Abstract
Since its discovery in 1981, the Ku complex has been extensively studied under multiple cellular contexts, with most work focusing on Ku in terms of its essential role in non-homologous end-joining (NHEJ). In this process, Ku is well-known as the DNA-binding subunit for DNA-PK, which is central to the NHEJ repair process. However, in addition to the extensive study of Ku’s role in DNA repair, Ku has also been implicated in various other cellular processes including transcription, the DNA damage response, DNA replication, telomere maintenance, and has since been studied in multiple contexts, growing into a multidisciplinary point of research across various fields. Some advances have been driven by clarification of Ku’s structure, including the original Ku crystal structure and the more recent Ku–DNA-PKcs crystallography, cryogenic electron microscopy (cryoEM) studies, and the identification of various post-translational modifications. Here, we focus on the advances made in understanding the Ku heterodimer outside of non-homologous end-joining, and across a variety of model organisms. We explore unique structural and functional aspects, detail Ku expression, conservation, and essentiality in different species, discuss the evidence for its involvement in a diverse range of cellular functions, highlight Ku protein interactions and recent work concerning Ku-binding motifs, and finally, we summarize the clinical Ku-related research to date.
Keywords: Ku heterodimer, Ku-binding motifs, Telomere maintenance, Genome stability, Non-homologous end-joining, Cancer
Introduction
Ku has been implicated in a diverse range of functions including transcription, DNA replication, innate immunity, the DNA damage response, and telomere maintenance, many of which are mediated through Ku-protein and Ku–RNA interactions [1]. Ku has also been researched in less spotlighted functions such as nucleolar transcript regulation [2], Bax-mediated apoptosis [3], and viral double-stranded DNA detection [4]. Our understanding of Ku is constantly expanding, with new research advances suggesting the heterodimer is doing more than what we currently understand. Incidentally, Ku is highly abundant in human cells, having multiple processed pseudogenes and a Ku80 isoform, all of which imply that Ku functions in processes or regulatory capacities we have yet to identify.
While most proteins are first identified in model organisms prior to the identification of human homologs, Ku was first identified as an autoantigen in the serum of a Japanese patient [5]. Many of the advances made in Ku understanding followed the characterization of Ku70/80 knockout mice, which were radiosensitive, immunocompromised, and proportional dwarves [6]. Curiously, while Ku70/80 knockouts are viable in some species, the genes appear to be indispensable in humans and human cell lines [7, 8]. The essentiality of Ku in some species over others is still unclear.
Acquisition of the Ku crystal structure represented a significant milestone in our mechanistic understanding of Ku [9], including our knowledge on Ku–protein interactions. Structural studies also led to the recent identification of the Ku-binding motif (KBM), which allows proteins to interact directly with Ku [10, 11]. Related to the identification of Ku protein interactors, our lab recently used the new, high-throughput proximity-dependent biotin identification (BioID) technique [12] to comprehensively identify in vivo Ku protein interactors in human cells [13].
In this review, we provide an interdisciplinary discourse on the Ku heterodimer outside of its traditional role as a DNA repair protein involved in non-homologous end-joining (NHEJ) and V(D)J recombination. We focus on unifying the different aspects of Ku research that will be critical to further understand the heterodimer including its structure, function, cellular localization, expression levels, conservation, and essentiality.
Ku composition: structure, domains, and function
Ku, originally named after a Japanese patient with an autoimmune disease that recognized Ku as an autoantigen, is a protein heterodimer [5]. In humans, Ku is composed of two subunits, Ku70 (~ 70 kDa, 609 amino acids) and Ku80 (~ 80 kDa, 732 amino acids), which are encoded by the XRCC6 (X-ray repair cross-complementing protein 6, located on chromosome 22) and XRCC5 (chromosome 2) genes, respectively. Eukaryotic subunits Ku70 and Ku80 undergo domain swapping to form the stable, Ku heterodimer complex, resistant to separation unless subjected to high salt (i.e. 1.5 M NaCl, 1 M KCl) or strong, ionic detergents (i.e. 0.5% SDS, 1% sodium deoxycholate) [14, 15]. Dimerization appears to be important for the stability of both Ku70 and Ku80. In mice, the deletion of one subunit leads to severely reduced expression of the other subunit, implying that most Ku exists as an obligate heterodimer [6, 16]. Heterodimerization also seems to be a necessary prerequisite for successful Ku80 purification in bacteria [17]. However, researchers have been able to isolate Ku70, independent of Ku80 [17, 18], supporting the notion that Ku70 can form a homodimer. Indeed, Tadi et al. (2016) were able to produce a stable Ku70 homodimer in vitro using purified protein from both insect and bacterial cells [18]. However, while a fraction of cellular Ku70 may exist as a homodimer, to date, the Ku70 homodimer has yet to be identified or studied in an intracellular context.
The Ku heterodimer has been implicated in binding double-stranded DNA (dsDNA) [19] and facilitating the repair of double-stranded breaks (DSBs) through the NHEJ repair pathway [reviewed in [20]]. In 2001, the X-ray crystallography structures of Ku unbound and bound to DNA were obtained [9], representing a major milestone that enhanced our understanding of Ku70/80 dimerization and DNA association (Fig. 1). Ku70 and Ku80 share structural homology and dimerize to form a pseudo-symmetrical basket structure that encircles the DNA duplex [9]. Both subunits show a common topology consisting of three domains: a N-terminal α/β domain, a central β-barrel domain, and a subunit-specific helical C-terminal domain (CTD).
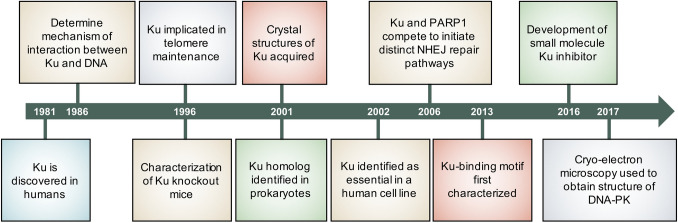
Fig. 1.
Timeline of Ku research milestones. Notable developments over the last 40 years pertaining to various research aspects about the Ku70/80 heterodimer
The N-terminal α/β domain of each Ku subunit is a divergent member of the von Willebrand factor A (vWA) family of domains [21], composed of a six-stranded β-sheet in a Rossman fold [9]. Disrupting the vWA domain in yeast Ku70/80 has been found to impair DNA repair and telomere regulation [22]. Although the amino edge of the vWA domain lies proximal to the DNA-binding groove, the vWA domain is not required for DNA binding. Instead, consistent with vWA domains in other proteins, the Ku vWA domains may mediate protein–protein interactions. For example, the Ku80 vWA domain has been shown to interact with APLF, a NHEJ repair protein essential for the recruitment of other repair factors [23].
Meanwhile, the central DNA-binding domain, composed of a seven-stranded antiparallel β-barrel, is essential for DNA binding and heterodimerization [9]. Heterodimerization results in a positively charged DNA-binding ring which fits sterically around the minor and major DNA grooves. Ku threads inwards on DNA similar to a nut threaded onto a bolt, with Ku70 positioned proximal and Ku80 distal to the DNA end [21, 24]. Ku slides onto DNA ends using an energy-free mechanism that is still poorly understood. No direct base contacts are made with DNA, implying that Ku associates with DNA in a sequence-independent manner [9]. Ku has a high affinity (Kd = 10–9 M) for dsDNA ends, 5′ and 3′ overhangs or blunt ends, and a significantly lower affinity for circular DNA and single-stranded DNA ends [1].
Finally, the C-terminal Ku regions are unique to each subunit, however, both contain a flexible linker and a globular structural domain (Fig. 2). The Ku70 C-terminal region contains a putative DNA-binding domain called the SAP (SAF-A/B, Acinus, and PIAS) domain (5 kDa, residues 559–609) [9]. The C-terminal structure was determined both in isolation using NMR [25] and as part of the unbound Ku70/80 crystal structure [9], and these studies discovered a common helix-extended loop-helix structural motif. There is evidence that SAP domains can bind DNA from studies on other SAP domain proteins [26, 27]. In the context of Ku, some studies suggest there could be interactions between Ku70-SAP and DNA [28, 29]. During DNA binding, the domain undergoes displacement that positions it in close proximity to the DNA-binding region of the Ku heterodimer [30, 31]. It is conceivable that Ku70-SAP may stabilize the interaction between Ku and DNA, though its exact function has yet to be conclusively elucidated.
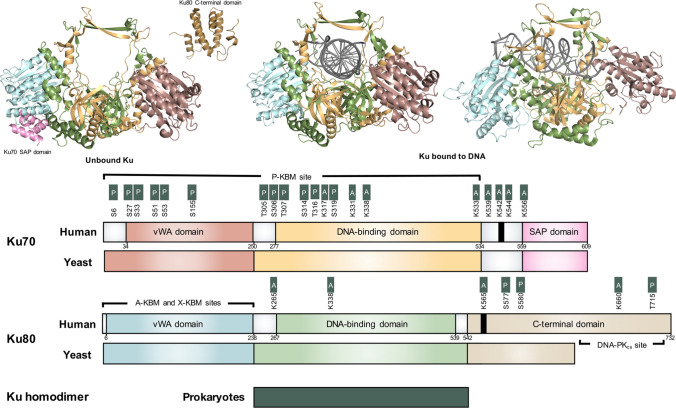
Fig. 2.
Ribbon diagram and corresponding schematic of Ku heterodimer domains. Figure adapted from Fell and Schild-Poulter (2015) review [1]. Ku ribbon diagram bound and unbound by DNA. Crystal structure data of human Ku (PDB: IJEY) is colored in a domain-specific manner. Ku80 C-terminus domain is portrayed in isolation using NMR structural data (PDB: IQ2Z). In the Ku schematic, human, yeast, and prokaryote Ku subunits are compared while the Ku von Willebrand A (vWA) domain, DNA-binding domain, and SAF-A/B, Acinus and PIAS (SAP) or C-terminal domain, are color-matched to the ribbon diagram. Nuclear Localization Signal (NLS) is indicated as a thick black line. DNA-PKcs and Ku-binding motif (KBM) interaction sites are indicated, as are key post-translational modification (PTM) sites for phosphorylation and acetylation
Initial crystallography studies were conducted with truncated Ku80 that was missing the CTD [9]. The Ku80-CTD structure (19 kDa, residues 542–732) was later determined using NMR by two different groups [32, 33]. The Ku80-CTD contains a globular domain composed of six α-helices (residues 592–709) that is flanked by disordered N- and C-termini. Single-particle electron microscopy and small angle X-ray scattering data show that in the presence of DNA, the Ku80-CTD undergoes displacement to form a flexible arm that extends from the DNA-binding core to interact with the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) [30, 34].
Although the Ku crystal structure was obtained in 2001, important regions of each subunit were not mapped out due to experimental difficulties. These regions have since been resolved in the context of the crystal structure of DNA-PKcs bound to the Ku80-CTD [35], and in two cryogenic electron microscopy (cryoEM) studies of DNA-PK containing full length Ku70/80 [36, 37]. CryoEM studies show DNA-PKcs and Ku both recognize and bind DNA separately and together result in a 30° kink in the DNA duplex in contrast to previous models that assumed the DNA is unbent [35, 38]. The role of DNA distortion in NHEJ remains unclear. CryoEM data also indicates that DNA-PKcs is activated allosterically by conformational changes initiated by DNA-PK holoenzyme (dsDNA, Ku70/80, and DNA-PKcs) formation [36, 37, 39]. CryoEM structures of Ku in isolation have not yet been reported but may be useful to further study Ku structure, protein interactions, and its role in NHEJ [39].
Ku conservation and essentiality
Conservation among organisms
Ku, an essential component of NHEJ, was first identified in humans [5], though functional homologs have since been identified in almost all eukaryotes including vertebrates, insects, fungi, and in some prokaryotic lineages [40–42]. Sequence homology analysis revealed that yeast and other lower eukaryotic Ku subunits may have arisen from a gene duplication event of a common ancestral gene [41, 43]. The eukaryotic Ku70 and Ku80 subunits have diverged in primary sequence but share similar secondary and tertiary structure [9]. Ku homologs in different organisms also have little sequence similarity but universally share similar structure and function [41].
In eukaryotes, Ku and DNA ligase IV form the core components of the NHEJ repair complex [41]. Many eukaryotes also have additional NHEJ factors such as DNA-PKcs. It was previously thought that DNA-PKcs was only found in higher eukaryotes, however, recently DNA-PKcs has been identified in invertebrates, fungi, plants, and protists [44]. In organisms that have functional DNA-PKcs, the Ku80 CTD binds with DNA-PKcs, and the subsequently formed DNA-PK holoenzyme recruits downstream NHEJ factors [45]. Notably, DNA-PKcs is absent in yeast and Caenorhabditis elegans [45, 46], organisms that rely primarily on homologous recombination (HR) as their main mechanism of DSB repair [47, 48]. Accordingly, yeast Ku80 contains a unique CTD, distinct from higher eukaryotes, capable of binding DNA ligase IV (Dnl4) instead of DNA-PKcs [48].
Prokaryotes were initially thought to rely solely on HR, however, Ku homologs have been found in multiple bacterial genera, including Mycobacterium, Pseudomonas, Bacillus, and Agrobacterium [40]. The core bacterial NHEJ complex is composed of homodimeric Ku and LigD, a multifunctional ligase with multiple catalytic domains [49]. Bacterial Ku subunits are smaller than eukaryotic Ku (approximately 30–40 kDa) yet share related structural homology. The β-barrel ring domain responsible for heterodimerization and binding dsDNA in eukaryotes is structurally and functionally conserved in bacterial Ku, but the N-terminal vWA domain and the CTD are not [50].
Homodimeric Ku is also present in a limited number of Archaean species, implying that NHEJ repair is rare in these organisms [51]. Only one species has been found with NHEJ complex components, and these components are closely related to their bacterial homologs [52]. Ku homologs are widespread across the kingdom of life. The Gam protein, found in bacteriophage Mu, is a homodimer capable of binding dsDNA ends. Gam has considerable sequence homology with prokaryotic and eukaryotic Ku and appears to be an ortholog [53]. However, there are notable examples of organisms missing Ku and NHEJ capability. For example, NHEJ machinery (Ku and LigD) is absent in E. coli [40]. Interestingly, Gam, in conjunction with native E. coli LigA, has been shown to activate NHEJ in E. coli [54]. Multiple other NHEJ factors have also been independently lost in several species of parasitic protists [55].
Essentiality among organisms
To date, there is no published viable Ku homozygous knockout human cell line, although notable knockdown and knockout attempts have been made in various mammalian cell lines (Table 1). In human HCT116 cells, homozygous Ku80 knockouts were reportedly not viable after a limited number of cell doublings [7]. Similarly, for Ku70, a genome-wide knockout study found that deletion was lethal in near-haploid human cell lines, HAP1 and KBM7 [8]. Ku has been proposed to be essential in humans due to its role in telomere maintenance [7, 56]. Ku knockout was also lethal in Ustilago maydis, a fungal species, due to unprotected telomeres triggering cell cycle arrest [57].
Table 1.
Mammalian cell lines with Ku70 or Ku80 deficiencies
| Subunit | Species | Cell line/tissue | Type of cell line | Type of deficiency | Viability | Reference(s) |
|---|---|---|---|---|---|---|
| Ku70 | Human | RERF-LC-A1 | Immortal | Knockdown | Viable | [58] |
| Nalm-6 | Immortal | Heterozygous | Viable | [59] | ||
| HCT116 | Immortal | Heterozygous | Viable | [60] | ||
| Knockout | Inviable | |||||
| HAP1 | Immortal | Knockout | Inviable | [8] | ||
| KBM7 | ||||||
| Mouse | Mouse Embryonic Fibroblasts (MEFs) | Immortal | Knockout | Viable | [61] | |
| Embryonic Stem (ES) cells | Primary | Heterozygous | Viable | [16] | ||
| Knockout | ||||||
| Fibroblasts | Primary | Heterozygous | Viable | [62] | ||
| Knockout | ||||||
| B and T lymphocytes, thymocytes, MEFs, spleen, bone marrow | Primary | Knockout | Viable | [63] | ||
| Ku80 | Hamster | sxi-2, sxi-3 | Immortal | Knockout | Viable | [64] |
| CHO-xrs-6 or xrs6a | Immortal | Knockout | Viable | [65, 66] | ||
| CHO-xrs-4 or xrs4a | ||||||
| CHO-xrs-5 or xrs5a | Immortal | Knockdown | Viable | [66] | ||
| XR-V15B | Immortal | Knockout | Viable | [67] | ||
| XR-V9B | Immortal | Knockout | Viable | [68] | ||
| Human | MRC5V1 | Immortal | Knockdown | Viable | [69] | |
| HCT116 | Immortal | Heterozygous | Viable | [7] | ||
| Knockout | Inviable | |||||
| HeLa | Immortal | Knockdown | Reduced viability | [70] | ||
| U2OS | ||||||
| SAOS | ||||||
| Nalm-6 | Immortal | Heterozygous | Viable | [59] | ||
| Mouse | MEFs | Immortal | Knockout | Viable | [6] | |
| Bone marrow, B and T lymphocytes, MEFs, spleen | Primary | Knockout | Viable | [6, 71] | ||
| ES cells, fibroblasts, thymus | Primary | Knockout | Viable | [6] | ||
| Thymocytes | Primary | Knockout | Viable | [71] |
axrs hamster cell lines can be reverted to wild-type levels of Ku80 with azacytidine treatment suggesting they may not be true knockouts, but instead contain one silenced, but functional allele [66]
Despite the inability to produce a viable total knockout of Ku in human cells, non-lethal Ku knockdowns through heterozygous gene knockout or RNA-based silencing have been achieved in different human cell lines [72–74]. Heterozygous Ku70 and Ku80 knockouts in HCT116 displayed slower cell proliferation, shortened telomeres, and hypersensitivity to ionizing radiation (IR), which indicates decreased NHEJ repair function [7, 73, 74]. Several human disorders are linked to NHEJ factor mutations, though no genetic disease has thus far been associated with a mutation in Ku. This combined data suggest that Ku is likely essential in humans. Contrarily, in most species, Ku appears to be dispensable (Table 2).
Table 2.
Viable species in which Ku was knocked out
| Kingdom | Species | Reference(s) |
|---|---|---|
| Animalia | Bombyx mori | [82] |
| Caenorhabditis elegans | [83] | |
| Chicken cell line DT40 | [84] | |
| Chinese hamster ovary (CHO) mutant cell lines | [85] | |
| Mus musculus | [16] | |
| Fungi | Claviceps purpurea | [86] |
| Penicillium chrysogenum | [86] | |
| Penicillium decumbens | [86] | |
| Penicillium marneffei | [86] | |
| Trichoderma virens | [86] | |
| Saccharomyces cerevisiae | [48] | |
| Schizosaccharomyces pombe | [87] | |
| Plantae | Arabidopsis thaliana | [88] |
| Protozoa | Toxoplasma gondii | [89] |
Species or cell lines, grouped by Kingdom, that were viable after XRCC5 (Ku80) and/or XRCC6 (Ku70) genes were successfully knocked out
Unlike their human cell line counterparts, Ku knockout mice are viable and have been extensively studied [6, 75]. Despite Ku knockout mice being viable and capable of reproduction, Ku deficiency in mice is linked to a host of symptoms [71, 75–81] (Table 3). Ku70 or Ku80 knockout mice share many characteristic phenotypes, but there are a few notable differences including early aging for Ku80 deficient mice and early incidence of cancer in Ku70 knockouts.
Table 3.
Ku70 and Ku80 knockout phenotypes in mice
| Ku70 Knockout Mice | Ku80 Knockout Mice |
|---|---|
| Viable | Viable |
| Can reproduce | Can reproduce |
| Ku70 knockout mother unable to sustain pups | Ku80 knockout mother unable to sustain pups |
| K/O mice smaller than control mice | K/O mice smaller than control mice |
| Fewer proliferating cells | Early loss of proliferating cells for fibroblasts |
| Premature senescence of cells | Fibroblasts show prolonged doubling time |
| Cells sensitive to ionizing radiation | Cells are radiation sensitive |
| Lack mature B cells or serum immunoglobulin | Arrest in T and B lymphocyte development |
| V(D)J rearrangement deficiency | V(D)J rearrangement deficiency |
| Increased incidence of thymic tumors | Smaller spleen and lymph nodes |
| Chromosomal instability | Chromosomal instability |
| Early aging/shorter lifespan |
Ku localization and expression
Cellular localization
Ku is predominantly observed in the nucleus [90]. Following transcription and translation, Ku subunits can translocate into the nucleus together, as a heterodimer [90], or independently, as each subunit contains its own nuclear localization signal [91]. In the nucleus, Ku has been shown to bind DNA DSBs with a high affinity [41, 92]. Ku’s key roles associated with DNA repair [reviewed in [1]], telomere function [93], DNA replication [94, 95], and transcription [96, 97] validate its purpose as a nuclear protein.
Further investigations of the nuclear distribution pattern of Ku revealed its localization in both the nucleoplasm and nucleolus [98–100]. Ku is a dynamic, highly mobile protein complex and its nuclear mobility was shown to be regulated by inositol hexakisphosphate (InsP6), an enzyme cofactor for DNA-PK and a direct Ku interactor [101–103]. In the absence of DNA damage, both Ku subunits are localized to the nucleolus [98, 104, 105], where Ku has been shown to associate with ribosomal RNA and RNA-binding proteins [98, 100]. In response to cellular stress signals, certain nucleolar proteins are found to be relocated to initiate countermeasures. Ku is one definitive example of this phenomenon as nucleolar Ku shuttles back to the nucleoplasm to initiate DNA break repair upon UV- or IR-induced DNA damage [98, 99]. These findings speculate that a portion of the Ku population exists in the nucleolus in the absence of DNA damage.
Although Ku has been well established as a nuclear protein, there are several studies which have observed its presence in the cytoplasm [reviewed in [90]]. Bax, a cytoplasmic protein, has been found to interact with Ku70, and this interaction was suggested to inhibit Bax-mediated apoptosis [3]. In addition, cytosolic Ku has been shown to act as a pattern recognition receptor that recognizes viral DNA in human cells [4]. Many of the studies that claimed to detect Ku in the cytoplasm relied on subcellular fractionation techniques where Ku may have “leaked out” of the nucleus to contaminate the cytoplasmic fractions, so the presence of Ku in the cytoplasm remains controversial.
Mitochondria contain their own genetic information in the form of a circular plasmid of DNA, which warrants mitochondria as a possible destination for Ku as a NHEJ repair factor. An earlier study reported that mitochondrial extracts of Ku-deficient hamster cells showed DNA end-binding activity, implying that some other factor associates with broken DNA ends and that Ku is not needed in the mitochondria to repair DSBs [106]. This speculation was recently confirmed as another group found that NHEJ was undetectable from the mitochondrial extracts of rat tissue and human cells [107]. Instead, the study identified factors involved in microhomology-mediated end-joining, an alternative DSB repair pathway to NHEJ, and suggest this pathway may be predominantly responsible for maintaining genomic integrity in the mitochondria [107].
Protein expression
Ku70 and Ku80 are abundant proteins with approximately 400,000 units of Ku per cell in established human cell lines [108], and Ku levels appear to be ubiquitous and abundant regardless of the cell type or cell cycle phase. Some in vivo human tissues (i.e. skin, muscle, nerve, lung, ovary, kidney, etc.) were found to show heterogeneity in Ku70 expression [109]. Furthermore, tissue‐specific overexpression of Ku70 and Ku80 homologs has been demonstrated in the ovary and testes of Xenopus laevis [110]. In Drosophila melanogaster, expression levels are constant through all developmental stages except oogenesis and early embryogenesis when they increase 25-fold [111]. These findings suggest a role for Ku in development. In Arabidopsis thaliana, expression was constant, albeit low, across all plant tissues examined [112]. Although Ku is conserved in almost all eukaryotic and a number of prokaryotic lineages, Ku expression patterns appear to be highly variable.
Highly expressed genes, like XRCC6 (Ku70), are prone to producing processed pseudogenes (PP). PPs are created in germline cells through reverse transcription and random integration of mRNA into the genome. PPs are often non-functional and have lost the ability to produce proteins [113]. Ku70 has five PPs (XRCC6P1-5) in the human genome. While XRCC6 is located on chromosome 22, three PPs are found on chromosomes 1 (XRCC6P3), 8 (XRCC6P4), and 10 (XRCC6P1) and two are found on chromosome X (XRCC6P2 and XRCC6P5). It is unclear, at present, whether any of these pseudogenes can or have been reactivated in human cells. Notably, no Ku80 pseudogenes have been reported.
The human Ku80 locus produces two protein isoforms: Ku80 and an alternative splice variant named Ku86 autoantigen related protein-1 (KARP-1), that is transcribed from a different upstream promoter [114]. KARP-1 shares certain biochemical properties with Ku80. KARP-1 is able to bind, colocalize, and stabilize Ku70 in vivo, strongly indicating that KARP-1 can heterodimerize with Ku70 [115]. KARP-1 has been shown to positively regulate DNA-PK activity [114] and partially complement the radiosensitivity of Ku80-deficient cells [115]. Notably KARP-1, not Ku80, expression is strongly induced by DNA damage in a p53- and ATM-dependent manner [116]. Whether KARP-1 has a distinct role or functions in mammalian DNA repair is still unclear, though KARP-1 was reported to have a protective role against cell damage caused by oxidative stress in rats [117].
Identifying Ku protein interactors
Ku70, Ku80, and DNA-PKcs
Ku is often referred to as the DNA-binding subunit of the DNA-PK complex, which assembles in response to DSBs [1]. The extreme Ku80 C-terminus (residues 439–592) interacts with DNA-PKcs [43, 118, 119] and promotes DNA-PKcs autophosphorylation at DNA breaks. It is well recognized that Ku is important for recruiting and activating DNA-PKcs at DSBs, although one study observed DNA-PKcs could still be activated in Ku-deficient hamster cells [120], though this remains to be verified. Recently, another study found that the DNA sequence and end structure of the break may also play a role in DNA-PK assembly, specifically impacting the interaction between DNA-PKcs and the Ku80 C-terminus [121]. Aside from the broken DNA end composition, it is also possible that DNA-PKcs may interact differently with Ku depending on the NHEJ processing factors recruited to the break. DNA-PK acts as a hub for the rest of the DNA repair factors during NHEJ [reviewed in [122]]. Independent of Ku, DNA-PKcs function in other cellular processes has been reported [123].
Ku interactome studies
As highly abundant proteins in human cells, Ku70 and Ku80 are frequently identified in affinity purification and mass spectrometry datasets. In this sense, Ku is sometimes seen as a “contaminating” protein, as it is often observed outside of a logical biological context. However, at the same time, many diverse factors have been shown to interact with Ku [1, 13, 124], indicative of the fact that Ku has been implicated in multiple cellular processes: NHEJ [11, 18, 119, 125–127], telomere maintenance [128–131], DNA damage response [132–134], RNA biology [135–137], transcription [15, 138, 139], DNA replication [140, 141], V(D)J recombination [142, 143], and apoptosis [3, 144–146] (Fig. 3).
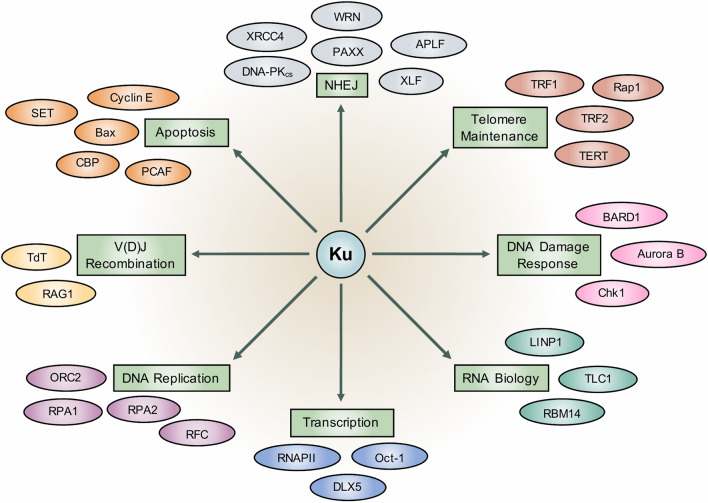
Fig. 3.
The Ku heterodimer functions in various cellular processes and interacts with multiple proteins. Figure adapted from Downs and Jackson (2004) [147] review. A selection of well-characterized cellular functions that have been shown to involve Ku, including key human Ku protein/RNA interactors
Originally, many studies identified candidate Ku interactors using high-throughput yeast-two-hybrid (Y2H) screens before validating interactions with low-throughput co-immunoprecipitation experiments and/or in vitro binding assays [10, 128, 148, 149]. Colocalization [150] and proximity ligation assays [151] are additional low-throughput techniques that can detect in vivo associations and have also been used previously to verify Ku interactors [132, 148].
We recently used proximity-dependent biotin identification (BioID) [12] to identify in vivo protein interaction candidates for Ku in human cells [13]. Using this technique with a second high-throughput proteomic technique known as affinity purification coupled to mass spectrometry (AP-MS), we were able to identify both known and novel Ku interactor candidates and create a comprehensive map of the Ku protein interactome [13]. Prior to our study, another group used tandem affinity purification tagging in human cells to identify Ku interactors and found 22 candidate proteins, many of which were shared with our study [124]. As technology continues to advance, more candidates can be identified using additional high-throughput techniques, allowing us to visualize the greater landscape and context of Ku functions throughout the cell.
The Ku-binding motif
Within the last decade, many studies identified a specific, 9-15 amino acid sequence referred to as a Ku-binding motif (KBM). The KBM is a conserved motif that mediates protein interactions with Ku [reviewed in [152]]. Thus far, only a handful of proteins have been found to contain a verified KBM, including Aprataxin PNKP-like factor (APLF) [125], Werner syndrome protein (WRN) [10], Modulator of retrovirus infection homologue (MRI, also known as CYREN) [10], XRCC4-like factor (XLF; also known as Cernunnos) [11], and Paralog of XRCC4 and XLF (PAXX) [11] (Fig. 4).
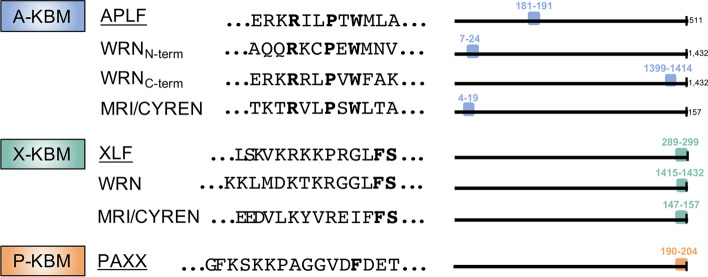
Fig. 4.
The human Ku-Binding Motifs (KBMs). Figure adapted from Frit et al. (2019) [152]. Different classes of KBM with known proteins containing verified KBMs, amino acid sequences, and approximate sequence location. Note that there is no clear consensus on the exact lengths and boundaries of the KBMs. Bolded amino acids are conserved. Each KBM was initially discovered in and named after the respective underlined protein
APLF, an intrinsically disordered protein that acts as an accessory factor for NHEJ, has been found to interact directly with Ku80, by binding to a hydrophobic pocket in the Ku80 vWA domain [11, 23]. Despite being predominantly nuclear, APLF does not contain its own nuclear localization signal [125]. Disrupting the interaction between Ku and APLF by mutating the APLF KBM (referred to as A-KBM in future studies) resulted in APLF becoming more cytoplasmic, suggesting that the A-KBM, and the interaction it mediates with Ku, is essential for retaining APLF in the nucleus [125].
Meanwhile, WRN was found to contain two KBMs: one at its N-terminus and another at its C-terminus [10]. While not a core NHEJ factor, WRN enhanced DSB repair using both KBMs [10]. The C-terminal KBM in WRN is adjacent to a novel, structurally similar but functionally distinct motif called the XLF-like motif [10]. This new motif was found at the extreme C-terminus of WRN, XLF (after which it was named), MRI/CYREN [11, 153], and PAXX [10].
Nemoz et al. (2018) proposed that this related XLF-like motif is also a true KBM and referred to it as X-KBM. Using crystallography, they showed that X-KBM also interacts with Ku80, albeit in a different region from the A-KBM. X-KBM seems to bind specific sites within the Ku80 vWA domain; specifically, X-KBM occupied an internal pocket formed when the Ku80 α/β domain underwent an outward rotation [11]. This unique interaction with the Ku80 vWA was also supported by a study conducted in X. laevis, where researchers suggested that the availability of this Ku80 vWA binding site may be conditional [154].
Nemoz et al. (2018) also identified a third sub-category of KBM, which they called P-KBM for the motif found in the C-terminus of PAXX. While PAXX was initially suspected to contain an X-KBM [10], implying an interaction with Ku80 in the same manner as XLF, the C-terminus of PAXX did not compete with XLF for Ku80 binding and instead bound Ku70 [11]. Another group had also previously deduced a PAXX-Ku70 interaction; they found that the C-terminus of PAXX was essential for Ku interaction, while the C-termini of both Ku70 and Ku80 were not essential for PAXX interaction [18]. Although the exact Ku70 binding site for P-KBM is unknown, the researchers noted that Ku binding to DNA results in a protrusion of the W148 residue in the Ku70 vWA, which they proposed could be an important contact for PAXX interaction [18].
Thus far, all the verified proteins containing KBMs are known or suspected to be involved in NHEJ [10]. At a double-stranded DNA break, Ku is threaded onto the ends of broken DNA and interaction with KBM-containing factors XLF or PAXX is only detectable in this specific context, with the inclusion of DNA [18]. In the future, it will be of interest to see if KBMs are identified in proteins outside of the NHEJ context. To this end, using the minimal KBM sequence (R-X-X-P-X-W), Grundy et al. (2016) identified over 600 putative KBM proteins [10]. Overall, KBMs represent a key motif that allows proteins to interact with either Ku80 or Ku70. Based on our current understanding, KBMs can be divided further into three sub-categories: A-KBMs, which dictate interaction with the Ku80 vWA hydrophobic pocket; X-KBMs, which also interact with the Ku80 vWA albeit distinct from A-KBMs; and finally P-KBMs, a third class that mediates interaction with Ku70 (Fig. 4).
Post-translational modifications
Ku functionality is modulated by several post-translational modifications (PTMs) including phosphorylation, acetylation, ubiquitination, and sumoylation. Some PTMs are associated with Ku function in NHEJ [155], DNA repair pathway selection [156], DNA damage response (DDR) [132], DNA replication [94], and Ku degradation [157], though many PTMs remain functionally uncharacterized.
Phosphorylation
Due to the close association of Ku with DNA-PKcs during NHEJ, initial studies focused on identifying potential phosphorylation sites of Ku by DNA-PKcs [158, 159]. Early in vitro studies identified several Ku80 residues (S577, S580, T715) that were phosphorylated by DNA-PK [158, 160, 161]. Similarly, Ku70 phosphorylation sites (S51, S53, S319) were also identified, and most were found as part of a DNA-PK consensus motif (S/T-Q) [159], with the exception of Ku70 S6 [158]. Phosphoablative mutation analysis of Ku80 S577, S580, T715 and Ku70 S6 demonstrated that phosphorylation of these sites was not needed for NHEJ repair function [160], raising the question of what functional role these phosphorylation events may play. A later study demonstrated that Ku70 phosphorylation by DNA-PKcs at other sites (T305, S306, T307, S314 and T316) displaced Ku from DNA ends, thereby preventing NHEJ and promoting HR repair during the S phase of the cell cycle [156]. Taken together, these results suggest some Ku phosphorylation may be involved in DNA repair pathway selection while the function of other sites is still unclear.
Aside from DNA-PKcs, cyclin-dependent kinases (CDKs) also phosphorylate Ku and other Ku70 phosphorylation sites have been identified using proteomic studies [162]. Cyclin A1-bound CDK2 was found to phosphorylate both Ku70 and Ku80 [163] and showed phosphorylation-mediated regulation of Ku70’s heterodimer DNA-binding ability in vitro [164]. One group demonstrated that DNA replication was regulated through Ku70 phosphorylation by cyclin/CDKs. Various cyclin/CDK proteins were suggested to phosphorylate Ku70 and inhibit its interaction with replication origins during S, G2, and M phases, while dephosphorylation of Ku70 during the G1 phase facilitated the assembly of the origin recognition complex [94]. These early results imply that the balance between phosphorylated and dephosphorylated Ku70 may function to prevent untimely replication initiation.
Previous research in our lab identified Ku70 S155 as a novel phosphorylation site that is phosphorylated in response to severe DNA damage, although the kinase protein responsible is unknown [132, 165]. The proposed functions of Ku70 S155 phosphorylation in cell cycle arrest and the DDR are discussed later in this review (see “DNA damage response”). Finally, proteomic screens have identified other potential Ku70 phosphorylation sites phosphorylated by checkpoint kinase 1 (Chk1) and polo-like kinase, though the functions mediated by these phosphorylation events remain elusive [166, 167]. In some human cancer cell lines, Ku phosphorylation has been shown to confer mechanisms for radiation-resistance and survival [155, 168]. For example, in certain chemoresistant leukemia cell lines, a novel form of Ku70 was identified in which residues S27 and S33 were phosphorylated, and this phosphorylation seemed to lead to a faster, but more unfaithful NHEJ repair process [155].
Acetylation
Both Ku70 and Ku80 have multiple lysine residues that have been identified as acetylation sites. Ku80 sites include K265, K338, K565, and K660 [169] while numerous Ku70 sites have also been identified (Fig. 2) [3, 170, 171]. Ku70 acetylation has been shown to impair NHEJ by modifying the Ku70 lysine residues needed for binding dsDNA ends: K539 and K542 [171, 172] and K317, K331, K338 [170]. Two histone acetyltransferase enzymes, CBP and PCAF, have been shown to be responsible for Ku acetylation [3]. CBP was capable of acetylating both Ku70 [171] and Ku80 [173] at various lysine residues. Meanwhile, histone deacetylases, such as SIRT1 [174] and SIRT6 [175], deacetylate Ku.
Current research suggests that Ku70 acetylation negatively affects NHEJ-mediated DNA repair. Knocking down CBP in neuroblastoma cells decreased the levels of acetylated Ku70 while showing increased DNA repair activity and cell survival [171]. Acetylated Ku70 (K539, K542) has also been implicated in the initiation of Bax-mediated apoptosis [3, 172], providing a possible explanation for why increased cell survival is observed in CBP knockdown neuroblastoma cells. Interestingly, there are cellular mechanisms preventing Ku70 acetylation. Certain Ku70 interactors (i.e. SET and SMAR1) have been shown to safeguard Ku70 from acetylation [144, 176]. Inhibition of Ku70 acetylation by INHAT subunit SET/TAF-1β regulates Ku70-mediated DNA damage response [144], while SMAR1 was shown to coordinate HDAC6-induced deacetylation of Ku70 and dictate cell fate upon irradiation [176].
Ubiquitination and sumoylation
Ubiquitination and addition of ubiquitin-like proteins (UBLs) (i.e. NEDD8, SUMO) play a vital role in regulating cellular responses to DNA repair pathways [reviewed in [177]]. Ubiquitination by E3 ligases RNF138 and RNF8 has been associated with freeing Ku80 from repaired dsDNA by targeting the subunit for degradation [177, 178]. However, little is known about the impact of these modifications on Ku70. Neddylation-dependent Ku70 ubiquitination has been shown to promote Ku70 removal from DNA damage sites after repair [157]. In yeast Ku70, sumoylation at the C-terminal lysine residues were found to be favorable for Ku70 and broken DNA end association [179]. Ubiquitination is one of the more understudied Ku PTMs and an area in need of further investigation.
Diversity of Ku function
Ku is best characterized for its indispensable role in NHEJ, one of the available pathways for repairing DNA DSBs. Ku is the first protein of the NHEJ repair complex to recognize and bind broken DNA ends, binding within 10 s of a DSB forming [180]. Ku threads onto both broken ends in a sequence-independent manner, with most models depicting a single heterodimer binding each end [1, 20]. Once bound, Ku recruits DNA-PKcs, which protects the DNA ends from degradation [181]. After autophosphorylation, DNA-PKcs undergoes a conformational change allowing other repair factors access to the ends [182]. Proteins involved in bridging and processing DSBs are also recruited, although the exact order of recruitment is still uncertain [183]. XRCC4 and XLF are recruited to DSBs and have been shown to form an alternating filament that resembles a mobile, “molecular sleeve” to bridge the broken ends [184]. Other factors recruited include DNA polymerases and endonucleases that may be needed to process the DNA ends to be compatible for rejoining. DNA end processing without a homologous template can introduce errors, hence NHEJ is known for being an error-prone process [185]. The extent of the DNA damage may dictate which processivity factors are recruited and their abundance. Finally, DNA ligase IV is responsible for ligating the DNA backbone and completing repair [186]. Ku in NHEJ has been studied and reviewed extensively by others in various contexts: DSB repair pathway choice [20, 156, 187], function and kinetics of NHEJ [1, 188], and Ku removal from repaired dsDNA [189].
Related to its function in repairing DSBs, Ku is also essential for the repair of intentional, programmed DNA breaks formed during V(D)J recombination and class switch recombination, the cellular lymphocyte-specific processes that generate antigen diversity and immunoglobulin types, respectively, in human immune cells [190, 191]. V(D)J recombination relies on many of the same factors needed in NHEJ, discussed in the previous paragraph (for more information, see review by [192]). Ku’s role in V(D)J recombination is further detailed later on in the review (see “Immune dysfunction”). Class switch recombination changes the type of immunoglobulin produced by a B cell through recombination of only the immunoglobulin constant region, modifying the function of the immunoglobin while leaving antigen specificity unchanged. In class switch recombination, intentional DSBs are introduced within the antibody gene locus and are typically repaired by Ku through the NHEJ pathway. However, unlike V(D)J recombination, if NHEJ factors are knocked out, class switch recombination breaks can also be repaired by the backup or alternative NHEJ repair process, microhomology-mediated end-joining [193].
Aside from NHEJ and immune system-related processes, Ku has been implicated in other cellular processes (Fig. 3). Here, we review research conducted about Ku function in the DNA damage response, DNA replication, transcription, telomere maintenance, and RNA biology.
DNA damage response
The DNA damage response (DDR) is a complex network of signaling pathways initiated upon detection of DNA damage [194]. DNA damage can arise internally from endogenous insults (DNA replication errors, reactive oxygen species, etc.) or externally from exogenous sources (ionizing radiation, chemicals, etc.) [195]. Numerous processes have been implicated in the DDR including cell cycle regulation, senescence, apoptosis, and DNA repair pathways including NHEJ [195, 196]. The DDR can be divided into three processes: scanning and detection, signal transduction and amplification, and repair and final appraisal. Here, we review the role of Ku in these processes in response to a DSB.
Various factors are responsible for scanning DNA for damage including the MRN (Mre11, Rad50, Nibrin/NBS1) [195, 197] and BASC (BRCA1-associated genome surveillance complex) [198] complexes. Some initially hypothesized that there was a specific “sensor” for each type of DNA lesion [199]. Conceivably, many suspected that Ku, an early responder to DSBs, played such a role in the DDR [197, 199]. However, Ku is not the only known sensor of DSBs, as MRN [200] and PARP1 [201] are also capable of recognizing and binding DSBs. By nature, not all DSBs are identical so there could be subtle differences governing the recognition of each DSB.
Once the damage has been detected, a “transducer” is needed to amplify the signal of damage and activate downstream proteins. Kinases such as ATM, ATR, and DNA-PKcs are the most commonly known transducer proteins responsible for signal amplification during the DDR [194, 202]. In response to DSBs, the ATM kinase phosphorylates multiple proteins including p53, 53BP1, BRCA1, CHK2, and histone H2AX at serine 139 [202, 203]. Aside from kinase proteins, our lab found that the S155 residue of Ku70, upon phosphorylation, appears to inhibit the Aurora B kinase and may be involved in promoting cell cycle arrest and the DDR [132]. Whether and how Ku interaction with Aurora B leads to cell cycle arrest is still unknown. Using irradiated murine Ku- and Ku70/ATM-knockout cell lines, another group demonstrated that Ku is needed for ATM-dependent ATR activation [204], providing further evidence that Ku may function in this aspect of the DDR.
In response to DSBs, phosphorylation of p53 leads to cell cycle checkpoint arrest, allowing the cell time to assess and repair the damage or trigger senescence/apoptosis if the damage is too great [202]. Ku is well established as being involved in the NHEJ repair aspect of the DDR. As Ku has been implicated in both DNA repair and apoptosis [3], it is tempting to speculate that Ku could also be appraising the repair attempt, ready to signal apoptosis if necessary [165]. The mechanism for deeming DNA damage beyond repair is still unclear though there are several possibilities (PTMs, cellular localization changes, retention time or occupancy levels at DSBs, etc.).
DNA replication
One of the less defined roles played by Ku is its function in DNA replication. In yeast, Ku was initially identified as being involved in the timing of the DNA replication cycle [205], and this was further supported by studies that established Ku as a modulator of activation time of replication origins in regions proximal to telomeric or sub-telomeric sequences [206]. Yeast Ku has also been shown to function at DNA replication forks. Upon replication fork arrest, Ku has been implicated in the cell recovery process, leading to the restart of replication [207]. The function of Ku at fork arrests may be a stabilizing action in yeast [208]. Ku may also function to limit DSB resection during DNA replication-associated damage, which could inhibit the initial steps leading to the HR repair pathway [209]. In support of Ku’s action as a regulator of DNA replication, one study characterized the removal of yeast Ku at arrested replication forks as integral to fork resection and replication restart [210].
Ku has also been implicated in regulating DNA replication in human cells. Early data suggested that there was a changing relationship between DNA-PKcs forming a complex with replication protein A during replication, to a complex of DNA-PKcs and Ku in response to DNA damage [140]. Through quantification of Ku binding in vivo at different cell cycle stages, Ku was identified as a protein that binds to mammalian sequences containing replication origins [211, 212]. Specifically, independent of DNA-PKcs, Ku has been found to directly bind to the A3/4 sequence found in mammalian replication origins [95]. Ku’s association with proteins involved in DNA repair may indicate a mechanism through which Ku is recruited to replication origins [141]. In the event of replication-induced DNA damage, Ku has been found to act as a protector of DNA replication by binding breaks and preventing the unloading of replication machinery from chromatin [213]. In advanced human metastatic breast cancer, higher levels of Ku were associated with chromatin, and Ku was found in the replication complexes formed at certain, more active origins of replication, implicating Ku’s role in DNA replication in breast tumorigenesis [211]. Phosphorylation of human Ku70 has also been linked to replication-related functions and was described earlier in this review (see “Post-translational modifications”). Finally, Ku80 knockdown in HeLa cells was directly associated with a decrease in the total rate of DNA replication [214].
Transcription
Ku is well-known for its sequence-independent association with broken DNA ends [9, 41, 108]. However, in the late 1990s, many groups reported Ku-DNA site-specific interaction [215–218], implying that Ku acts as a transcription factor to regulate gene expression. In support of this, the Ku70 SAP domain has been shown to have some DNA-binding capability independent of Ku80 [25, 28, 29], although it is currently unclear if this DNA-binding activity is sequence-specific. Ku was also found to associate with other DNA structures like hairpins or bubbles [reviewed in [41]], implying that rather than recognizing DNA by sequence, Ku may instead recognize and bind in vivo DNA structures that form at certain sites [147].
Several studies have suggested that Ku binds directly to specific DNA elements, such as the heat-shock element in the promoter of the hsp70 gene [219] and negative regulatory element 1 in vitro [216, 217], implicating Ku in the regulation of transcription. Using an in vitro transcription system with linear DNA, the displacement of transcription factors by Ku was observed [220]. However, Ku is not universally recognized as a bona fide transcription factor as its mode of DNA sequence-specific binding is not understood and the mechanisms through which it may regulate transcription remain unclear.
Ku has also been proposed to function more globally in transcription by modulating RNA polymerase II (RNAPII) expression or through protein–protein interactions with transcription-related proteins and factors. For instance, it has been reported that DSBs and DDR proteins may be a necessary component in regulating the release of paused RNAPII into active transcriptional elongation within human cells [221]. DNA-PK has been shown to phosphorylate the C-terminal domain of RNAPII [222] and Ku has been shown to interact with RNAPII [97, 138]. One group reported that Ku associates indirectly with RNAPII elongation sites via direct interaction between Ku80 and elongation proteins [97]. Recently, Ku was shown to bind the RNA hairpin structure of 7SK short nuclear RNA (snRNA), implicating Ku as a potential member of the 7SK short nuclear ribonucleoprotein (snRNP) complex that regulates transcriptional elongation [2]. Meanwhile, DNA-PK was shown to phosphorylate numerous transcription factors in vitro [reviewed in [223]]. Ku has also been found to work in tandem with other transcriptional proteins to initiate glucocorticoid receptor-dependent transcription [224].
While the connection between Ku and transcription is viable, Ku appears to regulate transcription under multiple contexts, making it a challenging and controversial area of study. Ku has also been implicated in the transcriptional regulation of gene expression in various diseases including breast cancer [225], Wilson disease [226], and HIV [227]. It is clear that a lot remains to be elucidated regarding Ku’s precise role in transcriptional processes relating to gene expression and implications of Ku in disease states, as well as identifying specific mechanisms of action.
Telomere maintenance
Telomeres are repetitive DNA sequences found at the ends of chromosomes that maintain genomic integrity. In mammals, telomeres protect chromosomal ends through the formation of a t-loop. In a t-loop, single-stranded DNA overhangs invade double-stranded telomeric TTAGGG repeats and associate with a six-member complex of proteins that are collectively known as the shelterin complex [228]. Both the shelterin complex and the t-loop form a cap that prevents DNA repair factors from recognizing the chromosome ends as potential DNA damage [228]. Telomeric DNA and associated protein complexes are involved in regulating gene expression through telomere position effects [229] and the three-dimensional genomic landscape [230], in which the proximity of genes to telomeric sequences and the conformation of chromatin can directly influence gene expression. Telomere biology, as well as telomeric dysregulation, have been extensively reviewed recently [229, 231], but despite this, some aspects of telomere regulation are still poorly understood.
One understudied area of research involves elucidating the network of proteins involved in telomere regulation. In the late 1990′s, researchers began to identify that Ku, which was classically recognized for its key role in DSB repair, was implicated in telomere regulation [129, 232–236]. Studies of mutant strains of yeast ku70 (yku70) and ku80 (yku80) revealed an essential role of Ku in telomere regulation, where a reduction or complete ablation of Ku function in yeast resulted in decreased telomere length [235, 236]. In contrast, in Drosophila, Ku deficiency results in abnormally long telomeres [237]. Ku-deficient mice displayed abnormal telomere lengths with telomeric fusions, indicating that Ku likely plays a role in preventing the occurrence of telomeric fusions [238, 239]. Depletion of a fungal form of Ku was found to cause cell cycle arrest, likely due to DNA damage signals from unprotected telomeres, although this needs further validation [57]. In mammalian cell lines, Ku depletion resulted in shortened telomeres and increased apoptosis [7, 74, 240]. These studies suggest Ku is required for proper telomere maintenance across different species, though the exact regulation mechanism is unclear. Ku has been shown to interact indirectly with telomeres through telomere-associated proteins in mice [238]. In mammals, Ku interacts with multiple members of the shelterin complex to protect chromosome ends [228]. In lower eukaryotes like yeast, Ku also functions in concert with other proteins to produce telomere position effects [241], but Ku’s function in telomere position effects of mammals still needs to be elucidated.
While Ku function in telomere biology has been reviewed [56, 242, 243], the precise mechanism of Ku at telomeres has yet to be fully elucidated. Human cells with a knockdown of Ku80 were reported to have shortened telomeres and increased cell lethality, though the timeline of telomere shortening upon Ku depletion was not characterized [74]. In human cells, it was further demonstrated that loss of Ku80 resulted in telomere loss through the formation of t-circles and subsequent cell death [244]. These findings suggest that Ku is essential in humans due to its role in telomere maintenance, but future studies will be needed to fully characterize Ku’s mechanism of action at telomeres that results in cell death.
RNA biology
Traditionally, RNA was understood for its central role as the template for protein synthesis, however, contemporary research has demonstrated that the majority of the human genome produces non-protein coding RNA species that can regulate gene expression or even impact disease progression [245]. Collectively, the interplay between RNA, proteins, and small molecules can also play an important role in the regulation of gene expression and protein function [246]. Recently, multiple DDR proteins have been found to bind RNA to regulate gene expression and modulate DDR signaling and repair [247]. Specifically, Ku has been found to associate with RNA structures to participate in multiple cellular functions.
Ku association with nuclear RNAs was identified in the 1990s as a means to modulate enzyme activity and potentially gene expression [248, 249]. In yeast, Ku associates with TLC1, the RNA component of telomerase, and acts to modulate telomerase activity [250, 251]. Yeast Ku recognizes and binds to a specific stem-loop hairpin structure of TLC1 [250, 252], and Ku binding to RNA and DNA was found to be mutually exclusive [253]. In human cells, a similar stem-loop structure within the RNA component of human telomerase was recognized and bound by Ku, suggesting that Ku interaction with telomerase RNA is conserved amongst species [254].
Ku’s association with RNA may also function to regulate transcription. Ku has been found to bind TAR RNA at the 5′ end of mRNA transcripts of the HIV-1-gene [248, 249]. Ku may directly affect HIV-1 expression at the transcriptional level and its latency in infected cells through RNA interactions [227]. As reviewed earlier, Ku has been implicated in the regulation of transcriptional elongation through its interaction with the RNA hairpin structure of 7SK snRNA, which acts as a scaffold for the formation of the 7SK snRNP complex [2].
Ku has been shown to interact with a long noncoding RNA known as LINP1, which acts as a scaffold for the Ku heterodimer and DNA-PKcs to function in response to damaged DNA [135, 255]. The inherent dsDNA repair function of Ku may play a role in resistance to cancer radiation therapies. The Ku-LINP1 interaction was shown to increase NHEJ repair efficiency. As this particular long noncoding RNA is overexpressed in triple-negative breast cancer, it was suggested that this Ku-RNA interaction may promote a radiation-resistance mechanism in tumor cells by enhancing NHEJ repair of DSBs [255]. Thapar et al. (2020) delve further into how LINP1 structurally functions in NHEJ and how it can effectively replace the NHEJ factor PAXX [135].
RNA:DNA hybrids, created during the formation of R-loops, appear to have varied and contrasting physiological effects that have been reviewed recently [256, 257]. While RNA:DNA hybrids are known to be a source of genomic instability, more recent research suggests that R-loops may play beneficial roles such as regulating gene expression and facilitating DNA double-stranded break repair [257, 258]. Evidence for this restorative action in DNA repair has been demonstrated in yeast [259], as well as in mammalian cells [260]. The removal of R-loops in human cells has been shown to reduce the efficiency of both HR and NHEJ, lending support that the RNA:DNA hybrids are critical for DNA repair, though mechanistic details have yet to be fully elucidated [260]. Recent data indicate that R-loops are generated at break sites and play a role in the establishment of DNA repair and the DDR [260, 261]. A number of DNA repair factors have been found to interact with R-loops, notably PARP1, DHX9, DDX5, and DNA-PK [258, 262, 263]. Specifically, DHX9 has been implicated in suppressing R-loops and maintaining genomic integrity in vivo [263]. Recently, PARP1, DHX9, and DDX5 were also identified as potential Ku interactors using both BioID and affinity purification coupled to mass spectrometry [13]. Ku70 and Ku80 were also found to have an affinity for RNA:DNA hybrid structures using pull-down assays followed by mass spectrometry [262]. Given Ku’s association with DNA repair factors that interact with RNA:DNA hybrids, as well as emerging evidence that Ku itself may interact with RNA:DNA hybrids, it will be interesting to explore and identify Ku’s role in maintaining genomic stability through R-loop interactions.
One study identified a Ku-mediated link between DNA repair and mRNA translation through Ku’s association with p53 mRNA [264]. The stem-loop structure in the untranslated region of the p53 transcript when bound by Ku was found to repress translation, and this repression was released as Ku relocated from the p53 mRNA to broken DNA ends in response to genotoxic stress [264]. In another study, Ku was found to move out of the nucleolus in response to sheared DNA that was added to live cells, suggesting that its association with RNA switches to DNA in the event of DSBs [98]. It was reported that in the nucleolus of mouse cells, Ku may affect rRNA processing, as the assembly of a catalytically defective or inactive DNA-PK by Ku resulted in an accumulation of unprocessed pre-rRNA precursors [100]. Overall, the context of Ku interaction with mammalian RNA within the nucleolus is still unclear. As a relatively new field, there is much to be learned regarding Ku and RNA biology.
Study of Ku in human diseases
Aging and telomere defects
Aging is the time-sensitive deterioration of physiological properties occurring at the cellular and organismal level. Accumulation of genetic defects over time and less efficient repair are contributing factors to aging [265].
Knocking out any of the DNA-PK subunits in mice caused premature aging [265, 266]. Yet the early understanding was that only deletions of Ku80 and/or DNA PKcs were responsible for aging phenotypes in mice whereas deletions of Ku70 correlated with a high incidence of cancer [265]. Later, these findings were found to be inaccurate as deletions of Ku70 and Ku80 subunits lead to identical aging phenotypes, highlighting the essentiality of both subunits [266]. Other than its substantial role in DSB repair, DNA-PK is found to be involved in metabolic dysregulation during aging. In aging smooth muscle cells, increasing DSBs caused constitutive DNA-PK activation leading to down regulation of 5′-AMP-activated protein kinase (AMPK) activity [267]. DNA-PK phosphorylation of HSP90 impaired chaperone functionality on AMPK leading to misfolding. The depleted AMPK levels caused reduced fitness and declined mitochondrial function in aging mice. These findings accentuate a novel role for DNA-PK in mediating DNA damage-induced metabolic imbalance during aging [267].
Ku function in telomere maintenance can also contribute to the aging phenotype. Telomere shortening leads to replicative senescence of somatic cells [265]. Loss of t-loop structure and/or the protein cap protection (shelterin complex proteins) due to telomere shortening triggers the DDR [265] leading to permanent cell cycle arrest and/or apoptosis, depending on the cell type [268]. Ku plays a protective role at telomeres by binding the shelterin complex [231]. As discussed earlier, yeast [232, 235], human cancer cells [74, 240] and certain mouse models deficient in Ku [238] have shown reduced telomere length which emphasizes Ku’s protective role against replicative senescence. Increased telomere associations and fusions were observed in aging Ku knockout mice [239, 266], and could be the result of defects in telomere cap protection.
A direct relationship between Ku and human aging disorders is uncertain, however, mutations to some potential Ku protein interactors may lead to premature aging. Hutchinson–Gilford progeria (HGPS) is a premature aging syndrome in humans caused by a single point mutation in the LMNA gene that codes for progerin, a truncated splice variant of lamin A [126, 269]. Ectopically-expressed DNA-PKcs was shown to co-immunoprecipitate with progerin in HGPS fibroblasts, implying the two may interact [269]. Compared to normal fibroblasts, HGPS fibroblasts showed reduced expression of Ku70, Ku80, and DNA-PKcs upon the accumulation of progerin and the loss of DNA-PK was found to contribute to the phenotypes observed in HGPS fibroblasts [269]. Another Ku protein interactor, WRN, enhances NHEJ repair [10, 270, 271], and loss of function mutations in WRN can result in Werner syndrome, an autosomal recessive disorder characterized by premature aging [271, 272]. Loss of function mutations in WRN impair its localization to the nucleus [272], and may negatively affect WRN and Ku interaction and NHEJ repair during aging.
Immune dysfunction: the Ku autoantigen
Ku was initially discovered as an autoantigen after the detection of anti-Ku antibodies in the sera of scleroderma–polymyositis overlap syndrome patients [5]. Since then, anti-Ku antibodies have been identified with varying prevalence (< 1–27%) in a wide spectrum of connective tissue diseases including systemic lupus erythematosus, Sjögren's syndrome, rheumatoid arthritis, and systemic sclerosis (SSc) [273–276]. The prevalence of anti-Ku autoantibodies in connective tissue diseases varies due to the detection immunoassay, type of autoimmune disorder, and the genetic and geographical background of the subjects studied [5, 273, 274, 277–280]. Anti-Ku antibody profiles have been associated with some clinical manifestations. A significant association was reported between anti-Ku antibodies and musculoskeletal manifestations of disease (myositis, arthritis, and joint contractures) in SSc patients [276]. Anti-Ku positive patients who also display elevated serum creatine kinase levels appear to be at significantly higher risk of developing interstitial lung disease [281, 282]. Antibodies against other NHEJ factors have also been found in the serum of connective tissue disease patients suggesting a concerted autoimmune response to DDR proteins and that DNA damage may be a factor in the development of connective tissue diseases [273, 283, 284].
Ku in innate immunity
The innate immune response relies on the detection of bacterial and viral pathogens by pattern-recognition receptors (PRR) that induce the production of cytokines and chemokines. Membrane-bound and cytoplasmic PRR detect foreign DNA using conserved, essential molecules of pathogens [285]. DNA-PK has been identified as a cytosolic PRR for DNA viruses in murine fibroblasts and mice. After infection from the vaccinia virus (VV), which has a linear dsDNA genome, DNA-PK was shown to co-localize with sites of viral DNA replication in the cytoplasm and was involved in the activation of the innate immune system. Accordingly, the innate immune response is impaired in mice lacking DNA-PK components [286]. Ku alone has also been shown to detect viruses in human cells. Ku70 specifically functions as a cytosolic PRR that recognizes dsDNA and induced interferon activation in HEK293 cells [4]. In several human cell lines, Ku70 overexpression inhibited expression of human T lymphotropic virus type 1 (HTLV-1), a retrovirus that can cause leukemia, whereas Ku70 knockdown promoted HTLV-1 expression [287]. Cytoplasmic Ku was able to promote hepatitis-associated chemokine secretion after the detection of cytosolic hepatitis B viral DNA in human liver-derived cells [288]. Ku70 also mediated the innate immune response in human macrophages to a viral infection by herpes simplex virus-2 [289]. Interestingly, and speaking to the importance of DNA-PK in restraining viral infection, viruses appear to have developed subversion mechanisms to counter DNA-PK-mediated host detection. Two VV proteins, C4 and C16, are able to bind Ku and block its DNA-binding capabilities, attenuating the host innate immune response [290, 291].
Impairment in V(D)J recombination
The immune system relies on T- and B-lymphocytes producing an adaptive immune response using antigen-specific T-cell receptors (TCRs) and immunoglobulin (Ig) antibodies, respectively. V(D)J recombination generates diversity in the variable regions of TCRs and Ig antibodies by the controlled creation and NHEJ-driven repair of DSBs [292, 293]. Animals lacking NHEJ are severely immunodeficient as evidenced by Ku70/80 knockout mice. Immunological defects include smaller immune system organs and arrested T- and B-lymphocyte development associated with impaired V(D)J recombination [6, 75]. Although mutations in Ku have not been identified in humans, mutations in other NHEJ factors have been associated with severe combined immunodeficiency (SCID), a group of genetic disorders that results in persistent and recurrent infections [294, 295].
Cancer
In healthy cells, cell division is carefully regulated and adheres to a strict process; cells that bypass these regulations and divide unchecked are considered cancerous. DNA replication stress or repair errors can lead to chromosomal aberrations and genomic instability, a hallmark of cancer [296]. In many cancers, the expression of DNA repair proteins is dysregulated [296]. Aside from expression, the localization, function, protein interactions, and PTMs of these proteins can also be dysregulated in cancer cells.
In the early 2000s, a prevailing hypothesis was that Ku expression could correlate with cancer treatment outcomes [297, 298]. Many treatments target cancerous cells by creating intentional DSBs using radiation or chemotherapy. Since Ku is essential for NHEJ, it was reasonable to hypothesize that lower Ku levels indicated increased sensitivity to DNA breaks. In support of this, Ku knockout mice demonstrate increased sensitivity to IR [63]. Similarly, siRNA downregulation of Ku70 in two cancerous cell lines also led to increased sensitivity to radiation and etoposide, a chemotherapy drug [299].
Ku levels were assessed in tumor biopsies using immunohistochemistry and were correlated to tumor radiosensitivity and the disease-free survival of patients. In two separate studies, Ku levels in colon cancer tumors were significantly reduced [298, 300]. Another group observed that in advanced rectal carcinoma tumors, tumors with a greater percentage of increased Ku-level cells were more likely to be radioresistant, while patients with a lower percentage tended to have better disease-free survival [297]. Another study assessed Ku levels in cervical cancer patients, though somewhat mixed results were observed as only some of the tumors showed correlation with Ku expression [301]. Contrary to the original hypothesis, some radiosensitive tumors actually had a higher percentage of cells showing increased Ku levels [301]. A second hypothesis proposed that Ku levels could be elevated in cancers, thus leading to an increased incidence of error-prone NHEJ and greater genomic instability. There was some evidence in support of this hypothesis too as some studies observed elevated Ku levels in various cancers [302, 303]. Elevated Ku expression has been correlated with increased resistance to both radiation [297] and cisplatin chemotherapy [304].
To a lesser degree, other studies have investigated possible Ku dysregulations at the functional level. For example, one study observed altered Ku DNA-binding activity in both breast and bladder tumors [305]. Another study found that EAF2, an androgen-responsive tumor suppressor, regulates NHEJ by recruiting and retaining Ku at sites of DNA damage in prostate cancer cells [306]. Finally, using chemotherapy-resistant lymphocytic leukemia cells, one group identified a novel phosphorylated form of Ku70 (residues S27 and S33) that conferred faster, more error-prone NHEJ [155]. Intriguingly, this study implies that some cancer cells adopt a Ku70-dependent chemotherapy resistance mechanism where NHEJ is enhanced.
Ku expression and NHEJ activity must be carefully regulated and balanced in human cells to preserve genomic stability, acting as an essential defense against oncogenesis. Directly connecting Ku to cancer in terms of diagnosis and prognosis has been challenging. In spite of some studies providing support for Ku levels as a prognostic tool, Ku expression does not appear to be a consistent parameter, possibly due to intra-tumor diversity, cancer progression, or variability among cancer types. However, Ku does merit further investigation as a direct therapeutic target. Recently, a small molecule Ku inhibitor capable of sensitizing human cells to radiation was published (Fig. 1). While some iteration of this molecule could be used in cancer treatments, problems such as tumor-specificity and toxicity still require resolution [307].
Conclusions
Recent research advances suggest Ku’s cellular role is far more wide-reaching than its long-established DNA repair function. The study of Ku is expanding to encompass numerous fields of research, many of them involving regulatory processes. Significant structural knowledge has been gained from the recently acquired cryoEM structures of DNA-PK [36, 37], while the function of other protein domains such as Ku70′s SAP domain remains under investigation. Another active field of Ku research is the identification of Ku protein interactors, particularly those containing the recently identified KBMs. Identifying and validating Ku protein interactors, including KBM proteins, will be a useful tool in the future to infer new cellular processes that implicate Ku function. Recent promising research investigating Ku’s role in innate immunity [289, 291] and the development of a small molecule Ku inhibitor [307] indicate that Ku’s clinical relevance should not be overlooked. Studies have shown that Ku appears to function as a sensor for viral dsDNA, a trait that could be exploited to enhance human immunity to dsDNA viruses. Already, there is evidence that viruses can develop strategies to overcome DNA-PK’s sensory capabilities as viral proteins capable of blocking DNA-PK’s recognition of viral dsDNA have been identified. Additionally, while Ku’s outlook as a tool for predicting cancer diagnosis and prognosis has yielded mixed results, increased cellular Ku levels seem to correlate with enhanced resistance to radiation, making Ku a worthy chemotherapy target to improve cancer treatment. The creation of viable Ku knockout cell lines in humans and a variety of other species would greatly assist in disease studies and elucidation of protein function. The multifunctional Ku protein has become a point of interdisciplinary research for many fields and further investigation could lead to more discoveries, at both the molecular and clinical level.
Acknowledgements
We are grateful to Mohamed Aly for reading and providing feedback on the review.
Authors’ contributions
S.A. and G.P. conceptualized the idea of the review. S.A., G.P., R.D.K., and N.B. wrote the original draft and S.A., G.P, and R.D.K. prepared the figures and tables. C.S.P. provided edits, suggestions, and critical feedback. The final version of the review was edited by all authors.
Funding
This work was supported by a Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant and a NSERC Discovery Accelerator Supplement to C.S.P.. S.A. and G.P. were supported by Ontario Graduate Scholarships.
Availability of data and material
Not applicable.
Code availability
Not applicable.
Declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
Not applicable.
Informed consent
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Fell VL, Schild-Poulter C. The Ku heterodimer: function in DNA repair and beyond. Mutat Res Rev Mutat Res. 2015;763:15–29. doi: 10.1016/j.mrrev.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 2.Shadrina O, Garanina I, Korolev S, et al. Analysis of RNA binding properties of human Ku protein reveals its interactions with 7SK snRNA and protein components of 7SK snRNP complex. Biochimie. 2020;171–172:110–123. doi: 10.1016/j.biochi.2020.02.016. [DOI] [PubMed] [Google Scholar]
- 3.Cohen HY, Lavu S, Bitterman KJ, et al. Acetylation of the C terminus of Ku70 by CBP and PCAF controls Bax-mediated apoptosis. Mol Cell. 2004;13:627–638. doi: 10.1016/s1097-2765(04)00094-2. [DOI] [PubMed] [Google Scholar]
- 4.Zhang X, Brann TW, Zhou M, et al. Cutting edge: Ku70 is a novel cytosolic DNA sensor that induces type III rather than type I IFN. J Immunol. 2011;186:4541–4545. doi: 10.4049/jimmunol.1003389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mimori T, Akizuki M, Yamagata H, et al. Characterization of a high molecular weight acidic nuclear protein recognized by autoantibodies in sera from patients with polymyositis-scleroderma overlap. J Clin Invest. 1981;68:611–620. doi: 10.1172/jci110295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nussenzweig A, Chen C, da Costa SV, et al. Requirement for Ku80 in growth and immunoglobulin V(D)J recombination. Nature. 1996;382:551–555. doi: 10.1038/382551a0. [DOI] [PubMed] [Google Scholar]
- 7.Li G, Nelsen C, Hendrickson EA. Ku86 is essential in human somatic cells. Proc Natl Acad Sci USA. 2002;99:832–837. doi: 10.1073/pnas.022649699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blomen VA, Májek P, Jae LT, et al. Gene essentiality and synthetic lethality in haploid human cells. Science. 2015;350:1092–1096. doi: 10.1126/science.aac7557. [DOI] [PubMed] [Google Scholar]
- 9.Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- 10.Grundy GJ, Rulten SL, Arribas-Bosacoma R, et al. The Ku-binding motif is a conserved module for recruitment and stimulation of non-homologous end-joining proteins. Nat Commun. 2016;7:11242. doi: 10.1038/ncomms11242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nemoz C, Ropars V, Frit P, et al. XLF and APLF bind Ku80 at two remote sites to ensure DNA repair by non-homologous end joining. Nat Struct Mol Biol. 2018;25:971–980. doi: 10.1038/s41594-018-0133-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roux KJ, Kim DI, Raida M, Burke B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol. 2012;196:801–810. doi: 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abbasi S, Schild-Poulter C. Mapping the Ku interactome using proximity-dependent biotin identification in human cells. J Proteome Res. 2019;18:1064–1077. doi: 10.1021/acs.jproteome.8b00771. [DOI] [PubMed] [Google Scholar]
- 14.Wang J, Satoh M, Pierani A, et al. Assembly and DNA binding of recombinant Ku (p70/p80) autoantigen defined by a novel monoclonal antibody specific for p70/p80 heterodimers. J Cell Sci. 1994;107(Pt 11):3223–3233. doi: 10.1242/jcs.107.11.3223. [DOI] [PubMed] [Google Scholar]
- 15.Schild-Poulter C, Pope L, Giffin W, et al. The binding of Ku antigen to homeodomain proteins promotes their phosphorylation by DNA-dependent protein kinase. J Biol Chem. 2001;276:16848–16856. doi: 10.1074/jbc.M100768200. [DOI] [PubMed] [Google Scholar]
- 16.Gu Y, Jin S, Gao Y, et al. Ku70-deficient embryonic stem cells have increased ionizing radiosensitivity, defective DNA end-binding activity, and inability to support V(D)J recombination. Proc Natl Acad Sci USA. 1997;94:8076–8081. doi: 10.1073/pnas.94.15.8076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanakahi LA. 2-Step purification of the Ku DNA repair protein expressed in Escherichia coli. Protein Expr Purif. 2007;52:139–145. doi: 10.1016/j.pep.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tadi SK, Tellier-Lebègue C, Nemoz C, et al. PAXX is an accessory c-NHEJ factor that associates with Ku70 and has overlapping functions with XLF. Cell Rep. 2016;17:541–555. doi: 10.1016/j.celrep.2016.09.026. [DOI] [PubMed] [Google Scholar]
- 19.Mimori T, Hardin JA. Mechanism of interaction between Ku protein and DNA. J Biol Chem. 1986;261:10375–10379. doi: 10.1016/S0021-9258(18)67534-9. [DOI] [PubMed] [Google Scholar]
- 20.Scully R, Panday A, Elango R, Willis NA. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat Rev Mol Cell Biol. 2019;20:698–714. doi: 10.1038/s41580-019-0152-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doherty AJ, Jackson SP. DNA repair: Topological analysis of chromatin-associated protein complexes using single affinity purification. Curr Biol. 2001;11:R920–R924. doi: 10.1016/S0960-9822(01)00555-3. [DOI] [PubMed] [Google Scholar]
- 22.Ribes-Zamora A, Mihalek I, Lichtarge O, Bertuch AA. Distinct faces of the Ku heterodimer mediate DNA repair and telomeric functions. Nat Struct Mol Biol. 2007;14:301–307. doi: 10.1038/nsmb1214. [DOI] [PubMed] [Google Scholar]
- 23.Grundy GJ, Rulten SL, Zeng Z, et al. APLF promotes the assembly and activity of non-homologous end joining protein complexes. EMBO J. 2013;32:112–125. doi: 10.1038/emboj.2012.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoo S, Kimzey A, Dynan WS. Photocross-linking of an oriented DNA repair complex. Ku bound at a single DNA end. J Biol Chem. 1999;274:20034–20039. doi: 10.1074/jbc.274.28.20034. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Z, Zhu L, Lin D, et al. The three-dimensional structure of the C-terminal DNA-binding domain of human Ku70. J Biol Chem. 2001;276:38231–38236. doi: 10.1074/jbc.M105238200. [DOI] [PubMed] [Google Scholar]
- 26.Göhring F, Schwab BL, Nicotera P, et al. The novel SAR-binding domain of scaffold attachment factor A (SAF-A) is a target in apoptotic nuclear breakdown. EMBO J. 1997;16:7361–7371. doi: 10.1093/emboj/16.24.7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suzuki R, Shindo H, Tase A, et al. Solution structures and DNA binding properties of the N-terminal SAP domains of SUMO E3 ligases from Saccharomyces cerevisiae and Oryza sativa. Proteins. 2009;75:336–347. doi: 10.1002/prot.22243. [DOI] [PubMed] [Google Scholar]
- 28.Lehman JA, Hoelz DJ, Turchi JJ. DNA-dependant conformational changes in the Ku heterodimer. Biochemistry. 2008;47:4359–4368. doi: 10.1021/bi702284c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu S, Pluth JM, Cucinotta FA. Putative binding modes of Ku70-SAP domain with double strand DNA: a molecular modeling study. J Mol Model. 2012;18:2163–2174. doi: 10.1007/s00894-011-1234-x. [DOI] [PubMed] [Google Scholar]
- 30.Rivera-Calzada A, Spagnolo L, Pearl LH, Llorca O. Structural model of full-length human Ku70–Ku80 heterodimer and its recognition of DNA and DNA-PKcs. EMBO Rep. 2007;8:56–62. doi: 10.1038/sj.embor.7400847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Makowski MM, Willems E, Jansen PWTC, Vermeulen M. Cross-linking immunoprecipitation-MS (xIP-MS): topological analysis of chromatin-associated protein complexes using single affinity purification. Mol Cell Proteomics. 2016;15:854–865. doi: 10.1074/mcp.M115.053082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harris R, Esposito D, Sankar A, et al. The 3D solution structure of the C-terminal region of Ku86 (Ku86CTR) J Mol Biol. 2004;335:573–582. doi: 10.1016/j.jmb.2003.10.047. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Z, Hu W, Cano L, et al. Solution structure of the C-terminal domain of Ku80 suggests important sites for protein-protein interactions. Structure. 2004;12:495–502. doi: 10.1016/j.str.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 34.Hammel M, Yu Y, Mahaney BL, et al. Ku and DNA-dependent protein kinase dynamic conformations and assembly regulate DNA binding and the initial non-homologous end joining complex. J Biol Chem. 2010;285:1414–1423. doi: 10.1074/jbc.M109.065615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sibanda BL, Chirgadze DY, Ascher DB, Blundell TL. DNA-PKcs structure suggests an allosteric mechanism modulating DNA double-strand break repair. Science. 2017;355:520–524. doi: 10.1126/science.aak9654. [DOI] [PubMed] [Google Scholar]
- 36.Sharif H, Li Y, Dong Y, et al. Cryo-EM structure of the DNA-PK holoenzyme. Proc Natl Acad Sci USA. 2017;114:7367–7372. doi: 10.1073/pnas.1707386114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yin X, Liu M, Tian Y, et al. Cryo-EM structure of human DNA-PK holoenzyme. Cell Res. 2017;27:1341–1350. doi: 10.1038/cr.2017.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spagnolo L, Rivera-Calzada A, Pearl LH, Llorca O. Three-dimensional structure of the human DNA-PKcs/Ku70/Ku80 complex assembled on DNA and its implications for DNA DSB repair. Mol Cell. 2006;22:511–519. doi: 10.1016/j.molcel.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 39.Chaplin AK, Hardwick SW, Liang S, et al. Dimers of DNA-PK create a stage for DNA double-strand break repair. Nat Struct Mol Biol. 2020 doi: 10.1038/s41594-020-00517-x. [DOI] [PubMed] [Google Scholar]
- 40.Bowater R, Doherty AJ. Making ends meet: repairing breaks in bacterial DNA by non-homologous end-joining. PLoS Genet. 2006;2:e8. doi: 10.1371/journal.pgen.0020008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dynan WS, Yoo S. Interaction of Ku protein and DNA-dependent protein kinase catalytic subunit with nucleic acids. Nucleic Acids Res. 1998;26:1551–1559. doi: 10.1093/nar/26.7.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aravind L, Koonin EV. Prokaryotic homologs of the eukaryotic DNA-end-binding protein Ku, novel domains in the Ku protein and prediction of a prokaryotic double-strand break repair system. Genome Res. 2001;11:1365–1374. doi: 10.1101/gr.181001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gell D, Jackson SP. Mapping of protein–protein interactions within the DNA-dependent protein kinase complex. Nucleic Acids Res. 1999;27:3494–3502. doi: 10.1093/nar/27.17.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lees-Miller JP, Cobban A, Katsonis P, et al. Uncovering DNA-PKcs ancient phylogeny, unique sequence motifs and insights for human disease. Prog Biophys Mol Biol. 2020 doi: 10.1016/j.pbiomolbio.2020.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith GCM, Jackson SP. The DNA-dependent protein kinase. Genes Dev. 1999;13:916–934. doi: 10.1101/gad.13.8.916. [DOI] [PubMed] [Google Scholar]
- 46.Lemmens BBLG, Tijsterman M. DNA double-strand break repair in Caenorhabditis elegans. Chromosoma. 2011;120:1–21. doi: 10.1007/s00412-010-0296-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clejan I, Boerckel J, Ahmed S. Developmental modulation of nonhomologous end joining in Caenorhabditis elegans. Genetics. 2006;173:1301–1317. doi: 10.1534/genetics.106.058628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emerson CH, Bertuch AA. Consider the workhorse: nonhomologous end joining in budding yeast. Biochem Cell Biol. 2016;94:396–406. doi: 10.1139/bcb-2016-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shuman S, Glickman MS. Bacterial DNA repair by non-homologous end joining. Nat Rev Microbiol. 2007;5:852–861. doi: 10.1038/nrmicro1768. [DOI] [PubMed] [Google Scholar]
- 50.Pitcher RS, Brissett NC, Doherty AJ. Nonhomologous end-joining in bacteria: a microbial perspective. Annu Rev Microbiol. 2007;61:259–282. doi: 10.1146/annurev.micro.61.080706.093354. [DOI] [PubMed] [Google Scholar]
- 51.White MF, Allers T. DNA repair in the archaea—an emerging picture. FEMS Microbiol Rev. 2018;42:514–526. doi: 10.1093/femsre/fuy020. [DOI] [PubMed] [Google Scholar]
- 52.Bartlett EJ, Brissett NC, Doherty AJ. Ribonucleolytic resection is required for repair of strand displaced nonhomologous end-joining intermediates. Proc Natl Acad Sci USA. 2013;110:E1984–E1991. doi: 10.1073/pnas.1302616110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.di Fagagna F, d’Adda WGR, Doherty AJ, Jackson SP. The Gam protein of bacteriophage Mu is an orthologue of eukaryotic Ku. EMBO Rep. 2003;4:47–52. doi: 10.1038/sj.embor.embor709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bhattacharyya S, Soniat MM, Walker D, et al. Phage Mu Gam protein promotes NHEJ in concert with Escherichia coli ligase. Proc Natl Acad Sci USA. 2018;115:E11614–E11622. doi: 10.1073/pnas.1816606115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nenarokova A, Záhonová K, Krasilnikova M, et al. Causes and effects of loss of classical nonhomologous end joining pathway in parasitic eukaryotes. MBio. 2019;10:e01541–e1619. doi: 10.1128/mBio.01541-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Indiviglio SM, Bertuch AA. Ku’s essential role in keeping telomeres intact. Proc Natl Acad Sci USA. 2009;106:12217–12218. doi: 10.1073/pnas.0906427106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.de Sena-Tomás C, Yu EY, Calzada A, et al. Fungal Ku prevents permanent cell cycle arrest by suppressing DNA damage signaling at telomeres. Nucleic Acids Res. 2015;43:2138–2151. doi: 10.1093/nar/gkv082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Omori S, Takiguchi Y, Suda A, et al. Suppression of a DNA double-strand break repair gene, Ku70, increases radio- and chemosensitivity in a human lung carcinoma cell line. DNA Repair. 2002;1:299–310. doi: 10.1016/s1568-7864(02)00006-x. [DOI] [PubMed] [Google Scholar]
- 59.Uegaki K, Adachi N, So S, et al. Heterozygous inactivation of human Ku70/Ku86 heterodimer does not affect cell growth, double-strand break repair, or genome integrity. DNA Repair (Amst) 2006;5:303–311. doi: 10.1016/j.dnarep.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 60.Fattah FJ, Lichter NF, Fattah KR, et al. Ku70, an essential gene, modulates the frequency of rAAV-mediated gene targeting in human somatic cells. PNAS. 2008;105:8703–8708. doi: 10.1073/pnas.0712060105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gama V, Gomez JA, Mayo LD, et al. Hdm2 is a ubiquitin ligase of Ku70-Akt promotes cell survival by inhibiting Hdm2-dependent Ku70 destabilization. Cell Death Differ. 2009;16:758–769. doi: 10.1038/cdd.2009.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li GC, Ouyang H, Li X, et al. Ku70: a candidate tumor suppressor gene for murine T cell lymphoma. Mol Cell. 1998;2:1–8. doi: 10.1016/s1097-2765(00)80108-2. [DOI] [PubMed] [Google Scholar]
- 63.Ouyang H, Nussenzweig A, Kurimasa A, et al. Ku70 is required for DNA repair but not for T cell antigen receptor gene recombination in vivo. J Exp Med. 1997;186:921–929. doi: 10.1084/jem.186.6.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee SE, Pulaski CR, He DM, et al. Isolation of mammalian cell mutants that are X-ray sensitive, impaired in DNA double-strand break repair and defective for V(D)J recombination. Mutat Res/DNA Repair. 1995;336:279–291. doi: 10.1016/0921-8777(95)00002-2. [DOI] [PubMed] [Google Scholar]
- 65.Finnie NJ, Gottlieb TM, Blunt T, et al. DNA-dependent protein kinase activity is absent in xrs-6 cells: implications for site-specific recombination and DNA double-strand break repair. PNAS. 1995;92:320–324. doi: 10.1073/pnas.92.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Singleton BK, Priestley A, Steingrimsdottir H, et al. Molecular and biochemical characterization of xrs mutants defective in Ku80. Mol Cell Biol. 1997;17:1264–1273. doi: 10.1128/MCB.17.3.1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zdzienicka MZ, Tran Q, van der Schans GP, Simons JWIM. Characterization of an X-ray-hypersensitive mutant of V79 Chinese hamster cells. Mutat Res/DNA Repair Rep. 1988;194:239–249. doi: 10.1016/0167-8817(88)90025-9. [DOI] [PubMed] [Google Scholar]
- 68.Zdzienicka MZ. Mammalian mutants defective in the response to ionizing radiation-induced DNA damage. Mutat Res/DNA Repair. 1995;336:203–213. doi: 10.1016/0921-8777(95)00003-3. [DOI] [PubMed] [Google Scholar]
- 69.Marangoni E, Le Romancer M, Foray N, et al. Transfer of Ku86 RNA antisense decreases the radioresistance of human fibroblasts. Cancer Gene Ther. 2000;7:339–346. doi: 10.1038/sj.cgt.7700111. [DOI] [PubMed] [Google Scholar]
- 70.Jaco I, Muñoz P, Blasco MA. Role of human Ku86 in telomere length maintenance and telomere capping. Cancer Res. 2004;64:7271–7278. doi: 10.1158/0008-5472.CAN-04-1381. [DOI] [PubMed] [Google Scholar]
- 71.Zhu C, Bogue MA, Lim D-S, et al. Ku86-deficient mice exhibit severe combined immunodeficiency and defective processing of V(D)J recombination intermediates. Cell. 1996;86:379–389. doi: 10.1016/S0092-8674(00)80111-7. [DOI] [PubMed] [Google Scholar]
- 72.Fan Y, Li J, Wei W, et al. Ku80 gene knockdown by the CRISPR/Cas9 technique affects the biological functions of human thyroid carcinoma cells. Oncol Rep. 2019;42:2486–2498. doi: 10.3892/or.2019.7348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fattah KR, Ruis BL, Hendrickson EA. Mutations to Ku reveal differences in human somatic cell lines. DNA Repair. 2008;7:762–774. doi: 10.1016/j.dnarep.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ghosh G, Li G, Myung K, Hendrickson EA. The lethality of Ku86 (XRCC5) loss-of-function mutations in human cells is independent of p53 (TP53) Radiat Res. 2007;167:66–79. doi: 10.1667/RR0692.1. [DOI] [PubMed] [Google Scholar]
- 75.Gu Y, Seidl KJ, Rathbun GA, et al. Growth retardation and leaky SCID phenotype of Ku70-deficient mice. Immunity. 1997;7:653–665. doi: 10.1016/S1074-7613(00)80386-6. [DOI] [PubMed] [Google Scholar]
- 76.Nussenzweig A, Sokol K, Burgman P, et al. Hypersensitivity of Ku80-deficient cell lines and mice to DNA damage: the effects of ionizing radiation on growth, survival, and development. Proc Natl Acad Sci U S A. 1997;94:13588–13593. doi: 10.1073/pnas.94.25.13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Karanjawala ZE, Grawunder U, Hsieh CL, Lieber MR. The nonhomologous DNA end joining pathway is important for chromosome stability in primary fibroblasts. Curr Biol. 1999;9:1501–1504. doi: 10.1016/s0960-9822(00)80123-2. [DOI] [PubMed] [Google Scholar]
- 78.Hasty P, Vijg J. Accelerating aging by mouse reverse genetics: a rational approach to understanding longevity. Aging Cell. 2004;3:55–65. doi: 10.1111/j.1474-9728.2004.00082.x. [DOI] [PubMed] [Google Scholar]
- 79.Vogel H, Lim D-S, Karsenty G, et al. Deletion of Ku86 causes early onset of senescence in mice. PNAS. 1999;96:10770–10775. doi: 10.1073/pnas.96.19.10770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Difilippantonio MJ, Zhu J, Chen HT, et al. DNA repair protein Ku80 suppresses chromosomal aberrations and malignant transformation. Nature. 2000;404:510–514. doi: 10.1038/35006670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ferguson DO, Sekiguchi JM, Chang S, et al. The nonhomologous end-joining pathway of DNA repair is required for genomic stability and the suppression of translocations. Proc Natl Acad Sci U S A. 2000;97:6630–6633. doi: 10.1073/pnas.110152897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ma S, Chang J, Wang X, et al. CRISPR/Cas9 mediated multiplex genome editing and heritable mutagenesis of BmKu70 in Bombyx mori. Sci Rep. 2014;4:4489. doi: 10.1038/srep04489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.C. elegans Deletion Mutant Consortium Large-scale screening for targeted knockouts in the Caenorhabditis elegans genome. G3 (Bethesda) 2012;2:1415–1425. doi: 10.1534/g3.112.003830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fukushima T, Takata M, Morrison C, et al. Genetic analysis of the DNA-dependent protein kinase reveals an inhibitory role of Ku in late S-G2 phase DNA double-strand break repair. J Biol Chem. 2001;276:44413–44418. doi: 10.1074/jbc.M106295200. [DOI] [PubMed] [Google Scholar]
- 85.Jeggo PA, Kemp LM. X-ray-sensitive mutants of Chinese hamster ovary cell line isolation and cross-sensitivity to other DNA-damaging agents. Mutat Res/DNA Repair Rep. 1983;112:313–327. doi: 10.1016/0167-8817(83)90026-3. [DOI] [PubMed] [Google Scholar]
- 86.Qiao Y-M, Yu R-L, Zhu P. Advances in targeting and heterologous expression of genes involved in the synthesis of fungal secondary metabolites. RSC Adv. 2019;9:35124–35134. doi: 10.1039/C9RA06908A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Baumann P, Cech TR. Protection of telomeres by the Ku protein in fission yeast. Mol Biol Cell. 2000;11:3265–3275. doi: 10.1091/mbc.11.10.3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.West CE, Waterworth WM, Story GW, et al. Disruption of the Arabidopsis AtKu80 gene demonstrates an essential role for AtKu80 protein in efficient repair of DNA double-strand breaks in vivo. Plant J. 2002;31:517–528. doi: 10.1046/j.1365-313X.2002.01370.x. [DOI] [PubMed] [Google Scholar]
- 89.Fox BA, Ristuccia JG, Gigley JP, Bzik DJ. Efficient gene replacements in Toxoplasma gondii strains deficient for nonhomologous end joining. Eukaryot Cell. 2009;8:520–529. doi: 10.1128/EC.00357-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Koike M. Dimerization, translocation and localization of Ku70 and Ku80 proteins. J Radiat Res. 2002;43:223–236. doi: 10.1269/jrr.43.223. [DOI] [PubMed] [Google Scholar]
- 91.Koike M, Shiomi T, Koike A. Ku70 can translocate to the nucleus independent of Ku80 translocation and DNA-PK autophosphorylation. Biochem Biophys Res Commun. 2000;276:1105–1111. doi: 10.1006/bbrc.2000.3567. [DOI] [PubMed] [Google Scholar]
- 92.Bertinato J, Schild-Poulter C, Haché RJG. Nuclear localization of Ku antigen is promoted independently by basic motifs in the Ku70 and Ku80 subunits. J Cell Sci. 2001;114:89–99. doi: 10.1242/jcs.114.1.89. [DOI] [PubMed] [Google Scholar]
- 93.Miyoshi T, Sadaie M, Kanoh J, Ishikawa F. Telomeric DNA ends are essential for the localization of Ku at telomeres in fission yeast. J Biol Chem. 2003;278:1924–1931. doi: 10.1074/jbc.M208813200. [DOI] [PubMed] [Google Scholar]
- 94.Mukherjee S, Chakraborty P, Saha P. Phosphorylation of Ku70 subunit by cell cycle kinases modulates the replication related function of Ku heterodimer. Nucleic Acids Res. 2016;44:7755–7765. doi: 10.1093/nar/gkw622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schild-Poulter C, Matheos D, Novac O, et al. Differential DNA binding of Ku antigen determines its involvement in DNA replication. DNA Cell Biol. 2003;22:65–78. doi: 10.1089/104454903321515887. [DOI] [PubMed] [Google Scholar]
- 96.Bertinato J, Tomlinson JJ, Schild-Poulter C, Haché RJG. Evidence implicating Ku antigen as a structural factor in RNA polymerase II-mediated transcription. Gene. 2003;302:53–64. doi: 10.1016/s0378111902010892. [DOI] [PubMed] [Google Scholar]
- 97.Mo X, Dynan WS. Subnuclear localization of Ku protein: functional association with RNA polymerase II elongation sites. Mol Cell Biol. 2002;22:8088–8099. doi: 10.1128/mcb.22.22.8088-8099.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Adelmant G, Calkins AS, Garg BK, et al. DNA ends alter the molecular composition and localization of Ku multicomponent complexes. Mol Cell Proteomics. 2012;11:411–421. doi: 10.1074/mcp.M111.013581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Moore HM, Bai B, Boisvert F-M, et al. Quantitative proteomics and dynamic imaging of the nucleolus reveal distinct responses to UV and ionizing radiation. Mol Cell Proteomics. 2011;10:009241. doi: 10.1074/mcp.M111.009241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shao Z, Flynn RA, Crowe JL, et al. DNA-PKcs has KU-dependent function in rRNA processing and haematopoiesis. Nature. 2020;579:291–296. doi: 10.1038/s41586-020-2041-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hanakahi LA, West SC. Specific interaction of IP6 with human Ku70/80, the DNA-binding subunit of DNA-PK. EMBO J. 2002;21:2038–2044. doi: 10.1093/emboj/21.8.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Byrum J, Jordan S, Safrany ST, Rodgers W. Visualization of inositol phosphate-dependent mobility of Ku: depletion of the DNA-PK cofactor InsP6 inhibits Ku mobility. Nucleic Acids Res. 2004;32:2776–2784. doi: 10.1093/nar/gkh592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cheung JCY, Salerno B, Hanakahi LA. Evidence for an inositol hexakisphosphate-dependent role for Ku in mammalian nonhomologous end joining that is independent of its role in the DNA-dependent protein kinase. Nucleic Acids Res. 2008;36:5713–5726. doi: 10.1093/nar/gkn572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dejmek J, Iglehart JD, Lazaro J-B. DNA-dependent protein kinase (DNA-PK)-dependent cisplatin-induced loss of nucleolar facilitator of chromatin transcription (FACT) and regulation of cisplatin sensitivity by DNA-PK and FACT. Mol Cancer Res. 2009;7:581–591. doi: 10.1158/1541-7786.MCR-08-0049. [DOI] [PubMed] [Google Scholar]
- 105.Higashiura M, Shimizu Y, Tanimoto M, et al. Immunolocalization of Ku-proteins (p80/p70): localization of p70 to nucleoli and periphery of both interphase nuclei and metaphase chromosomes. Exp Cell Res. 1992;201:444–451. doi: 10.1016/0014-4827(92)90293-h. [DOI] [PubMed] [Google Scholar]
- 106.Coffey G, Lakshmipathy U, Campbell C. Mammalian mitochondrial extracts possess DNA end-binding activity. Nucleic Acids Res. 1999;27:3348–3354. doi: 10.1093/nar/27.16.3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tadi SK, Sebastian R, Dahal S, et al. Microhomology-mediated end joining is the principal mediator of double-strand break repair during mitochondrial DNA lesions. Mol Biol Cell. 2016;27:223–235. doi: 10.1091/mbc.E15-05-0260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mimori T, Hardin JA, Steitz JA. Characterization of the DNA-binding protein antigen Ku recognized by autoantibodies from patients with rheumatic disorders. J Biol Chem. 1986;261:2274–2278. doi: 10.1016/S0021-9258(17)35929-X. [DOI] [PubMed] [Google Scholar]
- 109.Choi EK, Lee YH, Choi YS, et al. Heterogeneous expression of Ku70 in human tissues is associated with morphological and functional alterations of the nucleus. J Pathol. 2002;198:121–130. doi: 10.1002/path.1164. [DOI] [PubMed] [Google Scholar]
- 110.Yagura T, Sumi K. Molecular cloning and sequencing of cDNAs encoding homologues of human Ku70 and Ku80 autoantigen from Xenopus and their expression in various Xenopus tissues. Biochim Biophys Acta Gene Struct Expr. 1999;1445:160–164. doi: 10.1016/S0167-4781(99)00028-7. [DOI] [PubMed] [Google Scholar]
- 111.Jacoby DB, Wensink PC. Yolk protein factor 1 is a Drosophila homolog of Ku, the DNA-binding subunit of a DNA-dependent protein kinase from humans. J Biol Chem. 1994;269:11484–11491. doi: 10.1016/S0021-9258(19)78149-6. [DOI] [PubMed] [Google Scholar]
- 112.Tamura K, Adachi Y, Chiba K, et al. Identification of Ku70 and Ku80 homologues in Arabidopsis thaliana: evidence for a role in the repair of DNA double-strand breaks. Plant J. 2002;29:771–781. doi: 10.1046/j.1365-313x.2002.01258.x. [DOI] [PubMed] [Google Scholar]
- 113.Pink RC, Carter DRF. Pseudogenes as regulators of biological function. Essays Biochem. 2013;54:103–112. doi: 10.1042/bse0540103. [DOI] [PubMed] [Google Scholar]
- 114.Myung K, He DM, Lee SE, Hendrickson EA. KARP-1: a novel leucine zipper protein expressed from the Ku86 autoantigen locus is implicated in the control of DNA-dependent protein kinase activity. EMBO J. 1997;16:3172–3184. doi: 10.1093/emboj/16.11.3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Koike M, Yutoku Y, Koike A. KARP-1 works as a heterodimer with Ku70, but the function of KARP-1 cannot perfectly replace that of Ku80 in DSB repair. Exp Cell Res. 2011;317:2267–2275. doi: 10.1016/j.yexcr.2011.06.015. [DOI] [PubMed] [Google Scholar]
- 116.Myung K, Braastad C, He DM, Hendrickson EA. KARP-1 is induced by DNA damage in a p53- and ataxia telangiectasia mutated-dependent fashion. Proc Natl Acad Sci USA. 1998;95:7664–7669. doi: 10.1073/pnas.95.13.7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Do E, Taira E, Irie Y, et al. Molecular cloning and characterization of rKAB1, which interacts with KARP-1, localizes in the nucleus and protects cells against oxidative death. Mol Cell Biochem. 2003;248:77–83. doi: 10.1023/a:1024157515342. [DOI] [PubMed] [Google Scholar]
- 118.Davis AJ, Lee K-J, Chen DJ. The N-terminal region of the DNA-dependent protein kinase catalytic subunit is required for its DNA double-stranded break-mediated activation. J Biol Chem. 2013;288:7037–7046. doi: 10.1074/jbc.M112.434498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Singleton BK, Torres-Arzayus MI, Rottinghaus ST, et al. The C terminus of Ku80 activates the DNA-dependent protein kinase catalytic subunit. Mol Cell Biol. 1999;19:3267–3277. doi: 10.1128/mcb.19.5.3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Weterings E, Verkaik NS, Keijzers G, et al. The Ku80 carboxy terminus stimulates joining and artemis-mediated processing of DNA ends. Mol Cell Biol. 2009;29:1134–1142. doi: 10.1128/MCB.00971-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Radhakrishnan SK, Lees-Miller SP. DNA requirements for interaction of the C-terminal region of Ku80 with the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) DNA Repair. 2017;57:17–28. doi: 10.1016/j.dnarep.2017.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Jette N, Lees-Miller SP. The DNA-dependent protein kinase: a multifunctional protein kinase with roles in DNA double strand break repair and mitosis. Prog Biophys Mol Biol. 2015;117:194–205. doi: 10.1016/j.pbiomolbio.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Douglas P, Ye R, Radhamani S, et al. Nocodazole-induced expression and phosphorylation of Anillin and other mitotic proteins are decreased in DNA-dependent protein kinase catalytic subunit-deficient cells and rescued by inhibition of the anaphase-promoting complex/cyclosome with proTAME but not Apcin. Mol Cell Biol. 2020;40:e00191–e219. doi: 10.1128/MCB.00191-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bürckstümmer T, Bennett KL, Preradovic A, et al. An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nat Methods. 2006;3:1013–1019. doi: 10.1038/nmeth968. [DOI] [PubMed] [Google Scholar]
- 125.Shirodkar P, Fenton AL, Meng L, Koch CA. Identification and functional characterization of a Ku-binding motif in aprataxin polynucleotide kinase/phosphatase-like factor (APLF) J Biol Chem. 2013;288:19604–19613. doi: 10.1074/jbc.M112.440388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chung JH. The role of DNA-PK in aging and energy metabolism. FEBS J. 2018;285:1959–1972. doi: 10.1111/febs.14410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mari P-O, Florea BI, Persengiev SP, et al. Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proc Natl Acad Sci USA. 2006;103:18597–18602. doi: 10.1073/pnas.0609061103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Song K, Jung D, Jung Y, et al. Interaction of human Ku70 with TRF2. FEBS Lett. 2000;481:81–85. doi: 10.1016/s0014-5793(00)01958-x. [DOI] [PubMed] [Google Scholar]
- 129.Hsu H-L, Gilley D, Blackburn EH, Chen DJ. Ku is associated with the telomere in mammals. Proc Natl Acad Sci USA. 1999;96:12454–12458. doi: 10.1073/pnas.96.22.12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.O’Connor MS, Safari A, Liu D, et al. The human Rap1 protein complex and modulation of telomere length. J Biol Chem. 2004;279:28585–28591. doi: 10.1074/jbc.M312913200. [DOI] [PubMed] [Google Scholar]
- 131.Chai W, Ford LP, Lenertz L, et al. Human Ku70/80 associates physically with telomerase through interaction with hTERT. J Biol Chem. 2002;277:47242–47247. doi: 10.1074/jbc.M208542200. [DOI] [PubMed] [Google Scholar]
- 132.Fell VL, Walden EA, Hoffer SM, et al. Ku70 serine 155 mediates Aurora B inhibition and activation of the DNA damage response. Sci Rep. 2016;6:37194. doi: 10.1038/srep37194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Feki A, Jefford CE, Berardi P, et al. BARD1 induces apoptosis by catalysing phosphorylation of p53 by DNA-damage response kinase. Oncogene. 2005;24:3726–3736. doi: 10.1038/sj.onc.1208491. [DOI] [PubMed] [Google Scholar]
- 134.Goudelock DM, Jiang K, Pereira E, et al. Regulatory interactions between the checkpoint kinase Chk1 and the proteins of the DNA-dependent protein kinase complex. J Biol Chem. 2003;278:29940–29947. doi: 10.1074/jbc.M301765200. [DOI] [PubMed] [Google Scholar]
- 135.Thapar R, Wang JL, Hammel M, et al. Mechanism of efficient double-strand break repair by a long non-coding RNA. Nucleic Acids Res. 2020;48:10953–10972. doi: 10.1093/nar/gkaa784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gallardo F, Olivier C, Dandjinou AT, et al. TLC1 RNA nucleo-cytoplasmic trafficking links telomerase biogenesis to its recruitment to telomeres. EMBO J. 2008;27:748–757. doi: 10.1038/emboj.2008.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yuan M, Eberhart CG, Kai M. RNA binding protein RBM14 promotes radio-resistance in glioblastoma by regulating DNA repair and cell differentiation. Oncotarget. 2014;5:2820–2826. doi: 10.18632/oncotarget.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Maldonado E, Shiekhattar R, Sheldon M, et al. A human RNA polymerase II complex associated with SRB and DNA-repair proteins. Nature. 1996;381:86–89. doi: 10.1038/381086a0. [DOI] [PubMed] [Google Scholar]
- 139.Willis DM, Loewy AP, Charlton-Kachigian N, et al. Regulation of osteocalcin gene expression by a novel Ku antigen transcription factor complex. J Biol Chem. 2002;277:37280–37291. doi: 10.1074/jbc.M206482200. [DOI] [PubMed] [Google Scholar]
- 140.Shao RG, Cao CX, Zhang H, et al. Replication-mediated DNA damage by camptothecin induces phosphorylation of RPA by DNA-dependent protein kinase and dissociates RPA:DNA-PK complexes. EMBO J. 1999;18:1397–1406. doi: 10.1093/emboj/18.5.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Matheos D, Ruiz MT, Price GB, Zannis-Hadjopoulos M. Ku antigen, an origin-specific binding protein that associates with replication proteins, is required for mammalian DNA replication. Biochim Biophys Acta. 2002;1578:59–72. doi: 10.1016/s0167-4781(02)00497-9. [DOI] [PubMed] [Google Scholar]
- 142.Raval P, Kriatchko AN, Kumar S, Swanson PC. Evidence for Ku70/Ku80 association with full-length RAG1. Nucleic Acids Res. 2008;36:2060–2072. doi: 10.1093/nar/gkn049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Purugganan MM, Shah S, Kearney JF, Roth DB. Ku80 is required for addition of N nucleotides to V(D)J recombination junctions by terminal deoxynucleotidyl transferase. Nucleic Acids Res. 2001;29:1638–1646. doi: 10.1093/nar/29.7.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kim K-B, Kim D-W, Park JW, et al. Inhibition of Ku70 acetylation by INHAT subunit SET/TAF-Iβ regulates Ku70-mediated DNA damage response. Cell Mol Life Sci. 2014;71:2731–2745. doi: 10.1007/s00018-013-1525-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Gomez JA, Gama V, Yoshida T, et al. Bax-inhibiting peptides derived from Ku70 and cell-penetrating pentapeptides. Biochem Soc Trans. 2007;35:797–801. doi: 10.1042/BST0350797. [DOI] [PubMed] [Google Scholar]
- 146.Mazumder S, Plesca D, Kinter M, Almasan A. Interaction of a cyclin E fragment with Ku70 regulates Bax-mediated apoptosis. Mol Cell Biol. 2007;27:3511–3520. doi: 10.1128/MCB.01448-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Downs JA, Jackson SP. A means to a DNA end: the many roles of Ku. Nat Rev Mol Cell Biol. 2004;5:367–378. doi: 10.1038/nrm1367. [DOI] [PubMed] [Google Scholar]
- 148.Song K, Jung Y, Jung D, Lee I. Human Ku70 interacts with heterochromatin protein 1alpha. J Biol Chem. 2001;276:8321–8327. doi: 10.1074/jbc.M008779200. [DOI] [PubMed] [Google Scholar]
- 149.Yang CR, Yeh S, Leskov K, et al. Isolation of Ku70-binding proteins (KUBs) Nucleic Acids Res. 1999;27:2165–2174. doi: 10.1093/nar/27.10.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Costes SV, Daelemans D, Cho EH, et al. Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys J. 2004;86:3993–4003. doi: 10.1529/biophysj.103.038422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Fredriksson S, Gullberg M, Jarvius J, et al. Protein detection using proximity-dependent DNA ligation assays. Nat Biotechnol. 2002;20:473–477. doi: 10.1038/nbt0502-473. [DOI] [PubMed] [Google Scholar]
- 152.Frit P, Ropars V, Modesti M, et al. Plugged into the Ku-DNA hub: the NHEJ network. Prog Biophys Mol Biol. 2019;147:62–76. doi: 10.1016/j.pbiomolbio.2019.03.001. [DOI] [PubMed] [Google Scholar]
- 153.Hung PJ, Johnson B, Chen B-R, et al. MRI is a DNA damage response adaptor during classical non-homologous end joining. Mol Cell. 2018;71:332–342.e8. doi: 10.1016/j.molcel.2018.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kim K, Min J, Kirby TW, et al. Ligand binding characteristics of the Ku80 von Willebrand domain. DNA Repair (Amst) 2020;85:102739. doi: 10.1016/j.dnarep.2019.102739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bouley J, Saad L, Grall R, et al. A new phosphorylated form of Ku70 identified in resistant leukemic cells confers fast but unfaithful DNA repair in cancer cell lines. Oncotarget. 2015;6:27980–28000. doi: 10.18632/oncotarget.4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lee K-J, Saha J, Sun J, et al. Phosphorylation of Ku dictates DNA double-strand break (DSB) repair pathway choice in S phase. Nucleic Acids Res. 2016;44:1732–1745. doi: 10.1093/nar/gkv1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Brown JS, Lukashchuk N, Sczaniecka-Clift M, et al. Neddylation promotes ubiquitylation and release of Ku from DNA-damage sites. Cell Rep. 2015;11:704–714. doi: 10.1016/j.celrep.2015.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Chan DW, Ye R, Veillette CJ, Lees-Miller SP. DNA-dependent protein kinase phosphorylation sites in Ku 70/80 heterodimer. Biochemistry. 1999;38:1819–1828. doi: 10.1021/bi982584b. [DOI] [PubMed] [Google Scholar]
- 159.Jin S, Weaver DT. Double-strand break repair by Ku70 requires heterodimerization with Ku80 and DNA binding functions. EMBO J. 1997;16:6874–6885. doi: 10.1093/emboj/16.22.6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Douglas P, Gupta S, Morrice N, et al. DNA-PK-dependent phosphorylation of Ku70/80 is not required for non-homologous end joining. DNA Repair. 2005;4:1006–1018. doi: 10.1016/j.dnarep.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 161.Koike M, Koike A. Accumulation of Ku80 proteins at DNA double-strand breaks in living cells. Exp Cell Res. 2008;314:1061–1070. doi: 10.1016/j.yexcr.2007.11.014. [DOI] [PubMed] [Google Scholar]
- 162.Chi Y, Welcker M, Hizli AA, et al. Identification of CDK2 substrates in human cell lysates. Genome Biol. 2008;9:R149. doi: 10.1186/gb-2008-9-10-r149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Müller-Tidow C, Ji P, Diederichs S, et al. The cyclin A1-CDK2 complex regulates DNA double-strand break repair. Mol Cell Biol. 2004;24:8917–8928. doi: 10.1128/MCB.24.20.8917-8928.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ji P, Bäumer N, Yin T, et al. DNA damage response involves modulation of Ku70 and Rb functions by cyclin A1 in leukemia cells. Int J Cancer. 2007;121:706–713. doi: 10.1002/ijc.22634. [DOI] [PubMed] [Google Scholar]
- 165.Fell VL, Schild-Poulter C. Ku regulates signaling to DNA damage response pathways through the Ku70 von Willebrand A domain. Mol Cell Biol. 2012;32:76–87. doi: 10.1128/MCB.05661-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Blasius M, Forment JV, Thakkar N, et al. A phospho-proteomic screen identifies substrates of the checkpoint kinase Chk1. Genome Biol. 2011;12:R78. doi: 10.1186/gb-2011-12-8-r78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kettenbach AN, Wang T, Faherty BK, et al. Rapid determination of multiple linear kinase substrate motifs by mass spectrometry. Chem Biol. 2012;19:608–618. doi: 10.1016/j.chembiol.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Pucci S, Mazzarelli P, Sesti F, et al. Interleukin-6 affects cell death escaping mechanisms acting on Bax–Ku70-Clusterin interactions in human colon cancer progression. Cell Cycle. 2009;8:473–481. doi: 10.4161/cc.8.3.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Koike M, Yutoku Y, Koike A. Cloning of canine Ku80 and its localization and accumulation at DNA damage sites. FEBS Open Bio. 2017;7:1854–1863. doi: 10.1002/2211-5463.12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Al-Emam A, Arbon D, Kysela B. Deacetylation of Ku70 regulates ionizing-radiation induced DNA damage responses in human cells. BMC Genomics. 2014;15:P24. doi: 10.1186/1471-2164-15-S2-P24. [DOI] [Google Scholar]
- 171.Subramanian C, Hada M, Opipari AW, et al. CREB-binding protein regulates Ku70 acetylation in response to ionization radiation in neuroblastoma. Mol Cancer Res. 2013;11:173–181. doi: 10.1158/1541-7786.MCR-12-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Subramanian C, Opipari AW, Bian X, et al. Ku70 acetylation mediates neuroblastoma cell death induced by histone deacetylase inhibitors. Proc Natl Acad Sci USA. 2005;102:4842–4847. doi: 10.1073/pnas.0408351102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Xiao Y, Wang J, Qin Y, et al. Ku80 cooperates with CBP to promote COX-2 expression and tumor growth. Oncotarget. 2015;6:8046–8061. doi: 10.18632/oncotarget.3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Li L, Ye S, Yang M, et al. SIRT1 downregulation enhances chemosensitivity and survival of adult T-cell leukemia-lymphoma cells by reducing DNA double-strand repair. Oncol Rep. 2015;34:2935–2942. doi: 10.3892/or.2015.4287. [DOI] [PubMed] [Google Scholar]
- 175.Tao N-N, Ren J-H, Tang H, et al. Deacetylation of Ku70 by SIRT6 attenuates Bax-mediated apoptosis in hepatocellular carcinoma. Biochem Biophys Res Commun. 2017;485:713–719. doi: 10.1016/j.bbrc.2017.02.111. [DOI] [PubMed] [Google Scholar]
- 176.Chaudhary N, Nakka KK, Chavali PL, et al. SMAR1 coordinates HDAC6-induced deacetylation of Ku70 and dictates cell fate upon irradiation. Cell Death Dis. 2014;5:e1447. doi: 10.1038/cddis.2014.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Schwertman P, Bekker-Jensen S, Mailand N. Regulation of DNA double-strand break repair by ubiquitin and ubiquitin-like modifiers. Nat Rev Mol Cell Biol. 2016;17:379–394. doi: 10.1038/nrm.2016.58. [DOI] [PubMed] [Google Scholar]
- 178.Postow L, Ghenoiu C, Woo EM, et al. Ku80 removal from DNA through double strand break–induced ubiquitylation. J Cell Biol. 2008;182:467–479. doi: 10.1083/jcb.200802146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Hang LE, Lopez CR, Liu X, et al. Regulation of Ku-DNA association by Yku70 C-terminal tail and SUMO modification. J Biol Chem. 2014;289:10308–10317. doi: 10.1074/jbc.M113.526178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Williams ES, Stap J, Essers J, et al. DNA double-strand breaks are not sufficient to initiate recruitment of TRF2. Nat Genet. 2007;39:696–699. doi: 10.1038/ng0607-696. [DOI] [PubMed] [Google Scholar]
- 181.Weterings E, Verkaik NS, Brüggenwirth HT, et al. The role of DNA dependent protein kinase in synapsis of DNA ends. Nucleic Acids Res. 2003;31:7238–7246. doi: 10.1093/nar/gkg889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Dobbs TA, Tainer JA, Lees-Miller SP. A structural model for regulation of NHEJ by DNA-PKcs autophosphorylation. DNA Repair (Amst) 2010;9:1307–1314. doi: 10.1016/j.dnarep.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Radhakrishnan SK, Jette N, Lees-Miller SP. Non-homologous end joining: emerging themes and unanswered questions. DNA Repair (Amst) 2014;17:2–8. doi: 10.1016/j.dnarep.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Brouwer I, Sitters G, Candelli A, et al. Sliding sleeves of XRCC4-XLF bridge DNA and connect fragments of broken DNA. Nature. 2016;535:566–569. doi: 10.1038/nature18643. [DOI] [PubMed] [Google Scholar]
- 185.Menon V, Povirk LF. End-processing nucleases and phosphodiesterases: an elite supporting cast for the non-homologous end joining pathway of DNA double-strand break repair. DNA Repair (Amst) 2016;43:57–68. doi: 10.1016/j.dnarep.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 186.Critchlow SE, Bowater RP, Jackson SP. Mammalian DNA double-strand break repair protein XRCC4 interacts with DNA ligase IV. Curr Biol. 1997;7:588–598. doi: 10.1016/s0960-9822(06)00258-2. [DOI] [PubMed] [Google Scholar]
- 187.Deshpande RA, Myler LR, Soniat MM, et al. DNA-dependent protein kinase promotes DNA end processing by MRN and CtIP. Sci Adv. 2020;6:eaay0922. doi: 10.1126/sciadv.aay0922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Deriano L, Roth DB. Modernizing the nonhomologous end-joining repertoire: alternative and classical NHEJ share the stage. Annu Rev Genet. 2013;47:433–455. doi: 10.1146/annurev-genet-110711-155540. [DOI] [PubMed] [Google Scholar]
- 189.Postow L. Destroying the ring: freeing DNA from Ku with ubiquitin. FEBS Lett. 2011;585:2876–2882. doi: 10.1016/j.febslet.2011.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Fugmann SD, Lee AI, Shockett PE, et al. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu Rev Immunol. 2000;18:495–527. doi: 10.1146/annurev.immunol.18.1.495. [DOI] [PubMed] [Google Scholar]
- 191.Jackson SP, Jeggo PA. DNA double-strand break repair and V(D)J recombination: involvement of DNA-PK. Trends Biochem Sci. 1995;20:412–415. doi: 10.1016/s0968-0004(00)89090-8. [DOI] [PubMed] [Google Scholar]
- 192.Bassing CH, Swat W, Alt FW. The mechanism and regulation of chromosomal V(D)J recombination. Cell. 2002;109:S45–S55. doi: 10.1016/s0092-8674(02)00675-x. [DOI] [PubMed] [Google Scholar]
- 193.Soulas-Sprauel P, Rivera-Munoz P, Malivert L, et al. V(D)J and immunoglobulin class switch recombinations: a paradigm to study the regulation of DNA end-joining. Oncogene. 2007;26:7780–7791. doi: 10.1038/sj.onc.1210875. [DOI] [PubMed] [Google Scholar]
- 194.Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 195.Nowsheen S, Yang ES. The intersection between DNA damage response and cell death pathways. Exp Oncol. 2012;34:243–254. [PMC free article] [PubMed] [Google Scholar]
- 196.Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 197.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Wang Y, Cortez D, Yazdi P, et al. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 2000;14:927–939. doi: 10.1101/gad.14.8.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Rouse J, Jackson SP. Interfaces between the detection, signaling, and repair of DNA damage. Science. 2002;297:547–551. doi: 10.1126/science.1074740. [DOI] [PubMed] [Google Scholar]
- 200.Lamarche BJ, Orazio NI, Weitzman MD. The MRN complex in double-strand break repair and telomere maintenance. FEBS Lett. 2010;584:3682–3695. doi: 10.1016/j.febslet.2010.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Wang M, Wu W, Wu W, et al. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 2006;34:6170–6182. doi: 10.1093/nar/gkl840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Blackford AN, Jackson SP. ATM, ATR, and DNA-PK: the trinity at the heart of the DNA damage response. Mol Cell. 2017;66:801–817. doi: 10.1016/j.molcel.2017.05.015. [DOI] [PubMed] [Google Scholar]
- 203.Burma S, Chen BP, Murphy M, et al. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J Biol Chem. 2001;276:42462–42467. doi: 10.1074/jbc.C100466200. [DOI] [PubMed] [Google Scholar]
- 204.Tomimatsu N, Tahimic CGT, Otsuki A, et al. Ku70/80 modulates ATM and ATR signaling pathways in response to DNA double strand breaks. J Biol Chem. 2007;282:10138–10145. doi: 10.1074/jbc.M611880200. [DOI] [PubMed] [Google Scholar]
- 205.Barnes G, Rio D. DNA double-strand-break sensitivity, DNA replication, and cell cycle arrest phenotypes of Ku-deficient Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1997;94:867–872. doi: 10.1073/pnas.94.3.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Cosgrove AJ, Nieduszynski CA, Donaldson AD. Ku complex controls the replication time of DNA in telomere regions. Genes Dev. 2002;16:2485–2490. doi: 10.1101/gad.231602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Miyoshi T, Kanoh J, Ishikawa F. Fission yeast Ku protein is required for recovery from DNA replication stress. Genes Cells. 2009;14:1091–1103. doi: 10.1111/j.1365-2443.2009.01337.x. [DOI] [PubMed] [Google Scholar]
- 208.Sánchez A, Russell P. Ku stabilizes replication forks in the absence of Brc1. PLoS ONE. 2015;10:e0126598. doi: 10.1371/journal.pone.0126598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Foster SS, Balestrini A, Petrini JHJ. Functional interplay of the Mre11 nuclease and Ku in the response to replication-associated DNA damage. Mol Cell Biol. 2011;31:4379–4389. doi: 10.1128/MCB.05854-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Teixeira-Silva A, Ait Saada A, Hardy J, et al. The end-joining factor Ku acts in the end-resection of double strand break-free arrested replication forks. Nat Commun. 2017;8:1982. doi: 10.1038/s41467-017-02144-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Abdelbaqi K, Di Paola D, Rampakakis E, Zannis-Hadjopoulos M. Ku protein levels, localization and association to replication origins in different stages of breast tumor progression. J Cancer. 2013;4:358–370. doi: 10.7150/jca.6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Novac O, Matheos D, Araujo FD, et al. In vivo association of Ku with mammalian origins of DNA replication. Mol Biol Cell. 2001;12:3386–3401. doi: 10.1091/mbc.12.11.3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Park S-J, Ciccone SLM, Freie B, et al. A positive role for the Ku complex in DNA replication following strand break damage in mammals. J Biol Chem. 2004;279:6046–6055. doi: 10.1074/jbc.M311054200. [DOI] [PubMed] [Google Scholar]
- 214.Rampakakis E, Di Paola D, Zannis-Hadjopoulos M. Ku is involved in cell growth, DNA replication and G1-S transition. J Cell Sci. 2008;121:590–600. doi: 10.1242/jcs.021352. [DOI] [PubMed] [Google Scholar]
- 215.Galande S, Kohwi-Shigematsu T. Poly(ADP-ribose) polymerase and Ku autoantigen form a complex and synergistically bind to matrix attachment sequences. J Biol Chem. 1999;274:20521–20528. doi: 10.1074/jbc.274.29.20521. [DOI] [PubMed] [Google Scholar]
- 216.Giffin W, Torrance H, Rodda DJ, et al. Sequence-specific DNA binding by Ku autoantigen and its effects on transcription. Nature. 1996;380:265–268. doi: 10.1038/380265a0. [DOI] [PubMed] [Google Scholar]
- 217.Giffin W, Kwast-Welfeld J, Rodda DJ, et al. Sequence-specific DNA binding and transcription factor phosphorylation by Ku autoantigen/DNA-dependent protein kinase. J Biol Chem. 1997;272:5647–5658. doi: 10.1074/jbc.272.9.5647. [DOI] [PubMed] [Google Scholar]
- 218.Ludwig DL, Chen F, Peterson SR, et al. Ku80 gene expression is Sp1-dependent and sensitive to CpG methylation within a novel cis element. Gene. 1997;199:181–194. doi: 10.1016/s0378-1119(97)00366-1. [DOI] [PubMed] [Google Scholar]
- 219.Li GC, Yang SH, Kim D, et al. Suppression of heat-induced hsp70 expression by the 70-kDa subunit of the human Ku autoantigen. Proc Natl Acad Sci USA. 1995;92:4512–4516. doi: 10.1073/pnas.92.10.4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Ono M, Tucker PW, Capra JD. Ku is a general inhibitor of DNA-protein complex formation and transcription. Mol Immunol. 1996;33:787–796. doi: 10.1016/0161-5890(96)00030-2. [DOI] [PubMed] [Google Scholar]
- 221.Bunch H, Lawney BP, Lin Y-F, et al. Transcriptional elongation requires DNA break-induced signalling. Nat Commun. 2015;6:10191. doi: 10.1038/ncomms10191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Dvir A, Stein LY, Calore BL, Dynan WS. Purification and characterization of a template-associated protein kinase that phosphorylates RNA polymerase II. J Biol Chem. 1993;268:10440–10447. doi: 10.1016/S0021-9258(18)82219-0. [DOI] [PubMed] [Google Scholar]
- 223.Anderson CW. DNA damage and the DNA-activated protein kinase. Trends Biochem Sci. 1993;18:433–437. doi: 10.1016/0968-0004(93)90144-c. [DOI] [PubMed] [Google Scholar]
- 224.Trotter KW, King HA, Archer TK. Glucocorticoid receptor transcriptional activation via the BRG1-dependent recruitment of TOP2β and Ku70/86. Mol Cell Biol. 2015;35:2799–2817. doi: 10.1128/MCB.00230-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Nolens G, Pignon J-C, Koopmansch B, et al. Ku proteins interact with activator protein-2 transcription factors and contribute to ERBB2 overexpression in breast cancer cell lines. Breast Cancer Res. 2009;11:R83. doi: 10.1186/bcr2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Oh WJ, Kim EK, Ko JH, et al. Nuclear proteins that bind to metal response element a (MREa) in the Wilson disease gene promoter are Ku autoantigens and the Ku-80 subunit is necessary for basal transcription of the WD gene. Eur J Biochem. 2002;269:2151–2161. doi: 10.1046/j.1432-1033.2002.02865.x. [DOI] [PubMed] [Google Scholar]
- 227.Manic G, Maurin-Marlin A, Laurent F, et al. Impact of the Ku complex on HIV-1 expression and latency. PLoS ONE. 2013;8:e69691. doi: 10.1371/journal.pone.0069691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 229.Shay JW, Wright WE. Telomeres and telomerase: three decades of progress. Nat Rev Genet. 2019;20:299–309. doi: 10.1038/s41576-019-0099-1. [DOI] [PubMed] [Google Scholar]
- 230.Wood AM, Laster K, Rice EL, Kosak ST. A beginning of the end: new insights into the functional organization of telomeres. Nucleus. 2015;6:172–178. doi: 10.1080/19491034.2015.1048407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Shay JW. Telomeres and aging. Curr Opin Cell Biol. 2018;52:1–7. doi: 10.1016/j.ceb.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 232.Boulton SJ, Jackson SP. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 1998;17:1819–1828. doi: 10.1093/emboj/17.6.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Gravel S, Larrivée M, Labrecque P, Wellinger RJ. Yeast Ku as a regulator of chromosomal DNA end structure. Science. 1998;280:741–744. doi: 10.1126/science.280.5364.741. [DOI] [PubMed] [Google Scholar]
- 234.Martin SG, Laroche T, Suka N, et al. Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell. 1999;97:621–633. doi: 10.1016/s0092-8674(00)80773-4. [DOI] [PubMed] [Google Scholar]
- 235.Porter SE, Greenwell PW, Ritchie KB, Petes TD. The DNA-binding protein Hdf1p (a putative Ku homologue) is required for maintaining normal telomere length in Saccharomyces cerevisiae. Nucleic Acids Res. 1996;24:582–585. doi: 10.1093/nar/24.4.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Boulton SJ, Jackson SP. Identification of a Saccharomyces cerevisiae Ku80 homologue: roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic Acids Res. 1996;24:4639–4648. doi: 10.1093/nar/24.23.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Melnikova L, Biessmann H, Georgiev P. The Ku protein complex is involved in length regulation of Drosophila telomeres. Genetics. 2005;170:221–235. doi: 10.1534/genetics.104.034538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Hsu HL, Gilley D, Galande SA, et al. Ku acts in a unique way at the mammalian telomere to prevent end joining. Genes Dev. 2000;14:2807–2812. doi: 10.1101/gad.844000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Samper E, Goytisolo FA, Slijepcevic P, et al. Mammalian Ku86 protein prevents telomeric fusions independently of the length of TTAGGG repeats and the G-strand overhang. EMBO Rep. 2000;1:244–252. doi: 10.1093/embo-reports/kvd051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Espejel S, Franco S, Rodríguez-Perales S, et al. Mammalian Ku86 mediates chromosomal fusions and apoptosis caused by critically short telomeres. EMBO J. 2002;21:2207–2219. doi: 10.1093/emboj/21.9.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Fisher TS, Zakian VA. Ku: a multifunctional protein involved in telomere maintenance. DNA Repair (Amst) 2005;4:1215–1226. doi: 10.1016/j.dnarep.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 242.Bertuch AA, Lundblad V. The Ku heterodimer performs separable activities at double-strand breaks and chromosome termini. Mol Cell Biol. 2003;23:8202–8215. doi: 10.1128/mcb.23.22.8202-8215.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Sui J, Zhang S, Chen BPC. DNA-dependent protein kinase in telomere maintenance and protection. Cell Mol Biol Lett. 2020;25:2. doi: 10.1186/s11658-020-0199-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Wang Y, Ghosh G, Hendrickson EA. Ku86 represses lethal telomere deletion events in human somatic cells. PNAS. 2009;106:12430–12435. doi: 10.1073/pnas.0903362106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Matsui M, Corey DR. Non-coding RNAs as drug targets. Nat Rev Drug Discov. 2017;16:167–179. doi: 10.1038/nrd.2016.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Donlic A, Hargrove AE. Targeting RNA in mammalian systems with small molecules. Wiley Interdiscip Rev: RNA. 2018;9:e1477. doi: 10.1002/wrna.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Dutertre M, Vagner S. DNA-damage response RNA-binding proteins (DDRBPs): perspectives from a new class of proteins and their RNA targets. J Mol Biol. 2017;429:3139–3145. doi: 10.1016/j.jmb.2016.09.019. [DOI] [PubMed] [Google Scholar]
- 248.Kaczmarski W, Khan SA. Lupus autoantigen Ku protein binds HIV-1 TAR RNA in vitro. Biochem Biophys Res Commun. 1993;196:935–942. doi: 10.1006/bbrc.1993.2339. [DOI] [PubMed] [Google Scholar]
- 249.Yoo S, Dynan WS. Characterization of the RNA binding properties of Ku protein. Biochemistry. 1998;37:1336–1343. doi: 10.1021/bi972100w. [DOI] [PubMed] [Google Scholar]
- 250.Peterson SE, Stellwagen AE, Diede SJ, et al. The function of a stem-loop in telomerase RNA is linked to the DNA repair protein Ku. Nat Genet. 2001;27:64–67. doi: 10.1038/83778. [DOI] [PubMed] [Google Scholar]
- 251.Stellwagen AE, Haimberger ZW, Veatch JR, Gottschling DE. Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends. Genes Dev. 2003;17:2384–2395. doi: 10.1101/gad.1125903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Chen H, Xue J, Churikov D, et al. Structural insights into yeast telomerase recruitment to telomeres. Cell. 2018;172:331–343. doi: 10.1016/j.cell.2017.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Pfingsten JS, Goodrich KJ, Taabazuing C, et al. Mutually exclusive binding of telomerase RNA and DNA by Ku alters telomerase recruitment model. Cell. 2012;148:922–932. doi: 10.1016/j.cell.2012.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Ting NSY, Yu Y, Pohorelic B, et al. Human Ku70/80 interacts directly with hTR, the RNA component of human telomerase. Nucleic Acids Res. 2005;33:2090–2098. doi: 10.1093/nar/gki342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Zhang Y, He Q, Hu Z, et al. Long noncoding RNA LINP1 regulates repair of DNA double-strand breaks in triple-negative breast cancer. Nat Struct Mol Biol. 2016;23:522–530. doi: 10.1038/nsmb.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Santos-Pereira JM, Aguilera A. R loops: new modulators of genome dynamics and function. Nat Rev Genet. 2015;16:583–597. doi: 10.1038/nrg3961. [DOI] [PubMed] [Google Scholar]
- 257.Crossley MP, Bocek M, Cimprich KA. R-loops as cellular regulators and genomic threats. Mol Cell. 2019;73:398–411. doi: 10.1016/j.molcel.2019.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Bader AS, Hawley BR, Wilczynska A, Bushell M. The roles of RNA in DNA double-strand break repair. Br J Cancer. 2020;122:613–623. doi: 10.1038/s41416-019-0624-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Ohle C, Tesorero R, Schermann G, et al. Transient RNA-DNA hybrids are required for efficient double-strand break repair. Cell. 2016;167:1001–1013.e7. doi: 10.1016/j.cell.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 260.Lu W-T, Hawley BR, Skalka GL, et al. Drosha drives the formation of DNA:RNA hybrids around DNA break sites to facilitate DNA repair. Nat Commun. 2018;9:532. doi: 10.1038/s41467-018-02893-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Britton S, Dernoncourt E, Delteil C, et al. DNA damage triggers SAF-A and RNA biogenesis factors exclusion from chromatin coupled to R-loops removal. Nucleic Acids Res. 2014;42:9047–9062. doi: 10.1093/nar/gku601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Wang IX, Grunseich C, Fox J, et al. Human proteins that interact with RNA/DNA hybrids. Genome Res. 2018;28:1405–1414. doi: 10.1101/gr.237362.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Cristini A, Groh M, Kristiansen MS, Gromak N. RNA/DNA hybrid interactome identifies DXH9 as a molecular player in transcriptional termination and R-loop-associated DNA damage. Cell Rep. 2018;23:1891–1905. doi: 10.1016/j.celrep.2018.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Lamaa A, Le Bras M, Skuli N, et al. A novel cytoprotective function for the DNA repair protein Ku in regulating p53 mRNA translation and function. EMBO Rep. 2016;17:508–518. doi: 10.15252/embr.201541181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Lombard DB, Chua KF, Mostoslavsky R, et al. DNA repair, genome stability, and aging. Cell. 2005;120:497–512. doi: 10.1016/j.cell.2005.01.028. [DOI] [PubMed] [Google Scholar]
- 266.Li H, Vogel H, Holcomb VB, et al. Deletion of Ku70, Ku80, or both causes early aging without substantially increased cancer. Mol Cell Biol. 2007;27:8205–8214. doi: 10.1128/MCB.00785-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Park S-J, Gavrilova O, Brown AL, et al. DNA-PK promotes the mitochondrial, metabolic, and physical decline that occurs during aging. Cell Metab. 2017;25:1135–1146.e7. doi: 10.1016/j.cmet.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Armanios M. Telomeres and age-related disease: how telomere biology informs clinical paradigms. J Clin Invest. 2013;123:996–1002. doi: 10.1172/JCI66370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Liu G-H, Barkho BZ, Ruiz S, et al. Recapitulation of premature ageing with iPSCs from Hutchinson–Gilford progeria syndrome. Nature. 2011;472:221–225. doi: 10.1038/nature09879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Karmakar P, Snowden CM, Ramsden DA, Bohr VA. Ku heterodimer binds to both ends of the Werner protein and functional interaction occurs at the Werner N-terminus. Nucleic Acids Res. 2002;30:3583–3591. doi: 10.1093/nar/gkf482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Shamanna RA, Croteau DL, Lee J-H, Bohr VA. Recent advances in understanding Werner syndrome. F1000Research. 2017;6:1779. doi: 10.12688/f1000research.12110.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Opresko PL, Cheng W-H, von Kobbe C, et al. Werner syndrome and the function of the Werner protein; what they can teach us about the molecular aging process. Carcinogenesis. 2003;24:791–802. doi: 10.1093/carcin/bgg034. [DOI] [PubMed] [Google Scholar]
- 273.Belizna C, Henrion D, Beucher A, et al. Anti-Ku antibodies: clinical, genetic and diagnostic insights. Autoimmun Rev. 2010;9:691–694. doi: 10.1016/j.autrev.2010.05.020. [DOI] [PubMed] [Google Scholar]
- 274.Cavazzana I, Ceribelli A, Quinzanini M, et al. Prevalence and clinical associations of anti-Ku antibodies in systemic autoimmune diseases. Lupus. 2008;17:727–732. doi: 10.1177/0961203308089442. [DOI] [PubMed] [Google Scholar]
- 275.Chang WSJ, Schollum J, White DHN, Solanki KK. A cross-sectional study of autoantibody profiles in the Waikato systemic sclerosis cohort, New Zealand. Clin Rheumatol. 2015;34:1921–1927. doi: 10.1007/s10067-015-2981-3. [DOI] [PubMed] [Google Scholar]
- 276.Rozman B, Cucnik S, Sodin-Semrl S, et al. Prevalence and clinical associations of anti-Ku antibodies in patients with systemic sclerosis: a European EUSTAR-initiated multi-centre case-control study. Ann Rheum Dis. 2008;67:1282–1286. doi: 10.1136/ard.2007.073981. [DOI] [PubMed] [Google Scholar]
- 277.Hoa S, Hudson M, Troyanov Y, et al. Single-specificity anti-Ku antibodies in an international cohort of 2140 systemic sclerosis subjects: clinical associations. Medicine. 2016;95:e4713. doi: 10.1097/MD.0000000000004713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Low AHL, Wong S, Thumboo J, et al. Evaluation of a new multi-parallel line immunoassay for systemic sclerosis-associated antibodies in an Asian population. Rheumatology (Oxford) 2012;51:1465–1470. doi: 10.1093/rheumatology/kes055. [DOI] [PubMed] [Google Scholar]
- 279.Patterson KA, Roberts-Thomson PJ, Lester S, et al. Interpretation of an extended autoantibody profile in a well-characterized Australian Systemic Sclerosis (Scleroderma) cohort using principal components analysis. Arthritis Rheumatol. 2015;67:3234–3244. doi: 10.1002/art.39316. [DOI] [PubMed] [Google Scholar]
- 280.Rodriguez-Reyna TS, Hinojosa-Azaola A, Martinez-Reyes C, et al. Distinctive autoantibody profile in Mexican Mestizo systemic sclerosis patients. Autoimmunity. 2011;44:576–584. doi: 10.3109/08916934.2011.592886. [DOI] [PubMed] [Google Scholar]
- 281.Ogawa-Momohara M, Muro Y, Akiyama M. Overlap of systemic lupus erythematosus and myositis is rare in anti-Ku antibody-positive patients. Ann Rheum Dis. 2019 doi: 10.1136/annrheumdis-2019-216375. [DOI] [PubMed] [Google Scholar]
- 282.Spielmann L, Nespola B, Séverac F, et al. Anti-Ku syndrome with elevated CK and anti-Ku syndrome with anti-dsDNA are two distinct entities with different outcomes. Ann Rheum Dis. 2019;78:1101–1106. doi: 10.1136/annrheumdis-2018-214439. [DOI] [PubMed] [Google Scholar]
- 283.Schild-Poulter C, Su A, Shih A, et al. Association of autoantibodies with Ku and DNA repair proteins in connective tissue diseases. Rheumatology (Oxford) 2008;47:165–171. doi: 10.1093/rheumatology/kem338. [DOI] [PubMed] [Google Scholar]
- 284.Takeda Y. Autoantibodies against DNA double-strand break repair proteins. Front Biosci. 2001;6:d1412. doi: 10.2741/Takeda. [DOI] [PubMed] [Google Scholar]
- 285.Pichlmair A, e Sousa CR. Innate recognition of viruses. Immunity. 2007;27:370–383. doi: 10.1016/j.immuni.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 286.Ferguson BJ, Mansur DS, Peters NE, et al. DNA-PK is a DNA sensor for IRF-3-dependent innate immunity. Elife. 2012;1:e00047. doi: 10.7554/eLife.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Wang J, Kang L, Song D, et al. Ku70 senses HTLV-1 DNA and modulates HTLV-1 replication. J Immunol. 2017;199:2475–2482. doi: 10.4049/jimmunol.1700111. [DOI] [PubMed] [Google Scholar]
- 288.Li Y, Wu Y, Zheng X, et al. Cytoplasm-translocated Ku70/80 complex sensing of HBV DNA induces hepatitis-associated chemokine secretion. Front Immunol. 2016;7:569. doi: 10.3389/fimmu.2016.00569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Sui H, Zhou M, Imamichi H, et al. STING is an essential mediator of the Ku70-mediated production of IFN-λ1 in response to exogenous DNA. Sci Signal. 2017;10:eaah5054. doi: 10.1126/scisignal.aah5054. [DOI] [PubMed] [Google Scholar]
- 290.Peters NE, Ferguson BJ, Mazzon M, et al. A mechanism for the inhibition of DNA-PK-mediated DNA sensing by a virus. PLOS Pathog. 2013;9:e1003649. doi: 10.1371/journal.ppat.1003649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Scutts SR, Ember SW, Ren H, et al. DNA-PK is targeted by multiple vaccinia virus proteins to inhibit DNA sensing. Cell Rep. 2018;25:1953–1965.e4. doi: 10.1016/j.celrep.2018.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Bonilla FA, Oettgen HC. Adaptive immunity. J Allergy Clin Immunol. 2010;125:S33–S40. doi: 10.1016/j.jaci.2009.09.017. [DOI] [PubMed] [Google Scholar]
- 293.Roth DB. V(D)J recombination: mechanism, errors, and fidelity. Microbiol Spectr. 2014 doi: 10.1128/microbiolspec.MDNA3-0041-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Fischer A. Severe combined immunodeficiencies (SCID) Clin Exp Immunol. 2000;122:143–149. doi: 10.1046/j.1365-2249.2000.01359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Woodbine L, Gennery AR, Jeggo PA. The clinical impact of deficiency in DNA non-homologous end-joining. DNA Repair. 2014;16:84–96. doi: 10.1016/j.dnarep.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 296.Sishc BJ, Davis AJ. The role of the core non-homologous end joining factors in carcinogenesis and cancer. Cancers. 2017;9:81. doi: 10.3390/cancers9070081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Komuro Y, Watanabe T, Hosoi Y, et al. The expression pattern of Ku correlates with tumor radiosensitivity and disease free survival in patients with rectal carcinoma. Cancer. 2002;95:1199–1205. doi: 10.1002/cncr.10807. [DOI] [PubMed] [Google Scholar]
- 298.Rigas B, Borgo S, Elhosseiny A, et al. Decreased expression of DNA-dependent protein kinase, a DNA repair protein, during human colon carcinogenesis. Cancer Res. 2001;61:8381–8384. [PubMed] [Google Scholar]
- 299.Ayene IS, Ford LP, Koch CJ. Ku protein targeting by Ku70 small interfering RNA enhances human cancer cell response to topoisomerase II inhibitor and gamma radiation. Mol Cancer Ther. 2005;4:529–536. doi: 10.1158/1535-7163.MCT-04-0130. [DOI] [PubMed] [Google Scholar]
- 300.Beggs AD, Domingo E, McGregor M, et al. Loss of expression of the double strand break repair protein ATM is associated with worse prognosis in colorectal cancer and loss of Ku70 expression is associated with CIN. Oncotarget. 2012;3:1348–1355. doi: 10.18632/oncotarget.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Wilson CR, Davidson SE, Margison GP, et al. Expression of Ku70 correlates with survival in carcinoma of the cervix. Br J Cancer. 2000;83:1702–1706. doi: 10.1054/bjoc.2000.1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Hu H, Zhang Y, Zou M, et al. Expression of TRF1, TRF2, TIN2, TERT, KU70, and BRCA1 proteins is associated with telomere shortening and may contribute to multistage carcinogenesis of gastric cancer. J Cancer Res Clin Oncol. 2010;136:1407–1414. doi: 10.1007/s00432-010-0795-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Mazzarelli P, Parrella P, Seripa D, et al. DNA end binding activity and Ku70/80 heterodimer expression in human colorectal tumor. World J Gastroenterol. 2005;11:6694–6700. doi: 10.3748/wjg.v11.i42.6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Ma Q, Li P, Xu M, et al. Ku80 is highly expressed in lung adenocarcinoma and promotes cisplatin resistance. J Exp Clin Cancer Res. 2012;31:99. doi: 10.1186/1756-9966-31-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Pucci S, Mazzarelli P, Rabitti C, et al. Tumor specific modulation of KU70/80 DNA binding activity in breast and bladder human tumor biopsies. Oncogene. 2001;20:739–747. doi: 10.1038/sj.onc.1204148. [DOI] [PubMed] [Google Scholar]
- 306.Ai J, Pascal LE, Wei L, et al. EAF2 regulates DNA repair through Ku70/Ku80 in the prostate. Oncogene. 2017;36:2054–2065. doi: 10.1038/onc.2016.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Weterings E, Gallegos AC, Dominick LN, et al. A novel small molecule inhibitor of the DNA repair protein Ku70/80. DNA Repair (Amst) 2016;43:98–106. doi: 10.1016/j.dnarep.2016.03.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.
Not applicable.