Abstract
A cDNA fragment encoding part of a DNA methyltransferase was isolated from maize. The putative amino acid sequence identically matched that deduced from a genomic sequence in the database (accession no. AF063403), and the corresponding gene was designated as ZmMET1. Bacterially expressed ZmMET1 actively methylated DNA in vitro. Transcripts of ZmMET1 could be shown to exclusively accumulate in actively proliferating cells of the meristems of mesocotyls and root apices, suggesting ZmMET1 expression to be associated with DNA replication. This was confirmed by simultaneous decrease of transcripts of ZmMET1 and histone H3, a marker for DNA replication, in seedlings exposed to wounding, desiccation and salinity, all of which suppress cell division. Cold stress also depressed both transcripts in root tissues. In contrast, however, accumulation of ZmMET1 transcripts in shoot mesocotyls was not affected by cold stress, whereas those for H3 sharply decreased. Such a differential accumulation of ZmMET1 transcripts was consistent with ZmMET1 protein levels as revealed by western blotting. Expression of ZmMET1 is thus coexistent, but not completely dependent on DNA replication. Southern hybridization analysis with a methylation-sensitive restriction enzyme revealed that cold treatment induced demethylation of DNA in the Ac/Ds transposon region, but not in other genes, and that such demethylation primarily occurred in roots. These results suggested that the methylation level was decreased selectively by cold treatment, and that ZmMET1 may, at least partly, prevent such demethylation.
INTRODUCTION
DNA of higher plants contains 5-methyl cytosine (m5C) as up to 30% of the total cytosine residues (1). It occurs predominantly in CG and CNG, and levels change in a tissue- and/or development-specific manner (2–4). The physiological functions in plants are not absolutely clear, but it is proposed to play an important role for modulating DNA compartmentalization, determining chromatin structure, genomic imprinting, regulation of tissue-specific gene expression, aging and gene silencing (1,4,5).
Methylation of cytosines is catalyzed by an enzyme, DNA (cytosine-5) methyltransferase (MET) (EC 2.1.1.37), which transfers a methyl group from S-adenosylmethionine (AdoMet) to the fifth position of a cytosine residue. Depending on the catalytic properties, isoforms are classified into two major groups; de novo MET and maintenance MET (6,7). Since cytosine methylation is a post replicative process (8), maintenance MET which preferentially methylates cytosine residues in hemi-methylated DNAs has been intensively studied (9–11). Corresponding genes have been successfully isolated from several species, including Arabidopsis thaliana, pea, carrot and tobacco plants (1,12–15), and the genomic sequence for maize has also been reported (6). A survey of polypeptides encoded by these genes revealed that they are classified into three groups, METI, METII and METIII (6,16). Among these proteins, METI has been most intensively studied, being of considerable size, ranging up to 170 kDa, and containing well conserved domains found not only in plant METs but also in the animal counterparts (2,3,6,17). Further analyses showed the polypeptides to comprise two domains, a C-terminal catalytic domain, which is similar in structure to prokaryote MET, and a regulatory N-terminal domain (1). The catalytic domain contains up to 10 conserved motifs, designated I to X, which are essentially organized in the same order. The motifs I, II, III, IV, V and X constitute the AdoMet-binding pocket and the highly conserved motif IV binds DNA. The subregion between motifs VIII and IX constitutes the domain that recognizes the target methylation site (18,19).
In contrast to this physico-chemical characterization, the physiological significance in plants has received only limited attention. In the carrot and pea, MET transcripts are mainly observed in meristematic tissues, suggesting them to be associated with cell division (12,13). However, no experiments have so far been carried out on the impact of exogenous factors on MET expression, despite increasing knowledge on the essential roles of DNA methylation, for example in gene silencing and/or self-defense (4,11). Transposable elements have been shown to be heavily methylated, probably for silencing by the host plants (11,20) and vernalization was proposed to be associated with demethylation of genomic DNA (21,22). In an attempt to understand MET roles in plants, we selected maize as an experimental material, because previous studies have relatively well characterized its genome silenced by hypermethylation and the included multi-copies of transposons and highly repetitive regions (23). In the present study, isolation and molecular characterization of a cDNA fragment encoding MET from maize were carried out, and possible associations not only with DNA replication but also with self-defense of genomic DNA explored.
MATERIALS AND METHODS
Plant material and stress treatments
Maize seeds (Zea mays L. cv. Golden arrow) were germinated and grown in a hydroponic system with 1/5 diluted Murashige and Skoog (MS) medium under continuous light for 10–15 days at 23°C and 70% relative humidity in a growth cabinet. Seedlings were dissected into leaf blades, leaf sheaths, mesocotyls, mature roots (without meristem) and root apices (with meristem), frozen in liquid nitrogen and stored at –85°C until use. To investigate wounding responses, excised segments of mesocotyl were incubated under the same light and temperature conditions in covered petri dishes containing 30 ml of MS medium. For drought treatment, MS medium was removed from the cultures allowing roots to dry in an incubator. Salt stress was applied by adding 0.2 M NaCl to the medium and chilling experiments accomplished by transferring seedlings to an incubator at 4°C with the same light and humidity conditions. Ethyl-methane sulfonate (EMS) treatments were performed by submerging entire seedlings in MS medium containing EMS at 0.5%. After incubation for 24 h under the same light and temperature conditions, samples were rinsed with 5% sodium thiosulfate solution to neutralize EMS, transferred back to the original hydroponic culture condition, and incubated further under the same light, humidity and temperature conditions.
Isolation of ZmMET1 cDNA, probe preparation and DNA sequencing
A cDNA library, constructed in a ZAP cloning vector (Stratagene, La Jolla, CA), was screened with a 32P-labeled cDNA fragment of NtMET1, a gene encoding a tobacco MET. After secondary screening, positive clones were purified and excised in vivo in accordance with the manufacturer’s instructions. A resulting cDNA fragment of 2646 bp was amplified from the λZAP clone with universal primer pairs of M13–20 and RV to produce a probe for DNA blot analysis. A DNA fragment of 456 bp between the catalytic motifs I and VI was amplified with primer pairs of P11 (forward: 5′-CGTCTAGCTACTCTTGACATTTTTG) and P12 (reverse: 5′-AAAGTTCCGAACATTTTCTAACAG) to produce the probe cDNA, P1 for in situ hybridization. A DNA fragment of 459 bp between the catalytic motif X and the 3′ untranslated region was amplified with primer pairs of P21 (forward: 5′-ATCACAGTCCGGGAATGTGC) and P22 (reverse: 5′-AAGTTAATCTCATGTTGTCATTAATCACCA) to produce the probe cDNA, P2 for RNA blot analysis. A 480 bp DNA fragment for maize histone H3 was amplified from a cDNA library with primer pairs of P31 (forward: 5′-AGCACCAAAGCTCACGATGG) and P32 (reverse: 5′-CTACAAGCAGGCCCGAAGC) to produce the probe cDNA, P3 for RNA blot analysis. DNA sequencing was performed using the dideoxynucleotide chain termination method with a Vistra system Thermo Sequenase™ pre-mixed cycle sequencing kit (Amersham Life Science, Buckinghamshire, UK and Molecular Dynamics, Sunnyvale, CA) and an automated laser fluorescence sequencing apparatus (Hitachi SQ5500). Sequence analysis and comparisons were made with the MacDNAsis software (Hitachi, Tokyo, Japan; Pharmacia, Tokyo, Japan) and the BLASTX and the BLASTN programs (www BLAST server of the National Center for Biotechnology Information). A 2546 bp cDNA fragment for Ac was amplified from a SK− plasmid construct (provided by K. Shimamoto) by using universal primer pairs of M13–20 and RV to produce a probe for DNA blot analysis.
GST-fusion proteins
The ZmME1 fragment encoding MET region was cloned in-flame into BamHI and XhoI sites of pGEX4T (Pharmacia) to create pZmMET1. The insert DNA of the recombinant plasmid was confirmed by direct sequencing. To generate GST-fusion proteins, those plasmids were introduced into Escherichia coli JM109 cells. The expressed proteins were purified using glutathione–Sepharose (Pharmacia) columns as described by the supplier.
Enzyme assays
MET activity for time course analysis was measured in a reaction mixture of 900 µl containing 10% glycerol, 50 mM Tris–HCl, pH 7.5, 20 mM EDTA, 3 mM DTT, 0.25 µg/µl RNase A, 35 ng/µl dcm– E.coli (strain ME7791) genomic DNA, 20 µCi (methyl-3H)AdoMet (72 Ci/mmol, Amersham Pharmacia Biotech) and 0.5 ng/µl or 1 ng/µl of GST-ZmMET1 protein purified as described above. Alternatively, 1 ng/µl boiled GST-ZmMET1 protein or 2.25 × 10–5 U/µl of M.HapII was used for negative and positive controls, respectively. Incubations were done at 37°C, and a 300 µl aliquot was sampled every 90 min and DNA was extracted with phenol/chloroform followed by ethanol precipitation. The DNA pellet was dried, dissolved in a 300 µl TE buffer, spotted on a Whatman GF/C filter paper and counted for radioactivity. Steady-state kinetics were performed in reaction mixtures of 300 µl as described above containing various concentrations of (methyl-3H)AdoMet and 1 ng/µl of GST-ZmMET1 protein. For background estimation, 1 ng/µl boiled GST-ZmMET1 protein was used. Reaction was performed at 37°C for 90 min and DNA was extracted as described above.
Western blotting
Antibody against ZmET1 was raised by immunizing rabbits with synthetic peptides corresponding to the C-terminal catalytic regions (Cys1186-Ser1201 and Cys1469-His1489). Total protein samples were prepared from 0.5 g maize tissues pulverized in liquid nitrogen, and suspended in 500 µl cold lysis solution containing 50% glycerol, 40 mM Tris–Cl, pH 7.5, 0.1% SDS, 200 mM DTT, 1 mM PMSF. The sample was subjected to SDS–PAGE, blotted to polyvinylidene difluoride membranes (Immobilon 0.45 µm pore size, Millipore, Bedford, MA). Immunoblotting was performed using 1:2000 diluted anti-MET, and the locations of the antigens were visualized by using diluted 1:5000 horseradish peroxydase-conjugated goat anti-rabbit secondary antibody (Bio-Rad, Tokyo, Japan) and the ECL reagent (Amersham Life Science), followed by exposure to X-ray film.
DNA and RNA blot hybridization
Maize genomic DNA extracted with the cetyltrimethylammonium bromide method (24) was digested with appropriate restriction endonucleases, fractionated (10 µg per lane) in 0.8% agarose gels and blotted onto nylon membranes (Hybond N+, Amersham) after denaturation. Hybridization was performed at 50 or 65°C with appropriate probes (25). Membranes were washed in a buffer solution containing 2× SSC and 0.1% SDS at 50°C three times for 30 min each (medium stringency), or in a buffer solution containing 0.5× SSC and 0.1% SDS at 65°C three times for 30 min each (high stringency), and subjected to autoradiography with X-ray film. Total RNA was isolated by the acid guanidinium–phenol–chloroform (AGPC) method (26) and aliquots of 20 µg were fractionated on denaturing agarose gels, transferred onto nylon membranes (Hybond N, Amersham), and after crosslinking by UV irradiation, subjected to hybridization with 32P-labeled probe. The membranes were washed in a solution containing 0.5× SSC and 0.1% (w/v) SDS at 65°C for 1 h twice and subjected to autoradiography with X-ray film.
In situ RNA hybridization
In situ hybridization was performed as described earlier (27), with modification. Tissues were cut into 0.5 cm segments and fixed at 4°C for 6 h in 50 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)] buffer solution (pH 7.5) containing 0.25% (v/v) glutaraldehyde and 4% (w/v) paraformaldehyde. Fixed tissues, dehydrated through a series of concentrated ethanol and t-butyl alcohol and embedded in Paraplast (Sigma, Tokyo, Japan), were sectioned at 10 µm with a microtome and transferred onto a slide glass coated with poly-l-lysine. Digoxigenin-UTP labeled RNA probes were prepared according to the manufacturer’s instructions (Boehringer, Mannheim, Germany). Sense and anti-sense probes were transcribed using T7 RNA polymerase from the P1 fragment of ZmMET1 cDNA prepared as described above and cloned in a pGEM-T vector (Promega, Tokyo, Japan). After hybridization, slide glasses were washed twice in 0.2× SSC at 55°C for 1 h, then incubated with 20 µg/ml RNase A at 37°C for 30 min. After washing in 0.2× SSC at 55°C for 1 h, immunological detection of hybridized probes was carried out following the manufacturer’s instructions (Boehringer) with slight modification. Samples were successively incubated in DIG buffer (100 mM Tris–HCl, pH 7.5 and 150 mM NaCl) containing 1.5% blocking agent (Boehringer) for 1 h, and in DIG buffer containing diluted antibody-conjugate 1:2000 for 1 h. After three washes with DIG buffer containing 0.1% Tween-20 for 30 min each, slide glasses were briefly washed with 100 mM Tris–HCl, pH 9.5, containing 100 mM NaCl, 50 mM MgCl2 and 0.1% Tween-20 (detection buffer) and incubated further for 1 h in a detection buffer containing 337.5 µg/ml nitroblue tetrazolium salt and 175 µg/ml 5-bromo-4-chloro-3-indoyl phosphate toluidinium salt. Color development was terminated with addition of TE buffer.
RESULTS
Isolation and structure of ZmMET1 cDNA
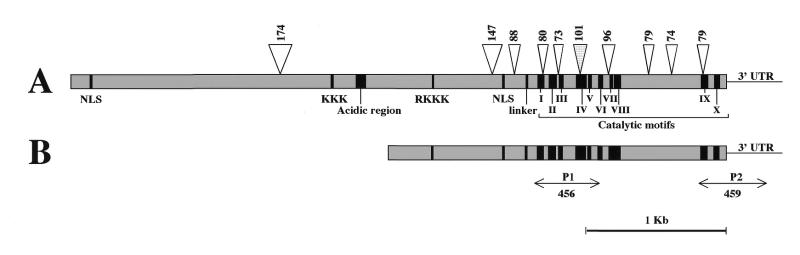
A maize cDNA library was screened with a cDNA fragment encoding tobacco MET (15) as the probe, and two clones, which differed in size but were identical in partial DNA sequence, were finally isolated. A cDNA clone of 2646 bp was further analyzed (EMBL accession no. AF229183) and found to be identical with the recently deposited sequence for the maize MET gene in the database (6; EMBL accession no. AF063403). Upon comparison of our cDNA fragment with the genomic sequence together with cDNAs from other plant species, the full-length cDNA was predicted to be 4573 bp in size containing an open reading frame for 1525 amino acids (171 kDa), and designated as ZmMET1 (Fig. 1). ZmMET1 is composed of two domains, the C-terminal catalytic domain which contains 10 conserved motifs of METs, and the N-terminal domain for regulatory functions. It should be noted that 9 out of 10 introns are localized in the N-terminal region.
Figure 1.
Schematic representation of ZmMET1 gene. Organization deduced from sequences of genome (AF063403) (A) and partial cDNA (B) is illustrated. The open reading frame is represented by boxes, the positions and numbers of nucleotide introns are indicated. Several functional domains based on homology with other known sequences were identified; the I–X catalytic motifs of eukaryote METase; NLS, nuclear localization signal; acidic domain; and other basic sites are also indicated. P1 and P2 indicate the DNA fragments amplified by PCR and used as templates for the in situ hybridization and mRNA blots analysis respectively.
MET activity
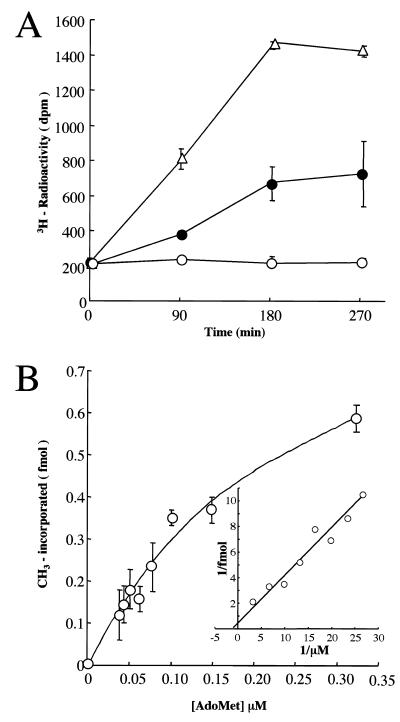
Bacterially expressed ZmMET1 was incubated with (methyl-3H)AdoMet and dcm– E.coli genomic DNA, and 3H-incorporation into DNA was measured at various time intervals (Fig. 2A). The reaction increased linearly up to 3 h, reaching a plateau thereafter. It is clear that this is due to enzymatic activity, because no increase in 3H-incorporation was observed with heat-inactivated GST-ZmMET1 protein. The observed reaction pattern was consistent with that of M.HapII. These observations clearly show that bacterially expressed protein indeed possesses MET activity. Velocity measurements at various AdoMet concentrations were then performed (Fig. 2B). The reaction velocity was concentration-dependent, showing a hyperbolic curve, and the double-reciprocal plots (Fig. 2B, insert) gave a Km value of 0.3 µM.
Figure 2.
Properties of bacterially expressed ZmMET1. (A) Time course assay. Bacterially expressed ZmMET1 at 1 ng/µl (closed circles) was incubated in a standard assay mixture with E.coli genomic DNA as the methyl acceptor. Aliquots were sampled every 90 min and the incorporated 3H-radioactivity into the DNA was measured in triplicate. As the positive control, 2.25 × 10–5 U/µl of M.HapII (open triangles) was assayed. The negative control was made of heat-inactivated ZmMET1 at 1 ng/µl (open circles). (B) Steady-state kinetics. Assays were performed in reaction mixtures as described in Materials and Methods containing indicated concentrations of (methyl-3H)AdoMet and 1 ng/µl of GST-ZmMET1 protein. Alternatively, 1 ng/µl heat-inactivated GST-ZmMET1 protein was used for background estimation. Incubations were done at 37°C for 90 min and the incorporated 3H-radioactivity into the DNA was measured in triplicate. The insert is the corresponding Lineweaver–Burk plot.
Molecular organization and tissue specificity
The molecular organization of the ZmMET1 in maize genome was determined by Southern hybridization analysis (Fig. 3). When genomic DNA was digested with BamHI, for which no site is present over at least 8 kb of the genomic region, a hybridization signal was only detected in the high molecular weight region. In contrast, EcoRI is predicted to produce three fragments of 1.5, >1.6 and >4.4 kb, all of which were identified. Similarly, EcoRV produced two fragments of 1.9 and 3.5 kb, and HindIII one fragment of 1 kb. These hybridization patterns were essentially the same under high and low stringency washing conditions, indicating ZmMET1 to be present as a single copy per haploid maize genome (Fig. 3A and B). However, under low stringency washing conditions, extra signals that appeared not to result from ZmMET1, were also observed (Fig. 3A). This indicates that another MET gene is possibly present in maize.
Figure 3.
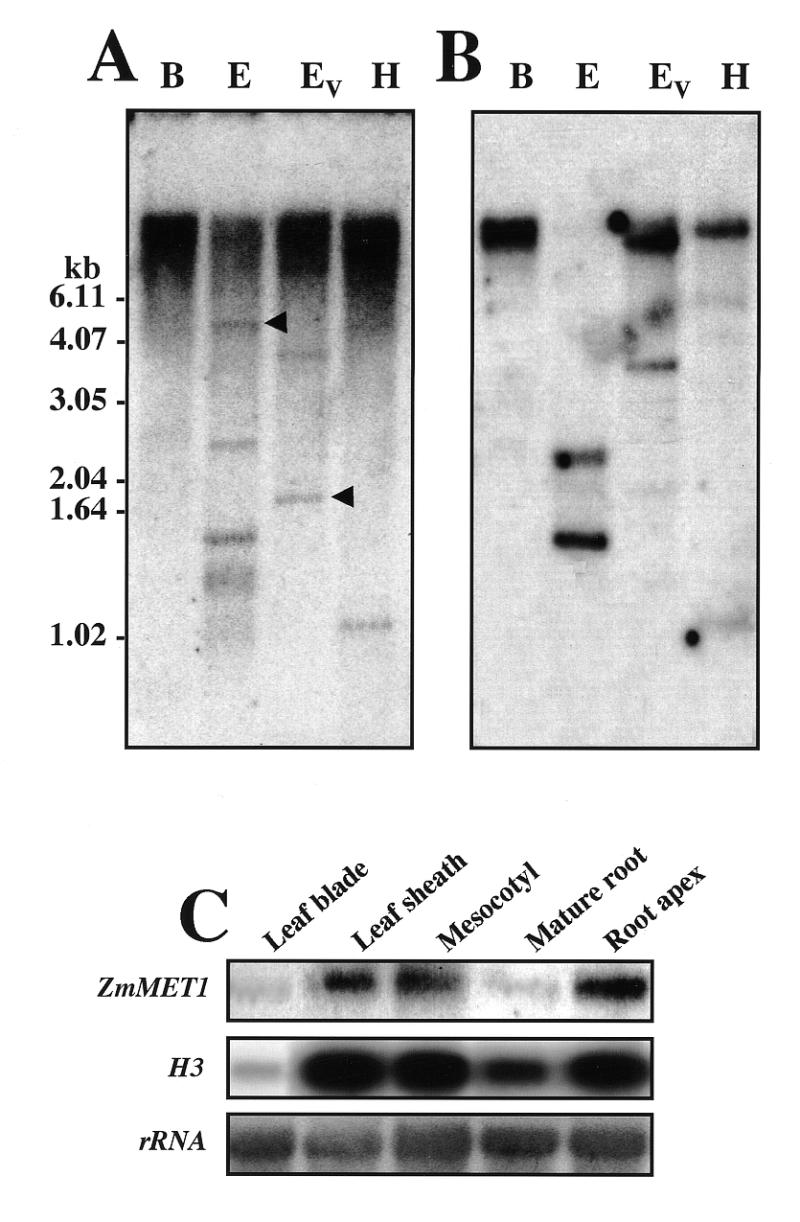
Southern and northern blot analyses. Genomic DNA (10 µg per lane) was digested with BamHI, EcoRI, EcoRV or HindIII, separated on an agarose gel, blotted and probed with a radioactively-labeled ZmMET1 cDNA fragment. The same membrane was washed at low (A) and high stringency (B). The size of the hybridizing signals is indicated along with the positions of DNA molecular markers. Hybridizing signals observed only in blots washed under low stringency are indicated with arrowheads. (C) RNA blot analysis of transcripts of ZmMET1 and H3. RNA blot analysis was carried out with 20 µg of total RNA extracted from leaf blades, leaf sheaths, mesocotyl, mature root region and root apices of 15 day old seedlings. Hybridization was performed successively with the P2 and P3 fragments of the ZmMET1 cDNA and H3 cDNA respectively.
Tissue-specific accumulation of ZmMET1 transcripts was examined by northern hybridization analysis (Fig. 3C). It is clearly shown that leaf sheaths, mesocotyls and root apices, but not leaf blades and mature root tissues, had large amounts. This accumulation pattern is essentially the same as that for the histone H3 gene, which is exclusively expressed in dividing cells of germinating maize seedlings (28), suggesting ZmMET1 to be preferentially switched on in dividing tissues.
In situ hybridization
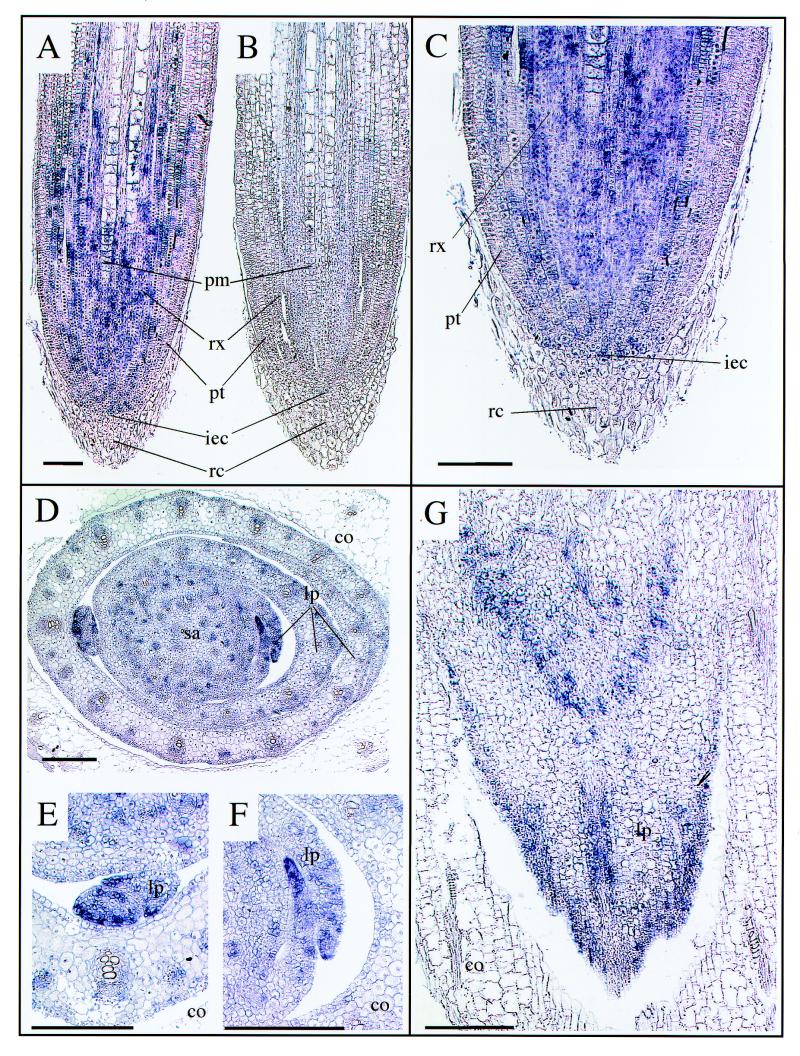
To further identify the particular cells in which ZmMET1 transcripts accumulate, in situ hybridization experiments were performed (Fig. 4). Leaf sheath node regions and root apices were analyzed because of their highest accumulation of ZmMET1 transcripts as shown by northern assay (Fig. 3C). In root apex sections, the digoxygenin-UTP labeled anti-sense probe was mainly localized in the initials of the epidermis and cortex, in the radicle cortex and potential metaxylem tissues, but not in the protoderm and root caps (Fig. 4A and C). Transcript levels appeared to decrease in all distal tissues as they progressively elongated and/or differentiated (Fig. 4A). In the leaf sheath node region, transcripts were accumulated at the leaf primordium edges as shown in transversal sections (Fig. 4D, E and F) and in longitudinal section (Fig. 4G). However, they were found not to be uniformly distributed among the cells, showing a blotchy pattern.
Figure 4.
Cell specific localization of ZmMET1 mRNA in maize root apex and leaf sheath node edge regions. In situ hybridization of ZmMET1 transcripts was performed with digoxigenin-UTP labeled ZmMET1 anti-sense (A, C, D, E, F, G) or sense (B) probes. Samples are from longitudinal sections of a 10 day old root apex (A, B); close-up of (A) showing the root cap and initials of procambium, epidermis and cortex (C); transversal sections of the leaf sheath node region (D); close-up of (D) showing leaf the primordium (E, F); longitudinal section of the leaf sheath node region (G). sa, shoot apex; lp, leaf primordia; co, coleoptile; pm, potential metaxylem; pt, protoderm; rx, radicle cortex; iec, initials of the epidermis and cortex; rc; root cap. Bars are 250 µm.
Stress response
The experiments detailed above suggested that ZmMET1 is predominantly expressed in proliferating cells. Subsequently we examined effects of various stresses that may interfere with cell division. As the control, the histone H3 gene (H3) was selected, because its transcripts are mainly encountered during the S phase of the cell cycle, so that it can be used as a marker gene for DNA replication (29,30). Changes of transcript level for ZmMET1 and H3 in coleptile node and mesocotyl regions in response to stress were examined by northern hybridization (Fig. 5). Mechanical wounding by direct cutting of mesocotyl tissues significantly inhibited transcript accumulation of both ZmMET1 and H3 (Fig. 5A). When seedlings were subjected to drought (Fig. 5B), or to salinity by being transferred to MS medium containing 0.5 M NaCl (Fig. 5C), levels of ZmMET1 and H3 transcripts began to sharply decrease after several hours. These observations indicate that expression of ZmMET1 and H3 is seriously affected when cell division or DNA replication is disturbed, and therefore that ZmMET1 may be involved in DNA replication complex. This conclusion, however, was countered by the finding that, when seedlings were treated with low temperature, no change was observed in ZmMET1 transcript levels, while H3 transcripts were significantly decreased (Fig. 5D).
Figure 5.
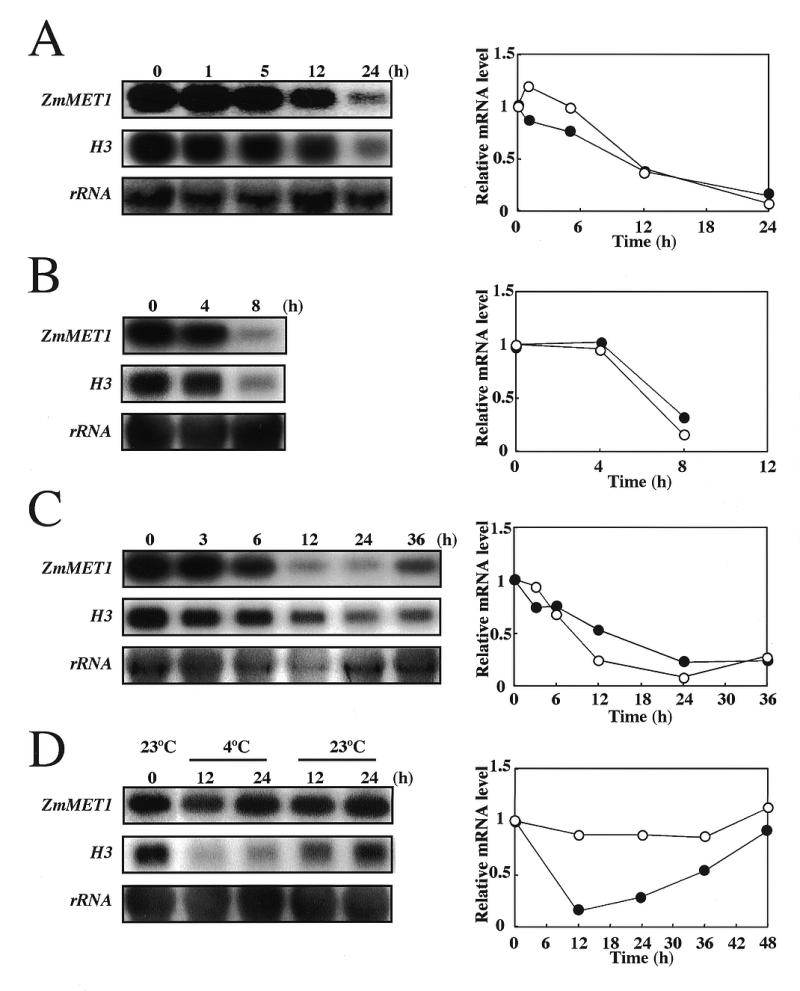
Effects of stresses on ZmMET1 and H3 transcript accumulation. Stress treatments were mechanical wounding (A), drought (B), salinity (C) and chilling (D). RNA analysis was carried out with 20 µg of total RNA extracted from the coleptile node and mesocotyl from 15 day old maize seedlings. After stress treatments as indicated, RNA blots were probed successively with the P2 and P3 fragments of the ZmMET1 cDNA and H3 cDNA, respectively. Results of a densitometric estimation of relative amounts of mRNA for ZmMET1 (open circles) and for H3 (closed circles) are graphically shown.
Cold stress and DNA damage
The resistance of ZmMET1 to cold was further investigated by subjecting seedlings to a temperature of 4°C for up to 4 days, during which period RNA was extracted from mesocotyl and root tissues. The level of ZmMET1 transcripts in the mesocotyl was weakly affected for up to 4 days (Fig. 6A), while that in root tissues was significantly decreased to ∼75% of the initial level (Fig. 6B). Transcripts for H3 were decreased in both tissues (Fig. 6A and B). This indicates that regulation of ZmMET1 transcript level is not always coupled with DNA replication in mesocotyl tissues. A possible explanation is that ZmMET1 is involved in DNA repair, which is required for restoration of damaged DNA under cold stress (31). In order to examine this possibility, maize seedlings were treated for 24 h with EMS which severely damages DNA, and allowed to recover for up to 24 h. Transcript levels of both ZmMET1 and H3 were then estimated by northern assay in mesocotyl and root tissues (Fig. 6C and D). Results showed both transcripts to be decreased upon DNA damage, indicating ZmMET1 not necessarily to function in repair system.
Figure 6.
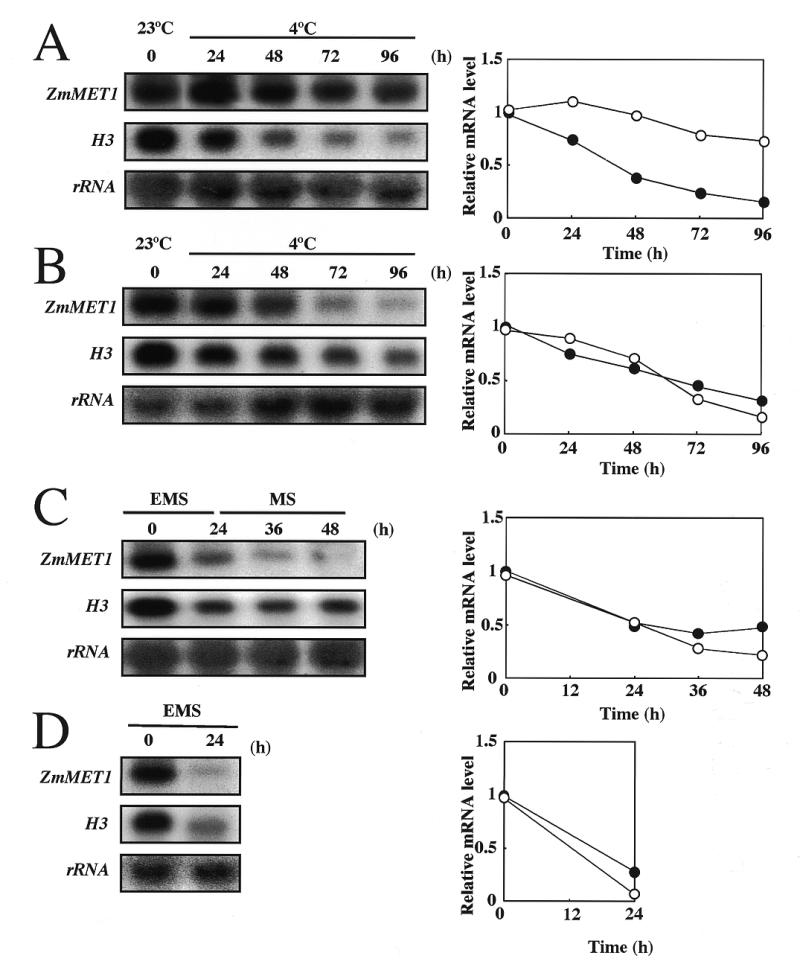
Effects of cold and EMS on ZmMET1 and H3 transcript accumulation. Treatments were by chilling (A, B) and EMS (C, D) in mecocotyl (A, C) and root tissues (B, D). RNA analysis was carried out with 20 µg of total RNA extracted from the coleptile node and mesocotyl from 15 day old maize seedlings. After stress treatments as indicated, RNA blots were probed successively with the P2 and P3 fragments of the ZmMET1 cDNA and H3 cDNA, respectively. Results of a densitometric estimation of relative amounts of mRNA for ZmMET1 (open circles) and for H3 (closed circles) are graphically shown.
In order to examine whether the differential accumulation of ZmMET1 transcripts reflects the protein level, western blotting was performed with extracts of tissues subjected to cold treatments (Fig. 7). In mesocotyl tissues, the protein level was not affected during transitory cold treatment (Fig. 7A), being consistent with unchanged transcript levels (Fig. 5D). Even under prolonged cold treatments, MET proteins in mesocotyl tissues were constitutively present, whereas those in root tissues rapidly declined (Fig. 7B). These differential profiles of MET1 protein are consistent with accumulation patterns of its transcript (Fig. 6A and B), indicating the correlation between transcription and translation of ZmMET1 in response to external stimulation.
Figure 7.
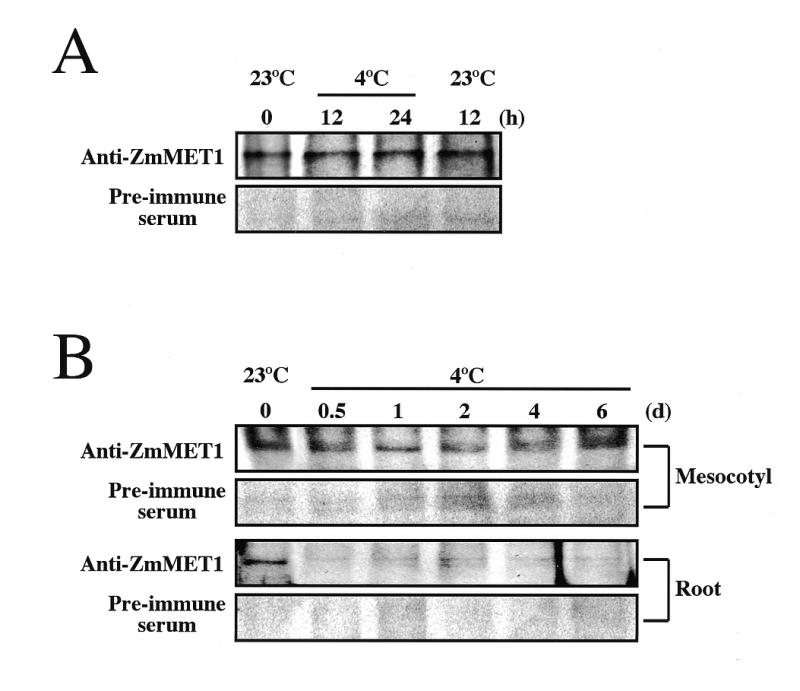
Immuno-blotting assay of ZmMET1. Cold treatment was given to mesocotyl tissues for transitory period (A) and to mesocotyl and root tissues for prolonged period (B). Total protein samples prepared from maize tissues treated with cold were separated by electrophoresis (110 µg per lane) and transferred to polyvinylidene difluoride membranes. After binding of rabbit anti-ZmMET1 antibodies or pre-immune serum, locations of the antigen were visualized by using horseradish peroxydase-conjugated goat anti-rabbit antibody and the ECL reagent (Amersham Life Science), followed by exposure to X-ray film.
Altered DNA methylation status under cold stress
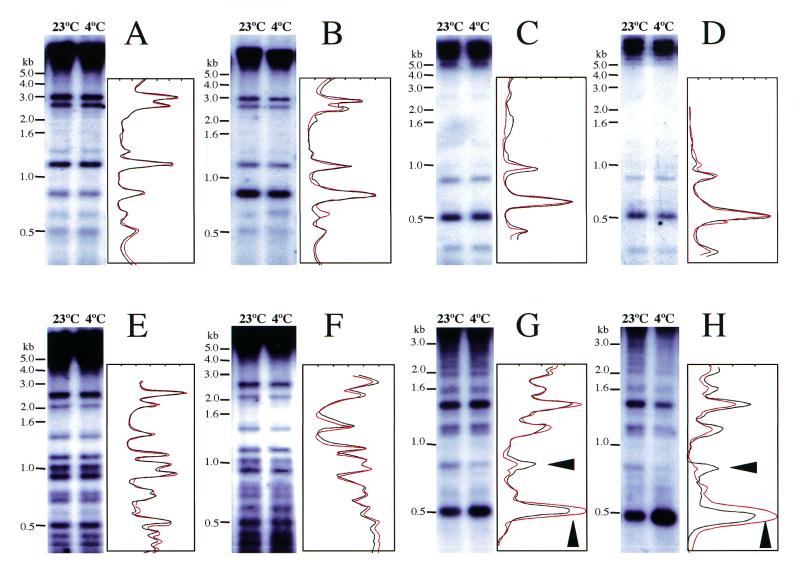
The possible reason for continuous expression of ZmMET1 in the mesocotyl under cold conditions is that ZmMET1 is required to continue methylation of DNA. To assess this question, genomic DNA was examined for methylation status by Southern hybridization analysis. DNA samples from mesocotyl and root seedlings treated or untreated by cold stress were digested with a restriction endonuclease, HpaII, which recognizes methylation of the second cytosine in CCGG sequence. The methylation pattern was examined at loci for ZmPK4, which is up-regulated by cold (32) (Fig. 8A and B), for ZmMET1 itself (Fig. 8C and D), for H3 which is down-regulated by cold (Fig. 8E and F) and for the Ac/Ds transposon (Fig. 8G and H). The hybridization for the ZmPK4 locus demonstrated the methylation pattern to be the same for both untreated and treated cases, although tissue-specific differences were distinct (Fig. 8A and B). Results for the ZmMET1 and H3 regions were completely the same among all samples regardless of the cold treatment (Fig. 8C, D, E and F). However, methylation of the Ac/Ds transposon locus showed distinct patterns. DNA isolated from cold-treated mesocotyl tissues showed essentially the same pattern, although slight decrease and increase of 0.8 and 0.5 kb fragments, respectively, was observed (Fig. 8G). However, in DNA isolated from cold-treated root tissues, a clear change in restriction pattern occurred, showing disappearance of several large-size fragments and increase in a 0.5 kb fragment (Fig. 8H). This indicates demethylation at CCGG sites had occurred. Similar experiments with another restriction endonuclease, MspI, which recognizes the first cytosine in CCGG, did not show any difference among these genes (data not shown), indicating the change in methylation to have occurred at the second cytosine. It should be noted that the observed difference in methylation pattern is analytical, since, due to a high copy number of Ac/Ds (23), data only suggest that the ratio of individual restriction fragment to the total population might be changed. Within these experimental limitations, however, it could be concluded that, while methylation pattern in individual genes did not vary, that in a transposon(s) did change upon cold stress. To determine whether or not the altered methylation pattern affected expression of the Ac/Ds transposon, northern hybridization was performed but no expression was detectable (data not shown).
Figure 8.
Effects of cold on DNA methylation. Southern blot analysis of genomic DNA extracted from mesocotyl (A, C, E, G) or root tissues (B, D, F, H) untreated (23°C) or treated by cold stress (4°C). The DNA extracts were digested with a restriction endonuclease, HpaII, separated on an agarose gel (10 µg per lane), blotted and probed with a radioactively labeled ZmPK4 cDNA fragment (A, B), ZmMET1 cDNA fragment (C, D), Histone H3 cDNA fragment (E, F) or Ac/Ds cDNA fragment (G, H). The membranes were washed at high stringency. The size of the hybridizing signals is indicated along with the positions of DNA molecular markers. The densitometric trace with black and red lines represents the relative intensity of signals for untreated (23°C) and treated (4°C) plants, respectively. Arrowheads indicate the positions of a 0.8 and a 0.5 kb fragment (G, H).
DISCUSSION
The present paper documents findings for properties of a gene encoding a MET from maize. The partial putative amino acid sequence, encoded by a 2.6 kb cDNA fragment (EMBL accession no. AF229183) obtained by screening of a cDNA library using a tobacco cDNA for MET (15) as the probe showed high similarity with those of other known plant DNA METs. It also matched well with the amino acid sequence predicted from the genomic sequence (EMBL accession no. AF063403), which was submitted to the database during the course of the present study. Comparison of the deduced polypeptide with other METs indicated that it is comprised of 1525 amino acids with a relative molecular mass of 171.5 kDa, having 10 conserved motifs characteristic of METs in the C-terminal region and several alternating clusters of lysine–arginine residues that separate the N- and C-terminal domains. DNA methyltransferase activity was confirmed by in vitro assay using bacterially expressed protein to efficiently methylate DNA from E.coli. These characteristics clearly indicate that it belongs to the MET1 family (1,2,3,6), and therefore it was designated as ZmMET1.
Northern blot analyses showed ZmMET1 to be essentially only expressed in tissues where the proportion of proliferating cells is substantial, as shown by active accumulation of H3 transcripts. This was corroborated by the spatial distribution revealed by in situ hybridization, ZmMET1 transcripts being predominantly accumulated in the apical meristem, where cells actively divide (12,13,17). However, the hybridization signal appears to be situated in clusters of cells disseminated in the youngest tissues with blotchy labeling patterns. A similar pattern was observed with carrot MET and it was interpreted as possibly reflecting cell-cycle regulation (13). Cell-cycle dependent transcription for MET has also been reported for mammals and bacteria (8,33). These observations strongly suggest that MET is closely associated with DNA replication not only in enzymatic activity, but also in its transcriptional regulation.
In order to examine whether expression of ZmMET1 is strictly linked with DNA replication, its transcript levels in response to physico-chemical stresses that affect cell proliferation, or DNA replication, were assayed by northern hybridization. Wounding, which suppressed cell division as shown by a decrease of H3 transcripts, also strongly inhibited ZmMET1 transcript accumulation. It is generally considered that methyl-jasmonate, ethylene and/or abscisic acid mediate wound signals (34,35). However, we found none of these chemicals to induce ZmMET1 transcript accumulation (unpublished observations), suggesting that signal transduction molecules arising from wounding are not directly involved in regulation of ZmMET1 expression. Water seizure caused by drought or salinity also simultaneously suppressed ZmMET1 transcript accumulation and cell proliferation, suggesting that expression of ZmMET1 is indeed dependent on DNA replication.
However, this conclusion was countered by the finding that cold treatment had only a limited impact on ZmMET1 transcript levels, despite strong repression of H3 transcript accumulation. This was confirmed by western blotting showing the levels of ZmMET1 protein to be associated with its transcript levels. Since it is established that cell division is inhibited by chilling (36), this discrepancy in transcript accumulation patterns suggests that cell division and ZmMET1 expression are probably normally coexistent but not completely associated. This is in line with previous observations of mature tissues in which, in spite of low rates of cell division, high expression of MET genes was apparent (2,17). One possible explanation is that cold stress may induce DNA damage (31), which activates DNA repair machinery requiring METs to maintain the initial methylation patterns. In order to assess this hypothesis, we examined the effects of DNA damage by using EMS and UV-C irradiation, but no increase of ZmMET1 transcript level was observed (unpublished observations).
A second explanation is that hypermethylation of genomic DNA occurs to silence genes that are not necessary in a cold environment. It has been proposed that, upon chilling, several hundreds of genes are up- or down-regulated to cope with severe stress (37), and it is conceivable that DNA methylation participates in such a regulation system. Indeed, recent work showed an altered methylation pattern with cold treatment in A.thaliana seedlings (21). In the present study, we found that, under cold stress, methylation patterns in individual genes appeared to be faithfully maintained in all tissues examined, whereas those in transposons did change in a tissue-specific manner. Since our experiments were only diagnostically performed by detecting methylation at limited cytosine residues in limited genomic regions, it is too early to conclude that overall methylation status has changed upon cold treatments. Nevertheless, the finding is of particular interest regarding biological significance of METs. First, demethylation appears to occur selectively in particular genome regions. To our knowledge, this is the first clear evidence of cold-induced demethylation of distinct gene(s), although global demethylation was observed in cold stressed chicory (22). Secondly, cold-induced change in methylation status may potentially change expression of related genes. This idea is consistent with a finding that vernalization is mediated through a gradual activation of relevant genes by altered methylation status during cold treatment (21). Thirdly, methylation blocks, or at least reduces expression of Ac/Ds transposons. Although we could not detect transcripts of Ac/Ds after cold treatment of root tissues, in which the Ac/Ds region was demethylated, it is highly probable that some of the Ac/Ds loci were activated upon cold stress. Indeed, Ac/Ds is known to be largely quiescent during normal growth, but activated upon stress treatments including wounding, pathogen attack and cell culture (11,20,38–41). Activation of Ac/Ds has also been reported to be inversely correlated with methylation status (42). These observations are consistent with the idea that methylation functions as a defense mechanism against invading DNAs (4,11). Fourthly, if the former assumption is correct, ZmMET1 may play a crucial role in preventing demethylation. This is supported by our finding that, in cold-stressed mesocotyl tissues, ZmMET1 remains largely expressed and demethylation does occur weakly, in contrast to the case in roots, where ZmMET1 is silenced. The reason for differential accumulation of ZmMET1 transcripts between the mesocotyl and roots is not clear, but it is conceivable that the self-defense system of the above-ground parts of plants, which suffer more severe temperature changes than roots, is more strongly constituted in comparison with that in other parts of the plant body. Although further evidence from enzyme activity measurement and quantitation of m5C is needed, the inverse relationship between ZmMET1 expression and DNA methylation status strongly supports the idea that the latter changes dynamically depending on the environmental conditions, and that such changes may be modulated by METs.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Dr K. Shimamoto (Nara Institute of Science and Technology) for a generous gift of the SK– plasmid construct containing Ac cDNA fragment, and Ms Y. Tatsumi and Ms C. Hata (CREST) for technical assistance. This work was supported by a grant to the Core Research for Evolutional Science and Technology (CREST) from the Japan Science and Technology Corporation (JST).
DDBJ/EMBL/GenBank accession no. AF229183
REFERENCES
- 1.Finnegan E.J. and Dennis,E.S. (1993) Nucleic Acids Res., 21, 2383–2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goto K., Numata,M., Komura,J.I., Ono,T., Bestor,T.H. and Kondo,H. (1994) Differentiation, 56, 39–44. [DOI] [PubMed] [Google Scholar]
- 3.Robertson K.D., Uzvolgyi,E., Liang,G., Talmadge,C., Sumegi,J., Gonzales,F.A. and Jones,P.A. (1999) Nucleic Acids Res., 27, 2291–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Razin A. and Cedar,H. (1991) Microbiol. Rev., 55, 451–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lewis J. and Bird,A. (1991) FEBS Lett., 285, 155–159. [DOI] [PubMed] [Google Scholar]
- 6.Cao X., Springer,N.M., Muszynski,M.G., Phillips,R.L., Kaeppler,S. and Jacobsen,S.E. (2000) Proc. Natl Acad. Sci. USA, 97, 4979–4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okano M. and Xie,S. (1998) Nature Genet., 19, 219–220. [DOI] [PubMed] [Google Scholar]
- 8.Leonhardt H., Page,A.W., Weier,H.U. and Bestor,T.H. (1992) Cell, 71, 865–873. [DOI] [PubMed] [Google Scholar]
- 9.Meyer P., Niedenhof,I. and Ten Lohuis,M. (1994) EMBO. J., 13, 2084–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flynn J., Glickman,J.F. and Reich,N.O. (1996) Biochemistry, 35, 7308–7315. [DOI] [PubMed] [Google Scholar]
- 11.Wang L., Heinlein,M. and Kunze,R. (1996) Plant Cell, 8, 747–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pradhan S., Cummings,M., Roberts,R.J. and Adams,R.L. (1998) Nucleic Acids Res., 26, 1214–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bernacchia G., Primo,A., Giorgetti,L., Pitto,L. and Cella,R. (1998) Plant J., 13, 317–329. [DOI] [PubMed] [Google Scholar]
- 14.Bernacchia G., Para,A., Pedrali-Noy,G. and Cella,R. (1998) Plant Physiol., 116, 446–446. [Google Scholar]
- 15.Nakano Y., Steward,N., Kusano,T. and Sano,H. (2000) Plant Cell Physiol., 41, 448–457. [DOI] [PubMed] [Google Scholar]
- 16.Genger R.K., Kovac,K.A., Dennis,E.S., Peacock,W.J. and Finnegan,E.J. (1999) Plant Mol. Biol., 41, 269–278. [DOI] [PubMed] [Google Scholar]
- 17.Kimura H., Takeda,T., Tanaka,S., Ogawa,T. and Shiota,K. (1998) Biochem. Biophys. Res. Com., 253, 495–501. [DOI] [PubMed] [Google Scholar]
- 18.Mi S. and Roberts,R.J. (1992) Nucleic Acids Res., 20, 4811–4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng X., Kumar,S., Posfai,J., Pflugrath,J.W. and Roberts,R.J. (1993) Cell, 74, 299–307. [DOI] [PubMed] [Google Scholar]
- 20.Dennis E.S. and Brettell,R.I. (1990) Philos. Trans. R. Soc. Lond. B. Biol. Sci., 326, 217–229. [DOI] [PubMed] [Google Scholar]
- 21.Finnegan E.J., Genger,R.K., Kovac,K., Peacock,W.J. and Dennis,E.S. (1998) Proc. Natl Acad. Sci. USA, 95, 5824–5829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Demeulemeester M.A.C., Van Stallen,N. and De Proft,M.P. (1999) Plant Sci., 142, 101–108. [Google Scholar]
- 23.Johns M.A. (1990) J. Mol. Evol., 30, 493–499. [Google Scholar]
- 24.Rogers O. and Bendich,A.J. (1994) In Gelvin,S.B. and Schilperoort,R.A. (eds), Extraction of Total Cellular DNA from Plants, Algae and Fungi. Plant Molecular Biology Manual. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp. D1:1–8.
- 25.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 26.Ullrich A., Shine,J., Chirgwin,J., Pictet,R., Tischer,E., Rutter,W.J. and Goodman,H.M. (1977) Science, 196, 1313–1319. [DOI] [PubMed] [Google Scholar]
- 27.Jackson D.P. (1991) In Bowles,D.J., Gurr,S.J. and McPhereson,M. (eds), Molecular Plant Pathology: A Practical Approach. In situ Hybridization in Plants. Oxford University Press, Oxford, UK, pp. 163–174.
- 28.Chaubet N., Clement,B., Philipps,G. and Gigot,C. (1991) Plant Mol. Biol., 17, 935–940. [DOI] [PubMed] [Google Scholar]
- 29.Reichheld J.P., Gigot.C. and Chaubet-Gigot,N. (1998) Nucleic Acids Res., 26, 3255–3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vogel M.C., Papadopoulos,T., Muller-Hermelink,H.K., Drahovsky,D. and Pfeifer,G.P. (1988) FEBS Lett., 236, 9–13. [DOI] [PubMed] [Google Scholar]
- 31.Koukalova B., Kovarik,A., Fajkus,J. and Siroky,J. (1997) FEBS Lett., 414, 289–292. [DOI] [PubMed] [Google Scholar]
- 32.Ohba H., Steward,N., Kawasaki,S., Ikeda,Y., Berberich,T., Koizumi,N., Kusano,T. and Sano,H. (2000) Mol. Gen. Genet., 263, 359–366. [DOI] [PubMed] [Google Scholar]
- 33.Reisenauer A., Quon,K. and Shapiro,L. (1999) J. Bacteriol., 181, 2430–2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seo S., Sano,H. and Ohashi,Y. (1997) Physiol. Plant., 101, 740–745. [Google Scholar]
- 35.Morgan P.W. and Drew Malcolm,C. (1997) Physiol. Plant., 100, 620–630. [Google Scholar]
- 36.Francis D. and Barlow,P.W. (1988) Symp. Soc. Exp. Biol., 42, 181–201. [PubMed] [Google Scholar]
- 37.Thomashow M.F. (1998) Plant Physiol., 118, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wessler S.R. (1996) Curr. Biol., 6, 959–961. [DOI] [PubMed] [Google Scholar]
- 39.Takeda S., Sugimoto,K., Otsuki,H. and Hirochika,H. (1998) Plant Mol. Biol., 36, 365–376. [DOI] [PubMed] [Google Scholar]
- 40.Schlappi M., Raina,R. and Fedoroff,N. (1994) Cell, 77, 427–437. [DOI] [PubMed] [Google Scholar]
- 41.Hirochika H., Sugimoto,K., Otsuki,Y., Tsugawa,H. and Kanda,M. (1996) Proc. Natl Acad. Sci. USA, 93, 7783–7788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brutnell T.P., May,P.B. and Dellaporta,S.L. (1997) Genetics, 147, 823–834. [DOI] [PMC free article] [PubMed] [Google Scholar]