Abstract
The two group IC3 pre-tRNA introns from Azoarcus and Synechococcus share very analogous secondary structures. They are small group I ribozymes that possess only two peripheral domains, P2 and P9. However, the 3′-splice site hydrolysis activity of the Synechococcus ribozyme critically depends on P2 whereas that of Azoarcus does not, indicating that the structure–function relationships of the two ribozymes are strikingly different despite their structural resemblance. To identify the element(s) that determines the catalytic properties of these ribozymes, we undertook analyses of chimeric ribozymes prepared by swapping their structural elements. We found that the difference can be attributed to a small number of nucleotides within the conserved core region. Further analysis by employing in vitro selection revealed that a base triple interaction (P4bp3 × J6/7-2) is a critical element for determining activity and suggests the existence of a novel base quintuple involving the base triple P4bp5 × J8/7-5.
INTRODUCTION
The group I self-splicing intron ribozymes share a phylogenetically conserved secondary structure consisting of two helical domains, P4–P6 and P8–P3–P7, that contain the active sites (Fig. 1; 1,2). In addition to this conserved region, they possess peripheral elements characteristic of each subgroup (2,3). However, in some cases these structurally similar ribozymes do not exhibit comparable activity, even those belonging to the same subgroup (4,5). One such example is a group IC1 intron from Physarum polycephalum that is 1500-fold less active than the Tetrahymena IC1 intron, one of its closest relatives (5). The basis of this difference likely resides within the limited number of sequence variations and/or minor alterations in their peripheral structures.
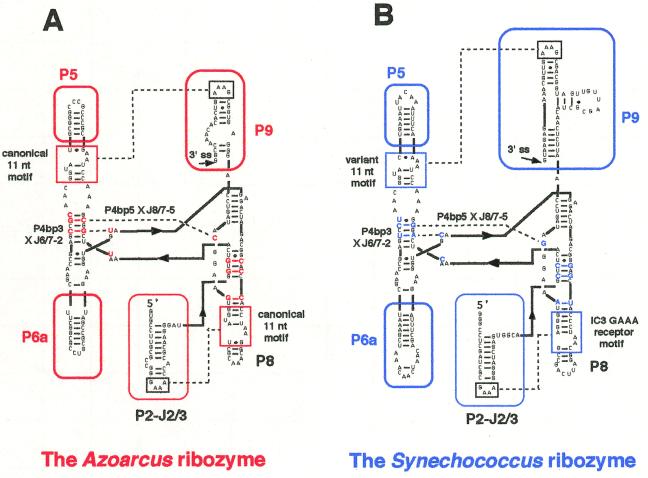
Figure 1.
The secondary structures of the two group IC3 introns and their conserved cores. (A) The secondary structure of the L-8 form of the group IC3 intron ribozyme from the tRNA precursor of Azoarcus sp. BH72. This form lacks the 5′-exon and the first 8 nt of the intron. The regions and nucleotides characteristic of this ribozyme are indicated in red. Broken lines indicate the tertiary interactions (GAAA loop × GAAA receptor, P4bp3 × J6/7-2 and P4bp5 × J8/7-5) discussed in the text. The ΔP2 form of this ribozyme lacks the P2–J2/3 region of the L-8 form. (B) The secondary structure of the L-8 form of the group IC3 intron ribozyme from the tRNA precursor of Synechococcus PCC6301. This also lacks the 5′-exon and the first 8 nt of the intron. The regions and nucleotides characteristic of this ribozyme are indicated in blue. Broken lines indicate the tertiary interactions (GAAA loop × GAAA receptor, P4bp3 × J6/7-2 and P4bp5 × J8/7-5) discussed in the text. The ΔP2 form of this ribozyme lacks the P2–J2/3 region of the L-8 form.
The IC3 introns, a subgroup of the group I introns, contain only two characteristic peripheral elements, P2 and P9, with GAAA tetraloops at their termini (Fig. 1; 1,6–12). Phylogenetic comparison of the IC3 introns strongly suggested that these GAAA loops in P2 and P9 interact with specific receptors in P8 and P5, respectively (Fig. 1). The IC3 introns from pre-tRNAIle of the purple bacterium Azoarcus sp. HB72 (termed the Azoarcus ribozyme; Fig. 1A) and pre-tRNALeu of the cyanobacterium Synechococcus PCC6301 (termed the Synechococcus ribozyme; Fig. 1B) share these features as well as highly similar secondary structures, with the exception of the P9 region (Fig. 1; 8,10). The Azoarcus ribozyme is particularly active at high temperatures (up to 75°C), high pH (up to 10.2) or in the presence of high concentrations of urea (up to 7.5 M), presumably due to its GC-rich secondary structure and the presence of two canonical 11 nt GAAA receptor motifs which can interact with GAAA loops more strongly than any other known GNRA loop receptors (13,14). We analyzed the structure–function relationships of these two group I introns at the nucleotide level by systematically constructing chimeric introns and by in vitro selection experiments.
MATERIALS AND METHODS
Ribozymes
Plasmids encoding derivatives of the Synechococcus IC3 intron involving the coreSyn variants were prepared from pTL3 using PCR and the resulting constructs were verified by DNA sequencing (10). Plasmids encoding the coreAzo variants were prepared using PCR from pAZIVS which was constructed by subcloning the Azoarcus intron into a pTZ18U vector. The L-8 and ΔP2 forms of the two ribozymes lack the first 8 nt of the introns and the P2-J2/3 regions (Fig. 1), respectively. Precursors of the intron ribozymes possess the 40 nt of the 5′-end of the 3′-exon with the sequence 5′-AAA AUC CGC UGA CUG UAA AGG UCG UGA GGG UUC GAG UCC C-3′ or 5′-AAU CCG UUG GUG CUG GGU UCG ACU CCC AGG GGG CCC ACC A-3′ in the case of the Synechococcus and the Azoarcus introns, respectively. The template DNAs for in vitro transcription of these precursor RNAs were generated by 20 cycles of PCR (94°C for 1 min, 55°C for 1 min, 72°C for 2 min) using ExTaq DNA polymerase (Takara Shuzo, Tokyo, Japan). For PCR, 1 ng template plasmid (an appropriate derivative of pTL3 or pAZIVS) and the primer set listed in the following section were employed.
Primers for PCR
Sense primers (the promoter sequence for T7 RNA polymerase is underlined). AL-8, 5′-TAA TAC GAC TCA CTA TAG GGC CTT GCG CCG GGA AAC CAC-3′; ADP2, 5′-TAA TAC GAC TCA CTA TAG GTG TCA AAT TCG GCG AAA CCT-3′; SL-8, 5′-TAA TAC GAC TCA CTA TAG GGC CTC GAT CGC GAA AGG-3′; SDP2, 5′-TAA TAC GAC TCA CTA TAG CTC TCA AAC TCA GGG AAA CCT-3′.
Antisense primers (the 3′-exon sequence is underlined). A3E-AP9, 5′-TGG TGG GCC CCC TGG GAG TCG AAC CCA GCA CCA ACG GAT TCC GGT TTG TGT GAC TTT CGC CAC TCC CTG GAC TAT-3′; A3E-SP9, 5′-TGG TGG GCC CCC TGG GAG TCG AAC CCA GCA CCA ACG GAT TCA TTC TCT TTG CAA CTT TCG CTG CCA TCA ACA-3′; S3E-SP9, 5′-GGG ACT CGA ACC CTC ACG ACC TTT ACA GTC AGC GGA TTT TCA TTC TCT TTG CAA CTT TCG CTG CCA TCA ACA-3′.
Primer sets for PCR
AL-8 and A3E-AP9 for the L-8 forms of the Azoarcus ribozyme, the coreAzo-4, coreAzo-6 and coreAzo-7 chimeric variants. ADP2 and A3E-AP9 for the ΔP2 forms of the Azoarcus ribozyme, the coreAzo-4, coreAzo-6 and coreAzo-7 chimeric variants. AL-8 and A3E-SP9 for the L-8 forms of the coreAzo-1, coreAzo-2, coreAzo-3 and coreAzo-5 chimeric variants. ADP2 and A3E-SP9 for the ΔP2 forms of the coreAzo-1, coreAzo-2, coreAzo-3 and coreAzo-5 chimeric variants. SL-8 and S3E-SP9 for the L-8 forms of the Synechococcus ribozyme and its 11nt-Syn variant. SDP2 and S3E-SP9 for the ΔP2 forms of the Synechococcus ribozyme and its 11nt-Syn variant. SL-8 and A3E-SP9 for the L-8 forms of the 3E-Syn and 11nt/3E-Syn variants and the coreSyn-2, coreSyn-3 and coreSyn-5 chimeric variants. SDP2 and A3E-SP9 for the ΔP2 forms of the 3E-Syn and 11nt/3E-Syn variants and the coreSyn-2, coreSyn-3 and coreSyn-5 chimeric variants. SL-8 and A3E-AP9 for the L-8 forms of the coreSyn-4, coreSyn-6, coreSyn-7 and coreSyn-8 chimeric variants. SDP2 and A3E-AP9 for the ΔP2 forms of the coreSyn-4, coreSyn-6, coreSyn-7 and coreSyn-8 chimeric variants.
Preparation of RNAs
All RNAs employed in this study were prepared by transcription in vitro with T7 RNA polymerase in the presence of [α-32P]GTP and purified by electrophoresis in 5% polyacrylamide denaturing gels as described (15).
Assay of the 3′-splice site hydrolysis reaction
Uniformly 32P-labeled precursor ribozymes possessing their 3′-exons were dissolved in distilled water followed by incubation at 80°C for 3 min. After cooling and incubation at the reaction temperature for 10 min, the reaction was initiated by adding 5× concentrated reaction buffer. The 3′-splice site hydrolysis reactions were assayed in a buffer containing 40 mM Tris–HCl, pH 8.3, 10 mM MgCl2 at 37°C. Aliquots were removed at specific times and quenched on ice by the addition of an equal volume of stop solution (150 mM EDTA, 70% formamide and 0.25% xylene cyanol). The products were electrophoresed in 5% polyacrylamide denaturing gels. The gels were dried and relative intensities of each band were quantitated using a Bio-Image Analyzer BAS-2500 (Fuji Film, Tokyo, Japan).
In vitro selection of active variant ribozymes
The DNA templates for the initial pool of coreMix variant ribozymes were prepared as follows. Oligonucleotides Mix-1 (5′-AAG CTA AAG CTA AAC AAC TAA CAG CTT TAG AAG GTG YAG AGA CTA GAC GGS ASC YAC CTA AGG GAT TCA GCC TAT G-3′, 76 mer, Y = C/T, S = G/C) and Mix-2 (5′-TTT CGC CAC TCC CTG GAC TAT SCC TTY ACC ATA GGC TGA ATC CCT TAG G-3′, 49 mer,Y = C/T, S = G/C) were hybridized and converted into double-stranded DNA by primer extension with ExTaq (Takara Shuzo). Using 1 pmol of this DNA as template, 10 cycles of PCR amplification (20 µl scale, 94°C for 1 min, 50°C for 1 min, 72°C for 1 min, denoted PCR1) were performed with two oligonucleotides (10 pmol each), Mix-3 (5′-GGT GAT TCG TCA TCT GGC AGS TST CAA AYT CRG SGA AAC CTA AGA CTT TAA ACA TAA AAG TTA TGG CAA YSC YGA GCC AAG CTA AAG CTA AAC AAC TA-3′, 98 mer,Y = C/T, S = G/C, R = A/G) and Mix-2. With the amplified fragment from PCR1 as template, an additional PCR (200 µl scale, 94°C for 1 min, 50°C for 1 min, 72°C for 1 min, denoted PCR2) was performed with primer Mix-4 (5′-TAA TAC GAC TCA CTA TAG GTG ATT CGT CAT CTG GCA-3′, promoter sequence for T7 RNA polymerase underlined) and primer A3E-AP9. The initial pool of the variant ribozyme was synthesized by in vitro transcription with T7 RNA polymerase on the resulting DNA templates.
The selection cycles were carried out by repeating the following steps. Step 1: the pool of uniformly 32P-labeled precursor RNAs of variant ribozymes was subjected to the 3′-splice site hydrolysis reaction for the specified time (90 min for the first to third cycles, 30 min for the fourth cycle and 10 min for the fifth and sixth cycles) under the assay conditions described above. Step 2: the reaction mixture was electrophoresed in a 5% acrylamide denaturing gel and active variants were isolated. Step 3: RT–PCR was performed with primers Mix-4 and A3E-AP9 for amplification of active variants and the regeneration of their 3′-exons and T7 promoter sequences. Step 4: precursors of the active variant ribozymes were transcribed using the DNA fragments amplified in Step 3 as templates.
For RT–PCR, RNA PCR Kit version 2.1 (Takara Shuzo) was used. After six rounds of selection, the resulting DNA fragments were cloned into pTZ18U, from which 28 clones were isolated and sequenced.
RESULTS
The structural elements
Effects of L2 × P8 interaction. Group IC3 introns from Synechococcus and Azoarcus tRNAs perform self-splicing reactions efficiently in the absence of protein factors (10,14). We have demonstrated that a shortened form of the Synechococcus ribozyme lacking the first 8 nt and 5′-exon (termed the L-8 form of the ribozyme) performs the site-specific hydrolysis reaction at the 3′-splice site, relying on a tertiary interaction between L2 and P8 (16; see also Fig. 2B).
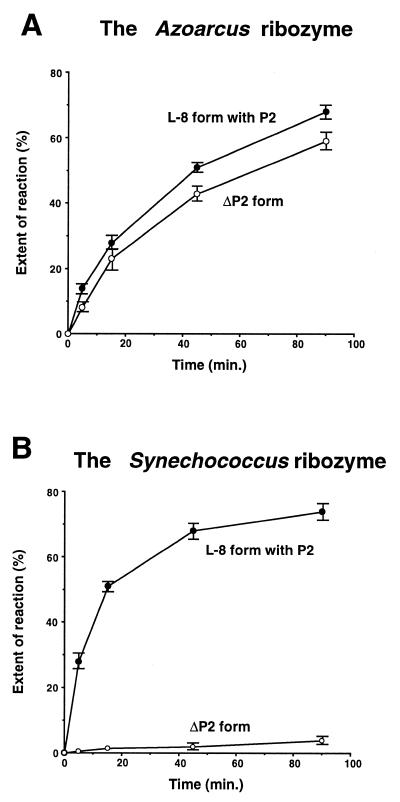
Figure 2.
The 3′-splice site-specific hydrolysis reactions of the Azoarcus and Synechococcus ribozyme in the presence or absence of the P2 element. (A) Time course of the 3′-splice site-specific hydrolysis reactions of the L-8 form and the ΔP2 form of the Azoarcus ribozyme. (B) Time course of the 3′-splice site-specific hydrolysis reactions of the L-8 form and the ΔP2 form of the Synechococcus ribozyme.
To determine whether the L2 × P8 interaction is also important for the Azoarcus ribozyme, we constructed and tested a mutant Azoarcus ribozyme lacking its P2 element (termed the ΔP2 form). We found that the ΔP2 form performs the 3′-splice site hydrolysis reaction with an efficiency comparable to that of its L-8 form possessing P2 under our assay conditions (40 mM Tris–HCl, pH 8.3, 10 mM MgCl2, 37°C) (Fig. 2A). This result indicates that unlike the Synechococcus ribozyme, the Azoarcus ribozyme is much less dependent on the L2 × P8 interaction.
To identify the factor(s) underlying this difference between the two ribozymes, we investigated their structure–function relationships in the absence of the P2 element. The overall secondary structures of the two ribozymes are closely related, but we were able to identify differences by comparing their secondary structures and primary sequences as follows. [1] The sequence of the 3′-exon (8,10; see also Materials and Methods); [2] the structure of the GAAA receptors in P5 and P8 (Fig. 1); [3] the secondary structure of P9 (Fig. 1); [4] the GC content of the P5 and P6a elements (Fig. 1); [5] identification of nucleotides at 14 positions in the conserved core region that differ between the two introns (Figs 1 and 5). (In addition, we found differences in the sequences and secondary structures of the P2 and J2/3 regions: This is irrelevant here because the ΔP2 forms of the ribozymes lack the P2 as well as the J2/3 region.)
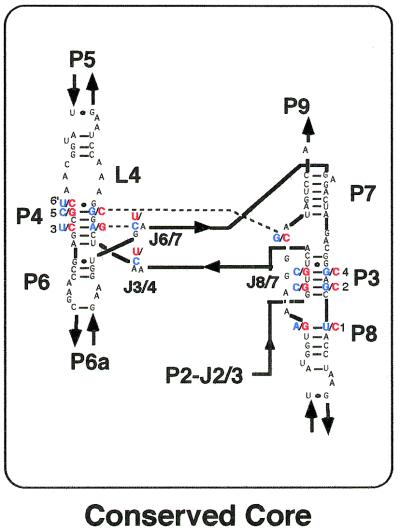
Figure 5.
The secondary structures of the conserved core regions of the 11nt/3E-Syn variant and the Azoarcus ribozyme. The core region consists of 92 nt involving P3, P4, L4, P6, P7 and part of P8. Between the Azoarcus ribozyme and the 11nt/3E-Syn variant (see text) 78 of the 92 nt in the core regions are identical. The 14 positions that differ between the two ribozymes are indicated in red or blue.
Effects of the 3′-exon and GAAA receptors. We examined the effects of the 3′-exon (factor [1]) and the two GAAA receptors (factor [2]) independently by domain substitution (Fig. 3). Replacement of the Synechococcus ribozyme 3′-exon with that of the Azoarcus ribozyme (termed the 3E-Syn variant) only slightly improved the activity of the ΔP2 form of the Synechococcus ribozyme (Fig. 3A).
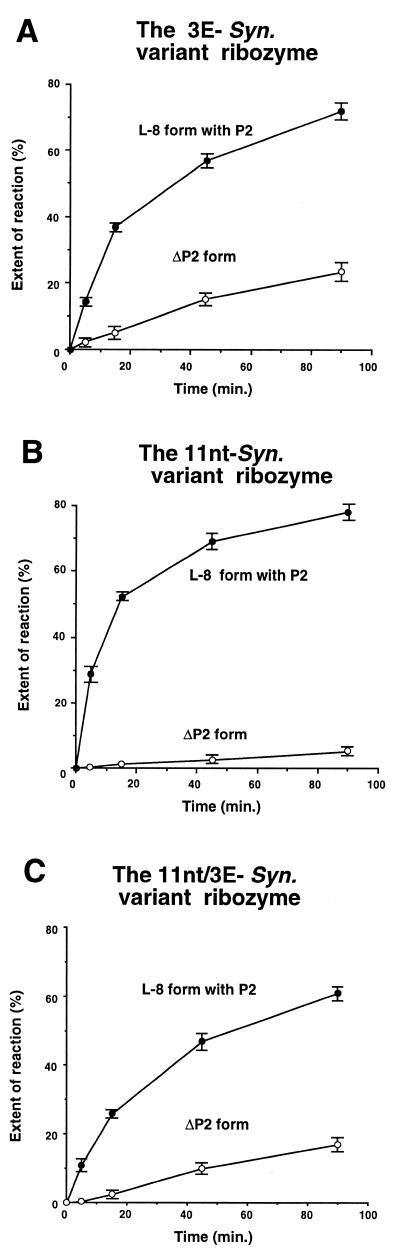
Figure 3.
The 3′-splice site-specific hydrolysis reactions of the variant Synechococcus ribozymes in the presence or absence of the P2 element. (A) Time course of the 3′-splice site-specific hydrolysis reactions of the L-8 and ΔP2 forms of the 3E-Syn variant ribozyme which possess the 3′-exon of the Azoarcus ribozyme. (B) Time course of the 3′-splice site-specific hydrolysis reactions of the L-8 and ΔP2 forms of the 11nt-Syn variant ribozyme which possess the canonical 11 nt motifs as GAAA receptors in P5 and P8 elements. (C) Time course of the 3′-splice site-specific hydrolysis reactions of the L-8 and ΔP2 forms of the 11nt/3E-Syn variant ribozyme which possess the 3′-exon of the Azoarcus ribozyme as well as the canonical 11 nt motifs as GAAA receptors in P5 and P8 elements.
The GAAA receptors in the Azoarcus ribozyme are canonical 11 nt GAAA receptor motifs while those of the Synechococcus ribozyme are not. These receptors in the P5 and P8 elements are predicted phylogenetically to interact with GAAA tetraloops in P9 and P2, respectively. Under our assay conditions, a mutant Synechococcus ribozyme possessing two canonical 11 nt receptor motifs (the 11nt-Syn variant) displayed almost identical activity to that of the original Synechococcus ribozyme either in the presence or absence of the P2 element (compare Fig. 3B with 2B).
A double mutant (the 11nt/3E-Syn variant) also exhibited a similar level of activity (Fig. 3C), indicating that neither the sequences of the 3′-exon nor the structures of the two GAAA receptors in P5 and P8 are critical determinants. On the basis of these results, we decided to employ the 11nt/3E-Syn variant ribozyme as the standard ribozyme for the following experiments.
Effects of the P5, P6a and P9 elements. The effects of P5, P6a and P9 (factors [3] and [4], respectively) were examined by employing the Azoarcus ribozyme and the 11nt/3E-Syn variant ribozyme. The secondary structures of P5 and P6a of the two ribozymes resemble each other; however, the stem regions of the Azoarcus ribozyme are GC-rich compared with those of the 11nt/3E-Syn variant (Fig. 1A and B). The P9 secondary structure of the Azoarcus ribozyme is small and structurally simple compared with that of the Synechococcus ribozyme (Fig. 1A and B).
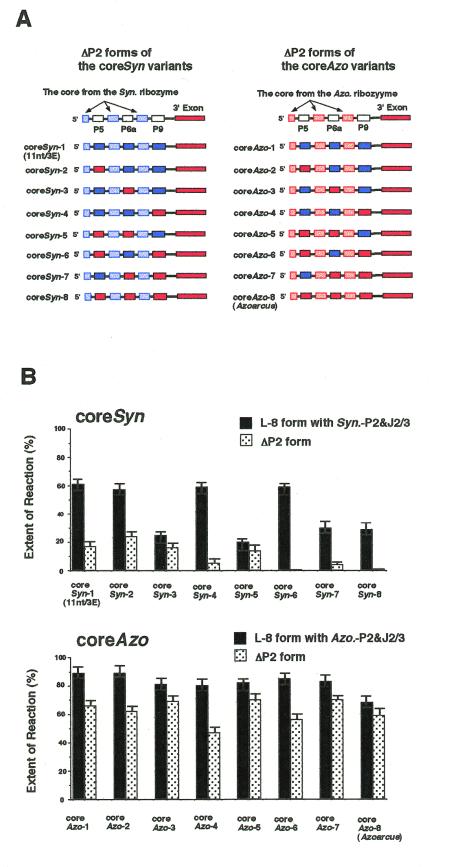
To investigate the relevance of these elements, we designed a series of chimeric ribozymes by swapping corresponding elements between the Azoarcus and the 11nt/3E-Syn variant ribozymes (Fig. 4A; 17). The coreSyn variants were created by swapping the P5, P6a and/or P9 elements of the 11nt/3E-Syn variant ribozyme with those of the Azoarcus ribozyme (Fig. 4A, left). Another series, termed coreAzo variants, was made by swapping the P5, P6 and/or P9 elements of the Azoarcus ribozyme with those of the 11nt/3E-Syn variant ribozyme (Fig. 4A, right). The activities of the coreSyn and coreAzo variants were examined and are summarized in Figure 4B.
Figure 4.
Structures and activities of the chimeric introns constructed from the Azoarcus ribozyme and the 11nt/3E-Syn variant. (A) Schematic representation of ΔP2 forms of the coreSyn series and coreAzo series of the chimeric ribozymes. Regions colored in red or blue indicate the structural element(s) from the Azoarcus ribozyme or the 11nt/3E-Syn variant, respectively. (B) Relative activity of the 3′-splice site hydrolysis reactions by the chimeric ribozyme with or without the P2 element. Uniformly 32P-labeled precursor ribozymes were incubated for 90 min. Activities of the L-8 or ΔP2 form variants are indicated with black or gray bars, respectively. The coreSyn and coreAzo variants with the same number share the same set of P5, P6a and/or P9 elements.
A coreSyn chimeric ribozyme should be activated by the P5, P6a and/or P9 peripheral elements from the Azoarcus ribozyme if they primarily contribute to activating the ΔP2 form of the Azoarcus ribozyme. [In this case, a coreAzo chimeric ribozyme should be inactivated when it carries the corresponding element(s) from the Synechococcus ribozyme.] As shown in Figure 4B, the elements modestly activated or inactivated the ΔP2 forms of the coreSyn ribozyme or those of the coreAzo ribozyme. However, the ΔP2 forms of the coreSyn variants having the Azoarcus P5, P6a and/or P9 peripheral elements were significantly less active than the corresponding coreAzo variants, indicating that neither the P5, P6a nor P9 region is the major determinant of the activity of the ΔP2 form of the Azoarcus ribozyme. The difference in activity thus appears to reside in the conserved core regions.
[Note: the activity was reduced for the L-8 forms of several coreSyn ribozymes having P6a of the Azoarcus ribozyme (Fig. 4B). For instance, replacement of the P6a element of the 11nt/3E-Syn (= coreSyn-1) variant with that from the Azoarcus ribozyme decreased activity in the presence of P2 (the L-8 form) (compare the activity of the L-8 form of coreSyn-3 with that of coreSyn-1; Fig. 4B, upper graph). A similar decrease in activity was also seen in the L-8 forms of the coreSyn variants having Azoarcus P6a (compare the activities of the L-8 forms of the coreSyn-5, coreSyn-7 and coreSyn-8 variants with those of the coreSyn-2, coreSyn-4 and coreSyn-6 variants, respectively; Fig. 4B, upper graph). However, the same replacement did not affect the ΔP2 form, indicating that the sequence and structure of the P6a region is not critical for activity of the ΔP2 form in coreSyn variants.]
Identification of the critical nucleotides in the core region
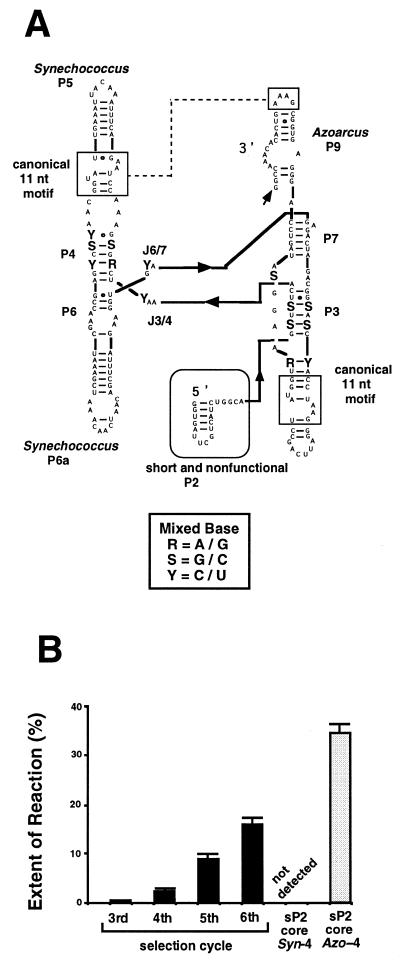
Comparison of the Azoarcus and 11nt/3E-Syn variant ribozymes lacking the P2 region revealed that the major determinant for the hydrolysis activity of the ΔP2 form of the Azoarcus ribozyme is neither its GAAA receptors, 3′-exon nor its peripheral region (P5, P6a or P9) (Figs 3 and 4). This indicates that the determinant should exist within the conserved core region (factor [5]), consisting of P3, P4, L4, P6, P7 and the proximal part of the P8 element (Fig. 5). Among the 92 nt in the core regions of the 11nt/3E-Syn variant and the Azoarcus ribozymes, 14 are different (Fig. 5). We evaluated the participation of these nucleotides in the mechanism of ribozyme activation by employing in vitro selection (18–20).
In vitro selection. We constructed a pool of variant ribozymes (termed the coreMix pool) having mixed bases at the 14 positions in the core of a chimeric ribozyme containing P5 and P6a of the Synechococcus ribozyme and P9 of the Azoarcus ribozyme (Fig. 6A; see also Fig. 4A). The organization of the peripheral elements of the coreMix pool variants is identical to that of the coreSyn-4 or coreAzo-4 variants and so we employed the two variants as the benchmark. Mixed bases consisting of the two original bases from the Synechococcus and Azoarcus ribozyme were introduced at the 14 sites (Figs 5 and 6A), yielding 214 different variants. The coreMix pool was designed to possess a short and non-functional P2 stem (termed the sP2 form) as the priming site for RT–PCR required during the selection cycles (Fig. 6A). [Note: the addition of a sP2 element moderately decreased the activity of the coreAzo-4 ribozyme: compare the activity of the ΔP2 form of the coreAzo-4 variant (Fig. 4B, lower graph) with that of its sP2 form (Fig. 6B) and the inactivated coreSyn-4 variant (Fig. 6B).]
Figure 6.
In vitro genetic analysis of the conserved core region of the two group IC3 intron ribozymes. (A) The secondary structure of the coreMix variant pool for the in vitro selection. Mixed nucleotides were introduced at the 14 positions indicated by R(A/G), S(G/C) or Y(C/U) in the core region. (B) The 3′-splice site-specific hydrolysis activity of the pool at each round of the selection cycle. Uniformly 32P-labeled precursor ribozymes from each round were incubated for 90 min. The activities of the sP2 form of coreSyn-4 (not detected) or coreAzo-4 (gray bar) are also indicated.
The activity of the coreMix pool, which was not detected prior to the third cycle, increased gradually through the fourth, fifth and sixth selection cycles (Fig. 6B). After six rounds, the activity of the pool became ∼40% of the activity of the sP2 form of the coreAzo-4 variant (Fig. 6B). The DNA fragments amplified after six cycles of selection were subcloned and 28 randomly picked clones were isolated, sequenced and assayed for hydrolysis activity (Fig. 7).
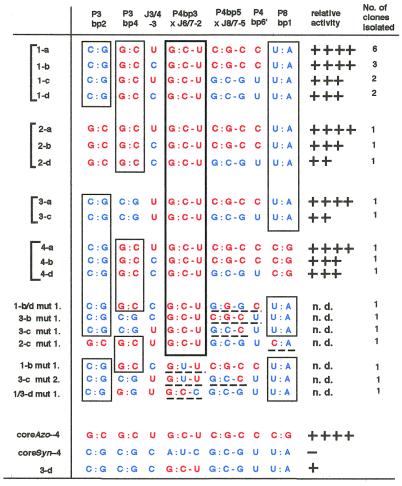
Figure 7.
Nucleotides at the 14 positions in the selected coreMix variants after six rounds of the selection cycle. Mixed bases were introduced at the positions in the initial pool. Nucleotides from the Azoarcus or Synechococcus ribozymes are indicated as red or blue, respectively. The following symbols indicate the relative activity: ++++, >70% activity of the sP2 form of coreAzo-4; +++, 40–70% activity of the sP2 form of coreAzo-4; ++, 10–40% activity of the sP2 form of the coreAzo-4; +, ∼10% activity of the sP2 form of coreAzo-4; –, no activity detected; n.d., not determined.
Identification of the critical base triples. Among the 14 positions containing mixed bases, three positions [a third base pair of P4 (P4bp3) and the second nucleotide of J6/7 (J6/7-2)] were nearly invariant (Fig. 7). In the wild-type, P4bp3 is a G:C pair in the Azoarcus ribozyme (Fig. 1A) whereas it is an A:U pair in the Synechococcus ribozyme (Fig. 1B). In the selected variants, 26 of 28 clones have the Azoarcus pair (G:C) at this position and the remaining two (1-b mut.1 and 3-c mut. 2) possess a G:U pair (Fig. 7). In the wild-type, J6/7-2 is a U in the Azoarcus ribozyme (Fig. 1A) and a C in the Synechococcus ribozyme (Fig. 1B). In the selected variants, 27 of the 28 clones possessed the Azoarcus base (U) at this position (Fig. 7). Because phylogenetic analyses indicated that P4bp3 and J6/7-2 co-vary to form a base triple (21), the Azoarcus and Synechococcus ribozyme are likely to form a G:C-U and A:U-C base triple at the sites, respectively. Our selection analysis strongly suggested that the IC3 ribozymes prefer the Azoarcus type (G:C-U) base triple to the Synechococcus type (A:U-C) to readily conduct the 3′-splice site hydrolysis reaction in the absence of the L2 × P8 interaction.
To see whether the G:C-U base-triple in the P4bp3 × J6/7-2 interaction is critical in activating the sP2 form of group IC3 ribozymes, we replaced the A:U-C base triple in the coreSyn-4 variant with the G:C-U base triple (class 3-d variant). The resulting class 3-d variant was active whereas the corresponding coreSyn-4 variant was inactive (Fig. 7), indicating that the identity of the P4bp3 × J6/7-2 base triple is critically important. However, the 3-d variant was ~10-fold less active than the coreAzo-4 variant (Fig. 7), suggesting that another element(s) in the core of the Azoarcus ribozyme assist further improvement in activity.
From sequence comparison of the selected variants, two positions were identified as likely candidates for the elements assisting improvement in the activity of IC3 ribozymes (Fig. 7). One is the fourth base pair of the P3 region (P3bp4), consisting of either a G:C pair (the Azoarcus ribozyme) or a C:G pair (the Synechococcus ribozyme). In the selected variants, 22 of 28 clones possessed a G:C base pair (the Azoarcus type) in P3bp4. However, the contribution of P3bp4 to the activity remains unclear (Fig. 7); the activities of class 1-a and class 3-a or class 1-c and class 3-c (note the difference between class 1-a and 3-a or between class 1-c and 3-c is only P3bp4) were comparable.
The second candidate was the base triple between the fifth base pair of P4 and the fifth nucleotide in J8/7 (denoted the P4bp5 × J8/7-5 base triple). This base triple has been shown to be critically important for ribozymic activity of IC3 introns (22,23). The P4bp5 × J8/7-5 base triple consists of C:G-C or G:C-G in the Azoarcus and Synechococcus ribozymes, respectively. In the selected variants, 16 of the 28 contain the C:G-C base triple (the Azoarcus type) whereas the remaining 12 clones possess the G:C-G base triple (the Synechococcus type).
Interestingly, sequence comparison indicated that this base triple also co-varies with the nucleotide in the sixth base pair in P4 (P4bp6′); this nucleotide is C or U in the Azoarcus and Synechococcus ribozymes, respectively. The base triple and the nucleotide co-vary in 24 of the clones (Fig. 7), suggesting the existence of an interaction between the two elements. Comparison of the activities of class 1-a, 1-b, 2-b or 3-a with class 1-c, 1-d, 2-d or 3-c, respectively, suggested that the variants having the Azoarcus type base triple (C:G-C) and P4bp6′ (C) are more active than those having the Synechococcus type base triple (G:C-G) and P4bp6′ (U).
The remaining three positions (two base pairs and one nucleotide) were also examined. It was found that they are not critical for hydrolytic activity of the selectants although they seem to prefer the Synechococcus type nucleotides to the Azoarcus type in the second base pair in P3 (P3bp2) and in the first base pair in P8 (P8bp1).
DISCUSSION
The Azoarcus and Synechococcus IC3 ribozymes are structurally very similar and equally active in vitro. We found that disruption of the long range interactions between L2 and P8 greatly affect the 3′-splice site hydrolysis reaction of the Synechococcus ribozyme, but not the Azoarcus ribozyme (Fig. 2A). Investigation of the factors determining the activity of these ribozymes revealed that the peripheral regions, such as the GC-rich P5 and P6a elements and the two 11 nt GAAA receptors, are not critical under our assay conditions (Figs 3 and 4). We therefore attempted to identify the crucial element within the conserved core region by using in vitro selection (Fig. 6). From a sequence comparison of the selected variants (Fig. 7) it was found that the base triple between P4bp3 and J6/7-2, G:C-U in the Azoarcus ribozyme and A:U-C in the Synechococcus ribozyme (Fig. 7), is critically important. The fact that most of the selectants (27 of 28 clones) possess G:C-U strongly suggests that the Azoarcus type base triple (G:C-U) is preferred over the Synechococcus type (A:U-C) for enhanced catalytic activity of the IC3 intron ribozymes.
One model for explaining the importance of this base triple can be proposed on the basis of the following results. It has been known that: (i) recognition of the conserved guanosine at the 3′-end of the group I intron is critical for its 3′-splice site hydrolysis reaction (24); (ii) the Azoarcus ribozyme can recognize the guanosine ~20- to 30-fold more strongly than other group I introns (25). This may indicate that the Azoarcus type base triple (G:C-U) accelerates the reaction by appropriately tuning binding of guanosine at the active site.
Previous analyses of base triples involving P4bp3 × J6/7-2 indicated that the P4 and J6/7 regions form two base triples, P4bp2 × J6/7-1 and P4bp3 × J6/7-2 (21). The P4bp2 × J6/7-1 base triple is C:G-G in both the Azoarcus and Synechococcus group IC3 ribozymes (Fig. 1). Mutational analyses of the base triples demonstrated that the Tetrahymena group IC1 ribozyme is most active when it possesses G:C-U/C:G-C [which represents the two base triples at P4bp2 × J6/7-1(G:C-U) and P4bp3 × J6/7-2(C:G-C)] whereas the T4 SunY group IA2 ribozyme is more active with C:G-C/G:C-U (21,26). Although these results suggest that the optimal set of base triples differs for the respective subgroup introns, it is also known that the Azoarcus type base triples (C:G-G/G:C-U) function efficiently both in the Tetrahymena (90% activity compared with G:C-U/C:G-C) and T4 SunY (∼70% activity compared with C:G-C/G:C-U) introns (21,26). Combining the previous and present results together, it is conceivable that the Azoarcus type base triples (C:G-G/G:C-U) are very suitable for the P4 × J6/7 position. This is supported by the fact that three phylogenetically divergent (subgroups IC1, IC3 and IA2) group I intron ribozymes can use this set of triples without sacrificing their activity and that the same triples are also found in the subgroup IA3, IB3 and IC2 introns (1). To determine the functional roles of the P4 × J6/7 base triples in the folding and reaction step of the group I ribozyme more precisely, we are currently examining mutational effects of the base triples quantitatively by performing kinetic analyses of the folding and reaction process of this class of ribozymes.
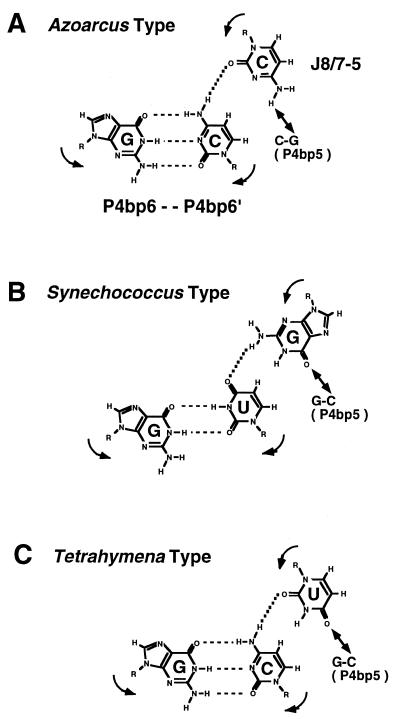
The selection analysis revealed that, in the active selectants, the base triple at P4bp5 × J8/7-5 often co-varies with P4bp6′ (Fig. 7). This base triple is functionally important in the group IC3 introns (22,23). The selection analysis showed that most of the selectants containing G:C-G (the Synechococcus type base triple) possess a U at P4bp6′ whereas those containing C:G-C (the Azoarcus type base triple) possess a C at P4bp6′ (Fig. 7). This co-variation can also be found among the group IC3 introns. The IC3 introns having the triple G:C-G also possess a U at P4bp6′ without exception (9–12) and the Azoarcus ribozyme having the C:G-C triple possesses a C at P4bp6′ (8; Fig. 1), suggesting the existence of an interaction between the P4bp5 × J8/7-5 base triple and P4bp6′. One model for this hypothetical interaction is an isosteric base quintuple consisting of two base pairs (P4bp5 and P4bp6) and one nucleotide (J8/7-5), as shown in Figure 8: the putative Azoarcus type (Fig. 8A) and Synechococcus type (Fig. 8B) base quintuples can be written as C:G-C-G:C and G:C-G-G:U, respectively. In this model, the base moiety of nucleotide J8/7-5 should be out of the plane of the P4bp5 or P4bp6 base pair and in an axial position when interacting with them simultaneously. The observation that the Tetrahymena group IC1 ribozyme having the P4bp5 × J8/7-5 base triple (G:C-U) is capable of forming a similar type of base quintuple (G:C-U-G:C) is consistent with the hypothesis shown in Figure 8C, although further biochemical analyses are necessary to confirm the existence of such a base quintuple.
Figure 8.
Possible base quintuple formed among P4bp5, P4bp6 and J8/7-5. Arrows indicate the polarity of the phosphodiester backbone. Solid lines with two arrowheads indicate the previously proposed tertiary interaction between P4bp5 and J8/7-5 (22,23). The combination of the possible base quintuples in (A) the Azoarcus IC3 ribozyme, (B) the Synechococcus IC3 ribozyme and (C) the Tetrahymena IC1 ribozyme.
The nature of the nucleotides in the core region of the Azoarcus ribozyme is critical to establishing its active structure. This is distinctively different from the strategies for activation in other group I intron ribozymes. For instance, the activity of the Tetrahymena ribozyme is significantly dependent on its large peripheral extension, P5abc (27,28), and the intron from mitochondrial rRNA of Neurospora functions efficiently only when it associates with CYT-18 protein (29,30). Thus at least three strategies for establishing a catalytically active structure exist for group I intron ribozymes, presumably due to selection pressure during the course of molecular evolution (31). We presume that similar strategies for establishing functionally active structures of RNAs will be found in other classes of ribozymes as well as in other RNA molecules which function depending on their higher order structures.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Drs D.A. Shub and J.M. Burke for the gift of plasmid pBGB1.1 encoding the Azoarcus intron ribozyme. We also thank Dr Ruth Yu and members of the T. Inoue Laboratory for critical reading of the the manuscript. This work was supported by Grants-in-Aids for Scientific Research on Priority areas from the Ministry of Education, Science, Sports and Culture, Japan.
REFERENCES
- 1.Michel F. and Westhof,E. (1990) J. Mol. Biol., 216, 585–610. [DOI] [PubMed] [Google Scholar]
- 2.Jaeger L., Michel,F. and Westhof,E. (1996) In Eckstein,F. and Lilley,D.M.J. (eds), Catalytic RNA. Springer, Berlin, Germany, pp. 33–52.
- 3.Lehnert V., Jaeger,L., Michel,F. and Westhof,E. (1996) Chem. Biol., 3, 993–1009. [DOI] [PubMed] [Google Scholar]
- 4.Davila-Aponte J.A., Huss,V.A., Sogin,M.L. and Cech,T.R. (1991) Nucleic Acids Res., 19, 4429–4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rocheleau G.A. and Woodson,S.A. (1994) Nucleic Acids Res., 22, 4315–4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu M.Q., Kathe,S.D., Goodrich-Blair,H., Nierzwicki-Bauer,S.A. and Shub,D.A. (1990) Science, 250, 1566–1570. [DOI] [PubMed] [Google Scholar]
- 7.Kuhsel M.G., Strickland,R. and Palmer,J.D. (1990) Science, 250, 1570–1573. [DOI] [PubMed] [Google Scholar]
- 8.Reinhold-Hurek B. and Shub,D.A. (1992) Nature, 357, 173–176. [DOI] [PubMed] [Google Scholar]
- 9.Biniszkiewicz D., Cesnaviciene,E. and Shub,D.A. (1994) EMBO J., 13, 4629–4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sugita M., Luo,L., Ohta,M., Itadani,H., Matsubayashi,T. and Sugiura,M. (1995) DNA Res., 2, 71–76. [DOI] [PubMed] [Google Scholar]
- 11.Paquin B., Kathe,S.T., Nierzwicki-Bauer,S.A. and Shub,D.A. (1997) J. Bacteriol., 179, 6798–6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paquin B., Heinfling,A. and Shub,D.A. (1999) J. Bacteriol., 181, 1049–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costa M. and Michel,F. (1995) EMBO J., 14, 1276–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanner M.A. and Cech,T.R. (1996) RNA, 2, 74–83. [PMC free article] [PubMed] [Google Scholar]
- 15.Milligan J.F., Groebe,D.R., Witherell,G.W. and Uhlenbeck,O.C. (1987) Nucleic Acids Res., 15, 8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ikawa Y., Naito,D., Aono,N., Shiraishi,H. and Inoue,T. (1999) Nucleic Acids Res., 27, 1859–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanner N.K. and Sargueil,B. (1995) J. Mol. Biol., 252, 583–595. [DOI] [PubMed] [Google Scholar]
- 18.Szostak J.W. and Ellington,A.D. (1993) In Gesteland,R.F. and Atkins,J.F. (eds), RNA World, 1st Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 511–534.
- 19.Lorsch J.R. and Szostak,J.W. (1996) Acc. Chem. Res., 29, 103–110. [DOI] [PubMed] [Google Scholar]
- 20.Williams K.P. and Bartel,D.P. (1996) In Eckstein,F. and Lilley,D.M.J. (eds), Catalytic RNA. Springer, Berlin, Germany, pp. 367–382.
- 21.Michel F., Ellington,A., Couture,S. and Szostak,J. (1990) Nature, 347, 578–580. [DOI] [PubMed] [Google Scholar]
- 22.Tanner M. and Cech,T. (1997) Science, 275, 847–849. [DOI] [PubMed] [Google Scholar]
- 23.Tanner M., Anderson,E., Gutell,R. and Cech,T. (1997) RNA, 3, 1037–1051. [PMC free article] [PubMed] [Google Scholar]
- 24.van der Horst G. and Inoue,T. (1993) J. Mol. Biol., 229, 685–694. [DOI] [PubMed] [Google Scholar]
- 25.Kuo L.Y., Davidson,L.A. and Pico,S. (1999) Biochim. Biophys. Acta, 1489, 281–292. [DOI] [PubMed] [Google Scholar]
- 26.Green R. and Szostak,J.W. (1994) J. Mol. Biol., 235, 140–155. [DOI] [PubMed] [Google Scholar]
- 27.Joyce G.F., van der Horst,G. and Inoue,T. (1989) Nucleic Acids Res., 17, 7879–7889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van der Horst G., Christian,A. and Inoue,T. (1991) Proc. Natl Acad. Sci. USA, 88, 184–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akins R.A. and Lambowitz,A.M. (1987) Cell, 50, 331–345. [DOI] [PubMed] [Google Scholar]
- 30.Lambowitz A.M., Caprara,M.G., Zimmerly,S. and Perlman,P. (1998) In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds), RNA World, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 451–485.
- 31.Inoue T. (1994) Nature, 370, 99–100. [DOI] [PubMed] [Google Scholar]