Abstract
BRCA1 expression is repressed by aberrant cytosine methylation in sporadic breast cancer. We hypothesized that aberrant cytosine methylation of the BRCA1 promoter was associated with the transcriptionally repressive effects of histone hypoacetylation and chromatin condensation. To address this question, we developed an in vitro model of study using normal cells and sporadic breast cancer cells with known levels of BRCA1 transcript to produce a 1.4 kb 5-methylcytosine map of the BRCA1 5′ CpG island. While all cell types were densely methylated upstream of –728 relative to BRCA1 transcription start, all normal and BRCA1 expressing cells were non-methylated downstream of –728 suggesting that this region contains the functional BRCA1 5′ regulatory region. In contrast, the non-BRCA1 expressing UACC3199 cells were completely methylated at all 75 CpGs. Chromatin immunoprecipitations showed that the UACC3199 cells were hypoacetylated at both histones H3 and H4 in the BRCA1 promoter compared to non-methylated BRCA1 expressing cells. The chromatin of the methylated UACC3199 BRCA1 promoter was inaccessible to DNA–protein interactions. These data indicate that the epigenetic effects of aberrant cytosine methylation, histone hypoacetylation and chromatin condensation act together in a discrete region of the BRCA1 5′ CpG island to repress BRCA1 transcription in sporadic breast cancer.
INTRODUCTION
CpG islands are GC-rich regions of DNA that have a higher than expected frequency of CpG dinucleotides. These islands are usually located at the 5′ end of genes and are associated with transcriptional promoters (1). Cytosines of CpG islands are non-methylated in normal tissues regardless of transcription status, whereas aberrant cytosine methylation of the 5′ CpG islands of genes is commonly associated with their transcriptional repression (2).
The aberrant cytosine methylation of CpG islands is associated with the alteration of chromatin structure to a protein-inaccessible state, which appears to participate in the transcriptional repression of the associated gene (3–5). A protein-inaccessible chromatin structure is also directly linked to the acetylation status of core histones in the nucleosomes of gene promoters (6,7). Hypoacetylated histones are associated with transcriptionally inert regions of heterochromatin, whereas acetylated histones are associated with transcriptionally active regions of euchromatin (8–10). Recent reports have identified a mechanistic pathway of epigenetic silencing by cytosine methylation, histone hypoacetylation and chromatin condensation suggesting that these mechanisms act together to inactivate gene transcription (11,12).
In this study we investigated the mechanisms of epigenetic silencing of the breast cancer susceptibility gene BRCA1 in sporadic breast cancer. BRCA1 is a tumor suppressor gene whose expression is repressed in a large portion of sporadic breast cancer patients and is associated with a malignant phenotype (13–15). Recent reports indicate that aberrant cytosine methylation of BRCA1 occurred in two of six and two of seven sporadic breast cancer specimens, respectively (16,17). In addition to these studies, we used high resolution bisulfite sequencing to identify aberrant cytosine methylation of the BRCA1 5′ CpG island in three of 21 sporadic breast cancer specimens. These three specimens also expressed the lowest levels of BRCA1 transcript by RT–PCR analysis (18). We hypothesized that the aberrant cytosine methylation of the BRCA1 promoter is associated with histone hypoacetylation, chromatin condensation and transcriptional repression of BRCA1 in sporadic breast cancer.
To test this hypothesis, we developed an in vitro model of study using normal cells and sporadic breast cancer cells with known levels of BRCA1 transcript to produce a 1.4 kb 5-methylcytosine map of the BRCA1 5′ CpG island. High resolution bisulfite sequence analysis showed that the non-methylated CpG island domain extends downstream of –728 relative to transcription start in normal and BRCA1 expressing cells. In contrast, the region upstream of –728, although still CpG rich, was methylated in both normal cells and breast cancer cell lines. The non-methylated domain contains maximal BRCA1 promoter activity and is the target region for aberrant cytosine methylation in breast cancer cells (17–20). We identified one BRCA1-negative sporadic breast cancer cell line that was aberrantly methylated at all 75 CpG dinucleotides analyzed. Chromatin immunoprecipitation assays revealed that the aberrantly methylated BRCA1 promoter of the BRCA1-negative breast cancer cells is associated with hypoacetylated histones H3 and H4 compared to the non-methylated BRCA1 promoter of normal and tumorigenic breast cells that express BRCA1. In addition, the aberrant cytosine methylation and histone hypoacetylation of the BRCA1 promoter coincides with a protein-inaccessible chromatin structure and transcriptional repression of BRCA1. These data indicate that the epigenetic effects of aberrant cytosine methylation, histone hypoacetylation and chromatin condensation act together in the discrete region of the BRCA1 promoter to repress BRCA1 transcription in sporadic breast cancer.
MATERIALS AND METHODS
Cell culture
MCF7 cells were obtained from the American Type Culture Collection (Rockville, MD) and cultured in RPMI 1640 supplemented with 5% fetal bovine serum (Gibco BRL, Grand Island, NY), 100 U/ml penicillin (Gibco BRL) and 1% glutamate (Gibco BRL) in a 5% CO2 atmosphere at 37°C. UACC3199 is an early passage sporadic breast cancer cell line derived from an infiltrating ductal carcinoma isolated from axillary lymph nodes. In this study, UACC3199 was passaged not more than 20 times and maintains the original genotype and phenotype of the primary tumor (21,22). Normal human mammary epithelial cells (HMEC) were purchased from Clonetics (San Diego, CA) and grown according to the manufacturer’s instructions. The peripheral blood lymphocytes (PBL) and human foreskin fibroblasts (HFF) are primary cells.
High resolution bisulfite sequencing of the BRCA1 5′ region
Genomic DNA was modified with sodium bisulfite as previously described (19). A 1.4 kb sequence of the BRCA1 5′ flanking region was amplified in two separate nested PCR reactions. A 656 bp nested PCR product from –591 to +66 relative to the BRCA1 transcription start site was amplified from the bisulfite-modified DNA with bisulfite specific primers derived from the reported BRCA1 sequence (GenBank accession no. U37574). The primer sets used were as follows:
primer 1 (nt 895–916), 5′-GGGGTTGGATGGGAATTGTAG-3′;
primer 2 (nt 1688–1792), 5′-CTCTACTACCTTTACCCAAAAACA-3′;
primer 3 (nt 989–1013), 5′-GTTTATAATTGTTGATAAGTATAAG-3′;
primer 4 (nt 1626–1646), 5′-AAAACCCCACAACCTATCCC-3′.
An 802 bp fragment of the BRCA1 upstream region from –1369 to –567 was also amplified from the bisulfite-modified DNA by nested PCR using the following primer sets:
primer 1 (nt 177–200), 5′-TTAGTTTAGAGAGGGGTTTTTATA-3′;
primer 2 (nt 1094–1119), 5′-CCACAATATTCCTTAAAAACTATAAT-3′;
primer 3 (nt 211–232), 5′-GGGTTGAAGGGTTTTTTTAGTA-3′;
primer 4 (nt 989–1013), 5′-CTTATACTTATCAACAATTATAAAC-3′.
Primers 1 and 2 were used in the first round of amplification and primers 3 and 4 were used in the second round of amplification under the following conditions: 95°C for 1 min followed by 35 cycles of 92°C for 1 min, 56°C for 3 min, 72°C for 1 min; ending with a final extension of 72°C for 5 min and a quick chill to 4°C. The resultant PCR products were cloned into pGEM-T-Easy TA (Promega, Madison, WI) according to the manufacturer’s instructions. Positive recombinants were isolated using the Qiaprep Spin Plasmid Miniprep Kit (Qiagen, Valencia, CA) and sequenced on an ABI automated DNA sequencer. Ten recombinants from each cell type were analyzed for the BRCA1 regulatory region. For the BRCA1 upstream region, ten recombinants were analyzed for PBL and HFF, eight recombinants for HMEC, and six recombinants for MCF7 and UACC3199. All non-methylated cytosines were successfully converted in the bisulfite reaction in all of the recombinants analyzed (Fig. S2).
Acetyl-histone H3 and H4 chromatin immunoprecipitations and PCR amplification of the BRCA1 and GAPDH promoters
Chromatin immunoprecipitations using the acetyl-histone H3 and H4 antibodies were performed according to the manufacturer’s instructions (Upstate Biotech, Lake Placid, NY). Cells were rinsed in 1× PBS and treated with 1% formaldehyde for 10 min at 37°C to form DNA–protein cross-links. The cells were rinsed in ice cold 1× PBS containing protease inhibitors (1 mM PMSF, 1 µg/ml aprotinin, 1 µg/ml pepstatin A), scraped and collected by centrifugation at 4°C. Cells were resuspended in an SDS lysis buffer containing protease inhibitors and incubated on ice for 10 min. The DNA–protein complexes were sonicated to lengths between 200 and 1000 bp as determined by gel electrophoresis (Fig. S3), isolated by centrifugation, and diluted with buffer containing protease inhibitors. The sample was divided equally into thirds: +Ab, –Ab and control sample. The +Ab and –Ab samples were pre-cleared with a Salmon Sperm DNA/Protein A Agarose Slurry. Following pre-clearing, the +Ab sample was exposed to the acetyl-histone H3 or H4 antibody and all of the samples were incubated overnight at 4°C with rotation. The chromatin–antibody complexes were collected using the Salmon Sperm DNA/Protein A Agarose Slurry and then sequentially washed with the manufacturer’s low salt buffer, high salt buffer, LiCl buffer, and twice with Tris–EDTA. The chromatin–antibody complexes were eluted and the DNA–protein cross-links were reversed with 5 M NaCl at 65°C for 4 h. All samples were treated with proteinase K, and the acetyl-histone H3 or H4 enriched fraction of genomic DNA was recovered by phenol/chloroform extractions and ethanol precipitations and quantitated using pico green (Molecular Probes, Eugene, OR). PCR amplification of the BRCA1 promoter (GenBank accession no. U37574) was performed using the following primers:
primer 1 (nt 1349–1369), 5′-GGCAGGCACTTTATGGCAAAC-3′,
primer 2 (nt 1757–1778), 5′-TTCGGAAATCCACTCTCCCACG-3′.
PCR amplification of the GAPDH promoter (GenBank accession no. J04038) was performed using the following primers:
primer 1 (nt 827–851), 5′-TAGTGTCCTGCTGCCCACAGTCCAG-3′,
primer 2 (nt 1168–1187), 5′-GGCGACGCAAAAGAAGATGC-3′.
Both PCRs were performed under the following conditions: 95°C for 4 min followed by 40 cycles of 95°C for 1 min, 68°C for 1 min and 72°C for 2 min and ending with a final extension of 72°C for 5 min. PCR products were size fractionated on a 3% TBE agarose gel, stained with ethidium bromide, and visualized on the Eagle Eye II Still Video System (Stratagene, La Jolla, CA).
Chromatin accessibility assays of the BRCA1 5′ region
Chromatin accessibility assays were performed as previously described (5) with minor modifications. Twenty million cells were washed twice with 1× PBS, gently scraped and collected by centrifugation. Nuclei were extracted by resuspension of cells in ice cold 1× RSB (10 mM Tris–HCl pH 8, 3 mM MgCl2, 10 mM NaCl, 0.05% NP-40). The nuclei were collected by centrifugation, resuspended in appropriate 1× restriction endonuclease buffer, and divided into four aliquots of 200 µl/aliquot. Zero, 25, 75, or 225 U of either EcoRI or SstI (Gibco BRL) was added to the nuclei and incubated at 37°C for 15 min. Genomic DNA was isolated using the QIAamp Tissue Kit (Qiagen) and 7.5 µg of this DNA was digested with 25 U of BamHI (Gibco BRL). Following phenol/chloroform extractions, the DNA was size-fractionated on a 1% TBE agarose gel and capillary transferred onto a 0.45 µm pore size Nytran Plus membrane (Schleicher and Schuell, Keene, NH). The membranes were hybridized with a probe generated from –564 to –204 of the BRCA1 5′ region and 32P-labeled using the random primer method. Membranes were washed once in 2× SSC–0.5% SDS for 30 min at room temperature followed by two washes at 62°C for 30 min in a 0.1% SSC–0.5% SDS solution. Results were visualized by autoradiography.
RESULTS
5-methylcytosine map of the BRCA1 5′ region
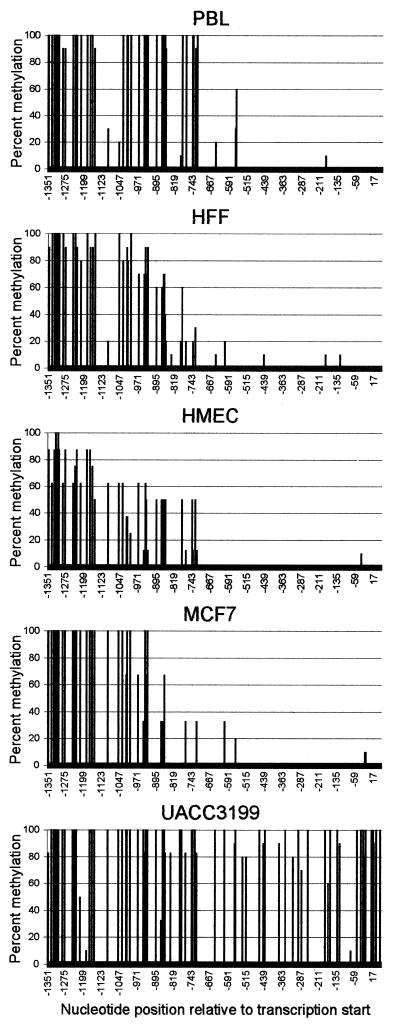
We developed an in vitro model of study using normal cells and sporadic breast cancer cells with known levels of BRCA1 transcript to produce a 1.4 kb 5-methylcytosine map of the BRCA1 5′ CpG island. The cell types used to generate this 5-methylcytosine map were the BRCA1-positive normal HMEC, BRCA1-negative normal human PBL, normal HFF and the BRCA1-positive MCF7 and BRCA1-negative UACC3199 human sporadic breast cancer cell lines (19). The presence of 5-methylcytosine in the 1.4 kb sequence of the BRCA1 5′ region was determined by bisulfite sequencing of two separate PCR amplicons. The first nested PCR amplification contained 30 CpG dinucleotides and extended from –591 to +66 relative to the BRCA1 transcription start site. The second nested PCR amplification was the BRCA1 5′ upstream region which contained 45 CpG dinucleotides and extended from –1369 to –567. Clones from each cell type were analyzed to create a high resolution 5-methylcytosine map of the BRCA1 5′ region as illustrated in Figure 1. The sequence information obtained for each cell type was compared to the known BRCA1 sequence to determine the frequency of 5-methylcytosine for each CpG dinucleotide.
Figure 1.
High resolution 5-methylcytosine map of the BRCA1 5′ region. Bisulfite modified DNA from PBL, HFF, HMEC and the sporadic breast cancer cell lines MCF7 and UACC3199 were PCR amplified, cloned and sequenced. Clones from each cell type were analyzed to obtain a percent methylation of the 75 CpG dinucleotides in the BRCA1 5′ region located on the y-axis. The x-axis represents the nucleotide position relative to the BRCA1 transcriptional start site (GenBank accession no. U37574).
The five cell types analyzed were densely methylated in the 5′ region upstream of –728. In contrast, 5-methylcytosine was absent in all normal cells and the BRCA1-positive MCF7 cells downstream of –728 suggesting that this non-methylated domain is the functional 5′ regulatory region of the BRCA1 CpG island. Unlike the other cell types, the BRCA1-negative UACC3199 cells were completely methylated downstream of –728. The aberrant cytosine methylation of the BRCA1 promoter in UACC3199 is associated with an observed 10-fold decrease in BRCA1 transcript compared to HMEC (19). These results indicate that aberrant cytosine methylation of the BRCA1 promoter is associated with transcriptional repression of BRCA1.
Aberrant cytosine methylation of the BRCA1 promoter is associated with histone H3 and H4 hypoacetylation
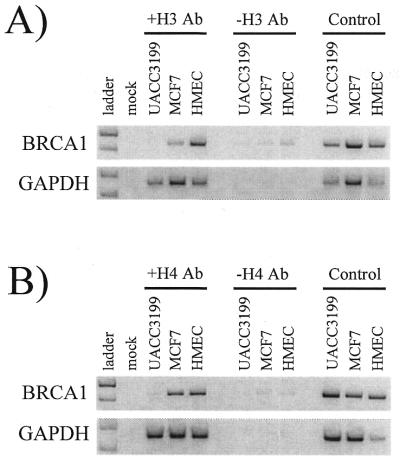
To determine if aberrant cytosine methylation of the BRCA1 promoter was associated with histone hypoacetylation, chromatin immunoprecipitations of UACC3199, MCF7 and HMEC were performed using acetyl-histone H3 and H4 antibodies. Following chromatin immunoprecipitations, the acetyl-histone H3 and H4 enriched fraction of genomic DNA was purified and isolated. The enriched DNA was analyzed by PCR for the presence of the BRCA1 promoter (from –232 to +198), as well as the constitutively active GAPDH promoter (from –311 to +50).
Figure 2A shows the results of one of the acetyl-histone H3 immunoprecipitation experiments. The acetyl-histone H3 enriched genomic DNA from the methylated BRCA1-negative UACC3199 cells failed to amplify the 429 bp BRCA1 PCR product compared to the non-methylated BRCA1-positive HMEC and MCF7 cells as seen in the +H3 Ab lanes. The BRCA1 PCR product was not amplified in the no acetyl-histone H3 antibody control (–H3 Ab lanes), but was amplified in the genomic DNA positive control in all cell types analyzed. As expected, each cell line had acetylated histone H3 associated with the constitutively expressed GAPDH CpG island promoter.
Figure 2.
Acetylation status of histones H3 and H4 in the BRCA1 and GAPDH promoters in UACC3199, MCF7 and HMEC. Chromatin immunoprecipitations using acetyl-histone H3 and H4 antibodies were performed on UACC3199, MCF7 and HMEC. Following isolation of the acetyl-histone H3 and H4 enriched fraction of genomic DNA, the BRCA1 or GAPDH promoters were PCR amplified. Presence of a PCR product indicates acetylation of the immunoprecipitated histone. Acetylation status of histone H3 (A) or histone H4 (B) for the BRCA1 (top) and GAPDH (bottom) promoters in UACC3199, MCF7 and HMEC is shown. The different cell types analyzed are shown across the top and are grouped according to their incubation with acetyl-histone H3 or H4 antibody (+H3 Ab or +H4 Ab), no acetyl-histone antibody (–H3 Ab or –H4 Ab), or control sample. These experiments were performed three times each with similar results.
Similar results were obtained from the acetyl-histone H4 immunoprecipitation experiments. Figure 2B shows that the acetyl-histone H4 enriched genomic DNA from the methylated BRCA1-negative UACC3199 cells failed to amplify the 429 bp BRCA1 PCR product compared to the non-methylated BRCA1-positive HMEC and MCF7 cells as seen in the +H4 Ab lanes. The BRCA1 PCR product was not amplified in the no acetyl-histone H4 antibody control (–H4 Ab lanes), but was amplified in the genomic DNA positive control in all cell types analyzed. Each cell line had acetylated histone H4 associated with the constitutively expressed GAPDH CpG island promoter.
The appearance of faint bands in the –Ab lanes of all the BRCA1 PCR samples and different band intensities of the positive control samples underscores that, based on the limitations of the assay, these experiments are purely qualitative in nature and that degrees of (hypo)acetylation of each histone cannot be quantitated. However, Figure 2 does show that the methylated, BRCA1-negative UACC3199 cells are hypoacetylated at histones H3 and H4 compared to the non-methylated, BRCA1-positive HMEC and MCF7 cells. These data indicate that active transcription of BRCA1 and GAPDH coincides with a non-methylated and histone acetylated promoter. In contrast, the aberrant cytosine methylation and histone hypoacetylation of the UACC3199 BRCA1 promoter is associated with the observed transcriptional repression of BRCA1.
Aberrant cytosine methylation and histone hypoacetylation prevent accessibility of proteins to the BRCA1 promoter
Chromatin accessibility assays were performed to determine if the aberrant cytosine methylation and histone H3 and H4 hypoacetylation of the BRCA1 promoter were associated with the remodeling of chromatin to a transcriptionally repressive state. We investigated two regions of the BRCA1 5′ region for protein accessibility: the methylated upstream region and the BRCA1 promoter. Intact nuclei were isolated from the non-methylated, histone acetylated, BRCA1-positive HMEC and MCF7 cells, and the methylated, histone hypoacetylated, BRCA1-negative UACC3199 cells. The nuclei were subjected to an in vivo restriction endonuclease digestion by EcoRI or SstI. Following the in vivo digestion and isolation of genomic DNA, an in vitro restriction endonuclease digestion was performed with BamHI to release DNA fragments of predictable sizes for Southern analysis.
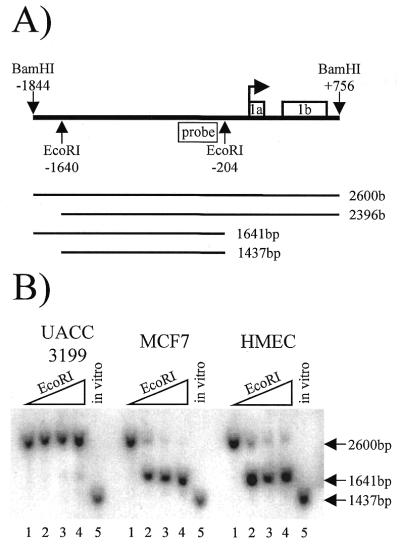
The BRCA1 5′ region has EcoRI restriction sites in both the methylated 5′ region at –1640 and in the BRCA1 promoter at –204 (Fig. 3A). Nuclei from HMEC, MCF7 and UACC3199 were exposed to 0, 25, 75 or 225 U of EcoRI for 15 min. Following isolation of genomic DNA, the samples were digested in vitro using BamHI, which cleaves at –1844 and +756 to yield a 2600 bp fragment. A radiolabeled probe was designed between the EcoRI cut sites such that a maximum of four possible products could be visualized by Southern analysis, as illustrated in Figure 3A. Inaccessibility of EcoRI at both restriction sites yields a 2600 bp fragment, which is represented by the no EcoRI control located in the first lane of each cell type. Accessibility of EcoRI in the upstream region only would result in a 2396 bp fragment, whereas accessibility of EcoRI in the promoter only would result in a 1641 bp fragment. Accessibility at both EcoRI sites would yield a 1437 bp fragment, as represented by the in vitro positive control shown in the fifth lane of each cell type.
Figure 3.
EcoRI chromatin accessibility assay of the BRCA1 5′ region. (A) A schematic showing the in vivo EcoRI and in vitro BamHI restriction sites relative to the BRCA1 transcription start site (bent arrow), the BRCA1 probe, exons 1a and 1b, and the four possible cleavage products and their predicted sizes. (B) A Southern blot that shows each cell type and the resultant in vivo EcoRI digest products. The lanes, from left to right, are 0, 25, 75 and 225 U of EcoRI; the in vitro control EcoRI digest is shown in the fifth lane for each cell type. This experiment was performed three times with similar results.
Figure 3B shows the results of one of the EcoRI chromatin accessibility experiments. The EcoRI site in the BRCA1 promoter was accessible to enzymatic cleavage in the non-methylated, histone acetylated, BRCA1-positive MCF7 and HMEC cells with as little as 25 U of EcoRI as illustrated by the appearance of a 1641 bp fragment in the second lane of these samples. In contrast, the BRCA1 promoter was inaccessible to EcoRI cleavage in the methylated, histone hypoacetylated, BRCA1-negative UACC3199 cells at all concentrations of EcoRI as demonstrated by the absence of the 1641 bp fragment or 1437 bp fragment in the second, third and forth lanes. The EcoRI site located at –1640 in the methylated upstream region flanking the BRCA1 5′ regulatory region was inaccessible to all concentrations of EcoRI in UACC3199, MCF7 and HMEC, as evidenced by the lack of either the 2396 or the 1437 bp fragment.
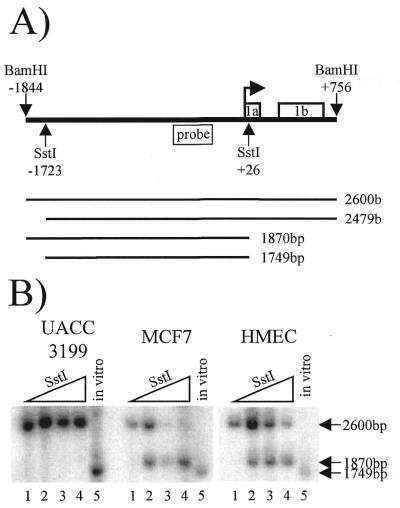
A similar chromatin accessibility assay was performed using the SstI restriction endonuclease as the in vivo restriction endonuclease (Fig. 4). The SstI restriction sites reside at –1723 in the methylated upstream region and at +26 in exon 1a. Using the same radiolabeled probe as the EcoRI experiments, Southern analysis yields four possible products as illustrated in Figure 4A. Inaccessibility of SstI at both restriction sites yields a 2600 bp fragment, which is represented by the no SstI control located in the first lane of each cell type (Fig. 4B). Accessibility of SstI in the methylated upstream region only would result in a 2479 bp fragment, whereas, accessibility of SstI in exon 1a only would result in a 1870 bp fragment. Accessibility at both SstI sites would yield a 1749 bp fragment, as represented by the in vitro positive control shown in the fifth lane of each cell type.
Figure 4.
SstI chromatin accessibility assay of the BRCA1 5′ region. (A) A schematic showing the in vivo SstI and in vitro BamHI restriction sites relative to the BRCA1 transcription start site (bent arrow), the BRCA1 probe, exons 1a and 1b, and the four possible in vivo SstI cleavage products and their predicted sizes. (B) A Southern blot that shows each cell type and the resultant in vivo SstI digest products. The lanes, from left to right, are 0, 25, 75 and 225 U of SstI; the in vitro control SstI digest is shown in the fifth lane of each cell type. This experiment was performed three times with similar results.
Figure 4B shows the results of one of the SstI chromatin accessibility experiments. The SstI site located near the BRCA1 transcription start site was accessible to as little as 25 U of SstI in the non-methylated, histone acetylated, BRCA1-positive MCF7 and HMEC cells as illustrated by the appearance of the 1870 bp fragment in the second lane of each of these samples. In contrast, the methylated, histone hypoacetylated, BRCA1-negative UACC3199 cells were inaccessible to all concentrations of SstI as demonstrated by the absence of the 1870 bp fragment or 1749 bp fragment in the second, third and fourth lanes. The SstI site located at –1723 in the methylated upstream region flanking the BRCA1 5′ regulatory region was inaccessible to all concentrations of SstI in UACC3199, MCF7 and HMEC, as evidenced by the lack of either the 2479 bp fragment or the 1749 bp fragment.
These data indicate that chromatin condensation of the BRCA1 promoter coincides with the aberrant cytosine methylation and histone hypoacetylation of this discrete region of DNA, and the transcriptional inactivation of BRCA1. A summary of these results is shown in Figure 5.
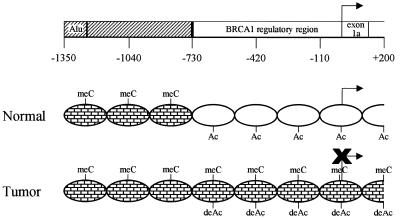
Figure 5.
Transcriptional repression of BRCA1 by aberrant cytosine methylation, histone hypoacetylation and chromatin condensation of the BRCA1 5′ regulatory region in sporadic breast cancer. Top, a schematic of the BRCA1 5′ flanking region. The BRCA1 upstream region (hashed box) contains a repetitive element (Alu) and extends to the beginning of the BRCA1 5′ regulatory region at –728 (black vertical line). The numbers represent the nucleotide position relative to the BRCA1 transcription start site (bent arrow). BRCA1-positive normal breast cells (middle) are non-methylated, contain acetylated histones H3 and H4 (Ac) and have a protein accessible chromatin conformation (open ovals) in the BRCA1 5′ regulatory region. In contrast, the BRCA1-negative tumor cells (bottom) have an aberrantly methylated BRCA15′ regulatory region (meC), hypoacetylated histones H3 and H4 (deAc), and condensed chromatin (bricked ovals) coincident with BRCA1 repression. Both normal and tumor cells have a methylated (meC) and chromatin condensed (bricked ovals) BRCA1 upstream region.
DISCUSSION
In this study we used an in vitro sporadic breast cancer cell line, UACC3199, with methylation-associated BRCA1 repression to analyze histone H3 and H4 acetylation status as well as chromatin structure of the BRCA1 promoter. Our findings show that aberrant cytosine methylation of the BRCA1 promoter coincides with the transcriptionally repressive effects of histone hypoacetylation and chromatin condensation. The aberrant cytosine methylation of the BRCA1 promoter observed in UACC3199 is a selective event as the 5′ CpG islands of the p15 and deoxycytosine kinase genes were found to be non-methylated (Fig. S1). The methylated BRCA1 promoter of the BRCA1-negative UACC3199 cells contain hypoacetylated histones H3 and H4 compared to the non-methylated, BRCA1-positive HMEC and MCF7 cells indicating that histone hypoacetylation occurs only in the aberrantly methylated BRCA1 promoter. In contrast to the BRCA1-positive HMEC and MCF7 cells, the methylated and hypoacetylated BRCA1 promoter of UACC3199 contains condensed chromatin that coincides with the observed transcriptional repression of BRCA1.
Taken together, our data indicates that aberrant cytosine methylation, histone hypoacetylation and chromatin condensation act together in the discrete region of the BRCA1 promoter to inactivate BRCA1 transcription in sporadic breast cancer. This is the first study to show the direct association of these three epigenetic events in the inactivation of a human tumor suppressor gene. Our findings are consistent with recent reports of the imprinted human fragile X mental retardation gene, FMR1. The FMR1 gene is inactivated in fragile X cells by cytosine methylation, hypoacetylation of histones H3 and H4 and chromatin condensation in its 5′ regulatory region compared to normal cells (23,24). Similarly, in an in vitro system, a methylated herpes simplex virus thymidine kinase reporter is transcriptionally repressed in stably transfected L-cells and is associated with hypoacetylated histone H4 and DNase I insensitivity (25). These studies directly indicate that cytosine methylation, histone hypoacetylation and chromatin condensation act together in mammalian gene promoters to suppress transcription.
In contrast to the non-methylated BRCA1 promoter, the region upstream of –728 is densely methylated in all normal cell types analyzed regardless of BRCA1 expression. In addition, the chromatin accessibility assays show that this methylated region is inaccessible to in vivo endonuclease digestion. Although we attempted to determine the acetylation status of the 5′ methylated region, the presence of a repetitive element upstream of –1273 blunted our efforts to successfully analyze this region. These data indicate that the methylated region upstream of –728 is inaccessible to DNA–protein interactions and suggests that the non-methylated and accessible region downstream of –728 represents the functional 5′ regulatory region of the BRCA1 CpG island. This conclusion is reinforced by BRCA1 promoter studies which show that reporter constructs lacking the region upstream of –728 have significantly higher promoter activities compared to reporter constructs that contain the region upstream of –728 (19,20).
The 5-methylcytosine pattern of the BRCA1 CpG island observed in the normal cells is typical of other human tumor suppressor gene CpG islands such as E-cadherin and VHL (26). While the 5′ regulatory region of these genes are non-methylated in normal cells, the upstream region contains a methylated repetitive element. Recent studies suggest that methylated repetitive elements may function as cis-acting ‘de novo methylation centers’ (27,28). In cultured human fibroblasts overexpressing DNA methyltransferase, methylation spread from the methylated repetitive element into the 5′ regulatory regions of both the E-cadherin and VHL genes (26). The mouse aprt gene is protected from its 5′ de novo methylation center by the presence of an Sp1 site located in the 5′ regulatory region (29). Deletion of this Sp1 site results in the aberrant cytosine methylation of the aprt regulatory region and transcriptional repression of aprt (30). It is possible that, like aprt, the BRCA1 5′ regulatory region is protected from aberrant cytosine methylation by DNA-binding proteins located in the region around –728. It is interesting to note that there is an Sp1 consensus sequence located at –687 between the last methylated CpG of the upstream methylated domain and the first non-methylated CpG dinucleotide of the non-methylated regulatory region.
Based on these results and the current scientific literature, we propose the following temporal sequence of epigenetic repression of BRCA1 in sporadic breast cancer. The aberrant cytosine methylation of the BRCA1 5′ regulatory region is, most likely, the first epigenetic event. Previous studies have shown that a methylated reporter construct transfected into mammalian cells was able to transcribe the reporter gene for 8 h (31,32). After 8 h, however, the methylated construct became transcriptionally inert which coincided with alterations in the nucleosomal array and an inability of RNA polymerase to bind the regulatory region. These data indicate that methylation of the regulatory region is not directly responsible for transcriptional repression, rather, methylation leads to a transcriptionally repressive chromatin state.
The aberrant cytosine methylation of the BRCA1 5′ regulatory region may be followed by the binding of methylation specific methyl binding proteins (33–35). One of these methyl binding proteins, MeCP2, has been shown to associate with a transcriptional repressor complex that includes histone deacetylases (11,12). We speculate that the methylated BRCA1 5′ regulatory region recruits MeCP2, or an analogous methyl binding protein, and a repressor complex that is capable of deacetylating histones H3 and H4, as observed. In turn, the deacetylation of histones, probably in concert with other chromatin remodeling proteins, results in the observed chromatin condensation and transcriptional repression of BRCA1.
This study shows that aberrant cytosine methylation, histone hypoacetylation, and chromatin condensation act together in a discrete region of the BRCA1 5′ CpG island to inactivate BRCA1 transcription. Our data suggests that the aberrant cytosine methylation observed in other human tumor suppressor gene CpG island promoters coincide with alterations in the composition and structure of the associated chromatin to a transcriptionally repressive state. These epigenetic alterations may reflect a common set of events necessary for the inappropriate transcriptional inactivation of human tumor suppressor genes and the progression of cancer.
SUPPLEMENTARY MATERIAL
See Supplementary Material available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Elliot Epner and David Bearss for critical review of the manuscript, Kathy Massey-Brown of the Arizona Cancer Center Cell Culture Core Facility for her assistance with cell culture, Skip Vaught and his sequencing crew at the Laboratory of Systematics and Evolution, University of Arizona, for the automated DNA sequencing, Nick Holtan for his assistance with the upstream 5-methylcytosine mapping and Jonathan Dodge for his assistance with the 5-methylcytosine mapping of p15 and dCK. This work was supported by NCI 65662 and DAMD 17-98-18279 to B.W.F. and 3P30CA23074-19 to the Arizona Cancer Center.
REFERENCES
- 1.Gardiner-Garden M. and Frommer,M. (1987) J. Mol. Biol., 196, 261–282. [DOI] [PubMed] [Google Scholar]
- 2.Bird A. (1992) Cell, 70, 5–8. [DOI] [PubMed] [Google Scholar]
- 3.Antequera F., Boyes,J. and Bird,A. (1990) Cell, 62, 503–514. [DOI] [PubMed] [Google Scholar]
- 4.Costello J.F., Futscher,B.W., Kroes,R.A. and Pieper,R.O. (1994) Mol. Cell. Biol., 14, 6515–6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watts G.S., Pieper,R.O., Costello,J.F., Peng,Y.M., Dalton,W.S. and Futscher,B.W. (1997) Mol. Cell. Biol., 17, 5612–5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hansen J.C., Tse,C. and Wolffe,A.P. (1998) Biochemistry, 37, 17637–17641. [DOI] [PubMed] [Google Scholar]
- 7.Ng H.H. and Bird,A. (1999) Curr. Opin. Genet. Dev., 9, 158–163. [DOI] [PubMed] [Google Scholar]
- 8.Ura K., Kurumizaka,H., Dimitrov,S., Almouzni,G. and Wolffe,A.P. (1997) EMBO J., 16, 2096–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wade P.A., Pruss,D. and Wolffe,A.P. (1997) Trends Biochem. Sci., 22, 128–132. [DOI] [PubMed] [Google Scholar]
- 10.Wolffe A.P. and Hayes,J.J. (1999) Nucleic Acids Res., 27, 711–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones P.L., Veenstra,G.J., Wade,P.A., Vermaak,D., Kass,S.U., Landsberger,N., Strouboulis,J. and Wolffe,A.P. (1998) Nature Genet., 19, 187–191. [DOI] [PubMed] [Google Scholar]
- 12.Nan X., Ng,H.H., Johnson,C.A., Laherty,C.D., Turner,B.M., Eisenman,R.N. and Bird,A. (1998) Nature, 393, 386–389. [DOI] [PubMed] [Google Scholar]
- 13.Holt J.T., Thompson,M.E., Szabo,C., Robinson-Benion,C., Arteaga,C.L., King,M.C. and Jensen,R.A. (1996) Nature Genet., 12, 298–302. [DOI] [PubMed] [Google Scholar]
- 14.Seery L.T., Knowlden,J.M., Gee,J.M., Robertson,J.F., Kenny,F.S., Ellis,I.O. and Nicholson,R.I. (1999) Int. J. Cancer, 84, 258–262. [DOI] [PubMed] [Google Scholar]
- 15.Yoshikawa K., Honda,K., Inamoto,T., Shinohara,H., Yamauchi,A., Suga,K., Okuyama,T., Shimada,T., Kodama,H., Noguchi,S., Gazdar,A.F., Yamaoka,Y. and Takahashi,R. (1999) Clin. Cancer Res., 5, 1249–1261. [PubMed] [Google Scholar]
- 16.Dobrovic A. and Simpfendorfer,D. (1997) Cancer Res., 57, 3347–3350. [PubMed] [Google Scholar]
- 17.Mancini D.N., Rodenhiser,D.I., Ainsworth,P.J., O’Malley,F.P., Singh,S.M., Xing,W. and Archer,T.K. (1998) Oncogene, 16, 1161–1169. [DOI] [PubMed] [Google Scholar]
- 18.Rice J.C., Ozcelik,H., Maxeiner,P., Andrulis,I. and Futscher,B.W. (2000) Carcinogenesis, 21, in press. [DOI] [PubMed] [Google Scholar]
- 19.Rice J.C., Massey-Brown,K.S. and Futscher,B.W. (1998) Oncogene, 17, 1807–1812. [DOI] [PubMed] [Google Scholar]
- 20.Xu C.F., Chambers,J.A. and Solomon,E. (1997) J. Biol. Chem., 272, 20994–20997. [DOI] [PubMed] [Google Scholar]
- 21.Thompson F., Emerson,J., Dalton,W., Yang,J.M., McGee,D., Villar,H., Knox,S., Massey,K., Weinstein,R., Bhattacharyya,A. et al. (1993) Genes Chromosomes Cancer, 7, 185–193. [DOI] [PubMed] [Google Scholar]
- 22.Trent J., Yang,J.M., Emerson,J., Dalton,W., McGee,D., Massey,K., Thompson,F. and Villar,H. (1993) Genes Chromosomes Cancer, 7, 194–203. [DOI] [PubMed] [Google Scholar]
- 23.Coffee B., Zhang,F., Warren,S.T. and Reines,D. (1999) Nature Genet., 22, 98–101. [DOI] [PubMed] [Google Scholar]
- 24.Godde J.S., Kass,S.U., Hirst,M.C. and Wolffe,A.P. (1996) J. Biol. Chem., 271, 24325–24328. [DOI] [PubMed] [Google Scholar]
- 25.Eden S., Hashimshony,T., Keshet,I., Cedar,H. and Thorne,A.W. (1998) Nature, 394, 842. [DOI] [PubMed] [Google Scholar]
- 26.Graff J.R., Herman,J.G., Myohanen,S., Baylin,S.B. and Vertino,P.M. (1997) J. Biol. Chem., 272, 22322–22329. [DOI] [PubMed] [Google Scholar]
- 27.Mummaneni P., Walker,K.A., Bishop,P.L. and Turker,M.S. (1995) J. Biol. Chem., 270, 788–792. [DOI] [PubMed] [Google Scholar]
- 28.Yates P.A., Burman,R.W., Mummaneni,P., Krussel,S. and Turker,M.S. (1999) J. Biol. Chem., 274, 36357–36361. [DOI] [PubMed] [Google Scholar]
- 29.Mummaneni P., Bishop,P.L. and Turker,M.S. (1993) J. Biol. Chem., 268, 552–528. [PubMed] [Google Scholar]
- 30.Mummaneni P., Yates,P., Simpson,J., Rose,J. and Turker,M.S. (1998) Nucleic Acids Res., 26, 5163–5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buschhausen G., Wittig,B., Graessmann,M. and Graessmann,A. (1987) Proc. Natl Acad. Sci. USA, 84, 1177–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kass S.U., Landsberger,N. and Wolffe,A.P. (1997) Curr. Biol., 7, 157–165. [DOI] [PubMed] [Google Scholar]
- 33.Boyes J. and Bird,A. (1991) Cell, 64, 1123–1134. [DOI] [PubMed] [Google Scholar]
- 34.Hendrich B. and Bird,A. (1998) Mol. Cell. Biol., 18, 6538–6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meehan R.R., Lewis,J.D. and Bird,A.P. (1992) Nucleic Acids Res., 20, 5085–5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.