Abstract
To expand the genetic code for specification of multiple non-natural amino acids, unique codons for these novel amino acids are needed. As part of a study of the potential of quadruplets as codons, the decoding of tandem UAGA quadruplets by an engineered tRNALeu with an eight-base anticodon loop, has been investigated. When GCC is the codon immediately 5′ of the first UAGA quadruplet, and release factor 1 is partially inactivated, the tandem UAGAs specify two leucines with an overall efficiency of at least 10%. The presence of a purine at anticodon loop position 32 of the tRNA decoding the codon 5′ to the first UAGA seems to influence translation of the following codon. Another finding is intraribosomal dissociation of anticodons from codons and their re-pairing to mRNA at overlapping or nearby codons. In one case where GCC is replaced by CGG, only a single Watson–Crick base pair can form upon re-pairing when decoding is resumed. This has implications for the mechanism of some cases of programmed frameshifting.
INTRODUCTION
This study is part of a broader investigation of codons suitable for expansion of the genetic code in order to create new protein functions and provide molecular beacons for structural studies (1). Translational incorporation of non-standard amino acids into proteins requires that codons be uniquely available to specify the new amino acids. One possibility is to use quadruplet codons for these special events, while the great majority of translation proceeds in a standard triplet mode. One approach is to engineer tRNAs with an extra base in their anticodon loops, eight instead of the nearly universal seven.
Hohsaka et al. (2) have demonstrated incorporation in vitro of two non-standard amino acids specified by non-adjacent ACCU and CGGG quadruplets. Our recent in vivo work, following an earlier precedent (3), has focused on single quadruplets (4) and this same system is extended here to tandem UAGA UAGA quadruplets. With the prospect of incorporating multiple non-standard amino acids (1), it is important to know if there are limitations on the functioning of tRNAs with enlarged anticodon loops functioning at adjacent ribosomal sites. Dramatic advances with aminoacylation of non-natural amino acids (1,5,6) makes it reasonable to focus on just the codon issue and in the present study standard amino acids are incorporated. The results, in addition to showing quadruplet decoding, also reveal unexpected facets of tRNA function pertinent to framing and the use of programmed frameshifting and bypassing for gene expression. The use of such frameshifting for expression of standard chromosomal genes may be much wider than expected (7).
A major concern with the use of quadruplet codons is the extent to which the tRNAs will misbehave during this special decoding; such as dissociation of anticodon pairing, of either peptidyl tRNA decoding the codon just prior to the quadruplet codon or of the engineered tRNA itself, and re-pairing to mRNA at other triplet or quadruplet sequences, respectively (4,8). Transient anticodon dissociation, by normal tRNAs with standard seven-membered anticodon loops, and re-pairing to mRNA at an overlapping codon is at the heart of most programmed frameshifting (9,10). Much consideration of the mechanisms involved in programmed frameshifting has centered on the premise that at least two anticodon bases have to be involved in re-pairing to mRNA at the codon in the new frame (11). The issue of whether one base pair might be adequate for re-pairing, at least in some circumstances, is addressed in the present work which has relevance for interpreting the mechanism of some cases of programmed frameshifting. However, not all programmed frameshifting involves detachment and re-pairing. In the +1 frameshifting used for expression of the yeast transposable element Ty3, ‘once only’ pairing is involved (12) and distinctive features of the wobble base of the P-site tRNA ensure that the next codon base is not available for pairing by the incoming tRNA (11). The present work addresses whether this ‘skipped base’ decoding is also relevant to work with enlarged tRNA anticodons and whether the identity of an anticodon loop base 5′ of the anticodon is relevant.
Some cases of quadruplet decoding in the presence of a mutant tRNA involve a near-cognate wild-type tRNA in the decoding event (13). However, in the present study, the engineered tRNA itself leads to amino acid incorporation as is essential for code expansion studies.
MATERIALS AND METHODS
Strains and growth conditions
Strain Su1675 ara, thi, Δlac-pro, recA/F’ lacIq, proAB+, KmR is a Rec– derivative (14) of Escherichia coli CSH26 and was used in our earlier experiments. MRA8 is a prfA1 derivative of E.coli MG1655 (15). Strains were grown in LB broth and antibiotics were added to 100 µg/ml for ampicillin, 50 µg/ml for kanamycin and 25 µg/ml for chloramphenicol. Unless stated otherwise, strains were grown at 37°C.
Construction of plasmids
Plasmids were constructed with a Su6-like tRNALeu gene with an extra base 5′ to the anticodon as described (4). Another set of plasmids were constructed in which a cassette with single or tandem UAGA quadruplets was inserted between glutathione S-transferase (GST) and lacZ in a GST–lacZ gene fusion for measurement of quadruplet decoding.
Construction of GST–malE vector, GM-1
The malE coding region was amplified from plasmid pMAL-C2 (New England Biolabs, Beverly, MA) by PCR using the following primers: ATATTAGTTAACTGAAAATCGAAGAAGGTAAACTGG and TTATTACTCGAGTTACGAGCTCGAATTAGTCTGCGCGT. HpaI and XhoI restriction endonuclease cleavage sites are in italics and malE sequences underlined. The 1110 bp PCR product was digested with HpaI and XhoI and ligated with vector pGEX-5X (Amersham Pharmacia Biotech, Piscataway, NJ) previously digested with SmaI and XhoI. The SmaI site was destroyed by ligation to the HpaI blunt end of the PCR fragment leaving the BamHI and EcoRI sites from the pGEX-5X polylinker available for cloning. Following electroporation into E.coli cells, several ampicillin resistant clones were selected and screened by PCR for the appropriately sized insert. One positive clone was selected and the malE sequence determined by DNA sequencing. There were two nucleotide changes compared to the wild-type malE sequence: one was a third position change of A→G that did not affect the protein sequence and the second was a C→T change that resulted in a valine substitution for an alanine residue. This single amino acid substitution does not interfere with MalE binding to amylose resin.
Construction of GST–malE gene fusion plasmids for protein characterization
Oligonucleotides containing UAGA quadruplets in various frames and with various 3′ and 5′ contexts (Table 1) were inserted into BamHI–EcoRI cut GM1 vector. Isolated plasmids were screened by sequencing to ensure correct insert and reading frame.
Table 1. mRNA sequence flanking quadruplets.
| Plasmid name | 5′-mRNA-3′ | Outgoing frame | |
|---|---|---|---|
| Assay constructs | pAGCC1 | AGC│UUCGCCUAGAAGA.UUG│GGC | + |
| pAGCC2 | AGC│UUCGCCUAGAUAGAUUG│GGC | – | |
| pAGCC2X | AGC│UUCGCCUAGAUACAUUG│GGC | – | |
| Protein characterization constructs | pMGCC2 | G│AUCCGCGCCUAGAUAGA│UUG | – |
| pMCGA2 | G│AUCCAUCGAUAGAUAGA│UUG | – | |
| pMCGG2 | G│AUCCAUCGGUAGAUAGA│UUG | – | |
| tRNA anticodon loop | 3′-AAAUCNUA-5′ |
The mRNA sequence of the area surrounding the tandem (or single) UAGA quadruplets is shown with the associated plasmid name. The sections of mRNA shown correspond to the DNA oligos used to generate the constructs. Italicized letters indicate bases that are part of the restriction sites used in cloning DNA oligos into the respective plasmid vectors. Vertical bars indicate incoming and outgoing reading frames relative to the 5′ and 3′ fusions, respectively. Sequences are aligned for ease of comparison within groups. The final column indicates the outgoing reading frame relative to the 5′ fusion. The bottom row shows the 3′-AUCN-5′ tRNALeu anticodon loop for comparison with the aligned UAGA mRNA sequences.
Plasmid isolation and DNA sequencing
Plasmids were isolated by a modified mini alkaline lysis/PEG precipitation method from the Perkin-Elmer Corporation. DNA sequencing was performed on an ABI 373 instrument.
Western blotting and protein quantification
Cell extracts were made by boiling cells in cracking buffer (6 M urea, 1% SDS, 125 mM Tris–HCl pH 7.2) and applied onto 10% (w/v) SDS–polyacrylamide gels. Separated proteins were blotted onto Immobilon-P membranes (Millipore Corp., Bedford, MA) using a Trans-Blot electrophoretic transfer cell (Bio-Rad Laboratories, Hercules, CA). The blotting buffer was 39 mM glycine, 48 mM Tris base, 0.05% SDS, 20% methanol and proteins were blotted at 0.5 A for 3 h. The GST termination products and GST–lacZ full-length products were visualized by chemiluminescence with a BM Chemiluminescence Western Blotting Kit (Roche Molecular Biochemicals, Indianapolis, IN) as per the manufacturer’s instructions. The primary antibody was anti-GST (Sigma, St Louis, MO). Chemiluminescence signals were quantified with a Lumi-Imager digital camera system (Roche Molecular Biochemicals).
Protein purification
MR8 cells transformed with various GST–malE fusions (Table 1) and pACYC184-based supP genes modified to have either a 3′-AUCU-5′ or 3′-AUCA-5′ extended anticodon loop, were grown in 1 l multiples in Terrific broth supplemented with 100 µg/ml ampicillin and 50 µg/ml chloramphenicol at 30°C for ∼2–3 h. Growth temperature was raised to 40°C and growth was continued for 6–8 h more. Cells were harvested and resuspended in 15 ml phosphate-buffered saline (PBS) per liter of culture and stored at –20°C overnight. Cells were disrupted by sonication and cell debris was removed by centrifugation at 10 000 r.p.m. for 20 min at 4°C in a Sorvall SS34 rotor. The supernatant was recovered and centrifuged at 45 000 r.p.m. for 2 h at 4°C in a Beckmann VTi50 rotor. Cleared lysate was applied to two Glutathione Sepharose™ 4B (Amersham Pharmacia Biotech) columns prepared with 2 ml bed volumes as per the manufacturer’s instructions. The columns were washed with 25 ml PBS and bound protein was eluted with 3 ml of 10 mM glutathione (Roche Molecular Biochemicals), 50 mM Tris pH 8.0. The eluate was diluted to 25 ml in maltose-binding protein buffer (MBPB; 20 mM Tris pH 7.4, 100 mM NaCl, 1 mM EDTA, 1 mM DTT) and applied to one amylose resin column (New England Biolabs) prepared with a 2 ml bed volume as per the manufacturer’s instructions. The column was washed with 25 ml MBPB and bound protein was eluted with 10 mM maltose in MBPB. Eluate was concentrated and washed with 6 ml HPLC-grade water in a Centricon 30 microconcentrator (Amicon, Beverly, MA).
Electrospray mass spectrometry
Molecular weights of proteins were determined using positive-ion electrospray mass spectrometry. The electrospray ionization process generates a series of multiply charged molecular ions from which very accurate (better than 0.01%) mass assignments are derived for each protein product. The amino acid composition of each protein was determined based on the exact molecular weight measured for the intact protein along with requirements dictated by the known sequence of the corresponding mRNA.
Following affinity purification, protein samples were prepared for electrospray ionization by removing salts, buffers, glutathione and other contaminants by trapping the proteins on a 1 mm C8 reverse-phase guard column (Optiguard, Optimize Technologies, Oregon City, OR) and washing the protein extensively with HPLC-grade water. The proteins were then eluted with a 67% methanol solution containing 0.9% formic acid directly into the electrospray interface of a Quattro-II mass spectrometer (Micromass, Beverly, MA). Samples were infused at a rate of 4 µl/min using a syringe pump for solvent delivery. Mass spectra were obtained from five to 50 accumulated spectra using continuum-data storage in the positive-ion mode while scanning 900–1400 Da in 4 s. A cone voltage of 60 eV and a spray voltage of 3.2 kV were used. Molecular mass spectra (Figs 3–5) show measured protein molecular weights that were generated by deconvolution of the multiply charged molecular-ion series using MaxEnt software (Micromass). Mass spectra were background subtracted prior to MaxEnt processing using a 3% threshold level. Relative normalization scales were not included in the figures of the molecular mass spectra, since protein purification procedures, electrospray ionization and processing of mass spectra do not necessarily represent accurate relative amounts of termination and full-length proteins.
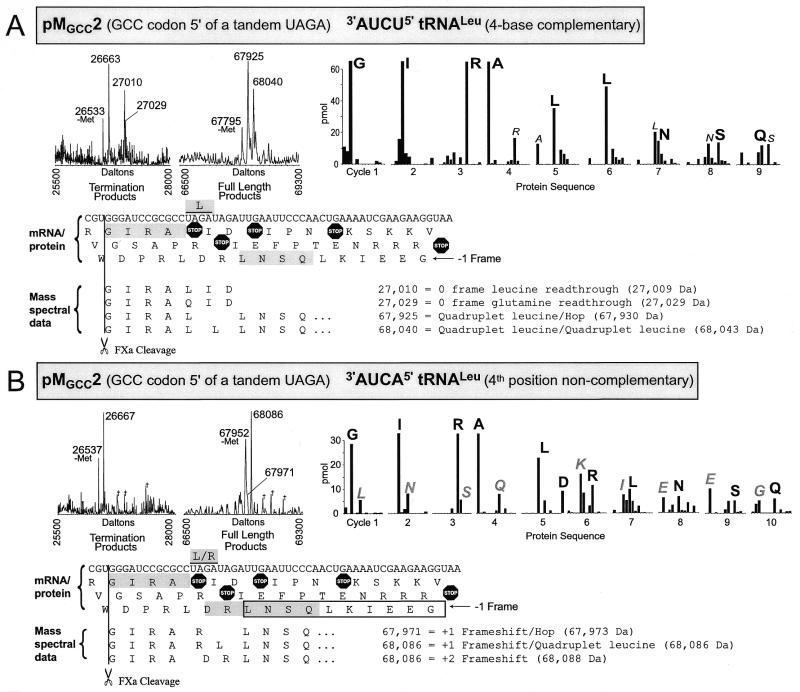
Figure 3.
Mass spectra and protein sequencing data shown for translation products of pMGCC2 translated in the presence of (A) 3′-AUCU-5′ tRNALeu with four base complementarity to the UAGA quadruplet or (B) 3′-AUCA-5′ tRNALeu which is non-complementary in the fourth quadruplet position. Molecular mass spectra show termination products and full-length fusion products. Peaks labeled ‘-Met’ are the result of in vivo N-terminal methionine removal from the protein ~131 Da higher in mass. Peaks labeled with daggers represent masses for which no protein interpretation was found. Interpretation of the products associated with the major peaks are shown by alignment of interpreted amino acid sequence below the mRNA sequence for the construct except for products terminating at UAG of the UAGA quadruplet which are not shown. Interpreted sequence for peaks representing standard termination and minus methionine proteins are not shown. Measured mass and the corresponding theoretical mass (in parentheses) are shown for each major peak. Theoretical mass values were calculated using average isotopes. For peptide N-terminal sequencing, vertical bars correspond to amino acid recovery for each cycle and are represented in the order, AEGIKLNQRSTVW, indicating all significant amino acids recovered. The protein sequence data for this figure only have an aspartic acid in cycle five and the amino acids represented by vertical bars for that cycle are in the order AEGIKLNQRSTVWD. The major amino acid recovered in each cycle is labeled in large black letters; minor amino acids are labeled with small italic letters; large gray letters label major secondary amino acid recovery where appropriate. Interpretations of the protein sequence data are shown superimposed on the mRNA 3-frame translated sequence with gray shaded boxes indicating the major sequence and outline boxes indicating the secondary sequences where appropriate. The plasmids generating the mRNA and the over-expressed modified tRNALeu are indicated in the figure.
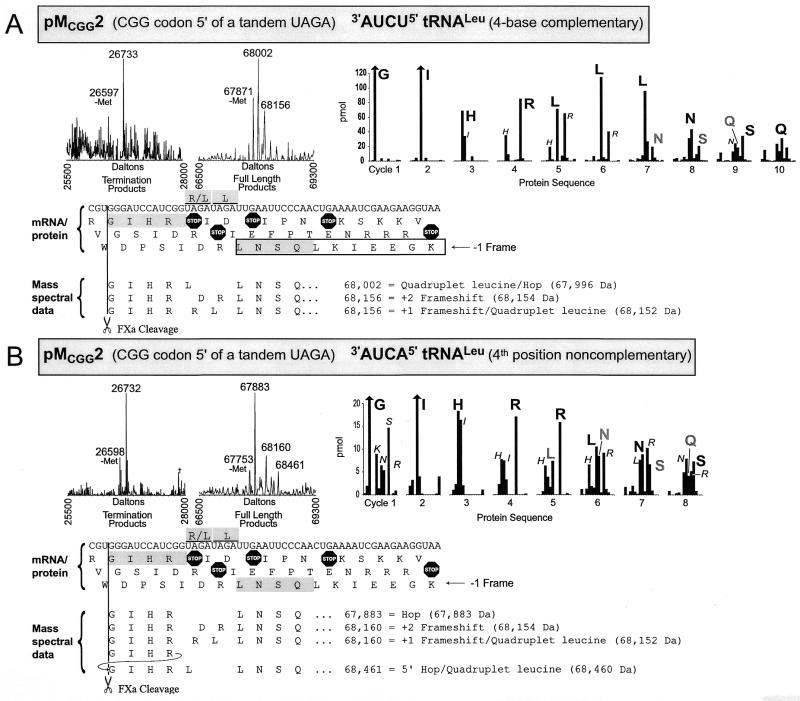
Figure 5.
Mass spectra and protein sequencing data shown for translation products of pMCGG2 translated in the presence of (A) 3′-AUCU-5′ tRNALeu with four base complementarity to the UAGA quadruplet or (B) 3′-AUCA-5′ tRNALeu which is non-complementary in the fourth quadruplet position. See legends to Figures 3 and 4 for further details.
Protein sequencing
Fusion protein intended for protein sequencing was digested with Factor Xa protease (New England Biolabs) as per the manufacturer’s recommended conditions and further purified by electrophoresis through 10% (w/v) SDS–polyacrylamide gels and electrophoretic transfer to PVDF membranes (Immobilon-P, Millipore Corp.) as previously described (16). N-terminal sequencing of peptides was carried out on either Applied Biosystems model 477 or model 492 (PE Biosystems, Foster City, CA). Lag corrected data were tabulated for figure graphs.
RESULTS
A modified tRNALeu and a UAGN quadruplet in mRNA to probe for quadruplet reading
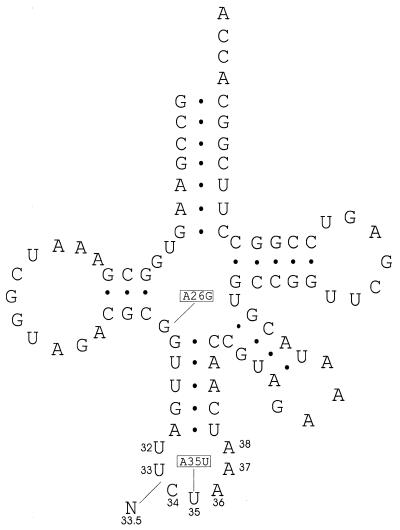
Wild-type leuX codes for a leucine tRNA with a 3′-AAC-5′ anticodon. Amber suppressor tRNA Su6 is encoded by the supP allele of the leuX gene (17) and differs from the wild-type gene by two mutations, A26G and A35U (Fig. 1). The latter mutation gives tRNA Su6 a 3′-AUC-5′ anticodon allowing it to suppress UAG stop codons. This tRNA has a 30–100% efficiency of suppression (18). After its anticodon loop had been modified to contain eight nucleotides, Su6 was found to translate a UAGA quadruplet with a 13–26% efficiency (4). In the current study, oligonucleotides designed to create variants of the Su6 tRNA each with one extra nucleotide either A or U inserted immediately 5′ of the anticodon were placed into pACYC184 under the control of the inducible tac promoter (Fig. 1). The mutant tRNA will be written as 3′-AUCN-5′ tRNALeu to readily identify the extra nucleotide and its position in relation to the anticodon of its parent.
Figure 1.
The cloverleaf structure of Su6 tRNA as deduced from the DNA sequence is shown. Boxed letters indicate locations where Su6 deviates from the product of the wild-type leuX gene. The ‘N’ shows the location of the nucleotide (designated as position 33.5) inserted to create an eight base anticodon loop. The figure is adapted from Yoshimura et al. (17), with tRNA nucleotide numbering as in Sprinzl et al. (33).
Assays to determine the efficiency of decoding tandem UAGA quadruplets by 3′-AUCU-5′ tRNALeu
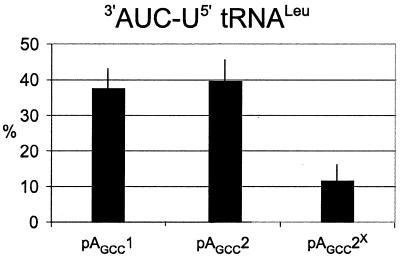
To measure reading by the mutant tRNA at single and tandem UAGA quadruplets, oligonucleotides (Table 1) were inserted between a gene encoding GST (GST) and β-galactosidase (lacZ) producing a GST–lacZ fusion as described (4). Escherichia coli strain MRA8 has a temperature sensitive release factor 1 at non-permissive temperatures (42°C) (15), and 3′-AUCN-5′ tRNALeu was found to have a two to four times higher level of quadruplet translation in this strain (4). The release factor mutant reduces the competition between release factor 1 mediated termination and quadruplet decoding at the UAGA quadruplet. Immunoblotting assays were done on proteins encoded by constructs with a single UAGA quadruplet (pAGCC1), a UAGA UAGA pair (pAGCC2) as well as a UAGA UACA pair (pAGCC2X; the superscripted X denotes the UAGA to UACA change) to determine the efficiency with which ribosomes traverse the two quadruplets in the presence of 3′-AUCU-5′ tRNALeu and a temperature sensitive release factor 1 at 42°C (Fig. 2). Both constructs with tandem quadruplets had the 3′ fusion in the –1 frame to capture the product of a +1 shift at each quadruplet, while the construct with a single quadruplet was framed with the 3′ fusion in the +1 frame. Surprisingly, these assays revealed that the efficiency of tandem UAGA quadruplets is similar (∼40%) to that found with a single UAGA quadruplet (4). Changing the second UAGA quadruplet to UACA, however, lowered the level of shifting to the –1 frame by 4-fold.
Figure 2.
Assays of the products of single, tandem, or modified tandem UAGA quadruplets in the mRNA in the presence of 3′-AUCU-5′ tRNALeu. Data represents percent of full-length fusion relative to full-length plus terminated fusion. Data points are means of four separate experiments. Error bars indicate average deviation from the mean. X-axis labels indicate plasmid used to generate mRNA.
Characterization of protein products
To investigate the specific products of translation of the tandem UAGA quadruplets, oligonucleotides were inserted between a gene encoding GST and maltose-binding protein (malE) in vector GM1. Proteins were isolated from strains with these constructs containing tandem UAGA quadruplets in different contexts and with either 3′-AUCU-5′ tRNALeu (four base complementarity) or 3′-AUCA-5′ tRNALeu (fourth base non-complementary). The products were analyzed by both mass spectrometry and protein sequencing.
Mass spectral measurements allowed interpretation of the specific amino acids incorporated at the quadruplet region. Based on prediction from the mRNA sequence, a predicted molecular weight is calculated for the proportion of the protein expected to be synthesized by standard triplet translation, the difference between this predicted mass and the measured molecular weight allows inference of the amino acid specified by the quadruplet codon. The measured molecular weights of proteins agreed with these inferred masses within 0.01%. The mass measurements of large molecules (e.g. 68 000 Da) does not permit unambiguous distinction of certain amino acid substitutions that differ in molecular weight by only a few daltons (i.e. leucine, 113 Da; asparagine, 114 Da; aspartic acid, 115 Da).
A GCC codon 5′ of tandem UAGA quadruplets. Protein characterization construct pMGCC2, has two tandem UAGA quadruplets with a GCC codon immediately 5′ of the first UAGA quadruplet (Fig. 3A). Here again, a shift to the –1 frame is required to allow for the possibility of a +1 frameshift at each of the two UAGA quadruplets. Mass spectrometric analysis shows three termination products. A 26 663 Da product whose mass is consistent with termination at the UAG of the first UAGA quadruplet (26 667 Da), a 27 010 Da product which corresponds to UAG readthrough with triplet specification of leucine plus two additional amino acids (27 009 Da) and a 27 029 Da product tentatively assigned as UAG readthrough with triplet specification of glutamine (27 029 Da). However, the latter may instead represent a sodium adduct of the 27 010 Da peak. Two major full-length proteins are evident in the molecular mass spectrum. The first peak at 67 925 Da corresponds to a full-length fusion that achieves the –1 frameshift by the two UAGA quadruplets together specifying a single leucine and continuing translation at the UUG leucine codon immediately 3′ of the second UAGA quadruplet, ‘GIRALLNSQ’ (67 930 Da). This could be accomplished by the first UAGA pairing with the anticodon of the 3′-AUCU-5′ tRNALeu, the mRNA then dissociating from the anticodon, slipping and re-pairing to the anticodon via the second UAGA—that is, by ribosomal hopping. Depending on whether there is a three or a four base interaction between the 3′-AUCU-5′ tRNALeu and the first UAGA quadruplet, either AUA (rare isoleucine codon) or UAG may stimulate this hop as seen previously with other slow-to-decode codons. Over-expressed tRNAIle which decodes AUA would be predicted to reduce this hopping if it is the stimulatory element. The mass of the second peak at 68 040 Da, corresponds to a full-length fusion with each UAGA quadruplet specifying a leucine, ‘GIRALLLNSQ’ (68 043 Da). The interpretation of these peaks is compatible with the protein sequence data, although the tandem leucine incorporation is not confirmed by the protein sequence data. Each of the two peaks in Figure 3A labeled 67 925 and 68 040, have similar smaller peaks on their right shoulder. The mass assignment for these small peaks corresponds to the sequences, ‘GIRARLNSQ’ and ‘GIRA(LR)LNSQ’, respectively. The order of the leucine and arginine in parentheses and the relative contribution of ArgLeu versus LeuArg is not determined since these minor products are not seen in the protein sequence data and the molecular mass data specifies composition, but not sequence order. Remarkably, no product was seen which corresponded to the first UAGA specifying leucine or arginine with subsequent termination at UAG of the second UAGA quadruplet.
In the presence of 3′-AUCA-5′ tRNALeu (Fig. 3B) the standard UAG termination product and a full-length protein of 68 086 Da are identified. The 68 086 Da peak is compatible with two sequence interpretations, ‘GIRALRLNSQ’ (68 086 Da) and ‘GIRADRLNSQ’ (68 088 Da). Protein sequencing shows that the predominant sequence begins ‘GIRA...’. In cycle five, there are two amino acids: leucine and aspartic acid. Leucine in cycle five has two possible origins. A minor sequence (Fig. 3B, boxed), ‘LNSQLKIEEG’ has a leucine in cycle five (underlined) and may result from Factor Xa cleavage of the sequence ‘GIRARLNSQLKIEEG’ as discussed previously (4). It is also possible that the fourth base non-complementary 3′-AUCA-5′ tRNALeu encodes leucine at the UAGA quadruplet to produce the sequence ‘GIRALRLNSQ’. The aspartic acid seen in cycle five must be encoded by the GAU (Asp) codon UAGAUAGA within the tandem quadruplets to produce the sequence ‘GIRADRLNSQ’. The mass data do not permit the distinction between leucine and aspartic acid and the result is discussed below. The peak at mass 67 952 Da, which represents the 68 086 Da protein without the N-terminal methionine, has a broad right shoulder. By increasing instrument resolution, the peak was better resolved, although at a lower signal intensity, and was assigned a molecular weight of 67 971 Da (higher resolution data not shown). This mass indicates that a single arginine is encoded by the tandem UAGA quadruplets to yield the sequence, ‘GIRARLNSQ’ (67 973 Da).
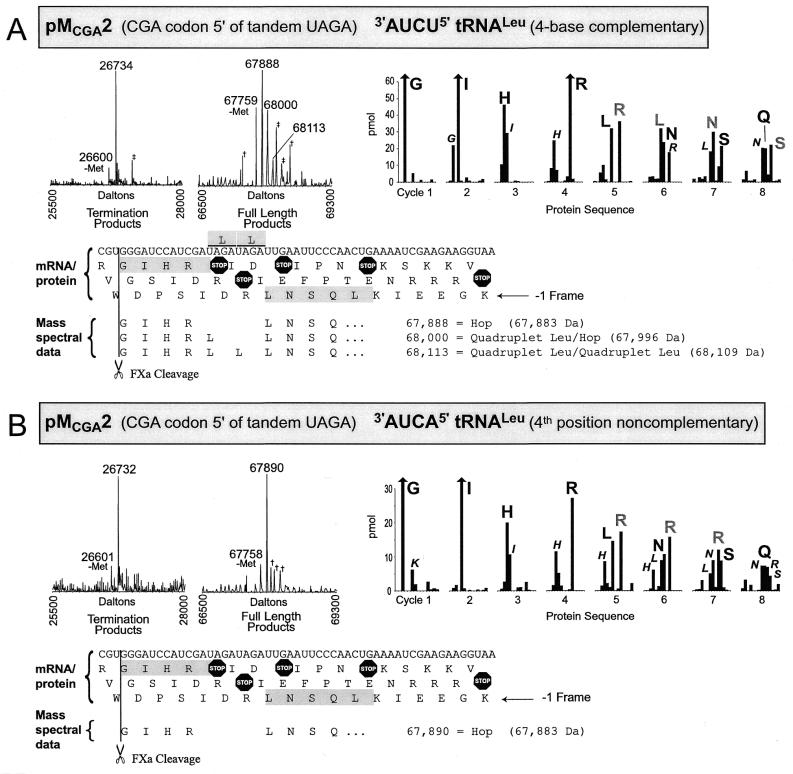
A CGA codon 5′ of tandem UAGA quadruplets. Is the encoding of aspartic acid specific for when peptidyl tRNAAla is decoding GCC 5′ of the UAGA quadruplet or is it also seen with tRNAArg decoding CGA as the 5′ codon? Construct pMCGA2 has two UAGA quadruplets with a CGA codon immediately 5′ of the first quadruplet. In the presence of 3′-AUCU-5′ tRNALeu the product of termination at UAG of the first UAGA quadruplet is observed (Fig. 4A). A number of full-length products are found of which the most intense, at 67 888 Da, corresponds to the sequence, ‘GIHRLNSQ’ (67 883 Da). This product could arise by tRNA![]() (anticodon 3′-GCI-5′) hopping forward four bases and skipping a U from CGA to AGA without pairing of the first base of the AGA, and with a net result that only one arginine is incorporated for both UAGA quadruplets. The next peak with a mass assignment of 68 000 Da corresponds to a single leucine inserted for the two UAGA quadruplets, ‘GIHRLLNSQ’ (67 996 Da). This leucine is likely encoded by the first UAGA quadruplet followed by hopping forward to pair with the second UAGA quadruplet as above. The next higher-mass protein is seen at 68 113 Da which matches the mass of a product with two leucines inserted, one for each of the two UAGA quadruplets ‘GIHRLLLNSQ’ (68 109 Da). While it is difficult to interpret protein sequence data alone for complex mixtures of products such as this, the sequence information is compatible with the predictions based on the mass spectrometric data. The remaining significant peaks in the mass spectrum represent glutathione derivatives or are unassigned as indicated in Figure 4A.
(anticodon 3′-GCI-5′) hopping forward four bases and skipping a U from CGA to AGA without pairing of the first base of the AGA, and with a net result that only one arginine is incorporated for both UAGA quadruplets. The next peak with a mass assignment of 68 000 Da corresponds to a single leucine inserted for the two UAGA quadruplets, ‘GIHRLLNSQ’ (67 996 Da). This leucine is likely encoded by the first UAGA quadruplet followed by hopping forward to pair with the second UAGA quadruplet as above. The next higher-mass protein is seen at 68 113 Da which matches the mass of a product with two leucines inserted, one for each of the two UAGA quadruplets ‘GIHRLLLNSQ’ (68 109 Da). While it is difficult to interpret protein sequence data alone for complex mixtures of products such as this, the sequence information is compatible with the predictions based on the mass spectrometric data. The remaining significant peaks in the mass spectrum represent glutathione derivatives or are unassigned as indicated in Figure 4A.
Figure 4.
Mass spectra and protein sequencing data shown for translation products of pMCGA2 translated in the presence of (A) 3′-AUCU-5′ tRNALeu with four base complementarity to the UAGA quadruplet or (B) 3′-AUCA-5′ tRNALeu which is non-complementary in the fourth quadruplet position. See Figure 3 legend for further details; in addition: double daggers indicate glutathione derivatives; vertical bars with upper arrows have been truncated due to very high amino acid recovery.
In the presence of 3′-AUCA-5′ tRNALeu (Fig. 4B) the major products are from termination at UAG of the first UAGA quadruplet and a 67 890 Da molecular weight protein derived from a CGA to AGA hop to yield the sequence, ‘GIHRLNSQ’ (67 883 Da), as described in the previous paragraph. This sequence is consistent with protein sequence data.
A CGG codon 5′ of tandem UAGA quadruplets. To further investigate the hopping discussed, studies were performed with construct pMCGG2 in which a CGG codon is located immediately 5′ of tandem UAGA quadruplets (Fig. 5A). This allows further comparison with the pMGCC2 construct, in which the aspartic acid was encoded, but reduces the possibility of hopping since with standard pairing only a single C-G pair would be possible in the AGA landing site. As anticipated, in the presence of 3′-AUCU-5′ tRNALeu, the product due to termination at UAG of the UAGA quadruplet is predominant. In contrast to pMCGA2, the peak at 68 002 Da indicates that the CGG design generates only one leucine for both UAGA quadruplets producing the sequence ‘GIHRLLNSQ’ (67 996 Da), with no tRNAArg hopping and no tandem leucine incorporation detected. The peak at 68 156 Da in this spectrum could be either a +2 frameshift due to ignoring the first two ribosomal A-site bases (UA) with the resulting sequence, ‘GIHRDRLNSQ’ (68 154 Da), or an arginine encoded by one UAGA quadruplet and a leucine encoded by the other to produce, ‘GIHR(RL)LNSQ’ (68 152 Da). The order of the L and R in parentheses cannot be determined from the mass spectral data. Based on the protein sequence data (cycle five) the +2 frameshift to the GAU codon is not occurring in this case. Instead, a leucine is encoded by one UAGA quadruplet and an arginine by the other. Separating the major and minor sequences in the protein sequencing data is complicated by the combination of leucines from the two products. Clarification, however, is provided by the mass spectrum, which gives no evidence for incorporation of two leucines to give the sequence, ‘GIHRLLLNSQ’.
In the presence of 3′-AUCA-5′ tRNALeu (Fig. 5B), three full-length products and the expected termination product were identified by mass spectrometry. The molecule at 67 883 Da corresponds to the sequence ‘GIHRLNSQ’ (67 883 Da) which suggests that tRNA![]() hops from CGG to AGA, consistent with the protein sequence. This hop is somewhat surprising because only one G-C pair is available for re-pairing, but no other explanation is apparent. The next peak at 68 160 Da corresponds to a protein in which the tandem UAGA quadruplets are negotiated either by a +2 frameshift due to ignoring the first two A-site bases (UA) with the sequence, ‘GIHRDRLNSQ’ (68 154 Da), or an arginine inserted for one UAGA quadruplet and a leucine inserted for the other as discussed in the previous paragraph, ‘GIHR(RL)LNSQ’ (68 152 Da). The protein sequence data supports the ‘GIHR(RL)LNSQ’ interpretation.
hops from CGG to AGA, consistent with the protein sequence. This hop is somewhat surprising because only one G-C pair is available for re-pairing, but no other explanation is apparent. The next peak at 68 160 Da corresponds to a protein in which the tandem UAGA quadruplets are negotiated either by a +2 frameshift due to ignoring the first two A-site bases (UA) with the sequence, ‘GIHRDRLNSQ’ (68 154 Da), or an arginine inserted for one UAGA quadruplet and a leucine inserted for the other as discussed in the previous paragraph, ‘GIHR(RL)LNSQ’ (68 152 Da). The protein sequence data supports the ‘GIHR(RL)LNSQ’ interpretation.
The signal for the third full-length protein of 68 461 Da is weak and a corresponding product was not detected in the protein sequencing data. Its mass is compatible with a glutathione derivative of the 68 160 Da protein, however the major protein seen in this spectrum (67 883 Da) does not have a corresponding glutathione derivative. The alternative involves a 12 nt 5′ hop. Since this is speculative, further work is necessary before this possibility merits discussion.
This final section reinforces the observation that four-base complementarity between the tRNA and the UAGA quadruplet is favorable for leucine incorporation while lack of complementarity allows translation to proceed by other events.
DISCUSSION
Recent work has provided promising evidence for the utility of quadruplet codons in code expansion studies (2,4). The present work adds to this evidence by showing that two tandem UAGA quadruplets in conjunction with an engineered tRNA having an eight base anticodon loop can encode two specified amino acids. The engineered tRNA is directly responsible for leucine being encoded by UAGA, an important consideration for efforts to get incorporation of a non-standard amino acid due to decoding of a specified codon.
Single base landing
All previously known cases of both +1 and –1 frameshifting (11) and, until now, all cases of bypassing suggest a requirement for two bases pairing in the landing site. Here, only a single base is involved in Watson–Crick pairing when a CGG decoding tRNAArg hops from CGG to AGA in the presence of 3′-AUCA-5′ tRNALeu. This suggests that it may be premature to eliminate consideration of one base re-pairing in cases of +1 and –1 frameshifting as well. In most cases of programmed –1 frameshifting that do not involve a stop or rare codon at the ribosomal A-site, two tRNAs shift in tandem (19), though in at least one case (20) only one tRNA shifts frame. In several other cases, trying to decide whether dissociation and re-pairing is involved hinges on knowing how many bases have to be involved in re-pairing for it to be a viable option, and it has long been realized (21,22), this has been a real issue. The present work suggests that caution is appropriate before accepting the conclusions of Sundararajan et al. (11) that two anticodon bases have to be involved in re-pairing. In concurrent work (A.J.Herr, J.F.Atkins and R.F.Gesteland, manuscript in preparation) we have extensively explored this significant issue.
Position 32 purine
Interestingly, with GCC UAGA UAGA U, the simplest interpretation for GAU functioning as a codon (to specify aspartic acid), is that decoding involves both re-pairing of tRNAAla to CCU and skipping of the next codon base, A. In their insightful studies of Saccharomyces cerevisiae Ty3 programmed frameshifting, Sundararajan et al. (11) showed an effect for a special interaction of the third codon base with anticodon base 34, in mediating skipping of the next mRNA base. In the present case, base 34 of the cognate tRNAAla for GCC is an unmodified G which can form a standard Watson–Crick pair with the third codon base C. Instead, a distinctive feature of this anticodon loop is the presence of a purine at position 32. Escherichia coli GCC decoding tRNAAla is one of only two wild-type E.coli tRNAs with a purine at position 32 (23). Of the 2726 known tRNAs from all organisms, only ~2% have a purine at position 32 and only 0.4% of known tRNAs have an A:U combination for positions 32:38 [A:U at these positions are thought to pair via a bifurcated hydrogen bond equivalent to a U:U pair at this position (24)]. Indeed it would seem likely that tRNAs with a purine at position 32 may play a special role and that the current base skipping following re-pairing to mRNA at CCU may be due to the purine at position 32 of tRNAAla discriminating against incoming scarce AGA decoding tRNAArg which has a wild-type seven-member anticodon loop. Previously, it was shown that a C at position 32 of a tRNAGly had a direct effect on the tRNA containing it to cause frameshifting (25). An alternate possibility is that a different mRNA structure, due to a purine run, is important. Interactions between rRNA, mostly A1492 and A1493, of the 30S subunit and the mRNA backbone decrease the dissociation of cognate tRNA in the A-site and may trigger significant conformational changes elsewhere (26). With base skipping in the A-site, subtle changes in mRNA conformation may affect either tRNA or rRNA contacts.
The experiments reported here utilized a derivative of a stop codon, but the goal of providing many new codons in a mixed triplet/quadruplet decoding system requires the use of quadruplets for which a normal tRNA is available to read the first three nucleotides as a triplet. The efficiency of relevant +1 frameshift mutant external suppressors gives some grounds for optimism, though the possible effect of signals known to stimulate frameshifting need to be explored. Some tRNAs with eight-member anticodon loops mediate quadruplet decoding by pairing of three anticodon nucleotides followed by a secondary pairing using two of those three and one flanking nucleotide. Though different from the detachment and re-pairing of the same anticodon in the programmed frameshifting in decoding E.coli release factor 2, it would not be surprising if this process was influenced by the same stimulatory signal, a Shine–Dalgarno sequence positioned 3 nt 5′ of the shift site (14). Despite early suggestions to the contrary, it has recently been proposed that four anticodon bases are never simultaneously involved in pairing to four codon bases (27). If true (28), this might mean that all quadruplet decoding mediated by tRNAs with eight-member anticodon loops would be responsive to appropriately positioned Shine–Dalgarno stimulation (a negative result would, of course, not be strong evidence for simultaneous quadruplet pairing). Experiments are needed to investigate the effects of different programmed frameshift stimulators on quadruplet decoding because of their potential impact on quadruplet translation efficiency and specificity and because of the potential for insights into understanding specification of codon and anticodon size and the interrelatedness of the two.
It is significant that the levels of translation of single and tandem UAGA quadruplets into the correct frame as seen in the immunoassays in Figure 2 are the same, ∼40%. However as seen throughout this study, multiple products can arise in the same reading frame because the ribosome employs various translation events to circumvent the UAGA quadruplet. The mass spectral data indicate how much of the single and tandem UAGA quadruplet decoding actually results from incorporation of leucine. At a single UAGA quadruplet, leucine was encoded two-thirds of the time (4). About one-third of ribosomes translating tandem UAGA quadruplets decode a leucine at both UAGAs (Leu-Leu). While this type of mass spectral data do not lend themselves to precise quantitative distinctions, it seems reasonable to conclude that decoding of two tandem UAGA quadruplets as leucine occurs at ∼50% or less of decoding of a single UAGA quadruplet as leucine (consistent with the 44% expected for independent events). When account is taken of termination and other translational events, tandem UAGA quadruplets specify two leucines at least 10% of the time.
In this study there is significant insertion of a single leucine for one UAGA quadruplet followed by hopping forward to the second UAGA quadruplet. This raises the question of whether there is a significant level of hopping from the single UAGA quadruplet in the earlier study (4) and, without the presence of a second UAGA to ‘capture’ them, these hopping ribosomes would land at dispersed sites and be lost to detection by our mass spectral analysis. This would affect the interpretation that the levels of the leucine insertion at single and tandem UAGA quadruplets occur at similar levels. However, it is not likely in the case of the single UAGA quadruplet that landing of such hopping ribosomes would be dispersed enough to avoid detection altogether. In this and other studies (A.J.Herr, J.F.Atkins and R.F.Gesteland, manuscript in preparation), we have seen hopping ribosomes utilize multiple landing sites. Yet even in multiple landing site cases, a restricted subset of possible codons is employed for landing. Among these sites, those with the greatest potential for pairing are used preferentially, leading to several discrete and detectable peaks. Despite the second UAGA of the GCC UAGA UAGA ‘capturing’ the ribosomes that took off from the first UAGA, we believe that direct comparison of single leucine specified by a single UAGA and double leucine specified by tandem UAGA is meaningful.
It is significant here that among the termination products no product is seen which corresponds to encoding of a single leucine by the first UAGA quadruplet and termination at UAG of the second UAGA quadruplet. If translation of each of the tandem UAGA quadruplets is an independent event, then we would expect to see a detectable amount of product corresponding to single leucine incorporation followed by termination. The presence of the expanded anticodon loop in the P-site tRNA may influence accessibility of release factor 1 to the A-site UAG(A). Isaksson and colleagues (29) have shown a functional interaction between peptidyl tRNA![]() , which decodes GGA and GGG, but not peptidyl tRNA
, which decodes GGA and GGG, but not peptidyl tRNA![]() , which decodes GGC and GGU, and release factor 1 during termination at UAG. We have not exploited the consequent poor termination at UAG following GGA (29) but in a concurrent study, Herr et al. (A.J.Herr, J.F.Atkins and R.F.Gesteland, manuscript in preparation) have been concerned with dissociation and re-pairing potential of tRNA
, which decodes GGC and GGU, and release factor 1 during termination at UAG. We have not exploited the consequent poor termination at UAG following GGA (29) but in a concurrent study, Herr et al. (A.J.Herr, J.F.Atkins and R.F.Gesteland, manuscript in preparation) have been concerned with dissociation and re-pairing potential of tRNA![]() because of its relevance to this project and also the 50 nt translational bypassing in decoding phage T4 gene 60 (30,31).
because of its relevance to this project and also the 50 nt translational bypassing in decoding phage T4 gene 60 (30,31).
This study has provided further evidence that quadruplet decoding has potential in schemes for expanding the genetic code. It focuses attention on the need to test programmed frameshifting stimulatory signals as a method of increasing the efficiency and specificity of quadruplet decoding, and it identifies detachment and re-pairing as an issue that will need to be addressed in potential quadruplet systems. Finally, it reports the involvement of only a single Watson–Crick base pair on re-pairing of a tRNA anticodon to mRNA, and provides further support for special functional properties of a purine at tRNA position 32.
NOTE ADDED IN PROOF
A discussion of the code expansion studies of Schultz and Romesberg and their colleagues, including quadruplet codons and new bases, has recently been published (32).
Acknowledgments
ACKNOWLEDGEMENTS
We thank Monica Rydén-Aulin (Department of Microbiology, Stockholm University, Sweden) for sending us the MRA8 strain. This work was supported by Department of Energy grant DEFG03-99ER62732 to R.F.G. and NIH grant RO1-GM48152 to J.F.A.
REFERENCES
- 1.Liu D. and Schultz,P. (1999) Proc. Natl Acad. Sci. USA, 96, 4780–4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hohsaka T., Ashizuka,Y., Sasaki,H., Murakami,H. and Sisido,M. (1999) J. Am. Chem. Soc., 121, 12194–12195. [Google Scholar]
- 3.Curran J.F. and Yarus,M. (1987) Science, 238, 1545–1550. [DOI] [PubMed] [Google Scholar]
- 4.Moore B., Persson,B.C., Nelson,C.C., Gesteland,R.F. and Atkins,J.F. (2000) J. Mol. Biol., 298, 195–209. [DOI] [PubMed] [Google Scholar]
- 5.Saks M.E., Sampson,J.R., Nowak,M.W., Kearney,P.C., Du,F., Abelson,J.N., Lester,H.A. and Dougherty,D.A. (1996) J. Biol. Chem., 271, 23169–23175. [DOI] [PubMed] [Google Scholar]
- 6.Liu D.R., Magliery,T.J., Pastrnak,M. and Schultz,P.G. (1997) Proc. Natl Acad. Sci. USA, 94, 10092–10097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ross-Macdonald P., Coelho,P.S.R., Roemer,T., Agarwal,S., Kumar,A., Jansen,R., Cheung,K.-H., Sheehan,A., Symoniatis,D., Umansky,L. et al., (1999) Nature, 402, 413–418. [DOI] [PubMed] [Google Scholar]
- 8.O’Connor M., Gesteland,R.F. and Atkins,J.F. (1989) EMBO J., 8, 4315–4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farabaugh P.J. (1996) Annu. Rev. Genet., 30, 507–528. [DOI] [PubMed] [Google Scholar]
- 10.Atkins J.F., Böck,A., Matsufuji,S. and Gesteland,R.F. (1999) In Gesteland,R.F, Cech,T.R. and Atkins,J.F. (eds), The RNA World. 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 637–673.
- 11.Sundararajan A., Michaud,W.A., Qian,Q., Stahl,G. and Farabaugh,P.J. (1999) Mol. Cell, 4, 1005–1015. [DOI] [PubMed] [Google Scholar]
- 12.Farabaugh P.J., Zhao,H. and Vimaladithan,A. (1993) Cell, 74, 93–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qian Q., Li,J.-N., Zhao,H., Hagervall,T.G., Farabaugh,P.J. and Björk,G.R. (1998) Mol. Cell, 1, 471–482. [DOI] [PubMed] [Google Scholar]
- 14.Weiss R.B., Dunn,D.M., Atkins,J.F. and Gesteland,R.F. (1987) Cold Spring Harb. Symp. Quant. Biol., 52, 687–693. [DOI] [PubMed] [Google Scholar]
- 15.Zhang S., Rydén-Aulin,M., Kirsebom,L.A. and Isaksson,L.A. (1994) J. Mol. Biol., 242, 614–618. [DOI] [PubMed] [Google Scholar]
- 16.Matsudaira P. (1987) J. Biol. Chem., 262, 10035–10038. [PubMed] [Google Scholar]
- 17.Yoshimura M., Inokuchi,H. and Ozeki,H. (1984) J. Mol. Biol., 177, 627–644. [DOI] [PubMed] [Google Scholar]
- 18.Miller J.H. and Albertini,A.M. (1983) J. Mol. Biol., 164, 59–71. [DOI] [PubMed] [Google Scholar]
- 19.Jacks T., Madhani,H.D., Masiarz,F.R. and Varmus,H.E. (1988) Cell, 55, 447–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mejlhede N., Atkins,J.F. and Neuhard,J. (1999) J. Bacteriol., 181, 2930–2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Condron B., Gesteland,R.F. and Atkins,J.F. (1991) Nucleic Acids Res., 19, 5607–5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brierley I., Jenner,A.J. and Inglis,S.C. (1992) J. Mol. Biol., 227, 463–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mims B., Prather,N. and Murgola,E.J. (1985) J. Bacteriol., 162, 837–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Auffinger P. and Westhof,E. (1999) J. Mol. Biol., 292, 467–483. [DOI] [PubMed] [Google Scholar]
- 25.O’Connor M. (1998) J. Mol. Biol., 279, 727–736. [DOI] [PubMed] [Google Scholar]
- 26.Yoshizawa S., Fourmy,D. and Puglisi,J. (1999) Science, 285, 1722–1725 [DOI] [PubMed] [Google Scholar]
- 27.Farabaugh P.J. and Björk,G.R. (1999) EMBO J., 18, 1427–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Atkins J.F., Herr,A.J., Massire,C., O’Connor,M., Ivanov,I. and Gesteland,R.F. (2000) In Garrett,R.A., Douthwaite,S.R., Liljas,A., Matheson,A.T., Moore,P.B. and Noller,H.F. (eds), The Ribosome: Structure, Function, Antibiotics and Cellular Interactions. ASM Press, Washington, DC, pp. 369–383.
- 29.Zhang S., Rydén-Aulin,M. and Isaksson,L.A. (1998) J. Mol. Biol., 284, 1243–1246. [DOI] [PubMed] [Google Scholar]
- 30.Weiss R.B., Huang,W.M. and Dunn,D.M. (1990) Cell, 62, 117–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herr A.J, Gesteland,R.F. and Atkins,J.F. (2000) EMBO J., 19, 2671–2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Service R.F. (2000) Science, 289, 232– 235. [DOI] [PubMed] [Google Scholar]
- 33.Sprinzl M., Horn,C., Brown,M., Ioudovitch,A. and Steinberg,S. (1998) Nucleic Acids Res., 26, 148–153. [DOI] [PMC free article] [PubMed] [Google Scholar]