Abstract
With the onset of Listeria monocytogenes resistance to the bacteriocin nisin, the search for alternative antimicrobial treatments is of fundamental importance. In this work, we set out to investigate proteins and lipids involved in the resistance mechanisms of L. monocytogenes against the antimicrobial peptides (AMPs) nisin and fengycin. The effect of sub-lethal concentrations of nisin and lipopeptide fengycin secreted by Bacillus velezensis P34 on L. monocytogenes was investigated by mass spectrometry-based lipidomics and proteomics. Both AMPs caused a differential regulation of biofilm formation, confirming the promotion of cell attachment and biofilm assembling after treatment with nisin, whereas growth inhibition was observed after fengycin treatment. Anteiso branched-chain fatty acids were detected in higher amounts in fengycin-treated samples (46.6%) as compared to nisin-treated and control samples (39.4% and 43.4%, respectively). In addition, a higher relative abundance of 30:0, 31:0 and 32:0 phosphatidylglycerol species was detected in fengycin-treated samples. The lipidomics data suggest the inhibition of biofilm formation by the fengycin treatment, while the proteomics data revealed downregulation of important cell wall proteins involved in the building of biofilms, such as the lipoteichoic acid backbone synthesis (Lmo0927) and the flagella-related (Lmo0718) proteins among others. Together, these results provide new insights into the modification of lipid and protein profiles and biofilm formation in L. monocytogenes upon exposure to antimicrobial peptides.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-022-04292-4.
Keywords: Antimicrobial peptides, Antimicrobial resistance, Nisin, Fengycin, Fatty acids, Membrane proteins
Introduction
Listeria monocytogenes is a foodborne pathogen causing listeriosis, a severe disease affecting primarily immunocompromised people, pregnant women and children, causing septicemia, meningitis and abortions [1]. The main route of L. monocytogenes transmission in cases of food contamination occurs during the production process [2]. The food industry uses antimicrobial compounds for extending shelf life and limiting the spread of foodborne pathogens, including L. monocytogenes [3]. Bacteria produce diverse types of antimicrobial peptides (AMPs), including those ribosomally synthesized, such as bacteriocins [4], and non-ribosomally synthesized AMPs, e.g., cyclic lipopeptides (CLPs) [5].
The lantibiotic nisin, a positively charged bacteriocin produced by Lactococcus lactis subsp. lactis, is widely used to control pathogens like L. monocytogenes in foods [6, 7]. Nisin inhibits bacterial growth through a dual mode of action, combining the generation of pores in the cell membrane and interruption of the cell wall biosynthesis mediated by specific interaction with the lipid II, which is recognized as a precursor component of bacterial cell wall [8, 9]. However, the acquisition of nisin tolerance or resistance in L. monocytogenes has already been observed. The antimicrobial resistance of L. monocytogenes to nisin can be influenced by environmental conditions and genetic characteristics [10, 11]. Some strains have high natural nisin resistance compared with others. As a consequence, the efficacy of this bacteriocin might be reduced [12, 13]. The upregulation of membrane proteins involved in biofilm formation was previously reported among the strategies employed by L. monocytogenes to evade nisin action [14].
Bacillus strains host multiple biosynthetic gene clusters that confer the ability to produce CLPs, collectively grouped into the surfactin, fengycin and iturin families [15]. Bacillus velezensis P34, a strain isolated from the intestine of an Amazonian fish, produces CLPs, mainly fengycin [16]. Although fengycin has been mostly reported as a potent antifungal lipopeptide [17], its antibacterial activity is also recognized [18]. Fengycin activity has been confirmed against Gram-positive bacteria, including the effect on reducing the presence of Staphylococcus aureus in human intestine by modulating quorum sensing [19].
The phospholipid phosphatidylglycerol (PG), along with cardiolipin, lipoteichoic acid (LTA) and phosphatidylethanolamine (PE) has been described to play a crucial role in membrane stability in Listeria [20, 21]. Surface charge modifications induced by changes in teichoic acids and membrane lipids have been included among the variety of resistance mechanisms to AMPs [22, 23]. Such modifications influence the interaction of AMPs with bacterial cells and consequently their bacteriolytic activity, often explained by two different mechanisms [24, 25]: (a) after binding to cell surface components, the peptide penetrates the membrane by forming channels, as in the case of nisin, or (b) the peptide aggregates on the membrane surface causing damage, a mechanism that may be exerted by fengycin [26, 27].
Membrane proteins should not be considered as isolated entities but part of a complex structure surrounded by lipid molecules, playing a pivotal role in maintaining the full structural and functional integrity of biological membranes [28]. The composition of bacterial cell surface also influences the biofilm formation capacity, already considered as one of the resistance strategies of L. monocytogenes. Biofilms formed by this foodborne pathogen contain teichoic acids as a major matrix component [29] and can be developed on different surfaces and inaccessible locations of food processing facilities, which is an important problem for the management of L. monocytogenes contaminations. Taking into account the biofilm formation process allows proposing better hygienic practices against this pathogen [30, 31].
The hypothesis we set out to test in this study is that L. monocytogenes is able to form biofilm as resistance strategy to sub-lethal concentration of nisin, and that the use of AMPs such as the CLP fengycin might overcome this concern. To test this hypothesis, L. monocytogenes was exposed to sub-lethal doses of nisin and fengycin and the molecular responses behind the bacterial resistance and biofilm modulation were investigated via mass spectrometry-based lipidomics and proteomics approaches.
Materials and methods
Chemicals
Reagents to perform the derivatization of lipid components into fatty acid methyl ester (FAME) derivatives, sodium methoxide (MeONa) and boron trifluoride-methanol solution (BF3, 14% in methanol) were acquired from Merck Life Science (Merck KGaA, Darmstadt, Germany). A reference standard solution of C4–C24 even carbon saturated FAMEs (1000 μg/mL) was acquired from Merck Life Science for determining experimental linear retention index (LRI) of fatty acid methyl esters (FAMEs).
Antimicrobials
Nisin (Chrisin®) was provided by Chr. Hansen A/S (Hørsholm, Denmark). In agreement with the manufacturer, the formulation contains 2.5% (w/w) pure nisin. Nisin stock solution (1 mg/mL) was prepared by diluting Chrisin® in 0.01 M HCl and stored at 4 °C. The working concentrations were obtained by dilution of the nisin stock solution in a 10 mM sodium phosphate buffer (pH 7.0). The sub-lethal nisin concentration of 0.1 µg/mL was previously determined through the study of inhibitory effects on microbial growth curves [14].
The production of fengycin peptides by B. velezensis P34 was carried out as described previously [16]. The obtained product showed 3,200 activity units (AU) per mL. Thus, dilutions in buffer phosphate pH 7 were performed to achieve the sub-lethal concentration of 100 AU/mL on L. monocytogenes, as demonstrated elsewhere [32].
Listeria monocytogenes cultivation
Listeria monocytogenes ATCC 7644 was pre-cultured in Brain Heart Infusion (BHI, Oxoid, Basingstoke, UK) agar to prepare a bacterial suspension in saline solution (8.5 g/L NaCl) with an OD600 of 0.150, corresponding to a 0.5 McFarland turbidity standard. This suspension (3 mL) was inoculated in 300 mL BHI broth to obtain a concentration of 106 colony forming units per mL (CFU/mL) and incubated in a rotary shaker at 37 °C and 150 rpm. After 6 h cultivation, the defined quantities of antimicrobials (fengycin or nisin) dissolved in 10 mM phosphate buffer pH 7.0 were added to the cultures and samples were incubated for additional 1 h. Control samples of untreated bacteria were prepared using phosphate buffer only. The cellular pellets were collected by centrifugation at 10,000g for 15 min at 4 °C, washed with PBS buffer pH 7.4 before protein and lipid extraction. Additional bacterial cultures were performed to determine L. monocytogenes growth curves for each treatment. All tests were performed using three biological replicates.
Extraction of lipid components
The lipid fraction of L. monocytogenes was initially extracted using Bligh and Dyer protocol as recently reported [33]. Briefly, 4 mL of a methanol/chloroform solution (2:1 v:v) was added to each cellular pellet contained in a 25 mL Falcon tube. The extraction mixture was homogenized using vortex mixing (5 min). After, 1 mL of chloroform and 2 mL of aqueous sodium chloride (NaCl) saturated solution were added to the mixture. The sample was vortexed and centrifuged for 10 min at 2000 g to separate methanol and chloroform layers. The lower organic phase containing lipid compounds was collected using a Pasteur pipette and transferred to a 2 mL vial. The chloroform solvent was removed under constant nitrogen flow.
Derivatization of lipid extract
The derivatization of lipid compounds into FAMEs for gas chromatography (GC) analysis was performed as reported earlier [34]. Briefly, the procedure involved the use of two different derivatizing agents: methanolic solution of MeONa (0.5% w/v) and methanolic solution of BF3 (14% w/v). An aliquot (500 µL) of the derivatizing agents was added to the lipid extract, and the reaction temperatures were maintained at 95 °C for 30 min. Subsequently, 300 µL of n-heptane and 200 µL of saturated NaCl solution were added to the mixture. The upper heptanic FAMEs layer was collected and directly injected into GC instrumentation for chromatographic separation in triplicate.
GC–MS and GC–FID analysis
The separation and identification of FAME derivatives were carried out using a GC/MS QP2010 Ultra (Shimadzu, Duisburg, Germany) instrument equipped with a split–split-less injector (280 °C) and an AOC-20i auto-sampler. A medium-polarity ionic liquid (IL) column, named SLB-IL60 30 m × 0.25 mm id × 0.25 µm df (Merck Life Science) was used for the separation of FAMEs. Quantitative analyses were performed with a GC-2010 instrument (Shimadzu) equipped with a split–split-less injector (280 °C), a flame ionization detector (FID) and AOC-20i auto-sampler. Chromatographic conditions included the volume injections of 3.0 μL, the following temperature program 50 °C–280 °C at 3.0 °C/min, the helium used as carrier gas at 30 cm/s linear velocity and a pressure of 26.6 kPa. MS parameters were as follows: mass range 40–550 amu, ion source temperature 220 °C, interface temperature 250 °C. The FID parameters include detector temperature settled at 300 °C (sampling rate 40 ms) and gas flows were 40 mL/min for hydrogen, 40 mL/min for make up (nitrogen) and 400 mL/min for air, respectively. Carrier gas was helium, at a constant linear velocity of 30.0 cm/s and a pressure of 99.4 kPa. Data collection and peak assignment were the same as those described in detail previously [34].
UHPLC–MS/MS analysis
The analyses were performed on a Shimadzu Ultra High-Performance Liquid Chromatograph-Nexera X-2 system (Shimadzu), including two LC-30 AD dual-plunger parallel-flow pumps, a DGU-20A5R degasser, a CTO-20AC column oven and a SIL-30AC auto-sampler. The UHPLC system was coupled to a LCMS-8060 triple quadrupole mass spectrometer equipped with ESI interface (Shimadzu). Mobile phases were: (A) 20 mM ammonium formate and (B) 2-propanol/acetonitrile/water (60:36:4 v/v/v) with 0.1% formic acid. The gradient program was: 0–6 min, 80–100% B (held for 16 min). The flow rate was 0.4 mL/min; column used was Ascentis Express C18, 100 × 2.1 mm, 2.7 μm dp (Merck Life Science, Darmstadt, Germany), the oven was set at 40 °C and the injection volume was 5 µL. MS and MS/MS acquisitions were performed using ESI source switching between positive ( +) and negative (-) ionization modes, with the following parameters: interface temperature, 450 °C; CDL temperature, 250 °C; heat block temperature, 200 °C; nebulizing gas flow (N2), 3 L/min; drying gas flow (N2), 5 L/min; acquisition range, 350–1250 m/z ( +) and 150–1250 m/z (-). Additional MS/MS experiments were optimized through the injection of single phospholipid (PL) standards, and the selected events are reported elsewhere [35].
Lipidomics data analysis
Data acquisition and processing were performed with the LabSolution ver. 5.95 software (Shimadzu Europa, Duisburg, Germany). The LIPID MAPS Structure Database (LMSD; https://www.lipidmaps.org/data/structure/) was employed for compound identification [36] by searching for the ion types reported in the Table S1, using a mass tolerance of ± 0.2 m/z. Additionally, raw data were converted to mzML format using MSconvert [37] and XIC was manually inspected via the GNPS Dashboard [38]. The urls for XIC visualizations through the GNPS dashboard are provided in the supplementary information.
Total protein extraction and processing
The preparation of protein samples and proteomic data acquisition were performed as reported previously [39, 40]. Briefly, bacterial cells were suspended in Tris–HCl buffer pH 7.5 containing 10 μL Halt™ protease inhibitor cocktail (Thermo Scientific, Rockford, IL, USA) and sonicated for 5 cycles of 30 s with intervals of 1 min in an ultrasonic cell disruptor in ice bath to maintain low temperatures. The lysed cell solutions were centrifuged at 10,000 g for 20 min and the supernatants were lyophilized before protein quantification by the Bradford method. Protein samples at a concentration of 10 μg/mL were digested following standard protocols [41], using 2 μg MS grade trypsin (Promega, Madison, WI, USA). Sample desalination was performed employing C18 stage tips, using 60% (v/v) acetonitrile and 0.1% (v/v) formic acid for peptides elution [42]. The samples were dried in a vacuum concentrator and stored at −20 °C for further analysis by nanoLC–MS/MS.
Proteomics mass spectrometry analysis
The dried samples were reconstituted in 10 μL formic acid (0.1% v/v). Then, an aliquot of 3 μL was analyzed using LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA) coupled to the EASY-nLC system (Proxeon Biosystem, West Palm Beach, FL, USA) through a Proxeon nanoelectrospray ion source. The peptide separation was carried out employing an acetonitrile gradient (2–90% v/v) containing 0.1% (v/v) formic acid, and using a PicoFrit Column (20 cm x ID75 μm, 5 μm particle size, New Objective) at a flow rate of 300 μL/min over 65 min. The instrument methods were set up in the data-dependent acquisition mode. Nano-electrospray voltage was set to 2.2 kV, and the source temperature was set to 275 °C. The DDA setting included a resolution r = 60,000 and the 20 peptides with higher intense peptide ions (top 20) with charge states ≥ 2 were sequentially isolated to a target value of 5000 and fragmented in the high-pressure linear ion trap by CID (collision-induced dissociation) with a normalized collision energy of 35%. Dynamic exclusion was enabled with an exclusion size list of 500 peptides, an exclusion duration of 60 s and a repetition count of 1. The data were processed using MaxQuant v1.3.0.3 software [43] and MS/MS where matched against UniProt database against L. monocytogenes (total of 2844 protein sequences), using the Andromeda search tool [44]. MaxQuant parameters were set as follows: trypsin was used as a protease, with maximum 2 missed cleavages and minimum peptide length of 7. Carbamidomethylation (C) was set as a fixed modification. Oxidation (M) and acetylation (Protein N-term) were set as variable modifications. Mass tolerance was set to 20 ppm, peptide and protein false discovery rate (FDR) cut-off was set to 0.01.
Biofilm assay
The influence of antimicrobials on biofilm formation by L. monocytogenes was evaluated by the crystal violet method [45]. Briefly, a fresh culture in BHI broth was prepared with an initial concentration of 106 CFU/mL L. monocytogenes and incubated for 6 h at 37 °C under aerobic condition. Aliquots were used to create respectively: (i) untreated samples, and cultures treated with (ii) 0.1 μg/mL nisin or (iii) 100 AU/mL fengycin. Ten wells of a 96-well microplate were prepared for each of these treatments and incubated at 37 °C for 24 h. After incubation, the wells were washed three times with sterile saline, fixed with methanol and the biofilm was dried overnight at room temperature. Crystal violet (2% w/v) was used for staining the fixed biofilm before washing the wells for residual dye elimination. Then, 95% (v/v) ethanol was added to the wells for dye extraction and the optical density of ethanol solutions was measured at 570 nm using a microplate reader (SpectraMax M2e). Optical density comparison was made for interpreting the results from wells containing different treatments [45].
Statistical analysis
The MetaboAnalyst 3.068 online software infrastructure was used for the statistical analysis of both the lipidomics and proteomics data. Principal component analysis (PCA) and partial least squares discriminate analysis (PLS-DA) were carried out to determine the variation between the control and the samples treated with nisin and fengycin. The cutoff value VIP (Variable Importance in Projection) ≥ 1 was used to prioritize important features from PLA-SDA statistical analysis, as established by Chong and Jun [46]. Proteins with VIP ≥ 1.0 were submitted to the String v11 Database for constructing protein–protein interactions [47], and the networks were visualized with Cytoscape (www.cytoscape.org/). One-way ANOVA followed by Student’s t test (95% confidence level) was used to evaluate the differences among treatments (control, nisin and fengycin) on lipids composition.
Results and discussion
Differential fatty acids profile
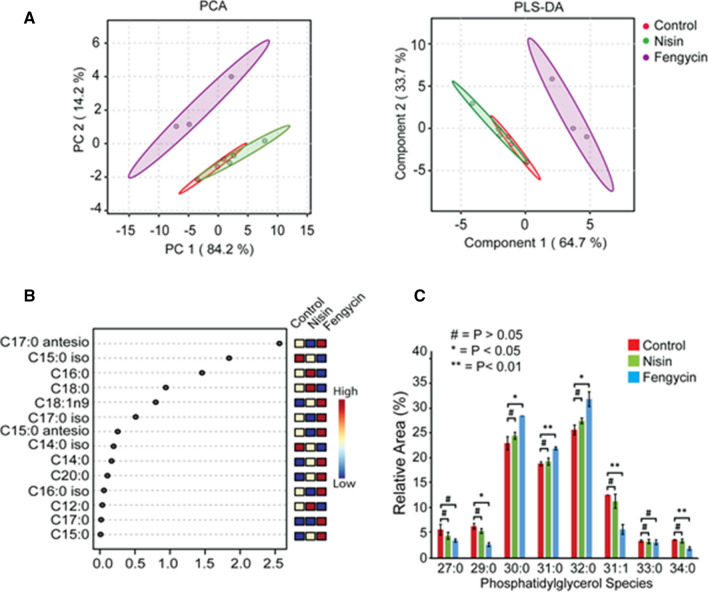
To investigate the changes in lipid profiles of L. monocytogenes after the treatments with nisin and fengycin, we performed a GC–MS analysis of fatty acids methyl esters (FAMEs) derivatives. In our analysis, we observed a total of 14 fatty acids, including the three most abundant FAs classes, namely straight FAs, iso-FAs and anteiso-FAs, and a mono-unsaturated fatty acid (MUFA). The GC profile of reported FAMEs can be seen in Fig. S1. According to the statistical analysis, the only class of FAs that showed significant differences (P < 0.05) was the anteiso-FA, whose values were significantly increased in the samples treated with fengycin (46.62 ± 3.55%) in comparison to those from nisin-treated samples (39.39 ± 2.47%) (Table 1). Accordingly, the additional multivariate statistical analysis including both PCA and PLS-DA, considering each fatty acid separately, revealed statistically significant differences (P < 0.05) for the fengycin-treated samples in comparison to the control and nisin-treated samples (Fig. 1A). Three fatty acids with variable importance in the projection (VIP) scores > 1.0 were detected (Fig. 1B). The FA anteiso-C17:0 showed the highest VIP score and higher concentration in samples treated with fengycin, and downregulated in the nisin treatment. The iso-C15:0 appeared downregulated by the treatment with fengycin, whereas the C16:0 was upregulated in the nisin-treated samples and downregulated by the treatment with fengycin.
Table 1.
List of FAME derivatives identified in Listeria monocytogenes treated with nisin, fengycin and untreated samples. The FAMEs were also grouped as saturated, branched and unsaturated compounds
| Compounds | MSSim | LRIref | LRIexp | Control | Nisin | Fengycin |
|---|---|---|---|---|---|---|
| C12:0 | 96 | 1200 | 1200 | 0.81 ± 0.15 | 1.04 ± 0.05 | 0.77 ± 0.15 |
| Iso-C14:0 | 93 | 1361 | 1361 | 0.82 ± 0.03 | 0.73 ± 0.07 | 0.60 ± 0.10 |
| C14:0 | 96 | 1400 | 1400 | 0.98 ± 0.07 | 1.12 ± 0.02 | 1.17 ± 0.09 |
| Iso-C15:0 | 93 | 1462 | 1461 | 17.24 ± 1.04 | 15.76 ± 0.98 | 15.14 ± 2.16 |
| Anteiso-C15:0 | 94 | 1476 | 1475 | 27.88 ± 0.89 | 25.01 ± 1.94 | 28.17 ± 2.49 |
| C15:0 | 90 | 1500 | 1500 | 0.19 ± 0.02 | 0.19 ± 0.01 | 0.20 ± 0.01 |
| Iso-C16:0 | 93 | 1560 | 1560 | 3.21 ± 0.13 | 3.11 ± 0.10 | 3.27 ± 0.35 |
| C16:0 | 96 | 1600 | 1600 | 16.13 ± 1.53 | 18.83 ± 1.87 | 14.46 ± 2.11 |
| Iso-C17:0 | 94 | 1660 | 1660 | 4.70 ± 0.27 | 4.43 ± 0.14 | 5.28 ± 0.33 |
| Anteiso-C17:0 | 96 | 1676 | 1676 | 15.53 ± 0.43 | 14.38 ± 0.54 | 18.45 ± 1.06 |
| C17:0 | 90 | 1700 | 1700 | 0.10 ± 0.01 | 0.10 ± 0.01 | 0.12 ± 0.02 |
| C18:0 | 96 | 1800 | 1800 | 11.87 ± 1.27 | 14.53 ± 1.69 | 10.79 ± 3.09 |
| C18:1n9 | 96 | 1811 | 1810 | 0.40 ± 0.12 | 0.61 ± 0.10 | 1.31 ± 0.71 |
| C20:0 | 90 | 2000 | 2000 | 0.14 ± 0.02 | 0.18 ± 0.02 | 0.27 ± 0.28 |
| Total | 100.00 | 100.00 | 100.00 | |||
| Straight-FA | 30.21 ± 3.07 | 35.99 ± 3.65 | 27.78 ± 5.75 | |||
| Iso-FA | 25.97 ± 1.46 | 24.03 ± 1.28 | 24.29 ± 2.93 | |||
| Anteiso-FA | 43.41 ± 1.32 | 39.39 ± 2.47 | 46.62 ± 3.55 | |||
| Unsaturated-FA | 0.40 ± 0.12 | 0.61 ± 0.10 | 1.31 ± 0.71 |
MSSim database spectral similarity, LRIref reference LRI, LRIexp experimental LRI, FAMEs profile is expressed as mean percentage (%) of total fatty acids ± standard deviation (SD)
Fig. 1.
Lipidomic analysis of Listeria monocytogenes exposed to nisin and fengycin. A Principal Component Analysis (PCA) and Partial Least Squares-Discriminant Analysis (PLS-DA) in fatty acids samples of L. monocytogenes treated with sub-lethal doses of fengycin (blue), nisin (green) and control (red). B Partial Least Squares-Discriminant Analysis (PLS-DA) with the variable importance projection (VIP) score. The boxes indicate relative concentration of the corresponding fatty acids in L. monocytogenes cultures for each specific group including the control and the treated with sub-lethal concentration of nisin and fengycin. C Comparison of relative quantification of individual PG molecular species in total lipid extracts (n = 3 biological replicates of treated bacteria) employing MS/MS using electrospray ionization in negative ionization modes (-), by precursor ion scan experiments, through selective monitoring of 153 Da fragment. The statistical analysis was performed using the two-tailed Student’s t test (#P > 0.05; *P < 0.05; **P < 0.01)
In previous studies, the characterization of membrane lipids in L. monocytogenes after exposure to some bacteriocins, such as nisin A, divergicin M35 and leucocin A, has been reported. These studies describe the modulation of the saturated–unsaturated membrane fatty acids ratio, including either reduction of the membrane fluidity coming from an improvement of the straight FAs concentrations [48, 49] or improving of the membrane fluidity through the increase of the branched FAs concentrations [50]. In this study, the nisin-treated samples showed increased amounts of long acyl chain straight saturated fatty acids (C16:0), in comparison with the control, suggesting increased thermodynamically stable interactions between acyl chains and thus increasing the membrane bilayer rigidity [51]. On contrary, the upregulation of branched fatty acid (anteiso-C17:0) improved the membrane fluidity on the fengycin-treated samples. Some studies showed that Gram-positive bacteria, such as Staphylococcus aureus and L. monocytogenes, increase the percentage of saturated fatty acids in bacterial cells forming biofilm, with a simultaneous decrease of iso-FAs and anteiso-FAs [52, 53].
LC–MS/MS-based lipidomic analysis
The intact polar lipid profiles detected in L. monocytogenes comprises mostly phosphatidylglycerols (PGs), diglycosyl-diacylglycerols (DGDGs) and diacylglycerols (DGs). Detected masses, class, partition number (PN) employed to simplify the process of identification, carbon number (CN), double bond number (DB) and retention time (RT) values are shown for each lipid species in Table S1. Sodium adducts were observed for DGDGs, while DGs were present as dehydrated protonated ions. On the other hand, LPGs and PGs were detected in negative ion mode, by precursor ion scan experiments, through selective monitoring of 153 Da fragments (Table S2). Elution of the lipid species occurred according to increasing PN (Fig. S2), derived from the equation PN = CN–2DB. The mass identification was carried out by comparing the obtained data with the information reported by Tatituri et al. [54] and further confirmed using the LIPID MAPS Structure Database [37]. Relative abundance percentages of all the reported PG compounds allowed us to detect significant differences among the treatments as shown in Fig. 1C. In particular, the most abundant PG species 30:0, 31:0 and 32:0 were found in higher amounts in fengycin-treated samples. MS/MS analysis confirmed the fatty acids 15:0/15:0, 15:0/16:0 and 15:0/17:0 as component of the PG species (Figure S3). The examination of the obtained MS data allowed us to correlate the relative abundance for some of the PG species in fengycin-treated samples to the higher quantification of anteiso branched FAs forms [54].
Several factors can influence the lipid profile of biological samples, including the culture medium and conditions, isolation and handling of lipid fraction, and even the activity of endogenous lipases ex vivo [55]. In this study, cardiolipin could not be identified due to a limitation of the analysis, while PE was detected in low amounts. Although these compounds are recognized as major lipids in Listeria, their abundances halved during elongation phase in Listeria innocua [56].
Proteomics analysis
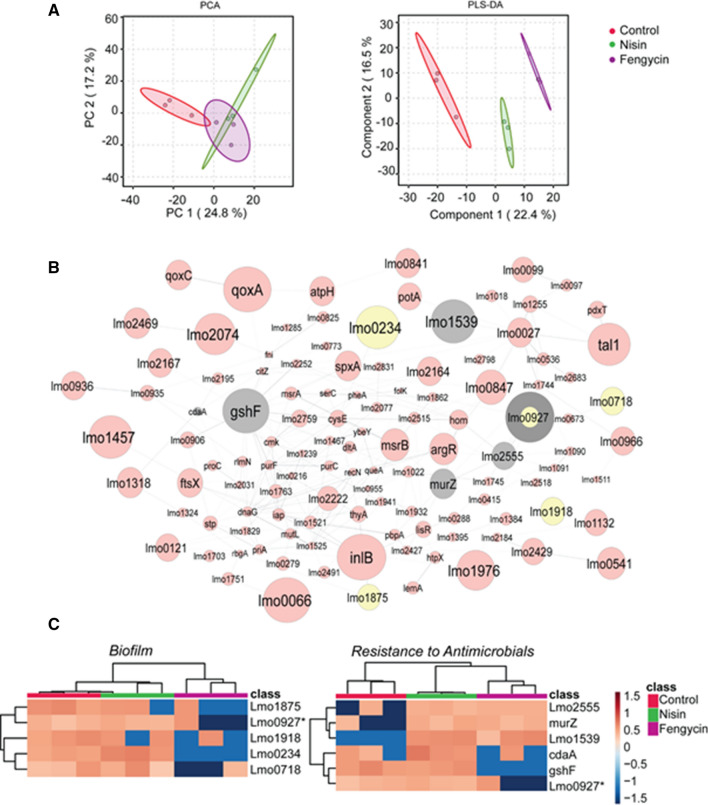
The changes of protein expression after the treatment with nisin and fengycin were investigated through a proteomics approach. A total of 951 LFQ (label-free quantified) proteins were detected, among the control and treated samples. After eliminating features that were not present in at least half of the samples, the database was reduced to 797 LFQ protein intensities. PCA showed discrimination between the control group and both treated sample groups, although no discrimination between nisin and fengycin groups was observed. Supervised PLS-DA analysis showed better discrimination among the three groups of samples (Fig. 2A) with 142 proteins being considered as drivers for the discrimination (VIP ≥ 1.0) (Table S3). This protein group was used to create a protein interaction network using functional enrichment analysis with STRING (LIT).With this network and according to their VIP scores, we prioritized protein families involved in biofilm regulation and the bacterial resistance process (Fig. 2B). Heatmaps with the proteins considered for their role on both resistance to antimicrobials and biofilm formation are shown in (Fig. 2C). The implications of these results are detailed in the next sections.
Fig. 2.
Proteomic analysis of Listeria monocytogenes exposed to nisin and fengycin. A Principal Component Analysis (PCA) and B Partial Least Squares-Discriminant Analysis (PLS-DA) in proteome samples of Listeria monocytogenes treated with fengycin (blue), nisin (green) and control (red). B Protein–protein interaction networks generated on STRING v11 database based on the identified proteins, all the proteins with VIP > 1 where reported in the figure, the node size represents the score obtained for each protein. The proteins involved in the biofilm process are colored in gray while those involved in the resistance process in yellow. C Heatmap of the proteins identified by PLS-DA statistical analysis. Samples are represented in columns, as the triplicate control samples (red), triplicate samples treated with sub-lethal nisin concentration (green) and triplicate sub-lethal fengycin concentration. Each colored cell on the map corresponds to the relative abundance of the proteins
Proteins associated with membrane stress resistance
The proteomic analysis also revealed a group of proteins associated with membrane resistance responses to AMPs (Table 2), including the LTA backbone synthase Lmo0927 protein, di-adenylate cyclase protein and glutathione synthase GshF protein that were downregulated exclusively in the fengycin-treated samples, while Lmo2555 protein, MurZ and Lmo1539 protein were upregulated in both treatments considered in this work.
Table 2.
Detected proteins involved in membrane stress response in Listeria monocytogenes, with the respective VIP score and gene name
| Protein name | VIP score | Regulation | Treatment | Gene name |
|---|---|---|---|---|
| Glutathione biosynthesis protein GshAB/(GshF) | 4.81 | Down | Fengycin | gshAB |
| Lmo0927 proteina | 2.76 | Down | Fengycin | lmo0927 |
| Diadenylate cyclase | 1.40 | Down | Fengycin | dacA/lmo2120 |
| Lmo2555 protein | 2.88 | Up | Fengycin/nisin | lmo2555 |
| UDP-N-acetylglucosamine 1-carboxyvinyltransferase 2 (MurZ) | 3.12 | Up | Fengycin/nisin | lmo2552 |
| Lmo1539 protein | 4.65 | Up | Fengycin/nisin | lmo1539 |
aProtein shared with the group of proteins associated with biofilm formation
The Lmo0927 protein is involved in LTA backbone synthesis, working also structurally on the bacterial cell membrane by extending the glycerol phosphate backbone chain. Besides Lmo0927, two glycosyltransferases, coded by lmo2555 and lmo2554 genes (the latter not differentially observed in this work), play a pivotal role to produce monoglucosyl-diacylglycerol and diglycosyl-diacylglycerols, respectively [57].
In general, the resistance strategy in Gram-positive bacteria includes modification of teichoic acids with D-alanine [58] and modification of negatively charged phosphatidylglycerol with positively charged lysine [59], to avoid interactions with cationic AMPs. In many Gram-positive bacteria, lipid species like monoglucosyl-diacylglycerols (MGDG) and diglycosyl-diacylglycerols (DGDG) act as membrane anchors for the LTA constituents of the cell wall [60]. The upregulation of Lmo2555 acting as MGDG synthase, observed in both nisin and fengycin treatments may act toward maintaining the membrane composition, as shown by the construction of deletion mutant, counteracting the complete loss of glycolipids with the consequent phenotypic changes [61]. The presence of MGDG in L. monocytogenes membrane guaranteed by Lmo2555 protein supports a protein-mediated adaptation against osmotic stressors like salts and sucrose [62], and maintaining the anionic lipid surface charge density as observed in Acholeplasma laidlawii [63]. Specifically, a simulation with bacterial MGDG synthetase demonstrated local enrichment of cardiolipin and phosphatidylglycerol in the immediate area of this protein through interaction with their positively charged residues [64]. In addition, the strong reduction of Lmo0927 could be related with the possible action of fengycin on the multi-drug resistance (MDR) transporters needed for full production of LTA, possibly via c-di-AMP efflux [65]. In support of this hypothesis, the proteomic results showed a downregulation of the related dacA protein (lmo2120 gene), a di-adenylate cyclase with c-di-AMP synthesis activity involved in type I interferon response within the host cell [66]. C-di-AMP showed a role on the osmotic homeostasis during environmental adaptation of Gram-positive bacteria [67], including resistance to acid and oxidative stress [68], resistance to extreme cell wall stress in S. aureus [69] and antibiotic tolerance associated with biofilm formation [70]. In this context, c-di-AMP has also been shown to play a role in Mycobacterium smegmatis for the negative regulation of fatty acid synthesis [71].
Another differentially expressed protein possibly associated with bacterial response to the membrane perturbation caused by antimicrobials was the glutathione (GSH) biosynthesis protein gshAB. In this study, gshAB was upregulated in nisin-treated samples, whereas the exposure to fengycin was related with the downregulation this protein in L. monocytogenes (Table 2). Although the knowledge about the physiological function of GSH in Gram-positive bacteria is limited, it has been shown to play important roles in cellular antioxidant mechanisms. Previous studies have demonstrated that bacterial cells containing GSH were capable of protecting the membrane structure when exposed to various environmental stresses, such as osmotic pressure, oxidative, and acid stress [72, 73]. Moreover, glutathione has emerged as a posttranslational regulator of protein function under conditions of oxidative stress, by the direct modification of proteins via glutathionylation.
Finally, both AMPs caused the upregulation of both UDP-N-acetylglucosamine 1-carboxyvinyltransferase (MurZ, lmo2552 gene) and putative glycerol uptake facilitator protein (GlpF-2, lmo1539 gene), which are associated to membrane stress resistance under the regulation of SigB. Lmo2552 protein has been associated with vancomycin tolerance in L. monocytogenes [74], whereas the Lmo1539 protein can be associated with the fact that a number of genes controlled by the key virulence factor PrfA are upregulated when L. monocytogenes growth in the presence of glycerol [75]. However, an alternative explanation for these results may come from dynamic simulations studies. Molecular dynamic simulation of the acyl-chain order in lipids around Escherichia coli aquaporins AqpZ and GlpF showed an increase in the acyl-chain order parameter. Yet the bilayer thickness decreases toward the protein and meets effectively the hydrophobic matching condition. The simulation also indicate that the lipid bilayer adapts to the channel by a hydrophobic matching condition reflecting the tendency of the lipid molecules for creating curved structures, and the transport function of the channel was modulated [76].
Differential biofilm organization after fengycin treatment
The fengycin treatment showed specific protein regulation toward the inhibition of biofilm formation, compared with the control and the nisin treatment. The proteomic analysis revealed a group of proteins suggesting the negative action on Listeria biofilm organization. Like other compounds acting against biofilm formation through inhibition of LTA synthesis [77], fengycin treatment strongly downregulated the Lmo0927 protein (LtaS). The LTA biosynthesis machinery represents an attractive target for the development of novel antimicrobial drugs against Gram-positive bacteria. The LTA deficiency in S. aureus altered the bacterial surface hydrophobicity and resulted in reduction of biofilm formation [78, 79]. Additional proteins with high VIP scores including putative pyruvate phosphate di-kinase regulatory protein, Lmo1875 protein, Lmo0718 protein, and Lmo1918 protein were observed (Table 3). All these proteins work on the biofilm promotion and were detected as downregulated in fengycin-treated samples.
Table 3.
Detected proteins involved in biofilm formation by Listeria monocytogenes, with the respective VIP score and gene name
| Protein name | VIP score | Regulation | Treatment | Gene name |
|---|---|---|---|---|
| Putative pyruvate phosphate dikinase regulatory protein | 4.54 | Down | Fengycin | lmo0234 |
| Lmo1875 protein | 2.71 | Down | Fengycin | lmo1875 |
| Lmo0718 protein | 2.92 | Down | Fengycin | lmo0718 |
| Lmo1918 protein | 2.90 | Down | Fengycin | lmo1918 |
| Lmo0927 proteina | 2.76 | Down | Fengycin | lmo0927 |
aProtein shared with the group of proteins associated with the membrane stress response
The putative pyruvate phosphate dikinase regulatory protein and Lmo1875 protein, have been previously reported to be among the upregulated proteins in sessile L. monocytogenes cells inside biofilms, when treated with lactocin AL705 in a comparative study with planktonic cells [80]. The flagellum contributes to the biofilm organization and its biosynthesis require dozens of genes from a large operon working on the bacterial motility. The operon also includes the protein Lmo0718, which was detected as downregulated in fengycin-treated samples. Thus, inhibition of flagella-related proteins may represent a possible loss on the biofilm building capacity [81]. Another downregulated protein in the fengycin-treated samples was the Lmo1918 protein with unknown function, although a deficient mutant effectively reduced the propensity of L. monocytogenes to form biofilms [82].
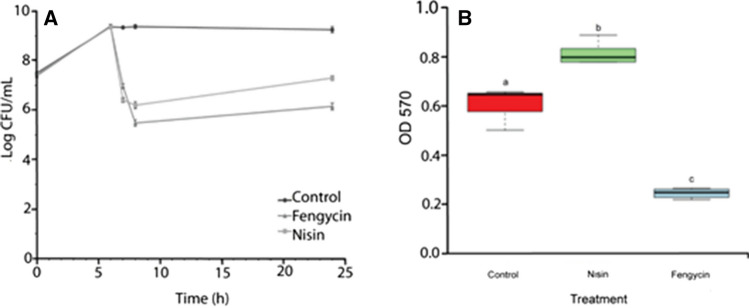
The growth curves of L. monocytogenes treated with AMPs showed a rapid reduction of viable cell numbers after the addition of nisin and fengycin (Fig. 3A). At 7 h cultivation, when the bacterial cells were collected for the omics analysis, a similar number of viable cells was revealed for both treatments. However, cell counts of L. monocytogenes treated with nisin were higher than fengycin treatment at 24 h. Moreover, experimental biofilm quantification assay permitted to detect significant differences among samples (Fig. 3B). Biofilm promotion was induced by the treatment with sub-lethal concentration of nisin (ODNis > ODCont), while a strong inhibition of biofilm was observed by the action of fengycin (ODFen < ODCont).
Fig. 3.
A Growth curves of Listeria monocytogenes in BHI broth during 24 h at 37 °C. The bacterial cultures were treated with antimicrobial peptides (nisin and fengycin) after 6 h cultivation. The data represent the mean of three experiments and the bars indicate standard deviations. Significant differences (P < 0.05) by Tukey’s test were observed among the microbial growth curves only after adding the antimicrobials. B Boxplot of spectrophotometry data showing the optical density (OD570) of L. monocytogenes biofilm after 24 h growth in BHI broth under different treatments: negative control, nisin (0.1 μg/mL) and fengycin (100 AU/mL), respectively. Different lower case letter over the plot of each treatment indicates significant differences (P < 0.05)
Concluding remarks
The exposure of L. monocytogenes to sub-lethal concentrations of nisin and fengycin resulted in different molecular responses. The proteomics analysis enabled us to illuminate possible mechanisms involved with membrane resistance in both AMP treatments, as well as the propensity of biofilm inhibition by the fengycin-treated samples.
Lipidomics analysis of the L. monocytogenes fatty acids profiles showed that membranes are enriched in branched FAs, as reported previously [83, 84]. Similarly, higher amounts of branched FAs have been reported in other Gram-positive bacteria, such as Staphylococcus and Bacillus species [85], conferring increased membrane fluidity as compared with other bacterial groups. Alterations in membrane lipid composition were observed after both nisin and fengycin treatments, suggesting an alteration of membrane fluidity as bacterial response to these AMPs. The variation of relative abundance of PG species in the fengycin-treated samples might be associated to the downregulation of Lmo0927 protein (LTA synthase) and with the related variation of available LTA [86].
Bacteria can randomly differentiate into persister cells during regular growth, but this phenotypic change can be also induced by environmental signals indicating potential threats for the bacteria. In bacterial populations, this phenomenon is largely controlled by stress signaling pathways, such as the general stress response or the SOS response. Thus, formation of persister cells is stimulated under conditions that favor the activation of these signaling pathways, such as biofilm formation, hostile environments, and response to injury caused by sub-lethal doses of antimicrobials [87]. Some studies indicate that fengycin auto-aggregates by inserting their acyl chains into model membrane bilayers, which is related to membrane perturbation and damage [26, 27]. Fengycin binding and aggregation disturb phospholipid bilayers, appearing to affect bacterial membranes in a dose-dependent manner [88], playing a key role in cell damage. Thus, we hypothesize that the exposure to sub-lethal concentration of fengycin induces a stress–response mechanism that results in altered membrane fluidity, allowing the surviving cells to face the potential damage caused by surface-active lipopeptides like fengycin. This mechanism could involve the regulation of proteins related to the synthesis of surface lipids, as suggested above, and mediated by general stress response regulons, such as σB, VirB or PrfA, which have been largely associated with the resistance of L. monocytogenes to environmental stresses [89].
One of the L. monocytogenes responses to the nisin exposure was the switch from planktonic to sessile organization, building biofilms to tolerate the environmental stress. In agreement, the treatment with sub-lethal nisin concentration induces the upregulation of proteins associated with biofilm formation in this species [14]. Contrarily, the treatment of L. monocytogenes with sub-lethal fengycin concentration showed an improvement of the BCFAs species with possible preference for a planktonic state, which can be related to the effective reduction in biofilm formation. Furthermore, the anteiso BCFAs were observed as a L. monocytogenes resistance mechanism against the stress produced by host defense mechanisms including both AMPs and peptidoglycan hydrolases inside the phagosome. This fatty acid modulation has also shown to compromise the production of the key virulence factor listeriolysin O, suggesting an influence on the virulence regulation [90].
The biofilm formation and membrane modification represent two of the several mechanisms involved in bacterial resistance. Considering the effect of differentially regulated proteins on the membrane structure, the proteins related with LTA metabolism should be emphasized. LTAs play a pivotal role in the ability of Gram-positive bacteria to grow, replicate and form biofilm, modulating the membrane structural stability and integrity under different physiological conditions [91, 92].
This study provides evidence that there is a link between the use of a sub-lethal dose of fengycin and inhibition of biofilm formation in L. monocytogenes. It was also confirmed that the bacterial physiology was reorganized to promote the biofilm formation when a sub-lethal concentration of nisin was employed. We hypothesize that several proteins contribute to increase the cell membrane stability through protein–lipid interactions. Together, our results provide new insights into the molecular mechanisms of biofilm inhibition in L. monocytogenes, which could be leveraged as possible new strategies to confront L. monocytogenes and other Gram-positive bacteria, including multi-resistant strains.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
Authors thank Dr. Adriana Franco Paes Leme, M.Sc. Romenia Ramos Domingues and Dr. Bianca Alves Pauletti, from Mass Spectrometry Laboratory of LNBio/CNPEM (Campinas, Brazil) for helping us to perform the LC-MS/MS procedures.
Author contributions
PS, AB, LM, and MdA contributed to the study conception and design. Material preparation, data collection, data analysis, and statistical analysis were performed by PS, FFV, GM, DD, DP and GVC. The manuscript was written by PS and critically revised by AB. All the authors read and approved the final manuscript.
Funding
This work received financial support from CNPq (Brasilia, Brazil) [grant 308880/2021–8]. PS was a former recipient of a PhD fellowship from CAPES. PS and DP were supported through the Deutsche Forschungsgemeinschaft through the CMFI Cluster of Excellence (EXC 2124).
Availability of data and material
The lipidomics and proteomics mass spectrometry data are publicly available through the MassIVE repository under the following accession numbers: MSV000088672 and MSV000088738.
Declarations
Competing interests
Authors declare no conflicts of interest.
Ethical approval
This research did not involve human participants.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Radoshevich L, Cossart P. Listeria monocytogenes: towards a complete picture of its physiology and pathogenesis. Nature Rev Microbiol. 2018;16:32–46. doi: 10.1038/nrmicro.2017.126. [DOI] [PubMed] [Google Scholar]
- 2.Duze ST, Marimani M, Patel M. Tolerance of Listeria monocytogenes to biocides used in food processing environments. Food Microbiol. 2021;97:103758. doi: 10.1016/j.fm.2021.103758. [DOI] [PubMed] [Google Scholar]
- 3.Zhao X, Kuipers OP. Identification and classification of known and putative antimicrobial compounds produced by a wide variety of Bacillales species. BMC Genomics. 2016;17:882. doi: 10.1186/s12864-016-3224-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chikindas ML, Weeks R, Drider D, Chistyakov VA, Dicks LM. Functions and emerging applications of bacteriocins. Curr Opin Biotechnol. 2018;49:23–28. doi: 10.1016/j.copbio.2017.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fira D, Dimkić I, Berić T, Lozo J, Stanković S. Biological control of plant pathogens by Bacillus species. J Biotechnol. 2018;285:44–55. doi: 10.1016/j.jbiotec.2018.07.044. [DOI] [PubMed] [Google Scholar]
- 6.Cotter PD, Hill C, Ross RP. Bacteriocins: developing innate immunity for food. Nature Rev Microbiol. 2005;3:777–788. doi: 10.1038/nrmicro1273. [DOI] [PubMed] [Google Scholar]
- 7.Alvarez-Sieiro P, Montalbán-López M, Mu D, Kuipers OP. Bacteriocins of lactic acid bacteria: extending the family. Appl Microbiol Biotechnol. 2016;100:2939–2951. doi: 10.1007/s00253-016-7343-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang W, Zhang Z, Han X, Wang J, Tang J, Dong S, Wang E. Concentration-dependent behavior of nisin interaction with supported bilayer lipid membrane. Biophys Chem. 2002;99:271–279. doi: 10.1016/S0301-4622(02)00227-2. [DOI] [PubMed] [Google Scholar]
- 9.Prince A, Sandhu P, Ror P, Dash E, Sharma S, Arakha M, Jha S, Akhter Y, Saleem M. Lipid-II independent antimicrobial mechanism of nisin depends on its crowding and degree of oligomerization. Sci Rep. 2016;6:37908. doi: 10.1038/srep37908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu S, Yu PL, Wheeler D, Flint S. Transcriptomic study on persistence and survival of Listeria monocytogenes following lethal treatment with nisin. J Global Antimicrob Resist. 2018;15:25–31. doi: 10.1016/j.jgar.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 11.Jiang X, Geng Y, Ren S, Yu T, Li Y, Liu G, Wang H, Meng H, Shi L. The VirAB-VirSR-AnrAB multicomponent system is involved in resistance of Listeria monocytogenes EGD-e to cephalosporins, bacitracin, nisin, benzalkonium chloride, and ethidium bromide. Appl Environ Microbiol. 2019;85:01470–1519. doi: 10.1128/AEM.01470-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malekmohammadi S, Kodjovi KK, Sherwood J, Bergholz TM. Genetic and environmental factors influence Listeria monocytogenes nisin resistance. J Appl Microbiol. 2017;123:262–270. doi: 10.1111/jam.13479. [DOI] [PubMed] [Google Scholar]
- 13.Wambui J, Eshwar AK, Aalto-Araneda M, Pöntinen A, Stevens MJ, Njage PM, Tasara T. The analysis of field strains isolated from food, animal and clinical sources uncovers natural mutations in Listeria monocytogenes nisin resistance genes. Front Microbiol. 2020;11:549531. doi: 10.3389/fmicb.2020.549531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stincone P, Miyamoto KN, Timbe PPR, Lieske I, Brandelli A. Nisin influence on the expression of Listeria monocytogenes surface proteins. J Proteom. 2020;226:103906. doi: 10.1016/j.jprot.2020.103906. [DOI] [PubMed] [Google Scholar]
- 15.Ongena M, Jacques P. Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol. 2008;16:115–125. doi: 10.1016/j.tim.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 16.Stincone P, Veras FF, Pereira JQ, Mayer FQ, Varela APM, Brandelli A. Diversity of cyclic antimicrobial lipopeptides from Bacillus P34 revealed by functional annotation and comparative genome analysis. Microbiol Res. 2020;238:126515. doi: 10.1016/j.micres.2020.126515. [DOI] [PubMed] [Google Scholar]
- 17.Vanittanakom N, Loeffler W, Koch U, Jung G. Fengycin-a novel antifungal lipopeptide antibiotic produced by Bacillus subtilis F-29-3. J Antibiot. 1986;39:888–901. doi: 10.7164/antibiotics.39.888. [DOI] [PubMed] [Google Scholar]
- 18.Medeot DB, Fernandez M, Morales GM, Jofré E. Fengycins from Bacillus amyloliquefaciens MEP218 exhibit antibacterial activity by producing alterations on the cell surface of the pathogens Xanthomonas axonopodis pv vesicatoria and Pseudomonas aeruginosa PA01. Front Microbiol. 2020 doi: 10.3389/fmicb.2019.03107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piewngam P, Zheng Y, Nguyen TH, Dickey SW, Joo HS, Villaruz AE, Glose KA, Fisher EL, Hunt RL, Li B, Chiou J, Pharkjaksu S, Khongthong S, Cheung GYC, Kiratisin P, Otto M. Pathogen elimination by probiotic Bacillus via signalling interference. Nature. 2018;562:532–537. doi: 10.1038/s41586-018-0616-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fischetti VA, Novick RP, Ferretti JJ, Portnoy DA, Braunstein M, Rood JI. Gram-Positive Pathogens. 3. Washington DC: ASM Press; 2019. [Google Scholar]
- 21.Crandall AD, Montville TJ. Nisin resistance in Listeria monocytogenes ATCC 700302 is a complex phenotype. Appl Environ Microbiol. 1998;64:231–237. doi: 10.1128/AEM.64.1.231-237.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sahl HG, Shai Y. Bacterial resistance to antimicrobial peptides. Biochim Biophys Acta. 2015;1848:3019–3020. doi: 10.1016/j.bbamem.2015.08.009. [DOI] [PubMed] [Google Scholar]
- 23.Omardien S, Brul S, Zaat SA. Antimicrobial activity of cationic antimicrobial peptides against gram-positives: current progress made in understanding the mode of action and the response of bacteria. Front Cell Dev Biol. 2016;4:111. doi: 10.3389/fcell.2016.00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee TH, Hall KN, Aguilar MI. Antimicrobial peptide structure and mechanism of action: a focus on the role of membrane structure. Curr Top Med Chem. 2016;16:25–39. doi: 10.2174/1568026615666150703121700. [DOI] [PubMed] [Google Scholar]
- 25.Sani MA, Separovic F. How membrane-active peptides get into lipid membranes. Acc Chem Res. 2016;49:1130–1138. doi: 10.1021/acs.accounts.6b00074. [DOI] [PubMed] [Google Scholar]
- 26.Horn JN, Cravens A, Grossfield A. Interactions between fengycin and model bilayers quantified by coarse-grained molecular dynamics. Biophys J. 2013;105:1612–1623. doi: 10.1016/j.bpj.2013.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sur S, Romo TD, Grossfield A. Selectivity and mechanism of fengycin, an antimicrobial lipopeptide, from molecular dynamics. J Phys Chem B. 2018;122:2219–2226. doi: 10.1021/acs.jpcb.7b11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Donnarumma D, Maestri C, Giammarinaro PI, Capriotti L, Bartolini E, Veggi D, Petracca R, Scarselli M, Norais N. Native state organization of outer membrane porins unraveled by HDx-MS. J Proteome Res. 2018;17:1794–1800. doi: 10.1021/acs.jproteome.7b00830. [DOI] [PubMed] [Google Scholar]
- 29.Brauge T, Sadovskaya I, Faille C, Benezech T, Maes E, Guerardel Y, Midelet-Bourdin G. Teichoic acid is the major polysaccharide present in the Listeria monocytogenes biofilm matrix. FEMS Microbiol Lett. 2016;363(2):229. doi: 10.1093/femsle/fnv229. [DOI] [PubMed] [Google Scholar]
- 30.Silva S, Teixeira P, Oliveira R, Azeredo J. Adhesion to and viability of Listeria monocytogenes on food contact surfaces. J Food Protec. 2008;71:1379–1385. doi: 10.4315/0362-028X-71.7.1379. [DOI] [PubMed] [Google Scholar]
- 31.Kocot AM, Olszewska MA. Biofilm formation and microscopic analysis of biofilms formed by Listeria monocytogenes in a food processing context. LWT Food Sci Technol. 2017;84:47–57. doi: 10.1016/j.lwt.2017.05.042. [DOI] [Google Scholar]
- 32.Motta AS, Flores FS, Souto AA, Brandelli A. Antibacterial activity of a bacteriocin-like substance produced by Bacillus sp. P34 that targets the bacterial cell envelope. Antonie Van Leeuwenhoek. 2008;93:275–284. doi: 10.1007/s10482-007-9202-2. [DOI] [PubMed] [Google Scholar]
- 33.Hines KM, Shen T, Ashford NK, Waalkes A, Penewit K, Holmes EA, McLean K, Salipante SJ, Werth BJ, Xu L. Occurrence of cross-resistance and β-lactam seesaw effect in glycopeptide-, lipopeptide-and lipoglycopeptide-resistant MRSA correlates with membrane phosphatidylglycerol levels. J Antimicrob Chemother. 2020;75:1182–1186. doi: 10.1093/jac/dkz562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kováčik J, Micalizzi G, Dresler S, Wójciak-Kosiord M, Ragosta E, Mondello L. The opposite nitric oxide modulators do not lead to the opposite changes of metabolites under cadmium excess. J Plant Physiol. 2020;252:153228. doi: 10.1016/j.jplph.2020.153228. [DOI] [PubMed] [Google Scholar]
- 35.Rigano F, Arena P, Mangraviti D, Donnarumma D, Dugo P, Donato P, Mondello L, Micalizzi G. Identification of high-value generating molecules from the wastes of tuna fishery industry by liquid chromatography and gas chromatography hyphenated techniques with automated sample preparation. J Sep Sci. 2021;44:1571. doi: 10.1002/jssc.202100108. [DOI] [PubMed] [Google Scholar]
- 36.Sud M, Fahy E, Cotter D, Brown A, Dennis EA, Glass CK, Merrill AH, Jr, Murphy RC, Raetz CRH, Russell DW, Subramaniam S. Lmsd: Lipid maps structure database. Nucleic Acids Res. 2007;35:D527–D532. doi: 10.1093/nar/gkl838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adusumilli R, Mallick P. Data conversion with ProteoWizard msConvert. In: Comai L, Katz J, Mallick P, editors. Proteomics: Methods in Enzymology. NY: Springer; 2017. pp. 339–368. [DOI] [PubMed] [Google Scholar]
- 38.Petras D, Phelan VV, Acharya D, Allen AE, Aron AT, Bandeira N, et al. GNPS Dashboard: collaborative exploration of mass spectrometry data in the web browser. Nature Meth. 2021;19:134–136. doi: 10.1038/s41592-021-01339-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pinilla CMB, Stincone P, Brandelli A. Proteomic analysis reveals differential responses of Listeria monocytogenes to free and nanoencapsulated nisin. Int J Food Microbiol. 2021;346:109170. doi: 10.1016/j.ijfoodmicro.2021.109170. [DOI] [PubMed] [Google Scholar]
- 40.Stincone P, Comerlato CB, Brandelli A. Proteomic analysis of Listeria monocytogenes exposed to free and nanostructured antimicrobial lipopeptides. Mol Omics. 2021;17:426–437. doi: 10.1039/D0MO00178C. [DOI] [PubMed] [Google Scholar]
- 41.Villén J, Gygi SP. The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nature Protoc. 2008;3:1630–1638. doi: 10.1038/nprot.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using stage tips. Nature Protoc. 2007;2:1896. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 43.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized ppb-range mass accuracies and proteome-wide protein quantification. Nature Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 44.Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 45.Stepanović S, Vuković D, Hola V, Bonaventura GD, Djukić S, Ćirković I, Ruzicka F. Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS. 2007;115:891–899. doi: 10.1111/j.1600-0463.2007.apm_630.x. [DOI] [PubMed] [Google Scholar]
- 46.Chong IG, Jun CH. Performance of some variable selection methods when multicollinearity is present. Chemom Intell Lab Syst. 2005;78:103–112. doi: 10.1016/j.chemolab.2004.12.011. [DOI] [Google Scholar]
- 47.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, von Mering C. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martínez B, Rodríguez A. Antimicrobial susceptibility of nisin resistant Listeria monocytogenes of dairy origin. FEMS Microbiol Lett. 2005;252:67–72. doi: 10.1016/j.femsle.2005.08.025. [DOI] [PubMed] [Google Scholar]
- 49.Naghmouchi K, Drider D, Hammami R, Fliss I. Effect of antimicrobial peptides divergicin M35 and nisin A on Listeria monocytogenes LSD530 potassium channels. Curr Microbiol. 2008;56:609–612. doi: 10.1007/s00284-008-9134-8. [DOI] [PubMed] [Google Scholar]
- 50.Vadyvaloo V, Hastings JW, van der Merwe MJ, Rautenbach M. Membranes of class IIa bacteriocin-resistant Listeria monocytogenes cells contain increased levels of desaturated and short-acyl-chain phosphatidylglycerols. Appl Environ Microbiol. 2002;68:5223–5230. doi: 10.1128/AEM.68.11.5223-5230.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Denich TJ, Beaudette LA, Lee H, Trevors JT. Effect of selected environmental and physico-chemical factors on bacterial cytoplasmic membranes. J Microbiol Meth. 2003;52:149–182. doi: 10.1016/S0167-7012(02)00155-0. [DOI] [PubMed] [Google Scholar]
- 52.Gianotti A, Serrazanetti D, Kamdem SS, Guerzoni ME. Involvement of cell fatty acid composition and lipid metabolism in adhesion mechanism of Listeria monocytogenes. Int J Food Microbiol. 2008;123:9–17. doi: 10.1016/j.ijfoodmicro.2007.11.039. [DOI] [PubMed] [Google Scholar]
- 53.Dubois-Brissonnet F, Trotier E, Briandet R. The biofilm lifestyle involves an increase in bacterial membrane saturated fatty acids. Front Microbiol. 2016;7:1673. doi: 10.3389/fmicb.2016.01673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tatituri RV, Wolf BJ, Brenner MB, Turk J, Hsu FF. Characterization of polar lipids of Listeria monocytogenes by HCD and low-energy CAD linear ion-trap mass spectrometry with electrospray ionization. Anal Bioanal Chem. 2015;407:2519–2528. doi: 10.1007/s00216-015-8480-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Furse S, Egmond MR, Killian JA. Isolation of lipids from biological samples. Mol Membr Biol. 2015;32:55–64. doi: 10.3109/09687688.2015.1050468. [DOI] [PubMed] [Google Scholar]
- 56.Furse S, Jakubec M, Rise F, Williams HE, Rees CED, Halskau Ø. Evidence that Listeria innocua modulates its membrane’s stored curvature elastic stress, but not fluidity, through the cell cycle. Sci Rep. 2017;7:8012. doi: 10.1038/s41598-017-06855-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Webb AJ, Karatsa-Dodgson M, Gründling A. Two-enzyme systems for glycolipid and polyglycerolphosphate lipoteichoic acid synthesis in Listeria monocytogenes. Mol Microbiol. 2009;74:299–314. doi: 10.1111/j.1365-2958.2009.06829.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abachin E, Poyart C, Pellegrini E, Milohanic E, Fiedler F, Berche P, Trieu-Cuot P. Formation of D-alanyl-lipoteichoic acid is required for adhesion and virulence of Listeria monocytogenes. Mol Microbiol. 2002;43:1–14. doi: 10.1046/j.1365-2958.2002.02723.x. [DOI] [PubMed] [Google Scholar]
- 59.Thedieck K, Hain T, Mohamed W, Tindall BJ, Nimtz M, Chakraborty T, Wehland J, Jänsch L. The MprF protein is required for lysinylation of phospholipids in listerial membranes and confers resistance to cationic antimicrobial peptides (CAMPs) on Listeria monocytogenes. Mol Microbiol. 2006;62:1325–1339. doi: 10.1111/j.1365-2958.2006.05452.x. [DOI] [PubMed] [Google Scholar]
- 60.Neuhaus FC, Baddiley J. A continuum of anionic charge: Structures and functions of D-alanyl-teichoic acids in Gram-positive bacteria. Microbiol Mol Biol Rev. 2003;67:686–723. doi: 10.1128/MMBR.67.4.686-723.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Theilacker C, Sava I, Sanchez-Carballo P, Bao Y, Kropec A, Grohmann E, Holst O, Huebner J. Deletion of the glycosyltransferase bgsB of Enterococcus faecalis leads to a complete loss of glycolipids from the cell membrane and to impaired biofilm formation. BMC Microbiol. 2011;11:67. doi: 10.1186/1471-2180-11-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wikström M, Xie J, Bogdanov M, Mileykovskaya E, Heacock P, Wieslander Å, Dowhan W. Monoglucosyldiacylglycerol, a foreign lipid, can substitute for phosphatidylethanolamine in essential membrane-associated functions in Escherichia coli. J Biol Chem. 2004;279:10484–10493. doi: 10.1074/jbc.M310183200. [DOI] [PubMed] [Google Scholar]
- 63.Edman M, Berg S, Storm P, Wikström M, Vikström S, Öhman A, Wieslander Å. Structural features of glycosyltransferases synthesizing major bilayer and nonbilayer-prone membrane lipids in Acholeplasma laidlawii and Streptococcus pneumoniae. J Biol Chem. 2003;278:8420–8428. doi: 10.1074/jbc.M211492200. [DOI] [PubMed] [Google Scholar]
- 64.Ge C, Gómez-Llobregat J, Skwark MJ, Ruysschaert JM, Wieslander Å, Lindén M. Membrane remodeling capacity of a vesicle-inducing glycosyltransferase. FEBS J. 2014;281:3667–3684. doi: 10.1111/febs.12889. [DOI] [PubMed] [Google Scholar]
- 65.Tadmor K, Pozniak Y, Burg Golani T, Lobel L, Brenner M, Sigal N, Herskovits AA. Listeria monocytogenes MDR transporters are involved in LTA synthesis and triggering of innate immunity during infection. Front Cell Infect Microbiol. 2014;4:16. doi: 10.3389/fcimb.2014.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Woodward JJ, Iavarone AT, Portnoy DA. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science. 2010;328:1703–1705. doi: 10.1126/science.1189801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Devaux L, Sleiman D, Mazzuoli MV, Gominet M, Lanotte P, Trieu-Cuot P, Kaminski PA, Firon A. Cyclic di-AMP regulation of osmotic homeostasis is essential in Group B Streptococcus. PLoS Genet. 2018;14:e1007342. doi: 10.1371/journal.pgen.1007342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rallu F, Gruss A, Ehrlich SD, Maguin E. Acid-and multistress-resistant mutants of Lactococcus lactis: identification of intracellular stress signals. Mol Microbiol. 2000;35:517–528. doi: 10.1046/j.1365-2958.2000.01711.x. [DOI] [PubMed] [Google Scholar]
- 69.Corrigan RM, Abbott JC, Burhenne H, Kaever V, Gründling A. c-di-AMP is a new second messenger in Staphylococcus aureus with a role in controlling cell size and envelope stress. PLoS Pathog. 2011;7:e1002217. doi: 10.1371/journal.ppat.1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Römling U, Balsalobre C. Biofilm infections, their resilience to therapy and innovative treatment strategies. J Int Med. 2012;272:541–561. doi: 10.1111/joim.12004. [DOI] [PubMed] [Google Scholar]
- 71.Zhang L, Li W, He ZG. DarR, a TetR-like transcriptional factor, is a cyclic di-AMP-responsive repressor in Mycobacterium smegmatis. J Biol Chem. 2013;288:3085–3096. doi: 10.1074/jbc.M112.428110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang J, Du GC, Zhang Y, Liao XY, Wang M, Li Y, Chen J. Glutathione protects Lactobacillus sanfranciscensis against freeze-thawing, freeze-drying, and cold treatment. Appl Environ Microbiol. 2010;76:2989–2996. doi: 10.1128/AEM.00026-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Masip L, Veeravalli K, Georgiou G. The many faces of glutathione in bacteria. Antioxid Redox Signal. 2006;8:753–762. doi: 10.1089/ars.2006.8.753. [DOI] [PubMed] [Google Scholar]
- 74.Shin JH, Kim J, Kim SM, Kim S, Lee JC, Ahn JM, Cho JY. σB-dependent protein induction in Listeria monocytogenes during vancomycin stress. FEMS Microbiol Lett. 2010;308:94–100. doi: 10.1111/j.1574-6968.2010.01998.x. [DOI] [PubMed] [Google Scholar]
- 75.Joseph B, Mertins s, Stoll R, Schar J, Umesha KR, Luo Q, Muller-Altrock S, Goebel W, Glycerol metabolism and PrfA activity in Listeria monocytogenes. J Bacteriol. 2008;190:5412–5430. doi: 10.1128/JB.00259-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jensen MØ, Mouritsen OG. Lipids do influence protein function-the hydrophobic matching hypothesis revisited. Biochim Biophys Acta-Biomembranes. 2004;1666:205–226. doi: 10.1016/j.bbamem.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 77.Naclerio GA, Onyedibe KI, Sintim HO. Lipoteichoic acid biosynthesis inhibitors as potent inhibitors of S. aureus and E. faecalis growth and biofilm formation. Molecules. 2020;25:2277. doi: 10.3390/molecules25102277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fedtke I, Mader D, Kohler T, Moll H, Nicholson G, Biswas R, Henseler K, Götz F, Zähringer U, Peschel A. A Staphylococcus aureus ypfP mutant with strongly reduced lipoteichoic acid (LTA) content: LTA governs bacterial surface properties and autolysin activity. Mol Microbiol. 2007;65:1078–1091. doi: 10.1111/j.1365-2958.2007.05854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Percy MG, Grundling A. Lipoteichoic acid synthesis and function in gram-positive bacteria. Annu Rev Microbiol. 2014;68:81–100. doi: 10.1146/annurev-micro-091213-112949. [DOI] [PubMed] [Google Scholar]
- 80.Melian C, Castellano P, Segli F, Mendoza LM, Vignolo GM. Proteomic analysis of Listeria monocytogenes FBUNT during biofilm formation at 10°C in response to lactocin AL705. Front Microbiol. 2021;12:43. doi: 10.3389/fmicb.2021.604126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Toledo-Arana A, Lasa I. Advances in bacterial transcriptome understanding: from overlapping transcription to the excludon concept. Mol Microbiol. 2020;113:593–602. doi: 10.1111/mmi.14456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chang Y, Gu W, Fischer N, McLandsborough L. Identification of genes involved in Listeria monocytogenes biofilm formation by mariner-based transposon mutagenesis. Appl Microbiol Biotechnol. 2012;93:2051–2062. doi: 10.1007/s00253-011-3719-z. [DOI] [PubMed] [Google Scholar]
- 83.Mastronicolis SK, German JB, Smith GM. Diversity of the polar lipids of the food-borne pathogen Listeria monocytogenes. Lipids. 1996;31:635–640. doi: 10.1007/BF02523834. [DOI] [PubMed] [Google Scholar]
- 84.Mastronicolis SK, German JB, Smith GM. Isolation and fatty acid analysis of neutral and polar lipids of the food bacterium Listeria monocytogenes. Food Chem. 1996;57:451–456. doi: 10.1016/0308-8146(95)00241-3. [DOI] [Google Scholar]
- 85.Whittaker P, Fry FS, Curtis SK, Al-Khaldi SF, Mossoba MM, Yurawecz MP, Dunkel VC. Use of fatty acid profiles to identify food-borne bacterial pathogens and aerobic endospore-forming bacilli. J Agric Food Chem. 2005;53:3735–3742. doi: 10.1021/jf040458a. [DOI] [PubMed] [Google Scholar]
- 86.Seki T, Furumi T, Hashimoto M, Hara H, Matsuoka S. Activation of extracytoplasmic function sigma factors upon removal of glucolipids and reduction of phosphatidylglycerol content in Bacillus subtilis cells lacking lipoteichoic acid. Genes Genet Syst. 2019;94:71–90. doi: 10.1266/ggs.18-00046. [DOI] [PubMed] [Google Scholar]
- 87.Harms A, Maisonneuve E, Gerdes K. Mechanisms of bacterial persistence during stress and antibiotic exposure. Science. 2016;354:4268. doi: 10.1126/science.aaf4268. [DOI] [PubMed] [Google Scholar]
- 88.Ma Z, Wang N, Hu J, Wang S. Isolation and characterization of a new iturinic lipopeptide, mojavensin A produced by a marine-derived bacterium Bacillus mojavensis B0621A. J Antibiot. 2012;65:317–322. doi: 10.1038/ja.2012.19. [DOI] [PubMed] [Google Scholar]
- 89.Krawczyk-Balska A, Markiewicz Z. The intrinsic cephalosporin resistome of Listeria monocytogenes in the context of stress response, gene regulation, pathogenesis and therapeutics. J Appl Microbiol. 2016;120:251–265. doi: 10.1111/jam.12989. [DOI] [PubMed] [Google Scholar]
- 90.Sun Y, Wilkinson BJ, Standiford TJ, Akinbi HT, O'Riordan MX. Fatty acids regulate stress resistance and virulence factor production for Listeria monocytogenes. J Bacteriol. 2012;194:5274–5284. doi: 10.1128/JB.00045-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bharatiya B, Wang G, Rogers SE, Pedersen JS, Mann S, Briscoe WH. Mixed liposomes containing gram-positive bacteria lipids: lipoteichoic acid (LTA) induced structural changes. Colloids Surf B. 2021;199:111551. doi: 10.1016/j.colsurfb.2020.111551. [DOI] [PubMed] [Google Scholar]
- 92.Hesser AR, Schaefer K, Lee W, Walker S. Lipoteichoic acid polymer length is determined by competition between free starter units. Proc Natl Acad Sci USA. 2020;117:29669–29676. doi: 10.1073/pnas.2008929117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The lipidomics and proteomics mass spectrometry data are publicly available through the MassIVE repository under the following accession numbers: MSV000088672 and MSV000088738.