Abstract
Although damaged cells can be repaired, cells that are considered unlikely to be repaired are eliminated through apoptosis, a type of predicted cell death found in multicellular organisms. Apoptosis is a structured cell death involving alterations to the cell morphology and internal biochemical changes. This process involves the expansion and cracking of cells, changes in cell membranes, nuclear fragmentation, chromatin condensation, and chromosome cleavage, culminating in the damaged cells being eaten and processed by other cells. The ubiquitin–proteasome system (UPS) is a major cellular pathway that regulates the protein levels through proteasomal degradation. This review proposes that apoptotic proteins are regulated through the UPS and describes a unique direction for cancer treatment by controlling proteasomal degradation of apoptotic proteins, and small molecules targeted to enzymes associated with UPS.
Keywords: Cancer, Cell death, DUB, UPS
Introduction
Apoptosis, an essential process for development and homeostasis in all multicellular organs, is divided into the intrinsic and extrinsic pathways [1] (Fig. 1). Problems with apoptosis give rise to numerous diseases, including cancer [1]. Rheumatoid arthritis and cancers are the well-known examples of under-apoptosis, whereas ischemic cardiomyopathy, acquired immune deficiency syndrome (AIDS), Alzheimer’s and Parkinson’s disease are some of the known examples of excessive-apoptosis [2, 3]. For the treatment of diseases caused by insufficient or excessive apoptosis, the protein levels of apoptotic proteins need to be controlled. A typical process that regulates protein levels is called the ubiquitin–proteasome system (UPS). Ubiquitination is a process that involves degradation of a target protein [4]. The ubiquitin–proteasome system proceeds through E1 (Ubiquitin-activating enzyme), E2 (Ubiquitin-conjugating enzyme), and E3 (Ubiquitin-protein ligase enzyme) [5]. E1, E2, and E3 enzymes induce the target protein to undergo proteasomal degradation, by forming the ubiquitin chain on the target protein [5]. Deubiquitinating enzymes (DUBs) cut off the ubiquitin chain formed on the target protein, reducing the amount of ubiquitination of the target protein [5] (Fig. 2).
Fig. 1.
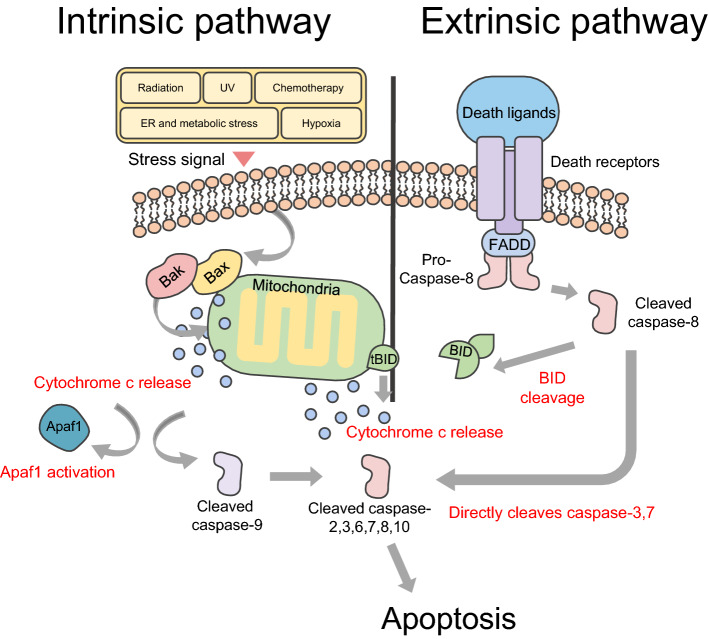
Extrinsic and intrinsic pathways in apoptosis. Internal stimuli, including DNA damage, activate apoptotic proteins belonging to the Bcl-2 family Bax and Bak. And this induces formation of Bax-Bak complexes in the mitochondrial outer membrane. The complexes release cytochrome c and enhance levels of apoptotic protease activator 1 (Apaf1). Released cytochrome c promotes proteolytic maturation of the activator caspase-9. Caspase-9 then cleaves and activates other effectors, caspase-2, 3, 6–8, and 10, and consequently induces apoptosis, and this series of processes is called the intrinsic pathway in apoptosis. As the death receptor and ligand bind, FAS-associated death domain protein (FADD) and pro-caspase-8 line up to bind to the intracellular portion of the death receptor. At this time, the recruited caspase 8 can directly cleave and activate caspase-3 and caspase-7 and proteolytically activate the BH3-only protein BH3-interacting domain death agonist (BID). Truncated BID (tBID) promotes mitochondrial membrane permeability through activation of the Bax-Bak complex of the outer mitochondrial membrane, and then induces apoptosis through cleavage and activation of caspases, and this series of processes is called the extrinsic pathway in apoptosis
Fig. 2.
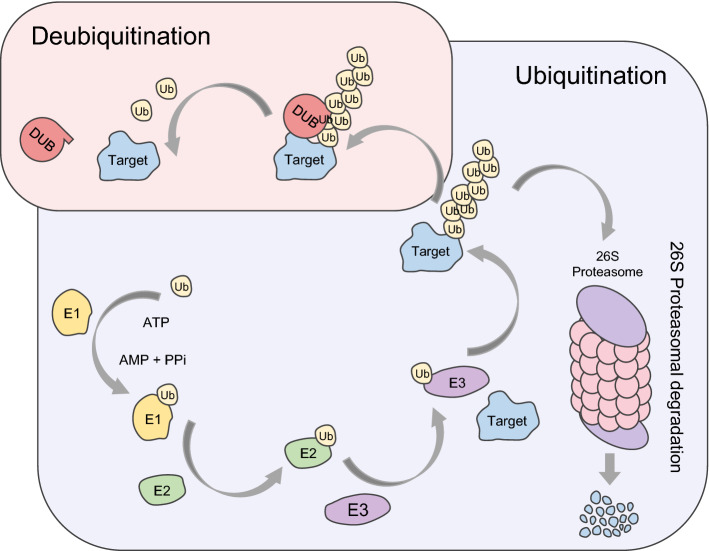
A pathway of protein proteasomal degradation through UPS and counteraction by DUBs. UPS requires a series of processes with involvement of E1, E2, and E3 enzymes. The result of ubiquitination is degradation of the target protein. (blue = E1 enzyme, green = E2 enzyme, purple = E3 enzyme, gray = target protein, red = DUB)
Apoptosis
Nucleated animal cells contain substances that induce cell destruction in an inactive form [1]. The onset of apoptosis is very tightly controlled by an activation mechanism [1]. This is because once apoptosis is initiated, it irreversibly leads to cell death. Therefore, it is essential that the onset is strictly regulated, until apoptosis is required. The intrinsic pathway is activated by intracellular signals released when cells are stressed, and by proteins released from the mitochondrial intermembrane space [6]. Mitochondria are important organelles for eukaryotes; without mitochondria, cell respiration is inhibited, leading to rapid cell death [6]. This is an important factor to be considered in cell death. Apoptotic proteins that target cells affect them in a variety of ways [6]. These proteins make passages on the mitochondrial membrane to expand the mitochondria or increase the permeability of the mitochondrial membrane to release apoptotic substances in the mitochondrial intermembrane space [6]. Nitric oxide is also involved in cell death [7]. Nitrogen monoxide increases the permeability of the mitochondrial membrane by decreasing the concentration gradient formed in the mitochondria, thereby facilitating apoptosis [7]. It is well known that p53 is responsible for cell cycle regulation and tumor suppression, and studies associating p53 and apoptosis have been identified [8]. Apoptosis by p53 involves the regulation of transcription-dependent and transcription-independent functions of p53. When enabled, p53 transactivates numerous apoptosis-related genes, including Bax, Noxa, PUMA, PIG3, p53AIP1, Killer/DR5, CD95 (Fas), and Prep [8, 9]. Most of these targets are members of the apoptosis-promoting Bcl-2 family genes [8]. In addition to these nuclear activities, p53 is also capable of inducing cytoplasmic death by mediating transcription-independent cytoplasm activation [10]. In particular, p53 is rapidly restricted to the mitochondria in response to various cell death signals, including ionizing radiation [11]. In mitochondria, p53 induces outer mitochondrial permeation (MOMP) to jointly release pro-apoptotic factors between mitochondrial membranes [10]. It is suggested that p53 interacts with Bak, Bcl-2, and Bcl-xl in the mitochondria, and acts as a BH3-specific protein and a direct activator or inhibitor of Bax or Bak [12]. The mitochondrial outer membrane permeabilization pore (MAC), which creates channels that increase the permeability of the outer mitochondrial membrane, is controlled by the Bcl-2 protein. The Bcl-2 protein also plays a key role in apoptosis. A second mitochondrial-derived protein, the mitochondrial-derived activator of caspase (SMAC), is released into the mitochondrial substrate [13]. SMAC inhibits the activity of these proteins by binding to proteins (IAPs) that inhibit apoptosis [13]. This inhibits the prevention of apoptosis by IAPs and makes it possible to initiate apoptosis. In addition, IAPs block the activity of caspase (cysteine protease/proteinase), and SMAC indirectly increases caspase activity and is involved in apoptosis [13]. The increased permeability of the mitochondrial membrane directly activates the enzyme that causes apoptosis [14]. One of the outcomes of increased caspase activity is cell death by mitochondrial cytochrome c [15]. Cytochrome c is present in the inner mitochondrial membrane as an enzyme involved in oxidative phosphorylation [15]. Apoptosis causes the outflow of cytochrome c into the cytoplasm, and is activated to break down the DNA of the cell and promote apoptosis [15]. Briefly, cytochrome c is released into the cytoplasm due to a channel formed in the outer mitochondrial membrane (MAC: mitochondrial apoptosis-induced channel) [6]. The released cytochrome c binds to the apoptosis protease activating factor-1 (Apaf-1), and subsequently to pro-caspase-9, to form a protein complex [6]. This complex activates pro-caspase-9 to form caspase-9 for the generation of cascades that activate other substances involved in apoptosis [6]. Anti-apoptotic Bcl-2 family members such as Bcl-2, BCL2L1, and MCL1 inhibit the action of BH3-only proteins, thereby inhibiting the progression of intrinsic apoptosis [16]. The death ligands (DRs) such as tumor necrosis factor-related apoptosis-inducing ligand (TRAIL), Fas ligand (FasL), and tumor necrosis factor-α (TNF-α) initiate the extrinsic apoptosis pathway [17–19]. They belong to the TNF receptor family, characterized by a cysteine-rich extracellular domain and a cytoplasmic death domain [20]. There are eight types of DRs and they are divided into two groups [21]. The first group includes the receptors Fas (DR2), TRAILR1 (DR4), and TRAIL2 (DR5) that can be activated by FasL and TRAIL [22]. TRAIL ligands bind to DR and TRAIL receptors (TRAIL-R), trigger apoptosis signals, and induce the formation and activation of death-inducing signaling complex (DISC). The second group includes the receptors TNFR1 (DR1), TRAMP (DR3), DR6, and EDAR. DR recruits the TNF-associated death domain (TRADD) as an adapter protein and binds to TNF-2,5 receptor-associated factor (TRAF2,5), the receptor-interacting protein kinase (RIP1 or RIPK1), and cellular inhibitors of apoptosis protein (cIAP). They bind to death receptors, and together with pro-caspase-8, FAS-associated death domain protein (FADD) or TNF receptor-associated death domain (TRADD) is aligned to associate with the intracellular region of death receptors [19]. In addition, the formation of DISC by caspase-8 binding to FADD can be inhibited when a protein called FLICE-like inhibitory protein (c-FLIP) is bound to FADD [23]. They bind to death receptors, and either FAS-associated death domain protein (FADD) or TNF receptor-associated death domain (TRADD) along with pro-caspase-8 is lined up to bind the intracellular region of death receptors [19]. Recruited caspase-8 directly cleaves and activates caspase-3 and 7 [24], and it proteolytically activates a BH3-only protein, the BH3-interacting domain death agonist (BID) [17]. Truncated BID (tBID) promotes mitochondrial membrane permeability through activation of the Bax-Bak complex in the outer mitochondrial membrane, and then induces apoptosis through cleavage and activation of caspases, which are involved in an extrinsic pathway of apoptosis [17].
UPS
Ubiquitin, a small polypeptide of 76 amino acids, is attached to the target protein. After several repetitions, many ubiquitins form a polyubiquitin chain on the target protein [25]. This action has many functions, including protein degradation, DNA damage response, stress response, and translation [26].
Ubiquitin-specific peptidase (USP) is a DUB that specifically recognizes and removes ubiquitin from proteins that belong to a large family of cysteine proteases [27]. USP is reported to be involved in many diseases, including cancer proliferation, inflammation, and neurodegenerative disorders [28, 29]. DUBs are divided into nine families. The subfamilies are divided into two protease groups according to the type of enzyme cleavage. Particularly, the cysteine protease class includes monocyte chemotactic protein-induced proteins (MCPIP), MIU-containing novel DUB family (MINDY), Machado-Joseph disease protein domain protease (MJD), ovarian cancer protease (OTU), after permuted papain fold peptidases of dsRNA viruses and eukaryotes (PPPDE) ubiquitin-specific protease (USP), ubiquitin C-terminal hydrolase (UCH), and ZUFSP families (Fig. 3). The metalloprotease type contains the Jab1/Pab1/MPN metal enzyme motif protease (JAMM) family [30].
Fig. 3.
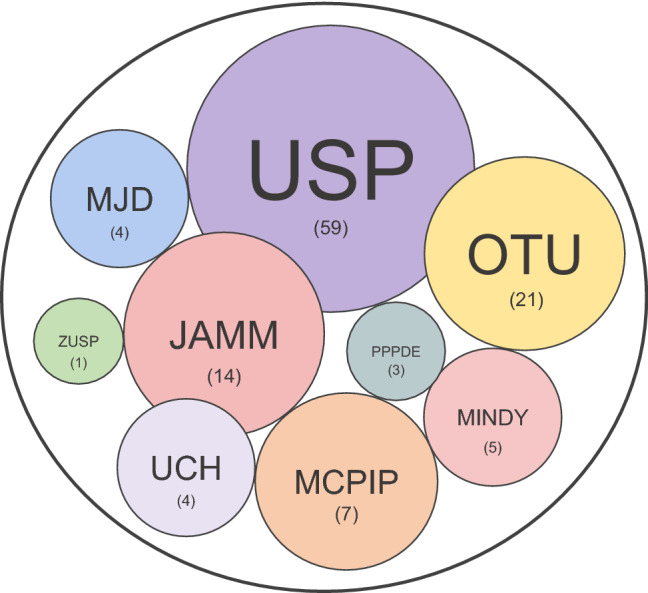
Types of DUB families. A total of nine families of DUBs have been identified including ubiquitin-specific protease (USP), ubiquitin C-terminal hydrolases protease (UCH), Machado–Joseph disease protein domain protease (MJD), ovarian tumor protease (OTU), Jab1/Pab1/MPN metallo-enzyme motif protease (JAMM), monocyte chemotactic protein-induced protease (MCPIP), permuted papain fold peptidase of dsDNA viruses and eukaryotes (PPPDE), motif interacting with Ub-containing novel DUB family (MINDY), and zinc finger with UFM1-specific peptidase domain protein (ZUFSP)
DUBs for apoptotic regulation
Pro-apoptotic proteins mediate apoptosis to destroy the damaged cells that cannot be recovered. DUBs that increase apoptosis by inhibiting proteasomal degradation of pro-apoptotic proteins through deubiquitination have been introduced. This process can be used to control the proliferation of cancer cells [31]. Conversely, E3 ligase, which increases the ubiquitination of pro-apoptotic proteins, promotes the degradation of apoptotic proteins. Understanding this process is important in cancer therapy [32].
CYLD
CYLD was identified as a mutable gene from familial cylindromatosis to develop cutaneous appendage tumors [33]. The N-terminal domain of CYLD comprises three cytoskeleton-related protein-glycine-rich (CAP-Gly) domains; the first two domains of CYLD mediate microtubule binding and the last is the CAP-Gly domain. This last domain couples to inhibitors of κB (IκB) kinase (IKK) adapter protein NF-κB essential modulator (NEMO) [34, 35]. CYLD has been widely studied in relation to NF-κB signaling [34]. It has been reported that the C-terminal portion of CYLD mediates M1 and K63 polyubiquitination, and the USP domain mediates cleavage of K11 and K48 polyubiquitin bonds in vitro. CYLD also has two stored proline-rich (PR) motifs and a TRAF2-binding motif (PVQES) that can interact with the SH3 domain of other proteins [36]. Additional studies have shown how the specificity of the K63 and M1 polyubiquitin chains are achieved [37–39]. IKKβ induces ubiquitination and degradation through IκBα phosphorylation, translocating NF-κB to the nucleus. CYLD is a DUB that ablates K63 and M1 polyubiquitin chains from various NF-κB signaling proteins [40]. The K48-K63 branch is shown to inhibit the K63-linked deubiquitination from CYLD [41]. Consistent with its role in deubiquitination of several key NF-κB signaling proteins, CYLD deficiency leads to constitutive NF-κB activation and subsequent inflammatory gene expression [42–44]. CYLD is known as a tumor suppressor protein in DUB [45], and this function of CYLD has also been studied using mouse models. CYLD knockout mice are not naturally tumor-causing but are more susceptible to chemically induced skin tumors than wild type mice [46].
OTUB1
OTU deubiquitinase, ubiquitin aldehyde binding 1 (OTUB1) known as otubain 1, belongs to the ovarian tumor domain (OTU) family of DUB. p53 is a well-known tumor suppressor [47]. OTUB1 is known as a regulator of p53, and regulates the mouse double minute 2 homolog (MDM2) modulating ubiquitination on p53 [48]. OTUB1 binds to the p53-MDM2-UbcH5 complex. It was confirmed that OTUB1 mediates the stabilization and activation of p53 by regulating the ubiquitination of p53 by MDM2 [48]. In addition, a previous study has shown that OTUB1 induces stabilization of murine mouse double minute 4 (MDM4) protein (MDMX) by inhibiting the MDM2-mediated MDMX ubiquitination, as well as MDMX accumulation in the mitochondria and cytoplasm, p53 (S46) phosphorylation, and mitochondrial-mediated apoptosis [49]. OTUB1 is known to negatively regulate the innate antiviral immune response and MHC-II antigen presentation and participates in the maturation and apoptosis of lymphocytes. OTUB1 plays an important role in apoptosis through autophagy related 5 (ATG5), which is necessary for the formation of autophagy vesicles, and abnormal expression of ATG5 thereby affects the initiation of autophagy [50]. A study has shown that ATG5 regulation by OTUB1 indirectly modulates autophagy and the immune processes [50].
UCHL1
Ubiquitin C-terminal hydrolase L1 (UCHL1) is an important member of the UCH family. UCHL1 protects against proteasomal degradation by removing the K48 polyubiquitin chain of phorbol-12-myristate-13-acetate-induced protein 1 (NOXA/PMAIP1), which is a known important mediator of DNA damage-induced cell death [51]. NOXA expression was enhanced in chemotherapy-resistant tumor samples, and UCHL1 deficiency reduced the DNA-damage-related apoptosis [51]. According to bioinformatics data of a previous study, the expression level of UCHL1 was suppressed in nasopharyngeal carcinoma cell lines [52]. It has been demonstrated that UCHL1 has dual functions due to its property to deubiquitinate p53 and p14ARF, and ubiquitinate MDM2 [52]. This means that UCHL1 can be applied as a factor that induces apoptosis and is involved in the pathogenesis of nasopharyngeal carcinoma [52]. In addition, the expression level of UCHL1 is decreased in hepatocellular carcinoma (HCC) and other digestive tumors [53]. Restoration of UCHL1 expression inhibits cell proliferation and induces apoptosis through the caspase pathway [53].
UCHL5/UCH37
Ubiquitin C-terminal hydrolase L5 (UCHL5/UCH37) is a DUB belonging to the cysteine protease family. UCHL5 is related to the 26S proteasome through Rpn132 and appears to play a role in interfering with the proteasome-associated degradation of substrates by detaching the polyubiquitin chain from substrates [54]. UCHL5 promotes the breakdown of inducible nitric oxide synthase and IκBα in the proteasome [54]. UCHL5 and the ubiquitin-specific protease 14 (USP14) inhibitor (b-AP15) induces cancer cell death by regulating UCHL5 levels in ovarian cancer [55]. b-AP15 inhibits phosphorylation of Smad2 and inhibits its invasive ability during TGF-β signaling [55].
USP2
MDM4 tumor protein inhibits tumor formation by regulating the apoptotic mediator p53. Ubiquitin-specific protease 2a (USP2a) is a DUB that protects MDM4 from degradation. Therefore, the USP2a-MDM4 interaction can be positioned as one of the key factors determining the malignant potential of human cancers [56]. It has been reported that isoforms of USP2, USP2a and USP2c mediate cell death by targeting the RIP1 protein [57]. In the same study, TRAF2 was determined to be the second target for USP2a and USP2c [57]. In addition, it was confirmed that knockdown of USP2c, but not USP2a, induces apoptosis through siRNA. The difference between the results of USP2a and USP2c is that the two proteins are homologous, but differ in the range of target substrates depending on the N terminal domain [57]. Because it has been demonstrated that USP2a mediates the TNF signaling pathway, a deubiquitination assay with factors in the intrinsic apoptosis pathway was performed [58]. It was found that PIP1 protein was accumulated by USP2a and USP2c [57]. An increase in the level of RIP1 can induce apoptosis in MCF7 cells [57]. The ubiquitin ligase TRAF2 plays a role in attaching K63 polyubiquitin chain to RIP1 through the activation of the transcription factor NF-κB, which is inducible by TNF. On the other hand, TRAF2 itself is also ubiquitinated at K48 and K63, and conjugation with the K63 ubiquitin chain is required for RIP1 activity. The K48 polyubiquitin chain of TRAF2 is removed by USP2a and USP2c, but the K63 chain was not [57]. Downregulation of USP2a through siRNA was confirmed to inhibit TNF-induced apoptosis [57].
USP4
Ubiquitin-specific protease 4 (USP4) contains domains including USP (DUSP), ubiquitin-like (UBL), UCHD1, and UCDH2. The DUSP and UBL domains of USP4 and USP15 share high homology [59]. It was found that the catalytic efficiency of the enzyme activity of USP4 is related to the DUSP-UBL domain [60]. Tumor suppressor protein retinoblastoma protein (pRb) is the well-known substrate of USP4 [61]. In head and neck squamous cell carcinoma (HNSCC), USP4 is upregulated and causes TNF-α-induced apoptosis through RIP1 targeting deubiquitination [62]. This finding indicates that the USP4 has a tumor suppressor role [62].
USP7
Ubiquitin-specific protease 7 (USP7) appears to regulate the kinetics of p53 and MDM2 pathway by p53 and its E3 ligase MDM2. Other tumor-related factors such as FOXO, phosphatase and tensin homolog (PTEN), and cis pin are consequently involved in cell cycle control, DNA damage response, and apoptosis. Consistently, abnormal expression and activity of USP7 have both been linked to numerous cancer types and it is considered as powerful cancer treatment targets [63, 64]. In a previous study, the decreased expressions of USP7 or p53 mutation was confirmed in 71% of the 131 patients identified with non-small-cell lung carcinoma (NSCLC); in particular, regulation of the p53 pathway by HAUSP expression in adenocarcinoma of NSCLC cancer plays an important role in cancer prognosis [65]. It was confirmed that the mRNA levels of apoptotic proteins (such as p21 and Bax) were lower in patients with reduced USP7 expression or p53 mutation, than levels obtained in the control group [65]. Regulation of p53 expression through USP7 has been identified as a treatment method for NSCLC [65]. In the absence of DNA damage, both upregulation and downregulation of USP7 in the human colon cancer cell xenograft model stabilizes the level of p53, induces apoptosis, and inhibits tumor growth [66].
USP9X
Ubiquitin-specific protease 9X (USP9X) plays a role in both cancer cell proliferation and death, depending on the tumor type. It has been shown that in colon cancer, USP9X acts as an indirect regulator that directly regulates the protein stability of FBW7, leading to cancer cell death [67]. In addition, USP9X showed high mutation levels in pancreatic ductal adenocarcinoma (PDA), thereby confirming that conditional deletion of USP9X interacts with KrasG12D to accelerate PDA tumorigenesis [68]. USP9X is a tumor suppressor gene with prognostic and therapeutic relevance in PDA [68]. Myeloid cell leukemia-1 (MCL1) is a pro-survival Bcl-2 family member that promotes cell survival [69]. High levels of MCL1 expression are confirmed in B- and mantle-cell lymphomas [70]. It acts as a factor in chemical resistance and disease recurrence. USP9X cleaves the K48 polyubiquitin chain of MCL1 and regulates the protein stability of MCL1 by regulating proteasomal degradation [70]. Patients with multiple myeloma presenting with high expression of USP9X have a poor prognosis, and knockdown of USP9X enhances ubiquitination of MCL1 to enhance apoptosis [70].
USP10
Ubiquitin-specific protease 10 (USP10) was found to regulate PTEN in lung and breast cancer cell lines [71, 72]. It was confirmed that the expression of USP10 in lung cancer tissue was lower than in normal cells, and knockdown of USP10 increased the tumor growth or invasion in mice [71]. PTEN and USP10 interact directly, and the metastatic effect of USP10 knockdown was negated by PTEN transfection [71]. p53 is known to be an important tumor suppressor [73]. A mutation or downregulation of p53 occurs in about 50% human cancers [74]. In a previous study, it has been demonstrated that miRNA can negatively regulate protein expression through direct labeling, or positively regulate cellular functions through inhibition of negative protein regulators [75]. USP10 has DUB activity against p53 [76]. A recent paper reported that miRNA-138 inhibits USP10 expression by directly binding to 3'-UTR of USP10 mRNA, a positive regulator of p53 [73]. This in turn downregulates the protein level of p53, resulting in decreased apoptosis and defects in cell cycle arrest [73].
USP14
IU1, a small molecule inhibitor of USP14, is known to increase apoptosis [77]. Treatment with a DUB inhibitor of USP14 and UCHL5 b-AP15 inhibits the DUB activities, mediates cell apoptosis, and decreases migration in GCB- and ABC-DLBCL, by activating the caspase and mitochondrial mediated apoptosis [78].
USP24
In U2OS cells, overexpression of ubiquitin-specific protease 24 (USP24) causes cell death and increases caspase-3 cleavage [79]. USP24 stabilizes the protein level of Bax by binding, followed by deubiquitination. In addition, yeast two-hybrid screening identified a protein that can interact with USP24, and a nonhomologous end joining factor, Ku70, was selected as a protein binding partner for USP24. USP24 is known to be involved in DNA repair, telomere maintenance, and V(D)J recombination [79]. In a previous study, it was also demonstrated that USP24 stabilizes the p300 protein, induces the acetylation of ku70, and reduces the interaction between ku70 and Bax [79].
USP28
Ubiquitin-specific protease 28 (USP28) is required for c-Myc stability in human tumor cells. USP28 binds to c-Myc through interaction with the F-box protein FBW7α, which is a part of the SCF type ubiquitin ligase. Therefore, the stabilizing c-Myc exists in the nucleus, and not in the nucleolus, which is decomposed by FBW7γ. High expression levels of USP28 are essential for colon and breast cancers, and stabilization of c-Myc by USP28 is essential for tumor cell proliferation [80]. [1–3]triazolo[4,5-d]pyrimidine, one of inhibitors for USP28, downregulates cancer cell proliferation in gastric cancer cell line GES-1 [81], and USP28 is also highly expressed in diverse cancers; bladder cancer [82], colorectal cancer [83], and non-small cell lung cancer [84].
USP30
Parkin levels are limited in most cells, and when high levels of heterozygous parkin are overexpressed, a representative mass clearance of mitochondria is observed. The researchers demonstrated that high levels of Parkin resulted in sensitization of hTERT-RPE1 cells to CCCP-induced, PINK1-dependent cell death. They showed that a unique MOM DUB, the ubiquitin-specific protease 30 (USP30), directly interferes with this expression of parkin activity. Importantly, data suggest that USP30 is associated with the apoptosis pathway, and its depletion also sensitizes cancer cells to cell death induced by the BH3 mimic [85]. USP30 inhibitors MF-094, MF-095, and FT385 can therefore be used in the treatment of Parkinson’s disease by inducing apoptosis [86, 87].
USP33
In a previous study, the clinical significance of ubiquitin-specific protease 33 (USP33) expression in tumor tissues of papillary thyroid carcinoma (PTC) patients was investigated. It was confirmed that the knockdown of USP33 in the PTC cell lines (TPC-1 and BCPAP) increases the cell viability and invasion ability [88]. USP33 interacts with Robo1 (Roundabout homolog 1), a major receptor for Slit that normally acts as a tumor suppressor in PTC cells [88]. The apoptotic effect on the PTC cells is confirmed by the interaction between Robo1 and USP33 [88].
DUBs for anti-apoptotic regulation
DUB has been reported to reduce apoptosis by inhibiting proteasomal degradation of anti-apoptotic proteins through deubiquitination. This process can be used to control excessive killing of normal cells. Conversely, E3 ligase, which increases the ubiquitination of anti-apoptotic proteins, may promote the degradation of anti-apoptotic proteins [89].
A20
A20 (also known as TNFAIP3) is a member of the OTU family, and contains an N-terminal OTU domain and zinc-finger motifs on the C-terminal region [90]. A20 is known to deconjugate K11, K48, and K63 polyubiquitin chains in vitro, but it was confirmed that only the K63 polyubiquitin chain is attached to the NF-κB protein [91]. In vitro and in vivo experimental data show that loss of A20 induces PIPK1 kinase-dependent and independent cell death upon single TNF stimulation [92]. In the absence of linear ubiquitination, A20 is recruited to a complex via the ZF4 and ZF7 domains, but in this scenario, it protects against cell death via the DUB activity [92].
FAM188B
Anoikis is a type of cell death induced by cell detachment. Family with sequence similarity 188 member B (FAM188B) is expected to be a member of the new DUB, but its function remains to be studied. It has been shown that knockdown of FAM188B increases anoikis in A549 and H1299 cell lines expressing WT-EGFR, and in H1975 cell line expressing TKI-resistant EGFR mutations [93]. FAM188B induces tumor proliferation by maintaining EGFR and carcinogenic protein activity levels [93].
JOSD1
Myeloid cell leukemia 1 (MCL1) is an important anti-apoptotic member of the Bcl-2 family. S63845, is a known inhibitor of MCL1, effective in hematologic cancer but not in solid cancer [94–97]. According to a recent study, Josephin domain containing 1 (JOSD1) was found to be one of highly expressed DUBs in cervical cancer [97]. Moreover, depletion of JOSD1 is reported to result in the death of gynecological cancer cell lines [97]. MCL1 has a short half-life and is sensitive to changes in protein synthesis or degradation. Therefore, the regulation of MCL1 is an important factor in determining apoptosis. USP9X, USP13, USP24, DUB3, JOSD1 and Ku70 reverses MCL1 K48-linked ubiquitination and prevents the proteasomal degradation. This leads to an increase in the half-life of MCL1 resulting in anti-apoptosis [98].
OTUB1
In a previous study, the expression of apoptosis-related proteins after OTUB1 silencing was investigated to explore the mechanism of OTUB1-mediated growth of HCC cells. Results revealed that compared to the control, decrease in the expression of OTUB1 by shOTUB1 in HepG2 and LM3 cell lines resulted in increased expressions of the apoptotic proteins caspase-3, caspase-9, and Bax, and decreased expression of the anti-apoptotic protein Bcl-2. These results indicate that OTUB1-silencing enhances apoptosis in HCC cells [99].
PPPDE1
The PPPDE family members are a newly discovered type of DUB, but their functions are not yet to be elucidated. It has been demonstrated that PPPDE1 is a DUB of ribosomal protein S7 and that it regulates the activity of ubiquitin via K48 and K63 binding [100]. PPPDE1 is a critical regulator of p53 protein and its downstream apoptosis pathway [101]. The knockdown of PPPDE1 results in reduced tumorigenesis in hepatocellular carcinoma cells [101].
UCHL5/UCH37
In the ABC- and GCB-subtypes of diffuse large B-cell lymphoma (DLBCL), the b-AP15 downregulates migration and induces apoptosis by regulating UCHL5 and USP14 [78]. In multiple myeloma (MM), UCHL5 and USP14 are highly expressed, compared to normal cells. b-AP15 also mediates the cell viability in MM cells by regulating UCHL5 and USP14, without inhibition of proteasome activity [102]. In the enterochromaffin (EC) cell line, the expression of UCHL5 is high, and one study reported that the expression of UCHL5 decreases the survival rate [103]. On the other hand, overexpression of UCHL5 promotes the cell cycle and proliferation, and reduces apoptosis in EC cells [103].
USP1
Ubiquitin-specific protease 1 (USP1) deubiquitinates the K48-linked polyubiquitin chain of histone demethylase lysine-specific demethylase 4 a (KDM4A), which is upregulated in prostate cancer (PC) [104]. The protein expression of KDM4A is stabilized by USP1 in vitro and in vivo [104]. Inhibition of USP1 by ML323 mediates reduction of cell proliferation in PC. ML323 is potentially a new treatment method for patients who are resistant to existing PC anticancer drugs [104]. USP1 promotes stem cell maintenance and radiation resistance in glioblastoma multiforme (GBM) tumors via stabilization of inhibitor of DNA binding 1 (ID1) and checkpoint kinase 1 (CHEK1) [105]. Targeting and lowering the level of USP1 may therefore reduce the survival of GBM, making it an effective treatment for GBM [105]. In addition, inhibitors of USP1 (including pimozide) suppress the DUB activity of USP1 and promote the degradation of ID1 [106]. These small molecules are known to induce apoptosis in acute myeloid leukemia (AML) and K562 [106].
USP2
Ubiquitin-specific protease 2 (USP2) is a multifunctional DUB. USP2 regulates cell cycle progression, and thus carcinogenesis through deubiquitination of cyclins and Aurora-A. Other tumorigenic molecules, including epidermal growth factor and fatty acid synthase, are also targets for USP2. USP2 further prevents the p53 signal [107]. Downregulation of USP2 in the triple negative breast cancer (TNBC) cell line causes apoptosis by regulating FAS and Cyclin D1. Treatment with the USP2 inhibitor ML364 results in apoptosis in the TNBC cell line [108], and downregulation of cyclin D1 by ML364 blocks the G0/G1 phase and disrupts the cell cycle progression in HCT116 cell and mino cell lines [109].
USP4
Ubiquitin-specific protease 4 (USP4) deubiquitinates and stabilizes TβRI (which plays a tumor-promoting role in liver cancer), and subsequently activates the TβRI/pSmad2 signaling pathway (which potentiates cell migration and invasion capacity) in vivo and in vitro [110]. Using quantitative proteomics analysis, cyclophilin A (CypA) was selected as a second potential target of USP4 in liver cancer, which was further confirmed by Co-IP analysis. CypA has been demonstrated to mediate malignant biological behaviors such as cell proliferation and metastasis by USP4 expression [111]. Similar to breast cancer, USP4 negatively regulates miR-148a in liver cancer, suggesting a potential therapeutic role for miR-148a in liver cancer [112]. High expression of USP4 is associated with poor clinical outcomes in lung cancer patients. USP4 is a novel deubiquitinating enzyme for Twist Family BHLH Transcription Factor 1 (Twist 1) that stabilizes Twist 1 and mediates the growth of lung cancer stem cells [113].
USP5
Activity of ubiquitin-specific protease 5 (USP5) is suppressed by the BRAF inhibitor (vemurafenib) in sensitive, but not in acquired or intrinsically resistant cells. USP5 knockdown overcomes acquired vemurafenib resistance and sensitizes BRAF and NRAS mutant melanoma cells to apoptosis initiated by MEK inhibitors, cytokines, or DNA-damaging agents. Knockdown and overexpression studies have demonstrated that USP5 regulates p53 levels and alters cell growth and cell cycle distribution associated with p21 induction. USP5 also regulates the intrinsic apoptotic pathway by modulating the p53-dependent FAS expression [114, 115]. USP5 is overexpressed in colorectal cancer tissue and promotes colorectal cancer cell proliferation and resistance to chemotherapy [116]. USP5 is known to promote tumorigenesis and drug resistance by inhibiting p14ARF-p53 signaling in hepatocellular carcinoma [117], and it induces epithelial–mesenchymal transition (EMT) in hepatocellular carcinoma cells by targeting SLUG, a known transcription factor [118]. A previous study confirmed that β-catenin signaling activation, which plays an important role in EMT, is inhibited in USP5 knockdown NSCLC cells [119]. It has been shown that the mRNA and protein levels of USP5 increased in clinical samples of liver cancer and HCC cells compared to the control [120]. Interestingly, MDM2 is downregulated by siUSP5, and p14ARF-p53 signaling was activated to inhibit cell proliferation and lead to apoptosis [120]. On the other hand, USP5 overexpression suppressed the expression of p14ARF and p53, promoted MDM2 expression, and enhanced cell proliferation [120].
USP7
USP7 is a known DUB of the tumor suppressor protein PTEN [121]. However, the tumor suppressor function of PTEN is achieved by the nuclear location of PTEN [121]. Deubiquitination of specific lysine of PTEN regulates the intracellular localization of PTEN protein, and deubiquitination of PTEN through USP7 controls the localization in the cytoplasm rather than the nucleus, which may inhibit the tumor suppression of PTEN [121]. In cervical cancer, USP7 mediates the binding and interaction of MRE11-RAD50-NBS1 (MRN) complex with DNA damage checkpoint protein 1 (MDC1) through deubiquitination of MDC1, and this process mediates DNA damage response (DDR) [122]. siUSP7 consequently inhibits the binding of MRN complex with MDC1, and it impairs the recruitment of p53 binding protein 1 (53BP1) and breast cancer protein 1 (BRCA1) in HeLa and MCF7 cells [122]. It is of interest that the high level of USP7 has been found in cervical cancer patients [122].
USP8
Ubiquitin-specific protease 8 (USP8) knockdown inhibits lung cancer cell proliferation and promotes apoptosis [123]. It may reduce the expression of RTK, thereby reducing the viability of gefitinib-resistant and sensitive NSCLC cells [123]. Knockdown of USP8 downregulated the expression of p-AKT, indicating that USP8 knockdown inhibits cell proliferation by inhibiting the PI3K/AKT pathway [123]. USP8 directly deubiquitinates FLIP, improves protein stability, and inhibits DR-induced apoptosis [124].
USP9X
Ubiquitin-specific protease 9X (USP9X) preserves the MCL1 expression by removing the polyubiquitin chain [125]. Inhibition of USP9X through WP1130 causes apoptosis in non-small lung cancer cells [126]. WP1130 enhances TRAIL-induced apoptosis through USP9X-dependent miR-708-mediated downregulation of c-FLIP [127]. Inhibition of the DUB USP9X induces pre-B cell homeobox 1 (PBX1) degradation, thereby stimulating prostate cancer cell apoptosis [128]. In breast cancer cells such as MCF7 and T47D, USP9X is shown to be a DUB for BRCA1, that plays an important role in DNA double-strand break repair [129]. Depletion of USP9X levels decreases the protein levels and half-life, along with increased ubiquitination of BRCA1 [129]. Knockdown of USP9X significantly reduces the homologous recombination (HR) efficiency. Taken together, these results indicate that USP9X plays a role in HR repair, and modulates the sensitivity of cancer cells to DNA damaging factors [129].
USP10
Analysis of USP10 expression revealed significantly higher levels in breast cancer patients, which is correlated with tumor progression and lower overall survival rates [72]. Interestingly, upregulation of USP10 protein level and additional depletion of USP10 in PI3K inhibitor-resistant breast cancer cells, when compared to the parental control group, resensitized these cells to PI3K inhibitors [72]. In addition, a patient-derived xenograft (PDX) model of breast cancer patients performed after administration of PI3K inhibitors showed an amount correlation between USP10 and PTEN protein levels [72]. The mRNA expression of USP10 is upregulated in endometriosis patients, and USP10 increases the protein stability of Raf-1 through deubiquitination to Raf-1 [130]. It induces the migration and proliferation of ectopic endometrial stromal cells and reduces apoptosis by mediating the activation of Raf-1/MEK/ERK signaling pathway [130]. The reversed result was confirmed by downregulation of USP10 through siUSP10 [130].
USP11
Ubiquitin-specific protease 11 (USP11) regulates the ubiquitination of cellular IAP2 (cIAP2), and increased levels of cIAP2 cause apoptosis in UACC-62 cells [131]. In a previous study, cIAP2, which protects cancer cells from apoptosis, is stabilized by USP11 and inhibits apoptosis [132]. It was confirmed that cIAP1 is expressed at high levels in almost all carcinomas; SMAC-mimicking treatment results in rapid reduction in the expression and function of cIAP1. Conversely, low expression levels of cIAP2 show lesser alterations in the expression after SMAC-mimicking treatment [132]. Knockdown of cIAP2 further allows cancer cells to reach apoptosis, and depletion of USP11 could decrease the expression levels of cIAP2 without affecting the expression levels of other IAPs such as cIAP1, XIAP, and ML-IAP [132].
USP13
MCL1 is an anti-apoptotic Bcl-family protein. Ubiquitin-specific protease 13 (USP13) regulates MCL1 stability in lung and ovarian cancer cells. Using the CRISPR/Cas9 system, it was confirmed that tumor growth was suppressed in USP13 KO nude mice [125]. Spautin-1 acts as an inhibitor of USP13 in MDCK and HeLa cells [133, 134].
USP14
The expression of USP14 is known to be associated with p53 deficiency diseases [135], colorectal cancer [136], intrahepatic bile duct cancer [137], lung cancer [138, 139], and ovarian cancer [140]. According to a previous study, the dysregulation of USP14 expression leads to cancer cell death by altering the expression level of anti-apoptotic protein Bcl-xl in epithelial ovarian cancer (EOC) cells. When the level of USP14 was reduced by shUSP14, the level of Bcl-xl protein also decreased, whereas high expression of USP14 was pathologically associated with poor prognosis in ovarian cancer patients [140]. In addition, knockdown of USP14 in SKOV3 cells contributed to the inhibition of cell proliferation [140]. Aurora-B is ubiquitinated and degraded during cell apoptosis induced by chemotherapeutic drugs for leukemia [141]. FBXW7 mediates Aurora-B ubiquitination breakdown during apoptosis induced by chemotherapy drugs [141]. Meanwhile, USP14 prevents protein degradation by deubiquitination of Aurora-B and inhibits chemotherapeutic drug apoptosis in leukemic cells [141]. Conversely, administration of b-AP15, an inhibitor of USP14, significantly increases the death rate of leukemic cells, in a dose-dependent manner [141]. Knockdown of USP14 in metastatic melanoma patients reduces the viability of melanoma cells [142]. It has been confirmed that inhibition of USP14 rapidly triggers ROS generation leading to apoptosis, including accumulation of polyubiquitinated proteins and chaperones, and mitochondrial dysfunction [142]. Moreover, in the lung carcinoma cell line A549, treatment with siRNA or DUB inhibitor IU1-47 of USP14 significantly reduces the proliferation rate and induces cell cycle arrest [143].
USP15
Ubiquitin-specific protease 15 (USP15) is an up-regulator of apoptosis in degenerative nucleus pulposus (NP) cells. Expression of USP15 decreases the phosphorylation of AKT, and upregulation of USP15 promotes cell apoptosis [144]. A previous study demonstrated that USP15 is a component of FKBP5/AKT signaling in NP cells [144]. It is well known that the mRNA levels of USP15 are upregulated in patients with MM [145]. Silencing of USP15 induces MM cell antiproliferative cell death as well as inhibits the expression of NF-κBp65 in the nucleus and cytoplasm, whereas overexpression of USP15 exerts an adverse effect [145]. Moreover, in vivo experiments have shown that silencing of USP15 inhibits MM tumor growth and NF-κBp65 expression [145]. Overexpression of USP15 promotes NF-κBp65 expression through inhibition of ubiquitination [145]. USP15 mediates enhancement of the TGF-β pathway by deubiquitination of the TGF-β receptor (TβR-I) in carcinomas such as glioblastoma, breast cancer, and ovarian cancer [146]. Higher expression of USP15 is obtained in these carcinomas [146]. In addition, in the mouse model derived from glioblastoma patients, downregulation of USP15 reduces the expression level of TGF-β and this depletion reduces the carcinogenic potential of cells [146].
USP18
Cervical cancer is a disease that causes cancer at the entrance of the uterus and is the second largest female cancer in the world after breast cancer [147]. In cervical cancer cell progression, the activation level of PI3K/AKT signaling is an important factor. A previous study reported that overexpression of ubiquitin-specific protease 18 (USP18) inhibits apoptosis and promotes cell proliferation in cervical cancer cell lines (Caski and SiHa) by regulating AKT phosphorylation [148], while USP18 silencing by siUSP18 mediates apoptosis in cervical cancer cell lines (Caski and SiHa) [148]. The expression of USP18 is upregulated in HCC [149]. Knockdown of USP18 inhibits HCC cell growth and induces cell cycle arrest and premature apoptosis [149]. And this regulation is through a component of the p53 apoptotic pathway and member of anti-apoptotic Bcl-2 family, BCL2L1 [149]. It was confirmed that BCL2L1 decreased with the decrease of USP18 [149].
USP21
Ubiquitin-specific protease 21 (USP21) is reported to deubiquitinate the H2A protein and nonhistone proteins such as the GLI family zinc finger 1 (GLI1) [150] and retinoic-acid-inducible-gene I (RIG-I) [151]. In previous studies, USP21 was determined to mediate cancer proliferation. USP21 is highly expressed in bladder cancer (BC) [152], and it has been confirmed that the high expression of USP21 is closely related to tumor size, epithelial-middle lobe metastasis (EMT), and poor prognosis [152]. USP21 directly regulates the protein level of enhancer of zeste 2 polycomb repressive complex 2 subunit (EZH2), which is known to be one of the cancer-inducing factors, and is highly expressed in numerous cancers [152]. The down-regulation of USP21 by siRNA inhibits the invasion and proliferation in renal cell carcinomas (RCC) [153]. USP21 has the potential to be an RCC therapeutic target associated with inhibition of interleukin 8 (IL-8) [153]. It has been reported that USP21 deubiquitinates Fos-related antigen 1 (FRA1), increasing the FRA-1 stability and improving the expression of FRA-1 target genes in colon cancer cells [154]. USP21 improves FRA-1 stability and AP-1 target gene expression by deubiquitinating Fra-1. Considering all the above, USP21 can be an attractive treatment target in metastatic colorectal carcinomas (mCRCs) with high Fra-1 expression [154].
USP22
Ubiquitin-specific protease 22 (USP22) is a protein that functions to induce apoptosis and plays an oncogenic role. In gastric cancer, the mRNA and protein expression levels of USP22 are high, and knockdown of USP22 increases cell viability and proliferation in vitro [155]. In addition, it has been determined that the USP22 activating c-Myc/NAMPT/SIRT adjusts the FOXO1 and YAP signaling pathways. [155]. In pancreatic ductal adenocarcinoma cells, USP22 reduces cell apoptosis by regulation of DYRK1A, and knock-down of USP22 shows opposing outcomes [156]. USP22 overexpression induces enhanced resistance to apoptosis and treatment resistance in multiple cancer cell lines [157, 158]. It has been demonstrated that knockdown of USP22 using miRNA inhibits the growth of colorectal cancer [159].
USP51
Zinc‑finger E‑box binding homeobox 1 (ZEB1) contains two zinc finger clusters in the N-terminal and C-terminal regions, which bind to the E-Box sequence (CACCT) or similar sequence (CACCG) to regulate downstream target gene expressions. ZEB1 promotes tumor cell metastasis, invasion, and resistance to treatment. ZEB1 expression is associated with treatment resistance in several cancers, and inhibition of ZEB1 has been shown to reverse chemical resistance in docetaxel-resistant human lung cancer cells [160]. Overexpression of USP51 causes ubiquitination of ZEB1 in MM cells and lowers the protein stability of ZEB1 through proteasomal degradation. Downstream ZEB1 in MM cells results in apoptosis and decreased cell proliferation [160]. It has also been demonstrated that knockdown of ZEB1 reduces cell proliferation and causes apoptosis in metanephric MM cells and mK3 cells [161]. Overexpression of ZEB1 mediates the proliferation and migration and reduces the cell apoptosis in MM cells. ZEB1 regulates cell proliferation and apoptosis with Six2 in MM cell lines [161].
Small molecular inhibitors for DUBs
As mentioned in previous sections, regulation of DUBs is important in the death of cancer cells. In fact, numerous studies have demonstrated the effectiveness of DUB inhibitors in regulating apoptosis (Table 1) [77, 78, 81, 86, 108, 109, 126, 127, 143, 162–173]. These inhibitors are newly made through continuous research, and further studies are required to prove their effects in vivo and in vitro, for further application in clinical trials. The well-known DUB inhibitor b-AP15 blocks the DUB activity by regulating 19S regulatory particles (19S RP) [163, 174]. b-AP15 is known as an inhibitor of UCHL5 and USP14 [78, 163]. Proteolysis-targeting chimera (PROTAC), which was first designed in 2001, is a new drug development technology with a concept of inducing proteasomal degradation by inducing protein ubiquitination with a compound that connects E3 ligase and target protein using a linker connecting the E3 ligase ligand module and the target protein ligand-module [175]. In 2020, Arvinas, the first PROTAC company established across the globe, announced the result of a phase 1 clinical trial showing that PROTAC had an actual therapeutic effect. PROTACs that are used in clinical trials are ARV-471 and ARV-110 from Arvinas. A number of multinational pharmaceutical companies are preparing for clinical trials and focusing on diverse researches, hence making this a trendy and powerful new drug development platform. Recently, various developments (including folate-caged PROTAC [176], p-PROTAC [177], and antibody-PROTAC) [178] have been developed. DUBTAC, which includes ligands that can link DUB and target proteins, has also been reported [179]. In future DUB research, methods to control the degradation of target proteins by grafting DUB to new technologies, including DUBTAC, need to be studied.
Table 1.
Small molecules for DUBs that regulate pro-apoptotic and anti-apoptotic pathways
| Small molecules | Proteins | References |
|---|---|---|
| [1,2,3]triazolo[4,5-d]pyrimidine | USP28 | [81] |
| AZ1 | USP25 | [162] |
| USP28 | ||
| b-AP15 | UCHL5/UCHL5 | [78, 163] |
| USP14 | ||
| DUBs-IN-2 | USP8 | [164] |
| FT385 | USP30 | [86] |
| IU1 | USP14 | [77] |
| IU1-47 | USP14 | [143] |
| LDN57444 | UCHL1 | [165] |
| ML324 | USP1/UAF1 | [173] |
| ML364 | USP2 | [108, 109] |
| MF-094 | USP30 | [86] |
| MF-095 | USP30 | [86] |
| NCI677397 | USP24 | [166] |
| NSC632839 | USP2 | [168] |
| USP7 | ||
| P22077 | USP7 | [167] |
| USP47 | ||
| Spautin-1 | USP13 | [169] |
| Subquinocin | CYLD | [170] |
| Vialinin A | USP4 | [171, 172] |
| USP5 | ||
| WP1130 | USP9X | [126, 127, 166] |
| USP24 |
Conclusion
Apoptosis is an indispensable process from the viewpoint of cell growth, development, and life maintenance, and when a problem occurs in this system, diseases inevitably occur in the human body. These diseases include representative diseases among human death factors. The UPS is known to be a very important process that regulates the breakdown of proteins (Table 2). Controlling the degree of ubiquitination of apoptotic proteins may be effective in the treatment of diseases related to apoptosis control due to failure to quantitatively control apoptosis proteins. Regulation of the expression of apoptotic proteins has been attempted in many papers and studies. More care should be taken to control cell apoptosis by proteins that are involved in both pro-apoptosis and anti-apoptosis processes, such as OTUB1, UCHL1, UCHL5/UCH37, USP2, USP4, USP7, USP9X, USP10, and USP14. This is because regulation of these protein levels can trigger or inhibit cell death, and hence results may vary from cell lines. However, since application of these proteins can affect both pro-apoptosis and anti-apoptosis, and different results can be obtained depending on whether the protein is overexpressed or knocked down, it can be difficult and highly useful at the same time. Since the action of these proteins for each type of cells and tissues is continuously required, it is necessary to pay attention to setting them as therapeutic targets. It has been suggested that the action of DUBs with ‘double roles’ can be classified by cancer types. Therefore, the direction of local cancer treatment can be attempted either in perturbing or activating the normal cellular functions. In addition, modulating anti-apoptotic proteins by regulating protein expression using E3 ligases and DUBs can be used in disease treatment as a way to induce or inhibit apoptosis. In addition, the recently developed PROTAC technology can be applied to degrade or increase the expression of a target protein using the ubiquitin–proteasome system, proposing great potential as a therapeutic tool for diverse cancers and diseases.
Table 2.
Pro-apoptotic vs anti-apoptotic roles of DUBs
| DUBs | Roles | References |
|---|---|---|
| A20 | Anti-apoptotic | [90–92] |
| CYLD | Pro-apoptotic | [34, 36–40] |
| FAM188B | Anti-apoptotic | [93] |
| JOSD1 | Anti-apoptotic | [94, 97, 98] |
| OTUB1 | Pro-apoptotic | [49, 50] |
| Anti-apoptotic | [99] | |
| PPPDE1 | Anti-apoptotic | [100, 101] |
| UCHL1 | Pro-apoptotic | [52, 53] |
| Anti-apoptotic | [180, 181] | |
| UCHL5/UCH37 | Pro-apoptotic | [54, 55] |
| Anti-apoptotic | [78, 103] | |
| USP1 | Anti-apoptotic | [105, 106] |
| USP2 | Pro-apoptotic | [56–58] |
| Anti-apoptotic | [107–109] | |
| USP4 | Pro-apoptotic | [61, 62] |
| Anti-apoptotic | [111–113] | |
| USP5 | Anti-apoptotic | [114–119] |
| USP7 | Pro-apoptotic | [65, 66] |
| Anti-apoptotic | [121, 122] | |
| USP8 | Anti-apoptotic | [123, 124] |
| USP9X | Pro-apoptotic | [67–70] |
| Anti-apoptotic | [127–129] | |
| USP10 | Pro-apoptotic | [71, 73, 76] |
| Anti-apoptotic | [72, 130] | |
| USP11 | Anti-apoptotic | [131, 132] |
| USP13 | Anti-apoptotic | [125] |
| USP14 | Pro-apoptotic | [78] |
| Anti-apoptotic | [140] | |
| USP15 | Anti-apoptotic | [144–146] |
| USP18 | Anti-apoptotic | [148, 149] |
| USP21 | Anti-apoptotic | [152–154] |
| USP22 | Anti-apoptotic | [155–159] |
| USP24 | Pro-apoptotic | [79] |
| USP28 | Pro-apoptotic | [80–84] |
| USP30 | Pro-apoptotic | [85–87] |
| USP33 | Pro-apoptotic | [88] |
| USP51 | Anti-apoptotic | [160, 161] |
Acknowledgements
We would like to thank members of Baek’s laboratory for their comments on the manuscript.
Author contributions
Both HSC and KHB contributed to the writing and design of the article. HSC prepared the figures and tables. KHB obtained the funding.
Funding
This research was supported by Basic Science Program Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2020R1I1A207500311).
Availability of data and materials
As this is a review article, there is no requirement for data availability.
Declarations
Conflict of interest
The authors have no competing interests to declare.
Ethical approval and consent to participate
As this is a review article, no ethical approval was necessary.
Consent for publication
All authors have confirmed consent for publication.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Elmore S. Apoptosis: a review of programmed cell death. Toxicol Pathol. 2007;35:495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Famularo G, De Simone C, Marcellini S. Apoptosis: mechanisms and relation to AIDS. Med Hypotheses. 1997;48:423–429. doi: 10.1016/s0306-9877(97)90041-4. [DOI] [PubMed] [Google Scholar]
- 3.Erekat NS. Apoptosis and its role in Parkinson’s disease. In: Stoker TB, Greenland JC, editors. Parkinson’s disease: pathogenesis and clinical aspects. Brisbane, Australia: Codon Publications; 2018. pp. 65–82. [Google Scholar]
- 4.Ciechanover A. Intracellular protein degradation: From a vague idea thru the lysosome and the ubiquitin-proteasome system and onto human diseases and drug targeting. Best Pract Res Clin Haematol. 2017;30:341–355. doi: 10.1016/j.beha.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 5.Deng L, Meng T, Chen L, Wei W, Wang P. The role of ubiquitination in tumorigenesis and targeted drug discovery. Signal Transduct Target Ther. 2020;5:11. doi: 10.1038/s41392-020-0107-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tait SW, Green DR. Mitochondrial regulation of cell death. Cold Spring Harb Perspect Biol. 2013;5:a008706. doi: 10.1101/cshperspect.a008706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murphy MP. Nitric oxide and cell death. Biochim Biophys Acta. 1999;1411:401–414. doi: 10.1016/s0005-2728(99)00029-8. [DOI] [PubMed] [Google Scholar]
- 8.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 9.Attardi LD, et al. PERP, an apoptosis-associated target of p53, is a novel member of the PMP-22/gas3 family. Genes Dev. 2000;14:704–718. doi: 10.1101/GAD.14.6.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Green DR, Kroemer G. Cytoplasmic functions of the tumour suppressor p53. Nature. 2009;458:1127–1130. doi: 10.1038/nature07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee CL, Blum JM, Kirsch DG. Role of p53 in regulating tissue response to radiation by mechanisms independent of apoptosis. Transl Cancer Res. 2013;2:412–421. doi: 10.3978/j.issn.2218-676X.2013.09.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chipuk JE, Green DR. How do BCL-2 proteins induce mitochondrial outer membrane permeabilization? Trends Cell Biol. 2008;18:157–164. doi: 10.1016/j.tcb.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adrain C, Creagh EM, Martin SJ. Apoptosis-associated release of Smac/DIABLO from mitochondria requires active caspases and is blocked by Bcl-2. EMBO J. 2001;20:6627–6636. doi: 10.1093/emboj/20.23.6627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ceballos-Cancino G, Espinosa M, Maldonado V, Melendez-Zajgla J. Regulation of mitochondrial Smac/DIABLO-selective release by survivin. Oncogene. 2007;26:7569–7575. doi: 10.1038/sj.onc.1210560. [DOI] [PubMed] [Google Scholar]
- 15.Wang C, Youle RJ. The role of mitochondria in apoptosis. Annu Rev Genet. 2009;43:95–118. doi: 10.1146/annurev-genet-102108-134850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cavalcante GC, et al. A cell’s fate: an overview of the molecular biology and genetics of apoptosis. Int J Mol Sci. 2019;20:4133. doi: 10.3390/ijms20174133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ashkenazi A. Targeting the extrinsic apoptosis pathway in cancer. Cytokine Growth Factor Rev. 2008;19:325–331. doi: 10.1016/j.cytogfr.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 18.Villa-Morales M, Fernandez-Piqueras J. Targeting the Fas/FasL signaling pathway in cancer therapy. Expert Opin Ther Targets. 2012;16:85–101. doi: 10.1517/14728222.2011.628937. [DOI] [PubMed] [Google Scholar]
- 19.Li K, et al. The involvement of TNF-alpha and TNF-beta as proinflammatory cytokines in lymphocyte-mediated adaptive immunity of Nile tilapia by initiating apoptosis. Dev Comp Immunol. 2021;115:103884. doi: 10.1016/j.dci.2020.103884. [DOI] [PubMed] [Google Scholar]
- 20.Ashkenazi A, Dixit VM. Death receptors: signaling and modulation. Science. 1998;281:1305–1308. doi: 10.1126/science.281.5381.1305. [DOI] [PubMed] [Google Scholar]
- 21.Papenfuss K, Cordier SM, Walczak H. Death receptors as targets for anti-cancer therapy. J Cell Mol Med. 2008;12:2566–2585. doi: 10.1111/j.1582-4934.2008.00514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kretz AL, et al. TRAILblazing strategies for cancer treatment. Cancers. 2019;11:456. doi: 10.3390/cancers11040456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee EW, Seo J, Jeong M, Lee S, Song J. The roles of FADD in extrinsic apoptosis and necroptosis. BMB Rep. 2012;45:496–508. doi: 10.5483/bmbrep.2012.45.9.186. [DOI] [PubMed] [Google Scholar]
- 24.Beaudouin J, Liesche C, Aschenbrenner S, Horner M, Eils R. Caspase-8 cleaves its substrates from the plasma membrane upon CD95-induced apoptosis. Cell Death Differ. 2013;20:599–610. doi: 10.1038/cdd.2012.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lilienbaum A. Relationship between the proteasomal system and autophagy. Int J Biochem Mol Biol. 2013;4:1–26. [PMC free article] [PubMed] [Google Scholar]
- 26.Flick K, Kaiser P. Protein degradation and the stress response. Semin Cell Dev Biol. 2012;23:515–522. doi: 10.1016/j.semcdb.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heideker J, Wertz IE. DUBs, the regulation of cell identity and disease. Biochem J. 2015;467:191. doi: 10.1042/bj4670191. [DOI] [PubMed] [Google Scholar]
- 28.Garg AV, Ahmed M, Vallejo AN, Ma A, Gaffen SL. The deubiquitinase A20 mediates feedback inhibition of interleukin-17 receptor signaling. Sci Signal. 2013;6:ra44. doi: 10.1126/scisignal.2003699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, et al. Deubiquitinating enzymes regulate PARK2-mediated mitophagy. Autophagy. 2015;11:595–606. doi: 10.1080/15548627.2015.1034408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park HB, Kim JW, Baek KH. Regulation of Wnt signaling through ubiquitination and deubiquitination in cancers. Int J Mol Sci. 2020;21:3904. doi: 10.3390/ijms21113904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He M, Zhou Z, Wu G, Chen Q, Wan Y. Emerging role of DUBs in tumor metastasis and apoptosis: Therapeutic implication. Pharmacol Ther. 2017;177:96–107. doi: 10.1016/j.pharmthera.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bader M, Steller H. Regulation of cell death by the ubiquitin-proteasome system. Curr Opin Cell Biol. 2009;21:878–884. doi: 10.1016/j.ceb.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun SC. CYLD: a tumor suppressor deubiquitinase regulating NF-kappaB activation and diverse biological processes. Cell Death Differ. 2010;17:25–34. doi: 10.1038/cdd.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harhaj EW, Dixit VM. Regulation of NF-kappaB by deubiquitinases. Immunol Rev. 2012;246:107–124. doi: 10.1111/j.1600-065X.2012.01100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saito K, et al. The CAP-Gly domain of CYLD associates with the proline-rich sequence in NEMO/IKKgamma. Structure. 2004;12:1719–1728. doi: 10.1016/j.str.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 36.Ritorto MS, et al. Screening of DUB activity and specificity by MALDI-TOF mass spectrometry. Nat Commun. 2014;5:4763. doi: 10.1038/ncomms5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Komander D, et al. The structure of the CYLD USP domain explains its specificity for Lys63-linked polyubiquitin and reveals a B box module. Mol Cell. 2008;29:451–464. doi: 10.1016/j.molcel.2007.12.018. [DOI] [PubMed] [Google Scholar]
- 38.Komander D, et al. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep. 2009;10:466–473. doi: 10.1038/embor.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sato Y, et al. Structures of CYLD USP with Met1- or Lys63-linked diubiquitin reveal mechanisms for dual specificity. Nat Struct Mol Biol. 2015;22:222–229. doi: 10.1038/nsmb.2970. [DOI] [PubMed] [Google Scholar]
- 40.Lork M, Verhelst K, Beyaert R. CYLD, A20 and OTULIN deubiquitinases in NF-kappaB signaling and cell death: so similar, yet so different. Cell Death Differ. 2017;24:1172–1183. doi: 10.1038/cdd.2017.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ohtake F, Saeki Y, Ishido S, Kanno J, Tanaka K. The K48–K63 branched ubiquitin chain regulates NF-kappaB signaling. Mol Cell. 2016;64:251–266. doi: 10.1016/j.molcel.2016.09.014. [DOI] [PubMed] [Google Scholar]
- 42.Brummelkamp TR, Nijman SM, Dirac AM, Bernards R. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-kappaB. Nature. 2003;424:797–801. doi: 10.1038/nature01811. [DOI] [PubMed] [Google Scholar]
- 43.Kovalenko A, et al. The tumour suppressor CYLD negatively regulates NF-kappaB signalling by deubiquitination. Nature. 2003;424:801–805. doi: 10.1038/nature01802. [DOI] [PubMed] [Google Scholar]
- 44.Trompouki E, et al. CYLD is a deubiquitinating enzyme that negatively regulates NF-kappaB activation by TNFR family members. Nature. 2003;424:793–796. doi: 10.1038/nature01803. [DOI] [PubMed] [Google Scholar]
- 45.Fernandez-Majada V, et al. The tumour suppressor CYLD regulates the p53 DNA damage response. Nat Commun. 2016;7:12508. doi: 10.1038/ncomms12508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Massoumi R, Chmielarska K, Hennecke K, Pfeifer A, Fassler R. Cyld inhibits tumor cell proliferation by blocking Bcl-3-dependent NF-kappaB signaling. Cell. 2006;125:665–677. doi: 10.1016/j.cell.2006.03.041. [DOI] [PubMed] [Google Scholar]
- 47.Sparks A, et al. The degradation of p53 and its major E3 ligase Mdm2 is differentially dependent on the proteasomal ubiquitin receptor S5a. Oncogene. 2014;33:4685–4696. doi: 10.1038/onc.2013.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun XX, Challagundla KB, Dai MS. Positive regulation of p53 stability and activity by the deubiquitinating enzyme Otubain 1. EMBO J. 2012;31:576–592. doi: 10.1038/emboj.2011.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen Y, Wang YG, Li Y, Sun XX, Dai MS. Otub1 stabilizes MDMX and promotes its proapoptotic function at the mitochondria. Oncotarget. 2017;8:11053–11062. doi: 10.18632/oncotarget.14278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pierdominici M, et al. Role of autophagy in immunity and autoimmunity, with a special focus on systemic lupus erythematosus. FASEB J. 2012;26:1400–1412. doi: 10.1096/fj.11-194175. [DOI] [PubMed] [Google Scholar]
- 51.Brinkmann K, et al. Ubiquitin C-terminal hydrolase-L1 potentiates cancer chemosensitivity by stabilizing NOXA. Cell Rep. 2013;3:881–891. doi: 10.1016/j.celrep.2013.02.014. [DOI] [PubMed] [Google Scholar]
- 52.Li L, et al. The tumor suppressor UCHL1 forms a complex with p53/MDM2/ARF to promote p53 signaling and is frequently silenced in nasopharyngeal carcinoma. Clin Cancer Res. 2010;16:2949–2958. doi: 10.1158/1078-0432.CCR-09-3178. [DOI] [PubMed] [Google Scholar]
- 53.Yu J, et al. Epigenetic identification of ubiquitin carboxyl-terminal hydrolase L1 as a functional tumor suppressor and biomarker for hepatocellular carcinoma and other digestive tumors. Hepatology. 2008;48:508–518. doi: 10.1002/hep.22343. [DOI] [PubMed] [Google Scholar]
- 54.Mazumdar T, et al. Regulation of NF-kappaB activity and inducible nitric oxide synthase by regulatory particle non-ATPase subunit 13 (Rpn13) Proc Natl Acad Sci U S A. 2010;107:13854–13859. doi: 10.1073/pnas.0913495107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fukui S, et al. The proteasome deubiquitinase inhibitor bAP15 downregulates TGF-beta/Smad signaling and induces apoptosis via UCHL5 inhibition in ovarian cancer. Oncotarget. 2019;10:5932–5948. doi: 10.18632/oncotarget.27219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang CL, et al. Ubiquitin-specific protease 2a stabilizes MDM4 and facilitates the p53-mediated intrinsic apoptotic pathway in glioblastoma. Carcinogenesis. 2014;35:1500–1509. doi: 10.1093/carcin/bgu015. [DOI] [PubMed] [Google Scholar]
- 57.Mahul-Mellier AL, et al. De-ubiquitinating proteases USP2a and USP2c cause apoptosis by stabilising RIP1. Biochim Biophys Acta. 2012;1823:1353–1365. doi: 10.1016/j.bbamcr.2012.05.022. [DOI] [PubMed] [Google Scholar]
- 58.Mahul-Mellier AL, et al. De-ubiquitinating protease USP2a targets RIP1 and TRAF2 to mediate cell death by TNF. Cell Death Differ. 2012;19:891–899. doi: 10.1038/cdd.2011.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Park JK, Das T, Song EJ, Kim EE. Structural basis for recruiting and shuttling of the spliceosomal deubiquitinase USP4 by SART3. Nucleic Acids Res. 2016;44:5424–5437. doi: 10.1093/nar/gkw218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Clerici M, Luna-Vargas MP, Faesen AC, Sixma TK. The DUSP-Ubl domain of USP4 enhances its catalytic efficiency by promoting ubiquitin exchange. Nat Commun. 2014;5:5399. doi: 10.1038/ncomms6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hu B, et al. Spotlight on USP4: structure, function, and regulation. Front Cell Dev Biol. 2021;9:595159. doi: 10.3389/fcell.2021.595159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hou X, Wang L, Zhang L, Pan X, Zhao W. Ubiquitin-specific protease 4 promotes TNF-alpha-induced apoptosis by deubiquitination of RIP1 in head and neck squamous cell carcinoma. FEBS Lett. 2013;587:311–316. doi: 10.1016/j.febslet.2012.12.016. [DOI] [PubMed] [Google Scholar]
- 63.Zhou J, et al. USP7: target validation and drug discovery for cancer therapy. Med Chem. 2018;14:3–18. doi: 10.2174/1573406413666171020115539. [DOI] [PubMed] [Google Scholar]
- 64.Schauer NJ, et al. Selective USP7 inhibition elicits cancer cell killing through a p53-dependent mechanism. Sci Rep. 2020;10:5324. doi: 10.1038/s41598-020-62076-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Masuya D, et al. The HAUSP gene plays an important role in non-small cell lung carcinogenesis through p53-dependent pathways. J Pathol. 2006;208:724–732. doi: 10.1002/path.1931. [DOI] [PubMed] [Google Scholar]
- 66.Becker K, Marchenko ND, Palacios G, Moll UM. A role of HAUSP in tumor suppression in a human colon carcinoma xenograft model. Cell Cycle. 2008;7:1205–1213. doi: 10.4161/cc.7.9.5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Khan OM, et al. The deubiquitinase USP9X regulates FBW7 stability and suppresses colorectal cancer. J Clin Invest. 2018;128:1326–1337. doi: 10.1172/JCI97325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Perez-Mancera PA, et al. The deubiquitinase USP9X suppresses pancreatic ductal adenocarcinoma. Nature. 2012;486:266–270. doi: 10.1038/nature11114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bolomsky A, et al. MCL-1 inhibitors, fast-lane development of a new class of anti-cancer agents. J Hematol Oncol. 2020;13:173. doi: 10.1186/s13045-020-01007-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schwickart M, et al. Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature. 2010;463:103–107. doi: 10.1038/nature08646. [DOI] [PubMed] [Google Scholar]
- 71.Sun J, et al. USP10 inhibits lung cancer cell growth and invasion by upregulating PTEN. Mol Cell Biochem. 2018;441:1–7. doi: 10.1007/s11010-017-3170-2. [DOI] [PubMed] [Google Scholar]
- 72.Bhattacharya U, Neizer-Ashun F, Mukherjee P, Bhattacharya R. When the chains do not break: the role of USP10 in physiology and pathology. Cell Death Dis. 2020;11:1033. doi: 10.1038/s41419-020-03246-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Luo Z, et al. A negative feedback regulatory loop between miR-138 and TP53 is mediated by USP10. Oncotarget. 2019;10:6288–6296. doi: 10.18632/oncotarget.27275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol. 2010;2:a001008. doi: 10.1101/cshperspect.a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 76.Yuan J, Luo K, Zhang L, Cheville JC, Lou Z. USP10 regulates p53 localization and stability by deubiquitinating p53. Cell. 2010;140:384–396. doi: 10.1016/j.cell.2009.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Boselli M, et al. An inhibitor of the proteasomal deubiquitinating enzyme USP14 induces tau elimination in cultured neurons. J Biol Chem. 2017;292:19209–19225. doi: 10.1074/jbc.M117.815126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jiang L, et al. Proteasomal cysteine deubiquitinase inhibitor b-AP15 suppresses migration and induces apoptosis in diffuse large B cell lymphoma. J Exp Clin Cancer Res. 2019;38:453. doi: 10.1186/s13046-019-1446-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang SA, Hung JJ. Ubiquitin-specific protease 24 regulates apoptosis through Deubiquinating Bax and Mediating Ku70 Acetylation. FASEB J. 2015;29:569. doi: 10.1096/fasebj.29.1_supplement.569.9. [DOI] [Google Scholar]
- 80.Popov N, et al. The ubiquitin-specific protease USP28 is required for MYC stability. Nat Cell Biol. 2007;9:765–774. doi: 10.1038/ncb1601. [DOI] [PubMed] [Google Scholar]
- 81.Liu Z, Zhao T, et al. Discovery of [1,2,3]triazolo[4,5-d]pyrimidine derivatives as highly potent, selective, and cellularly active USP28 inhibitors. Acta Pharm Sin B. 2020;10:1476–1491. doi: 10.1016/j.apsb.2019.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Guo G, Xu Y, Gong M, Cao Y, An R. USP28 is a potential prognostic marker for bladder cancer. Tumour Biol. 2014;35:4017–4022. doi: 10.1007/s13277-013-1525-1. [DOI] [PubMed] [Google Scholar]
- 83.Diefenbacher ME, et al. The deubiquitinase USP28 controls intestinal homeostasis and promotes colorectal cancer. J Clin Invest. 2014;124:3407–3418. doi: 10.1172/JCI73733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang L, et al. Overexpression of deubiquitinating enzyme USP28 promoted non-small cell lung cancer growth. J Cell Mol Med. 2015;19:799–805. doi: 10.1111/jcmm.12426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liang JR, et al. USP30 deubiquitylates mitochondrial Parkin substrates and restricts apoptotic cell death. EMBO Rep. 2015;16:618–627. doi: 10.15252/embr.201439820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rusilowicz-Jones EV, et al. USP30 sets a trigger threshold for PINK1-PARKIN amplification of mitochondrial ubiquitylation. Life Sci Alliance. 2020;3:e202000768. doi: 10.26508/lsa.202000768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kluge AF, et al. Novel highly selective inhibitors of ubiquitin specific protease 30 (USP30) accelerate mitophagy. Bioorg Med Chem Lett. 2018;28:2655–2659. doi: 10.1016/j.bmcl.2018.05.013. [DOI] [PubMed] [Google Scholar]
- 88.Jia M, Guo Y, Lu X. USP33 is a biomarker of disease recurrence in papillary thyroid carcinoma. Cell Physiol Biochem. 2018;45:2044–2053. doi: 10.1159/000488041. [DOI] [PubMed] [Google Scholar]
- 89.Lee JC, Peter ME. Regulation of apoptosis by ubiquitination. Immunol Rev. 2003;193:39–47. doi: 10.1034/j.1600-065x.2003.00043.x. [DOI] [PubMed] [Google Scholar]
- 90.Verhelst K, van Loo G, Beyaert R. A20: attractive without showing cleavage. EMBO Rep. 2014;15:734–735. doi: 10.15252/embr.201439014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wertz IE, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 92.Priem D, et al. A20 protects cells from TNF-induced apoptosis through linear ubiquitin-dependent and -independent mechanisms. Cell Death Dis. 2019;10:692. doi: 10.1038/s41419-019-1937-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jang EJ, et al. FAM188B downregulation sensitizes lung cancer cells to anoikis via EGFR downregulation and inhibits tumor metastasis In vivo. Cancers. 2021;13:247. doi: 10.3390/cancers13020247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kotschy A, et al. The MCL1 inhibitor S63845 is tolerable and effective in diverse cancer models. Nature. 2016;538:477–482. doi: 10.1038/nature19830. [DOI] [PubMed] [Google Scholar]
- 95.Moujalled DM, et al. Combining BH3-mimetics to target both BCL-2 and MCL1 has potent activity in pre-clinical models of acute myeloid leukemia. Leukemia. 2019;33:905–917. doi: 10.1038/s41375-018-0261-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li Z, He S, Look AT. The MCL1-specific inhibitor S63845 acts synergistically with venetoclax/ABT-199 to induce apoptosis in T-cell acute lymphoblastic leukemia cells. Leukemia. 2019;33:262–266. doi: 10.1038/s41375-018-0201-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wu X, Luo Q, et al. JOSD1 inhibits mitochondrial apoptotic signalling to drive acquired chemoresistance in gynaecological cancer by stabilizing MCL1. Cell Death Differ. 2020;27:55–70. doi: 10.1038/s41418-019-0339-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Senichkin VV, Streletskaia AY, Gorbunova AS, Zhivotovsky B, Kopeina GS. Saga of Mcl-1: regulation from transcription to degradation. Cell Death Differ. 2020;27:405–419. doi: 10.1038/s41418-019-0486-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ni Q, Chen J, et al. Expression of OTUB1 in hepatocellular carcinoma and its effects on HCC cell migration and invasion. Acta Biochim Biophys Sin. 2017;49:680–688. doi: 10.1093/abbs/gmx056. [DOI] [PubMed] [Google Scholar]
- 100.Xie X, Wang X, et al. PPPDE1 is a novel deubiquitinase belonging to a cysteine isopeptidase family. Biochem Biophys Res Commun. 2017;488:291–296. doi: 10.1016/j.bbrc.2017.04.161. [DOI] [PubMed] [Google Scholar]
- 101.Xie X, Wang X, et al. PPPDE1 promotes hepatocellular carcinoma development by negatively regulate p53 and apoptosis. Apoptosis. 2019;24:135–144. doi: 10.1007/s10495-018-1491-6. [DOI] [PubMed] [Google Scholar]
- 102.Tian Z, et al. A novel small molecule inhibitor of deubiquitylating enzyme USP14 and UCHL5 induces apoptosis in multiple myeloma and overcomes bortezomib resistance. Blood. 2014;123:706–716. doi: 10.1182/blood-2013-05-500033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liu D, Song Z, Wang X, Ouyang L. Ubiquitin C-Terminal Hydrolase L5 (UCHL5) accelerates the growth of endometrial cancer via activating the Wnt/beta-Catenin signaling pathway. Front Oncol. 2020;10:865. doi: 10.3389/fonc.2020.00865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cui SZ, et al. Targeting USP1-dependent KDM4A protein stability as a potential prostate cancer therapy. Cancer Sci. 2020;111:1567–1581. doi: 10.1111/cas.14375. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 105.Lee JK, et al. USP1 targeting impedes GBM growth by inhibiting stem cell maintenance and radioresistance. Neuro Oncol. 2016;18:37–47. doi: 10.1093/neuonc/nov091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mistry H, et al. Small-molecule inhibitors of USP1 target ID1 degradation in leukemic cells. Mol Cancer Ther. 2013;12:2651–2662. doi: 10.1158/1535-7163.MCT-13-0103-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kitamura H, Hashimoto M. USP2-related cellular signaling and consequent pathophysiological outcomes. Int J Mol Sci. 2021;22:1209. doi: 10.3390/ijms22031209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nida S, Ambe L, Farha I, Shamshad Z, Zehra H. Inhibition of USP2 Induces apoptosis through down regulation of fatty acid synthase and Cyclin D1 in triple negative breast cancer. Curr Proteomics. 2020;17:425. doi: 10.2174/157016461766619100809352. [DOI] [Google Scholar]
- 109.Davis MI, et al. Small molecule inhibition of the ubiquitin-specific protease USP2 accelerates cyclin D1 degradation and leads to cell cycle arrest in colorectal cancer and mantle cell lymphoma models. J Biol Chem. 2016;291:24628–24640. doi: 10.1074/jbc.M116.738567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Qiu C, et al. Correction for: Ubiquitin-specific protease 4 promotes metastasis of hepatocellular carcinoma by increasing TGF-beta signaling-induced epithelial-mesenchymal transition. Aging. 2019;11:3408–3409. doi: 10.18632/aging.102010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Li T, et al. Ubiquitin-specific protease 4 promotes hepatocellular carcinoma progression via cyclophilin A stabilization and deubiquitination. Cell Death Dis. 2018;9:148. doi: 10.1038/s41419-017-0182-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Heo MJ, et al. microRNA-148a dysregulation discriminates poor prognosis of hepatocellular carcinoma in association with USP4 overexpression. Oncotarget. 2014;5:2792–2806. doi: 10.18632/oncotarget.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li F, et al. The deubiquitinase USP4 stabilizes Twist1 protein to promote lung cancer cell stemness. Cancers. 2020;12:1582. doi: 10.3390/cancers12061582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Potu H, et al. Usp5 links suppression of p53 and FAS levels in melanoma to the BRAF pathway. Oncotarget. 2014;5:5559–5569. doi: 10.18632/oncotarget.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dayal S, et al. Suppression of the deubiquitinating enzyme USP5 causes the accumulation of unanchored polyubiquitin and the activation of p53. J Biol Chem. 2009;284:5030–5041. doi: 10.1074/jbc.M805871200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Xu X, et al. Ubiquitin specific peptidase 5 regulates colorectal cancer cell growth by stabilizing Tu translation elongation factor. Theranostics. 2019;9:4208–4220. doi: 10.7150/thno.33803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liu Y, et al. Ubiquitin specific peptidase 5 mediates Histidine-rich protein Hpn induced cell apoptosis in hepatocellular carcinoma through P14–P53 signaling. Proteomics. 2017;17:1600350. doi: 10.1002/pmic.201600350. [DOI] [PubMed] [Google Scholar]
- 118.Meng J, et al. USP5 promotes epithelial-mesenchymal transition by stabilizing SLUG in hepatocellular carcinoma. Theranostics. 2019;9:573–587. doi: 10.7150/thno.27654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Xue S, et al. USP5 promotes metastasis in non-small cell lung cancer by inducing epithelial-mesenchymal transition via Wnt/beta-catenin pathway. Front Pharmacol. 2020;11:668. doi: 10.3389/fphar.2020.00668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Liu Y, et al. Usp5 functions as an oncogene for stimulating tumorigenesis in hepatocellular carcinoma. Oncotarget. 2017;8:50655–50664. doi: 10.18632/oncotarget.16901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Song MS, et al. The deubiquitinylation and localization of PTEN are regulated by a HAUSP-PML network. Nature. 2008;455:813–817. doi: 10.1038/nature07290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Su D, et al. Ubiquitin-specific protease 7 sustains DNA damage response and promotes cervical carcinogenesis. J Clin Invest. 2018;128:4280–4296. doi: 10.1172/JCI120518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rong Z, Zhu Z, Cai S, Zhang B. Knockdown of USP8 inhibits the growth of lung cancer cells. Cancer Manag Res. 2020;12:12415–12422. doi: 10.2147/IJN.S259191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Jeong M, et al. USP8 suppresses death receptor-mediated apoptosis by enhancing FLIPL stability. Oncogene. 2017;36:458–470. doi: 10.1038/onc.2016.215. [DOI] [PubMed] [Google Scholar]
- 125.Zhang S, et al. Deubiquitinase USP13 dictates MCL1 stability and sensitivity to BH3 mimetic inhibitors. Nat Commun. 2018;9:215. doi: 10.1038/s41467-017-02693-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kushwaha D, et al. USP9X inhibition promotes radiation-induced apoptosis in non-small cell lung cancer cells expressing mid-to-high MCL1. Cancer Biol Ther. 2015;16:392–401. doi: 10.1080/15384047.2014.1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kim S, et al. WP1130 enhances TRAIL-induced apoptosis through USP9X-dependent miR-708-mediated downregulation of c-FLIP. Cancers (Basel) 2019;11:344. doi: 10.3390/cancers11030344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Liu Y, et al. Inhibition of the deubiquitinase USP9x induces pre-B cell homeobox 1 (PBX1) degradation and thereby stimulates prostate cancer cell apoptosis. J Biol Chem. 2019;294:4572–4582. doi: 10.1074/jbc.RA118.006057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lu Q, Zhang FL, Lu DY, Shao ZM, Li DQ. USP9X stabilizes BRCA1 and confers resistance to DNA-damaging agents in human cancer cells. Cancer Med. 2019;8:6730–6740. doi: 10.1002/cam4.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chen Q, et al. USP10 promotes proliferation and migration and inhibits apoptosis of endometrial stromal cells in endometriosis through activating the Raf-1/MEK/ERK pathway. Am J Physiol Cell Physiol. 2018;315:C863–C872. doi: 10.1152/ajpcell.00272.2018. [DOI] [PubMed] [Google Scholar]
- 131.Lee EW, et al. USP11-dependent selective cIAP2 deubiquitylation and stabilization determine sensitivity to Smac mimetics. Cell Death Differ. 2015;22:1463–1476. doi: 10.1038/cdd.2014.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lee EW, Song J. USP11: A key regulator of cIAP2 stability and sensitivity to SMAC mimetics. Mol Cell Oncol. 2016;3:e1029829. doi: 10.1080/23723556.2015.1029829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Liu J, et al. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell. 2011;147:223–234. doi: 10.1016/j.cell.2011.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Morgan EL, et al. The deubiquitinase (DUB) USP13 promotes Mcl-1 stabilisation in cervical cancer. Oncogene. 2021;40:2112–2129. doi: 10.1038/s41388-021-01679-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ma YS, et al. Inhibition of USP14 deubiquitinating activity as a potential therapy for tumors with p53 deficiency. Mol Ther Oncolytics. 2020;16:147–157. doi: 10.1016/j.omto.2019.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Shinji S, et al. Ubiquitin-specific protease 14 expression in colorectal cancer is associated with liver and lymph node metastases. Oncol Rep. 2006;15:539–543. [PubMed] [Google Scholar]
- 137.Chuensumran U, et al. Ubiquitin-specific protease 14 expression associated with intrahepatic cholangiocarcinoma cell differentiation. Asian Pac J Cancer Prev. 2011;12:775–779. [PubMed] [Google Scholar]
- 138.Wu N, et al. Over-expression of deubiquitinating enzyme USP14 in lung adenocarcinoma promotes proliferation through the accumulation of beta-catenin. Int J Mol Sci. 2013;14:10749–10760. doi: 10.3390/ijms140610749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wu N, Zhang C, Bai C, Han YP, Li Q. MiR-4782-3p inhibited non-small cell lung cancer growth via USP14. Cell Physiol Biochem. 2014;33:457–467. doi: 10.1159/000358626. [DOI] [PubMed] [Google Scholar]
- 140.Wang Y, et al. Ubiquitin-specific protease 14 (USP14) regulates cellular proliferation and apoptosis in epithelial ovarian cancer. Med Oncol. 2015;32:379. doi: 10.1007/s12032-014-0379-8. [DOI] [PubMed] [Google Scholar]
- 141.Song C, Ma R, Yang X, Pang S. The deubiquitinating enzyme USP14 regulates leukemic chemotherapy drugs-induced cell apoptosis by suppressing ubiquitination of aurora kinase B. Cell Physiol Biochem. 2017;42:965–973. doi: 10.1159/000478679. [DOI] [PubMed] [Google Scholar]
- 142.Didier R, et al. Targeting the proteasome-associated deubiquitinating enzyme USP14 impairs melanoma cell survival and overcomes resistance to MAPK-targeting therapies. Mol Cancer Ther. 2018;17:1416–1429. doi: 10.1158/1535-7163.MCT-17-0919. [DOI] [PubMed] [Google Scholar]
- 143.Moghadami AA, et al. Inhibition of USP14 induces ER stress-mediated autophagy without apoptosis in lung cancer cell line A549. Cell Stress Chaperones. 2020;25:909–917. doi: 10.1007/s12192-020-01125-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Yu B, et al. USP15 promotes the apoptosis of degenerative nucleus pulposus cells by suppressing the PI3K/AKT signalling pathway. J Cell Mol Med. 2020;24:13813–13823. doi: 10.1111/jcmm.15971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zhou L, et al. USP15 inhibits multiple myeloma cell apoptosis through activating a feedback loop with the transcription factor NF-kappaBp65. Exp Mol Med. 2018;50:1–12. doi: 10.1038/s12276-018-0180-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Eichhorn PJ, et al. USP15 stabilizes TGF-beta receptor I and promotes oncogenesis through the activation of TGF-beta signaling in glioblastoma. Nat Med. 2012;18:429–435. doi: 10.1038/nm.2619. [DOI] [PubMed] [Google Scholar]
- 147.Francies FZ, Bassa S, Chatziioannou A, Kaufmann AM, Dlamini Z. Splicing genomics events in cervical cancer: insights for phenotypic stratification and biomarker potency. Genes. 2021;12:130. doi: 10.3390/genes12020130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Diao W, et al. USP18 promotes cell proliferation and suppressed apoptosis in cervical cancer cells via activating AKT signaling pathway. BMC Cancer. 2020;20:741. doi: 10.1186/s12885-020-07241-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Cai J, et al. Downregulation of USP18 inhibits growth and induces apoptosis in hepatitis B virus-related hepatocellular carcinoma cells by suppressing BCL2L1. Exp Cell Res. 2017;358:315–322. doi: 10.1016/j.yexcr.2017.07.006. [DOI] [PubMed] [Google Scholar]
- 150.Heride C, et al. The centrosomal deubiquitylase USP21 regulates Gli1 transcriptional activity and stability. J Cell Sci. 2016;129:4001–4013. doi: 10.1242/jcs.188516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Fan Y, et al. USP21 negatively regulates antiviral response by acting as a RIG-I deubiquitinase. J Exp Med. 2014;211:313–328. doi: 10.1084/jem.20122844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Chen Y, Zhou B, Chen D. USP21 promotes cell proliferation and metastasis through suppressing EZH2 ubiquitination in bladder carcinoma. Onco Targets Ther. 2017;10:681–689. doi: 10.2147/OTT.S124795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Peng L, Hu Y, Chen D, Jiao S, Sun S. Ubiquitin specific peptidase 21 regulates interleukin-8 expression, stem-cell like property of human renal cell carcinoma. Oncotarget. 2016;7:42007–42016. doi: 10.18632/oncotarget.9751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Yun SI, et al. Ubiquitin-specific protease 21 promotes colorectal cancer metastasis by acting as a Fra-1 deubiquitinase. Cancers. 2020;12:207. doi: 10.3390/cancers12010207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Liu H, et al. Oncogenic USP22 supports gastric cancer growth and metastasis by activating c-Myc/NAMPT/SIRT1-dependent FOXO1 and YAP signaling. Aging. 2019;11:9643–9660. doi: 10.18632/aging.102410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Bai Z, Du Y, Cong L, Cheng Y. The USP22 promotes the growth of cancer cells through the DYRK1A in pancreatic ductal adenocarcinoma. Gene. 2020;758:144960. doi: 10.1016/j.gene.2020.144960. [DOI] [PubMed] [Google Scholar]
- 157.Lin Z, et al. USP22 antagonizes p53 transcriptional activation by deubiquitinating Sirt1 to suppress cell apoptosis and is required for mouse embryonic development. Mol Cell. 2012;46:484–494. doi: 10.1016/j.molcel.2012.03.024. [DOI] [PubMed] [Google Scholar]
- 158.Armour SM, et al. A high-confidence interaction map identifies SIRT1 as a mediator of acetylation of USP22 and the SAGA coactivator complex. Mol Cell Biol. 2013;33:1487–1502. doi: 10.1128/MCB.00971-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Xu H, Liu YL, Yang YM, Dong XS. Knock-down of ubiquitin-specific protease 22 by micro-RNA interference inhibits colorectal cancer growth. Int J Colorectal Dis. 2012;27:21–30. doi: 10.1007/s00384-011-1275-8. [DOI] [PubMed] [Google Scholar]
- 160.Zhou F, et al. Knockdown of ubiquitinspecific protease 51 attenuates cisplatin resistance in lung cancer through ubiquitination of zincfinger Ebox binding homeobox 1. Mol Med Rep. 2020;22:1382–1390. doi: 10.3892/mmr.2020.11188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Gu Y, et al. Zeb1 Is a potential regulator of Six2 in the proliferation, apoptosis and migration of metanephric mesenchyme cells. Int J Mol Sci. 2016;17:1283. doi: 10.3390/ijms17081283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Wrigley JD, et al. Identification and characterization of dual inhibitors of the USP25/28 deubiquitinating enzyme subfamily. ACS Chem Biol. 2017;12:3113–3125. doi: 10.1021/acschembio.7b00334. [DOI] [PubMed] [Google Scholar]
- 163.Oh YT, Deng L, Deng J, Sun SY. The proteasome deubiquitinase inhibitor b-AP15 enhances DR5 activation-induced apoptosis through stabilizing DR5. Sci Rep. 2017;7:8027. doi: 10.1038/s41598-017-08424-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kageyama K, Asari Y, Sugimoto Y, Niioka K, Daimon M. Ubiquitin-specific protease 8 inhibitor suppresses adrenocorticotropic hormone production and corticotroph tumor cell proliferation. Endocr J. 2020;67:177–184. doi: 10.1507/endocrj.EJ19-0239. [DOI] [PubMed] [Google Scholar]
- 165.Bi HL, et al. Inhibition of UCHL1 by LDN-57444 attenuates Ang II-Induced atrial fibrillation in mice. Hypertens Res. 2020;43:168–177. doi: 10.1038/s41440-019-0354-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wang SA, et al. USP24 promotes drug resistance during cancer therapy. Cell Death Differ. 2021;28:2690–2707. doi: 10.1038/s41418-021-00778-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Antao AM, Tyagi A, Kim KS, Ramakrishna S. Advances in deubiquitinating enzyme inhibition and applications in cancer therapeutics. Cancers. 2020;12:1579. doi: 10.3390/cancers12061579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Pozhidaeva A, et al. USP7-specific inhibitors target and modify the enzyme’s active site via distinct chemical mechanisms. Cell Chem Biol. 2017;24:1501–1512.e5. doi: 10.1016/j.chembiol.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 169.Pesce E, et al. The autophagy inhibitor Spautin-1 antagonizes rescue of mutant CFTR through an autophagy-independent and USP13-mediated mechanism. Front Pharmacol. 2018;9:1464. doi: 10.3389/fphar.2018.01464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Yamanaka S, et al. Subquinocin, a small molecule inhibitor of CYLD and USP-family deubiquitinating enzymes, promotes NF-kappaB signaling. Biochem Biophys Res Commun. 2020;524:1–7. doi: 10.1016/j.bbrc.2019.12.049. [DOI] [PubMed] [Google Scholar]
- 171.Okada K, et al. Vialinin A is a ubiquitin-specific peptidase inhibitor. Bioorg Med Chem Lett. 2013;23:4328–4331. doi: 10.1016/j.bmcl.2013.05.093. [DOI] [PubMed] [Google Scholar]
- 172.Yoshioka Y, et al. Ubiquitin-specific peptidase 5, a target molecule of vialinin A, is a key molecule of TNF-alpha production in RBL-2H3 cells. PLoS ONE. 2013;8:e80931. doi: 10.1371/journal.pone.0080931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Dexheimer TS et al (2010) Discovery of ML323 as a novel Inhibitor of the USP1/UAF1 deubiquitinase complex. In: Probe reports from the NIH molecular libraries program. Bethesda (MD)
- 174.Wang X, et al. Synthesis and evaluation of derivatives of the proteasome deubiquitinase inhibitor b-AP15. Chem Biol Drug Des. 2015;86:1036–1048. doi: 10.1111/cbdd.12571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Sakamoto KM, et al. Protacs: chimeric molecules that target proteins to the Skp1-Cullin-F box complex for ubiquitination and degradation. Proc Natl Acad Sci U S A. 2001;98:8554–8559. doi: 10.1073/pnas.141230798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Liu J, et al. Cancer selective target degradation by folate-caged PROTACs. J Am Chem Soc. 2021;143:7380–7387. doi: 10.1021/jacs.1c00451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Jin J, et al. The peptide PROTAC modality: a novel strategy for targeted protein ubiquitination. Theranostics. 2020;10:10141–10153. doi: 10.7150/thno.46985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Maneiro MA, et al. Antibody-PROTAC conjugates enable HER2-dependent targeted protein degradation of BRD4. ACS Chem Biol. 2020;15:1306–1312. doi: 10.1021/acschembio.0c00285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Henning NJ, et al. Deubiquitinase-targeting chimeras for targeted protein stabilization. bioRxiv. 2021;44:959. doi: 10.1101/2021.04.30.441959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Davies CW, et al. The co-crystal structure of ubiquitin carboxy-terminal hydrolase L1 (UCHL1) with a tripeptide fluoromethyl ketone (Z-VAE(OMe)-FMK) Bioorg Med Chem Lett. 2012;22:3900–3904. doi: 10.1016/j.bmcl.2012.04.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Gu Y, et al. The deubiquitinating enzyme UCHL1 negatively regulates the immunosuppressive capacity and survival of multipotent mesenchymal stromal cells. Cell Death Dis. 2018;9:459. doi: 10.1038/s41419-018-0532-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
As this is a review article, there is no requirement for data availability.