Abstract
The poly(A) tail of eukaryotic mRNAs regulates translation and RNA stability through an association with the poly(A)-binding protein (PABP). The role of PABP in selective polyadenylation/deadenylation and translational recruitment/repression of maternal mRNAs that occurs in early development is not fully understood. Here, we report studies including UV-crosslinking and immunoblotting assays to characterise PABP in the early developmental stages of the clam Spisula solidissima. A single, 70 kDa PABP, whose sequence is highly homologous to vertebrate, yeast and plant PABPs, is detected in oocytes. The levels of clam PABP are constant in early embryogenesis, although its ability to crosslink labelled poly(A) is ‘masked’ shortly after fertilisation and remains so until the larval stage. Full RNA-binding potential of PABP in embryo lysates was achieved by brief denaturation with guanidinium hydrochloride followed by dilution for binding and crosslinking or by controlled treatment of lysates with Ca2+-dependent micrococcal nuclease. Masking of PABP, which accompanies cytoplasmic polyadenylation in maturing oocytes and in in vitro activated oocyte lysates, is very likely due to an association with mRNAs that bear new PABP target binding sites and thus prevent protein binding to the labelled A-rich probe. Functional implications of these findings as well as the potential application of this unmasking method to other RNA-binding proteins is discussed.
INTRODUCTION
The poly(A)-binding protein (PABP), by binding the 3′ poly(A) tail of eukaryotic mRNA, plays important roles in translational efficiency and message stabilisation in the cytoplasm and 3′-end formation in the nucleus. In eukaryotes, during the initiation phase of protein synthesis the 5′ cap structure m7GpppG binds the eIF4F complex composed of the cap-binding protein eIF4E, the adaptor protein eIF4G and the RNA helicase eIF4A to recruit the small ribosomal subunit (reviewed in 1). The synergistic effect of the cap and the poly(A) tail on translation observed in vivo (2) and in vitro (3,4) is mediated by eIF4G bridging a 5′–3′ interaction between eIF4E and PABP (5–8). Indeed, atomic force microscopy studies strikingly illustrate the ability of these three proteins to circularise capped and polyadenylated mRNA (9). It is thought that such bringing together of the distal ends of transcripts enhances the ability of full-length mRNAs to undergo re-initiation of protein synthesis.
PABP protects the poly(A) tail from deadenylases in vertebrates (10,11) and thus serves to stabilise mRNA by preventing the initial stage of a common pathway of eukaryotic mRNA decay, prior to decapping and 5′→3′ exonucleolytic digestion (reviewed in 12). Recent work shows that mRNA stabilisation is an intrinsic property of PABP that is independent of poly(A) (13). Saccharomyces cerevisiae PABP also functions to regulate poly(A) tail length of pre-mRNA, by interacting with CFI, the cleavage and polyadenylation factor, and through an apparent inhibition of poly(A) polymerase (14,15).
PABP, which is essential in yeast, contains four tandem RNA recognition motif domains (RRM 1–4) at the N-terminus and a much less conserved C-terminal region. Binding of PABP to poly(A) is principally promoted by conserved RNP-1 aromatic residues in RRMs 2+4 (16,17). RRM 2 also contains the residues specifying the functional interaction with eIF4G in yeast and man (4,6,18). The C-terminal, non-RNA-binding portion of the protein contributes to multimerise PABP molecules in the presence of poly(A) to form a higher order structure with regularly spaced PABPs on a single RNA molecule (16). Intriguingly, a recent report suggests that the specific interaction observed between the C-teminus of PABP and eRF3 (polypeptide chain releasing factor) prevents this repeated structure (19).
Levels of PABP in somatic cells appear to be tightly controlled by a proposed autoregulatory translation mechanism involving the 5′-untranslated region (UTR) of PABP mRNA, which contains an A-rich tract capable of binding PABP. PABP present in large excess over 3′ poly(A) binding sites was suggested to associate with the leader of its own mRNA and thus repress its synthesis. Derepression could take place when either the level of polyadenylated transcripts increased or if pre-existing poly(A) tails were lengthened, both scenarios providing additional 3′ target sites (20). For example, in resting cells growth stimulated by serum, PABP synthesis is increased, in the absence of changes in PABP mRNA levels (21), while in terminally differentiated reticulocytes, which are transcriptionally inert and contain stores of PABP, PABP mRNA is largely repressed (22). Subsequent studies demonstrated directly that the PABP 5′-UTR A-rich tract is responsible for autoregulation of PABP mRNA translation and thus determining PABP levels, in vitro and in vivo (20,23–25). Strikingly, ectopically expressed PABP in HeLa or NIH 3T3 cells specifically reduces synthesis of the cognate host cell protein (24,25). The maintenance of an optimal PABP to polyadenylated mRNA ratio appears critical for mediation of the important PABP functions.
Stored maternal mRNAs undergo regulated changes in poly(A) tail length during oocyte maturation and early embryonic development. Investigations in a variety of organisms indicate the critical impact that these mRNA modifications have on their translation: deadenylation silences the mRNAs, while poly(A) extension triggers their expression (reviewed in 26,27). These processes have been best characterised in Xenopus and mouse (28–32), though similar studies in the fruit fly (33) and clam (34,35) point to their evolutionary conservation (36).
In Xenopus, cytoplasmic polyadenylation of maternal mRNA was first observed during oocyte maturation. mRNAs possessing certain U-rich sequences, called CPEs for cytoplasmic polyadenylation elements, in their 3′-UTR have short poly(A) tails in immature oocytes which are substantially elongated during maturation. The CPEs, composed of U4-6A1-2U, are usually found in relatively close proximity upstream of the ubiquitous nuclear polyadenylation signal AAUAAA, also required for cytoplasmic polyadenylation (26,27). Absence of CPEs in the 3′-UTR results in mRNA deadenylation upon maturation, with concomitant release from polysomes (26,27). While the specific CPE-binding factor (CPEB) has been characterised in Xenopus and clam (37–40), little in detail is yet understood about its dual roles as a translational repressor in the oocyte (35,41) or as a cytoplasmic polyadenylation factor in the egg (35,37).
The mechanism by which the extension of a 3′ poly(A) tail on a message influences ribosome binding remains far from clear, but is assumed to potentially involve poly(A)-binding proteins and cap-binding factors. Few studies have investigated the presence and role of PABP in mediating poly(A) functions during the early stages of development. In sea urchin eggs and early embryos, two different PABPs of 66 and 80 kDa have been identified as equally abundant proteins which are readily purified by affinity chromatography (42). These are present at about 50-fold excess over poly(A) binding sites, based on a binding stoichiometry of one PABP per 25 A residues (42). In clear contrast, in Xenopus laevis, immunoblotting experiments do not detect any significant amounts of PABP until the neurula stage (43). More recent evidence indicates that PABP levels in Xenopus oocytes are indeed very low, with less than one PABP molecule per poly(A) binding site (44). This low level of PABP might be required for normal development by allowing the specific deadenylation of selected mRNAs. Poly(A) bound to PABP is resistant to the activity of the Xenopus default deadenylase which acts upon the mRNAs lacking CPEs during meiotic maturation (45).
The purpose of our work was to characterise PABPs in the early stages of development of the surf clam Spisula solidissima, which has been the subject of previous studies of the control of translation and polyadenylation (35,46–48). The technique of UV-crosslinking with a 32P-labelled A-rich RNA probe allowed us to detect a single 70 kDa PABP in clam oocyte and egg lysates, subsequently confirmed by western blotting. We show that while the overall levels of PABP and its mRNA are constant in early embryogenesis, the amount of free PABP declines rapidly upon fertilisation concomitant with maternal mRNA polyadenylation. Thus in eggs and early embryos, PABP and polyadenylated RNA levels are tightly coupled, unlike in oocytes which contain an excess of PABP over poly(A) binding sites. The implications from these observations are contrasted with those reported in other invertebrates and vertebrates.
MATERIALS AND METHODS
Clam oocyte and activated egg extracts
Oocytes were harvested from mature female clams obtained by the Marine Resource Centre at the Woods Hole Marine Biological Laboratory, USA. Fertilisation of oocytes was achieved by diluting concentrated sperm 100-fold in filtered sea water and adding a 1/200 volume of this dilution to the oocyte suspension. Parthenogenetically activated oocytes (eggs) were obtained by incubation of oocytes with 40 mM KCl (final concentration) in filtered sea water for 60 min (40,48). All incubations were performed at 18°C.
Extracts from either oocytes or eggs were prepared in dilute or concentrated versions as described (40,48). Briefly, cells were lysed in 2 vol of T buffer (pH 6.8 or 7.2) in the presence of a 1/1000 dilution of a protease inhibitor solution (10 mg/ml leupeptin, pepstatin and chymostatin in DMSO), RNAguard (1/1000 dilution of stock 30 U/l) and a 1 mM final concentration of DTT. Dilute extracts were used solely for UV-crosslinking reactions. Concentrated extracts were prepared likewise except that oocytes and eggs were washed with T buffer followed by a quick spin and removal of excess buffer by aspiration before lysis. Concentrated oocyte extracts competent for protein synthesis and polyadenylation were assayed as in Walker et al. (48) and Minshall et al. (35), respectively.
UV-crosslinking
To assay for PABP, an A-rich RNA probe was transcribed using T7 RNA polymerase and [32P]ATP (PB 10160; Amersham) as the labelled nucleotide from the pTZ18-based vector described previously (20). The sequence of the A-rich region derived from the 5′-UTR of human PABP mRNA was 5′-CTAGCAGGCCTA6TCCA8TCTA7TCTTTTA6CCCCA7TTTACA6T-3′. Detection of p82/CPEB was performed using the 3′-UTR masking element sequence of S.solidissima ribonucleotide reductase mRNA as probe (48).
Standard UV-crosslinking conditions were as previously described (20). For the phosphatase treatment, oocyte/egg extracts were incubated with potato acid phosphatase at 0.5 µg/µl extract (48). Samples of 2 µl were then used for UV-crosslinking for identification of PABP and p82. Denaturing/renaturing of extracts with guanidinium hydrochloride was performed by adding 2 µl of a saturated solution of guanidinium chloride (GuHCl) to 8 or 10 µl of oocyte/egg extracts (final concentration >1.5 M). After an incubation period of 5 min at room temperature they were then diluted in the UV-crosslinking reaction (0.5–10 µl final reaction volume; final concentration of GuHCl <100 mM). Treatment with micrococcal nuclease was performed by incubating lysates with increasing concentrations of micrococcal nuclease (0, 25 or 250 U for 10 µl extract) with 1–2.5 mM CaCl2 at 20°C for 18 min followed by inactivation with 2–5 mM EGTA and UV-crosslinking.
To immunoprecipitate UV crosslinked PABP, crosslinking reactions were carried out as described above, except that heparin was omitted. Following RNase treatment, five 10 µl reactions were pooled together and diluted to 200 µl with NETS buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 5 mM EDTA, 0.05% NP40). An aliquot of 5 µl of anti-Dm PABP serum was added. The rabbit polyclonal serum was raised against recombinant Drosophila PABP, made in Escherichia coli (generously supplied by V. Lefrère; 49). The reactions were left at 4°C on a rotary shaker for 1 h. After this time, 10 µl of slurried protein A–Sepharose 6MB beads (Pharmacia) were added and incubation continued for another hour. The beads were collected by brief centrifugation in a microcentrifuge, washed three times with 1 ml NETS buffer and finally boiled in 20 µl of 2× SDS sample buffer, prior to gel electrophoresis.
Western blotting
Western blotting was carried out essentially as described previously (40). Blots were blocked with Tris-buffered saline (TBST = 50 mM Tris–HCl pH 8.0, 150 mM NaCl, 0.1% Tween-20) containing 5% non-fat dried milk. The membrane was incubated for 2 h at room temperature with the anti-Dm PABP serum diluted in TBST containing 5% Marvel (1:2000). The blots were developed with polyclonal goat anti-rabbit alkaline phosphatase-conjugated secondary antibody (dilution 1:6000; Sigma). Drosophila melanogaster embryo proteins were a gift of Dr Stephan Grunert (Wellcome/CRC).
Cloning of clam oocyte PABP by RT–PCR
Reverse transcription (RT) of clam oocyte RNA was performed by denaturing 5–10 µg RNA at 65°C for 3 min and incubating with reverse transcriptase (Promega), as suggested by the manufacturer, in the presence of 1 mM dNTP, 0.75 U/µl RNase inhibitor, 10 mM DTT and 0.1 µg/µl primer solution at 42°C for 2 h. The primer solution contained a mix of oligos containing 20 T residues followed by either A, G or C, to allow anchoring to the 5′-end of the poly(A) tail.
PCR reactions were performed using two sets of degenerate oligonucleotides designed to anneal to conserved regions of PABP as primers. The 5′ primer of each set corresponds to the most conserved region of PABP, positioned between RRMs 1 and 2 [primer 4370, 5′-CGGGATCCATGTGGTCTCA(G/A)CGTGA-3′]. The first of the two alternative 3′ primers anneals to another conserved region positioned at the end of RRM 4 [primer 4371, 5′-CGGGATCC(G/T)(T/C)TG(A/G)GC(T/C)A(T/A)(T/G)GC(T/A)AC-3′] whilst the second 3′ primer anneals to the conserved motif located in the C-terminus of PABP [primer 4372, 5′-CGGGATCCAT(G/T/A)CC(A/G)GT(G/A)AT(C/T)TT(A/G)CC-3′]. All primers included sites for BamHI (underlined) at their 5′-end. Nested PCR, using Taq DNA polymerase (Boehringer) with 2.5 mM MgCl2 and a 50°C annealing temperature for 30 cycles, was initially performed with primers 4370/4372 and the product of expected size was re-amplified with primers 4370/4371. This second PCR reaction produced only one DNA fragment which was digested with BamHI, subcloned into the BamHI site of plasmid pGEM3zf+ (Promega), sequenced and used as a probe in northern blots (accession no. AF255335).
Northern blotting
Total RNA from Spisula oocytes/eggs at different time points was prepared using TriReagent (Molecular Research Center, OH) according to the manufacturer’s instructions. Aliquots of 20 µg RNA for each time point were glyoxylated according to Ausubel et al. (50) and run on a vertical 1.5% (w/v) agarose gel. Radiolabelled DNA markers were HindIII fragments of λ DNA labelled with [35S]dATP using DNA polymerase I Klenow fragment. The labelled DNA was boiled for 3 min before being treated with glyoxal as for RNA. After electrophoresis, RNA was transferred to Hybond-N (Amersham) in 20× SSC and crosslinked to the membrane by irradiation on a UV transilluminator (302 nm) for 10 min followed by baking at 80°C for 2 h. Membranes were prehybridised in prehybridisation solution for 2 h at 65°C and with the PABP DNA probe (labelled by random priming using the PrimeIt kit; Stratagene) at ∼5 × 105 c.p.m./ml. Hybridisation was performed at 65°C overnight before membranes were removed and washed twice for 45 min at 60°C in 0.1× SSC, 0.1% SDS. Membranes were air dried and subjected to autoradiography.
RESULTS
Specific poly(A)-binding activity can be detected in clam oocyte extracts but not in those from activated eggs
To identify S.solidissima proteins which preferentially bind poly(A), UV-crosslinking assays of clam oocyte and egg extracts were performed using an A-rich 32P-labelled probe. This RNA probe, which corresponds to a portion of the 5′-UTR of human PABP mRNA, is 73 nt long and consists of stretches of 6–8 A residues interspersed by pyrimidine residues. Similar A-rich regions are present in many PABP mRNAs cloned to date (20,51). The A-rich probe crosslinks to rabbit reticulocyte and wheatgerm PABP in a specific manner; the crosslinking is competed out by poly(A) but not by poly(U) or poly(G) competitor homopolymers (20; Fig. 1A). Use of this probe, rather than an equivalent length of pure A tract, is preferred in UV-crosslinking due to the ease with which the covalently linked RNA, following UV irradiation, can be digested with RNase A and/or T1.
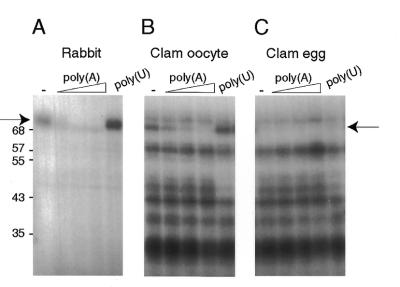
Figure 1.
UV-crosslinking analysis of PABPs from rabbit reticulocytes, clam oocytes and eggs. Extracts were pre-incubated in the absence (–) or presence of competitor poly(A) (increasing concentrations of 1, 4 and 16 µg/ml) and poly(U) (4 µg/ml) for 5 min at 30°C and mixed with the 32P-labelled A-rich oligonucleotide probe. Samples were then used for UV-crosslinking, RNase treatment and SDS–PAGE followed by autoradiography. The arrow on the left indicates rabbit PABP and that on the right indicates the putative clam PABP band. The numbers on the left indicate the molecular weight markers (kDa).
In contrast to reticulocyte lysate, which contains a unique PABP, several clam oocyte proteins were crosslinked to the A-rich probe, including one the size of rabbit PABP. However, only the 70 kDa clam polypeptide binds poly(A) specifically, as its binding can be competed by added poly(A) but not by poly(U) (Fig. 1A and B). Based on its size and specific poly(A) binding this oocyte protein is presumed to be the clam PABP homologue. When egg extracts were assayed by the same method essentially all the bands detected in oocytes were also present in eggs, with the significant and striking exception of the 70 kDa protein (Fig. 1C). We concluded that either PABP protein levels or the ability of PABP to bind RNA was regulated in maturing Spisula oocytes.
Identification of clam PABP
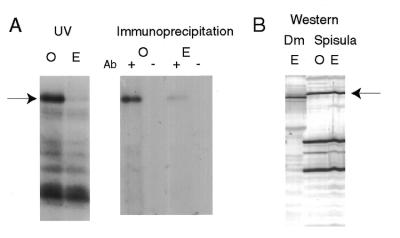
First we needed to verify that the 70 kDa crosslinked band from clam oocyte lysates which exhibited poly(A)-specific binding was indeed clam PABP. To do so, we used antibodies raised against recombinant D.melanogaster PABP (49) in immunoprecipitation assays and western blots. UV reactions of both oocyte and egg lysates irradiated in the presence of the labelled A-rich probe were immunoprecipitated with anti-Dm PABP antibodies and the complexes captured on protein A–Sepharose beads. Control reactions were performed in the absence of antibody. The results show that the 70 kDa protein labelled by UV-crosslinking in oocyte lysates (and present at very reduced levels in egg lysates) is recognised by anti-Dm PABP antibodies, confirming its initial identification as PABP (Fig. 2A). Moreover, using the same serum, but this time in western blots of total clam oocyte and egg lysates, we show that clam PABP is present in oocytes and eggs at equivalent levels. While several non-specific bands are recognised by the antibodies on the clam protein blots, a 70 kDa polypeptide corresponding to the immunoprecipitated PABP, and migrating close to Drosophila PABP, is evident in both clam oocytes and eggs (Fig. 2B). The 70 kDa clam PABP is found essentially exclusively in the oocyte cytoplasm (not shown). Taken together, the approaches of RNA binding (Fig. 1) and immunological detection (Fig. 2) led us to conclude that there was a significant difference in the levels of available PABP before and after oocyte activation. While in oocytes there is a significant amount of PABP free to bind RNA, in eggs this protein is either inactive or inaccessible.
Figure 2.
Identification of the oocyte 70 kDa polypeptide as PABP. (A) UV-crosslinking samples of clam oocyte (O) and eggs (E) (see Fig. 1) were immunoprecipitated with (+) or without (–) rabbit antibodies raised against D.melanogaster PABP and protein A–Sepharose. (B) Western blot of D.melanogaster embryo (E) and Spisula oocyte (O) and egg (E) protein samples, probed with anti-Dm PABP antibodies and alkaline phosphatase-conjugated goat anti-rabbit antibodies. The arrows indicate the PABP band.
Unmasking the full RNA-binding potential of clam PABP
We reasoned that the RNA probe may only detect free protein but cannot compete for PABP complexed to RNA and/or protein. We first tested whether phosphorylation of PABP (or of a putative regulatory factor) was involved in its differential RNA binding by treating oocyte and egg lysates with potato acid phosphatase and then UV-crosslinking. The efficacy of the phosphatase in dephosphorylating RNA-binding proteins was checked in control reactions using clam p82/CPEB, a protein that binds CPE-like motifs in clam maternal mRNAs and whose phosphorylation in maturing oocytes results in a discernible size shift in denaturing SDS–PAGE gels (40,48). While p82/CPEB was dephosphorylated, the same treatment did not reveal PABP poly(A) binding in eggs (Fig. 3A).
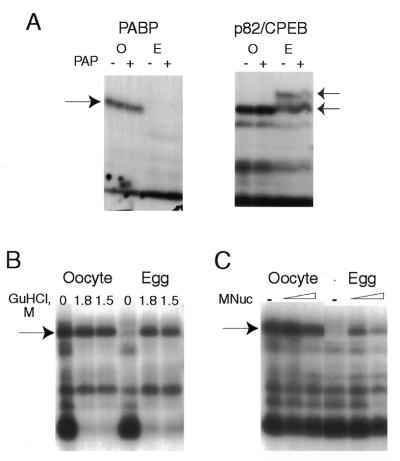
Figure 3.
Unmasking of poly(A)-binding activity in Spisula egg lysates. Clam oocyte and egg extracts were incubated under different conditions prior to UV-crosslinking. (A) Aliquots were incubated with potato acid phosphatase (+) or water (–) as control for 30 min at 30°C. The samples were then used for UV-crosslinking with either the A-rich RNA probe, for PABP detection, or the 3′-UTR of ribonucleotide reductase, for detection of p82/CPEB. The dark arrow on the left indicates PABP; the two lighter arrows on the right point to the unmodified and phosphorylated forms of p82/CPEB. (B) Aliquots were incubated with a saturated GuHCl solution for 5 min at room temperature, to a final concentration of 1.8 or 1.5 M, and subsequently diluted and submitted to the UV-crosslinking protocol. (C) Aliquots were incubated with increasing concentrations of micrococcal nuclease (0, 2500 and 25 000 U/ml) before EGTA treatment and UV-crosslinking assays.
To test whether egg PABP was unavailable to bind the RNA probe due to its participation in protein–protein or RNA–protein complexes, extracts from both oocytes and eggs were briefly denatured with GuHCl and subsequently rapidly diluted in the presence of the labelled probe followed by UV-crosslinking (Fig. 3B). In practice, denaturing/renaturing of extracts with GuHCl was achieved by incubating aliquots of lysates with a saturated solution of GuHCl (∼9 M) at a final concentration of ∼1.5 M. After 5 min, the samples were diluted in the UV-crosslinking mix to reduce the concentration of GuHCl to <100 mM (see Materials and Methods). Following this treatment, not only is clam PABP clearly present in egg extracts, but its levels are equivalent to those in oocytes (Fig. 3B).
Figure 3C shows the results of an experiment that tested whether the absence of free PABP in eggs is due to it binding endogenous mRNA. Oocyte and egg extracts were treated with micrococcal nuclease (0, 25 or 250 U for 10 µl extract) in the presence of CaCl2 to degrade lysate RNA, followed by subsequent enzyme inactivation with EGTA and UV-crosslinking. Again, PABP previously undetectable in egg extracts can now be identified by UV-crosslinking (Fig. 3C). (The higher levels of nuclease used in this experiment may have been incompletely inactivated by EGTA, leading to probe digestion and loss of PABP crosslinking.) These results suggest that the absence of poly(A)-binding activity in egg extracts is due to PABP being unavailable for binding the labelled RNA probe as it is already tightly associated with endogenous poly(A)+ mRNA. Conditions which result in disruption of the native mRNP (GuHCl) or which destroy mRNA (micrococcal nuclease) unmask the RNA-binding potential of the egg PABP.
Next, we employed a gel filtration method to independently assess the proportion of PABP in large RNP complexes and in the cytosol. Clam oocyte and egg lysates were fractionated on Sepharose CL6B large pore columns in low salt buffer (48) and fractions were analysed by western blotting using anti-PABP antibodies. Densitometric scanning revealed an ∼2-fold increase in complexed PABP in eggs, compared to oocytes (data not shown), supporting our conclusions from the UV-crosslinking assays regarding the differential availability of PABP in these stages. In contrast, clam p82/CPEB and its phosphorylated form p92 are found exclusively in RNP in both stages (N.Minshall and N.Standart, unpublished results).
Cloning of clam PABP
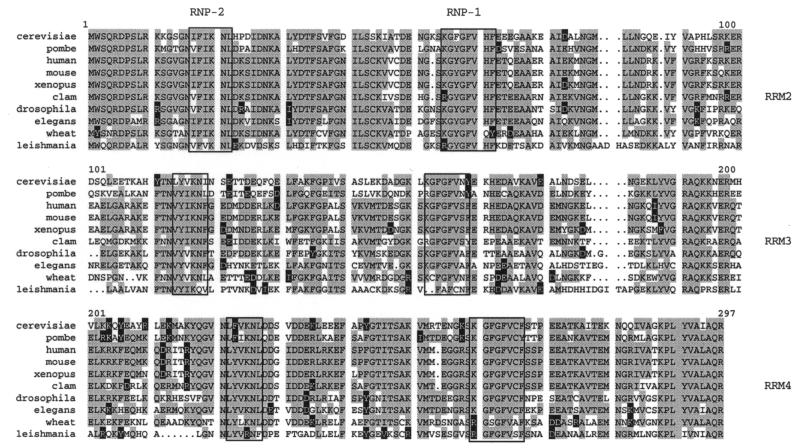
In order to explore in detail the expression of PABP and PABP mRNA in early clam development, we needed to clone the Spisula protein, as no PABP sequence from any marine invertebrate was available. A partial clam PABP cDNA was obtained using degenerate primers that anneal to highly conserved regions of PABP. These primers amplified fragments of predicted size from rabbit reticulocyte and Xenopus egg total RNA (not shown). Two sets of nested PCR reactions were performed with clam oocyte RNA to amplify a 858 bp cDNA fragment, corresponding to RRMs 2–4 of PABP (see Materials and Methods for details). The sequenced clone revealed a high degree of homology to PABPs from a variety of organisms, ranging from fern to man (Fig. 4). For example, 85% of clam residues were identical/homologous to human PABP. The distinct sequences of the individual RRM regions are reflected in their different functions. In particular, RRMs 2+4 showed high sequence conservation between clam and, notably, the other vertebrate PABPs. RRM 2 contains the high affinity poly(A) binding site while RRM 4 promotes non-specific polypyrimidine RNA binding (16,17). RRM 2 and RRM 4 are required for poly(A) and cap-dependent stimulation of translation. In addition to its role in binding RNA, RRM 2 of yeast PABP is required for its functional interaction with eukaryotic translation initiation factor 4G (18). The sequence of clam PABP fully supported our previous conclusions of the presence of a ‘conventional’ PABP in Spisula, based on the size of a specific PABP (Fig. 1) and immunological detection (Fig. 2).
Figure 4.
Sequence comparison between clam PABP and PABPs from other organisms. Clam PABP cDNA containing the RRM 2–RRM 4 region (AF255335) is aligned with PABP sequences from S.cerevisiae (AAA34838), Schizosaccharomyces pombe (P31209), man (P11940), mouse (AAB70164.1), X.laevis (P20965), D.melanogaster (S30887), Caenorhabditis elegans (T26427), wheat (T06979) and Leishmania major (AF093062). The RNP-2 and RNP-1 motifs are boxed; four or more identical amino acids are highlighted in grey while homologous amino acids are highlighted in black.
Developmental expression of PABP and PABP mRNA in clam embryos
During the first few hours of embryonic development of most organisms any changes in the pattern of protein synthesis depend on regulation at the post-transcriptional level. In clam embryos, no substantial de novo mRNA synthesis occurs until around 3–4 h of development (52,53).
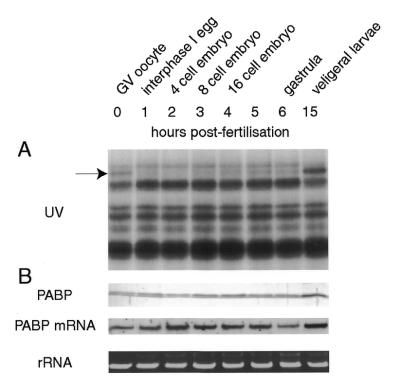
To determine whether and when free PABP levels can be detected in early embryogenesis, extracts were prepared at different time points after fertilisation and assayed by UV-crosslinking (Fig. 5A). The drop in the level of free PABP that occurs early after fertilisation (Figs 2 and 3) is not reversed in the first hours of development. Only after embryonic mRNA synthesis resumes at ∼3–4 h after fertilisation is an increase in free PABP levels first detected and only 15 h after fertilisation, when the larvae reach the swimming stage, does its level approach that seen in the oocyte. Western blots of the same extracts show that total PABP levels remain constant throughout this period except for an increase at the 15 h time point, confirming that the changes seen by UV-crosslinking are largely at the level of free protein (Fig. 5B).
Figure 5.
Levels of free PABP in developing clam embryos. (A) Fertilised oocytes were cultured in sea water at 18°C and their embryonic development monitored by microscopic examination. At intervals, as indicated, aliquots were taken and extracts prepared for UV-crosslinking with the A-rich probe to detect PABP (arrow). (B) The same timed samples were also analysed by western blotting with anti-PABP antibodies (upper), by northern blotting probed with a clam PABP cDNA (middle) and by agarose gel electrophoresis and EtBr staining for rRNA loading control (lower).
As discussed earlier, PABP synthesis in somatic cells is presumed to be regulated at the translational level by binding of free PABP to the 5′-UTR of its mRNA, which contains an A-rich tract (20,23). According to this model, a reduction in free protein would relieve the inhibition of translation and allow a rapid recovery of free protein levels, unless other control mechanisms were acting upon PABP. Since more than 6 h are necessary for the recovery of free PABP levels, we investigated whether any changes in the level of PABP mRNA might be responsible for this behaviour. The PABP cDNA was used as a probe in northern blots with RNA samples prepared from different time points. Equal loading of samples was monitored by agarose gel electrophoresis and EtBr staining of rRNA. PABP mRNA levels remain constant throughout early Spisula development and only at the swimming stage (15 h) is an increase in its concentration observed, presumably responsible for the increase in PABP seen at this stage (Fig. 5B). It appears from the lack of de novo synthesis of PABP after the drop in free protein levels that mechanisms other than the somatic autoregulatory one regulate the translation of this mRNA during early clam development. Due to the large size of clam PABP mRNA (estimated to be ∼3.3 kb) we could not determine directly from the northern analysis whether this mRNA was subject to polyadenylation and/or deadenylation during development.
PABP masking coincides with cytoplasmic polyadenylation of clam maternal mRNAs
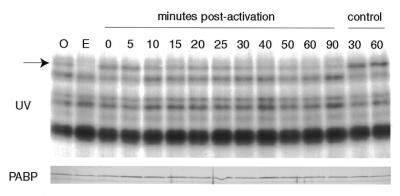
In Spisula, as in higher eukaryotes, cytoplasmic polyadenylation of maternal mRNAs is temporally and functionally correlated with the major changes seen in the pattern of protein synthesis that follows meiotic maturation. Polyadenylation of cyclin A, one of the most abundant translationally regulated maternal mRNAs, occurs ∼20–25 min after fertilisation (34). We set out to define more precisely at what time after activation the drop in free PABP levels occurs, to see if a correlation can be made with the timing of cytoplasmic polyadenylation of maternal mRNAs. As shown in Figure 6, aliquots of KCl (parthenogenetically)-activated oocytes were taken at frequent intervals after oocyte activation and the behaviour of free PABP was monitored by UV-crosslinking. The intensity of labelled PABP decreases gradually from ∼10 to 30 min after activation in the absence of significant changes in PABP protein levels as shown by western blotting. Thus the drop in free PABP levels coincides with the period of maximum cytoplasmic polyadenylation of maternal mRNA.
Figure 6.
Free PABP levels decline ∼30 min after oocyte activation. Parthenogenetically (KCl) activated oocytes were cultured at 18°C and frequent samples, as indicated, were withdrawn for UV-crosslinking to detect PABP (arrow). Samples of oocytes (O) and 1 h egg (E) as well as control samples (no KCl addition) were analysed alongside. The lower panel is a western blot of the same samples developed with anti-PABP antibodies.
pH-induced activation of oocyte lysates reduces available PABP
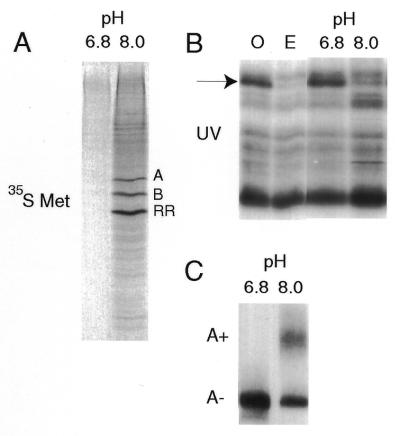
We previously described an in vitro activation system from clam oocytes based on the natural permanent rise in pH that accompanies their maturation. The intracellular pH of clam oocytes, pH 6.8–6.9, rises rapidly in the fertilised egg (48,54; and references therein). Concentrated lysates were prepared from clam oocytes in T buffer, pH 6.8. To mimic activation, an equal volume of either pH 6.8 (control) or 8.0 (final pH ∼7.2, activated) was added to the concentrated lysates, followed by incubation at 18°C for 2 h. As reported earlier, raising the pH increases the rate and pattern of protein synthesis, including selective stimulation of translation of the abundant mRNAs encoding cyclins A and B and the small subunit of ribonucleotide reductase (48; Fig. 7A). We then tested whether the pH-activated oocyte extracts could produce the changes seen in free PABP levels observed after fertilisation in vivo. Indeed, a clear reduction in PABP-binding activity, similar to that observed in the extracts prepared from eggs, occurs in the in vitro activated samples (Fig. 7B). This reduction in available PABP correlated with the pH-induced activation of polyadenylation, as assayed with the 3′-UTR of ribonucleotide reductase mRNA as substrate (35; Fig. 7C). Thus both in vivo (Fig. 6) and in vitro (Fig. 7), newly polyadenylated RNA sequesters available PABP.
Figure 7.
Reduction in free PABP levels on activation in vitro. Concentrated clam oocyte extracts were activated by mixing with an equal volume of buffer at pH 6.8 (control) or 8.0 (activated) and incubated for 2 h at 18°C. (A) Protein synthesis profile of the unactivated and activated samples assayed in the presence of [35S]methionine. The three major translationally up-regulated mRNAs encode cyclins A (A) and B (B) and the small subunit of ribonucleotide reductase (RR). (B) UV-crosslinking reactions of oocyte (O) and 1 h egg (E) lysates and the pH 6.8/8.0 samples assayed with the 32P-labelled A-rich RNA probe. The arrow indicates PABP. (C) Polyadenylation assays of the pH 6.8/8.0 samples assayed with the 32P-labelled ribonucleotide reductase 3′-UTR probe. A–, the non-adenylated labelled RNA probe; A+, migration of the RNA probe extended by ~150 A residues.
DISCUSSION
We report two major findings in this study of PABP in the early embryogenesis of the clam Spisula. First, we show that while levels of PABP in oocytes and early embryos are essentially constant, the amount of PABP available to bind added poly(A) declines dramatically during meiotic maturation, at a time when maternal mRNAs undergo cytoplasmic polyadenylation, and does not recover until well after zygotic transcription. Thus in oocytes there appears to be a significant excess of PABP over poly(A) binding sites, while in maturing eggs and early embryos this excess is sequestered by newly polyadenylated RNA. The functional implications of our observations in Spisula will be contrasted below with those arising from previous studies of PABP in developing sea urchins and Xenopus. Secondly, and more generally, the simple methodology we developed for unmasking the full RNA-binding potential of egg PABP, by either brief GuHCl denaturation or controlled micrococcal nuclease digestion, should in principle be applicable to any study of RNA-binding proteins. In particular, we envisage that high affinity RNA-binding proteins that are part of a large RNP complex or ones that undergo regulation in development or differentiation may be inaccessible at different stages to probing with labelled RNA until the complexes and/or target RNA is destroyed.
Mature Xenopus oocytes accumulate an enormous pool of polyadenylated mRNA during oogenesis, but the overall poly(A) content subsequently decreases upon progesterone-stimulated oocyte maturation, from 1.8 ng/oocyte to 1.0 ng/egg. Poly(A)+ RNA contains two distinct size classes of poly(A) in oocytes (mean size 60 and 20 nt long) but three in eggs and embryos (100, 80 and 20 nt) (55). In the light of more recent studies these results can be interpreted as arising from a relatively large scale deadenylation of housekeeping messages that lack a CPE accompanied by extension of poly(A) tails on a specific subset of CPE-containing mRNAs. These processes are largely temporally separated; different mRNAs are polyadenylated at varying times during maturation leading up to and following germinal vesicle breakdown (GVBD) while deadenylation occurs abruptly at GVBD (26,27). Quantitative immunoblotting analysis indicates that there is <5 pg PABP/embryo at the cleavage stage and that PABP is undetectable earlier. In view of these estimates, oocytes, eggs and early embryos contain <1 PABP per 25 nt binding site (44). It is likely that Xenopus oocytes and eggs contain the same very low level of PABP and, therefore, that maturation results in higher occupancy of the remaining poly(A+) RNA by PABP. The PABP:mRNA ratio is critical for the process of deadenylation; overexpression (8–10 times) of PABP stabilises and prevents translational inactivation of non-CPE-containing mRNAs, implying a primary role for PABP in poly(A+) RNA stabilisation in metazoan oocytes (45). It is important to note that our current understanding of Xenopus PABP is based on the assumption of a single polypeptide species capable of binding poly(A)+ RNA, but it is possible that there exist additional PABPs in Xenopus (56).
Indeed, in sea urchins, there are two forms of PABP, of 66 and 80 kDa, found in approximately equal ratios in nuclear and cytoplasmic fractions. Sea urchin PABPs are, in striking contrast to Xenopus, surprisingly abundant proteins, comprising ∼0.6% of total cellular protein; approximately 50 times more than required to bind all poly(A) in eggs. In this invertebrate, the 3-fold increase in poly(A) content after fertilisation is accompanied by an increase in bound PABP, nevertheless, PABPs are still in 15 times excess over binding sites. Thus in eggs and two-cell embryos >95% of PABP appears uncomplexed to mRNA (42). While this apparent difference between Xenopus and sea urchins is surprising, in the absence of sequence information it is unclear whether one or both sea urchin proteins are in fact members of the classic well-characterised PABP family.
The single 70 kDa Spisula PABP is highly homologous to human, yeast and plant PABPs. The overall poly(A) content in clam oocytes increases ∼2–3-fold after fertilisation (57). Deadenylation and polyadenylation of different maternal mRNAs occurs rapidly after fertilisation and at the same time, ∼25–35 min after fertilisation (34). The results presented in this study indicate that the free concentration of the PABP in the cytoplasm varies as maturation proceeds. We also provide evidence that the process of cytoplasmic polyadenylation may be responsible for the reduction in free poly(A)-binding activity during maturation. As in Xenopus then, levels of PABP are limiting at the time of polyadenylation/deadenylation of maternal mRNAs. The participation of PABP in the polyadenylation process itself and the consequences of increased PABP complexed to polyadenylated RNAs in early developmental stages is still very much unknown, but the dramatic changes seen in its binding activity following fertilisation indicate that this protein might also have an important direct or indirect role in controlling poly(A) metabolism and protein synthesis during early development.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Valerie Lefrere for the Drosophila anti-PABP antibody, Stephan Grunert for Drosophila embryo proteins and Richard Jackson and Mike Wormington for stimulating suggestions and discussions. O.P.M.N. was funded by the Brazilian Research Council CNPq, its program RHAE and the British Council. J.A.W. acknowledges funding from the James A. and Faith Miller Fellowship and MBL Associates Fellowship 1996, Marine Biological Laboratory, Woods Hole.
DDBJ/EMBL/GenBank accession no. AF255335
REFERENCES
- 1.Hentze M.W. (1997) Science, 275, 500–501. [DOI] [PubMed] [Google Scholar]
- 2.Gallie D.R. (1991) Genes Dev., 5, 2108–2116. [DOI] [PubMed] [Google Scholar]
- 3.Tarun S.Z. and Sachs,A.B. (1995) Genes Dev., 9, 2997–3007. [DOI] [PubMed] [Google Scholar]
- 4.Otero L.J., Ashe,M.P. and Sachs,A.B. (1999) EMBO J., 18, 3153–3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tarun S.Z. Jr and Sachs,A.B. (1996) EMBO J., 15, 7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 6.Imataka H., Gradi,A. and Sonenberg,N. (1998) EMBO J., 17, 7480–7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Piron M., Vende,P., Cohen,J. and Poncet,D. (1998) EMBO J., 17, 5811–5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fraser C.S., Pain,V.M. and Morley,S.J. (1999) J. Biol. Chem., 274, 196–204. [DOI] [PubMed] [Google Scholar]
- 9.Wells S.E., Hillner,P.E., Vale,R.D. and Sachs,A.B. (1998) Mol. Cell, 2, 135–140. [DOI] [PubMed] [Google Scholar]
- 10.Bernstein P., Peltz,S.W. and Ross,J. (1989) Mol. Cell. Biol., 9, 659–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ford L., Bagga,P. and Wilusz,J. (1997) Mol. Cell. Biol., 17, 398–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caponigro G. and Parker,R. (1996) Microbiol. Rev., 60, 233–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coller J.M., Gray,N.K. and Wickens,M.P. (1998) Genes Dev., 12, 3226–3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amrani N., Minet,M., Le Gouar,M., Lacroute,F. and Wyers,F. (1997) Mol. Cell. Biol., 17, 3694–3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Minvielle-Sebastia L., Preker,P.J., Wiederkehr,T., Strahm,Y. and Keller,W. (1997) Proc. Natl Acad. Sci. USA, 94, 7897–7902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kühn U. and Pieler,T. (1996) J. Mol. Biol., 256, 20–30. [DOI] [PubMed] [Google Scholar]
- 17.Deardoff J.A. and Sachs,A.B. (1997) J. Mol. Biol., 269, 67–81. [DOI] [PubMed] [Google Scholar]
- 18.Kessler S.H. and Sachs,A.B. (1998) Mol. Cell. Biol., 18, 51–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoshino S.-I., Imai,M., Kobayashi,T., Uchida,N. and Katada,T. (1999) J. Biol. Chem., 274, 16677–16680. [DOI] [PubMed] [Google Scholar]
- 20.de Melo Neto O.P., Standart,N. and de Sa,C.M. (1995) Nucleic Acids Res., 23, 2198–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomas G. and Thomas,G. (1986) J. Cell Biol., 103, 2137–2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maundrell K., Imaizumi-Scherrer,M.T., Maxwell,E.S., Civelli,O. and Scherrer,K. (1983) J. Biol. Chem., 258, 1387–1390. [PubMed] [Google Scholar]
- 23.Bag J. and Wu,J. (1996) Eur. J. Biochem., 237, 143–152. [DOI] [PubMed] [Google Scholar]
- 24.Wu J. and Bag,J. (1998) J. Biol. Chem., 273, 34535–34542. [DOI] [PubMed] [Google Scholar]
- 25.Hornstein E., Harel,H., Levy,G. and Meyuhas,O. (1999) FEBS Lett., 457, 209–213. [DOI] [PubMed] [Google Scholar]
- 26.Wickens M., Kimble,J. and Strickland,S. (1996). In Hershey,J.W.B., Mathews,M.B. and Sonenberg,N. (eds), Translational Control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 410–450.
- 27.Richter J.D. (1996) In Hershey,J.W.B., Mathews,M.B. and Sonenberg,N. (eds), Translational Control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 481–503.
- 28.Huarte J., Stutz,A., O’Connell,M.L., Gubler,P., Belin,D., Darrow,A.L., Strickland,S. and Vassali,J.-D. (1992) Cell, 69, 1021–1030. [DOI] [PubMed] [Google Scholar]
- 29.Varnum S.M. and Wormington,W.M. (1990) Genes Dev., 4, 2278–2286. [DOI] [PubMed] [Google Scholar]
- 30.Sheets M., Fox,C., Hunt,T., Van de Woude,G. and Wickens,M. (1994) Genes Dev., 8, 926–938. [DOI] [PubMed] [Google Scholar]
- 31.Gebauer F., Xu,W., Cooper,G. and Richter,J. (1994) EMBO J., 13, 5712–5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sheets M.D., Wu,M. and Wickens,M. (1995) Nature, 374, 511–516. [DOI] [PubMed] [Google Scholar]
- 33.Sallés F.J., Lieberfarb,M.E., Wreden,C., Gergen,J.P. and Strickland,S. (1994) Science, 266, 1996–1999. [DOI] [PubMed] [Google Scholar]
- 34.Rosenthal E.T. and Ruderman,J.V. (1987) Dev. Biol., 121, 237–246. [DOI] [PubMed] [Google Scholar]
- 35.Minshall N., Walker,J., Dale,M. and Standart,N. (1999) RNA, 5, 27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verrotti A., Thompson,S., Wreden,C., Strickland,S. and Wickens,M. (1996) Proc. Natl Acad. Sci. USA, 93, 9027–9032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hake L.E. and Richter,J.D. (1994) Cell, 79, 617–627. [DOI] [PubMed] [Google Scholar]
- 38.Stebbins-Boaz B., Hake,L.E. and Richter,J.D. (1996) EMBO J., 15, 2582–2592. [PMC free article] [PubMed] [Google Scholar]
- 39.Hake L.E., Mendez,R. and Richter,J.D. (1998) Mol. Cell. Biol., 18, 685–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walker J., Minshall,C., Hake,L., Richter,J. and Standart,N. (1999) RNA, 5, 14–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Moor C. and Richter,J.D. (1999) EMBO J., 18, 2294–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Drawbridge J., Grainger,J.L. and Winkler,M.M. (1990) Mol. Cell. Biol., 10, 3994–4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zelus B.D., Giebelhaus,D.H., Eib,D.W., Kenner,K.A. and Moon,R.T. (1989) Mol. Cell. Biol., 9, 2756–2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stambuk R. and Moon,R. (1992) Biochem. J., 287, 761–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wormington M., Searfoss,A. and Hurney,C. (1996) EMBO J., 15, 900–909. [PMC free article] [PubMed] [Google Scholar]
- 46.Standart N., Dale,M., Stewart,E. and Hunt,T. (1990) Genes Dev., 4, 2157–2168. [DOI] [PubMed] [Google Scholar]
- 47.Standart N. and Dale,M. (1993) Dev. Genet., 14, 492–499. [DOI] [PubMed] [Google Scholar]
- 48.Walker J., Dale,M. and Standart,N. (1996) Dev. Biol., 173, 292–305. [DOI] [PubMed] [Google Scholar]
- 49.Lefrère V. and Duncan,R. (1994) Nucleic Acids Res., 22, 1640–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (1995) Short Protocols in Molecular Biology. John Wiley & Sons.
- 51.Hotchkiss T.L., Nerantzakis,G.E., Dills,S.C., Shang,L. and Read,L.K. (1999) Mol. Biochem. Parasitol., 98, 117–129. [DOI] [PubMed] [Google Scholar]
- 52.Rosenthal E.T., Hunt,T. and Ruderman,J.V. (1980) Cell, 20, 487–494. [DOI] [PubMed] [Google Scholar]
- 53.Westendorf J.M. (1988) Molecular characterization of cyclin B: a protein that induces M-phase in the early clam embryo. Harvard University, Cambridge, MA.
- 54.Katsu Y., Minshall,N., Nagahama,Y. and Standart,N. (1999) Dev. Biol., 209, 186–199. [DOI] [PubMed] [Google Scholar]
- 55.Sagata N., Shiokawa,K. and Yamana,K. (1980) Dev. Biol., 77, 431–448. [DOI] [PubMed] [Google Scholar]
- 56.Swiderski R. and Richter,J. (1988) Dev. Biol., 128, 349–358. [DOI] [PubMed] [Google Scholar]
- 57.Rosenthal E.T., Tansey,T.R. and Ruderman,J.V. (1983) J. Mol. Biol., 166, 309–327. [DOI] [PubMed] [Google Scholar]