Abstract
Four years after the original sequence submission, we have re-annotated the genome of Mycoplasma pneumoniae to incorporate novel data. The total number of ORFss has been increased from 677 to 688 (10 new proteins were predicted in intergenic regions, two further were newly identified by mass spectrometry and one protein ORF was dismissed) and the number of RNAs from 39 to 42 genes. For 19 of the now 35 tRNAs and for six other functional RNAs the exact genome positions were re-annotated and two new tRNALeu and a small 200 nt RNA were identified. Sixteen protein reading frames were extended and eight shortened. For each ORF a consistent annotation vocabulary has been introduced. Annotation reasoning, annotation categories and comparisons to other published data on M.pneumoniae functional assignments are given. Experimental evidence includes 2-dimensional gel electrophoresis in combination with mass spectrometry as well as gene expression data from this study. Compared to the original annotation, we increased the number of proteins with predicted functional features from 349 to 458. The increase includes 36 new predictions and 73 protein assignments confirmed by the published literature. Furthermore, there are 23 reductions and 30 additions with respect to the previous annotation. mRNA expression data support transcription of 184 of the functionally unassigned reading frames.
INTRODUCTION
This study presents a re-annotation of the Mycoplasma pneumoniae genome, updating the original published annotation by Himmelreich et al. (1; deposited in GenBank) through further sequence analysis, incorporation of knowledge from the literature and new experimental data. There are inherent difficulties in genome annotation, even if the genome considered is small (the M.pneumoniae genome has a size of only 816 kb). In the original annotation 328 proteins (48%) from M.pneumoniae had no functional assignment. Comparisons and contradictory results with the genome annotation of the closely related Mycoplasma genitalium (2–7) illustrate that functional annotation is a continuing effort.
With these difficulties in mind, we have tried to approach the re-annotation in a more formal way. First, we re-examine gene contents and reading frame lengths (Table 1) and define the semantics used for the re-annotation (Table 3). Second, important steps in the annotation reasoning and the programs used are given, allowing reproducibility. Third, new experimental genome analysis data from M.pneumoniae support our effort.
Table 1. Identification of genes and reading frame lengtha.
|
677 previously annotated reading frames | |
| +, 6 | Intergenic hits to a hypothetical protein |
| MPN605(MP236–237), MPN482(MP359–360), MPN418(MP421–422), MPN388(MP450–451 MPN270(MP564–565), MPN254(MP579.1), | |
| (MP313–314)b, (MP365–366)b, (MP383–384)b, (MP384–385)b | |
| +, 4 | New complete protein genes |
| MPN069(MP085–086) 50S ribosomal protein L33 | |
| MPN495(MP346.1) PTS pentitol phosphotransferase EIIB | |
| MPN296(MP540–541) 30S ribosomal protein S21 | |
| MPN242(MP590–591) SecG | |
| +, 2 | Short hypothetical proteins |
| –, 1 | the original MP237 was a too short, different reading frame and was deleted |
| 688 protein reading frames (after our re-annotation) | |
| Re-examination of protein reading frame lengthsc | |
| +, 12 | N-terminal extensionsd |
| MPN118(MP037), MPN077(MP078), MPN033(MP121), MPN661(MP181), MPN651(MP191), MPN475(MP365), MPN448(MP392), MPN396(MP443), MPN395(MP444), MPN345(MP492), MPN336(MP501), MPN306(MP531) | |
| For MPN033(MP121) (uracil phosphotransferase; P75081) and MPN395(MP444) (adenine phosphoribosyltransferase) the 2-dimensional gel molecular weights confirm the predicted extension | |
| +, 4 | C-terminal extensionsd |
| MPN111(MP044), MPN108(MP047), MPN032(MP122), MPN520(MP322) | |
| –, 8 | Proteins shortened at the N-terminus |
| The following protein reading frames are shorter (N-terminus begins later) than the previously annotated M.pneumoniae GenBank annotation | |
| MPN073(MP082), MPN643(MP199), MPN639(MP203), MPN611(MP231), MPN444(MP395), MPN432(MP408), MPN320(MP517), MPN170(MP662) | |
aAll intergenic regions between any of the previously annotated protein reading frames were re-screened applying sequence analysis to identify hitherto overlooked reading frames (top). Similarly, previously unrecognized extensions became apparent by sequence comparison as well as shortened reading frames (bottom).
bThese four reading frames contain in-frame stops and are not counted.
cData of these reading frame modifications were shared with SwissProt and either are or will very soon be updated in SwissProt.
dThe C-terminal extensions are supported by sequence alignment to related protein reading frames from other organisms. However, they are only possible with frame shifting or mutation of stop codons. This indicates either pseudogenes or sequencing errors in these regions. In addition to the N- and C-terminal extensions listed, there is a potential intergenic extension. Adjacent ORFs MPN347(MP490) and MPN345(MP492) may be connected with MPN346(MP491) to form one gene via the intergenic regions, but this would again require some frame shifting [hsdR restriction enzyme (pseudo)gene, sequencing error or gene fragments].
Table 3. Re-annotation of protein function: the different re-annotation categories.
| Category | Cases | |
|---|---|---|
| Proteins originally annotated in GenBank | 677 | |
| Conf | Re-annotation is consistent to the functional annotation of GenBank (‘confirmed’; only if a function is assigned, otherwise labeled as a hypothetical protein, next category); e.g. MPN125(MP030) excinuclease ABC, subunit C | 297 |
| hypotheticala | The function of the protein seems to be unknown (even if stated otherwise in GenBank) and there is no orthologous protein in any other species so far; e.g. MPN376(MP460) (no similar sequences found) | 45 |
| conserved hypothetical | Class additional to original GenBank annotation; the function of the protein is treated as unknown (even if stated otherwise in GenBank), but there are homologous proteins in other species, for example in M.genitalium; e.g. MPN239(MP593) with its homolog MG101 | 178 |
| wrong | The functional annotation in GenBank has been completely replaced or the protein reading frame deleted. It is followed by an explanation; e.g. MP237 original reading frame was deleted | 4 |
| less | The functional annotation in GenBank cannot be justified by database searches according to our knowledge, for example MPN007(MP147) originally annotated as DNA polymerase subunit δ′ is now modified to ‘similar to DNA-polymerase subunits’ | 18 |
| more_ | This study adds some new, additional feature to the functional annotation of the protein; e.g. MPN324(MP513), described as ribonucleoside diphosphate reductase but we added that this is the α-chain | 30 |
| new_conf | No functional prediction was annotated in GenBank M.pneumoniae sequence, but latest versions of other genomes and databanks [GenBank M.genitalium (revised), SwissProt, etc.] or recent literature indicate similar functional features and are confirmed in this study; e.g. MPN549(MP293) previously annotated ‘hypothetical protein’ is now predicted as a phophodiesterase with a DHH domain, as also described by Fukuda et al. (32) (however, the paper mislabeled this orf4 of the P1 operon as P1 itself) | 73 |
| new | The functional prediction is new. No other source of this information is available, at least to our knowledge; e.g. MPN435(MP405), ‘MG306 homolog’ before, now annotated by sequence similarity as amino acid permease | 32 |
| New protein reading frames (see Table 1; four new functional annotated reading frames; five conserved hypothetical; two hypothetical) | 11 | |
| Total protein reading framesb | 688 | |
| RNA genesc | 42 | |
| RNA and protein genes in the complete genome | 730 | |
aOnly two hypothetical entries remained unchanged, only 297 from a new total of 688 protein reading frames stay as before (43%).
bTotal number of proteins unassigned: 223 + 7 = 230.
c35 tRNA, 19 re-annotated tRNA positions, two new tRNALeu (codons TTG and CTC); six other functional RNAs (positions re-annotated); one new RNA of 200 nt (27).
The protein and RNA inventory of M.pneumoniae is made much more complete by the re-annotation, as shown by examples from all annotation categories discussed below.
MATERIALS AND METHODS
Computational genome and sequence analysis techniques
The complete genome of M.pneumoniae was extensively compared to available completely sequenced genomes (in particular to M.genitalium) to better assign and identify the encoded proteins therein. Furthermore, iterative sequence analysis searches (PSI-BLAST; 8) compared M.pneumoniae sequences to other organisms and public databases. The statistical expectancy value for reporting hits by chance was generally set at a conservative threshold of an expected value E of 10–6.
To independently check and test these results, we applied not only other programs with similar function, such as HMM and fasta searches, but also complementary tools and methods, such as domain analysis, phylogenetic analysis, analysis of context and clusters of orthologous genes. This also included analysis of gene duplications, replacement by unrelated sequences (non-orthologous displacement; 9) and gene neighborhood to determine orthology (10). Furthermore, we applied the different tools using extensive sequence analysis protocols as described and reviewed previously (11). Amongst other tests, this included verification of detected similarities by reciprocal searches from identified sequences and determination of the exact region where the sequence similarity was actually found. In particular, the multidomain architecture of many proteins has been taken into account. Functional assignments were tested and confirmed, including sequence searches from sequences with experimentally determined functions (12). Significant links to experimentally determined functions were established.
Phylogenetic analysis was applied to analyze gene duplication events and clarify the substrate specificity of the encoded enzymes.
Detailed data for each reading frame, including annotation reasoning and programs used, are available on our web site (www.bork.embl-heidelberg.de/Annot/MP/ ). The updated annotation data are furthermore deposited with GenBank (update of accession no. U00089; 1).
A number of standard features are included in the web table: gene numbering (original GenBank number and new revised numbering from the putative origin of replication in accordance with widely used numbering schemes for prokaryotic genomes), GenBank identifier and accession no.; original GenBank annotation and revised annotation; where applicable and of interest, proteins with similar sequence with known 3-dimensional folds (13); metabolic pathway assignment (14); MG orthologs and MP homologs; intrinsic features (transmembrane domains, protein export signals, low complexity regions and coiled coils); domain annotations according to the SMART program suite (15); characterizing comments on reading frames.
Experimental genome analysis techniques
Mycoplasma pneumoniae culture, treatment of cells and protein extraction are described in Proft and Herrmann (16).
2-Dimensional gel electrophoresis followed standard procedures (17). The pH gradient in the first dimension was from pH 3 to pH 10 and in the second dimension vertical slab gels were used.
Protein identification by mass spectrometry (details in 18). Colloidal Coomassie Blue stained protein spots were cut out and tryptic gel digests were done. The tryptic peptides were eluted, concentrated and analysed by on-line micro-HPLC and ion trap mass spectrometry (MS/MS). Ion trap mass spectrometry permitted identification of the protein by comparing the masses of tryptic peptides and their fragmentation pattern to a protein database directly translated from the DNA sequence. An in-depth analysis of the 2-dimensional gel and mass spectrometry data for M.pneumoniae will be published elsewhere (J.T.Regula, B.Ueberle, G.Boguth, A.Görg, M.Schnölzer, R.Herrmann and R.Frank, submitted for publication).
mRNA expression. Measurement of mRNA expression of the different M.pneumoniae genes (comparing different growth temperatures) followed standard techniques using DNA arrays (19; H.W.H.Göhlmann, C.U.Zimmermann and R.Herrmann, unpublished data).
Other techniques. Standard molecular biology techniques for genome sequencing, cloning, northern hybridization and protein analysis were applied according to Sambrook et al. (20).
RESULTS AND DISCUSSION
Identification of genes and reading frame length
RNA. Encoded RNAs and RNA genes were identified by systematic sequence comparison to orthologous RNAs from different prokaryotic and eukaryotic species and to GenBank and to available RNA databases for specific RNA types (21–25). We did not consider other completely novel or non-consensus RNA variants (26,27). Two new tRNALeu were added and the positions of 19 of the original 33 tRNA annotations were revised. Furthermore, re-annotation of the positions of three rRNA genes (5S rRNA, 16S rRNA and 23S rRNA) and of three other functional RNA molecules (RNase P, 10Sa RNA and 4.5S RNA) were included, as well as a description of a new 200 nt RNA. The 200 nt RNA, named MP200 RNA, was further analyzed in detail, including northern analysis. It is highly abundant. Its rich stem–loop structure and the potential to encode cysteine-rich peptides is conserved between M.pneumoniae and M.genitalium, however, its specific function is still unclear (28).
Proteins. The intergenic regions were re-analyzed by sequence comparisons to identify unrecognized reading frames (Table 1, top). This yielded a total of 12 new proteins (two unassigned short proteins identified by mass spectrometry, six hypothetical proteins and four with predicted functional features) (Fig. 1). Furthermore, one of the original reading frames was dismissed and four with sequence similarity to proteins were discarded as they contain frameshifts and are likely pseudogenes (Table 1). Apart from PSI-BLAST searches these results were checked by extensive protein family alignments and other techniques as explained in Materials and Methods. As a result, the current number of protein genes we report here is 688, an increase of 11 from the previous annotation.
Figure 1.
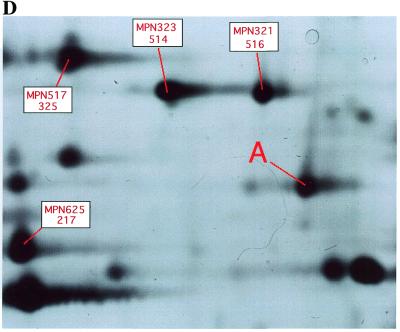
(A) Peptides identified by mass spectrometry of the protein MPN033(MP121) (see Materials and Methods). Those peptides matching the genome-derived sequence are shown in bold. The protein reading frame sequence not covered by these peptides is shown in plain text. Extension of the MPN033(MP121) sequence respective to its original annotation could be confirmed. The methionine at the start is shown in italic. The start position given in the original annotation is underlined. The exact start sequence is predicted (as shown) at the methionine directly before the furthest N-terminal peptide determined. (B) Identification of three new short proteins by mass spectrometry. These proteins are shorter than 100 amino acids. The methionine at the start is shown in italic. The first protein shows high similarity to a pentitol phosphotransferase IIB subunit. This peptide was also predicted from screening intergenic regions and by Reizer et al. (30). The other two are hypothetical (show no similarities). The short proteins are given here as maximal extensions between two stop codons according to the peptides sequenced. A detailed analysis of the peptide data derived for these proteins as well as the proteome of M.pneumoniae will be published elsewhere (J.T.Regula, B.Ueberle, G.Boguth, A.Görg, M.Schnölzer, R.Herrmann and R.Frank, submitted for publication). (C) Sequence of the reading frame MPN254(MP579.1) (hypothetical) predicted between MPN255(MP579) and MPN253(MP580) according to the mass spectrometric data. Peptides matching the genome-derived sequence identified by mass spectrometry are shown in bold. Protein sequence not covered by these peptides is shown in plain text (protein coverage 40.8% by amino acid count, 40.1% by mass). (D) Mycoplasma pneumoniae proteins were separated by 2-dimensional gel electrophoresis in a pH gradient from 3 to 10, in a vertical 12.5% slab gel and stained with silver. A part of the 2-dimensional gel showing the presence of the product of gene MPN254(MP579.1) (labeled A, sequence as shown in C). Previously known MP proteins surrounding MPN254(MP579.1) in the 2-dimensional gel are labeled in red, with the MPN number given at the top and the number according to Himmelreich et al. (1) at the bottom.
All protein reading frames were consistently renumbered (MPN numbers; see our web page) from the origin of replication as in other prokaryotic genome efforts. Genome identifiers for the proteins discussed in the paper, sorted according to MPN number, are summarized with their alternative identifiers in Table 2 [the new number, old identifier according to Himmelreich et al. (1), PID and ORF identifier are listed]. In the following the MP numbers according to the original numbering system after Himmelreich et al. (1) are given as subscripts in parentheses for reference to previous papers. These MP numbers are not identical to the subsequent GenBank numbering.
Table 2. Genome identifiers for the proteins discussed (sorted according to MPN).
| MPN | MP | PID | orf |
|---|---|---|---|
| MPN007 |
147.0 |
PID:g1673807 |
D12_orf253 |
| MPN032 |
122.0 |
PID:g1673781 |
B01_orf108 |
| MPN033 |
121.0 |
PID:g1673780 |
B01_orf178 |
| MPN047 |
107.0 |
PID:g1673764 |
D09_orf451 |
| MPN051 |
103.0 |
PID:g1673759 |
D09_orf384 |
| MPN068 |
086.0 |
PID:g1673741 |
D09_orf125 |
| MPN069 |
085.0–086.0 |
50S rp L33 |
48 aa |
| MPN073 |
082.0 |
PID:g1673736 |
D09_orf388 |
| MPN077 |
078.0 |
PID:g1673732 |
R02_orf469 |
| MPN078 |
077.0 |
PID:g1673731 |
R02_orf694 |
| MPN079 |
076.0 |
PID:g1673730 |
R02_orf300 |
| MPN095 |
060.0 |
PID:g1673710 |
R02_orf254 |
| MPN096 |
059.0 |
PID:g1673709 |
R02_orf264 |
| MPN108 |
047.0 |
PID:g1673696 |
C09_orf404 |
| MPN111 |
044.0 |
PID:g1673692 |
C09_orf422 |
| MPN113 |
042.0 |
PID:g1673690 |
C09_orf223 |
| MPN118 |
037.0 |
PID:g1673685 |
C09_orf143b |
| MPN125 |
030.0 |
PID:g1673677 |
C09_orf586L |
| MPN158 |
674.0 |
PID:g1674379 |
VXpSPT7_orf269 |
| MPN170 |
662.0 |
PID:g1674366 |
VXpSPT7_orf184 |
| MPN210 |
622.0 |
PID:g1674324 |
G07_orf808 |
| MPN223 |
609.0 |
PID:g1674310 |
G07_orf312 |
| MPN237 |
595.0 |
PID:g1674296 |
G07_orf478V |
| MPN239 |
593.0 |
PID:g1674294 |
K04_orf222 |
| MPN242 |
590.0–591.0 |
SecG |
76 aa |
| MPN243 |
590.0 |
PID:g1674290 |
K04_orf726 |
| MPN254 |
579.1 |
|
A65_orf157 |
| MPN270 |
564.0–565.0 |
hypothetical |
95 aa |
| MPN272 |
563.1 |
E1553 |
A65_orf94 |
| MPN274 |
562.0 |
PID:g1674260 |
A65_orf266 |
| MPN280 |
556.0 |
PID:g1674253 |
A65_orf569 |
| MPN294 |
542.0 |
PID:g1674238 |
H10_orf206 |
| MPN296 |
540.0–541.0 |
30S rp S21 |
60 aa |
| MPN298 |
539.0 |
PID:g1674234 |
H10_orf119 |
| MPN304 |
533.0 |
PID:g1674228 |
H10_orf238 |
| MPN305 |
532.0 |
PID:g1674227 |
H10_orf198 |
| MPN306 |
531.0 |
PID:g1674226 |
H10_orf273o |
| MPN308 |
529.0 |
PID:g1674224 |
F10_orf565 |
| MPN320 |
517.0 |
PID:g1674210 |
F10_orf328 |
| MPN321 |
516.0 |
PID:g1674209 |
F10_orf160 |
| MPN323 |
514.0 |
PID:g1674207 |
F10_orf153 |
| MPN324 |
513.0 |
PID:g1674206 |
F10_orf721 |
| MPN345 |
492.0 |
PID:g1674183 |
H91_orf206 |
| MPN346 |
491.0 |
PID:g1674182 |
H91_orf115 |
| MPN347 |
490.0 |
PID:g1674181 |
H91_orf376 |
| MPN372 |
465.0 |
PID:g1674154 |
A19_orf591 |
| MPN376 |
461.0 |
PID:g1674149 |
A19_orf1140 |
| MPN377 |
460.1 |
E3366 |
A19_orf74 |
| MPN386 |
452.0 |
PID:g1674139 |
F11_orf229 |
| MPN388 |
450.0–451.0 |
hypothetical |
42 aa |
| MPN395 |
444.0 |
PID:g1674131 |
F11_orf133 |
| MPN396 |
443.0 |
PID:g1674129 |
F11_orf887 |
| MPN397 |
442.0 |
PID:g1674128 |
F11_orf733 |
| MPN407 |
432.0 |
PID:g1674117 |
F11_orf879 |
| MPN418 |
421.0–422.0 |
hypothetical |
140 aa |
| MPN427 |
413.0 |
PID:g1674098 |
A05_orf290 |
| MPN431 |
409.0 |
PID:g1674094 |
A05_orf317 |
| MPN432 |
408.0 |
PID:g1674093 |
A05_orf382 |
| MPN435 |
405.0 |
PID:g1674089 |
A05_orf395 |
| MPN444 |
395.0 |
PID:g1674078 |
H08_orf289 |
| MPN448 |
392.0 |
PID:g1674075 |
H08_orf263 |
| MPN455 |
385.0 |
PID:g1674067 |
H08_orf287 |
| MPN456 |
384.0 |
PID:g1674066 |
H08_orf1005 |
| MPN457 |
383.0 |
PID:g1674064 |
H08_orf329V |
| MPN474 |
366.0 |
PID:g1674046 |
P01_orf1033 |
| MPN475 |
365.0 |
PID:g1674045 |
P01_orf292 |
| MPN479 |
361.0 |
PID:g1674040 |
P01_orf197 |
| MPN482 |
358.0–359.0 |
hypothetical |
64 aa |
| MPN491 |
350.0 |
PID:g1674028 |
P02_orf474 |
| MPN492 |
349.0 |
PID:g1674027 |
P02_orf305 |
| MPN493 |
348.0 |
PID:g1674026 |
P02_orf218 |
| MPN494 |
347.0 |
PID:g1674025 |
P02_orf159 |
| MPN495 |
346.1 |
C5841 |
P02_orf95 |
| MPN496 |
346.0 |
PID:g1674024 |
P02_orf660 |
| MPN508 |
334.0 |
PID:g1674009 |
P02_orf509 |
| MPN509 |
333.0 |
PID:g1674008 |
P02_orf427 |
| MPN510 |
332.0 |
PID:g1674007 |
P02_orf458 |
| MPN511 |
331.0 |
PID:g1674006 |
F04_orf260V |
| MPN512 |
330.0 |
PID:g1674005 |
F04_orf154 |
| MPN517 |
325.0 |
PID:g1673999 |
G12_orf166a |
| MPN520 |
322.0 |
PID:g1673995 |
G12_orf861 |
| MPN527 |
315.0 |
PID:g1673988 |
G12_orf225 |
| MPN528 |
314.0 |
PID:g1673987 |
G12_orf184 |
| MPN529 |
313.0 |
PID:g1673985 |
G12_orf109 |
| MPN547 |
295.0 |
PID:g1673966 |
G12_orf558 |
| MPN548 |
294.0 |
PID:g1673965 |
G12_orf326 |
| MPN549 |
293.0 |
PID:g1673964 |
G12_orf325 |
| MPN558 |
284.0 |
PID:g1673955 |
H03_orf191 |
| MPN557 |
285.0 |
PID:g1673956 |
H03_orf612 |
| MPN562 |
280.0 |
PID:g1673951 |
H03_orf248 |
| MPN571 |
271.0 |
PID:g1673942 |
D02_orf660 |
| MPN605 |
237.0 |
PID:g1673905 |
C12_orf157La |
| MPN608 |
234.0 |
PID:g1673901 |
C12_orf225 |
| MPN609 |
233.0 |
PID:g1673900 |
C12_orf329 |
| MPN610 |
232.0 |
PID:g1673899 |
C12_orf651V |
| MPN611 |
231.0 |
PID:g1673898 |
C12_orf385 |
| MPN625 |
217.0 |
PID:g1673883 |
C12_orf141 |
| MPN639 |
203.0 |
PID:g1673867 |
E09_orf287o |
| MPN643 |
199.0 |
PID:g1673863 |
E09_orf302 |
| MPN651 |
191.0 |
PID:g1673855 |
E09_orf379 |
| MPN652 |
190.0 |
PID:g1673854 |
E09_orf364 |
| MPN653 |
189.0 |
PID:g1673853 |
E09_orf143V |
| MPN661 | 181.0 | PID:g1673844 | K05_orf499 |
1The original GenBank MP237 here was another, too short reading frame and has been deleted; instead there is a new reading frame stretching into the previously intergenic region.
MPN, updated genome numbering scheme; MP, original numbering after Himmelreich et al. (1). PID numbering and ORF accession codes are given in addition. The full table is available on our web site.
The reading frame lengths were also re-examined. Previously unrecognized extensions of different MP proteins became apparent and are summarized in Table 1 (bottom). The eight re-annotated proteins that have been shortened at the N-terminus are already included in the SwissProt sequence database. Protein fragments and overlaps were also identified. For example, MPN305(MP532) and MPN304(MP533) are N- and C-terminal fragments of arginine deiminase. Re-sequencing suggests that the separating frameshift is real, while intact MPN560(MP282) provides arginine deiminase activity.
As a further validation of the results derived by sequence comparison and theoretical analysis, two of the predicted N-terminal extensions (Table 1, bottom) were directly confirmed using 2-dimensional gel electrophoresis and mass spectrometry. Applying this combination, 350 protein spots were resolved and analyzed in a systematic effort to study the proteome of M.pneumoniae. Figure 1A shows peptides of the protein MPN033(MP121) identified by mass spectroscopy in bold. Protein reading frame sequences not covered by these peptides are shown in plain text. The other predictions are currently being examined by the same techniques. In Figure 1B mass spectrometry data for three new, short proteins in M.pneumoniae are shown. Two of these short proteins show no homology to any known sequences (also not in HMM and SMART searches), while the third reading frame has significant similarity to a small subunit of the PTS system (expected E value applying PSI-BLAST of 10–36). This confirmed experimentally an ORF between P02_orf660 and P02_orf159 already suggested by Reizer et al. (29), as well as by our screen for proteins in previously intergenic regions (MPN495(MP346.1); Table 1). Furthermore, the hypothetical protein MPN254(MP579.1) predicted from the intergenic screen was confirmed by the same technique (Fig. 1C). The localization of the 2-dimensional gel spot for this protein before tryptic digestion for mass spectrometry is shown in Figure 1D.
Re-annotation of protein function
We considered a functional feature to be predicted for the product of a reading frame if either its molecular function could be predicted (e.g. ‘methyltransferase’) or the biological context has become clear. Thus, a transmembrane domain (predicted as an intrinsic feature) is not considered specific enough for a functional annotation, however, ‘permease’ (indicating the biological activity) is. Similarly, a non-specific description regarding an external stimulus (such as ‘glucose-inhibited protein’) was not considered to be sufficient for a functional annotation, whereas the cellular role (i.e. ‘cell division protein’) is. Different functional re-assignment categories are given together with an example for each category in Table 3. Apart from the first group of 297 proteins for which the annotation could be confirmed (‘conf‘; Table 3, top; 43% from a total of 688 proteins), modifications of the original annotations were made. These included semantic modifications (mainly in the classification of hypothetical proteins) and modified functional assignments (in all protein categories).
In the following only a few examples for each re-annotation category (hypothetical, conserved hypothetical, wrong, less, more_, new_conf and new) are discussed in the order they appear in Table 3. More data are summarized in the tables and each reading frame annotation for the whole genome can be found at http://www.bork.embl-heidelberg.de/Annot/MP/
Proteins of unknown function
The original GenBank annotation of M.pneumoniae does not provide a known functional feature for 328 protein reading frames. These protein reading frames are listed in Table 4 under four different categories. Part of our effort was motivated by the goal to add functional information to these entries. For example, 42 proteins were previously assigned as ‘putative lipoproteins’ only and four putatitive lipoproteins which were given a defined functional assignment (1). For these proteins, the prokaryotic lipoprotein motif (prosite PS00013) is present [lipobox, Met++, more or less hydrophobic leader region Leu(Ala/Ser)(Gly/Ala)Cys; the leader region is very short in MPN561(MP281) and MPN051(MP103)]. Palmitylation assays indicate that the number of proteins with lipid attachment sites in M.pneumoniae should be 25–30 (Pyrowolakis and Herrmann, unpublished results), but so far only the subunit b of the F0F1-type ATPase has been identified experimentally as a lipoprotein (30). A reliable, homology-based prediction requires the identification of a related sequence with a domain confirmed to be involved in lipid binding. This was the case for only six of the 42 putative lipoproteins. Another two were found to have a distinct function. The other 34 sequences were re-annotated more conservatively as ‘hypothetical’ or ‘conserved hypothetical’ (the next two categories in Table 3; conserved hypothetical if there was a related protein sequence in another species).
Table 4. Proteins with unknown function.
| Old |
|
New |
|
|||
|---|---|---|---|---|---|---|
| Total | Percentb | Total | Percent | |||
| ‘MG[…] homolog’a | 137 | 20% | not used | not used | ||
| ‘orf’ | 94 | 14% | not used | not used | ||
| ‘hypothetical’ | 63 | 9% | 47 | 7% | ||
| ‘putative lipoprotein’c | 34 | 5% | not used | not used | ||
| ‘conserved hypothetical’d | not used | not used | 183 | 27% | ||
| Unknown function | 328 | 48% | 230 | 33% | ||
aAdditional 36 proteins with a homolog in M.genitalium were also classified as putative lipoproteins (see that category).
bPercentages values are rounded to the nearest integer.
cThe proteins counted here contain only a lipoprotein prosite motif, but could not be linked by sequence analysis to a known, experimentally characterized lipoprotein. Six further putative lipoproteins could be confirmed and were annotated as lipoproteins (see text for details). Four other proteins are lipoproteins already apparent in the original annotation but not counted here as they did not have the keyword ‘putative lipoprotein’.
dIncluding five new ones found in intergenic regions.
Expression of mRNA (Table 4) was confirmed by gene expression data for 184 of the (conserved) hypothetical proteins using macroarrays (19). The macroarray data are given for individual reading frames in our complete genome annotation table (see Materials and Methods; presence of an mRNA for an individual reading frame is labeled ‘mRNA expressed’ in the web table).
Re-annotation of functional assigned proteins
Four annotations were completely replaced (wrong; example in Table 3). In several cases the original annotation was too broad and a less specific one (keyword ‘less’; Table 3, middle) had to be chosen. MPN007(MP147) is an example. It was originally annotated as DNA polymerase III subunit δ′. However, there is not enough sequence similarity to confirm that functional assignment. The sequence similarities in PSI-BLAST runs to other subunits such as γ and τ have similarly high E values (ranging from 10–7 to 10–4 for each of them; protein length is well covered; similar results are apparent from phylogenetic analyses or analyzing the domain architecture) and only similarity to an unspecified subunit of DNA polymerase III is annotated by us.
New functional features compared to GenBank were annotated in 109 cases, including predictions for four completely new reading frames. Each of these adds some information to the predicted protein and enzymatic repertoire of M.pneumoniae (Table 5). We defined three categories: novel functional features, novel annotation integrating public knowledge and novel prediction (more_, new_conf and new; Table 3, bottom).
Table 5. Re-annotated molecular functions encoded in M.pneumoniae reading frames (selected examples).
| Transport | |
| MPN274(MP562) | Sulfate/molybden ABC transporter subunit |
| MPN113(MP042) | G3P transporter |
| MPN068(MP086) | secE protein transport system |
| MPN096(MP059) | Amino acid permeases |
| MPN095(MP060) | Amino acid permeases |
| MPN396(MP443) | Similar to secD |
| MPN435(MP405 ) | Amino acid permeases |
| MPN431(MP409) | Amino acid permeases |
| MPN308(MP529) | Amino acid permease |
| MPN625(MP217) | Osmotically inducible protein C family |
| MPN611(MP231) | Phosphate assimilation |
| MPN571(MP271) | Hemolysin like protein |
| MPN508(MP334) | Membrane translocator |
| MPN509(MP333) | Membrane translocator |
| MPN510(MP332) | Membrane translocator |
| MPN511(MP331) | Membrane translocator |
| MPN512(MP330) | Membrane translocator |
| MPN474(MP366) | Structural protein |
| Metabolism | |
| MPN047(MP107) | Nicotinate phosphoribosyl-transferase |
| MPN032(MP122) | Hydrolase |
| MPN558(MP284) | Methyltransferase |
| MPN557(MP285) | NADH oxidoreductase |
| MPN548(MP294) | Pseudouridine synthase |
| MPN547(MP295) | Dihydroxyacetone kinase |
| MPN527(MP315) | Membrane-integrated oxidoreductase |
| MPN491(MP350) | Membrane nuclease |
| MPN427(MP413) | Hydrolase/phosphatase |
| MPN407(MP432) | Lipase |
| MPN336(MP501) | Nucleotidyl transferase |
| MPN298(MP539) | Acyl carrier protein synthase |
| MPN294(MP542) | Protease |
| MPN280(MP556) | Similar to single-stranded RNA(DNA) processing enzyme |
| MPN243(MP590) | RNase R |
| Pathogenicity | |
| MPN372(MP465) | Similar to pertussis toxin subunit 1 |
| Regulation | |
| MPN223(MP609) | HPr(Ser) kinase |
Examples for new annotations are shown. Italics indicate that this functional assignment seems to be completely new and is not backed up by previously published literature or databases (new, Table 3), otherwise it is new_conf (Table 3). Details on these shortened, abbreviated annotations are given in the text; the complete list, detailed comments and additional information can be found at http://www.bork.embl-heidelberg.de/Annot/MP/ . New data are also included at this site if they shed new light on already known annotations; for example, in the hsdS restriction enzyme system recent data from Mycoplasma pulmonis enzymes similar in sequence suggest rapid evolution (42).
Novel functional features. In 30 cases we could add functional features to the functional annotation present (more_; Table 3). An example is MPN237(MP595), which was originally annotated as an amidase homolog. Sequence analysis using PSI-BLAST shows that one can be more precise about this finding; this sequence is similar to glutamine-tRNA amidotransferase subunit A (this is also evident from the family alignment). There is high and significant homology over the full sequence length to, for example, the recently experimentally characterized sequence from Bacillus subtilis (31). This similarity has also been included in the recent update of the homologous M.genitalium sequence by GenBank.
Novel annotation integrating public knowledge. Since the release of the original GenBank annotation (1), new data on the sequence entries have become available and the sequence analysis software has been enhanced (see for example 8). To integrate these new data in an unbiased and systematic fashion, first all sequence entries were re-analyzed with the latest sequence analysis software (see Materials and Methods). The old annotation was also extensively compared to the results from a survey of recent literature and public database updates such as the SwissProt sequence database. The complete M.genitalium sequence has been recently updated and a number of papers (see for example 32–34) have described novel predictions and experiments for many of the M.pneumoniae genes. Inconsistencies with the original annotation found by our own sequence analysis can be resolved with higher certainty by systematically retrieving and critically comparing this public data from different sources.
MPN558(MP284) and MPN557(MP285) provide typical examples (Table 5). Originally annotated as glucose-inhibited cell division proteins B and A, detailed sequence comparisons, including PSI-BLAST, domain architecture and complementary sequence analysis methods (such as predicting protein 3-dimensional structures based on homologous sequence searches; http://www.bork.embl-heidelberg.de ) show that their actual molecular functions seem to be a methyltransferase [MPN558(MP284)] and an NADH oxidoreductase [MPN557(MP285)], including homologs with known structure (1BHJ.brk and 1FEA.brk, respectively). Specific queries for these findings revealed that this information has already been noted by others, for example regarding the latest version of clusters of orthologous genes (COG0357 and COG0445, respectively; 35). However, these novel predictions were not considered in the last GenBank update of M.genitalium and in the recent literature (see for example 36).
Novel prediction. There are 36 cases where (at least to our knowledge) the functional assignment is completely new (Table 5). An example is the protein secretion system in M.pneumoniae. The system has been well characterized in Escherichia coli (35). Cytosolic chaperones or regulators (trigger factor, SecB, DnaK, bacterial signal recognition particle and FtsY) deliver the protein to a membrane transporter (SecA). The receptor should also function as a motor to push the protein across the membrane via specific protein channels (SecY, SecG, SecE, SecD and SecF). Himmelreich et al. (1) noted that they had identified trigger factor, DnaK, SRP and FtsY as well as SecA, whereas of the channel-forming proteins only SecY could be assigned, leaving the secretion pathway incomplete.
We have now annotated protein reading frames similar to SecD, SecE and SecG, yielding a new, more complete picture of this secretory pathway in M.pneumoniae. As several pathogenicity factors (e.g. re-annotated hydrolases and lipases; Table 5) are secreted, the respective protein channels are potential drug targets.
SecE and SecG were annotated by integrating public knowledge. MPN068(MP086) is a SecE homolog (new_conf, updated COG0690; 35). MPN242, a region previously annotated as intergenic, is the missing SecG homolog. The YvaL homology has also been reported by Bellgard and Gojobori (38). YvaL has in the meantime been experimentally verified to be a SecG homolog (39).
However, MPN396(MP443), with its similarity to secD, provides an example of a novel prediction (Fig. 2). This protein had been annotated before as a conserved hypothetical protein, the MG277 homolog from M.genitalium (1,35,36 and in the SwissProt update of M.genitalium). PSI-BLAST searches indicate similarity to the secDF protein from B.subtilis after the second iteration.
Figure 2.
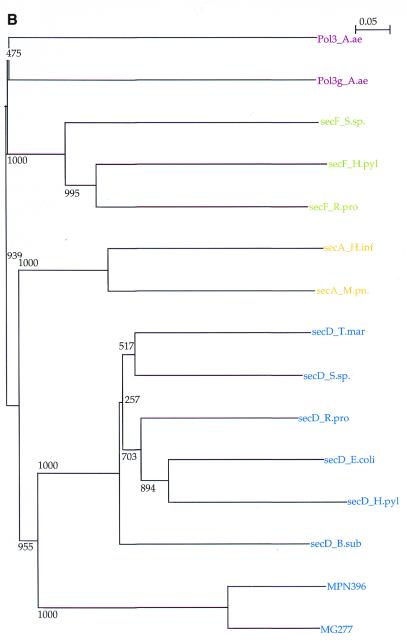
(A) Sequence alignment of MPN280(MP555) with related secD sequences. Only the central part (140 amino acid positions) of the alignment is given. After the M.pneumoniae sequence the M.genitalium homolog is shown (MG277), aligned with secD proteins from various species (top to bottom) (SwissProt identifier/accession no.): Mycobacterium leprae (SECD_MYCLE); Mycobacterium tuberculosis (SECD_MYCTU); Bacillus subtilis (accession no. AAC31122; the secD domain from the fusion protein secDF only); Treponema pallidum (SECD_TREPA); Thermotoga maritima (accession no. Q9WZW4); Borrelia burgdorferi (accession no. AAC66993); Helicobacter pylori (SECD_ECOLI); secD from Campylobacter jejuni (accession no. CAB73348); Escherichia coli (SECD_ECOLI); Haemophilus influenzae (SECD_HAEIN); Rickettsia prowazecki (SECD_RICPR); Streptomyces coelicolor (SECD_STRCO); Synechocystes PCC6803 (SECD_SYNY3). (B) Phylogenetic tree with bootstrap values (1000 trials) comparing certified secD and secF domains (T.mar, Thermotoga maritima; S.sp., Synechocystes PCC6803; R.pro, Rickettsia prowazecki; H.pyl, Helicobacter pylori; E.col, E.coli, B.sub, Bacillus subtilis) with MPN280(MP555) and its homolog MG277, secA from H.influenzae and MPN210(MP622) from M.pneumoniae and polymerase III subunits (Aquifex aeolicus).
Further analysis re-tested this suggestion and showed that protein MPN396(MP443) contains a domain similar to secD and a second part (which may perhaps be another domain involved in secretion, such as a fusion with the related secF as in B.subtilis secDF). The similarity of the secD-like domain in MPN396(MP443) was confirmed by PSI-BLAST searches from established secD proteins [finding MPN396(MP443) with expected values well below 10–6 in the second iteration]. Moreover, clusters of orthologous genes and gene neighborhoods (both available using the STRING tool at http://www.bork.EMBL-Heidelberg.DE/C-GOD ) back this prediction by independent methods. Detailed sequence alignment (the central portion is displayed in Fig. 2A) shows clear homology to other secD domains but indicates also that the Mycoplasma sequences are only secD-like. A phylogenetic tree of established secD and secF sequences including MPN396 and MG277 gives a similar result (Fig. 2B). We suggest that MPN396 with its secD-like domain should further complete the secretory repertoire in M.pneumoniae; however, experiments and analyses have now to better determine its exact relation to the established members of the sec family characterized to date.
No homologous sequence has been found for SPase I in the secretory pathway in M.pneumoniae. SPase I would cleave the signal peptide before secretion. However, suitable cleavage sites have been identified for several M.pneumoniae proteins (1) and one of the proteases identified may contain this function, such as the new annotated intracellular protease MPN386(MP542) (new_conf, COG 0693).
Re-annotated molecular functions enable predictions on higher levels
The re-annotation of molecular functions may in addition provide some answers regarding higher levels of cellular interactions such as transport (several new annotated permeases and transporters are listed in Table 5), secretion (example above) and pathogenicity factors. Metabolism, multiple substrate use and existing operons are also better described.
Metabolism. As an example, MPN547(MP295) was previously annotated as a homolog of MG369, which in the recent update of M.genitalium (December 1999) is still given as a conserved hypothetical protein. Detailed sequence analysis (see Materials and Methods) shows, for example, similarity to experimentally characterized dihydroxyacetone kinases from different bacteria and fungi in PSI-BLAST searches of the N-terminal 300 amino acids with significant E values below 10–7, also apparent from the latest COG table (35). The dihydroxyacetone kinase domain could yield ATP by transforming dihydroxyacetone phosphate and ADP into dihydroxyacetone and ATP. The predicted activity can be metabolically connected to phospholipid metabolism in M.pneumoniae and the necessary supply of dihydroxyacetone phosphate via MPN051(MP103) (glycerol 3-phosphate dehydrogenase reading frame, confirmed in re-annotation). The remaining sequence of MPN547(MP295) (total length 558 amino acids) may regulate or add further to this predicted enzyme activity.
Multiple substrates. There seem to be M.pneumoniae enzymes which can interact with several substrates, for example MPN158(MP674). As already indicated in the first annotation and in SwissProt (P22990), given its clear and high sequence similarity over the full length to biochemically well-characterized enzymes from Gram-positive homologs, the encoded enzyme can act as both a riboflavin kinase and an FMN adenylyltransferase using one substrate binding site (according to biochemical data for the Corynebacterium ammoniagenes enzyme; 40). However, considering that MPN047(MP107) is now re-annotated as nicotinate phosphoribosyltransferase (by sequence similarity, including biochemically well-characterized family members) and that MPN562(MP280) is and was annotated as an NH3-dependent NAD synthase, it is tempting to speculate that MPN158(MP674) also has nicotinate-nucleotide adenylyltransferase activity besides FMN adenylyltransferase activity. This capability would complete the synthesis of NAD from imported nicotinic acid, a pathway so far incomplete. The reaction mechanism and substrate seem to be sufficiently similar to suggest this, but, as further experimental evidence is lacking, we have kept the original annotation and suggest this further activity of the reading frame product only as a comment.
Apparent operons. The phosphate uptake system was more completely annotated. It is composed of MPN611(MP231) (new assignment, similar to phosphate-binding protein PTS, for example from E.coli, previously annotated as ‘conserved with MG412’), MPN610(MP232), MPN609(MP233) and MPN608(MP234). It is probably regulated by MPN397(MP442) (ppGpp 3′-pyrophosphorylase).
A ribulose uptake operon is apparent. Small operons were known previously for fructose (MPN078(MP077) and MPN079(MP076)) and mannitol (MPN651(MP191)–MPN653(MP189)). Ribulose is now found to be transported (MPN496(MP346), MPN494(MP347)) and channeled via d-arabinose 6-hexulose 3-phosphate synthase (MPN493(MP348)) and d-arabinose 6-hexulose 3-phosphate isomerase MPN492(MP349) into fructose 6-phosphate and glycolysis. Of these proteins, MPN496 and MPN493 were not functionally annotated before and MPN494 had been annotated as a hypothetical phosphotransferase. These new functional assignments also became apparent on integrating data from SwissProt annotations with further direct experimental data published and realized for homologous proteins. Furthermore, we have now added a description of and data on the pentitol BC subunit of the ribulose transporter (MPN495(MP346.1); see Table 1 and data in Fig. 1B), not annotated before.
Lessons for genome annotation
The re-annotation presented here is only our current interpretation of the genome sequence. There remains a substantial fraction of proteins unassigned (230 of 688 or 33%) and even this prototype of a small or even minimal genome (34,41) is far from being completely understood. To reduce the level of errors, close cooperation, regular updates and deposition of the findings in databases such as SwissProt and GenBank is required. We support calls for concerted efforts in re-annotation and a consistent nomenclature (3,42,43).
Regular, well-documented further updates of genome sequences will yield a considerable gain in information. We have focused mainly on the molecular functions of the proteins because these can be directly deduced from the protein sequence and/or simple experimental tests. Furthermore, we approached the re-annotation in a more formal way, including semantics, re-annotation categories and inclusion of programs and reasoning to allow reproducibility. New experimental data were integrated, including data from this study on mRNA expression and proteome analysis. In this way, three new RNAs and 12 new proteins were identified, protein lengths (24 cases) and RNA positions (25 cases) were corrected and several new operons predicted. On the next level of re-annotation, the increase of 31% in functional assignments obtained (from 349 to 458) was not only quantitative but improved our overall knowledge regarding pathogenicity factors, secretion, transporters and metabolism of M.pneumoniae.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Amos Bairoch for information on recent SwissProt annotation efforts on Mycoplasma and all our collaboration partners at the European Multimedia Laboratory (Heidelberg) and Lion Biosciences AG (Heidelberg), as well as Richard Roberts and Janos Posfai (New England Biolabs, Beverly, MA), Kerstin Lühn and Jürgen Brosius (University of Münster), Mitsuhiro Itaya (Mitsubishi Kasei Institute, Tokyo), Warren C. Lathe (EMBL), Ake Wieslander (Stockholm University), Robert Turner (Scripps Institute, La Jolla, CA) and Jonathan Reizer (University of California at San Diego, La Jolla, CA) for discussions, comments and making unpublished material available to us. This research was supported by the BMBF (grants 0312212 and ‘Genominfo’), DFG (SFB544/B2, He780/10-1 and SPP ‘Informatikmethoden zur Analyse grosser genomischer Datenmengen’), the Graduiertenkolleg ‘Pathogene Mikroorganismen: Molekulare Mechanismen und Genome’ and by the Fonds der Chemischen Industrie.
DDBJ/EMBL/GenBank accession no. U00089
REFERENCES
- 1.Himmelreich R., Hilbert,H., Plagens,H., Pirkl,E., Li,B.-C. and Herrmann,R. (1996) Nucleic Acids Res., 24, 4420–4449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Himmelreich R., Plagens,H., Hilbert,H., Reiner,B. and Herrmann,R. (1997) Nucleic Acids Res., 25, 701–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brenner S.E. (1999) Trends Genet., 15, 132–133. [DOI] [PubMed] [Google Scholar]
- 4.Koonin E.V., Mushegian,A.R. and Rudd,K.E. (1996) Curr. Biol., 6, 404–416 [DOI] [PubMed] [Google Scholar]
- 5.Ouzounis C., Casari,G., Valencia,A. and Sander,C. (1996) Mol. Microbiol., 20, 898–900. [DOI] [PubMed] [Google Scholar]
- 6.Fraser C.M., Gocayne,J.D., White,O. et al. (1995) Science, 270, 397–403. [DOI] [PubMed] [Google Scholar]
- 7.Pennisi E. (1999) Science, 286, 447–450. [DOI] [PubMed] [Google Scholar]
- 8.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koonin E.V., Mushegian,A.R. and Bork,P. (1996) Trends Genet., 12, 334–336. [PubMed] [Google Scholar]
- 10.Huynen M.A. and Bork,P. (1998) Proc. Natl Acad. Sci. USA, 95, 5849–5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bork P., Dandekar,T., Diaz-Lazcoz,Y., Eisenhaber,F., Huynen,M. and Yuan,Y. (1998) J. Mol. Biol., 283, 707–725. [DOI] [PubMed] [Google Scholar]
- 12.Bork P. and Gibson,T.J. (1996) Methods Enzymol., 266, 162–184. [DOI] [PubMed] [Google Scholar]
- 13.Huynen M., Doerks,T., Eisenhaber,F., Orengo,C., Sunyaev,S., Yuan,Y. and Bork,P. (1998) J. Mol. Biol., 280, 323–326. [DOI] [PubMed] [Google Scholar]
- 14.Dandekar T., Schuster,S., Snel,B., Huynen,M. and Bork,P. (1999) Biochem. J., 343, 115–124. [PMC free article] [PubMed] [Google Scholar]
- 15.Schultz J., Copley,R.R., Doerks,T., Ponting,C.P. and Bork,P. (2000) Nucleic Acids Res., 28, 231–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Proft T. and Herrmann,R. (1994) Mol. Microbiol., 13, 337–348. [DOI] [PubMed] [Google Scholar]
- 17.Görg A., Obermaier,C., Boguth,G., Harder,A., Scheibe,B., Wildgruber,R. and Weiss,W. (2000) Electrophoresis, 21, 1037–1053. [DOI] [PubMed] [Google Scholar]
- 18.Eng J.K., McCormack,A.L. and Yates,J.R. (1994) J. Am. Soc. Mass Spectrom., 5, 976–989. [DOI] [PubMed] [Google Scholar]
- 19.Southern E.M. (1996) Trends Genet., 12, 110–115. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 21.Brown J.W. (1999) Nucleic Acids Res., 27, 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Rijk P., Robbrecht,E., de Hoog,S., Caers,A., Van de Peer,Y. and De Wachter,R. (1999) Nucleic Acids Res., 27, 174–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szymanski M., Barciszewska,M.Z., Barciszewski,J. and Erdmann,V.A (1999) Nucleic Acids Res., 27, 158–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van de Peer Y., Robbrecht,E., de Hoog,S., Caers,A., De Rijk,P. and De Wachter,R. (1999) Nucleic Acids Res., 27, 179–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williams K.P. (1999) Nucleic Acids Res., 27, 165–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guigo R. (1997) Comput. Chem., 21, 215–222. [DOI] [PubMed] [Google Scholar]
- 27.Dandekar T., Beyer,K., Bork,P., Kenealy,M.R., Pantopoulos,K., Hentze,M., Sonntag-Buck,V., Flouriot,G., Gannon,F. and Schreiber,S. (1998) Bioinformatics, 14, 271–278. [DOI] [PubMed] [Google Scholar]
- 28.Göhlmann H.W.H, Weiner,J., Schön,A. and Herrmann,R. (2000) J. Bacteriol., 182, 3281–3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reizer J., Paulsen,I.T., Reizer,A., Titgemeyer,F., Saier,M.H.Jr (1996) Microb. Comp. Genomics, 1, 151–164. [DOI] [PubMed] [Google Scholar]
- 30.Pyrowolakis G., Hoffmann,D. and Herrmann,R. (1998) J. Biol. Chem., 273, 24792–24796. [DOI] [PubMed] [Google Scholar]
- 31.Curnow A.W., Hong,K.W., Yuan,R., Kim,S., Martins,O. and Winkler,W. (1997) Proc. Natl Acad. Sci. USA, 94, 11819–11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fukuda Y.,Washio,T. and Tomita,M. (1999) Nucleic Acids Res., 27, 1847–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aravind L. and Koonin,E.V. (1998) Trends Biochem.Sci., 23, 17–19. [DOI] [PubMed] [Google Scholar]
- 34.Hutchison C.A., Peterson,S.N., Gill,S.R., Cline,R.T., White,O., Fraser,C.M., Smith,H.O. and Venter,J.C. (1999) Science, 286, 2165–2169. [DOI] [PubMed] [Google Scholar]
- 35.Tatusov R.L., Galperin,M.Y., Natale,D.A. and Koonin,E.V. (2000) Nucleic Acids Res., 28, 33–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Müller A., MacCallum,R.M. and Sternberg,M.J. (1999) J. Mol. Biol., 293, 1257–1271. [DOI] [PubMed] [Google Scholar]
- 37.Schatz G. and Dobberstein,B. (1996) Science, 271, 1519–1526. [DOI] [PubMed] [Google Scholar]
- 38.Bellgard M.I. and Gojobori,T. (1999) Gene, 238, 33–37. [DOI] [PubMed] [Google Scholar]
- 39.Van Wely K.H., Swaving,J., Brockhulzen,C.F., Rose,M., Quax,W.J. and Driessen,A.J. (1999) J. Bacteriol., 181, 1786–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Efimov I., Kuusk,V., Zhang,X. and McIntire,W.S. (1998) Biochemistry, 37, 9716–9723. [DOI] [PubMed] [Google Scholar]
- 41.Mushegain A.R. and Koonin,E.V. (1996) Proc. Natl Acad. Sci. USA, 93, 10268–10273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kyrpides N.C. and Ouzounis,C.A. (1998) Science, 281, 1457. [DOI] [PubMed] [Google Scholar]
- 43.Kyrpides N.C. and Ouzounis,C.A. (1999) Mol. Microbiol., 32, 886–887. [DOI] [PubMed] [Google Scholar]
- 44.Dybvig K., Sitaraman,R. and French,C.T. (1998) Proc. Natl Acad. Sci. USA, 95, 13923–13928. [DOI] [PMC free article] [PubMed] [Google Scholar]