Abstract
Putative RNA-binding proteins (RBPs), zygote arrested-1 (ZAR1), and ZAR2 (also known as ZAR1L), have been identified as maternal factors that mainly function in oogenesis and embryogenesis. Despite divergence in their spatio-temporal expression among species, the CxxC structure of the C-terminus of ZAR proteins is highly conserved and is reported to be the functional domain for the activity of the RBPs of ZAR proteins. In oocytes from Xenopus laevis and zebrafish, ZAR proteins have been reported to bind to maternal transcripts and inhibit translation in immature growing oocytes, whereas in fully grown mouse oocytes, they promote the translation during meiotic maturation. Thus, ZAR1 and ZAR2 may be required for the maternal-to-zygotic transition by stabilizing the maternal transcriptome in oocytes with partial functional redundancy. In addition, recent studies have suggested non-ovarian expression and function of ZAR proteins, particularly their involvement in tumorigenesis. ZAR proteins are potentially associated with tumor suppressors and can serve as epigenetically inactivated cancer biomarkers. In this review, studies on Zar1/2 are systematically summarized, and some issues that require discussion and further investigation are introduced as perspectives.
Keywords: RNA-binding proteins, Meiosis, mRNA homeostasis, Oocyte, Zygote, Maternal-to-zygotic transition, Tumorigenesis
Introduction
RNA-binding proteins (RBPs)
RBPs interact with RNAs by recognizing specific structures or motifs and forming ribonucleoprotein complexes [1–3]. To date, many biological events involving RNA have been confirmed to be regulated by RBP interactions, such as processing [4], alternative splicing, translation, translocation, and degradation [4–9], for which the specificity and affinity of the binding are vital.
To accurately recognize and bind to target transcripts, divergent functional domains are acquired, which are generally known as RNA-binding domains. An RBP commonly comprises more than one RNA-binding domain [10], some of which interact with other proteins to regulate the fate of transcripts. Zinc finger domains have been widely identified in many RBPs, which function via a Zn2+-dependent mechanism. Zinc finger domains may bind to nucleic acids or proteins owing to their divergent structures [11, 12], with the arrangement of cysteine residues being a crucial contributor [10, 12].
Oogenic meiosis is vital for the reproduction of animals, during which transcription is silenced, and physiological activities are determined by accumulated maternal transcripts [13, 14] thus, oocytes are an ideal model to explore the post-transcriptional regulatory mechanisms of RBPs. Many RBPs have been identified in oocytes [13, 14], some of which are known as oocyte-specific maternal RBPs [7, 8]. As these RBPs appear to be functionally important for maintaining homeostasis of the maternal transcriptome, in-depth research on their mechanisms is needed.
Oocyte meiosis
In mammals, the biogenesis of germ cells between males and females is remarkably different. After sex differentiation, female germ cells undergo mitotic proliferation and enter meiosis until they are arrested at the diploid stage of prophase before birth [15, 16]. However, the male germ cell line maintains the ability to renew after birth [17]. Thus, owing to the finiteness of oocyte quantity in postnatal females, the quality of oocytes is considered one of a determining factors of fertility in female mammals [13, 14].
Prolonged and discontinuous meiosis occurs throughout oogenesis [13, 16], and oocyte development arrested at the prophase of the first meiotic division for decades. Thereafter, the meiotic-resumed oocytes continuously develop into metaphase of the second meiotic division and are arrested for the second time, but are not reactivated until fertilized [17]. Thus, the normal process of meiosis is one of the criteria for determining the developmental quality of oogenesis [18, 19].
As oocytes resume meiosis I at the beginning of ovulation, ensuring normal oocyte development is important. At this stage, with the completion of genomic DNA methylation [20], transcriptional silencing events occur in meiotic oocytes; thus, the accumulation of maternal transcripts in growing oocytes is vital for subsequent developmental competence [17]. Accordingly, genes that are specifically expressed and functional for maternal transcriptome homeostasis at this stage have been widely studied.
Maternal-to-zygotic transition (MZT)
The destination of oogenesis is fertilization; thus, the potency of embryogenesis is an important criterion for the quality of oocytes [17]. As transcription is silenced in meiotic oocytes until zygotic genome activation (ZGA) after fertilization, some of the maternal mRNAs transcribed in growing oocytes are also vital for the early stage of embryogenesis [13, 21, 22]. Thus, the balance between translation and degradation of maternal transcripts is essential for the transition of development control from maternal to zygotic genes, a process called the maternal-to-zygotic transition (MZT).
As RBPs are closely related to the post-transcriptional regulation of RNA, they play important roles in MZT [23]. Zygote arrested-1 (Zar1) was first identified in Mus musculus as one of the earliest discovered maternal factors during MZT [3, 7, 24], and Zar2 (also known as Zar1-like, Zar1l) was found to be homologous to Zar1 [2, 3, 25]. Both Zar1 and Zar2 are important for MZT with partial functional redundancy, which may be related to their RNA-binding capacity [8]. ZAR proteins do not conform to the general definition of an RBP or a maternal factor for the following reasons.
As RBPs, ZAR1 and ZAR2 lack typical RNA-binding domains and possess an atypical zinc finger domain, of which the structures are highly conserved among species [2, 7, 8, 24–27] (Fig. 1).
As maternal factors, Zar1 and Zar2 are expressed as early as the growing stage of oogenesis, that is, oocytes in primary, secondary, and early antral follicles, and play partially redundant roles in maintaining the stability of the maternal transcriptome, subsequently influencing embryogenesis [2, 8, 28]. In comparison, the typical maternal factors regulating MZT, such as BTG4, CNOT6L, and PABPN1L, are stored in growing oocytes as mature mRNAs and are transiently translated into proteins after meiotic resumption, which is the final stage of oogenesis.
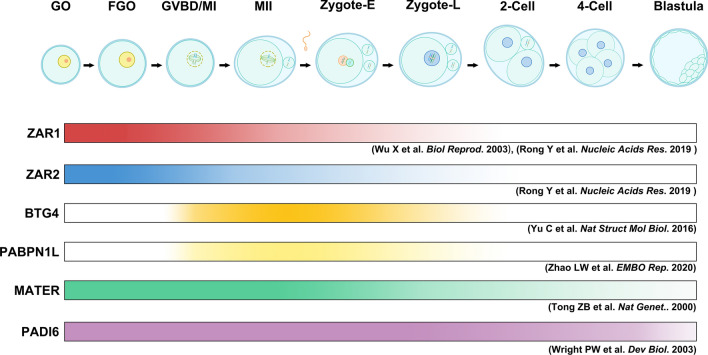
The temporal expression of Zar1 and Zar2 in murine oocytes is highest during oocyte growth and gradually decreases until disappearance in 2-cell stage, which is unique compared to other typical maternal effectors, such as MSY2, BTG4, PABPN1L, or PADI6 [29–31] (Fig. 2). As discussed later, new genetic evidence reveals that ZAR1/2 are required as early as in growing oocytes instead of functioning after fertilization. This is consistent with their expression pattern but is remarkably different from other well-established maternal factors.
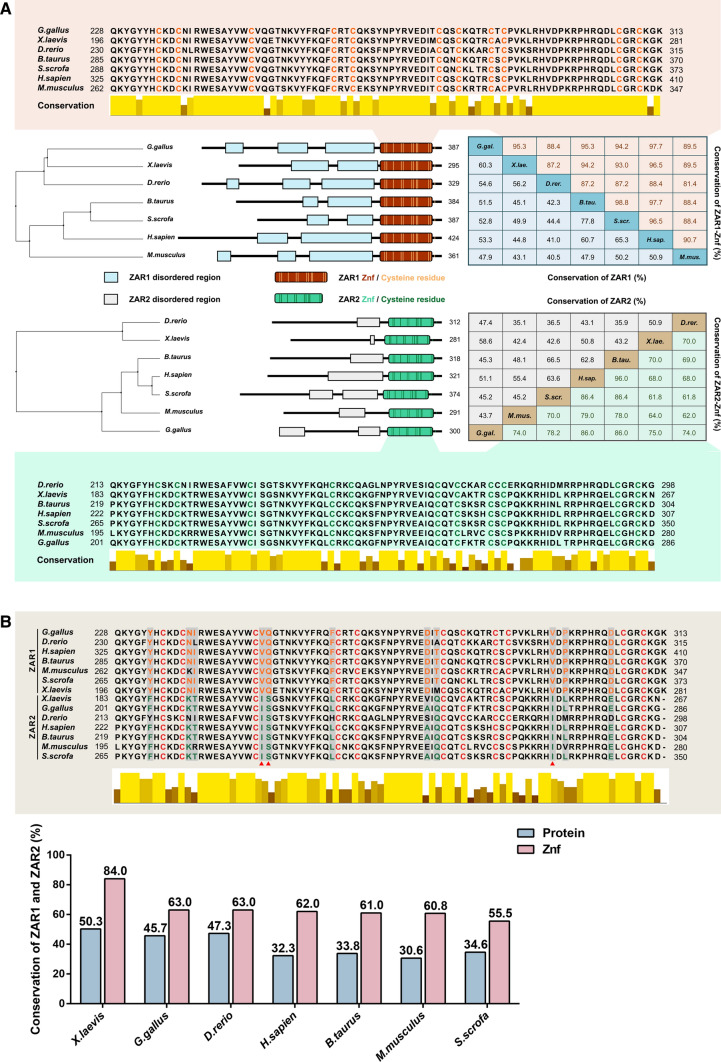
Fig. 1.
Of ZAR1 and ZAR2 among species. A Phylogenetic tree, schematic, amino acid identity (%), and sequence alignment of ZAR1 and ZAR2 among species. B Sequence alignment to display the amino acid similarity of the C-terminus between ZAR1 and ZAR2. The interleaved bar plot shows the conservation of amino acids and zinc-finger domains between ZAR1 and ZAR2. The grey boxes highlight the distinguishing residues, and the red deltas indicate the completely conserved residues among the chosen species. The protein sequences of Zar1/2 were collected from NCBI using the following reference sequences: Homo sapiens (NP_783318.1/NP_001130043.1), Mus musculus (NP_777366.1/NP_001153165.1), Xenopus laevis (NP_001083958.1/NP_001153159.1), Bos taurus (NP_001069671.1/NP_001120912.1), Sus scrofa (NP_001123428.1/ P_020920840.1), Danio rerio (NP_919362.2/NP_001186296.1), and Gallus gallus (XP_003641256.4/NP_001165014.2). The phylogenetic tree and sequence alignment were generated in Jalview using protein sequences analyzed by ClustalX. A schematic was scaled in proportion to the position of amino acids, and domain information was obtained from UniProt (Znf refers to zinc finger domain). Amino acid identity was calculated using Vector NTI 10
Fig. 2.
Temporal expression of Zar1/2 compared to other maternal effectors in mice. The pattern of Zar1/2 and some key factors of oogenesis and maternal-to-zygotic transition in mice. The gradient ribbons show the protein expression patterns. The references are labelled below the corresponding ribbon. Zygote-E refers to early zygotes, and Zygote-L refers to late zygote
Thus, the function of ZAR proteins needs to be clarified. In this review, we summarize the research progress of Zar1 and Zar2 using existing studies and propose prospects for further studies that have not been elucidated.
Of note, another ‘ZAR1’ (HOPZ-ACTIVATED RESISTANCE 1) was discovered in Arabidopsis, which is reported to recognize foreign pathogens and activate immunity to resist invasion [32]. ‘ZAR1’ in plants completely differs from the vertebrate Zar1 reviewed in this article [32].
Zar1 and Zar2
History of Zar1 and Zar2
Zar1 was discovered by Wu in 2003 [24] and was defined as one of the first discovered oocyte-specific maternal factors in mice, because the deficiency of Zar1 has no effect on the development of mice but causes infertility in female mice, resulting in the arrest of zygotes at the two-cell stage [8, 24].
The orthologs of Zar1 have been successively identified among species [7, 24, 26, 27, 33, 34]. ZAR1 may function as a maternal RBP, which causes translation repression when deleted in Xenopus and zebrafish [25, 35]. According to Rong et al. [8], the transcriptome is dysregulated in Zar1/2 knockout oocytes, thereby further confirming the role of ZAR1 as an RBP. Recently, two novel rare SNPs of ZAR1, a homolog of Zar1 in humans, were reported to be potentially associated with human zygote arrest [36].
Zar2 was first identified in bovine by Sangiorgio [25]. Owing to the similarity in structure between Zar1 and Zar2, but difference in localization on chromosome of Zar2 as Zar1, Zar2 was first called Zar1l. In 2013, Yamamoto et al. [26] formally distinguished Zar1 and Zar2 by identifying 12 conserved amino acid differences at the C-terminus (Fig. 1B) and synteny [26]. Studies on Zar2 have been conducted on various species [2, 3, 25]. In 2010, the function of Zar2 in mice was first reported by Hu et al., which indicated that Zar2 is vital for embryogenesis [2]. Subsequently, the functional similarity between Zar2 and Zar1 in Xenopus revealed that Zar2 bound to mRNA and repressed translation in early oocytes [3, 26]. However, the results of in vivo experiments performed by Rong showed that Zar2 gene knockout led to a slight downregulation of the oocyte development rate in mice [8].
By establishing Zar1/2 double knockout in mice, the in vivo functions of Zar1 and Zar2 were systematically studied, revealing that the oogenesis of Zar1/2−/− oocytes was impaired with abnormal spindle assembly, delayed meiosis resumption, and dysregulated transcriptome [8]. Thus, the primary notion that Zar1 is a maternal effector that functions after fertilization was revealed to be inaccurate.
Identity of Zar1 and Zar2
Conservation of Zar1 and Zar2
Soon after the discovery of Zar1 and Zar2, orthologs of these two genes were successively identified among species, and their evolutionary conservatism was comprehensively analyzed [2, 3, 24–27, 33–35]. Multiple sequence alignment analysis revealed that the conservation of Zar1 and Zar2 among species was limited, which may be due to divergent ancestral lineages (Fig. 1). However, the C-terminus of the homolog of these two proteins is highly conserved among species [7, 24, 26], suggesting that the C-terminus of ZAR proteins is functionally important (Fig. 1).
Although ZAR1 and ZAR2 are homologs, the conservation between these two proteins in species is not high, with the C-terminus being slightly higher than the full-length proteins. These similarities and differences may explain why ZAR1 and ZAR2 are partially redundant.
Expression pattern of ZAR1 and ZAR2
To identify the molecular functions of ZAR1 and ZAR2 in vivo, knowledge of their spatial and temporal distributions is important. There are similarities and differences in the distribution of ZAR proteins in different species, but among them all, ZAR proteins were found to have a conserved ovary localization [7, 8]. In human tissue samples, ZAR proteins were found to be expressed in many other non-ovarian organs, such as the lung, spleen, and heart [25, 27, 33, 34]. Several studies have also indicated that ZAR proteins are expressed in some cancer cells [37–40], suggesting that they may have non-ovarian functions.
High levels of ZAR1 and ZAR2 expression are conserved in oocytes and embryos of many mammalian and non-mammalian species [2, 7, 8, 24–28, 33, 34]. In murine oocytes, ZAR1 is preferentially located in the cortical region, whereas ZAR2 is evenly distributed [8, 24]. Notably, ZAR proteins showed the highest abundance in mice at the early stage of oogenesis, with a continuous decline, and ceased to be expressed before ZGA (Fig. 2). In contrast, the expression of ZAR proteins can persist in the blastosphere in cattle and pigs [8, 24], which is far beyond ZGA [21]. The temporal difference may be attributed to the divergent time nodes of ZGA among different species [33]. Furthermore, the diverse localizations suggest that ZAR proteins may not be completely redundant, which requires in-depth studies.
In general, the highest expression in immature mouse oocytes, which is unusual among many maternal factors, such as Btg4 and Pabpn1l [15, 29], suggests that ZAR proteins may play important roles in oogenesis (Fig. 2), which has been confirmed by Rong et al. [8].
Structure of ZAR1 and ZAR2
ZAR proteins have a conserved C-terminus in all homologs, which contain 12 conserved cysteine residues [8]. In 2003, the C-terminus of ZAR1 was initially found to contain an unconventional plant homeodomain (PHD), known as a zinc finger domain, with an H to C substitution, which is potentially associated with transcriptional regulation [24]. Thereafter, the RNA-binding competence of ZAR1 and ZAR2 was reported to be significantly downregulated with the mutation of the C-terminus [8, 26, 35], along with the Zn2+ dependency of the binding activity. ZAR1 and ZAR2 putatively contain a CxxC zinc finger domain. However, as the CxxC domain of ZAR1 lacks a conclusive correlation with typical zinc finger domains, ZAR proteins may contain an undescribed ZAR-specific zinc finger domain [3] (Fig. 1).
In addition, in the zinc finger domains in the C-terminus of ZAR proteins, there are 11 conserved residues that distinguish ZAR proteins into ZAR1 and ZAR2 (Fig. 1B). Although the residue differences between ZAR1 and ZAR2 are significant, the biochemical properties of these alternative amino acids have not been investigated. Thus, further studies are needed to elucidate the relationship between the function and differences of residues.
The N-terminus of ZAR1 and ZAR2 is less conserved among species [3, 26, 35]. In 2010, Hu reported that two-cell embryos overexpressing ZAR2 C-terminus-EGFP could be arrested by dominant-negative effects [2]. Thus, structures other than the C-terminus of ZAR2 may also have important biofunctions. However, the functional domains and mechanisms of the N-terminus of ZAR proteins remain unknown.
ZAR1 was reported to contain disordered domains that are generally found in proteins that tend to form hydrogels or amyloid-like aggregates [35] (Fig. 1A). In cells, some non-membrane organelles consisting of RNA and RBPs are vital for post-transcriptional and translational regulation, creating a separate chemical environment by phase separation in the cytoplasm [41]. The amyloid-like structure caused by phase-separated-protein-forming hydrogels is known to repress target mRNA translation during gametogenesis [42]. Zar2 can also form foci in the cytoplasm when overexpressed in late 2-cell-stage embryos [2]. Thus, the structure of ZAR proteins indicates their phase separation properties, despite the need for more evidence.
Function of Zar1 and Zar2
Since the discovery of ZAR1 in 2003, studies on the functions of Zar1 and Zar2 have been conducted among species [24]. To date, these studies have reported that ZAR proteins are potentially involved in the regulation of the transcriptome and translation during MZT, as described below.
ZAR1/ZAR2 regulate the transcriptome in oocytes
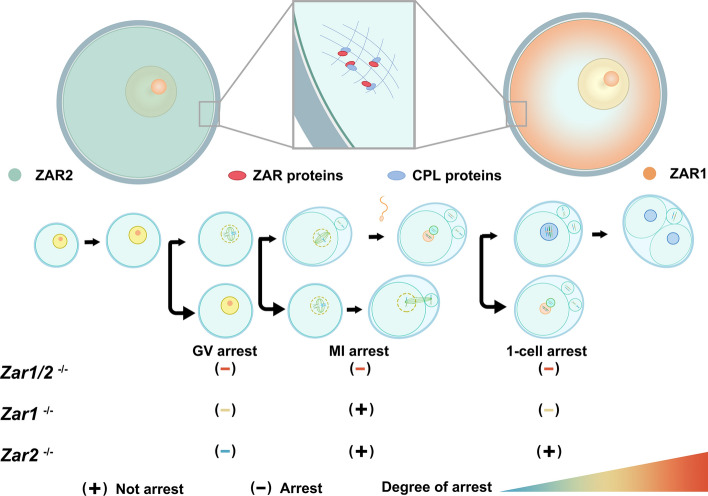
As early as 2003, it was hypothesized that Zar1 regulates maternal mRNA in oocytes because of the atypical zinc finger domain in its C-terminus [24]. In 2019, Rong et al. reported that numerous differentially expressed genes were observed in Zar1/2−/− oocytes, in addition to their binding and co-localization to PADI6, MATER, and MSY2 in the cytoplasm of mouse oocytes, which are RBPs that maintain transcriptome homeostasis by assembling cytoplasmic lattices (CPLs) [8, 43, 44] (Fig. 3). Accordingly, the importance of Zar1 and Zar2 in maintaining homeostasis of the transcriptome has been indicated [8].
Fig. 3.
Localization and role of Zar1/2 in mice oocytes. In Mus musculus, ZAR1 and ZAR2 are distributed differently as ZAR1 is located in cortical areas, whereas ZAR2 is evenly distributed in the cytoplasm. ZAR proteins putatively interact with cytoplasmic lattice (CPL) proteins to maintain oogenesis. In Zar1−/− or Zar2−/− oocytes, meiosis is abnormal, and Zar2−/− mice are fertile. However, in Zar1/2−/− oocytes, meiotic maturation defects were much more serious than in Zar1−/− and Zar2−/− oocytes, with some oocytes arrested at meiosis I (MI) and exhibiting abnormal spindles
Germline-specific cytoplasmic chromatoid bodies (C-bodies) have a structure similar to that of the processing body (P-body) in male germ cells [45], which may engage in chromosome modification and genome stability. In 2010, Zar1 and Zar2 were reported to be co-localized with components of the C-body and P-body in murine somatic cells, suggesting that ZAR proteins may be associated with the stability of mRNAs.
The detailed mechanisms by which ZAR proteins influence the maternal transcriptome remain unknown, and ZAR-binding transcripts need to be identified. A recently developed technique, LACE-seq [46], suitable for a small number of oocytes, can be used to resolve this issue and to analyze the binding motif of ZAR proteins.
Regulation of translation
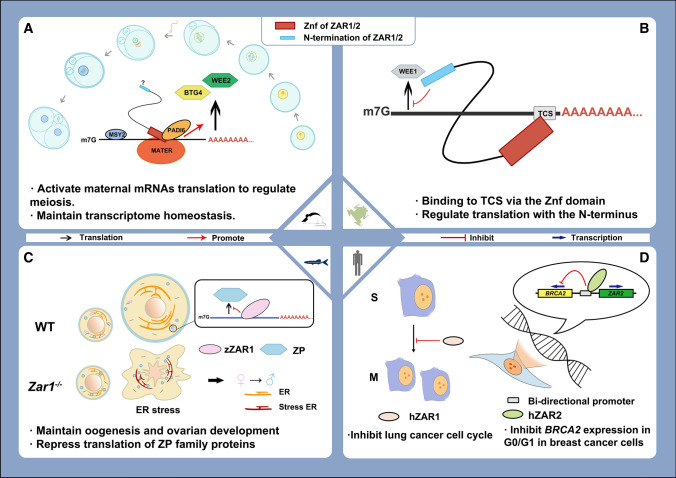
The 3ʹ-untranslated region (3ʹ-UTR) of mRNA is commonly known to regulate mRNA-related processing, including translation [47] (Fig. 4). Some cis-regulatory elements are located on the 3ʹ-UTR of mRNA and are associated with spatio-temporal translation, such as cytoplasmic polyadenylation elements [48, 49], polyadenylation signals [5, 9], and translational control sequence (TCSs) [50]. TCSs were first identified in the 3ʹ-UTR of Wee1 mRNA in oocytes of Xenopus laevis [50], which functions dually to repress translation in immature oocytes while activating translation in meiotic oocytes [51]. In 2012, Xenopus Zar2 was first identified as a trans-acting factor binding to TCSs through its C-terminus and was found to repress the translation of maternal transcripts depending on its N-terminus [3]. Later in 2013, the same function of Xenopus ZAR1 was revealed, with a markedly higher affinity than Xenopus ZAR2 to the TCS [26]. In 2016, the translationally repressive function of ZAR1 was reported in zebrafish oocytes, in which the expression of zona pellucida mRNA was suppressed to maintain oogenesis and ovarian development [35]. Some translational regulatory elements were also reported to be co-immunoprecipitated with ZAR1 in zebrafish, such as CPEB1 [35, 49], suggesting that ZAR1 may interact with other proteins to repress translation in immature oocytes. However, the detailed mechanisms involved have not yet been elucidated.
Fig. 4.
Function model of Zar1/2 among species. The function of Zar1/2 among species reported to date. A In Mus musculus, ZAR proteins were reported to activate translation and maintain homeostasis of the maternal transcriptome by binding to mRNAs and interacting with other proteins, such as MSY2 and PADI6, in oocytes with partially functional complementarity. The C-terminus is functionally involved in the binding of RNAs and proteins. B In Xenopus laevis oocytes, ZAR proteins were reported to specifically recognize the TCS in the 3ʹ UTR of Wee1 mRNA, which represses translation. C In zebrafish, ZAR1 was reported to bind mRNAs of zona pellucida (Zp) and repress their translation in early stage oocytes. Deletion of ZAR1 induces endoplasmic reticulum (ER) stress, which leads to oocyte apoptosis and female-to-male sex transition. zZAR1 refers to ZAR1 in zebrafish. D In humans, ZAR1 and ZAR2 were found to function in repressing the development of cancer cells, whereby ZAR1 represses cell cycles and causes lung cancer cells to arrest at the S phase. ZAR2 binds to the bi-promoter of BRCA2 and ZAR2, thereby repressing the transcription of BRCA2 to repress breast cancer cells. hZAR1 and hZAR2 refer to ZAR1 and ZAR2 in humans, respectively
In murine oocytes, ZAR1/ZAR2 was reported to activate the translation of some maternal transcripts, such as Btg4 and Wee2 [35, 49]. The contrasting results may be due to the different stage oocytes used in the studies. In studies on X. laevis and zebrafish, immature growing oocytes were used, and the phenomenon of promoting translation was mainly observed after meiotic resumption, characterized by germinal vesicle breakdown (GVBD) in mice. According to the significant reduction in maternal mRNA levels in Zar1/2−/− growing oocytes and fully grown oocytes, it is possible that some transcripts of translation-related proteins fail to accumulate normally, thereby, decreasing the translational level. However, this hypothesis requires more experimental data.
In conclusion, although the involvement of ZAR1 and ZAR2 in translational regulation has been confirmed, the underlying mechanisms require further study.
ZAR1/ZAR2 regulates meiotic maturation of oocytes
As Zar1 was first identified as a maternal effect gene in mammals, it was reported to have no effect on oogenesis [24]. However, an in-depth study by Rong et al. found that the exact functions of Zar1 and Zar2 were executed during meiosis rather than post-fertilization, which aligns with the expression window of ZAR proteins [8]. Zar2−/− female mice were fertile, but their GVBD rate was slightly downregulated and their polar body-1 (PB1) emission was delayed. Subsequently, the same phenotypes were found in Zar1−/− oocytes with more severe effects, in addition to mutated female infertility. However, in Zar1/2−/− oocytes, the process of meiosis was significantly abnormal, with a much lower rate of GVBD and the emission rate of PB1, in addition to disordered spindle assembly [8]. Some maternal transcripts, such as Tpx2 and Wee2, which are associated with spindle assembly and the progression of meiosis, respectively, failed to accumulate in Zar1/2 null oocytes [8]. Thus, Zar1 and Zar2 showed partial functional redundancy; the offset by Zar2 may explain why Zar1 null mice were first reported to have normal oogenesis [8].
In contrast to Zar1−/− murine oocytes that can develop to be fertilized, the mutation of Zar1 in zebrafish was reported to cause severe developmental arrest of oocytes, eventually causing a female to male sex reversal [35]. Thus, Zar1 is functionally divergent among species, even within the same cell type. Further studies are needed to determine whether there are other mechanisms of Zar1 in other species and whether Zar2 has similar functions.
ZAR1/ZAR2 regulate embryogenesis
The development of Zar1-deficient murine oocytes was initially reported to be normal during meiosis until arrest in two-cell stage [7, 8]. Thus, early studies on ZAR proteins were mainly focused on preimplantation embryogenesis, where failed pronuclei fusion and ZGA were observed [7, 8], while some of the totipotency genes critical for ZGA, such as MuERV-L [52], are not activated in mouse Zar1/2♀−/♂+ 1-cell zygotes [8]. However, whether the zygotic abnormalities caused by the aftershock of the deficiency in early oogenesis or ZAR proteins function directly in preimplantation embryogenesis remains unknown.
Histone methylation modifications, such as H3K4 [53] and H3K9 [54], have divergent functions in gene expression by activating and repressing transcription, respectively. In 2010, Hu et al. [2] found that when C-terminus-mutated Zar2 was overexpressed in mouse zygotes by mRNA microinjection, H3K4me2/3 was significantly downregulated, whereas H3K9me3 was upregulated. In addition, the expression levels of Dppa2 and Dppa4 [55], which are chromatin modification components that might function in transcriptional activation [56], were significantly downregulated in early embryos overexpressing mutated Zar2. In addition, transcription activity was also found to be significantly downregulated in embryos that were arrested at the 2-cell stage. These results suggest that ZAR2 may maintain histone–demethylation-associated modifications to ensure ZGA [2].
Whether the same effects described above exist in Zar1 null oocytes and whether in vivo histone modifications are influenced by ZAR proteins in oocytes remains unclear. Thus, further research is necessary to determine whether ZAR proteins influence DNA and histone methylation in oocytes. During oogenesis, prolonged transcriptional silencing events occur in fully grown oocytes within preovulatory follicles, for which de novo methylation of the genome is necessary. Thus, oocytes are the desired models for identifying chromatin modifications [17].
The initiation of ZGA requires the degradation of maternal transcripts as the premise [13]; thus, the regulation of maternal transcriptome in early embryos is important. The P-body and C-body are structures that might be specific for storing and degrading transcripts [57], of which the C-body is germ-line specific [58]. EIF2C1 (AGO1), EIF2C2 (AGO2), DDX6, and LSM14A are known components of the P-body [58], and PIWIL1, PIWIL2, and LIN28 are components of the C-body [56], which have been reported to co-localize or interact with ZAR2 when ectopically expressed in somatic cells [2]. Thus, ZAR1 and ZAR2 may engage in the formation of germline-specific C-bodies in embryos to maintain embryo development, but the detailed mechanism needs to be clarified.
Furthermore, PIWI and AGO are both subfamily members of the argonaute protein, and are functionally associated with maintaining the stability of the transcriptome by regulating transposons [46, 59, 60]. Some transposable elements, such as MT and MuERV-L, are reported to be associated with the initiation of transcription and acquisition of totipotency [53, 61, 62] in ZGA, which are important for MZT. Recently, AGO2 was confirmed to repress LTR-driven activation of transcription in mouse oocytes [46], whereas ZAR1 and ZAR2 were found to co-localize with AGO1, AGO2, and PIWIL2 when expressed in murine somatic cells and human 293 T cells, and the mRNA level of piwil2 was also found to be significantly downregulated in Zar2 mutated 2-cell embryos [2], suggesting that ZAR1 and ZAR2 may play a role in the regulation of transposable elements in both oocytes and embryos, which needs further evidence to confirm.
ZAR1 regulates human preimplantation embryogenesis
ZAR1 is the human homolog of mouse Zar1, which was also detected in human ovaries [24, 38, 63] thus, the functions of ZAR1 were considered. In a recent etiological analysis of patients with recurrent zygote arrest in artificial insemination, two single nucleotide polymorphism sites (SNPs) in ZAR1 were detected with significant statistical differences compared to controls, which are both synonymous variations in ZAR1 [63]. Synonymous variations have been reported to be related to many human diseases by regulating the fate of mRNAs [63, 64]. Thus, these two SNPs of ZAR1 may be partially associated with zygote arrest, suggesting that ZAR1 may maintain human preimplantation embryogenesis. However, as research on human oocytes has not focused on specific genes, the mechanisms of ZAR1 and its potential involvement in human fertility require further elucidation.
Zar1 in non-oocyte tissues
According to previous studies, homologs of Zar1 among species were found to be expressed in other non-ovarian organs [24, 27, 28] (Fig. 4), which suggests that the functions of ZAR proteins in cell lines other than germ cells need to be elucidated.
ZAR1 was first reported to be specifically expressed in the ovaries and testes [7, 24]. However, in recent studies, the expression of ZAR1 was found in cancer cell lines [37–40]. More abnormal hypermethylation of the promoter or non-promoter regions of ZAR1 has been reported in many cancer cell lines, such as melanoma [37], brain tumors [40] and lung cancers [38], compared to their paracancer tissues or benign tumors.
ZAR1 is more highly expressed in normal lung cells than in other non-ovarian organs [38]. However, in lung cancer cell lines, ZAR1 was confirmed to be inactivated by a hypermethylated promoter [38]. The demethylation treatment could effectively recover the expression of ZAR1 and inhibit tumorigenesis, and when ZAR1 was overexpressed in cancer cell lines, the cell cycle of cancer cell lines was inhibited [38] (Fig. 4D). Moreover, in the primary tissues of lung cancers, ZAR1 was also aberrantly methylated compared to the para-carcinoma tissues, of which the methylation level was significantly higher in the primary tissues of non-small cell lung carcinoma than in small cell lung carcinoma [38].Thus, ZAR1 may not only be an epigenetically repressed growth inhibitory factor of lung cancer cells but also a hypermethylated biomarker to detect lung cancer cells and distinguish non-small cell lung carcinoma from small cell lung carcinoma.
In addition, in malignant melanomas and diffuse brain tumors, such as glioblastomas and pituitary adenomas, the off-promoters of the ZAR1 intergenic regions are frequently aberrantly methylated [37, 38]. However, contrary to be inhibited in lung cancer cells, the expression of ZAR1 is abnormally activated in melanomas and remains undetectable in glioblastomas [37, 38]. In addition, the overexpression or inhibition of ZAR1 in melanoma cells would not affect carcinogenesis; thus, there was no evidence that the extensive aberrant methylation in non-promoter regions was correlated with ZAR1 expression and tumorigenesis in these cancer cell lines [37, 38]. Nonetheless, as the abnormally high levels of methylation in non-promoter-intergenic regions were found to occur prevalently, they could be used as a biomarker to identify cancers mentioned above.
The transcription of ZAR2 and BRCA2 is initiated by a bidirectional promoter, and ZAR2 was found to repress breast cancer cells by binding to the bidirectional promoter, thereby partly silencing the expression of BRCA2 in the G0/G1 stage of the cell cycle [39] (Fig. 4D). In general, ZAR proteins are potentially associated with tumor suppressors and can serve as epigenetically inactivated cancer biomarkers [37, 38, 40]. However, whether ZAR proteins are vital for repressing tumorigenesis remains unknown and requires further investigation.
Perspectives
Zar1 was initially identified in mouse oocytes as a maternal factor that causes the arrest of 2-cell embryos when mutated, whereas Zar2 was first identified as a homolog of Zar1. ZAR proteins have been confirmed to be expressed in many other species with partially conserved protein structures. Accordingly, the functions of these two proteins were further studied in mice, X. laevis, and zebrafish (Fig. 4). However, many gaps in our understanding of these functions remain, which are as follows:
Maternal factors are defined as genes whose products are accumulated in growing oocytes, but have little effect on oogenesis and fertilization, and specifically function in embryogenesis. However, it has been demonstrated in both mouse and zebrafish that ZAR proteins play an earlier and more important role in meiotic maturation than in preimplantation embryogenesis, which coincides with their expression window. Thus, it is inaccurate to identify Zar1 as a maternal effector. Further research is needed to determine the detailed mechanisms by which ZAR proteins regulate oogenesis.
Zar1 and Zar2 are conserved among vertebrate species (Fig. 1). However, the spatiotemporal distribution of ZAR proteins among the species studied showed divergence. Non-oogenesis developmental functions in non-ovarian tissues and different mechanisms of translational regulation have been reported. Thus, comparative studies of ZAR proteins among species and cell types are needed to further understand their functions.
The structures of ZAR1 and ZAR2 are noteworthy, because their C-termini are highly conserved. Although the CxxC domain of ZAR proteins has been reported to be functionally Zn2+-dependent, there are no typical zinc finger structures to define it, which is potentially a ZAR-protein-specific zinc finger domain with a unique function. In addition, the N-terminus of ZAR proteins is partially conserved among species; however, the existence of specific functional domains remains to be determined. Thus, the protein structures of ZAR proteins need to be clearly identified for in-depth functional elucidation.
Transcriptome regulation at the early stage of oogenesis and the RNA-binding activity of Zar1/2 have been reported. However, whether the stability of the transcriptome is directly or indirectly influenced by Zar1/2 remains unknown. Thus, the transcripts that directly interact with ZAR1 need to be identified. ZAR1/2 in X. laevis prefers to bind the TCS of transcripts; however, whether there are binding specificities of ZAR1 and ZAR2 in other animals remains unknown. To address this, techniques for identifying RBP-interacting RNAs, such as RIP-seq and LACE-seq [46], should be used in future investigations.
Zar1 and Zar2 are reported to be partially redundant in mouse oocytes as double knockout of these two genes causes more serious phenotypes than single knockouts. In addition, ZAR1 was reported to be specifically located in the cortical cytoplasm beneath the cell membrane in mouse oocytes, which co-localizes with MSY2, a known component of CPL, whereas ZAR2 was reported to be distributed evenly in the oocytes (Fig. 3). Furthermore, in combination with the large protein amount and specific expression frame of ZAR1/2 in growing and fully grown oocytes, ZAR proteins potentially act as RNA translocators by binding transcripts in a one-to-one ratio, which also implies that these two proteins have different assignments. Thus, further research is needed to determine the functional differences between Zar1 and Zar2.
In summary, Zar1 and Zar2 synergistically play roles in both oogenesis and preimplantation embryogenesis as atypical maternal factors while maintaining meiosis via partial redundancy. Thus, Zar1 and Zar2 might be maternal factors that function simultaneously as maternal RBPs, which maintain homeostasis of the transcriptome during oogenesis by interacting with other proteins to form complexes and bind to maternal mRNAs, ultimately influencing oocytes and embryos. Oocytes are one of the most ideal models for determining the fate of mRNAs owing to the existence of transcriptional silencing and recovery transition during MZT, coupled with highly regulated and synchronized meiotic and mitotic cell cycles. Thus, further research using oocytes and early stage embryos is required to clarify the detailed functional mechanisms of ZAR proteins.
Acknowledgements
This work was supported by the National Key Research and Development Program of China (2021YFC2700100), the National Natural Science Foundation of China (Award Numbers: 31930031 and 31890781), the National Ten-Thousands Talents Program of China, the Natural Science Foundation of Zhejiang Province, China [D22C68649], and the Key Research and Development Program of Zhejiang Province (2021C03098 and 2021C03100). We thank Drs Shu-Yan Ji and Yan Rong for their contributions to the ZAR1/2 studies and helpful discussions. We thank Editage (www.editage.cn) for English language editing.
Author contributions
WYK wrote the manuscript and prepared the figures. FHY conceived and revised the manuscript.
Funding
This work was funded by the National Natural Science Foundation of China (Award Numbers: 31930031 and 31890781).
Declarations
Conflict of interest
The authors declare no conflict of interest.
Ethics approval and consent to participate
Not applicable.
Consent for publication
The authors declare that they have permission to publish this review and that it has not previously been published elsewhere.
Availability of data and materials
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Glisovic T, Bachorik JL, Yong J, Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008;582:1977–1986. doi: 10.1016/j.febslet.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu J, Wang F, Zhu X, Yuan Y, Ding M, Gao S. Mouse ZAR1-like (XM_359149) colocalizes with mRNA processing components and its dominant-negative mutant caused two-cell-stage embryonic arrest. Dev Dyn. 2010;239:407–424. doi: 10.1002/dvdy.22170. [DOI] [PubMed] [Google Scholar]
- 3.Charlesworth A, Yamamoto TM, Cook JM, Silva KD, Kotter CV, Carter GS, et al. Xenopus laevis zygote arrest 2 (zar2) encodes a zinc finger RNA-binding protein that binds to the translational control sequence in the maternal Wee1 mRNA and regulates translation. Dev Biol. 2012;369:177–190. doi: 10.1016/j.ydbio.2012.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Nostrand EL, Pratt GA, Yee BA, Wheeler EC, Blue SM, Mueller J, et al. Principles of RNA processing from analysis of enhanced CLIP maps for 150 RNA binding proteins. Genome Biol. 2020;21:90. doi: 10.1186/s13059-020-01982-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang J, Zhang H, Cao L, Dai X, Zhao L, Liu H, et al. Oocyte meiosis-coupled poly(A) polymerase α phosphorylation and activation trigger maternal mRNA translation in mice. Nucleic Acids Res. 2021;49:5867–5880. doi: 10.1093/nar/gkab431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sha Q, Zheng W, Wu Y, Li S, Guo L, Zhang S, et al. Dynamics and clinical relevance of maternal mRNA clearance during the oocyte-to-embryo transition in humans. Nat Commun. 2020;11:4917. doi: 10.1038/s41467-020-18680-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu X, Wang P, Brown CA, Zilinski CA, Matzuk MM. Zygote Arrest 1 (Zar1) is an evolutionarily conserved gene expressed in vertebrate Ovaries1. Biol Reprod. 2003;69:861–867. doi: 10.1095/biolreprod.103.016022. [DOI] [PubMed] [Google Scholar]
- 8.Rong Y, Ji S, Zhu Y, Wu Y, Shen L, Fan H. ZAR1 and ZAR2 are required for oocyte meiotic maturation by regulating the maternal transcriptome and mRNA translational activation. Nucleic Acids Res. 2019;47:11387–11402. doi: 10.1093/nar/gkz863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao LW, Fan HY. Revisiting poly(A)-binding proteins: multifaceted regulators during gametogenesis and early embryogenesis. BioEssays. 2021;43:2000335. doi: 10.1002/bies.202000335. [DOI] [PubMed] [Google Scholar]
- 10.Corley M, Burns MC, Yeo GW. How RNA-binding proteins interact with RNA: molecules and mechanisms. Mol Cell. 2020;78:9–29. doi: 10.1016/j.molcel.2020.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bestor TH. Activation of mammalian DNA methyltransferase by cleavage of a Zn binding regulatory domain. EMBO J. 1992;11:2611–2617. doi: 10.1002/j.1460-2075.1992.tb05326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chuang LSH, Ng H, Chia J, Li BFL. Characterisation of independent DNA and multiple Zn-binding domains at the N terminus of human DNA-(Cytosine-5) methyltransferase: modulating the property of a DNA-binding domain by contiguous Zn-binding motifs. J Mol Biol. 1996;257:935–948. doi: 10.1006/jmbi.1996.0213. [DOI] [PubMed] [Google Scholar]
- 13.Sha Q, Zhang J, Fan H. A story of birth and death: mRNA translation and clearance at the onset of maternal-to-zygotic transition in mammals†. Biol Reprod. 2019;101:579–590. doi: 10.1093/biolre/ioz012. [DOI] [PubMed] [Google Scholar]
- 14.Vastenhouw NL, Cao WX, Lipshitz HD. The maternal-to-zygotic transition revisited. Development. 2019;146:v161471. doi: 10.1242/dev.161471. [DOI] [PubMed] [Google Scholar]
- 15.Zhao LW, Zhu YZ, Chen H, Wu YW, Pi SB, Chen L, et al. PABPN1L mediates cytoplasmic mRNA decay as a placeholder during the maternal-to-zygotic transition. EMBO Rep. 2020;21:e49956. doi: 10.15252/embr.201949956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sánchez F, Smitz J. Molecular control of oogenesis. Biochimica et Biophysica Acta (BBA) Mol Basis Dis. 2012;1822:1896–1912. doi: 10.1016/j.bbadis.2012.05.013. [DOI] [PubMed] [Google Scholar]
- 17.Saitou M, Yamaji M (2012) Primordial Germ Cells in Mice. Cold Spring Harbor Perspectives in Biology 4:a8375 [DOI] [PMC free article] [PubMed]
- 18.Coticchio G, Sereni E, Serrao L, Mazzone S, Iadarola I, Borini A. What criteria for the definition of oocyte quality? Ann N Y Acad Sci. 2004;1034:132–144. doi: 10.1196/annals.1335.016. [DOI] [PubMed] [Google Scholar]
- 19.Gilchrist RBLMTJ. Oocyte-secreted factors: regulators of cumulus cell function and oocyte quality. Hum Reprod Update. 2008;14:159–177. doi: 10.1093/humupd/dmm040. [DOI] [PubMed] [Google Scholar]
- 20.Stewart KR, Veselovska L, Kim J, Huang J, Saadeh H, Tomizawa S, et al. Dynamic changes in histone modifications precede de novo DNA methylation in oocytes. Genes Dev. 2015;29:2449–2462. doi: 10.1101/gad.271353.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamatani T, Carter MG, Sharov AA, Ko MSH. Dynamics of global gene expression changes during mouse preimplantation development. Dev Cell. 2004;6:117–131. doi: 10.1016/s1534-5807(03)00373-3. [DOI] [PubMed] [Google Scholar]
- 22.Peaston AE, Evsikov AV, Graber JH, de Vries WN, Holbrook AE, Solter D, et al. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev Cell. 2004;7:597–606. doi: 10.1016/j.devcel.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 23.Christou-Kent M, Dhellemmes M, Lambert E, Ray PF, Arnoult C. Diversity of RNA-binding proteins modulating post-transcriptional regulation of protein expression in the maturing mammalian oocyte. Cells. 2020;9:662. doi: 10.3390/cells9030662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu X, Viveiros MM, Eppig JJ, Bai Y, Fitzpatrick SL, Matzuk MM. Zygote arrest 1 (Zar1) is a novel maternal-effect gene critical for the oocyte-to-embryo transition. Nat Genet. 2003;33:187–191. doi: 10.1038/ng1079. [DOI] [PubMed] [Google Scholar]
- 25.Sangiorgio L, Strumbo B, Brevini TAL, Ronchi S, Simonic T. A putative protein structurally related to zygote arrest 1 (Zar1), Zar1-like, is encoded by a novel gene conserved in the vertebrate lineage. Comp Biochem Physiol B Biochem Mol Biol. 2008;150:233–239. doi: 10.1016/j.cbpb.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 26.Yamamoto TM, Cook JM, Kotter CV, Khat T, Silva KD, Ferreyros M, et al. Zar1 represses translation in Xenopus oocytes and binds to the TCS in maternal mRNAs with different characteristics than Zar2. Biochimica et Biophysica Acta (BBA) Gene Regul Mech. 2013;1829:1034–1046. doi: 10.1016/j.bbagrm.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uzbekova S, Roy-Sabau M, Dalbiès-Tran R, Perreau C, Papillier P, Mompart F, et al. Zygote arrest 1 gene in pig, cattle and human: evidence of different transcript variants in male and female germ cells. Reprod Biol Endocrinol. 2006;4:12. doi: 10.1186/1477-7827-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michailidis G, Argiriou A, Avdi M. Expression of chicken zygote arrest 1 (Zar1) and Zar1-like genes during sexual maturation and embryogenesis. Vet Res Commun. 2010;34:173–184. doi: 10.1007/s11259-010-9343-z. [DOI] [PubMed] [Google Scholar]
- 29.Yu C, Ji S, Sha Q, Dang Y, Zhou J, Zhang Y, et al. BTG4 is a meiotic cell cycle–coupled maternal-zygotic-transition licensing factor in oocytes. Nat Struct Mol Biol. 2016;23:387–394. doi: 10.1038/nsmb.3204. [DOI] [PubMed] [Google Scholar]
- 30.Yu J, Hecht NB, Schultz RM. Expression of MSY2 in mouse oocytes and preimplantation Embryos1. Biol Reprod. 2001;65:1260–1270. doi: 10.1095/biolreprod65.4.1260. [DOI] [PubMed] [Google Scholar]
- 31.Wright PW, Bolling LC, Calvert ME, Sarmento OF, Berkeley EV, Shea MC, et al. ePAD, an oocyte and early embryo-abundant peptidylarginine deiminase-like protein that localizes to egg cytoplasmic sheets. Dev Biol. 2003;256:74–89. doi: 10.1016/s0012-1606(02)00126-4. [DOI] [PubMed] [Google Scholar]
- 32.Hu M, Qi J, Bi G, Zhou J. Bacterial effectors induce oligomerization of immune receptor ZAR1 in vivo. Mol Plant. 2020;13:793–801. doi: 10.1016/j.molp.2020.03.004. [DOI] [PubMed] [Google Scholar]
- 33.Brevini TAL, Cillo F, Colleoni S, Lazzari G, Galli C, Gandolfi F. Expression pattern of the maternal factor zygote arrest 1 (Zar1) in bovine tissues, oocytes, and embryos. Mol Reprod Dev. 2004;69:375–380. doi: 10.1002/mrd.20140. [DOI] [PubMed] [Google Scholar]
- 34.Pennetier S, Uzbekova S, Perreau C, Papillier P, Mermillod P, Dalbiès-Tran R. Spatio-temporal expression of the germ cell marker genes MATER, ZAR1, GDF9, BMP15, andVASA in adult bovine tissues, oocytes, and preimplantation Embryos1. Biol Reprod. 2004;71:1359–1366. doi: 10.1095/biolreprod.104.030288. [DOI] [PubMed] [Google Scholar]
- 35.Miao L, Yuan Y, Cheng F, Fang J, Zhou F, Ma W, et al. Translation repression by maternal RNA binding protein zar1 is essential for early oogenesis in zebrafish. Development. 2016;144:128–138. doi: 10.1242/dev.144642. [DOI] [PubMed] [Google Scholar]
- 36.Tian Y, Yang J, Peng Y, Chen T, Huang T, Zhang C, et al. Variation screening of zygote arrest 1(ZAR1) in women with recurrent zygote arrest during IVF/ICSI programs. Reprod Sci. 2020;27:2265–2270. doi: 10.1007/s43032-020-00246-y. [DOI] [PubMed] [Google Scholar]
- 37.Shinojima Y, Terui T, Hara H, Kimura M, Igarashi J, Wang X, et al. Identification and analysis of an early diagnostic marker for malignant melanoma: ZAR1 intra-genic differential methylation. J Dermatol Sci. 2010;59:98–106. doi: 10.1016/j.jdermsci.2010.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Richter AM, Kiehl S, Köger N, Breuer J, Stiewe T, Dammann RH. ZAR1 is a novel epigenetically inactivated tumour suppressor in lung cancer. Clin Epigenet. 2017;9:60. doi: 10.1186/s13148-017-0360-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Misra S, Sharma S, Agarwal A, Khedkar SV, Tripathi MK, Mittal MK, et al. Cell cycle-dependent regulation of the bi-directional overlapping promoter of human BRCA2/ZAR2 genes in breast cancer cells. Mol Cancer. 2010;9:50. doi: 10.1186/1476-4598-9-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watanabe T, Yachi K, Ohta T, Fukushima T, Yoshino A, Katayama Y, et al. Aberrant hypermethylation of non-promoter zygote Arrest 1 (Zar1) in human brain tumors. Neurol Med Chir. 2010;50:1062–1069. doi: 10.2176/nmc.50.1062. [DOI] [PubMed] [Google Scholar]
- 41.Courchaine EMLA. Droplet organelles? EMBO J. 2016;35:1603–1612. doi: 10.15252/embj.201593517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yurttas P, Vitale AM, Fitzhenry RJ, Cohen-Gould L, Wu W, Gossen JA, et al. Role for PADI6 and the cytoplasmic lattices in ribosomal storage in oocytes and translational control in the early mouse embryo. Development. 2008;135:2627–2636. doi: 10.1242/dev.016329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang J, Morales CR, Medvedev S, Schultz RM, Hecht NB. In the absence of the mouse DNA/RNA-binding protein MSY2, messenger RNA instability leads to spermatogenic Arrest1. Biol Reprod. 2007;76:48–54. doi: 10.1095/biolreprod.106.055095. [DOI] [PubMed] [Google Scholar]
- 45.Ramat A, Simonelig M. Functions of PIWI proteins in gene regulation: new arrows added to the piRNA Quiver. Trends Genet. 2021;37:188–200. doi: 10.1016/j.tig.2020.08.011. [DOI] [PubMed] [Google Scholar]
- 46.Su R, Fan L, Cao C, Wang L, Du Z, Cai Z, et al. Global profiling of RNA-binding protein target sites by LACE-seq. Nat Cell Biol. 2021;23:664–675. doi: 10.1038/s41556-021-00696-9. [DOI] [PubMed] [Google Scholar]
- 47.Mayr C. What Are 3′ UTRs doing? Cold Spring Harbor Perspect Biol. 2019;11:a34728. doi: 10.1101/cshperspect.a034728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sha Q, Dai X, Dang Y, Tang F, Liu J, Zhang Y, et al. A MAPK cascade couples maternal mRNA translation and degradation to meiotic cell cycle progression in mouse oocyte. Development. 2016;144:452–463. doi: 10.1242/dev.144410. [DOI] [PubMed] [Google Scholar]
- 49.Dai X, Jiang J, Sha Q, Jiang Y, Ou X, Fan H. A combinatorial code for mRNA 3′-UTR-mediated translational control in the mouse oocyte. Nucleic Acids Res. 2019;47:328–340. doi: 10.1093/nar/gky971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Charlesworth A, Welk J, MacNicol AM. The temporal control of Wee1 mRNA translation during xenopus oocyte maturation is regulated by cytoplasmic polyadenylation elements within the 3′-untranslated region. Dev Biol. 2000;227:706–719. doi: 10.1006/dbio.2000.9922. [DOI] [PubMed] [Google Scholar]
- 51.Wang YY, Charlesworth A, Byrd SM, Gregerson R, MacNicol MC, MacNicol AM. A novel mRNA 3′ untranslated region translational control sequence regulates Xenopus Wee1 mRNA translation. Dev Biol. 2008;317:454–466. doi: 10.1016/j.ydbio.2008.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, et al. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012;487:57–63. doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.VerMilyea MD, O'Neill LP, Turner BM. Transcription-independent heritability of induced histone modifications in the mouse preimplantation embryo. PLoS ONE. 2009;4:e6086. doi: 10.1371/journal.pone.0006086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grewal SIS, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 55.De Iaco A, Coudray A, Duc J, Trono D. DPPA2 and DPPA4 are necessary to establish a 2C-like state in mouse embryonic stem cells. EMBO Rep. 2019;20(5):e47382. doi: 10.15252/embr.201847382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Masaki H, Nishida T, Kitajima S, Asahina K, Teraoka H. Developmental pluripotency-associated 4 (DPPA4) localized in active chromatin inhibits mouse embryonic stem cell differentiation into a primitive ectoderm lineage. J Biol Chem. 2007;282:33034–33042. doi: 10.1074/jbc.M703245200. [DOI] [PubMed] [Google Scholar]
- 57.Kotaja NSP. The chromatoid body: a germ-cell-specific RNA-processing centre. Nat Rev Mol. 2007;1:85–90. doi: 10.1038/nrm2081. [DOI] [PubMed] [Google Scholar]
- 58.Pressman S, Bei Y, Carthew R. SnapShot: posttranscriptional gene silencing. Cell. 2007;130:570–571. doi: 10.1016/j.cell.2007.07.042. [DOI] [PubMed] [Google Scholar]
- 59.Lim AK, Lorthongpanich C, Chew TG, Tan CWG, Shue YT, Balu S, et al. The nuage mediates retrotransposon silencing in mouse primordial ovarian follicles. Development. 2013;140:3819–3825. doi: 10.1242/dev.099184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 61.Taborska E, Pasulka J, Malik R, Horvat F, Jenickova I, Jelić Matošević Z, et al. Restricted and non-essential redundancy of RNAi and piRNA pathways in mouse oocytes. PLOS Genet. 2019;15:e1008261. doi: 10.1371/journal.pgen.1008261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Franke V, Ganesh S, Karlic R, Malik R, Pasulka J, Horvat F, et al. Long terminal repeats power evolution of genes and gene expression programs in mammalian oocytes and zygotes. Genome Res. 2017;27:1384–1394. doi: 10.1101/gr.216150.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bai D, Sun J, Jia Y, Yin J, Zhang Y, Li Y, et al. Genome transfer for the prevention of female infertility caused by maternal gene mutation. J Genet Genom. 2020;47:311–319. doi: 10.1016/j.jgg.2020.06.002. [DOI] [PubMed] [Google Scholar]
- 64.Sauna ZE, Kimchi-Sarfaty C. Understanding the contribution of synonymous mutations to human disease. Nat Rev Genet. 2011;12:683–691. doi: 10.1038/nrg3051. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.