Dear Editor,
Multiple myeloma (MM) is a plasma cell neoplasm (PCN) [1]. Sensitive and accurate identification of genetic abnormalities is critical for patient risk stratification and therapeutic decision-making as well as understanding pathogenesis. Recurring genetic abnormalities include structural variants (SVs), such as rearrangements involving IGH and MYC genes; copy number variants (CNVs), such as 1q+, del(17p), del(13q); hyperdiploidy/hypodiploidy; and various gene mutations, such as mutations in the RAS pathway, BRAF, FAM46C, DIS3, and TP53 [2]. Trisomies/hyperdiploidy and IGH rearrangements/fusions are considered primary abnormalities present at disease initiation while 1q+, del(17p)/TP53 mutations and MYC rearrangements/fusions are considered secondary abnormalities that develop during disease progression [3]. High-risk genetic abnormalities including del(17p), t(4;14)/IGH::FGFR3/NSD2, and t(14;16)/IGH::MAF have been used along with serum markers for MM risk stratification in the Revised International Stage System [4]. Other genetic abnormalities including t(14;20)/IGH::MAFB, and MYC rearrangements/fusions are considered potential high-risk biomarkers in MM [2]. Recently whole-genome sequencing (WGS) studies have revealed novel candidate driver genes and catastrophic complex rearrangements (chromoanagenesis) associated with poor clinical outcomes [5]. Furthermore, the presence of some genetic abnormalities can clearly impact response to specific therapies, as exemplified by t(11;14)/IGH::CCND1 and response to BCL2 inhibition [2, 3, 6]. Therefore, detection of genome-wide genetic abnormalities is essential.
Detection of genetic abnormalities has routinely relied on karyotyping, fluorescence in-situ hybridization (FISH), and next-generation sequencing (NGS). Karyotyping offers a low-resolution whole-genome analysis and requires the presence of actively dividing cancer cells [7]. FISH is more sensitive than karyotyping within targeted areas covered by probes but still leaves most of the genome unexamined [7]. Furthermore, complex gene rearrangements can make interpretation and analysis of FISH difficult. For example, FISH failed to detect ~70% of all MYC SV subtypes reported by NGS [8] and variant IGH rearrangements may mask IGH rearrangements or cause equivocal results [9]. Chromosomal microarray (CMA) testing has been reported to provide a whole-genome analysis and improve diagnostic yield for CNVs compared to FISH/karyotyping [10]. However, CMA is not capable of detecting balanced SVs (e.g., IGH or MYC rearrangements/fusions), which limits its clinical utility. Currently, RNA-sequencing and WGS show promising genetic data in MM [5]. These approaches require complex bioinformatics pipelines for the detection of SVs/CNVs, an abundance of CD138+ plasma cells, and large capital equipment requirements, which may be challenging to establish in a clinical laboratory.
Optical genome mapping (OGM) is an emerging technology that uses fluorescently labeled ultra-high molecular weight genomic DNA to restructure genome-wide SV and CNV maps. It has a simple workflow and straightforward bioinformatics analysis pipelines. It has been shown to detect genome-wide SVs/CNVs in hematologic malignancies [11–13]. To date, two pilot studies of OGM on CD138+ plasma cells have been reported: one study compared OGM data between 4 extramedullary and 7 intramedullary MM [14] and the other compared OGM and FISH results in 20 MM cases with plasma cell percentages of ≥10% [15]. Although both studies showed promising OGM results in MM for detection of genome-wide SVs and CNVs, there is no multi-center study reporting the clinical utility of combined OGM and NGS for pathogenesis/prognostication of MM and other PCN (with plasma cell percentages of <10%) in clinical practice.
This study includes concurrent karyotyping, FISH, OGM, and NGS analyses on 45 PCN patients from The Johns Hopkins Hospital and The University of Texas MD Anderson Cancer Center from January 2022 to November 2023 (Table 1, see Materials and Methods in the supplementary file for details). It includes 35 MM, 6 smoldering myeloma, 2 monoclonal gammopathy of undetermined/renal significance, and 2 amyloid light chain (AL) amyloidosis (Supplementary Table S1). One patient has simultaneous diagnosis of both MM and myelodysplastic syndrome (MDS) (case #30). OGM was performed on CD138+ plasma cells for cases #1–30 with median 25% plasma cells by morphology, and on fresh biopsy/aspirate for cases #31–45 without CD138-enrichment having >50% plasma cells. All FISH and NGS were performed on CD138+ uncultured specimens. The specimen was considered abnormal based on laboratory-established cutoffs/criteria. Concordance of karyotyping/FISH and OGM results was determined by a systematic review of the loci of interest for each sample. This study was approved by the Institutional Review Board and performed in accordance with the Declaration of Helsinki.
Table 1.
Genetic abnormalities by karyotyping, FISH, OGM, and NGS.
| IDa | Sex | Age | PC % core/diff (flow)b | Karyotype | FISH abnormality | OGM for FISH loci | OGM additional CNVs & SVs (Tier 1/2) | Pathogenic gene mutation by NGS |
|---|---|---|---|---|---|---|---|---|
| 1 | F | 55 | 25% (5.3%) | Normal | IGH::CCND1 | Yes | −X, −13, and 11q+ | No pathogenic mutations |
| 2 | M | 77 | 13% (2.5%) | Normal | 1q22+ | Yes | Hyperdiploidy, -X, −20, 2p+d | IKZF3 p.H474P |
| 3 | M | 65 | 13% (0.5%) | Normal (−Y) | IGH::MAF | Yes | −Y | No pathogenic mutations |
| 4 | F | 68 | 55% (7.3%) | N/A | 1p36.3−, 1q22+ | Yes | Hypodiploidy, 9q+ | No pathogenic mutations |
| 5 | F | 48 | 75% (5.6%) | Abnormal | 1q22+, IGH sep., MAF−, MAFB− | Yes (IGH::MAFAc) | 13q−d | No pathogenic mutations |
| 6 | F | 67 | 90% (46.3%) | Abnormal | 1q22+, IGH::CCND1 | Yes | −13, +15, chromoanagenesis, 1p−d | KRAS p.G12A, NFKBIZ p.E122fs |
| 7 | M | 76 | 25% (17.8%) | Normal | IGH::CCND1, TP53− | Yes | −Y, −13, 11q+, 16q−, CD38− | IDH1 p.R132L, KRAS p.Q61H, DIS3 p.G948a, TP53 p.K373fs |
| 8 | M | 76 | 5% (1.5%) | Normal | 1q22+, IGH::MAF, CCND1+ | Yes | Hyperdiploidy, −13, 20p−, −22 | N/A |
| 9 | M | 72 | 5% (0.6%) | Normal | Normal | Yes | Hyperdiploidy, −13 | N/A |
| 10 | F | 51 | 50% (11.5%) | N/A | Equivocal IGH sep., MAF− | Yes (IGH::MYCc) | Hyperdiplody, −X, −13, 16q− | KRAS p.G60D, TENT5C p.K82fs & p.H96fs, PTPN11 p.N308D |
| 11 | F | 69 | 55% (0.8%) | Normal | TP53− | Yes | IGH::MAFB, hypodiploidy, 1p−, 5p−, 5q−, 22q− | N/A |
| 12 | F | 66 | 13% (0.3%) | Normal | IGH::MAF | Yes | −X, +4, +9, +16, +21, TNFRSF17+ | No pathogenic mutations |
| 13 | M | 74 | 65% (31.1%) | N/A | 1q22+, TP53− | Yes | Hyperdiploidy, −Y, 10p−, 17q+, TNFRSF17+, chromoanagenesis | No pathogenic mutations |
| 14 | M | 63 | 45% (4.8%) | Normal | IGH rearrangement | Yes (IGH::CCND3c) | MYC::IGL, Xq+, 1p−, 14q−, 15q+ | No pathogenic mutations |
| 15 | M | 75 | 15% (1.3%) | Normal (−Y) | MYC+, TP53− | Yes | Hyperdiploidy, −Y, 6q−, 8p−, 8q+, 17q+ | KRAS p.Q61H |
| 16 | F | 63 | 50% (20.8%) | Normal | MYC rearrangement | Yes (inverted MYC) | Hyperdiploidy, −X, −18, chromoanagenesis, CD38+d | NRAS p.Q61R |
| 17 | F | 75 | 8% (0.7%) | N/A | 1q22+ | Yes | −X, −13, +15, 1p− | IDH2 p.R140Q |
| 18 | M | 73 | <10% (0.3%) | Normal | IGH::CCND1 | Yes | None | No pathogenic mutations |
| 19 | M | 65 | 85% (21%) | Normal | IGH::CCND1 | Yes | Hyperdiploidy, −13, 10q+, 12p−, 16q− | KRAS p.Q61H & p.G12C, PIK3R2 p.P121S, RB1 p.E440K & p.N505T, CCND1 p.I132M |
| 20 | M | 67 | 45% (N/A) | N/A | 1q22+, IGH::CCND1 | Yes | Hyperdiploidy, −Y, −13, 3q+, 11q+, TNFRSF17+, MYC sep. | KRAS p.Q61H, IKZF3 p.Y202D |
| 21 | M | 74 | 35% (N/A) | N/A | 1q22+, IGH− | Yes | −Y, −13, −14, bi-allelic RB1−d | No pathogenic mutations |
| 22 | F | 85 | 80% (32.8%) | N/A | 1q22+, IGH::MAF | Yes | −13, −22, 1p−d | N/A |
| 23 | M | 63 | 25% (4.0%) | Normal (−Y) | IGH− | Yes | Hyperdiploidy, Yq+, 14q− | N/A |
| 24 | M | 87 | 20% (11.1%) | Normal | Normal | Yes | Hyperdiploidy, chromoanagenesis 1, 11, 1p−d | No pathogenic mutations |
| 25 | F | 39 | 8% (1.6%) | Normal | IGH− | Yes | Hypodiploidy | No pathogenic mutations |
| 26 | M | 89 | 65% (19.1%) | Normal | 1q22+, IGH::CCND1 | Yes | −Y, 6q−, chromoanagenesis | No pathogenic mutations |
| 27 | M | 71 | 13% (0.6%) | Normal | Normal | Yes | Hyperdiploidy, 1p−, 6p+ | DIS3 p.D487H |
| 28 | M | 57 | 20% (4.2%) | Normal | Normal | Yes | Hyperdiploidy, 6q−, 16q−, MYC::IGL, chromoanagenesis | NRAS p.Q61L |
| 29 | M | 57 | 60% (14%) | N/A | Normal | Yes | Hyperdiploidy, Xq+ | N/A |
| 30 | M | 86 | 20% (0.9%) | Abnormal | IGH::CCNDI | Yes | −7, +8, −5q, dic (12; 17) (p13.32; p13.3) | TP53 p.R213G |
| 31 | M | 41 | 55% (4.6%) | Abnormal | 1q22+, MYC− | Yes (except 1q+) | IGH::FGFR3/NSD2 | No pathogenic mutations |
| 32 | M | 66 | 62% (14%) | Normal | CCND+, +9 | Yes | Hyperdiploidy, −Y, 6q− | N/A |
| 33 | M | 70 | 67% (23%) | Abnormal | CKS1B+, RB1− | Yes | +3, +7, +22, MYC::PECAM1 | N/A |
| 34 | F | 66 | 93% (21%) | Abnormal | CKS1B+, FGFR3+, IGH+, MYC+, +9, CCND1+, MAF::IGH, TP53− | Yes | −(X, 7, 11, 13, 16, 22), +(3, 6, 8, 9, 18, 20), chromoanagenesis 1, 2, 12 | DNMT3A c.2711C > T |
| 35 | M | 74 | 70% (34%) | Abnormal | FGFR3+, MYC sep., +9, CCND1+, MAF− | Yes (IGL::MYCc) | Hyperdiploidy, 18q+, chromoanagenesis 13, 16, 18 | N/A |
| 36 | F | 57 | 75% (32%) | Normal | IGH::CCND1, CKS1B+, −13 | Yes | +15, −X, MYC::TENT5C | BCL7A c.92+1G > C, NRAS p.Q61H |
| 37 | F | 74 | 57% (5%) | Normal | IGH::MAF, TP53−, −13 | Yes (except TP53−) | −X, TMEM200C sep., ARIH2 sep. | ATM p.Q2637a & p.K2440E, BRAF p.D594N, TP53 p.R337C, KLHL6 p.E17a, KRAS p.G13D, PIK3CA p.H665Y, PTPN1 c.494C > T, PLCG2 p.S1029C |
| 38 | M | 79 | 94% (40%) | Abnormal | IGH::CCND1, 5′MYC amp | Yes | 9p− | N/A |
| 39 | M | 63 | 74% (28%) | Abnormal | IGH::CCND1, CDKN2C−, CKS1B+, MAF−, RB1− | Yes | None | N/A |
| 40 | M | 78 | 75% (81%) | Abnormal | CKS1B+, MYC+, CDKN2A−, +9, CCND+, −13, MAF− | Yes | Hyperdiploidy, IGH::HDAC9, STK11 sep., 12p−d | DH2 p.V305M, KLF2 p.P125H, NRAS p.A59D |
| 41 | M | 70 | 52% (26%) | Normal | IGH::CCND1 | Yes | None | ASXL1 p.G646fs, CCND1 p.K46Q, CDKN2B c.158_ 159dupTC, IGLL5 p.S47G, p.A32D, c.1A > G (<5) & p.S59N, PLCG2 p.K381M, TP53 p.A276G |
| 42 | F | 68 | 71% (40%) | Abnormal | FGFR3−, IGH−, CCND+, +9 | Yes | Hyperdiploidy, 1p−, PTGFR::CD53, AL 606923.2 sep., KIAA1109::FOXN3, AP3B1::EDIL3, ELP4::AC131571.1, DIAPH2 sep., MACC1::ETV4d | IRAK1 p.N345S, NF1 p.A706V, RBMX p.A66V, p.R298a & p.R6C, TENT5C p.L142P |
| 43 | F | 76 | 78% (89%) | Abnormal | CKS1B+, IGH::FGFR3/ NSD2, MYC sep., CDKN2A−, −13, IGH+ | Yes (MYC::NBEAc) | None | N/A |
| 44 | M | 49 | 79% (35%) | Abnormal | −1, TP53−, MYC−, −13, IGH−, MAF− | Yes | Hypodiploidy, 6q− | N/A |
| 45 | M | 56 | 58% (53%) | Abnormal | CKS1B+, CCND1::IGH, −13 | Yes | 1p−d, numerous intra- and inter-chromosomal rearrangement | DNMT3A p.R729W & p.L566a, GNA13 p.C18S, IGLL5 p.A54V, SOX11 p.D233E |
NGS performed in CLIA/CAP-certified molecular diagnostics labs with coverage (>250×) and mutant allele frequency (>5%).
sep. rearrangement, + gain, − deletion/loss, N/A not available.
aOGM performed on CD138+ cells for sample IDs 1–30 and on fresh biopsy/aspirate without CD138+ for samples IDs 31–45.
bFor sample IDs 1–31, % PC reported for core biopsy from surgical pathology report. For sample IDs 32–45, % PC reported based on differential results.
cOGM revealed translocation partners.
d≥5 additional copy number variants (gains >10 Mb & losses >5 Mb) besides monosomy or trisomy.
In this cohort, 14 cases (38%) had abnormal karyotypes and a total of 233 genomic loci were tested by FISH with an average of 5 FISH loci per specimen (Supplementary Table S1). Ninety-eight percent of these loci (229 out of 233) displayed concordance between FISH and OGM (Table 1). Four discordant loci demonstrated the following: equivocal IGH rearrangement by IGH break-apart FISH had IGH::MYC fusion by OGM (case #10); MYC rearrangement by FISH had complex nested inversions within 8q by OGM (case #16); and two low-level CNVs by FISH were undetected by OGM due to low levels of plasma cells (cases #31 and #37 – both unselected cases with <5% plasma cells by flow) (Supplementary Fig. S1). Compared with FISH loci, OGM achieved 100% sensitivity, specificity, and accuracy in CD138+ cases and 96.6% sensitivity, 100% specificity, and 98.3% accuracy in unselected cases. Furthermore, OGM identified potential translocation partners in 5 cases, which supported IGH and/or MYC rearrangements by FISH (IGH::MAFA in case #5, IGH::MYC in case #10, IGH::CCND3 in case #14, IGL::MYC in case #35, MYC::NBEA in case #43). OGM identified two t(4; 14) samples with IGH::FGFR3/NSD2 fusions (cases #31 and #43).
Compared to limited FISH loci, OGM provided a genome-wide profile of SVs and CNVs. OGM revealed 18 hyperdiploidy, 4 hypodiploidy, and 9 IGH or MYC rearrangements that were not tested by FISH, as well as 8 cases (18%) with chromoanagenesis (complex genomic rearrangements and copy number alterations) (Table 1). OGM changed the prognostication beyond standard cytogenetics/FISH testing in eight cases (18% of cases in this study). Most samples displayed additional SVs/CNVs (Tier 3) by OGM: on average, an additional 7 intra- and inter-chromosomal translocation events (ranging from 0 to 90 translocations with a median of 4), an additional 0–5 CNV gain events (>1 Mb), and 0–14 CNV loss events (>1 Mb) per sample (Supplementary Table S2).
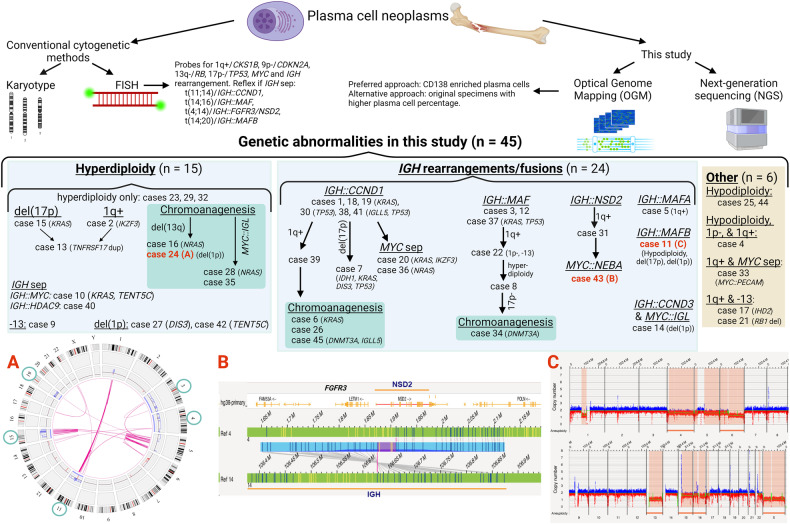
OGM’s comprehensive genome-wide SVs/CNVs profile led to classifying genetic subtypes in MM (Fig. 1). All cases had known MM abnormalities including 24 with IGH gene fusions, 15 with hyperdiploidy, and 6 had other abnormalities. These cases can be further classified according to additional MM abnormalities. Chromoanagenesis was frequent in cases with hyperdiploidy (n = 4, 27% of all hyperdiploidy cases) and in cases with IGH::CCND1 (n = 3, 23% of all IGH::CCND1 fusion cases). MYC::IGL fusions and NRAS mutations were found in half of the cases with chromoanagenesis and hyperdiploidy (cases #28, #35, and cases #16, #28, respectively). Gain of 1q was found in all cases with chromoanagenesis and IGH rearrangements/fusions, which might be a novel molecular subtype. Concurrent myeloma NGS results detected pathogenic mutations in 29 cancer genes, with recurrent mutations in KRAS (27%), IGLL5, NRAS, TENT5C, TP53 (10%) and CCND1, CDKN2B, DIS3, DNMT3A, IDH1/2, IKZF3, and PLCG2 (7%).
Fig. 1. Potential genetic testing workflow to classify PCN genetic subtypes in a clinical setting.
OGM and NGS on CD138+ plasma cells are preferred, although alternative approaches include OGM and NGS on original specimens with a higher plasma cell burden. A Circos plot of case #24 showing chromoanagenesis (red lines in the center) and hyperdiploidy (gain of chromosomes 3, 4, 11, 15, and 19, green circles). B Breakpoints of case #43 showing an IGH::FGFR3/NDS2 fusion. C Whole-genome view of case #11 shows hypodiploidy with losses of chromosomes 4, 6, 13, 15, 16, X, del(17p), and partial del(1p). Common pathogenic mutations in this cohort are listed for each case.
Given that only approximately one-third of cases had abnormal karyotypes and that FISH included assessment for a limited number of loci, OGM allowed for a more comprehensive definition of the plasma cell genome, not only for well-established FISH targets/regions, but also for hyperdiploidy/hypodiploidy, chromoanagenesis, atypical IGH and/or MYC translocation partners, and 366 novel SVs/CNVs that might be important in the formation and development of MM (Supplementary Tables S1–2). For case #30 with both MM and MDS, MM FISH on CD138+ plasma cells detected an IGH::CCND1 fusion while MDS FISH panel and karyotype on the fresh bone marrow detected del(5q), −7, +8, and dic(12p; 17p). OGM was able to detect both the MM and MDS abnormalities.
Genome-wide SVs/CNVs by OGM may be therapeutically helpful. For example, gain of BCMA/TNFRSF17 on 16p13.13 was found in patients #12 (+16) and #13 and #20 (BCMA/TNFRSF17+), detecting BCMA CNVs, such as gain/amplification or bi-allelic loss, may be associated with the effectiveness or resistance of BCMA-targeted monoclonal antibodies or CAR T-cell therapies. Patients #19 and #40 had a loss of GPRC5D on 12p13.1, who may not be ideal candidates for GPRC5D-targeted therapies.
In this study, genome-wide OGM analysis facilitated classification of genetic subtypes in PCN. We propose a potential clinical workflow for diagnostic testing for PCN (Fig. 1). We advise to perform OGM and NGS on CD138+ plasma cells. Genome-wide OGM results will not only provide comprehensive SVs/CNVs landscape for clarifying cytogenetic risks, but also may lead to the discovery of novel genetic biomarkers. In the absence of complex bioinformatics pipelines, OGM is an emerging method for genome-wide detection of SVs/CNVs in PCN.
In conclusion, this is the first and largest study (performed at two academic centers) reporting the value of combined OGM and NGS for PCN pathogenesis in clinical practice. A combination of OGM for genome-wide SVs/CNVs and NGS to interrogate for gene mutations may become an essential approach for evaluating genetic abnormalities in MM in the clinical setting. Future multi-center studies that incorporate larger numbers of MM cases, obtain comprehensive clinical data, and follow various treatment strategies will shed light on how genetic subtypes of MM are related to treatment response rates, survival, and overall prognosis.
Supplementary information
Supplementary Methods and Supplementary Figure S1
Acknowledgements
We thank all personnel of the Cytogenomic Laboratories in the Johns Hopkins and the University of Texas MD Anderson Cancer Center, and Johns Hopkins Genomics who involved in genetic testing. We also thank Amanda Dossat, Michael Gallagher, Manasi Pimpley, Erika Headrick, Stephanie Burke, and Natacha Diaz-Meyer from Bionano for a validation of OGM analysis pipelines. This study was supported by the Johns Hopkins School of Medicine Department of Pathology. RZO, the Florence Maude Thomas Cancer Research Professor, would like to acknowledge support from the Leukemia & Lymphoma Society (SCOR-12206-17), the National Cancer Institute (R01 CA266612), the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation, and the Riney Family Multiple Myeloma Research Fund at MD Anderson from the Paula and Rodger Riney Foundation.
Author contributions
YZ, CBK, CDK, and GT performed study concept and design; YZ and MK performed writing; YZ, MK, VS, JG, LM, PL, KB, CY, CDK, RX, and GT provided acquisition, analysis, and interpretation of data; MK, VS, JG, KT, JM, and KB provided technical and materials support. YZ, MK, VS, JG, LM, PL, JM, KB, CY, CDK, RX, KT, RO, CH, SA, PI, CBK, and GT contributed to the review, revision, and approval of the final paper.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41408-024-01059-x.
References
- 1.Palumbo A, Anderson K. Multiple myeloma. N Engl J Med. 2011;364:1046–60. doi: 10.1056/NEJMra1011442. [DOI] [PubMed] [Google Scholar]
- 2.Cowan AJ, Green DJ, Kwok M, Lee S, Coffey DG, Holmberg LA, et al. Diagnosis and management of multiple myeloma: a review. JAMA. 2022;327:464–77. doi: 10.1001/jama.2022.0003. [DOI] [PubMed] [Google Scholar]
- 3.Rajkumar SV. Multiple myeloma: 2022 update on diagnosis, risk stratification, and management. Am J Hematol. 2022;97:1086–107. doi: 10.1002/ajh.26590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palumbo A, Avet-Loiseau H, Oliva S, Lokhorst H, Goldschmidt H, Rosinol L, et al. Revised international staging system for multiple myeloma: a report from International Myeloma Working Group. J Clin Oncol. 2015;33:2863–9. doi: 10.1200/JCO.2015.61.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rustad EH, Yellapantula VD, Glodzik D, Maclachlan KH, Diamond B, Boyle EM, et al. Revealing the impact of structural variants in multiple myeloma. Blood Cancer Discov. 2020;1:258–73. doi: 10.1158/2643-3230.BCD-20-0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cardona-Benavides IJ, de Ramon C, Gutierrez NC. Genetic abnormalities in multiple myeloma: prognostic and therapeutic implications. Cells. 2021;10:336. doi: 10.3390/cells10020336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saxe D, Seo EJ, Bergeron MB, Han JY. Recent advances in cytogenetic characterization of multiple myeloma. Int J Lab Hematol. 2019;41:5–14. doi: 10.1111/ijlh.12882. [DOI] [PubMed] [Google Scholar]
- 8.Sharma N, Smadbeck JB, Abdallah N, Zepeda-Mendoza C, Binder M, Pearce KE, et al. The prognostic role of MYC structural variants identified by NGS and FISH in multiple myeloma. Clin Cancer Res. 2021;27:5430–9. doi: 10.1158/1078-0432.CCR-21-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rabani H, Ziv M, Lavi N, Aviv A, Suriu C, Shalata A, et al. Deletions and amplifications of the IGH variable and constant regions: a novel prognostic parameter in patients with multiple myeloma. Leuk Res. 2020;99:106476. doi: 10.1016/j.leukres.2020.106476. [DOI] [PubMed] [Google Scholar]
- 10.Christensen T, Deng W, McMahill B, Schappert J, Liu W, Saleki R, et al. Utilization of magnetic-activated cell sorting and high-density single nucleotide polymorphism microarrays improves diagnostic yield and prognostic value in clinical testing for patients with multiple myeloma and normal routine chromosome study. Acta Haematol. 2014;132:233–6. doi: 10.1159/000361074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang H, Garcia-Manero G, Sasaki K, Montalban-Bravo G, Tang Z, Wei Y, et al. High-resolution structural variant profiling of myelodysplastic syndromes by optical genome mapping uncovers cryptic aberrations of prognostic and therapeutic significance. Leukemia. 2022;36:2306–16. doi: 10.1038/s41375-022-01652-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levy B, Baughn LB, Akkari Y, Chartrand S, LaBarge B, Claxton D, et al. Optical genome mapping in acute myeloid leukemia: a multicenter evaluation. Blood Adv. 2023;7:1297–307. doi: 10.1182/bloodadvances.2022007583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luhmann JL, Stelter M, Wolter M, Kater J, Lentes J, Bergmann AK, et al. The clinical utility of optical genome mapping for the assessment of genomic aberrations in acute lymphoblastic leukemia. Cancers. 2021;13:4388. doi: 10.3390/cancers13174388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kriegova E, Fillerova R, Minarik J, Savara J, Manakova J, Petrackova A, et al. Whole-genome optical mapping of bone-marrow myeloma cells reveals association of extramedullary multiple myeloma with chromosome 1 abnormalities. Sci Rep. 2021;11:14671. doi: 10.1038/s41598-021-93835-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giguere A, Raymond-Bouchard I, Collin V, Claveau JS, Hebert J, LeBlanc R. Optical genome mapping reveals the complex genetic landscape of myeloma. Cancers. 2023;15:4687. doi: 10.3390/cancers15194687. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Methods and Supplementary Figure S1
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.