Abstract
Acute promyelocytic leukemia (APL) is a hematological malignancy driven by the oncoprotein PML-RARα, which can be treated with arsenic trioxide (As2O3) or/and all-trans retinoic acid. The protein arginine methyltransferase 5 (PRMT5) is involved in tumorigenesis. However, little is known about the biological function and therapeutic potential of PRMT5 in APL. Here, we show that PRMT5 is highly expressed in APL patients. PRMT5 promotes APL by interacting with PML-RARα and suppressing its ubiquitination and degradation. Mechanistically, PRMT5 attenuates the interaction between PML-RARα and its ubiquitin E3 ligase RNF4 by methylating RNF4 at Arg164. Notably, As2O3 treatment triggers the dissociation of PRMT5 from PML nuclear bodies, attenuating RNF4 methylation and promoting RNF4-mediated PML-RARα ubiquitination and degradation. Moreover, knockdown of PRMT5 and pharmacological inhibition of PRMT5 with the specific inhibitor EPZ015666 significantly inhibit APL cells growth. The combination of EPZ015666 with As2O3 shows synergistic effects on As2O3-induced differentiation of bone marrow cells from APL mice, as well as on apoptosis and differentiation of primary APL cells from APL patients. These findings provide mechanistic insight into the function of PRMT5 in APL pathogenesis and demonstrate that inhibition of PRMT5, alone or in combination with As2O3, might be a promising therapeutic strategy against APL.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-022-04358-3.
Keywords: PRMT5, PML-RARα, RNF4, Methylation, Ubiquitination, Acute promyelocytic leukemia
Introduction
Acute promyelocytic leukemia (APL) is a hematological malignancy that is a subtype of acute myeloid leukemia (AML) [1]. APL is mainly driven by an oncoprotein encoded by a fusion gene consisting of the N terminus of promyelocytic leukemia protein (PML) and the C terminus of retinoic acid receptor α (RARα) [2, 3]. PML-RARα functions as a transcriptional repressor of related genes and attenuates the assembly and function of PML nuclear bodies (PML-NBs), which results in deregulation of transcription, differentiation arrest, and enhances self-renewal of leukemia-initiating blast cells [4–6]. All-trans retinoic acid (ATRA) and arsenic trioxide (As2O3) are currently used for the clinical treatment of APL [7, 8]. The combination of ATRA with As2O3 or chemotherapy dramatically improves the prognosis of APL patients [9]. Mechanistically, these combination therapies have produced better APL patient outcomes by promoting PML-RARα degradation and thus inducing the differentiation of APL blasts and decreasing the abundance of leukemia-initiating cells [10, 11]. PML-RARα degradation is a SUMO-triggered and RNF4/ubiquitination-mediated process [12, 13]. However, this therapeutic scheme may cause serious side events such as systemic infection and secondary leukemia [14, 15]. In addition, some APL patients fail to respond to therapy targeting PML-RARα or relapse after a complete remission [16–18].
Recent studies have reported that protein arginine methyltransferases (PRMTs) function in several cancers [19, 20]. PRMTs are a group of proteins with 11 known members that can catalyze the addition of 1 or 2 methyl groups to the guanidine nitrogen atoms of an arginine residue [21, 22]. Among the members of the PRMT family, PRMT5 has been a focus of significant attention in cancer research [23–25]. Overexpression of PRMT5 has been reported in hematologic malignancies, and PRMT5 has been shown to regulate proliferation and self-renewal of chronic myeloid leukemia (CML) stem cells [26]. However, the role of PRMT5 in APL has not been defined.
In this study, we investigated the expression and roles of PRMT5 in APL cell lines and primary APL cells from patients. We found that PRMT5 promotes the growth of APL cells by inhibiting PML-RARα ubiquitination. By methylating RNF4 at R164, PRMT5 inhibits the interaction between PML-RARα and RNF4. Inhibition of PRMT5 activity in combination with As2O3 promotes differentiation of bone marrow (BM) cells from APL mice and primary APL cells from APL patients. These results demonstrate the oncogenic role of PRMT5 in APL pathogenesis, and provide a rationale for a potential therapeutic strategy targeting PRMT5 in APL treatment.
Materials and methods
Animal studies
A mouse model of APL was established by intravenously injecting NOD-SCID mice with NB4 cells [27]. All mice were housed in specific pathogen-free barrier facilities. BM cells were isolated from mice and analyzed by CD11b staining to determine their differentiation rate. Animals were handled following the ‘Principles for the Utilization and Care of Vertebrate Animals’ and the ‘Guide for the Care and Use of Laboratory Animals’. Animal studies were approved by the IACUC of the Center for Experimental Animal Research (China) and Peking University Laboratory Animal Center (IACUC No. LSCZhengX-2-1).
Primary human leukemia samples
Primary APL and normal mononuclear cells were freshly obtained from the bone marrow (BM) tissue of seven APL patients and six healthy donors at the Institute of Hematology and Blood Diseases Hospital of Peking Union Medical College (PUMC). Informed consent was obtained from all participants in accordance with the Declaration of Helsinki. The study was approved by the Ethics Committee of Peking University (IRB00001052-16020).
His-ubiquitin pulldown assay
His-ubiquitin pulldown assays were performed following a method described in a previous study [28]. Briefly, HEK293T cells were transfected with his-ubiquitin and the indicated plasmids. After 48 h, the cells were harvested and lysed in His-pulldown buffer. The lysate was incubated with 60 µL of Ni2+ beads for 4 h and then washed 4 times with wash buffer. The beads were denatured with 2 × SDS loading buffer and ubiquitin was assessed using the indicated antibodies.
Clonogenic survival assay
First, 150–750 cells were seeded in solid agarose medium in 6-well plates in triplicate. After 24 h, cells were cultured in medium containing a different concentration of EPZ015666. After 12 days, the number of clones was counted. The survival fraction was normalized to the number of untreated cells.
Duolink proximity ligation assay (PLA)
To detect the interaction between PRMT5 and PML-RARα/RNF4, we used the Duolink® In Situ PLA® kit (DUO92101, Sigma-Aldrich). The Duolink proximity ligation assay was performed following the manufacturer’s method [29]. Fluorescence images were obtained under a confocal laser scanning microscope (Zeiss LSM 710) using a 63 × oil objective lens.
His-ubiquitin immunoprecipitation assay
HEK293T cells were transfected with his-ubiquitin and the other indicated plasmids. After 48 h, the cells were harvested, lysed in RIPA buffer and sonicated. The supernatants were incubated with the indicated antibodies at 4 °C for 4 h or overnight, followed by incubation with 30 μL protein G beads for 3 h. Finally, the beads were washed three times using RIPA buffer and denatured with 2 × SDS loading buffer. Ubiquitin was assessed using anti-his or anti-ubiquitin antibodies.
In vitro methylation assay
Recombinant GST-fusion protein RNF4 WT and RNF4 R164K were expressed in E. coli BL21 and purified using a glutathione-Sepharose 4B column (GE Healthcare). Myc-tagged PRMT5 was immunoprecipitated from transfected HEK293T cells. GST-RNF4 WT/R164K fusion protein and immunoprecipitated PRMT5 protein were incubated in 30 μL of a solution consisting of 5 mM MgCl2, 20 mM HEPES, 1 mM ethylenediaminetetraacetic acid, 1 mM DTT, 10% glycerol, and 100 μM S-(5′-adenosyl)-l-methionine iodide (pH 7.9) (Sigma Aldrich) at 37 °C for 1 h. The reaction was stopped with SDS sample buffer and the sample was subjected to SDS-PAGE, followed by immunoblotting with anti-SDMe and anti-RNF4 antibodies.
In vitro ubiquitination assay
HEK293T cells were co-transfected with Flag-PML-RARα and His-SUMO2. After 48 h, cells were harvested and precipitated using Flag beads. Flag-PML-RARα was eluted using Flag peptide. SUMOylated Flag-PML-RARα was precipitated using Ni2+ beads. His-SUMO2-Flag-PML-RARα, GST-RNF4 WT/R164K, 10 μM UBE1 (R&D Systems), 1 μM UbcH5a (R&D Systems) and 1 mM ubiquitin were incubated together in reaction buffer (50 mM Tris–HCl, 5 mM MgCl2, 2 mM ATP, and 1 mM DTT; pH 7.5) at 37 °C for 4 h. RNF4 ubiquitination was detected by western blotting with anti-ubiquitin antibodies.
Statistical analysis
Statistical analysis was performed using Student’s t test. All the results are presented as mean ± SEM (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
Additional information regarding the materials and methods is included in the Supplementary Information.
Results
PRMT5 is highly expressed in APL cells and interacts with PML-RARα
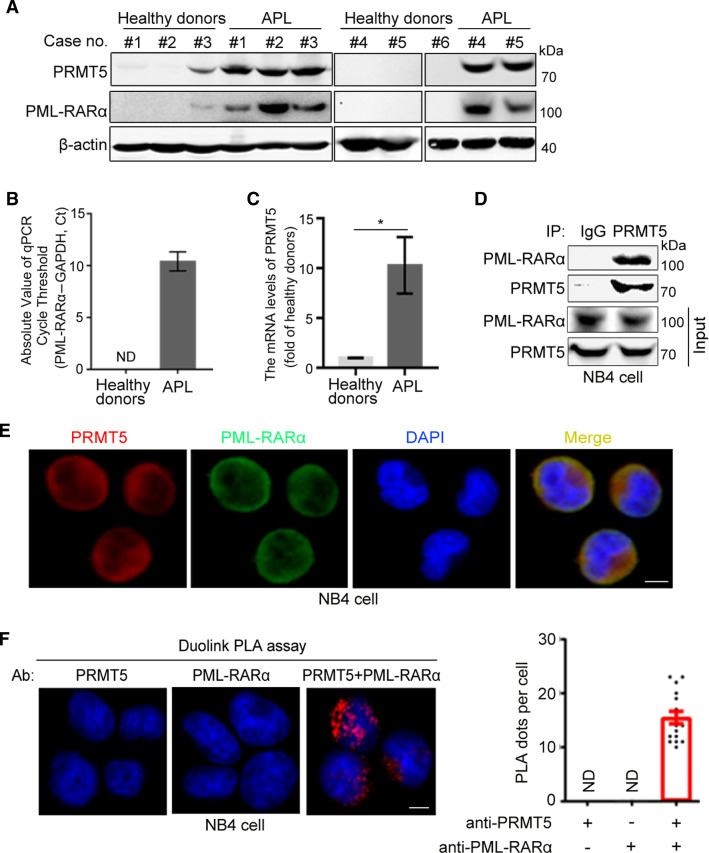
Previous studies have shown that the oncogenic fusion protein PML-RARα plays an important role in the process of APL development [30, 31]. The arginine methyltransferase PRMT5 has attracted significant attention for its clinical effect on tumorigenesis and its exceptional levels of accumulation in colon, breast and blood cancers [32]. To examine whether PRMT5 is involved in the pathogenesis of APL, we analyzed the protein levels of PRMT5 in APL patient samples. We found that the abundance of PRMT5 in BM from APL patients was significantly higher than that of normal cells (Fig. 1A), and PRMT5 expression was positively correlated with the level of PML-RARα (Fig. 1A). Furthermore, BM from APL patients expressed higher mRNA levels of PML-RARα and PRMT5 in comparison with BM from healthy donors (Fig. 1B, C). To illustrate the regulatory mechanism of PRMT5 in APL, we determined whether PRMT5 could interact with PML-RARα. Co-immunoprecipitation (co-IP) assays showed that Flag-tagged PML-RARα was associated with endogenous PRMT5 (Supplementary Fig. S1A). Moreover, the endogenous interaction between PRMT5 and PML-RARα was confirmed in human leukemia NB4 cells, an APL cell line in which PML-RARα is expressed endogenously (Fig. 1D). The immunofluorescence experiments also showed that endogenous PML-RARα and PRMT5 co-localized in NB4 cells (Fig. 1E), and this observation was verified by a PLA, which allowed us to detect in situ protein interaction with high specificity and sensitivity (Fig. 1F). Taken together, these findings demonstrate that PRMT5 is elevated in APL and is a novel partner of PML-RARα.
Fig. 1.
PRMT5 is highly expressed in APL cells and interacts with PML-RARα. A Immunoblotting to assess PML-RARα and PRMT5 expression in BM from healthy people (#1–#6) and APL patients (#1–#5). The qRT-PCR analyses of PML-RARα (B) and PRMT5 (C) mRNA expression in BM from healthy donors and APL patients. D The endogenous interaction between PML-RARα and PRMT5 was confirmed by co-IP experiments in NB4 cells. E Representative images of NB4 cells immunostained for PRMT5 (red), PML-RARα (green), and DAPI (blue). Scale bar, 10 μm. F Duolink PLA assays were performed to confirm the in situ interaction between PRMT5 and PML-RARα (red dots) in NB4 cells. Scale bar, 10 μm. Quantification results for the PLA dots indicating PRMT5–PML-RARα interaction are shown as mean ± SEM
PRMT5 promotes the stability of PML-RARα
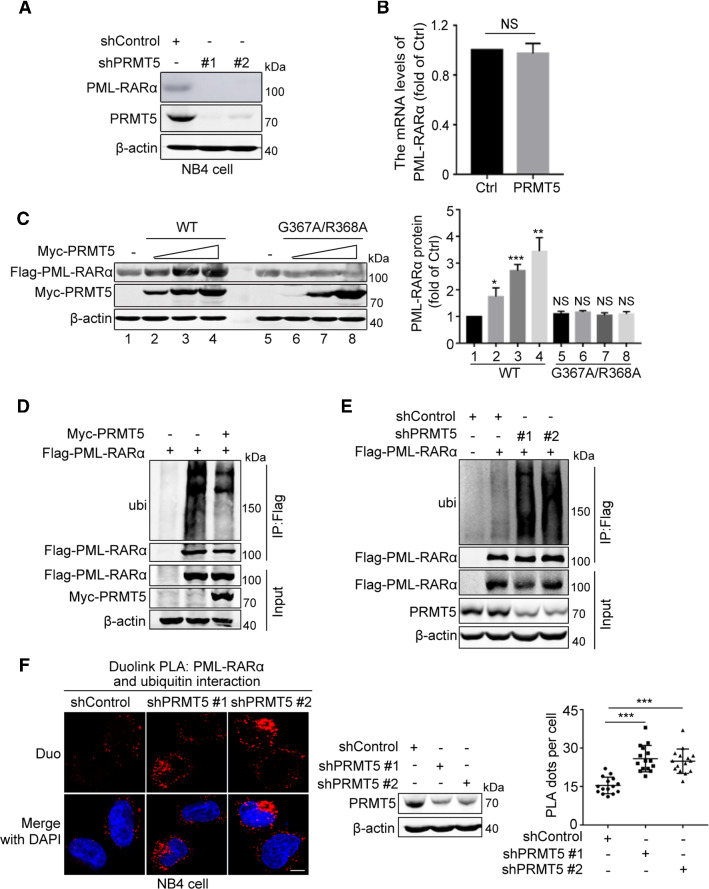
Targeting PML-RARα for degradation is a conventional strategy for APL treatment [27, 33]. Because PRMT5 is associated with PML-RARα, we next explored the effect of PRMT5 on the expression level of PML-RARα. We found that knockdown of PRMT5 in HEK293T cells decreased the protein abundance of PML-RARα (Supplementary Fig. S1B). Moreover, we assessed the effect of PRMT5 on the protein stability of endogenous PML-RARα in NB4 cells. As expected, knockdown of PRMT5 decreased the abundance of PML-RARα in NB4 cells (Fig. 2A). However, overexpression of PRMT5 had no influence on the mRNA abundance of PML-RARα (Fig. 2B), suggesting that PRMT5 regulates PML-RARα at the protein level rather than promoting its transcription.
Fig. 2.
PRMT5 increases the stability of PML-RARα by reducing its ubiquitination. A The effect of depleted PRMT5 on PML-RARα stability was analyzed by immunoblotting in PRMT5 knockdown and control NB4 cells. B The qRT-PCR analyses of PML-RARα mRNA expression in HEK293T cell transfected with Myc-PRMT5 and Flag-PML-RARα. Statistical analysis was performed using Student’s t-test. Data are shown as mean ± SEM (n = 3). NS, not significant. C The effect of PRMT5 enzymatic activity on PML-RARα expression was analyzed by immunoblotting in HEK293T cells transfected with Flag-PML-RARα and increasing amounts of PRMT5 wild-type or the enzymatically inactive PRMT5 G367A/R368A mutant (left). Quantification results are shown as the mean ± SEM (n = 3) (right). D HEK293T cells transfected with different plasmids were treated with 10 μM MG132 for 8 h and then subjected to IP ubiquitination analyses using the indicated antibodies. E The shControl- and shPRMT5-HEK293T cells transfected with Flag-empty or Flag-PML-RARα were treated with 10 μM MG132 for 8 h and then subjected to IP ubiquitination analyses using the indicated antibodies. F The in situ binding between PML-RARα and ubiquitin in NB4 cells was detected by Duolink PLA immunofluorescence using anti-PML-RARα and anti-ubiquitin antibodies. Scale bar, 10 μm. Quantification results for the PLA dots indicating PML-RARα-ubiquitin interaction are shown as mean ± SEM
Because PRMT5 is a methyltransferase, we assessed whether the methyltransferase activity of PRMT5 plays a role in its effect on the stability of PML-RARα. We thus constructed an enzymatically inactive mutant form of PRMT5, PRMT5 G367A/R368A, in which the enzymatic sites Gly367 and Arg368 were mutated to Ala. In contrast to wild-type PRMT5, which promoted PML-RARα stability, the PRMT5 G367A/R368A mutant did not affect the stability of PML-RARα (Fig. 2C), suggesting that the methyltransferase activity of PRMT5 is critical for its effect on PML-RARα. Collectively, these results indicate that PRMT5 enhances the stability of PML-RARα.
PRMT5 inhibits ubiquitination of PML-RARα by interacting with RNF4
Previous studies revealed that degradation of PML-RARα induced by As2O3 treatment is achieved through a SUMO-dependent, ubiquitin-mediated process [12, 13]. SUMOylation of PML-RARα triggers RNF4-mediated ubiquitination and degradation; therefore, the stability of PML-RARα is regulated by SUMOylation and ubiquitination. We examined whether PRMT5 affected the stability of PML-RARα by modulating its SUMOylation and ubiquitination in cells treated with or without As2O3. The results of His-pull down assays showed that PRMT5 inhibited ubiquitination of PML-RARα (Supplementary Fig. S2A), but it had no effect on its SUMOylation (Supplementary Fig. S2B). Moreover, the inhibitory effect of PRMT5 on PML-RARα ubiquitination was confirmed by immunoprecipitation assays under denatured condition (Fig. 2D). In contrast, knockdown of PRMT5 significantly increased PML-RARα ubiquitination (Fig. 2E). Consistently, Duolink PLA assays revealed that knockdown of PRMT5 promoted ubiquitination of PML-RARα in NB4 cells (Fig. 2F). Next, using His-pull down assays, we showed that PRMT5 inhibited K48-linked ubiquitination, but it had no effect on K63-linked ubiquitination of PML-RARα (Supplementary Fig. S2C).
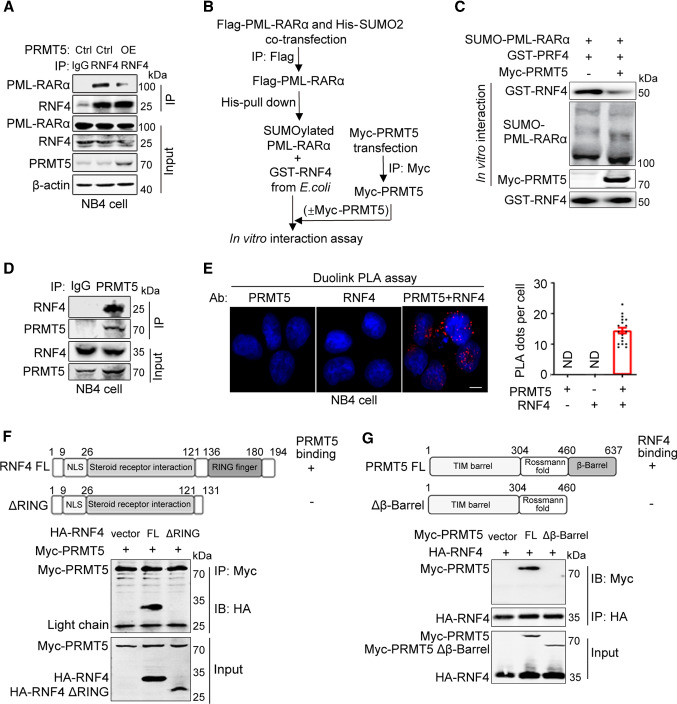
To clarify how PRMT5 regulates RNF4-mediated PML-RARα ubiquitination, we generated PRMT5-overexpressed NB4 cells and investigated whether PRMT5 functions by affecting the interaction between RNF4 and its substrate PML-RARα. The results of co-IP experiments showed that overexpression of PRMT5 attenuated the endogenous PML-RARα–RNF4 interaction (Fig. 3A). Moreover, because RNF4 is a poly-SUMO-specific ubiquitin E3 ligase, which binds to the poly-SUMO chain of PML-RARα to induce PML-RARα degradation [12], we assessed whether PRMT5 inhibited the interaction between RNF4 and SUMO-modified PML-RARα by performing an in vitro interaction assay (Fig. 3B). As expected, PRMT5 inhibited the interaction between RNF4 and SUMOylated PML-RARα (Fig. 3C). Based on these findings, we hypothesized that PRMT5 likely interacted with RNF4. Indeed, the results of co-IP and PLA assays showed that PRMT5 interacted with both exogenous and endogenous RNF4 (Fig. 3D, E; Supplementary Fig. S2D). Furthermore, identification of the critical domain responsible for the binding of PRMT5 to RNF4 revealed that the RING finger domain of RNF4 and the β-Barrel domain of PRMT5 were necessary for PRMT5–RNF4 binding (Fig. 3F, G). Collectively, our findings demonstrate that PRMT5 represses PML-RARα ubiquitination by blocking the interaction between RNF4 and SUMOylated PML-RARα.
Fig. 3.
PRMT5 represses RNF4–PML-RARα binding by interacting with RNF4. A The effect of PRMT5 on the interaction between endogenous PML-RARα and RNF4 was examined by co-IP experiments in NB4 cells with stably overexpressed PRMT5. B Diagram of the in vitro interaction assay used for C. HEK293T cells were transfected with Flag-PML-RARα and His-SUMO2. SUMOylated PML-RARα was immunoprecipitated with Flag beads, eluted by Flag peptide, and then purified by His-pulldown. Myc-PRMT5 was purified from transfected HEK293T cells by immunoprecipitation using anti-Myc antibodies. SUMOylated PML-RARα was incubated with purified GST-RNF4 in the absence and presence of Myc-PRMT5. The interaction between RNF4 and SUMOylated PML-RARα was assessed by immunoblotting. C The effect of PRMT5 on the RNF4-SUMOylated PML-RARα interaction was examined by in vitro interaction assay. D The endogenous interaction between PRMT5 and RNF4 was confirmed in NB4 cells by co-IP experiments. E Duolink PLA assays were performed to confirm the in situ interaction between PRMT5 and RNF4 (red dots) in NB4 cells. Scale bar, 10 μm. Quantification results for the PLA dots indicating PRMT5-RNF4 interaction are shown as mean ± SEM. F Schematic representation of full-length RNF4 and the RNF4 ΔRING mutation construct. The interactions between Myc-PRMT5 and HA-RNF4 full-length or ΔRING were detected by Co-IP assays. G Schematic representation of full-length PRMT5 and the PRMT5 Δβ-Barrel mutation construct. The interactions between HA-RNF4 and Myc-PRMT5 full-length or Δβ-Barrel were detected by Co-IP assays
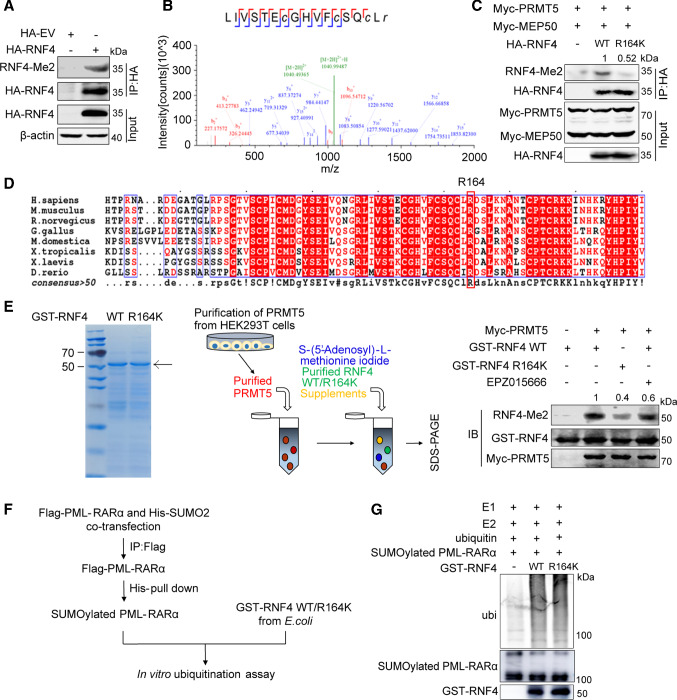
RNF4 is methylated at R164 by PRMT5
PRMT5 is a type II arginine methyltransferase that adds two symmetric methyl groups to the arginine residue of its substrates [34]. Since PRMT5 is associated with RNF4, we speculated that RNF4 is a substrate of PRMT5. To test this hypothesis, we first examined whether RNF4 is methylated with an antibody against symmetric dimethylated arginine. Indeed, RNF4 showed a methylation signal at the expected position (Fig. 4A). Next, using mass spectrometry, we identified arginine 164 as the methylation site of RNF4 (Fig. 4B). Mutation of Arg164 to Lys obviously reduced the level of PRMT5/MEP50-mediated RNF4 methylation (Fig. 4C), and RNF4 Arg164 is highly conserved in various vertebrate species (Fig. 4D). Moreover, in vitro methylation assay also indicated that PRMT5 catalyzed RNF4 methylation (Fig. 4E). As Arg164 is located in the RING finger domain of RNF4, to further investigate the effect of RNF4 R164 methylation on its regulatory function, we examined whether RNF4 R164 methylation affects PML-RARα ubiquitination by in vitro ubiquitination assays using SUMOylated PML-RARα proteins (prepared as shown in Fig. 4F). The RNF4 WT and R164K mutant were capable of ubiquitinating PML-RARα (Fig. 4G), suggesting that RNF4 methylation did not attenuate its ubiquitin E3 ligase activity. These results demonstrate that the R164 site of RNF4 is methylated by PRMT5, and this methylation has no effect on its E3 enzyme activity.
Fig. 4.
RNF4 is methylated at R164 by PRMT5. A HA-tagged RNF4 was immunoprecipitated with the HA antibody from transfected HEK293T cells and methylation of RNF4 was detected using anti-SDMe-Arginine antibodies. B Mass spectrometric (MS) analysis of RNF4 methylation sites. HEK293T cells were transfected with HA-RNF4. HA-RNF4 was immunopurified with anti-HA affinity beads followed by MS. C Immunoblotting of the methylation of RNF4 R164. HEK293T cells were transfected with the indicated plasmids for 48 h. D Amino acid sequences from the indicated species were aligned using the ESPript 3.0 website. Conserved sequences are highlighted. E Assay for in vitro methylation of RNF4 by PRMT5 using purified proteins. Left panel, Coomassie blue staining of GST-fused RNF4 WT or the RNF4 R164K mutant purified from E. coli. Middle panel, schematic representation of the in vitro methylation assay. Right panel, purified Myc-PRMT5 proteins were incubated with purified GST-RNF4 WT or GST-RNF4 R164K mutant proteins. The reaction mixtures were separated by SDS-PAGE and immunoblotted using the indicated antibodies. F Diagram of the in vitro ubiquitination assay used for G. G HEK293T cells were transfected with Flag-PML-RARα and His-SUMO2. Flag-tagged PML-RARα was purified from transfected HEK293T cells by immunoprecipitation using anti-Flag antibodies, after which His-SUMO2-Flag-PML-RARα was purified by Ni2+ pulldown and incubated with purified GST-RNF4 WT or GST-RNF4 R164K mutant proteins at 37 °C for 4 h. The effect of R164 methylation on its E3 enzyme activity was analyzed by measuring the ubiquitination level of PML-RARα
RNF4 methylation inhibits PML-RARα ubiquitination and degradation by blocking RNF4–PML-RARα interaction
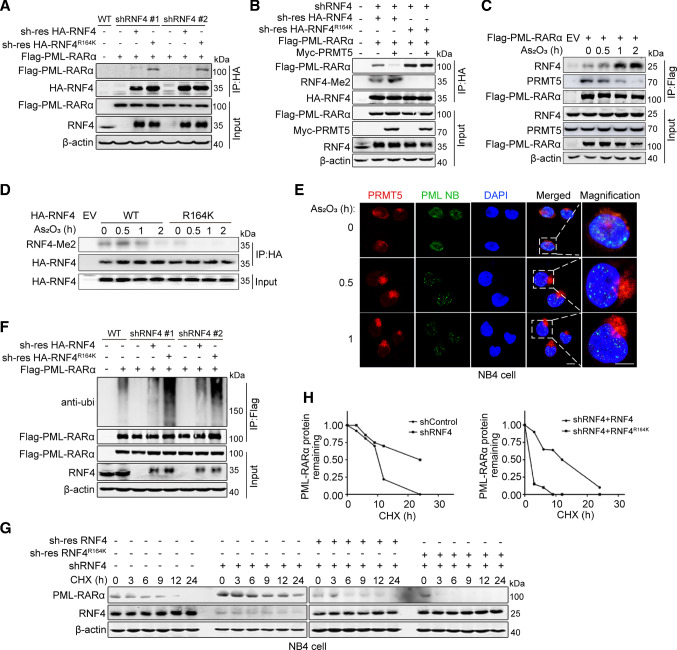
The results described above suggest that RNF4 is a substrate for PRMT5, but methylation of RNF R164 does not affect its ubiquitin E3 ligase activity. To explore whether methylation of RNF4 influences binding between RNF4 and PML-RARα, we re-introduced shRNA-resistant RNF4 WT or RNF R164K in RNF4-knockdown 293 T cells, and detected the association of PML-RARα with RNF4 WT or the RNF R164K mutant. The RNF4 R164K mutant showed a stronger interaction with PML-RARα in comparison with RNF4 WT (Fig. 5A). We also examined the influence of PRMT5-mediated RNF4 methylation on the interaction between RNF4 and PML-RARα. As shown in Fig. 5B, the RNF4 R164K mutant showed a stronger interaction with PML-RARα in comparison with RNF4 WT; upon overexpression of PRMT5, RNF4 WT disassociated from PML-RARα, while the RNF4 R164K mutant was still bound to PML-RARα. In addition, Co-IP assays showed that treatment with As2O3 disturbed the interaction of PRMT5 with PML-RARα (Fig. 5C) and RNF4 (Supplementary Fig. S3A), while promoting RNF4–PML-RARα interaction (Fig. 5C and Supplementary Fig. S3B). Next, we performed in vitro pulldown assays, which showed that PRMT5 directly interacted with RNF4 but not with PML-RARα (Supplementary Fig. S3C and S3D). These results indicate that PRMT5 directly interacts with RNF4 and inhibits the interaction between RNF4 and PML-RARα by competitively binding to RNF4. Consistently, we found that the abundance of WT RNF4 methylated by PRMT5 gradually decreased following As2O3 treatment (Fig. 5D). It has been reported that As2O3 triggers the recruitment of RNF4 to PML nuclear bodies and promotes PML or PML-RARα ubiquitination and proteasome-dependent degradation [13]. Therefore, we determined whether As2O3 could inhibit the interaction between PRMT5 and RNF4 by affecting the localization of PRMT5 within PML nuclear bodies. As we predicted, IF assays showed that the localization of PRMT5 in PML nuclear bodies was reduced significantly at 30 min after As2O3 treatment (Fig. 5E). These results indicate that As2O3 treatment results in the dissociation of PRMT5 from PML nuclear bodies, and thus PRMT5 no longer interacts with and methylates RNF4. Given our findings that PRMT5 modulates PML-RARα turnover (Fig. 2) and the RNF4 R164K mutant has a stronger interaction with PML-RARα (Fig. 5A), we determined whether PRMT5 suppresses PML-RARα ubiquitination and promotes PML-RARα abundance by regulating RNF4 methylation. We separately measured the effects of RNF4 WT and the R164K mutant on PML-RARα ubiquitination, as well as the half-life of the PML-RARα protein. As shown in Fig. 5F and Supplementary Fig. S4, the RNF4 R164K mutant led to increased PML-RARα ubiquitination in the absence and presence of As2O3. Consistently, the RNF4 R164K mutant promoted PML-RARα degradation (Fig. 5G, H). Taken together, our results indicate that PRMT5 stabilizes PML-RARα by catalyzing RNF4 methylation which inhibits the interaction between RNF4 and PML-RARα.
Fig. 5.
RNF4 methylation inhibits PML-RARα ubiquitination and degradation by blocking RNF4–PML-RARα interaction. A The interaction between PML-RARα and RNF4 WT/R164K was examined by co-IP assay in shRNF4 HEK293T cells transfected with shRNA-resistant RNF4 WT or RNF4 R164K mutant plasmids. B The effect of RNF4 methylation on RNF4–PML-RARα interaction was examined by co-IP assay in shRNF4 HEK293T cells transfected with shRNA-resistant RNF4 WT or R164K with/without Myc-PRMT5 plasmids. C The effects of As2O3 on the PRMT5–PML-RARα and RNF4–PML-RARα interactions were analyzed by immunoblotting. HEK293T cells were transfected with the indicated plasmids and treated with As2O3 (1 μM) for 0 h, 0.5 h, 1 h or 2 h. D Immunoblotting to measure the level of RNF4 methylation. HEK293T cells transfected with the indicated plasmids were treated with As2O3 (1 μM) for 0 h, 0.5 h, 1 h or 2 h. E Representative images of NB4 cells immunostained for PML (green), PRMT5 (red) and DAPI (blue). NB4 cells were treated with As2O3 (2 μM) for 0, 0.5 or 1.0 h to examine changes in the co-localization of PRMT5 and PML nuclear bodies. Scale bar, 10 μm. F Denatured IP assays of PML-RARα ubiquitination in shRNF4 HEK293T cells transfected with shRNA-resistant RNF4 WT or RNF4 R164K mutant plasmids. G Immunoblotting to measure the abundance of PML-RARα. The shRNF4 NB4 cells were infected with lentiviruses stably expressing shRNA-resistant RNF4 WT or the RNF4 R164K mutant, after which they were treated with CHX (10 μg/mL). H Semi-quantification of PML-RARα levels, with β-actin used as a loading control. The relative PML-RARα level at time 0 was set as 1
Inhibition of PRMT5 together with As2O3 synergistically restrains the proliferation and differentiation of APL cells
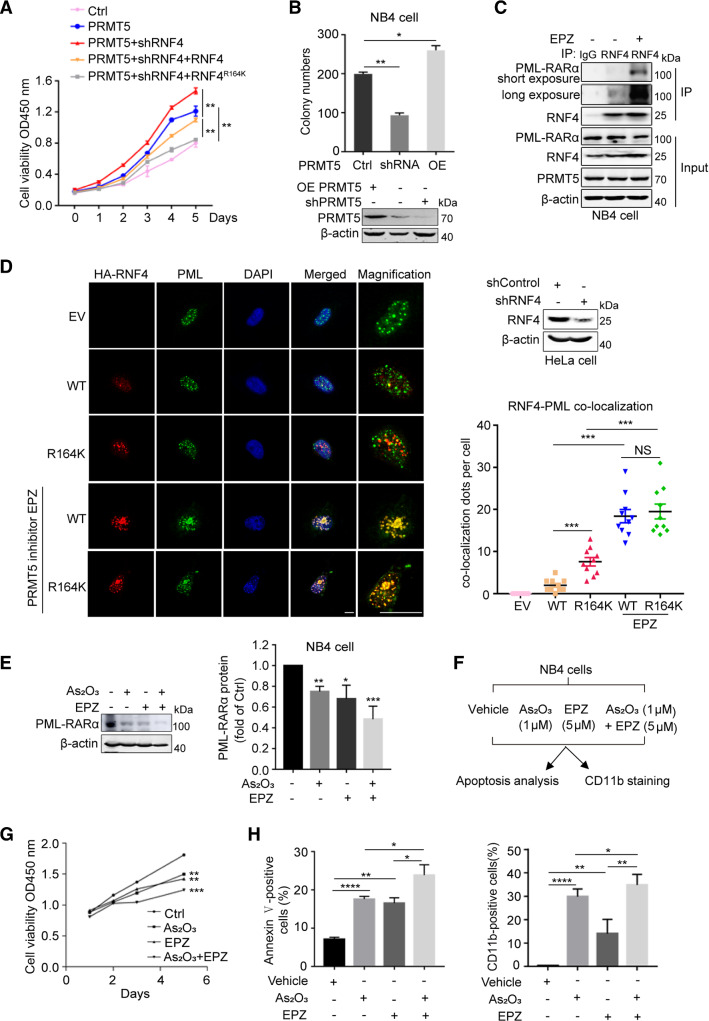
The results described above suggest that PRMT5 might regulate the progression of APL by modulating PML-RARα stability. We generated a NB4 cell line that stably expressed PRMT5 and RNF4 shRNA, and re-expressed sh-res RNF4 WT or RNF4 R164K, after which we performed cell viability assays (Supplementary Fig. S5A). The results revealed that PRMT5 overexpression enhanced NB4 cell viability, and knockdown of RNF4 in NB4 cells with overexpressed PRMT5 clearly increased cell viability. In comparison with re-expression of RNF4 WT, re-introduction of RNF4 R164K significantly inhibited the improvement in cell viability induced by RNF4 knockdown in PRMT5-overexpressed NB4 cells (Fig. 6A). We also examined the influence of PRMT5 on NB4 cell proliferation using colony formation assays. The number of NB4 clones was reduced by depletion of PRMT5, but it was increased by PRMT5 overexpression (Fig. 6B). As PRMT5 stabilized PML-RARα by methylating RNF4, we assumed that chemically inhibition of PRMT5 might affect RNF4–PML-RARα interaction. Indeed, the results of co-IP assays in NB4 cells treated with or without PRMT5 inhibitor EPZ015666 showed that inhibition of PRMT5 promoted the interaction between RNF4 and PML-RARα (Fig. 6C). Furthermore, we performed nuclear cytoplasmic protein separation and IF assays to investigate the effects of PRMT5 inhibition on the subcellular localization of RNF4 WT and the RNF4 R164K mutant. The results of nuclear and cytoplasmic protein separation assays showed that chemical inhibition of PRMT5 did not affect the nuclear localization of RNF4 WT or R164K, and their nucleoplasmic levels remained unchanged (Supplementary Fig. S5B). The results of IF assays showed that in comparison with RNF4 WT, more of the RNF4 R164K mutant aggregated in PML nuclear bodies. In contrast, in cells treated with PRMT5 inhibitor EPZ015666, both RNF4 WT and the RNF4 R164K mutant showed increased co-localization with PML nuclear bodies (Fig. 6D). These data indicate that PRMT5 inhibitor EPZ015666 promotes RNF4–PML-RARα interaction.
Fig. 6.
Inhibition of PRMT5 enhances PML-RARα degradation and the therapeutic efficacy of As2O3 in NB4 cells. A sh-res RNF4 WT or RNF4 R164K was re-expressed in NB4 cells that stably expressed PRMT5 and RNF4 shRNA, and then cell viability was measured with the CCK8 kit. B Colony formation of NB4 cells with stable PRMT5 knockdown or PRMT5 overexpression (OE PRMT5) cultured in methylcellulose, wild-type NB4 cell was used as a control. The numbers of cell clones were counted. The PRMT5 protein level was confirmed by immunoblotting. C The interaction between endogenous RNF4 and PML-RARα was confirmed by co-IP in NB4 cells treated with or without PRMT5 inhibitor EPZ015666 (EPZ). D shRNF4 HeLa cells transfected with sh-res HA-RNF4 WT or sh-res HA-RNF4 R164K were treated with or without PRMT5 inhibitor EPZ, and subjected to immunofluorescence assay using antibodies against HA (red) and PML (green). Nuclei were stained with DAPI (blue). Immunofluorescence images and the number of co-localization dots per cell are shown. Scale bar, 20 μm. E Effects of EPZ (5 μM, 60 h) or/and As2O3 (2 μM, 2 h) on endogenous PML-RARα protein levels in NB4 cells were detected by western blotting. Quantification results are shown as the mean ± SEM (n = 3). F Experimental design for evaluating the growth, differentiation and apoptosis rate of NB4 cells exposed to the indicated treatment. G Cell viability analyses of NB4 cells cultured in 96-well plates with 5 μM EPZ, or 1 μM As2O3, or 5 μM EPZ in combination with 1 μM As2O3. Cell viability was measured with the CCK8 kit. H The proportions of apoptotic cells (left) and differentiated cells (right) were determined by flow cytometry. The results of A, B, D, E, G and H are shown as the mean ± SEM (n = 3). Statistical analysis was performed using Student’s t test, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. NS, not significant
Next, we determined whether the combination of the PRMT5 inhibitor EPZ015666 with As2O3 had synergistic effects. As expected, in comparison with the effect of As2O3 or EPZ015666 alone, the combination of As2O3 with EPZ015666 more efficiently destabilized PML-RARα in NB4 cells (Fig. 6E). Consistently, we found that EPZ015666 inhibited the growth of NB4 cells, and the combination of EPZ015666 with As2O3 further suppressed the growth of APL cells (Fig. 6F, G). Moreover, EPZ015666 potently increased the apoptosis and differentiation percentages of NB4 cells induced by As2O3 treatment (Fig. 6H).
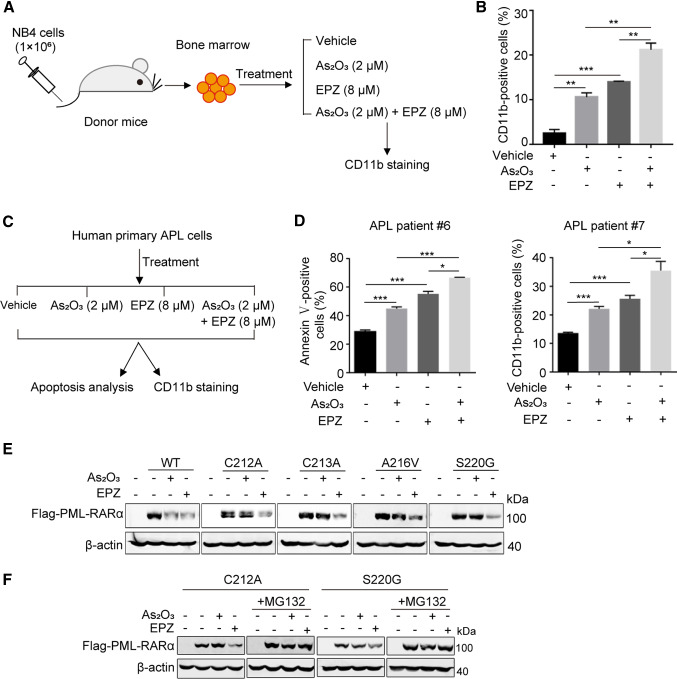
Moreover, we explored the biological significance of PRMT5 inhibition using an APL mouse model. BM cells were isolated from APL mice and treated with As2O3 or/and EPZ015666, after which the differentiation rate was examined (Fig. 7A). Consistent with the observations in NB4 cells, the combination of EPZ015666 with As2O3 showed synergistic effects on the differentiation of BM cells from APL mice (Fig. 7B). We also isolated primary APL cells from bone marrow samples collected from APL patients, after which the cells were treated with EPZ015666, As2O3, or EPZ015666 together with As2O3, and rates of cell apoptosis and differentiation were examined (Fig. 7C). The results further confirmed the synergistic effects of EPZ015666 and As2O3 (Fig. 7D). Together, these results suggest that PRMT5 plays a critical role in APL pathogenesis, and the combination of PRMT5 inhibitor EPZ015666 with As2O3 enhances the anti-APL efficacy of As2O3.
Fig. 7.
The combination of PRMT5 inhibitor EPZ015666 with As2O3 impedes proliferation and differentiation of APL cells. A Experimental design for studying differentiation of BM from APL mice in vitro with the indicated treatment. B The proportion of differentiated cells was determined by flow cytometry. C The strategy for studying apoptosis and differentiation of human primary APL cells from APL patients with the indicated treatment. D The proportions of apoptotic cells (left) and differentiated cells (right) in primary human APL cells (APL patients #6 and #7) were determined by flow cytometry. The results of B and D are shown as the mean ± SEM (n = 3). Statistical analysis was performed using Student’s t test, *p < 0.05, **p < 0.01, ***p < 0.001. E EPZ induced degradation of PML-RARα in cells that were resistant to As2O3. HeLa cells transfected with the indicated plasmids were treated with As2O3 (1 μM, 16 h) or EPZ (5 μM, 60 h) and the protein abundance of PML-RARα was detected by western blotting. F Determination of the PML-RARα protein degradation pathway by EPZ. HeLa cells transfected with the indicated plasmids were treated with As2O3 (1 μM, 16 h) or EPZ (10 μM, 20 h), and 10 μM MG132 was added to culture for an additional 12 h, after which, the abundance of PML-RARα protein was determined by western blotting
Because drug resistance due to PML-RARα mutations is an intractable problem in APL therapy, we investigated the effects of PRMT5 inhibitor EPZ015666 on clinically reported As2O3-resistant mutants including C212A, C213A, A216V, and S220G [35]. EPZ015666 treatment reduced the abundance of PML-RARα in cells harboring As2O3-resistant PML-RARα mutations (Fig. 7E). In addition, we found that the proteasome inhibitor MG132 reduced EPZ015666-induced PML-RARα degradation of As2O3-resistant mutants C212A and S220G, suggesting that EPZ015666 promotes PML-RARα degradation by the proteasome degradation pathway (Fig. 7F). These data indicate that inhibition of PRMT5 promotes the degradation of PML-RARα, and this mechanism has the potential to be exploited to treat As2O3-resistant APL patients.
Discussion
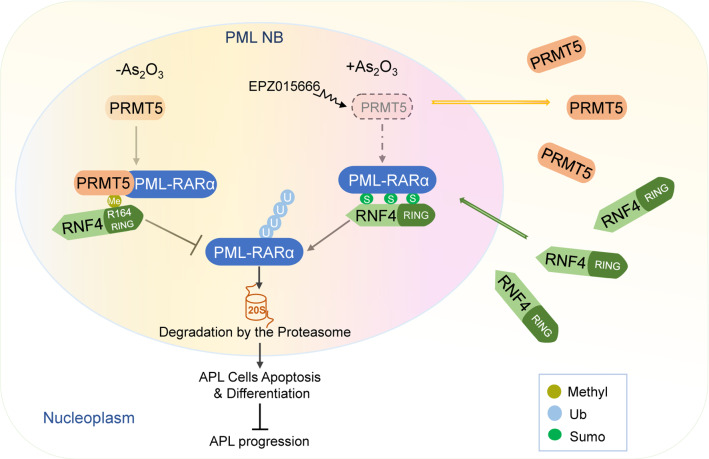
In this study, we demonstrated the regulatory role of arginine methyltransferase PRMT5 in PML-RARα degradation and APL development. We found that PRMT5 promotes PML-RARα stability by methylating RNF4 at R164, which blocks the interaction of RNF4 with PML-RARα and subsequently inhibits ubiquitination of PML-RARα and enhances the stability of PML-RARα, thus inhibiting apoptosis and differentiation of APL cells. Following As2O3 treatment, PRMT5 dissociates from PML nuclear bodies, while RNF4 shuttles into PML nuclear bodies, which promotes the interaction of RNF4 with PML-RARα and accelerates PML-RARα degradation. Furthermore, inhibition of PRMT5 by administration of its inhibitor EPZ015666 eradicates APL cells and enhances the sensitivity of APL cells to As2O3 by accelerating PML-RARα degradation (Fig. 8).
Fig. 8.
A working model for regulation of PML-RARα stability and APL pathogenesis by PRMT5. PRMT5 associates with and stabilizes PML-RARα to promote APL pathogenesis by methylating RNF4 at R164. PRMT5 inhibitor EPZ015666 in combination with As2O3 increases APL cells apoptosis and differentiation by disrupting PRMT5/RNF4 interaction and enhancing RNF4/PML-RARα interaction, thus suppressing APL progression
Previous studies demonstrated that PML-RARα degradation is mediated by SUMOylation and ubiquitination [12, 13]. SUMOylation is critically important for enhanced PML/PML-RARα degradation during the early drug treatment stage. The S214L mutation of PML-RARα can disrupt the organization of PML nuclear bodies and changes in dynamics, producing resistance to both As2O3 and ATRA [36, 37]. PCGF2, a Polycomb group protein, can form the PCGF2–UBE2I complex with UBE2I to impede PML-RARα degradation. However, As2O3 facilitates disruption of the PCGF2–UBE2I complex, and UBE2I interacts with PML-RARα, which induces PML-RARα SUMOylation and subsequent degradation [38]. Interestingly, here we found that PRMT5 inhibited ubiquitination, rather than SUMOylation, of PML-RARα (Fig. 2D, E; Supplementary Fig. S3A and S3B), indicating that PRMT5 regulates PML-RARα abundance by influencing the ubiquitination process. Notably, PRMT5 methyltransferase activity is an essential regulator of PML-RARα stability. PRMT5 and PRMT9 are type II PRMTs that catalyze mono- and symmetric di-methylation and function similarly in several biological processes [39]. We explored whether PRMT9 affected the stability of PML-RARα, as was shown for PRMT5. No influence of PRMT9 on PML-RARα stability was observed in cells transfected with different amounts of PRMT9 (Supplementary Fig. S6A and S6B), suggesting that the stability of PML-RARα is affected by PRMT5 specifically. PRMT5-catalyzed methylation of RNF4 blocked its interaction with PML-RARα, thus stabilizing PML-RARα. In addition, we found that in comparison with RNF4 WT, the RNF4 R164K mutant promoted degradation of PML but not RARα in HeLa cells (non-APL cells) without PML-RARα (Supplementary Fig. S7). This finding is consistent with a previous study reporting that PML and PML-RARα are degraded through a RNF4/ubiquitin-mediated pathway [13]. In addition, we showed that PRMT5 did not methylate PML-RARα (Supplementary Fig. S8). This is the first study showing that the abundance of PML-RARα is regulated by methyltransferase PRMT5 through methylation of RNF4. Identification of the PRMT5–RNF4–PML-RARα axis by this study revealed a novel mechanism underlying APL pathogenesis. However, further studies are necessary to investigate whether methylation of RNF4 causes conformational changes and subsequently inhibits PML-RARα binding.
PRMT5 has been reported to function in several hematological malignancies, such as acute lymphoblastic leukemia (ALL) [40], acute myeloid leukemia (AML) [41, 42], chronic myelogenous leukemia (CML) and multiple myeloma (MM) [26]. PRMT5 regulates the binding of the splicing regulator SRSF1 to mRNAs and proteins by methylating SRSF1, which promotes the survival of AML cells and provides potential biomarkers for the treatment response to PRMT5 inhibitors [42]. PRMT5-mediated H4R3sme2 inhibits cell differentiation in pediatric ALL [40]. The regulatory mechanisms underlying these effects of PRMT5 on hematological malignancies generally invovle regulation of transcription, either by catalyzing symmetric di-methylation of histone protein to regulate gene transcription or by methylating transcription factors to alter their transcriptional activities [43]. In this study, we demonstrated that PRMT5 promotes APL progression by regulating the protein level of oncogene PML-RARα. Ours and previous studies indicate that PRMT5 regulates blood cancers through distinct mechanisms.
Our study demonstrates the critical role of PRMT5 in APL pathogenesis and suggests that inhibition of PRMT5 might be a potential therapeutic option against APL. Indeed, we found that EPZ015666, a recently identified selective and orally bioavailable PRMT5 inhibitor [44], inhibited APL cell proliferation and increased APL cell differentiation. Notably, the combination of EPZ015666 with As2O3 enhanced the anti-APL efficacy of As2O3 in BM from APL mice, as well as in primary APL cells from APL patients.
Maimaitiyiming et al. recently reported that hyperthermia destabilized PML/RARα protein in clinically identified As2O3-resistant mutants via the SIAH2 E3 ligase, which is a mechanism distinct from that of arsenic therapy. Moreover, hyperthermia and As2O3 treatment were shown to act synergistically in destabilizing PML-RARα [35]. Here, we found that EPZ015666 induced degradation of As2O3-resistant PML-RARα mutants by the proteasome degradation pathway. However, the clinical therapeutic efficacy of the combination of EPZ015666 with As2O3 for refractory or relapsed APL patients remains to be explored.
In conclusion, our study reveals the critical role of the PRMT5/RNF4/PML-RARα axis in APL cells and proposes a potential approach involving the combination of As2O3 with a PRMT5 inhibitor for the treatment of patients with APL.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant nos. 82130081 and 81730080), Natural Science Foundation of Beijing Municipality (5212008). We thank the National Center for Protein Sciences and the Core Facilities of Life Sciences at Peking University, particularly Guilan Li, Dong Liu, Yinghua Guo, Liying Du, Hongxia Lv, Siying Qin, and Xiaochen Li for technical help.
Author contributions:
YY, XH and XZ designed this study. XH, YY, DZ, YZ, and MW performed the experiments. XH and YY analyzed the data and wrote the manuscript. KL and HHZ provided NOD-SCID mice and APL patient samples. XZ supervised this study and wrote the manuscript.
Availability of data and materials
The data and materials are available by contacting the corresponding author upon reasonable request.
Declarations
Conflict of interest
The authors declare no potential conflicts of interest.
Ethical approval
Animal studies were performed with ethics approval authorized by the IACUC of the Center for Experimental Animal Research (China) and Peking University Laboratory Animal Center (IACUC No. LSCZhengX-2-1). The study of APL patients was approved by the Ethics Committee of Peking University (IRB00001052-16020) and was conducted in accordance with the principles of the Declaration of Helsinki.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xinping Huang and Yongfeng Yang contributed equally to this work.
References
- 1.de The H, Chen Z. Acute promyelocytic leukaemia: novel insights into the mechanisms of cure. Nat Rev Cancer. 2010;10(11):775–783. doi: 10.1038/nrc2943. [DOI] [PubMed] [Google Scholar]
- 2.Burnett AK, Russell NH, Hills RK, Bowen D, Kell J, Knapper S, et al. Arsenic trioxide and all-trans retinoic acid treatment for acute promyelocytic leukaemia in all risk groups (AML17): results of a randomised, controlled, phase 3 trial. Lancet Oncol. 2015;16(13):1295–1305. doi: 10.1016/S1470-2045(15)00193-X. [DOI] [PubMed] [Google Scholar]
- 3.Strocchio L, Gurnari C, Santoro N, Putti MC, Micalizzi C, Zecca M, et al. Arsenic trioxide and all-trans retinoic acid treatment for childhood acute promyelocytic leukaemia. Br J Haematol. 2019;185(2):360–363. doi: 10.1111/bjh.15507. [DOI] [PubMed] [Google Scholar]
- 4.Lallemand-Breitenbach V, de The H. PML nuclear bodies. Cold Spring Harb Perspect Biol. 2010;2(5):a000661. doi: 10.1101/cshperspect.a000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kwok C, Zeisig BB, Dong S, So CW. Forced homo-oligomerization of RARalpha leads to transformation of primary hematopoietic cells. Cancer Cell. 2006;9(2):95–108. doi: 10.1016/j.ccr.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 6.Martens JH, Brinkman AB, Simmer F, Francoijs KJ, Nebbioso A, Ferrara F, et al. PML-RARalpha/RXR alters the epigenetic landscape in acute promyelocytic leukemia. Cancer Cell. 2010;17(2):173–185. doi: 10.1016/j.ccr.2009.12.042. [DOI] [PubMed] [Google Scholar]
- 7.Kayser S, Schlenk RF, Platzbecker U. Management of patients with acute promyelocytic leukemia. Leukemia. 2018;32(6):1277–1294. doi: 10.1038/s41375-018-0139-4. [DOI] [PubMed] [Google Scholar]
- 8.Coombs CC, Tavakkoli M, Tallman MS. Acute promyelocytic leukemia: where did we start, where are we now, and the future. Blood Cancer J. 2015;5:e304. doi: 10.1038/bcj.2015.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lo-Coco F, Avvisati G, Vignetti M, Thiede C, Orlando SM, Iacobelli S, et al. Retinoic acid and arsenic trioxide for acute promyelocytic leukemia. N Engl J Med. 2013;369(2):111–121. doi: 10.1056/NEJMoa1300874. [DOI] [PubMed] [Google Scholar]
- 10.Lallemand-Breitenbach V, Zhu J, Chen Z, de The H. Curing APL through PML/RARA degradation by As2O3. Trends Mol Med. 2012;18(1):36–42. doi: 10.1016/j.molmed.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 11.Dos Santos GA, Kats L, Pandolfi PP. Synergy against PML-RARa: targeting transcription, proteolysis, differentiation, and self-renewal in acute promyelocytic leukemia. J Exp Med. 2013;210(13):2793–2802. doi: 10.1084/jem.20131121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tatham MH, Geoffroy MC, Shen L, Plechanovova A, Hattersley N, Jaffray EG, et al. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat Cell Biol. 2008;10(5):538–546. doi: 10.1038/ncb1716. [DOI] [PubMed] [Google Scholar]
- 13.Lallemand-Breitenbach V, Jeanne M, Benhenda S, Nasr R, Lei M, Peres L, et al. Arsenic degrades PML or PML-RARalpha through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat Cell Biol. 2008;10(5):547–555. doi: 10.1038/ncb1717. [DOI] [PubMed] [Google Scholar]
- 14.Lu Y, Yan JS, Xia L, Qin K, Yin QQ, Xu HT, et al. 2-Bromopalmitate targets retinoic acid receptor alpha and overcomes all-trans retinoic acid resistance of acute promyelocytic leukemia. Haematologica. 2019;104(1):102–112. doi: 10.3324/haematol.2018.191916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaut D, Sasine J, Schiller G. Secondary clonal hematologic neoplasia following successful therapy for acute promyelocytic leukemia (APL): a report of two cases and review of the literature. Leuk Res Rep. 2018;9:65–71. doi: 10.1016/j.lrr.2018.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lehmann-Che J, Bally C, de The H. Resistance to therapy in acute promyelocytic leukemia. N Engl J Med. 2014;371(12):1170–1172. doi: 10.1056/NEJMc1409040. [DOI] [PubMed] [Google Scholar]
- 17.Zhang X, Pan J. Resistance to arsenic trioxide and retinoic acid therapy in acute promyelocytic leukemia. Ann Hematol. 2017;96(4):707–708. doi: 10.1007/s00277-017-2923-z. [DOI] [PubMed] [Google Scholar]
- 18.Zhu HH, Qin YZ, Huang XJ. Resistance to arsenic therapy in acute promyelocytic leukemia. N Engl J Med. 2014;370(19):1864–1866. doi: 10.1056/NEJMc1316382. [DOI] [PubMed] [Google Scholar]
- 19.Smith E, Zhou W, Shindiapina P, Sif S, Li C, Baiocchi RA. Recent advances in targeting protein arginine methyltransferase enzymes in cancer therapy. Expert Opin Ther Targets. 2018;22(6):527–545. doi: 10.1080/14728222.2018.1474203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jarrold J, Davies CC. PRMTs and arginine methylation: cancer’s best-kept secret? Trends Mol Med. 2019;25(11):993–1009. doi: 10.1016/j.molmed.2019.05.007. [DOI] [PubMed] [Google Scholar]
- 21.Pal S, Sif S. Interplay between chromatin remodelers and protein arginine methyltransferases. J Cell Physiol. 2007;213(2):306–315. doi: 10.1002/jcp.21180. [DOI] [PubMed] [Google Scholar]
- 22.Blanc RS, Richard S. Arginine methylation: the coming of age. Mol Cell. 2017;65(1):8–24. doi: 10.1016/j.molcel.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 23.Yan F, Alinari L, Lustberg ME, Martin LK, Cordero-Nieves HM, Banasavadi-Siddegowda Y, et al. Genetic validation of the protein arginine methyltransferase PRMT5 as a candidate therapeutic target in glioblastoma. Cancer Res. 2014;74(6):1752–1765. doi: 10.1158/0008-5472.CAN-13-0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu D, Gur M, Zhou Z, Gamper A, Hung MC, Fujita N, et al. Interplay between arginine methylation and ubiquitylation regulates KLF4-mediated genome stability and carcinogenesis. Nat Commun. 2015;6:8419. doi: 10.1038/ncomms9419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanda M, Shimizu D, Fujii T, Tanaka H, Shibata M, Iwata N, et al. Protein arginine methyltransferase 5 is associated with malignant phenotype and peritoneal metastasis in gastric cancer. Int J Oncol. 2016;49(3):1195–1202. doi: 10.3892/ijo.2016.3584. [DOI] [PubMed] [Google Scholar]
- 26.Jin Y, Zhou J, Xu F, Jin B, Cui L, Wang Y, et al. Targeting methyltransferase PRMT5 eliminates leukemia stem cells in chronic myelogenous leukemia. J Clin Investig. 2016;126(10):3961–3980. doi: 10.1172/JCI85239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li K, Wang F, Cao WB, Lv XX, Hua F, Cui B, et al. TRIB3 Promotes APL progression through stabilization of the oncoprotein PML-RARalpha and inhibition of p53-mediated senescence. Cancer Cell. 2017;31(5):697–710 e7. doi: 10.1016/j.ccell.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 28.Li T, Guan J, Huang Z, Hu X, Zheng X. RNF168-mediated H2A neddylation antagonizes ubiquitylation of H2A and regulates DNA damage repair. J Cell Sci. 2014;127(Pt 10):2238–2248. doi: 10.1242/jcs.138891. [DOI] [PubMed] [Google Scholar]
- 29.Zhu D, Xu R, Huang X, Tang Z, Tian Y, Zhang J, et al. Deubiquitinating enzyme OTUB1 promotes cancer cell immunosuppression via preventing ER-associated degradation of immune checkpoint protein PD-L1. Cell Death Differ. 2021;28(6):1773–1789. doi: 10.1038/s41418-020-00700-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de The H, Pandolfi PP, Chen Z. Acute promyelocytic leukemia: a paradigm for oncoprotein-targeted cure. Cancer Cell. 2017;32(5):552–560. doi: 10.1016/j.ccell.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 31.Cicconi L, Fenaux P, Kantarjian H, Tallman M, Sanz MA, Lo-Coco F. Molecular remission as a therapeutic objective in acute promyelocytic leukemia. Leukemia. 2018;32(8):1671–1678. doi: 10.1038/s41375-018-0219-5. [DOI] [PubMed] [Google Scholar]
- 32.Yang Y, Bedford MT. Protein arginine methyltransferases and cancer. Nat Rev Cancer. 2013;13(1):37–50. doi: 10.1038/nrc3409. [DOI] [PubMed] [Google Scholar]
- 33.Nasr R, Lallemand-Breitenbach V, Zhu J, Guillemin MC, de The H. Therapy-induced PML/RARA proteolysis and acute promyelocytic leukemia cure. Clin Cancer Res. 2009;15(20):6321–6326. doi: 10.1158/1078-0432.CCR-09-0209. [DOI] [PubMed] [Google Scholar]
- 34.Karkhanis V, Hu YJ, Baiocchi RA, Imbalzano AN, Sif S. Versatility of PRMT5-induced methylation in growth control and development. Trends Biochem Sci. 2011;36(12):633–641. doi: 10.1016/j.tibs.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maimaitiyiming Y, Wang QQ, Yang C, Ogra Y, Lou YJ, Smith CA, et al. Hyperthermia selectively destabilizes oncogenic fusion proteins. Blood Cancer Discov. 2021;2(4):388–401. doi: 10.1158/2643-3230.BCD-20-0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao S, Shi P, Zhong Q, Shao S, Huang Y, Sun Y, et al. Identification of a point mutation PML(S214L)-RARalpha that alters PML body organization, dynamics and SUMOylation. Biochem Biophys Res Commun. 2019;511(3):518–523. doi: 10.1016/j.bbrc.2019.02.101. [DOI] [PubMed] [Google Scholar]
- 37.Bai DM, Zheng XF. PML-RARA mutations confer varying arsenic trioxide resistance. Protein Cell. 2017;8(4):296–301. doi: 10.1007/s13238-016-0356-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jo S, Lee YL, Kim S, Lee H, Chung H. PCGF2 negatively regulates arsenic trioxide-induced PML-RARA protein degradation via UBE2I inhibition in NB4 cells. Biochim Biophys Acta. 2016;1863(7 Pt A):1499–1509. doi: 10.1016/j.bbamcr.2016.03.019. [DOI] [PubMed] [Google Scholar]
- 39.Li ASM, Li F, Eram MS, Bolotokova A, Dela Sena CC, Vedadi M. Chemical probes for protein arginine methyltransferases. Methods. 2020;175:30–43. doi: 10.1016/j.ymeth.2019.11.017. [DOI] [PubMed] [Google Scholar]
- 40.Mei M, Zhang R, Zhou ZW, Ying Z, Wang J, Zhang H, et al. PRMT5-mediated H4R3sme2 confers cell differentiation in pediatric B-cell precursor acute lymphoblastic leukemia. Clin Cancer Res. 2019;25(8):2633–2643. doi: 10.1158/1078-0432.CCR-18-2342. [DOI] [PubMed] [Google Scholar]
- 41.Tarighat SS, Santhanam R, Frankhouser D, Radomska HS, Lai H, Anghelina M, et al. The dual epigenetic role of PRMT5 in acute myeloid leukemia: gene activation and repression via histone arginine methylation. Leukemia. 2016;30(4):789–799. doi: 10.1038/leu.2015.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Radzisheuskaya A, Shliaha PV, Grinev V, Lorenzini E, Kovalchuk S, Shlyueva D, et al. PRMT5 methylome profiling uncovers a direct link to splicing regulation in acute myeloid leukemia. Nat Struct Mol Biol. 2019;26(11):999–1012. doi: 10.1038/s41594-019-0313-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu F, Rui L. PRMT5 in gene regulation and hematologic malignancies. Genes Dis. 2019;6(3):247–257. doi: 10.1016/j.gendis.2019.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chan-Penebre E, Kuplast KG, Majer CR, Boriack-Sjodin PA, Wigle TJ, Johnston LD, et al. A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models. Nat Chem Biol. 2015;11(6):432–437. doi: 10.1038/nchembio.1810. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data and materials are available by contacting the corresponding author upon reasonable request.