Abstract
The conventional phytopathogen Pseudomonas syringae reportedly possesses several virulence determinants against Caenorhabditis elegans; however, their action mechanisms remain elusive. This study reports the nematicidal activity and action receptor of a methyl-accepting chemotaxis protein (MCP03) of a wild-type P. syringae MB03 against C. elegans. Purified MCP03 exhibited nematicidal toxicity against C. elegans at a half-lethal concentration of 124.4 μg mL−1, alongside detrimental effects on the growth and brood size of C. elegans. Additionally, MCP03-treated worms exhibited severe pathological destruction of the intestine and depressed wrinkles of the cuticle. Yeast two-hybrid assays identified a subunit of COP9 signalosome, namely CSN-5, which functioned as an MCP03 action receptor. In vitro pull-down verified the binding interaction between MCP03 and CSN-5. RNA interference assays confirmed that MCP03 antagonizes CSN-5, thereby adversely affecting the brood size and cuticle integrity of C. elegans. Following MCP03 infection, the expression of genes related to reproduction, growth, and cuticle formation, such as kgb-1, unc-98, and col-117, was considerably downregulated, indicating pathological changes in MCP03-treated nematodes. Therefore, we proposed that MCP03 antagonizes CSN-5, causing lethality as well as detrimental effects on the fertility, growth, and morphogenesis of C. elegans, which can provide new insights into the signaling pathways and mechanisms underlying the nematicidal action of MCP03 toward C. elegans.
Keywords: Pseudomonas syringae, Methyl-accepting chemotaxis protein, Caenorhabditis elegans, Receptor protein, Nematicidal activity
1. Introduction
Plant-parasitic nematodes (PPNs) are a large group of soil-borne roundworms that mainly attack underground plant parts and cause stunted growth, thereby endangering nutrient supply from the soil and causing a first-hand global annual yield loss of >$157 billion [1]. Additionally, these nematodes increase the susceptibility of plant roots to other pathogenic fungal and bacterial infections and serve as vectors for certain plant-pathogenic viruses [2]. Although various chemical nematicides and agricultural management have been the typical implements for controlling PPNs for a long time, integrated biological control has become a prioritized approach in combating PPNs owing to its sustainability and eco-friendly nature [3,4].
Various soil-borne bacteria belonging to several genera, such as Bacillus [5,6], Pseudomonas [7,8], Pasteuria [9], and Burkholderia [10], reportedly exhibit potent nematicidal activity. These bacteria have developed various strategies, including the production of external toxins, invasive enzymes, or other active substances, to trap and kill nematodes and act as external parasites by utilizing extracellular proteases to digest the nematode cuticle [11]. In addition, they can produce nematode-toxic metabolites after entering the nematode body; for instance, the Cry proteins from the nematicidal bacterium Bacillus thuringiensis formed lytic pores in the cell membrane of intestinal epithelial cells upon ingestion [12].
Chemotaxis is a conventional bacterial behavior that allows them to sense chemical cues in their surroundings, thereby enabling them to relocate to favorable niches away from toxic substances. Methyl-accepting chemotaxis proteins (MCPs) are a family of chemoreceptors responsible for the chemotactic behaviors of numerous bacteria [13]. Moreover, chemotaxis is involved in the pathogenicity of various bacterial pathogens, including Vibrio cholera [14], Coronobacter sakazakii [15], Campylobacter jejuni [16], Pseudomonas aeruginosa [16], and Pseudomonas syringae [17]. The connection between chemotaxis and pathogenicity relies on the detection of pathogenicity-related signal molecules in the hosts by MCPs, which play a critical role in regulating certain cellular activities, such as biofilm formation, toxin production, exopolysaccharide production, flagellum biosynthesis, cell survival, motility, pathogenicity, and antibiotic resistance [18].
Recently, the conventional phytopathogen P. syringae has been reported to exhibit nematicidal activity against the free-living model nematode Caenorhabditis elegans [19,20]. In addition, a recent genome-wide prediction analysis also identified several potent nematicidal factors against C. elegans in a P. syringae wild-type strain MB03, including an MCP (namely MCP03) [21]. The current study aims to elucidate the nematicidal activity and action mechanism of MCP03. Because of the current technical limitations of molecular genetic studies of PPNs, C. elegans was employed as a target nematode owing to its well-characterized genetic background [22]. We identified for the first time that a subunit of a COP9 signalosome (CSN) functioned as a receptor protein (namely CSN-5) of MCP03 in C. elegans, which may associatively exert destructive effects on the intestinal tract, exhibited lethal and detrimental effects on egg-laying and growth, and surface cuticles by downregulating the expression of genes related to these activities. Therefore, a putative outline mechanism underlying the nematicidal action of MCP03 following its binding with CSN-5 has been proposed.
2. Materials and methods
2.1. Bacterial and nematode strains, plasmids, and culture conditions
The bacterial strains and plasmids used in this study are listed in Table 1. A wild-type strain of P. syringae MB03 [23] (CCTCC No. M2014114, China Center for Type Culture Collection) provided the gene resource for mcp03. The Escherichia coli strains DH5α, BL21(DE3), and HT115(DE3) were employed for cloning or expressing certain genes using different plasmid vectors, whereas E. coli OP50 cells were used as food for feeding C. elegans unless specified. P. syringae and E. coli cells were routinely cultured at 28 °C and 37 °C in Luria–Bertani (LB) medium [24] supplemented with 100 μg mL−1 ampicillin (Amp), 50 μg mL−1 kanamycin (Kan), or 20 μg mL−1 tetracycline (Tet) when required, respectively. The C. elegans strains used in this study include the wild-type Bristol strain N2, green fluorescence protein (GFP)-labeled transgenic strain FT63 (dlg::gfp), which were obtained from Caenorhabditis Genetics Center (College of Biological Sciences, University of Minnesota, MN55108, USA). The csn-5 silenced strain was obtained via RNA interference (RNAi) assay. All C. elegans strains were cultivated at 25 °C under standard conditions on a nematode growth medium [20] using E. coli OP50 as food. The synchronized worms were prepared according to the standard protocol [25].
Table 1.
Bacterial, yeast and nematode strains, plasmids and oligonucleotide primers used in this study.
| Strains/plasmids/primers | Phenotypes/sequences a | Sources |
|---|---|---|
| Bacterial strains | ||
| P. syringae | ||
| MB03 | A wild-type strain with high ice-nucleating activity | Li et al.23 |
| E. coli | ||
| DH5α | supE44ΔlacU169(Φ80 lacZΔM15) hdsR17 recA1endA1 gyrA96 thi-1 relA1 | Laboratory stock |
| BL21(DE3) | F−ompT hsdS (rB–mB–) dcm + Tetr galλ(DE3) endA The [argU proL Camr] [argU ileY leuW Strep/Specr] | Invitrogen |
| OP50 | Ampr, uracil auxotroph, Food source used for C. elegans | Laboratory stock |
| HT115(DE3) | Tetr, an RNase III-deficient E. coli strain | CGC b |
| MB830 | E. coli BL21(DE3) harboring pGEX-6P-1 | This study |
| MB831 | E. coli BL21(DE3) harboring pMB831 | This study |
| MB833 | E. coli BL21(DE3) harboring pMB833 | This study |
| MB834 | E. coli HT115(DE3) harboring pMB834 | This study |
| Yeasts | ||
| Y2Hgold | MATa, trp1, leu2, ura3, his3, gal4Δ, gal80Δ | Clontech |
| Y187 | MATа, ura3, his3, ade2, trp1, Leu2, gal4Δ, met, gal80Δ | Clontech |
| MB832 | Y2Hgold harboring pMB832 | This study |
| Nematodes | ||
| N2 | A wild-type C. elegans | CGC |
| FT63 (dlg::gfp) | A GFP-labeled transgenic C. elegans expressed (dlg::gfp) in epithelial cell junction | CGC |
| LG333 (skn-1b::gfp) | A GFP-labeled transgenic C. elegans expressed (skn-1b::gfp) | CGC |
| Plasmids | ||
| pET-28a | Kanr, E. coli expression vector, 5369 bp | Novagen |
| pMB831 | Kanr, pET28a derivative harboring mcp03 gene at EcoR I/Xho I site, 7138 bp | This study |
| pGBKT7 | Ampr, pGEX-6P-1 derivative harboring csn-5 gene at EcoR I site, 6000 bp | TaKaRa, Inc. |
| pMB832 | Kanr, pGBKT7 derivative harboring mcp03 gene at Nde I/BamH I sites, 9100 bp | This study |
| pGADT7-csn-5 | Ampr, Y2H expressing csn-5, 9100 bp | This study |
| pGBKT7-53 | Kanr, Y2H expression vector, positive control plasmid, 8300 bp | TaKaRa, Inc. |
| pGBKT7-lam | Kanr, Y2H expression vector, negative control plasmid, 7900 bp | TaKaRa, Inc. |
| pGEX-6P-1 | Ampr, E. coli expression vector, 4900 bp | Novagen |
| pMB833 | Ampr, pGEX-6P-1 derivative harboring csn5 gene at EcoR I site, 6000 bp | This study |
| pL4440 | Ampr, E. coli expression vector, 2800 bp | Takara |
| pMB834 | Ampr, pL4440 derivative harboring csn-5 gene at Hind III site, 3900 bp | This study |
| Oligonucleotide primers c | ||
| F-pET28a-mcp03 | 5c−CCGGAATTCATGCAGACGTTAAAGGC−3′ | |
| R-pET28a-mcp03 | 5′–CCGCTCGAGTTATTTACCCAACAGCAGC–3′ | |
| F-pGBKT7-mcp03 | 5′–CGCCATATGATGCAGACGTTAAAGGCTTTG–3′ | |
| R-pGBKT7-mcp03 | 5′–CGCGGATCCTTATTTACCCAACAGCAGCGC–3′ | |
| F-pGEX-6P-1-csn5 | 5′–CCCCTGGGATCCCCGGAAATGGAAGTTGATACGTC–3′ | |
| R-pGEX-6P-1-csn5 | 5′–CCGCTCGAGTCGACCCGGGTTAAGCATCGGCCATCTC–3′ | |
| F-pL4440-csn5 | 5′–CAGGAATTCGATATCAAGATGGAAGTTGATAACGTC–3′ | |
| R-pL4440-csn5 | 5′–TCGACGGTATCGATAAGTTAAGCATCGGCCATCTC–3′ | |
| For qRT-PCR | ||
| F-GAPDH | 5′–TAACTTCGGTATCATCGAAGGACTC–3′ | |
| R-GAPDH | 5′–GACGGAAACATCTGGTGTAGGGA–3′ | |
| F-csn5 | 5′–TTCGCTCTTCCAGTTGAGGG–3′ | |
| R-csn5 | 5′–CGACCTTCCGTATCGCACAT–3′ | |
| F-col-117 | 5′–GGTGTGCGTCACTCTTCCAA–3′ | |
| R-col-117 | 5′–ACGGTTTCCAGATGGGATGG–3′ | |
| F-mpk-1 | 5′–TGGACTGGGTGCAAGTGAAG–3′ | |
| R-mpk-1 | 5′–GCAACGAGCCATCTGAAACC–3′ | |
| F-kgb-1 | 5′–GACGATGAGGTAAACGCCCC–3′ | |
| R-kgb-1 | 5′–GTGAAAATGTCGTGGTCGGC–3′ | |
| F-unc-98 | 5′–AGCAAGAAGCGAGTCCTACC–3′ | |
| R-unc-98 | 5′–TGCTTCGGCTCGTATCCTTC–3′ | |
Note.
Ampr, ampicillin resistance, Kanr, kanamycin resistance, Tetr, tetracycline resistance..
CGC, Caenorhabditis Genetics Center, College of Biological Sciences, University of Minnesota..
The underlined sequences indicate the restriction enzyme sites..
2.2. Cloning, expression, purification, and labeling of MCP03 protein
The oligonucleotide primers used in this study are listed in Table 1. To construct the recombinant plasmid pMB831 (Fig. 1B), the mcp03 gene was amplified via polymerase chain reaction (PCR) from the P. syringae MB03 genome (GenBank accession No. NZ_LAGV00000000.1) using the primers F-pET28a-mcp03 and R-pET28a-mcp03. The amplified fragment was inserted into the Xho I and EcoR I sites of the E. coli expression vector pET28a to generate pMB831. This recombinant plasmid was subsequently transformed into E. coli BL21(DE3) to yield the recombinant strain MB831.
Fig. 1.
Expression, structural organization, and nematicidal activity of MCP03. (A) Conserved domain analysis of MCP03. (B) Schematic representation of the construction of plasmid pMB831 expressing the mcp03 gene. (C) SDS–PAGE analysis of whole-cell lysate of recombinant E. coli MB831, E. coli BL21(DE3) was used as the negative control. (D) Whole-cell nematicidal activity of E. coli strain MB831 against C. elegans. P. syringae MB03 and E. coli BL21(DE3) were used as the positive and negative control, respectively. (*** indicate p < 0.001). (E) SDS–PAGE and (F) Western blot analysis of purified MCP03 protein. (G) Nematicidal activity assay of purified MCP03 protein against C. elegans. Error bars represent the standard deviations from the mean of three independent experiments.
To induce MCP03 expression, E. coli MB831 cells were cultured in LB broth with Kan at 37 °C until the cell optical density at 600 nm (OD600) reached 0.6, following which 0.2 mmol L−1 isopropyl-β-d-thiogalactopyranoside (IPTG) was added to the cultures, which were further incubated at 30 °C for 6 h. The cells were harvested through centrifugation (5000 rpm for 10 min), resuspended in phosphate-buffered saline (PBS, pH 7.4), and homogenized using a high-pressure homogenizer (NS1001L 2 K, Niro Soavi, Germany) at 1000 psi. The supernatants were collected through centrifugation (12,000 rpm for 10 min at 4 °C). The MCP03 protein was purified from the supernatants using a nickel nitrilotriacetic acid spin column (Qiagen). The protein samples were subsequently solubilized using 20 mmol L−1 HEPES (pH 6.0) and stored at −80 °C following quantification [26]. Finally, the purified MCP03 was labeled using N-hydroxysuccinimide-rhodamine (Rho) (Pierce 46,102) [27].
2.3. Nematicidal bioassays
The whole-cell bioassay was performed to identify the nematicidal potential of the MCP03 protein [20]. The expression of the MCP03 protein in the recombinant E. coli MB831 cells was induced according to the protocol described in Section 2.2. In each well of a 96-well microtiter plate, 40 μL of recombinant E. coli cells with an OD600 of 0.7, 5 μL of a worm solution (30–50 worms), 5 μL of 8 mmol L−1 FUDR (5-fluorodeoxyuridine), 0.6 μL of 10 mg mL−1 chloramphenicol, and S medium were added up to a total volume of 200 μL. The assay plates were then incubated at 25 °C for 5 days. Three replicates were used for each tested sample, and a minimum of three independent experiments were performed. The E. coli BL21(DE3) and P. syringae MB03 cells were used as negative and positive controls, respectively, in this assay. Experimental nematodes were scored five days after worm deposition and were considered dead when they did not respond to prodding with a platinum wire under a stereomicroscope (Olympus IX73, Tokyo, Japan).
The nematicidal bioassays and the LC50 determination of the purified MCP03 protein against C. elegans were performed following a previously established method [20]. Briefly, the purified MCP03 sample was tested at different concentrations (45, 90, 135, 180, and 225 μg mL−1). In each well, we added 10 μL of purified MCP03 protein, 5 μL of a worm solution (30–50 worms), 5 μL of 8 mmol L−1 FUDR (5-fluorodeoxyuridine), 1 μL of 50 mg mL−1 Kan, and S medium supplemented with E. coli OP50 as a food source up to a total volume of 200 μL. Each concentration was tested in triplicate, with a minimum of three repetitions. The microtiter plates were sealed with parafilm to maintain humidity, and the assay was conducted at 25 °C. Furthermore, the death of the worms was recorded after 5 days.
2.4. Growth inhibition and brood size bioassays
The growth inhibition assay was performed using synchronized L1-stage C. elegans [20]. Briefly, 10 μL E. coli OP50, MCP03 protein at different concentrations (40, 60, 80, 100, and 120 μg mL−1), 1 μL 50 μg mL−1 Kanamycin, 5 μL L1 stage larvae of C. elegans (30–50 worms), and S medium were added to each well of a 96-well microtiter plate. For a negative control, protein solution was substituted with S medium. Four independent wells and three biological replicates were used for each protein concentration. The worms were consequently incubated for 3 days at 25 °C. First, the nematodes were anesthetized using 15 mmol L−1 sodium azide. Then, a minimum of 20 worms were photographed at 100 × magnification using an Olympus digital camera on a microscope (Olympus IX73, Tokyo, Japan) for each concentration. The lengths of these worms were subsequently determined using NIH Image J1.33 software. Finally, we plotted a comparative graph representing the average worm lengths against those of the control group.
The brood size of C. elegans was measured in a liquid-based 96-well microtiter plate assay. Each well contained 5 μL E. coli OP50 suspension in S medium at an OD600 of 0.2, purified MCP03 protein at different concentrations (20, 40, 60, 80, 100, and 120 μg mL−1), a single L4 stage worm, and S medium up to the final volume of 120 μL. Each protein concentration was assayed in four wells with a minimum of three repetitions. Finally, the eggs in each well were counted under an inverted microscope (Olympus IX73, Tokyo, Japan).
2.5. Analysis of C. elegans intestinal pathology
The wild-type C. elegans N2 and the transgenic C. elegans FT63 (DLG::GFP) were fed with Rho-labeled MCP03 and unlabeled MCP03, respectively. Bovine serum albumin (BSA) was used as the negative control. After 72 h of incubation, the worms were rendered unconscious using a 15 mmol L−1 sodium azide and fixed on a microscopic slide coated with 2 % agarose for imaging. The worms were examined using a fluorescence microscope (80i, Nikon, Japan) in two different modes: green fluorescence (GFP) and red fluorescence (RFP).
2.6. Yeast two-hybrid assay
The yeast two-hybrid (Y2H) assays were performed to screen the potential receptor of MCP03 following the standard protocol [28], as shown in Supplementary Fig. S1. A cDNA library from C. elegans was constructed using the plasmid vector pGADT7 and transformed into the yeast strain Y187. Full-length MCP03 was fused to the GAL4 DNA binding domain in the vector pGBKT7 to yield the recombinant plasmid pMB832 (Supplementary Fig. S2) and was transformed into the yeast host strain Y2H Gold to yield the recombinant MB832. Two-hybrid interactions were screened by mating MB832 with the A.D. fusion C. elegans cDNA library. Blue colonies on SD/-Trp-Leu medium supplemented with X-α Gal (X-alpha galactosidase) and AbA (Aureobasidin A) were further analyzed on SD/-Trp-Leu-His-Ade/X-α Gal/AbA (QDO/X/A) medium. The resultant clones that appeared on the QDO/X/A medium were subjected to colony PCR using specific primers to identify the interacting partners. The resultant csn-5 was cloned into the vector pGADT7, and the interaction with MCP03 was tested in a Y2H X- α gal assay [28].
2.7. Expression and purification of recombinant protein from C. elegans
Total RNA was extracted from the wild-type strain N2 of C. elegans using the Trizol method [29]. Subsequently, cDNA synthesis was performed using the TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen Biotech). Furthermore, csn-5 was amplified using the F-pGEX-6P-1-csn-5 and R-pGEX-6P-1-csn-5 primers (Table 1) and then cloned into the expression vector pGEX-6P-1 using the Gibson one-step assembly method [30] to generate the recombinant pMB833 (Supplementary Fig. S3), which was transformed into E. coli BL21(DE3) to yield recombinant E. coli MB833. For the induction of CSN-5 expression, MB833 cells were cultured in LB broth until an OD600 of 0.6 at 37 °C was reached, following which 0.2 mmol L−1 IPTG was added, and the cultures were further incubated for 4 h at 30 °C. The recombinant protein was purified using affinity chromatography with glutathione Sepharose 4 B. The eluted protein was subsequently dialyzed and stored at −80 °C.
2.8. Scanning electron microscope
The worm surface morphologies of C. elegans N2, MCP03-treated N2, and N2 CSN-5 RNAi were observed under a scanning electron microscope (SEM, JSM-6390/LV, JEOL, Japan) according to the previously described sample preparation and fixation procedures [31].
2.9. Pull-down assay
E. coli MB831 and MB833 expressing His-tag fused MCP03 and glutathione S-transferase (GST) tagged CSN-5, respectively, and E. coli MB830 harboring plasmid pGEX-6P-1 (as the negative control) were IPTG-induced according to the above protocols. The cells were harvested via centrifugation at 12,000 rpm for 10 min at 4 °C, resuspended with PBS (pH 7.4), and homogenized using a high-pressure homogenizer (NS1001L 2 K, Niro Soavi, Germany) at 1000 psi. Following centrifugation of the disrupted suspensions, the supernatants from MB831/MB833 and MB831/MB830 (the negative control) were grouped and incubated with glutathione-coupled Sepharose beads in a rotative incubator for 3 h at 4 °C, following which they were centrifuged. The beads were washed with precooled (4 °C) PBS (pH 7.4) at least five times to elute unbound proteins. The bead-bound proteins were detected using 12.5 % sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and Western blot analysis using an anti-His-tag antibody, according to the standard protocols [24].
2.10. Western blot analysis
To analyze the expression of MCP03 in E. coli MB831 and the binding interaction between MCP03 and CSN-5, after the separation of total proteins via 10 % and 12.5 % SDS–PAGE, respectively, the gels were electro-transferred to a nitrocellulose membrane and incubated overnight with a 1:11,000 dilution of anti-His-tag antibody, followed by incubation with an HRP-conjugated secondary antibody at a 1:1500 dilution. The membrane was then visualized using an enhanced chemiluminescence substrate as described by the manufacturer (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
2.11. Construction of csn-5 RNAi strain
An RNAi strain (namely E. coli MB834) was constructed and used to silence csn-5 expression in C. elegans N2 following a previously described protocol [32]. The csn-5 fragment was PCR-amplified from the cDNA of C. elegans N2 using the primers F-RNAi.csn-5 and R-RNAi.csn-5 (Table 1). The amplified fragment was subsequently digested using Hind III and cloned into an RNAi plasmid vector pL4440 to yield recombinant pMB834 (Supplementary Fig. S4A), which was transformed into E. coli HT115 to generate E. coli MB834. The expression of dsRNA in MB834 was induced using 1 mmol L−1 IPTG at the final concentration (Supplementary Fig. S4B).
2.12. Real-time quantitative PCR
Synchronized C. elegans N2 L4 stage worms were fed with purified MCP03 or BSA (negative control). The worms were collected 24 h after feeding and used for extracting total RNAs according to a previously reported protocol [29]. Real-time quantitative PCR (RT-qPCR) was performed to determine the expression levels of selected C. elegans genes using the comparative cycle threshold method (2–△△C.T. method) [33]. The primers used for RT-qPCR analysis of the selected genes are listed in Table 1. GAPDH was used as an internal reference gene, while RNAs from BSA-fed C. elegans N2 were used as the negative control.
2.13. Data analysis
Statistical analysis was performed using GraphPad Prism 8.3 (GraphPad Software, LLC, Boston, MA, USA), and data were derived from at least three biological replicates unless otherwise indicated. Statistical significance was defined as p < 0.05.
3. Results
3.1. Molecular characterization, expression and purification, and nematicidal activity of MCP03
The gene mcp03 (gene locus “VT47_10,710”) from the genome of P. syringae MB03 was characterized using the online tool BLASTn in the GenBank nucleotide sequence database at the National Center for Biotechnology Information server (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The MCP03 protein was predicted to encode a 598 amino acid (AA) protein with a theoretical molecular weight (Mw) of ∼66 kDa. Conserved domain architecture analysis revealed two domains of MCP03: a HAMP domain, spanning AAs 93–149, and an MCP signal domain, spanning AAs 154–424 (Fig. 1A). The gene mcp03 was amplified from the P. syringae MB03 genome and was expressed in E. coli MB831 via constructing an expression plasmid pMB831 (Fig. 1B). Fig. 1C shows that an additional band with a molecular mass equal to that of the predicted MCP03 was present in the E. coli MB831 profile (indicated by the arrow). However, this was not found in the BL21(DE3) profile (the negative control), indicating the successful expression of MCP03 in E. coli MB831 cells. Subsequently, a liquid-based bioassay was performed to evaluate the virulence potential of E. coli MB831 intact cells toward C. elegans. MB831 exhibited significant (p < 0.001) nematicidal activity against C. elegans compared with that shown by the negative control E. coli BL21(DE3) cells (P. syringae MB03 intact cells were used as a positive control) (Fig. 1D). Therefore, the MCP03 protein expressed in MB831 was purified, and SDS–PAGE and Western blot analyses confirmed that MCP03 was purified as a single protein component at high purity (Fig. 1E & F). We further evaluated the nematicidal activity of the purified MCP03 against C. elegans. Fig. 1G shows that the purified MCP03 exhibited pronounced nematicidal activity against C. elegans in a dose-dependent pattern, with a calculated LC50 value of 124.47 (99.22−147.47) μg mL−1, indicating the high toxicity of this protein against C. elegans.
3.2. MCP03 affects different phenotypes of C. elegans
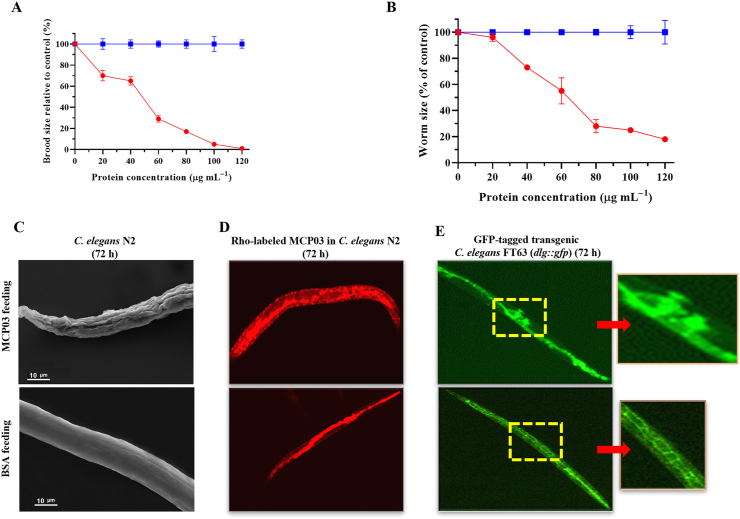
The virulence of purified MCP03 protein was assessed by investigating its effects on the spawning number and growth of C. elegans N2, using BSA as the negative control. Fig. 2A shows the detrimental effect of MCP03 on the brood size of worms following a dose increase from 0 to 120 μg mL−1, and the spawning rate declined to 0 at a dose of 120 μg mL−1, indicating the pathogen-induced impairment of the reproductive system of C. elegans N2 when fed MCP03. Moreover, Fig. 2B shows that low MCP03 concentrations (<20 μg mL−1) exerted only a slight repressive effect on the size of the synchronized L1-stage C. elegans worms; however, higher MCP03 concentrations induced a greater decrease in worm size, and at 120 μg mL−1 of MCP03, the area of the worm was only 20 % compared to the control BSA, thereby indicating a dose-dependent detrimental effect of MCP03 on the growth of C. elegans.
Fig. 2.
Detrimental effects of MCP03 on C. elegans N2. (A) Brood size assay of C. elegans under different concentrations of MCP03. (B) Worm size assay of C. elegans treated with different doses of MCP03. (C) SEM micrograph of the surface morphology of C. elegans after feeding with MCP03 or BSA. (D) Fluorescence micrograph of C. elegans N2 fed with Rho-labeled MCP03 (Magnification: 400×). Rho-labeled BSA was used as the negative control. In the MCP03-treated worm, red fluorescence was distributed throughout the entire body, while the control N2 fed with Rho-labeled BSA displayed red fluorescence confined to only the intestinal tract. (E) Fluorescence micrograph of GFP-tagged C. elegans FT63 (dlg::gfp) fed with MCP03 (Magnification: 400×). The yellow-dotted rectangle in the treated nematode micrograph highlights the area of intestinal tissue damage, indicating blurry and fragmented GFP fluorescence. Conversely, the intestine of the control worm fed with BSA exhibited an integrated, intact bamboo-shaped intestinal tract. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Interestingly, MCP03 also demonstrated disruptive impairment on the surface cuticles of C. elegans N2, as severe depressed dents and wrinkles of the cuticles were clearly visualized under SEM on the surface phenotype of worms fed with MCP03 in 72 h, which strongly contrasted with the smooth surface of worms fed with BSA (Fig. 2C).
To investigate whether MCP03 acted on the intestinal tract of worms, we examined the intestinal morphology of two C. elegans worms that were fed with Rho-labeled or un-labeled MCP03 after 72 h: the wild-type N2 and GFP-tagged FT63 (dlg::gfp) respectively. Fig. 2D shows that the intestinal tract of N2 treated with Rho-labeled MCP03 appeared to be pathologically altered under the upper CLSM micrography, as disseminative red fluorescence was distributed throughout the entire worm body, while the control N2 fed with Rho-labeled BSA displayed red fluorescence confined to only the intestinal tract. Moreover, the destructive effect of MCP03 on the integrity of the epithelial junctions of worms was confirmed using MCP03-treated FT63(dlg::gfp) worms as substantial disruption of epithelial junctions was visible via upper CLSM micrography (Fig. 2E) (indicated by the yellow dotted–rectangle region). Conversely, the control worm fed with BSA exhibited an integrated, intact, bamboo-shaped intestinal tract.
3.3. Identification of CSN-5 as a putative receptor of MCP03
To identify the receptor protein interacting with MCP03 to approximately actuate its toxicity against C. elegans N2, we performed an N2 genome-wide Y2H screening using MCP03 as the bait. We first performed the control experiments; the positive controls, pGBKT7-53 and pGADT7-T, exhibited interaction on the DDO/X/A plate (Supplementary Fig. S5A), validating the assay's sensitivity. Meanwhile, the negative control, involving pGADT7-T and pGBKT7-Lam, demonstrated the absence of interaction, confirmed by the lack of growth on the DDO/X/A plate (Supplementary Fig. S5B). Fig. 3A shows a typical trilobite pattern of mating yeast cells, indicating the successful mating of MB832 and the Y187 Y2H library cells carrying various fractions of C. elegans cDNA. Fig. 3B shows blue colonies on QDO/X/A plates, indicating the presence of potential prey proteins interacting with MCP03 in these colonies. Therefore, these four colonies were subjected to PCR amplification of their harbored prey cDNAs, revealing that all four prey cDNA fragments were of similar size in the electrophoresis profiles (∼1100 bps) (Fig. 3C). These fragments were sequenced and found to be an identical ORF comprising 1107-bp nucleotide sequences, which was further identified as the csn-5 gene of C. elegans by searching the WormBase database (https://wormbase.org//). CSN-5 was the exclusive candidate receptor in our yeast two-hybrid screen, signifying a specific and substantial interaction within our experimental framework. Notably, the limited number of hits may result from variations in screening conditions or bait selection. The predicted protein CSN-5 comprises 369 AA with a theoretical Mw of ∼67 kDa and is a key subunit of the COP9 signalosome. This conserved protein complex plays a critical role in regulating gene expression, cell proliferation, and eukaryotic cell cycle [34]. Conserved domain architecture analysis revealed that CSN-5 possesses an MPN superfamily domain spanning AA45–AA292. Three motifs within this domain were identified, namely Mov34/MPN/JAMM, as well as a zinc-binding site with metalloproteinase activity (Supplementary Fig. S6A). The three-dimensional structure of CSN-5 exhibits a compact architecture characterized by multiple α-helices and β-sheets that form a barrel fold (Supplementary Fig. S6B).
Fig. 3.
Yeast two-hybrid assay for the identification of the MCP03-interacting receptor. (A) Micrograph of the mating yeast cells. The red arrow indicates a typically successful trilobite mating pattern. In (B), (a)–(d) indicate positive colonies in the preliminary screening on the QDO/X/A medium plate. (C) Electrophoresis of PCR-amplified plasmid cDNAs from positive colonies. Lane M, DNA Mw marker. Lane (a)–(d), PCR-amplified products from the colony a–d on (B). (D) Yeast response validation on CSN-5 by cotransformation of bait plasmid pMB832 and prey plasmid pGADT7-csn-5 into Y187. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.4. Binding interaction of MCP03 with CSN-5
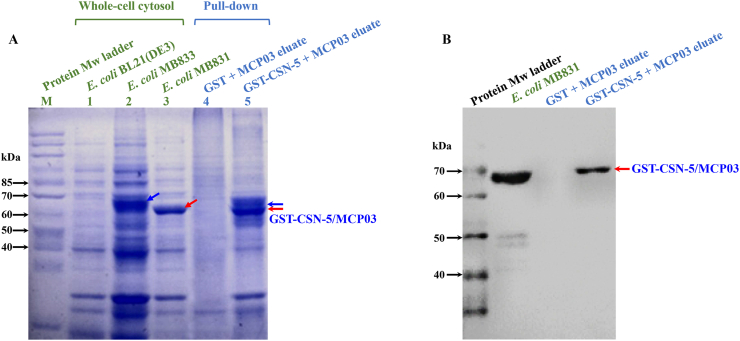
An in vivo yeast response validation of CSN-5 via the cotransformation of bait plasmid pMB832 and prey plasmid pGADT7-csn-5 into Y187 revealed a positive colony of CSN-5 (Fig. 3D), thereby verifying the binding between CSN-5 and MCP03. In vitro, pull-down affinity chromatography assay and Western blot analysis were also performed to confirm the binding activity of the purified GST-tagged CSN-5 protein (GST–CSN-5) and MCP03. Fig. 4A shows that GST–CSN-5 was successfully expressed in E. coli MB833, with the predicted Mw of ∼67 kDa (Fig. 4A, lane 2 indicated by blue arrow), while His-tagged MCP03 was expressed in MB831 at the Mw of ∼66 kDa (Fig. 4A, lane 3 indicated by red arrow). Following coincubation of the purified GST–CSN-5 and MCP03, they formed a binding complex that was identified in the SDS–PAGE profile (Fig. 4A, lane 5). Conversely, no binding complex was detected in the coincubation of GST and MCP03 alone (Fig. 4A, lane 4). Western blot analysis using a specific anti-His antibody further visualized a ∼67 kDa positive band in the GST–CSN-5/MCP03 complex, slightly higher than that of the control MCP03 at ∼66 kDa, which appeared to be caused by the complex formed with GST–CSN-5 at a relatively higher Mw (Fig. 4B). Overall, these results confirm the binding activity between MCP03 and CSN-5.
Fig. 4.
Binding interaction between MCP03 and CSN-5. (A) SDS–PAGE analysis of recombinant E. coli cells expressing CSN-5 and in vitro pull-down assay of MCP03 and CSN-5 interaction. (B) Western blot analysis of the pull-down fractions.
3.5. Effects of silencing csn-5 by RNAi on brood size, growth size, and cuticle integrity
RNAi experiments were conducted to silence csn-5 in C. elegans N2 (the RNAi-treated C. elegans N2 was named “N2-CSN-5-RNAi”). The growth, brood size, and cuticle morphology of N2-CSN-5-RNAi were then compared with N2. The csn-5 gene of N2 was cloned to pL4440 to construct pMB834, and a 1273-bp dsRNA specific for csn-5 RNAi was synthesized upon IPTG induction in E. coli MB834 cells harboring pMB834 (Supplementary Fig. S4A). The C. elegans N2-CSN-5-RNAi worms were yielded by feeding the dsRNA to N2. Fig. 5 shows only a limited effect of csn-5 RNAi on the growth of N2 (Fig. 5A) and the average individual size of twenty worms (Fig. 5B), indicating that CSN-5 did not significantly regulate the growth of C. elegans (p > 0.05). However, the RNAi on csn-5 caused a significant inhibitory effect on the brood size of N2-CSN-5-RNAi compared to N2, as the N2-CSN-5-RNAi worms laid approximately 65 % fewer eggs compared with that of the wild-type N2 (p < 0.05) (Fig. 5C), indicating that CSN-5 served as the action receptor of MCP03 in suppressing the reproduction system of C. elegans. Moreover, the csn-5 RNAi caused morphological changes on the surface cuticles of worms, as the crumples and depressed dents of the cuticles were visible in the N2-CSN-5 RNAi worms (Fig. 5D, indicated by the red arrow), consistent with the similar pathological pattern of MCP03 treatment (Fig. 5D), further identifying that CSN-5 is a functional action receptor of MCP03.
Fig. 5.
Effects of csn-5 RNAi on the worm size, brood size, and surface cuticle of C. elegans N2. (A) Microscopic examination of the wild-type and csn-5 RNAi-treated C. elegans N2 worms (Magnification: 40×). (B) Worm size quantification of the wild-type and csn-5 RNAi-treated C. elegans. CSN-5 did not significantly (p > 0.05) regulate the growth of C. elegans. (C) Quantification of the average spawning number of the wild-type and csn-5 RNAi-treated C. elegans. CSN-5 caused a significant (p < 0.05) inhibitory effect on the brood size of N2-CSN-5-RNAi compared to N2. (D) SEM micrograph of the csn-5 RNAi-C. elegans worms. The wild-type N2 worms fed with BSA and MCP03 were used as the negative and positive controls, respectively. The red arrow indicates the depressed dent of the worm surface. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.6. RT-qPCR analysis of selected genes of C. elegans N2
To investigate the effects of MCP03 on C. elegans at the molecular level, we conducted RT-qPCR analysis on a subset of genes associated with growth, brood size, and cuticle formation of the worm. Specifically, we analyzed the expression levels of col-117, which encodes collagen belonging to the Col-3 family and serves as the main component of stratum corneum on the C. elegans surface [35], kgb-1, a gene involved in regulating the reproductive function of C. elegans [36], unc-98, a gene involved in the assembly and maintenance of myofibrils [37,38], and mpk-1, a gene involved in regulating the resveratrol-mediated longevity of C. elegans through MPK-1 signaling pathways [39]. Fig. 6 shows that after feeding MCP03 for 24 h, the expression levels of col-117, kgb-1, and unc-98 in C. elegans N2 were substantially downregulated, those of mpk-1 were significantly upregulated, and those of csn-5 remained unchanged. The downregulation of col-117 expression is consistent with the observed depressed morphology of stratum corneum on the worm surface after feeding MCP03 (Fig. 5D); the downregulation of kgb-1 affected the fertility of worms, as identified previously because KGB-1 and CSN-5 interact with GLH-1 and regulate the reproductive function of nematodes [38]. In addition, the downregulation of unc-98 affected the formation of C. elegans muscle tissue [40]. These results suggest that MCP03 has a significant impact on the expression levels of key genes associated with C. elegans cuticle formation, reproduction, growth, and muscle tissue development, elucidating the molecular mechanisms underlying its effects on these physiological processes.
Fig. 6.
RT-qPCR of expression activities of several selected genes related to fertility, growth, and cuticle formation of C. elegans.
4. Discussion
This study aimed to investigate the nematicidal activity and underlying action mechanism of MCP03 in the P. syringae MB03-C. elegans infection model. We evaluated the direct nematicidal activity of MCP03 against C. elegans and its detrimental effects on the growth, brood size, and external and internal morphology of C. elegans. Furthermore, we identified CSN-5 as an MCP03-binding protein in C. elegans. Although the role of MCPs as key players in chemotaxis and pathogenicity has been well-characterized in different bacteria [41], to the best of our knowledge, this study was the first to provide new insights into the nematicidal activity and action mechanism of a bacterial MCP against nematodes.
In the P. syringae MB03 genome, 46 MCP-encoding genes (chemoreceptor genes) were annotated. Notably, the genome-wide prediction of nematicidal genes of P. syringae MB03 using VirulentPred and a liquid fast-killing assay using recombinant E. coli cells revealed the nematicidal potential of MCP03 among these MCPs [21]. Consistent with this prediction, the bioassays of heterologously expressed MCP03 exhibited lethal activity against C. elegans, with an LC50 of 124.47 (99.22–147.47) μg mL−1, and multiple detrimental effects on the growth, reproduction, and morphology of C. elegans. Although the activity is relatively lower than that of Cry5Da1, a well-known eminent bacterial nematicidal toxin of B. thuringiensis with an LC50 of 36.69 μg mL−1 against C. elegans [42], given the multifaceted nematode-toxic activities, MCP03 still holds promise as a potential nematode-pest control agent for agricultural, horticultural, or forestry applications.
CSN-5 is a gene in C. elegans that encodes a component of the COP9 signalosome (CSN) complex. C. elegans contains seven CSN subunits (CSN 1, 2, 3, 4, 5, 6, and 7), which possess proteasome component domains that promote protein–protein interactions and have nucleic acid–binding properties [43]. Conversely, CSN-5 and CSN-6 possess an MPN domain with a JAMM (Jab/MPN/Mov34) sequence and exhibit metalloproteinase activity [44]. The three-dimensional structure of CSN-5 exhibits a compact architecture characterized by multiple α-helices and β-sheets that form a barrel fold responsible for mediating protein–protein interactions (Supplementary Fig. S6B) [45]. Notably, CSN-5 is also involved in ubiquitin-dependent protein degradation, cell cycle regulation, and multiple signal transduction processes [46]. This suggests its potential role in mediating crucial protein–protein interactions central to these cellular functions. Herein, CSN-5 was identified as the binding receptor of MCP03 through two distinct in vitro experiments. Owing to the unavailability of the CSN-5:GFP strain of C. elegans, we were unable to identify the in vivo interaction between MCP03 and CSN-5 proteins in this study as constructing such a strain was not feasible within the constraints of our laboratory resources and it was also not available through CGC. The data shown in Fig. 2 indicate that MCP03 targeted the intestinal tissues of N2 and caused severe destructive impairment of the integrity of epithelial junctions. We believe that these pathological changes could be the main lethal determinants of MCP03 treatment as these changes are associated with other stepwise pathological processes, such as decreased food intake, substance, and energy metabolism collapse, and septicemia, which ultimately lead to the death of C. elegans. Evaluating the impact of MCP03 treatment on C. elegans intestinal permeability at sequential time points would provide valuable insights into anatomical changes of the intestine in correlation with variations in worm size. However, whether these pathological processes were related to the binding of MCP03 and CSN-5 remains unknown. Therefore, further studies are warranted to include the expression of CSN5 in RNAi knockdown C. elegans to demonstrate if the nematicidal activity of MCP03 is restored. Additionally, the changes in the structure and permeability of the intestines after the suppression of CSN5 could be tested to explore the role and action mechanism of the MCP03-CSN-5 binding complex in the pathogenicity of MCP03.
Several investigations have demonstrated that CSN-5 interacts with UNC-98 and UNC-96, which are involved in the assembly and maintenance of myofibers [37]. The outer surface of MCP03-treated N2 severely shrank (Fig. 2C), col-117 and unc-98 expressions were significantly downregulated (Fig. 6), and the cuticle of worms subjected to CSN-5 RNAi silencing exhibits a pathological pattern consistent with the effects observed in worms treated with MCP03 (Fig. 5D). These results suggest that MCP03 interacts with CSN-5 and downregulates the expression of collagen and certain muscle-related proteins, causing severely depressed wrinkles on the surface of the worms. In addition, Fig. 2A illustrates that MCP03 had a substantial inhibitory effect on the reproductive capacity of N2, leading to a significant reduction in brood size. RNAi with csn-5 significantly reduced the spawning rate of N2 (Fig. 5C), confirming the critical role of CSN-5 in regulating the fertility activity of C. elegans. This could be attributed to the downregulation of kgb-1 expression, as CSN-5 and KGB-1 can regulate the expression of GLH-1, which is crucial for the fertility of C. elegans [36]. Notably, the expression of csn-5 in N2 remained unaltered following MCP03 treatment. We speculate that MCP03 was bound to CSN-5 and inhibited its activity to some extent without significantly reducing the expression of the protein. However, MCP03 may interact with other undetected receptors in C. elegans, potentially because of limitations in the coverage of the cDNA library and screening processes employed in this study.
Based on the results obtained in this study, we propose a few outlines for the pathogenicity of MCP03 against C. elegans (Fig. 7). When MCP03 infects C. elegans worms, it impacts their epidermal layer, reducing collagen expression and deteriorating the stratum corneum. DAF-16 is positively regulated when worms are infected, and the epidermis is damaged [47]. Consequently, the nematodes detect external threats, which trigger their immune response and upregulate the expression of MPK-1, which promotes resveratrol-mediated nematode lifespan in a non-dependent manner through the SIR-2.1/DAF-16 pathway (closely related to regulating nematode lifespan and immunity) by regulating SKN-1 accumulation in the nucleus [39]. Furthermore, MCP03 interacts with CSN-5 gene within the nematode, which exerts an inhibitory effect on CSN-5 activity, which subsequently impacts various aspects of nematode physiology, notably the fertility-related gene glh-1 that interacts with CSN-5 and the unc-98 and unc-96 genes that are associated with myofibril function. Consequently, the egg-laying rate of nematodes decreases, and their outer surface undergoes shrinkage. It is important to highlight the involvement of CSN-5 within nematodes in mediating ubiquitin-dependent protein degradation. Cellular protein degradation predominantly relies on the ubiquitin–proteasome system. The disruption of this system can result in severe consequences, including cell cancer or necrosis. After feeding on MCP03, nematodes also experience certain intestinal damage, which may be linked to the CSN-5 receptor protein. However, the precise underlying mechanisms of this intestinal damage warrant further research for a comprehensive understanding.
Fig. 7.
The proposed action mechanism underlying MCP03 pathogenicity against C. elegans.
5. Conclusions
The present study demonstrates the nematicidal activity of MCP03 against C. elegans and the detrimental effects on intestinal tissues, fertility capability, growth, and surface cuticle morphology. Y2H assays identified a subunit of the CSN signaling complex, CSN-5, as an action receptor of MCP03. The binding interaction between MCP03 and CSN-5 was confirmed through two distinct in vitro experiments such as Yeast two-hybrid and pull-down assays. Following the MCP03 treatment, several genes related to fertility, growth, and cuticle formation were found to be downregulated in N2 worms. Owing to its relatively high nematicidal virulence and multiple detrimental activities, MCP03 has the potential to be pursued for developing a bionematicide.
Data availability statement
Data will be made available upon request.
CRediT authorship contribution statement
Jiaoqing Li: Writing – original draft, Validation, Methodology, Investigation, Funding acquisition, Data curation, Conceptualization. Haiyan Dai: Writing – original draft, Validation, Methodology, Investigation, Formal analysis, Data curation. Anum Bashir: Writing – original draft, Methodology, Investigation, Formal analysis, Data curation. Zhiyong Wang: Methodology, Investigation. Yimin An: Formal analysis, Data curation. Xun Yu: Writing – review & editing, Investigation. Liangzheng Xu: Supervision, Conceptualization. Lin Li: Writing – review & editing, Supervision, Funding acquisition, Data curation, Conceptualization.
Declaration of competing interest
The authors declare no competing financial interests.
Acknowledgments
This work was funded by a grant from Meizhou City's 2021 Guangdong Provincial Rural Revitalization Strategy Special Fund (Grant no. 2021A0305002), a grant from Guangdong Provincial Science and Technology Innovation Strategy Special Fund (Grant no. KTP20210213), and a grant from Guangdong Pomelo Engineering Technology Development Center, Guangdong Provincial Key Scientific Research Platform Construction Project of Regular University (Grant no. 2019GCZX007).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e30366.
Contributor Information
Liangzheng Xu, Email: xlzheng@jyu.edu.cn.
Lin Li, Email: lilin@mail.hzau.edu.cn.
Appendix B. Supplementary data
The following are the Supplementary data to this article.
References
- 1.Abad P., Gouzy J., Aury J.M., Castagnone-Sereno P., Danchin E.G.J., Deleury E., Perfus-Barbeoch L., Anthouard V., Artiguenave F., Blok V.C., Caillaud M.C., Coutinho P.M., Dasilva C., De Luca F., Deau F., Esquibet M., Flutre T., Goldstone J.V., Hamamouch N., Hewezi T., Jaillon O., Jubin C., Leonetti P., Magliano M., Maier T.R., Markov G.V., McVeigh P., Pesole G., Poulain J., Robinson-Rechavi M., Sallet E., Ségurens B., Steinbach D., Tytgat T., Ugarte E., van Ghelder C., Veronico P., Baum T.J., Blaxter M., Bleve-Zacheo T., Davis E.L., Ewbank J.J., Favery B., Grenier E., Henrissat B., Jones J.T., Laudet V., Maule A.G., Quesneville H., Rosso M.N., Schiex T., Smant G., Weissenbach J., Wincker P. Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nat. Biotechnol. 2017;26:909–915. doi: 10.1038/nbt.1482. [DOI] [PubMed] [Google Scholar]
- 2.Topalović O., Geisen S. Nematodes as suppressors and facilitators of plant performance. New Phytol. 2023;238:2305–2312. doi: 10.1111/nph.18925. [DOI] [PubMed] [Google Scholar]
- 3.Castillo P., Navas-Cortés J.A., Landa B.B., Jiménez-Díaz R.M., Vovlas N. Plant-parasitic nematodes attacking chickpea and their in planta interactions with rhizobia and phytopathogenic fungi. Plant Dis. 2008;92:840–853. doi: 10.1094/PDIS-92-6-0840. [DOI] [PubMed] [Google Scholar]
- 4.Sabbahi R., Hock V., Azzaoui K., Saoiabi S., Hammouti B. A global perspective of entomopathogens as microbial biocontrol agents of insect pests. J. Agric. Food Res. 2022;10 doi: 10.1016/j.jafr.2022.100376. [DOI] [Google Scholar]
- 5.Luo H., Xiong J., Zhou Q., Xia L., Yu Z. The effects of Bacillus thuringiensis Cry6A on the survival, growth, reproduction, locomotion, and behavioral response of Caenorhabditis elegans. Appl. Microbiol. Biotechnol. 2013;97:10135–10142. doi: 10.1007/s00253-013-5249-3. [DOI] [PubMed] [Google Scholar]
- 6.Peng D., Lin J., Huang Q., Zheng W., Liu G., Zheng J., Zhu L., Sun M. A novel metalloproteinase virulence factor is involved in Bacillus thuringiensis pathogenesis in nematodes and insects. Environ. Microbiol. 2016;18:846–862. doi: 10.1111/1462-2920.13069. [DOI] [PubMed] [Google Scholar]
- 7.Dubern J.F., Cigana C., De Simone M., Lazenby J., Juhas M., Schwager S., Bianconi I., Döring G., Eberl L., Williams P., Bragonzi A., Cámara M. Integrated whole‐genome screening for Pseudomonas aeruginosa virulence genes using multiple disease models reveals that pathogenicity is host specific. Environ. Microbiol. 2015;17:4379–4393. doi: 10.1111/1462-2920.12863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun X., Zhang R., Ding M., Liu Y., Li L. Biocontrol of the root-knot nematode Meloidogyne incognita by a nematicidal bacterium Pseudomonas simiae MB751 with cyclic dipeptide. Pest Manag. Sci. 2021;77:4365–4374. doi: 10.1002/ps.6470. [DOI] [PubMed] [Google Scholar]
- 9.Phani V., Shivakumara T.N., Davies K.G., Rao U. Meloidogyne incognita fatty acid- and retinol-binding protein (Mi-FAR-1) affects nematode infection of plant roots and the attachment of Pasteuria penetrans endospores. Front. Microbiol. 2017;8:2122. doi: 10.3389/fmicb.2017.02122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tedesco P., Di Schiavi E., Esposito F.P., de Pascale D. Evaluation of Burkholderia cepacia complex bacteria pathogenicity using Caenorhabditis elegans. Bio. Protoc. 2016;6 doi: 10.21769/BioProtoc.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang X., Tian B., Niu Q., Yang J., Zhang L., Zhang K. An extracellular protease from Brevibacillus laterosporus G4 without parasporal crystals can serve as a pathogenic factor in infection of nematodes. Res. Microbiol. 2005;156:719–727. doi: 10.1016/j.resmic.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 12.Crickmore N. Using worms to better understand how Bacillus thuringiensis kills insects. Trends Microbiol. 2005;13:347–350. doi: 10.1016/j.tim.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 13.Wadhams G.H., Armitage J.P. Making sense of it all: bacterial chemotaxis. Nat. Rev. Mol. Cell Biol. 2004;5:1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- 14.Nishiyama S., Suzuki D., Itoh Y., Suzuki K., Tajima H., Hyakutake A., Homma M., Butler-Wu S.M., Camilli A., Kawagishi I. Mlp24 (McpX) of Vibrio cholerae implicated in pathogenicity functions as a chemoreceptor for multiple amino acids. Infect. Immun. 2012;80:3170–3178. doi: 10.1128/IAI.00039-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi Y., Kim S., Hwang H., Kim K.P., Kang D.H., Ryu S. Plasmid-encoded MCP is involved in virulence, motility, and biofilm formation of Cronobacter sakazakii ATCC 29544. Infect. Immun. 2015;83:197–204. doi: 10.1128/IAI.02633-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McLaughlin H.P., Caly D.L., McCarthy Y., Ryan R.P., Dow J.M. An orphan chemotaxis sensor regulates virulence and antibiotic tolerance in the human pathogen Pseudomonas aeruginosa. PLoS One. 2012;7 doi: 10.1371/journal.pone.0042205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu X., Lund S.P., Scott R.A., Greenwald J.W., Records A.H., Nettleton D., Lindow S.E., Gross D.C., Beattie G.A. Transcriptional responses of Pseudomonas syringae to growth in epiphytic versus apoplastic leaf sites. Proc. Natl. Acad. Sci. U.S.A. 2013;110:E425–E434. doi: 10.1073/pnas.1221892110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matilla M.A., Krell T. The effect of bacterial chemotaxis on host infection and pathogenicity. FEMS Microbiol. Rev. 2018;42 doi: 10.1093/femsre/fux052. [DOI] [PubMed] [Google Scholar]
- 19.Ali M., Sun Y., Xie L., Yu H., Bashir A., Li L. The pathogenicity of Pseudomonas syringae MB03 against Caenorhabditis elegans and the transcriptional response of nematicidal genes upon different nutritional conditions. Front. Microbiol. 2016;7:805. doi: 10.3389/fmicb.2016.00805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bashir A., Sun Y., Yu X., Sun X., Li L. Nematicidal effects of 2-methyl-aconitate isomerase from the phytopathogen Pseudomonas syringae MB03 on the model nematode Caenorhabditis elegans. J. Invertebr. Pathol. 2021;185 doi: 10.1016/j.jip.2021.107669. [DOI] [PubMed] [Google Scholar]
- 21.Ali M., Gu T., Yu X., Bashir A., Wang Z., Sun X., Ashraf N.M., Li L. Identification of the genes of the plant pathogen Pseudomonas syringae MB03 required for the nematicidal activity against Caenorhabditis elegans through an integrated approach. Front. Microbiol. 2022;13 doi: 10.3389/fmicb.2022.826962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaletta T., Hengartner M.O. Finding function in novel targets: C. elegans as a model organism. Nat. Rev. Drug Discov. 2006;5:387–398. doi: 10.1038/nrd2031. [DOI] [PubMed] [Google Scholar]
- 23.Li Q., Yan Q., Chen J., He Y., Wang J., Zhang H., Yu Z., Li L. Molecular characterization of an ice nucleation protein variant (InaQ) from Pseudomonas syringae and the analysis of its transmembrane transport activity in Escherichia coli. Int. J. Biol. Sci. 2012;8:1097–1108. doi: 10.7150/ijbs.4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook J., Russell D.W. third ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y: 2001. Molecular Cloning: a Laboratory Manual. [Google Scholar]
- 25.Porta-de-la-Riva M., Fontrodona L., Villanueva A., Cerón J. Basic Caenorhabditis elegans methods: synchronization and observation. J. Vis. Exp. 2012;64 doi: 10.3791/4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 27.Griffitts J.S., Whitacre J.L., Stevens D.E., Aroian R.V. Bt toxin resistance from loss of a putative carbohydrate-modifying enzyme. Science. 2001;293:860–864. doi: 10.1126/science.1062441. [DOI] [PubMed] [Google Scholar]
- 28.Paiano A., Margiotta A., De Luca M., Bucci C. Yeast two-hybrid assay to identify interacting proteins. Curr Protoc Protein Sci. 2019;95:e70. doi: 10.1002/cpps.70. [DOI] [PubMed] [Google Scholar]
- 29.Green M.R., Sambrook J. J. Total RNA extraction from Caenorhabditis elegans. Cold Spring Harb. Protoc. 2020;2020 doi: 10.1101/pdb.prot101683. [DOI] [PubMed] [Google Scholar]
- 30.Gibson D.G., Young L., Chuang R.Y., Venter J.C., Hutchison C.A., 3rd, Smith H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 31.Zhu J., Cai X., Harris T.L., Gooyit M., Wood M., Lardy M., Janda K.D. Disarming Pseudomonas aeruginosa virulence factor LasB by leveraging a Caenorhabditis elegans infection model. Chem. Biol. 2015;22:483–491. doi: 10.1016/j.chembiol.2015.03.012. [DOI] [PubMed] [Google Scholar]
- 32.Conte D., Jr., MacNeil L.T., Walhout A.J.M., Mello C.C. RNA interference in Caenorhabditis elegans. Curr. Protoc. Mol. Biol. 2015;109:26.23.21–26.23.30. doi: 10.1002/0471142727.mb2603s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 34.Qin N., Xu D., Li J., Deng X.W. COP9 signalosome: discovery, conservation, activity, and function. J. Integr. Plant Biol. 2020;62:90–103. doi: 10.1111/jipb.12903. [DOI] [PubMed] [Google Scholar]
- 35.Broday L., Hauser C.A., Kolotuev I., Ronai Z. Muscle-epidermis interactions affect exoskeleton patterning in Caenorhabditis elegans. Dev. Dyn. 2007;236:3129–3136. doi: 10.1002/dvdy.21341. [DOI] [PubMed] [Google Scholar]
- 36.Gerke P., Keshet A., Mertenskötter A., Paul R.J. The JNK-like MAPK KGB-1 of Caenorhabditis elegans promotes reproduction, lifespan, and gene expressions for protein biosynthesis and germline homeostasis but interferes with hyperosmotic stress tolerance. Cell. Physiol. Biochem. 2014;34:1951–1973. doi: 10.1159/000366392. [DOI] [PubMed] [Google Scholar]
- 37.Gieseler K., Qadota H., Benian G.M. vol. 2017. WormBook; 2017. pp. 1–59. (Development, Structure, and Maintenance of C. elegans Body Wall Muscle). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith P., Leung-Chiu W.M., Montgomery R., Orsborn A., Kuznicki K., Gressman-Coberly E., Mutapcic L., Bennett K. The GLH proteins, Caenorhabditis elegans P granule components, associate with CSN-5 and KGB-1, proteins necessary for fertility, and with ZYX-1, a predicted cytoskeletal protein. Dev. Biol. 2002;251:333–347. doi: 10.1006/dbio.2002.0832. [DOI] [PubMed] [Google Scholar]
- 39.Yoon D.S., Cha D.S., Choi Y., Lee J.W., Lee M.H. MPK-1/ERK is required for the full activity of resveratrol in extended lifespan and reproduction. Aging Cell. 2019;18 doi: 10.1111/acel.12867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller R.K., Qadota H., Stark T.J., Mercer K.B., Wortham T.S., Anyanful A., Benian G.M. CSN-5, a component of the COP9 signalosome complex, regulates the levels of UNC-96 and UNC-98, two components of M-lines in Caenorhabditis elegans muscle. Mol. Biol. Cell. 2009;20:3608–3616. doi: 10.1091/mbc.e09-03-0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salah Ud-Din A.I.M., Roujeinikova A. Methyl-accepting chemotaxis proteins: a core sensing element in prokaryotes and archaea. Cell. Mol. Life Sci. 2017;74:3293–3303. doi: 10.1007/s00018-017-2514-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Geng C., Liu Y., Li M., Tang Z., Muhammad S., Zheng J., Wan D., Peng D., Ruan L., Sun M. Dissimilar crystal proteins Cry5Ca1 and Cry5Da1 synergistically act against Meloidogyne incognita and delay Cry5Ba-Based nematode resistance. Appl. Environ. Microbiol. 2017;83:e03505–e03516. doi: 10.1128/AEM.03505-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim T., Hofmann K., von Arnim A.G., Chamovitz D.A. PCI complexes: pretty complex interactions in diverse signaling pathways. Trends Plant Sci. 2001;6:379–386. doi: 10.1016/s1360-1385(01)02015-5. [DOI] [PubMed] [Google Scholar]
- 44.Zhang S.N., Pei D.S., Zheng J.N. The COP9 signalosome subunit 6 (CSN6): a potential oncogene. Cell Div. 2013;8:14. doi: 10.1186/1747-1028-8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vijayabaskar M.S., Vishveshwara S. Insights into the fold organization of TIM barrel from interaction energy based structure networks. PLoS Comput. Biol. 2012;8 doi: 10.1371/journal.pcbi.1002505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Braus G.H., Irniger S., Bayram O. Fungal development and the COP9 signalosome. Curr. Opin. Microbiol. 2010;13:672–676. doi: 10.1016/j.mib.2010.09.011. [DOI] [PubMed] [Google Scholar]
- 47.Sandhu A., Badal D., Sheokand R., Tyagi S., Singh V. Specific collagens maintain the cuticle permeability barrier in Caenorhabditis elegans. Genetics. 2021;217 doi: 10.1093/genetics/iyaa047. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available upon request.