Abstract
Faithful chromosome segregation requires correct attachment of kinetochores with the spindle microtubules. Erroneously-attached kinetochores recruit proteins to activate Spindle assembly checkpoint (SAC), which senses the errors and signals cells to delay anaphase progression for error correction. Temporal control of the levels of SAC activating-proteins is critical for checkpoint activation and silencing, but its mechanism is not fully understood. Here, we show that E3 ubiquitin ligase, SCF-FBXW7 targets BubR1 for ubiquitin-mediated degradation and thereby controls SAC in human cells. Depletion of FBXW7 results in prolonged metaphase arrest with increased stabilization of BubR1 at kinetochores. Similar kinetochore stabilization is also observed for BubR1-interacting protein, CENP-E. FBXW7 induced ubiquitination of both BubR1 and the BubR1-interacting kinetochore-targeting domain of CENP-E, but CENP-E domain degradation is dependent on BubR1. Interestingly, Cdk1 inhibition disrupts FBXW7-mediated BubR1 targeting and further, phospho-resistant mutation of Cdk1-targeted phosphorylation site, Thr 620 impairs BubR1-FBXW7 interaction and FBXW7-mediated BubR1 ubiquitination, supporting its role as a phosphodegron for FBXW7. The results demonstrate SCF-FBXW7 as a key regulator of spindle assembly checkpoint that controls stability of BubR1 and its associated CENP-E at kinetochores. They also support that upstream Cdk1 specific BubR1 phosphorylation signals the ligase to activate the process.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-023-05019-9.
Keywords: Kinetochore, Mitosis, Checkpoint, BubR1, FBXW7, CENP-E, SCF complex
Introduction
Establishment of correct attachment of chromosomal kinetochore (KT) with the microtubules (MTs) is essential for error-free chromosome segregation, failure of which leads to birth defects, embryonic lethality, and aneuploidy [1–3]. Cells use dedicated surveillance machinery, called spindle assembly checkpoint (SAC) (also termed as mitotic checkpoint), which senses the attachment errors and relays signals for error correction by delaying metaphase to anaphase progression. Defective mitotic checkpoint substantially contributes to chromosomal instability and has been commonly found in many human cancers [4]. Genetic analyses of mitotic checkpoint proteins in human cancers earlier showed that there are minimal alterations in their gene or mRNA levels [4, 5]. Subsequent studies demonstrated that the epigenetic control of the mitotic checkpoint components at the protein level plays a key role in the mitotic progression and maintenance of chromosomal stability[6].
Activation of SAC involves both formation of the mitotic checkpoint complex (MCC) and recruitment of the checkpoint protein complexes consisting of Bub3, Bub1, BubR1, and Mad1-Mad2 to the unattached KTs [7]. Specifically, the initial activation of SAC signaling involves recruitment of the Bub3-Bub1/BubR1 (Bub) complex to the unattached KTs. Upon recruitment of the Bub complex to KTs, Bub1 gets phosphorylated and aids in recruitment of Mad1-Mad2 complex together with Cdc20 to the unattached site to further activate the signaling. Following KT recruitment of Mad1-Mad2 and Cdc20, Mad2 undergoes a conformational change causing the release of the Mad2-Cdc20 complex from the KTs. The KT-released Cdc20-Mad2 then associates with the cytosolic Bub3-BubR1 to form the MCC [8–11]. Upon establishment of correct KT-MT attachment, MCC is disassembled leading to the release of Cdc20 and thereby, activation of APC to trigger chromosome segregation and anaphase progression [12].
Centromere Protein E (CENP-E/KIF10) is a kinesin motor from the Kinesin-7 family, which localizes to the unattached KTs and utilizes plus-end directed microtubule motility to slide the mono-oriented chromosomes to the spindle equator [13]. CENP-E-depletion causes chromosomal misalignment and mitotic arrest defects [14]. Similar to BubR1, CENP-E level is up-regulated during G2 phase and peaks during mitosis before its proteolytic degradation on mitotic exit [15, 16]. CENP-E level remains high prior to metaphase and localizes as a large crescent structure in the outer kinetochore including the outermost fibrous corona layer [17]. Upon attainment of KT-MT attachment at metaphase, its kinetochore level is reduced to a substantial level, though a residual amount is still to be associated for maintaining the attachment [18]. BubR1 aids in recruitment of CENP-E to the KTs and further, the persistent maintenance of CENP-E association with KT requires BubR1 [13, 15, 16, 19]. BubR1 forms a stable complex with CENP-E by binding to the KT-targeting C-terminal domain of CENP-E [19, 20]. However, the mechanism of how CENP-E level at KT is differentially maintained remains less clear.
The genetic mutations and expression level of BubR1 are strongly associated with chromosomal instability and cancer. Over-expression of BubR1 is found in mitotic cells with chromosomal instability and it correlates with increased proliferation of cancer cells [4, 21, 22]. Suppression of BubR1 expression also leads to premature mitotic exit and a high percentage of unaligned kinetochores without the formation of stable attachment of chromosomes with the spindle microtubules [23, 24]. BubR1 is involved in mitotic checkpoint activation through its binding in the form of the Bub complex to the outer kinetochore protein, Knl1 [25, 26]. Further, Cdk1-mediated phosphorylation of BubR1 at Thr 620 facilitates its binding to Polo-like kinase 1(Plk1), which further phosphorylates BubR1 at additional sites that are necessary for the KT recruitment of PP2A- B56 and subsequent dephosphorylation of Knl1 to silence the checkpoint [27, 28]. Regulation of BubR1 level and its phosphorylation to the optimal level is therefore critical for balancing SAC activation vs. silencing and timely progression to anaphase. However, the mechanism underlying such molecular control has been less defined.
Though the role of APC in mitosis has been well established, there are indications of the roles of other ubiquitin ligases in regulating mitosis. E3 ubiquitin ligase, UBR5 promotes dissociation of Bub3-BubR1 sub-complex from the MCC by ubiquitylating BubR1 and thereby, facilitates APC activation and anaphase progression [29]. SCF (Skp1-Cullin-FBox), a multi-subunit complex belonging to a large family of ubiquitin ligases, plays critical roles in cell cycle control via ubiquitin-mediated degradation of key cellular proteins. While the Skp1 and Cullin 1 units form the core structure of the ligase machinery, the F-box component serves as the E3 ligase that targets the substrates for activation of ubiquitination [30–32]. There are about 69 members of the F-box proteins in humans [33]. SCFFBXW7 with its substrate targeting subunit FBXW7 is a highly conserved E3 ligase complex that controls the turnover of several critical cell cycle and proliferation-controlling proteins including c-Myc, c-Jun, Notch, and Cyclin E [34–39]. FBXW7 is mutated in numerous cancers including breast, ovarian, endometrial, and colon; and it functions as a tumor suppressor [40, 41]. FBXW7-deficient human cells are chromosomally unstable with high abundance of micronuclei and aneuploidy [42]. FBXW7 abrogation has also been shown to induce mitotic defects such as multipolar spindles, chromosome misalignments, and centrosome amplification [43]. However, understanding of the regulatory mechanisms of FBXW7 in mitosis has been limited so far. Here, we show FBXW7 regulates spindle assembly checkpoint and anaphase progression by targeting BubR1 and its binding partner, CENP-E in human cells. Partial FBXW7 depletion leads to increased levels of BubR1, and its binding partner, CENP-E at kinetochores in mitotic synchronized cells and similarly, its over-expression significantly reduces their levels. Biochemical results show that FBXW7 regulates stability of the mitotic checkpoint complex by controlling the cellular levels of these two proteins via ubiquitin-mediated degradation. Pharmacological inhibition of Cyclin-dependent kinase 1(Cdk1) suppresses FBXW7-mediated BubR1 degradation and further, phospho-deficient mutation of the Cdk1-targeting site of BubR1 mimics such effect, whereas the phospho-mimetic mutation stimulates BubR1 degradation.
Results
FBXW7 regulates mitotic progression
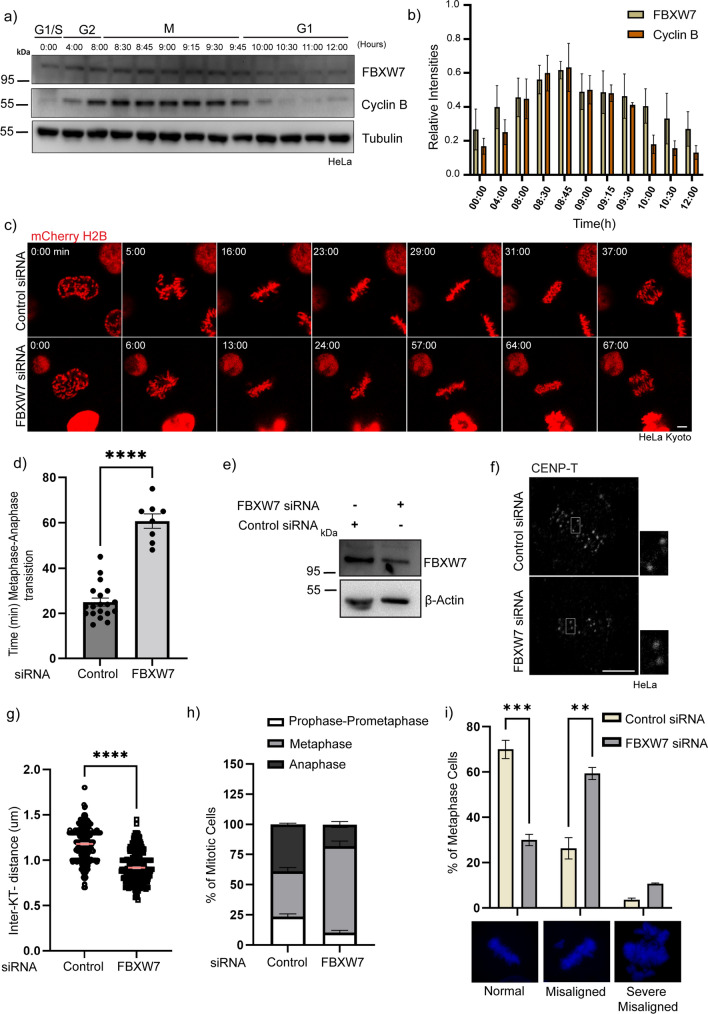
We checked the cellular expression levels of FBXW7 during the cell cycle. HeLa cells were synchronized using double thymidine and the cell lysates were probed for FBXW7 after collecting the cells at constant intervals post-release of thymidine. The levels of FBXW7 expression showed a steep increase during the M phase. Cyclin B was probed as a mitotic marker (Fig. 1a, b). Since FBXW7 levels were high during mitosis, we sought to investigate its role in the progression of mitosis. FBXW7 was depleted using siRNA in H2B mCherry stably expressed HeLa Kyoto cells and mitotic progression was monitored by time-lapse imaging in live cells that were synchronized by double thymidine and then released from thymidine block. The thymidine-released cells were imaged from the time soon after the nuclear envelope breakdown till the anaphase. The level of FBXW7-depletion was about 60% by FBXW7 siRNA (Fig. 1e). Mitotic cells with such partial depletion of FBXW7 showed a significant number of chromosomes that are loosely congressed and took prolonged time to proceed to anaphase (Fig. 1c). Specifically, metaphase to anaphase transition was significantly delayed. While the control cells completed metaphase to anaphase progression in ~ 25 min, the FBXW7 depleted cells took ~ 60 min for the same (Fig. 1d). Consistent with such delay, the percentage of metaphase cells was higher in FBXW7 depleted cells as compared to control cells (Fig. 1h). It was also observed that the inter-kinetochore distances were reduced in the FBXW7-depleted cells as compared to control, implying defects in achieving required tension at the KT-MT attachment site (Fig. 1f,g). The partial FBXW7-depleted cells also showed an increased number of misaligned chromosomes as compared to control cells (Fig. 1i). The results indicated that FBXW7 has an essential role in anaphase progression.
Fig. 1.
FBXW7 depletion leads to a delay in metaphase to anaphase transition a Expression analysis of FBXW7 during the cell cycle. HeLa cells were synchronized with double thymidine treatment and then released to enter the S phase. Cells were collected at constant intervals and then probed for endogenous FBXW7 levels. b Histogram of FBXW7/cyclin B levels normalized to tubulin showed peaks during mitosis. Data = mean ± SEM from three independent experiments. c Selected frames of mCherry-tagged HeLa H2B cells after 48 h post-transfection of FBXW7 siRNA (lower panel) or control siRNA (upper panel). Cells were monitored upon entry to mitosis and mitotic timing was checked by live cell imaging. Images were acquired with a time interval of 1 min for 120 min. scale bar: 5 µm d Plot of mitotic timing FBXW7-depleted cells showed a delay of 45—65 min in metaphase to anaphase transition, whereas control cells enter anaphase within 15–25 min.****p < 0.0001 by student’s t-test. Each dot represent a single cell. e Lysate of HeLa cells showing FBXW7 levels in FBXW7 siRNA vs control siRNA. f Representative image of HeLa cells treated with FBXW7 siRNA (left panel) or control siRNA (right panel) probed with inner-kinetochore protein CENP -T as a marker. scale bar 5 µm. g Individual distances are marked on the dot plot Data = mean ± SEM ****p < 0.0001 by t-test h Percentage of mitotic cells in asynchronous HeLa cells were plotted in FBXW7 siRNA v/s Control siRNA condition. FBXW7 siRNA-treated cells showed increased metaphase cells as compared to Control siRNA. ~ 300–350 cells were analyzed. Data = mean ± SEM. i Graph showing the percentage of HeLa cells having congression defects in FBXW7 siRNA v/s Control siRNA condition. Data = mean ± SEM. ***p = 0.0001 by 2-way ANOVA
FBXW7 regulates BubR1 levels at metaphase kinetochores
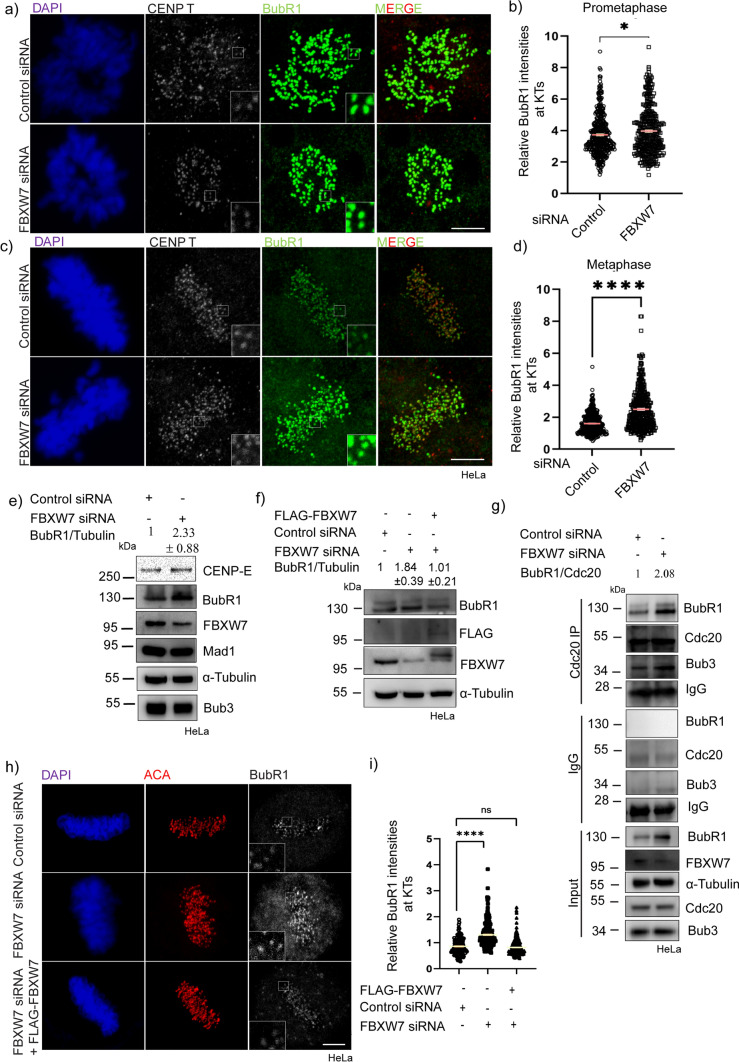
Since the partial FBXW7 depletion results in a delay of metaphase to anaphase transition and reduced inter-kinetochore tension, we next investigated how FBXW7 regulates the localization of proteins associated with mitotic checkpoint signaling. HeLa cells depleted of FBXW7 by siRNA were synchronized using double thymidine and then imaged for the spindle assembly checkpoint proteins after releasing thymidine and fixing the cells at prometaphase and metaphase. Immunofluorescence imaging showed a robust increase of BubR1 localization at the kinetochores in the partially FBXW7-depleted metaphase cells as compared to the control (Fig. 2c). Intensity analysis of individual kinetochores showed an increase of 1.5 times in FBXW7-depleted cells as compared to control (Fig. 2d). Under the similar condition, the KT intensity of MAD1 was not visibly affected, but MAD2 showed a significant increase in response to partial FBXW7 depletion (Fig. S1a–c). KT intensity of BubR1 also showed an increase in prometaphase cells under the partial FBXW7 depleted condition; however, the effect was less pronounced than metaphase cells (Fig. 2 a, b). KT localization of BubR1 showed similar levels to the control cells, when exogenous FLAG-FBXW7 was expressed under FBXW7-depleted background (Fig. 2h, i).
Fig. 2.
BubR1 levels were increased substantially in FBXW7-depleted cells. a, c Representative images of HeLa cells after 18 h of FBXW7 siRNA or control siRNA treatment followed by synchronization at prometaphase and metaphase by treating with thymidine followed by its release at appropriate times prior to stain for BubR1 and inner kinetochore marker, CENP T. b, d Dot Plots representing intensities of kinetochore-localized BubR1 in prometaphase and metaphase cells in FBXW7 siRNA vs. control HeLa cells. BubR1 intensities were normalized with respect to that of CENP T in all cases. ~ 300–400 kinetochores were analyzed from three independent experiments. Data = mean ± SEM. ****p < 0.0001, *p = 0.0178 by unpaired t-test e Lysates of mitotic synchronized HeLa cells after 18 h of FBXW7 siRNA or control siRNA were assessed for the levels of BubR1. Tubulin was probed as a control. BubR1 showed 2.33 ± 0.88 times increased overall levels in FBXW7-depleted cells as compared to control cells. f Lysates of mitotic synchronized HeLa cells after 18 h of FBXW7 siRNA or control siRNA or FBXW7 siRNA treatment rescued with FLAG-FBXW7 expression were assessed for the overall levels of BubR1. Tubulin was probed as a control. g Lysates of mitotic synchronized HeLa cells transfected with control siRNA or FBXW7 siRNA were subjected to Cdc20 immunoprecipitation and probed for BubR1 and Bub3 proteins. h Images of HeLa cells transfected with FBXW7 siRNA or control siRNA or FBXW7 siRNA rescued with FLAG-FBXW7 followed by synchronization at metaphase using double thymidine treatment followed by its release and fixed at the appropriate time. Cells were probed for BubR1 and inner kinetochore marker ACA. i Dot plot of intensities of kinetochore-localized BubR1 normalized with corresponding ACA intensity at the metaphase plate. ~ 300–400 kinetochores were analyzed. Data = mean ± SEM. ****p < 0.0001, and ns = 0.4774 from one-way ANOVA
We next checked how FBXW7 depletion affects the cellular levels of BubR1 and other checkpoint proteins. The level of BubR1 showed a robust increase in the partially FBXW7-depleted mitotic synchronized cells as compared to control mitotic HeLa cells (Fig. 2e). Further, expression of exogenous FLAG-FBXW7 reversed the increase of BubR1 level to the level of control cells in the partial FBXW7-depleted condition (Fig. 2f). To verify if the FBXW7 depletion-induced stimulation of BubR1 level is associated with the activity of mitotic checkpoint complex (MCC), we checked the levels of MCC proteins including BubR1 and Bub3 associated with Cdc20 by immunoprecipitation by using Cdc20 antibody from the cell lysates of FBXW7-depleted mitotic synchronized HeLa cells. The IP of Cdc20 showed a significant (~ two fold) increase of BubR1 in the FBXW7 depleted condition as compared to the control (Fig. 2g). The amount of Bub3 was also increased in the IP, however, its cellular level was not considerably increased upon partial depletion of FBXW7 (Fig. 2g) suggesting that the assembly of the MCC complex proteins was induced presumably due to increased stabilization of BubR1. Interestingly, outer-kinetochore-associated BubR1 interacting protein CENP-E also showed a significant increase in its cellular level, nearly similar to BubR1 in the partial FBXW7-depleted conditions in HeLa cells (Fig. 2e). Similar BubR1 and CENP-E regulation were apparent in U2OS cells (Fig. S1f).
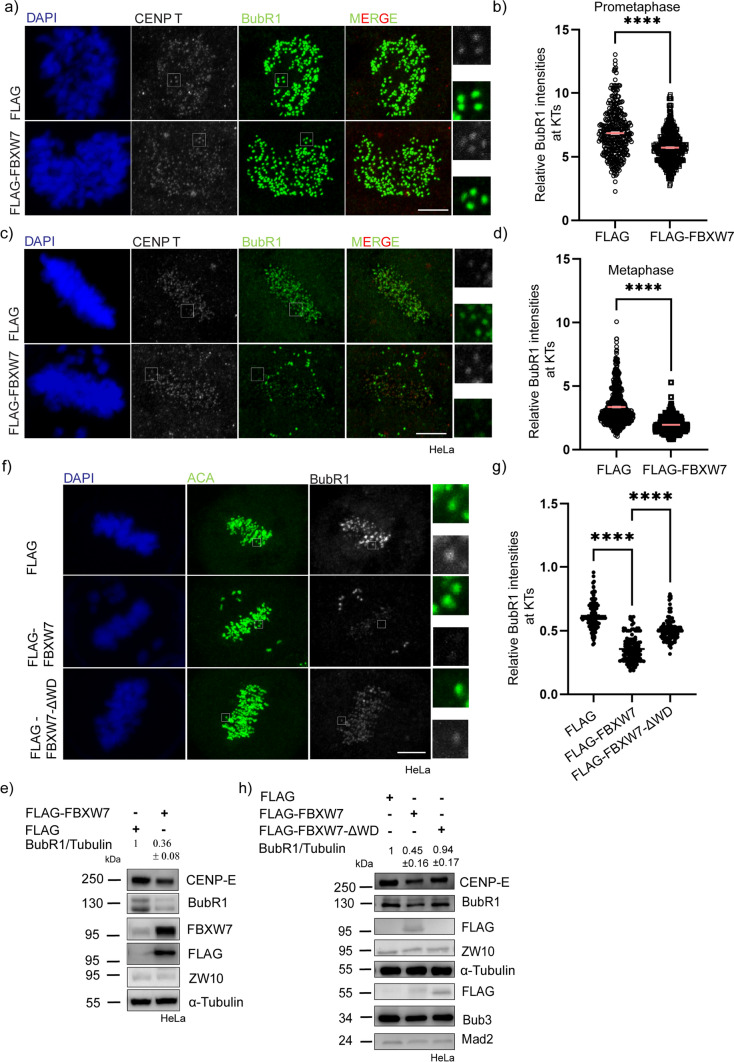
Next, we examined the FBXW7-mediated effects on BubR1 by overexpressing exogenous FBXW7 plasmid in HeLa cells. Mitotically synchronized FLAG-FBXW7-expressed prometaphase and metaphase HeLa cells showed significantly reduced KT localization of BubR1 (Fig. 3a, c). However, the effect was more pronounced at metaphase KT. Intensity analysis showed about 17% and 54% reduction of BubR1 levels at the prometaphase and metaphase kinetochores, respectively (Fig. 3b, d). Expression of a substrate-binding WD domain-deleted form of FBXW7, FLAG-FBXW7-ΔWD rescued KT localization of BubR1 supporting BubR1 as a substrate of the ligase (Fig. 3f, g). Consistent with the KT localization effect, the cellular levels of BubR1 was substantially reduced upon FLAG-FBXW7 expression in HeLa and U2OS cells (Figs. 3e, S1g). Similar results were observed in the case of CENP-E as well (Figs. 3e, S1g). The deletion of the WD domain of FBXW7 showed a loss of function of these effects on CENP-E (Fig. 3h). Furthermore, treatment of proteasomal inhibitor MG132 suppressed the effect of FLAG-FBXW7-mediated BubR1 and CENP-E degradation, suggesting proteasomal degradation of these proteins induced by the ligase (Fig. S1h). The levels of Aurora A and cyclin E were also checked as positive controls in both partial FBXW7-depleted as well as overexpressed condition (Fig. S1i, j). Staining of endogenous FBXW7 also showed its localization at the mitotic kinetochores (Fig. S1k).
Fig. 3.
FBXW7 over-expression reduces overall levels of BubR1 a, c Representative images of HeLa cells after 12 h of FLAG-tagged FBXW7α or control FLAG plasmid expression followed by synchronization at prometaphase and metaphase by treating with thymidine followed by its release and fixed at appropriate times prior to stain for BubR1 and inner kinetochore marker, CENP-T. b, d Plots of intensities of kinetochore-localized BubR1 at prometaphase and metaphase cells in FLAG-FBXW7-expressed vs. control cells. BubR1 intensities were normalized with respect to that of CENP-T in all cases. ~ 300–400 kinetochores were analyzed from three independent experiments. Data = mean ± SEM.****p < 0.0001 from unpaired t-test. e Lysates of mitotic synchronized HeLa cells expressed with control FLAG plasmid or FLAG-tagged FBXW7, were assessed for the CENP-E and BubR1 levels. Tubulin was probed as a control. BubR1 and CENP-E levels were reduced substantially in FBXW7-overexpressed cells. f Confocal images of HeLa cells after 12 h of FLAG-tagged FBXW7 or control FLAG or FLAG-FBXW7-ΔWD expression followed by synchronization at metaphase by treating with thymidine followed by its release and fixed at appropriate times prior to stain for BubR1 and inner kinetochore marker, ACA g Plot of intensities of kinetochore localized BubR1 at metaphase in control vs FLAG-FBXW7 expressed vs FLAG-FBXW7-ΔWD-expressed cells. BubR1 intensities were normalized with respect to that of ACA in all cases. ~ 300 kinetochores were analyzed from three independent experiments. Data = mean ± SEM. ****p < 0.0001 from one-way ANOVA h Lysates of mitotic synchronized cells expressed with Control FLAG vs FLAG-FBXW7 vs FLAG-FBXW7-ΔWD were assessed for overall BubR1 and CENP-E levels. Tubulin was probed as a control
To rule out the possibility of BubR1 and CENP-E degradation during prolonged mitotic arrest under normal condition, we checked BubR1 and CENP-E levels in control cells during prolonged mitotic arrest up to 5 h by treating the cells with Nocodazole. Levels of both BubR1 and CENP-E were nearly unchanged during that time span (Fig. S2a). In order to assess whether BubR1 and CENP-E are degraded after metaphase, control and FLAG-FBXW7-expressed cells were treated with Monastrol (an inhibitor for prometaphase arrest) and the levels of BubR1 and CENP-E levels were compared in the Monastrol-treated and wash-out conditions. The BubR1 and CENP-E levels were considerably reduced (~ 70%) in the Monastrol-washout cells expressed with Flag-FBXW7 as compared to control, whereas such reduction was much less (~ 30%) pronounced in the Monastrol-treated condition (Fig. S2b). These results suggest that BubR1 and CENP-E are degraded as the cells exit mitosis. To verify if BubR1 and CENP-E are synthesized during mitosis, we assessed the FBXW7 depletion-mediated effect on BubR1 and CENP-E levels in the presence of Cyclohexamide (CHX), an inhibitor of protein synthesis. While the control cells showed time-dependent decrease of both BubR1 and CENP-E levels, they were considerably more stabilized in the partial FBXW7-depleted cells (Fig. S2c) supporting their rescue. The results suggest that FBXW7 plays an important role in regulating the homeostasis of BubR1 and CENP-E during mitosis.
FBXW7-mediated CENP-E degradation is BubR1 dependent
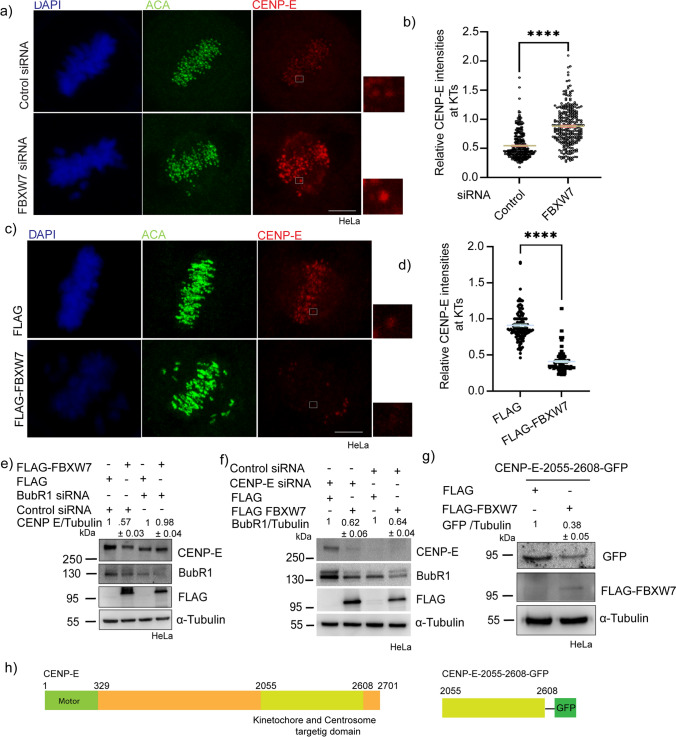
Since FBXW7 regulates the cellular level of CENP-E, we first assessed how it affects KT localization of CENP-E. We showed that FBXW7 partial depletion led to a significant increase in CENP-E localization at the KTs of metaphase-synchronized HeLa cells (Fig. 4a). Similarly, the expression of exogenous FLAG-FBXW7 showed reduced KT localization of CENP-E (Fig. 4c). Intensity analysis showed about 48% increase and 55% reduction of CENP-E localization in FBXW7-depleted and over-expressed cells, respectively (Fig. 4b, d). Since CENP-E binds to BubR1 and is recruited to kinetochores in a BubR1-dependent manner [44], we assessed whether FBXW7-mediated CENP-E regulation requires BubR1. The effect of FLAG-FBXW7 expression on CENP-E level was examined in HeLa cells that were depleted of BubR1 by siRNA. Unlike in control cells, FLAG-FBXW7 expression in BubR1-depleted cells showed no change in CENP-E level (Fig. 4e). Interestingly, CENP-E depletion did not cause any effect on FLAG-FBXW7 expression-mediated BubR1 degradation in HeLa cells (Fig. 4f). BubR1 interacts with CENP-E within its kinetochore-targeting domain (2055–2608) [44]. We, therefore, verified whether the kinetochore-targeting domain of CENP-E, which consists of the BubR1-binding site, is degraded by FBXW7. HeLa cells were co-transfected with CENP-E-2055–2608-GFP and FLAG-FBXW7 and the level of CENP-E-2055–2608-GFP was assessed in comparison to control cells expressed only with CENP-E-2055–2608-GFP. The level of CENP-E-2055–2608-GFP showed a marked reduction in the FLAG-FBXW7-expressed cells (Fig. 4g, h). The results indicate that the FBXW7 mediates CENP-E degradation in its BubR1-bound form. Further to check if depletion of BubR1 could rescue the FBXW7 depletion-caused mitotic delay, we depleted BubR1 partially (about 50%) along with FBXW7 depletion using corresponding siRNAs in H2B mCherry stably-expressed HeLa Kyoto cells and monitored mitotic progression by time-lapse imaging in live cells that were synchronized by double thymidine and then released from thymidine block. The thymidine-released cells were imaged from the time soon after the nuclear envelope breakdown till the anaphase. The levels of BubR1 and FBXW7-depletion by siRNA are shown in Fig. S2d. Consistent with the previous observation, the partially FBXW7-depleted mitotic cells showed a significant number of chromosomes that are loosely congressed and the cells took prolonged time to proceed to anaphase as compared to control cells (Fig. S2e, Movie M3, M4). While the FBXW7-depleted cells completed metaphase to anaphase progression in ~ 60 min, the cells with partially depleted BubR1 along with FBXW7 did so in ~ 25 min, nearly similar to control cells (~ 24 min) (Fig. S2f, Movie M3–M5). This supports that FBXW7-depletion induced metaphase to anaphase delay is due to increased BubR1.
Fig. 4.
FBXW7-mediated CENP-E degradation is BubR1 dependent. a Representative images of HeLa cells after 18 h of FBXW7 siRNA or control siRNA treatment followed by synchronization by treating with thymidine and released at the appropriate time prior to stain for CENP-E and inner kinetochore marker, ACA. b Dot Plots representing intensities of kinetochore localized BubR1 at metaphase cells in FBXW7 siRNA vs. control cells. CENP-E intensities were normalized with respect to that of ACA in all cases. ~ 300–400 kinetochores were analyzed from three independent experiments. Data = mean ± SEM. ****p < 0.0001 unpaired t-test. c Representative images of HeLa cells after 12 h of FLAG-tagged FBXW7α or control FLAG plasmid expression followed by synchronization at prometaphase and metaphase by treating with thymidine followed by its release and fixed at appropriate times prior to stain for BubR1 and inner kinetochore marker, CENP-T. d Plots of intensities of kinetochore localized CENP-E at metaphase cells in FLAG-FBXW7 expressed vs. control cells. CENP-E intensities were normalized with respect to that of ACA in all cases. ~ 300–400 kinetochores were analyzed. Data = mean ± SEM.****p < 0.0001 from unpaired t-test. e Overall levels of CENP-E were unaffected in cells depleted of BubR1 followed by FBXW7 overexpression vs control siRNA followed by FBXW7 overexpression. f Overall BubR1 levels were significantly reduced in CENP-E-depleted cells overexpressed with FLAG-FBXW7 with respect to control cells. g Overall levels of CENP-E-2055–2608-GFP showed a significant reduction in the FLAG-FBXW7-expressed cells with respect to control cells h Schematic representation of the amino acid regions of CENP-E-WT and CENP-E-2055–2608-GFP
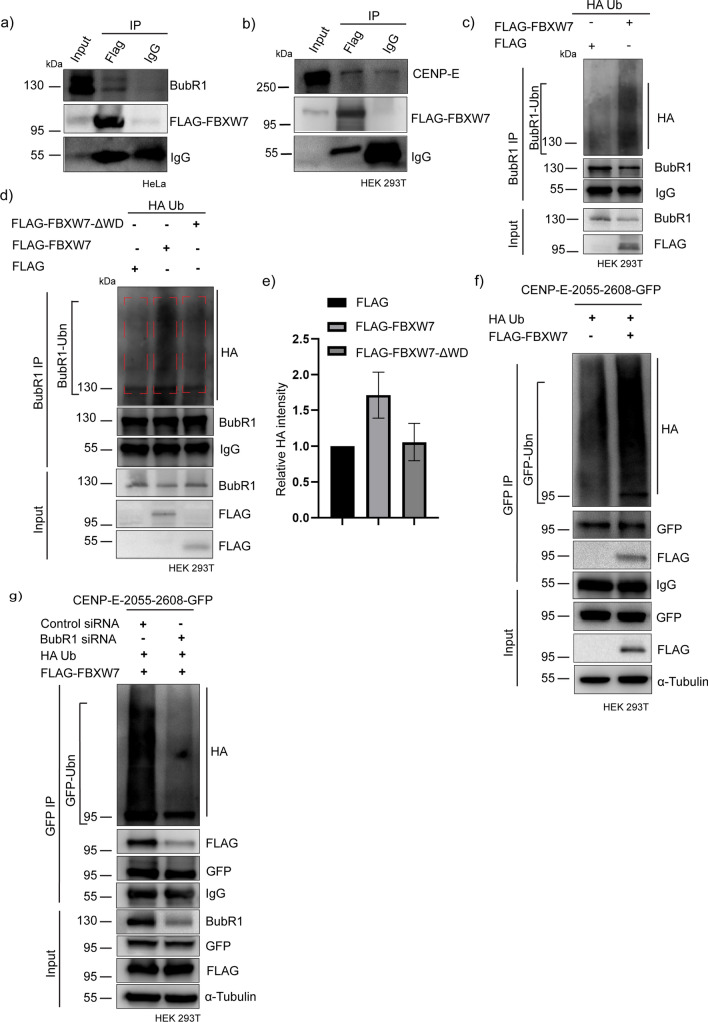
FBXW7 interacts with BubR1 and ubiquitinates BubR1 and CENP-E
We next sought to determine whether FBXW7 interacts with BubR1 and CENP-E. Pull-down of FLAG-FBXW7 from mitotic synchronized FLAG-FBXW7-expressed HeLa cells showed the presence of endogenous BubR1 (Fig. 5a). Similarly, co-immunoprecipitation of BubR1 showed its association with FLAG-FBXW7 (Fig. S2h). Pull-down of FLAG-FBXW7 from mitotic synchronized FLAG-FBXW7-expressed HEK293T cells also showed the presence of CENP-E (Fig. 5b). Similarly, FLAG-FBXW7 was present in the co-immunoprecipitate of CENP-E (Fig. S2g). Next, we checked whether FBXW7 induces ubiquitination of these proteins. Ubiquitination was analyzed from the lysates of mitotic synchronized HEK293T cells that were expressed with FLAG-FBXW7 and HA-Ub followed by treatment with proteasome inhibitor, MG132. BubR1 showed a strong ubiquitination pattern spanning over a region above its molecular size in the FLAG-FBXW7-overexpressed cells as compared to control FLAG empty vector plus HA-Ub- transfected cells (Fig. 5c). BubR1 ubiquitination level was substantially reduced in cells expressed with FLAG-FBXW7-ΔWD, substantiating the conclusion that BubR1 is a substrate for ubiquitination by SCFFBXW7 (Fig. 5d). Intensity analysis of ubiquitinated BubR1 level (BubR1-Ubn) showed 1.7 times increased ubiquitination in FLAG-FBXW7 expressed cells as compared to control. Cells overexpressed with FLAG-FBXW7-ΔWD showed BubR1 ubiquitination comparable to the control cells (Fig. 5e).
Fig. 5.
BubR1 is a novel substrate of FBXW7. a HeLa cells overexpressed with FLAG-FBXW7 were synchronized at mitosis followed by the addition of MG132 treatment and then cell lysate was subjected to FLAG pull down by using FLAG antibody and then probed for BubR1 b HEK293T cells expressed with FLAG-FBXW7 were synchronized at mitosis followed by the addition of MG132, were subjected to FLAG pull down by using FLAG antibody and then probed for CENP-E. c HEK293T cells were expressed with FLAG-FBXW7 and HA-tagged Ubiquitin (HA-Ub) for 18 h followed by mitotic synchronization using thymidine and addition of MG132 and BubR1 was immunoprecipitated from the cell lysate. The BubR1 immunoprecipitate from the lysates of FLAG-FBXW7-expressed cells were probed for HA-Ub and compared to the BubR1 immunoprecipitate from control cell lysate. d HEK293T cells were expressed with FLAG-FBXW7 or FLAG-FBXW7-ΔWD together with HA-Followed by mitotic synchronization using thymidine and MG132. BubR1 was co immunoprecipitated and its ubiquitination was compared by probing with an HA antibody. e Plot of BubR1 ubiquitination levels in different condition as of d. f GFP immunoprecipitate of mitotic synchronized HEK293T cells expressed with FLAG-FBXW7 and HA-Ub along with CENP-E-2055–2608-GFP was probed for HA-Ub and the levels of ubiquitinated proteins were compared with respect to control CENP-E-2055–2608-GFP cells expressed with FLAG control plasmid. g HEK293T cells were co-expressed with CENP-E-2055–2608-GFP together with FLAG-FBXW7 and HA-Ub in BubR1 siRNA or Control siRNA transfected condition for 18 h followed by mitotic synchronization using thymidine followed by the addition of MG132. Then CENP-E-2055–2608-GFP was immunoprecipitated by GFP antibody and probed for HA
Then, we assessed the ubiquitination of CENP-E-2055–2608-GFP by FLAG-FBXW7. Immunoprecipitate of CENP-E-2055–2608-GFP from mitotic synchronized HEK 293T cells upon expression of FLAG-FBXW7 and HA-Ub followed by treatment with proteasome inhibitor, MG132 showed an increased ubiquitination pattern in the FLAG-FBXW7-overexpressed cells as compared to control (Fig. 5f). We examined whether FBXW7-mediated CENP-E-2055–2608-GFP ubiquitination is dependent on BubR1. Ubiquitination levels of CENP-E-2055–2608-GFP in control vs BubR1-depleted cells were compared. HEK293T cells depleted of BubR1 by siRNA were expressed with FLAG-FBXW7 and HA-Ub and then ubiquitination was analyzed in the GFP immunoprecipitate from the lysates of mitotic synchronized MG132-treated cells. CENP-E-2055–2608-GFP immunoprecipitation from BubR1-depleted cells showed substantially reduced ubiquitination as compared to that of control siRNA-treated cells (Fig. 5g). Consistent with reduced ubiquitination, the interaction between FLAG-FBXW7 and CENP-E-2055–2608-GFP was markedly reduced in BubR1-depleted condition as compared to control as evident from the much-reduced amount of FLAG-FBXW7 associated with the CENP-E-2055–2608-GFP immunoprecipitate in the BubR1 depleted cell lysate (Fig. 5g).
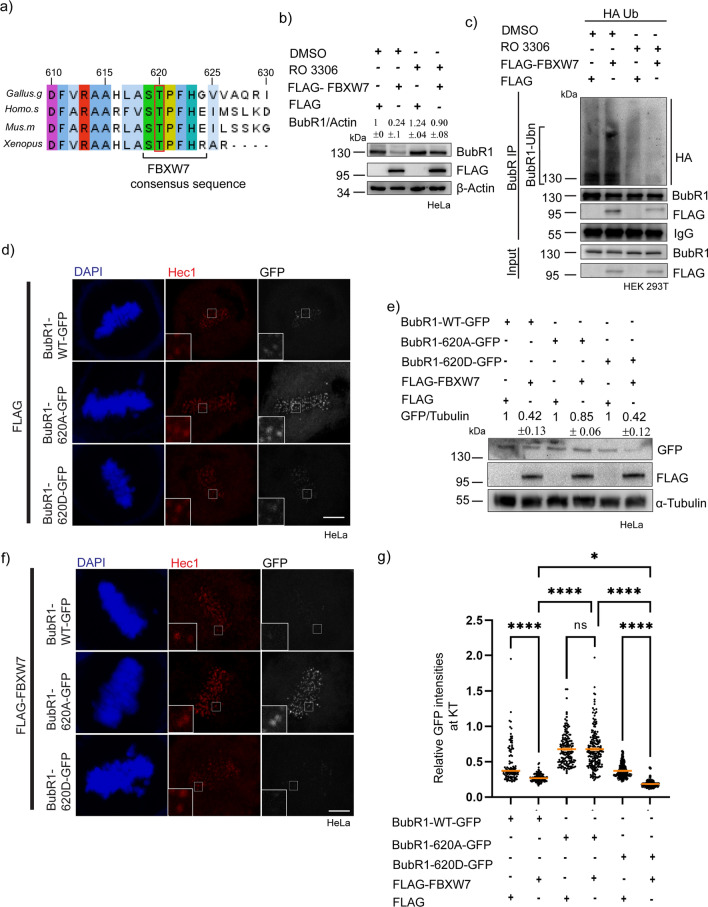
Cdk1 phosphorylation facilitates FBXW7-mediated BubR1 ubiquitination
Previous studies have shown that Cdk1 mediates BubR1 phosphorylation at Thr 620 [27, 45]. Amino acid sequence analysis also showed the presence of a consensus sequence specific to FBXW7 targeting phospho-degron sequence (STPFHE) that includes Thr 620 and the same is conserved in several higher vertebrates [27, 41, 46] (Fig. 6a). Therefore, we first checked the role of Cdk1 in FBXW7-mediated BubR1 degradation. HeLa cells expressed with FLAG-FBXW7 were treated with Cdk1 inhibitor, RO3306 [47] and BubR1 levels were compared with the untreated control cells. RO3306 treatment in FLAG-FBXW7-expressed cells rescued BubR1 to a level comparable to control cells, supporting the involvement of Cdk1 in this regulation (Fig. 6b). The level of BubR1 ubiquitination was also substantially reduced in RO3306-treated cells suggesting that Cdk1-mediated phosphorylation could be involved in FBXW7-mediated BubR1 ubiquitination (Fig. 6c). To determine the role of Cdk1-mediated Thr 620 phosphorylation in BubR1 regulation by FBXW7, HeLa cells co-expressed with wild-type (BubR1-WT-GFP) or phosphomimetic (BubR1-T620D-GFP) or phophoresistant (BubR1-T620A-GFP) and FLAG-FBXW7 were analyzed for BubR1 intensity at the kinetochores. While in the BubR1-WT-GFP and BubR1-T620D-GFP cells, KT localization of both the proteins was substantially reduced in the FLAG-FBXW7-overexpressed condition as compared to control cells expressed with those BubR1 constructs, but not FLAG-FBXW7, under similar condition, the BubR1-T620A-GFP localization did not show any change (Fig. 6d, f). Intensity analysis of individual KT-localized BubR1 showed similar nature (Fig. 6g). The total cellular level of BubR1-T620A-GFP also did not show any change in control vs. FLAG-FBXW7 expressed condition, whereas the BubR1-WT-GFP and BubR1-T620D-GFP showed a marked reduction in the FLAG-FBXW7 expressed cells as compared to control (Fig. 6e).
Fig. 6.
Cdk1-mediated phosphorylation at Thr 620in BubR1 acts as phosphodegron for FBXW7-mediated degradation. a Conservation of Thr620 (T620) of BubR1 among different species. b Overall levels of BubR1 were unaffected in Cdk1 inhibitor-treated cells in control vs FLAG-FBXW7 overexpressed cells. c Levels of ubiquitinated BubR1were reduced in Cdk1 inhibitor-treated HEK293T cells expressed with FLAG-FBXW7 and HA-Ub as compared to control conditions. d, f Representative images of Mitotic synchronized HeLa cells expressed with BubR1-WT-GFP or BubR1-T620A-GFP or BubR1 T620D-GFP together with control FLAG-plasmid or FLAG-FBXW7 g Dot plot of intensities of GFP-tagged BubR1 WT and mutants at kinetochores normalized to Hec1 as marker in metaphase cells expressed with BubR1-WT-GFP or BubR1-T620A-GFP or BubR1-T620D-GFP together with FLAG-FBXW7 vs. FLAG empty vector. ~ 200–300 kinetochores were analyzed. Data = mean ± SEM. ****p < 0.0001, *p = 0.211, ns > 0.9999 by one-way ANOVA e The overall cellular levels of BubR1-T620D-GFP showed increased degradation, whereas the BubR1-T620A-GFP showed reduced degradation in FLAG-FBXW7 expressed cells
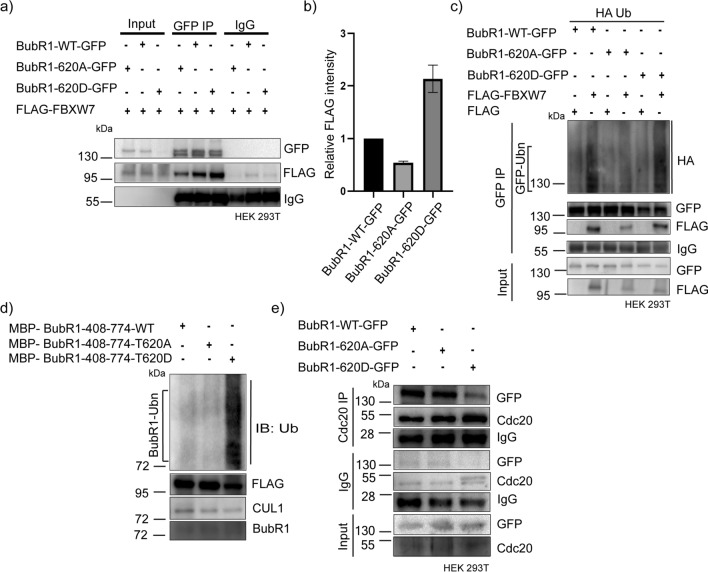
To verify if the interaction of BubR1 with FBXW7 is different in the T620A vs.T620D condition, the immunoprecipitation of BubR1-T620A-GFP and BubR1-T620D-GFP from the lysates of HEK293T cells expressed with the respective mutants together with FLAG-FBXW7 were compared for the presence of FLAG-FBXW7. Amount of FLAG-FBXW7 was substantially higher in the BubR1-T620D-GFP IP as compared to BubR1-T620A-GFP IP (Fig. 7a). Plot of FLAG-FBXW7 levels with respect to BubR1 immunoprecipitate level (WT and mutants) shows about ~ four fold increase of FLAG-FBXW7 in the BubR1-T620D-GFP IP as compared to BubR1-T620A-GFP IP (Fig. 7b). FLAG-FBXW7 in BubR1-WT-GFP IP also appeared to be slightly higher than BubR1-T620A-GFP condition (Fig. 7b). These results indicate that FBXW7-BubR1 interaction is facilitated in the BubR1 Thr 620-phosphorylated condition. We then assessed how the Thr 620 phosphorylation affects FBXW7-mediated BubR1 ubiquitination by analyzing the levels of ubiquitinated BubR1-T620D-GFP vs. BubR1-T620A-GFP from the IP of the respective BubR1-GFP proteins in the HEK293T cells expressed with the BubR1 mutants together with FLAG-FBXW7 and HA-Ub. Ubiquitination of BubR1-WT-GFP was also analysed for comparison. While BubR1-T620D-GFP exhibited a high level of ubiquitination, even higher than BubR1-WT-GFP form, BubR1-T620A-GFP did not show any detectable ubiquitination (Fig. 7c). We also assessed ubiquitination of purified MBP-tagged recombinant BubR1-408–774-T620D protein. All the SCFFBXW7 protein components (Skp1, Cul1, Rbx, FBXW7) were purified by FLAG-FBXW7 pulldown in HEK293T cells and the pulldown sample mixture was added to increasing amounts of MBP-BubR1-408–774-T620D. MBP-BubR1-408–774-T620D showed increased ubiquitination in a dose-dependent manner (Fig. S2i, ). In-vitro ubiquitination assay (see Methods) also showed increased ubiquitination only in the phosphomimetic mutant of BubR1, whereas the wild-type (MBP-BubR1-408–774-WT) and phopho-resistant mutant (MBP-BubR1-408–774-T620A) showed negligible ubiquitination supporting that BubR1 620 phosphorylation is required for ubiquitination of BubR1 (Fig. 7d). Further to check how BubR1 phosphorylation regulates MCC stability, mitotic synchronized HeLa cells expressed with BubR1-WT-GFP/BubR1-T620A-GFP/BubR1-T620D-GFP and treated with MG132 were assayed for Cdc20 pull-down to assess the levels of BubR1 wildtype vs. the mutants. Interestingly, while BubR1-WT-GFP or BubR1-T620A-GFP could interact strongly with Cdc20, the same was substantially impaired in the case of BubR1-T620D-GFP (Fig. 7e). These data suggest that Cdk1 -targeted phosphorylation of BubR1 at Thr 620 induces BubR1 dissociation from the MCC complex.
Fig. 7.
BubR1 phosphorylation at Thr 620 promotes its ubiquitination by FBXW7 a HEK293T cells were co-expressed withBubR1-WT-GFP or BubR1-T620A-GFP or BubR1-T620D-GFP together with FLAG-FBXW7 for 18 h followed by mitotic synchronization using thymidine followed by the addition of MG132. Then GFP-tagged BubR1 was immunoprecipitated with GFP antibody and levels of FLAG-FBXW7 were compared by immunoblotting with FLAG antibody. b The histogram plot shows the relative intensity of FLAG-FBXW7 normalized to corresponding BubR1-WT-GFP or BubR1-T620A-GFP or BubR1-T620D-GFP level. c HEK293T cells were co-expressed with BubR1-WT-GFP or BubR1-T620A-GFP or BubR1-T620D-GFP together with FLAG-FBXW7 and HA-Ub and the lysates of cells after mitotic synchronization by thymidine release followed by MG132 were probed for HA-Ub. d Recombinant MBP-tagged BubR1-408–774-WT or MBP-tagged BubR1-408–774-T620A or MBP-tagged BubR1-408–774-T620D bound with amylose resin was incubated separately with a mixture of SCFFBXW7 components, Myc-Skp-1, Myc-Cul-1, HA-Rbx-1, and FLAG-FBXW7, followed by E1, E2, Ub, and ATP. SCFFBXW7component were isolated from HEK293T cells as described in Methods. Samples were immunoblotted for BubR1 and ubiquitin by antibodies. e Lysates of mitotic synchronized HEK293T cells transfected with BubR1-WT-GFP or BubR1-T620A-GFP or BubR1-T620D-GFP for 18 h followed by mitotic synchronization by thymidine release followed by MG132 addition were subjected to Cdc20 immunoprecipitation and probed for GFP-tagged BubR1 protein levels
Discussion
Increased expression of FBXW7 during mitosis prompted us to investigate the mitotic-specific function of SCFFBXW7. Our results revealed that loss of FBXW7 induced lagging chromosomes and reduced inter-kinetochore tension in both metaphase plate-congressed and uncongressed chromosomes, suggesting defects in microtubule attachments and/or tension between the sister kinetochore pairs. This was accompanied by increased recruitment of MCC components to the kinetochores, BubR1 and Mad2 (Fig. 2, S1). Interestingly the overall cellular levels of BubR1 but not Bub3 showed a significant increase in response to partial FBXW7 depletion (Fig. 2). FBXW7 overexpression led to a reduction in BubR1 levels, which substantiated the possibility that FBXW7 directly regulates the stability of BubR1 during mitosis (Fig. 2). Consistently, we have demonstrated that FBXW7 interacts with and induces ubiquitination of BubR1 leading to its degradation (Fig. 5, S2). Taken together, the findings are in support of the notion that FBXW7-mediated direct control of BubR1 stability plays an important role in mitotic progression.
It has been shown previously that Cdk1 targeting BubR1 phosphorylation at Thr 620 is essential for the timely progression of mitosis as T620A mutation led to prolonged metaphase arrest [27]. BubR1 T620 phosphorylation is required for recruitment of Plk1 to BubR1 and the recruited Plk1 phosphorylates further to other sites in BubR1. This follows PP2A-B56-recruitment to kinetochore, which is required for timely mitotic progression [27]. Here, we find that the same mutation resulted in resistance against FBXW7-mediated BubR1 ubiquitination and degradation (Figs. 6, 7). As FBXW7 partial depletion also showed a similar metaphase to anaphase onset delay and increased BubR1 stabilization (Fig. 1), a reasonable possibility could be that FBXW7-mediated BubR1 degradation regulates timely mitotic progression. Thr620 phosphorylation could also play a role in the regulation of MCC assembly and associated SAC activation as we showed that the T620A mutant interacted with other MCC proteins relatively more strongly than the T620D mutant (Fig. 7). Earlier Cdc20 phosphorylation by Cdk1 has also been reported to destabilize MCC by promoting the release of BubR1 from the complex [48]. We found that Cdc20 interaction was substantially reduced in the BubR1T620D mutant form as compared to the BubR1 T620A form (Fig. 7). On the other hand, Cdc20 IP showed an increased level of BubR1 in FBXW7 partial-depleted condition (Fig. 2), suggesting that both Cdk1-mediated phosphorylation and FBXW7-mediated ubiquitination of BubR1 play an additional role in MCC disassembly and this mechanism could be independent of Cdc20 phosphorylation-mediated MCC disassembly shown earlier [12]. Our results here highlight the additional molecular control of MCC stability via Cdk1 site-specific phosphorylation of BubR1 and its stability by FBXW7.
Previous studies have shown that CENP-E level is associated with fibrous corona structure spanning a larger area allowing efficient capture of microtubules laterally by the kinetochores during early mitosis till prometaphase [17]. However, such lateral attachments generate various attachment errors, which are eventually corrected by favoring end-on attachment during metaphase [49]. Conversion of lateral to end-on attachments has been shown to be associated with the loss of larger fibrous corona structures including CENP-E organization to constricted smaller dot-like structures [17, 50, 51]. Therefore, CENP-E levels at the kinetochores need to be reduced in a mitotic stage-specific manner and this process needs to be molecularly controlled. Here, we show that FBXW7 is a key regulator in controlling CENP-E levels during mitosis [52]. FBXW7-depletion leads to increased accumulation of CENP-E at the kinetochores and also increased its cellular levels (Fig. 4). This is likely due to the abrogation of FBXW7-mediated ubiquitination followed by CENP-E degradation as we have shown that FBXW7 induces ubiquitination and degradation of the kinetochore targeting domain of CENP-E (Fig. 5). We also observed that this regulation is dependent on BubR1 as in the BubR1-depleted cells, FBXW7 did not exert any effect on CENP-E level (Fig. 4). BubR1 depletion also impaired FBXW7-mediated ubiquitination of the kinetochore binding domain of CENP-E (Fig. 5). It is possible that the kinetochore-recruited BubR1-bound CENP-E is targeted for degradation by FBXW7. BubR1 has been shown to bind to CENP-E within the kinetochore targeting domain of CENP-E and it is also required to recruit CENP-E to the kinetochore [15]. The persistent presence of some amount of CENP-E at the kinetochores during metaphase is also needed for stabilization of microtubule-kinetochore attachment [18], suggesting that kinetochore-localized CENP-E level needs to be correctly balanced. Such a balance could be maintained by SCFFBXW7 through BubR1 and CENP-E degradation. In conclusion, this study reveals the key role of an SCF family ligase, FBXW7 in the control of mitotic progression by regulating protein BubR1 and its binding partner, CENP-E. Previously, a few other F-box family E3 ligases such as FBXO42 and FBXO6 have been shown to play roles in mitotic progression [53, 54]. Therefore, it remains to be understood in the future the functional link of FBXW7-mediated BubR1 and CENP-E regulation to the mitotic-specific roles of those ligases. It is possible that the coordination of multiple SCF E3 ligases orchestrates the temporal control of various other molecular regulators of mitosis.
Materials and methods
Cell lines, siRNAs, reagents
HeLa, U2OS, or HEK293T cells originally obtained from ATCC were cultured in DMEM media, supplemented with 10% FBS, 2 mM L-glutamine,1.5 mg/ml sodium bicarbonate, 100 mg/ml penicillin, 100 mg/ml streptomycin. Cells were synchronized by 2 mM thymidine (SigmaAldrich, U.S.A.) and collected/fixed after 9 h of thymidine release. Final concentration 9 μM RO3306 (SigmaAldrich, U.S.A.) was used for CDK1 inhibition experiments. We used either single siRNA or ribonuclease III-prepared siRNA pools (esiRNA) to suppress protein expression. esiRNA (Cat no: EHU075951, Sigma-Aldrich) targeted against the 1218–1625 nucleotide region of FBXW7 (NM_033632) was used. For transfection by single FBXW7 siRNA, RNA sequence of 5′ACCUUCUCUGGAGAGAGAAAUGC3′ was used. For depletion of endogenous BubR1 with simultaneous expression of exogenous BubR1 plasmids, BubR1 siRNA targeted to the 3’-UTR region (5’-GTCTCACAGATTGCTGCCT-3′) (Dharmacon, U.S.A.) was used. Control siRNA (5’-UCUAUAUCAUGGCCGACAA-3′) or Luciferase esiRNA (Cat no: EHUFLUC-SigmaAldrich, U.S.A.) was used as control. Lipofectamine RNAiMAX (Invitrogen, U.S.A.) was used as a vehicle for the transfection of siRNA and esiRNA; and lipofectamine 3000 or 2000 (Invitrogen, U.S.A.) was used for plasmid DNA transfection.
Antibodies
Rabbit anti-FBXW7 (A301-720A) antibody from Bethyl laboratories, U. S. A. was used. Anti-mouse α-tubulin, anti-rabbit HA (H6908), and mouse Flag M2 (F1804) antibodies were procured from Sigma Aldrich. Mouse monoclonal Actin, mouse monoclonal BubR1 and mouse monoclonal Mad2 (610,678) antibodies were obtained from BD Biosciences. Mouse monoclonal CENP E (sc-376685), mouse monoclonal GFP antibody (sc-9996), mouse monoclonal Hec1 antibody (sc-135934), mouse monoclonal Mad1 (sc-47746) and mouse IgG (sc-2025) were obtained from Santa Cruz Biotechnology. Anti-rabbit polyclonal CENP T (ab-220280), anti-rabbit polyclonal ZW10 (ab-21582) anti-rabbit polyclonal Knl1 (ab222055) and anti-rabbit polyclonal Bub3 (ab133699) were obtained from Abcam. Anti-centromere antibody (ACA) was obtained from Antibodies Incorporated, CA. All HRP-conjugated, Alexa 488-, and Alexa 647-conjugated secondary antibodies were purchased from Jackson ImmunoResearch, PA, U.S.A. Alexa 568-conjugated anti-rabbit secondary antibody was obtained from ThermoFisher Scientific, MA, U.S.A.
Plasmids and proteins
pcDNA5-eGFP-AID-BubR1 were obtained from Addgene. pcDNA5-eGFP-AID- BubR1(T620A), pcDNA5-eGFP-AID-BubR1(T620D) were generated using site-directed mutagenesis techniques. MBP-BubR1-408–778 and MBP-CENP-E-2055–2608 plasmids were obtained from Julie Welburn, University of Edinburgh. MBP-BubR1-408–778 (T620D), MBP-BubR1-408–778 (T620A) construct was generated using site-directed mutagenesis. The recombinant MBP-tagged BubR1 proteins were purified. FLAG-tagged human FBXW7-ΔWD plasmid was generated using FLAG-tagged human FBXW7(alpha form) as a template originally obtained from Bruce E. Clurman, Fred Hutchinson Cancer Research Center, U.S.A. MBP-CENP-E-2055–2608 (obtained from Julie Welburn, University of Edinburgh) construct was sub-cloned to generate CENP-E-2055–2608-GFP. Recombinant human proteins E1, E2 and ubiquitin were obtained from Boston Chemicals, U.S.A. Myc-Cul1, Myc-Skp-1 and HA-RBX-1 were obtained from wenyi wei, Beth Israel Medical centre, U.S.A.
Cell synchronization and Co-immunoprecipitation (co-IP)
HEK293T cells were mitotic synchronized by double thymidine treatment followed by thymidine release. MG132 were added to the cells after 7 h of thymidine release and incubated for the next 6 h. For immunoprecipitation, cells were lysed using lysis buffer (20 mM Tris–HCl, pH 7.4, 0.1% Triton X-100, 50 mM NaCl, 10 mM EGTA) supplemented with phosphatase inhibitor 2 and 3 and protease inhibitor (Sigma). Cell lysates were incubated with corresponding antibodies as specified for IP then incubated with A/G agarose beads.
MG132 (25 μM) (Sigma Aldrich, U.S.A.), whenever used for relevant experiments, was added for 2 h after 7 h of thymidine release. For prolonged mitotic arrest, the double-thymidine synchronized cells were released from thymidine and then were arrested at mitosis by Nocodazole (0.33 μM). The mitotic arrested cells were collected by shake-off and then were treated with Nocodazole (0.33 μM) or 100 nM cycloheximide (CHX) plus Nocodazole (0.33 μM) for the time as indicated.
Immunofluorescence confocal microscopy and image analysis
Metaphase or prometaphase-arrested cells were fixed using ice cold methanol or 4% PFA. For PFA fixation, cells were pre-extracted for 5 min using 0.1% triton X 100 in PHEM buffer (60 mM PIPES, 25 mM HEPES, 10 mM EGTA, and 2 mM MgCl2) [54]. The cells were then fixed using 4% PFA in PHEM for 15 min at room temperature followed by blocking with 1% PBSAT (1X PBS, 1% BSA, 0.5% Triton X 100). Methanol-fixed cells were incubated with primary antibodies Knl1 (1:200), CENP-T (1:200), ACA (1:700), α-Tubulin (1:700), BubR1(1:200), Hec1(1:100) for 3 to 4 h at room temperature. PFA fixed cells were incubated with primary antibodies, Knl1 (1:400), CENP-T (1:400), Mad1 (1:200), and Mad2 (1:200) for 4 h or overnight at room temperature. DNA was stained using DAPI and then the cells were mounted with ProLong Gold. Images were acquired in Zeiss LSM 880 confocal microscope and Leica SP5 inverted confocal microscope with a 63X oil immersion objective. Image contrast was adjusted by changing the LUT values for better visibility. Raw confocal images were quantified for individual kinetochore intensity across Z or from projection images using ZEN blue software and ImageJ Fiji. Background intensity was measured from a region adjacent to kinetochores and subtracted from the kinetochore intensities. All graphs were plotted and analyzed for statistical significance using GraphPad Prism9.
Ubiquitination assays
In vivo ubiquitination assay- HEK293T cells were transfected with HA-Ub with either control FLAG or FLAG-FBXW7-WT or FLAG-FBXW7-ΔWD and synchronized by single thymidine treatment followed by its release and treating with MG132 (25 μM) during the last 6 h before collecting cells for lysis. BubR1 or CENP-E ubiquitination was assessed in mitotic synchronized cells [42]
In vitro ubiquitination assay- SCFFBXW7 complex components were purified as described [42].Briefly, they were purified from HEK293T cells by transfecting Flag-FBXW7 WT, HA-Rbx-1, Myc-Skp-1, and Myc-Cul-1 followed by FLAG-FBXW7 pull-down. Recombinant MBP-tagged BubR1-408–774 WT or MBP-tagged BubR1-408–774-620A or MBP-tagged BubR1-408–774-620D was pulled down using amylose resin from the BL21 DE3 cell lysate after expressing the plasmids. For ubiquitination reaction, the SCFFBXW7 complex proteins and MBP-BubR1 proteins (WT or T620A or T620D mutant) were mixed together and then purified E1 (100 ng), E2 (100 ng) (UBCH3), Ubiquitin (5 μg) together with ATP (2 mM) were added to the mixture.
Western blot analysis
Raw data quantification was performed using Quantity one software and the corresponding mean intensity values of the protein bands were obtained. Background intensities were obtained from the regions adjacent to the protein bands and were subtracted from the total intensities of protein bands to obtain the net intensities of the bands. The net intensity values were normalised with respect to net intensities of the corresponding loading controls (tubulin or actin).
Statistical analysis
Data are presented as mean ± SEM. The normally distributed data were analyzed with modifiedStudent’s t-test at the 99% confidence level. Wherever applicable, one-way ANOVAfollowed by Tukey’s multiple comparison tests was performed. The data were plotted and analysed using GraphPad Prism 9 software. The figures were organized using Adobe Photoshop and Adobe Illustrator.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Dr. Don Cleveland, UCSD for the CENP-E wild type plasmid and Dr. Julie Welburn, University of Edinburgh for the MBP-BubR1-408-778 and MBP-CENP-E-2055-2608 plasmids. We also thank Dr. Bruce E Clurman, Fred Hutchinson Cancer Research Center, U. S. A. for FBXW7 (alpha) plasmid. Financial support from DBT, Govt. of India, and DST-SERB, Govt. of India to TKM are thankfully acknowledged.
Funding
Department of Biotechnology, Ministry of Science and Technology, India.
Data availability statement
All relevant data are available.
Declarations
Conflict of interest
The authors declare no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Holland AJ, Cleveland DW. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol. 2009;10:478–487. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ricke RM, Van Ree JH, Van Deursen JM. Whole chromosome instability and cancer: a complex relationship. Trends Genet. 2008;24:457–466. doi: 10.1016/j.tig.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schvartzman JM, Sotillo R, Benezra R. Mitotic chromosomal instability and cancer: mouse modelling of the human disease. Nat Rev Cancer. 2010;10:102–115. doi: 10.1038/nrc2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daniel P, Cahill CL, Jian Yu, Riggins GJ, Willson JKV, Markowitz SD, Kinzler KW, Vogelstein B. Mutations of mitotic checkpoint genes in human cancers. Nature. 1998;392:300–303. doi: 10.1038/32688. [DOI] [PubMed] [Google Scholar]
- 5.Masuda A, Takahashi T. Chromosome instability in human lung cancers: possible underlying mechanisms and potential consequences in the pathogenesis. Oncogene. 2002;21:6884–6897. doi: 10.1038/sj.onc.1205566. [DOI] [PubMed] [Google Scholar]
- 6.Nezi L, Musacchio A. Sister chromatid tension and the spindle assembly checkpoint. Curr Opin Cell Biol. 2009;21:785–795. doi: 10.1016/j.ceb.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 7.Shepperd LA, Meadows JC, Sochaj AM, et al. Phosphodependent recruitment of Bub1 and Bub3 to Spc7/KNL1 by Mph1 kinase maintains the spindle checkpoint. Curr Biol. 2012;22:891–899. doi: 10.1016/j.cub.2012.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lara-Gonzalez P, Pines J, Desai A. Spindle assembly checkpoint activation and silencing at kinetochores. Semin Cell Dev Biol. 2021;117:86–98. doi: 10.1016/j.semcdb.2021.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu ST, Zhang H. The mitotic checkpoint complex (MCC): looking back and forth after 15 years AIMS. Mol Sci. 2016;3:597–634. doi: 10.3934/molsci.2016.4.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang G, Kruse T, Lopez-Mendez B, et al. Bub1 positions Mad1 close to KNL1 MELT repeats to promote checkpoint signalling. Nat Commun. 2017;8:15822. doi: 10.1038/ncomms15822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Piano V, Alex A, Stege P, et al. CDC20 assists its catalytic incorporation in the mitotic checkpoint complex. Science. 2021;371:67–71. doi: 10.1126/science.abc1152. [DOI] [PubMed] [Google Scholar]
- 12.Lischetti T, Nilsson J. Regulation of mitotic progression by the spindle assembly checkpoint Mol. Cell Oncol. 2015;2:e970484. doi: 10.4161/23723548.2014.970484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Craske B, Welburn JPI. Leaving no-one behind: how CENP-E facilitates chromosome alignment. Essays Biochem. 2020;64:313–324. doi: 10.1042/EBC20190073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schaar BT, Chan GKT, Maddox P, Salmon ED, Yen TJ. CENP-E function at kinetochores is essential for chromosome alignment. JCB. 1997;139:1373–1382. doi: 10.1083/jcb.139.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yen TJ, Li G, Schaar BT, et al. CENP-E is a putative kinetochore motor that accumulates just before mitosis. Nature. 1992;359:536–539. doi: 10.1038/359536a0. [DOI] [PubMed] [Google Scholar]
- 16.Brown KD, Coulson RM, Yen TJ, et al. Cyclin-like accumulation and loss of the putative kinetochore motor CENP-E results from coupling continuous synthesis with specific degradation at the end of mitosis. J Cell Biol. 1994;125:1303–1312. doi: 10.1083/jcb.125.6.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Magidson V, Paul R, Yang N, et al. Adaptive changes in the kinetochore architecture facilitate proper spindle assembly. Nat Cell Biol. 2015;17:1134–1144. doi: 10.1038/ncb3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu M, Chang Y, Hu H, et al. LUBAC controls chromosome alignment by targeting CENP-E to attached kinetochores. Nat Commun. 2019;10:273. doi: 10.1038/s41467-018-08043-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ciossani G, Overlack K, Petrovic A, et al. The kinetochore proteins CENP-E and CENP-F directly and specifically interact with distinct BUB mitotic checkpoint Ser/Thr kinases. J Biol Chem. 2018;293:10084–10101. doi: 10.1074/jbc.RA118.003154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan GK, Schaar BT, Yen TJ. Characterization of the kinetochore binding domain of CENP-E reveals interactions with the kinetochore proteins CENP-F and hBUBR1. J Cell Biol. 1998;143:49–63. doi: 10.1083/jcb.143.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grabsch H, Takeno S, Parsons WJ, et al. Overexpression of the mitotic checkpoint genes BUB1, BUBR1, and BUB3 in gastric cancer–association with tumour cell proliferation. J Pathol. 2003;200:16–22. doi: 10.1002/path.1324. [DOI] [PubMed] [Google Scholar]
- 22.Yamamoto Y, Matsuyama H, Chochi Y, et al. Overexpression of BUBR1 is associated with chromosomal instability in bladder cancer. Cancer Genet Cytogenet. 2007;174:42–47. doi: 10.1016/j.cancergencyto.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 23.Kops GJ, Foltz DR, Cleveland DW. Lethality to human cancer cells through massive chromosome loss by inhibition of the mitotic checkpoint. Proc Natl Acad Sci U S A. 2004;101:8699–8704. doi: 10.1073/pnas.0401142101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lampson MA, Renduchitala K, Khodjakov A, et al. Correcting improper chromosome-spindle attachments during cell division. Nat Cell Biol. 2004;6:232–237. doi: 10.1038/ncb1102. [DOI] [PubMed] [Google Scholar]
- 25.Elowe S. Bub1 and BubR1: at the interface between chromosome attachment and the spindle checkpoint. Mol Cell Biol. 2011;31:3085–3093. doi: 10.1128/MCB.05326-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson VL, Scott MI, Holt SV, et al. Bub1 is required for kinetochore localization of BubR1, Cenp-E, Cenp-F and Mad2, and chromosome congression. J Cell Sci. 2004;117:1577–1589. doi: 10.1242/jcs.01006. [DOI] [PubMed] [Google Scholar]
- 27.Elowe S, Hummer S, Uldschmid A, et al. Tension-sensitive Plk1 phosphorylation on BubR1 regulates the stability of kinetochore microtubule interactions. Genes Dev. 2007;21:2205–2219. doi: 10.1101/gad.436007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suijkerbuijk SJ, Vleugel M, Teixeira A, et al. Integration of kinase and phosphatase activities by BUBR1 ensures formation of stable kinetochore-microtubule attachments. Dev Cell. 2012;23:745–755. doi: 10.1016/j.devcel.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 29.Kaisari S, Miniowitz-Shemtov S, Sitry-Shevah D, et al. Role of ubiquitin-protein ligase UBR5 in the disassembly of mitotic checkpoint complexes. Proc Natl Acad Sci U S A. 2022 doi: 10.1073/pnas.2121478119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 31.Lee EK, Diehl JA. SCFs in the new millennium. Oncogene. 2014;33:2011–2018. doi: 10.1038/onc.2013.144. [DOI] [PubMed] [Google Scholar]
- 32.Skaar JR, Pagan JK, Pagano M. Mechanisms and function of substrate recruitment by F-box proteins. Nat Rev Mol Cell Biol. 2013;14:369–381. doi: 10.1038/nrm3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stephenson SEM, Costain G, Blok LER, et al. Germline variants in tumor suppressor FBXW7 lead to impaired ubiquitination and a neurodevelopmental syndrome. Am J Hum Genet. 2022;109:601–617. doi: 10.1016/j.ajhg.2022.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davis RJ, Welcker M, Clurman BE. Tumor suppression by the Fbw7 ubiquitin ligase: mechanisms and opportunities. Cancer Cell. 2014;26:455–464. doi: 10.1016/j.ccell.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grim JE, Gustafson MP, Hirata RK, et al. Isoform- and cell cycle-dependent substrate degradation by the Fbw7 ubiquitin ligase. J Cell Biol. 2008;181:913–920. doi: 10.1083/jcb.200802076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malyukova A, Dohda T, Von Der Lehr N, et al. The tumor suppressor gene hCDC4 is frequently mutated in human T-cell acute lymphoblastic leukemia with functional consequences for Notch signalling. Cancer Res. 2007;67:5611–5616. doi: 10.1158/0008-5472.CAN-06-4381. [DOI] [PubMed] [Google Scholar]
- 37.Maser RS, Choudhury B, Campbell PJ, et al. Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature. 2007;447:966–971. doi: 10.1038/nature05886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’neil J, Grim J, Strack P, et al. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. J Exp Med. 2007;204:1813–1824. doi: 10.1084/jem.20070876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Welcker M, Orian A, Grim JE, et al. A nucleolar isoform of the Fbw7 ubiquitin ligase regulates c-Myc and cell size. Curr Biol. 2004;14:1852–1857. doi: 10.1016/j.cub.2004.09.083. [DOI] [PubMed] [Google Scholar]
- 40.Fan J, Bellon M, Ju M, et al. Clinical significance of FBXW7 loss of function in human cancers. Mol Cancer. 2022;21:87. doi: 10.1186/s12943-022-01548-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yeh CH, Bellon M, Nicot C. FBXW7: a critical tumor suppressor of human cancers. Mol Cancer. 2018;17:115. doi: 10.1186/s12943-018-0857-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rajagopalan H, Jallepalli PV, Rago C, et al. Inactivation of hCDC4 can cause chromosomal instability. Nature. 2004;428:77–81. doi: 10.1038/nature02313. [DOI] [PubMed] [Google Scholar]
- 43.Badarudeen B, Gupta R, Nair SV, et al. The ubiquitin ligase FBXW7 targets the centriolar assembly protein HsSAS-6 for degradation and thereby regulates centriole duplication. J Biol Chem. 2020;295:4428–4437. doi: 10.1074/jbc.AC119.012178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Legal T, Hayward D, Gluszek-Kustusz A, et al. The C-terminal helix of BubR1 is essential for CENP-E-dependent chromosome alignment. J Cell Sci. 2020 doi: 10.1242/jcs.246025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singh P, Pesenti ME, Maffini S, et al. BUB1 and CENP-U, primed by CDK1, are the main PLK1 kinetochore receptors in mitosis. Mol Cell. 2021;81(67–87):e69. doi: 10.1016/j.molcel.2020.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lan H, Tan M, Zhang Q, et al. LSD1 destabilizes FBXW7 and abrogates FBXW7 functions independent of its demethylase activity. Proc Natl Acad Sci U S A. 2019;116:12311–12320. doi: 10.1073/pnas.1902012116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vassilev LT, Tovar C, Chen S, et al. Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc Natl Acad Sci U S A. 2006;103:10660–10665. doi: 10.1073/pnas.0600447103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miniowitz-Shemtov S, Eytan E, Ganoth D, et al. Role of phosphorylation of Cdc20 in p31(comet)-stimulated disassembly of the mitotic checkpoint complex. Proc Natl Acad Sci U S A. 2012;109:8056–8060. doi: 10.1073/pnas.1204081109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Itoh G, Ikeda M, Iemura K, et al. Lateral attachment of kinetochores to microtubules is enriched in prometaphase rosette and facilitates chromosome alignment and bi-orientation establishment. Sci Rep. 2018;8:3888. doi: 10.1038/s41598-018-22164-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rodriguez-Rodriguez JA, Lewis C, Mckinley KL, et al. Distinct roles of RZZ and Bub1-KNL1 in mitotic checkpoint signaling and kinetochore expansion. Curr Biol. 2018;28(3422–3429):e3425. doi: 10.1016/j.cub.2018.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pereira C, Reis RM, Gama JB, et al. Self-Assembly of the RZZ complex into filaments drives kinetochore expansion in the absence of microtubule attachment. Curr Biol. 2018;28(3408–3421):e3408. doi: 10.1016/j.cub.2018.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sacristan C, Ahmad MUD, Keller J, et al. Dynamic kinetochore size regulation promotes microtubule capture and chromosome biorientation in mitosis. Nat Cell Biol. 2018;20:800–810. doi: 10.1038/s41556-018-0130-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu HZ, Wang ZQ, Shan HZ, et al. Overexpression of Fbxo6 inactivates spindle checkpoint by interacting with Mad2 and BubR1. Cell Cycle. 2018;17:2779–2789. doi: 10.1080/15384101.2018.1557488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang Y, Yao Y, Xu HZ, et al. Defects in chromosome congression and mitotic progression in KIF18A-deficient cells are partly mediated through impaired functions of CENP-E. Cell Cycle. 2009;8:2643–2649. doi: 10.4161/cc.8.16.9366. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are available.