Abstract
The 8-aza-7-deazaadenine (pyrazolo[3,4-d]pyrimidin-4-amine) N8-(2′-deoxyribonucleoside) (2) which has an unusual glycosylation position was introduced as a universal nucleoside in oligonucleotide duplexes. These oligonucleotides were prepared by solid-phase synthesis employing phosphoramidite chemistry. Oligonucleotides incorporating the universal nucleoside 2 are capable of forming base pairs with the four normal DNA nucleosides without significant structural discrimination. The thermal stabilities of those duplexes are very similar and are only moderately reduced compared to those with regular Watson–Crick base pairs. The universal nucleoside 2 belongs to a new class of compounds that form bidentate base pairs with all four natural DNA constituents through hydrogen bonding. The base pair motifs follow the Watson–Crick or the Hoogsteen mode. Also an uncommon motif is suggested for the base pair of 2 and dG. All of the new base pairs have a different shape compared to those of the natural DNA but fit well into the DNA duplex as the distance of the anomeric carbons approximates those of the common DNA base pairs.
INTRODUCTION
The degeneracy of the genetic code leads to a multitude of possible DNA sequences coding for a particular protein. Oligonucleotides that take this redundancy into account must be synthesized and used for the screening of the potential DNA or RNA candidates. This has led to the concept of mixed primers or probes. An alternative approach uses less discriminatory base analogs. Such bases are described as universal or ambiguous. A true universal base should hybridize with all four natural bases without discrimination and significant destabilization of the DNA duplex.
Hypoxanthine is one of the most widely used universal bases. Its nucleoside 2′-deoxyinosine forms base pairs with 2′-deoxyadenosine, 2′-deoxycytidine, 2′-deoxyguanosine and 2′-deoxythymidine; however, it does not bind with equal affinity to these nucleosides. Studies of 9mer oligonucleotides show a wide range of Tm values (15°C) when 2′-deoxyinosine is inserted in duplexes opposite each of the four natural nucleosides (1,2). Thus, it cannot be considered as a true universal nucleoside. In PCR and sequencing applications, hypoxanthine has been shown to behave like guanine (3). Recently, the base pairing properties of 7-deaza-2′-deoxyinosine were investigated and found to be similar to those of 2′-deoxyinosine (4). Other ambiguous nucleosides are represented by 1-(2′-deoxy-β-d-ribofuranosyl)imidazole-4-carboxamide (5) and the compounds P and K (6,7). The latter have the ability to act as amino and imino tautomers. Thus, the pyrimidine analog P pairs with dA and dG while the purine analog K forms base pairs with dC and dT. The pairing modes were confirmed by NMR spectroscopy (8).
Apart from the nucleosides forming base pairs by hydrogen bonding there are a number of universal residues that do not show such capabilities. Heterocyclic bases, such as 5-nitroindole (9,10), 3-nitropyrrole (11) and difluorotoluene (12) can form strong base stacks when they replace the natural bases. As hydrogen bonding is abolished, the specificity of base recognition is lost. Nevertheless, specific base recognition is still possible when the shape of the universal base fits well in a base pair that can be considered as isomorphous. Abasic sugar derivatives have also been evaluated as universal residues which are neither able to form hydrogen bonds nor stabilize a base pair by stacking interactions (13).
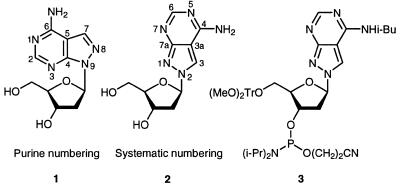
This manuscript describes a novel type of a universal base or, more precisely, a universal nucleoside which is derived from 8-aza-7-deazaadenine (pyrazolo[3,4-d]pyrimidin-4-amine). The base is linked to the 2′-deoxyribose moiety in an unusual way (Scheme 1). The nitrogen-8 (purine numbering is used throughout Results and Discussion) is the glycosylation site but not the usual position-9 (see 1 and 2). Oligonucleotides incorporating the unusually linked nucleoside 2 are prepared by solid-phase synthesis (14), thereby employing the phosphoramidite 3 as monomeric building block. Hybridization experiments will demonstrate the ambiguous character of the nucleoside 2, which still forms specific hydrogen bonds towards the four canonical DNA constituents.
Scheme 1.
RESULTS AND DISCUSSION
Monomers
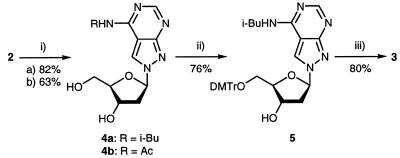
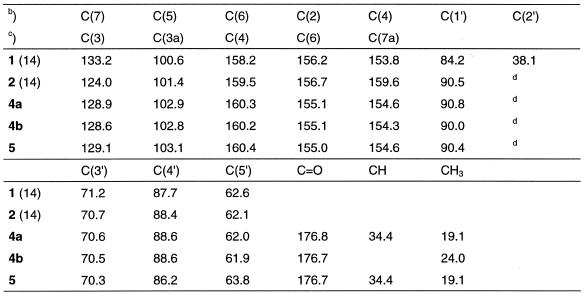
Oligonucleotides containing compound 2 have been recently described, and its base pairing properties with dT was studied in duplexes with parallel and antiparallel chain orientation (14,15). During these studies the amino group of nucleoside 2 was protected with a benzoyl residue. Due to the particular structure of compound 2 the benzoyl-protecting group was extraordinarily stable under the regular deprotection conditions (concentrated ammonia, elevated temperature) and not orthogonal to the protecting groups of the other four regular DNA constituents. Thus, other protecting groups were evaluated. The acetyl and the isobutyryl group were chosen for this purpose (Scheme 2). Acetylation as well as isobutyrylation was performed by using the corresponding anhydrides and employing the protocol of transient protection (16). Both derivatives (4a,b) were isolated as monoacyl derivatives. Next, the half-life values (τ) of deprotection were measured in 25% aqueous ammonia. The reaction was followed UV-spectrophotometrically at 20 and 40°C (Table 1). While the benzoyl derivative showed a τ of almost 5 h, the acetyl as well as the isobutyryl derivatives were quickly removed. As the isobutyryl derivative 4a was easier to handle and was isolated in higher yield (82%) it was used for further manipulations. Then, compound 4a was converted into its 4,4′-dimethoxytrityl derivative (5, 76%) and was treated with chloro(2-cyanoethoxy)-N,N-diisopropylamino-phosphine to give the phosphoramidite 3 in 80% yield. All monomeric compounds were characterized by elemental analysis as well as by 1H- and 13C-NMR spectra (see Materials and Methods and Table 6). The 13C-NMR signals were assigned by gated-decoupled 13C-NMR or heteronuclear 1H/13C-NMR correlation spectra. The 13C-NMR indicated a significant upfield shift of the C7 signal when the glycosylic position is changed from N9 to N8.
Scheme 2. (i) (a) (i-Bu)2O, pyridine, r.t., 3 h. (b) Ac2O, pyridine, r.t., 3 h. (ii) DMT-Cl, pyridine, r.t., 3 h. (iii) (i-Pr)2NP(Cl)O(CH2)2CN, CH2Cl2, r.t., 20 min.
Table 1. Half-life values (τ) of the protected z8c7Ad* derivatives measured UV-spectrophotometrically in 25% aq. ammonia.
aMeasured at 40°C.
bMeasured at 20°C.
Table 6. 13C-NMR chemical shifts of the 8-aza-7-deazaadenine 2′-deoxyribofuranosidesa.
aMeasured in (d6)DMSO.
bPurine numbering.
cSystematic numbering.
dSuperimposed by DMSO.
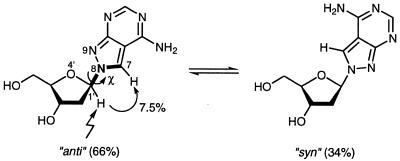
Regarding the base pairing, it was of interest to calculate the syn-anti equilibrium of the N8-linked nucleoside 2. The population of the conformers was determined by 1D-NOE difference spectroscopy. The distance between H1′ and H7 of compound 2 is nearly the same as in the case of the N9-linked nucleosides between H1′ and H8, so the syn-anti population ratio was calculated by using a calibration graph published earlier for regularly linked nucleosides (17). Because of the undefined syn and anti orientation of N8-glycosylated nucleobases, the torsion angle χ used throughout this manuscript is formed by the bonds O4′-C1′-N9-C7. The orientation of the base is ‘anti’ when the distance between H1′ and H7 is a minimum and ‘syn’ when this distance is a maximum (Scheme 3). This is opposite to the definition as in the case of N9-linked nucleosides. Here, the orientation of the base is syn when the distance between H1′ and H8 is a minimum and anti when this distance is a maximum. Nevertheless, this opposite definition is used as the conformation of the nucleoside 2 correlates to that of the purine nucleosides when incorporated in a DNA-duplex. As a nuclear Overhauser enhancement of 7.5% was determined for H7 upon irradiation of H1′ the nucleoside 2 adopts a 66% population of ‘anti’ conformers and a 34% population in the ‘syn’ state. Thus, compound 2 shows similar conformer equilibrium as found for 2′-deoxyadenosine (60% anti) (17).
Scheme 3. Two-state ‘syn-anti’ equilibrium of N8-8-aza-7-deaza-2′-deoxyadenosine.
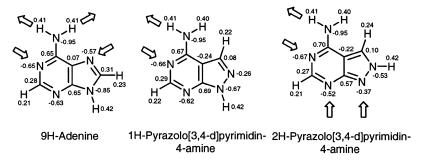
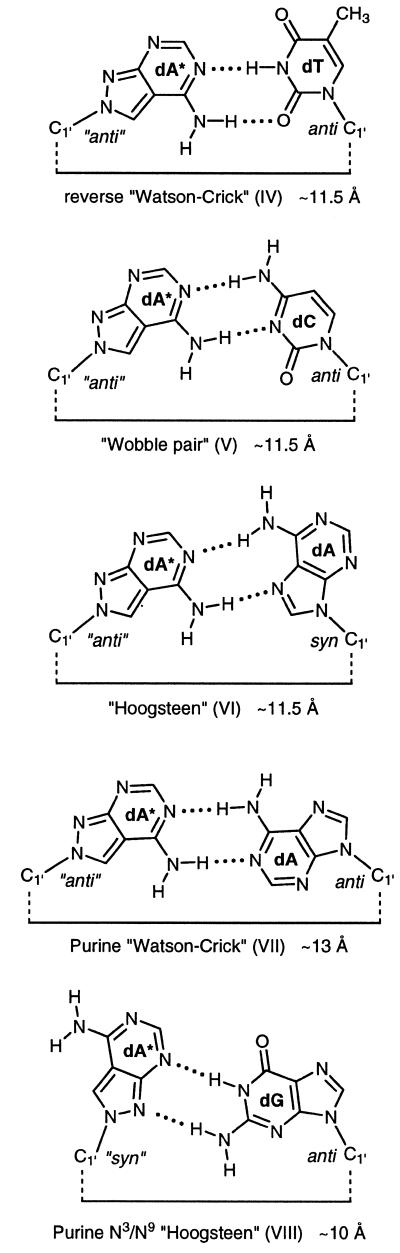
The interaction energies of base pairs depend on the charge densities of the atoms. Thus, a comparison of the charge densities of adenine (18), 1H-pyrazolo[3,4-d]pyrimidin-4-amine and 2H-pyrazolo[3,4-d]pyrimidin-4-amine was made using ab initio calculations at the 6–31G* level (HyperChem Release 5, Hypercube Inc., Gainesville, FL). A geometry optimization on the STO-3G level was made before. This led to the pattern of the proton donor and acceptor sites shown in Scheme 4. The 1H- and 2H-tautomeric forms of the base are used as models for the identically positioned 2′-deoxyribonucleosides. The charge densities of the Watson–Crick recognition sites are similar for all three nucleosides. However, the direct linking of the two hetero atoms (nitrogen-8 and nitrogen-9) in the five-membered ring of the pyrazolo[3,4-d]pyrimidine system has a very significantly marked base-weakening effect (compare the N9-tautomers of adenine with 8-aza-7-deazaadenine). The 2H-tautomer which represents a model for compound 2 shows also a reduced charge density for the nitrogen-9 as it is directly linked to nitrogen-8. Nevertheless, nitrogen-9 of compound 2 is a better acceptor than nitrogen-8 in compound 1. According to this the nucleoside 2 is able to form Watson–Crick or reverse Watson–Crick base pairs. The ‘Hoogsteen’ site of 2 can involve nitrogens 3 and 9 as acceptors within DNA duplexes being different from that of regular Hoogsteen base pairs.
Scheme 4. Partial atomic charges (in electron units) of adenine, 1H-pyrazolo[3,4-d]pyrimidin-4-amine and 2H-pyrazolo[3,4-d]pyrimidin-4-amine calculated at the 6-31G* level.
Oligonucleotides
Oligonucleotides containing compound 2 were prepared by solid-phase synthesis using the standard protocol of an automated DNA synthesizer (Applied Biosystems, Weiterstadt, Germany) (19). The coupling efficiency of the phosphoramidite was always higher than 98%. Deprotection was performed with 25% aqueous NH3, and the oligonucleotides were purified by OPC-cartridges (20) or by reverse-phase HPLC (see Materials and Methods). The homogenity of the oligonucleotides was proved by reverse-phase HPLC. MALDI-TOF mass spectra of the oligonucleotides were measured (15) and are in agreement with the calculated data. The nucleoside composition was determined after oligonucleotide digestion with snake-venom phosphodiesterase followed by alkaline phosphatase (21).
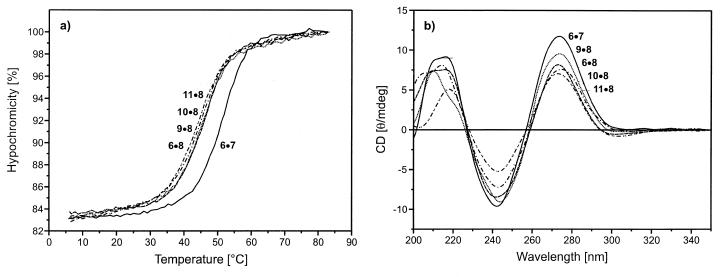
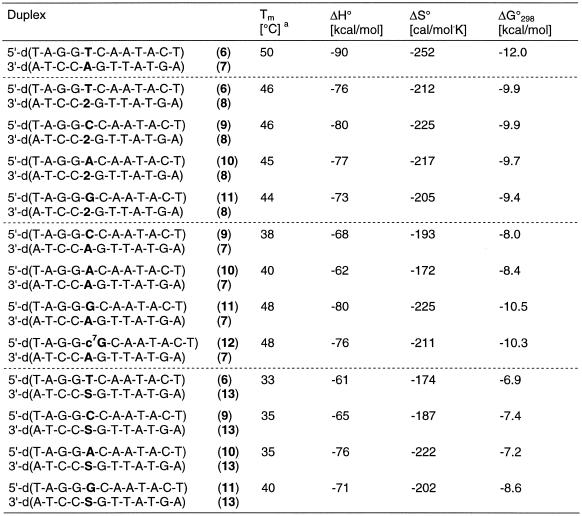
For the hybridization experiments the oligonucleotide duplex 5′-d(TAGGTCAATACT)•3′-d(ATCCAGTTATGA) (6•7) was used as a reference to study the influence of base-modified nucleosides on the duplex stability. First, the central dA residue was replaced by compound 2 leading to the oligonucleotide 8. The latter was hybridized with the second strand containing the canonical nucleosides dT, dC, dA and dG (6, 9, 10, 11) opposite to compound 2. Melting curves were measured and CD-spectra were taken (Fig. 1 ). The melting curves were also used to determine the thermodynamic data that were obtained by curve shape analysis using the program Meltwin 3.0 (22). In the cases of the duplexes incorporating compound 2 only moderate Tm decreases (4–6°C) were observed with regard to the reference duplex 6•7 (Table 2). Within the series of modified duplexes the Tm difference was small (ΔTm = 2°C). The CD-spectra of the various duplexes showed the typical shape of a B-DNA (Fig. 1).
Figure 1.
(a) Melting curves and (b) CD spectra of the oligonucleotides 6•7, 6•8, 9•8, 10•8 and 11•8. Measured in 1 M NaCl, 100 mM MgCl2, 60 mM Na-cacodylate (pH 7.0) with 5 µM single-strand concentration.
Table 2. Tm values of antiparallel-stranded oligonucleotides containing one N8-8-aza-7-deaza-2′-deoxyadenosine (2, dA*) residue opposite to each of the four natural nucleosides dA, dC, dG and dT.
aMeasured at 260 nm in 1 M NaCl, 100 mM MgCl2 and 60 mM Na-cacodylate (pH 7.0) with 5 µM single-strand concentration.
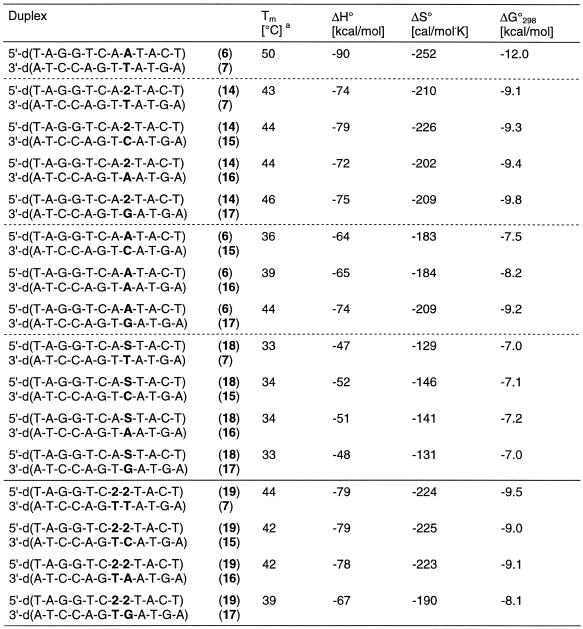
When the three canonical nucleosides dA, dG and dC were positioned opposite to 2′-deoxyadenosine, the Tm value decreased by 2–12°C (Table 2) while the dA analog 2 opposite all of the four canonical nucleosides results in a Tm decrease of only 4–6°C. We have also introduced 1,2-dideoxyribose (dS) as an abasic sugar residue opposite the four canonical bases. These experiments were performed in order to demonstrate the Tm decrease in the absence of a heterocyclic base residue. Compared to compound 2 (ΔTm = –4 to –6°C) the abasic residues resulted in a much higher Tm decrease (ΔTm = –10 to –17°C). This indicates a rather large contribution of the base of compound 2 to the duplex stability. In this concern it should be noted that the duplex 11•13 with three dG residues in a row and with an abasic residue opposite to the dG tract gives a significantly higher Tm value than the others. This might be explained by the strong stacking interactions of the consecutive dG residues, which can stabilize the helix even when only one of the opposite bases (dC) is absent. From these experiments it can be concluded that compound 2 is a universal nucleoside as it base pairs almost equally well with all of the four DNA constituents. In order to test the universal base pairing properties of 2 further we have selected another position within the oligonucleotide duplex. Also in these series of experiments a single substitution was made. The oligonucleotide duplexes carrying an abasic site (dS) were used for comparison. Also in this case compound 2 behaves like a universal nucleoside (Table 3). The duplexes in which two of the universal bases are substituted were investigated next. In these sequences with compound 2 residues as nearest neighbors one universal residue base pairs with dT while the second is located opposite the four regular nucleosides. In these cases the ambiguous base pairing properties are also obvious, and the differences in the Tm values were not significantly different from those of duplexes containing only one universal nucleoside residue (Table 3).
Table 3. Tm values of antiparallel-stranded oligonucleotides containing N8-8-aza-7-deaza-2′-deoxyadenosine (2, dA*) residues opposite to each of the four natural nucleosides dA, dC, dG and dT.
aMeasured at 260 nm in 1 M NaCl, 100 mM MgCl2 and 60 mM Na-cacodylate (pH 7.0) with 5 µM single-strand concentration.
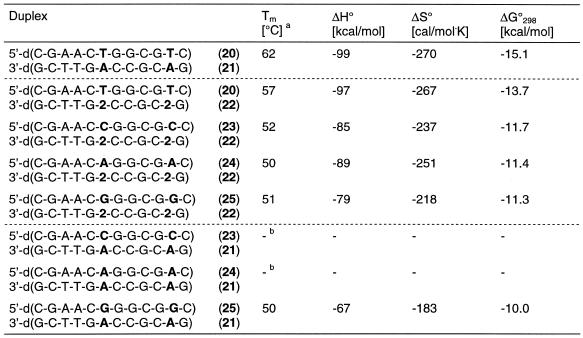
We have also investigated the effect of the universal nucleoside 2 on the stability of a dodecamer duplex having a higher dC-dG content and showing a Tm value of 62°C (duplex 20•21, Table 4) compared to 50°C of the duplex 6•7. In this duplex two compound 2 residues were incorporated and two regular nucleosides were placed opposite them. This resulted in ΔTm values varying by 7°C. This Tm range is higher than that of oligonucleotides containing two compound 2 residues in a row (ΔTm = 5°C). Nevertheless, duplexes containing two mismatches do not even hybridize, except those incorporating dG-dA base pairs (Table 4).
Table 4. Tm values of antiparallel-stranded oligonucleotides containing two N8-8-aza-7-deaza-2′-deoxyadenosine (2, dA*) residues opposite each of the four natural nucleosides dA, dC, dG and dT.
aMeasured at 260 nm in 1 M NaCl, 100 mM MgCl2 and 60 mM Na-cacodylate (pH 7.0) with 5 µM single strand concentration.
bNo sigmoidal melting curve.
Base pair motifs
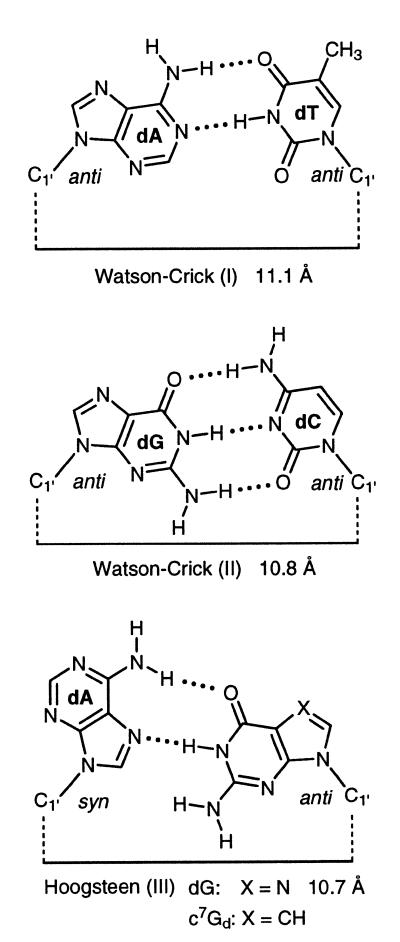
The universal nucleoside 2 belongs to the class of universal nucleosides forming specific base pairs by hydrogen bonding and stacking interaction and not by the base stacking alone. Universal nucleosides with properties of non-specific base stackers have been realized with 5-nitroindole or 3-nitropyrrole 2′-deoxyribonucleosides. Despite the fact that compound 2 forms hydrogen-bonded base pairs, the base pair structures have to be different from those of regular nucleosides. The regular DNA base pairs that are used for comparison form bidentate dA-dT or tridentate dG-dC Watson–Crick motifs (23). The Hoogsteen pairs are usually bidentate, as shown for the dG-dA pair (Scheme 5). The regular Watson–Crick base pairs are isomorphous leading to very particular base pair overlaps within the DNA helix. The base pairs formed by the nucleoside 2 with the four canonical nucleosides are not isomorphous, neither among each other nor in comparison to the natural base pairs. According to that the base pair overlaps are different in the case of those with regular or universal nucleosides. Due to almost identical Tm values of duplexes containing compound 2 and very similar stabilities with regard to the dA-dT pair (see reference duplex 6•7) the novel base pairs have bidentate structures. This is underlined by the fact that the ΔG-values of compound 2 when interacting with the normal nucleosides are in the stability range of a dA-dT but not of a dG-dC pair.
Scheme 5.
The new base pairs were constructed under the following assumptions:
‘Watson–Crick’ and ‘Hoogsteen’ base pairs are allowed as well as those with the reversed modes.
The nucleoside 2 can act as acceptor in a new Hoogsteen-like mode via nitrogen-9 and nitrogen-1.
The distances between the anomeric carbons of the universal nucleoside 2 and its natural counterparts should approximate those of the Watson–Crick base pairs (11 Å) (23). The length of the H-bonds was arbitrarily set to 2 Å.
When these factors are taken into account the following base pair motifs (IV–VIII) are suggested (Scheme 6): motif IV (2 pairs with dT): in this case the 2-oxo group of dT and the 6-amino group of 2 are the H-donors and nitrogen-3 of dT and nitrogen-1 of compound 2 are the acceptor atoms forming a reverse Watson–Crick base pair (15). Motif V (2 pairs with dC): the amino groups of dC or 2 are the proton donors and nitrogen-3 of dC and nitrogen-1 of 2 are the acceptor sites. This motif is related to a ‘wobble’ pair. Motifs VI and VII (2 pairs with dA): in this case two base pairs can be formed. However, the base pair VI is preferred as only in this case the anomeric carbons approximate the distance of normal B-DNA base pairs. The motif follows a ‘Hoogsteen’ mode. Motif VIII (2 pairs with dG): for this base pair a rather unusual motif (VIII) is suggested. Two hydrogen bonds are formed with dG as the H-donor and nitrogen-3 and nitrogen-9 of compound 2 as acceptor sites. Compared to the other motifs the distance between the anomeric carbons is reduced. The motif can be described as purine N3/N9-‘Hoogsteen’ pair.
Scheme 6.
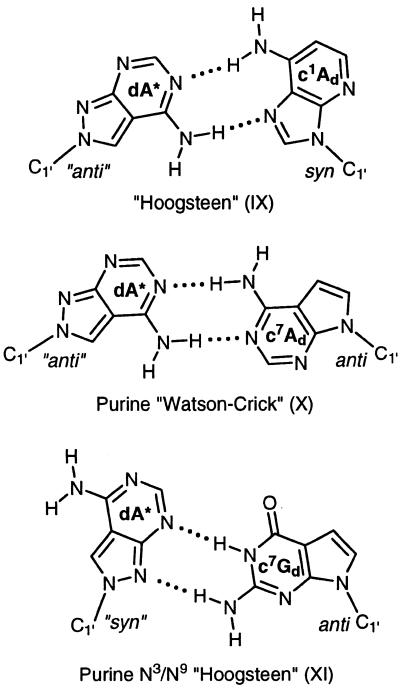
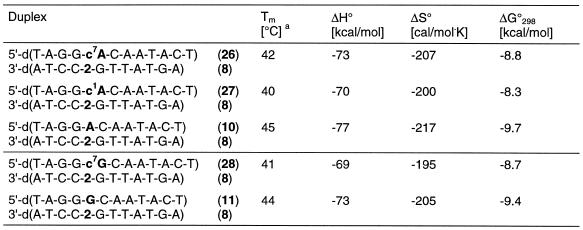
These base pair motifs are supported by some preliminary experiments performed with base-modified nucleosides located opposite to 2 (Table 5). Replacement of the dA residue opposite to compound 2 by c7Ad (duplex 26•8) or c1Ad (duplex 27•8) resulted in only moderate Tm changes suggesting that the base pair motifs (IX and X) can be formed. A similar experiment has been performed by the replacement of dG residues opposite to compound 2 by 7-deaza-2′-deoxyguanosine (c7Gd; duplex 28•8). Also in this case the Tm decrease was only 3°C supporting the structure of the base pair motif XI (Scheme 7).
Table 5. Tm values of antiparallel-stranded oligonucleotides containing one N8-8-aza-7-deaza-2′-deoxyadenosine (2, dA*) residue opposite to either c7Ad, c1Ad or c7Gd.
aMeasured at 260 nm in 1 M NaCl, 100 mM MgCl2 and 60 mM Na-cacodylate (pH 7.0) with 5 µM single strand concentration.
Scheme 7.
CONCLUSION
The pyrazolo[3,4-d]pyrimidine nucleoside 2 represents a new class of universal nucleosides which hybridize almost equally well with the four canonical DNA-constituents dA, dT, dG and dC. From model building studies it is obvious that the ambiguous character of compound 2 is controlled by bidentate base pairs. Thus, hydrogen bonding and not stacking interactions play the crucial role. The base pairs of 2 with the four canonical nucleosides are different in shape to those of the regular base pairs of B-DNA. However, as the distances of the anomeric carbons approximate those of a regular Watson–Crick base pair they fit well in a B-DNA-duplex. Nucleoside 2 might not be the only new universal nucleoside of this type. It is expected that other ‘adenine’ derivatives having the sugar attached to the 8-position show a similar behavior.
MATERIALS AND METHODS
General
All chemicals were purchased from Aldrich, Sigma (Sigma–Aldrich Chemie GmbH, Deisenhofen, Germany) or Fluka (Buchs, Switzerland). Solvents were of laboratory grade. Elemental analyses were performed by Mikroanalytisches Laboratorium Beller (Göttingen, Germany). NMR spectra were measured on Avance DPX 250 or AMX 500 spectrometers (Bruker, Karlsruhe, Germany) operating at proton resonance frequencies of 250.13 and 500.14 MHz (125.13 MHz for 13C), respectively. Chemical shifts are in p.p.m. relative to TMS as internal standard. J-values are given in Hz. M.p.’s were measured with a Büchi SMP-20 apparatus (Büchi, Flawil, Switzerland) and are uncorrected. UV spectra were recorded on a U 3200 spectrometer (Hitachi, Tokyo, Japan). Thin-layer chromatography (TLC) was performed on aluminum sheets, silica gel 60 F254, 0.2 mm layer (Merck, Darmstadt, Germany) and column flash chromatography (FC) on silica gel 60 (Merck) at 0.4 bar (4 × 104 Pa). The 13C-NMR data are shown in Table 6.
Measurement of the protecting group stabilities
The stabilities of the amino protecting groups were determined UV-spectrophotometrically with a U 3200 spectrometer (Hitachi) heated by a RC 6 CP thermostat (Lauda, Lauda-Königshofen, Germany) at two temperatures (20 and 40°C). UV-overlay spectra of compounds 4a and 4b were measured followed by time scans at that wavelength where the highest absorbance change was observed (Table 1).
2-(2-Deoxy-β-d-erythro-pentofuranosyl)-4-(isobutyrylamino)-2H-pyrazolo[3,4-d]pyrimidine (4a)
Compound 2 (1.0 g, 4.0 mmol) was dried by co-evaporation with pyridine (2 × 25 ml). The residue was suspended in pyridine (25 ml), and Me3SiCl (2.5 ml, 19.7 mmol) was added at room temperature (r.t.). After 15 min of stirring, isobutyric anhydride (3.26 ml, 19.7 mmol) was introduced in the solution, and the stirring was continued for 3 h. The mixture was cooled (ice bath), water (5 ml) and subsequently 25% aq. NH3 (5 ml) were added, and the stirring was continued for another 15 min. The solvent was evaporated and the residue dissolved in H2O (25 ml). The aq. solution was washed with ether (2 × 15 ml), separated and evaporated. Co-evaporation with toluene (2 × 20 ml), and then with MeOH (2 × 20 ml) furnished a colorless foam. This was applied to FC (column 12 × 2 cm, CH2Cl2-MeOH 8:2) to yield a colorless foam (1.05 g, 82%). Rf (CH2Cl2-MeOH 8:2) 0.25; λmax(MeOH)/nm 239 (ɛ/dm3 mol–1 cm–1 8100), 273 (8900), 304 (6500); δH[250 MHz; (d6)DMSO] 1.15 (6 H, s, Me2), 2.40 (1 H, m, 2′-Hα), 2.67 (1 H, m, 2′-Hβ), 2.91 (1 H, h, J 6.7, CH), 3.60 (2 H, m, 5′-H2), 3.93 (1 H, m, 4′-H), 4.46 (1 H, m, 3′-H), 4.90 (1 H, t, J 4.3, 5′-OH), 5.36 (1 H, d, J 3.1, 3′-OH), 6.48 (1 H, ‘t’, J 5.5, 1′-H), 8.61 (1 H, s, 6-H), 9.00 (1 H, s, 3-H), 11.20 (1 H, s, NH). (Found: C, 52.48; H, 6.05; N, 21.70. C14H19N5O4 requires C, 52.33; H, 5.96; N, 21.79%.)
4-(Acetylamino)-2-(2-deoxy-β-d-erythro-pentofuranosyl)-2H-pyrazolo[3,4-d]pyrimidine (4b)
Compound 2 (12) (150 mg, 0.6 mol) was dried by co-evaporation with anh. pyridine and then dissolved in pyridine (4 ml). Me3SiCl (0.4 ml, 3.2 mmol). The solution was treated with acetic anhydride (0.28 ml, 3.0 mmol) under stirring for 3 h at r.t. The mixture was cooled (ice bath) and water (1 ml) was added. After 15 min, the solution was treated with ice-cold 10% aq. NH3 (1 ml) with stirring. This was continued for 15 min, the solvent was evaporated, and the residue was co-evaporated with toluene (2 × 10 ml). The solid material was dissolved in MeOH and applied to FC (column 3 × 10 cm, CH2Cl2-MeOH 8:2). Compound 4b was isolated as a colorless powder (110 mg, 63%). Rf (CH2Cl2-MeOH 8:2) 0.2; λmax(MeOH)/nm 239 (ɛ/dm3 mol–1 cm–1 8400), 273 (9300), 302 (6700); δH[250 MHz; (d6)DMSO] 2.24 (3 H, s, Me), 2.41 (1 H, m, 2′-Hα), 2.65 (1 H, m, 2′-Hβ), 3.55 (2 H, m, 5′-H2), 3.94 (1 H, m, 4′-H), 4.43 (1 H, m, 3′-H), 4.91 (1 H, t, J 5.3, 5′-OH), 5.37 (1 H, d, J 4.4, 3′-OH), 6.46 (1 H, ‘t’, J 6.1, 1′-H), 8.60 (1 H, s, 6-H), 8.95 (1 H, s, 3-H), 11.22 (1 H, s, NH). (Found: C, 48.99; H, 5.25; N, 23.68. C12H15N5O4 requires C, 49.14; H, 5.16; N, 23.88%.)
2-[2-Deoxy-5-O-(4,4′-dimethoxytriphenylmethyl)-β-d-erythro-pentofuranosyl]-4-(isobutyrylamino)-2H-pyrazolo[3,4-d]pyrimidine (5)
Compound 4a (1.0 g, 3.1 mmol) was dried by repeated co-evaporation with anhydrous pyridine (2 × 20 ml) and dissolved in dry pyridine (25 ml). Then, 4,4′-dimethoxytriphenylmethyl chloride (1.22 g, 3.6 mmol) was added, and the solution was stirred for 3 h under argon at r.t. Then MeOH (5 ml) was added, and the mixture was stirred for another 30 min. The solution was then reduced to half the volume, dissolved in CH2Cl2 (100 ml), and extracted twice with a 5% aq. NaHCO3 solution (50 ml) followed by saturated brine (50 ml). The organic layer was dried (Na2SO4), filtered, evaporated and co-evaporated twice with toluene (30 ml). The residue was purified by FC (silica gel, 4 × 10 cm, solvent CH2Cl2-acetone 8:2). Compound 5 was obtained as a colorless foam (1.46 g, 76%). Rf (CH2Cl2-acetone 8:2) 0.3; λmax(MeOH)/nm 236 (ɛ/dm3 mol–1 cm–1 28 500), 273 (11 400), 305 (6700); δH[250 MHz; (d6)DMSO] 1.11 [6 H, dd, J 6.9, J 10.4, (CH3)2], 2.43 (1 H, m, 2′-Hα), 2.81 (1 H, m, 2′-Hβ), 2.90 (1 H, h, J 6.8, CH), 3.14 (2 H, m, 5′-H2), 3.66 and 3.69 (6 H, 2 s, OMe), 4.04 (1 H, m, 4′-H), 4.52 (1 H, m, 3′-H), 5.42 (1 H, d, J 5.0, 3′-OH), 6.54 (1 H, dd, J 6.4, J 2.7, 1′-H), 6.74 (4 H, m, ArH), 7.11–7.29 (9 H, m, ArH), 8.62 (1 H, s, 6-H), 9.05 (1 H, s, 3-H), 11.22 (1 H, s, NH). (Found: C, 67.36; H, 5.98; N, 11.30. C35H37N5O6 requires C, 67.40; H, 5.98; N, 11.23%.)
2-[2-Deoxy-5-O-(4,4′-dimethoxytriphenylmethyl)-β-d-erythro-pentofuranosy]-4-(isobutyrylamino)-2H-pyrazolo[3,4-d]pyrimidin-3′-[(2-cyanoethyl)-N,N-diisopropyl-phosphoramidite] (3)
A stirred solution of compound 5 (800 mg, 1.28 mmol) in anh. CH2Cl2 (40 ml) was pre-flushed with Ar. Then, (i-Pr)2EtN (0.42 ml, 2.41 mmol) and 2-cyanoethyl diisopropylchlorophosphoramidite (0.42 ml, 1.88 mmol) were added under Ar. The reaction was monitored by TLC. After stirring for 20 min, the reaction mixture was diluted with CH2Cl2 (40 ml) and washed with a 5% aq. NaHCO3 solution (2 × 20 ml) followed by saturated brine (20 ml). The organic layer was dried (Na2SO4), evaporated and co-evaporated with CH2Cl2 (2 × 20 ml). The residue was purified by FC (silica gel, 4 × 10 cm, solvent: CH2Cl2-acetone 9:1) to yield compound 3 as a colorless foam (840 mg, 80%). Rf (CH2Cl2-acetone 9:1) 0.5 and 0.6; δP [101 MHz; CDCl3] 149.5, 149.7.
Synthesis and purification of the oligonucleotides 6–28
The oligonucleotide synthesis was performed on an ABI 392-08 DNA synthesizer (Applied Biosystems, Weiterstadt, Germany) in a 1 µmol scale using the phosphoramidite 3 and those of the regular 2′-deoxynucleosides (Applied Biosystems) following the synthesis protocol for 3′-phosphoramidites (19). The crude oligonucleotides were purified and detritylated on oligonucleotide purification cartridges (Applied Biosystems) following the standard protocol for purification (20). The oligonucleotides were lyophilized on a Speed Vac evaporator to yield colorless solids which were stored frozen at –18°C. The MALDI-TOF MS data were determined and found to be in agreement with the calculated values. The enzymatic hydrolysis of the oligomers was performed as described by Seela and Lampe (21). Quantification of the constituents was made on the basis of the peak areas, which were divided by the extinction coefficients of the nucleoside (ɛ260 values: dA 15 400, dC 7300, dG 11 400, dT 8800, 2 6600). Snake venom phophodiesterase (EC 3.1.15.1., Crotallus durissus) and alkaline phosphatase (EC 3.1.3.1., Escherichia coli) were generous gifts of the Roche Diagnostics GmbH (Penzberg, Germany). Oligonucleotide analysis was carried out on reverse phase HPLC (RP-18 HPLC) with a Merck-Hitachi-HPLC: 250 × 4 mm RP-18 column; gradients of 0.1 M (Et3NH)OAc (pH 7.0)/ MeCN 95:5 (A) and MeCN (B); gradient I: 50 min 0–50% B in A, flow rate 1 ml/min; gradient II: 20 min 0–25% B in A, flow rate 0.7 ml/min; 30 min 25–40% B in A, flow rate 1 ml/min; gradient III: 20 min 0–25% B in A, flow rate 1 ml/min.
Determination of Tm values and thermodynamic data
Absorbance versus temperature profiles were measured on a Cary-1/1E UV/VIS spectrophotometer (Varian, Mulgrave, Australia) equipped with a Cary thermoelectrical controller. The temperature values were measured in the reference cell with a Pt-100 resistor. The heating and cooling rate was 1.0°C/min. The thermodynamic data (ΔH°, ΔS°, ΔG°298) were calculated using the ‘MeltWin 3.0’ program package (22). The CD spectra were recorded with a Jasco-600 (Jasco, Tokyo, Japan) spectropolarimeter with a thermostatically (Lauda RCS-6 bath) controlled 1 cm cuvette.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Mr Yang He for the NMR spectra, Dr Helmut Rosemeyer for the PSEUROT calculations, Mrs Elisabeth Feiling for the oligonucleotide syntheses and Dr J. Gross (Universität Heidelberg, Germany) for the MALDI-TOF spectra. Financial support by the Deutsche Forschungsgemeinschaft and Roche Diagnotics GmbH (Penzberg, Germany) is gratefully acknowledged.
REFERENCES
- 1.Martin F.H. and Castro,M.M. (1985) Nucleic Acids Res., 13, 8927–8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kawase Y., Iwai,S., Inoue,H., Miura,K. and Ohtsuka,E. (1986) Nucleic Acids Res., 14, 7727–7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kamiya H., Sakaguchi,T., Murata,N., Fujimuro,M., Miura,H., Ishikawa,H., Shimizu,M., Inoue,H., Nishimura,S., Matsukage,A., Masutani,C., Hanaoka,F. and Ohtsuka,E. (1992) Chem. Pharm. Bull. (Tokyo), 40, 2792–2795. [DOI] [PubMed] [Google Scholar]
- 4.Seela F. and Mittelbach,C. (1999) Nucl. Nucl., 18, 425–441. [Google Scholar]
- 5.Johnson W.T., Zhang,P. and Bergstrom,D.E. (1997) Nucleic Acids Res., 25, 559–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kong Thoo Lin P. and Brown,D.M. (1989) Nucleic Acids Res., 17, 10373–10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown D.M. and Kong Thoo Lin,P. (1991) Carbohydr. Res., 216, 129–139. [DOI] [PubMed] [Google Scholar]
- 8.Nederman A.N.R., Stone,M.J., Kong Thoo Lin,P., Brown,D.M. and Williams,D.H. (1991) J. Chem. Soc. Chem. Commun., 19, 1357–1359. [Google Scholar]
- 9.Loakes D., Brown,D.M., Linde,S. and Hill,F. (1995) Nucleic Acids Res., 23, 2361–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loakes D. and Brown,D.M. (1994) Nucleic Acids Res., 22, 4039–4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nichols R., Andrews,P.C., Zhang,P. and Bergstrom,D.E. (1994) Nature, 369, 492–493. [DOI] [PubMed] [Google Scholar]
- 12.Moran S., Ren,R.X.-F. and Kool,E.T. (1997) Proc. Natl Acad. Sci. USA, 94, 10506–10511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takeshita M., Chang,C.-N., Johnson,F., Will,S. and Grollman,A.P. (1987) J. Biol. Chem., 262, 10171–10179. [PubMed] [Google Scholar]
- 14.Seela F. and Kaiser,K. (1988) Helv. Chim. Acta, 71, 1813–1823. [Google Scholar]
- 15.Seela F., Zulauf,M. and Debelak,H. (2000) Helv. Chim. Acta, 83, 1437–1453. [Google Scholar]
- 16.Ti G.S., Gaffney,B.L. and Jones,R.A. (1982) J. Am. Chem. Soc., 104, 1316–1319. [Google Scholar]
- 17.Rosemeyer H., Toth,G., Golankiewicz,B., Kazimierczuk,Z., Bourgeois,W., Kretschmer,U., Muth,H.P. and Seela,F. (1990) J. Org. Chem., 55, 5784–5790. [Google Scholar]
- 18.Jiang S.-P., Raghunathan,G., Ting,K.-L., Xuan,J.C. and Jernigan,R.L. (1994) J. Biomol. Struct. Dyn., 12, 367–382. [DOI] [PubMed] [Google Scholar]
- 19.Users Manual of the DNA synthesizer. Applied Biosystems, Weiterstadt, Germany, p. 392.
- 20.Manual for the Oligonucleotide Purification Cartridges. Applied Biosystems, Weiterstadt, Germany.
- 21.Seela F. and Lampe,S. (1991) Helv. Chim. Acta, 74, 1790–1800. [Google Scholar]
- 22.McDowell J.A. and Turner,D.H. (1996) Biochemistry, 35, 14077–14089. [DOI] [PubMed] [Google Scholar]
- 23.Kennard O. (1987) In Eckstein,F. and Lilley,D.M.J. (eds), Nucleic Acids and Molecular Biology. Springer-Verlag, Heidelberg, Germany, pp. 25–52.