Abstract
Small extracellular vesicle (sEV)-mediated intercellular communication regulates multiple aspects of growth and development in multicellular organisms. However, the mechanism underlying cargo recruitment into sEVs is currently unclear. We show that the key nucleo-cytoplasmic transport (NCT) protein—RanGTPase, in its GTP-bound form (RanGTP), is enriched in sEVs secreted by mammalian cells. This recruitment of RanGTP into sEVs depends on the export receptor CRM1 (also called XPO1). The recruitment of GAPDH, a candidate cargo protein, into sEVs is regulated by the RanGTP–CRM1axis in a nuclear export signal (NES)-dependent manner. Perturbation of NCT through overexpression or depletion of nuclear transport components affected the recruitment of Ran, CRM1 and GAPDH into sEVs. Our studies, thus, suggest a link between NCT, particularly the Ran–CRM1 axis, and recruitment of NES-containing cargoes into the sEVs. Collectively, these findings implicate RanGTPase as a link between NCT and sEV mediated intercellular communication.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-022-04422-y.
Keywords: Small extracellular vesicles, RanGTPase, Exportin1/CRM1, Nuclear export signal, Exosome, Intercellular communication
Introduction
RanGTPase (referred as ‘Ran’ hereafter) is a highly conserved protein involved in the nucleo-cytoplasmic transport (NCT) of macromolecules across the nuclear envelope (NE) [1]. The asymmetric distribution of Ran’s regulator proteins, the GTPase activating protein RanGAP1 in the cytoplasm and the guanine nucleotide exchange factor RCC1 in the nucleus, creates a steep gradient of RanGTP across the NE, wherein the concentration of RanGTP is higher in the nucleus and RanGDP in the cytoplasm. This gradient determines the directionality of transport by enabling the import complex assembly in the cytoplasm and its disassembly in the nucleus, and the export complex assembly in the nucleus and its dissociation in the cytoplasm [2, 3]. NCT of cargoes requires specific transport adapters/receptors called karyopherins; importins are receptors involved in nuclear import and exportins are involved in nuclear export [3]. Based on the current model, an export complex consisting of the protein cargo containing a nuclear export signal (NES), RanGTP and CRM1 (also called Exportin-1 or XPO-1) assembles in the nucleus [4]. This complex travels through the nuclear pore complex (NPC) to the cytoplasm, where its disassembly is mediated by RanBP1/2-assisted, RanGAP1-mediated hydrolysis of RanGTP [2]. Conversely, import complexes consisting of the protein cargoes possessing nuclear localization signal (NLS), importin-α and importin-β form in the cytoplasm. Once in the nucleus, RanGTP binds importin-β, thus dissociating the complex and releasing the cargo. In addition to its role in NCT, Ran is also implicated in cell division, post mitotic nuclear envelope assembly and cell cycle progression [2].
Multicellular organisms have also evolved various intercellular communication mechanisms to coordinate processes during growth and homeostasis. Although ligand–receptor signaling is a well-established mode of intercellular communication, recent studies have highlighted the involvement of small extracellular vesicles (sEVs) and tunneling nanotubes (TNTs) in cell-to-cell signaling [5, 6]. Two of the sEVs are exosomes and microvesicles [7, 8]. Exosomes are 30–150 nm sized membrane vesicles of endocytic origin formed in the multivesicular bodies (MVBs), which get released into the extracellular milieu when the MVBs fuse with the plasma membrane [9–12]. Microvesicles, on the other hand, are generated by budding of the plasma membrane [9, 13]. Different cell types, including dendritic cells, B cells, T cells, macrophages, reticulocytes and tumor cells, are known to secrete exosomes [13]. Exosomes contain proteins, mRNAs and small RNA species including miRNAs, which have been shown to be functional in the recipient cells [5, 8, 12]. Studies indicated a role for hnRNPA2B1, Ras and YBX1 in the recruitment of subsets of miRNAs into exosomes [14–17]. The proteins targeted into exosomes include members of ESCRT machinery such as Alix and TSG101, tetraspanins such as CD63 and CD81, heat-shock proteins, cytoskeletal proteins and regulators of intracellular trafficking such as Rab GTPases [18]. There are also many soluble proteins including metabolic enzymes such as GAPDH present in the exosomes [13, 19]. However, the signals and mechanisms involved in recruiting soluble proteins into sEVs (exosomes or microvesicles) are unclear.
We had reported earlier that RanGTPase possesses the ability to move from one cell to another in a GTP- and CRM1-dependent manner [20]. Given that Ran, a critical regulator of NCT, and other components of NCT such as CRM1/XPO-1, are present in sEVs [21, 22], we wished to explore a possible connection between NCT and intercellular communication through sEVs. Here, we confirmed that Ran is present in the sEVs secreted by different cell lines. Interestingly, the recruitment of Ran into sEVs was found to be GTP-dependent, and requires CRM1. Furthermore, we show that RanGTP–CRM1 axis regulates recruitment of a cargo, GAPDH, into sEVs in an NES-dependent manner, thus linking NCT with recruitment of a subset of cargoes into sEVs.
Results
Ran gets recruited into small extracellular vesicles (sEVs)
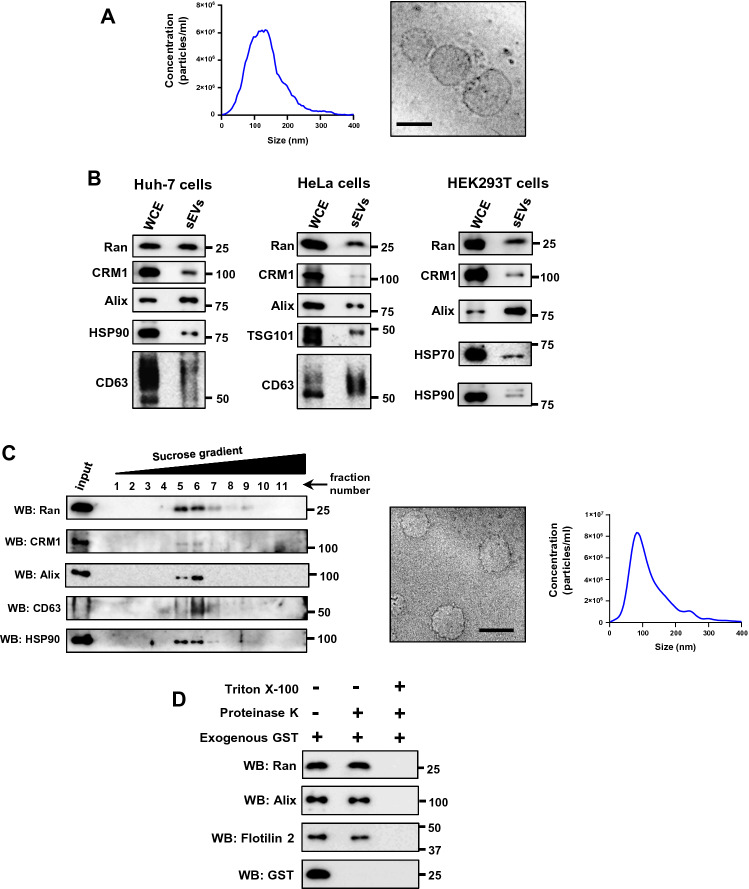
A previous study reported that intercellular movement of Ran occurs in a GTP- and CRM1-dependent manner [20]. Protein profiling studies have identified Ran as a component of sEVs [23–28]. We, therefore, investigated if sEVs serve as a means for the intercellular transfer of Ran. sEVs were isolated from conditioned medium using a previously described ultracentrifugation-based method [29]. The 100,000g pellet obtained from HeLa cell conditioned medium, considered as sEVs, was assessed by nano-particle tracking analysis (NTA) and Cryo-transmission electron microscopy (Cryo-TEM) (Fig. 1A). Cryo-EM images showed this fraction to be enriched with vesicles and the size of these vesicles ranged from 30 to 150 nm (Fig. 1A). Moreover, western blotting analysis confirmed the presence of exosome markers such as Alix, HSP90, TSG101 and CD63 in sEVs isolated from Huh-7, HeLa and HEK293T cells (Fig. 1B). Interestingly, Ran and CRM1 proteins were also detected in the sEVs derived from different cell lines (Fig. 1B). These results indicate that sEVs are enriched in exosomes, and Ran and CRM1 may be present in sEVs.
Fig. 1.
Ran and CRM1 are present in small extracellular vesicles. A Left panel: nanoparticle tracking analysis (NTA) of small extracellular vesicles (sEVs) isolated from HeLa cells. Right panel: cryo-transmission electron microscopic (Cryo-TEM) image of HeLa sEVs (100,000g pellet). Scale bar—100 nm. B sEVs were isolated from Huh-7, HeLa or HEK293T cells as indicated. The respective whole cell extracts (WCE) and small extracelluar vesicles (sEVs) were probed with indicated antibodies by western blotting. Alix, HSP90, HSP70, CD63 and TSG101 were used as sEVs (exosome) markers. Molecular weight markers (in numbers) are as indicated. C Left panel: sEVs isolated from HeLa cells (100,000g pellet) were further subjected to density-based separation using sucrose gradient (0–60%). The fractions were probed for the presence of indicated proteins using western blotting (WB). Alix, CD63 and HSP90 were used as (sEV exosome) markers. Middle panel: cryo-transmission electron microscopic (Cryo-TEM) image of vesicles isolated from fractions (5–7) corresponding to exosomes. Scale bar, 100 nm. Right panel: particle distribution plot (NTA) of purified sEVs/exosomes. D HeLa sEVs (100,000g pellet) were added with purified glutathione S-transferase (exogenous GST), left untreated (−) or treated (+) with proteinase K in the absence (−) or presence (+) of triton X-100 as indicated. The samples were subjected to western blot (WB) analysis using specific GST, Ran, Alix or Flotilin-2 antibodies. Alix and Flotilin-2 were used as sEV markers. GST was used as a control to monitor Proteinase K activity in the absence of triton X-100. Molecular weight markers (in numbers) are as indicated
However, owing to the heterogeneity of secreted vesicles [31–33] and lack of specific isolation protocols, it is not possible to rule out the presence of other vesicle types in the preparations [8]. Therefore, throughout the manuscript, the isolated vesicles will be referred to as ‘small extracellular vesicles (sEVs)’ and at specific instances as ‘exosomes’.
To verify if Ran and CRM1 are present in the exosomal fractions, the 100,000g pellet was resuspended and separated on a sucrose density gradient [17, 29]. Exosomes are known to float at a density of ~ 1.08–1.18 g/ml (fractions 5–7) in a continuous sucrose gradient [12, 29]. Consequently, Ran and CRM1 co-fractionated with the exosomal markers Alix, CD63 and HSP90 (Fig. 1C), indicating that these proteins are /may be present in the vesicles, particularly exosomes, secreted by cells. Ran was also detected in sEVs positive for CD63 [an exosome marker [10]] that were isolated by an immunoaffinity-based method, further confirming that Ran is present in exosomes (Fig. S1). To test if Ran is present inside the isolated vesicles, the vesicles were treated with proteinase K in the presence or absence of triton X-100 [34]. In the absence of the detergent triton X-100, intravesicular proteins are shielded from proteinase K activity, by the intact vesicular membrane. On the contrary, triton X-100 dissolves the membrane, thus making the proteins inside the vesicle accessible to proteinase K mediated degradation. Our results showed that Ran and the sEV markers Alix and Flotilin-2 were degraded in the presence of triton X-100, and remained intact in its absence (Fig. 1D), indicating that Ran is present inside the vesicles. To ensure that Proteinase K was active in the absence of the detergent, purified glutathione S-transferase (GST) was exogenously added to the sEVs and the Proteinase K reaction was performed. GST was degraded in the presence and absence of the detergent (Fig. 1D), indicating that Proteinase K was active even in the absence of the detergent.
Next, we tested whether the recruitment of Ran into sEVs was sensitive to extracellular signaling cues. It is already known that stimulation of macrophages by lipopolysaccharide (LPS) increases secretion of bioactive molecules through sEVs [35–37]. Treatment of the mouse macrophage cell line RAW 264.7 with LPS significantly increased the relative level of Ran in sEVs as compared to the exosome marker Alix (Fig. S2). Taken together, the results showed that Ran is present in sEVs derived from different cell lines, and its recruitment might be regulated by extracellular signaling.
Recruitment of Ran into sEVs is GTP dependent
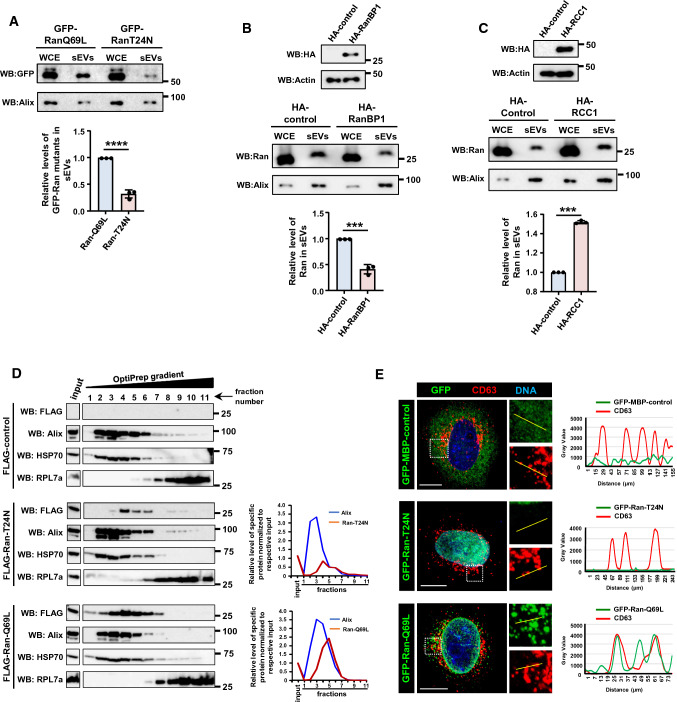
Previous studies have shown that the GTP-bound form of Ran can move from one cell to another [20]. As Ran is present in sEVs, we reasoned that Ran might move between cells through sEVs, thus predicting that GTP-bound Ran is preferably recruited into sEVs. Consistent with this, the GTP-locked mutant Ran-Q69L [38] was recruited into sEVs at significantly higher levels as compared to the nucleotide-free mutant Ran-T24N [39] (Fig. 2A). Further, the cellular levels of RanGTP were altered using different approaches, and the recruitment of Ran to sEVs was monitored. Overexpression of RanBP1, a protein that fosters RanGTP hydrolysis and hence decreases cellular RanGTP, significantly reduced the level of Ran in sEVs secreted by HEK293T cells, as compared to the control (Fig. 2B). On the contrary, increased cellular levels of RanGTP, achieved via ectopic expression of the guanine nucleotide exchange factor for Ran-RCC1 in HEK293T cells, resulted in increased recruitment of Ran into sEVs (Fig. 2C). These results support the conclusion that recruitment of Ran into sEVs occurs in its GTP-bound state.
Fig. 2.
Recruitment of Ran into sEVs is GTP-dependent. A Huh-7 cells were transfected with GFP-Ran-Q69L or GFP-Ran-T24N mutant for 16 h. Post 48 h, sEVs were isolated and the levels of Ran and Alix were analyzed in the whole cell extract (WCE) and sEVs (100,000g pellet) (top panel). Relative recruitment of Ran into sEVs, in comparison to Alix, was calculated from three independent sets and plotted (right panel). Data are presented as mean ± SEM, P‐value was calculated using Student's t test and ****P < 0.0001. B HEK293T cells were transfected with pcDNA-empty-vector (HA-control) or HA-RanBP1 construct for 16 h and sEVs were isolated after 48 h. The expression of HA-RanBP1 was analyzed by western blotting (top panel). sEVs were isolated and analyzed for levels of Ran and Alix (middle panel). Relative recruitment of Ran into sEVs, with respect to Alix, was calculated from three independent sets and plotted (bottom panel). Data are presented as mean ± SEM, P value was calculated using Student's t test and ***P < 0.001. C HEK293T cells were transfected with pcDNA-empty-vector (HA-control) or HA-RCC1 construct for 16 h and sEVs were isolated after 48 h. The expression of HA-RCC1 was analyzed by western blotting (top panel). sEVs were isolated and analyzed for levels of Ran and Alix (middle panel). Relative recruitment of Ran into sEVs, with respect to Alix, was calculated from three independent experiments and plotted (bottom panel). Data are presented as mean ± SEM, P‐value was calculated using Student's t test and ***P < 0.001. D Huh-7 cells were transfected with FLAG-empty vector (FLAG-control), FLAG-Ran-T24N or FLAG-Ran-Q69L. The cell lysates were separated on OptiPrep density gradient (3–30%). Fractions collected and analyzed for the presence of indicated proteins using western blotting (WB). Graph plots (right) indicate the amount of Ran and Alix (sEV-exosome marker) across the gradient fractions normalized to respective input level. E HeLa cells were transfected with GFP-MBP-control, GFP-Ran-Q69L or GFP-Ran-T24N construct (green) for 21 h. Cells were fixed and immunostained for endogenous CD63 [marker for multi-vesicular body (MVB); red]. DNA was stained with Hoechst 33,342 (blue). The intensity line plots (right) indicate spatial distribution of GFP (green) and CD63 (red) shown in the zoomed region. Scale bar, 10 µm
Exosomes are derived from MVBs [5]. Given that RanGTP is preferably recruited to sEVs (and possibly to exosomes), the association of the two Ran mutants (Ran-Q69L and Ran-T24N) with MVBs within the cells was assessed via two approaches, cellular fractionation and immunocytochemistry. Huh-7 cells expressing FLAG-control (empty vector), FLAG-Ran-Q69L or FLAG-Ran-T24N were fractionated using OptiPrep (iodixanol) gradient centrifugation [40]. FLAG-Ran-Q69L co-fractionated with MVB markers such as Alix and HSP70 (Fig. 2D). FLAG-Ran-T24N was also detected in MVB fractions; however, its levels were substantially reduced as compared to Ran-Q69L (Fig. 2D). This is consistent with the observation that RanT24N gets recruited to the sEVs to a lesser extent than Ran-Q69L (Fig. 2A). To validate these observations, we transfected GFP-MBP-control, GFP-Ran-Q69L or GFP-Ran-T24N in HeLa cells and assessed the co-localization between the Ran mutants and endogenous CD63 (MVB marker) using specific antibodies. GFP-RanQ69L predominantly localized to the NE and cytoplasmic punctate structures, whereas GFP-Ran-T24N primarily localized to the nucleus and was diffusely present in the cytoplasm (Fig. 2E). Interestingly, the cytoplasmic Ran-Q69L puncta often associated with CD63, whereas Ran-T24N or GFP-MBP-control showed no discernible association with MVBs (Fig. 2E). Collectively, these results showed that the GTP-bound form of Ran preferably associates with MVBs, supporting the conclusion that RanGTP is targeted to exosomes.
Enrichment of Ran in sEVs is dependent on CRM1
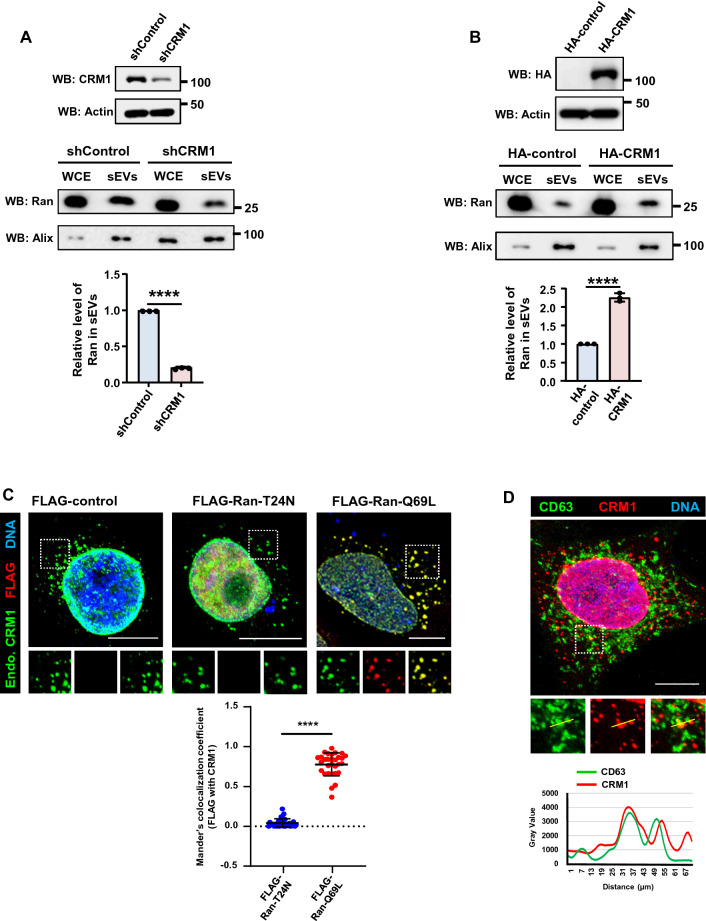
During nuclear export, the cooperative binding of RanGTP with the CRM1–NES-containing cargo (CRM1–NES*Cargo) complex creates a stable trimolecular export complex [4, 41, 42]. Earlier studies have shown that treatment of the cells with LMB, an inhibitor of CRM1 that interferes with RanGTP–CRM1–NES*Cargo complex formation, reduced the intercellular transfer of Ran [4, 20]. To examine whether CRM1 is required for the recruitment of Ran into the sEVs, we depleted endogenous CRM1 from HeLa cells using inducible CRM1 shRNA. CRM1 depletion led to substantially decreased levels of Ran in sEVs as compared to control cells (Fig. 3A). On the contrary, overexpression of CRM1 in HEK293T led to enhanced recruitment of Ran into sEVs (Fig. 3B). Moreover, the GTP-bound mutant of Ran (Q69L), but not the nucleotide-free Ran mutant (T24N), colocalized with endogenous CRM1 (Fig. 3C). Endogenous CRM1 also associated with CD63 (Fig. 3D). These results suggest that recruitment of Ran into the sEVs depends on the export receptor CRM1. The data also indicate that RanGTP and CRM1 may be recruited to the exosomes as part of the trimolecular export complexes. RanBP3, a cofactor that facilitates the export complex formation [43, 44], was also detected in sEVs (Fig. S3).
Fig. 3.
Recruitment of Ran into sEVs depends on CRM1. A HeLa cell line harboring inducible control shRNA (shControl) or CRM1 shRNA (shCRM1) was induced with doxycycline for 36 h. The sEVs were isolated from the conditioned medium after a second round of doxycycline induction for 36 h. Depletion of CRM1 was assessed by western blotting (WB) (top panel). sEVs were isolated and analyzed for levels of Ran and Alix (middle panel). Relative recruitment of Ran into sEVs, with respect to Alix, was calculated from three independent experiments and plotted (bottom panel). Data are presented as mean ± SEM, P‐value was calculated using Student's t test and ****P < 0.0001. B HEK293T cells were transfected with pCI-neo-HA-empty-vector (HA-control) or HA-CRM1 construct for 16 h and sEVs were isolated after 48 h. The expression of HA-CRM1 was analyzed by western blotting (top panel). sEVs were isolated and analyzed for levels of Ran and Alix (middle panel). Relative recruitment of Ran into sEVs, with respect to Alix, was calculated from three independent sets and plotted (bottom panel). Data are presented as mean ± SEM, P‐value was calculated using Student's t test and ****P < 0.0001. C HeLa cells were transfected with FLAG-empty vector (FLAG-control), FLAG-Ran-T24N or FLAG-Ran-Q69L construct for 21 h. Top panel: cells were fixed and immunostained for FLAG (red) and endogenous CRM1 (Endo. CRM1; green). DNA was stained with Hoechst 33342 (blue). Scale bar, 10 µm. Bottom panel: quantitative representation of Manders’ colocalization coefficients obtained by analyzing the red fluorescence intensity (FLAG) overlapping with green fluorescence intensity (CRM1) from 30 cells derived out of three independent experiments (10 cells/per experiment). Mann–Whitney test was performed for statistical analysis. Data represented as mean ± SD, ****P < 0.0001. D Top panel; HeLa cells were co-immunostained for endogenous CRM1 (red) and CD63 (green) using specific antibodies. DNA was stained with Hoechst 33342 (blue). The intensity line plots (bottom panel) indicate spatial distribution of CD63 (green) in relation to CRM1 (red) shown in the zoomed region. Scale bar, 10 µm
Recruitment of GAPDH to sEVs is dependent on NES
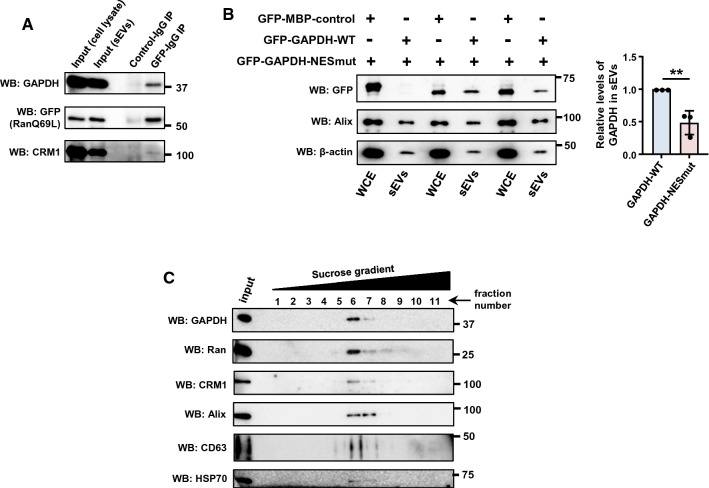
The interaction between RanGTP and CRM1 is stabilized when CRM1 is bound to an NES-containing export cargo (NES*Cargo) [45]. Our data show that RanGTP is recruited to sEVs in a CRM1-dependent manner, thus allowing us to hypothesize that RanGTP–CRM1–NES*Cargo complexes might be directed to the sEVs. To test this, sEVs isolated from Huh-7 cells expressing GFP-Ran-Q69L were subjected to co-immunoprecipitation using GFP-specific antibody. Mass-spectrometric analysis of the immunoprecipitates enabled identification of GFP-Ran-Q69L-interacting proteins (Table S1). GAPDH being one of them, was selected as the candidate NES*cargo for further studies as its presence in exosomes is well characterized [19, 28], and its export from the nucleus is known to be mediated by Ran and CRM1 [46].
Co-immunoprecipitation analysis confirmed that GAPDH is present in complex with RanGTP and CRM1 in sEVs (Fig. 4A). Interestingly, as compared to wild-type GAPDH, the recruitment of the GAPDH NES-mutant K259N into the sEVs was significantly reduced (Fig. 4B), thus suggesting GAPDH is sorted into sEVs in an NES-dependent manner. Additionally, GAPDH, along with Ran and CRM1, was present in sEVs isolated through sucrose density gradient separation (Fig. 4C). Collectively, the data suggest that RanGTP and CRM1 may mediate the delivery of NES containing cargo proteins into sEVs.
Fig. 4.
GAPDH is a cargo recruited to sEVs using RanGTP–CRM1 axis. A sEVs isolated from Huh-7 cells expressing GFP-Ran-Q69L were lysed and subjected to immunoprecipitation (IP) using rabbit IgG (control IgG) or GFP-specific antibody (GFP-IgG). The immunoprecipitates were probed for the presence of GAPDH, GFP (Ran-Q69L) and CRM1 using specific antibodies. Molecular weight markers (in numbers) are as indicated. B Dependence of GAPDH for recruitment into sEVs on nuclear export signal (NES). Left panel: sEVs were purified from Huh-7 cells expressing GFP-MBP-control, GFP-GAPDH-wild type (WT) or GFP-GAPDH NES mutant (K259N) and subjected to western blotting to detect the presence of indicated proteins. Alix was used as sEV (exosome marker). Right panel: quantitative data depicting the relative level of GFP-tagged proteins in the sEVs normalized to the levels of Alix in the respective sample. Data are presented as mean ± SEM from there independent experiments, P value was calculated using Student's t test and **P < 0.01. C sEVs (100,000g pellet) isolated from HeLa cells were further separated on sucrose density gradient (0–60%). Individual fractions were collected and subjected to western blotting (WB) using indicated antibodies. Alix, CD63 and HSP70 were used as sEV (exosome) markers
Recruitment of GAPDH into sEVs is linked with the NCT
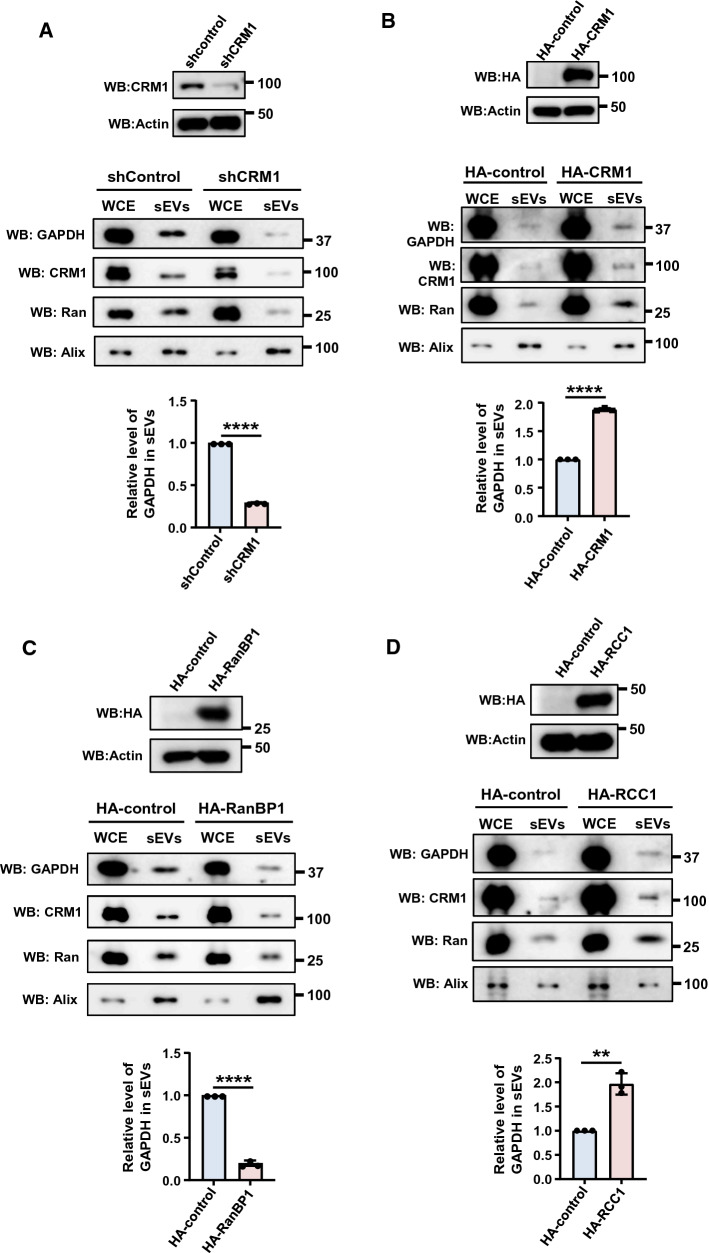
To verify the recruitment of GAPDH into sEVs is dependent on NCT, the NCT process was altered by overexpressing or depleting critical proteins of the pathway. Depletion of CRM1, which potentially reduces the nuclear export of NES-dependent cargoes, resulted in a significant reduction in the level of GAPDH in the sEVs as compared to control cells (Fig. 5A). However, overexpression of CRM1 that would enhance nuclear export resulted in increased levels of GAPDH in the sEVs (Fig. 5B). On the contrary, overexpression of RanBP1 that enhances disassembly of RanGTP–CRM1–NES*Cargo complexes, reduced the GAPDH level in the sEVs (Fig. 5C). Similarly, ectopic expression of RCC1, the guanine nucleotide exchange factor for Ran, that is expected to increase RanGTP levels, led to substantially increased recruitment of GAPDH into sEVs (Fig. 5D). Collectively, these results suggest that recruitment of cargo such as GAPDH depends on NCT, particularly on the export of the RanGTP–CRM1–NES*Cargo complex.
Fig. 5.
NCT is linked with recruitment of cargoes into sEVs. A HeLa cells harboring Inducible control shRNA (shControl) or CRM1 (shCRM1) were induced with doxycycline for 36 h. sEVs were isolated from the conditioned medium after a second round of doxycycline induction for 36 h. The depletion of CRM1 was analyzed by western blotting (top panel). The isolated sEVs were subjected to western blotting (WB) to analyze the relative levels of indicated proteins. Alix was used as sEV (exosome) marker (middle panel). Quantitative representation of relative levels of GAPDH normalized to that of Alix in the respective sEV samples (bottom panel). Data are presented as mean ± SEM from three independent experiments, P‐value was calculated using Student's t test and **P < 0.01. B HEK293T cells were transfected with pCI-neo-HA-empty-vector (HA-control) or HA-CRM1 construct for 16 h and forty eight hours later the sEVs were isolated. The expression of HA-CRM1 was analyzed by western blotting (top panel). sEVs were isolated and analyzed for levels of GAPDH, Ran and Alix (middle panel). Relative recruitment of GAPDH into sEVs, with respect to Alix, was calculated from three independent sets and plotted (bottom panel). Data are presented as mean ± SEM, P value was calculated using Student's t test and ****P < 0.0001. C HEK293T cells were transfected with pcDNA-empty-vector (HA-control) or HA-RanBP1 construct for 16 h and sEVs were isolated after 48 h. The expression of HA-RanBP1 was analyzed by western blotting (top panel). The sEVs isolated were analyzed for levels of GAPDH, Ran and Alix by western blotting (WB) (middle panel). Relative recruitment of GAPDH into sEVs, as compared to Alix, was calculated from three independent experiments and plotted (bottom panel). Data are presented as mean ± SEM, P value was calculated using Student's t test and ****P < 0.0001. D HEK293T cells were transfected with pcDNA-empty-vector (HA-control) or HA-RCC1 construct for 16 h and sEVs were isolated after 48 h. The expression of HA-RCC1 was analyzed by western blotting (top panel). The isolated sEVs were analyzed for levels of GAPDH, Ran and Alix by western blotting (WB) (middle panel). Relative recruitment of GAPDH into sEVs, as compared to Alix, was calculated from three independent experiments and plotted (bottom panel). Data are presented as mean ± SEM, P value was calculated using Student's t test and **P < 0.01
Intercellular transport of Ran depends on exosome biogenesis pathway and NCT
Given that Ran gets recruited into sEVs in a GTP- and CRM1-dependent manner and can undergo intercellular transport, it seems plausible that the intercellular transfer of Ran [20] may occur through sEVs. It has been shown that components of the endosomal sorting complex required for transport (ESCRT) machinery participates in the biogenesis of extracellular vesicles, and depletion of TSG101 significantly reduced the biogenesis of exosomes [47]. To test whether intercellular transport of Ran is mediated through exosomes, TSG101 was depleted from HeLa cells using specific siRNA, and cell-to-cell transfer of Ran was monitored using a previously described transient transfection assay using mCherry-α-tubulin as a co-transfection marker as described earlier [20]. Transfer of Ran between cells was significantly reduced in TSG101 depleted cells as compared to controls (Fig. S4), suggesting that intercellular transport of Ran occurs through sEVs, and particularly via exosomes. Even though tubulin was identified as a component of sEVs [5], there was no detectable level of transferred tubulin in the neighboring cells observed within the time frame (10 h) of the assay (Fig. S4). However, within this time, transferred Ran-Q69L was visible in the neighboring cells (Fig. S4). To confirm the specificity and robustness of intercellular Ran transfer, we also used mCherry-histone H2B (a nuclear protein) as a co-transfection marker (Fig. S5). Similar to mCherry-α-tubulin, the intercellular transfer of mCherry-H2B was also not observed within the time and conditions of the assay (Fig. S5), but Ran-Q69L transfer was distinctly detectable. Under the same conditions, GFP-MBP-control was also not detected in the neighboring mCherry-H2B-negative cells.
The prominent role of Ran in NCT suggests that NCT might regulate the transfer of Ran between cells. To confirm this, NCT was manipulated by depletion of RanBP1 and CRM1. Interestingly, absence of RanBP1 increased (Fig. S6A) and CRM1 decreased (Fig. S6B) the intercellular transfer of Ran, indicating that transport of Ran between cells and NCT are interlinked.
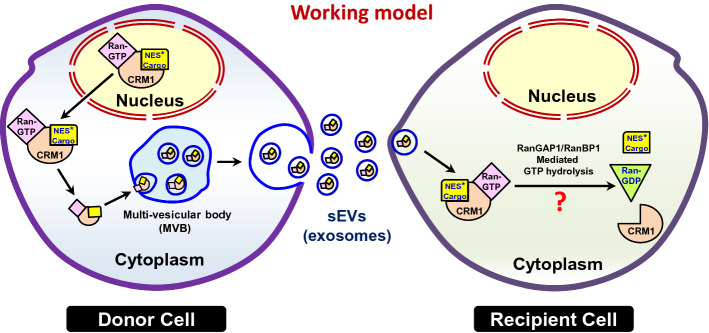
Based on our findings, we propose a working model (Fig. 6). Some of the nuclear export complexes may escape GTP-hydrolysis-dependent disassembly in the cytoplasm, and alternatively get recruited into the sEVs formed through the MVBs. Recipient cells may take up the sEVs, and the export complexes may be disassembled in the recipient cells through the cytoplasmic RanGAP1-mediated hydrolysis of GTP on Ran.
Fig. 6.
Working model for Ran–CRM1-mediated recruitment of cargoes into sEVs. Some of the RanGTP–CRM1–NES*Cargo export complexes may escape disassembly in the cytoplasm, and get recruited into the intra-luminal vesicles of the multi-vesicular bodies (MVBs) generated by inward budding. The MVBs eventually fuse with the plasma membrane to release the intra-luminal vesicles, called exosomes, a class of sEVs. This implies a role for the NCT machinery in the sorting of a subset of cargoes into sEVs released by the donor cells. In the recipient cells, this complex may be disassembled, thus releasing the cargo due to cytoplasmic RanGAP1-mediated hydrolysis of GTP on Ran
Discussion
Conventionally believed to be a means for cellular waste disposal, the sEVs are now emerging as a key player in intercellular communication. Research over the past decade has highlighted a role for EVs in cancer and neurodegenerative diseases. Moreover, the cargo content of the sEVs appears to be condition specific, thereby suggesting that the cells may have regulatory mechanisms that determine the same. However, in general, the cellular mechanism(s) governing the recruitment of cargo into the vesicles remains unclear.
Here we show that RanGTPase, a crucial player in NCT, is important for the sorting of a subset of protein cargoes into sEVs, particularly into exosomes. Our results further show that this recruitment depends on a RanGTP–CRM1 axis and the presence of a nuclear export signal (NES) in the cargo—which enables the association of the cargo with RanGTP and CRM1. Here, we have used GAPDH a candidate cargo. It is important to identify additional cargoes and validate their dependence on this mechanism for recruitment into sEVs, to generalize our findings. Nevertheless, these findings define a possible mechanism for the recruitment of cargo into sEVs, by which any nuclear export cargo can potentially be recruited into sEVs. Recently, it has been shown that CRM1/Ran complex is involved in recruitment of specific m7G-modifed RNAs into EVs, which was found to be restricted by CD47 [22]. Our results supporting a role for Ran/CRM1 in the recruitment of soluble protein cargo such as GAPDH extends the repertoire of molecules dependent on Ran/CRM1 for recruitment into sEVs. Additionally, the findings suggest an interplay between NCT, particularly the nuclear export, and intercellular communication through sEVs.
It is important to note, however, that the molecular machinery involved in recruitment of RanGTP–CRM1–NES*Cargo into sEVs remains to be elucidated. This may require the interaction of RanGTP–CRM1–NES*Cargo with sEV/exosome biogenesis machinery. Additionally, the cellular or physiological factors determining the disassembly of the export complex or its escape from hydrolysis and recruitment into exosomes within the donor cell remain to be studied.
The new findings reported here also provide a possible mechanistic explanation for the release of cargo in the recipient cells. RanBP1/RanGAP1-mediated hydrolysis of GTP on Ran in the cytoplasm of the recipient cells may dissociate the complex, thus releasing the cargo in recipient cells (Fig. 6). However, further studies are warranted for exploring this mechanism.
What is the possibility that cells use sEVs to remove excess RanGTP when it builds up beyond a threshold level, rather than serving any function in intercellular communication? Our results, however, suggest that this is unlikely the case, as we find GTP-bound Ran being taken up by the neighboring cells (Fig. S4, S5 and S6) [20]. Moreover, RanGTP, along with CRM1/XPO-1, recruits specific cargoes such as GAPDH in an NES-dependent manner into sEVs. Based on this, we conclude that the recruitment of RanGTP–CRM1–NES*Cargo into the sEVs has specific role in intercellular communication.
NCT and sEV-mediated intercellular communication are impaired in multiple cancers and neurodegenerative diseases [48–60]. Interestingly, both Ran and CRM1 are overexpressed in many cancers [48, 61, 62]. Although the role of Ran and CRM1 in cancer development and progression is not mechanistically well understood, several attempts to interfere with their functions have yielded encouraging results in restricting the tumor size and metastasis [63–66]. The interconnection between NCT and sEVs that we identified may provide additional avenues to explore the contribution of this cross-talk in the development and persistence of multiple diseases where both NCT and sEV compositions are altered.
Materials and methods
Cells and transfections
HeLa S3, HEK293T and Huh-7 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS). RAW 264.7 cells were grown in RPMI-1640 with 10% FBS. The cells were maintained with antibiotics and routinely tested for mycoplasma contamination. The serum used for exosome experiments was depleted of exosomes by a prior ultracentrifugation at 100,000g for 3 h.
For transient expression of proteins, the cells were transfected 12 h after plating using polyethyleneimine, linear (PEI, MW-25000, Polysciences, Inc.). The siRNA transfections were performed using Lipofectamine RNAiMax (Invitrogen). Depletion of TSG101 by siRNA was achieved by initial transfection of 40 nM control or TSG101 siRNA for 48 h, followed by a second round of transfection of 40 nM siRNA for further 24 h.
Expression vectors and siRNA
The constructs GFP-MBP, GFP-RanQ69L and GFP-RanT24N were described earlier [20, 67]. The constructs pcDNA-HA-RCC1, pcDNA4-His-Xpress-CRM1 and pGEX-RanBP1 were generous gifts from Mary Dasso (NICHD, NIH, Bethesda, USA). FLAG-Ran-Q69L and FLAG-Ran-T24N were generated by subcloning respective ORFs into a modified phCMV1-FLAG (Geneatlantis) vector. The CRM1 open reading frame PCR amplified using pcDNA4-His-Xpress-CRM1 as a template and was cloned at ApaI/SalI sites of pCI-neo-HA vector (Promega) to generate HA-CRM1. For obtaining HA-RanBP1 construct, the ORF was amplified from pGEX-RanBP1 using specific primers and was cloned at the EcoRI/XhoI sites of pcDNA-HA vector (Invitrogen). pCMV-GAPDH-Flag was generously provided by Wei Liu (Zhejiang University School of Medicine, China). The GAPDH ORF was subcloned into BglII/EcoRI sites of pEGFP-C1 vector (Clontech). The GAPDH NES mutant (K259N) was made by a PCR-based method using appropriate primers. The clones were confirmed by sequencing. For knockdown studies, control siRNA was described earlier [67], and siRNA against TSG101 [target sequence 5′ CGUCCUAUUUCGGCAUCCU 3′] was obtained from Dharmacon.
Generation of inducible shRNA stable cell lines
Stable inducible shRNA HeLa cell lines for RanBP1 and CRM1 were obtained by lentivirus transduction. The shRNA target sequences for non-specific control (5′ GUGGACUCUUGAAAGUACUAU 3′), RanBP1 (5′ GCGAGGCACUGGUGACGUCAA 3′) and CRM1 (5′ GUGGUGAAUUGCUUAUACCAU 3′) were cloned into Tet-pLKO-puro vector (Addgene #21915). For generating lentivirus, HEK293T cells were co-transfected with the packaging vectors psPAX2 (Addgene #12260) and pMD2.G (Addgene #12259) and specific Tet-pLKO-puro constructs. Tet-pLKO-puro vector containing control shRNA sequence was used to generate shRNA control cells. The virus containing conditioned medium was collected 48 h after transfection. For transduction, the virus containing medium was diluted 1:1 with DMEM, mixed with polybrene (Sigma) to a final concentration of 8 µg/ml, and added to HeLa cells for 48 h. A pool of HeLa cells harboring each inducible shRNA was selected by puromycin (Invitrogen) treatment (2 µg/ml).
Isolation of sEVs by ultracentrifugation
Small extracellular vesicles (sEVs) were isolated from different cell lines (HeLa S3, HEK293T, Huh-7 and RAW 264.7 cells) using a series of ultracentrifugation and micro-filtration steps as previously described [29]. Briefly, cells were grown in DMEM or RPMI-1640 with 10% exosome-depleted FBS and after specific time points, the culture medium was collected. The sEVs were isolated from the conditioned medium by three sequential centrifugation steps at 4 °C: 500g for 15 min; 10,000g for 30 min; followed by micro-filtration through 0.22 µm filter; and a spin at 100,000g for 3 h. The pellets were washed once with 1 × PBS and further centrifuged at 100,000g for 2 h. The sEV pellets obtained were resuspended in 1 × PBS, NP-40 lysis buffer (20 mM tris–HCl, pH 8, 137 mM NaCl, 10% Glycerol, 1% NP40 and 2 mM EDTA) or RIPA (50 mM Tris–HCl, pH 8, 150 mM NaCl, 1% NP40, 0.1% SDS, 0.5% sodium deoxycholate and 1 mM EDTA) supplemented with protease inhibitor cocktail (Roche), 10 mM NaF (Sigma-Aldrich) and 2 mM PMSF (Sigma). The 100,000g pellet obtained was considered as sEVs and used in most of the studies.
Purification of sEVs by sucrose gradient centrifugation
Purification of sEVs through sucrose density gradient was performed as described earlier with some modifications [17]. Briefly, 4 × 106 HeLa cells (per 100 mm plate) were grown for 48 h. Conditioned medium from twenty 100 mm plates was collected and pooled. sEVs were isolated using ultracentrifugation as described above, and the 100,000g pellet was resuspended in 1 × PBS. The sucrose gradient was prepared by diluting a stock solution of 60% (w/v) sucrose in a gradient buffer (20 mM Tris–HCl, pH 7.4 and 137 mM NaCl). The sEV suspension was diluted with 1.1 ml of 60% sucrose solution to obtain a final concentration of 55% sucrose, and dispensed at the bottom of a 5 ml centrifuge tube. This was then sequentially overlaid with 40% sucrose (1100 µl), 20% sucrose (1100 µl), and finally with the gradient buffer (without sucrose, 1100 µl), and subjected to ultracentrifugation at 150,000g for 16 h in MLS-50 rotor (Optima MAX-XP Benchtop Ultracentrifuge Beckman Coulter). Eleven fractions of 400 µl each were collected from top of the gradient, diluted with 3.6 ml 1 × PBS and pelleted at 100,000g for 2 h. Each fraction was resuspended in 1 × PBS for Cryo-TEM or RIPA buffer for western blotting. Fractions (number 5, 6, and 7) were pooled and used for Cryo-TEM.
Purification of CD63-positive exosomes
CD63-positive exosomes were isolated using Exosome-Human CD63 isolation kit (Invitrogen) according to the manufacturer’s instructions. Briefly, pre-enriched exosomes (100,000g pellet) isolated from eight 100 mm HeLa culture plates (~ 50 ml culture media), were first resuspended in 100 µl of 1 × PBS. This solution was then incubated with anti-CD63 antibodies bound to dynabeads (Invitrogen) (100 µl) for 18 h at 4 °C with constant mixing. Later, the bead-bound exosomes were separated using a magnet, washed with 1 × PBS and lysed in 25 µl of RIPA buffer. The supernatant was then removed, added with SDS-loading dye, heated and analyzed by western blotting.
Nanoparticle tracking analysis (NTA)
NTA for determination of the size distribution of the isolated sEVs was performed using Zetaview (Particle Metrix, Germany) as per manufacturer’s instructions. The data were analyzed using Zetaview software (version 8.05.11 SP4).
Cryo-transmission electron microscopy (Cryo-TEM)
Samples were prepared for Cryo-TEM by the frozen hydrated vitrified technique [68] using a Vitrobot Mark IV (FEI Thermo-Fisher, Hillsboro, Oregon) semi-automated sample preparation system. About 4 µl of sEV sample was taken on a holey formvar carbon film 200-mesh Cu (Cu-200HFC Pacific Grid Tech San Francisco, CA) kept in an environmental chamber at a temperature 22 °C and humidity 95%. The sample was made into a thin film by blotting once for one second with a blot force setting of one, and then plunged into liquid ethane at its freezing point. The resultant vitrified grid was transferred under liquid nitrogen to a Gatan Model 655 cryo holder (Gatan, Pleasanton, CA, USA) and then cryo-transferred into the TEM goniometer whilst maintaining the cold chain throughout. Imaging was done in a JEOL 2100 HRTEM (Akishima, Tokyo, Japan) operating at 120 keV while maintaining the sample holder at about − 172 to − 174 °C as measured by a Gatan Smartset model 900 (Gatan, Pleasanton, CA, USA) cold stage controller. The anti-contamination device of the TEM was filled with liquid nitrogen to prevent the vacuum deterioration during analysis. Images were captured by Orius Camera (Gatan, Pleasanton, CA, USA) controlled by Gatan Digital Micrograph software saved in native format as dm3 and then converted to JPEG/TIFF formats.
Protease protection assay
Protease protection assay was performed as described earlier with minor modifications [34]. HeLa-derived sEVs were resuspended in 120 µl of 1 × PBS and added with 100 ng of purified glutathione S-transferase (GST) and were equally separated into three vials. One vial was maintained as untreated control, the second was treated with proteinase K (25 µg/ml final concentration) and the third one with proteinase K (25 µg/ml final concentration) and 0.1% triton X-100 (final concentration). These were incubated for 30 min at 37 °C. The reactions were stopped by the addition of 5 mM PMSF and 5 × SDS-loading dye (for a final concentration of 1.5 ×) and heated at 95 °C for 7 min before analyzing by western blotting.
MVB fractionation
Subcellular fractionation was performed with 3–30% continuous gradient prepared using OptiPrep (Sigma) as described previously [40], with minor modifications. Gradients were made in a 50 mM HEPES (pH 7.0)-based buffer containing 78 mM KCl, 4 mM MgCl2 and 10 mM ethylene glycol tetra-acetic acid (EGTA). Cells were washed, scraped and lysed with a Dounce homogenizer in HEPES buffer (50 mM, pH 7.0) containing 0.25 M sucrose, 78 mM KCl, 4 mM MgCl2, 8.4 mM CaCl2 and 10 mM EGTA, supplemented with complete protease inhibitor cocktail (Roche), 10 mM NaF (Sigma-Aldrich) and 2 mM PMSF (Sigma-Aldrich), 0.5 mM DTT. The lysate was centrifuged twice at 1000g for 5 min and layered on top of the gradient and centrifuged at 133,000g for 5 h. Eleven fractions were collected from top to bottom, and the proteins were precipitated using 100% trichloroacetic acid and analyzed for the presence of specific proteins by western blotting.
Antibodies
The following antibodies were used for western blotting with the indicated dilutions: mouse anti-Ran (1:10,000, BD biosciences 610340), mouse anti-CRM1 (1:2,000, BD 611832), rabbit anti-CRM1(1:3000, Sigma) mouse anti-HSP70 (1:2000, Santa Cruz sc-66048), mouse anti-HSP90 (1:3000, R&D), mouse anti-GAPDH (1:3000), goat anti-TSG101 (1:1500, Santa Cruz sc-6037), mouse anti-α-tubulin (1:10,000, Sigma T5168), mouse anti-HA (1:1000, Covance Research Products MMS-101R), mouse anti-Alix (1:2000, Santa Cruz sc-271975), mouse anti-GFP (1:5000, Santa Cruz sc-9996), mouse anti-β-actin (1:5000, Sigma), rabbit anti-RFLP7a (1:2,000, CST), rabbit anti-CD63 (1:1000, in-house, this paper), rabbit anti-GFP (1:5000, in-house) [69], mouse anti- FLAG (1:3000, Sigma, F1804), mouse anti-RanBP3 (1:1000, BD 612,050), mouse anti-GST (1:3000, Santa Cruz sc-138), mouse anti-Flotilin-2 (1:3000, BD biosciences 610383), donkey anti-rabbit IgG-HRP (1:10,000, GE Healthcare NA-934) and sheep anti-mouse IgG-HRP (1:10,000, GE Healthcare NA-931).
The following antibodies were used for indirect immunofluorescence with indicated dilutions: mouse anti-CD63 (1:300, Santa Cruz, sc-5275), rabbit anti-XPO1/CRM1 (1:500, Sigma, HPA042933), mouse anti- FLAG (1:300, Sigma, F1804), rabbit anti-GFP (1:250, in-house) [69], Alexa Fluor donkey anti-rabbit 488 (Invitrogen, 1:500, A21206), Alexa Fluor donkey anti-rabbit 594 (Invitrogen, 1:500, A21207), Alexa Fluor donkey anti-mouse 488 (Invitrogen, 1:500, A21202), and Alexa Fluor donkey anti-mouse 594 (Invitrogen, 1:500, A21203).
Production of antibodies against human CD63
A coding region corresponding to the extracellular domain of human CD63 (108–200 aa) was PCR amplified from pEGFP-CD63 (a kind gift from Frederik Vilhardt, University of Copenhagen, Denmark) and cloned at the EcoRI/XhoI sites of pET30a. Recombinant protein was expressed in BL21 (DE3) RIL (Novagen) and lysed in denaturation buffer (8 M urea, 100 mM NaH2PO4, 10 mM tris, 1 mM DTT, pH 8.0). The lysate was incubated with Ni–NTA beads and sequentially washed with the wash buffer (100 mM NaH2PO4, 10 mM tris, pH 6.3, containing 8 M, 4 M, and 2 M urea). The last wash was performed with TBS containing 20 mM imidazole. The protein was eluted with TBS containing 250 mM imidazole. The eluted protein was dialysed with 100 mM HEPES buffer, pH 7.6, with 5% glycerol. The recombinant protein was used for raising polyclonal antibodies in rabbit and for affinity purification of antibodies.
Cell lysis, co-immunoprecipitation and mass spectrometric analysis
For preparation of whole cell extract (WCE), cells were washed once with 1 × PBS, scrapped in 1 × PBS on ice, resuspended in NP-40 or RIPA lysis buffer, sonicated and cleared by centrifugation at 15,300g for 20 min at 4 °C. For isolation of proteins from sEVs, the pellets were resuspended in NP-40 or RIPA lysis buffer. Protein estimation was performed using Bradford reagent (Bio-Rad) for both WCE and sEVs and a known amount of protein was analyzed by western blotting.
For co-immunoprecipitation (co-IP) experiments, sEVs were isolated from the Huh-7 cells transfected with GFP-RanQ69L and lysed in cold NP40 lysis buffer containing protease inhibitor cocktail (Roche), 1 mM NaF and 200 mM PMSF. The lysate was centrifuged at 15,300g for 2 min. The supernatant was removed and starting material was taken out and added with 3 × SDS-PAGE loading dye and heated at 95 °C for 5 min. Protein-A sepharose beads (Invitrogen) were bound with anti-GFP antibodies or control rabbit IgG (Vector labs) and the lysate was added to these beads and incubated overnight, at 4 °C, with continuous mixing. The immunoprecipitates were then washed twice with lysis buffer and once with TBS before eluting it in SDS-PAGE loading dye with heating for 5 min at 95 °C. The samples were separated by SDS-PAGE and then transferred Semi-dry transfer onto a PVDF membrane (Merck/Millipore). The membrane was then analyzed by western blotting using specific antibodies and developed using ECL-Plus Western Detection Kit (Thermo Scientific, Cyanagen) following the manufacturer’s instructions. Images of the blots were acquired using Image-Quant LAS 4000 (GE Healthcare).
For all the relative recruitment of proteins into sEVs (Figs. 2A–C, 3A, B, 4C, 5A–D, and S2), initially the level of specific protein in sEVs was normalized to that in the WCE, which was further normalized to a similar value obtained for the sEV marker such as Alix.
For identification of GFP-Ran-Q69L-sepcific interacting proteins, the immunoprecipitates were then washed twice with lysis buffer and twice with tris-buffer (25 mM, pH 7.2). Proteins were eluted in 0.1% RapiGest SF (Waters, 186001861), further subjected to in-solution digestion and the peptides were analyzed using Orbitrap fusion mass spectrometer (Thermo Scientific). The protein identification was performed using Proteome Discoverer software (version 2.2, Thermo Scientific) by applying the Sequest HT database search engine with the cut-off criteria of 1% false discovery rate (FDR).
Immunofluorescence and confocal microscopy
Cells were fixed with 4% Paraformaldehyde (PFA) for 15 min or Methanol for 5 min at room temperature (RT) as per the required experimental condition after 18–20 h of transfection. PFA or methanol fixed coverslips were permeabilized with 0.2% triton X-100 for 15 min (for PFA fixation) and for 30 s (for methanol fixation). Coverslips were then incubated with primary and secondary antibodies in 2% normal horse serum (NHS) blocking solution for 45 min each at RT, with respective antibody dilutions. DNA was stained with Hoechst 33342 during secondary antibody incubation. After each incubation step the coverslips were washed thrice with 1 × PBS. Coverslips were then mounted in VectaShield anti-fade mounting media for confocal microscopy.
The images were obtained using a confocal laser scanning microscope (Olympus FV3000) with 100 × objective (oil, 1.45 NA), which were processed using CellSens and Adobe Photoshop. All the quantitative analysis of the immunofluorescence experiments was performed using data obtained from at least three independent repeats, with the aid of FIJI ImageJ and Microsoft Excel software.
Transient transfection assay for Ran transfer
Intercellular transfer of Ran was performed as described earlier [20]. Briefly, HeLa cells were co-transfected with GFP-Ran-Q69L and mCherry-α-tubulin or mCherry-Histone H2B (transfection marker) for 10 h. Cells were fixed and immunostained using GFP-specific rabbit polyclonal antibodies (in house). The number of GFP-positive (recipient) cells surrounding mCherry-α-tubulin or mCherry-Histone H2B expressing cells (donor) was counted. GFP-MBP was used as a control in case of Fig. S5. The transfer of Ran from at least 25 donor cells from three experiments was assessed for each condition mentioned.
Statistical analysis
The experiments were independently repeated at least three times, and the values are expressed as mean ± SEM or mean ± SD, as indicated in figure legends. P values were calculated using Student’s t test or Mann–Whitney test (Graph pad prism). Graphs were plotted using Graph pad prism software. P values are indicated by asterisks (*). P value between 0.05 and 0.01 is indicated by single asterisk (*), between 0.01 and 0.001 is indicated by double asterisk (**) and between 0.001 and 0.0001 is indicated by triple asterisk (***), and less than 0.0001 is indicated by four asterisk (****).
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Mary Dasso (NICHD, NIH, USA), Frederik Vilhardt (University of Copenhagen, Denmark) and Wei Liu (Zhejiang University School of Medicine, China) for sharing the reagents. The initial help from Joseph lab members for this study is gratefully acknowledged. We thank NCCS Bio-imaging facility and the staff for help with microscopy, and Lizanne Oliveira for the editorial help. We are grateful to Joseph and Seshadri lab members for scientific discussions and helpful suggestions. We thank Mahesh Kulkarni (NCL, Pune) and Atanu Basu (NIV, Pune) for their initial help and insightful discussions on the project. The authors acknowledge support of IIT Bombay Cryo-HRTEM Central Facility in Chemical Engineering Department, and technical help by Shobha Ramagiri. The authors also acknowledge the Department of Biotechnology, Government of India (BT/PR10855/BRB/10/1330/2014) for funding Orbitrap mass spectrometer to NCCS. This work was supported by funding from the Department of Biotechnology (DBT), Ministry of Science and Technology (grant number BT/PR32331/BRB/10/1774/2019) to J.J. and intramural funding from NCCS. Financial support, in the form of fellowships, from the Department of Biotechnology (DBT, Govt. of India) to S.C and S.P., from the University Grants Commission (UGC, Govt. of India) to A.L., and from Council of Scientific and Industrial Research (CSIR, Govt. of India) to D.K., is gratefully acknowledged.
Author contributions
SC, DK, AL and SP performed experiments and analyzed the data. JB and SR helped with EM and proteomics studies, respectively. VS helped in experimental design and critical assessment of the project. JJ conceived the study, supervised the project, and wrote the article. All authors were involved in editing the manuscript.
Funding
This work was supported by funding from the Department of Biotechnology (DBT), Ministry of Science and Technology (grant number BT/PR32331/BRB/10/1774/2019) to J.J. and intramural funding from NCCS.
Data availability
Not Applicable.
Declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent to publish
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Takai Y, Sasaki T, Matozaki T. Small GTP-binding proteins. Physiol Rev. 2001;81:153–208. doi: 10.1152/physrev.2001.81.1.153. [DOI] [PubMed] [Google Scholar]
- 2.Joseph J. Ran at glance. J Cell Sci. 2006;119:3481–3484. doi: 10.1242/jcs.03071. [DOI] [PubMed] [Google Scholar]
- 3.Macara IG. Transport into and out of the nucleus. Microbiol Mol Biol Rev. 2001;65:570–594. doi: 10.1128/MMBR.65.4.570-594.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fung HYJ, Chook YM. Atomic basis of CRM1-cargo recognition, release and inhibition. Semin Cancer Biol. 2014;27:52–61. doi: 10.1016/j.semcancer.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 6.Zurzolo C. Tunneling nanotubes: reshaping connectivity. Curr Opin Cell Biol. 2021;71:139–147. doi: 10.1016/j.ceb.2021.03.003. [DOI] [PubMed] [Google Scholar]
- 7.Lo Cicero A, Stahl PD, Raposo G. Extracellular vesicles shuffling intercellular messages: for good or for bad. Curr Opin Cell Biol. 2015;35:69–77. doi: 10.1016/j.ceb.2015.04.013. [DOI] [PubMed] [Google Scholar]
- 8.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Latifkar A, Hur YH, Sanchez JC, et al. New insights into extracellular vesicle biogenesis and function. J Cell Sci. 2019;132:jcs222406. doi: 10.1242/jcs.222406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hessvik NP, Llorente A. Current knowledge on exosome biogenesis and release. Cell Mol Life Sci. 2018;75:193–208. doi: 10.1007/s00018-017-2595-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pettinato G, Manivel JC, Ravetto C, et al. Papillary cystic tumor of the pancreas: a clinicopathologic study of 20 cases with cytologic, immunohistochemical, ultrastructural, and flow cytometric observations, and a review of the literature. Am J Clin Pathol. 1992;98:478–488. doi: 10.1093/ajcp/98.5.478. [DOI] [PubMed] [Google Scholar]
- 12.Pegtel DM, Gould SJ. Exosomes. Annu Rev Biochem. 2019;88:487–514. doi: 10.1146/annurev-biochem-013118-111902. [DOI] [PubMed] [Google Scholar]
- 13.Meldolesi J. Exosomes and ectosomes in intercellular communication. Curr Biol. 2018;28:R435–R444. doi: 10.1016/j.cub.2018.01.059. [DOI] [PubMed] [Google Scholar]
- 14.Villarroya-Beltri C, Gutiérrez-Vázquez C, Sánchez-Cabo F, et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. 2013;4:2980. doi: 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tourrière H, Chebli K, Zekri L, et al. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J Cell Biol. 2003;160:823–831. doi: 10.1083/jcb.200212128. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Sexton RE, Mpilla G, Kim S, et al. Ras and exosome signaling. Semin Cancer Biol. 2019;54:131–137. doi: 10.1016/j.semcancer.2019.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shurtleff MJ, Temoche-Diaz MM, Karfilis KV, et al. Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. Elife. 2016;5:e19276. doi: 10.7554/eLife.19276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simons M, Raposo G. Exosomes - vesicular carriers for intercellular communication. Curr Opin Cell Biol. 2009;21:575–581. doi: 10.1016/j.ceb.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 19.Mathivanan S, Ji H, Simpson RJ. Exosomes: Extracellular organelles important in intercellular communication. J Proteomics. 2010;73:1907–1920. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 20.Khuperkar D, Helen M, Magre I, Joseph J. Inter-cellular transport of Ran GTPase. PLoS ONE. 2015;10:e0125506. doi: 10.1371/journal.pone.0125506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duijvesz D, Burnum-Johnson KE, Gritsenko MA, et al. Proteomic profiling of exosomes leads to the identification of novel biomarkers for prostate cancer. PLoS ONE. 2013;8:e82589. doi: 10.1371/journal.pone.0082589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaur S, Saldana AC, Elkahloun AG, et al. CD47 interactions with exportin-1 limit the targeting of m7G-modified RNAs to extracellular vesicles. J Cell Commun Signal. 2021 doi: 10.1007/s12079-021-00646-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi DS, Lee JM, Gun WP, et al. Proteomic analysis of microvesicles derived from human colorectal cancer cells. J Proteome Res. 2007;6:4646–4655. doi: 10.1021/pr070192y. [DOI] [PubMed] [Google Scholar]
- 24.Guescini M, Guidolin D, Vallorani L, et al. C2C12 myoblasts release micro-vesicles containing mtDNA and proteins involved in signal transduction. Exp Cell Res. 2010;316:1977–1984. doi: 10.1016/j.yexcr.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 25.Koppen T, Weckmann A, Müller S, et al. Proteomics analyses of microvesicles released by Drosophila Kc167 and S2 cells. Proteomics. 2011;11:4397–4410. doi: 10.1002/pmic.201000774. [DOI] [PubMed] [Google Scholar]
- 26.Mears R, Craven RA, Hanrahan S, et al. Proteomic analysis of melanoma-derived exosomes by two-dimensional polyacrylamide gel electrophoresis and mass spectrometry. Proteomics. 2004;4:4019–4031. doi: 10.1002/pmic.200400876. [DOI] [PubMed] [Google Scholar]
- 27.Miguet L, Pacaud K, Felden C, et al. Proteomic analysis of malignant lymphocyte membrane microparticles using double ionization coverage optimization. Proteomics. 2006;6:153–171. doi: 10.1002/pmic.200500133. [DOI] [PubMed] [Google Scholar]
- 28.Kugeratski FG, Hodge K, Lilla S, et al. Quantitative proteomics identifies the core proteome of exosomes with syntenin-1 as the highest abundant protein and a putative universal biomarker. Nat Cell Biol. 2021;23:631–641. doi: 10.1038/s41556-021-00693-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Théry C, Amigorena S, Raposo G, Clayton A (2006) Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol 30:Chapter 3:Unit 3.22. 10.1002/0471143030.cb0322s30 [DOI] [PubMed]
- 30.Mathieu M, Martin-Jaular L, Lavieu G, Théry C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat Cell Biol. 2019;21:9–17. doi: 10.1038/s41556-018-0250-9. [DOI] [PubMed] [Google Scholar]
- 31.Pluchino S, Smith JA. Explicating exosomes: reclassifying the rising stars of intercellular communication. Cell. 2019;177:225–227. doi: 10.1016/j.cell.2019.03.020. [DOI] [PubMed] [Google Scholar]
- 32.Jeppesen DK, Fenix AM, Franklin JL, et al. Reassessment of exosome composition. Cell. 2019;177:428–445.e18. doi: 10.1016/j.cell.2019.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beltraminelli T, Perez CR, De Palma M (2021) Disentangling the complexity of tumor-derived extracellular vesicles. Cell Rep 35 [DOI] [PubMed]
- 34.Rajagopalan V, Canals D, Luberto C, et al. Critical determinants of mitochondria-associated neutral sphingomyelinase (MA-nSMase) for mitochondrial localization. Biochim Biophys Acta Gen Subj. 2015;1850:628–639. doi: 10.1016/j.bbagen.2014.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang G, Jin S, Huang W, et al. LPS-induced macrophage HMGB1-loaded extracellular vesicles trigger hepatocyte pyroptosis by activating the NLRP3 inflammasome. Cell Death Discov. 2021;7:1–11. doi: 10.1038/s41420-021-00729-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y, Zhao M, Liu S, et al. Macrophage-derived extracellular vesicles: diverse mediators of pathology and therapeutics in multiple diseases. Cell Death Dis. 2020;11:1–18. doi: 10.1038/s41419-020-03127-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McDonald MK, Tian Y, Qureshi RA, et al. Functional significance of macrophage-derived exosomes in inflammation and pain. Pain. 2014;155:1527–1539. doi: 10.1016/j.pain.2014.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bischoff FR, Klebe C, Kretschmer J, et al. RanGAP1 induces GTPase activity of nuclear Ras-related Ran. Proc Natl Acad Sci U S A. 1994;91:2587–2591. doi: 10.1073/pnas.91.7.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klebe C, Bischoff FR, Ponstingl H, Wittinghofer A. Interaction of the nuclear GTP-binding protein ran with its regulatory proteins RCC1 and RanGAP1. Biochemistry. 1995;34:639–647. doi: 10.1021/bi00002a031. [DOI] [PubMed] [Google Scholar]
- 40.Ghosh S, Bose M, Ray A, Bhattacharyya SN. Polysome arrest restricts miRNA turnover by preventing exosomal export of miRNA in growth-retarded mammalian cells. Mol Biol Cell. 2015;26:1072–1083. doi: 10.1091/mbc.E14-11-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Monecke T, Dickmanns A, Ficner R. Allosteric control of the exportin CRM1 unraveled by crystal structure analysis. FEBS J. 2014;281:4179–4194. doi: 10.1111/febs.12842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Monecke T, Haselbach D, Voß B, et al. Structural basis for cooperativity of CRM1 export complex formation. Proc Natl Acad Sci USA. 2013;110:960–965. doi: 10.1073/pnas.1215214110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Englmeier L, Fornerod M, Bischoff FR, et al. RanBP3 influences interactions between CRM1 and its nuclear protein export substrates. EMBO Rep. 2001;2:926–932. doi: 10.1093/embo-reports/kve200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nemergut ME, Lindsay ME, Brownawell AM, Macara IG. Ran-binding protein 3 links Crm1 to the Ran guanine nucleotide exchange factor. J Biol Chem. 2002;277:17385–17388. doi: 10.1074/jbc.C100620200. [DOI] [PubMed] [Google Scholar]
- 45.Petosa C, Schoehn G, Askjaer P, et al. Architecture of CRM1/Exportin1 suggests how cooperativity is achieved during formation of a nuclear export complex. Mol Cell. 2004;16:761–775. doi: 10.1016/j.molcel.2004.11.018. [DOI] [PubMed] [Google Scholar]
- 46.Brown VM, Krynetski EY, Krynetskaia NF, et al. A novel CRM1-mediated nuclear export signal governs nuclear accumulation of glyceraldehyde-3-phosphate dehydrogenase following genotoxic stress. J Biol Chem. 2004;279:5984–5992. doi: 10.1074/jbc.M307071200. [DOI] [PubMed] [Google Scholar]
- 47.Colombo M, Moita C, Van Niel G, et al. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J Cell Sci. 2013;126:5553–5565. doi: 10.1242/jcs.128868. [DOI] [PubMed] [Google Scholar]
- 48.Boudhraa Z, Carmona E, Provencher D, Mes-Masson AM. Ran GTPase: a key player in tumor progression and metastasis. Front Cell Dev Biol. 2020;8:345. doi: 10.3389/fcell.2020.00345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim HJ, Taylor JP. Lost in transportation: nucleocytoplasmic transport defects in ALS and other neurodegenerative diseases. Neuron. 2017;96:285–297. doi: 10.1016/j.neuron.2017.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hachiya N, Sochocka M, Brzecka A, et al. Nuclear envelope and nuclear pore complexes in neurodegenerative diseases—new perspectives for therapeutic interventions. Mol Neurobiol. 2021;58:983–995. doi: 10.1007/s12035-020-02168-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mortimore GE, Pösö AR. Intracellular protein catabolism and its control during nutrient deprivation and supply. Annu Rev Nutr. 1987;7:539–564. doi: 10.1146/annurev.nu.07.070187.002543. [DOI] [PubMed] [Google Scholar]
- 52.Bitetto G, Di Fonzo A. Nucleo-cytoplasmic transport defects and protein aggregates in neurodegeneration. Transl Neurodegener. 2020;9:1–16. doi: 10.1186/s40035-020-00205-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Becker A, Thakur BK, Weiss JM, et al. Extracellular vesicles in cancer: cell-to-cell mediators of metastasis. Cancer Cell. 2016;30:836–848. doi: 10.1016/j.ccell.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thompson AG, Gray E, Heman-Ackah SM, et al. Extracellular vesicles in neurodegenerative disease-pathogenesis to biomarkers. Nat Rev Neurol. 2016;12:346–357. doi: 10.1038/nrneurol.2016.68. [DOI] [PubMed] [Google Scholar]
- 55.Conlan RS, Pisano S, Oliveira MI, et al. Exosomes as reconfigurable therapeutic systems. Trends Mol Med. 2017;23:636–650. doi: 10.1016/j.molmed.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Candelario KM, Steindler DA. The role of extracellular vesicles in the progression of neurodegenerative disease and cancer. Trends Mol Med. 2014;20:368–374. doi: 10.1016/j.molmed.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sabri N, Roth P, Xylourgidis N, et al. Distinct functions of the Drosophila Nup153 and Nup214 FG domains in nuclear protein transport. J Cell Biol. 2007;178:557–565. doi: 10.1083/jcb.200612135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barrès V, Ouellet V, Lafontaine J, et al. An essential role for Ran GTPase in epithelial ovarian cancer cell survival. Mol Cancer. 2010;9:272. doi: 10.1186/1476-4598-9-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kurisetty VV, Johnston PG, Johnston N, et al. RAN GTPase is an effector of the invasive/metastatic phenotype induced by osteopontin. Oncogene. 2008;27:7139–7149. doi: 10.1038/onc.2008.325. [DOI] [PubMed] [Google Scholar]
- 60.Matchett KB, McFarlane S, Hamilton SE, et al. Ran GTPase in nuclear envelope formation and cancer metastasis. Adv Exp Med Biol. 2014;773:323–351. doi: 10.1007/978-1-4899-8032-8_15. [DOI] [PubMed] [Google Scholar]
- 61.Rensen WM, Mangiacasale R, Ciciarello M, Lavia P. The GTPase Ran: regulation of cell life and potential roles in cell transformation. Front Biosci. 2008;13:4097–4121. doi: 10.2741/2996. [DOI] [PubMed] [Google Scholar]
- 62.Çağatay T, Chook YM. Karyopherins in cancer. Curr Opin Cell Biol. 2018;52:30–42. doi: 10.1016/j.ceb.2018.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dakir E-H, Pickard A, Srivastava K, et al. The anti-psychotic drug pimozide is a novel chemotherapeutic for breast cancer. Oncotarget. 2018;9:34889–34910. doi: 10.18632/oncotarget.26175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Haggag YA, Matchett KB, Falconer RA, et al. Novel ran-RCC1 inhibitory peptide-loaded nanoparticles have anti-cancer efficacy in vitro and in vivo. Cancers (Basel) 2019;11:222. doi: 10.3390/cancers11020222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Azmi AS, Uddin MH, Mohammad RM. The nuclear export protein XPO1—from biology to targeted therapy. Nat Rev Clin Oncol. 2021;18:152–169. doi: 10.1038/s41571-020-00442-4. [DOI] [PubMed] [Google Scholar]
- 66.Azizian NG, Azizian NG, Li Y, Li Y. XPO1-dependent nuclear export as a target for cancer therapy. J Hematol Oncol. 2020;13:61. doi: 10.1186/s13045-020-00903-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sahoo MR, Gaikwad S, Khuperkar D, et al. Nup358 binds to AGO proteins through its SUMO -interacting motifs and promotes the association of target mRNA with miRISC. EMBO Rep. 2017;18:241–263. doi: 10.15252/embr.201642386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bellare JR, Davis HT, Scriven LE, Talmon Y. Controlled environment vitrification system: an improved sample preparation technique. J Electron Microsc Tech. 1988;10:87–111. doi: 10.1002/jemt.1060100111. [DOI] [PubMed] [Google Scholar]
- 69.Murawala P, Tripathi MM, Vyas P, et al. Nup358 interacts with APC and plays a role in cell polarization. J Cell Sci. 2009;122:3113–3122. doi: 10.1242/jcs.037523. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not Applicable.