Abstract
Many tumors are now understood to be heterogenous cell populations arising from a minority of epithelial-like cancer stem cells (CSCs). CSCs demonstrate distinctive metabolic signatures from the more differentiated surrounding tumor bulk that confer resistance to traditional chemotherapeutic regimens and potential for tumor relapse. Many CSC phenotypes including metabolism, epithelial-to-mesenchymal transition, cellular signaling pathway activity, and others, arise from altered mitochondrial function and turnover, which are regulated by constant cycles of mitochondrial fusion and fission. Further, recycling of mitochondria through mitophagy in CSCs is associated with maintenance of reactive oxygen species levels that dictate gene expression. The protein machinery that drives mitochondrial dynamics is surprisingly simple and may represent attractive new therapeutic avenues to target CSC metabolism and selectively eradicate tumor-generating cells to reduce the risks of metastasis and relapse for a variety of tumor types.
Keywords: Mitochondrial dynamics, Mitochondrial morphology, Cancer stem cells, Metabolism, Therapeutic resistance, Signaling, EMT
Introduction
Death from cancer currently ranks as second only to heart disease in number of deaths per year within the United States [1]. This burden reflects an urgent need for a greater understanding of the diversity of tumor biology to propel the development of effective therapeutic tools or combinations of currently existing medicines to target cancer in novel ways. To this end, recent research in cancer biology has reshaped the way that the cellular makeup and physiology of many solid and liquid tumors are understood by offering a hierarchical model of tumor organization to better explain cancer outcomes in both the laboratory and clinic. This interpretation, termed the cancer stem cell model, has been reviewed extensively elsewhere [2, 3], and states that for numerous tumors, there exists substantial cellular heterogeneity established from a minority of long-lived stem cells that slowly and asymmetrically divide to self-replicate and give rise to daughter cells. Daughter cells have impressive short-term proliferative capacity to form the bulk of tumors and to differentiate into more mature cell types, but lack significant tumor-initiating capability. Experimental evidence for this model of cancer has so-far been most robust in studies of glioblastoma [4, 5], leukemia [6, 7], breast [8, 9], lung [10, 11], and colorectal cancers [12]. Cancer stem cells are also known to be inherently resistant to conventional chemotherapeutic regimens and are believed to be the source of relapse after clinical remission. Therefore, increased understanding and appreciation for cancer stem cell biology are paramount to the development of new therapeutic regimens that aim to destroy this population of cells and to eradicate tumors in a manner that minimizes risk of future relapse.
Cancer stem cells often represent a small minority of cells within a tumor, thus identification and isolation of these cells away from non-CSCs is crucial for efforts aimed at characterizing their biology. To this end, groups typically rely on the expression of cell surface markers that suggest stemness based on either their expression in non-cancer stem cells or coexpression with stemness markers in cancer cell subpopulations. Single surface epitope expression is often used for these means, but combinations of gene expression are also frequently employed in different systems. For example, CD133 positivity is used extensively to mark cancer stem cells in colon [13], lung [14], and brain [15] CSCs, whereas a CD24−CD44+ population is often purified as breast cancer stem cells [9]. Stemness is also frequently measured by functional behaviors such as capacity of cells to form tumor spheres, termed spherogenicity, colony formation, limiting dilution assays, and serial transplantation assay performance.
Traditional chemotherapeutic regimens capitalize on destroying vastly proliferative cells, and can be quite successful in shrinking primary tumors. Unfortunately, these tumors often relapse, potentially due to CSC quiescence-driven therapeutic evasion and continued proliferation of daughter cells that repopulate tumors. This process is similar to the regrowth of epithelia in the body after initial insult, where basal somatic stem cells are tasked with regeneration of the cellular differentiation hierarchy that makes up that particular tissue. These intrinsic properties permit survival of CSCs which then resume the process of tumor expansion; however, it is becoming increasingly clear that external signals from the tumor microenvironment also drive cancer stem cell phenotypes and must be considered as well, as reviewed by Prager et al. [16]. Similar to non-cancer stem cells, it appears that cancer stem cell phenotypes can be markedly plastic and informed by signals from the niche they inhabit. It has been observed that upon selective ablation of LGR5+ colorectal cancer stem cells, differentiated daughter cells migrate into the CSC niche and dedifferentiate into LGR5+ CSCs to drive further tumor growth after ablation cessation [12, 17]. In contrast, Chen et al. found ablation of glioma cancer stem cells does not result in restoration of CSCs by differentiated progeny [4]. These results suggest that this phenomenon may be tissue-specific and make imperative our consideration of the tumor microenvironment on cancer stem cell physiology and therapeutic development.
Each of the cellular behaviors that distinguish CSCs from the tumor bulk and surrounding non-transformed stroma have been intimately linked with altered mitochondrial function. Mitochondria perform an array of vital functions including orchestration of cellular metabolism, sequestration of calcium, production of reactive oxygen species, synthesis of metabolic intermediates that remodel chromatin, and the primary hubs of apoptotic regulation [18]. These operations receive input from extracellular and intracellular signals to allow cells to adapt to stress and environmental change. Research in mitochondrial biology in the past two decades has begun to elucidate how mitochondrial dynamics, or the process of mitochondrial remodeling through constant cycles of organellar fusion and fission, contributes to normal cellular physiology and how it is co-opted to drive tumorigenesis [19]. Because cancer stem cells and mitochondria in cancer have been summarized thoroughly elsewhere [2, 3, 18], this review will focus primarily on how mitochondrial dynamics in particular empowers cancer stem cell behavior and how it may serve as a new target of therapeutic focus.
Mitochondrial dynamics
In contrast to the classical electron micrograph images of mitochondria as bean-shaped and discrete organelles in the cytoplasm, mitochondrial shape and size are incredibly dynamic and can be impressively heterogeneous even within a single cell. Continuous processes of mitochondrial fusion that merge shorter mitochondrial fragments into larger, more interconnected structures are balanced by fission, which severs longer mitochondrial networks into smaller particles [20]. Perhaps unexpectedly, the protein machinery that drives mitochondrial fusion and fission is strikingly simple. Fission is generally initiated upon mitochondrial membrane constriction by endoplasmic reticulum contacts [21]. Next, the large cytoplasmic GTPase dynamin-related protein 1, or DRP1, is recruited to mitochondria where it oligomerizes and spiralizes on the outer mitochondrial membrane before constricting upon GTP hydrolysis to cleave a mitochondrion into two [22]. Currently, there exists conflicting evidence of whether other proteins such as dynamin 2 are required to completely sever mitochondria, but the role of DRP1 in this process is paramount [23–25]. DRP1 also possesses multiple amino acid residues subject to post-translational modification that modulate its activity. Phosphorylation at serine 616 (S616) has been extensively demonstrated as a fission-activating modification [26, 27], whereas phosphorylation at S637 decreases fission activity of DRP1[28].
Mitochondrial fusion relies primarily on three other GTPases. Outer mitochondrial membrane fusion is accomplished by outer membrane-bound mitofusin 1 (MFN1) and mitofusin 2 (MFN2) [29]. These proteins form both heterodimers and homodimers that then hydrolyze GTP to bring together and fuse the outer membranes of two mitochondria [30]. Fusion of inner mitochondrial membranes relies on the action of optic atrophy 1 (OPA1) which lines the inner mitochondrial membrane to integrate two membranes into one and also serves to maintain mitochondrial cristae structural fidelity [31, 32]. Interestingly, OPA1 is cleaved from a fusiogenic long form (L-OPA1) into a pro-fission short form (S-OPA1) by proteases including YME1L and OMA1, with the ratio between the long and short forms dictating OPA1 action and mitochondrial morphology [33–35]. Mitofusins 1 and 2 offer functional redundancy in that both must be ablated to achieve significant impairment in mitochondrial fusion whereas removal of functional OPA1 alone is sufficient to accomplish similar means.
One consequence of fission–fusion cycling is the mixing of internal contents upon inner and outer mitochondrial membrane fusion, allowing areas of local dysfunction to be compensated for by intact neighboring regions in the newly formed mitochondrion [36]. Regions of mitochondria that harbor severely dysfunctional machinery such as electron transport chain complexes can be purified from the population by mitochondrial fission and targeted for degradation through a number of mitochondria-specific autophagy pathways, termed mitophagy [37]. Immediately following fission, one sister mitochondrion remains polarized with its mitochondrial membrane potential (MMP) intact whereas the other sister mitochondrion is depolarized. This depolarization acts as a functional challenge to newly formed fragments that must successfully repolarize or else face mitophagic degradation and elimination from the cellular mitochondrial pool [38]. Mitophagy therefore serves in mitochondrial quality control and is primarily carried about by either the PINK1-Parkin pathway or the BNIP3L/NIX and FUNDC1 pathway. Briefly, the PINK1-Parkin pathway works through stabilization of the kinase PINK1 on the mitochondrial outer membrane upon mitochondrion depolarization. PINK1 then phosphorylates ubiquitin chains present on mitochondrial outer membrane proteins, which in turn promotes recruitment of the E3 ubiquitin ligase Parkin. Parkin recruitment leads to additional ubiquitination of outer membrane proteins leading to the subsequent recruitment of autophagy receptors and the targeted removal of these depolarized mitochondria [39]. NIX and FUNDC1 are mitochondrial outer membrane proteins that can directly act as mitophagy receptors that interact with LC3 to promote autophagosome formation and facilitate mitochondrial degradation [40, 41]. The interaction between these receptors and LC3 can be induced by a variety of stimuli and is regulated through post-translational modifications.
Apart from content mixing and quality control, mitochondrial dynamics also dictate mitochondrial size and morphology. Both the average size and distribution of mitochondrial sizes within any given cell depends on dominance of fission, which favors smaller mitochondria, or fusion, which generates larger networks. Mitochondrial size affects function, as highly fused tubular mitochondrial networks demonstrate increased ATP synthase dimerization and ATP synthesis, and decreased degradation compared to fragmented mitochondria [42]. Conversely, smaller mitochondria are frequently observed in so-called “immature” cell states such as those in somatic stem cells and many tumor cells that have high-fission activity [43–46]. This fragmented morphology has been largely associated with heightened rates of glycolytic flux, decreased oxidative phosphorylation activity, increased reactive oxygen species (ROS) formation, and increased efficiency of both elimination through mitophagy as well as intracellular mitochondrial mobility [19]. Although these generalizations do not faithfully describe all cell types and contexts, the effects of mitochondrial dynamics on stem cells in the context of health and disease are beginning to yield intriguing patterns and a base of knowledge from which to grow.
The sensitivity of mitochondrial dynamics to intracellular and extracellular signaling coupled with the functional changes that mitochondrial dynamics can impose within a cell place this process as a key mediator of phenotypic change in response to stimuli. Given the myriad challenges that cancer cells must overcome to form primary tumors and then metastasize, it is unsurprising that the field of cancer is beginning to uncover critical ways in which modulating mitochondrial dynamics aids cells in their journey through transformation to metastasis.
Signaling pathway effects on mitochondrial dynamics
A primary function of mitochondria is to allow cells to adjust to cues originating from both extracellular and intracellular environments. To accomplish this, mitochondria must be sensitive to changes in signaling pathway activations through alterations in metabolism and morphology which then feed back to modulate activity of other signaling pathways. Because the effects of various signaling pathways on mitochondrial dynamics have been reviewed extensively elsewhere [47], we will provide only a brief discussion to establish some structure for how these pathways may fit into a cancer stem cell framework.
The RAS-MAPK axis is perhaps the most well-annotated pathway with regard to its effects on mitochondrial dynamics and spans many cellular model systems. Multiple groups including our own have identified that ERK directly phosphorylates DRP1 on serine 616 to potentiate DRP1-mediated mitochondrial fission [26, 48]. Since ERK activation lies downstream of RAS, RAF, and MEK activity, increased flux through any part of this cascade increases DRP1 fission activity and decreases mitochondrial size. ERK-mediated phosphorylation of DRP1 has been demonstrated by our group and others to be required for RAS-mediated tumorigenesis in multiple cancer types including pancreatic ductal adenocarcinoma [26] and melanoma [48], as well as for nuclear reprogramming of MEFs into epithelial-like colonies through overexpression of pluripotency factors [49]. Moreover, ERK kinase has been found to directly phosphorylate T562 of MFN1, decreasing its ability to tether mitochondria together and therefore preventing mitochondrial fusion [50].
Multiple groups have implicated the PI3K-AKT pathway as a pro-mitochondrial fission pathway. Tondera et al. found that in PC3 prostate cancer cells, the mitochondrial protein MTP18 (MTFP1) promotes mitochondrial fission and cellular proliferation, and that its expression is upregulated with increased PI3K activity, independent of AKT activity [51]. They also describe an anti-apoptotic role for MTP18 in HaCaT keratinocytes as evidenced by increased cleaved PARP following UVB exposure in MTP18 knockdown conditions [51]. Conversely, in neurons treated with Amyloid β, calcium influx stimulates CaMKII-mediated phosphorylation and activation of AKT which itself directly phosphorylates DRP1 S616 to increase mitochondrial fission [52]. In this system, Kim et al. observed an increase in apoptosis following mitochondrial fragmentation, in contrast to the findings in PC3 cells. It therefore seems that PI3K-AKT governance of mitochondrial morphology stems from multiple nodes in this pathway and demonstrates the complexity of signal integration on mitochondrial dynamics.
MYC is a transcription factor proto-oncogene that has been extensively studied across multiple cancer types and is associated with promotion of cell proliferation and growth. Effects of MYC on mitochondrial dynamics are variable depending on cellular context, but in Burkitt lymphoma, MYC directly occupies the DRP1 promoter to increase DRP1 transcription and mitochondrial fission [53]. A pro-fragmentation function of MYC has also been reported in mouse embryonic fibroblasts where c-Myc overexpression alone increases DRP1 localization to mitochondria and increased phosphorylation at S579 of DRP1, a post-translational modification that is fissiogenic as evidenced by increased mitochondrial fragmentation in phosphomimetic mutants at this residue [54]. Conversely, in breast mammary epithelia, c-Myc is an important driver of mitochondrial fusion through PLD6 activity and inhibition of YAP/TAZ function which is crucial in maintenance of stemness phenotypes [55].
Execution of mitochondrial fission is also achieved through activation of the cellular stress sensor AMP-activated protein kinase (AMPK). In studies of U2OS cells, AMPK expression is necessary for mitochondrial fragmentation following introduction of mitochondrial stressors antimycin-A and rotenone [56]. Further, chemical activation of AMPK in the absence of these stressors produces significant mitochondrial fission mediated by direct phosphorylation at S155 and S172 of Mitochondrial Fission Factor (MFF) which serves as a receptor to recruit DRP1 to mitochondria [56]. Additionally, signaling pathway strength, including that of AMPK, can be influenced by mitochondrial dynamics as well. In a model of glioblastoma, Xie et al. discovered that knockdown of DRP1 expression decreases oxygen consumption rate and increases cell stress, activating AMPK [57]. These reports exemplify how mitochondrial function is altered by incoming signals and how mitochondria then affect signaling strength of pathways that are sensitive to mitochondrial outputs.
Mitochondrial dynamics in cancer stem cell metabolism
Reorganization of mitochondrial structure through mitochondrial fusion and fission is known to be associated with a host of functional outcomes in the context of non-transformed stem cells as well as cancer stem cells. A wide variety of metabolic phenotypes are associated with mitochondrial morphologies that are highly dependent on tissue type and differentiation status. Classically, immature cell types such as embryonic stem cells and many tumor cells demonstrate highly fragmented mitochondria that rely on aerobic glycolysis for energy production [45, 46]. Elevated rates of glycolysis, even in the presence of oxygen, permit use of glycolytic intermediates in anabolic metabolism to fuel cell growth and proliferation through increased synthesis of essential building blocks such as nucleotides and lipids [58]. Additionally, metabolic phenotypes coordinated by mitochondria have been well described to affect cellular identity reprogramming through modulation of chromatin accessibility [59, 60].
Mitochondria orchestrate gene expression through synthesis of metabolites that form retrograde signals by serving as cofactors and substrates for gene expression-modifying enzymes. The majority of research on mitochondrial intermediate-driven gene expression changes has focused on production of acetyl-CoA [61, 62], alpha-ketoglutarate [63], succinate [64], and reactive oxygen species [65, 66]. Included within this group of enzymes are the Ten-eleven translocation (TET) and PHD families of DNA and histone demethylases, respectively, which are both activated by high levels of alpha-ketoglutarate and inhibited by succinate and fumarate [67]. Additionally, Jumonji domain-containing histone demethylase activity is also reliant on alpha-ketoglutarate. Acetyl-CoA synthesized in the cytoplasm from mitochondrial-derived citrate is the substrate for acetylation of histones, DNA, and cytosolic enzymes. Alterations in levels of reactive oxygen species synthesized within mitochondria can both directly modify enzymes such as p38 MAPK [68] and DUSP6 [69], and activate antioxidant programs such as the NRF2 pathway that modulate gene expression through direct binding of ROS-sensitive transcription factors [70]. Additionally, mitochondrial content affects mitochondrial capacity to buffer cytosolic calcium, which modulates activity of calcium-sensitive cellular signaling such as that mediated by Calcineurin [71], as discussed in greater detail in the therapeutic resistance section. Retrograde signaling networks can form significant interactions in which one mitochondrial metabolite may influence activation of another mitochondrial retrograde signaling program, as exemplified by work in the Avadhani lab demonstrating mitochondrial ROS activation of calcineurin signaling [72].
Mitochondrial metabolite-sensitive signaling is regulated by mitochondrial dynamics and affects cellular metabolism. In elegant studies of mouse embryonic cortex development, Khacho et al. demonstrated that neural stem cell (NSC) mitochondrial morphology exhibits a transient fragmentation phase critical to differentiation [73]. In this work, highly glycolytic SOX2+ NSCs with elongated mitochondria differentiate into SOX2− TBR2+ neural progenitors that exhibit increased oxidative phosphorylation and fragmented mitochondrial morphology. When progenitors then differentiate further into DCX+ neurons, their mitochondria revert to a fused network morphology. Genetic inhibition of mitochondrial fragmentation through depletion of DRP1 blocks the conversion of neural stem cells into the more differentiated progenitor cells in vivo, indicating decreased differentiation potential and increased self-renewal, a finding also supported by in vitro neurosphere formation assays. Conversely, knockout of fusion components MFN1/2 or OPA1 knockdown decreases self-renewal in vivo and in vitro. Interestingly, knockout of either DRP1 or MFN1/2 increases oxygen consumption rate but does not impair mitochondrial function. This group also found that a highly interconnected mitochondrial morphology is linked with decreased reactive oxygen species formation and increased transcription of self-renewal genes. Upon mitochondrial fragmentation, mitochondrial ROS levels rise and stimulate the ROS-protection transcription factor NRF2 which inhibits self-renewal genes and promotes transcription of differentiation genes [73].
In contrast, Folmes et al. found that mouse embryonic fibroblasts (MEFs) demonstrate well-connected mitochondrial networks with abundant cristae that are converted to highly fragmented and cristae-poor populations when they are reprogrammed into induced pluripotent stem cells (iPSCs) [74]. This morphological change is coupled with a departure from oxidative phosphorylation-predominant metabolism in differentiated MEFs into a highly glycolytic metabolism in iPSCs. Increases in glycolytic gene expression profiles precede activation of pluripotency genes, adding a temporal relationship between engagement of glycolytic metabolism and nuclear reprogramming [74]. Additionally, dedifferentiation is severely diminished upon inhibition of glycolytic capacity [74]. Their findings support the notion that mitochondrial dynamics are intimately linked with metabolic phenotypes that inform differentiation status and plasticity. These studies and those of Khacho also demonstrate how different tissue types display varying dependencies on both mitochondrial morphology and metabolism to support stem versus more differentiated cellular phenotypes. This is a critical concept to justify future investigations of the influence that mitochondrial dynamics holds on metabolism of different populations of stem cells throughout the body.
Focusing on stem cells in the context of cancer, Civenni et al. discovered that metastatic and hormone treatment-refractory prostate cancer cells upregulate mitochondrial fission factor (MFF) compared to primary tumor and normal prostate counterparts [75]. MFF functions as a mitochondrial receptor to which DRP1 binds to initiate constriction and mitochondrial fission. Knockdown of MFF decreases cancer stem cell proliferation in in vitro tumor sphere formation assays and significantly decreases in vivo tumor growth. Interestingly, MFF knockdown is associated with significant reductions in both oxygen consumption rate and spare respiratory capacity of tumor stem cells in tumor sphere assays, but not in adherent cells representative of the tumor bulk [75]. This indicates that even within a tumor, the stem-like cell populations can exhibit differential metabolic dependencies on mitochondrial dynamics machinery from the more differentiated non-stem tumor bulk.
The results in prostate cancer stem cells were supported by investigations in a patient-derived xenograph model of glioblastoma. Xie et al. found that brain tumor-initiating cells (BTICs), the cancer stem cell equivalent, display a significantly more fragmented mitochondrial morphology compared to the more differentiated non-BTIC tumor cells [57]. Additionally, even though the total levels of DRP1 are comparable between BTIC and non-BTIC cells, levels of activating phospho-S616 are elevated in BTIC versus non-BTIC. Conversely, the expression of the DRP1-inactivating phopho-S637 is reduced in the stem cell population versus the more differentiated tumor bulk. This observation was tested functionally by differentiating BTIC into non-BTIC, which over the course of differentiation leads to decreases in phospho-S616 and increases in phospho-S637 of DRP1, suggesting significant inactivation. To further complement the functional importance of activating and inactivating phosphorylations of DRP1 in glioblastoma differentiation and stemness, the group overexpressed phosphomimetic S616 and phospho-dead S637 DRP1 in differentiated non-BTICs and observed decreased expression of glioblastoma differentiation markers GFAP and MAP2 and concomitant increased expression of stemness markers such as NES, NANOG, SSEA-1 (FUT4), and Oct4, among others. Metabolically, shRNA knockdown of DRP1 in BTIC decreases oxygen consumption rate, with basal OCR falling to half of control knockdown and maximal OCR reaching only one-third of that in control knockdown. DRP1 knockdown cells also demonstrate decreased in vitro proliferation and tumor sphere formation, as well as inhibition of in vivo tumor growth [57]. These studies highlight the importance of mitochondrial dynamics in glioblastoma metabolism and stemness gene expression, and suggest that future therapy for this disease may benefit from drugs that affect mitochondrial dynamics.
Two publications in the last six years from the Jordan lab have shed light on the relationship between mitochondrial morphology, metabolism, and cancer stem cell identity in acute myeloid leukemia. Leukemic stem cells (LSCs) in primary AML samples are purified from non-LSCs based on low levels of reactive oxygen species in LSCs compared to ROS-high non-LSCs [76]. These leukemic stem cells are metabolically dormant compared to non-LSCs and exhibit lower levels of oxidative phosphorylation, glycolysis, and ATP, although their diminished metabolic needs were met primarily through oxidative phosphorylation. LSCs display fewer and highly fragmented mitochondria compared to non-LSCs that results from increased expression of pro-fission FIS1 following AMPK activation [77]. Treatment with mitochondrial stressor valinomycin causes significant accumulation of mitochondria in non-LSCs with more fused mitochondria compared to LSCs, indicating that FIS1-mediated mitochondrial fission is critical to clearing damaged mitochondria in LSCs and that non-LSCs were unable to do so. Further, knockdown of FIS1 in primary AML inactivates the myeloid anti-differentiation gene GSK3, therefore promoting cellular differentiation and reducing maintenance of cancer stem cell identity. In all, this group delineated a mechanism by which AML leukemic stem cells are characterized by low levels of ROS and preserve stem cell identity through mitochondrial fragmentation-dependent elimination of stressed mitochondria [76, 77].
Mitochondrial dynamics in cancer stem cell metastasis
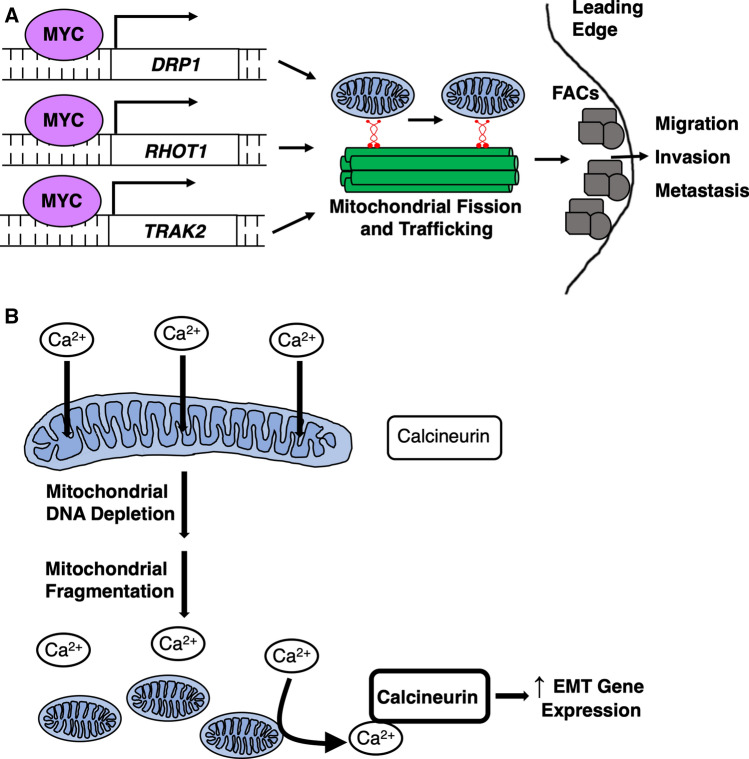
Most deaths from cancer occur following metastasis of the primary tumor to a distant site and not due to stress of the primary tumor itself. Cancer stem cells are well-equipped to make the epithelial-to-mesenchymal transition (EMT) to facilitate physical transit from tissue of origin to site of metastasis through unique and numerous mitochondrial processes [78]. Induction of EMT drives highly glycolytic metabolism [79–81], which is dependent on mitochondrial dynamics, as MFN1 knockdown promotes fragmented mitochondrial morphology, increases expression of glycolysis genes, and increases cancer cell migration and metastasis [82]. EMT also involves the downregulation of epithelial gene expression such as cell–cell adhesion marker E-cadherin and upregulation of motility and polarization genes necessary for cells to transit across tissues and into circulatory systems. This process is regulated by signaling networks such as AMPK and linked with marked changes in mitochondrial intracellular localization as mitochondria are trafficked to the cellular leading edge to fuel local reactions around structures involved in cellular motility such as focal adhesions [83, 84]. In fact, the oncogenic transcription factor MYC directly binds to the promoters of DRP1 and mitochondrial trafficking genes including RHOT1, RHOT2, and TRAK2 to promote mitochondrial fission and motility within cells (Fig. 1a) [53]. Mitochondrial fission permits mitochondrial redistribution along the cortical actin cytoskeleton which is inhibited upon knockdown of MYC or any of the trafficking genes. Further, cells with decreased mitochondrial fragmentation and trafficking demonstrate significantly impaired focal adhesion dynamics, cellular motility, and invasion capacity [53]. In agreement with these results, Desai et al. found that destabilizing mitochondrial dynamics to favor fusion in breast cancer cells by overexpressing OPA1 or a dominant-negative DRP1 inhibits the anterior localization of mitochondria between the nucleus and leading edge of migrating cells [85]. Failure to properly traffic mitochondria to the anterior compartment causes slower cellular migration and decreases directional persistence [85]. Both of the above studies speak to the critical role that proper mitochondrial dynamics plays in intracellular motility of mitochondria to sustain reorganization and function of molecular structures that propel cancer cells movement.
Fig. 1.
Mechanisms of mitochondrial dynamics-mediated EMT. a Myc upregulates expression of DRP1 and mitochondrial trafficking genes RHOT1 and TRAK2 by binding their promoter regions. Upregulation of these components increases mitochondrial motility to the cellular leading edge to promote focal adhesion complex (FAC) dynamics important for cellular migration, invasion, and metastasis [53]. b Depletion of mitochondrial DNA leads to mitochondrial fragmentation, which decreases mitochondrial calcium buffering. Increased cytosolic calcium activates calcineurin signaling to upregulate EMT gene expression [71]
Apart from the effects of mitochondrial dynamics on intracellular redistribution, changes in mitochondrial morphology can direct EMT through gene expression alterations as well. Numerous tumors, including breast cancer, colorectal cancer, and lung cancer, have inherently low copy numbers of the mitochondrial genome (mtDNA) [86]. Experimental reduction in mtDNA copy number in breast cancer leads to cells with fewer and more fragmented mitochondria, and greater spherogenic potential and proportion of CD44high/CD24low cells, both markers of stemness, that are reverted upon mtDNA restoration [71]. Depletion of mtDNA also reduces mitochondrial calcium sequestration within the cell which then stimulates Calcineurin-dependent upregulation of EMT gene expression and increases cellular movement velocity and in vitro invasion (Fig. 1b) [71].
Another focus of mitochondrial function in cancer stem cell EMT comes from mitochondrial production of reactive oxygen species. In breast cancer and colon adenocarcinoma tumor spheres, cancer stem cells isolated for high levels of reactive oxygen species (RH-TS) were more successful in establishing metastases than were the ROS-low cells (RL-TS) from the same parental spheres [65]. By transmission electron microscopy, RH-TS harbor swollen and cristae-poor mitochondria whereas RL-TS mitochondria are denser with well-formed cristae. Additionally, ROS-high cells demonstrate a gene expression signature of increased fatty acid beta-oxidation and ROS detoxification programs compared to ROS-low cells. This pattern is paralleled by increased MAPK pathway activity through oxidation of ROS-sensitive p38 MAPK and upregulation of classic EMT gene expression such as MMP1 and LOX [65]. Conversely, in studies of normal human mammary epithelial cells, forced EMT through overexpression of EMT master transcription factor SNAIL or treatment with TGFβ increases mitochondrial fusion by upregulating MFN1 [87]. Interestingly, the MFN1-PKCς complex is required in this system to tether fused mitochondria to the cortical membrane, which upon asymmetric cell division, ensures that mitochondria are unequally segregated between the two daughter cells. Daughter cells that retain fused mitochondrial networks exhibit increased glutathione synthesis and protection from reactive oxygen species, which drives stem cell self-renewal whereas cells that receive fewer mitochondria progress through luminal differentiation. Knockdown of MFN1 decreases stemness measured by spherogenic potential in both normal mammary epithelial cells and in BT549 and MDA-MB-231 stem cell-rich breast cancer cell lines [87]. Because the role of MFN1 in EMT somewhat conflicts between studies assessing MFN1-driven glycolysis in EMT and these studies, it is important to explore the relationships between mitochondrial dynamics components and EMT across multiple systems and tumor types. In general, these studies demonstrate the critical role of mitochondrial structure in determining mitochondrial segregation and function, such as ROS synthesis, and highlight how structure can act both upstream and downstream of EMT signaling activation. This complexity is also exemplified by studies in esophageal squamous cell carcinoma (ESCC) in which activation of EMT through TGFβ stimulation increases expression of CSC marker CD44 in a parkin-dependent manner [88]. Knockdown of parkin suppresses mitophagy, increases levels of mitochondrial ROS, and decreases proportion of CD44high cells. Cellular turnover of mitochondria to control concentrations of mitochondrial metabolites like ROS is an emerging theme that appears to affect many cellular phenotypes such as identity, EMT, and therapeutic resistance as we will explore in the next section.
Mitochondrial dynamics in cancer stem cell therapeutic resistance
Much of the rationale for research on cancer stem cells lies in the historical failure of traditional therapeutic regimens to achieve tumor remission without relapse. Therefore, investigation into how CSCs may be efficiently targeted while sparing non-transformed host tissue is critical to therapy development and treatment success. The identification of cancer stem cells and tumor heterogeneity driving tumor formation and relapse is itself monumental, but insufficient to achieve these ambitious goals. Further inquiry into how cancer stem cells evade traditional therapies and what vulnerabilities they demonstrate for potential targeting is crucial. Because identified mechanisms of therapeutic resistance are often tissue- or tumor-specific, this section is subdivided by tumor type.
Pancreatic ductal adenocarcinoma (PDAC) CSCs:
Studies by Viale et al. strongly support the notion that targeting the tumor bulk is often unsuccessful at eliminating cancer stem cells [89]. In their work, spontaneous PDAC tumor cells surviving complete KRASG12D ablation are characterized as pancreatic cancer stem cells by CD133+ and CD44high expression and demonstrate upregulation of mitochondrial biogenesis transcription factor PGC1A and increased mitochondrial mass. Surviving cells also display significant differences in mitochondrial morphology, increased dependency on oxygen consumption, decreased glycolytic flux, and increased sensitivity to OXPHOS inhibition. ATP synthase inhibitor oligomycin significantly diminishes in vitro spherogenic potential of these cells and extends lifespan of mice subject to tumor relapse compared to vehicle control [89]. These studies nicely support both the existence of differential metabolic profiles of the tumor bulk versus CSCs and how pharmacotherapy aimed at mitochondrial function in CSCs may be most effective in preventing relapse.
Gynecological CSCs:
In cervical and ovarian cancers, Kong et al. found that mitochondrial fusion may endow therapeutic resistance [90]. Cisplatin treatment in chemosensitive cells with an intermediate mitochondrial morphology phenotype induces massive mitochondrial fragmentation and apoptosis whereas chemoresistant cells exhibit very interconnected and tubular mitochondrial networks both before and after treatment. One proposed mechanism for maintenance of fused morphology is decreased OMA1 protease-mediated processing of OPA1. Cisplatin treatment did not affect the ratio of the pro-fusion long form L-OPA1 to the fissiogenic S-OPA1 in chemoresistant cells, but led to decreased relative L-OPA1 and increased S-OPA1 in chemosensitive cells [90]. Alternatively, Zampieri et al. find that mitochondrial morphology in cisplatin-sensitive versus cisplatin-resistant cells is cell line dependent, where resistant COV-362 cells demonstrate an abundance of networked mitochondria compared to sensitive cells, but SKOV-3 cells demonstrate no such difference [91]. Both cell lines display upregulation of mitophagy and dependence on mitophagy to maintain CSC phenotypes like clonogenicity in the context of cisplatin resistance, indicating that regardless of mitochondrial morphology, targeting mitophagy may be efficacious.
Breast CSCs:
Therapeutic resistance in breast cancer stem cells (BCSCs) is mediated by increased fatty acid oxidation through upregulation of CPT1B [92]. CPT1B is the long chain fatty acid transporter localized to mitochondria, and serves as the rate-limiting step in fatty acid oxidation. Consistent with this, BCSCs exhibit a significant decrease in cell proliferation, tumor sphere formation, and proportion of tumor-initiating cells upon pharmacologic inhibition of CPT1B. Additionally, non-CSCs demonstrate perinuclear mitochondrial distribution whereas BCSCs exhibit a branched network phenotype [92]. Similarly, mitochondrial metabolism through ROS synthesis was discovered to drive breast CSC identity and therapeutic resistance through increased ROS-sensitive HIF1A activity [93]. Generation of chemotherapy-resistant breast cancer cell lines induces upregulation of MYC and the anti-apoptotic and Bcl-2 family protein MCL1. This is accompanied by increases in proportion of breast CSCs, mammosphere formation, oxidative phosphorylation, and mitochondrial ROS. Morphologically, knockdown of MCL1 leads to fragmentation of elongated mitochondrial networks into smaller and rounder mitochondria. Similarly, MYC knockdown decreases the number of mitochondria per cell as measured by transmission electron microscopy and mtDNA content. MYC and MCL1 were found to cooperate in maintenance of breast CSC identity by increasing mitochondrial biogenesis and ROS-mediated HIF1A stabilization. HIF1A activates stem cell genes, including NANOG [93]. These groups present strong evidence for BCSC chemotherapeutic resistance driven by metabolic transitions accompanied by elongated mitochondrial network morphology and points to fatty acid oxidation inhibition as a new therapeutic avenue to eradicate breast cancer stem cells.
Lung CSCs:
Studies of lung adenocarcinoma present another interesting mechanism of therapeutic resistance in CSCs through alteration of one-carbon metabolism. One-carbon metabolism comprises the transfer of one-carbon units between molecules and is mediated by various species of folate, most of which are unable to traverse the mitochondrial membranes, so compartment-specific enzymes exist to restore levels of necessary forms of folate for reactions in the cytosol and mitochondria. These reactions are critical for many anabolic pathways including nucleotide synthesis, amino acid homeostasis, oxidation–reduction protection, and epigenetic manipulation [94]. Mitochondrial methylenetetrahydrofolate dehydrogenase 2 (MTHFD2) activity generates folate intermediates used for the synthesis of purine nucleotides. Overexpression of this enzyme has been previously identified in a meta-analysis of human tumor gene expression, but its significance to cancer stem cell biology in particular has been largely unexplored [95]. An exception comes from studies of lung adenocarcinoma cancer stem cells in which expression of MTHFD2 is crucial for tumorigenesis and expression of stem cell genes. Knockdown of MTHFD2 in this system decreases tumor-initiating cell frequency, tumor sphere formation, and expression of stem cell transcription factor SOX2. Additionally, lung adenocarcinoma cells resistant to the EGFR inhibitor gefitinib demonstrate significant upregulation of MTHFD2 compared to the chemosensitive parental cell line and a marked sensitivity to MTHFD2 knockdown. This work implicates one-carbon metabolism in cancer stem cell gene expression signatures and development of therapeutic resistance; however, it is yet to be determined how mitochondrial dynamics itself may influence one-carbon metabolism enzyme activity or substrate availability. It is possible that mitochondrial morphology or turnover may affect efficiency of one-carbon metabolism, but further studies directly manipulating mitochondrial dynamics machinery will be required to address this.
Glioblastoma CSCs:
Another mechanism by which mitochondrial dynamics appears to dictate drug resistance in cancer stem cells is through mitophagy-mediated mitochondrial quality control. The mitophagy receptor NIX is upregulated in glioblastoma stem cells (GSCs) residing in the hypoxic tumor niche [96]. Knockdown of NIX in GSCs decreases the expression of stemness markers CD133, Oct4, and SOX2, and attenuates GSC cell viability [96]. Further, NIX knockdown increases survival compared to control in a xenograft model. The NIX promoter contains both antioxidant response and hypoxia response elements, causing NIX upregulation in the presence of either hypoxia or elevated levels of reactive oxygen species. In hypoxia, there is a time-dependent decrease in mitochondrial mass in GSCs upon hypoxia treatment, which is prevented by NIX knockdown. Interestingly, NIX also promotes activity of the mTOR pathway, leading to stimulation of hypoxia signaling which itself has been shown to upregulate expression of the multidrug resistance (MDR1) gene [97]. It is therefore feasible that maintenance of hypoxia signaling through mitophagy confers chemotherapeutic resistance.
Colorectal CSCs:
Studies in colorectal cancer agree with those in glioblastoma in describing chemoresistance of CSCs by means of mitophagy [98]. In this system, expression of NIX/BNIP3L and mitophagy activity is upregulated in colorectal CSCs compared to non-CSCs. Treatment with mitochondrial ROS-inducing doxorubicin causes mitochondrial mass to decrease as BNIP3L expression increases and autophagy components colocalize with mitochondria. This occurs to a greater extent in CSCs and leads to decreased levels of mitochondrial superoxide in CSCs than non-CSCs [98]. Mitophagy therefore appears to be a strong defense against doxorubicin-induced mitochondrial ROS. Although the authors do not comment on mitochondrial morphology per se, it would be worthwhile to explore how mitochondrial fusion and fission are affected in the context of mitophagy-mediated chemotherapy resistance and whether manipulation of fusion-fission components affects cellular resistance and mitophagy phenotypes.
Acute myeloid leukemia CSCs:
Leukemic stem cells (LSC) in acute myeloid leukemia appear to also rely on low levels of reactive oxygen species to potentiate chemoresistance [76]. Primary AML stem cells purified by selection for low levels of ROS demonstrate an increase in mitochondrial respiration, upregulation of anti-apoptotic protein BCL-2, and exquisite resistance to the standard AML chemotherapy agent daunorubicin. This group did not explore changes in mitochondrial dynamics specifically in LSCs versus non-stem AML cells, but high FIS1 expression has been reported as a risk factor for poor clinical response in AML [7]. FIS1 plays a role in mitochondrial fission and mitophagy, so it is tempting to draw links between studies in AML and those in colorectal CSCs to hypothesize that FIS1 increases the rate of mitophagy to depress mitochondrial ROS and maintain stemness and resistance mechanisms in AML.
Interestingly, radiotherapy exposure has also been found to promote cancer stem cell phenotypes including EMT and metastatic potential, CSC marker expression, and metabolic changes in many tumor types such as prostate, breast, and lung cancer [99]. One mechanism by which this occurs is through radiotherapy-induced mitochondrial production of reactive oxygen species that then activate transcription factors critical to these processes such as SNAIL for EMT and HIF1A for numerous CSC phenotypes. Although data specifically on mitochondrial dynamics in the context of irradiation effects on cancer stem cells are lacking, Patten et al. have found that upon irradiation, mesenchymal stem cells exhibit elevated mitochondrial oxidative phosphorylation, increased electron transport chain supercomplex formation, and slight mitochondrial elongation [100]. These changes occur as adaptations to decrease DNA damage after an initial priming dose of radiation before treatment with a full dose. Additionally, this priming behavior is dependent on mitochondrial dynamics, as OPA1 knockout MEFs are unable to exhibit priming and demonstrate no significant difference in DNA damage after a full dose regardless of priming irradiation [64]. It is therefore possible that similar changes in mitochondrial morphology occur within cancer stem cells after radiotherapy given the mitochondrial metabolic response that CSCs exhibit following radiotherapy and that chemotherapy administration often leads to a similar metabolic profile associated with increased mitochondrial mass and fusion into extended networks.
Conclusions
Although the field of mitochondrial dynamics is still in its adolescence, it is becoming increasingly evident that mitochondrial fusion–fission cycling and its command of mitochondrial size are powerful dictators of mitochondrial metabolism in health and disease. The molecular mechanisms that drive mitochondrial fission and fusion are surprisingly simple, with, as far as we know, only a few proteins being absolutely instrumental in these processes. This knowledge coupled with the power that mitochondrial dynamics machinery exerts over essential mitochondrial functions like coordination of metabolism, formation of reactive oxygen species, and the rate of mitophagy makes the development of specific and potent pharmacologic inhibitors, and perhaps potentiators, of proteins such as DRP1, MFN1, MFN2, and OPA1 imperative. These tools would enable investigators and clinicians to bypass the complex cell signaling effects that modulate mitochondrial dynamics proteins upstream of their action and to enforce certain mitochondrial phenotypes in cells that may be most sensitive to these changes.
Although the details of how mitochondrial dynamics affect mitochondrial metabolism and chemotherapeutic resistance in cancer stem cells are heterogeneous across tumor types, in general, it appears that CSCs rely heavily on oxidative phosphorylation and mitophagy for maintenance of stem cell character and for survival under chemotherapeutic challenge (Fig. 2). Because many traditional therapies have historically targeted features of the tumor bulk, often characterized by high rate of proliferation and growth, achieving remission has remained an enormous challenge as cancer stem cells may repopulate the tumor niche after treatment cessation. Further insights into how mitochondrial dynamics affect mitochondrial function in cancer stem cells will inform future treatment; however, it is also critical to gain a deeper understanding of tumor heterogeneity to appreciate how therapy affects cancer stem cell populations and tumors more broadly. For instance, how well do CSC markers perform in identifying tumor initiation capacity between histologically similar tumors from different patients? How does culturing technique affect CSC proportions and phenotypes? How do different subpopulations of CSCs within the same tumor compare to each other based on CSC marker expression and metabolic signatures? Although groups have made some progress on this front [101], much work remains. Identifying the extent and patterns of tumor heterogeneity and how it associates with cellular metabolism is vital to therapy aimed at CSC destruction. Approaching these questions will rely on new techniques to assess complex CSC phenotypes in vivo and may benefit from combining lineage tracing approaches with single-cell metabolomics and single-cell gene expression analysis.
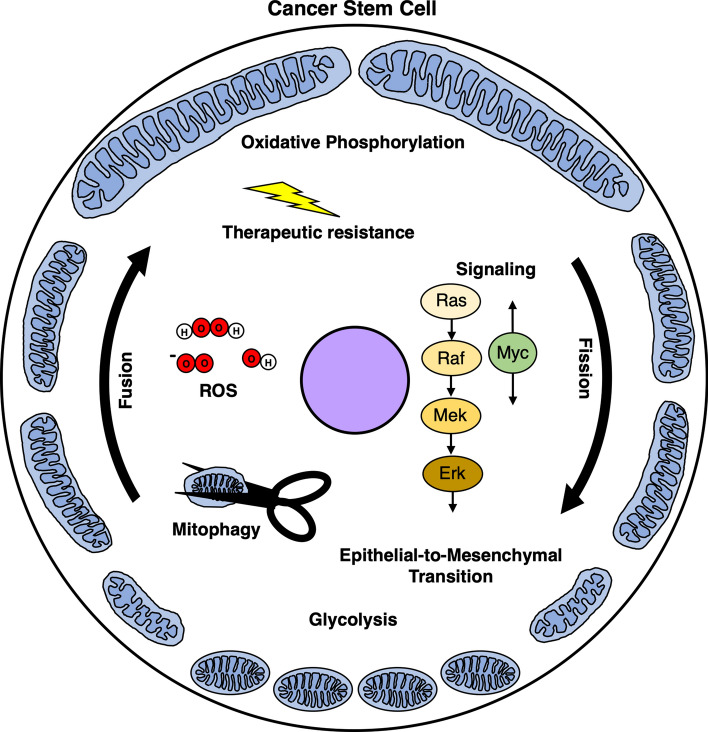
Fig. 2.
General phenotypes associated with mitochondrial fission or fusion in cancer stem cells (CSCs). Oncogenic signaling pathways affect mitochondrial dynamics and vice versa; for example, MAPK signaling increases mitochondrial fission whereas Myc has mixed effects. Mitochondrial fission is generally associated with increased rates of mitophagy, EMT, and glycolysis. Conversely, mitochondrial fusion is frequently associated with increased oxidative phosphorylation and therapeutic resistance
Novel therapeutic avenues will likely come from development of new chemotherapeutics and adapting currently available and FDA-approved medicines toward CSC destruction. Experimental evidence that the latter is possible comes from studies of colorectal cancer stem cells. Treatment of colorectal cancer cell lines with standard chemotherapeutic agent oxaliplatin demonstrates a dose-dependent enrichment of colorectal CSCs [102]. These cells exhibit standard CSC behaviors such as slow rates of cell proliferation and elevated sphere-forming potential, as well as increased activation of autophagy and lysosomal pathways. Treatment with the antimalarial lysosomal inhibitor mefloquine vastly inhibits the expression of key lysosomal proteins LAMP1 and LAMP2, as well as mitophagy components PINK1 and Parkin, and leads to mitochondrial ballooning and dissolution of cristae structure in CSCs, but not in healthy tissue. Combination therapy of mefloquine with oxaliplatin almost completely attenuates CSC growth in three xenograft mouse models [102]. These results encourage the discovery of new combinations of current medicines that capitalize on the unique metabolic and morphologic signatures of mitochondria in cancer stem cells. Additionally, although the manner through which mitochondrial-derived ROS affect cancer stem cells is still controversial, pharmacologic modulation of ROS levels in cancer may be worth exploring, especially because of the availability of clinically approved mitochondrial-targeted antioxidants like MitoQ [103], which is currently being tested in clinical trials for inflammatory and vascular diseases.
Ultimately, it will take a concerted effort to further delineate patterns of mitochondrial dynamics and metabolism that differentiate cancer stem cells from the tumor bulk and healthy host tissue. This will demand a new focus and understanding of the role cancer stem cells specifically play in the formation and maintenance of tumors, as well as the variety of mitochondrial phenotypes that they demonstrate, which render them sensitive to treatments that target mitochondrial functions. To gain the most insight, future work should also focus on directly perturbing mitochondrial dynamics components (Table 1) as opposed to relying solely on proposed inhibitors that may have off-target effects.
Table 1.
Studies with direct manipulation of mitochondrial dynamics machinery
| Model system | Mitochondrial dynamics perturbation | Major findings | References |
|---|---|---|---|
| Human mammary epithelial cells |
Mdivi-1 Drp1 K38A overexpression |
Drp1 inhibition decreases YAP/TAZ-dependent clonogenicity | [55] |
| Human primary AML and AML cell lines | Fis1 shRNA knockdown | Fis1 knockdown inhibits mitophagy, colony formation, and engraftment potential | [77] |
| Human glioblastoma PDX |
Drp1 S616E S637A overexpression Drp1 shRNA knockdown |
Drp1 S616E S637A overexpression inhibits differentiation and upregulates stemness gene expression Drp1 knockdown decreases OCR, in vitro proliferation, spherogenesis, and in vivo tumor growth |
[57] |
| Mouse embryonic cortex development |
Drp1 or Mfn1/2 knockout Opa1 shRNA knockdown |
Inhibition of mitochondrial fission decreases ROS production and neural stem cell differentiation Inhibition of mitochondrial fusion promotes ROS production and differentiation |
[73] |
| Human prostate cancer cell lines | Mff shRNA knockdown | Mff knockdown decreases OCR, cell proliferation, spherogenesis, and in vivo tumor growth | [75] |
|
Human mammary epithelial cells Human breast cancer cell lines |
Mfn1 knockdown | Mfn1 knockdown decreases stemness and tumor sphere formation | [87] |
| Mouse embryonic fibroblasts | Opa1 knockout | Opa1 knockout inhibits protective priming to irradiation exposure | [100] |
Acknowledgements
We thank the members of the Kashatus lab for review of the manuscript.
Funding
This work is supported by NIH grant CA200755 (to D.F.K.).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References:
- 1.Murphy SL, Kochanek KD, Xu J, Arias E (2015) Mortality in the United States, 2014. NCHS Data Brief No. 229 [PubMed]
- 2.Batlle E, Clevers H. Cancer stem cells revisited. Nat Med. 2017;23:1124–1134. doi: 10.1038/nm.4409. [DOI] [PubMed] [Google Scholar]
- 3.Beck B, Blanpain C. Unravelling cancer stem cell potential. Nat Rev Cancer. 2013;13:727–738. doi: 10.1038/nrc3597. [DOI] [PubMed] [Google Scholar]
- 4.Chen J, Li Y, Yu TS, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu J, Yuan X, Xu Q, et al. Cancer stem cells in glioblastoma. Stem cells cancer stem cells, vol 1 stem cells cancer stem cells. Ther Appl Dis Inj. 2012;1:113–120. doi: 10.1007/978-94-007-1709-1_14. [DOI] [Google Scholar]
- 6.Dick JE, Bonnet D. Human Acute Myeloid Leukaemia is organised as a heirarchy that originates from a primitive haematopoetic cell. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 7.Tian Y, Huang Z, Wang Z, et al. Identification of novel molecular markers for prognosis estimation of acute myeloid leukemia: over-expression of PDCD7, FIS1 and Ang2 may indicate poor prognosis in pretreatment patients with acute myeloid leukemia. PLoS ONE. 2014;9:5–10. doi: 10.1371/journal.pone.0084150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fillmore CM, Kuperwasser C. Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Res. 2008;10:1–13. doi: 10.1186/bcr1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Al-Hajj M, Wicha M, Benito-Hernandez A, et al. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci. 2003;100(7):3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masciale V, Grisendi G, Banchelli F, et al. Isolation and identification of cancer stem-like cells in adenocarcinoma and squamous cell carcinoma of the lung: a pilot study. Front Oncol. 2019;9:1–12. doi: 10.3389/fonc.2019.01394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herreros-Pomares A, de-Maya-GironesCalabuig-Fariñas JDS, et al. Lung tumorspheres reveal cancer stem cell-like properties and a score with prognostic impact in resected non-small-cell lung cancer. Cell Death Dis. 2019;10:1–14. doi: 10.1038/s41419-019-1898-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Sousa E, Melo F, Kurtova AV, Harnoss JM, et al. A distinct role for Lgr5 + stem cells in primary and metastatic colon cancer. Nature. 2017;543:676–680. doi: 10.1038/nature21713. [DOI] [PubMed] [Google Scholar]
- 13.O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 14.Bertolini G, Roz L, Perego P, et al. Highly tumorigenic lung cancer CD133+ cells display stem-like features and are spared by cisplatin treatment. Proc Natl Acad Sci USA. 2009;106:16281–16286. doi: 10.1073/pnas.0905653106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 16.Prager BC, Xie Q, Bao S, Rich JN. Cancer stem cells: the architects of the tumor ecosystem. Cell Stem Cell. 2019;24:41–53. doi: 10.1016/j.stem.2018.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shimokawa M, Ohta Y, Nishikori S, et al. Visualization and targeting of LGR5 + human colon cancer stem cells. Nature. 2017;545:187–192. doi: 10.1038/nature22081. [DOI] [PubMed] [Google Scholar]
- 18.Vyas S, Zaganjor E, Haigis MC. Mitochondria and Cancer. Cell. 2016;166:555–566. doi: 10.1016/j.cell.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen H, Chan DC. Mitochondrial dynamics in regulating the unique phenotypes of cancer and stem cells. Cell Metab. 2017;26:39–48. doi: 10.1016/j.cmet.2017.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bereiter-Hahn J, Vöth M. Dynamics of mitochondria in living cells: shape changes, dislocations, fusion, and fission of mitochondria. Microsc Res Tech. 1994;27:198–219. doi: 10.1002/jemt.1070270303. [DOI] [PubMed] [Google Scholar]
- 21.Friedman JR, Lackner LL, West M, et al. ER tubules mark sites of mitochondrial division. Science (80-) 2011;334:358–362. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ingerman E, Perkins EM, Marino M, et al. Dnm1 forms spirals that are structurally tailored to fit mitochondria. J Cell Biol. 2005;170:1021–1027. doi: 10.1083/jcb.200506078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamerkar SC, Kraus F, Sharpe AJ, et al. Dynamin-related protein 1 has membrane constricting and severing abilities sufficient for mitochondrial and peroxisomal fission. Nat Commun. 2018 doi: 10.1038/s41467-018-07543-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fonseca TB, Sánchez-Guerrero Á, Milosevic I, Raimundo N. Mitochondrial fission requires DRP1 but not dynamins. Nature. 2019;570:E34–E42. doi: 10.1038/s41586-019-1296-y. [DOI] [PubMed] [Google Scholar]
- 25.Lee JE, Westrate LM, Wu H, et al. Multiple dynamin family members collaborate to drive mitochondrial division. Nature. 2016;540:139–143. doi: 10.1038/nature20555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kashatus JA, Nascimento A, Myers LJ, et al. Erk2 phosphorylation of Drp1 promotes mitochondrial fission and MAPK-driven tumor growth. Mol Cell. 2015;57:537–551. doi: 10.1016/j.molcel.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taguchi N, Ishihara N, Jofuku A, et al. Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J Biol Chem. 2007;282:11521–11529. doi: 10.1074/jbc.M607279200. [DOI] [PubMed] [Google Scholar]
- 28.Chang CR, Blackstone C. Cyclic AMP-dependent protein kinase phosphorylation of Drp1 regulates its GTPase activity and mitochondrial morphology. J Biol Chem. 2007;282:21583–21587. doi: 10.1074/jbc.C700083200. [DOI] [PubMed] [Google Scholar]
- 29.Santel A, Fuller MT. Control of mitochondrial morphology by a human mitofusin. J Cell Sci. 2001;114:867–874. doi: 10.1242/jcs.114.5.867. [DOI] [PubMed] [Google Scholar]
- 30.Chen H, Detmer SA, Ewald AJ, et al. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olichon A, Emorine LJ, Descoins E, et al. The human dynamin-related protein OPA1 is anchored to the mitochondrial inner membrane facing the inter-membrane space. FEBS Lett. 2002;523:171–176. doi: 10.1016/S0014-5793(02)02985-X. [DOI] [PubMed] [Google Scholar]
- 32.Olichon A, Baricault L, Gas N, et al. Loss of OPA1 perturbates the mitochondrial inner membrane structure and integrity, leading to cytochrome c release and apoptosis. J Biol Chem. 2003;278:7743–7746. doi: 10.1074/jbc.C200677200. [DOI] [PubMed] [Google Scholar]
- 33.Ehses S, Raschke I, Mancuso G, et al. Regulation of OPA1 processing and mitochondrial fusion by m-AAA protease isoenzymes and OMA1. J Cell Biol. 2009;187:1023–1036. doi: 10.1083/jcb.200906084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ishihara N, Fujita Y, Oka T, Mihara K. Regulation of mitochondrial morphology through proteolytic cleavage of OPA1. EMBO J. 2006;25:2966–2977. doi: 10.1038/sj.emboj.7601184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anand R, Wai T, Baker MJ, et al. The i-AAA protease YME1L and OMA1 cleave OPA1 to balance mitochondrial fusion and fission. J Cell Biol. 2014;204:919–929. doi: 10.1083/jcb.201308006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rolland SG, Motori E, Memar N, et al. Impaired complex IV activity in response to loss of LRPPRC function can be compensated by mitochondrial hyperfusion. Proc Natl Acad Sci USA. 2013 doi: 10.1073/pnas.1303872110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pickles S, Vigié P, Youle RJ. Mitophagy and quality control mechanisms in mitochondrial maintenance. Curr Biol. 2018;28:R170–R185. doi: 10.1016/j.cub.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cho HM, Ryu JR, Jo Y, et al. Drp1-Zip1 interaction regulates mitochondrial quality surveillance system. Mol Cell. 2019;73:364–376.e8. doi: 10.1016/j.molcel.2018.11.009. [DOI] [PubMed] [Google Scholar]
- 39.Lazarou M, Sliter DA, Kane LA, et al. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524:309–314. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu L, Feng D, Chen G, et al. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat Cell Biol. 2012;14:177–185. doi: 10.1038/ncb2422. [DOI] [PubMed] [Google Scholar]
- 41.Novak I, Kirkin V, McEwan DG, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gomes LC, Di BG, Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat Cell Biol. 2011;13:589–598. doi: 10.1038/ncb2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.St. JohnRamalho-SantosGray JCJHL, et al. The expression of mitochondrial DNA transcription factors during early cardiomyocyte in vitro differentiation from human embryonic stemn cells. Clon Stem Cells. 2005;7:141–153. doi: 10.1089/clo.2005.7.141. [DOI] [PubMed] [Google Scholar]
- 44.Lee WTY, John JS. The control of mitochondrial DNA replication during development and tumorigenesis. Ann N Y Acad Sci. 2015;1350:95–106. doi: 10.1111/nyas.12873. [DOI] [PubMed] [Google Scholar]
- 45.Prigione A, Fauler B, Lurz R, et al. The senescence-related mitochondrial/oxidative stress pathway is repressed in human induced pluripotent stem cells. Stem Cells. 2010;28:721–733. doi: 10.1002/stem.404. [DOI] [PubMed] [Google Scholar]
- 46.Zhang J, Khvorostov I, Hong JS, et al. UCP2 regulates energy metabolism and differentiation potential of human pluripotent stem cells. EMBO J. 2011;30:4860–4873. doi: 10.1038/emboj.2011.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nagdas S, Kashatus DF. The interplay between oncogenic signaling networks and mitochondrial dynamics. Antioxidants. 2017 doi: 10.3390/antiox6020033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Serasinghe MN, Wieder SY, Renault TT, et al. Mitochondrial division is requisite to RAS-induced transformation and targeted by oncogenic MAPK pathway inhibitors. Mol Cell. 2015;57:521–536. doi: 10.1016/j.molcel.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prieto J, León M, Ponsoda X, et al. Early ERK1/2 activation promotes DRP1-dependent mitochondrial fission necessary for cell reprogramming. Nat Commun. 2016 doi: 10.1038/ncomms11124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pyakurel A, Savoia C, Hess D, Scorrano L. Extracellular regulated kinase phosphorylates mitofusin 1 to control mitochondrial morphology and apoptosis. Mol Cell. 2015;58:244–254. doi: 10.1016/j.molcel.2015.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tondera D, Santel A, Schwarzer R, et al. Knockdown of MTP18, a novel phosphatidylinositol 3-kinase-dependent protein, affects mitochondrial morphology and induces apoptosis. J Biol Chem. 2004;279:31544–31555. doi: 10.1074/jbc.M404704200. [DOI] [PubMed] [Google Scholar]
- 52.Kim DI, Lee KH, Gabr AA, et al. Aβ-Induced Drp1 phosphorylation through Akt activation promotes excessive mitochondrial fission leading to neuronal apoptosis. Biochim Biophys Acta Mol Cell Res. 2016;1863:2820–2834. doi: 10.1016/j.bbamcr.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 53.Agarwal E, Altman BJ, Ho Seo J, et al. Myc regulation of a mitochondrial trafficking network mediates tumor cell invasion and metastasis. Mol Cell Biol. 2019 doi: 10.1128/mcb.00109-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Prieto J, Seo AY, León M, et al. MYC induces a hybrid energetics program early in cell reprogramming. Stem Cell Rep. 2018;11:1479–1492. doi: 10.1016/j.stemcr.2018.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.von Eyss B, Jaenicke LA, Kortlever RM, et al. A MYC-driven change in mitochondrial dynamics limits YAP/TAZ function in mammary epithelial cells and breast cancer. Cancer Cell. 2015;28:743–757. doi: 10.1016/j.ccell.2015.10.013. [DOI] [PubMed] [Google Scholar]
- 56.Toyama EQ, Herzig S, Courchet J, et al. Metabolism: AMP-activated protein kinase mediates mitochondrial fission in response to energy stress. Science (80-) 2016;351:275–281. doi: 10.1126/science.aab4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xie Q, Wu Q, Horbinski CM, et al. Mitochondrial control by DRP1 in brain tumor initiating cells. Nat Neurosci. 2015;18:501–510. doi: 10.1038/nn.3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.DeBerardinis RJ, Chandel NS. We need to talk about the Warburg effect. Nat Metab. 2020;2:127–129. doi: 10.1038/s42255-020-0172-2. [DOI] [PubMed] [Google Scholar]
- 59.Sperber H, Mathieu J, Wang Y, et al. The metabolome regulates the epigenetic landscape during naive-to-primed human embryonic stem cell transition. Nat Cell Biol. 2015;17:1523–1535. doi: 10.1038/ncb3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kaelin WG, McKnight SL. Influence of metabolism on epigenetics and disease. Cell. 2013;153:56–69. doi: 10.1016/j.cell.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee JV, Carrer A, Shah S, et al. Akt-dependent metabolic reprogramming regulates tumor cell Histone acetylation. Cell Metab. 2014;20:306–319. doi: 10.1016/j.cmet.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wellen KE, Hatzivassiliou G, Sachdeva UM, et al. ATP-citrate lyase links cellular metabolism to histone acetylation. Science (80-) 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Carey BW, Finley LWS, Cross JR, et al. Intracellular α-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature. 2015;518:413–416. doi: 10.1038/nature13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Selak MA, Armour SM, MacKenzie ED, et al. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-α prolyl hydroxylase. Cancer Cell. 2005;7:77–85. doi: 10.1016/j.ccr.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 65.Wang C, Shao L, Pan C, et al. Elevated level of mitochondrial reactive oxygen species via fatty acid β-oxidation in cancer stem cells promotes cancer metastasis by inducing epithelial-mesenchymal transition. Stem Cell Res Ther. 2019 doi: 10.1186/s13287-019-1265-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hamanaka RB, Glasauer A, Hoover P, et al. Mitochondrial reactive oxygen species promote epidermal differentiation and hair follicle. Development. 2013;6:1–10. doi: 10.1126/scisignal.2003638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.MacKenzie ED, Selak MA, Tennant DA, et al. Cell-permeating α-ketoglutarate derivatives alleviate pseudohypoxia in succinate dehydrogenase-deficient cells. Mol Cell Biol. 2007;27:3282–3289. doi: 10.1128/mcb.01927-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rodríguez-Colman MJ, Schewe M, Meerlo M, et al. Interplay between metabolic identities in the intestinal crypt supports stem cell function. Nature. 2017;543:424–427. doi: 10.1038/nature21673. [DOI] [PubMed] [Google Scholar]
- 69.Ma R, Ma L, Weng W, et al. DUSP6 SUMOylation protects cells from oxidative damage via direct regulation of Drp1 dephosphorylation. Sci Adv. 2020 doi: 10.1126/sciadv.aaz0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xue D, Zhou X, Qiu J. Emerging role of NRF2 in ROS-mediated tumor chemoresistance. Biomed Pharmacother. 2020;131:110676. doi: 10.1016/j.biopha.2020.110676. [DOI] [PubMed] [Google Scholar]
- 71.Guha M, Srinivasan S, Ruthel G, et al. Mitochondrial retrograde signaling induces epithelial-mesenchymal transition and generates breast cancer stem cells. Oncogene. 2014;33:5238–5250. doi: 10.1038/onc.2013.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Srinivasan S, Koenigstein A, Joseph J, et al. Role of mitochondrial reactive oxygen species in osteoclast differentiation. Ann N Y Acad Sci. 2010;1192:245–252. doi: 10.1111/j.1749-6632.2009.05377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Khacho M, Clark A, Svoboda DS, et al. Mitochondrial dynamics impacts stem cell identity and fate decisions by regulating a nuclear transcriptional program. Cell Stem Cell. 2016;19:232–247. doi: 10.1016/j.stem.2016.04.015. [DOI] [PubMed] [Google Scholar]
- 74.Folmes CDL, Nelson TJ, Martinez-Fernandez A, et al. Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metab. 2011;14:264–271. doi: 10.1016/j.cmet.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Civenni G, Bosotti R, Timpanaro A, et al. Epigenetic control of mitochondrial fission enables self-renewal of stem-like tumor cells in human prostate cancer. Cell Metab. 2019;30:303–318.e6. doi: 10.1016/j.cmet.2019.05.004. [DOI] [PubMed] [Google Scholar]
- 76.Lagadinou ED, Sach A, Callahan K, et al. BCL-2 inhibition targets oxidative phosphorylation and selectively eradicates quiescent human leukemia stem cells. Cell Stem Cell. 2013;12:329–341. doi: 10.1016/j.stem.2012.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pei S, Minhajuddin M, Adane B, et al. AMPK/FIS1-mediated mitophagy is required for self-renewal of human AML stem cells. Cell Stem Cell. 2018;23:86–100.e6. doi: 10.1016/j.stem.2018.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ye X, Tam WL, Shibue T, et al. Distinct EMT programs control normal mammary stem cells and tumour-initiating cells. Nature. 2015;525:256–260. doi: 10.1038/nature14897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rodríguez-García A, Samsó P, Fontova P, et al. TGF-β1 targets Smad, p38 MAPK, and PI3K/Akt signaling pathways to induce PFKFB3 gene expression and glycolysis in glioblastoma cells. FEBS J. 2017;284:3437–3454. doi: 10.1111/febs.14201. [DOI] [PubMed] [Google Scholar]
- 80.Masin M, Vazquez J, Rossi S, et al. GLUT3 is induced during epithelial-mesenchymal transition and promotes tumor cell proliferation in non-small cell lung cancer. Cancer Metab. 2014;2:11. doi: 10.1186/2049-3002-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu M, Quek L-E, Sultani G, Turner N. Epithelial-mesenchymal transition induction is associated with augmented glucose uptake and lactate production in pancreatic ductal adenocarcinoma. Cancer Metab. 2016;4:1–13. doi: 10.1186/s40170-016-0160-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang Z, Li TE, Chen M, et al. MFN1-dependent alteration of mitochondrial dynamics drives hepatocellular carcinoma metastasis by glucose metabolic reprogramming. Br J Cancer. 2020;122:209–220. doi: 10.1038/s41416-019-0658-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cunniff B, McKenzie AJ, Heintz NH, Howe AK. AMPK activity regulates trafficking of Mitochondria to the leading edge during cell migration and matrix invasion. Mol Biol Cell. 2016;27:2662–2674. doi: 10.1091/mbc.E16-05-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Caino MC, Ghosh JC, Chae YC, et al. PI3K therapy reprograms mitochondrial trafficking to fuel tumor cell invasion. Proc Natl Acad Sci USA. 2015;112:8638–8643. doi: 10.1073/pnas.1500722112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Desai SP, Bhatia SN, Toner M, Irimia D. Mitochondrial localization and the persistent migration of epithelial cancer cells. Biophys J. 2013;104:2077–2088. doi: 10.1016/j.bpj.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lee HC, Yin PH, Lin JC, et al. Mitochondrial genome instability and mtDNA depletion in human cancers. Ann N Y Acad Sci. 2005;1042:109–122. doi: 10.1196/annals.1338.011. [DOI] [PubMed] [Google Scholar]
- 87.Wu MJ, Chen YS, Kim MR, et al. Epithelial-mesenchymal transition directs stem cell polarity via regulation of mitofusin. Cell Metab. 2019;29:993–1002.e6. doi: 10.1016/j.cmet.2018.11.004. [DOI] [PubMed] [Google Scholar]
- 88.Whelan KA, Chandramouleeswaran PM, Tanaka K, et al. Autophagy supports generation of cells with high CD44 expression via modulation of oxidative stress and Parkin-mediated mitochondrial clearance. Oncogene. 2017 doi: 10.1038/onc.2017.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Viale A, Pettazzoni P, Lyssiotis CA, et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature. 2014;514:628–632. doi: 10.1038/nature13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kong B, Tsuyoshi H, Orisaka M, et al. Mitochondrial dynamics regulating chemoresistance in gynecological cancers. Ann N Y Acad Sci. 2015;1350:1–16. doi: 10.1111/nyas.12883. [DOI] [PubMed] [Google Scholar]
- 91.Zampieri LX, Grasso D, Bouzin C et al (2020) Mitochondria participate in chemoresistance to cisplatin in human ovarian cancer cells. Mol Cancer Res molcanres.1145.2019. 10.1158/1541-7786.mcr-19-1145 [DOI] [PubMed]
- 92.Wang T, Fahrmann JF, Lee H, et al. JAK/STAT3-regulated fatty acid β-oxidation is critical for breast cancer stem cell self-renewal and chemoresistance. Cell Metab. 2018;27:136–150.e5. doi: 10.1016/j.cmet.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lee K, GiltnaneBalko JMJM, et al. MYC and MCL1 cooperatively promote chemotherapy-resistant breast cancer stem cells via regulation of mitochondrial oxidative phosphorylation. Cell Metab. 2017;26:633–647.e7. doi: 10.1016/j.cmet.2017.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ducker GS, Rabinowitz JD. Cell metabolism review one-carbon metabolism in health and disease. Cell Metab. 2017;25:27–42. doi: 10.1016/j.cmet.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nilsson R, Jain M, Madhusudhan N, et al. Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nat Commun. 2014 doi: 10.1038/ncomms4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jung J, Zhang Y, Celiku O et al (2019) Mitochondrial NIX promotes tumor survival in the hypoxic niche of glioblastoma. Cancer Res canres.0198.2019. 10.1158/0008-5472.can-19-0198 [DOI] [PMC free article] [PubMed]
- 97.Comerford KM, Wallace TJ, Karhausen J, et al. Hypoxia-inducible factor-1-dependent regulation of the multidrug resistance (MDR1) gene. Cancer Res. 2002;62:3387–3394. [PubMed] [Google Scholar]
- 98.Yan C, Luo L, Guo CY, et al. Doxorubicin-induced mitophagy contributes to drug resistance in cancer stem cells from HCT8 human colorectal cancer cells. Cancer Lett. 2017;388:34–42. doi: 10.1016/j.canlet.2016.11.018. [DOI] [PubMed] [Google Scholar]
- 99.Lee SY, Jeong EK, Ju MK, et al. Induction of metastasis, cancer stem cell phenotype, and oncogenic metabolism in cancer cells by ionizing radiation. Mol Cancer. 2017;16:1–25. doi: 10.1186/s12943-016-0577-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Patten DA, Ouellet M, Allan DS, et al. Mitochondrial adaptation in human mesenchymal stem cells following ionizing radiation. FASEB J. 2019;33:9263–9278. doi: 10.1096/fj.201801483rr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Stewart JM, Shaw PA, Gedye C, et al. Phenotypic heterogeneity and instability of human ovarian tumor-initiating cells. Proc Natl Acad Sci USA. 2011;108:6468–6473. doi: 10.1073/pnas.1005529108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Takeda M, Koseki J, Takahashi H, et al. Disruption of endolysosomal Rab5/7 efficiently eliminates colorectal cancer stem cells. Cancer Res. 2019;79:1426–1437. doi: 10.1158/0008-5472.CAN-18-2192. [DOI] [PubMed] [Google Scholar]
- 103.Adlam VJ, Harrison JC, Porteous CM, et al. Targeting an antioxidant to mitochondria decreases cardiac ischemia-reperfusion injury. FASEB J. 2005;19:1088–1095. doi: 10.1096/fj.05-3718com. [DOI] [PubMed] [Google Scholar]