Summary
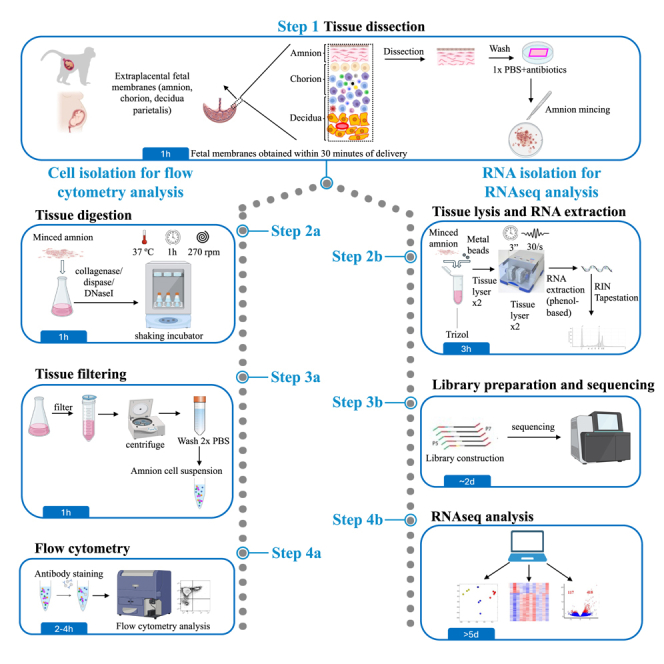
The amnion is a thin layer of fetal origin in contact with the amniotic fluid which plays a key role at the feto-maternal interface during pregnancy. Here, we present a protocol for isolation of human and Rhesusmacaque amnion cells. We describe steps for tissue dissection, cell isolation for flow cytometry analysis, and RNA isolation for RNA sequencing library preparation and analysis. This protocol can provide insights into altered immunological pathways during intrauterine infections to develop new therapeutic strategies.
For complete details on the use and execution of this protocol, please refer to Presicce et al.1
Subject areas: Sequence analysis, Flow Cytometry, Immunology, Model Organisms
Graphical abstract
Highlights
-
•
Instructions for optimal amnion tissue digestion and amnion cell isolation
-
•
Protocol to characterize amnion cells by flow cytometry
-
•
Detailed steps for amnion cell RNA-seq library preparation and analysis
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
The amnion is a thin layer of fetal origin in contact with the amniotic fluid which plays a key role at the feto-maternal interface during pregnancy. Here, we present a protocol for isolation of human and rhesus macaque amnion cells. We describe steps for tissue dissection, cell isolation for flow cytometry analysis, and RNA isolation for RNA sequencing library preparation and analysis. This protocol can provide insights into altered immunological pathways during intrauterine infections to develop new therapeutic strategies.
Before you begin
Institutional permissions
The use of human samples for laboratory experiments requires approved institutional review board (IRB). Similarly, all animal procedures must be approved by the Institutional Animal Care and Use Committees (IACUC). Human placentas were collected under UCLA IRB# 20–000579 and non-human primate under the IACUC (protocol # 22121) at the University of California Davis at the time of birth.
Your institution may require additional and/or region-specific permission as well as restrictions with some materials/reagents. Therefore, do not start working before the right approvals have been obtained.
Preparation of items for placenta collection
Timing: 4 h
-
1.
Sterile container to collect the placenta at room temperature (RT) (20°C–25°C) at the time of delivery. Placentas from both vaginal delivery and cesarean section can be used.
-
2.
Decontaminate a class II biosafety (BSL2) cabinet with UV light.
-
3.Sterilize:
-
a.Two tweezers and one pair of scissors.
-
b.Two metal trays.
-
a.
-
4.In the BSL2 cabinet, place:
-
a.A container with 10% (v/v) bleach solution to collect liquid waste.
-
b.Another container with a biohazard bag to discard the unused tissue and contaminated plastics.
-
a.
-
5.
Place absorbent pads, tube racks, sterile 50 mL conical tubes, sterile PBS, sterile large Petri dishes, and disposable scalpels in the BSL2 cabinet.
-
6.
Prepare 500 mL of sterile PBS + antibiotics (penicillin 10,000 unit/mL; streptomycin 10,000 μg/mL).
-
7.
Pre-warm DMEM/F-12 media (37°C).
-
8.
Pre-warm shaking incubator (37°C).
Preparation of reagent stocks
Timing: 30 min
-
9.
Reconstitute collagenase A powder in DMEM/F-12 [50 mg/mL] in sterile conditions.
-
10.
Reconstitute dispase II powder in DMEM/F-12 [12.5 mg/mL] in sterile conditions.
-
11.
Reconstitute DNaseI powder in sterile PBS [2 mg/mL] in sterile conditions. Filter with syringe filter (PES 0.2 μm), aliquot 200 μL/vial, and store at −20°C up to one year.
CRITICAL: Collagenase A and dispase II powders are very hard to dissolve. Before reconstitution, powders should be at RT.
For 2.5 g of collagenase A, slowly add 20 mL of pre-warmed DMEM/F-12 and incubate at RT for 10 min without resuspending the mixture. Add 30 mL of pre-warmed DMEM/F-12 and with a motorized pipet filler slowly pipette up and down avoiding bubbles and until the powder is completely dissolved. Therefore, the final concentration of the stock solution will be 50 mg/mL and it will be enough to digest 50 samples. Aliquot this stock solution in sterile 1.5 mL tubes (1 mL/tube) and keep it at 4°C up to one month. Make sure to gently resuspend the stock solution before adding it to the sample into the Erlenmeyer flask.
For dispase II, weigh 125 mg of powder/sample and transfer it into a 50 mL conical tube (up to four samples). Slowly add 10 mL/sample of pre-warmed DMEM/F-12 and incubate at RT without resuspending the mixture. After 10 min, gently resuspend the mix avoiding bubbles. Keep it at 4°C up to one week. Make sure to gently resuspend the stock solution before adding it to the sample into the Erlenmeyer flask.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Live/Dead Fixable Aqua Dead Cell Stain Kit (human/rhesus) (1:200) | Thermo Fisher Scientific | Cat# L34957 |
| Anti-non-human primate CD45 (clone D058-1283) (1:20) | BD Biosciences | Cat# 562394; RRID:AB_756078 |
| Anti-human CD45 (clone HI30) (1:20) | BD Biosciences | Cat# 562279; RRID:AB_11154577 |
| Stabilizing fixative | BD Biosciences | Cat# 338036 |
| Biological samples | ||
| Human placenta | University of California, Los Angeles | |
| Non-human primate placenta | University of California, Davis | |
| Chemicals, peptides, and recombinant proteins | ||
| Dulbecco’s phosphate-buffered saline (DPBS) w/o Ca2+/Mg2+ | Thermo Fisher Scientific | Cat# 14190144 |
| Penicillin-Streptomycin | Thermo Fisher Scientific | Cat# 15140122 |
| Collagenase A | Roche | Cat# 11088793001 |
| DNaseI | Roche | Cat# 10104159001 |
| Dispase II | Life Technologies | Cat# 11320-033 |
| DMEM/F-12 | Gibco | Cat# 11320-033 |
| ACK lysing buffer | Gibco | Cat# A1049-01 |
| Trypan blue stain | Gibco | Cat# 15250-061 |
| Fetal calf serum | Thermo Fisher Scientific | Cat# 16000-044 |
| Human IgG | Sigma | Cat# I2511 |
| Trizol | Sigma | Cat# T8424 |
| Critical commercial assays | ||
| NEBNext rRNA Depletion kit with sample purification beads | New England Biolabs | Cat# E6350 |
| NEBNext Ultra II RNA kit with sample purification beads | New England Biolabs | Cat# E7765 |
| NEBNext Multiplex oligos for Illumina (96 unique dual index primer pairs) | New England Biolabs | Cat# E6440 |
| Illumina TruSeq RNA Exome Enrichment | Illumina | Cat# #20020490 |
| Illumina Exome Panel – Enrichment Oligos | Illumina | Cat# 20020183 |
| SPRIselect | Beckman Coulter | Cat# B23317 |
| Qubit RNA BR Assay Kit | Thermo Fisher Scientific | Cat# Q10210 |
| Qubit dsDNA BR Assay Kit | Thermo Fisher Scientific | Cat# Q32850 |
| Qubit 1× dsDNA HS Assay Kit | Thermo Fisher Scientific | Cat# Q33230 |
| RNA ScreenTape | Agilent | Cat# 5067-5576 |
| RNA sample buffer | Agilent | Cat# 5067-5577 |
| D1000 ScreenTape | Agilent | Cat# 5067-5582 |
| D1000 sample buffer | Agilent | Cat# 5067-5602 |
| RNeasy Mini kit (optional) | QIAGEN | Cat# 74104 |
| Deposited data | ||
| Bulk RNA-seq, human amnion | GEO | GSE243830 |
| Bulk RNA-seq, rhesus amnion | GEO | GSE243830 |
| Experimental models: Organisms/strains | ||
|
Macaca mulatta Females, 8–10 years old |
University of California, Davis | |
| Software and algorithms | ||
| BD FACSDiva | BD Biosciences | http://www.bdbiosciences.com/us/instruments/research/software/flowcytometry-acquisition/bdfacsdivasoftware/m/111112/features |
| FlowJo, Version 10 | FlowJo | https://www.flowjo.com |
| Other | ||
| Cell lyser/homogenizer | ||
| Cell strainer 70 μm | Fisherbrand | Cat# 22363548 |
| 50 mL polypropylene conical tubes | Falcon | Cat# 352070 |
| 2 mL sterile tubes | Eppendorf | Cat# 022600044 |
| 1.5 mL sterile tubes | Fisherbrand | Cat# 05-408-129 |
| 1.5 mL Phasemaker tubes | Invitrogen | Cat# A33248 |
| 2.4 mm metal beads | Omni International | Cat# 19-640-3 |
| 5 mL serological pipet | Falcon | Cat# 357543 |
| 10 mL serological pipet | Falcon | Cat# 357551 |
| 25 mL serological pipet | Falcon | Cat# 357535 |
| 5 mL sterile disposable syringes | BD | Cat# 309646 |
| Syringe filters, PES (0.2 μm), Sterile | Basix | Cat# 13-1001-06 |
| Sterile specimen containers | Repligen | Cat# 725500 |
| Parafilm wrapping film | Fisher Scientific | Cat# 13-374-12 |
| Cell scraper | Falcon | Cat# 353085 |
| Scissors | Fisherbrand | Cat# 08-940 |
| Tissue forceps | Fisherbrand (Stoelting) | Cat# 10-001-274 |
| Flow cytometry analyzer | BD Biosciences | Fortessa |
| Shaking incubator | New Brunswick Scientific | Excella E24 |
| Centrifuge | Beckman Coulter | Allegra X-I4R |
| Mini centrifuge | Beckman Coulter | Microfuge 20R |
| Small FACS tubes | USA Scientific | Cat# 1412-101000 |
| Non-tissue culture-treated plates, 96 well, U-bottom | Falcon | Cat# 351177 |
| Disposable scalpels | Feather | Cat# 5200030 |
| −80°C freezer | Thermo Fisher Scientific | Cat# TDE40086FA |
| Qubit 4 fluorometer (or earlier versions) | Thermo Fisher Scientific | Q33238 |
| Qubit assay tubes | Thermo Fisher Scientific | Q32856 |
| 4200 TapeStation System | Agilent | G2991BA |
| Optical tube strip caps (8× strip) | Agilent | Cat# 401425 |
| Optical tube strips (8× strip) | Agilent | Cat# 401428 |
| Loading tips, 1 Pk | Agilent | Cat# 5067-5153 |
Materials and equipment
Washing buffer with antibiotics
-
•
1× Pen/Strep: add 50 mL of antibiotics in 500 mL 1× sterile PBS.
Can be stored for up to 1 week at 4°C.
Digestion buffer
| Reagent | Stock concentration | Volume of stock solutions | Final concentration |
|---|---|---|---|
| Collagenase A | 50 mg/mL | 1 mL | 0.5 mg/mL |
| Dispase II | 12.5 mg/mL | 10 mL | 1.25 mg/mL |
| DMEM/F12 | 89 mL | ||
| Total | 100 mL |
Should be made fresh in the Erlenmeyer after adding the minced tissue.
Flow cytometry wash buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| 1 × PBS | 9.8 mL | |
| Fetal calf serum (FCS) | 2% | 200 μL |
| Total | 10 mL |
Keep on ice. Can be stored for up to 1 week at 4°C.
Flow cytometry staining buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Flow cytometry staining buffer | 9.8 mL | |
| Human IgG | 20 μg/mL | 200 μL |
| Total | 10 mL |
Should be made fresh on the day of use and kept on ice.
Surface marker master mix (per tube/well)
| Reagent | Final concentration | Amount |
|---|---|---|
| Flow cytometry staining buffer | 44.75 μL | |
| Live/Dead Fixable Aqua Dead Cell Stain Kit | 1:200 | 0.25 μL |
| Anti-NHP or -human CD45 | 1:10 | 5 μL |
| Total | 50 μL |
Should be made fresh on the day of use and kept at 4°C protected from light.
Step-by-step method details
Amnion collection
Timing: 1 h
In this section, we describe in detail how to dissect the amnion from the amnion-chorion-decidua membranes (Figure 1). Figure 1 shows human term placenta. However, the same protocol has been validated for both human and non-human primate preterm placentas.2,3,4
-
1.Place the placenta on a sterile tray. Find the point of rupture in the membranes and lift the fetal membranes (i.e., amnion-chorion-decidua parietalis).
-
a.With sterile scissors, cut the membranes and place them on the tray with the decidua parietalis on the top and the amnion on the bottom. For a correct orientation: the amnion is smooth and shiny while the decidua parietalis is rough (Figure 1A).
-
b.With sterile tweezers, remove the blood clots (Figure 1B).
-
c.Rinse the fetal membranes with washing buffer with antibiotics (Figure 1C).
-
d.Pre-wet a second sterile tray with washing buffer with antibiotics and transfer the fetal membranes (see the troubleshooting section – problem 1).
-
a.
-
2.
With a cell scraper, gently remove the chorion-decidua and collect the amnion (Figures 1D and 1E).
-
3.
Weigh and record the amnion weight.
-
4.
Place the amnion in a pre-wet sterile Petri dish with washing buffer with antibiotics (Figure 1F).
-
5.
Finely mince the amnion using opposing scalpels (Figure 1G).
-
6.
Proceed with either tissue digestion to obtain amnion cell suspension for flow cytometry analysis or RNA extraction for RNA-seq experiments.
-
7.After amnion collection, follow:
-
a.Steps 8–29 for cell isolation and flow cytometry analysis.
-
b.Steps 30–71 for RNA extraction and RNA-seq library preparation.
-
a.
CRITICAL: placenta must be collected and processed within 30 min after delivery.
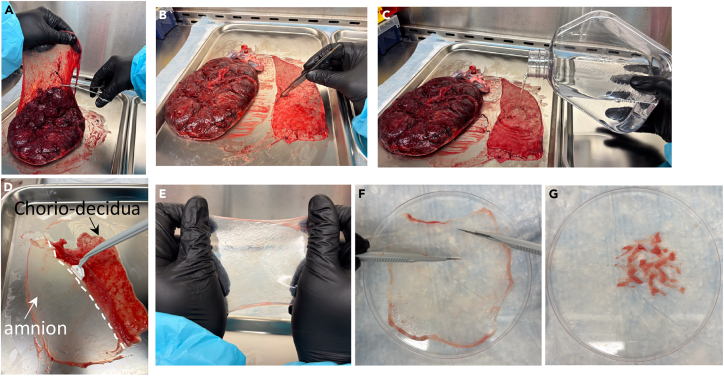
Figure 1.
Amnion collection
(A) Find the rupture of membrane site, lift the chorioamnion-decidua layers (i.e., fetal membranes) and separate them from the placenta with sterile scissors.
(B) Place the fetal membranes with the decidua parietalis facing upward. Remove any blood clots with sterile tweezer.
(C) Rinse the fetal membranes with washing buffer with antibiotics.
(D) Amnion collection is done by gently scraping the decidua parietalis and the chorion using a cell scraper.
(E) The isolated amnion is very thin, clear, and elastic.
(F) Place the amnion on a sterile Petri dish.
(G) Finely mince the amnion.
Amnion digestion, cell isolation, and cell staining for flow cytometry
Timing: 3 h
In this section, we describe in detail the steps for optimal amnion tissue digestion and for amnion cell isolation. We recommend beginning with fresh tissues for amnion cell isolation
-
8.Place the minced amnion in a sterile Erlenmeyer flask and add the digestion buffer (see “Digestion buffer” table above) consisting of:
-
a.1 mL of stock solution of collagenase A.
-
b.10 mL of stock solution of dispase II.
-
c.Pre-warmed medium at 37°C (89 mL).
-
a.
-
9.
Seal the flask with parafilm wrapping film and incubate for 30 min at 37°C with rotation (225 RPM) (Figure 2A).
-
10.
Move the flask to the safety cabinet and add 200 μL of stock solution of DNaseI (final concentration 4 μg/mL).
-
11.
Seal the flask and incubate for 30 min at 37°C with rotation (225 RPM).
-
12.
A successful digestion results in the complete amnion dissociation (Figure 2B).
-
13.
Filter the digested amnion through a sterile 70-μm nylon mesh to remove cell clumps of residuals of undigested amnion (Figure 2C).
-
14.
Pellet the cell suspension for 7 min at 600 × g (RT) (Figure 2D).
-
15.
Gently discard the supernatant (see the troubleshooting section – problem 2).
-
16.
Resuspend the cell pellet in 50 mL of sterile PBS 1× (Figure 2E).
-
17.
Pellet the cell solution for 7 min at 600 × g (RT).
-
18.
Repeat steps 16 and 17.
-
19.
Proceed with cell count with trypan blue exclusion test and record the cell viability.
-
20.
In FACS tubes or in wells of rounded 96-well, resuspend 1 × 106 cells in 50 μL of blocking buffer/tube or well and incubate for 10 min at 4°C.
-
21.
Add antibodies: Live/Dead Fixable Aqua Dead Cell Stain Kit and CD45.
-
22.
Protect the tubes/plates from the light and incubate for 20 min at 4°C.
-
23.
Add 500 μL/tube or 150 μL/well of cold FACS washing buffer.
-
24.
Spin the tubes or plates for 4 min at 930 × g at 4°C.
-
25.
Carefully aspirate the supernatant without disturbing the cell pellet.
-
26.
Repeat steps 23→25.
-
27.
Resuspend the cell pellet in 200 μL of BD stabilizing solution.
-
28.
Where a plate was used for staining, transfer the cell solution in a FACS tube. If not, proceed directly to step 29.
-
29.
Acquire at least 500,000 event/tube. Representative flow cytometry gating strategy is shown in Figure 3.
CRITICAL: After each centrifugation, carefully aspirate the supernatant because cell pellet may be loose.
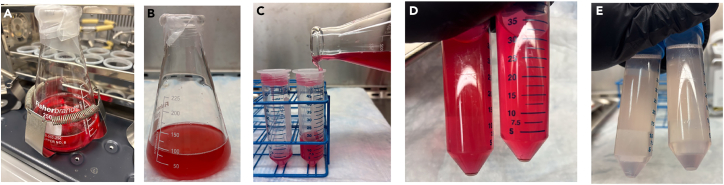
Figure 2.
Amnion digestion
(A) The minced amnion is placed in a sterile flask with digestion medium and incubated at 37°C in agitation (225 rpm) for 30’.
(B) With a complete digestion, the tissue is totally lysed and no floating pieces are visible.
(C) Filter the digested amnion through a sterile 70-μm nylon mesh to remove cell clumps of residuals of undigested amnion.
(D) Pellet the cell solution (7 min at 600 × g, at RT).
(E) Gently discard the supernatant and wash the cells in sterile PBS 1×.
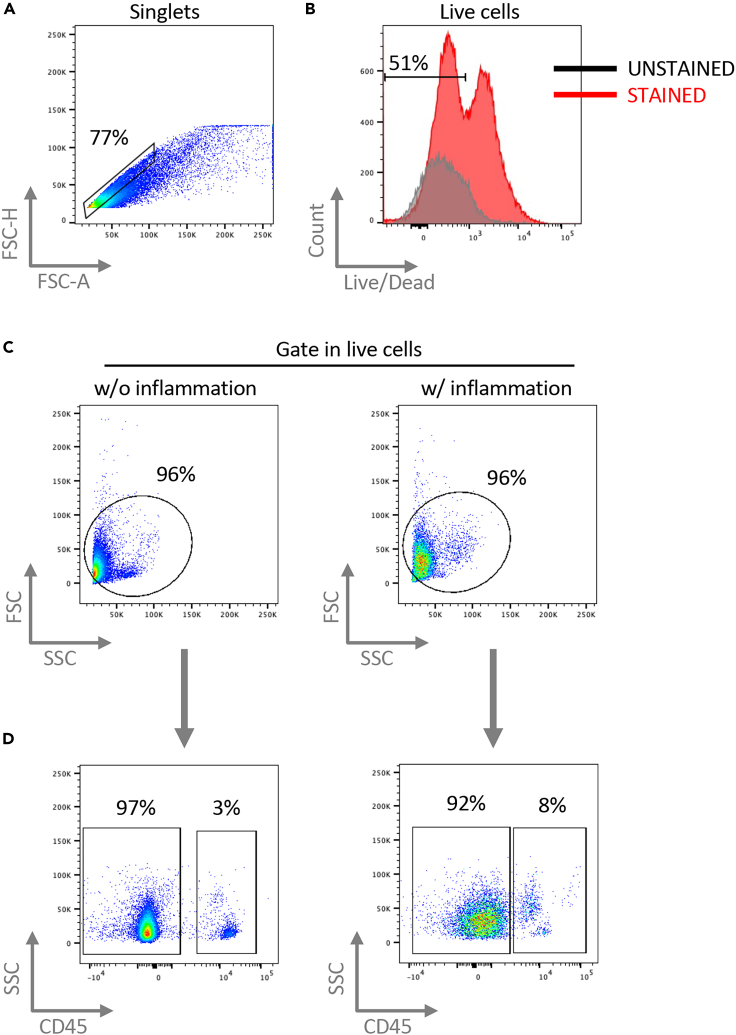
Figure 3.
Gating strategy for flow cytometry phenotype analysis of amnion cells
Representative density plots of Rhesus samples. The inflammation was induced by intra-amniotic injection of LPS in Rhesus macaque at ∼80% gestation (corresponding to 30–32 weeks of the human gestation). Age-matched controls were injected at the same gestational age with saline and use as controls (i.e., w/o inflammation). Note: the same gating strategy can be used for human preterm and term samples (not shown).
(A) Singlets gating to exclude doublets.
(B) Live cells positive gate.
(C) Forward (FSC) and side (SSC) scatter gating on cells to exclude cellular debris.
(D) CD45− cells negative gate representing amnion epithelial and mesenchymal cells and CD45+ cells positive gating representative infiltrating leukocytes. Note that in the inflamed sample the frequency of infiltrating neutrophils is higher compared to the non-inflamed sample.
RNA extraction, library preparation, and RNA-seq
Timing: varies (>3 days)
In this section, we describe in detail the steps for optimal RNA extraction (downloadable from https://tools.thermofisher.com/content/sfs/manuals/MAN0016162_100045875_PhasemakerTubes_QR.pdf) from amnion tissue and library construction for Next Generation Sequencing (NGS) (Figure 4A). Starting with freshly minced amnion is preferable. However, getting multiple samples at the same time may be challenging. Therefore, minced amnion can be snap frozen in liquid N2 and kept at −80°C up to ten years.
-
30.Pre-label 3 sets of sterile and nuclease-free tubes:
-
a.2 mL tube.
-
b.2 mL Phasemaker tube.
-
c.1.5 mL tube.
-
a.
-
31.
Pre-cool the minicentrifuge at 4°C.
-
32.
Centrifuge the Phasemaker tubes for 30 s at 14,000 × g prior to use.
-
33.
Using sterile tweezers, transfer 50–100 mg of minced amnion in a sterile 2 mL tube.
-
34.
Add 2 sterile metal beads/tube.
-
35.
Add 1 mL of Trizol reagent/tube (Figure S1A).
-
36.
Transfer the tube/s into the tissue lyser.
-
37.
After 3 min at 30/s check if the amnion is completely lysed (Figure S1B). If not, repeat step 37.
-
38.
Transfer each sample in one Phasemaker tube (Figure S1C).
-
39.
Incubate for 5 min at RT to allow complete dissociation of the nucleoproteins complex.
-
40.
Add 0.2 mL of chloroform and shake the tube/s vigorously by hand for 15 s.
-
41.
Incubate for 15 min.
-
42.
Centrifuge the tube/s for 5 min at 14,000 × g at 4°C.
-
43.
The solution will be separated into three phases: a colorless upper phase containing RNA, an interphase, and a lower red phenol-chloroform phase (Figure S1D).
-
44.
Carefully transfer the aqueous upper phase to a new 1.5 mL tube. Proceed to RNA precipitation (refer to Step 2 of RNA isolation protocol using Phasemaker tubes; downloadable from https://tools.thermofisher.com/content/sfs/manuals/MAN0016162_100045875_PhasemakerTubes_QR.pdf). Total RNA precipitate forms a white gel-like pellet at the bottom of the tube (Figure S1E). Wash the RNA (refer to Step 3 of RNA isolation protocol using Phasemaker tubes; downloadable from https://tools.thermofisher.com/content/sfs/manuals/MAN0016162_100045875_PhasemakerTubes_QR.pdf) and solubilize the RNA (refer to Step 4 of RNA isolation protocol using Phasemaker tubes; downloadable from https://tools.thermofisher.com/content/sfs/manuals/MAN0016162_100045875_PhasemakerTubes_QR.pdf).
-
45.
Measure the concentration of extracted RNA using the Qubit RNA BR Assay, according to the manufacturer’s protocol (see the troubleshooting section – problem 3).
-
46.
Evaluate the quality (RNA integrity – RINe, and the DV200) of the extracted RNA using the Agilent TapeStation Eukaryotic RNA Assay (Figure 4B). If the concentration measured in the previous step exceeds 500 ng/μL, then dilute the RNA accordingly (between 20 to 500 ng/μL). Alternatively, equivalent methods such as BioAnalyzer, QIAxcel, LabChip GX, etc., can be used (see the troubleshooting section – problem 4).
-
47.
Both human and non-human primate RNA samples are processed according to NEBNext rRNA Depletion Kit (Human/Mouse/Rat). We suggest starting with at least 200 ng of total RNA (see the troubleshooting section – problem 5).
-
48.
Referring to the Section 2 of the NEB manual E6350 (downloadable from https://zenodo.org/records/10691497), perform the following steps: hybridization of probes to the RNA; RNase H digestion; DNase I digestion; RNA purification using NEBNext RNA Purification beads. Purified RNA has a volume of 5 μL.
-
49.
Proceed with the library preparation using the NEBNext Ultra II Directional RNA kit. For partially degraded samples (RINe 2-7) continue with the fragmentation and priming step (steps 21a→d Section 2 of the NEB manual E6350; downloadable from https://zenodo.org/records/10691497), while for highly degraded RNA samples (RINe <2, usually Rhesus samples in our experience) skip to the priming step (steps 22a→d Section 2 of the NEB manual E6350; downloadable from https://zenodo.org/records/10691497).
-
50.PARTIALLY DEGRADED (RIN 2-7) SAMPLES ONLY:
-
a.Mix the rRNA-depleted sample (5 μL) with the 1st strand reaction buffer (4 μL) and random primers (1 μL).
-
b.Incubate the 10 μL reaction for 8 min at 94°C, then immediately transfer the samples on ice.
-
c.Add 8 μL of strand specificity reagent and 2 μL of 1st strand synthesis enzyme mix. Incubate the 20 μL reaction for 10 min at 25°C; 30 min at 42°C; 15 min at 70°C; hold at 4°C (heated lid > 85°C).
-
d.Proceed to 2nd strand synthesis (step 23 Section 2 of the NEB manual E6350; downloadable from https://zenodo.org/records/10691497).
-
a.
-
51.HIGHLY DEGRADED (RIN <2) SAMPLES ONLY:
-
a.Prime the rRNA-depleted RNA by adding 1 μL of random primers and incubate for 5 min at 65°C (heated lid > 90°C).
-
b.Immediately transfer on ice and add 8 μL of Strand Specificity Reagent, 4 μL of 1st strand reaction buffer and 2 μL of 1st strand synthesis enzyme mix.
-
c.Mix by pipetting and incubate the 20 μL reaction for 10 min at 25°C; 30 min at 42°C; 15 min at 70°C; hold at 4°C (heated lid > 85°C).
-
d.Proceed to second strand synthesis step 23 (Section 2 of the NEB manual E6350)
-
a.
-
52.
Proceed with the second strand synthesis, the following purification and EndPrep reactions according to the NEB manual (section 2, steps: 2.7, 2.8, 2.9; downloadable from https://zenodo.org/records/10691497).
-
53.
Ligate the End-Repaired and dA-tailed cDNA fragments (60 μL) using 2.5 μL of a 10-fold dilution of the NEBNext Adaptor (dilution obtained using the Adaptor Dilution Buffer; kit NEBNext Multiplex Oligos for Illumina - 96 Unique Dual Index Primer Pairs), 1 μL of the ligation enhancer and 30 μL of the Ligation Master Mix.
-
54.
Incubate for 15 min at 20°C in a thermocycler with heated lid off (or with open lid).
-
55.
Add 3 μL of the USERenzyme and incubate for 15 min at 37°C (heated lid > 50°C).
-
56.
Proceed to the purification using 0.9 volumes (87 μL) of the Sample Purification Beads.
-
57.Enrich purified adapter-ligated DNA fragments (15 μL) via PCR by mixing in the following components: 25 μL of NEBNext Ultra II Q5 Master Mix; 10 μL of dual index primer pair. Although discouraged, it is also possible to use:
-
a.Combinatorial dual indexed primers;
-
b.Single indexed i7 primer and universal i5 primer.
-
a.
-
58.
Incubate the reaction for 30 s at 98°C for the initial denaturation; followed by 12 cycles of: 98°C for 10 s; 65°C for 75 s. The final extension is performed at 65°C for 5 min, then hold at 4°C.
-
59.
Purify the amplified libraries with 0.9 volumes of Sample Purification beads according to manufacturer’s guidelines, resulting in 20 μL of purified final library.
-
60.
Quantify the final libraries using Qubit BR DNA Assay and the fragment distribution checked using the Agilent TapeStation D1000 Assay (see the troubleshooting section – problems 6 and 7).
-
61.
The final libraries are now ready to be enriched for the coding transcriptome using the Illumina TruSeq RNA Exome kit.
-
62.
Mix approximately 200 ng for each of four libraries in one pool (4-plex pool). If the number of samples is lower or if a different plexity is desired, please check the reference guide.
-
63.
Perform two rounds of hybridization/capture according to the TruSeq RNA Exome Reference Guide (1000000039582; downloadable from https://zenodo.org/records/10691497).
-
64.
After the second elution from streptavidin beads is complete, purify libraries using 1.8 volumes (45 μL) of SPRIselect beads.
-
65.
Mix the eluted enriched library pool (25 μL) with the Illumina PCR Primer cocktail (5 μL) and Enhanced PCR Mix (20 μL).
-
66.
Amplify the 50 μL reaction using the following program: initial denaturation for 30 s at 98°C; 10 cycles of: 98°C for 10 s, 60°C for 30 s, 72°C for 30 s; final elongation at 72°C for 5 min, then hold at 6°C.
-
67.
Purify amplified library pools using 1.8 volumes (90 μL) of SPRIselect beads and elute them in 30 μL.
-
68.
Quantify purified final library pools using Qubit HS DNA Assay and the fragment distribution checked using the Agilent TapeStation D1000 Assay (Figure 4C) (see the troubleshooting section – problem 7).
-
69.
Dilute library pools and submit them for Next-Generation Sequencing (Illumina) according to the service provider recommendations. The recommended read length for sequencing is 50 paired-end, but other options are allowed (not shorter than 50 single-end).
-
70.
Obtained fastQ files are then analyzed using any of the popular RNA-seq pipelines (such as STAR with -quantMode GeneCounts; HiSat2 followed by featureCounts, or others).
-
71.
A few examples of the resulting alignments are shown in Figures 5A–5C for both Rhesus and Human samples.
CRITICAL: After each centrifugation, carefully aspirate the supernatant because cell pellet may be loose.
Note: RNA purification is not needed because the polymer in the Phasemaker Tubes is inert and does not interfere with standard RNA applications such as RT-PCR, dot blot hybridization, poly(A)+ selection, RNase protection assay, etc. However, a commercial kit (e.g., RNeasy Kit by QIAGEN) can be used to improve the RNA quality according to the manufacturer’s instructions (downloadable from: https://www.qiagen.com/us/resources/resourcedetail?id=0e32fbb1-c307-4603-ac81-a5e98490ed23&lang=en, https://www.qiagen.com/us/resources/resourcedetail?id=f9b2e5ef-9456-431a-85ed-2a2b9fbd503d&lang=en).
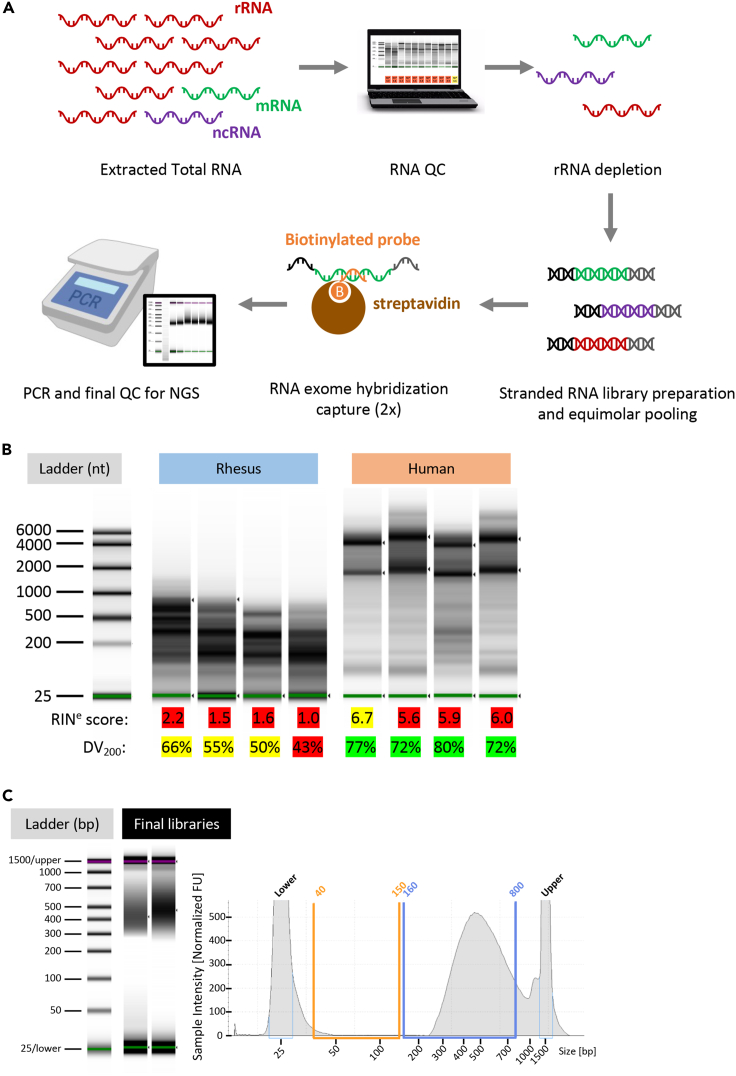
Figure 4.
RNA library preparation
(A) RNA library preparation workflow. Total extracted RNA was depleted from rRNA after initial quality assessment. Depleted RNA was then used as input for stranded library preparation. Two rounds of capture hybridization using biotinylated probes were performed before the final amplification, purification, and QC, then submit for Next-Generation Sequencing.
(B) Images from the TapeStation RNA QC. The RINe score is automatically calculated by Eukaryotic RNA Assay, and the values can range from 1 (extremely degraded) to 10 (intact). RINe scores are highlighted in red (degraded RNA), yellow (mild/moderate degradation), and green (intact/minimal degradation). The DV200 represents the amount (in percent) of total RNA that is above the 300 nt. DV200 values are highlighted in red (< 50%), yellow (between 50% and 70%), and green (> 70%). The green line at the bottom of each lane represents the lower marker.
(C) Example of final libraries (pool of samples) post hybridization capture. In the left panel, two libraries are visualized as gel view. The green and purple lines in each lane represent the lower marker, and upper marker, respectively. In the right panel, one library is visualized as electropherogram/region view. If present, adapter-dimers usually lie in the orange-highlighted region, while insert-containing fragments are usually found above 160–200 bp (blue region).
Figure 5.
RNA sequencing reads distribution
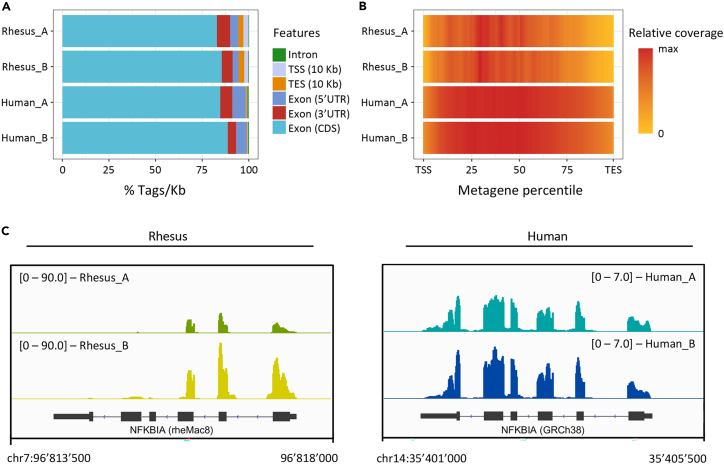
(A) Distribution of aligned reads on annotation features for two Rhesus and two Human samples. Genes: CDS Exons, 5′UTR Exons, 3′UTR Exons, Introns; Intragenic: 10 Kb upstream of the TSS, 10 Kb downstream of the TES. CDS = coding sequence; UTR = untranslated region; TSS = transcriptional start site; TES = transcript end site. Data obtained from the read_distribution.py module of RSeQC; plot generated with ggplot2 (v 3.4.4) in R (v 4.3.2).
(B) Distribution of reads over the metagene body for two Rhesus and two Human samples. Data obtained from the geneBody_coverage2.py module of RSeQC using CPM-normalized bigwig files, generated with deepTools bamCoverage (v 3.5.4.post1) using a --binSize = 10; plot generated with ggplot2 (v 3.4.4) in R (v 4.3.2) from scaled data (set max = 1).
(C) Genome browser screenshots for the Rhesus (left – genome rheMac8) and Human (right – genome GRCh38) NFKBIA gene showing the coverage of RNA-seq data (same bigwig files used in panel B).
Expected outcomes
The protocol is optimized for the isolation of amnion cells as well as RNA extraction from amnion tissues for NGS from both human and non-human primate placentas regardless of the gestational age. Cell yield depends on the amount of amnion tissues. Approximately, 1 g of tissue will result in 0.5–1 × 106 cells. However, the inflammation may increase cell yield because of the leukocyte infiltration as shown in Figure 3.
Limitations
The placentas should be stored at RT and the protocol works best when using fresh amnion tissues obtained within 30 min after delivery. Placentas up to 5 h after delivery can be used, but cell yield and quality dramatically decrease. When using frozen samples for RNA-seq, samples must be snap frozen very quickly after delivery in liquid nitrogen to ensure a good RNA quality.
Troubleshooting
Problem 1
The amnion gets dry very quickly and tends to adhere firmly to the tray and/or Petri dish (related to Step 1d).
Potential solution
Pre-wet both the tray and/or Petri dish with washing buffer with antibiotics and work as fast as possible.
Problem 2
The pellet is bloody (related to step 15).
Potential solution
-
•
Add 10 mL of ACK lysis buffer, incubate at RT for 7 min;
-
•
Fill the tube with sterile PBS, pellet the cell solution for 7 min at 600 × g (RT);
-
•
Discard the supernatant;
-
•
Wash with PBS and pellet the cell solution for 7 min at 600 × g (RT);
-
•
Gently discard the supernatant.
Problem 3
The concentration of the RNA is too low to be measured by the Qubit BR RNA assay (related to step 45).
Potential solution
If the starting material was less than the suggested 50 mg, the efficiency can be lower, impacting the final yield. If the concentration is too low to be measured with the Qubit RNA BR Assay, it can still be assessed with the High-Sensitivity counterpart Qubit RNA HS Assay (Thermo Fisher Scientific, cat# Q32852). However, an extremely low yield suggests problems during the RNA extraction, hence re-extraction from a new sample of tissue is highly recommended.
Problem 4
The quality of the RNA in terms of degradation is extremely low (related to step 46).
Potential solution
If the RINe score is below 3, then consider the DV200 method. This method measures the percentage of the RNA fragments larger than 200 bases. If the DV200 < 30%, extraction of RNA from a new tissue chunk is highly recommended.
Problem 5
The concentration of the RNA is too low to reach 200 ng in 12 μL (< 16 ng/μL).
Potential solution
The protocol for rRNA depletion allows total RNA input as low as 5 ng (related to step 47). However, this should be avoided since it will be necessary to increase the number of PCR cycles during the final amplification of the library prep prior to RNA exome probe hybridization, leading to a reduced complexity.
Please do not proceed with RNA/cDNA amplification attempts prior to rRNA depletion, as it will most likely not be compatible with this depletion protocol.
If a total amount of 200 ng of RNA has been extracted, but the concentration is too low, reduce the volume of the extracted RNA using SpeedVac at low temperatures (<50°C).
Problem 6
The total yield of the library prep is less than 200 ng (related to step 60).
Potential solution
Perform the last PCR (steps 57 → 60) on PCR amplified DNA for a total of 3-5 cycles. Use the same PCR barcoded primers or use universal Illumina primers (P5/P7). e.g., IDT xGen Library Amplification Primer Mix, 16 rxn, cat# 1077675; or Primer1: 5′ AATGATACGGCGACCACCGAGAT 3′ and Primer2: 5′ CAAGCAGAAGACGGCATACGA 3′.
Do not consider primers/adapter dimers (visible with the TapeStation assay) when measuring the concentration of the libraries. E.g. a sample has a contamination of adapter dimers of 13%: correct the Qubit-measured final library concentration to 87% (100-13%).
Problem 7
There are traces of adapter/primer dimers on the TapeStation report (related to steps 60 and 68).
Potential solution
If the contamination with primer/adapter dimers occurs after the rRNA-depleted library prep (step 60), it is usually not an issue, as the following steps (probe hybridization and capture) will remove it.
If the contamination (primer/adapter dimers > 1%) occurs post hybridization capture (step 68), perform the purification with the sample beads an additional time (step 67).
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact Pietro Presicce. Email: (ppresicce@mednet.ucla.edu).
Technical contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the technical contact, Monica Cappelletti (mcappelletti@mednet.ucla.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
This study did not generate or analyze dataset.
Acknowledgments
We would like to thank Dr. Claire A. Chougnet in the Division of Immunobiology at Cincinnati Children’s Hospital Medical Center for providing non-human primates used in this study. This study was supported by National Insitute of Health (NIH) grants R01 HD98389 (S.G.K.), K12 HD000849 (Y.A.), and the Burroughs Wellcome Fund (Y.A.). The graphical abstract was created with BioRender.com (https://BioRender.com).
Author contributions
M.C., D.S., M.M., and P.P. participated in data generation. M.C., D.S., M.M., M.P., M.R.J., Y.A., S.G.K., and P.P. participated in analysis and interpretation of data. S.G.K. and P.P. participated in the conception and design of the study and S.G.K. and Y.A. obtained the funding. M.C., M.M., and P.P. wrote the manuscript. All authors have reviewed the manuscript and approve the final version.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2024.103044.
Supplemental information
References
- 1.Presicce P., Cappelletti M., Morselli M., Ma F., Senthamaraikannan P., Protti G., Nadel B.B., Aryan L., Eghbali M., Salwinski L., et al. Amnion responses to intrauterine inflammation and effects of inhibition of TNF signaling in preterm Rhesus macaque. iScience. 2023;26 doi: 10.1016/j.isci.2023.108118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Presicce P., Cappelletti M., Senthamaraikannan P., Ma F., Morselli M., Jackson C.M., Mukherjee S., Miller L.A., Pellegrini M., Jobe A.H., et al. TNF-Signaling Modulates Neutrophil-Mediated Immunity at the Feto-Maternal Interface During LPS-Induced Intrauterine Inflammation. Front. Immunol. 2020;11:558. doi: 10.3389/fimmu.2020.00558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Presicce P., Park C.W., Senthamaraikannan P., Bhattacharyya S., Jackson C., Kong F., Rueda C.M., DeFranco E., Miller L.A., Hildeman D.A., et al. IL-1 signaling mediates intrauterine inflammation and chorio-decidua neutrophil recruitment and activation. JCI Insight. 2018;3 doi: 10.1172/jci.insight.98306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Presicce P., Senthamaraikannan P., Alvarez M., Rueda C.M., Cappelletti M., Miller L.A., Jobe A.H., Chougnet C.A., Kallapur S.G. Neutrophil recruitment and activation in decidua with intra-amniotic IL-1beta in the preterm rhesus macaque. Biol. Reprod. 2015;92:56. doi: 10.1095/biolreprod.114.124420. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate or analyze dataset.