Abstract
The human cerebral cortex is a uniquely complex structure encompassing an unparalleled diversity of neuronal types and subtypes. These arise during development through a series of evolutionary conserved processes, such as progenitor cell proliferation, migration and differentiation, incorporating human-associated adaptations including a protracted neurogenesis and the emergence of novel highly heterogeneous progenitor populations. Disentangling the unique features of human cortical development involves elucidation of the intricate developmental cell transitions orchestrated by progressive molecular events. Crucially, developmental timing controls the fine balance between cell cycle progression/exit and the neurogenic competence of precursor cells, which undergo morphological transitions coupled to transcriptome-defined temporal states. Recent advances in bulk and single-cell transcriptomic technologies suggest that alongside protein-coding genes, non-coding RNAs exert important regulatory roles in these processes. Interestingly, a considerable number of novel long non-coding RNAs (lncRNAs) and microRNAs (miRNAs) have appeared in human and non-human primates suggesting an evolutionary role in shaping cortical development. Here, we present an overview of human cortical development and highlight the marked diversification and complexity of human neuronal progenitors. We further discuss how lncRNAs and miRNAs constitute critical components of the extended epigenetic regulatory network defining intermediate states of progenitors and controlling cell cycle dynamics and fate choices with spatiotemporal precision, during human neurodevelopment.
Keywords: Long non-coding RNAs, miRNAs, Neurogenesis, Single-cell RNA sequencing, Epitranscriptomics, Evolution
Introduction
The human cerebral cortex is one of the most complex structures in the brain responsible for high-order cognitive functions and social behavior as well as sensory, motor and emotional control, while its impairment results in neurodevelopmental, neuropsychiatric and neurodegenerative disorders. The cerebral cortex is a laminated structure organized into six layers comprising a large number of neurons that are classically divided into two major types: the glutamatergic excitatory pyramidal projection neurons and the GABAergic inhibitory interneurons. Pyramidal projection neurons account for approximately 80% of all cortical neurons and represent the only output system relaying information from one cortical area to another or to other brain regions, but also the largest input system receiving information from other cortical areas or brain regions. Interneurons, on the other hand, comprise 20% of the cortical neuronal population and form local synaptic connections that play critical roles in shaping network activity. Cortical neurons are further subdivided into a multitude of subtypes characterized by different molecular and electrophysiological properties as well as by distinct patterns of synaptic connections. Understanding how this remarkable neuronal diversity is generated and how neural circuits are formed during cortical development remains a challenge, particularly in humans.
Cortical development encompasses intricate cell transitions orchestrated by progressive molecular events. These include evolutionary conserved processes, such as progenitor cell proliferation, neuronal birth, migration and differentiation, synapse formation and maturation, synaptic pruning and cell death. Evolutionary adaptations in humans have led to protracted neurogenesis and the generation of an unprecedented diversity of cell types and subtypes from a uniquely heterogeneous population of human progenitors [1, 2]. Cortical projection neurons develop through waves of neurogenesis from radial glial progenitors residing in the ventricular zone and from intermediate progenitors of an enlarged subventricular zone. The six-layered neocortex is thus generated in an inside-out fashion, with the earliest-born neurons populating the deepest layers, and the last-born neurons populating the most superficial layers. Human-specific mechanisms include the emergence of novel radial glia populations and other unique features of human progenitors. Crucially, developmental timing controls the fine balance between cell cycle progression/exit and the neurogenic competence of precursor cells which undergo morphological transitions and related transcriptome-defined temporal states [3].
A body of research accumulating over the last 15 years has uncovered a set of key transcriptional regulators that control mammalian cortical development. However, identification of the exact changes and the precise molecular innovations that distinguish the human brain has been elusive. Data from large-scale consortia investigating functional genomic elements such as ENCODE and FANTOM [4, 5], revolutionized our understanding of mammalian genomes in terms of architecture, activity and regulation. It was thus revealed that the human genome encodes only ~ 20,000 protein-coding genes, which represent less than 2% of the total genome sequence while the majority is transcribed into non-coding RNAs. Notably, a large fraction of these (~ 40%) are expressed in the brain, with characteristic spatiotemporal patterns arguing for functional implications. Indeed, long non-coding RNAs (lncRNAs) and microRNAs (miRNAs), the most numerous members of the non-coding family, participate in regulatory networks during cortical development [6, 7]. In sharp contrast to the highly conserved repertoire of protein-coding genes, the extent of non-protein-coding intronic and intergenic sequences does scale with increased developmental complexity. Particularly, the emergence of primate and human-specific non-coding RNAs in combination with their capacity to modulate large gene-regulatory networks render them ideal candidates as drivers of human brain complexity and evolution [8, 9]. Reconstruction of human gene-regulatory networks uncovered that both lncRNAs and miRNAs are involved in acquisition of cell-type identities while they undergo dynamic transitions even among closely related cell types during neuronal commitment. It thus seems that non-coding RNAs endow neural cells with an additional level of control for the precise spatiotemporal deployment of genes, essential for accomplishment of complex neurobiological traits.
Here, we present an overview of human cortical development and highlight exciting new discoveries regarding the marked diversification and complexity of human neuronal progenitors and their neuronal progeny. We present gene expression data and cellular differences underpinning the divergent evolution of human neuronal circuits and present emerging evidence supporting a critical role of lncRNAs and miRNAs in regulating progenitor turnover and cell-fate choice during human neural development. In particular, we discuss how non-coding RNAs exert spatiotemporal control of cell cycle dynamics and fate acquisition through their association with key molecular networks and signaling pathways. Finally, we illustrate the evolving expression patterns of lncRNAs and miRNAs in distinct human progenitor types, including radial glia and intermediate progenitors, and their relevance to the human neurogenic program.
Species-specific diversity of the brain’s cellular composition: the evolution of complexity
A challenging scope of contemporary neuroscience concerns reconstruction of the unique elements that build the human brain. Efforts are directed towards elucidation of species-specific cellular, molecular and biochemical features that shape neuronal connectivity and dictate high-order cognitive functions and social behavior. The basic principles of neurogenesis are conserved across species and include proliferation and diversification of neural progenitor cells that give rise to an extensive array of distinct neuronal and glial phenotypes [10–12].
Brain evolution has favored expansion of the cortical surface area, along with increase in the radial thickness of the cortical plate [13]. This structural innovation is instructive for brain enlargement and involves an expanded primate germinal zone, a region where progenitors reside and proliferate [14]. Consequent evolutionary adaptations incorporate a protracted period of human neurogenesis, and accompanying changes in the morphology and abundance of both excitatory and inhibitory neurons, as well as glial cells, resulting in more complex patterns of synaptogenesis and neuronal connectivity [13].
In the last decade, the characterization of neural and glial diversity has become possible on a global scale thanks to revolutionary methodological advances. In particular, single-cell RNA-sequencing technologies provide a platform to track transcriptional changes and identify molecular cell-type specifications along neurodevelopmental processes as well as among species. A number of studies have exploited these technologies to highlight the extent of cell-type diversity in the nervous system of different species including drosophila, zebrafish mouse, non-human and human primates [15–22]. Transcriptional heterogeneity of neuronal and non-neuronal cells, including astrocytes and oligodendrocytes, has thus been ascribed with molecularly distinct and regionally restricted populations linked to specific neural circuits [23, 24]. Similarly, analysis of the adult human brain allowed an unprecedented transcriptome-level resolution of cell types and subtypes. One study remarkably revealed 35 distinct cellular clusters, including excitatory and inhibitory neuronal subtypes in the cortex and cerebellum, as well as non-neuronal cells such as astrocytes and oligodendrocytes, while it illustrated substantial regional heterogeneity among particular excitatory neuronal classes [25]. Further investigations in the mouse and human brain demonstrated that the adult cortex consists of at least 8 to 19 distinct excitatory neuronal subtypes distributed across six layers [10, 18, 26, 27]. Recently, a more elaborate analysis in two areas of the mouse neocortex, the primary visual cortex and the lateral motor cortex, uncovered 133 functionally distinct transcriptomic cell types [20]. Interestingly, it was shown that most glutamatergic types are area-specific while most GABAergic types are shared across cortical areas, a dichotomy that correlates well with their developmental origins and connectivity patterns. Most glutamatergic types are born locally from the ventricular and subventricular zones of the developing cortex and project to other regions, whereas nearly all GABAergic interneurons migrate from the ganglionic eminences to reach the cortex and form local connections [20].
As expected, comparison of single-cell transcriptomic data across species revealed divergent features of the human brain at cellular resolution. Assessment of mouse and human datasets demonstrated a surprisingly well-conserved cellular architecture that enables matching of homologous types and allows predictions of the properties of human cortical types. Beyond fundamental similarities, extensive differences between homologous human and mouse cell types were identified, including marked alterations in proportions, laminar distribution and morphology. In addition, divergent genes appear functionally relevant as they are associated with connectivity and signaling [21]. Human-specific neuronal signatures were also detected in relation to non-human apes, and specifically in 7 out of the 33 brain regions investigated, including the primary somatosensory cortex, internal capsule, and cerebellar white matter [22].
Origins of diversity: evolutionary adaptations of neuronal progenitors
Elucidation of the human-specific mechanisms through which neuronal complexity arises involves interrogation of the molecular specifications that regulate developmental transitions and commitment of progenitor cells. The human cortex not only presents the greatest expansion across evolution, but also associates with high-order cognitive functions and serious neurodevelopmental, psychiatric and neurodegenerative disorders. Cortical neurogenesis integrates a sequence of proliferation-differentiation events which are reproduced in a timely fashion and with remarkable fidelity to yield a six-layered structure in an inside–out mode, whereby early-born neurons occupy the deeper layers and later-born neurons are sequentially positioned in upper layers [28–34]. The principles and mechanisms that underlie the timely production of adequate classes of cells remain only partially understood and are still under intense investigation [35]. In fact, we are only at the beginning of defining aspects of cortical development that are shared across mammals and those that are human specific. The ultimate goal is to reveal differences that distinguish humans from other species and understand how these features translate into human behavior and disease.
Unique features and competence of human neural progenitors
In general terms during development, neural stem cells or neural progenitors initially undergo a phase of exponential amplification via symmetric divisions, and then switch to a neurogenic phase via asymmetric divisions producing a self-renewed progenitor and a sister cell committed to differentiation. Later on, terminal symmetric divisions yield two post-mitotic neuronal cells exhausting the pool of proliferating progenitors. The prevailing hypothesis for cortical neurogenesis is that of a progressive restriction of progenitor potential over time [36] (Fig. 1), against the alternative view which presumes the existence of distinct, fate-restricted progenitors already present before the onset of neurogenesis [37, 38].
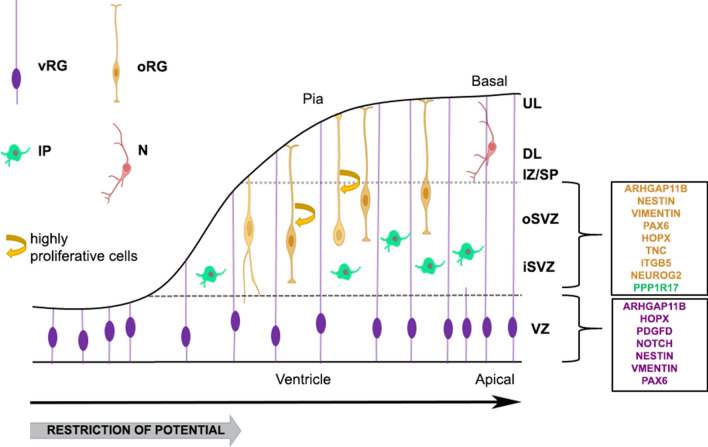
Fig. 1.
Schematic illustration of the laminar and cellular organization of the developing cortex, including the location and morphology of different progenitor cell populations. The two main proliferative regions, the ventricular (VZ) and subventricular (SVZ) zones, are depicted. vRG apical progenitors residing in the VZ give rise to intermediate progenitors (IPs) and outer radial glial cells (oRGs) that populate a secondary proliferative region, the subventricular zone (SVZ). Pia-contacting oRGs, delaminate from the ventricular zone (VZ) and translocate to the enlarged outer subventricular zone (oSVZ), thus subdividing the SVZ into inner (iSVZ) and outer (oSVZ) zones. oRGs form contact with the basal lamina and are highly proliferative cells with diverse morphologies. IP cells also exhibit variable phenotypes. Divergent gene expression of human-associated cortical progenitor types is highlighted in the boxes on the right. The black arrow below the ventricle denotes the progress of neurogenesis, which starts during the 6–7th postconceptional week. Multipotential proliferating progenitors progressively become more fate-restricted, as indicated by the thick gray arrow, to generate the diversity of neurons occupying the deep (DL) and upper layers (UL) of the cortical pate. Neurons (N); Intermediate Zone/Subplate (IZ/SP)
Human cortical neurogenesis starts around the seventh postconceptional week [39], when neuroepithelial (NE) cells lining the ventricles gradually transform into radial glia (ventricular radial glia; vRG) to compose the proliferative ventricular zone (VZ). NE cells display a symmetric dynamic pattern of cell division that serves to expand the progenitor cell pool, while Unlike RG cells, they retract their basal fibers before division [40]. Characterization of RG cells at cellular, molecular and physiological levels has revealed considerable species‐specific differences [18, 41, 42]. Although rodent RGs are an excellent model for the study of basic aspects of cortical development, their counterparts in primates, including humans, are considerably different with respect to their location, developmental potential and molecular signature. For example, outer radial glia (oRGs), also called basal radial glia, are a novel type of glial progenitors which constitute an evolutionary adaptation observed in mammals with a large brain, including human and non-human primates [14, 43–45]. Thus in primates including humans, vRG cells produce two types of basal progenitors, the intermediate progenitors (IP) and the outer radial glia (oRG) both of which move out of the ventricular zone to populate a secondary proliferative region, the subventricular (SVZ). oRGs, in particular, delaminate from the ventricular zone (VZ) and translocate to the outer subventricular zone (oSVZ), thus subdividing the SVZ into inner and outer zones [14, 43–45]. Therefore, the inner SVZ (iSVZ) is populated mostly by IPs and the outer SVZ is composed mainly of oRG cells [46].
To add to the diversification of primate progenitor populations, at later stages of neurodevelopment during a period coinciding with the transition from neurogenesis to gliogenesis, vRGs in humans give rise to a morphologically distinct type called truncated radial glial cell that remains in the VZ [43]. Although topologically divergent, the mouse neocortex presents an oSVZ-like germinal zone [44, 47, 48]. Still, the volume of the oSVZ in the macaque and human brain far exceeds that of the iSVZ, underlining its importance in evolution as a whole, and particularly in brain expansion [14]. Therefore, the generation of a greater number of neurons in the human brain appears in part due to the establishment of an enlarged proliferative zone and to the vast increase in the number of highly proliferative oRG cells [3, 45, 46]. In particular the oSVZ, which is the principal proliferative region in the dorsal cortex during upper layer neurogenesis, is directly linked to the increased neuronal number and complexity associated with the upper cortical layer in primates [49]. It is recognized that the temporally restricted expression of specific transcription factors in the VZ matches with the timely generation of different classes of neurons, suggesting a temporal orchestration of progenitor competence [29, 50]. For example, Ngn1 is required for specifying the fates of lower layer excitatory neurons, but not later during upper layer formation [51]. Consistently, genes that are restricted to either upper or deep-layer neurons are also enriched in subsets of progenitors during development. Moreover, early cortical RGCs express markers of lower layer neurons, including Emx2, Fezf2, and Sox2 [52–55], while genes specific for upper layer neurons, such as Satb2, are expressed at high levels in IPCs in the SVZ during middle and late stages of neocortical development [56]. These studies suggest that the mechanisms governing the production of early-born versus late-born neurons involve the timely expression of specific transcription factors. However, other studies have challenged this idea reporting upper layer Cux2 progenitors to be present in the VZ during deep-layer neuron generation [38]. Therefore, although the correlation between sequential transcription factor expression in progenitor cells and neuron generation is generally accepted, a concrete elucidation of this mechanism at work is still lacking.
In the absence of distinct types of pre-committed progenitors in proliferating germinal zones, the question arises if fate restriction in single progenitor types is temporally regulated in an irreversible manner. In this respect, heterochronic transplantation studies demonstrated that early progenitors, like radial glial cells, are temporally plastic and can re-enter past neurogenic states when exposed to an earlier-stage environment, while more progressed intermediate progenitors lose this fate plasticity, suggesting a dual effect of timing and environmental influence [57, 58]. Moreover, the reproducible pattern of cortical neurogenesis argues that the precise timing of differentiation may be somehow inherently encoded within lineages of individual progenitor cells [59, 60]. New single-cell RNA-sequencing data generated at high temporal resolution to trace the molecular identities of successive generations of apical progenitors, indicated that dynamically expressed genes drive apical progenitors from being internally directed to more exteroceptive states [50] (Fig. 1). Notably, while at early stages, cell cycle-related and nucleus/chromatin-related processes are prominent, later in development the susceptibility of apical progenitors to environmental signals increases, as highlighted by higher expression of membrane receptors, cell–cell signaling-related proteins, and excitability-related proteins [50].
The fact that a given neuronal cell type can be specified during a very limited time window and that multipotential progenitors progressively become more fate-restricted, dictates that characterization of the spectrum of temporal progenitor states is essential for constructing specific lineage trees that predict developmental timing of fate acquisition. The concept of cell identity, therefore, has been suggested to integrate three major aspects: phenotypic characterization including functionality, allocation to a lineage through knowledge of developmental history, and description of cell state in response to diverse stimuli [61].
Molecular profile, morphological characteristics and cell cycle regulation of human progenitors
Pioneering studies have laid the ground for defining broad categories of neural progenitors according to their molecular profile. However, more recent single-cell transcriptomic approaches produced data that add to the diversity of neural progenitor states but also highlight species-specific deviations of restricted identities. For example, vRG and oRG cells share expression of characteristic markers of radial glia including nestin, vimentin, and PAX6 [3, 62]. However, their molecular profiles are highly distinct as oRG preferentially express genes related to extracellular matrix formation, migration, and stemness [63]. Moreover, oRG cells express several newly discovered markers, including HOPX, TNC and ITGB5, giving them a unique transcriptional signature [63, 64] (Fig. 1). With similar approaches, it was identified that human Neurog2 is a critical factor in delamination and expansion of oRGs as well as in regulation of their neurogenic potential [65] (Fig. 1). Unbiased single-cell transcriptome analysis revealed two mouse subtypes of IP cells, expressing general IP markers such as Tbr2 and Afap1, with one subtype enriched in more mature neuronal differentiation markers, such as Neurod1 and Mgat5b (Fig. 1). In human, a distinct species-specific IP cell population was identified with oRG-like morphology and characteristic expression of a novel cytoplasmic marker, PPP1R17 [63, 66] (Fig. 1). Taken together, single-cell transcriptomics illuminated an array of specific and comparative aspects shaping human and mouse cortical development and shed light at the diversity of progenitor cell types and their individual differentiation paths. The vast dataset produced was systematically organized into Shiny Cortex, a publicly available resource [67].
It is noteworthy, that while developmental atlases and cell trajectories have revealed a wealth of information on molecular profiles and can help identify cellular differentiation paths, a full understanding of cell-type specification requires further lineage tracing experiments and correlations with specific morphotypes. Morphology is an important component characterizing the phenotype and state of progenitor cells, further relating to their proliferation or migration status. This notion is enhanced by recent findings showing that cell shape changes early in development during the neuroepithelial (NE) to radial glia (RG) transition are delayed in humans, extending the proliferative NE stage and thus contributing to human-specific neocortical expansion [68].
Regarding later stages, rodent intermediate progenitors have distinct multipolar morphology [69, 70], whereas human and non-human primate neural progenitors have diverse morphologies suggesting an expanded potential for transitional states [71] (Fig. 1). Currently, the defining morphology of oRG cells is controversial [71–73]. Studies in the macaque showed that oSVZ progenitor cells may display a mixture of bipolar, unipolar, and multipolar morphologies, and may alternate between morphologies during a given cell cycle [71]. More complex phenotypes exist also for human IPs. For example, IPs within the human oSVZ may be multipolar, unipolar, or bipolar, with radially or tangentially oriented processes [63]. Diversification between species was reported on the basis of morphology, localization and the expression of the IP-specific marker TBR2, demonstrating that there may be a greater proportion of IP-like oSVZ progenitors compared to radial glia-like progenitors in the macaque as compared to human [71].
Another important attribute of progenitor state is cell cycle properties. In general, neural progenitors undergo several modes of cell division. As already discussed, these include symmetrical proliferative divisions that expand the pool of neuronal precursor cells, followed by asymmetrical neurogenic divisions and, finally, terminal symmetrical divisions that produce two neurons and deplete the pool of proliferative cells [72, 74]. The mode of division was shown to be characteristic of distinct species-specific progenitor populations providing some explanation for the increase of brain size across evolutionary time. Particularly in the mouse, vRG cell divisions result in vRG self-renewal, direct neurogenesis, or production of intermediate progenitor cells that divide once to produce two neurons [28, 46, 69]. In contrast, only a small proportion of human oRG cell divisions produce IP cells, while the vast majority of divisions appear to lead to oRG self-renewal [75]. IP cells similarly self-renew many times before producing neurons [3], while oRG cells appear less restricted than multipolar IPs in their lineage potential [63]. These observations illustrate the increased proliferative capacity of human progenitors, underlying brain enlargement.
Human progenitors, including oRG cells, exhibit species distinct gene expression patterns while cell cycle genes among those are characteristic of their divergent properties. For example, PDGFD gene encodes a platelet-derived growth factor acting through the PDGFRB receptor, which is expressed in RG cells and regulates cell cycle progression and cell expansion in human but not mouse progenitors [76] (Fig. 1). In addition, the human-specific gene ARHGAP11B, which is highly enriched in human RG cells, favors oRG generation and amplification [77, 78], while Notch signaling in particular has been linked to the proliferative capacity of human cortical progenitors [79] as it has been implicated in the clonal expansion of human RGs [80–82] (Fig. 1).
The plane of divisions as well as cell cycle length also appears to play a role in progenitor proliferative aptitude and neurogenic competency, baring multiple evolutionary implications. It has been proposed that, an evolutionary shift from a vertical cleavage plane angle to a mix of vertical and horizontal divisions favored self-renewal of progenitors and generation of oRGs, and may have contributed to an increased oRG cell population and the development of an oSVZ in humans [83, 84]. The “cell cycle length” hypothesis suggests that the length of the cell cycle, particularly G1 phase, is a critical determinant for the onset of neurogenesis, by allowing the accumulation of differentiation-driving factors. Indeed, it has been shown that during mouse corticogenesis, G1 lengthening is both necessary and sufficient to switch neural progenitors to neurogenesis [11, 85–88]. The length of G1 phase changes dramatically in both VZ and oSVZ during primate development, which may be associated with a decreased rate of cell cycle exit and an increase in proliferative divisions in the oSVZ [71]. In line, comparison between human and chimpanzee cerebral organoids, revealed that human RG cells display a longer mitotic phase [41], which has been linked to proliferative divisions rather than asymmetric neurogenic divisions. This again suggests that human RGs may favor proliferative over neurogenic divisions for a longer period, which is consistent with a larger brain [89] (Fig. 1). Recent studies provided a breakthrough in explaining the slower tempo of human neurogenesis by attributing temporal differences during development between species, a phenomenon termed allochrony, to cell-autonomous differences in biochemical reaction speeds within neural progenitors [90].
Dynamic gene expression profiles underscore the heterogeneity of human neuronal progenitors
In an effort to distinguish intermediate progenitor cell states, recent transcriptomic data revealed the remarkable heterogeneity of these populations [71]. Single-cell sequencing demonstrated the previously unappreciated diversity of transcriptional states within both human apical RG and oRG cells, which by contrast appears markedly simplified in mouse [65, 91]. A number of studies attest the enhanced diversity of the human progenitor populations, including previously unidentified transient NPC populations [92, 93] (Fig. 1). Of note, single-cell resolution of the human forebrain during development revealed new intermediate progenitor markers that enabled identification of diverse neuron subtypes [19]. In addition, single-cell RNA sequencing of human, chimpanzee and macaque cerebral organoids complemented by previous data from fetal brain tissue, revealed gain of human-specific gene expression which associates with proliferation of radial glia [89]. Indeed, human progenitor diversity is characterized by unique expression profiles compared to mouse [1], but also, importantly, human radial glial progenitors differ most strikingly from mouse in their graded transition from neuroepithelial radial glia to delaminated, multipolar, neuronal lineage-committed intermediate progenitors [65]. This diversity of neural precursors includes multiple cell types with transcriptional profiles of mixed identity displaying co-expression of genes which are typically thought to define separate types of stem, progenitor or differentiated cells [1]. For example, the mRNAs of neuronal markers, such as Cux1 and Tle4, are expressed in early apical progenitors, while the mRNAs of Ctip2 and Neurod2 are readily detectable in newborn IPs [50]. This phenomenon, whereby the mRNAs for proteins that will be expressed in progeny are already present but not translated in the parent cell, is called transcriptional priming or pre-patterning, and has been linked to precursor competence. According to this mechanism, progenitors express a broad profile of mRNAs important for both pluripotency and differentiation, allowing for post-transcriptional control in progenitors and newborn progeny alike, through rapid expression of the appropriate proteins. In line, transcriptional priming of the transcription factor Eomes (Tbr2) was suggested to be a driver of precursor lineage diversity [1]. Transcriptional priming is logical from a kinetic standpoint to ensure rapid changes in protein expression, as transcription can persist over 10 min, whereas post-transcriptional regulation is faster, with an average translation rate of about 1 min.
It is now being recognized that the dynamic transcriptomic states of progenitors are most probably instructive of their competence, which is gradually restricted over time. It becomes clear that to accomplish a precise and predictable differentiation program, precursor cells require rapid, tightly controlled changes in gene expression. Small regulatory RNAs, including increasing numbers of miRNAs are widely acknowledged as fine regulators of spatiotemporal gene expression via control of mRNA translation and degradation [7]. High developmental complexity, on the other hand, requires an expansion of regulatory information which has been proposed to reside mainly in the non-protein-coding portion of the genome [94]. Indeed, multiple epigenetic regulatory systems, including DNA methylation, histone modifications and non-coding RNAs work in harmony to guide sequential lineage specification of neural progenitors [95]. Next, we focus on the effects of long non-coding RNAs and miRNAs in human progenitor diversity and function. We will examine and discuss recent evidence that link the functional precision of non-coding RNAs to intricate transitions specific of the human neurogenic program. We will highlight the implication of lncRNAs and miRNAs in establishing distinct molecular signatures, in regulation of the proliferative capacity and neurogenic potential, as well as the transcriptional heterogeneity exhibited by human cortical progenitors.
Non-coding RNAs in human neurogenesis
While only a very small fraction of the human genome codes for proteins, corresponding to approximately 20,000 protein-coding genes, non-coding RNAs (ncRNAs) make up roughly 80% of the human transcriptome [96]. The most numerous classes of ncRNAs include lncRNAs and miRNAs which both have been linked to brain development.
Biogenesis and mode of function of lncRNAs and miRNAs
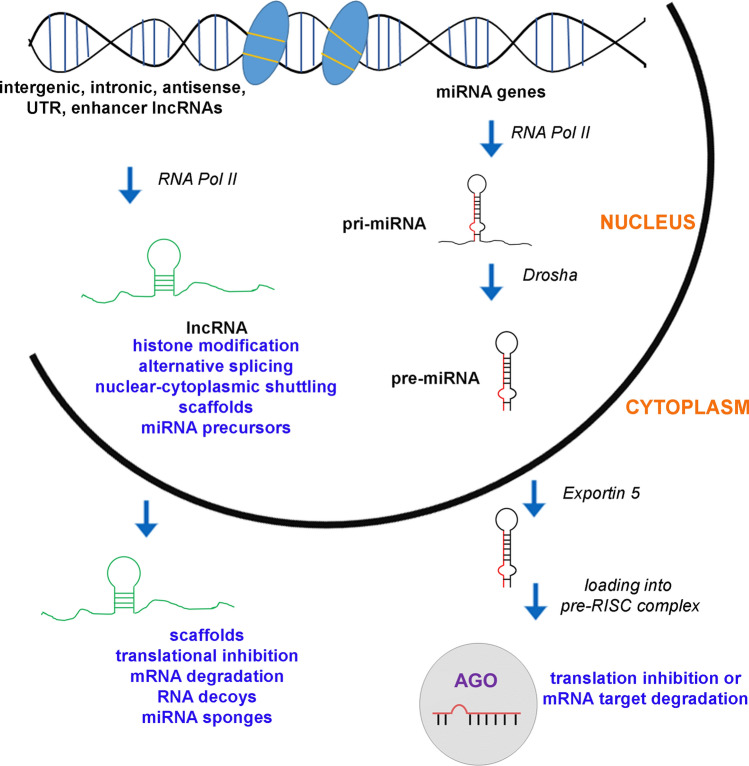
LncRNAs can range from 200 nucleotides up to several kilobases in length and can be categorized according to their genomic location and function. As with protein-coding genes, lncRNAs undergo 5’ capping, 3’ polyadenylation and splicing modifications [97] and are transcribed by RNA polymerase II. Regarding their genomic location, this may vary and includes intergenic regions, introns of protein-coding genes, and also gene-regulatory regions such as UTRs [98], promoters [99] and enhancers [100]. Presenting an amazing diversity, lncRNAs fall into several mechanistic categories [101–103]. Globally, they act as scaffolds, providing a site for other interactions, thus recruiting protein complexes based on their sequence specificity. LncRNAs have been implicated in transcriptional modulation, post-transcriptional control (alternative splicing), nuclear-cytoplasmic shuttling, translational inhibition, mRNA degradation, formation of RNA decoys preventing proteins accessing other RNAs (RBP decoy) and regulation of protein activity [101] (Fig. 2). LncRNA–mRNA interactions can regulate mRNA levels by increasing or decreasing their stability. They can also act as precursors for small ncRNAs, such as miRNAs and small nucleolar RNAs (snoRNAs) [102] (Fig. 2). Alternatively, lncRNAs can, through specific base-pairing, sponge miRNAs from other binding events (Fig. 2). Their functional diversity relies on the inherent properties of RNA molecules, like their modular organization and the ability to fold into different structures. Of note, lncRNAs can modulate transcription of protein-coding genes at either their own locus (cis-regulation) or distant loci (trans-regulation) by altering the chromatin state, possibly by providing a scaffold for chromatin-modifying complexes or by guiding ribonucleoprotein complexes to target loci [103]. The majority of research over the last 10 years has focused on those lncRNAs that remain in the nucleus, but sequencing RNAs in the cytoplasm has revealed that many lncRNAs are also present in the cytoplasm.
Fig. 2.
Schematic representation of lncRNA and miRNA biosynthesis and function. Briefly, lncRNAs are transcribed by RNA polymerase II, while they fall into several mechanistic categories: they act as scaffolds, as miRNA sponges, and precursors for miRNAs. They are also involved in alternative splicing, nuclear-cytoplasmic shuttling, translational inhibition, mRNA degradation, and formation of RNA decoys. The production of mature miRNA begins with the transcription of double-stranded primary miRNA (pri-miRNA) by RNA polymerase II. Following a series of cleavages, the mature ‘‘guide’’ miRNA strand remains which is loaded into a pre-miRNA-induced silencing complex (pre-RISC) containing Argonaute (Ago) and other proteins. Mature miRNAs silence their mRNA targets either by direct degradation and/or by suppression of translation
Contrary to the highly conserved protein-coding genes, thousands of new lncRNAs have emerged during primate nervous system evolution. About a third of the lncRNAs are unique to the primate lineage [104], and only ~ 12% of human lncRNAs appear to be conserved in other vertebrate species [105, 106]. Interestingly, only 72% of human lncRNAs are also expressed in macaque, compared to 98% of human protein-coding genes in all primates [107].
microRNAs (miRNAs) are another class of non-coding RNAs of smaller size, approximately ∼22 nucleotides in length, that function as post-transcriptional regulators. Mature miRNAs silence their mRNA targets either by direct degradation and/or by suppression of translation (Fig. 2). The miRNA-mRNA interaction assumes partial sequence complementarity and occurs between the mRNA’s 3’UTR and a 6–8 nucleotides long ‘‘seed’’ sequence at the 5’ end of the microRNA. The production of mature miRNA begins with the transcription of double-stranded primary miRNA (pri-miRNA) by RNA polymerase II. The pri-miRNA is then cleaved by the RNase-III enzyme, Drosha, producing ∼70-bp pre-miRNAs that are exported from the nucleus into the cytoplasm by Exportin 5 (Exp5). miRNA double-stranded duplexes of 21–23 nucleotides are the product of cleavage of pre-miRNA sequences by the Dicer enzyme. These duplexes are loaded into a pre-miRNA-induced silencing complex (pre-RISC) containing Argonaute (Ago) and other proteins, where the complementary ‘‘passenger’’ strand is removed so that just the mature ‘‘guide’’ miRNA strand remains (Fig. 2). Association with mRNA targets may enhance miRNA stability, a phenomenon known as target-mediated miRNA protection, while the introduction of additional target sites can also promote miRNA accumulation [108]. On the other hand, miRNA–mRNA interactions can also destabilize the miRNA and promote its degradation through a process known as target RNA directed miRNA degradation [109]. The degree of sequence complementarity infers the outcome of the miRNA–mRNA interaction, with higher complementarity favoring miRNA degradation and lower complementarity favoring miRNA stabilization [110]. A single microRNA can target multiple mRNAs simultaneously, while a single mRNA may be regulated by different microRNAs [111] underlining the vast regulatory potential of the miRNA network.
As an estimated 70% is expressed in the nervous system [112], miRNAs have emerged as important post-transcriptional regulators of gene expression involved in neurogenesis and neural function in mammalian species [113, 114].
The evolving role of lncRNAs in the proliferation and differentiation of neuronal progenitors
Implying widespread functional implications, about 40% of the identified human lncRNAs are specifically expressed in the brain [104, 115], corresponding to 4000–20,000 lncRNA genes, a remarkably high number compared to the approximately 20,000–25,000 protein-coding genes [116]. Generally, lncRNAs exhibit tissue-specific expression, including the brain where they show regionally segregated expression patterns [9]. They are dynamically expressed during neuronal development demonstrating higher spatiotemporal, cell type and tissue specificity, compared to coding genes [6, 98, 117, 118]. Therefore, many lncRNAs are enriched in specific sub-populations of the mouse [118] and human [65] cortex. This property along with their functional diversity and the emergence of a significant number of primate and human-specific lncRNAs, renders them attractive candidates as drivers of human brain complexity and evolution [119].
Studies in rodents identified conserved lncRNAs as key regulators of neural lineage entry [120] which act by directing transcription factors or chromatin remodeling machineries to specific lineage-specifying genes. For example, the transcription factor REST that acts as a neuronal gene repressor, induces the expression of the lncRNA Rmst, which in turn drives neural differentiation by recruiting the neural transcription factor Sox2 to key neurogenesis-promoting genes, such as Dlx1, Ascl1, and Hey2 [121], thereby acting in trans (Table 1). Sox2 expression is also regulated by another lncRNA called TUNA (Tcl1 upstream neuron-associated lncRNA), which forms a complex with three RNA-binding proteins (RBPs): PTBP1, hnRNP-K, and NCL. The TUNA-RBP complex was detected at the promoters of Nanog, Sox2, and Fgf4, and knockdown of TUNA or the individual RBPs inhibited neural differentiation of mouse embryonic stem cells [122]. In addition, the lncRNA Paupar regulates in cis the expression of the transcription factor Pax6 [123], a critical factor for neural progenitor generation, while the lncRNAs Riken‐201 and Riken‐203 were shown to modulate the expression of Sox6 and regulate neural differentiation by repressing the function of miR-96 and miR-467a-3p [124].
Table 1.
Specification of the role of lncRNAs and miRNAs in progenitor function as identified in human experimental setups
| lncRNA/miRNA | Target/function | Species-specificity | Ref |
|---|---|---|---|
| Cell cycle/proliferation | |||
| LncND | Maintains the neural progenitor pool; acts as a miRNA sponge for miR-143-3p to regulate the Notch signaling pathway | Primate | [112] |
| FMR4 | Promotes proliferation of human neural precursor cells | Primate | [113] |
| miR-2115 | Enriched in radial glia; regulates their proliferation by fine-tuning the expression of ORC4, a known regulator of DNA replication | Primate | [97] |
| miR4673 | Involved through the NOTCH pathway in amplification of the proliferative capacity of human neural progenitors and the delay of their differentiation | Hominin specific | [142] |
| miR-9 | One of its key targets is Stathmin, a protein that increases microtubule instability; miR-9 coordinates the proliferation and migration of human neural progenitor cells | Conserved | [136–138] |
| let-7 | Link between Sox2 and LIN28/let-7 pathway in maintaining NPC proliferation and their neurogenic potential | Conserved | [136–138] |
| miR-1301-3p | Targets the mRNA for the histone-lysine N-methyltransferases mll1 and mll2 that function towards induction of neurogenesis | Primate | [97] |
| Differentiation | |||
| Rmst | Drives neural differentiation by recruiting the neural transcription factor Sox2 | Conserved | [104] |
| lncRNA_N1 | Associates with the REST/coREST complex to regulate gene expression and neural cell-fate specification | Conserved | [104] |
| lncRNA_N2 | Promotes neurogenesis by maintaining MIR-125B and LET7A levels in neural progenitors | Conserved | [104] |
| Pnky | Controls the balance between self-renewal and neuronal differentiation in dividing neural stem cells by interacting with the mRNA splicing regulator PTBP1 | Conserved | [108] |
| miR-125 | Promotes exit form pluripotency and potentiation of neural specification by targeting SMAD4 | Conserved | [136–138] |
| miR-1290 | Targets crucial cell cycle genes and acts as an upstream regulator during neuronal differentiation | Primate | [143, 144] |
| miR-197 | Forms together with MeCP2, ADAM10 and NOTCH a regulatory axis for human progenitor differentiation | Conserved | [141] |
| miR-934 | Controls progenitor to neuroblast transition and impacts on differentiation of newborn neurons, including neurite growth | Primate | [145] |
Another two lncRNAs, namely linc-Brn1b and Pnky are located in proximity and co-expressed with Brn1 or Brn2, which are implicated in the proliferation and neuronal commitment of progenitors. Linc-Brn1b mutants were found to display distinct abnormalities in the generation of upper layer II–IV neurons in the neocortex [125]. The neural-specific lncRNA Pnky, on the other hand, is expressed in the nucleus of dividing neural stem cells (NSCs) in the developing mouse brain and detected in human VZ area (Fig. 3). It controls the balance of self-renewal and neuronal differentiation in dividing NSCs by interacting with the mRNA splicing regulator PTBP1, which is expressed in NSCs and represses the inclusion of neural exons in non-neural cells [126].
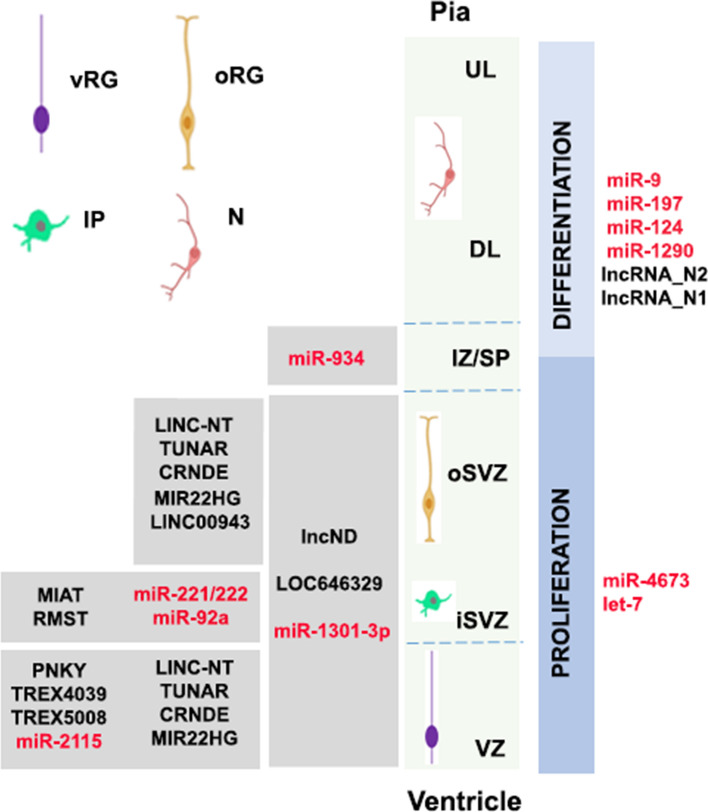
Fig. 3.
A series of sequencing studies, the majority at single-cell resolution, have reported non-coding RNA expression characterizing the consecutive states of progenitor cells during human cortical neurogenesis. The schematic diagram depicts the different zones of the developing human cortex indicating the main regions of cell proliferation or differentiation. A number of lncRNAs (black fonts) and miRNAs (red fonts) that associate with progenitor populations within distinct regions of the developing cortex are shown on the left. Non-coding RNAs that associate with progenitor cell proliferation or differentiation are shown on the right. Ventricular radial glia (vRG); intermediate progenitors (IP); outer radial glia (oRGs); neurons (N); ventricular zone (VZ); inner subventricular zone (iSVZ); outer subventricular zone (oSVZ); deep layer (DL); upper layer (UL), Intermediate Zone/Subplate (IZ/SP)
Considering the emergence of a significant number of human-specific lncRNAs across evolution, subsequent research in human models added substantially to our knowledge on the impact of lncRNAs in fundamental functions of human neural progenitors. For this purpose, different systems were examined, including human fetal brain tissue, human embryonic stem or precursor cell lines and induced pluripotent stem cells along with brain organoids. These endeavors were accompanied by appropriate differentiation protocols and some also incorporated state-of-the-art sequencing methodologies at single-cell resolution. Initial studies showed lncRNAs to be necessary components of human neural developmental gene networks, and it was next demonstrated that certain lncRNAs were linked to radial glia proliferation or were found to be enriched in oRG cells [65]. Moreover, specific lncRNAs have been associated with distinct cellular identities or transient neurodevelopmental cell states in the fetal human cortex, inferring their participation in the intricate transitions of early developmental events [6]. Importantly, it was recently shown that human-specific lncRNAs contribute to the phylogenetic divergence of the striatum [127].
The first direct demonstration that lncRNAs are indispensable for human neurogenesis was based on microarray expression profiling of differentially expressed lncRNAs during differentiation of human embryonic stem cells. This work further identified 36 novel lncRNAs that play a role in progenitor function, including lncRNA_N1 which associates with the REST/coREST complex to regulate gene expression and neural cell-fate specification, and the lncRNA_N2 which promotes neurogenesis by maintaining MIR-125B and LET7A levels in neural progenitors [121] (Table 1) (Fig. 3).
Adding to previous data, a recent study used an optimized protocol to generate human progenitors and identify several lncRNAs with dynamic expression during neurogenesis [128], while others explored publicly available sequencing datasets towards examination of novel lncRNAs affecting progenitor differentiation [129]. In agreement, combination of epigenetics, network and GO analysis complemented by experimental validation showed that divergent lncRNAs are more specifically associated with neuronal but not astrocytic differentiation [129].
A series of publications reported the involvement of lncRNAs in human progenitor proliferation. LncND (long non-coding neuro-development), which appeared in the Catarrhini branch of primates, is enriched in radial glia cells in the VZ and SVZ of the developing human brain and declines in neurons (Fig. 3). LncND acts as a miRNA sponge for miR-143-3p to regulate the Notch signaling pathway. Inhibition of lncND released miR-143-3p resulting in downregulation of Notch, which reduced cell proliferation and induced neuronal differentiation. Remarkably, gain-of-function of lncND in the developing mouse cortex led to an expansion of the PAX6 + radial glia cell population, supporting its role in maintaining the neural progenitor pool by regulating the Notch signaling pathway (Table 1) [130]. FMR4, an antisense lncRNA was shown to promote proliferation of progenitors isolated from human fetal brains, possibly through interaction with chromatin and forward affecting global histone methylation and mRNA expression [131] (Table 1). Single-cell transcriptome analyses of fetal brain from gestational weeks 13 to 23 detected known while uncovering many novel lncRNAs that are specific to distinct cell types and are abundantly expressed in individual cells. Among those, LOC646329 demonstrated high expression in the radial glia subpopulation and was shown to regulate their proliferation [132] (Fig. 3), while LINC00943 was found to be specifically expressed in oRG cells [92] (Fig. 3).
Additional single-cell sequencing studies of human fetal tissue enhanced the observation that lncRNAs display dynamic patterns of expression during human neurodevelopment while matching their expression profiles with the amazing transcriptional heterogeneity exhibited by human cortical progenitors. Non-conserved lncRNAs were connected to critical neural developmental processes such as progenitor pluripotency, neurogenesis and epithelial–mesenchymal transition. On the other hand, certain conserved lncRNAs, including LINC-NT, TUNAR, CRNDE and MIR22HG, that are absent in mouse RG were found enriched in human apical and outer RG, suggesting potentially distinct functions in cortical development (Fig. 3). Of the 15 known mouse IP-enriched lncRNAs [6], only two, MIAT and RMST, showed considerable expression in human fetal cortex (Fig. 3), and even these showed cell-type enrichment patterns distinct from those in mouse, while, the human RG subtype-specific transcripts that were identified include many lncRNAs that lack homologous mouse transcripts. Finally, long non-coding transcripts were especially enriched in human oRG cells determining their molecular identity and influencing their function [65].
Work on 3D brain models strengthened the concept that lncRNAs establish transient developmental cell states. Time-coupled transcriptomics at single-cell resolution of cerebral cortex organoids from human, chimpanzee, orangutan, and rhesus revealed 386 transiently expressed lncRNAs (TrEx lncRNAs) in human that have a remarkably conserved expression pattern in other great ape species as well. During early cortical neuron differentiation, TrEx lncRNAs were found to associate with short-lived cell-type intermediates. To mention a few, TREX108 and TREX8168 overlapping with MIR219-2 are highly expressed in neuroepithelial cells and absent later, TREX4039 is slightly expressed in neuroepithelial cells and RGs, and TREX5008 is restricted to the large RG cluster at only a particular time point [133] (Fig. 3).
The evolving role of miRNAs in the proliferation and differentiation of neuronal progenitors
miRNAs have properties that make them particularly suitable for driving developmental and evolutionary changes. These include the apparent ease by which miRNAs arise de novo in the genome and the fact that small changes in their sequence or expression levels can have broad consequences over a network of molecular targets. miRNA expression during development has been studied in several species, including rodents [134], non-human primates, and humans [135]. It has been shown that miRNA profiles change as cells pass through lineage stages [136], suggesting that miRNAs are tuning gene expression in a manner that establishes an identity boundary for a cell. This spatiotemporal mode of function of miRNAs matches with the requirement for precision of the molecular mechanisms that couple proliferation with competence in neural progenitors. Indeed, as shown in mouse brain development, not only miRNAs mediate developmental timing by driving cell-type transitions [137, 138], but their ablation leads to complete disorganization of cell-fate generation in the cortex [138–140]. This observation is further strengthened by the requirement for expression of gradients of certain miRNAs by neural progenitors to ensure temporal patterning of the embryonic neocortex and allow for the sequential generation of layer-specific neurons at precise times [141].
Studies on experimental animals have uncovered a number of miRNAs that seem to intervene at the margin between cell cycle exit and fate decision. Let-7 is a miRNA that controls cell cycle dynamics to drive neural differentiation by repressing genes that promote proliferation and cell cycle progression. Specifically, in the cortex it has been shown to repress the epigenetic regulator Hmga2 and the proliferation-promoting nuclear receptor TLX, which controls neural stem cell proliferation in the developing brain [142].
MiR-9 together with miR-124, two of the most abundant miRNAs in the brain, target REST which, as mentioned above, opposes neuronal differentiation [143, 144], while REST itself acts as an inhibitor of miR-124 expression. MiR-9 is also involved in a negative feedback loop with TLX (Fig. 3). Further, miR-210 was demonstrated to be required for normal cell cycle progression of mouse neural progenitors. It has been implicated in radial glia proliferation during neocortical development through regulation of the cyclin-dependent kinase CDK7 [145]. Other miRNAs involved in radial glia proliferation are miR-30e alongside miR-181d [146], miR-7 [147], and miR-34/449 [148, 149]. miR-92, on the other hand, targets EOMES (TBR2), the T-box transcription factor that is preferentially expressed in cortical intermediate progenitors, and regulates cortical neuron production [150] (Fig. 3). Specification of intermediate progenitor cells engages several more miRNAs, including miR-214 and miR-15b [151, 152], as well as miR-20a/b and miR-23a [153].
miRNA research on human embryonic stem cell differentiation models accompanied by miRNA profiling underscored the effects of known miRNAs, such as let-7, miR-9, and miR-125 in human progenitor proliferation, and neuronal commitment [154–156]. Interestingly, similar work also uncovered new targets and roles for certain miRNAs, stressing the need to investigate miRNA function in human experimental setups (Table 1). For example, miR-125 with a previously reported neuronal function was shown to act at earlier stages to promote exit from pluripotency and potentiate neural specification by targeting SMAD4.
Access to human tissue in combination with high-throughput sequencing techniques provided important insight into the significance of the miRNA-driven transcriptome changes across brain development and evolution [7]. In a comprehensive study aiming to characterize the landscape of miRNA–mRNA interactions during human brain development, Nowakovski et al. [114] established an innovative single-cell approach for combined mRNA and miRNA profiling at single-cell resolution using human fetal tissue samples corresponding to peak neurogenesis [Gestational week (GW) 15 and 16.5] and early gliogenesis (GW 19–20.5). The vast majority of miRNAs analyzed were enriched in at least one cell type, demonstrating significant specificity. For example, miR-221/222 and miR-92a were found enriched in cortical IPs, in line with other reports and consistent with their proposed roles in controlling proliferation (Fig. 3). Most interestingly, dynamic changes in miRNA abundance were revealed across closely related cells and in concordance with neuronal differentiation and maturation, suggesting that miRNA-mRNA interactions operate as functional modules in acquisition of cell-type identities and undergo dynamic transitions during brain development. For instance, targets of miR-92b-3p, let-7–5p, miR-421, and miR-137 were enriched alongside radial glial markers but also in excitatory neurons, but only at early stages.
Along with highly conserved miRNAs, a number of new brain miRNAs have emerged. Following evolutionary adaptations, more than 100 primate (including human) and 14 human-specific miRNAs have been identified in the developing brain [157]. Integration of novel miRNAs into ancient gene circuitry is suggested to exert additional regulation over proliferation of neural progenitors in cortical germinal areas, a region that demonstrates significant expansion across brain evolution [158]. Indeed, data so far emphasize the involvement of primate-specific miRNAs in the regulation of cell cycle dynamics operating during progenitor proliferation and neuronal differentiation. The list of related miRNAs is growing and includes a great ape-specific miRNA, miR-2115, which is enriched in radial glia (Fig. 3). It regulates their proliferation by fine-tuning the expression of ORC4, a known regulator of DNA replication [114] (Table 1). Another primate-specific miRNA, miR-1301-3p was detected in the germinal zones of the visual cortex of the macaque developing brain. miR-1301-3p targets the mRNA for histone-lysine N-methyltransferases mll1 and mll2 (MixedLineage, Leukemia) that functions towards induction of neurogenesis (Table 1, Fig. 3). miR-197 and the hominin-specific miRNA-4673 are involved in the regulation of NOTCH, a signaling pathway pivotal for proliferation (Table 1). miR-197 forms together with MeCP2, ADAM10, and NOTCH a regulatory axis for human progenitor differentiation [159]. miR4673, on the other hand, is encoded in intron 4 of human Notch-1 (Fig. 3). This miRNA has a remarkable impact on the regulation of the developmental clock as it instructs bimodal reprogramming of the cell cycle, leading to initial synchronization of neural precursors at the G0 phase of the cell cycle followed by accelerated progression through interphase. The end result is the amplification of the proliferative capacity of human neural progenitors and the delay of their differentiation [160].
miR-934 and miR-1290 are also found only in great apes, including human, and exert their function during early neurodevelopmental events. miR-1290 targets crucial cell cycle genes and acts as an upstream regulator during neuronal differentiation (Table 1), while miR-934 displays a stage-specific expression pattern during progenitor expansion and early neuron generation (Fig. 3). Interestingly, we have shown that miR-934 directly controls progenitor to neuroblast transition and impacts on neurite growth of newborn neurons (Table 1), further affecting the expression of genes associated with the subplate zone (Table 1), a transient compartment most prominent in primates that emerges during early corticogenesis [161–163]. Finally, a recent study provided for the first-time direct evidence for an evolutionary emerged miRNA that mediates fate specialization in mouse cortex. The authors demonstrated that miR-409-3p diversifies between generation of corticospinal motor neurons and callosal projection neurons both of which are born at the same developmental time and share common progenitors [164]. miR-409-3p is enriched in corticospinal motor neurons and belongs to the 12qF1 cluster that during evolution co-appeared with the motor cortex and the corpus callosum. It functions to promote acquisition of the corticospinal motor neuron fate by repressing the callosal projection neuron transcriptional regulator LMO4, which is required for identity establishment of callosal neurons [165].
Challenges and future perspectives
A series of observations suggest that both lncRNAs and miRNAs constitute critical components of the extended gene-regulatory network controlling cell cycle dynamics and fate choices during human neurodevelopment. Recent advances in high-throughput RNA-sequencing technologies, applied at bulk and at single-cell level, have started to provide a more comprehensive insight of precursor diversity. Such methodologies facilitated also profiling non-coding RNA expression patterns during neurogenesis. Accumulating evidence highlighted their spatiotemporal expression and cell-type specificity, and their role in mediating progenitor transitions emphasizing their involvement in sculpting the expanded repertoire of cortical neurons.
To evaluate the functional significance of lncRNAs and miRNAs as drivers of human brain complexity and evolution, relevant studies need to be complemented by lineage tracing experiments, while a consensus has to be reached regarding definition of subtypes and molecular signatures across developmental time. Clearly the technological advancements of RNA sequencing along with the formulation of appropriate algorithms that enable the automatic, high-throughput single-cell capture, and transcriptome analysis of cortical tissue is invaluable for the resolution of regional cell type landscapes and reconstruction of developmental trajectories. Still, to link molecular cell types to morphological and physiological correlates, the resolution of transcriptomic data should be combined with positional information of individual cells in three-dimensional space. Coming to fill this gap, spatial transcriptomics allow capturing single-cell transcriptomic data from a given location while retaining spatial information [166]. However, improvements of this nascent methodology are required to assure single-cell resolution and provide increase of the extent of sequenced mRNAs.
Gaining insight into the mechanisms generating particular cell types, further requires determining the developmental history of otherwise seemingly equivalent neuronal subtypes. Clonal history has traditionally been explored by microscopic tracing of cells during development, monitoring the heritable expression of genetically encoded chromogenic or fluorescent proteins and, more recently, using next-generation sequencing. Pseudotemporal analysis algorithms may produce trajectories from a static landscape of the brain, or when examining different time points. However, several innovative approaches have recently been formulated for simultaneous lineage tracing and transcriptome profiling in thousands of single cells in the zebra fish [167–169]. The challenge ahead is to apply such strategies in mouse and human cells.
The functional diversity of lncRNAs and the fact that they can modulate transcription of protein-coding genes either at their own locus (cis-regulation) or at distant loci (trans-regulation), adds additional levels of difficulty in delineating the mode of action of individual lncRNAs. The emergence of a considerable number of primate and human-specific lncRNAs and miRNAs in combination with the complexity of the human brain poses the necessity to use human-based models for research purposes. In line, 3D brain organoids appear to embody human cellular diversity and spatial architecture and recapitulate the organization of neural progenitor zones to a considerable degree, reflecting developmental events during embryonic stages in vivo [170, 171]. Particularly, the transcriptome of dorsal forebrain organoids grown for 40–100 days correlated best with fetal cortex tissue at ages 8–16 post-conception week, indicating similar pace of development, while single-cell analysis of telencephalic organoids and fetal human cortex showed that they contain very similar cell types [172–175].
Such systems can be invaluable to understand the critical input of non-coding RNAs in dictating intricate neurogenic decisions. However, these models require systematic benchmarking against primary tissue to validate their fidelity [176].
Recently, important advancements in cortical organoid protocols allow long-term culture with decreased necrosis, improved nutrient access and attainment of morphological and functional maturation [177–179]. An important milestone is the long-term maintenance of human cortical organoids reaching developmental progression and maturation remarkably corresponding to postnatal stages between 250 and 300 days [180]. Finally, the generation and fusion of region-specific organoids, termed assembloids, offer the opportunity to study how different brain regions communicate with each other, opening new avenues for understanding inter-regional brain connectivity and function during development and disease. Examples are assembloids of dorsal and ventral forebrain, as well as more complex systems integrating the components of a cortico-motor circuit [181–184].
Taken together, combining knowledge of well-resolved lineage trees coupled to transcriptional coding and non-coding information and other epigenetic events in association with morphology, cell positioning and formation of connectivity is a demanding venture that will provide insight to the central developmental neurobiology question of how neuronal diversity arises and importantly might explain how neural circuits are affected in particular disorders.
Acknowledgements
We thank the members of the Matsas Lab for stimulating discussions and comments.
Author contributions
KP conceived and wrote the review that was critically revised and edited by RM.
Funding
We acknowledge funding from: Stavros Niarchos Foundation to the Hellenic Pasteur Institute as part of the Foundation’s initiative to support the Greek Research Center Ecosystem; the Hellenic Foundation for Research and Innovation (HFRI) under the “1st Call for HFRI Research Projects to support Postdoctoral Researchers” (Project PARKINSynapse 899) and the “1st Call for HFRI Research Projects to support Faculty members and Researchers and the procurement of high-cost research equipment” (Project 1019-DiseasePHENOTarget); the Hellenic General Secretariat for Research and Innovation Flagship Action for Neurodegenerative Diseases on the basis of Personalized Medicine (2018ΣΕ01300001).
Declarations
Conflict interests
The authors declare that they have no conflict of interest.
Ethics approval and consent to participate
Not applicable.
Consent for publication
The authors have given their consent for publication.
Data availability
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Li Z, Tyler WA, Zeldich E, Santpere Baro G, Okamoto M, Gao T, et al. Transcriptional priming as a conserved mechanism of lineage diversification in the developing mouse and human neocortex. Sci Adv. 2020;6:45. doi: 10.1126/sciadv.abd2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Le Bail R, Bonafina A, Espuny-Camacho I, Nguyen L. Learning about cell lineage, cellular diversity and evolution of the human brain through stem cell models. Curr Opin Neurobiol. 2020;66:166–177. doi: 10.1016/j.conb.2020.10.018. [DOI] [PubMed] [Google Scholar]
- 3.Lui JH, Hansen DV, Kriegstein AR. Development and evolution of the human neocortex. Cell. 2011;146(1):18–36. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309(5740):1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 5.Consortium EP. Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447(7146):799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aprea J, Prenninger S, Dori M, Ghosh T, Monasor LS, Wessendorf E, et al. Transcriptome sequencing during mouse brain development identifies long non-coding RNAs functionally involved in neurogenic commitment. EMBO J. 2013;32(24):3145–3160. doi: 10.1038/emboj.2013.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prodromidou K, Matsas R. Species-specific miRNAs in human brain development and disease. Front Cell Neurosci. 2019;13:559. doi: 10.3389/fncel.2019.00559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qureshi IA, Mattick JS, Mehler MF. Long non-coding RNAs in nervous system function and disease. Brain Res. 2010;1338:20–35. doi: 10.1016/j.brainres.2010.03.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci USA. 2008;105(2):716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lake BB, Ai R, Kaeser GE, Salathia NS, Yung YC, Liu R, et al. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science. 2016;352(6293):1586–1590. doi: 10.1126/science.aaf1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hardwick LJ, Ali FR, Azzarelli R, Philpott A. Cell cycle regulation of proliferation versus differentiation in the central nervous system. Cell Tissue Res. 2015;359(1):187–200. doi: 10.1007/s00441-014-1895-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lodato S, Arlotta P. Generating neuronal diversity in the mammalian cerebral cortex. Annu Rev Cell Dev Biol. 2015;31:699–720. doi: 10.1146/annurev-cellbio-100814-125353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hill RS, Walsh CA. Molecular insights into human brain evolution. Nature. 2005;437(7055):64–67. doi: 10.1038/nature04103. [DOI] [PubMed] [Google Scholar]
- 14.Smart IH, Dehay C, Giroud P, Berland M, Kennedy H. Unique morphological features of the proliferative zones and postmitotic compartments of the neural epithelium giving rise to striate and extrastriate cortex in the monkey. Cereb Cortex. 2002;12(1):37–53. doi: 10.1093/cercor/12.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosenberg AB, Roco CM, Muscat RA, Kuchina A, Sample P, Yao Z, et al. Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science. 2018;360(6385):176–182. doi: 10.1126/science.aam8999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paul A, Crow M, Raudales R, He M, Gillis J, Huang ZJ. Transcriptional architecture of synaptic communication delineates GABAergic neuron identity. Cell. 2017;171(3):522–39e20. doi: 10.1016/j.cell.2017.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farnsworth DR, Saunders LM, Miller AC. A single-cell transcriptome atlas for zebrafish development. Dev Biol. 2020;459(2):100–108. doi: 10.1016/j.ydbio.2019.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nowakowski TJ, Bhaduri A, Pollen AA, Alvarado B, Mostajo-Radji MA, Di Lullo E, et al. Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science. 2017;358(6368):1318–1323. doi: 10.1126/science.aap8809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhong S, Zhang S, Fan X, Wu Q, Yan L, Dong J, et al. A single-cell RNA-seq survey of the developmental landscape of the human prefrontal cortex. Nature. 2018;555(7697):524–528. doi: 10.1038/nature25980. [DOI] [PubMed] [Google Scholar]
- 20.Tasic B, Yao Z, Graybuck LT, Smith KA, Nguyen TN, Bertagnolli D, et al. Shared and distinct transcriptomic cell types across neocortical areas. Nature. 2018;563(7729):72–78. doi: 10.1038/s41586-018-0654-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hodge RD, Bakken TE, Miller JA, Smith KA, Barkan ER, Graybuck LT, et al. Conserved cell types with divergent features in human versus mouse cortex. Nature. 2019;573(7772):61–68. doi: 10.1038/s41586-019-1506-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khrameeva E, Kurochkin I, Han D, Guijarro P, Kanton S, Santel M, et al. Single-cell-resolution transcriptome map of human, chimpanzee, bonobo, and macaque brains. Genome Res. 2020;30(5):776–789. doi: 10.1101/gr.256958.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bayraktar OA, Bartels T, Holmqvist S, Kleshchevnikov V, Martirosyan A, Polioudakis D, et al. Astrocyte layers in the mammalian cerebral cortex revealed by a single-cell in situ transcriptomic map. Nat Neurosci. 2020;23(4):500–509. doi: 10.1038/s41593-020-0602-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marques S, Zeisel A, Codeluppi S, van Bruggen D, Mendanha Falcao A, Xiao L, et al. Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science. 2016;352(6291):1326–1329. doi: 10.1126/science.aaf6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lake BB, Chen S, Sos BC, Fan J, Kaeser GE, Yung YC, et al. Integrative single-cell analysis of transcriptional and epigenetic states in the human adult brain. Nat Biotechnol. 2018;36(1):70–80. doi: 10.1038/nbt.4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeisel A, Munoz-Manchado AB, Codeluppi S, Lonnerberg P, La Manno G, Jureus A, et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347(6226):1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- 27.Tasic B, Menon V, Nguyen TN, Kim TK, Jarsky T, Yao Z, et al. Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat Neurosci. 2016;19(2):335–346. doi: 10.1038/nn.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noctor SC, Flint AC, Weissman TA, Dammerman RS, Kriegstein AR. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 2001;409(6821):714–720. doi: 10.1038/35055553. [DOI] [PubMed] [Google Scholar]
- 29.Frantz GD, Weimann JM, Levin ME, McConnell SK. Otx1 and Otx2 define layers and regions in developing cerebral cortex and cerebellum. J Neurosci. 1994;14(10):5725–5740. doi: 10.1523/JNEUROSCI.14-10-05725.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anderson SA, Kaznowski CE, Horn C, Rubenstein JL, McConnell SK. Distinct origins of neocortical projection neurons and interneurons in vivo. Cereb Cortex. 2002;12(7):702–709. doi: 10.1093/cercor/12.7.702. [DOI] [PubMed] [Google Scholar]
- 31.Angevine JB, Jr, Sidman RL. Autoradiographic study of cell migration during histogenesis of cerebral cortex in the mouse. Nature. 1961;192:766–768. doi: 10.1038/192766b0. [DOI] [PubMed] [Google Scholar]
- 32.Arimatsu Y, Ishida M. Distinct neuronal populations specified to form corticocortical and corticothalamic projections from layer VI of developing cerebral cortex. Neuroscience. 2002;114(4):1033–1045. doi: 10.1016/S0306-4522(02)00201-4. [DOI] [PubMed] [Google Scholar]
- 33.Rakic P. Neurons in rhesus monkey visual cortex: systematic relation between time of origin and eventual disposition. Science. 1974;183(4123):425–427. doi: 10.1126/science.183.4123.425. [DOI] [PubMed] [Google Scholar]
- 34.Takahashi T, Goto T, Miyama S, Nowakowski RS, Caviness VS., Jr Sequence of neuron origin and neocortical laminar fate: relation to cell cycle of origin in the developing murine cerebral wall. J Neurosci. 1999;19(23):10357–10371. doi: 10.1523/JNEUROSCI.19-23-10357.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uzquiano A, Gladwyn-Ng I, Nguyen L, Reiner O, Gotz M, Matsuzaki F, et al. Cortical progenitor biology: key features mediating proliferation versus differentiation. J Neurochem. 2018;146(5):500–525. doi: 10.1111/jnc.14338. [DOI] [PubMed] [Google Scholar]
- 36.Guo C, Eckler MJ, McKenna WL, McKinsey GL, Rubenstein JL, Chen B. Fezf2 expression identifies a multipotent progenitor for neocortical projection neurons, astrocytes, and oligodendrocytes. Neuron. 2013;80(5):1167–1174. doi: 10.1016/j.neuron.2013.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han W, Sestan N. Cortical projection neurons: sprung from the same root. Neuron. 2013;80(5):1103–1105. doi: 10.1016/j.neuron.2013.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Franco SJ, Gil-Sanz C, Martinez-Garay I, Espinosa A, Harkins-Perry SR, Ramos C, et al. Fate-restricted neural progenitors in the mammalian cerebral cortex. Science. 2012;337(6095):746–749. doi: 10.1126/science.1223616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Silbereis JC, Pochareddy S, Zhu Y, Li M, Sestan N. The cellular and molecular landscapes of the developing human central nervous system. Neuron. 2016;89(2):248–268. doi: 10.1016/j.neuron.2015.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Subramanian L, Bershteyn M, Paredes MF, Kriegstein AR. Dynamic behaviour of human neuroepithelial cells in the developing forebrain. Nat Commun. 2017;8:14167. doi: 10.1038/ncomms14167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mora-Bermudez F, Badsha F, Kanton S, Camp JG, Vernot B, Kohler K, et al. Differences and similarities between human and chimpanzee neural progenitors during cerebral cortex development. Elife. 2016;2016:5. doi: 10.7554/eLife.18683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu Y, Sousa AMM, Gao T, Skarica M, Li M, Santpere G, et al. Spatiotemporal transcriptomic divergence across human and macaque brain development. Science. 2018;362:6420. doi: 10.1126/science.aat8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nowakowski TJ, Pollen AA, Sandoval-Espinosa C, Kriegstein AR. Transformation of the radial glia scaffold demarcates two stages of human cerebral cortex development. Neuron. 2016;91(6):1219–1227. doi: 10.1016/j.neuron.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vaid S, Camp JG, Hersemann L, Eugster Oegema C, Heninger AK, Winkler S, et al. A novel population of Hopx-dependent basal radial glial cells in the developing mouse neocortex. Development. 2018;145:20. doi: 10.1242/dev.169276. [DOI] [PubMed] [Google Scholar]
- 45.Fietz SA, Kelava I, Vogt J, Wilsch-Brauninger M, Stenzel D, Fish JL, et al. OSVZ progenitors of human and ferret neocortex are epithelial-like and expand by integrin signaling. Nat Neurosci. 2010;13(6):690–699. doi: 10.1038/nn.2553. [DOI] [PubMed] [Google Scholar]
- 46.Hansen DV, Lui JH, Parker PR, Kriegstein AR. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature. 2010;464(7288):554–561. doi: 10.1038/nature08845. [DOI] [PubMed] [Google Scholar]
- 47.Shitamukai A, Konno D, Matsuzaki F. Oblique radial glial divisions in the developing mouse neocortex induce self-renewing progenitors outside the germinal zone that resemble primate outer subventricular zone progenitors. J Neurosci. 2011;31(10):3683–3695. doi: 10.1523/JNEUROSCI.4773-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martinez-Cerdeno V, Cunningham CL, Camacho J, Antczak JL, Prakash AN, Cziep ME, et al. Comparative analysis of the subventricular zone in rat, ferret and macaque: evidence for an outer subventricular zone in rodents. PLoS ONE. 2012;7(1):E30178. doi: 10.1371/journal.pone.0030178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lukaszewicz A, Savatier P, Cortay V, Giroud P, Huissoud C, Berland M, et al. G1 phase regulation, area-specific cell cycle control, and cytoarchitectonics in the primate cortex. Neuron. 2005;47(3):353–364. doi: 10.1016/j.neuron.2005.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Telley L, Agirman G, Prados J, Amberg N, Fievre S, Oberst P, et al. Temporal patterning of apical progenitors and their daughter neurons in the developing neocortex. Science. 2019;364:6440. doi: 10.1126/science.aav2522. [DOI] [PubMed] [Google Scholar]
- 51.Schuurmans C, Armant O, Nieto M, Stenman JM, Britz O, Klenin N, et al. Sequential phases of cortical specification involve Neurogenin-dependent and -independent pathways. EMBO J. 2004;23(14):2892–2902. doi: 10.1038/sj.emboj.7600278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leingartner A, Richards LJ, Dyck RH, Akazawa C, O'Leary DD. Cloning and cortical expression of rat Emx2 and adenovirus-mediated overexpression to assess its regulation of area-specific targeting of thalamocortical axons. Cereb Cortex. 2003;13(6):648–660. doi: 10.1093/cercor/13.6.648. [DOI] [PubMed] [Google Scholar]
- 53.Chen B, Schaevitz LR, McConnell SK. Fezl regulates the differentiation and axon targeting of layer 5 subcortical projection neurons in cerebral cortex. Proc Natl Acad Sci USA. 2005;102(47):17184–17189. doi: 10.1073/pnas.0508732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen B, Wang SS, Hattox AM, Rayburn H, Nelson SB, McConnell SK. The Fezf2-Ctip2 genetic pathway regulates the fate choice of subcortical projection neurons in the developing cerebral cortex. Proc Natl Acad Sci USA. 2008;105(32):11382–11387. doi: 10.1073/pnas.0804918105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bani-Yaghoub M, Tremblay RG, Lei JX, Zhang D, Zurakowski B, Sandhu JK, et al. Role of Sox2 in the development of the mouse neocortex. Dev Biol. 2006;295(1):52–66. doi: 10.1016/j.ydbio.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 56.Britanova O, Akopov S, Lukyanov S, Gruss P, Tarabykin V. Novel transcription factor Satb2 interacts with matrix attachment region DNA elements in a tissue-specific manner and demonstrates cell-type-dependent expression in the developing mouse CNS. Eur J Neurosci. 2005;21(3):658–668. doi: 10.1111/j.1460-9568.2005.03897.x. [DOI] [PubMed] [Google Scholar]
- 57.Oberst P, Fievre S, Baumann N, Concetti C, Bartolini G, Jabaudon D. Temporal plasticity of apical progenitors in the developing mouse neocortex. Nature. 2019;573(7774):370–374. doi: 10.1038/s41586-019-1515-6. [DOI] [PubMed] [Google Scholar]
- 58.Okamoto M, Miyata T, Konno D, Ueda HR, Kasukawa T, Hashimoto M, et al. Cell-cycle-independent transitions in temporal identity of mammalian neural progenitor cells. Nat Commun. 2016;7:11349. doi: 10.1038/ncomms11349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gaspard N, Bouschet T, Hourez R, Dimidschstein J, Naeije G, van den Ameele J, et al. An intrinsic mechanism of corticogenesis from embryonic stem cells. Nature. 2008;455(7211):351–357. doi: 10.1038/nature07287. [DOI] [PubMed] [Google Scholar]
- 60.Shen Q, Wang Y, Dimos JT, Fasano CA, Phoenix TN, Lemischka IR, et al. The timing of cortical neurogenesis is encoded within lineages of individual progenitor cells. Nat Neurosci. 2006;9(6):743–751. doi: 10.1038/nn1694. [DOI] [PubMed] [Google Scholar]
- 61.Morris SA. The evolving concept of cell identity in the single cell era. Development. 2019;146:12. doi: 10.1242/dev.169748. [DOI] [PubMed] [Google Scholar]
- 62.Estivill-Torrus G, Pearson H, van Heyningen V, Price DJ, Rashbass P. Pax6 is required to regulate the cell cycle and the rate of progression from symmetrical to asymmetrical division in mammalian cortical progenitors. Development. 2002;129(2):455–466. doi: 10.1242/dev.129.2.455. [DOI] [PubMed] [Google Scholar]
- 63.Pollen AA, Nowakowski TJ, Chen J, Retallack H, Sandoval-Espinosa C, Nicholas CR, et al. Molecular identity of human outer radial glia during cortical development. Cell. 2015;163(1):55–67. doi: 10.1016/j.cell.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thomsen ER, Mich JK, Yao Z, Hodge RD, Doyle AM, Jang S, et al. Fixed single-cell transcriptomic characterization of human radial glial diversity. Nat Methods. 2016;13(1):87–93. doi: 10.1038/nmeth.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Johnson MB, Wang PP, Atabay KD, Murphy EA, Doan RN, Hecht JL, et al. Single-cell analysis reveals transcriptional heterogeneity of neural progenitors in human cortex. Nat Neurosci. 2015;18(5):637–646. doi: 10.1038/nn.3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kawaguchi A, Ikawa T, Kasukawa T, Ueda HR, Kurimoto K, Saitou M, et al. Single-cell gene profiling defines differential progenitor subclasses in mammalian neurogenesis. Development. 2008;135(18):3113–3124. doi: 10.1242/dev.022616. [DOI] [PubMed] [Google Scholar]
- 67.Kageyama J, Wollny D, Treutlein B, Camp JG. ShinyCortex: exploring single-cell transcriptome data from the developing human cortex. Front Neurosci. 2018;12:315. doi: 10.3389/fnins.2018.00315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Benito-Kwiecinski S, Giandomenico SL, Sutcliffe M, Riis ES, Freire-Pritchett P, Kelava I et al (2021) An early cell shape transition drives evolutionary expansion of the human forebrain. Cell [DOI] [PMC free article] [PubMed]
- 69.Noctor SC, Martinez-Cerdeno V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci. 2004;7(2):136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- 70.Nelson BR, Hodge RD, Bedogni F, Hevner RF. Dynamic interactions between intermediate neurogenic progenitors and radial glia in embryonic mouse neocortex: potential role in Dll1-Notch signaling. J Neurosci. 2013;33(21):9122–9139. doi: 10.1523/JNEUROSCI.0791-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Betizeau M, Cortay V, Patti D, Pfister S, Gautier E, Bellemin-Menard A, et al. Precursor diversity and complexity of lineage relationships in the outer subventricular zone of the primate. Neuron. 2013;80(2):442–457. doi: 10.1016/j.neuron.2013.09.032. [DOI] [PubMed] [Google Scholar]
- 72.Takahashi T, Nowakowski RS, Caviness VS., Jr The leaving or Q fraction of the murine cerebral proliferative epithelium: a general model of neocortical neuronogenesis. J Neurosci. 1996;16(19):6183–6196. doi: 10.1523/JNEUROSCI.16-19-06183.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.De Juan RC, Borrell V. Coevolution of radial glial cells and the cerebral cortex. Glia. 2015;63(8):1303–1319. doi: 10.1002/glia.22827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cai L, Hayes NL, Takahashi T, Caviness VS, Jr, Nowakowski RS. Size distribution of retrovirally marked lineages matches prediction from population measurements of cell cycle behavior. J Neurosci Res. 2002;69(6):731–744. doi: 10.1002/jnr.10398. [DOI] [PubMed] [Google Scholar]
- 75.Nonaka-Kinoshita M, Reillo I, Artegiani B, Martinez-Martinez MA, Nelson M, Borrell V, et al. Regulation of cerebral cortex size and folding by expansion of basal progenitors. EMBO J. 2013;32(13):1817–1828. doi: 10.1038/emboj.2013.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lui JH, Nowakowski TJ, Pollen AA, Javaherian A, Kriegstein AR, Oldham MC. Radial glia require PDGFD-PDGFRbeta signalling in human but not mouse neocortex. Nature. 2014;515(7526):264–268. doi: 10.1038/nature13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Florio M, Albert M, Taverna E, Namba T, Brandl H, Lewitus E, et al. Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science. 2015;347(6229):1465–1470. doi: 10.1126/science.aaa1975. [DOI] [PubMed] [Google Scholar]
- 78.Namba T, Dóczi J, Pinson A, Xing L, Kalebic N, Wilsch-Bräuninger M, et al. Human-specific ARHGAP11B acts in mitochondria to expand neocortical progenitors by glutainolysis. Neuron. 2020;105(5):867–81.e9. doi: 10.1016/j.neuron.2019.11.027. [DOI] [PubMed] [Google Scholar]
- 79.Florio M, Heide M, Pinson A, Brandl H, Albert M, Winkler S, et al. Evolution and cell-type specificity of human-specific genes preferentially expressed in progenitors of fetal neocortex. Elife. 2018;2018:7. doi: 10.7554/eLife.32332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Visel A, Minovitsky S, Dubchak I, Pennacchio LA. VISTA enhancer browser–a database of tissue-specific human enhancers. Nucleic Acids Res. 2007;35:D88–92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gregorio I, Braghetta P, Bonaldo P, Cescon M. Collagen VI in healthy and diseased nervous system. Dis Models Mech. 2018;11:6. doi: 10.1242/dmm.032946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schep AN, Wu B, Buenrostro JD, Greenleaf WJ. chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat Methods. 2017;14(10):975–978. doi: 10.1038/nmeth.4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.LaMonica BE, Lui JH, Hansen DV, Kriegstein AR. Mitotic spindle orientation predicts outer radial glial cell generation in human neocortex. Nat Commun. 2013;4:1665. doi: 10.1038/ncomms2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chenn A, McConnell SK. Cleavage orientation and the asymmetric inheritance of Notch1 immunoreactivity in mammalian neurogenesis. Cell. 1995;82(4):631–641. doi: 10.1016/0092-8674(95)90035-7. [DOI] [PubMed] [Google Scholar]
- 85.Dehay C, Kennedy H. Cell-cycle control and cortical development. Nat Rev Neurosci. 2007;8(6):438–450. doi: 10.1038/nrn2097. [DOI] [PubMed] [Google Scholar]
- 86.Caviness VS, Jr, Takahashi T, Nowakowski RS. The G1 restriction point as critical regulator of neocortical neuronogenesis. Neurochem Res. 1999;24(4):497–506. doi: 10.1023/A:1022579712262. [DOI] [PubMed] [Google Scholar]
- 87.Salomoni P, Calegari F. Cell cycle control of mammalian neural stem cells: putting a speed limit on G1. Trends Cell Biol. 2010;20(5):233–243. doi: 10.1016/j.tcb.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 88.Lange C, Huttner WB, Calegari F. Cdk4/cyclinD1 overexpression in neural stem cells shortens G1, delays neurogenesis, and promotes the generation and expansion of basal progenitors. Cell Stem Cell. 2009;5(3):320–331. doi: 10.1016/j.stem.2009.05.026. [DOI] [PubMed] [Google Scholar]
- 89.Kanton S, Boyle MJ, He Z, Santel M, Weigert A, Sanchis-Calleja F, et al. Organoid single-cell genomic atlas uncovers human-specific features of brain development. Nature. 2019;574(7778):418–422. doi: 10.1038/s41586-019-1654-9. [DOI] [PubMed] [Google Scholar]
- 90.Matsuda M, Hayashi H, Garcia-Ojalvo J, Yoshioka-Kobayashi K, Kageyama R, Yamanaka Y, et al. Species-specific segmentation clock periods are due to differential biochemical reaction speeds. Science. 2020;369(6510):1450–1455. doi: 10.1126/science.aba7668. [DOI] [PubMed] [Google Scholar]
- 91.Fietz SA, Lachmann R, Brandl H, Kircher M, Samusik N, Schroder R, et al. Transcriptomes of germinal zones of human and mouse fetal neocortex suggest a role of extracellular matrix in progenitor self-renewal. Proc Natl Acad Sci USA. 2012;109(29):11836–11841. doi: 10.1073/pnas.1209647109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fan X, Fu Y, Zhou X, Sun L, Yang M, Wang M, et al. Single-cell transcriptome analysis reveals cell lineage specification in temporal-spatial patterns in human cortical development. Sci Adv. 2020;6(34):eaaz2978. doi: 10.1126/sciadv.aaz2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Eze UC, Bhaduri A, Haeussler M, Nowakowski TJ, Kriegstein AR. Single-cell atlas of early human brain development highlights heterogeneity of human neuroepithelial cells and early radial glia. Nat Neurosci. 2021;24(4):584–594. doi: 10.1038/s41593-020-00794-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Taft RJ, Pheasant M, Mattick JS. The relationship between non-protein-coding DNA and eukaryotic complexity. BioEssays. 2007;29(3):288–299. doi: 10.1002/bies.20544. [DOI] [PubMed] [Google Scholar]
- 95.Yoon KJ, Vissers C, Ming GL, Song H. Epigenetics and epitranscriptomics in temporal patterning of cortical neural progenitor competence. J Cell Biol. 2018;217(6):1901–1914. doi: 10.1083/jcb.201802117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et al. Landscape of transcription in human cells. Nature. 2012;489(7414):101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15(1):7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 98.Mercer TR, Qureshi IA, Gokhan S, Dinger ME, Li G, Mattick JS, et al. Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci. 2010;11:14. doi: 10.1186/1471-2202-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011;43(7):621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143(1):46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yoon JH, Abdelmohsen K, Gorospe M. Posttranscriptional gene regulation by long noncoding RNA. J Mol Biol. 2013;425(19):3723–3730. doi: 10.1016/j.jmb.2012.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhang X, Wang W, Zhu W, Dong J, Cheng Y, Yin Z, et al. Mechanisms and functions of long non-coding RNAs at multiple regulatory levels. Int J Mol Sci. 2019;20:22. doi: 10.3390/ijms20225573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43(6):904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154(1):26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25(18):1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Necsulea A, Soumillon M, Warnefors M, Liechti A, Daish T, Zeller U, et al. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature. 2014;505(7485):635–640. doi: 10.1038/nature12943. [DOI] [PubMed] [Google Scholar]
- 108.Chatterjee S, Fasler M, Bussing I, Grosshans H. Target-mediated protection of endogenous microRNAs in C. elegans. Dev Cell. 2011;20(3):388–396. doi: 10.1016/j.devcel.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 109.Fuchs Wightman F, Giono LE, Fededa JP, de la Mata M. Target RNAs strike back on microRNAs. Front Genet. 2018;9:435. doi: 10.3389/fgene.2018.00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Marcinowski L, Tanguy M, Krmpotic A, Radle B, Lisnic VJ, Tuddenham L, et al. Degradation of cellular mir-27 by a novel, highly abundant viral transcript is important for efficient virus replication in vivo. PLoS Pathog. 2012;8(2):e1002510. doi: 10.1371/journal.ppat.1002510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Saliminejad K, Khorram Khorshid HR, Soleymani Fard S, Ghaffari SH. An overview of microRNAs: biology, functions, therapeutics, and analysis methods. J Cell Physiol. 2019;234(5):5451–5465. doi: 10.1002/jcp.27486. [DOI] [PubMed] [Google Scholar]
- 112.Adlakha YK, Saini N. Brain microRNAs and insights into biological functions and therapeutic potential of brain enriched miRNA-128. Mol Cancer. 2014;13:33. doi: 10.1186/1476-4598-13-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Davis GM, Haas MA, Pocock R. MicroRNAs: not "fine-tuners" but key regulators of neuronal development and function. Front Neurol. 2015;6:245. doi: 10.3389/fneur.2015.00245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Nowakowski TJ, Rani N, Golkaram M, Zhou HR, Alvarado B, Huch K, et al. Regulation of cell-type-specific transcriptomes by microRNA networks during human brain development. Nat Neurosci. 2018;21(12):1784–1792. doi: 10.1038/s41593-018-0265-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 116.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22(9):1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Belgard TG, Marques AC, Oliver PL, Abaan HO, Sirey TM, Hoerder-Suabedissen A, et al. A transcriptomic atlas of mouse neocortical layers. Neuron. 2011;71(4):605–616. doi: 10.1016/j.neuron.2011.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Molyneaux BJ, Goff LA, Brettler AC, Chen HH, Hrvatin S, Rinn JL, et al. DeCoN: genome-wide analysis of in vivo transcriptional dynamics during pyramidal neuron fate selection in neocortex. Neuron. 2015;85(2):275–288. doi: 10.1016/j.neuron.2014.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Briggs JA, Wolvetang EJ, Mattick JS, Rinn JL, Barry G. Mechanisms of long non-coding RNAs in mammalian nervous system development, plasticity, disease, and evolution. Neuron. 2015;88(5):861–877. doi: 10.1016/j.neuron.2015.09.045. [DOI] [PubMed] [Google Scholar]
- 120.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477(7364):295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ng SY, Johnson R, Stanton LW. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 2012;31(3):522–533. doi: 10.1038/emboj.2011.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lin N, Chang KY, Li Z, Gates K, Rana ZA, Dang J, et al. An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Mol Cell. 2014;53(6):1005–1019. doi: 10.1016/j.molcel.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Vance KW, Sansom SN, Lee S, Chalei V, Kong L, Cooper SE, et al. The long non-coding RNA Paupar regulates the expression of both local and distal genes. EMBO J. 2014;33(4):296–311. doi: 10.1002/embj.201386225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhang L, Xue Z, Yan J, Wang J, Liu Q, Jiang H. LncRNA Riken-201 and Riken-203 modulates neural development by regulating the Sox6 through sequestering miRNAs. Cell Prolif. 2019;52(3):e12573. doi: 10.1111/cpr.12573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sauvageau M, Goff LA, Lodato S, Bonev B, Groff AF, Gerhardinger C, et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife. 2013;2:e01749. doi: 10.7554/eLife.01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ramos AD, Andersen RE, Liu SJ, Nowakowski TJ, Hong SJ, Gertz C, et al. The long noncoding RNA Pnky regulates neuronal differentiation of embryonic and postnatal neural stem cells. Cell Stem Cell. 2015;16(4):439–447. doi: 10.1016/j.stem.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bocchi VD, Conforti P, Vezzoli E, Besusso D, Cappadona C, Lischetti T, et al. The coding and long noncoding single-cell atlas of the developing human fetal striatum. Science. 2021;372:6542. doi: 10.1126/science.abf5759. [DOI] [PubMed] [Google Scholar]
- 128.Grassi DA, Brattas PL, Jonsson ME, Atacho D, Karlsson O, Nolbrant S, et al. Profiling of lincRNAs in human pluripotent stem cell derived forebrain neural progenitor cells. Heliyon. 2020;6(1):e03067. doi: 10.1016/j.heliyon.2019.e03067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Prajapati B, Fatma M, Maddhesiya P, Sodhi MK, Fatima M, Dargar T, et al. Identification and epigenetic analysis of divergent long non-coding RNAs in multilineage differentiation of human Neural Progenitor Cells. RNA Biol. 2019;16(1):13–24. doi: 10.1080/15476286.2018.1553482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Rani N, Nowakowski TJ, Zhou H, Godshalk SE, Lisi V, Kriegstein AR, et al. A primate lncRNA mediates notch signaling during neuronal development by sequestering miRNA. Neuron. 2016;90(6):1174–1188. doi: 10.1016/j.neuron.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Peschansky VJ, Pastori C, Zeier Z, Wentzel K, Velmeshev D, Magistri M, et al. The long non-coding RNA FMR4 promotes proliferation of human neural precursor cells and epigenetic regulation of gene expression in trans. Mol Cell Neurosci. 2016;74:49–57. doi: 10.1016/j.mcn.2016.03.008. [DOI] [PubMed] [Google Scholar]
- 132.Liu SJ, Nowakowski TJ, Pollen AA, Lui JH, Horlbeck MA, Attenello FJ, et al. Single-cell analysis of long non-coding RNAs in the developing human neocortex. Genome Biol. 2016;17:67. doi: 10.1186/s13059-016-0932-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Field AR, Jacobs FMJ, Fiddes IT, Phillips APR, Reyes-Ortiz AM, LaMontagne E, et al. Structurally conserved primate LncRNAs are transiently expressed during human cortical differentiation and influence cell-type-specific genes. Stem Cell Rep. 2019;12(2):245–257. doi: 10.1016/j.stemcr.2018.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Nielsen JA, Lau P, Maric D, Barker JL, Hudson LD. Integrating microRNA and mRNA expression profiles of neuronal progenitors to identify regulatory networks underlying the onset of cortical neurogenesis. BMC Neurosci. 2009;10:98. doi: 10.1186/1471-2202-10-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Somel M, Liu X, Tang L, Yan Z, Hu H, Guo S, et al. MicroRNA-driven developmental remodeling in the brain distinguishes humans from other primates. PLoS Biol. 2011;9(12):e1001214. doi: 10.1371/journal.pbio.1001214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kosik KS. MicroRNAs tell an evo-devo story. Nat Rev Neurosci. 2009;10(10):754–759. doi: 10.1038/nrn2713. [DOI] [PubMed] [Google Scholar]
- 137.Volvert ML, Rogister F, Moonen G, Malgrange B, Nguyen L. MicroRNAs tune cerebral cortical neurogenesis. Cell Death Differ. 2012;19(10):1573–1581. doi: 10.1038/cdd.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Saurat N, Andersson T, Vasistha NA, Molnar Z, Livesey FJ. Dicer is required for neural stem cell multipotency and lineage progression during cerebral cortex development. Neural Dev. 2013;8:14. doi: 10.1186/1749-8104-8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kawase-Koga Y, Otaegi G, Sun T. Different timings of Dicer deletion affect neurogenesis and gliogenesis in the developing mouse central nervous system. Dev Dyn. 2009;238(11):2800–2812. doi: 10.1002/dvdy.22109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.De Pietri TD, Pulvers JN, Haffner C, Murchison EP, Hannon GJ, Huttner WB. miRNAs are essential for survival and differentiation of newborn neurons but not for expansion of neural progenitors during early neurogenesis in the mouse embryonic neocortex. Development. 2008;135(23):3911–3921. doi: 10.1242/dev.025080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Shu P, Wu C, Ruan X, Liu W, Hou L, Fu H, et al. Opposing gradients of microRNA expression temporally pattern layer formation in the developing neocortex. Dev Cell. 2019;49(5):764–854. doi: 10.1016/j.devcel.2019.04.017. [DOI] [PubMed] [Google Scholar]
- 142.Fairchild CLA, Cheema SK, Wong J, Hino K, Simo S, La Torre A. Let-7 regulates cell cycle dynamics in the developing cerebral cortex and retina. Sci Rep. 2019;9(1):15336. doi: 10.1038/s41598-019-51703-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Conaco C, Otto S, Han JJ, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc Natl Acad Sci USA. 2006;103(7):2422–2427. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Visvanathan J, Lee S, Lee B, Lee JW, Lee SK. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 2007;21(7):744–749. doi: 10.1101/gad.1519107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Abdullah AI, Zhang H, Nie Y, Tang W, Sun T. CDK7 and miR-210 co-regulate cell-cycle progression of neural progenitors in the developing neocortex. Stem Cell Rep. 2016;7(1):69–79. doi: 10.1016/j.stemcr.2016.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Nigro A, Menon R, Bergamaschi A, Clovis YM, Baldi A, Ehrmann M, et al. MiR-30e and miR-181d control radial glia cell proliferation via HtrA1 modulation. Cell Death Disease. 2012;3:e360. doi: 10.1038/cddis.2012.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Pollock A, Bian S, Zhang C, Chen Z, Sun T. Growth of the developing cerebral cortex is controlled by microRNA-7 through the p53 pathway. Cell Rep. 2014;7(4):1184–1196. doi: 10.1016/j.celrep.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Fededa JP, Esk C, Mierzwa B, Stanyte R, Yuan S, Zheng H, et al. MicroRNA-34/449 controls mitotic spindle orientation during mammalian cortex development. EMBO J. 2016;35(22):2386–2398. doi: 10.15252/embj.201694056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Fineberg SK, Kosik KS, Davidson BL. MicroRNAs potentiate neural development. Neuron. 2009;64(3):303–309. doi: 10.1016/j.neuron.2009.10.020. [DOI] [PubMed] [Google Scholar]
- 150.Nowakowski TJ, Fotaki V, Pollock A, Sun T, Pratt T, Price DJ. MicroRNA-92b regulates the development of intermediate cortical progenitors in embryonic mouse brain. Proc Natl Acad Sci USA. 2013;110(17):7056–7061. doi: 10.1073/pnas.1219385110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Shu P, Fu H, Zhao X, Wu C, Ruan X, Zeng Y, et al. MicroRNA-214 modulates neural progenitor cell differentiation by targeting Quaking during cerebral cortex development. Sci Rep. 2017;7(1):8014. doi: 10.1038/s41598-017-08450-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lv X, Jiang H, Liu Y, Lei X, Jiao J. MicroRNA-15b promotes neurogenesis and inhibits neural progenitor proliferation by directly repressing TET3 during early neocortical development. EMBO Rep. 2014;15(12):1305–1314. doi: 10.15252/embr.201438923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ghosh T, Aprea J, Nardelli J, Engel H, Selinger C, Mombereau C, et al. MicroRNAs establish robustness and adaptability of a critical gene network to regulate progenitor fate decisions during cortical neurogenesis. Cell Rep. 2014;7(6):1779–1788. doi: 10.1016/j.celrep.2014.05.029. [DOI] [PubMed] [Google Scholar]
- 154.Delaloy C, Liu L, Lee JA, Su H, Shen F, Yang GY, et al. MicroRNA-9 coordinates proliferation and migration of human embryonic stem cell-derived neural progenitors. Cell Stem Cell. 2010;6(4):323–335. doi: 10.1016/j.stem.2010.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Boissart C, Nissan X, Giraud-Triboult K, Peschanski M, Benchoua A. miR-125 potentiates early neural specification of human embryonic stem cells. Development. 2012;139(7):1247–1257. doi: 10.1242/dev.073627. [DOI] [PubMed] [Google Scholar]
- 156.Cimadamore F, Amador-Arjona A, Chen C, Huang CT, Terskikh AV. SOX2-LIN28/let-7 pathway regulates proliferation and neurogenesis in neural precursors. Proc Natl Acad Sci USA. 2013;110(32):E3017–E3026. doi: 10.1073/pnas.1220176110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Berezikov E. Evolution of microRNA diversity and regulation in animals. Nat Rev Genet. 2011;12(12):846–860. doi: 10.1038/nrg3079. [DOI] [PubMed] [Google Scholar]
- 158.Arcila ML, Betizeau M, Cambronne XA, Guzman E, Doerflinger N, Bouhallier F, et al. Novel primate miRNAs coevolved with ancient target genes in germinal zone-specific expression patterns. Neuron. 2014;81(6):1255–1262. doi: 10.1016/j.neuron.2014.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wang YM, Zheng YF, Yang SY, Yang ZM, Zhang LN, He YQ, et al. MicroRNA-197 controls ADAM10 expression to mediate MeCP2's role in the differentiation of neuronal progenitors. Cell Death Differ. 2019;26(10):1863–1879. doi: 10.1038/s41418-018-0257-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Farahani R, Rezaei-Lotfi S, Simonian M, Hunter N. Bi-modal reprogramming of cell cycle by MiRNA-4673 amplifies human neurogenic capacity. Cell Cycle. 2019;18(8):848–868. doi: 10.1080/15384101.2019.1595873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Moore D, Meays BM, Madduri LSV, Shahjin F, Chand S, Niu M, et al. Downregulation of an evolutionary young mir-1290 in an ipsc-derived neural stem cell model of autism spectrum disorder. Stem Cells Int. 2019;2019:8710180. doi: 10.1155/2019/8710180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Yelamanchili SV, Morsey B, Harrison EB, Rennard DA, Emanuel K, Thapa I, et al. The evolutionary young miR-1290 favors mitotic exit and differentiation of human neural progenitors through altering the cell cycle proteins. Cell Death Dis. 2014;5:e982. doi: 10.1038/cddis.2013.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Prodromidou K, Vlachos IS, Gaitanou M, Kouroupi G, Hatzigeorgiou AG, Matsas R. MicroRNA-934 is a novel primate-specific small non-coding RNA with neurogenic function during early development. Elife. 2020;2020:9. doi: 10.7554/eLife.50561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Greig LC, Woodworth MB, Galazo MJ, Padmanabhan H, Macklis JD. Molecular logic of neocortical projection neuron specification, development and diversity. Nat Rev Neurosci. 2013;14(11):755–769. doi: 10.1038/nrn3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Diaz JL, Siththanandan VB, Lu V, Gonzalez-Nava N, Pasquina L, MacDonald JL, et al. An evolutionarily acquired microRNA shapes development of mammalian cortical projections. Proc Natl Acad Sci USA. 2020;117(46):29113–29122. doi: 10.1073/pnas.2006700117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Rodriques SG, Stickels RR, Goeva A, Martin CA, Murray E, Vanderburg CR, et al. Slide-seq: A scalable technology for measuring genome-wide expression at high spatial resolution. Science. 2019;363(6434):1463–1467. doi: 10.1126/science.aaw1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Spanjaard B, Hu B, Mitic N, Olivares-Chauvet P, Janjuha S, Ninov N, et al. Simultaneous lineage tracing and cell-type identification using CRISPR-Cas9-induced genetic scars. Nat Biotechnol. 2018;36(5):469–473. doi: 10.1038/nbt.4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Alemany A, Florescu M, Baron CS, Peterson-Maduro J, van Oudenaarden A. Whole-organism clone tracing using single-cell sequencing. Nature. 2018;556(7699):108–112. doi: 10.1038/nature25969. [DOI] [PubMed] [Google Scholar]
- 169.Frieda KL, Linton JM, Hormoz S, Choi J, Chow KK, Singer ZS, et al. Synthetic recording and in situ readout of lineage information in single cells. Nature. 2017;541(7635):107–111. doi: 10.1038/nature20777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501(7467):373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Qian X, Nguyen HN, Song MM, Hadiono C, Ogden SC, Hammack C, et al. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell. 2016;165(5):1238–1254. doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Amiri A, Coppola G, Scuderi S, Wu F, Roychowdhury T, Liu F, et al. Transcriptome and epigenome landscape of human cortical development modeled in organoids. Science. 2018;362:6420. doi: 10.1126/science.aat6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Luo C, Lancaster MA, Castanon R, Nery JR, Knoblich JA, Ecker JR. Cerebral organoids recapitulate epigenomic signatures of the human fetal brain. Cell Rep. 2016;17(12):3369–3384. doi: 10.1016/j.celrep.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Mariani J, Coppola G, Zhang P, Abyzov A, Provini L, Tomasini L, et al. FOXG1-dependent dysregulation of GABA/glutamate neuron differentiation in autism spectrum disorders. Cell. 2015;162(2):375–390. doi: 10.1016/j.cell.2015.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Velasco S, Kedaigle AJ, Simmons SK, Nash A, Rocha M, Quadrato G, et al. Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature. 2019;570(7762):523–527. doi: 10.1038/s41586-019-1289-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Bhaduri A, Andrews MG, Kriegstein AR, Nowakowski TJ. Are organoids ready for prime time? Cell Stem Cell. 2020;27(3):361–365. doi: 10.1016/j.stem.2020.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Bhaduri A, Andrews MG, Mancia Leon W, Jung D, Shin D, Allen D, et al. Cell stress in cortical organoids impairs molecular subtype specification. Nature. 2020;578(7793):142–148. doi: 10.1038/s41586-020-1962-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Qian X, Su Y, Adam CD, Deutschmann AU, Pather SR, Goldberg EM, et al. Sliced human cortical organoids for modeling distinct cortical layer formation. Cell Stem Cell. 2020;26(5):766–81e9. doi: 10.1016/j.stem.2020.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Giandomenico SL, Sutcliffe M, Lancaster MA. Generation and long-term culture of advanced cerebral organoids for studying later stages of neural development. Nat Protoc. 2021;16(2):579–602. doi: 10.1038/s41596-020-00433-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Gordon A, Yoon SJ, Tran SS, Makinson CD, Park JY, Andersen J et al (2021) Long-term maturation of human cortical organoids matches key early postnatal transitions. Nat Neurosci [DOI] [PMC free article] [PubMed]
- 181.Xiang Y, Tanaka Y, Cakir B, Patterson B, Kim KY, Sun P, et al. hESC-derived thalamic organoids form reciprocal projections when fused with cortical organoids. Cell Stem Cell. 2019;24(3):487–97e7. doi: 10.1016/j.stem.2018.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Birey F, Andersen J, Makinson CD, Islam S, Wei W, Huber N, et al. Assembly of functionally integrated human forebrain spheroids. Nature. 2017;545(7652):54–59. doi: 10.1038/nature22330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Xiang Y, Tanaka Y, Patterson B, Kang YJ, Govindaiah G, Roselaar N, et al. Fusion of regionally specified hPSC-derived organoids models human brain development and interneuron migration. Cell Stem Cell. 2017;21(3):383–98e7. doi: 10.1016/j.stem.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Andersen J, Revah O, Miura Y, Thom N, Amin ND, Kelley KW, et al. Generation of functional human 3D cortico-motor assembloids. Cell. 2020;183(7):1913–29e26. doi: 10.1016/j.cell.2020.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.