Abstract
The chloroplast ribosomal protein CS1, the homolog of the bacterial ribosomal protein S1, is believed to be involved in the process of ribosome binding to mRNA during translation. Since translation control is an important step in chloroplast gene expression, and in order to study initiation complex formation, we studied the RNA-binding properties of CS1 protein. We found that most of the CS1 protein in spinach chloroplast co-purified with the 30S ribosomal subunit. The relative binding affinity of RNA to CS1 was determined using the UV-crosslinking competition assay. CS1 protein binds the ribohomopolymer poly(U) with a relatively high binding affinity. Very low binding affinities were obtained for the other ribohomopolymers, poly(G), poly(A) and poly(C). In addition, no specific binding of CS1, either in the 30S complex or as a recombinant purified protein, was obtained to the 5′-untranslated region of the mRNA in comparison to the other parts. RNA-binding experiments, in which the N- and C-termini of the protein were analyzed, revealed that the RNA-binding site is located in the C-terminus half of the protein. These results suggest that CS1 does not direct the 30S complex to the initiation codon of the translation site by specific binding to the 5′-untranslated region. In bacteria, specific binding is derived by base pairing between 16S rRNA and the Shine–Dalagarno sequences. In the chloroplast, nuclear encoded and gene-specific translation factors may be involved in the determination of specific binding of the 30S subunit to the initiator codon.
INTRODUCTION
Chloroplasts are photosynthetic organelles that contain their own DNA and gene expression machinery. The control of chloroplast gene expression is performed at the levels of transcription, post-transcription, translation and protein degradation (1–4). For example, translation of the psbA mRNA, which encodes the D1 protein of the photosystem II reaction center, is activated by light (5). In the green alga Chlamydomonas reinhardti, this translation activation was found to be correlated with the binding of a complex of several proteins to the 5′-untranslated region (UTR) of the mRNA in redox- and ADP-phosphorylation-dependent processes (6,7). The chloroplast translation machinery resembles that of bacteria in many respects. It is composed of 70S ribosomes and many chloroplast genes contain Shine–Dalagarno (SD) sequences in their 5′-UTR (8). However, while in Escherichia coli the SD sequence base pairs with 16S rRNA, the necessity of this sequence for translation initiation in the chloroplast is much less restricted (8). This might be explained by the observation that nuclear encoded proteins activate chloroplast translation of specific genes (1). One can therefore hypothesize that instead of SD sequences, chloroplast 5′-UTRs contain sequence elements which function as binding sites for nuclear encoded proteins, which interact with the ribosomes to initiate translation (9,10). A similar situation was described for yeast mitochondria (11).
The bacterial ribosomal protein S1 was implicated as a good candidate for involvement in the process of mRNA recognition and binding by the 30S ribosomal subunit to the translation initiation site (9,12,13). The protein is an acidic RNA-binding protein, consisting of a C-terminal part with four repeat motifs that bind RNA and an N-terminus double domain that binds the ribosome (13). The S1 protein has been shown to bind poly(U) sequences with high affinity (14). High binding affinity for poly(A) sequences was also reported (15). The chloroplast homolog of bacterial S1 protein is CS1. It is encoded in the nucleus, much shorter than the bacterial protein and composed of three RNA-binding domains (16). It was suggested that the ribosomal-binding repeats located in the C-terminal part of the bacterial protein are superimposed on the RNA-binding domains in the middle of the chloroplast protein (16). The chloroplast CS1 protein was reported to specifically bind RNA sequences resembling the 5′-UTR of chloroplast genes, as well as poly(A) sequence (16,17). A protein that shows a striking similarity to spinach CS1 was isolated from cyanobacteria (18).
Here we have analyzed spinach CS1 protein in terms of its association with the small subunit of the ribosome and binding specificity to RNA molecules. We found that most of the CS1 population is associated with the 30S complex and that the protein, either embedded in the 30S complex or as a purified recombinant protein, does not display a higher binding affinity for specific RNA sequences. Similar to bacterial S1, CS1 binds the ribohomopolimer poly(U) with a higher affinity than other ribohomopolymers. The RNA-binding domain was localized in the C-terminal half of CS1. The results suggest that binding of 30S to the translation initiation site is not due to the high affinity of CS1 for a specific nucleotide sequence in the 5′-UTR.
MATERIALS AND METHODS
Purification of the 30S ribosomal subunit complex
Chloroplast protein-soluble extract was prepared from young leaves of hydroponically grown spinach (Spinacia oleracea cv Viroflay) plants as previously described (19). The 30S fraction was isolated from this extract by applying 70 mg protein to a Superdex 200 (Pharmacia) size exclusion column in a buffer containing 20 mM HEPES pH 7.9, 60 mM KCl, 12.5 mM MgCl2, 0.1 mM EDTA, 2 mM DTT and 5% glycerol (buffer E). The fractions of 600–700 kDa were pooled and applied to a heparin (Hi-trap 1 ml; Pharmacia) column that was developed with a KCl gradient in buffer E. Fractions eluted at 0.3–0.4 M KCl were dialyzed and applied to a resource Q column (Pharmacia) that was developed with a KCl gradient in buffer E. The 30S complex was eluted as a sharp peak at 0.5 M KCl. It was characterized as 30S by the identification of several proteins known to be associated in the 30S complex (Fig. 1) and 16S RNA by RNA blot (not shown).
Figure 1.
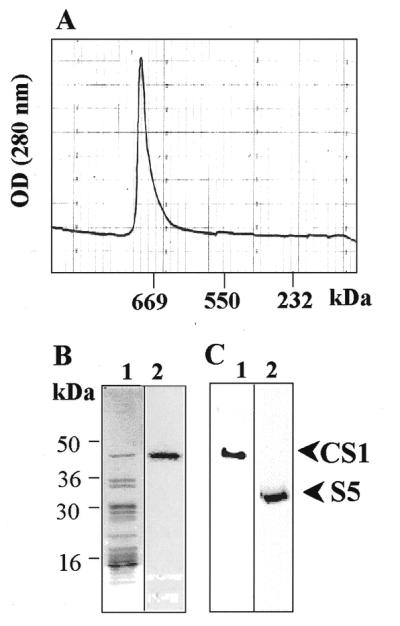
Isolation of CS1 protein with the 30S ribosomal subunit. The 30S ribosomal small subunit was isolated as described in Materials and Methods. The isolated complex was fractionated on a Superdex 200 size exclusion column and the elution profile monitored by absorption at 280 nm (A). The protein profile of the high molecular weight complex was analyzed by SDS–PAGE and CS1 protein by UV-crosslinking to psbA 5′-UTR RNA (B, lanes 1 and 2, respectively). The CS1 and S5 ribosomal proteins were identified by an immunoblot of the fraction with the corresponding specific antibodies (C, lanes 1 and 2, respectively).
Preparation of Escherichia coli-produced CS1
An oligo(dT) primed cDNA was synthesized from total spinach leaf RNA as previously described (20). The CS1 cDNA lacking the transit peptide that mediates transport into the chloroplast was PCR amplified using primers corresponding to the mature protein (without the transit peptide directing it into the chloroplast) and oligo(dT). For expression in E.coli, the CS1 cDNA was cloned into the BamHI and SalI sites of the expression vector pQE30 (Qiagen). Thus, six histidines and several additional residues were added to the N-terminus of the mature protein. The recombinant protein was produced in SG13009[pREP4] cells by growing to an OD600 of 0.9 on LB medium (100 ml) containing ampicilin (100 µg/ml) and kanamycin (25 µg/ml), followed by the addition of IPTG (2 mM) for 3 h. Bacterial cells were collected by centrifugation, resuspended in 5 ml buffer containing 0.1 M sodium phosphate, 0.01 M Tris–Cl pH 8.0 and 8 M urea, and broken using a French press operating at 20 000 p.s.i. The non-soluble fraction was collected by centrifugation for 20 min at 13 000 g and the CS1 or N-CS1 proteins were solubilized as previously described (21). The C-CS1 protein was produced in bacteria as a soluble protein and was therefore directly purified with Ni–NTA–agarose. The solubilized protein was incubated in buffer E containing 8.0 M urea and Ni–NTA–agarose (Bio-Labs) for 1 h at 4°C with gentle mixing. The material was applied to a column, washed with 10 vol buffer containing 8.0 M urea, 0.1 M sodium phosphate and 0.01 M Tris–HCl, pH 8.0, and 10 vol washing buffer (8.0 M urea, 0.1 M sodium phosphate, 0.01 M Tris–HCl, pH 6.3). The protein was eluted using a linear gradient to 0.5 M imidazole in a buffer containing 8.0 M urea, 0.1 M sodium phosphate, 0.01 M Tris–HCl, pH 4.5, followed by dialysis against buffer E. Additional purification included fractionation on a heparin column (Hitrap; Pharmacia). Protein concentration was determined using the Bio-Rad protein assay kit (Bio-Rad). To verify that the RNA-binding properties of the recombinant protein were not altered by the histidine residues, other proteins were expressed using the same vector system and checked for RNA binding. No RNA binding was observed using several proteins that were overexpressed in this way (data not shown). Following Ni2+ affinity and heparin column purification, fractionation of the recombinant protein by SDS–PAGE revealed a single protein species; this preparation lacked detectable RNase activity when incubated with radiolabeled RNAs (data not shown).
Preparation of CS1 deletion mutants
The PstI site located at amino acid number 180 of the CS1 cDNA was used to subclone the DNA fragments for expression of the first 180 (N-CS1) or last 191 amino acids (C-CS1) in the same expression vector. The N-CS1 and C-CS1 proteins were expressed in bacteria and purified as described above for CS1. Since C-CS1 accumulated in the soluble fraction of the bacteria, this fraction was collected and applied to a Ni2+ affinity column.
Preparation of synthetic RNAs
Construction of the plasmid used to transcribe the psbA 5′- and 3′-ends and the amino acid coding region RNAs, as well as the petD 3′-end and RNA derived from the KS plasmid (Stratagene) linearized with BamHI and transcribed with T3 RNA polymerase, was described previously (22). For the experiments described here, the insert was transferred into Bluescript KS+ (Stratagene) so that transcription with T7 or T3 RNA polymerase generated the mRNA-like strand. RNA for UV-crosslinking experiments was prepared with 2.5 µM [α-32P]UTP and 25 µM non-radioactive UTP (22). Competitor RNA for the competition experiments was trace-labeled with 0.0025 µM [α-32P]UTP and 0.5 mM non-radioactive UTP (22).
UV-crosslinking
UV-crosslinking of protein to [α-32P]UTP-labeled RNAs was carried out as previously described (23). Essentially, 3 fmol RNA (240 000 c.p.m.), or the amount indicated in the figure legends, were incubated in 15.5 µl with 50 ng protein in a buffer containing 10 mM HEPES pH 7.9, 30 mM KCl, 6 mM MgCl2, 0.05 mM EDTA, 1 mM DTT and 8% glycerol. Following 1.8 J UV irradiation in a UV-crosslinking apparatus (Hoefer), the RNA was digested with 1 µg RNase A at 37°C for 30 min and the proteins fractionated by SDS–PAGE. The label transferred from the RNA to the proteins was detected by autoradiography.
Gel mobility shift assay
Gel mobility shift assays coupled with RNase T1 protection assays were performed as previously described (24). Proteins were mixed with 32P-labeled RNA. Following 5 min incubation at room temperature, 10 U of RNase T1 was added and the reaction was incubated for an additional 5 min before electrophoresis of the sample on a 1× TBE, 5% native polyacrylamide gel. RNA–protein complexes were detected by autoradiography.
Size exclusion chromatography and immunoblots
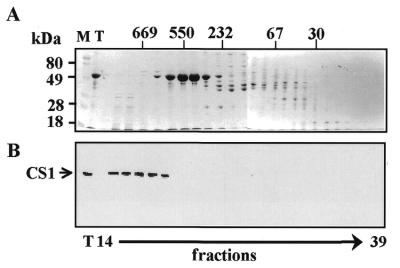
Size exclusion chromatography was performed by applying 4 mg chloroplast protein extract to a Superdex-200 column (Pharmacia). The column was developed in buffer E at a flow rate of 1 ml/min. Fractions of 0.5 ml were collected, precipitated with 80% cold acetone and analyzed by SDS–PAGE and immunoblots were decorated with specific antibodies. The protein standards for size estimation were as follows: thyroglobulin, 669 kDa; RubP-carboxylase, 550 kDa; catalase, 232 kDa; aldolase, 158 kDa; bovine serum albumin, 67 kDa; casein, 30 kDa. Specific antibodies to the CS1 and S5 proteins were kindly provided by Dr Silva Lerbs-Mache.
RESULTS
Association of the CS1 protein with the ribosomal small subunit (30S)
In order to characterize the RNA-binding properties of CS1 protein in the chloroplast, we first asked whether it is associated with the 30S fraction (the small subunit of the ribosome). In such a case, it would be important to study the RNA-binding properties of the protein within this complex. The 30S subunit was isolated through successive fractionation using size exclusion, heparin affinity and anion exchange columns (see Materials and Methods). The isolated protein–RNA complex, when fractionated on a size exclusion column, was eluted as a single sharp peak of ∼650–700 kDa (Fig. 1A). Using an RNA blot, we found a major RNA molecule that hybridized to the 16S small subunit ribosomal RNA probe. No signal was obtained with a probe corresponding to the 23S ribosomal RNA of the large subunit (50S) (data not shown). Analyzing the protein profile of this complex disclosed ∼15–20 polypeptides. Two of these proteins with molecular weights of 47 and 33 kDa were identified using specific antibodies as the ribosomal 30S proteins CS1 and S5, respectively (Fig. 1B and C; 16,25). In addition, the 47 kDa protein bound RNA corresponding to the 5′-UTR of psbA in a UV-crosslinking assay (Fig. 1B). We therefore concluded that the isolated high molecular weight complex is the ribosomal 30S small subunit.
Although CS1 protein was isolated as a subunit of the 30S protein–RNA complex, there is also a possibility that it exists in the chloroplast as a ‘free’ protein that is not associated with the 30S subunit, as was reported before for the S1 bacterial protein (13,15). In order to analyze what proportion of the CS1 protein population was associated with the 30S complex, soluble chloroplast protein extract was fractionated on a size exclusion column and presence of the CS1 protein in each fraction was monitored by immunoblot. The results presented in Figure 2 show that CS1 protein was detected mainly in fractions eluting in the 650–700 kDa region (Fig. 2). Only trace amounts of signal were obtained in the 50 kDa region when the immunoblot was overexposed (not shown). We concluded that under our experimental conditions most of the CS1 population is associated with the 30S ribosomal subunit. Therefore, we next asked what are the binding properties of this protein to different RNA molecules, either embedded in the 30S ribosomal complex or as a purified recombinant protein?
Figure 2.
The CS1 population is associated with the 30S ribosomal subunit. Soluble chloroplast proteins were fractionated on a Superdex 200 size exclusion column. The protein profile of each fraction was analyzed by SDS–PAGE and silver staining (A). M, molecular weight markers; T, total proteins. The following proteins were fractionated on the same column as size markers and their elution peaks are indicated at the top of the figure: thyroglobulin, 669 kDa; RuBP bisphosphate carboxylase, 550 kDa; catalase, 232 kDa; aldolase, 158 kDa; bovine serum albumin, 67 kDa; casein, 30 kDa. The location of CS1 was monitored by an immunoblot using specific antibodies (B).
RNA-binding properties of the CS1 protein
Since the 30S ribosomal small subunit binds the mRNA during the translation initiation process, the binding site for this complex is expected to be located in the 5′-UTR of the mRNA. We therefore used the 5′-UTR of psbA mRNA, which encodes the photosystem II reaction center protein D1, as the target RNA for the binding assays. Due to light-dependent rapid turnover of the D1 protein, psbA mRNA is one of the most abundant mRNAs in the chloroplast and is actively translated in light. A synthetic [32P]RNA corresponding to the 5′-UTR of psbA was UV-crosslinked to the 30S ribosomal subunit complex. The major signal was obtained with the 47 kDa polypeptide, which was identified as CS1 using specific antibodies (Figs 1 and 3–5). In addition, recombinant bacterially expressed CS1 protein binds the same RNA in the UV-crosslinking assay (Fig. 4). We have previously shown that the relative strength of the UV-crosslinking signal cannot directly be accounted for by the affinity of the RNA for the protein (22,23). Therefore, we monitored the relative affinity of CS1 protein for synthetic RNA molecules using the UV-crosslinking competition method (23). In this method, the UV-crosslinking signal is competed for by addition of extra non-labeled RNA. RNA that competes with the radioactive signal at a lower concentration binds the protein with a higher affinity (23).
Figure 3.
CS1 binds poly(U) with a higher affinity than other ribohomopolymers. UV-crosslinking assay of the 30S fraction with synthetic [32P]RNA representing the psbA 5′-UTR. The ribohomopolymers poly(G), poly(A), poly(C) and poly(U) were added 5 min before addition of the radioactive RNA. –, no competitor added. The radioactive signal of the 47 kDa CS1 protein is indicated.
Figure 5.
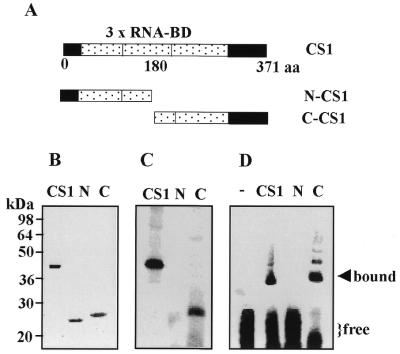
The RNA-binding site is located in the N-terminal half of the CS1 protein. The recombinant CS1 protein, as well as the N- (N-CS1) and the C-termini (C-CS1) parts, were overexpressed and purified from E.coli cells. (A) Schematic representation of the CS1, N-CS1 and C-CS1 proteins. The three RNA-binding domains are indicated by dashed boxes. The number of amino acids, starting from the first ATG, are indicated. (B) Stained SDS–PAGE of the recombinant bacterially expressed CS1, N-CS1 (N) and C-CS1 (C). (C) UV-crosslinking analysis of the recombinant protein to psbA 5′-UTR RNA. Each protein was mixed with [32P]RNA, UV irradiated following digestion of the RNA with RNase and subjected to SDS–PAGE and autoradiography. (D) Gel mobility shift assay of binding of the proteins to psbA 5′-UTR RNA. Proteins were incubated with RNA and digested with RNases. The protein–RNA complexes were analyzed by non-denaturing PAGE and autoradiography.
Figure 4.
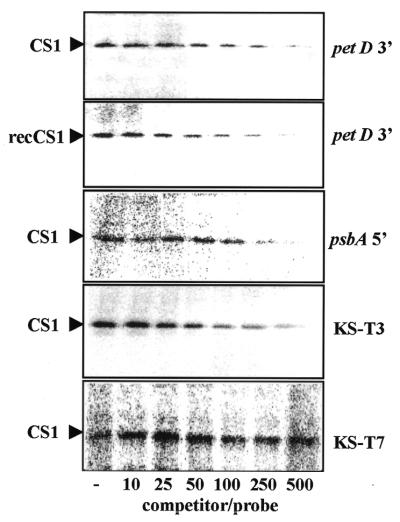
RNA-binding characteristics of the CS1 protein. RNA binding was analyzed using the UV-crosslinking competition assay. The 30S fraction (CS1) or the recombinant CS1 protein (recCS1) were mixed with synthetic [32P]RNA representing the psbA 5′-UTR in the presence of non-radioactive competitor RNA, as indicated in the figure. No competitor was added in the lane marked –. Following incubation, the reaction mixture was UV-crosslinked, digested with RNase and analyzed by SDS–PAGE and autoradiography. KS-T3 indicates RNA containing 14 uridines that was transcribed using T3 RNA polymerase from plasmid Bluescript linearized with BamHI. KS-T7 represents RNA containing eight uridines that was transcribed from the same plasmid using T7 RNA polymerase.
Binding to ribohomopolymers
The 5′-UTR of chloroplast genes are often enriched with adenosines and uridines. For example, if one compares the 70 nt upstream and downstream of the first ATG of the psbA gene of spinach chloroplast, there are 27 uridines, in comparison to 15 in the amino acid coding region. For rbcL and petB, the numbers are 27 and 31, as compared to 20 and 20, respectively. These differences are even more pronounced when counting only stretches of uridines that are more than 3 nt: 4 compared to 1, 3 compared to 1 and 6 compared to 0, for psbA, rbcL and petB, respectively. In order to see whether the CS1 protein displays a high binding affinity for a sequence enriched in adenosines or uridines, we analyzed the affinity of CS1 protein in the 30S complex for ribohomopolymers. As described in the Introduction, spinach CS1 protein was characterized as binding specifically to the 5′-UTR of chloroplast RNAs and as displaying a high binding affinity for poly(A) but not for other ribohomopolymers (16). However, bacterial S1 protein was found to bind poly(U) sequences with high affinity (14). The results presented in Figure 3 show that when CS1 embedded in the 30S ribosomal subunit was tested in the UV-crosslinking competition assay, poly(U) competed for the UV-crosslinking signal very well. The addition of a 50-fold excess of the competitor reduced the UV-crosslinking signal by >50%, which is typical of a high binding affinity (22,23). However, poly(A), poly(G) and poly(C) did not display any (or only very low) affinity for CS1. Only a slight reduction in the UV-crosslinking signal could be obtained at 500-fold excess of ribohomopolymer to radioactive RNA. This binding activity is characteristic of proteins and RNA molecules that do not bind to each other under physiological conditions. Analyzing purified recombinant CS1 produced similar results (not shown). In addition, manipulating the buffer contents and the conditions of the binding assay did not result in binding of another ribohomopolymer. We therefore concluded that CS1 protein, either as a purified recombinant protein or embedded within the 30S ribosomal subunit, displays a high binding affinity exclusively for poly(U).
Binding to different RNA molecules
We then analyzed synthetic RNA molecules corresponding to different parts of several chloroplast mRNAs. We found that all, not only those representing the 5′-UTR, exhibited the UV-crosslinking signal, indicating RNA binding. However, since the UV-crosslinking signal does not represent binding affinity, the possibility still existed that even though CS1 bound every RNA molecule that was tested, it bound the 5′-UTR sequence with a higher affinity. We first asked whether CS1 in the 30S ribosomal complex, in comparison to the isolated recombinant protein, would display a similar or different affinity for RNA. To this end, a competition UV-crosslinking experiment was performed in which non-radioactive RNA corresponding to the 3′-end of petD mRNA competed for binding of CS1, either in the 30S ribosomal complex or as a single recombinant protein. The results demonstrated similar binding affinities for the CS1 protein under both conditions (Fig. 4, upper two panels). We then tested other synthetic RNA molecules that similarly represented the 5′-UTR, the amino acid coding region or the 3′-UTR of several chloroplasts. All of them revealed similar affinities in which an ∼50-fold excess of the competitor was required to reduce the UV-crosslinking signal by 50%. This is a similar affinity to that obtained when poly(U) was the competitor (Fig. 3). Several examples in which the competitors were RNAs derived from transcription of the petD 3′-UTR, psbA 5′-UTR and the Bluescript plasmid are shown in Figure 4. Moreover, higher binding affinities than those representing the 5′-UTRs were not obtained and we reproducibly determined that the binding affinity of CS1 for the 5′-UTR of psbA RNA is lower than the affinity for RNA representing the 3′-UTR of petD (Fig. 4). These results demonstrate that CS1, either as an isolated recombinant protein or embedded in the 30S ribosomal complex, binds different RNA molecules, as well as poly(U), with similar affinities and does not display a higher binding affinity for nucleotide sequences in the 5′-UTR. Together with the observation that no binding was obtained with poly(A), poly(G) and poly(C), the results suggest that the affinity of CS1 for RNA molecules is derived from the number of uridine stretches. A similar suggestion was made for bacterial S1 protein (14). In order to test this hypothesis, we compared the binding affinity of CS1 for RNAs derived from transcription of the vector Bluescript linearized with BamHI and transcribed either by T3 or T7 RNA polymerase. The RNA transcribed by T3 RNA polymerase contained 14 uridines (including two pairs), whereas the RNA transcribed using T7 RNA polymerase contained eight (including one pair). The results revealed that while the T3-transcribed RNA showed a similar affinity to the other RNA molecules, the T7-transcribed RNA displayed a very low binding affinity (Fig. 4, bottom two panels). The fact that while 14 uridine residues in the RNA molecule were sufficient to result in a high affinity for CS1 while eight uridines were not strengthens the hypothesis that the affinity of CS1 for an RNA molecule is derived from the number of uridines.
Mapping the RNA-binding site of CS1 protein
By sequence comparison to bacterial S1 protein, as well as to other RNA-binding proteins, it was suggested that the CS1 amino acid sequence contains three putative RNA-binding domains (Fig. 5A; 16). In order to define the RNA-binding site of CS1, we used the PstI restriction site located approximately in the middle of the cDNA to express the N- or C-terminal halves of the protein in bacteria. The two parts, referred to as N-CS1 and C-CS1 as shown in Figure 5A, were purified to homogeneity and UV-crosslinked to psbA 5′-UTR RNA (Fig. 5B and C). Binding was detected with C-CS1 while no crosslinking signal was obtained with N-CS1. This result suggests that the RNA-binding site is located in the C-terminal half of the protein. Nevertheless, the lack of a UV-crosslinking signal did not necessarily imply that this part of the protein does not bind RNA. If the labeled nucleotide of the RNA is not close enough to a crosslinked amino acid in the protein and is not protected from RNase digestion, a UV-crosslinking signal will not be obtained even though the protein binds the RNA. To verify whether or not the 5′ protein binds RNA, we used the RNase T1 gel mobility shift assay (24,26). In this assay, the RNA and protein were mixed and allowed to form a complex. The mixture was then treated with RNase T1 in order to digest any RNA that is not protected by binding to a protein. Following digestion, the mixture was fractionated on a non-denaturing polyacrylamide gel, where the RNA–protein complex was retarded compared to unbound RNA. As shown in Figure 5D, retarded RNA–protein complexes were obtained with full-length CS1 and C-CS1, but not with N-CS1. Taken together, we conclude that the RNA-binding site of CS1 protein is localized in the C-terminal half of the protein.
DISCUSSION
This paper describes the characterization of CS1 in terms of its association with the ribosomal small subunit and its RNA-binding properties. In E.coli, S1 protein plays a role in mRNA binding to the 30S ribosomal complex (13,14). This protein is required for translation of most, if not all, mRNAs (9). The N-terminus of this protein is associated with the 30S ribosomal subunit by protein–protein interactions, but its long and flexible C-terminal domain has a high affinity for pyrimidine sequences (12–14). Indeed, many E.coli mRNAs have pyrimidine-rich sequences upstream of the SD sequences (13). The suggested model of initiation complex formation implies S1-dependent formation of a 30S–mRNA binary complex with the SD sequences followed by an interaction of S1 with the downstream pyrimidine stretch. In bacteria, specificity is derived by base pairing between 16S rRNA and the SD sequence. In chloroplasts, the involvement of nuclear encoded proteins that bind the 5′-UTR of chloroplast mRNAs and modulate their translation has been postulated (1,7,27). For example, three such proteins were identified in Chlamydomonas chloroplasts as being involved in the light-dependent translation of D1 protein (27). Nuclear encoded proteins binding the 5′-UTR and modulating translation were also identified for the mRNAs of Chlamydomonas and higher plants (4,26,28–35). Therefore, a clear difference between the prokaryotic translation initiation mechanism and that of the chloroplast is that in the chloroplast additional translation regulators encoded in the nucleus are involved.
RNA-binding studies of bacterial S1 protein disclosed a low binding affinity for SD sequences but a high binding affinity for pseudoknot-containing RNA ligands (12). Other studies found, similar to our results, a high binding affinity of S1 for oligo(U) sequences and suggested that its in vivo target is the oligo(U) sequences preceding the SD domain (14). However, specific poly(A) binding by S1 and CS1 were also reported (15,16). The solution structure of the S1 motif from the enzyme polynucleotide phosphorylase (PNPase) using NMR methods displayed a structural motif that is common to several proteins, including RNases, translation initiation factors and cold shock-induced proteins, suggesting that this motif was derived from an ancient nucleic acid-binding protein (36). Interestingly, the bacterial and chloroplast PNPases were recently characterized as poly(U) and poly(A) high affinity binding proteins (37,38). CS1 harbors three copies of this S1 RNA-binding motif. However, the result of the experiment presented in Figure 5, in which the N-terminal half of the protein does not bind RNA, suggests that each one of the S1 motifs displays a different affinity for RNA and that the one near the N-terminus does not bind RNA by itself.
Unlike previous reports that CS1 differs from S1 in its affinity for ribohomopolymers and that it displays a specific nucleotide sequence binding affinity (16,17), we show here that both proteins bind the same ribohomopolymer, poly(U), with high binding affinity and bind every RNA molecule that is not a ribohomopolymer of G, A or C with similar affinity. We suggest that previous studies in which specific binding to the 5′-UTR was observed were the result of a lack of uridine stretches in the molecules that did not bind the protein. Therefore, we suggest that CS1 does not direct specific binding the 30S ribosomal subunit to the 5′-UTR. The nuclear encoded translation modulators, described above for chloroplast translation initiation control, determine the specificity of formation of the initiation translation complex around the first ATG.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Dr Silva Lerbs-Mache for the CS1 and S5 antibodies and helpful advice and Dr Avihai Danon for critical reading of the manuscript. This work was supported by the Israel Science Foundation, administered by the Israel Academy of Science and Humanities, by a grant from the Niedersachsen Foundation for Science and Art and by a grant from the Israel–Japan Scientific Cooperation Research Foundation.
REFERENCES
- 1.Stern D.B., Higgs,D.C. and Yang,J. (1997) Trends Plant Sci., 2, 308–315. [Google Scholar]
- 2.Schuster G., Lisitsky,I. and Klaff,P. (1999) Plant Physiol., 120, 937–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hayes R., Kudla,J. and Gruissem,W. (1999) Trends Biochem. Sci., 24, 199–202. [DOI] [PubMed] [Google Scholar]
- 4.Goldschmidt-Clermont M. (1998) Int. Rev. Cytol., 177, 115–180. [DOI] [PubMed] [Google Scholar]
- 5.Barber J. and Andersson,B. (1992) Trends Biochem. Sci., 17, 61–66. [DOI] [PubMed] [Google Scholar]
- 6.Mayfield S.P., Yohn,C.B., Cohen,A. and Danon,A. (1995) Annu. Rev. Plant Physiol. Plant Mol. Biol., 46, 147–166. [Google Scholar]
- 7.Danon A. (1997) Plant Physiol., 115, 1293–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hauser C.R., Gillham,N.W. and Boynton,J.E. (1998) In Rochaix,J.-D., Goldschmidt-Cleremont,M. and Merchant,S. (eds), The Molecular Biology of Chloroplasts and Mitochondria in Chlamydomonas, Kluwer Academic, Dordrecht, The Netherlands, Vol. 7, pp. 197–217.
- 9.Sorensen M.A., Fricke,J. and Pedersen,S. (1998) J. Mol. Biol., 280, 561–569. [DOI] [PubMed] [Google Scholar]
- 10.Subramanian A.R. (1993) Trends Biochem. Sci., 18, 177–181. [DOI] [PubMed] [Google Scholar]
- 11.Costanzo M.C. and Fox,T.D. (1990) Annu. Rev. Genet., 24, 91–113. [DOI] [PubMed] [Google Scholar]
- 12.Ringquist S., Jones,T., Snyder,E.E., Gibson,T., Boni,I. and Gold,L. (1995) Biochemistry, 34, 3640–3648. [DOI] [PubMed] [Google Scholar]
- 13.Subramanian A.R. (1983) Prog. Nucleic Acid Res., 28, 101–143. [DOI] [PubMed] [Google Scholar]
- 14.Boni I.V., Isaeva,D.M., Musychenko,M.L. and Tzareva,N.V. (1991) Nucleic Acids Res., 19, 155–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalapos M.P., Paulus,H. and Sarkar,N. (1997) Biochimie, 79, 493–502. [DOI] [PubMed] [Google Scholar]
- 16.Franzetti B., Carol,P. and Mache,R. (1992) J. Biol. Chem., 267, 19075–19081. [PubMed] [Google Scholar]
- 17.Alexander C., Faber,N. and Klaff,P. (1998) Nucleic Acids Res., 26, 2265–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sugita M., Sugita,C. and Sugiura,M. (1995) Mol. Gen. Genet., 246, 142–147. [DOI] [PubMed] [Google Scholar]
- 19.Schuster G. and Gruissem,W. (1991) EMBO J., 10, 1493–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lisitsky I., Klaff,P. and Schuster,G. (1996) Proc. Natl Acad. Sci. USA, 93, 13398–13403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Citovsky V., Knorr,D., Schuster,G. and Zambryski,P. (1990) Cell, 60, 637–647. [DOI] [PubMed] [Google Scholar]
- 22.Lisitsky I., Liveanu,V. and Schuster,G. (1994) Nucleic Acids Res., 22, 4719–4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lisitsky I., Liveanu,V. and Schuster,G. (1995) Plant Physiol., 107, 933–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Q., Adams,C.C., Usack,L., Yang,J., Monde,R. and Stern,D.B. (1995) Mol. Cell. Biol., 15, 2010–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou D.-X. and Mache,R. (1989) Mol. Gen. Genet., 219, 204–208. [DOI] [PubMed] [Google Scholar]
- 26.Danon A. and Mayfield,S.P. (1994) Science, 266, 1717–1719. [DOI] [PubMed] [Google Scholar]
- 27.Somanchi A. and Mayfield,S.P. (1999) Curr. Opin. Plant Biol., 2, 404–409. [DOI] [PubMed] [Google Scholar]
- 28.Rochaix J.-D., Kuchka,M., Mayfield,S., Schirmer-Rahire,M., Girard-Bascou,J. and Bennoun,P. (1989) EMBO J., 8, 1013–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stampacchia O., Girard-Bascou,J., Zanasco,J.-L., Zerges,W., Bennoun,P. and Rochaix,J.-D. (1997) Plant Cell, 9, 773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zerges W., Girard-Bascou,J. and Rochaix,J.D. (1997) Mol. Cell. Biol., 17, 3440–3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barkan A., Voelker,R., Mendel-Hartvig,J., Johnson,D. and Walker,M. (1995) Physiol. Plant., 93, 163–170. [Google Scholar]
- 32.McCormac D.J. and Barkan,A. (1999) Plant Cell, 11, 1709–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Drager R.G. and Stern,D.B. (1998) In Rochaix,J.-D., Goldschmidt-Clermont,M. and Merchant,S. (eds), Molecular Biology of Chlamydomonas: Chloroplasts and Mitochondria. Kluwer Academic, Dordrecht, The Netherlands, Vol. 7, pp. 165–140.
- 34.Higgs D.C., Shapiro,R.S., Kindle,K.L. and Stern,D.B. (1999) Mol. Cell. Biol., 19, 8479–8491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trebitsh T., Levitan,A., Sofer,A. and Danon,A. (2000) Mol. Cell. Biol., 20, 1116–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bycroft M., Hubbard,T.J., Proctor,M., Freund,S.M. and Murzin,A.G. (1997) Cell, 88, 235–242. [DOI] [PubMed] [Google Scholar]
- 37.Lisitsky I., Kotler,A. and Schuster,G. (1997) J. Biol. Chem., 272, 17648–17653. [DOI] [PubMed] [Google Scholar]
- 38.Lisitsky I. and Schuster,G. (1999) Eur. J. Biochem., 261, 468–474. [DOI] [PubMed] [Google Scholar]