Abstract
Numerous mechanisms involved in promoting cancer cell survival under nutrient starvation have been described. Long noncoding RNAs (lncRNAs) have emerged as critical players in colorectal cancer (CRC) progression, but the role of lncRNAs in the progression of CRC under nutrient starvation has not been well clarified. Here, we identified a lncRNA, LINC01615, that was significantly upregulated in response to serum starvation. LINC01615 can contribute to the adaptation of CRC cells to serum-deprived conditions and enhance cell survival under similar conditions. LINC01615 activated the pentose phosphate pathway (PPP) under serum starvation, manifested as decreased ROS production and enhanced nucleotide and lipid synthesis. Glucose-6-phosphate dehydrogenase (G6PD) is a key rate-limiting enzyme of the PPP, and LINC01615 promoted G6PD expression by competitively binding with hnRNPA1 and facilitating G6PD pre-mRNA splicing. Moreover, we also found that serum starvation led to METTL3 degradation by inducing autophagy, which further increased the stability and level of LINC01615 in a m6A-dependent manner. LINC01615 knockdown combined with oxaliplatin achieved remarkable antitumor effects in PDO and PDX models. Collectively, our results demonstrated a novel adaptive survival mechanism permitting tumor cells to survive under limiting nutrient supplies and provided a potential therapeutic target for CRC.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-022-04675-7.
Keywords: LINC01615, Pentose phosphate pathway, Serum starvation, Cell survival, Autophagy
Introduction
Cancer cells rely on the availability of nutrients to maintain cell growth and survival. In rapidly growing solid tumors, including colorectal cancer (CRC), delayed tumor angiogenesis usually leads to a shortage of many nutritional components, including glucose, amino acids, and lipids [1, 2]. To survive, cancer cells have evolved complex regulatory networks to withstand nutritional deficiency. Understanding the tolerance mechanism may provide new strategies for cancer therapy [3].
Long noncoding RNAs (lncRNAs) are a class of noncoding RNAs > 200 nucleotides (nt) in length. Abnormal expression of lncRNAs has been reported in various types of cancers and plays important roles in tumorigenesis and progression, including in the adaptation to various nutrient absence environments [4]. Studies have reported that the lncRNA GAS5 can be induced by the absence of growth factors or serum starvation to inhibit cell proliferation and promote apoptosis [5]. Moreover, under glucose starvation conditions, the lncRNA TRINGS can be induced by P53 and protects cancer cells from necrosis [6]. These studies suggest that lncRNAs may be involved in helping cells adapt to nutrient starvation. However, more specific mechanisms regarding the role of lncRNAs in resistance to nutrient starvation still need to be characterized.
The pentose phosphate pathway (PPP) has been demonstrated to be an important pathway for cell adaptation to various stresses, including nutrient deficiency [7]. The PPP starts from the branch point of glycolysis, glucose-6-phosphate (G6P). G6P dehydrogenase (G6PD) is the first rate-limiting enzyme that catalyzes the initial step of the PPP shunt, converting G6P to 6-phosphoglucolactone [8]. The PPP plays a vital role in meeting the cellular demands of biosynthesis and antioxidant defense [9]. It generates ribose-5-phosphate (R5P) for de novo synthesis of nucleotides and produces NADPH for biosynthesis of lipids and detoxification of reactive oxygen species (ROS).
In our study, we used serum-free medium to simulate nutritional deficiency [10, 11] and treat CRC cells. LINC01615 was identified as a nutrient starvation-associated lncRNA. Further studies confirmed that LINC01615 can inhibit apoptosis and promote cell survival under starvation conditions by activating G6PD-induced PPP flux. The autophagy-reduced m6A writer METTL3 can lead to upregulated expression of LINC01615. Intriguingly, we also demonstrated that LINC01615 depletion has strong synergistic antitumor activity when combined with oxaliplatin. Overall, this lncRNA renders cells capable of withstanding nutrient starvation and may be a novel target for CRC treatment.
Material and methods
Patient and tissue specimens
This study was reviewed and approved by the ethics committee of Xuzhou Medical University (XYFY2019-KL221-01). Written informed consent was obtained from all patients. The study included 76 patients with CRC who were 42–84 years old, all of whom underwent surgery from 2019.12 to 2020.05 at the Department of General Surgery, Affiliated Hospital of Xuzhou Medical University. Clinicopathological characteristics were also examined, such as age, sex, tumor size, depth of invasion, tumor differentiation, lymph node invasion, TNM stage, and metastasis. Tumors were classified and graded based on the pTNM classification advocated by the International Union against Cancer.
Cell lines and cell culture
The human normal colonic epithelial cell line FHC and the CRC cell lines SW480, HT29, SW620, DLD1, HCT116, and LoVo were purchased from the Chinese Academy of Science (Shanghai, China). HCT116 cells were cultured in McCoy’s 5A medium (HyClone, USA) containing 10% fetal bovine serum (Gibco, USA), 100 U/ml penicillin, and 100 μg/ml streptomycin (Vicmed, China), while FHC, SW480, and 293T cells were cultured in DMEM, and HT29, SW620, DLD1, and LoVo cells were cultured in RPMI 1640 medium. All cells were cultured at 37 °C in a humidified incubator with 5% CO2. Serum-free medium was used to replace complete medium 6 h before collecting cells, and the cells were treated with bafilomycin A1 at a concentration of 0.5 µM for 6 h.
Short interfering RNA (siRNA) and plasmid construction, plasmid transfection and RNA interference
Full-length LINC01615 (NR_132622, 647 bp), full-length G6PD (NM_000402, 2223 bp), full-length hnRNPA1 (NM_031157, 3744 bp) and LINC01615 antisense were also synthesized by GENEWIZ and cloned into the pcDNA3.1 vector (Invitrogen). F2: LINC01615/1–249, F3: LINC01615/250–399, F4: LINC01615/400–549, and F5: LINC01615/500–647 were generated by polymerase chain reaction (PCR) and subcloned into the pcDNA3.1 vector. P1: hnRNPA1/1–97, P2: hnRNPA1/97–184, P3: hnRNPA1/184–240, P4: hnRNPA1/240–372 and P5: hnRNPA1/1–372 constructs were cloned into a pcDNA3.1-Flag vector.
CRC cells were transfected with siRNA (GenePharma, China) using siLentFect lipid reagent (Bio-Rad, USA) and were infected with plasmid (Genechem, China) using Lipofectamine 3000 transfection reagent (ThermoFisher, USA) according to the manufacturer’s protocol. The transfection effects were verified by real-time quantitative reverse transcription PCR (qRT‒PCR) and Western blotting (WB). All sequences are provided in the Supplementary Materials and Methods.
Northern blot assay
Northern blotting was performed with a Northern Blot Kit (Short Chain) (Bersinbio, China) according to the manufacturer’s protocol. Total RNA was extracted from cells, electrophoresed in a formaldehyde denaturing agarose gel and transferred to a nylon membrane. After ultraviolet cross-linking, the membrane was prehybridized and incubated with a DIG-labeled probe. Finally, the membrane was incubated with anti-DIG and detected with CSPD.
Oxidative PPP flux analysis
The flux through the oxidative branch of the PPP was measured as described by Mancuso et al. [12]. Briefly, cells were incubated for 9 h with either complete medium or serum-free medium, both containing 10 mM 13C-2-d-glucose (CortecNet, CC855P1). Media were then collected and centrifuged at 900 × g for 5 min, and supernatant fractions were incubated with 70% ethanol. After centrifugation at 20,000 × g for 30 min, the supernatant fractions were dried and then resuspended in 100% D2O. PPP flux was analyzed based on the rate of glucose consumption and the ratio of 13C incorporated into carbon 2 (generated by glycolysis) and carbon 3 (generated by the PPP) of lactate determined by NMR spectroscopy using a Bruker Avance 400 MHz spectrometer.
NADP+/NADPH detection assay
NADP+/NADPH was tested using WST-8 according to the manufacturer’s protocol of the NADP+/NADPH Assay Kit with WST-8 (Beyotime, China).
ROS evaluation
ROS levels were analyzed as previously described [13]. Briefly, cells were incubated with 50 µM H2DCF-DA at 37 °C for 30 min. Cells were then washed twice with phosphate buffered saline (PBS), resuspended in ice-cold PBS, and collected. Fluorescence was immediately measured using a FACScan Flow Cytometer (Becton Dickinson, San Jose, CA).
Chromatin isolation by RNA purification-mass spectrometry (ChIRP-MS)
A ChIRP kit (Bersinbio, China) was used to test the protein-binding lncRNAs according to the manufacturer’s protocol. Cells were cross-linked with paraformaldehyde and lysed by sonication, and then the lysis solution was incubated with biotin-labeled probes and magnetic beads. Finally, proteins were extracted and detected by mass spectrometry (Aksomics, China).
RNA immunoprecipitation (RIP) and m6A MeRIP
A RIP Kit (Merck, USA) was used to test the RNA-binding protein according to the manufacturer’s protocol. The antibody was prebound to magnetic beads and incubated with cell lysates, and then RNA was eluted from magnetic beads and detected by qPCR. Similar to the RIP experiment, m6A MeRIP was performed using a riboMeRIP m6A Transcriptome Profiling Kit (RIBOBIO, China) according to the manufacturer’s protocol. Total RNA was fragmented and purified, and then the m6A antibody was prebound to magnetic beads and incubated with the RNA prepared above. Finally, the specific RNA was eluted and detected by qPCR.
m6A dot blot assay
Total RNA was extracted and loaded onto an Amersham Hybond-N + membrane (GE, USA). Then, the membrane was blocked with 5% skimmed milk and incubated with anti-m6A overnight at 4 °C after cross-linking with UV radiation. On day 2, the membrane was incubated with HRP-conjugated goat anti-rabbit IgG and developed with an ECL detection system.
Patient-derived organoids
The generation of patient-derived organoids (PDOs) was performed as described in a previous study [14]. Briefly, tumor tissue was obtained from surgical resections of patients with CRC, cut into 1–3 mm3 pieces, and digested with 200 U/ml collagenase (Sigma, USA) and 100 U/ml hyaluronidase (Sigma, USA). The PDO was mixed with growth factor-reduced Matrigel and cultured in 24-well plates.
PDO was transiently transfected with a lentivirus expressing LINC01615, sh-LINC01615 (GenePharma, China), or was incubated with oxaliplatin (10 μM) for 4 days [15]. Paired DIC images for each PDO were acquired on day 1 and day 4 after treatment [16] and used to analyze the growth of organoids. The evaluation of PDO growth was performed following the method described previously [17]. Briefly, the surface area at each time point was outlined manually in Fiji, and a growth fold change was calculated as the ratio of a surface area from a given time point (4 days) over day 1. For each experiment, the growth fold change was normalized to the median of the control, which was set to have a value of 1.
Patient-derived xenograft models
Patient-derived xenograft models (PDX models) were established as described in a previous study [18]. Briefly, tumor tissue was obtained from surgical resections of patients with CRC and cut into 2 × 2 × 3 mm3 pieces, and then fresh RPMI 1640 was used to wash the tissues twice. NOD/SCID mice were chosen as the first-generation PDX mice that carried patient tissues. Subsequent generations of PDX mice were BALB/c-nude mice. When each xenografted tumor grew to 1–2 cm3, we harvested the xenografted tumor and immediately transplanted them into next-generation mice three times. When the tumors had grown to an appropriate volume in the third-generation PDX mice, we randomly divided the mice into four groups. For the control group, we injected 200 μl PBS every 4 days. For the sh-LINC01615 group, we injected lentivirus expressing LINC01615 shRNA#1 into tumor tissues every 4 days. For the oxaliplatin group, we delivered oxaliplatin by intraperitoneal (i.p.) injection (5 mg/kg) every 4 days [19]. For the combination group, we injected lentivirus expressing LINC01615 shRNA#1 into tumor tissues and delivered oxaliplatin by intraperitoneal (i.p.) injection (5 mg/kg) every 4 days.
Gene set enrichment analysis (GSEA)
GSEA was performed on the normalized data using the GSEA v2.0 tool (http://www.broad.mit.edu/gsea/). We compared the gene expression between cells with high and low LINC01615 expression. One gene set was used for GSEA (KEGG_PENTOSE_PHOSPHATE_PATHWAY), and the detailed genes in the gene sets can be found in MSigDB (http://software.broadinstitute.org/gsea/msigdb/genesets.jsp). The p values of the differences between the two gene sets were analyzed with the Kolmogorov–Smirnov test.
Statistical analysis
All values are expressed as the mean ± standard deviation (SD). The significance of the differences was determined by one-way ANOVA or Student’s t test. The Chi-square test was used to evaluate the relationship between expression and clinicopathological characteristics. Spearman’s correlation coefficient was used to calculate correlations between two groups. Kaplan–Meier analysis was employed for survival analysis, and the differences in survival probabilities were estimated using the log-rank test. p < 0.05 was considered significant. Statistical analyses were performed using SPSS version 20.0 (SPSS, Inc., USA).
More procedures are available in the Supplemental Materials and Methods.
Results
LINC01615 is upregulated in response to serum deprivation and predicts poor survival in CRC
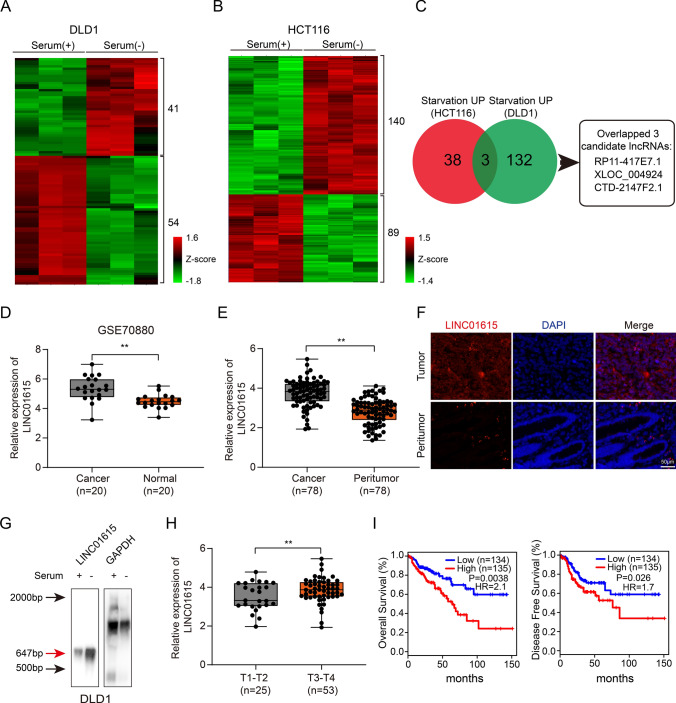
To assess special lncRNAs induced by nutritional deficiency in CRC, we carried out lncRNA microarray analysis in DLD1 and HCT116 cells grown in normal medium or serum-free medium, which is the most common nutrient deficiency inducer [20]. A total of 54 downregulated lncRNAs and 41 upregulated lncRNAs (p < 0.05, FC ≥ 2 or ≤ − 2) were identified in DLD1 cells (Fig. 1A), and 89 downregulated lncRNAs and 140 upregulated lncRNAs (p < 0.05, FC ≥ 2 or ≤ − 2) were identified in HCT116 cells. (Fig. 1B). We overlapped the upregulated lncRNAs and the downregulated lncRNAs upon starvation in both cell lines, and three candidate upregulated lncRNAs and one downregulated lncRNA were finally found (Figs. 1C, S1A). We focused more on the three upregulated lncRNAs. Since the length of XLOC_004924 was less than 400 bp and CTD-2147F2.1 had no predictive value for prognosis (p = 0.6, Fig. S1B) based on the data from TCGA, we excluded the above two lncRNAs and chose RP11-417E7.1 (LINC01615) for further study. A coding potential algorithm (PhyloCSF) showed that LINC01615 had no protein coding capacity (Fig. S1C). We examined the expression of LINC01615 in various types of cancers in TCGA database, demonstrating that LINC01615 was overexpressed in most types of cancers including CRC (Fig. S1D). We further assessed LINC01615 expression in the GEO database (GSE115856 and GSE70880) and 78 CRC tissues compared with peritumor tissues, confirming that LINC01615 was significantly upregulated in CRC tissues (Figs. 1D, E, S1E). This trend was further verified by FISH (Fig. 1F). Northern blot analysis with a specific RNA probe for LINC01615 showed that LINC01615 could be detected at its expected size of 647 bp, the expression of LINC01615 was increased under serum deprivation (Fig. 1G).
Fig. 1.
LINC01615 is upregulated in response to serum deprivation and predicts poor survival in CRC. A Hierarchical clustering showing differentially expressed lncRNAs of DLD1 cells cultured in normal medium or serum-free medium (FC ≥ 2 or ≤ − 2, p < 0.05). B Hierarchical clustering showing differentially expressed lncRNAs of HCT116 cells cultured in normal medium or serum-free medium (FC ≥ 2 or ≤ − 2, p < 0.05). C Venn diagram showing the numbers of overlapping lncRNAs between the differentially upregulated lncRNAs in HCT116 and DLD1 cells after serum-free medium treatment. D The relative expression of LINC01615 in tumor tissue and corresponding peritumor tissue based on data obtained from the GEO database (GSE70880). E The relative expression levels of LINC01615 were assessed by qRT–PCR in 78 paired CRC tissues and peritumor tissues. F FISH analysis of LINC01615 expression in CRC tissues and peritumor tissues. G Northern blot analysis of LINC01615 expression in DLD1 cells with or without serum supplementation. H LINC01615 expression in CRC tissues from T1–T2 stage CRC patients and T3–T4 stage CRC patients. I Kaplan–Meier analysis of the OS rate and DFS rate in CRC patients in the TCGA database with high or low expression of LINC01615. Data are the means ± SDs (n = 3 independent experiments), **p < 0.01
We evaluated the potential correlation between LINC01615 expression and clinicopathological features and found that LINC01615 was significantly upregulated in T3-T4 CRC patients compared with T1-T2 CRC patients (Fig. 1H). Kaplan–Meier analysis revealed that higher expression of LINC01615 was associated with lower overall survival (OS) rates (p = 0.0038, hazard ratio = 2.1) and lower disease-free survival (DFS) rates (p = 0.026, hazard ratio = 1.7) in 269 CRC cases from TCGA (Fig. 1I).
LINC01615 preserves cell survival under starvation stress
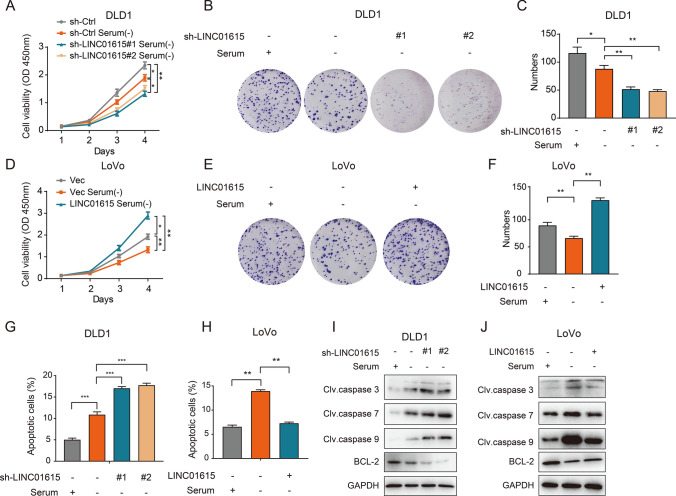
To investigate the role of LINC01615 in CRC progression, we tested the expression of LINC01615 in the normal intestinal epithelial cell line FHC and CRC cell lines and found that the expression of LINC01615 was relatively high in DLD1 and HCT116 cells and was relatively low in LoVo cells (Fig. S1F). Subsequently, we silenced LINC01615 in DLD1 and HCT116 cells and overexpressed it in LoVo cells (Fig. S1G–H). CCK-8 and colony formation assays demonstrated that the proliferative ability of cells was significantly inhibited by LINC01615 knockdown without serum provision and that overexpressing LINC01615 substantially reversed the inhibitory effect of serum deprivation on cell proliferation (Figs. 2A–F, S1I–K). However, knockdown of LINC01615 only slightly altered cell proliferation in normal medium (Fig. S1L–M). Then, we explored the effects of LINC01615 on cell death under starvation conditions. Annexin V/PI assays showed that knockdown of LINC01615 increased the ratio of apoptotic cells, while overexpression of LINC01615 reversed serum deprivation-induced apoptosis (Figs. 2G, H, S1N–P). We also explored the expression of apoptosis-related proteins under starvation. Increased expression of cleaved caspase 3, caspase 7, and caspase 9 and decreased expression of BCL-2 were observed after LINC01615 depletion, whereas the opposite expression of apoptosis-related proteins was detected after LINC01615 overexpression (Figs. 2I, J, S1Q). Together, these findings confirmed that LINC01615 sustains cell survival by promoting cell proliferation and inhibiting apoptosis under starvation.
Fig. 2.
LINC01615 preserves cell survival under starvation stress. A DLD1 cells were transfected with either a nontargeting shRNA (sh-Ctrl) or a shRNA targeting LINC01615 (sh-LINC01615), and LoVo cells were transfected with either an empty vector or a plasmid containing LINC01615. Twenty-four hours after transfection, the cells were starved for 6 h. The CCK-8 assay showed the effects of LINC01615 knockdown on proliferation in DLD1 cells without serum supplementation. B Colony formation assays were performed to examine the effects of LINC01615 knockdown on proliferation in DLD1 cells without serum supplementation. C Statistical analysis of colony formation numbers in (B). D CCK-8 assay showed the effects of LINC01615 overexpression on proliferation in LoVo cells without serum supplementation. E Colony formation assays were performed to examine the effects of LINC01615 overexpression on proliferation in LoVo cells without serum supplementation. F Statistical analysis of colony formation numbers in E. G Flow cytometry data presented as histograms showing the effects of LINC01615 knockdown on the apoptotic rate in DLD1 cells without serum supplementation. H Flow cytometry data presented as histograms showing the effects of LINC01615 overexpression on the apoptotic rate in LoVo cells without serum supplementation. I WB was used to examine the effects of LINC01615 knockdown on the expression of cleaved caspase 3, caspase 7, caspase 9 and BCL-2 in DLD1 cells without serum supplementation. J WB was used to examine the effects of LINC01615 overexpression on the expression of cleaved caspase 3, caspase 7, caspase 9 and BCL-2 in LoVo cells without serum supplementation. Data are the means ± SDs (n = 3 independent experiments), *p < 0.05, **p < 0.01
LINC01615 regulates the PPP to sustain cell survival under serum starvation
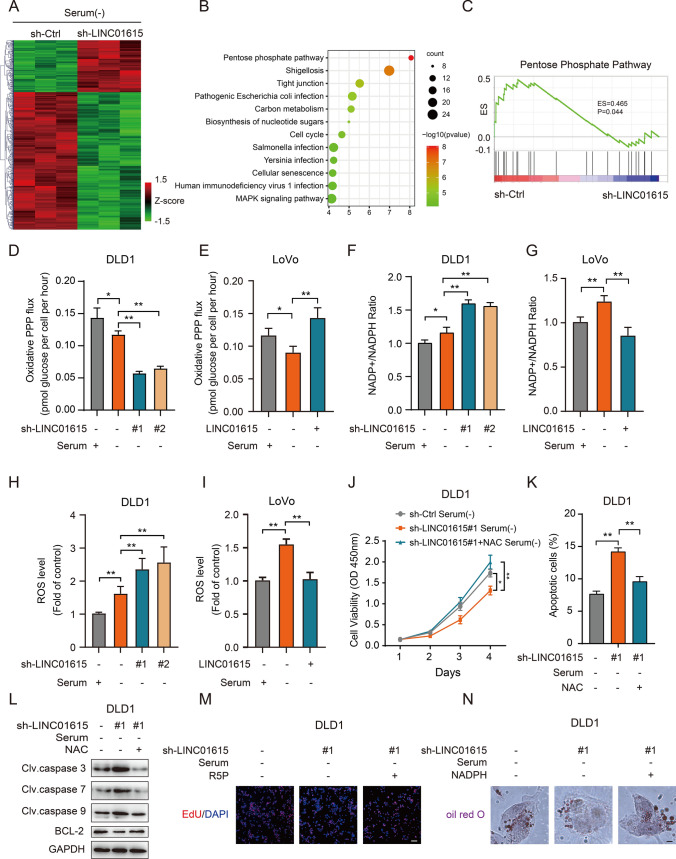
To gain further insight into the biological pathways of LINC01615 involved in the regulation of cell survival, RNA-seq analysis was performed in DLD1 cells transfected with LINC01615 shRNA and control shRNA transfectants under starvation conditions. A total of 895 dysregulated genes (p < 0.05, FC ≥ 2 or ≤ − 2) were detected (Fig. 3A). KEGG enrichment revealed that the significantly enriched pathways included the PPP, shigellosis, tight junction and carbon metabolism (Fig. 3B). Among the above-mentioned pathways, the PPP is a well-recognized pathway that promotes cell survival under environmental stress by providing raw materials for biosynthesis [21] and clearing ROS [22]. Gene set enrichment analyses (GSEA) indicated a positive correlation between LNC01615 and the PPP (Fig. 3C). We then assessed whether LINC01615 exerts its function though the PPP. We traced labeled [2−13C] glucose for the indicated times and examined the PPP flux by quantifying the amount of labeled glucose entering the PPP. The results showed that knockdown of LINC01615 significantly reduced the PPP flux by over 50% without serum provision, while overexpression of LINC01615 significantly increased the PPP flux (Figs. 3D–E, S2A). Increased PPP flux is a major source of cellular NADPH, which provides the reducing power to support reductive biosynthetic reactions and to maintain cellular redox homeostasis [23]. We found that knockdown of LINC01615 significantly increased NADP+/NADPH ratios and ROS levels, while overexpression of LINC01615 had the opposite effects (Figs. 3F–I, S2B–D). We then used the ROS scavenger N-acetyl-L-cysteine (NAC) to eliminate ROS in LINC01615-silenced cells under starvation conditions and found that NAC reversed the induction of apoptosis and the suppression of cell growth in LINC01615-silenced cells (Figs. 3J–L, S2E–H).
Fig. 3.
LINC01615 promotes cell survival through the PPP. A Hierarchical clustering showing differentially expressed mRNAs in LINC01615 shRNA- and control-transfected cells (fold change > 2 or fold change < 0.5, p < 0.05). Red and green reflect the max and min levels, respectively. B, C Overview of KEGG (B) and GSEA (C) of LINC01615-regulated gene expression events. Fisher’s exact P values for each enriched functional category are shown. D, E DLD1 cells were transfected with LINC01615 shRNA or control shRNA. LoVo cells were transfected with LINC01615 overexpression plasmid or control vector. Then, the cells were cultured in medium containing [2-13C] glucose and without serum supplementation. Oxidative PPP flux was measured based on the rate of glucose consumption and the ratio of 13C incorporated into carbon 2 (indicating glycolysis) and carbon 3 (indicating PPP) of lactate by nuclear magnetic resonance (NMR) spectroscopy. F, G NADP+/NADPH ratios were measured in the cells from D, E. H, I ROS levels were measured in the cells from D, E. J CCK-8 assay showed the effect of LINC01615 inhibition on proliferation in DLD1 cells treated with or without NAC under serum-free starvation conditions. K Flow cytometry data presented as histograms show the effects of LINC01615 inhibition on the apoptotic rate in DLD1 cells treated with or without NAC under serum-free starvation conditions. L WB was used to examine the effects of LINC01615 inhibition on cleaved caspase 3, caspase 7, caspase 9 and BCL-2 expression in DLD1 cells treated with or without NAC under serum-free starvation conditions. M The EdU staining assay showed the effect of LINC01615 inhibition on nucleotide synthesis in DLD1 cells treated with or without R5P under serum-free starvation conditions (100 × magnification). N Oil red O staining assay showed the effect of LINC01615 inhibition on lipid synthesis in DLD1 cells treated with or without NADPH under serum-free starvation conditions (1000 × magnification). Data are the means ± SDs (n = 3 independent experiments), *p < 0.05, **p < 0.01
PPP branches have also been reported to provide ribulose-5-phosphate (R5P) and NADPH for nucleotide and lipid synthesis [9]. A 5-ethynyl-2’-deoxyuridine (EdU) incorporation assay demonstrated that LINC01615 silencing suppressed de novo DNA synthesis, and the addition of R5P increased DNA synthesis and cell viability in LINC01615-depleted cells (Figs. 3M, S2I-M). Consistently, LINC01615 silencing suppressed lipid accumulation stained by oil Red O, and the addition of NADPH restored lipid accumulation and cell viability in LINC01615-depleted cells (Figs. 3N, S2N–P). These results suggest that LINC01615 regulates PPP flux to promote cell survival in response to starvation.
LINC01615 regulates the PPP through the key rate-limiting enzyme G6PD
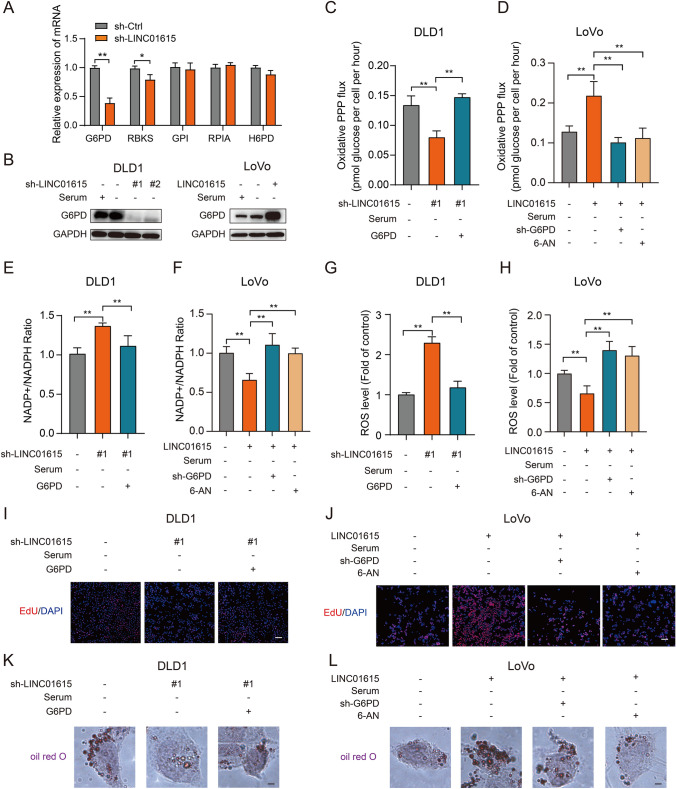
To further investigate the key gene involved in LINC01615 regulating the PPP, we detected the expression of the PPP gene set in DLD1 cells. Interestingly, we found that G6PD, the rate-limiting enzyme of the PPP, was the most significantly changed gene in the PPP gene set after LINC01615 knockdown in DLD1 cells (Figs. 4A, S3A). We speculated that LINC01615 might activate the PPP through G6PD. The Western blot results further confirmed that knockdown of LINC01615 led to a marked reduction in G6PD protein levels without serum provision, while overexpression of LINC01615 upregulated G6PD expression (Fig. 4B). By analyzing TCGA sequencing data, we found that G6PD was significantly upregulated in CRC tissues and was positively correlated with LINC01615 expression (Fig. S3B–C). Similarly, the IHC results confirmed the higher expression of G6PD in CRC tissues (Fig. S3D), and patients with high levels of LINC01615 showed higher levels of G6PD (Fig. 6D).
Fig. 4.
LINC01615 regulates the PPP through the key rate-limiting enzyme G6PD. A The effects of LINC01615 knockdown on the expression of PPP genes in DLD1 cells. B WB was used to examine the effects of LINC01615 knockdown and overexpression on the expression of G6PD in DLD1 and LoVo cells under serum-free starvation conditions. C, D DLD1 cells were cotransfected with LINC01615 shRNA, control shRNA or G6PD overexpression plasmid. LoVo cells were cotransfected with LINC01615 overexpression plasmid, control vector or G6PD shRNA and treated with or without 6-AN. Then, the cells were cultured in medium containing [2-13C] glucose and without serum supplementation. Oxidative PPP flux was measured based on the rate of glucose consumption and the ratio of 13C incorporated into carbon 2 (indicating glycolysis) and carbon 3 (indicating PPP) of lactate by NMR spectroscopy. E–L NADP+/NADPH ratios and ROS levels and nucleotide and lipid synthesis were measured in the cells from C, D. Data are the means ± SDs (n = 3 independent experiments), * p < 0.05, ** p < 0.01
Fig. 6.
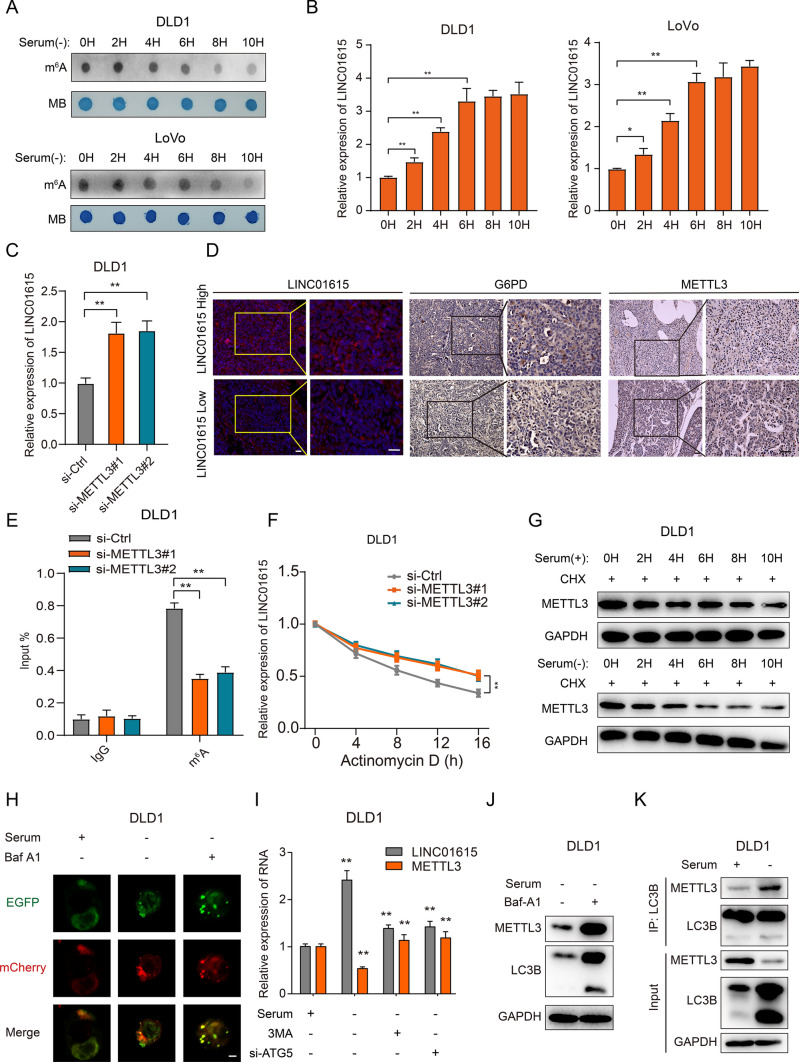
Autophagy-reduced METTL3 upregulates LINC01615 in a m6A modification-dependent manner. A A dot blot assay was used to analyze the m6A content of total RNA in DLD1 and LoVo cells cultured without serum supplementation for different times. Methylene blue staining was used as a loading control. B qRT–PCR analysis of LINC01615 expression in DLD1 and LoVo cells cultured without serum supplementation for different times. C qRT–PCR analysis of LINC01615 expression in DLD1 cells transfected with METTL3 siRNA or control siRNA. D Representative images of LINC01615, METTL3 and G6PD analyzed by FISH or IHC in LINC01615 high-expression and LINC01615 low-expression tumors. E MeRIP-qPCR showing the m6A modification enrichment of LINC01615 in DLD1 cells after METTL3 depletion. F qRT–PCR analysis of the stability of LINC01615 in DLD1 cells with or without METTL3 deletion. G WB analysis of METTL3 protein stability in DLD1 cells cultured with or without serum supplementation for different times. H Representative confocal microscopy images of LC3B puncta distribution in DLD1 cells under normal or serum-free starvation conditions and treated with or without Baf-A1 (1000 × magnification). I Analysis of the expression of METTL3 and LINC01615 in DLD1 cells transfected with ATG5 siRNA, METTL3 overexpression plasmid, control vector or treated with 3-MA 30 min before the end of each experimental time. J WB analysis of METTL3 expression in DLD1 cells treated with or without Baf-A1 under serum-free starvation conditions. K Co-IP analysis was performed with specific antibodies against METTL3 and LC3B in DLD1 cells under normal or serum-free starvation conditions. WB analysis of the interaction between METTL3 and LC3B. Data are the means ± SDs (n = 3 independent experiments), *p < 0.05, **p < 0.01
To further ascertain whether LINC01615 mediates the regulation of PPP flux by G6PD, we silenced G6PD with shRNA in LINC01615-overexpressing LoVo cells under starvation conditions and overexpressed G6PD in LINC01615-knockdown DLD1 cells. As expected, knockdown of G6PD or treatment with 6-aminonicotinamide (6-AN, an inhibitor of G6PD) in LoVo cells abolished the effects of LINC01615 overexpression on increasing PPP flux and nucleotide and lipid synthesis and decreasing the NADP + /NADPH ratio and ROS level (Figs. 4C–L, S3E–F). In contrast, overexpression of G6PD in LINC01615-knockdown DLD1 cells showed the opposite effects (Figs. 4C–L, S3E–F). These results suggest that LINC01615 activates the PPP by upregulating G6PD expression.
LINC01615 regulates the splicing of G6PD pre-mRNA by recruiting hnRNPA1
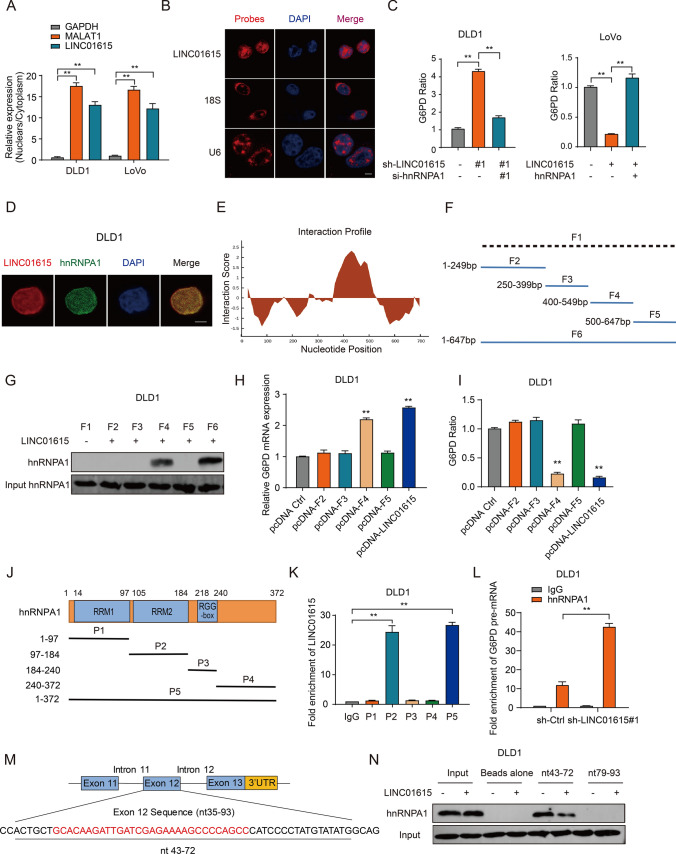
To explore the molecular mechanism by which LINC01615 regulates G6PD expression, we first predicted the competing endogenous RNAs (ceRNAs) of G6PD through the StarBase website. The results showed that LINC01615 was not a ceRNA of G6PD (data not shown). In addition to acting as ceRNAs, accumulating evidence suggests that lncRNAs can also bind with different proteins to exert their function [24]. We then conducted ChIRP assays followed by MS analyses to search for proteins that might interact with LINC01615. Our results revealed 19 potential proteins that were pulled down by LINC01615 probes (Table S1). Of these, hnRNPA1, with a higher interacting rank, was reported to be a nuclear protein, which was consistent with the location of LINC01615 (Fig. 5A, B). In addition, as a member of the hnRNP family, hnRNPA1 possesses RNA chaperone activity [25] and functions as a splicing repressor [26]. Intriguingly, LINC01615 could also negatively regulate the splicing of G6PD, as reflected by changes in the G6PD pre-mRNA/mature mRNA ratio upon LINC01615 knockdown or overexpression (Fig. 5C). We speculated that LINC01615 regulates the splicing of G6PD through the recruitment of hnRNPA1.
Fig. 5.
LINC01615 regulates the splicing of G6PD pre-mRNA by recruiting hnRNPA1. A The nuclear and cytoplasmic fractions of DLD1 and LoVo cells were subjected to qRT–PCR. MALAT1 was the nuclear positive control; GAPDH was the cytoplasmic positive control. B RNA-FISH performed in DLD1 cells. LINC01615 probes are red, and nuclei are stained with DAPI (1000 × magnification). C The left histogram shows the G6PD pre-mRNA/mature mRNA ratio in DLD1 cells transfected with control shRNA, LINC01615 shRNA, or cotransfected LINC01615 shRNA and hnRNPA1 siRNA. The histogram on the right shows the G6PD pre-mRNA/mature mRNA ratio in LoVo cells transfected with control vector, LINC01615 overexpression plasmid, or cotransfected LINC01615 overexpression plasmid and hnRNPA1 overexpression plasmid. D Colocalization analysis was performed with specific probes against LINC01615 and a specific antibody against hnRNPA1. Nuclei are stained with DAPI. LINC01615 probes are red; hnRNPA1 is green (1000 × magnification). E Nucleotides 401–452 of LINC01615 are in the main region that interacts with hnRNPA1. F Sequence mapping of full-length (F6) and truncated LINC01615 (F1–F5). G WB of hnRNPA1 in samples pulled down by full-length (F6) or truncated LINC01615 (F1–F5). H G6PD mRNA expression in samples pulled down by full-length (F6) or truncated LINC01615 (F1–F5). I G6PD mRNA ratio in samples pulled down by full-length (F6) or truncated LINC01615 (F1–F5). J Domain mapping of Flag-labeled full-length hnRNPA1 (P5) or truncated hnRNPA1 (P1–P4). K Antibodies against Flag were used for RIP, followed by LINC01615 qRT–PCR in DLD1 cells. L Antibodies against hnRNPA1 were used for RIP, followed by G6PD pre-mRNA qRT–PCR in DLD1 cells transfected with LINC01615 shRNA or control shRNA. M The structures of the G6PD and oligonucleotide sequences used in the pulldown experiments. The sequence from nt 35 to nt 93 of exon 12 is shown diagrammatically. Oligonucleotides 43–72 nt and 79–93 nt were used in pulldown assays. N Oligonucleotides 43–72 nt and 79–93 nt were covalently linked to adipic acid beads and incubated with nuclear extracts (150 μg of protein) from DLD1 cells overexpressing LINC01615 or control cells. The proteins were eluted and analyzed with hnRNPA1 antibody. Data are the means ± SDs (n = 3 independent experiments), *p < 0.05, **p < 0.01
We further verified the interaction between LINC01615 and hnRNPA1. Immunofluorescence colocalization assays showed that LINC01615 and hnRNPA1 colocalized in the nucleus in CRC cells (Fig. 5D). RIP assays with hnRNPA1 antibody showed that hnRNPA1 bound specifically to LINC01615 (Fig. S3G), and this interaction was enhanced by nutrient deprivation (Fig. S3H). Then, we predicted the binding sites of LINC01615 with hnRNPA1 by CatRAPID, which revealed that nt 401–452 of LINC01615 constituted the main region that interacted with hnRNPA1 (Fig. 5E). We constructed one biotinylated full-length LINC01615 and five biotinylated fragments of LINC01615 (F1: full-length antisense LINC01615, F2: 1–249 bp, F3: 250–399 bp, F4: 400–549 bp, F5: 500–647 bp, and F6: full-length sense LINC01615) to pull down hnRNPA1 in DLD1 cell lysates (Fig. 5F). We found that the fragment between 400 and 549 bp of LINC01615 was crucial for the interaction with hnRNPA1 (Fig. 5G). In addition, ectopic expression of LINC01615 fragment 4 (400–549 bp) resulted in increased G6PD mRNA levels and decreased G6PD pre-mRNA/mature mRNA ratios, whereas the other fragments did not exert these effects (Fig. 5H–I). As previously reported, hnRNPA1 contains two RNA recognition motifs (RRM1 and RRM2 domains) and one RNA-binding domain (Fig. 5J). To identify the binding domain of hnRNPA1 with LINC01615, we cloned a series of Flag-tagged hnRNPA1 constructs (P1: 1–97 aa, P2: 97–184 aa, P3: 184–240 aa, P4: 240–372 aa, and P5: 1–372). RIP assays were performed with an anti-Flag antibody and demonstrated that both P2 and P5 interacted with LINC01615 (Fig. 5K), suggesting that the RRM2 domain of hnRNPA1 is required for the interaction with LINC01615.
Next, we investigated how the interaction of LINC01615 and hnRNPA1 affected G6PD pre-mRNA splicing. The RIP assay indicated that hnRNPA1 could also bind specifically to G6PD pre-mRNA (Fig. S3I), and knockdown of LINC01615 significantly increased the binding of hnRNPA1 with G6PD pre-mRNA, whereas overexpression of LINC01615 inhibited their binding (Figs. 5L, S3J). Then, we knocked down hnRNPA1 in DLD1 cells and overexpressed hnRNPA1 in LoVo cells (Fig. S3K-L). The results showed that knockdown of hnRNPA1 in LINC01615-silenced cells decreased the ratio of G6PD pre-mRNA/mature mRNA. In contrast, overexpression of hnRNPA1 in LINC01615-overexpressing cells increased the ratio of G6PD pre-mRNA/mature mRNA (Fig. 5C). This suggests that LINC01615 abolishes the hnRNPA1-induced suppression of G6PD pre-mRNA splicing.
Next, we investigated the potential binding sites between hnRNPA1 and G6PD pre-mRNA. The Flag-hnRNPA1 truncated plasmids were transfected into CRC cells. Similar to the results on the binding domain of hnRNPA1 with LINC01615, the RIP assay demonstrated that the RRM2 domain of hnRNPA1 was also responsible for the interaction with G6PD pre-mRNA (Fig. S3M). Previous studies reported that hnRNP proteins could inhibit G6PD pre-mRNA splicing by binding to nt 43–72 of exon 12 of G6PD [27]. To confirm the binding sites of G6PD pre-mRNA with hnRNPA1, we designed two RNA oligonucleotides (nt 43–72 and nt 79–93) of exon 12 to pull down hnRNPA1. We found that overexpression of LINC01615 decreased the binding of hnRNPA1 to the nt 43–72 oligonucleotide, while no protein bound to the nt 79–93 oligonucleotide (Fig. 5M, N). We also treated LINC01615-overexpressing cells with hnRNPA1 plasmids with or without RRM2 domain. and found that hnRNPA1 plasmids with the RRM2 domain can inhibit LINC01615-mediated CRC cell survival. The inhibitory function was abrogated when transfected with truncated plasmids without the RRM2 domain (Fig. S3N-Q). These data suggest that LINC01615 can cover the RRM2 domain of hnRNPA1 and inhibit hnRNPA1 binding to exon 12 of G6PD pre-mRNA, leading to increased G6PD pre-mRNA splicing.
Starvation-reduced METTL3 upregulates LINC01615 in a m6A modification-dependent manner
m6A modification modulates all stages of the RNA life cycle and thereby regulates the expression and functions of RNAs. Previous studies have reported that starvation can influence m6A modification of RNA through m6A demethylases [28]. SRAMP (http://www.cuilab.cn/sramp/) prediction revealed that several m6A sites are dispersed throughout LINC01615 (Fig. S4A). Therefore, we elucidated whether the upregulation of LINC01615 under nutrient deprivation is dependent on m6A demethylase-induced m6A modification. As expected, nutrient deprivation increased LINC01615 expression and decreased the total m6A level in a time-dependent manner (Fig. 6A, B). Then, we determined whether the decreased level of m6A modification was mediated by m6A methylase or demethylase, and the effects of nutrient deprivation on the m6A methylases METTL3 and METTL14 and the m6A demethylase FTO were evaluated. The results showed that nutrient deprivation decreased the expression of METTL3 but had no effects on METTL14 and FTO (Fig. S4B), suggesting that METTL3 may be involved in the m6A level of LINC01615 under nutrient deprivation. Then, we silenced or upregulated METTL3 with siRNA or overexpression plasmids, respectively (Fig. S4C-D). METTL3 depletion clearly induced LINC01615 levels, while METTL3 overexpression decreased LINC01615 levels (Figs. 6C, S4E). Overexpression of METTL3 markedly reduced the induction of LINC01615 by nutrient deprivation (Fig. S4F), suggesting that the upregulation of LINC01615 upon starvation was induced by METTL3. Analysis of the TCGA-COAD and GTEx datasets showed that METTL3 expression was negatively correlated with LINC01615 expression (R = − 0.23, Fig. S4G). The IHC results further confirmed that high LINC01615 expression was correlated with low METTL3 expression (Figs. 6D, S4H). Then, we investigated whether METTL3-mediated m6A modification regulated LINC01615 expression. MeRIP assays revealed that silencing METTL3 decreased the m6A modification of LINC01615 in DLD1 cells, while overexpressing METTL3 had the opposite effects in LoVo cells (Figs. 6E, S4I). Furthermore, we assessed LINC01615 stability in CRC cells upon METTL3 inhibition or overexpression. After treating the cells with actinomycin D to block the de novo synthesis of RNA, the depletion of METTL3 resulted in increased stability of LINC01615, whereas overexpression of METTL3 had the opposite effects (Figs. 6F, S4J). These data suggest that nutrient deprivation decreases METTL3, which enhances the stability of LINC01615 in a m6A-dependent manner.
Starvation reduces METTL3 stability by inducing autophagy-mediated protein degradation
To determine the mechanism by which starvation reduces METTL3, we assessed the stability of METTL3 protein under starvation. Compared with cells in normal medium, serum deprivation reduced METTL3 stability (Figs. 6G, S5A). The autophagy‒lysosome pathway is one of the main protein degradation mechanisms [29]. Studies have indicated that nutrient starvation can induce autophagy to degrade proteins [30], so we investigated whether the decreased stability of METTL3 under serum starvation was mediated by autophagic degradation. TEM and EGFP-mCherry-LC3 staining assays revealed that autophagic flux was increased under serum starvation (Figs. 6H, S5B–E). Then, we knocked down the essential autophagy gene ATG5 or treated cells with the autophagy inhibitor 3-MA and found that starvation-reduced METTL3 was clearly upregulated and LINC01615 was reduced (Figs. 6I, S5F–G). In addition, we used the lysosome inhibitor bafilomycin A1 (Baf-A1) to inhibit lysosomes, and cells treated with Baf-A1 showed reduced autophagy, increased accumulation of METTL3 and reduced accumulation of LINC01615 (Figs. 6J, S5G–H). To further assess the relationship between METTL3 and autophagosomes, we validated the interaction between METTL3 and LC3B by Co-IP, and serum deprivation increased the interaction between METTL3 and LC3B (Figs. 6K, S5I). IF assay revealed that METTL3 colocalized with LC3B and LAMP1 in CRC cells under starvation (Fig. S5J), quantitative colocalization level in cytoplasm was measured using Pearson's correlation analysis (Rr), relative Rr value between METTL3 and LAMP1 was 0.537 ± 0.048 and relative Rr value between METTL3 and LC3B was 0.695 ± 0.052. These results suggested that serum starvation degraded METTL3 through the autophagy‒lysosome pathway.
LINC01615 knockdown enhances oxaliplatin efficacy in PDO and PDX models
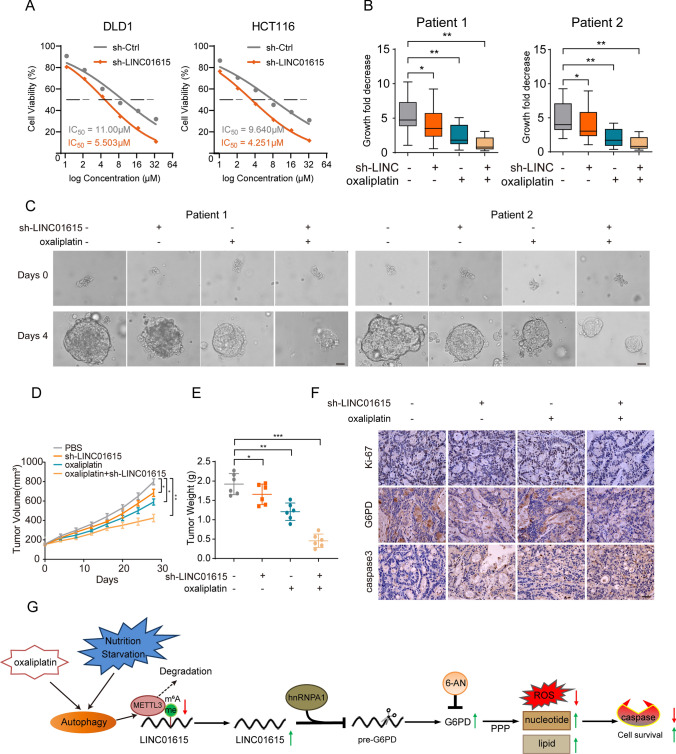
Previous studies have suggested that chemotherapy, which functions as a significant autophagy inducer, could inhibit nucleotide synthesis, induce apoptosis, elevate cellular ROS levels and show synergistic anticancer effects in colorectal cancer cells [31–33]. Therefore, we investigated whether knockdown of LINC01615 affects the antitumor effect of chemotherapy drugs, such as oxaliplatin. MTT assays confirmed that LINC01615 knockdown increased oxaliplatin sensitivity in CRC cells, and the IC50 values for the cells with LINC01615 knockdown were decreased compared with those of the control (Fig. 7A). Then, we built PDOs from two human CRC patient tissues and found that the expression level of LINC01615 was higher in cancer tissues than in adjacent normal tissues (Fig. S6A). We found that LINC01615 knockdown or oxaliplatin treatment (10 μM) caused a substantial reduction in PDO growth (Fig. 7B–C). The combination of LINC01615 knockdown and oxaliplatin treatment (10 μM) caused a more significant reduction in PDO growth than any individual treatment (Fig. 7B–C).
Fig. 7.
Knockdown of LINC01615 enhances oxaliplatin efficacy in PDO and PDX models. A The sensitivities of DLD1 and HCT116 cells infected with LINC01615 shRNA or not under different concentrations of oxaliplatin were determined by MTT assay. B Measurement of the growth of PDO in response to knockdown LINC01615 and oxaliplatin. C Effects of sh-LINC01615 and oxaliplatin on the viability of PDO cells (scale bar, 100 μm). D The tumor growth curves of the PDX mice were measured and recorded for the control group, sh-LINC01615 group, oxaliplatin group, and sh-LINC01615 combined with oxaliplatin group throughout the experiment (n = 6). E The tumor weights of the PDX mice were measured and recorded for the control group, sh-LINC01615 group, oxaliplatin group, and sh-LINC01615 combined with oxaliplatin group throughout the experiment (n = 6). F The expression levels of Ki-67, G6PD and caspase 3 were observed in PDX tumor tissues of the control group, sh-LINC01615 group, oxaliplatin group, and sh-LINC01615 combined with oxaliplatin group by IHC. G Schematic representation showing that nutrient deprivation causes activation of autophagy and decreases METTL3 expression. Reduced METTL3 decreases the m6A modification of LINC01615 and upregulates the expression of LINC01615. Overexpression of LINC01615 promotes G6PD pre-mRNA splicing and upregulates the expression of G6PD by competitively binding with hnRNPA1. Overexpression of G6PD promoted cell survival under nutrient starvation and oxaliplatin treatment by promoting PPP flux, increasing nucleotide and lipid synthesis and decreasing ROS production to minimize oxidative damage. Data are the means ± SDs (n = 3 independent experiments), *p < 0.05, **p < 0.01, ***p < 0.01
Considering that PDX has been proven to be a more realistic experimental model [34], we constructed PDX models and injected poly(amino-co-ester) polyplexes of LINC01615 shRNA intratumorally into PDX mice to further assess the clinical benefits of targeting LINC01615. Similar to the results of PDO, we found that LINC01615 knockdown or oxaliplatin treatment (5 mg/kg) in vivo suppressed tumor growth relative to the negative control group (Figs. 7D–E, S6B). The combination of LINC01615 knockdown and oxaliplatin treatment (5 mg/kg) resulted in a more significant reduction in tumor growth than LINC01615 knockdown or oxaliplatin treatment alone (Figs. 7D–E, S6B). Furthermore, qRT–PCR was performed in PDX tumors, and we found decreased levels of LINC01615 in the LINC01615 knockdown group and sh-LINC01615 combined with oxaliplatin group relative to the negative control group (Fig. S6C). The expression levels of Ki-67 and G6PD were downregulated in the sh-LINC01615 group and sh-LINC01615 combined with oxaliplatin group (Fig. 7F). Caspase 3 was upregulated in all treatment groups, especially in the combined treatment group (Figs. 7F, S6D). We also examined the autophagy degradation status (LC3B) and METTL3 level in PDX tumors, oxaliplatin treatment increased LC3B-II/I and decreased METTL3 expression (Fig. S6E). The aforementioned results strongly suggest the potential of LINC01615 as a therapeutic target for CRC treatment and enhance oxaliplatin efficacy.
In summary, this study illustrates the mechanism by which nutrient starvation-reduced METTL3 upregulates the level of LINC01615 and LINC01615 recruits hnRNPA1 to modulate G6PD splicing and PPP activity, which regulates cell survival in adaptation to nutrient deprivation and chemosensitivity to oxaliplatin (Fig. 7G).
Discussion
In response to nutrient stress, cancer cells have developed strategies to overcome nutrient starvation, and these strategies, such as alterations in cell metabolism, aim to maximize the usage of nutrients and mitigate stress damage [13, 35]. To discover new mechanisms of adaptation to nutritional stress, we compared differentially expressed lncRNAs under nutrient starvation in two CRC cell lines and identified LINC01615 (Fig. 1). Further experiments demonstrated that CRC cells counteract nutrient-limited conditions by means of LINC01615-mediated apoptosis inhibition (Fig. 2). Reports on whether lncRNAs are involved in adaptation to nutrient starvation are still lacking, and this is an in-depth study showing that lncRNAs play an important role in the response to adverse nutritional conditions.
Several lncRNAs have been reported to be associated with the PPP in cancers. In multiple myeloma, the lncRNA PDIA3P interacts with c-Myc to enhance the transcription of G6PD, leading to an increase in PPP flux and promoting the proliferation and drug resistance of multiple myeloma [36]. In osteosarcoma, the lncRNA OR3A4 increased G6PD expression by sponging miR-1207-5p, leading to the activation of the PPP and the increased production of NADPH, thus promoting the growth of cancer cells [37]. Accordingly, in our study, GSEA showed that the differentially expressed gene sets upon LINC01615 knockdown were significantly related to the PPP. To test whether the PPP was involved in LINC01615-mediated regulation of cell survival, we treated cells with a competitive inhibitor of G6PD, 6-AN, and found that the function of LINC01615 could be abolished by 6-AN. Oxidized and nonoxidized PPPs have been identified to be excessively activated in multiple cancers, including breast cancer, pancreatic ductal adenocarcinoma and CRC [38–40], and PPP-derived NADPH and R5P function as cytoplasmic free radical scavengers and crucial sources of lipid and nucleotide synthesis [8, 41, 42]. Then, we treated LINC01615-depleted cells with the ROS scavenger NAC and found that NAC significantly reduced apoptosis and enhanced cell survival, suggesting that PPP-dependent ROS production eventually led to the promotion of apoptosis and suppression of cell survival, which is consistent with previous findings that overproduction of free radicals, such as ROS and reactive nitrogen species (RNS), can induce cellular apoptosis, senescence, autophagy, and necrosis [43, 44]. Meanwhile, we cultured LINC01615-depleted cells in medium with additional NADPH or R5P supplementation and found that the synthesis of lipids or nucleotides was enhanced, which provide required intermediates for cell growth necessitating biosynthesis [21] (Fig. 3). These findings suggest that LINC01615 maintains cell growth activity under nutrient-limited conditions.
We also demonstrated that LINC01615 promotes PPP flux by modulating G6PD splicing under starvation conditions. Splicing is a process in which introns are removed and exons are joined to produce a mature mRNA. The regulation of G6PD expression is controlled by changes in the rate of pre-mRNA splicing [27]. A previous study showed that G6PD pre-mRNA contained an exonic splicing silencer that localized to nt 43–72 of exon 12, and the binding of hnRNPs to the silencer inhibited the splicing of G6PD pre-mRNA [45]. Our results identified hnRNPA1 as a novel protein that interacts with LINC01615 and G6PD pre-mRNA. RNA pulldown demonstrated that hnRNPA1 can bind to exon 12, suggesting that LINC01615 acts as a blocker to inhibit the binding of hnRNPA1 to G6PD pre-mRNA and promote G6PD splicing. Previous research also reports that the enzymatic activity and expression levels of G6PD are regulated by nutritional signals, including a high-carbohydrate diet, polyunsaturated fatty acids, and hormonal signals [46]. Notably, we demonstrated for the first time that serum deprivation altered the expression and activity of G6PD via a lncRNA, which improves our understanding of the complex relationship between nutritional signals and G6PD. More mechanistic research underlying the changes in G6PD “sensing” nutrition signals needs to be carried out in the future.
As a member of the hnRNP protein family [47], hnRNPA1 is a splicing factor and is localized in the nucleus but can shuttle between the nucleus and cytoplasm [48]. The structure of hnRNPA1 contains two conserved RRMs and a flexible glycine-rich C-terminal region consisting of Arg-Gly-Gly tripeptide repeats (RGGs), which participate in both general and specific interactions with proteins or RNAs [49]. Studies have indicated that hnRNPA1 is also essential for controlling the splicing of various gene transcripts, such as CCDC50 [50], PKM [51] and Tau [52]. In addition to the above gene transcripts, our study identified G6PD as a new splicing substrate of hnRNPA1. By constructing a truncated protein, we demonstrated that RRM2 of hnRNPA1 was the functional region for splicing inhibition. LINC01615 promoted G6PD pre-mRNA splicing by competitively suppressing the binding of G6PD pre-mRNA with RRM2 (Figs. 4, 5). These findings enrich the research on the regulatory mechanism of G6PD splicing.
We also investigated the mechanism leading to LINC01615 upregulation in response to serum starvation. It has been reported that m6A modification can be found on almost all types of RNAs and can affect RNA transcription, processing, translation, and metabolism [53]. Indeed, we found several m6A sites dispersed throughout LINC01615, and nutrient starvation decreased the m6A level in CRC cells. This suggests that m6A modification may be involved in serum starvation-induced upregulation of LINC01615. Recent findings have suggested that nutrient deprivation could induce the expression of m6A demethylases [54]. We detected the expression of several demethylases and methylases, and the results revealed that the reduction in METTL3 is responsible for nutrient stress-induced upregulation of LINC01615. METTL3 was identified as the first RNA methylase; it can bind to multiple types of RNAs, including mRNA, tRNA, and snRNA, and mainly methylate m6A modification [55]. We demonstrated that METTL3-induced m6A modification targeted and destabilized LINC01615, which is consistent with the notion that m6A enrichment leads to RNA decay [56]. Autophagy is a conserved process of intracellular lysosomal degradation, and the substrates of autophagy include damaged organelles and proteins [57]. Starvation and other stressors can dramatically induce autophagy [58]. In this study, we found that knockdown of the autophagy-related gene ATG5 or 3-MA treatment enhanced starvation-reduced METTL3 and induced LINC01615 expression, demonstrating that LINC01615 is induced by starvation stress through the autophagy pathway. Further study showed that METTL3 can interact with LC3, and this interaction can be enhanced by starvation, demonstrating that METTL3 is an autophagy substrate (Fig. 6). Our findings are indicative of an autophagy-METTL3-LINC01615 axis that is a critical adaptation mechanism under nutrient starvation.
Oxaliplatin is one of the main chemotherapy drugs used as the first-line therapy for CRC, and previous research has indicated that oxaliplatin-related autophagy induction could lead to chemotherapy resistance [59]. The antitumor effect of oxaliplatin is closely related to ROS accumulation [60]. As we expected, our results indicated that LINC01615 knockdown enhanced oxaliplatin efficacy in PDO and PDX models (Fig. 7). The possible reason is that the high expression of LINC01615 induced by oxaliplatin-related autophagy was inhibited, which led to decreased cell proliferation and increased apoptosis. These findings may provide a useful strategy for sensitizing CRC to chemotherapy.
Taken together, our data suggest that LINC01615 can promote cell survival under starvation conditions. Mechanistically, LINC01615 promotes PPP flux to decrease ROS production and increase nucleotide and lipid synthesis by regulating G6PD pre-mRNA splicing. Furthermore, starvation-induced degradation of METTL3 through autophagy led to increased stability and levels of LINC01615 in a m6 A-dependent manner. We also demonstrated that LINC01615 knockdown combined with oxaliplatin has a strong synergistic effect against CRC. Our findings could be helpful for understanding the mechanism of adaptation to nutrient starvation and developing therapeutic strategies for CRC.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file2 Supplementary Figure 1 (A) Venn diagram showing the numbers of overlapping lncRNAs between the differentially downregulated lncRNAs in HCT116 and DLD1 cells after serum-free medium treatment. (B) Kaplan–Meier analysis of the OS rate in CRC patients in the TCGA database with high or low expression of CTD-2147F2.1. (C) The non-coding nature of LINC01615 was predicted by coding-potential analysis (PhyloCSF). (D) The relative expression of LINC01615 in various types of tumor tissues and corresponding peritumor tissues based on data obtained from TCGA database. (E) The relative expression of LINC01615 in tumor tissue and corresponding peritumor tissue based on data obtained from the GEO database (GSE115856). (F) qRT-PCR showed the expression of LINC01615 in normal intestinal epithelial cell line FHC and CRC cell lines (DLD1, HT-29, SW620, SW480, LoVo and HT29). (G) qRT-PCR examined the expression of LINC01615 after knockdown in DLD1 and HCT116 cells. (H) qRT-PCR examined the expression of LINC01615 after overexpression in LoVo cells. (I) CCK-8 assay showed the effects of LINC01615 knockdown on proliferation in HCT116 cells without serum supplementation. (J) Colony formation assays were performed to examine the effects of LINC01615 knockdown on proliferation in HCT116 cells without serum supplementation. (K) Statistical analysis of colony formation numbers in (J). (L-M) CCK-8 assay showed the effect of LINC01615 knockdown on proliferation in DLD1 and HCT116 cells under normal conditions. (N-O) Images of flow cytometry analysis showing the effects of LINC01615 knockdown and LINC01615 overexpression on the apoptotic rate in DLD1, HCT116 and LoVo cells without serum supplementation. (P) Flow cytometry data presented as histograms showing the effects of LINC01615 knockdown on the apoptotic rate in HCT116 cells without serum supplementation. (Q) WB was used to examine the effects of LINC01615 knockdown on the expression of cleaved caspase 3, caspase 7, caspase 9 and BCL-2 in HCT116 cells without serum supplementation. GAPDH was used as a loading control. Data are shown as the mean ± S.E. * p<0.05, ** p<0.01. (TIF 6432 kb)
Supplementary file3 Supplementary Figure 2 (A) HCT116 cells were transfected with LINC01615 shRNA or control shRNA. Then, the cells were cultured in medium containing [2-13C] glucose and without serum supplementation. Oxidative PPP flux was measured based on the rate of glucose consumption and the ratio of 13C incorporated into carbon 2 (indicating glycolysis) and carbon 3 (indicating PPP) of lactate by nuclear magnetic resonance (NMR) spectroscopy. (B) NADP+/NADPH ratios were measured in the cells from (A). (C) ROS levels were measured in the cells from (A). (D) Images of flow cytometry analysis showing the effects of LINC01615 knockdown and LINC01615 overexpression on the ROS level in DLD1 and LoVo cells without serum supplementation. (E) CCK-8 assay showed the effects of LINC01615 inhibition on proliferation in HCT116 cells treated with or without NAC under serum-free starvation conditions. (F) Images of flow cytometry analysis showing the effects of LINC01615 inhibition on the apoptotic rate in DLD1 and HCT116 cells treated with or without NAC under serum-free starvation conditions. (G) Flow cytometry data presented as histograms show the effects of LINC01615 inhibition on the apoptotic rate in HCT116 cells treated with or without NAC under serum-free starvation conditions. (H) WB was used to examine the effects of LINC01615 inhibition on cleaved caspase 3, caspase 7, caspase 9 and BCL-2 expression in HCT116 cells treated with or without NAC under serum-free starvation conditions. (I-J) CCK-8 assay showed the effects of LINC01615 inhibition on proliferation in DLD1 and HCT116 cells treated with or without R5P under serum-free starvation conditions. (K) Statistical analysis of EdU (+) cells in (Figure 3M). (L) EdU staining assay showed the effect of LINC01615 inhibition on nucleotide synthesis in HCT116 cells treated with or without R5P under serum-free starvation conditions. (M) Statistical analysis of EdU (+) cells in (L). (N-O) CCK-8 assay showed the effects of LINC01615 inhibition on proliferation in DLD1 and HCT116 cells treated with or without NADPH under serum-free starvation conditions. (P) Oil red O staining assay showed the effect of LINC01615 inhibition on lipid synthesis in HCT116 cells treated with or without NADPH under serum-free starvation conditions. Data are shown as the mean ± S.E. * p<0.05, ** p<0.01. (TIF 5630 kb)
Supplementary file4 Supplementary Figure 3 (A) The top-scoring genes recurring in the PPP gene sets, including G6PD. (B) The statistical analysis of G6PD expression in 275 colon cancer tissues and 41 normal tissues; paired t-test. In the boxplots, the middle line depicts the median and the whiskers are the min to max range. (C) The correlation between LINC01615 and G6PD expression in 275 colon cancer tissues. (D) The expression of G6PD in normal tissues and CRC tissues was assessed by IHC. (E-F) Statistical analysis of EdU (+) cells in (Figure 4I-J). (G) Antibodies against hnRNPA1 were used for RIP, followed by LINC01615 qRT–PCR in DLD1 and LoVo cells. (H) Antibodies against hnRNPA1 were used for RIP, followed by LINC01615 qRT–PCR, in DLD1 cells with or without serum supplementation. (I) Antibodies against hnRNPA1 were used for RIP, followed by G6PD pre-mRNA qRT–PCR in DLD1 and LoVo cells. (J) Antibodies against hnRNPA1 were used for RIP, followed by G6PD pre-mRNA qRT–PCR in LoVo cells transfected with LINC01615 overexpression plasmid or control vector. (K-L) WB was used to examine the effects of hnRNPA1 knockdown and overexpression on the expression of hnRNPA1 in DLD1, HCT116 and LoVo cells. (M) Antibodies against Flag were used for RIP, followed by G6PD pre-mRNA qRT–PCR in DLD1 cells. (N) Colony formation assay was conducted to examine the effects of hnRNPA1 truncated plasmids with or without RRM2 domain on proliferation in DLD1 cells which overexpressing LINC01615 and without serum supplementation. (O) Statistical analysis of colony formation numbers in (N). (P) Images of flow cytometry analysis showing the effects of hnRNPA1 truncated plasmid with or without RRM2 domain on the apoptotic rate in DLD1 cells overexpressing LINC01615 under serum-free starvation conditions. (Q) Flow cytometry data presented as histograms showing the effects of hnRNPA1 truncated plasmid with or without RRM2 domain on the apoptotic rate in DLD1 cells overexpressing LINC01615 and without serum supplementation. Data are the means ± SD (n = 3 independent experiments), * p<0.05, ** p<0.01. (TIF 7126 kb)
Supplementary file5 Supplementary Figure 4 (A) SRAMP (http://www.cuilab.cn/sramp/) prediction revealed several m6A sites distributed in LINC01615. (B) WB analysis of the expression of METTL3, METTL14 and FTO in DLD1 cells cultured with or without serum supplementation. (C-D) WB was used to examine the effects of METTL3 knockdown and overexpression on the expression of METTL3 in DLD1 and LoVo cells. (E) qRT–PCR analysis of the expression of LINC01615 in LoVo cells transfected with METTL3 overexpression plasmid or control vector. (F) qRT–PCR analysis of LINC01615 expression in LoVo cells cultured with or without serum supplementation and transfected with METTL3 overexpression plasmid or control vector. (G) The correlation between METTL3 and LINC01615 expression in colon tissues. (H) Statistical analysis of IHC staining scores in (Figure 6D). (I) meRIP-qPCR showing the m6A modification enrichment of LINC01615 in LoVo cells after METTL3 overexpression. (J) qRT–PCR analysis of the stability of LINC01615 in LoVo cells with or without METTL3 overexpression. Data are the means ± SD (n = 3 independent experiments), * p<0.05, ** p<0.01. (TIF 2502 kb)
Supplementary file6 Supplementary Figure 5 (A) WB analysis of METTL3 protein stability in LoVo cells cultured with or without serum supplementation for different times. (B) Transmission electron microscopy images of autophagic vacuoles in DLD1 and LoVo cells under normal or serum-free starvation conditions. Arrows indicate autophagosomes; scale bar, 500 nm. (C) Representative confocal microscopy images of LC3B puncta distribution in LoVo cells under normal or serum-free starvation conditions and treated with or without Baf A1 (1000× magnification). (D-E) Statistical analysis of fluorescence puncta of autolysosome (free red) and autophagosome (yellow) in (Figure 6H) and (C). (F) WB analysis of ATG5, METTL3 and LC3B expression after ATG5 knockdown in DLD1 cells without serum supplementation. (G) The expression of LINC01615 after 3-MA, ATG5 knockdown and Baf A1 treatment in DLD1 cells without serum supplementation. (H) WB analysis of METTL3 and LC3B expression in LoVo cells treated with or without Baf-A1 under serum-free starvation conditions. (I) Co-IP analysis was performed with specific antibodies against HA and Flag in 293T cells under normal or serum-free starvation conditions after transfection of HA-METTL3 and Flag-LC3B plasmids. WB analysis of the interaction between METTL3 and LC3B. (J) Representative confocal microscopy images of METTL3, LC3B and LAMP1 distribution in DLD1 cells under serum-free starvation conditions (1000× magnification). Data are the means ± SD (n = 3 independent experiments), * p<0.05, ** p<0.01. (TIF 11211 kb)
Supplementary file7 Supplementary Figure 6 (A) qRT-PCR was used to detect the expression of LINC01615 in two donor patients’ tumor tissues. (B) PDXs images of control group, sh-LINC01615 group, oxaliplatin group, and sh-LINC01615 combined with oxaliplatin group. (C) The expression levels of LINC01615 in PDX tumors of control group, sh-LINC01615 group, oxaliplatin group, and sh-LINC01615 combined with oxaliplatin group through qRT–PCR. (D) Statistical analysis of IHC staining scores in (Figure 7F). (E) WB analysis of LC3B and METTL3 expression in sh-Ctrl and sh-LINC01615 group treated with or without oxaliplatin. Data are the means ± SD (n = 3 independent experiments), * p<0.05, ** p<0.01. (TIF 3063 kb)
Acknowledgements
We thank all individuals who participated in this work.
Abbreviations
- CRC
Colorectal cancer
- PPP
Pentose phosphate pathway
- G6PD
G6P dehydrogenase
- lncRNAs
Long noncoding RNAs
- ROS
Reactive oxygen species
- 3-MA
3-Methyladenine
- Baf-A1
Bafilomycin
- shRNA
Short hairpin RNA
- siRNA
Short interfering RNA
- qRT–PCR
Real-time quantitative reverse transcription PCR
- WB
Western blot
- ChIRP-MS
Chromatin isolation by RNA purification-mass spectrometry
- RIP
RNA immunoprecipitation
- PDO
Patient-derived organoid
- PDX
Patient-derived xenograft
- GSEA
Gene set enrichment analysis
- OS
Overall survival
- DFS
Disease-free survival
- ceRNA
Competing endogenous RNA
- Co-IP
Co-immunoprecipitation
- IHC
Immunohistochemistry
- 6-AN
6-Aminonicotinamide
- NAC
N-Acetyl-L-cysteine
- RGG
Arg-Gly-Gly tripeptide repeats
- CCK-8
Cell counting kit-8
- NMR
Nuclear magnetic resonance
- FISH
Fluorescence in situ hybridization
- meRIP
Methylated RNA immunoprecipitation
- TEM
Transmission electron microscopy
Author contributions
YZ and LX conceived and designed the experiment, performed experiment, and wrote the manuscript. ZR, XL, JS, PZ, CZ, SG, NW, and XZ performed experiment, collected data and analyzed data. CX, ZL, MM, YZ, and YC interpreted data and constructed figures. CL conceptualized the study, designed the experiment, interpreted data and reviewed the manuscript. All authors read and approved the final version of the manuscript. YZ and CL have accessed and verified the data, and CL was responsible for the decision to submit the manuscript.
Funding
This research was supported by grants from the National Natural Science Foundation of China (No.82072729, No.81602568, No.81773130), China Postdoctoral Science Foundation (No.2018M643009), the Natural Science Foundation of Jiangsu (BK20211606), the National Natural Science Foundation of Hunan Province (No. 2019JJ50906) and Xuzhou Key R&D Program (KC20064).
Availability of data and materials
The data generated in this study are available upon request from the corresponding author.
Declarations
Conflict of interest
The authors declare no potential conflicts of interest.
Ethical approval and consent to participate
This study was performed according to the ethical standards of Declaration of Helsinki and was approved by the ethics committee of the Xuzhou Medical University (XYFY2019-KL221-01). All animal experiments were approved by the Animal Care Committee of Xuzhou Medical University.
Consent for publication
We have obtained consents to publish this paper from all the participants of this study.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yi Zhang, Lei Xu and Zeqiang Ren contributed equally to this work.
References
- 1.Cederholm T, et al. ESPEN guidelines on definitions and terminology of clinical nutrition. Clin Nutr. 2017;36:49–64. doi: 10.1016/j.clnu.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 2.Payton M, et al. Antagonism of Ang-Tie2 and Dll4-Notch signaling has opposing effects on tumor endothelial cell proliferation, evidenced by a new flow cytometry method. Lab Invest. 2014;94:1296–1308. doi: 10.1038/labinvest.2014.116. [DOI] [PubMed] [Google Scholar]
- 3.Izuishi K, Kato K, Ogura T, Kinoshita T, Esumi H. Remarkable tolerance of tumor cells to nutrient deprivation: possible new biochemical target for cancer therapy. Cancer Res. 2000;60:6201–6207. [PubMed] [Google Scholar]
- 4.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3:ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khan MR, Xiang S, Song Z, Wu M. The p53-inducible long noncoding RNA TRINGS protects cancer cells from necrosis under glucose starvation. EMBO J. 2017;36:3483–3500. doi: 10.15252/embj.201696239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li TY, et al. ULK1/2 constitute a bifurcate node controlling glucose metabolic fluxes in addition to autophagy. Mol Cell. 2016;62:359–370. doi: 10.1016/j.molcel.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 8.Yang HC, et al. The redox role of G6PD in Cell growth, cell death, and cancer. Cells. 2019 doi: 10.3390/cells8091055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patra KC, Hay N. The pentose phosphate pathway and cancer. Trends Biochem Sci. 2014;39:347–354. doi: 10.1016/j.tibs.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernard M, et al. Autophagy drives fibroblast senescence through MTORC2 regulation. Autophagy. 2020;16:2004–2016. doi: 10.1080/15548627.2020.1713640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang S, et al. Sirtuin 1 promotes autophagy and proliferation of endometrial cancer cells by reducing acetylation level of LC3. Cell Biol Int. 2021;45:1050–1059. doi: 10.1002/cbin.11549. [DOI] [PubMed] [Google Scholar]
- 12.Mancuso A, Sharfstein ST, Tucker SN, Clark DS, Blanch HW. Examination of primary metabolic pathways in a murine hybridoma with carbon-13 nuclear magnetic resonance spectroscopy. Biotechnol Bioeng. 1994;44:563–585. doi: 10.1002/bit.260440504. [DOI] [PubMed] [Google Scholar]
- 13.Desideri E, et al. MAPK14/p38alpha-dependent modulation of glucose metabolism affects ROS levels and autophagy during starvation. Autophagy. 2014;10:1652–1665. doi: 10.4161/auto.29456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Driehuis E, Kretzschmar K, Clevers H. Establishment of patient-derived cancer organoids for drug-screening applications. Nat Protoc. 2020;15:3380–3409. doi: 10.1038/s41596-020-0379-4. [DOI] [PubMed] [Google Scholar]
- 15.Arena S, et al. A subset of colorectal cancers with cross-sensitivity to olaparib and oxaliplatin. Clin Cancer Res. 2020;26:1372–1384. doi: 10.1158/1078-0432.CCR-19-2409. [DOI] [PubMed] [Google Scholar]
- 16.Xiao L, et al. Long noncoding RNA uc173 promotes renewal of the intestinal mucosa by inducing degradation of MicroRNA 195. Gastroenterology. 2018;154:599–611. doi: 10.1053/j.gastro.2017.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiao Z, et al. Statin-induced GGPP depletion blocks macropinocytosis and starves cells with oncogenic defects. Proc Natl Acad Sci USA. 2020;117:4158–4168. doi: 10.1073/pnas.1917938117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X, et al. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol Cancer. 2019;18:20. doi: 10.1186/s12943-018-0935-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ju HQ, et al. Disrupting G6PD-mediated Redox homeostasis enhances chemosensitivity in colorectal cancer. Oncogene. 2017;36:6282–6292. doi: 10.1038/onc.2017.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang X, et al. Roles for VEGF-C/NRP-2 axis in regulating renal tubular epithelial cell survival and autophagy during serum deprivation. Cell Biochem Funct. 2019;37:290–300. doi: 10.1002/cbf.3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stincone A, et al. The return of metabolism: biochemistry and physiology of the pentose phosphate pathway. Biol Rev Camb Philos Soc. 2015;90:927–963. doi: 10.1111/brv.12140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bensaad K, Cheung EC, Vousden KH. Modulation of intracellular ROS levels by TIGAR controls autophagy. EMBO J. 2009;28:3015–3026. doi: 10.1038/emboj.2009.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu J, et al. Overexpression of ULK1 Represents a potential diagnostic marker for clear cell renal carcinoma and the antitumor effects of SBI-0206965. EBioMedicine. 2018;34:85–93. doi: 10.1016/j.ebiom.2018.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu S, et al. Ai-lncRNA EGOT enhancing autophagy sensitizes paclitaxel cytotoxicity via upregulation of ITPR1 expression by RNA-RNA and RNA-protein interactions in human cancer. Mol Cancer. 2019;18:89. doi: 10.1186/s12943-019-1017-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee YJ, Wang Q, Rio DC. Coordinate regulation of alternative pre-mRNA splicing events by the human RNA chaperone proteins hnRNPA1 and DDX5. Genes Dev. 2018;32:1060–1074. doi: 10.1101/gad.316034.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kashima T, Rao N, David CJ, Manley JL. hnRNP A1 functions with specificity in repression of SMN2 exon 7 splicing. Hum Mol Genet. 2007;16:3149–3159. doi: 10.1093/hmg/ddm276. [DOI] [PubMed] [Google Scholar]
- 27.Hong X, et al. PTEN antagonises Tcl1/hnRNPK-mediated G6PD pre-mRNA splicing which contributes to hepatocarcinogenesis. Gut. 2014;63:1635–1647. doi: 10.1136/gutjnl-2013-305302. [DOI] [PubMed] [Google Scholar]
- 28.Zhou J, et al. N(6)-methyladenosine guides mRNA alternative translation during integrated stress response. Mol Cell. 2018;69:636–647.e637. doi: 10.1016/j.molcel.2018.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, Le WD. Autophagy and ubiquitin-proteasome system. Adv Exp Med Biol. 2019;1206:527–550. doi: 10.1007/978-981-15-0602-4_25. [DOI] [PubMed] [Google Scholar]
- 30.He C. Balancing nutrient and energy demand and supply via autophagy. Curr Biol. 2022;32:R684–r696. doi: 10.1016/j.cub.2022.04.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen W, Lian W, Yuan Y, Li M. The synergistic effects of oxaliplatin and piperlongumine on colorectal cancer are mediated by oxidative stress. Cell Death Dis. 2019;10:600. doi: 10.1038/s41419-019-1824-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McQuade RM, Stojanovska V, Bornstein JC, Nurgali K. Colorectal cancer chemotherapy: the evolution of treatment and new approaches. Curr Med Chem. 2017;24:1537–1557. doi: 10.2174/0929867324666170111152436. [DOI] [PubMed] [Google Scholar]
- 33.Ahmad S. Platinum-DNA interactions and subsequent cellular processes controlling sensitivity to anticancer platinum complexes. Chem Biodivers. 2010;7:543–566. doi: 10.1002/cbdv.200800340. [DOI] [PubMed] [Google Scholar]
- 34.Zhao M, et al. Combining neratinib with CDK4/6, mTOR, and MEK inhibitors in models of HER2-positive cancer. Clin Cancer Res. 2021;27:1681–1694. doi: 10.1158/1078-0432.CCR-20-3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chikashige Y, et al. Gcn2 eIF2α kinase mediates combinatorial translational regulation through nucleotide motifs and uORFs in target mRNAs. Nucleic Acids Res. 2020;48:8977–8992. doi: 10.1093/nar/gkaa608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang X, et al. LncRNA PDIA3P interacts with c-Myc to regulate cell proliferation via induction of pentose phosphate pathway in multiple myeloma. Biochem Biophys Res Commun. 2018;498:207–213. doi: 10.1016/j.bbrc.2018.02.211. [DOI] [PubMed] [Google Scholar]
- 37.Wang X, Chen K, Zhao Z. LncRNA OR3A4 regulated the growth of osteosarcoma cells by modulating the miR-1207-5p/G6PD signaling. Onco Targets Ther. 2020;13:3117–3128. doi: 10.2147/OTT.S234514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Q, et al. Rac1 activates non-oxidative pentose phosphate pathway to induce chemoresistance of breast cancer. Nat Commun. 2020;11:1456. doi: 10.1038/s41467-020-15308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen S, et al. Enhanced pentose phosphate pathway activity promotes pancreatic ductal adenocarcinoma progression via activating YAP/MMP1 axis under chronic acidosis. Int J Biol Sci. 2022;18:2304–2316. doi: 10.7150/ijbs.69526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin J, et al. The POU2F1-ALDOA axis promotes the proliferation and chemoresistance of colon cancer cells by enhancing glycolysis and the pentose phosphate pathway activity. Oncogene. 2022;41:1024–1039. doi: 10.1038/s41388-021-02148-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ju HQ, Lin JF, Tian T, Xie D, Xu RH. NADPH homeostasis in cancer: functions, mechanisms and therapeutic implications. Signal Transduct Target Ther. 2020;5:231. doi: 10.1038/s41392-020-00326-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ge T, et al. The Role of the Pentose Phosphate Pathway in Diabetes and Cancer. Front Endocrinol. 2020;11:365. doi: 10.3389/fendo.2020.00365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dreher D, Junod AF. Role of oxygen free radicals in cancer development. Eur J Cancer (Oxf, Engl 1990) 1996;32A:30–38. doi: 10.1016/0959-8049(95)00531-5. [DOI] [PubMed] [Google Scholar]
- 44.Moloney JN, Cotter TG. ROS signalling in the biology of cancer. Semin Cell Dev Biol. 2018;80:50–64. doi: 10.1016/j.semcdb.2017.05.023. [DOI] [PubMed] [Google Scholar]
- 45.Szeszel-Fedorowicz W, Talukdar I, Griffith BN, Walsh CM, Salati LM. An exonic splicing silencer is involved in the regulated splicing of glucose 6-phosphate dehydrogenase mRNA. J Biol Chem. 2006;281:34146–34158. doi: 10.1074/jbc.M603825200. [DOI] [PubMed] [Google Scholar]
- 46.Triantaphyllopoulos KA, Laliotis GP, Bizelis IA. Energy balance-dependent regulation of ovine glucose 6-phosphate dehydrogenase protein isoform expression. Adipocyte. 2014;3:30–38. doi: 10.4161/adip.26437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roy R, et al. hnRNPA1 couples nuclear export and translation of specific mRNAs downstream of FGF-2/S6K2 signalling. Nucleic Acids Res. 2014;42:12483–12497. doi: 10.1093/nar/gku953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu W, et al. Heterogeneous nuclear ribonucleoprotein A1 exerts protective role in intracerebral hemorrhage-induced secondary brain injury in rats. Brain Res Bull. 2020;165:169–177. doi: 10.1016/j.brainresbull.2020.09.023. [DOI] [PubMed] [Google Scholar]
- 49.Roy R, Huang Y, Seckl MJ, Pardo OE. Emerging roles of hnRNPA1 in modulating malignant transformation. Wiley Interdiscip Rev RNA. 2017 doi: 10.1002/wrna.1431. [DOI] [PubMed] [Google Scholar]
- 50.Sun G, et al. HnRNP A1 - mediated alternative splicing of CCDC50 contributes to cancer progression of clear cell renal cell carcinoma via ZNF395. J Exp Clin Cancer Res. 2020;39:116. doi: 10.1186/s13046-020-01606-x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 51.Lan Z, et al. The interaction between lncRNA SNHG6 and hnRNPA1 contributes to the growth of colorectal cancer by enhancing aerobic glycolysis through the regulation of alternative splicing of PKM. Front Oncol. 2020;10:363. doi: 10.3389/fonc.2020.00363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu Y, et al. hnRNP A1 regulates alternative splicing of tau exon 10 by targeting 3' splice sites. Cells. 2020 doi: 10.3390/cells9040936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen XY, Zhang J, Zhu JS. The role of m(6)A RNA methylation in human cancer. Mol Cancer. 2019;18:103. doi: 10.1186/s12943-019-1033-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang S, et al. m(6)A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat Commun. 2019;10:2782. doi: 10.1038/s41467-019-10669-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Niu Y, et al. N6-methyl-adenosine (m6A) in RNA: an old modification with a novel epigenetic function. Genom Proteom Bioinform. 2013;11:8–17. doi: 10.1016/j.gpb.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou Z, et al. Mechanism of RNA modification N6-methyladenosine in human cancer. Mol Cancer. 2020 doi: 10.1186/s12943-020-01216-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jiao L, et al. Regulation of glycolytic metabolism by autophagy in liver cancer involves selective autophagic degradation of HK2 (hexokinase 2) Autophagy. 2018;14:671–684. doi: 10.1080/15548627.2017.1381804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Amaravadi R, Kimmelman AC, White E. Recent insights into the function of autophagy in cancer. Genes Dev. 2016;30:1913–1930. doi: 10.1101/gad.287524.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu W, et al. HMGB1-mediated autophagy modulates sensitivity of colorectal cancer cells to oxaliplatin via MEK/ERK signaling pathway. Cancer Biol Ther. 2015;16:511–517. doi: 10.1080/15384047.2015.1017691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yin H, et al. Chiauranib selectively inhibits colorectal cancer with KRAS wild-type by modulation of ROS through activating the p53 signaling pathway. Am J Cancer Res. 2020;10:3666–3685. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary file2 Supplementary Figure 1 (A) Venn diagram showing the numbers of overlapping lncRNAs between the differentially downregulated lncRNAs in HCT116 and DLD1 cells after serum-free medium treatment. (B) Kaplan–Meier analysis of the OS rate in CRC patients in the TCGA database with high or low expression of CTD-2147F2.1. (C) The non-coding nature of LINC01615 was predicted by coding-potential analysis (PhyloCSF). (D) The relative expression of LINC01615 in various types of tumor tissues and corresponding peritumor tissues based on data obtained from TCGA database. (E) The relative expression of LINC01615 in tumor tissue and corresponding peritumor tissue based on data obtained from the GEO database (GSE115856). (F) qRT-PCR showed the expression of LINC01615 in normal intestinal epithelial cell line FHC and CRC cell lines (DLD1, HT-29, SW620, SW480, LoVo and HT29). (G) qRT-PCR examined the expression of LINC01615 after knockdown in DLD1 and HCT116 cells. (H) qRT-PCR examined the expression of LINC01615 after overexpression in LoVo cells. (I) CCK-8 assay showed the effects of LINC01615 knockdown on proliferation in HCT116 cells without serum supplementation. (J) Colony formation assays were performed to examine the effects of LINC01615 knockdown on proliferation in HCT116 cells without serum supplementation. (K) Statistical analysis of colony formation numbers in (J). (L-M) CCK-8 assay showed the effect of LINC01615 knockdown on proliferation in DLD1 and HCT116 cells under normal conditions. (N-O) Images of flow cytometry analysis showing the effects of LINC01615 knockdown and LINC01615 overexpression on the apoptotic rate in DLD1, HCT116 and LoVo cells without serum supplementation. (P) Flow cytometry data presented as histograms showing the effects of LINC01615 knockdown on the apoptotic rate in HCT116 cells without serum supplementation. (Q) WB was used to examine the effects of LINC01615 knockdown on the expression of cleaved caspase 3, caspase 7, caspase 9 and BCL-2 in HCT116 cells without serum supplementation. GAPDH was used as a loading control. Data are shown as the mean ± S.E. * p<0.05, ** p<0.01. (TIF 6432 kb)
Supplementary file3 Supplementary Figure 2 (A) HCT116 cells were transfected with LINC01615 shRNA or control shRNA. Then, the cells were cultured in medium containing [2-13C] glucose and without serum supplementation. Oxidative PPP flux was measured based on the rate of glucose consumption and the ratio of 13C incorporated into carbon 2 (indicating glycolysis) and carbon 3 (indicating PPP) of lactate by nuclear magnetic resonance (NMR) spectroscopy. (B) NADP+/NADPH ratios were measured in the cells from (A). (C) ROS levels were measured in the cells from (A). (D) Images of flow cytometry analysis showing the effects of LINC01615 knockdown and LINC01615 overexpression on the ROS level in DLD1 and LoVo cells without serum supplementation. (E) CCK-8 assay showed the effects of LINC01615 inhibition on proliferation in HCT116 cells treated with or without NAC under serum-free starvation conditions. (F) Images of flow cytometry analysis showing the effects of LINC01615 inhibition on the apoptotic rate in DLD1 and HCT116 cells treated with or without NAC under serum-free starvation conditions. (G) Flow cytometry data presented as histograms show the effects of LINC01615 inhibition on the apoptotic rate in HCT116 cells treated with or without NAC under serum-free starvation conditions. (H) WB was used to examine the effects of LINC01615 inhibition on cleaved caspase 3, caspase 7, caspase 9 and BCL-2 expression in HCT116 cells treated with or without NAC under serum-free starvation conditions. (I-J) CCK-8 assay showed the effects of LINC01615 inhibition on proliferation in DLD1 and HCT116 cells treated with or without R5P under serum-free starvation conditions. (K) Statistical analysis of EdU (+) cells in (Figure 3M). (L) EdU staining assay showed the effect of LINC01615 inhibition on nucleotide synthesis in HCT116 cells treated with or without R5P under serum-free starvation conditions. (M) Statistical analysis of EdU (+) cells in (L). (N-O) CCK-8 assay showed the effects of LINC01615 inhibition on proliferation in DLD1 and HCT116 cells treated with or without NADPH under serum-free starvation conditions. (P) Oil red O staining assay showed the effect of LINC01615 inhibition on lipid synthesis in HCT116 cells treated with or without NADPH under serum-free starvation conditions. Data are shown as the mean ± S.E. * p<0.05, ** p<0.01. (TIF 5630 kb)
Supplementary file4 Supplementary Figure 3 (A) The top-scoring genes recurring in the PPP gene sets, including G6PD. (B) The statistical analysis of G6PD expression in 275 colon cancer tissues and 41 normal tissues; paired t-test. In the boxplots, the middle line depicts the median and the whiskers are the min to max range. (C) The correlation between LINC01615 and G6PD expression in 275 colon cancer tissues. (D) The expression of G6PD in normal tissues and CRC tissues was assessed by IHC. (E-F) Statistical analysis of EdU (+) cells in (Figure 4I-J). (G) Antibodies against hnRNPA1 were used for RIP, followed by LINC01615 qRT–PCR in DLD1 and LoVo cells. (H) Antibodies against hnRNPA1 were used for RIP, followed by LINC01615 qRT–PCR, in DLD1 cells with or without serum supplementation. (I) Antibodies against hnRNPA1 were used for RIP, followed by G6PD pre-mRNA qRT–PCR in DLD1 and LoVo cells. (J) Antibodies against hnRNPA1 were used for RIP, followed by G6PD pre-mRNA qRT–PCR in LoVo cells transfected with LINC01615 overexpression plasmid or control vector. (K-L) WB was used to examine the effects of hnRNPA1 knockdown and overexpression on the expression of hnRNPA1 in DLD1, HCT116 and LoVo cells. (M) Antibodies against Flag were used for RIP, followed by G6PD pre-mRNA qRT–PCR in DLD1 cells. (N) Colony formation assay was conducted to examine the effects of hnRNPA1 truncated plasmids with or without RRM2 domain on proliferation in DLD1 cells which overexpressing LINC01615 and without serum supplementation. (O) Statistical analysis of colony formation numbers in (N). (P) Images of flow cytometry analysis showing the effects of hnRNPA1 truncated plasmid with or without RRM2 domain on the apoptotic rate in DLD1 cells overexpressing LINC01615 under serum-free starvation conditions. (Q) Flow cytometry data presented as histograms showing the effects of hnRNPA1 truncated plasmid with or without RRM2 domain on the apoptotic rate in DLD1 cells overexpressing LINC01615 and without serum supplementation. Data are the means ± SD (n = 3 independent experiments), * p<0.05, ** p<0.01. (TIF 7126 kb)
Supplementary file5 Supplementary Figure 4 (A) SRAMP (http://www.cuilab.cn/sramp/) prediction revealed several m6A sites distributed in LINC01615. (B) WB analysis of the expression of METTL3, METTL14 and FTO in DLD1 cells cultured with or without serum supplementation. (C-D) WB was used to examine the effects of METTL3 knockdown and overexpression on the expression of METTL3 in DLD1 and LoVo cells. (E) qRT–PCR analysis of the expression of LINC01615 in LoVo cells transfected with METTL3 overexpression plasmid or control vector. (F) qRT–PCR analysis of LINC01615 expression in LoVo cells cultured with or without serum supplementation and transfected with METTL3 overexpression plasmid or control vector. (G) The correlation between METTL3 and LINC01615 expression in colon tissues. (H) Statistical analysis of IHC staining scores in (Figure 6D). (I) meRIP-qPCR showing the m6A modification enrichment of LINC01615 in LoVo cells after METTL3 overexpression. (J) qRT–PCR analysis of the stability of LINC01615 in LoVo cells with or without METTL3 overexpression. Data are the means ± SD (n = 3 independent experiments), * p<0.05, ** p<0.01. (TIF 2502 kb)
Supplementary file6 Supplementary Figure 5 (A) WB analysis of METTL3 protein stability in LoVo cells cultured with or without serum supplementation for different times. (B) Transmission electron microscopy images of autophagic vacuoles in DLD1 and LoVo cells under normal or serum-free starvation conditions. Arrows indicate autophagosomes; scale bar, 500 nm. (C) Representative confocal microscopy images of LC3B puncta distribution in LoVo cells under normal or serum-free starvation conditions and treated with or without Baf A1 (1000× magnification). (D-E) Statistical analysis of fluorescence puncta of autolysosome (free red) and autophagosome (yellow) in (Figure 6H) and (C). (F) WB analysis of ATG5, METTL3 and LC3B expression after ATG5 knockdown in DLD1 cells without serum supplementation. (G) The expression of LINC01615 after 3-MA, ATG5 knockdown and Baf A1 treatment in DLD1 cells without serum supplementation. (H) WB analysis of METTL3 and LC3B expression in LoVo cells treated with or without Baf-A1 under serum-free starvation conditions. (I) Co-IP analysis was performed with specific antibodies against HA and Flag in 293T cells under normal or serum-free starvation conditions after transfection of HA-METTL3 and Flag-LC3B plasmids. WB analysis of the interaction between METTL3 and LC3B. (J) Representative confocal microscopy images of METTL3, LC3B and LAMP1 distribution in DLD1 cells under serum-free starvation conditions (1000× magnification). Data are the means ± SD (n = 3 independent experiments), * p<0.05, ** p<0.01. (TIF 11211 kb)
Supplementary file7 Supplementary Figure 6 (A) qRT-PCR was used to detect the expression of LINC01615 in two donor patients’ tumor tissues. (B) PDXs images of control group, sh-LINC01615 group, oxaliplatin group, and sh-LINC01615 combined with oxaliplatin group. (C) The expression levels of LINC01615 in PDX tumors of control group, sh-LINC01615 group, oxaliplatin group, and sh-LINC01615 combined with oxaliplatin group through qRT–PCR. (D) Statistical analysis of IHC staining scores in (Figure 7F). (E) WB analysis of LC3B and METTL3 expression in sh-Ctrl and sh-LINC01615 group treated with or without oxaliplatin. Data are the means ± SD (n = 3 independent experiments), * p<0.05, ** p<0.01. (TIF 3063 kb)
Data Availability Statement
The data generated in this study are available upon request from the corresponding author.