Abstract
A strategy employing gene trap mutagenesis and site-specific recombination (Cre/loxP) has been used to identify genes that are transiently expressed during early mouse development. Embryonic stem cells expressing a reporter plasmid that codes for neomycin phosphotransferase and Escherichia coli LacZ were infected with a retroviral gene trap vector (U3Cre) carrying coding sequences for Cre recombinase (Cre) in the U3 region. Activation of Cre expression from integrations into active genes resulted in a permanent switching between the two selectable marker genes and consequently the expression of β-galactosidase (β-Gal). As a result, clones in which U3Cre had disrupted genes that were only transiently expressed could be selected. Moreover, U3Cre-activating cells acquired a cell autonomous marker that could be traced to cells and tissues of the developing embryo. Thus, when two of the clones with inducible U3Cre integrations were passaged in the germ line, they generated spatial patterns of β-Gal expression.
Fusions between promoterless genes that encode an easily assayable gene product and the controlling elements of cellular genes have proved valuable tools for studying gene function and regulation in mammalian cells. Several types of vectors, referred to as “gene traps,” that insert a promoterless reporter gene into mostly random chromosomal sites, including transcriptionally active regions, have been developed. By selecting for gene expression, recombinants in which the reporter gene is fused to the regulatory elements of an endogenous gene are obtained. Transcripts generated by these fusions faithfully reflect the activity of a disrupted cellular gene and serve as a molecular tag to clone any gene linked to a specific function (for reviews, see references 9, 13, 14, and 37). Particularly useful in this regard have been mouse embryonic stem (ES) cell lines which can pass genes introduced in vitro to transgenic offspring in vivo (1, 15, 21, 34).
Approximately 50% of genes disrupted in ES cell lines after infection or electroporation of gene trap vectors generated recessive phenotypes in mice (7, 32, 35). Based on this high efficiency of gene inactivation, it appears possible to isolate cell lines with integrations in most expressed genes (2 × 104 to 4 × 104). However, it would not be practical to pass all mutations to the germ line since many mutations will involve genes of lesser significance. Moreover, genes that are not expressed in ES cells generally go unobserved. Therefore, it would be useful to prescreen mutagenized ES-cell clones for mutations that affect genes involved in significant biological processes. To this end, three types of screens have been developed.
One approach involves sequencing DNA regions immediately adjacent to a gene trap vector. Comparison of the flanking sequences with nucleic acid databases reveals instances in which the gene trap has disrupted known genes (12, 37). A second approach uses a marker gene which can be easily visualized in the developing embryo. Analysis of pattern formation in chimeric or transgenic mice identifies mutations in developmentally regulated genes (6, 7, 39). A third approach relies on a reporter gene that requires an N-terminal signal sequence for expression. This effectively selects for mutations in genes that encode secreted proteins (31).
However, none of these strategies specifically selects for mutations in genes that are only transiently expressed. Since during mouse development most genes are expressed in a temporally and spatially restricted manner, it would be advantageous to develop a gene trap approach that enables the recovery of integrations into transiently expressed genes. To this end, a gene trap expressing the site-specific recombinase Cre was used to induce a permanent switching between two selectable marker genes expressed from a constitutive promoter. Since recombination uncouples the expression of the marker gene from the Cre-activating cellular promoter, genes that are expressed in a temporally restricted manner can be identified. Moreover, promoter uncoupling results in the acquisition of a cell autonomous marker which facilitates the analysis of cell fate and migration in the developing embryo.
In this paper we describe five ES-cell clones with gene trap integrations in genes that are induced during differentiation. Two of these clones generated spatial patterns of gene expression in transgenic embryos.
MATERIALS AND METHODS
Plasmids.
The ppgklxLacZ vector was derived from ppgklxtkneoIL3 (24) by replacing the genes encoding tkneo and interleukin 3 (IL-3) with coding sequences for Neo and LacZ, respectively. Briefly, a loxPLacZ fragment was assembled in pGEM30 by blunt-end ligation of the LacZ gene (derived from U3LacZ [20]) into the EcoRI site of the loxP flanking polylinker (24). loxPLacZ was then cloned as a SalI/XhoI fragment into the XhoI site of pSBC-2 (3) upstream of the simian virus 40 polyadenylation site. A fragment extending from the XmnI site of ppgklxtkneoIL3 to the NheI site of the downstream loxP site was isolated by a partial digestion and cloned into the respective sites of pSBC-2-loxLacZ to obtain ppgklxtkneoLacZ. Finally, ppgklxneoLacZ was obtained by replacing the tkneo fusion gene with neo by using blunt-end cloning and the RsrII and BglII restriction sites of ppgklxtkneoLacZ. The gene trap construct pGgU3Creen(−) was obtained as described previously (24).
Cells and viruses.
D3 ES cells were grown on irradiated (32 Gy) mouse embryo fibroblast feeder layers in Dulbecco’s modified Eagle medium supplemented with 15% (vol/vol) preselected and heat-inactivated fetal calf serum (Linaris, Bettingen, Germany), 100 mM nonessential amino acids (Gibco), 0.1 mM β-mercaptoethanol (Sigma), 1,000 U of leukemia inhibitory factor (LIF) (ESGRO; Gibco/BRL) per ml, and 5 mg each of penicillin and streptomycin per ml. In some experiments differentiation was induced by withdrawing LIF and feeder layers whereas in others differentiation was allowed to proceed in bacterial plates with Dulbecco’s modified Eagle medium supplemented with 20% fetal calf serum, 0.3 mM β-mercaptoethanol, and 100 mM nonessential amino acids.
ppgklxneoLacZ was transduced into D3 cells by electroporation with a Bio-Rad Gene Pulser at 240 V and 500 μF. Following incubation for 24 h, cells were selected for 7 days in 320 μg of active G418 (Gibco/BRL) per ml.
Infection of ES cells with the U3Cre gene trap virus was performed by incubating 150 ES cells with 50 μl of virus-containing supernatant for 2 h as described previously (24).
Amplification and cloning of upstream sequences.
Inverse PCR from genomic DNAs was performed with the Cre-specific primers described previously (24). 5′ rapid amplification of cDNA ends (RACE) was performed with 1 μg of total RNA by using the 5′ RACE kit from Gibco/BRL according to the manufacturer’s instructions. The specific Cre reverse primers were as follows: 5′-GGTATGCTCAGAAAACGCCTG-3′, 5′-CGAACCTCATCACTCGTTGCATC-3′ (nested), and 5′-CGGTCAGTAAATTGGACACCTTCC-3′ (nested). Amplification reactions were performed in a Perkin-Elmer thermocycler, and amplified DNA was cloned into the pGEMT vector (Promega) as described previously (24). Inserts were sequenced with an ABI 310 genetic analyzer (Perkin-Elmer).
Nucleic acid hybridization analyses.
DNA and RNA hybridizations were performed on Hybond N or Hybond N+ membranes (Amersham) by using [32P]dCTP-labeled probes and a random priming labeling kit (Rediprime; Amersham). Blots either were scanned with a PhosphorImager (Molecular Dynamics) and analyzed with IPLabGel software (Molecular Dynamics) or were exposed to Kodak-Biomax autoradiography film.
Histochemical analysis of LacZ expression.
Cells were stained for 2 h at 37°C with a solution of 1 mg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) per ml in phosphate-buffered saline after fixation for 15 min in a solution of 2% formaldehyde and 0.2% glutaraldehyde.
Embryos and fetuses were stained overnight at 37°C after fixation for 60 min and preincubation for 30 min in phosphate-buffered saline solution containing 0.15 mg of chloroquin per ml, 10 mM potassium ferricyanide, 10 mM potassium ferrocyanide, 2 mM magnesium chloride, and 0.2% Triton X-100 (pH 7.4) to reduce endogenous background, i.e., β-Gal. Stained embryos were embedded in glycol methacrylate (JB-4; Polysciences) and blocks were cut to 3-μm thicknesses with glass knives on an ultramicrotome (LKB) as described recently (16). Sections were counterstained with nuclear fast red, mounted on clean glass slides, and analyzed by standard microscopy.
Nucleotide sequence accession numbers.
GenBank accession numbers for the gene trap flanking sequences are as follows: 1C7a, emb|AJ2237718; 1C7b, emb|AJ223719; 2C2a, emb|AJ223720; 2C2b, emb|AJ223721; 2E12, emb|AJ223722; 7G4, gb|L20255; and 8G3, emb|AJ223723.
RESULTS
Construction of an ES cell line amenable to selectable marker switching by Cre recombinase.
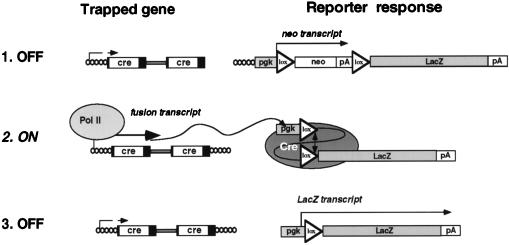
To obtain an ES cell line that would allow Cre recombinase to switch two selectable marker genes, we electroporated D3 ES cells with the expression plasmid ppgklxneoLacZ. ppgklxneoLacZ consists of two tandemly arrayed selectable marker genes that are expressed from a pgk promoter (Fig. 1). The 5′ gene codes for neomycin phosphotransferase and is flanked by two loxP recombination targets in identical orientation. The 3′ gene codes for β-galactosidase (β-Gal) (20) and terminates in a simian virus 40 polyadenylation sequence. To suppress LacZ translation from dicistronic transcripts, two tandem copies of bovine growth hormone polyadenylation sequence were inserted downstream of the neo gene. Thus, cells expressing ppgklxneoLacZ, albeit neomycin resistant, should not express LacZ. Since Cre recombinase excises the sequences flanked by loxP (25, 26), Cre expression was expected to delete the neo gene and thereby place the lacZ gene just downstream of the pgk promoter. As a result, cells would lose neomycin resistance and start synthesizing β-Gal (Fig. 1).
FIG. 1.
Mechanism of LacZ activation by U3Cre integrations into transiently expressed genes. Cre expressed from cell-provirus fusion transcripts (left) excises neo from ppgklxneoLacZ (right) and places lacZ downstream of the constitutive pgk promoter. This induces synthesis of β-Gal which is independent of U3Cre expression. For further explanation see the text.
Several ppgklxneoLacZ-expressing cell lines were isolated in G418 and assayed individually for background β-Gal activity by X-Gal staining. Four of 10 cell lines tested consistently failed to stain with X-Gal (LacZ−) even after prolonged periods in culture. When induced to differentiate by withdrawing of LIF and feeder layers, the cells continued to stain negative, indicating that the LacZ− phenotype is stable (data not shown). One cell line expressing a single copy of ppgklxneoLacZ (plnLacZ13) was selected for further analysis. When transfected with a plasmid expressing Cre recombinase (pMCCre− [10]), the plnLacZ13 cells stained positive for β-Gal and harbored recombined reporter plasmids (data not shown).
Recombined plnLacZ13 expresses LacZ ubiquitously in the early embryo.
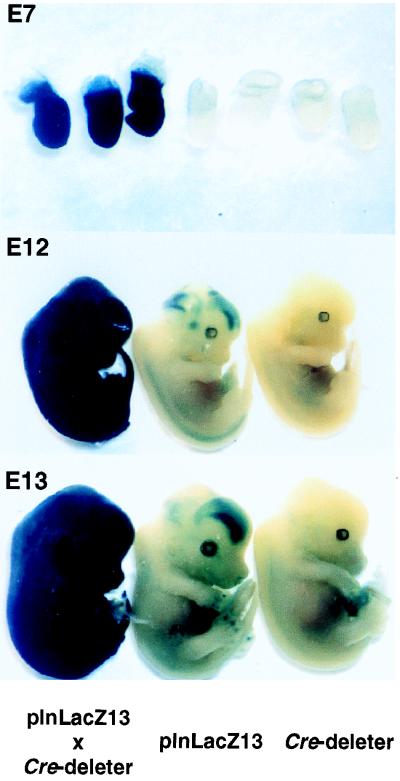
Since one goal of this study was to devise a system that would enable analysis of cell fate and migration in the developing embryo, it was important to investigate whether a recombined reporter plasmid would express LacZ in all embryonic tissues. Therefore, we injected the ppgklxneoLacZ-expressing plnLacZ13 cells into C57BL/6 blastocysts and transferred these into the uteri of NMRI mouse recipients. Chimeric males were mated with C57BL/6 females and agouti offspring carrying the transgene were identified by tail blotting. Mice heterozygous for the transgene were crossed to a transgenic Cre deleter strain which has been shown to express the recombinase ubiquitously (29). X-Gal staining of the developing embryos showed that in doubly transgenic mice β-Gal was expressed ubiquitously. However, virtually no β-Gal expression was observed in embryos carrying only one transgene (Fig. 2). Although plnLacZ13 mice showed some β-Gal expression within a small circumscribed area of the forebrain, presumably due to a slight leakiness of the reporter plasmid, this background was clearly distinguishable from positive controls on microscopic tissue sections (data not shown).
FIG. 2.
In vivo recombination of ppgklxLacZ in transgenic plnLacZ13 mice. Embryos resulting from crosses between heterozygous plnLacZ13 and Cre deleter strains were recovered at the indicated stages of gestation (E = day) and stained with X-Gal as described in Materials and Methods. Embryos in each panel are littermates.
Ubiquitous expression of β-Gal was confirmed for plnLacZ13mice by staining embryonic tissue sections, including those from the liver, kidney, intestine, heart, limb, and rib cartilage (data not shown). This suggested that ppglxneoLacZ had integrated into a chromosomal locus that does not significantly restrict expression. Moreover, no obvious loss-of-function abnormalities were observed in homozygous plnLacZ13 mice, indicating that the integration did not disrupt any vital genes.
A U3Cre/plnLacZ13 integration library preselected in G418 contains clones with inducible LacZ expression.
A library of approximately 1 × 105 to 2 × 105 independent proviral integrations was constructed in microtiter plates by infecting plnLacZ13 cells (≈150 cells per well) with the retroviral gene trap vector U3Cre, which contains a promoterless Cre recombinase gene in the U3 region of the long terminal repeat (LTR). As has been shown in previous studies, activation of this type of gene trap occurs from integrations in or near 5′ exons and does not require in-frame fusions with the coding sequences of cellular genes (12, 24, 36, 38).
Since activation of Cre expression from integrations into transcribed genes results in loss of the neomycin resistance gene (Fig. 1), the library was first selected in G418. This was expected to eliminate all integrations into genes that express Cre to levels high enough to cause recombination. Thus, surviving cells were likely to have provirus integrations in transcriptionally inactive chromosomal regions (including silent genes) or in genes expressed too weakly to cause recombination.
To identify cell pools with inducible U3Cre integrations, aliquots removed from each well were allowed to differentiate for 4 days without LIF or feeder layers. This period was chosen because we have found in earlier experiments that by this time over 50% of the cells express the mesodermal marker brachyury, which is indicative of gastrulation (11, 33). Since gastrulation corresponds to cellular diversification, we expected enhanced gene activity and therefore a higher fraction of genes that would be accessible to gene trap mutagenesis. Of 960 pools, 44 stained positive for β-Gal. Upon retesting, nine pools expressed β-Gal even before differentiation, indicating that G418 selection did not completely eliminate the integrations into expressed genes. Five pools with inducible β-Gal expression and one pool with constitutive β-Gal expression were selected to isolate the cell lines 2E12, 1C7, 8G3, 2H7, 2C2, and 7G4, respectively.
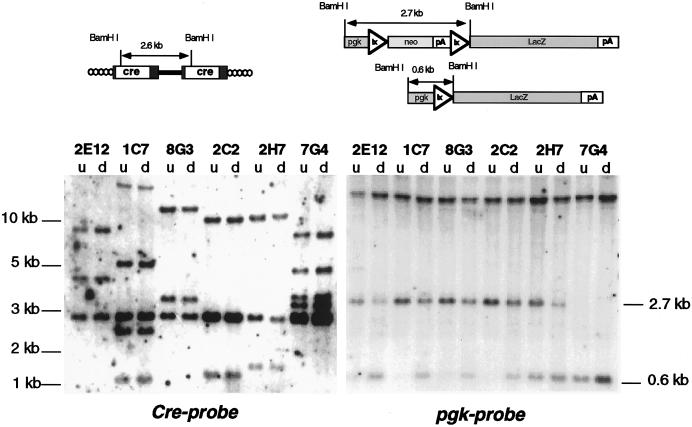
To verify clonality, genomic DNAs were cleaved with BamHI and hybridized on Southern blots to a Cre-specific probe. Since BamHI cleaves within the 5′ third of Cre, each nonrearranged provirus should generate three hybridizing fragments, i.e., a constant fragment of 2.6 kb derived from the provirus and two variable fragments representing sequences extending from the BamHI sites in the LTRs to the BamHI sites in the 5′ and 3′ flanking cellular DNA, respectively. Figure 3 (left panel) shows that each clone contained one to three nonrearranged proviruses, which is consistent with a multiplicity of infection of ∼1.5.
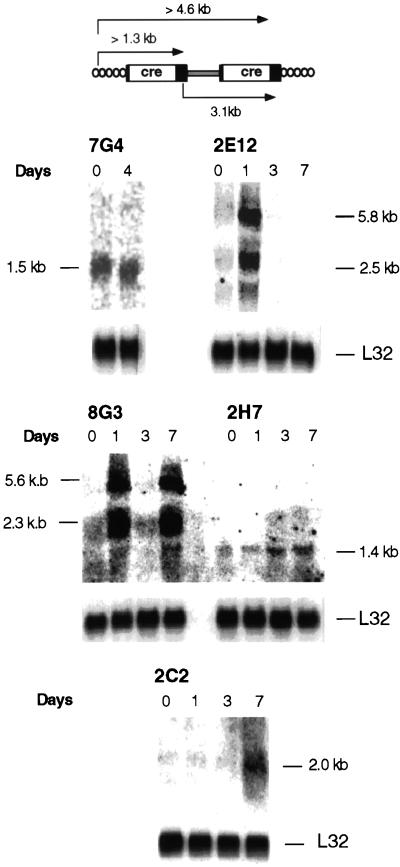
FIG. 3.
U3Cre provirus integrations and site-specific recombination in clones isolated after induction of differentiation. (Upper panel) Structure of U3Cre proviruses (left) and reporter plasmids (right) before and after recombination. (Lower panel) Southern blot analysis of clones recovered from the plnLacZ13 integration library following induction of differentiation. Genomic DNAs extracted from individual clones before (u, undifferentiated) and after (d, differentiated) differentiation were cleaved with BamHI, fractionated on agarose gels, blotted onto nylon filters, and hybridized to 32P-labeled Cre-specific (left) or pgk-specific (right) probes. The large-molecular-size bands hybridizing to pgk represent the endogenous pgk promoter.
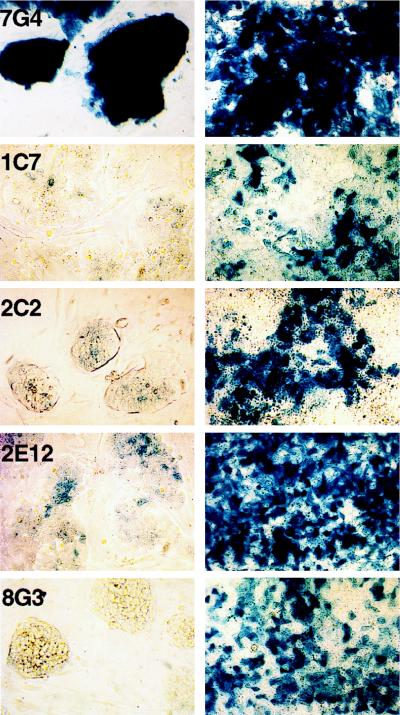
All clones generated differentiated progeny that stained positive for β-Gal, indicating that U3Cre expression was induced. However, the fraction of LacZ+ cells among the progeny of each clone varied from as high as 100% in 7G4 cells to 25% in 1C7 cells (Fig. 4). Assuming that differentiating cells gradually lose their proliferative potential, this staining heterogeneity is likely to reflect gene activation in distinct precursor cells. Thus, a less mature progenitor would yield higher numbers of LacZ+ cells than any of its more differentiated progeny.
FIG. 4.
Induction of β-Gal expression in differentiating U3Cre-expressing clones. Cells were induced to differentiate by the withdrawal of LIF and feeder layers for 4 days. Cells were stained with X-Gal before (left panels) and after (right panels) differentiation and examined by light microscopy at a ×100 magnification. Samples are representative of four independent inductions.
To confirm recombination, genomic DNAs were cleaved with BamHI and hybridized on Southern blots to a pgk-specific probe. As shown in Fig. 3 (right panel, top), BamHI cuts within the 5′ ends of pgk and lacZ, such that nonrecombined cells generate an internal hybridizing fragment of 2.7 kb. Since this fragment accommodates all sequences flanked by loxP, its deletion should generate a residual fragment of 0.6 kb. Figure 3 (right panel, bottom) shows that each differentiated clone contained this fragment, indicating that ppgklxneoLacZ had recombined. However, as expected from the heterogeneous staining patterns, recombination was mostly incomplete.
Recombination of ppgklxneoLacZ is caused by inducible cell-provirus fusion transcripts.
To ascertain that Cre was expressed from cell-provirus fusion transcripts, Northern blots with RNAs from differentiating clones were hybridized to a Cre-specific probe. U3 gene trap integrations expressed in ES cells typically generate cell-provirus fusion transcripts of variable sizes that initiate in a nearby cellular promoter and terminate in the polyadenylation site of the 5′ LTR. In some cases, a second transcript which initiates at the same cellular promoter but terminates in the polyadenylation site of the 3′ LTR is also seen (Fig. 5, upper panel) (2, 35).
FIG. 5.
Analysis of cell-provirus fusion transcripts in clones with inducible LacZ expression. (Upper panel) Predicted transcripts from U3Cre integrations into expressed genes. (Lower panels) Northern blot analysis of cell-provirus fusion transcripts in differentiating clones. RNAs were extracted from individual clones after incubation of the cells for various intervals in bacterial plates without LIF or feeder layers. Twenty micrograms of total RNA (7G4, 2E12, 8G3, and 2H7) and 5 μg of polyadenylated RNA (2C2) were fractionated on formaldehyde-agarose gels, blotted onto nylon filters, and hybridized to 32P-labeled Cre or L32 (19) probes.
As expected, each clone expressed cell DNA-provirus fusion transcripts which, in all but the 7G4 cells, were induced by differentiation. More importantly, clones 2E12 and 8G3 expressed the fusion transcripts only transiently (Fig. 5, lower panel). Interestingly, U3Cre expression in clone 8G3 was induced twice, on days 1 and 7. This pattern is reminiscent of the expression of certain cytokine genes in differentiating ES cells (28).
Sequences flanking the U3Cre proviruses hybridize to single-copy genes.
To investigate whether U3Cre proviruses had disrupted single-copy genes, upstream proviral sequences were isolated by inverse PCR or 5′ RACE and sequenced (8, 36). All sequences showed typical cell DNA-provirus junctions and varied in size from 4 to 500 nucleotides (Table 1). While the sequences from inducible integrations showed no homology to known genes or expressed sequence tags, the 7G4 flanking sequence was 100% identical with the 5′ end of the stathmin cDNA (Table 1 and data not shown). Stathmin is a cell cycle-regulated, tubulin binding protein which has been previously shown to be highly expressed in proliferating cells (4, 17, 23).
TABLE 1.
Summary of results obtained with ES-cell clonesa
| Cell line | Proviruses (n) | No. of nucleotides in provirus flanking DNA (method) | Hybridization to single-copy genes | Homology to known genes | Nature of induction of fusion transcript | Fusion gene inheritance |
|---|---|---|---|---|---|---|
| 1C7 | 2 | 217 (IP) | Yes | No | ND | No |
| 154 (IP) | Yes | No | ||||
| 2C2 | 2b | 120 (IP) | Yes | No | Gradual | Yes |
| 147 (IP) | Yes | No | ||||
| 2E12 | 1 | 498 (IP) | Yes | No | Transient | No |
| No | ||||||
| 7G4 | 3c | 39 (5′ RACE) | NT | Stathmin | Constitutive | Yes |
| 8G3 | 1 | 177 (IP) | Yes | No | Transient | Yes |
| 2H7 | 1 | 4 (IP) | NT | NT | Gradual | No |
The number of proviruses per cell was determined by Southern blotting as described in the legend to Fig. 3. For inverse PCR, genomic DNAs were digested with MseI and religated to obtain circular molecules. After cleavage with PvuII, 1 μg of DNA from each sample was used for PCR as described in Materials and Methods. 5′ RACE was performed with 10 μg of total cellular RNA as described in Materials and Methods. Fusion transcripts were visualized on Northern blots as described in the legend to Fig. 5. IP, inverse PCR; ND, not detected; NT, not tested.
Provirus integrations were segregated by breeding into C57BL/6 mice. Integration sites were identified by tail blotting and hybridization to 32P-labeled flanking sequence probes. The Cre-activating integration (2C2b) was identified by mating either transgenic mouse to a plnLaZ13 reporter mouse and by staining the developing embryos with X-Gal.
Provirus integrations were segregated by breeding into C57BL/6 mice. The U3Cre-expressing integration was identified by tail blotting and hybridization to a 32P-labeled HindIII/SpeI restriction fragment derived from a stathmin cDNA clone (5).
Each of the cloned flanking sequences hybridized to a single BamHI restriction fragment in wild-type DNA, indicating that they were derived from single-copy genes. Additional hybridizing fragments representing the allele occupied by the provirus were generated in each corresponding parental clone (data not shown).
β-Gal expression patterns in transgenic embryos.
To test whether the selected clones were still totipotent, we individually injected each into blastocysts and mated the resulting chimeras with C57BL/6 mice. Analysis of the offspring indicated that three clones had contributed to the germ line despite extensive in vitro manipulation. Moreover, the transgenes were inherited in each case in an autosomal Mendelian fashion (Table 1).
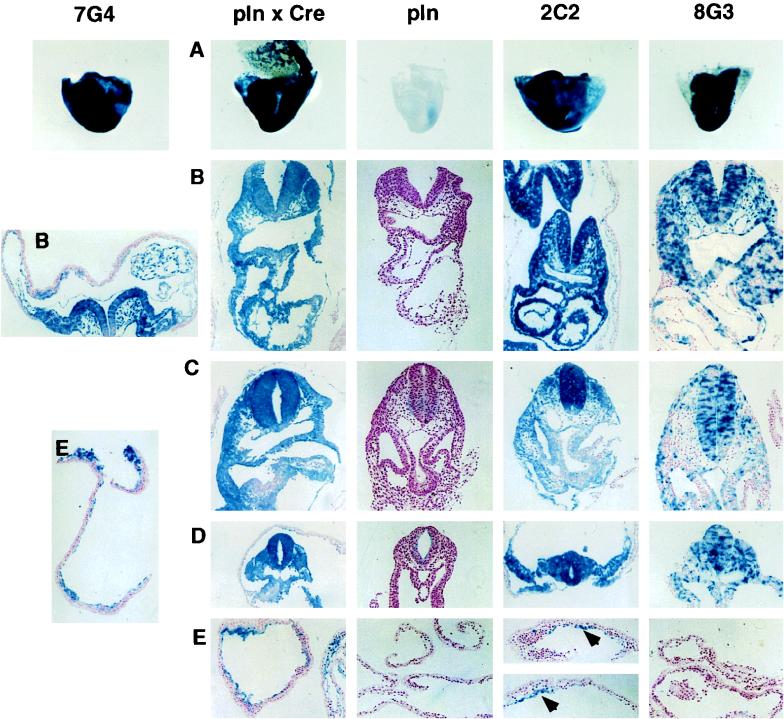
To examine whether the inducible gene trap integrations were also inducible in vivo, mice bearing the U3Cre but not the ppgklxneoLacZ transgene were mated with homozygous plnLacZ13 mice. Developing embryos were recovered after 8.5 days of gestation and stained for β-Gal expression. Two different lines of transgenic mice were used as positive controls. One strain originated from a doubly transgenic plnLacZ13/Cre deleter mouse from which it received a recombined reporter plasmid through the germ line. The other strain was derived from clone 7G4, which expresses Cre constitutively. In both cases, embryos stained completely with X-Gal, indicating ubiquitous recombination of ppgklxneoLacZ. In contrast, 8G3 embryos did not express β-Gal in extraembryonic tissues (Fig. 6). On stained embryo sections, LacZ+ cells were missing from the yolk sac and the amnion, indicating that Cre was activated after extraembryonic lineage segregation (Fig. 6). However, the overall staining pattern was a mosaic between LacZ+ and LacZ− cells. Since no mosaic was detected in the positive controls, the pattern may have been caused by an independent activation of Cre in different lineages. Alternatively, the short-lived fusion transcripts expressed in 8G3 cells (Fig. 5) may have prevented ubiquitous recombination of ppgklxneoLacZ. As has been shown previously, Cre recombinase seems to require at least 72 h to recombine 80% of its targets (33a).
FIG. 6.
β-Gal expression patterns in transgenic embryos. Embryos resulting from crosses between heterozygous 7G4, 2C2, or 8G3 mice and homozygous plnLacZ13 reporter mice were recovered at 8.5 days of gestation and stained with X-Gal. A, whole mounts; B, heart; C, intestinal stalk; D, somites; E, yolk sac. pln, plnLacZ13; Cre, Cre deleter. The whole mounts and tissue sections were examined by light microscopy at ×20 and ×100 magnifications, respectively. Cell clusters in 2C2 are marked by arrowheads. Note that despite the presence of recombined reporter plasmids, the endodermal component of the extraembryonic membranes does not express LacZ.
In 2C2 embryos, LacZ+ cells were seen in both the embryo proper and extraembryonic tissues (Fig. 6). However, unlike the positive controls, the LacZ+ cells appeared in clusters throughout the extraembryonic membranes. Although suggestive of a progenitor-progeny relationship, these clusters most likely reflect Cre activation in distinct embryonic cell lineages.
DISCUSSION
This study exploited the combined features of gene trap mutagenesis and site-specific recombination to identify genes that are induced during early mouse development. ES cells expressing a specific reporter construct were infected with gene trap retroviruses containing coding sequences for Cre recombinase in the U3 region. By using the β-Gal gene as a conditionally expressed marker gene, cell clones in which Cre was expressed from transcripts initiating in the flanking cellular DNA could be identified. Five inducible U3Cre integrations were selected during ES-cell differentiation, two of which were passed to the germ line. Since U3Cre expression was also induced during embryonic development, the results confirm previous studies showing that genes regulated in vitro are also regulated in vivo (18, 20, 22, 27, 30, 32).
Analysis of provirus flanking sequences derived from the inducible clones identified integrations into as-yet-unidentified single-copy genes. Although recent results seem to suggest that up to 60% of all gene trap integrations are integrations into previously characterized genes or expressed sequence tags (12), we believe that the present strategy does not favor recovery of integrations into known genes. This is because selection is applied for integrations into 5′ exons of transiently expressed genes, both of which are underrepresented in conventional cDNA libraries.
The gene trap approach described here has several unique advantages. Unlike previously described strategies in which gene trap insertions are restricted to expressed genes, switching between two selectable marker genes by means of Cre recombinase allows enrichment for integrations into silent genes. By first eliminating integrations into active genes, it is possible to screen large pools of recombinant clones for integrations into regulated genes. This circumvents the laborious analysis of individual clones (6, 20, 35, 39).
The most important feature of the system is the recombinase-induced uncoupling between β-Gal and Cre expression. As a result, LacZ continues to be expressed even in the absence of Cre recombinase, thereby providing the means for the recovery of transiently expressed genes. In line with this, two of the five fusion transcripts analyzed in this study were present only transiently in differentiating cells. Since most developmentally regulated genes are expressed in a temporally and spatially restricted manner, the strategy is ideal for the molecular analysis of mouse development.
A further consequence of promoter uncoupling is the acquisition of an autonomous marker by the gene trap-activating cells. Such a genetic marker could be useful for demarcating cell lineages and for tracking cell fate and migration in the developing embryo. Although the LacZ expression patterns described in this study are reminiscent of gene trap activations in many different lineages, activations at later stages of differentiation are likely to be more restricted and thus earmark the progenitors of more specialized cells and tissues.
Staining patterns will also provide valuable clues as to which tissues or organs should be examined for loss-of-function abnormalities induced by gene disruptions. This feature is particularly helpful in cases where mutations generate only subtle phenotypes.
Finally, the strategy used here is prone to yield strains of mice in which Cre expression is controlled by tissue-specific promoters. Such strains should be a valuable tool for targeted gene disruptions in special cells or tissues.
ACKNOWLEDGMENTS
We thank Frieder Schwenk and Klaus Rajewsky for providing the Cre deleter mouse, André Sobel for the stathmin cDNA, and Christina Friedel and Sabine Reindel for technical assistance.
This work was supported by grants from the Deutsche Forschungsgemeinschaft, Bonn, Germany, and the VW Foundation, Wolfsburg, Germany, to H.V.M.
REFERENCES
- 1.Bradley A, Evans M, Kaufman M H, Robertson E. Formation of germ-line chimeras from embryo derived teratocarcinoma cell lines. Nature. 1984;309:255–256. doi: 10.1038/309255a0. [DOI] [PubMed] [Google Scholar]
- 2.DeGregori J, Russ A, von Melchner H, Rayburn H, Priyaranjan P, Jenkins N A, Copeland N G, Ruley H E. A murine homolog of the yeast RNA1 gene is required for postimplantation development. Genes Dev. 1994;8:265–276. doi: 10.1101/gad.8.3.265. [DOI] [PubMed] [Google Scholar]
- 3.Dirks W, Wirth M, Hauser H. Dicistronic transcription units for gene expression in mammalian cells. Gene. 1993;128:247–249. doi: 10.1016/0378-1119(93)90569-o. [DOI] [PubMed] [Google Scholar]
- 4.Doye V, Kellermann O, Buc-Caron M H, Sobel A. High expression of stathmin in multipotential teratocarcinoma and normal embryonic cells versus their early differentiated derivatives. Differentiation. 1992;50:89–96. doi: 10.1111/j.1432-0436.1992.tb00489.x. [DOI] [PubMed] [Google Scholar]
- 5.Doye V, Soubrier F, Bauw G, Boutterin M C, Beretta L, Koppel J, Vandekerckhove J, Sobel A. A single cDNA encodes two isoforms of stathmin, a developmentally regulated neuron-enriched phosphoprotein. J Biol Chem. 1989;264:12134–12137. [PubMed] [Google Scholar]
- 6.Forrester L, Nagy A, Sam M, Watt A, Stevenson L, Bernstein A, Joyner A, Wurst W. An induction gene trap screen in embryonic stem cells: identification of genes that respond to retinoic acid in vitro. Proc Natl Acad Sci USA. 1996;93:1677–1682. doi: 10.1073/pnas.93.4.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedrich G, Soriano P. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 1991;5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- 8.Frohman M A, Dush M K, Martin G R. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gossler A, Zachgo J. Gene and enhancer trap screens in ES cell chimeras. In: Joyner A L, editor. Gene targeting: a practical approach. Oxford, England: IRL Press at Oxford University; 1993. pp. 181–227. [Google Scholar]
- 10.Gu H, Zou Y, Rajewsky K. Independent control of immunoglobulin switch recombination at individual switch regions evidenced through Cre-IoxP-mediated gene targeting. Cell. 1993;73:1155–1164. doi: 10.1016/0092-8674(93)90644-6. [DOI] [PubMed] [Google Scholar]
- 11.Herrmann B G, Labeit S, Poustka A, King T R, Lehrach H. Cloning of the T gene required in mesoderm formation in the mouse. Nature. 1990;343:617–622. doi: 10.1038/343617a0. [DOI] [PubMed] [Google Scholar]
- 12.Hicks G G, Shi E, Li X M, Li C H, Pawlak M, Ruley H E. Functional genomics in mice by tagged sequence mutagenesis. Nat Genet. 1997;16:338–344. doi: 10.1038/ng0897-338. [DOI] [PubMed] [Google Scholar]
- 13.Hill D H P, Wurst W. Mutagenic strategies to dissect mammalian development. Curr Top Dev Biol. 1993;28:181–206. doi: 10.1016/s0070-2153(08)60213-6. [DOI] [PubMed] [Google Scholar]
- 14.Hill D H P, Wurst W. Screening for pattern formation in mouse using gene trap technology. Methods Enzymol. 1993;255:663–681. doi: 10.1016/0076-6879(93)25043-2. [DOI] [PubMed] [Google Scholar]
- 15.Hooper M, Hardy K, Handyside A, Hunter S, Monk M. HPRT-deficient (Lesch-Nyhan) mouse embryos derived from germline colonization by cultured cells. Nature. 1987;326:292–295. doi: 10.1038/326292a0. [DOI] [PubMed] [Google Scholar]
- 16.Lazik A, Liu Y, Bringas P, Sangiorgi F, Maxson R. A sensitive method for analyzing b-galactosidase reporter gene expression in tissue sections of mouse embryos. Trends Genet. 1996;12:445–446. doi: 10.1016/0168-9525(96)99999-0. [DOI] [PubMed] [Google Scholar]
- 17.Marklund U, Osterman Ö, Melander H, Bergh A, Gullberg M. The phenotype of a “Cdc2 kinase target site-deficient” mutant of oncoprotein 18 reveals a role of this protein in cell cycle control. J Biol Chem. 1994;48:30626–30635. [PubMed] [Google Scholar]
- 18.Muthuchamy M, Pajak L, Howles P, Doetschman T, Wieczorek D. Developmental analysis of tropomyosin gene expression in embryonic stem cells and mouse embryos. Mol Cell Biol. 1993;13:3311–3323. doi: 10.1128/mcb.13.6.3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neznanov N, Thorey I S, Ceceña G, Oshima R G. Transcriptional insulation of the human keratin 18 gene in transgenic mice. Mol Cell Biol. 1993;13:2214–2223. doi: 10.1128/mcb.13.4.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reddy S, Rayburn H, von Melchner H, Ruley H E. Fluorescence activated sorting of totipotent embryonic stem cells expressing developmentally regulated lacZ fusion genes. Proc Natl Acad Sci USA. 1992;89:6721–6725. doi: 10.1073/pnas.89.15.6721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robertson E, Bradley A, Kuehn M, Evans M. Germ-line transmission of genes introduced into cultured pluripotential cells by retroviral vector. Nature. 1986;323:445–448. doi: 10.1038/323445a0. [DOI] [PubMed] [Google Scholar]
- 22.Rogers M B, Hosler B A, Gudas L J. Specific expression of a retinoic acid-regulated, zinc-finger gene, Rex-1, in preimplantation embryos, trophoblasts and spermatocytes. Development. 1991;113:815–824. doi: 10.1242/dev.113.3.815. [DOI] [PubMed] [Google Scholar]
- 23.Rowlands D C, Williams A, Jones N A, Guest S S, Reynolds G M, Barber P C, Brow G. Stathmin expression is a feature of proliferating cells of most, if not all, cell lineages. Lab Investig. 1995;72:100–113. [PubMed] [Google Scholar]
- 24.Russ A P, Friedel C, Ballas K, Kalina U, Zahn D, Strebhardt K, von Melchner H. Identification of genes induced by factor deprivation in hematopoietic cells undergoing apoptosis using gene-trap mutagenesis and site-specific recombination. Proc Natl Acad Sci USA. 1996;93:15279–15284. doi: 10.1073/pnas.93.26.15279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sauer B. Functional expression of the cre-lox site-specific recombination system in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:2087–2096. doi: 10.1128/mcb.7.6.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sauer B, Henderson N. Site-specific recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc Natl Acad Sci USA. 1988;85:5166–5170. doi: 10.1073/pnas.85.14.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scherer C A, Chen J, Nachabeh A, Hopkins N, Ruley H E. Transcriptional specificity of the pluripotent embryonic stem cell. Cell Growth Differ. 1996;7:1393–1401. [PubMed] [Google Scholar]
- 28.Schmitt R M, Bruyns E, Snodgrass H R. Hematopoietic development of embryonic stem cells in vitro: cytokine and receptor gene expression. Genes Dev. 1991;5:728–740. doi: 10.1101/gad.5.5.728. [DOI] [PubMed] [Google Scholar]
- 29.Schwenk F, Baron U, Rajewsky K. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 1995;23:5080–5081. doi: 10.1093/nar/23.24.5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharpe N G, Williams D G, Latchman D S. Regulated expression of the small nuclear ribonucleoprotein particle protein SmN in embryonic stem cell differentiation. Mol Cell Biol. 1990;10:6817–6820. doi: 10.1128/mcb.10.12.6817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skarnes W, Moss J, Hurtley S, Beddington R. Capturing genes encoding membrane and secreted proteins important for mouse development. Proc Natl Acad Sci USA. 1995;92:6592–6596. doi: 10.1073/pnas.92.14.6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skarnes W C, Auerbach B A, Joyner A L. A gene trap approach in mouse embryonic stem cells: the lacZ reporter is activated by splicing, reflects endogenous gene expression, and is mutagenic in mice. Genes Dev. 1992;6:903–918. doi: 10.1101/gad.6.6.903. [DOI] [PubMed] [Google Scholar]
- 33.Smith J C, Price B M J, Green J B A, Weigel D, Herrmann B G. Expression of a Xenopus homolog of brachyury (T) is an immediate-early response to mesoderm induction. Cell. 1991;67:79–87. doi: 10.1016/0092-8674(91)90573-h. [DOI] [PubMed] [Google Scholar]
- 33a.Stewart, F. Personal communication.
- 34.Thompson S, Clarke A R, Pow A M, Hooper M L, Melton D W. Germ line transmission and expression of a corrected HPRT gene produced by gene targeting in embryonic stem cells. Cell. 1989;56:313–321. doi: 10.1016/0092-8674(89)90905-7. [DOI] [PubMed] [Google Scholar]
- 35.von Melchner H, DeGregori J V, Rayburn H, Reddy S, Friedel C, Ruley H E. Selective disruption of genes expressed in totipotent embryonal stem cells. Genes Dev. 1992;6:919–927. doi: 10.1101/gad.6.6.919. [DOI] [PubMed] [Google Scholar]
- 36.von Melchner H, Reddy S, Ruley H E. Isolation of cellular promoters by using a retrovirus promoter trap. Proc Natl Acad Sci USA. 1990;87:3733–3737. doi: 10.1073/pnas.87.10.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.von Melchner H, Ruley H E. Functional analysis of the mammalian genome by gene entrapment. In: Farzane F, Cooper D N, editors. Functional analysis of the human genome. Oxford, England: Bios Scientific Publishers; 1995. pp. 109–129. [Google Scholar]
- 38.von Melchner H, Ruley H E. Identification of cellular promoters by using a retrovirus promoter trap. J Virol. 1989;63:3227–3233. doi: 10.1128/jvi.63.8.3227-3233.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wurst W, Rossant J, Prideaux V, Konacka M, Joyner A L, Guillemont F, Gasca S, Auerbach A, Ang S L. Genetic screen for novel pattern formation genes in mouse using gene trap approaches in ES cells. Genetics. 1995;139:889–899. doi: 10.1093/genetics/139.2.889. [DOI] [PMC free article] [PubMed] [Google Scholar]