Abstract
To aid in the prioritization of deubiquitinases (DUBs) as anticancer targets, we developed an approach combining activity-based protein profiling (ABPP) with mass spectrometry in both non-small cell lung cancer (NSCLC) tumor tissues and cell lines along with analysis of available RNA interference and CRISPR screens. We identified 67 DUBs in NSCLC tissues, 17 of which were overexpressed in adenocarcinoma or squamous cell histologies and 12 of which scored as affecting lung cancer cell viability in RNAi or CRISPR screens. We used the CSN5 inhibitor, which targets COPS5/CSN5, as a tool to understand the biological significance of one of these 12 DUBs, COPS6, in lung cancer. Our study provides a powerful resource to interrogate the role of DUB signaling biology and nominates druggable targets for the treatment of lung cancer subtypes.
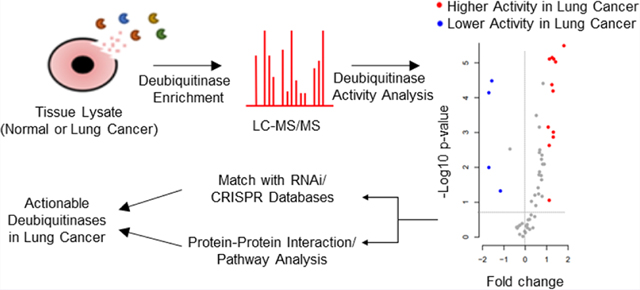
Graphical Abstract
INTRODUCTION
The ubiquitin (Ub) post-translational modification plays a critical role in the control of numerous cellular processes and is frequently implicated in various human diseases. Deubiquitinases (DUBs) that remove ubiquitin (Ub) from the modified substrates have been implicated in regulating transcription factors, cell cycle checkpoints, DNA damage repair, chromatin remodeling, and kinase signaling.1,2 The human genome encodes ~100 DUBs and controls a number of key cancer pathways including RAS signaling, the epithelial−mesenchymal transition (EMT), deregulation of p53 activity, NF-κB signaling, and interferon receptor signaling.3−6 There is growing interest in developing potential DUB inhibitors (DUBi) to attack vulnerabilities in Ub biology as novel therapeutics, based on the biological roles enumerated above and aberrant DUB expression in a number of cancers.7,8
A key prerequisite to accelerate the drug discovery and understanding of Ub biology in cancer can be facilitated by evaluating key Ub-regulating enzymes as drug targets in the context of cancer subtypes. While numerous atlases using DNA or mRNA sequencing have been reported, these can be limited given the absence of functional activity or clear knowledge of genetic mutation on protein function. We hypothesized that an activity-based landscape of DUBs would accelerate efforts toward targeting DUBs. To this end, we employed activity-based protein profiling (ABPP) using a DUB substrate mimic as an activity-based probe (ABP) to screen for DUB enzymes and provide a snapshot of the active enzymes within a tumor sample. Previously described, HA-tagged DUB-ABPs are comprised of full length Ub that has the C-terminal glycine replaced with a cysteine-reactive electrophilic group that covalently modifies the DUB after binding.9 Different HA-tagged ubiquitin probes such as HA-UbVME, HA-UbC2Br, and HA-UbPA show marginally different reactivity profiles for DUBs.9,10 In order to maximize DUB labeling, we compared probe-captured fractions from labeling using individual probes from commercial source and in-house (Moffitt) synthesized batches, as well as different ratios of two HA-tagged probes;: HA-UbVME and HA-UbPA in NSCLC cell lysates. Both Western blot and mass spectrometry data indicate broader ranges of DUBs were captured by using a 1:1 ratio of the in-house synthesized HA-UbVME and HA-UbPA probes. Hence, we employed a combination of HA-UbVME and HA-UbPA (1:1) probes to evaluate DUBs using ABPP in NSCLC tumors.
RESULTS AND DISCUSSION
Differential Expression of DUBs in Lung Tumors.
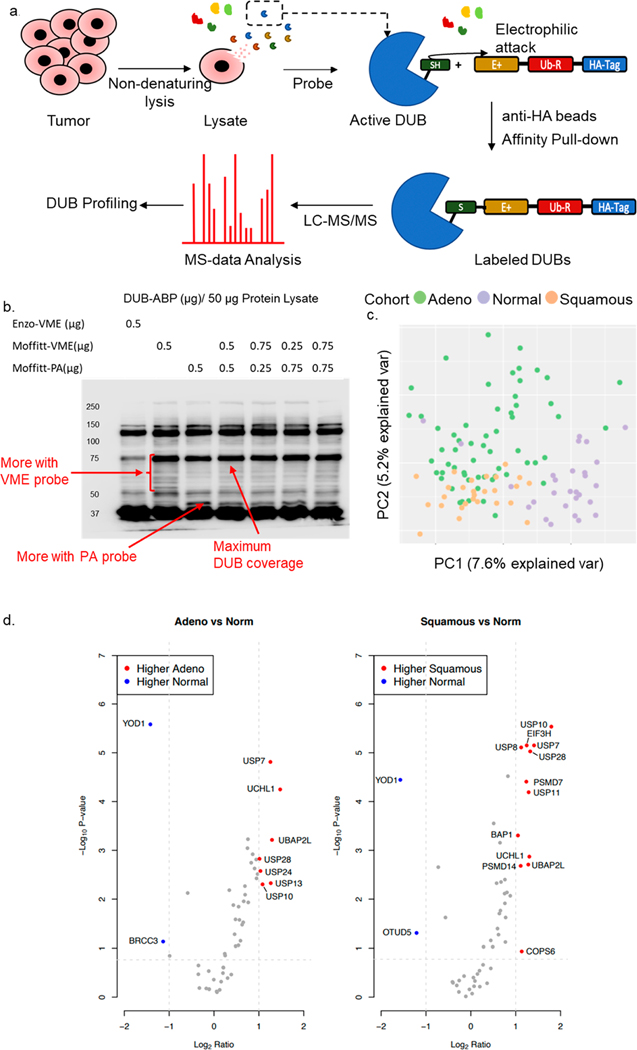
We engaged active DUB site-directed activity-based probes9,10 to capture DUB-ABP protein complexes from tumor samples (Figure 1a). We designed a cohort of 90 surgically resected lung cancer tumor tissues along with 30 unmatched normal tumor-adjacent (NTA) lung tissues. We selected 60 adenocarcinomas, 30 with oncogenic KRAS mutations, given the high unmet medical need to address this class,11 and 30 KRAS wild type while 30 consisted of squamous cell carcinoma, representing the two major histological subtypes of NSCLC. Equal distributions for gender, smoking status, and pathological stages were considered for determining the cohort of tumor samples (Supplementary Table 1). We optimized the recovery of the DUB proteins in lysates from lung cancer cell lines using in house generated DUB-ABP in 1 mg of homogenized tumor proteome (Supplementary Figure 1). In our hands, probe-enriched samples show that the combination of HA-UbVME and HA-UbPA probes (1:1) provides broader coverage of DUBs in protein lysate (Figure 1b). Proteins were subjected to in-gel digestion and quantified using liquid chromatography−tandem mass spectrometry (LC-MS/MS). We first assessed all identified proteins and observed a clear division in principal component analysis (PCA) within NTA and tumor samples (Figure 1c).
Figure 1.
Design and outcome for DUB-ABPP in NSCLC tumors: (a) Schematic diagram for the DUB-ABPP protocol. (b) Optimization of DUB-ABP using commercially available (Enzo-VME) and Moffitt generated (Moffitt-VME ad Moffitt-PA) probes. (c) PCA of tissue specimens using all proteins identified in tumor-ABPP. The graph indicates a clear separation between adenocarcinoma, squamous cell carcinoma, and NTA samples. (d) Volcano plot for the differentially active DUBs in adenocarcinoma and squamous cell carcinoma versus NTA. The significance cutoff is marked by dotted lines. Labeled DUBs indicate significantly higher activity (red) and significantly lower activity (blue) in tumors versus NTA.
Additional statistical analysis is described in the Supporting Information (Supplementary Figure 2). We identified a total of 67 DUBs from all 6 DUB families (Supplementary Table 2) and removed 14 DUBS from further analysis due to the rarity of their detection in the data set (Supplementary Figure 3). The remaining 53 DUBs were analyzed to evaluate differentially expressed DUBs (|log2 ratio| ≥ 1 and a Benjamini−Hochberg adjusted p-value ≤ 0.25 for two subtypes) in different subtypes of NSCLC. We identified 9 differentially expressed DUBs (UCHL1, UBP2L, USP13, USP7, USP10, USP24, USP28, BRCC3, and YOD1) in adenocarcinoma, and 14 differentially expressed DUBs (USP10, USP7, USP28, UCHL1, USP11, UBP2L, EIF3H, PSMD7, COPS6, USP8, PSMD14, BAP1, OTUD5, and YOD1) in squamous cell carcinoma compared to NTA (Figure 1d, Supplementary Table 3). Among these UCHL1, UBP2L, USP10, USP28, and USP7 were upregulated while YOD1 was downregulated in both adenocarcinoma and squamous cell carcinoma compared to NTA. We performed the Western blots for a subset of DUBs including USP10 and USP7 on the lysates derived from the leftover tissue samples used in the DUB-ABPP to look at the expression in tissues. The Western blots confirmed the differential expression for USP10 and USP7 and matched with the DUB-ABPP data showing higher expression in both lung adenocarcinoma and squamous tissue samples (Supplementary Figure 4a). We also examined the dose dependent effect on the cell viability in 12 NSCLC cell lines for inhibition of two upregulated DUBs identified through DUB-ABPP in tumor samples. Highly selective and potent DUB inhibitor targeting USP10 (spautin-1) resulted in significant loss of cell viability in 4 NSCLC cell lines tested while USP28 inhibitor (AZ-1) significantly decreased the cell viability of all 12 cell lines tested (Supplemetary Figure 4c). Although USP7 was also significantly upregulated, USP-7 inhibition did not affect the cell viability. This may be due to the lower potency of the available USP7 inhibitor (data not shown). Both DUBs, USP10 and USP28, are highly active in both lung adenocarcinoma and squamous carcinoma in DUB-ABPP data as compared to NTA and can be exploited to understand the previously unknown roles of these DUBs in lung cancer.12,13 Further, we assessed the absence of 14 DUBs not consistently detected across the cohort. OTUD1 was enriched in squamous (present in 67% of samples) compared to the tumor-adjacent (present in 17% of samples) by a Fisher’s exact test (p = 1.81 × 10−4, OR = 9.55 (Supplementary Figure 3). We identified differentially detected DUBs in different NSCLC histologies compared to NTA, but no DUBs were differentially expressed between KRAS mutant and KRAS wild type adenocarcinomas.
Analysis of DUB Interacting Proteins (DIPs).
As the affinity purification in this study involves nondenaturing conditions for the enrichment of ABP-labeled DUBs, proteins with noncovalent interactions with the ABP-labeled DUB proteins (DIPs) were also captured. We hypothesized that DIPs can provide insights into specific DUB related pathways that may differ across lung cancer subtypes. An earlier study has categorized high-confidence candidate interacting proteins of DUBs including components of large macromolecular regulatory complexes, such as the proteasome, COP9 signalosome, spliceosome, EIF3 complex, and transcriptional complexes.14 This study has reported substantial interconnectivity among DIPs. To understand the role of DUBs and DIPs in lung cancer subtypes, we next analyzed the DIPs associated with differentially expressed DUBs in particular subtypes of NSCLC using BioGRID.15 In the tumor-ABPP data, we identified DUBs that interact with the above-mentioned regulatory complexes such as USP4, USP15, USP39 (spliceosome), PSMD7, PSMD14 (proteasome), EIF3H, EIF3F (EIF3 complex), and COPS5, COPS6 (COP9 signalosome) and observed a significant enrichment of members of these protein complexes in samples enriched by DUB-ABP capture (Figure 2a). This result is consistent with the enrichment of DIPs along with DUBs in DUB-ABP pulldown samples with reproducible interactor proteins in NSCLC cell lines (Supplementary Figure 5, Supplementary Table 4), and therefore, ABPP data can also be used to explore differentially expressed DIPs. We performed the Western blots for COPS6 and DIPs of COPS6 including EGFR and FBXO22 which were enriched in squamous cell carcinoma. Western blot results showed they appeared to be higher in squamous tissue samples compared to normal adjacent tissue in accordance with the observations from the DUB-ABPP data. (Supplementary Figure 4b)
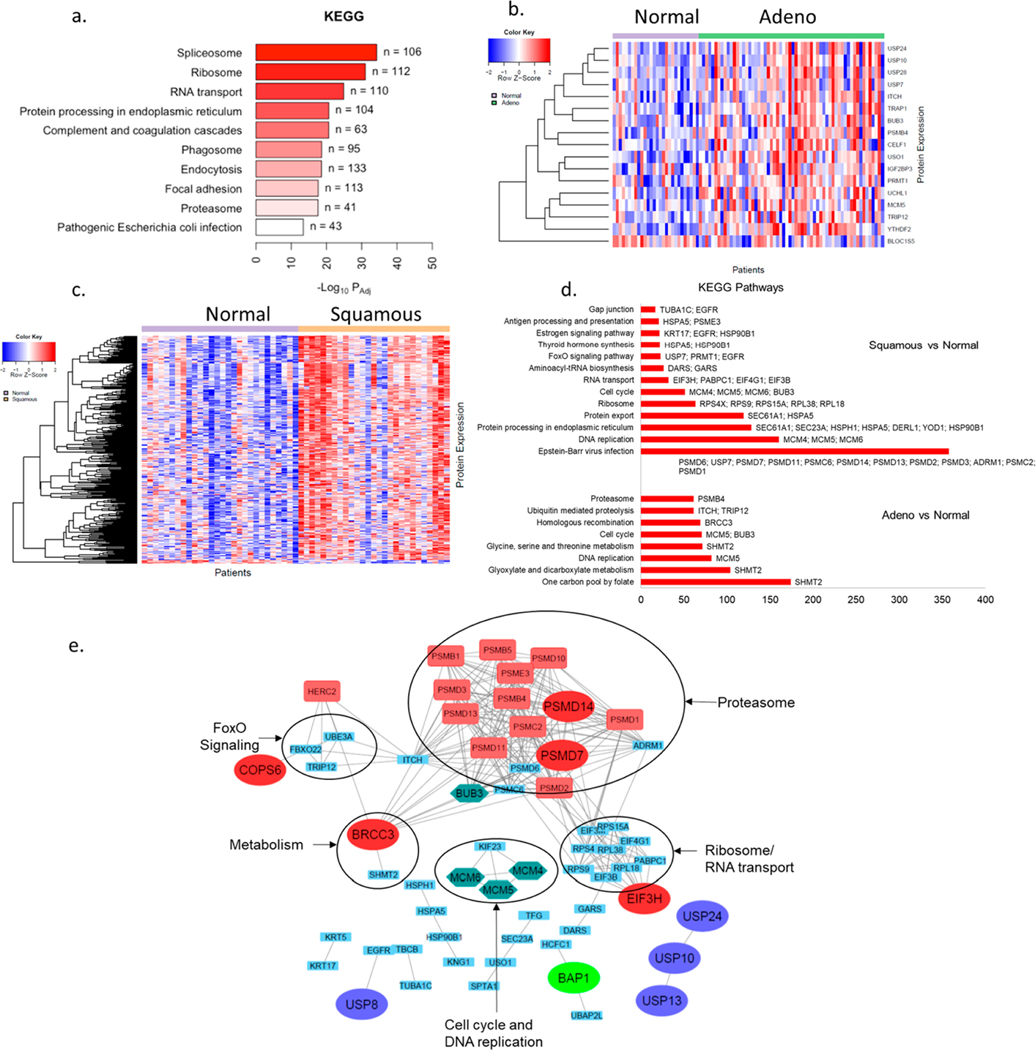
Figure 2.
Pathway analysis for DUBs and DIPs in NSCLC tumors: (a) KEGG pathway analysis of all proteins identified in tumor ABPP. (b) Heat map for 19 differentially expressed DIPs of differentially active DUBs in adenocarcinoma versus NTA. (c) Heat map for 75 differentially expressed DIPs of differentially active DUBs in squamous carcinoma versus NTA. (d) Major pathways identified using KEGG pathway analysis of differentially expressed DIPs in adenocarcinoma and squamous carcinoma versus NTA. (e) STRING database association between differentially active DUBs in either of the tumor subtypes vs NTA and their differentially expressed DIPs. DUBs belonging to different subfamilies are represented by enlarged ovals of different colors. Proteins in the green hexagon are involved in cell cycle and DNA repair pathways. Proteins in red rectangles represent contributors to the G2/M cell cycle checkpoint.
We next focused on analyzing the levels of DIPs recovered in the pulldowns for known interactors of the differentially expressed DUBs in our data, which can yield altered DUB-associated pathways in lung cancer subtypes. We explored known DIPs of differentially expressed DUBs in each NSCLC tumor subtype. We identified 168 DIPs for the nine differentially expressed DUBs in adenocarcinoma versus NTA using BioGRID. Of these, 19 DIPs were differentially expressed in adenocarcinoma vs NTA (Figure 2b, Supplementary Table 5). Similarly, we identified 370 DIPs for the 14 differentially expressed DUBs in squamous cell carcinoma versus NTA. Of these, 75 DIPs were differentially expressed in squamous cell carcinoma versus NTA (Figure 2c, Supplementary Table 5). KEGG pathway analysis of the differentially expressed DUBs and DIPs in adenocarcinoma vs NTA revealed the differentially expressed DIPs involved in cell cycle, DNA replication (MCM5; BUB3), and multiple metabolic pathways (SHMT2). Similar analysis in squamous cell carcinoma versus NTA revealed the differentially expressed DIPs involved in antigen, immune system (HSPA5; PSME3), protein processing in endoplasmic reticulum (SEC61A1; SEC23A; HSPH1; HSPA5; DERL1; YOD1; HSP90B1), cell cycle, and DNA replication (MCM4; MCM5; MCM6; BUB3), protein export (SEC61A1; HSPA5), and RNA transport (EIF3H; PABPC1; EIF4G1; EIF3B) (Figure 2d). Differentially expressed DUBs in one of the tumor subtypes versus NTA and their differentially expressed DIPs identified via the BioGRID database were further visualized by STRING16 network analysis. The STRING analysis for the association of differentially expressed DUBs and DIPs in different subtypes of NSCLC supports that these proteins are extensively interconnected as part of large molecular complexes, such as the proteasome and ribosome (Figure 2e), and further analysis can provide insights into the role of these DUBs in NSCLC.
Comaprison of DUB Profile between Lung Cancer Cell Lines and Tumors.
Having developed an activity landscape along with nominating differentially active DUBs in lung cancer subtypes using tumors and NTA, we next turned our attention to delineating possible vulnerabilities related to these DUBs. To create a link between the tissue specimens and cell lines that can be probed for vulnerabilities, we compared the DUB profiles of tumor samples with the DUB profiles from 25 adenocarcinoma LC cell lines (Supplementary Table 6). We identified 53 DUBs in LC cell lines; 51 out of 53 were included in the group of 67 DUBs identified in tumor samples (Supplementary Figure 6). Among the additional DUBs identified exclusively in tumor samples versus cell line samples, five of the DUBs, namely, MINDY4, MINDY2, JOSD2, USP38, and USP43 were identified in more than 70% of the tumor samples but none of the cell line samples. Also, USP24 was identified in 100% of the tumor samples but only 2/25 cell line samples. These six DUBs, therefore, may be specific to tumor-associated cells, such as tumor resident immune cells or fibroblasts or endothelial cells. We compared the DIPs from 25 lung adenocarcinoma cell lines to the DIPs from lung adenocarcinoma tumor samples. Among the 1081 DIPs of DUBs identified in lung adenocarcinoma tumor samples and 1131 DIPs of DUBs identified in the cell line samples, 841 DIPs matched between the tumor and cell lines (Supplementary Figure 5, Supplementary Table 4).
DUB Activity from ABPP Compared to DUB-Gene Essentiality.
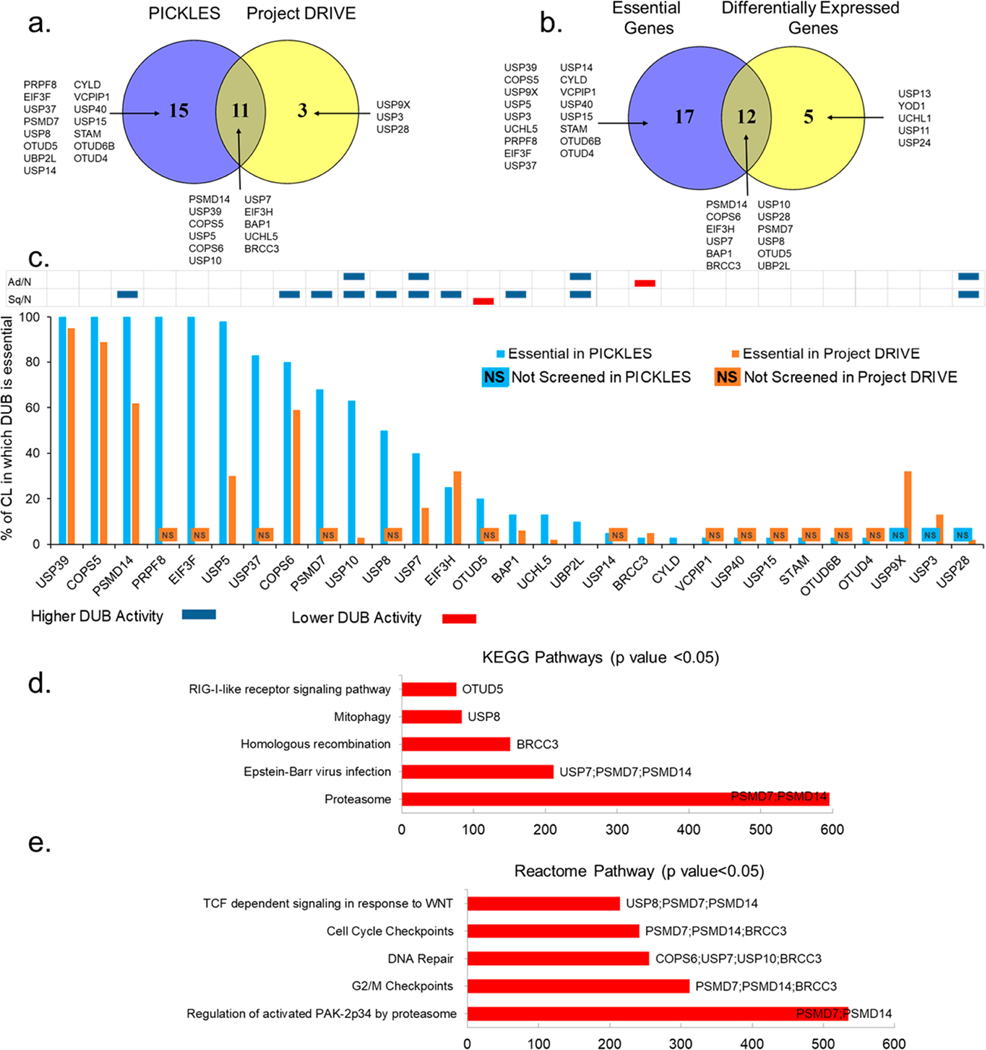
Based on this high degree of overlap, this cell line panel can serve as a model to generate leads from tumor profiling data and RNAi or CRISPR screens. Next, we searched the DUBs identified through our tumor ABPP analyses for their effects on cell viability. We leveraged essentiality through viability effects of mRNA knockdown from two resources: (i) Project DRIVE, a large scale RNAi screen of 7,837 genes including 35 DUBs in 67 lung cancer cell lines,17 and (ii) PICKLES, a pooled in vitro CRISPR knockout library for 18,000 protein-coding genes including 92 DUBs in 40 lung cancer cell lines.18 We explored both databases to achieve the maximum coverage of DUBs. Sensitivity score cut offs for statistically significant loss of viability were employed to nominate essential genes from Project DRIVE and the PICKLES database.17,18 Fourteen DUBs in Project DRIVE and 26 DUBs in PICKLES were identified as essential in one or more LC cell lines tested (Figure 3a). Out of these, 11 DUBs were essential in both screenings. Therefore, a total of 29 DUBs scored as essential in at least one LC cell line tested in either vulnerability screen. We then matched these 29 essential DUBs with the 17 differentially expressed DUBs in the ABPP data in either of the tumor subtypes compared to NTA. Twelve out of 17 differentially expressed DUBs overlapped with 29 essential genes from the screenings (Figure 3b, Supplementary Table 7). The DUB gene essentiality score was calculated based on the percentage of LC cell lines tested in which a specific DUB is essential (Figure 3c). The data revealed that most of the upregulated DUBs were essential in a large number of cell lines tested.
Figure 3.
DUBs essentiality in lung cancer: (a) Overlap between the 26 DUBs (PICKLES) and 14 DUBs (Project DRIVE) identified as essential in viability screens. (b) Overlap between all essential DUBs and differentially active DUBs in ABPP for either NSCLC tumor subtype. (c) DUB essentiality score. The y-axis represents the percentage of cell lines in which specific DUB is essential in PICKLES (blue column) or Project DRIVE (orange column) screening. No column corresponding to any DUB on the x-axis indicates that the specific DUB was not screened in PICKLES (NS in blue box) and/or Project DRIVE (NS in orange box). The table above the columns compares DUB activity for LC subtypes, Adeno vs NTA and Squamous vs NTA in blue (higher DUB activity) and red (lower DUB activity), respectively. (d) KEGG pathway analysis of 12 DUBs that are differentially active in adenocarcinoma and squamous carcinoma vs NTA and are essential in at least one of the two gene essentiality screens. (e) Reactome pathway analysis of 12 DUBs that are differentially active in adenocarcinoma and squamous carcinoma vs NTA and are essential in at least one of the two gene essentiality screens.
Directed by these results, we next focused our analysis on these 12 DUBs (PSMD14, COPS6, PSMD7, USP10, USP8, USP7, EIF3H, OTUD5, BAP1, UBP2L, BRCC3, and USP28), which are differentially active in LC tumor subtypes and score as essential genes in viability screens. KEGG and Reactome pathway analysis indicates these DUBs are involved in vital cellular processes such as DNA repair, cell cycle and G2/M checkpoints, DNA damage recognition in GG-NER, and innate immune response regulation (Figure 3d and e). Among these hits, BAP1, BRCC3, USP10, USP28, USP7, and USP8 control functionally interconnected DNA damage response and cell cycle checkpoints and are frequently deregulated in tumorigenesis.19−23 This subset of DUBs contains attractive drug candidates. COPS6, a component of the COP9 signalosome complex (CSN complex), appears to be extremely intriguing, because it scores as essential in 80% and 60% of the LC cell lines tested in PICKLES and Project DRIVE, respectively. In our data, COPS6 is highly upregulated in squamous cell carcinoma, a tumor type with high unmet medical need. COPS6 is also involved in degradation of several cancer related proteins and pathways such as EGFR, p53, c-myc and c-Jun, and, hence, an interesting candidate in cancer therapeutics.24,25
Potential Role of COPS5/COPS6 in Lung Cancer.
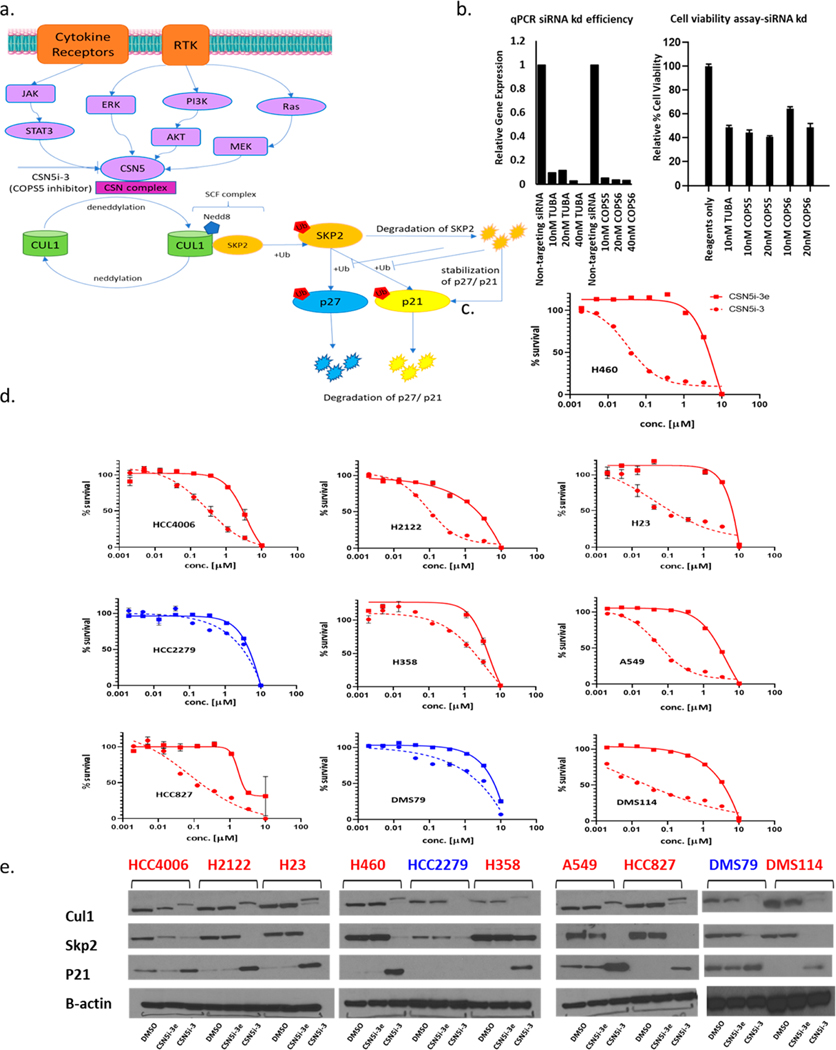
We next took advantage of recent developments in small molecules that target the CSN complex. First, we validated essentiality for COPS6 and COPS5 in our own hands using siRNA knockdown in H460 KRAS mutant lung adenocarcinoma cells. COPS5 is the active DUB component associated with COPS6 and involved in the enzymatic activity of the COP9 signalosome complex,26 specifically targeting the ubiquitin-like molecule Nedd8 (Figure 4a). We observed significant suppression of cell growth in H460 cells in accordance with the observations from the previously mentioned public databases (Figure 4b). To further extend our understanding of the role of COPS5/COPS6 in lung tumorigenesis, we studied inhibition of cell growth using a known small molecule inhibitor of COPS5, CSN5i-3, which was revealed as a selective and potent inhibitor of COPS5 through a lead optimization approach,27 along with the inactive enantiomer CSN5i-3e as a control. CSN5i-3 resulted in cell death in H460 LC cells with an IC50 < 100 nM, while the inactive enantiomer has no effect on cell viability up to 1 μM (Figure 4c). We analyzed the effect of CSN5i-3 and CSN5i-3e on cell viability in a more diverse set of lung cancer cell lines, including those with EGFR or KRAS mutations, and small cell lung cancers. Most cell lines were similar in response to H460; CSN5i-3 was much more effective than CSN5i-3e. However, HCC2279 (NSCLC, EGFR mutant lung adenocarcinoma) and DMS79 (SCLC) were resistant to CSN5i-3 until treated with concentration > 1 μM (Figure 4d). COPS5 mediates the deneddylation of the cullin subunits of the SCF-type E3 ligase complexes.24 CSN5i-3 inhibition of COPS5 resulted in inhibition of the deneddylation process, and hence, accumulation of Cul1 in all the cell lines tested. Interestingly, treatment of all the cell lines sensitive to CSN5i-3 resulted in significant accumulation of p21, a substrate of CUL1-SKP2,28 whereas the two resistant cell lines, HCC2279 and DMS79, did not show any significant change in p21 expression after CSN5i-3 treatment (Figure 4e). The lack of response in these two cell lines does not appear to be related to lack of target engagement, as CSN5i-3 effectively inhibited Cul1 deneddylation and stimulated Skp2 degradation in all cell lines tested. Therefore, it appears that the COPS5-SKP2-p21 axis may have cell line specific roles in controlling cell viability.
Figure 4.
Effect of COPS5 inhibition using CSN5i-3 and CSN5i-3e on NSCLC and SCLC cell lines. (a) Schematic representation of regulation of cullin neddylation and corresponding substrates by COPS5. (b) Effect of COPS5/COPS6 siRNA knockdown on cell viability in H460 cells. α-tubulin (TUBA) is used as a housekeeping gene control. (c) Effect of CSN5i-3 (active enantiomer) and CSN5i-3e (inactive enantiomer) on cell viability in H460 cells. (d) Effect of CSN5i-3 and CSN5i-3e on cell viability in a panel of NSCLC and SCLC cell lines. (e) Effect on substrates of Cul1 by CSN5i-3 and CSN5i-3e. In parts c−e the cell lines showing loss of viability due to CSN5i-3 treatment are shown in red and the cell lines unaffected by CSN5i-3 treatment are shown in blue.
Starting with surgically resected lung tumors and NTA, we identified active DUBs using mass spectrometry, found differentially active DUBs by comparing NTA in two major NSCLC subtypes (adenocarcinoma and squamous cell carcinoma), and were able to nominate DUBs with roles in controlling cell viability by integrating our ABPP results with publicly available RNAi and CRISPR screens. Highly selective and potent DUB inhibitors targeting USP10 and USP28 were used to validate the nominated targets from our ABPP data. Both DUBs, USP28 and USP10, are highly active in both lung adenocarcinoma and squamous carcinoma in DUB-ABPP data as compared to NTA and can be exploited to understand the previously unknown roles of these DUBs in lung cancer.
Our results with COPS6/COPS5 inhibition are particularly important for targeting squamous cell lung cancer, which has few successes in targeted agents and high unmet medical need. Further validation of the pathways associated with COPS5/COPS6 in the ABPP data for these particular cell lines or subtypes of LC may provide information to understand why some cell lines respond to this DUB inhibition while others do not. While previous studies have provided thelandscape of DUBs and the DUBome in cell lines,9,10 our study presents the most comprehensive evaluation of DUBs in human NSCLC tumor samples using ABPP. The 12 active DUBs nominated from our data, which are differentially active in LC tumor subtypes and are essential genes in viability screens, have been associated with vital cellular processes. These DUBs can present potential therapeutic candidates for novel drugs, and further studies using RNAi knockdown or selective inhibitors can provide further insights into the associated pathways.
This study involving DUB-ABPs and mass spectrometry in human tumor samples is the largest study to date to our knowledge using ABPP in tumors and mass spectrometry. Our data can serve as a powerful resource for exploring the pathways associated with DUBs in lung cancer, and our approach combining tumor profiling with RNAi and CRISPR viability screens may enable development of new therapeutic strategies targeting DUBs or DUB interacting proteins.
METHODS
Detailed experimental procedures for the production of ABP-probes and for the analysis of LC-MS/MS data are provided in the Supporting Information. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifiers PXD028802 and 10.6019/PXD028802
Supplementary Material
ACKNOWLEDGMENTS
We thank B. Kessler from University of Oxford for his generous gift of plasmid for probe synthesis at Moffitt Cancer Center. This work has been supported in part by R50CA211447 (HRL), Proteomics & Metabolomics Core, Chemical Biology Core, and Biostatistics and Bioinformatics Shared Resource at Moffitt Cancer Center, an NCI-designated Comprehensive Cancer Center (P30-CA076292).
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acschembio.2c00018.
Optimization for probe/ protein lysate ratio for HA-UbVME probes in two NSCLC cell lines, statistical analysis of DUB-ABPP data, Western blots for DUBs, DUBs eliminated from ABPP data after evaluation of missingness in tumor profiling data, comparison of DUB interacting proteins in cell lines and tumors, MALDI MS of Moffitt HA-UbVME and HA-UbPA probes, and methods for probe synthesis and LC-MS/MS sample preparation (PDF)
LC-MS/MS raw data files and LC-MS/MS data analysis parameters (XLSX)
Complete contact information is available at: https://pubs.acs.org/10.1021/acschembio.2c00018
The authors declare no competing financial interest.
Contributor Information
Shikha Mahajan, Department of Thoracic Oncology, H. Lee Moffitt Cancer Center and Research Institute, Tampa, Florida 33612, United States.
Anurima Majumder, Department of Thoracic Oncology, H. Lee Moffitt Cancer Center and Research Institute, Tampa, Florida 33612, United States.
Paul A. Stewart, Biostatistics and Bioinformatics Shared Resource, H. Lee Moffitt Cancer Center and Research Institute, Tampa, Florida 33612, United States
Yian Ann Chen, Department of Biostatistics and Bioinformatics, H. Lee Moffitt Cancer Center and Research Institute, Tampa, Florida 33612, United States.
Emma Adhikari, Department of Tumor Biology, H. Lee Moffitt Cancer Center and Research Institute, Tampa, Florida 33612, United States.
Bin Fang, Proteomics & Metabolomics Core, H. Lee Moffitt Cancer Center and Research Institute, Tampa, Florida 33612, United States.
Yan Yang, Chemical Biology Core, H. Lee Moffitt Cancer Center and Research Institute, Tampa, Florida 33612, United States.
Harshani Lawrence, Chemical Biology Core, H. Lee Moffitt Cancer Center and Research Institute, Tampa, Florida 33612, United States.
Fumi Kinose, Department of Thoracic Oncology, H. Lee Moffitt Cancer Center and Research Institute, Tampa, Florida 33612, United States.
John M. Koomen, Department of Molecular Oncology, H. Lee Moffitt Cancer Center and Research Institute, Tampa, Florida 33612, United States
Eric B. Haura, Department of Thoracic Oncology, H. Lee Moffitt Cancer Center and Research Institute, Tampa, Florida 33612, United States
REFERENCES
- (1).Pinto-Fernandez A; Kessler BM DUBbing Cancer: Deubiquitylating Enzymes Involved in Epigenetics, DNA Damage and the Cell Cycle As Therapeutic Targets. Front Genet. 2016, 7, 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Skaug B; Jiang X; Chen ZJ The role of ubiquitin in NF-kappaB regulatory pathways. Annu. Rev. Biochem. 2009, 78, 769–96. [DOI] [PubMed] [Google Scholar]
- (3).Mustachio LM; Lu Y; Tafe LJ; Memoli V; Rodriguez-Canales J; Mino B; Villalobos PA; Wistuba I; Katayama H; Hanash SM; et al. Deubiquitinase USP18 Loss Mislocalizes and Destabilizes KRAS in Lung Cancer. Mol. Cancer Res. 2017, 15 (7), 905–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Saldana M; VanderVorst K; Berg AL; Lee H; Carraway KL Otubain 1: a non-canonical deubiquitinase with an emerging role in cancer. Endocr Relat Cancer. 2019, 26 (1), R1–R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Wertz I; Dixit V. A20–a bipartite ubiquitin editing enzyme with immunoregulatory potential. Adv. Exp. Med. Biol. 2014, 809, 1–12. [DOI] [PubMed] [Google Scholar]
- (6).Zheng H; Gupta V; Patterson-Fortin J; Bhattacharya S; Katlinski K; Wu J; Varghese B; Carbone CJ; Aressy B; Fuchs SY; et al. A BRISC-SHMT complex deubiquitinates IFNAR1 and regulates interferon responses. Cell Rep. 2013, 5 (1), 180–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Cremer A; Stegmaier K. Targeting DUBs to degrade oncogenic proteins. Br J. Cancer. 2020, 122, 1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Wertz IE; Murray JM Structurally-defined deubiquitinase inhibitors provide opportunities to investigate disease mechanisms. Drug Discov Today Technol. 2019, 31, 109–123. [DOI] [PubMed] [Google Scholar]
- (9).Borodovsky A; Ovaa H; Kolli N; Gan-Erdene T; Wilkinson KD; Ploegh HL; Kessler BM Chemistry-based functional proteomics reveals novel members of the deubiquitinating enzyme family. Chem. Biol. 2002, 9 (10), 1149–59. [DOI] [PubMed] [Google Scholar]
- (10).Ekkebus R; van Kasteren SI; Kulathu Y; Scholten A; Berlin I; Geurink PP; de Jong A; Goerdayal S; Neefjes J; Heck AJ; et al. On terminal alkynes that can react with active-site cysteine nucleophiles in proteases. J. Am. Chem. Soc. 2013, 135 (8), 2867–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Román M; Baraibar I; López I; Nadal E; Rolfo C; Vicent S; Gil-Bazo I. KRAS oncogene in non-small cell lung cancer: clinical perspectives on the treatment of an old target. Mol. Cancer. 2018, 17 (1), 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Hu C; Zhang M; Moses N; Hu CL; Polin L; Chen W; Jang H; Heyza J; Malysa A; Caruso JA; et al. The USP10-HDAC6 axis confers cisplatin resistance in non-small cell lung cancer lacking wild-type p53. Cell Death Dis. 2020, 11 (5), 328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Wrigley JD; Gavory G; Simpson I; Preston M; Plant H; Bradley J; Goeppert AU; Rozycka E; Davies G; Walsh J; et al. Identification and Characterization of Dual Inhibitors of the USP25/28 Deubiquitinating Enzyme Subfamily. ACS Chem. Biol. 2017, 12 (12), 3113–3125. [DOI] [PubMed] [Google Scholar]
- (14).Sowa ME; Bennett EJ; Gygi SP; Harper JW Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009, 138 (2), 389–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Oughtred R; Stark C; Breitkreutz BJ; Rust J; Boucher L; Chang C; Kolas N; O’Donnell L; Leung G; McAdam R; et al. The BioGRID interaction database: 2019 update. Nucleic Acids Res. 2019, 47 (D1), D529–D541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Szklarczyk D; Gable AL; Lyon D; Junge A; Wyder S; Huerta-Cepas J; Simonovic M; Doncheva NT; Morris JH; Bork P; et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47 (D1), D607–D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).McDonald ER 3rd; de Weck A; Schlabach MR; Billy E; Mavrakis KJ; Hoffman GR; Belur D; Castelletti D; Frias E; Gampa K; et al. Project DRIVE: A Compendium of Cancer Dependencies and Synthetic Lethal Relationships Uncovered by Large-Scale, Deep RNAi Screening. Cell. 2017, 170 (3), 577–592.e10. [DOI] [PubMed] [Google Scholar]
- (18).Lenoir WF; Lim TL; Hart T. PICKLES: the database of pooled in-vitro CRISPR knockout library essentiality screens. Nucleic Acids Res. 2018, 46 (D1), D776–D780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Atanassov BS; Koutelou E; Dent SY The role of deubiquitinating enzymes in chromatin regulation. FEBS Lett. 2011, 585 (13), 2016–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Popov N; Wanzel M; Madiredjo M; Zhang D; Beijersbergen R; Bernards R; Moll R; Elledge SJ; Eilers M. The ubiquitin-specific protease USP28 is required for MYC stability. Nat. Cell Biol. 2007, 9 (7), 765–74. [DOI] [PubMed] [Google Scholar]
- (21).Zhang D; Zaugg K; Mak TW; Elledge SJ A role for the deubiquitinating enzyme USP28 in control of the DNA-damage response. Cell. 2006, 126 (3), 529–42. [DOI] [PubMed] [Google Scholar]
- (22).Cheung M; Testa JR BAP1, a tumor suppressor gene driving malignant mesothelioma. Transl Lung Cancer Res. 2017, 6 (3), 270–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Deng J; Hou G; Fang Z; Liu J; Lv XD Distinct expression and prognostic value of OTU domain-containing proteins in non-small-cell lung cancer. Oncol Lett. 2019, 18 (5), 5417–5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Fang L; Lu W; Choi HH; Yeung SC; Tung JY; Hsiao CD; Fuentes-Mattei E; Menter D; Chen C; Wang L; et al. ERK2-Dependent Phosphorylation of CSN6 Is Critical in Colorectal Cancer Development. Cancer Cell. 2015, 28 (2), 183–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Hou J; Deng Q; Zhou J; Zou J; Zhang Y; Tan P; Zhang W; Cui H. CSN6 controls the proliferation and metastasis of glioblastoma by CHIP-mediated degradation of EGFR. Oncogene 2017, 36 (8), 1134–1144. [DOI] [PubMed] [Google Scholar]
- (26).Lingaraju GM; Bunker RD; Cavadini S; Hess D; Hassiepen U; Renatus M; et al. Crystal structure of the human COP9 signalosome. Nature 2014, 512, 161–165. [DOI] [PubMed] [Google Scholar]
- (27).Schlierf A; Altmann E; Quancard J; Jefferson AB; Assenberg R; Renatus M; Jones M; Hassiepen U; Schaefer M; Kiffe M; et al. Targeted inhibition of the COP9 signalosome for treatment of cancer. Nat. Commun. 2016, 7, 13166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Cope GA; Deshaies RJ Targeted silencing of Jab1/Csn5 in human cells downregulates SCF activity through reduction of F-box protein levels. BMC Biochem. 2006, 7, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.