Summary
Biallelic loss-of-function variants in the MUSK gene result in two allelic disorders: (1) congenital myasthenic syndrome (CMS; OMIM: 616325), a neuromuscular disorder that has a range of severity from severe neonatal-onset weakness to mild adult-onset weakness, and (2) fetal akinesia deformation sequence (OMIM: 208150), a form of pregnancy loss characterized by severe muscle weakness in the fetus. The MUSK gene codes for muscle-specific kinase (MuSK), a receptor tyrosine kinase involved in the development of the neuromuscular junction. Here, we report a case of neonatal-onset MUSK-related CMS in a patient harboring compound heterozygous deletions in the MUSK gene, including (1) a deletion of exons 2–3 leading to an in-frame MuSK protein lacking the immunoglobulin 1 (Ig1) domain and (2) a deletion of exons 7–11 leading to an out-of-frame, truncated MuSK protein. Individual domains of the MuSK protein have been elucidated structurally; however, a complete MuSK structure generated by machine learning algorithms has clear inaccuracies. We modify a predicted AlphaFold structure and integrate previously reported domain-specific structural data to suggest a MuSK protein that dimerizes in two locations (Ig1 and the transmembrane domain). We analyze known pathogenic variants in MUSK to discover domain-specific genotype-phenotype correlations; variants that lead to a loss of protein expression, disruption of the Ig1 domain, or Dok-7 binding are associated with the most severe phenotypes. A conceptual model is provided to explain the severe phenotypes seen in Ig1 variants and the poor response of our patient to pyridostigmine.
Keywords: MUSK gene, Congenital myasthenic syndrome (CMS), Fetal akinesia deformation sequence (FADS), Neuromuscular junction (NMJ), MuSK protein structure, Domain-specific genotype-phenotype correlations, Pyridostigmine, β2-adrenergic receptor agonist, Ig1 domain, Precision medicine
MUSK loss-of-function variants lead to highly variable clinical severity, from fetal loss to adult-onset weakness. Here we present a case of neonatal-onset congenital myasthenic syndrome due to loss of the Ig1 domain. We provide an improved AlphaFold structure and suggest a protein domain-based logic to the widely variable disease onset.
Introduction
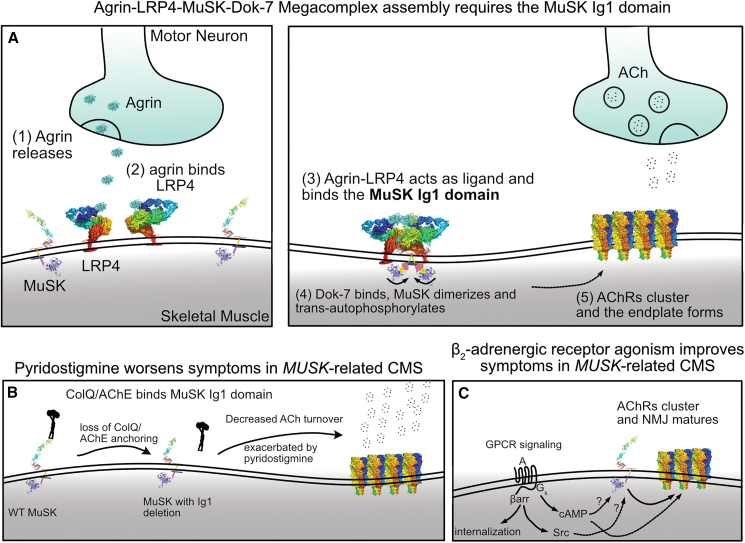
Congenital myasthenic syndrome (CMS) is a neuromuscular disorder characterized by neuromuscular junction (NMJ) abnormalities.1 CMS has been linked to variants in 35 genes and can be categorized as presynaptic, synaptic, postsynaptic, or mixed pre- and postsynaptic with glycosylation defects based on which gene is mutated.2 Muscle-specific kinase (MuSK) is a receptor tyrosine kinase located postsynaptically, and rare biallelic variants underlie MUSK-related CMS.3,4 MuSK contains seven domains.5 The first immunoglobulin domain (Ig1) allows MuSK to homodimerize when activated by its ligand agrin/low-density lipoprotein receptor-related protein 4 (LRP4) and is important for binding to a collagen-like tail subunit of asymmetric acetylcholinesterase (ColQ) and acetylcholinesterase (AChE) for clearance of ACh from the synapse.6,7 Early in development, Wnt signaling through the MuSK frizzled (Frz) domain plays a role in guiding growing motor neuron axons to muscle fibers.8 Later in development, MuSK signaling in response to neuronally released agrin bound to LRP4 leads to activation of MuSK through an agrin-LRP4-MuSK-Dok-7 megacomplex that leads to proper acetylcholine receptor (AChR) clustering and endplate formation.6,7 Intracellularly, MuSK contains a protein kinase domain, which trans-autophosphorylates for its own activation as well as phosphorylating key downstream effectors, such as Dok-7.9,10
Biallelic pathogenic loss-of-function variants in the MUSK gene, including missense variants and deletions, lead to two genetic conditions characterized by NMJ abnormalities. The first condition, fetal akinesia deformation sequence (FADS), is a form of in utero demise characterized by a sequence of defects related to decreased fetal movement.11 Movement of the fetus is important for the proper development of a number of organ systems. FADS often results in respiratory insufficiency due to pulmonary hypoplasia, skeletal muscle disease with joint contractures, and intrauterine growth retardation (IUGR). Characteristic dysmorphic features can also result due to the FADS and IUGR, including micrognathia, single palmar creases, hypertelorism, and posteriorly rotated ears.
The second disorder, CMS, presents with neuromuscular weakness ranging from neonatal- to adult-onset limb-girdle weakness or even isolated vocal cord paralysis.4 Though CMS and FADS are considered two different disease entities, both are associated with a recessive inheritance pattern and caused by biallelic loss-of-function variants; therefore, they represent a continuum of manifestations of the same disease process and can be more generically described as MUSK-related disorders.
Here, we report a case of a neonatal onset MUSK-related CMS in a patient harboring compound heterozygous MUSK deletions, including (1) an in-frame deletion of exons 2–3 leading to shortened MuSK protein lacking an Ig1 domain and (2) a deletion of exons 7–11 leading to an out-of-frame, truncated MuSK protein. We performed a literature review of known MUSK variants, identifying 24 cases of CMS3,4,12,13,14,15,16,17,18,19,20,21,22 and 21 cases of FADS.11,23,24,25 We provide a protein domain-specific genotype-phenotype correlation based on the structure and function of the MuSK protein. We find that pathogenic variants within the Ig1 domain are associated with a more severe phenotype. Finally, given that our patient experienced clinical deterioration with a short trial of pyridostigmine, an AChE inhibitor used to treat myasthenia gravis, and subsequently improved with oral albuterol, a short-acting β2 adrenergic receptor agonist, we review our understanding of current treatment paradigms and agree with current recommendations that discourage the use of cholinesterase inhibitors in MUSK-related CMS.26
Subjects and methods
Genetic testing
The genetic evaluation of the patient was performed during an inpatient consult (D.GJ. and K.G.Q.T.) and continued in an outpatient setting (B.T.C. and M.T.M.) at Duke University Hospital. Written informed consent was obtained for reporting of clinical information. Per the Duke University Health System IRB policy, the review of medical records for publication of a single case report is not considered by the IRB to be research involving human subjects, and therefore such a report of medical cases is considered exempt.
SNP microarray analysis
Copy-number variant analysis was performed on DNA derived from uncultured peripheral blood leukocytes of the patient using the Affymetrix Cytoscan HD array and Affymetrix ChAS software 4.3 (Thermo Fisher Scientific).
NextGen sequencing panel
Genomic DNA was isolated from peripheral blood leukocytes by the Duke Molecular and Cytogenetics Lab and then submitted to Invitae (San Francisco, CA, USA) for Illumina next-generation sequencing technology to interrogate 143 genes associated with neuromuscular disorders. Both MUSK deletions identified by NGS were subsequently confirmed by array comparative genomic hybridization (MUSK transcript NM_005592.3.). Parental testing was performed to confirm the phase of the variants.
Protein modeling
The MuSK protein structure was predicted with AlphaFold and obtained from the Membranome 3.0 database.27,28,29,30 Custom adjustments to the structure were made in pymol to present it in a more biologically plausible orientation (The PyMOL Molecular Graphics System, v.2.0 Schrödinger). The following structures were obtained from the Protein Data Bank: dimer of Ig1 and Ig2 domains (PDB: 2EIP),31 Frz-like cysteine rich domain (PDB: 3HKL),6 unphosphorylated, autoinhibited tyrosine kinase (PDB: 1LUF),9 and peptide fragment with DOK7 (PDB: 3ML4).10 The AlphaFold structure (MUSK_HUMAN) was obtained from Membranome 3.0 (ID MUSK_HUMAN) as a base.29,30 Custom alignment in pymol was performed to generate models using existing structures as an anchor point. Two models were developed. In model 1, dimerization was modeled in both the Ig1 and transmembrane domain using the TMDOCK model. In model 2, dimerization was modeled in the Ig1 domain alone using prior crystal structures of the Ig1 and Ig2 domains as a starting point. Given the physical constraints required for kinase trans-autophosphorylation (i.e., the need for the two kinase domains to be close enough in physical space to rotate and phosphorylate one another), the model with the transmembrane domains homodimerizing was selected as the most plausible structure (though further experimental validation is warranted).
Results
Case presentation
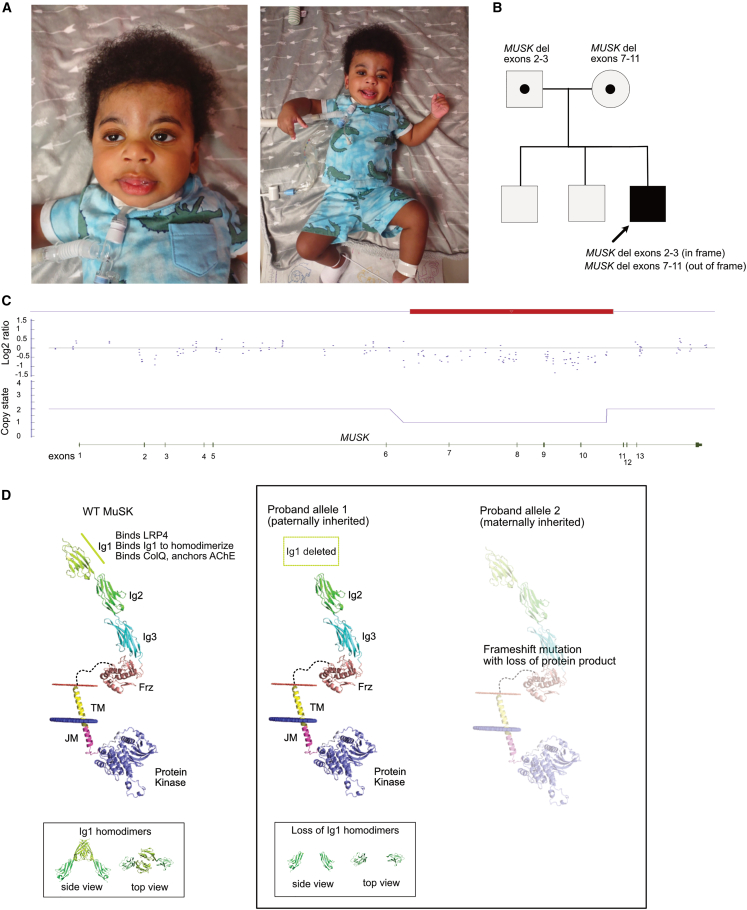
The patient was born at 38 weeks gestational age and presented at birth with generalized hypotonia, bilateral ptosis, muscle weakness, and respiratory failure requiring neonatal intensive care. There were no prenatal concerns, and he was born symmetric and appropriate for gestational age (birth weight: 3.17 kg [44th percentile]; head circumference: 35 cm [71st percentile]; length: 53 cm [92nd percentile]). APGAR scores were 2 (1 min) and 7 (5 min), and he required intubation at the 15 h of life for respiratory insufficiency and acidosis. Spinal muscular atrophy testing was negative both on the North Carolina newborn screen and on follow-up testing. An SNP chromosomal microarray revealed (1) a 104 kb duplication of 17q25.3 with a breakpoint within the SEPT9 gene with chromosomal coordinates chr17:75,447,984–75,551,771 (hg build 19) and (2) an interstitial deletion of 43 kilobases on 9p31.3 involving MUSK with chromosomal coordinates chr9:113,501,707–113,544,998 (hg build 19). Duplications and missense variants in SEPT9 have been associated with autosomal dominant hereditary neuralgic amyotrophy, characterized by severe upper extremity neuropathic pain and dysmorphic features (hypotelorism, cleft palate, blepharophimosis, and unusual skin fold creases) not observed in our patient.32,33,34,35 Our patient did exhibit trigonocephaly (Figure 1).
Figure 1.
Loss of the Ig1 domain leads to neonatal-onset MUSK-related CMS
(A) Clinical photographs taken at 10 months of age show ptosis and myopathic facies, which are typical of congenital myasthenic syndrome. He has low tone in his mouth, but he does not have macroglossia. Additionally, he has evidence of trigonocephaly, which is not a typical dysmorphic feature of MUSK-related CMS and may be related to a separate unidentified genetic or environmental modifier. At 10 months of age, he exhibited severe hypotonia and was unable to hold his head unassisted for more than a few seconds. Antigravity movements of his extremities were present.
(B) The patient inherited two intragenic deletions in MUSK in trans.
(C) SNP chromosome microarray analysis revealed an intronic deletion encompassing exons 7–11. The smaller loss of material encompassing exons 2–3 was below detection for his initial microarray, although in retrospect, there are probes in this region with signal intensity suggesting a deletion.
(D) The paternally inherited allele with the in-frame deletion is predicted to result in a MuSK protein with the Ig1 domain deleted but the remainder of the protein intact. His maternally inherited allele is an out-of-frame deletion predicted to result in loss of protein expression.
A neuromuscular gene panel covering 143 genes was performed (Invitae Comprehensive Neuromuscular Disorders Panel). The gene panel confirmed the pathogenic MUSK deletion detected on microarray, providing better resolution of exons involved. The panel also revealed a second smaller pathogenic deletion involving exons 2–3 of MUSK, which was below the SNP microarray detectable range. Subsequent parental testing demonstrated that each parent was heterozygous for one of the deletions, thus confirming a diagnosis of MUSK-related CMS. In addition to both deletions, a paternally inherited missense variant in MUSK was also observed (c.667G>A; p.Val223Ile), which is obligated to be present on the paternally inherited allele with the deletions of exons 2–3. Variants of uncertain significance in genes CHRND, HNRNPDL, and SPEG and a pseudodeficiency allele in GAA were observed (Table S1).
While the patient was in the neonatal intensive care unit after the diagnosis of CMS, a trial of pyridostigmine was performed at an oral dose of 1 mg/kg every 6 h for 2 days. The patient experienced acute worsening with bradycardia, respiratory distress, and increased secretions while receiving pyridostigmine, and the medication was discontinued. Two days following this pyridostigmine trial, the patient was noted to have a new-onset choreiform movements. An electroencephalogram revealed generalized slowing but no epileptiform activity, and neuroimaging (brain MRI) was normal. His choreiform movements ceased, and he returned to normal baseline movements 3 days after they were first noted.
Oral albuterol therapy was then initiated at 1 mg/kg every 8 h. Improvement in strength and tone were noted with decreased ventilator settings, and strength improved from 3/5 (antigravity) to 4/5 (strength against light resistance) on the MRC scale in bilateral upper and lower extremities. Ptosis improved, though it was still present, and proximal muscle strength improved from 3/5 to 4/5. As albuterol was well tolerated, it was increased to a maintenance dose of 2 mg/kg every 8 h.
The patient was 10 months of age at his last evaluation in the Medical Genetics clinic (Figure 1). At that time, he was showing normal cognitive development. He continued to have significant weakness, including considerable head lag, but he was able to sit unassisted for a few seconds when placed in a seated position. He remains on 24 h mechanical ventilation for respiratory support but is currently working toward weaning down his ventilatory needs.
Structural modeling of MuSK using previous experimental and predicted structures
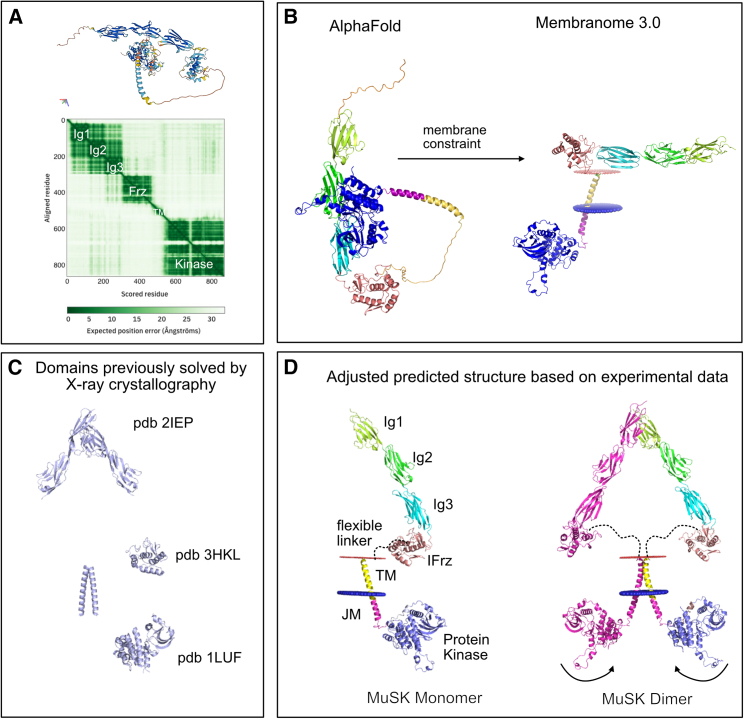
X-ray crystallography data exist for the MuSK Ig1 and Ig2 domain (PDB: 2IEP), the Frz-like domain (PDB: 3HKL), the kinase domain (PDB: 1LUF), and a 2:2 complex between a portion of the juxtamembrane domain of MuSK and Dok7 (PDB: 3ML4).6,9,10,31 An experimental structure for the complete protein has not been solved by X-ray crystallography or cryoelectron microscopy (cryo-EM). An in-silico-predicted model generated by AlphaFold exists, but this structure does not consider the presence of the membrane, which separates the intracellular and extracellular portions of the protein (Figure 2).27,28 The AlphaFold structure loops the extracellular Ig domains adjacent to the intracellular kinase domain (Figure 2A). Membranome provides transmembrane information to AlphaFold structures to improve structure prediction, but this automated tool does not consider additional experimental evidence for individual proteins.29,30 For the MuSK structure in the Membranome 3.0 database, the linker region is removed, and the extracellular domain is tilted horizontally along the membrane, which would not allow for homodimerization of the Ig1 domain or the transmembrane domain or trans-autophosphorylation of the kinase domain (Figure 2B).
Figure 2.
Modeling the structure of MuSK using experimental and predicted data suggests dimerization occurs at both the Ig1 domain and the transmembrane domain
(A) The MUSK_HUMAN structure from the AlphaFold database (https://alphafold.ebi.ac.uk/entry/O15146) has a biologically unlikely orientation with the intracellular kinase domain interacting closely with the extracellular Ig2 domain.
(B) Membranome removes disordered regions and forces a membrane constraint to correctly separate the intracellular and extracellular domains; however, without the flexible linker, which is present between the extracellular domain and transmembrane domain, the extracellular portion of the protein is positioned horizontally along the membrane, which would not allow for Ig1 dimerization concurrently with kinase trans-autophosphorylation.
(C) Previously solved domains by X-ray crystallography were obtained from the RCSB Protein Data Bank (https://rcsb.org).
(D) Allowing for the flexible linker between the Frz-like and transmembrane domain to modify the position of the extracellular domain allows for an orientation of the MuSK protein that is biologically plausible and allows for dimerization at the Ig1 domain, within the transmembrane domain, and within reach for trans-autophosphorylation of the kinase domains.
To overcome the challenges, we combined available information from crystallography, functional data, and the AlphaFold model to generate an improved predicted structural representation of the MuSK protein (Figure 2). This structure aligns previously solved domains into a logical structure that is able to homodimerize at both the Ig1 and transmembrane domains. The Ig1 domains adopt the orientation observed in the previously solved structure,31 and the positioning of the transmembrane domain is consistent with reports that replacement of the transmembrane domain with a constitutively homodimerizing form led to constitutive clustering and MuSK activation.36 The flexible linker between the transmembrane and the Frz-like domain has sufficient length for the structure to be energetically plausible. The kinase domains also have sufficient flexibility to turn inwards to trans-autophosphorylate upon agrin/LRP4 activation.
Two regions of the protein, the disordered linker regions and the Dok-7-binding juxtamembrane domain, remain poorly resolved and require further refinement; this is typical of disordered regions in AlphaFold-predicted structures.37 This juxtamembrane domain that connects the transmembrane domain with the kinase domain has been shown to be involved in Dok-7 binding and is in a region of the AlphaFold structure that has “low” to “confident” ratings (between 50 and 90 per-residue confidence score predicted local distance difference test [pLDDT]). This low confidence is consistent with a degree of flexibility of the linker region to accommodate confirmations with binding partners.
Genotype-phenotype correlation suggests domain-specific symptom severity
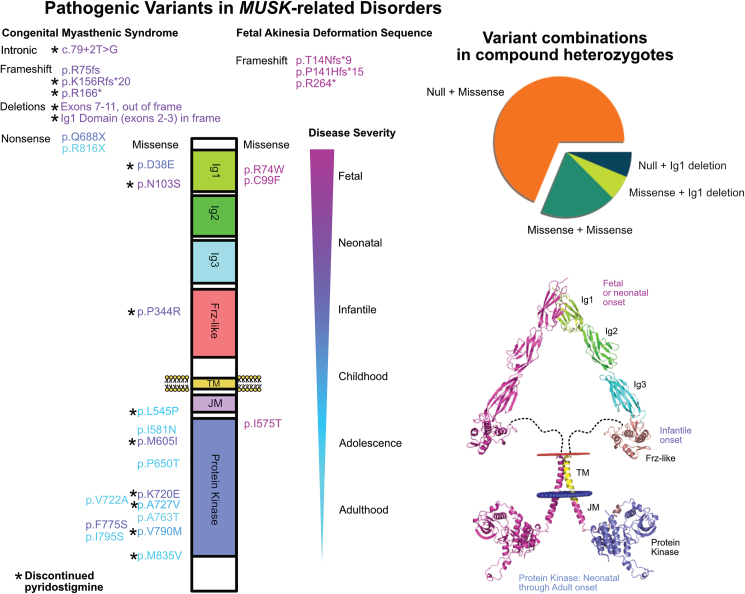
The primarily expressed isoform of MuSK is composed of seven domains and one flexible, disordered linker region (Figure 3). Extracellularly, MuSK is composed of three Ig-like domains (Ig1, Ig2, and Ig3), a Frz-like cysteine rich domain, and a flexible disordered linker region connecting the extracellular domains to the transmembrane domain.38 MuSK contains a single-pass transmembrane domain. The intracellular domain is composed of a short juxtamembrane domain with one phosphorylated residue important for DOK7 interactions and a protein kinase domain.5,9,39
Figure 3.
Missense pathogenic variants in the MUSK gene are most severe in the Ig1 domain and most frequently found in the kinase domain
Pathogenic variants that have been observed in patients to date are plotted along their position within the MuSK protein and separated by CMS (on the left) and FADS (on the right). Variant color is given based the disease severity scale from fetal (purple) to adulthood (cyan) onset of symptoms. Null alleles consistently resulted in the most severe phenotype. For missense variants, those that impact the extracellular domain (Ig1 or Frz-like) were associated with earlier onset of symptoms. Variants were most frequent in the kinase domain (which is also the largest domain). The kinase domain variants had the largest range of disease onset. Variants that were observed in patients who trialed and discontinued pyridostigmine are marked with an asterisk (∗); all patients with extracellular missense variants discontinued pyridostigmine, but so did many patients with kinase variants. The most frequent combination of variants in compound heterozygotes involves one null allele and one missense allele.
We reviewed the literature for previously reported variants in MUSK. Forty-six patients, including the case we present here, with twenty-nine unique disease-related alleles were observed3,4,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25 (Table S2). The variants appeared in four of the seven MuSK domains, including the Ig1, Frz-like, juxtamembrane, and kinase domains (Figure 3). Variants leading to loss of the full-length MuSK protein (nonsense, frameshift, and large deletions) represented 62% of the alleles, and 38% were missense. Homozygous loss-of-function variants resulted in the most severe phenotype (FADS). Missense variants were frequently observed in compound heterozygosity with a loss-of-function allele (55% of unique disease combinations). The patient reported in this study is unique in his combination of a loss-of-function allele and an allele with a deletion of the Ig1 domain.
The Ig1 domain performs multiple functions integral to known MuSK function, including binding between MuSK monomers for homodimerization and binding to the AChE COLQ to anchor AChE in the NMJ.6,7 Four missense variants were observed in the Ig1 domain; two resulted in FADS, and two were observed in neonatal/infantile-onset CMS. Complete loss of the Ig1 domain through deletions involving exons 2–3 has been previously reported. The previous case reported with a deletion of exons 2–3 had a second allele with a p.D38E variant also impacting the Ig1 domain. Of note, our patient’s deletion of exons 2–3 was inherited from an unaffected parent, providing evidence that the variant does not have a dominant-negative effect.
The p.N103S allele was observed both in a homozygous state and in heterozygosity with a null allele.4,15,40,41 The allele frequency of the p.N103S allele is 2.41e−5 on gnomAD and is present in Latino/admixed American and European/non-Finnish populations. The variant was also described in a Chinese family.40 The only described individual that is heterozygous for a loss-of-function variant plus the p.N103S variant presented with neonatal-onset CMS. Of the four patients reported so far with homozygous p.N103S variants, significant variability has been observed.40 One presented in the neonatal period, one in childhood, one at 13 years of age, and one at 27 years of age. Apart from the 27-year-old who was not on treatment, all the patients responded poorly to AChE inhibitor therapy and improved with a β2-receptor agonist.40
One missense variant (p.P344R) was observed in the Frz-like domain. The variant presented in a homozygous state in a patient with infantile-onset CMS. The Frz-like domain interacts with Wnt morphogens, and recent work in a Frz-like-domain knockout mouse revealed defects in muscle prepatterning and NMJ synapse differentiation.42
The intracellular domain is comprised of a juxtamembrane domain, which contains a critical phosphorylated residue (Y554) within an NXPY binding site43 (note that amino acid numbering has been adjusted since the original publication from Y553 to Y554). Phosphorylation at Y554 following agrin/LRP4 activation leads to Dok-7 binding and ultimately AChR clustering. No pathogenic variants were observed at this residue, but functional data have shown loss of AChR clustering that we predict would cause disease if observed in patients.
Two phosphorylated regions involving five tyrosine residues are involved in agrin/LRP4-mediated activation of the MuSK kinase.43,44 Y554 and Y577 are found in, and adjacent to, the juxtamembrane domain. The activation loop within the tyrosine kinase is composed of Y751, Y755, and Y756. Interestingly, the most severe missense homozygous variant, which resulted in FADS, results in the substitution of threonine, a residue that can also be phosphorylated two amino acids upstream of the typically phosphorylated Y577 residue (Figure 4).11 Functional data from fetal tissue and myocytes with the p.I575T variant show loss of MuSK autoactivation, loss of kinase activity, and loss of AChR clustering.11 The mechanism of this substitution has not been completely elucidated; whether the severity of this change is related to improper phosphorylation of the substituted threonine or loss of normal phosphorylation of the Y577 or Y554 residues remains an open question. Rare missense variants at amino acid position 575 appear in gnomAD at a low frequency. p.I575V appears at an allele frequency of 3.1e−5 in the European (non-Finnish) population data and p.I575N at an allele frequency of 6.4e−5 in the African/African American population. Variants resulting in substitutions at any of the five phosphorylated residues are not present in gnomAD but would presumably be deleterious.
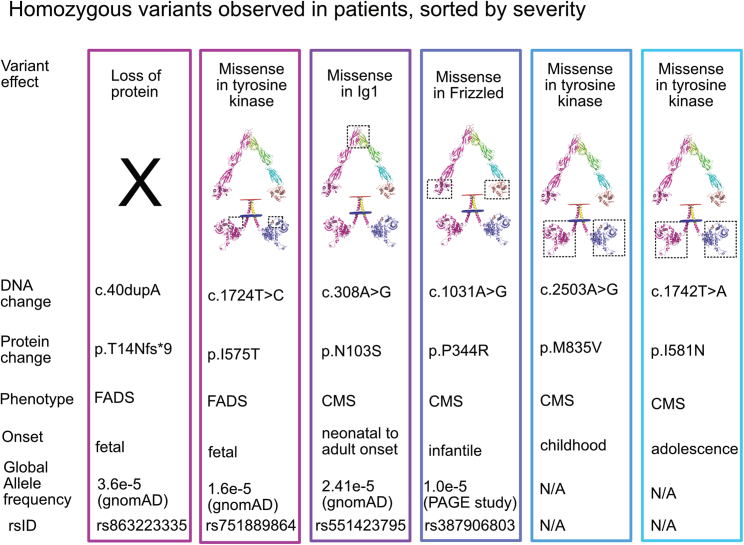
Figure 4.
Homozygous MUSK variants previously reported in patients
Six variants have been observed in a homozygous state. Two of these variants result in FADS (c.40dupA and c.1724T>C). The c.40dupA allele leads to a complete loss of protein expression. It is the only combination that leads to a complete loss of protein expression observed in patients to date, though presumably, any other combination that leads to complete loss of MuSK will be lethal in utero. The c.1724T>C allele leads to a p.I575T missense variant. This variant leads to the placement of a residue that is capable of being phosphorylated within two amino acids of the prototypically phosphorylated tyrosine at position 577. Further experimental validation of the p.I575T variant is needed to determine if it alters phosphorylation at Y577 or leads to disease through another mechanism. The remaining four variants result in CMS and are located within the Ig1, Frz-like, and kinase domains. The p.N103S variant is the most frequent of the homozygous variants and results in a range of symptom severity, from neonatal onset to adult onset.
The tyrosine kinase domain, which is the largest MuSk domain, contained the most pathogenic variants (12/18 missense alleles observed). Except for the p.I575T variant described above, no other kinase variants were observed in FADS. The kinase variants were spread along the length of the domain (Figure 3). Two variants, p.I581N and p.M835V, were observed in a homozygous state (Figure 4). These variants resulted in less severe disease with childhood or adolescent onset of symptoms. The remaining missense kinase variants appeared as compound heterozygous with the second allele containing a loss-of-function variant (7/10) or separate missense variants in the kinase domain (3/10) (Figure 5).
Figure 5.
Conceptual models suggest the importance of the Ig1 domain for MuSK protein function
(A) The formation of the agrin-LRP4-MuSK-Dok-7 megacomplex occurs stepwise. Unlike most receptor tyrosine kinases, which bind directly to a ligand, agrin first binds LRP4, and the agrin-LRP4 complex serves as a ligand for MuSK. Following agrin-LRP4-MuSK binding on the extracellular side, Dok-7 binds intracellularly, leading to MuSK activation by trans-autophosphorylation (i.e., the kinase domains from opposite MuSK monomers phosphorylate each other). Activated MuSK phosphorylates downstream targets to result in AChR clustering and endplate formation.
(B) The Ig1 domain is the site of binding for ColQ, allowing for proper localization of AChE in the synapse. This suggests a model to explain why our patient experienced severe worsening in the setting of a trial of pyridostigmine (an AChE inhibitor).
(C) Systemic β-agonist therapy is the current mainstay of therapy, though the mechanism of signaling through the β-adrenergic receptor is not completely known. Whether G-protein signaling, β-arrestin signaling, or both are involved is an open question.
Compound heterozygous variants with one kinase missense change and a loss-of-function variant resulted in neonatal (4/7 observations) or congenital vocal cord paralysis and/or adult-onset limb-girdle weakness (3/7 observations). The p.V790M variant in compound heterozygosity with a loss-of-function allele was observed in two separate cases of neonatal-onset CMS.3,17 Reminiscent of observations in patients, a knockin mouse model of the p.V790M variant (p.V789M in the mouse) recapitulated the CMS phenotype when the p.V790M variant was heterozygous with a loss-of-function allele.45 No phenotype was observed in the p.V790M homozygous state. Consistent with its frequent appearance in our cohort, p.V790M is found in almost all populations in gnomAD and appears at a global allele frequency of 2.10e−4, making it the most common allele among pathogenic or likely pathogenic missense variants.
Discussion
Here, we report a case of neonatal MUSK-related CMS and review previously reported cases of MUSK-related disease. We use the atlas of cases developed here to extract correlations between the genotype and phenotype and propose that grouping cases by the impacted protein domain(s) predicts disease severity. Combining functional evidence with machine-learning-based predictions and experimental structural data, we develop a complete MuSK structure for illustrative purposes. We use this structure to highlight domain-specific changes among previously reported cases.
Our patient’s combination of genetic changes is unique for cases of MUSK-related disease described to date. Previously reported cases either resulted in absent MuSK protein expression (typically resulting in FADS) or MuSK expression with a single amino acid change. One previous case presented with one allele producing a MuSK protein with a missense change and the other allele producing MuSK with a loss of the Ig1 domain.14 In our patient, the only MuSK protein present is a form with the Ig1 domain missing. Given the importance of the Ig1 domain to core MuSK functions—dimerization, agrin/LRP4 response, and AChE anchoring—it is not surprising that our patient presented with severe symptoms.6,7 One limitation of this study is the use of SNP microarray and next-generation sequencing to characterize our patient’s deletions without further confirmation by RNA and western blotting methods. Further experimental investigation of this Ig1-deficient variant is warranted to determine its impact on cell signaling.
Protein structure determination is key to our mechanistic understanding of protein function. X-ray crystallography and cryo-EM are mainstays to solving protein structures, but in recent years, computational modeling has shown increasing promise in successful prediction.27,28 The model of MuSK proposed here was informed by both computational models and the prior crystallographic determination of Ig1 domains and the juxtamembrane domain interaction with Dok-7.6,9,10,31 Our model integrates disparate sources of data, and additional experimental data are important to validate these predictions. Tools for refining protein models like extensive molecular dynamics simulation, as recently done for the tyrosine-protein kinase KIT, could be a direction of future research to better understand the conformations and dynamics of the linker and other low-confidence and flexible regions.46
As with prior reports of postsynaptic CMS, our patient had a poor response to a trial of the AChE-inhibiting medication pyridostigmine. By reviewing cases of MUSK-related CMS with variants affecting the Ig1 domain, we find that they all have exhibited a poor response to pyridostigmine (this paper and Gallenmüller et al.14). Given the role of Ig1 in anchoring AChE to the NMJ, it would be informative to investigate AChE localization in patients with Ig1 variants. Regardless of the mechanism, we further highlight existing recommendations that pyridostigmine should not be used in patients with MUSK-related CMS,2 particularly when their variant impacts the Ig1 domain.
Our patient exhibited a good response to therapy with an oral β2-adrenergic receptor agonist. Previous reports suggested that β2-adrenergic receptor agonists appeared to have the greatest therapeutic effect for variants that disrupt kinase function4; however, our patient does not fit that pattern, suggesting that a β2-adrenergic receptor agonist may be a reasonable first-line therapy for all MUSK-related CMS.2
The mechanism by which activation of the β2-adrenergic receptor, a prototypical G-protein-coupled receptor (GPCR), improves symptoms in MUSK-related CMS, a disorder of a receptor tyrosine kinase, is not clear. GPCR signaling occurs both through G proteins and β-arrestins. Activation of the Gs alpha subunit, for example, increases cAMP, which may directly or indirectly activate MuSK or may lead through a separate pathway to MuSK effector activation (Figure 5C). β-Arrestin signaling has been shown to activate the non-receptor tyrosine kinase Src,47 and activated Src has been shown to act within the MuSK/agrin pathway for AChR clustering.48 Further elucidation of this pathway is important, as biased agonists for G-protein or β-arrestin activation exist that may form the basis of a more effective precision medicine approach to therapy.
Recently, agonist antibodies directed against MuSK have been proposed as a potential therapy for postsynaptic CMS.49 The treatment showed promise in a mutant Dok-7 mouse model of CMS. The antibodies target the Frz-like domain of MuSK to induce MuSK activity, proposed to induce MuSK dimerization independent of Dok-7 or other adaptor proteins. However, the exact mechanism of MuSK activation by these antibodies is unclear. In a separate study it was shown that full-length antibody could induce MuSK activation and AChR clustering, while a fragment antigen binding region was insufficient, leading them to suggest that dimerization of MuSK was required for antibody activity.50 In contrast, previous literature found that single-chain variable fragments (scFVs) against MuSK could induce MuSK activation, but the authors could not rule out the possibility of scFV dimers.51 As one of our patient’s MuSK variants contains a functional Frz-like domain and a functional kinase domain, he would likely be an excellent candidate for such a therapy.
A review of cases here provides some logic to the variant profile typically observed in MUSK-related disorders. Twenty-one variant combinations were observed, with 28% occurring in homozygosity. Of the homozygous variants, only one, to our knowledge, leads to complete loss of protein expression (c.40dupA). We hypothesize that any other allele combination that leads to complete loss of protein expression will result in FADS. Of the compound heterozygous variants, 73% occurred with one allele that resulted in complete loss of protein expression (frameshift, nonsense, or out-of-frame deletion) and the other allele with a missense variant. This suggests a mild deleterious effect of haploinsufficiency that is further worsened by a loss-of-function missense variant in compound heterozygous cases. Further evidence for this pattern of inheritance comes from a mouse model of the p.V790M variant, which does not have a phenotype in the p.V790 homozygous state but has a striking neuromuscular phenotype in the compound heterozygous setting of one p.V790 allele and one MUSK-deleted allele.45
Conclusion
In conclusion, we provide the detailed clinical phenotype of a case of MUSK-related CMS in the setting of a MuSK protein lacking the Ig1 domain. Our patient presented with severe neonatal respiratory distress and a poor response to pyridostigmine. Therapy with a systemic β2-adrenergic receptor agonist has been beneficial, though the mechanism of action remains unclear. We use previous structural data to improve machine-learning-predicted models of the MuSK protein. We review prior reports to search for general rules, which may assist with variant classification and prediction. We find that the Ig1 and kinase domains were the most frequently impacted by pathogenic missense variants and that most cases of MUSK-related CMS were in compound heterozygous individuals with one null allele and one missense allele. Treatments that increase expression of a functional allele or act downstream of the LRP4/MUSK/agrin/Dok-7 pathway may prove efficacious.
Data and code availability
The MUSK AlphaFold2-generated structure is available for download from the AlphaFold Protein Structure Database (https://alphafold.ebi.ac.uk/entry/O15146). The Membranome transmembrane-corrected structure is available from the Membranome database (https://membranome.org/proteins/2668).
Acknowledgments
We wish to thank the patient reported here and his family for their participation in this work. Research reported in this publication was supported by National Heart, Lung, and Blood Institute (NHLBI) of the National Institutes of Health under award number 1R38HL143612 and the Duke Resident Physician-Scientist Program to B.T.C.
Author contributions
Conceptualization, B.T.C. and M.T.M.; methodology, B.T.C., S.W.L., and L.F.; software, S.W.L.; formal analysis, B.T.C., S.W.L., L.F., and K.L.D.; resources, B.T.C., D.G.J., C.D.H., M.K.E., C.E.P., E.C.S., K.G.Q.T., and M.T.M.; data curation, B.T.C., S.W.L., and A.B.; writing – original draft, B.T.C., S.W.L., and L.F.; writing – review and editing, B.T.C., S.W.L., L.F., A.B., D.G.J., M.A., K.L.D., C.H.D., M.K.E., C.E.P., E.C.S., K.G.Q.T., and M.T.M.; visualization, B.T.C., S.W.L., and L.F.; supervision, M.T.M.; project administration, B.T.C.; funding acquisition, B.T.C. and M.T.M.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xhgg.2024.100288.
Contributor Information
Benjamin T. Cocanougher, Email: benjamin.cocanougher@duke.edu.
Marie T. McDonald, Email: marie.mcdonald@duke.edu.
Supplemental information
References
- 1.Vanhaesebrouck A.E., Beeson D. The congenital myasthenic syndromes: expanding genetic and phenotypic spectrums and refining treatment strategies. Curr. Opin. Neurol. 2019;32:696. doi: 10.1097/WCO.0000000000000736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abicht A., Müller J.S., Lochmüller H. Mirzaa G.M., editors, GeneReviews® [Internet] University of Washington; Seattle: 2021. Congenital Myasthenic Syndromes Overview; pp. 1993–2023.https://www.ncbi.nlm.nih.gov/books/NBK1168/ [PubMed] [Google Scholar]
- 3.Chevessier F., Faraut B., Ravel-Chapuis A., Richard P., Gaudon K., Bauche S., et al. MUSK, a new target for mutations causing congenital myasthenic syndrome. Hum. Mol. Genet. 2004;13:3229–3240. doi: 10.1093/hmg/ddh333. [DOI] [PubMed] [Google Scholar]
- 4.Owen D., Töpf A., Preethish-Kumar V., Lorenzoni P.J., Vroling B., Scola R.H., et al. Recessive variants of MuSK are associated with late onset CMS and predominant limb girdle weakness. Am. J. Med. Genet. 2018;176(7):1594–1601. doi: 10.1002/ajmg.a.38707. [DOI] [PubMed] [Google Scholar]
- 5.Valenzuela D.M., Stitt T.N., DiStefano P.S., Rojas E., Mattsson K., Compton D.L., et al. Receptor tyrosine kinase specific for the skeletal muscle lineage: expression in embryonic muscle, at the neuromuscular junction, and after injury. Neuron. 1995;15:573–584. doi: 10.1016/0896-6273(95)90146-9. [DOI] [PubMed] [Google Scholar]
- 6.Stiegler A.L., Burden S.J., Hubbard S.R. Crystal structure of the frizzled-like cysteine-rich domain of the receptor tyrosine kinase MuSK. J. Mol. Biol. 2009;393:1–9. doi: 10.1016/j.jmb.2009.07.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang W., Coldefy A.S., Hubbard S.R., Burden S.J. Agrin binds to the N-terminal region of Lrp4 protein and stimulates association between Lrp4 and the first immunoglobulin-like domain in muscle-specific kinase (MuSK) J. Biol. Chem. 2011;286:40624–40630. doi: 10.1074/jbc.M111.279307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jing L., Lefebvre J.L., Gordon L.R., Granato M. Wnt signals organize synaptic prepattern and axon guidance through the zebrafish unplugged/MuSK receptor. Neuron. 2009;61:721–733. doi: 10.1016/j.neuron.2008.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Till J.H., Becerra M., Watty A., Lu Y., Ma Y., Neubert T.A., et al. Crystal structure of the MuSK tyrosine kinase: insights into receptor autoregulation. Structure. 2002;10:1187–1196. doi: 10.1016/s0969-2126(02)00814-6. [DOI] [PubMed] [Google Scholar]
- 10.Bergamin E., Hallock P.T., Burden S.J., Hubbard S.R. The cytoplasmic adaptor protein Dok7 activates the receptor tyrosine kinase MuSK via dimerization. Mol. Cell. 2010;39:100–109. doi: 10.1016/j.molcel.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan-Sindhunata M.B., Mathijssen I.B., Smit M., Baas F., De Vries J.I., Van Der Voorn J.P., et al. Identification of a Dutch founder mutation in MUSK causing fetal akinesia deformation sequence. Eur. J. Hum. Genet. 2015;23:1151–1157. doi: 10.1038/ejhg.2014.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Al-Shahoumi R., Brady L.I., Schwartzentruber J., Tarnopolsky M.A. Two cases of congenital myasthenic syndrome with vocal cord paralysis. Neurology. 2015;84:1281–1282. doi: 10.1212/WNL.0000000000001396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ben Ammar A., Soltanzadeh P., Bauché S., Richard P., Goillot E., Herbst R., et al. A mutation causes MuSK reduced sensitivity to agrin and congenital myasthenia. PLoS One. 2013;8 doi: 10.1371/journal.pone.0053826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gallenmüller C., Müller-Felber W., Dusl M., Stucka R., Guergueltcheva V., Blaschek A., et al. Salbutamol-responsive limb-girdle congenital myasthenic syndrome due to a novel missense mutation and heteroallelic deletion in MUSK. Neuromuscul. Disord. 2014;24:31–35. doi: 10.1016/j.nmd.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giarrana M.L., Joset P., Sticht H., Robb S., Steindl K., Rauch A., Klein A. A severe congenital myasthenic syndrome with “dropped head” caused by novel MUSK mutations. Muscle Nerve. 2015;52:668–673. doi: 10.1002/mus.24687. [DOI] [PubMed] [Google Scholar]
- 16.Luan X., Tian W., Cao L. Limb-girdle congenital myasthenic syndrome in a Chinese family with novel mutations in MUSK gene and literature review. Clin. Neurol. Neurosurg. 2016;150:41–45. doi: 10.1016/j.clineuro.2016.08.021. [DOI] [PubMed] [Google Scholar]
- 17.Maggi L., Brugnoni R., Scaioli V., Winden T.L., Morandi L., Engel A.G., et al. Marked phenotypic variability in two siblings with congenital myasthenic syndrome due to mutations in MUSK. J. Neurol. 2013;260:2894–2896. doi: 10.1007/s00415-013-7118-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maselli R.A., Arredondo J., Cagney O., Ng J.J., Anderson J.A., Williams C., et al. Mutations in MUSK causing congenital myasthenic syndrome impair MuSK–Dok-7 interaction. Hum. Mol. Genet. 2010;19:2370–2379. doi: 10.1093/hmg/ddq110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mihaylova V., Salih M.A.M., Mukhtar M.M., Abuzeid H.A., El-Sadig S.M., Von Der Hagen M., et al. Refinement of the clinical phenotype in musk-related congenital myasthenic syndromes. Neurology. 2009;73:1926–1928. doi: 10.1212/WNL.0b013e3181c3fce9. [DOI] [PubMed] [Google Scholar]
- 20.Murali C., Li D., Grand K., Hakonarson H., Bhoj E. Isolated vocal cord paralysis in two siblings with compound heterozygous variants in MUSK: expanding the phenotypic spectrum. Am. J. Med. Genet. 2019;179:655–658. doi: 10.1002/ajmg.a.61060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pinto M.V., Saw J.L., Milone M. Congenital Vocal Cord Paralysis and Late-Onset Limb-Girdle Weakness in MuSK–Congenital Myasthenic Syndrome. Front. Neurol. 2019;10:1300. doi: 10.3389/fneur.2019.01300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen Y., Wang B., Zheng X., Zhang W., Wu H., Hei M. A Neonate With MuSK Congenital Myasthenic Syndrome Presenting With Refractory Respiratory Failure. Frontiers in pediatrics. 2020;8:166. doi: 10.3389/fped.2020.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilbe M., Ekvall S., Eurenius K., Ericson K., Casar-Borota O., Klar J., et al. MuSK: a new target for lethal fetal akinesia deformation sequence (FADS) J. Med. Genet. 2015;52:195–202. doi: 10.1136/jmedgenet-2014-102730. [DOI] [PubMed] [Google Scholar]
- 24.Li N., Qiao C., Lv Y., Yang T., Liu H., Yu W.Q., Liu C.X. Compound heterozygous mutation of MUSK causing fetal akinesia deformation sequence syndrome: A case report. World journal of clinical cases. 2019;7:3655. doi: 10.12998/wjcc.v7.i21.3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ding L., Xie Y., Yuan T., Sui Z., Liu H., Li Z. Fetal Akinesia Deformation Sequence (FADS) with Compound Heterozygous Variants in MuSK: A Case Report. Authorea. 2020;1 doi: 10.22541/au.160097884.45196854. Preprint at. [DOI] [Google Scholar]
- 26.Dowling J.J.D., Gonorazky H., Cohn R.D., Campbell C. Treating pediatric neuromuscular disorders: The future is now. Am. J. Med. Genet. 2018;176:804–841. doi: 10.1002/ajmg.a.38418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Varadi M., Anyango S., Deshpande M., Nair S., Natassia C., Yordanova G., et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022;50:D439–D444. doi: 10.1093/nar/gkab1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lomize A.L., Pogozheva I.D. TMDOCK: an energy-based method for modeling α-helical dimers in membranes. J. Mol. Biol. 2017;429:390–398. doi: 10.1016/j.jmb.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 30.Lomize A.L., Hage J.M., Pogozheva I.D. Membranome 2.0: database for proteome-wide profiling of bitopic proteins and their dimers. Bioinformatics. 2018;34:1061–1062. doi: 10.1093/bioinformatics/btx720. [DOI] [PubMed] [Google Scholar]
- 31.Stiegler A.L., Burden S.J., Hubbard S.R. Crystal structure of the agrin-responsive immunoglobulin-like domains 1 and 2 of the receptor tyrosine kinase MuSK. J. Mol. Biol. 2006;364:424–433. doi: 10.1016/j.jmb.2006.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeannet P.Y., Watts G.D., Bird T.D., Chance P.F. Craniofacial and cutaneous findings expand the phenotype of hereditary neuralgic amyotrophy. Neurology. 2001;57:1963–1968. doi: 10.1212/wnl.57.11.1963. [DOI] [PubMed] [Google Scholar]
- 33.Laccone F., Hannibal M.C., Neesen J., Grisold W., Chance P.F., Rehder H. Dysmorphic syndrome of hereditary neuralgic amyotrophy associated with a SEPT9 gene mutation–a family study. Clin. Genet. 2008;74:279–283. doi: 10.1111/j.1399-0004.2008.01022.x. [DOI] [PubMed] [Google Scholar]
- 34.Collie A.M., Landsverk M.L., Ruzzo E., Mefford H.C., Buysse K., Adkins J.R., et al. Non-recurrent SEPT9 duplications cause hereditary neuralgic amyotrophy. J. Med. Genet. 2010;47:601–607. doi: 10.1136/jmg.2009.072348. [DOI] [PubMed] [Google Scholar]
- 35.Grosse G.M., Bauer C., Kopp B., Schrader C., Osmanovic A. Identification of a rare SEPT9 variant in a family with autosomal dominant Charcot-Marie-Tooth disease. BMC Med. Genet. 2020;21:1–7. doi: 10.1186/s12881-020-0984-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones G., Moore C., Hashemolhosseini S., Brenner H.R. Constitutively active MuSK is clustered in the absence of agrin and induces ectopic postsynaptic-like membranes in skeletal muscle fibers. J. Neurosci. 1999;19:3376–3383. doi: 10.1523/JNEUROSCI.19-09-03376.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruff K.M., Pappu R.V. AlphaFold and implications for intrinsically disordered proteins. J. Mol. Biol. 2021;433 doi: 10.1016/j.jmb.2021.167208. [DOI] [PubMed] [Google Scholar]
- 38.Herbst R. MuSk function during health and disease. Neurosci. Lett. 2020;716 doi: 10.1016/j.neulet.2019.134676. [DOI] [PubMed] [Google Scholar]
- 39.Jennings C.G., Dyer S.M., Burden S.J. Muscle-specific trk-related receptor with a kringle domain defines a distinct class of receptor tyrosine kinases. Proc. Natl. Acad. Sci. USA. 1993;90:2895–2899. doi: 10.1073/pnas.90.7.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Y., Qiao K., Yan C., Song J., Huan X., Luo S., et al. Congenital myasthenia syndrome in a Chinese family with mutations in MUSK: A hotspot mutation and literature review. J. Clin. Neurosci. 2020;76:161–165. doi: 10.1016/j.jocn.2020.03.036. [DOI] [PubMed] [Google Scholar]
- 41.Wadwekar V., Nair S.S., Tandon V., Kuruvilla A., Nair M. Congenital myasthenic syndrome: ten years clinical experience from a quaternary care south-Indian hospital. J. Clin. Neurosci. 2020;72:238–243. doi: 10.1016/j.jocn.2019.11.030. [DOI] [PubMed] [Google Scholar]
- 42.Messéant J., Dobbertin A., Girard E., Delers P., Manuel M., Mangione F., et al. MuSK frizzled-like domain is critical for mammalian neuromuscular junction formation and maintenance. J. Neurosci. 2015;35:4926–4941. doi: 10.1523/JNEUROSCI.3381-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Herbst R., Burden S.J. The juxtamembrane region of MuSK has a critical role in agrin-mediated signaling. EMBO J. 2000;19:67–77. doi: 10.1093/emboj/19.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zong Y., Jin R. Structural mechanisms of the agrin–LRP4–MuSK signaling pathway in neuromuscular junction differentiation. Cell. Mol. Life Sci. 2013;70:3077–3088. doi: 10.1007/s00018-012-1209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chevessier F., Girard E., Molgó J., Bartling S., Koenig J., Hantaï D., Witzemann V. A mouse model for congenital myasthenic syndrome due to MuSK mutations reveals defects in structure and function of neuromuscular junctions. Hum. Mol. Genet. 2008;17:3577–3595. doi: 10.1093/hmg/ddn251. [DOI] [PubMed] [Google Scholar]
- 46.Inizan F., Hanna M., Stolyarchuk M., Chauvot de Beauchêne I., Tchertanov L. The first 3D model of the full-length KIT cytoplasmic domain reveals a new look for an old receptor. Sci. Rep. 2020;10:1–13. doi: 10.1038/s41598-020-62460-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pakharukova N., Masoudi A., Pani B., Staus D.P., Lefkowitz R.J. Allosteric activation of proto-oncogene kinase Src by GPCR–beta-arrestin complexes. J. Biol. Chem. 2020;295:16773–16784. doi: 10.1074/jbc.RA120.015400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mohamed A.S., Rivas-Plata K.A., Kraas J.R., Saleh S.M., Swope S.L. Src-class kinases act within the agrin/MuSK pathway to regulate acetylcholine receptor phosphorylation, cytoskeletal anchoring, and clustering. J. Neurosci. 2001;21:3806–3818. doi: 10.1523/JNEUROSCI.21-11-03806.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oury J., Zhang W., Leloup N., Koide A., Corrado A.D., Ketavarapu G., et al. Mechanism of disease and therapeutic rescue of Dok7 congenital myasthenia. Nature. 2021;595:404–408. doi: 10.1038/s41586-021-03672-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cantor S., Zhang W., Delestree N., Remedio L., Mentis G.Z., Burden S.J. Preserving neuromuscular synapses in ALS by stimulating MuSK with a therapeutic agonist antibody. Elife. 2018;7 doi: 10.7554/eLife.34375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xie M.H., Yuan J., Adams C., Gurney A. Direct demonstration of MuSK involvement in acetylcholine receptor clustering through identification of agonist ScFv. Nat. Biotechnol. 1997;15:768–771. doi: 10.1038/nbt0897-768. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MUSK AlphaFold2-generated structure is available for download from the AlphaFold Protein Structure Database (https://alphafold.ebi.ac.uk/entry/O15146). The Membranome transmembrane-corrected structure is available from the Membranome database (https://membranome.org/proteins/2668).