Abstract
Purα is a ubiquitous, sequence-specific DNA- and RNA-binding protein which is highly conserved in eukaryotic cells. Purα has been implicated in diverse cellular functions, including transcriptional activation and repression, translation and cell growth. Moreover, this protein has been shown to be involved in regulating several human viruses which replicate in the central nervous system (CNS), including human immunodeficiency virus type I (HIV-1) and JC virus (JCV). Purα exerts part of its activity by interacting with cellular proteins, including pRb, E2F, cyclin A, Sp1 and members of the Y-box family of proteins, including YB-1 and MSY1, as well as viral proteins such as polyomavirus large T-antigen and HIV-1 Tat. The ability of Purα to interact with its target DNA sequence and to associate with several viral and cellular proteins is modulated by RNA. Purα has also been shown to be involved in cell growth and proliferation. Its association with pRb, E2F and cyclin A coupled with its fluctuating levels throughout the cell cycle, position Purα as a crucial factor in the cell cycle. Moreover, microinjection studies demonstrate that Purα causes either a G1 or G2 arrest depending on the cell cycle time of injection. The gene encoding Purα has been localized to a human locus which is frequently deleted in myelogenous leukemias and other cancers and Purα gene deletions have been detected in many cases of lymphoid cancers. The following review details the structural characteristics of Purα, its family members and the involvement of this protein in regulating various cellular and viral genes, viral replication and cell growth.
PURα: STRUCTURE, GENE FAMILY AND EVOLUTIONARY CONSERVATION
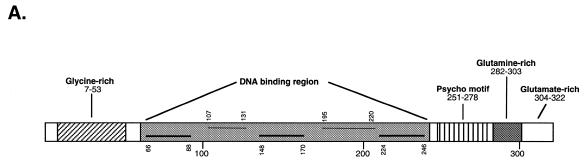
Since the initial cloning of human Purα (GenBank accession no. M96684) in 1992 (1), several laboratories have cloned genes encoding proteins homologous to Purα (Table 1). Analysis of the predicted 322 amino acid human Purα protein reveals a modular structure with a central region composed of three 23 amino acid class I repeats interspersed with two 26 amino acid class II repeats (1; Fig. 1A). Although not completely identical, each repeat preserves several identical and conservatively substituted amino acid residues (Fig. 1B). The central repeat region of Purα is important for binding to its single-stranded DNA target sequence (2,3). This region also contains sequences important for its interaction with several regulatory proteins, including T-antigen and YB-1, as well as its oligomerization domain (4–6). The class II motifs have been shown to be involved in the interaction between Purα and the HIV-1 Tat protein (7).
Table 1. Proteins identical to Purα.
| Factor | Reference |
|---|---|
| Myelin enhancing factor 1 (MEF-1) | (13) |
| Single-stranded cAMP response element-binding protein (ssCRE-BP) | (45) |
| Specific single-stranded DNA-binding factor 1 (SPSF1) | (22) |
| BI and BII FE65 promoter complexes | (14) |
| Neuronal nicotinic acetylcholine receptor promoter-binding protein 43 (NARP43) | (19) |
| Vascular actin single-stranded DNA-binding factor 2 (VACssBF2), p46 component | (15) |
| (CAG)-element-recognizing protein 1 (CAGER-1) | (16) |
Figure 1.
Structural organization of human Purα protein. (A) Graphical representation of the domains and motifs of Purα. The three basic aromatic class I repeats are indicated by heavy horizontal lower lines in the central repeat region and the two acidic leucine-rich regions are indicated by light horizontal upper lines in the central repeat region. (B) Class I motifs are aligned at the top and class II repeat motifs are aligned at the bottom. Solid boxes indicate identical amino acid residues and dotted boxes indicate conservative changes.
In addition to the repeat modules located in the central region of the protein, Purα contains several other notable structural features. The N-terminus contains a glycine-rich region including a stretch of 18 glycine residues broken only by a single serine residue. The region from residue 261 to 274 contains a potential amphipathic α-helix with opposing basic and aromatic side chains. Within this amphipathic helix of Purα is a region of limited homology to the large tumor antigens of several polyomaviruses, including simian virus 40 (SV40), JC virus (JCV), BK virus (BKV) and several other viral as well as yeast proteins (8). Since the consensus sequence derived from alignment of Purα and these various other proteins contains PSY and C, this domain has been termed the ‘psycho’ motif (8). Interestingly, the homology of Purα to T-antigen spans a region of T-antigen necessary for its interaction with the product of the human retinoblastoma tumor suppressor gene, pRb (9). Moreover, studies have demonstrated that Purα binds the hypophosphorylated form of pRb and that this interaction involves the region of Purα encompassing the psycho motif (3).
The region from Lys203 to Lys229 contains a characteristic PEST (proline–glutamate–serine–threonine) sequence which is thought to facilitate protein turnover in cells (10). The C-terminus of Purα contains a glutamine/glutamate-rich domain with half of the residues from amino acid 276 to 321 being either glutamine or glutamate. Within this region, there is a stretch of seven glutamine residues as well as a sequence of five glutamate resides broken by a single glycine. Glutamine-rich sequences are thought to function as transcriptional activation domains (11) and Purα has been shown to be a transcriptional transactivator of several genes (12–14).
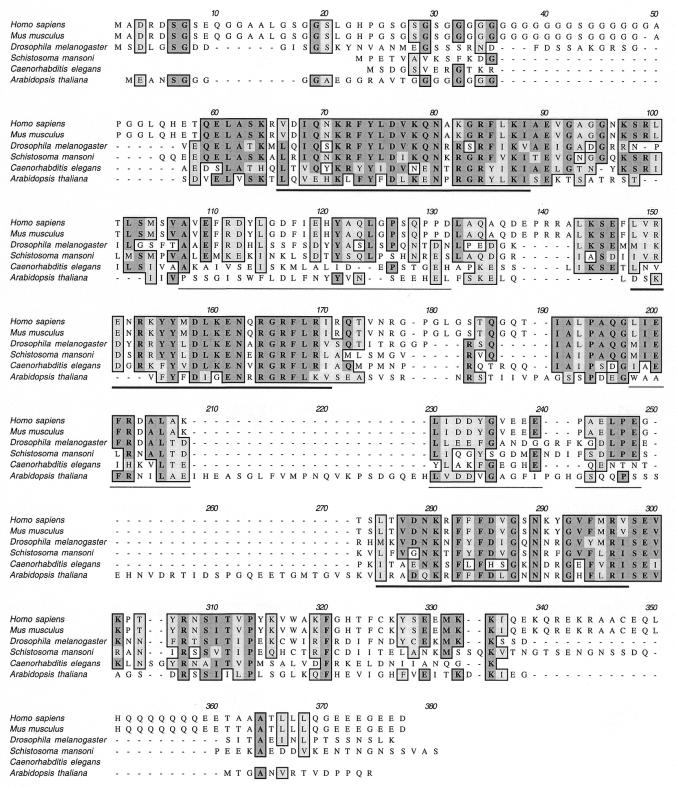
The sequence of Purα is highly conserved among species. Amino acid sequence alignment of human and murine (U02098) Purα reveals only two amino acid differences (8). Mouse Purα lacks Gly49 and includes an Ala306→Thr substitution. In addition to mammalian cells, Purα sequences have also been cloned from Drosophila melanogaster (AF02159), Caenorhabditis elegans (U70852), Schistosoma mansoni (AF254148) and Arabidopsis thaliana (AF136152). The various protein sequences share a high degree of homology, particularly in the class I and, to a lesser degree, class II repeats (Fig. 2).
Figure 2.
Alignment of Purα sequences from Homo sapiens, Mus musculus, Drosophila melanogaster, Schistosoma mansoni, Caenorhabditis elegans and Arabidopsis thaliana. Dark gray boxed amino acids indicate sequence identity; light gray boxed amino acids indicate conserved amino acids. The heavy underline and thin underline indicate class I and class II repeats of human Purα, respectively.
Purα is one member of the Pur protein family; Purβ and Purγ are other family members which have been identified. Murine Purα and Purβ (AF017630) possess 71% sequence identity (15). Mouse Purβ exhibits the same modular structure as Purα and the basic 23 amino acid class I repeats and acidic 26 amino acid class II repeats are highly conserved between murine Purα and Purβ. There are, however, several notable differences between these two Pur proteins. More specifically, Purβ possesses a glycine-rich stretch which interrupts the second class II repeat and lacks the potential casein kinase II phosphorylation site present in the C-terminal region of the ‘psycho’ motif of Purα. Other striking differences between Purα and Purβ lie in the N- and C-terminal ends of the proteins. While the N-terminus of Purα exhibits an almost uninterrupted stretch of glycine residues (17 of 18), the glycine-rich stretch of Purβ (19 of 26) is interrupted by a six amino acid sequence (23FQPAPR28). Moreover, the C-terminal glutamine-rich stretch of Purα is absent in Purβ.
A sequence for the Pur family member Purγ has been entered in the GenBank database (AF195513). Human Purγ possesses 54% sequence identity to human Purα. The class I and II repeats are conserved between Purα and Purγ. Notable differences between these two human Pur family members include a dissimilar N-terminus, a 23 amino acid insertion within the first class II repeat of Purγ and the absence of the C-terminal glutamine-rich stretch in Purγ. The gene encoding Purγ is located head-to-head with the gene encoding the Werner syndrome helicase, WRN, at chromosome band 8p11. PURG had previously gone undetected in a sequence for the promoter region of WRN (AB003173). The first exon of WRN is separated from PURG by ∼1 kb, raising the prospect of genetic co-regulation.
It is interesting that there are three members of the Pur family identified to date. Although there is little functional evidence addressing the question of whether these protein family members are functionally redundant or biologically unique, several structural features suggest that they likely possess different functions. While the central repeat region is highly conserved between these Pur family members, there are various structural differences in other regions of the proteins. For example, a potential casein kinase phosphorylation site which is present in the Rb-binding domain of Purα is absent in the other family members. There are many other structural differences between these Pur family members. Moreover, it is currently unclear if these proteins are differentially expressed. Additional studies are necessary to address these questions and elucidate the cellular necessity of each family member.
In 1999, Yano et al. (16) sequenced two proteins that were reported as binding to DNA CAG or CGG triplet repeat elements. The sequences corresponded to Purα and Purβ. These workers named their proteins CAGER-1 and CAGER-2, claiming that the original cDNA clones for Purα and Purβ were ‘misassigned’ and that Purα actually encodes a 28 kDa protein. Both of those claims are in error, however, and it is very likely that the CAGERs are Purα and Purβ. Purα was originally identified as a protein of 322 amino acids, the cDNA of which was cloned from both HeLa cells and fetal liver libraries (1). That paper was the first instance in which the terms ‘Purα’ and ‘Purβ’ were used. The name Purα was never used to refer to a 28 kDa protein as erroneously stated by Yano et al. (16). In an earlier paper a protein had been identified as a Pur factor based on its ability to bind to a purine-rich single-stranded DNA sequence (17). When UV crosslinked and bound to an oligonucleotide, that protein migrated as a band of 28 kDa. That protein was never referred to as Purα. Nor was it ever claimed that 28 kDa would be an accurate molecular weight for that protein. Based on its properties, it is most likely a Pur family member or a breakdown product of a Pur family member, but it was never given the name Purα. Yano et al. (16) wished to distinguish the putative CAGER proteins from Purα and Purβ on the basis of tissue distribution, noting that CAGER levels are particularly high in postnatal mouse brain. The observation has been published that Purα levels are high in postnatal mouse brain (18). This is not inconsistent with their observations of putative CAGERs. Considerable evidence indicates, however, that Purα is virtually ubiquitous, in contrast to the claim for CAGERS. Many other workers concur. The sequence for human Purα is unchanged today from its originally published version in 1992 (1). Various workers have sequenced true Purα cDNA clones, as distinct from Purβ or Purγ, from the following human cells and tissues: HeLa cells (M96684); fetal liver (M96684); leiomyosarcoma (AW452746); germ cell tumors (AI63208); testis (AI223140); prostate (AI417720); kidney tumor (AI672340); B cells from a chronic lymphocytic leukemia patient (AI082370); carcinoid tumor (AA976072); lung tumor (AI829930); parathyroid tumor (AI022564); colon adenocarcinoma (AW084098); ovarian tissue (AW298527). Purα cDNA has also been sequenced from the following mouse tissues: vascular smooth muscle (AF02159); spinal cord (AI85069); pineal gland (AI845412); mammary gland (AI647004); bone (AW744672).
PURα AND NUCLEIC ACIDS
Purα binds both single-stranded and double-stranded DNA in a sequence-specific fashion, but has a preference for the purine-rich single-stranded form of its recognition sequence which is composed of repeats of (GGN) (1,2,5,7,13–15,19–26). Moreover, various studies have demonstrated that the interaction between Purα and its target PUR sequence elements results in the formation of multimeric complexes (3,5,7,26). The interaction between Purα and its target sequence is modulated by several different proteins, including pRb (3), YB-1 (5), T-antigen (19) and MyEF-2 (26).
Purα also interacts with RNA and RNA molecules have been shown to modulate the activity of Purα (18,27,28). A Purα-associated cellular RNA, named PU-RNA, with significant homology to 7SL RNA has been shown to inhibit the interaction between Purα and its target DNA sequence within the myelin basic protein gene promoter (18). In contrast, other studies have demonstrated that the DNA-binding activity of a Pur factor is RNA-dependent (29). This Pur factor, however, while binding to a GC-rich sequence, had a molecular mass (29 kDa) different from that of Purα. Purα has also been shown to interact with the HIV-1 TAR RNA and this interaction modulates HIV-1 gene transcription via a TAR-dependent mechanism (27). Another Purα-associated cellular RNA has been shown to mediate the interaction between Purα and the HIV-1 Tat protein and enhance transcriptional activation of the HIV-1 promoter in the presence of Purα and Tat (28). RNA is also important in the self-association of Purα (6).
PURα AND GENE TRANSCRIPTION
Purα has been implicated in the transcriptional control of many cellular genes, including myelin basic protein (MBP), murine vascular smooth muscle (VSM) α-actin, neuron-specific FE65 gene promoter, neuronal nicotinic acetylcholine (nAch) receptor and the single-stranded cAMP response element. In addition, this protein has been implicated in the control of several viral promoters, including the JCV early gene promoter and the HIV-1 long terminal repeat.
MBP represents the first cellular gene which was shown to be transcriptionally regulated by Purα. Expression of MBP, which is restricted to glial cells within the CNS, is differentially regulated during brain development. The cell type and developmental expression of MBP is regulated at the transcriptional level (30) and a proximal element, termed MB1, which contains the Purα binding motif, dictates cell type-specific expression of MBP (31,32). Purification of a 39 kDa MB1-binding protein from mouse brain at the peak of myelination identified MEF-1 (myelin enhancing factor 1), with amino acid identity to Purα (13). Interestingly, the expression of MEF-1/Purα and its interaction with the MB1 sequence occurs in a developmental stage-specific manner which coincide with the pattern of MBP gene expression (18,22). Functionally, Purα stimulates transcription of the MBP promoter in glial cells (13,33), and the GC/GA-rich domain within the MB1 region which is necessary for binding of Purα is required for this activity.
Purα interacts with other cellular regulatory proteins, including MyEF-2 and Sp1, which regulate MBP gene transcription. MyEF-2 is a single-stranded DNA-binding protein which decreases transcription of the MBP gene (34). It was shown that the interaction bewteen Purα and MyEF-2 can determine the binding of these proteins to their target DNA sequences within the MB1 motif (26). Evidently, both proteins can exert a negative effect on each other’s DNA binding activity. Based on the programmed expression of Purα and MyEF-2 during myelination and the interplay between these two proteins, a model for their involvement in the transcriptional regulation of the MBP gene during the course of brain development has been proposed (26). According to this model, the association of MyEF-1 with MB1 in the early phase of brain development when the level of Purα is minimum suppresses MBP gene transcription. In the later stages, when the level of Purα increases, the interaction between Purα and MB1 displaces MyEF-2 from the MBP promoter and results in stimulation of the MBP gene.
Purα also associates with the ubiquitous transcription factor Sp1, which has been shown to regulate MBP gene expression (33). Sp1 interacts with the MB1 DNA motif at a region that partially overlaps with the Purα binding site. Sp1 enhances the interaction between Purα and MB1 without the formation of a Purα–MB1–Sp1 complex. Functionally, overexpression of Purα and Sp1 in CNS cells results in synergistic stimulation of the MBP promoter.
Mouse VSM α-actin is another target gene regulated by Purα (15). Transcriptional regulation of the VSM α-actin promoter in both fibroblasts and undifferentiated myoblasts is mediated, in part, by an asymmetrical polypurine/polypyrimidine tract containing an inverted muscle-specific MCAT (AGGAATG) enhancer motif which has been shown to interact with at least three distinct DNA binding activities. The double-stranded form of this sequence serves as a binding site for a transcription enhancer factor 1-related protein which has been implicated in transactivation (35,36). Two distinct single-stranded DNA binding activities, called vascular actin single-strand binding factors, VACssBF 1 and 2, have been implicated in transcriptional repression (37). VACssBF 2, which consists of two distinct polypeptides (p44 and p46), was shown to be necessary and sufficient for repression (38). Further experiments revealed the identity of the p46 and p44 components of VACssBF2 as Purα and Purβ, respectively (15). This group also demonstrated that Purα and Purβ interact with each other and can bind as hetero- and homodimers to the purine-rich strand of the MCAT enhancer and that both Pur proteins interact with the mouse Y-box protein MSY1 (39). MSY1 is highly homologous to the human protein YB-1, previously reported to interact with Purα (5). These Y-box proteins bind to pyrimidine-rich elements that could be the complementary sequence of many PUR elements. The finding that Purα associates with Y-box proteins in two different systems highlights the potential significance of this interaction in gene regulation.
Interestingly, Purα, Purβ and MSY1 also interact with an mRNA sequence derived from the coding region of VSM α-actin which has similarity to the MCAT enhancer in the 5′ promoter of the gene (40). Furthermore, this sequence suppressed mRNA translation when placed in the 5′-untranslated region of a reporter mRNA. Translational efficiency was restored by mutations within this sequence which impaired the binding of Purα, Purβ and MSY1, suggesting that these proteins may also participate in translation.
Purα has been suggested to play a role in the transcriptional regulation of the neuronal nAch receptor gene, specifically the β4 subunit gene (21). A sequence element in the promoter region of the rat β4 subunit gene (termed E1) was used in a purification scheme to isolate nuclear proteins which interact with this element. Four polypeptides with apparent molecular masses of 31, 43, 65 and 114 kDa were detected. Peptide sequence analysis of the 43 kDa polypeptide indicated that it was the bovine homolog of Purα. Functionally, a mutation within the E1 sequence, which abrogated the interaction with Purα, resulted in a 70% reduction in promoter activity, suggesting a functional role of Purα in transcriptional control of the nAch receptor gene (41). Additionally, Sp1 and Sp3 have been shown to be involved in β4 gene expression via their interactions with E2, which is located immediately downstream of E1 (42). As Purα both functionally and structurally interacts with Sp1 (33), the interaction between these two proteins may regulate expression from the β4 subunit of the nAch receptor gene as well.
Purα has also been implicated in transcriptional regulation of the FE65 gene. FE65 is a 90 kDa adaptor protein that interacts with the Alzheimer β-amyloid precursor protein. The FE65 gene has a TATA-less promoter that drives expression mainly in neurons. The minimal promoter region which is functional in neural cells forms three major DNA–protein complexes, termed BI, BII and BIII, when incubated with rat brain nuclear extract. Purification of BI and BII revealed that both proteins were Purα (14). In Chinese hamster ovary cells, where the activity of the FE65 promoter is very low, transient overexpression of Purα increased expression from the FE65 minimal promoter. Interestingly, purification of the BIII protein revealed identity to the YYI transcription factor. Although YY1 also activated the FE65 minimal promoter, no cooperation was observed between Purα and YY1 (14).
Purα has also been implicated in chronic morphine dependence. Osugi et al. (43,44) demonstrated that the DNA binding activity of a nuclear single-stranded cyclic AMP response element (ssCRE)-binding protein (ssCRE-BP) is decreased after chronic morphine administration. Subsequent purification of murine ssCRE-BP revealed identity to mouse Purα (45). Interestingly, the levels of ssCRE-BP/Purα mRNA or protein were not changed by chronic morphine treatment (45). The DNA binding activity of Purα, however, was shown to be enhanced by the addition of a heat stable activator (46). Purification of this activator revealed identity to calmodulin (CaM) and CaM was shown to enhance the interaction between Purα and various PUR elements derived from the 5′-non-coding regions of various genes, including the myelin basic protein gene, the nAch receptor β4 subunit gene, the somatostatin ssCRE and the CaM response element of the neuropeptide Y gene (47). These data suggest that Purα may regulate transcription of various genes through Ca/CaM and subsequent characterization of the physiological role of Purα and Ca/CaM in control of transcription and replication via PUR elements represents exciting prospects in signaling mechanisms involved in Purα function.
Purα has been implicated in control of expression of various other cellular genes, including the transforming growth factor β-1 (TGFβ-1) promoter (24) and the A.thaliana translation elongation factor eEF1A via its interaction with interstitial telomere sequences (25). Other studies suggest that Purα may be increased in allergic airway inflammation (48). Additionally, a Pur element has been shown to function as an enhancer for the clusterin gene in Rous sarcoma virus-infected avian fibroblasts (v-src) (49).
In considering mechanisms by which Purα family members may influence transcription through binding to DNA promoter elements, parallels may be found in the far upstream element (FUSE)-binding protein (FBP) family of transcription factors (50). As do the Pur proteins, FBP proteins bind specifically to a single strand of their DNA recognition elements, FUSE. Although there is little homology between Pur and FBP proteins, there are certain functionally significant similarities. Both FBP proteins and Purα possess a prominent poly(G) motif near their N-termini, as do several other single-stranded DNA-binding proteins. In both protein families, the most conserved sequence is in the central DNA-binding region. FBP proteins and Purα also possess sequences of known transcriptional activation capacity near their C-termini. The ability of FBP to bind its melted DNA element upstream of the c-myc gene in vivo has been related to the degree of c-myc transcription (51). It has been proposed that the single-stranded nature of FUSE is created by supercoiling resulting from transcription and that by binding the torsionally strained DNA, FBP can directly measure promoter activity. The melted DNA at FUSE can serve as a flexible hinge, facilitating interaction between FBP and other protein factors (52).
As mentioned earlier, in addition to its role in regulating varied and diverse cellular genes, Purα is also involved in regulating several human viruses which replicate in the CNS, including JCV and HIV-1. Expression of JCV is tissue-specific and is determined primarily at the level of viral gene transcription (reviewed in 53). The transcriptional control region of this virus is composed of a bidirectional 98 bp promoter/enhancer repeat which has a pentanucleotide repeat sequence (AGGGAAGGGA) designated the lytic control element (LCE). This motif, which affects viral gene expression and replication, has been shown to interact with a nuclear factor termed the lytic control element binding protein (LCP-1) which bears remarkable similarity to Purα (54). Subsequent studies demonstrated that this PUR sequence element does indeed interact with Purα (2).
Purα has been shown to stimulate JCV early gene transcription and decrease the ability of the viral regulatory protein, T-antigen, to increase the level of JCV late gene transcription (12). The functional antagonism between Purα and T-antigen is mitigated by their physical interaction with one another (4). A mutant Purα which is unable to interact with T-antigen is incapable of abrogating T-antigen-mediated transcriptional activation. The complementary strand of the LCE and similar PUR sequences in JC variant strains contain a polypyrimidine stretch which has been shown to interact with the Y-box binding protein YB-1 (2,5). Moreover, Purα interacts with YB-1 and this interaction regulates JC viral early and late gene transcription (5). This is reminiscent of the interaction between Purα and MSY1, suggesting that members of the Pur and Y-box families can combinatorally regulate various cellular and viral genes.
Interestingly, there is a high incidence of the JCV-induced demyelinating disease progressive multifocal leukoencephalopathy (PML) among individuals with acquired immunodeficiency syndrome (AIDS), suggesting that the presence of HIV-1 in the brains of infected individuals induces JCV gene transcription and replication. In support of this synergism, the transregulatory protein of HIV-1, Tat, has been shown to activate the JCV late gene promoter (55,56). Tat is a transcription activator that interacts with a cis-acting RNA sequence called TAR and several DNA-binding transcription factors to stimulate transcription of the HIV genome (57). Interestingly, the Tat-responsive sequence of JCV contains a PUR element and Purα and Tat synergistically activate the JCV late gene promoter (7). Results from protein–protein interaction studies revealed that Purα has the ability to interact with the HIV-1 Tat protein (7,28). This interaction is dependent on RNA molecules, as treatment with RNase abrogates their interaction. Several RNA species involved in the interaction between HIV-1 Tat and Purα have been cloned and these molecules specifically reconstitute the interaction between these two proteins. Furthermore, co-expression of the RNA in the sense orientation results in increased transcriptional activity of Purα and Tat on the HIV-1 LTR (28).
PURα AND CONTROL OF CELL GROWTH
There is mounting evidence that Purα is integrally involved in cell growth. Purα was originally cloned by its affinity for the PUR element adjacent to a region of stably bent DNA 1.6 kb upstream of the human c-myc gene (1). This element is near the center of an initiation zone of chromosomal replication. Moreover, PUR elements are present at eukaryotic origins of DNA replication. Purα has also been shown to interact with viral origins of DNA replication, including the JCV and bovine papillomavirus origins (19,23). The first functional indication of Purα’s role in replication emerged from studies utilizing the JC viral origin of replication (19). These studies demonstrated that Purα suppresses JC viral DNA replication in human glial cells. Interestingly, when an antisense RNA was expressed, JCV DNA replication was stimulated, suggesting that the endogenous Purα exerts a negative effect on replication. In addition to the location of PUR elements and the effect of Purα on viral DNA replication, additional evidence supporting a role for Purα in the control of cellular growth and proliferation is derived from alterations in Purα levels and intracellular localization during the cell cycle, the association of Purα with key cell cycle regulatory proteins and viral oncoproteins, protein microinjection studies and identification of gene deletions in human tumors (3,20,58–63).
Alterations in the intracellular levels of Purα have been demonstrated in CV-1 cells (58). Levels of Purα decline precipitously in the G1 phase of the cell cycle, just prior to the onset of S phase, and remain low in early S phase. Levels subsequently recover throughout the late S and G2 phases of the cell cycle to peak at mitosis. Purα levels remain maximal through cytokinesis and re-entry into early G1. Earlier studies demonstrated that Purα interacts with the hypophosphorylated form of the retinoblastoma tumor suppressor gene product, pRb (3). pRb is an integral protein involved in progression of the cell cycle (reviewed in 64). The hypophosphorylated form of pRb complexes with a variety of proteins in the G0 and G1 phases of the cell cycle, including transcription factors such as E2F. Hyperphosphorylation of pRb in late G1 results in the release of these transcription factors from pRb and allows E2F to activate genes necessary for progression through G1 and entry into the S phase of the cell cycle. Co-immunoprecipitation studies have demonstrated that the association between Purα and pRb is restricted to the G1 phase of the cell cycle (58). Moreover, during the late S and G2 phases of the cell cycle, Purα co-immunoprecipitates with cyclin A and co-localizes with cyclin A in replication foci in the nucleus. These observations suggest a role for Purα in mediating cell cycle events in the late S and G2 phases of the cell cycle. In addition, the decline in Purα levels at the beginning of S phase could itself play a regulatory role representing a positive signal for the onset of DNA replication. These studies were performed in CV-1 cells. Additional studies in other cell lineages investigating the cell cycle-dependent alteration in intracellular levels are necessary to determine if this trend occurs in other cell systems.
Interestingly, Purα has also been shown to interact with E2F-1 and this interaction decreases the ability of E2F-1 to exert its transcriptional activity upon the dihydrofolate reductase gene (DHFR) promoter (20). This suppression is due to the ability of Purα to inhibit the interaction of E2F-1 with its target sequence. Interestingly, Purα and E2F-1 bind to the same region of pRb, suggesting that the association of Purα with pRb may liberate E2F-1. These observations indicate that Purα may play an important role in the activity of E2F-1 during the cell cycle. Future studies detailing the phase(s) during which Purα interacts with E2F-1 will help clarify Purα’s emerging relationship with these key cell cycle regulators.
Evidence from protein microinjection studies provides perhaps the most direct evidence of Purα’s role in cell cycle progression. Stacey et al. (59) microinjected Purα into NIH 3T3 cells and employed a video time-lapse technique to determine the cell cycle position. Approximately 80% of cells injected with Purα were inhibited from passing through mitosis, with most cells blocked in the G2 phase, although a lesser block was seen in G1. In this study, cells were also injected with a mutant Purα protein which contains the first 215 amino acids. This mutant contained two of the three DNA-binding repeats, but importantly lacked the Rb-binding and glutamine-rich domains. Interestingly, microinjection of this mutant had no effect upon cell cycle progression. These observations provide substantial evidence that Purα is involved in cell cycle progression. Additional experiments are necessary to fully decipher the molecular mechanism(s) involved in this process.
As noted above, Purα also interacts with large T-antigen from several polyomaviruses (4,19,65). One well-characterized function of viral oncoproteins, including T-antigens, is cellular transformation. One mechanism by which these proteins are able to cause cellular transformation is via their interaction with the products of tumor suppressor genes, such as p53 and pRb. The interaction between Purα and these viral oncoproteins raises interesting questions as to the cellular role of Purα. This is particularly noteworthy in the light of several observations regarding Purα. PURA, the gene encoding Purα, has been localized to human chromosome band 5q31 (60). Loss of heterozygosity at this locus is frequently associated with hematological malignancies, particularly myelodysplastic syndromes and myeloid leukemia (61,62). Moreover, PURA gene deletions have been demonstrated in many cases of myelogenous leukemia and myelodysplastic syndrome (63).
CONCLUSION
Since its first description 8 years ago, much has been learned about Purα. This sequence-specific DNA- and RNA-binding protein is involved in diverse cellular functions, including transcription, translation and cell growth. Additionally, it is involved in regulating HIV-1 and JC viruses. Although the exact mechanism of its involvement in the cell cycle is as yet undescribed, Purα plays a role in control of the cell cycle. Moreover, the fact that Purα is highly conserved among various eukaryotic organisms suggests that Purα is a critical protein. Future experiments dissecting the role of this founding member of the Pur family as well as other family members will ultimately unravel the diverse functions of this protein.
Acknowledgments
ACKNOWLEDGEMENTS
The authors wish to thank past and present members of the Center for NeuroVirology and Cancer Biology for their insightful discussions, sharing of ideas and reagents. We also wish to thank C. Schriver for preparation of this manuscript. This work was made possible by grants awarded by the NIH to K.K. and E.J.
REFERENCES
- 1.Bergemann A.D., Ma,Z.W. and Johnson,E.M. (1992) Sequence of cDNA comprising the human pur gene and sequence-specific single-stranded-DNA-binding properties of the encoded protein. Mol. Cell. Biol., 12, 5673–5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen N.N., Chang,C.F., Gallia,G.L., Kerr,D.A., Johnson,E.M., Krachmarov,C.P., Barr,S.M., Frisque,R.J., Bollag,B. and Khalili,K. (1995) Cooperative action of cellular proteins YB-1 and Purα with the tumor antigen of the human JC polyomavirus determines their interaction with the viral lytic control element. Proc. Natl Acad. Sci. USA, 92, 1087–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson E.M., Chen,P.L., Krachmarov,C.P., Barr,S.M., Kanovsky,M., Ma,Z.W. and Lee,W.H. (1995) Association of human Purα with the retinoblastoma protein, Rb, regulates binding to the single-stranded DNA Purα recognition element. J. Biol. Chem., 270, 24352–24360. [DOI] [PubMed] [Google Scholar]
- 4.Gallia G.L., Safak,M. and Khalili,K. (1998) Interaction of the single-stranded DNA-binding protein Purα with the human polyomavirus JC virus early protein T-antigen. J. Biol. Chem., 273, 32662–32669. [DOI] [PubMed] [Google Scholar]
- 5.Safak M., Gallia,G.L. and Khalili,K. (1999) Reciprocal interaction between two cellular proteins, Purα and YB-1, modulates transcriptional activity of JCVCY in glial cells. Mol. Cell. Biol., 19, 2712–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gallia G.L., Darbinian,N., Johnson,E.M. and Khalili,K. (1999) Self-association of Purα is mediated by RNA. J. Cell. Biochem., 75, 1–15. [PubMed] [Google Scholar]
- 7.Krachmarov C.P., Chepenik,L.G., Barr-Vagell,S. and Khalili,K. (1996) Activation of the JC virus Tat-responsive transcriptional control element by association of the Tat protein of human immunodeficiency virus 1 with cellular protein Purα. Proc. Natl Acad. Sci. USA, 93, 14112–14117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma Z.W., Bergemann,A.D. and Johnson,E.M. (1994) Conservation in human and mouse Purα of a motif common to several proteins involved in initiation of DNA replication. Gene, 149, 311–314. [DOI] [PubMed] [Google Scholar]
- 9.DeCaprio J.A., Ludlow,J.W., Figge,J., Shew,J.Y., Huang,C.M., Lee,W.H., Marsilio,E., Paucha,E. and Livingston,D.M. (1988) SV40 large tumor antigen forms a specific complex with the product of the retinoblastoma susceptibility gene. Cell, 54, 275–283. [DOI] [PubMed] [Google Scholar]
- 10.Rogers S., Wells,R. and Rechsteiner,M. (1986) Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science, 234, 364–368. [DOI] [PubMed] [Google Scholar]
- 11.Tjian R. and Maniatis,T. (1994) Transcriptional activation: a complex puzzle with few easy pieces. Cell, 77, 5–8. [DOI] [PubMed] [Google Scholar]
- 12.Chen N.N. and Khalili,K. (1995) Transcriptional regulation of human JC polyomavirus promoters by cellular proteins YB-1 and Purα in glial cells. J. Virol., 69, 5843–5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haas S., Thatikunta,P., Steplewski,A., Johnson,E.M., Khalili,K. and Amini,S. (1995) A 39-kDa DNA-binding protein from mouse brain stimulates transcription of myelin basic protein gene in oligodendrocytes. J. Cell Biol., 130, 1171–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zambrano N., DeRenzis,S., Minopoli,G., Faraonio,R., Donini,V., Scaloni,A., Cimino,F. and Russo,T. (1997) DNA-binding protein Purα and transcription factor YY1 function as transcription activators of the neuron-specific FE65 gene promoter. Biochem. J., 328, 293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kelm R.J. Jr, Elder,P.K., Strauch,A.R. and Getz,M.J. (1997) Sequence of cDNAs encoding components of vascular actin single-stranded DNA-binding factor 2 establish identity to Purα and Purβ. J. Biol. Chem., 272, 26727–26733. [DOI] [PubMed] [Google Scholar]
- 16.Yano H., Wang,B.E., Ahmad,I., Zhang,J., Abo,T., Nakayama,J., Krempen,K. and Kohwi,Y. (1999) Identification of (CAG)n and (CGG)n repeat-binding proteins, CAGERs expressed in mature neurons of the mouse brain. Exp. Cell Res., 251, 388–400. [DOI] [PubMed] [Google Scholar]
- 17.Bergemann A.D. and Johnson,E.M. (1992) The HeLa pur factor binds single-stranded DNA at a specific element conserved in gene flanking regions and origins of DNA replication. Mol. Cell. Biol., 12, 1257–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tretiakova A., Gallia,G.L., Shcherbik,N., Jameson,B., Johnson,E.M., Amini,S. and Khalili,K. (1998) Association of Purα with RNAs homologous to 7SL determines its binding ability to the myelin basic protein promoter DNA sequence. J. Biol. Chem., 273, 22241–22247. [DOI] [PubMed] [Google Scholar]
- 19.Chang C.F., Gallia,G.L., Muralidharan,V., Chen,N.N., Zoltick,P., Johnson,E. and Khalili,K. (1996) Evidence that replication of human neurotropic JC virus DNA in glial cells is regulated by the sequence-specific single-stranded DNA-binding protein Purα. J. Virol., 7, 4150–4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Darbinian N., Gallia,G.L., Kundu,M., Shcherbik,N., Tretiakova,A., Giordano,A. and Khalili,K. (1999) Association of Purα and E2F-1 suppresses transcriptional activity of E2F-1. Oncogene, 18, 6398–6402. [DOI] [PubMed] [Google Scholar]
- 21.Du Q., Tomkinson,A.E. and Gardner,P.D. (1997) Transcriptional regulation of neuronal nicotinic acetylcholine receptor genes. A possible role for the DNA-binding protein Purα. J. Biol. Chem., 272, 14990–14995. [DOI] [PubMed] [Google Scholar]
- 22.Haas S., Gordon,J. and Khalili,K. (1993) A developmentally regulated DNA-binding protein from mouse brain stimulates myelin basic protein gene expression. Mol. Cell. Biol., 13, 3103–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jurk M., Weissinger,F., Lottspeich,F., Schwarz,U. and Winnacker,E.L. (1996) Characterization of the single-strand-specific BPV-1 origin binding protein, SPSF I, as the HeLa Purα factor. Nucleic Acids Res., 24, 2799–2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thatikunta P., Sawaya,B.E., Denisova,L., Cole,C., Yusibova,G., Johnson,E.M., Khalili,K. and Amini,S. (1997) Identification of a cellular protein that binds to Tat-responsive element of TGFβ-1 promoter in glial cells. J. Cell. Biochem., 67, 466–477. [PubMed] [Google Scholar]
- 25.Tremousaygue D., Manevski,A., Bardet,C., Lescure,N. and Lescure,B. (1999) Plant interstitial telomere motifs participate in the control of gene expression in root meristems. Plant J., 20, 553–561. [DOI] [PubMed] [Google Scholar]
- 26.Muralidharan V., Tretiakova,A., Steplewski,A., Haas,S., Amini,S., Johnson,E. and Khalili,K. (1997) Evidence for inhibition of MyEF-2 binding to MBP promoter by MEF-1/Purα. J. Cell. Biochem., 66, 524–531. [DOI] [PubMed] [Google Scholar]
- 27.Chepenik L.G., Tretiakova,A.P., Krachmarov,C.P., Johnson,E.M. and Khalili,K. (1998) The single-stranded DNA binding protein, Pur-α, binds HIV-1 TAR RNA and activates HIV-1 transcription. Gene, 210, 37–44. [DOI] [PubMed] [Google Scholar]
- 28.Gallia G.L., Darbinian,N., Tretiakova,A., Ansari,S.A., Rappaport,J., Brady,J., Wortman,M.J., Johnson,E.M. and Khalili,K. (1999) Association of HIV-1 Tat with the cellular protein, Purα, is mediated by RNA. Proc. Natl Acad. Sci. USA, 96, 11572–11577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herault Y., Chatelain,G., Brum,G. and Michel,D. (1995) RNA-dependent DNA binding activity of the Pur factor, potentially involved in DNA replication and gene transcription. Gene Expr., 4, 85–93. [PMC free article] [PubMed] [Google Scholar]
- 30.Campagnoni A.T. and Macklin,W.B. (1988) Cellular and molecular aspects of myelin protein gene expression. Mol. Neurobiol., 2, 41–89. [DOI] [PubMed] [Google Scholar]
- 31.Devine-Beach K.A., Haas,S. and Khalili,K. (1992) Analysis of the proximal transcriptional element of the myelin basic protein gene. Nucleic Acids Res., 20, 545–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Devine-Beach K.A., Lashgari,M.S. and Khalili,K. (1990) Myelin basic protein gene transcription: identification of proximal and distal cis-acting regulatory elements. J. Biol. Chem., 265, 13830–13835. [PubMed] [Google Scholar]
- 33.Tretiakova A., Steplewski,A., Johnson,E.M., Khalili,K. and Amini,S. (1999) Regulation of myelin basic protein gene transcription by Sp1 and Purα: evidence for association of Sp1 and Purα in brain. J. Cell. Physiol., 181, 160–168. [DOI] [PubMed] [Google Scholar]
- 34.Haas S., Steplewski,A., Siracusa,L.D., Amini,S. and Khalili,K. (1995) Identification of a sequence-specific single-stranded DNA binding protein that suppresses transcription of the mouse myelin basic protein gene. J. Biol. Chem., 270, 12503–12510. [DOI] [PubMed] [Google Scholar]
- 35.Cogan J.G., Sun,S., Stoflet,E.S., Schmide,L.H.J., Getz,M.J. and Strauch,A.R. (1995) Plasticity of vascular smooth muscle alpha-actin gene transcription. Characterization of multiple, single- and double-strand specific DNA-binding proteins in myoblasts and fibroblasts. J. Biol. Chem., 270, 11310–11321. [DOI] [PubMed] [Google Scholar]
- 36.Stoflet E.S., Schmidt,L.J., Elder,P.K., Korf,G.M., Foster,D.N., Strauch,A.R. and Getz,M.J. (1992) Activation of muscle-specific actin gene promoter in serum-stimulated fibroblasts. Mol. Biol. Cell, 3, 1073–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun S., Stoflet,E.S., Cogan,J.G., Strauch,A.R. and Getz,M.J. (1995) Negative regulation of the vascular smooth muscle alpha-actin gene in fibroblasts and myoblasts: disruption of enhancer function by sequence-specific single-stranded-DNA-binding proteins. Mol. Cell. Biol., 15, 2429–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kelm R.J. Jr, Sun,S., Strauch,A.R. and Getz,M.J. (1996) Repression of transcriptional enhancer factor-1 and activator protein-1-dependent enhancer activity by vascular actin single-stranded DNA binding factor 2. J. Biol. Chem., 271, 24278–24285. [DOI] [PubMed] [Google Scholar]
- 39.Kelm R.J. Jr, Cogan,J.G., Elder,P.K., Strauch,A.R. and Getz,M.J. (1999) Molecular interactions between single-stranded DNA-binding proteins associated with an essential MCAT element in the mouse smooth muscle α-actin promoter. J. Biol. Chem., 274, 14238–14245. [DOI] [PubMed] [Google Scholar]
- 40.Kelm R.J. Jr, Elder,P.K. and Getz,M.J. (1999) The single-stranded DNA-binding proteins, Purα, Purβ and MSY1 specifically interact with an exon 3-derived mouse vascular smooth muscle α-actin messenger RNA sequence. J. Biol. Chem., 274, 38268–38275. [DOI] [PubMed] [Google Scholar]
- 41.Hu M., Bigger,C.B. and Gardner,P.D. (1995) A novel regulatory element of a nicotinic acetylcholine receptor gene interacts with a DNA binding activity enriched in rat brain. J. Biol. Chem., 270, 4497–4502. [DOI] [PubMed] [Google Scholar]
- 42.Bigger C.B., Casanova,E.A. and Gardner,P.D. (1996) Transcriptional regulation of neuronal nicotinic acetylcholine receptor genes. Functional interactions between Sp1 and the rat beta 4 subunit gene promoter. J. Biol. Chem., 271, 32842–32848. [DOI] [PubMed] [Google Scholar]
- 43.Osugi T., Taniura,H., Ikemoto,M. and Miki,N. (1991) Effects of chronic exposure of NG108-15 cells to morphine or ethanol on binding of nuclear factors to cAMP-response element. Biochem. Biophys. Res. Commun., 174, 25–31 [DOI] [PubMed] [Google Scholar]
- 44.Osugi T., Ikemoto,M., Tanaka,H., Wang,X.B. and Miki,N. (1994) Modulation by chronic morphine administration of single-stranded cAMP response element (ssCRE) binding proteins in the mouse cerebellum. Mol. Brain Res., 21, 256–262. [DOI] [PubMed] [Google Scholar]
- 45.Osugi T., Ding,Y., Tanaka,H., Kuo,C.H., Do,E., Irie,Y. and Miki,N. (1996) Involvement of a single-stranded DNA binding protein, ssCRE-BP/Purα, in morphine dependence. FEBS Lett., 391, 11–16. [DOI] [PubMed] [Google Scholar]
- 46.Ding Y., Osugi,T., Kuo,C.H., Tanaka,H., Do,E., Irie,Y. and Miki,N. (1997) Characterization of a nuclear factor that enhances DNA binding activity of ssCRE-BP/Purα, a single-stranded DNA binding protein. Neurochem. Int., 31, 45–54. [DOI] [PubMed] [Google Scholar]
- 47.Kuo C.H., Nishikawa,E., Ichikawa,H., Sadakata,T., Niu,S.Y. and Miki,N. (1999) Calmodulin functions as an activator of Purα binding to single-stranded purine-rich DNA elements (PUR elements). Biochem. Biophys. Res. Commun., 255, 406–411. [DOI] [PubMed] [Google Scholar]
- 48.Wei E.Q., Irie,Y., Kuo,C.H., Ding,Y., Niu,S.Y., Do,E. and Miki,N. (1998) A single stranded DNA-binding protein, ssCRE-BP/Purα, in rat lung and its increase in allergic airway inflammation. Jpn. J. Pharmacol., 78, 419–427. [DOI] [PubMed] [Google Scholar]
- 49.Herault Y., Chatelain,G., Brum,G. and Michel,D. (1993) The PUR element stimulates transcription and is a target for single strand-specific binding factors conserved among vertebrate classes. Cell Mol. Biol. Res., 39, 717–725. [PubMed] [Google Scholar]
- 50.Davis-Smyth T., Duncan,R.C., Zheng,T., Michelotti,G. and Levens,D. (1996) The far upstream element-binding proteins comprise an ancient family of single-strand DNA-binding transactivators. J. Biol. Chem., 271, 31679–31687. [DOI] [PubMed] [Google Scholar]
- 51.He L., Liu,J., Collins,I., Sanford,S., O’Connell,B., Benham,C.J. and Levens,D. (2000) Loss of FBP function arrests cellular proliferation and extinguishes c-myc expression. EMBO J., 19, 1034–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tomonaga T., Michelotti,G.A., Libutti,D., Uy,A., Sauer,B. and Levens,D. (1998) Unrestraining genetic processes with a protein-DNA hinge. Mol. Cell, 1, 759–764. [DOI] [PubMed] [Google Scholar]
- 53.Raj G.V. and Khalili,K. (1995) Transcriptional regulation: lessons from the human neurotropic polyomavirus, JCV. Virology, 213, 283–291. [DOI] [PubMed] [Google Scholar]
- 54.Tada H. and Khalili,K. (1992) A novel sequence-specific DNA-binding protein, LCP-1, interacts with single-stranded DNA and differentially regulates early gene expression of the human neurotropic JC virus. J. Virol., 66, 6885–6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chowdhury M., Taylor,J.P., Chang,C.-F., Rappaport,J. and Khalili,K. (1992) Evidence that a sequence similar to TAR is important for induction of the JC virus late promoter by human immunodeficiency virus type 1 Tat. J. Virol., 66, 7355–7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chowdhury M., Kundu,M. and Khalili,K. (1993) GC/GA rich sequence confers Tat-responsiveness to human neurotropic virus promoter, JCVL, in cells derived from CNS. Oncogene, 8, 887–892. [PubMed] [Google Scholar]
- 57.Jones K.A. (1997) Taking a new TAK on Tat transactivation. Genes Dev., 11, 2593–2599. [DOI] [PubMed] [Google Scholar]
- 58.Itoh H., Wortman,M.J., Kanovsky,M., Uson,R.R., Gordon,R.E., Alfano,N. and Johnson,E.M. (1998) Alterations in Purα levels and intracellular localization in the CV-1 cell cycle. Cell Growth Differ., 9, 651–665. [PubMed] [Google Scholar]
- 59.Stacey D.W., Hitomi,M., Kanovsky,M., Gan,L. and Johnson,E.M. (1999) Cell cycle arrest and morphological alterations following microinjection of NIH3T3 cells with Purα. Oncogene, 18, 4254–4261. [DOI] [PubMed] [Google Scholar]
- 60.Ma Z.W., Pejovic,T., Najfeld,V., Ward,D.C. and Johnson,E.M. (1995) Localization of PURA, the gene encoding the sequence-specific single-stranded-DNA-binding protein Purα, to chromosome band 5q31. Cytogenet. Cell Genet., 71, 64–67. [DOI] [PubMed] [Google Scholar]
- 61.LeBeau M.M., Espinosa,R., Neuman,W.L., Stock,W., Roulston,D., Larson,R.A., Keinanen,M. and Westbrook,C.A. (1993) Cytogenetic and molecular delineation of the smallest commonly deleted region of chromosome 5 in malignant myeloid diseases. Proc. Natl Acad. Sci. USA, 90, 5484–5488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pedersen B. (1993) 5q–: pathogenic importance of the common deleted region and clinical consequences of the entire deleted segment. Anticancer Res., 13, 1913–1916. [PubMed] [Google Scholar]
- 63.Lezon-Geyda K.A., Najfeld,V. and Johnson,E.M. (1997) The PURA gene, encoding the single-stranded DNA-binding protein Purα, as a marker for 5q31 alteration in myeloproliferative disorders, a potentially early step in induction of AML. FASEB J., 11, A100. [Google Scholar]
- 64.Weinberg RA. (1995) The retinoblastoma protein and cell cycle control. Cell, 81, 323–330. [DOI] [PubMed] [Google Scholar]
- 65.Tretiakova A., Otte,J., Croul,S.E., Kim,J.H., Johnson,E.M., Amini,S. and Khalili,K. (1999) Association of JC virus large T-antigen with myelin basic protein transcription factor (MEF-1/Purα) in hypomyelinated brains of mice transgenically expressing T antigen. J. Virol., 73, 6076–6084. [DOI] [PMC free article] [PubMed] [Google Scholar]