Summary
Background
Both defects in mismatch repair (dMMR) and high microsatellite instability (MSI-H) have been recognised as crucial biomarkers that guide treatment strategies and disease management in colorectal cancer (CRC). As MMR and MSI tests are being widely conducted, an increasing number of MSI-H tumours have been identified in CRCs with mismatch repair proficiency (pMMR). The objective of this study was to assess the clinical features of patients with pMMR/MSI-H CRC and elucidate the underlying molecular mechanism in these cases.
Methods
From January 2015 to December 2018, 1684 cases of pMMR and 401 dMMR CRCs were enrolled. Of those patients, 93 pMMR/MSI-H were identified. The clinical phenotypes and prognosis were analysed. Frozen and paraffin-embedded tissue were available in 35 patients with pMMR/MSI-H, for which comprehensive genomic and transcriptomic analyses were performed.
Findings
In comparison to pMMR/MSS CRCs, pMMR/MSI-H CRCs exhibited significantly less tumour progression and better long-term prognosis. The pMMR/MSI-H cohorts displayed a higher presence of CD8+ T cells and NK cells when compared to the pMMR/MSS group. Mutational signature analysis revealed that nearly all samples exhibited deficiencies in MMR genes, and we also identified deleterious mutations in MSH3-K383fs.
Interpretation
This study revealed pMMR/MSI-H CRC as a distinct subgroup within CRC, which manifests diverse clinicopathological features and long-term prognostic outcomes. Distinct features in the tumour immune-microenvironment were observed in pMMR/MSI-H CRCs. Pathogenic deleterious mutations in MSH3-K383fs were frequently detected, suggesting another potential biomarker for identifying MSI-H.
Funding
This work was supported by the Science and Technology Commission of Shanghai Municipality (20DZ1100101).
Keywords: Mismatch repair, Microsatellite instability, Colorectal cancer, Diagnostic biomarker, Predictive modelling
Research in context.
Evidence before this study
-
1.
Defects in mismatch repair (dMMR) and high microsatellite instability (MSI-H) have been recognised as crucial biomarkers directing treatment strategies and disease management in colorectal cancer (CRC).
-
2.
With the widespread use of MMR and MSI tests, an increasing number of MSI-H tumours have been identified in mismatch repair proficient (pMMR) CRCs, estimated to occur in a range of 1%–10%. Given that MSI tests have historically been administered to a relatively limited subset of pMMR CRCs, the actual proportion of MSI-H occurrences within pMMR CRCs could potentially be higher than current estimates suggest.
-
3.
pMMR/MSI-H CRC has been recognised as a distinct subgroup, but previous studies rarely analysed the clinical phenotype and underlying genotype, nor did those investigations provide an exhaustive explanation, except for technical bias of IHC and MSI testing methods, MLH1 methylation, and somatic MMR gene mutation.
Added value of this study
-
1.
A relatively high prevalence of MSI-H CRC was identified within pMMR CRCs (5.5%, 93/1684).
-
2.
Compared with pMMR/MSS CRCs, pMMR/MSI-H CRCs presented significantly reduced tumour progression and better long-term prognosis.
-
3.
pMMR/MSI-H cohorts exhibited a higher presence of CD8+ T cells and NK cells in comparison to the pMMR/MSS group.
-
4.
Mutational signature analysis revealed that nearly all samples exhibited deficiencies in MMR genes and deleterious mutations in MSH3-K383fs were also identified.
-
5.
An MSI prediction model was constructed for screening pMMR cases.
Implications of all the available evidence
-
1.
pMMR/MSI-H CRC represents a distinct subgroup of CRC, which manifests diverse clinicopathological features and long-term prognostic outcomes.
-
2.
Distinct features of the tumour immune-microenvironment were found to be inherent in pMMR/MSI-H CRCs.
-
3.
Pathogenic deleterious mutations in MSH3-K383fs were frequently detected, suggesting it as a potential biomarker for MSI-H.
-
4.
The nomogram serves as a valuable tool for physicians to facilitate the screening of patients with MSI-H tumours through pMMR test results.
Introduction
Colorectal cancer (CRC) is one of the most common malignant tumours worldwide, and the fifth leading cause of cancer-related death globally.1 CRC tumours manifest heterogeneous phenotypes yielded of different molecular mechanisms, therefore presenting varied heterogeneous outcomes and drug sensitivities.2,3 One well-established malignant transformation mechanism in CRC is the induction of microsatellite instability (MSI) due to defects in mismatch repair (dMMR), which culminates in the accumulation of deleterious mutations.3, 4, 5 Tumours characterised by microsatellite instability-high (MSI-H) had demonstrated an enhanced responsiveness to programmed cell death 1 (PD-1) inhibitor therapies,6,7 which significantly improved long-term prognostic outcomes of those patients.8 Consequently, several PD-1 inhibitors have received endorsement in current guidelines as the first-line of treatment for unresectable or metastatic MSI-H/dMMR CRC.9,10
Expression levels of mismatch repair (MMR) genes, including MLH1, MSH2, MSH6, and PMS2, assessed using immunohistochemistry (IHC) staining, are routinely employed for Lynch syndrome (LS) screening. Furthermore, MSI testing is recommended for tumours showcasing deficiencies in any of these MMR proteins.11 Given the MSI-H and dMMR recognised as critical biomarkers directing treatment strategies and disease management in CRC, the emphasis on MSI status screening has substantially expanded. Over the past decade, MSI tests were also carried out based on empirical risk factors such as a family history of cancer, early-onset CRC, and the presence of multiple primary CRCs. As a result, an increasing number of MSI-H tumours were identified in mismatch repair proficient (pMMR) CRCs, estimated to range between 1% and 10%.12,13 Given that MSI tests have historically been administered to a relatively limited subset of pMMR CRCs, the actual proportion of MSI-H occurrences within pMMR CRCs could potentially be higher than current estimates suggest.
Even though researchers have recognised the pMMR/MSI-H CRC as a distinct group, rarely studies analysed the clinical phenotype and underlying genotype, nor did those investigations provide an exhaustive explanation, except for technical bias of IHC and MSI testing methods, MLH1 methylation, and somatic MMR gene mutation.14,15 The lack of systematic study on clinical features and molecular mechanisms of this distinctive groups not only restricts further exploration of MSI-H biomarkers in pMMR CRCs, but also obstructs the identification of MSI-H tumours in clinical work.
This retrospective study recruited 1684 patients with pMMR CRC and 401 patients with dMMR CRC from a cohort of consecutive 8216 patients with CRC spanning from 2015 to 2018. Of these, 93 pMMR/MSI-H CRC cases were identified, accounting for 5.5% (93/1684) patient with pMMR CRC. The relatively high proportion of MSI-H tumours was identified in pMMR CRC group, indicating MSI-H tumours may harbour other molecular mechanisms except for deficiency of mismatch repair function. In this study, we meticulously assessed the clinical features of pMMR/MSI-H CRC patients and utilised both whole exome sequencing (WES) and transcriptomic analysis to reveal the underlying molecular mechanism from these cases. Our findings indicated CRC patients exhibiting certain clinical hallmarks possess a heightened likelihood of presenting with MSI-H, which lends support to introducing MSI testing for those patients. Simultaneously, our data pointed to the deleterious mutation of MSH3-K383fs as a biomarker that's indicative of MSI-H. Finally, we constructed a prediction model for MSI-H to facilitate the identification of MSI-H CRC in pMMR cases.
Methods
Participant and sample collection
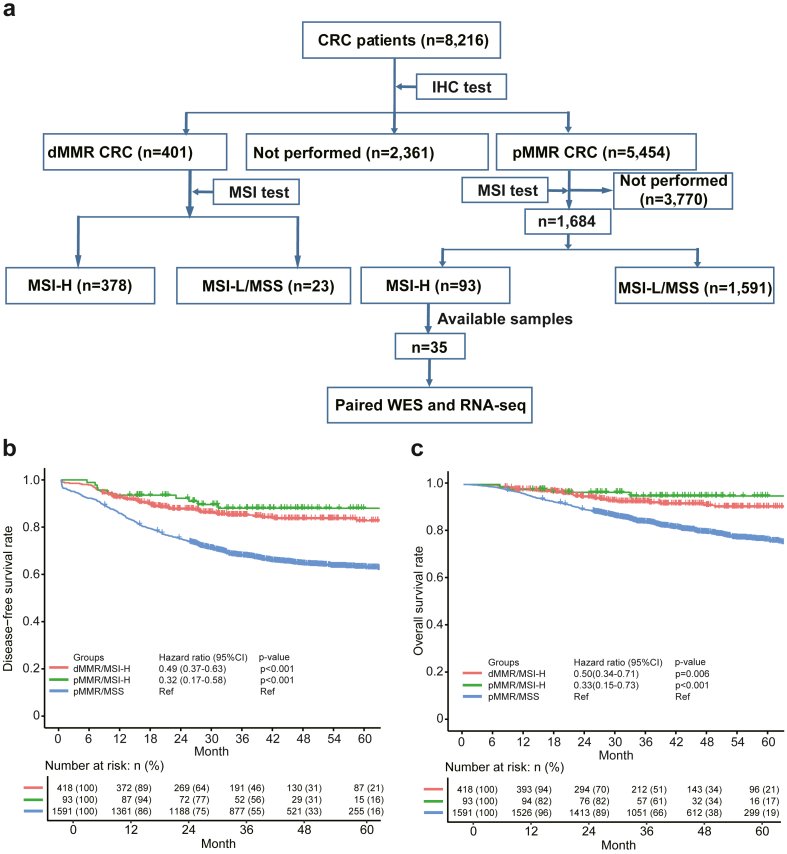
A total of 8216 patients with CRC who received treatment at the Fudan University Shanghai Cancer Center between January 1 2015 and December 31 2018 were retrospectively included in this study. A total of 401 cases of dMMR and 5454 of pMMR CRCs were identified by IHC. MSI tests were performed for all dMMR tumours and 1684 pMMR tumours, the flow chart of patients’ selection was illustrated in Fig. 1a.
Fig. 1.
Flowchart of samples selection and clinical features of pMMR/MSI-H samples. a, Flowchart detailing the enrolment process for patients with colorectal cancer. b, Disease-free survival curves comparing different patient groups. c, Overall survival curves comparing different patient groups. Differences in OS and DFS were assessed using stratified log-rank tests and Cox regression for hazard ratios, with dMMR/MSI-H as the reference. Sample size: dMMR/MSI-H, n = 418; pMMR/MSI-H, n = 93; pMMR/MSS, n = 1591.
Data including demographic information, family cancer history, medical history and pathological results were extracted from the electronic medical record, which were valid and complete. All patients were followed up as of June 30th, 2023.
IHC and MSI Analysis System
Tumour representative blocks were carefully selected for analysis with normal–tumour junctions in order to assess staining results properly. VENTANA MMR RxDx Panel was use for IHC. IHC of MMR proteins (MLH1, PMS2, MSH2, and MSH6) was performed on 4-μm thick paraffin tissue sections using monoclonal antibodies against the following proteins: MLH1 [VENTANA® anti-MLH1 (M1) Mouse Monoclonal Primary Antibody, cat. 790-5091/07862237001, Ready-to-use, Roche], PMS2 [VENTANA® anti-PMS2 (A16-4) Mouse Monoclonal Primary Antibody, cat. 790-5094/07862261001, Roche], MSH2 [VENTANA® anti-MSH2 (G219-1129) Mouse Monoclonal Primary Antibody, cat. 760-5093/08033684001, Ready-to-use, Roche], and MSH6 [VENTANA® anti-MSH6 (SP93) Rabbit Monoclonal Primary Antibody, cat. 790-5092/07862245001, Ready-to-use, Roche]. Staining was performed on the autostainer Benchmark XT/Ultra (Ventana, Medical Systems, Tucson, AZ, USA) using an OptiView universal DAB IHC detection and amplification kit (cat. 760-099/06396518001, Roche), according to the manufacturer's instructions.
To ensure accuracy, we re-checked the previously saved HE stained sections and conducted repeat IHC experiments on these available samples. The sections were deparaffinized at 65 °C for 1–2 h, followed by three washes with xylene, each lasting 10 min, and subsequently rehydrated through graded alcohols to distilled water. Next, the slides underwent antigen unmasking by heating with Sodium citrate-EDTA antigen repair solution (cat, No. P0086, Beyotime, China) and were cooled to room temperature for 1.5 h. After rinsing in distilled water and TBS, the sections were incubated with primary monoclonal antibodies overnight at 4 °C. The rabbit-anti-MLH1 (EPR3894) (1:300 dilution, Abcam, cat.ab92312, UK), rabbit-anti-MSH2 (EPR21017-123) (1:300 dilution, Abcam, cat.ab227941, UK), rabbit-anti-MSH6 (EPR3945) (1:200 dilution, Abcam, cat.ab92471, UK), and rabbit-anti-PMS2 (EPR3947) (1:100 dilution, Abcam, cat.ab110638, UK) antibodies were used as primary antibodies. Subsequently, the sections were incubated with horseradish peroxidase-conjugated secondary antibody (cat. No. GK500705, GeneTech) at room temperature for 30–60 min, followed by incubation with 3′-diaminobenzidine (cat. No.GK500705, GeneTech, China) for 5 min. The slides were then counterstained with haematoxylin, dehydrated with a graded series of alcohols, and mounted with coverslips and mounting medium. The staining density was measured using a Leica CCD camera DFC420 connected to a Leica DM IRE2 microscope (Leica Microsystems Imaging Solutions Ltd.). Non-cancerous colonic mucosa, stromal cells, infiltrating lymphocytes, or the centres of lymphoid follicles served as internal positive controls, while known dMMR CRC samples served as external negative controls.
Nuclear expression of all the four MMR markers (MLH1, PMS2, MSH2, and MSH6) indicated pMMR, which was derived from a pMMR/MSI-H case (Figure S1). Complete loss of nuclear expression of any of the four MMR markers in the neoplastic cells was considered as deficient MMR expression or dMMR. Each result was confirmed independently by a minimum of two seasoned pathologists.
The Promega™ MSI Analysis System v1.2 (RUO) was utilised to determine MSI status. The protocol provided by the manufacturer was strictly adhered to. The classification into MSI-H and Microsatellite stable (MSS) states was performed in alignment with the directives provided in the manufacturer's guidelines.
Whole exome sequencing of paired normal-tumour samples
In order to perform comprehensive genomic and transcriptomic analyses, we carefully reviewed the tissue specimens from the Tissue Bank of our hospital. Frozen and paraffin-embedded tissue were available in 35 pMMR/MSI-H cases, for paired normal-tumour WES alongside RNA sequencing (Figure S2a).
Genomic DNA was extracted from tumour tissues and lymphocytes using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany). The DNA concentration was subsequently quantified using the Qubit 3.0 (Thermo Fisher Scientific, Inc., Waltham, MA, USA), following the manufacturer's instructions. Library preparation was achieved using the SureSelect Human All Exon Kit V6 (Agilent Technologies, Santa Clara, California, USA), adhering to the manufacturer's protocol. The quality of the captured libraries was assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies) before being sequenced on the NovaSeq 6000 system (Illumina, San Diego, California, USA), according to the manufacturer's guidelines.
Raw sequencing reads were preprocessed by trim_galore (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) for subsequent analysis: (1) adapter trimming: (2) remove the reads in which the N base has reached a certain percentage (default length of 8 bp); (3) remove the reads which contain low-quality bases (default quality threshold value ≤ 20) above a certain portion (default 40%); (4) sliding window trimming: the bases in the sliding window (default is 4 bp) with mean quality below cutting quality (default is 20) will be cut. The cleaned reads were aligned to the reference human genome (build hg38, https://hgdownload.soe.ucsc.edu/goldenPath/hg38/bigZips/) using Sentieon bwa-mem.16 Subsequent processing including sorting reads and marking duplicates were performed according to best practices of the GATK Toolkit v417,18 (https://gatk.broadinstitute.org/hc/en-us). Sequence depth and coverage were obtained using qualimap.19 To identify all the variants, we used two somatic mutation callers for single nucleotide variants (SNVs) and indels: Mutect220 and Strelka2.21 To improve specificity, a panel of normal sample filtration was used to remove background germline variations and artifacts. Mutect2 was based on bam files which were processed by quality score recalibration that was performed using GATK4 (v 4.1.1.0). Somatic mutations were then annotated using VEP.22 To obtain reliable mutation calls, we used a two-step approach. First, chose mutations that were identified in both of the two callers (Mutect2 and Strelka2). Second, additional filtering with three criteria was performed: (1) variant allele frequency (VAF) ≥ 8%; (2) sequencing depth in the region ≥ 8; (3) sequence reads in support of the variant call ≥ 2. Tumour mutation burden (TMB) was defined as the number of somatic mutations per Mb by pyTMB (https://github.com/bioinfo-pf-curie/TMB). Samples with over 10 muts/Mb were labelled as TMB-H.
RNA sequencing for tumour
We used 100 ng total RNA from all subjects to prepare sequencing libraries using the TruSeq stranded total RNA sample preparation kit (Illumina). The quality of the resulting complementary DNA libraries was evaluated with the Agilent 2100 Bioanalyzer (Agilent Technologies). Quantification was achieved with the KAPA library quantification kit (Kapa Biosystems, Massachusetts, USA) according to the manufacturer's library quantification protocol. After cluster amplification of the denatured templates, sequencing was conducted in a paired-end format (2 × 101 bp) on the Illumina NovaSeq 6000 platform.
Raw RNA-seq reads were first subjected to quality control and adapter trimming using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and Cutadapt (https://github.com/marcelm/cutadapt), respectively. The high-quality reads were then aligned to the reference genome using a splice-aware aligner, HISAT2.23 The aligned reads were further processed and sorted according to best practices of the GATK Toolkit v4. To detect genetic variants, including SNVs and small indels, we utilised a workflow similar to that used for whole exon sequencing based variant calling process for subsequent analysis. The variant calling process involved local realignment around indels, base quality score recalibration, and variant quality score recalibration. The variant calls generated by Strelka2 and Mutect2 were then compared, and only mutations that were concordantly identified by both algorithms were retained. This stringent filtering criterion ensured that only high-confidence mutations were included for further analysis. Variant sites were filtered based on various criteria, including read depth, mapping quality, variant allele frequency, and annotation databases, to ensure the accuracy and reliability of the identified variants. To annotate the detected variants, functional impact analysis was performed using VEP.
Classification of MMR gene missense mutations
Mutations in MMR genes (MLH1, MSH2, MSH6, PMS2) were primarily annotated through the VEP and categorized in line with the guidelines established by the American College of Medical Genetics and Genomics and the Association for Molecular Pathology (ACMG-AMP). Missense mutations that were initially designated as variants of uncertain significance (VUS) were further reclassified using both the Evidence-based Variant Classification (evolutionary experimental data-based method) and REVEL (ensemble method) system24 (Figure S2b). When both methods concurrently reclassify a VUS variant as pathogenic, it is then considered a potential pathogenic variant. However, if only one method identifies it as pathogenic, we continue to classify it as a VUS. This meticulous annotation and classification approach ensures an accurate assessment of the clinical relevance of MMR gene mutations.
Copy number variation and tumour purity
Copy number variants were determined using the cnvkit software (https://github.com/etal/cnvkit) which deploys a sophisticated algorithm to accurately discern genomic alterations within tumour samples. Oncogenic copy number alterations were further annotated for their oncogenic relevance using the OncoKB Annotator,25 a comprehensive oncology knowledgebase designed to elucidate the effects and potential clinical implications of cancer-related variations (http://oncokb.org). To assess tumour purity, RNA sequencing data were utilised. The raw RNA-seq reads were mapped to the reference transcriptome using HISAT2.23 The mapped reads were then quantified to estimate gene expression levels. The tumour purity was inferred by analysing the expression levels of tumour-specific genes and normal tissue-specific genes using established computational approaches (e.g., ESTIMATE algorithm26).
Mutational signature analysis
Mutational signatures were identified using the R package MutSigCV and NMF.27,28 The normalization method was set to ‘exome2genome’. This approach organised sample information in the form of the fraction of mutations in each of the 96 trinucleotides and determined the weighted combination of the COSMIC signatures (https://cancer.sanger.ac.uk/signatures/) that most closely reconstructed the mutational profile.
Clonal decomposition
We employed TIMER29 (Tumor Immune Estimation Resource), a computational tool specifically designed for tumour immune microenvironment analysis. Utilising the gene expression profiles obtained from RNA-seq data, TIMER applied deconvolution algorithms to infer the proportions of distinct immune cell populations within the tumour microenvironment. Furthermore, the TCGA-COAD dataset obtained from the TCGA Research Network underwent tumour immune microenvironment analysis using the same methods. By leveraging TIMER's capabilities, we estimated the relative abundance of various immune cell types, such as T cells, B cells, natural killer cells, macrophages, and dendritic cells, contributing to the clonal composition of the tumour.
TCGA-COAD dataset analysis
The gene expression RNA-seq and clinical files for Colonic Adenocarcinoma from The Cancer Genome Atlas (TCGA) were extracted from the GDC portal (https://portal.gdc.cancer.gov/). The clinical annotation file containing MSI information can be downloaded from https://www.linkedomics.org/data_download/TCGA-COADREAD/. Based on the MSI status, we categorized the TCGA-COAD dataset into pMMR/MSS (n = 369) and dMMR/MSI-H groups (n = 81). To ensure consistency in tumour purity analysis, we applied the ESTIMATE algorithm to analyse the RNA-seq data of the TCGA-COAD dataset. For a more in-depth analysis of the immune microenvironment, we utilised the immune cell proportion annotation file provided by TIMER for TCGA-COAD. This enabled us to conduct further comparative analyses.
Predictive modelling
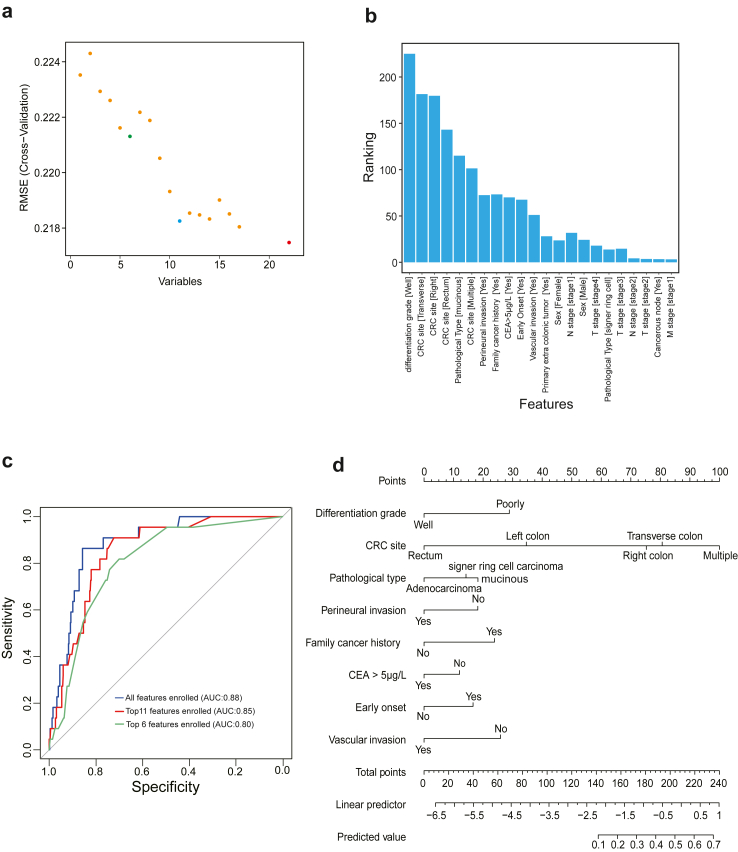
We used the createDataPartition function from the caret package (v.6.0–94) for random splitting of the sample, dividing 1684 pMMR patients into a training cohort (n = 1179) and a validation cohort (n = 505) in a 7:3 ratio. Features selection for the logistic regression model in the training cohort was performed using recursive feature elimination by rfe function, repeated 10 times with 10-fold cross-validation, to understand the impact of various feature combinations on the model's performance. We also evaluated the individual impact of each feature. Out of 22 total features, we observed that the top 6 and top 11 feature combinations corresponded to the first and second lowest RMSE values, respectively, while the inclusion of all features resulted in the lowest overall RMSE. Considering each feature's contribution ranking to the model, logistic regression models were constructed with sets of top-6, top-11, and all features, utilising the lrm function for model development on the training dataset. Predictions for the validation dataset were made using the predict function from the stats package (v.4.2.0). Model performance was evaluated using the roc function from the pROC package (v.1.18.5). Calibration residuals were calculated using the calibrate function from the rms package (v.6.7–1), were combined with AUC to provide a comprehensive evaluation of the models' performance. Based on this combined assessment, we selected the logistic regression model with 11 features for subsequence analysis. Finally, we constructed a nomogram using the nomogram function, employing a 0.5 cut-off value to identify potential MSI-H tumours in patients with pMMR tumour.
Statistics
All statistics analysis in this study were performed by experienced professional statistical experts. All statistical analyses were performed using R (version 4.0.2) in Rstudio v.1.2 software. When comparing clinical features between pMMR/MSI-H and pMMR/MSS, we focused on categorical variables and calculated proportions to assess differences between the two groups. T-tests were used for hypothesis testing of each variable between the two groups. For comparisons involving pMMR/MSI-H and other groups (pMMR/MSS, dMMR/MSI-H) in terms of immune infiltration, tumour purity, tumour heterogeneity, and MMR gene expression, we employed the Wilcoxon test to assess differences between two groups and the Kruskal–Wallis H test for overall differences among three or more groups. Linear regression analysis was used to validate the correlation between MMR genes and MSI-score, with the statistical differences in this correlation assessed using the Spearman correlation coefficient. Survival analysis primarily utilised the “survival” package (v.2.11–4), with the “surv” and “survfit” functions. Kaplan–Meier curves were generated using the “ggsurvplot” function from the “survminer” package (v0.4.9). Overall or paired differences in overall survival (OS) and disease-free survival (DFS) were evaluated with a stratified log-rank test. Hazard ratios and confidence intervals were estimated through Cox regression analysis, using dMMR/MSI-H as the reference group. The coxph function was employed to assess pairwise survival differences between different groups. Moreover, we used the cox.zph function from the survival package (v.3.5–7), which performs a global test of the proportional hazards assumption. A statistically non-significant p-value from this test suggests that the proportional hazards assumption holds. All statistical analyses with p-value <0.05 were considered statistically significant (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, N.S., not significant).
Ethics
All procedures were conducted at the Fudan University Shanghai Cancer Center and adhered to the Declaration of Helsinki. This study was approved by the Ethics Committee of our hospital (2005-ZZK-29), and written informed consents were obtained from all participants.
Role of funders
The funders did not have any role in study design, data collection, data analyses, interpretation, or writing of report.
Results
Clinical features and long-term prognostic of patients with pMMR/MSI-H CRC
We retrospectively collected 8216 patients from 2015 to 2018, a total of 1684 cases of pMMR CRCs were enrolled. Among them, 1591 patients with pMMR/MSS tumours and 93 (5.5%, 93/1684) cases of pMMR/MSI-H CRCs were identified. The clinical features of pMMR/MSI-H cases and their comparison with pMMR/MSS cases have been summarized in Table 1. Compared with pMMR/MSS CRCs, a higher proportion of multiple primary CRCs (t-test, p < 0.001), right colon cancers (t-test, p < 0.001), mucinous (t-test, p < 0.001), and poorly differentiation grade (t-test, p < 0.001) tumours were observed; but less cancerous node (t-test, p = 0.002), vascular invasion (t-test, p = 0.02), perineural invasion (t-test, p = 0.002) and stage III/IV CRCs (t-test, p < 0.001) were observed.
Table 1.
Demographic and clinical characteristics of patients with pMMR/MSI-H and pMMR/MSS tumors.
| Variables | pMMR/MSI-H (n = 93) | pMMR/MSS (n = 1591) | p-value |
|---|---|---|---|
| Male (%) | 59 (63.4) | 891 (56.0) | 0.194 |
| Early onset | 56 (60.2) | 549 (35.5) | <0.001 |
| CEA >5 μg/L | 34 (36.6) | 687 (43.2) | 0.252 |
| Metachronous CRC (%) | 6 (6.5) | 35 (2.2) | 0.025 |
| CRC site (%) | <0.001 | ||
| Right | 39 (41.9) | 251 (15.8) | |
| Transverse | 17 (18.3) | 75 (4.7) | |
| Left | 18 (19.4) | 460 (28.9) | |
| Rectum | 8 (8.6) | 763 (48.0) | |
| Multiple | 11 (11.8) | 42 (2.6) | |
| Pathological type (%) | <0.001 | ||
| Adenocarcinoma | 63 (67.7) | 1388 (87.2) | |
| Mucinous | 25 (26.9) | 143 (9.0) | |
| Signer ring cell | 5 (5.4) | 60 (3.8) | |
| Differentiation grade (%) | <0.001 | ||
| Well | 46 (49.5) | 1153 (72.5) | |
| Poorly | 47 (50.5) | 438 (27.5) | |
| Vascular invasion (%) | 17 (18.3) | 480 (30.2) | 0.02 |
| Perineural invasion (%) | 15 (16.1) | 511 (32.1) | 0.002 |
| Cancerous node (%) | 4 (4.3) | 286 (18.0) | 0.001 |
| TNM stage (%) | <0.001 | ||
| I | 16 (17.2) | 315 (19.8) | |
| II | 52 (55.9) | 426 (26.8) | |
| III | 24 (25.8) | 634 (39.8) | |
| IV | 1 (1.1) | 216 (13.6) | |
| Family cancer history (%) | 48 (51.6) | 621 (39.0) | 0.021 |
| Extra-colonic cancer (%) | 19 (20.4) | 169 (10.6) | 0.006 |
Survival analysis revealed that 5-year OS and DFS of pMMR/MSI-H cases were 95% and 88%, which were comparable with those of 94% and 83% for dMMR/MSI-H (CoxPH test, p = 0.44 for DFS, p = 0.66 for OS). These findings suggest that patients with pMMR/MSI-H CRC had a prognosis similar to dMMR/MSI-H CRC. The 5-year OS and DFS of pMMR/MSI-H CRC cases were significantly higher than those of 77% and 63% for pMMR/MSS cases (CoxPH test, p < 0.0001 for DFS, p = 0.0033 for OS) (Fig. 1b and c). Thus, patients with pMMR/MSI-H CRC manifested significantly better prognostic outcome, compared with those of patients with pMMR/MSS CRC.
Distinctive tumour microenvironment features between pMMR/MSI-H and dMMR/MSI-H tumours
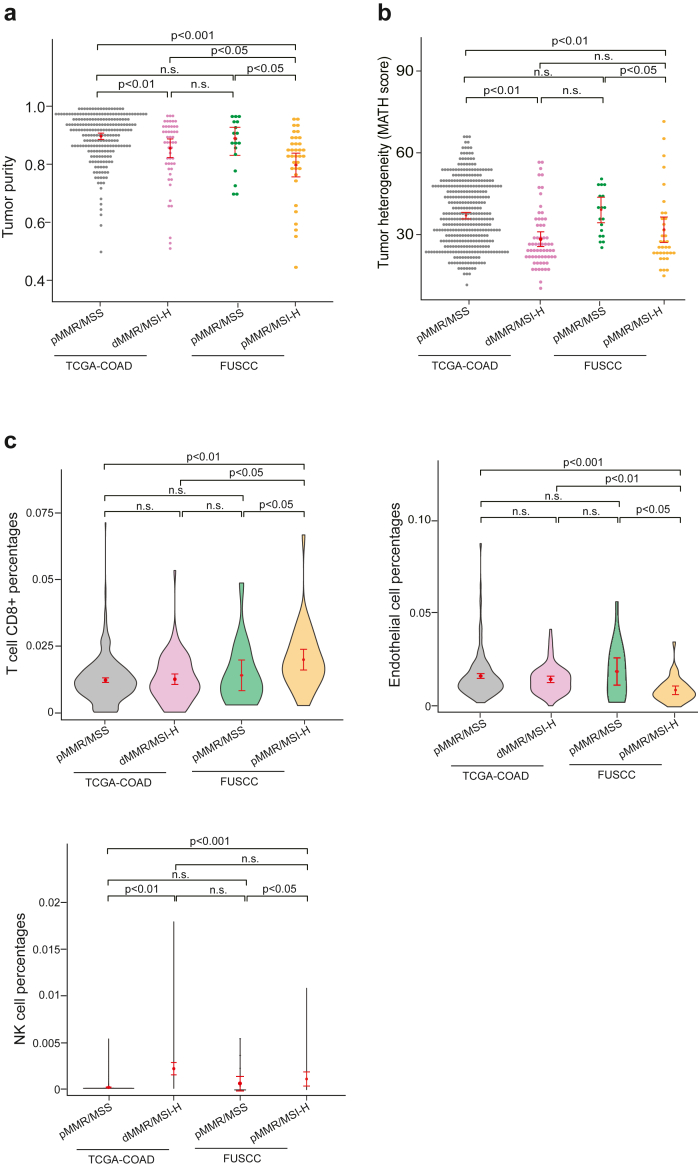
To investigate the differences in the tumour microenvironment between pMMR/MSI-H and other tumour types. We utilised the TCGA-COAD dataset to compare tumour purity and tumour heterogeneity. By dividing the TCGA-COAD dataset into pMMR/MSS and dMMR/MSI groups and comparing them with our cohort, we found that our cohort displayed diminished levels of both tumour purity and tumour heterogeneity (Fig. 2a and b). Furthermore, we explored the tumour microenvironment indicators and found that the discordance samples exhibited specific tumour immune-microenvironment characteristics. While there wasn't a marked difference in terms of the overall immune score, the microenvironment score, or the population of specific immune cells across the groups, it was evident that the pMMR/MSI-H cohorts exhibited a higher presence of CD8+ T cells and NK cells, along with a lower proportion of endothelial cells, B cells and macrophage cells comparison to the pMMR/MSS group (Fig. 2c and Figure S3). These findings highlight distinct tumour immune-microenvironment features inherent to pMMR/MSI-H CRCs.
Fig. 2.
Comprehensive genomic and transcriptomic insights into pMMR/MSI-H samples. a, Comparative analysis of tumour purity. Sample size: TCGA-pMMR/MSS, n = 323; TCGA-dMMR/MSI-H, n = 50; FUSCC-pMMR/MSS, n = 18; FUSCC-pMMR/MSI-H, n = 35. b, Investigation of tumour heterogeneity. Sample size: TCGA-pMMR/MSS, n = 360; TCGA-dMMR/MSI-H, n = 63; FUSCC-pMMR/MSS, n = 18; FUSCC-pMMR/MSI-H, n = 35. c, Tumour immune microenvironment analysis depicting proportions of CD8+ T cells, NK cells, and endothelial cells, in sequential order from left to right. Pairwise comparisons between samples were performed using the Wilcoxon test, with p < 0.05 indicating significant differences. Sample size: TCGA-pMMR/MSS, n = 353; TCGA-dMMR/MSI-H, n = 78; FUSCC-pMMR/MSS, n = 18; FUSCC-pMMR/MSI-H, n = 35.
Mutations profiling in the patients with pMMR/MSI-H tumours
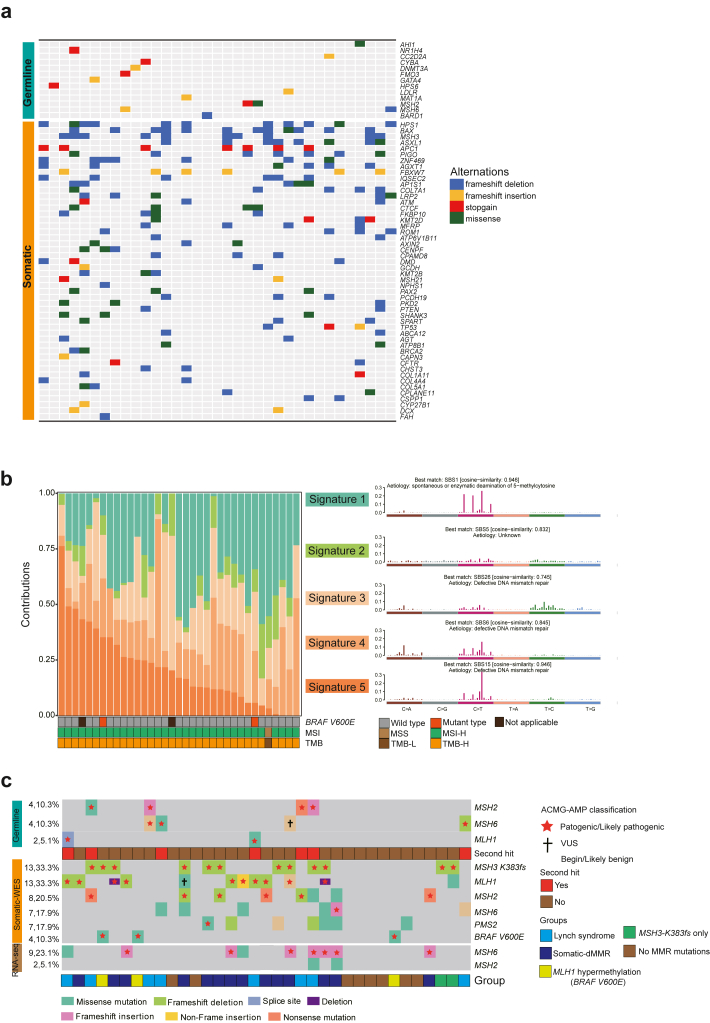
To investigate the specific mutational landscape of pMMR/MSI-H tumours, we analysed both germline and somatic pathogenic mutations in these patients. In our study of germline mutations, we identified a total of 14 pathogenic mutations across 13 genes. Notably, two mutations were found in MSH2 and another two in MSH6, with an additional mutation identified in the cancer susceptibility gene, BARD1. Within the somatic mutations, genes related to genomic stability and transcription factors showed the highest mutation frequencies (Fig. 3a).
Fig. 3.
Analysis of Mutational Signatures and Distribution of MMR Gene Mutations. a, The heatmap illustrates the distribution of pathogenic mutations, with germline pathogenic mutations presented on the top and somatic pathogenic mutations depicted at the bottom, pMMR/MSI-H (n = 35). b, Identification of tumour mutational signatures and their representation across samples, featuring the mutational signatures on the right and their distribution across samples on the left, pMMR/MSI-H (n = 35). c, Distribution of MMR and BRAF-V600E gene mutations in pMMR/MSI-H samples, pMMR/MSI-H (n = 35).
Mutational signature analysis uncovered DNA mismatch repair deficiency as the predominant cause of discordant results
Mutational signatures provide invaluable insights into the molecular intricacies of tumours, acting as definitive markers for their classification and stratification. The heterogeneity observed in these signatures across various tumour subtypes sheds light on their origin, clinical evolution, and potential therapeutic sensitivities. We conducted a comprehensive analysis of pMMR/MSI-H samples, revealing the presence of five distinct mutational signatures. One of the mutational signatures showed a high similarity to SBS1, while three mutational signatures were similar to those associated with DNA mismatch repair deficiency. Additionally, signature 2 displayed similarity to SBS5. In a broader analysis of mutational signature composition across all 35 samples, expanding our analysis to encompass all 35 samples, it became evident that all of them carried mutational signatures suggesting DNA mismatch repair deficiency (Fig. 3b). These mutational signature findings provide direct evidence that nearly all 35 inconsistent samples exhibit deficiencies in MMR genes. This pivotal observation highlights the widespread occurrence of DNA mismatch repair deficiencies in these samples.
MMR genes mutations profiling in the patients with pMMR/MSI-H tumours
Prompted by the findings from the mutational signature analysis, we initially assessed the germline MMR gene mutation in all 35 patients. Analysis of MMR pathway-related genes, including MLH1, MSH2, MSH6, PMS2, MLH3 and MSH3, revealed 3 MLH1 variants (SNPs), 99 MSH2 variants (2 insertions and 97 SNPs), 27 MSH6 variants (1 insertions, 1 deletion and 25 SNPs), 56 PMS2 variants (SNPs), 1 MLH3 variant (SNP), and 42 MSH3 variants (SNPs). Among these, pathogenic variants classified based on ACMG-AMP guidelines were identified in 6 patients, including 1 MLH1 variant, 3 MSH2 variants, and 2 MSH6 variants. Considering the possibility that missense mutations might affect protein function without impacting protein expression, we reclassified 14 VUS missense mutations in MMR genes by functional prediction tools. Ultimately, this led to identifying 2 pathogenic variants, 1 in MLH1 and 1 in MSH2, while the remaining variants remained as VUS. In summary, among 35 patients, we identified 8 patients who carried pathogenic MMR genes mutation, suggestive of LS (Fig. 3c). Seven patients with LS have second hits in their tumours.
Using WES-based somatic mutation analysis, we identified 10 non-Lynch patients carrying pathogenic MMR mutations and 2 patients with MLH1 copy number deletion. RNAseq-based somatic MMR gene mutation analysis identified 2 patients with pathogenic MMR mutations. Additionally, we detected 2 cases with pathogenic mutations in the non-classical MMR gene, including 2 MSH3-K383fs deleterious mutations. In total, we identified 16 patients exhibiting somatic dMMR, this being unrelated to MLH1 hypermethylation (Fig. 3c). Among those patients with somatic-dMMR, three patients with MMR gene mutations exhibited a variant allele VAF less than 15%, indicating a low proportion of dMMR cell components within the tumour, potentially due to intratumorally heterogeneity (Figure S4) leading to failure in tissue sampling or IHC experiments. The remaining 4 MMR mutations had VAFs ranging from 30% to 45% (Table S1).
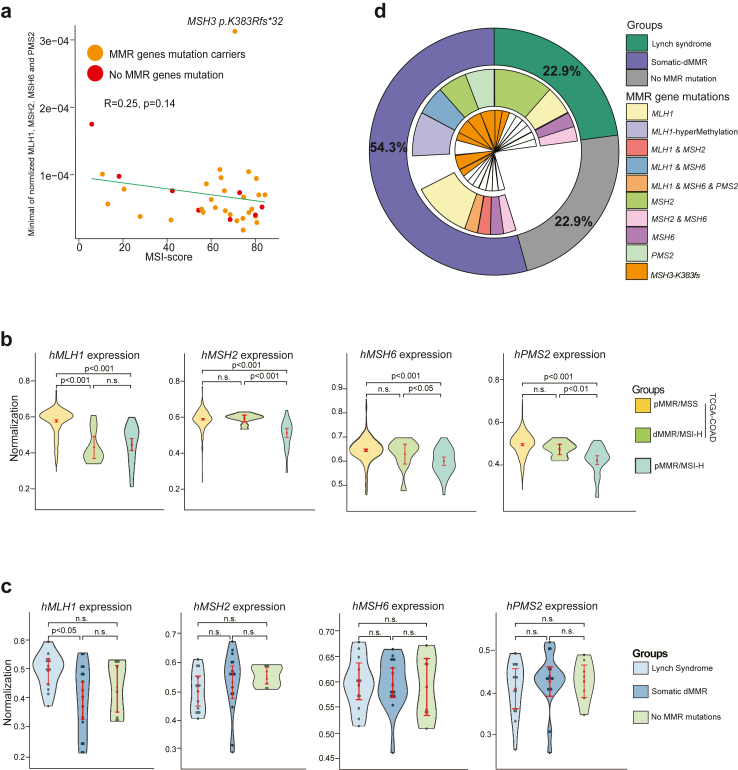
Inexplicable pMMR/MSI-H tumours: absence of germline or somatic MMR mutations
Following our comprehensive assessment of MMR mutations across 35 samples, we discerned that 8 pMMR/MSI-H samples lacked identifiable sources of MSI-H based on pathogenic MMR mutations. Therefore, we meticulously investigated the clinical features of these patients. Notably, a significant number of them presented with CMS3 and CMS4 subtypes. Additionally, two samples exhibited a tumour purity of less than 70% (Table S1). RNA-seq expression profiling of these samples manifested a notably reduced expression of MMR genes. A pronounced negative correlation was observed between MMR gene expression and MSI, underscoring the influence of diminished MMR gene expression on MSI status (Fig. 4a). Compared to the TCGA-COAD dataset, the expression levels of all four MMR genes in our cohort were significantly lower in the pMMR/MSS group. When compared to the dMMR/MSI-H group, our cohort still exhibited lower expression levels of MMR genes, except for MLH1 (Fig. 4b). However, when we stratified our cohort into three groups based on germline and somatic MMR gene status and compared the expression of MMR genes, we found that, apart from MLH1, there were no significant differences in gene expression among the groups. Specifically, the somatic-dMMR group showed significantly reduced expression of hMLH1 (Fig. 4c). Consequently, we cannot exclude the possibility that MLH1 hypermethylation might be responsible for the dMMR status observed in these samples.
Fig. 4.
Regulation of MMR gene expression may partly account for the occurrence of pMMR/MSI-H. a, Correlation of MMR gene expression with MSI score (calculated by MSIsensor) across pMMR/MSI-H samples. Analysed with Spearman's correlation; p < 0.05 indicates significance. b, Comparison of MMR gene expression in TCGA-COAD data and pMMR/MSI-H samples. Pairwise comparisons between samples were performed using the Wilcoxon test, with p < 0.05 indicating significant differences. Sample size: TCGA-pMMR/MSS, n = 360; TCGA-dMMR/MSI-H, n = 78; FUSCC-pMMR/MSS, n = 18; pMMR/MSI-H, n = 35. c, Comparison of MMR gene expression across different pMMR/MSI-H categorizations. Pairwise comparisons between samples were performed using the Wilcoxon test, with p < 0.05 indicating significant differences. d, A summary of the molecular characteristics for pMMR/MSI-H: the outer circle categorises patients according to the statuses of germline and somatic mutations in MMR genes; the middle layer circle represents the mutation statuses of MMR genes; and the inner circle details the detection of somatic MSH3-K383fs variants.
Overall, among all pMMR/MSI-H cases, LS accounted for 22.9% (8/35), including 2 cases of MLH1, 4 of MSH2, 1 of MSH6, and 1 of MSH2 and MSH6 mutation; somatic dMMR counted for 54.3% (19/35), including 7 cases of MLH1, 3 of MSH2, 1 of MSH6, 2 of PMS2, 1 case with co-mutations in MLH1 and MSH2, 3 of MLH1 hypermethylation, and 2 of MSH3-K383fs; 8 cases of Inexplicable pMMR/MSI-H tumours (Fig. 4d).
Risk prediction models for screening potential pMMR/MSI-H
Given it is crucial to identify potential MSI-H from patients with pMMR CRC, we sought to develop a clinically user-friendly nomogram model for clinical screening. Initially, we divided all 1684 patients into a training cohort and a validation cohort in a 7:3 ratio, with their clinical features compared in Table S2. Using a logistic regression model, we assessed the contribution of features and their combinations to the model, with results indicating improved model performance with the addition of more features (Fig. 5a). However, considering the need for simplicity and practicality in clinic, we selected three feature combination sets for further model building: top 6, top 11, and all features. The ranking of features based on their contribution to model performance guided our selection decision-making (Fig. 5b). Consequently, we constructed three logistic regression models with varying features. Based on the validation set evaluation, the models with decreasing number of features demonstrated AUCs of 0.88, 0.85, and 0.80, respectively (Fig. 5c). Therefore, considering the balance of performance, simplicity, and practicality, we selected the logistic regression model with 11 features for subsequent use. This model, presented as a nomogram, includes features such as tumour differentiation, CRC site, pathological type, perineural invasion, family cancer history, CEA level, early onset, and vascular invasion. Patients whose scores exceed the threshold of 0.5 on our model are recommended to proceed with MSI testing (Fig. 5d).
Fig. 5.
Development of a Nomogram for Identifying Potential MSI-H cases within the pMMR results. a, A scatter plot illustrating the relationship between the number of features and model performance. b, Distribution of feature rankings based on their contribution to model performance. c, AUC performance evaluation of logistic regression models constructed with different feature selections. d, Nomogram constructed from the top 11 feature logistic regression model.
Discussion
The treatment landscape for CRC has transitioned into the era of immunotherapy, dMMR and MSI have been endorsed as paramount biomarkers for deploying PD-1 blockade and other immunotherapeutic strategies.7,8 MSI-H serves not only as the hallmark but also the consequence of dMMR, and has traditionally been recommended for tumours exhibiting dMMR. Multiple studies have underscored the sensitivity of MSI-H and dMMR as predictive biomarkers for the efficacy of immunotherapies.9, 10, 11,30 Therefore, the indication of MSI testing has been much expanded, and a negligible proportion of MSI-H CRCs were identified in pMMR cases.
In this study, we discerned that pMMR/MSI-H CRC was a distinct group of CRCs that manifested heterogeneity in clinicopathologic features and long-term prognostic outcomes. Predominantly, these patients exhibited mucinous and poorly differentiated tumours, presented with multiple primary CRCs, and had a familial cancer history. Compared with pMMR/MSS tumours, those pMMR/MSI-H CRC have significantly better prognostic outcomes, which were compatible with patients with dMMR/MSI-H. Combining WES and RNA sequencing, pathogenic deleterious in MSH3 were commonly observed, with some samples harbouring mutations in MSH3 alone and others exhibiting co-occurrence with mutations in other MMR genes, lending as another potential molecular mechanism for MSI-H. At last, an MSI-H prediction model was constructed for screening on patients with pMMR tumour. Both the highlighted clinical and molecular biomarkers, coupled with our predictive model, promise to streamline the identification of potential MSI-H.
As IHC is widely accessible, it has been regularly implemented in pathological analysis for CRC tumours. MSI testing is typically conducted via PCR. Discrepancies between these two methods can arise due to lower tumour purity, as reported in previous studies,14,15 and as observed in our research. These findings implicated the importance of rigorous quality control for the samples before confirming MSI-H status.
Except for methodological bias, a notable proportion of MSI-H exists within pMMR tumours. Upon examining clinical features, we found that these CRC tumours exhibited similar characteristics of LS-associated CRCs, such as a higher proportion of poorly differentiated and mucinous adenocarcinoma.31 These observations suggested that the phenotypes of MSI-H were approximately identical regardless of the causes of MSI-H. Thus, MSI-H may be derived from strong driving factors, such as dMMR, which results in homogenized characteristics. Given that MSI-H indicates specific features within the tumour microenvironment, influencing therapeutic choices in the clinic, we also analysed the immune microenvironment using transcriptome data. The pMMR/MSI-H CRCs displayed an elevated presence of CD8+ T cells and NK cells, suggesting an active tumour immune response, as reported in dMMR/MSI-H tumours.32 Therefore, our findings imply that MSI-H may serve as a more encompassing and sensitive biomarker for immunotherapies compared to dMMR alone. In addition, the distinctive tumour immune environment could potentially hinder tumour progression, leading to fewer instances of cancerous nodes, vascular invasion, perineural invasion, and metastasis, which in turn could translate to higher PFS and OS rates.
To elucidate the underlying mechanisms of pMMR/MSI-H CRCs, we conducted paired normal-tumour WES for these tumours. Mutational signature analysis pinpointed MMR deficiencies in all 35 patients with pMMR/MSI-H, establishing that the dMMR status is the predominant contributor to MSI-H. Thus, we conducted a systematic analysis of MMR gene mutation in tumours, based on DNA and RNA sequencing results. Pathogenic deleterious mutations in MMR genes were detected in at least 14 patients. Additionally, 3 patients exhibited MLH1 hypermethylation and 2 patients exhibited unique somatic MSH3 mutations (MSH3-K383fs). We discerned a subset of pMMR/MSI-H samples harbouring missense mutations in MMR genes, which might result in function impairment of the MMR proteins without affecting expression levels. Meanwhile, frameshift mutations in MSH6 at the transcript level were frequently observed in our cohort, possibly arising from post-transcriptional editing or RNA modifications. While the mutated MSH6 transcripts can still be translated into proteins, their functional integrity is compromised, inducing MSI-H. Therefore, it's imperative to screen for both somatic and germline mutations of MMR genes, given that somatic mutations can equally precipitate MSI-H.
Some samples exhibited low VAF for somatic MMR gene mutations, indicating a minority presence of dMMR tumour cells. This observation can be ascribed to tumour heterogeneity and potential shortcomings in IHC procedures. In a previous study conducted at our centre, our researchers reported several cases of pMMR/MSI-H, in which tumour heterogeneity was observed within the intratumoral region.33 In this study, tumour heterogeneity of MMR protein expression was also observed. In these cases, a portion of glands exhibited dMMR, with MLH1/PMS2 or MSH2/MSH6 proteins being simultaneously absent in the same region. This finding indicates that tumour heterogenous MMR protein expression should also be considered when MSI-H is detected in pMMR tumours. In another subset of samples, despite the absence of recognisable pathogenic mutations in MMR genes, there was diminished expression of these genes. This hints at the role of expression regulation mechanisms disrupting DNA mismatch repair signalling. Such regulatory mechanisms might encompass heightened methylation heterogeneity, silencer elements, and mutations within promoters.
The MSH3 is a DNA MMR gene implicated in tumorigenesis of colon cancer exhibiting MSI-H.34 Certain research links biallelic germline mutations in MSH3 to a recessive subtype of colorectal adenomatous polyposis.35 In our study, pathogenic deleterious in MSH3 were commonly observed, with some samples harbouring mutations in MSH3 alone and co-occurrence with mutations in other MMR genes. Previous studies have reported that homozygous loss of MSH3 can predispose individuals to CRC and MSI-H status.35, 36, 37 While the association between heterozygous loss of MSH3 and MSI-H status in human tumours has not been well studied, our study provides clinical validation of the association between heterozygous loss of MSH3 and MSI. Even if the MSH3 mutation is incidental, its potential as a biomarker of MSI cannot be overlooked, given its pronounced mutation frequency in MSI-H CRCs.
Finally, we developed a prediction model for MSI-H using clinical and pathological characteristics. Importantly, the aim of the prediction model was not to replace MSI testing, nor was it capable of taking it instead. Instead, its outcomes offer a guideline for MSI-H screening, directing clinicians' attention to key risk factors of pMMR/MSI-H CRCs. Even in cases where IHC indicates pMMR, MSI-H testing may be recommended for those identified as high-risk by the prediction model.
There are still some limitations to consider. Firstly, we did not perform MLH1 methylation testing. Despite the known accuracy constraints of methylation testing, and its potential inability to discern subclonal MLH1 hypermethylation, its absence might lead to an underestimation of MLH1 methylation heterogeneity. Secondly, the intrinsic limitations of WES impeded a detailed evaluation of copy number variations, possibly underestimation of MMR gene copy number variations. At last, the study of the immune microenvironment was based on RNA-seq data, incorporating dMMR/MSI data from TCGA for comparison. On one hand, the estimation of immune cell composition using RNA-seq might be less accurate and not as reliable as IHC for detecting immune markers. On the other hand, inherent systematic variations in TCGA-COAD dataset may introduce biases, potentially challenge the robustness of our immune microenvironment research.
In conclusion, we highlight the relatively increased prevalence of MSI-H in pMMR CRCs. This study revealed pMMR/MSI-H CRC as a distinct subgroup of CRC, which manifests diverse clinicopathological features and long-term prognostic outcomes. In comparison with pMMR/MSS tumours, pMMR/MSI-H CRCs exhibited significantly better Prognosis, which were compatible with patients with dMMR/MSI-H. Distinct tumour immune-microenvironment features were found to be inherent in pMMR/MSI-H CRCs. Notably, pathogenic deleterious mutations in MSH3-K383fs were frequently detected, suggesting another potential biomarker for MSI-H. At last, an MSI prediction model was constructed for screening pMMR cases. Understanding the molecular mechanisms underlying pMMR/MSI-H may provide theoretical basis for management of immunotherapy strategies for such CRC patients.
Contributors
Conceptualization: Ye Xu, K.L., F.Q.L., S.W. and Yun Xu; Methodology: K.L., Yun Xu, C.L., and F.Q.L.; Data curation: K.L., Yun Xu, C.L., M.H.L., X.Y. Z. and M.H.S.; Investigation: K.L., Yun Xu, C.L., M.H.L., and F.Q.L.; Formal analysis: Yun Xu, K.L., F.Q.L., S.W. and Ye Xu.; Validation: Yun Xu., K.L., F.Q.L., S.W. and Ye Xu; Writing original draft: K.L. and Yun Xu; Review and editing: L.Y.Z., S.W., F.Q.L. and Ye Xu; Accessed and verified the underlying data: Ye Xu, Yun Xu and K. L.; Supervision: L.Y.Z., S.W., F.Q.L. and Ye Xu; Project administration: L.Y.Z., S.W., F.Q.L. and Ye Xu; Funding acquisition: Ye Xu; All authors have read, edited, and approved the final manuscript.
Data sharing statement
The raw sequencing data of germline reported in this paper have been deposited in the Genome Sequence Archive in the National Genomics Data Center, China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences that are temporarily controlled. That germline raw data is available from the corresponding author upon reasonable request. All somatic variant sequencing data for this study have been uploaded to the SRA and stored the corresponding MAF files for germline and somatic sequencing on GitHub (https://github.com/kyle-lk/Mismatch_between_MMR_and_MSI_in_CRC). Desensitized clinical features and codes are also available on GitHub. Ye Xu, Yun Xu and Kai Liu were responsible for accessing and verifying the underlying data associated with this study.
Declaration of interests
The authors declare no competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We acknowledge all authors participating in this study. And this study was funded by the Science and Technology Commission of Shanghai Municipality (Grant No. 20DZ1100101; Recipient of the grant: Ye Xu).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2024.105142.
Contributor Information
Sheng Wang, Email: wangs601@163.com.
Fangqi Liu, Email: liufq021@163.com.
Ye Xu, Email: yexu@shmu.edu.cn.
Appendix A. Supplementary data
The IHC expression of pMMR tumours. All four IHC images are derived from a specimen and these sections were consecutively cut from the same paraffin-embedded block. MMR proteins (hMLH1, hMSH2, hMSH6, and hPMS2) are proficiently expressed. Original magnification × 40, scale bar: 625 μm.
Workflow of functional annotation of MMR gene mutations.
Comparative analysis of tumour microenvironment characteristics in dMMR/MSI-H and pMMR/MSS data sourced from TCGA, along with our pMMR/MSS and pMMR/MSI-H data. Study compared utilising Wilcoxon test, p < 0.05 indicating significance. Sample size: TCGA-pMMR/MSS, n = 353; TCGA-dMMR/MSI-H, n = 78; FUSCC-pMMR/MSS, n = 18; FUSCC-pMMR/MSI-H, n = 35.
The IHC of low proportion of dMMR cell components within the pMMR tumours. IHC images are derived from a specimen and these sections were consecutively cut from the same paraffin-embedded block. Intratumoral regions in red boxes were identified as pMMR (MSH2/MSH6 proficiency), and regions in blue boxes were identified as dMMR (MSH2/MSH6 deficiency). Original magnification × 200, scale bar: 100 μm.
References
- 1.Sung H., Ferlay J., Siegel R.L., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Komor M.A., Bosch L.J., Bounova G., et al. Consensus molecular subtype classification of colorectal adenomas. J Pathol. 2018;246(3):266–276. doi: 10.1002/path.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guinney J., Dienstmann R., Wang X., et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21(11):1350–1356. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Stratton M.R. Exploring the genomes of cancer cells: progress and promise. Science. 2011;331(6024):1553–1558. doi: 10.1126/science.1204040. [DOI] [PubMed] [Google Scholar]
- 6.van Ginkel J., Tomlinson I., Soriano I. The evolutionary landscape of colorectal tumorigenesis: recent paradigms, models, and hypotheses. Gastroenterology. 2023;164(5):841–846. doi: 10.1053/j.gastro.2022.11.049. [DOI] [PubMed] [Google Scholar]
- 7.Amodio V., Lamba S., Chila R., et al. Genetic and pharmacological modulation of DNA mismatch repair heterogeneous tumors promotes immune surveillance. Cancer Cell. 2023;41(1):196–209.e5. doi: 10.1016/j.ccell.2022.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andre T., Shiu K.K., Kim T.W., et al. Pembrolizumab in microsatellite-instability-high advanced colorectal cancer. N Engl J Med. 2020;383(23):2207–2218. doi: 10.1056/NEJMoa2017699. [DOI] [PubMed] [Google Scholar]
- 9.Biller L.H., Schrag D. Diagnosis and treatment of metastatic colorectal cancer: a review. JAMA. 2021;325(7):669–685. doi: 10.1001/jama.2021.0106. [DOI] [PubMed] [Google Scholar]
- 10.Cervantes A., Adam R., Rosello S., et al. Metastatic colorectal cancer: ESMO clinical practice guideline for diagnosis, treatment and follow-up. Ann Oncol. 2023;34(1):10–32. doi: 10.1016/j.annonc.2022.10.003. [DOI] [PubMed] [Google Scholar]
- 11.Vikas P., Messersmith H., Compton C., et al. Mismatch repair and microsatellite instability testing for immune checkpoint inhibitor therapy: ASCO endorsement of College of American pathologists guideline. J Clin Oncol. 2023;41(10):1943–1948. doi: 10.1200/JCO.22.02462. [DOI] [PubMed] [Google Scholar]
- 12.Quintanilha J.C.F., Graf R.P., Fisher V.A., et al. Comparative effectiveness of immune checkpoint inhibitors vs chemotherapy in patients with metastatic colorectal cancer with measures of microsatellite instability, mismatch repair, or tumor mutational burden. JAMA Netw Open. 2023;6(1) doi: 10.1001/jamanetworkopen.2022.52244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sui Q., Zhang X., Chen C., et al. Inflammation promotes resistance to immune checkpoint inhibitors in high microsatellite instability colorectal cancer. Nat Commun. 2022;13(1):7316. doi: 10.1038/s41467-022-35096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Salins A.G.D., Tachon G., Cohen R., et al. Discordance between immunochemistry of mismatch repair proteins and molecular testing of microsatellite instability in colorectal cancer. Esmo Open. 2021;6(3):100120. doi: 10.1016/j.esmoop.2021.100120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shia J. Immunohistochemistry versus microsatellite instability testing for screening colorectal cancer patients at risk for hereditary nonpolyposis colorectal cancer syndrome. Part I. The utility of immunohistochemistry. J Mol Diagn. 2008;10(4):293–300. doi: 10.2353/jmoldx.2008.080031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H., Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26(5):589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiao W., Ren L., Chen Z., et al. Toward best practice in cancer mutation detection with whole-genome and whole-exome sequencing. Nat Biotechnol. 2021;39(9):1141–1150. doi: 10.1038/s41587-021-00994-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKenna A., Hanna M., Banks E., et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia-Alcalde F., Okonechnikov K., Carbonell J., et al. Qualimap: evaluating next-generation sequencing alignment data. Bioinformatics. 2012;28(20):2678–2679. doi: 10.1093/bioinformatics/bts503. [DOI] [PubMed] [Google Scholar]
- 20.Cibulskis K., Lawrence M.S., Carter S.L., et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31(3):213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim S., Scheffler K., Halpern A.L., et al. Strelka2: fast and accurate calling of germline and somatic variants. Nat Methods. 2018;15(8):591–594. doi: 10.1038/s41592-018-0051-x. [DOI] [PubMed] [Google Scholar]
- 22.McLaren W., Gil L., Hunt S.E., et al. The ensembl variant effect predictor. Genome Biol. 2016;17(1):122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019;37(8):907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frazer J., Notin P., Dias M., et al. Disease variant prediction with deep generative models of evolutionary data. Nature. 2021;599(7883):91–95. doi: 10.1038/s41586-021-04043-8. [DOI] [PubMed] [Google Scholar]
- 25.Chakravarty D., Gao J., Phillips S.M., et al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol. 2017;2017 doi: 10.1200/PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshihara K., Shahmoradgoli M., Martinez E., et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alexandrov L.B., Nik-Zainal S., Wedge D.C., Campbell P.J., Stratton M.R. Deciphering signatures of mutational processes operative in human cancer. Cell Rep. 2013;3(1):246–259. doi: 10.1016/j.celrep.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Helleday T., Eshtad S., Nik-Zainal S. Mechanisms underlying mutational signatures in human cancers. Nat Rev Genet. 2014;15(9):585–598. doi: 10.1038/nrg3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li T., Fan J., Wang B., et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77(21):e108–e110. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ganesh K., Stadler Z.K., Cercek A., et al. Immunotherapy in colorectal cancer: rationale, challenges and potential. Nat Rev Gastroenterol Hepatol. 2019;16(6):361–375. doi: 10.1038/s41575-019-0126-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sinicrope F.A. Lynch syndrome-associated colorectal cancer. N Engl J Med. 2018;379(8):764–773. doi: 10.1056/NEJMcp1714533. [DOI] [PubMed] [Google Scholar]
- 32.Hale V.L., Jeraldo P., Chen J., et al. Distinct microbes, metabolites, and ecologies define the microbiome in deficient and proficient mismatch repair colorectal cancers. Genome Med. 2018;10(1):78. doi: 10.1186/s13073-018-0586-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang J., Zhang X., Wang Q., et al. Histomorphological and molecular genetic characterization of different intratumoral regions and matched metastatic lymph nodes of colorectal cancer with heterogenous mismatch repair protein expression. J Cancer Res Clin Oncol. 2023;149(7):3423–3434. doi: 10.1007/s00432-022-04261-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ikeda M., Orimo H., Moriyama H., et al. Close correlation between mutations of E2F4 and hMSH3 genes in colorectal cancers with microsatellite instability. Cancer Res. 1998;58(4):594–598. [PubMed] [Google Scholar]
- 35.Adam R., Spier I., Zhao B., et al. Exome sequencing identifies biallelic MSH3 germline mutations as a recessive subtype of colorectal adenomatous polyposis. Am J Hum Genet. 2016;99(2):337–351. doi: 10.1016/j.ajhg.2016.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Plaschke J., Kruger S., Jeske B., et al. Loss of MSH3 protein expression is frequent in MLH1-deficient colorectal cancer and is associated with disease progression. Cancer Res. 2004;64(3):864–870. doi: 10.1158/0008-5472.can-03-2807. [DOI] [PubMed] [Google Scholar]
- 37.Villy M.C., Masliah-Planchon J., Schnitzler A., et al. MSH3: a confirmed predisposing gene for adenomatous polyposis. J Med Genet. 2023;60(12):1198–1205. doi: 10.1136/jmg-2023-109341. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The IHC expression of pMMR tumours. All four IHC images are derived from a specimen and these sections were consecutively cut from the same paraffin-embedded block. MMR proteins (hMLH1, hMSH2, hMSH6, and hPMS2) are proficiently expressed. Original magnification × 40, scale bar: 625 μm.
Workflow of functional annotation of MMR gene mutations.
Comparative analysis of tumour microenvironment characteristics in dMMR/MSI-H and pMMR/MSS data sourced from TCGA, along with our pMMR/MSS and pMMR/MSI-H data. Study compared utilising Wilcoxon test, p < 0.05 indicating significance. Sample size: TCGA-pMMR/MSS, n = 353; TCGA-dMMR/MSI-H, n = 78; FUSCC-pMMR/MSS, n = 18; FUSCC-pMMR/MSI-H, n = 35.
The IHC of low proportion of dMMR cell components within the pMMR tumours. IHC images are derived from a specimen and these sections were consecutively cut from the same paraffin-embedded block. Intratumoral regions in red boxes were identified as pMMR (MSH2/MSH6 proficiency), and regions in blue boxes were identified as dMMR (MSH2/MSH6 deficiency). Original magnification × 200, scale bar: 100 μm.