To the Editor:
Bioinformatics software comes in a variety of programming languages and requires diverse installation methods. This heterogeneity makes management of a software stack complicated, error-prone, and inordinately time-consuming. Whereas software deployment has traditionally been handled by administrators, ensuring the reproducibility of data analyses1–3 requires that the researcher be able to maintain full control of the software environment, rapidly modify it without administrative privileges, and reproduce the same software stack on different machines.
The Conda package manager (https://conda.io) has become an increasingly popular means to overcome these challenges for all major operating systems. Conda normalizes software installations across language ecosystems by describing each software with a human readable ‘recipe’ that defines meta-information and dependencies, as well as a simple ‘build script’ that performs the steps necessary to build and install the software. Conda builds software packages in an isolated environment, transforming them into relocatable binaries. Importantly, it obviates reliance on system-wide administration privileges by allowing users to generate isolated software environments in which they can manage software versions by project, without generating incompatibilities and side effects (Supplementary Results). These environments support reproducibility, as they can be rapidly exchanged via files that describe their installation state. Conda is tightly integrated into popular solutions for reproducible data analysis such as Galaxy4, bcbio-nextgen (https://github.com/chapmanb/bcbio-nextgen), and Snakemake5. To further enhance reproducibility guarantees, Conda can be combined with container or virtual machine-based approaches and archive facilities such as Zenodo (Supplementary Results). Finally, although Conda provides many commonly used packages by default, it also allows users to optionally include additional, community-managed repositories of packages (termed channels).
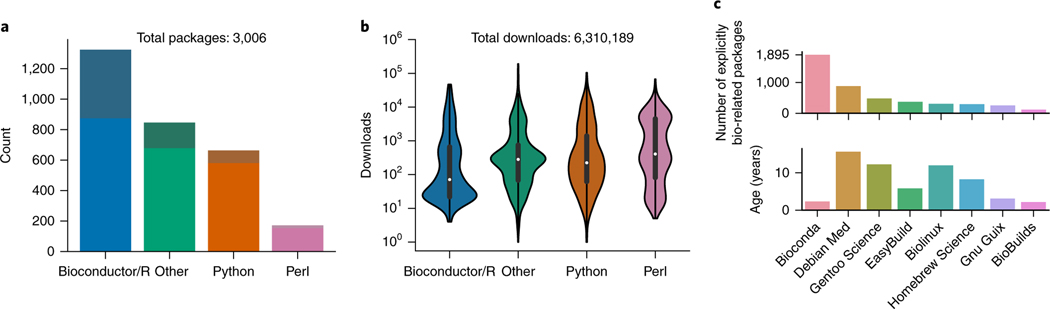
To unlock the benefits of Conda for the life sciences, we present the Bioconda project (https://bioconda.github.io). The Bioconda project provides over 3,000 Conda software packages for Linux and macOS. Rapid turnaround times (Supplementary Results) and extensive documentation (https://bioconda.github.io/contributing.html) have led to a growing community of over 200 international scientists working in the project (Supplementary Results). The project is led by a core team, which is complemented by interest groups for particular language ecosystems. Unlimited (in time and space) storage for generated packages is donated by Anaconda Inc. All other used infrastructure is free of charge. Bioconda provides packages from various language ecosystems such as Python, R (CRAN and Bioconductor), Perl, Haskell, Java, and C/C++ (Fig. 1a). Many of the packages have complex dependency structures that require various manual steps for installation when not relying on a package manager like Conda (Supplementary Results). With over 6.3 million downloads, Bioconda has become a backbone of bioinformatics infrastructure that is used heavily across all language ecosystems (Fig. 1b). It is complemented by the conda-forge project (https://condaforge.github.io), which hosts software not specifically related to the biological sciences. This separation has proven beneficial, because the focused nature of the Bioconda community allows for fast turnaround times and support when a user needs to contribute packages or fix problems. Nevertheless, the two projects collaborate closely, and the Bioconda team maintains over 500 packages hosted by conda-forge.
Fig. 1 |. Package numbers and usage.
a, Package count per language ecosystem (saturated colors on the lower portions of the bars represent explicitly life-science-related packages). b, Distribution of per-package downloads, separated by language ecosystem. The term “other” encompasses all packages that do not fall into one of the specific categories named. White dots represent the mean; dark bars represent the interval between upper and lower quartiles. c, Comparison of the number of explicitly life-science-related packages in Bioconda with that in Debian Med (https://www.debian.org/devel/debian-med), Gentoo Science Overlay (category sci-biology; https://github.com/gentoo/sci), EasyBuild (module bio; https://easybuilders.github.io/easybuild), Biolinux6, Homebrew Science (tag bioinformatics; https://brew.sh), GNU Guix (category bioinformatics; https://www.gnu.org/s/guix), and BioBuilds (https://biobuilds.org). The lower graph shows the project age since the first release or commit. Statistics obtained 25 October 2017.
Bioconda is not the only effort to distribute bioinformatics software (Fig. 1c). The alternatives can be categorized into system-wide (Debian-Med, Genotoo Science, Biolinux, and Homebrew) and per-user (EasyBuild, GNU Guix, and BioBuilds) installation mechanisms. The system-wide approaches lack the ability to put the scientist in control of the installed software stack, and thus do not meet the requirements for reproducibility outlined above. All per-user-based approaches provide a similar feature set (BioBuilds is also using the Conda package manager). However, among all available approaches, Bioconda, despite being the most recent, is by far the most comprehensive, with thousands of software libraries and tools that are maintained by hundreds of international contributors (Fig. 1c).
For reproducible data science, it is crucial that software libraries and tools be provided via an easy-to-use, unified interface, so that they can be easily deployed and sustainably managed. With its ability to maintain isolated software environments, integration into major workflow management systems, and lack of requirement for any administration privileges for use, the Conda package manager is the ideal tool to ensure sustainable and reproducible software management. Bioconda packages have been well received by the community, with over six million downloads so far. We invite everybody to join the Bioconda community, participate in maintaining or publishing new software, and work toward the goal of a central, comprehensive, and language-agnostic collection of easily installable software for the life sciences.
Supplementary Material
Acknowledgements
We thank all contributors, the conda-forge team, and Anaconda Inc. for excellent cooperation. Further, we thank Travis CI (https://travis-ci.com) and Circle CI (https://circleci.com) for providing free Linux and macOS computing capacity. Finally, we thank ELIXIR (https://www.elixir-europe.org) for constant support and donation of staff. This work was supported by the Intramural Program of the National Institute of Diabetes and Digestive and Kidney Diseases, US National Institutes of Health (R.D.), the Netherlands Organisation for Scientific Research (NWO) (VENI grant 016. Veni.173.076 to J.K.), the German Research Foundation (SFB 876 to J.K.), and the NYU Abu Dhabi Research Institute for the NYU Abu Dhabi Center for Genomics and Systems Biology, program number CGSB1 (grant to J.R. and A. Yousif).
Footnotes
Reporting Summary. Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Competing interests
The authors declare no competing interests.
Additional Information
Supplementary information is available for this paper at https://doi.org/10.1038/s41592-018-0046-7.
A full list of authors and affiliations is available as Supplementary Table 1.
Data availability.
Data and code underlying the presented results are enclosed in a Snakemake workflow archive available at https://doi.org/10.5281/zenodo.1068297. The archive can also be used to automatically reproduce all results and figures presented in this paper.
References
- 1.Mesirov JP Science 327, 415–416 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker M. Nature 533, 452–454 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Munafò MR et al. Nat. Hum. Behav. 1, 0021 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Afgan E. et al. Nucleic Acids Res. 44, W3–W10 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Köster J. & Rahmann S. Bioinformatics 28, 2520–2522 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Field D. et al. Nat. Biotechnol. 24, 801–803 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data and code underlying the presented results are enclosed in a Snakemake workflow archive available at https://doi.org/10.5281/zenodo.1068297. The archive can also be used to automatically reproduce all results and figures presented in this paper.