Abstract
Background and aim
Most patients with hepatocellular carcinoma (HCC) in China have been diagnosed with spleen deficiency syndrome (SDS), which accelerates the progression of HCC by disrupting the tumor microenvironment homeostasis. This study aimed to investigate the intercellular crosstalk in HCC with SDS.
Experimental procedure
An HCC-SDS mouse model was established using orthotopic HCC transplantation based on reserpine-induced SDS. Single-cell data analysis and cancer cell prediction were conducted using Seurat and CopyKAT package, respectively. Intercellular interactions were explored using CellPhoneDB and CellChat and subsequently validated using co-culture assays, ELISA and histological staining. We performed pathway activity analysis using gene set variation analysis and the Seurat package. The extracellular matrix (ECM) remodeling was assessed using a gel contraction assay, atomic force microscopy, and Sirius red staining. The deconvolution of the spatial transcriptomics data using the “CARD” package based on single-cell data.
Results and conclusion
We successfully established the HCC-SDS mouse model. Twenty-nine clusters were identified. The interactions between cancer cells and cancer-associated fibroblasts (CAFs) were significantly enhanced via platelet-derived growth factor (PDGF) signaling in HCC-SDS. CAFs recruited in HCC-SDS lead to ECM remodeling and the activation of TGF-β signaling pathway. Deconvolution of the spatial transcriptome data revealed that CAFs physically surround cancer cells in HCC-SDS. This study reveals that the crosstalk of CAFs-cancer cells is crucial for the tumor-promoting effect of SDS. CAFs recruited by HCC via PDGFA may lead to ECM remodeling through activation of the TGF-β pathway, thereby forming a physical barrier to block immune cell infiltration under SDS.
Keywords: Hepatocellular carcinoma, Cancer-associated fibroblasts, Spleen-deficiency syndrome, Single-cell RNA sequencing, Spatial transcriptomics, Cell crosstalk, Tumor microenvironment, Traditional Chinese medicine
Graphical abstract

Highlights
-
•
The interactions between cancer cells and CAFs were enhanced via PDGF signaling in HCC with HCC-SDS.
-
•
CAFs recruited in HCC-SDS lead to ECM remodeling through activation of the TGF-β pathway.
-
•
CAFs recruited in HCC-SDS form a physical barrier surrounding cancer cells that may block immune cell infiltration.
List of abbreviations
- BSA
Bovine serum albumin
- CAF
Cancer-associated fibroblast
- DAPI
4′,6-Diamidino-2-phenylindole
- DMEM
Dulbecco's Modified Eagle Medium
- ECM
Extracellular matrix
- ELISA
Enzyme linked immunosorbent assay
- FBS
Fetal bovine serum
- GO
Gene Ontology
- GSVA
Gene set variation analysis
- HCC
Hepatocellular carcinoma
- iCAFs
Inflammatory CAFs
- IF
Immunofluorescence
- IHC
Immunohistochemistry
- AFM
Atomic force microscopy
- IL-1
Interleukin-1
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- myCAFs
Myofibroblastic CAFs
- PBS
Phosphate-buffered saline
- PC
Principal component
- PDGF
Platelet-derived growth factor
- PDGFA
PDGF subunit A
- PDGFB
PDGF subunit B
- PDGFR
PDGF receptor
- PDGFRA
PDGF receptor A
- PDGFRB
Platelet Derived Growth Factor Receptor Beta
- PVDF
Polyvinylidene fluoride
- p-SMAD2
Phospho-SMAD2
- RIPA
Radioimmunoprecipitation assay
- scRNA-seq
Single-cell RNA sequencing
- SDS
Spleen deficiency syndrome
- SDS-PAGE
Sodium Dodecyl Sulphate-PolyAcrylamide Gel Electrophoresis
- SPF
Specific pathogen-free
- ST
Spatial transcriptome
- TBST
Tris-buffered saline with Tween® 20
- TCM
Traditional Chinese Medicine
- TGF-β
Transforming growth factor-β
- TME
Tumor microenvironment
- UMAP
Uniform Manifold Approximation and Projection
- α-SMA
α-Smooth muscle actin
1. Introduction
Hepatocellular carcinoma (HCC) is one of the most common malignancies with poor prognosis and has developed into a significant public health issue.1 Traditional Chinese Medicine (TCM) has had remarkable success in treating HCC, based on its distinctive therapeutic approaches and a wide range of herbs.2,3 Interestingly, the majority of patients with HCC in China receive a clinical diagnosis of spleen deficiency syndrome (SDS), characterized by loss of appetite, weakness, pallor, weight loss, and loose stools.4 According to TCM, SDS is considered the core pathogenic factor in the HCC tumor microenvironment (TME) and is closely associated with the progression of HCC. For instance, the degree of malignancy was more severe, and the survival time was shorter in HCC rats with SDS, and SDS may accelerate HCC development by interfering with the TME homeostasis and by decreasing the infiltration of cytotoxic cells.5,6 Meanwhile, data from both laboratory and clinical studies tend to support the notion that Chinese herbal medicine can ameliorate the SDS and limit HCC progression by improving the pre-tumorigenic microenvironment.7,8 However, the underlying mechanism by which SDS regulates HCC progression remains to be explored.
The interactions among the cellular components in TME collectively drive tumor progression and response to immunotherapy.9 As the most prevalent cell type in the tumor stroma in TME, cancer-associated fibroblasts (CAFs) have recently been widely shown to be linked to poor prognosis and survival in a range of cancers.10 In the TME, several factors, such as interleukin-1 (IL-1), platelet-derived growth factor (PDGF), and transforming growth factor-β (TGF-β), contribute to the recruitment and activation of CAFs.11 Functionally, CAFs play a crucial role in accelerating tumor progression. CAFs produce components of the extracellular matrix (ECM) that can form a physical barrier that hinders drug delivery and restricts infiltration of cytotoxic T cells into the tumor; both events contribute to the establishment of an immunosuppressive environment.12,13 In addition, CAFs promote immune escape by directly suppressing the activity of immune effector cells and recruiting immunosuppressive cells.14 With the help of cytokines, chemokines, growth factors, and mechanical cues, CAFs are also crucial for promoting tumor growth.15 However, further research is needed to completely understand the specific mechanisms of crosstalk between cancer cells and CAFs in HCC, particularly with SDS.
Exploring the characteristics of the TME and spatial intratumor architecture in HCC is a crucial step in improving patient prognosis. Traditional research strategies, mainly at the bulk-tumor level, have inherent limitations in providing precise information about individual cells in a highly mixed TME.16 Single-cell RNA sequencing (scRNA-seq) is a technological innovation that solves the problem of scarce information collection regarding cellular heterogeneity by enabling the analysis of single-cell transcriptomes.17 Based on scRNA-seq technology, the cell types and characteristics of the TME in HCC have been refined.18 The spatial transcriptome (ST) platform can quantify the whole-transcriptome mRNA expression of thousands of genes superimposed on histological information from the same tissue slice.19 Gene expression profiles are mapped back to their original locations, enabling a direct link between histology and gene expression.20 The integration of scRNA-seq with ST improves sequencing capabilities and enables the in-situ visualization of signatures to be combined with the mapping of more cell types than ST alone.21
In this study, we aimed to dissect the crosstalk between cancer cells and CAFs in HCC with SDS by integrating scRNA-seq and ST, potentially assisting in preventing SDS-related immunosuppression, and further guiding immunotherapy for targeted CAFs combined with spleen-strengthening herbal therapy.
2. Materials and methods
2.1. Animals and experimental groups
Forty C57BL/6 mice (male; 4–6 weeks old; weighing 14–19 g) and four nude mice (4 weeks old, weighing approximately 20 g) were purchased from the Animal Center of the First Affiliated Hospital of Sun Yat-sen University (production license: SCXK [Guangdong] 2020–0056). The animals were kept in a specific pathogen-free (SPF) environment at the Animal Center of the First Hospital of Sun Yat-sen University (animal use permit: SYXK [Guangdong] 2020–0108). The use of mice in this experiment was approved by the Clinical Experimentation and Animal Ethics Committee of the First Hospital of Sun Yat-sen University (approval number 2020:402). The C57BL/6 mice were randomly divided into two groups (20 mice/group): HCC and HCC with SDS (HCC-SDS).
2.2. Establishment of the HCC-SDS model
SDS of the mice in the HCC-SDS group was induced by subcutaneous injection of reserpine (0.1 mg/kg/day) (purity ≥98.0 %, Macklin Biomedical Co., LTD, Shanghai, China) once a day for 14 days.4,6 The mice in the HCC group were injected subcutaneously with saline, following the same dosing scheme. The body weights of all mice and the amount of food consumed in each cage were recorded daily. The odor, mental state, body temperature, respiratory status, fur smoothness, food intake, and stools of the mice were also monitored. At day 14 post-reserpine injection, the severity of SDS in the HCC-SDS group was evaluated using the SDS rating scale (Table 1). Three days after injection of reserpine, HCC was established in mice by orthotopic tumor implantation.6 Approximately 1 × 106 Hepa1-6 cells in the logarithmic growth period were injected subcutaneously into the axilla of nude mice. After the tumors grew subcutaneously, they were collected and divided into fragments of approximately 1 mm × 1 mm and transplanted into the liver of C57BL/6 mice after sodium pentobarbital anesthesia. After transplantation, the mice were closely observed daily, and their body weight and feed consumption were recorded. Twenty-two days later, blood was collected from the eye using sterile forceps, and tumor tissue samples were collected and preserved for subsequent experiments.
Table 1.
Spleen deficiency syndrome rating scale.
| Index/Score | Grading standards |
|||
|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |
| Body odor | Odor-free | Mild odor | Medium odor | Severe odor |
| Mental state | Stable | Irritable | Fatigue | Somnolence |
| Chill & fever | Normal | Cowered | Chill | Arched back &trembling |
| Respiration | Normal | Panting | Tachypnea | Faint |
| Fur | Gloss | Matted | Fluffy & erect | Brown & erect |
| Defecate | Normal | Wet | Wet rotten | Mucous stool |
| Appetite | Normal | Reduced to 50 % | Reduced to 25 % | Not at all |
The calculated sum of the scores of each item (total score ≤3, no SD; 4 ≤ total scores≤ 7, mild SD; 8≤ total scores ≤14, typical SD; 15≤ total scores ≤21, severe SD).
2.3. Cell culture
The human hepatoma cell line MHCC97H and the mouse hepatoma cell line Hepa1-6 were obtained from the State Key Laboratory of Oncology in South China (Sun Yat-sen University Cancer Center, China). The human liver fibroblast cell line LX-2 was purchased from the Institute of Cell Resource Center, Chinese Academy of Sciences (Shanghai, China). All reagents used for cell culture were purchased from Gibco (USA). The cells were incubated at 37 °C and 5 % CO2 with Dulbecco's Modified Eagle Medium (DMED) supplemented with 10 % fetal bovine serum (FBS) and 1 % penicillin/streptomycin. Subcultures were conducted every 2–3 days.
2.4. Cell co-culture assay
The LX-2 and Hepa1-6 cells were digested and resuspended for counting. LX-2 cells (5 × 104) were seeded in the central chamber of a cell co-culture slide (ibidi GmbH, Germany), and equal numbers of Hepa1-6 cells were seeded in the other eight chambers. After the cells adhered, the medium was replaced with one containing 10 % of serum from HCC and HCC-SDS mice; for the control group, the same amount of phosphate-buffered saline (PBS) was used in place of the serum, and cells were co-cultured at 37 °C with 5 % CO2 for 4 h. The morphological changes of LX-2 cells were observed under a brightfield microscope (Olympus, Japan).
2.5. Transwell co-culture assay
Briefly, 1 × 105 LX-2 cells were seeded in the top chamber (Corning, 8 μm, USA) with 200 μL containing serum-free medium, and the lower chamber (Corning) was filled with a 600 μL mixture of hepatoma cells and medium with 10 % mice serum from HCC and HCC-SDS mice, and HCC-SDS mice serum plus a PDGF receptor B (PDGFRB) inhibitor (imatinib, 2 μM, MedChemExpress, USA) respectively; for the control group, the same amount of PBS was used in place of the serum. After incubation at 37 °C, 5 % CO2 for 24 h, the cells were fixed in 4 % paraformaldehyde and stained with 0.5 % crystal violet for 20 min. The cells in the top chamber were gently scraped using a cotton swab, and the migrated cells were imaged under a brightfield microscope.
2.6. Wound-healing assay
LX-2 and hepatoma cells were seeded on both sides of the wound healing culture insert (ibidi), and cell density was determined to be close to 100 % after adhesion. After stimulation with a medium containing 10 % serum from HCC and HCC-SDS mice or equal amount of PBS for the control group, the cells were photographed under a brightfield microscope to observe the relative motility at 0 and 24 h.
2.7. Gel contraction assays
The contractility of CAFs was assessed using a gel contraction assay. Co-culture was performed using the Transwell system. 500 μL of type I collagen (Corning, USA) was added to the 24-well plate, yielding a final collagen concentration of 2 mg/mL. Once the gel polymerized (1 h), 5 × 104 Hepa1-6 cells were seeded in the upper chamber, and the same number of LX-2 were seeded on the gel in the lower chamber. After the cells adhered, the medium was replaced with one containing 10 % of serum from HCC, HCC-SDS mice or HCC-SDS mice serum plus a TGF-β inhibitor (SB431542, 10 μM, MedChemExpress), the control group was replaced by PBS. Gel contraction was monitored at 0 and 48 h.
2.8. Evaluation of PDGF subunit A (PDGFA) levels in medium
LX-2 and Hepa1-6 cells were transwell co-cultured for 24 h at 37 °C as described above, and treated with 10 % serum from HCC and HCC-SDS mice; for the control group, the same amount of PBS was used in place of the serum. To measure the concentration of the PDGFA in medium, culture media were collected and centrifuged at 1000×g for 20 min. The levels of PDGFA in the supernatant was examined using a PDGFA enzyme linked immunosorbent assay (ELISA) kit (ELK Biotechnology, China) according to the manufacturer's instruction.
2.9. Western blotting
Western blotting analysis was performed to detect the protein levels of α-smooth muscle actin (α-SMA), PDGFA, PDGFRB, SMAD2, phospho-SMAD2 (p-SMAD2), Collagen I and Collagen III. Total proteins from cell and tissue samples were extracted using a radioimmunoprecipitation assay (RIPA) lysis buffer (Bioss, USA) containing proteinase inhibitors (Bioss) and a cocktail of phosphatase inhibitors (Bioss). The proteins were separated via 10 % sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto a polyvinylidene fluoride (PVDF) membrane. After blocking with 5 % skim milk for 1 h, PVDF membranes were incubated with specific primary antibodies (Zen BioScience, China) overnight at 4 °C. The next day, the membranes were washed thrice and incubated with a second antibody (Bioss) for 2 h at room temperature. Blots were imaged using a ChemiDoc XRS system (Bio-Rad, USA).
2.10. Immunofluorescence (IF) staining
IF analysis was performed using frozen sections of tumor tissue from HCC and HCC-SDS mice and cell slides after co-culture. The slides were first sealed with 5 % bovine serum albumin (BSA) for 30 min, and then incubated overnight with primary antibodies against α-SMA (Zen BioScience) or p-SMAD2 (Cell Signaling Technology, USA) at 4 °C. After washing thrice with tris-buffered saline with Tween® 20 (TBST), the slides were incubated with a second antibody (1:500; Bioss) combined with appropriate Alexa Fluor 594, protected from light, and then sealed with neutral gum containing 4′,6-diamidino-2-phenylindole (DAPI). Fields were randomly selected in each section to observe the staining condition under an inverted fluorescence microscope (Olympus, Japan).
2.11. Immunohistochemistry (IHC)
The HCC tissues were fixed in 4 % paraformaldehyde, embedded in paraffin, and sectioned. After deparaffinization and rehydration, antigen retrieval was performed using a citrate buffer (pH 6.0). The sections were then blocked with a goat serum solution for 45 min and incubated with primary antibodies at 4 °C overnight. The tissue sections were then incubated with secondary antibodies for 30 min at room temperature, followed by incubation with DAB for 5 min. Finally, the sections were stained with hematoxylin and mounted. Three fields were randomly selected for each section to observe the expression of target proteins under appropriate magnifications.
2.12. Sirius red staining
Sirius red staining was performed to evaluate the tumor tissue fibrosis. Briefly, paraffin-embedded tumor sections were dewaxed and incubated with Sirius red solution (Solarbio, China) for 1 h at 26 °C. After washing with running water for 10 min, the sections were dehydrated with ethanol and sealed with neutral gum. Fields were randomly selected in each section to observe the staining condition under an inverted microscope (Olympus).
2.13. Atomic force microscopy (AFM) measurements
AFM was used to measure the stiffness of tumor tissue. Briefly, frozen sections of tumor tissues from HCC and HCC-SDS mice were incubated in PBS containing protease inhibitors (1:100, Bioss) and 20 mg/mL propidium iodide (Sigma Aldrich, Germany) before the examination. A BIO-AFM (MFP-3D BIO, Asylum Research, USA) mounted on a fluorescence microscope (Olympus) was used to conduct AFM indentation. With a 10 μm diameter borosilicate glass spherical tip covering, the silicon nitride cantilevers were adjusted with a spring constant of 0.03 N/m. Before each measurement, the cantilever underwent a thermal oscillation calibration. Young's elastic modulus was calculated using a Hertz model with a Poisson's ratio of 0.5 based on the force-distance curves acquired by cantilever tapping the tumor matrix.
2.14. Single-cell RNA-sequencing and data analysis
2.14.1. Transcriptome sequencing and library preparation
Transcriptional sequencing and library preparation of one normal murine liver tissue sample and two HCC and three HCC-SDS murine tumor tissues was performed by Hangzhou Lianchuan Biotechnology Co., Ltd.
2.14.2. Quality control, cell-type clustering, and major cell-type annotation
Quality control and cell-type clustering were performed using the Seurat package22 of the R 3.6.2 software, and major cell-type annotation was performed manually. Standard quality control steps were used to filter unreliable cells. For each cell, two quality measurements were calculated: cells that had higher than 25 % expression of mitochondrial genes or expressed less than 500 genes were removed. In addition, genes were filtered to retain those expressed in at least one more cell. The normalized data were subjected to dimensionality analysis using principal component (PC) analysis. The top 45 PCs were selected for clustering, and the clustering parameter resolution was set to 0.7. After the clustering was completed, the cells were manually annotated based on the CellMarker23 and PanglaoDB24 databases and combined with the differential genes of each cluster.
2.14.3. Cancer cell prediction
We used the CopyKAT package25 to predict cancer cells. Using endothelial cells from the healthy group as a reference, fibroblasts, hepatocytes, and endothelial cells were extracted for prediction. Cells that undergo considerable aneuploidy mutations are considered cancer cells. The cancer cells we predicted were deemed to be annotated for downstream analysis as a new cell type.
2.14.4. Intercellular interaction analyses
We conducted intercellular interaction analysis using CellPhoneDB26 and CellChat.27 CellPhoneDB was used to calculate the number of interactions between cell–cell and receptor–ligand pairs, and CellChat was used to calculate the communication status between cells in terms of signaling pathways.
2.14.5. Functional and pathway activity analyses
To further clarify the major functions of active CAFs, we performed Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses. After that, the gene set variation analysis (GSVA) algorithm28 and the AddModuleScore function in Seurat were used to calculate pathway activity. Based on the GSVA algorithm, the Hallmark pathway dataset in the msigdbr package for mouse species was employed, with the data matrix as input, and the results were visualized using the pheatmap package.29 Based on the AddModuleScore function, Seurat objects and gene sets were selected and run, and finally, the results were mapped using Uniform Manifold Approximation and Projection (UMAP) plots.
2.15. Spatial transcriptomics validation
To further investigate the spatial co-localization of single cell clusters in HCC with SDS, we used the “CARD” package30 to deconvolute the ST database of single-cell data. Tumor tissues from HCC and HCC-SDS mice were cryosectioned at 10 μm thickness. The sections were mounted on Visium Spatial Gene Expression Slides. General morphological analysis and sequencing data were spatially aligned, stained with hematoxylin and eosin, and processed according to the Visium Spatial Gene Expression User Guide. Tissue sections were treated for permeability for 12 min, and libraries were prepared according to Visium user guidelines for spatial gene expression. Finally, the samples were sequenced on a NovaSeq 6000 (illumine, USA). We used the “CARD” package and created the “CARD” object using the “CreateCARDObject” function. The result was calculated using the “CARD_deconvolution” function with default parameters.
2.16. Statistical analysis
All data were analyzed using the GraphPad Prism 8.3.0 software and expressed as the mean ± SD. Differences between the two groups were analyzed using a two-tailed Student's t-test, and one-way ANOVA was used for multiple-group comparisons. Statistical significance was set at p < 0.05.
3. Results
3.1. Successful establishment of the HCC-SDS model
First, the experimental scheme for SDS induction in mice with transplanted HCC is presented in Fig. 1A. The average body weight and food intake of the HCC-SDS group mice decreased significantly (p < 0.0001) during reserpine treatment (Fig. 1B and C). On the other hand, the HCC group showed a moderate and stable increase in mean body weight, and food intake was similar to the initial level. As expected, SDS assessment revealed that the mice in the HCC-SDS group had significantly (p < 0.0001) higher scores than those in the HCC group (Fig. 1D). Consistently, the body weight and food intake of mice in the SDS-HCC group were significantly (p < 0.0001) lower than those in the HCC group after orthotopic HCC transplantation (Fig. 1E and F), and the postoperative HCC-SDS group maintained the SDS state until the mice were killed (Fig. 1G). The mice were dissected 22 days after transplantation. Tumors grew well in the livers of both groups of mice, and the tumors in the HCC-SDS group were noticeably larger than those in the HCC group (Fig. 1H), which suggests that, to some degree, HCC was more malignant in the HCC-SDS group. These results indicated that the HCC-SDS model was successfully established.
Fig. 1.
Establishment of the HCC-SDS model using reserpine and orthotopic transplantation. (A) Flow chart of the experimental protocol for establishment of the HCC-SDS model. (B) Changes in body weight during reserpine injection. (C) Food consumption per cage during reserpine injection, 5 mice per cage. (D) SDS score statistics following 14-day reserpine injection. (E) Changes in body weights following transplantation. (F) Food consumption per cage following transplantation, 5 mice per cage. (G) SDS score statistics before euthanasia. (H) Representative images of tumors. n = 10, ****p < 0.0001.
3.2. Single-cell transcriptomes define heterogeneity in HCC with SDS
To assemble a cell atlas of the TME in HCC-SDS, six samples (three HCC-SDS tissues, two HCC tissues, and one normal liver tissue) were sequenced using scRNA-seq technology. Fig. 2A shows the results after quality control, standardization, dimensionality reduction, clustering, and annotation of the sequencing data. A total of 58835 cells—27405 cells from HCC-SDS, 24366 cells from HCC, and 7064 cells from normal liver tissue, comprising immune cells, hepatocytes, and stromal cells—made up our single-cell atlas. Fig. 2B and C shows the distribution of the three groups and six tissue samples in UMAP. The cells formed 29 clusters, which we annotated based on known marker genes; the annotation methodology is shown in Fig. 2D. Next, we calculated the proportion of each cell type to characterize the changes in each cell type in the three groups. Interestingly, we observed some differences in the proportion of cell composition between the different clusters in the three groups (Fig. 2E), suggesting that the TME between HCC-SDS and HCC tissues is somewhat heterogeneous.
Fig. 2.
Identification of cell clusters and annotation. (A) UMAP plot of 29 cell clusters based on 58,835 single-cell transcriptomes. UMAP plot visualizations of (B) the three groups and (C) six tissue samples. (D) Dot plot depicting the cell-type-specific markers. (E) Proportion bar graph representing cluster frequency in the three groups. In each group, the sum of each cluster proportion is 100 %.
3.3. The proportion of cancer cells increases in HCC with SDS
Known marker genes alone cannot determine the malignancy of cells; therefore, we used the CopyKAT package to infer copy number variants from single-cell transcriptome data to identify cancer cells. Here, we used the endothelial cells of the healthy group as a reference and identified potential cancer cells (Fig. 3A). We predicted cancer cells in hepatocytes and fibroblasts, and 13,663 cells were predicted to be malignant; these were mostly composed of hepatocytes-C1, proliferating hepatocytes-C1, hepatocytes-C2, and proliferating hepatocytes-C3 (Fig. 3B and C). The distribution of cancer cells in the two groups is illustrated in Fig. 3D. Of the 13,663 cells, 9631 belonged to the HCC-SDS group (35 % of the total number of cells in the HCC-SDS group) and 4032 to the HCC group (16 % of the total number of cells in the HCC group) (Fig. 3E).
Fig. 3.
Prediction of cancer cells. (A) Heatmap of the predicted cancer cells. The upper markers are chromosomes 1 to 22 and X. The color markers on the left represent the predicted cell types, where green are normal cells and red are predicted cancer cells. Blue in the heat map indicates loss and red indicates amplification; a more obvious color change indicates a more obvious aneuploidy variation. (B) Cell types used for CopyKAT analysis. (C) UMAP plot of predicted cancer cells using CopyKAT analysis. Red indicates predicted cancer cells; light gray, non-cancer cells; and dark gray, filtered cells. (D) Distribution of cancer cells in the two groups. (E) Proportion bar graph representing the percentage of cancer cells.
3.4. Cancer cells–CAFs interaction was associated with PDGF signaling in HCC with SDS
To explore the differences in intercellular interactions between the TME of the HCC-SDS and HCC groups, we performed cell–cell interaction analyses. The results from CellPhoneDB showed that cancer cells and CAF–C1 interacted predominately in the HCC group (Fig. 4A), while the predominant interactions in the HCC-SDS group were CAF–C1, CAF–C2, CAF–C3, and CAF–C4 (Fig. 4B). Furthermore, we observed that the interactions between cancer cells and CAFs were significantly enhanced in the HCC-SDS group (Fig. 4C), while those between cancer cells and T cells were significantly reduced (Fig. 4D). Based on the CellChat package, compared to the HCC group, cancer cells in the HCC-SDS group exhibited a higher frequency of incoming and outgoing signaling (Fig. 4E). To further investigate and quantify the ligand–receptor interaction between cancer cells and CAFs, we investigated the signaling pathways involved in the interactions between cancer cells and CAFs. We found that in the HCC-SDS group, the PDGF pathway and related ligand–receptor interactions, including PDGFA–PDGFRB, PDGFA–PDGF receptor A (PDGFRA), PDGFB subunit B (PDGFB)–PDGFRB, and PDGFB–PDGFRA, affected cancer cells–CAFs interaction, with PDGFA–PDGFRB presenting the highest communication contribution (Fig. 4F and G). The above results suggest that cancer cells–CAFs interactions were associated with PDGF signaling in the HCC with SDS.
Fig. 4.
Cell–cell interactions. The interactions among 29 cell types in the (A) HCC group and (B) HCC-SDS group. (C) Frequency of interaction between cancer cells and CAFs. (D) Frequency of interaction between cancer cells and T cells. (E) Incoming and outgoing interaction strength between cancer cells and CAFs. (F) Heatmap of the predicted PDGF pathway. (G) Contribution of each receptor–ligand pair to cancer cells–CAFs interactions in the PDGF pathway in the HCC-SDS group.
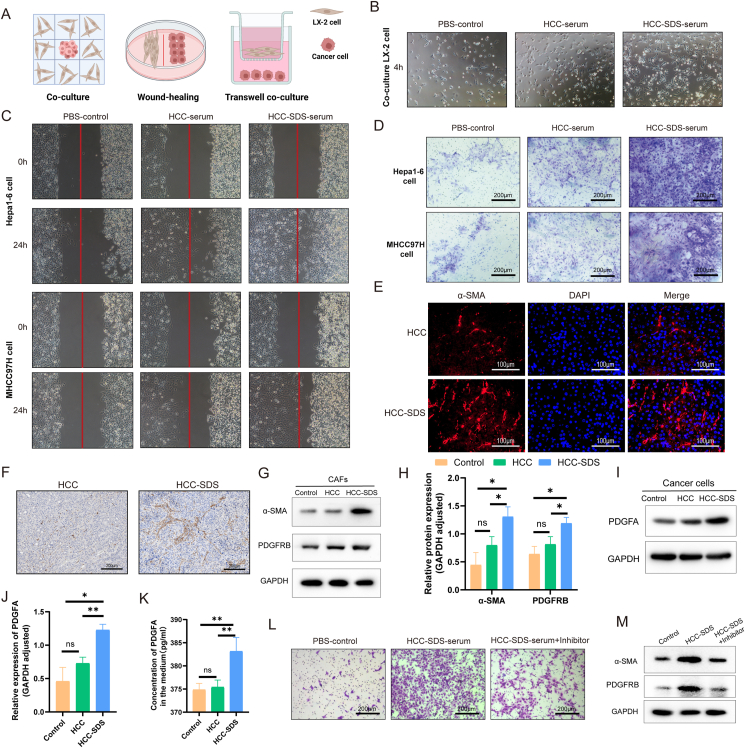
3.5. Cancer cells recruit CAFs by secreting PDGFA in HCC with SDS
To investigate whether SDS promoted the interaction between cancer cells and CAFs and the potential role of SDS in this event, we used three co-culture systems to verify the recruitment of CAFs by hepatoma cells in vitro (Fig. 5A). CAFs appeared bipolar or multipolar with elongated shapes after co-culture with Hepa1-6 cells stimulated with HCC-SDS serum for 4 h (Fig. 5B), exhibiting typically activated fibroblast morphology.31 The wound-healing assay showed that the capacity of hepatoma cells incubated in the HCC-SDS serum to recruit CAFs was enhanced compared to that of cells in the HCC serum and PBS (Fig. 5C). Similarly, as illustrated in Fig. 5D, the stronger capacity of hepatoma cells to recruit CAFs was observed in the Transwell assay in both cell lines in the HCC-SDS serum. Furthermore, The IF and IHC staining showed that the protein level of α-SMA in tumor tissues increased in the HCC-SDS group compared with that in the HCC group (Fig. 5E and F). CAFs were identified as activated fibroblasts using western blotting analysis, and the α-SMA expression of CAFs in the HCC-SDS group significantly (p < 0.5) increased after co-culturing (Fig. 5G and H). Additionally, compared with the other groups, the expression of PDGFA in HCC cells treated with the HCC-SDS serum was up-regulated, which corresponded to an increased expression of PDGFRB in CAFs (Fig. 5G, H, I, J). Meanwhile, ELISA ‘s results showed that more PDGFA was detected in the supernatant of the co-culture medium treated with serum from mice in the SDS-HCC group (Fig. 5K). The recruitment of CAFs in the serum of SDS-HCC mice was attenuated by PDGFRB inhibitors (imatinib)32,33 to inhibit PDGFA-PDGFRB receptor-ligand pair communication (Fig. 5L and M). These results are in line with research on cell–cell interactions, suggesting that cancer cells may recruit more CAFs via PDGFA in HCC-SDS.
Fig. 5.
Recruitment of CAFs by cancer cells. (A) Schematic representation of the experimental design of the co-culture assay. (B) Morphological changes of CAFs after 4-h co-culture with cancer cells. (C) Relative motility of CAFs after co-culturing with cancer cells in the wound-healing assay. CAFs are noted on the left and hepatoma cells on the right. (D) Representative photographs of CAFs recruited by cancer cells in the Transwell assay; scale bar: 200 μm. (E) Immunofluorescence analysis of α-SMA expression in HCC and HCC-SDS tissues; scale bar: 100 μm. (F) Immunohistochemical analysis of α-SMA expression in HCC and HCC-SDS tissue; scale bar: 200 μm. Western blotting analysis (G) and quantification (H) of α-SMA and PDGFRB in CAFs after co-culturing. Western blotting analysis (I) and quantification (J) of PDGFA in the cancer cells after co-culturing. (K) Levels of PDGFA in the medium were measured by ELISA. (L) Representative photographs of CAFs recruited by cancer cells treated with or without PDGFRB inhibitor in the Transwell assay; scale bar: 200 μm. (M) Western blotting analysis of α-SMA and PDGFRB in CAFs treated with or without PDGFRB inhibitor. *p < 0.5, **p < 0.01.
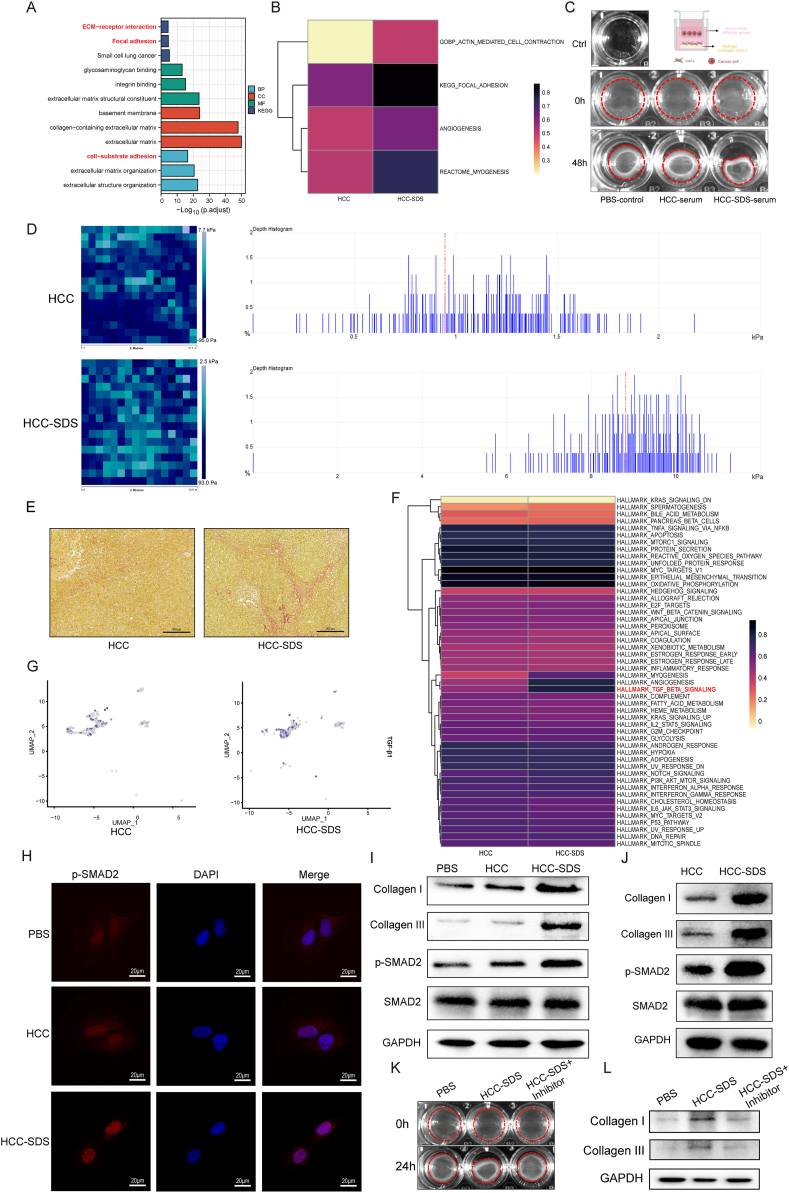
3.6. Enhanced ECM remodeling and activation of the TGF-β pathways in CAFs under SDS
We further analyzed the pathway enrichment and pathway activity of CAF differential genes to verify the contribution of SDS to CAF recruitment by HCC cells. As shown in Fig. 6A, KEGG enrichment annotation suggested that the genes with upregulated expression were significantly enriched in the pathway related to EMC–receptor interaction and focal adhesion. GO enrichment analysis of the biological process also suggested that the genes with upregulated expression were enriched in cell–substrate adhesion signaling. In agreement with the results of KEGG and GO enrichment analyses, GSVA indicated enhanced activation of ECM remodeling-related pathways in the HCC-SDS group samples (Fig. 6B). Fibroblasts promote matrix remodeling through active contraction, leading to matrix stiffening and desmoplasia.34 We observed that CAFs cultured in HCC-SDS serum contracted the gel to a greater extent than those in HCC serum and PBS (Fig. 6C). By calculating the elastic modulus of tumor tissues using AFM, we found that the tumor of the HCC-SDS mouse had a significantly stiffer matrix; the majority of the surface stiffness from the HCC-SDS group were gathered in 8–10 kPa, while those in the HCC group were gathered in 0.5–1.5 kPa (Fig. 6D). Moreover, Sirius red staining showed that the level of collagen was significantly increased in HCC-SDS group compared with HCC group (Fig. 6E). Therefore, we proposed that CAFs activated in HCC-SDS might lead to ECM remodeling. To explore the underlying molecular regulatory mechanism, we investigated the potential signaling pathways associated with ECM remodeling in CAFs using GSVA; we observed that the activity of the TGF-β pathway was significantly enhanced in the HCC-SDS group (Fig. 6F). The results of the Seurat AddModuleScore algorithm also showed that, compared with the HCC group, the expression of TGF-β1 was relatively upregulated in CAFs in the HCC-SDS group (Fig. 6G). TGF signaling pathway has long been identified with its strategic role in ECM assembly and remodeling during cancer progression, by promoting SMAD phosphorylation and translocation into the nucleus, thus regulating the transcription of target genes.35 Western blotting analysis revealed greatly elevated phosphorylation of SMAD2 and the IF also exhibited an increase in nuclear localization p-SMAD2, in the CAFs of the HCC-SDS group relative to that in the HCC group and PBS group after co-culture (Fig. 6H and I). Of note, TGF-β is a key contributor to shaping the ECM by stimulating the production and remodeling of collagens in CAFs.31,36 To further investigate whether TGF-β activation in CAFs regulates ECM-associated collagens, we have detected the expression of Collagen I and Collagen III. We found that, with the increase of SMAD2 phosphorylation, the expression of ECM-remodeling related proteins Collagen I and Collagen III were up-regulated under HCC-SDS (Fig. 6I). This differential expression pattern of the TGF-β signaling pathway between the SD-HCC and HCC groups was further confirmed at the protein level in the liver tumor tissues by Western blotting (Fig. 6J). Meanwhile, the ECM-remodeling effect of the CAFs under HCC-SDS was largely reversed by the specific inhibitor of the TGF-β signaling pathway in vitro (Fig. 6K and L). Thus, based on the abovementioned results, we speculated that CAFs recruited by cancer cells under SDS enhance contractility, resulting in ECM remodeling by activation of the TGF-β pathway.
Fig. 6.
Signal pathway analysis and biological function validations of activated CAFs. (A) The KEGG pathway and GO enrichment analyses of differentially expressed genes in CAFs. (B) GSVA heatmap of the ECM remodeling-related pathway in activated CAFs. (C) Representative images of gel contraction assay of CAFs, contracted gel within the red line. (D) Representative images of Young's modulus heatmap (left) and stiffness distribution (right) of the tumor matrix measured by AFM. (E) Representative images of Sirius red staining in HCC and HCC-SDS tissue; scale bar: 200 μm. (F) GSVA heatmap of Cancer Hallmark pathways in activated CAFs. (G) UMAP plot of Hallmark-TGFβ1 score. (H) Representative immunofluorescence images of p-SMAD2 in CAFs after co-culture; scale bar: 20 μm. Western blot analysis of p-SMAD, Collagen I and Collagen III protein expression in CAFs after co-culture (I) and tumor tissues of both groups (J). (K) Representative images of gel contraction assay of CAFs treated with or without TGF-β inhibitor, contracted gel within the red line. (L) Western blot analysis of Collagen I and Collagen III protein expression in CAFs with or without TGF-β inhibitor.
3.7. Spatial co-localization between cancer cells and CAFs
As our scRNA-seq analysis only investigated the function of CAFs, we sought to validate the co-localization of cancer cells and CAFs using ST analysis. Interestingly, as demonstrated in Fig. 7, the deconvolution results of HCC spatial transcriptomic data showed that CAFs physically surround cancer cells in the TME under SDS, while no such co-localization was observed in the HCC group.
Fig. 7.
Spatial transcriptomic analyses of co-localization of cancer cells and CAFs. The red circles are the spatial barrier structure composed of CAFs located near the tumor boundary.
4. Discussion
In China, most patients with HCC are clinically diagnosed with SDS, which is considered one of the major physiological factors contributing to the formation of the pro-tumorigenic microenvironment in HCC.4,6 In our study, consistent with previous reports,4,6 the HCC-SDS mice showed symptoms typical of SDS, including loss of appetite, mental depression, weight loss, laziness and pile-ups, withered sparse hair, and loose stools (perianal uncleanliness), which were not observed in the HCC mice, indicating that model of HCC-SDS was successfully established. Moreover, the tumors in the HCC-SDS group were substantially larger than those in the HCC group, suggesting that SDS promoted the malignant progression of HCC to some extent. However, it is still difficult to comprehensively assess the characteristics of the HCC-SDS model based only on the changes in symptoms and indicators, and an in-depth analysis of the changes in the TME may be beneficial for understanding the influence of SDS on HCC progression.
The interactions among the cellular components of TME collectively determine tumor progression and response to immunotherapy.9 Here, we mapped a single-cell atlas of HCC with SDS to dissect its TME characteristics and intercellular interactions. We first focused on the proportion of each cell type and found that the proportion of cells in the HCC TME was altered by SDS. Previous studies have also found that SDS interferes with TME homeostasis in HCC by decreasing the proportion and activity of cytotoxic cells.5,37 Notably, the number of cancer cells was significantly higher in SDS. The SDS-induced immunosuppressive microenvironment avoids the killing of cytotoxic cells37; thus, more cancer cells can be observed under SDS, indicating that SDS promoted HCC progression at the TME level. Prior studies have demonstrated that the most abundant cells in TME are CAFs, which are mainly derived from mesenchymal fibroblasts and regulate tumor biological behavior directly or indirectly by promoting angiogenesis, mesenchymal remodeling, drug resistance, immunosuppression, and invasive metastasis.10,38 In our study, cellular interactions between cancer cells and CAFs were significantly enhanced under SDS, revealing an underlying correlation between CAFs and the tumor-promoting effects of SDS. Consistent with the above results, cancer cells were found to recruit and activate more CAFs in SDS serum in in vitro co-cultures. IHC and IF staining of HCC-SDS tissues also revealed a similar trend. Much evidence suggests that the function of CAFs is not always pro-tumorigenic.11 Several clinical studies have found a positive correlation between some specific phenotypes of CAFs and patient survival in lung and pancreatic cancer.39,40 Furthermore, the depletion of α-SMA + CAFs accelerated the development of pancreatic ductal adenocarcinoma and helped polarize tumor-infiltrating T cells.40,41 In liver metastasis and pancreatic cancer, the depletion of myCAF results in the downregulation of its derivative COL1A, promoting tumor growth in the absence of mechanical constraints.42
Subsequently, we investigated the mode of communication between cancer cells and CAFs and found that PDGF-related ligand–receptor interactions were highly activated in the HCC-SDS group and that PDGFA–PDGFRB was the primary contributor pair. Paracrine signaling of cancer cell-derived PDGF has been identified as a major driver of CAF recruitment in lung43 and breast carcinomas,44 malignant melanoma,45 and cholangiocarcinoma.46 PDGF secreted by cancer cells induces fibroblast migration by interacting with the PDGF receptor (PDGFR); this process leads to CAF recruitment by activating Rho/ROCK, which in turn exerts pro-tumor growth and pro-angiogenic effects.45,46 Therefore, we hypothesized that cancer cells may recruit more CAFs through paracrine PDGFA signaling under HCC-SDS. This conjecture was confirmed in the in vitro co-culture experiment. These results provide important evidence that the cancer cells–CAFs interaction may be the source of the tumor-promoting effects of SDS.
CAFs are emerging as central players in shaping the TME phenotype toward immunosuppression.47 CAFs promote tumor immunosuppression through multiple mechanisms, including the secretion of multiple cytokines and chemokines, as well as mediating the recruitment and affecting the functional differentiation of innate and adaptive immune cells.48 Moreover, CAFs directly disrupt the function of cytotoxic lymphocytes and inhibit their activity.48 They also create a physical barrier around the tumor through ECM remodeling, blocking the infiltration of immune cells. Interestingly, we found that the expression pattern of recruited CAFs was associated with the ECM remodeling-related pathways. Tumor stiffness and desmoplasia is governed by fibroblasts active contraction, which contribute significantly to ECM remodeling.34 We also observed enhanced contractility of CAFs, increased desmoplasia and greater tumor stromal stiffness under SDS, suggesting that CAFs activated in HCC-SDS might lead to ECM remodeling. Meanwhile, the TGF-β signaling pathway was significantly activated and regulated the expression of Collagen I and Collagen III in CAFs in HCC-SDS. A hallmark of CAFs in the TME is the overproduction of collagen components, which leads to ECM remodeling and affects tumor progression.49 Of note, previous studies have proposed that CAF-derived TGF-β1 is one of the most crucial regulators of ECM, regulating ECM rigidity and fibrosis.35 Increased ECM remodeling resulting from CAFs contributes to the creation of a physical barrier to block immune cell infiltration, thereby promoting the formation of an immunosuppressive microenvironment.50 Even with infiltration of cells with anti-tumor immune responses, CAFs hinder the communication between immune and cancer cells, disrupting anti-tumor effects. In our study, ST analysis showed that CAFs formed a physical barrier around cancer cells in the HCC-SDS group, while cellular interaction analysis showed reduced interactions between cancer cells and T cells. Therefore, we speculated that CAFs recruited by HCC-SDS may participate in ECM remodeling by activating the TGF-β pathway, thus forming a physical barrier to block immune cell infiltration.
To our knowledge, this is the first study to focus on cell crosstalk in HCC-SDS based on scRNA-seq and ST. Our study revealed the potential implication of the crosstalk between CAFs and cancer cells in immunosuppression in HCC-SDS. Moreover, our study suggests that the activation and recruitment of CAFs likely constitutes a key part of the underlying mechanism, which may serve as a potential novel immunotherapeutic target to modulate the HCC stroma by targeting PDGFA and PDGFRB. However, our study had some noteworthy limitations. First, based on the expression of α-SMA, cytokines, and chemokines, CAFs can be divided into myofibroblastic (myCAFs) and inflammatory CAFs (iCAFs).51 Thus, it is necessary to explore the mechanism of different CAF subsets in HCC-SDS. In addition, we did not validate the genetic depletion of CAFs in vivo. So far, nearly all in vivo evidence on CAF depletion is based on studies that used specific genetically engineered mice to block the accumulation of CAFs. The accumulation of CAFs was inhibited by crossing abnormal α-SMA expression transgenic mice with spontaneous tumor mice. In our study, we used an in situ transplanted HCC mice model, unlike the spontaneous tumor mouse model, where the possibility exists to perform CAF-specific genetic manipulation.
5. Conclusions
The present study revealed the crucial role of the crosstalk between CAFs and cancer cells in the pro-tumor effect of SDS by integrating single-cell and ST. We speculated that CAFs recruited by HCC via PDGFA may lead to ECM remodeling through activation of the TGF-β pathway, thus forming a physical barrier to block immune cell infiltration in the TME of SDS. These findings provide important insights into the link between intercellular crosstalk and immunosuppression in SDS that would contribute to our better understanding of the role of SDS in HCC progression in TCM.
Funding
This work was supported by the National Natural Science Foundation of China (No.82174173, No.81903967, No.82104962, No.81873248, and No.82304944), Guangdong Basic and Applied Basic Research Foundation (NO. 2020A1515011003).
Availability of data and materials
The datasets used and/or analyzed during the current study have been deposited in Zenodo (https://doi.org/10.5281/zenodo.10162501)
Ethics approval and consent to participate
Animal experiments were approved by the Clinical Experimentation and Animal Ethics Committee of the First Affiliated Hospital of Sun Yat-sen University (approval number 2020:402).
Declaration of competing interest
The authors declare no conflicts of interest.
Footnotes
Peer review under responsibility of The Center for Food and Biomolecules, National Taiwan University.
Contributor Information
Yan Chen, Email: cheny556@mail.sysu.edu.cn.
Shijun Zhang, Email: zhshjun@mail.sysu.edu.cn.
Zhuomao Mo, Email: anchormok@zju.edu.cn.
References
- 1.Singal A.G., Lampertico P., Nahon P. Epidemiology and surveillance for hepatocellular carcinoma: new trends. J Hepatol. 2020;72(2):250–261. doi: 10.1016/j.jhep.2019.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu X., Li M., Wang X., et al. Effects of adjuvant traditional Chinese medicine therapy on long-term survival in patients with hepatocellular carcinoma. Phytomedicine : international journal of phytotherapy and phytopharmacology. 2019;62 doi: 10.1016/j.phymed.2019.152930. [DOI] [PubMed] [Google Scholar]
- 3.Liao X., Bu Y., Jia Q. Traditional Chinese medicine as supportive care for the management of liver cancer: past, present, and future. Genes & diseases. 2020;7(3):370–379. doi: 10.1016/j.gendis.2019.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luo H., Chen Y., Sun B., Xiang T., Zhang S. Establishment and evaluation of orthotopic hepatocellular carcinoma and drug-induced hepatocellular carcinoma in mice with spleen-deficiency syndrome in traditional Chinese medicine. Afr J Tradit, Complementary Altern Med : AJTCAM. 2017;14(1):165–173. doi: 10.21010/ajtcam.v14i1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun B., Meng J., Xiang T., et al. Effect of the herbal formulation Jianpijiedu on the TCRVβCDR3 repertoire in rats with hepatocellular carcinoma and subjected to food restriction combined with laxative. Exp Ther Med. 2016;11(3):818–826. doi: 10.3892/etm.2015.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y., Li P., Mao S., et al. Exosome CTLA-4 regulates PTEN/CD44 signal pathway in spleen deficiency internal environment to promote invasion and metastasis of hepatocellular carcinoma. Front Pharmacol. 2021;12 doi: 10.3389/fphar.2021.757194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mo Z., Cao Z., Yu L., et al. An integrative analysis reveals the potential mechanism between herbal medicine yinchen and immunoregulation in hepatocellular carcinoma. BioMed Res Int. 2020;2020 doi: 10.1155/2020/8886914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yao Z., Li Y., Wang Z., et al. Research on anti-hepatocellular carcinoma activity and mechanism of Polygala fallax Hemsl. J Ethnopharmacol. 2020;260 doi: 10.1016/j.jep.2020.113062. [DOI] [PubMed] [Google Scholar]
- 9.Binnewies M., Roberts E.W., Kersten K., et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med. 2018;24(5):541–550. doi: 10.1038/s41591-018-0014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mao X., Xu J., Wang W., et al. Crosstalk between cancer-associated fibroblasts and immune cells in the tumor microenvironment: new findings and future perspectives. Mol Cancer. 2021;20(1):131. doi: 10.1186/s12943-021-01428-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biffi G., Tuveson D.A. Diversity and biology of cancer-associated fibroblasts. Physiol Rev. 2021;101(1):147–176. doi: 10.1152/physrev.00048.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang H., Hegde S., Knolhoff B.L., et al. Targeting focal adhesion kinase renders pancreatic cancers responsive to checkpoint immunotherapy. Nat Med. 2016;22(8):851–860. doi: 10.1038/nm.4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Provenzano P.P., Cuevas C., Chang A.E., Goel V.K., Von Hoff D.D., Hingorani S.R. Enzymatic targeting of the stroma ablates physical barriers to treatment of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;21(3):418–429. doi: 10.1016/j.ccr.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feig C., Jones J.O., Kraman M., et al. vol. 110. 2013. Targeting CXCL12 from FAP-expressing carcinoma-associated fibroblasts synergizes with anti-PD-L1 immunotherapy in pancreatic cancer; pp. 20212–20217. (Proceedings of the National Academy of Sciences of the United States of America). 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pickup M.W., Mouw J.K., Weaver V.M. The extracellular matrix modulates the hallmarks of cancer. EMBO Rep. 2014;15(12):1243–1253. doi: 10.15252/embr.201439246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suvà M.L., Tirosh I. Single-cell RNA sequencing in cancer: lessons learned and emerging challenges. Mol Cell. 2019;75(1):7–12. doi: 10.1016/j.molcel.2019.05.003. [DOI] [PubMed] [Google Scholar]
- 17.Baslan T., Hicks J. Unravelling biology and shifting paradigms in cancer with single-cell sequencing. Nat Rev Cancer. 2017;17(9):557–569. doi: 10.1038/nrc.2017.58. [DOI] [PubMed] [Google Scholar]
- 18.Sun Y., Wu L., Zhong Y., et al. Single-cell landscape of the ecosystem in early-relapse hepatocellular carcinoma. Cell. 2021;184(2):404–421.e416. doi: 10.1016/j.cell.2020.11.041. [DOI] [PubMed] [Google Scholar]
- 19.Rao A., Barkley D., França G.S., Yanai I. Exploring tissue architecture using spatial transcriptomics. Nature. 2021;596(7871):211–220. doi: 10.1038/s41586-021-03634-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ståhl P.L., Salmén F., Vickovic S., et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science (New York, N.Y.) 2016;353(6294):78–82. doi: 10.1126/science.aaf2403. [DOI] [PubMed] [Google Scholar]
- 21.Zhu Q., Shah S., Dries R., Cai L., Yuan G.C. Identification of spatially associated subpopulations by combining scRNAseq and sequential fluorescence in situ hybridization data. Nat Biotechnol. 2018 doi: 10.1038/nbt.4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hao Y., Hao S., Andersen-Nissen E., et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184(13):3573–3587.e3529. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X., Lan Y., Xu J., et al. CellMarker: a manually curated resource of cell markers in human and mouse. Nucleic Acids Res. 2019;47(D1):D721–d728. doi: 10.1093/nar/gky900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Franzén O., Gan L.M., Björkegren J.L.M. PanglaoDB: a web server for exploration of mouse and human single-cell RNA sequencing data. Database : the journal of biological databases and curation. 2019;2019 doi: 10.1093/database/baz046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao R., Bai S., Henderson Y.C., et al. Delineating copy number and clonal substructure in human tumors from single-cell transcriptomes. Nat Biotechnol. 2021;39(5):599–608. doi: 10.1038/s41587-020-00795-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Efremova M., Vento-Tormo M., Teichmann S.A., Vento-Tormo R. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat Protoc. 2020;15(4):1484–1506. doi: 10.1038/s41596-020-0292-x. [DOI] [PubMed] [Google Scholar]
- 27.Jin S., Guerrero-Juarez C.F., Zhang L., et al. Inference and analysis of cell-cell communication using CellChat. Nat Commun. 2021;12(1):1088. doi: 10.1038/s41467-021-21246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hänzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilk A.J., Rustagi A., Zhao N.Q., et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat Med. 2020;26(7):1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma Y., Zhou X. Spatially informed cell-type deconvolution for spatial transcriptomics. Nat Biotechnol. 2022;40(9):1349–1359. doi: 10.1038/s41587-022-01273-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu X., Lu Y., Huang J., et al. CD16(+) fibroblasts foster a trastuzumab-refractory microenvironment that is reversed by VAV2 inhibition. Cancer Cell. 2022;40(11):1341–1357.e1313. doi: 10.1016/j.ccell.2022.10.015. [DOI] [PubMed] [Google Scholar]
- 32.Cadamuro M., Brivio S., Mertens J., et al. Platelet-derived growth factor-D enables liver myofibroblasts to promote tumor lymphangiogenesis in cholangiocarcinoma. J Hepatol. 2019;70(4):700–709. doi: 10.1016/j.jhep.2018.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guan Y., Enejder A., Wang M., et al. A human multi-lineage hepatic organoid model for liver fibrosis. Nat Commun. 2021;12(1):6138. doi: 10.1038/s41467-021-26410-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen Y., Wang X., Lu J., et al. Reduction of liver metastasis stiffness improves response to bevacizumab in metastatic colorectal cancer. Cancer Cell. 2020;37(6):800–817.e807. doi: 10.1016/j.ccell.2020.05.005. [DOI] [PubMed] [Google Scholar]
- 35.Fullár A., Dudás J., Oláh L., et al. Remodeling of extracellular matrix by normal and tumor-associated fibroblasts promotes cervical cancer progression. BMC Cancer. 2015;15:256. doi: 10.1186/s12885-015-1272-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horn L.A., Chariou P.L., Gameiro S.R., et al. Remodeling the tumor microenvironment via blockade of LAIR-1 and TGF-β signaling enables PD-L1-mediated tumor eradication. J Clin Invest. 2022;132(8) doi: 10.1172/JCI155148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao N. Evidence of spleen yang deficiency and spleen qi deficiency in rats with spleen deficiency due to reserpine. J Tradit Chin Med. 2008;5:449–452. [Google Scholar]
- 38.Kashima H., Noma K., Ohara T., et al. Cancer-associated fibroblasts (CAFs) promote the lymph node metastasis of esophageal squamous cell carcinoma. Int J Cancer. 2019;144(4):828–840. doi: 10.1002/ijc.31953. [DOI] [PubMed] [Google Scholar]
- 39.Ono S., Ishii G., Nagai K., et al. Podoplanin-positive cancer-associated fibroblasts could have prognostic value independent of cancer cell phenotype in stage I lung squamous cell carcinoma: usefulness of combining analysis of both cancer cell phenotype and cancer-associated fibroblast phenotype. Chest. 2013;143(4):963–970. doi: 10.1378/chest.12-0913. [DOI] [PubMed] [Google Scholar]
- 40.Özdemir B.C., Pentcheva-Hoang T., Carstens J.L., et al. Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer Cell. 2014;25(6):719–734. doi: 10.1016/j.ccr.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McAndrews K.M., Chen Y., Darpolor J.K., et al. Identification of functional heterogeneity of carcinoma-associated fibroblasts with distinct IL6-mediated therapy resistance in pancreatic cancer. Cancer Discov. 2022;12(6):1580–1597. doi: 10.1158/2159-8290.CD-20-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhattacharjee S., Hamberger F., Ravichandra A., et al. Tumor restriction by type I collagen opposes tumor-promoting effects of cancer-associated fibroblasts. J Clin Invest. 2021;131(11) doi: 10.1172/JCI146987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tejada M.L., Yu L., Dong J., et al. Tumor-driven paracrine platelet-derived growth factor receptor alpha signaling is a key determinant of stromal cell recruitment in a model of human lung carcinoma. Clin Cancer Res : an official journal of the American Association for Cancer Research. 2006;12(9):2676–2688. doi: 10.1158/1078-0432.CCR-05-1770. [DOI] [PubMed] [Google Scholar]
- 44.Shao Z.M., Nguyen M., Barsky S.H. Human breast carcinoma desmoplasia is PDGF initiated. Oncogene. 2000;19(38):4337–4345. doi: 10.1038/sj.onc.1203785. [DOI] [PubMed] [Google Scholar]
- 45.Anderberg C., Li H., Fredriksson L., et al. Paracrine signaling by platelet-derived growth factor-CC promotes tumor growth by recruitment of cancer-associated fibroblasts. Cancer Res. 2009;69(1):369–378. doi: 10.1158/0008-5472.CAN-08-2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berasain C., Avila M.A. Platelet-derived growth factor D: a new player in the complex cross-talk between cholangiocarcinoma cells and cancer-associated fibroblasts. Hepatology (Baltimore, Md. 2013;58(3):853–855. doi: 10.1002/hep.26476. [DOI] [PubMed] [Google Scholar]
- 47.Ruhland M.K., Loza A.J., Capietto A.H., et al. Stromal senescence establishes an immunosuppressive microenvironment that drives tumorigenesis. Nat Commun. 2016;7 doi: 10.1038/ncomms11762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mhaidly R., Mechta-Grigoriou F. Fibroblast heterogeneity in tumor micro-environment: role in immunosuppression and new therapies. Semin Immunol. 2020;48 doi: 10.1016/j.smim.2020.101417. [DOI] [PubMed] [Google Scholar]
- 49.Chan J.S., Tan M.J., Sng M.K., et al. Cancer-associated fibroblasts enact field cancerization by promoting extratumoral oxidative stress. Cell Death Dis. 2017;8(1) doi: 10.1038/cddis.2016.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sorokin L. The impact of the extracellular matrix on inflammation. Nat Rev Immunol. 2010;10(10):712–723. doi: 10.1038/nri2852. [DOI] [PubMed] [Google Scholar]
- 51.Elyada E., Bolisetty M., Laise P., et al. Cross-species single-cell analysis of pancreatic ductal adenocarcinoma reveals antigen-presenting cancer-associated fibroblasts. Cancer Discov. 2019;9(8):1102–1123. doi: 10.1158/2159-8290.CD-19-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study have been deposited in Zenodo (https://doi.org/10.5281/zenodo.10162501)