Abstract
Protein misfolding is involved in a large number of diseases, among which cystic fibrosis. Complex intra- and inter-domain folding defects associated with mutations in the cystic fibrosis transmembrane regulator (CFTR) gene, among which p.Phe508del (F508del), have recently become a therapeutical target. Clinically approved correctors such as VX-809, VX-661, and VX-445, rescue mutant protein. However, their binding sites and mechanisms of action are still incompletely understood. Blind docking onto the 3D structures of both the first membrane-spanning domain (MSD1) and the first nucleotide-binding domain (NBD1), followed by molecular dynamics simulations, revealed the presence of two potential VX-809 corrector binding sites which, when mutated, abrogated rescue. Network of amino acids in the lasso helix 2 and the intracellular loops ICL1 and ICL4 allosterically coupled MSD1 and NBD1. Corrector VX-445 also occupied two potential binding sites on MSD1 and NBD1, the latter being shared with VX-809. Binding of both correctors on MSD1 enhanced the allostery between MSD1 and NBD1, hence the increased efficacy of the corrector combination. These correctors improve both intra-domain folding by stabilizing fragile protein–lipid interfaces and inter-domain assembly via distant allosteric couplings. These results provide novel mechanistic insights into the rescue of misfolded proteins by small molecules.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-021-03994-5.
Keywords: Chemical chaperone, Cystic fibrosis, Protein folding, Binding site, Molecular dynamics
Introduction
Protein misfolding is a key feature of many pathological conditions known as conformational diseases. Pharmacological chaperones offer attractive therapeutical solutions, by providing molecular scaffolds enabling misfolded proteins to recover their native fold and hence their activity [1]. Multi-domain and multi-pass membrane proteins, which undergo complex co- and post-translational folding in the endoplasmic reticulum [2, 3], are particularly challenging targets for the development of pharmacological chaperones. A prominent case is the cystic fibrosis transmembrane regulator (CFTR) protein, encoded by the CFTR gene, with numerous mutations identified in patients affected by cystic fibrosis (CF), such as the most frequent mutation p.Phe508del (F508del), leading to protein misfolding [4].
CFTR folding occurs co-translationally and includes both intra- and inter-domain folding steps [5–7]. The first stages consist in the insertion of transmembrane helices (TMs) of the first membrane-spanning domain (MSD1) within the membrane via dimer hairpins [2]. Then MSD1 TMs pack tightly and its N- and C-termini assemble with its first intracellular loop (ICL1). This assembly is strengthened by interactions with the downstream nucleotide-binding domain 1 (NBD1) [8]. NBD1-located F508del was shown to alter both domain stability and inter-domain assembly, mainly involving interactions with intracellular loop ICL4 within the second MSD (MSD2) [9, 10].
The screening of small molecules enhancing proper folding of mutant CFTR led to the identification of drugs named correctors, such as VX-809 or its structurally closely related analogue VX-661. While very effective in vitro, their activity was suboptimal in clinical trials even in combination with the CFTR potentiator VX-770 [11, 12], this latter enhanced channel function [13]. Corrector combinations targeting defects associated with different folding steps showed synergistic rescue on CFTR-F508del [14]. A corrector combination (VX-661 and VX-445) in association with the potentiator VX-770 (triple combination therapy) showed great clinical benefits to CF patients carrying at least one F508del allele [15]. Nonetheless, as these molecules have been identified using high throughput screening, their mechanisms of action and binding sites remain largely elusive, especially for the recently described VX-445 [15, 16].
VX-809 binding to native-like 3D structures—rather than unfolded/misfolded intermediates—is supported by the fact that the drug binds to purified CFTR and stabilizes F508del CFTR after its rescue at the cell surface [17]. Protease-susceptibility assays have, moreover, shown that early MSD1 packing may well resemble a native-like structure [8]. VX-809 stabilizes truncated CFTR fragments that contain complete MSD1 [18, 19] and strengthens MSD1 assembly [8]. Corrective conformational changes or stabilization of MSD1 by VX-809 appears to be communicated to other domains, including NBD1, MSD2, and NDB2. VX-809 is highly effective at correcting MSD1-specific disease-causing mutations (e.g. P67L and L206W) [14, 19–24] with the exception of G85E [24] which causes a severe defect in TM insertion and packing [25]. Several mutants form a hot spot around a groove predicted to stably bind phospholipids [26]. This groove is lined by TM1, TM2, TM3, and TM6, a four-helix block that would stay unstable until the swapped TM10–TM11 pair from MSD2 joins it in the CFTR full-length assembly, as observed in its cryo-EM 3D structure [27]. This groove could be considered as the MSD1 Achilles heel and is a potential binding site for small molecules that stabilize MSD1 during the folding process. This potential binding site differs from a composite, multi-domain binding pocket [28] encompassing F374, L375, and amino acids from the lasso, NBD1, MSD2, and ICL4.
In addition to these data on MSD1, NMR spectroscopy, differential scanning calorimetry, and statistical analysis revealed direct binding of VX-809 to NBD1 and allosteric coupling between the binding site and the distant F508 position, at the NBD1:ICL4 interface [29].
In this study, we explored the potential VX-809 binding sites in both MSD1 and NBD1, by combining blind docking and molecular dynamics simulations with biochemical experiments. We especially considered secondary mutations identified in cis of F508del (on the same allele), which have been shown to preclude the response to correction [30, 31]. The lack of response could be associated with additional folding defects adding up with the F508del folding defect and rendering the protein non-responsive to treatment (e.g. L467F located in NBD1) [30] or with the alteration of corrector binding sites (e.g. F87L located in MSD1) [31].
We showed that VX-809 can be stably accommodated in an MSD1 groove and described an allosteric coupling linking this site to the F508del region on NBD1. In addition, we highlighted another potential VX-809 binding site within NBD1, consistent with previous experimental evidence [29]. While MSD1 mutants prevented both F508del and L206W rescue by VX-809, NBD1 mutants prevented the rescue of only F508del, indicating that mutants differ in their sensitivity to corrector single or double occupancy. The sensitivity of CFTR-F508del to the novel corrector VX-445 and VX-809/VX-445 combination was also consistent with the presence of two binding sites located on MSD1 and NBD1 and the need for an allosteric coupling between CFTR domains to fully correct F508del.
Materials and methods
Plasmids and mutagenesis
The cDNA of wild-type (WT) CFTR (M470) was subcloned in pTracer as previously published [32]. Mutagenesis of all the indicated mutants was performed using the QuickChange XL II mutagenesis kit (Agilent) following the manufacturer’s instructions. Obtained mutants were fully sequenced, amplified, and purified (Macherey–Nagel). Plasmid concentrations were measured using a Nanodrop (Thermo Fisher Scientific).
Cell culture and transfection
HEK293 cells were purchased from ATCC and cultivated in DMEM medium supplemented with 10% fetal calf serum (Thermo Fisher Scientific). Cells were maintained at 37 °C, 5% CO2. For Western blot analysis, cells were seeded in six-well plates and transfected with CFTR plasmids using Lipofectamine 3000 (Thermo Fisher Scientific).
Western blot analysis
Transfected cells were treated with 3 µM of VX-809 (Selleckchem) for 24 h before being lysed in RIPA buffer containing protease inhibitors and protein concentration assessed using RcDc assay (BioRad). In some experiments cells were treated with 3 µM of VX-445 (racemic mixture, Selleckchem), alone and in combination with VX-809. Western blot analysis was performed using 60 µg of protein from each sample separated on a 7% acrylamide gel. After transfer onto nitrocellulose membranes, CFTR was probed using antibody 660 (NACF Foundation) and alpha-tubulin probed with antibody DM1A (Santa Cruz). Finally, membranes were incubated with appropriate secondary antibodies coupled to fluorochromes and signal detection was performed with the Odyssey scanner (Li-Cor). Quantification of the Western blots was performed using ImageJ software as described in Fig. S1.
Statistical analysis
Quantitative variables were described as mean (± SEM). Comparisons to WT conditions and between treated and untreated conditions were made with one-way ANOVA followed by Fischer test for p evaluation.
Visualization of 3D structures, molecular docking, and molecular dynamics simulations
3D structures were visualized with the UCSF Chimera package [33]. 2D and 3D structures of the drugs were extracted from the ZINC [34] or PubChem [35] databases (VX-809, VX-661, VX-445 (4S)-methyl, and (4R)-methyl enantiomers in the trimethylpyrrolidinyl group). Blind docking was performed using the EADock DSS tool provided by the online SwissDock server [36, 37]. The cryo-EM 3D structure of human CFTR in ATP-bound conformation (pdb:6MSM) was considered for exploring the MSD1 binding sites, while for exploring the NBD1 binding sites, we used the crystal structure of human F508del CFTR NBD1 (pdb:4WZ6), from which the regulatory extension (aa 648–671) was removed. Additional docking experiments were done using the cryo-EM 3D structure of human CFTR in ATP-free conformation (pdb:5UAK).
For molecular dynamics (MD) simulations, the 3D structure of human CFTR in the ATP-bound conformation was embedded in a 1-palmitoyl-2-oleoyl-phosphatidylcholine (POPC) bilayer and solvated in a 150 mM NaCl solution. The CHARMM36 force field [38] was used for the protein, lipids and ions, and the TIP3P model for water. Minimization, equilibration, and production steps were performed on the occigen/cines supercomputer using Gromacs 2019.1 [39]. The standard CHARMM-GUI inputs [40] were used for the minimization and equilibration of the systems. During these steps, harmonic restraints applied to the protein-heavy atoms and the lipid heads and were gradually released during 1.2 ns. The production dynamics were then performed in the NPT ensemble without any restraints. Nose–Hoover thermostat [41] and Parrinello–Rahman barostat [42] were used to keep the temperature and the pressure constant at 310 K and 1 bar. Periodic boundary conditions were used and the particle mesh Ewald algorithm was applied to treat long-range electrostatic interactions [43]. A switching function was applied between 10 and 12 Å for the non-bonded interactions. LINCS [44] was applied to constrain the bond lengths involving hydrogen atoms. The integration time step was set to 2 fs and the overall length of the trajectory was 1 μs for the simulation of apo CFTR.
MD simulations were also performed of the CFTR protein in complex with the correctors in the proposed binding sites, to evaluate the stability of the predicted conformations. Corrector parameters were generated using the CgenFF module in CHARMM-GUI server [45]. These MD simulations were run for at least 125 ns, with calculations of Root Mean Square Deviations (RMSD) done for estimating corrector stability within the binding sites. Binding poses at 125 ns were selected for illustration in the different CFTR-drug complexes.
The creation and analysis of CFTR networks (Fig. S6) were performed using the R package Bio3D [46, 47]. In this method, MD simulations are used as an input that first undergoes a correlation analysis. Hence, a matrix of residue pairs dynamical cross-correlations is calculated and the output is then used to generate a correlation network with residues as nodes that are linked by weighted edges that are proportional to their degree of correlated motion. The Girvan–Newman clustering method [48] is finally used to identify communities of highly correlated residues. The full residue network and coarse-grained community network were visualized and analyzed with VMD.
Results
A potential VX-809 binding site highlighted within MSD1 by molecular docking
We evaluated the effect of L53V on CFTR correction, a mutation located in the N-terminal part of CFTR, called the lasso motif [27]. In transiently transfected HEK293 cells, CFTR-L53V showed a maturation profile comparable to wild-type (WT) with the presence of both fully glycosylated band C and partially glycosylated band B at a similar ratio (Figs. 1A, B, S1). In combination with F508del, L53V prevented VX-809 rescue (Fig. 1A, B). These results were very similar to those obtained with mutation F87L or L467F (Fig. 1B). Nonetheless, the underlying mechanism involved could differ, with L467F inducing an additional folding defect leading to the production of non-rescuable proteins while L53V and F87L could alter a potential VX-809 binding site. These results led us to further evaluate the region in the vicinity of amino acids L53 and F87 for the presence of potential VX-809 binding sites.
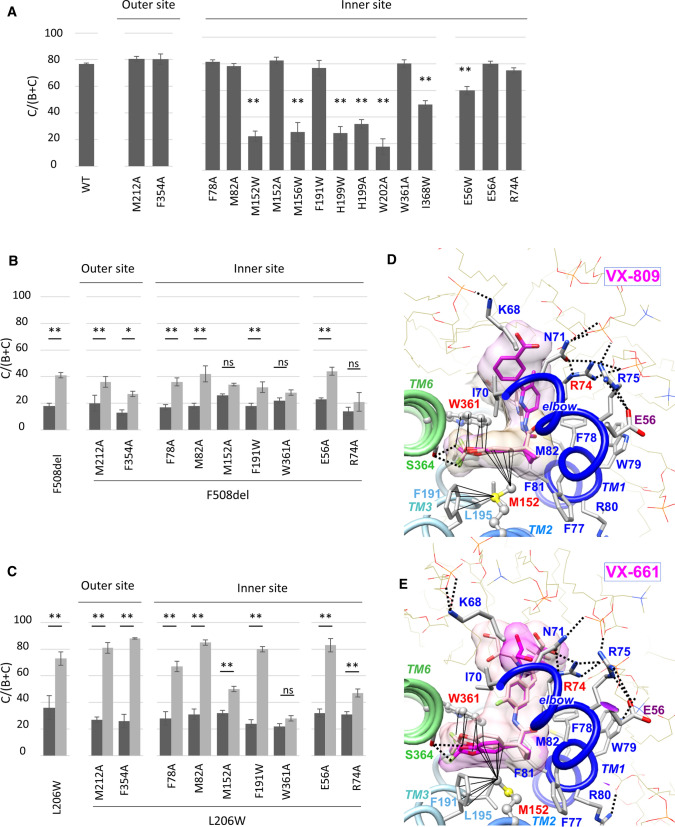
Fig. 1.
Identification of a potential VX-809 binding site within MSD1. A Western blot analysis of WT and mutant CFTR. CFTR fully glycosylated (band C) and core glycosylated (band B) are indicated. Cells were treated with VX-809 (3 µM, 24 h) when indicated. Tubulin (Tub) was probed to assess equal loading amounts. B Quantification of the C/(B + C) maturation ratio of the indicated mutants. Dark grey represents control conditions and light grey VX-809 treatments. Measures are means ± SEM of n = 3–20, with * indicating p < 0.05, ** indicating p < 0.01 and ns indicating no statistical differences compared to untreated condition (one-way ANOVA followed by Fischer test for p evaluation). C Two main pockets were identified at the level of the membrane inner and outer leaflets on the 3D structure of human CFTR MSD1, extracted from the cryo-EM 3D structure of the full-length protein (pdb: 6MSM). The membrane inner leaflet site corresponds to the best docking scores obtained after blind docking using SwissDock, with the surface envelope of VX-809 (representative members of the six first clusters) shaded in pink. The second binding site was identified at the level of the membrane outer leaflet, with fewer clusters (surface envelope (representative members of three clusters) shaded in purple) and less favorable docking scores. The lasso is shown in purple, MSD1 TM1 to TM3 in blue, MDS1 TM4 to TM6 in green. F508 and L206 are shown with pink spheres, L53 and F87 with yellow spheres. D Focus on amino acids within the potential VX-809 binding sites at the level of the membrane inner (bottom) and outer (top) leaflets. The conformations of VX-809 corresponding to the best docking scores in the two sites are shown (inner site (cluster 0): SwissDock FullFitness score of −1938; outer site (cluster 7): SwissDock FullFitness score of −1924). Tested amino acids are labeled in magenta
Blind docking of VX-809 was performed on the experimental 3D structure of the human ATP-bound CFTR (PDB 6MSM [27]). This structure appears more adapted than the ATP-free one for the analysis of the full allosteric networks associated with inter-domain assembly, after VX-809 binding to MSD1. Only the MSD1 (aa 1–390) was considered as a target for docking with default parameters. Clusters gathering similar conformations (or binding modes) of VX-809 within sites were ranked according to the average FullFitness of their elements [49]. After having excluded one unauthorized site in the full-length CFTR, two binding pockets were identified on MSD1, totalizing 17 and 11 clusters (pink and purple surfaces in Fig. 1C). The first pocket was located at the level of the membrane inner leaflet and was formed by amino acids from the elbow (L73), TM1 (F78, F81, M82), TM3 (F191, H199, W202), TM2 (M152, M156) and TM6 (W361, S364, I368) (Fig. 1D—bottom, Fig. S2A). The 43 conformations of the six first clusters are characterized by a unique position of the benzodioxolyl core of VX-809, deeply embedded in the pocket, as it is also the case for the amide group, while the methylpyridine and benzoic acid portions adopt more variable positions, outside the pocket (Fig. S2B—left). Blind docking of VX-661 highlighted the same binding pocket in the five best clusters, with a very similar position of the common benzodioxolyl core, whereas variable positions are observed outside the pocket for the second half of the drug (Fig. S2B—right). Hence the difluorobenzodioxolyl group is the key anchor element of the two correctors in the binding pocket. This site is embedded within a hot spot for class II mutations associated with either mild or severe folding defects (Fig. S3) [26, 50], consistent with fragility in the folding and/or packing of the transmembrane helices (TM) in this region.
The second site, highlighted at the level of the membrane outer leaflet, involves amino acids of TM3 (V208, A209, M212), TM4 (F224, I231, A234, L235), and TM6 (V350, F354) (Fig. 1D—top).
Exploring key residues of the MSD1 VX-809 binding pockets
We further explored these potential pockets by mutating key amino acids implicated in VX-809 binding.
The site located at the level of the membrane outer leaflet (Fig. 1D) was evaluated by mutants M212A and F354A, opposite to each other. These mutations did not alter protein maturation in a WT background (Fig. 2A) nor prevent VX-809 correction in F508del CFTR (Fig. 2B), which is partially rescued, and in L206W CFTR, which can be fully rescued (Fig. 2C).
Fig. 2.
Exploring MSD1 inner membrane leaflet binding site. A–C Quantification of the C/(B + C) maturation ratio of the indicated mutants. Dark grey represents control conditions and light grey VX-809 treatments. Measures are means ± SEM of n = 3–20, with * indicating p < 0.05, ** indicating p < 0.01 and ns indicating no statistical differences compared to WT or untreated condition (one-way ANOVA followed by Fischer test for p evaluation). D Focus on the VX-809 position within the membrane inner leaflet binding site, before (light pink) and after (magenta) MD simulation (125 ns), highlighting the stable position of the drug in the binding site and interactions with lipids (POPC, thin sticks) covering the site. The tight contacts established with M152 and W361 are represented with black lines. Two H-bonds (dashed lines) are present between S364 OG and VX-809 F1 (3.08 Å) and F2 (3.02 Å). Color code is the same as in Fig. 1C, D, with amino acids labels colored accordingly. E Similar view for VX-661, before (light pink) and after (magenta) MD simulation (125 ns). Two H-bonds are present between S364 OG and VX-661 F3 (2.98 Å) and O3 (3.02 Å)
The pocket located at the level of the membrane inner leaflet was evaluated by mutating a set of nine amino acids. Six of the eleven mutations first tested in a WT background (M152W, M156W, H199W, H199A, W202A, and I368W, Fig. 2A) decreased maturation, confirming the vulnerability of this region. Single mutants M156W, H199A, and I368W could be rescued by VX-809 while M152W, H199W, and W202A showed only a partial or no response (Fig. S4A), consistent with these amino acids being implicated in VX-809 induced correction.
The effect of mutants F78A, M82A, M152A, F191W, and W361A that were not deleterious in the WT context was then tested on the rescue of F508del (Fig. 2B) and L206W (Fig. 2C). While F78A, M82A, and F191W had no effect, M152A and W361A reduced or prevented correction by VX-809. The importance of M152 and W361 in VX-809 binding was further strengthened by their position on both sides of the difluorobenzodioxolyl group, which is the key anchor point of the drug (Fig. 1D). Molecular dynamics (MD) simulation indicated a very stable position of VX-809 within the site, with H-bonds between fluorine atoms and S364 hydroxyl, as well as a particular involvement of lipids covering the VX-809 methylpyridine and benzoic acid portion and establishing, through their phosphate groups, key interactions with several basic amino acids (Fig. 2D). A similar situation is observed for the MD simulation of the CFTR:VX-661 complex (Fig. 2E). To assess if M152A and W361A altered VX-809 binding affinity, higher VX-809 concentrations were used. While CFTR-M152A-F508del was partially rescued at high concentrations, CFTR-W361A-F508del remained not responsive (Fig. S5 (A, B and C)). For secondary mutations which induced CFTR misfolding (M152W, M156W, H199W/A, and I368W, Fig. S3B), both F508del and L206W correction was prevented (Fig. S4B), probably due to additive folding defects leading to the production of non-rescuable channels, as previously demonstrated for L467F [30] (Fig. 1B).
We then targeted more distant amino acids implicated in the architecture of the membrane inner leaflet pocket. Amino acids E56 and R74 are located at the cytoplasmic side of the pocket, either at its edge (R74) or at the interface between the elbow and the lasso (E56 which forms a salt bridge with R75) (Fig. 1D). Single mutation E56A and R74A did not alter CFTR maturation (Fig. 2A). While E56A had no apparent effect, R74A prevented F508del correction and mitigated L206W correction (Fig. 2B, C), even at high concentrations (Fig. S5D).
Taken together, these results confirmed the sensitivity of this region to mutations and showed that amino acids located in the MSD1 membrane inner leaflet pocket (M152, W361) or involved in its architecture (R74) influence VX-809 efficacy towards both F508del and L206W, supporting thereby its relevance as a VX-809 binding site.
Allosteric coupling with NBD1 F508del region
To understand the impact of an MSD1 binding site on the NBD1:ICL4 interface affected by the F508del mutation, we searched for allosteric coupling between the two regions (Fig. 3A). A network of connected amino acids could be highlighted, including a salt bridge formed between K162 (in ICL1, at the end of TM2) and E1075 (in ICL4, at the beginning of TM11), the latter being connected to the NBD1 F508 region (Fig. 3A). This salt bridge not present on the cryo-EM 3D structure (Fig. S6A—top), appeared quickly in a 1 µs long MD simulation and remained stable over time (Fig. S6A—middle). Adding up to the salt-bridges linking K163/K166 (ICL1) to E379 (at the end of TM6) (Fig. S6A—top and middle) reinforces the network linking the MSD1 core helices to MSD1 ICLs. After MD simulation in the presence of VX-809, the K162-E1075 salt bridge remained very stable as well (Fig. 3A and Fig. S6A—bottom). Remarkably, the occupancy of the binding site by VX-809 is accompanied by a movement of the lasso Lh2 relative to ICL1–ICL4, leading K162 to shift an additional H-bond from E54 to D47. This movement of Lh2 (aa 45–63) could be seen in the light of the VX-809-induced protection of the MSD1 N-ter (aa 49) from proteolytic cleavage [8]. Analysis of the 1 µs long MD simulation run on CFTR in the absence of VX-809 indicated a community of highly correlated residues in the vicinity of the MSD1 VX-809 binding site and ICL4 (in contact with NBD1 F508) via the lasso and ICL1, that is disrupted by double-mutant K162A-E1075A (Fig. S6B). Altogether, these results strengthen the importance of the lasso Lh2 for allosteric coupling between MSD1 and NBD1, displaying a tight network of salt-bridges involving the potential MSD1 VX-809 binding site (through the E56-R75 bond) and ICLs (through the E54/D47-K162-E1075 network). This in turn impacts the ICL4:NBD1 interface involving F1074 (Figs. 3A, S6A). The VX-809 binding site is also connected to ICL1 through M152 (on the same TM2 helix than K162) and the bond network linking the end of TM6 to K163/K166. Allosteric coupling was experimentally tested by disrupting the K162-E1075 salt bridge. Single mutant K162A showed a reduced maturation whereas E1075A was not altered (Fig. 3B). Both K162A and E1075A prevented F508del rescue while not affecting L206W correction (Fig. 3C). CFTR-F508del-E1075A maturation was not rescued by higher VX-809 concentrations (Fig. S5E) but swapping amino acids K162E-E1075K of the salt bridge restored VX-809 rescue (Fig. 3C). These results demonstrated the necessity of allosteric coupling between MSD1 and NBD1 to rescue F508del, contrary to L206W. Allosteric coupling towards NBD1 could also explain the differential effect of mutations L53V and F87L in combination with either F508del (Fig. 1B) or L206W (Fig. 3C).
Fig. 3.
VX-809 MSD1 binding site and F508 region are allosterically coupled. A Path linking the potential VX-809 binding site in MSD1 and the F508 position at the interface between NBD1 (light pink) and ICL4 (orange) and involving lasso Lh2 (purple) and ICL1 (light blue) (3D structure after MD simulation in the presence of VX-809). The surface envelope of VX-809 is pink-shaded. The K162-E1075 bond network is highlighted in the inset. Color code is the same as in Fig. 1C, D, with amino acids labels colored accordingly. B, C Quantification of the C/(B + C) maturation ratio of the indicated mutants. Dark grey represents control conditions and light grey VX-809 treatments. Measures are means ± SEM of n = 3–20, with * indicating p < 0.05, ** indicating p < 0.01 and ns indicating no statistical differences compared to WT or untreated condition (one-way ANOVA followed by Fischer test for p evaluation)
A second potential VX-809 binding site on NBD1
As a VX-809 binding site was also described on NBD1 [29], we performed blind docking of VX-809 on the crystal structure of human F508del CFTR NBD1 (pdb:4WZ6) [51], from which the regulatory extension (aa 648–671) has been removed. We did not consider NBD1 extracted from the complete CFTR assembly (or the corresponding NBD1:NBD2 dimer) as this 3D structure lacks helix H9, which is a critical feature of the possible binding site [29]. While the two first docking binding modes are located near the F508del loop (blue in Fig. 4A) and ATP-binding site (green), respectively, the following seven clusters (orange, 65 conformations), focused on a single region, involving strands S3, S9, S10, and helices H3, H8, and H9 (Fig. 4A). The binding site, similar to the one proposed by Hudson and colleagues [29], is oriented towards the NBD1:NBD2 interface, in a region critical for CFTR trafficking [52]. MD simulation of the VX809:NDB1 complex, considering the best docking score (cluster 2), led to refine the position of the drug in the site, and highlight the critical role of H620, Y625, and K643 (Fig. 4A). A strong H-bond is observed between VX-809 O5 and Y625 hydroxyl group, while significant interactions also exist between the two VX-809 fluorine atoms and the S623 hydroxyl group. A similar docking of VX-661 led to highlighting the same binding pocket in six of the ten best clusters (the first four ones corresponding to the F508del loop), supporting the relevance of this site (Fig. S7).
Fig. 4.
Intact NBD1 binding site is required for F508 rescue. A Left: View of the human CFTR F508del NBD1, extracted from a crystal structure (pdb: 4WZ6) from which the regulatory extension (RE) has been removed and on which VX-809 has been docked (blind docking). Three main pockets were identified in the high-ranking clusters, the third one (orange) gathering seven clusters out of nine (SwissDock FullFitness scores of −1274 to −1270). Middle: Representative conformations of each of the seven clusters of the third pocket, with its main amino acids highlighted in ball and stick. The positions of G622 and G628, identified as critical for CFTR trafficking [52], are indicated with brown circles. Right: Position of VX-809 (cluster 2) in the NBD1 binding site, before (light orange—lower position) and after (dark orange—upper position) MD simulation, highlighting the refined position of the drug and tight contacts established with H620 and Y625, as well as with S623 and basic amino acids belonging to H8 and H9 (K643). B–D. Quantification of the C/(B + C) maturation ratio of the indicated mutants. Dark grey represents control conditions and light grey VX-809 treatments. Measures are means ± SEM of n = 3–20, with * indicating p < 0.05, ** indicating p < 0.01 and ns indicating no statistical differences compared to WT or untreated condition (one-way ANOVA followed by Fischer test for p evaluation)
A set of mutations was introduced within this potential VX-809 binding site (labeled in pink Fig. 4A). H620A, Y625A, and K643A did not alter CFTR processing while S459W reduced maturation without preventing VX-809 rescue (Figs. 4B and S4C). Strikingly, VX-809 rescue of F508del and L206W were differently affected by these mutations. Indeed, while S459W, H620A, Y625A, and K643A prevented F508del correction, they did not affect L206W restoration (Figs. 4C, D and S4D). Treatment of CFTR-F508del-H620A with higher VX-809 concentrations did not rescue maturation (Fig. S5F). These results indicate again that the two mutants are not corrected by the same mechanism with mutations located within NBD1 affecting F508del correction, contrary to L206W. These differences could be explained by distinct folding defects associated with each mutation, with F508del destabilizing NBD1, contrary to L206W [53].
Some of the highlighted sites can also accommodate corrector VX-445
CFTR-F508del correction is greatly enhanced by the combination of two correctors, e.g. VX-809/VX-445 or VX-661/VX-445 with the latter showing clear clinical benefits [15]. We evaluated the sensitivity of our mutants to VX-445 and the VX-445/VX-809 combination. VX-445 enhanced CFTR-F508del maturation alone, reaching comparable rescue to VX-809, and up to near WT levels when in combination with VX-809 (Fig. 5A). Mutation L467F prevented response to both treatments, contrary to L53V and F87L (Fig. S8A).
Fig. 5.
Rescue by VX-445 and VX-809/VX-445 requires both MSD1 and NBD1 binding sites integrity. A Quantification of the C/(B + C) maturation ratio of the indicated mutants. Dark grey represents control conditions, light grey VX-445 treatments and black VX-809/VX-445 treatments. Measures are means ± SEM of n = 3–20, with * indicating p < 0.05, ** indicating p < 0.01 and ns indicating no statistical differences compared to untreated condition (one-way ANOVA followed by Fischer test for p evaluation). B, C. Blind docking of VX-445 (4S enantiomer) on NBD1 (B) and on MSD1 (C), followed by MD simulations. For NBD1, three main pockets were identified in the ten high-ranking clusters, as for VX-809, the third one (orange) gathering five clusters out of ten (SwissDock FullFitness scores: −1662 to −1656), whereas for MSD1, only one pocket (purple) was allowed (first eight clusters shown, SwissDock FullFitness scores: −2324 to −2313). Color code is the same as in Fig. 1C, D. Representative conformations of the first clusters in MSD1 and NBD1 (orange site) are given in Fig. S9 for both the 4S and 4R enantiomer, highlighting the critical role of the 2,2,4-trimethylpyrrolidinyl group in pocket binding. In the boxes are shown the positions of VX-445 (4S enantiomer) in the NBD1 binding site (cluster 2) and the MSD1 binding site (cluster 8), before (light orange and light purple, respectively) and after (dark orange and dark purple, respectively) MD simulation (125 ns), highlighting the refined positions of the drug within the sites. An intermediate position of VX-445 after 60 ns (2), between 0 ns (1) and 125 ns (3) was indicated for the MSD1 binding site. The orange circle in the MSD1 box highlights the interaction of W361 with the 3,3,3-trifluoro-2,2-dimethylpropoxy extremity of VX-445.
Regarding secondary mutations located within NBD1, H620A prevented VX-445 and blunted VX-809/VX-445 correction while Y625A had no effect (Fig. 5A). This is consistent with a VX-445 binding site in the vicinity of H620, a hypothesis supported by blind-docking of VX-445 on NDB1, highlighting a preferential binding site similar to that targeted by VX-809 (Fig. 5B). Docking was performed with the 4S enantiomer (asymmetry present in the 2,2,4-trimethylpyrrolidinyl group, which exhibits greater efficacy than the 4R enantiomer in the presence of VX-661 [16]), and strikingly, for the vast majority of the clusters, the 2,2,4-trimethylpyrrolidinyl group acts as the critical anchor in the pocket (the 4-methyl C and H atoms are highlighted in blue in Fig. S9B—top, dark green). The same is true for the 4R enantiomer (Fig. S9B—top, light green), with docking scores similar to that observed for the 4S one. As for VX-809, MD simulation led to refining the position of VX-445 in the site and underlined the critical role of H620 (Fig. 5B).
Among the MSD1 secondary mutations tested, W361A abrogated VX-445 and markedly reduce VX-445/VX-809 correction while R74A and M152A had no effect (Fig. 5A). These results suggested that W361A may affect a VX-445 binding site within MSD1 possibly distinct from the VX-809 site embedded in the inner leaflet as neither R74A nor M152A altered correction. Blind-docking of VX-445 on MSD1 identified a unique potential VX-445 binding site (Fig. 5C), corresponding to the outer membrane leaflet site described in Fig. 1C for VX-809. As for the NBD1 binding site, the 2,2,4-trimethylpyrrolidinyl group acts as the critical anchor in the pocket characterizing this site, whatever the VX-445 enantiomer (the 4-methyl C and H atoms are highlighted in blue in Fig. S9B—bottom), in particular interacting with the side chain of F354. Docking scores are also similar between the two enantiomers. Concordantly, while M212A and F354A located within this site did not prevent VX-809 correction (Fig. 2A), they prevented VX-445 and reduced VX-445/VX-809 correction (Fig. 5A).
The fact that double mutants could be partially or completely rescued with VX-445/VX-809 combination (Fig. 5A) or VX-445/VX-809 combination and incubation of the cells at low temperature (Figure S8B) indicated that the absence of response to single correctors was not caused by irreversible misfolding of the protein, leading to non-correctable channels.
MD simulations of CFTR with VX-445 bound to this site showed that one binding pose, for which the 3,3,3-trifluoro-2,2-dimethylpropoxy extremity is in contact with the inner leaflet binding site, evolved towards a stable position after ~ 100 ns (Fig. 5C). This position implied a shift towards W361, with which VX-445 tightly interacts, however, without reaching the VX-809 binding site. MD simulation considering another pose, orientated along the same axis but in an opposite way, led to a rapid escape of the drug out of the binding site. We also ran MD simulations with correctors in both sites (i.e. VX-445 in the outer membrane leaflet site and VX-809 or VX-661 in the inner membrane leaflet site) and observed in this case a stable binding of VX-809/VX-661, as previously observed, together with a stable position of VX-445 (Figs. 6 and S10). Correctors interacted with each other through their extremities, consistent with synergic behavior. The VX-445/VX-661 couple showed a qualitatively better complementarity to the protein surface, with strong interaction between VX-661 and R74 (Fig. S10). E1075A, which interrupted the allosteric coupling between MSD1 and NBD1 (Figs. 3 and S4), prevented VX-445 and VX-445/VX-809 correction while reconstitution of the K162-E1075 salt bridge (K162E-E1075K) restored full correction (Fig. 5A). As for VX-809 and VX-661 alone, slight local conformational changes were observed (Figs. 3 and S6), in particular in the elbow helix and the lasso Lh2, allowing D47 to bind K162 in ICL1, which forms a salt bridge with E1075 in ICL4.
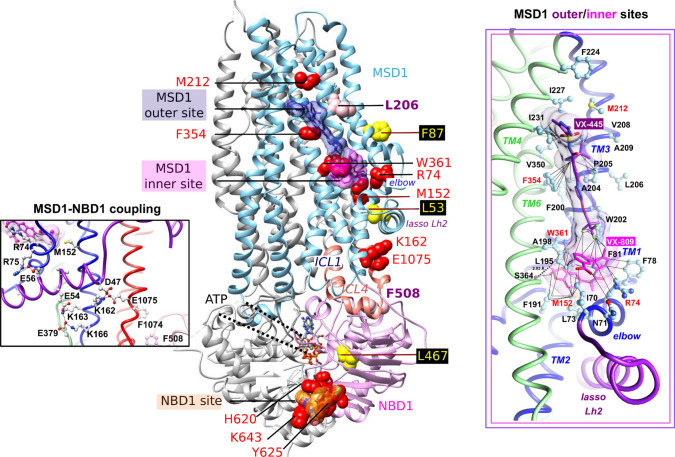
Fig. 6.
Potential corrector binding sites on the CFTR 3D structure. View of the 3D structure of human CFTR (ribbon representation, pdb 6MSM), on which the correctors are highlighted in their stable positions (after 125 ns MD simulations) in their potential binding sites using grids (magenta and orange for VX-809/VX-661 and purple for VX-445). Ribbons are represented in blue for MSD1 and pink for NBD1. Amino acids L206 and F508 are indicated in pink. Mutations identified in the context of complex alleles which prevent VX-809 rescue of CFTR-F508del are represented in yellow. Secondary mutations preventing the correction of CFTR-F508del or both CFTR-L206W and CFTR-F508del are in red. The 3D structure of human F508del NBD1 (pdb:4WZ6) has been considered for NBD1 helices H8/H9, including the K643 position. At right are shown with atomic details the positions and contacts of VX-809 and VX-445, considered simultaneously, into the inner and outer membrane sites, respectively (also see Fig. S9 for detailed comments and comparison with the VX-661/VX-445 complex). Color code for the TM helices is as indicated in Fig. 1. At left is represented the bond network involved in the coupling between MSD1 and NBD1 (illustrated in the CFTR-VX-661/VX-445 complex, also see Fig. S10 for a view of the network in the different complexes)
Taken together, results obtained with point mutations and modeling suggest the presence of two binding sites on MSD1, one for each of the correctors, and a shared site on NBD1. The MSD1 sites are allosterically coupled to the F508del region via the K162-E1075 salt bridge, allostery which is reinforced by the simultaneous occupancy of the sites, hence their enhanced efficacy (Fig. 6).
Discussion
Several small molecules targeting CFTR have been identified in the past decade, enabling to enhance channel activity, protein stability or folding, highlighting the possibility to correct misfolded CFTR using such a strategy. Corrector VX-809 has been developed first as it partially restores trafficking of F508del CFTR [54]. While VX-809 does not improve isolated NBD1 stability [18], it acts by stabilizing the NBD1:ICL4 interface [9, 10, 55]. Some studies have suggested a possible binding site at the level of this interface (e.g. [56, 57].), which is supported here by the fact that the best scoring pose of VX-809 on the F508del NBD1 3D structure indeed fills this interstitial area (Fig. 4A). However, this seems specific to F508del CFTR, as VX-809 was also shown to be efficient on a wide range of class II mutations which are located on other CFTR domains [14, 19–24], but preserve the NBD1:ICL4 interface. Moreover, the various locations of the VX-809 rescuable mutations on the whole CFTR protein, in particular in MSD1, are in favor of allosteric effects and/or multiple binding sites for this small drug.
Seminal works have shown that VX-809 bind to two isolated domains: MSD1 [18, 19] and NBD1 [29], inducing different effects. Binding to MSD1 was suggested by the stabilization of truncated CFTR forms, encompassing amino acids 1–373 [19]. This allowed a strong rescue of mutants located in the N-terminal part of CFTR, such as E56K, P67L, L69H, L206W or W361R [14, 19, 22, 24, 26], indicating that initial targeting of MSD1 folding and assembly accounted for the bulk VX-809 rescue. Molinski and colleagues have proposed a potential binding site located close to the cytoplasm and involving interfaces with other domains [28]. Nonetheless, mutants in this site (K166E/Q) showed a strong response to VX-809 [8]. We followed here a strategy consisting in using MSD1 secondary mutations identified in patients carrying F508del to identify amino acids modulating the VX-809 response (Figs. 1, 6). Both L53V and F87L prevented F508del rescue and are located in the vicinity of a potential binding pocket, which was also identified by blind-docking experiments at the level of the membrane inner leaflet. Both, VX-809 and VX-661 are particularly well accommodated in the pocket, as the 2,2-difluoro-1,3-benzodioxolyl portion of VX-809/VX-661, common to the VX-809 derivatives ALK-809 and SUL-809 [58], especially interacts with W361, S364, F81, F191 and M152, while the other portion of the drugs contacts membrane lipids and/or amino acids at the periphery of the site (Fig. S2).
Mutants discussed above (L69H, E56K, P67L, L206W, and W361R) are also in close vicinity or within this pocket. MD simulations indicate that L206W established strong interactions with P205 and W202, thereby moving the side chain of this last amino acid away from its position within the pocket in the WT situation, with W361 and W202 interacting with the same lipid (Fig. S12). Importantly, MD simulations of the WT protein indicated that lipids can be stably accommodated in the putative VX-809 binding site studied here [26], suggesting that VX-809 may mimic such molecules for stabilizing these fragile areas. Such modified interactions with lipids might account for a modified architecture of the pocket that can be rescued by displacement of lipids by the corrector. This feature could represent a key step in the corrector’s mode of action leading to global stabilization of protein–lipid interactions favoring protein folding. Further work is now needed to investigate more complex lipid compositions of the membranes. Indeed, like other groups [59, 60], we have performed our simulations with simple systems currently used for mimicking cell membranes, made of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), but future simulations should consider more complex mixtures containing POPC, phosphatidylethanolamine, sphingomyelin and cholesterol.
The VX-809/VX-661 binding site was also found in the ATP-free 3D structure of human CFTR [61], with superimposable positions of the amino acid side chains participating in the pocket. The MSD1 VX-809/VX-661 binding site could, therefore, be conformation independent. The R74 side chain, located at the edge of the pocket, is, however, oriented differently in the two structures, preventing access to the drug in the docking experiments in the ATP-free structure. The side chain of this amino acid appears, therefore, to be mobile, while also playing a critical direct role in corrector binding, as supported by our modeling and site-directed mutagenesis. In contrast, the MSD1 VX-445 binding site could be conformation-dependent, as this one includes the TM4 helix, which adopts different positions relative to the MSD1 TM1–TM2–TM3–TM6 block depending on the conformation of the channel [27, 61]. However, this hypothesis should be qualified as the critical interactions observed after MD in the complex are made with amino acids from TM3 and TM6, with a particular role of the 2,2,4-trimethylpyrrolidinyl group in pocket anchoring.
Besides the action on a native-like structure, the mode of action of correctors might also be linked to enhanced TM hairpin insertion within the membrane. This was illustrated for V232D, associated with unfavorable insertion of the TM3/4 hairpin that remained within the inner leaflet of the membrane [62]. V232D is located at the level of an MSD1 hydrophobic pocket [63], which corresponds to the VX-445 binding site highlighted here (Fig. 6). The V232D side-chain orientation towards lipids, however, does not preclude packing of TMs after their insertion in the membrane, consistent with the rescue of this mutant by VX-809 [63]. This is not the case for mutant G85E which was associated with a shortening of the TM1 span, leading to defective integration within the membrane [25] and the production of a non-rescuable CFTR [24]. Indeed, in this case, the packing of the TMs would also be directly affected, as G85 is in direct contact with the TM3 H199 side chain near the VX-809 binding site. Thus, rescuing partially folded intermediates by enhanced insertion of TM hairpins could also represent a key step in the mode of action of correctors, favoring the packing of MSD domains when key actors of this packing are not affected.
In the context of full-length CFTR-F508del, correction levels achieved by VX-809 are, however, more modest [12, 19]. This limitation has been associated with additional NBD1 instability specific for the F508del mutation, which alters inter-domain interactions of NBD1 occurring in particular through the MSD2 ICL4 loop [64]. NBD1 binders correcting the instability of the domain have been intensively searched and have led to identifying molecules that bind different sites on NBD1, such as c407 [65], crotoxin [66], nanobodies [67], BIA (5-bromoindole-3-acetic acid), and the dual corrector-potentiator CFFT-001 [68–70]. Direct binding of VX-809 to NBD1 was demonstrated more recently [29]. NMR data suggest that VX-809 occupies a site similar to that targeted by CFFT-001, and that allosteric coupling occurs between this site and the NBD1:ICL4 interface [29, 69, 71]. While NBD1 thermo-instability was still detectable in the presence of VX-809, enhanced association to MSD1 via ICL4 favored protein assembly and CFTR-F508del rescue. Our results are consistent with this binding pocket being targeted by both VX-809/VX-661 and VX-445 correctors, with the latter recently shown to directly bind and stabilize NBD1 [16]. The 2,2,4-trimethylpyrrolidinyl group of VX-445 appeared to be a key element for drug anchorage in the NBD1 pocket, while the 2,2-difluoro-1,3-benzodioxolyl portion of VX-809/VX-661 appeared to play this role in the MSD1 membrane inner leaflet site. Of note, the conformation of the NBD1 VX-445 binding site evolved alongside the MD simulation with a displacement of helix H9 while the 2,2,4-trimethylpyrrolidinyl group was maintained buried within an inner pocket (Fig. S13). It should be noted here that another binding site was recently proposed for VX-809 and aminoarylthiazole-VX-809 hybrid derivatives, next to the ATP-binding site, in a location similar to the second scoring pose highlighted here by blind docking on the F508del NBD1 3D structure [72]. Whether or not this binding site is also relevant for increasing NBD1 stability and CFTR rescue deserves further investigation.
Coupling between the MSD1 VX-809 binding site and the F508 region in NBD1 involves a tight network of amino acids from the elbow, lasso Lh2, ICL1, and ICL4 supported here with secondary mutations. This network includes D47, E54, E56, R74, M152, K162, K163, K166, and E1075 (Fig. 6—left box). One key feature of allostery may be linked to the shift of the lasso Lh2 upon VX-809 binding and the associated interaction of D47 with K162, allowing a synergy between three communication pathways leading from MSD1 to K162 (the TM2 way: M152K162; two elbow-Lh2 ways: (i) R74R75E56D47K162, (ii) R74R75E56E54K163K162) (Table S1). The critical role played by this pivotal interface in CFTR assembly and pharmacological repair, is also highlighted by other recent studies focused on ICL1 and Lh2 [8, 73, 74]. These results presented here in the F508del background indicate the necessity of stabilizing inter-domain folding steps to efficiently rescue misfolded F508del, contrary to L206W (Fig. 6).
Overall, the rationale of our work was to consider correction of double mutants in the F508del or L206W background to support the implication of the mutated residue in the corrector binding site or the corrector's effect.
One can, however, not rule out the possibility that even in the absence of folding defects induced by a single mutation, additional folding defects may be associated with either F508del and L206W, leading to the production of a non-rescuable CFTR protein. However, this appeared not to be the case since rescue of double mutants was achieved, for mutants in the MSD1 VX-445 binding site by VX-809 (Fig. 2A) and for mutants in the MSD1 VX-809 binding site by VX-445 (R74A and M152A, Fig. 5A). Also, high levels of correction were obtained for all double mutants with the VX445 + VX809 combination (Fig. 5A) alone and with low-temperature incubation of the cells (Fig. S8B). These results reveal the possibility to correct double mutants and argue in favor of the implication of amino acids targeted by secondary mutations in the binding site.
Interpretation of the effect of point mutations relative to the participation of the corresponding amino acids in a binding site should also always be carried out with particular care as mutations could affect corrector binding but also allosteric changes induced by corrector binding within the site, which may be distant. Nonetheless, the results presented here for the MSD1 VX-809/VX-661 binding site are supported by recent cryo-EM results posted in a preprint server (bioRxiv 2021.06.18.449063) after deposition of our study (bioRxiv 2021.05.04.442442) and showing the presence of VX-809/VX-661 within the inner leaflet binding site, with the same orientations and contacts. The present study brings additional information on another possible binding site on NBD1 and on the yet less explored VX-445 ones, as well as on their interplay with lipids.
To conclude, it appears that VX-809 affects CFTR differently depending on the binding site, with the MSD1 site inducing a global stabilization of the initial folding steps while the NBD1 site stabilizes NBD1-ICL4 interaction. As VX-445 was shown to affect NBD1 folding [16] and binding of both VX-445 and VX-809 to MSD1 leads to increased allosteric coupling with NBD1, it can be suggested that both mechanisms contribute to the enhanced rescue observed with the VX-445/VX-809 combination. These results shed light on the mechanism of action of these pharmacological chaperones which target both intra-domain and inter-domain folding steps to achieve full correction. These also give useful considerations for understanding key elements in the folding of multi-domain membrane proteins and for ongoing drug discovery related to conformational diseases.
Supplementary Information
Below is the link to the electronic supplementary material.
Author contributions
NB conceived the protocols, performed experiments and analyzed data. AhE conceived the protocols and performed MD simulations, as well as data curation and analysis. JPM was involved in the conceptualization, design of the study, and data analysis. BC, IP, and NS performed experiments and statistical analysis. RZ and JLD participated in the analysis of the protein–drug interactions. AlE and ISG participated in the writing of the manuscript. IC and AH conceived and coordinated the project, conceived the protocols, performed experiments, analyzed data, and wrote the manuscript. All authors have reviewed the manuscript and approved its submission.
Funding
This work was performed using HPC resources from GENCI-[CINES] (Grants 2017-A0020707206, 2018-A0040707206, 2019-A0060707206 and 2020-A0080707206) and benefited from the financial support of the French Association Vaincre la Mucoviscidose and Association pour l’Aide à la Recherche contre la Mucoviscidose (AARM).
Availability of data and material
Data and material are available upon request.
Code availability
Not applicable.
Declarations
Conflict of interest
NB, AhE, JPM, BC, IP, NS, RZ, JLD, AlE, IC, and AH declare no competing interests. ISG reports grants from Vertex Therapeutics, Eloxx Pharmaceuticals and participation in the scientific board of Proteostasis Therapeutics; outside the submitted work.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Nesrine Baatallah and Ahmad Elbahnsi contributed equally to this work and should be considered as first authors.
Isabelle Callebaut and Alexandre Hinzpeter should be considered as co-last authors.
Contributor Information
Isabelle Callebaut, Email: isabelle.callebaut@sorbonne-universite.fr.
Alexandre Hinzpeter, Email: alexandre.hinzpeter@inserm.fr.
References
- 1.Convertino M, Das J, Dokholyan N. Pharmacological chaperones: design and development of new therapeutic strategies for the treatment of conformational diseases. ACS Chem Biol. 2016;11:1471–1489. doi: 10.1021/acschembio.6b00195. [DOI] [PubMed] [Google Scholar]
- 2.Kim SJ, Skach WR. Mechanisms of CFTR folding at the endoplasmic reticulum. Front Pharmacol. 2012;3:201. doi: 10.3389/fphar.2012.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marinko JT, Huang H, Penn WD, Capra JA, Schlebach JP, Sanders CR. Folding and misfolding of human membrane proteins in health and disease: from single molecules to cellular proteostasis. Chem Rev. 2019;119:5537–5606. doi: 10.1021/acs.chemrev.8b00532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Veit G, Avramescu RG, Chiang AN, Houck SA, Cai Z, Peters KW, et al. From CFTR biology toward combinatorial pharmacotherapy: expanded classification of cystic fibrosis mutations. Mol Biol Cell. 2016;27:424–433. doi: 10.1091/mbc.e14-04-0935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cui L, Aleksandrov L, Chang XB, Hou YX, He L, Hegedus T, et al. Domain interdependence in the biosynthetic assembly of CFTR. J Mol Biol. 2007;365:981–994. doi: 10.1016/j.jmb.2006.10.086. [DOI] [PubMed] [Google Scholar]
- 6.Du K, Lukacs GL. Cooperative assembly and misfolding of CFTR domains in vivo. Mol Biol Cell. 2009;20:1903–1915. doi: 10.1091/mbc.e08-09-0950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kleizen B, van Vlijmen T, de Jonge HR, Braakman I. Folding of CFTR is predominantly cotranslational. Mol Cell. 2005;20:277–287. doi: 10.1016/j.molcel.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 8.Kleizen B, van Willigen M, Mijnders M, Peters F, Grudniewska M, Hillenaar T, et al. Co-translational folding of the first transmembrane domain of ABC-transporter CFTR is supported by assembly with the first cytosolic domain. J Mol Biol. 2021;433:166955. doi: 10.1016/j.jmb.2021.166955. [DOI] [PubMed] [Google Scholar]
- 9.Mendoza JL, Schmidt A, Li Q, Nuvaga E, Barrett T, Bridges RJ, et al. Requirements for efficient correction of DF508 CFTR revealed by analyses of evolved sequences. Cell. 2012;148:164–174. doi: 10.1016/j.cell.2011.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rabeh WM, Bossard F, Xu H, Okiyoneda T, Bagdany M, Mulvihill CM, et al. Correction of both NBD1 energetics and domain interface is required to restore ΔF508 CFTR folding and function. Cell. 2012;148:150–163. doi: 10.1016/j.cell.2011.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taylor-Cousar JL, Munck A, McKone EF, van der Ent CK, Moeller A, Simard C, et al. Tezacaftor-ivacaftor in patients with cystic fibrosis homozygous for Phe508del. N Engl J Med. 2017;377:2013–2023. doi: 10.1056/NEJMoa1709846. [DOI] [PubMed] [Google Scholar]
- 12.Wainwright CE, Elborn JS, Ramsey BW, Marigowda G, Huang X, Cipolli M, et al. Lumacaftor-ivacaftor in patients with cystic fibrosis homozygous for Phe508del CFTR. N Engl J Med. 2015;373:220–231. doi: 10.1056/NEJMoa1409547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Goor F, Hadida S, Grootenhuis PD, Burton B, Cao D, Neuberger T, et al. Rescue of CF airway epithelial cell function in vitro by a CFTR potentiator, VX-770. Proc Natl Acad Sci USA. 2009;106:18825–18830. doi: 10.1073/pnas.0904709106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Veit G, Xu H, Dreano E, Avramescu RG, Bagdany M, Beitel LK, et al. Structure-guided combination therapy to potently improve the function of mutant CFTRs. Nat Med. 2018;24:1732–1742. doi: 10.1038/s41591-018-0200-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Middleton PG, Mall MA, Dřevínek P, Lands LC, McKone EF, Polinemi D, et al. Elexacaftor–tezacaftor–ivacaftor for cystic fibrosis with a single Phe508del allele. N Engl J Med. 2019;381:1809–1819. doi: 10.1056/NEJMoa1908639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Veit G, Roldan A, Hancock MA, Da Fonte DF, Xu H, Hussein M, et al. Allosteric folding correction of F508del and rare CFTR mutants by elexacaftor-tezacaftor-ivacaftor (Trikafta) combination. JCI Insight. 2020;5(18):e139983. doi: 10.1172/jci.insight.139983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eckford PD, Ramjeesingh M, Molinski S, Pasyk S, Dekkers JF, Li C, et al. VX-809 and related corrector compounds exhibit secondary activity stabilizing active F508del-CFTR after its partial rescue to the cell surface. Chem Biol. 2014;21:666–678. doi: 10.1016/j.chembiol.2014.02.021. [DOI] [PubMed] [Google Scholar]
- 18.Loo TW, Bartlett MC, Clarke DM. Corrector VX-809 stabilizes the first transmembrane domain of CFTR. Biochem Pharmacol. 2013;86:612–619. doi: 10.1016/j.bcp.2013.06.028. [DOI] [PubMed] [Google Scholar]
- 19.Ren HY, Grove DE, Houck SA, Sopha P, van Goor F, Hoffman BJ, et al. VX-809 corrects folding defects in cystic fibrosis transmembrane conductance regulator protein through action on membrane-spanning domain 1. Mol Biol Cell. 2013;24:3016–3024. doi: 10.1091/mbc.e13-05-0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Avramescu RG, Kai Y, Xu H, Bidaud-Meynard A, Schnúr A, Frenkiel S, et al. Mutation-specific downregulation of CFTR2 variants by gating potentiators. Hum Mol Genet. 2017;26:4873–4885. doi: 10.1093/hmg/ddx367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sabusap CM, Wang W, McNicholas CM, Chung WJ, Fu L, Wen H, et al. Analysis of cystic fibrosis-associated P67L CFTR illustrates barriers to personalized therapeutics for orphan diseases. JCI Insight. 2016;1:e86581. doi: 10.1172/jci.insight.86581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharma H, Jollivet Souchet M, Callebaut I, Prasad R, Becq F. Function, pharmacological correction and maturation of new Indian CFTR gene mutations. J Cyst Fibros. 2015;14:34–41. doi: 10.1016/j.jcf.2014.06.008. [DOI] [PubMed] [Google Scholar]
- 23.Han ST, Rab A, Pellicore MJ, Davis EF, McCague AF, Evans TA, et al. Residual function of cystic fibrosis mutants predicts response to small molecule CFTR modulators. JCI Insight. 2018;3:e121159. doi: 10.1172/jci.insight.121159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oliver KE, Rauscher R, Mijnders M, Wang W, Wolpert MJ, J M,, et al. Slowing ribosome velocity restores folding and function of mutant CFTR. J Clin Invest. 2019;129:5236–5253. doi: 10.1172/JCI124282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patrick AE, Karamyshev AL, Millen L, Thomas PJ. Alteration of CFTR transmembrane span integration by disease-causing mutations. Mol Biol Cell. 2011;22:4461–4471. doi: 10.1091/mbc.e11-05-0396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Billet A, Elbahnsi A, Jollivet-Souchet M, Hoffmann B, Mornon JP, Callebaut I, et al. Functional and pharmacological characterization of the rare CFTR mutation W361R. Front Pharmacol. 2020;17:295. doi: 10.3389/fphar.2020.00295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Z, Liu F, Chen J. Molecular structure of the ATP-bound, phosphorylated human CFTR. Proc Natl Acad Sci USA. 2018;115:12757–12762. doi: 10.1073/pnas.1815287115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Molinski SV, Shahani VM, Subramanian AS, MacKinnon SS, Woollard G, Laforet M, et al. Comprehensive mapping of cystic fibrosis mutations to CFTR protein identifies mutation clusters and molecular docking predicts corrector binding site. Proteins. 2018;86:833–843. doi: 10.1002/prot.25496. [DOI] [PubMed] [Google Scholar]
- 29.Hudson RP, Dawson JE, Chong PA, Yang Z, Millen L, Thomas PJ, et al. Direct binding of the corrector VX-809 to human CFTR NBD1: evidence of an allosteric coupling between the binding site and the NBD1:CL4 interface. Mol Pharmacol. 2017;92:124–135. doi: 10.1124/mol.117.108373. [DOI] [PubMed] [Google Scholar]
- 30.Baatallah N, Bitam S, Martin N, Servel N, Costes B, Mekki C, et al. Cis variants identified in F508del complex alleles modulate CFTR channel rescue by small molecules. Hum Mutat. 2018;39:506–514. doi: 10.1002/humu.23389. [DOI] [PubMed] [Google Scholar]
- 31.Masson A, Schneider-Futschik EK, Baatallah N, Nguyen-Khoa T, Girodon E, Hatton A, et al. Predictive factors for lumacaftor/ivacaftor clinical response. J Cyst Fibros. 2019;18:368–374. doi: 10.1016/j.jcf.2018.12.011. [DOI] [PubMed] [Google Scholar]
- 32.Fanen P, Clain J, Labarthe R, Hulin P, Girodon E, Pagesy P, et al. Structure-function analysis of a double-mutant cystic fibrosis transmembrane conductance regulator protein occurring in disorders related to cystic fibrosis. FEBS Lett. 1999;452:371–374. doi: 10.1016/S0014-5793(99)00647-X. [DOI] [PubMed] [Google Scholar]
- 33.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 34.Sterling T, Irwin JJ. ZINC 15—ligand discovery for everyone. J Chem Inf Model. 2015;55:2324–2337. doi: 10.1021/acs.jcim.5b00559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim S, Chen J, Cheng T, Gindulyte A, He J, He S, et al. PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res. 2021;49:D1388–D1395. doi: 10.1093/nar/gkaa971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grosdidier A, Zoete V, Michielin O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucl Acids Res. 2011;39:W270–W277. doi: 10.1093/nar/gkr366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grosdidier A, Zoete V, Michielin O. Fast docking using the CHARMM force field with EADock DSS. J Comput Chem. 2011;32:2149–2159. doi: 10.1002/jcc.21797. [DOI] [PubMed] [Google Scholar]
- 38.MacKerell AJ, Feig M, Brooks CI. Extending the treatment of backbone energetics in protein force fields: Limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comp Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 39.Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, et al. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. doi: 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- 40.Jo S, Kim T, Iyer VG, Im W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J Comput Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 41.Nosé S. A unified formulation of the constant temperature molecular dynamics methods. J Chem Phys. 1984;81:511–519. doi: 10.1063/1.447334. [DOI] [Google Scholar]
- 42.Parrinello M, Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J Appl Phys. 1981;52:7182–7190. doi: 10.1063/1.328693. [DOI] [Google Scholar]
- 43.Darden T, York D, Pedersen L. Particle mesh Ewald: an N⋅log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. doi: 10.1063/1.464397. [DOI] [Google Scholar]
- 44.Hess B, Bekker H, Berendsen HJ, Fraaije JG. LINCS: a linear constraint solver for molecular simulations. J Comput Chem. 1997;18:1463–1472. doi: 10.1002/(SICI)1096-987X(199709)18:12<1463::AID-JCC4>3.0.CO;2-H. [DOI] [Google Scholar]
- 45.Kim S, Lee J, Jo S, Brooks CL, Lee HS, Im W. CHARMM-GUI ligand reader and modeler for CHARMM force field generation of small molecules. J Comput Chem. 2017;38:1879–1886. doi: 10.1002/jcc.24829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sethi A, Eargle J, Black AA, Luthey-Schulten Z. Dynamical networks in tRNA: protein complexes. Proc Natl Acad Sci USA. 2009;106:6620–6625. doi: 10.1073/pnas.0810961106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skjærven L, Yao X-Q, Scarabelli G, Grant BJ. Integrating protein structural dynamics and evolutionary analysis with Bio3D. BMC Bioinformatics. 2014;15:399. doi: 10.1186/s12859-014-0399-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Girvan M, Newman MEJ. Community structure in social and biological networks. Proc Natl Acad Sci USA. 2002;99:7821–7826. doi: 10.1073/pnas.122653799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grosdidier A, Zoete V, Michielin O. EADock: docking of small molecules into protein active sites with a multiobjective evolutionary optimization. Proteins. 2007;67:1010–1025. doi: 10.1002/prot.21367. [DOI] [PubMed] [Google Scholar]
- 50.Estabrooks S, Brodsky JL. Regulation of CFTR biogenesis by the proteostatic network and pharmacological modulators. Int J Mol Sci. 2020;21:452. doi: 10.3390/ijms21020452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hall JD, Wang H, Byrnes LJ, Shanker S, Wang K, Efremov IV, et al. Binding screen for cystic fibrosis transmembrane conductance regulator correctors finds new chemical matter and yields insights into cystic fibrosis therapeutic strategy. Protein Sci. 2016;25:360–373. doi: 10.1002/pro.2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Billet A, Melin P, Jollivet M, Mornon J-P, Callebaut I, Becq F. C terminus of nucleotide binding domain 1 contains critical features for cystic fibrosis transmembrane conductance regulator trafficking and activation. J Biol Chem. 2010;285:22132–22140. doi: 10.1074/jbc.M110.120683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Protasevich I, Yang Z, Wang C, Atwell S, Zhao X, Emtage S, et al. Thermal unfolding studies show the disease causing F508del mutation in CFTR thermodynamically destabilizes nucleotide-binding domain 1. Protein Sci. 2010;19:1917–1931. doi: 10.1002/pro.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van Goor F, Hadida S, Grootenhuis PD, Burton B, Stack JH, Straley KS, et al. Correction of the F508del-CFTR protein processing defect in vitro by the investigational drug VX-809. Proc Natl Acad Sci USA. 2011;108:18843–18848. doi: 10.1073/pnas.1105787108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.He L, Kota P, Aleksandrov AA, Cui L, Jensen T, Dokholyan NV, et al. Correctors of ΔF508 CFTR restore global conformational maturation without thermally stabilizing the mutant protein. FASEB J. 2013;27:536–545. doi: 10.1096/fj.12-216119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Farinha CM, King-Underwood J, Sousa M, Correia AR, Henriques BJ, Roxo-Rosa M, et al. Revertants, low temperature, and correctors reveal the mechanism of F508del-CFTR rescue by VX-809 and suggest multiple agents for full correction. Chem Biol. 2013;20:943–955. doi: 10.1016/j.chembiol.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 57.Okiyoneda T, Veit G, Dekkers JF, Bagdany M, Soya N, Xu H, et al. Mechanism-based corrector combination restores DF508-CFTR folding and function. Nat Chem Biol. 2013;9:444–454. doi: 10.1038/nchembio.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sinha C, Zhang W, Moon CS, Actis M, Yarlagadda S, Arora K, et al. Capturing the direct binding of CFTR correctors to CFTR by using click chemistry. ChemBioChem. 2015;16:2017–2022. doi: 10.1002/cbic.201500123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Corradi V, Gu R-X, Vergani P, Tieleman D. Structure of transmembrane helix 8 and possible membrane defects in CFTR. Biophys J. 2018;114:1751–1754. doi: 10.1016/j.bpj.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Farkas B, Tordai H, Padányi R, Tordai A, Gera J, Paragi G, et al. Discovering the chloride pathway in the CFTR channel. Cell Mol Life Sci. 2020;77:765–778. doi: 10.1007/s00018-019-03211-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu F, Zhang Z, Csanády L, Gadsby DC, Chen J. Molecular structure of the human CFTR ion channel. Cell. 2017;169:85–95. doi: 10.1016/j.cell.2017.02.024. [DOI] [PubMed] [Google Scholar]
- 62.Krainer G, Treff A, Hartmann A, Stone TA, Schenkel M, Keller S, et al. A minimal helical-hairpin motif provides molecular-level insights into misfolding and pharmacological rescue of CFTR. Commun Biol. 2018;1:154. doi: 10.1038/s42003-018-0153-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Loo TW, Clarke DM. The cystic fibrosis V232D mutation inhibits CFTR maturation by disrupting a hydrophobic pocket rather than formation of aberrant interhelical hydrogen bonds. Biochem Pharmacol. 2014;88:46–57. doi: 10.1016/j.bcp.2013.12.027. [DOI] [PubMed] [Google Scholar]
- 64.He L, Aleksandrov LA, Cui L, Jensen TJ, Nesbitt KL, Riordan JR. Restoration of domain folding and interdomain assembly by second-site suppressors of the DeltaF508 mutation in CFTR. FASEB J. 2010;24:3103–3112. doi: 10.1096/fj.09-141788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Odolczyk N, Fritsch J, Norez C, Servel N, da Cunha MF, Bitam S, et al. Discovery of novel potent ΔF508-CFTR correctors that target the nucleotide binding domain. EMBO Mol Med. 2013;5:1484–1501. doi: 10.1002/emmm.201302699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Faure G, Bakouh N, Lourdel S, Odolczyk N, Premchandar A, Servel N, et al. Rattlesnake phospholipase A2 increases CFTR-chloride channel current and corrects ∆F508CFTR dysfunction: impact in cystic fibrosis. J Mol Biol. 2016;428:2898–2915. doi: 10.1016/j.jmb.2016.05.016. [DOI] [PubMed] [Google Scholar]
- 67.Sigoillot M, Overtus M, Grodecka M, Scholl D, Garcia-Pino A, Laeremans T, et al. Domain-interface dynamics of CFTR revealed by stabilizing nanobodies. Nat Commun. 2019;10:2636. doi: 10.1038/s41467-019-10714-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.He L, Aleksandrov AA, An J, Cui L, Yang Z, Brouillette CG, et al. Restoration of NBD1 thermal stability is necessary and sufficient to correct DF508 CFTR folding and assembly. J Mol Biol. 2015;427:106–120. doi: 10.1016/j.jmb.2014.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hudson RP, Chong PA, Protasevich II, Vernon R, Noy E, Bihler H, et al. Conformational changes relevant to channel activity and folding within the first nucleotide binding domain of the cystic fibrosis transmembrane conductance regulator. J Biol Chem. 2012;287:28480–28494. doi: 10.1074/jbc.M112.371138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhenin M, Noy E, Senderowitz H. REMD simulations reveal the dynamic profile and mechanism of action of deleterious, rescuing, and stabilizing perturbations to NBD1 from CFTR. J Chem Inf Model. 2015;55:2349–2364. doi: 10.1021/acs.jcim.5b00312. [DOI] [PubMed] [Google Scholar]
- 71.Dawson J, Farber P, Forman-Kay J. Allosteric coupling between the intracellular coupling helix 4 and regulatory sites of the first nucleotide binding domain of CFTR. PLoS One. 2013;8:e74347. doi: 10.1371/journal.pone.0074347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Righetti G, Casale M, Liessi N, Tasso B, Salis A, Tonelli M, et al. Molecular docking and QSAR studies as computational tools exploring the rescue ability of F508del CFTR correctors. Int J Mol Sci. 2020;21:8084. doi: 10.3390/ijms21218084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Loo TW, Clarke DM. Corrector VX-809 promotes interactions between cytoplasmic loop one and the first nucleotide-binding domain of CFTR. Biochem Pharmacol. 2017;136:24–31. doi: 10.1016/j.bcp.2017.03.020. [DOI] [PubMed] [Google Scholar]
- 74.Pomperada Sabusap CM, Joshi D, Simhaev L, Oliver KE, Senderowitz H, van Willigen M, et al. The CFTR P67L variant reveals a key role for N-terminal lasso helices in channel folding, maturation, and pharmacologic rescue. J Biol Chem. 2021;296:100598. doi: 10.1016/j.jbc.2021.100598. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data and material are available upon request.
Not applicable.