Abstract
The base excision repair (BER) pathway is essential for maintaining the stability of DNA in all organisms and defects in this process are associated with life-threatening diseases. It is involved in removing specific types of DNA lesions that are induced by both exogenous and endogenous genotoxic substances. BER is a multi-step mechanism that is often initiated by the removal of a damaged base leading to a genotoxic intermediate that is further processed before the reinsertion of the correct nucleotide and the restoration of the genome to a stable structure. Studies in human and yeast cells, as well as fruit fly and nematode worms, have played important roles in identifying the components of this conserved DNA repair pathway that maintains the integrity of the eukaryotic genome. This review will focus on the components of base excision repair, namely, the DNA glycosylases, the apurinic/apyrimidinic endonucleases, the DNA polymerase, and the ligases, as well as other protein cofactors. Functional insights into these conserved proteins will be provided from humans, Saccharomyces cerevisiae, Drosophila melanogaster, and Caenorhabditis elegans, and the implications of genetic polymorphisms and knockouts of the corresponding genes.
Keywords: Oxidative DNA damage and repair, Sub-pathways, Genome instability, Organismal differences, Neurodegenerative diseases, Cancers
Introduction
The structure of DNA is chemically unstable and vulnerable to various alterations at a high frequency. Approximately, 70,000 different DNA lesions or modifications are generated in a human cell every 24 h (Fig. 1) [1]. The endogenous sources of reactive oxygen species (ROS), e.g., hydrogen peroxide, superoxide anion, hydroxyl radical can damage the DNA primarily at nucleobase. If these lesions are not removed, they lead to various types of mutations in both prokaryotes and eukaryotes (Fig. 2) [2]. Several reactions including, hydrolysis, alkylation, oxidation, methylation, and deamination, can destabilize the nuclear bases and alter the base-pairing property resulting in severe genetic mutations [3]. Several DNA damages are generated by replication errors, such as incorporating 8-oxoguanine into the genome via DNA polymerases [4]. Besides the intrinsic sources of DNA damage, ionizing radiation can produce a barrage of oxidized base lesions and DNA single- and double-strand breaks in the genome. In addition, various chemotherapeutic agents such as temozolomide, bleomycin, anthracyclines, and melphalan produce several types of genotoxic DNA lesions that block the DNA replication and transcription processes [5]. Both healthy and tumor cells must process these destructive DNA lesions if the cells were to survive and maintain physiological functions [6]. Thus, understanding the detailed mechanism of how various cells process DNA lesions has been a benchmark for the selective destruction of cancer cells while protecting the normal cells.
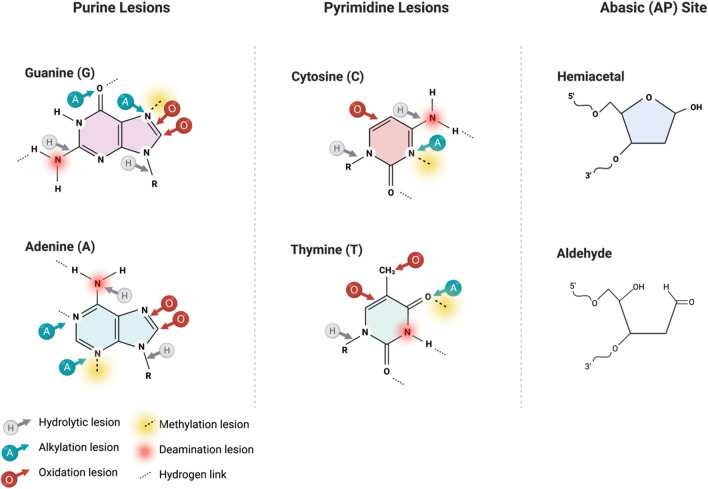
Fig. 1.
Summary of DNA base lesions. Common sites of spontaneous hydrolysis (grey arrow marked H), alkylation (blue arrow marked A), oxidation (red arrow marked O), methylation (yellow shaded dashed line), and deamination (red shaded circle) within guanine, adenine, cytosine, and thymine. DNA glycosylases and various BER proteins recognize these damaged bases and the subsequent abasic site lesion. The figure was generated with a license from BioRender.com
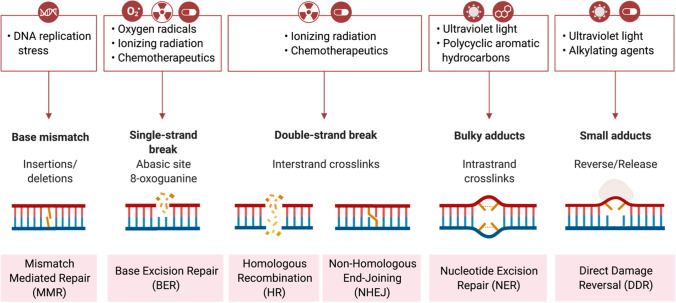
Fig. 2.
Common causes and repair mechanisms of DNA damage in eukaryotes. Several DNA damaging agents may lead to different types of DNA lesions. Each can be corrected by a particular genome repair mechanism, including mismatch repair, base-excision repair, homologous recombination repair, non-homologous end-joining, nucleotide excision repair, or direct damage reversal. The figure was generated with a license from BioRender.com
Several fundamental DNA repair mechanisms exist in eukaryotes that show specificity towards different types of DNA lesions (Fig. 2). These DNA repair mechanisms include (1) mismatch mediated repair (MMR), (2) base excision repair (BER), (3) homologous and non-homologous recombinational repair (HR and NHEJ), (4) nucleotide excision repair (NER), and (5) direct damage reversal (DDR) [7]. This review will focus entirely on the BER pathway, as outline in more detail below. This pathway is responsible for repairing non-bulky DNA damaged bases generated, for example, by endogenous ROS, ionizing radiation, and chemotherapeutic agents [2]. Components of the BER pathway perform five sequential enzymatic reactions requiring a DNA glycosylase, an apurinic/apyrimidinic (AP) endonuclease, DNA polymerase β, and its associated 5′-deoxyribophosphodiesterase (5′-dRPase) activity, as well as a DNA ligase and the scaffold protein, XRCC1, to coordinate the reactions (Fig. 3) [7]. This multistage process is coordinated by the BER enzymes and protein factors, which maintain DNA repair efficiency and genome stability [8, 9].
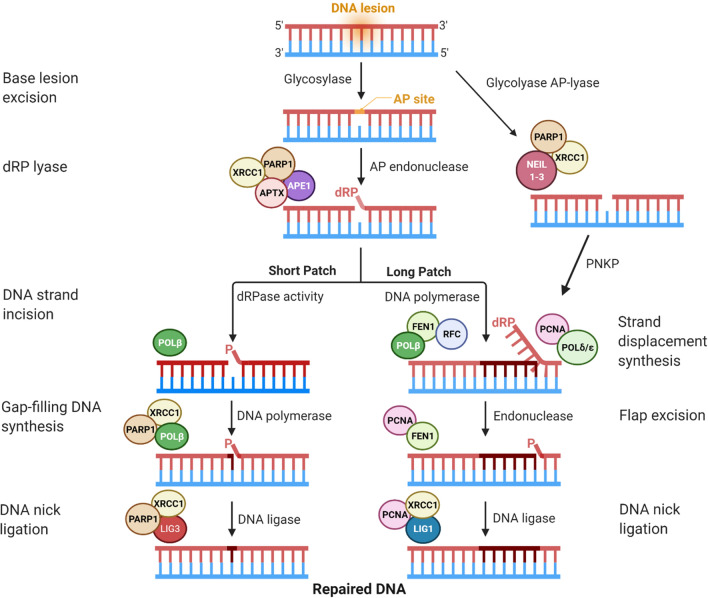
Fig. 3.
A simplified scheme illustrates the Short Patch and Long Patch base excision repair (BER) pathways in eukaryotes. Lesion-specific DNA glycosylases (e.g., UNG1) recognize and remove the damaged base resulting in an abasic site. An APE1 incision follows this to create a single-strand break with 3′-hydroxyl and 5′-dRP ends. The POLβ-dRP lyase activity excises the latter, and POLβ simultaneously fills the gap with either single-nucleotide (left BER, Short Patch) or 2 to 11 nucleotides with FEN1/RFC coupling (right BER, Long Patch). The choice between Short Patch BER or Long Patch BER depends on the state of the 5′dRP end. NEIL-DNA glycosylases (NEIL1-3) contain a β,δ-elimination activity that results in a single-nucleotide gap with a 3′-phosphate end. PNKP will then remove the 3′-phosphate, and the pathway may proceed via Long Patch Repair. Finally, ligation of the DNA-strand nicks is performed through a LIG3-XRCC1-PARP1 complex to complete the Short Patch BER. The polymerase activity will be switched to POLδ/ε when the 5′-dRP termini resist POLβ activity, which can add 2 to 11 nucleotides in the gap. This pathway leaves a flap recognized and removed via FEN1 endonuclease activity that forms a complex with PCNA (right branch). The Long Patch BER is completed when the remaining DNA backbone nick is sealed by DNA LIGI, also associated with PCNA and XRCC1. Orange haze depicts the modified base, and nascent nucleotide(s) are shown in dark red. AP site apurinic/apyrimidinic site, APE1 AP-endonuclease 1, dRP 5′-deoxyribose phosphate, POL DNA polymerase, XRCC1 X-ray cross-complementing protein 1, LIG1-3 DNA ligase I and 3, PCNA proliferating cell nuclear antigen, RFC replication factor C, FEN1 Flap endonuclease, PARP1 poly (ADP-ribose) polymerase, PNKP polynucleotide kinase phosphatase, APTX aprataxin, NEIL1-3 endonuclease VIII-like glycosylases 1, 2, and 3. The figure was generated with a license from BioRender.com
Studying the BER pathway in mammalian systems is complicated by the enzymatic redundancy, substrate overlap of some enzymes, and lethality caused by single-gene deletion such as TDG encoding thymine DNA glycosylase and APE1 encoding the major AP endonuclease [10]. As such, many contributions towards our understanding of the cellular mechanism of the eukaryotic BER process have been derived from studies performed in the budding yeast Saccharomyces cerevisiae (S. cerevisiae), as well as from Caenorhabditis elegans (C. elegans) and Drosophila melanogaster (D. melanogaster) [6, 11]. In this review, we describe the functions of the various components of the BER pathway in human cells and draw a comparison with those of S. cerevisiae, C. elegans, and D. melanogaster, as depicted in Tables 1 and 2. Despite the high conservation of the components of the BER pathway across these organisms, there are several distinct differences highlighted below. Herein, we also summarize the defective BER components leading to pathophysiological conditions such as neurodegenerative diseases and tumorigenesis. Finally, we will provide some insights on the knowledge gap and the likely direction for future studies in the field.
Table 1.
The proteins involved in BER in humans, yeast, drosophila, and C. elegans: Consequences of defective BER
| BER Enzyme | Human | S. cerevisiae | D. melanogaster | C. elegans | Damaged base type | Interactions* | Substrates | Diseases associated with BER defect | Localization | References |
|---|---|---|---|---|---|---|---|---|---|---|
| DNA glycosylase | ||||||||||
| Uracil DNA glycosylase | UNG | Ung1 | – | UNG-1 | Deaminated base | APE1 enhances glycosylase activity; Associated with RPA, PCNA, and XRCC1 | U, U:A, U:G, 5-FU in ssDNA and dsDNA | Immunological defects (hyper-IgM syndrome), colorectal cancer | Nuclei and Mitochondria | [11, 24, 32] |
| SMUG1 | – | Dmel | – | APE1 enhances glycosylase activity | dU, 5-fU, 5-hmU, U:G, U:A | Cervical squamous cell carcinoma, colorectal cancer | Nuclei | [27, 73] | ||
| MBD4 | – | MBD-R2 | – | Associated with MLH1 | U or T in U/TpG:5-meCpG, 5-fU, 5hmU/ssDNA, Tg:G | Colorectal cancer, myeloid neoplasia, immunological defects | [24, 73] | |||
| G-T-mismatch DNA glycosylase | TDG | – | THD1 | – | APE1 enhances glycosylase activity; Associated with RXR/RAR, RAD9, NEIL1/NEIL2, and SIRT1 | U:G, EthenoC:G, T:G, 8-oxoA:C, G, and T, 5-OHC, 5-fU, 5-caC | Spinocerebellar ataxia with axonal neuropathy, colorectal, liver, breast, thyroid, and lung cancer | [44, 50] | ||
| 3-methyl adenine DNA glycosylase | AAG/MPG | Mag1 | – | – | Alkylated base | Association with hHR23 and XRCC1 | 3-MeA, 7-MeA, 3-MeG, 7-MeG, N7-mG, N3-mG, 1-mG Etheno A, m6A, hypoxanthine from dsDNA | Ischemic stroke, inflammatory diseases, lung, and intestinal cancer | Nuclei and MPG Mitochondria | [85] |
| 8-oxoguanine DNA glycosylase/AP lyase | OGG1 | Ogg1 | OGG1 | – | Oxidized base | APE1 enhances glycosylase activity; Associated with XRCC1 and SIRT3 | 8-oxoG:C, faPyG, FaPyC, 8-oxoA, and AP site | Bladder cancer, depression, intestinal inflammation, rheumatoid arthritis | Nuclei and Mitochondria | [60, 61] |
| A-G-mismatch DNA glycosylase/AP lyase | MUTYH | – | – | – | Associated with APE1, PCNA, and RPA | A:8-oxoG, A:G, A:C, 2-hA, 2-oxoA | Depression, Alzheimer’s disease, head and neck cancer | [68] | ||
| Nth Endonuclease III-Like 1 | NTHL1 | Ntg1/ Ogg2 (Eth1) | CG9272 | NTH-1 | Oxidized base | APE1 enhances glycosylase activity; Associated with XPG and XRCC1 | faPyG, Tg/dsDNA, Cg, 5-DHU, 5-ohU and 5-ohC in dsDNA, 5,6-HT, and AP site | Polyposis, colorectal cancer | Nuclei and Mitochondria | [23, 56, 73] |
| – | Ntg2 | – | – | Associated with MLH1 | – | Nuclei | [58] | |||
| AP endonuclease | ||||||||||
| Endonuclease III | APE1 (HAP1/REF1) | Apn1 | APEX1 | APN-1 | Oxidized base | UNG1 enhances glycosylase activity; Associated with SMUG, NTHL1, OGG1, TDG, MUTYH, XRCC1, PCNA, LIG1, and FEN1 | AP sites, 3'-oxo-C | Breast, bladder, lung, liver, ovarian, and uterine cancer | Nuclei and Mitochondria | [44, 73] |
| APE2 | Apn2 | EXO-3 | Associated with PCNA and POL30 | AP sites; A in A:8-oxoG | Breast, uterus, kidney, lung, liver, and skin cancer | Nuclei and APE2 Mitochondria | ||||
| Endonuclease IV | NEIL1 | – | – | – | Oxidized base | Associated with FEN1, XRCC1, PCNA, POLδ, RPA, PARP1, and TDG | Tg, 5-ohU, 5-ohC, faPyA/G, Urea, 8-oxoG, Sp, Gh in ss and dsDNA | Depression, colorectal cancer, immunodeficiency and neurodegenerative diseases | Nuclei and Mitochondria | [81] |
| NEIL2 | Interacts with XRCC1, TDG, and POLδ | Overlap with NTH1 and NEIL1 | Colorectal cancer | Nuclei | [82] | |||||
| NEIL3 | Associated with RPA and POLδ | Oxidized purines, faPyG, faPyA, Sp and Gh in ssDNA | Immunodeficiency and neurodegenerative diseases | [81] | ||||||
| DNA polymerase | ||||||||||
| DNA polymerase | POLε | POLε | – | – | Alkylated base and ultraviolet radiation | Associated with POLβ, RFC1, and PCNA | SSB with a 1 nt-gap, abasic sites with 3′OH and 5′dRP | Colorectal cancer | Nuclei and Mitochondria | [108, 119] |
| POLδ | POLδ | – | – | Associated with NEIL1-3 | ||||||
| POLβ | TRF4 | – | – | Associated with PCNA, POLε, PARP1, XRCC1, and FEN1 | Alzheimer’s disease, thymic hyperplasia, breast, and colon cancer | |||||
| DNA ligase | ||||||||||
| DNA ligase | LIG1 | Cdc9 | – | LIG-1 | Oxidized base and ionizing radiation | Associated with PCNA, POLα, FEN1, and APE1 | DNA with 3'-OH/5"-P nick | Immunodeficiency and cancer predisposition | Nuclei | [23, 132, 133] |
| LIG3 | – | – | – | Associated with PARP1 and XRCC1 | Nuclei and Mitochondria | |||||
| LIG4 | – | LIG4 | LIG-4 | Nuclei | ||||||
Fapy 2,6-diamino-4-oxo-5-formamidopyrimidine, fU fluorouracil, hmU 5-(hydroxymethyl) uracil, hoU Deoxyuridine, dU, 5-hydroxyuracil, Tg thymine glycol, 3-meA 3-methyl-adenine, oxo- oxidized base, OH hydroxyl termini, deoxyribose phosphate dRP; P phosphorylated end/molecule, guanidinohydantoin Gh, spiroiminodihydantoin Sp, ssDNA single strand-DNA, dsDNA double strand-DNA, A adenine, C cytosine, T Thymine, G guanine, U uracil, Ig immunoglobulin
Table 2.
Additional factors involved in BER in humans, yeast, drosophila, and C. elegans: Consequences of defective BER
| BER Enzyme | Human | S. cerevisiae | D. melanogaster | C. elegans | Damaged base type | Interactions | Substrates | Diseases associated with BER defect | Localization | References |
|---|---|---|---|---|---|---|---|---|---|---|
| Accessory proteins | ||||||||||
| X-ray repair cross-complementing group | XRCC1 | – | XRCC4/XLF | – | Oxidized base | Associated with OGG1, NTHL1, NEIL1, NEIL2, MPG1, POLβ, LIG3, PARP1, PARP2, UNG2, PNK, ATPX, PCNA, and APE1 | Abasic sites with 3′-OH and 5′-dRP and other nucleotide gaps | Breast, lung, liver, and uterine cancer, rheumatoid arthritis, Alzheimer’s disease, and nonsyndromic cerebellar ataxias | Nuclei | [135, 154] |
| Poly (ADP-ribose) polymerase | PARP1 | – | PARP | PME-1 | Alkylated base | ROS enhances PARP activity; Associated with LIG3, XRCC1, POL3, OGG1, BRCA1/2, SIRT1, SIRT6, TET1, and several non-BER proteins | Alzheimer’s disease, breast, lung, ovarian, liver, and uterine cancer, lung inflammatory disorders, cardiovascular disease, diabetes, asthma, sepsis, arthritis, atherosclerosis, and nonsyndromic cerebellar ataxias | Nuclei | [138, 149, 150] | |
| PME-2 | ||||||||||
| Flap endonuclease | FEN1 | Rth1/Rad27 | CG8648 | CRN-1 | Oxidized base, ionizing radiation, and AP site | Associated with PCNA, POL4, NEIL1, LIG1, APE1, and PNCA | Alzheimer’s, Huntington, Amyotrophic lateral sclerosis, Parkinson, Hereditary spastic paraplegia Friedreich ataxia, and cancer | Nuclei and Mitochondria | [121] | |
| CG10670 | ||||||||||
| Proliferating cell nuclear antigen | PCNA | Pol30/2 | POLE | POLε | Alkylated base and oxidized base | Associated with APE1, MUTYH, POLβ, XRCC1, FEN1, LIG1, and several other proteins | Multinucleated gap | Hodgkin's disease, autoimmune and neurodegenerative diseases | Nuclei | [73, 115, 116] |
| POLη | Pol3 | POLD | POLH-1 | Associated LIG1, FEN1, PCNA, POLε, and PARP1 | – | |||||
| POLk | Pol IV | POLB | POLK-1 | |||||||
APE1 AP-endonuclease 1, dRP 5′-deoxyribose phosphate, POL DNA polymerase, XRCC1 X-ray cross-complementing protein 1, LIG1-3 DNA ligase I and 3, PCNA proliferating cell nuclear antigen, RFC replication factor C, FEN1 Flap endonuclease, PARP1 poly (ADP-ribose) polymerase, PNKP polynucleotide kinase phosphatase, APTX aprataxin, NEIL1-3 endonuclease VIII-like glycosylases 1 and 2, ROS reactive oxygen species
Base excision repair—a genome maintenance process
The first functional enzymes of the BER process were isolated and characterized from the Escherichia coli (E. coli) model in 1960. During the 1970s to 1980s, several BER encoding genes and recombinant proteins have been cloned and characterized using E. coli [12, 13]. This establishment allows the studying of BER mechanisms in eukaryotes including yeast and mammalian cells. By the end of the 1990s, the BER process was primarily known in mammalian cells [14–16]. In vitro, about 70 different modified bases have been created and installed in oligonucleotide substrates. More than 15 of these lesions have been accurately quantified in the cellular genome using various techniques such as mass spectrometry (Fig. 1). The modified bases include oxidized purines and pyrimidines that, when processed, lead to apurinic/apyrimidinic (AP) sites and subsequently DNA single-strand breaks [6]. However, the efficiency of the BER recombinant DNA glycosylases involved in the first step of the damaged base removal could not be accurately determined as a fraction of the proteins were inactive. These enzymes showed significantly low enzymatic activities in vitro, which raised the issue of how the base excision repair factors maintain the DNA integrity in mammalian cells when enormous levels of spontaneous lesions are produced daily [17, 18].
The expression of BER pathway proteins is increased and activated in response to DNA damage or during the G1 phase of the cell cycle in both nuclei and mitochondria [19]. Thus, the primary function of BER is to correct the frequently produced non-helix minor distorting nucleobase mutations in the DNA sequences [19]. Generally, BER comprises five distinct steps (Fig. 3). This process is initiated by the recognition and excision of the damaged base in the DNA helix by a DNA glycosylase leaving an apurinic/apyrimidinic (AP) site [17, 18, 20, 21]. The abasic site has two different cyclic organic structures in the genome, a main hemiacetal molecule and an open-ring aldehyde molecule (Fig. 1). An AP endonuclease then incises the resulting AP site on the 5′-side of the sugar-phosphate backbone creating a nick or a break bearing a 3′-hydroxyl terminus and 5′-deoxyribose phosphate (dRp) structure [21]. This allows the entry of DNA polymerase β to replace the damaged base with the correct single nucleotide, in a process referred to as Short Patch Repair, while simultaneously removing the dRp with its intrinsic dRp lyase activity [16]. Alternatively, the damaged base can be replaced by at least 2–11 nucleotides that entail other factors, including PCNA and FEN1 endonuclease, in a process referred to as Long Patch Repair. The resulting nick left by DNA polymerase β is closed by DNA ligase 3 [20].
DNA glycosylases of BER in eukaryotes
DNA glycosylases (DNGs) are a group of highly conserved DNA repair enzymes responsible for recognizing and initiating the repair process of damaged DNA bases. The eukaryotic genome encodes several BER DNGs that seem to share a phylogenic origin. The genome of humans, yeast, fruit fly, and the nematode worm encodes 11, 5, 6, and 2 DNGs, respectively [22–24]. While some of the DNGs are highly substrate-specific, others show substrate redundancy and overlap. The DNGs belong to two categories: (1) monofunctional that catalyzes the cleavage of the N-glycosylic bond and removal of the damaged bases leaving an abasic site, and (2) bifunctional that removes oxidized bases with an associated AP lyase activity on the 3′-baseless termini [25]. The monofunctional DNGs include uracil-DNA glycosylase-1 and -2 (UNG-1, UNG-2), single-strand selective monofunctional uracil DNA glycosylase-1 (SMUG1), thymine DNA glycosylase (TDG), methyl CpG binding domain-4 (MBD4) in mammalian cells; thymine DNA glycosylase-1 (THD1), and MBD-R2 in D. melanogaster; UNG-1 in C. elegans; Ung1 and methyladenine DNA glycosylase (Mag1) in S. cerevisiae [22–24].
The bifunctional DNGs, following removal of the damaged base, cleave the DNA backbone 3′-phosphate to the lesion using an activated amino moiety as a nucleophile in a β-elimination reaction [25]. This enzymatic action results in a single-strand DNA break with 3′-blocking unsaturated aldehyde and a 5′-phosphate terminal. Moreover, some bifunctional DNGs incise the DNA 3′ and 5′ in a β, δ-elimination reaction that leaves a 3′ and a 5′-phosphate end [25]. Bifunctional DNGs proteins include endonuclease III-like protein 1 (NTHL1) and 8-Oxo-Guanine DNA glycosylase (OGG1) in humans; NTH1 in D. melanogaster and C. elegans, and the N-glycosylase-1 and -2 (Ntg1 and Ntg2) and Ogg1 in S. cerevisiae [22–24]. Depending on the chemical structure, members of DNGs are sorted into four prominent families, including uracil-DNA glycosylases (UDG), helix-hairpin-helix glycosylases (HhH), 3-methyl-purine glycosylases (MPG), and endonuclease VIII-like glycosylases (NEIL) [6].
Uracil DNA N-glycosylases superfamily
While uracil in RNA is benign, its presence in DNA is premutagenic. Uracil in DNA results either from cytosine deamination or dUTP misincorporation opposite adenine during replication. BLASTP database search for UDGs identified six families that include UNG, SMUG, TDG, and MBD4 proteins [26]. This superfamily seems to share the same α/β fold of several conserved principal elements that correspond to three motifs. The lateral motifs are located near the N- and C-termini and are in the substrate-binding pocket. However, the third motif is positioned centrally and is essential for the conformational structure of UDGs to support its binding to the DNA strand [26]. UNG, SMUG, and TDG belong to different DNA glycosylase families and show structural and functional differences. For example, these differences might explain the observation that the single-strand-selective monofunctional uracil-DNA glycosylase I (SMUG-1) can successfully complement UNG1 function in S. cerevisiae, but not that of E. coli mutants [27, 28].
Uracil DNA glycosylase excises uracil bases in both single- and double-stranded DNA and shows greater activity towards U in single-stranded DNA followed by U:G and U:A in double-stranded DNA [29]. It is highly specific for uracil bases and belongs to the family I of DNGs [30]. It has been demonstrated that overexpression of UNG1 can significantly enhance cellular resistance against oxidative stress [31]. It is believed that oxidative stress spares UNG1 degradation via protection by a disulfide link with Peroxiredoxin 3, an essential antioxidant protein in the mitochondria, such that UNG1 protects the mitochondrial genome from oxidation [31]. In humans, alternative splicing produces two isoforms of UNG: mitochondrial UNG-1 and nuclear UNG-2. Both enzymes share a widely conserved C-terminus involved in uracil removal. However, the N-terminus often varies among species as it seems to be involved in protein regulation, localization, and interprotein interactions [30]. Sequence analysis indicates that UNG homologs vary slightly in size and share moderate homology among organisms. For instance, the human UNG2 shares 58.2% similarity with C. elegans UNG1, while it shares 54% amino acid identity with its homolog in S. cerevisiae [11, 32]. As a result, the protein size varies slightly among organisms, e.g., 34.6 kDa in humans versus 40.5 kDa in S. cerevisiae [11].
Interestingly, the genome of D. melanogaster does not encode the uracil DNA glycosylase. Therefore, it accumulates significant levels of uracil into the larval DNA [22]. In addition, the downregulation of deoxyuracil triphosphatase (dUTPase) during DNA replication increases dUTP in the nucleotide pool, thereby further elevating the level of uracil in the larval genome [22]. Although the mechanism that limits uracil into the genome of D. melanogaster is not yet fully understood, it is noteworthy that this organism possesses the enzyme Dme1 to remove uracil (see below). Of note, a specific base repair enzyme named “Uracil-DNA Degrading Factor” also exists in fruit flies that might limit uracil incorporation into the genome [33].
SMUG1 is another monofunctional member of the DNA glycosylase family that is responsible for removing uracil and its derivatives from single- and double-stranded DNA in nuclear chromatin [34]. It was also recently shown to have a critical role in regulating transcription, cell cycle, apoptosis, and the maturation of telomerase [35, 36]. Interestingly, deleting SMUG1 reduces the human RNA telomerase gene levels and consequently results in severe telomere depletion [36]. A TBLASTN search for sequence homology of human SMUG1 confirmed its preservation in prokaryotes and eukaryotes [37]. However, in contrast to vertebrates, non-vertebrates and insects (except sea urchins) seem to encode either a UNG or a SMUG uracil DNA glycosylase. Examples are C. elegans and S. cerevisiae, which only encode uracil DNA glycosylase [23, 27]. Furthermore, homology to human SMUG1 appears to increase in higher eukaryotes. For instance, the D. melanogaster Dmel shares about 39% identity and 61% similarity with SMUG-1, while M. musculus shares 89% identity and 96% similarity with its human homolog [27]. Although SMUG1 recognizes uracil in both single- and double-stranded DNA, like UNG [38], it can also remove several oxidation products of uracil, such as 5-hydroxyuracil, 5-hydroxymethyluracil, and 5-formyl uracil. Both UNG1 and SMUG1 appear to coordinate binding to the damaged base and execute the BER pathway via distinct mechanisms. In contrast to the human nuclear UNG2, the catalytic activity of SMUG1 is not under the control of the cell cycle; however, it is influenced by the nature of the base pairs flanking uracil [38]. Further, the SMUG1 protein shows a particular preference to A:T base pairs and has a lower contribution to U:G repair [39]. This is congruent with the structure of the conserved DNA wedge motif of SMUGs, which accommodates a more invasive interaction with dsDNA compared to UNG, allowing for contact with adjacent nucleotides [38].
Deletion of SMUG1 results in a normal phenotype, fertility, and survival rate in mice, although the 5-hmU and uracil activities are abolished [40]. The data revealed that SMUG1 serves as the major 5-hmU excision activity and an auxiliary function for UNG. However, SMUG1/UNG-double deletion is not lethal and the mice survive, suggesting that another redundant enzyme(s) exists to process the U:G and T:G mismatches left by the deamination of cytosine, 5-methylcytosine, and 5-hydroxymethylcytosine to produce uracil, thymine, and 5-hmU, respectively. A combination of the double SMUG1/UNG-deletion mice with mice lacking the mismatch repair protein Msh2 increased the cancer predisposition of the triple deleted mice [40]. A previous study showed that SMUG1 deficiency is associated with aggressive breast cancer [41], and a more recent study indicated that SMUG1 is among 25 genes whose expression is significantly associated with the risk of pancreatic cancers [42]. Thus, the inability to remove specific DNA lesions by SMUG1 absence may lead to a more unstable genome in particular cancers.
TDG is the founding member of the widely conserved mismatch uracil DNA glycosylases. These BER proteins have two vital activities: epigenetic regulation and maintenance of the genome [43]. They share a commonly conserved central motif that harbors the catalytic activity but vary markedly in their N- and C- termini; the reason why most orthologues share between 37 and 52% identity only [44]. TDG is best known for its ability to repair U:G mismatches, but it also removes 5-derivatives of U and C such as 5-methyluracil, 5-hydroxyuracil, 5-carboxylcytosine, 5-hydroxymethylcytosine, and 5-formylcytosine [45]. It is also essential for maintaining the epigenetic integrity of CpG (or CG) islands by removing thymine in T:G mismatches that arise from 5-methylcytosine deamination [46]. During epigenetic control of gene expression, 5-methylcytosine is demethylated in a process that requires three oxidation steps by the ten-eleven translocation (TET) dioxygenases to produce 5-hydroxymethylcytosine, 5-formylcytosine, and 5-carboxycytosine, and these latter two oxidized cytosines can be excised by TDG followed by the insertion of unmodified cytosines by the BER process [47]. Thus, TDG cooperates with TET to erase methylated cytosines at CpG, through an oxidation-excision process, and there is evidence for similar roles in non-CG regions of the genome [47].
Like other UDGs, TDG employs a nucleotide flipping mechanism to recognize its substrates. However, TDGs uniquely identify the target base through interacting with the complementary base, which explains the strict preference for guanine-mismatched base pairs such as T:G and U:G [48]. This mechanistic model enables the identification of mismatched base pairs that distinguish the removal of epigenetically modified bases such as non-damaged 5-methylcytosine. The mismatch recognizing uracil DNA glycosylases can form an enormous catalytic cavity around the target base allowing the enzymes to accommodate various bulky derivatives of purines and pyrimidines [45]. However, substrate specificity seems to be affected by the phylogenic origin of different TDGs. Gene deletion of TDG in mice results in embryonic lethality associated with impaired epigenetic regulation impacting developmental gene expression [49]. The TDGs of humans, chickens, and fruit flies efficiently excise thymine and its derivatives; however, orthologs in Schizosaccharomyces pombe (Thp1p) and E. coli cannot do the same [50, 51]. Likewise, only vertebrate TDG can remove derivatives of 5mC [44]. So far, C. elegans does not seem to possess a TDG [23].
Helix–Hairpin–helix DNA glycosylases superfamily
NTH-1 belongs to the helix–hairpin–helix superfamily of DNA endonuclease III-like N-glycosylases, a bifunctional DNA enzyme that removes oxidatively damaged pyrimidine bases as well as AP sites [52]. The enzyme uses a β-elimination reaction to incise DNA strands on the 3′-side of the abasic site leaving a single-strand DNA break terminated with 3′-α, β unsaturated aldehyde. Phylogenic analysis revealed that almost every eukaryotic genome encodes a NTH enzyme [53]. Blast analysis of the C. elegans database identified a homologous protein NTH-1 exhibiting similar enzymatic activities like the human NTHL1 [54]. The genome of Arabidopsis thaliana (A. thaliana), S. pombe, and S. cerevisiae encodes two homologs of human NTHL1 attributed to lineage-specific duplication events. In yeast, the two homologs known as Ntg1 and Ntg2, both isozymes vary significantly in the spectrum of lesions process in addition to their cellular localization [52]. The yeast Ntg1, like its human ortholog NTH1, localizes to both the nucleus and mitochondria. Ntg1 and NTH1 possess both nuclear and mitochondrial targeting signals to regulate the metabolism of the genome under oxidative damage [52].
In contrast, Ntg2 is localized strictly in the nucleus and shares a conserved endonuclease-III iron-sulfur center that is not detected in Ntg1. The iron-sulfur cluster is inserted into Ntg2 through the aid of the cytosolic iron-sulfur assembly machinery, and defects in components of this machinery such as MMS19 lead to genomic instability [55]. The NTHL1 ortholog in other eukaryotes such as D. melanogaster has yet to be examined for regulatory responses [27].
Oxidizing agents can produce many DNA base lesions including a ring-opened formamido-pyrimidine derivative of guanine and adenine (faPyG and faPyA), as well as several modified pyrimidines such as thymine glycol, 5,6-dihydrothymine, 5-hydroxyuracil, 5-hydroxycytosine, and dihydroxy-uracil [56]. Both Ntg1 and Ntg2 are responsible for excising these oxidized base lesions. However, several oxidants can stimulate and regulate the catalytic activity of Ntg1 and not Ntg2 [52]. In addition, Ntg1 can also excise 8-oxoguanine (8-oxoG) opposite guanine but not Ntg2 [57]. Deletion of Ntg1 and Ntg2 have not shown any changes in the toxicity effect of MMS, radiation, and hydrogen peroxide nor resulted in any striking spontaneous mutation phenotypes. The results suggest that oxidized bases and the subsequent abasic sites can be repaired through alternative mechanisms in S. cerevisiae [58]. Recently, the NTHL1 variation, p.Q90*, has been associated with a rare colorectal polyposis known as NTHL1-associated polyposis (NAP), which is believed to be due to an inability to remove oxidative pyrimidine DNA lesions, such as 5-hydroxyuracil [56, 59].
OGG1 is present in most eukaryotic organisms, including mammals, yeast, and fruit fly [60, 61]. However, it is not present in the nematodes C. elegans nor C. briggsae [23]. It has been established that OGG1 efficiently removes, for example, 7,8-dihydro-8-oxoguanine and -8-oxoadenine in the DNA produced by various oxidative agents, including normal aerobic metabolism and exposure to gamma-irradiation [62]. Similar to Ntg1 and Ntg2, OGG1 can incise abasic sites opposite cytosine in dsDNA [62]. The bi-functional N-glycosylase and AP lyase activities of OGG1 are greatly affected by the identity of the base corresponding to the lesion with a noticeable preference for cytosine and G-derived formamidopyrimidine [4]. The active site lysine residue of OGG1 attacks the 8-oxoguanine at the C1′ position and forms a protein-deoxyribose intermediate, which cleaves the N-glycosidic bond and releases free 8-oxoguanine. The AP-lyase activity of OGG1 then acts at the AP site to produce a single DNA strand break terminated with a 3′-phosphate group [4]. Several studies have observed that the AP lyases activity of OGG1 is not efficient compared to its glycosylase catalytic activity and can be replaced by more robust HhH enzymes such as NEIL1 [63]. Alkylating agents and antioxidants could upregulate the expression of OGG1 as well as the AP endonuclease APE1, which could enhance the AP lyase activity of OGG1 by several folds [62]. OGG1 is present in various isoforms with its prominent localization in the mitochondria, except for the 1a-isoform with a potent nuclear targeting signal. The observation that OGG1 is localized to both organelles underscores its importance in maintaining the stability of the mitochondrial and nuclear genome [64].
Recently, new findings revealed that the Cut like homeobox 1 gene, residing in a highly amplified chromosomal region in glioblastoma, encoding the CUX1 transcription factor, contains distinct domains referred to as CUT domains that serve as additional factors for the BER pathway [65, 66]. These CUT domains can interact and stimulate the activity of several enzymes, including OGG1 and APE1 and, more recently, the polymerase and 5′-dRP lyase activities of DNA polymerase β of the BER pathway. CUX1 knockdown in glioblastoma cells displayed sensitivity to temozolomide, which indirectly induces AP sites. This response correlates with diminishing OGG1 and APE1 activities. In contrast, glioblastoma cells expressing high levels of CUX1 are resistant to temozolomide, which correlates with increasing OGG1 and APE1 activities [66]. Both enzymes generate highly genotoxic DNA lesions, blocked 3′-ends, and DNA strand breaks. However, these toxic lesions apparently do not accumulate as the CUT domains also stimulate the activities of DNA polymerase β to ensure the completion of the BER pathway [65]. Besides the CUT domains, the histone deacetylase HDAC1 can interact and stimulate OGG1 activity [67]. Mice lacking HDAC1 showed enrichment of 8-oxoG lesions in the guanine-rich sequence present in the promoter regions of many downregulated genes in aged brains. Activation of HDAC1 decreased 8-OxoG lesions in the aged brain by stimulating OGG1 activity and improving cognitive deficiencies. This finding suggests a key role for OGG1 in forestalling brain aging and leading to neurodegeneration [67].
MYH/MUTYH is a monofunctional guanine mispaired-specific adenine–DNA glycosylase localized in both mitochondria and nucleus [68]. MUTHY is responsible for removing adenine and oxidized adenine such as 2-hydroxyadenine when misincorporated opposite to 7,8-dihydro-8-oxoguanine or 2,6-diamino-4-hydroxy-5-formamidopyrimidine (FaPyG); thereby preventing the accumulations of G:C to T:A mutations. MUTYH has an iron-sulfur center, like Ntg2 and NTH1. It is mainly upregulated in the S phase as well as by several proteins, including APE1, proliferating cell nuclear antigen (PCNA), and MSH2/6 mismatch repair complex [68]. So far, no AP lyase activity has been assigned to MUTYH, as shown for several other DNA glycosylases. In response to oxidative damage, MUTYH is recruited by the catalytic activity of the histone/protein deacetylase of SIRT6 to repair oxidative lesions on the telomeres [69].
Deleting the MUTYH gene causes a mutator phenotype as its deficiency elevates intestinal and lung cancer risk by accumulating 8-oxoguanine lesions [70]. Furthermore, the deletion of MYTH in mice manifested an elevated rate of mutation associated with spontaneous tumorigenesis [71]. More recently, genome-wide studies identified MUTYH as one of the additional significantly mutated genes in colorectal cancer. MUTYH was also identified as a pathogenic variant following whole-exome sequencing of germline DNA derived from women at high risk for hereditary breast and ovarian cancers and are not carriers of BRCA1, BRCA2, nor TP53 mutation [72]. MUTYH plays a critical role in preventing mutations that are the result of adenine mispairing with 8-oxoG lesions. Although orthologs of MUTYH are conserved in several eukaryotes, including S. pombe, a MUTYH-like homolog has not been reported in S. cerevisiae, C. elegans, or D. melanogaster [23, 73, 74].
MBD4, also known as MED1, is a nuclear monofunctional mismatch-specific DNA glycosylase with a HhH type DNA glycosylase domain and methyl-CpG-binding domain. The encoding gene of MBD4 has been detected in different organisms such as MIG in E. coli; however, it is not found in D. melanogaster or C. elegans [23, 73]. Mammalian MBD4 can recognize and excise uracil, 5-hydroxymethyluracil, thymine, and thymine oxidized derivatives (e.g., thymine glycol, 5-formyluracil) when mispaired to guanine. These lesions are usually caused by the spontaneous deamination of 5-methylcytosine (5-mC) and cytosine to thymine and uracil, which resulted in T:G or U:G mismatches, respectively, with preference to CpG dinucleotides [75]. Several other activities are associated with MBD4, such as modulating gene expression. However, the complete glycosylase functionality is not entirely characterized and requires further exploration. The deamination of 5mC remains the primary source of age-related somatic mutations. The mutation of the MBD4 gene in mammals has been found to promote the incidence of colorectal carcinoma induced by intestinal inflammation, which leads to microsatellite instability [76]. Additional studies revealed that the germline deficiency of MBD4 could enhance cancer susceptibility by predisposing affected individuals to acute myeloid leukemia, breast cancer, as well as uveal melanoma [77–79].
Endonuclease VIII-like DNA glycosylases superfamily
The endonuclease VIII-like enzymes NEIL1 (nei-like 1), NEIL2, and NEIL3 are bifunctional DNA glycosylases that belong to the Fpg/Nei superfamily of DNA glycosylases with the structural fold of hairpin-2-turn-hairpin [80]. They can remove a broad spectrum of oxidative lesions, many shared with the structurally distinct enzymes OGG1 and NTH1. The NEIL DNA glycosylases could efficiently remove damaged purines and pyrimidines such as faPyA, faPyG, spiroiminodihydantoin, guanidinohydantoin, hydroxyuracil, and urea. NEIL1 and NEIL2 are well-characterized in mammalian systems and seem to possess a unique preference towards oxidative lesions in bubble DNA structures over duplex DNA structures. BER initiated by NEIL1 or NEIL2 typically involves β, δ-elimination cleavage at the AP site to produce a one-base gap with a 3′-phosphate, which is not an effective substrate for DNA polymerases and, thus the 3′-end requires further processing [80]. NEIL1 is associated with replicative proteins and is involved in DNA repair during the S-phase [81]. NEIL2 is associated with RNA polymerase II and displayed preferential repair of oxidized DNA lesions in the transcribed strand. It prefers cytosine oxidation products and adopts a unique conformation upon binding to its substrate that is not observed for NEIL1 and NEIL3 [82]. NEIL3 could excise oxidized pyrimidines and formamidopyrimidines with a higher preference towards ssDNA and bubble DNA structure. This enzyme is also involved in processing specific types of DNA inter-strand cross-link lesions [83].
Despite their different substrate preference, NEILs, like other members of the Fpg/Nei family of DNGs, share several conserved structural motifs. Some of the most prominent motifs include a helix-two-turns-helix motif, a zinc finger motif, a N-terminal proline residue, as in NEIL1 and NEIL2 [80]. The zinc finger motif of NEIL1 lacks the appropriate residues and loops to coordinate a zinc atom; for this, it is identified as a "zinc-less-finger" motif. This motif contains a highly conserved Arg227 residue required for the DNG activity of NEIL1 [81]. In contrast, NEIL3 contains additional valine residues and two zinc-finger-Gly-Arg-Phe motifs (Zf-GRF) in its extreme C-terminal end to serve as a nucleophilic center [83]. These motifs promote interactions with the replication factor PCNA and the AP endonuclease APE1 and not APE2. The evidence to date suggests that NEIL3 Zf-GRF prevents APE1 activity on ssDNA to avoid excess strand breakage [84]. So far, neither the genome of C. elegans, D. melanogaster, nor S. cerevisiae encodes homologs of this family [6, 23, 73].
Methyl-purine glycosylases superfamily
The sole member of the alkylpurine-DNA-N-glycosylase (APNG), also known as methylpurine-DNA-N-glycosylase (MPG), superfamily is the 3-alkyladenine DNA glycosylase (AAG) or N-methylpurine DNA glycosylase (MDG) [85]. APNG can excise alkylated DNA base lesions including N3-methyladenine, N7-methylguanine, deaminated purines (e.g., oxanine, xanthine, hypoxanthine), and purine exocyclic adducts (e.g., 1, N6-ethenoadenine), as well as adenine opposite to cytosine to create an AP site. However, it has no activity towards oxidized DNA base lesions [86]. It can repair the methylation damage rapidly from either single- or double-stranded DNA. APNG is present in mitochondria and nuclei, and its regulation depends on the cell cycle [85]. High-level expression of APNG has been shown to be an independent prognostic factor for the overall survival of glioblastoma patients treated with the chemotherapeutic agent temozolomide [87]. This anticancer alkylating agent produces mainly N3-methyladenine and N7-methylguanine, nearly 80% of the total lesions. It is believed that the high expression of APNG, which lacks AP lyase activity, creates an abundance of toxic AP sites from the temozolomide-induced lesions that can be further processed to produce additional toxic lesions, such as DNA single-strand breaks [87].
During the repair process, APE1 and PCNA can interact with APNG, and apparently, this complex can enhance the turnover of the enzyme and the recruitment of XRCC1 to displace it from the AP site [85]. As such, it seems more likely that the accumulation of toxic AP sites might lead to lethality, as brain tumor cells with decreased levels of APE1 are sensitive to temozolomide [66]. APNG counterpart exists in other organisms such as the MAG1 enzymes in S. cerevisiae and S. pombe and alkA in E. coli. In general, these enzymes have different structural characteristics from the other three DNG families [86]. So far, the APNG ortholog has not been found in C. elegans nor D. melanogaster raising the question of how do these organisms process alkylation DNA damage [23, 73].
Apurinic/apyrimidinic endonucleases
The next step in the BER pathway after the action of the DNA glycosylase is the processing of the AP site by the main AP endonuclease enzymes, APE1/APEX1/Ref1/HAP1. In mammalian tissues, the AP site is a typical genomic lesion that accumulates in excess of 10,000 lesions within a day for a given cell [88]. AP sites are cytotoxic and carcinogenic, as well as lead to DNA–protein cross-links. Therefore, it is essential to prevent the accumulation of AP sites to maintain genome integrity. APE1 is regulated by phosphorylation, acetylation, and ubiquitylation. It is the principal AP endonuclease facilitating the removal of AP sites in eukaryotic cells as it performs almost 95% of the DNA repair activity [89]. This enzyme has a vital role in the proper functioning of the BER pathway to suppress carcinogenesis [90]. In addition to APE1, a second AP endonuclease APE2 exists in mammalian cells, and both are localized to the nucleus and mitochondria [91].
APE1 has a robust AP endonuclease activity and a weaker 3′- to 5′-exonuclease activity, while APE2 is a poor AP endonuclease activity, but displayed very strong 3′ to 5′-exonuclease and 3′-diesterase activities [88]. Recent evidence demonstrated that APE2 forms an interaction with PCNA, which stimulates APE2 3′-5′-exonuclease activity, and together the APE2-PCNA complex resects single-strand DNA breaks in the 3′ to 5′-direction creating a signal to activate the ATR-Chk1 DNA damage response pathway [92]. Another study revealed that downregulation of APE2, or FEN1, in either BRCA1- and BRCA2-deficient tumor cells caused synthetic lethality [93]. The authors proposed that the APE2-PCNA complex is required to process AP sites at replication forks, preventing replication-induced double-strand breaks at unrepaired AP sites that would necessitate the homologous recombination DNA repair functions, BRAC1 and BRAC2 [93]. Thus, in the absence of APE2 and BRAC2, cells cannot process the AP sites or the DNA strand breaks arising from stalled replication at AP sites leading to the lethal phenotype. It is not clear why the predominant AP endonuclease activity of APE1 cannot substitute for APE2 deficiency to avoid its synthetic lethality with BRAC2. However, the study suggests that APE2 might be a valuable chemotherapeutic target against refractory and metastatic BRCA-inactivated cancer cells in combination with PARP inhibitors.
APE1, and not APE2, has a nucleotide incision repair (NIR) activity that can cleave directly on the 5′-side of several oxidized base lesions such as 5,6-dihydro-2′-deoxyuridine and alpha-2′-deoxynucleosides to produce a 3′-hydroxyl group and leaving the 5′-end with the oxidized base that is then removed by the FEN1 (flap structure-specific endonuclease 1) [89, 94]. Besides APE1 roles in processing damaged single- and double-stranded DNA, it is also endowed with the ability to: (1) bind and cleave AP site on single-stranded RNA and to process microRNA such as miR-221/222 to regulate gene expression [95, 96], (2) utilize its redox cysteine Cys65 to reduce several transcription factors such as STAT3, AP-1, p53, and NF-κB to activate their transcriptional functions, and (3) interact with many proteins via its N-terminal 1–127 residues [97, 98]. One notable APE1 interacting proteins is nucleophosmin (NPM1), a nucleolar protein that performs many roles, such as ribosome biogenesis [99]. A variant of NPM1, NPM1c + , which causes the protein to relocalize to the cytoplasm, is associated with nearly a third of acute myeloid leukemia. This NPM1 variant also caused cytoplasmic localization of APE1 and leading to defects in BER [99].
Several factors, including XRCC1, RAD9-RAD1-HUS1 checkpoint complex, and HSP70, can potentially enhance the activity of APE1 and modulate its function [100]. SIRT1 and SIRT6, members of a sirtuin protein family, can also regulate APE1 activity and other BER enzymes such as MUTYH, OGG1, NEIL1, NEIL2, and LIG3 through their deacetylation activity [100]. Proteins that interact with APE1 can also control its redox state. For example, Peroxiredoxin I, PRDX1, which functions to decompose H2O2 and serves as a chaperone to protect proteins from oxidation-induced destabilization, can sequester APE1 from activating NFkB to prevent stimulated expression of pro-inflammatory genes, such as IL-8 [97]. Recent studies have shown that PRDX1 is bound to the telomere likely to prevent the accumulation of H2O2 that can cause oxidative DNA lesions [101]. Alternatively, PRDX1 could recruit APE1 to process the AP sites left following NEIL3 removal of spontaneous 8-oxoguanine lesions produced in the repetitive TTAGGG sequence in the telomere [102].
APE1-functionally related AP endonucleases exist in other species. In C. elegans, two genes apn-1 and exo-3, have been isolated and characterized. These encode the AP endonuclease APN-1 and EXO-3 that share 44% and 27% identity with the E. coli endonuclease IV and exonuclease III, respectively [103]. Although C. elegans EXO-3 shares a significant identity with APE1, it lacks the NIR activity, but this activity is inherited by the C. elegans APN-1 [103]. The deletion of both apn-1 and exo-3 genes is not lethal, but the resulting animals showed decreased progeny size [32]. The observation that the double deletion animal is not lethal may indicate that C. elegans possesses an additional enzyme(s) to combat AP sites [32]. In D. melanogaster, only a single AP endonuclease called the recombination repair protein 1 (Rrp1) exists. Rrp1 is an orthologue of APE1, but it also encompasses another function involved in recombinational DNA repair [104]. Although D. melanogaster does not appear to possess a member of the endo IV family of AP endonucleases, it contains the ribosomal protein S3 that can cleave AP sites and serve as the auxiliary enzyme [105]. We have recently isolated the related S3 protein from C. elegans and are in the process of determining if it can function as an AP endonuclease.
As in E. coli and C. elegans, S. cerevisiae possesses two well-characterized AP endonucleases, Apn1 and Apn2 [106]. A striking difference is noted in the structure and function of the major AP endonucleases in humans and yeast. The E. coli endonuclease IV counterpart Apn1 is responsible for nearly 95% of all AP endonuclease and 3’-diesterase activities in yeast. In contrast, the E. coli exonuclease III counterpart in human cells APE1 accounts for most AP endonuclease activity, but this enzyme has a weaker 3′-diesterase [106]. The yeast Apn2 also possesses strong AP endonuclease and 3′-diesterase activities, yet defects in these enzymes lead to different phenotypes. For example, yeast mutants lacking Apn1 are very sensitive to methyl methanesulfonate, which indirectly produces AP sites, but not yeast mutants lacking Apn2. Yeast lacking Apn1 and Apn2 are viable, while deletion of APE1 leads to embryonic lethality [107].
DNA polymerases/lyases
The multifunctional enzyme DNA polymerase β is recruited into the BER process following the AP endonuclease activity [108]. DNA polymerase β has two essential catalytic functions (1) the dRP lyase, residing within the 8 kDa N-terminal domain, which catalyzes the removal of 5′-dRP leaving a 5′-phosphate [16], and (2) the magnesium-dependent nucleotidyl-transferase activity of the 31 kDa C-terminal domain that inserts a single nucleotide at the 3′-hydroxyl end to replace the damaged base [109]. Removal of the 5′-dRP occurs significantly fast, such that it allows the polymerase to efficiently incorporate the correct complementary nucleotide [108]. DNA synthesis activity of the polymerase β can fill the one-base gap left after cleavage of the AP site, in a process known as a Short Patch Repair (Fig. 3). This one base gap filling will result in a nicked DNA or short (nt-1) gapped-site that will be efficiently ligated to restore the intact DNA by the gap-filling activity of polymerase β and DNA ligase [109, 110]. Mutations such as Arg137Glu that lower the polymerase β activity led to embryonic abnormalities in mice and a range of additional phenotypes, including the accumulation of DNA double-strand breaks, increased apoptosis, and sensitivity to methyl methanesulfonate (MMS), as well as enhance tumor progression and migration [111, 112].
It is noteworthy that the 5′-dRP left at the AP site can also be removed by other protein activities, such as the 5′-flap endonuclease activity of FEN1 that has the ability to create the gap to stimulate the DNA polymerase activity as observed in vitro [16, 113]. In this mode of repair, termed Long Patch Repair, the replicative DNA polymerase β and/or possibly polymerases δ and ε in conjunction with PCNA and the replication factor RFC insert multiple nucleotides at the single nucleotide gap, at least 2 to 11 nucleotides (Fig. 3) [113]. At the same time, the 5′-dRP strand is displaced in the form of a flap, which is then cleaved by FEN1 [16]. An alternative to this Long Patch Repair process is unwinding the nicked DNA in the 3′- to 5′-direction by the DNA helicase RECQ1 requiring the roles of the endonuclease ERCC1-XPF, PARP1 (poly (ADP-ribose) polymerase-1), and RPA leading to the incision of the flap. Thus, human cells have developed multiple ways to process base lesions via the BER pathway [114].
So far, no ortholog of DNA polymerase β has been found in the metazoan invertebrate genomes, including C. elegans, D. melanogaster, and few other insects such as Diptera [73, 115]. It is possible that the alternative PCNA-dependent pathway, including the use of the FEN1-like enzyme, could be operational in the BER pathway in these organisms [116]. In D. melanogaster, the 5′-dRP moieties at AP sites generated by AP endonuclease are removed through the FEN1-like flap enzymes (CG8648 and CG10670), suggesting that there is no requirement for the 5′-dRPase β-lyase activity of polymerase β [73]. In C. elegans, POLQ-1 has been identified as the DNA polymerase required to insert a single nucleotide in the BER pathway [115]. In the absence of any treatment, C. elegans polq-1 mutants showed elevated spontaneous germ cell apoptosis as in the case of polymerase deficient mice [32]. The C. elegans POLQ-1 is functionally related to the human DNA polymerase theta, which is responsible for repairing DNA double-strand breaks in the alternative NHEJ pathway. In fact, C. elegans POLQ-1 deficient mutants are sensitive to aristolochic acid, a genotoxic food contaminant that generates DNA double-strand breaks [117].
Computational analysis revealed that the S. cerevisiae has two DNA polymerases of the X family, DNA polymerase IV (pol IV), the homolog of human polymerase β, and TRF4. Yeast pol IV can fill short gaps and extend the primer termini with a high rate of errors [118]. In addition, pol IV activity is enhanced by AP sites, and it has been shown to possess an intrinsic 5′-dRP lyase activity, as human polymerase β [16, 118]. The evidence to date indicates that pol IV has an established role in the yeast non-homologous end-joining pathway to repair DNA double-strand breaks. Mutants lacking pol IV showed mild or no sensitivity to the classical monofunctional alkylating agent methylmethane sulfonate used to monitor defects in the BER pathway, excluding a major role for pol IV in this pathway [16]. Instead, the DNA resynthesis in BER is performed mainly by the yeast DNA polymerase ε and δ, homologs of the human polymerase ε and δ, with a possible role also for polymerase α in the pathway [119].
It is noteworthy that the 5′-end left by the AP endonucleases Apn1 and Apn2 is primarily removed by the Rad27/FEN1 flap endonuclease [120]. The lack of the 5′-dRP removal can lead to abortive ligation as the ligase catalyzes the adenylation of the 5′-dRP to produce a (5′-AMP)-dRP group [110]. This abortive ligation product can also be formed by inserting mismatched or oxidized nucleotides into a DNA gap by DNA pol IV. While yeast can remove the 5′-AMP-dRP using Rad27, the Hnt3 enzyme, an ortholog of human aprataxin (APTX) can also excise this 5′-block [121]. Since pol IV also possesses a 5′-dRP lyase activity, it is believed that it is part of a larger family of enzymes that act to remove the 5′-AMP-dRP group from the BER intermediate [120].
DNA ligases
The base excision repair culminates through a DNA ligase action that seals the break in the DNA backbone by inducing a phosphodiester bond formation between the 3′-hydroxyl end of the new nucleotide and the corresponding 5′-phosphate terminus [122]. Further, this enzyme is also essential in the DNA replication of the Okazaki fragments [123]. DNA ligase III (LIG3) is responsible for sealing the nick following the single nucleotide insertion by the Short Patch BER pathway. In contrast, DNA ligase I (LIG1) is operational in the Long Patch pathway [114]. However, LIG3 is the sole enzyme in the mitochondria [122, 124]. Both DNA LIG1 and LIG3 are associated with the presence of a nuclear scaffold protein X-ray repair cross-complementing protein 1 (XRCC1) and used ATP to activate the 5′-phosphate by the addition of adenosine monophosphate and releasing of the pyrophosphate [122]. The roles of these ligases are not limited to the BER pathway, as they performed functions in other DNA repair pathways, such as the DNA end-joining pathway during the class switching recombination [125, 126]. A third DNA ligase, LIG4, is essentially involved in the non-homologous end-joining and has no function in the BER pathway [127].
The insertion of mismatched or oxidized nucleotides into a DNA gap by DNA polymerase β could directly lead to the sealing of the 3′-nicked with the defective base through DNA LIG3 [128]. Such incorrectly matched or damaged base pair will fail the nick ligation process through DNA LIG1, which would generate products of abortive ligation with a 5′-adenylated dRP group. The resulting 5′-AMP with modified nucleotides are highly cytotoxic, leading to DNA replication errors and DNA-strands break. As such, it is crucial to correct the 5′-AMP BER lesions to maintain DNA and cell stability. Different DNA enzymes are involved in the repairing of the 5′-AMP abortive products that generate a 5′-phosphate terminal at the DNA break and enhance the ligation process [128]. These enzymes include FEN1 and the protein partner of XRCC1, APTX. Thus, APTX has the potential to prevent the abortive events of DNA ligation from inhibiting the Short Patch Repair process [129, 130]. In general, the mechanism of the ligases involved three steps (1) adenylation at the lysine active site, (2) transfer of the adenylyl group to the 5′- phosphate end, and (3) attacking the 3′-hydroxyl group to ligate the DNA ends and remove the AMP group [122].
The DNA ligase responsible for completing the BER process in C. elegans is controversial, although the animal possesses LIG1 and LIG4 and lacks the XRCC1 scaffold protein to coordinate the ligation process [131]. However, in D. melanogaster, it appears that ligation of the nicked DNA is performed by a complex of proteins including XRCC4, DNA LIG4, and XLF (a factor involved in NHEJ)[132].
In S. cerevisiae, the CDC9 gene encodes the essential DNA ligase, LIG1, which plays a more general function in the mitochondrial and nuclear genome in sealing nicked DNA following the removal of the damaged base [133]. In contrast to DNA LIGI, which is conserved among all eukaryotes, DNA LIG3 is only present in vertebrates [133]. Knockout studies in vertebrate cells showed that DNA LIG3 acts as a backup polymerase upon loss of LIGI and rescues DNA replication via an alternative Okazaki fragment-ligation mechanism [134]. Thus, LIG1 and LIG3 might share selective functions.
Co-factor proteins
In addition to the proteins mentioned above, cofactors are also involved in the BER pathway and do not participate directly in the DNA processing reactions (Table 2). These molecules are considered accessory factors such as PARP and XRCC1, which provide a significant opportunity to modulate the BER reactions for clinical applications. These factors stabilize the DNA strands until the repair process is completed; however, the exact mechanism is still not fully understood [135]. In eukaryotes, PARP and XRCC1 are potentially involved in different cellular processes such as gene regulation and transcription, genome repair, cell signaling, and apoptosis [136]. These factors can modulate several DNA repair proteins such as APE1, polymerase β, SIRT1, and DNA LIG1 by protein–protein interactions with no detectable enzymatic activities (Fig. 3; Table 2) [137]. The deacetylation activity of SIRT1 maintains PARP1 function through its binding protein DBC1. Both complexes, SIRT1/DBC1 and PARP1/DBC1 can be modulated by nicotinamide adenine dinucleotide (NAD+) levels [138]. Interestingly, the involvement of these cofactors in the BER pathway is highly specific; for instance, pyrimidine base damage requires XRCC1 activity but does not require PARP [136].
In humans, 17 proteins in the PARP family share similarities to PARP1 [137]. The PARP proteins have essential roles in guiding the repair of DNA lesions and preventing cellular apoptosis caused by cytotoxic DNA-strand breaks. PARP1 can abruptly detect the strand breaks and activate its poly-(ADP) ribosylation activity to transfer the ADP-ribose from the redox cofactor NAD+ to itself and other target proteins, such as PARP2 in the nuclei [138]. Furthermore, PARP1 could direct, for example, the repair of 8-oxoguanine lesions via the Long-Patch BER pathway [136]. In Long Patch repair, the activities of FEN1 and polymerase β can be inhibited by PARP1, but it has a minor impact on Short Patch BER, although it can compete for AP sites and inhibit APE1 activity. It is noteworthy that PARP1 may also exhibit abasic and dRP lyase actions upon the deficiency of APE1 as a backup excision repair mechanism [139]. PARP2 is another protein of the PARP family that has a crucial role in preventing mutagenesis, particularly in the PARylation of polynucleotide kinase (PNKP) and XRCC1 [140]. Recent evidence indicates that PARP1 and PAPR2 perform distinct roles to regulate the base-excision DNA repair process [140, 141]. For example, while PARP1 plays a more significant role in the upstream part of the BER pathway, such as stimulating the release of APE1, PARP2 enhances the downstream components such as LIG3 [140, 141]. Mice lacking PARP1 have a compromised DNA repair process, and the deletion of both PARP1 and PARP2 delays the ligation of DNA-strand breaks, resulting in a lethal phenotype [140]. Another study has found that the deficiency of both PARP1 and PARP2 proteins improved the efficacy of cytotoxic drugs and reduced the tumor burden [142].
The cytotoxic effect of many chemotherapies could stimulate and upregulate the protein expression of PARP1 in both normal and malignant mammalian cells [143]. Besides BER, PARP1 also has roles in other DNA repair pathways, including single- and double-strand break repair and nucleotide excision repair [144]. Presently, inhibitors of PARP1 are being exploited to use as single or in combination with chemotherapeutic agents for treating various cancers, including breast and ovarian [145]. For instance, the Food and Drug Administration has approved the use of PARP inhibitors in cases of BRCA1 and BRCA2 mutations and metastatic breast cancer due to the potent synthetic lethal effect [146]. Thus, defining the exact mechanism of the PARP family in DNA repair may help develop highly selective PARP-targeted medications that could treat the tumor within its microenvironment.
Two nematode PARP factors have been found, known as PME1 and PME2 [147]. PME-1 and PME-2 share 31 and 24% identity at the amino acid level with the human PARP1 and PARP2, respectively. PME-1 has a C-terminal with the same basic structure of PARP as well as an N-terminal that contains two zinc fingers. PME knockout C. elegans are sensitive to cisplatin and ionizing radiation, but not to manganese-induced oxidative DNA lesions, as assessed by growth and survival rates [147–149]. The D. melanogaster genome contains only one gene, PARPS-1, encoding a homolog of human PARP1 [150]. It has similar conserved domains as the human PARP1 and performs roles in DNA repair, regulates the nuclear chromatin structure, and maintains the organism survival [138]. In the absence of PARPS-1, the expression of at least 600 genes is altered, suggesting the importance of PARPS-1 function in various physiological pathways [151]. The variation observed in BER components between yeast and humans is also expected to influence the nature of the interactions during the process. Indeed, a significant distinction between yeast and human BER lies in the absence of a PARP to exert regulatory control on the BER pathway in yeast [152].
In the case of the BER cofactor XRCC1, no enzymatic activity has been assigned to this protein. However, it plays a role in coordinating components of the BER pathway [153]. In the Short Patch Repair pathway, the human XRCC1 forms a complex with DNA polymerase β and LIG3 to facilitate enzyme ligation after the inserted base [154]. In the Long Patch Repair pathway, a complex consisting of LIG1, PCNA, and XRCC1 is responsible for sealing the DNA-strand breaks that follow the action of the DNA polymerase and FEN1. At DNA single-strand break, PARP1 and or PARP2 can recognize the lesion and recruit XRCC1 [143]. This nuclear protein stabilizes and enhances various BER components in mammalian cells. XRCC1 can generate protein–protein interactions with UNG2, NTH1, NEIL1, NEIL2, OGG1, PNKP, MPG, APE1, and DNA polymerase β, using three distinct domains, a N-terminal and two independent BRCT domains (Fig. 3; Tables 1, 2). These XRCC1-complexes serve to coordinately regulate the activity of the BER proteins [155].
The downregulation of the XRCC1 gene during the early development of mice resulted in embryonic mortality [156]. Genetic mutation in XRCC1 can enhance the risk of tumorigeneses such as head, neck, esophageal, and breast cancers and decrease the survival rate [157, 158]. The deficiency of XRCC1 showed elevated levels of DNA strand lesions and brain tissue damage associated with the death of mouse embryos [159]. Furthermore, genomic deletion of XRCC1 displayed sensitivities to various genotoxic agents in eukaryotes [160, 161]. However, there is still a need to study the cellular response of PARP1 and XRCC1 deficiency to DNA single strand break and different chemotherapies. So far, the gene ortholog of the scaffold protein XRCC1 has not been found in C. elegans nor D. melanogaster, and it is not clear how these organisms coordinate the BER process [73, 131]. On the other hand, XRCC1 of the fission yeast S. pombe appears to play a more prominent role in cell cycle regulation than DNA repair, and S. cerevisiae seems to lack XRCC1 [162, 163]. It remains unclear how the final steps of BER are coordinated in yeast in the absence of XRCC1.
BER pathway and disease
The disruption and deregulation of the DNA damage tolerance and response and defects in DNA repair reactions promote the genomic instability that leads to various diseases. Oxidative stress-induced lesions or polymorphisms in genes encoding BER proteins are associated with the progression of many pathologies, including neuropathogenesis (e.g., Parkinson’s, Huntington’s, and Alzheimer’s diseases), as well as cardiovascular diseases, diabetes, and cancers (Tables 1, 2) [164]. Globally, cancer is the second major cause of mortality and is responsible for an estimated 10 million cancer deaths in 2020 [165]. Indeed, genetic analyses of these patients detected the presence of base lesions that are characteristics of defects and altered regulation of the BER process [164]. Genetic polymorphisms of MPG could enhance the progression of different pathologies, mainly ischemic stroke, lung cancer, and inflammatory diseases [166, 167]. Mice devoid of the MPG DNA glycosylase showed high levels of DNA lesions with an elevated probability of intestinal cancer under oxidative conditions [168]. In addition, a comprehensive genotyping analysis uncovered a significant association between cervical squamous cell carcinoma and several SMUG1 genetic polymorphisms, including rs3087404(A/G) and rs2029167(A/G) [169]. This association could help predict the occurrence of several types of cervical carcinomas, such as cervical intraepithelial neoplasia.
The deficiency of OGG1, NEIL1-3, APE1, APE2, MDB4, and MUTYH was found to promote the development and progression of neurodegenerative disorders, aging, depression, as well as many types of tumors, including colon, bladder, liver, head, neck, breast, kidney, and lung [170–173]. For instance, breast cancer has been observed to be associated with mutation of various excision proteins, such as APE1, APE2, POL β, XRCC1, and PARP1. Resistance to chemotherapies and tumor progression has been associated with the upregulation of these proteins [174]. Patients with Alzheimer, Parkinson, rheumatoid arthritis, immunological dysfunction, and intestinal inflammatory diseases are predicted to have a low level of UNG, OGG1, MUTYH, FEN1, PARP1, and XRCC1 proteins with an elevated level of 8-oxoguanine lesion, underscoring the possibility of using OGG1 and 8-oxoguanine lesions as a diagnostic tool for these diseases [175–179]. Recently, studies have reported a strong association between TDG function and the incidence of many tumorigeneses including lung, breast, liver, and thyroid cancers [180–182]. In addition, the inactivation of TDG protein promotes the development of hepatic cancer, which is associated with bile acid accumulation and diabetic status [181]. The deficiencies or genetic variants in NTHL1 glycosylase protein are associated with aging and many types of cancer, such as invasive ductal carcinoma bladder, intestinal, and squamous cell carcinomas [59, 168]. Likewise, the altered regulation of LIG1 and LIG3 is associated with tumor proliferation [183].
The protein levels of PARP1 are elevated in several pathophysiological conditions such as human cancers and inflammatory disorders. Hence, the suppression of PARP1 levels can enhance the sensitivity of cells towards radiotherapy and chemotherapy by inhibiting the correction of DNA-stand breaks and base lesions [184]. A study performed in mice under atherosclerosis conditions showed that the cerebral infarct volume directly relates to the PARP activity and that inhibiting its activity can significantly decrease arterial inflammation [185]. As observed for many BER proteins, mutations of the vital scaffold factor, XRCC1, are associated with an elevated risk of breast cancer and cancer progression [186]. In addition, XRCC1 has been recently shown to govern the repair of DNA single-strand breaks at specific sites such as enhancers and demethylated regions in the genome of neurons. It is believed that defects in XRCC1 or its protein partners PNKP and APTX lead to the accumulation of DNA single-strand breaks in neurons and, as such, are the cause of neurodegenerative diseases [187, 188].
APE1 also plays a central role in combating the onset and progression of tumors by regulating the oxidative stress response and genes involved in chemoresistance [10]. The abnormal cytoplasmic localization of APE1 and its strongly interacting partner NPM1 is linked to tumor progression and chemoresistance of high-grade serous ovarian cancer [189]. There is growing evidence that APE1 can be used as a specific biomarker and a potential treatment target for various pathologies by disrupting its interactions with other partners [10]. In addition to APE1, variants of DNA polymerase β serve as specific base mutators leading to genomic instability, mutagenesis, aging, and neurodegenerative diseases. It is noteworthy that at least 30% of colorectal cancer patients have mutations within the DNA polymerase β gene [190, 191]. Furthermore, this impaired DNA enzyme has been associated with different solid cancers, enhanced cell apoptosis, and higher sensitivity to chemotherapy [192]. In general, the BER process is crucial to suppress genomic instability, thereby preventing numerous anomalies, including the onset and progression of tumors and age-related neurodegenerative disorders [10, 172].
Conclusions and future directions
The genomic and biochemical analyses of the BER process have progressed tremendously since the first discovery of the uracil DNA glycosylase by Lindahl in 1974. The BER repair process is carried out by the handover of essential proteins to the next that together function in coordination to maintain DNA stability, which otherwise would lead to various diseases. Coordination of these steps is obtained through an overlapping interaction of enzyme-product and protein–protein that maintains the intermediates flow during the repair process. Several aspects of the pathway remain poorly understood; for example, how are the enzymes (1) precisely coordinated at the DNA lesion intermediates? (2) regulated to promote the association and disassociation at the DNA lesion site? (3) maintained following the completion of the repair process? Moreover, (4) reassembled before reengaging with the removal of the subsequent DNA lesion? Besides these shortcomings in the BER mechanism, other advancements in the genomic and precision medicine field have led to enormous contributions whereby single nucleotide polymorphisms within the BER genes are directly linked to specific types of diseases. However, it remains a challenge to correct these BER gene mutations by gene therapy to forestall disease initiation and progression. The ease of genetic manipulation of some eukaryotic organisms such as S. cerevisiae, C. elegans, and D. melanogaster has extensively revealed complementary functions of the BER enzymes. The repair mechanisms identified in these organisms were intuitive in understanding the role of the conserved BER process. However, it is noteworthy that D. melanogaster and C. elegans lack some proteins known to be essential in human cells. We believe that both D. melanogaster and C. elegans could provide new insights into the alternative mechanism(s) of processing base lesions that may help unravel auxiliary pathways in human cells.
Acknowledgements
We thank the College of Health and Life Sciences, Hamad Bin Khalifa University, for providing scholarships to both N.N.H and N.E. The schematic representations were created using Biorender.com.
Author contributions
NNH, wrote the entire manuscript using a draft provided by NE; NNH, prepared the Tables and Figures and completed the reference list; DR, extensively revised the complete manuscript and both DR and NNH finalized the manuscript.
Funding
Qatar Foundation to the College of Health and Life Sciences, Hamad Bin Khalifa University, Education City, Doha, Qatar.
Declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
All authors gave their consent for publication.
Availability of data and material
Not applicable.
Conflict of interest
All the authors declared that there are no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Tubbs A, Nussenzweig A. Endogenous DNA damage as a source of genomic instability in cancer. Cell. 2017;168(4):644–656. doi: 10.1016/j.cell.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matkarimov BT, Saparbaev MK. DNA repair and mutagenesis in vertebrate mitochondria: evidence for asymmetric DNA strand inheritance. Adv Exp Med Biol. 2020;1241:77–100. doi: 10.1007/978-3-030-41283-8_6. [DOI] [PubMed] [Google Scholar]
- 3.Tremblay S, Wagner JR. Dehydration, deamination and enzymatic repair of cytosine glycols from oxidized poly(dG-dC) and poly(dI-dC) Nucleic Acids Res. 2008;36(1):284–293. doi: 10.1093/nar/gkm1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boiteux S, Coste F, Castaing B. Repair of 8-oxo-7,8-dihydroguanine in prokaryotic and eukaryotic cells: Properties and biological roles of the Fpg and OGG1 DNA N-glycosylases. Free Radic Biol Med. 2017;107:179–201. doi: 10.1016/j.freeradbiomed.2016.11.042. [DOI] [PubMed] [Google Scholar]
- 5.Slyskova J, et al. Base and nucleotide excision repair facilitate resolution of platinum drugs-induced transcription blockage. Nucleic Acids Res. 2018;46(18):9537–9549. doi: 10.1093/nar/gky764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bauer NC, Corbett AH, Doetsch PW. The current state of eukaryotic DNA base damage and repair. Nucleic Acids Res. 2015;43(21):10083–10101. doi: 10.1093/nar/gkv1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chatterjee N, Walker GC. Mechanisms of DNA damage, repair, and mutagenesis. Environ Mol Mutagen. 2017;58(5):235–263. doi: 10.1002/em.22087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moor NA, Lavrik OI. Protein–protein interactions in DNA base excision repair. Biochemistry (Mosc) 2018;83(4):411–422. doi: 10.1134/S0006297918040120. [DOI] [PubMed] [Google Scholar]
- 9.Beard WA, et al. Eukaryotic base excision repair: new approaches shine light on mechanism. Annu Rev Biochem. 2019;88:137–162. doi: 10.1146/annurev-biochem-013118-111315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ayyildiz D, et al. Architecture of the human ape1 interactome defines novel cancers signatures. Sci Rep. 2020;10(1):28. doi: 10.1038/s41598-019-56981-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boiteux S, Jinks-Robertson S. DNA repair mechanisms and the bypass of DNA damage in Saccharomyces cerevisiae. Genetics. 2013;193(4):1025–1064. doi: 10.1534/genetics.112.145219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ljungquist S, Andersson A, Lindahl T. A mammalian endonuclease specific for apurinic sites in double-stranded deoxyribonucleic acid. II. Further studies on the substrate specificity. J Biol Chem. 1974;249(5):1536–1540. doi: 10.1016/S0021-9258(19)42916-5. [DOI] [PubMed] [Google Scholar]
- 13.Muench KF, Misra RP, Humayun MZ. Sequence specificity in aflatoxin B1–DNA interactions. Proc Natl Acad Sci USA. 1983;80(1):6–10. doi: 10.1073/pnas.80.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362(6422):709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 15.Ruf A, de Murcia G, Schulz GE. Inhibitor and NAD+ binding to poly(ADP-ribose) polymerase as derived from crystal structures and homology modeling. Biochemistry. 1998;37(11):3893–3900. doi: 10.1021/bi972383s. [DOI] [PubMed] [Google Scholar]
- 16.Sobol RW, et al. The lyase activity of the DNA repair protein beta-polymerase protects from DNA-damage-induced cytotoxicity. Nature. 2000;405(6788):807–810. doi: 10.1038/35015598. [DOI] [PubMed] [Google Scholar]
- 17.Mitra S, et al. Choreography of oxidative damage repair in mammalian genomes. Free Radic Biol Med. 2002;33(1):15–28. doi: 10.1016/S0891-5849(02)00819-5. [DOI] [PubMed] [Google Scholar]
- 18.Caldecott KW. Mammalian DNA base excision repair: Dancing in the moonlight. DNA Repair (Amst) 2020;93:102921. doi: 10.1016/j.dnarep.2020.102921. [DOI] [PubMed] [Google Scholar]
- 19.Drohat AC, Coey CT. Role of base excision "Repair" enzymes in erasing epigenetic marks from DNA. Chem Rev. 2016;116(20):12711–12729. doi: 10.1021/acs.chemrev.6b00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boldinova EO, et al. Isoforms of base excision repair enzymes produced by alternative splicing. Int J Mol Sci. 2019;20(13):3279. doi: 10.3390/ijms20133279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Demple B, Harrison L. Repair of oxidative damage to DNA: enzymology and biology. Annu Rev Biochem. 1994;63:915–948. doi: 10.1146/annurev.bi.63.070194.004411. [DOI] [PubMed] [Google Scholar]
- 22.Muha V, et al. Uracil-containing DNA in Drosophila: stability, stage-specific accumulation, and developmental involvement. PLoS Genet. 2012;8(6):e1002738. doi: 10.1371/journal.pgen.1002738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elsakrmy N, Zhang-Akiyama Q-M, Ramotar D (2020) The base excision repair pathway in the nematode caenorhabditis elegans. Front Cell Dev Biol 8(1447) [DOI] [PMC free article] [PubMed]
- 24.Lucas-Lledo JI, Maddamsetti R, Lynch M. Phylogenomic analysis of the uracil-DNA glycosylase superfamily. Mol Biol Evol. 2011;28(3):1307–1317. doi: 10.1093/molbev/msq318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobs AL, Schar P. DNA glycosylases: in DNA repair and beyond. Chromosoma. 2012;121(1):1–20. doi: 10.1007/s00412-011-0347-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarno A, et al. Uracil-DNA glycosylase UNG1 isoform variant supports class switch recombination and repairs nuclear genomic uracil. Nucleic Acids Res. 2019;47(9):4569–4585. doi: 10.1093/nar/gkz145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pettersen HS, et al. Uracil-DNA glycosylases SMUG1 and UNG2 coordinate the initial steps of base excision repair by distinct mechanisms. Nucleic Acids Res. 2007;35(12):3879–3892. doi: 10.1093/nar/gkm372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elateri I, et al. hSMUG1 can functionally compensate for Ung1 in the yeast Saccharomyces cerevisiae. DNA Repair (Amst) 2003;2(3):315–323. doi: 10.1016/S1568-7864(02)00221-5. [DOI] [PubMed] [Google Scholar]
- 29.Schormann N, Ricciardi R, Chattopadhyay D. Uracil-DNA glycosylases-structural and functional perspectives on an essential family of DNA repair enzymes. Protein Sci. 2014;23(12):1667–1685. doi: 10.1002/pro.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weiser BP, et al. N-terminal domain of human uracil DNA glycosylase (hUNG2) promotes targeting to uracil sites adjacent to ssDNA-dsDNA junctions. Nucleic Acids Res. 2018;46(14):7169–7178. doi: 10.1093/nar/gky525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Z, et al. Hydrogen peroxide mediated mitochondrial UNG1-PRDX3 interaction and UNG1 degradation. Free Radic Biol Med. 2016;99:54–62. doi: 10.1016/j.freeradbiomed.2016.07.030. [DOI] [PubMed] [Google Scholar]
- 32.Papaluca A, et al. UNG-1 and APN-1 are the major enzymes to efficiently repair 5-hydroxymethyluracil DNA lesions in C elegans. Sci Rep. 2018;8(1):6860. doi: 10.1038/s41598-018-25124-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bekesi A, et al. A novel fruitfly protein under developmental control degrades uracil-DNA. Biochem Biophys Res Commun. 2007;355(3):643–648. doi: 10.1016/j.bbrc.2007.01.196. [DOI] [PubMed] [Google Scholar]
- 34.Kuznetsova AA, et al. Pre-steady-state kinetic analysis of damage recognition by human single-strand selective monofunctional uracil-DNA glycosylase SMUG1. Mol Biosyst. 2017;13(12):2638–2649. doi: 10.1039/C7MB00457E. [DOI] [PubMed] [Google Scholar]
- 35.An MJ et al (2021) Ablation of SMUG1 Reduces Cell Viability and Increases UVC-Mediated Apoptosis in Hepatocarcinoma HepG2 Cells. Genes (Basel) 12(2) [DOI] [PMC free article] [PubMed]
- 36.Kroustallaki P, et al. (2019) SMUG1 promotes telomere maintenance through telomerase RNA Processing. Cell Rep. 2019;28(7):1690–1702. doi: 10.1016/j.celrep.2019.07.040. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Z, et al. Structural basis of substrate specificity in geobacter metallireducens SMUG1. ACS Chem Biol. 2016;11(6):1729–1736. doi: 10.1021/acschembio.6b00164. [DOI] [PubMed] [Google Scholar]
- 38.Alsoe L, et al. Uracil accumulation and mutagenesis dominated by cytosine deamination in CpG dinucleotides in mice lacking UNG and SMUG1. Sci Rep. 2017;7(1):7199. doi: 10.1038/s41598-017-07314-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dingler FA, et al. Uracil excision by endogenous SMUG1 glycosylase promotes efficient Ig class switching and impacts on A: T substitutions during somatic mutation. Eur J Immunol. 2014;44(7):1925–1935. doi: 10.1002/eji.201444482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kemmerich K, et al. Germline ablation of SMUG1 DNA glycosylase causes loss of 5-hydroxymethyluracil- and UNG-backup uracil-excision activities and increases cancer predisposition of Ung-/-Msh2-/- mice. Nucleic Acids Res. 2012;40(13):6016–6025. doi: 10.1093/nar/gks259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abdel-Fatah TM, et al. Single-strand selective monofunctional uracil-DNA glycosylase (SMUG1) deficiency is linked to aggressive breast cancer and predicts response to adjuvant therapy. Breast Cancer Res Treat. 2013;142(3):515–527. doi: 10.1007/s10549-013-2769-6. [DOI] [PubMed] [Google Scholar]
- 42.Zhong J, et al. A Transcriptome-wide association study identifies novel candidate susceptibility genes for pancreatic cancer. J Natl Cancer Inst. 2020;112(10):1003–1012. doi: 10.1093/jnci/djz246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dodd T, et al. Uncovering universal rules governing the selectivity of the archetypal DNA glycosylase TDG. Proc Natl Acad Sci USA. 2018;115(23):5974–5979. doi: 10.1073/pnas.1803323115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Popov AV et al (2019) Reading targeted DNA damage in the active demethylation pathway: role of accessory domains of eukaryotic AP endonucleases and thymine-DNA glycosylases. J Mol Biol [DOI] [PubMed]
- 45.Pidugu LS, et al. Excision of 5-carboxylcytosine by thymine DNA glycosylase. J Am Chem Soc. 2019;141(47):18851–18861. doi: 10.1021/jacs.9b10376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bellacosa A, Drohat AC. Role of base excision repair in maintaining the genetic and epigenetic integrity of CpG sites. DNA Repair (Amst) 2015;32:33–42. doi: 10.1016/j.dnarep.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.DeNizio JE, et al. TET-TDG active DNA demethylation at CpG and non-CpG sites. J Mol Biol. 2021;433(8):166877. doi: 10.1016/j.jmb.2021.166877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deckard CE, Banerjee DR, Sczepanski JT. Chromatin structure and the pioneering transcription factor FOXA1 regulate TDG-mediated removal of 5-formylcytosine from DNA. J Am Chem Soc. 2019;141(36):14110–14114. doi: 10.1021/jacs.9b07576. [DOI] [PubMed] [Google Scholar]
- 49.Cortazar D, et al. Embryonic lethal phenotype reveals a function of TDG in maintaining epigenetic stability. Nature. 2011;470(7334):419–423. doi: 10.1038/nature09672. [DOI] [PubMed] [Google Scholar]
- 50.Hardeland U, et al. The versatile thymine DNA-glycosylase: a comparative characterization of the human, Drosophila and fission yeast orthologs. Nucleic Acids Res. 2003;31(9):2261–2271. doi: 10.1093/nar/gkg344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Porat N, et al. Direct detection of chicken genomic DNA for gender determination by thymine-DNA glycosylase. Br Poult Sci. 2011;52(1):58–65. doi: 10.1080/00071668.2010.549665. [DOI] [PubMed] [Google Scholar]
- 52.Williams, S.C. and J.L. Parsons, NTH1 Is a New Target for Ubiquitylation-Dependent Regulation by TRIM26 Required for the Cellular Response to Oxidative Stress. Mol Cell Biol, 2018. 38(12). [DOI] [PMC free article] [PubMed]
- 53.Denver DR, Swenson SL, Lynch M. An evolutionary analysis of the helix-hairpin-helix superfamily of DNA repair glycosylases. Mol Biol Evol. 2003;20(10):1603–1611. doi: 10.1093/molbev/msg177. [DOI] [PubMed] [Google Scholar]
- 54.Morinaga H, et al. Purification and characterization of Caenorhabditis elegans NTH, a homolog of human endonuclease III: essential role of N-terminal region. DNA Repair (Amst) 2009;8(7):844–851. doi: 10.1016/j.dnarep.2009.04.020. [DOI] [PubMed] [Google Scholar]
- 55.Stehling O, et al. MMS19 assembles iron-sulfur proteins required for DNA metabolism and genomic integrity. Science. 2012;337(6091):195–199. doi: 10.1126/science.1219723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shinmura K, et al. Defective repair capacity of variant proteins of the DNA glycosylase NTHL1 for 5-hydroxyuracil, an oxidation product of cytosine. Free Radic Biol Med. 2019;131:264–273. doi: 10.1016/j.freeradbiomed.2018.12.010. [DOI] [PubMed] [Google Scholar]
- 57.Swartzlander DB, et al. Identification of SUMO modification sites in the base excision repair protein, Ntg1. DNA Repair (Amst) 2016;48:51–62. doi: 10.1016/j.dnarep.2016.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gellon L, et al. Synergism between base excision repair, mediated by the DNA glycosylases Ntg1 and Ntg2, and nucleotide excision repair in the removal of oxidatively damaged DNA bases in Saccharomyces cerevisiae. Mol Genet Genomics. 2001;265(6):1087–1096. doi: 10.1007/s004380100507. [DOI] [PubMed] [Google Scholar]
- 59.Das, L., V.G. Quintana, and J.B. Sweasy, NTHL1 in genomic integrity, aging and cancer. DNA Repair (Amst), 2020. 93: p. 102920. [DOI] [PMC free article] [PubMed]
- 60.Lu J, Liu Y. Deletion of Ogg1 DNA glycosylase results in telomere base damage and length alteration in yeast. EMBO J. 2010;29(2):398–409. doi: 10.1038/emboj.2009.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yasukawa T, et al. Drosophila Ogg1 is required to suppress 8-oxo-guanine accumulation following oxidative stress. Genes Genet Syst. 2015;90(1):11–20. doi: 10.1266/ggs.90.11. [DOI] [PubMed] [Google Scholar]
- 62.Zhou X, et al. OGG1 is essential in oxidative stress induced DNA demethylation. Cell Signal. 2016;28(9):1163–1171. doi: 10.1016/j.cellsig.2016.05.021. [DOI] [PubMed] [Google Scholar]
- 63.Ferino, A. and L.E. Xodo, Effect of DNA Glycosylases OGG1 and Neil1 on Oxidized G-Rich Motif in the KRAS Promoter. Int J Mol Sci, 2021. 22(3). [DOI] [PMC free article] [PubMed]
- 64.Ogawa A, et al. Enzyme kinetics of an alternative splicing isoform of mitochondrial 8-oxoguanine DNA glycosylase, ogg1-1b, and compared with the nuclear ogg1-1a. J Biochem Mol Toxicol. 2015;29(2):49–56. doi: 10.1002/jbt.21605. [DOI] [PubMed] [Google Scholar]
- 65.Ramdzan, Z.M., et al., CUT Domains Stimulate Pol beta Enzymatic Activities to Accelerate Completion of Base Excision Repair. J Mol Biol, 2021. 433(4): p. 166806. [DOI] [PubMed]
- 66.Kaur S, et al. CUX1 stimulates APE1 enzymatic activity and increases the resistance of glioblastoma cells to the mono-alkylating agent temozolomide. Neuro Oncol. 2018;20(4):484–493. doi: 10.1093/neuonc/nox178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pao PC, et al. HDAC1 modulates OGG1-initiated oxidative DNA damage repair in the aging brain and Alzheimer's disease. Nat Commun. 2020;11(1):2484. doi: 10.1038/s41467-020-16361-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Koger N, et al. Analysis of MUTYH alternative transcript expression, promoter function, and the effect of human genetic variants. Hum Mutat. 2019;40(4):472–482. doi: 10.1002/humu.23709. [DOI] [PubMed] [Google Scholar]
- 69.Tan J, et al. An ordered assembly of MYH glycosylase, SIRT6 protein deacetylase, and Rad9-Rad1-Hus1 checkpoint clamp at oxidatively damaged telomeres. Aging (Albany NY) 2020;12(18):17761–17785. doi: 10.18632/aging.103934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thibodeau, M.L., et al., Base excision repair deficiency signatures implicate germline and somatic MUTYH aberrations in pancreatic ductal adenocarcinoma and breast cancer oncogenesis. Cold Spring Harb Mol Case Stud, 2019. 5(2). [DOI] [PMC free article] [PubMed]
- 71.Isoda T, et al. Abnormality in Wnt signaling is causatively associated with oxidative stress-induced intestinal tumorigenesis in MUTYH-null mice. Int J Biol Sci. 2014;10(8):940–947. doi: 10.7150/ijbs.9241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Felicio PS, et al. Whole-exome sequencing of non-BRCA1/BRCA2 mutation carrier cases at high-risk for hereditary breast/ovarian cancer. Hum Mutat. 2021;42(3):290–299. doi: 10.1002/humu.24158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sekelsky J. DNA Repair in Drosophila: Mutagens, Models, and Missing Genes. Genetics. 2017;205(2):471–490. doi: 10.1534/genetics.116.186759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ishchenko AA, et al. The 3'->5' exonuclease of Apn1 provides an alternative pathway to repair 7,8-dihydro-8-oxodeoxyguanosine in Saccharomyces cerevisiae. Mol Cell Biol. 2005;25(15):6380–6390. doi: 10.1128/MCB.25.15.6380-6390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yakovlev DA, et al. Search for Modified DNA Sites with the Human Methyl-CpG-Binding Enzyme MBD4. Acta Naturae. 2017;9(1):88–98. doi: 10.32607/20758251-2017-9-1-88-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yu AM, et al. The Mbd4 DNA glycosylase protects mice from inflammation-driven colon cancer and tissue injury. Oncotarget. 2016;7(19):28624–28636. doi: 10.18632/oncotarget.8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang Y, et al. Epigenetic silencing of RNF144A expression in breast cancer cells through promoter hypermethylation and MBD4. Cancer Med. 2018;7(4):1317–1325. doi: 10.1002/cam4.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Repo P, et al. Germline loss-of-function variants in MBD4 are rare in Finnish patients with uveal melanoma. Pigment Cell Melanoma Res. 2020;33(5):756–762. doi: 10.1111/pcmr.12892. [DOI] [PubMed] [Google Scholar]
- 79.Sanders MA, et al. MBD4 guards against methylation damage and germ line deficiency predisposes to clonal hematopoiesis and early-onset AML. Blood. 2018;132(14):1526–1534. doi: 10.1182/blood-2018-05-852566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fleming AM, Burrows CJ. Formation and processing of DNA damage substrates for the hNEIL enzymes. Free Radic Biol Med. 2017;107:35–52. doi: 10.1016/j.freeradbiomed.2016.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Albelazi, M.S., et al., The Biochemical Role of the Human NEIL1 and NEIL3 DNA Glycosylases on Model DNA Replication Forks. Genes (Basel), 2019. 10(4). [DOI] [PMC free article] [PubMed]
- 82.Eckenroth, B.E., et al., Unique Structural Features of Mammalian NEIL2 DNA Glycosylase Prime Its Activity for Diverse DNA Substrates and Environments. Structure, 2021. 29(1): p. 29–42 e4. [DOI] [PMC free article] [PubMed]
- 83.Li N, et al. Cooperation of the NEIL3 and Fanconi anemia/BRCA pathways in interstrand crosslink repair. Nucleic Acids Res. 2020;48(6):3014–3028. doi: 10.1093/nar/gkaa038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rodriguez AA, et al. An autoinhibitory role for the GRF zinc finger domain of DNA glycosylase NEIL3. J Biol Chem. 2020;295(46):15566–15575. doi: 10.1074/jbc.RA120.015541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fu D, Samson LD. Direct repair of 3, N(4)-ethenocytosine by the human ALKBH2 dioxygenase is blocked by the AAG/MPG glycosylase. DNA Repair (Amst) 2012;11(1):46–52. doi: 10.1016/j.dnarep.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Troll, C.J., et al., Interplay between base excision repair activity and toxicity of 3-methyladenine DNA glycosylases in an E coli complementation system. Mutat Res, 2014. 763: p. 64–73. [DOI] [PMC free article] [PubMed]
- 87.Fosmark S, et al. APNG as a prognostic marker in patients with glioblastoma. PLoS ONE. 2017;12(6):e0178693. doi: 10.1371/journal.pone.0178693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Maher RL, Wallace SS, Pederson DS. The lyase activity of bifunctional DNA glycosylases and the 3'-diesterase activity of APE1 contribute to the repair of oxidized bases in nucleosomes. Nucleic Acids Res. 2019;47(6):2922–2931. doi: 10.1093/nar/gky1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kuznetsova AA, Fedorova OS, Kuznetsov NA. Kinetic features of 3'-5' exonuclease activity of human AP-endonuclease APE1. Molecules. 2018;23(9):2101. doi: 10.3390/molecules23092101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ma X, et al. Downregulation of APE1 potentiates breast cancer cells to olaparib by inhibiting PARP-1 expression. Breast Cancer Res Treat. 2019;176(1):109–117. doi: 10.1007/s10549-019-05189-w. [DOI] [PubMed] [Google Scholar]
- 91.Kladova OA, et al. Modulation of the apurinic/apyrimidinic endonuclease activity of human APE1 and of its natural polymorphic variants by base excision repair proteins. Int J Mol Sci. 2020;21(19):7147. doi: 10.3390/ijms21197147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lin Y, et al. APE2 promotes DNA damage response pathway from a single-strand break. Nucleic Acids Res. 2018;46(5):2479–2494. doi: 10.1093/nar/gky020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mengwasser KE, et al. Genetic screens reveal FEN1 and APEX2 as BRCA2 synthetic lethal targets. Mol Cell. 2019;73(5):885–899. doi: 10.1016/j.molcel.2018.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gros L, et al. The major human AP endonuclease (Ape1) is involved in the nucleotide incision repair pathway. Nucleic Acids Res. 2004;32(1):73–81. doi: 10.1093/nar/gkh165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kuznetsova AA, et al. Effect of the substrate structure and metal ions on the hydrolysis of undamaged RNA by human AP endonuclease APE1. Acta Naturae. 2020;12(2):74–85. doi: 10.32607/actanaturae.10864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chohan M, et al. Human apurinic/apyrimidinic endonuclease 1 (APE1) has 3' RNA phosphatase and 3' exoribonuclease activities. J Mol Biol. 2015;427(2):298–311. doi: 10.1016/j.jmb.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 97.Nassour H, et al. Peroxiredoxin 1 interacts with and blocks the redox factor APE1 from activating interleukin-8 expression. Sci Rep. 2016;6:29389. doi: 10.1038/srep29389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tell G, Fantini D, Quadrifoglio F. Understanding different functions of mammalian AP endonuclease (APE1) as a promising tool for cancer treatment. Cell Mol Life Sci. 2010;67(21):3589–3608. doi: 10.1007/s00018-010-0486-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vascotto C, et al. Functional regulation of the apurinic/apyrimidinic endonuclease 1 by nucleophosmin: impact on tumor biology. Oncogene. 2015;34(26):3482. doi: 10.1038/onc.2015.191. [DOI] [PubMed] [Google Scholar]
- 100.Kabzinski J, et al. Sirt3 regulates the level of mitochondrial DNA repair activity through deacetylation of NEIL1, NEIL2, OGG1, MUTYH, APE1 and LIG3 in colorectal cancer. Pol Przegl Chir. 2019;92(1):1–4. doi: 10.5604/01.3001.0013.5539. [DOI] [PubMed] [Google Scholar]
- 101.Aeby E, et al. Peroxiredoxin 1 protects telomeres from oxidative damage and preserves telomeric DNA for extension by telomerase. Cell Rep. 2016;17(12):3107–3114. doi: 10.1016/j.celrep.2016.11.071. [DOI] [PubMed] [Google Scholar]
- 102.Zhou J, et al. NEIL3 repairs telomere damage during s phase to secure chromosome segregation at mitosis. Cell Rep. 2017;20(9):2044–2056. doi: 10.1016/j.celrep.2017.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shatilla A, et al. Identification of two apurinic/apyrimidinic endonucleases from Caenorhabditis elegans by cross-species complementation. DNA Repair (Amst) 2005;4(6):655–670. doi: 10.1016/j.dnarep.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 104.Sander M, Lowenhaupt K, Rich A. Drosophila Rrp1 protein: an apurinic endonuclease with homologous recombination activities. Proc Natl Acad Sci USA. 1991;88(15):6780–6784. doi: 10.1073/pnas.88.15.6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Graifer D, Karpova G. Ribosomal protein uS3 in cell biology and human disease: Latest insights and prospects. BioEssays. 2020;42(12):e200124. doi: 10.1002/bies.202000124. [DOI] [PubMed] [Google Scholar]
- 106.Ramotar D, et al. Cellular role of yeast Apn1 apurinic endonuclease/3'-diesterase: repair of oxidative and alkylation DNA damage and control of spontaneous mutation. Mol Cell Biol. 1991;11(9):4537–4544. doi: 10.1128/mcb.11.9.4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Johnson RE, et al. Identification of APN2, the Saccharomyces cerevisiae homolog of the major human AP endonuclease HAP1, and its role in the repair of abasic sites. Genes Dev. 1998;12(19):3137–3143. doi: 10.1101/gad.12.19.3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Beard WA. DNA polymerase beta: Closing the gap between structure and function. DNA Repair (Amst) 2020;93:102910. doi: 10.1016/j.dnarep.2020.102910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Karimi-Busheri F, et al. Repair of DNA strand gaps and nicks containing 3'-phosphate and 5'-hydroxyl termini by purified mammalian enzymes. Nucleic Acids Res. 1998;26(19):4395–4400. doi: 10.1093/nar/26.19.4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Whitaker AM, et al. Base excision repair of oxidative DNA damage: from mechanism to disease. Front Biosci (Landmark Ed) 2017;22:1493–1522. doi: 10.2741/4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhao S, et al. Mutation in DNA polymerase beta causes spontaneous chromosomal instability and inflammation-associated carcinogenesis in mice. Cancers (Basel) 2019;11(8):1160. doi: 10.3390/cancers11081160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang M, et al. DNA polymerase beta modulates cancer progression via enhancing CDH13 expression by promoter demethylation. Oncogene. 2020;39(33):5507–5519. doi: 10.1038/s41388-020-1386-1. [DOI] [PubMed] [Google Scholar]
- 113.Liu Y, et al. DNA polymerase beta and flap endonuclease 1 enzymatic specificities sustain DNA synthesis for long patch base excision repair. J Biol Chem. 2005;280(5):3665–3674. doi: 10.1074/jbc.M412922200. [DOI] [PubMed] [Google Scholar]
- 114.Woodrick J, et al. A new sub-pathway of long-patch base excision repair involving 5' gap formation. EMBO J. 2017;36(11):1605–1622. doi: 10.15252/embj.201694920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Asagoshi K, et al. Single-nucleotide base excision repair DNA polymerase activity in C elegans in the absence of DNA polymerase beta. Nucleic Acids Res. 2012;40(2):670–681. doi: 10.1093/nar/gkr727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Boehm EM, Gildenberg MS, Washington MT. The Many roles of PCNA in eukaryotic DNA REPLICATION. Enzymes. 2016;39:231–254. doi: 10.1016/bs.enz.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Volkova NV, et al. Mutational signatures are jointly shaped by DNA damage and repair. Nat Commun. 2020;11(1):2169. doi: 10.1038/s41467-020-15912-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bebenek K, et al. Biochemical properties of Saccharomyces cerevisiae DNA polymerase IV. J Biol Chem. 2005;280(20):20051–20058. doi: 10.1074/jbc.M501981200. [DOI] [PubMed] [Google Scholar]
- 119.Jain R, Aggarwal AK, Rechkoblit O. Eukaryotic DNA polymerases. Curr Opin Struct Biol. 2018;53:77–87. doi: 10.1016/j.sbi.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 120.Gellon L, et al. Intrinsic 5'-deoxyribose-5-phosphate lyase activity in Saccharomyces cerevisiae Trf4 protein with a possible role in base excision DNA repair. DNA Repair (Amst) 2008;7(2):187–198. doi: 10.1016/j.dnarep.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Daley JM, Wilson TE, Ramotar D. Genetic interactions between HNT3/Aprataxin and RAD27/FEN1 suggest parallel pathways for 5' end processing during base excision repair. DNA Repair (Amst) 2010;9(6):690–699. doi: 10.1016/j.dnarep.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sallmyr A, et al. Human DNA ligases in replication and repair. DNA Repair (Amst) 2020;93:102908. doi: 10.1016/j.dnarep.2020.102908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Williams JS, et al. High-fidelity DNA ligation enforces accurate Okazaki fragment maturation during DNA replication. Nat Commun. 2021;12(1):482. doi: 10.1038/s41467-020-20800-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bauer RJ, et al. Comparative analysis of the end-joining activity of several DNA ligases. PLoS ONE. 2017;12(12):190062. doi: 10.1371/journal.pone.0190062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Pergolizzi G, Wagner GK, Bowater RP. Biochemical and Structural characterisation of DNA ligases from bacteria and archaea. Biosci Rep. 2016;36(5):00391. doi: 10.1042/BSR20160003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lu G, et al. Ligase I and ligase III mediate the DNA double-strand break ligation in alternative end-joining. Proc Natl Acad Sci U S A. 2016;113(5):1256–1260. doi: 10.1073/pnas.1521597113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kurosawa A, Kuboshima H, Adachi N. Complex genetic interactions between DNA polymerase beta and the NHEJ ligase. FEBS J. 2020;287(2):377–385. doi: 10.1111/febs.15012. [DOI] [PubMed] [Google Scholar]
- 128.Caglayan M, Wilson SH. Oxidant and environmental toxicant-induced effects compromise DNA ligation during base excision DNA repair. DNA Repair (Amst) 2015;35:85–89. doi: 10.1016/j.dnarep.2015.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ismail IH, et al. The RNF138 E3 ligase displaces Ku to promote DNA end resection and regulate DNA repair pathway choice. Nat Cell Biol. 2015;17(11):1446–1457. doi: 10.1038/ncb3259. [DOI] [PubMed] [Google Scholar]
- 130.Horton JK, et al. XRCC1 phosphorylation affects aprataxin recruitment and DNA deadenylation activity. DNA Repair (Amst) 2018;64:26–33. doi: 10.1016/j.dnarep.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wilson DM, 3rd, et al. Systematic analysis of DNA crosslink repair pathways during development and aging in Caenorhabditis elegans. Nucleic Acids Res. 2017;45(16):9467–9480. doi: 10.1093/nar/gkx660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gorski MM, et al. The Drosophila melanogaster DNA Ligase IV gene plays a crucial role in the repair of radiation-induced DNA double-strand breaks and acts synergistically with Rad54. Genetics. 2003;165(4):1929–1941. doi: 10.1093/genetics/165.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Donahue SL, et al. Mitochondrial DNA ligase function in Saccharomyces cerevisiae. Nucleic Acids Res. 2001;29(7):1582–1589. doi: 10.1093/nar/29.7.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Arakawa H, et al. Functional redundancy between DNA ligases I and III in DNA replication in vertebrate cells. Nucleic Acids Res. 2012;40(6):2599–2610. doi: 10.1093/nar/gkr1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Polo LM, et al. Efficient single-strand break repair requires binding to both poly(ADP-Ribose) and DNA by the central BRCT domain of XRCC1. Cell Rep. 2019;26(3):573–581. doi: 10.1016/j.celrep.2018.12.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Reynolds P, et al. Disruption of PARP1 function inhibits base excision repair of a sub-set of DNA lesions. Nucleic Acids Res. 2015;43(8):4028–4038. doi: 10.1093/nar/gkv250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lavrik OI. PARPs' impact on base excision DNA repair. DNA Repair (Amst) 2020;93:102911. doi: 10.1016/j.dnarep.2020.102911. [DOI] [PubMed] [Google Scholar]
- 138.Wilk A, et al. Extracellular NAD(+) enhances PARP-dependent DNA repair capacity independently of CD73 activity. Sci Rep. 2020;10(1):651. doi: 10.1038/s41598-020-57506-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sukhanova M, Khodyreva S, Lavrik O. Poly(ADP-ribose) polymerase 1 regulates activity of DNA polymerase beta in long patch base excision repair. Mutat Res. 2010;685(1–2):80–89. doi: 10.1016/j.mrfmmm.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 140.Hanzlikova H, et al. Overlapping roles for PARP1 and PARP2 in the recruitment of endogenous XRCC1 and PNKP into oxidized chromatin. Nucleic Acids Res. 2017;45(5):2546–2557. doi: 10.1093/nar/gkw1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kutuzov MM, et al. The contribution of PARP1, PARP2 and poly(ADP-ribosyl)ation to base excision repair in the nucleosomal context. Sci Rep. 2021;11(1):4849. doi: 10.1038/s41598-021-84351-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Mateu-Jimenez M, et al. Reduced tumor burden through increased oxidative stress in lung adenocarcinoma cells of PARP-1 and PARP-2 knockout mice. Biochimie. 2016;121:278–286. doi: 10.1016/j.biochi.2015.11.030. [DOI] [PubMed] [Google Scholar]
- 143.Abbotts R, Wilson DM., 3rd Coordination of DNA single strand break repair. Free Radic Biol Med. 2017;107:228–244. doi: 10.1016/j.freeradbiomed.2016.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ray Chaudhuri A. Nussenzweig A (2017) The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat Rev Mol Cell Biol. 2017;18(10):610–621. doi: 10.1038/nrm.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Velagapudi UK, et al. Recent development in the discovery of PARP inhibitors as anticancer agents: a patent update (2016–2020) Expert Opin Ther Pat. 2021;31:1–15. doi: 10.1080/13543776.2021.1886275. [DOI] [PubMed] [Google Scholar]
- 146.Gelmon KA, et al. Clinical effectiveness of olaparib monotherapy in germline BRCA-mutated, HER2-negative metastatic breast cancer in a real-world setting: phase IIIb LUCY interim analysis. Eur J Cancer. 2021;152:68–77. doi: 10.1016/j.ejca.2021.03.029. [DOI] [PubMed] [Google Scholar]
- 147.Dequen F, Gagnon SN, Desnoyers S. Ionizing radiations in Caenorhabditis elegans induce poly(ADP-ribosyl)ation, a conserved DNA-damage response essential for survival. DNA Repair (Amst) 2005;4(7):814–825. doi: 10.1016/j.dnarep.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 148.Crone B, et al. Elemental bioimaging of cisplatin in caenorhabditis elegans by LA-ICP-MS. Metallomics. 2015;7(7):1189–1195. doi: 10.1039/C5MT00096C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Neumann C, et al. The role of poly(ADP-ribose) polymerases in manganese exposed Caenorhabditis elegans. J Trace Elem Med Biol. 2020;57:21–27. doi: 10.1016/j.jtemb.2019.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Boamah EK, et al. Poly(ADP-Ribose) polymerase 1 (PARP-1) regulates ribosomal biogenesis in Drosophila nucleoli. PLoS Genet. 2012;8(1):1002442. doi: 10.1371/journal.pgen.1002442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bordet G, et al. Poly(ADP-ribose) polymerase 1 in genome-wide expression control in Drosophila. Sci Rep. 2020;10(1):21151. doi: 10.1038/s41598-020-78116-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Tao Z, Gao P, Liu HW. Studies of the expression of human poly(ADP-ribose) polymerase-1 in Saccharomyces cerevisiae and identification of PARP-1 substrates by yeast proteome microarray screening. Biochemistry. 2009;48(49):11745–11754. doi: 10.1021/bi901387k. [DOI] [PubMed] [Google Scholar]
- 153.Kirby TW, et al. Nuclear localization of the DNA repair scaffold XRCC1: uncovering the functional role of a bipartite NLS. Sci Rep. 2015;5:13405. doi: 10.1038/srep13405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Cannan WJ, et al. The human ligase IIIalpha-XRCC1 protein complex performs DNA nick repair after transient unwrapping of nucleosomal DNA. J Biol Chem. 2017;292(13):5227–5238. doi: 10.1074/jbc.M116.736728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Vasil'eva IA, Moor NA, Lavrik OI. Effect of human XRCC1 protein oxidation on the functional activity of its complexes with the key enzymes of DNA base excision repair. Biochemistry (Mosc) 2020;85(3):288–299. doi: 10.1134/S0006297920030049. [DOI] [PubMed] [Google Scholar]
- 156.Xu C, et al. Deficiency of X-ray repair cross-complementing group 1 in primordial germ cells contributes to male infertility. FASEB J. 2019;33(6):7427–7436. doi: 10.1096/fj.201801962RR. [DOI] [PubMed] [Google Scholar]
- 157.Dutta D, et al. Effect of Arg399Gln single-nucleotide polymorphism in XRCC1 gene on survival rate of Indian squamous cell head-and-neck cancer patients. J Cancer Res Ther. 2020;16(3):551–558. doi: 10.4103/jcrt.JCRT_476_18. [DOI] [PubMed] [Google Scholar]
- 158.Kaur J, et al. Association of XRCC1, XRCC2 and XRCC3 gene polymorphism with Esophageal cancer risk. Clin Exp Gastroenterol. 2020;13:73–86. doi: 10.2147/CEG.S232961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ghosh S, et al. Partial loss of the DNA repair scaffolding protein, Xrcc1, results in increased brain damage and reduced recovery from ischemic stroke in mice. Neurobiol Aging. 2015;36(7):2319–2330. doi: 10.1016/j.neurobiolaging.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Puerta-Garcia E, et al. Effect of DPYD, MTHFR, ABCB1, XRCC1, ERCC1 and GSTP1 on chemotherapy related toxicity in colorectal carcinoma. Surg Oncol. 2020;35:388–398. doi: 10.1016/j.suronc.2020.09.016. [DOI] [PubMed] [Google Scholar]
- 161.Soliman AHM, et al. Genetic polymorphisms in XRCC1, OGG1, and XRCC3 DNA repair genes and DNA damage in radiotherapy workers. Environ Sci Pollut Res Int. 2020;27(35):43786–43799. doi: 10.1007/s11356-020-10270-9. [DOI] [PubMed] [Google Scholar]
- 162.Fenech M, et al. Cloning and characterization of the rad4 gene of Schizosaccharomyces pombe; a gene showing short regions of sequence similarity to the human XRCC1 gene. Nucleic Acids Res. 1991;19(24):6737–6741. doi: 10.1093/nar/19.24.6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Marintchev A, et al. Domain specific interaction in the XRCC1-DNA polymerase beta complex. Nucleic Acids Res. 2000;28(10):2049–2059. doi: 10.1093/nar/28.10.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Malfatti MC, et al. New perspectives in cancer biology from a study of canonical and non-canonical functions of base excision repair proteins with a focus on early steps. Mutagenesis. 2020;35(1):129–149. doi: 10.1093/mutage/gez051. [DOI] [PubMed] [Google Scholar]
- 165.Sung H, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 166.Ryu CS, et al. MPG and NPRL3 polymorphisms are associated with ischemic stroke susceptibility and post-stroke mortality. Diagnostics (Basel) 2020;10(11):947. doi: 10.3390/diagnostics10110947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.de Barrios O, et al. ZEB1 promotes inflammation and progression towards inflammation-driven carcinoma through repression of the DNA repair glycosylase MPG in epithelial cells. Gut. 2019;68(12):2129–2141. doi: 10.1136/gutjnl-2018-317294. [DOI] [PubMed] [Google Scholar]
- 168.Broderick P, et al. Evaluation of NTHL1, NEIL1, NEIL2, MPG, TDG, UNG and SMUG1 genes in familial colorectal cancer predisposition. BMC Cancer. 2006;6:243. doi: 10.1186/1471-2407-6-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ye F, et al. Association of SMUG1 SNPs in intron region and linkage disequilibrium with occurrence of cervical carcinoma and HPV infection in Chinese population. J Cancer. 2019;10(1):238–248. doi: 10.7150/jca.27103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Jiang S et al. (2021) Associations of APE1 and OGG1 polymorphisms with onset and prognosis of bladder cancer. Panminerva Med [DOI] [PubMed]
- 171.Czarny P, et al. Association between single nucleotide polymorphisms of MUTYH, hOGG1 and NEIL1 genes, and depression. J Affect Disord. 2015;184:90–96. doi: 10.1016/j.jad.2015.05.044. [DOI] [PubMed] [Google Scholar]
- 172.Sliwinski T, et al. Polymorphisms of the DNA base excision repair gene MUTYH in head and neck cancer. Exp Oncol. 2009;31(1):57–59. [PubMed] [Google Scholar]
- 173.Jensen KA, Shi X, Yan S. Genomic alterations and abnormal expression of APE2 in multiple cancers. Sci Rep. 2020;10(1):3758. doi: 10.1038/s41598-020-60656-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Wright G, Gassman NR. Transcriptional dysregulation of base excision repair proteins in breast cancer. DNA Repair (Amst) 2020;93:102922. doi: 10.1016/j.dnarep.2020.102922. [DOI] [PubMed] [Google Scholar]
- 175.Simon H, et al. OGG1 deficiency alters the intestinal microbiome and increases intestinal inflammation in a mouse model. PLoS ONE. 2020;15(1):e0227501. doi: 10.1371/journal.pone.0227501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Mohamed RH, et al. Association of XRCC1 and OGG1 DNA repair gene polymorphisms with rheumatoid arthritis in Egyptian patients. Gene. 2016;578(1):112–116. doi: 10.1016/j.gene.2015.12.021. [DOI] [PubMed] [Google Scholar]
- 177.Kwiatkowski D, et al. Variants of base excision repair genes MUTYH, PARP1 and XRCC1 in Alzheimer's disease risk. Neuropsychobiology. 2015;71(3):176–186. doi: 10.1159/000381985. [DOI] [PubMed] [Google Scholar]
- 178.Imai K, et al. Human uracil-DNA glycosylase deficiency associated with profoundly impaired immunoglobulin class-switch recombination. Nat Immunol. 2003;4(10):1023–1028. doi: 10.1038/ni974. [DOI] [PubMed] [Google Scholar]
- 179.Zheng L, et al. Fen1 mutations result in autoimmunity, chronic inflammation and cancers. Nat Med. 2007;13(7):812–819. doi: 10.1038/nm1599. [DOI] [PubMed] [Google Scholar]
- 180.Zhu Y, et al. LINC00467 is up-regulated by TDG-mediated acetylation in non-small cell lung cancer and promotes tumor progression. Oncogene. 2020;39(38):6071–6084. doi: 10.1038/s41388-020-01421-w. [DOI] [PubMed] [Google Scholar]
- 181.Yan JB, et al. Insulin and metformin control cell proliferation by regulating TDG-mediated DNA demethylation in liver and breast cancer cells. Mol Ther Oncolytics. 2020;18:282–294. doi: 10.1016/j.omto.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Li D, et al. The HOTAIRM1/miR-107/TDG axis regulates papillary thyroid cancer cell proliferation and invasion. Cell Death Dis. 2020;11(4):227. doi: 10.1038/s41419-020-2416-1. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 183.Tomkinson AE, Naila T, Khattri BS. Altered DNA ligase activity in human disease. Mutagenesis. 2020;35(1):51–60. doi: 10.1093/mutage/gez026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Ke Y, et al. The role of PARPs in inflammation-and metabolic-related diseases: molecular mechanisms and beyond. Cells. 2019;8(9):1047. doi: 10.3390/cells8091047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Pazzaglia S, Pioli C. Multifaceted role of PARP-1 in DNA repair and inflammation: pathological and therapeutic implications in cancer and non-cancer diseases. Cells. 2019;9(1):41. doi: 10.3390/cells9010041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Li Q, Ma R, Zhang M. XRCC1 rs1799782 (C194T) polymorphism correlated with tumor metastasis and molecular subtypes in breast cancer. Onco Targets Ther. 2018;11:8435–8444. doi: 10.2147/OTT.S154746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Wu W et al (2021) Neuronal enhancers are hotspots for DNA single-strand break repair. Nature [DOI] [PMC free article] [PubMed]
- 188.Yoon G, Caldecott KW. Nonsyndromic cerebellar ataxias associated with disorders of DNA single-strand break repair. Handb Clin Neurol. 2018;155:105–115. doi: 10.1016/B978-0-444-64189-2.00007-X. [DOI] [PubMed] [Google Scholar]
- 189.Londero AP, et al. Expression and prognostic significance of APE1/Ref-1 and NPM1 proteins in high-grade ovarian serous cancer. Am J Clin Pathol. 2014;141(3):404–414. doi: 10.1309/AJCPIDKDLSGE26CX. [DOI] [PubMed] [Google Scholar]
- 190.Meng L, et al. 2',3'-dideoxycytidine, a DNA polymerase-beta inhibitor, reverses memory deficits in a mouse model of Alzheimer's disease. J Alzheimers Dis. 2019;67(2):515–525. doi: 10.3233/JAD-180798. [DOI] [PubMed] [Google Scholar]
- 191.Wu Q, et al. Two polymorphic mutations in promoter region of DNA polymerase beta in relatively higher percentage of thymic hyperplasia patients. Thorac Cancer. 2020;12:588–592. doi: 10.1111/1759-7714.13773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Murphy DL, et al. The E288K colon tumor variant of DNA polymerase beta is a sequence specific mutator. Biochemistry. 2012;51(26):5269–5275. doi: 10.1021/bi3003583. [DOI] [PMC free article] [PubMed] [Google Scholar]