Abstract
Regulatory T (Treg) cells that infiltrate human tumors exhibit stronger immunosuppressive activity compared to peripheral blood Treg cells (PBTRs), thus hindering the induction of effective antitumor immunity. Previous transcriptome studies have identified a set of genes that are conserved in tumor-infiltrating Treg cells (TITRs). However, epigenetic profiles of TITRs have not yet been completely deciphered. Here, we employed ATAC-seq and CUT&Tag assays to integrate transcriptome profiles and identify functional regulatory elements in TITRs. We observed a global difference in chromatin accessibility and enhancer landscapes between TITRs and PBTRs. We identified two types of active enhancer formation in TITRs. The H3K4me1-predetermined enhancers are poised to be activated in response to tumor microenvironmental stimuli. We found that AP-1 family motifs are enriched at the enhancer regions of TITRs. Finally, we validated that c-Jun binds at regulatory regions to regulate signature genes of TITRs and AP-1 is required for Treg cells activation in vitro. High c-Jun expression is correlated with poor survival in human HCC. Overall, our results provide insights into the mechanism of AP-1-mediated activation of TITRs and can hopefully be used to develop new therapeutic strategies targeting TITRs in liver cancer treatment.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-023-04746-3.
Keywords: Tumor-infiltrating Treg cells, Epigenetic profiles, Chromatin accessibility, Enhancer, AP-1, c-Jun
Introduction
Hepatocellular carcinoma (HCC) is the major cause of cancer-related death globally and its incidence and cancer-specific mortality are increasing rapidly worldwide [1]. Over the past several years, multiple immunotherapeutic modalities, such as immune-checkpoint inhibition therapy, have been used to treat HCC, but only a minority of patients could significantly benefit from the therapies [2]. Clinical data have revealed a highly immunosuppressive intratumor environment alongside dysfunctional tumor-infiltrating lymphocytes (TILs) in HCC [3]. Several studies have confirmed that the existence of an immunosuppressive tumor microenvironment (TME) caused mainly by the accumulation of regulatory T (Treg) cells [4]. Determination of molecular mechanism of Treg cells infiltrating the tumor tissues could facilitate the development of novel strategies in liver cancer treatment.
Treg cells that express the master transcription factor FOXP3 in peripheral lymphoid and non-lymphoid organs play an essential role in negative regulation of the immune response in mice and humans [5, 6]. These cells are able to migrate into inflammatory sites throughout the body, restrain inflammatory responses to self-antigens [7], and exert immunosuppressive functions via various mechanisms [8].
Abundant Treg cells have been observed in different experimental mouse tumor models and human cancers, constituting 10–50% of CD4+ T cells in tumor tissues compared with 2–5% of CD4+ T cells in the peripheral blood of healthy individuals. Treg cells can migrate to the TME by different chemokine receptors sensing multiple chemokines in various cancers [9]. The high frequency of TITRs and the increased ratios of FOXP3+ Treg cells to CD8+ T cells are related to poor prognoses of patients with various types of cancer [4]. Moreover, specific depletion of Treg cells is able to enhance tumor immunity effectively in murine models [10, 11]. Notably, clinical evidence indicates that the success of CTLA-4 blockade immunotherapy relies on Fc-mediated depletion of tumor-infiltrating CTLA-4+ Treg cells [12–14]. Thus, targeting TITRs may be a promising anti-tumor therapy, and gaining a deeper fundamental understanding of the unique properties of TITRs is warranted.
Several studies have revealed that the phenotype of TITRs is differed from the phenotype of Treg cells in peripheral blood [15, 16]. These TME-localized Treg cells, which are predominantly FOXP3high effector Treg cells and have a strong immunosuppressive activity, exhibit highly activated features characterized by high expression of T cell activation-associated markers, such as CTLA-4, OX40, GITR and TIM3 [9]. Transcriptome analysis studies have shown that TITRs are different from those in peripheral blood and matched non-cancerous tissues [17–19]. Recent single-cell transcriptome analyses have revealed the preferential enrichment for CTLA-4high Treg cells in HCC and NSCLC [20, 21], and the epigenetic landscapes of Treg cells in blood have been well investigated [22–25]. However, the epigenetic profiles of Treg cells localized in human tumors remain largely unexplored, which might provide useful insights into the mechanisms of gene regulation in TITRs.
In this study, we sorted TITRs and PBTRs from treatment-naïve primary HCC patients. ATAC-seq (Assay for Transposase Accessible Chromatin using Sequencing) [26], RNA-seq and CUT&Tag (Cleavage Under Targets and Tagmentation) [27] were performed to characterize the chromatin accessibility profiles, transcriptomic features, histone modifications and transcription factors in Treg cells. We observed that the chromatin accessibility landscape of TITRs is distinct from that of PBTRs. Functional annotation analysis revealed that the hyper-accessible regions in TITRs were largely gene distal, which often contains enhancer elements. Furthermore, we identified two types of active enhancer formation in TITRs. Significantly, H3K4me1-predetermined enhancers give the TITRs a set of transcriptional options to rapidly deploy in response to tumor microenvironmental stimuli. Finally, we found that c-Jun binds at regulatory regions to regulate activated genes of TITRs. AP-1 is required for activation of Treg cells in vitro. High c-Jun expression is associated with poor survival in human HCC. Our findings could help understand the mechanisms underlying specific expression patterns of TITRs, thereby accelerating the emergence of new therapeutic strategies targeting TITRs in liver cancer treatment.
Materials and methods
Patients
All human liver samples and blood were collected preoperatively and were obtained from the Department of General Surgery, Nanfang Hospital, Southern Medical University, Guangzhou, China. All clinical samples and information were collected with informed consent of the patients. The study was approved by the Medical Ethical Committee of Nanfang Hospital of Southern Medical University.
Tumor specimens
Tumor specimens and peripheral blood samples were obtained prospectively after surgical resection from treatment-naive HCC patients. Tumor specimens were preserved in the Tissue Storage Solution (Miltenyi). Tumor tissues and blood samples were processed within 12 h after isolation. For preparation of single-cell suspensions, fresh tumor specimens were cut into small pieces with scissors and digested by Liberase TL (Sigma) at 37 °C for 30 min under continuous rotation. Peripheral blood mononuclear cells were isolated using Ficoll-Pague Plus (GE).
Flow cytometry
Treg cells were purified by flow cytometry sorting including the antibodies: anti-CD3 FITC (clone OKT3), anti-CD4 PerCP-Cy5.5 (clone OKT4), anti-CD25 PE (clone BC96), and anti-CD127 PE/CF594 (clone A019D5). Cell sorting was performed on Aria II instrument (BD) by the Medical Research Institute. Antibodies for flow cytometric analyses were as follows: anti-CD4 BV421 (clone OKT4), anti-FOXP3 AF647 (clone 206D), anti-CD25 PE/dazzle594 (clone M-A251), anti-CTLA4 BV605 (clone BNI3), anti-ICOS APC/Fire750 (clone C398.4A), anti-PD1 PE (clone EH12.2H7), anti-GITR BV605 (clone 108-17), anti-TIM3 APC/Fire750 (clone F38-2E2).
ATAC-seq library preparation
We used TruePrep DNA Library Prep Kit (Vazyme, TD501) to construct the ATAC-seq library. Detailed procedures refer to Supplemental materials.
CUT&Tag library preparation
We used CUT&Tag High-Sensitivity Kit (Novoprotein) to construct the CUT&Tag library. Antibodies for this experiment were as follows: anti-H3K4me1 (Abcam, ab8895), anti-H3K27ac (Abcam, ab4729), anti-c-Jun (CST, 9165). Detailed procedures refer to Supplemental materials.
RNA-seq library preparation
The mRNA Amplification Kit (Vazyme, N712) was used to build the RNA-seq library. Approximately 1 ng full-length DNA was used per tagmentation reaction using TruePrep DNA Library Prep Kit (Vazyme, TD503), on the basis of the manufacturer’s protocol. Detailed procedures refer to Supplemental materials.
ATAC-seq and CUT&Tag data analysis
Sequencing reads were evaluated by FastQC and trimmed by Trim Galore. Clipped reads were aligned to the hg19 reference genome with Bowtie2 using the following options: bowtie2 -x hg19 –very-sensitive -X 1000 -p 10 -1 Reads1.fq.gz -2 Reads2.fq.gz. Aligned files were filtered using Samtools. PCR duplicated reads and reads aligning to the mitochondrial genome were filtered using Picards. Peaks were called using MACS2 with options: macs2 callpeak -t sample.bed -n sample -shift -100 -extsize 200 -nomodel -B -SPMR -g hs -keep-dup all. The output Bedgraph files from MACS2 were converted to bigWig format for peak visualization on the IGV Genome Browser. Using consensus peak sets, the number of fragments within peaks were counted using featureCounts. The differential analysis was performed using DESeq2. Peaks with absolute log2 fold change ≥ 1 and adjusted p value < 0.05 were considered significantly different between tumor-infiltrating and PBTRs. Peak annotation was performed with R package ChIPseeker. GO and GSEA analysis were performed with R package clusterProfiler. TFs motif analysis was carried out using HOMER tool. Genomic signal density plots were calculated by R package EnrichedHeatmap. TF regulatory networks were drawn using Cytoscape.
RNA-seq data analysis
Sequencing reads were evaluated by FastQC and trimmed by Trim Galore. Trimmed reads were aligned to the hg19 reference genome with HISAT2. Aligned files were filtered using Samtools. PCR duplicated reads were filtered using Picards. Transcripts were quantified at a gene level using featureCounts based on the GENECODE annotation. The differential analysis was analyzed by DESeq2. Genes with absolute log2 fold change ≥ 1 and adjusted p value < 0.05 were considered significantly different between tumor-infiltrating and PBTRs.
Transcription factor footprinting analysis
TF motifs were collected from Homo sapiens Comprehensive Model Collection database. TF footprinting analysis was carried out using TOBIAS tools with the principal workflow including the following steps. ATACorrect tool was used to correct the bias of ATAC-seq reads (TOBIAS ATACorrect -bam sample.bam -genome hg19genome.fa.gz -peaks merged_peaks.bed -blacklist hg19_blacklist.bed). Then, ScoreBigwig tool was used to calculate footprint scores from corrected cutsites (TOBIAS FootprintScores -signal corrected.bw -regions merged_peaks.bed). The BINDetect tool was used to estimate differentially bound motifs (TOBIAS BINDetect -motifs HOCOMOCOv11_full_HUMAN_mono_meme_format.meme -signals tumor_Tregs_footprints.bw blood_Tregs_footprints.bw -genome hg19genome.fa.gz -peaks merged_peaks_annotated.bed -peak_header merged_peaks_annotated_header.txt -outdir BINDetect_output -cond_names Tumor_versus_Blood -cores 8). The PlotAggregate tool was used to visualize TF footprints based on ATAC-seq signals (TOBIAS PlotAggregate -TFBS FOXP3_all.bed -signals tumor_Tregs_corrected.bw blood_Tregs_footprints.bw -output FOXP3_footprint_comparison_all.pdf -share_y both -plot_boundaries -signal-on-x).
TCGA data analysis
The TCGA bladder urothelial carcinoma (BLCA), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), esophageal carcinoma (ESCA), brain lower grade glioma (LGG), hepatocellular carcinoma (LIHC), thymoma (THYM) data were used to evaluate the correlation of gene expression and patient survival. The statistical analysis was performed by GEPIA.
Results
The chromatin accessibility landscape of TITRs is distinct from those of PBTRs
Treg cells heavily infiltrate tumors and are highly immunosuppressive in the TME [9, 28, 29]. Several studies have found that TITRs are phenotypically distinct from Treg cells in peripheral blood [17–20], suggesting that TME components have a significant impact on the phenotype and function of TITRs.
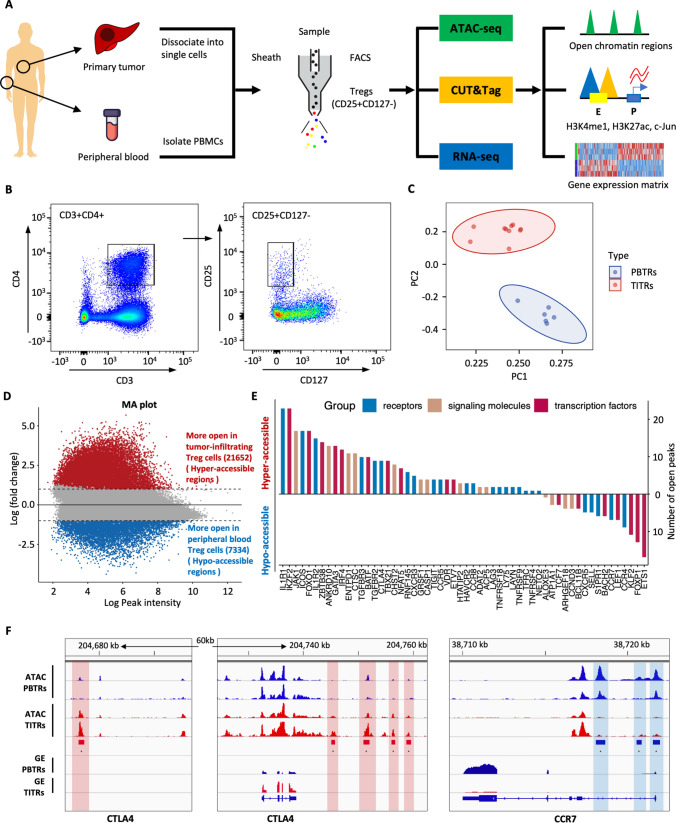
To elucidate the dynamic and precise mechanisms of transcriptional regulation of TITRs, we investigated the chromatin accessibility landscape and gene expression profiling of Treg cells using ATAC-seq and RNA-seq. The overall scheme of this study is presented in Fig. 1A. TITRs and PBTRs were sorted by flow cytometry (Fig. 1B). Cells were stained with anti-FOXP3 and the purity of cells was higher than 95% (Fig. S1A). The insert size distribution has obvious periodicity, as expected (Fig. S1B). A high enrichment of read counts around TSS regions showed that the datasets satisfy quality control (Fig. S1C). Pearson correlation analysis demonstrated that our datasets were highly reproducible between biological replicates (Fig. S1D). RNA-seq profiling was used for determining the differentially expressed genes in TITRs compared to PBTRs (Fig. S2, Supplementary Table 1).
Fig. 1.
Distinct chromatin accessibility landscape of tumor-infiltrating Treg cells (TITRs) and peripheral blood Treg cells (PBTRs). A Schematic representation of the overall experimental study design. ATAC-seq, CUT&Tag and RNA-seq were applied to Treg cells with biological replicates purified from tumor tissues and peripheral blood, and the sequence data set was used for downstream bioinformatical analyses. B Representative flow cytometry plots showed the sorting strategy of Treg cells based on CD25 and CD127 expression from tumor tissues and peripheral blood. C Principal component analysis (PCA) plot showed that TITRs were clearly differentiated from PBTRs based on normalized read counts of chromatin accessibility. TITRs are indicated in red, and PBTRs are indicated in blue. D MA plot shows the differential chromatin accessibility between TITRs and PBTRs. The red (blue) dots represent the significantly accessible peaks between the groups (absolute log2 fold change ≥ 1, adjusted p value < 0.05). E The number of open peaks associated with genes encoding receptors, signaling molecules and transcription factors in TITRs and PBTRs. F Genome browser snapshot of ATAC signals and gene expression (GE) levels at the CTLA4 and CCR7 gene locus in TITRs and PBTRs. Red rectangles and vertical shaded boxes (red) indicate hyper-accessible chromatin peaks in TITRs. Blue rectangles and vertical shaded boxes (blue) indicate hypo-accessible chromatin peaks
Principal component analysis (PCA) on chromatin accessibility demonstrated that TITRs clustered together and were separated from PBTRs (Figs. 1C, S3A), indicating distinct differences existed in Treg cells between tumor and peripheral blood. To study chromatin opening and closing in TITRs and PBTRs, we performed differential chromatin accessibility analysis between these two groups. In TITRs, 21652 peaks were identified with increased accessibility (defined as “hyper-accessible” regions) and 7334 peaks with decreased accessibility (defined as “hypo-accessible” regions) (Fig. 1D). Genes with differential accessible chromatin peaks that have been previously involved in regulation of Treg cell suppression and function are listed in Fig. 1E and Supplementary Table 2. These genes included signaling molecules, surface receptors and transcription factors. For example, CTLA-4, a key regulator for controlling suppressive function of Treg cells [30], has multiple hyper-accessible peaks at the distal-intergenic regions in TITRs. RNA-seq data also found the upregulated expression of CTLA4 in TITRs, which is consistent with previous reports [13, 31]. Treg cells expressing homing receptors can migrate to a range of tissues and inflammation sites. Several chemokine receptors, including CCR1, CCR5, CCR8 and CXCR3, were highly expressed by TITRs, and emerged in the more open chromatin sites. However, CCR7, a chemokine receptor associated with early resting or quiescence of T cells [32], has three hypo-accessible peaks at the promoter and intronic regions, and one hypo-accessible peak at the distal-intergenic region (Fig. 1F). These findings indicated that TITRs not only have specific gene expression patterns, as shown in previous studies [17–19], but also have distinct chromatin accessibility features compared to PBTRs, which motivated us to elucidate what the intrinsic mechanism of the open chromatin signatures might be.
Positive correlation between the changes of chromatin accessibility and the expression of genes
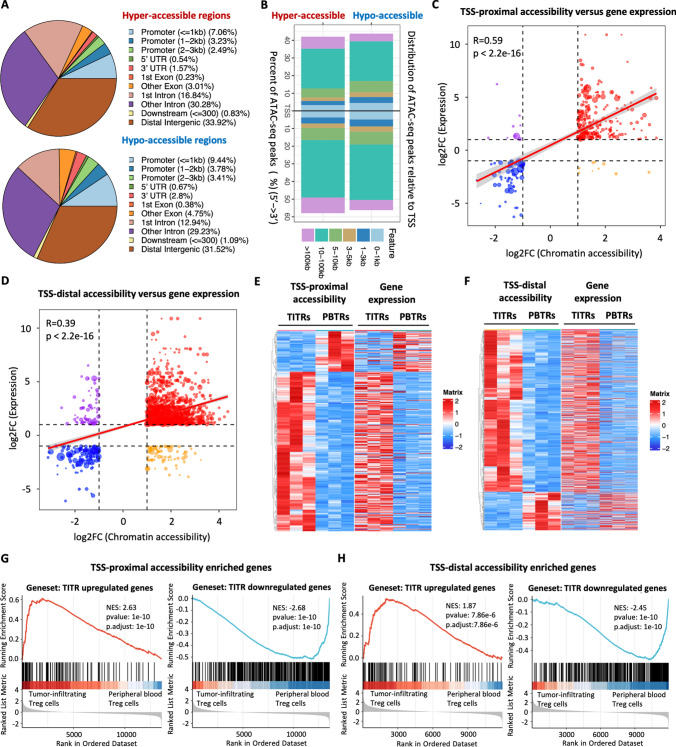
To understand the potential regulatory function that may influence the gene expression of TITRs, we segmented the genome into promoters, introns, untranslated regions (UTRs), and distal intergenic (> 10 kb from known TSSs) features based on GENCODE annotation. Functional annotation analysis revealed that the differential accessible chromatin regions were mainly distributed in gene distal elements (Fig. 2A). Among all samples, the great majority of regions were distributed in intron or intergenic regions (Fig. S3B). Next, we analyzed the locations of differential accessible chromatin regions relative to annotated genes in the human genome. Approximately 24% of hypo-accessible chromatin regions localize within 5 kilobases (kb) of a TSS, while more than 65% of hypo-accessible chromatin regions localized 5–100 kb from the TSS (Fig. 2B). Approximately 18% of hyper-accessible chromatin regions localize within 5 kb of a TSS, while the remaining 80% of hyper-accessible chromatin regions localized 5–100 kb from the TSS, with some even further than 100 kb (Fig. 2B).
Fig. 2.
Positive concordance between changes of chromatin accessibility occurring in gene distal regulatory regions and gene expression. A The pie charts show the percentage of hyper-accessible and hypo-accessible chromatin regions with respect to different genomic features. B Distance distribution of hyper-accessible and hypo-accessible chromatin regions relative to transcription start site (TSS). C Positive correlation of differential chromatin accessibility peaks at TSS-proximal regions with differential expression genes. The horizontal and vertical axis represent the log2 foldchange values derived from differential accessibility and differential gene expression analysis, respectively. Correlation was calculated using the Pearson correlation coefficient analysis. D Positive correlation of differential chromatin accessibility peaks at TSS-distal regulatory regions with differentially expressed genes. The horizontal and vertical axis represent the log2 foldchange values derived from differential accessibility and differential gene expression analysis, respectively. Correlation was calculated using the Pearson correlation coefficient analysis. E Heatmap shows significant changes of chromatin accessibility at TSS-proximal regions and expression of corresponding genes. Rows represent significantly different ATAC-seq peaks and the corresponding genes, respectively. Columns represent each sample. F Heatmap shows significant changes of chromatin accessibility at TSS-distal regions and expression of the nearest gene. Rows represent significantly different ATAC-seq peaks and nearest genes, respectively. Column represents each sample. G GSEA plot shows enrichment levels of indicated gene sets in TITRs versus PBTRs. Genes associated with the hyper-accessible peaks at TSS-proximal regions are ranked at the left side and genes associated with the hypo-accessible peaks at TSS-proximal regions are ranked at the right side. Left panel: enrichment plots generated by GSEA using published TITR upregulated genes (identified by Magnuson et al. [19]). Right panel: enrichment plots generated by GSEA using TITRs downregulated gene sets identified from this study. H GSEA plots show enrichment levels of indicated gene sets in TITRs versus PBTRs. Genes associated with the hyper-accessible peaks at TSS-distal regulatory regions are ranked at the left side and genes associated with the hypo-accessible peaks at TSS-distal regulatory regions are ranked at the right side. Left panel: enrichment plots generated by GSEA using published TITR upregulated genes (identified by Magnuson et al. [19]). Right panel: enrichment plots generated by GSEA using TITRs downregulated gene sets identified from this study
We postulated that the differential chromatin accessibility we observed might mark a combined activation of promoter and distal regulatory elements at specific developmental stages to drive target gene expression. We then divided the differential chromatin accessibility peaks into TSS-proximal (± 5 kb) and TSS-distal (> 5 kb) peaks based on distance to the nearest TSS for further analyses. We found that fold changes of gene expression were significantly positively correlated with changes of chromatin accessibility at both TSS-proximal peaks (Pearson’s correlation r = 0.59, p < 2.2e−16) (Fig. 2C) and TSS-distal peaks (Pearson’s correlation r = 0.39, p < 2.2e−16) (Fig. 2D), although there was a subset of peaks with discordant changes. The possible reason for the weak correlation between TSS-distal peaks and gene expression might lie in the fact that the association of distal regulatory regions to target genes is more vulnerable to multiple factors, such as chromatin loops and topologically associating domains. We also observed that normalized read counts of chromatin accessibility at TSS-proximal and TSS-distal regions were consistent with the transcriptional levels of proximal target genes (Fig. 2E, F). We found that genes with higher chromatin accessibility levels tended to show higher expression (Figure S3C). Further pathway analyses indicated that genes associated with TSS-proximal regions are involved in the cytokine-cytokine receptor interaction pathway, whereas genes associated with TSS-distal regions are involved in the T cell receptor signaling pathway (Figure S3D).
For hyper-accessible chromatin regions, gene set enrichment plots showed that upregulated genes of TITRs across species and tumor types [19] were significantly enriched among genes adjacent to the TSS-proximal and TSS-distal regions in TITRs compared to the PBTRs (Fig. 2G, H). For hypo-accessible chromatin regions, genes downregulated in TITRs identified by RNA-seq analysis were significantly enriched among genes adjacent to the TSS-proximal and TSS-distal peaks in PBTRs compared to the TITRs (Fig. 2G, H). Gene ontology (GO) analysis showed that the genes adjacent to hyper-accessible chromatin regions were highly enriched in those involved in the activation of immune responses, leukocyte differentiation, cell migration, and immune response-regulating signaling pathways, whereas terms related to DNA repair and mRNA processing were found in the hypo-accessible chromatin region-associated genes (Figure S3D).
Overall, the results indicate that extensive chromatin remodeling occurs mainly in intronic and distal-intergenic regions and that changes of gene expression are highly associated with chromatin remodeling in TITRs.
De novo enhancer activation and poised enhancer activation in TITRs
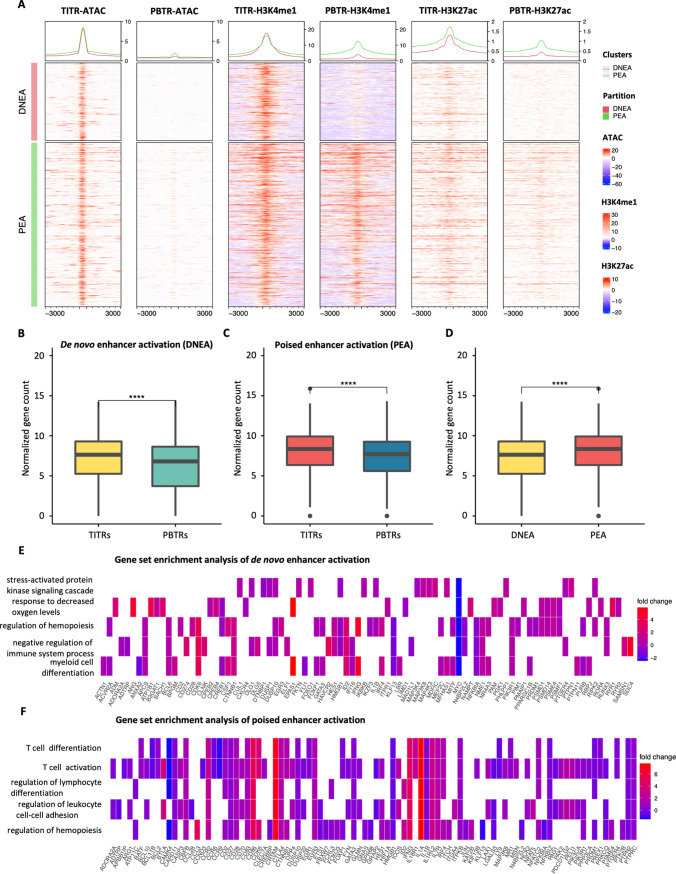
Our ATAC-seq data analysis found that a large portion of hyper-accessible regions were largely located in introns and distal intergenic regions (Fig. 2A), which often contains enhancer elements. To comprehensively evaluate alteration of chromatin modifications and identify putative enhancers in TITRs, we performed CUT&Tag using antibodies to H3K4me1 and H3K27ac in the sorted Treg cells. Following integration of ATAC-seq and CUT&Tag data, we found that a portion of the hyper-accessible chromatin regions (3259 peaks) that displayed de novo establishment of open chromatin also showed de novo establishment of H3K4me1 deposition in TITRs. We hereafter refer to these regions as “de novo enhancer activation” (DNEA) regions (Fig. 3A). Interestingly, the remaining large portion of hyper-accessible chromatin regions (6885 peaks) have high H3K4me1 deposition signal in both TITRs and PBTRs, and thus were designated as “poised enhancer activation” (PEA) regions (Fig. 3A). We also found counterparts in the hypo-accessible chromatin regions, defined as “enhancer deactivation” (EDE) and “poised enhancer deactivation” (PEDE) regions (Fig. S4). There are some shared epigenetic features between TITRs and PBTRs, which we defined as “typical enhancers” (TEs) and “poised enhancers” (PEs) (Fig. S5).
Fig. 3.
De novo enhancer activation and poised enhancer activation in TITRs. A Categorical heatmaps show activation of de novo enhancers and poised enhancers. The enhancers were defined by H3K4me1 peaks over 3 kb distant from TSSs. The first two heatmaps show the normalized read counts of ATAC-seq at each peak (± 3 kb from the peaks) in TITRs and PBTRs samples. The middle two heatmaps show the normalized read counts of H3K4me1 signal at each peak (± 3 kb from the peaks) in TITRs and PBTRs samples. The last two heatmaps show the normalized read counts of H3K27ac signal at each peak (± 3 kb from the peaks) in TITRs and PBTRs samples. H3K4me1 peaks are divided into equal windows. Metagene plot showing the average reads density of chromatin accessibility, with H3K4me1 and H3K27ac modification signals at each peak (± 3 kb from the peaks), respectively. B The average gene expression levels of de novo enhancer activation regions adjacent genes in TITRs were significantly higher than PBTRs. Y‐axis represents the normalized read count. Significance was checked by t test: ****, p value < 0.0001. C The average gene expression levels of poised enhancer activation regions adjacent genes in TITRs were significantly higher than PBTRs. Y‐axis represents the normalized read count. Significance was checked by t test: ****, p value < 0.0001. D The average gene expression levels of poised enhancer activation regions adjacent genes were significantly higher than the average gene expression levels of de novo enhancer activation nearest genes in TITRs. Y‐axis represents the normalized read count. Significance was checked by t test: ****, p value < 0.0001. E Heatmap plot displays the significantly enriched biological process of de novo enhancer activation regions adjacent genes and representative genes are displayed below. F Heatmap plot displays the significantly enriched biological process of poised enhancer activation regions adjacent genes and representative genes are displayed below
Previous studies showed that enhancers tend to interact with adjacent genes through a loop mechanism to activate gene transcription [33]. We found that the average expression level of genes affected by the DNEA and PEA regions in TITRs is significantly higher than genes unaffected by the DNEA or PEA regions in PBTRs (Fig. 3B, C), indicating that formation of an active enhancer has a positive effect on targeted gene expression. Furthermore, we found that the proximal genes of PEA regions show a higher average expression level (Fig. 3D). GO analysis of DNEA-adjacent genes revealed significantly enriched terms, such as negative regulation of the immune system, and included such known genes as HAVCR2 (Fig. 3E, S6A). HAVCR2 has been reported negatively regulate the immune responses against viral infection. Recent studies have shown that HAVCR2 is also expressed by Treg cells and TIM3 + Treg cells are the predominant Treg cell population in tumors. The increased chromatin accessibility and de novo accumulation of H3K4me1 and H3K27ac marks was observed at the intronic regions of HAVCR2 (also known as TIM3) in TITRs (Figure S6B). GO analysis of PEA-adjacent genes revealed significantly enriched biological process terms, such as T cell differentiation, T cell activation and cytokine receptor activity and included such known genes as BATF, BCL11B, CBFB and IRF4 (Fig. 3F). For example, the active enhancer signature of BATF, instructing the suppressive activity of TITRs, is shown by the induction of open chromatin peaks, and enriched H3K4me1 and H3K27ac modifications at the intronic regions (Figure S6C), suggesting that BATF uses the putative enhancer to enhance gene expression in TITRs.
AP-1 family motifs are enriched at the enhancer regions of TITRs
The chromatin accessibility landscape resulting from the binding of transcription factors (TFs) in combination with histone modifications acting on cis-regulatory elements coordinately regulate gene expression [34]. Therefore, to identify potential transcription factors involved in driving these dramatic differences in accessibility, we implemented enrichment analysis of TF binding motif using Homer suite within the differentially accessible chromatin regions. Motif analysis showed that some motifs were highly enriched in hyper-accessible chromatin regions, such as Fra1, JunB, Fos, and BATF motifs (Fig. 4A and Supplementary Document 1), and the majority of these transcription factors belong to AP-1 family [35]. In hypo-accessible chromatin regions, some motifs, such as ETV1, ERG, ETS1, and Etv2, were more enriched in PBTRs (Fig. S7A and Supplementary Document 2).
Fig. 4.
AP-1 family binding motifs are enriched in the enhancer regions of TITRs. A Heatmap shows the percentage of motifs enriched in hyper-accessible chromatin regions compared to the background sites harboring the same motifs. Top 40 overrepresented motifs were selected with a p value < 10–100. B Volcano plot displays differential TF binding activity estimated from TOBIAS between TITRs and PBTRs. Each dot represents one motif obtained from JASPAR database. Tumor-infiltrating Treg cell-specific TFs are highlighted in red and peripheral blood Treg cell-specific TFs are highlighted in blue. AP-1 family members are selected and presented. Clustering tree represents TFs with more than 50% overlap. Complete TFs activity are listed in Supplementary Table 2. C Accumulated footprint plot of JUN. The accumulated plots are centered at the total possible JUN binding sites between TITRs and PBTRs. D Heatmap shows genome-wide differential JUN footprint activity and differential corresponding gene expression between TITRs and PBTRs. The column of binding prediction represents whether the site will be bound/unbound by JUN in the TITRs. E JUN transcription factor regulatory network. Nodes indicate gene colored based on differential gene expression between TITRs and PBTRs. JUN node is colored based on footprint fold change between TITRs and PBTRs. Node sizes represent network degree
Recently, computational analysis of footprints has emerged as a more effective and accurate technique to infer multiple TF occupancy genome-wide compared with chromatin immunoprecipitation sequencing (ChIP-seq) [36, 37]. Hence, the differential TF binding plots were generated (Fig. 4B and Supplementary Document 3) by comparing the TOBIAS-based footprinting scores between TITRs and PBTRs. The differential TOBIAS scores of each TF footprint are shown in Supplementary Table 3. FOXP3 creates footprints both in TITRs and PBTRs, as expected (Figure S8A). We observed a group of TFs with significant activity shifts (Fig. 4B), all belonging to the AP-1 family [35]. We found that this group of motifs from the AP-1 family cluster together in Treg cells (Fig. 4B). Consistent with the gene expression, footprint score of JUN was much higher in TITRs compared with PBTRs (Fig. 4C), suggesting that JUN has higher binding activity in TITRs. There was a significantly positive correlation between the gain of JUN binding and the targeted gene expression (Fig. 4D). By integrating the TF footprint scores and gene expression, we constructed a cross-regulatory network around JUN in TITRs (Fig. 4E).
Several TFs known to be involved in effector Treg cell differentiation and gene expression regulation were enriched in TITRs (Figs. S8A, S9A). We also observed a shift of ETS1, Fli1, ERG, ETV2 and other TF activity in PBTRs (Figs. S7B, S9B). ETS-1 controls the development of natural regulatory T cells [38], which has been found to bind the FOXP3 enhancer region CNS2 [39]. An aggregated footprint of ETS-1 was clearly visible in PBTRs (Fig. S7C), which supports the previous notion[38]. Overall, the results described above suggest that AP-1 family members have important functions for the maintenance of open chromatin in TITRs.
c-Jun binds at regulatory regions to regulate signature genes of TITRs
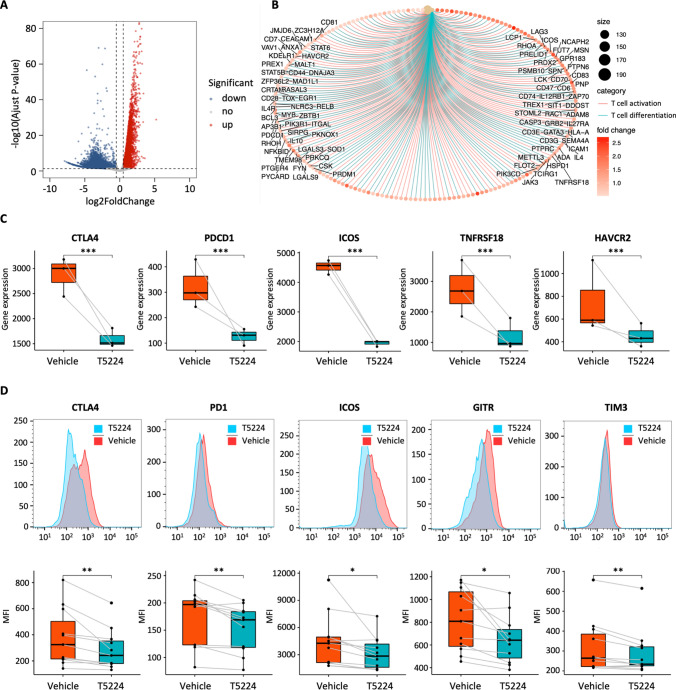
To further validate the predicted transcription factors in regulating specific gene expression of Treg cells in the TME, we performed CUT&Tag assay for c-Jun on tumor-infiltrating Treg cells. Our CUT&Tag analysis identified approximately 18,000 peaks significantly bound by c-Jun (Fig. 5A). Interestingly, functional annotation analysis revealed that the nearly half of c-Jun bound peaks were located in promoter regions, which suggest that c-Jun mediates interactions between promoters and enhancers in close proximity (Fig. 5A). We then sought to identify enriched DNA sequence motifs within the c-Jun bound peaks and found that Jun-AP1 binding motif was significantly enriched (Fig. 5B), which further confirmed the quality and specificity of our c-Jun CUT&Tag experiment.
Fig. 5.
Identification of c-Jun directly target genes in TITRs. A The pie chart shows the percentage of c-Jun binding cites in TITRs with respect to different genomic signatures. B Motif enrichment analysis was performed for c-Jun binding sites in TITRs. C The overlap between c-Jun target genes and genes defined by differential ATAC-seq peaks. D GO analysis shows the biological processes associated with overlapped genes. E List of c-Jun target genes encoding receptors, signaling molecules and transcription factors in TITRs. F Categorical heatmaps show chromatin accessibility, H3K4me1, H3K27ac modification and c-Jun CUT&Tag signals at DENA and PEA regions. Metagene plot showing the average reads density of chromatin accessibility, with H3K4me1 and H3K27ac modification and c-Jun CUT&Tag signals at each peak (± 3 kb from the peaks), respectively. G Genome browser snapshot of ATAC signals and gene expression (GE) levels at the TNFRSF18, HAVCR2 and CCR8 gene locus in TITRs and PBTRs. Vertical shaded boxes (red) indicate c-Jun binding sites in TITRs
To better understand the role of c-Jun in regulating epigenetic changes underlying alterations in gene expression of TITRs, we analyzed the colocalization of c-Jun binding sites with the differential accessible chromatin regions identified by ATAC-seq. We observed 5801 c-Jun target genes overlap with genes defined by differential ATAC-seq peaks (Fig. 5C), which indicated that these genes were not only directly bound by c-Jun, but also were differential regulated on the epigenetic level. We found that functional enrichment of these common genes was highly enriched in the T cell activation and T cell differentiation (Fig. 5D), whereas enrichment terms related to ribonucleoprotein complex biogenesis and RNA catabolic process were found in the c-Jun specific target genes. Similarly, c-Jun target genes with differential ATAC-seq peaks previously involved in regulation of Treg cell suppression and function are listed in Fig. 5E and Supplementary Table 4, and included receptors, signaling molecules and transcription factors.
Next, to gain additional molecular mechanism of how c-Jun regulates the transcription, we determined whether the c-Jun might be involved in the regulation of enhancer activation of TITRs. We found both “poised enhancers activation” and “de novo enhancers activation” regions have a clear enrichment of c-Jun occupancy in TITRs (Fig. 5F). For example, the HAVCR2 (Tim3) gene locus had two c-Jun binding sites (Fig. 5G). One of them bound to the promoter region of HAVCR2 locus. Additional binding site became accessible only in TITRs, suggesting that c-Jun promote differential expression of HAVCR2 on TITRs by acting on this enhancer. These data indicate that c-Jun directly binds at enhancer regions which chromatin has been opened and can drive activated gene expression in TITRs.
Inhibition of AP-1 interfered with Treg cells activation in vitro
To assess whether AP-1 family members can be induced from stimulated Treg cells in vitro, we reanalyzed the GSE138603 dataset from the NCBI GEO public database. This data set includes the transcriptomes of expanded human Treg cells that were either unstimulated or treated with anti-CD3/anti-CD28 for 24h. Transcriptome analysis revealed in vitro stimulated Treg cells exhibit an activated phenotype with high expression of Treg signature genes compared to the unstimulated Treg cells (data not shown). We found that several AP-1 family members including c-Jun were significantly elevated in Treg cells following stimulation. Several co-inhibitory and co-stimulatory molecules were also included.
To interrogate the requirement of AP-1 expression to promote the activation of PBTRs, we utilized the chemical inhibitor T5224, which specifically inhibited the DNA binding activity of AP-1 [40]. CD25 + CD127− Treg cells from peripheral blood were sorted and stimulated with anti-CD3/anti-CD28 in the presence of T5224 or vehicle (DMSO) for 24h. Transcriptome profiling assays on treated and untreated Treg cells were performed. Our transcriptome sequencing data demonstrated considerable changes took place in gene expression after T5224 treatment (Fig. 6A). A total of 5773 genes were significantly upregulated and 9171 downregulated in response to treatment with vehicle for 24h, compared with T5224 treated Treg cells. GO analysis revealed enrichment in genes related to T cell activation and differentiation (Fig. 6B). Interestingly, our analysis revealed that core Treg cells signature genes such as CTLA4, PDCD1 (PD1) and several immune checkpoints including TNFRSF18 (GITR), TNFRSF4(OX-40), ICOS were significantly reduced in T5224-treated Treg cells (Fig. 6C), compared to vehicle-treated Treg cells. Among them, HAVCR2 and PD-L1 were selected for qPCR, and the results were consistent with transcriptome sequencing (Figure S10A). At the protein level, inhibiting AP-1 caused downregulation of immune checkpoints such as CTLA4, ICOS, and GITR (Fig. 6D). To further clarify whether Treg function was impaired after T5224 treatment, we performed a CFSE-based suppression assay of Treg cells. We found that T5224-treated Treg cells displayed attenuated suppressive functions, compared to vehicle-treated Treg cells (Fig. S10B). These data suggested that AP-1 is required for activation of Treg cells in vitro and plays an essential role in regulating activated genes in TITRs.
Fig. 6.
AP-1 is required for Treg cells activation in vitro. A Differential gene expression analysis between vehicle and T5224 treated Treg cells. Significantly upregulated genes of vehicle treated cells are labeled in red, and significantly downregulated genes of T5224 treated cells are labeled in blue. B GO analysis of upregulated genes in vehicle treated cells revealed a significant enrichment of T cell activation related genes. C Boxplots show fold change of gene expression between vehicle and T5224 treated Treg cells. Significance was determined using DESeq2 analysis. ***, p value < 0.001. D Mean fluorescence intensity plots (MFI; top) and histogram plots (bottom) of selected proteins gated on CD25 + FOXP3 + Treg cells. Significance was checked by paired t test. *, p value < 0.05, **, p value < 0.01
c-Jun expression was associated with worse patient outcome in multiple cancers
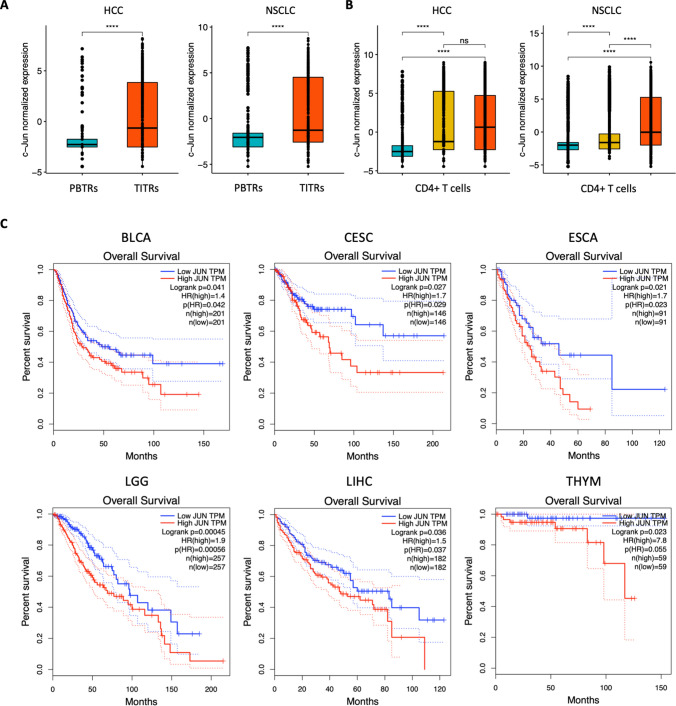
Next, we reanalyzed published single-cell datasets of tumor-infiltrating lymphocytes from HCC and NSCLC patients. We observed that TITRs exhibited significantly higher level of c-Jun than PBTRs isolated from peripheral blood, tumor, and adjacent normal tissues from HCC and NSCLC patients. (Fig. 7A). Higher expression of c-Jun was also detected in CD4+ T cells isolated from tumors, compared to peripheral blood and normal tissues (Fig. 7B), which is consistent with the findings that tumors contained a higher ratio of effector Treg cells. To investigate the potential clinical relevance of our findings, we asked whether the expression of c-Jun correlate with the clinical outcome of cancer patients. We then employed a web tool GEPIA (Gene Expression Profiling Interactive Analysis) for TCGA survival analysis. We found that high expression of c-Jun is clinically associated with poor prognosis in a variety of cancers including HCC (Fig. 7C).
Fig. 7.
c-Jun expression was associated with worse patient outcome in multiple cancers. A Box plots show the normalized gene expression of c-Jun in PBTRs and TITRs isolated from peripheral blood and tumors of HCC and NSCLC patients. (peripheral blood, cyan; tumors, red). Significance was checked by t test: ****, p value < 0.0001. B Box plots show the normalized gene expression of c-Jun in CD4+ T cells isolated from peripheral blood, normal tissues and tumors of HCC and NSCLC patients. (peripheral blood, cyan; normal tissues, yellow; tumors, red). Significance determined by t test: ****, p value < 0.0001. C Overall survival of patients with low and high expression of c-Jun in multiple cancers from TCGA dataset.
Discussion
As discussed in the introduction, the TME is highly immunosuppressive via a range of mechanisms, which may impair tumor cells clearance. It has been recognized that human tumors often harbor large numbers of CD4+CD25+ Treg cells with highly activated effector features and superior suppressive functions. Therefore, it is valuable to comprehensively profile the epigenome, which could extensively elucidate the mechanism of gene regulation of TITRs and offer potentially therapeutic strategies targeting TITRs.
We compared the chromatin accessibility landscape of TITRs and PBTRs from the same individual using ATAC-seq in a genome-wide scale. TITRs and PBTRs were derived from the same individual, thus precluding the effect of genetic variation, and provided a perfect model to account for the gene regulation mechanism. Howard Y. Chang et al. have applied single-cell ATAC-seq to predict cell type-specific enhancers and transcription factors of human intratumoral T cells [41]. However, they mainly focused on the regulatory trajectory of exhausted CD8+ T cells, rather than on Treg cells. Recently, CCR4+ Treg cells with expression of PD-1 and TCF1 exhibited stem-like properties as have been reported in HCC. ATAC-seq data showed plentiful chromatin remodeling between CCR4+ and CCR4− Treg cells, and revealed increased accessibility at the promoters of CCR4 in CCR4+ Treg cells [42].
We report a comprehensive analysis of changes in chromatin accessibility in Treg cells and observe more open chromatin regions in the TITRs than those in PBTRs, which is consistent with the transcriptomic findings that there are more significantly up-expressed genes in TITRs. Significantly, we found that fold changes of gene expression were positively correlated with changes of chromatin accessibility. GO analysis showed that the hyper-accessible chromatin regions adjacent genes were related to T cell differentiation process. Many differentially expressed TFs involved in regulating differentiated Treg cells, including BATF, VDR, IRF4, and PRDM1, have elevated chromatin accessibility and corresponding gene expression. These findings suggest that changes of chromatin accessibility could explain the inherent reason for the differential gene expression between TITRs and PBTRs.
The cytokine milieu and specific TFs polarizing CD4+ T helper cell differentiation can drive the functional specialization of Treg cells in parallel to suppressing the corresponding T helper cell responses in inflammation [43]. We also observed that the elevated expression of the Th1 core factor T-bet, CXCR3 and Th2 master factor GATA3 is accompanied by a greater degree of chromatin accessibility, as opposed to the Th17 TF RORγt (RORC) and Th17-associated chemokine receptor CCR6. However, a few exceptions have been observed. For example, CCR4, known to be mainly expressed on effector Treg cells in tumor tissues, was associated with hypo-accessible peaks in PBTRs. The causes of this discrepancy may lie in additional regulatory mechanisms, such as DNA methylation, involved in its transcription. Single-cell RNA sequencing of infiltrating lymphocytes from HCC revealed CCR4 was expressed at lower levels on tumor-infiltrating Treg cells compared with peripheral blood Treg cells. These results suggest that Treg cells comprise a complex and heterogeneous population with diverse functions to regulate the mixed immune response in the TME.
Secondly, we provided comprehensive putative enhancer profiles using H3K4me1 and H3K27ac modifications in the TITRs and PBTRs and found massive remodeling of the enhancer landscape between TITRs and PBTRs. Mijnheer et.al found that super-enhancers of TBX21 are increased in synovial fluid-derived Treg cells in juvenile idiopathic arthritis patients. Moreover, they found VDR as a novel marker related to effector Treg cell differentiation [44]. We also found enrichment of these TFs in our tumor-derived Treg cells, which may be the reason for the similar gene expression profile of Treg cells from SF and the TME [44].
In mammalian cells, active and poised enhancers are commonly marked by H3K4me1 in a cell-type-specific manner [45, 46]. Our previous studies demonstrated that the poised chromatin state can predict the gene transcription potential [47, 48]. It has long been known that poised chromatin is an important element in gene regulation during the development of the immune system [49, 50]. Given that not all differentially upregulated genes conform to the same regulatory patterns by enhancers, we hypothesized that the transcription of induced genes may be separately regulated by diverse enhancer activation. In fact, we found a large portion of hyper-accessible chromatin regions have a high H3K4me1 deposition signal in both TITRs and PBTRs, and were designated as PEA regions in TITRs. Furthermore, the poised enhancer activation was accompanied with the upregulation of adjacent genes associated with T cell activation and T cell differentiation processes in TITRs. Comparative analysis of TCR repertoires of Treg and conventional T cells revealed that the most dominant Treg cells in metastatic melanoma and gastrointestinal tumors were recruited from the peripheral blood, after which their phenotypic signatures were influenced by the local TME [18, 51]. These findings reconfirmed our speculation that these H3K4me1-predetermined enhancers give the TITRs a set of transcriptional options to deploy rapidly in response to TME stimuli. In addition to PENA, DENA regions may have played a significant role in regulating gene expression of TITRs. Because these DENA regions that displayed de novo establishment of open chromatin and H3K4me1 deposition in TITRs occurred only in tumor microenvironment, but not in peripheral environment. We speculate that DENA may not only require the prolonged exposure of TITRs to tumor antigens, but also may require signals from cytokines and other signaling molecules present in the TME. Therefore, DENA regions are more specific regulatory elements for controlling gene expression of TITRs with more immunosuppressive activity.
Lastly, we predict a series of TFs that might be involved in establishing tumor-infiltrating Treg cell-specific gene expression programs using an unbiased method called digital genomic footprinting [37, 52]. Interestingly, enrichment of transcription factor motifs showed distinct patterns between hyper-accessible and hypo-accessible chromatin regions. We observed a shift of AP-1 family member activity in TITRs. AP-1 participated in a broad range of biological processes, including cellular survival, proliferation and differentiation [35]. AP-1 facilitates enhancer remodeling of precursor-activated T cells, and is essential for Treg cell differentiation at the stage of thymocyte maturation [53]. AP-1 establishes the open chromatin at the effector genomic loci during the early stages of primary naive CD4+ T cell activation in vitro [54]. These findings led us to postulate that AP-1 family might have a positive role in tumor-infiltrating Treg cells differentiation process.
CUT&Tag assay of c-Jun performed on TITRs and integrated multi-omics data suggest that c-Jun binds at regulatory regions of genes associated T cell activation and differentiation process in tumor-infiltrating Treg cells. The TF footprint analysis results provide a rich resource for future exploration of the regulatory network in TITRs. Identification of the upstream transcriptional regulators of c-Jun is helpful to construct the gene regulatory network. We found that the motif of RELB and NR1H2 existed in the c-Jun promoter region, and the gene expression level of RELB and NR1H2 were higher in the TITRs. Therefore, we predicted that RELB and NR1H2 might are responsible for c-Jun expression in TITRs, adding a layer of information on the transcriptional hierarchy governing TITRs gene activation.
To further demonstrate the positive function of AP-1 in the activation of Treg cells, we performed in vitro functional experiments. We took advantage of small molecule AP-1 inhibitor to study the effect on the gene transcriptional and protein levels of Treg cells. Interestingly, the most striking impact of AP-1 inhibition was the marked downregulation of several immune checkpoint molecules (e.g., CTLA4, TNFRSF18, HAVCR2), which involved in controlling T cell activation. Additionally, despite significant decreases in Helios and GATA3 transcript level, no significant decreases in IKZF2 and GATA3 protein were detected in T5224-treated Treg cells. Results from CUT&Tag and in vitro assay provide further support that the AP-1 has a positive role for the activation of TITRs.
In conclusion, the epigenetic landscape of TITRs is clearly distinct from that of PBTRs, including chromatin accessibility and enhancer landscapes. Transcriptional changes of TITRs are reflected at the epigenetic level via global regulation of the activation of enhancers; in particular, AP-1 is necessary for the activation of TITRs. Our findings could help in better understanding of the mechanism of specific gene expression patterns of TITRs and facilitate the development of novel and potential therapeutic strategies targeting TITRs in liver cancer treatment.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Dr. Jianming Zeng (University of Macau), and all the members of his bioinformatics team, biotrainee, for generously sharing their experience and codes.
Author contributions
BZ designed and finished the main experiments, performed the bioinformatics analyses, analyzed the data, and wrote the manuscript. QZ coordinated clinical contributions, collected clinical samples, and discussed the results. TX analyzed the data, assisted with figure presentation, and wrote the manuscript. YW assisted with figure presentation and flow cytometry cell sorting. DZ and ZC designed and supervised the project, interpreted the results and edited the manuscript. BG designed and supervised the project, analyzed the data, and wrote the manuscript.
Funding
This work was supported by grants to B.G. from the National Natural Science Foundation of China (31870875); to Z.C. from the National Natural Science Foundation of China (82171745); and to D.Z. from the National Natural Science Foundation of China (82071781 and 82271782). This work was also supported by Grants to Q.Z. from the Natural Science Foundation of Guangdong Province (2021A1515011015) and to Y.W. from the Basic Research Project of Baoan District (2020JD471).
Availability of data and material
All sequence data generated in this study are deposited in Sequence Read Archive (SRA) under NCBI database: PRJNA824422.
Declarations
Conflict of interest
The authors declare no competing interests.
Ethics approval and consent to participate
All clinical samples and information were collected with informed consent of the patients. The study was approved by the Medical Ethical Committee of Nanfang Hospital of Southern Medical University.
Consent for publication
All authors agree for publication.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Baowen Zhuo, Qifan Zhang and Tingyan Xie: contributed equally.
Bo Guo: Lead Contact.
Contributor Information
Zhengliang Chen, Email: zhlchen@smu.edu.cn.
Daming Zuo, Email: zdaming@smu.edu.cn.
Bo Guo, Email: guobomail@gmail.com.
References
- 1.Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391:1301–1314. doi: 10.1016/S0140-6736(18)30010-2. [DOI] [PubMed] [Google Scholar]
- 2.Feng GS, Hanley KL, Liang Y, Lin X. Improving the efficacy of liver cancer immunotherapy: the power of combined preclinical and clinical studies. Hepatology. 2021;73(Suppl 1):104–114. doi: 10.1002/hep.31479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang XH, Yamagiwa S, Ichida T, Matsuda Y, Sugahara S, Watanabe H, Sato Y, Abo T, Horwitz DA, Aoyagi Y. Increase of CD4+ CD25+ regulatory T-cells in the liver of patients with hepatocellular carcinoma. J Hepatol. 2006;45:254–262. doi: 10.1016/j.jhep.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 4.Tanaka A, Sakaguchi S. Regulatory T cells in cancer immunotherapy. Cell Res. 2017;27:109–118. doi: 10.1038/cr.2016.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ohkura N, Kitagawa Y, Sakaguchi S. Development and maintenance of regulatory T cells. Immunity. 2013;38:414–423. doi: 10.1016/j.immuni.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 6.Josefowicz SZ, Lu LF, Rudensky AY. Regulatory T cells: mechanisms of differentiation and function. Annu Rev Immunol. 2012;30:531–564. doi: 10.1146/annurev.immunol.25.022106.141623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kitagawa Y, Sakaguchi S. Molecular control of regulatory T cell development and function. Curr Opin Immunol. 2017;49:64–70. doi: 10.1016/j.coi.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 8.Vignali DA, Collison LW, Workman CJ. How regulatory T cells work. Nat Rev Immunol. 2008;8:523–532. doi: 10.1038/nri2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Togashi Y, Shitara K, Nishikawa H. Regulatory T cells in cancer immunosuppression—implications for anticancer therapy. Nat Rev Clin Oncol. 2019;16:356–371. doi: 10.1038/s41571-019-0175-7. [DOI] [PubMed] [Google Scholar]
- 10.Shimizu J, Yamazaki S, Sakaguchi S. Induction of tumor immunity by removing CD25+CD4+ T cells: a common basis between tumor immunity and autoimmunity. J Immunol. 1999;163:5211–5218. doi: 10.4049/jimmunol.163.10.5211. [DOI] [PubMed] [Google Scholar]
- 11.Onizuka S, Tawara I, Shimizu J, Sakaguchi S, Fujita T, Nakayama E. Tumor rejection by in vivo administration of anti-CD25 (interleukin-2 receptor alpha) monoclonal antibody. Cancer Res. 1999;59:3128–3133. [PubMed] [Google Scholar]
- 12.Simpson TR, Li F, Montalvo-Ortiz W, Sepulveda MA, Bergerhoff K, Arce F, Roddie C, Henry JY, Yagita H, Wolchok JD, et al. Fc-dependent depletion of tumor-infiltrating regulatory T cells co-defines the efficacy of anti-CTLA-4 therapy against melanoma. J Exp Med. 2013;210:1695–1710. doi: 10.1084/jem.20130579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arce Vargas F, Furness AJS, Litchfield K, Joshi K, Rosenthal R, Ghorani E, Solomon I, Lesko MH, Ruef N, Roddie C, et al. Fc effector function contributes to the activity of human anti-CTLA-4 antibodies. Cancer Cell. 2018;33:649–663e644. doi: 10.1016/j.ccell.2018.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ha D, Tanaka A, Kibayashi T, Tanemura A, Sugiyama D, Wing JB, Lim EL, Teng KWW, Adeegbe D, Newell EW, et al. Differential control of human Treg and effector T cells in tumor immunity by Fc-engineered anti-CTLA-4 antibody. Proc Natl Acad Sci U S A. 2019;116:609–618. doi: 10.1073/pnas.1812186116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Azizi E, Carr AJ, Plitas G, Cornish AE, Konopacki C, Prabhakaran S, Nainys J, Wu K, Kiseliovas V, Setty M, et al. Single-cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell. 2018;174:1293–1308e1236. doi: 10.1016/j.cell.2018.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toker A, Nguyen LT, Stone SC, Yang SYC, Katz SR, Shaw PA, Clarke BA, Ghazarian D, Al-Habeeb A, Easson A, et al. Regulatory T cells in ovarian cancer are characterized by a highly activated phenotype distinct from that in melanoma. Clin Cancer Res. 2018;24:5685–5696. doi: 10.1158/1078-0432.CCR-18-0554. [DOI] [PubMed] [Google Scholar]
- 17.De Simone M, Arrigoni A, Rossetti G, Gruarin P, Ranzani V, Politano C, Bonnal RJP, Provasi E, Sarnicola ML, Panzeri I, et al. Transcriptional landscape of human tissue lymphocytes unveils uniqueness of tumor-infiltrating T regulatory cells. Immunity. 2016;45:1135–1147. doi: 10.1016/j.immuni.2016.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Plitas G, Konopacki C, Wu K, Bos PD, Morrow M, Putintseva EV, Chudakov DM, Rudensky AY. Regulatory T cells exhibit distinct features in human breast cancer. Immunity. 2016;45:1122–1134. doi: 10.1016/j.immuni.2016.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Magnuson AM, Kiner E, Ergun A, Park JS, Asinovski N, Ortiz-Lopez A, Kilcoyne A, Paoluzzi-Tomada E, Weissleder R, Mathis D, Benoist C. Identification and validation of a tumor-infiltrating Treg transcriptional signature conserved across species and tumor types. Proc Natl Acad Sci USA. 2018;115:E10672–E10681. doi: 10.1073/pnas.1810580115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng C, Zheng L, Yoo JK, Guo H, Zhang Y, Guo X, Kang B, Hu R, Huang JY, Zhang Q, et al. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell. 2017;169:1342–1356e1316. doi: 10.1016/j.cell.2017.05.035. [DOI] [PubMed] [Google Scholar]
- 21.Guo X, Zhang Y, Zheng L, Zheng C, Song J, Zhang Q, Kang B, Liu Z, Jin L, Xing R, et al. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat Med. 2018;24:978–985. doi: 10.1038/s41591-018-0045-3. [DOI] [PubMed] [Google Scholar]
- 22.Schmidl C, Hansmann L, Lassmann T, Balwierz PJ, Kawaji H, Itoh M, Kawai J, Nagao-Sato S, Suzuki H, Andreesen R, et al. The enhancer and promoter landscape of human regulatory and conventional T-cell subpopulations. Blood. 2014;123:e68–78. doi: 10.1182/blood-2013-02-486944. [DOI] [PubMed] [Google Scholar]
- 23.Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, Cui K, Kanno Y, Roh TY, Watford WT, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–167. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohkura N, Yasumizu Y, Kitagawa Y, Tanaka A, Nakamura Y, Motooka D, Nakamura S, Okada Y, Sakaguchi S. Regulatory T cell-specific epigenomic region variants are a key determinant of susceptibility to common autoimmune diseases. Immunity. 2020;52:1119–1132e1114. doi: 10.1016/j.immuni.2020.04.006. [DOI] [PubMed] [Google Scholar]
- 25.Giles JR, Manne S, Freilich E, Oldridge DA, Baxter AE, George S, Chen Z, Huang H, Chilukuri L, Carberry M, et al. Human epigenetic and transcriptional T cell differentiation atlas for identifying functional T cell-specific enhancers. Immunity. 2022;55:557–574e557. doi: 10.1016/j.immuni.2022.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaya-Okur HS, Wu SJ, Codomo CA, Pledger ES, Bryson TD, Henikoff JG, Ahmad K, Henikoff S. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat Commun. 1930;2019:10. doi: 10.1038/s41467-019-09982-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wing JB, Tanaka A, Sakaguchi S. Human FOXP3(+) regulatory T cell heterogeneity and function in autoimmunity and cancer. Immunity. 2019;50:302–316. doi: 10.1016/j.immuni.2019.01.020. [DOI] [PubMed] [Google Scholar]
- 29.Kim HR, Park HJ, Son J, Lee JG, Chung KY, Cho NH, Shim HS, Park S, Kim G, In Yoon H, et al. Tumor microenvironment dictates regulatory T cell phenotype: Upregulated immune checkpoints reinforce suppressive function. J Immunother Cancer. 2019;7:339. doi: 10.1186/s40425-019-0785-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wing K, Onishi Y, Prieto-Martin P, Yamaguchi T, Miyara M, Fehervari Z, Nomura T, Sakaguchi S. CTLA-4 control over Foxp3+ regulatory T cell function. Science. 2008;322:271–275. doi: 10.1126/science.1160062. [DOI] [PubMed] [Google Scholar]
- 31.Alvisi G, Brummelman J, Puccio S, Mazza EM, Tomada EP, Losurdo A, Zanon V, Peano C, Colombo FS, Scarpa A, et al. IRF4 instructs effector Treg differentiation and immune suppression in human cancer. J Clin Invest. 2020;130:3137–3150. doi: 10.1172/JCI130426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations (*) Annu Rev Immunol. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gibcus JH, Dekker J. The hierarchy of the 3D genome. Mol Cell. 2013;49:773–782. doi: 10.1016/j.molcel.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klemm SL, Shipony Z, Greenleaf WJ. Chromatin accessibility and the regulatory epigenome. Nat Rev Genet. 2019;20:207–220. doi: 10.1038/s41576-018-0089-8. [DOI] [PubMed] [Google Scholar]
- 35.Shaulian E, Karin M. AP-1 as a regulator of cell life and death. Nat Cell Biol. 2002;4:E131–136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- 36.Li Z, Schulz MH, Look T, Begemann M, Zenke M, Costa IG. Identification of transcription factor binding sites using ATAC-seq. Genome Biol. 2019;20:45. doi: 10.1186/s13059-019-1642-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bentsen M, Goymann P, Schultheis H, Klee K, Petrova A, Wiegandt R, Fust A, Preussner J, Kuenne C, Braun T, et al. ATAC-seq footprinting unravels kinetics of transcription factor binding during zygotic genome activation. Nat Commun. 2020;11:4267. doi: 10.1038/s41467-020-18035-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mouly E, Chemin K, Nguyen HV, Chopin M, Mesnard L, Leite-de-Moraes M, Burlen-defranoux O, Bandeira A, Bories JC. The Ets-1 transcription factor controls the development and function of natural regulatory T cells. J Exp Med. 2010;207:2113–2125. doi: 10.1084/jem.20092153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Polansky JK, Schreiber L, Thelemann C, Ludwig L, Kruger M, Baumgrass R, Cording S, Floess S, Hamann A, Huehn J. Methylation matters: binding of Ets-1 to the demethylated Foxp3 gene contributes to the stabilization of Foxp3 expression in regulatory T cells. J Mol Med (Berl) 2010;88:1029–1040. doi: 10.1007/s00109-010-0642-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aikawa Y, Morimoto K, Yamamoto T, Chaki H, Hashiramoto A, Narita H, Hirono S, Shiozawa S. Treatment of arthritis with a selective inhibitor of c-Fos/activator protein-1. Nat Biotechnol. 2008;26:817–823. doi: 10.1038/nbt1412. [DOI] [PubMed] [Google Scholar]
- 41.Satpathy AT, Granja JM, Yost KE, Qi Y, Meschi F, McDermott GP, Olsen BN, Mumbach MR, Pierce SE, Corces MR, et al. Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. Nat Biotechnol. 2019;37:925–936. doi: 10.1038/s41587-019-0206-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao Y, You M, Fu J, Tian M, Zhong X, Du C, Hong Z, Zhu Z, Liu J, Markowitz GJ, et al. Intratumoral stem-like CCR4+ regulatory T cells orchestrate the immunosuppressive microenvironment in HCC associated with hepatitis B. J Hepatol. 2022;76:148–159. doi: 10.1016/j.jhep.2021.08.029. [DOI] [PubMed] [Google Scholar]
- 43.Chaudhry A, Rudensky AY. Control of inflammation by integration of environmental cues by regulatory T cells. J Clin Invest. 2013;123:939–944. doi: 10.1172/JCI57175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mijnheer G, Lutter L, Mokry M, van der Wal M, Scholman R, Fleskens V, Pandit A, Tao W, Wekking M, Vervoort S, et al. Conserved human effector Treg cell transcriptomic and epigenetic signature in arthritic joint inflammation. Nat Commun. 2021;12:2710. doi: 10.1038/s41467-021-22975-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhuo B, Yu J, Chang L, Lei J, Wen Z, Liu C, Mao G, Wang K, Shen J, Xu X. Quantitative analysis of chromatin accessibility in mouse embryonic fibroblasts. Biochem Biophys Res Commun. 2017;493:814–820. doi: 10.1016/j.bbrc.2017.08.065. [DOI] [PubMed] [Google Scholar]
- 48.Yu J, Xiong C, Zhuo B, Wen Z, Shen J, Liu C, Chang L, Wang K, Wang M, Wu C, et al. Analysis of local chromatin states reveals gene transcription potential during mouse neural progenitor cell differentiation. Cell Rep. 2020;32:107953. doi: 10.1016/j.celrep.2020.107953. [DOI] [PubMed] [Google Scholar]
- 49.Smale ST, Fisher AG. Chromatin structure and gene regulation in the immune system. Annu Rev Immunol. 2002;20:427–462. doi: 10.1146/annurev.immunol.20.100301.064739. [DOI] [PubMed] [Google Scholar]
- 50.Lau CM, Adams NM, Geary CD, Weizman OE, Rapp M, Pritykin Y, Leslie CS, Sun JC. Epigenetic control of innate and adaptive immune memory. Nat Immunol. 2018;19:963–972. doi: 10.1038/s41590-018-0176-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ahmadzadeh M, Pasetto A, Jia L, Deniger DC, Stevanovic S, Robbins PF, Rosenberg SA. Tumor-infiltrating human CD4(+) regulatory T cells display a distinct TCR repertoire and exhibit tumor and neoantigen reactivity. Sci Immunol. 2019 doi: 10.1126/sciimmunol.aao4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Galas DJ, Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978;5:3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Samstein RM, Arvey A, Josefowicz SZ, Peng X, Reynolds A, Sandstrom R, Neph S, Sabo P, Kim JM, Liao W, et al. Foxp3 exploits a pre-existent enhancer landscape for regulatory T cell lineage specification. Cell. 2012;151:153–166. doi: 10.1016/j.cell.2012.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yukawa M, Jagannathan S, Vallabh S, Kartashov AV, Chen X, Weirauch MT, Barski A. AP-1 activity induced by co-stimulation is required for chromatin opening during T cell activation. J Exp Med. 2020 doi: 10.1084/jem.20182009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequence data generated in this study are deposited in Sequence Read Archive (SRA) under NCBI database: PRJNA824422.