Abstract
Objectives
To describe the epidemiology of Pneumocystis jirovecii pneumonia and colonization diagnosed by next-generation sequencing (NGS) and explore the usefulness of the number of P. jirovecii sequence reads for the diagnosis of P. jirovecii pneumonia.
Methods
We examined the NGS results for P. jirovecii in respiratory samples collected from patients and analysed their clinical, radiological and microbiological characteristics.
Results
Among 285 respiratory samples collected over a 12-month period (January to December 2022), P. jirovecii sequences were detected in 56 samples from 53 patients. Fifty (94.3%) of the 53 patients were HIV-negative. Following our case definitions, 37 (69.8%) and 16 (30.2%) of the 53 patients had P. jirovecii infection and colonization respectively. P. jirovecii infection was associated with presence of underlying disease with immunosuppression (94.6% vs 18.8%, P < 0.05), positive serum 1,3-β-D-glucan (41.2% vs 0%, P < 0.01) and higher number of P. jirovecii sequence reads (P < 0.005). In contrast, P. jirovecii colonization was associated with the male sex (93.8% vs 54.1%, P < 0.01), another definitive infectious disease diagnosis of the respiratory tract (43.8% vs 2.7%, P < 0.001) and higher survival (100% vs 67.6%, P < 0.01). Although P. jirovecii pneumonia was associated with higher number of P. jirovecii reads in respiratory samples, only a sensitivity of 82.14% and a specificity of 68.75% could be achieved.
Conclusion
Detection of P. jirovecii sequences in respiratory samples has to be interpreted discreetly. A combination of clinical, radiological and laboratory findings is still the most crucial in determining whether a particular case is genuine P. jirovecii pneumonia.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11046-024-00849-y.
Keywords: Pneumocystis jirovecii, Pneumonia, Respiratory tract, Colonization, Next-generation sequencing
Introduction
Pneumocystis jirovecii is a fungus that causes pneumonia in immunocompromised patients. Clinically P. jirovecii pneumonia is characteristically associated with fever, shortness of breath and hypoxia and radiologically ground glass opacities are often observed. In general, it causes a relatively milder disease with lower (10–12%) mortality in human immunodeficiency virus (HIV)-positive patients but more severe disease with higher (30–50%) mortality in other immunocompromised patients who are HIV-negative [1]. Traditionally, laboratory diagnosis of P. jirovecii pneumonia was achieved by direct detection of P. jirovecii asci in respiratory tract specimens by microscopic examination after Grocott-Gomori methenamine silver (GMS) or immunofluorescence staining [2]. In recent years, polymerase chain reaction (PCR) has also been used for the detection of P. jirovecii [3–6]. Although it has improved the sensitivity of detection, it is not able to distinguish between genuine P. jirovecii pneumonia and P. jirovecii colonization of the respiratory tract [7, 8].
In the last few years, next-generation sequencing (NGS) has emerged as a technology for laboratory diagnosis of many culture-negative infections [9, 10]. We have recently reported its application in confirming the first case of listeria meningitis in a patient with autoantibody against interferon gamma as well as understanding the spectrum of Q fever, fungal infections and culture-negative meningitis and encephalitis [10–13]. It is notable that we have shown, in our recent review, that in fact P. jirovecii is the commonest fungal organism detected by NGS in clinical specimens [10]. Furthermore, others have also shown that NGS is much more sensitive than direct GMS staining and microscopy for the detection of P. jirovecii in respiratory samples [14–16]. In this study, we describe the epidemiology of P. jirovecii infection and colonization diagnosed by NGS in our hospital and discuss the reasons that may account for such phenomena. In addition, we also explore the usefulness of the number of P. jirovecii sequence reads for the diagnosis of P. jirovecii pneumonia and discuss how to interpret NGS results.
Materials and Methods
Ethical Statement
This study was approved by the Institutional Review Board of The University of Hong Kong—Shenzhen Hospital ([2022]120), and the requirement of obtaining informed consent was exempted.
Patients
This study was conducted over a 12-month period (January to December 2022) in The University of Hong Kong—Shenzhen Hospital, Shenzhen, China. This 1,400-bed multi-specialty hospital was established in 2012 and provides primary to tertiary medical services to the residents of Shenzhen city in both inpatient and outpatient settings. Supported through the policy from the government of Shenzhen, the hospital is established as a reform model medical institution in China, and many new medical technologies can be introduced to the hospital first. The laboratory reports of all respiratory samples submitted for NGS were examined. The clinical details, laboratory data and radiological findings of all patients with P. jirovecii sequence reads detected in their respiratory samples were retrieved from the hospital electronic record system and analysed.
Case Definitions
According to the Consensus Definitions of Invasive Fungal Disease of the European Organization for Research and Treatment of Cancer and the Mycoses Study Group [17], a case of P. jirovecii infection is defined as a P. jirovecii NGS-positive patient, whom after careful consideration of his/her clinical, radiological and laboratory findings, the clinician-in-charge has decided to prescribe specific anti-P. jirovecii treatment. A case of P. jirovecii colonization is defined as a P. jirovecii NGS-positive patient, whom after careful consideration of his/her clinical, radiological and laboratory findings, the clinician-in-charge has decided not to prescribe specific anti-P. jirovecii treatment.
Microbiological and Other Laboratory Tests
Clinical specimens were collected and handled according to standard protocols [18]. Direct detection of P. jirovecii and acid-fast bacilli were performed by GMS stain and Ziehl–Neelsen stain, respectively. The identities of bacterial and fungal isolates were confirmed by matrix-assisted laser desorption ionization–time-of-flight mass spectrometry [19]. Cryptococcal antigen detection was performed using lateral flow assay (Norman, USA). 1,3-β-D-glucan detection was performed using Test Kit for the Detection of Fungus 1,3-β-D-Glucan (Photometric Assay) (A & C Biological Ltd, Zhanjiang, China). Real-time PCR for Mycobacterium tuberculosis was performed using M. tuberculosis DNA Fluorescence Diagnostic Kit (PCR-Fluorescence Probing) (Sansure Biotech, Hunan, China); and real-time RT-PCR for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was performed using 2019-nCoV Nucleic Acid Test Kit (Biogerm, Shanghai, China). Real-time RT-PCR for influenza virus A and real-time PCR for herpes simplex virus (HSV) were performed by KingMed Diagnostics company.
NGS
The sputum and bronchoalveolar lavage (BAL) samples were sent to KingMed Diagnostics company, Sagene company, Vision Medicals company, Dinfectome company or GensKey company for targeted NGS (tNGS) or metagenomics NGS (mNGS) analysis.
Statistical Analysis
A comparison of characteristics between the P. jirovecii infection and colonization groups was performed. Chi-square test was used for categorical variables and unpaired Student’s t-test or Mann–Whitney U test was used for continuous variables. P < 0.05 was considered as statistically significant.
Results
Patients
During the 12-month study period, a total of 285 respiratory samples from 241 patients were submitted for tNGS or mNGS analyses. Among these 285 samples, P. jirovecii sequence reads were detected in 56 samples from 53 patients. For these 53 patients, the male to female ratio was 35:18. The median age was 61 (range 30 to 85) years. Thirty-eight (71.7%) of the 53 patients had underlying immunocompromised conditions, the commonest being malignancies (n = 19), followed by connective tissue and autoimmune diseases on corticosteroid and/or other immunosuppressive treatment (n = 15), solid organ transplant recipients on corticosteroid and/or other immunosuppressive treatment (n = 4) and HIV infection (n = 3) (Table 1).
Table 1.
Demographic and clinical characteristics of patients in whom Pneumocystis jirovecii was detected by NGS
| Case no | Sex/age | Underlying disease(s) | Key clinical manifestation(s) | Immunosuppressive and/or chemotherapy of underlying disease(s) | Colonization/infection of P. jirovecii | Anti-P. jirovecii treatment | Outcome |
|---|---|---|---|---|---|---|---|
| 1 | F/51 | Dermatomyositis, ILD | Fever, skin rash and rupture, SOB | Methylprednisolone, hydroxychloroquine, tacrolimus | Infection | TMP-SMX | Improved |
| 2 | M/72 | Renal transplantation, DM | Cough, fever | Cyclosporin A, prednisone, MMF | Colonization | None | Improved |
| 3 | M/44 | Renal transplantation | Cough, SOB, fever | Tacrolimus, prednisone | Infection | TMP-SMX | Improved |
| 4 | M/66 | Hepatocellular carcinoma, chronic HBV infection, immune pneumonitis | Fever, diarrhea, SOB | Prednisone, MMF, sorafenib, camrelizumab, apatinib, radiotherapy | Infection | TMP-SMX | Succumbed |
| 5 | F/69 | Brest carcinoma | SOB | Abemaciclib | Infection | TMP-SMX | Succumbed |
| 6 | M/67 | Lung transplantation, DM, COPD, pulmonary heart disease, CKD | SOB | Tacrolimus, prednisone, MMF | Infection | TMP-SMX, caspofungin, clindamycin | Succumbed |
| 7 | F/45 | Breast carcinoma | SOB | Doxorubicin, cyclophosphamide, paclitaxel, dexamethasone | Infection | TMP-SMX | Improved |
| 8 | M/40 | Hyperthyroidism, gout | Fever, headache | None | Colonization | None | Improved |
| 9 | F/49 | Neuromyelitis optica spectrum disorders, Hashimoto's thyroiditis | Fever, headache, limb spasticity, SOB | MMF, prednisone | Infection | TMP-SMX | Improved |
| 10 | F/48 | Breast carcinoma, adult Still's disease | SOB, fever | Radiotherapy, methylprednisolone, cyclosporin A | Infection | TMP-SMX | Improved |
| 11 | M/58 | MDS | Cough, SOB | None | Infection | TMP-SMX | Improved |
| 12 | M/47 | Chronic HBV infection, AIDS | Cough, fatigue, fever | None | Infection | TMP-SMX | Improved |
| 13 | M/51 | Nasopharyngeal carcinoma, renal transplantation, chronic HBV infection | Facial edema, SOB, cough, sore throat | Tacrolimus, MMF, paclitaxel, cisplatin, capecitabine, tegafur, gimeracil, oteracil, gemcitabine, cetuximab, docetaxel, nimotuzumab, radiotherapy, anlotinib | Infection | Caspofungin, clindamycin | Succumbed |
| 14 | M/67 | Hypertension, antisynthetase syndrome | Cough, fever | None | Infection | TMP-SMX | Improved |
| 15 | M/47 | Chronic HBV infection, bronchiectasis, hamartoma of left lung | Cough, fever | None | Colonization | None | Improved |
| 16 | M/54 | Meningioma, ANCA-associated small-vessel vasculitis, COPD | Cough | None | Infection | TMP-SMX | Improved |
| 17 | F/69 | Lung carcinoma | Fever, SOB | Almonertinib, osimertinib, dexamethasone | Infection | TMP-SMX | Improved |
| 18 | F/80 | RA, ILD, pulmonary arterial hypertension | SOB, fever | Leflunomide, hydroxychloroquine, prednisone, dexamethasone, iguratimod, tripterygium glycosides, tofacitinib, denosumab | Infection | TMP-SMX | Improved |
| 19 | M/63 | COPD | SOB, cough, hemoptysis | None | Colonization | None | Improved |
| 20 | F/66 | Overlap syndrome | Cough, recurrent SOB | Prednisone, hydroxychloroquine, azathioprine | Colonization | None | Improved |
| 21 | M/71 | Membranous nephropathy, DM, hypertension, CHD | General edema | None | Infection | TMP-SMX | Improved |
| 22 | F/35 | Myasthenia gravis, Ekbom syndrome, xerophthalmia, post-resection of thymoma | SOB | Prednisone, azathioprine | Infection | TMP-SMX, caspofungin, clindamycin | Improved |
| 23 | M/62 | Asthma, ABPA | SOB, fever | None | Colonization | None | Improved |
| 24 | M/82 | CKD | Cough, SOB | None | Colonization | None | Improved |
| 25 | F/59 | Breast carcinoma, radiation pneumonitis | Cough, SOB | Pharmorubicin, cyclophosphamide, docetaxel, abemaciclib | Infection | TMP-SMX | Improved |
| 26 | M/42 | Aplastic anemia, community acquired pneumonia | Cough, sore throat, fever | None | Colonization | None | Improved |
| 27 | M/63 | Subacute combined degeneration | Fatigue, myalgia | Methylprednisolone | Infection | TMP-SMX | Improved |
| 28 | F/53 | T cell lymphoma | SOB | None | Infection | TMP-SMX | Improved |
| 29 | F/52 | Breast carcinoma | None | Doxorubicin, cyclophosphamide, paclitaxel, dexamethasone | Infection | TMP-SMX | Improved |
| 30 | F/46 | Breast carcinoma | Fever, cough | Doxorubicin, cyclophosphamide, paclitaxel, dexamethasone | Infection | TMP-SMX | Improved |
| 31 | M/61 | Mantle cell lymphoma | Erythema, desquamation, fever | Zanubrutinib, prednisone | Infection | TMP-SMX | Improved |
| 32 | M/52 | Chronic HBV infection, mediastinal solitary fibrous tumor | Fever, cough | None | Colonization | None | Improved |
| 33 | M/46 | Thymoma, myasthenia gravis, bronchiectasis | Cough | Methylprednisolone | Infection | TMP-SMX, caspofungin, clindamycin | Improved |
| 34 | M/73 | T cell lymphoma, DM, liver cirrhosis | Cough, SOB, fever | Chidamide, thalidomide, lenalidomide, cisplatin, gemcitabine, L-asparaginase, ifosfamide, etoposide, vincristine, dexamethasone, pomalidomide, cytarabine | Infection | TMP-SMX | Succumbed |
| 35 | M/75 | Gastric carcinoma | None | Oxaliplatin, capecitabine | Infection | None | Succumbed |
| 36 | M/65 | ILD | Palpitation, cough, hemoptysis, chest distress | Prednisone, nintendanib, methylprednisolone, pirfenidone | Infection | TMP-SMX | Succumbed |
| 37 | F/36 | Breast carcinoma | Chest distress, SOB | Doxorubicin, cyclophosphamide, paclitaxel | Infection | TMP-SMX | Improved |
| 38 [20] | F/71 | Hemophagocytic lymphohistiocytosis, Still's disease | Fever, fatigue, chest distress, SOB | Dexamethasone, prednisone, cyclosporine, tocilizumab | Infection | TMP-SMX | Succumbed |
| 39 | M/66 | DM | Fever | None | Colonization | None | Improved |
| 40 | F/55 | Anti-MDA5 antibody dermatomyositis | Cough, palpitation | None | Infection | TMP-SMX | Succumbed |
| 41 | M/30 | AIDS | Fever, SOB | None | Infection | TMP-SMX | Improved |
| 42 | F/55 | Breast carcinoma | Fever | Doxorubicin, cyclophosphamide | Infection | TMP-SMX | Improved |
| 43 | M/85 | DM | Fever, cough | None | Colonization | None | Improved |
| 44 | M/73 | COPD, CHD, gout, hypertension, renal calculi, BPH | Fever, cough, coma | None | Infection | TMP-SMX | Succumbed |
| 45 | M/44 | AIDS | SOB, cough, diarrhea | None | Infection | TMP-SMX | Improved |
| 46 | M/66 | DM | None | None | Colonization | None | Improved |
| 47 | M/66 | None | Cough, fever | None | Colonization | None | Improved |
| 48 | M/84 | DM | Cough, sputum | None | Colonization | None | Improved |
| 49 | F/75 | Ovarian malignant teratoma, RA | SOB, fever | Bleomycin, etoposide, cisplatin | Infection | TMP-SMX | Succumbed |
| 50 | M/67 | COPD | Fever, cough | None | Colonization | None | Improved |
| 51 | M/55 | None | SOB, cough, fever | None | Colonization | None | Improved |
| 52 | M/73 | RA, ILD | SOB, fever | Methotrexate, hydroxychloroquine, sulfasalazine, iguratimod | Infection | TMP-SMX | Improved |
| 53 | M/53 | Dermatomyositis, ILD | SOB, cough | Prednisone, cyclophosphamide, pirfenidone | Infection | TMP-SMX | Succumbed |
F, Female; M, Male; ILD, Interstitial lung disease; SOB, Shortness of breath; TMP-SMX, Trimethoprim-sulfamethoxazole; DM, Diabetes mellitus; MMF, Mycophenolate mofetil; HBV, Hepatitis B virus; COPD, Chronic obstructive pulmonary disease; CKD, Chronic kidney disease; MDS, Myelodysplastic syndrome; AIDS, Acquired immune deficiency syndrome; ANCA, Anti-neutrophil cytoplasmic antibodies; RA, Rheumatoid arthritis; CHD, Coronary heart disease; ABPA, Allergic bronchopulmonary aspergillosis; MDA5, Melanoma differentiation-associated protein 5; BPH, Benign prostatic hyperplasia
NGS Analysis
In 44 of the 53 patients, P. jirovecii was detected by tNGS; whereas in nine patients, it was detected by mNGS. In samples collected from 31 (58.5%) of the 53 patients, sequence reads of other potential respiratory pathogens were detected (Table 2). These included bacteria (Acinetobacter baumannii, Bordetella pertussis, Chlamydia psittaci, Haemophilus influenzae, Klebsiella pneumoniae, Legionella pneumophilia, Pseudomonas aeruginosa, Staphylococcus aureus, Stenotrophomonas maltophilia and Streptococcus pneumoniae), mycobacteria (Mycobacterium abscessus, Mycobacterium chelonae, Mycobacterium intracellulare, Mycobacterium kansasii and M. tuberculosis), viruses (adenovirus, influenza virus and rhinovirus) and fungi (Aspergillus fumigatus, Cryptococcus neoformans and Trichosporon asahii). In samples collected from 26 (49.1%) of the 53 patients, sequences that were considered as contaminants or colonizers were present. Most of them were bacteria and yeasts present in the oral cavity of immunocompetent or immunocompromised hosts (Supplementary Table 1). In samples collected from eight (cases 5, 17, 29, 30, 31, 32, 37 and 51) of the 53 patients, Tropheryma whipplei, a bacterium of doubtful clinical significance in the respiratory tract [21], was detected (Table 2).
Table 2.
NGS analysis and other key laboratory results of patients in the present cohort
| Case no | Gomori methenamine silver staining | NGS | 1,3-β-D-glucan (pg/mL) | Other positive microbiological tests | ||
|---|---|---|---|---|---|---|
| Specimen | tNGS/mNGS | Sequencing result (number of reads) | ||||
| 1 | Negative | BAL | mNGS | Pneumocystis jirovecii (5), Prevotella melaninogenica (81), Veillonella parvula (5), Mycobacterium intracellulare (1) | 201.61 | None |
| 2 | Not done | Sputum | tNGS | Enterobacter cloacae complex (9), P. jirovecii (19), EBV (17,782), CMV (50), HHV-7 (49), Ureaplasma urealyticum (2) | < 37.5 | None |
| 3 | Negative | BAL | tNGS | P. jirovecii (1886) | < 37.5 | None |
| 4 | Negative | BAL | mNGS | Rothia mucilaginosa (45,448), Streptococcus mitis (13,051), Streptococcus pneumoniae (11,961), P. jirovecii (168), HSV-1 (17), Olsenella uli (18,072), P. melaninogenica (8702), Staphylococcus haemolyticus (6259), V. parvula (3530), Parvimonas micra (3037), Corynebacterium simulans (1984), Cryptobacterium curtum (1390), Filifactor alocis (379), Leptotrichia buccalis (329), Corynebacterium striatum (269), Atopobium parvulum (261), Peptostreptococcus anaerobius (152), Clostridioides difficile (136), Actinomyces oris (82) | > 600 | None |
| 5 | Negative | Sputum | tNGS | Tropheryma whipplei (19,095), Haemophilus influenzae (387), Staphylococcus aureus (26), P. jirovecii (14), rhinovirus C (16,789), EBV (7569) | < 37.5 | None |
| 6 | Negative | BAL | tNGS | Candida albicans (27,428), EBV (3357), Enterococcus faecalis (1925), Stenotrophomonas maltophilia (321), P. jirovecii (141), HHV-7 (136), Trichosporon asahii (34) | < 37.5 | None |
| 7 | Negative | BAL | mNGS | S. aureus (3), P. jirovecii (2), Neisseria flavescens (40), Haemophilus parainfluenzae (15), P. melaninogenica (15), R. mucilaginosa (12), Porphyromonas gingivalis (11), Fusobacterium nucleatum (6), Capnocytophaga granulosa (4), Peptostreptococcus stomatis (4), Aggregatibacter segnis (4), F. alocis (4), Treponema denticola (4), Veillonella dispar (4), Streptococcus pseudopneumoniae (3) | < 37.5 | None |
| 8 | Not done | Sputum | tNGS | Chlamydia psittaci (49), Acinetobacter baumannii (177), H. influenzae (33), Pseudomonas aeruginosa (22), S. maltophilia (14), HHV-7 (32), P. jirovecii (11), HHV-6 (6) | Not done | None |
| 9 | Not done | BAL | mNGS | P. jirovecii (830), Streptococcus species (23), R. mucilaginosa (10), Abiotrophia defectiva (7), Granulicatella adiacens (5), Staphylococcus epidermidis (3), Tannerella forsythia (2), Prevotella denticola (1), EBV (1), Torque teno virus (1) | > 600 | None |
| 10 | Positive | BAL | tNGS | P. jirovecii (48,609), EBV (3) | 394.01 | None |
| 11 | Negative | BAL | mNGS | S. aureus (79), C. albicans (1757), P. jirovecii (2), human polyomavirus 5 (4), Mycobacterium chelonae (3), S. haemolyticus (264,865), Lactobacillus rhamnosus (12,454), Mogibacterium timidum (72), Corynebacterium tuberculostearicum (9) | < 37.5 | None |
| 12 | Positive | BAL | tNGS | P. jirovecii (84,000), Bordetella pertussis (1705), rhinovirus A (44) | 49.72 | None |
| 13 | Not done | Sputum | tNGS | Klebsiella pneumoniae (44,975), A. baumannii (15,986), S. maltophilia (11,464), E. faecalis (11,480), EBV (826), P. jirovecii (87), E. cloacae complex (32), HSV-1 (9) | < 37.5 | None |
| 14 | Not done | BAL | mNGS | P. jirovecii (15), P. melaninogenica (1405), R. mucilaginosa (930), Campylobacter concisus (432), Streptococcus infantis (363), Gemella sanguinis (269), Veillonella atypica (189), Eikenella corrodens (123), Actinomyces graevenitzii (115), Solobacterium moorei (104), Capnocytophaga sputigena (101), H. parainfluenzae (83), A. defectiva (66), L. buccalis (54), P. stomatis (51), Oribacterium sinus (45) | < 37.5 | None |
| 15 | Negative | Sputum | tNGS | EBV (1215), HHV-7 (741), P. jirovecii (72) | < 37.5 | None |
| 16 | Negative | BAL | tNGS | P. jirovecii (4476), C. albicans (4447), Legionella pneumophilia (17), HHV-7 (83), CMV (5) | < 37.5 | None |
| Negative | BAL | tNGS | P. jirovecii (322), C. albicans (178), CMV (12), HHV-7 (10) | < 37.5 | None | |
| 17 | Negative | BAL | tNGS | C. albicans (424), P. jirovecii (135), CMV (135), H. influenzae (19), T. whipplei (54), HHV-7 (6) | 61.96 | None |
| 18 | Negative | BAL | tNGS | Rhinovirus (192), P. aeruginosa (278), P. jirovecii (105), S. pneumoniae (5), EBV (7) | 183.52 | Sputum culture: P. aeruginosa |
| 19 | Not done | Sputum | tNGS | P. aeruginosa (40,038), EBV (882), CMV (23), HHV-7 (12), P. jirovecii (7) | < 37.5 | Sputum culture: P. aeruginosa |
| 20 | Not done | Sputum | tNGS | P. aeruginosa (48,178), Mycobacterium kansasii (150), S. maltophilia (603), EBV (274), P. jirovecii (166), CMV (115), C. albicans (27), HHV-7 (19) | < 37.5 | Sputum for AFB smear: positive; sputum culture: P. aeruginosa |
| 21 | Not done | Sputum | tNGS | S. aureus (6472), P. jirovecii (184), HHV-7 (136), Haemophilus haemolyticus (113) | < 37.5 | None |
| 22 | Not done | Sputum | tNGS | Mycobacterium abscessus (3907), H. influenzae (46,294), H. haemolyticus (355), P. jirovecii (212), EBV (129), HSV-1 (97), CMV (32), HHV-7 (30) | Not done | None |
| 23 | Negative | Sputum | tNGS | Influenza virus A (38,696), Aspergillus fumigatus (76), P. jirovecii (1) | < 37.5 | NPS for Influenza A virus RNA: positive |
| 24 | Not done | Sputum | tNGS | K. pneumoniae (54,414), A. baumannii (10,292), E. faecalis (8694), EBV (1517), CMV (93), S. aureus (88), HHV-7 (46), P. jirovecii (13) | Not done | None |
| 25 | Negative | BAL | tNGS | P. jirovecii (172) | < 37.5 | None |
| 26 | Not done | Sputum | tNGS | A. baumannii (173), H. haemolyticus (51), S. aureus (21), P. jirovecii (8) | < 37.5 | None |
| 27 | Negative | BAL | mNGS | Escherichia coli (61,948), K. pneumoniae (1424), E. cloacae complex (3289), P. jirovecii (1104), HSV (1,095,105), EBV (53), HHV-7 (26), HHV-6B (4), S. epidermidis (494,451), V. parvula (215,980), Actinomyces dentalis (22,892), S. infantis (46,135), Campylobacter curvus (32,006), Neisseria bacilliformis (27,509), Corynebacterium matruchotii (12,641), Cutibacterium acnes (11,078), E. corrodens (12,967), C. albicans (4452), Trichomonas tenax (723) | 128.64 | BAL culture: K. pneumoniae, C. albicans |
| 28 | Not done | Sputum | tNGS | P. jirovecii (117), HHV-7(11) | < 37.5 | None |
| 29 | Negative | BAL | tNGS | P. jirovecii (4137), P. aeruginosa (27), E. coli (14), T. whipplei (749) | Not done | BAL culture: P. aeruginosa |
| 30 | Negative | BAL | tNGS | P. jirovecii (655), T. whipplei (14) | < 37.5 | None |
| 31 | Negative | BAL | mNGS | P. jirovecii (878), T. whipplei (234), Torque teno virus (510), CMV (200), S. pseudopneumoniae (280), Streptococcus oralis (57), H. parainfluenzae (73), H. haemolyticus (10), G. adiacens (14), R. mucilaginosa (13) | 87.61 | None |
| 32 | Not done | Sputum | tNGS | K. pneumoniae (6055), H. haemolyticus (1738), EBV (330), S. aureus (94), T. whipplei (366), HHV-7 (77), P. jirovecii (32), HHV-6 (11), CMV (5) | Not done | None |
| 33 | Not done | Sputum | tNGS | H. haemolyticus (1960), HHV-7 (365), P. jirovecii (274), T. asahii (1), EBV (34), C. albicans (7) | 87.6 | None |
| Not done | Sputum | tNGS | H. haemolyticus (1385), H. influenzae (14,635), HHV-7 (548), EBV (77), P. jirovecii (45), C. albicans (20) | None | ||
| Not done | BAL | tNGS | P. jirovecii (189), C. albicans (23), CMV (6), HHV-7 (4) | None | ||
| 34 | Not done | Sputum | tNGS | C. albicans (20,746), A. baumannii (1112), K. pneumoniae (340), P. jirovecii (260) | 76.97 | Sputum culture: A. baumannii, C. albicans |
| 35 | Not done | BAL | tNGS | Nontuberculosis mycobacteria (185), P. jirovecii (52) | Not done | None |
| 36 | Negative | Sputum | tNGS | K. pneumoniae (24,020), H. influenzae (8038), EBV (199), C. albicans (163), A. baumannii (41), HHV-7 (36), P. jirovecii (27) | < 37.5 | BAL culture: K. pneumoniae |
| 37 | Negative | BAL | tNGS | P. jirovecii (9600), T. whipplei (8) | 225.8 | None |
| 38 [20] | Positive | BAL | tNGS | P. jirovecii (34,019), CMV (2975), EBV (8), K. pneumoniae (21), C. albicans (2) | 293.99 | Blood and pleural effusion culture: Nocardia kroppenstedtii; BAL culture: K. pneumoniae, N. kroppenstedtii |
| 39 | Negative | BAL | tNGS | Mycobacterium tuberculosis complex (43,834), P. jirovecii (927) | < 37.5 | BAL for AFB smear: positive; BAL for M. tuberculosis DNA: positive |
| 40 | Positive | BAL | tNGS | P. jirovecii (25,284), EBV (17) | < 37.5 | None |
| 41 | Positive | BAL | tNGS | P. jirovecii (58,385), EBV (374), E. faecalis (199) | 93.8 | None |
| 42 | Negative | BAL | tNGS | P. jirovecii (2470) | < 37.5 | None |
| 43 | Negative | BAL | tNGS | HSV-1 (580), E. coli (11), HHV-7 (4), C. albicans (10,441), P. jirovecii (49) | < 37.5 | BAL for HSV DNA: positive |
| 44 | Negative | BAL | tNGS | P. aeruginosa (3638), K. pneumoniae (58), CMV (61), P. jirovecii (47) | < 37.5 | BAL culture: P. aeruginosa |
| 45 | Positive | BAL | tNGS | P. jirovecii (64,629), CMV (81) | < 37.5 | None |
| 46 | Not done | BAL | tNGS | Cryptococcus neoformans (73,023), P. jirovecii (796) | < 37.5 | Serum for cryptococcal antigen: positive |
| 47 | Not done | Sputum | tNGS | P. aeruginosa (6493), H. haemolyticus (80), adenovirus C (12), C. albicans (102), P. jirovecii (62), EBV (23), HHV-7 (4) | < 37.5 | BAL for M. tuberculosis complex tNGS: 11 reads |
| 48 | Not done | BAL | tNGS | K. pneumoniae (56,021), P. aeruginosa (18,635), S. aureus (2661), P. jirovecii (1108), EBV (792) | Not done | BAL culture: K. pneumoniae, P. aeruginosa |
| 49 | Not done | Sputum | tNGS | K. pneumoniae (37,575), S. aureus (13,401), P. aeruginosa (13,123), H. haemolyticus (25), C. albicans (6927), P. jirovecii (201) | < 37.5 | None |
| 50 | Not done | BAL | tNGS | K. pneumoniae (73,975), P. jirovecii (56), CMV (12) | < 37.5 | BAL culture: K. pneumoniae |
| 51 | Not done | Sputum | tNGS | H. haemolyticus (25), T. whipplei (1373), P. jirovecii (202) | Not done | Throat swab for SARS-CoV-2 RNA: positive |
| 52 | Negative | BAL | mNGS | K. pneumoniae (177), P. jirovecii (11), HHV-6B (1), Corynebacterium propinquum (442,826), Dolosigranulum pigrum (140,682), V. parvula (76,864), C. acnes (39,845), Prevotella salivae (32,519), A. dentalis (2687), Megasphaera micronuciformis (6162), Moraxella nonliquefaciens (3916), C. concisus (4279), S. epidermidis (2934), Streptococcus anginosus (855), S. salivarius (588), C. albicans (5555) | < 37.5 | None |
| 53 | Negative | BAL | tNGS | P. jirovecii (27) | < 37.5 | None |
BAL, Bronchoalveolar lavage; NGS, Next-generation sequencing; tNGS, Targeted NGS; mNGS, Metagenomics NGS; EBV, Epstein-Barr virus; CMV, Cytomegalovirus; HHV, Human herpes virus; HSV, Herpes simplex virus; AFB, Anti-fast bacilli; NPS, Nasopharyngeal swab; SARS-CoV-2, Severe acute respiratory syndrome coronavirus 2
Other Microbiology Tests
Serum cryptococcal antigen was positive in one patient (case 46), and acid-fast bacilli were detected in two patients (cases 20 and 39) and M. tuberculosis DNA in one patient (case 39) (Table 2). HSV DNA was positive in the BAL of one patient (case 43). Influenza A virus and SARS-CoV-2 RNA were each positive in the nasopharyngeal swab of two patients respectively (cases 23 and 51) (Table 2).
P. jirovecii Infection and Colonization
According to our case definitions, 37 (69.8%) of the 53 patients with P. jirovecii sequence reads detected in their respiratory samples had P. jirovecii infection, whereas the other 16 (30.2%) of the 53 were considered as colonization. P. jirovecii infection was associated with the presence of underlying disease with immunosuppression (35/37, 94.6%) compared to P. jirovecii colonization (3/16, 18.8%) (P < 0.05) (Table 3). Only two patients with P. jirovecii pneumonia did not have major immunosuppression. The first one (case 21, Table 1) was a 71-year-old man with membranous nephropathy, hypertension, diabetes mellitus and coronary heart disease. He refused to receive corticosteroid and other immunosuppressive treatment for his membranous glomerulonephritis. The second one (case 44, Table 1) was a 73-year-old man with chronic obstructive pulmonary disease, coronary heart disease, gout, hypertension, renal calculi and benign prostatic hyperplasia. In addition to underlying diseases, patients with P. jirovecii infection were associated with higher number of P. jirovecii sequence reads in their respiratory samples than those with P. jirovecii colonization (P < 0.005) (Table 3). Furthermore, there were significantly more patients with positive serum 1,3-β-D-glucan results in the infection (14/34, 41.2%) than in the colonization group (0/11) (P < 0.01) (Table 3). On the other hand, P. jirovecii colonization was associated with a predominance of the male sex (15/16, 93.8%) compared to P. jirovecii infection (20/37, 54.1%) (P < 0.01) (Table 3). Patients with P. jirovecii colonization was also associated with another definitive infectious disease diagnosis of the respiratory tract (7/16, 43.8%) compared to P. jirovecii infection (1/37) (P < 0.001) (Table 3). The definitive diagnoses of these seven patients were psittacosis (case 8), M. kansasii pulmonary infection (case 20), cryptococcosis (case 46), tuberculosis (case 39 and 47), influenza (case 23) and COVID-19 (case 51) (Table 2). Moreover, a significantly higher proportion of patients with P. jirovecii colonization (by definition did not receive specific anti-P. jirovecii treatment, 16/16, 100%) had improved compared to those with P. jirovecii infection (by definition received specific anti-P. jirovecii treatment, 25/37, 67.6%) (P < 0.01) (Table 3).
Table 3.
Comparison of characteristics in patients with Pneumocystis jirovecii infection and colonization
| Patient characteristics | Infection (n = 37) | Colonization (n = 16) | P-value | |
|---|---|---|---|---|
| Age (years) | 57.5 ± 12.7 | 63.4 ± 13.7 | 0.1356 | |
| Sex | Female | 17 | 1 | 0.0051 |
| Male | 20 | 15 | ||
| Underlying immunocompromised condition | All | 35 | 3 | < 0.0001 |
| HIV | 3 | 0 | ||
| Solid tumour on chemotherapy | 18 | 0 | ||
| Haematological malignancy | 3 | 0 | ||
| Connective tissue disease/autoimmune disease | 14 | 2 | ||
| Solid organ transplant | 3 | 1 | ||
| Clinical manifestations | Fever | 19 | 11 | 0.2407 |
| Cough | 15 | 12 | 0.0212 | |
| Shortness of breath | 24 | 6 | 0.065 | |
| Laboratory test | aMedian number of P. jirovecii sequence reads detected via tNGS in 44 of 53 patients (interquartile range), /100 K original reads | 236 (108, 8319) | 52.50 (11.5, 193) | 0.002 |
| Positive GMS staining respiratory samples in 31 of 53 patients | 6/27 | 0/4 | 0.5614 | |
| Positive serum 1,3-β-D-glucan in 45 of 53 patients | 14/34 | 0/11 | 0.0098 | |
| Definitive diagnosis of other infectious diseases | 1 | 7 | 0.0005 | |
| Outcome | Improved | 25 | 16 | 0.0096 |
| Succumbed | 12 | 0 | ||
tNGS, Targeted next-generation sequencing; GMS, Gomori methenamine silver
aAll the other nine patients with P. jirovecii detected via metagenomics NGS had P. jirovecii infection
Discussion
In this study, 53 patients in our hospital with P. jirovecii sequences in their respiratory samples were detected by tNGS or mNGS analysis. Among these 53 patients, only three were HIV-positive (cases 12, 41 and 45, Table 1), whereas the other 50 were HIV-negative. This is very different from the general epidemiology of P. jirovecii infections, of which HIV infection is the single most important risk factor. The common reasons for immunosuppression in the HIV-negative patients in this cohort were solid tumour or haematological malignancies on chemotherapy and autoimmune diseases or solid organ transplant recipients on corticosteroid and/or other immunosuppressive treatment, which is consistent with the changing epidemiological profile of P. jirovecii infection in the past decades [22]. For HIV-positive patients with P. jirovecii infections, the fungal loads in their respiratory tracts are usually high and direct microscopic examination after GMS staining, sometimes even using induced sputum samples, is often sufficient for making a diagnosis. In contrast, for the other immunocompromised patients, the fungal load is usually low and bronchoscopic examination has to be performed to collect BAL samples so as to improve the yield. In fact, for all the three HIV-positive patients in the present cohort, their BAL samples were also positive for P. jirovecii by direct microscopic examination after GMS staining, whereas for the 29 BAL samples obtained from the HIV-negative patients that were submitted for microscopic examination after GMS staining, only three were positive for P. jirovecii (P < 0.001 by Fisher’s Exact test). This is in line with the high number of P. jirovecii sequence reads (84,000 for cases 12, 58,385 for case 41 and 64,629 for case 45) (Table 2) observed in the three BAL samples collected from the three HIV-positive patients examined by tNGS, which is significantly higher than the number of P. jirovecii sequence reads (median 655, range 27 to 48,609) in the BAL samples collected from the 21 HIV-negative patients examined by tNGS (P < 0.001 by Mann–Whitney U test).
Detection of P. jirovecii sequence reads in respiratory samples has to be interpreted discreetly. Traditionally, P. jirovecii infection was diagnosed in the laboratory by direct detection of P. jirovecii asci in respiratory samples after GMS staining in immunocompromised patients with suspected clinico-radiological features, such as shortness of breath, hypoxia and ground glass infiltrates on chest radiographs. In the past decades, a number of PCR assays have been developed for the detection of P. jirovecii in respiratory tract specimens [3–6]. In some of these studies, colonization of P. jirovecii in the respiratory tract has been suggested [5–8]. In the present cohort, 16 (29.6%) of the 53 patients with P. jirovecii sequences in their respiratory samples detected by NGS analysis recovered without receiving specific anti-P. jirovecii therapy. In some of these 16 patients, other respiratory pathogens were present. For example, C. psittaci was detected from the sputum of Case 8 and he responded to doxycycline well; and in Cases 23 and 51, influenza A virus and SARS-CoV-2 RNA were detected in their nasopharyngeal and throat swabs respectively (Table 2). All of the 16 patients improved after receiving specific antimicrobial therapy to the other pathogens identified or just symptomatic treatment. In these 16 patients, P. jirovecii was considered as colonizers of the respiratory tract, rather than pathogens; and they were associated with the male gender, absence of underlying disease, negative serum 1,3-β-D-glucan, and a lower number of P. jirovecii sequence reads (Table 3). It is interesting to note that in our recent study on the detection of T. whipplei in respiratory samples by NGS, Whipple disease was never suspected to be a diagnosis in any of the patients before detection of the bacterium; and the presence of T. whipplei in the respiratory specimens of these patients was still elusive [21].
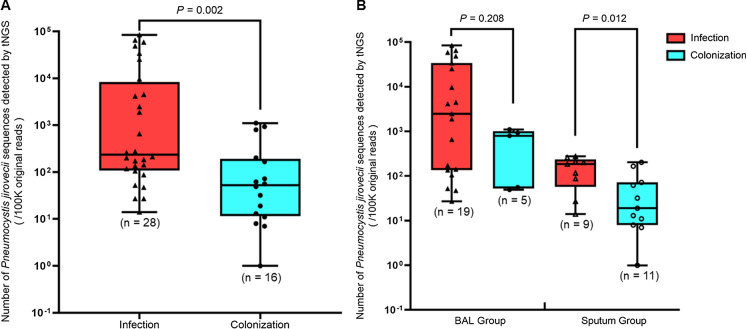
Although infection is associated with a significantly higher number of P. jirovecii sequence reads as compared to colonization, clinical judgement is still the most crucial in determining whether a particular case is genuine P. jirovecii pneumonia. When the number of P. jirovecii sequence reads in all respiratory (sputum and BAL) samples between the infection and colonization groups were compared, it was observed that the number of sequence reads was significantly higher in the infection than the colonization group (P < 0.005) (Fig. 1A). However, for example, if 79.5 reads were used as the cutoff for distinguishing between P. jirovecii pneumonia and colonization, only a sensitivity of 82.14% and a specificity of 68.75% could be achieved. Furthermore, when the analysis was performed for the sputum group, the number of sequence reads was still significantly higher in the infection than the colonization group (P < 0.05); but when the analysis was performed for the BAL group, there was no difference between the number of reads in the two groups, although there was still a trend towards a higher number of reads in the infection than the colonization group (Fig. 1B). All these showed that the number of sequence reads is not a reliable parameter to indicate whether a particular patient has P. jirovecii pneumonia or just P. jirovecii colonization. In contrast to the number of P. jirovecii sequence reads, it was shown in the present cohort that positive serum 1,3-β-D-glucan and direct GMS staining of respiratory samples were highly specific, although not sensitive, for P. jirovecii infection (Table 3); and hence would be useful for the prediction of P. jirovecii pneumonia if these results were positive. The final diagnosis of P. jirovecii pneumonia should be made using a combination of clinical, radiological and laboratory findings.
Fig. 1.
Distribution in number of P. jirovecii sequence reads in respiratory samples from patients in the present cohort detected by targeted next-generation sequencing (tNGS). Panel A: boxplot showing number of P. jirovecii sequence reads distribution in all respiratory samples from patients with P. jirovecii infection and colonization. Panel B: boxplot showing number of P. jirovecii sequence reads distribution in bronchoalveolar lavage (BAL) and sputum samples from patients with P. jirovecii infection and colonization
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We are grateful to the staff at the Department of Clinical Microbiology and Infection Control, The University of Hong Kong—Shenzhen Hospital for their technical support and assistance.
Author contributions
Fanfan Xing, Susanna K. P. Lau and Patrick C. Y. Woo designed the conceptualization; Fanfan Xing, Chaowen Deng, Zhendong Luo, Min Liu, Haiyan Ye, Linlin Sun and Patrick C. Y. Woo did data collection and curation; Fanfan Xing, Chaowen Deng and Patrick C. Y. Woo performed the data analysis; Fanfan Xing, Simon K. F. Lo, Susanna K. P. Lau and Patrick C. Y. Woo audited the methodology; Simon K. F. Lo supervised the laboratory administration and resources; Susanna K. P. Lau and Patrick C. Y. Woo supervised the research; Fanfan Xing and Patrick C. Y. Woo wrote the original draft; Fanfan Xing, Chaowen Deng, Zhendong Luo, Min Liu, Haiyan Ye, Linlin Sun, Chi-Ching Tsang, Simon K. F. Lo, Susanna K. P. Lau and Patrick C. Y. Woo reviewed, edited and approved the draft.
Funding
This work was partly supported by the Health Commission of Shenzhen Municipality under the Sanming Project of Medicine [SZSM201911014] in Shenzhen; the Ministry of Education in Taiwan under the framework of the Higher Education Sprout Project (MOE-112-S-023-A).
Declarations
Transparency
Patrick C. Y. Woo has provided scientific advisory/laboratory services for Gilead Sciences, Incorporated; International Health Management Associates, Incorporated; Merck & Corporation, Incorporated; Micología Molecular S.L. and Pfizer, Incorporated. The other authors report no conflict of interest. The funding sources had no role in study design, data collection, analysis, interpretation, or writing of the report. The authors alone are responsible for the content and the writing of the manuscript.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Susanna K. P. Lau, Email: skplau@hku.hk
Patrick C. Y. Woo, Email: pcywoo@hku.hk
References
- 1.Morris A, Norris KA. Colonization by P. jirovecii and its role in disease. Clin Microbiol Rev. 2012;25(2):297–317. doi: 10.1128/CMR.00013-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Desoubeaux G, Franck-Martel C, Caille A, Drillaud N, Carluer L, de Kyvon MA, Bailly É, Chandenier J. Use of calcofluor-blue brightener for the diagnosis of P. jirovecii pneumonia in bronchial-alveolar lavage fluids: a single-center prospective study. Med Mycol. 2017;55(3):295–301. doi: 10.1093/mmy/myw068. [DOI] [PubMed] [Google Scholar]
- 3.Wakefield AE, Pixley FJ, Banerji S, Sinclair K, Miller RF, Moxon ER, Hopkin JM. Detection of Pneumocystis carinii with DNA amplification. Lancet. 1990;336(8713):451–453. doi: 10.1016/0140-6736(90)92008-6. [DOI] [PubMed] [Google Scholar]
- 4.Robberts FJ, Liebowitz LD, Chalkley LJ. Polymerase chain reaction detection of Pneumocystis jiroveci: evaluation of 9 assays. Diagn Microbiol Infect Dis. 2007;58(4):385–392. doi: 10.1016/j.diagmicrobio.2007.02.014. [DOI] [PubMed] [Google Scholar]
- 5.Huggett JF, Taylor MS, Kocjan G, Evans HE, Morris-Jones S, Gant V, Novak T, Costello AM, Zumla A, Miller RF. Development and evaluation of a real-time PCR assay for detection of P. jirovecii DNA in bronchoalveolar lavage fluid of HIV-infected patients. Thorax. 2008;63(2):154–159. doi: 10.1136/thx.2007.081687. [DOI] [PubMed] [Google Scholar]
- 6.To KK, Wong SC, Xu T, Poon RW, Mok KY, Chan JF, Cheng VC, Chan KH, Hung IF, Yuen KY. Use of nasopharyngeal aspirate for diagnosis of pneumocystis pneumonia. J Clin Microbiol. 2013;51(5):1570–1574. doi: 10.1128/JCM.03264-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aguilar YA, Rueda ZV, Maya MA, Vera C, Rodiño J, Muskus C, Vélez LA. Is it possible to differentiate P. jirovecii pneumonia and colonization in the immunocompromised patients with pneumonia? J Fungi. 2021;7(12):1036. doi: 10.3390/jof7121036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fan LC, Lu HW, Cheng KB, Li HP, Xu JF. Evaluation of PCR in bronchoalveolar lavage fluid for diagnosis of P. jirovecii pneumonia: a bivariate meta-analysis and systematic review. PLoS ONE. 2013;8(9):e73099. doi: 10.1371/journal.pone.0073099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilson MR, Naccache SN, Samayoa E, Biagtan M, Bashir H, Yu G, Salamat SM, Somasekar S, Federman S, Miller S, Sokolic R, Garabedian E, Candotti F, Buckley RH, Reed KD, Meyer TL, Seroogy CM, Galloway R, Henderson SL, Gern JE, DeRisi JL, Chiu CY. Actionable diagnosis of neuroleptospirosis by next-generation sequencing. N Engl J Med. 2014;370(25):2408–2417. doi: 10.1056/NEJMoa1401268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsang CC, Teng JLL, Lau SKP, Woo PCY. Rapid genomic diagnosis of fungal infections in the age of next-generation sequencing. J Fungi. 2021;7(8):636. doi: 10.3390/jof7080636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xing F, Hung DLL, Lo SKF, Chen S, Lau SKP, Woo PCY. Next-generation sequencing-based diagnosis of bacteremic Listeria monocytogenes meningitis in a patient with anti-interferon gamma autoantibodies: a case report. Infect Microb Dis. 2022;4(1):44–46. doi: 10.1097/IM9.0000000000000080. [DOI] [Google Scholar]
- 12.Xing F, Ye H, Deng C, Sun L, Yuan Y, Lu Q, Yang J, Lo SKF, Zhang R, Chen JHK, Chan JFW, Lau SKP, Woo PCY. Diverse and atypical manifestations of Q fever in a metropolitan city hospital: Emerging role of next-generation sequencing for laboratory diagnosis of Coxiella burnetii. PLoS Negl Trop Dis. 2022;16(4):e0010364. doi: 10.1371/journal.pntd.0010364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xing F, Yang Q, Deng C, Sun L, Luo Z, Ye H, Yang J, Lo SKF, Lau SKP, Woo PCY. Clinical impact of next-generation sequencing on laboratory diagnosis of suspected culture-negative meningitis and encephalitis. J Infect. 2022;85(5):573–607. doi: 10.1016/j.jinf.2022.08.026. [DOI] [PubMed] [Google Scholar]
- 14.Jiang J, Bai L, Yang W, Peng W, An J, Wu Y, Pan P, Li Y. Metagenomic next-generation sequencing for the diagnosis of P. jirovecii pneumonia in Non-HIV-infected patients: a retrospective study. Infect Dis Ther. 2021;10(3):1733–1745. doi: 10.1007/s40121-021-00482-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang D, Fang S, Hu X, Xu Q, Chu X, Mei X, Xie W. Metagenomic Next-Generation Sequencing Is Highly Efficient in Diagnosing P. jirovecii Pneumonia in the Immunocompromised Patients. Front Microbiol. 2022;17(13):913405. doi: 10.3389/fmicb.2022.913405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu X, Zhang J, Ma W, Xing L, Ning H, Yao M. Pneumocystis Jirovecii pneumonia diagnosis via metagenomic next-generation sequencing. Front Med. 2022;9(9):812005. doi: 10.3389/fmed.2022.812005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donnelly JP, Chen SC, Kauffman CA, Steinbach WJ, Baddley JW, Verweij PE, Clancy CJ, Wingard JR, Lockhart SR, Groll AH, Sorrell TC, Bassetti M, Akan H, Alexander BD, Andes D, Azoulay E, Bialek R, Bradsher RW, Bretagne S, Calandra T, Caliendo AM, Castagnola E, Cruciani M, Cuenca-Estrella M, Decker CF, Desai SR, Fisher B, Harrison T, Heussel CP, Jensen HE, Kibbler CC, Kontoyiannis DP, Kullberg BJ, Lagrou K, Lamoth F, Lehrnbecher T, Loeffler J, Lortholary O, Maertens J, Marchetti O, Marr KA, Masur H, Meis JF, Morrisey CO, Nucci M, Ostrosky-Zeichner L, Pagano L, Patterson TF, Perfect JR, Racil Z, Roilides E, Ruhnke M, Prokop CS, Shoham S, Slavin MA, Stevens DA, Thompson GR, Vazquez JA, Viscoli C, Walsh TJ, Warris A, Wheat LJ, White PL, Zaoutis TE, Pappas PG. Revision and update of the consensus definitions of invasive fungal disease from the European organization for research and treatment of cancer and the mycoses study group education and research consortium. Clin Infect Dis. 2020;71(6):1367–1376. doi: 10.1093/cid/ciz1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carroll KC, Pfaller MA. Manual of clinical microbiology. 12. UK: ASM Press; 2019. [Google Scholar]
- 19.Lau SK, Tang BS, Teng JL, Chan TM, Curreem SO, Fan RY, Ng RH, Chan JF, Yuen KY, Woo PC. Matrix-assisted laser desorption ionisation time-of-flight mass spectrometry for identification of clinically significant bacteria that are difficult to identify in clinical laboratories. J Clin Pathol. 2014;67(4):361–366. doi: 10.1136/jclinpath-2013-201818. [DOI] [PubMed] [Google Scholar]
- 20.Xing F, Xia Y, Lu Q, Lo SKF, Lau SKP, Woo PCY. Rapid diagnosis of fatal Nocardia kroppenstedtii bacteremic pneumonia and empyema thoracis by next-generation sequencing: a case report. Front Med. 2023;18(10):1226126. doi: 10.3389/fmed.2023.1226126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xing F, Lo SW, Liu M, Deng C, Ye H, Sun L, Yang J, Lo SKF, Lau SKP, Woo PCY. Emergence of Tropheryma whipplei detection in respiratory samples by next-generation sequencing: pathogen or innocent bystander? J Infect. 2023;86(2):154–225. doi: 10.1016/j.jinf.2022.12.004. [DOI] [PubMed] [Google Scholar]
- 22.Cillóniz C, Dominedò C, Álvarez-Martínez MJ, Moreno A, García F, Torres A, Miro JM. Pneumocystis pneumonia in the twenty-first century: HIV-infected versus HIV-uninfected patients. Expert Rev Anti Infect Ther. 2019;17(10):787–801. doi: 10.1080/14787210.2019.1671823. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.