Abstract
Studies of the monogenic autoimmune disease immunodysregulation polyendocrinopathy enteropathy X-linked syndrome (IPEX) have elucidated the essential function of the transcription factor FOXP3 and thymic-derived regulatory T cells (Tregs) in controlling peripheral tolerance. However, the presence and the source of autoreactive T cells in IPEX remain undetermined. Here, we investigated how FOXP3 deficiency affects the T cell receptor (TCR) repertoire and Treg stability in vivo and compared T cell abnormalities in patients with IPEX to those in patients with autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy syndrome (APECED). To study Tregs independently of their phenotype and to analyze T cell autoreactivity, we combined Treg-specific demethylation region analyses, single-cell multi-omic profiling, and bulk TCR sequencing. We found that patients with IPEX, unlike patients with APECED, have expanded autoreactive T cells originating from both autoreactive effector T cells (Teffs) and Tregs. In addition, a fraction of the expanded Tregs from patients with IPEX lost their phenotypic and functional markers including CD25 and FOXP3. Functional experiments with CRISPR/Cas9-mediated FOXP3 knock-out Tregs and Tregs from patients with IPEX indicated that the patients’ Tregs gain a Th2 skewed Teff-like function, which is consistent with immune dysregulation observed in these patients. Analyses of FOXP3 mutation-carrier mothers and a patient with IPEX after hematopoietic stem cell transplantation, indicated that Tregs expressing non-mutated FOXP3 prevent the accumulation of autoreactive Teffs and unstable Tregs. These findings could be directly used for diagnostic and prognostic purposes and for monitoring the effects of immunomodulatory treatments.
One Sentence Summary:
Autoreactive T cells in patients with IPEX syndrome originate from both effector T cells and unstable, Th2-skewed regulatory T cells.
INTRODUCTION
Autoimmunity occurs when immune cells fail to distinguish “self” from “non-self” and initiate immune reactions against their autologous tissues. To limit T cell autoreactivity, developing T cells are selected in the thymus by medullary thymic epithelial cells (mTECs), B cells, and dendritic cells, which express and present self-antigens to T cells (1), a mechanism referred as central T cell tolerance. The expression of tissue-restricted self-antigens (TRAs) in mTECs is regulated by the autoimmune regulator (AIRE). Autoreactive T cells that recognize self-antigens with high affinity are eliminated (negative selection), whereas T cells that recognize self-peptides presented by major histocompatibility complexes (MHCs) with low affinity are positively selected and become effector T cells (Teffs). Alternatively, some T cells with intermediate to high affinity to self-antigens may be positively selected and adopt a unique developmental fate by differentiating into regulatory T cells (Tregs). Hence, the T cell receptor (TCR) repertoire of Tregs is physiologically self-reactive (2–4). However, the process of negative selection is not 100% accurate. Some autoreactive Teffs escape negative selection and enter the periphery (5–7). To prevent autoimmunity, these autoreactive T cells are controlled by various regulatory mechanisms collectively referred to as peripheral tolerance.
Two human monogenic diseases with autoimmunity exemplify the importance of central and peripheral tolerance. Mutations in AIRE result in failed T cell selection and development of autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy syndrome (APECED), manifested by progressive autoimmune targeting of various tissues (8). Analyses of the Treg and Teff TCR repertoires of these patients indicated that some of the common Treg TCR clones were present within the Teff compartment, indicating an altered distribution of the self-reactive T cells as a consequence of aberrant selection (9). Mutations in FOXP3, a transcription factor essential for Treg function, lead to the prototypic example of Treg deficiency, called immune dysregulation polyendocrinopathy enteropathy X-linked syndrome (IPEX), a severe, early-onset, multiorgan, and life-threatening autoimmunity (10). However, the presence of autoreactive T cells and their lineage origin in patients with IPEX has not been characterized, representing an important gap in understanding the role of FOXP3 and Tregs in controlling autoreactive T cells in humans.
Evaluation of the epigenetic landscape of the Treg-specific demethylated region (TSDR) is the best approach to define thymus-derived Tregs since demethylation of the TSDR is unique to FOXP3-expressing Tregs and distinguishes them from Teffs that can transiently express FOXP3 upon activation (11). Surprisingly, patients with IPEX have an increased frequency of TSDR-demethylated cells relative to CD4+ T cells in whole blood samples (12, 13), although the frequency of Tregs measured by their standard phenotypic markers CD3+CD4+CD25high and FOXP3+, or CD127−, or both FOXP3+ CD127− ranges from low to normal (14–17). This discrepancy led us to hypothesize that Tregs from patients with IPEX are unstable or plastic and expand beyond the phenotypic Treg compartment. Indeed, data mainly from murine studies revealed that defects during Treg development or inflammatory conditions may give rise to unstable or plastic Tregs often referred as “wannabe” or “ex” Tregs, which originate from physiologically self-reactive Tregs but may acquire Teff-like phenotype (18–24). These data called for human studies of Treg plasticity in samples from patients with IPEX to unravel whether some phenotypical Teffs have a Treg origin, what Teff phenotype Tregs may acquire, and how Treg plasticity or instability may affect TCR repertoire autoreactivity.
Here, we show that the Teff compartment of patients with IPEX, unlike that of patients with APECED or healthy donors (HDs), contains TSDR-demethylated cells demonstrating the presence of a population that displays a Treg lineage marker but does not express CD25, FOXP3, or both CD25 and FOXP3, two molecules normally expressed by Tregs. We named this population “loss-of-identity” Tregs and characterized, at the single cell-level, their transcriptome, TCR repertoire, and protein expression. We show that the presence of loss-of-identity Tregs in vivo is prevented by healthy Tregs expressing unmutated FOXP3. Moreover, Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/Cas9-mediated FOXP3 knockout (FOXP3KO) HD Tregs and IPEX Tregs showed proinflammatory, and Th2-effector functions, recapitulating the immune dysregulation observed in patients with IPEX. Together with the TCR repertoire analyses in phenotypically and epigenetically defined T cell subsets, our data indicate that loss of FOXP3 function in humans leads to increased autoreactivity in Teffs and expansion of Tregs with Teff-like phenotype, thereby suggesting an expansion of two sources of autoreactive T cells in patients with IPEX.
RESULTS
The Teff compartment of patients with IPEX contains TSDR-demethylated cells and a fraction of the TSDR-demethylated cells is FOXP3 negative.
This study was performed on samples from patients with IPEX, mutation carrier mothers, and patients with APECED. Mutations, age, and sex of participants included in the study are summarized in table S1. To investigate Treg plasticity in patients with IPEX, we measured the frequency of TSDR-demethylated cells in sorted CD3+CD4+ T cell subsets with different expression of CD25 and CD127, the relevant receptors for interleukin (IL)-2 and IL-7 differentially regulating Treg and Teff homeostasis (25). To this aim, two subpopulations of Teffs were sorted as CD25−CD127low and CD25-/dimCD127high, hereafter named Teff1 and Teff2, respectively (Fig. 1A). In parallel, CD25highCD127low Tregs were isolated, and shortly expanded in vitro to obtain a sufficient amount of DNA for the analyses. The total frequencies of Teff1s, Teff2s, and Tregs were not different between HDs and patients with IPEX (Fig. 1B), and neither were frequencies of CD4+ T cells (Fig. 1C). To assess TSDR demethylation, we used a finely optimized quantitative polymerase chain reaction (qPCR) assay with primers and probes specific for the demethylated TSDR sequence, and thus, instead of analyzing the degree of demethylation in bulk population, this assay determines the frequency of the cells with fully or largely demethylated TSDRs (11, 26). Results show that, unlike in HDs, a significant (p<0.0001) fraction of IPEX Teff1s was TSDR-demethylated (Fig. 1D, fig. S1). The presence of TSDR-demethylated cells in Teff2 did not reach statistical significance (Fig. 1D, p=0.0582) As expected, the sorted CD25highCD127low Tregs of both patients with IPEX and HDs contained almost entirely TSDR-demethylated cells (Fig. 1D). These results indicate that T cells with this Treg epigenetic marker were found among the CD25-negative cell population in samples from patients with IPEX.
Figure 1. The Teff compartment of patients with IPEX contains TSDR-demethylated cells and a fraction of them are FOXP3−.
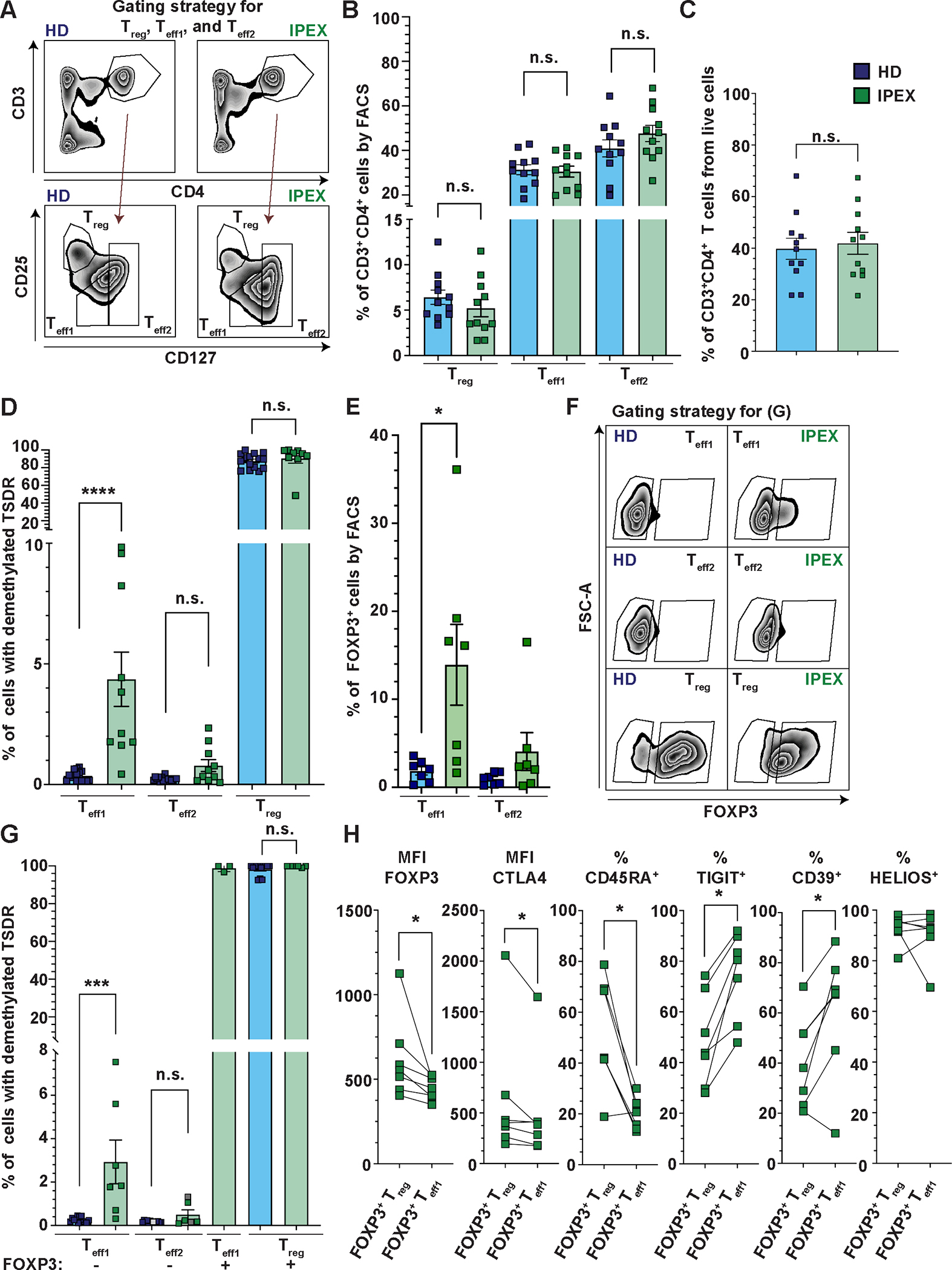
(A) Representative image of a gating strategy to sort and analyze the frequencies of Teff1, Teff2, and Treg subpopulations. (B and C) Quantification of the frequencies of Teff1s, Teff2s, and Tregs (B) and CD3+CD4+ T cells (C) from PBMC of 11 patients with IPEX (green) and 11 HDs (blue). (D) Analysis of the frequencies of TSDR-demethylated cells in sorted Teff1s, Teff2s, and Tregs of 10 patients with IPEX and 16 HDs. Cells were sorted as shown in (A). (E) Analyses by FACS of FOXP3 expression in Teff1 and Teff2 populations in 7 patients with IPEX. The FACS gating strategy is shown in fig. S2A. (F) Representative FACS plots show the gating strategy used to sort FOXP3− Teff1s and Teff2s and FOXP3+ Teff1s and Tregs from patients with IPEX and HDs for subsequent TSDR demethylation analysis. The Teff1s, Teff2s, and Tregs were gated as shown in (A). (G) Quantification of the frequency of TSDR-demethylated cells in FOXP3− Teff1s and Teff2s, and FOXP3+ Teff1s and Tregs in 7 (3 for FOXP3+ Teff1s) patients with IPEX and 13 HDs. Data points shown in gray (IPEX Teff2s) represent samples with a low cell number or genome copies, where the values may be partially imprecise. (H) FACS analyses (Median Fluorescence Intensity, MFI, and percentages, %) of FOXP3, CTLA4, CD45RA, TIGIT, CD39, and HELIOS expression in FOXP3+ Tregs and FOXP3+ Teff1s in 7 patients with IPEX. The data are analyzed in a pairwise fashion comparing the respective populations within the same patients. Data are presented as mean ± standard error of the mean (SEM). Significance was evaluated using the Mann-Whitney test (B to G) or Wilcoxon matched-pairs test (H). n.s., not significant; *p<0.05, ***p<0.001, ****p<0.0001.
The vast majority of IPEX-causing mutations do not abolish FOXP3 expression but diminish FOXP3 function (13, 15). However, FOXP3+ cells were detected mainly in Teff1 compartment of patients with IPEX (Fig. 1E and fig. S2A). The results from this fluorescence-activated cell sorting (FACS) analysis were an important part of the initial rationale for further characterizing the Teff1 and Teff2 compartments (fig. S2B), and to test if i) the FOXP3+ Teff1s are TSDR demethylated and ii) the TSDR-demethylated cells are present also among FOXP3− Teffs. To this aim, we analyzed the frequency of TSDR-demethylated cells in the sorted FOXP3− Teff1, FOXP3− Teff2, FOXP3+ Teff1, and FOXP3+ Treg subsets (patients with IPEX: n=7 and HDs: n=13) (Fig. 1F and G, fig. S2C). We found that FOXP3+ Teff1s are comprised almost exclusively of TSDR-demethylated cells and that TSDR-demethylated cells are also present among FOXP3− Teff1s in patients with IPEX, but not in HDs. Compared with phenotypic Tregs, the FOXP3+ Teff1s are predominantly CD45RA−, express lower FOXP3 and cytotoxic T-lymphocyte-associated protein 4 (CTLA4), but express more CD39 and TIGIT (Fig. 1H, fig. S2A and D). Both phenotypic Tregs and FOXP3+ Teffs expressed Helios. In summary, these analyses suggest that the FOXP3+ Teff1s are antigen-experienced Tregs that downregulated FOXP3 expression. Collectively, our data show that in patients with IPEX, a fraction of the epigenetically-defined Tregs do not express CD25, have reduced or no expression of FOXP3, and some gain CD127 expression.
Characterization of Treg subpopulations using single-cell multi-omic profiling of CD4+ T cells from patients with IPEX revealed a disease-specific population of Tregs that is dominated by CD25− cells.
To gain further insight into Treg heterogeneity in samples from patients with IPEX, we performed single-cell multi-omic RNA, protein, and TCR profiling using the 10X genomics platform. We assayed CD4+ T cells isolated from three HDs and three patients with IPEX. After data filtering, we obtained 17,865 T cells from HDs and 17,566 T cells from patients with IPEX, relatively evenly distributed amongst the samples (Fig. 2A). Cells were clustered and visualized using uniform manifold approximation and projection (UMAP) according to their transcriptomics (Fig. 2B). Clusters were annotated according to their differential expression of key genes (Fig. 2C) and the expression of CD45RA protein (based on sequencing of antibody derived tags [ADT]) (fig. S3A). We identified the following populations: cytotoxic (GZMK+, GZMA+), Th1/Cytotoxic (TBX21+, GZMB+), Th2 (GATA3+), Th17 (RORC+, CCR6+), naïve (CD45RA+ [ADT]), memory (adt_CD45RA−), Treg naïve (FOXP3+, IKZF2+, CD45RA+ [ADT]), and two populations of memory Treg (FOXP3+, IKZF2+, CD45RA−) (Fig. 2B and C, fig. S3A). The Treg memory 1 population included cells from both HDs and patients with IPEX, whereas the Treg memory 2 population was almost exclusively composed of cells isolated from patients with IPEX (Fig. 2D). The Treg memory 2 population was also among the most frequent memory T cell populations in these patients. TCR analyses demonstrated that Treg memory 1 and Treg memory 2 were clonally related, suggesting a common origin of these populations (Fig. 2E and fig. S3B). Compared with Treg memory 1, Treg memory 2 cells demonstrated reduced expression of FOXP3 and IL2RA, but increased expression of IL7R, CD69, and ZPF36L2, which was previously shown to be a negative regulator of IKZF2 expression (27) (Fig. 2F, data file S1). In addition, the Treg memory 2 population showed differential expression of genes associated with tumor necrosis factor (TNF)-α signaling. Indeed, the enrichment in expression of TNF-α signaling target genes was confirmed by pathway enrichment analysis (Fig. 2G and fig. S3C). These data demonstrate that patients with IPEX have both typical (Treg memory 1) and atypical (Treg memory 2) memory Tregs. We observed that the disease-specific Treg memory 2 are clonally most related to typical Tregs, they display a gene expression signature indicative of TNF-α exposure, and demonstrate reduced mRNA expression of prototypical Treg markers.
Figure 2. Characterization of Treg subpopulations using single-cell multi-omic profiling of CD4+ T cells revealed a population of Tregs specific for patients with IPEX.
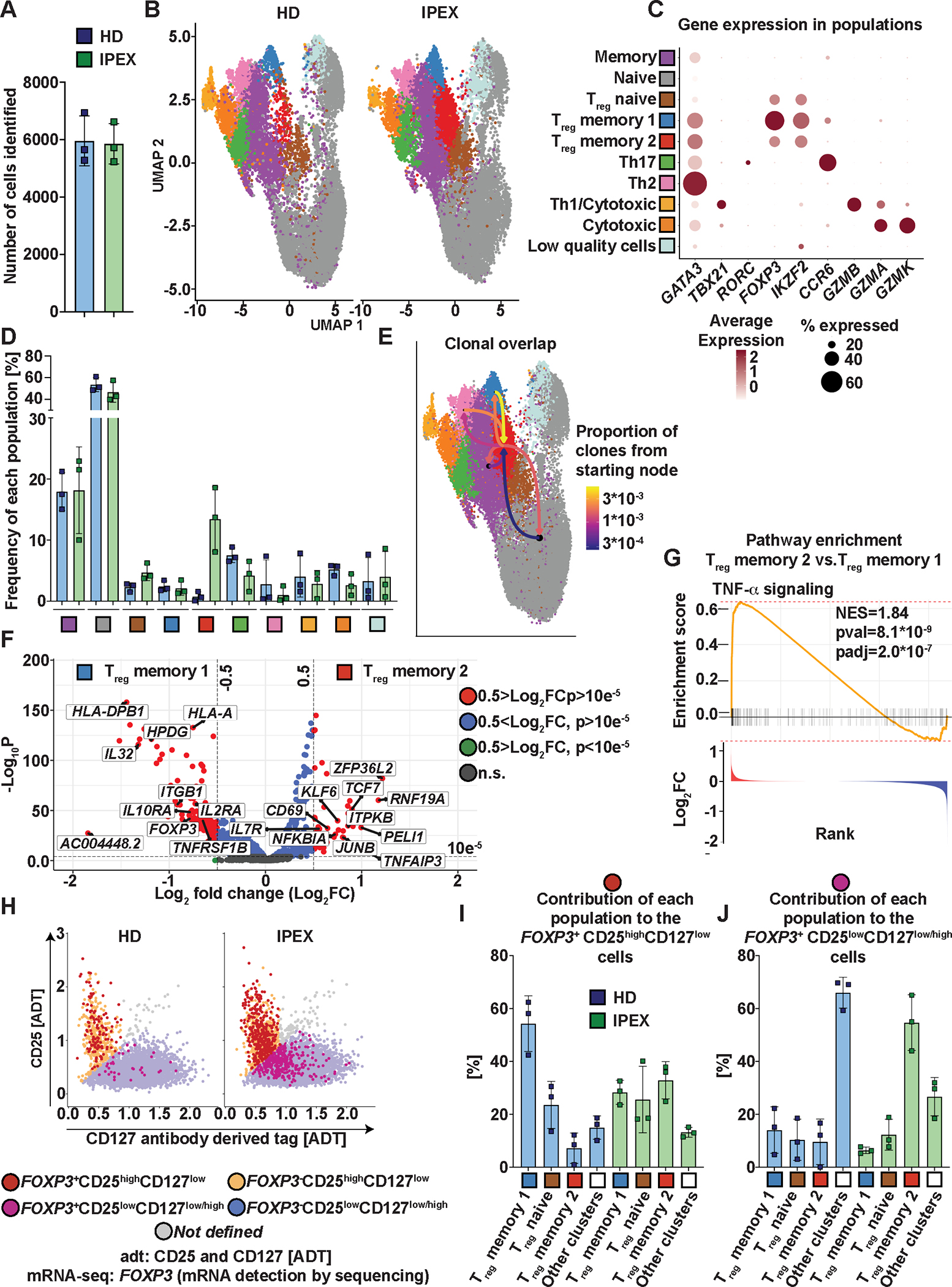
(A) Number of cells identified per donor. (B) UMAP projection of the T cell populations identified in HDs’ samples (left) and samples from patients with IPEX (right). (C) Expression of selected genes in the different populations identified by the specific color. The dot color intensity indicates the amount of expression and the size of the dot indicates the number of positive cells for a given gene in the population. (D) Distribution of the identified populations in the CD4+ T cells from HDs and patients with IPEX. The frequency of each population is defined as a percentage of cells present in the indicated populations from the total number of cells in a given sample. (E) TCR repertoire overlaps between Treg memory 2 populations and all other populations identified. The color of an arrow indicates the proportion of the overlapping clones between the populations. Arrows always start from a population to which the proportion is indicated. (F) Differential expression analyses between Treg memory 2 and 1. The relevant genes are indicated by a label. The genes with positive fold change (FC) are upregulated in the Treg memory 2 compared with the Treg memory 1. P values were calculated using the Wilcoxon rank sum test. n.s, not significant. (G) TNF-α signaling was identified as the most significantly differentially expressed gene set by fast gene set enrichment analysis (FGSEA) comparing Treg memory 2 and 1. Genes were ranked based on fold changes; the top panel shows the enrichment score and the bottom panel the fold changes of the genes. (H) Representative FeatureScatter plot of CD25 and CD127 expression using antibody-derived tag data [ADT]. The plot shows one IPEX and HD sample. The FOXP3 positivity was determined based on the mRNA expression. (I and J) Contribution of each T cell population from (B to D) to the total number of FOXP3+CD25highCD127low (I) and FOXP3+CD25lowCD127low/high (J) cells. FOXP3+CD25highCD127low and FOXP3+CD25lowCD127low/high cells were identified as shown in (H). “Other clusters” are pooled non-Treg populations from (B to D). Data are presented as mean ± standard deviation (SD).
To validate our cytometric findings in Fig. 1, we assessed CD127 and CD25 expression as determined using ADT in conjunction with FOXP3 transcript expression (Fig. 2H). In both HDs and patients with IPEX, we observed a high frequency of FOXP3+ cells among the CD25highCD127low cells (fig. S3D). In contrast to HDs, where the FOXP3+ cells made up a minor fraction of the CD25lowCD127low/high, about 6% of all CD25lowCD127low/high cells in patients with IPEX were FOXP3+ (fig. S3E), recapitulating our cytometric findings. Next, we assessed how the FOXP3+ CD25lowCD127low/high and the FOXP3+ CD25highCD127low cells were distributed across the different transcriptionally-defined Treg populations. The FOXP3+ CD25highCD127low cells from both HDs and patients with IPEX associated primarily with the Treg memory 1 and Treg naive population (Fig. 2I). However, about a third of the FOXP3+ CD25highCD127low cells of patients with IPEX were also present in the Treg memory 2 populations. FOXP3+ CD25lowCD127low/high cells, although infrequent in HDs (fig. S3E), were spread among the non-Treg populations in HDs, and in patients with IPEX were primarily associated with the Treg memory 2 population (Fig. 2J).
These data show that the atypical Treg memory 2 population is mainly composed of Tregs that have lost CD25 and gained CD127 expression. Consistent with the FOXP3+CD25lowCD127low/high cells displaying a Treg memory 2 transcriptional signature, when comparing FOXP3+CD25highCD127low versus FOXP3+CD25lowCD127low/high from patients with IPEX we observed a similar set of differentially expressed genes (fig. S3F, data file S2) and enrichment pathways (fig. S3G and H).
TCR repertoire analyses indicate increased autoreactivity in CD4+ T cells from patients with IPEX.
We analyzed the TCR repertoires in CD4+ T cells from patients with IPEX to potentially illustrate the FOXP3-dependent role of Tregs in controlling autoreactive T cell expansion in humans in vivo. We performed DNA sequencing of TCRB of Teff1s, Teff2s, and Tregs from seven patients with IPEX and ten HDs. In total, we identified 5.4×105 and 7.4×105 Teff1, 5.1 ×105 and 6.9×105 Teff2, and 1.7 ×105 and 2.3 ×105 Treg TCR sequences from patients with IPEX and HDs, respectively. The number of sequences identified per sample for each subpopulation was well-balanced between the HDs and patients with IPEX (Fig. 3A). The fraction of productive rearrangement did not show a significant difference in all three cell populations of patients with IPEX as compared with those of HDs (Tregs p=0.2698, Teff1s p=0.3148, Teff2s p=0.4747), and the overall Simpson clonality index was largely normal (Fig. 3B and C). We did not find any major bias in TCRB gene usage (fig. S4). Alteration of the TCR repertoires of Teff1s and especially Teff2s from patients with IPEX were evidenced by their different distributions of CDR3ß length compared with their counterparts in HDs. Indeed, whereas Tregs from both patients with IPEX and HDs display a similar CDR3ß length distribution, Teff1s and Teff2s in patients with IPEX were enriched in clones with longer CDR3ß loops (Fig. 3D). To investigate potential TCR autoreactivity, we analyzed the presence of autoreactivity promoting and limiting amino acid (AA) doublets at positions 6 and 7 of the CDR3ß loop. These AA doublets are TCR features associated with negative and positive T cell selection, respectively, and were suggested to be a mean for distinguishing normal versus autoimmunity-prone T cell repertoires (3, 4, 28, 29). We found increased frequencies of the autoreactivity promoting AA doublets in the TCR repertoires of both Teff1s and Teff2s from patients with IPEX (Fig. 3E). Consistently, we found a significantly reduced frequency of autoreactivity limiting AA doublets in Teff2s (p=0.0431, Fig. 3F). We conclude that the differential length usage of CDR3ß, increased frequency of autoreactivity AA promoting doublets, and reduced frequency of autoreactivity limiting AA doublets in Teffs from patients with FOXP3 mutation indicate increased autoreactivity in the patients’ Teff compartments. Although Teff1 is composed of both TSDR-demethylated and bona fide Teff, Teff2 contains only a negligible number of TSDR-demethylated cells. Therefore, the abnormal TCR repertoire in Teff2 is consistent with the idea that increased autoreactivity originates from bona fide Teffs.
Figure 3. TCR analyses indicate increased TCR repertoire autoreactivity in patients with IPEX.
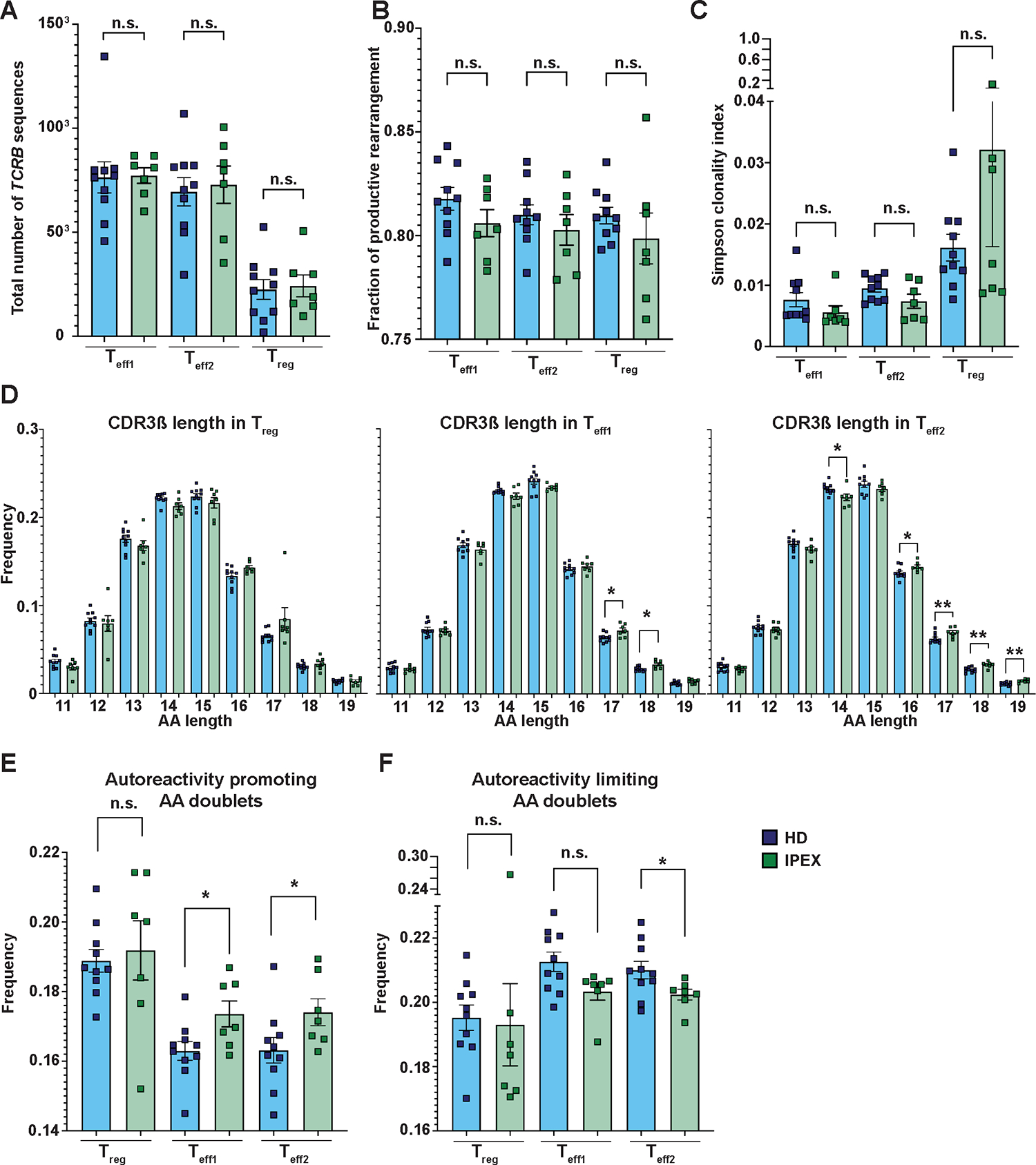
(A) Shown is the number of TCRB sequences that were identified in Teff1s, Teff2s, and Tregs from patients with IPEX (green) and HDs (blue). (B) Shown is the fraction of TCRB productive rearrangements for Teff1s, Teff2s, and Tregs from patients and HDs. (C) Shown is the Simpson clonality index for Teff1s, Teff2s, and Tregs from patients with IPEX and HDs. (D) Analysis of CDR3ß length of Tregs (left), Teff1s (middle), and Teff2s (right) from patients with IPEX and HDs. (E and F) Frequency of autoreactivity promoting (E) and autoreactivity limiting (F) AA doublets in CDR3ß sequences in Teff1, Teff2, and Treg populations from patients with IPEX and HDs. TCRB sequencing was performed in all three T cell subsets of 10 HD and 7 IPEX patients (A to F). Analyses in (C to G) were performed from productive rearrangements. Data are presented as mean ± SEM. Statistical significance was evaluated using the Mann-Whitney test; n.s., not significant; *p<0.05, **p<0.01.
In addition, we found a positive correlation between the frequency of TSDR-demethylated cells and the frequency of autoreactivity promoting AA doublets in Teff1s, suggesting that TSDR-demethylated cells in the Teff1 compartment contribute to changes in Teff1 TCR repertoire, indicative of increased autoreactivity (Fig. 4A). There was no significant correlation between the frequency of TSDR demethylated cells in Teff2 compartment and frequency of autoreactivity promoting AA doublets in Teff1 compartment (p=0.2357, Fig. 4A), and neither between the frequency of TSDR-demethylated cells and the presence of autoreactivity promoting AA doublets within Teff2 compartment (p=0.1667, Fig. 4B). However, we found a positive correlation between the presence of TSDR-demethylated cells in Teff1 and the frequency of autoreactivity promoting AA doublets in Teff2 (p=0.0341, Fig. 4B). These data suggest that the presence of TSDR-demethylated cells in Teff1 is predictive of an abnormal TCR repertoire that may be associated with increased autoreactivity of the Teff2 compartment composed mostly of bona fide Teffs. Hence, the loss-of-identity Treg population may favor the expansion of autoreactive Teffs.
Figure 4. Frequencies of TSDR-demethylated cells correlate with increased frequencies of autoreactivity promoting AA doublets in Teff compartments of patients with IPEX.
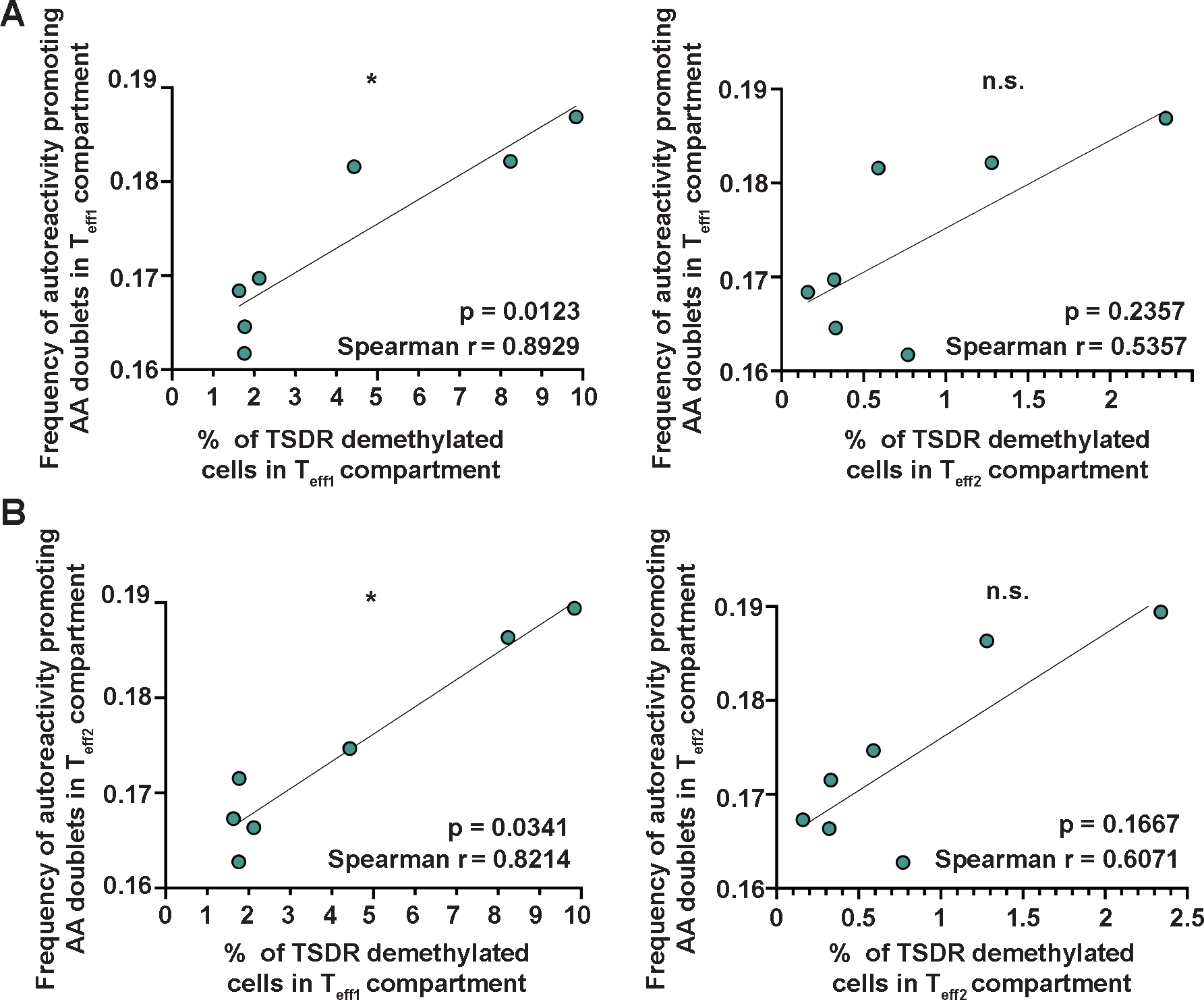
(A) Correlation between % of TSDR-demethylated cells in Teff1s (left) and Teff2s (right), and frequency of autoreactivity promoting AA doublets in Teff1s. (B) Correlation between % of TSDR-demethylated cells in Teff1s (left) and Teff2s (right), and the frequency of autoreactivity promoting AA doublets in Teff2s. Data were analyzed using Spearman correlation; n.s., not significant; *p<0.05. The p-values, Spearman r, and a simple linear regression are shown (n=7).
Mothers of patients with IPEX carrying FOXP3 mutations have a normal distribution of TSDR-demethylated cells and a normal TCR repertoire.
The FOXP3 gene is located in the X chromosome. As a consequence, in carrier mothers of FOXP3 mutations, random chromosome X inactivation results in the expression of mutated FOXP3 in 50% of Teffs (30, 31). In contrast to Teffs, the majority of carrier mothers’ CD25high Tregs express the non-mutated FOXP3 allele (30, 32). Therefore, unlike in patients with IPEX, the Tregs in the healthy mothers are functional and able to control the partially mutated Teffs. However, it is unknown if i) some FOXP3-mutated TSDR-demethylated cells are present in Teff compartment, and ii) carrier mothers have abnormal TCR repertoires indicative of increased autoreactivity in the Teffs. We sorted Teff1s, Teff2s, and Tregs from five carrier mothers and from five sex-matched HDs. We did not find TSDR-demethylated cells in any of the Teff cell subsets tested (Fig. 5A). These results demonstrate that in carrier mothers, as well as in HDs, in which functional Tregs are present, TSDR-demethylated cells are restricted to Treg compartment.
Figure 5. IPEX carrier mothers have a normal distribution of TSDR-demethylated cells and TCR repertoires of Teff2, and the presence of donor-derived cells prevents expansion and normalizes the distribution of TSDR-demethylated cells in a patient with IPEX post HSCT.
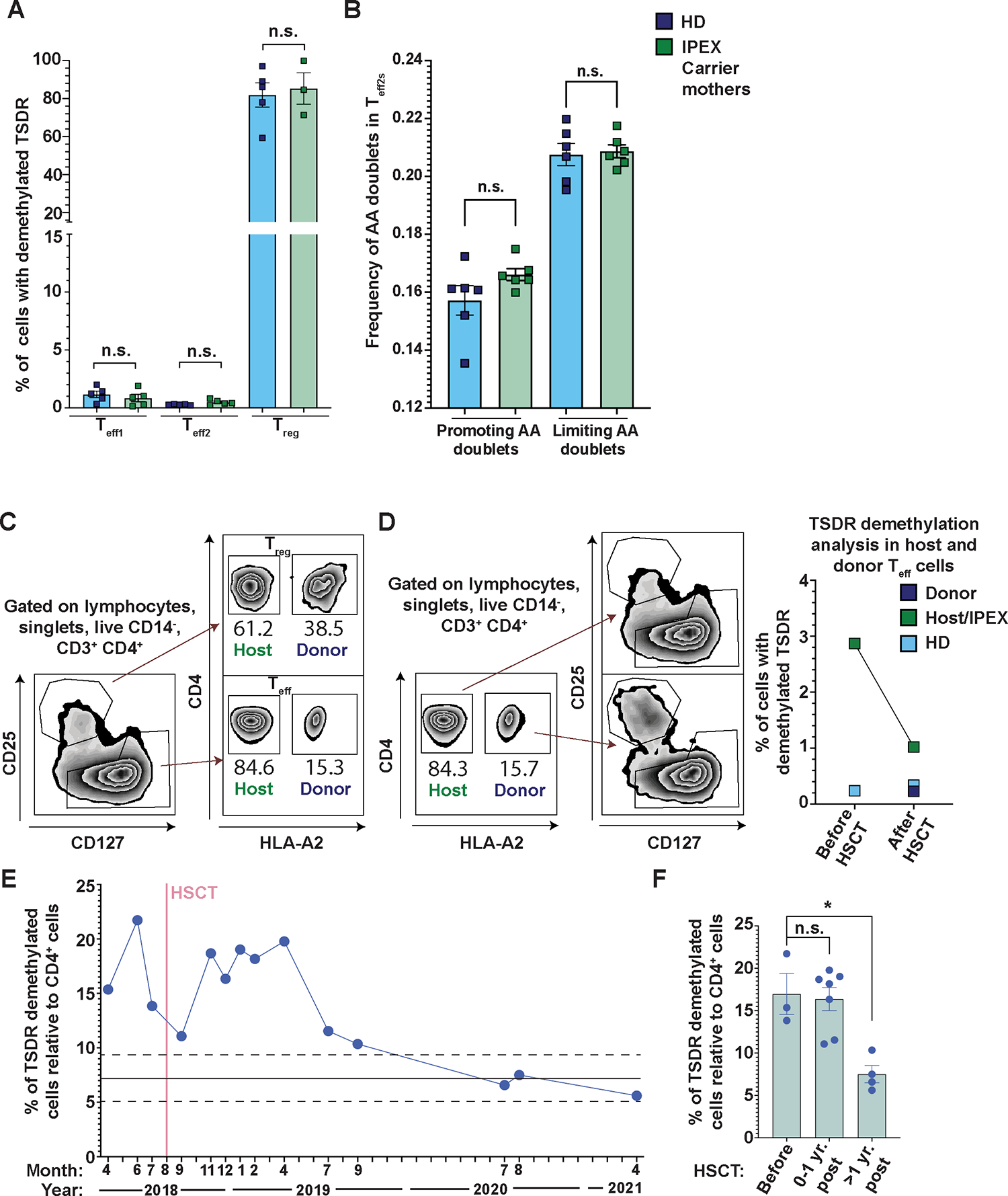
(A) Shown are the frequencies of TSDR-demethylated cells from sorted Teff1s, Teff2s, and Tregs from 5 FOXP3 mutation carrier mothers (green) and 5 sex-matched HDs (blue). The sorting strategy was the same as in Fig. 1A. (B) Frequency of autoreactivity promoting and limiting AA doublets in CDR3ß repertoire of Teff2 cells from 6 FOXP3 carrier mothers and 6 sex-matched HDs. Analysis in (B) was performed from productive rearrangements. (C) FACS analysis of chimerism 2.5 years post-transplantation in a patient with IPEX shows a competitive advantage of healthy donor-derived Tregs over patient-derived Tregs. Donor-derived cells are HLA-A2+ and host-derived cells are HLA-A2−. (D) TSDR demethylation analysis of sorted host- and donor-derived Teffs from the patient with IPEX before and 2.5 years post-transplantation. In addition, HD Teffs were sorted in parallel as a control (light blue). (Left panel) Gating strategy. (Right panel) Quantification of the TSDR-demethylated cell frequencies in the sorted populations. (E) TSDR demethylation analysis from whole blood at various time points pre- and post-transplantation. The frequencies of TSDR-demethylated cells were normalized to % of CD4+ T cells also determined by epigenetic analyses. The mean and SD of HD (previously published data (13); for each value see data file S4) are depicted as full and dashed lines, respectively. (F) Quantification of TSDR:CD4 ratio before transplantation, less than one year post- transplantation, and more than one year post-transplantation. Data are presented as mean ± SEM. Data in (F) were analyzed using Dunn’s multiple comparisons test and in (A and B) using Mann-Whitney test; n.s., not significant; *p<0.05.
We sequenced TCRB repertoire of Teff2s from six carrier mothers and six sex-matched HDs (Fig. 5B). We did not find any differences in the frequencies of autoreactivity promoting and limiting AA doublets in carrier mothers as compared with HDs. These data further support that the aberrant TCR repertoire in Teffs of patients with IPEX mainly originates from the expansion of autoreactive TCR clones owing to a loss of Treg function, rather than resulting from a FOXP3 cell-intrinsic effect in Teffs.
Decreased frequency of TSDR-demethylated cells in Teff were observed in a patient with IPEX after hematopoietic stem cell transplantation, despite low donor chimerism.
We analyzed the frequency of TSDR-demethylated cells in the Teffs of a patient with IPEX prior to and up to 2.5 years post-hematopoietic stem cell transplantation (HSCT), which resulted in mixed donor chimerism. Patient- and the donor-derived cells could be distinguished using FACS by the differential expression of human leukocyte antigen (HLA)-A2. The patient donor chimerism 2.5 years post-transplantation was only about 15% in Teff compartment and about 38% in Treg compartment (Fig. 5C). Despite the relatively low chimerism at this time post-HSCT in both Tregs and Teffs, we observed a three-fold reduction in the frequency of TSDR-demethylated cells in the host-derived Teffs as compared with the pre-HSCT sample (Fig. 5D). In addition, the kinetics of the TSDR demethylation in whole blood showed that the proportion of TSDR-demethylated cells relative to the frequencies of CD4+ T cells significantly decreased over time (p=0.0498), reaching normal values one-year post-HSCT (Fig. 5E and F). These data suggest that the presence of even a small proportion (about 40%) of the donor-derived Tregs prevents the expansion of FOXP3 mutated Tregs and the accumulation of TSDR-demethylated cells in Teff compartment.
In patients with APECED, TSDR-demethylated cells are restricted to the Treg cell subset but TCR repertoire analyses indicate increased autoreactivity.
To get insight into the potential thymic origin of TSDR-demethylated cells in Teffs, we analyzed TSDR demethylation in Tregs and Teffs from patients with APECED, in which a T cell-extrinsic defect in thymic selection leads to impaired Treg commitment and presence of clones that were meant to be Tregs within the Teff pool (9). We sorted Teff1s, Teff2s, and Tregs from three patients with APECED and five HD or carrier relatives, if available. In contrast to patients with IPEX, TSDR-demethylated cells were not detected in the Teffs of patients with APECED, whereas APECED Tregs were exclusively comprised of TSDR-demethylated cells (Fig. 6A). In line with the aberrant Teff and Treg selection, analyses of previously reported TCR sequencing data (9) together with newly generated TCRB sequencing data (total six patients with APECED and six HD) showed an increased frequency of autoreactivity promoting AA doublets in both Teffs and Tregs of patients with APECED as compared with the HDs (Fig. 6B). The frequency of autoreactivity limiting AA doublets in Teffs was also reduced (Fig. 6B). Hence, although patients with APECED present with abnormal TCR repertoire consistent with increased Teff autoreactivity, the Treg lineage commitment in AIRE deficiency does not result in the aberrant presence of TSDR-demethylated cells in the Teff compartment. These results, therefore, suggest that the presence of TSDR-demethylated cells in Teff compartment of patients with IPEX specifically results from defective FOXP3 function.
Figure 6. Patients with APECED have a normal distribution of TSDR-demethylated cells, but an increased and reduced frequency of autoreactivity promoting and limiting AA doublets, respectively, in Treg and Teff compartments.
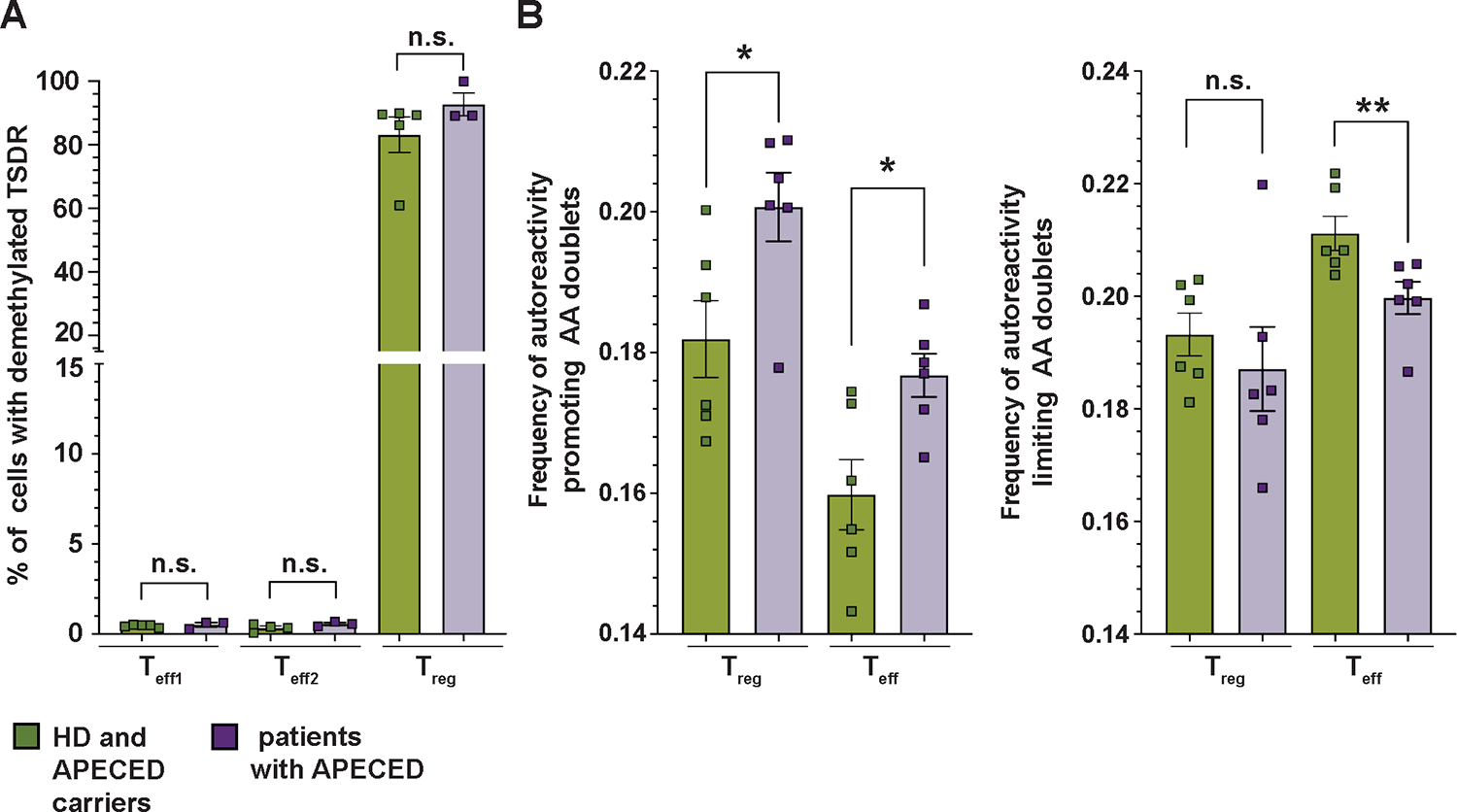
(A) Frequencies of TSDR-demethylated cells in sorted Teff1, Teff2, and Treg populations from 3 patients with APECED (purple) and 5 HDs (green), including 4 parents and 1 HD. (B) Analysis of TCR receptor autoreactivity from previously reported TCR sequencing data of four patients with APECED and four HDs (9) together with newly obtain CDR3ß sequences from two patients with APECED syndrome and two HDs. (Left graph) Frequency of autoreactivity promoting AA doublets in CDR3ß of Tregs and Teffs. (Right graph) Frequency of autoreactivity limiting AA doublets in CDR3ß of Tregs and Teffs. Analyses were done from productive rearrangements. Data are presented as mean ± SEM. Data were analyzed using the Mann-Whitney test. n.s., not significant; *p<0.05, **p<0.01.
TCR-stimulated FOXP3KO Tregs from HDs and Tregs from patients with IPEX showed greater production of proinflammatory cytokines compared to controls.
Since patients with IPEX have TSDR-demethylated cells present in Teff compartment and a fraction of these cells do not express FOXP3, we investigated the Teff function of Tregs following the loss of FOXP3 expression. These experiments serve as a model to study FOXP3-deficient Tregs and to identify cytokines that are the most affected by the loss of FOXP3 expression. We generated FOXP3KO Tregs from HD Tregs as described previously (33). Briefly, we used CRISPR/Cas9 technology combined with Adeno-Associated Virus Type 6 (AAV6) delivery of FOXP3 homologous template containing truncated nerve grow factor receptor (NGFR) reporter gene under the control of PGK promoter. FOXP3KO Tregs were purified using NGFR (Fig. 7A). Control Tregs were treated as the FOXP3KO but electroporated in the absence of GuideRNA/Cas9 and not treated with AAV6. Upon TCR-mediated activation, we observed an enhanced expansion of FOXP3KO Tregs (Fig. 7B) and a slight but significant increase in the proportions of TSDR-demethylated cells in FOXP3KO over control Tregs (p=0.0244, Fig. 7C), which is in line with the increased proliferative capacity in the absence of FOXP3 and confirms high purity of the FOXP3KO Tregs. Of note, analysis of CD25 and CD127 expression is not reliable in Treg cultures due to IL-2 media supplementation. To assess Teff-like function in FOXP3KO Tregs, we collected the supernatant after the 3-day expansion and analyzed the production of cytokines using a multiplex Luminex assay. Data were normalized to the control:FOXP3KO cell ratio at the end of the experiment, taking into account the higher expansion of the FOXP3KO Tregs. This normalization may favor the control Tregs. We observed increased production of IL-13, IL-17, TNF-α and granulocyte-macrophage colony-stimulating factor (GM-CSF) by the FOXP3KO Tregs (Fig. 7D and fig. S5), suggesting that FOXP3-deficient Tregs can confer Teff-like function, better expand in response to TCR stimulation, and produce Th2 and Th17 cytokines. To exclude non-specific effects of AAV6 or GuideRNA/Cas9, we repeated the experiments with control Tregs and Tregs treated with AAV6 and scramble GuideRNA/Cas9. The data indicate that the above-described differences between FOXP3KO Tregs and control Tregs were specifically caused by loss of FOXP3 expression (fig. S6). Next, we examined ex vivo stimulated Tregs and Teffs from patients with IPEX and HDs for the production of IL-13, IL-17, TNF-α, and GM-CSF, which we found to be controlled by FOXP3. Unlike HDs, phenotypic FOXP3+CD25+CD127− Tregs from patients with IPEX produced GM-CSF and IL-13 (Fig. 7E and F and fig. S7). These data indicate that, in contrast to HDs, even the phenotypic Tregs from patients with IPEX can acquire a Teff phenotype to some extent and participate in the inflammation and polarization of immune response towards a Th2 phenotype.
Figure 7. FOXP3KO Tregs exhibit greater expansion and increased production of proinflammatory cytokines in response to TCR stimulation in vitro, and TCR stimulated Tregs from patients with IPEX exhibit increased proinflammatory cytokine production ex vivo.
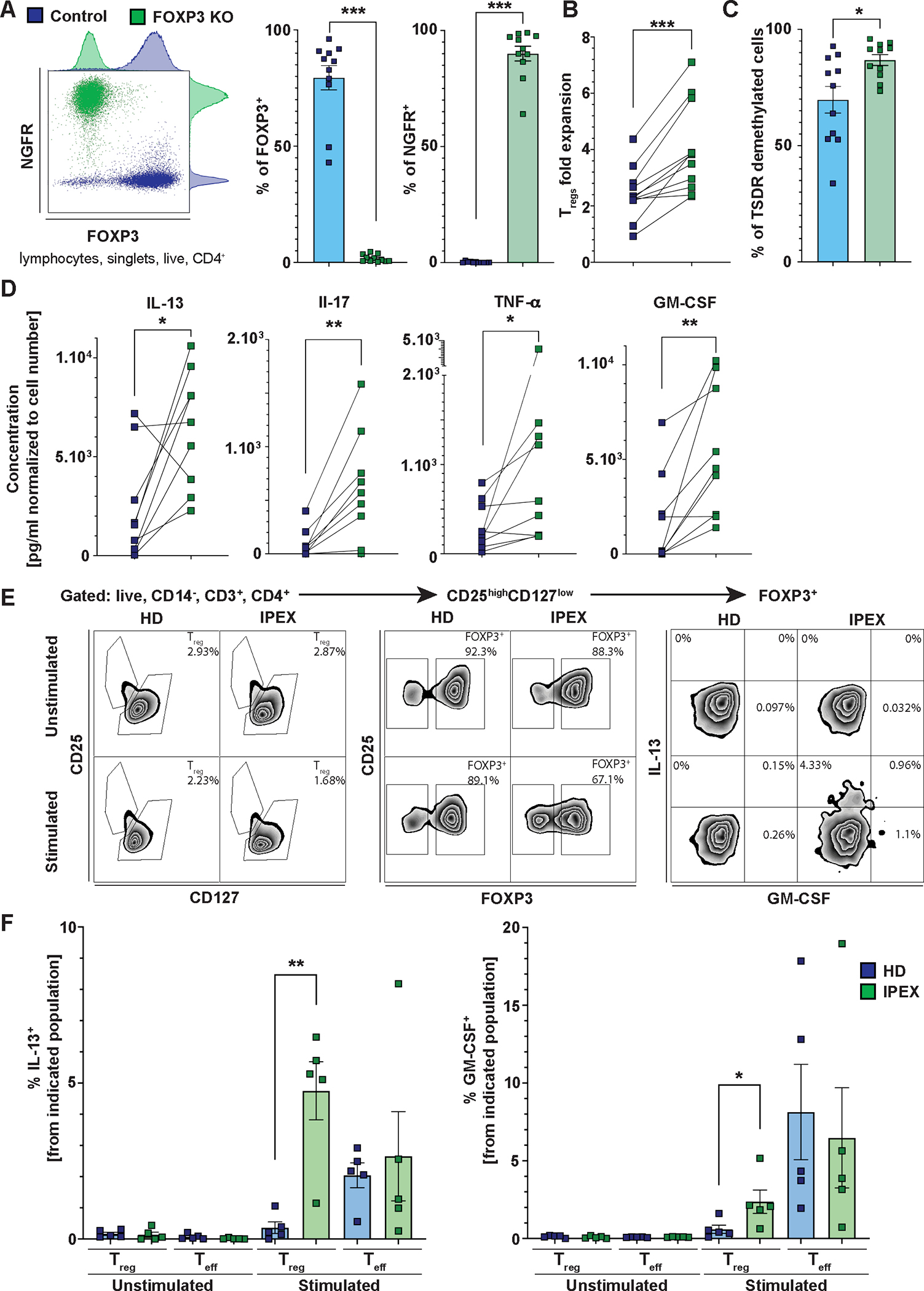
(A) Representative FACS plot (left), quantification of FOXP3 expression (middle), and quantification of NGFR expression (right) in FOXP3KO and control Tregs from 11 HDs. (B) Treg expansion upon CD3/CD28 stimulation. Cells were counted 3 days post anti-CD3 and anti-CD28 stimulation. The data are plotted as a fold increase of the cell number from day 0. Lines connect control and FOXP3KO Tregs from the same donor. (C) Frequency of TSDR-demethylated cells in control and FOXP3KO Tregs. (D) Analysis of cytokine production from supernatants of control and FOXP3KO Tregs using a Luminex-based multiplexed assay. Only significantly upregulated or downregulated cytokines are shown. The cytokine concentrations were normalized to the ratio of control:FOXP3KO Treg numbers at the time of supernatant collection. (E) FACS analysis of proinflammatory cytokine production by Tregs from patients with IPEX and HDs. The representative gating strategy shows Brefeldin A- and Monensin-treated PBMCs from patients with IPEX and HDs unstimulated or stimulated with PMA and ionomycin for six hours. (F) Quantification of IL-13 and GM-CSF production by stimulated or unstimulated Tregs and Teffs from five HDs and five patients with IPEX. Representative gating strategy is shown in (E). Data are presented as mean ± SEM. Data in (A to D) were analyzed using the Wilcoxon matched-pairs test. Statistical significance in (F) was evaluated using the Mann-Whitney test. *p<0.05, **p<0.01, ***p<0.001.
Treg subpopulations isolated from patients with IPEX were characterized after TCR stimulation using single-cell multi-omic profiling.
We performed the RNA, protein, and TCR profiling in CD4+ T cells from the same samples as in Fig. 2, but we first stimulated the cells for 6 hours with anti-CD3 and anti-CD28 antibody-coated beads. We obtained data for 16,458 T cells from HD and 18,018 T cells from patients with IPEX (Fig. 8A). Once again, cells were clustered and visualized using UMAP according to their transcriptomics (Fig. 8B). Clusters were annotated based on mRNA expression of key genes (Fig. 8C), and CD45RA protein expression (based on ADT) (fig. S8A). We identified the following populations: Th1/Cytotoxic (TBX21+, GZMB+, GZMK+, GZMA+), Th2 (GATA3+), Th1/Th17 (RORC+, TBX21+), naïve (CD45RA+ [ADT]), memory (CD45RA− [ADT]), Treg naïve (FOXP3+, IKZF2+, CD45RA+ [ADT]), Treg memory 1 (FOXP3+, IKZF2+), Treg memory 2 (FOXP3+, IKZF2+), and Treg/Teff population (FOXP3low, IKZF2low) (Fig. 8B and C, fig. S8A). Similarly, to ex vivo sequenced cells, the TCR-stimulated Treg memory 2 cells were specific to patients with IPEX (Fig. 8D). To confirm that this stimulated population was analogous to the Treg memory 2 population in our ex vivo experiment, we analyzed the TCR overlap between ex vivo and stimulated populations (Fig. 8E). Indeed, we detected the highest TCR overlap between TCR-stimulated Treg memory 1 and ex vivo Treg memory 1 as well as between TCR-stimulated Treg memory 2 and ex vivo Treg memory 2 populations, thereby demonstrating the correspondence between the ex vivo and TCR stimulated populations. (Fig. 8E). Consistent with shared clonality between the Treg memory 1 and Treg memory 2 in the ex vivo analysis, we observed shared clonality between TCR-stimulated and ex vivo Treg memory 1 and Treg memory 2, and vice versa (Fig. 8E). For the TCR overlap between non-Treg populations, see fig. S8B. Differential expression analyses between Treg memory 1 and 2 showed downregulation of IL2RA, FOXP3 and CTLA4, and upregulation of GATA3, suggesting preferential polarization of the atypical Tregs to Th2-like phenotype (Fig. 8F, data file S3). Pathway analyses showed an increased expression of cMYC target genes in Treg memory 2 (Fig. 8G and fig. S8C). cMYC is known to be upregulated upon TCR stimulation in T cells(34, 35), thus the relative increase in expression of cMYC target genes in Treg memory 2 over Treg memory 1 may suggest higher responsiveness to TCR stimulation, which may potentially explain the superior in vitro expansion of FOXP3KO Tregs compared with FOXP3-competent Tregs (Fig. 7B). Consistent with the in vitro FOXP3KO Treg and ex vivo Treg data from patients with IPEX, we observed IL13 expressing Tregs in both Treg memory 1 and 2 compartments of patients with IPEX, and only a very limited number of IL13 expressing Tregs in HDs (Fig. 8H and I, fig. S8D). Importantly, Tregs from patients with IPEX accounted for 7 to 20% of all IL13 expressing CD4+ T cells, suggesting a substantial contribution of patients’ Treg to the IL-13 producing cell pool (Fig. 8J). We did not observe an increase in the production of other proinflammatory cytokines from our single-cell multi-omics experiments (fig. S8D and E); however, these data may be partially ascribed to differences in cytokine kinetics or limitations to sequencing approaches.
Figure 8. Characterization of Treg subpopulations post-TCR stimulation using single-cell multi-omic profiling revealed increased IL13 expression in Treg populations from patients with IPEX.
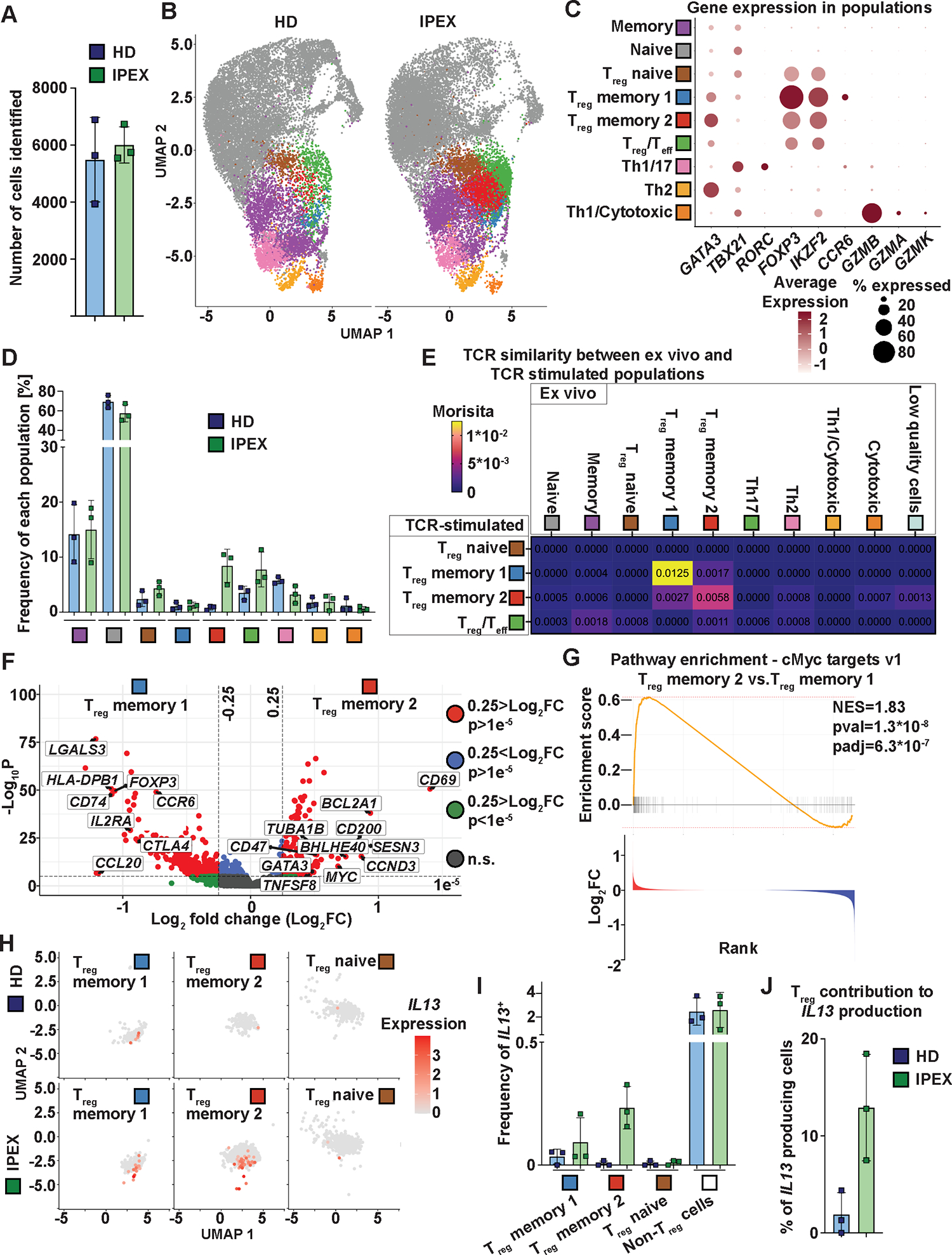
(A) Number of cells identified per donor. (B) UMAP projection of the T cell populations identified in HDs (left) and patients with IPEX (right). (C) Expression of selected genes in the populations. Color intensity indicates the amount of expression and the size of the dot indicates the percentage of positive cells. (D) Distribution of CD4+ T cells from HD and patients with IPEX in the identified populations. Frequency of each population is defined as a percentage of cells present in the indicated population from the total number of cells in a given sample. The same color code as in (B) and (C) indicates the population. (E) Morisita overlaps of TCR repertoires between TCR-stimulated populations: Treg naïve, Treg memory 1, Treg memory 2, and Treg/Teff populations, as well as all ex vivo populations identified in Fig. 2. (F) Differential expression analyses between Treg memory 2 and 1. The relevant genes are indicated by a label. The genes with positive fold change (FC) are upregulated in Treg memory 2 compared with Treg memory 1. HLA-DRB1 is not shown (p-value = 3.9E−162, log2 fold change = −2.6). n.s., not significant. (G) cMYC target genes were identified as the most significantly differentially expressed gene set by FGSEA comparing Treg memory 2 and Treg memory 1 populations. Genes were ranked based on fold changes; the top panel shows the enrichment score and the bottom panel the fold change of the genes. (H) Feature dot plot shows expression of IL13 in Treg populations in patients with IPEX and HDs. (I) Quantification of IL13-expressing cells in patients with IPEX and HDs. The frequency on x axis is defined as in (D) and all other populations are presented as “Non-Treg cells”. (J) % of IL13-expressing cells from the Treg populations from all IL13.
To provide further evidence of TCR autoreactivity in Treg and Teff populations from patients with IPEX, we analyzed the presence of autoreactivity promoting and limiting AA doublets from the scRNA and TCR sequencing data. To obtain enough CDR3ß sequences for the analyses, we pooled the data from ex vivo and TCR-stimulated cells, and from HDs and patients with IPEX in Treg populations. We observed that the Treg memory 2 compartment was enriched in the frequency of autoreactivity promoting AA doublets and depleted of autoreactivity limiting AA doublets (fig. S8F), again supporting that the Treg memory 2 population, composted in majority by FOXP3+ TSDR-demethylated CD25low/high cells, is enriched for autoreactive cells.
DISCUSSION
In the present work, we elucidated two concomitant and potentially related phenomena contributing to the etiology of IPEX, a distinctive example of monogenic autoimmunity and loss of immune regulation. First, CD4+ T cells from patients with IPEX with TSDR demethylation, the epigenetic lineage marker of Tregs, were present in the Teff compartment. Second, the TCR repertoire of these patients was enriched in autoreactive sequences, resembling those of Tregs.
The TSDR demethylated cells in the CD4+ Teff compartment of patients with IPEX comprised both FOXP3+ and FOXP3− cells. Since FOXP3+ Teffs from patients with IPEX were exclusively TSDR demethylated, we were able to identify and study them. FOXP3+ Teffs displayed a memory phenotype and had reduced expression of molecules linked to suppressive function, such as FOXP3 and CTLA4, compared with phenotypic Tregs. Consistent with their memory phenotype, they expressed high abundance of CD39 and TIGIT (36). Using a combination of mRNA, protein, and TCR profiling, we provided evidence that these FOXP3+ Teffs correspond to an IPEX-specific, transcriptionally distinct, Treg population that we called Treg memory 2. The mRNA analyses of the Treg memory 2 population revealed an upregulation of IL7R and ZPF36L2, a negative regulator of IKZF2 expression (27), as well as an enrichment of TNF-α signaling target genes. Yet, the TCR repertoire of the Treg memory 2 population was the most similar to that of classical memory Tregs (Treg memory 1). Hence, the FOXP3+ TSDR demethylated cells present in the Teff compartment of patients with IPEX are memory Tregs with altered expression of key Treg molecules, with a gene expression signature indicative of exposure to an inflammatory environment, and a TCR repertoire clonally related with prototypical Tregs that are potentially their precursors. We, therefore, propose that these TSDR-demethylated cells, which expanded beyond the CD25highCD127low phenotypical Treg compartment, are unstable Tregs, which we consider loss-of-identity Tregs. Although we cannot exclude the possibility that some Tregs exit the thymus with loss-of-identity Treg phenotype, the evidence presented above suggest otherwise. These data provide important insights into the heterogeneity of FOXP3-deficient Tregs observed by Zemmour et al. (37) They reported two populations of Tregs in FOXP3-deficient patients, one of which was only present in the disease. Based on our TSDR data, TCR data, and the simultaneous profiling of Treg protein and transcriptional markers, we demonstrated that FOXP3 deficiency gives rise to two different subsets of memory Tregs, one of which loses its phenotypical characteristics.
The presence of loss-of-identity Tregs can well explain the overall increased proportion of TSDR-demethylated cells in whole blood of patients with IPEX, recently deemed diagnostically relevant and associated with disease activity (12, 13), as well as its discrepancy with the frequency of Tregs as determined by FACS (13–17). The absence of the loss-of-identity Tregs not only in carrier mothers and healthy donors, but also in patients with APECED, supports dysfunctional mutated FOXP3 as a primary cause of Treg instability. Therefore, the loss-of-identity Tregs might represent a human counterpart of the “ex” and unstable Tregs described in mice (18–24). However, although loss-of-identity Tregs in patients with IPEX correlated with an increased T cell autoreactivity, the latter occurred in patients with APECED independently from the Treg expansion, likely caused by direct impaired thymic deletion of autoreactive T cells (13). Importantly, we provide evidence that the presence of functional Tregs in vivo can prevent or control the accumulation or the expansion of the loss-of-identity Tregs, as observed in carrier mothers or in the transplanted patient with IPEX in which functional donor-derived Tregs had a selective advantage.
The phenotype analyses of the loss-of-identity Tregs showed reduced expression or even complete absence of crucial suppressive molecules such as FOXP3, CD25, and CTLA4. As a consequence, these cells likely lose regulatory functions. However, they are still derived from the self-antigen specific selected Tregs, as indicated by the TCR analysis, and are therefore likely to play a pathogenic role in patients with IPEX. In agreement with this scenario, the Teff2 compartment of patients with IPEX, which was composed mainly of bona fide Teffs, had an increased frequency of autoreactivity promoting AA doublets, as well as a reduced frequency of autoreactivity limiting AA doublets, thus indicating expansion of autoreactive bona fide Teffs in the absence of functional Tregs. Considering that the increase in the frequency of autoreactivity promoting AA doublets in Teff2 correlated with the frequency of TSDR-demethylated cells in Teff1, we speculate that the autoreactive Teffs expand freely because of the lack of functional Tregs in patients with FOXP3 mutations (15). Consistent with this hypothesis, IPEX carrier mothers had a normal TCR repertoire, indicating that the expansion of autoreactive Teffs in patients with IPEX is largely caused by a lack of FOXP3-dependent Treg function.
A clinically relevant question is to what extent the unstable Tregs are capable of effector functions. We showed that, upon TCR-mediated activation, FOXP3KO Tregs from HDs upregulated the production of the proinflammatory cytokine TNF-α and the Th2 polarizing cytokine IL-13, both of which we found among the most upregulated cytokines in plasma from patients with IPEX(13). Indeed, and in contrast to HDs, even the CD25highCD127low Tregs from patients with IPEX produced IL-13. Similarly, we detected IL-13 production by TCR-stimulated Treg memory 2 cells using scRNA-seq. These findings are in line with data from a murine model where IPEX-like disease was caused by attenuated FOXP3 expression; in this model, the Tregs with low FOXP3 expression acquired a Th2 Teff-like phenotype (23). As expected, we also observed increased production of IL-17 by FOXP3KO Tregs, which reflects the Treg/Th17 imbalance in the absence of functional FOXP3 (38–40). However, we did not observe IL-17 production in ex vivo experiments, possibly because Tregs isolated from peripheral blood of patients with IPEX and of HDs are preferentially Th2 polarized as indicated by GATA3 expression in the scRNA-seq. Unlike in Lam et al., we did not observe a broad upregulation of cytokine secretion in FOXP3KO Tregs but rather an increase in specific cytokines, including GM-CSF (41). Interestingly, we found GM-CSF to be produced also by phenotypic CD25highCD127low Tregs from patients with IPEX. GM-CSF produced by T cells can act in a paracrine fashion to induce phagocyte activation and production of proinflammatory cytokines and chemokines such as IL-1β, TNF-α, CCL17, and CCL22, a mechanism associated with the pathogenesis of T cell mediated autoimmune diseases, such as rheumatoid arthritis (RA) and multiple sclerosis (MS) (42–45). Moreover, we observed increased plasma concentrations of cytokines and chemokines associated with dysregulation of the phagocytic compartment in patients with IPEX (13), including TNF-α, CCL17, and CCL22. Although further work is necessary to fully unravel the role of GM-CSF in the pathogenesis of IPEX disease, GM-CSF neutralizing antibodies might be an attractive treatment option, as they are currently in clinical trials for RA and MS (42). Overall, our in vitro and ex vivo data indicated that, upon activation, FOXP3 fully- or partially- deficient Tregs upregulate the expression of effector cytokines, specifically those that we described being increased in patients with IPEX. These data might provide at least a partial explanation for the observed Th2 polarization in patients with IPEX and their Th2- and Th17-associated pathologies (such as eczema and enteropathy, respectively).
As IPEX is a rare disease, experimental studies are limited by the amount of material per patient and the number of patients available. In addition, shipment of the collected samples made some of them less suitable for certain types of experiments, such as ex vivo scRNA-seq. Our study missed assessments of multiple time points during the course of the disease prior and after immunosuppressive treatment. Such data could provide a more definitive picture of the temporal dynamic of the unstable Tregs and autoreactive Teffs, which could be associated to the clinical manifestations. Similarly, tissue samples, including from the thymus and target organs, could provide additional information about the development, homing, and function of the unstable Tregs and autoreactive Teffs in patients with IPEX. Finally, Treg instability analyzed in a larger number of patients would likely allow us to dissect the mutation-specific effects.
Collectively, these data contribute to the basic knowledge about the role of FOXP3 as a gatekeeper of Treg identity and quiescence as well as in preventing autoreactive Teff expansion in humans. In addition, these data are of interest from a translational perspective, such as in the context of Treg-based therapies, not only in patients with IPEX, but also in patients with other autoimmune diseases in which the loss-of-identity Tregs and expansion of autoreactive T cells are still poorly defined or where similar Treg instability has been suggested (46, 47).
MATERIALS AND METHODS
Study Design
The research objective was to determine the presence and the distribution of autoreactive T cells in patients with IPEX. More specifically, we aimed to unravel how FOXP3 deficiency affects the T cell repertoire autoreactivity and Treg stability in vivo. The data obtained in IPEX patients were compared with those in APECED patients and HD. The inclusion criteria for patients with IPEX consisted of detected FOXP3 mutation by gene sequencing and the presence of clinical symptoms or signs of autoimmunity. The inclusion criteria for patients with APECED consisted of detected homozygous AIRE mutation by gene sequencing. In patient 63, the AIRE mutation was confirmed with certainty only in one allele, but the patient clinically presented with APECED manifestations including autoimmune adrenal insufficiency, autoimmune hypoparathyroidism, and hypothyroidism, and thus was included in the study. Patient and healthy donor details are summarized in table S1. Patients’ and HDs’ samples were collected upon informed consent on an institutional review board approved protocol (IRB #34131) of the Center for Genetic Immune Diseases, Stanford, CA. In addition, the samples from patients with APECED were collected as described previously (9) and under an institutional review board approved protocol (#0906005336). HDs were sex-matched siblings, parents, and pediatric healthy individuals. In addition, samples from adult HDs were purchased from Stanford Blood Center. For TSDR-demethylation analysis in patients with APECED, parents of patients with APECED were used as a control if available (4 out of 5 samples, non-sex matched), and for TCR analyses, sex-matched HDs were used as controls.
FACS staining, sorting, and analysis
Ficoll-Paque PLUS media (Fisher Scientific)-isolated peripheral blood mononuclear cells (PBMCs) from whole blood obtained from a standard blood draw, buffy coat, or Leukoreduction System Chambers were stained for 30 minutes on ice, washed with phosphate-buffered saline (PBS), analyzed, and sorted using BD FACSAria II Cell Sorter (BD Biosciences). The high cell purities after the cell sorting were confirmed by reanalysis of the sorted samples (when enough cells were available) and by TSDR demethylation analysis. Staining of intracellular antigens was done using Foxp3 / Transcription Factor Staining Buffer Set (eBioscience) according to manufacturer recommendations. FACS data were analysed using FlowJo software (FlowJo LLC). If multiple brilliant violet or ultraviolet dyes were used, cells were stained in Brilliant stain buffer or brilliant staining buffer plus (BD Biosciences). Antibodies used in this study are summarized in table S2.
TSDR demethylation analyses
Sorted CD14−CD3+CD4+CD25−CD127low Teff1 and CD14−CD3+CD4+CD25-/dimCD127high Teff2 pellets were frozen in PBS and CD14−CD3+CD4+CD25highCD127low Tregs were expanded using Dynabeads Human T-Activator CD3/CD28 (1:1 beads:Treg ratio, Gibco), and cultured in X-VIVO 15 with Gentamicin, L-Gln, Phenol Red (Lonza), supplemented with 5% human serum (Millipore Sigma) and 300 IU/ml IL-2 (Peprotech) for 5 to 10 days to obtain a sufficient number of cells for TSDR demethylation. TSDR demethylation was analyzed using qPCR as described previously (26). Briefly, cell pellets were thawed and DNA was isolated, bisulfite converted, and analyzed using qPCR. Data were analyzed by Epimune GmbH. TSDR-demethylation data obtained from the whole blood samples of transplanted patients were normalized to the number of CD4+ T cells, also determined by epigenetic analysis (13). The % of cells with demethylated TSDR is defined as follows: (TSDR demethylated copies)/((GAPDH copies)/2/100), and thus was independent of sex.
Single cell RNA, protein, and TCR profiling sample preparation and analysis
PBMCs were thawed and CD4+ T cells were isolated using a CD4+ T Cell Isolation Kit, human (Miltenyi Biotec). High CD4+ T cell purity was confirmed by FACS (fig. S8G). For TCR stimulation, 5e5 T cells (except one donor where only 3.3e5 where available) were plated in 200 μl of X-VIVO15 with Gentamicin, L-Gln, Phenol Red (Lonza), supplemented with 5% human serum (Millipore Sigma), and stimulated for 6 hours with Dynabeads Human T-Activator CD3/CD28 (1:1 beads:cell ratio, Gibco). Cells and beads were centrifuged 300g for 2 min at the start of the activation. Both stimulated and ex vivo cells were stained with oligo-nucleotide tag-conjugated CD127, CD25, and CD45RA antibodies; and cells from patients with IPEX were stained with hashtag antibody 1 and HD cells with hashtag antibody 2 (for details about the antibodies, see table S2). The antibody premix was centrifuged at 14e3 g for 8 min and staining was performed in PBS with 1% bovine serum albumin (BSA; Miltenyi Biotec) for 30 min on ice. Samples were washed three times, cell concentration was adjusted to 1e6 per ml, and samples from HDs and patients with IPEX were mixed in pairs (always HD and IPEX sample) right before the mixed samples were loaded onto a chromium controller (10x Genomics). Super loading was performed to target 20,000 cells for each pair of samples (48).
Preparation of transcriptome, repertoire (TCR), and protein (ADT and hashtag) cDNA libraries was performed according to the 10x Genomics protocol: “Chromium Next GEM Single Cell 5’ Reagent Kits v2” (CG000330 Rev D). cDNA libraries were sequenced at high-depth using the NovaSeq X Plus Platform. The 10x Genomics Cloud Analysis platform was used to process and align raw reads to the human genome (GRCh38) and to generate count matrices and TCR annotations for single cells.
The analyses were performed in R using standard Seurat workflow (49). Briefly, low-quality cells were removed based on low read counts and high mitochondrial DNA content, RNA and ADT data were normalized and de-hashtagged, and doublets were removed based on hashtag staining. Contaminating CD79A and CD14 double positive and CD3E and CD247 double negative clusters and cells were removed from analyses. Data were scaled, TCR genes were removed from variable genes used for principal component analysis using Trex package (50), and data were integrated using the harmony package (51). Ex vivo data were analyzed separately, and TCR-stimulated data were analyzed together with ex vivo data and separated post-scaling. TCR repertoires were analyzed using scRepertoire package and, for the TCR overlap analyses, the AA sequence of TCRα and β chains were used (52). CD25 and CD127 gating was performed in pairs in which the libraries were prepared, where each pair always contained HD and IPEX samples. Differential gene expression analyses between the populations were performed using the FindMarkers function (Seurat) and the results were plotted using EnhanceVulcano package (53). Pathway analyses were performed using fgsea package; all genes were used for the analyses and their differential expression was ranked based on fold changes (54). In addition, scCustomize package and ColorBrewer were used for visualization (55, 56).
TCR repertoire analyses
Sorted cell pellets were frozen in PBS. Upon thawing, DNA was isolated using DNeasy Blood and Tissue Kit (Qiagen). The TCRB was sequenced by Adaptive biotechnologies. Data were processed by Adaptive and initial analysis was done using immunoSEQ analyzer online tool (Adaptive biotechnologies). The analysis of the frequency of autoreactivity promoting and limiting doublets was performed as described previously (29). Briefly, CDR3ß sequences shorter than 8 amino acids were removed because of the conserved amino acid residues at the end of the CDR3ß loop. The frequency of autoreactivity promoting and limiting doublets defined by B. D. Stadinski et al., were calculated as a sum of frequencies of productive rearrangements of a given set of doublets (3, 29). TCRB sequences are available online at (https://clients.adaptivebiotech.com/). The previously published TCR data from patients with APECED (9) and respective HDs are available under immuneACCESS DOI https://doi.org/10.21417/B7V926 (https://clients.adaptivebiotech.com/pub/sng-2019-sciimmunol).
FOXP3 knock out in primary Tregs
To isolate CD3+CD4+CD25highCD127− Tregs, we used PBMCs from Leukoreduction System (LRS) chambers and enriched Tregs using the CD4+CD25+CD127dim/− Regulatory T Cell Isolation Kit II, human (Miltenyi Biotec) followed by FACS sorting of the CD4+CD25+CD127dim/− cells to obtain a pure population. Tregs were stimulated for 7 to 8 days with Dynabeads Human T-Activator CD3/CD28 (1:1 beads:Treg cell ratio, Gibco), and cultured in X-VIVO 15 with Gentamicin, L-Gln, and Phenol Red (Lonza), supplemented with 5% human serum (Millipore Sigma) and 300 IU/mL IL-2 (Peprotech). We subsequently edited the cells using Cas9 (Integrated DNA Technologies), chemically modified guide RNA (Synthego corporation), and AAV6 virus (Signagen laboratories, and in house produced)-mediated delivery of homologous template containing NGFR reporter gene under control of PGK promoter as described previously (33). Control Tregs were treated the same as FOXP3KO Tregs but were electroporated in the absence of CRISPR/Cas9 and not treated with AAV6. Scramble Tregs were nucleofected in the presence of ScrambleGuide/Cas9 (Negative Control Scramble #1, mod, Synthego) and treated with the same batch of the AAV6 virus used to generate the FOXP3KO Tregs. Cells were expanded for 7 to 10 days and NGFR+ cells were purified using MACSelect LNGFR MicroBeads and autoMACS Pro Separator (Miltenyi Biotec). Two donors were expanded for an additional 9 days to obtain sufficient cell numbers for analyses. Subsequently, 1×105 cells were plated in technical duplicates or triplicates, stimulated with Dynabeads Human T-Activator CD3/CD28 (1:1 beads:Treg cell ratio, Gibco), and analyzed for cell count, FOXP3 expression (in Fig. 7, FOXP3+ cells were determined as double positive for two antibody clones 259D and 150D; in fig. S7 only clone 259D was used), and cytokine production 3 days post stimulation. Samples with less than 30% of TSDR-demethylated cells were removed from analyses. For the cytokine production, the technical duplicates were averaged and for each donor, the values were normalized to control:FOXP3 KO cell number ratio at the time of supernatant collection with the following formula: (Technical duplicate average)/((FOXP3 KO)/control [Treg number])).
In the case of control and scramble Tregs, the averaged cytokine concentrations were not normalized to cell number ratio because scramble and control did not show any difference in proliferation.
Cytokine production analyses
For cytokine production by FOXP3KO Tregs, supernatants were collected 3 days post stimulation as described above. Cytokines were measured using a Luminex-based multiplexed assay (Th1/2/9/17 18-plex Human ProcartaPlex Panel, Thermo Fisher) per manufacturer recommendation. For cytokine production by ex vivo stimulated PBMCs from patients with IPEX and HDs, cells were thawed and 2 ×106 cells per ml were incubated in X-VIVO 15 with Gentamicin, L-Gln, and Phenol Red (Lonza), supplemented with 5% human serum (Millipore Sigma), Brefeldin A (BioLegend) and Monensin (BioLegend) for 6 hours. Cells were left untreated or treated with PMA and ionomycin (Cell Activation Cocktail (without Brefeldin A), BioLegend). Next, cells were washed and stained as indicated in the FACS staining, sorting, and analysis section above.
Statistical analysis
All raw, individual-level data for experiments where n<20 are presented in data file S4. Statistical analyses were performed using GraphPad Prism software version 9 (GraphPad Software, Inc.) or R studio software packages as indicated in methods. Normal distribution was not assumed, and the number of observations did not allow for proper testing of normality, therefore, non-parametric two-tailed test Mann-Whitney for non-paired values and non-parametric two-tailed Wilcoxon test for paired values were used. In addition, two-tailed Spearman correlation and Dunn’s multiple comparisons test were used to evaluate significance. Adjustments to alpha levels for multiple comparisons were made using Bonferroni correction for differential gene expression analyses between the clusters (adjusted p values are reported in data file S1 to S3) and using Benjamini-Hochberg method for the FGSEA. Statistical tests are also indicated in the figure legend. Significance is indicated as follows: *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. N represents the number of donors or values per group.
Supplementary Material
Acknowledgments:
We would like to thank the patients and families who made this study possible by kindly donating their blood. In addition, we would like to thank Robert A. Freeborn and Benjamin Thomas and Rhonda Perriman for English corrections, Colin Scott Waichler for help with R code, Veronica Maria Tagi for pediatric HD collection, Laura Passerini for some of the carrier mother sample collection, Kenneth Weinberg for providing the Aria II instrument, and Catherine Carswell-Crumpton and Cheng Pan from the FACS core at the Institute for Stem Cell Biology and Regenerative Medicine for kind technical support.
Funding:
RB is an Anne T. and Robert M. Bass Faculty Scholar, Maternal and Child Research Institute (MCHRI), Department of Pediatrics, Stanford University. This work was supported by MCHRI grant new idea (to RB), MCHRI grant postdoctoral support (to SB), the Bonnie Uytengsu and family endowment for the Center for Genetic Immune Diseases (CGID) and the Center for Definitive and Curative Medicine (CDCM) (to RB and MGR), and by NIH/NIAID grant AI-061093 (to EM). MAC was supported by Center for Pediatric Immunology at St. Louis Children’s Hospital and Washington University. MM was supported by a St. Baldrick’s Foundation Scholar award and by K08 HL151809. EL was supported by the Agency for Science Technology & Research, Singapore.
Competing Interests:
JS was employed by Epimune GmbH, a company using epigenetic technology. SO is employed by and is a shareholder of Precision for Medicine and is a shareholder of Epimune GmbH. SO is inventor of FOXP3 related patents: “Detection and quality control of regulatory T cells through DNA-methylation analysis of the Foxp3 gene (US-10876163-B2)”, “Assay for Determining the Type and/or Status of a Cell Based on the Epigenetic Pattern and the Chromatin Structure (US-20220145390-A1), and “Epigenetic modification of the loci for CAMTA1 and/or FOXP3 as a marker for cancer treatment“ (US-20070243161-A1). RB is co-inventor with SO of the patent US-20230183804-A1 entitled “Epigenetic Method To Detect And Distinguish IPEX And IPEX-Like Syndromes, In Particular In Newborns”. MK consults for Pharming Healthcare and Horizon Therapeutics. The other authors declared no competing interests.
Data availability:
All data associated with this study are in the paper or supplementary materials. In addition, TCR sequencing data are deposited at https://clients.adaptivebiotech.com/, and single-cell multiomics data are available at GEO under record GSE247274.
References
- 1.Klein L, Kyewski B, Allen PM, Hogquist KA, Positive and negative selection of the T cell repertoire: what thymocytes see (and don’t see). Nat Rev Immunol 14, 377–391 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li MO, Rudensky AY, T cell receptor signalling in the control of regulatory T cell differentiation and function. Nat Rev Immunol 16, 220–233 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stadinski BD, Shekhar K, Gomez-Tourino I, Jung J, Sasaki K, Sewell AK, Peakman M, Chakraborty AK, Huseby ES, Hydrophobic CDR3 residues promote the development of self-reactive T cells. Nat Immunol 17, 946–955 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lagattuta KA, Kang JB, Nathan A, Pauken KE, Jonsson AH, Rao DA, Sharpe AH, Ishigaki K, Raychaudhuri S, Repertoire analyses reveal T cell antigen receptor sequence features that influence T cell fate. Nat Immunol 23, 446–457 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Danke NA, Koelle DM, Yee C, Beheray S, Kwok WW, Autoreactive T cells in healthy individuals. J Immunol 172, 5967–5972 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Filion MC, Proulx C, Bradley AJ, Devine DV, Sekaly RP, Decary F, Chartrand P, Presence in peripheral blood of healthy individuals of autoreactive T cells to a membrane antigen present on bone marrow-derived cells. Blood 88, 2144–2150 (1996). [PubMed] [Google Scholar]
- 7.Boehncke WH, Brembilla NC, Autoreactive T-Lymphocytes in Inflammatory Skin Diseases. Front Immunol 10, 1198 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Martino L, Capalbo D, Improda N, D’Elia F, Di Mase R, D’Assante R, D’Acunzo I, Pignata C, Salerno M, APECED: A Paradigm of Complex Interactions between Genetic Background and Susceptibility Factors. Front Immunol 4, 331 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sng J, Ayoglu B, Chen JW, Schickel JN, Ferre EMN, Glauzy S, Romberg N, Hoenig M, Cunningham-Rundles C, Utz PJ, Lionakis MS, Meffre E, AIRE expression controls the peripheral selection of autoreactive B cells. Sci Immunol 4, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bacchetta R, Barzaghi F, Roncarolo MG, From IPEX syndrome to FOXP3 mutation: a lesson on immune dysregulation. Ann N Y Acad Sci 1417, 5–22 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Baron U, Floess S, Wieczorek G, Baumann K, Grutzkau A, Dong J, Thiel A, Boeld TJ, Hoffmann P, Edinger M, Turbachova I, Hamann A, Olek S, Huehn J, DNA demethylation in the human FOXP3 locus discriminates regulatory T cells from activated FOXP3(+) conventional T cells. Eur J Immunol 37, 2378–2389 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Barzaghi F, Passerini L, Gambineri E, Ciullini Mannurita S, Cornu T, Kang ES, Choe YH, Cancrini C, Corrente S, Ciccocioppo R, Cecconi M, Zuin G, Discepolo V, Sartirana C, Schmidtko J, Ikinciogullari A, Ambrosi A, Roncarolo MG, Olek S, Bacchetta R, Demethylation analysis of the FOXP3 locus shows quantitative defects of regulatory T cells in IPEX-like syndrome. J Autoimmun 38, 49–58 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Narula M, Lakshmanan U, Borna S, Schulze JJ, Holmes TH, Harre N, Kirkey M, Ramachandran A, Tagi VM, Barzaghi F, Grunebaum E, Upton JEM, Hong-Diep Kim V, Wysocki C, Dimitriades VR, Weinberg K, Weinacht KG, Gernez Y, Sathi BK, Schelotto M, Johnson M, Olek S, Sachsenmaier C, Roncarolo M-G, Bacchetta R, Epigenetic and Immunological Indicators of IPEX Disease in subjects with FOXP3 gene mutation. Journal of Allergy and Clinical Immunology. [DOI] [PubMed] [Google Scholar]
- 14.Gambineri E, Ciullini Mannurita S, Hagin D, Vignoli M, Anover-Sombke S, DeBoer S, Segundo GRS, Allenspach EJ, Favre C, Ochs HD, Torgerson TR, Clinical, Immunological, and Molecular Heterogeneity of 173 Patients With the Phenotype of Immune Dysregulation, Polyendocrinopathy, Enteropathy, X-Linked (IPEX) Syndrome. Front Immunol 9, 2411 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bacchetta R, Passerini L, Gambineri E, Dai M, Allan SE, Perroni L, Dagna-Bricarelli F, Sartirana C, Matthes-Martin S, Lawitschka A, Azzari C, Ziegler SF, Levings MK, Roncarolo MG, Defective regulatory and effector T cell functions in patients with FOXP3 mutations. J Clin Invest 116, 1713–1722 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kinnunen T, Chamberlain N, Morbach H, Choi J, Kim S, Craft J, Mayer L, Cancrini C, Passerini L, Bacchetta R, Ochs HD, Torgerson TR, Meffre E, Accumulation of peripheral autoreactive B cells in the absence of functional human regulatory T cells. Blood 121, 1595–1603 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barzaghi F, Amaya Hernandez LC, Neven B, Ricci S, Kucuk ZY, Bleesing JJ, Nademi Z, Slatter MA, Ulloa ER, Shcherbina A, Roppelt A, Worth A, Silva J, Aiuti A, Murguia-Favela L, Speckmann C, Carneiro-Sampaio M, Fernandes JF, Baris S, Ozen A, Karakoc-Aydiner E, Kiykim A, Schulz A, Steinmann S, Notarangelo LD, Gambineri E, Lionetti P, Shearer WT, Forbes LR, Martinez C, Moshous D, Blanche S, Fisher A, Ruemmele FM, Tissandier C, Ouachee-Chardin M, Rieux-Laucat F, Cavazzana M, Qasim W, Lucarelli B, Albert MH, Kobayashi I, Alonso L, Diaz De Heredia C, Kanegane H, Lawitschka A, Seo JJ, Gonzalez-Vicent M, Diaz MA, Goyal RK, Sauer MG, Yesilipek A, Kim M, Yilmaz-Demirdag Y, Bhatia M, Khlevner J, Richmond Padilla EJ, Martino S, Montin D, Neth O, Molinos-Quintana A, Valverde-Fernandez J, Broides A, Pinsk V, Ballauf A, Haerynck F, Bordon V, Dhooge C, Garcia-Lloret ML, Bredius RG, Kalwak K, Haddad E, Seidel MG, Duckers G, Pai SY, Dvorak CC, Ehl S, Locatelli F, Goldman F, Gennery AR, Cowan MJ, Roncarolo MG, Bacchetta R, Primary C Immune Deficiency Treatment, B. the Inborn Errors Working Party of the European Society for, T. Marrow, Long-term follow-up of IPEX syndrome patients after different therapeutic strategies: An international multicenter retrospective study. J Allergy Clin Immunol 141, 1036–1049 e1035 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Komatsu N, Okamoto K, Sawa S, Nakashima T, Oh-hora M, Kodama T, Tanaka S, Bluestone JA, Takayanagi H, Pathogenic conversion of Foxp3+ T cells into TH17 cells in autoimmune arthritis. Nat Med 20, 62–68 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Saxena V, Lakhan R, Iyyathurai J, Bromberg JS, Mechanisms of exTreg induction. Eur J Immunol 51, 1956–1967 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dominguez-Villar M, Hafler DA, Regulatory T cells in autoimmune disease. Nat Immunol 19, 665–673 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Charbonnier LM, Cui Y, Stephen-Victor E, Harb H, Lopez D, Bleesing JJ, Garcia-Lloret MI, Chen K, Ozen A, Carmeliet P, Li MO, Pellegrini M, Chatila TA, Functional reprogramming of regulatory T cells in the absence of Foxp3. Nat Immunol 20, 1208–1219 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gavin MA, Rasmussen JP, Fontenot JD, Vasta V, Manganiello VC, Beavo JA, Rudensky AY, Foxp3-dependent programme of regulatory T-cell differentiation. Nature 445, 771–775 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Wan YY, Flavell RA, Regulatory T-cell functions are subverted and converted owing to attenuated Foxp3 expression. Nature 445, 766–770 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Van Gool F, Nguyen MLT, Mumbach MR, Satpathy AT, Rosenthal WL, Giacometti S, Le DT, Liu W, Brusko TM, Anderson MS, Rudensky AY, Marson A, Chang HY, Bluestone JA, A Mutation in the Transcription Factor Foxp3 Drives T Helper 2 Effector Function in Regulatory T Cells. Immunity 50, 362–377 e366 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katzman SD, Hoyer KK, Dooms H, Gratz IK, Rosenblum MD, Paw JS, Isakson SH, Abbas AK, Opposing functions of IL-2 and IL-7 in the regulation of immune responses. Cytokine 56, 116–121 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baron U, Werner J, Schildknecht K, Schulze JJ, Mulu A, Liebert UG, Sack U, Speckmann C, Gossen M, Wong RJ, Stevenson DK, Babel N, Schurmann D, Baldinger T, Bacchetta R, Grutzkau A, Borte S, Olek S, Epigenetic immune cell counting in human blood samples for immunodiagnostics. Sci Transl Med 10, (2018). [DOI] [PubMed] [Google Scholar]
- 27.Makita S, Takatori H, Iwata A, Tanaka S, Furuta S, Ikeda K, Suto A, Suzuki K, Ramos SBV, Nakajima H, RNA-Binding Protein ZFP36L2 Downregulates Helios Expression and Suppresses the Function of Regulatory T Cells. Front Immunol 11, 1291 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rowe JH, Delmonte OM, Keles S, Stadinski BD, Dobbs AK, Henderson LA, Yamazaki Y, Allende LM, Bonilla FA, Gonzalez-Granado LI, Celikbilek Celik S, Guner SN, Kapakli H, Yee C, Pai SY, Huseby ES, Reisli I, Regueiro JR, Notarangelo LD, Patients with CD3G mutations reveal a role for human CD3gamma in Treg diversity and suppressive function. Blood 131, 2335–2344 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Daley SR, Koay HF, Dobbs K, Bosticardo M, Wirasinha RC, Pala F, Castagnoli R, Rowe JH, Ott de Bruin LM, Keles S, Lee YN, Somech R, Holland SM, Delmonte OM, Draper D, Maxwell S, Niemela J, Stoddard J, Rosenzweig SD, Poliani PL, Capo V, Villa A, Godfrey DI, Notarangelo LD, Cysteine and hydrophobic residues in CDR3 serve as distinct T-cell self-reactivity indices. J Allergy Clin Immunol 144, 333–336 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Di Nunzio S, Cecconi M, Passerini L, McMurchy AN, Baron U, Turbachova I, Vignola S, Valencic E, Tommasini A, Junker A, Cazzola G, Olek S, Levings MK, Perroni L, Roncarolo MG, Bacchetta R, Wild-type FOXP3 is selectively active in CD4+CD25(hi) regulatory T cells of healthy female carriers of different FOXP3 mutations. Blood 114, 4138–4141 (2009). [DOI] [PubMed] [Google Scholar]
- 31.Tommasini A, Ferrari S, Moratto D, Badolato R, Boniotto M, Pirulli D, Notarangelo LD, Andolina M, X-chromosome inactivation analysis in a female carrier of FOXP3 mutation. Clin Exp Immunol 130, 127–130 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seitz C, Joly AL, Fang F, Frith K, Gray P, Andersson J, The FOXP3 full-length isoform controls the lineage-stability of CD4(+)FOXP3(+) regulatory T cells. Clin Immunol 237, 108957 (2022). [DOI] [PubMed] [Google Scholar]
- 33.Goodwin M, Lee E, Lakshmanan U, Shipp S, Froessl L, Barzaghi F, Passerini L, Narula M, Sheikali A, Lee CM, Bao G, Bauer CS, Miller HK, Garcia-Lloret M, Butte MJ, Bertaina A, Shah A, Pavel-Dinu M, Hendel A, Porteus M, Roncarolo MG, Bacchetta R, CRISPR-based gene editing enables FOXP3 gene repair in IPEX patient cells. Sci Adv 6, eaaz0571 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lindsten T, June CH, Thompson CB, Multiple mechanisms regulate c-myc gene expression during normal T cell activation. Embo j 7, 2787–2794 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Preston GC, Sinclair LV, Kaskar A, Hukelmann JL, Navarro MN, Ferrero I, MacDonald HR, Cowling VH, Cantrell DA, Single cell tuning of Myc expression by antigen receptor signal strength and interleukin-2 in T lymphocytes. Embo j 34, 2008–2024 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dwyer KM, Hanidziar D, Putheti P, Hill PA, Pommey S, McRae JL, Winterhalter A, Doherty G, Deaglio S, Koulmanda M, Gao W, Robson SC, Strom TB, Expression of CD39 by human peripheral blood CD4+ CD25+ T cells denotes a regulatory memory phenotype. Am J Transplant 10, 2410–2420 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zemmour D, Charbonnier LM, Leon J, Six E, Keles S, Delville M, Benamar M, Baris S, Zuber J, Chen K, Neven B, Garcia-Lloret MI, Ruemmele FM, Brugnara C, Cerf-Bensussan N, Rieux-Laucat F, Cavazzana M, Andre I, Chatila TA, Mathis D, Benoist C, Single-cell analysis of FOXP3 deficiencies in humans and mice unmasks intrinsic and extrinsic CD4(+) T cell perturbations. Nat Immunol 22, 607–619 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eisenstein EM, Williams CB, The Treg/Th17 Cell Balance: A New Paradigm for Autoimmunity. Pediatric Research 65, 26–31 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Passerini L, Olek S, Di Nunzio S, Barzaghi F, Hambleton S, Abinun M, Tommasini A, Vignola S, Cipolli M, Amendola M, Naldini L, Guidi L, Cecconi M, Roncarolo MG, Bacchetta R, Forkhead box protein 3 (FOXP3) mutations lead to increased TH17 cell numbers and regulatory T-cell instability. J Allergy Clin Immunol 128, 1376–1379 e1371 (2011). [DOI] [PubMed] [Google Scholar]
- 40.Zhou L, Lopes JE, Chong MM, Ivanov II, Min R, Victora GD, Shen Y, Du J, Rubtsov YP, Rudensky AY, Ziegler SF, Littman DR, TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature 453, 236–240 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lam AJ, Lin DTS, Gillies JK, Uday P, Pesenacker AM, Kobor MS, Levings MK, Optimized CRISPR-mediated gene knockin reveals FOXP3-independent maintenance of human Treg identity. Cell Rep 36, 109494 (2021). [DOI] [PubMed] [Google Scholar]
- 42.Lotfi N, Thome R, Rezaei N, Zhang GX, Rezaei A, Rostami A, Esmaeil N, Roles of GM-CSF in the Pathogenesis of Autoimmune Diseases: An Update. Front Immunol 10, 1265 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Achuthan A, Cook AD, Lee MC, Saleh R, Khiew HW, Chang MW, Louis C, Fleetwood AJ, Lacey DC, Christensen AD, Frye AT, Lam PY, Kusano H, Nomura K, Steiner N, Förster I, Nutt SL, Olshansky M, Turner SJ, Hamilton JA, Granulocyte macrophage colony-stimulating factor induces CCL17 production via IRF4 to mediate inflammation. J Clin Invest 126, 3453–3466 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hamilton JA, GM-CSF-Dependent Inflammatory Pathways. Front Immunol 10, 2055 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Piseddu I, Röhrle N, Knott MML, Moder S, Eiber S, Schnell K, Vetter V, Meyer B, Layritz P, Kühnemuth B, Wiedemann GM, Gruen J, Perleberg C, Rapp M, Endres S, Anz D, Constitutive Expression of CCL22 Is Mediated by T Cell-Derived GM-CSF. J Immunol 205, 2056–2065 (2020). [DOI] [PubMed] [Google Scholar]
- 46.Ferreira RC, Simons HZ, Thompson WS, Rainbow DB, Yang X, Cutler AJ, Oliveira J, Castro Dopico X, Smyth DJ, Savinykh N, Mashar M, Vyse TJ, Dunger DB, Baxendale H, Chandra A, Wallace C, Todd JA, Wicker LS, Pekalski ML, Cells with Treg-specific FOXP3 demethylation but low CD25 are prevalent in autoimmunity. J Autoimmun 84, 75–86 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ju B, Zhu L, Wang J, Zheng J, Hao Z, Luo J, Zhang J, Hu N, An Q, Feng X, Huo Y, He L, The proportion and phenotypic changes of CD4(+)CD25(−)Foxp3(+) T cells in patients with untreated rheumatoid arthritis. BMC Immunol 23, 41 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stoeckius M, Zheng S, Houck-Loomis B, Hao S, Yeung BZ, Mauck WM 3rd, Smibert P, Satija R, Cell Hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol 19, 224 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hao Y, Hao S, Andersen-Nissen E, Mauck WM 3rd, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, Hoffman P, Stoeckius M, Papalexi E, Mimitou EP, Jain J, Srivastava A, Stuart T, Fleming LM, Yeung B, Rogers AJ, McElrath JM, Blish CA, Gottardo R, Smibert P, Satija R, Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e3529 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.V. D. Borcherding N. (https://github.com/ncborcherding/Trex, 2022), vol. R package version 0.99.6. [Google Scholar]
- 51.Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, Baglaenko Y, Brenner M, Loh PR, Raychaudhuri S, Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods 16, 1289–1296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Borcherding N, Bormann NL, Kraus G, scRepertoire: An R-based toolkit for single-cell immune receptor analysis. F1000Res 9, 47 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blighe K, Rana S, and Lewis M,. (https://github.com/kevinblighe/EnhancedVolcano., 2018).
- 54.Korotkevich G, Sukhov V, Budin N, Shpak B, Artyomov MN, Sergushichev A, Fast gene set enrichment analysis. bioRxiv, 060012 (2021). [Google Scholar]
- 55.Marsh S (https://CRAN.R-project.org/package=scCustomize, 2021).
- 56.Neuwirth E. (https://CRAN.R-project.org/package=RColorBrewer, 2022).
- 57.Sullivan S, Gordon S, Ujhazi B, Zegarra W, Bacchetta R, Csomos K, Walter J, LATE ONSET IPEX SYNDROME IN AN ADOLESCENT MALE. Annals of Allergy, Asthma & Immunology 129, S131 (2022). [Google Scholar]
- 58.Kobayashi I, Shiari R, Yamada M, Kawamura N, Okano M, Yara A, Iguchi A, Ishikawa N, Ariga T, Sakiyama Y, Ochs HD, Kobayashi K, Novel mutations of FOXP3 in two Japanese patients with immune dysregulation, polyendocrinopathy, enteropathy, X linked syndrome (IPEX). J Med Genet 38, 874–876 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsuda M, Torgerson TR, Selmi C, Gambineri E, Carneiro-Sampaio M, Mannurita SC, Leung PS, Norman GL, Gershwin ME, The spectrum of autoantibodies in IPEX syndrome is broad and includes anti-mitochondrial autoantibodies. J Autoimmun 35, 265–268 (2010). [DOI] [PubMed] [Google Scholar]
- 60.Gambineri E, Perroni L, Passerini L, Bianchi L, Doglioni C, Meschi F, Bonfanti R, Sznajer Y, Tommasini A, Lawitschka A, Junker A, Dunstheimer D, Heidemann PH, Cazzola G, Cipolli M, Friedrich W, Janic D, Azzi N, Richmond E, Vignola S, Barabino A, Chiumello G, Azzari C, Roncarolo MG, Bacchetta R, Clinical and molecular profile of a new series of patients with immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome: inconsistent correlation between forkhead box protein 3 expression and disease severity. J Allergy Clin Immunol 122, 1105–1112.e1101 (2008). [DOI] [PubMed] [Google Scholar]
- 61.Halonen M, Eskelin P, Myhre AG, Perheentupa J, Husebye ES, Kämpe O, Rorsman F, Peltonen L, Ulmanen I, Partanen J, AIRE mutations and human leukocyte antigen genotypes as determinants of the autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy phenotype. J Clin Endocrinol Metab 87, 2568–2574 (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data associated with this study are in the paper or supplementary materials. In addition, TCR sequencing data are deposited at https://clients.adaptivebiotech.com/, and single-cell multiomics data are available at GEO under record GSE247274.


