Abstract
Background
Cholangiocarcinoma (CCA) is a class of malignant tumors originating from bile duct epithelial cells. Due to difficult early diagnosis and limited treatment, the prognosis of CCA is extremely poor. BMI1 is dysregulated in many human malignancies. However, the prognostic significance and oncogenic role of BMI1 in cholangiocarcinoma (CCA) are not well elucidated.
Methods
In the present study, we investigated its clinical importance and the potential mechanisms in the progression of CCA. We detected BMI1 expression in a large CCA cohort. We demonstrated that BMI1 was substantially upregulated in CCA tissues and was identified as an independent prognostic biomarker of CCA. Moreover, overexpression of BMI1 promoted CCA proliferation, migration, and invasion. And BMI1 knockdown could inhibit proliferation and metastases of CCA in vitro and in vitro/vivo validation. Interestingly, we found that CCA-derived exosomes contain BMI1 proteins, which can transfer BMI1 between CCA cells. The unique BMI1-containing exosomes promote CCA proliferation and metastasis through autocrine/paracrine mechanisms. In addition, we demonstrated that BMI1 inhibits CD8+T cell-recruiting chemokines by promoting repressive H2A ubiquitination in CCA cells.
Conclusions
BMI1 is an unfavorable prognostic biomarker of CCA. Our data depict a novel function of BMI1 in CCA tumorigenesis and metastasis mediated by exosomes. Besides, BMI1 inhibition may augment immune checkpoint blockade to inhibit tumor progression by activating cell-intrinsic immunity of CCA.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-022-04500-1.
Keywords: Cholangiocarcinoma, BMI1, Progression, prognosis, Exosomes, Antitumor immunity
Introduction
Cholangiocarcinoma (CCA) is a highly malignant tumor with an extremely poor prognosis [1, 2], and its etiology and pathogenesis remain largely unclear [3]. According to the anatomical location, CCA is further divided into three subtypes, including intrahepatic CCA (iCCA), perihilar CCA (pCCA), and distal CCA (dCCA) [4, 5]. The global incidence of CCA has been rising over the past decades [6]. East and Southeast Asia, especially Thailand and China, had the highest prevalence of CCA [7]. Surgical resection is the only curative option for the treatment of CCA. Unfortunately, most patients are diagnosed at advanced stages with lymph node invasion and distant metastasis, and the surgical resection rate is only 30–40%[8]. Furthermore, high postoperative recurrence rates and chemical resistance attribute to the dismal prognosis, and the 5-year overall survival (OS) rate is less than 30% after radical resection [9]. The prognosis of CCA is poor due to the difficulties of early diagnosis and the resistance to chemotherapy and radiotherapy [10]. There are few recognized target drugs for CCA, but the effects of most targeted drugs are not satisfactory. Therefore, it is urgently necessary to identify novel biomarkers or target therapies to improve clinical outcomes in CCA patients.
The polycomb group (PcG) proteins are transcriptional repressors that regulate lineage choices during development and differentiation, while they play an essential role in cancer [11]. As a core member of the PcG gene family, B-cell-specific Moloney murine leukemia virus integration site 1 (BMI1) contains an E3 ubiquitin ligase resulting in gene silencing through ubiquitination of histone H2A at lysine 119 [12]. BMI1 was first identified as an oncogene in lymphoma collaboration with Myc [13]. Recently, many studies, including ours, have reported that the expression of BMI1 is increased in various human cancers, and it is correlated with poor prognoses [14–16]. Targeting BMI1 significantly decreases proliferation and metastasis of cancers in different ways, including inducing autophagy-mediated necroptosis and dampening epithelial–mesenchymal transition [17, 18]. In CCA, previous studies show that BMI1 drives proliferation and suppresses senescence [19–21]. However, how BMI1 functions in CCA tumorigenesis and progression is still largely obscure.
Exosomes are defined as a class of bilayer membrane vesicles with 40–150 nm diameter. They originate from multivesicular bodies (MVBs) and are released into the extracellular environment by many mammalian cell types [22]. The exosomes carrying a series of bioactive substances, such as proteins, RNA, and DNA, can dock and fuse to the membrane of target cells [23]. Recently, exosomes have emerged as a major player in intercellular communication through various mechanisms [22, 24], specifically in cancer development and progression [25]. Secreted exosomes by cancer cells can be internalized by the same and other cell types through autocrine, paracrine, and endocrine mechanisms to regulate cell proliferation and metastasis [26]. Proteins loaded in these exosomes reflect the abnormal protein distribution between cancer cells, which can be transferred to recipient cells to regulate tumor behaviors. Notably, our previous studies have shown that the expression of BMI1 is increased in plasma/serum of cancer patients [27, 28]. These findings suggest that BMI1 may exist in an extracellular pattern, such as exosomes.
Currently, immune checkpoint blocking (ICB) and combined molecular targeted therapy have become an important strategy for CCA treatment. However, even though patients with CCA usually express programmed cell death protein 1 (PD-1) and its ligand (PD-L1) [29, 30], their sensitivity to immune checkpoint inhibitors (ICI) is very low. Therefore, it is crucial to find out the key targets of drug resistance in immunotherapy. Chemokines are an essential component of cytokines. Many tumors have one or more chemokine networks, which further affect tumor growth and metastasis, as well as the immune infiltration of tumor-related macrophages and lymphocytes [31]. Studies have showed that inhibition of histone lysine-specific demethylase 1 can enhance the binding of CD8+T cells with chemokines (CCL5, CXCL9, CXCL10), and PD-L1 develop an antitumor role in breast cancer [32]. Dangaj D et al. showed that CCL5 and CXCL9 overexpression was associated with CD8+T cell infiltration in solid tumors, and their coexpression revealed immunoreactive tumors with prolonged survival and response to checkpoint blockade [33]. Similarly, high expression of CXCL9 induces tumor immune infiltration of CD8+T cells, causing an antitumor immune response [34, 35].
PD1 blockade combined with chemotherapy has been approved for treating recurrent or metastatic CCA. Moreover, the defensive functions of immune cells are suppressed in CCA, and blocking immune checkpoint molecules expressed on tumor-infiltrating T cells can enhance the defensive functions of T cells [36]. Unexpectedly, part of the patient with lymph node metastasis showed vast resistance to antibody-mediated checkpoint blockade [37], indicating that CCA may be intrinsically resistant to PD1 blockade.
In the present study, we systematically investigated the expressions and prognostic significance of BMI1 in a large CCA cohort consisting of 58 iCCAs, 123 pCCAs, and 101 dCCAs, and we showed that BMI1 predicted the worst prognosis. Results from in vitro and in vivo experiments confirmed the critical role of BMI1 in the proliferation, migration, and invasion of CCA. Notably, we demonstrated that CCA cell-derived BMI1 or exosomal BMI1 led to CCA proliferation and metastasis for the first time. Finally, we identified that inhibition of BMI1 enhances immune checkpoint blockade by activating tumor cell-intrinsic immunity, resulting in the tumorigenesis and metastasis of CCA.
Materials and methods
Ethics statement
The study was approved by the Ethics Committee of Qilu Hospital of Shandong University, and written informed consent was obtained from each patient.
Cells and reagents
Human CCA cell lines QBC-939, HUCCT1 and RBE, and biliary epithelial cell line HIBEpiC were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The human CCA cell line HCCC-9810 was obtained from the American Type Culture Collection (Manassas, VA, USA). All cell lines were authenticated using short tandem repeat (STR) analysis, and the databases of the Chinese Academy of Sciences or ATCC were used as references. All cells were maintained in Dulbecco's Modified Eagle medium (DMEM, Gibico, New York, USA). In contrast, RBE and HuCCT1 cells were cultured in Roswell Park Memorial Institute-1640 medium (RPMI-1640, Gibico) supplemented with 10% fetal bovine serum (FBS, Gibico) and penicillin/streptomycin (HyClone, Utah, USA) in a humidified atmosphere containing 5% CO2. The information of reagents and antibodies was summarized in Supplementary Table 1. Other reagents without special instruction are purchased from Sigma-Aldrich (Burlington, MA, USA).
Patients and tissue microarray (TMA)
A total of 930 CCA patients (258 iCCA, 412 pCCA and 260 dCCA)) who underwent surgical resection from 2010 to 2020 in Qilu Hospital of Shandong University were included in the primary cohort. The validation cohort consisted of 58 iCCA patients, 123 pCCA patients, and 101 dCCA patients (Supplementary Table 2) who were selected from the primary cohort according to the criteria as follows: (I) patients who underwent radical resection with a clear surgical margin; (II) patients with available formalin-fixed tumor tissues, follow-up and complete medical records, (III) patients with post-operational survival time more than one month, and (IV) patients without a history of other malignancies. The tumors were classified and staged according to the 8th edition of the AJCC/UICC TNM classification system.
The representative paraffin-embedded sections of CCA tissues were used for TMA construction and IHC analysis. Before IHC analysis, hematoxylin and eosin staining was performed to confirm the histological features of all samples. For TMA construction, core biopsies of 1.5 mm in diameter were taken from each sample and arranged into TMA slides.
IHC and scoring
IHC was performed according to the standard protocol [4, 38]. For TMA, the slides were submerged in EDTA buffer (pH 9.0) for optimal antigen retrieval. For the slides of xenografts, sodium citrate buffer (pH 6.0) was used for antigen retrieval. The primary anti-BMI1 antibody, anti-ki67 antibody, anti-E-cadherin antibody, anti-CD8 antibody, anti-CCL5 antibody, anti-CXCL9 antibody, or ant-CD8A was applied.
Stained slides were screened using a slides scanner (Pannoramic MIDI; 3DHISTECH, Budapest, Hungary). The tumor area was selected by a senior pathologist and evaluated by Quant Center software. The staining intensity was stratified as weak, moderate, or strong, and the area of each staining was calculated. IHC score = (percentage of cells of weak intensity × 1) + (percentage of cells of moderate intensity × 2) + (percentage of cells of strong intensity × 3), according to previous studies. The cohort was divided into different groups according to the cut-offs of IHC scores, which was confirmed as the point with the highest sum of specificity and sensitivity in the receiver operating characteristic (ROC) curves.
Quantitative real-time PCR
Total mRNA was extracted from frozen CCA tissues or cells using TRIzol reagent (Invitrogen, USA). Purified RNA was reversely transcribed into cDNA using High Capacity cDNA Reverse Transcription Kit (Takara, Dalian, China), and qRT-PCR was conducted using SYBR Premix Ex Taq ™ (Takara). GAPDH was adopted as an internal control, and target gene relative expression was calculated using the 2−ΔΔCt method. All primers were designed using Primer Express version 5.0 software (Applied Biosystems). The primers used for qPCR are listed in Supplementary Table 3.
Western blotting
Total proteins were extracted from frozen CCA tissues or cells using RIPA lysis buffer (Beyotime, Shanghai, China) with 1% PMSF (Beyotime) on ice for 30 min. Protein concentration was determined using a bicinchoninic acid (BCA) assay kit (Beyotime). Proteins were denatured and separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) on 10% gels and then transferred to polyvinylidene difluoride (PVDF) membranes. Then, proteins were incubated with primary antibodies overnight at 4 °C, incubated with secondary antibodies, washed, and subjected to detection using an enhanced chemiluminescence kit (Millipore, Bedford, MA, USA). Quantitative analysis of western blotting was performed using Image J.
Lentivirus transduction and stable cell lines
To enhance the expression levels of BMI1, the human BMI1 gene was PCR-amplified from cDNA and cloned into a pLenti-EF1a lentiviral vector (GeneChem, China). To silence BMI1, HEK293T cells were transiently transfected with two short hairpin RNAs (shRNAs) and packaging plasmids GV, pHelper1.0, and pHelper2.0. QBC-939 cells were plated at 2 × 106 cells per 6-well plate and transfected with 10 µg of the indicated plasmids. After 48 h, lentiviral supernatants were collected and used to infect QBC-939 cells in the presence of 8 μg/ml of polybrene (Sigma-Aldrich). Stable cell lines were selected with media containing 2 μg/ml of puromycin for 14 days. The knockdown and overexpression efficiency levels were validated by western blotting and quantitative real-time PCR. The related sequences are listed in Supplementary Table 4.
CCK8 proliferation assay
Cells transfected with the indicated shRNAs or vector were plated into 96-well plates (5 × 103 cells/well) and treated with PTC-209 (10 μM) if indicated. After incubation for the indicated times, CCK8 reagent (Dojindo, Japan) was added, and cells were incubated at 37 °C for 1 h. The optical density at 450 nm was detected using a spectrophotometer (Molecular Devices Company, San Jose, CA, USA).
Colony formation assay
Cells were seeded into 6-well plates (500 cells/well) and cultured for 2 weeks at 37 °C. After fixation with methanol for 15 min, cells were stained with 0.1% crystal violet (Sigma-Aldrich) for 20 min. The number of colonies (more than 50 cells) was counted under a microscope at 10 × magnification.
Wound healing assay
Cells were seeded in 6-well plates (2 × 105 cells/well) and cultured at 37 °C. After attachment, a wound was created in the cell monolayer using a sterile pipette tip. Cells were washed twice with cold phosphate-buffered saline (PBS), and the initial wound size was measured using a microscope. Then, cells were cultured with serum-free medium for 24 h at 37 °C, and the wound size was measured again. The wound closure percentage = (1 − [final wound size/initial wound size]) × 100.
Transwell assay
The 8.0 μm pore transwell chamber was purchased from BD Biosciences (Franklin Lakes, NJ, USA). The upper chamber was pre-coated with Matrigel (BD Biosciences) for the invasion experiments. Cells were starved of serum-free medium for 6 h before the experiment. 2 × 105 cells were suspended and plated into each upper chamber. The lower chambers contained media with 10% FBS to facilitate the migration or invasion of cells. After incubation for 12 h, cells adhering to the lower surface were fixed with methanol and stained with 0.1% crystal violet (Sigma-Aldrich) for 20 min. Migrated or invaded cell number was counted from three randomly selected visual fields with a microscope.
Conditioned medium (CM) preparation
CCA cells at 80% confluence were maintained in DMEM supplemented with 10% exosome-depleted FBS for 48 h, the medium of cells was collected and concentrated for 1000×g to discard the pellets. The supernatant was filtered with a low-protein-binding filter (0.2 µm) (Millipore) and concentrated ten times with Amicon Ultra 15 ml filters at 4000g.
Exosome isolation and identification
Exosomes were isolated from CM using the ultracentrifugation method. Briefly, CM was sequentially centrifuged at 300g for 30 min, 3000g for 30 min, 20,000g for 30 min, and 100,000g for 80 min at 4 °C. The pellets were washed with cold PBS and purified by another centrifugation at 100,000g for 80 min at 4 °C. Subsequently, the purified exosomes were resuspended in PBS for protein detection and functional assays. Transmission electron microscopy (JEM-1-11 microscope, Japan) was used to image exosomes at 100 keV. The quality of exosomes was determined by the Nanosight NS300 instrument equipped with ZetaView PMX 110 (Particle Metrix, Meerbusch, Germany). The protein concentration of exosomes was determined using a BCA Protein Assay Kit (Beyotime).
Exosome labeling and tracing
According to the manufacturer's instructions, isolated exosomes were labeled with a PKH26Red or PKH67 Green Fluorescent Cell Linker Kit (Sigma-Aldrich). QBC-939 or RBE cells were exposed to labeled exosomes at 37 °C for 6 h. Cells were washed with cold PBS, then fixed with 4% paraformaldehyde for 30 min, and stained with DAPI for 20 min at room temperature. Finally, cells were observed under a confocal microscope (Leica Microsystems AG, Germany).
ELISA
Human CCL5 (RANTES) ELISA Kit (Cat. No. RAB0076) and CXCL9 (MIG) ELISA Kit (Cat. No. RAB0138) were obtained from Sigma-Aldrich. Concentrations of CCL5 or CXCL9 in the CCA cells medium were measured according to the manufacturer's instructions. To detect the protein levels of CCL5 or CXCL9 of supernatant, QBC-939 or RBE cells were treated with PTC-209 or shBMI1 for 48 h. The collected medium was concentrated to 1 ml. For ELISA detection, 100 µl of standard or sample was added to the appropriate wells and incubated for 2 h at room temperature. After washing, 200 µl of CCL5 or CXCL9 conjugate was added and incubated for 2 h at room temperature. Substrate solution was added for 30 min at room temperature. After adding 50 µl of stop solution, the ELISA absorbance values could be read on the BIOTEK spectrophotometer (Vermont, USA) at 450 nm.
ChIP-qPCR
The enzymatic ChIP kit was obtained from Cell Signaling Technology (Cat. No. 9003), and ChIP-qPCR assays were performed according to the manufacturer's instructions. Briefly, QBC-939 cells were sequentially treated with DTBP solution (Sigma-Aldrich Cat. No. D2338) and formaldehyde and harvested with a cell scraper. Then, the cell pellet was lysed with ChIP lysis buffer and sonicated to generate fragmented chromatin samples. Chromatin samples were incubated with anti-BMI1 or anti-Ubiquityl-Histone H2A antibodies overnight at 4 °C. The rabbit IgG (Cell Signaling Technology, Cat. No. 2729) was used as the negative control, and a non-immunoprecipitated sample (2%) was used as the input control. The purified DNA levels were quantified by qPCR. The primers used for ChIP-qPCR are listed in Supplementary Table 5.
Tumor xenograft models
All animal experiments were approved by the Medical Ethics Committee of Shandong University. Female BALB/c nude mice (5–6 weeks old, 16–18 g) were purchased from GemPharmatech Co., Ltd. (Nanjing, China). Mice were randomly divided into three groups (n = 6). QBC-939 cells were transfected with EGFP-shBMI1 by the lentivirus (GeneChem, China). After screening with puromycin, stable high EGFP-expressing QBC-939 cells were observed by fluorescence microscopy and selected by flow cytometry. Stable clones of QBC-939 cells (5 × 106 cells), transfected with BMI1 knockdown, were subcutaneously injected into the right flanks of nude mice. The tumor was monitored by a live imaging system (IVIS Spectrum). Radiant efficiency was measured to quantify the tumor burden of mice. Tumor diameters were measured with an external caliper every 2 days. Tumor volume was calculated according to the formula as follows: V = (L × W2)/2, where V is the volume (mm3), L is the length (mm), and W is the width (mm). The tumors were excised, photographed, and weighed after 28 days. For the in vivo metastasis assay, stable clones of QBC-939 cells (2 × 106 cells) with BMI1 knockdown were injected into the tail vein of BALB/c nude mice (n = 6). Mice weights were measured every week. After 8 weeks, the mice were sacrificed, and the weights of the livers and lungs were calculated to assess the actual tumor burden. The number of nodules on the livers and lungs was confirmed by histological staining and counted.
To explore the effects of exosomes on tumor progression, 100 µl isolated exosomes were injected into the tail vein weekly after subcutaneous or tail vein injection of QBC-939 cells.
Statistical analysis
All values were represented as mean ± standard deviation (SD). Statistical analyses were carried out using SPSS 26.0 and GraphPad prism 8.1 software. The Chi-square test assessed the correlation between BMI1 and clinicopathologic characteristics. Survival curves were plotted using the Kaplan–Meier method and compared using the log-rank test. The independent prognostic significance of clinicopathological characteristics was analyzed using multivariate analysis with the Cox proportional hazards regression model. Linear correlations were analyzed with Pearson correlation analysis. In in vitro and in vivo experiments, one-way or two-way ANOVA was used to compare the statistical difference between groups. P values less than 0.05 were considered statistically significant.
Results
BMI1 expression is significantly upregulated in CCA and is correlated with poor prognosis
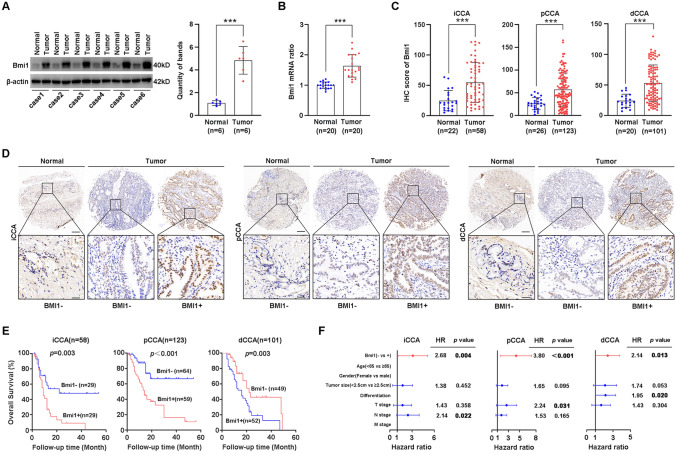
To illustrate the expression of BMI1 in CCA, we first determined the expression pattern of BMI1 in six pairs of CCA tissues and their adjacent normal tissues using WB. The expression of BMI1 in CCA samples was significantly higher than in adjacent normal tissues (Fig. 1A). Similar results were also noticed in 20 paired CCA tissues and their adjacent normal tissues by qPCR (Fig. 1B). Moreover, we investigated the expression of BMI1 with IHC in 68 tumor-adjacent normal tissues and 282 CCAs (58 iCCAs, 123 pCCAs, and 101 dCCAs). IHC scores of BMI1 in CCAs were significantly higher than the scores of tumor-adjacent normal tissues (Fig. 1C).
Fig. 1.
BMI1 expression is upregulated in CCA tissues and predicts a poor clinical outcome. A BMI1 expression was detected in 6 pairs of CCA tissues and adjacent normal duct tissues with WB (left), and the bands were quantified (right). BMI1 protein level was normalized to β-actin. B The mRNAs of BMI1 in 20 pairs of CCA tissues (6 iCCAs, 7 pCCAs, and 7 dCCAs) and adjacent normal tissues were detected with qPCR. C The IHC score of BMI1 in iCCA, pCCA, and dCCA was calculated between tumor and normal tissues. D Representative IHC images of BMI1 expression in iCCA, pCCA, and dCCA and adjacent normal tissues (top: × 100 magnification, scale bar, 200 μm; bottom: × 400 magnification, scale bar, 50 μm). E Patients with different expression patterns of BMI1 had distinct prognoses. Patients with high BMI1 expression had significantly poorer overall survival than those with low BMI1 expression (Kaplan–Meier method). F Forest plot showing the association between BMI1 expression and CCA survival using multivariate analyses. *, ** and *** represents P < 0.05, 0.01 and 0.001 respectively. Statistical significance was analyzed with paired t test (A, B), one-way ANOVA (C) or log-rank test (E)
Moreover, the CCA patients were categorized into subsets with low/high BMI1 expression based on the IHC score (Fig. 1D and Supplementary Table 2). The correlation between the BMI1 expression and corresponding clinical follow-up information was analyzed by the Kaplan–Meier method. High expression of BMI1 was remarkably associated with a lower OS in all CCA subtypes (Fig. 1E). These results show that BMI1 is a prognostic indicator of CCA.
Prognostic significance of BMI1 in CCA
Univariate analysis with a log-rank test was performed to analyze the prognostic value of BMI1 and other clinicopathological factors (Supplementary Tables 6–8). In iCCA patients, larger tumor size (P = 0.016), advanced T stage (P = 0.028), and advanced N stage (P = 0.022) were all associated with a lower 3-year OS rate. In pCCA patients, larger tumor size (P = 0.039), advanced T stage (P = 0.012), advanced N stage (P = 0.025), and advanced tumor–lymph node–metastasis (TNM) stage (P = 0.011) indicated a worse prognosis. In dCCA patients, larger tumor size (P = 0.013), poor differentiation (P = 0.015), and advanced T stage (P = 0.008) are associated with a poor outcome.
Multivariate analysis was also performed to identify the independent prognostic factors using the Cox regression model (Fig. 1F and Supplementary Tables 6–8). All factors with a P value ≤ 0.05 in univariate analysis were enrolled in multivariate analysis, except the TNM stage. As a result, BMI1 was identified as an independent prognostic biomarker in all CCA subtypes (iCCA, P = 0.004; pCCA, P < 0.001; dCCA, P = 0.013). All the results suggest an independent prognostic significance of BMI1 in CCA.
BMI1 was required for proliferation, migration, and invasion of CCA cells
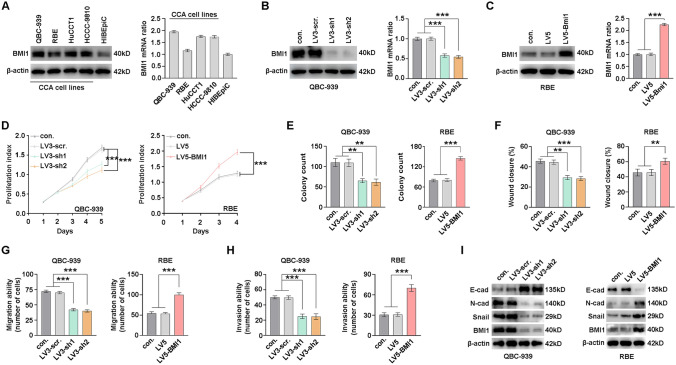
The above-mentioned results urged us to explore the function of BMI1 in human CCA tumorigenesis and/or progression. The expression of BMI1 was detected in a series of CCA cell lines using WB and qPCR, including normal biliary epithelium cell line HIBEpiC, CCA cell lines QBC-939, RBE, HuCCT1, and HCCC-9810. HIBEpiC had a lower BMI1 expression compared with CCA cell lines (Fig. 2A). BMI1 was knocked down or overexpressed with lentivirus in QBC-939 and RBE cells, respectively, and the expression efficiency was verified with WB and qPCR (Fig. 2B, C). Both CCK-8 and colony formation assays indicated that BMI1 knockdown remarkably impaired the proliferation of QBC-939 cells, and overexpression of BMI1 prompted the proliferation of RBE cells (Fig. 2D, E, and Supplementary Fig. 1A). Meanwhile, CCA cells migratory and invasive abilities were evaluated with wound-healing assay and transwell assay. Notably, BMI1 knockdown decreased the migration and invasion of QBC-939 cells, whereas BMI1 overexpression enhanced the migration and invasion in RBE cells (Fig. 2F–H and Supplementary Fig. 1B–D). Therefore, we investigated the regulation of BMI1 on EMT biomarkers, including E-cadherin, N-cadherin, and Snail. BMI1 knockdown increased E-cadherin expression and decreased expression of N-cadherin and Snail. In contrast, BMI1 overexpression had opposite regulation on the expression of EMT biomarkers (Fig. 2I), indicating that BMI1 was able to promote EMT. Taken together, our results show that BMI1 plays an essential role in the proliferation and metastasis of CCA cells.
Fig. 2.
BMI1 promotes CCA cells proliferation, migration, and invasion in vitro. A BMI1 expressions in different CCA cell lines and biliary epithelial cell lines (HIBEpiC) were detected with WB (left) and qPCR (right). B Knockdown of BMI1 in QBC-939 cells with two different shRNAs (LV3-sh1 and LV3-sh2) and scramble (LV3-scr.) were verified by WB (left) and qPCR (right). C Overexpression of BMI1 in RBE cells with lentivirus encoding LV5-BMI1 was verified by WB (left) and qPCR (right). CCA cell proliferation was determined by CCK8 assays (D) and colony formation assays (E) after silencing or overexpressing BMI1 in QBC939 and RBE cells. BMI1 was silenced or overexpressed in QBC939 and RBE cells, and the migration (F, G) or invasion (H) were detected by wound-healing assay or transwell assay. I EMT biomarkers were detected after BMI1 knockdown in QBC-939 cells and overexpression in RBE cells. *, ** and *** represents P < 0.05, 0.01 and 0.001 respectively. Statistical significance was analyzed with one-way ANOVA (B, C, E–H) or two-way ANOVA (D). Data were from at least three independent experiments and shown as mean ± SEM
BMI1 facilitated tumor proliferation and metastasis of CCA
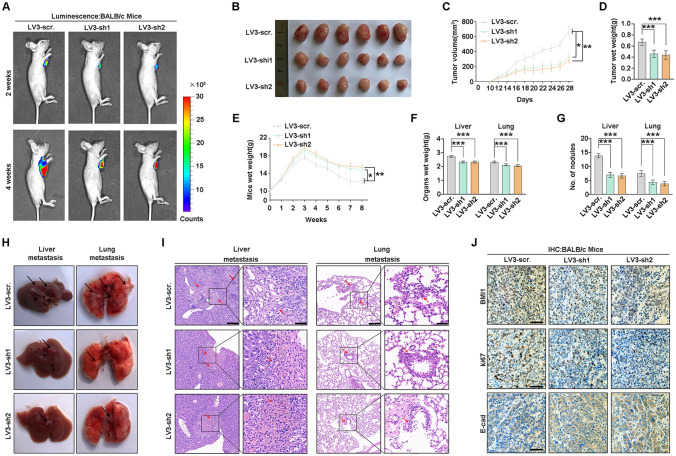
Experiments in vivo were performed to verify the function of BMI1 on tumor proliferation and metastasis. Stable BMI1-silenced (LV3-sh1 and LV3-sh2) or control QBC-939 cells (LV3-scr.) were subcutaneously injected into BALB/c nude mice. Radiant efficiency and diameters of xenografts with BMI1 knockdown were lower than the control group (Fig. 3A, B, Supplementary Fig. 2). Similarly, the volume and weight of xenografts with BMI1 depletion were notably lower than the control group (Fig. 3C, D).
Fig. 3.
BMI1 promotes CCA proliferation and metastasis in vivo. A Subcutaneous xenograft models in nude mice were established with BMI1-silenced stable QBC-939 cells and control cells. A live imaging system monitored the tumors, and the representative bioluminescent images were shown after 2 and 4 weeks. B The xenograft tumors were detected in the three groups after 28 days. C The volume of xenograft tumors was recorded from day 10 to day 28. D Weights of xenograft tumors were measured after 28 days. E Stable BMI1-silenced QBC-939 cells and control cells were injected via the tail vein of nude mice. The weights of mice were measured weekly. F Livers and lungs were weighed to assess the tumor burden of metastatic foci. G The number of metastatic nodules in livers and lungs of nude mice. H Representative gross specimens of metastasis lesions in livers and lungs. I Representative H&E staining of metastasis lesions in livers and lungs. Arrows indicate metastatic lesions (left: scale bars, 200 μm; right: scale bars, 50 μm). J Representative IHC images of BMI1, Ki67, and E-cadherin expression in xenograft tumors (scale bars, 100 μm). *, ** and *** represents P < 0.05, 0.01 and 0.001 respectively. Statistical significance was analyzed with one-way ANOVA (D, F, G) or two-way ANOVA (C, E). Data were from at least three independent experiments and shown as mean ± SEM
Stable BMI1-silenced or control QBC-939 cells were injected into BALB/c nude mice via the tail vein. The mice injected with BMI1-silenced QBC-939 cells had heavier body weight, lower organs weight, and fewer metastatic nodules in livers and lungs (Fig. 3E–G), reflecting that BMI1 knockdown attenuated the systemic metastasis of CCA. Intrahepatic and lung metastases were rarely detected in the BMI1-depleted group but frequently found in the control group (Fig. 3H). H&E and IHC staining with anti-human mitochondria antibody of the metastatic nodules on livers and lungs was further performed to confirm that the nodules were formed by CCA cells, and BMI1 knockdown strikingly decreased the number of metastatic lesions (Fig. 3I and Supplementary Fig. 3). IHC and qPCR further verified the expression of BMI1, Ki67, and E-cadherin in xenografts (Fig. 3J and Supplementary Fig. 4). These results suggested the potency of BMI1 to promote CCA proliferation and metastasis.
Secreted BMI1 but not rhBMI1 promotes CCA proliferation and metastasis
Our previous study showed that the expression of BMI1 is increased in plasma/serum of cancer patients [27]. Therefore, we examined whether BMI1 protein is secreted into the medium from CCA cells. QBC-939 and RBE cells used for BMI1 overexpression were subjected to standardized conditions to collect CM. The secreted BMI1 proteins were both detectable in the CM derived from BMI1-overexpressing cells (CM/LV5-BMI1), vector cells (CM/LV5), and control cells (CM/con.) (Fig. 4A), suggesting that BMI1 can be secreted into CM. To evaluate the function of secreted BMI1 in CCA cell proliferation and metastasis, we used the transcriptional inhibitor PTC-209 of BMI1 to eliminate the effect of endogenous BMI1 in CCA cells. PTC-209 reduced BMI1 expression of CCA cells in a dose-dependent manner, and PTC-209 at 10 µM significantly inhibited BMI1 levels (Fig. 4B). Moreover, QBC-939 and RBE cells were incubated with CM in the presence of PTC-209. An increase in BMI1 was observed in recipient CCA cells following treatment with CM/LV5-BMI1 (Fig. 4C). Both CCK-8 and colony formation assays showed that CM/LV5-BMI1 significantly promoted the proliferation of CCA cells compared to CM/vector (Fig. 4D, E, and Supplemental Fig. 5A). As expected, CM/LV5-BMI1 enhanced the migration and invasion of CCA cells (Fig. 4F–H and Supplementary Fig. 5B–D). It suggests the BMI1 proteins could be transported from the extracellular into the cells and promote CCA progression.
Fig. 4.
Secreted BMI1 but not rhBMI1 promotes CCA proliferation, migration, and invasion. A BMI1 protein in the CM derived from BMI1-overexpressing cells (CM/LV5-BMI1), vector cells (CM/LV5), and control cells (CM/con.) were detected by WB. B QBC-939 and RBE cells were incubated in BMI1 inhibitor PTC-209 at different concentrations for 24 h, and the BMI1 expression was detected with WB. C QBC-939 and RBE cells were incubated with CM in the presence of 10 μM PTC-209 for 24 h, and the BMI1 expression was detected with WB. QBC-939 and RBE cells were incubated with CM with 10 μM PTC-209 for indicated time or 2 weeks, and the proliferation was detected with CCK8 (D) and colony formation assay (E). F QBC-939 and RBE cells were incubated with CM with 10 μM PTC-209 for 24 h, and the migration was detected with the wound-healing assay. QBC-939 and RBE cells were incubated with CM with 10 μM PTC-209 for 12 h, and the migration (G) and invasion (H) was detected with transwell assays. Cell proliferation was determined in QBC-939 and RBE cells upon treatment with different concentrations of rhBMI1 according to the CCK8 assay. Cell migration (J) and invasion (K) was determined in QBC-939 and RBE cells upon treatment with different concentrations of rhBMI1 according to transwell assay. N.S. represents not significant. *, ** and *** represents P < 0.05, 0.01 and 0.001 respectively. Statistical significance was analyzed with one-way ANOVA (E–H, J, K) or two-way ANOVA (D, I). Data were from at least three independent experiments and shown as mean ± SEM
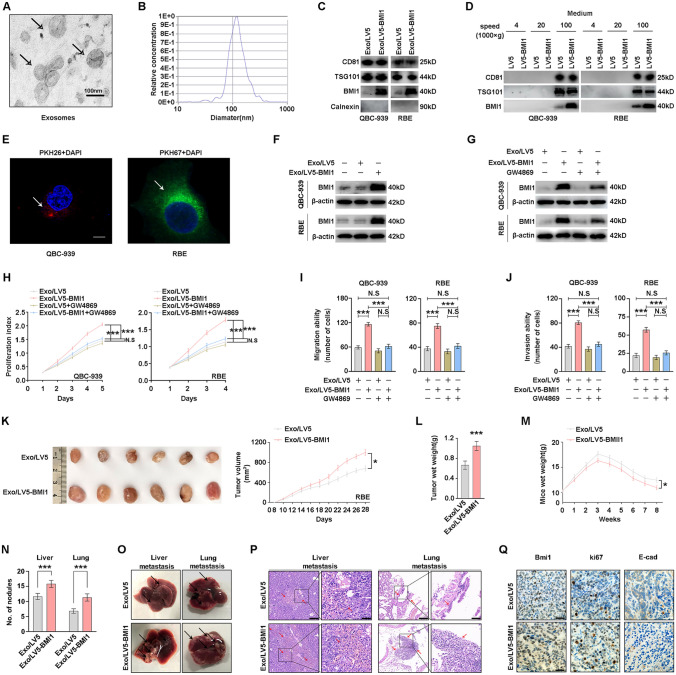
Fig. 5.
Intercellular transfer of BMI1 by exosomes through autocrine/paracrine promotes CCA proliferation, migration, and invasion. A The purified exosomes derived from the CCA medium were analyzed under electron microscopy (scale bar, 100 nm). B The diameter distribution and relative concentration of purified exosomes were analyzed by nanoparticle tracking analysis. C Exosomal markers (CD81, TSG101, and Calnexin) and BMI1 in exosomes purified from BMI1-overexpressing cells (Exo/LV5-BMI1) and vector cells (Exo/LV5) were detected by WB. D Exosomal markers (CD81and TSG101) and BMI1 in the CCA medium were detected by WB under differential ultracentrifugation. E Parental CCA cells were incubated with PKH26/PKH67-labeled exosomes for 2 h (red, PKH26; green, PKH67; blue: nucleus), and the arrows indicate exosomes incorporated into CCA cells (scale bar, 100 μm). F BMI1 in recipient CCA cells incubated with Exo/LV5-BMI1 and Exo/LV5 was detected by WB. G Recipient CCA cells were treated with exosomes with or without 5 μM GW4869 for 72 h, and the BMI1was detected by WB. H Recipient CCA cells were treated with exosomes with or without 5 μM GW4869 for the indicated time, and the proliferation was detected with CCK8 assay. Recipient CCA cells were treated with exosomes with or without 5 μM GW4869 for 12 h, and the migration (I) and invasion (J) were detected with transwell assays. K Subcutaneous xenograft models in nude mice were established with QBC-939 cells, and purified exosomes (Exo/LV5-BMI1 and Exo/LV5) were injected into the tail vein of mice weekly. The xenograft tumors were observed after 28 days (left), and the volume of xenograft tumors was recorded from day 8 to day 28(right). L Weights of xenograft tumors were measured. M QBC-939 cells were injected into the tail vein of nude mice, and purified exosomes were injected into the tail vein of mice weekly. The wet weights of mice were measured. N The number of metastatic nodules in livers and lungs of nude mice. O Representative gross specimens of metastatic lesions in livers and lungs. P Representative H&E staining of metastasis lesions in livers and lungs. Arrows indicate metastasis lesions (left: scale bars, 200 μm; right: scale bars, 50 μm). Q Representative IHC images of BMI1, Ki67, and E-cadherin expression in xenograft tumors (scale bars, 100 μm). N.S. represents not significant. *, ** and *** represents P < 0.05, 0.01 and 0.001 respectively. Statistical significance was analyzed with one-way ANOVA (I, J, L, N) or two-way ANOVA (H, K, M). Data were from at least three independent experiments and shown as mean ± SEM
To investigate whether secreted BMI1 directly promotes CCA proliferation and metastasis, CCA cells were incubated with different concentrations of recombinant human BMI1 (rhBMI1). Interestingly, the proliferation, migration, or invasion of QBC-939 and RBE cells was not affected by rhBMI1 (Fig. 4I–K and Supplementary Fig. 6). The results suggest that intracellular BMI1 promoted CCA cells progression rather than extracellular BMI1. Besides, the secreted BMI1 in CM may enter CCA cells independent of a cell membrane-anchored receptor. So, we speculated that secreted BMI1 was likely to be present in a particle-associated form and could be internalized by CCA cells to function intracellularly.
Exosome-mediated intercellular transfer of BMI1 between CCA cells to promote CCA proliferation and metastasis
Based on the above results, we found that BMI1 has both intracellular and extracellular forms. Hence, we can reasonably suspect that BMI1 may exist in extracellular vesicles, such as exosomes. To uncover this conjecture, exosomes were purified from CCA cells culture supernatant by graded centrifugation. According to the typical cup-shaped morphology and a diameter range (40–150 nm), the isolated vesicles were identified as exosomes using electron microscopy and nanoparticle tracking analysis (Fig. 5A, B). WB further confirmed these vesicles by detecting the exosomal markers CD81, TSG101, and Calnexin (Fig. 5C). Besides, we only showed that 100,000×g pellets derived from CCA cells contained BMI1 proteins (Fig. 5D). Above results reveal that exosomal fractions from CCA cells contained BMI1 proteins.
Moreover, we further explored whether BMI1-containing exosomes can be internalized by CCA cells to affect their biological behavior. For this purpose, parental CCA cells were incubated with exosomes labeled with PKH26/PKH67. As measured by laser scanning confocal microscopy, the recipient cells exhibited high uptake efficiency for exosomes (Fig. 5E). Then, we measured the BMI1 proteins levels in recipient CCA cells treated with exosomes derived from BMI1-overexpressing cells (Exo/LV5-BMI1) and vector cells (Exo/LV5). Moreover, BMI1 expression was increased in the recipient cells after treatment with Exo/LV5-BMI1 (Fig. 5F). To further confirm this conclusion, GW4869 was used to block exosomes generation and release from CCA cells. BMI1 expression was significantly reduced in recipient CCA cells treated with Exo/LV5-BMI1 and GW4869 (Fig. 5G). The above observations suggested that CCA-secreted BMI1 can be transferred between CCA cells via exosomes. This result urged us to investigate whether exosomes containing BMI1 are sufficient to induce the progression of CCA cells. CCK-8 and colony formation assays showed that the proliferation of CCA cells was significantly stimulated by Exo/LV5-BMI1 compared with Exo/LV5, whereas eliminated with GW4869 (Fig. 5H). Besides, Exo/LV5-BMI1 led to increased cell migration and invasion which can be blocked by GW4869 (Fig. 5I, J, and Supplementary Fig. 7). On the contrary, Exo/shBMI1 attenuated tumor proliferation, migration and invasion compared with Exo/LV3 control cells (Supplementary Fig. 8A–C). Moreover, exosomes from CCA cells were used to incubate BMI1-silencing CCA as recipient cells, using Exo/shBMI1 as a control. Similarly, exosomes from control CCA cells could induce the proliferation, migration and invasion of BMI1-silencing CCA cells(Supplementary Fig. 8D–F). Above results showed that BMI1 can regulate CCA progression in an exosome-dependent manner.
Furthermore, subcutaneous xenografts in nude mice were established with QBC-939 cells, and the purified exosomes into the tail vein of mice weekly. After 28 days, the Exo/LV5-BMI1 group exhibited larger tumor sizes and weights than the Exo/LV5 group (Fig. 5K, L). As to tumor metastasis, QBC-939 cells were injected into the tail vein of nude mice, and purified exosomes were injected into the tail vein of mice weekly. The mice injected with Exo/LV5-BMI1 had lower body weight, heavier organs weight, and more metastatic nodules in the livers and lungs (Fig. 5M, N, and Supplementary Fig. 9). Liver and lung metastases were more frequently found in the Exo/LV5-BMI1 group (Fig. 5O). H&E staining of the metastatic nodules on livers and lungs revealed that Exo/LV5-BMI1 increased the number of metastatic lesions (Fig. 5P and Supplementary Fig. 10). Similarly, IHC further verified the expression of BMI1, Ki67, and E-cadherin in the xenografts (Fig. 5Q and Supplementary Fig. 11). Taken together, these results show that the unique BMI1-containing exosomes secreted by CCA have progression-promoting functions in CCA cells.
BMI1 attenuates tumor cell-intrinsic antitumor immunity in CCA cells
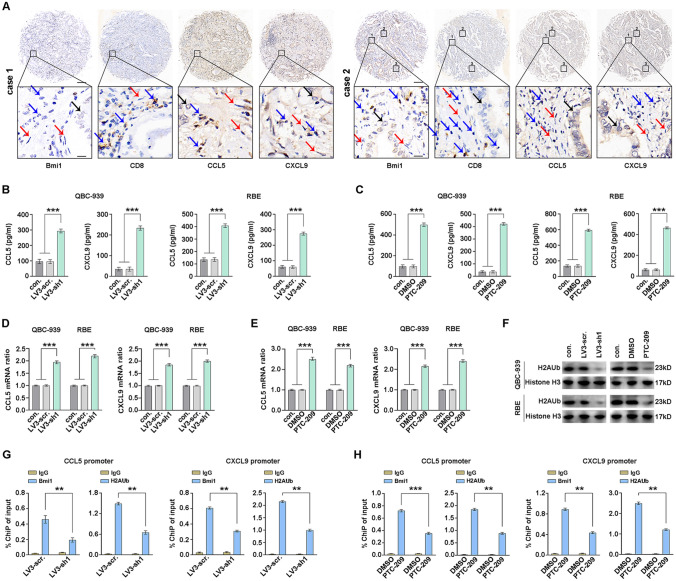
It has been found that BMI1 inhibition augments PD1 blockade by activating tumor cell-intrinsic immunity in neck squamous cell carcinoma [39]. Inspired by this conclusion, we explored the levels of chemokines related to CD8+T cells recruitment in CCA tissues. CCL5 and CXCL9, as essential members of the chemokine family, were confirmed to promote the recruitment of CD8+T lymphocytes into tumor sites. We performed IHC to examine the expression of BMI1, CD8, CCL5, and CXCL9 in human CCA samples (Fig. 6A and Supplementary Fig. 12). And the IHC demonstrated that there was a negative correlation between BMI1 and CD8 in human CCA tissues (Supplementary Fig. 13). Next, we found BMI1 knockdown or PTC-209 remarkably increased the CCL5 and CXCL9 in both QBC-939 and RBE cells as determined by ELISA and qPCR (Fig. 6B–E).
Fig. 6.
BMI1 reduces the expression of CD8+T cell-attracting chemokines in CCA cells. A Representative IHC images of BMI1, CD8, CCL5, and CXCL9 in human CCA samples. The black arrows indicate tumor cells, the blue arrows indicate lymphocytes cells, and the red arrows indicate stromal cells (top: × 100 magnification, scale bar, 200 μm; bottom: × 400 magnification, scale bar, 25 μm). ELISA showed that the protein levels of CCL5 or CXCL9 in the medium of QBC-939 and RBE cells were induced by BMI1 knockdown (B) or PTC-209 (C). QBC-939 and RBE cells were treated with BMI1 knockdown (D) or PTC-209 (E), and the CCL5 or CXCL9 mRNA levels were analyzed by qPCR. F QBC-939 and RBE cells were treated with BMI1 knockdown or PTC-209, and the H2AUb was detected by WB. G The CHiP-qPCR assay showed that BMI1 occupied on the promoters of CCL5 and CXCL9 regions were reduced by BMI1 knockdown. H The CHiP-qPCR assay showed that PTC209 reduced H2AUb levels on the promoters of CCL5 and CXCL9 regions. *, ** and *** represents P < 0.05, 0.01 and 0.001 respectively. Statistical significance was analyzed with one-way ANOVA. Data were from at least three independent experiments and shown as mean ± SEM
BMI1 is required for the H2AUb to exert a transcriptional inhibitory effect. The level of H2AUb was reduced with BMI1 knockdown or PTC-209 in CCA cells (Fig. 6F). We explored whether BMI1 inhibited the transcription of chemokines by H2AUb in QBC-939 cells. ChIP-qPCR revealed that BMI1 specifically occupied on the promoters of CCL5 and CXCL9, and the knockdown of BMI1 reduced the levels of H2AUb on their promoters (Figs. 6G). Consistently, PTC209 treatment significantly reduced the levels of BMI1 on the promoters of CCL5 and CXCL9, and levels of H2AUb on the promoters were significantly decreased by PTC209 (Figs. 6H). The ChIP-qPCR revealed that BMI1 specifically occupied on the promoters of CCL5 and CXCL9, and BMI1 inhibition could derepress chemokine expression intrinsically. Our results suggest that targeting BMI1 may be a novel combination therapy strategy, and anti-PD1 plus BMI1 inhibitor is expected to be a new combination treatment for CCA.
Discussion
Due to ineffective adjuvant therapy, CCA patients usually have poor prognoses. Comprehensive treatment with surgery and chemotherapy or targeted drugs has become the international consensus for CCA treatment. Unfortunately, treatment selection for CCA has stagnated in recent decades. In combination with cisplatin, gemcitabine is the main chemotherapeutic regimen for CCA. However, the benefits of chemotherapy or radiotherapy in CCA patients are very limited [40]. The development of new drug targets and therapies is typically based on identifying novel biomarkers, which largely rely on cohort studies, including large numbers of patients. However, lower surgical resection rates increased the difficulty of obtaining specimens and establishing the cohorts. Here, we established a cohort consisting of 282 CCA patients with available tissues and follow-ups, which is the largest retrospective cohort known to study CCA. Our study confirmed BMI1 as an unfavorable prognostic biomarker of all CCA subtypes. Our results suggest that BMI1 detection could help stratify high-risk patients and guide more severe post-operational supervision.
At present, there are many studies on the molecular pathways of BMI1 in tumors. For example, BMI1 is first reported to regulate tumor cell proliferation through the Ink4A/Arf locus, encoding tumor suppressors p16 and p19Arf [16, 41]. BMI1 also inhibits autophagy-mediated necroptosis via the PINK1–PARK2-dependent mitochondrial pathway [17]. Guo J et al. have found that BMI1 drives the formation and progression of intrahepatic CCA [42]. However, the detailed mechanism of BMI1 in CCA has not been elucidated. In our studies, we found that BMI1 significantly promoted cell proliferation, migration, and invasion, strongly suggesting that BMI1 was a key determinant of cancer cell proliferation and metastasis.
Our studies have reported that BMI1 mRNA is detectable in serum/plasma of tumor patients, which is a potential non-invasive biomarker for cancer diagnosis and prognosis [27, 28]. BMI1 mRNA is fragmented in blood, so BMI1 detection is difficult. Intensive research has described the role of tumor-derived exosomes in tumor progression [43, 44]. Exosomes can protect cellular contents, like DNA, RNA, and protein, from degradation in circulation [45]. The exchange of materials between cells through autocrine, paracrine, and endocrine mechanisms can be mediated by exosomes [22, 23, 46, 47]. This better explains why BMI1 is stable in the peripheral blood. For the first time, we demonstrated that its protein could also be packaged in exosomes shed by CCA cells and transferred to the recipient cells. As expected, the BMI1-containing exosomes promote CCA proliferation and metastasis in vitro and in vivo. These data suggest that BMI1 may exert an oncogenic effect through the autocrine/paracrine mechanism. As we know, CCA is a highly heterogeneous malignancy [2]. A proportion of CCA cells had high BMI1 expression, but some cells expressed low BMI1. In our study, BMI1 was mainly expressed in CCA nucleus, but also expressed in the cytoplasm (Fig. 1D), and CCA nucleus usually had stronger IHC staining than cytoplasm. The intrinsic expression of BMI1, and the imported BMI1 by exosome, which is the dominant pathway for CCA cells to possess BMI1, is an interesting scientific question and requires further investigation. Maybe high expression of BMI1 in the nucleus mainly depend on BMI1 transcription, while exosomal import is an important compensatory mechanism to cells with low BMI transcription.
Currently, treating and preventing CCA metastasis remains a significant challenge. Although PD1 blockade-based comprehensive therapies have achieved some efficacy in clinical trials, the objective response rates towards CCA are not very high, indicating that immune evasion plays a critical role in CCA progression and metastasis. Increasing evidence suggests that targeting tumor cell-intrinsic antitumor immunity can inhibit tumor progression [48]. As known, BMI1 mediates gene silencing via H2AUb and represses gene expression [49]. Here, we showed that BMI1 inhibition induced transcription of chemokines (CCL5 and CXCL9) in the CCA cells by erasing repressive H2AUb markers epigenetically. Therefore, BMI1 inhibition in these tumor cells should also intrinsically activate their immune response and recruit CD8+T cells. As an analog of PTC-209, PTC-596 has been in clinical trials to treat solid tumors [49, 50]. In advanced CCA patients with high BMI1 expression, anti-PD1 combined with PTC596 may achieve better therapeutic effects.
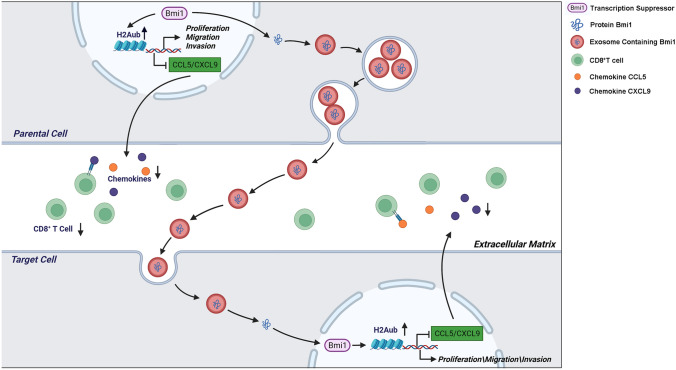
In conclusion, the present study indicates that BMI1 is a prognostic biomarker of iCCA, pCCA, and dCCA and predicts poor outcomes. By in vitro and in vivo experiments, BMI1 plays a regulatory role in CCA proliferation and metastasis. Mechanistically, we show exosomal BMI1 serves as an intercellular messenger for proliferation and metastasis of CCA cells for the first time. More importantly, we point out that BMI1 leads to tumor progression by inhibiting the internal immunity of CCA cells (Fig. 7). Thus, our study demonstrates an underlying mechanism of CCA development and provides a new combination therapy strategy suppressing CCA proliferation and metastasis.
Fig. 7.
Schematic illustration of the mechanisms underlying BMI1 in CCA cells. BMI1 promotes CCA progression in two interrelated mechanisms: (1) The BMI1-containing exosomes from CCA cells promote CCA proliferation and metastasis through the autocrine/paracrine pathway; (2) BMI1 attenuates CCL5 and CXCL9 expression, which could inhibit the recruitment of CD8+T cells and further depress cell-intrinsic antitumor immunity
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Dr. Xiaoqing Yang from the Department of Pathology, the Qianfoshan Hospital of Shandong University, for evaluating the IHC results and distinguish different cell types in the HE staining.
Abbreviations
- CCA
Cholangiocarcinoma
- iCCA
Intrahepatic cholangiocarcinoma
- pCCA
Perihilar cholangiocarcinoma
- dCCA
Distal cholangiocarcinoma
- TMA
Tissue microarray
- IHC
Immunohistochemistry
- qRT-PCR
Quantitative real-time polymerase chain reaction
- WB
Western blot
- IHC
Immunohistochemistry
- ELISA
Enzyme-linked immunosorbent assays
- ChIP-qPCR
Chromatin immunoprecipitation-qPCR
- OS
Overall survival rate
- DMEM
Dulbecco's Modified Eagle Medium
- FBS
Fetal bovine serum
- SDS-PAGE
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- PVDF
Polyvinylidene difluoride
- PMSF
Phenylmethanesulfonyl fluoride
- PBS
Phosphate buffer saline
- CCK-8
Cell counting kit-8
- DMSO
Dimethyl sulfoxide
- CM
Conditioned medium
- BMI1
B-cell-specific Moloney leukemia virus insertion site 1
- E-cad
E-cadherin
- N-cad
N-cadherin
- EMT
Epithelial-mesenchymal transition
- MVBs
Multivesicular bodies
- shRNA
Short hairpin RNA
- TNM
Tumor–node–metastasis
- rhBMI1
Recombinant human BMI1
- exoBMI1
Exosomal BMI1
- CD8A
CD8 antigen
- CCL5
C–C motif chemokine ligand 5
- CXCL9
C-X-C motif chemokine ligand 9
- PD1
Programmed cell death protein 1
- ICB
Immune checkpoint blocking
- ICI
Immune checkpoint inhibitors
Author contributions
ZL, CH, LJ, YW, and JL performed the experiments. ZL designed the experiments and wrote the paper. ZL, LZ, KL, and BQ collected the clinical samples. XL, YW, WM, TC, and AS perform the follow-up. ZL, CH, and LZ participated in data analysis and interpretation. XZ and YX designed the study, YX and ZZ contributed to study supervision and revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant nos. 81900728, 82072676, 82172791), Shandong Province Natural Science Foundation (Grant nos. ZR2019MH008, ZR2020MH238), Shandong Province Key R&D Program (Major Scientific Innovation Projects, 2021CXGC011105), Shandong Medical and Health Technology Development Project (Grant no. 2018WSB20002), Clinical Research Foundation of Shandong University (Grant no. 2020SDUCRCA018), Key Research and Development Program of Shandong Province (Grant no. 2019GSF108254). The funders had no role in study design, data collection, analysis, interpretation, and manuscript writing.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Conflict of interest
There are no relevant conflicts of interest.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of Qilu Hospital of Shandong University, and written informed consent was obtained from each patient. The Laboratory Animal Care and Use Committees of the hospital approved all experimental procedures.
Consent for publication
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Zengli Liu, Chunxiao Hu, and Lijie Zheng have contributed equally.
Contributor Information
Zongli Zhang, Email: zzlzzl1900@163.com.
Yunfei Xu, Email: xuyunfei1988@126.com.
References
- 1.Rizvi S, Khan SA, Hallemeier CL, Kelley RK, Gores GJ. Cholangiocarcinoma—evolving concepts and therapeutic strategies. Nat Rev Clin Oncol. 2018;15(2):95–111. doi: 10.1038/nrclinonc.2017.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu J, Ren G, Li K, Liu Z, Wang Y, Chen T, Mu W, Yang X, Li X, Shi A, et al. The Smad4-MYO18A-PP1A complex regulates beta-catenin phosphorylation and pemigatinib resistance by inhibiting PAK1 in cholangiocarcinoma. Cell Death Differ. 2021;29:818–831. doi: 10.1038/s41418-021-00897-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banales JM, Marin JJG, Lamarca A, Rodrigues PM, Khan SA, Roberts LR, Cardinale V, Carpino G, Andersen JB, Braconi C, et al. Cholangiocarcinoma 2020: the next horizon in mechanisms and management. Nat Rev Gastroenterol Hepatol. 2020;17(9):557–588. doi: 10.1038/s41575-020-0310-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen T, Li K, Liu Z, Liu J, Wang Y, Sun R, Li Z, Qiu B, Zhang X, Ren G, et al. WDR5 facilitates EMT and metastasis of CCA by increasing HIF-1alpha accumulation in Myc-dependent and independent pathways. Mol Ther. 2021;29(6):2134–2150. doi: 10.1016/j.ymthe.2021.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Z, Liu J, Chen T, Sun R, Liu Z, Qiu B, Xu Y, Zhang Z. HMGA1-TRIP13 axis promotes stemness and epithelial mesenchymal transition of perihilar cholangiocarcinoma in a positive feedback loop dependent on c-Myc. J Exp Clin Cancer Res. 2021;40(1):86. doi: 10.1186/s13046-021-01890-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Florio AA, Ferlay J, Znaor A, Ruggieri D, Alvarez CS, Laversanne M, Bray F, McGlynn KA, Petrick JL. Global trends in intrahepatic and extrahepatic cholangiocarcinoma incidence from 1993 to 2012. Cancer. 2020;126(11):2666–2678. doi: 10.1002/cncr.32803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valle JW, Kelley RK, Nervi B, Oh DY, Zhu AX. Biliary tract cancer. Lancet. 2021;397(10272):428–444. doi: 10.1016/S0140-6736(21)00153-7. [DOI] [PubMed] [Google Scholar]
- 8.Blechacz B. Cholangiocarcinoma: current knowledge and new developments. Gut Liver. 2017;11(1):13–26. doi: 10.5009/gnl15568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Razumilava N, Gores GJ. Cholangiocarcinoma. Lancet. 2014;383(9935):2168–2179. doi: 10.1016/S0140-6736(13)61903-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu YF, Liu ZL, Pan C, Yang XQ, Ning SL, Liu HD, Guo S, Yu JM, Zhang ZL. HMGB1 correlates with angiogenesis and poor prognosis of perihilar cholangiocarcinoma via elevating VEGFR2 of vessel endothelium. Oncogene. 2019;38(6):868–880. doi: 10.1038/s41388-018-0485-8. [DOI] [PubMed] [Google Scholar]
- 11.Bracken AP, Helin K. Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat Rev Cancer. 2009;9(11):773–784. doi: 10.1038/nrc2736. [DOI] [PubMed] [Google Scholar]
- 12.Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell. 2005;20(6):845–854. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 13.Haupt Y, Bath ML, Harris AW, Adams JM. bmi-1 transgene induces lymphomas and collaborates with myc in tumorigenesis. Oncogene. 1993;8(11):3161–3164. [PubMed] [Google Scholar]
- 14.Zhang X, Wang CX, Zhu CB, Zhang J, Kan SF, Du LT, Li W, Wang LL, Wang S. Overexpression of Bmi-1 in uterine cervical cancer: correlation with clinicopathology and prognosis. Int J Gynecol Cancer. 2010;20(9):1597–1603. [PubMed] [Google Scholar]
- 15.Song LB, Zeng MS, Liao WT, Zhang L, Mo HY, Liu WL, Shao JY, Wu QL, Li MZ, Xia YF, et al. Bmi-1 is a novel molecular marker of nasopharyngeal carcinoma progression and immortalizes primary human nasopharyngeal epithelial cells. Cancer Res. 2006;66(12):6225–6232. doi: 10.1158/0008-5472.CAN-06-0094. [DOI] [PubMed] [Google Scholar]
- 16.Kim JH, Yoon SY, Kim CN, Joo JH, Moon SK, Choe IS, Choe YK, Kim JW. The Bmi-1 oncoprotein is overexpressed in human colorectal cancer and correlates with the reduced p16INK4a/p14ARF proteins. Cancer Lett. 2004;203(2):217–224. doi: 10.1016/j.canlet.2003.07.009. [DOI] [PubMed] [Google Scholar]
- 17.Dey A, Mustafi SB, Saha S, Kumar Dhar Dwivedi S, Mukherjee P, Bhattacharya R. Inhibition of BMI1 induces autophagy-mediated necroptosis. Autophagy. 2016;12(4):659–670. doi: 10.1080/15548627.2016.1147670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ye K, Chen QW, Sun YF, Lin JA, Xu JH. Loss of BMI-1 dampens migration and EMT of colorectal cancer in inflammatory microenvironment through TLR4/MD-2/MyD88-mediated NF-kappaB signaling. J Cell Biochem. 2018;119(2):1922–1930. doi: 10.1002/jcb.26353. [DOI] [PubMed] [Google Scholar]
- 19.Sasaki M, Yamaguchi J, Ikeda H, Itatsu K, Nakanuma Y. Polycomb group protein Bmi1 is overexpressed and essential in anchorage-independent colony formation, cell proliferation and repression of cellular senescence in cholangiocarcinoma: tissue and culture studies. Hum Pathol. 2009;40(12):1723–1730. doi: 10.1016/j.humpath.2009.01.027. [DOI] [PubMed] [Google Scholar]
- 20.Fan L, Xu C, Wang C, Tao J, Ho C, Jiang L, Gui B, Huang S, Evert M, Calvisi DF, et al. Bmi1 is required for hepatic progenitor cell expansion and liver tumor development. PLoS ONE. 2012;7(9):e46472. doi: 10.1371/journal.pone.0046472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Correnti M, Cappon A, Pastore M, Piombanti B, Lori G, Oliveira D, Munoz-Garrido P, Lewinska M, Andersen JB, Coulouarn C, et al. The protease-inhibitor SerpinB3 as a critical modulator of the stem-like subset in human cholangiocarcinoma. Liver Int. 2022;42(1):233–248. doi: 10.1111/liv.15049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simons M, Raposo G. Exosomes–vesicular carriers for intercellular communication. Curr Opin Cell Biol. 2009;21(4):575–581. doi: 10.1016/j.ceb.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 23.Ela S, Mager I, Breakefield XO, Wood MJ. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov. 2013;12(5):347–357. doi: 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- 24.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 25.McAndrews KM, Kalluri R. Mechanisms associated with biogenesis of exosomes in cancer. Mol Cancer. 2019;18(1):52. doi: 10.1186/s12943-019-0963-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, Molina H, Kohsaka S, Di Giannatale A, Ceder S, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527(7578):329–335. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang X, Yang X, Zhang Y, Liu X, Zheng G, Yang Y, Wang L, Du L, Wang C. Direct serum assay for cell-free bmi-1 mRNA and its potential diagnostic and prognostic value for colorectal cancer. Clin Cancer Res Off J Am Assoc Cancer Res. 2015;21(5):1225–1233. doi: 10.1158/1078-0432.CCR-14-1761. [DOI] [PubMed] [Google Scholar]
- 28.Zhang X, Wang C, Wang L, Du L, Wang S, Zheng G, Li W, Zhuang X, Zhang X, Dong Z. Detection of circulating Bmi-1 mRNA in plasma and its potential diagnostic and prognostic value for uterine cervical cancer. Int J Cancer. 2012;131(1):165–172. doi: 10.1002/ijc.26360. [DOI] [PubMed] [Google Scholar]
- 29.Sabbatino F, Villani V, Yearley JH, Deshpande V, Cai L, Konstantinidis IT, Moon C, Nota S, Wang Y, Al-Sukaini A, et al. PD-L1 and HLA Class I antigen expression and clinical course of the disease in intrahepatic cholangiocarcinoma. Clin Cancer Res. 2016;22(2):470–478. doi: 10.1158/1078-0432.CCR-15-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loeuillard E, Yang J, Buckarma E, Wang J, Liu Y, Conboy C, Pavelko KD, Li Y, O'Brien D, Wang C, et al. Targeting tumor-associated macrophages and granulocytic myeloid-derived suppressor cells augments PD-1 blockade in cholangiocarcinoma. J Clin Investig. 2020;130(10):5380–5396. doi: 10.1172/JCI137110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Balkwill F. Cancer and the chemokine network. Nat Rev Cancer. 2004;4(7):540–550. doi: 10.1038/nrc1388. [DOI] [PubMed] [Google Scholar]
- 32.Qin Y, Vasilatos SN, Chen L, Wu H, Cao Z, Fu Y, Huang M, Vlad AM, Lu B, Oesterreich S, et al. Inhibition of histone lysine-specific demethylase 1 elicits breast tumor immunity and enhances antitumor efficacy of immune checkpoint blockade. Oncogene. 2019;38(3):390–405. doi: 10.1038/s41388-018-0451-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dangaj D, Bruand M, Grimm AJ, Ronet C, Barras D, Duttagupta PA, Lanitis E, Duraiswamy J, Tanyi JL, Benencia F, et al. Cooperation between constitutive and inducible chemokines enables T cell engraftment and immune attack in solid tumors. Cancer Cell. 2019;35(6):885–900 e810. doi: 10.1016/j.ccell.2019.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pascual-Garcia M, Bonfill-Teixidor E, Planas-Rigol E, Rubio-Perez C, Iurlaro R, Arias A, Cuartas I, Sala-Hojman A, Escudero L, Martinez-Ricarte F, et al. LIF regulates CXCL9 in tumor-associated macrophages and prevents CD8(+) T cell tumor-infiltration impairing anti-PD1 therapy. Nat Commun. 2019;10(1):2416. doi: 10.1038/s41467-019-10369-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fukuda Y, Asaoka T, Eguchi H, Yokota Y, Kubo M, Kinoshita M, Urakawa S, Iwagami Y, Tomimaru Y, Akita H, et al. Endogenous CXCL9 affects prognosis by regulating tumor-infiltrating natural killer cells in intrahepatic cholangiocarcinoma. Cancer Sci. 2020;111(2):323–333. doi: 10.1111/cas.14267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou G, Sprengers D, Mancham S, Erkens R, Boor PPC, van Beek AA, Doukas M, Noordam L, Campos Carrascosa L, de Ruiter V, et al. Reduction of immunosuppressive tumor microenvironment in cholangiocarcinoma by ex vivo targeting immune checkpoint molecules. J Hepatol. 2019;71(4):753–762. doi: 10.1016/j.jhep.2019.05.026. [DOI] [PubMed] [Google Scholar]
- 37.Sadeghlar F, Vogt A, Mohr RU, Mahn R, van Beekum K, Kornek M, Weismuller TJ, Branchi V, Matthaei H, Toma M, et al. Induction of cytotoxic effector cells towards cholangiocellular, pancreatic, and colorectal tumor cells by activation of the immune checkpoint CD40/CD40L on dendritic cells. Cancer Immunol Immunother. 2021;70(5):1451–1464. doi: 10.1007/s00262-020-02746-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Z, Sun R, Zhang X, Qiu B, Chen T, Li Z, Xu Y, Zhang Z. Transcription factor 7 promotes the progression of perihilar cholangiocarcinoma by inducing the transcription of c-Myc and FOS-like antigen 1. EBioMedicine. 2019;45:181–191. doi: 10.1016/j.ebiom.2019.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jia L, Zhang W, Wang CY. BMI1 inhibition eliminates residual cancer stem cells after PD1 blockade and activates antitumor immunity to prevent metastasis and relapse. Cell Stem Cell. 2020;27(2):238–253 e236. doi: 10.1016/j.stem.2020.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hezel AF, Deshpande V, Zhu AX. Genetics of biliary tract cancers and emerging targeted therapies. J Clin Oncol. 2010;28(21):3531–3540. doi: 10.1200/JCO.2009.27.4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397(6715):164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 42.Guo J, Deng N, Xu Y, Li L, Kuang D, Li M, Li X, Xu Z, Xiang M, Xu C. Bmi1 drives the formation and development of intrahepatic cholangiocarcinoma independent of Ink4A/Arf repression. Pharmacol Res. 2021;164:105365. doi: 10.1016/j.phrs.2020.105365. [DOI] [PubMed] [Google Scholar]
- 43.Kamerkar S, LeBleu VS, Sugimoto H, Yang S, Ruivo CF, Melo SA, Lee JJ, Kalluri R. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 2017;546(7659):498–503. doi: 10.1038/nature22341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martins VR, Dias MS, Hainaut P. Tumor-cell-derived microvesicles as carriers of molecular information in cancer. Curr Opin Oncol. 2013;25(1):66–75. doi: 10.1097/CCO.0b013e32835b7c81. [DOI] [PubMed] [Google Scholar]
- 45.Zhao R, Zhang Y, Zhang X, Yang Y, Zheng X, Li X, Liu Y, Zhang Y. Exosomal long noncoding RNA HOTTIP as potential novel diagnostic and prognostic biomarker test for gastric cancer. Mol Cancer. 2018;17(1):68. doi: 10.1186/s12943-018-0817-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu X, Liang Y, Li X, Ouyang K, Wang M, Cao T, Li W, Liu J, Xiong J, Li B, et al. Exosome-mediated delivery of kartogenin for chondrogenesis of synovial fluid-derived mesenchymal stem cells and cartilage regeneration. Biomaterials. 2021;269:120539. doi: 10.1016/j.biomaterials.2020.120539. [DOI] [PubMed] [Google Scholar]
- 47.Nieland L, Morsett LM, Broekman MLD, Breakefield XO, Abels ER. Extracellular vesicle-mediated bilateral communication between glioblastoma and astrocytes. Trends Neurosci. 2021;44(3):215–226. doi: 10.1016/j.tins.2020.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sen T, Rodriguez BL, Chen L, Corte CMD, Morikawa N, Fujimoto J, Cristea S, Nguyen T, Diao L, Li L, et al. Targeting DNA damage response promotes antitumor immunity through STING-mediated T-cell activation in small cell lung cancer. Cancer Discov. 2019;9(5):646–661. doi: 10.1158/2159-8290.CD-18-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blackledge NP, Rose NR, Klose RJ. Targeting Polycomb systems to regulate gene expression: modifications to a complex story. Nat Rev Mol Cell Biol. 2015;16(11):643–649. doi: 10.1038/nrm4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Infante R, Bedard L, Shapiro G, Bauer TM, Prawira A, Laskin O, et al. Phase 1 results of PTC596, a novel small molecule targeting cancer stem cells (CSCs) by reducing levels of BMI1 protein. J Clin Oncol. 2017;35(15):2574–2574. doi: 10.1200/JCO.2017.35.15_suppl.2574. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.