Abstract
Here we report a transgenic mouse line that exhibits significant deviations from a classic pattern of parental imprinting. When the transgene is passed through the female germline, it is completely silenced in some offspring while in others expression is reduced. This variable expressivity does not appear to be the result of differences in the presence of unlinked modifiers. Female transmission of the transgene is associated with hypermethylation. The transgene is generally reactivated on passage through the male germline. Extended pedigrees reveal complex patterns of inheritance of the phenotype. The most likely explanation for this result is that the imprint is not completely erased and reset when passed through the germline of either sex. FISH analysis reveals that the transgene has integrated into chromosome 3 band E3, a region not known to carry imprinted genes, and the integration site shows no sign of allele-specific differential methylation. These findings, in conjunction with other recent work, raise the possibility that the introduction of foreign DNA into the mammalian genome, either through retrotransposition or transgenesis, may be associated with parental imprinting that is not always erased and reset during meiosis.
INTRODUCTION
A number of mammalian autosomal genes display monoallelic expression (reviewed in 1,2). This inactivation of an allele depending on the gamete of origin has been termed parental, genomic or gametic imprinting. Parental imprinting with differential DNA modification usually results in complete silencing of one parental allele with gene expression limited to the other.
Despite extensive study of a number of human genes and their murine homologues and an increasing number of mouse transgenes which have been shown to exhibit parental imprinting, the mechanisms responsible for this phenomenon are still unknown. In mice, two types of transgene imprinting or parent-of-origin effects are seen: one present only with mixed genetic backgrounds (3–6), which is thought to be due to the presence of strain-specific modifers (7), and classic genomic imprinting, which occurs in inbred strains (8,9). It is generally thought that in classic genomic imprinting the imprint: (i) arises during gametogenesis; (ii) is stably inherited through mitosis; (iii) is reset in the germline after the sex of the embryo is determined, so that it can be reversed in subsequent generations (10–12).
DNA methylation is the strongest candidate for the epigenetic modification responsible for imprinting, because it correlates with transcriptional inactivity of certain genes and is stably maintained through cell division and because the degree of methylation is different in spermatogenesis and oogenesis (1). More recently, chromatin packaging has been implicated as a possible epigenetic modifier of gene expression involved in imprinting (13–16).
Whatever epigenetic modification is responsible for imprinting, it is clear that it can be reversed when the allele is transmitted through the opposite parental germline in a subsequent generation. This has led to the idea that the imprint is erased or neutralised in the germ cells and reset according to sex (8,11,17). This is consistent with the notion that most epigenetic modifications are erased during meiosis in order to re-establish the totipotency of the genome in each generation. However, in some species, including plants (reviewed in 18), yeast (19,20) and Drosophila (21–23), epigenetic modifications at some loci can be inherited through the germline. The evidence for this in mammals has, until recently, been restricted to situations involving either transgenes (3,24,25) or nuclear transfer (26). We have recently shown (27) that epigenetic modifications at an endogenous locus (agouti viable yellow) in the mouse can be inherited through meiosis. Interestingly, the viable yellow allele at the agouti locus is the result of insertion of a retroviral element (28) which results in the locus displaying variable expressivity and incomplete parental imprinting (29).
Here we report a transgenic mouse line that exhibits incomplete parental imprinting and what appears to be germline inheritance of the imprint. When the transgene is transmitted through the female, expression is reduced in some offspring and completely silenced in others, i.e. the transgene displays variable expressivity. This variable expressivity does not appear to be the result of unlinked genetic modifiers. Furthermore, the phenotype of the mother (whether she carries an active or an inactive locus) correlates with the probability of silencing in her offspring. In general the imprint is reversed following passage through the male germline, but if the male inherits the transgene from a female carrying a completely silenced transgene, then reversal of the imprint does not occur in all offspring. These unusual grandparental effects suggest that the imprint is not always completely erased and reset on passage through either the female or the male germline. The transgene array is more methylated following female transmission than male transmission. The transgene has been localised by FISH to a region on chromosome 3 which does not contain any known imprinted genes. The integration site has been cloned and shows no signs of allele-specific differential methylation. These results suggest that in some instances epigenetic modifications associated with parental imprinting are not completely erased and reset in the germline. It may be that transgenes and retroviruses are particularly prone to these types of effects. Such mechanisms would result in unusual patterns of inheritance of a particular phenotype and may be operating at a number of loci in the mammalian genome.
MATERIALS AND METHODS
Transgene construct and the generation of transgenic mice
The transgene construct DM2 contains the human ζ-globin promoter from –127 to +6 linked to an oligonucleotide (SDK) containing the Shine–Dalgarno and Kozak sequences, the lacZ reporter gene with the SV40 polyadenylation signal and a 4.1 kb fragment containing the human α-globin locus enhancer region (HS-40) previously shown to direct high levels of expression in erythroid cells (30). Three transgenic lines were produced by microinjection of DNA into the pronuclei of fertilised mouse oocytes from the P.O. (Pathology Oxford) mouse strain (31). The P.O. strain has been maintained as a closed colony for the past 20 years; the mice used in this study are from a P.O. colony formed 6 years ago from three mating pairs. Initial studies of three lines produced with this construct have been reported previously (31). In that study transgene expression was only recorded following transmission through the male. One of these three lines, 239B, is the focus of this study. The copy number of line 239B was estimated to be 5–8 (31). Southern blotting of tail DNA was used to identify transgenic mice and to determine the copy number.
β-Galactosidase activity in whole cells and lysates
Transgene expression was analysed at the single cell level in erythroid cells from mice at 12.5 days post coitum (d.p.c.) or 4 weeks after birth as described previously (31). Cells were fixed and then stained with 5′-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) at 37°C for up to 24 h. A minimum of 1 × 105 erythrocytes were inspected for detectable blue stain; non-expressors are those mice having no stained cells in >105 erythrocytes. For positively staining mice, at least 500 erythroid cells were counted to determine the percentage of stained cells.
The activity of β-galactosidase (β-gal) in erythroid cells was assayed in lysates of blood obtained from 12.5 d.p.c. embryos. Embryos were bled into phosphate-buffered saline. A cell count was determined with a hemocytometer. The cells were collected by centrifugation, resuspended in 250 mM Tris (pH 7.4), then lysed by freeze–thawing and the cellular debris removed. The lysate was assayed for β-gal activity using the substrate o-nitrophenyl-β-d-galactopyranoside (ONPG) as previously described (31). β-gal activity per expressing cell was calculated by correcting the activity obtained per erythroid cell with the percentage of expressing (blue staining) cells.
Methylation analysis of the transgene array and the 3′ genomic flank at the site of integration
DNA was purified from circulating erythroid cells of 12.5 d.p.c. transgenic embryos or whole embryos. Aliquots of 2 µg DNA were digested with BamHI alone or BamHI with either MspI, HpaII, HinP1I or HhaI. The resulting fragments were analysed by Southern transfer and hybridisation following separation on a 1.2% agarose gel. The membranes were probed with either a 4.1 kb fragment containing the HS-40 region, a 3.5 kb SDK lacZ fragment, a 1.4 kb mouse α-globin fragment or a 301 bp fragment containing the genomic sequence at the 3′ integration site [obtained by ligation-mediated PCR (LMPCR)]. The probes were labelled by random priming using [α-32P]dCTP (Bresatec) and the Megaprime Labelling Kit (Amersham).
Culturing of mouse embryonic fibroblasts
Embryos resulting from the cross between a homozygous transgenic male and a wild-type P.O. female were dissected at 12.5 d.p.c. and bled. After removing the head and liver the remainder of two embryos were pooled and then homogenised using a 19G needle and syringe in Amnio-max medium (Life Technologies). The emulsion was then treated with collagenase (2000 U) for 4 h and then incubated at 37°C in 5% CO2. Twenty-four hours prior to harvesting, the confluent cultures were passaged. Mouse metaphase cells were prepared using standard colcemid and harvesting procedures.
Fluorescent in situ hybridisation (FISH)
Plasmid clone DM2, representing the entire transgene construct, was random-prime labelled with biotin (High-Prime kit; Boehringer Mannheim) and hybridised without repeat suppression to denatured transgenic mouse metaphase chromosomes spread on microscope slides using methods previously described (32). Slides were incubated overnight at 37°C and washed to a final stringency of 0.1× SSC at 60°C before signal amplification with FITC–avidin and biotinylated anti-avidin (Vector Laboratories). Slides were mounted in glycerol containing antifade and DAPI as counterstain for G-band visualisation before analysis under a Leitz Aristoplan microscope. Selected fields were transferred for image processing to a Power Macintosh 8100-80 computer via a Photometrics cooled CCD camera using appropriate wavelength filters, IPLab Spectrum and Multiprobe extension software. G-band nomenclature for description of mouse karyotypes and probe localisation was according to Nesbitt and Francke (33).
Cloning the 3′ integration site by LMPCR
The method and linker sequences were modified from Steigerwald et al. (34). Genomic DNA from line 239B was digested with one of five enzymes (EcoRV, ScaI, DraI, SspI or XmnI). This was followed by ligation of the linkers. Nested PCR was performed with transgene-specific primers, homologues to the 3′-end of the HS-40 fragment in the transgene construct and a primer to the linker. The PCR product was gel purified, cloned into pGem-T Easy (Promega) cloning vector and sequenced.
RESULTS
Inheritance of the transgenic phenotype following female transmission
Three transgenic lines (278A, 268B and 239B) were established following microinjection of a construct, DM2, containing the human ζ-globin gene promoter, the reporter gene lacZ and an erythroid-specific enhancer (HS-40). All three lines exhibit transgene expression in erythroid cells as reported previously (31). Further studies revealed that transgene expression was inherited in a Mendelian pattern in two of these lines (278A and 268B), but that the third (239B) exhibited a complex non-Mendelian pattern of inheritance of transgene expression. Initially this was not reported, as the line was maintained by breeding transgenic males to wild-type females (31). Line 239B is the focus of this study. When the transgene was inherited through the female only 76% of the transgenic offspring expressed the transgene, which is significantly different from the 100% seen when the transgene was inherited through the male (Table 1). This indicates that approximately one-quarter of the genotypically positive offspring have a silenced transgene following female transmission. This silencing of the transgene was not observed when it was inherited from the father: with male transmission the genotype of the offspring always correlated with the phenotype. These breeding experiments were repeated after backcrossing the transgene for three generations into two inbred strains, C57BL/6 and CBA/Ca. The results were essentially the same (data not shown): the transgene was silenced in some offspring following female transmission but never following male transmission. This parent of origin-dependent silencing is similar to parental imprinting, but in this case it is incomplete in that it does not occur in all the transgenic offspring resulting from female transmission.
Table 1. Penetrance of the phenotype following maternal and paternal transmission.
| Female × male | Phenotypically positive (%) | Genotypically positive (%) | Transgenic offspring phenotypically positive (%) |
|---|---|---|---|
| WT × 239B | 47.9 (n = 242) | 47.9 (n = 242) | 100 (n = 116) |
| 239B × WT | 37.0 (n = 265) | 48.7 (n = 265) | 76 (n = 129) |
Matings were set up between phenotypically positive hemizygous transgenic males and wild-type females and the reciprocal cross between wild-type male and phenotypically positive hemizygous transgenic females. Phenotypic analysis was performed on the offspring of such crosses by staining erythrocytes with X-Gal to detect the presence of an active transgene. For a sample to score phenotypically negative, no expressing cells were detected in >105 erythroid cells. Genotypic analysis was performed by Southern transfer and hybridisation using mouse genomic DNA and the lacZ gene as probe.
Reduced transgene expression with female transmission
Variegated expression in erythrocytes is typical of transgenes consisting of globin regulatory elements linked to the reporter gene lacZ (31,35,36). Each transgenic line expresses the transgene at a characteristic level determined by the number of erythrocytes producing β-gal. The proportion of erythrocytes expressing the transgene is different in each transgenic line and is, in general, stably inherited through subsequent generations (31,35–37). When the transgene in line 239B was inherited from the male, ∼22% of the circulating erythrocytes expressed the transgene at 12.5 d.p.c. When the transgene was inherited from the female it was completely silenced in some individuals (see Table 1) and in the others the expression was limited to ∼4% of the circulating erythrocytes (Table 2). Thus female transmission of the transgene increases the extent of variegation. A similar result was seen when hemizygote crosses were performed using transgenic mice backcrossed for three generations into two inbred strain backgrounds (C57BL/6 and CBA/Ca) (data not shown). The sensitivity of the β-gal/X-Gal staining assay used in this study has allowed us to detect those offspring with reduced transgene activity, which might otherwise have been classified as non-expressing offspring.
Table 2. Transgene expression in phenotypically positive offspring following maternal and paternal transmission.
| Female × male | Percentage of erythrocytes expressing lacZ (%) | Transgene activity per 1010 expressing cells |
|---|---|---|
| WT × 239B | 21.6 ± 5.9 (n = 50) | 175.0 ± 56 (n = 16) |
| 239B × WT | 4.1 ± 2.0 (n = 47) | 99.3 ± 26 (n = 11) |
Circulating erythrocytes from 12.5 d.p.c. embryos were collected and assayed for the number of cells expressing the transgene by X-Gal staining. Only samples showing some transgene expression were used in this instance. Transgene activity was determined by incubating the whole cell lysates with ONPG followed by spectrophotometric analysis and correcting for the number of expressing cells. Data presented are the means ± SD and n represents the number of individual pups analysed.
The decrease in transgene expression seen following female transmission was not limited to the number of erythrocytes expressing the transgene. The level of transgene activity in each expressing cell can be calculated by assaying lacZ activity in cell lysates and correcting for the number of cells expressing the transgene. Following female transmission, this level was approximately half the level found in expressing cells following male transmission (Table 2). It appears that in those offspring which show incomplete imprinting following female transmission transgene expression is reduced ∼10-fold and that the majority of this effect is achieved by a reduction in the percentage of cells expressing the transgene. There is a small (2-fold) effect on the level of transgene activity in an expressing cell.
The imprint is reversed through the male germline
The inheritance of transgene expression was followed for a number of generations (Fig. 1 and Table 3). When a phenotypically positive female resulting from male inheritance of the transgene was crossed to a wild-type male, 76% of the transgenic offspring carried an active transgene (Fig. 1A and Tables 1 and 3A). The remaining 24% of offspring carried a silenced transgene. Genotypically positive but phenotypically negative male offspring were then bred to wild-type females. All the resulting 55 transgenic offspring expressed the transgene (Fig. 1B and Table 3B). This demonstrates that the imprint is reversed through the male germline. After reversal of the imprint, the number of erythrocytes expressing the transgene was the same as that observed following transmission from phenotypically positive males: ∼22% of erythrocytes expressed the transgene (data not shown). A similar cross was performed with the transgene in the CBA/Ca background with similar results (data not shown). This reversal of the imprint through the opposite sex is consistent with classic models of genomic imprinting.
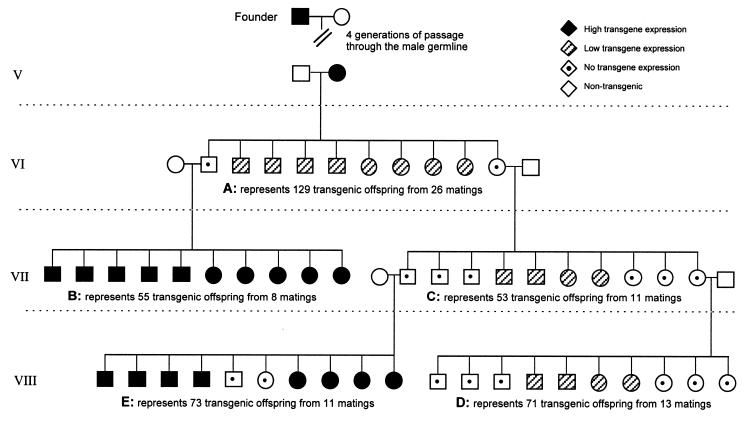
Figure 1.
Representative pedigree of 239B transmission. The phenotypes depicted represent the percentages obtained for the total mice analysed for each of the five matings (A–E) adapted to a litter size of 10 pups. Genotyping was performed by Southern transfer and hybridisation using genomic mouse DNA probed with the lacZ gene and phenotypic analysis was determined by X-Gal staining of erythrocytes. All crosses were performed using P.O. heterozygote transgenic mice and P.O. wild-type mice of the opposite sex. Following convention, circles represent females and males are depicted by squares. It can be seen that the phenotype of the offspring depends not only on the phenotype of the transgenic parent but also grandparental effects are evident with male transmission. When the grandmother (female parent cross C) has a silenced transgene the silencing modification is not always reversed through the subsequent male germline (male parent cross E), resulting in some progeny with no transgene expression.
Table 3. Penetrance of the transgene is reduced when the female parent or grandparent carries a silenced transgene.
| Mating: Transgenic parent × wild-type | Transgenic offspring with an active transgene (%) |
|---|---|
| A: Expressing female (from an expressing male) | 76 (n = 129) |
| B: Non-expressing male (from an expressing female) | 100 (n = 53) |
| C: Non-expressing female (from an expressing female) | 38a (n = 55) |
| D: Non-expressing female from a non-expressing female | 42 (n = 71) |
| E: Non-expressing male from a non-expressing female | 79b (n = 73) |
Heterozygous transgenic mice were mated to wild-type mice of the opposite sex and the litters resulting from such crosses analysed for the presence of the transgene both phenotypically and genotypically as previously described. The letter indicates where this cross lies on the pedigree (Fig. 1).
aThe value 38% is statistically different from 76% in A with P = 4.8 × 10–6.
bThe value 79% is statistically different from 100% in B with P = 0.001.
Variable expressivity of the transgene
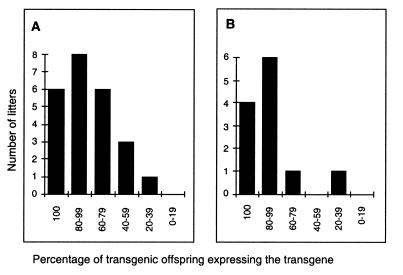
The presence of both completely silenced and expressing offspring following female transmission of the transgene could arise from segregation of a genetic modifier or be due to some intrinsic variable expressivity of the transgene, which is independent of unlinked modifiers. We believe that the former explanation is unlikely since inheritance of a modifier is inconsistent with the extended pedigree. Furthermore, analysis of the percentage of expressing offspring in individual litters (Fig. 2A and data not shown) reveals a normal distribution rather than the bimodal distribution that one would expect if a sub-population of the colony carried such a modifier.
Figure 2.
Distribution of litters based on the percentage of expressing offspring within a litter. Individual litters were grouped into categories depending on the percentage of expressing offspring within the litter. The numbers of litters obtained for each category are plotted on the y-axis. (A) Litters resulting from crosses between wild-type males and non-imprinted females. (B) Litters resulting from crosses between selected wild-type males and non-imprinted females. These selected wild-type males were generated by crossing two silenced parents.
In an attempt to clarify this further we carried out the following experiment. If a modifier (associated with silencing) is present in a sub-population of the P.O. strain then by crossing a silenced transgenic male to a silenced transgenic female, the resulting wild-type mice would have a higher probability of carrying the putative modifier than normal wild-type mice. Wild-type males selected in this way were crossed to non-imprinted females to examine the frequency of silenced offspring in the subsequent litters (Fig. 2B). The distribution of frequencies of expressing offspring per litter was not significantly different in these litters from that obtained with non-selected wild-type males. If the variable expressivity was the result of a modifier one would have expected the litters from selected wild-type males to have contained more silent offspring. This was not the case. These results are inconsistent with the existence of such a modifier. It therefore seems more likely that at this locus the transgene is susceptible to variable expressivity. Variable expressivity of this type has been reported at a number of murine genes: agouti viable yellow (38) and axin-fused (39). In these cases, the variability is seen in inbred strains where all individuals are theoretically genetically identical. The variability is set up by stochastic epigenetic processes, which are not well understood, and appears to coincide with retroviral insertions.
Our observations of transgene expression following crosses into the inbred strains C57BL/6 and CBA/Ca also argue against the idea of a genetic modifier being the cause of the variable expressivity. If the variable expressivity was due to a modifier then it would have to be present in a sub-population of both the C57BL/6 and CBA/Ca strains, as there was no dilution of the effect when the transgene was crossed into these strains for three generations (data not shown). Since C57BL/6 and CBA/Ca are both inbred strains, maintained by brother/sister matings, sub-populations should not exist: all animals should be genetically identical.
Grandparental effects through the female germline
When a phenotypically negative transgenic female was crossed with a wild-type male, only 38% of her transgenic offspring carried an active transgene (Fig. 1C and Table 3C). This is significantly lower than the 76% of transgenic offspring which we found when the female parent was phenotypically positive (Fig. 1A and Table 3A). This suggests that the imprint is not completely erased and then reset through the female germline but that some form of the imprint is inherited following female transmission. The probability that any offspring will express the transgene is affected by whether or not its transgenic mother actually expressed the transgene. Decreased penetrance of the transgene was also seen when the transgene was backcrossed for three generations into the inbred strains (data not shown). This maintenance or enhancement of the imprint through the female gamete differs from classic models of gametic imprinting.
The imprinted transgene was then followed for a further generation. When a silenced female who had received the transgene from a silenced female parent was crossed with a wild-type male (Fig. 1D and Table 3D), 42% of the offspring contain an active transgene. This once again supports the notion that some memory of the imprint is inherited through the female germline without erasure and resetting and that imposition of the imprint on germ cells is once again variable.
Grandparental effects through the male germline
When a silenced male who received the transgene from a silenced female was mated to a wild-type female (Fig. 1E and Table 3E) the imprint was not completely reversed in his offspring; 79% carried an active transgene. This contrasts with the complete reversal (reversal in 100% of offspring) of the imprint following male transmission when the male had received the transgene from an expressing female (Fig. 1B and Table 3B). Some compounding effect of two generations of imprinting may prevent complete erasure in the male germline. Thus the probability that offspring will express the transgene is affected by the phenotype of the grandmother, despite the fact that the phenotype of the father was identical in both cases.
Methylation state of the imprinted transgene
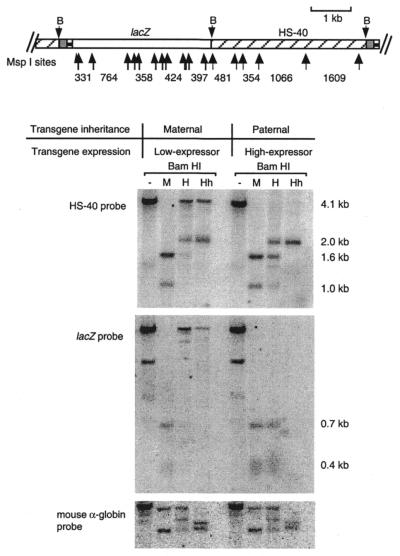
As methylation has been reported to correlate with imprinting of a number of transgenes and endogenous genes, we investigated the methylation state of the imprinted transgene. The methylation state of ∼50 CpG sites in the lacZ and αHS-40 regions was analysed with methylation-sensitive restriction enzymes, using DNA extracted from circulating erythrocytes of 12.5 d.p.c. embryos (Fig. 3). The degree of methylation of the transgene following female transmission was greater than that following male transmission, and this was true for both the lacZ portion, containing 14 MspI/HpaII and 28 HhaI sites, and the αHS-40 portion of the transgene. Not all sites within the transgene were methylated following female transmission, as some digestion of the transgene was still evident (Fig. 3).
Figure 3.
Schematic representation of the transgene construct and the methylation state of the 239B transgene following male and female transmission. DNA was extracted from circulating erythrocytes collected from 12.5 d.p.c. embryos, then digested with either BamHI (B) alone or a combination of BamHI and either MspI (M), HpaII (H) or HhaI (Hh). Following separation on a 1.2% agarose gel, Southern blotting and hybridisation was performed using either a 4.1 kb HS-40 fragment or the lacZ gene. The membrane was also hybridised with a 1.4 kb fragment from the mouse α-globin gene to check for complete digestion of the enzymes. Not all band sizes are given as the difference in patterning is indicative of the differential methylation states of the transgene following male and female transmission.
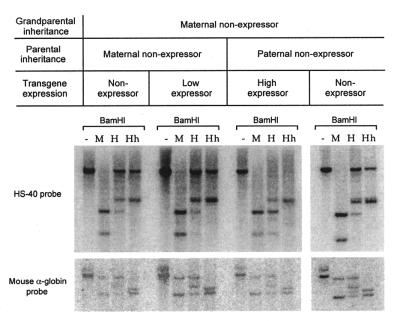
In light of our finding that there were grandmaternal effects on phenotype following male transmission, we decided to investigate the methylation pattern of those offspring in which the transgene was silent despite passage through the male gamete. These were all individuals who had inherited the transgene from a non-expressing male, who had himself inherited the transgene from a non-expressing female. The methylation pattern of the transgene in these individuals is similar to that found following passage directly from a non-expressing female (Fig. 4). This is consistent with the idea that the imprint in these individuals has not been erased on passage through the male germline. Nevertheless, it should be noted that the methylation state of the transgene does not correlate tightly with expression, since we see no difference in the methylation state of non-expressors compared with low expressors (Fig. 4). Preliminary analysis of the sperm of non-expressing males reveals that the transgene remains heavily methylated despite the fact that the transgene is less methylated in his progeny. This suggests that the methylation is erased at some stage after fertilisation (data not shown).
Figure 4.
Methylation state of the transgene array following two generations of imprinting. DNA was extracted from circulating erythrocytes and digested as described in Figure 3. Expressing and non-expressing individuals were collected from litters represented in Figure 1 (D, offspring with a non-expressing mother and grandmother; E, offspring with a non-expressing father and a non-expressing grandmother). A banding pattern, suggestive of extensive methylation, was obtained following female transmission (as seen in Fig. 3) and with the non-expressing individual resulting from inheritance through a non-expressing female then a non-expressing male.
Mapping of the transgene integration site to chromosome 3
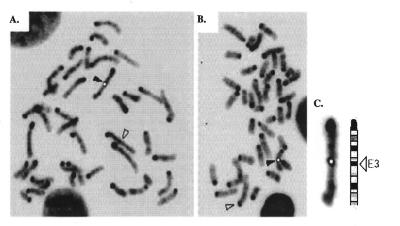
Since 239B was one of three transgenic lines that contained the same construct and the other two exhibited normal Mendelian inheritance, the complex pattern of inheritance is unlikely to be the result of some factor intrinsic to the transgene construct. We therefore attempted to determine whether the transgene array in 239B had integrated into a previously identified imprinted region in the mouse genome. A number of mouse chromosomes have been found to contain imprinted genes (for reviews see 2,5,33). FISH studies identified a clear signal proximal on one chromosome 3 in 30/30 metaphase cells analysed from the cultured transgenic mouse tissue. The mice used in this study were hemizygous for the transgene. The biotinylated DM2 plasmid probe mapped specifically to chromosome 3 band E3 in all cells, marking this as the likely integration site of the transgene (Fig. 5). This region has not previously been reported to contain any endogenous genes which display parental imprinting.
Figure 5.
(A and B) Representative metaphase cells of transgenic mouse line 239B showing a fluorescent signal (black arrowhead) on one mouse chromosome 3 band E3 after FISH with the DM2 transgene construct within p-127zetalacZHS-40. The chromosome 3 homologue is indicated with an open arrowhead. (C) Chromosome 3 from (A) aligned with the ideogram showing precise location of the probe signal in band E3. The signal has been painted white to increase clarity in black and white.
Methylation at the site of integration
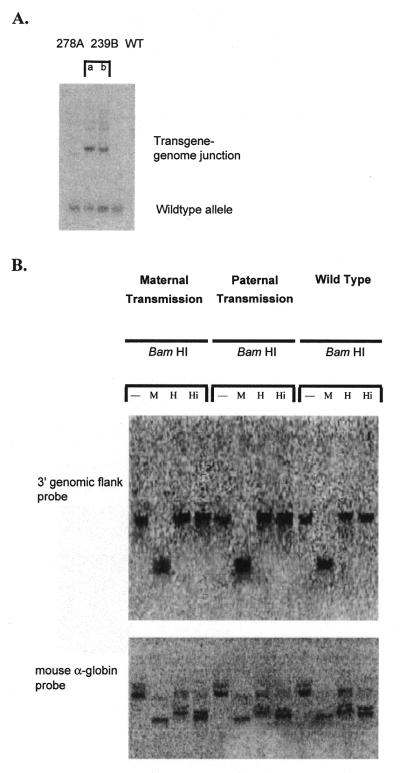
Using LMPCR, the 3′ genomic flank at the site of integration of the transgene was cloned. We obtained 426 bp of novel DNA and PCR experiments confirmed that this DNA was adjacent to the 3′-end of the transgene array (data not shown). The sequence showed no special features; in particular it was not especially rich in CpG dinuclotides. This 426 bp fragment was cloned into the pGem-T Easy cloning vector. Southern analysis of genomic DNA, using 301 bp of this sequence as probe, revealed that the integration site was unique in the genome (Fig. 6A). A single band was obtained with wild-type DNA and with DNA from another transgenic line carrying the same transgene (278A), while two bands were obtained with 239B DNA. The two bands correspond to the different alleles in the hemizygous transgenic mouse; the wild-type allele and the transgene–genome junction at the 3′ integration site of the other allele.
Figure 6.
(A) The 3′ genomic flank sequence is unique in the genome. Southern blotting of DraI-digested genomic DNA and hybridisation with a 301 bp radiolabelled HindIII–EcoRI fragment of the 3′ genomic sequence from the site of integration (obtained by LMPCR) generated a single band in DNA samples from line 278A and wild-type (WT) mice and two bands in DNA from line 239B. The 239B DNA was obtained from a hemizygous mouse so that one allele generated a wild-type band and the other band represents the transgene–genome junction. Samples a and b are independent samples from line 239B. (B) Susceptibility of the 3′ site of integration of the transgene to digestion by methylation-sensitive restriction enzymes. Transgenic embryos were collected following either maternal or paternal transmission of the transgene as well as their wild-type littermates. DNA extracted from these 12.5 d.p.c. embryos was digested with BamHI alone or in combination with either MspI (M) (methylation-insensitive), HpaII (H) (methylation-sensitive) or HinP1I (Hi) (methylation-sensitive). The membrane was hybridised using a radiolabelled 301 bp HindIII–EcoRI fragment of the 3′ genomic DNA from the site of integration (obtained by LMPCR). The membrane was stripped and reprobed with the radiolabelled mouse α-globin gene to check for complete digestion of the DNA in all samples.
Methylation-sensitive restriction enzyme analysis revealed that the 3′ flanking region of the 239B integration site was methylated regardless of germline transmission or the presence of the transgene. DNA samples were prepared from mice which had inherited the transgene from their mother (female transmission), from those who inherited the transgene from their father (male transmission) and from non-transgenic littermates. Following restriction enzyme digestion and hybridisation with the 3′ flank DNA, no differential methylation patterns could be seen (Fig. 6B).
DISCUSSION
It is estimated that ∼10–20% of all transgenes exhibit some form of parental imprinting (5). In many of these cases the imprinting is lost when the transgene is introduced into an inbred strain (3–6,40), whereas others retain the imprint on an inbred background (8,9). In the latter case, true germline transgene imprinting, the imprint is thought to be completely erased in both male and female primordial germ cells and reset during maturation of the gametes. Here we report a mouse transgene that is parentally imprinted and displays incomplete penetrance and variable expressivity. Detailed breeding studies suggest that the imprint is not completely erased in the germline, resulting in unusual patterns of inheritance of transgene expression.
Incomplete imprinting
In many early studies of imprinted genes, both endogenous and transgenes, it appeared that the parent of origin imprint was complete, with no expression from the imprinted locus. However, recent studies with more sensitive assays have found that, in at least some cases, silencing of the imprinted locus is incomplete (41–43). The mouse Grf1 gene is expressed in adult brain tissue mainly from the paternal allele with a small amount (<10%) of maternal expression as determined by PCR (42). The human p57KIP2 gene is imprinted with preferential expression from the maternal allele but low levels of expression are seen from the paternal allele (44). The mouse H19 gene is mainly, but not exclusively, expressed from the maternal allele: the paternal allele expresses H19 at ∼5% the level produced from the maternal allele (45). IGF-2 is incompletely imprinted in the human (46) and in the mouse (47). In these cases it is not clear whether every cell in a population is repressed to the same extent or whether repression is stochastic and a small number of cells in a population continue to show biallelic expression.
In the transgenic line 239B described here, repression of transgene expression associated with imprinting is incomplete in a significant proportion of the offspring who have inherited the transgene from the mother. In these cases, a single cell assay of transgene expression reveals that the transgene is expressed at close to normal levels but in significantly fewer cells than when the transgene is inherited through the male. This decrease in the number of cells expressing the transgene is reminiscent of the variegated or heterocellular gene silencing observed in many situations: position–effect variegation in Drosophila (48,49), as well as telomeric and repeat-induced gene silencing in yeast (50,51), Drosophila (52,53) and mammals (54,55). Cellular mosaicism has been observed in many cases of epigenetic silencing of both transgenes and endogenous genes (4,31,37,56). In many of these cases heterochromatin is implicated as a causative factor and it is worth noting that in Drosophila and yeast these phenomena occur in the absence of DNA methylation.
We have found that when the transgene is inherited from the female not every offspring in a single litter is imprinted to the same extent. The offspring fall into two categories: in some the transgene is completely silenced and in others the transgene is incompletely silenced. We believe that these differences in phenotype are not the result of underlying genetic differences at unlinked loci. The transgenic line has been made and maintained in the P.O. background. The P.O. strain has been maintained as a closed colony for the past 20 years and the mice used in this study are derived from a P.O. colony formed 6 years ago from three mating pairs. There have been a number of reports of variable expressivity of transgenes in mixed genetic backgrounds (3,4). Similar observations have been reported at the IGF2R locus in humans (57,58). Xu and colleagues (58) reported sporadic monoallelic expression in three of 14 fetuses, raising the possibility that the locus was subject to polymorphic imprinting. Imprinting at the WT1 gene in human placentae is observed in some but not all fetuses (59). Of course, in the case of these human studies genetic differences at other loci are thought to be the explanation for the variability. However, variable expression and methylation of a transgene locus have been reported among genetically identical mice (25,60) and a number of endogenous loci display variable expressivity in inbred strains in the mouse: such as the agouti mutants Avy, Aiapy, Ahvy (38), the Disorganisation mutant Ds (61) and the axin-fused mutant Fu (39,62). The mechanism underlying this variation is unknown, although in Avy mice the phenotype correlates with the methylation state of the locus (63). Interestingly, both the Avy and Fu mutations display parental imprinting effects: expressivity and penetrance are dependent on the parent of origin of the mutant allele (29,62).
In this study we have found that the transgene is more methylated following female transmission than following male transmission and methylation state thus parallels the activity of the transgene to some degree. Normal patterns of expression of the imprinted mouse endogenous genes Igf2, H19 and Igf2r are altered in mice deficient in DNA methyltransferase (64). When both expression and methylation state of imprinted transgenes have been reported there has been a strong correlation between the two: usually manifest as silencing or repression associated with hypermethylation (4–6,8,9,24,65). As the mechanisms of establishment and maintenance of the epigenetic modification have not been determined, it is not known whether the correlation between methylation and imprinting is causative or consequential.
Incomplete erasure of the imprint during meiosis
Use of the sensitive β-gal/X-Gal assay has not only enabled the detection of transgene activity in a small percentage of a population of erythrocytes, but also enabled the transmission of transgene activity to be analysed more thoroughly than is permitted by the more common assays of transgene expression. It has been possible to show that as the transgene is passed through subsequent generations, the percentage of offspring which carry the imprint is affected by the phenotype of the parent or grandparent. When the mother carries an active transgene, it will be completely silenced in 24% of her offspring and partially silenced in the remainder. However, when the mother carries a silent transgene, significantly more of her offspring (62%) will inherit a completely silent transgene. With male transmission where there has been only one round of imprinting (the transgene is silent in the male but active in his mother), the imprint is reversed in all offspring. However, where there have been two rounds of imprinting (the transgene is silent in the male and silent in his mother), the reversal through the male germline is incomplete and occurs in only 79% of the offspring. Classic models of imprinting, which describe erasure through both germlines and then resetting of the imprint depending on sex, would predict that all transgenic offspring resulting from male transmission would express the transgene. The grandparental effects do not appear to be the result of genetic differences at other loci, but rather suggest that the imprint is itself not erased in some cases when passed through the germline.
Epigenetic modifications associated with gene silencing are generally considered to be inherited during mitosis but cleared during meiosis, enabling the genome to return to the totipotent state in each newly developing embryo. There have been a number of reports of particular loci in yeast (19,20), Drosophila (21,22) and mice (3,25) in which epigenetic modifications are not erased in meiosis. The recent report of meiotic inheritance of epigenetic modifications at the agouti viable yellow (Avy) locus is of particular relevance (27). Agouti viable yellow displays variable expressivity and incomplete parental imprinting. These effects are not observed at the wild-type agouti locus and are presumably a result of the retroviral insertion associated with the mutation. The results reported here are strikingly similar, suggesting that the introduction of DNA into the mammalian genome, either by retroviral insertion or transgenesis, can result in complex grandparental effects on inheritance of the phenotype.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Linda Weaving for technical assistance and David Martin and Hugh Morgan for critical reading of the manuscript. This work was supported by grants from the National Health and Medical Research Council of Australia to E.W. and the Cancer Society of New Zealand and Lottery Health New Zealand to C.M. M.K. was supported by an Australian Postgraduate Award.
REFERENCES
- 1.Barlow D. (1995) Science, 270, 1610–1613. [DOI] [PubMed] [Google Scholar]
- 2.Morison I.M. and Reeve,A.E. (1998) Hum. Mol. Genet., 7, 1599–1609. [DOI] [PubMed] [Google Scholar]
- 3.Allen N.D., Norris,M.L. and Surani,M.A. (1990) Cell, 61, 853–861. [DOI] [PubMed] [Google Scholar]
- 4.McGowan R., Campbell,R., Peterson,A. and Sapienza,C. (1989) Genes Dev., 3, 1669–1676. [DOI] [PubMed] [Google Scholar]
- 5.Reik W., Howlett,S.K. and Surani,M.A. (1990) Development, (suppl.), 99–106. [Google Scholar]
- 6.Sapienza C., Peterson,A.C., Rossant,J. and Balling, R (1987) Nature, 328, 251–254. [DOI] [PubMed] [Google Scholar]
- 7.Engler P., Haasch,D., Pinkert,C.A., Doglio,L., Glymour,M., Brinster,R. and Storb,U. (1991) Cell, 65, 939–947. [DOI] [PubMed] [Google Scholar]
- 8.Chaillet J.R., Vogt,T.F., Beier,D.R. and Leder,P. (1991) Cell, 66, 77–83. [DOI] [PubMed] [Google Scholar]
- 9.Sasaki H., Hamada,T., Ueda,T., Seki,R., Higashinakagawa,T. and Sakaki,Y. (1991) Development, 111, 573–581. [DOI] [PubMed] [Google Scholar]
- 10.Chaillet J.R. (1994) Mutat. Res., 307, 441–449. [DOI] [PubMed] [Google Scholar]
- 11.Szabó P.E. and Mann,J.R. (1995) Genes Dev., 9, 1857–1868. [DOI] [PubMed] [Google Scholar]
- 12.Villar A.J., Eddy,E.M. and Pederson,R.A. (1995) Dev. Biol., 172, 264–271. [DOI] [PubMed] [Google Scholar]
- 13.Hark A.T. and Tilghman,S.M. (1998) Hum. Mol. Genet., 7, 1979–1985. [DOI] [PubMed] [Google Scholar]
- 14.Svensson K., Mattsson,R., James,T.C., Wentzel,P., Pilartz,M., MacLaughin,J., Miller,S.J., Olsson,T., Eriksson,U.J. and Ohlsson,R. (1998) Development, 125, 61–69. [DOI] [PubMed] [Google Scholar]
- 15.Szabó P.E., Pfeifer,G.P. and Mann,J.R. (1998) Mol. Cell. Biol., 18, 6767–6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khosla S., Aitchison,A., Gregory,R., Allen,N.D. and Feil,R. (1999) Mol. Cell. Biol., 19, 2556–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Surani M.A. (1991) Curr. Opin. Genet. Dev., 1, 241–246. [DOI] [PubMed] [Google Scholar]
- 18.Matzke M. and Matzke,A.J.M. (1993) Annu. Rev. Plant Physiol. Plant Mol. Biol., 44, 53–76. [Google Scholar]
- 19.Grewal S.I.S. and Klar,A.J.S. (1996) Cell, 86, 95–101. [DOI] [PubMed] [Google Scholar]
- 20.Klar A.J.S. (1998) Trends Genet., 14, 299–301. [DOI] [PubMed] [Google Scholar]
- 21.Cavalli G. and Paro,R. (1998) Cell, 93, 505–518. [DOI] [PubMed] [Google Scholar]
- 22.Dorn R., Krauss,V., Reuter,G. and Saumweber,H. (1993) Proc. Natl Acad. Sci. USA, 90, 11376–11380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dorn R., Szidonya,J., Korge,G., Sehnert,M., Taubert,H., Archoukieh,E., Tschiersch,B., Morawietz,H., Wustmann,G., Hoffmann,G. and Reuter,G. (1993) Genetics, 133, 279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hadchouel M., Farza,H., Simon,D., Tiollais,P. and Pourcel,C. (1987) Nature, 329, 454–456. [DOI] [PubMed] [Google Scholar]
- 25.Sutherland H.G.E., Morgan,H.D., Kearns,M.M., Headley,A.P., Morris,C., Martin,D.I.K. and Whitelaw,E. (2000) Mamm. Genome, 11, 347–355. [DOI] [PubMed] [Google Scholar]
- 26.Roemer I., Reik,W., Dean,W. and Klose,J. (1997) Curr. Biol., 7, 277–280. [DOI] [PubMed] [Google Scholar]
- 27.Morgan H.D., Sutherland,H.G.E., Martin,D.I.K. and Whitelaw,E. (1999) Nature Genet., 23, 314–318. [DOI] [PubMed] [Google Scholar]
- 28.Duhl D.M.J., Vrieling,H., Miller,K.A., Wolff,G.L. and Barsh,G.S. (1994) Nature Genet., 8, 59–64. [DOI] [PubMed] [Google Scholar]
- 29.Wolff G.L. (1978) Genetics, 88, 529–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharpe J.A., Chan-Thomas,P.S., Lida,J., Ayyub,H., Wood,W.G. and Higgs,D.R. (1992) EMBO J., 11, 4565–4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robertson G., Garrick,D., Wu,W., Kearns,M., Martin,D. and Whitelaw,E. (1995) Proc. Natl Acad. Sci. USA, 92, 5371–5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morris C.M., Courtay,C., Guerts van Kessel,A., Heisterkamp,N. and Groffen,J. (1993) Hum. Genet., 91, 31–36. [DOI] [PubMed] [Google Scholar]
- 33.Nesbitt M.N. and Francke,U. (1973) Chromosoma, 41, 145–158. [DOI] [PubMed] [Google Scholar]
- 34.Steigerwald S.D., Pfeifer,G.P. and Riggs,A.D. (1990) Nucleic Acids Res., 18, 1435–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garrick D., Sutherland,H., Robertson,G. and Whitelaw,E. (1996) Nucleic Acids Res., 24, 4902–4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sutherland H.G.E., Martin,D.I.K. and Whitelaw,E. (1997) Mol. Cell. Biol., 17, 1607–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Graubert T.A., Hug,B.A., Wesselschmidt,R., Hsieh,C.-L., Ryan,T.M., Townes,T.M. and Ley,T.J. (1998) Nucleic Acids Res., 26, 2849–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perry W.L., Copeland,N.G. and Jenkins,N.A. (1994) Bioessays, 16, 705–707. [DOI] [PubMed] [Google Scholar]
- 39.Zeng L., Fagotto,F., Zhang,T., Hsu,W., Vasicek,T.J., Perry,W.L., III, Lee,J.J., Tilghman,S.M., Gumbiner,B.M. and Costantini,F. (1997) Cell, 90, 181–192. [DOI] [PubMed] [Google Scholar]
- 40.Sapienza C., Paquette,J., Tran,T.H. and Peterson,A. (1989) Development, 107, 165–168. [DOI] [PubMed] [Google Scholar]
- 41.Barlow D.P. (1997) EMBO J., 16, 6899–6905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Plass C., Shibata,H., Kalcheva,I., Mullins,L., Kotelevtseva,N., Mullins,J., Kato,R., Sasaki,H., Hirotsune,S., Okazaki,Y., Held,W.A., Hayashizaki,Y. and Chapman,V.M. (1996) Nature Genet., 14, 106–109. [DOI] [PubMed] [Google Scholar]
- 43.Schuster-Gossler K., Chazottes,D.S., Guénet,J.-L., Zachgo,J. and Gossler,A. (1996) Mamm. Genome, 7, 20–24. [DOI] [PubMed] [Google Scholar]
- 44.Matsuoka S., Thompson,J.S., Edwards,M.C., Barletta,J.M., Grundy,P., Kalikin,L.M., Harper,J.W., Elledge,S.J. and Feinberg,A.P. (1996) Proc. Natl Acad. Sci. USA, 97, 3026–3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leighton P.A., Ingram,R.S., Eggenschwiler,J., Efstratiadis,A. and Tilghman,S.M. (1995) Nature, 375, 34–39. [DOI] [PubMed] [Google Scholar]
- 46.Kalscheuer V.M., Mariman,E.C., Schepens,M.T., Rehder,H. and Ropers,H.H. (1993) Nature Genet., 5, 74–78. [DOI] [PubMed] [Google Scholar]
- 47.Sasaki H., Jones,P.A., Chaillet,J.R., Ferguson-Smith,A.C., Barton,S.C., Reik,W. and Surani,M.A. (1992) Genes Dev., 6, 1843–1856. [DOI] [PubMed] [Google Scholar]
- 48.Henikoff S. (1981) Chromosoma, 83, 381–393. [DOI] [PubMed] [Google Scholar]
- 49.Henikoff S. (1992) Curr. Opin. Genet. Dev., 2, 907–912. [DOI] [PubMed] [Google Scholar]
- 50.Gottschling D.E., Aparicio,O.M., Billington,B.L. and Zakian,V.A. (1990) Cell, 63, 751–762. [DOI] [PubMed] [Google Scholar]
- 51.Renauld H., Aparicio,O.M., Zierath,P.D., Billington,B.L., Chablani,S.K. and Gottschling,D.E. (1993) Genes Dev., 7, 1133–1145. [DOI] [PubMed] [Google Scholar]
- 52.Dorer D.R. and Henikoff,S. (1994) Cell, 77, 1–20. [DOI] [PubMed] [Google Scholar]
- 53.Sabl J.F. and Henikoff,S. (1996) Genetics, 142, 447–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Davis B.P. and MacDonald,R.J. (1988) Genes Dev., 2, 13–22. [DOI] [PubMed] [Google Scholar]
- 55.Garrick D., Fierring,S., Martin,D.I.K. and Whitelaw,E. (1998) Nature Genet., 18, 56–59. [DOI] [PubMed] [Google Scholar]
- 56.Rubin D.C., Ong,D.E. and Gordon,J.I. (1989) Proc. Natl Acad. Sci. USA, 86, 799–806. [Google Scholar]
- 57.Smrzka O.W., Fae,I., Stöger,R., Kurzbauer,R., Fischer,G.F., Henn,T., Weith,A. and Barlow,D.P. (1995) Hum. Mol. Genet., 4, 1945–1952. [DOI] [PubMed] [Google Scholar]
- 58.Xu Y., Goodyer,C.G., Deal,C. and Polychronakos,C. (1993) Biochem. Biophys. Res. Commun., 197, 747–754. [DOI] [PubMed] [Google Scholar]
- 59.Jinno Y., Yun,K., Nishiwaki,K., Kubota,T., Ogawa,O., Reeve,A.E. and Nikawa,N. (1994) Nature Genet., 6, 305–309. [DOI] [PubMed] [Google Scholar]
- 60.Weichman K. and Chaillet,J.R. (1997) Mol. Cell. Biol., 17, 5269–5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hummel K.P. (1958) J. Exp. Zool., 137, 389–423. [DOI] [PubMed] [Google Scholar]
- 62.Reed S.C. (1937) Genetics, 22, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Michaud E.J., van Vugt,M.J., Bultman,S.J., Sweet,H.O., Davisson,M.T. and Woychik,R.P. (1994) Genes Dev., 8, 1463–1472. [DOI] [PubMed] [Google Scholar]
- 64.Li E., Beard,C. and Jaenisch,R. (1993) Nature, 366, 362–365. [DOI] [PubMed] [Google Scholar]
- 65.Swain J.L., Stewart,T.A. and Leder,P. (1987) Cell, 50, 719–727. [DOI] [PubMed] [Google Scholar]