Abstract
The epithelial-to-mesenchymal transition (EMT) is a reversible process that may interact with tumour immunity through multiple approaches. There is increasing evidence demonstrating the interconnections among EMT-related processes, the tumour microenvironment, and immune activity, as well as its potential influence on the immunotherapy response. Long non-coding RNAs (lncRNAs) are emerging as critical modulators of gene expression. They play fundamental roles in tumour immunity and act as promising biomarkers of immunotherapy response. However, the potential roles of lncRNA in the crosstalk of EMT and tumour immunity are still unclear in sarcoma. We obtained multi-omics profiling of 1440 pan-sarcoma patients from 19 datasets. Through an unsupervised consensus clustering approach, we categorised EMT molecular subtypes. We subsequently identified 26 EMT molecular subtype and tumour immune-related lncRNAs (EILncRNA) across pan-sarcoma types and developed an EILncRNA signature-based weighted scoring model (EILncSig). The EILncSig exhibited favourable performance in predicting the prognosis of sarcoma, and a high-EILncSig was associated with exclusive tumour microenvironment (TME) characteristics with desert-like infiltration of immune cells. Multiple altered pathways, somatically-mutated genes and recurrent CNV regions associated with EILncSig were identified. Notably, the EILncSig was associated with the efficacy of immune checkpoint inhibition (ICI) therapy. Using a computational drug-genomic approach, we identified compounds, such as Irinotecan that may have the potential to convert the EILncSig phenotype. By integrative analysis on multi-omics profiling, our findings provide a comprehensive resource for understanding the functional role of lncRNA-mediated immune regulation in sarcomas, which may advance the understanding of tumour immune response and the development of lncRNA-based immunotherapeutic strategies for sarcoma.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-022-04462-4.
Keywords: Sarcoma, Epithelial-to-mesenchymal transition, LncRNA, Tumour immunity, Prognostic risk model, Machine learning
Introduction
Sarcomas are a heterogeneous group of primary mesenchymal tumours, derived from bone, cartilage, muscle and other connective tissues. More than 100 different sarcoma subtypes varying in pathology, clinical presentation, molecular characteristics, and response to therapy have been identified, 80% of which are soft tissue sarcomas (STS), while 15% are bone sarcomas and 5% are gastrointestinal stromal tumours [1]. Although relatively rare, sarcomas are often fatal and are responsible for significant mortality as the most aggressive childhood cancers [2]. The clinical management of sarcomas is highly challenging due to misdiagnosis and late diagnosis, as well as their heterogeneity, aggressive nature and resistance to conventional treatments such as surgery, radiation and chemotherapy [3]. Consequently, novel therapeutic strategies are urgently needed for sarcomas. Recently, immunotherapy has been successfully applied in several cancers [4]. As a promising treatment strategy, several clinical trials on immunotherapy (such as immune checkpoint inhibitor (ICI) therapy) for sarcoma patients have shown profound beneficial effects on patient survival [5, 6]. However, some refractory patients still have disproportionate responses to immunotherapy [7]. Thus, it is imperative to explore biomarkers that can function as molecular targets or modulators in the aspect of tumour immunology for sarcomas.
Epithelial-to-mesenchymal transition (EMT) is a complex process in which epithelial cells lose their apical–basal polarity and acquire mesenchymal characteristics including a fibroblast-like morphology and increased migratory capacity [8]. The reverse process, described as mesenchymal-to-epithelial transition (MET), has also been reported [9]. EMT constitutes a critical characteristic in the tumour microenvironment (TME), which has been identified as playing crucial roles in cancer metastasis and immune escape in several carcinomas [10]. In contrast to carcinomas, a variable degree of epithelial/mesenchymal differentiation has been observed in various sarcoma histological subtypes, which can be either more epithelial-like (such as Ewing sarcoma, synovial sarcomas) or more mesenchymal-like (such as osteosarcoma, chondrosarcoma), while the existence of sarcoma subtypes presenting both extreme phenotypes within one tumour has also been reported [11]. Accumulating evidence indicates that many sarcomas undergo EMT- and MET-related processes to take advantage of both biological features leading to high aggressiveness and unfavourable clinical outcomes [11, 12]. However, few studies have reported any association among EMT, TME and tumour immunity in sarcomas or any potential regulators.
The long non-coding RNAs (lncRNAs), which are more than 200 nucleotides in length, play a pivotal role in various biological processes including epigenetic and transcriptional regulation, interaction with protein complexes and cell communication. They are highly-conserved molecules with potential abilities to regulate cell proliferation, development and differentiation, as well as pathogenesis [13]. Although a large number of lncRNAs have been identified as tumour suppressor genes and oncogenes, most of their functions and mechanisms are still unclear. Some of the well-studied lncRNAs such as XIST, ZEB2-AS1 and NORAD have been demonstrated to play a putative role in the EMT regulation of various carcinomas [14]. During the past decade, a number of lncRNAs have emerged as critical elements in the regulation of diverse biological processes including the EMT, where they promote or attenuate the oncogenesis of sarcomas [15]. Owing to the highly-conserved characteristics of lncRNAs which are often expressed in a tumour-specific manner, they are thought to be promising therapeutic targets and biomarkers for cancer diagnosis or prognosis assessment. Most recently, researchers have been attempting to systematically identify the functions of lncRNAs in the processes of EMT and MET through the use of efficient gene editing tools [14]. Increasing evidence reveals that lncRNAs function as communicators and mediators, being directly or indirectly involved in the crosstalk between tumour cells and infiltrating immune cells within the tumour immune microenvironment (TIME), where they participate in cancer onset and progression [16, 17]. For example, Huang et al. reported that the lncRNA NKILA promotes tumour immune evasion by sensitising T cells to activation-induced cell death [18]. Hu et al. identified the oncogenic lncRNA LINK-A that regulates cancer cell antigen presentation and intrinsic tumour suppression [19].
In this study, we integrated large-size pan-sarcoma datasets with multi-omics profiling. Through a machine learning approach, we identified pan-sarcoma EMT molecular subtypes and identified lncRNAs in the crosstalk of EMT and immune microenvironment across sarcomas. Based on the results, we constructed an lncRNA-based computational model and demonstrated it as a predictive biomarker for assessing the prognosis of patients with sarcomas, as well as a comprehensive resource for understanding the functional role of lncRNA-mediated immune regulation and developing potential clinical implications of lncRNA-based immunotherapeutic strategies in precision medicine for sarcomas.
Methods
Pan-sarcoma data collection
Overall, we collected 19 public sarcoma datasets from the National Cancer Institute Genomic Data Commons—The Cancer Genome Atlas (NCI GDC TCGA)—Therapeutically Applicable Research to Generate Effective Treatments (NCI GDC TARGET), the Gene Expression Omnibus (GEO) and the European Molecular Biology Laboratory's European Bioinformatics Institute (EMBL-EBI) databases. Accessions for the datasets used in the present study are as follows: phs000178 (TCGA-SARC Sarcoma), phs000468 (TARGET-OS Osteosarcoma), GSE13433, GSE142162, GSE14827, GSE17618, GSE20196, GSE20559, GSE23980, GSE34620, GSE34800, GSE37371, GSE66533, GSE71118, GSE87437, E-MEXP-1922, E-MEXP-3628, E-MEXP-964, E-TABM-1202.
For the TCGA-SARC dataset, RNA-Sequencing (RNA-Seq) data of raw count format and FPKM (Fragments Per Kilobase of transcript per Million mapped reads) format, masked somatic mutation data (mutect2), masked copy number segment data and survival follow-up data with clinicopathological characteristics were obtained from the TCGA data portal using the TCGAbiolinks [20] R package (version 2.20.1). TCGA-SARC molecular subtype data and other characteristics of patients were obtained from Lazar et al.’s study [21]. TCGA-SARC immune subtype data were curated from Thorsson et al.’s study [22]. For the TARGET-OS dataset, RNA-Seq data of raw count format and TPM (Transcripts Per Million) format and latest clinical information were obtained from TARGET data matrix. The Homo sapiens GRCh38.104 annotation file was downloaded from Ensembl [23] for gene symbol and biotype annotations corresponding to Ensembl identification. DESeq2 [24] R package (version 1.32.0) was applied to filter out low-abundance genes, normalize RNA-Seq counts data and perform variance stabilizing transformation. RNA-Seq data of FPKM format was transformed to TPM format using a previously described method [25].
For microarray datasets, raw or processed data and the available clinical information were downloaded from GEO [26] and EMBL-EBI [27]. When possible, available Affymetrix CEL files within each dataset were re-processed and re-normalized individually into expression matrix through the robust multi-array average expression measure using the affyPLM [28] R package (version 1.68.0). The arrayQualityMetrics [29] R package (version 3.48.0) was applied to exclude low-quality and outlier samples of microarray datasets. All microarray data used in this study was based on Affymetrix Human Genome U133 Plus 2.0 Array. We utilized the Combat method of sva [30] R Package (version 3.40.0) to correct the batch effect caused by technical variation and differences across the 17 microarray datasets, and combine them into a pan-sarcoma microarray dataset of 1085 samples. The hgu133plus2.db [31] R package (version 3.13.0) was applied to map probes into gene symbols, in which the probe with the highest mean values was selected when multiple probes were mapped to one gene. In total, 1440 sarcoma patients were included in this study. Detailed information for all datasets and patients were documented in Supplementary file 1.
Immunotherapy data collection
RNA-Seq data and clinical information from patients with tumours treated with anti-programmed death-1 (PD-1) or anti-PD-ligand-1 (PD-L1) immune checkpoint inhibitor (ICI) therapy were obtained from Kim et al.’s study (GSE176307) [32], including overall survival, progression-free survival and treatment response of 89 urothelial cancer patients.
Clustering molecular pattern of EMT signature expression
We collected curated EMT-related gene lists reported by 5 pan-cancer studies via EMTome [33–38], and combined them into an EMT signature (Supplementary file 1). To cluster EMT molecular pattern of sarcoma patients, we utilized the ConsensusClusterPlus [39] R package (version 1.56.0) to perform an unsupervised consensus clustering on expression of EMT signature in 1085 pan-sarcoma samples based on K-means algorithm. The resampling was set to be 1000 repetitions to ensure the clustering stability. Distance matrix of consensus clustering was extracted, and a silhouette analysis was applied to assess how similar an individual was matched to its assigned cluster as compared to other clusters using the CancerSubtypes [40] R package (version 1.18.0).
Computation of the EMT score
EMT gene signatures with annotation of epithelial and mesenchymal markers from Tuan et al.’s and Hollern et al.’s studies were separately used to compute the EMT score [34, 38]. The EMT score for each sample was calculated as , in which M and E respectively represent the normalized expression of the mesenchymal maker genes and epithelial maker genes, and respectively represent the number of corresponding genes, as described in a previous study[41].
Functional enrichment analysis
The clusterProfiler [42] R package (version 4.0.5) was used for over representation analysis and pre-ranked gene set enrichment analysis (GSEA). The non-parametric gene set variation analysis (GSVA) was conducted using the GSVA [43] R package (version 1.40.1). A | normalized enrichment score (NES) |≥ 1.0 and adjust P value < 0.05 was considered with significance for the pre-ranked GSEA. The GSVA enrichment scores were applied to the limma [44] R package (version 3.48.3) to fit a linear model, and the alteration was considered with significance when the | log2FoldChange |≥ 0.2 and adjust P value < 0.05. Gene sets of Gene Ontology (GO) [45] Biological Process section (c5.go.bp.v7.3), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway (c2.cp.kegg.v7.3) [46], WikiPathways (c2.cp.wikipathways.v7.3) [47] and Reactome (c2.cp.reactome.v7.3) [48] pathway were downloaded from the Molecular Signatures Database (MSigDB) [49].
Evaluation of TME cell infiltration abundance
The CIBERSORTx [50] algorithm with the LM22 signature matrix (a signature matrix containing 22 functionally defined human immune subsets profiled by microarrays) was utilized to quantify the abundance of 22 types of TME infiltrating cells. We set parameters of CIBERSORTx as follows: 100 times for permutation test, batch correction—bulk mode, absolute mode of output scores, and RNA-Seq expression data without quantile normalization, while microarray expression data with quantile normalization. The overall fraction of stromal and immune cells infiltration in the sarcoma samples was calculated by using the xCell [51] via the immunedeconv [52] R package (version 2.0.4).
Weighted gene co-expression network analysis
Weighted gene co-expression network analysis (WGCNA) is commonly used for mining gene co-expression networks and hub genes based on pairwise correlations in genomic applications [53]. In the present study, to identify lncRNAs in gene modules that were most relevant to EMT molecular subtype, we applied the WGCNA [54] R package (version 1.70-3) to construct weighted gene co-expression modules and module–trait relationship from the pan-sarcoma samples. The threshold of scale-free topology fitting index (R2) was set as 0.90. The minimum module size was set as 30 and the threshold for merging modules was set as of 30%. Intramodular analysis was performed by calculating the correlation of module membership and gene significance for EMT molecular subtype.
Identification of immune-related lncRNAs in sarcoma
We downloaded curated human immune gene list with function and Gene Ontology term from the ImmPort project [55], and mapped gene symbols to Ensembl IDs. In total, we obtained 1752 immune genes in 17 immune functional pathways in subsequent analyses (Supplementary file 3). To identify potential immune-related lncRNA modifiers, we proposed a computational method that integrates a gene expression-based immunology framework as follows: (1) All lncRNAs were ranked based on their co-expression relationship with immune marker genes; (2) Infiltrations of immune cells were estimated through CIBERSORTx with absolute score mode, all lncRNAs were ranked based on the correlation between their expression and the abundance of a given infiltrating immune cell component; (3) GSVA enrichment score of the 17 immune functional pathways were computed for each sample, all lncRNAs were ranked based on the correlation between their expression and the GSVA enrichment score of a given immune functional pathway. Pearson's correlation coefficients (PCC) were calculated for each step, where a lncRNA with a PCC ≥ 0.3 and adjusted p value < 0.05 was considered as candidate immune-related lncRNAs.
Development of an EMT- and tumour immune-related lncRNA signature scoring model
We identified lncRNAs that concurrently correlates to EMT molecular subtype and tumour immune in sarcoma. An EMT- and tumour immune-related lncRNA signature scoring model (EILncSig) was constructed by a method similar to a previous study [56]: (1) The prognostic value of each candidate lncRNA was firstly evaluated by univariate Cox proportional hazards regression analysis; (2) A weighted combination was applied by using the regression coefficients in the multivariate Cox regression analysis. The EILncSig score for each patient was defined as , where Exp and Beta represent the normalized expression and regression coefficient of candidate lncRNA and K represent the number of lncRNAs in the EILncSig scoring model. We applied time-dependent ROC curves analysis and Kaplan–Meier survival analysis to evaluate the prognostic prediction value of EILncSig scoring model through the survivalROC [57] and survminer [58] R packages (version 1.0.3 and 0.4.9). The optimal cutpoint for dividing patients into high- and low- EILncSig levels was defined by the surv_cutpoint function of the survminer R package, where the parameter—minimal proportion of observations per group was set to 30% to avoid the occurrence of too few patients in a certain group. We additionally performed the ROC curves analysis to evaluate the prediction value of EILncSig scoring model on sarcoma subtypes through the pROC [59] R package (version 1.18.0). Univariate and multivariate Cox regression analyses were performed on EILncSig and available clinicopathological characteristics. When conducting model training and validating, we used completely independent dataset/cohort.
Clustering analysis of expression pattern based on pan-cancer TME signatures
The categorizing method for pan-cancer TME patterns and 29 sets of gene expression signatures describing pan-cancer TME characteristics were obtained from Bagaev et al.’s study [60] (Supplementary file 5). After performing GSVA on all the TME signatures for each patient, the GSVA enrichment scores were robustly standardized (median-centered and scaled by median absolute deviation) within each cohort. By using ConsensusClusterplus [39] R package (version 1.56.0), we applied an unsupervised clustering algorithm to analysis the standardized GSVA scores of TME signatures. K-means clustering algorithm was used and resampling was set to be 1000 repetitions. An analysis of t-distributed stochastic neighbour embedding (t-SNE) by using the Rtsne [61] R package (version 0.15) was further conducted and visualized on a 3D map with the scatterplot3d [62] R package (version 0.3-41).
Analysis of somatic mutation and recurrent regions of somatic copy number alteration
Analysis and visualization of somatic mutations of TCGA-SARC dataset was performed through the Maftools [63] R package (version 2.8.05). To determine significantly amplified or deleted regions of SCNA, we applied GISTIC 2.0 [64] to analyze DNA copy number segmentation profiles. The analytic process of GISTIC 2.0 was completed on the GenePattern platform [65]. Parameters of GISTIC 2.0 were set as follows: noise threshold—0.3, focal length cutoff—0.5, confidence level—90%, q value threshold—0.25, copy-ratio cap—1.5 and arm-level peel-off mode enabled. We applied GenomicRanges [66] R package (version 1.44.0) to determine genes that overlapped within any “wide peak” region identified by GISTIC 2.0 with a residual q value less than 0.05.
Analysis of differentially expressed genes
The DESeq2 R package was applied to process RNA-Seq counts data and then identify differentially expressed genes (DEG) between two groups. The differential expression threshold was defined with a fold-change of threshold at 1.5 and an adjusted p value < 0.05. The DEG results were presented in volcano plots and heatmaps by EnhancedVolcano [67] and pheatmap [68] R packages (version 1.10.0 and 1.0.12).
Discovery of potential drugs based on CMAP database
The Connectivity Map (CMAP) database [69] provides large-scale pharmacogenomic data including systematic drug-induced perturbation. We downloaded the curated CMAP perturbation dataset (version 2016) via the PharmacoGx [70] R package (version 2.4.0). Then we ranked and selected the top 500 DEGs to represent the transcriptomic alteration for EILncSig and utilized PharmacoGx R package to measure the concordance of transcriptomic difference and drug induced cellular molecular alterations. The GSEA method was implemented for connectivity scores calculation and permutation testing was set as 100 times to detect the significance.
Statistical analysis
Statistical tests in this study were conducted by using the R software (version 4.1.2, https://www.r-project.org). The ggplot2 R package (version 3.3.5) and extensions [71] were used for data analysis and visualization. The Wilcoxon signed-rank test and Kruskal–Wallis test were applied to compare continuous variables for two groups and three or more groups, respectively. Categorical data was tested by the chi-square test. The Kaplan–Meier method, log-rank test and Cox proportional hazards regression analysis were used in prognostic analysis. Correlation analysis of continuous variables was performed by using the Pearson correlation test, while the Spearman correlation test was performed instead considering the influence of outliers when necessary. A statistical test is considered with statistical significance at two-sided p < 0.05. When necessary, the Benjamini–Hochberg method was applied for p value adjustment.
Results
Derivation of de novo pan-sarcoma EMT molecular subtypes from the perspective of EMT signature
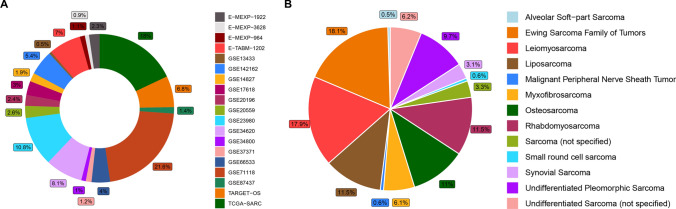
First, we collected EMT process-related genes that were curated by Tuan et al. [34], Rokavec et al. [35], Kandimalla et al. [36], Koplev et al. [37] and Hollern et al. [38] in their pan-cancer studies. In total, 630 genes were annotated and combined into a merged EMT signature. Detailed gene symbols and gene types (epithelial/mesenchymal marker) are delineated in Supplementary file 1. As shown in Fig. 1A, B, a total of 1440 sarcoma patients of various histological subtypes were included in the present study, for whom RNA-Seq expression data were contained in TCGA-SARC and TARGET-OS and microarray expression data based on the same platform were contained in the other datasets. To obtain a comprehensive understanding of the pan-sarcoma EMT molecular subtypes, we combined transcriptomic profiling and available clinical information of 17 datasets that were tested on the same platform (GSE13433, GSE142162, GSE14827, GSE17618, GSE20196, GSE20559, GSE23980, GSE34620, GSE34800, GSE37371, GSE66533, GSE71118, GSE87437, E-MEXP-1922, E-MEXP-3628, E-MEXP-964, E-TABM-1202) (Fig. S1A). A large pan-sarcoma expression dataset containing 1,085 samples with over 12 subtypes was involved in further clustering analysis.
Fig. 1.
Overview of pan-sarcoma data in the present study. A Sample size of enrolled datasets. B Sample size of involved sarcoma histology subtypes
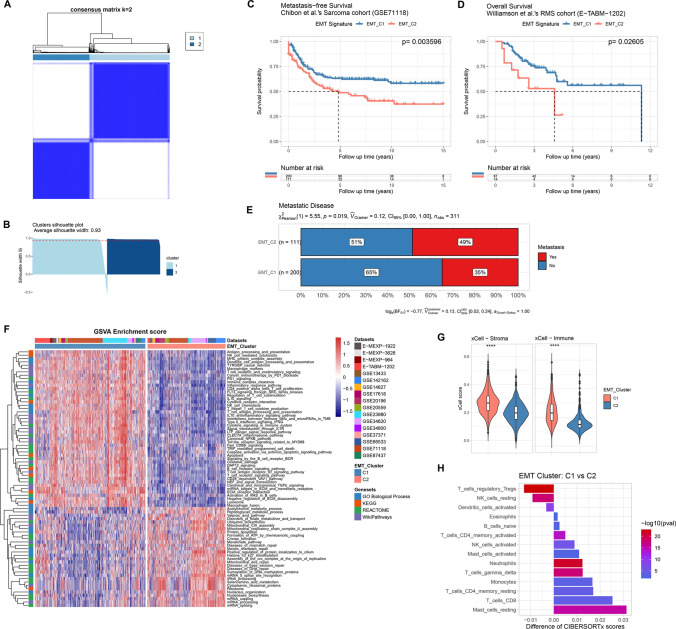
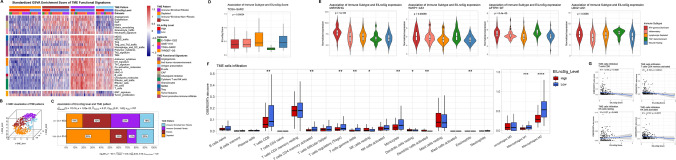
Through an unsupervised consensus clustering of the expression pattern of the merged EMT signature, we classified sarcoma patients into distinct EMT molecular subtypes, where 636 patients were assigned to EMT Cluster_1 (EMT_C1) and 449 patients were assigned to Cluster_2 (Supplementary file 2). Consensus matrix and silhouette analysis (average width: 0.93) showed satisfactory clustering results (Fig. 2A, B). To reveal the association of EMT molecular subtypes and prognosis of sarcoma patients, we performed Kaplan–Meier survival analysis on patients with matched expression profiling and clinical information. For the sarcoma cohort from Chibon et al. (GSE71118), we obtained a p value of 0.003596 from the log-rank test, indicating that patients of EMT_C2 had significantly worse metastasis-free survival (MFS) (Fig. 2C). A consistent result was also found as shown in Fig. 2D that patients of EMT_C2 had significantly worse overall survival (OS) in the rhabdomyosarcoma cohort of Williamson et al. (E-TABM-1202, log-rank p = 0.02605). Furthermore, more patients with metastatic disease were found in EMT_C2 (Fig. 2E, 49% vs 35%, p = 0.019). To analyse the biological processes and pathway variations of the distinct EMT molecular subtypes, we implemented gene set variation analysis (GSVA). As shown in Fig. 2F, oxidative damage, TGF-β signalling and several immune-related pathways including interleukin-10 (IL-10) signalling, type II-interferon (IFNG) signalling, T/B cell receptor signalling pathway and NK cell chemotaxis/cytotoxicity were significantly enriched in the EMT_C1 group while mRNA capping/processing/splicing, nucleolus organisation and several DNA damage repair-related pathways including mismatch repair and base excision repair were significantly enriched in the EMT_C2 group. Accumulating studies have reported a potential association between TME-infiltrating immune cells and dysregulated EMT/MET in the tumour. Thus, we applied the xCell tool, a novel gene signature-based ssGSEA method to estimate the overall TME infiltration status and found that both stromal and immune scores of EMT_C1 were significantly higher than those of EMT_C2 (Fig. 2G). Moreover, CIBERSORTx, a deconvolution algorithm, was applied to assess the infiltrating abundance of various immune cell types between EMT subgroups (Fig. 2H and Fig. S1B, C). Activated memory CD4+ T cells, activated NK cells, γδ-T cells and CD8+ T cells showed high infiltration in the EMT_C1 group, whereas regulatory T cells (Tregs), resting NK cells and activated dendritic cells were more abundant in the EMT_C2 group. In addition, we found that patients of EMT_C1 possessed higher EMT scores, which indicated a tendency to the mesenchymal phenotype (Fig. S1D). We further examined whether there existed any over-representation of sarcoma subtypes in the EMT molecular classification. As shown in Fig. S1E, samples of each sarcoma subtype were segregated to the EMT C1 and C2 clusters in different proportions. We observed that the EMT C1 cluster involved more myxofibrosarcoma and undifferentiated pleomorphic sarcoma patients, but fewer synovial sarcoma and Ewing sarcoma patients. However, liposarcoma, osteosarcoma, leiomyosarcoma and rhabdomyosarcoma were not found to be enriched in either the EMT C1 or C2 clusters (Fig. S1F).
Fig. 2.
Unsupervised consensus clustering of pan-sarcoma EMT molecular subtypes base on expression pattern of EMT signature. A The consensus matrix heatmap showing clustering result of EMT signature expression. B Assessment of the consensus clustering by silhouette analysis. C, D Kaplan–Meier survival analysis of MFS and OS for patients between EMT molecular subtypes in corresponding cohorts, respectively. E Distribution of sarcoma with metastatic disease between EMT molecular subtypes. F.A Heatmap showing GSVA enrichment scores of differentially variated biological processes and pathways. G Comparison of the overall TME infiltration status (stromal and immune scores) between EMT_C1 and EMT_C2 via the xCell tool. H Comparison of the infiltrating abundance of various immune cells between EMT_C1 and EMT_C2 through CIBERSORTx
WGCNA and identification of lncRNAs associated with EMT molecular subtypes
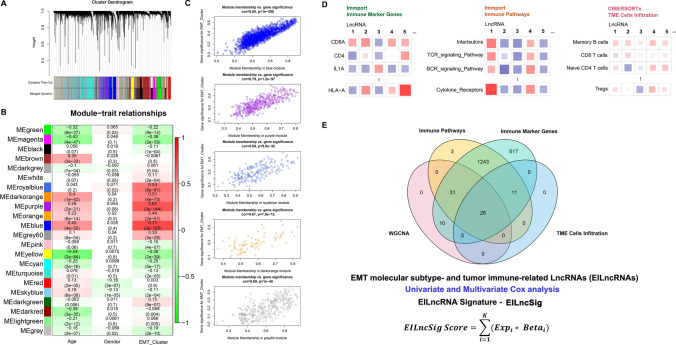
We used variance-stability-transformed expression data via DESeq2 as the input data for WGCNA. The best β value in the co-expression network was calculated to be 7 (Fig. S2A-C). A total of 21 gene modules were finally identified after dynamic tree cutting and module merging processes (Figs. 3A, S2E and Supplementary file 3). As shown in the module–trait relationship, many modules were found to be significantly correlated (p value < 0.05) with the EMT clusters (Fig. 3B). We screened modules with relatively high correlation coefficients (≥ 0.3). Furthermore, after the intramodular analysis, we finally identified five gene modules which showed a good correlation of module membership and gene significance for the EMT molecular subtype (Figs. 3C and S2F). According to gene biotype annotation of Ensemble GRCh38.104, 72 lncRNAs in the five gene modules were identified as EMT molecular subtype-associated lncRNAs (Supplementary file 3).
Fig. 3.
Identification of EMT molecular subtype and tumour immune-related lncRNA (EILncRNA) across pan-sarcoma types. A Cluster Dendrogram showing the merged dynamic gene modules in the WGCNA process. B A heatmap for WGCNA module–trait relationship. C Intramodular analysis on the correlation of module membership and gene significance for EMT molecular subtype (showing the five modules with high correlation). D A schematic diagram showing the parallelly computational process for identifying immune-related lncRNAs. E Intersection of EMT molecular subtype and tumour immune-related lncRNAs (EILncRNA) across pan-sarcoma types. An EILncRNA signature-based scoring model (EILncSig) was constructed by combination of the normalized expression of prognostic EILncRNAs weighted by their corresponding multivariate Cox regression coefficients
Identification of immune-related lncRNAs across pan-sarcoma types
To identify candidate lncRNA modifiers that are relevant to tumour immunity across pan-sarcoma types, we proposed a three-line parallel computational approach, which involves correlations of lncRNA expression to (1) immune marker gene expression, (2) immune-related pathway activity and (3) abundance of TME-infiltrating immune cells. Briefly, the Pearson correlation test on normalised lncRNA expression and corresponding terms was performed for each step as shown in the schematic diagram (Fig. 3D). LncRNAs in the correlation pairs with a Pearson correlation coefficient ≥ 0.3 and an adjusted p value < 0.05 were selected. A total of 37 lncRNAs were identified as robust candidates involved in tumour immunity across pan-sarcoma types (Supplementary file 3).
Construction and validation of a pan-sarcoma EILncRNA signature scoring model
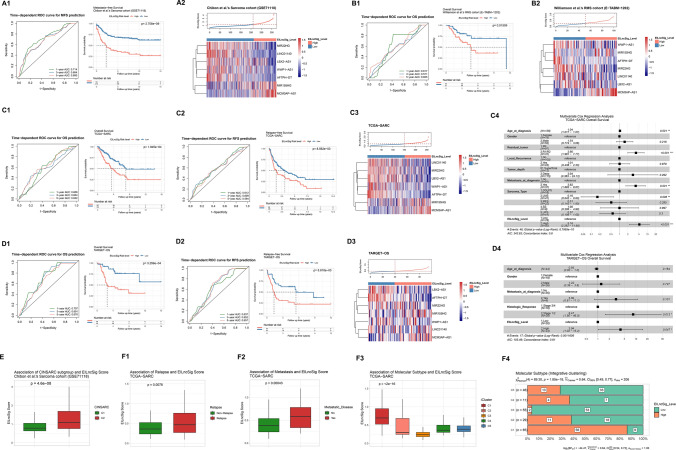
As shown in Fig. 3E, we finally identified 26 lncRNAs that showed a concurrent relationship to EMT molecular subtype and tumour immunity across pan-sarcoma types (EILncRNA). Considering the heterogeneity of sarcoma subtypes and the complexity of interactions between EMT and tumour immunity, we proposed to develop an EILncRNA signature-based scoring model (EILncSig) to quantitatively estimate the crosstalk characteristics of EMT, the tumour immune microenvironment (TIME) and tumour immunity for individual sarcoma patients. When conducting model training and validating, we used completely independent cohort without duplicated samples. We selected the sarcoma dataset of Chibon et al. (GSE71118) as the training cohort, which has the largest sample size (n = 311) with clinical information (MFS) in the present study. We performed univariate Cox proportional hazards regression analysis to clarify the prognostic significance of the 26 EILncRNAs. A total of seven EILncRNAs (MIR22HG, LINC01140, LBX2-AS1, WWP1-AS1, AFTPH-DT, MIR155HG and MCM3AP-AS1) were then selected to construct the EILncRNA signature-based scoring model. EILncSig score was computed as the sum of the normalised expression of the seven EILncRNAs weighted by corresponding multivariate Cox regression coefficients (Supplementary file 4).
As shown in the time-dependent receiver operating characteristic (ROC) curve analysis for MFS prediction, areas under the curve (AUC) were 0.714, 0.684 and 0.680 for 1, 3 and 5 years, respectively. By using the optimal cutoff value of EILncSig score, patients in the training cohort were stratified to high- and low-EILncSig groups. Kaplan–Meier survival analysis showed that patients of the high-EILncSig group had significantly worse MFS (log-rank p = 2.708e–9) (Fig. 4A1). The distribution of the EILncSig score and the seven-EILncRNA expression between high- and low-EILncSig groups is shown in Fig. 4A2.
Fig. 4.
The EILncSig is associated with prognosis and molecular subtypes in sarcoma (construction and validation). A, B (1) Time-dependent ROC curve analysis of EILncSig score for predicting the MFS/OS probability and Kaplan–Meier analysis on MFS/OS of high- and low-EILncSig groups stratified by optimal cutoff point. (2) Ranked distribution of EILncSig scores and a heatmap of LncRNA expression pattern (z score) (Training cohort: GSE71118 and Validation cohort: E-TABM-1202). C, D (1) Time-dependent ROC curve analysis of EILncSig score for predicting the OS probability and Kaplan–Meier analysis on OS of high- and low-EILncSig groups stratified by optimal cutoff point. (2) Time-dependent ROC curve analysis of EILncSig score for predicting the RFS probability and Kaplan–Meier analysis on RFS of high- and low-EILncSig groups stratified by optimal cutoff point. (3) Ranked distribution of EILncSig scores and a heatmap of LncRNA expression pattern (z score). (4) A forest plot of multivariate Cox regression analysis of EILncSig levels and clinicopathological characteristics on the overall survival. (c1–c4): TCGA-SARC and (d1-d4): TARGET-OS). E The association of EILncSig and CINSARC subtypes (GSE71118). F The association of EILncSig and (1) Relapse, (2) Metastasis and (3 & 4) Integrative molecular subtypes (TCGA-SARC)
To validate whether the EILncSig scoring model demonstrates robust effectiveness across pan-sarcoma patients, we included three independent datasets as testing cohorts (the rhabdomyosarcoma cohort from Williamson et al., E-TABM-1202, n = 101; TCGA-SARC sarcoma, n = 259; and TARGET-OS osteosarcoma, n = 95) for further validation. The risk score for each patient was calculated and all patients were stratified into high- and low-risk groups. For the rhabdomyosarcoma cohort of Williamson et al., the time-dependent ROC curve analysis indicated EILncSig as a prognostic predictor for OS. Kaplan–Meier survival analysis showed significantly worse OS of patients in the high-risk group (log-rank p = 0.01509, Fig. 4B). Consistent results from the time-dependent ROC curve and Kaplan–Meier survival analyses on both OS and relapse-free survival (RFS) were also successfully validated in the other two validation cohorts (TCGA-SARC and TARGET-OS) as shown in Fig. 4C, D (C1: TCGA-SARC OS, C2: TCGA-SARC RFS, D1: TARGET-OS OS and D2: TARGET-OS RFS). We also investigated whether there was any enrichment of sarcoma subtypes between the high- and low-EILncSig groups. We examined the sarcoma dataset of Chibon et al. (GSE71118) and TCGA-SARC dataset which involved pan-sarcoma samples. As shown in Fig. S3A, B, several sarcoma subtypes were observed to be enriched in either the high- or low-EILncSig group. Consistently, leiomyosarcoma was obviously enriched in the high-EILncSig group, while myxofibrosarcoma was more enriched in the low-EILncSig group.
To confirm whether the EILncSig scoring stratification could be an independent prognostic factor of other clinical features, patients from TCGA-SARC and TARGET-OS with available clinicopathologic parameters were analysed by univariate and multivariate Cox regression analyses (Supplementary file 4) to test the performance of the EILncSig after being adjusted for clinicopathologic parameters including age, gender, tumour depth, tumour metastasis, residual tumour after surgery, local recurrence, tumour grade (histological response) and sarcoma subtype. As shown by the multivariate Cox regression analyses in Fig. 4C4, 4D4, the hazard ratios (HRs) of high-EILncSig versus low-EILncSig for OS were 5.163 (p = 0.00008; 95% CI 2.282–11.680) in TCGA-SARC testing cohort and 3.938 (p = 0.04687; 95% CI 1.019–15.217) in the TARGET-OS testing cohort. Therefore, the EILncSig was identified as an independent factor for the OS prediction. Taken together, the results of the training and testing cohorts indicated that the EILncSig scoring model could be an excellent model for predicting the prognosis of sarcoma patients, which may aid in formulating precise therapeutic strategies for patients with sarcoma. In addition, we preliminarily evaluated whether the EILncSig scoring possessed predictive value on the sarcoma subtypes. In the combined pan-sarcoma expression dataset (n = 1085), the mean EILncSig of each sarcoma subtype varied (Fig. S3C). We applied ROC curve analysis to assess the predictive value. As demonstrated in Fig. S3D, the EILncSig scoring may predict sarcoma subtypes to a certain extent—a high EILncSig score may predict Ewing sarcoma (AUC = 0.747) and a low EILncSig score may predict liposarcoma or myxofibrosarcoma (AUC = 0.711 and 0.761, respectively).
We further examined the associations of EILncSig scores with multiple tumour characteristics across pan-sarcoma patients. Chibon et al. established a prognostic gene expression signature, complexity model in sarcomas (CINSARC), to improve sarcoma patient grading. As shown in Fig. 4E, higher EILncSig scores were found in the CINSARC_C2 group (p = 4.6e-8). As for TCGA-SARC cohort, relapse patients and patients with metastasis were found with higher EILncSig scores (p = 0.0078 and 0.00043, Fig. 4F1-2). A congruent result was also found in the integrative clustering (iCluster) molecular subtypes of sarcoma identified by Alexander et al. The iCluster_C1 group in which patients have the worst prognosis, possessed higher EILncSig scores, whereas the iCluster_C3 group had the lowest EILncSig scores (p < 2e–16, Fig. 4F3). In addition, we found that EILncSig scores were positively correlated with EMT scores and the EMT_C2 cluster had higher EILncSig scores in the combined pan-sarcoma dataset (Fig. S3E, F), which demonstrated a significant association between EILncSig and EMT molecular phenotype across pan-sarcoma patients.
TME and immune patterns associated with EILncSig in sarcoma
Bagaev et al. developed 29 sets of gene expression signatures describing pan-cancer TME characteristics and applied them in exploring TME patterns in pan-cancer patients. Four TME subtypes (immune-enriched, fibrotic (IE/F); immune-enriched, non-fibrotic (IE); fibrotic (F); and depleted (D)) were defined to demonstrate the role of TME in cancer progression and metastasis. We selected sarcoma datasets (TCGA-SARC, TARGET-OS, GSE71118 and E-TABM-1202) to analyse the characteristics of TME across pan-sarcoma patients. After computing EILncSig scores and assigning patients to high- and low-EILncSig levels within each cohort, all patients were included in the clustering analysis of the TME pattern. We utilised an unsupervised clustering method to assign the pan-sarcoma patients to one of four groups by using robustly standardised GSVA enrichment scores of the 29 functional gene expression signature (FGES) sets (Supplementary file 5 and Fig. S3G). As shown in the heatmap (Fig. 5A), sarcoma patients with distinct FGES characteristics along with high- and low-EILncSig stratifications were distributed among the four TME patterns. We utilised the t-SNE analysis to demonstrate the definite diversity of sarcoma patients with each TME pattern (Fig. 5B). Furthermore, high- and low-EILncSig stratifications and four TME patterns presented significant concordant relationships among sarcoma patients (Fig. 5C). Consistent with the previous results, the TME-depleted pattern with the worst prognosis covered 50% of the high-EILncSig group whereas TME-IE and IE/F patterns representing better prognosis were more enriched in the low-EILncSig group.
Fig. 5.
The EILncSig associates distinct TME and immune patterns in sarcoma. A A heatmap of the robustly standardized GSVA enrichment scores for patients assigned into four distinct TME patterns based on unsupervised consensus clustering of the TME-pattern signatures in combined sarcoma dataset. B A 3D t-sne distribution of sarcoma patients corresponding to each TME pattern. C Distinct distribution of TME patterns in high and low EILncSig groups. D Comparison of the EILncSig scores among TCGA-SARC immune subtypes. E Varied expression pattern of four LncRNAs of the EILncSig among TCGA-SARC immune subtypes (with significance). F Distribution and Comparison of TME-infiltrating cells (CIBERSORTx absolute score) between high- and low-EILncSig groups. G Significant correlations between EILncSig scores and infiltrations of CD8 T cells, CD4 memory activated T cells, resting NK cells and activated NK cells
Thorsson et al. identified immune subtypes (wound healing, IFN-γ dominant, inflammatory, lymphocyte depleted and TGF-β dominant) to define pan-cancer immune response patterns that impact prognosis and tumour-immune interactions. We collected information on the immune subtype of TCGA-SARC samples (five immune subtypes involved in total) (Supplementary file 5). As shown in Fig. 5D, there was a significant difference in EILncSig scores among the five immune subtypes (p = 0.00034), with extremely low EILncSig scores in the TGFβ-dominant immune subtype. Additionally, further analysis revealed that expression levels of five EILncRNAs (lncRNAs WWP1-AS1, AFTPH-DT, LBX2-AS1, MCM3AP-AS1 and miR155HG) were also significantly different among the five immune subtypes (Fig. 5E and Fig. S3H). In the aspect of TME-infiltrating immune cells estimated by CIBERSORTx (Supplementary file 5 and Fig. S3I), CD8+ T cells, activated memory CD4+ T cells, Tregs, γδ-T cells, monocytes and macrophages (M1 and M2) showed high infiltration in the low-EILncSig group of better prognoses, whereas resting NK cells and dendritic cells (resting and activated) were more abundant in the high-EILncSig group (Fig. 5F). Furthermore, Spearman correlation analysis showed that the EILncSig score was negatively correlated with CD8+ T cells, activated memory CD4+ T cells, and activated NK cells, but positively correlated with resting NK cells (Fig. 5G).
The transcriptomic alteration, SNV and sCNA associated with EILncSig in sarcoma
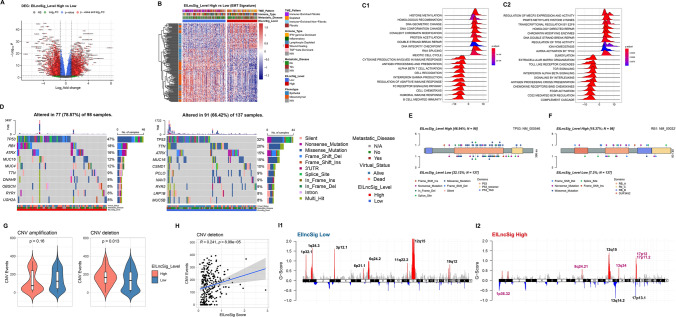
Given that the EILncSig developed from the lncRNA modulation in the pan-sarcoma crosstalk of EMT molecular and tumour immune characteristics, we further assessed the potential value of EILncSig in the perception of transcriptomic genomic alterations in sarcoma. First, we performed DEG analysis on the 259 samples of TCGA-SARC dataset via DESeq2 and found that 6,621 genes (3,384 upregulated and 3,237 downregulated) were significantly differentially expressed in the high-EILncSig group (Fig. 6A and S4A). As shown in the heatmap of Fig. 6B, 186 EMT-related genes belonged to the DEGs set, in which a major subset of mesenchymal marker genes were upregulated in the low-EILncSig group. This result is also consistent with the positive correlation between EILncSig scores and EMT scores in the combined pan-sarcoma microarray dataset. We further used the DESeq2 Wald statistic as a rank list for pre-ranked gene set enrichment analysis (GSEA). As shown in Fig. 6C, ridge plots of GSEA revealed that several gene sets, including DNA damage repair, TP53 activity regulation, histone methylation and protein acetylation, were enriched in the high-EILncSig group, whereas tumour immune activity-related gene sets, such as immune response regulation, cytokine production, interferon and interleukin signalling, were enriched in the low-EILncSig group.
Fig. 6.
Integrative analysis of EILncSig involved in transcriptomic and genomic characteristics of sarcoma. A A volcano plot of DEGs between high- and low-EILncSig groups. B A heatmap showing expression pattern of EMT-related genes in the DEG set. C Ridge plots of selected GSEA results in the gene sets of 1) Gene Ontology biological process and 2) REACTOME pathways. D Somatic mutation landscape and top mutated genes of sarcoma patients in high- and low-EILncSig groups. E, F Lollipop plot showing mutation sites of TP53 and RB1 genes corresponding to high- and low-EILncSig groups. G Comparison of total CNV events (amplification and deletion) between EILncSig groups. H Positive correlation between EILncSig scores and CNV deletion events. I Recurrent somatic CNV regions identified by GISTIC 2.0 in sarcoma patients of high- and low-EILncSig groups (Distinct focal peaks identified in the high EILncSig group are highlighted in purple colour)
We analysed the somatic mutation data of samples with matched EILncSig scores from TCGA-SARC, with 98 and 137 patients in the high- and low-EILncSig groups, respectively (Figs. S4B, C and 6D). TP53 mutation was found as top1 mutation both in the high- and low-EILncSig groups. However, a higher mutation frequency (47% vs. 32%) was observed in the high-EILncSig group. The mutation frequency of RB1, a well-known tumour suppressor gene, was much higher in the high-EILncSig group (ranking 2nd) than that in the low-EILncSig group. Another widely studied cancer-related gene, TTN, was also found to be mutated with relatively high differential frequencies in the low-EILncSig group. We identified specific mutation sites of TP53, RB1 and TTN corresponding to their amino acid location between the high- and low-EILncSig groups (Figs. 6E, F and S4D).
As for the somatic copy number alteration (sCNA), we evaluated its divergence associated with EILncSig by using GISTIC 2.0, which involved 258 samples with matched EILncSig scores in TCGA-SARC (Fig. S4E). As shown in Fig. 6G, higher copy number deletion events were found in the high-EILncSig group while no significant difference of amplification was observed. In addition, we found a significantly positive correlation between copy number deletion events and EILncSig scores (R = 0.241, p = 8.99e–05) (Figs. S4F and 6H). As the previous GSEA showed that DDR-related pathways were found to be activated in the EILncSig-high group, these results indicated that the EILncSig might potentially reflect genome instability in sarcoma. Moreover, we implemented functions of GISTIC 2.0 to identify recurrent focal sCNA regions. As shown in Fig. 6I, there were multiple obvious amplification peaks in the low-EILncSig group, while amplifications on chromosomes 8, 13 and 17 and deletions on chromosomes 1, 13 and 17 were found with higher absolute G-scores in the high-EILncSig group. We identified several distinct sCNA peaks in the high-EILncSig group, such as focal amplification peaks, including the well-studied cancer-driven gene MYC (8q.24.21), several oncogenic genes TFDP1, CUL4A, GAS6 (13q34), DNA damage response related genes TOP3A, ALKBH5 (17p11.2), along with focal deletion peaks including the tumour suppressor gene TP73 (1p36.32) (Supplementary file 6).
EILncSig as a potential predictor of immunotherapy response
Accumulating studies are focusing on identifying robust indicators of immunotherapy response in cancer patients. Predictive efficacy of biomarkers such as expression of certain immune checkpoint inhibitors (ICI), tumour neoantigen burden (TNB) and microsatellite instability (MSI) have been studied in specific cancer types [72–74]. The clinical development of cancer immunotherapy and advances in genomic analysis have also validated the important role of the TME in response to ICI therapy. Considering the association of EILncSig with immune-infiltrating cells and immune process activation, we evaluated the potential capacity of EILncSig as a predictor of immunotherapy response. Previous studies have demonstrated that complex crosstalk exists among tumour immune response, immune infiltration and expression of ICI genes.
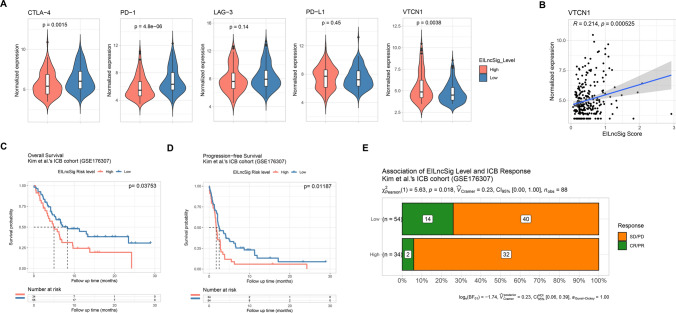
Herein, we first compared the expression of several common ICI genes between patients stratified by EILncSig in TCGA-SARC dataset as shown in Fig. 7A and Fig. S5. We found that the expressions of multiple ICI genes including CTLA-4 and PD-1 were significantly higher in the low-EILncSig group. When considering the globally high level of immune infiltration of the low-EILncSig group, ICI genes that are highly expressed in immune cells are considered to be abundantly expressed. However, we found that expressions of PD-L1, LAG-3, SIGLEC6 and IDO2 did not differ between EILncSig groups and the expression of VTCN1 was even significantly higher in the high-EILncSig group. In addition, VTCN1 expression was positively correlated to the EILncSig scores (Fig. 7B).
Fig. 7.
Potential predictive value of the EILncSig scoring model on response to immunotherapy. A Comparison of normalized expression of ICI genes between sarcoma patients of EILncSig groups. B Positive correlation between normalized expression of VTCN1 and EILncSig scores. C, D Kaplan–Meier analysis on OS and PFS of high- and low-EILncSig groups stratified by optimal cutoff value in the GSE176307 ICI therapy cohort. E Proportions of ICI therapy response corresponding to high- and low-EILncSig groups. (CR complete response, PR partial response, SD stable disease, PD progressive disease)
Next, we examined the capacity of the EILncSig to predict the ICI therapy response in an independent clinical cohort. The cohort of Kim et al. (GSE176307), a publicly accessible PD1/PD-L1 therapy dataset with RNA-Seq and follow-up data, was used in this study. Patients were stratified to high- and low-EILncSig groups using the same method (Supplementary file 7). Time-dependent ROC curve analysis showed that EILncSig scores could be used to predict patients’ PFS and OS. Kaplan–Meier survival analysis revealed that patients in the high-EILncSig group had worse OS and PFS after ICI therapy (log-rank p = 0.03753 and 0.01187, Fig. 7C, D). Moreover, a lower percentage of high-EILncSig patients achieved complete/partial response (CR/PR) while a higher percentage suffered from stable/progressive disease (SD/PD) as compared to the low-EILncSig group (p = 0.018, Fig. 7E). Taken together, these data show that low-EILncSig patients experienced significant clinical benefits, better therapeutic responses and remarkably prolonged survival after ICI therapy.
Discovery of potential drugs that target EILncSig in sarcoma
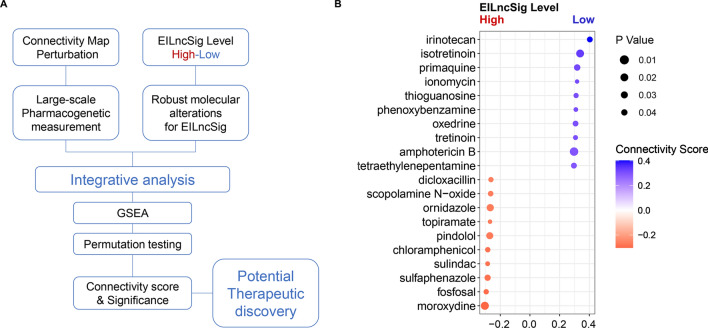
Exploring the complex molecular interactions and regulatory mechanisms of tumour immunity is indeed the exact route to improving immunotherapeutic efficacy. However, it is noteworthy that the combination of immunotherapy and classical chemotherapeutic drugs could be an achievable approach to promote the effectiveness of immunotherapy [75, 76]. Herein, we mined the CMAP database and interactively analysed large-scale pharmacogenetic data with molecular characteristics of EILncSig, to discover drugs that may have the potential capacity to convert sarcoma from high-EILncSig into low-EILncSig status (Fig. 8A and Supplementary file 7).
Fig. 8.
Screen of compounds that have potential capacity to convert EILncSig phenotype based on integrative analysis of pharmacogenetic perturbation database. A A schematic diagram displays the workflow for interactively analysis of large-scale pharmacogenetic data with molecular characteristics of EILncSig. A connectivity score represents the correlation of a compound perturbation with the transcriptomic characteristics of EILncSig. P value is computed by permutation testing and adjusted to the determine significance of the connectivity. B A bubble plot presents potential compounds that have potential capacity to convert EILncSig phenotype. Blue: favourable compounds that may activate transcriptomic alternation from high- to low- EILncSig phenotype. Red: adverse to Blue
As shown in Fig. 8B, promising drugs with positive connective scores were predicted, such as the topoisomerase I inhibitor irinotecan, the retinoid drug isotretinoin, the Ca2+ ionophore ionomycin and the antimetabolite drug tioguanine. Although these drugs have different molecular targets, an increasing number of recent publications have validated the potential of these drugs in immune modulation. For example, He et al. developed a PD-L1-targeting immune liposome (P-Lipo) for co-delivering irinotecan and JQ1, which successfully elicited antitumour immunity in colorectal cancer through induction of immunogenic cell death (ICD) by irinotecan and interference in the immunosuppressive PD-1/PD-L1 pathway by JQ1 [77]. The antitumour immunity or immune-enhancing effect of specific compounds still need to be further validated in sarcoma, while we surmised that these results may be supportive to expanding novel combination strategies of classic drugs with immunotherapy for sarcoma patients and will provide a fundamental basis for further experiments and clinical trials.
Discussion
Sarcoma is a highly heterogeneous malignant tumour, with a highly aggressive clinical phenotype and unfavourable clinical outcomes. Owing to the complex molecular profiling and varying clinicopathological characteristics across sarcoma types, only a limited number of patients obtain satisfactory clinical benefits from common therapeutic strategies [78]. Immunotherapy has become a hotspot in cancer research and takes cancer treatment into a new era. Although immunotherapy for sarcoma has been successful in some cases, its application prospect and effectiveness are still unclear across heterogeneous sarcomas as compared to specific well-studied cancers such as leukaemia [6]. Notably, emerging evidence has presented a boosted therapeutic efficacy by combining immunotherapy with modulation of specific functional targets [79]. To explore the potential application of a combined immunotherapy strategy for sarcoma, it is worthwhile to identify biomarkers that function as molecular targets or critical regulators in tumour immunity across sarcoma types.
EMT is a reversible process that may interact with tumour immunity through multiple approaches such as affecting the TIME. Recent studies have demonstrated the interconnections among EMT-related processes, TME, and immune activity, as well as the potential influence on immunotherapy response. It is notable that increasing evidence shows that certain sarcomas reside in an intermediate EMT/MET-related state, such as the metastable phenotype, which allows tumour cells to switch between epithelial and mesenchymal differentiation [11]. The combined presence of epithelial and mesenchymal features likely plays an indispensable role in the aggressiveness of such sarcomas. To precisely define the regulators of EMT/MET-related processes in sarcomas, we defined two distinct EMT-related molecular subtypes based on a combined EMT signature, identified 26 EILncRNAs, and then constructed a 7-lncRNA signature scoring model (EILncSig) that can stratify sarcoma patients with distinct prognoses, immune microenvironment characteristics, as well as genomic and transcriptomic variations. In the current study, the EILncSig was validated as a robust evaluating tool for the prognosis of patients with sarcoma through the examination of multiple independent datasets incorporating various sarcoma types.
Over the past several decades, accumulating studies have revealed the important roles of the TME in sarcoma genesis, as well as in predicting the prognosis of sarcoma patients [80]. An increased understanding of TME patterns in sarcoma is essential for improving patient outcomes and quality of life. Bagaev et al. developed 29 sets of gene expression signatures describing pan-cancer TME characteristics and defined four TME subtypes to uncover the bidirectional interaction between sarcoma cells and TME [60]. Accordingly, the high-EILncSig group was mainly composed of the TME-depleted pattern whereas TME-IE and IE/F patterns were more enriched in the low-EILncSig group in our current study. Moreover, the EILncSig score was negatively correlated with infiltrations of CD8 + T cells, activated memory CD4 + T cells, and activated NK cells. It is generally accepted that cytotoxic CD8 + T cells, following successful priming, recognise tumour-specific (neoantigens) or tumour-associated antigens and exert anti-tumour function primarily via the release of cytotoxic molecules such as perforin and granzymes [81]. Taken together, our findings indicate that the EILncSig is closely associated with TIME characteristics across pan-sarcoma patients.
The EILncSig also reflects changes in the expression of genes involved in multiple vital hallmarks in sarcomas. Based on the GSVA, we found that several pathways involved in proliferation and metabolism were enriched in the high-risk group whereas tumour immune activity-related gene sets were enriched in the low-risk group. We also found that the somatic mutational profile and sCNA landscape also differed significantly between the high- and low-risk groups. The high-risk group had significantly higher mutational frequency, especially when it came to the well-known tumour suppressor genes TP53 and RB-1. Consistent with the GSVA results, copy number deletion events were markedly enriched with increased EILncSig scores, indicating the potential crosstalk between EILncSig and the genome instability of sarcoma. The sCNA analysis revealed that the high-risk group had multiple recurrent focal amplification peaks covering genomic regions of MYC (8q24.21), TFDP1, CUL4A, and GAS6 (13q34), along with focal deletion peaks including the tumour suppressor gene TP73 (1p36.32). The c-MYC proto-oncogene plays a crucial role in various stages of tumourigeneses, such as proliferation, growth, apoptosis, metabolism, DNA replication and angiogenesis, which can also induce radio- and chemo-resistance of sarcoma cells by suppressing radiation-induced apoptosis and DNA damage, promoting radiation-induced DNA repair and transcriptional regulation of ABC transporter family genes [82, 83]. The transcription factor p73 is a structural and functional homolog of TP53 and can mimic and/or substitute for p53 onco-suppressive functions and has attracted considerable attention for therapeutic cancer management because of the rare mutation [84]. Galtsidis et al. demonstrated that p73 regulated the miR-3158-containing network involved in EMT, thus modulating the cell migration in osteosarcoma [85].
Drug resistance to conventional chemotherapy is one of the most challenging problems in the clinical management of sarcomas. Immune checkpoint inhibitor therapy has recently achieved substantial advances in clinical care for many cancer types including sarcoma [5, 6]. An early assessment of ICI therapy response by predictive biomarkers is crucial for the selection of patients who are most likely to benefit from ICI therapy. Although ICI genes were supposed to be highly expressed in the low-EILncSig group with higher immune infiltration, we still found that the expressions of PD-L1 and LAG-3 showed no significant difference between EILncSig groups and the expression of VTCN1 was significantly higher in the high-EILncSig group. These findings suggest that high-EILncSig sarcoma patients may potentially benefit from ICI therapy against PD-L1, LAG3 and VTCN1. Furthermore, the cohort of Kim et al. (GSE176307) [32] was used to compare the survival distributions of patients stratified by EILncSig. The low-EILncSig patients were found to experience significant clinical benefits, better therapeutic responses and markedly prolonged survival after ICI therapy, indicating that the complex interplay between immune infiltration and ICI genes in the TME has an impact on sarcoma patients’ survival. In addition, we identified multiple drugs that may possess the potential to improve the immunotherapeutic response, which may guide the development of novel chemo-immunotherapy strategies for sarcoma patients. Irinotecan is a first-line chemo-drug in colorectal and pancreatic cancer and other solid tumours, which functions as a topoisomerase I inhibitor; thereby, inducing double-stranded DNA breakage and cell death [86]. Accumulating evidence has recently demonstrated that irinotecan can induce ICD and upregulate tumour-specific antigens, thus triggering an anti-tumour immune response [87]. He et al. and Liu et al. have validated the superior anti-tumour effect and enhanced patient survival of chemo-immunotherapy by combining delivery of anti-PD-L1 and irinotecan [77, 88]. However, further validation of the immune-enhancing effects of specific drugs combined with immunotherapy are warranted in sarcoma.
Although lncRNAs lack protein-coding capability, they are emerging as critical regulators of gene expression in diverse biological processes and play pivotal roles in the tumourigenesis and development of cancer. Some components of the EILncSig have been reported to be dysregulated and function as imperative regulators in several cancers including specific sarcomas. LncRNA MIR155HG, also referred to as the B‑cell integration cluster, has been identified as an oncogene which could play a promotional role in EMT regulation [89]. Notably, a recent study showed that a 17-amino acid micro-peptide encoded by MIR155HG regulates antigen presentation and suppresses autoimmune inflammation [90]. MCM3AP-AS1 was found to be dysregulated in a variety of cancers. A recent study revealed that MCM3AP-AS1 regulates the abundance of M2 macrophage infiltration within the tumour immune microenvironment and may be a potential target to treat bone metastasis of prostate cancer [91]. LINC01140 has recently been reported to participate in the regulation of immune response and EMT [92]. In particular, Hu et al. found that LINC01140 is downregulated in metastatic sarcoma and low LINC01140 expression is associated with unfavourable prognosis of sarcoma [93]. LncRNA MIR22HG is located in 17p13.3, a chromosomal region which is frequently hypermethylated or deleted, and the existing studies demonstrated that MIR22HG functions as either a tumour suppressor or a tumour promoter in numerous cancer types, the regulatory mechanism of which involves Wnt/β-catenin, Notch, EMT and STAT3 signalling pathways [94]. A study by Xu et al. showed that overexpression of MIR22HG triggers T cell infiltration and consequently promotes immune response in colorectal cancer [95]. In addition, a recent study revealed that MIR22HG plays an anti-tumour role in osteosarcoma by acting as a competing endogenous RNA (ceRNA) to the miR-629-5p/TET3 axis [96]. These experimentally-validated findings provide further support to interpret the role of EILncSig in the crosstalk of EMT and tumour immunity.
Conclusion
In summary, we identified lncRNAs which play roles in the crosstalk of EMT and tumour immunity across pan-sarcoma types and constructed a lncRNA-based computational model. Our findings provide a comprehensive resource for understanding the functional role of lncRNA-mediated immune regulation in sarcomas. The constructed EILncSig in our study may serve as a robust predictor of prognosis for patients with sarcomas, as well as a potential biomarker of ICI therapy response that facilitates a more accurate selection of sarcoma patients who may benefit from immunotherapy. The present study established the groundwork for developing potential clinical applications of lncRNA-based immunotherapeutic strategies in precision medicine.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary figure 1 Clustering analysis of pan-sarcoma EMT molecular subtypes. (a). Principle component analysis and datasets combination (before batch correction: left, after batch correction: right) via Combat function of the sva R package. (b). A stack plot showing overall TME-infiltering cells estimated by CIBERSORTx between EMT molecular subtypes. (c). Comparison of absolute scores of each TME-infiltering cell type estimated by CIBERSORTx. (d). Comparison of EMT scores between EMT molecular subtypes.(e). A stack plot showing the composition of sarcoma types between EMT molecular subtypes. (f). For each sarcoma type, proportions of patients with and without specific sarcoma between EMT molecular subtypes
Supplementary figure 2 WGCNA for the identification of EMT molecular subtype related gene modules.(a). Preliminary sample clustering for the detection of outliers. (b). Sample dendrogram and trait heatmap of enrolled sample (not available information in grey). (c). Analysis of network topology for various soft-thresholding powers. Left: the scale-free fit index. Right: the mean connectivity. (d). A histogram and visual assessment of scale-free network topology. (e). Clustering of module eigengenes to quantify co-expression similarity of entire modules for modules merging. (f). Intramodular analysis on the correlation of module membership and gene significance for EMT molecular subtype (showing the five modules with relatively low correlation)
Supplementary figure 3. Evaluation of EILncSig in aspects of EMT, immune subtypes and TME characteristics. (a). In the GSE71118 dataset, a stack plot showing the composition of sarcoma types and the proportions of patients with/without specific sarcoma types between EILncSig groups. (b). In the TCGA-SARC dataset, a stack plot showing the composition of sarcoma types and the proportions of patients with/without specific sarcoma types between EILncSig groups. (c). Comparison of EILncSig scores among various sarcoma types in the combined pan-sarcoma expression dataset. (d). ROC analysis and corresponding AUC for evaluating whether EILncSig score could predict certain sarcoma types. (e). Correlation of EILncSig scores and EMT scores. (f). Comparison of EILncSig scores between EMT molecular subtypes identified by consensus clustering. (g). Consensus matrix of the unsupervised consensus clustering on TME pattern in the combined sarcoma dataset. (h). Expression pattern of lncRNAs of the EILncSig among TCGA-SARC immune subtypes. (i). A stack plot showing overall TME-infiltering cells estimated by CIBERSORTx between EILncSig groups of TCGA-SARC
Supplementary figure 4. Supplementary results of analysis of transcriptomic and genomic characteristics of sarcoma. (a). A heatmap showing DEGs expression of sarcoma patients between EILncSig groups of TCGA-SARC. (b). Somatic mutation landscape of TCGA-SARC patients. (c). Waterfall plot showing top mutated genes of TCGA-SARC patients (n=237). (d). Lollipop Plot showing mutation sites of TTN genes corresponding to high- and low-EILncSig groups. (e). Total CNV events (amplification and deletion) of TCGA-SARC patients. (f). No correlation between EILncSig scores and CNV amplification event
Supplementary figure 5. Supplementary results of analysis of transcriptomic and genomic characteristics of sarcoma. Comparison of normalized expression of ICI genes between sarcoma patients of EILncSig groups (TIM-3, SIGLEC6, IDO1 and IDO2)
Supplementary file 1. Sheet 1-3. Clinical information of patients included in the present study from 17 microarray datasets, TCGA-SARC and TARGET-OS RNA-Seq dataset. Sheet 4. Sample size for each dataset and overview of sarcoma histology subtypes. Sheet 5. The combined EMT signature.
Supplementary file 2. Sheet 1. Result of consensus clustering for pan-sarcoma EMT molecular subtypes based on EMT signature expression pattern. Sheet 2-5. Comparisons of GSVA enrichment scores of patients in different EMT molecular subtypes. Sheet 6. Immune Score, Stromal Score and Microenvironment Score estimated by xCell.Sheet 7. Absolute score of each TME-infiltrating cell type estimated by CIBERSORTx. Sheet 8. EMT scores of patients of EMT molecular subtypes.
Supplementary file 3. Sheet 1. Detailed WGCNA results. Sheet 2. The 72 lncRNAs identified to be associated to the EMT molecular subtypes. Sheet 3. Immune genes of Immport database with annotation. Sheet 4. GSVA scores of immune pathways. Sheet 5. Correlation test of lncRNA–immune pathway pairs. Sheet 6. Correlation test of lncRNA–immune genes pairs. Sheet 7. Correlation test of lncRNA–TME-infiltrating cell pairs. Sheet 8. The 37 robust lncRNA candidates involved in tumour immunity across pan-sarcoma type
Supplementary file 4. Sheet 1. Gene list of EILncRNA. Sheet 2. Univariate and multivariate Cox regression analysis of EILncRNA on training cohort GSE71118. Sheet 3. EILncSig scores and stratification of EILncSig groups of training and validation cohorts. Sheet 4-5. Detailed clinical information use in the Cox regression analysis of EILncSig and clinical characteristics (TARGET-OS and TCGA-SARC). Sheet 6. Supporting clinical information and molecular subtype of TCGA-SARC obtained from Alexander et al.'s study (https://doi.org/10.1016/j.cell.2017.10.014). Sheet 7. Cox regression analysis results of EILncSig with clinicopathological characteristics in TARGET-OS and TCGA-SARC
Supplementary file 5. Sheet 1. TME Signatures of Bagaev et al.’s study (https://doi.org/10.1016/j.ccell.2021.04.014). Sheet 2. Standardized GSVA enrichment scores of the 29 functional TME gene expression signatures. Sheet 3. Consensus clustering of TME pattern. Sheet 4. Supporting immune subtype of TCGA-SARC obtained from Thorsson et al.'s study (https://doi.org/10.1016/j.immuni.2018.03.023). Sheet 5. Absolute scores of TME-infiltrating cells estimated by CIBERSORTx (TCGA-SARC)
Supplementary file 6. Sheet 1. DEG analysis result of patients in high- and low-EILncSig groups. Sheet 2-3. GESA result based on the gene sets of GOBP and REACTOME pathways. Sheet 4. Putative copy-number variation events from GISTIC 2.0 (TCGA-SARC). Sheet 5-6. Identification of genes within recurrent CNV regions
Supplementary file 7. Sheet 1. EILncSig and clinical information of an ICI therapy cohort GSE176307. Sheet 2. Connectivity scores and significance test for drug prediction based on CMAP database
Acknowledgements
The results here are based on the data generated by the TCGA Research Network and the Therapeutically Applicable Research to Generate Effective Treatments. The study reported herein fully satisfies the TCGA and TARGET publication requirements (https://www.cancer.gov/tcga; https://ocg.cancer.gov/programs/target). The authors would like to thank the TCGA, TARGET and GEO developed by National Institutes of Health and the ArrayExpress developed by the European Bioinformatics Institute.
Abbreviations
- EMT
Epithelial-to-mesenchymal transition
- TME
Tumour microenvironment
- LncRNA
Long non-coding RNAs
- EILncRNA
EMT and tumour Immune-related lncRNAs
- EILncSig
EILncRNA signature-based scoring model
- CNV
Copy number variation
- ceRNA
Competitive endogenous RNA
- STS
Soft tissue sarcomas
- ICI
Immune checkpoint inhibitor
- UCA1
Urothelial carcinoma-associated 1
- PD1
Programmed cell death 1
- LIMIT
LncRNA Inducing IFN-γ, MHC-I and Immunogenicity of tumour
- MFS
Metastasis-free survival
- RFS
Relapse-free survival
- OS
Overall survival
- GSVA
Gene set variation analysis
- IL-10
Interleukin-10
- IFNG
Interferon
- TIME
Tumour immune microenvironment
- ROC
Receiver operating characteristic
- AUC
Areas under the curve
- TME subtypes-IE/F
Immune-enriched, fibrotic form
- TME subtypes-IE
Immune-enriched, non-fibrotic form
- TME subtypes-F
Fibrotic form
- TME subtypes-D
Depleted form
- FGES
Functional gene expression signatures
- sCNA
Somatic copy number alteration
- TNB
Tumour neoantigen burden
- MSI
Microsatellite instability
- CR/PR
Complete/partial response
- SD/PD
Stable/progressed disease
- P-Lipo
PD-L1-targeting immune liposome
- ICD
Immunogenic cell death
- PDL-1
PD-ligand-1
- GSEA
Gene set enrichment analysis
- NES
Normalized enrichment score
- GO
Gene Ontology
- KEGG
Kyoto Encyclopaedia of Genes and Genomes
- MSigDB
Molecular Signatures Database
- WGCNA
Weighted gene co-expression network analysis
- PCC
Pearson's correlation coefficients
- DEG
Differentially-expressed genes
- Sarcoma DDLPS
Dedifferentiated liposarcoma
- Sarcoma LMS
Leiomyosarcoma
- Sarcoma MFS
Myxofibrosarcoma
- Sarcoma SS
Synovial sarcoma
- Sarcoma UPS
Undifferentiated pleomorphic sarcoma
Author contributions
DS and SM contributed equally to this work. DS, SM, FP, BZ and BH collected data. DS, SM, FP, MM and WT analysed data and conducted statistical analysis. All authors contributed to data interpretation. DS, SM, FP, MM, WT, ZZ, ZS and JL drafted and revised the manuscript. DS, MS, MM and WT organized the R scripts and processed data. DS, ZZ and JL jointly conceived and supervised the study.
Funding
This work was supported by grants from the National Natural Science Foundation of China (grant No. 82072978 to JL, No. 82072979 to ZZ) and the Natural Science Foundation of Hubei Province (Grant No. 2020CFB861 to JL).
Availability of data and materials
The accession IDs, web links for publicly available datasets analysed in this study are described in method section. All software and R packages used in our study are publicly available and denoted in the method section. Processed results of the present study are available in Supplementary files. R scripts and processed datasets for data analysis and visualisation are available on the GitHub (https://github.com/dyshi9/CMLS-D-22-00823).
Declarations
Competing interests
The authors declare that they do not have any competing conflicts of interest.
Consent for publication
All authors reviewed and approved the final manuscript for publication.
Ethics approval and consent to participate
The patient cohorts we used were publicly available datasets that were collected with patients’ informed consent.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Deyao Shi, Shidai Mu have contributed equally to this work.
Contributor Information
Deyao Shi, Email: shideyao@hust.edu.cn.
Zhicai Zhang, Email: zhicaizhang@126.com.
Jianxiang Liu, Email: liujianxiang@hust.edu.cn.
References
- 1.Ferrari A, Dirksen U, Bielack S. Sarcomas of Soft Tissue and Bone. Prog Tumor Res. 2016;43:128–141. doi: 10.1159/000447083. [DOI] [PubMed] [Google Scholar]
- 2.Damerell V, Pepper MS, Prince S. Molecular mechanisms underpinning sarcomas and implications for current and future therapy. Signal Transduct Target Ther. 2021;6:246. doi: 10.1038/s41392-021-00647-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kasper B. The challenge of finding new therapeutic avenues in soft tissue sarcomas. Clin Sarcoma Res. 2019;9:5. doi: 10.1186/s13569-019-0115-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waldman AD, Fritz JM, Lenardo MJ. A guide to cancer immunotherapy: from T cell basic science to clinical practice. Nat Rev Immunol. 2020;20:651–668. doi: 10.1038/s41577-020-0306-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grünewald TG, Alonso M, Avnet S, Banito A, Burdach S, Cidre-Aranaz F, Di Pompo G, Distel M, Dorado-Garcia H, Garcia-Castro J, et al. Sarcoma treatment in the era of molecular medicine. EMBO Mol Med. 2020;12:e11131. doi: 10.15252/emmm.201911131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Groisberg R, Hong DS, Behrang A, Hess K, Janku F, Piha-Paul S, Naing A, Fu S, Benjamin R, Patel S, et al. Characteristics and outcomes of patients with advanced sarcoma enrolled in early phase immunotherapy trials. J Immunother Cancer. 2017;5:100. doi: 10.1186/s40425-017-0301-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hegde PS, Chen DS. Top 10 Challenges in Cancer Immunotherapy. Immunity. 2020;52:17–35. doi: 10.1016/j.immuni.2019.12.011. [DOI] [PubMed] [Google Scholar]
- 8.Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- 9.Migault M, Sapkota S, Bracken CP. Transcriptional and post-transcriptional control of epithelial-mesenchymal plasticity: why so many regulators? Cell Mol Life Sci. 2022;79:182. doi: 10.1007/s00018-022-04199-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Terry S, Savagner P, Ortiz-Cuaran S, Mahjoubi L, Saintigny P, Thiery JP, Chouaib S. New insights into the role of EMT in tumor immune escape. Mol Oncol. 2017;11:824–846. doi: 10.1002/1878-0261.12093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sannino G, Marchetto A, Kirchner T, Grünewald TGP. Epithelial-to-mesenchymal and mesenchymal-to-epithelial transition in mesenchymal tumors: a paradox in sarcomas? Cancer Res. 2017;77:4556–4561. doi: 10.1158/0008-5472.CAN-17-0032. [DOI] [PubMed] [Google Scholar]
- 12.Kahlert UD, Joseph JV, Kruyt FAE. EMT- and MET-related processes in nonepithelial tumors: importance for disease progression, prognosis, and therapeutic opportunities. Mol Oncol. 2017;11:860–877. doi: 10.1002/1878-0261.12085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mishra S, Verma SS, Rai V, Awasthee N, Chava S, Hui KM, Kumar AP, Challagundla KB, Sethi G, Gupta SC. Long non-coding RNAs are emerging targets of phytochemicals for cancer and other chronic diseases. Cell Mol Life Sci. 2019;76:1947–1966. doi: 10.1007/s00018-019-03053-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohammadinejad R, Biagioni A, Arunkumar G, Shapiro R, Chang KC, Sedeeq M, Taiyab A, Hashemabadi M, Pardakhty A, Mandegary A, et al. EMT signaling: potential contribution of CRISPR/Cas gene editing. Cell Mol Life Sci. 2020;77:2701–2722. doi: 10.1007/s00018-020-03449-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Min L, Garbutt C, Tu C, Hornicek F, Duan Z. Potentials of long noncoding RNAs (LncRNAs) in sarcoma: from biomarkers to therapeutic targets. Int J Mol Sci. 2017;18(4):731. doi: 10.3390/ijms18040731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu QL, Zhang Z, Wei X, Zhou ZG. Noncoding RNAs in tumor metastasis: molecular and clinical perspectives. Cell Mol Life Sci. 2021;78:6823–6850. doi: 10.1007/s00018-021-03929-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang M, Zhou L, Yu F, Zhang Y, Li P, Wang K. The functional roles of exosomal long non-coding RNAs in cancer. Cell Mol Life Sci. 2019;76:2059–2076. doi: 10.1007/s00018-019-03018-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang D, Chen J, Yang L, Ouyang Q, Li J, Lao L, Zhao J, Liu J, Lu Y, Xing Y, et al. NKILA lncRNA promotes tumor immune evasion by sensitizing T cells to activation-induced cell death. Nat Immunol. 2018;19:1112–1125. doi: 10.1038/s41590-018-0207-y. [DOI] [PubMed] [Google Scholar]
- 19.Hu Q, Ye Y, Chan LC, Li Y, Liang K, Lin A, Egranov SD, Zhang Y, Xia W, Gong J, et al. Oncogenic lncRNA downregulates cancer cell antigen presentation and intrinsic tumor suppression. Nat Immunol. 2019;20:835–851. doi: 10.1038/s41590-019-0400-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D, Sabedot TS, Malta TM, Pagnotta SM, Castiglioni I, et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44:e71. doi: 10.1093/nar/gkv1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Network CGAR. Comprehensive and integrated genomic characterization of adult soft tissue sarcomas. Cell. 2017;171:950–965.e928. doi: 10.1016/j.cell.2017.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH, Porta-Pardo E, Gao GF, Plaisier CL, Eddy JA, et al. The immune landscape of cancer. Immunity. 2018;48:812–830.e814. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Howe KL, Achuthan P, Allen J, Allen J, Alvarez-Jarreta J, Amode MR, Armean IM, Azov AG, Bennett R, Bhai J, et al. Ensembl 2021. Nucleic Acids Res. 2021;49:D884–d891. doi: 10.1093/nar/gkaa942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pachter L (2011) Models for transcript quantification from RNA-Seq. arXiv preprint, arXiv:1104.3889
- 26.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013;41:D991–995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, Lopez R. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47:W636–w641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heber S, Sick B. Quality assessment of Affymetrix GeneChip data. OMICS. 2006;10:358–368. doi: 10.1089/omi.2006.10.358. [DOI] [PubMed] [Google Scholar]
- 29.Kauffmann A, Gentleman R, Huber W. arrayQualityMetrics—a bioconductor package for quality assessment of microarray data. Bioinformatics. 2009;25:415–416. doi: 10.1093/bioinformatics/btn647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carlson M (2021) hgu133plus2.db: Affymetrix Human Genome U133 Plus 2.0 Array annotation data. (R package version 3.2.3)
- 32.Rose TL, Weir WH, Mayhew GM, Shibata Y, Eulitt P, Uronis JM, Zhou M, Nielsen M, Smith AB, Woods M, et al. Fibroblast growth factor receptor 3 alterations and response to immune checkpoint inhibition in metastatic urothelial cancer: a real world experience. Br J Cancer. 2021;125:1251–1260. doi: 10.1038/s41416-021-01488-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Busuioc C, Ciocan-Cartita CA, Braicu C, Zanoaga O, Raduly L, Trif M, Muresan MS, Ionescu C, Stefan C, Crivii C, Al Hajjar N, Mǎrgǎrit S, Berindan-Neagoe I. Epithelial-mesenchymal transition gene signature related to prognostic in colon adenocarcinoma. J Pers Med. 2021;11(6):476. doi: 10.3390/jpm11060476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tan TZ, Miow QH, Miki Y, Noda T, Mori S, Huang RY, Thiery JP. Epithelial-mesenchymal transition spectrum quantification and its efficacy in deciphering survival and drug responses of cancer patients. EMBO Mol Med. 2014;6:1279–1293. doi: 10.15252/emmm.201404208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rokavec M, Kaller M, Horst D, Hermeking H. Pan-cancer EMT-signature identifies RBM47 down-regulation during colorectal cancer progression. Sci Rep. 2017;7:4687. doi: 10.1038/s41598-017-04234-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kandimalla R, Gao F, Li Y, Huang H, Ke J, Deng X, Zhao L, Zhou S, Goel A, Wang X. RNAMethyPro: a biologically conserved signature of N6-methyladenosine regulators for predicting survival at pan-cancer level. NPJ Precis Oncol. 2019;3:13. doi: 10.1038/s41698-019-0085-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koplev S, Lin K, Dohlman AB, Ma'ayan A. Integration of pan-cancer transcriptomics with RPPA proteomics reveals mechanisms of epithelial-mesenchymal transition. PLoS Comput Biol. 2018;14:e1005911. doi: 10.1371/journal.pcbi.1005911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hollern DP, Swiatnicki MR, Andrechek ER. Histological subtypes of mouse mammary tumors reveal conserved relationships to human cancers. PLoS Genet. 2018;14:e1007135. doi: 10.1371/journal.pgen.1007135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26:1572–1573. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu T, Le TD, Liu L, Su N, Wang R, Sun B, Colaprico A, Bontempi G, Li J. CancerSubtypes: an R/Bioconductor package for molecular cancer subtype identification, validation and visualization. Bioinformatics. 2017;33:3131–3133. doi: 10.1093/bioinformatics/btx378. [DOI] [PubMed] [Google Scholar]
- 41.Mak MP, Tong P, Diao L, Cardnell RJ, Gibbons DL, William WN, Skoulidis F, Parra ER, Rodriguez-Canales J, Wistuba II, et al. A patient-derived, pan-cancer EMT signature identifies global molecular alterations and immune target enrichment following epithelial-to-mesenchymal transition. Clin Cancer Res. 2016;22:609–620. doi: 10.1158/1078-0432.CCR-15-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Consortium GO The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019;47:D330–d338. doi: 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kutmon M, Riutta A, Nunes N, Hanspers K, Willighagen EL, Bohler A, Mélius J, Waagmeester A, Sinha SR, Miller R, et al. WikiPathways: capturing the full diversity of pathway knowledge. Nucleic Acids Res. 2016;44:D488–494. doi: 10.1093/nar/gkv1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jassal B, Matthews L, Viteri G, Gong C, Lorente P, Fabregat A, Sidiropoulos K, Cook J, Gillespie M, Haw R, et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2020;48:D498–d503. doi: 10.1093/nar/gkz1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Newman AM, Steen CB, Liu CL, Gentles AJ, Chaudhuri AA, Scherer F, Khodadoust MS, Esfahani MS, Luca BA, Steiner D, et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol. 2019;37:773–782. doi: 10.1038/s41587-019-0114-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18:220. doi: 10.1186/s13059-017-1349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sturm G, Finotello F, Petitprez F, Zhang JD, Baumbach J, Fridman WH, List M, Aneichyk T. Comprehensive evaluation of transcriptome-based cell-type quantification methods for immuno-oncology. Bioinformatics. 2019;35:i436–i445. doi: 10.1093/bioinformatics/btz363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4:17. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 54.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinform. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bhattacharya S, Dunn P, Thomas CG, Smith B, Schaefer H, Chen J, Hu Z, Zalocusky KA, Shankar RD, Shen-Orr SS, et al. ImmPort, toward repurposing of open access immunological assay data for translational and clinical research. Sci Data. 2018;5:180015. doi: 10.1038/sdata.2018.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun J, Zhang Z, Bao S, Yan C, Hou P, Wu N, Su J, Xu L, Zhou M. Identification of tumor immune infiltration-associated lncRNAs for improving prognosis and immunotherapy response of patients with non-small cell lung cancer. J Immunother Cancer. 2020;8(1):e000110. doi: 10.1136/jitc-2019-000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heagerty PJ, Saha-Chaudhuri. P, Saha-Chaudhuri MP (2013) survivalROC: time-dependent ROC curve estimation from censored survival data. (R package version 1.0.3)
- 58.Kassambara A, Kosinski M, Biecek P, Fabian S (2021) survminer: Drawing Survival Curves using ‘ggplot2’ (R package version 0.4.9)
- 59.Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, Müller M. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bagaev A, Kotlov N, Nomie K, Svekolkin V, Gafurov A, Isaeva O, Osokin N, Kozlov I, Frenkel F, Gancharova O, et al. Conserved pan-cancer microenvironment subtypes predict response to immunotherapy. Cancer Cell. 2021;39(6):845–865.e7. doi: 10.1016/j.ccell.2021.04.014. [DOI] [PubMed] [Google Scholar]
- 61.Van Der Maaten L. Accelerating t-SNE using tree-based algorithms. J Mach Learn Res. 2014;15:3221–3245. [Google Scholar]
- 62.Ligges U, Maechler M. scatterplot3d—an R package for visualizing multivariate data. J Stat Softw. 2003;8:1–20. doi: 10.18637/jss.v008.i11. [DOI] [Google Scholar]
- 63.Mayakonda A, Lin DC, Assenov Y, Plass C, Koeffler HP. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28:1747–1756. doi: 10.1101/gr.239244.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 66.Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, Carey VJ. Software for computing and annotating genomic ranges. PLoS Comput Biol. 2013;9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Blighe K, Rana S, Lewis M (2022) EnhancedVolcano: publication-ready volcano plots with enhanced colouring and labeling. (R package version 1.14.0)
- 68.Kolde R (2019) Pheatmap: pretty heatmaps. . (R package version 1.0.12)
- 69.Subramanian A, Narayan R, Corsello SM, Peck DD, Natoli TE, Lu X, Gould J, Davis JF, Tubelli AA, Asiedu JK, et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell. 2017;171:1437–1452.e1417. doi: 10.1016/j.cell.2017.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Smirnov P, Safikhani Z, El-Hachem N, Wang D, She A, Olsen C, Freeman M, Selby H, Gendoo DM, Grossmann P, et al. PharmacoGx: an R package for analysis of large pharmacogenomic datasets. Bioinformatics. 2016;32:1244–1246. doi: 10.1093/bioinformatics/btv723. [DOI] [PubMed] [Google Scholar]
- 71.Wickham H. GGPLOT2: elegant graphics for data analysis 2016. New York: Springer-Verlag; 2016. [Google Scholar]
- 72.Robert C. A decade of immune-checkpoint inhibitors in cancer therapy. Nat Commun. 2020;11:3801. doi: 10.1038/s41467-020-17670-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim K, Kim HS, Kim JY, Jung H, Sun JM, Ahn JS, Ahn MJ, Park K, Lee SH, Choi JK. Predicting clinical benefit of immunotherapy by antigenic or functional mutations affecting tumour immunogenicity. Nat Commun. 2020;11:951. doi: 10.1038/s41467-020-14562-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhao P, Li L, Jiang X, Li Q. Mismatch repair deficiency/microsatellite instability-high as a predictor for anti-PD-1/PD-L1 immunotherapy efficacy. J Hematol Oncol. 2019;12:54. doi: 10.1186/s13045-019-0738-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Galon J, Bruni D. Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat Rev Drug Discov. 2019;18:197–218. doi: 10.1038/s41573-018-0007-y. [DOI] [PubMed] [Google Scholar]
- 76.Zheng H, Zhao W, Yan C, Watson CC, Massengill M, Xie M, Massengill C, Noyes DR, Martinez GV, Afzal R, et al. HDAC inhibitors enhance T-Cell chemokine expression and augment response to PD-1 immunotherapy in lung adenocarcinoma. Clin Cancer Res. 2016;22:4119–4132. doi: 10.1158/1078-0432.CCR-15-2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.He ZD, Zhang M, Wang YH, He Y, Wang HR, Chen BF, Tu B, Zhu SQ, Huang YZ. Anti-PD-L1 mediating tumor-targeted codelivery of liposomal irinotecan/JQ1 for chemo-immunotherapy. Acta Pharmacol Sin. 2021;42:1516–1523. doi: 10.1038/s41401-020-00570-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pingping B, Yuhong Z, Weiqi L, Chunxiao W, Chunfang W, Yuanjue S, Chenping Z, Jianru X, Jiade L, Lin K, et al. Incidence and mortality of sarcomas in Shanghai, cHina, during 2002–2014. Front Oncol. 2019;9:662. doi: 10.3389/fonc.2019.00662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li G, Kryczek I, Nam J, Li X, Li S, Li J, Wei S, Grove S, Vatan L, Zhou J, et al. LIMIT is an immunogenic lncRNA in cancer immunity and immunotherapy. Nat Cell Biol. 2021;23:526–537. doi: 10.1038/s41556-021-00672-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ehnman M, Chaabane W, Haglund F, Tsagkozis P. The tumor microenvironment of pediatric sarcoma: mesenchymal mechanisms regulating cell migration and metastasis. Curr Oncol Rep. 2019;21:90. doi: 10.1007/s11912-019-0839-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhu N, Hou J. Assessing immune infiltration and the tumor microenvironment for the diagnosis and prognosis of sarcoma. Cancer Cell Int. 2020;20:577. doi: 10.1186/s12935-020-01672-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gravina GL, Festuccia C, Popov VM, Di Rocco A, Colapietro A, Sanità P, Monache SD, Musio D, De Felice F, Di Cesare E, et al. c-Myc sustains transformed phenotype and promotes radioresistance of embryonal rhabdomyosarcoma cell lines. Radiat Res. 2016;185:411–422. doi: 10.1667/RR14237.1. [DOI] [PubMed] [Google Scholar]
- 83.Xu BS, Chen HY, Que Y, Xiao W, Zeng MS, Zhang X. ALK(ATI) interacts with c-Myc and promotes cancer stem cell-like properties in sarcoma. Oncogene. 2020;39:151–163. doi: 10.1038/s41388-019-0973-5. [DOI] [PubMed] [Google Scholar]
- 84.Logotheti S, Richter C, Murr N, Spitschak A, Marquardt S, Pützer BM. Mechanisms of functional pleiotropy of p73 in cancer and beyond. Front Cell Dev Biol. 2021;9:737735. doi: 10.3389/fcell.2021.737735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Galtsidis S, Logotheti S, Pavlopoulou A, Zampetidis CP, Papachristopoulou G, Scorilas A, Vojtesek B, Gorgoulis V, Zoumpourlis V. Unravelling a p73-regulated network: the role of a novel p73-dependent target, MIR3158, in cancer cell migration and invasiveness. Cancer Lett. 2017;388:96–106. doi: 10.1016/j.canlet.2016.11.036. [DOI] [PubMed] [Google Scholar]
- 86.Del Rio M, Mollevi C, Bibeau F, Vie N, Selves J, Emile JF, Roger P, Gongora C, Robert J, Tubiana-Mathieu N, et al. Molecular subtypes of metastatic colorectal cancer are associated with patient response to irinotecan-based therapies. Eur J Cancer. 2017;76:68–75. doi: 10.1016/j.ejca.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 87.McKenzie JA, Mbofung RM, Malu S, Zhang M, Ashkin E, Devi S, Williams L, Tieu T, Peng W, Pradeep S, et al. The effect of topoisomerase i inhibitors on the efficacy of T-cell-based cancer immunotherapy. J Natl Cancer Inst. 2018;110:777–786. doi: 10.1093/jnci/djx257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu X, Jiang J, Liao YP, Tang I, Zheng E, Qiu W, Lin M, Wang X, Ji Y, Mei KC, et al. Combination chemo-immunotherapy for pancreatic cancer using the immunogenic effects of an irinotecan silicasome nanocarrier plus anti-PD-1. Adv Sci (Weinh) 2021;8:2002147. doi: 10.1002/advs.202002147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cui W, Meng W, Zhao L, Cao H, Chi W, Wang B. TGF-β-induced long non-coding RNA MIR155HG promotes the progression and EMT of laryngeal squamous cell carcinoma by regulating the miR-155-5p/SOX10 axis. Int J Oncol. 2019;54:2005–2018. doi: 10.3892/ijo.2019.4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Niu L, Lou F, Sun Y, Sun L, Cai X, Liu Z, Zhou H, Wang H, Wang Z, Bai J, et al. A micropeptide encoded by lncRNA MIR155HG suppresses autoimmune inflammation via modulating antigen presentation. Sci Adv. 2020;6:eaaz2059. doi: 10.1126/sciadv.aaz2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen Y, Chen Z, Mo J, Pang M, Chen Z, Feng F, Xie P, Yang B. Identification of HCG18 and MCM3AP-AS1 that associate with bone metastasis, poor prognosis and increased abundance of M2 macrophage infiltration in prostate cancer. Technol Cancer Res Treat. 2021;20:1533033821990064. doi: 10.1177/1533033821990064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xia R, Geng G, Yu X, Xu Z, Guo J, Liu H, Li N, Li Z, Li Y, Dai X, et al. LINC01140 promotes the progression and tumor immune escape in lung cancer by sponging multiple microRNAs. J Immunother Cancer. 2021;9(8):e002746. doi: 10.1136/jitc-2021-002746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hu X, Han W, Lou N. High levels of LINC01140 expression predict a good prognosis and improve radiotherapy in sarcoma patients. Crit Rev Eukaryot Gene Expr. 2021;31:9–20. doi: 10.1615/CritRevEukaryotGeneExpr.2021038597. [DOI] [PubMed] [Google Scholar]
- 94.Zhang L, Li C, Su X. Emerging impact of the long noncoding RNA MIR22HG on proliferation and apoptosis in multiple human cancers. J Exp Clin Cancer Res. 2020;39:271. doi: 10.1186/s13046-020-01784-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Xu J, Shao T, Song M, Xie Y, Zhou J, Yin J, Ding N, Zou H, Li Y, Zhang J. MIR22HG acts as a tumor suppressor via TGFβ/SMAD signaling and facilitates immunotherapy in colorectal cancer. Mol Cancer. 2020;19:51. doi: 10.1186/s12943-020-01174-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhao H, Zhang M, Yang X, Song D. Overexpression of long non-coding RNA MIR22HG represses proliferation and enhances apoptosis via miR-629-5p/TET3 axis in osteosarcoma cells. J Microbiol Biotechnol. 2021;31:1331–1342. doi: 10.4014/jmb.2106.06028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure 1 Clustering analysis of pan-sarcoma EMT molecular subtypes. (a). Principle component analysis and datasets combination (before batch correction: left, after batch correction: right) via Combat function of the sva R package. (b). A stack plot showing overall TME-infiltering cells estimated by CIBERSORTx between EMT molecular subtypes. (c). Comparison of absolute scores of each TME-infiltering cell type estimated by CIBERSORTx. (d). Comparison of EMT scores between EMT molecular subtypes.(e). A stack plot showing the composition of sarcoma types between EMT molecular subtypes. (f). For each sarcoma type, proportions of patients with and without specific sarcoma between EMT molecular subtypes
Supplementary figure 2 WGCNA for the identification of EMT molecular subtype related gene modules.(a). Preliminary sample clustering for the detection of outliers. (b). Sample dendrogram and trait heatmap of enrolled sample (not available information in grey). (c). Analysis of network topology for various soft-thresholding powers. Left: the scale-free fit index. Right: the mean connectivity. (d). A histogram and visual assessment of scale-free network topology. (e). Clustering of module eigengenes to quantify co-expression similarity of entire modules for modules merging. (f). Intramodular analysis on the correlation of module membership and gene significance for EMT molecular subtype (showing the five modules with relatively low correlation)
Supplementary figure 3. Evaluation of EILncSig in aspects of EMT, immune subtypes and TME characteristics. (a). In the GSE71118 dataset, a stack plot showing the composition of sarcoma types and the proportions of patients with/without specific sarcoma types between EILncSig groups. (b). In the TCGA-SARC dataset, a stack plot showing the composition of sarcoma types and the proportions of patients with/without specific sarcoma types between EILncSig groups. (c). Comparison of EILncSig scores among various sarcoma types in the combined pan-sarcoma expression dataset. (d). ROC analysis and corresponding AUC for evaluating whether EILncSig score could predict certain sarcoma types. (e). Correlation of EILncSig scores and EMT scores. (f). Comparison of EILncSig scores between EMT molecular subtypes identified by consensus clustering. (g). Consensus matrix of the unsupervised consensus clustering on TME pattern in the combined sarcoma dataset. (h). Expression pattern of lncRNAs of the EILncSig among TCGA-SARC immune subtypes. (i). A stack plot showing overall TME-infiltering cells estimated by CIBERSORTx between EILncSig groups of TCGA-SARC
Supplementary figure 4. Supplementary results of analysis of transcriptomic and genomic characteristics of sarcoma. (a). A heatmap showing DEGs expression of sarcoma patients between EILncSig groups of TCGA-SARC. (b). Somatic mutation landscape of TCGA-SARC patients. (c). Waterfall plot showing top mutated genes of TCGA-SARC patients (n=237). (d). Lollipop Plot showing mutation sites of TTN genes corresponding to high- and low-EILncSig groups. (e). Total CNV events (amplification and deletion) of TCGA-SARC patients. (f). No correlation between EILncSig scores and CNV amplification event
Supplementary figure 5. Supplementary results of analysis of transcriptomic and genomic characteristics of sarcoma. Comparison of normalized expression of ICI genes between sarcoma patients of EILncSig groups (TIM-3, SIGLEC6, IDO1 and IDO2)
Supplementary file 1. Sheet 1-3. Clinical information of patients included in the present study from 17 microarray datasets, TCGA-SARC and TARGET-OS RNA-Seq dataset. Sheet 4. Sample size for each dataset and overview of sarcoma histology subtypes. Sheet 5. The combined EMT signature.
Supplementary file 2. Sheet 1. Result of consensus clustering for pan-sarcoma EMT molecular subtypes based on EMT signature expression pattern. Sheet 2-5. Comparisons of GSVA enrichment scores of patients in different EMT molecular subtypes. Sheet 6. Immune Score, Stromal Score and Microenvironment Score estimated by xCell.Sheet 7. Absolute score of each TME-infiltrating cell type estimated by CIBERSORTx. Sheet 8. EMT scores of patients of EMT molecular subtypes.
Supplementary file 3. Sheet 1. Detailed WGCNA results. Sheet 2. The 72 lncRNAs identified to be associated to the EMT molecular subtypes. Sheet 3. Immune genes of Immport database with annotation. Sheet 4. GSVA scores of immune pathways. Sheet 5. Correlation test of lncRNA–immune pathway pairs. Sheet 6. Correlation test of lncRNA–immune genes pairs. Sheet 7. Correlation test of lncRNA–TME-infiltrating cell pairs. Sheet 8. The 37 robust lncRNA candidates involved in tumour immunity across pan-sarcoma type
Supplementary file 4. Sheet 1. Gene list of EILncRNA. Sheet 2. Univariate and multivariate Cox regression analysis of EILncRNA on training cohort GSE71118. Sheet 3. EILncSig scores and stratification of EILncSig groups of training and validation cohorts. Sheet 4-5. Detailed clinical information use in the Cox regression analysis of EILncSig and clinical characteristics (TARGET-OS and TCGA-SARC). Sheet 6. Supporting clinical information and molecular subtype of TCGA-SARC obtained from Alexander et al.'s study (https://doi.org/10.1016/j.cell.2017.10.014). Sheet 7. Cox regression analysis results of EILncSig with clinicopathological characteristics in TARGET-OS and TCGA-SARC
Supplementary file 5. Sheet 1. TME Signatures of Bagaev et al.’s study (https://doi.org/10.1016/j.ccell.2021.04.014). Sheet 2. Standardized GSVA enrichment scores of the 29 functional TME gene expression signatures. Sheet 3. Consensus clustering of TME pattern. Sheet 4. Supporting immune subtype of TCGA-SARC obtained from Thorsson et al.'s study (https://doi.org/10.1016/j.immuni.2018.03.023). Sheet 5. Absolute scores of TME-infiltrating cells estimated by CIBERSORTx (TCGA-SARC)
Supplementary file 6. Sheet 1. DEG analysis result of patients in high- and low-EILncSig groups. Sheet 2-3. GESA result based on the gene sets of GOBP and REACTOME pathways. Sheet 4. Putative copy-number variation events from GISTIC 2.0 (TCGA-SARC). Sheet 5-6. Identification of genes within recurrent CNV regions
Supplementary file 7. Sheet 1. EILncSig and clinical information of an ICI therapy cohort GSE176307. Sheet 2. Connectivity scores and significance test for drug prediction based on CMAP database
Data Availability Statement
The accession IDs, web links for publicly available datasets analysed in this study are described in method section. All software and R packages used in our study are publicly available and denoted in the method section. Processed results of the present study are available in Supplementary files. R scripts and processed datasets for data analysis and visualisation are available on the GitHub (https://github.com/dyshi9/CMLS-D-22-00823).