Abstract
We have modified the tetracycline-regulatable system so that all components are present on a stable extrachromosomal vector that can replicate in a wide variety of mammalian cells. An EBV/human ori vector is used to carry the system, overcoming the species specificity of conventional Epstein–Barr virus vectors. By placing the transcriptional transactivator gene under autoregulation, better induction characteristics are obtained. This system offers greater speed and sensitivity than previously reported methods. It can be applied within 3–4 weeks and produces an induction range of several hundred-fold with a low background.
INTRODUCTION
The tetracycline-inducible gene expression system developed by Gossen and Bujard (1) has proven useful for inducing or repressing at will the expression of a particular gene in mammalian cells. This system relies on the presence or absence of tetracycline (Tc) or a commonly used analog, doxycycline (Dox), to control gene expression. Key components of the Tc-system are of prokaryotic origin and independent of mammalian host cell regulation. Because gene expression is highly specific and tightly controlled, the Tc-inducible gene expression system has become widely used.
Variants of the system have been developed to actively turn on or turn off a gene (2). The ‘Tet-On’ system includes two main components. The first is a fusion gene between the N-terminal region of a reverse tetracycline repressor and the C-terminal domain of the Herpes simplex virus VP16 activation domain (2). The product of the fusion gene, called the reverse transcription transactivator (rtTA), selectively activates transcription from its cis-acting binding site, TetP, in the presence of Tc or Dox. The TetP element supplies promoter activity and is positioned upstream of the gene that is to be controlled by Tc or Dox. It consists of a series of tetracycline operators directly upstream of the cytomegalovirus (CMV) immediate-early minimal promoter, providing binding sites that are recognized by an activated rtTA protein. When the inducer is added to the system, rtTA binds to the TetP element, and the gene under its control is transcribed.
To use the system, each component has to be sequentially integrated into the chromosomes, then functionally tested. Typical methods for generating regulatable cell lines rely on the random integration of each component into the host chromosomes. Because genomic DNA flanking the point of integration can have a dramatic influence on the expression of introduced genes, it is imperative to screen for clones that provide desirable regulatory characteristics. While it is possible to obtain clones that produce low background levels while maintaining the potential to achieve high levels of gene expression upon induction, creating these cell lines is a time consuming and laborious process. To make the procedure more efficient, an extrachromosomal Epstein–Barr virus (EBV) vector can be used to carry one or both of the components of the Tc-regulatable system, reducing the need to screen many different clones. Because EBV vectors remain outside the chromosomes, the problems associated with random integration are eliminated.
In the original protocol two rounds of transfection and screening are required, to integrate the Tc-responsive transactivator and to introduce the TetP cassette controlling the gene of interest. By including the TetP element driving the expression of a gene on an EBV vector (3), the second screening process becomes unnecessary. All cells carrying the EBV vector will have a similar, high level of expression of the gene of interest upon induction. This system is especially advantageous when a variety of genes are under study. Once a cell line properly expressing the transactivator element has been produced, different genes can readily be cloned on the EBV vector and introduced into the cell line without further screening. This development provides a more efficient means to generate Tc-regulatable cell lines, but still requires prior production of a cell line properly expressing the transactivator element.
By including both elements necessary for Tc-regulation on an EBV vector (4), the position effects of chromosomal DNA do not influence gene expression of either component. It is thus unnecessary to screen clones for optimal expression. However, the high constitutive level of the transactivator produced by this system due to the multicopy, well-expressed EBV vector results in only low levels of induction, even in the presence of near toxic levels of Tc. Furthermore, efficient EBV replication occurs only in primate and canine cells, restricting the host range of both of these EBV-based systems.
To address these problems, we have created a new system with the following features. In place of the EBV viral origin of replication, we have used a human genomic sequence that has been shown to mediate efficient replication in a wide variety of mammalian cells, including rodent cells (5,6). This feature eliminates the species restriction associated with conventional EBV vectors, but preserves the favorable extrachromosomal retention and replication properties. In addition, our system places the rtTA gene under autoregulation, so high amounts of rtTA are made only when desired, during induction. This feature creates a desirable background to induction ratio. With this one-plasmid system, inducible cell lines can be generated in ~3 weeks, versus the original system, which required a minimum of 5–6 months for creation of such lines.
MATERIALS AND METHODS
Plasmid construction
A bi-directional TetP cassette that regulates the expression of the firefly luciferase gene and the reverse tetracycline-responsive transcriptional activator gene (rtTA) was constructed using the pBI-L plasmid from Clontech (Palo Alto, CA) as a base. The 1-kb rtTA gene was removed from pTet-On (Clontech) with restriction enzymes EcoRI and BamHI. This fragment was ligated into the EcoRV site of pBI-L, positioning the bi-directional TetP immediately upstream of the rtTA start codon. To accommodate the blunt ends generated by EcoRV, the EcoRI–BamHI rtTA fragment was made blunt by using T4 polymerase. The completed bi-directional rtTA-tetP-luciferase cassette was removed with BsrBI and placed into the SmaI site of pEGFP-1 (Clontech), resulting in pG-BON, which was used as a control vector. The EBV human-ori vector pDYAL has been described (5) and was used to create pDYAL-BON. A linker containing the restriction sites AgeI and XhoI was placed in the NruI site of pDYAL resulting in the plasmid pDYAL-AX. The 5.4-kb AgeI–XhoI fragment containing the bi-directional rtTA-luciferase expression cassette was removed from pG-BON and ligated into pDYAL-AX digested with AgeI and XhoI, creating pDYAL-BON. All plasmids were propagated in the Escherichia coli strain DH10B.
Cloning into larger plasmids such as pDYAL-AX, which is ~25 kb in length, is more difficult than cloning into smaller plasmids. However, we have routinely cloned into such plasmids, making use of the following tips. Because competing ligation reactions with a smaller plasmid backbone will occur more easily, contaminating vector backbone present with the insert fragment is likely to produce background colonies. To avoid this problem, a pure source of the insert fragment is helpful. In particular, if the insert fragment is purified from a plasmid backbone that bears a different antibiotic resistance from that of pDYAL, one can eliminate background colonies derived from contaminating plasmid backbone. We also find it helpful to use in-gel ligation rather than gel extraction for the large fragment, in order to improve recovery. We run fragments on 0.75% SeaPlaque agarose (FMC, Rockland, MN) for such ligations. We have been able to clone successfully into a large plasmid with no other special provisions, and routinely obtain the desired clone simply by screening a small number of individual mini-preps derived from candidate colonies. We have not found rearrangements to be a problem with pDYAL or its derivatives.
Tissue culture
Experiments were performed in C127I and C57/MG mouse cell lines (ATCC). C127 is a non-virally transformed clonal cell line derived from mammary tumor tissue of an RIII mouse. Cell line C57 is a non-tumorogenic, spontaneously transformed epithelial cell line generated from mammary tissue. Cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum, penicillin and streptomycin. Both cell lines were grown at 37°C in 5% CO2.
Transfections and experimental design
Cell lines were transfected using the Lipofectamine Plus Reagent from Gibco BRL (Gaithersburg, MD) according to the manufacturer’s guidelines. Briefly, 100-mm plates were transfected with 5 µg DNA, 20 µl Plus Reagent and 30 µl Lipofectamine using OPTI-MEM I medium (Gibco BRL) for 3.5 h. DNA:lipid complexes were then removed and cells were gently washed once and growth medium replaced. One or two 100-mm plates were transfected with the pG-BON control vector or pDYAL-BON. Because pG-BON carries the neomycin resistance gene and pDYAL-BON possesses the hygromycin resistance gene, selection was carried out using growth media supplemented with either 1.2 mg/ml G418 or 600 µg/ml hygromycin, respectively.
After transfection, cells were allowed to recover for 36–48 h, then selective medium was added. Medium was generally changed every 3–4 days until the cells approached confluence. Cells transfected with pG-BON grew faster than cells transfected with pDYAL-BON. As a result, pG-BON transfectants were split twice at 1:5 during the 3-week selection period, while pDYAL-BON transfectants often required only one split at 1:2.
At ~3 weeks post-transfection, cells were trypsinized from the 100-mm plates and aliquoted to 60-mm plates for gene expression analysis under a range of doxycycline (Sigma, St Louis, MO) doses. Since the cells transfected with pG-BON grew faster, less of the suspension was used in comparison to the pDYAL-BON group. For pG-BON transfections, 300 µl of 10 ml of cell suspension were aliquoted to 60-mm plates, whereas 500 µl of the pDYAL-BON cell suspension was used to achieve similar cell numbers between the two groups. Two milliliters of the remaining cells were plated on a 100-mm plate to continue propagation. For gene expression analysis, the 60-mm plates were left overnight to recover, then the medium was removed and replaced with selective medium supplemented with doxycycline at concentrations of 0, 1 or 5 µg/ml. For each concentration of doxycycline tested, duplicates of each plate were made to provide samples for 24 and 48 h time points.
Luciferase assays
Cells cultured with different amounts of doxycycline were harvested at 24 and 48 h and gently washed twice with 1 ml of calcium/magnesium-free phosphate buffered saline. Samples were collected by adding 300 µl of a lysing reagent containing 25 mM Tris–HCl (pH 7.8), 2 mM EDTA (pH 8.0), 5% glycerol (v/v) and 0.5% Triton X-100 (v/v). After 2 min, cells were scraped into tubes using a rubber policeman. Tubes were spun briefly for 2–3 min at ~10 000 g to separate the aqueous fraction from cell debris. A portion of the sample was removed, aliquoted to several tubes, and quickly frozen with N2 (L) and stored at –80°C.
Levels of firefly luciferase were determined using the Luciferase Assay System of Promega (Madison, WI). For each sample, 10 µl of cell extract and 100 µl of Luciferase Assay Reagent (Promega) were mixed and assayed in a Turner TD-20e luminometer. The luminometer integration period was set at 15 s with a delay time of 3 s. Samples with high levels of luciferase were diluted with the lysing reagent supplemented with 1 mg/ml BSA. The amount of luciferase in each sample was determined from a standard curve generated by using recombinant luciferase.
The protein level in each sample was determined with the DC Protein Assay from Bio-Rad (Hercules, CA). Extracts were diluted 5-fold with water to prevent interference. Samples with high levels of protein were further diluted to obtain results that coincided with the standard curve. These subsequent dilutions were done with a 5-fold dilution of the lysing reagent so that all samples maintained a similar chemical composition.
RESULTS AND DISCUSSION
The C127 and C57 mouse cell lines were transfected with two Tc-regulatable plasmids, pG-BON and pDYAL-BON (Fig. 1). Both plasmids carried all components of the Tc-regulatable system within a cassette controlled by a bi-directional TetP promoter. This promoter arrangement consists of the tetracycline-responsive element comprising seven copies of the 42-bp tet operator sequence. This element is positioned between two minimal CMV promoters that lack the enhancer that is part of the complete CMV promoter. The promoter is minimally expressed in the absence of binding of rtTA to the tet operator sequences. The promoter controls the luciferase gene and the rtTA reverse tetracycline transcriptional activator gene.
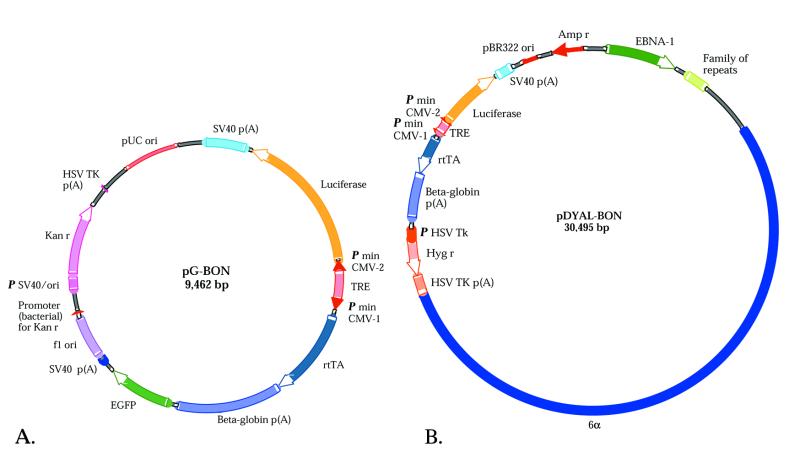
Figure 1.
Tetracycline-inducible vectors. A tetracycline regulatable bidirectional cassette is carried by (A) a conventional plasmid, pG-BON and (B) an extrachromosomally retained, autonomously replicating EBV/human ori vector, pDYAL-BON. Amp r, ampicillin resistance; EGFP, enhanced green fluorescent protein; HSV TK, herpes simplex virus thymidine kinase; Hyg r, hygromycin resistance; Kan r, kanamycin and neomycin resistance; P, promoter; p(A); polyA addition site; TRE, tetracycline responsive element; 6α, 16.2 kb fragment of alphoid repeat DNA from human chromosome 17.
The pG-BON plasmid lacks the ability to replicate extrachromosomally, whereas pDYAL-BON can replicate extrachromosomally and is retained long-term in mammalian cells. pDYAL-BON contains the origin of replication, oriP, from EBV, in which the dyad symmetry element needed for replication has been replaced with a 16.2 kb fragment of human alphoid DNA derived from the centromere of chromosome 17. This fragment confers the ability to replicate once per cell cycle in a variety of mammalian cells, including human, monkey and rodent cells (5,6).
Selection for the neomycin or hygromycin resistance markers on the plasmids was carried out for 3 weeks. Populations of resistant cells were expected to consist of randomly integrated plasmid DNA in the case of pG-BON and extrachromosomally replicating plasmid in the case of pDYAL-BON. To confirm this expectation, plasmid DNA was purified from the cell populations by performing Hirt extraction (7) and showing, in the case of pDYAL-BON-transfected mouse cells, the presence of plasmid DNA, all of which was DpnI-resistant. In the case of pG-BON-transfected mouse cells, no plasmid DNA was detected. The former result is consistent with the presence of extrachromosomal plasmid DNA that has undergone at least two rounds of replication in the mammalian cells. In practice, extrachromosomal plasmid DNA is present at this time after transfection only if the DNA has the ability to be retained and replicated in mammalian cells (8).
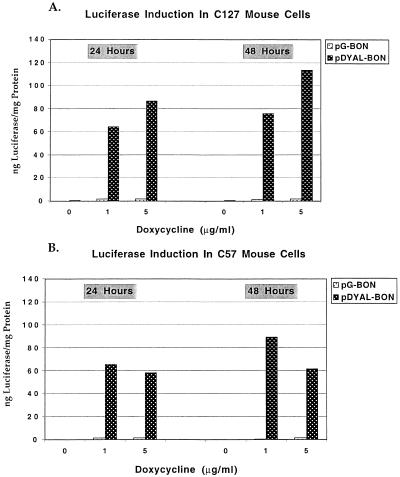
The populations of resistant cells were used for experiments demonstrating doxycycline induction. Dox was added to culture medium at concentrations of 1 and 5 µg/ml, and luciferase induction was measured 24, 48 and 72 h later. Luciferase was maximally expressed in C127 and C57 cells within 48 h of doxycycline induction. Luciferase values are presented in Table 1 and graphed in Figure 2. Measurements were also taken at 72 h, with the result that luciferase levels were stable, but not higher than at the earlier time points. It is typical of Tc-controlled gene expression systems to respond with similar kinetics. However, considering that expression of luciferase in our system is dependent on the prior production of rtTA through a positive feedback mechanism, reaching near maximum levels of luciferase by 24 h was a particularly favorable feature of this system.
Table 1. Luciferase induction measurements.
| pG-BON | pDYAL-BON | ||||
|---|---|---|---|---|---|
| Harvest | Dox (µg/ml) | ng Luc/mg protein | Fold-induction | ng Luc/mg protein | Fold-induction |
| C127 mouse cells | |||||
| 24 h | 0 | 0.15 | ∅ | 0.42 | ∅ |
| 24 h | 1 | 1.74 | 11.60 | 64.51 | 153.60 |
| 24 h | 5 | 1.93 | 12.87 | 86.87 | 206.83 |
| 48 h | 0 | 0.12 | ∅ | 0.34 | ∅ |
| 48 h | 1 | 1.38 | 11.50 | 75.97 | 223.44 |
| 48 h | 5 | 1.94 | 16.17 | 113.57 | 334.03 |
| C57 mouse cells | |||||
| 24 h | 0 | 0.03 | ∅ | 0.16 | ∅ |
| 24 h | 1 | 1.66 | 55.33 | 65.64 | 410.25 |
| 24 h | 5 | 1.52 | 50.67 | 58.31 | 364.44 |
| 48 h | 0 | 0.05 | ∅ | 0.25 | ∅ |
| 48 h | 1 | 0.58 | 11.60 | 89.40 | 357.60 |
| 48 h | 5 | 1.77 | 65.40 | 61.50 | 246.00 |
Values of luciferase were determined by using a standard curve generated with recombinant luciferase. Luciferase values were normalized to protein concentrations in the extracts. The fold-inductions in mouse cell lines C127 and C57 are also presented. The data represent the average of two independent experiments, which gave closely similar results.
Figure 2.
Doxycyline-mediated luciferase induction. The data in Table 1 are graphed here. Luciferase levels were evaluated 24 and 48 h after introduction of Dox. Experiments were performed in (A) C127 and (B) C57 mouse cell lines.
The bi-directional tetP promoter cassette was able to mediate Dox-controlled gene expression whether the regulating elements were carried on the conventional pG-BON plasmid or on the EBV/human ori pDYAL-BON vector. However, the EBV/human ori vector was consistently more effective. When the bi-directional unit was introduced into C57 mouse cells on the EBV/human ori vector, it was possible to induce luciferase expression >400-fold within 24 h of being exposed to 1 µg/ml Dox. In contrast, mouse cells transfected with the conventional pG-BON plasmid bearing the same bi-directional cassette induced expression of luciferase at a much lower level, in the range of 10–50-fold. The difference probably reflects the consistent gene expression observed on the extrachromosomal vector, as opposed to the randomly integrated vector, in which many of the integration sites may not induce well due to position effects.
The extrachromosomally replicating EBV/human ori pDYAL-BON vector is generally present in multiple copies, ~1–100 per cell. The copy number tends to decrease slowly over time, even when selection is maintained (9). Using the same cell lines that were assayed above at 3 weeks, we also induced luciferase expression with Dox at 1 month and obtained essentially identical results. For more prolonged studies, freezing of the transfected cells during the first month and subsequent thawing before use is recommended. We tested the freezing and thawing protocol on these transfected cells, frozen ~1 month after transfection. After thawing of the cells, Dox-induced luciferase expression had the same characteristics as that observed for the cells before freezing.
There are two advantages to including the Tc-controlled regulatable cassette on an EBV/human ori vector. First, all the components involved in controlled regulation remain extrachromosomal. Gene expression from such extrachromosomal vectors has proven to be stable and consistent (9), independent of the position effects and silencing that are routinely observed with chromosomally integrated transgenes. Therefore, each cell receiving the vector will generate approximately the same level of stable gene expression. It is not necessary to isolate clones, nor to screen such clones for optimal activity. This feature saves months of time and labor.
By using an EBV/human ori vector instead of a conventional EBV vector, we gain the broad host range uniquely displayed by these vectors. Inclusion of a human genomic fragment to mediate replication in place of the dyad symmetry element present in the EBV oriP viral origin frees the replication origin from the species specificity typical of viral origins of replication (10). We have demonstrated replication function and long-term retention of such vectors in human, monkey, mouse and hamster cells (6,11). No restrictions on vector replication have been observed, so it is likely that the vectors will also replicate in many other mammalian cell types.
A previous problem with inclusion of both the transactivator and the target gene on a multicopy extrachromosomal vector was an elevated supply of the transactivator, resulting in a poor induction ratio. To correct this problem, we created a situation in which the rtTA gene was highly expressed only when induced gene expression was desired, rather than constitutively expressing the transactivator. By placing the rtTA gene under control of its own gene product, presence of the inducer is required to produce a significant amount of rtTA. In this way, the induction signal is amplified upon provision of the inducer, leading to a rapid and dramatic induction of gene expression of the regulated gene of interest. A similar idea has been applied previously in the context of a retroviral system (12), but here the simplicity and convenience of a plasmid system is preserved.
This system should prove useful for rapidly creating mammalian cell lines carrying an inducible gene that is under tight control. This feature is especially valuable when creation of a large panel of inducible cell lines is required.
Acknowledgments
ACKNOWLEDGEMENTS
C.R.S. was supported by a pre-doctoral fellowship from Public Health Service National Research Service Award T32 CA09302 from the National Cancer Institute. This work was supported by National Institutes of Health grant DK51834 to M.P.C.
REFERENCES
- 1.Gossen M. and Bujard,H. (1992) Proc. Natl Acad. Sci. USA, 89, 5547–5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kistner A., Gossen,M., Zimmermann,F., Jerecic,J., Ullmer,C., Lubbert,H. and Bujard,H. (1996) Proc. Natl Acad. Sci. USA, 93, 10933–10938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jost M., Kari,C. and Rodeck,U. (1997) Nucleic Acids Res., 25, 3131–3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lang Z. and Feingold,J.M. (1996) Gene, 168, 169–171. [DOI] [PubMed] [Google Scholar]
- 5.Haase S.B. and Calos,M.P. (1991) Nucleic Acids Res., 19, 5053–5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krysan P.J. and Calos,M.P. (1993) Gene, 136, 137–143. [DOI] [PubMed] [Google Scholar]
- 7.Hirt B. (1967) J. Mol. Biol., 26, 365–369. [DOI] [PubMed] [Google Scholar]
- 8.Krysan P.J., Haase,S.B. and Calos,M.P. (1989) Mol. Cell. Biol., 9, 1026–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wohlgemuth J.G., Kang,S.H., Bulboaca,G.H., Nawotka,K.A. and Calos,M.P. (1996) Gene Ther., 3, 503–512. [PubMed] [Google Scholar]
- 10.Calos M.P. (1996) Trends Genet., 12, 463–466. [DOI] [PubMed] [Google Scholar]
- 11.Heinzel S.S., Krysan,P.J., Tran,C.T. and Calos,M.P. (1991) Mol. Cell. Biol., 11, 2263–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoffman A., Nolan,G.P. and Blau,H.M. (1996) Proc. Natl Acad. Sci. USA, 93, 5185–5190. [DOI] [PMC free article] [PubMed] [Google Scholar]