Abstract
The phytohormone auxin plays crucial roles in nearly every aspect of plant growth and development. Auxin signaling is activated through the phytohormone-induced proteasomal degradation of the Auxin/INDOLE-3-ACETIC ACID (Aux/IAA) family of transcriptional repressors. Notably, many auxin-modulated physiological processes are also regulated by nitric oxide (NO) that executes its biological effects predominantly through protein S-nitrosylation at specific cysteine residues. However, little is known about the molecular mechanisms in regulating the interactive NO and auxin networks. Here, we show that NO represses auxin signaling by inhibiting IAA17 protein degradation. NO induces the S-nitrosylation of Cys-70 located in the intrinsically disordered region of IAA17, which inhibits the TIR1–IAA17 interaction and consequently the proteasomal degradation of IAA17. The accumulation of a higher level of IAA17 attenuates auxin response. Moreover, an IAA17C70W nitrosomimetic mutation renders the accumulation of a higher level of the mutated protein, thereby causing partial resistance to auxin and defective lateral root development. Taken together, these results suggest that S-nitrosylation of IAA17 at Cys-70 inhibits its interaction with TIR1, thereby negatively regulating auxin signaling. This study provides unique molecular insights into the redox-based auxin signaling in regulating plant growth and development.
Keywords: Arabidopsis thaliana, Auxin, Aux/IAA, Nitric oxide, S-nitrosylation, Intrinsically disordered region
Introduction
The phytohormone auxin plays a pivotal role in numerous aspects of plant growth and development. The predominant nuclear auxin signaling pathways are comprised by three classes of major components, TRANSPORT INHIBITOR RESPONSE1/AUXIN SIGNALING F-BOX PROTEIN (TIR1/AFB) receptors, AUXIN RESPONSE FACTOR (ARF) transcription factors, and Auxin/INDOLE-3-ACETIC ACID (Aux/IAA) transcriptional repressor proteins (Morffy and Strader, 2022). Aux/IAA proteins directly repress ARF transcriptional activity under low auxin conditions. With an increased level, auxin promotes interaction between the SCFTIR1/AFB and Aux/IAAs, leading to polyubiquitylation of the repressor proteins and their subsequent degradation by the 26S proteasome. The degradation of Aux/IAA proteins relieves ARF transcriptional repression, allowing ARF-mediated regulation of auxin-responsive genes (Jing and Strader, 2018). Aux/IAA repressor proteins thus tightly control the transcription of auxin-responsive genes. In Arabidopsis, 29 Aux/IAA proteins are found to interact with other auxin signaling components to regulate downstream auxin responses (Figueiredo and Strader, 2022). Most Aux/IAA proteins contain three conserved domains, namely an N-terminal domain I (DI) for recruiting co-repressors, a degron domain (DII) that determines protein stability, and a C-terminal type I/II Phox and Bem1p (PB1) protein-protein interaction domain (Powers and Strader, 2020).
In addition to its dominant roles in regulating growth and development, auxin signaling actively interacts with other signaling pathways, including those mediated by other phytohormones and signaling molecules, to modulate a wide range of physiological processes. Among those, nitric oxide (NO) has been implied to play a role in regulating auxin signaling. A major physiological role of NO is executed by protein post-translational modifications (PTMs). PTMs are critical for maintaining the fundamental cell functions through regulating protein stability, conformation, subcellular localization, and interactions (Gupta et al., 2020). Aux/IAA proteins are modified by multiple PTMs including ubiquitylation, cis-trans isomerization, and phosphorylation (Figueiredo and Strader, 2022). As a small gaseous signaling molecule, NO plays myriad roles in plant growth and development, including hormone regulation, signaling transduction, biotic and abiotic stress responses (Feng et al., 2019; Kolbert et al., 2019; Gupta et al., 2022). PTMs by NO take place primarily through three types of modifications: S-nitrosylation (the formation of a nitrosothiol group on cysteine residues), metal nitrosylation (interaction of NO with metalloproteins), and tyrosine nitration (covalently addition of NO to the tyrosine residues) (Pande et al., 2022). Current evidence indicates that NO predominantly acts via protein S-nitrosylation, in which an NO moiety is covalently added to a reactive cysteine (Cys) thiol (SH) forming an S-nitrosothiol (SNO) (Gupta et al., 2022). S-nitrosylation is a highly conserved redox-based PTM that plays crucial roles in various physiological processes via regulating protein activities (Fernando et al., 2019). In plants, S-nitrosylation of target proteins is involved in regulating various aspects of phytohormonal networks, including synthesis, transport, degradation, and signal transduction (Zhang et al., 2019; Pande et al., 2022). Recent studies suggest that NO plays an important role in regulating auxin responses, polar transport, and signal transduction (Fernandez-Marcos et al., 2011; Terrile et al., 2012; Shi et al., 2015; Iglesiasa et al., 2018). NO reduces root meristem size and inhibits root growth by reducing PIN-dependent auxin transport in Arabidopsis (Fernandez-Marcos et al., 2011; Ni et al., 2017). S-nitrosylation of auxin receptor TIR1 at Cys-140 is important for TIR1 function (Terrile et al., 2012). ARABIDOPSIS SKP1-LIKE1 (ASK1), an adaptor in the subunits of the SKP1–Cullin–F-box (SCF) complex, is S-nitrosylated and involved in SCFTIR1/AFBs assembly (Iglesiasa et al., 2018). These findings suggest that NO might be fine-tuning SCFTIR1/AFBs activity and auxin signaling transduction. Yet, the molecular mechanisms connecting NO and auxin networks remain largely unknown.
Here, we show that NO represses auxin signaling by inhibiting the interaction between the auxin co-receptor TIR1 and IAA17 repressor protein through S-nitrosylation of IAA17 at Cys-70 in the intrinsically disordered region (IDR), thus revealing a mechanism modulating the complex redox-auxin interaction in regulating plant growth and development.
Results
Nitric oxide inhibits IAA17 protein degradation
The intracellular level of S-nitrosoglutathione (GSNO), a major bioactive NO species, is tightly controlled by GSNO reductase (GSNOR) that catalyzes the turnover of GSNO (Feechan et al., 2005; Lee et al., 2008; Chen et al., 2009). Mutations in the single-copied Arabidopsis GSNOR1 gene cause a severe growth phenotype, mimicking defective signaling of cytokinin and auxin (Lee et al., 2008; Chen et al., 2009; Kwon et al., 2012). To explore possible regulatory roles of NO in auxin signaling, we investigate its effects on the stability of Aux/IAA proteins, which is key to auxin-mediated growth and development. To this end, we used the Arabidopsis HS::IAA17NT-GUS marker line, in which the N-terminal domains I and II of IAA17/AXR3 (IAA17NT) were fused to the β-glucuronidase encoding gene (GUS) reporter protein under the control of a heat-shock inducible promoter (HS) (Gray et al., 2001). After heat shock treatment at 37°C for 2 h, seedlings were then treated with IAA, the proteasome inhibitor MG132, or one of two NO donors, GSNO or sodium nitroprusside (SNP). As shown previously (Gray et al., 2001), auxin rapidly induced the degradation of the IAA17 fusion protein (Figs. 1A, S1, S2). However, the NO donors GSNO and SNP strongly blocked this IAA-enhanced degradation over the equivalent time course, stabilizing the fusion protein to a similar extent as that observed with MG132 (Figs. 1A, S1, S2). These results demonstrate that NO inhibits IAA17 protein degradation, which is consistent with the observations previously reported (Shi et al., 2015). To further validate the results, we quantified the total GUS protein abundance from HS::IAA17NT-GUS plants grown under different treatments by immunoblot analysis. Consistent with the staining results, NO significantly inhibited IAA17 protein degradation (Figs. 1B, 1C, S2). Moreover, the GUS enzymatic activity of the HS::IAA17NT-GUS reporter line was significantly increased after GSNO treatment in a concentration-dependent manner when compared with IAA treatment (Fig. 1D). Taken together, these results suggest that NO regulates IAA17 protein degradation.
Fig. 1.
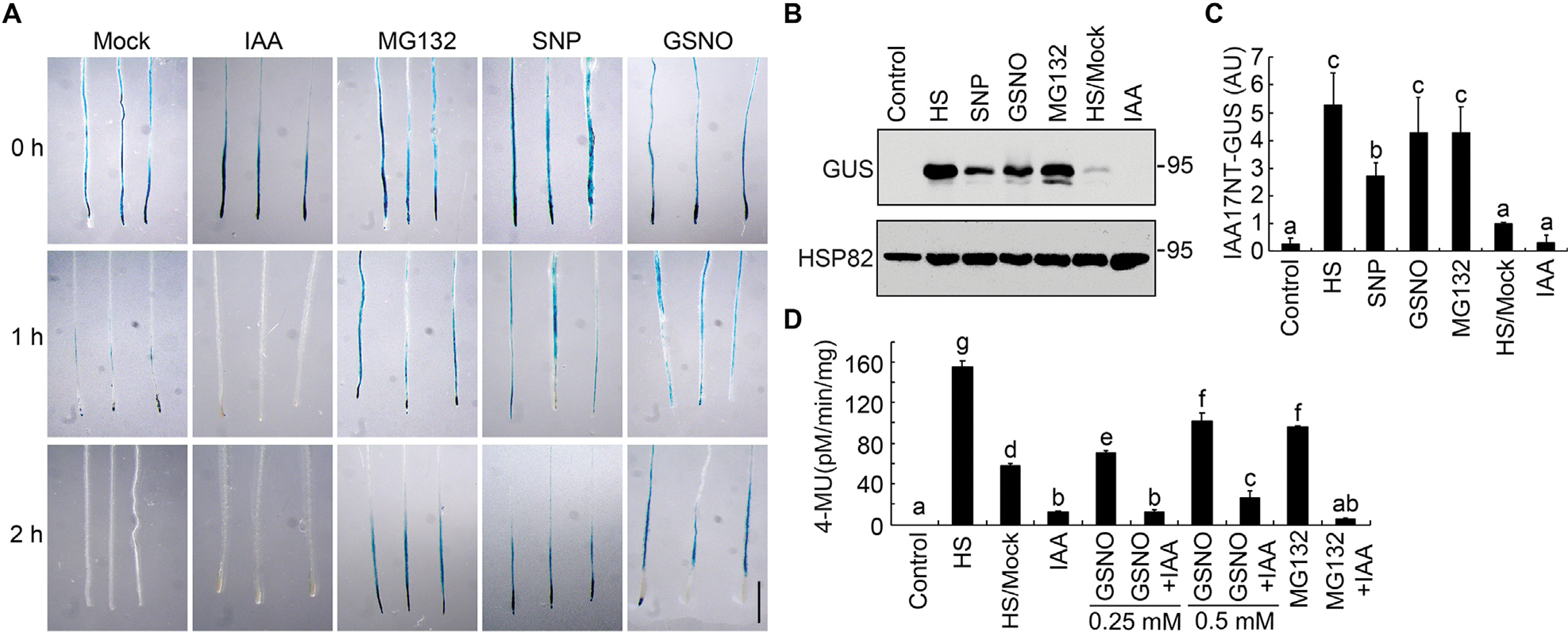
Nitric oxide stabilizes IAA17-GUS protein. A: Histochemical staining of the GUS activity in 7-day-old seedlings carrying an HS::IAA17NT-GUS reporter. After heat shock at 37°C for 2 h, the seedlings were transferred to media in the absence (mock) or the presence of 10 μM IAA, 50 μM MG132, 200 μM SNP, and 200 μM GSNO and incubated at 22°C for the indicated time prior to GUS staining. B and C: Immunoblotting analysis and quantification of IAA17NT-GUS levels in seedlings shown in (A). Anti-HSP82 is used as a loading control. D: Analysis of the GUS activity in 7-day-old HS::IAA17NT-GUS seedlings treated with the indicated chemicals and concentrations. HS/Mock and other columns, HS samples treated with DMSO (HS/Mock) and various chemicals at 22°C for 2 h. Data are mean ± SD of three independent experiments. Different letters indicate individual groups for multiple comparisons with significant differences (one-way ANOVA, Duncan, P < 0.05). HS, heat shock. Scale bar, 1 cm (A).
We next examined IAA17 protein accumulation in Arabidopsis NO-overproducing mutants NO overproducer1 (nox1) GSNO reductase1 (gsnor1–3) and repressor of gsnor1 (gsnor1 rog1–1) After crossing these NO-overproducing mutants to the HS::IAA17NT-GUS background and applying a heat shock treatment at 37°C for 2 h, all of the mutants displayed higher accumulation of IAA17NT-GUS fusion protein when compared with mock treatment throughout the time course (Fig. 2A), further supporting that NO plays a role in reducing IAA17 degradation (Figs. 1A, S1, S2). To further investigate IAA17 protein accumulation in the NO-overproducing mutants, IAA17 antibodies were generated in mice using His-IAA17 recombinant protein as an antigen (Fig. S3). We found that the accumulation of IAA17 was increased in nox1, gsnor1–3, and gsnor1 rog1–1, similarly in the IAA17 gain-of-function mutant (axr3–1 and axr3–3) and overexpression lines (35S::IAA17-FLAG) (Fig. 2B). A similar phenotype was also observed in the overexpression lines treated with the NO donors GSNO or SNP (Fig. 2C and 2D). These results suggest that NO positively regulates the accumulation of IAA17 protein.
Fig. 2.
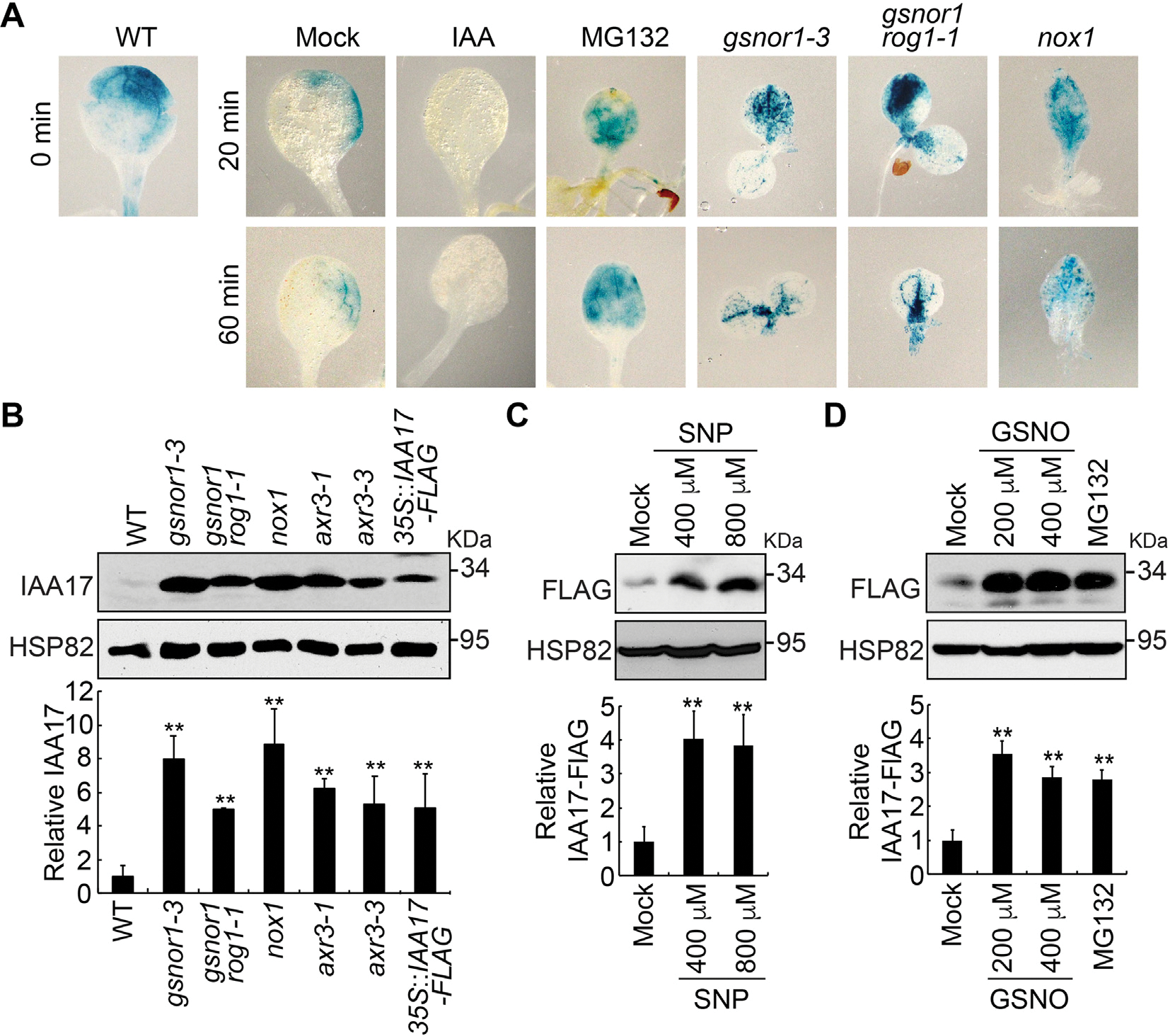
Nitric oxide promotes IAA17 protein accumulation. A: GUS staining of 7-day-old seedlings carrying an HS::IAA17NT-GUS reporter in Col-0 (WT) or NO-overproducing mutants (gsnor1–3, gsnor1 rog1–1, and nox1) treated with heat shock at 37°C for 2 h. B: Immunoblotting analysis (top) and quantification (bottom) of IAA17 protein levels in NO-overproducing mutants. IAA17 gain-of-function mutants of axr3–1 and axr3–3, and 35S::IAA17-FLAG transgenics are used as controls. C and D: Immunoblotting analysis (top) and quantification (bottom) of IAA17 protein levels treated with the nitric oxide donors SNP and GSNO. Treatment with MG132 is used as a control. Data are mean ± SD of three independent experiments. The statistical significance is determined by a two-sided Student’s t-test (Paired two sample for means). **, P < 0.01 when compared to the mock.
IAA17 is S-nitrosylated at Cys-15 and Cys-70
Since the major physiological activity of NO is executed by S-nitrosylation, we speculated that the NO-mediated inhibition of IAA17 protein degradation is likely via S-nitrosylation of the transcriptional repressor. Therefore, we employed a biotin-switch method to determine if IAA17 recombinant protein is S-nitrosylated. In this method, the nitrosothiol groups are replaced with a more stable biotin moiety via chemical reduction by ascorbate, and then identified using immunoblot analysis. We found that His- or GST4CS-tagged IAA17 recombinant proteins were S-nitrosylated by GSNO, but not by the reduced glutathione (GSH; a negative control) (Fig. 3A and 3B). Moreover, S-nitrosylation of the transgenic IAA17-FLAG protein was specifically detected in planta under normal growth conditions by an in vivo biotin-switch method (Fig. 3C).
Fig. 3.
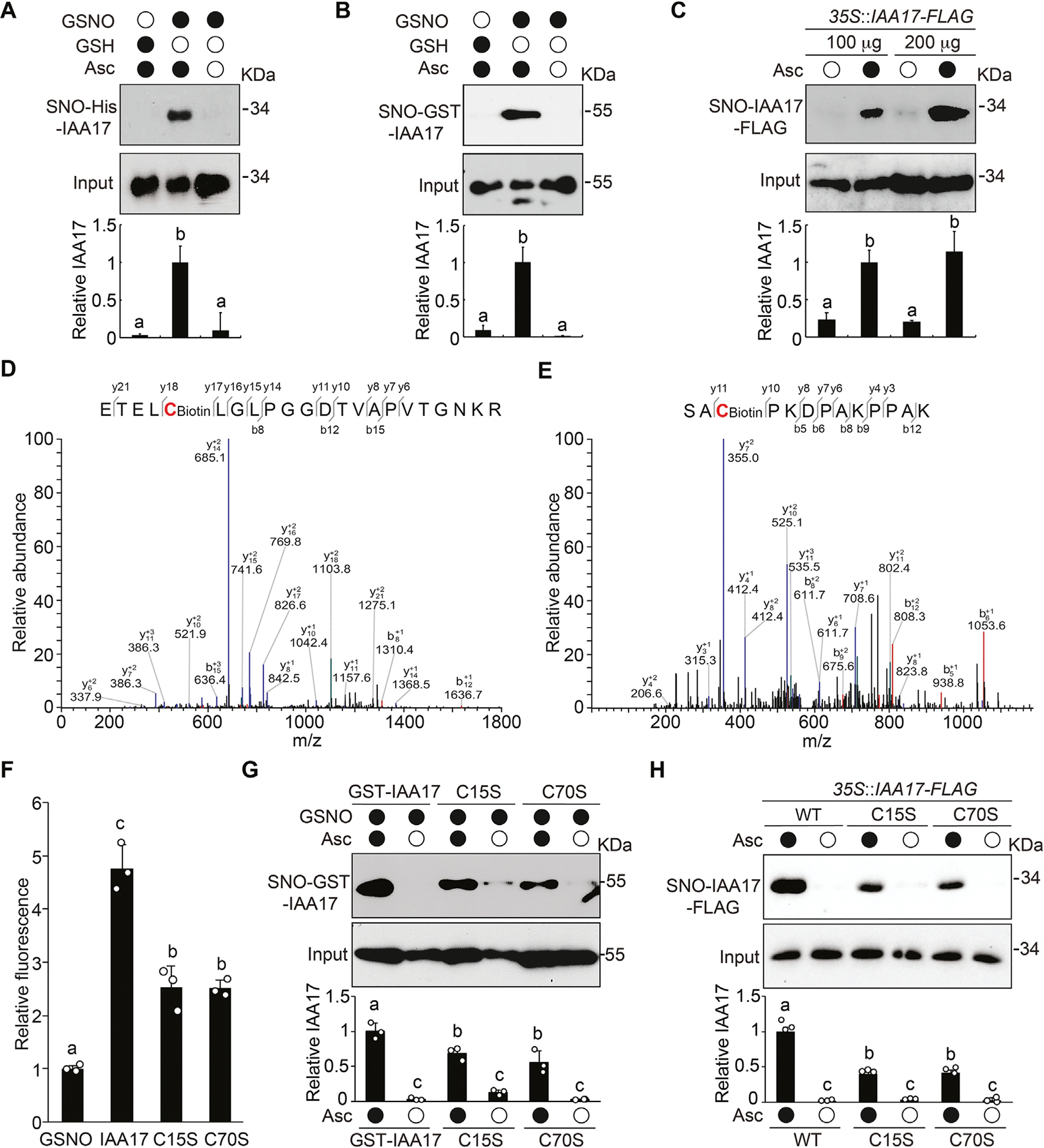
IAA17 is S-nitrosylated at Cys-15 and Cys-70. A and B: Immunoblotting analysis (top) and quantification (bottom) of S-nitrosylated recombinant His-IAA17 (A) and GST4CS-IAA17 (B) proteins (SNO) in vitro in the absence or presence of the reductant ascorbate sodium (Asc). Treatment with GSH is used as a negative control. C: In vivo analysis of S-nitrosylation of IAA17-FLAG protein (SNO) in extracts prepared from 35S::IAA17-FLAG seedlings. Quantitative analysis of the data is shown below the blot. D and E: Analysis of the S-nitrosylation sites of IAA17 using mass spectrometry from a biotin-charged IAA17 peptide. The b- and y-type product ions are indicated. Cys-15 and Cys-70 are identified as two S-nitrosylated residues. F: DAN analysis of S-nitrosylated residues in GSNO tripeptide and His-IAA17 recombinant protein. G: Analysis of S-nitrosylated GST4CS-IAA17, GST4CS-IAA17C15S, and GST4CS-IAA17C70S recombinant proteins. Quantitative analysis is shown below the blot. H: In vivo analysis of S-nitrosylated IAA17-FLAG protein (SNO) in extracts prepared from 35S::IAA17-FLAG, 35S::IAA17C15S-FLAG, and 35S::IAA17C70S-FLAG seedlings. Quantitative analysis is shown below the blot. Data are mean ± SD of three independent experiments. Different letters indicate individual groups for multiple comparisons with significant differences (one-way ANOVA, Duncan, P < 0.05). DAN, 2,3-diaminonaphthalene.
To identify the S-nitrosylated cysteine residues in IAA17, we utilized liquid chromatography-tandem mass spectrometry to analyze the tryptic fragments derived from GSNO-treated IAA17 recombinant protein. We found that Cys-15 and Cys-70 were S-nitrosylated in IAA17 (Fig. 3D and 3E). In addition, an experiment that measures the conversion ratio of 2,3-diaminonaphthalene (DAN) into fluorescent 2, 3-naphthyltriazole (NAT) catalyzed by NO released from the thiol group, was performed to determine the number of S-nitrosylated cysteine residues in IAA17. We compared the fluorescence from a peptide derived from GSNO that contains a single Cys residue with IAA17, IAA17C15S, and IAA17C70S proteins. IAA17 recombinant protein displayed approximately a 4-fold higher DAN-NAT conversion ratio than GSNO, whereas both IAA17C15S and IAA17C70S mutant proteins showed significantly reduced conversion ratio, suggesting that IAA17 contains at least two S-nitrosylated Cys residues (Fig. 3F). Consistently, IAA17C15S and IAA17C70S mutants showed a significant reduction in the S-nitrosylation levels compared with the wild-type IAA17 both in vitro (Fig. 3G) and in vivo (Fig. 3H). Taken together, these results indicate that Cys-15 and Cys-70 in IAA17 are S-nitrosylated sites.
Cys-70 in IDR of IAA17 modulates the TIR1–IAA17 interaction
Data presented above suggest that NO regulates the accumulation of IAA17 protein. Since Aux/IAA proteins are degraded by SCFTIR1-Aux/IAA receptor complex through the 26S proteasome, we explored the possible regulatory effects of NO on this pathway. First, we evaluated whether NO affects the proteasome. We examined the stability of several proteasome-dependent degradation proteins after treatment by NO donors GSNO or proteasome inhibitor MG132. We found that NO did not affect the stability of ARF1, ARF7, ARF19, or TIR1, whereas their degradation was inhibited by MG132 (Fig. S4). In addition, NO did not have detective effects on the protein level of protein arginine methyltransferase 5 (PRMT5) (Hu et al., 2017), but induced the ABA INSENSITIVE 5 (ABI5) protein degradation (Albertos et al., 2015), both of which are degraded through the proteasome (Stone et al., 2006; Zhang et al., 2016). These results suggest that NO may specifically regulate the stability of IAA17.
Next, we sought to determine whether Cys-15 or Cys-70 of IAA17 directly interacts with TIR1/ASK1. Sequence analysis predicts that the N-terminus of IAA17 is an intrinsically disordered region (IDR) encompassing residues 1 to 105, which contains both Cys-15 and Cys-70 of IAA17 (Fig. S5A). To investigate the roles of Cys-15and Cys70 in TIR1–IAA17 complex formation, we performed one bead-per-residue coarse-grained simulations of IAA17 with TIR1/ASK1 using the Mpipi model (Joseph et al., 2022) (Fig. S5B and S5C). In line with previous experiments, our simulations identified the KR and GWPPV motif in the IAA17 N-terminal IDR as the primary source of IAA17–TIR1/ASK1 interaction (Tan et al., 2007; Niemeyer et al., 2020) (Figs. 4A, S6A, S6C, S8). As expected, upon replacing the IAA17 N-terminal IDR with a length-matched poly-glycine-serine (poly-GS) dipeptide repeat, the IAA17–TIR1/ASK1 interaction was lost (Figs. S6B, S6D, S7B, S7D). These results suggest that the Mpipi model qualitatively recapitulates key features of this interaction. Our simulations of wild-type IAA17 suggest that Cys-70 interacts more frequently with TIR1/ASK1 than Cys-15, likely due to the proximity of Cys-70 to the GWPPV motif (Figs. 4A, S6A, S6C, S8). As a corollary, the TIR1/ASK1 residues that interact most frequently with Cys-70 are in the surface pocket of TIR1-LRR domain, adjacent to the TIR1 residues that interact with the GWPPV motif (Figs. 4B, S5D–S5F, S7A, S7C, S9). Taken together, these data suggest that Cys-70 in the IDR of IAA17 may directly interact with TIR1 upon IAA17–TIR1/ASK1 interaction.
Fig. 4.
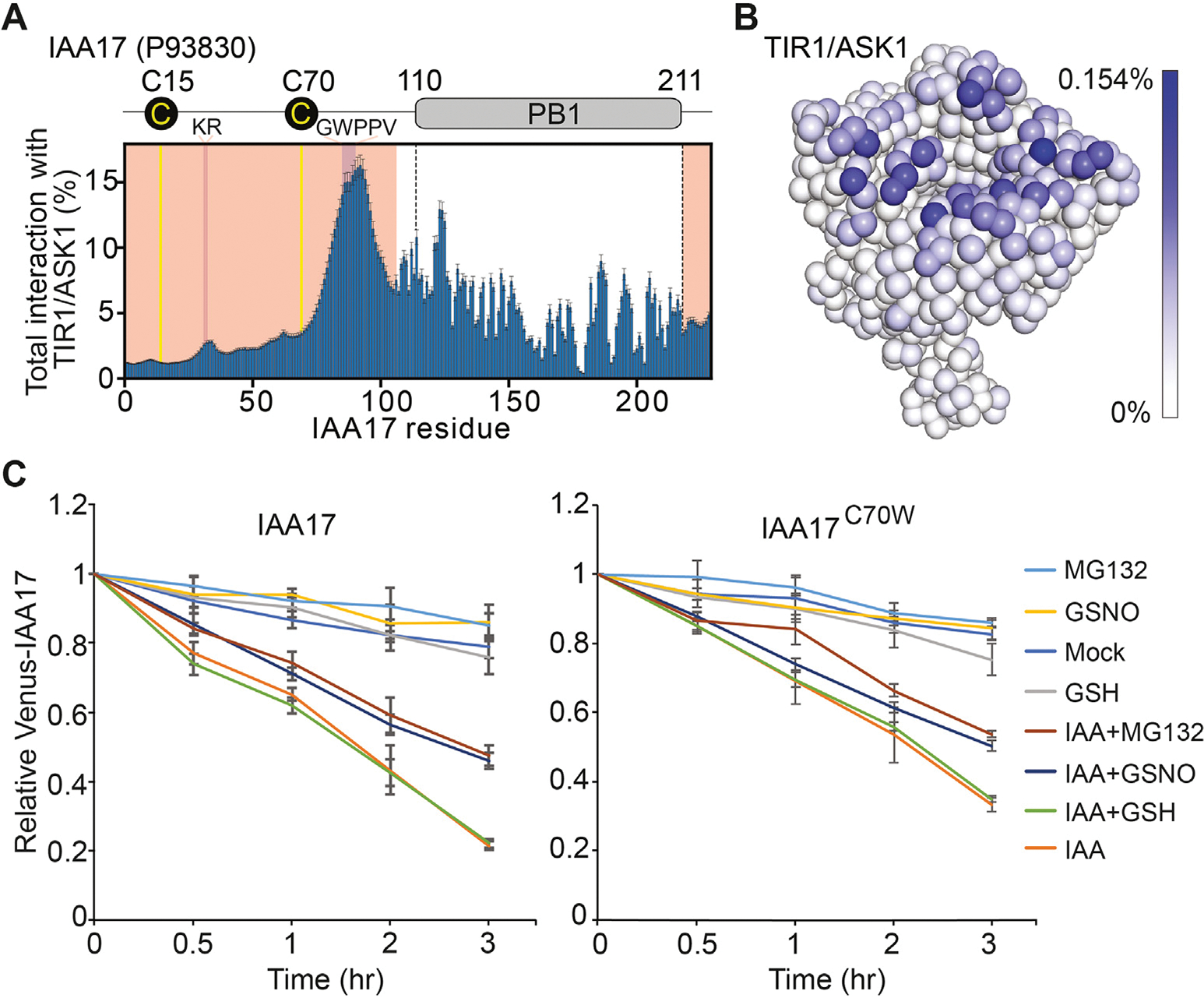
Cys-70 in the IDR of IAA17 contributes the interaction between TIR1 and IAA17. A: Plot showing the total percent interaction for each amino acid in IAA17 with every amino acid in the TIR1/ASK1 complex. Yellow lines denote the locations of Cys-15 and Cys70. Majenta indicates the location of the KR and GWPPV motif. Light red shows the location of the IAA17 IDR. B: The coarse-grained simulations display the frequency interactions between Cys-70 of IAA17 and the TIR1/ASK1 complex. The TIR1/ASK1 complex is colored with a gradient from white to blue, where the intensity of blue reflects the frequency of contacts between Cys-70 of IAA17 and individual residues of the TIR1/ASK1 complex. C: The IAA17 and IAA17C70W degradation dynamics were analyzed in the synthetic ratiometric yeast system by flow cytometry. Mean ratiometric value, relative to initial measurements at the time of treatments, is shown for each strain and treatment, with error bars representing ± SD of three biological replicates. IDR, intrinsically disordered region.
To test possibilities of the Cys-70 in regulating IAA17 degradation, a synthetic ratiometric Aux/IAA degradation yeast (Saccharomyces cerevisiae) system was used (Havens et al., 2012; Chaisupa et al., 2023). In this system, the Venus-tagged IAA17, and bicistronic mScarlet-I internal control, as well as the TIR1 were integrated into the yeast genome. Various mutations within IAA17 were also integrated, specifically of IAA17 at Cys-70 to serine (S), tryptophan (W), phenylalanine (F), or tyrosine (Y). Serine (S) is structurally similar to cysteine but cannot be nitrosylated, whereas tryptophan (W) has been shown to mimic an S-nitrosylated cysteine (Feng et al., 2013; Zhan et al., 2018). We found that the GSNO substantially inhibited the IAA17 degradation in the presence of IAA, which was similar to that of the MG132 treatment (Fig. 4C). However, the degradation of IAA17C70W mutation was reduced when treated with the GSNO and IAA at the same time, comparing to IAA17 wild-type protein (Fig. 4C). Other mutations of IAA17 (C70S, C70F, and C70Y) did not show detectable effects compared to IAA17 wild-type protein (Fig. S10). Taken together, these results suggest that S-nitrosylation of Cys-70 in the IDR of IAA17 plays a role in regulating IAA17 degradation.
S-nitrosylation of IAA17 inhibits its proteasomal degradation
Data presented above suggest that NO may regulate the TIR1-Aux/IAA interaction. To further test this possibility, a coimmunoprecipitation (Co-IP) assay was performed. To exclude the effects of S-nitrosylation of TIR1 and ASK1 (Terrile et al., 2012; Iglesiasa et al., 2018) in the assay and only evaluate the effects of NO on IAA17 stability, His-IAA17 recombinant protein was incubated with GSH (a negative control) or GSNO for 2 h, precipitated and washed with cold acetone to remove GSNO, and then the GSNO-free samples were used for the following assays. TIR1-Myc protein purified by immunoprecipitation from transgenic plants was incubated with or without GSNO-treated His-IAA17 recombinant proteins and the TIR1–IAA17 interaction was examined by a Co-IP experiment. The interaction between TIR1-Myc and His-IAA17 was significantly enhanced by auxin (IAA), similar to previous observations (Dharmasiri et al., 2005) (Fig. 5A). However, when incubated with IAA and the GSNO-treated His-IAA17, the interaction between TIR1-Myc and His-IAA17 was significantly reduced compared to when IAA was incubated with GSH-treated His-IAA17, similar to that of the control in the absence of auxin (Fig. 5A). These results suggest that NO negatively regulates the interaction of TIR1–IAA17 co-receptor in auxin signaling.
Fig. 5.
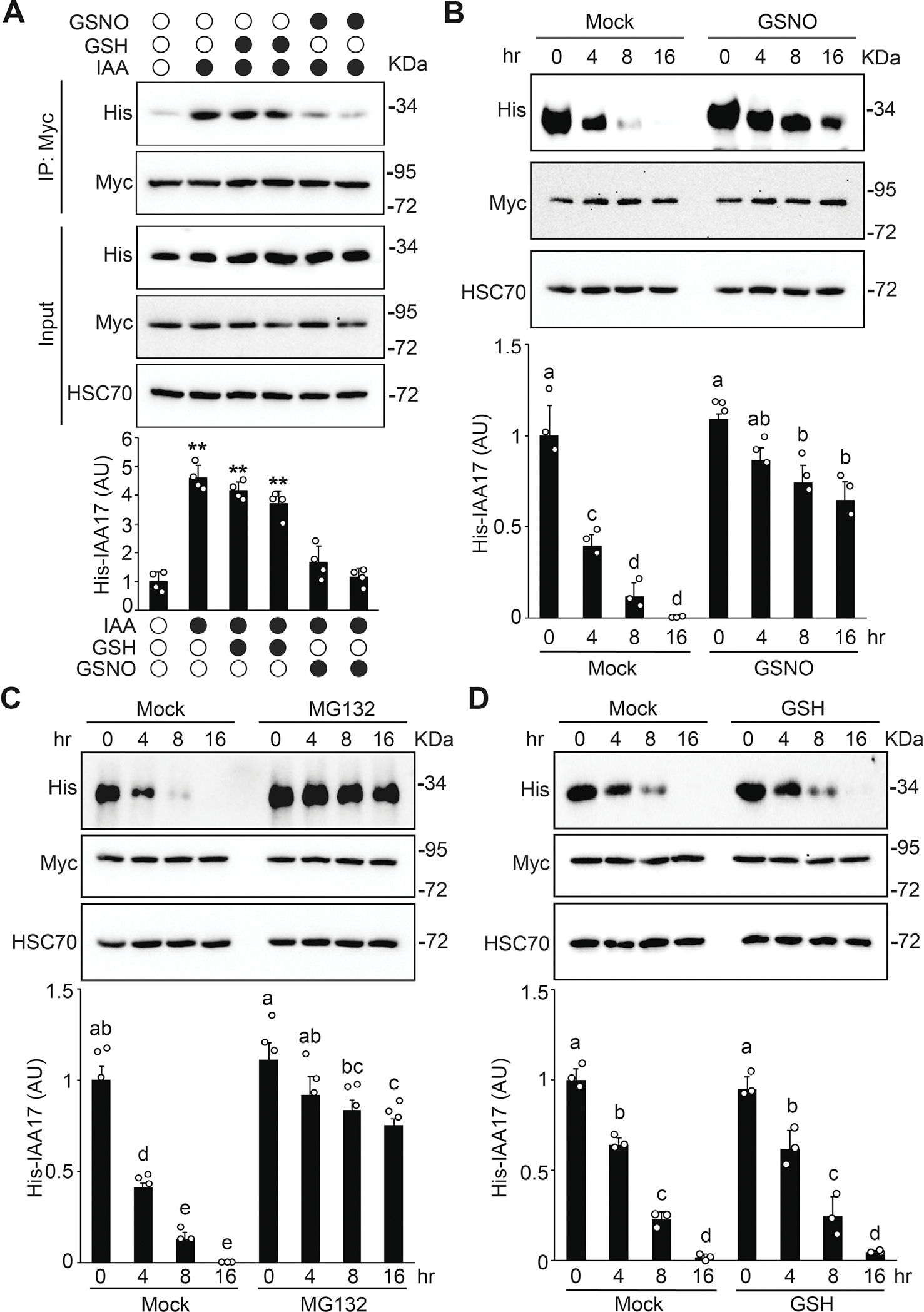
S-nitrosylation negatively regulates IAA17 protein degradation. A: GSNO negatively regulates the auxin-induced TIR1–IAA17 interaction. His-IAA17 recombinant protein was incubated with GSH (negative control) or GSNO for 2 h and then free GSNO was removed by precipitation. TIR1-Myc protein purified from TIR1-Myc transgenic plants by immunoprecipitation was incubated with His-IAA17 recombinant proteins treated with the indicated chemicals. The immunoprecipitated samples were analyzed by immunoblotting using antibodies as indicated. Quantitative analysis of the relative level of His-IAA17 is presented below the blots. Data are mean ± SD of three independent experiments. The statistical significance is determined by a two-sided Student’s t-test (Paired two sample for means). **, P < 0.01 when compared to the mock. B and C: In vitro degradation assay of His-IAA17 recombinant protein. His-IAA17 recombinant protein was incubated with GSNO for 2 h and free GSNO was removed by precipitation. Total protein extracts prepared from TIR1-Myc transgenic plants were incubated with GSNO-treated His-IAA17 recombinant protein for the indicated times in the absence (B) or the presence (C) of 50 μM MG132 for the indicated times. Immunoblotting analysis (top) and quantification (bottom) of His-IAA17 are shown. Anti-HSC70 is used as a loading control. D: In vitro His-IAA17 recombinant protein degradation treated with the negative control GSH as described in (B). Data are mean ± SD of three independent experiments. Different letters indicate individual groups for multiple comparisons with significant differences (one-way ANOVA, Duncan, P < 0.05).
To further explore the functional effects of NO on the SCFTIR1–Aux/IAA co-receptor formation, we performed an in vitro degradation assay to examine the stability of IAA17. When the transgenic plant lysate of TIR1-Myc was incubated with the His-IAA17 recombinant proteins, GSNO reduced the degradation of His-IAA17 compared with the mock over the time course, similar to the treatment with MG132 but not by GSH (Fig. 5B–5D). These results suggest that S-nitrosylation of IAA17 negatively regulates the TIR1–IAA17 co-receptor formation, thereby reducing the degradation of the IAA17 transcriptional repressors through the proteasomal pathway.
S-nitrosylation of IAA17 at Cys-70 attenuates auxin responsiveness
Data presented above indicate that IAA17 is S-nitrosylated at Cys-15 and Cys-70. To explore the regulatory roles of Cys-15 and Cys-70 in auxin responsiveness, we created transgenic lines driven by the IAA17 native promoter, including IAA17::IAA17-FLAG, IAA17::IAA17C15S-FLAG, IAA17::IAA17C15W-FLAG, IAA17::IAA17C70S-FLAG, and IAA17::IAA17C70W-FLAG, in an iaa17 (SALK_011820) T-DNA insertion mutant background. Transgenic plants were screened by quantitative reverse transcription-PCR (qRT-PCR) to identify those displaying comparable expression levels as that of wild-type (Fig. S11). Analysis of root elongation showed that only IAA17::IAA17C70W-FLAG transgenic plants were resistance to the inhibitory effects of the synthetic auxin 2,4-D (Figs. 6A, 6B, S12). In addition, only IAA17::IAA17C70W-FLAG transgenic plants displayed reduced lateral root density when treated with 2,4-D (Fig. 6C). Moreover, the 35S::IAA17C70W-FLAG transgenic plants showed defective growth and development phenotype compared with wild-type plants (Fig. S13). Taken together, these results suggest that S-nitrosylation of IAA17 at Cys-70 plays a critical role in regulating auxin responses.
Fig. 6.
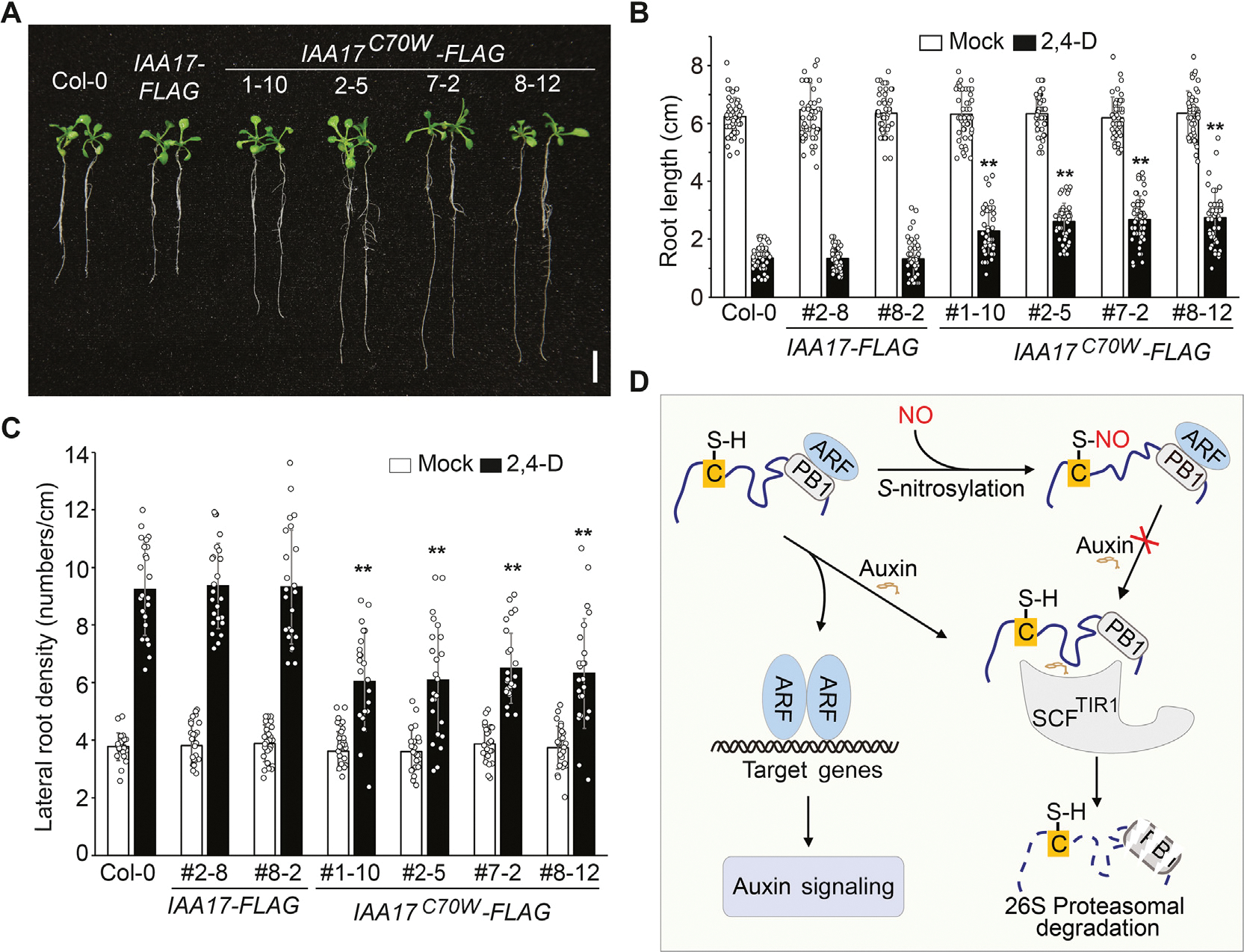
S-nitrosylation of IAA17 at Cys-70 attenuates auxin responsiveness. A: Seeds with the indicated genotypes were germinated and grown on 1/2 MS media with sucrose for 5 days. Seedlings were then transferred onto media containing 10 nM 2,4-D and cultured for additional 6 days. Representative seedlings were photographed. B and C: The primary root length and the lateral root density of seedlings shown in (A). n = 50 and n = 20 seedlings are used for the analysis of the root length and the lateral root density, respectively. Data are mean ± SD from three independent experiments and gray dots represent the individual values. The statistical significance is determined by a two-sided Student’s t-test (Paired two samples for means). **, P < 0.01 when compared to Col-0. D: A proposed model of NO-regulated auxin signaling. NO-mediated S-nitrosylation of IAA17 at Cys-70 may cause its structural alterations, resulting in the inhibition of its interaction with the SCFTIR1 complex and consequently its proteasomal degradation induced by auxin. The increased accumulation of IAA17 protein thus attenuates auxin responsiveness. Scale bar, 1 cm (A).
In summary, these results collectively support a model (Fig. 6D) in which S-nitrosylation of Cys-70 in the IDR modulates IAA17 protein accumulation by repressing the TIR1–IAA17 coreceptor interaction, thereby regulating auxin responsiveness to affect plant growth and development.
Discussion
NO is an important signal that regulates diverse biological processes in plants. As a dominant route to mediate NO bioactivity, S-nitrosylation plays crucial roles in regulating protein activities including stability (Feng et al., 2019). In this study, we find that S-nitrosylation of IAA17 by NO directly inhibits IAA17 protein degradation. Genetic and biochemical evidence demonstrates that NO regulates the proteasomal degradation of IAA17 by S-nitrosylation residue Cys-70 in the IDR that directly contacts the TIR1 receptor, thereby repressing the formation of the TIR1–IAA17 co-receptor to attenuate auxin responsiveness. The altered stability of the S-nitrosylated IAA17 protein is likely caused by its reduced interaction with TIR1, thus less susceptible to degrade in the proteasome.
Arabidopsis IAA17 contains five cysteine residues and two of them (Cys-15 and Cys-70) are identified as S-nitrosylation sites by LC-MS/MS and mutagenesis studies, of which Cys-70 may interact more frequently with TIR1 than Cys-15 as implied by molecular simulations. This result is supported by the observation that S-nitrosylation of IAA17 at Cys-70 plays a critical role in regulating auxin responses. While no apparent phenotype is observed in transgenic plants carrying an IAA17C15S gene, we cannot rule out the possibility that Cys-15 may play a role in regulating plant growth under stress conditions or in response to environmental changes. We notice that Cys-70 is not conserved in other Aux/IAA proteins in Arabidopsis, raising the possibility that S-nitrosylation at Cys-70 may specifically regulate IAA17 protein stability, which may imply functional divergence of Aux/IAA proteins as previously discovered in other members of this family (Ding and Friml, 2010; Cao et al., 2019; Lv et al., 2020; Yu et al., 2022).
The NO-auxin interplay plays an important role in regulating plant growth and responses to environmental stresses (Pagnussat et al., 2002, 2003; Otvos et al., 2005; Zhang et al., 2019; Pande et al., 2022). NO regulates auxin metabolism, transport, and signaling transduction, and auxin in turn also modulates NO accumulation and function (Campos et al., 2019; Gupta et al., 2022; Kohli et al., 2022). In this study, we find that S-nitrosylation of IAA17 at Cys-70 inhibits the interaction between TIR1 and IAA17, suggesting that NO plays an inhibitory role in regulating auxin signaling. We notice that our results are somewhat different from that of a previous study that shows a positive regulatory role of NO in auxin signaling by S-nitrosylation of TIR1 (Terrile et al., 2012). One explanation for these inconsistences would be that different auxin signaling components are tested under different assay conditions in these studies. Terrile et al. (2012) perform the binding assay of TIR1-Myc and GST-IAA3 in the presence of free GSNO. In the current study, we remove free GSNO prior to mixing TIR1-Myc with His-IAA17. Moreover, our previous study shows that GST contains four Cys residues that are S-nitrosylated (Feng et al., 2013), and we therefore use a mutant version of GST4CS in all our assays, while a wild-type GST is used by Terrile et al. (2012). Taking these considerations, while highly reactive free GSNO and the S-nitrosylation of the GST may contribute to the discrepancy between the two studies, we do not rule out the possibility that tissue- or development-specific regulation of auxin signaling by NO, which may explain the discrepancy between two studies. Moreover, we also notice that NO, either endogenous sources such as in the gsnor1 and nox1 mutants or externally supplied, generally promotes stress responses but inhibits growth and development (He et al., 2004; Lee et al., 2008; Chen et al., 2009; Kwon et al., 2012). NO has been shown to positively regulate both biotic and abiotic responses (Feng et al., 2019; Kolbert et al., 2019; Pande et al., 2022; Shang et al., 2022). On the other hand, while NO promotes seed germination in several species (Albertos et al., 2015; Li et al., 2016, 2018; Signorelli and Considine, 2018), the signaling molecule inhibits root elongation, shoot growth, leaf development, and reproductive development in most cases (Lee et al., 2008; Tada et al., 2008; Wang et al., 2009, 2015; Yun et al., 2011; Kwon et al., 2012). Consistent with this notion, NO-mediated protein S-nitrosylation has been shown to positively modulate stress responses connecting to the so-called stress phytohormones, including salicylic acid, abscisic acid, ethylene, and jasmonic acid (Lindermayr et al., 2006; Tada et al., 2008; Wang et al., 2009; Albertos et al., 2015; Ayyar, 2016; Pande et al., 2022). By contrast, this regulatory scheme appears to negatively regulate signaling mediated by the so-called growth-promoting phytohormones, including cytokinin, gibberellic acid, strigolactone, and brassinosteroid (Feng et al., 2013; Wang et al., 2015; Kolbert, 2019; Chen et al., 2022; Pande et al., 2022). Data presented in this study, consistent with that obtained from a previous study (Shi et al., 2015), support a negative regulatory role of NO in auxin signaling.
Finally, it is interesting to note that the S-nitrosylated Cys-70 of IAA17 is located in the IDR, which has been implied to play important roles in regulating the structure and functionality of these transcriptional repressors (Niemeyer et al., 2020; Figueiredo and Strader, 2022). In particular, PTMs likely act as key mechanisms to determine the spatial and temporal specificity of a given Aux/IAA protein (Jing et al., 2015; Winkler et al., 2017; Figueiredo and Strader, 2022). In this regard, the redox-based S-nitrosylation of Cys-70 may represent a unique mechanism to regulate the activity of IAA17 in yet unidentified physiological processes.
Materials and methods
Plant materials
All Arabidopsis (Arabidopsis thaliana) lines used in this study were in the Columbia (Col-0) background. The HS::IAA17NT-GUS (Gray et al., 2001) and TIR1-Myc (Gray et al., 1999) seeds were provided by Mark Estelle. The gsnor1–3 (Feechan et al., 2005) and nox1 (He et al., 2004) seeds were provided by Gary Loake and Yikun He, respectively. The gsnor1 rog1–1 mutant was described previously (Chen et al., 2020). The IAA17 gain-of-function mutants (axr3–1 and axr3–3) and T-DNA insertion mutants (SALK_011820 and SALK_065697) were obtained from ABRC.
Phenotypic analyses and chemical treatment
Seeds were surface sterilized for 15 min with 20% (v/v) bleach containing 0.01% (v/v) Triton X-100, then rinsed 4 times with sterile water. Sterilized seeds were stratified for 2 days at 4°C to promote uniform germination. After stratification, seeds were plated on 1/2 MS media solidified with 1% (w/v) sucrose at 22°C under continuous illumination.
To examine root elongation in Col-0 and transgenic plants, seedlings were grown as described above for 5 days of vertical growth at 22°C, and seedlings showing similar root length were transferred to the media supplemented with mock (EtOH) or the indicated concentrations of 2,4-D. After culturing for additional six days, the samples were photographed and the root phenotypes (the root length and lateral root density) were analyzed.
For other chemical treatments, seeds were germinated and grown on plant nutrient (PN) media solidified with 0.6% (w/v) agar and supplemented with 0.5% (w/v) sucrose (PNS) for 4 days, in the absence or the presence of various chemicals (GSH, Sigma-Aldrich, G4251; GSNO, Sigma-Aldrich, N4148; MG132, Sigma-Aldrich, M8699) with the indicated combinations and concentrations. The GVG::TIR1-Myc transgenic seeds were germinated and grown on PNS media for 7 days and then treated with 30 μM dexamethasone by spraying for 8 h to induce the TIR1-Myc expression. The samples were then transferred to liquid PN supplemented with either DMSO (Mock), 200 μmM GSH, 200 μM GSNO or 50 μM MG132 and cultured for 16 h. The samples were collected for the preparation of protein extracts for immunoblotting analysis.
Plasmid construction and genetic transformation
To create the pHis-IAA17 and pGST4CS-IAA17 expression vectors, the coding sequences of IAA17 were amplified from Arabidopsis cDNA and the stop codons were eliminated during PCR. The PCR fragments with appropriate restriction sites were cloned into the BamHI and SalI sites of pET28a (Novagen) and pGST4CS. The pGST4CS vector was generated from pGST4T-1 (Amersham Biosciences), which substitute of 4 S-nitrosylated Cys residues with Ser (Feng et al., 2013). The pGST4CS-IAA17C15S and pGST4CS-IAA17C70S plasmids were generated in a similar approach.
To construct 35S::IAA17-FLAG vector, the coding sequences of IAA17 were amplified from Arabidopsis cDNA and the stop codons was eliminated during PCR. The PCR fragments with appropriate restriction sites were digested with SmaI and cloned into the SmaI site of a pSK-FLAG vector to generate the IAA17-FLAG fusion gene, which was then cloned into the KpnHI and SalI sites of pWM101 under the control of the 35S promoter. The 35S::IAA17C70W-FLAG vector was generated in a similar approach.
To generate the IAA17::IAA17-FLAG vector, the genomic fragment of IAA17 including the putative promoter sequences and the coding regions was amplified by PCR using wild-type Arabidopsis genomic DNA as a template. The stop codon was eliminated and appropriate restriction sites were introduced during PCR. The PCR fragment digested with SmaI was cloned into the same site of a pSK-FLAG vector to generate an IAA17::IAA17-FLAG fusion gene. The fusion gene released by SalI and BamHI digestion was then cloned into the same restriction sites of pBI121. The IAA17::IAA17C15S-FLAG, IAA17::IAA17C15W-FLAG, IAA17::IAA17C70S-FLAG, and IAA17::IAA17C70W-FLAG vectors were generated in a similar approach.
Site-directed mutagenesis was performed using the Easy Mutagenesis System (TransGen Biotech, Beijing) following the manufacturer’s instructions. All PCR-amplified fragments were verified by extensive restriction digests and DNA sequencing analysis. Binary expression vectors were introduced into Agrobacterium tumefaciens strain GV3101.
The generation of transgenic Arabidopsis plants was performed via the Agrobacterium-mediated genetic transformation (Bechtold and Pelletier, 1998).
All primers used for plasmid construction are listed in Table S1.
Heat shock treatment and GUS activity assay
HS::IAA17NT-GUS plants were crossed into the gsnor1–3, nox1, and gsnor1 rog1–1 mutant background. Seven-day-old seedlings were submerged in liquid 1/2 MS media and heat shocked for 2 h at 37°C. After the heat shock, seedlings were transferred to media in the absence (Mock) or the presence of various chemicals with the indicated concentrations and incubated for 0–2 h at 22°C prior to GUS staining. For the GUS enzyme activity assay (Gray et al., 2001), seedlings were collected after 1 h treatment and stored in liquid nitrogen until protein extraction. The fluorometric assay was performed by incubating protein extracts in 2 mM 4-methylumbelliferyl-β-D-glucoronide (MUG), 50 mM KPO4 (pH 7.0), 0.1% SDS, 0.1% Triton X-100, 10 mM β-mercaptoethanol, and 10 mM EDTA for 16 h followed by analysis with a Microplate Reader. Protein extracts were prepared from 10 seedlings and data were normalized against total protein levels.
Antibody preparation and immunoblot analysis
To prepare anti-IAA17 antibodies, a full-length cDNA fragment of IAA17 was used to produce recombinant proteins tagged with 6× His. The purified His-IAA17 recombinant protein was used to immunize mice and rabbits, respectively. Immunoblotting analysis was performed as described previously (Jing et al., 2015, 2022). Total plant proteins are prepared by grinding seedlings in liquid nitrogen and then extracted in lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% [v/v] Nonidet P-40, 0.5% [w/v] sodium deoxycholate, 0.1% [w/v] SDS, 1 mM phenylmethylsulfonyl fluoride [PMSF], and 1% [v/v] protease inhibitor cocktail [Sigma-Aldrich, P9599]). After centrifugation twice at 13,000 g for 10 min at 4°C, the supernatant was collected and then subjected to SDS-PAGE. The proteins were electrically transferred onto a PVDF membrane and then detected with a primary antibody of indicated (usually at 1:5000 dilutions). The blot is incubated with a secondary antibody (HRP-conjugated goat anti-rabbit IgG or HRP-conjugated goat anti-mouse IgG; Beijing Dingguo Changsheng Biotechnology) at 1: 50000 dilutions. The signal was detected using a SuperSignal Western Femto Maximum Sensitivity Substrate kit (Thermo Scientific, Cat #34096) according to the manufacturer’s instructions. Arabidopsis HSP82 (Beijing Protein Innovation, Cat #K2010) or HSC70 (Enzo Life Sciences, ADI-SPA-817-D) was used as loading controls. The target or loading control bands were quantified using FIJI (ImageJ) and the mean values of 3–5 independent experiments were presented with statistical analysis (one-way ANOVA or Student’s t-test) of significant differences when applicable.
In vitro S-nitrosylation assay
Analysis of the in vitro S-nitrosylation modification was performed by a biotin-switch method as described previously (Chen et al., 2020). In brief, approximately 5 μg of recombinant protein was incubated with GSH or GSNO (200 μM) in the dark at room temperature for 30 min. The sample was precipitated with three volumes of cold acetone, and then washed with acetone three times and resuspended in 300 μL blocking buffer (250 mM HEPES, pH 7.7, 4 mM EDTA, 1 mM neocuproine, 2.5% [w/v] SDS, 200 mM S-methylmethane thiosulfonate). The reaction was carried out at 50°C for 1 h with frequent vortex. Then the sample was precipitated using cold acetone and dissolved in 48 μL HENS buffer (250 mM HEPES, pH 7.7, 4 mM EDTA, 1 mM neocuproine, 1% [w/v] SDS), followed by an addition of 6 μL 20 mM sodium ascorbate and 6 μL 4 mM biotin-HPDP. The reaction was incubated at room temperature for 1 h. All the above steps were carried out in a darkroom. Aliquots of each sample was analyzed by SDS-PAGE without boiling, followed by immunoblotting using an anti-biotin antibody (Cell Signaling Technology, Cat #7075). The blot was stripped using strip buffer (15 g/L glycine, 1% SDS, 1% Tween-20, pH 2.2) at room temperature for 30 min and re-incubated using an anti-His (Santa Cruz Biotechnology, Cat #SC-8036) or anti-GST antibody (Santa Cruz Biotechnology, Cat #SC-138) to analyze the input.
In vivo S-nitrosylation assay
In vivo S-nitrosylation assay was performed as previously described (Zhan et al., 2018). Total protein extracts were prepared by grinding seedlings in liquid nitrogen and then extracted in cold HEN buffer (250 mM HEPES, pH 7.7, 1 mM EDTA, 0.1 mM neocuproine, and 1% [v/v] protease inhibitor cocktail). After centrifugation at 13,000 rpm at 4°C for 20 min, the supernatant was collected and incubated with blocking buffer (250 mM HEPES, pH 7.7, 4 mM EDTA, 1 mM neocuproine, 2.5% [w/v] SDS, 200 mM S-methylmethane thiosulfonate) at 50°C for 1 h with frequent vortex. The samples were precipitated with cold acetone, followed by washing with acetone three times, and then dissolved in 240 μL of HEN buffer supplemented with 1% (w/v) SDS. After adding 20 μL of 500 mM sodium ascorbate and 20 μL of 4 mM biotin-HPDP, the samples were incubated at room temperature for 1 h, followed by precipitation and washing with cold acetone. The pellet was dissolved in 300 mL 250 mM HEPES buffer and neutralized with 800 μL of neutralization buffer (25 mM HEPES, pH 7.7, 100 mM NaCl, 1 mM EDTA, 0.5% Triton X-100), followed by adding 30 μL streptavidin beads (Thermo Scientific, Cat #29202) and incubating at 4°C for 16 h. After washing the beads three times with the buffer (25 mM HEPES, pH 7.7, 600 mM NaCl, 1 mM EDTA, 0.5% Triton X-100), the samples were subjected to SDS-PAGE and analyzed by immunoblotting.
DAN-NAT assay
The number of S-nitrosylated cysteine residues was measured by the 2,3-diaminonaphthalene (DAN) assay as described previously (Yang et al., 2015). In brief, recombinant His-IAA17 protein (approximately 200 μg) was treated with 200 μM GSNO at room temperature in a darkroom for 1 h. The samples were washed twice with 1 mL PBS to remove free GSNO, followed by adding 300 μL of 200 mM HgCl2 and 200 mM DAN for 30 min in the dark, allowing to converse DAN into fluorescent 2,3-naphthyltriazole (NAT). To stop the reaction, 15 3bcxL of 2.8 M NaOH was added and the reaction was incubated for 5 min at room temperature. The excitation spectrum of UV (365 ± 10 nm) and emission wavelength (450 nm), emitted from the NAT fluorescent signal, were measured using a fluorospectrometer (Nano Drop 3300, Thermo Scientific) and the relative DAN-NAT conversion rate was calculated as the fluorescent intensity of NAT per μM protein.
Mass spectrometry
Identification of S-nitrosylated cysteine residues was performed by mass spectrometric using the SNOSID method with minor modifications (Zhan et al., 2018). Briefly, approximately 100 μg His-IAA17 recombinant proteins were treated with 200 μM GSNO and then labeled with biotin-maleimide (Sigma-Aldrich, Cat #B1267). The biotinylated proteins were precipitated and washed with cold acetone, resuspended in 400 μL dissolved buffer (20 mM Tris-HCl, pH 7.7, 1 mM EDTA, 0.4% Triton X-100), and then digested by trypsin (Promega, Cat #V5280) at a final concentration of 10 μg/mL at 37°C for 16 h. The reaction was terminated by adding PMSF and followed by incubating with 50 μL streptavidin-agarose beads (Thermo Scientific, Cat #29202) at room temperature for 2 h. The streptavidin beads were washed three times in 1 mL washing buffer (20 mM Tris-HCl, pH 7.7, 1 mM EDTA, 0.4% Triton X-100, 600 mM NaCl), followed by washing three times with 1 mL buffer (5 mM ammonium bicarbonate and 20% acetonitrile). The streptavidin-bound peptides were eluted by adding 100 μL of 100 mM β-mercaptoethanol and then concentrated by a SpeedVac vacuum concentrators. The supernatants were concentrated to a final volume 20 μL–30 μL, and then stored at −20°C for subsequent analysis.
The trypsin-digested peptides were analyzed by LC-MS/MS using a BioBasic-18 column (0.18 × 150 mm2, Surveyor System, Thermo Inc.) coupled on-line with the ion trap mass spectrometer (LCQ Deca Xp Plus, Thermo Finnigan). The instrument was run in the data-dependent mode with cycling between one full MS scan from m/z 400–2000 and MS/MS scans of five most abundant ions using dynamic exclusion. The normalized collision energy was set to 35% for precursor ions fragmentation. Analysis of MS/MS spectra for peptide identification was performed by protein database searching engine with pFIND software as described previously (Li et al., 2005). The His-IAA17 sequence was used as a database for searching. The key searching parameters were setting as follows: (1) a precursor mass tolerance of 2.0 Da; (2) a fragment mass tolerance of 0.5 Da; (3) two miscleavages were allowed in the search; (4) variable modifications: +15.995 amu on methionine for oxidation, +451.540 amu on cysteine for biotinylation; and (5) show spectra or peptides when False Discovery Rate (FDR) ≤ 1%. Choose 2*Y/(X + Y), where X indicates the number of hits to target sequences, and Y indicates the number of hits to decoy sequences, respectively.
Co-immunoprecipitation assay
The co-immunoprecipitation (Co-IP) experiments were performed as previously described with minor modifications (Jing et al., 2022). Briefly, His-IAA17 recombinant protein (approximately 20 μg) was incubated with 200 μM GSH (negative control) or 200 μM GSNO at room temperature in the dark for 2 h and precipitated with three volumes of cold acetone for 20 min at −20°C. The samples were washed with cold acetone three times to remove free GSNO, dried at room temperature for 2 min, and resuspended in dissolved buffer (20 mM Tris-HCl, pH 7.7, 1 mM EDTA, 0.4% Triton X-100). To prepare TIR1-Myc protein, 7-day-old TIR1-Myc transgenic seedlings were ground in liquid nitrogen and extracted in lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10 mM MgCl2, 10% [v/v] glycerol, 0.1% [v/v] Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride, 1% [v/v] protease inhibitor cocktail [Sigma-Aldrich, P9599]). The extracts were cleared by centrifugation at 14,000 g for 15 min and supernatants containing 1.0 mg–2.0 mg total proteins were incubated with anti-Myc magnetic beads (ThermoFisher, 88842) at 4°C for 1 h with gentle shaking. The beads were collected, washed, and then mixed with the GSH-treated or GSNO-treated His-IAA17 recombinant proteins as described above, with or without 10 μM IAA as indicated. The reaction was run at 4°C for 2 h–3 h with gentle shaking. The beads were washed three times with 1 mL washing buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10 mM MgCl2, 0.1% [v/v] Nonidet P-40) and then subsequently used for immunoblotting.
In vitro protein degradation assay
The analysis of His-IAA17 protein degradation in vitro was performed as previously described with minor modifications (Jing et al., 2022). The preparation of GSH- or GSNO-treated His-IAA17 recombinant protein and TIR1-Myc transgenic protein was performed as described above (see “Co-immunoprecipitation assay” section). The TIR1-Myc-containing protein extracts (equivalent to approximately 1 mg total proteins) were mixed with His-IAA17 protein (treated with GSH or GSNO) or 50 μM MG132 in a total volume of 600 μL. The mixture was incubated at 4°C with gentle agitation, and 100 μL of each sample was collected at the indicated time points and then analyzed by immunoblotting.
Synthetic yeast (Saccharomyces cerevisiae) system assay
The synthetic yeast system assay of IAA17 degradation dynamics was performed as previously described with minor modification (Havens et al., 2012; Chaisupa et al., 2023). Yeast transformation was performed using a lithium acetate protocol (Gietz and Woods, 2002). All the vectors were digested by PmeI enzyme to linearize prior to transformation. The vector pGP8G containing TIR1 was transformed into the W303–1A (MATα leu2–3,112 trp1–1 can1–100 ura3–1 ade2–1 his3–11,15) strain. The pGP4G vector, which contains a radiometric sensor Venus in-frame fused to IAA17, followed by a 2A self-cleaving peptide and mScarlet, was transformed into the W814–29B (MATα ade2–1 trp1–1 can1–100 ura3–1 leu2–3,112 his3–11,15) strain. Various mutations of IAA17 at Cys-70 to serine (S), tryptophan (W), phenylalanine (F), and tyrosine (Y) were created using Site-Directed Mutagenesis System (Thermo Fisher, SuperFi II). The empty vector pGP4G-Venus-mScarlet was used as a control.
Flow cytometry measurements
The flow cytometry measurements were performed as previously described with modifications (Pierre-Jerome et al., 2017). Briefly, after mating W814–29B strains into W303–1A, a single colony was suspended in synthetic complete (SC) media. The initial cell concentration of each strain was measured by flow cytometry (CytoFLEX S, Beckman Coulter) and then diluted to 1 event/μL. The cultures were grown in 125 mL Erlenmeyer flasks at 30°C for 16 h in a shaker at 250 rpm and then filtered through a 40-μm cell strainer. The filtered cultures were aliquoted into 2-mL Eppendorf tubes and mixed with various chemicals (1 μM IAA, 200 μM GSH, 200 μM GSNO, and 50 μM MG132) and different combinations (IAA with GSH, IAA with GSNO, and IAA with MG132). The samples were cultured at 30°C in a shaker at 300 rpm. The samples were measured by flow cytometry (CytoFLEX S, Beckman Coulter) every 30 min for 3 h. The program was set to 10,000 events per well and the flowrate was at 130 μL/mL. The excitation and emission settings were selected as 488 nm and 525 nm for Venus, 561 nm and 585 nm for mScarlet. Deionized water of 100 μL was added between each sample to eliminate crossover events. Data were processed using FlowJo™ (v10.8.2 Software, BD Life Sciences). The values of each treatment were determined and normalized as a ratio of Venus divided by mScarlet.
Quantitative reverse transcription-PCR (qRT-PCR)
Total RNA was prepared using the RNeasy Plant Mini Kit (Qiagen). Quantitative reverse transcription-PCR (qRT-PCR) was performed using the iTaqTM Universal SYBR® Green Supermix (Bio-Rad) according to the manufacturer’s instructions. The reaction was run in a CFX96 REAL-Time PCR Detection System (Bio-Rad). The relative expression level of a target gene was analyzed with the delta-delta Ct method and normalized to the expression level of ACT7. All experiments were repeated at least twice (two biological repeats with three technical repeats for each experiment). The primers used for qRT-PCR are listed in Table S1.
Coarse-grained simulations
Molecular simulation was performed using the LAMMPS simulation engine (Thompson et al., 2022) with the default Mpipi parameters (Joseph et al., 2022). Disordered regions were modeled as fully flexible chains, while folded domains were modeled as fixed rigid bodies as described previously (Joseph et al., 2022; Cubuk et al., 2023). The relative position of beads in folded domains was determined using the location of the alpha carbon atoms for each amino acid, while disordered region starting configurations were generated as a non-overlapping starting configuration. For the TIR1/ASK1 complex, the crystal structure (Tan et al., 2007) (PDB ID: 2P1M) was used for generating the coarse-grained structure used for all simulations. This structure represents the holo (auxin bound) TIR1/ASK1 complex, because we lack forcefield parameters for small molecules auxin that is not included in the receptor. The PB1 domain of IAA17 spanned residues 105 to 218 and all residues outside the PB1 domain were modeled as flexible beads. The PB1/IDR definitions are largely consistent with predictions from the disorder predictor metapredict (V2) (Emenecker et al., 2021). The model for the IAA17 PB1 domain was acquired using ColabFold (Jumper et al., 2021; Mirdita et al., 2022), where the full-length IAA17 was used to generate the structure. In ColabFold, structures were ranked by pLDDT score, max_msa was set to 512:1024, use_turbo was set to True, num_models was set to 5, use_ptm was set to True, num_ensembles was set to 8, max_recycles was set to 3, training was set to False, and num_samples was set to 1. All simulations were run in a box with a box size of 300 Å3 with periodic boundary conditions. All simulations ran for 20,000,000 steps with 10 fs per step. At the beginning of each simulation, the first 5,000,000 steps (i.e., 20% of the simulation) were discarded as equilibration and were not used for downstream analysis. The coordinates of the amino acids in the simulation were recorded every 5000 steps. For all simulations, the temperature was set to 300 K. Thirty replicates were used for each simulated protein or protein variant (including IAA17 poly-GS IDR) to reduce the bias inherent to each simulation due to the random placement of the TIR1/ASK1 complex and IAA17 within the box. The simulations were analyzed using the Python package SOURSOP HICH based in part on MDTraj (McGibbon et al., 2015; Lalmansingh et al., 2023). For considering IAA17 either ‘bound’ or ‘unbound’ to TIR1/ASK1 during analysis, IAA17 was considered bound to the TIR1/ASK1 complex if at least one residue of IAA17 was in contact with the TIR1/ASK1 complex. Residues were considered in contact if their inter-residue distance was less than 7 Å, as determined using the get_interchain_distance function in SOURSOP. This was also used to calculate the frequency at which Cys-70 of IAA17 was in contact with the TIR1/ASK1 complex. For all images or videos of the simulations, the programs NGLview (Rose et al., 2018) or VMD (Humphrey et al., 1996) were used.
Supplementary Material
Acknowledgments
We thank Dr. Mark Estelle, Dr. Gary Loake, Dr. Yikun He, and the Arabidopsis Biological Resource Center (ABRC) for seeds; Jeffrey R. Allen, Joseph Cammarata, Katelyn Sageman-Furnas, Sunita Pathak, and Edward G. Wilkinson for critical comments on the manuscript; members of Zuo lab and Strader lab for helpful discussion. This work is supported by grants from the National Natural Science Foundation of China (31830017), Chinese Academy of Sciences (XDB27030207), the Hainan Excellent Talent Team, and State Key Laboratory of Plant Genomics (SKLPG2023–22).
Footnotes
Conflict of interest
The authors declare no competing interests.
CRediT authorship contribution statement
Hongwei Jing, Xiaolu Yang: Conceptualization, Experimentation, Data analysis, Writing - Original draft, Writing - Review & Editing. Ryan J. Emeneckere: Experimentation, Data analysis, Writing - Review & Editing. Jian Feng: Conceptualization, Experimentation. Jian Zhang, Marcelo Rodrigues Alves de Figueiredoc, Patarasuda Chaisupag: Experimentation. Clay Wright, Alex S. Holehouse, Lucia C. Strader: Conceptualization, Supervision, Writing - Review & Editing. Jianru Zuo: Conceptualization, Supervision, Writing - Original draft, Writing - Review & Editing, Funding acquisition.
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jgg.2023.05.001.
References
- Albertos P, Romero-Puertas MC, Tatematsu K, Mateos I, Sanchez-Vicente I, Nambara E, Lorenzo O, 2015. S-nitrosylation triggers ABI5 degradation to promote seed germination and seedling growth. Nat. Commun. 6, 8669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayyar PV, 2016. Uncovering the role of S-nitrosylation in jasmonic acid signalling during the plant immune response (PhD thesis). IMPS, The University of Edinburgh, Edinburgh. [Google Scholar]
- Bechtold N, Pelletier G, 1998. In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 82, 259–266. [DOI] [PubMed] [Google Scholar]
- Campos FV, Oliveira JA, Pereira MG, Farnese FS, 2019. Nitric oxide and phytohormone interactions in the response of Lactuca sativa to salinity stress. Planta 250, 1475–1489. [DOI] [PubMed] [Google Scholar]
- Cao M, Chen R, Li P, Yu Y, Zheng R, Ge D, Zheng W, Wang X, Gu Y, Gelova Z, et al. , 2019. TMK1-mediated auxin signalling regulates differential growth of the apical hook. Nature 568, 240–243. [DOI] [PubMed] [Google Scholar]
- Chaisupa P, Rahman MM, Hildreth SB, Moseley S, Gatling C, Helm RF, Wright RC, 2023. Genetically encoded, noise-tolerant, auxin biosensors in yeast facilitate metabolic engineering and directed evolution. bioRxiv. 10.1101/2023.03.21.533585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Sun S, Song CP, Zhou JM, Li J, Zuo J, 2022. Nitric oxide negatively regulates gibberellin signaling to coordinate growth and salt tolerance in Arabidopsis. J. Genet. Genomics 49, 756–765. [DOI] [PubMed] [Google Scholar]
- Chen L, Wu R, Feng J, Feng T, Wang C, Hu J, Zhan N, Li Y, Ma X, Ren B, et al. , 2020. Transnitrosylation mediated by the non-canonical catalase ROG1 regulates nitric oxide signaling in plants. Dev. Cell 53, 444–457. [DOI] [PubMed] [Google Scholar]
- Chen R, Sun S, Wang C, Li Y, Liang Y, An F, Li C, Dong H, Yang X, Zhang J, et al. , 2009. The Arabidopsis PARAQUAT RESISTANT2 gene encodes an S-nitrosoglutathione reductase that is a key regulator of cell death. Cell Res. 19, 1377–1387. [DOI] [PubMed] [Google Scholar]
- Cubuk J, Alston JJ, Incicco JJ, Holehouse AS, Hall KB, Stuchell-Brereton MD, Soranno A, 2023. The disordered N-terminal tail of SARS CoV-2 Nucleocapsid protein forms a dynamic complex with RNA. bioRxiv. 10.1101/2023.02.10.527914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M, 2005. The F-box protein TIR1 is an auxin receptor. Nature 435, 441–445. [DOI] [PubMed] [Google Scholar]
- Ding Z, Friml J, 2010. Auxin regulates distal stem cell differentiation in Arabidopsis roots. Proc. Natl. Acad. Sci. U. S. A. 107, 12046–12051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emenecker RJ, Griffith D, Holehouse AS, 2021. Metapredict: a fast, accurate, and easy-to-use predictor of consensus disorder and structure. Biophys. J. 120, 4312–4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feechan A, Kwon E, Yun BW, Wang Y, Pallas JA, Loake GJ, 2005. A central role for S-nitrosothiols in plant disease resistance. Proc. Natl. Acad. Sci. U. S. A. 102, 8054–8059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Chen L, Zuo J, 2019. Protein S-nitrosylation in plants: current progresses and challenges. J. Integr. Plant Biol. 61, 1206–1223. [DOI] [PubMed] [Google Scholar]
- Feng J, Wang C, Chen Q, Chen H, Ren B, Li X, Zuo J, 2013. S-nitrosylation of phosphotransfer proteins represses cytokinin signaling. Nat. Commun. 4, 1529. [DOI] [PubMed] [Google Scholar]
- Fernandez-Marcos M, Sanz L, Lewis DR, Muday GK, Lorenzo O, 2011. Nitric oxide causes root apical meristem defects and growth inhibition while reducing PIN-FORMED (PIN1)-dependent acropetal auxin transport. Proc. Natl. Acad. Sci. U. S. A. 108, 18506–18511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando V, Zheng X, Walia Y, Sharma V, Letson J, Furuta S, 2019. S-nitrosylation: an emerging paradigm of redox signaling. Antioxidants 8, 404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo MRA, Strader LC, 2022. Intrinsic and extrinsic regulators of Aux/IAA protein degradation dynamics. Trends Biochem. Sci. 47, 865–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Woods RA, 2002. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350, 87–96. [DOI] [PubMed] [Google Scholar]
- Gray WM, del Pozo JC, Walker L, Hobbie L, Risseeuw E, Banks T, Crosby WL, Yang M, Ma H, Estelle M, 1999. Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev. 13, 1678–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M, 2001. Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 414, 271–276. [DOI] [PubMed] [Google Scholar]
- Gupta KJ, Kaladhar VC, Fitzpatrick TB, Fernie AR, Moller IM, Loake GJ, 2022. Nitric oxide regulation of plant metabolism. Mol. Plant 15, 228–242. [DOI] [PubMed] [Google Scholar]
- Gupta KJ, Kolbert Z, Durner J, Lindermayr C, Corpas FJ, Brouquisse R, Barroso JB, Umbreen S, Palma JM, Hancock JT, et al. , 2020. Regulating the regulator: nitric oxide control of post-translational modifications. New Phytol. 227, 1319–1325. [DOI] [PubMed] [Google Scholar]
- Havens KA, Guseman JM, Jang SS, Pierre-Jerome E, Bolten N, Klavins E, Nemhauser JL, 2012. A synthetic approach reveals extensive tunability of auxin signaling. Plant Physiol. 160, 135–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Tang RH, Hao Y, Stevens RD, Cook CW, Ahn SM, Jing L, Yang Z, Chen L, Guo F, et al. , 2004. Nitric oxide represses the Arabidopsis floral transition. Science 305, 1968–1971. [DOI] [PubMed] [Google Scholar]
- Hu J, Yang H, Mu J, Lu T, Peng J, Deng X, Kong Z, Bao S, Cao X, Zuo J, 2017. Nitric oxide regulates protein methylation during stress responses in plants. Mol. Cell 67, 702–710. [DOI] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K, 1996. Vmd: Visual molecular dynamics. J. Mol. Graph. 14, 33–38. [DOI] [PubMed] [Google Scholar]
- Iglesiasa MJ, Terrile MC, Correa-Aragunde N, Colman SL, Izquierdo-Alvarez A, Fiol DF, Paris R, Sanchez-Lopez N, Marina A, Villalobos LIAC, et al. , 2018. Regulation of SCFTIR1/AFBs E3 ligase assembly by S-nitrosylation of Arabidopsis SKP1-like1 impacts on auxin signaling. Redox Biol. 18, 200–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing H, Korasick DA, Emenecker RJ, Morffy N, Wilkinson EG, Powers SK, Strader LC, 2022. Regulation of AUXIN RESPONSE FACTOR condensation and nucleo-cytoplasmic partitioning. Nat. Commun. 13, 4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing H, Strader LC, 2018. Structural biology of auxin signal transduction. In: Hejatko J (Ed.), Plant Structural Biology: Hormonal Regulations, 66th, p. 49. [Google Scholar]
- Jing H, Yang X, Zhang J, Liu X, Zheng H, Dong G, Nian J, Feng J, Xia B, Qian Q, et al. , 2015. Peptidyl-prolyl isomerization targets rice Aux/IAAs for proteasomal degradation during auxin signalling. Nat. Commun. 6, 7395. [DOI] [PubMed] [Google Scholar]
- Joseph JA, Reinhardt A, Aguirre A, Chew PY, Russell KO, Espinosa JR, Garaizar A, Collepardo-Guevara R, 2022. Physics-driven coarse-grained model for biomolecular phase separation with near-quantitative accuracy. Nat. Comput. Sci. 1, 732–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Zidek A, Potapenko A, et al. , 2021. Highly accurate protein structure prediction with AlphFold. Nature 596, 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli SK, Khanna K, Bhardwaj R, Corpas FJ, Ahmad P, 2022. Nitric oxide, salicylic acid and oxidative stress: is it a perfect equilateral triangle? Plant Physiol. Biochem. 184, 56–64. [DOI] [PubMed] [Google Scholar]
- Kolbert Z, 2019. Strigolactone-nitric oxide interplay in plants: the story has just begun. Physiol. Plant 165, 487–497. [DOI] [PubMed] [Google Scholar]
- Kolbert Z, Barroso JB, Brouquisse R, Corpas FJ, Gupta KJ, Lindermayr C, Loake GJ, Palma JM, Petrivalsky M, Wendehenne D, et al. , 2019. A forty year journey: the generation and roles of NO in plants. Nitric Oxide 93, 53–70. [DOI] [PubMed] [Google Scholar]
- Kwon E, Feechan A, Yun BW, Hwang BH, Pallas JA, Kang JG, Loake GJ, 2012. AtGSNOR1 function is required for multiple developmental programs in Arabidopsis. Planta 236, 887–900. [DOI] [PubMed] [Google Scholar]
- Lalmansingh JM, Keeley AT, Ruff KM, Pappu RV, Holehouse AS, 2023. SOURSOP: a Python package for the analysis of simulations of intrinsically disordered proteins. bioRxiv. 10.1101/2023.02.16.528879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee U, Wie C, Fernandez BO, Feelisch M, Vierling E, 2008. Modulation of nitrosative stress by S-nitrosoglutathione reductase is critical for thermotolerance and plant growth in Arabidopsis. Plant Cell 20, 786–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Fu Y, Sun R, Ling CX, Wei Y, Zhou H, Zeng R, Yang Q, He S, Gao W, 2005. pFind: a novel database-searching software system for automated peptide and protein identification via tandem mass spectrometry. Bioinformatics 21, 3049–3050. [DOI] [PubMed] [Google Scholar]
- Li R, Jia Y, Yu L, Yang W, Chen Z, Chen H, Hu X, 2018. Nitric oxide promotes light-initiated seed germination by repressing PIF1 expression and stabilizing HFR1. Plant Physiol. Biochem. 123, 204–212. [DOI] [PubMed] [Google Scholar]
- Li X, Pan Y, Chang B, Wang Y, Tang Z, 2016. NO promotes seed germination and seedling growth under high salt may depend on EIN3 protein in Arabidopsis. Front. Plant Sci. 6, 1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindermayr C, Saalbach G, Bahnweg G, Durner J, 2006. Differential inhibition of Arabidopsis methionine adenosyltransferases by protein S-nitrosylation. J. Biol. Chem. 281, 4285–4291. [DOI] [PubMed] [Google Scholar]
- Lv B, Yu Q, Liu J, Wen X, Yan Z, Hu K, Li H, Kong X, Li C, Tian H, et al. , 2020. Non-canonical AUX/IAA protein IAA33 competes with canonical AUX/IAA repressor IAA5 to negatively regulate auxin signaling. EMBO J. 39, e101515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGibbon RT, Beauchamp KA, Harrigan MP, Klein C, Swails JM, Hernandez CX, Schwantes CR, Wang LP, Lane TJ, Pande VS, 2015. MDTraj: a modern open library for the analysis of molecular dynamics trajectories. Biophys. J. 109, 1528–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirdita M, Schutze K, Moriwaki Y, Heo L, Ovchinnikov S, Steinegger M, 2022. ColabFold: making protein folding accessible to all. Nat. Methods 19, 679–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morffy N, Strader LC, 2022. Structural aspects of auxin signaling. Cold Spring Harb. Perspect. Biol. 14, a039883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M, Zhang L, Shi YF, Wang C, Lu Y, Pan J, Liu JZ, 2017. Excessive cellular S-nitrosothiol impairs endocytosis of auxin efflux transporter PIN2. Front. Plant Sci. 8, 1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemeyer M, Castillo EM, Ihling CH, Iacobucci C, Wilde V, Hellmuth A, Hoehenwarter W, Samodelov SL, Zurbriggen MD, Kastritis PL, et al. , 2020. Flexibility of intrinsically disordered degrons in AUX/IAA proteins reinforces auxin co-receptor assemblies. Nat. Commun. 11, 2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otvos K, Pasternak TP, Miskolczi P, Domoki M, Dorjgotov D, Szucs A, Bottka S, Dudits D, Feher A, 2005. Nitric oxide is required for, and promotes auxin-mediated activation of, cell division and embryogenic cell formation but does not influence cell cycle progression in alfalfa cell cultures. Plant J. 43, 849–860. [DOI] [PubMed] [Google Scholar]
- Pagnussat GC, Lanteri ML, Lamattina L, 2003. Nitric oxide and cyclic GMP are messengers in the indole acetic acid-induced adventitious rooting process. Plant Physiol. 132, 1241–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnussat GC, Simontacchi M, Puntarulo S, Lamattina L, 2002. Nitric oxide is required for root organogenesis. Plant Physiol. 129, 954–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pande A, Mun BG, Rahim W, Khan M, Lee DS, Lee GM, Al Azzawi TNI, Hussain A, Kim CK, Yun BW, 2022. Phytohormonal regulation through protein S-nitrosylation under stress. Front. Plant Sci. 13, 865542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre-Jerome E, Wright RC, Nemhauser JL, 2017. Characterizing auxin response circuits in Saccharomyces cerevisiae by flow cytometry. Methods Mol. Biol. 1497, 271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers SK, Strader LC, 2020. Regulation of auxin transcriptional responses. Dev. Dyn. 249, 483–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose AS, Bradley AR, Valasatava Y, Duarte JM, Prlic A, Rose PW, 2018. NGL viewer: web-based molecular graphics for large complexes. Bioinformatics 34, 3755–3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang JX, Li X, Li C, Zhao L, 2022. The role of nitric oxide in plant responses to salt stress. Int. J. Mol. Sci. 23, 6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi YF, Wang DL, Wang C, Culler AH, Kreiser MA, Suresh J, Cohen JD, Pan J, Baker B, Liu JZ, 2015. Loss of GSNOR1 function leads to compromised auxin signaling and polar auxin transport. Mol. Plant 8, 1350–1365. [DOI] [PubMed] [Google Scholar]
- Signorelli S, Considine MJ, 2018. Nitric oxide enables germination by a four-pronged attack on ABA-induced seed dormancy. Front. Plant Sci. 9, 296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Williams LA, Farmer LM, Vierstra RD, Callis J, 2006. KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell 18, 3415–3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada Y, Spoel SH, Pajerowska-Mukhtar K, Mou Z, Song J, Wang C, Zuo J, Dong X, 2008. Plant immunity requires conformational charges of NPR1 via S-nitrosylation and thioredoxins. Science 321, 952–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Calderon-Villalobos LIA, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N, 2007. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446, 640–645. [DOI] [PubMed] [Google Scholar]
- Terrile MC, Paris R, Calderon-Villalobos LIA, Iglesias MJ, Lamattina L, Estelle M, Casalongue CA, 2012. Nitric oxide influences auxin signaling through S-nitrosylation of the Arabidopsis TRANSPORT INHIBITOR RESPONSE 1 auxin receptor. Plant J. 70, 492–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AP, Aktulga HM, Berger R, Bolintineanu DS, Brown WM, Crozier PS, Veld PJI, Kohlmeyer A, Moore SG, Nguyen TD, et al. , 2022. LAMMPS-a flexible simulation tool for particle-based materials modeling at the atomic, meso, and continuum scales. Comput. Phys. Commun. 271, 108171. [Google Scholar]
- Wang P, Du Y, Hou YJ, Zhao Y, Hsu CC, Yuan F, Zhu X, Tao WA, Song CP, Zhu JK, 2015. Nitric oxide negatively regulates abscisic acid signaling in guard cells by S-nitrosylation of OST1. Proc. Natl. Acad. Sci. U. S. A. 112, 613–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YQ, Feechan A, Yun BW, Shafiei R, Hofmann A, Taylor P, Xue P, Yang FQ, Xie ZS, Pallas JA, et al. , 2009. S-nitrosylation of AtSABP3 antagonizes the expression of plant immunity. J. Biol. Chem. 284, 2131–2137. [DOI] [PubMed] [Google Scholar]
- Winkler M, Niemeyer M, Hellmuth A, Janitza P, Christ G, Samodelov SL, Wilde V, Majovsky P, Trujillo M, Zurbriggen MD, et al. , 2017. Variation in auxin sensing guides AUX/IAA transcriptional repressor ubiquitylation and destruction. Nat. Commun. 8, 15706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Mu J, Chen L, Feng J, Hu J, Li L, Zhou JM, Zuo J, 2015. S-nitrosylation positively regulates ascorbate peroxidase activity during plant stress responses. Plant Physiol. 167, 1604–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Zhang F, Friml J, Ding Z, 2022. Auxin signaling: research advances over the past 30 years. J. Integr. Plant Biol. 64, 371–392. [DOI] [PubMed] [Google Scholar]
- Yun BW, Feechan A, Yin M, Saidi NBB, Le Bihan T, Yu M, Moore JW, Kang JG, Kwon E, Spoel SH, et al. , 2011. S-nitrosylation of NADPH oxidase regulates cell death in plant immunity. Nature 478, 264–268. [DOI] [PubMed] [Google Scholar]
- Zhan N, Wang C, Chen L, Yang H, Feng J, Gong X, Ren B, Wu R, Mu J, Li Y, et al. , 2018. S-nitrosylation targets GSNO reductase for selective autophagy during hypoxia responses in plants. Mol. Cell 71, 142–154. [DOI] [PubMed] [Google Scholar]
- Zhang HT, Zeng LF, He QY, Tao WA, Zha ZG, Hu CD, 2016. The E3 ubiquitin ligase CHIP mediates ubiquitination and proteasomal degradation of PRMT5. Biochim. Biophys. Acta 1863, 335–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Huang D, Wang C, Wang B, Fang H, Huo J, Liao W, 2019. Recent progress in protein S-nitrosylation in phytohormone signaling. Plant Cell Physiol. 60, 494–502. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


