Abstract
The high mobility group A (HMGA) proteins are found to be aberrantly expressed in several tumors. Studies (in vitro and in vivo) have shown that HMGA protein overexpression has a causative role in carcinogenesis process. HMGA proteins regulate cell cycle progression through distinct mechanisms which strongly influence its normal dynamics along malignant transformation. Tumor protein p53 (TP53) is the most frequently altered gene in cancer. The loss of its activity is recognized as the fall of a barrier that enables neoplastic transformation. Among the different functions, TP53 signaling pathway is tightly involved in control of cell cycle, with cell cycle arrest being the main biological outcome observed upon p53 activation, which prevents accumulation of damaged DNA, as well as genomic instability. Therefore, the interaction and opposing effects of HMGA and p53 proteins on regulation of cell cycle in normal and tumor cells are discussed in this review. HMGA proteins and p53 may reciprocally regulate the expression and/or activity of each other, leading to the counteraction of their regulation mechanisms at different stages of the cell cycle. The existence of a functional crosstalk between these proteins in the control of cell cycle could open the possibility of targeting HMGA and p53 in combination with other therapeutic strategies, particularly those that target cell cycle regulation, to improve the management and prognosis of cancer patients.
Keywords: HMGA, TP53, Cell cycle, Cancer, Cell cycle-directed anti-cancer therapies
Introduction
HMGA proteins
The high mobility group A (HMGA) protein family is composed of HMGA1a, HMGA1b, and HMGA2. HMGA1a and HMGA1b proteins are encoded by the same gene, HMGA1, which is located at the chromosome band 6p21, whereas HMGA2 is generated from HMGA2 gene located at the chromosome band 12q13-15 [1]. The HMGA proteins possess an N-terminus region harboring three basic domains known as AT-hooks, which are able to bind the AT-rich sequences in the minor groove of the DNA. They also possess a C-terminus region harboring the so-called acidic carboxyl-terminal domain, whose function still remains unclear. Indeed, the presence of many negatively charged amino acid residues makes the C-terminal domain suitable to contribute to protein–protein interactions rather than binding to DNA [2].
The HMGA proteins share the same structure and are well-conserved throughout the evolution, accounting for their ability to regulate common targets [3]. Although HMGA proteins do not have an intrinsic transcriptional activity, they function as architectural chromatinic proteins that bind to AT-rich sequences of DNA, without possessing any specific consensus sequence. In this regard, recent studies that performed ChIP-seq experiments confirmed the preference of HMGA proteins for AT-rich genome [4, 5]. Although HMGA proteins do not possess intrinsic transcriptional activity, they are involved in several biological pathways through orchestration of the assemblage of transcriptional complexes. By directly interacting with DNA and transcriptional factors, HMGA proteins are able to modulate the expression of several human genes [1, 6–8]. Notably, most of the HMGA-regulated genes (such as E2F1, c-Myc and CCNA) are involved in cell proliferation and invasion [9–11].
Therefore, due to their pivotal roles, HMGA proteins are subject to several post-translational modifications including arginine/lysine methylation, lysine acetylation, and serine/threonine phosphorylation, all of which modulate their interaction with DNA and other proteins [12–14].
The physiologic role of HMGA proteins is mainly implicated during embryogenesis, where they are highly expressed. The characterization of knocked-out mouse models of both Hmga1 (Hmga1-null) and Hmga2 (Hmga2-null) genes clearly revealed the involvement of these proteins in several aspects of development [1]. Interestingly, Hmga1-null and heterozygous mice developed type 2 diabetes and cardiac hypertrophy, respectively [1, 15]. Conversely, a pygmy phenotype was found in Hmga2-null and heterozygous mice, with a body size reduction of 60% and 25%, respectively, and a substantial impairment of body fat tissue [15, 16]. Furthermore, the Hmga1/Hmga2 double knock-out mice model exhibited a “superpygmy” phenotype, 80% loss in body size, which was probably induced by a strongly downregulation of E2F1 activity [17].
HMGA oncogenic activity
HMGA protein expression is low or absent (mainly as it concerns HMGA2) in adult tissues, whereas HMGA protein overexpression is a feature of malignant neoplasms [1], thus representing a marker for malignancy [18] and a poor prognostic index, since their upregulation is frequently correlated with a diminished patient survival and the occurrence of distant metastases [19].
Interestingly, the upregulation of HMGA proteins is not only a malignancy marker; however, it is well established by several in vitro and in vivo studies that HMGA overexpression has a causative role in carcinogenesis process. The abrogation of HMGA expression impaired the neoplastic transformation of rat thyroid-derived cell lines induced by murine transforming retroviruses [1]. Moreover, HMGA1 silencing, which is achieved by transfecting a HMGA1 cDNA antisense construct, causes apoptotic cell death in human anaplastic thyroid carcinoma-derived cell lines, but not in normal thyroid cells [20]. As a corroboration of HMGA role in carcinogenesis, engineered mice overexpressing both Hmga1 or Hmga2 were reported to develop several neoplasms such as lipomas [21], hematopoietic tumors [22, 23], and pituitary adenomas [24].
Finally, recent studies have demonstrated that different microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) regulate HMGA expression [25–33], with the most studied being miRNA-let7 [25–27], lnc SNHG16, lnc RPSAP52, and HMGA1 pseudogenes [28–33].
TP53-general aspects
TP53 (tumor protein p53) was first identified in the late 1970 s, and, during the first following years of its discovery, it was assumed to be an oncogene [34, 35]. However, a decade after its identification, TP53 was established as a tumor suppressor gene [36–38]. This represented a milestone on the understanding of TP53 and the molecular basis of cancer. TP53 was initially observed in 1989, when it was described as highly mutated in a wide variety of distinct tumors [39, 40]. In 1990, it was demonstrated that mutations in TP53 is associated with Li-Fraumeni syndrome, an inherited familial predisposition to a wide range of cancers [40, 41]. Till date, TP53 is the most studied human gene [42], as well as the most frequently altered gene in cancer [43, 44], with loss of its activity recognized as the fall of a barrier that enables neoplastic transformation and tumor development [44–46].
In humans, the TP53 gene is located on the short arm of chromosome 17 (17p13.1) and encodes the p53 protein [47]. p53 is a 393 amino acid protein divided into three main functional domains: N-terminal domain, DNA binding domain (DBD), and C-terminal domain. The N-terminal domain is required for transcriptional activation; the DBD represents the central core through which the interaction between p53 and its target proteins occurs, while the C-terminal is responsible for p53 tetramerization ability and regulation of DNA-binding domain, and contains a nuclear export and nuclear localization signals [48, 49].
p53 plays an important role in multicellular organisms through regulating cell cycle, as well as through its function as a tumor suppressor. In normal non-stressed cells, the functional p53 protein has a short half-life and is hardly detected [50]. However, under stress signal such as oncogene assaults and DNA damage, among others, the protein accumulates and triggers the transcription of p53 target genes, leading to cell cycle arrest/DNA repair or apoptosis in extreme cases [51, 52]. In this regard, p53 has been considered “the guardian of the genome”, reflecting its importance in ensuring the proper functioning of cells [53].
TP53 in cancer
Based on its anti-cancer function, p53 acts as a transcription factor and is involved in several cellular processes including DNA repair, cell cycle arrest, senescence, and apoptosis, among others [54]. Therefore, it is not surprising that TP53 signaling pathway is virtually inactivated in all types of cancer, since approximately half of cancer patients contain an inactivating mutation in TP53 and the other half present disrupted p53 function as a result of defective signaling pathways or effector molecules that regulate its activity [55–57].
Cellular levels of p53 protein are key determinant of its function. The expression of p53 is precisely controlled by E3 ubiquitin ligase murine double minute 2 (MDM2), which targets p53 toward degradation by 26S proteasome, thus maintaining a basal level of the protein [58–60]. Nevertheless, MDM2 itself is a transcriptional target of p53, thus characterizing a regulatory feedback loop [61]. Contrarily, upon cellular stress, the kinases including ataxia telangiectasia mutated (ATM) and ataxia telangiectasia and Rad3 related (ATR) act as DNA damage sensors and trigger a cascade of phosphorylation that leads to the phosphorylation of p53, preventing its interaction with MDM2. Therefore, p53 is stabilized and its levels are increased in cells, enabling the transactivation of its target genes and execution of its crucial cellular functions [56, 62, 63].
As a result of the crucial role of p53, its cellular levels/activity must be also tightly controlled or regulated. Post-translational regulation of p53 accounts for most of the mechanisms that control p53 activity during stress conditions. The post-translational regulation includes phosphorylation, acetylation, ubiquitination, sumoylation, neddylation, methylation, and glycosylation [49, 64–72]. Moreover, it is now known that p53 regulation takes place at many levels. In addition to the post-translational regulation of p53, transcriptional, post-transcriptional and translation regulation mechanisms have been comprehensively reviewed by Niazi and colleagues [73].
The frequency of TP53 mutations in different tumor types greatly varies, nevertheless, it is especially high (> 80%) in tumors that are very difficult to treat such as triple-negative breast cancer [74], high-grade serous ovarian cancer [75], esophageal squamous cell carcinoma [76, 77], squamous and small cell type lung cancer [78, 79]. Missense mutations in DBD are the most frequent ones, accounting for about 90% of all mutations [80–82]. Notably, mutant p53 proteins accumulate to a greater extent, since these proteins are incapable of inducing the transcription of their negative regulator MDM2 [83, 84]. Moreover, certain types of tumor do not harbor p53 mutation, but present an impaired p53 function due to overexpression of its inhibitor MDM2 [55–57].
HMGA regulates TP53 expression and function
Interestingly, it has been reported that HMGA proteins regulate p53 at different levels. The first mechanism of this regulation is represented by HMGA-dependent p53 regulation through protein–protein interaction. The silencing of HMGA1 exerted an increased activation of p53 functions in some thyroid carcinoma-derived cell lines, suggesting the inhibitory activity of HMGA1 protein on p53 protein. Furthermore, co-immunoprecipitation data suggested that HMGA1 directly interact with p53 via the C-terminal tetramerization domain [85]. Moreover, HMGA1 negatively regulates p53 protein functions by decreasing the transcription of several p53 effectors including Bax and p21Cip1, and enhancing the expression of p53 repressor MDM2 [1]. Interestingly, by Wang et al. [86] demonstrated that HMGA2 promotes cell cycle progression and inhibition of apoptosis by directly binding to both p53 tetramerization domain and zinc finger domains of MDM2, which increased the MDM2-induced ubiquitination on p53 and, consequently, its degradation.
The second mechanism involves HMGA-dependent p53 regulation at transcriptional level. Through chromatin immunoprecipitation (ChIP) and luciferase assays, Puca et al. [87] demonstrated that HMGA1 binds p53 promoter, repressing its expression in a dose-dependent manner.
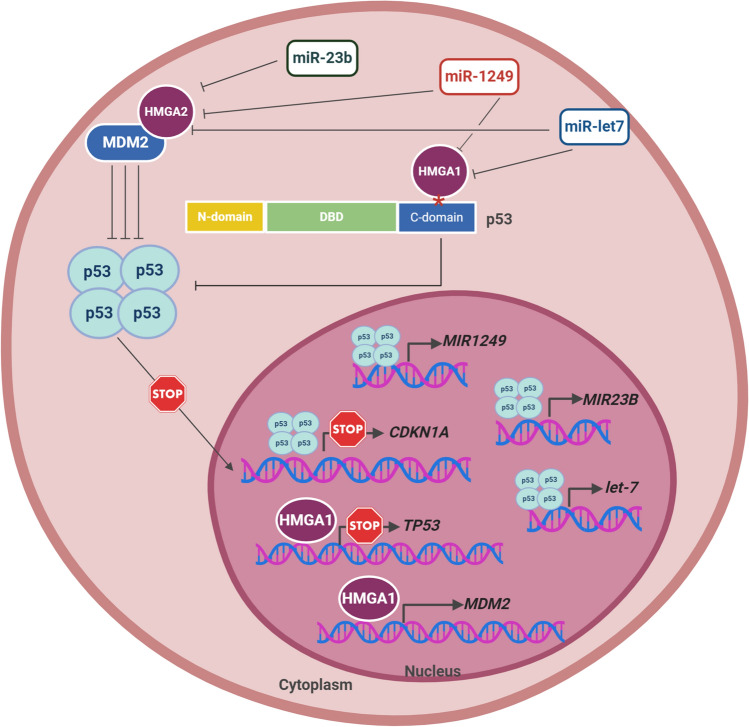
Surprisingly, p53 also seem to regulate the expression of HMGA proteins. Through induction of tumor suppressor miRNA (miR-1249), p53 suppressed colorectal cancer proliferation and metastasis by targeting HMGA2 [88]. In addition, activation of p53 in different cancer cell lines enhances the expression of other tumor suppressor miRNAs that target HMGA1 and/or 2 such as miR-23-b [89, 90] and miR-Let7 [91]. This highlights an important involvement of HMGA and TP53 interplay in cancer (Fig. 1).
Fig. 1.
TP53 and HMGA expression and/or function reciprocal regulation. Schematic representation of the mechanism through which TP53 and HMGA are capable of regulating each other expression and/or function. Specifically, to impair TP53 expression and function, HMGA1 is able to directly interact with p53 at the C-terminal oligomerization domain, blocking its tetramerization and, therefore, preventing its binding to DNA and consequent activation of its transcriptional targets, for example CDKN1A. Additionally, HMGA1 is capable of binding to TP53 promoter, inhibiting its transcription. Finally, HMGA1 also binds to the promoter of mouse double minute 2 (MDM2) gene, a repressor of p53, inducing its transcription. The mechanism through which HMGA2 decreases p53 expression consists in direct binding to the zinc finger domains of MDM2, thus increasing MDM2-induced ubiquitination of p53 and, consequently, its degradation. On the other hand, TP53 transcriptionally induces microRNA (miR) let-7, miRNA-1249, that target HMGA1 and HMGA2 for degradation, and miRNA-23b that target HMGA2 for degradation, depleting their cellular levels. *represents p53 oligomerization domain. DBD DNA binding domain
The role of HMGA and TP53 in cancer cell cycle control
Not disregarding the fact that HMGA and TP53 are involved in the regulation of several critical processes including cell proliferation, apoptosis, DNA repair, among others, thus triggering contrasting outcomes, herein we focus on their effects on the regulation of cell cycle. The cell cycle represents a key event that directly relates to tissue homeostasis, and alteration in the mechanisms involved in cell cycle regulation is highly associated with cancer development [92, 93]. HMGA proteins regulate the cell cycle through distinct mechanisms which strongly alter and directs its normal dynamics toward carcinogenesis [94]. TP53 is also tightly involved in cell cycle control, and cell cycle arrest is the main biological outcome observed upon p53 activation, thus preventing accumulation of damaged DNA and genomic instability, which presents a major barrier for tumor development [45, 46]. In the following sections, specific cell cycle mechanisms controlled by HMGA and TP53 are discussed according to the phases of cell cycle.
Early cell cycle phases-G1/S transition
p53 plays a crucial role in this phase of the cell cycle. Indeed, the activation of p53 leads to cell cycle arrest mainly through its capacity to regulate gene transcription [95]. Once p53 is activated in response to a wide variety of cellular stress, it induces cyclin-dependent kinase inhibitor 1A (CDKN1A) transcription, increasing the level of its product, p21Cip1 [96–99]. p21Cip1 then binds to cyclin E/cyclin-dependent kinase (CDK)2 and cyclin E/Cdk4 complexes, inactivating them and, therefore, blocking the phosphorylation of retinoblastoma (Rb) protein. Therefore, Rb remains bound to E2F1, preventing it from trans-activating its targets, resulting to cell cycle arrest at the G1 phase [97]. Moreover, p53 can induce G1/S cell cycle arrest by transcriptionally downregulating cell division cycle (CDC)25A in a p21Cip1-independent mechanism, as demonstrated in colorectal adenocarcinoma cells [100], and by repressing CCND1, as demonstrated in human non-small cell lung carcinoma and osteosarcoma cell lines. The inhibition of CCND1 occurs by switching its regulatory complex. p53 inhibits B cell lymphoma (Bcl)-3 expression, resulting to the reduced presence of CCND1 transcriptional activator p53/Bcl-3 complex, whereas its increment causes the association between p52 and histone deacetylases (HDAC)1, which enhances the presence of the transcriptional repressor complex [101]. Furthermore, it has been demonstrated that p53 acts a transcriptional inhibitor of CCNE2 via a p21Cip1-dependent mechanism in glioma cells [102].
Although the key mechanism of p53-mediated cell cycle arrest is the transcriptional repression of its cell cycle target genes, p53 can directly bind to the promoters of less than 5% of these genes [103], suggesting that the regulation of most of them occurs through indirect mechanisms. In this context, the modulation of DREAM (dimerization partner, RB-like, E2F and multi-vulval class B—MuvB) complex by p53 seems to play a crucial role in cell cycle control mechanisms. DREAM protein complex represses cell cycle genes during quiescence (G0), and orchestrates their expression at G1/S and G2/M phases in a time-coordinated manner, as a result of the shift from its transcriptional repression to activation assembly along cell cycle [104, 105]. p21Cip1 activation by p53 blocks the formation of cyclin-CDK complexes, thus maintaining pRB-like proteins in a hypophosphorylated state [106]. This enables them to associate with proteins to form the repressive DREAM complex configuration and hampers the assembly of the transcriptional activator complexes [104]. This characterizes the p53-DREAM pathway which has been previously demonstrated to regulate the expression of several cell cycle genes [98, 107, 108], leading to cell cycle arrest at G1 phase.
Interestingly, several evidences have indicated that one of the mechanisms by which p53 controls cell cycle and tumor progression is through miR-34 transcriptional regulation [109–115]. It was demonstrated that the induction of miR-34 expression leads to the inhibition of several cell cycle genes and proteins including cyclins E2 [116, 117], D2 [118], D3 [119], CDK4 and CDK6 [114, 116, 117, 119], E2F [114, 119], E2F3 [117, 118], Myc [117, 118] and Kras [118]. Furthermore, overexpression of miR-34 enhances the expression of the cyclin-dependent kinase inhibitor 2C (CDKN2C) [112]. Therefore, induction of miR-34 expression and modulation of its targets result to G1/S cell cycle arrest [113], as a p53-dependent mechanism. Downregulation of mir-34 is associated with TP53 mutation, as demonstrated in ovarian cancer, and a more malignant phenotype is associated with a worse overall survival [120, 121], making it a negative independent prognostic marker for breast [122, 123] and gastric carcinomas [124–127]. Consistently, ectopic expression of miR-34 was able to restore the p53 tumor suppressor functions in p53-deficient human pancreatic cancer cells by inhibiting cancer stem cell self-renewal and/or determining the cell fate [128].
The induction of miR-34 by TP53 presents an excellent demonstration of the interplay between TP53 and HMGA in regulation of cell cycle, since miR-34 has been reported to target HMGA2 expression, thus inducing arrest of gastric tumor cells in G1 phase, as well as decreasing the number of cells in S phase [129]. In addition, another demonstration revealed that HMGA genes may be involved in controlling the DREAM complex, thus hampering its assembly. Moreover, it was previously reported that HMGA2 gene silencing impacts on the expression of a central component of DREAM complex (that is, E2F4) in retinoblastoma cells [130]. Furthermore, p130, which is a RB-like protein and an essential component of DREAM complex, seems to be regulated by HMGA proteins. Moreover, HMGA1 was implicated as a driver in p130-negative human and murine retinoblastomas [131].
HMGA proteins also control the expression of regulatory proteins involved in the initial steps of cell cycle progression [94]. Thus, to elucidate the regulatory mechanisms of HMGA1 transcriptional network that is potentially related with lymphoma malignant transformation, Schuldenfrei and colleagues, using a HMGA1 transgenic mice model, demonstrated that HMGA1 overexpression is involved in the positive regulation of cyclin E [7, 132]. The upregulation of cyclin D and E1 expression by HMGA1 is associated with the activation of distinct mechanisms including the activation of Notch pathway and deregulation of Hippo signaling, which is represented by the nuclear localization of Yes-associated protein (YAP) [133–135]. Notch1 activation has been associated with tumor development [136, 137] and, interestingly, the expression of Notch 1 receptor and its ligand, delta-like canonical Notch ligand 1 (DLL1), are both inhibited by miR-34a [138]. Furthermore, miR-34 expression leads to diminished E2F3 mRNA levels [139]. Therefore, it seems that p53 tumor suppressor role counteracts the oncogenic effects of HMGA proteins in cell cycle control. To substantiate this, a growing body of evidence has been put forward to demonstrate a highly complex and context-dependent crosstalk between TP53 and Hippo pathways, since the deregulation of both signaling pathways are connected and associated with tumor progression [140].
HMGA1 also induces the expression of cyclins D1 and E1, triggering G1/S phase transition in cervical cancer cell progression. Furthermore, HMGA1 enhances cervical tumor cell invasiveness and proliferation by increasing the expression of miR-221/222 that target and, consequently, downregulate the tissue inhibitor of metalloproteinases 3 (TIMP3) [135] and p27, which plays a critical role in controlling G1/S transition [141]. HMGA2 also activates E2F1 through a mechanism that involves HDAC1 [9]. The relevance of the association between HMGA2 and histone deacetylases in the regulation of cell cycle was previously observed, though in a different context. It was reported that HMGA2 physically prevents the binding of HDAC1 to pRB/E2F1 complex, resulting in over-activity of E2F1 due to the increase in its acetylation which, in turn, promotes the transcription of regulatory genes involved in G1 phase progression [9]. The blockage of the physical interaction between HDAC1 and pRB/E2F1 complex caused by HMGA2, in addition to allowing G1 progression, it is also capable of preventing the activation of p53. This results due to the observation that HDAC1, when not bound to pRB/E2F1, deacetylates p53 in synergy with sirtuin (SIRT)1, preventing its overactivation [142] and transactivation of the target genes [143]. Therefore, the deacetylation of p53 suppresses its ability to trigger G1/S cell cycle arrest and increase miR-Let7a levels [91], which is one of the main epigenetic regulators of HMGA1 and HMGA2 [25–27], as well as the mechanism through which p53 counteracts the effects of HMGA on the cell cycle. Furthermore, HMGA1 positively regulates E2F1 [9] by directly increasing E2F1 expression and activity, promoting E2F1 release, due to its association with Rb [144, 145], and triggering G1/S progression. On the contrary, p53 activation induces the expression of miR-17-5p [91] which inhibits E2F1 [146], thus reinforcing the p53-mediated G1/S cell cycle arrest. Finally, the p53-transcriptional target, miR-34, causes a decrease in HDAC1 and SIRT1 levels [117], resulting in a positive feedback loop in which p53 triggers a cascade of events that amplifies its activation. Therefore, it is clear that the mechanisms governing cell cycle progression depend on the fundamental balance between the expression and/or activity of HMGA and p53 proteins.
Late cell cycle phases—G2/M progression
HMGA and p53 proteins also participate in the regulation of late cell cycle progression, that is, G2 to M phase transition. It was previously shown that HMGA2 counteracts the suppressive effect of the transcription factor p120E4F in cell cycle progression by displacing p120E4F from CCNA promoter, thus preventing its repressive effects over cyclin A expression. Following the displacement of p120E4F by HMGA, it binds onto the CCNA promoter, inducing the expression of cyclin A [11]. The association of cyclin A with CDKs such as CDK2 induces the entry into S phase, as well as progression to G2 phase [92, 93]. It is reasonable to hypothesize that the positive regulation of this protein by HMGA2 is associated with malignant transformation and/or progression. Interestingly, in line with this HMGA2 cell cycle control mechanism, ectopic expression of p120E4F in mouse embryo cells resulted in cell cycle arrest at G1/S phase, as well as a significant decrease in cyclins A, E, and D1, and CDK 4/6 and CDK2 activities associated with a marked increase in p21Cip1 expression [147]. Also in this case, the interplay between HMGA and p53 proteins is evident, since p53 interacts with p120E4F in human and murine cell lines, leading to cell cycle arrest, and that this association with p53 is required for p120E4F to exert its growth suppression activity [148]. Therefore, these data indicate that p120E4F may be an important p53 partner in the complex checkpoint network functions, and that p53 may indirectly regulate cyclins and CDKs activities via interaction with p120E4F. Furthermore, it was demonstrated that p120E4F simultaneously interacts with p14ARF and p53, forming a ternary complex in vivo that promotes G2 cell cycle arrest in a p53-dependent manner [149], thus reinforcing the importance of the association between p120E4F and p53 in cell cycle regulation.
Another evidence of the counteracting effects of HMGA and TP53 on cell cycle regulations was demonstrated with miR-23b and miR-130b. These miRNAs are able to target and downregulate HMGA2 in pituitary adenomas, promoting cell cycle arrest at G1 and G2 phases [150]. Therefore, the downregulation of miR-23b and miR-130b leads to an increase in the expression of both HMGA2 and cyclin A2, which also targets HMGA2 [150]. Interestingly, p53 is involved in miR-23b-modulated functions, since miR-23b levels are augmented in different cell lines following p53-induced expression, thus indicating that this miRNA represents a direct or indirect p53 target [89, 90]. Therefore, it could be suggested that the regulation exerted by miR-23b over HMGA2 and cyclin A2 levels resulted in cell cycle arrest which may be, at least in part, influenced by p53. Furthermore, the downregulation of miR-150 was associated with an increased HMGA2 and cyclin A expression in colorectal tumor cells [151], and, conversely, with a decreased activity of TP53 in human colorectal cancer cells [116].
Moreover, the circuits that govern HMGA2 expression during cell cycle appear to be complex. It was revealed that lncRNAs exhibited a regulatory effect on the expression of HMGA genes [30, 31]. In line with this, using a hepatocellular carcinoma cell model, it was demonstrated by Li and colleagues, that lncRNA SNHG16 acts as a decoy to mir-Let7b‐5p, which, besides inducing HMGA2 levels, also promotes the transition through G2/M phase via the enhancement of CDC25B expression [152]. However, although the authors did not demonstrate that the deregulation of cell cycle was a direct consequence of HMGA2 overexpression, it seems quite plausible that the aberrant expression of HMGA2 might have worked in synergy with CDC25B to elicit the G2/M transition.
One could point out the participation of TP53 in this regulatory network through which HMGA2 guides G2/M transition, since miR-Let7 expression is induced upon p53 activation [91]. Therefore, the lncRNA SNHG16 may operate to counteract the p53 cell cycle arrest effect in cells by overexpressing HMGA2 and contributing to cell cycle progression. In addition, expression of miR-17-5p, which is also induced upon p53 activation [91], was shown to be positively correlated with the expression of lncRNA SNHG16 [153]. E2F1 is negatively regulated by miR-17-5p [146]. Since lncRNA SNHG16 functions as a sponge for miR-17-5p, preventing the inhibition of its target genes [154], it could be suggested that the stabilization of E2F1 levels may represent another mechanism through which lncRNA SNHG16 restrains p53-mediated cell cycle arrest, thus favoring cell cycle progression. lncRNA SNHG16 is also capable of blocking one of the main mechanisms by which p53 controls cell cycle progression, which has to do with the induction of CDKN1A expression. SNHG16 enriches the histone methyltransferase Enhancer of Zeste Homolog 2 (EZH2) and recruits it to CDKN1A promoter [155], where it enhances H3K27me3 activity, resulting in CDKN1A repression [156]. Furthermore, it was reported that the overexpression of lncRNA RPSAP52 prevents HMGA2 degradation by competing with miR-15a, miR-15b, and miR-16 which are redirected to CDKN1A, causing its depletion [28]. Interestingly, in addition to CCNA transcriptional regulation by HMGA2, the circuit involved in the control of HMGA2 expression also influences cyclin A activity, since cyclin A/CDK2 complex is inhibited by p21Cip1 [93, 94], thus indicating the existence of a feedback loop mechanism in which HMGA2 directly or indirectly operates as a central element during cell cycle progression. In addition, it shows a significant conflict with TP53 cell cycle regulation, since p21Cip1 induction is the key mechanism for p53-mediated cell cycle control. Nevertheless, this is controversial following the reported miRNA-15a, 15b and 16 upregulation upon p53 activation [91], and their capacity of triggering apoptosis by targeting Bcl-2 [157]. One could hypothesize that this discrepancy could be due to the observation that miRNAs are promiscuous, that is, they possess multiple mRNA targets [158], and that lncRNA RPSAP52 might be interfering in the complex and highly coordinated cell cycle control network in which p53 and HMGA play crucial roles, shifting the effect from cell cycle arrest/apoptosis to cell cycle progression.
HMGA2 has also been reported as capable of regulating other important elements that are particularly involved in progression through G2 phase, as well as G2/M transition. HMGA proteins transcriptionally regulate the expression of cyclin A [1], cyclin B [94] and cyclin B2, apart from cooperating with p27 during pituitary tumorigenesis [94, 159]. p53 also controls the late stages of cell cycle progression, triggering G2 arrest under stressful cellular conditions [160]. The binding of cyclin-dependent kinase 1 (CDK1) to cyclin B1 is essential for its activation and to ensure G2/M transition. In this regard, p53 transcriptionally activates p21Cip1, as well as GADD45 (Growth Arrest and DNA-Damage-Inducible 45 Alpha) and 14-3-3 both of which simultaneously inhibits CDK1 activation [161, 162]. In addition, p53 transcriptionally represses CCNB1 and CCNB2 expression, resulting in G2 arrest [163–166]. p53 also inhibits CDK1 expression by inducing CDKN1A expression. Once expressed, p21Cip1 inhibits the cyclin-dependent kinase activity that promotes p130 and E2F4 which bind onto CDK1 promoter, causing its repression [160, 166, 167]. p53 also transcriptionally inhibits CCNA2 in a p21Cip1-dependent mechanism [168]. By modulating the DREAM complex, p53 regulates the expression of CCNB1, CDK1, CCNB2, CDC25C, among others [98, 107, 108]. Considering the potential role of HMGA in controlling DREAM complex assembly, it is important to consider the interference of HMGA on the expression of G2 phase genes through this mechanism. It is, therefore, evident that p53 and HMGA regulate several proteins involved in G2 phase of the cell cycle and, in most cases, exhibits an opposite effect, resulting in cell cycle arrest or progression, depending on the cellular state.
M phase and genomic stability
In addition to G1 and G2 checkpoints events, the phenomena occurring during M phase are also associated with tumor development and/or progression, and are regulated by HMGA and TP53. It has been observed that aberrant expression of HMGA proteins per se could affect genomic integrity during cancer progression, since it was reported that HMGA1 was associated with mitotic aberrations, such as chromosomes misalignment, besides promoting a significant alteration of the duration of metaphasis/anaphase due to the regulation of spindle assembly checkpoints (SAC) genes [169]. Also, the interplay between HMGA2 and Nek2 plays a crucial role on chromatin condensation during spermatocytes meiosis [170]. Therefore, these data suggest that cell cycle effects mediated by HMGA1 may only occur from a molecular instability threshold, which is probably represented by the loss of function of genes associated with the maintenance of genome integrity. These data interestingly reveal a scenario where HMGA proteins seems to be the “poison and antidote” at the same time, since cell cycle deregulation mediated by HMGA proteins in carcinogenesis is dependent on early alterations promoted by this same protein.
This strengths the suggested interplay between TP53 and HMGA in cell cycle controlling, and its consequences in tumor development and/or progression. Genomic integrity and tightly regulated cell cycle control mechanisms are crucial for maintaining tumor suppression. As discussed above, wild-type, active p53 controls virtually all the mechanisms that ensure proper cell cycle regulation. For instance, the modulation of DREAM regulatory complex by p53 is a compelling demonstration of the broad control exerted by p53 throughout cell cycle. The p53-DREAM pathway controls a great variety of cell cycle genes that act from G1 phase to the end of mitosis anaphase, indicating that p53 controls all the checkpoints present in cell cycle progression (G1/S, G2/M and SAC). Therefore, p53 controls SAC, chromosomal segregation, and mitotic spindle assembly [108, 171–180], and it could be stated that the deregulation caused by TP53 mutations may contribute to a general loss of cell cycle checkpoint control, resulting in aneuploidy and chromosomal instability.
In this regard, the proper functioning of p53 ensures genomic stability and tumor suppression. Nevertheless, since the maintenance of genomic integrity by p53 is lost in virtually all tumors [56–58], it could be stated that this may account for the genomic instability that drives cells toward tumorigenesis. The genomic instability caused by p53 loss may represent the molecular instability threshold needed for HMGA to function as an oncogene in cells. Moreover, HMGA overexpression is a very common feature in several tumors [1, 18, 19] and could also interfere with the tumor suppressor mechanisms exerted by the active p53. For instance, HMGA is also capable of intervening in the formation of the DREAM repressive complex through the modulation of E2F4 and p-130 expression [130, 131], representing a major intrusion in the broad control exerted by p53 throughout cell cycle.
Clinical perspectives of the interplay between HMGA and TP53 in cell cycle control
The inhibition of cell cycle progression has been described as a prominent therapeutic approach in the management of cancer [181]. In this regard, drugs that specifically inhibit the activity of target proteins involved in the control of cell cycle have been developed and tested. These drugs constitute two class of cell cycle inhibitors: CDK inhibitors [182] and cell cycle checkpoint inhibitors [183]. However, on the basis of the effects of HMGA and TP53 on cell cycle regulation, it is reasonable to envisage them as specific cell cycle target drugs in the future.
Although there is a possibility to block HMGA protein function to regulate the expression and activity of crucial molecules involved in cell cycle progression such as cyclins A, B, D, and E, as well as Rb and p53 [1], it, however, seems that the biological relevance of HMGA proteins cannot be neglected during the implementation of cell cycle-directed therapies. It was previously reported that several synthetic, semi-synthetic, and natural compounds that inhibit HMGA1 and HMGA2 function have been developed. The main approach employed is to prevent the binding of HMGA proteins to the AT-rich DNA regions [184]. However, most of these compounds present adverse side effects that could be associated with the lack of specificity of these drugs [185, 186]. In addition, the high expression levels of HMGA family members in adult stem cells [187] reinforce the need for a specific strategy that would be deviant of undesired systemic effects. The possibility to inhibit the accumulation of HMGA protein through the restoration of microRNAs that target HMGA proteins may be envisaged with future advancement in technology.
It is also crucial to consider the impact of TP53 mutational status when envisaging cell cycle-targeted cancer therapy. Loss of TP53 activity is widely spread among different types of tumors [56–58]. Therefore, in addition to considering the lack of p53 functions in tumors when proposing a cell cycle-directed therapy, p53 itself represents a very attractive target for cancer treatment, specifically the mutant p53 protein. Indeed, mutant p53 proteins lose their ability to bind DNA, accumulate in the cell, and are ready to be turned in its active form. It has been reported that the restoration of p53 wild-type functions results in regression of tumor [188–195].
Several compounds aimed at restoring wild-type p53 from mutant p53 have been produced and tested in pre-clinical studies, showing encouraging results [196–202]. Among the small p53 wild-type reactivating molecules, APR-246 is the most promising. APR-246 covalently bind to residues 277 and 124 of p53 protein [203, 204], restoring the wild-type conformation and re-establishing its transcriptional function [205, 206] in a wide range of p53 mutants [207]. This molecule was tested in a phase I/II clinical trial comprising 22 patients with hormone-refractory prostate cancer (n = 54) and hematological malignancies (n = 22), demonstrating its clinical effects in two patients and confirming its biological effects in tumor cells in vivo [208]. Since then, different phase I and II clinical trials are ongoing or just concluded (results not yet published) on the evaluation of the biological effects of APR-246 in combination with other drugs in esophageal cancer (NCT02999893), myeloid neoplasm (NCT03072043), and high-grade serous ovarian cancer (NCT02098343 and NCT03268382) patients.
In the cases where TP53 is not mutated but its function is hampered by overexpression of MDM2 that leads to p53 degradation, small molecule and peptide drugs have been developed to inhibit the interacting binding sites of p53 and MDM2, thus preventing the degradation of p53. A small peptide known as Idasanutlin is currently being tested through phases I, II and III clinical trials (NCT03850535 and NCT02545283) in patients with acute myeloid leukemia, since 75% of these patients possess a wild-type p53 [209]. It is important to consider some possible side effects of MDM2 antagonist treatment which includes: stabilization of p53 in normal cells, leading to undesired cell death; possible stabilization of mutant p53 in pre-malignant lesions, leading to increased risk of cancer progression; and possible increase in the expression of MDM2 degradation targets such as hormone receptors [198]. Therefore, the long-term use of peptides blocking p53-MDM2 interaction should be critically evaluated.
In addition, inducing synthetic lethality together with p53 mutation has also been envisaged as an anti-cancer therapeutic approach. Therefore, among the several p53 mutant synthetic lethal genes identified [210–214], some of them are part of p53-related pathways, since they are involved in G1 and G2/M checkpoints, DNA damage, as well as in the Rac and Rho pathway [199]. For this purpose, a Wee1 (involved in G2/M checkpoints) inhibitor (AZD1775) has been evaluated as a synthetic lethal agent in the TP53-mutated human cancers in phase I and II clinical trials, demonstrating that its use together with other chemotherapeutic drugs is efficient in the treatment of solid tumors and ovarian cancer patients [215, 216].
Another p53-targeted therapy is the introduction of a wild-type p53 in cancer cells using a defective adenovirus, followed by irradiation to cause DNA damage and induce p53-mediated apoptosis of head and neck tumors [217]. Although there are lots of progress required for the full implementation of p53-target therapies in tumors, this intervention strategy promises to be a powerful tool for improving cancer treatment, given the extremely encouraging results already obtained.
Conclusion
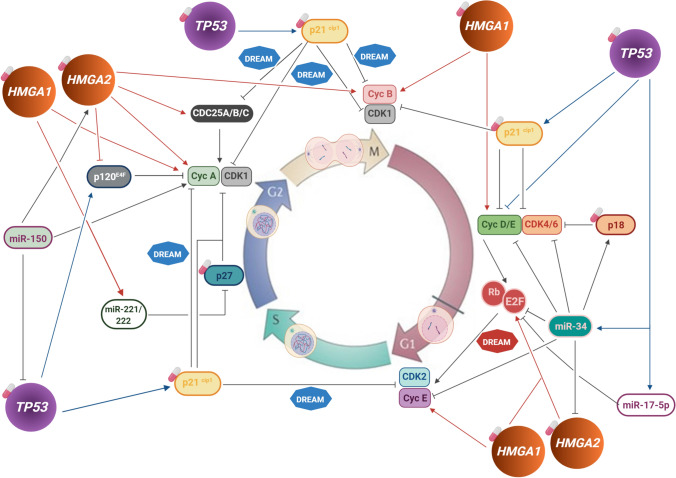
In the past years, it has been undeniably demonstrated that HMGA family members and TP53 play crucial roles in carcinogenesis. In addition, published data unveiled a functional interplay between HMGA proteins and TP53 in the regulation of crucial cellular processes including cell cycle (Fig. 2). In this regard, HMGA- and p53- targeted approaches, in combination with other therapeutic strategies, including those affecting cell cycle regulation, may greatly improve patients’ treatment response and prognosis. So far, the precise coordination of HMGA and p53 in controlling cell cycle and contributing to tumor development and/or progression is still not clear. Nevertheless, they are major cell cycle regulators with many common cell cycle control mechanisms, and are frequently altered in tumors. Therefore, it may be very promising to consider the interplay between HMGA and p53 in cell cycle control and tumor progression to improve therapeutic strategies, especially via targeting the cell cycle control.
Fig. 2.
Circuit representing TP53 and HMGA cell cycle control mechanisms in cancer. Schematic representation of the direct or indirect targets of TP53, HMGA1 and HMGA2 involved in the different cell cycle checkpoints and their effect exerted over the targets (Induction or inhibition). The involvement of DREAM complex in this circuit is also represented: transcriptional repression of p53 target genes which occurs through the modulation of DREAM complex towards its repressive assembly is indicated by the blue DREAM complex, whereas the upregulation of E2F4 by HMGA2 favoring DREAM transcriptional activator configuration and its potential effects is indicated by the red DREAM complex. It is possible to observe the overlap between TP53 and HMGA target molecules along cell cycle, nevertheless, leading to opposing outcomes. The white and pink pillules indicate the potential cell cycle druggable targets, i.e. cyclin-dependent kinase (CDK) inhibitors, TP53 and HMGA. The mechanistic details of the interplay between TP53 and HMGA in cell cycle regulation in cancer, as well as their potential use as therapeutic approach are described in the text
Funding
We would like to thank Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq—Brazil), Fundação de Amparo à Pesquisa Carlos Chagas Filho (FAPERJ—Brazil), Swiss Bridge Foundation and Associazione Italiana Ricerca sul Cancro (AIRC—Italy) for the financial support for this study.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Nathalia Meireles Da Costa, Email: nathalia.meireles@inca.gov.br.
Luiz Eurico Nasciutti, Email: luiz.nasciutti@histo.ufrj.br.
References
- 1.Fusco A, Fedele M. Roles of HMGA proteins in cancer. Nat Rev Cancer. 2007;7(12):899–910. doi: 10.1038/nrc2271. [DOI] [PubMed] [Google Scholar]
- 2.Ozturk N, Singh I, Mehta A, Braun T, Barreto G. HMGA proteins as modulators of chromatin structure during transcriptional activation. Front Cell Dev Biol. 2014;2:5. doi: 10.3389/fcell.2014.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vignali R, Marracci S. HMGA genes and proteins in development and evolution. Int J Mol Sci. 2020;21(2):1–39. doi: 10.3390/ijms21020654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang K, Guo W, Ren T, Huang Y, Han Y, Zhang H, et al. Knockdown of HMGA2 regulates the level of autophagy via interactions between MSI2 and Beclin1 to inhibit NF1-associated malignant peripheral nerve sheath tumour growth. J Exp Clin Cancer Res. 2019;38(1):185. doi: 10.1186/s13046-019-1183-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colombo DF, Burger L, Baubec T, Schübeler D. Binding of high mobility group A proteins to the mammalian genome occurs as a function of AT-content. PLoS Genet. 2017;13(12):e1007102. doi: 10.1371/journal.pgen.1007102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thanos D, Maniatis T. The high mobility group protein HMG I(Y) is required for NF-kappa B-dependent virus induction of the human IFN-beta gene. Cell. 1992;71(5):777–789. doi: 10.1016/0092-8674(92)90554-P. [DOI] [PubMed] [Google Scholar]
- 7.Forzati F, Federico A, Pallante P, Abbate A, Esposito F, Malapelle U, et al. CBX7 is a tumor suppressor in mice and humans. J Clin Invest. 2012;122(2):612–623. doi: 10.1172/JCI58620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Battista S, Fedele M, Martinez Hoyos J, Pentimalli F, Pierantoni GM, Visone R, et al. High-mobility-group A1 (HMGA1) proteins down-regulate the expression of the recombination activating gene 2 (RAG2) Biochem J. 2005;389:91–97. doi: 10.1042/BJ20041607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fedele M, Visone R, De Martino I, Troncone G, Palmieri D, Battista S, et al. HMGA2 induces pituitary tumorigenesis by enhancing E2F1 activity. Cancer Cell. 2006;9(6):459–471. doi: 10.1016/j.ccr.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 10.Cao XP, Cao Y, Zhao H, Yin J, Hou P. HMGA1 promoting gastric cancer oncogenic and glycolytic phenotypes by regulating c-myc expression. Biochem Biophys Res Commun. 2019;516(2):457–465. doi: 10.1016/j.bbrc.2019.06.071. [DOI] [PubMed] [Google Scholar]
- 11.Tessari MA, Gostissa M, Altamura S, Sgarra R, Rustighi A, Salvagno C, et al. Transcriptional activation of the cyclin A gene by the architectural transcription factor HMGA2. Mol Cell Biol. 2003;23(24):9104–9116. doi: 10.1128/MCB.23.24.9104-9116.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Q, Wang Y. HMG modifications and nuclear function. Biochim Biophys Acta. 2010;1799(1–2):28–36. doi: 10.1016/j.bbagrm.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sgarra R, Diana F, Rustighi A, Manfioletti G, Giancotti V. Increase of HMGA1a protein methylation is a distinctive characteristic of leukaemic cells induced to undergo apoptosis. Cell Death Differ. 2003;10(3):386–389. doi: 10.1038/sj.cdd.4401184. [DOI] [PubMed] [Google Scholar]
- 14.Sgarra R, Diana F, Bellarosa C, Dekleva V, Rustighi A, Toller M, et al. During apoptosis of tumor cells HMGA1a protein undergoes methylation: identification of the modification site by mass spectrometry. Biochemistry. 2003;42(12):3575–3585. doi: 10.1021/bi027338l. [DOI] [PubMed] [Google Scholar]
- 15.Foti D, Chiefari E, Fedele M, Iuliano R, Brunetti L, Paonessa F, Manfioletti G, Barbetti F, Brunetti A, Croce CM, Fusco A, Brunetti A. Lack of the architectural factor HMGA1 causes insulin resistance and diabetes in humans and mice. Nat Med. 2005;11(7):765–773. doi: 10.1038/nm1254. [DOI] [PubMed] [Google Scholar]
- 16.Anand A, Chada K. In vivo modulation of Hmgic reduces obesity. Nat Genet. 2000;24(4):377–380. doi: 10.1038/74207. [DOI] [PubMed] [Google Scholar]
- 17.Federico A, Forzati F, Esposito F, Arra C, Palma G, Barbieri A, et al. Hmga1/Hmga2 double knock-out mice display a “superpygmy” phenotype. Biol Open. 2014;3(5):372–378. doi: 10.1242/bio.20146759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pallante P, Sepe R, Puca F, Fusco A. High mobility group A proteins as tumor markers. Front Med (Lausanne) 2015;2:15–22. doi: 10.3389/fmed.2015.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X, Liu X, Li AY, Chen L, Lai L, Lin HH, et al. Overexpression of HMGA2 promotes metastasis and impacts survival of colorectal cancers. Clin Cancer Res. 2011;17(8):2570–2580. doi: 10.1158/1078-0432.CCR-10-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berlingieri MT, Pierantoni GM, Giancotti V, Santoro M, Fusco A. Thyroid cell transformation requires the expression of the HMGA1 proteins. Oncogene. 2002;21(19):2971–2980. doi: 10.1038/sj.onc.1205368. [DOI] [PubMed] [Google Scholar]
- 21.Arlotta P, Tai AK, Manfioletti G, Clifford C, Jay G, Ono SJ. Transgenic mice expressing a truncated form of the high mobility group I-C protein develop adiposity and an abnormally high prevalence of lipomas. J Biol Chem. 2000;275(19):14394–14400. doi: 10.1074/jbc.M000564200. [DOI] [PubMed] [Google Scholar]
- 22.Baldassarre G, Fedele M, Battista S, Vecchione A, Klein-Szanto AJ, Santoro M, et al. Onset of natural killer cell lymphomas in transgenic mice carrying a truncated HMGI-C gene by the chronic stimulation of the IL-2 and IL-15 pathway. Proc Natl Acad Sci USA. 2001;98(14):7970–7975. doi: 10.1073/pnas.141224998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu Y, Sumter TF, Bhattacharya R, Tesfaye A, Fuchs EJ, Wood LJ, et al. The HMG-I oncogene causes highly penetrant, aggressive lymphoid malignancy in transgenic mice and is overexpressed in human leukemia. Cancer Res. 2004;64(10):3371–3375. doi: 10.1158/0008-5472.CAN-04-0044. [DOI] [PubMed] [Google Scholar]
- 24.Fedele M, Pentimalli F, Baldassarre G, Battista S, Klein-Szanto AJ, Kenyon L, et al. Transgenic mice overexpressing the wild-type form of the HMGA1 gene develop mixed growth hormone/prolactin cell pituitary adenomas and natural killer cell lymphomas. Oncogene. 2005;24(21):3427–3435. doi: 10.1038/sj.onc.1208501. [DOI] [PubMed] [Google Scholar]
- 25.Oliveira-Mateos C, Sánchez-Castillo A, Soler M, Obiols-Guardia A, Piñeyro D, Boque-Sastre R, et al. The transcribed pseudogene RPSAP52 enhances the oncofetal HMGA2-IGF2BP2-RAS axis through LIN28B-dependent and independent let-7 inhibition. Nat Commun. 2019;10(1):3979–4036. doi: 10.1038/s41467-019-11910-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X, Cao L, Wang Y, Wang X, Liu N, You Y. Regulation of let-7 and its target oncogenes (Review) Oncol Lett. 2012;3(5):955–960. doi: 10.3892/ol.2012.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Martino M, Esposito F, Pellecchia S, Penha RCC, Botti G, Fusco A, et al. HMGA1-regulating microRNAs Let-7a and miR-26a are downregulated in human seminomas. Int J Mol Sci. 2020;21:3014–3023. doi: 10.3390/ijms21083014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D’Angelo D, Arra C, Fusco A. RPSAP52 lncRNA inhibits p21Waf1/CIP expression by interacting with the RNA binding protein HuR. Oncol Res. 2020;28(2):191–201. doi: 10.3727/096504019X15761465603129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ros G, Pegoraro S, De Angelis P, Sgarra R, Zucchelli S, Gustincich S, et al. HMGA2 antisense long non-coding RNAs as new players in the regulation of HMGA2 expression and pancreatic cancer promotion. Front Oncol. 2019;9:1526–1531. doi: 10.3389/fonc.2019.01526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.D’Angelo D, Mussnich P, Sepe R, Raia M, Del Vecchio L, Cappabianca P, et al. RPSAP52 lncRNA is overexpressed in pituitary tumors and promotes cell proliferation by acting as miRNA sponge for HMGA proteins. J Mol Med (Berl) 2019;97(7):1019–1032. doi: 10.1007/s00109-019-01789-7. [DOI] [PubMed] [Google Scholar]
- 31.Wang Z, Wang P, Cao L, Li F, Duan S, Yuan G, et al. Long intergenic non-coding RNA 01121 promotes breast cancer cell proliferation, migration, and invasion via the miR-150-5p/HMGA2 axis. Cancer Manag Res. 2019;11:10859–10870. doi: 10.2147/CMAR.S230367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Martino M, Forzati F, Arra C, Fusco A, Esposito F. HMGA1-pseudogenes and cancer. Oncotarget. 2016;7(19):28724–28735. doi: 10.18632/oncotarget.7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Esposito F, De Martino M, Petti MG, Forzati F, Tornincasa M, Federico A, et al. HMGA1 pseudogenes as candidate proto-oncogenic competitive endogenous RNAs. Oncotarget. 2014;5(18):8341–8354. doi: 10.18632/oncotarget.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lane DP, Crawford LV. T antigen is bound to a host protein in SV40-transformed cells. Nature. 1979;278:261–263. doi: 10.1038/278261a0. [DOI] [PubMed] [Google Scholar]
- 35.Linzer DI, Levine AJ. Characterization of a 54 K dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells. Cell. 1979;17:43–52. doi: 10.1016/0092-8674(79)90293-9. [DOI] [PubMed] [Google Scholar]
- 36.Eliyahu D, Michalovitz D, Eliyahu S, Pinhasi-Kimhi O, Oren M. Wild-type p53 can inhibit oncogene-mediated focus formation. Proc Natl Acad Sci USA. 1989;86:8763–8767. doi: 10.1073/pnas.86.22.8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Finlay CA, Hinds PW, Levine AJ. The p53 proto-oncogene can act as a suppressor of transformation. Cell. 1989;57:1083–1093. doi: 10.1016/0092-8674(89)90045-7. [DOI] [PubMed] [Google Scholar]
- 38.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 39.Baker SJ, Fearon ER, Nigro JM, Hamilton SR, Preisinger AC, Jessup JM, et al. Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science. 1989;244:217–221. doi: 10.1126/science.2649981. [DOI] [PubMed] [Google Scholar]
- 40.Nigro JM, Baker SJ, Preisinger AC, Jessup JP, Hosteller R, Cleary K, et al. Mutations in the p53 gene occur in diverse human tumour types. Nature. 1989;342:705–708. doi: 10.1038/342705a0. [DOI] [PubMed] [Google Scholar]
- 41.Malkin D, Li FP, Strong LC, Fraumeni JF, Jr, Nelson CE, Kim DH, et al. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science. 1990;250:1233–1238. doi: 10.1126/science.1978757. [DOI] [PubMed] [Google Scholar]
- 42.Dolgin E. The most popular genes in the human genome. Nature. 2017;551:427–431. doi: 10.1038/d41586-017-07291-9. [DOI] [PubMed] [Google Scholar]
- 43.Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rivlin N, Brosh R, Oren M, Rotter V. Mutations in the p53 tumor suppressor gene: important milestones at the various steps of tumorigenesis. Genes Cancer. 2011;2(4):466–474. doi: 10.1177/1947601911408889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mantovani F, Collavin L, Del Sal G. Mutant p53 as a guardian of the cancer cell. Cell Death Differ. 2019;26(2):199–212. doi: 10.1038/s41418-018-0246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hainaut P, Hollstein M. p53 and human cancer: the first ten thousand mutations. Adv Cancer Res. 2000;77:81–137. doi: 10.1016/S0065-230X(08)60785-X. [DOI] [PubMed] [Google Scholar]
- 48.Stommel JM, Marchenko ND, Jimenez GS, Moll UM, Hope TJ, Wahl GM. A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J. 1999;18:1660–1672. doi: 10.1093/emboj/18.6.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bode AM, Dong Z. Post-translational modification of p53 in tumorigenesis. Nat Rev Cancer. 2004;4(10):793–805. doi: 10.1038/nrc1455. [DOI] [PubMed] [Google Scholar]
- 50.Hernandez-Boussard T, Montesano R, Hainaut P. Sources of bias in the detection and reporting of p53 mutations in human cancer: analysis of the IARC p53 mutation database. Genet Anal. 1999;14(5–6):229–233. doi: 10.1016/S1050-3862(98)00030-8. [DOI] [PubMed] [Google Scholar]
- 51.Gudkov AV, Komarova EA. Dangerous habits of a security guard: the 2 faces of p53 as a drug target. Hum Mol Genet. 2007;16(Spec No 1):R67–R72. doi: 10.1093/hmg/ddm052. [DOI] [PubMed] [Google Scholar]
- 52.Lu C, El-Deiry WS. Targeting p53 for enhanced radio- and chemo-sensitivity. Apoptosis. 2009;14:597–606. doi: 10.1007/s10495-009-0330-1. [DOI] [PubMed] [Google Scholar]
- 53.Bykov VJ, Selivanova G, Wiman KG. Small molecules that reactivate mutant p53. Eur J Cancer. 2003;39:1828–1834. doi: 10.1016/S0959-8049(03)00454-4. [DOI] [PubMed] [Google Scholar]
- 54.Read AP, Strachan T. Cancer genetics. Human molecular genetics 2. New York: Wiley; 1999. [Google Scholar]
- 55.Bieging KT, Mello SS, Attardi LD. Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer. 2014;14:359–370. doi: 10.1038/nrc3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kruiswijk F, Labuschagne CF, Vousden KH. p53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nat Rev Mol Cell Biol. 2015;16:393–405. doi: 10.1038/nrm4007. [DOI] [PubMed] [Google Scholar]
- 57.Muller PA, Vousden KH. Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell. 2014;25:304–317. doi: 10.1016/j.ccr.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 59.Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor supressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/S0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 60.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 61.Barak Y, Juven T, Haffner R, Oren M. MDM2 expression is induced by wild type p53 activity. EMBO J. 1993;12:461–468. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hager KM, Gu W. Understanding the noncanonical pathways involved in p53-mediated tumor suppression. Carcinogenesis. 2014;35:740–746. doi: 10.1093/carcin/bgt487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kang R, Kroemer G, Tang D. The tumor suppressor protein p53 and the ferroptosis network. Free Radic Biol Med. 2019;133:162–168. doi: 10.1016/j.freeradbiomed.2018.05.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hupp TR, Lane DP. Allosteric activation of latent p53 tetramers. Curr Biol. 1994;4:865–875. doi: 10.1016/S0960-9822(00)00195-0. [DOI] [PubMed] [Google Scholar]
- 65.Prives C, Hall PA. The p53 pathway. J Pathol. 1999;187:112–126. doi: 10.1002/(SICI)1096-9896(199901)187:1<112::AID-PATH250>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 66.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 67.Brooks CL, Gu W. Ubiquitination, phosphorylation and acetylation: the molecular basis for p53 regulation. Curr Opin Cell Biol. 2003;15:164–171. doi: 10.1016/S0955-0674(03)00003-6. [DOI] [PubMed] [Google Scholar]
- 68.Michael D, Oren M. The p53-Mdm2 module and the ubiquitin system. Semin Cancer Biol. 2003;13:49–58. doi: 10.1016/S1044-579X(02)00099-8. [DOI] [PubMed] [Google Scholar]
- 69.Brooks CL, Gu W. p53 ubiquitination: mdm2 and Beyond. Mol Cell. 2006;21:307–315. doi: 10.1016/j.molcel.2006.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypothesis, in vivo veritas. Nat Rev Cancer. 2006;6:909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- 71.Horn HF, Vousden KH. Coping with stress: multiple ways to activate p53. Oncogene. 2007;26:1306–1316. doi: 10.1038/sj.onc.1210263. [DOI] [PubMed] [Google Scholar]
- 72.Tang Y, Zhao W, Chen Y, Zhao Y, Gu W. Actetylation is indispensable for p53 activation. Cell. 2008;133:612–626. doi: 10.1016/j.cell.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Niazi S, Purohit M, Niazi JH. Role of p53 circuitry in tumorigenesis: a brief review. Eur J Med Chem. 2018;158:7–24. doi: 10.1016/j.ejmech.2018.08.099. [DOI] [PubMed] [Google Scholar]
- 74.The Cancer Genome Atlas Research Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.The Cancer Genome Atlas Research Network Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Song Y, Li L, Ou Y, Gao Z, Li E, Li X, et al. Identification of genomic alterations in oesophageal squamous cell cancer. Nature. 2014;509:91–95. doi: 10.1038/nature13176. [DOI] [PubMed] [Google Scholar]
- 77.Souza-Santos PT, Soares Lima SC, Nicolau-Neto P, Boroni M, Meireles Da Costa N, Brewer L, et al. Mutations, differential gene expression, and chimeric transcripts in esophageal squamous cell carcinoma show high heterogeneity. Transl Oncol. 2018;11(6):1283–1291. doi: 10.1016/j.tranon.2018.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cancer Genome Atlas Research Network Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pfeifer M, Fernández-Cuesta L, Sos ML, George J, Seidel D, Kasper LH, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet. 2012;44:1104–1110. doi: 10.1038/ng.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.https://cancer.sanger.ac.uk/cosmic. COSMIC (Catalogue of Somatic Mutations in Cancer). Accessed Mar 2020
- 81.https://p53.iarc.fr/. TP53 database, IARC (International Agency for Research on Cancer). Accessed Mar 2020
- 82.Bouaoun L, Sonkin D, Ardin M, Hollstein M, Byrnes G, Zavadil J, et al. TP53 variations in human cancers: new lessons from the IARC TP53 database and genomics data. Hum Mutat. 2016;7(9):865–876. doi: 10.1002/humu.23035. [DOI] [PubMed] [Google Scholar]
- 83.Brosh R, Rotter V. When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer. 2009;9:701–713. doi: 10.1038/nrc2693. [DOI] [PubMed] [Google Scholar]
- 84.Terzian T, Suh YA, Iwakuma T, Post SM, Neumann M, Lang GA, et al. The inherent instability of mutant p53 is alleviated by Mdm2 or p16INK4a loss. Genes Dev. 2008;22:1337–1344. doi: 10.1101/gad.1662908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Frasca F, Rustighi A, Malaguarnera R, Altamura S, Vigneri P, Del Sal G, et al. HMGA1 inhibits the function of p53 family members in thyroid cancer cells. Cancer Res. 2006;66(6):2980–2989. doi: 10.1158/0008-5472.CAN-05-2637. [DOI] [PubMed] [Google Scholar]
- 86.Wang Y, Hu L, Wang J, Li X, Sahengbieke S, Wu J, et al. HMGA2 promotes intestinal tumorigenesis by facilitating MDM2-mediated ubiquitination and degradation of p53. J Pathol. 2018;246(4):508–518. doi: 10.1002/path.5164. [DOI] [PubMed] [Google Scholar]
- 87.Puca F, Colamaio M, Federico A, Gemei M, Tosti N, Bastos AU, Del Vecchio L, Pece S, Battista S, Fusco A. HMGA1 silencing restores normal stem cell characteristics in colon cancer stem cells by increasing p53 levels. Oncotarget. 2014;5(10):3234–3245. doi: 10.18632/oncotarget.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen X, Zeng K, Xu M, Liu X, Hu X, Xu T, et al. p53-induced miR-1249 inhibits tumor growth, metastasis, and angiogenesis by targeting VEGFA and HMGA2. Cell Death Dis. 2019;10(2):131–156. doi: 10.1038/s41419-018-1188-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.He L, Zhao X, He L. LINC01140 alleviates the oxidized low-density lipoprotein-induced inflammatory response in macrophages via suppressing miR-23b. Inflammation. 2020;43(1):66–73. doi: 10.1007/s10753-019-01094-y. [DOI] [PubMed] [Google Scholar]
- 90.Blume CJ, Hotz-Wagenblatt A, Hüllein J, Sellner L, Jethwa A, Stolz T, et al. p53-dependent non-coding RNA networks in chronic lymphocytic leukemia. Leukemia. 2015;29(10):2015–2023. doi: 10.1038/leu.2015.119. [DOI] [PubMed] [Google Scholar]
- 91.Tarasov V, Jung P, Verdoodt B, Lodygin D, Epanchintsev A, Menssen A, et al. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6(13):1586–1593. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- 92.Dai L, Zhao T, Bisteau X, Sun W, Prabhu N, Lim YT, et al. Modulation of protein-interaction states through the cell cycle. Cell. 2018;173(6):1481–1494. doi: 10.1016/j.cell.2018.03.065. [DOI] [PubMed] [Google Scholar]
- 93.Ingham M, Schwartz GK. Cell-cycle therapeutics come of age. J Clin Oncol. 2017;35(25):2949–2959. doi: 10.1200/JCO.2016.69.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fedele M, Fusco A. Role of the high mobility group A proteins in the regulation of pituitary cell cycle. J Mol Endocrinol. 2010;44(6):309–318. doi: 10.1677/JME-09-0178. [DOI] [PubMed] [Google Scholar]
- 95.Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137(3):413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 96.El-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75(4):817–825. doi: 10.1016/0092-8674(93)90500-P. [DOI] [PubMed] [Google Scholar]
- 97.Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potente inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-G. [DOI] [PubMed] [Google Scholar]
- 98.Quaas M, Müller GA, Engeland K. p53 can repress transcription of cell cycle genes through a p21(WAF1/CIP1)-dependent switch from MMB to DREAM protein complex binding at CHR promoter elements. Cell Cycle. 2012;11:4661–4672. doi: 10.4161/cc.22917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer. 2009;9:400–414. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rother K, Kirschner R, Sänger K, Böhlig L, Mössner J, Engeland K. p53 downregulates expression of the G1/S cell cycle phosphatase Cdc25A. Oncogene. 2007;26(13):1949–1953. doi: 10.1038/sj.onc.1209989. [DOI] [PubMed] [Google Scholar]
- 101.Rocha S, Martin AM, Meek DW, Perkins ND. p53 represses cyclin D1 transcription through down regulation of Bcl-3 and inducing increased association of the p52 NF-kappaB subunit with histone deacetylase 1. Mol Cell Biol. 2003;23(13):4713–4727. doi: 10.1128/MCB.23.13.4713-4727.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gorjala P, Cairncross JG, Gary RK. p53-dependent up-regulation of CDKN1A and down-regulation of CCNE2 in response to beryllium. Cell Prolif. 2016;49(6):698–709. doi: 10.1111/cpr.12291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fischer M, Steiner L, Engeland K. The transcription factor p53: not a repressor, solely an activator. Cell Cycle. 2014;13:3037–3058. doi: 10.4161/15384101.2014.949083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Engeland K. Cell cycle arrest through indirect transcriptional repression by p53: i have a DREAM. Cell Death Differ. 2018;25(1):114–132. doi: 10.1038/cdd.2017.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sadasivam S, DeCaprio JA. The DREAM complex: master coordinator of cell cycle-dependent gene expression. Nat Rev Cancer. 2013;13(8):585–595. doi: 10.1038/nrc3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rippin TM, Bykov VJ, Freund SM, Selivanova G, Wiman KG, Fersht AR. Characterization of the p53-rescue drug CP-31398 in vitro and in living cells. Oncogene. 2002;21:2119–2129. doi: 10.1038/sj.onc.1205362. [DOI] [PubMed] [Google Scholar]
- 107.Mannefeld M, Klassen E, Gaubatz S. B-MYB is required for recovery from the DNA damage-induced G2 checkpoint in p53 mutant cells. Cancer Res. 2009;69(9):4073–4080. doi: 10.1158/0008-5472.CAN-08-4156. [DOI] [PubMed] [Google Scholar]
- 108.Fischer M, Quaas M, Nickel A, Engeland K. Indirect p53-dependent transcriptional repression of Survivin, CDC25C, and PLK1 genes requires the cyclin-dependent kinase inhibitor p21/CDKN1A and CDE/CHR promoter sites binding the DREAM complex. Oncotarget. 2015;6(39):41402–41417. doi: 10.18632/oncotarget.6356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Christoffersen NR, Shalgi R, Frankel LB, Leucci E, Lees M, Klausen M, et al. p53-independent upregulation of miR-34a during oncogene-induced senescence represses MYC. Cell Death Differ. 2010;17(2):236–245. doi: 10.1038/cdd.2009.109. [DOI] [PubMed] [Google Scholar]
- 110.Wong MY, Yu Y, Walsh WR, Yang JL. microRNA-34 family and treatment of cancers with mutant or wild-type p53. Int J Oncol. 2011;38(5):1189–1195. doi: 10.3892/ijo.2011.970. [DOI] [PubMed] [Google Scholar]
- 111.He X, He L, Hannon GJ. The guardian’s little helper: microRNAs in the p53 tumor suppressor network. Cancer Res. 2007;67(23):11099–11101. doi: 10.1158/0008-5472.CAN-07-2672. [DOI] [PubMed] [Google Scholar]
- 112.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26(5):745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, et al. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol. 2007;17(15):1298–1307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- 114.Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010;17(2):193–199. doi: 10.1038/cdd.2009.56. [DOI] [PubMed] [Google Scholar]
- 115.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447(7148):1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Slattery ML, Mullany LE, Wolff RK, Sakoda LC, Samowitz WS, Herrick JS. The p53-signaling pathway and colorectal cancer: interactions between downstream p53 target genes and miRNAs. Genomics. 2019;111(4):762–771. doi: 10.1016/j.ygeno.2018.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kaller M, Liffers ST, Oeljeklaus S, Kuhlmann K, Röh S, Hoffmann R et al (2011) Genome-wide characterization of miR-34a induced changes in protein and mRNA expression by a combined pulsed SILAC and microarray analysis. Mol Cell Proteomics 10(8):M111.010462 [DOI] [PMC free article] [PubMed]
- 118.Zhu H, Dougherty U, Robinson V, Mustafi R, Pekow J, Kupfer S, et al. EGFR signals downregulate tumor suppressors miR-143 and miR-145 in Western diet-promoted murine colon cancer: role of G1 regulators. Mol Cancer Res. 2011;9(7):960–975. doi: 10.1158/1541-7786.MCR-10-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lal A, Thomas MP, Altschuler G, Navarro F, O’Day E, Li XL, et al. Capture of microRNA-bound mRNAs identifies the tumor suppressor miR-34a as a regulator of growth factor signaling. PLoS Genet. 2011;7(11):e1002363. doi: 10.1371/journal.pgen.1002363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Welponer H, Tsibulak I, Wieser V, Degasper C, Shivalingaiah G, Wenzel S, et al. The miR-34 family and its clinical significance in ovarian cancer. J Cancer. 2020;11(6):1446–1456. doi: 10.7150/jca.33831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Schmid G, Notaro S, Reimer D, Abdel-Azim S, Duggan-Peer M, Holly J, et al. Expression and promotor hypermethylation of miR-34a in the various histological subtypes of ovarian cancer. BMC Cancer. 2016;16:102–110. doi: 10.1186/s12885-016-2135-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bonetti P, Climent M, Panebianco F, Tordonato C, Santoro A, Marzi MJ, et al. Dual role for miR-34a in the control of early progenitor proliferation and commitment in the mammary gland and in breast cancer. Oncogene. 2019;38(3):360–374. doi: 10.1038/s41388-018-0445-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Park EY, Chang E, Lee EJ, Lee HW, Kang HG, Chun KH, et al. Targeting of miR34a-NOTCH1 axis reduced breast cancer stemness and chemoresistance. Cancer Res. 2014;74(24):7573–7582. doi: 10.1158/0008-5472.CAN-14-1140. [DOI] [PubMed] [Google Scholar]
- 124.Hui WT, Ma XB, Zan Y, Wang XJ, Dong L. Prognostic significance of miR-34a expression in patients with gastric cancer after radical gastrectomy. Chin Med J (Engl) 2015;128:2632–2637. doi: 10.4103/0366-6999.166019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kim CH, Kim HK, Rettig RL, Kim J, Lee ET, Aprelikova O, et al. miRNA signature associated with outcome of gastric cancer patients following chemotherapy. BMC Med Genomics. 2011;4:79–86. doi: 10.1186/1755-8794-4-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Katada T, Ishiguro H, Kuwabara Y, Kimura M, Mitui A, Mori Y, et al. microRNA expression profile in undifferentiated gastric cancer. Int J Oncol. 2009;34:537–542. [PubMed] [Google Scholar]
- 127.Zhang H, Li S, Yang J, Liu S, Gong X, Yu X. The prognostic value of miR-34a expression in completely resected gastric cancer: tumor recurrence and overall survival. Int J Clin Exp Med. 2015;8:2635–2641. [PMC free article] [PubMed] [Google Scholar]
- 128.Ji Q, Hao X, Zhang M, Tang W, Yang M, Li L, et al. MicroRNA miR-34 inhibits human pancreatic cancer tumor-initiating cells. PLoS ONE. 2009;4(8):e6816. doi: 10.1371/journal.pone.0006816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ji Q, Hao X, Meng Y, Zhang M, Desano J, Fan D, et al. Restoration of tumor suppressor miR-34 inhibits human p53-mutant gastric cancer tumorspheres. BMC Cancer. 2008;8:266–278. doi: 10.1186/1471-2407-8-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Venkatesan N, Krishnakumar S, Deepa PR, Deepa M, Khetan V, Reddy MA. Molecular deregulation induced by silencing of the high mobility group protein A2 gene in retinoblastoma cells. Mol Vis. 2012;18:2420–3247. [PMC free article] [PubMed] [Google Scholar]
- 131.Kooi IE, van Mil SE, MacPherson D, Mol BM, Moll AC, Meijers-Heijboer H, et al. Genomic landscape of retinoblastoma in Rb -/- p130 -/- mice resembles human retinoblastoma. Genes Chromosomes Cancer. 2017;56(3):231. doi: 10.1002/gcc.22429. [DOI] [PubMed] [Google Scholar]
- 132.Schuldenfrei A, Belton A, Kowalski J, Talbot CC, Jr, Di Cello F, Poh W, et al. HMGA1 drives stem cell, inflammatory pathway, and cell cycle progression genes during lymphoid tumorigenesis. BMC Genomics. 2011;12:549–585. doi: 10.1186/1471-2164-12-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Xi Y, Li YS, Tang HB. High mobility group A1 protein acts as a new target of Notch1 signaling and regulates cell proliferation in T leukemia cells. Mol Cell Biochem. 2013;374(1–2):173–180. doi: 10.1007/s11010-012-1517-2. [DOI] [PubMed] [Google Scholar]
- 134.Pegoraro S, Ros G, Ciani Y, Sgarra R, Piazza S, Manfioletti G. A novel HMGA1-CCNE2-YAP axis regulates breast cancer aggressiveness. Oncotarget. 2015;6(22):19087–19101. doi: 10.18632/oncotarget.4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Fu F, Wang T, Wu Z, Feng Y, Wang W, Zhou S, et al. HMGA1 exacerbates tumor growth through regulating the cell cycle and accelerates migration/invasion via targeting miR-221/222 in cervical cancer. Cell Death Dis. 2018;9(6):594–611. doi: 10.1038/s41419-018-0683-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Balint K, Xiao M, Pinnix CC, Soma A, Veres I, Juhasz I, et al. Activation of Notch1 signaling is required for beta-catenin-mediated human primary melanoma progression. J Clin Invest. 2005;115:3166–3176. doi: 10.1172/JCI25001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Grabher C, von Boehmer H, Look AT. Notch 1 activation in the molecular pathogenesis of T-cell acute lymphoblastic leukaemia. Nat Rev Cancer. 2006;6:347–359. doi: 10.1038/nrc1880. [DOI] [PubMed] [Google Scholar]
- 138.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/S0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 139.Welch C, Chen Y, Stallings RL. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene. 2007;26(34):5017–5022. doi: 10.1038/sj.onc.1210293. [DOI] [PubMed] [Google Scholar]
- 140.Furth N, Aylon Y, Oren M. p53 shades of Hippo. Cell Death Differ. 2018;25(1):81–92. doi: 10.1038/cdd.2017.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Visone R, Russo L, Pallante P, De Martino I, Ferraro A, Leone V, et al. MicroRNAs (miR)-221 and miR-222, both overexpressed in human thyroid papillary carcinomas, regulate p27Kip1 protein levels and cell cycle. Endocr Relat Cancer. 2007;14(3):791–798. doi: 10.1677/ERC-07-0129. [DOI] [PubMed] [Google Scholar]
- 142.Brooks CL, Gu W. The impact of acetylation and deacetylation on the p53 pathway. Protein Cell. 2011;2(6):456–462. doi: 10.1007/s13238-011-1063-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Brochier C, Dennis G, Rivieccio MA, McLaughlin K, Coppola G, Ratan RR, et al. Specific acetylation of p53 by HDAC inhibition prevents DNA damage-induced apoptosis in neurons. J Neurosci. 2013;33(20):8621–8632. doi: 10.1523/JNEUROSCI.5214-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ueda Y, Watanabe S, Tei S, Saitoh N, Kuratsu J, Nakao M. High mobility group protein HMGA1 inhibits retinoblastoma protein-mediated cellular G0 arrest. Cancer Sci. 2007;98(12):1893–1901. doi: 10.1111/j.1349-7006.2007.00608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Pierantoni GM, Battista S, Pentimalli F, Fedele M, Visone R, Federico A, et al. A truncated HMGA1 gene induces proliferation of the 3T3-L1 pre-adipocytic cells: a model of human lipomas. Carcinogenesis. 2003;24(12):1861–1869. doi: 10.1093/carcin/bgg149. [DOI] [PubMed] [Google Scholar]
- 146.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435(7043):839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 147.Fernandes ER, Zhang JY, Rooney RJ. Adenovirus E1A-regulated transcription factor p120E4F inhibits cell growth and induces the stabilization of the cdk inhibitor p21WAF1. Mol Cell Biol. 1998;18(1):459–467. doi: 10.1128/MCB.18.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sandy P, Gostissa M, Fogal V, Cecco LD, Szalay K, Rooney RJ, et al. p53 is involved in the p120E4F-mediated growth arrest. Oncogene. 2000;19(2):188–199. doi: 10.1038/sj.onc.1203250. [DOI] [PubMed] [Google Scholar]
- 149.Rizos H, Diefenbach E, Badhwar P, Woodruff S, Becker TM, Rooney RJ, et al. Association of p14ARF with the p120E4F transcriptional repressor enhances cell cycle inhibition. J Biol Chem. 2003;278(7):4981–4989. doi: 10.1074/jbc.M210978200. [DOI] [PubMed] [Google Scholar]
- 150.Leone V, Langella C, D’Angelo D, Mussnich P, Wierinckx A, Terracciano L, et al. Mir-23b and miR-130b expression is downregulated in pituitary adenomas. Mol Cell Endocrinol. 2014;390(1–2):1–7. doi: 10.1016/j.mce.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 151.Zhang ZC, Wang GP, Yin LM, Li M, Wu LL. Increasing miR-150 and lowering HMGA2 inhibit proliferation and cycle progression of colon cancer in SW480 cells. Eur Rev Med Pharmacol Sci. 2018;22(20):6793–6800. doi: 10.26355/eurrev_201810_16147. [DOI] [PubMed] [Google Scholar]
- 152.Li S, Peng F, Ning Y, Jiang P, Peng J, Ding X, et al. SNHG16 as the miRNA let-7b-5p sponge facilitates the G2/M and epithelial-mesenchymal transition by regulating CDC25B and HMGA2 expression in hepatocellular carcinoma. J Cell Biochem. 2020;121(3):2543–2558. doi: 10.1002/jcb.29477. [DOI] [PubMed] [Google Scholar]
- 153.Peng H, Li H. The encouraging role of long noncoding RNA small nuclear RNA host gene 16 in epithelial-mesenchymal transition of bladder cancer via directly acting on miR-17-5p/metalloproteinases 3 axis. Mol Carcinog. 2019;58(8):1465–1480. doi: 10.1002/mc.23028. [DOI] [PubMed] [Google Scholar]
- 154.Zhong JH, Xiang X, Wang YY, Liu X, Qi LN, Luo CP, et al. The lncRNA SNHG16 affects prognosis in hepatocellular carcinoma by regulating p62 expression. J Cell Physiol. 2020;235(2):1090–1102. doi: 10.1002/jcp.29023. [DOI] [PubMed] [Google Scholar]
- 155.Yang M, Wei W. SNHG16: a novel long-non coding RNA in human cancers. Onco Targets Ther. 2019;12:11679–11690. doi: 10.2147/OTT.S231630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Mu X, Chen M, Xiao B, Yang B, Singh S, Zhang B. EZH2 confers sensitivity of breast cancer cells to taxol by attenuating p21 expression epigenetically. DNA Cell Biol. 2019;38(7):651–659. doi: 10.1089/dna.2019.4699. [DOI] [PubMed] [Google Scholar]
- 157.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102(39):13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Liufu Z, Zhao Y, Guo L, Miao G, Xiao J, Lyu Y, et al. Redundant and incoherent regulations of multiple phenotypes suggest microRNAs’ role in stability control. Genome Res. 2017;27(10):1665–1673. doi: 10.1101/gr.222505.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Fedele M, Paciello O, De Biase D, Monaco M, Chiappetta G, Vitiello M, et al. HMGA2 cooperates with either p27kip1 deficiency or Cdk4R24C mutation in pituitary tumorigenesis. Cell Cycle. 2018;17(5):580–588. doi: 10.1080/15384101.2017.1403682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Taylor WR, Stark GR. Regulation of the G2/M transition by p53. Oncogene. 2001;20(15):1803–1815. doi: 10.1038/sj.onc.1204252. [DOI] [PubMed] [Google Scholar]
- 161.Hermeking H, Lengauer C, Polyak K, He TC, Zhang L, Thiagalingam S, et al. 14-3-3 sigma is a p53-regulated inhibitor of G2/M progression. Mol Cell. 1997;1:3–11. doi: 10.1016/S1097-2765(00)80002-7. [DOI] [PubMed] [Google Scholar]
- 162.Zhan Q, Chen IT, Antinore MJ, Fornace AJ., Jr Tumor suppressor p53 can participate in transcriptional induction of the GADD45 promoter in the absence of direct DNA binding. Mol Cell Biol. 1998;18:2768–2778. doi: 10.1128/MCB.18.5.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Krause K, Wasner M, Reinhard W, Haugwitz U, Dohna CL, Mössner J, et al. The tumour suppressor protein p53 can repress transcription of cyclin B. Nucleic Acids Res. 2000;28(22):4410–4418. doi: 10.1093/nar/28.22.4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Fischer M, Quaas M, Steiner L, Engeland K. The p53-p21-DREAM-CDE/CHR pathway regulates G2/M cell cycle genes. Nucleic Acids Res. 2016;44(1):164–174. doi: 10.1093/nar/gkv927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Innocente SA, Abrahamson JL, Cogswell JP, Lee JM. p53 regulates a G2 checkpoint through cyclin B1. Proc Natl Acad Sci USA. 1999;96(5):2147–2152. doi: 10.1073/pnas.96.5.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Taylor WR, DePrimo SE, Agarwal A, Agarwal ML, Schönthal AH, Katula KS, et al. Mechanisms of G2 arrest in response to overexpression of p53. Mol Biol Cell. 1999;10(11):3607–3622. doi: 10.1091/mbc.10.11.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Taylor WR, Schonthal AH, Galante J, Stark GR. p130/E2F4 binds to and represses the cdc2 promoter in response to p53. J Biol Chem. 2001;276(3):1998–2006. doi: 10.1074/jbc.M005101200. [DOI] [PubMed] [Google Scholar]
- 168.Müller GA, Engeland K. The central role of CDE/CHR promoter elements in the regulation of cell cycle-dependent gene transcription. FEBS J. 2010;277(4):877–893. doi: 10.1111/j.1742-4658.2009.07508.x. [DOI] [PubMed] [Google Scholar]
- 169.Pierantoni GM, Conte A, Rinaldo C, Tornincasa M, Gerlini R, Federico A, et al. Deregulation of HMGA1 expression induces chromosome instability through regulation of spindle assembly checkpoint genes. Oncotarget. 2015;6(19):17342–17353. doi: 10.18632/oncotarget.3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Di Agostino S, Fedele M, Chieffi P, Fusco A, Rossi P, Geremia R, et al. Phosphorylation of high-mobility group protein A2 by Nek2 kinase during the first meiotic division in mouse spermatocytes. Mol Biol Cell. 2004;15(3):1224–1232. doi: 10.1091/mbc.e03-09-0638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Ver Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 172.Rao CV, Yamada HY, Yao Y, Dai W. Enhanced genomic instabilities caused by deregulated microtubule dynamics and chromosome segregation: a perspective from genetic studies in mice. Carcinogenesis. 2009;30:1469–1474. doi: 10.1093/carcin/bgp081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Nam HJ, Naylor RM, van Deursen JM. Centrosome dynamics as a source of chromosomal instability. Trends Cell Biol. 2015;25:65–73. doi: 10.1016/j.tcb.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Funk LC, Zasadil LM, Weaver BA. Living in CIN: mitotic infidelity and its consequences for tumor promotion and suppression. Dev Cell. 2016;39:638–652. doi: 10.1016/j.devcel.2016.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Thompson SL, Compton DA. Proliferation of aneuploid human cells is limited by a p53- dependent mechanism. J Cell Biol. 2010;188:369–381. doi: 10.1083/jcb.200905057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Wolter P, Hanselmann S, Pattschull G, Schruf E, Gaubatz S. Central spindle proteins and mitotic kinesins are direct transcriptional targets of MuvB, B-MYB and FOXM1 in breast cancer cell lines and are potential targets for therapy. Oncotarget. 2017;8:11160–11172. doi: 10.18632/oncotarget.14466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Wolter P, Schmitt K, Fackler M, Kremling H, Probst L, Hauser S, et al. GAS2L3, a target gene of the DREAM complex, is required for proper cytokinesis and genomic stability. J Cell Sci. 2012;125:2393–2406. doi: 10.1242/jcs.097253. [DOI] [PubMed] [Google Scholar]
- 178.Li C, Lin M, Liu J. Identification of PRC1 as the p53 target gene uncovers a novel function of p53 in the regulation of cytokinesis. Oncogene. 2004;23:9336–9347. doi: 10.1038/sj.onc.1208114. [DOI] [PubMed] [Google Scholar]
- 179.Muller S, Almouzni G. Chromatin dynamics during the cell cycle at centromeres. Nat Ver Genet. 2017;18:192–208. doi: 10.1038/nrg.2016.157. [DOI] [PubMed] [Google Scholar]
- 180.Filipescu D, Naughtin M, Podsypanina K, Lejour V, Wilson L, Gurard-Levin ZA, et al. Essential role for centromeric factors following p53 loss and oncogenic transformation. Genes Dev. 2017;31:463–480. doi: 10.1101/gad.290924.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Schwartz GK, Shah MA. Targeting the cell cycle: a new approach to cancer therapy. J Clin Oncol. 2005;23(36):9408–9421. doi: 10.1200/JCO.2005.01.5594. [DOI] [PubMed] [Google Scholar]
- 182.Law ME, Corsino PE, Narayan S, Law BK. Cyclin-dependent kinase inhibitors as anticancer therapeutics. Mol Pharmacol. 2015;88(5):846–852. doi: 10.1124/mol.115.099325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Luserna Ghelli, di Rora’ A, Iacobucci I, Martinelli G. The cell cycle checkpoint inhibitors in the treatment of leukemias. J Hematol Oncol. 2017;10(1):77–91. doi: 10.1186/s13045-017-0443-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Huso TH, Resar LM. The high mobility group A1 molecular switch: turning on cancer—can we turn it off? Expert Opin Ther Targets. 2014;18(5):541–553. doi: 10.1517/14728222.2014.900045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Baluna R, Vitetta ES (1997) Vascular leak syndrome: a side effect of immunotherapy. Immunopharmacology 37(2–3):117–132 [DOI] [PubMed]
- 186.Beckerbauer L, Tepe JJ, Eastman RA, Mixter PF, Williams RM, Reeves R. Differential effects of FR900482 and FK317 onapoptosis, IL-2 gene expression, and induction of vascular leak syndrome. Chem Biol. 2002;9(4):427–441. doi: 10.1016/S1074-5521(02)00122-9. [DOI] [PubMed] [Google Scholar]
- 187.Parisi S, Piscitelli S, Passaro F, Russo T (2020) HMGA proteins in stemness and differentiation of embryonic and adult stem cells. Int J Mol Sci 21(1):E362 [DOI] [PMC free article] [PubMed]
- 188.Martins CP, Brown-Swigart L, Evan GI. Modeling the therapeutic efficacy of p53 restoration in tumors. Cell. 2006;127:1323–1334. doi: 10.1016/j.cell.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 189.Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656–660. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, et al. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 191.Christophorou MA, Martin-Zanca D, Soucek L, Lawlor ER, Brown-Swigart L, Verschuren EW, et al. Temporal dissection of p53 function in vitro and in vivo. Nat Genet. 2005;37:718–726. doi: 10.1038/ng1572. [DOI] [PubMed] [Google Scholar]
- 192.Jackson JG, Lozano G. The mutant p53 mouse as a pre-clinical model. Oncogene. 2013;32:4325–4330. doi: 10.1038/onc.2012.610. [DOI] [PubMed] [Google Scholar]
- 193.Xue C, Haber M, Flemming C, Marshall GM, Lock RB, MacKenzie KL, et al. p53 determines multidrug sensitivity of childhood neuroblastoma. Cancer Res. 2007;67:10351–10360. doi: 10.1158/0008-5472.CAN-06-4345. [DOI] [PubMed] [Google Scholar]
- 194.Kenzelmann Broz D, Attardi LD. In vivo analysis of p53 tumor suppressor function using genetically engineered mouse models. Carcinogenesis. 2010;31:1311–1318. doi: 10.1093/carcin/bgp331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Wang Y, Suh YA, Fuller MY, Jackson JG, Xiong S, Terzian T, et al. Restoring expression of wild-type p53 suppresses tumor growth but does not cause tumor regression in mice with a p53 missense mutation. J Clin Invest. 2011;121:893–904. doi: 10.1172/JCI44504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Bykov VJN, Eriksson SE, Bianchi J, Wiman KG. Targeting mutant p53 for efficient cancer therapy. Nat Rev Cancer. 2018;18(2):89–102. doi: 10.1038/nrc.2017.109. [DOI] [PubMed] [Google Scholar]
- 197.Duffy MJ, Synnott NC, Crown J. Mutant p53 as a target for cancer treatment. Eur J Cancer. 2017;83:258–265. doi: 10.1016/j.ejca.2017.06.023. [DOI] [PubMed] [Google Scholar]
- 198.Duffy MJ, Synnott NC, McGowan PM, Crown J, O’Connor D, Gallagher WM. p53 as a target for the treatment of cancer. Cancer Treat Rev. 2014;40(10):1153–1160. doi: 10.1016/j.ctrv.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 199.Levine AJ. Targeting therapies for the p53 protein in cancer treatments. Annu Rev Cancer Biol. 2019;3:21–34. doi: 10.1146/annurev-cancerbio-030518-055455. [DOI] [Google Scholar]
- 200.Joerger AC, Fersht AR. The p53 pathway: origins, inactivation in cancer, and emerging therapeutic approaches. Annu Rev Biochem. 2016;85:375–404. doi: 10.1146/annurev-biochem-060815-014710. [DOI] [PubMed] [Google Scholar]
- 201.Zhou X, Hao Q, Lu H. Mutant p53 in cancer therapy-the barrier or the path. J Mol Cell Biol. 2019;11(4):293–305. doi: 10.1093/jmcb/mjy072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Mantovani F, Walerych D, Sal GD. Targeting mutant p53 in cancer: a long road to precision therapy. FEBS J. 2017;284(6):837–850. doi: 10.1111/febs.13948. [DOI] [PubMed] [Google Scholar]
- 203.Lambert JM, Gorzov P, Veprintsev DB, Söderqvist M, Segerbäck D. PRIMA-1 reactivates mutant p53 by covalent binding to the core domain. Cancer Cell. 2009;15(5):376–388. doi: 10.1016/j.ccr.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 204.Zhang Q, Bykov VJN, Wiman KG, Zawacka-Pankau J. APR-246 reactivates mutant p53 by targeting cysteines 124 and 277. Cell Death Dis. 2018;9:439–451. doi: 10.1038/s41419-018-0463-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Bou-Hanna C, Jarry A, Lode L, Schmitz I, Schulze-Osthoff K, Kury S, et al. Acute cytotoxicity of MIRA-1/NSC19630, a mutant p53-reactivating small molecule, against human normal and cancer cells via a caspase-9-dependent apoptosis. Cancer Lett. 2015;359:211–217. doi: 10.1016/j.canlet.2015.01.014. [DOI] [PubMed] [Google Scholar]
- 206.Wang T, Lee K, Rehman A, Daoud SS. PRIMA-1 induces apoptosis by inhibiting JNK signaling but promoting the activation of Bax. Biochem Biophys Res Commun. 2007;352:203–212. doi: 10.1016/j.bbrc.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 207.Bykov VJ, Wiman KG. Mutant p53 reactivation by small molecules makes its way to the clinic. FEBS Lett. 2014;588(16):2622–2627. doi: 10.1016/j.febslet.2014.04.017. [DOI] [PubMed] [Google Scholar]
- 208.Lehmann S, Bykov VJ, Ali D, Andren O, Cherif H, Tidefelt U, et al. Targeting p53 in vivo: a first-in-human study with p53-targeting compound APR-246 in refractory hematologic malignancies and prostate cancer. J Clin Oncol. 2012;30:3633–3639. doi: 10.1200/JCO.2011.40.7783. [DOI] [PubMed] [Google Scholar]
- 209.Prokocimer M, Molchadsky A, Rotter V. Dysfunctional diversity of p53 proteins in adult acute myeloid leukemia: projections on diagnostic workup and therapy. Blood. 2017;130(6):699–712. doi: 10.1182/blood-2017-02-763086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Maiuri MC, Galluzzi L, Morselli E, Kepp O, Malik SA, Kroemer G. Autophagy regulation by p53. Curr Opin Cell Biol. 2010;2:181–185. doi: 10.1016/j.ceb.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 211.Wang X, Simon R. Identification of potential synthetic lethal genes to p53 using a computational biology approach. BMC Med Genom. 2013;6:30–40. doi: 10.1186/1755-8794-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Ma CX, Cai S, Li S, Ryan CE, Guo Z, Schaiff WT, et al. Targeting Chk1 in p53-deficient triple-negative breast cancer is therapeutically beneficial in human-in-mouse tumor models. J Clin Investig. 2012;122(4):1541–1552. doi: 10.1172/JCI58765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Baldwin A, Grueneberg DA, Hellner K, Sawyer J, Grace M, Li W et al (2010) Kinase requirements in human cells: V. Synthetic lethal interactions between p53 and the protein kinases SGK2 and PAK3. PNAS 107(28):12463–12468 [DOI] [PMC free article] [PubMed]
- 214.Shalem O, Sanjana NE, Zhang F. High-throughput functional genomics using CRISPR-Cas9. Nat Rev Genet. 2015;16(5):299–311. doi: 10.1038/nrg3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Leijen S, van Geel RM, Pavlick AC, Tibes R, Rosen L, Razak AR, et al. Phase I study evaluating WEE1 inhibitor AZD1775 as monotherapy and in combination with gemcitabine, cisplatin, or carboplatin in patients with advanced solid tumors. J Clin Oncol. 2016;34(36):4371–4380. doi: 10.1200/JCO.2016.67.5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Leijen S, van Geel RM, Sonke GS, de Jong D, Rosenberg EH, Marchetti S, et al. Phase II study of WEE1 inhibitor AZD1775 plus carboplatin in patients with TP53-mutated ovarian cancer refractory or resistant to first-line therapy within 3 months. J Clin Oncol. 2016;34(36):4354–4361. doi: 10.1200/JCO.2016.67.5942. [DOI] [PubMed] [Google Scholar]
- 217.Peng Z, Yu Q, Bao L. The application of gene therapy in China. IDrugs. 2008;11(5):346–350. [PubMed] [Google Scholar]