Abstract
The mammalian salivary gland develops as a highly branched structure designed to produce and secrete saliva. This review focuses on research conducted on mammalian salivary gland development, particularly on the differentiation of acinar, ductal, and myoepithelial cells. We discuss recent studies that provide conceptual advances in the understanding of the molecular mechanisms of salivary gland development. In addition, we describe the organogenesis of submandibular glands (SMGs), model systems used for the study of SMG development, and the key signaling pathways as well as cellular processes involved in salivary gland development. The findings from the recent studies elucidating the identity of stem/progenitor cells in the SMGs, and the process by which they are directed along a series of cell fate decisions to form functional glands, are also discussed. Advances in genetic tools and tissue engineering strategies will significantly increase our knowledge about the mechanisms by which signaling pathways and cells establish tissue architecture and function during salivary gland development, which may also be conserved in the growth and development of other organ systems. An increased knowledge of organ development mechanisms will have profound implications in the design of therapies for the regrowth or repair of injured tissues. In addition, understanding how the processes of cell survival, expansion, specification, movement, and communication with neighboring cells are regulated under physiological and pathological conditions is critical to the development of future treatments.
Keywords: Salivary gland, Morphogenesis, Development, Cell signaling, Submandibular glands
Introduction
Mammalian salivary glands comprise three pairs of major glands—the submandibular glands (SMGs), sublingual glands (SLGs), and parotid glands (PGs)—and several minor glands such as the lingual, buccal, palatal, and von Ebner’s glands in the circumvallate papillae. There are multiple cell types in the salivary glands, including acinar, ductal, myoepithelial, neuronal, lymphatic, and endothelial cells [1] (Fig. 1). Complex interactions among these cell types occur during salivary gland development; however, the precise timing and mechanism of these events are not fully understood. The most lateral and cranial point of the buccal groove (the labiogingival sulcus) gives rise to PGs at the 5 − 6th weeks of gestation in humans [2], and the medial and lateral paralingual groove (the linguogingival sulcus) appears at the ectoderm-derived oral epithelial condensation at the floor of the mouth, and is separated by the basement membrane surrounding neural crest-derived mesenchymal tissue at 5.5 − 6th weeks and 7 − 8th weeks of gestation in humans, respectively. In addition, the posterior one quarter of the medial paralingual groove gives rise to the anlage of the SMGs, and the anterior three quarter gives rise to the Wharton's duct, whereas the lateral paralingual originates the SLGs [3–5].
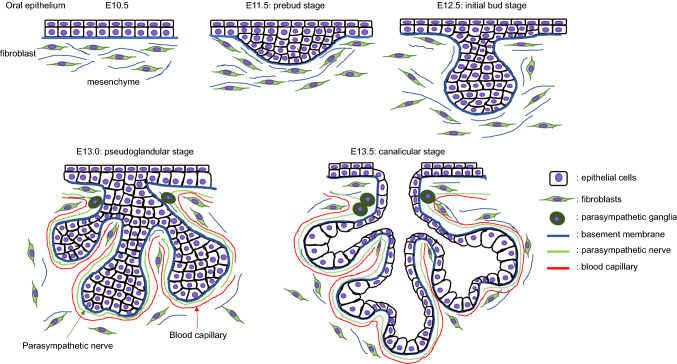
Fig. 1.
Diagrams of salivary gland development. Oral epithelium is thickened (pre-bud stage) and invaginates into the mesenchyme. The initial bud further proliferates and elongates (initial bud stage) and then stars branching to form multi-end buds (pseudoglandular stage). At the ducts, lumens are formed at the center of bilayers and differentiates into terminal buds (canalicular stage and terminal bud stage)
Mice are established models used in the study of SMG development through cytological and molecular analyses. SMG development is initiated with the formation of the primitive knot and thickening of the oral epithelium at embryonic day 11.5 (E11.5) in mice, and at the 6th week of gestation in humans (the prebud stage); this primitive knot invaginates into the condensed mesenchyme containing an endothelial plexus at E12.5 in mice, and at 7–8th week of gestation in humans (the initial bud stage) [6, 7]. This single epithelial bud then undergoes several rounds of branching morphogenesis at E13.5–E14.5 in mice (the pseudoglandular stage), which is defined by multiple cycles of cleft formation, expansion of the end buds, and duct tubulogenesis [7, 8]. At E13.5, the ductal cells differentiate into two layers, the basal cell layer and luminal cell layer, and a ductal lumen is formed at the midline of keratin 19 (K19)-expressing luminal cells at E13.5 and expanded by E16.5 (the canalicular stage). This luminal formation is regulated by parasympathetic nerve transmitter vasoactive intestinal peptide (VIP) and its receptor VIPR1, but not by acetylcholine (ACh) and its muscarinic receptor 1 (M1), which are required for bud formation from the oral epithelium and epithelial morphogenesis [9, 10] (Fig. 1).
The development of a lumen within the branched epithelium occurs in the following order: (1) the distal end of the main cord and the branch cords, (2) the proximal end of the main cords, and (3) the central portion of the main cord. The lumen is initially formed within the ducts before they develop into the terminal buds. In vitro studies using mammary gland epithelial cell lines and mouse SMG organ cultures suggested that apoptotic cell death in the central portion of the lumen is involved in the formation of a hollow lumen [11, 12]. Terminal differentiation of the end buds into secretory acini is apparent at the 19–24th week of gestation in humans and E17.5 in mice; this is followed by further growth and differentiation until a mature organ capable of nerve-stimulated secretion is formed (the terminal bud stage) [7, 13, 14]. Terminal differentiation of the gland then continues postnatally. These complex series of morphogenic steps suggest that multiple intrinsic and extrinsic signaling pathways are regulated in a spatiotemporal fashion. This review discusses the mechanism(s) by which salivary gland formation is affected through alterations in several cell signaling pathways.
Stem/progenitor cells in the SMGs
Multiple progenitor populations exist in both embryonic and adult salivary glands, and various nuclear, cytoplasmic, and cell surface markers have been used to characterize the salivary progenitor cells, including ASCL3, KIT, K5, K14, SOX2, and SOX9 [14–16]. c-KIT is a receptor tyrosine kinase type III that binds to a cytokine or stem cell factor to regulate cell proliferation, differentiation, and protection from apoptosis [17, 18]. It is expressed in end bud epithelial cells, but not in ductal cells, in the developing embryonic salivary glands [19], and c-KIT-expressing bud cells with high proliferation activity are subpopulated into K5-coexpressing proximal and K14-coexpressing distal progenitor cells [19]. Moreover, K5-expressing proximal progenitor cells coexpress K19 (K5+;K19+) during differentiation into the luminal cell layer and lose K5 expression (K5−;K19+) in well-developed luminal cells [9]. Interestingly, parasympathetic ACh/M1 signaling induces cell proliferation and epithelial differentiation in K5+ progenitor cells via epidermal growth factor (EGF) signaling [9]. On the other hand, K14+ distal progenitor cells are especially expanded by a combination of c-KIT and fibroblast growth factor (FGF) signaling mediated by receptor FGFR2b [19]. Previous transplantation studies demonstrate that c-KIT-expressing adult cells in intercalated, striated, and excretory ducts can accelerate repair after irradiation [20, 21], and a recent study indicates that these cells do not expand in cultured salivary gland organoid whereas all salivary gland cells in the organoid express K14 [22].
Progenitor cells expressing the mammalian achaete-scute homolog 3 (ASCL3), a transcription factor, exist in the striated and excretory duct cells of adult mouse salivary glands [23]. Three members of the Ascl gene family have been identified, and they are all expressed in tissue-specific progenitor cells and implicated in cell fate determination and differentiation events. Progenitor cells characterized by Ascl3 expression are present in all three major salivary glands in mice [24], and lineage tracing experiments show that ASCL3+ cells generate a subset of adult ductal and acinar cells in cultured sphere organoids [24–26]. Mice with a deletion of Ascl3 (Ascl3EGFP−Cre/EGFP−Cre) and elimination of Ascl3-expressing cells (Ascl3EGFP−Cre/+;R26RDTA/+) show hypoplastic salivary glands [26]; after ligation-induced trauma, damaged acinar cells are repopulated at the same extent as in control mice. Therefore, multiple progenitor-like cell populations that do not express Ascl3 are present and functionally compensate for Ascl3 absence in adult salivary glands [26].
SOX2, i.e., SRY (sex-determining region Y)-box 2, is a transcription factor that plays critical roles in embryonic development and cell fate determination and a marker of adult stem/progenitor cells [27–29]. In embryonic salivary gland development, SOX2-expressing bud cells can give rise to both acinar and ductal cells, but they are critical for acinar cell development [30]. Deletion of Sox2 in K14-expressing epithelial progenitor cells (K14-CreERT2;Sox2F/F induced at E10.5) results in severely hypoplastic SMGs and SLGs without affecting mesenchymal, neuronal, and ductal cell differentiation [30, 31]. Furthermore, the lack of Sox2-expressing progenitor cells causes failure to regenerate acinar cells after irradiation [31]. SOX9 is also expressed in epithelial progenitor cells, which give rise to both acinar and ductal cells [32], and K14-Cre;Sox9F/F conditional knockout mice fail to generate branches of end buds and exhibit developmental arrest at the bud stage in all three major salivary glands [32]. Moreover, prominin-1 (PROM1, a.k.a. CD133) is a transmembrane protein involved in stem cell capacity that is expressed in several stem/progenitor cells such as hematopoietic stem cells, neuronal/glial stem cells, endothelial progenitor cells, and cancer stem cells [33, 34]. PROM1-positive cells isolated from mouse SMGs can differentiate into acinar, ductal, and myoepithelial cells through regulation of Sox9 expression [35].
It is critical to recognize the difference between stem cells and progenitor cells, which, despite being frequently mentioned interchangeably, are not equivalent and exhibit distinct properties [36]. Stem cells can replicate indefinitely and produce both undifferentiated and differentiated progeny, whereas progenitor cells undergo only a finite number of cell divisions, do not self-renew, and are often limited in the number of cell types they can produce [37]. Long-term self-renewal and multipotent differentiation capacity are functional properties that require rigorous analysis of the cells within their native tissue niche [37]. It is known that the salivary glands contain multipotent stem cells that differentiate into either acinar cells or duct cells [38, 39], but evidence showing that differentiated cells can display cellular plasticity, particularly under stressful conditions, may help reconciling the various proposed stem cell populations in these structures [37].
Differentiation of mucous and serous acinar cells
The human salivary glands produce 0.5 − 1.5 L of saliva daily, which contains 99.5% of water, 0.3% of proteins, and 0.2% of both inorganic and organic substances [40]. Acinar cells are responsible for the production and secretion of α-amylase, mucins, and immunoglobulins essential for the digestion and taste of foods, lubrication, buffering, and prevention of dental and oral diseases [41]. There are two types of acinar cells in the salivary glands: serous and mucous acinar. Serous acinar cells are typically 8 − 12 pyramid-shaped cells with a well-developed rough endoplasmic reticulum (ER), round nuclei in the middle of the cytoplasm, and secretory granules at the apical cytoplasm; serous saliva contains a large amount of water, ions, and digestive α-amylase (AMY1A), which is crucial for food digestion [13, 42]. In contrast, mucous acinar cells have flat nuclei towards the basal cell surface with aggregated granules in the apical cytoplasm [13, 42] and secrete saliva containing mucins essential for lubrication and oral health [13, 42]. In humans, the PGs include only serous acini, but the SMGs and SLGs are a mixture of both serous and mucous acini (more serous acini in the SMGs, and predominantly mucous acini in the SLGs). In contrast, mouse PGs and SMGs are composed of pure serous acini, whereas the SLGs includes only mucous acini. Minor glands contain either mixed or mucous acini in both humans and mice [43].
The duct system and secretory duct
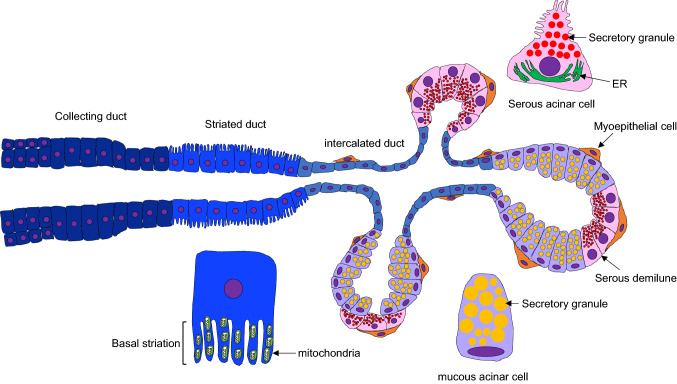
The mammalian duct system consists of intercalated ducts (IDs), striated ducts (SDs, a.k.a. intralobular ducts), and excretory ducts (EDs, a.k.a. interlobular ducts). Saliva produced by acinar cells in the glandular body flows sequentially through the IDs, SDs, and EDs. The IDs are lined by cuboidal to flattened single-layered epithelial cells between the acini and SDs and modify primary saliva components [44, 45]. The SDs are lined by single columnar epithelial cells, with vertical accumulation of mitochondria at the basal holding and numerous invaginations of the apical membrane, and also contain secretory granules and a smooth ER at the apical cytoplasm. They play a role in modifying the ion components of primary saliva through the secretion and resorption of ions transported bi-directionally between the ductal lumen and extracellular spaces [43, 46]. The EDs are lined by stratified epithelial cells, inner columnar to squamous cells, and basal cuboidal cells (Fig. 2).
Fig. 2.
A diagram of salivary glands. The salivary glands include collecting, striated, and intercalated duct cells in the duct and acinar (serous and mucous) and myoepithelial cells in the acini. Striated duct exhibit basal striation containing numerous mitochondria. Serous and mucous acinar cells contain numerous secretory granules in the cells
The PGs secrete saliva via the Stensen’s (a.k.a. Stenon’s) duct at the parotid papillae, the SMGs via the Wharton’s duct at the sublingual caruncle, and the SLGs through the Bartholin’s duct, which further connects with the Wharton’s duct at the sublingual caruncle or the ducts of Rivinus at the sublingual caruncle or sublingual fold [43]. In rodent males, granular convoluted tubules (GCTs), which are located between the IDs and SDs, develop in submandibular glands after sexual maturation, at 4 weeks old, in an androgen-dependent manner [43, 47, 48]. The GCTs are composed of simple columnar epithelia containing many eosinophilic secretory granules in the supranuclear cytoplasm [43]. These secretory granules contain various biologically active polypeptides such as cell growth factors EGF and nerve growth factor (NGF) and hormones, and undergo exocytosis in response to neural and hormonal stimuli [43]. Previous studies have demonstrated the role of testosterone in the development and maintenance of GCTs. For example, mice with a deficiency for the androgen receptor (Ar) (ArF/F; CAG-Cre mice) exhibit GCT maturation defects [49], and castrated male mice have underdeveloped GCTs that resemble those in female mice [50–54]. Moreover, GCTs in male mice with the testicular feminization mutation (Tfm), a spontaneous single-base deletion in the androgen receptor (Ar) gene, have fewer secretory granules and increased cytoplasmic vacuoles [55]. In addition, excessive cholesterol synthesis results in failed autophagy, specifically in the duct cells of the salivary glands, followed by the accumulation of NF-E2-related factor 2 (NRF2), a transcription factor known as one of the specific substrates for autophagy [56]. The accumulation of NRF2 suppresses Forkhead box protein a1 (Foxa1), which forms a transcriptional complex with the androgen receptor to regulate target genes, and is crucial for GCT differentiation [56].
Signaling pathways in the regulation of salivary gland formation
Fibroblast growth factor (FGF) family
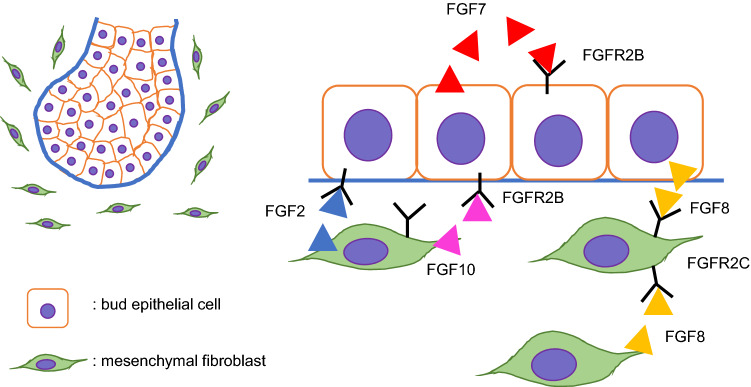
FGF signaling is transduced by 23 FGF ligands and four receptors and is essential for multiple branching organs, including the lungs, pancreas, prostate, and salivary glands [57]. Fgfr1b/2b and Fgf1/8/13 are expressed in the epithelium, and Fgfr1c/2c/3/4 and Fgf1/2/3/7/8/10/13 are expressed at the mesenchyme of SMG buds in mice [58] (Fig. 3). FGF10 is a major ligand for FGFR2B during development, and mice with loss of either Fgf10 or Fgfr2IIIb (Fgf10−/− or Fgfr2bLacZ/LacZ) exhibit similar phenotypes of dysgenesis or agenesis of the lungs, limbs, pancreas, kidney, and salivary glands that are more severe than those seen in Fgfr2bLacZ/+ or Fgf10± heterozygous mice [59–63]. Fgf10± mice exhibit delayed, hypoplastic SMG development, leading to reduced saliva secretion (a.k.a. xerostomia) [59]. Mice with a cranial neural crest (CNC) cell-specific deletion of Fgf10 (Wnt1-Cre;Fgf10F/F conditional knockout) exhibit agenesis of the salivary and lacrimal glands, which is similar to the phenotype of Fgf10 null mice [64]. In addition, FGF signaling can induce putative progenitor cells marked with the myelocytomatosis oncogene (MYC; a transcription factor), sex-determining region Y-box 9 (SOX9; a transcription factor), and KIT proto-oncogene receptor tyrosine kinase (KIT; a receptor tyrosine kinase) in the end buds of ex vivo SMG explants [19]. In mouse embryos with loss of either Fgf10 or Ffgr2b, loss of FGF signaling leads to failure of epithelial invagination and downgrowth into the surrounding mesenchymal tissue; thus, an initial SMG placode is detectable at E12, but as a hypoplastic end bud, and then the glands are no longer detectable at E13.5 [63, 65, 66]. In addition, double heterozygous mice for the Fgfr2c and Fgf10 deletions (Fgfr2c+/Δ;Fgf10±) exhibit more severe hypoplastic SMGs than single heterozygous Fgfr2c+/Δ mice [63, 67]. These phenotypic similarities seen in mice with loss of either Fgf10, Fgfr2b, or Fgfr2c suggest that FGF signaling is critical for exocrine gland development.
Fig. 3.
Schematic images of FGF signaling during salivary gland development
In humans, heterozygous mutations in genes related to FGF signaling are associated with autosomal dominant aplasia of the lacrimal and salivary glands (ALSG) syndrome and autosomal dominant lacrimo-auriculo-dento-digital (LADD) syndrome. ALSG is known to be caused by mutations in FGF10 [68–71], and LADD has been associated with mutations in FGF10, FGFR2, and FGFR3 [72–74]. Both syndromes are characterized by lacrimal and salivary glands aplasia/hypoplasia, dryness of the eye and mouth, dental caries, and oral infections. In addition, patients with LADD often display defects in the craniofacial region, in the digits, and the genitourinary system [69, 74]. FGF10/FGFR2B signaling via the mitogen-activated kinase-like protein (MAPK) and the phosphoinositide 3-kinase (PI3K) pathways is known to be involved in epithelial cell proliferation and expansion of epithelial end buds [75, 76]. In fact, the FGF10/FGFR2B complex binds to the heparan sulfate (HS) domain of extracellular matrix HS proteoglycans, such as perlecan; heparanase releases FGF10 from perlecan HS, increasing MAPK signaling activity, which is crucial for branching morphogenesis [77, 78]. LADD-related FGF10 mutations reduce the binding affinity to FGFR2B or stability of FGF10, resulting in reduced FGF10/FGFR2B signaling [79], and LADD-causing mutations in FGFR2 reduce receptor tyrosine kinase activity, resulting in impaired tyrosine autophosphorylation for FGF10/FGFR2B signaling [79, 80].
Another FGF ligand, FGF8, signals through FGFR2c. Mice with a conditional deletion of Fgf8 in the 1st pharyngeal arch ectoderm (Tfap2aIRESCre/+;Fgf8F/Null) and mice with a hypomorphic form of FGF8 (Fgf8H/Null) exhibit SMG agenesis with arrest at initial epithelial gland bud formation and hypoplastic SMGs with a few branched end buds with narrow luminae, respectively [81]. The branching defect caused by Fgf8 deficiency can be rescued with exogenous supplementation of FGF10 [81]. Altogether, these results indicate that FGF10/FGFR2b and FGF8/FGFR2c signaling share common downstream targets critical for SMG development. Interestingly, mice with mouse mammary tumor virus overexpressing Fgf8 or Fgf8b (MMTV-Fgf8 and MMTV-Fgf8b) develop ductal hyperplasia, as well as salivary and mammary adenocarcinomas [82].
FGF7 is explicitly expressed in epithelial tissues and signals via FGFR2. The supplementation of both FGF7 and FGF1 accelerates acinar cell differentiation and branching in cultured SMG explants [83]. Interestingly, mice expressing Fgf7 under the Krt14 promoter (K14-Fgf7 mice) exhibit small salivary glands with failure of duct differentiation [84]; however, Fgf7 null mice do not have any abnormality in the salivary glands [85]. In addition, mice with an ablation of antagonists of FGF signaling Sprouty (Spry) 1 and 2 in epithelial cells (Spry1/2 double null mice, Shh-Cre;K14-Cre;Spry1F/−;Spry2F/− in which approximately 80% of the Spry1/2 gene is deleted in epithelial cells) exhibit wide primary duct formation and abnormal branching due to reduction of K5+ duct progenitor cells and lack of parasympathetic ganglia (PSGs) and innervation in the SMGs; however, Wnt1-Cre;Spry1/2F/− conditional knockout mice do not have any defect in neither SMG morphogenesis nor gangliogenesis/innervation [86]. K5+ progenitor cells are known to produce and secrete WNT ligands (Wnt4, Wnt5b, Wnt7b, and Wnt10a), which activate WNT signaling in neurons of PSGs associated with neuronal survival, gangliogenesis, and innervation [86].
FGF2 is expressed in the SMG mesenchyme during development, and knockdown of Fgf2 in mesenchymal–epithelial co-cultures isolated from gland buds reveals that expression of acinar cell marker aquaporin 5 (AQP5), a water channel crucial for saliva secretion, in the epithelial cluster and epithelial cell survival is suppressed compared with the control. Moreover, supplementation of FGF2 in epithelial clusters promotes organogenesis and expression of AQP5 [87]. These studies strongly suggest that a balance of FGF signaling activity is crucial for proper epithelial morphogenesis and progenitor cell maintenance.
Epidermal growth factor (EGF) family
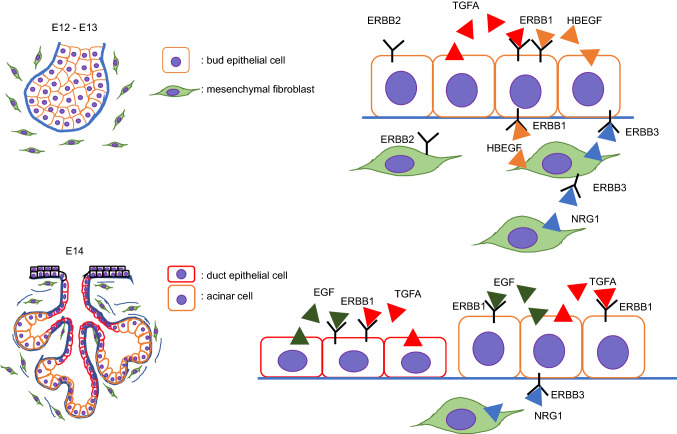
The EGF family of receptor tyrosine kinases (EGFR/ErbB1, ErbB2, ErbB2, and ErbB4) and their ligands play crucial roles in cell proliferation, migration, differentiation, survival, and apoptosis in the development and maintenance of various tissues [88, 89]. During salivary gland development, all four receptors are expressed in gland buds from E12 to E15. ErbB1 is expressed only in epithelial cells, ErbB2 and ErbB3 in epithelial cells and in a small population of surrounding mesenchymal cells and their ligands, and Egf/EGF in epithelial cells of ducts and end buds after E14, but not at E12 and E13. In addition, Tgfa/TGFA is expressed in epithelial cells of ducts and end buds after E12, Hbegf in both epithelial cells and the mesenchyme at E12 and E13, and Nrg1 only in the surrounding mesenchyme from E12 to E15 [90–92]. EGF is a ligand for ERBB1 and an inducer of branching of end buds, and increases production of integrin alpha 6 (ITGA6) at the basal cell membrane via activation of extracellular signal-regulated kinase (ERK) 1/2 signaling [93–97]. Heparin-binding EGF-like growth factor (HBEGF) and transforming growth factor-alpha (TGFα) are also ligands for ERBB1 that induce branching [97, 98]. In addition, stimulation of the parasympathetic nervous system via ACh/M1 signaling increases the proliferation of K5+ progenitor cells in the initial buds via ERRB1/HBEGF signaling, which in turn increases differentiation of K19+ luminal cells [9]. Finally, mesenchyme-specific neuregulin-1 (NRG1) is a ligand for both ERBB3 and ERBB4 and induces branching [90]. Mice with deletion of ErbB1 (ErbB1−/− mice) exhibit slightly small-sized SMGs due to impaired terminal bud branching and reduction of epithelial cell proliferation [92, 99] (Fig. 4).
Fig. 4.
Schematic images of EGF signaling during salivary gland development
NOTCH signaling
Notch signaling is a conserved intercellular signaling pathway that regulates cell proliferation, differentiation, and cell fate determination in various tissues during development and regeneration [100–104]. Notch signaling is activated by binding of ligands Jagged 1 and 2 (JAG1 and JAG2), delta-like canonical Notch ligand 1, 3, and 4 (DLL1, DLL3, and DLL4), or delta-like noncanonical Notch ligand 1 and 2 (DLK1 and DLK2) to their receptors NOTCH1-4. In human SMGs, NOTCH1-4, JAG1/2, and DLL1 are expressed more abundantly in duct cells compared to acinar cells [105]. Moreover, ductal ligation of the rat parotid gland demonstrates that expression of NOTCH1-4, JAG1/2, and DLL1 is induced in acinar cells during regeneration [105]. DLK1 and DLK2, transmembrane proteins containing six EGF-like repeats, are secreted as soluble forms that are cleaved by a disintegrin and metalloproteinase (ADAM) 17. DLK1 and DLK2, which act as negative NOTCH signaling regulators, play roles in cell growth and differentiation in various organs, including the brain, pituitary gland, skeletal muscles, cartilages, pancreas, lung, and SMGs [106–108]. At the bud stage, DLK1 is expressed in the surrounding mesenchyme, whereas DLK2 is expressed in the epithelial cells of the end buds. At the pseudoglandular and canalicular stages, DLK1 expression is detectable in both mesenchymal cells and distal-end bud epithelial cells, but not in ductal cells, whereas DLK2 is strongly expressed in duct cells and weakly expressed in the end buds. At the terminal bud stage, the expression of DLK1 is strongly detected in myoepithelial cells [109]. NOTCH signaling is activated in the epithelial cells of the buds and ducts, the surrounding mesenchymal cells, and the parasympathetic ganglion [109]. Inhibition of NOTCH signaling by DLK1 results in suppression of branching morphogenesis and innervation of parasympathetic nerve toward the end buds, without cell proliferation defects in bud cells [109]. As predicted, Dlk1 null mice (Dlk1−/−) exhibit small SMGs, SLGs, and PGs, resulting in reduced stimulating salivary flow without any morphological changes. However, the number of K14-expressing epithelial cells, but not K5-expressing cells, is increased in the developing SMGs of Dlk1−/− embryos [110].
WNT/β-catenin signaling
WNT signaling contributes to embryonic patterning, cell proliferation, cell fate, migration, polarization, and differentiation in multiple organisms and organ systems [111–113]. This signaling pathway is categorized into the β-catenin-dependent canonical pathway, and the independent noncanonical planar cell polarity pathway and WNT-Ca2+ pathway; the canonical pathway is activated by ligands WNT1, 2, 3, 3a, 7a/b, 8a, 9a/b, and 10a/b, and the noncanonical WNT/PCP and/or WNT/calcium pathways are activated by WNT4, 5a/b, 6, 7a, 11, and 16 [114–116]. These ligands bind to the receptor complex of LRP5/6 and Frizzled proteins (Fzd1 − Fzd10) [117]. Despite this abundance of ligands, WNT signaling itself is strictly regulated in a spatiotemporal manner during SMG development. Canonical WNT activity is first detectable at E11.5–E12.5 in mice at the condensed mesenchyme near the end buds and within the parasympathetic ganglia, and is restricted to ductal epithelial cells only after E14. WNT ligands are suggested to originate from K5+ ductal progenitor cells [86, 99, 118, 119]. Constitutively activated canonical WNT/β-catenin signaling inhibits branching morphogenesis while inhibited canonical WNT/β-catenin signaling accelerates branching morphogenesis in SMG tissue culture at the bud stage [118].
The interactions between the FGF and WNT pathways suggest that loss- or gain-of-function mutations in the FGF genes result in altered WNT signaling, leading to changes in ductal and ganglia morphogenesis [86]; for example, stimulation of FGF signaling suppresses expression of WNT ligands. The deficient gangliogenesis and branching in the SMGs of Spry1/2 mutant mice (Spry1/2 double null) can be rescued by haploinsufficiency of Fgf10 in the Spry1/2 mutant background (Spry1−/−;Spry2−/−;Fgf10±) combined with the Wnt activator [86]. Mice with a mesenchyme-specific deletion of β-catenin (Dermo1-Cre;Ctnnb1F/−) exhibit small SMGs with reduced branching due to suppressed expression of the gene coding for ectodysplasin A (EDA), but exogenous supplementation of EDA can rescue the branching defect caused by WNT signaling inhibition in these mice [119]. Moreover, mice with constitutively activated canonical WNT/β-catenin signaling (Rosa26-CreERT2;Ctnnb1Ex3F/F) show defects in the differentiation of acinar cells and luminal formation through suppression of KIT expression [83]. In addition, WNT3A (a ligand for canonical WNT/β-catenin signaling), but not WNT5A (a ligand for noncanonical WNT pathway), suppresses acinar cell differentiation and maintains progenitor cell status, suggesting that canonical WNT/β-catenin signaling regulates end bud differentiation [83]. In adult SMGs, a few cells among basal excretory cells co-expressing high epithelial cell adhesion molecule (EpCAM) and nuclear β-catenin are thought to constitute a stem cell population. These cells are in fact capable of forming spheres following treatment with both WNT3A and R-Spondin1. Moreover, the transplantation of these cells pretreated with WNT regenerates both acinar and ductal tissues in irradiated SMGs [120].
Hedgehog (HH) signaling
Congenital brain, limb, cochlear, neural crest, and craniofacial defects are present in individuals harboring mutations in hedgehog (HH) ligands (Sonic hedgehog, Desert hedgehog, Indian hedgehog), HH receptors [Patched (Ptch1) and Patched 2 (Ptch2), and Smoothened (Smo)], and mediators/transcription factors GLI1–3 [121, 122]. HH ligands bind to 12-transmembrane spanning receptor Ptch1 at the primary cilia, which is an antenna-like structure (described in detail below); in the absence of HH ligands, Ptch1 inhibits the activity of effector SMO, a 7 transmembrane-spanning G protein-coupled protein, in the primary cilia. Phosphorylation of SMO by the HH-Ptch1 complex activates SMO and induces translocation and accumulation of SMO in the primary cilia, where activated SMO processes GLI, and the GLI proteins then translocate into the nucleus to regulate target gene expression [121]. In general, GLI1 and GLI2 act as activators, but GLI3 has a dual function, acting as activator and suppressor depending on the presence and absence of HH ligands, respectively [122–125]. Heteromeric motor molecules (i.e., kinesins and dyneins) move in an anterograde and retrograde manner along the axoneme microtubules; intraflagellar transport (IFT) proteins bind to kinesins or dyneins to carry molecules between the cytosol and primary cilium, a microtubule-based cellular protrusion in nearly every cell except blood cells. Primary cilia play crucial role in the regulation of HH signaling; therefore, any failure in primary cilium formation and function disrupts HH signaling [126].
In the developing murine SMGs, Shh/SHH, its receptor Ptch1 and SMO, GLI1, and GLI3 are expressed at the ductal- and terminal-bud epithelial cells, whereas Dhh is expressed in the surrounding mesenchyme [119, 127, 128]. SMG development in Shh null mice (Shh−/−) is arrested before the pseudoglandular stage, with a few undifferentiated initial buds, and a similar branching defect is seen in SMG explants treated with cyclopamine, a HH signaling inhibitor. On the other hand, exogenous SHH increases branching [128, 129]. SHH also induces ErbB1, −2, and −3 expression in the epithelium and Egf, Nrg1, and Tgfα expression in the mesenchyme in cultured mouse SMG explants, which play a role in branching morphogenesis [129]. Ectopic expression of Gli1 in K5-Gli1 and MMTV-Gli1 transgenic mice exhibit compromised acinar cell differentiation, hyperplastic ductal structure, and cyst formation, suggesting that GLI1 plays a role in the regulation of both duct cell proliferation and acinar cell differentiation [130]. CNC cell-specific deletion of Shh effector Smo or Gli2/3 (Wnt1-Cre;SmoF/F, Wnt1-Cre;Gli2F/F;Gli3F/F, or Wnt1-Cre;Gli2F/F;Gli3Δ699/Δ699) exhibit SMG agenesis due to developmental arrest at the initial bud stage. On the other hand, transgenic mice overexpressing Gli3 (Wnt1-Cre;RosaGli3TFlag) display slightly hypoplastic SMGs [131]. These results suggest that SHH signaling in the surrounding mesenchyme is critical for differentiation and branching of epithelial cells in the SMGs. Moreover, mice with loss of Kif3a (a kinesin-2 motor protein subunit) in CNC cells (Wnt1-Cre;Kif3aF/F) exhibit SMG agenesis, which is caused by arrest in development at the initial bud stage, as seen in Smo and Gli2/3 mutant mice [131]. Although a relationship between SMG morphogenesis and primary cilium remains elusive, Ift88Orpk/Orpk transgenic mice (expressing a hypomorphic allele that results in reduced protein levels) display reduced duct extension and branching morphogenesis in the mammary glands [132].
Bone morphogenetic proteins (BMPs)/transforming growth factor β (TGFβ) signaling
The TGFβ superfamily is known to be involved in the development and morphogenesis of various organs [133, 134]. Ligands of the TGFβ superfamily include TGFβ, BMP, growth differentiation factor (GDF), Activin, Inhibin, Myostatin, and Nodal. The TGFβ superfamily receptors are serine/threonine kinase receptors categorized into type I, type II, and type III; type I comprises seven subtypes and type II five, where type III consist only of TGFBR3 (beta-glycan). Ligands bind to type II receptors and then recruit type I receptors to form a ligand-type II receptor complex, which phosphorylates/activates the type I receptor; this phosphorylated type I receptor then phosphorylates receptor-regulated SMAD (R-SMAD; SMAD1/5/8, or SMAD1/5/9 for BMPs, SMAD2/3 for TGFβs and activin/inhibin), which then binds to common partner SMAD (Co-SMAD; SMAD4). The SMAD complex then translocates into the nucleus to regulate target gene expression [135–138]. In the developing salivary glands, various BMP signaling molecules [e.g., ligands Bmp1-4, 6, and 7; receptors Bmpr1a, 1b, 2, and Acvr2b; antagonists Noggin (Nog) and Gremlin1 (Grem1)] are expressed in the SMGs and SLGs in a spatial–temporal-specific manner, whereas Bmp8b is expressed only in the SLGs [expression data provided by the Salivary Gland Molecular Anatomy project at NIDCR (https://sgmap.nidcr.nih.gov/sgmap/sgexp.html)] [58, 67, 127, 139–141]. For example, Bmp1, 2, 3, 4, and 6 and Bmpr1b are expressed at high levels in mesenchymal cells at E13.5, whereas Bmp7 is expressed at higher levels in the epithelium compared to the mesenchyme. Consistent with the role of BMP7 in epithelial organ development [142], Bmp7 null mice (Bmp7−/−) exhibit aberrant salivary gland morphogenesis with fewer end buds and luminal ducts as compared to wild-type controls [67], suggesting that BMP7 regulates branching morphogenesis.
FAM20C, a Golgi kinase, phosphorylates various proteins, including BMP4. The SMGs in Mmtv-Cre;Fam20cF/F mutant mice exhibit decreased acinar lobes and defects in maturation and secretion in GCTs due to suppression of canonical BMP signaling in duct cells [143]. Interestingly, patients with Sjögren’s syndrome and disease-onset NOD mice (a mouse model for Sjögren’s syndrome; disease-onset after the age of 20 weeks) highly express BMP6 in the salivary glands [144, 145]. Ectopic expression of BMP6 is known to induce hypofunction in the salivary glands and lacrimal glands with increased extracellular matrix and decreased AQP5 expression. Inhibition of BMP signaling by an ALK2/3 inhibitor can restore defective salivary gland function in NOD-Aecl1Aecl2 mice (also a Sjögren’s syndrome mouse model) [144, 145].
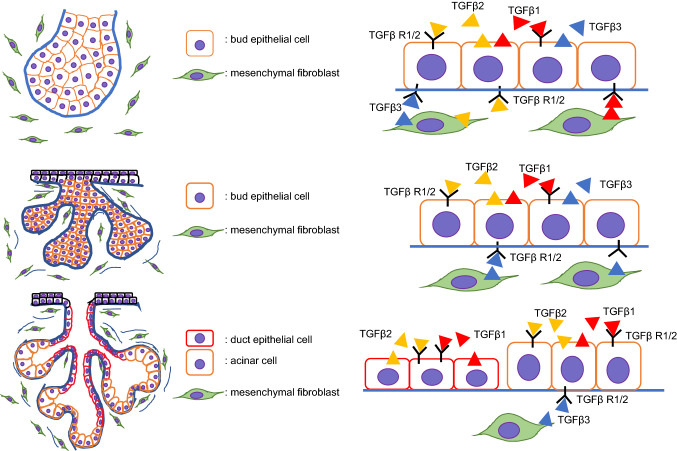
TGFβ1, -β2, and -β3 are also expressed in the developing salivary glands. All three ligands are present in both the oral epithelium and the adjacent mesenchyme, but TGFBR1 and TGFBR2 are expressed mainly in epithelial cells at the initial bud stage. TGFβ1, TGFβ2, TGFBR1, and TGFBR2 are expressed only in the branching bud epithelium, whereas TGFβ3 is present in both the epithelium and the surrounding mesenchyme at the pseudoglandular stage. At the canalicular stage, the expression of TGFβ1, TGFβ2, TGFBR1, and TGFBR2 is restricted to the terminal buds, luminal cells, and ducts, whereas TGFβ3 is expressed only in the surrounding mesenchyme. At the terminal bud stage, TGFBR2 expression shifts to the ductal epithelial cells only [92, 146, 147]. However, Tgfb1, Tgfb2, or Tgfb3 null mice (Tgfb1−/−, Tgfb2−/−, and Tgfb3−/−) show normal SMG development, suggesting that TGFβ ligands are functionally redundant during SMG development [92, 148] (Fig. 5). Exogenous supplementation of TGFβ1 to mouse SMG explants can suppress branching morphogenesis as well as acinar cell differentiation in a dose-dependent manner [149], and mice overexpressing Tgfb1 (MMTV-Cre;Tgfb1glo) develop severely hypoplastic SMGs with acinar cell atrophy and fibrosis in the adult phase [150]. Interestingly, Tgfb1−/− mice exhibit inflammatory cell infiltration in the SMGs, similarly to Sjögren’s syndrome, after postnatal day 10 due to failure in the suppression of the inflammatory response [148, 151].
Fig. 5.
Schematic images of TGFβ signaling during salivary gland development
Ectodysplasin a (EDA) signaling
EDA, a tumor necrosis factor family transmembrane protein, and its receptor EDAR play essential roles in the development of ectodermal tissues such as the teeth, skin, mammary glands, and salivary glands [152–155]. Mutations in EDA, EDAR, or EDARADD (an EDAR-associated death domain) cause autosomal recessive, autosomal dominant, or X-linked hypohidrotic ectodermal dysplasia (HED or XLHED), a syndrome variably characterized by tooth agenesis, hypoplastic teeth, sparse hair, dysfunction in exocrine glands, such as sweat glands, sebaceous glands, lacrimal glands, mammary glands, and mucous glands of the bronchi, and salivary glands [156–159]. Both loss- and gain-of-function studies in genetic mouse models have revealed the critical role of the EDA pathway in salivary gland ductal and acinar development. Eda null mice (Eda−/−, Tabby) and Edar null mice (Edar−/−) display defects in branching morphogenesis, resulting in hypoplastic salivary glands with a reduced number of ductal and acinar structures in the adult phase [119, 160–164]. In contrast, mouse embryos with specific overexpression of Eda in the epithelium under the K14 promoter (K14-Eda) and Edar transgenic mice (EdarTg951/Tg951) display increased branching and number of end buds in the SMGs [119, 165]. EDA signaling is known to be mediated by transcription factor nuclear factor kappa B (NF-κB) in tooth, hair follicle, and salivary gland development [119, 166–169], and mutations in the EDAR gene fail to activate NF-κB signaling [170]. Previous studies demonstrated that HH and WNT signaling pathways are downstream of EDA signaling in the developing salivary glands [119, 161, 171], and the expression of Shh is in fact suppressed in Eda mutant mice and upregulated in K14-Eda mice when compared with wild-type controls. The branching defects in the SMG of Eda and Edar mutant mice (Eda−/− and EdardlJ/dlJ) can be rescued by treatment with SHH ex vivo [119, 163], but the excessive branching phenotype in the SMGs of K14-Eda mice is restored by inhibition of HH signaling with cyclopamine [119]. Despite the fact that there are interactions between the EDA and WNT signaling pathways in early SMG development (earlier than E13 in mice) (e.g., the suppression of WNT signaling in the mesenchyme reduces Eda expression), these two pathways appear to function independently after E13 [119]. For example, the activity of WNT and EDA/NF-κB signaling does not co-localize during duct formation/lumenization of the SMGs. WNT activity is detectable in the surrounding mesenchyme before E15 and in the ducts after E15, whereas NF-κB signals are in fact localized only at the end buds, not the ducts [118, 172]. The mechanism of how the EDA pathway controls acinar formation in the SMGs, and whether the SMGs of individuals with HED show aberrant duct or acinar morphogenesis, remain to be determined.
Neurotrophic factors secreted by sympathetic and parasympathetic nerves
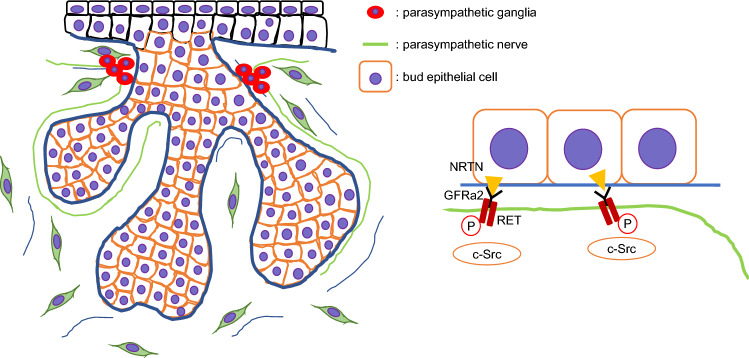
The glial cell line-derived neurotrophic factor (GDNF) ligand families (GLFs) belong to the TGFβ superfamily. GLFs comprise GDNF, neurturin (NRTN), artemin (ARTN), and persephin (PSPN), which play roles in the survival and differentiation of neurons through activation of RET, a receptor tyrosine kinase for members of GDNF family [173–175]. GLFs bind to the GDNF family receptor alpha (GFRα; GFRα1 for GDNF, GFRα2 for NRTN, GFRα3 for ARTN, and GFRα4 for PSPN), and then the complex forms a dimer with RET, resulting in the activation of tyrosine kinase for intracellular signal transduction. GDNF treatment of SMGs in irradiated mice promotes cell proliferation through activation of the GDNF-RET signaling pathway [176, 177]. Mice deficient for either Gfra2 or Ret (Gfra2−/− and Ret−/−) display hypoplastic submandibular ganglia and absence of parasympathetic innervation in the SLGs at birth, whereas Gfra1 null mice show no developmental defects [178, 179]. In the adult phase, the submandibular, otic, and sphenopalatine ganglia in Gfra2−/− mice are severely atrophic, resulting in absent innervation in the lacrimal, parotid, and sublingual glands [178, 180]. A recent ex vivo study suggests that NRTN is necessary for functional innervation of the developing salivary epithelium [10]. NRTN and GFRa2 are expressed in the SMG bud epithelium and parasympathetic nerve fibers, respectively [181]. The expression of Ret is detectable in the parasympathetic ganglia adjacent to the ducts and is required for guiding GFRα2-expressing parasympathetic nerves toward the developing end buds [181]. NRTN treatment induces neuronal outgrowth and synapse formation in SMG explants and promotes end bud regeneration and parasympathetic nerve innervation in irradiated mice [181, 182] (Fig. 6). Therefore, bidirectional communication between the branching epithelium and the ganglia are crucial for organogenesis.
Fig. 6.
Interaction of parasympathetic nerve system and salivary gland development
Diseases related to cranial nerve dysfunction are known to cause salivary gland dysfunction. For example, hereditary gelsolin amyloidosis, which is caused by autosomal dominant mutations in gelsolin, presents gelsolin amyloid deposition and xerostomia [183, 184]. Treacher Collins syndrome (TCS), which is caused by autosomal dominant mutations in TCOF1, POLR1C, and POLR1D (genes necessary for neural crest cell survival and migration), is characterized by craniofacial abnormalities such as underdeveloped zygomatic bones, micrognathia, cleft palate, and eyelid coloboma [185, 186]. Some studies suggest that salivary gland dysplasia and dysfunction are also associated with TCS [187].
The autonomic nervous system, comprising both sympathetic and parasympathetic nerves, regulates almost all organs’ functions with opposite outcomes (the frequently called fight-or-flight response). In the salivary glands, the activation of the sympathetic nervous system accelerates mucous saliva secretion from mucous acini; on the other hand, the activation of the parasympathetic nervous system stimulates serous saliva secretion from serous acini. ACh binds to receptors M1 − M5, which are G-protein coupled receptors. The M3 receptor plays a role in saliva flow, whereas the M1 and M5 receptors play a role in secretion [188–190]. Moreover, the activation of M1 and M3 receptors induces the translocation of AQP5 from the apical cytosol to lipid rafts on the apical membrane via increased intracellular Ca2+ concentration in acinar cells of the salivary glands [191–193]. Parasympathetic nerve denervation causes salivary gland atrophy similar to that seen in irradiated salivary glands. For instance, parasympathetic nerve denervation in rat SMGs upregulates expression of M1 and M3, resulting in increased resting saliva flow, but decreased stimulating saliva flow [194]. The parasympathetic nervous system, but not the sympathetic nervous system, is essential for the maintenance of epithelial progenitor cells in an undifferentiated state required for organogenesis [9]. On the other hand, VIP, another neurotransmitter from the parasympathetic nervous system, but not ACh/M1, regulates lumen formation during duct development [10].
Conclusion
Salivary gland development involves the interactions of multiple cell types, including epithelial, mesenchymal, endothelial, and neuronal cells. Various pathways cooperate to establish acinar and ductal growth, development of the ganglia, and progenitor cell survival/proliferation. Several mouse genetic studies indicate that these molecular pathways act within complex signaling networks, which require a systematic approach to elucidate how they affect the different morphogenic processes. Further advances in human genetics and the ever-increasing number of mouse models generated will significantly increase our knowledge of the mechanisms by which signaling pathways and cells establish the tissue architecture and function during salivary gland formation.
Acknowledgments
This study was supported by grants from the National Institute of Dental and Craniofacial Research, NIH (DE026509, DE026767, DE028340, and DE029818), and UTHealth School of Dentistry faculty funds to JI.
Abbreviations
- SOX9
Sex-determining region Y-box 9
- KIT
KIT proto-oncogene, receptor tyrosine kinase
- FGF
Fibroblast growth factor
- TGF-β
Transforming growth factor-β
- SMGs
Submandibular glands
- SLGs
Sublingual glands
- PGs
Parotid glands
- KRT19+
Keratin 19 positive
- MYC
Myelocytomatosis oncogene
- KIT
KIT proto-oncogene, receptor tyrosine kinase
- AKT
AKT serine/threonine kinase
- PI3K
Phosphatidylinositol 3-kinase
- EDA/EDAR
Ectodysplasin-A/ectodysplasin-A receptor
- MAPK
Mitogen-activated kinase-like protein
- HS
Heparan sulfate
- ECM
Extracellular matrix
- SHH
Sonic hedgehog
- NRG
Neuregulin
- HH
Hedgehog
- SMO
Smoothened
- PTCH
Patched
- BMP
Bone morphogenetic proteins
- GDNF
Glial cell line-derived neurotrophic growth factor
- NRTN
Neurturin
- Ach
Acetylcholine
- VIP
Vasoactive intestinal peptide
- Ascl3
Achaete-scute homolog 3
- SCF
Stem cell factor
- AQP
Aquaporin
- ID
Intercalated duct
- SD
Striated duct
- ED
Excretory duct
- NRF2
NF-E2-related factor 2
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Rothova M, Thompson H, Lickert H, Tucker AS. Lineage tracing of the endoderm during oral development. Dev Dyn. 2012;241:1183–1191. doi: 10.1002/dvdy.23804. [DOI] [PubMed] [Google Scholar]
- 2.Guizetti B, Radlanski RJ. Development of the parotid gland and its closer neighboring structures in human embryos and fetuses of 19–67 mm CRL. Ann Anat. 1996;178:503–508. doi: 10.1016/S0940-9602(96)80105-1. [DOI] [PubMed] [Google Scholar]
- 3.Quiros-Terron L, Arraez-Aybar LA, Murillo-Gonzalez J, De-la-Cuadra-Blanco C, Martinez-Alvarez MC, et al. Initial stages of development of the submandibular gland (human embryos at 5.5-8 weeks of development) J Anat. 2019;234:700–708. doi: 10.1111/joa.12955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Merida-Velasco JA, Sanchez-Montesinos I, Espin-Ferra J, Garcia-Garcia JD, Garcia-Gomez S, et al. Development of the human submandibular salivary gland. J Dent Res. 1993;72:1227–1232. doi: 10.1177/00220345930720081101. [DOI] [PubMed] [Google Scholar]
- 5.Guizetti B, Radlanski RJ. Development of the submandibular gland and its closer neighboring structures in human embryos and fetuses of 19–67 mm CRL. Ann Anat. 1996;178:509–514. doi: 10.1016/S0940-9602(96)80107-5. [DOI] [PubMed] [Google Scholar]
- 6.Lourenço SV, Kapas S. Integrin expression in developing human salivary glands. Histochem Cell Biol. 2005;124:391–399. doi: 10.1007/s00418-005-0784-3. [DOI] [PubMed] [Google Scholar]
- 7.Patel VN, Rebustini IT, Hoffman MP. Salivary gland branching morphogenesis. Differentiation. 2006;74:349–364. doi: 10.1111/j.1432-0436.2006.00088.x. [DOI] [PubMed] [Google Scholar]
- 8.Borghese E. The development in vitro of the submandibular and sublingual glands of Mus musculus. J Anat. 1950;84:287–302. [PMC free article] [PubMed] [Google Scholar]
- 9.Knox SM, Lombaert IM, Reed X, Vitale-Cross L, Gutkind JS, et al. Parasympathetic innervation maintains epithelial progenitor cells during salivary organogenesis. Science. 2010;329:1645–1647. doi: 10.1126/science.1192046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nedvetsky PI, Emmerson E, Finley JK, Ettinger A, Cruz-Pacheco N, et al. Parasympathetic innervation regulates tubulogenesis in the developing salivary gland. Dev Cell. 2014;30:449–462. doi: 10.1016/j.devcel.2014.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Debnath J, Mills KR, Collins NL, Reginato MJ, Muthuswamy SK, et al. The role of apoptosis in creating and maintaining luminal space within normal and oncogene-expressing mammary acini. Cell. 2002;111:29–40. doi: 10.1016/S0092-8674(02)01001-2. [DOI] [PubMed] [Google Scholar]
- 12.Teshima TH, Wells KL, Lourenco SV, Tucker AS. Apoptosis in early salivary gland duct morphogenesis and lumen formation. J Dent Res. 2016;95:277–283. doi: 10.1177/0022034515619581. [DOI] [PubMed] [Google Scholar]
- 13.Tucker AS. Salivary gland development. Semin Cell Dev Biol. 2007;18:237–244. doi: 10.1016/j.semcdb.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 14.Aure MH, Symonds JM, Mays JW, Hoffman MP. Epithelial cell lineage and signaling in murine salivary glands. J Dent Res. 2019;98:1186–1194. doi: 10.1177/0022034519864592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Emmerson E, Knox SM. Salivary gland stem cells: a review of development, regeneration and cancer. Genesis. 2018;56:e23211. doi: 10.1002/dvg.23211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwak M, Alston N, Ghazizadeh S. Identification of stem cells in the secretory complex of salivary glands. J Dent Res. 2016;95:776–783. doi: 10.1177/0022034516634664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lennartsson J, Ronnstrand L. Stem cell factor receptor/c-Kit: from basic science to clinical implications. Physiol Rev. 2012;92:1619–1649. doi: 10.1152/physrev.00046.2011. [DOI] [PubMed] [Google Scholar]
- 18.Ronnstrand L. Signal transduction via the stem cell factor receptor/c-Kit. Cell Mol Life Sci. 2004;61:2535–2548. doi: 10.1007/s00018-004-4189-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lombaert IM, Abrams SR, Li L, Eswarakumar VP, Sethi AJ, et al. Combined KIT and FGFR2b signaling regulates epithelial progenitor expansion during organogenesis. Stem Cell Reports. 2013;1:604–619. doi: 10.1016/j.stemcr.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lombaert IM, Brunsting JF, Wierenga PK, Faber H, Stokman MA, et al. Rescue of salivary gland function after stem cell transplantation in irradiated glands. PLoS ONE. 2008;3:e2063. doi: 10.1371/journal.pone.0002063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pringle S, Maimets M, van der Zwaag M, Stokman MA, van Gosliga D, et al. Human salivary gland stem cells functionally restore radiation damaged salivary glands. Stem Cells. 2016;34:640–652. doi: 10.1002/stem.2278. [DOI] [PubMed] [Google Scholar]
- 22.Kwak M, Ninche N, Klein S, Saur D, Ghazizadeh S. c-Kit(+) Cells in adult salivary glands do not function as tissue stem cells. Sci Rep. 2018;8:14193. doi: 10.1038/s41598-018-32557-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoshida S, Ohbo K, Takakura A, Takebayashi H, Okada T, et al. Sgn1, a basic helix-loop-helix transcription factor delineates the salivary gland duct cell lineage in mice. Dev Biol. 2001;240:517–530. doi: 10.1006/dbio.2001.0473. [DOI] [PubMed] [Google Scholar]
- 24.Rugel-Stahl A, Elliott ME, Ovitt CE. Ascl3 marks adult progenitor cells of the mouse salivary gland. Stem Cell Res. 2012;8:379–387. doi: 10.1016/j.scr.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bullard T, Koek L, Roztocil E, Kingsley PD, Mirels L, et al. Ascl3 expression marks a progenitor population of both acinar and ductal cells in mouse salivary glands. Dev Biol. 2008;320:72–78. doi: 10.1016/j.ydbio.2008.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arany S, Catalan MA, Roztocil E, Ovitt CE. Ascl3 knockout and cell ablation models reveal complexity of salivary gland maintenance and regeneration. Dev Biol. 2011;353:186–193. doi: 10.1016/j.ydbio.2011.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arnold K, Sarkar A, Yram MA, Polo JM, Bronson R, et al. Sox2(+) adult stem and progenitor cells are important for tissue regeneration and survival of mice. Cell Stem Cell. 2011;9:317–329. doi: 10.1016/j.stem.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou HY, Katsman Y, Dhaliwal NK, Davidson S, Macpherson NN, et al. A Sox2 distal enhancer cluster regulates embryonic stem cell differentiation potential. Genes Dev. 2014;28:2699–2711. doi: 10.1101/gad.248526.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang ZX, Teh CH, Kueh JL, Lufkin T, Robson P, et al. Oct4 and Sox2 directly regulate expression of another pluripotency transcription factor, Zfp206, in embryonic stem cells. J Biol Chem. 2007;282:12822–12830. doi: 10.1074/jbc.M611814200. [DOI] [PubMed] [Google Scholar]
- 30.Emmerson E, May AJ, Nathan S, Cruz-Pacheco N, Lizama CO, et al. SOX2 regulates acinar cell development in the salivary gland. Elife. 2017;6:8. doi: 10.7554/eLife.26620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Emmerson E, May AJ, Berthoin L, Cruz-Pacheco N, Nathan S, et al. Salivary glands regenerate after radiation injury through SOX2-mediated secretory cell replacement. EMBO Mol Med. 2018;3:10–24. doi: 10.15252/emmm.201708051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chatzeli L, Gaete M, Tucker AS. Fgf10 and Sox9 are essential for the establishment of distal progenitor cells during mouse salivary gland development. Development. 2017;144:2294–2305. doi: 10.1242/dev.146019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Z. CD133: a stem cell biomarker and beyond. Exp Hematol Oncol. 2013;2:17. doi: 10.1186/2162-3619-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mizrak D, Brittan M, Alison M. CD133: molecule of the moment. J Pathol. 2008;214:3–9. doi: 10.1002/path.2283. [DOI] [PubMed] [Google Scholar]
- 35.Tanaka J, Mabuchi Y, Hata K, Yasuhara R, Takamatsu K, et al. Sox9 regulates the luminal stem/progenitor cell properties of salivary glands. Exp Cell Res. 2019;382:111449. doi: 10.1016/j.yexcr.2019.05.030. [DOI] [PubMed] [Google Scholar]
- 36.Seaberg RM, van der Kooy D. Stem and progenitor cells: the premature desertion of rigorous definitions. Trends Neurosci. 2003;26:125–131. doi: 10.1016/S0166-2236(03)00031-6. [DOI] [PubMed] [Google Scholar]
- 37.Weng PL, Aure MH, Ovitt CE. Concise Review: a critical evaluation of criteria used to define salivary gland stem cells. Stem Cells. 2019;37:1144–1150. doi: 10.1002/stem.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okumura K, Nakamura K, Hisatomi Y, Nagano K, Tanaka Y, et al. Salivary gland progenitor cells induced by duct ligation differentiate into hepatic and pancreatic lineages. Hepatology. 2003;38:104–113. doi: 10.1053/jhep.2003.50259. [DOI] [PubMed] [Google Scholar]
- 39.Hisatomi Y, Okumura K, Nakamura K, Matsumoto S, Satoh A, et al. Flow cytometric isolation of endodermal progenitors from mouse salivary gland differentiate into hepatic and pancreatic lineages. Hepatology. 2004;39:667–675. doi: 10.1002/hep.20063. [DOI] [PubMed] [Google Scholar]
- 40.Liu J, Duan Y. Saliva: a potential media for disease diagnostics and monitoring. Oral Oncol. 2012;48:569–577. doi: 10.1016/j.oraloncology.2012.01.021. [DOI] [PubMed] [Google Scholar]
- 41.Wilmarth PA, Riviere MA, Rustvold DL, Lauten JD, Madden TE, et al. Two-dimensional liquid chromatography study of the human whole saliva proteome. J Proteome Res. 2004;3:1017–1023. doi: 10.1021/pr049911o. [DOI] [PubMed] [Google Scholar]
- 42.Hauser BR, Hoffman MP. Regulatory mechanisms driving salivary gland organogenesis. Curr Top Dev Biol. 2015;115:111–130. doi: 10.1016/bs.ctdb.2015.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Amano O, Mizobe K, Bando Y, Sakiyama K. Anatomy and histology of rodent and human major salivary glands: -overview of the Japan salivary gland society-sponsored workshop. Acta Histochem Cytochem. 2012;45:241–250. doi: 10.1267/ahc.12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dardick I, Naiberg J, Leung R, Ramjohn S, Christensen H, et al. Ultrastructural study of acinar and intercalated duct organization of submandibular and parotid salivary gland. Lab Invest. 1990;63:394–404. [PubMed] [Google Scholar]
- 45.Tandler B, Nagato T, Toyoshima K, Phillips CJ. Comparative ultrastructure of intercalated ducts in major salivary glands: a review. Anat Rec. 1998;252:64–91. doi: 10.1002/(SICI)1097-0185(199809)252:1<64::AID-AR7>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 46.Harunaga J, Hsu JC, Yamada KM. Dynamics of salivary gland morphogenesis. J Dent Res. 2011;90:1070–1077. doi: 10.1177/0022034511405330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dunn JF, Wilson JD. Developmental study of androgen responsiveness in the submandibular gland of the mouse. Endocrinology. 1975;96:1571–1578. doi: 10.1210/endo-96-6-1571. [DOI] [PubMed] [Google Scholar]
- 48.Berkman MD, Kronman JH. A histochemical study of the effects of castration and testosterone administration on the major salivary glands of Swiss mice. Acta Anat (Basel) 1970;76:200–219. doi: 10.1159/000143492. [DOI] [PubMed] [Google Scholar]
- 49.Adthapanyawanich K, Kumchantuek T, Nakata H, Yamamoto M, Wakayama T, et al. Morphology and gene expression profile of the submandibular gland of androgen-receptor-deficient mice. Arch Oral Biol. 2015;60:320–332. doi: 10.1016/j.archoralbio.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 50.Bhoola KD, Dorey G, Jones CW. The influence of androgens on enzymes (chymotrypsin-and trypsin-like proteases, renin, kallikrein and amylase) and on cellular structure of the mouse submaxillary gland. J Physiol. 1973;235:503–522. doi: 10.1113/jphysiol.1973.sp010400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Caramia F. Ultrastructure of mouse submaxillary gland. II. Effect of castration in the male. J Ultrastruct Res. 1966;16:524–536. doi: 10.1016/S0022-5320(66)80004-7. [DOI] [PubMed] [Google Scholar]
- 52.Chretien M. Action of testosterone on the differentiation and secretory activity of a target organ: the submaxillary gland of the mouse. Int Rev Cytol. 1977;50:333–396. doi: 10.1016/S0074-7696(08)60101-1. [DOI] [PubMed] [Google Scholar]
- 53.Kaiho M, Nakamura T, Kumegawa M. Morphological studies on the synthesis of secretory granules in convoluted tubules of mouse submandibular gland. Anat Rec. 1975;183:405–419. doi: 10.1002/ar.1091830305. [DOI] [PubMed] [Google Scholar]
- 54.Rogers AW, Brown-Grant K. The effects of castration on the ultrastructure and the iodide-concentrating ability of mouse submaxillary salivary glands. J Anat. 1971;109:51–62. [PMC free article] [PubMed] [Google Scholar]
- 55.Matsuura S, Sahara N, Suzuki K. Fine structure of submandibular glands of mice with testicular feminization (Tfm/Y) Cell Tissue Res. 1984;235:295–301. doi: 10.1007/BF00217853. [DOI] [PubMed] [Google Scholar]
- 56.Suzuki A, Shim J, Ogata K, Yoshioka H. Cholesterol metabolism plays a crucial role in the regulation of autophagy for cell differentiation of granular convoluted tubules in male mouse submandibular glands. Development. 2019;146(20):178335. doi: 10.1242/dev.178335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Iber D, Menshykau D. The control of branching morphogenesis. Open Biol. 2013;3:130088. doi: 10.1098/rsob.130088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hoffman MP, Kidder BL, Steinberg ZL, Lakhani S, Ho S, et al. Gene expression profiles of mouse submandibular gland development: FGFR1 regulates branching morphogenesis in vitro through BMP- and FGF-dependent mechanisms. Development. 2002;129:5767–5778. doi: 10.1242/dev.00172. [DOI] [PubMed] [Google Scholar]
- 59.May AJ, Chatzeli L, Proctor GB, Tucker AS. Salivary gland dysplasia in Fgf10 heterozygous mice: a new mouse model of xerostomia. Curr Mol Med. 2015;15:674–682. doi: 10.2174/1566524015666150831141307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.De Moerlooze L, Spencer-Dene B, Revest JM, Hajihosseini M, Rosewell I, et al. An important role for the IIIb isoform of fibroblast growth factor receptor 2 (FGFR2) in mesenchymal-epithelial signalling during mouse organogenesis. Development. 2000;127:483–492. doi: 10.1242/dev.127.3.483. [DOI] [PubMed] [Google Scholar]
- 61.Ohuchi H, Hori Y, Yamasaki M, Harada H, Sekine K, et al. FGF10 acts as a major ligand for FGF receptor 2 IIIb in mouse multi-organ development. Biochem Biophys Res Commun. 2000;277:643–649. doi: 10.1006/bbrc.2000.3721. [DOI] [PubMed] [Google Scholar]
- 62.Revest JM, Spencer-Dene B, Kerr K, De Moerlooze L, Rosewell I, et al. Fibroblast growth factor receptor 2-IIIb acts upstream of Shh and Fgf4 and is required for limb bud maintenance but not for the induction of Fgf8, Fgf10, Msx1, or Bmp4. Dev Biol. 2001;231:47–62. doi: 10.1006/dbio.2000.0144. [DOI] [PubMed] [Google Scholar]
- 63.Jaskoll T, Abichaker G, Witcher D, Sala FG, Bellusci S, et al. FGF10/FGFR2b signaling plays essential roles during in vivo embryonic submandibular salivary gland morphogenesis. BMC Dev Biol. 2005;5:11. doi: 10.1186/1471-213X-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Teshima TH, Lourenco SV, Tucker AS. Multiple cranial organ defects after conditionally knocking out Fgf10 in the neural crest. Front Physiol. 2016;7:488. doi: 10.3389/fphys.2016.00488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wells KL, Gaete M, Matalova E, Deutsch D, Rice D, et al. Dynamic relationship of the epithelium and mesenchyme during salivary gland initiation: the role of Fgf10. Biol Open. 2013;2:981–989. doi: 10.1242/bio.20135306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Makarenkova HP, Hoffman MP, Beenken A, Eliseenkova AV, Meech R, et al. Differential interactions of FGFs with heparan sulfate control gradient formation and branching morphogenesis. Sci Signal. 2009;2:ra55. doi: 10.1126/scisignal.2000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jaskoll T, Zhou YM, Chai Y, Makarenkova HP, Collinson JM, et al. Embryonic submandibular gland morphogenesis: stage-specific protein localization of FGFs, BMPs, Pax6 and Pax9 in normal mice and abnormal SMG phenotypes in FgfR2-IIIc(+/Delta), BMP7(-/-) and Pax6(-/-) mice. Cells Tissues Organs. 2002;170:83–98. doi: 10.1159/000046183. [DOI] [PubMed] [Google Scholar]
- 68.Chapman DB, Shashi V, Kirse DJ. Case report: aplasia of the lacrimal and major salivary glands (ALSG) Int J Pediatr Otorhinolaryngol. 2009;73:899–901. doi: 10.1016/j.ijporl.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 69.Milunsky JM, Zhao G, Maher TA, Colby R, Everman DB. LADD syndrome is caused by FGF10 mutations. Clin Genet. 2006;69:349–354. doi: 10.1111/j.1399-0004.2006.00597.x. [DOI] [PubMed] [Google Scholar]
- 70.Seymen F, Koruyucu M, Toptanci IR, Balsak S, Dedeoglu S, et al. Novel FGF10 mutation in autosomal dominant aplasia of lacrimal and salivary glands. Clin Oral Investig. 2017;21:167–172. doi: 10.1007/s00784-016-1771-x. [DOI] [PubMed] [Google Scholar]
- 71.Rodrigo MJ, Idoipe M, Izquierdo S, Satue M, Mateo A, et al. New pathogenic variant in the FGF10 gene in the agenesis of lacrimal and salivary gland syndrome: ophthalmological and genetic study. Ophthalmic Genet. 2018;39:125–128. doi: 10.1080/13816810.2017.1381976. [DOI] [PubMed] [Google Scholar]
- 72.Entesarian M, Matsson H, Klar J, Bergendal B, Olson L, et al. Mutations in the gene encoding fibroblast growth factor 10 are associated with aplasia of lacrimal and salivary glands. Nat Genet. 2005;37:125–127. doi: 10.1038/ng1507. [DOI] [PubMed] [Google Scholar]
- 73.Scheckenbach K, Balz V, Wagenmann M, Hoffmann TK. An intronic alteration of the fibroblast growth factor 10 gene causing ALSG-(aplasia of lacrimal and salivary glands) syndrome. BMC Med Genet. 2008;9:114. doi: 10.1186/1471-2350-9-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rohmann E, Brunner HG, Kayserili H, Uyguner O, Nurnberg G, et al. Mutations in different components of FGF signaling in LADD syndrome. Nat Genet. 2006;38:414–417. doi: 10.1038/ng1757. [DOI] [PubMed] [Google Scholar]
- 75.Carraro G, El-Hashash A, Guidolin D, Tiozzo C, Turcatel G, et al. miR-17 family of microRNAs controls FGF10-mediated embryonic lung epithelial branching morphogenesis through MAPK14 and STAT3 regulation of E-Cadherin distribution. Dev Biol. 2009;333:238–250. doi: 10.1016/j.ydbio.2009.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Boucherat O, Nadeau V, Berube-Simard FA, Charron J, Jeannotte L. Crucial requirement of ERK/MAPK signaling in respiratory tract development. Development. 2015;142:3801. doi: 10.1242/dev.131821. [DOI] [PubMed] [Google Scholar]
- 77.Patel VN, Knox SM, Likar KM, Lathrop CA, Hossain R, et al. Heparanase cleavage of perlecan heparan sulfate modulates FGF10 activity during ex vivo submandibular gland branching morphogenesis. Development. 2007;134:4177–4186. doi: 10.1242/dev.011171. [DOI] [PubMed] [Google Scholar]
- 78.Patel VN, Likar KM, Zisman-Rozen S, Cowherd SN, Lassiter KS, et al. Specific heparan sulfate structures modulate FGF10-mediated submandibular gland epithelial morphogenesis and differentiation. J Biol Chem. 2008;283:9308–9317. doi: 10.1074/jbc.M709995200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shams I, Rohmann E, Eswarakumar VP, Lew ED, Yuzawa S, et al. Lacrimo-auriculo-dento-digital syndrome is caused by reduced activity of the fibroblast growth factor 10 (FGF10)-FGF receptor 2 signaling pathway. Mol Cell Biol. 2007;27:6903–6912. doi: 10.1128/MCB.00544-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lew ED, Bae JH, Rohmann E, Wollnik B, Schlessinger J. Structural basis for reduced FGFR2 activity in LADD syndrome: Implications for FGFR autoinhibition and activation. Proc Natl Acad Sci U S A. 2007;104:19802–19807. doi: 10.1073/pnas.0709905104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jaskoll T, Witcher D, Toreno L, Bringas P, Moon AM, et al. FGF8 dose-dependent regulation of embryonic submandibular salivary gland morphogenesis. Dev Biol. 2004;268:457–469. doi: 10.1016/j.ydbio.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 82.Daphna-Iken D, Shankar DB, Lawshe A, Ornitz DM, Shackleford GM, et al. MMTV-Fgf8 transgenic mice develop mammary and salivary gland neoplasia and ovarian stromal hyperplasia. Oncogene. 1998;17:2711–2717. doi: 10.1038/sj.onc.1202212. [DOI] [PubMed] [Google Scholar]
- 83.Matsumoto S, Kurimoto T, Taketo MM, Fujii S, Kikuchi A. The WNT/MYB pathway suppresses KIT expression to control the timing of salivary proacinar differentiation and duct formation. Development. 2016;143:2311–2324. doi: 10.1242/dev.134486. [DOI] [PubMed] [Google Scholar]
- 84.Guo L, Yu QC, Fuchs E. Targeting expression of keratinocyte growth factor to keratinocytes elicits striking changes in epithelial differentiation in transgenic mice. EMBO J. 1993;12:973–986. doi: 10.1002/j.1460-2075.1993.tb05738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guo L, Degenstein L, Fuchs E. Keratinocyte growth factor is required for hair development but not for wound healing. Genes Dev. 1996;10:165–175. doi: 10.1101/gad.10.2.165. [DOI] [PubMed] [Google Scholar]
- 86.Knosp WM, Knox SM, Lombaert IM, Haddox CL, Patel VN, et al. Submandibular parasympathetic gangliogenesis requires sprouty-dependent Wnt signals from epithelial progenitors. Dev Cell. 2015;32:667–677. doi: 10.1016/j.devcel.2015.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hosseini ZF, Nelson DA, Moskwa N, Sfakis LM, Castracane J, et al. FGF2-dependent mesenchyme and laminin-111 are niche factors in salivary gland organoids. J Cell Sci. 2018;1:131. doi: 10.1242/jcs.208728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wieduwilt MJ, Moasser MM. The epidermal growth factor receptor family: biology driving targeted therapeutics. Cell Mol Life Sci. 2008;65:1566–1584. doi: 10.1007/s00018-008-7440-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sisto M, Lorusso L, Ingravallo G, Lisi S. Exocrine gland morphogenesis: insights into the role of amphiregulin from development to disease. Arch Immunol Ther Exp (Warsz) 2017;65:477–499. doi: 10.1007/s00005-017-0478-2. [DOI] [PubMed] [Google Scholar]
- 90.Miyazaki Y, Nakanishi Y, Hieda Y. Tissue interaction mediated by neuregulin-1 and ErbB receptors regulates epithelial morphogenesis of mouse embryonic submandibular gland. Dev Dyn. 2004;230:591–596. doi: 10.1002/dvdy.20078. [DOI] [PubMed] [Google Scholar]
- 91.Nitta M, Kume T, Nogawa H. FGF alters epithelial competence for EGF at the initiation of branching morphogenesis of mouse submandibular gland. Dev Dyn. 2009;238:315–323. doi: 10.1002/dvdy.21780. [DOI] [PubMed] [Google Scholar]
- 92.Jaskoll T, Melnick M. Submandibular gland morphogenesis: stage-specific expression of TGF-alpha/EGF, IGF, TGF-beta, TNF, and IL-6 signal transduction in normal embryonic mice and the phenotypic effects of TGF-beta2, TGF-beta3, and EGF-r null mutations. Anat Rec. 1999;256:252–268. doi: 10.1002/(SICI)1097-0185(19991101)256:3<252::AID-AR5>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 93.Nogawa H, Takahashi Y. Substitution for mesenchyme by basement-membrane-like substratum and epidermal growth factor in inducing branching morphogenesis of mouse salivary epithelium. Development. 1991;112:855–861. doi: 10.1242/dev.112.3.855. [DOI] [PubMed] [Google Scholar]
- 94.Koyama N, Kashimata M, Sakashita H, Sakagami H, Gresik EW. EGF-stimulated signaling by means of PI3K, PLCgamma1, and PKC isozymes regulates branching morphogenesis of the fetal mouse submandibular gland. Dev Dyn. 2003;227:216–226. doi: 10.1002/dvdy.10309. [DOI] [PubMed] [Google Scholar]
- 95.Koyama N, Hayashi T, Mizukoshi K, Matsumoto T, Gresik EW, et al. Extracellular regulated kinase5 is expressed in fetal mouse submandibular glands and is phosphorylated in response to epidermal growth factor and other ligands of the ErbB family of receptors. Dev Growth Differ. 2012;54:801–808. doi: 10.1111/dgd.12008. [DOI] [PubMed] [Google Scholar]
- 96.Kashimata M, Sayeed S, Ka A, Onetti-Muda A, Sakagami H, et al. The ERK-1/2 signaling pathway is involved in the stimulation of branching morphogenesis of fetal mouse submandibular glands by EGF. Dev Biol. 2000;220:183–196. doi: 10.1006/dbio.2000.9639. [DOI] [PubMed] [Google Scholar]
- 97.Kashimata M, Gresik EW. Epidermal growth factor system is a physiological regulator of development of the mouse fetal submandibular gland and regulates expression of the alpha6-integrin subunit. Dev Dyn. 1997;208:149–161. doi: 10.1002/(SICI)1097-0177(199702)208:2<149::AID-AJA2>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 98.Umeda Y, Miyazaki Y, Shiinoki H, Higashiyama S, Nakanishi Y, et al. Involvement of heparin-binding EGF-like growth factor and its processing by metalloproteinases in early epithelial morphogenesis of the submandibular gland. Dev Biol. 2001;237:202–211. doi: 10.1006/dbio.2001.0351. [DOI] [PubMed] [Google Scholar]
- 99.Haara O, Koivisto T, Miettinen PJ. EGF-receptor regulates salivary gland branching morphogenesis by supporting proliferation and maturation of epithelial cells and survival of mesenchymal cells. Differentiation. 2009;77:298–306. doi: 10.1016/j.diff.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 100.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 101.Chiba S. Notch signaling in stem cell systems. Stem Cells. 2006;24:2437–2447. doi: 10.1634/stemcells.2005-0661. [DOI] [PubMed] [Google Scholar]
- 102.Lai EC. Notch signaling: control of cell communication and cell fate. Development. 2004;131:965–973. doi: 10.1242/dev.01074. [DOI] [PubMed] [Google Scholar]
- 103.Luo Z, Shang X, Zhang H, Wang G, Massey PA, et al. Notch signaling in osteogenesis, osteoclastogenesis, and angiogenesis. Am J Pathol. 2019;189:1495–1500. doi: 10.1016/j.ajpath.2019.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lloyd-Lewis B, Mourikis P, Fre S. Notch signalling: sensor and instructor of the microenvironment to coordinate cell fate and organ morphogenesis. Curr Opin Cell Biol. 2019;61:16–23. doi: 10.1016/j.ceb.2019.06.003. [DOI] [PubMed] [Google Scholar]
- 105.Dang H, Lin AL, Zhang B, Zhang HM, Katz MS, et al. Role for Notch signaling in salivary acinar cell growth and differentiation. Dev Dyn. 2009;238:724–731. doi: 10.1002/dvdy.21875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Baladron V, Ruiz-Hidalgo MJ, Nueda ML, Diaz-Guerra MJ, Garcia-Ramirez JJ, et al. dlk acts as a negative regulator of Notch1 activation through interactions with specific EGF-like repeats. Exp Cell Res. 2005;303:343–359. doi: 10.1016/j.yexcr.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 107.Yevtodiyenko A, Schmidt JV. Dlk1 expression marks developing endothelium and sites of branching morphogenesis in the mouse embryo and placenta. Dev Dyn. 2006;235:1115–1123. doi: 10.1002/dvdy.20705. [DOI] [PubMed] [Google Scholar]
- 108.Garcia-Gallastegi P, Ruiz-Garcia A, Ibarretxe G, Rivero-Hinojosa S, Gonzalez-Siccha AD, et al. Similarities and differences in tissue distribution of DLK1 and DLK2 during E16.5 mouse embryogenesis. Histochem Cell Biol. 2019;152:47–60. doi: 10.1007/s00418-019-01778-4. [DOI] [PubMed] [Google Scholar]
- 109.Garcia-Gallastegui P, Ibarretxe G, Garcia-Ramirez JJ, Baladron V, Aurrekoetxea M, et al. DLK1 regulates branching morphogenesis and parasympathetic innervation of salivary glands through inhibition of NOTCH signalling. Biol Cell. 2014;106:237–253. doi: 10.1111/boc.201300086. [DOI] [PubMed] [Google Scholar]
- 110.Garcia-Gallastegui P, Luzuriaga J, Aurrekoetxea M, Baladron V, Ruiz-Hidalgo MJ, et al. Reduced salivary gland size and increased presence of epithelial progenitor cells in DLK1-deficient mice. Cell Tissue Res. 2016;364:513–525. doi: 10.1007/s00441-015-2344-z. [DOI] [PubMed] [Google Scholar]
- 111.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 112.Wang J, Sinha T, Wynshaw-Boris A. Wnt signaling in mammalian development: lessons from mouse genetics. Cold Spring Harb Perspect Biol. 2012;4:3. doi: 10.1101/cshperspect.a007963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Steinhart Z, Angers S. Wnt signaling in development and tissue homeostasis. Development. 2018;1:145. doi: 10.1242/dev.146589. [DOI] [PubMed] [Google Scholar]
- 114.Sugimura R, Li L. Noncanonical Wnt signaling in vertebrate development, stem cells, and diseases. Birth Defects Res C Embryo Today. 2010;90:243–256. doi: 10.1002/bdrc.20195. [DOI] [PubMed] [Google Scholar]
- 115.Wang HY, Malbon CC. Wnt signaling, Ca2+, and cyclic GMP: visualizing frizzled functions. Science. 2003;300:1529–1530. doi: 10.1126/science.1085259. [DOI] [PubMed] [Google Scholar]
- 116.Miller JR. The Wnts. Genome Biol. 2002;3:REVIEW3001. doi: 10.1186/gb-2001-3-1-reviews3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Niehrs C. The complex world of WNT receptor signalling. Nat Rev Mol Cell Biol. 2012;13:767–779. doi: 10.1038/nrm3470. [DOI] [PubMed] [Google Scholar]
- 118.Patel N, Sharpe PT, Miletich I. Coordination of epithelial branching and salivary gland lumen formation by Wnt and FGF signals. Dev Biol. 2011;358:156–167. doi: 10.1016/j.ydbio.2011.07.023. [DOI] [PubMed] [Google Scholar]
- 119.Haara O, Fujimori S, Schmidt-Ullrich R, Hartmann C, Thesleff I, et al. Ectodysplasin and Wnt pathways are required for salivary gland branching morphogenesis. Development. 2011;138:2681–2691. doi: 10.1242/dev.057711. [DOI] [PubMed] [Google Scholar]
- 120.Maimets M, Rocchi C, Bron R, Pringle S, Kuipers J, et al. Long-term in vitro expansion of salivary gland stem cells driven by wnt signals. Stem Cell Rep. 2016;6:150–162. doi: 10.1016/j.stemcr.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kopinke D, Norris AM, Mukhopadhyay S. Developmental and regenerative paradigms of cilia regulated hedgehog signaling. Semin Cell Dev Biol. 2020;5:3. doi: 10.1016/j.semcdb.2020.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mukhopadhyay S, Rohatgi R. G-protein-coupled receptors, Hedgehog signaling and primary cilia. Semin Cell Dev Biol. 2014;33:63–72. doi: 10.1016/j.semcdb.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Carballo GB, Honorato JR, de Lopes GPF, Spohr T. A highlight on Sonic hedgehog pathway. Cell Commun Signal. 2018;16:11. doi: 10.1186/s12964-018-0220-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sasai N, Toriyama M, Kondo T. Hedgehog signal and genetic disorders. Front Genet. 2019;10:1103. doi: 10.3389/fgene.2019.01103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lee RT, Zhao Z, Ingham PW. Hedgehog signalling. Development. 2016;143:367–372. doi: 10.1242/dev.120154. [DOI] [PubMed] [Google Scholar]
- 126.Kong JH, Siebold C, Rohatgi R. Biochemical mechanisms of vertebrate hedgehog signaling. Development. 2019;1:146. doi: 10.1242/dev.166892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Guan Z, Dong B, Huang C, Hu X, Zhang Y, et al. Expression patterns of genes critical for SHH, BMP, and FGF pathways during the lumen formation of human salivary glands. J Mol Histol. 2019;50:217–227. doi: 10.1007/s10735-019-09819-x. [DOI] [PubMed] [Google Scholar]
- 128.Jaskoll T, Leo T, Witcher D, Ormestad M, Astorga J, et al. Sonic hedgehog signaling plays an essential role during embryonic salivary gland epithelial branching morphogenesis. Dev Dyn. 2004;229:722–732. doi: 10.1002/dvdy.10472. [DOI] [PubMed] [Google Scholar]
- 129.Mizukoshi K, Koyama N, Hayashi T, Zheng L, Matsuura S, et al. Shh/Ptch and EGF/ErbB cooperatively regulate branching morphogenesis of fetal mouse submandibular glands. Dev Biol. 2016;412:278–287. doi: 10.1016/j.ydbio.2016.02.018. [DOI] [PubMed] [Google Scholar]
- 130.Fiaschi M, Kolterud A, Nilsson M, Toftgard R, Rozell B. Targeted expression of GLI1 in the salivary glands results in an altered differentiation program and hyperplasia. Am J Pathol. 2011;179:2569–2579. doi: 10.1016/j.ajpath.2011.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Elliott KH, Millington G, Brugmann SA. A novel role for cilia-dependent sonic hedgehog signaling during submandibular gland development. Dev Dyn. 2018;247:818–831. doi: 10.1002/dvdy.24627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.McDermott KM, Liu BY, Tlsty TD, Pazour GJ. Primary cilia regulate branching morphogenesis during mammary gland development. Curr Biol. 2010;20:731–737. doi: 10.1016/j.cub.2010.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zinski J, Tajer B, Mullins MC. TGF-beta family signaling in early vertebrate development. Cold Spring Harb Perspect Biol. 2018;1:10. doi: 10.1101/cshperspect.a033274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sieber C, Kopf J, Hiepen C, Knaus P. Recent advances in BMP receptor signaling. Cytokine Growth Factor Rev. 2009;20:343–355. doi: 10.1016/j.cytogfr.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 135.Wrana JL, Attisano L, Wieser R, Ventura F, Massague J. Mechanism of activation of the TGF-beta receptor. Nature. 1994;370:341–347. doi: 10.1038/370341a0. [DOI] [PubMed] [Google Scholar]
- 136.Katagiri T, Watabe T. Bone morphogenetic proteins. Cold Spring Harb Perspect Biol. 2016;1:8. doi: 10.1101/cshperspect.a021899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/S0092-8674(03)00432-X. [DOI] [PubMed] [Google Scholar]
- 138.Heldin CH, Moustakas A. Signaling receptors for TGF-beta family members. Cold Spring Harb Perspect Biol. 2016;3:8–21. doi: 10.1101/cshperspect.a022053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Takahashi H, Ikeda T. Transcripts for two members of the transforming growth factor-beta superfamily BMP-3 and BMP-7 are expressed in developing rat embryos. Dev Dyn. 1996;207:439–449. doi: 10.1002/(SICI)1097-0177(199612)207:4<439::AID-AJA8>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 140.Heikinheimo KA, Laine MA, Ritvos OV, Voutilainen RJ, Hogan BL, et al. Bone morphogenetic protein-6 is a marker of serous acinar cell differentiation in normal and neoplastic human salivary gland. Cancer Res. 1999;59:5815–5821. [PubMed] [Google Scholar]
- 141.Thomadakis G, Ramoshebi LN, Crooks J, Rueger DC, Ripamonti U. Immunolocalization of bone morphogenetic protein-2 and -3 and osteogenic protein-1 during murine tooth root morphogenesis and in other craniofacial structures. Eur J Oral Sci. 1999;107:368–377. doi: 10.1046/j.0909-8836.1999.eos107508.x. [DOI] [PubMed] [Google Scholar]
- 142.Zouvelou V, Luder HU, Mitsiadis TA, Graf D. Deletion of BMP7 affects the development of bones, teeth, and other ectodermal appendages of the orofacial complex. J Exp Zool B Mol Dev Evol. 2009;312B:361–374. doi: 10.1002/jez.b.21262. [DOI] [PubMed] [Google Scholar]
- 143.Miao N, Zhan Y, Xu Y, Yuan H, Qin C, et al. Loss of Fam20c causes defects in the acinar and duct structure of salivary glands in mice. Int J Mol Med. 2019;43:2103–2117. doi: 10.3892/ijmm.2019.4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Yin H, Cabrera-Perez J, Lai Z, Michael D, Weller M, et al. Association of bone morphogenetic protein 6 with exocrine gland dysfunction in patients with Sjogren’s syndrome and in mice. Arthritis Rheum. 2013;65:3228–3238. doi: 10.1002/art.38123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Yin H, Kalra L, Lai Z, Guimaro MC, Aber L, et al. Inhibition of bone morphogenetic protein 6 receptors ameliorates Sjogren's syndrome in mice. Sci Rep. 2020;10:2967. doi: 10.1038/s41598-020-59443-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kizu Y, Sakurai H, Katagiri S, Shinozaki N, Ono M, et al. Immunohistological analysis of tumour growth factor beta 1 expression in normal and inflamed salivary glands. J Clin Pathol. 1996;49:728. doi: 10.1136/jcp.49.9.728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Jaskoll T, Choy HA, Melnick M. Glucocorticoids, TGF-beta, and embryonic mouse salivary gland morphogenesis. J Craniofac Genet Dev Biol. 1994;14:217–230. [PubMed] [Google Scholar]
- 148.Shull MM, Ormsby I, Kier AB, Pawlowski S, Diebold RJ, et al. Targeted disruption of the mouse transforming growth factor-beta 1 gene results in multifocal inflammatory disease. Nature. 1992;359:693–699. doi: 10.1038/359693a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Janebodin K, Buranaphatthana W, Ieronimakis N, Hays AL, Reyes M. An in vitro culture system for long-term expansion of epithelial and mesenchymal salivary gland cells: role of TGF-beta1 in salivary gland epithelial and mesenchymal differentiation. Biomed Res Int. 2013;3:815895. doi: 10.1155/2013/815895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hall BE, Zheng C, Swaim WD, Cho A, Nagineni CN, et al. Conditional overexpression of TGF-beta1 disrupts mouse salivary gland development and function. Lab Invest. 2010;90:543–555. doi: 10.1038/labinvest.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Boivin GP, O'Toole BA, Orsmby IE, Diebold RJ, Eis MJ, et al. Onset and progression of pathological lesions in transforming growth factor-beta 1-deficient mice. Am J Pathol. 1995;146:276–288. [PMC free article] [PubMed] [Google Scholar]
- 152.Mikkola ML, Thesleff I. Ectodysplasin signaling in development. Cytokine Growth Factor Rev. 2003;14:211–224. doi: 10.1016/S1359-6101(03)00020-0. [DOI] [PubMed] [Google Scholar]
- 153.Mikkola ML. Molecular aspects of hypohidrotic ectodermal dysplasia. Am J Med Genet A. 2009;149A:2031–2036. doi: 10.1002/ajmg.a.32855. [DOI] [PubMed] [Google Scholar]
- 154.Cui CY, Schlessinger D. EDA signaling and skin appendage development. Cell Cycle. 2006;5:2477–2483. doi: 10.4161/cc.5.21.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Botchkarev VA, Fessing MY. Edar signaling in the control of hair follicle development. J Investig Dermatol Symp Proc. 2005;10:247–251. doi: 10.1111/j.1087-0024.2005.10129.x. [DOI] [PubMed] [Google Scholar]
- 156.Nordgarden H, Johannessen S, Storhaug K, Jensen JL. Salivary gland involvement in hypohidrotic ectodermal dysplasia. Oral Dis. 1998;4:152–154. doi: 10.1111/j.1601-0825.1998.tb00270.x. [DOI] [PubMed] [Google Scholar]
- 157.Bergendal B. Orodental manifestations in ectodermal dysplasia-a review. Am J Med Genet A. 2014;164A:2465–2471. doi: 10.1002/ajmg.a.36571. [DOI] [PubMed] [Google Scholar]
- 158.Schneider H, Hammersen J, Preisler-Adams S, Huttner K, Rascher W, et al. Sweating ability and genotype in individuals with X-linked hypohidrotic ectodermal dysplasia. J Med Genet. 2011;48:426–432. doi: 10.1136/jmg.2010.084012. [DOI] [PubMed] [Google Scholar]
- 159.Cluzeau C, Hadj-Rabia S, Jambou M, Mansour S, Guigue P, et al. Only four genes (EDA1, EDAR, EDARADD, and WNT10A) account for 90% of hypohidrotic/anhidrotic ectodermal dysplasia cases. Hum Mutat. 2011;32:70–72. doi: 10.1002/humu.21384. [DOI] [PubMed] [Google Scholar]
- 160.Blecher SR, Debertin M, Murphy JS. Pleiotropic effect of Tabby gene on epidermal growth factor-containing cells of mouse submandibular gland. Anat Rec. 1983;207:25–29. doi: 10.1002/ar.1092070104. [DOI] [PubMed] [Google Scholar]
- 161.Wells KL, Mou C, Headon DJ, Tucker AS. Defects and rescue of the minor salivary glands in Eda pathway mutants. Dev Biol. 2011;349:137–146. doi: 10.1016/j.ydbio.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 162.Jaskoll T, Zhou YM, Trump G, Melnick M. Ectodysplasin receptor-mediated signaling is essential for embryonic submandibular salivary gland development. Anat Rec A Discov Mol Cell Evol Biol. 2003;271:322–331. doi: 10.1002/ar.a.10045. [DOI] [PubMed] [Google Scholar]
- 163.Wells KL, Mou C, Headon DJ, Tucker AS. Recombinant EDA or Sonic Hedgehog rescue the branching defect in Ectodysplasin A pathway mutant salivary glands in vitro. Dev Dyn. 2010;239:2674–2684. doi: 10.1002/dvdy.22406. [DOI] [PubMed] [Google Scholar]
- 164.Mukaibo T, Munemasa T, Masaki C, Cui CY, Melvin JE. Defective NaCl reabsorption in salivary glands of Eda-null X-LHED mice. J Dent Res. 2018;97:1244–1251. doi: 10.1177/0022034518782461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Chang SH, Jobling S, Brennan K, Headon DJ. Enhanced Edar signalling has pleiotropic effects on craniofacial and cutaneous glands. PLoS ONE. 2009;4:e7591. doi: 10.1371/journal.pone.0007591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Voutilainen M, Lindfors PH, Trela E, Lonnblad D, Shirokova V, et al. Ectodysplasin/NF-kappaB promotes mammary cell fate via Wnt/beta-catenin pathway. PLoS Genet. 2015;11:e1005676. doi: 10.1371/journal.pgen.1005676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Lindfors PH, Voutilainen M, Mikkola ML. Ectodysplasin/NF-kappaB signaling in embryonic mammary gland development. J Mammary Gland Biol Neoplasia. 2013;18:165–169. doi: 10.1007/s10911-013-9277-5. [DOI] [PubMed] [Google Scholar]
- 168.Lippens S, Lefebvre S, Gilbert B, Sze M, Devos M, et al. Keratinocyte-specific ablation of the NF-kappaB regulatory protein A20 (TNFAIP3) reveals a role in the control of epidermal homeostasis. Cell Death Differ. 2011;18:1845–1853. doi: 10.1038/cdd.2011.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Schmidt-Ullrich R, Tobin DJ, Lenhard D, Schneider P, Paus R, et al. NF-kappaB transmits Eda A1/EdaR signalling to activate Shh and cyclin D1 expression, and controls post-initiation hair placode down growth. Development. 2006;133:1045–1057. doi: 10.1242/dev.02278. [DOI] [PubMed] [Google Scholar]
- 170.Okita T, Asano N, Yasuno S, Shimomura Y. Functional studies for a dominant mutation in the EDAR gene responsible for hypohidrotic ectodermal dysplasia. J Dermatol. 2019;46:710–715. doi: 10.1111/1346-8138.14983. [DOI] [PubMed] [Google Scholar]
- 171.Pummila M, Fliniaux I, Jaatinen R, James MJ, Laurikkala J, et al. Ectodysplasin has a dual role in ectodermal organogenesis: inhibition of Bmp activity and induction of Shh expression. Development. 2007;134:117–125. doi: 10.1242/dev.02708. [DOI] [PubMed] [Google Scholar]
- 172.Häärä O, Fujimori S, Schmidt-Ullrich R, Hartmann C, Thesleff I, et al. Ectodysplasin and Wnt pathways are required for salivary gland branching morphogenesis. Development. 2011;138:2681. doi: 10.1242/dev.057711. [DOI] [PubMed] [Google Scholar]
- 173.Airaksinen MS, Saarma M. The GDNF family: signalling, biological functions and therapeutic value. Nat Rev Neurosci. 2002;3:383–394. doi: 10.1038/nrn812. [DOI] [PubMed] [Google Scholar]
- 174.Poyhonen S, Er S, Domanskyi A, Airavaara M. Effects of neurotrophic factors in glial cells in the central nervous system: expression and properties in neurodegeneration and injury. Front Physiol. 2019;10:486. doi: 10.3389/fphys.2019.00486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Paratcha G, Ledda F. GDNF and GFRalpha: a versatile molecular complex for developing neurons. Trends Neurosci. 2008;31:384–391. doi: 10.1016/j.tins.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 176.Peng X, Varendi K, Maimets M, Andressoo JO, Coppes RP. Role of glial-cell-derived neurotrophic factor in salivary gland stem cell response to irradiation. Radiother Oncol. 2017;124:448–454. doi: 10.1016/j.radonc.2017.07.008. [DOI] [PubMed] [Google Scholar]
- 177.Xiao N, Lin Y, Cao H, Sirjani D, Giaccia AJ, et al. Neurotrophic factor GDNF promotes survival of salivary stem cells. J Clin Invest. 2014;124:3364–3377. doi: 10.1172/JCI74096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Rossi J, Tomac A, Saarma M, Airaksinen MS. Distinct roles for GFRalpha1 and GFRalpha2 signalling in different cranial parasympathetic ganglia in vivo. Eur J Neurosci. 2000;12:3944–3952. doi: 10.1046/j.1460-9568.2000.00292.x. [DOI] [PubMed] [Google Scholar]
- 179.Enomoto H, Heuckeroth RO, Golden JP, Johnson EM, Milbrandt J. Development of cranial parasympathetic ganglia requires sequential actions of GDNF and neurturin. Development. 2000;127:4877–4889. doi: 10.1242/dev.127.22.4877. [DOI] [PubMed] [Google Scholar]
- 180.Rossi J, Luukko K, Poteryaev D, Laurikainen A, Sun YF, et al. Retarded growth and deficits in the enteric and parasympathetic nervous system in mice lacking GFR alpha2, a functional neurturin receptor. Neuron. 1999;22:243–252. doi: 10.1016/S0896-6273(00)81086-7. [DOI] [PubMed] [Google Scholar]
- 181.Knox SM, Lombaert IM, Haddox CL, Abrams SR, Cotrim A, et al. Parasympathetic stimulation improves epithelial organ regeneration. Nat Commun. 2013;4:1494. doi: 10.1038/ncomms2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Ferreira JNA, Zheng C, Lombaert IMA, Goldsmith CM, Cotrim AP, et al. Neurturin gene therapy protects parasympathetic function to prevent irradiation-induced murine salivary gland hypofunction. Mol Ther Methods Clin Dev. 2018;9:172–180. doi: 10.1016/j.omtm.2018.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Juusela P, Tanskanen M, Nieminen A, Kari K, Suominen L, et al. Xerostomia in hereditary gelsolin amyloidosis. Amyloid. 2013;20:39–44. doi: 10.3109/13506129.2013.764284. [DOI] [PubMed] [Google Scholar]
- 184.Juusela P, Tanskanen M, Nieminen A, Uitto VJ, Blafield H, et al. Hereditary gelsolin amyloidosis mimicking Sjogren’s syndrome. Clin Rheumatol. 2009;28:1351–1354. doi: 10.1007/s10067-009-1260-6. [DOI] [PubMed] [Google Scholar]
- 185.Trainor PA. Craniofacial birth defects: The role of neural crest cells in the etiology and pathogenesis of Treacher Collins syndrome and the potential for prevention. Am J Med Genet A. 2010;152A:2984–2994. doi: 10.1002/ajmg.a.33454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Dixon J, Trainor P, Dixon MJ. Treacher collins syndrome. Orthod Craniofac Res. 2007;10:88–95. doi: 10.1111/j.1601-6343.2007.00388.x. [DOI] [PubMed] [Google Scholar]
- 187.Osterhus IN, Skogedal N, Akre H, Johnsen UL, Nordgarden H, et al. Salivary gland pathology as a new finding in Treacher Collins syndrome. Am J Med Genet A. 2012;158A:1320–1325. doi: 10.1002/ajmg.a.35331. [DOI] [PubMed] [Google Scholar]
- 188.Matsui M, Motomura D, Karasawa H, Fujikawa T, Jiang J, et al. Multiple functional defects in peripheral autonomic organs in mice lacking muscarinic acetylcholine receptor gene for the M3 subtype. Proc Natl Acad Sci USA. 2000;97:9579–9584. doi: 10.1073/pnas.97.17.9579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Gautam D, Heard TS, Cui Y, Miller G, Bloodworth L, et al. Cholinergic stimulation of salivary secretion studied with M1 and M3 muscarinic receptor single- and double-knockout mice. Mol Pharmacol. 2004;66:260–267. doi: 10.1124/mol.66.2.260. [DOI] [PubMed] [Google Scholar]
- 190.Tobin G, Giglio D, Gotrick B. Studies of muscarinic receptor subtypes in salivary gland function in anaesthetized rats. Auton Neurosci. 2002;100:1–9. doi: 10.1016/S1566-0702(02)00139-X. [DOI] [PubMed] [Google Scholar]
- 191.Cho G, Bragiel AM, Wang D, Pieczonka TD, Skowronski MT, et al. Activation of muscarinic receptors in rat parotid acinar cells induces AQP5 trafficking to nuclei and apical plasma membrane. Biochim Biophys Acta. 2015;1850:784–793. doi: 10.1016/j.bbagen.2015.01.009. [DOI] [PubMed] [Google Scholar]
- 192.Ishikawa Y, Eguchi T, Skowronski MT, Ishida H. Acetylcholine acts on M3 muscarinic receptors and induces the translocation of aquaporin5 water channel via cytosolic Ca2+ elevation in rat parotid glands. Biochem Biophys Res Commun. 1998;245:835–840. doi: 10.1006/bbrc.1998.8395. [DOI] [PubMed] [Google Scholar]
- 193.Ishikawa Y, Skowronski MT, Ishida H. Persistent increase in the amount of aquaporin-5 in the apical plasma membrane of rat parotid acinar cells induced by a muscarinic agonist SNI-2011. FEBS Lett. 2000;477:253–257. doi: 10.1016/S0014-5793(00)01763-4. [DOI] [PubMed] [Google Scholar]
- 194.Qi W, Cong X, Zhang XM, Wang ZY, Yang NY, et al. Parasympathectomy increases resting salivary secretion in normal and irradiated submandibular glands of rats. Eur J Oral Sci. 2017;125:110–118. doi: 10.1111/eos.12330. [DOI] [PubMed] [Google Scholar]