Abstract
Little is known about the impact of metabolic stimuli on brain tissue at a molecular level. The ketone body beta-hydroxybutyrate (BHB) can be a signaling molecule regulating gene transcription. Thus, we assessed lysine beta-hydroxybutyrylation (K-bhb) levels in proteins extracted from the cerebral cortex of mice undergoing a ketogenic metabolic challenge (48 h fasting). We found that fasting enhanced K-bhb in a variety of proteins including histone H3. ChIP-seq experiments showed that K9 beta-hydroxybutyrylation of H3 (H3K9-bhb) was significantly enriched by fasting on more than 8000 DNA loci. Transcriptomic analysis showed that H3K9-bhb on enhancers and promoters correlated with active gene expression. One of the most enriched functional annotations both at the epigenetic and transcriptional level was “circadian rhythms''. Indeed, we found that the diurnal oscillation of specific transcripts was modulated by fasting at distinct zeitgeber times both in the cortex and suprachiasmatic nucleus. Moreover, specific changes in locomotor activity daily features were observed during re-feeding after 48-h fasting. Thus, our results suggest that fasting remarkably impinges on the cerebral cortex transcriptional and epigenetic landscape, and BHB acts as a powerful epigenetic molecule in the brain through direct and specific histone marks remodeling in neural tissue cells.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-022-04673-9.
Keywords: Fasting, Cerebral cortex, Beta-hydroxybutyrylation, Transcriptome, Epigenome
Introduction
Nutrition is a key regulator of whole-body physiology and health. Nutrients impinge on tissue functions through modulation of metabolism, which results in adaptive changes in cellular biochemical/molecular processes, and ultimately tissue homeostasis. Notably, besides the well-documented influence on peripheral organs’ physiology/pathology, mounting evidence shows that the metabolic status can also affect neuronal function and metabolism, and influence brain physiology and ultimately cognitive processes, emotions and behavior [1–4]. Indeed, the high fat/high sugar western diet negatively affects brain health, leading to alterations in cognitive functions, anxiety, depression, and in general a higher incidence of emotional disorders [5, 6]. On the other hand, caloric restriction has been shown to prevent age-related brain damage and to be neuroprotective by influencing free radical metabolism, and the cellular stress response system [7, 8]. In 1920’s very low carbohydrate/high fat ketogenic diets (KD) were introduced in the clinic to treat refractory epilepsy, mimicking the increase in the blood level of ketone bodies [i.e., beta-hydroxybutyrate (BHB)], which were thought to mediate the effect of fasting on seizure events [9]. Notably, epilepsy has high comorbidity with neurodevelopmental disorders such as autism spectrum disorders (ASD), and KD is one of the therapeutic approaches proposed to treat ASD patients [10, 11]. KD seems also to be beneficial in the treatment of neurodegenerative and psychiatric diseases [12–14]. Finally, fasting and aerobic exercise may improve cognitive performance and protect against depression and anxiety-like behavior through the enhancement of hippocampal neurogenesis [15–17]. Thus, in short, diet robustly influences neurological outcomes, yet the molecular and biochemical underpinnings of these effects are poorly understood [18].
KD, fasting or starvation, and prolonged aerobic exercise, all promote the depletion of liver glycogen stores, favoring the decrease of blood glucose concentration. Due to its scarcity, glucose cannot be used to respond to the energy request of organismal tissues. Thus, to maintain metabolic homeostasis, our body switches to an alternative fuel: ketone bodies, produced via the activation of the so-called ketogenic pathway. Mainly in the liver, the acetyl-CoA produced by fatty acid oxidation is converted into three ketone bodies: acetoacetate, acetone, and BHB. The latter is the most abundant circulating ketone body and, under ketogenic conditions, the major energy source for metabolically active tissues, such as the brain [19]. Intriguingly, BHB is not just an energy carrier as it was believed in the past. Emerging evidence suggests that BHB is also a signaling molecule, capable of binding to the HCAR2 and FFAR3 receptors [20–22], and acting as an endogenous epigenetic regulator. Indeed, BHB selectively acts as a class 1 Histone deacetylase inhibitor, increasing histone acetylation and active gene expression [23, 24]. Finally, BHB is the chemical donor for a new epigenetic mark so far investigated in detail only in the liver: lysine beta-hydroxy-butyrylation (K-bhb). Liver K-bhb contributes to active gene expression and regulates genes involved in the hepatic metabolic response to starvation [25].
Despite the high ketone bodies demand of the brain, the impact of BHB on neural tissue epigenome and transcriptome has never been explored. To address this important issue, which could underline and shed light on the adaptation capability of neuronal circuits to nutritional changes, we analyzed in vivo neural tissue responses to a specific metabolic challenge consisting of 48-h fasting. Specifically, we discovered that fasting enhanced K-bhb in a variety of proteins. Then, we explored the genome-wide distribution of K9-bhb on histone H3 (H3K9-bhb) through chromatin immunoprecipitation, followed by sequencing (ChIP-seq) in the occipital cortex (an area corresponding to the visual cortex) of fasted (F) mice, and parallel measurement of the transcriptional response through RNA-seq. Dataset crossing pointed toward H3K9-bhb being an epigenetic mark related to fasting-induced gene expression, therefore, suggesting that fasting-driven increase in BHB could directly affect brain cells through the remodeling of the chromatin landscape and transcriptional responses.
Results
Fasting-driven increase in BHB causes a burst in protein lysine beta-hydroxybutyrylation in the brain
To assess the molecular response of brain tissue to fasting-induced ketosis, we adopted a protocol of prolonged fasting (Fig. 1a) and we assessed the biochemical action of a major metabolite produced upon fasting: BHB. Postnatal day (P) 33 mice were subjected to fasting (F) condition, corresponding to a total absence of food for 48 h (48 h), but ad libitum (AL) access to water. Their body weight and glycemia were monitored before and after fasting. F mice displayed a significant decrease in their body weight and blood glucose concentration after 48 h, while AL control mice did not show any significant difference (Figs. 1 b, c and S1a). Notably, BHB levels were strongly increased not only in F mice plasma and liver (Fig. 1d, S1b), but also in the cerebral cortex (Fig. 1e). Free fatty acids (FFA) were also quantified in plasma, however only a trend to increase was observed in F mice with respect to AL (Fig. S1c). On the contrary, total triglycerides were significantly increased in the liver of F mice (Fig. S1d), in line with previous literature reports [26]. Moreover, fat mass was analyzed by weighting subcutaneous, interscapular brown adipose tissue (BAT), epididymal fat (in male mice), and lean mass by measuring gastrocnemius and soleus muscle. The fat mass was significantly decreased in F mice (Fig. S1e) while the muscle mass was not altered (Fig. S1e), suggesting that 48 h fasting did not induce muscle degradation.
Fig. 1.
Fasting affects blood glucose concentration, locomotor activity and brain BHB. a Experimental timeline. b Weight change (%) before and after 48 h fasting (N = 13 AL, N = 14 F, unpaired 2-tailed t-test, t25 = 19.40 p < 0.0001); c Blood glucose concentration (mg/dL) before and after 48 h fasting (AL N = 9, F N = 10, two-way RM ANOVA time*treatment, interaction F1,17 = 64.19 p < 0.0001, post-hoc Sidak, AL vs F (post FASTING) t34 = 9.203 p < 0.0001); d plasma beta-hydroxybutyrate concentration in AL and F mice (mmol/L) (N = 6 AL, N = 6 F, unpaired 2-tailed t-test, t10 = 11.30 p < 0.0001). e Cortical beta-hydroxybutyrate concentration (pmol/mg) (N = 5 AL, N = 6 F, unpaired 2-tailed t-test, t9 = 5.958 p = 0.0002). f Mean spontaneous locomotor activity throughout the daily cycle before and during fasting (gray square) (N = 14 AL, N = 10 F, two-way RM ANOVA time*treatment, interaction F5,110 = 6.461 p < 0.0001, post-hoc Sidak, AL vs F (FASTING light day2) t132 = 5.612 p < 0.0001). White and black squares below the x axis represent day-time and night-time respectively. g Spontaneous locomotor activity trace throughout the daily cycle before and during fasting (gray square) (N = 14 AL, N = 10 F). One data point every 20 min. Error bars represent SEM
Finally, during the 48 h of food deprivation, F animals displayed a significant increase in spontaneous locomotor activity both during night and day time (Fig. 1f, g), showing that our protocol did not cause lethargy in the subjects and confirming behavioral changes reflecting the search for food [27].
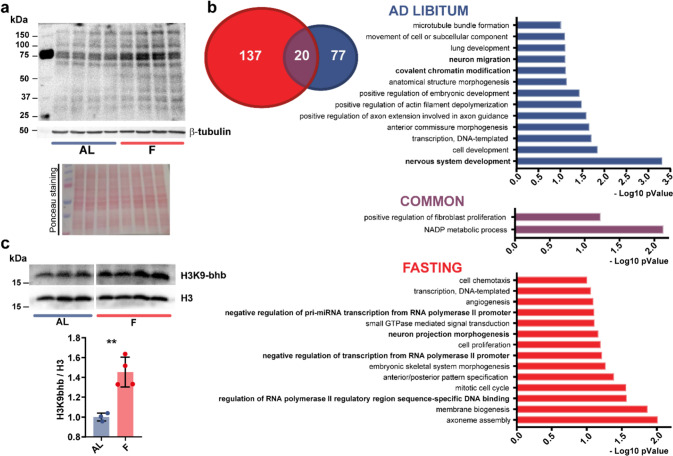
Since BHB has been demonstrated to be the chemical moiety for a new post-translational modification (PTM) on K residues [25, 28], we performed a western-blot analysis of protein extract from the occipital cortex (corresponding to the mouse visual cortex) of F and AL mice using a pan-K-bhb antibody. Strikingly, K-bhb was significantly increased in the occipital cortex of F mice with respect to AL (Fig. 2a), suggesting that the increased cortical BHB after fasting could be exploited, not only to produce energy, but also as a chemical donor for PTMs. As a control, we analysed the pan-K-bhb in the liver of the same mice and confirmed a significant upregulation in F mice with respect to AL (Fig. S1f).
Fig. 2.
Fasting increases protein Lysine beta-hydroxybutyrylation and modifies the beta-hydroxybutyrylation profile of the total proteome in the cerebral cortex. a Western blot analysis of total lysine beta-hydroxybutyrylation in the occipital cortex of F or AL mice using a pan-K-bhb antibody (top panel), a control beta-tubulin antibody (middle panel) and the Ponceau staining (bottom panel). b Left: Venn diagram of K-bhb proteins from the occipital cortex of AL (blue) and F mice (red). In purple the K-bhb proteins found in both conditions. Right: GO analysis (biological processes) of K-bhb proteins found in AL, F and in both groups (N = 3 AL, N = 3 F). c Western blot analysis of histone lysine beta-hydroxybutyrylation in the occipital cortex of F and AL mice using a specific H3K9-bhb antibody (top panel) or a control H3 antibody (middle panel). Histograms representing the normalized ratio of H3K9-bhb/H3 in F condition compared to AL (N = 3 AL, N = 4 F, unpaired 2-tailed t-test, t5 = 4.973 p = 0.0042). Error bars represent SEM
HPLC–MS analysis of K-bhb residues in protein extract from the occipital cortex highlighted the presence of 234 beta-hydroxybutyrilated proteins. In total, 137 proteins were beta-hydroxybutyrilated exclusively in F mice, 77 proteins exclusively in AL mice, and 20 proteins were beta-hydroxybutyrilated in both groups (Fig. 2b). The 20 common beta-hydroxybutyrilated proteins were enriched in annotations such as “NADP metabolic processes” and “positive regulation of fibroblast proliferation”'(Fig. 2b and Dataset S1). On the other hand, the F proteins displaying K-bhb clustered in specific GO annotations mainly related to “regulation of transcription, regulation of transcription-dependent on RNA pol II and morphogenesis” (Fig. 2b and Dataset S2). Also, the GO biological processes analysis of the proteins beta-hydroxybutyrilated in AL indicated an enrichment in “transcription, and chromatin modification” annotations (Fig. 2b, and Dataset S3). Immunoprecipitation (IP) of F mice cerebral cortex protein extract using the pan-K-bhb antibody and western blot confirmed HDAC2 beta-hydroxybutyrylation (Fig. S1g), in line with the MS data.
These findings suggest that BHB may have a more complex function related to transcriptional regulation, not just limited to HDAC inhibition or being a chemical moiety for histone PTMs, but also associated with the modulation of epigenetic (e.g. KDM1B, HDAC2), transcription factors (e.g. E2F4, HOXB3) or other chromatin-related proteins (e.g. SKOR1, CHD5) functions.
BHB has been demonstrated to modify K-bhb on histones with consequent alteration of the liver transcriptional program [25]. Thus, we studied H3K9-bhb in the brain using a specific antibody. H3K9-bhb was significantly more abundant in the cortex of F mice (Figs. 2c and S2a) with respect to the AL group. As expected, H3K9-bhb increased also in the liver of F mice (Fig. S2b). In contrast, the acetylation on K9-14 of histone H3 was not altered in the cortex of F mice (Fig. S2c), suggesting that this epigenetic mark might be less sensitive to the higher levels of BHB associated with prolonged fasting in the cerebral cortex.
Overall, these data demonstrate that beta-hydroxybutyrylation is conspicuously enhanced by fasting in neural tissue.
BHB impacts the chromatin state of the cerebral cortex through direct epigenetic action
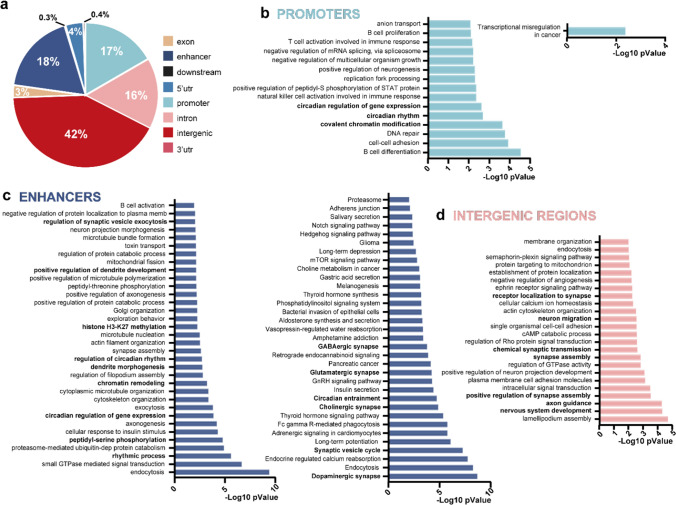
To further explore the role of H3K9-bhb in the CNS, we performed a ChIP-seq experiment and studied the genome-wide distribution of H3K9-bhb in the occipital cortex of mice subjected to 48 h fasting or fed AL. The ChIP-seq analysis revealed a striking increase in H3K9-bhb enriched loci in F with respect to AL. Indeed, the differential analysis between the two experimental conditions detected about 8400 enriched peaks (p < 0.05) in F vs AL (Dataset S4) distributed in intergenic (42%), promoter (17%), and enhancer regions (18%, Fig. 3a). Importantly, only one peak was significantly enriched in AL with respect to F (Dataset S4), suggesting that the increase in BHB observed during fasting is responsible for the dramatic increase in neural tissue H3K9-bhb.
Fig. 3.
Fasting induces robust changes in H3K9 beta-hydroxybutyrylation in the mouse cerebral cortex. a Pie chart of genome-wide distribution of H3K9-bhb enriched loci in F with respect to AL (Cyber-T p < 0.05). b GO Biological processes (left graph) and KEGG pathway (right graph) analysis of the genes in proximity of promoters. c Same as b for enhancer regions. d Same as b for introns. No significant KEGG pathway was found for gene introns
To gain further insight into the specific epigenetic effects of BHB in neural tissue, we crossed our H3K9-bhb ChIP-seq data with the already published H3K9-bhb ChIP-seq data in the liver [25]. The analysis of the data from Xie et al., according to our pipeline (see materials and methods) revealed that the total number of H3K9-bhb peaks in the brain was lower than in the liver (8404 brain loci vs 15,416 liver loci). Peak category distribution demonstrated that H3K9-bhb was principally present in promoter and enhancer regions in the mouse liver after 48 h fasting (Fig. S2d), while enhancer, promoter, and especially intergenic regions were the most prominent category in the brain (Fig. 3a). In the cerebral cortex, the GO “biological processes” analysis of the genes in proximity to promoter and enhancer regions (Datasets S5 and S6) highlighted a specific signature related to circadian rhythms, and pathways involving histone PTM and chromatin remodeling (Fig. 3b, c). Also, synaptic transmission, dendrite morphogenesis, and synapse assembly came up as enriched GO terms associated with the enhancer regions. Brain neurogenesis, brain and dendrite morphogenesis, and axon guidance terms were dominant among the genes associated with the promoter regions (Dataset S5). The KEGG pathway analysis in the enhancers confirmed the “circadian rhythms” and “synaptic transmission” categories (Fig. 3c, Dataset S6). Finally, intergenic regions showed enriched annotations about synapse regulation, transmission and plasticity (Fig. 3d, Dataset S7).
Data crossing revealed that H3K9-bhb was present in 547 promoter regions and 681 enhancers common to brain and liver (Dataset S8). KEGG pathway analysis highlighted several enriched annotations related to metabolism for the common enhancers, among them “Insulin signaling”, “mTOR signaling”, “FoxO signaling” pathway, and “circadian entrainment” and “circadian rhythms” (Dataset S9). “FoxO signaling” pathway was present also in the common promoter list (Dataset S10).
Overall, these data demonstrate that 48 h fasting is able to remodel the neural chromatin landscape, inducing a dynamic and robust H3K9-bhb in the cerebral cortex, preferentially targeting enhancers, promoters, and intergenic regions.
H3K9-bhb is linked to active gene expression in neural tissue
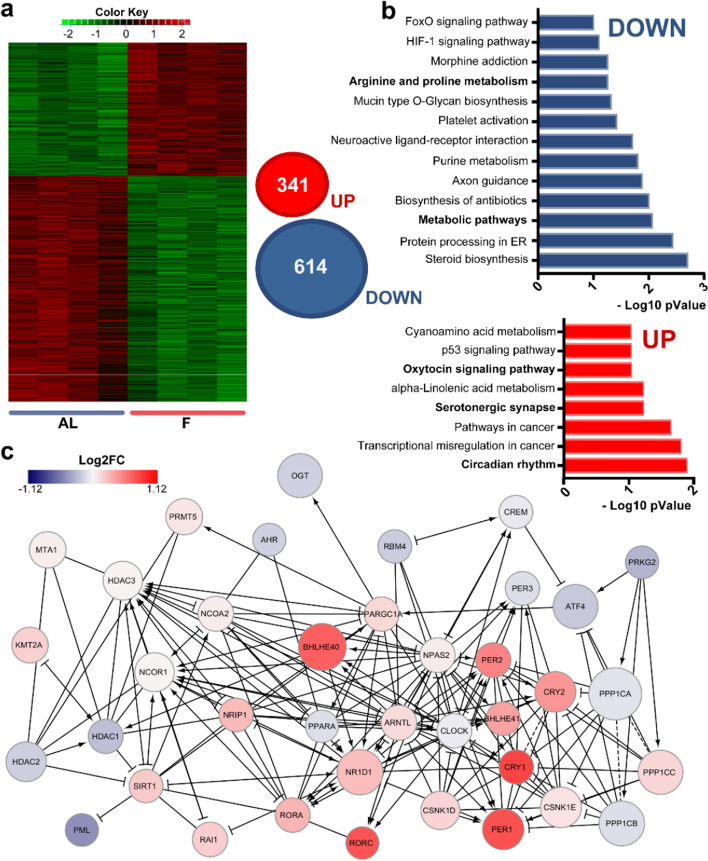
To investigate the impact of H3K9-bhb on gene expression in the visual cortex, we performed an RNA-seq experiment using the same cortical samples used for the ChIP-seq experiment. Fasting dramatically remodeled the transcriptome of the cortex altering the expression of 955 transcripts (Benjamini–Hochberg < 0.01. For the complete gene list see Dataset S11).
In particular, 341 genes were upregulated and 614 genes were downregulated in F mice (Fig. 4a). The effect was highly consistent in the different samples, as shown by the heatmap obtained from every biological replicate (Fig. 4a). The most overrepresented KEGG pathways among the downregulated genes in fasting were “steroid biosynthesis”, “protein processing in endoplasmic reticulum”, “metabolic pathway”, “purine metabolism” etc. (Fig. 4b, Dataset S12a); categories mainly related to metabolism, thus suggesting the existence of a brain adaptive response to conserve energy. KEGG pathways characterizing the UP in fasting transcripts were “serotonergic synapses”, “oxytocin signaling pathway”, and “p53 signaling pathway” (Fig. 4b, Dataset S12b). Notably, in the neocortex the most significant KEGG pathway in the fasting upregulated genes was “circadian rhythms'' (Fig. 4b); this is of interest since core-clock genes have been involved in the control of critical period onset in the mouse visual cortex [29]. Finally, the interaction network of the 955 differentially expressed genes revealed a variety of interactomes including “FoxO” “mTOR” “Insulin” and “PPAR” signaling pathways which are well-known biochemical cascades influenced by fasting; “Glycosaminoglycan metabolism”, “Chondroitin sulfate metabolism and biosynthesis” which are components of the perineuronal nets, important for ocular dominance plasticity and critical period timing [30]; and again “circadian rhythms” (Fig. S3; for a complete list see Dataset S13).
Fig. 4.
Fasting-driven transcriptional changes in the cerebral cortex. a Left: Heat map of RNA-seq analysis performed in the occipital cortex of F and AL mice (AL N = 4, F N = 4, p < 0.05). Right: Venn diagrams representing the differential expressed genes (DEGs) between AL and F in the RNA-seq dataset (n = 4, Benjamini–Hochberg < 0.01). b KEGG pathways analysis of DEGs in the occipital cortex of F mice. Upper panel: KEGG analysis of DEGs downregulated in the F condition. Lower panel: KEGG analysis of DEGs upregulated in the F condition. c Reactome pathway of DEG enriched in “circadian rhythms” annotation. Colors: Log2FC. Circle dimensions: Fasting means
Finally, gene set enrichment analysis on the whole RNA-seq data set using CAMERA [31] confirmed that “circadian rhythms” is significantly enriched in both GO and KEGG pathways (Fig. S4a, b). Intriguingly, the reactome pathway displayed a relationship between “circadian rhythms” terms and a variety of proteins related to epigenetic remodeling (Fig. 4c), suggesting that an interplay between circadian and epigenetic mechanisms could be responsible for the brain adaptive response to prolonged fasting.
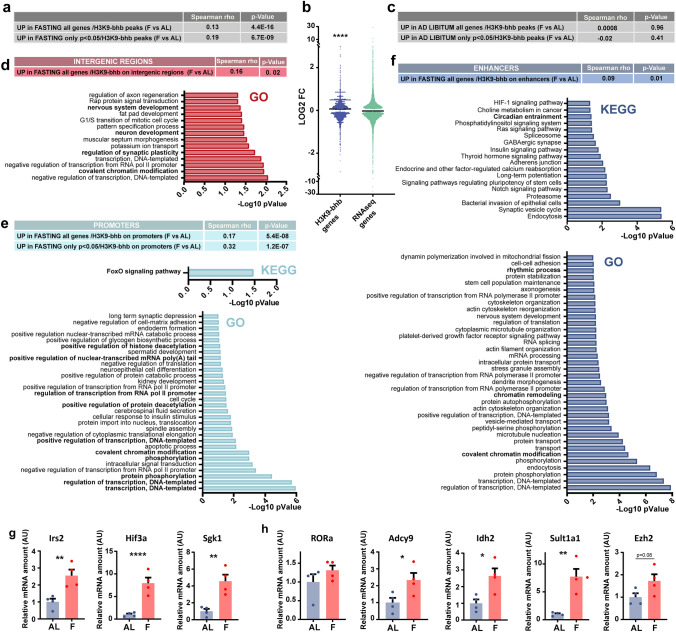
To understand the role played by H3K9-bhb on the cerebral cortex transcriptome, we crossed our RNA-seq and ChIP-seq data. We found a significant positive correlation between differential H3K9-bhb peaks (F vs AL) and the fold change of the genes that are upregulated in F (Fig. 5a, all the genes of the RNA-seq up in fasting, Spearman correlation: ρ = 0.13, p value = 4.4E−16, only genes up in fasting with p value < 0.05, Spearman correlation: ρ = 0.19, p value = 6.7E−09). Furthermore, the expression of genes located in close proximity to loci that are enriched in H3K9-bhb peaks was selectively upregulated by fasting with respect to all expressed genes (Fig. 5b, Mann–Whitney test: U = 19,670,177, p < 0.0001). It is worth noting that there was no significant correlation between the differential H3K9-bhb peaks and the genes upregulated in AL condition (Fig. 5c, all the genes of the RNA-seq up in AL, Spearman correlation: ρ = 0.0008, p value = 0.96. Figure 5c only genes up in AL with p value < 0.05, Spearman correlation: ρ = − 0.02, p value = 0.41). To further validate our RNA-seq and ChIP-seq datasets crossing, we performed qPCR analysis of selected transcripts that are significantly upregulated in F condition and whose genomic loci were associated with a significant enrichment in H3K9-bhb (Fig. 5g, h). Finally, we validated the expression of transcripts involved in the ketogenic response, such as Hmgcs2, the rate limiting enzyme of the ketogenic pathway, and Pdk4, a kinase which inhibits glucose utilization and increases fat metabolism in response to prolonged fasting and starvation (Fig. S4c). The qPCR data confirmed the sequencing experiment results.
Fig. 5.
H3K9-bhb is linked to active gene expression in the cortex of fasted mice. a Correlation between F-induced H3K9-bhb peak enrichment and transcript fold change revealed by RNA-seq (F vs AL, all the genes of the RNA-seq up in fasting, Spearman correlation: ρ = 0.13, p value = 4.4E−16; only genes up in fasting with p value < 0.05, Spearman correlation: ρ = 0.19, p value = 6.7E−09). b Fold change expression induced by fasting is significantly higher for genes in proximity to H3K9-bhb peaks than for all genes. Horizontal bar represents median, Mann–Whitney test: U = 19,670,177, p < 0.0001. c Lack of correlation between AL-induced H3K9-bhb peak enrichment and transcript fold changes revealed by RNA-seq (all the genes of the RNA-seq up in AL, Spearman correlation: ρ = 0.0008, p = 0.96; only genes up in AL with p < 0.05, Spearman correlation: ρ = − 0.02, p = 0.41). d Top: Correlation between H3K9-bhb enriched intergenic regions and transcripts upregulated in fasting (only genes up in fasting with p < 0.05, Spearman correlation: ρ = 0.16, p = 0.02). e Top: Correlation between H3K9-bhb enrichment on promoters and transcript fold-change for genes upregulated in fasting (all the genes of the RNA-seq up in fasting, Spearman correlation: ρ = 0.17, p = 5.4E−08; only genes up in fasting with p value < 0.05, Spearman correlation: ρ = 0.32, p = 1.2E−07). Bottom: KEGG pathways analysis and GO analysis of the H3K9-bhb promoters of genes upregulated in the fasting condition. f Top: Correlation between H3K9-bhb enrichment on enhancers and transcript fold-change for genes upregulated in fasting (all the genes of the RNA-seq up in fasting, Spearman correlation: ρ = 0.09, p = 0.01). Bottom: KEGG pathways analysis and GO analysis of the H3K9-bhb enhancers of genes upregulated in the fasting condition. g, h qPCR analysis of genes upregulated in the occipital cortex of fasted mice and associated with a significant enrichment in H3K9-bhb in (g) promoters (Irs2, Hif3a, Sgk1) and h enhancers (RORa, Adcy9, Idh2, Sult1a1, Ezh2) after 48 h fasting. Irs2 (insulin receptor substrate 2): N = 4 AL, N = 4 F unpaired 2-tailed t-test, t6 = 3.832 p = 0.0086; Hif3a: N = 4 AL, N = 4 F unpaired 2-tailed t-test, t6 = 5.474 p = 0.0016; Sgk1 (serum/glucocorticoid regulated kinase 1): N = 4 AL, N = 4 F unpaired 2-tailed t-test, t6 = 4.275 p = 0.0052; Rora (RAR-related orphan receptor alpha): N = 4 AL, N = 4 F unpaired 2-tailed t-test, t6 = 1.277 p = 0.2489; Adcy9 (adenylate cyclase 9): N = 4 AL, N = 4 F unpaired 2-tailed t-test, t6 = 2.620 p = 0.0396; Idh2 (isocitrate dehydrogenase 2): N = 4 AL, N = 4 F unpaired 2-tailed t-test, t6 = 3.206 p = 0.0185; Sult1a1 (sulfotransferase family 1A): N = 4 AL, N = 4 F unpaired 2-tailed t-test, t6 = 4.754 p = 0.0031; Ezh2 (Enhancer of zeste homolog 2): N = 4 AL, N = 4 F unpaired 2-tailed t-test, t6 = 2.088 p = 0.0818. Error bars represent SEM
Altogether, these results suggest that H3K9-bhb could be related to fasting-induced gene expression in the cerebral cortex of mice.
To gain a better insight into the transcriptional regulatory role of cortical K-bhb after 48 h fasting, we analyzed the list of differential and significantly enriched H3K9-bhb DNA loci (Dataset S4) separately considering promoters, enhancers and intergenic regions, and crossed them with the correspondent transcript levels obtained through the RNA-seq. H3K9-bhb was particularly abundant in intergenic regions (Fig. 3a). Epigenetic marks present in intergenic regions of brain chromatin have been shown to be sensitive to changes in neuronal activity, such as DNA methylation [32]. Fasting-driven changes in H3K9-bhb in intergenic regions positively correlated with the expression of correspondent upregulated transcripts in fasting (Fig. 5d, genes up in fasting with p value < 0.05, Spearman correlation: ρ = 0.16, p value = 0.02). GO biological processes analysis revealed the association with genes involved in regulation of transcription, chromatin modifications, synaptic plasticity and neurodevelopment (Fig. 5d). Promoters and enhancers are key elements for controlling transcript levels, and they were especially influenced by H3K9-bhb in F mice (Fig. 3a). We found a significant correlation between promoter hits and transcripts that are upregulated in fasting (Fig. 5e, all the genes of the RNA-seq up in fasting, Spearman correlation: ρ = 0.17, p value = 5.4E−08, only genes up in fasting with p value < 0.05, Spearman correlation: ρ = 0.32, p value = 1.2E−07). The genes clustered in the “FoxO signaling pathway” (Fig. 5e). Moreover, GO analysis of the term “Biological Process” showed a variety of annotations principally related to transcriptional regulation and PTMs (Dataset S14).
The correlation of RNA-seq genes with H3K9-bhb enhancers gave again a positive value (Fig. 5f, all the genes of the RNA-seq up in fasting, Spearman correlation: ρ = 0.09, p value = 0.01). Interestingly, pathways and terms concerning “circadian rhythms” came up both in the KEGG (Fig. 5f and Dataset S15) analysis and GO annotation (Fig. 5f and Dataset S15). Furthermore, GO biological processes related to “chromatin remodeling and modifications” were significant terms. Again and in keeping with the above analysis, we observed the presence of a signature specifically related to circadian clock and epigenetic mechanisms, strongly suggesting that those could be the molecular underpinning of the cerebral cortex adaptation to a fasting challenge.
Finally, to explore additional potential regulatory mechanisms in fasting-driven transcriptional reprogramming, we performed a transcription factor binding site (TFBS) analysis in the promoter, enhancer, intergenic regions of the genes obtained from the crossing of RNA-seq and ChIP-seq datasets (Complete list in Dataset S16). TFBS analysis in promoters highlighted GATA-2 and GATA-1 binding motif, transcription factors involved in depressive-like behavior, regulation of hippocampal synapse-related genes and spine density [33], and NRF-1 binding site as top hits (Fig. S4d). Intriguingly, NRF-1 is a regulator of oxidative phosphorylation, mitochondrial biogenesis genes and also of GABA-A receptor B1 subunit gene, suggesting a potential coupling between energy metabolism and neurotransmission [34]. In intergenic regions, TFBS analysis revealed sites related to several transcription factors (Fig. S4e). The top hits were E2F and Ezh2 sites, transcription factors whose roles are particularly relevant in the developing brain [35, 36]. Notably, Ezh2 gene expression was significantly upregulated in the cerebral cortex of F mice (Fig. 5h); and E2F4 was also present in the pool of beta-hydroxybutyrilated protein in F mice (Dataset S2), thus we could speculate a link between H3K9-bhb and E2F4 beta-hydroxybutyrylation in transcriptional regulation. Ultimately, Egr1 motif, involved in brain plasticity, learning and memory [37], was highly enriched (Fig. S4f) in enhancers. Notably, the core-clock regulators CLOCK:BMAL, and Bhlhe40 (also enriched in intergenic regions (Fig. S4e)) binding motifs were also significantly present in enhancer regions (Fig. S4f), again pointing toward the cortical clock involvement in neural adaptation mechanisms to a fasting challenge.
Fasting influences diurnal gene expression of core-clock genes and daily rhythms in locomotor activity
Protein synthesis regulation and circadian rhythms are important mechanisms in neural function and plasticity [38, 39]. Since GO categories associated with these mechanisms consistently emerged in our RNA-seq and ChIP-seq data, we selected these two pathways for in depth analysis of fasting regulation. First, we studied the “insulin and mTOR pathways” by assessing two key biochemical steps: serine 235–236 phosphorylation of S6 (Fig. S5a) and Serine 473 phosphorylation of AKT (Fig. S5b). Confirming the hypothesis indicated by the RNA- and ChIP-seq data, both PTMs were significantly reduced in fasting mice, suggesting that protein synthesis could be altered in the cerebral cortex of F mice, in line with a condition of energy saving. Second, we studied “circadian rhythms”, a process representing one of the top hits in the RNA-seq data (Fig. 4b) and in the differential ChIP-seq enhancer analysis (Fig. 5f). We analyzed the expression of core-clock genes in the visual cortex (Fig. 6a) of F and AL mice at four different time points [Zeitgeber time (ZT)] every 6 h throughout the daily cycle (Fig. 6b). The results demonstrated that several clock genes related to transcriptional inhibition—Per1, Cry1, Cry2, and Nr1d1 (Rev-erb alpha) —displayed similar changes in their diurnal profile upon fasting, and were significantly upregulated at ZT4 in F with respect to AL mice, validating the RNA-seq result which was performed at this ZT (Fig. 6c). On the other hand, Bmal1, Clock and Rora, belonging to the transcriptional activation limb of the core-clock machinery, were not significantly altered at any ZT in F mice (Fig. 6d), suggesting a certain degree of specificity of fasting effects on the inhibitory limb of the molecular clock. Prompted by these findings obtained in the visual cortex, we investigated whether circadian clock gene expression was also changed by fasting in the key brain structure for circadian rhythms, i.e., the suprachiasmatic nucleus (SCN) (Fig. S6a,b). We found that, as in the cortex, transcripts belonging to the negative limb of the core-clock were mainly impacted, with a significant increase at ZT22 (Fig. S6c). Moreover, Bmal1 oscillation was strongly dampened (Fig. S6d).
Fig. 6.
H3K9-bhb influences the expression of core-clock genes in the cortex of mice. a Schematic coronal section of the visual cortex showing the site of gene expression analysis. b Experimental timeline. The expression of core-clock genes of F and AL mice was analyzed at 4 different time points (ZT) every 6 h throughout the daily cycle. Mice included in a specific ZT started the fasting time specifically 2 days before (48 h before) at the corresponding ZT. c Quantitative real-time PCR analysis of genes related to transcriptional inhibition of the core- clock machinery (N = 4 per condition, per time-point. Per1:, two-way ANOVA time*treatment, interaction F3,24 = 3.234 p = 0.04, post-hoc Sidak, AL vs F (ZT4) t24 = 3.209 p = 0.015; Cry1: two-way ANOVA time*treatment, interaction F3,24 = 4.836 p = 0.009, post-hoc Sidak, AL vs F (ZT4) t24 = 3.317 p = 0.0115; Cry2: F, two-way ANOVA time*treatment, interaction F3,24 = 3.717 p = 0.0251, post-hoc Sidak, AL vs F (ZT4) t24 = 2.999 p = 0.0247, (ZT22) t24 = 2.769 p = 0.042; Rev-erb alpha, two-way ANOVA time*treatment, interaction F3,24 = 4.162 p = 0.0165, post-hoc Sidak, AL vs F (ZT4) t24 = 3.144 p = 0.0175). d Quantitative real-time PCR analysis of genes related to the transcriptional activation limb of the core clock machinery (N = 4 per condition, per time-point. Bmal1: F, two-way ANOVA time*treatment, interaction F3,24 = 1.167 p = 0.1804; Clock: two-way RM ANOVA time*treatment, interaction F3,24 = 0.94 p = 0.436; Rora: F, two-way RM ANOVA time*treatment, interaction F3,24 = 1.635 p = 0.3869). Error bars represent SEM
All these findings, together with the possible body-wide effect of fasting on circadian rhythmicity [40, 41], led us to explore whether fasting could alter the daily rhythmicity in spontaneous locomotor activity. Diurnal rhythmicity in locomotion was investigated by selecting three different epochs of 5 days each: the 5 days before fasting (pre-fasting), the 5 days after refeeding (T1) and the subsequent 5 days (T2) (Fig. 7a). Body weight was analyzed before, after fasting, at T1 and T2 (Fig. 7b). For every epoch, we computed the relative power spectrum at different oscillation frequencies and we integrated the power between 0.9 and 1.1 cycles/day as a measure of diurnal rhythmicity (Fig. 7c, see materials and methods and Fig. S7a). The results demonstrated that before fasting there were no differences between AL and F groups (Fig. 7d). However, both at T1 and T2 (after fasting), F mice displayed a decreased power at the diurnal frequency (Fig. 7d), suggesting that acute fasting for 48 h affects the normal development of rhythmicity in locomotion. Notably, this effect was persistent for at least 10 days after refeeding, suggesting a long-lasting trace of fasting effects on behavioral rhythmicity. The mechanisms underlying this fasting influence on behavior is not known, however, total locomotor activity did not change at T1 and T2 (Fig. S7b), and the body weight went back to AL level by T1, although a certain difference was observed at T2 (Figs. 7b and S7c). Moreover, the food intake was significantly increased 3, 4, and 5 days (T1) after refeeding with respect to the AL controls (Fig. S7c). However, in the subsequent days, it was comparable to AL mice (Fig. S7c). Also, gastrocnemius and soleus muscle mass did not change after refeeding (Fig. S7d, e), while subcutaneous fat and BAT were increased in F mice at T1 (Fig. S7d), and subcutaneous fat and epididymal fat were significantly higher at T2 (Fig. S7e). Finally, cortical expression changes in specific genes altered by 48 h fasting (Figs. 5g, h, 6c) were not observed at T1 and T2 (Fig. S8), indicating that at least part of the fasting-induced brain transcriptional reprogramming is not persistent after re-feeding.
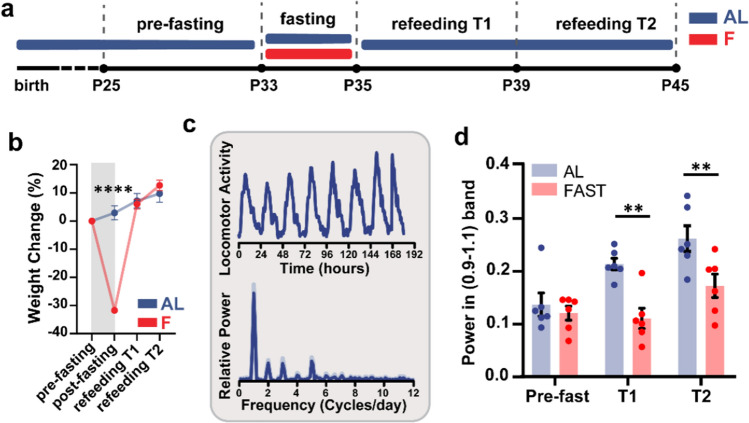
Fig. 7.
Fasting has long-lasting effects on the daily rhythmicity in spontaneous locomotor activity. a Experimental timeline. b Weight change (%) before (pre-fasting) and after 48 h fasting (post-fasting), and during the refeeding phase at two different time points: 5 days after the refeeding (refeeding T1), 10 days after the refeeding (refeeding T2) (AL N = 6, F N = 6, two-way RM ANOVA time*treatment, interaction F3,30 = 78.35 p < 0.0001, post-hoc Sidak, AL vs F (post-fasting) t40 = 12.7 p < 0.0001. Gray square represents the fasting period. c Representative trace of the locomotor activity of a single mouse in the time domain (top) and in the frequency domain (bottom). c Relative power of the circadian oscillation (total power between 0.9 and 1.1 cycles per day) in AL and FAST mice in three epochs: before fasting (Pre-fast), first 5 days of refeeding (T1) and second 5 days of refeeding (T2). (N = 6 per condition, two-way RM ANOVA time*treatment, interaction F2,20 = 4.11 p = 0.032, post-hoc Holm Sidak, AL vs FAST (Pre-fast) T30 = 0.598 p = 0.555, AL vs FAST (T1) T30 = 3.768 p = 0.002, AL vs FAST (T2) T30 = 3.274 p = 0.005). Error bars represent SEM
Discussion
Metabolic influence on the brain has been observed for years, however, little is known about the molecular mechanisms activated by metabolic stimuli in brain tissue. In particular caloric restriction or intermittent fasting have been found to ameliorate brain resilience to aging processes [42], but its biochemical and transcriptional consequences on brain tissue in vivo are poorly understood. BHB, a metabolic fuel for the brain mainly produced by the liver during fasting, has been shown to be beneficial in models of neurological diseases [43], epilepsy [44] and stroke [45]. These effects have been attributed to BDNF [46, 47], HCAR2 [48], and promotion of SIRT3 function [49]. However, our study shows that a surprisingly high number of proteins can undergo K-bhb, raising the possibility that multiple unknown pathways can mediate BHB effects in the brain with potential therapeutic implications for brain disorders. To our knowledge, this is the first proteome-wide report of beta-hydroxybutyrylation in the cerebral cortex. A recent report investigated K-bhb in the liver after 48 h fasting and found 267 beta-hydroxybutyrylated proteins, involved in several metabolic networks, energy and detoxification pathways [28]. In the brain, the proteins displaying K-bhb belonged to specific GO categories which included regulation of transcription and chromatin modifications, raising the possibility of a cross-talk among beta-hydroxybutyrylation and other epigenetic mechanisms. Further studies are needed to investigate the BHB action on specific proteins and in the distinct cell types present in the neural tissue.
Our work also reveals transcriptomic changes induced by fasting in the cerebral cortex, and demonstrates that BHB is an epigenetic molecule capable of remodeling the neural tissue chromatin landscape by post-translationally modifying the histone H3. Fasting-induced transcriptional changes and the enrichment of H3K9-bhb on a genome-wide scale were significantly correlated suggesting a regulatory role for H3K9-bhb in neural tissue. Previous work studied H3K9-bhb response to the same fasting period used here in the liver [25]. Our ChIP-seq analysis in cerebrocortical tissue revealed a number of DNA loci displaying differential H3K9-bhb after fasting summing up to about half of the DNA loci regulated in the liver, indicating that histone beta-hydroxybutyrylation may be a more pervasive PTM in liver than in the brain. Importantly, the data crossing of liver and cortex H3K9-bhb ChIP-seq results indicated several common promoters and enhancers among the two tissues. The KEGG pathways of the common enhancers were associated with “circadian rhythms” terms. The effects of fasting on the liver clock are well-known [40, 41], as well as the effects of H3K9-bhb on core-clock gene expression [25]. However, how food deprivation can impinge on the cerebral cortex clock is still obscure. Our transcriptome and ChIP-seq data converge into a possible involvement of circadian processes in the brain response to a prolonged fasting (see below).
Based on genomic localization analysis, H3K9-bhb was mainly enriched in the cerebral cortex intergenic, enhancer and promoter regions of fasting mice. The genes neighboring those sites were positively correlated with the RNA-seq results of genes UP in fasting, extending and confirming previous findings in the liver [25], and suggesting that H3K9-bhb is a new PTM linked to active gene expression also in the brain. Although our understanding of intergenic regions function in the brain is still limited, H3K9-bhb in these genomic loci might help regulate the expression of associated transcribed regions, as already observed for DNA methylation in the human brain [50].
The functional annotation analysis of transcriptome, beta-hydroxybutyrylome and epigenome suggests that fasting could impinge on specific pathways, which could cooperate in setting up the best response of brain cells to a condition of scarce access to food. In particular, “circadian rhythms” is the most significantly enriched KEGG pathway among the upregulated genes, and “circadian entrainment”, “circadian regulation of gene expression”, “rhythmic process”, and again “circadian rhythms'', come up in the ChIP-seq functional annotation analysis of enhancer and promoter regions. Thus, diurnal rhythms seem to represent biological processes particularly impacted by fasting in the cerebral cortex. The circadian clock is an inner oscillator system that ensures appropriate physiology and fitness to a variety of organisms [51, 52]. Changes in core-clock gene expression after 48-h fasting might help to optimize the energetic cycle in neuronal cells, which consume large amount of energy for electrical transmission and membrane potential maintenance; and/or non-neuronal cells, which need adequate fuel for surveillance/defense function (e.g., microglia), metabolism, tropism, and secretion/absorption of neurotransmitters, etc. (e.g., astrocytes). Intriguingly, our reactome analysis of the cortical transcriptome displayed the existence of a link between circadian rhythms related genes and epigenetic related transcripts (Fig. 4c). Epigenetic mechanisms participate in the formation of circadian oscillation of gene expression, intricately regulating the core-clock machinery [53]. The fasting challenge could directly control core-clock gene expression through a complex network of epigenetic proteins, possibly fine tuning H3K9-bhb and other histone PTMs. Furthermore, the circadian clock controls the onset of critical period plasticity in the visual cortex [29]. Thus, we speculate that the molecular/epigenetic/metabolic changes driven by fasting could contribute to altering the plasticity potential of the neural circuits through modulation of the endogenous 24-h oscillator. An effect of fasting on plasticity is also in line with fasting modulation of protein synthesis, a regulatory mechanism involved in plasticity [54], and the impact of the gut-brain axis on adult ocular dominance plasticity [55]. This hypothesis will be tested in future experiments.
Core-clock genes were also modulated in a time-of-the-day dependent manner in the SCN, suggesting the master clock could be sensitive to changes in the metabolic status [3]. As such, we found that fasting could interfere with normal rhythmicity in spontaneous locomotor activity (Fig. 7). The changes in the rhythmicity of activity were observed both 5 days and 10 days after refeeding suggesting a long-lasting “memory trace” related to locomotion in F mice with respect to AL.
“Oxytocin signaling” and “serotonergic synapses” are categories present in our transcriptome functional annotation, and they seem particularly relevant for brain function. Oxytocin and serotonin are neuromodulators involved in human affects and socialization [56]. For instance they have been shown to be implicated in ASD and depression [57, 58]. Prolonged fasting could promote social behavior to cooperate and increase the probability to survive during periods of food scarcity [59]. Based on our GO results, intermittent fasting, caloric restriction, KD or ketone supplements might be alternative strategies or adjuvant to traditional treatments for ASD, depression or other neuropsychiatric diseases. This is an intriguing hypothesis which needs further validation in preclinical models to rationalize the use of nutrition to improve brain disorders, and cognitive function in general.
An important future step would be to deeply investigate the functional relevance of H3K9-bhb in the brain fasting response, and to causally link H3K9-bhb to fasting-induced gene expression. At this time, these experiments are particularly challenging since we have only little information about the mechanisms underlying the beta-hydroxybutyrylation chemical reaction. Recently, human SIRT3 was shown to display class-selective histone de-betahydroxybutyrylase activities on H3K9-bhb [60]. SIRT3 seems to be preferentially a mitochondrial sirtuin [61], while its nuclear localization is controversial [62–64]. Thus, genetically or pharmacologically modulating SIRT3 in the cortex to target H3K9-bhb during fasting would not be an appropriate approach considering SIRT3 uncertain subcellular localization and the unspecific effects caused by its deacetylase activity. Furthermore, Huang et al. discovered that the acetyltransferase p300 can catalyze the enzymatic addition of BHB to K, while histone deacetylase 1 (HDAC1) and HDAC2 enzymatically remove K-bhb in cells in vitro [65]. Intriguingly, as HDAC2 was a hit in our K betahydroxybutyrilome analysis (Figs. 2b and S1g) in the cerebral cortex of F mice, a deeper investigation to understand the role of K-bhb on HDAC2 enzymatic activity might reveal a novel BHB-driven regulatory loop for PTMs levels during ketosis and beyond. Finally, since p300 is an acetyl-transferase, blocking its activity would be highly unspecific. A desirable approach could be to directly target H3K9-bhb in vivo for example using an intrabody against this specific PTM [66], however this type of tool is not available yet.
Before concluding, a reflection regarding the distinct effects of fasting-driven metabolic changes and ketone bodies should be made. The ability of BHB to substitute for blood glucose in energy production is essential for survival during prolonged starvation, particularly regarding brain function. Fasting is able to alter the subject metabolic status by impacting lipid, glucose and protein metabolism [67]. Fasting influences the levels of several metabolites, among which glucose, BHB, acetoacetate; hormones, such as insulin, leptin and FGF21. Some of these molecules have been shown to affect brain function or plasticity [4, 68–70]. Although our specific analysis of K-bhb and H3K9-bhb point toward a direct effect of BHB on neural tissue chromatin remodeling, as BHB is the only chemical donor for K-bhb identified so far, we cannot exclude the contribution of other players in the observed transcriptional reprogramming. Even more complicated to dissect is the mechanism underlying behavioral alteration in daily locomotor rhythmicity. Fasting can impinge on the endogenous clock in peripheral tissues [40, 41], thus we cannot exclude a pure-metabolic dependent effect on the SCN after a 48 h fasting challenge, or an interplay between ketone bodies as signaling/epigenetic molecules and other metabolic signals.
In summary, our work shows that fasting is a powerful regulator of the transcriptional status of neural tissue modifying the chromatin epigenetic landscape, potentially leaving a trace on circadian rhythms which could persist at the behavioral level. The regulated genes are not only involved in brain tissue metabolism, but they also include pathways that are relevant for synaptic transmission and plasticity, raising the hypothesis that the metabolic status of the individual might exert an important regulation on brain function.
Materials and methods
Animals and feeding
All experiments were carried out in accordance with the European Directives (2010/63/EU) and were approved by the Italian Ministry of Health (authorization number 354/2020-PR).
Male and female C57BL/6J mice were used in this study. Mice were housed in conventional cages (365 × 207 × 140 mm, 2–3 animals per cage) with nesting material. Mice were kept under a 12-h dark: 12-h light cycle, with food (standard diet mucedola 4RF25) and water ad libitum.
Mice were weaned on postnatal day (P) 21. At P33 a group of mice was subjected to fasting (F group) for 2 days, until P35, through the removal of the food from the cage and ad libitum access to water. Age-matched control mice (AL group) continued to have ad libitum access to food and water. After 2 days, mice were sacrificed by decapitation. Liver and occipital cortices (corresponding to the visual cortex) were harvested, snapped frozen in liquid nitrogen and stored at − 80 °C until use. The visual cortex was chosen as this area represents a gold standard to investigate plasticity processes in the brain and neuronal adaptation to nutritional challenges [4]. Subcutaneous fat, interscapular brown adipose tissue (BAT), epididymal fat, gastrocnemius, and soleus muscle were collected and weighted using a precision scale. For the day vs night sample analysis, mice were sacrificed every 6 h along the diurnal cycle (from ZT0 to ZT24). For every group of mice, the food was precisely removed 48 h before the ZT of the sacrifice.
For the refeeding experiments, mice were sacrificed 5 (T1, P40) or 10 days (T2, P45) after refeeding at ZT4. The harvesting and lean and fat mass measurements were performed as described above.
For the daily food intake analysis, mice were single housed and starting from the day after fasting, food pellets weight were measured using a precision scale every 24 h, until T2, corresponding to 10 days after refeeding.
Metabolomic analysis
About 20 mg of liver and cortex samples were homogenized with 300 µl cold methanol on Precellys Evolution Homogenizer (Bertin Instruments, Frankfurt, Germany) at 4 °C: 3 cycles of 30 s at 5500 rpm, with 10 s pause between each cycle for liver and 3 cycles of 30 s at 6800 rpm, with 10 s pause between each cycle for cortex. Samples were then centrifuged at 13,300 rpm for 20 min at 4 °C for proteins precipitation and extracted with modified Folch’s method: 900 µl of chloroform:methanol (2:1, v:v) and 200 µl of water and centrifuged at 13,300 rpm for 20 min at 4 °C for two times. Down phase was dried under nitrogen flux, reconstituted with chloroform:methanol (1:14, v:v) for triglyceride (TGs) and analyzed by LC–MS-QTOF (UHPLC 1290 infinity coupled with 6545 QTOF, Agilent, Santa Clara, CA) equipped ZORBAX Eclipse Plus C18 column 2.1 × 100 mm 1.8 µm (Agilent, Santa Clara, CA). Quantification was obtained using TG (15:0/15:0/15:0) (Larodan, Solna, SE) as an internal standard. For the analysis of beta-Hydroxybutyric acid (BHB) in tissue and plasma, polar phase was extracted with modified Folch’s method described above; the up-phase was dried under nitrogen flux, reconstituted with acetonitrile:water (8:2, v:v) and analyzed by LC–MS-QTOF equipped with a Poroshell 120 HILIC-Z column 2.1 × 150 mm 2.7 µm (Agilent, Santa Clara, CA). Quantification was performed using 13C-BHB (Cambridge Isotope Laboratories, Tewksbury, MA) as an internal standard. Plasma FFA composition was evaluated by gas chromatography-mass spectrometry (GC–MS, GC7890-MS5975, Agilent Technology, Santa Clara, CA) equipped with a capillary column (DB-5MS J&W, l 30 m; i.d. 0.25 mm; film thickness 0.25 µm, J&W, Agilent, Santa Clara, CA). After adding the internal standard, i.e., heptadecanoic acid (Sigma-Aldrich), plasma samples (40 µl) were deproteinized with 300 µl of cold methanol, centrifuged at 13,300 rpm for 20 min at 4 °C and the extracted with modified Folch’s method. Down phase was dried under nitrogen flux, reconstituted with 80 μL of acetonitrile, derivatized with 20 μL of N, O-Bis(trimethylsilyl) trifluoroacetamide with 1% trimethylchlorosilane (Sigma-Aldrich) for 40 min at 75 °C and injected into the GC–MS. Quantification was performed using heptadecanoic acid (Merck, Darmstadt, Germany), as an internal standard.
Protein extraction and Western Blot
For total protein extracts, liver and cerebral cortex samples were homogenized in modified RIPA buffer (50 mM Tris pH8, 150 mM NaCl, 5 mm EDTA, 15 mM MgCl2, 1% NP40) plus protease inhibitors. The samples were sonicated for 1 min on ice (10 s on/10 s off) and centrifuged for 15 min at 14,000×g, at 4 °C. The supernatant was recovered and the protein concentration was determined by Bradford assay (Biorad #5,000,006) using a Nanodrop Spectrophotometer (Thermoscientific 2000 C).
8% SDS-PAGE was performed to check Lysine-bhb, phospho(Ser235-236)S6, total S6, phospho(Ser473)AKT and total AKT levels. 15% SDS-PAGE was performed to analyze H3 and total H3K9-bhb. The samples were blotted onto nitrocellulose membranes (Biorad) and blocked in 5% BSA in Tris-buffered saline (TBS) for 1 h at RT. The nitrocellulose membrane was incubated at 4 °C overnight with the following antibodies: anti-phospho(Ser235-236)S6 (Cell Signalling Technology #2211S) 1:1000, anti-S6 (Cell Signalling Technology #2217) 1:1000, and anti-phospho (Ser473) AKT 1:1000 (Cell signalling technology #9271), AKT 1:1000 (Cell Signalling technology #9272), bhb-Lysine 1:2000 (PTM Biolabs #1201), H3K9-bhb 1:2000 (PTM Biolabs #1250), H3 1:5000 (Abcam #1791), beta-tubulin 1:3000 (Sigma #T4026) H3K9-K14ac 1:1000 (Millipore #06–599). Blots were then washed 3 times in TTBS for 30 min, incubated in HRP conjugated anti-mouse or anti-rabbit diluted (1:8000) in 2.5% BSA in TTBS for 1 h at RT. The membranes were then rinsed three times in TTBS and incubated in enhanced chemiluminescent substrate (Millipore) and acquired through a Chemidoc XRS instrument. Bands densitometry was analyzed through ImageJ software.
RNA extraction
Occipital cortex samples were homogenized in phenol/guanidine-based QIAzol Lysis Reagent (Qiagen #79,306). Chloroform was added and the samples were shaken for 15 s. The samples were left at 20–24 °C for 3 min and then centrifuged (12,000×g, 20 min, 4 °C). The upper phase aqueous solution, containing RNA, was collected in a fresh tube and the RNA was precipitated by the addition of isopropanol. Samples were mixed by vortexing, left at RT for 10 min and then centrifuged (12,000×g, 15 min, 4 °C). The supernatant was discarded and the RNA pellet was washed in 75% ethanol by centrifugation (7500×g, 5 min, 4 °C). The supernatant was discarded and the pellet was left to dry for a minimum of 15 min before resuspending in RNAse free water.
Due to the small amounts of tissue, samples containing the suprachiasmatic nucleus of the hypothalamus were homogenized in phenol/guanidine-based QIAzol Lysis Reagent (Qiagen #79,306) and then processed using the miRNeasy Mini Kit (Qiagen # 217,004).
RNA concentration was determined by Nanodrop Spectrophotometer (Thermoscientific 2000 C). RNA quality was analyzed via agarose gel electrophoresis (1% agarose). Total RNA was reverse transcribed using QuantiTech Reverse Transcription Kit (Qiagen # 205,311).Gene expression was analyzed by real-time PCR (Step one, Applied Biosystems), using PowerUp SYBR Green Master Mix (Thermo Fisher #A25742). The primers for gene expression were designed by Primer 3 software (v. 0.4.0) and all the sequences reported in Supplementary Materials.
Quantitative values for cDNA amplification were calculated from the threshold cycle number (Ct) obtained during the exponential growth of the PCR products. Threshold was set automatically by the Step one software. Data were analyzed by the ΔΔCt methods using 18S rRNA to normalize the cDNA levels of the transcripts under investigation.
RNA sequencing and data analysis
N = 4 biological replicates per experimental group were used in the RNA-seq experiment. Total RNA was extracted as described above, libraries were prepared and sequenced on Illumina HiSeq2500 instrument during a pair-end read 125 bp sequencing, producing sequencing results in FastQ format. The FastQ files were processed through the standard Tuxedo protocol, using Tophat and Cufflinks. Tophat was used to align the RNA-seq reads to the reference genome assembly mm10 and Cufflinks was used to calculate gene expression levels. This protocol outputs the FPKM values for each gene of each replicate. The differential analysis of the FPKM values across all experiments and control groups was conducted with Cyber-T, a differential analysis program using a Bayesian-regularized t-test [71, 72]. The p-value threshold used for determining differential expression was 0.05 for all groups. The statistic was corrected using the Benjamini–Hochberg (BH) test for significance.
ChIP-sequencing and analysis
N = 4 biological replicates per experimental group were used for the sequencing. IPs and controls were processed in parallel, and libraries were prepared at the Institute of Applied Genomics (Udine, Italy). The sequencing was performed on Hiseq2500 in 125 bp paired-end mode. The sequencing reads were aligned to the mouse genome assembly mm10 with Bowtie2. Enriched genome areas and peaks were identified with the MACS2 algorithm. Peak annotation was performed with in-house annotating tools. Peaks whose start to end position cover regions including promoter, enhancer, utr3, utr5, exon, intron, 2000 base pair downstream of promoter, or intergenic region were annotated and the percentages of the peak’s coverage of those regions were calculated. The closest gene and TSS to each peak were identified and their distances to the peak were calculated.
Analysis of locomotor activity
Locomotor activity was measured using Opto M3 multi-channel activity monitors (Columbus Instruments, OH, USA). Monitors were placed in the colony area and testing was conducted in the same conditions of animal facility housing with the exception of animals being individually housed in (33 × 15 × 13) cm (length × width × height) clear plastic cages. Locomotor activity was measured by integrating the number of infrared beam breaks over 1-min epochs for many days.
Total activity over an epoch was calculated as the sum of all beam brakes in that epoch. Diurnal rhythmicity was calculated by analyzing 5 days-long epochs in the frequency domain. For each mouse, we removed the non-oscillatory component of the activity trace by subtracting the mean activity, then we computed the periodogram power spectral density estimate at frequencies between 0.1 and 12 cycles/day in increments of 0.1. For each mouse, we normalized the power spectrum so that total power was equal to 1. The power of the circadian oscillation was defined as the sum of the normalized power between 0.9 and 1.1 cycles/day.
Statistical analysis
The majority of statistical analyses were performed using GraphPad Prism version 7 (GraphPad Software, San Diego, CA, USA).
Western blot: differences between groups were tested for significance using unpaired t-tests.
qPCR: Differences between groups were tested for significance using two-way ANOVA. Holm–Sidak's multiple comparisons post hoc tests were performed, when appropriate.
Correlation between RNA-seq and ChIP-seq datasets: Spearman correlation was run on the datasets. The results were considered significant when p was < 0.05.
Locomotor activity: differences between total activity and circadian rhythmicity were tested using a two-way RM ANOVA.
The effect of sex was tested in all the analysis, when it was absent, male and female mice were analyzed as one group, otherwise a two-way ANOVA was performed, and the male and female data were reported in distinct graphs. All data are represented as the mean ± SEM unless otherwise stated. N refers to single animals unless otherwise stated. In the figures *p < 0.05; **p < 0.001, ***p < 0.0001.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Manuel Tongiani, Andrea Tognozzi, Matteo Alberti and Maria Grazia Giuliano for their help with the experiments. Special thanks to Vania Liverani and Antonella Calvello (Scuola Normale Superiore) for technical assistance in the lab. We thank Prof. Concetta Morrone and Prof. Paola Binda (University of Pisa) for their insightful comments.
Author contributions
SC performed the experiments, analyzed the data, and prepared the figures. SCh performed the ChIP-seq and RNA-seq analysis. LL performed the daily activity analysis. FF and SR performed the Beta-hydroxybutyrilome analysis. FC, SF and AG performed the LC–MS-QTOF experiments and analysis. MS performed the RNA seq analysis. MC and FD helped with tissue harvesting and performed qPCR. FR helped with GO analysis. RM helped with correlations between ChIP-seq and RNA-seq. CM helped with ChIP-seq data alignment and analysis. PB supervised the RNA-seq and ChiP-seq data analysis. PT conceived and supervised the project, performed the experiments, and wrote the manuscript.
Funding
This research was supported by H2020-MSCA-IF-2016 749697 GaMePLAY, University of Pisa PRA-2020, Italian Ministry of University and Research PNRR Tuscany Health Ecosystem Milestone 8.9.1 and PNRR young MSCA_0000081 iNsPIReD to PT, and funded in part by EFSD-Lilly 2019 to AG and PT. The work of SCh, MS, CM, and PB was in part supported by NIH Grant GM123558 to PB. RM was supported by Fondazione Umberto Veronesi.
Data availability
RNA-seq and ChIP-seq datasets generated and analyzed during this study are available on the GEO database: accession number GSE168725.
Declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Ethics approval
All experiments were carried out in accordance with the European Directives (2010/63/EU) and were approved by the Italian Ministry of Health (authorization number 354/2020-PR).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Sara Cornuti, Siwei Chen have contributed equally to this work.
References
- 1.Gómez-Pinilla F. Brain foods: the effects of nutrients on brain function. Nat Rev Neurosci. 2008;9:568–578. doi: 10.1038/nrn2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mattson MP, Moehl K, Ghena N, et al. Intermittent metabolic switching, neuroplasticity and brain health. Nat Rev Neurosci. 2018;19:80–80. doi: 10.1038/nrn.2017.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tognini P, Samad M, Kinouchi K, et al. Reshaping circadian metabolism in the suprachiasmatic nucleus and prefrontal cortex by nutritional challenge. Proc Natl Acad Sci USA. 2020;117:29904–29913. doi: 10.1073/pnas.2016589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Padamsey Z, Katsanevaki D, Dupuy N, Rochefort NL. Neocortex saves energy by reducing coding precision during food scarcity. Neuron. 2022;110:280–296.e10. doi: 10.1016/j.neuron.2021.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dixon JB, Browne JL, Lambert GW, et al. Severely obese people with diabetes experience impaired emotional well-being associated with socioeconomic disadvantage: results from diabetes MILES—Australia. Diabetes Res Clin Pract. 2013;101:131–140. doi: 10.1016/j.diabres.2013.05.017. [DOI] [PubMed] [Google Scholar]
- 6.Dutheil S, Ota KT, Wohleb ES, et al. High-fat diet induced anxiety and anhedonia: impact on brain homeostasis and inflammation. Neuropsychopharmacology. 2016;41:1874–1887. doi: 10.1038/npp.2015.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walsh ME, Shi Y, Van Remmen H. The effects of dietary restriction on oxidative stress in rodents. Free Radical Biol Med. 2014;66:88–99. doi: 10.1016/j.freeradbiomed.2013.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo H, Chiang H-H, Louw M, et al. Nutrient sensing and the oxidative stress response. Trends Endocrinol Metab. 2017;28:449–460. doi: 10.1016/j.tem.2017.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lutas A, Yellen G. The ketogenic diet: metabolic influences on brain excitability and epilepsy. Trends Neurosci. 2013;36:32–40. doi: 10.1016/j.tins.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evangeliou A, Vlachonikolis I, Mihailidou H, et al. Application of a ketogenic diet in children with autistic behavior: pilot study. J Child Neurol. 2003;18:113–118. doi: 10.1177/08830738030180020501. [DOI] [PubMed] [Google Scholar]
- 11.Li Q, Liang J, Fu N, et al. A ketogenic diet and the treatment of autism spectrum disorder. Front Pediatr. 2021;9:650624. doi: 10.3389/fped.2021.650624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Louw E, van der Louw E, van den Hurk D, et al. Ketogenic diet guidelines for infants with refractory epilepsy. Eur J Paediatr Neurol. 2016;20:798–809. doi: 10.1016/j.ejpn.2016.07.009. [DOI] [PubMed] [Google Scholar]
- 13.Paoli A, Rubini A, Volek JS, Grimaldi KA. Beyond weight loss: a review of the therapeutic uses of very-low-carbohydrate (ketogenic) diets. Eur J Clin Nutr. 2013;67:789–796. doi: 10.1038/ejcn.2013.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murakami M, Tognini P. Molecular mechanisms underlying the bioactive properties of a ketogenic diet. Nutrients. 2022 doi: 10.3390/nu14040782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee J, Duan W, Mattson MP. Evidence that brain-derived neurotrophic factor is required for basal neurogenesis and mediates, in part, the enhancement of neurogenesis by dietary restriction in the hippocampus of adult mice. J Neurochem. 2002;82:1367–1375. doi: 10.1046/j.1471-4159.2002.01085.x. [DOI] [PubMed] [Google Scholar]
- 16.Vivar C, Potter MC, Choi J, et al. Monosynaptic inputs to new neurons in the dentate gyrus. Nat Commun. 2012;3:1107. doi: 10.1038/ncomms2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landry T, Huang H. Mini review: The relationship between energy status and adult hippocampal neurogenesis. Neurosci Lett. 2021;765:136261. doi: 10.1016/j.neulet.2021.136261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pizzorusso T, Tognini P. Interplay between metabolism, nutrition and epigenetics in shaping brain DNA methylation, neural function and behavior. Genes. 2020 doi: 10.3390/genes11070742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newman JC, Verdin E. Ketone bodies as signaling metabolites. Trends Endocrinol Metab. 2014;25:42–52. doi: 10.1016/j.tem.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kimura I, Inoue D, Maeda T, et al. Short-chain fatty acids and ketones directly regulate sympathetic nervous system via G protein-coupled receptor 41 (GPR41) Proc Natl Acad Sci. 2011;108:8030–8035. doi: 10.1073/pnas.1016088108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taggart AKP, Kero J, Gan X, et al. (d)-β-Hydroxybutyrate inhibits adipocyte lipolysis via the nicotinic acid receptor PUMA-G. J Biol Chem. 2005;280:26649–26652. doi: 10.1074/jbc.C500213200. [DOI] [PubMed] [Google Scholar]
- 22.Won Y-J, Lu VB, Puhl HL, 3rd, Ikeda SR. β-Hydroxybutyrate modulates N-type calcium channels in rat sympathetic neurons by acting as an agonist for the G-protein-coupled receptor FFA3. J Neurosci. 2013;33:19314–19325. doi: 10.1523/JNEUROSCI.3102-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shimazu T, Hirschey MD, Newman J, et al. Suppression of oxidative stress by β-hydroxybutyrate, an endogenous histone deacetylase inhibitor. Science. 2013;339:211–214. doi: 10.1126/science.1227166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tognini P, Murakami M, Liu Y, et al. Distinct circadian signatures in liver and gut clocks revealed by ketogenic diet. Cell Metab. 2017;26:523–538.e5. doi: 10.1016/j.cmet.2017.08.015. [DOI] [PubMed] [Google Scholar]
- 25.Xie Z, Zhang D, Chung D, et al. Metabolic regulation of gene expression by histone lysine β-hydroxybutyrylation. Mol Cell. 2016;62:194–206. doi: 10.1016/j.molcel.2016.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hashimoto T, Cook WS, Qi C, et al. Defect in peroxisome proliferator-activated receptor alpha-inducible fatty acid oxidation determines the severity of hepatic steatosis in response to fasting. J Biol Chem. 2000;275:28918–28928. doi: 10.1074/jbc.M910350199. [DOI] [PubMed] [Google Scholar]
- 27.Koubi HE, Robin JP, Dewasmes G, et al. Fasting-induced rise in locomotor activity in rats coincides with increased protein utilization. Physiol Behav. 1991;50:337–343. doi: 10.1016/0031-9384(91)90075-Y. [DOI] [PubMed] [Google Scholar]
- 28.Koronowski KB, Greco CM, Huang H, et al. Ketogenesis impact on liver metabolism revealed by proteomics of lysine β-hydroxybutyrylation. Cell Rep. 2021;36:109487. doi: 10.1016/j.celrep.2021.109487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kobayashi Y, Ye Z, Hensch TK. Clock genes control cortical critical period timing. Neuron. 2015;86:264–275. doi: 10.1016/j.neuron.2015.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pizzorusso T, Medini P, Berardi N, et al. Reactivation of ocular dominance plasticity in the adult visual cortex. Science. 2002;298:1248–1251. doi: 10.1126/science.1072699. [DOI] [PubMed] [Google Scholar]
- 31.Wu D, Smyth GK. Camera: a competitive gene set test accounting for inter-gene correlation. Nucleic Acids Res. 2012;40:e133–e133. doi: 10.1093/nar/gks461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo JU, Ma DK, Mo H, et al. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat Neurosci. 2011;14:1345–1351. doi: 10.1038/nn.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choi M, Wang SE, Ko SY, et al. Overexpression of human GATA-1 and GATA-2 interferes with spine formation and produces depressive behavior in rats. PLoS ONE. 2014;9:e109253. doi: 10.1371/journal.pone.0109253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li M, Cogswell SE, Hixson K, et al. Nuclear Respiratory Factor 1 (NRF-1) controls the activity dependent transcription of the GABA-A receptor beta 1 subunit gene in neurons. Front Mol Neurosci. 2018;11:285. doi: 10.3389/fnmol.2018.00285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pereira JD, Sansom SN, Smith J, et al. Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proc Natl Acad Sci U S A. 2010;107:15957–15962. doi: 10.1073/pnas.1002530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swiss VA, Casaccia P. Cell-context specific role of the E2F/Rb pathway in development and disease. Glia. 2010;58:377–390. doi: 10.1002/glia.20933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Veyrac A, Besnard A, Caboche J, et al. The transcription factor Zif268/Egr1, brain plasticity, and memory. Prog Mol Biol Transl Sci. 2014;122:89–129. doi: 10.1016/B978-0-12-420170-5.00004-0. [DOI] [PubMed] [Google Scholar]
- 38.Martin KC, Barad M, Kandel ER. Local protein synthesis and its role in synapse-specific plasticity. Curr Opin Neurobiol. 2000;10:587–592. doi: 10.1016/S0959-4388(00)00128-8. [DOI] [PubMed] [Google Scholar]
- 39.Hartsock MJ, Spencer RL. Memory and the circadian system: identifying candidate mechanisms by which local clocks in the brain may regulate synaptic plasticity. Neurosci Biobehav Rev. 2020;118:134–162. doi: 10.1016/j.neubiorev.2020.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kinouchi K, Magnan C, Ceglia N, et al. Fasting imparts a switch to alternative daily pathways in liver and muscle. Cell Rep. 2018;25:3299–3314.e6. doi: 10.1016/j.celrep.2018.11.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vollmers C, Gill S, DiTacchio L, et al. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc Natl Acad Sci USA. 2009;106:21453–21458. doi: 10.1073/pnas.0909591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mujica-Parodi LR, Amgalan A, Sultan SF, et al. Diet modulates brain network stability, a biomarker for brain aging, in young adults. Proc Natl Acad Sci USA. 2020;117:6170–6177. doi: 10.1073/pnas.1913042117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang H, Shan W, Zhu F, et al. Ketone bodies in neurological diseases: focus on neuroprotection and underlying mechanisms. Front Neurol. 2019;10:585. doi: 10.3389/fneur.2019.00585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simeone TA, Simeone KA, Rho JM. Ketone bodies as anti-seizure agents. Neurochem Res. 2017;42:2011–2018. doi: 10.1007/s11064-017-2253-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stephan JS, Sleiman SF. Exercise factors released by the liver, muscle, and bones have promising therapeutic potential for stroke. Front Neurol. 2021;12:600365. doi: 10.3389/fneur.2021.600365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sleiman SF, Henry J, Al-Haddad R, et al. Exercise promotes the expression of brain derived neurotrophic factor (BDNF) through the action of the ketone body β-hydroxybutyrate. Elife. 2016 doi: 10.7554/eLife.15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen L, Miao Z, Xu X. β-hydroxybutyrate alleviates depressive behaviors in mice possibly by increasing the histone3-lysine9-β-hydroxybutyrylation. Biochem Biophys Res Commun. 2017;490:117–122. doi: 10.1016/j.bbrc.2017.05.184. [DOI] [PubMed] [Google Scholar]
- 48.Rahman M, Muhammad S, Khan MA, et al. The β-hydroxybutyrate receptor HCA2 activates a neuroprotective subset of macrophages. Nat Commun. 2014;5:3944. doi: 10.1038/ncomms4944. [DOI] [PubMed] [Google Scholar]
- 49.Yin J, Han P, Tang Z, et al. Sirtuin 3 mediates neuroprotection of ketones against ischemic stroke. J Cereb Blood Flow Metab. 2015;35:1783–1789. doi: 10.1038/jcbfm.2015.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Telese F, Gamliel A, Skowronska-Krawczyk D, et al. “Seq-ing” insights into the epigenetics of neuronal gene regulation. Neuron. 2013;77:606–623. doi: 10.1016/j.neuron.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zarrinpar A, Chaix A, Panda S. Daily eating patterns and their impact on health and disease. Trends Endocrinol Metab. 2016;27:69–83. doi: 10.1016/j.tem.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tognini P, Murakami M, Sassone-Corsi P. Interplay between microbes and the circadian clock. Cold Spring Harb Perspect Biol. 2018 doi: 10.1101/cshperspect.a028365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eckel-Mahan K, Sassone-Corsi P. Epigenetic regulation of the molecular clockwork. Prog Mol Biol Transl Sci. 2013;119:29–50. doi: 10.1016/B978-0-12-396971-2.00002-6. [DOI] [PubMed] [Google Scholar]
- 54.Cajigas IJ, Will T, Schuman EM. Protein homeostasis and synaptic plasticity. EMBO J. 2010;29:2746–2752. doi: 10.1038/emboj.2010.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lupori L, Cornuti S, Mazziotti R, et al. The gut microbiota of environmentally enriched mice regulates visual cortical plasticity. Cell Rep. 2022;38:110212. doi: 10.1016/j.celrep.2021.110212. [DOI] [PubMed] [Google Scholar]
- 56.Mottolese R, Redouté J, Costes N, et al. Switching brain serotonin with oxytocin. Proc Natl Acad Sci U S A. 2014;111:8637–8642. doi: 10.1073/pnas.1319810111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamasue H, Domes G. Oxytocin and autism spectrum disorders. Curr Top Behav Neurosci. 2018;35:449–465. doi: 10.1007/7854_2017_24. [DOI] [PubMed] [Google Scholar]
- 58.Borroto-Escuela DO, Ambrogini P, Chruścicka B, et al. The role of central serotonin neurons and 5-HT heteroreceptor complexes in the pathophysiology of depression: a historical perspective and future prospects. Int J Mol Sci. 2021 doi: 10.3390/ijms22041927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weiss O, Dorfman A, Ram T, et al. Rats do not eat alone in public: Food-deprived rats socialize rather than competing for baits. PLoS ONE. 2017;12:e0173302. doi: 10.1371/journal.pone.0173302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang X, Cao R, Niu J, et al. Molecular basis for hierarchical histone de-β-hydroxybutyrylation by SIRT3. Cell Discov. 2019;5:35. doi: 10.1038/s41421-019-0103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Verdin E, Hirschey MD, Finley LWS, Haigis MC. Sirtuin regulation of mitochondria: energy production, apoptosis, and signaling. Trends Biochem Sci. 2010;35:669–675. doi: 10.1016/j.tibs.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bao J, Lu Z, Joseph JJ, et al. Characterization of the murine SIRT3 mitochondrial localization sequence and comparison of mitochondrial enrichment and deacetylase activity of long and short SIRT3 isoforms. J Cell Biochem. 2010;110:238–247. doi: 10.1002/jcb.22531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cooper HM, Spelbrink JN. The human SIRT3 protein deacetylase is exclusively mitochondrial. Biochem J. 2008;411:279–285. doi: 10.1042/BJ20071624. [DOI] [PubMed] [Google Scholar]
- 64.Hallows WC, Albaugh BN, Denu JM. Where in the cell is SIRT3?—functional localization of an NAD-dependent protein deacetylase. Biochem J. 2008;411:e11–e13. doi: 10.1042/BJ20080336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang H, Zhang D, Weng Y, et al. The regulatory enzymes and protein substrates for the lysine β-hydroxybutyrylation pathway. Sci Adv. 2021 doi: 10.1126/sciadv.abe2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chirichella M, Lisi S, Fantini M, et al. Post-translational selective intracellular silencing of acetylated proteins with de novo selected intrabodies. Nat Methods. 2017;14:279–282. doi: 10.1038/nmeth.4144. [DOI] [PubMed] [Google Scholar]
- 67.Patterson RE, Sears DD. Metabolic effects of intermittent fasting. Annu Rev Nutr. 2017;37:371–393. doi: 10.1146/annurev-nutr-071816-064634. [DOI] [PubMed] [Google Scholar]
- 68.Sa-Nguanmoo P, Tanajak P, Kerdphoo S, et al. FGF21 improves cognition by restored synaptic plasticity, dendritic spine density, brain mitochondrial function and cell apoptosis in obese-insulin resistant male rats. Horm Behav. 2016;85:86–95. doi: 10.1016/j.yhbeh.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 69.Ferrario CR, Reagan LP. Insulin-mediated synaptic plasticity in the CNS: anatomical, functional and temporal contexts. Neuropharmacology. 2018;136:182–191. doi: 10.1016/j.neuropharm.2017.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Silva B, Mantha OL, Schor J, et al. Glia fuel neurons with locally synthesized ketone bodies to sustain memory under starvation. Nat Metab. 2022;4:213–224. doi: 10.1038/s42255-022-00528-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Baldi P, Long AD. A Bayesian framework for the analysis of microarray expression data: regularized t -test and statistical inferences of gene changes. Bioinformatics. 2001;17:509–519. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- 72.Kayala MA, Baldi P. Cyber-T web server: differential analysis of high-throughput data. Nucleic Acids Res. 2012;40:W553–W559. doi: 10.1093/nar/gks420. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq and ChIP-seq datasets generated and analyzed during this study are available on the GEO database: accession number GSE168725.