Abstract
The efficiency of programmed ribosomal frameshifting in decoding antizyme mRNA is the sensor for an autoregulatory circuit that controls cellular polyamine levels in organisms ranging from the yeast Schizosaccharomyces pombe to Drosophila to mammals. Comparison of the frameshift sites and flanking stimulatory signals in many organisms now permits a reconstruction of the likely evolutionary path of the remarkably conserved mRNA sequences involved in the frameshifting.
mRNA SEQUENCES AND TRANSLATIONAL FRAMESHIFTING
When the general features of readout of the genetic code were solved in the 1960s, no one expected that the rules would be modifiable in an mRNA-specific manner. Several ways in which the readout process can be reprogrammed are known but the most prevalent type of such ‘recoding’ is site-specific 1 nt changes in reading frame. Some sequences are especially prone to ribosomal frameshifting in this manner and the efficiency with which the process occurs can be greatly augmented by signals contained within the mRNA. The signals can dictate a set ratio of frameshift to non-frameshift products, with the set level depending on the strength of the signals. Alternatively, the process can be responsive to external signals and serve a regulatory purpose. In some cases, up to half of ribosomes shift frame at a specific site, which is remarkably high compared to a 1 in 10 000 or less level of general frameshifting error. For instance, with Escherichia coli dnaX the frameshift product and the product of standard decoding function as distinct subunits, in a 1:1 ratio, in the major replicative polymerase, DNA polymerase III. This frameshifting is –1, like that occurring in the decoding of several bacterial, yeast, plant and animal viral genes and also bacterial insertion sequences of the IS3 family. The great majority of these cases involve slippage ‘backwards’ of mRNA relative to both P- and A-site codons. The ribosomal A-site is occupied by a codon specifying an abundant (or at least not a sparse) tRNA and the frameshifting is not used for regulatory purposes (reviewed in 1). In contrast, for many +1 frameshifts the A-site codon is either a stop codon or a rare codon. Such examples are seen in the expression of E.coli release factor 2 (2,3), the Saccharomyces cerevisiae transposable elements Ty1 and Ty3 (4), S.cerevisiae EST3 (ever shorter telomeres 3) (5) and S.cerevisiae ABP140, which encodes an actin filament-binding protein (6). For all of these cases except Ty3, the first base of the slow-to-decode A-site codon is utilized for re-pairing by the peptidyl tRNA as it realigns the reading frame following dissociation from the initial P-site codon. In Ty3 frameshifting, re-pairing is not involved but the nature of the codon–anticodon pairing of the third P-site codon base causes the first base of the A-site codon to be left unpaired (7). The incoming aminoacyl tRNA pairs with A-site bases 2–4 to effect the shift to the +1 frame. With Ty1, Ty3 and, by inference, EST3 and ABP140 +1 frameshifting, competition for 0 frame decoding of the original A-site codon permits responsiveness of the level of frameshifting to the concentration of the sparse tRNA. With the autoregulatory frameshifting required for release factor 2 synthesis, the equivalent competition is for release factor 2 itself.
For programmed frameshifting, mRNA signals that elevate the level of frameshifting above that found with the frameshifting sites on their own have been characterized. For +1 frameshifting to decode E.coli release factor 2, a Shine–Dalgarno interaction three bases 5′ of the shift with translating ribosomes is crucial (3,8,9), and for E.coli dnaX frameshifting a Shine–Dalgarno interaction 10 bases 5′ of the shift site is important (10). In addition, a stem–loop 3′ of the shift site is important for dnaX frameshifting and the efficiency of frameshifting is governed by the stability of the stem–loop (11). While a 3′ stem–loop is also important for HIV-1 gag–pol frameshifting (12,13), most known cases of animal virus frameshifting utilize a 3′ pseudoknot rather than a simple stem–loop as a stimulator (14,15). Atomic level structures are known for the pseudoknots that stimulate mouse mammary tumor virus gag–pol frameshifting (16,17) and a plant virus counterpart (18; reviewed in 19). For Ty3 +1 frameshifting, the 12 nt 3′ of the shift site that stimulate frameshifting appear to act without folding into a stem–loop or a pseudoknot (20).
Programmed frameshifting is widely known in decoding of viruses from bacteria (21), yeast (22,23), plants (24) and animals and also in decoding mobile chromosomal elements such as bacterial insertion sequences of the IS3 family (25) or various yeast Ty elements (4). Only a few examples of chromosomal non-transposon genes that utilize programmed frameshifting are known. These are: E.coli dnaX (11) and the gene for release factor 2; S.cerevisiae EST3 and ABP140; antizyme genes of yeast, nematodes, insects and vertebrates, as described below.
ANTIZYME AS A REGULATOR OF CELLULAR POLYAMINE LEVELS
Eukaryotes have evolved an autoregulatory mechanism whereby the efficiency of a +1 translational frameshifting event is used as a sensor of polyamine levels. The frameshift occurs in the translation of ornithine decarboxylase antizyme mRNA. Pioneering studies with a mammalian antizyme 1 gene showed that it is decoded from two partially overlapping open reading frames (ORFs) (26). A relatively short ORF1 is followed by a much longer overlapping ORF2. There is no evidence for independent translation initiation of ORF2. Instead, as initially shown by Matsufuji and colleagues, translation is initiated at a start codon for ORF1 and subsequent +1 translational frameshifting in the overlap between ORF1 and ORF2 is necessary for the synthesis of functional antizyme protein (reviewed in 27).
Antizyme was initially described as a biochemical activity that inhibits ornithine decarboxylase (ODC) (28–30) and is elevated in response to increased polyamine levels in cells (31). However, much skepticism prevailed until cDNA cloning (32) and monoclonal antibodies (33) permitted proper analysis of this normally extremely rare protein (2 p.p.m. of soluble protein). Antizyme protein binds to, and inhibits, ODC, a key enzyme in polyamine biosynthesis (32). Mammalian antizyme 1 can present its bound ODC (34,35) to the 26S proteosome for proteolytic degradation without ubiquitination (36,37). This is a catalytic reaction in which one antizyme molecule can destroy multiple molecules of ODC. In addition to its roles in inhibiting the function of ODC, mammalian antizyme 1 inhibits the cellular uptake of polyamines by both reducing import and also increasing excretion of polyamines (38–40). The regulatory roles of antizyme are summarized in Figure 1.
Figure 1.
Schematic representation of antizyme-dependent regulation of polyamines in the cell.
Polyamines, the product of ODC function, induce antizyme synthesis by increasing the efficiency of the obligatory +1 frameshifting (41,42). This closes an autoregulatory circuit in which antizyme has both the means to regulate polyamine levels and the translation elongation stage of its synthesis is a sensor to rapidly adjust antizyme expression in response to fluctuations in polyamine levels in the cell. As expected, perturbation of antizyme expression leads to significant alterations in cellular polyamine levels (43,44).
Several cis-acting mRNA sequences are known to be required for efficient +1 translational frameshifting on decoding the rat antizyme 1 mRNA (42,45). By inference, this same mechanism is employed by its orthologs in all mammals (discussed below). However, none of these cis-acting sequences appear to mediate polyamine-specific induction. Instead, the polyamine induction of frameshifting seems to be mediated directly by the translational machinery itself (the ribosome or some component of it) (42).
COMPILATION OF ANTIZYME SEQUENCES
Vertebrate antizyme genes
Subsequent to the cloning and characterization of rat antizyme 1, orthologs of this gene were cloned from mouse (46,47), human (48,49), a number of additional mammals, chick (50) and frog (51). Two recently described antizyme genes in zebrafish could also be classified as antizyme 1 orthologs (52). Members of the antizyme 1 gene subfamily share some features not present in other antizymes. In addition to the remarkable similarity of their frameshift sites, which is discussed below, all antizyme 1 orthologs share striking amino acid similarity in certain regions of their ORF1. ORF1 of rat antizyme 1 has two AUG initiation codons. In vitro and more recently in vivo studies have shown that both AUGs are used as initiators of translation (41,42,53). The same arrangement with similar positioning is seen in all orthologs of antizyme 1. Sequence analysis and subcellular localization experiments have shown that the polypeptide initiated from the first AUG codon of ORF1 contains a mitochondrial localization signal (53). Underlying the possible importance of this function, all orthologs of antizyme 1 share a high level of similarity within the first 20 amino acids of their ORF1 (85% identity compared to <45% identity for the rest of the protein). Initiation at the second AUG still leads to frameshifting downstream and results in a polypeptide that is ∼30 amino acids shorter. This shorter product lacks the mitochondrial localization signal present in the product from translation at the first AUG.
Apart from the mitochondrial localization sequence in antizyme 1, no biochemical function is known for the product of ORF1 of any antizyme gene, even though closely related antizymes usually share amino acid similarity in that region of the protein.
Antizymes 2 (54–56) and 3 (57,58) are recently discovered mammalian paralogs of antizyme 1. Antizyme 2 is more similar to antizyme 1 (55% amino acid identity) than is antizyme 3. The ORF1 of this gene is shorter than that of the antizyme 1 gene and the amino acid sequence of its product is not well conserved compared to antizyme 1 (54). In fact, only the region of ORF1 closest to the frameshift site is conserved at all and this is probably due to conservation of the nucleotide sequence (see below). Antizyme 3 is even more divergent (26 and 29% amino acid identity compared to antizymes 1 and 2, respectively, in the human). This antizyme paralog also has a short, non-conserved ORF1. A phylogenetic analysis of the three mammalian paralogs of antizyme based on their amino acid sequence (using invertebrate antizymes to root the tree) revealed the following evolutionary relationship. Antizyme 3 was the first to diverge from the other two, most likely early during vertebrate evolution (54). Antizymes 1 and 2 diverged later, perhaps shortly before the radiation of currently surviving members of the vertebrate phylum (Fig. 2). Antizyme 2 is under much tighter evolutionary pressure, as shown by the high level of similarity between the mouse and the human homologs (99.5% identity compared to only 84% identity between the mouse and human homologs of antizyme 1). This indicates that mammalian antizyme 2 has acquired a function that is both very important and non-overlapping with that of antizyme 1. Antizyme 3 is unique in that unlike all other antizymes tested so far, its expression is tissue and cell type specific. Antizyme 3 mRNA is detected only in post-meiotic male germline cells, where expression of antizyme 1 is down-regulated, strongly indicating that its function is not redundant with respect to antizyme 1 (57,58).
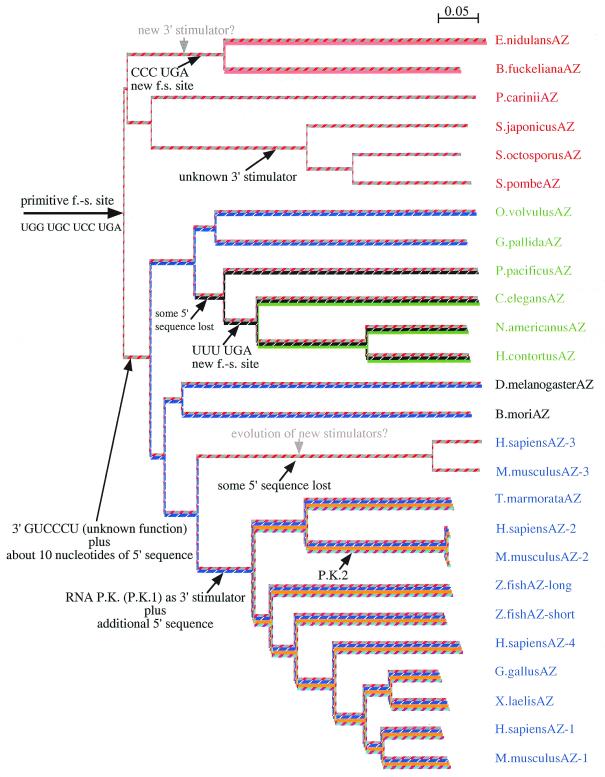
Figure 2.
Unrooted phylogenetic tree based on the amino acid sequence (both ORF1 and ORF2) of antizyme proteins. The full genus names are given in the legend to Figure 3. The tree was drawn using the neighbor-joining algorithm of the ClustalX program (88). The tree is overlaid with a schematic presentation of the most likely path of antizyme frameshift site evolution. Different colored lines indicate the evolution of specific cis-acting features of the frameshift site. Line colors are coordinated with coloration of the cis-acting elements shown in Figure 3B. Black arrows indicate the likely positions of known events in the evolution of antizyme frameshift sites. Grey arrows indicate possible evolutionary events. P.K., pseudoknot; f.-s., frameshift site.
An interesting antizyme homolog that shows characteristics intermediate between mammalian antizymes 1 and 2 was cloned from the marbled electric ray (Torpedo marmorata) (I.P.Ivanov, unpublished results). The primary sequence of this protein identifies it as a member of the antizyme 1 gene subfamily (i.e. ORF1 is longer and has two initiation codons at the appropriate positions). However, amino acid comparison between this protein and other antizymes reveals that it is more similar to antizyme 2 from humans and mice than it is to any of the true orthologs of antizyme 1 (66% identity, 81% similarity to mouse antizymes 2 and 1, respectively; 50% identity, 65% similarity to human antizymes 2 and 1, respectively). Analysis of the frameshift site of this gene also reveals intermediate characteristics. This finding demonstrates that antizyme 2 evolved from a gene that had the structural characteristics of antizyme 1 (and not the other way around).
Recently, we identified an EST sequence from a human brain library corresponding to an antizyme transcript different from the three known antizyme paralogs of humans. The complete cDNA sequence confirmed that this antizyme gene is different from the other three, even though it possesses features that clearly identify it as a member of the antizyme 1 subfamily (unpublished data). No additional entries corresponding to this gene have been placed in the EST databank since the original finding. An unsuccessful attempt was made to PCR amplify parts of this gene from human genomic DNA. Therefore, this gene is only tentatively designated human antizyme 4, pending confirmation of its origin.
Invertebrate antizyme genes
A single antizyme gene has been found in Drosophila. It was identified by positional cloning of the genes in the gutfeeling locus (59,60). This antizyme gene has served as a stepping stone to the identification of antizyme genes from other invertebrates. The next important step was the fortuitous identification of an antizyme gene (SPA) in the fission yeast Schizosaccharomyces pombe (44). This was done by an amino acid similarity search despite the only marginal similarity of SPA to fly and mammalian antizyme proteins (10% identity, 24% similarity to both Drosophila antizyme and human antizyme 1). The antizyme activity of SPA was confirmed by standard biochemical experiments (44). The discovery of antizyme in S.pombe and comparison of its sequence to that of antizymes from a vertebrate and fly has allowed the identification of homologs of this gene from numerous other organisms by searching the publicly available sequence databases [non-redundant (nr) or expressed sequence tags (dbEST)]. Three main criteria were used to identify new members of the antizyme gene family. First, sequence similarity (at the amino acid level) to one of the known antizymes has to be as high as or higher than that between SPA and the vertebrate homologs. Second, the highest similarity should be confined to two specific regions of the protein shown previously to be highly conserved in the known antizymes (see Fig. 3A). Third, the protein should be encoded by two overlapping reading frames, with the downstream ORF (in the +1 frame relative to ORF1) encoding the sequence with highest similarity to known antizymes. The last of these criteria is biased against antizymes that do not require frameshifting for expression but we have found that all candidates examined so far that fulfill the first two requirements also fulfill the last. Candidates were completely sequenced either as cDNA clones or genomic DNA. Applying this method, antizyme homologs were identified from Caenorhabditis elegans (worm), Necator americanus (worm), Haemonchus contortus (worm), Pristioncus pacificus (worm), Onchocerca volvulus (worm) (44), Globodera pallida (worm), Pneumocystsis carinii (fungus), Botryotinia fuckeliana (gray mold), Emericella nidulans (mold), T.marmorata (electric ray) and Bombyx mori (silk worm) (our unpublished results). (Some of these antizyme sequences have been confirmed by others; 61.) In two cases, Schizosaccharomyces octosporus and Schizosaccharomyces japonicus, the antizyme gene was identified using only molecular biology techniques (unpublished results).
Figure 3.
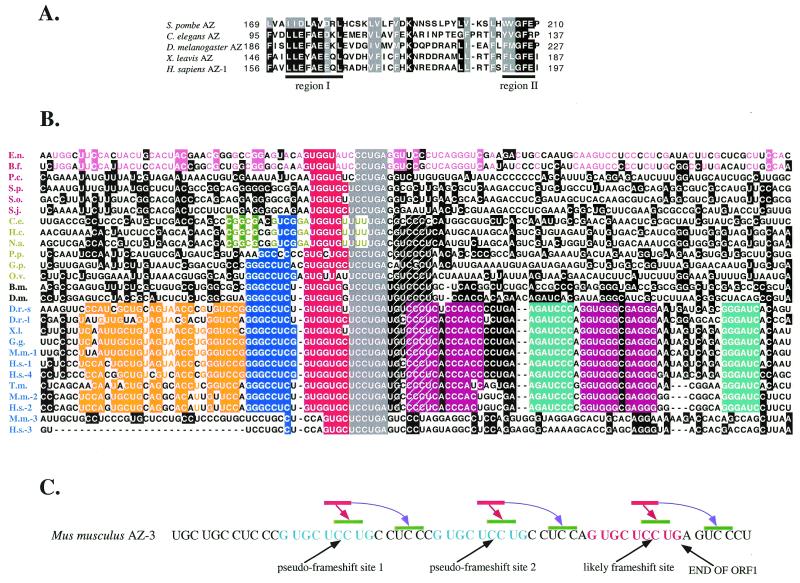
Sequence comparison and analysis of antizyme genes. (A) Partial amino acid alignment of five divergent antizyme proteins (yeast, nematode, insect, amphibian and mammalian). The two regions of antizymes showing the highest amino acid similarity among all known members of the gene family are underlined. Numbers indicate the amino acid position relative to the beginning of the protein. Black shading indicates at least four identical amino acids at a given position. Gray shading indicates at least five similar amino acids at a given position. (B) Nucleotide sequence comparison of the frameshift site region of various antizyme genes. The frameshift site is shown in gray shading. Dotted diagonal lines indicate the conserved 3′ sequence GYCCCY. The different cis-acting elements are colored separately (for additional explanation see main text). Species names are abbreviated as follows: B.f., Botryotinia fuckeliana; B.m. Bombyx mori; C.e., Caenorhabditis elegans; D.m., Drosophila melanogaster; D.r., Danio rerio; E.n., Emericella nidulans; G.g., Gallus gallus; G.p., Globodera pallida; H.c., Haemonchus contortus; H.s., Homo sapiens; M.m., Mus musculus; N.a., Necator americanus; O.v., Onchocerca volvulus; P.c., Pneumocystsis carinii; P.p., Pristioncus pacificus; S.j., Schizosaccharomyces japonicus; S.o., Schizosaccharomyces octosporus; S.p., Schizosaccharomyces pombe; T.m., Torpedo marmorata; X.l., Xenopus laevis. Yeast species names are in red, nematodes in green, insects in black and vertebrates in blue. (C) The frameshift site of mouse antizyme 3 mRNA. The sequence identical to the antizyme consensus is shown in bold and color (red for the ‘genuine’ frameshift site and blue for the ‘pseudo-frameshift’ sites). The authentic frameshift site is part of the red colored sequence. The nucleotides are grouped in triplets relative to the 0 frame (ORF1). The red bar indicates the P-site codon prior to frameshifting. Green bars indicate possible P-site codons after the shift event. Brown arrows indicate possible +1 ribosome shifts. Violet arrows indicate possible ribosome ‘hops’ on matching downstream codons in the +1 frame.
Conservation of amino acid domains
The functional domains of rat antizyme 1 responsible for binding to ODC, tagging ODC for degradation (62,63) and inhibition of polyamine transport (40) have been defined. The C-terminal half (corresponding to amino acids 121–227 of the human ortholog) is sufficient for binding to ODC and is also essential for inhibition of the polyamine transport function of antizyme. This is the domain that is most highly conserved among all the different antizymes and is the location of the two most highly conserved regions, corresponding to amino acids 159–167 and 192–196 of the human antizyme 1 protein (44). Amino acids 69–112 are necessary for antizyme-mediated destabilization of ODC and are sufficient to confer accelerated degradation to unstable heterologous proteins when linked to them covalently (64). This region shows no conservation between vertebrate and invertebrate antizymes. It is not clear if this is because invertebrate ODC is not destabilized following binding to antizyme or whether invertebrate antizymes have evolved different sequences for destabilizing ODC.
All vertebrate antizyme genes (except antizymes 3 and 4) plus the Drosophila homolog, but none of the other known invertebrate antizymes, end with two to four C-terminal negatively charged residues. Since the function of these negatively charged residues is not known (these amino acids are not necessary for binding to, or destabilization of, ODC), the significance of this difference is not apparent. However, these residues are reminiscent of the two negatively charged C-terminal residues of mammalian spermidine/spermine N1-acetyltransferase (SSAT), which are also conserved through great evolutionary distances (worms to humans). SSAT, like antizyme 1, has a short half-life. The two C-terminal glutamates of SSAT mediate the rapid turnover of that protein (65). It is tempting to speculate that the negatively charged C-terminal residues of most metazoan antizymes play a similar role.
Most of the recently discovered antizyme genes show only limited conservation of amino acid sequence (10–20% identity when compared to the previously known homologs), reflecting a large evolutionary divergence (as much as 1 000 000 000 years) and a relatively high rate of protein evolution. Phylogenetic analysis (Fig. 2) based on the amino acid sequences of the various antizyme genes indicates that they are products of divergent evolution (i.e. their phylogenetic relationship matches the phylogenetic relationship of the species that carry them). There is one exception. The phylogenetic analysis shows that Xenopus laevis antizyme 1 diverged from Gallus gallus antizyme 1 after the latter diverged from mouse/human antizyme 1. The reason for this anomaly is not known. This same analysis places the divergence of human antizyme 4 sometime before the radiation of the presently surviving tetrapods but after the divergence of zebrafish and tetrapods.
EVOLUTION OF CIS-ACTING SIGNALS IN ANTIZYME +1 RIBOSOMAL FRAMESHIFTING
For historical reasons the mRNA sequences that stimulate mammalian antizyme 1 +1 frameshifting have been studied in most detail (42) and have provided the paradigm for identifying the frameshift stimulators in all subsequently discovered antizyme genes. Three cis-acting elements are known to be necessary for efficient antizyme 1 frameshifting to levels up to 30%. The first, and least well understood, is a 50 nt sequence just 5′ of the frameshift site, which stimulates frameshifting 2.5–5-fold (42; S.Matsufuji, personal communication). The second signal is the UGA stop codon of ORF1, which stimulates frameshifting 15–20-fold. The third cis-acting element is a RNA pseudoknot that starts 3 nt 3′ of the stop codon of ORF1 and gives a 2.5–5-fold increase in frameshifting (42).
The 5′ signal was defined through serial deletions of ORF1 of antizyme 1. Since the decrease in frameshifting is stepwise with progressive loss of ORF1 sequence it appears likely that the ‘5′ element’ is modular. Analysis with this element indicates that it is the nucleotide and not the amino acid sequence that contains the recoding information (see below). Currently nothing is known about the mechanism through which this element works to stimulate +1 frameshifting. It is possible that the 5′ element of antizyme 1 frameshifting works through pairing with rRNA analogous to the Shine–Delgarno sequence that stimulates +1 and –1 frameshifting in prokaryotes. However, to date there has been no demonstration that rRNA of translocating eukaryotic ribosomes directly interacts with any mRNA sequence, although there are indications for rRNA–mRNA interaction in some initiation events (66–69).
Changing the UGA stop codon of antizyme 1 ORF1 to a sense codon dramatically reduces the efficiency of +1 frameshifting. Conversely, changing this UGA to the other two stop codons, UAA and UAG, leads to only a slight reduction in frameshifting efficiency (42). Early in vitro experiments showed that base pairing in stems 1 and 2 of the 3′ pseudoknot is essential for functioning of this 3′ element, however, maintaining the identity of base pairs in either stem is not necessary (42). Similar results were obtained from in vivo experiments (S.Matsufuji, personal communication). Disrupting either of the two stems of this pseudoknot results in a decrease in frameshifting equivalent to deleting the whole pseudoknot region. Antizyme decoding is the first, and so far the only, example of +1 frameshifting employing an RNA pseudoknot as a 3′ stimulator.
Fission yeast SPA is the only other antizyme gene for which analysis of frameshifting stimulators has been conducted. The observed +1 frameshifting efficiency in decoding this gene is 4–5%, more than 5-fold less than that of antizyme 1 in mammals (44). Despite the lower frameshift efficiency, SPA also has cis-acting frameshift stimulators. It contains only a very short 5′ stimulator corresponding to the last four sense codons of ORF1. Like mammalian antizyme 1, SPA contains an important 3′ frameshift stimulator (frameshift efficiency drops 7–10-fold without it) but its nature is very different from that of antizyme 1. The minimum 3′ sequence necessary for stimulating efficient frameshifting is more than 150 nt long (and may be as much as 180 nt) and the sequence cannot be obviously folded into a RNA pseudoknot analogous to that in antizyme 1. In fact, none of the predicted RNA secondary structures in this region are sufficient to induce efficient +1 frameshifting, suggesting that the 3′ stimulator of SPA frameshifting comprises an unusual structure or that it works via its primary sequence (44).
Nucleotide sequence comparison
Compilation and alignment of a comprehensive number of antizyme frameshift sites is shown in Figure 3B. These sequences include examples from vertebrates, insects, nematodes and fungi. This phylogenetic comparison of nucleotide sequences was combined with the experimental data to identify patterns in the evolution of antizyme frameshifting. Several regularities become apparent.
5′ Elements
As was suggested by the experimental data, phylogenetic comparison of the frameshift sites indicates that the mammalian 5′ element is modular. The different putative modules are colored orange, dark blue and red/gray (Fig. 3B). The orange colored stretch of nucleotides, which constitutes the 5′-half of the 5′ mammalian stimulator, is conserved among the vertebrate orthologs of antizymes 1 and 2 as well as human antizyme 4, but not antizyme 3 or any of the invertebrate genes. The dark blue colored block of 8–9 nt located downstream of the orange colored block but upstream of the red/gray block is conserved among vertebrate orthologs of antizymes 1, 2 and 4 and also the two insect homologs and at least two of the six known nematode homologs (the antizyme homologs in O.volvulus and G.pallida). The proximal 9 nt (indicated in red/gray) 5′ of the stop codon of ORF1 are largely conserved in all antizyme genes identified so far. 5′ Elements involved in recoding are known to work either through their nucleotide (release factor 2 and dnaX; 3,10) or amino acid (gene 60; 70,71) sequence. There are several lines of evidence, both phylogenetic and experimental, suggesting that the 5′ element of antizyme works through its nucleotide sequence. (i) Synonymous substitutions are no more frequent than non-synonymous ones within the 5′ element. (ii) The middle module (dark blue) is present in two different reading frames in the various metazoan antizymes. (iii) The first and second modules (orange and dark blue, respectively) stimulate +1 frameshifting 5-fold in S.pombe (44) even though the endogenous antizyme gene itself does not have these modules. Experiments in S.pombe showed that introducing either synonymous or antonymous changes in this sequence leads to equivalent reductions in frameshifting efficiency, approximately the same as deleting the entire region, indicating that at least in this heterologous system the first and second modules work through their nucleotide sequence (72). (iv) Site-directed mutagenesis experiments by Matsufuji and colleagues indicate that the 5′ element of mammalian antizyme 1 works through its nucleotide and not its amino acid sequence (personal communication).
Stop codon
The stop codon of ORF1 (shown in gray shading) of all antizyme genes identified to date is UGA. This finding is noteworthy because, as mentioned above, experiments with rat antizyme 1 have shown that substituting this codon with the other two stop codons UAA and UAG leads to only a marginal reduction in frameshifting efficiency. UGA is the rarest and, although context dependent (73), may be the least efficient of the three stop codons in translation termination in eukaryotes. This suggests the hypothesis that, like other examples of P-site ribosomal frameshifting, slow decoding of the UGA stop codon of antizyme 1 ORF1 favors frameshifting by allowing more time for the thermodynamically unfavorable reaction: decoding in the +1 frame. Perhaps this difference in translation termination efficiency leading to a small increase in frameshift efficiency is sufficient to have been subject to evolutionary selection.
3′ Elements
The region corresponding to the RNA pseudoknot of mammalian antizyme 1 mRNA is well conserved among all vertebrate orthologs of antizyme 1 plus human antizyme 4 and, to a lesser extent, electric ray antizyme and mammalian antizyme 2. The regions that base pair to form stems 1 and 2 of the pseudoknot, shown in Figure 3B in burgundy and light blue respectively, are especially highly conserved among these antizyme genes. These data clearly demonstrate that all vertebrate orthologs of antizymes 1 and 2 contain sequences that can fold to form an RNA pseudoknot. In fact, among all orthologs of antizyme 1 plus antizyme 4, not a single nucleotide has been altered in the base pairing regions. This still holds true even when all known orthologous sequences of vertebrate antizyme 1 are compared (Fig. 4A). Even though the regions involved in base pairing are absolutely conserved, the regions bordering the pseudoknot and the loops within the pseudoknot are only partially conserved. All but one nucleotide of loop 2 are changed in at least one ortholog of antizyme 1 and antizyme 4. The sequence of loop 1 is more conserved, with only one nucleotide variation among the vertebrate orthologs of antizyme 1. Similar results are seen when the pseudoknot sequences of antizymes 1 and 2 are compared. There is a single base pair of the antizyme 1 pseudoknot that is not absolutely conserved in the antizyme 2 counterpart; a C:G base pair has been changed to a U:G base pair. Also, the antizyme 2 pseudoknot contains additional possible G:C base pairs at the top and the bottom of stem 1 (Fig. 4B). For the counterpart pseudoknot involved in stimulating –1 frameshifting in decoding of the coronavirus, infectious bronchitis virus, a minimal length of 11 bp in stem 1 is important for efficient frameshifting, a reduction to 10 bp causing a 7-fold drop in activity (74). Whether the correspondence in this case to one turn of an A-helix is important is unresolved, however a similar topological requirement does not exist for antizyme +1 frameshifting. The pseudoknot stimulator of mouse mammary tumor virus (MMTV) gag–pol –1 frameshifting is characterized by a much shorter stem 1 than that of infectious bronchitis virus (IBV) or antizyme 1. Adding an extra base pair at the base of stem 1 would also alter the positioning of the pseudoknot relative to the UGA stop codon (moving it closer by 1 nt). Experiments with mammalian antizyme 1 pseudoknot sequences in reticulocyte lysate and S.cerevisiae have shown that changing the spacing between the stop codon and the beginning of the pseudoknot in blocks of 3 nt results in reduced frameshifting efficiency (45). More significantly, frameshifting with the mammalian antizyme 1 sequence in S.cerevisiae is almost exclusively –2 instead of +1, however, adding a 3 nt spacing between the stop codon and the beginning of the pseudoknot alters this ratio in favor of the +1 event (45). There is no discernable conservation of loop 2 sequences between the two antizyme paralogs. In fact, loop 2 of antizyme 2 is 3–4 nt shorter. Three of the six nucleotides of loop 1 are conserved between the two antizyme paralogs. Conservation of the A·C mismatch that breaks the base pairing of stem 1 is particularly noteworthy. Perhaps this mismatch creates a break in the stacking of stem 1 that is necessary for structural reasons. However, site-directed mutagenesis experiments have shown that changing the A to G, creating a G:C base pair, if anything increases frameshift efficiency under the conditions tested. Opening up the helix may allow protein recognition of the major groove that could perhaps be important for regulation or expression in certain tissues. It is also possible that this A·C forms some non-Watson–Crick base pairing in the context of stem 1 of the pseudoknot but this seems unlikely since, at least in one case, there is a naturally occurring U·C mismatch in that position. Either way, conservation of these two juxtaposed nucleotides over such evolutionary distances, >400 000 000 years, is a clear indication of their importance for some (most likely structural) aspect of antizyme pseudoknot function.
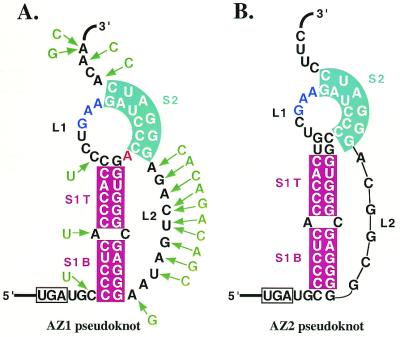
Figure 4.
The RNA pseudoknots of (A) human antizyme 1 and (B) human antizyme 2. The base pairs absolutely conserved between antizyme 1 and 2 RNA pseudoknots are shown as shaded boxes: burgundy for stem 1 and light blue for stem 2. The stop codon of ORF1 is indicated with a rectangle. The ‘wedged’ nucleotide between stems 1 and 2 of the antizyme 1 pseudoknot is in red. The only three nucleotides of loop 1 conserved between antizymes 1 and 2 are shown in dark blue. Nucleotide substitutions in the pseudoknot region of vertebrate orthologs of antizymes 1 and 4 are shown with green arrows. The substitution pattern is based on the antizyme sequences shown in Figure 3B plus the antizyme from Paralychtys olvaceus. L1, loop 1; L2, loop 2; S1B, stem 1 bottom; S1T, stem 1 top; S2, stem 2.
As noted above, the region 3′ of the frameshift site of electric ray antizyme shows characteristics intermediate between those of antizyme 1 and antizyme 2 counterparts. Like antizyme 1, stem 1 of the pseudoknot has 10, not 12, potential base pairs. Also, as in all antizyme 1 genes, the sequence immediately following stem 2 is ACA and not CUU, as it is in antizyme 2 mRNA. At the same time the sequence of loop 2 of electric ray antizyme is more similar to that of antizyme 2: it is 3 nt shorter and the deletion is followed by the sequence CGG, as in antizyme 2. This result affirms the phylogenetic relationship between the various vertebrate antizyme genes derived from amino acid comparisons but, even more importantly, suggests that the potential extra base pairs of stem 1 of antizyme 2 mRNA are functionally unrelated to the 3 nt deletion of loop 2. The electric ray antizyme pseudoknot has at least two features that are unique. A widely conserved C:G base pair in stem 1 is in this case U:G (this is at a different position to the variant seen in antizyme 2). Functional and structural experiments with the vpk derivative of the MMTV pseudoknot have shown that a ‘wedged’ base between stem 1 and stem 2 is required for efficient –1 frameshifting and that its identity, adenosine, is very important (75,76). However, this unpaired wedged adenosine makes only a modest contribution to the stability of the pseudoknot, suggesting that frameshifting efficiency is not strongly correlated with global stability of the RNA (77). Nevertheless, additional evidence has been obtained that a wedged base is important for some reason in pseudoknots of that general type. Conversion of a derivative of the IBV pseudoknot with a short ‘MMTV-like’ stem 1 to an active frameshift stimulator requires a wedged adenosine, absent in the wild-type pseudoknot, and the 3′ nucleotide in loop 2 closest to the junction also has to be an adenosine (78). The analogous base in the electric ray antizyme pseudoknot is G not A, as it is in antizymes 1 and 4. This finding is in agreement with previous site-directed mutagenesis experiments, which showed that the identity of this base has no influence on the +1 frameshifting efficiency of antizyme 1 (42).
The mechanism by which the 3′ RNA pseudoknot stimulates +1 frameshifting in antizyme is not well understood. The three most likely possibilities are: (i) the pseudoknot interferes with decoding of the stop codon of ORF1, thus extending a ribosomal pause during which the ribosome can switch reading frames; (ii) the pseudoknot is important for positioning of the first A-site codon in the new reading frame by preventing the sliding ribosome from slipping further downstream during the frameshift event; (iii) the pseudoknot is responsible for recruiting a cellular factor needed for efficent frameshifting.
The pattern of nucleotide conservation observed with the pseudoknot of vertebrate antizymes 1, 2 and 4 differs from the conventional wisdom about the pattern of conservation of RNA secondary structure. Previous phylogenetic analyses of RNA molecules with known secondary structures have shown, in general, that the identities of bases within helical regions are less conserved than those in non-helical regions (79). This is thought to be due to the fact that it is easier to form specific protein–RNA interactions with sequences that are single-stranded. In an RNA double helix most of the functional groups that can allow distinction between different bases are hidden deep in the narrow, inaccessible major groove of A-form RNA. This frees nucleotides within helices to vary as long as base pairing is maintained to preserve the overall secondary structure of the RNA molecule. In a recent phylogenetic analysis of RNA secondary structures necessary for mRNA editing reactions (80), a pattern of conservation similar to that of the antizyme pseudoknot was observed. The authors speculate that the reason for this unusual pattern of conservation in this case is that the specific substrate requirements of the enzymes catalyzing the editing reaction are part of the double-stranded regions, so mutations disrupting the helix are highly deleterious. It appears that the reason for the high frequency of co-variation within helical regions of previously studied RNA structures is that they are biased in favor of highly repeated RNA genes (mostly various rRNAs). Because of the redundancy of these RNA genes, mutations that lead to disruption of base pairing would be more easily tolerated than in genes with only two genomic copies, thus allowing more time for transition from one base pair to another. This will be true, however, only for genes whose function is under significant evolutionary pressure. The most likely reason for the higher conservation of loop regions of previously analyzed RNA structures is that these structures are interacting with proteins (in the case of rRNA dozens of proteins) or because the nucleotides in ‘loop’ regions are often involved in RNA secondary and tertiary structures. In the case of protein binding to loop regions the interactions require specific contacts between unpaired nucleotides and the protein and as a result these loop regions are under strong selection not to change. The same is true for ‘loop’ nucleotides involved in higher order RNA structures. Loops that are not required for the function of an RNA structure would be under little evolutionary constraints. One implication of this hypothesis is that perhaps the vertebrate antizyme RNA pseudoknot does not interact with proteins in a sequence-specific manner in order to perform its frameshift stimulatory function.
None of the invertebrate and fungal antizyme homologs have sequence conservation 3′ of the frameshift site that would indicate that RNA pseudoknots exist in these mRNAs. Site-directed mutagenesis and deletion experiments with a mammalian antizyme 1 gene have shown that there is no recoding information 3′ of the frameshift site outside the RNA pseudoknot context (42). Surprisingly, even though the invertebrate antizyme mRNAs lack a 3′ pseudoknot, some of them share sequence similarity in this region to their antizyme 1 and 2 homologs from vertebrates. This sequence (consensus GYCCCY) starts 2 nt after the UGA codon of ORF1 and is present in all metazoan homologs of antizyme identified except perhaps antizyme 3 and G.pallida antizyme (the molds B.fuckeliana and E.nidulans have partial conservation of this sequence but since this conservation is incomplete it is not clear if the sequence evolved before the emergence of metazoans). The role of this sequence, if any, in frameshifting is unclear. In the +1 frameshifting of yeast retrotransposon Ty3 a short sequence, up to 12 nt long immediately downstream of the frameshift site, which is not known to form secondary structure, is necessary for optimal frameshift efficiency (4).
There is an intriguing pattern of conservation for the two nucleotides immediately following the UGA stop codon of ORF1 from the different antizyme mRNAs. All fungal antizymes have G in the position just 3′ of the UGA codon, all nematodes have C and all vertebrate and insect antizymes identified so far, except antizyme 3, have U. Even more intriguing is conservation of the next 3′ nucleotide. In all antizyme genes except antizyme 3 and antizyme from G.pallida the nucleotide in that position is G. The sequence context 3′ of a stop codon is known to be important for the efficiency with which that stop codon is decoded by release factors. The efficiency of translation termination as a function of the identity of the adjacent 3′ nucleotide has been determined for UGA in both E.coli and mammals. In the two systems the requirements of 3′ nucleotides for efficient termination are different. In mammals the order of termination efficiency for the 3′ position is A > G >> C > U (73). In E.coli it is the opposite: U > C > G > A (81). The 3′ context specificity is probably species (phylum/kingdom) specific. We speculate that conservation of the nucleotide 3′ of UGA within a group of phylogenetically related antizyme mRNAs reflects the importance of this nucleotide for efficient frameshifting (inefficient translation termination) and the reason for its variation between groups is due to the different requirements for efficient termination in the different groups. The best indication that conservation of the first and second nucleotides 3′ of the UGA of antizyme is probably a result of their importance for efficient translation termination comes from analysis of the sequence from C.elegans. As mentioned above, in C.elegans (and all other nematodes) the identity of these two bases is CG. A comprehensive analysis of the 3′ termination context for most C.elegans ORFs shows that the bias for the nucleotide in the first 3′ position is G > A, U >> C, while it is U >> A > C > G for the second position (82). Assuming that selection is for efficient termination, the nucleotides least likely to facilitate efficient translational termination in C.elegans are those present just 3′ of the stop codon of antizyme ORF1. In fact, C.elegans exhibits preference for the following nine positions as well. In all but the last of these positions, worm antizyme has nucleotides that are least likely to support efficient termination of the stop codon. This is the region that contains the short 3′ sequence conserved among all metazoan antizymes. Therefore, it seems possible that, at least in worms, this conservation is due to the importance of the sequence for inefficient translation termination. There is a precedent: in E.coli efficient recognition of UGA by release factor 2 is dependant on the identity of nucleotides up to six bases 3′ of the stop codon (83,84), although it is not clear that release factor 2 interacts directly with more than the three bases of the stop codon (85). However, since mammalian genes do not show nucleotide bias so far 3′ of stop codons it seems unlikely that this conserved sequence has the same function there. Mammals, and vertebrates in general, however, show preference for the nucleotide immediately 3′ of the stop codon. As mentioned above, the least efficient nucleotide at that position is U. Uridine is seen at that position in all vertebrate antizymes except the orthologs of antizyme 3.
Frameshift site
In several antizymes there are alterations of the frameshift site itself. In three somewhat distantly related nematodes, C.elegans, N.americanus and H.contortus, it is UUU UGA rather than the usual UCC UGA. Similarly, in the two molds E.nidulans and B.fuckeliana and the fungus P.carinii the shift site is CCC UGA. These findings raise a question about the previous conclusion, based on site-directed mutational analysis, that P-site tRNA re-pairing is not part of the mechanism of +1 translational frameshifting of antizyme. A common mechanism for +1 frameshifting is re-pairing of the peptidyl tRNA to mRNA in the new reading frame. However, an alternative mechanism, in which peptidyl tRNA interferes with standard A-site decoding and causes 3′ bases 2–4 to constitute the next codon, has been documented for Ty3 frameshifting (4,7). Results of experiments with some mutants of the frameshift site of mammalian antizyme 1 indicated that the occlusion mechanism is likely (42). Subsequent experiments have challenged this conclusion. When tested in S.cerevisiae (45) and S.pombe (72), the mammalian antizyme 1 frameshift site can direct not only +1 but also a substantial fraction of –2 ribosomal frameshifting (although the ratio of the two products is different in the two yeasts). The –2 frameshift product is best explained as a result of peptidyl tRNA shifting and re-pairing with –2, with decoding resuming at the next available codon (CCU, proline). It seems unlikely that the same frameshift site can support two mechanistically different frameshift events: re-pairing and occlusion. A more likely explanation is that both +1 and –2 frameshifting, at least in yeast, result from peptidyl tRNA re-pairing (in the case of +1 re-pairing involving 1 or 2 bp in the new position). This case, however, may not be representative of events in the natural environment. In the case of UUU UGA and CCC UGA antizyme frameshift sites, the P-site tRNA decoding the last sense codon of ORF1 can slip in the +1 direction relative to the mRNA and form base pairing interactions that are as strong, or almost as strong, as in the original reading frame. More experiments will be needed to definitively determine the mechanism of +1 frameshifting in antizyme synthesis.
The frameshift sites of the two antizyme 3 genes stand in stark contrast to those in the other known antizyme genes in vertebrates. The sequence 3′ of the shift site cannot obviously fold into a RNA pseudoknot comparable to the 3′ structure of mammalian antizyme 1 mRNA. Furthermore, the 5′ sequence, which is highly conserved in all vertebrates (dark blue box) and in all mammals (orange box), is not conserved in these two mammalian genes (Fig. 3B). This is perhaps reflected in the low frameshifting efficiency observed with antizyme 3 (unpublished results). (It must be pointed out that, for technical reasons, none of these experiments were done in the cell type normally expressing antizyme 3, post-meiotic male germ cells.) However, an alternative is possible. A small triplication of five amino acids (15 nt) is observed immediately 5′ of the putative frameshift site of mouse antizyme 3. Closer inspection shows that the sequence that is triplicated includes 9 of the 10 nt of antizyme 3 ORF1 that are identical to the frameshift site consensus. The one difference changes the A of the UGA stop codon of ORF1 to a C to form a UGC cysteine codon. This triplication creates two ‘pseudo-frameshift sites’ (G-UGC-UCC-UG) such that +1 frameshifting on any one of the two UCC UGC sites would result in the synthesis of a protein product that is essentially identical to that resulting from authentic +1 frameshifting at the ‘normal’ UCC UGA frameshift site (Fig. 3C). Thus, any residual translational frameshifting at the two pseudo-frameshift sites would be additive to the total frameshifting efficiency. Yet another possibility is a short ribosome ‘hop’ from the UCC codon contained within the pseudo-frameshift site or the authentic frameshift site to the next available downstream UCC codon, which would result in a similar final outcome (Fig. 3C). In a ribosome ‘hop’ the P-site tRNA dissociates from the mRNA while still bound to the ribosome and ‘lands’ some distance downstream on a matching codon (70,71). A ribosome ‘hop’ event is mechanistically related to (compatible with) P-site +1 frameshifting (3). Whether these events occur during the translation of mouse antizyme 3 is still an open question. In this context it has to be noted that the human ortholog of antizyme 3 has only one (partial) copy of this pseudo-frameshift site.
In addition to the sequence similarities that are common to most antizyme frameshift sites, there are others that are peculiar to only one closely related subset of sequences. Alterations of the UCC UGA frameshift site to UUU UGA and CCC UGA (mentioned above) are two such examples, but there are several others. The frameshift site sequences of E.nidulans and B.fuckeliana antizymes share only a short region of similarity with the consensus (10 of the last 13 nt of ORF1). However, despite the long time of divergence between these two fungi (most likely >400 000 000 years), their antizyme genes share substantial similarity in the vicinity of the frameshift site in addition to the core region, shown in pink in Figure 3B. The importance of these conserved sequences for +1 frameshifting in the two molds has not been tested. However, since 50 nt in either direction of the frameshift site are the most highly conserved within the mRNA of these two genes, it appears that this conservation is likely due to the importance of these sequences for efficient +1 frameshifting. Extensive similarity between different fission yeasts is observed in the region 3′ of the frameshift site that has previously been shown experimentally to be necessary for efficient frameshifting. For example, a block of 35 nt starting 62 nt downstream of the UGA codon of ORF1 of SPA is absolutely conserved in all three known fission yeasts: S.pombe, S.octosporus and S.japonicus (striking conservation for sequences that likely diverged >300 000 000 years ago). The nematode antizyme genes that have a UUU UGA frameshift site share additional nucleotide conservation (shown in green in Fig. 3B) that is unique to just that subgroup. Comparison of antizyme genes from two distantly related insects (B.mori and D.melanogaster) shows that only a region of 29 nt in the vicinity of the frameshift site is conserved. When the sequences from two fruit fly species (D.melanogaster and Drosophila virilis) are compared, regions of high nucleotide conservation are significantly extended. A region of ∼100 nt surrounding the frameshift site is absolutely conserved between the two flies while the rest of the coding sequence is <75% conserved. This pattern of conservation, with some branches sharing unique features and the experimental data from SPA analysis showing the evolution of an apparently independent 3′ stimulator, strongly indicates that cis-acting frameshift stimulators may have evolved multiple times during the evolution of antizyme +1 frameshifting.
Evolution of the antizyme frameshift sequences
It seems logical to assume that the reason for the evolution of multiple cis-acting stimulators of frameshifting is that they have simply co-evolved with the translational machinery of the organism that carries them to give maximal frameshifting. This seemingly logical assumption is not supported by the experimental data. In fact, the opposite is sometimes the case. Experiments with antizyme frameshifting sequences tested in heterologous systems show that sometimes they support as high as or higher levels of frameshifting in a heterologous system. For example, a S.pombe frameshift cassette supports 4.4% +1 frameshifting in S.pombe (under conditions of high polyamines) but ∼5.5% in reticulocyte lysate (also under high polyamine conditions) (44). When the mammalian antizyme 1 cassette is tested in S.pombe it gives 9% +1 frameshifting (72), which is more than the frameshifting observed with the endogenous gene. When individual components (like the 5′ element and 3′ pseudoknot) of the antizyme 1 frameshift signal are tested in S.pombe they stimulate frameshifting as much as or more than they do in mammals, even though the endogenous fission yeast gene clearly does not contain these two sequences (except for the very end of the 5′ element) (72). These results suggest that the reason for the apparent variety of cis-acting frameshift stimulators in modern descendants of antizyme is that they have emerged rather late in the evolution of the gene and not because they have co-evolved with the translational apparatus (although that may turn out to be so in some particular cases). This variation probably also reflects the optimization of antizyme expression in each individual branch of evolution.
The combined knowledge gained from sequence analysis and standard molecular biology experiments allows us to discern the likely scenario of antizyme frameshift site evolution (Fig. 2). We postulate that the most ‘primitive’ version of the antizyme frameshift site was the sequence UGG UGC UCC UGA (red/gray lines). This sequence by itself is capable of inducing small but measurable levels of +1 frameshifting in mammals and in yeast (42,72). All antizyme genes identified so far have this sequence or some variation of it as part of their frameshift site. The genes for S.pombe SPA and human antizyme 1, two of the most divergent members of the antizyme gene family, share this exact sequence. We postulate that all other cis-acting mRNA frameshift stimulators are subsequent additions to this ‘primitive’ frameshift site.
In the fungal lineage the following sequence of events seems likely. Sometime during the radiation of Ascomycota some members of the phylum acquired CCC UGA instead of the original UCC UGA frameshift site. This event may have occurred twice, once in the lineage leading to E.nidulans and B.fuckeliana and a second time in the lineage leading to P.carinii (the phylogenetic relationship between P.carinii and the other fungi is currently not known). From the sequence conservation between the E.nidulans and B.fuckeliana antizyme genes we infer that this fungal branch may have evolved cis-acting frameshift stimulating sequences specific to this lineage (pink lines). Whether these cis-acting stimulators are working in a manner analogous to the 5′ and 3′ stimulators in mammals is not yet clear. It appears that sometime during the early evolution of fission yeasts a unique 3′ frameshift stimulator emerged. Initial characterization of this stimulator in S.pombe showed that it is responsible for up to a 10-fold increase in the efficiency of +1 frameshifting. The extensive conservation of this region among all three known fission yeasts suggests that this stimulator is likely present in all of them (unpublished data).
Sometime during the evolution of metazoa from protozoa two distinct additions were made to the ‘primitive’ antizyme frameshift site. First, a 6 nt sequence (GYCCCY) of unknown function evolved just 3′ of the stop codon of ORF1 (diagonally dotted lines). This sequence is present in almost all known members of the metazoan antizyme gene family. Second, a sequence of ∼10 nt was added to the already existing 5′ stimulator (dark blue lines). In the nematode lineage, members of clade V (86) appear to have lost this additional 10 nt sequence. The antizyme frameshift sites of C.elegans, N.americanus and H.contortus (all members of clade V) have undergone further transformation. Instead of the usual UCC UGA frameshift site, UUU UGA has emerged. In that same branch of nematodes a short region (shown with green lines) 5′ of the core frameshift site is absolutely conserved. Considering that the overall conservation of nucleotides among these three antizymes is rather limited, such conservation in this region becomes significant. Perhaps it is yet another version of the 5′ +1 frameshift stimulator.
Insect antizyme sequences do not appear to have acquired unique frameshift stimulators that are shared by both D.melanogaster (fruit fly) and B.mori (moth). The conservation pattern of the frameshift site of D.melanogaster and D.virilis suggests that additional 5′ and 3′ stimulators may have evolved in the Drosophila lineage.
Evolution of the frameshift sites of the various vertebrate paralogs of antizyme mRNAs has occurred in two separate directions. One is the direction taken by the two orthologs of antizyme 3 genes in mammals. These genes have lost some of the 5′ stimulatory sequences, including part of the ‘primitive’ frameshift site. This is perhaps reflected in the low level of +1 frameshifting seen with these coding sequences in vitro (unpublished results) and in vivo in tissue cultures (M.Howard, personal communication). It is possible that a specific factor(s) present in male post-meiotic germ cells recognizes other features of the antizyme frameshift region. As noted above, antizyme 3 may have evolved additional 5′ stimulators in the form of ‘pseudo-frameshift sites’, but no experimental data exist to support this hypothesis. A second direction in the evolution of vertebrate antizyme frameshift sites was taken by the vertebrate orthologs of the antizyme 1 and 2 genes and the human antizyme 4 gene. The common progenitor of all of them acquired two features not present in the frameshift site of any invertebrate antizymes or antizyme 3. A region of ∼30 nt (orange lines) immediately upstream of the already existing 5′ element was added. Deletion experiments have confirmed the importance of this region for efficient frameshifting (42,72; S.Matsufuji, personal communication). The most significant new feature is an RNA pseudoknot structure starting 3 nt 3′ of the frameshift site (burgundy/light blue lines). The existence of this structure and its importance for efficient +1 frameshifting in antizyme are supported both by site-directed mutagenesis (42,44,72) and by the phylogenetic data presented here. It appears that the antizyme 1 type of pseudoknot evolved first and the antizyme 2 variety evolved from it millions of years later.
CONCLUSION
Several conclusions can be made from the analysis of antizyme frameshift sites. First, recoding sites can be selected for over very long evolutionary periods (hundreds of millions of years). Second, some plasticity exists in the frameshift site, indicating a level of flexibility in setting up P-site recoding events. Third, setting a ‘primitive’ frameshift site is crucial. Once a +1 frameshift site is established by selection, cis-acting stimulators can evolve ‘at will’ to optimize the frameshift levels in any given system. More such analyses are essential if we are to better understand all requirements for +1 ribosome frameshift events.
The discovery of alternative decoding was not anticipated when the rules of the genetic code were worked out more than 30 years ago. Since the first examples were discovered, new cases have been reported each year. However, the vast majority of these are found in the compact genomes of ‘parasitic’ nucleic acid sequences. Examples of recoding required for expression of cellular genes are still very few. More importantly, all cellular genes requiring recoding were found ad hoc after detailed molecular analysis demonstrated that a major discrepancy existed between the observed and predicted gene product. As a result, even today we do not know the extent to which organisms employ recoding strategies for expression of their genes. Advances in genome research allow for the first time the possibility of a systematic search for such events. Some progress has been made in this regard with sequences that might support –1 frameshifting of the tandem slip variety (87). Unfortunately, this has not been possible for recoding events of the P-site frameshift kind, largely because of our incomplete understanding of the sequence requirements for such events. In this regard the molecular and phylogenetic analysis of antizyme frameshift sites provides one more step in the right direction.
Acknowledgments
ACKNOWLEDGEMENTS
Our work was dependent on the generosity of many in the provision of clones, cDNA libraries and yeast strains. We also highly appreciate discussions and collaborations with Dr S. Matsufuji (Jikei University School of Medicine, Tokyo) and thank him and Dr Brenda Bass for critical comments on the manuscript. We thank Jasmine Parvaz for technical help. This work was supported by Department of Energy grant DEFG03-99ER62732 to R.F.G. and NIH grant GM48152 to J.F.A. I.P.I. was supported by NIH Genetics Training Grant 5T32GM07464-23.
REFERENCES
- 1.Gesteland R.F. and Atkins,J.F. (1996) Annu. Rev. Biochem., 65, 741–768. [DOI] [PubMed] [Google Scholar]
- 2.Craigen W.J. and Caskey,C.T. (1986) Nature, 322, 273–275. [DOI] [PubMed] [Google Scholar]
- 3.Weiss R.B., Dunn,D.M., Atkins,J.F. and Gesteland,R.F. (1987) Cold Spring Harbor Symp. Quant. Biol., 52, 687–693. [DOI] [PubMed] [Google Scholar]
- 4.Farabaugh P.J., Zhao,H. and Vimaladithan,A. (1993) Cell, 74, 93–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morris D.K. and Lundblad,V. (1997) Curr. Biol., 7, 969–976. [DOI] [PubMed] [Google Scholar]
- 6.Asakura T., Sasaki,T., Nagano,F., Satoh,A., Obaishi,H., Nishioka,H., Imamura,H., Hotta,K., Tanaka,K., Nakanishi,H. and Takai,Y. (1998) Oncogene, 16, 121–130. [DOI] [PubMed] [Google Scholar]
- 7.Sundararajan A., Michaud,W.A., Qian,Q., Stahl,G. and Farabaugh,P.J. (1999) Mol. Cell, 4, 1005–1015. [DOI] [PubMed] [Google Scholar]
- 8.Weiss R.B., Dunn,D.M., Dahlberg,A.E., Atkins,J.F. and Gesteland,R.F. (1988) EMBO J., 7, 1503–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwartz R. and Curran,J.F. (1997) Nucleic Acids Res., 25, 2005–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larsen B., Wills,N.M., Gesteland,R.F. and Atkins,J.F. (1994) J. Bacteriol., 176, 6842–6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Larsen B., Gesteland,R.F. and Atkins,J.F. (1997) J. Mol. Biol., 271, 47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parkin N.T., Chamorro,M. and Varmus,H.E. (1992) J. Virol., 66, 5147–5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bidou L., Stahl,G., Grima,B., Liu,H., Cassan,M. and Rousset,J.-P. (1997) RNA, 3, 1153–1158. [PMC free article] [PubMed] [Google Scholar]
- 14.Brierley I., Digard,P. and Inglis,S.C. (1989) Cell, 57, 537–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.ten Dam E.B., Pleij,C.W.A. and Bosch,L. (1990) Virus Genes, 4, 121–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen L.X. and Tinoco,I. (1995) J. Mol. Biol., 247, 963–978. [DOI] [PubMed] [Google Scholar]
- 17.Gonzalez R.L. and Tinoco,I. (1999) J. Mol. Biol., 289, 1267–1282. [DOI] [PubMed] [Google Scholar]
- 18.Kim Y.-G., Su,L., O’Neill,A. and Rich,A. (1999) Proc. Natl Acad. Sci. USA, 96, 14234–14239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giedroc D.P., Theimer,C.A. and Nixon,P.L. (2000) J. Mol. Biol., 298, 167–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farabaugh P.J. (2000) Prog. Nucleic Acid Res. Mol. Biol., 64, 131–170. [DOI] [PubMed] [Google Scholar]
- 21.Levin M.E., Hendrix,R.W. and Casjens,S.R. (1993) J. Mol. Biol., 234, 124–139. [DOI] [PubMed] [Google Scholar]
- 22.Dinman J.D. (1995) Yeast, 11, 1115–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lopinski J.D., Dinman,J.D. and Bruenn,J.A. (2000) Mol. Cell. Biol., 20, 1095–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller W.A., Brown,C.M. and Wang,S. (1997) Semin. Virol., 8, 3–13. [Google Scholar]
- 25.Chandler M. and Fayet,O. (1993) Mol. Microbiol., 7, 497–503. [DOI] [PubMed] [Google Scholar]
- 26.Miyazaki Y., Matsufuji,S. and Hayashi,S. (1992) Gene, 113, 191–197. [DOI] [PubMed] [Google Scholar]
- 27.Gesteland R.F., Weiss,R.B. and Atkins,J.F. (1992) Science, 257, 1640–1641. [DOI] [PubMed] [Google Scholar]
- 28.Heller J.S., Fong,W.F. and Canellakis,E.S. (1976) Proc. Natl Acad. Sci. USA, 73, 1858–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kitani T. and Fujisawa,H. (1984) J. Biol. Chem., 259, 10036–10040. [PubMed] [Google Scholar]
- 30.Murakami Y. and Hayashi,S. (1985) Biochem. J., 226, 893–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hayashi S. and Canellakis,E.S. (1989) In Hayashi,S. (ed.), Ornithine Decarboxylase: Biology, Enzymology, and Molecular Genetics. Pergamon Press, New York, NY, pp. 47–58.
- 32.Matsufuji S., Miyazaki,Y., Kanamoto,R., Kameji,T., Murakami,Y., Baby,T.G., Fujita,K., Ohno,T. and Hayashi,S. (1990) J. Biochem., 108, 365–371. [DOI] [PubMed] [Google Scholar]
- 33.Matsufuji S., Kanamoto,R., Murakami,Y. and Hayashi,S. (1990) J. Biochem., 107, 87–91. [DOI] [PubMed] [Google Scholar]
- 34.Li X. and Coffino,P. (1992) Mol. Cell. Biol., 12, 3556–3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li X. and Coffino,P. (1993) Mol. Cell. Biol., 13, 2377–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murakami Y., Matsufuji,S., Kameji,T., Hayashi,S., Igarashi,K., Tamura,T., Tanaka,K. and Ichihara,A. (1992) Nature, 360, 597–599. [DOI] [PubMed] [Google Scholar]
- 37.Murakami Y., Matsufuji,S., Hayashi,S., Tanahashi,N. and Tanaka,K. (1999) Mol. Cell. Biol., 19, 7216–7227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitchell J.L.A., Judd,G.G., Bareyal-Leyser,A. and Ling,S.Y. (1994) Biochem. J., 299, 19–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suzuki T., He,Y., Kashiwagi,K., Murakami,Y., Hayashi,S. and Igarashi,K. (1994) Proc. Natl Acad. Sci. USA, 91, 8930–8934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sakata K., Kashiwagi,K. and Igarashi,K. (2000) Biochem. J., 347, 297–303. [PMC free article] [PubMed] [Google Scholar]
- 41.Rom E. and Kahana,C. (1994) Proc. Natl Acad. Sci. USA, 91, 3959–3963. [Erratum, Proc. Natl Acad. Sci. USA, 91, 9195.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matsufuji S., Matsufuji,T., Miyazaki,Y., Murakami,Y., Atkins,J.F., Gesteland,R.F. and Hayashi,S. (1995) Cell, 80, 51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murakami Y., Matsufuji,S., Miyazaki,Y. and Hayashi,S. (1994) Biochem. J., 304, 183–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ivanov I.P., Matsufuji,S., Murakami,Y., Gesteland,R.F. and Atkins,J.F. (2000) EMBO J., 19, 1907–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matsufuji S., Matsufuji,T., Wills,N.M., Gesteland,R.F. and Atkins,J.F. (1996) EMBO J., 15, 1360–1370. [PMC free article] [PubMed] [Google Scholar]
- 46.Kankare K., Uusi-Oukari,M. and Jänne,O.A. (1997) Biochem. J., 324, 807–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nilsson J., Koskiniemi,S., Persson,K., Grahn,B. and Holm,I. (1997) Eur. J. Biochem., 250, 223–231. [DOI] [PubMed] [Google Scholar]
- 48.Tewari D.S., Qian,Y., Thornton,R.D., Pieringer,J., Taub,R., Mochan,E. and Tewari,M. (1994) Biochim. Biophys. Acta, 1209, 293–295. [DOI] [PubMed] [Google Scholar]
- 49.Hayashi T., Matsufuji,S. and Hayashi,S. (1997) Gene, 203, 131–139. [DOI] [PubMed] [Google Scholar]
- 50.Drozdowski B., Gong,T.-W.L. and Lomax,M.I. (1998) Biochim. Biophys. Acta, 1396, 21–26. [DOI] [PubMed] [Google Scholar]
- 51.Ichiba T., Matsufuji,S., Miyazaki,Y. and Hayashi,S. (1995) Biochim. Biophys. Acta, 1262, 83–86. [DOI] [PubMed] [Google Scholar]
- 52.Saito T., Hascilowicz,T., Ohkido,I., Kikuchi,Y., Okamoto,H., Hayashi,S., Murakami,Y. and Matsufuji,S. (2000) Biochem. J., 345, 99–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mitchell J.L.A. and Judd,G.G. (1998) Biochem. J., 26, 591–595. [Google Scholar]
- 54.Ivanov I.P., Gesteland,R.F. and Atkins,J.F. (1998) Genomics, 52, 119–129. [DOI] [PubMed] [Google Scholar]
- 55.Zhou J., Atkins,J.F. and Gesteland,R.F. (1999) Gene, 232, 165–171. [DOI] [PubMed] [Google Scholar]
- 56.Zhu C., Lang,D.W. and Coffino,P. (1999) J. Biol. Chem., 274, 26425–26430. [DOI] [PubMed] [Google Scholar]
- 57.Ivanov I.P., Rohrwasser,A., Terreros,D.A., Gesteland,R.F. and Atkins,J.F. (2000) Proc. Natl Acad. Sci. USA, 97, 4808–4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tosaka Y., Tanaka,H., Yano,Y., Masai,K., Nozaki,M., Yomogida,K., Otani,S., Nojima,H. and Nishimune,Y. (2000) Genes Cells, 5, 265–276. [DOI] [PubMed] [Google Scholar]
- 59.Ivanov I.P., Simin,K., Letsou,A., Atkins,J.F. and Gesteland,R.F. (1998) Mol. Cell. Biol., 18, 1553–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Salzberg A., Golden,R., Bodmer,R. and Bellen,H.J. (1996) Genetics, 144, 183–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu C., Karplus,K., Grate,L. and Coffino,P. (2000) Bioinformatics, 16, 478–481. [DOI] [PubMed] [Google Scholar]
- 62.Ichiba T., Matsufuji,S., Miyazaki,Y., Murakami,Y., Tanaka,K., Ichihara,A. and Hayashi,S. (1994) Biochem. Biophys. Res. Commun., 200, 1721–1727. [DOI] [PubMed] [Google Scholar]
- 63.Li X. and Coffino,P. (1994) Mol. Cell. Biol., 14, 87–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li X., Stebbins,B., Hoffman,L., Pratt,G., Rechsteiner,M. and Coffino,P. (1996) J. Biol. Chem., 271, 4441–4446. [DOI] [PubMed] [Google Scholar]
- 65.Coleman C.S. and Pegg,A.E. (1997) J. Biol. Chem., 272, 12164–12169. [DOI] [PubMed] [Google Scholar]
- 66.Pestova T.V., Shatsky,I.N., Fletcher,S.P., Jackson,R.J. and Hellen,C.U.T. (1998) Genes Dev., 12, 67–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Verrier S.-B. and Jean-Jean,O. (2000) RNA, 6, 584–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hu M.C., Tranque,P., Edelman,G.M. and Mauro,V.P. (1999) Proc. Natl Acad. Sci. USA, 96, 1339–1344. [Google Scholar]
- 69.Yueh A. and Schneider,R.J. (2000) Genes Dev., 14, 414–421. [PMC free article] [PubMed] [Google Scholar]
- 70.Weiss R.B., Huang,W.M. and Dunn,D.M. (1990) Cell, 62, 117–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Herr A.J., Gesteland,R.F. and Atkins,J.F. (2000) EMBO J., 19, 2671–2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ivanov I.P., Gesteland,R.F., Matsufuji,S. and Atkins,J.F. (1998) RNA, 4, 1230–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McCaughan K.K., Brown,C.M., Dalphin,M.E., Berry,M.J. and Tate,W.P. (1995) Proc. Natl Acad. Sci. USA, 92, 5431–5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Napthine S., Liphardt,J., Bloys,A., Routledge,S. and Brierley,I. (1999) J. Mol. Biol., 288, 305–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen X., Kang,H., Shen,L.X., Chamorro,M., Varmus,H.E. and Tinoco,I. (1996) J. Mol. Biol., 260, 479–483. [DOI] [PubMed] [Google Scholar]
- 76.Kang H. and Tinoco,I. (1997) Nucleic Acids Res., 25, 1943–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Theimer C.A. and Giedroc,D.P. (1999) J. Mol. Biol., 289, 1283–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liphardt J., Napthine,S., Kontos,H. and Brierley,I. (1999) J. Mol. Biol., 288, 321–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gutell R.R., Larsen,N. and Woese,C.R. (1994) Microbiol. Rev., 58, 10–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Aruscavage P.J. and Bass,B.L. (2000) RNA, 6, 257–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Poole E.S., Brown,C.M. and Tate,W.P. (1995) EMBO J., 14, 151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jacobs G.H., Stockwell,P.A., Schrieber,M.J., Tate,W.P. and Brown,C.M. (2000) Nucleic Acids Res., 28, 293–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Major L.L., Poole,E.S., Dalphin,M.E., Mannering,S.A. and Tate,W.P. (1996) Nucleic Acids Res., 24, 2673–2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Poole E.S., Major,L.L., Mannering,S.A. and Tate,W.P. (1998) Nucleic Acids Res., 26, 954–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ito K., Uno,M. and Nakamura,Y. (2000) Nature, 403, 680–684. [DOI] [PubMed] [Google Scholar]
- 86.Blaxter M. (1998) Science, 282, 2041–2046. [DOI] [PubMed] [Google Scholar]
- 87.Hammell A.B., Taylor,R.C., Peltz,S.W. and Dinman,J.D. (1999) Genome Res., 9, 417–427. [PMC free article] [PubMed] [Google Scholar]
- 88.Higgins D.G., Thompson,J.D. and Gibson,T.J. (1996) Methods Enzymol., 266, 383–402. [DOI] [PubMed] [Google Scholar]