Abstract
Multiple morphological abnormalities of the flagella (MMAF) is a severe disease of male infertility, while the pathogenetic mechanisms of MMAF are still incompletely understood. Previously, we found that the deficiency of Ccdc38 might be associated with MMAF. To understand the underlying mechanism of this disease, we identified the potential partner of this protein and found that the coiled-coil domain containing 146 (CCDC146) can interact with CCDC38. It is predominantly expressed in the testes, and the knockout of this gene resulted in complete infertility in male mice but not in females. The knockout of Ccdc146 impaired spermiogenesis, mainly due to flagellum and manchette organization defects, finally led to MMAF-like phenotype. Furthermore, we demonstrated that CCDC146 could interact with both CCDC38 and CCDC42. It also interacts with intraflagellar transport (IFT) complexes IFT88 and IFT20. The knockout of this gene led to the decrease of ODF2, IFT88, and IFT20 protein levels, but did not affect CCDC38, CCDC42, or ODF1 expression. Additionally, we predicted and validated the detailed interactions between CCDC146 and CCDC38 or CCDC42, and built the interaction models at the atomic level. Our results suggest that the testis predominantly expressed gene Ccdc146 is essential for sperm flagellum biogenesis and male fertility, and its mutations might be associated with MMAF in some patients.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-023-05025-x.
Keywords: CCDC146, Male fertility, MMAF, Spermiogenesis, ODF transportation, Flagellum biogenesis
Background
Infertility is a widespread human health issue, affecting 10–15% of couples worldwide, and male factors account for around 50% of these cases [1, 2]. Male infertility is clinically diagnosed as absence of spermatozoa in the ejaculate (azoospermia), decreased sperm concentration (oligozoospermia), lower sperm motility (asthenozoospermia), or reduced percentage of morphologically normal sperm (teratozoospermia) [3–5]. MMAF is a subtype of asthenoteratozoospermia characterized by immotile spermatozoa presenting severe flagellar malformations [6]. Asthenozoospermia and teratozoospermia usually come from some defects of spermiogenesis.
Spermiogenesis is the final stage of development that involves complex and highly ordered differentiation of spermatids. This process is characterized by reshaping spermatids, including elongation and condensation of the nucleus, and the development of specific structures such as the acrosome, the transient manchette, the flagellum, and the shedding of the cytoplasm [7, 8]. Spermatozoa are highly specialized cells with a head containing genetic information and a tail that moves vigorously to reach eggs. The axoneme, a motility apparatus composed of “9 + 2” microtubules, is the core structure of tails. The sperm tail can be divided into three parts, including the midpiece, principal piece, and endpiece [9, 10] according to the accessory structures surrounding the axoneme. Mitochondrial sheath surrounds outer dense fibers (ODFs) in the midpiece, whereas fibrous sheath (FS) surrounds and replaces ODF 3 and ODF 8 in the principal piece, and the end piece consists of the axoneme only [11, 12].
Abnormalities of the axoneme and accessory structures mainly result in asthenozoospermia, associated with morphological flagellar defects such as abnormal tails, irregular mitochondrial sheaths, and irregular residual cytoplasm [13]. Previous studies have identified about 43 MMAF-related genes [14, 15], mainly from the DNAH protein family [16–21], coiled-coil domain containing (CCDC) protein family [22–24], cilia- and flagella-associated protein (CFAP) family [25–36] and other flagellum biogenesis and morphogenesis related proteins [37–54]. Mutations in these genes cause MMAF, characterized as sperm without flagella or with short, coiled, or irregular flagella [55]. Although an increasing number of studies explore the genetic etiology of MMAF, the pathogenetic mechanisms of MMAF are still not completely elucidated.
The coiled-coil domain-containing (CCDC) proteins have been investigated in the last decade for their multifunctional roles in general and male reproductive physiology [56–66]. CCDC proteins essentially contain the coiled-coil domain, a highly conserved superhelical protein motif characterized by one or more alpha-helical peptides wrapped around each other in a superhelical fashion [67, 68]. About 10% of an organism’s proteome is estimated to comprise the coiled-coil-forming sequences [69]. CCDC proteins are expressed in various tissues, their localizations in cells are varied, and they play functional roles in most physiological processes [70]. Most of the CCDC proteins in the male reproductive system are expressed in the testis, epididymis, seminal vesicle, or prostate [56]. In the testis, CCDC proteins are localized in the seminiferous duct cells, Leydig cells, and maturing spermatocytes [56]. Knockout of Ccdc42 causes sterility in male mice with malformation of the head-to-tail coupling apparatus (HTCA) and the sperm tail [71]. CCDC38 is also essential for spermiogenesis and flagellum biogenesis [72]. The knockout of Ccdc38 results in an MMAF-like phenotype in mice. Besides, CCDC42 and CCDC38 mediate ODF2 transport during flagellum biogenesis [72, 73]. Both proteins are essential for flagellum biogenesis and fertility in male mice, suggesting mutations of these two genes might be associated with male infertility in humans [71–73]. Understanding the molecular causes of MMAF requires identifying the components of these complex molecular machines in spermiogenesis.
Here, we identified CCDC146, which is highly expressed in the testis, can interact with CCDC38 and CCDC42. Importantly, Ccdc146−/− male mice show an abnormally elongated manchette and a MMAF-like phenotype. We found that the CCDC146 might interact with CCDC42 and CCDC38 to facilitate the transportation of some proteins via the IFT pathways during flagellum biogenesis. Additionally, all these proteins are essential for flagellum biogenesis and fertility in male mice, suggesting mutations of these genes might be associated with male infertility in humans.
Results
Ccdc146 is predominantly expressed in the testis
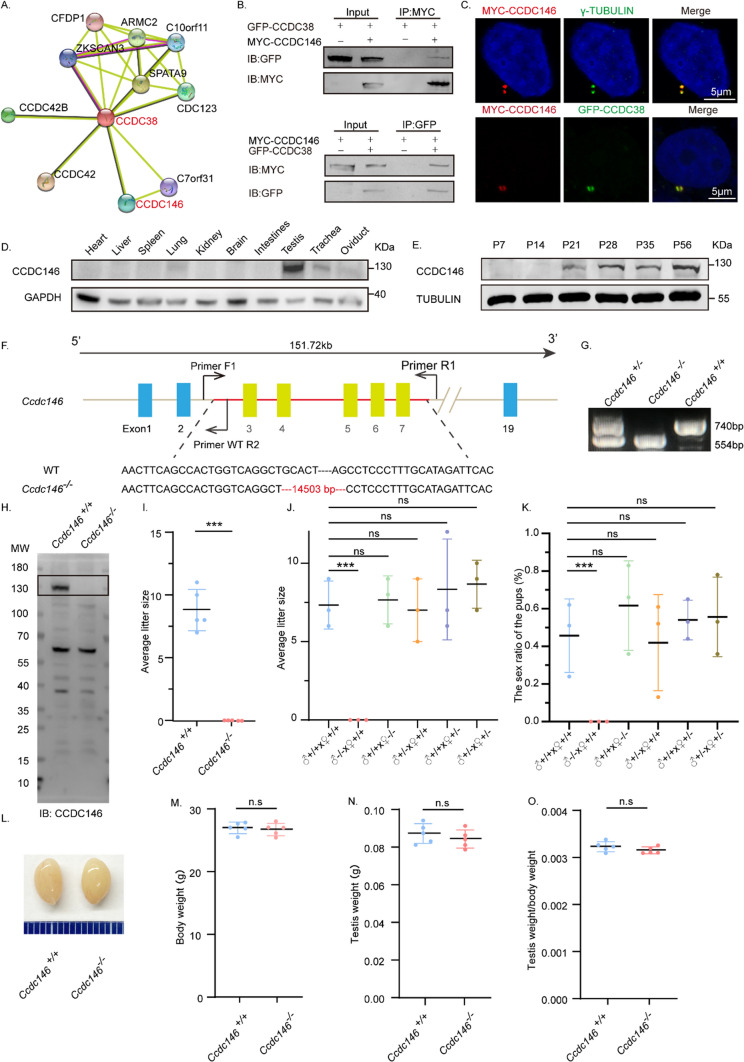
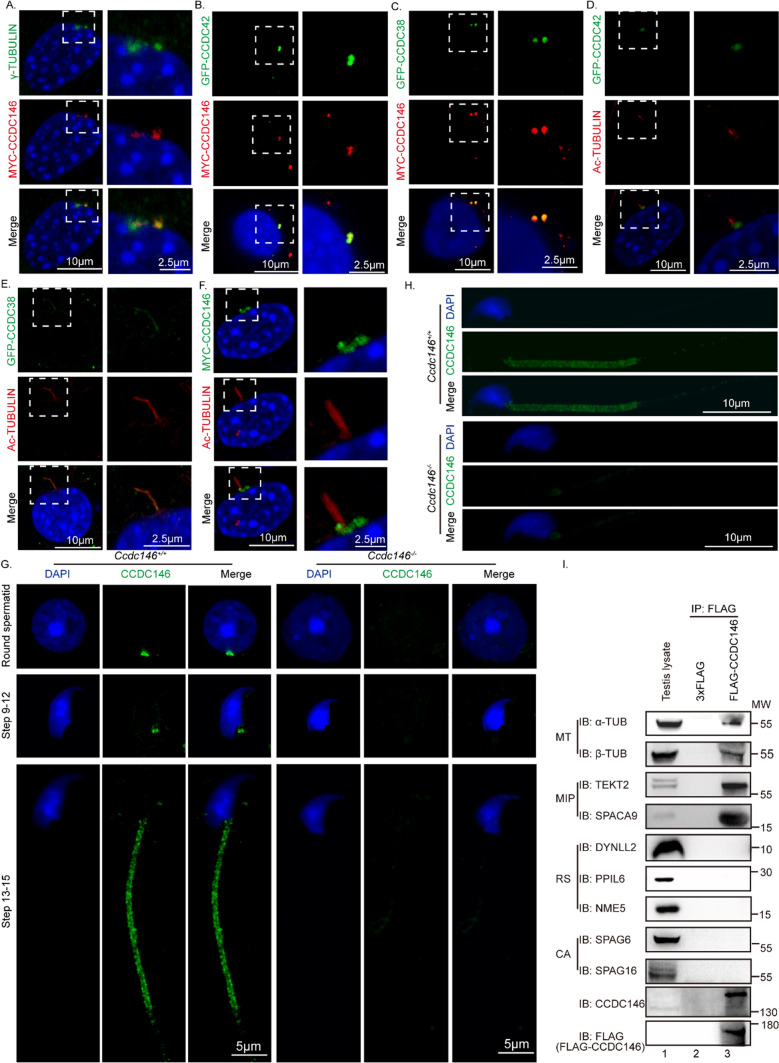
Recently, we found that the knockout of a testis-specific expressed gene Ccdc38 results in an MMAF-like phenotype [72]. To further understand the molecular mechanisms underlying MMAF, we tried to identify its potential interacting partners which might also be associated with this disease. We analyzed the potential interaction networks by the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING), and found that CCDC146 might be related to CCDC38 in human beings (Fig. 1A). Both of these two proteins were defined as centrosome-associated proteins during the assessment of bovine sperm samples [74]. To detect if there were real interaction or not, we constructed pEGFP-C1-Ccdc38 and pCSII-MYC-Ccdc146 plasmids. They were cotransfected into human embryonic kidney 293 expressing SV40 large T antigen (HEK293T) cells, then followed by reciprocal immunoprecipitation with either anti-MYC or anti-GFP antibody, and found that both of them can be immunoprecipitated by the other one (Fig. 1B). Additionally, pCSII-MYC-Ccdc146 and pEGFP-C1-Ccdc38 were transfected into HeLa cells respectively. After immunofluorescence with anti-MYC antibodies and anti-γ-TUBULIN or anti-GFP antibodies, we found that CCDC146 colocalized with γ-TUBULIN and CCDC38 (Fig. 1C), suggesting that CCDC146 indeed interacts with CCDC38. Then we examined its expression pattern in different tissues and found that it was predominantly expressed in the testis (Fig. 1D). Further immunoblotting of mouse testis lysates prepared from different days after birth was carried out. CCDC146 was detected in the testis at postnatal day 21 (P21), and the level increased continuously from postnatal P28 onward, with the highest levels detected in the adult testes (Fig. 1E). This time course corresponded with the onset of spermiogenesis, suggesting that CCDC146 might participate in this process.
Fig. 1.
The knockout of Ccdc146 leads to male infertility. A CCDC146 was related to CCDC38 predicted by the STRING database. B CCDC146 interacted with CCDC38. pCSII-MYC-Ccdc146 was transfected into HEK293T cells with pEGFP-C1-Ccdc38, 48 hours after transfection, cells were collected for immunoprecipitation with anti-MYC, and detected by anti-GFP or anti-MYC antibodies, respectively. C CCDC146 co-localized with CCDC38 and γ-TUBULIN in Hela cells. pCSII-MYC-Ccdc146 and pEGFP-C1-Ccdc38 were co-transfected into Hela cells. 24 h after transfection, cells were fixed and stained with anti-MYC and anti-γ-TUBULIN or anti-GFP antibodies, and the nucleus was stained with DAPI. Scale bar: 5 μm. D The expression of Ccdc146 in different tissues of adult mice. GAPDH was used as a loading control. E The expression of Ccdc146 on different days. TUBULIN was used as a loading control. F The schematic illustration of the knockout strategy for generating Ccdc146−/− mice lacking exon 3–7. Primer-F1, Primer-R1, and Primer-R2 were used as the genotyping primers. G Genotyping of Ccdc146−/− mice. H Western blotting of CCDC146 indicated the depletion efficiency in Ccdc146−/− male mice. I Ccdc146−/− male mice were infertile. The Y axis represents the average number of offspring per litter. Data are presented as means ± SEM. two-tailed Student’s t test; ***P < 0·001. J The number of pups per litter of the different crossing possibilities. Data are presented as means ± SEM. two-tailed Student’s t test; ns: no significance; ***P < 0.001. K The sex ratio of the pups of the different crossing possibilities. Data are presented as means ± SEM. two-tailed Student’s t test; ns: no significance; ***P < 0.001. L The size of the Ccdc146+/+ and Ccdc146−/− mice testes were unaffected. M Testis from Ccdc146+/+ and Ccdc146−/− male mice had no obvious difference in weight (n = 5). Data are presented as means ± SEM. two-tailed Student’s t-test; ns: no significance. N The body weight of Ccdc146+/+ and Ccdc146−/− male mice had no obvious difference (n = 5). Data are presented as means ± SEM. two-tailed Student’s t-test; ns: no significance. O The testis/body weight ratios in Ccdc146+/+ and Ccdc146−/− male mice were consistent (n = 5). Data are presented as means ± SEM. two-tailed Student’s t-test; ns no significance
Ccdc146 knockout leads to male infertility
To characterize the potential functions of CCDC146 during spermiogenesis, Ccdc146−/− mice were created using the CRISPR-Cas9 system from Cyagen Biosciences. Exon 3 to exon 7 of the Ccdc146 gene was selected as the target site (Fig. 1F). The founder animals were genotyped by genomic DNA sequencing and further confirmed by polymerase chain reaction. Two primers were designed to identify the Ccdc146−/− mice (Fig. 1F); the size of the Ccdc146 locus in Ccdc146+/+ mice was 740 bp, while the size of the locus in Ccdc146−/− mice was 554 bp (Fig. 1G). To confirm the knockout of this gene, we generated a rabbit anti-CCDC146 antibody following its purification, it was verified by staining the Hela cells, which were transfected with pCSII-MYC-Ccdc146 plasmid. We found that both anti-MYC and anti-CCDC146 antibodies colocalized to each other (Supplement Fig. S1A), suggesting the anti-CCDC146 antibody is specific to this protein. Furthermore, immunoblotting detection of CCDC146 indicated that the CCDC146 protein was successfully eliminated in the testis of Ccdc146−/− mice (Fig. 1H). Hence, we next examined the fertility of Ccdc146+/+ and Ccdc146−/− mice. Ccdc146−/− male mice exhibited normal mounting behaviors and produced coital plugs but failed to produce offspring after mating with WT adult female mice (Fig. 1I–K), while the fertility of Ccdc146 knockout female mice appeared normal (Fig. 1J, K). However, the knockout of Ccdc146 did not affect either testis size (Fig. 1L) or the ratio of testis weight to body weight (Fig. 1M–O). Taken together, the knockout of Ccdc146 leads to male infertility.
Ccdc146 knockout leads to MMAF in male mice
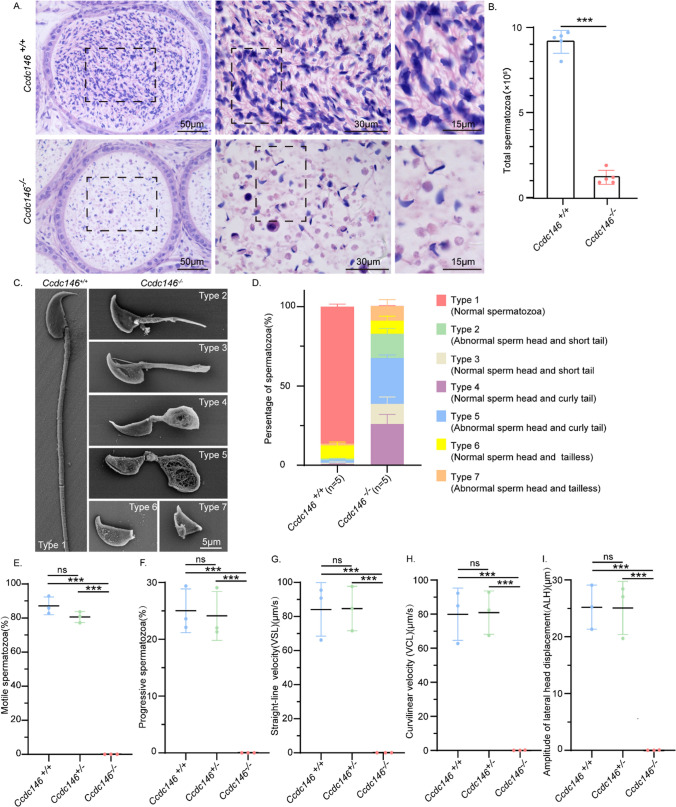
To further explore the cause of male infertility, we observed the transverse sections of the Ccdc146−/− cauda epididymis by hematoxylin and eosin (H&E) staining and found that there was a complete lack of spermatozoa or only a few spermatozoa in the epididymal lumen of Ccdc146−/− mice (Fig. 2A). We examined the spermatozoa released from the caudal epididymis and found that the sperm count in the Ccdc146−/− mice was significantly decreased compared with Ccdc146+/+ mice (Fig. 2B). To determine the morphological characteristics of the spermatozoa, we performed scanning electron microscopy (SEM) of single-sperm, further revealing the morphological abnormalities of Ccdc146−/− spermatozoa (Fig. 2C). The Ccdc146−/− caudal epididymis only contained malformed spermatozoa exhibiting the prominent MMAF phenotype of short, coiled, or absent flagella, compared with Ccdc146+/+ mice. In addition to the flagella abnormality, Ccdc146−/− mice had abnormal sperm heads as well (Fig. 2C, D). The ratio of spermatozoa with abnormal heads and flagella is shown in Fig. 2D. Abnormal sperm heads with short tails and normal sperm heads with curly tails were the major defect categories. Using the computer-assisted semen analysis (CASA) system, the Ccdc146 knockout male mice exhibited a significant decrease in sperm motility as compared to Ccdc146+/+ and Ccdc146+/- mice (Supplementary Movie 1–3). The percentage of motile spermatozoa of the Ccdc146+/+ mice was 87.67% and the progressive spermatozoa of the Ccdc146+/+ mice was 25%, while no motile spermatozoa were observed in the Ccdc146−/− mice (Fig. 2E, F). We further observed significant differences in sperm motility parameters, such as VSL, VCL, and ALH, between Ccdc146+/+ and Ccdc146−/− mice. (Fig. 2G–I). We also detected the cilia mobility of the respiratory tract cilia [26] and found that there was no difference in the beating of trachea cilia for Ccdc146+/+and Ccdc146−/− mice. The movies were provided in Supplementary Movie 4 and 5. These results indicate that Ccdc146 is important for sperm motility. Therefore, the knockout of Ccdc146 results in an MMAF together with an asthenospermia-like phenotype in mice.
Fig. 2.
Ccdc146 knockout results in MMAF. A H&E staining of the caudal epididymis of Ccdc146+/+ and Ccdc146−/− mice. B Sperm number obtained from Ccdc146+/+ and Ccdc146−/− mice (n = 5). Data are presented as means ± SEM. two-tailed Student’s t test; ***P < 0.001. C Scanning electron microscopy analysis of spermatozoa from the epididymis of Ccdc146+/+ and Ccdc146−/− mice. Six abnormal sperm phenotypes were observed: short tail, curly tail, and tailless with normal or abnormal head. D The percentage of different spermatozoa observed in Ccdc146+/+ and Ccdc146−/− caudal epididymis. E Motile sperm in Ccdc146+/+, Ccdc146+/- and Ccdc146−/− mice (n = 3). Data are presented as means ± SEM. two-tailed Student’s t test; ns: no significance; ***P < 0.001. F Progressive sperm in Ccdc146+/+, Ccdc146+/- and Ccdc146−/− mice (n = 3). Data are presented as means ± SEM. two-tailed Student’s t test; ns: no significance; ***P < 0.001. G The straight-line velocity of sperm from Ccdc146+/+, Ccdc146+/- and Ccdc146−/− mice (n = 3). Data are presented as means ± SEM. two-tailed Student’s t test; ns: no significance; ***P < 0.001. H The curvi linear velocity of sperm from Ccdc146+/+, Ccdc146+/- and Ccdc146−/− mice (n = 3). Data are presented as means ± SEM. two-tailed Student’s t test; ns: no significance; ***P < 0.001. I The amplitude of lateral head displacement of sperm from Ccdc146+/+, Ccdc146+/- and Ccdc146−/− mice. (n = 3). Data are presented as means ± SEM. two-tailed Student’s t test; ns: no significance; ***P < 0.001
CCDC146 is required for spermiogenesis
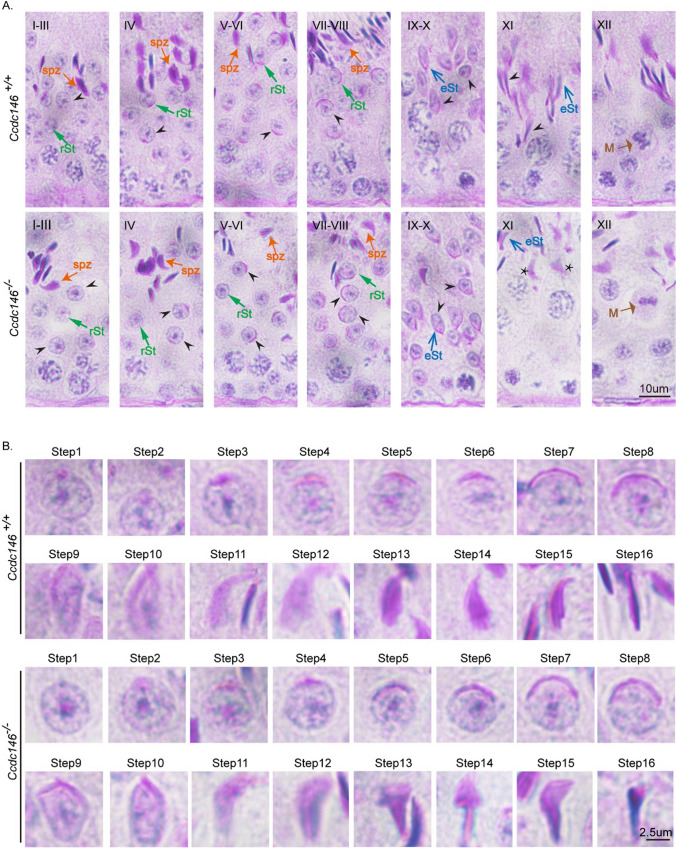
To address the rationale of knockout of Ccdc146 causing MMAF, we conducted Periodic acid–Schiff (PAS) staining to determine the stages of spermiogenesis in Ccdc146−/− and Ccdc146+/+ testes. The most prominent defects were observed in the spermatids at the stages of spermiogenesis, where abnormally elongated and constricted sperm head shapes were identified (Fig. 3A). Spermatid elongation is characterized by the formation of a transient microtubular structure, the manchette. The manchette microtubules (MTs) are connected to the perinuclear ring, which is closely associated with the acrosomal border, and extend progressively towards the caudal region, thus forming a skirt-like structure surrounding the nucleus [75, 76]. The manchette is essential for nuclear reshaping and is considered as a track for delivering cargos to assemble HTCA and sperm tail [77]. To clarify the detailed morphological effects of the Ccdc146 mutation on the structure of sperm heads, we analyzed the process of sperm head shaping between Ccdc146−/− and Ccdc146+/+ mice (Fig. 3B). Notably, the acrosome and nucleus morphology in Ccdc146−/− spermatids was normal compared with Ccdc146+/+ spermatids from step 1 to step 8, Head shaping started at step 9 to step 10, and the morphology of the elongated Ccdc146−/− spermatid heads was normal, compared with that of Ccdc146+/+ mice, whereas abnormal club-shaped heads were seen in step 11 spermatids in Ccdc146−/− mice. This phenomenon became more apparent between step 11 and step 16 (Fig. 3B). In conclusion, these results indicate that CCDC146 is required for normal spermiogenesis.
Fig. 3.
Impaired spermiogenesis in Ccdc146−/− mice. (A) PAS-hematoxylin staining of Ccdc146−/− testis sections showed abnormal sperm nuclear shape. rSt round spermatid, spt spermatozoa, M meiotic spermatocyte, eSt elongating spermatid. The asterisk indicates abnormally elongated spermatids at stage XI in Ccdc146−/− mice. (B) PAS-hematoxylin staining of spermatids at different steps from Ccdc146+/+ and Ccdc146−/− mice. Scale bars: 10 μm (A); 2·5 μm (B)
CCDC146 is required for sperm flagellum biogenesis and manchette function
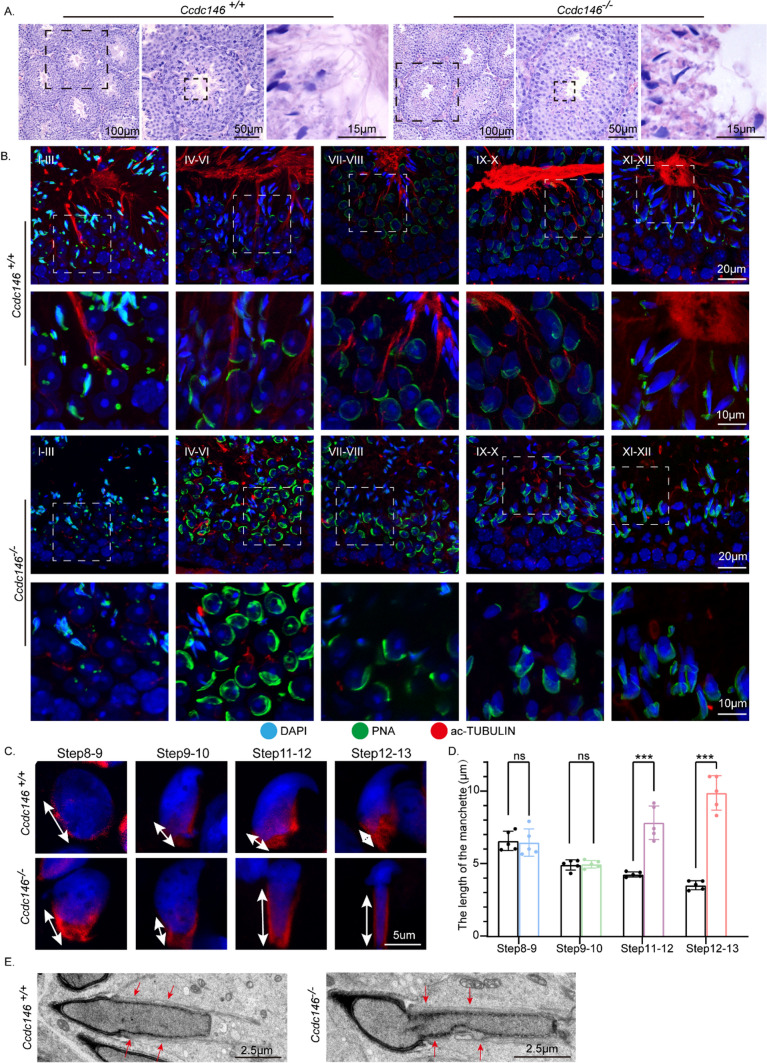
H&E staining was used to further observe the morphological changes of the seminiferous tubules. The seminiferous tubules of Ccdc146+/+ mice had a tubular lumen with flagella appearing from the developing spermatids. In contrast, the flagella were abnormal in the seminiferous tubules of Ccdc146−/− mice (Fig. 4A). These observations suggest that CCDC146 plays a vital role in flagellum biogenesis. To determine the causes of the flagellum defects in Ccdc146−/− mice, we investigated the effect of Ccdc146 knockout on flagellum biogenesis using the antibody against acetylated tubulin, a flagellum-specific marker [78]. Unlike the well-defined flagellum of the control group, the axoneme was absent in step 2–3 spermatids in Ccdc146−/− mice (Fig. 4B). In step 4–6, abnormally formed flagella were found in Ccdc146−/− testis sections (Fig. 4B). The presence of long and abnormal spermatid heads suggested defects in the function of the manchette, which is involved in sperm head shaping. Immunofluorescence staining for α/β-tubulin antibody showed that manchette formation was normal in step 8 to step 10 spermatids in Ccdc146−/− mice, while the manchettes of Ccdc146−/− mice spermatids were abnormally longer than WT controls during step 11 to step 13 (Fig. 4C, D). We performed transmission electron microscopy to study the organization of the sperm manchette in detail in Ccdc146−/− mice. During the chromatin condensation period starting from step 11 spermatids, the manchette of Ccdc146−/− mice appeared abnormally long, and the perinuclear ring constricted the sperm nucleus, causing severe defects in sperm head formation (Fig. 4C, D). The SOX9 staining [79] showed that the Sertoli cells were not affected by Ccdc146 knockout (Supplement Fig. S2A). We also detected the trachea cilia in Ccdc146−/− mice, and we found that it is similar to that of the Ccdc146+/+ mice (Supplement Fig. S3A). Thus, Ccdc146 knockout causes severe defects in sperm flagellum biogenesis and manchette function.
Fig. 4.
Disorganized flagellum and manchette in Ccdc146−/− spermatids. A The histology of the seminiferous tubules from Ccdc146+/+ and Ccdc146−/− male mice. B Immunofluorescence analysis of Ac-TUBULIN (red) and PNA lectin (green) to identify sperm flagellum biogenesis. C Abnormal manchette elongation in Ccdc146−/− spermatids. Spermatids from different manchette-containing steps were stained with α/β-TUBULIN antibody (red) to visualize the manchette. Ccdc146−/− spermatids displayed abnormal elongation of the manchette. D Statistical analysis of the length of the manchette. The color columns indicate the manchette length of the Ccdc146−/− spermatids in different steps, while the black columns indicate the manchette length of the Ccdc146+/+ spermatids. Data are presented as means ± SEM. two-tailed Student’s t test; ns no significance; ***P < 0.001. E TEM revealed that the manchette of elongating spermatids (steps 9–11) from Ccdc146.−/− mice were ectopically placed. The red arrowhead indicates the abnormal manchette. Scale bars: 100 μm (A); 50 μm (A); 20 μm (B); 15 μm (A); 10 μm (B); 5 μm (C); 2·5 μm (E)
ODFs and axonemes were abnormally organized in the Ccdc146−/− spermatozoa
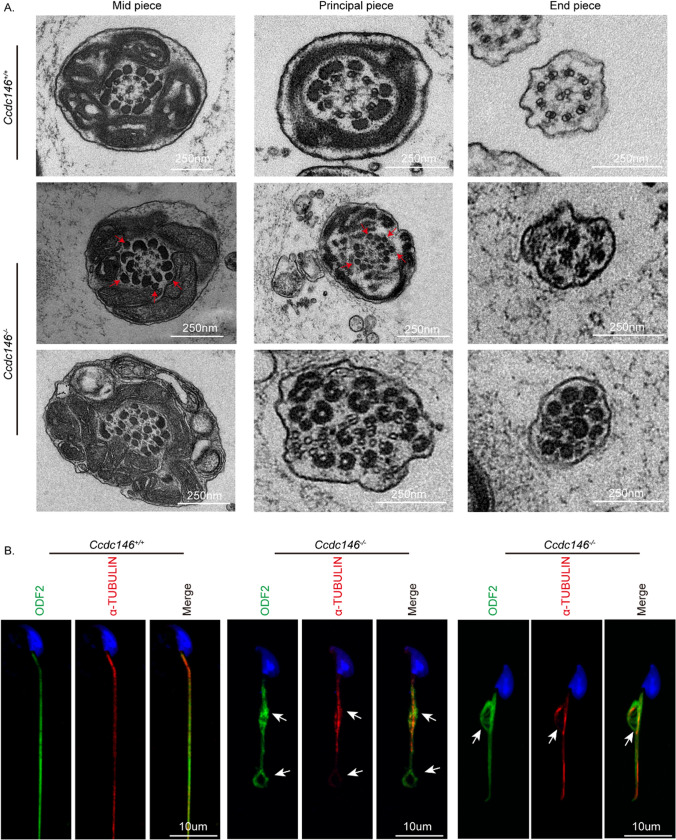
To dissect the mechanism underlying the defects of flagellum biogenesis, we sought to investigate whether the knockout of Ccdc146 affected the internal structure of mature sperm flagella. We performed transmission electron microscopy in the epididymis and observed that the ODFs and axonemes were abnormally organized in the Ccdc146−/− spermatozoa. Intriguingly, we noticed that there were several extra ODFs in the flagellum of Ccdc146−/− mice (Fig. 5A). Then, we used immunofluorescence to analyze the localization of ODF2 and tubulin in epididymal spermatozoa. ODF2 co-localized with α-tubulin in the midpiece and principal piece of Ccdc146+/+ sperm tails. In contrast, it displayed abnormal signals in the short tails of Ccdc146−/− spermatozoa (Fig. 5B, white arrow).
Fig. 5.
Impaired ODF transportation in Ccdc146−/− spermatids. A Cross-sections of Ccdc146−/− sperm tails revealed the disorganization of axonemal microtubules and tail accessory structures. Red arrowheads indicate an increase in outer dense fibers. B Immunofluorescence of ODF2 (green) and α-TUBULIN (red) in spermatids from Ccdc146+/+ and Ccdc146−/− mice. Nuclei were stained with DAPI (blue). Scale bars: 250 nm (A); 10 μm (B)
To reveal how CCDC146 affects ODF transport, we co-transfected with pCSII-MYC-Ccdc146, pEGFP-C1-Odf1, pCDNA-HA-Odf2, and pEGFP-C1-Odf4 in HEK293T cells. Interestingly, we found that ODF1 and ODF2 were not co-immunoprecipitated with CCDC146-, nor the ODF family protein ODF4 (Supplement Fig. S4A–D). The above findings reveal that CCDC146 may be involved in ODF assembly through indirect ways. This raises a fundamental question of whether CCDC146 indirectly causes ODF assembly abnormalities by affecting other proteins interacting with ODFs.
Previous studies have indicated that CCDC38 interacts with IFT88 and ODF2, and the knockout of Ccdc38 reduces the transport of ODF2 to the flagellum [59]. We also detected the interaction between the intraflagellar transport (IFT) complexes and CCDC146. Our findings indicated that CCDC146 can interact with IFT88 and IFT20, suggesting its potential involvement in the IFT pathways. (Supplement Fig. S4E, F). In addition, it has been reported that CCDC42 can interact with ODF1 and ODF2, and is involved in sperm flagellum biogenesis [71, 73]. We wondered whether CCDC146 affects ODF transportation by interacting with CCDC38 and CCDC42.
CCDC146 interacts with CCDC38 and CCDC42
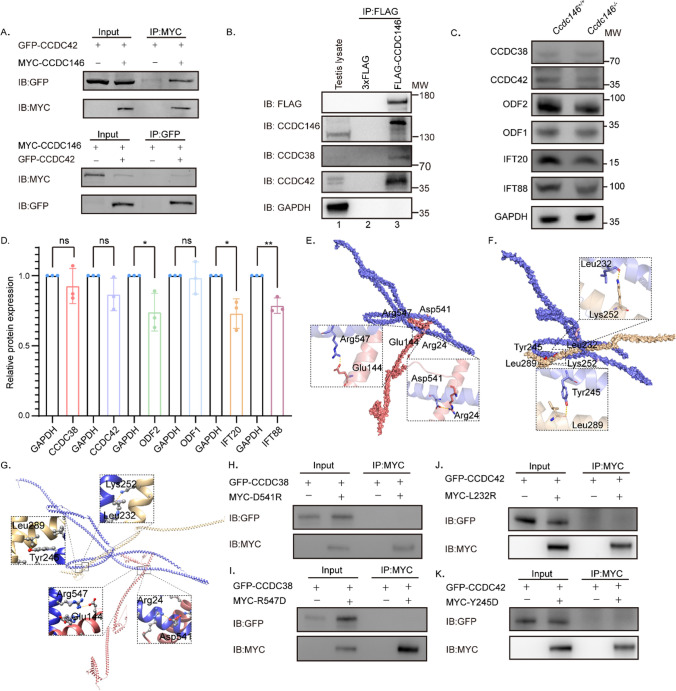
pCSII-MYC-Ccdc146 was then expressed in HEK293T cells and followed by immunoprecipitation experiments using the anti-MYC antibody, demonstrating that CCDC146 co-immunoprecipitated with CCDC38 and CCDC42 (Fig. 1B, Fig. 6A). The interaction between CCDC146 and CCDC38 or CCDC42 can also be detected in the mice testes (Fig. 6B). To further examine their relationship, we performed immunoblotting of CCDC42, CCDC38, ODF2, IFT88, IFT20, and ODF1 in the Ccdc146+/+ and Ccdc146−/− mice testes (Fig. 6C, Supplement Fig. S5).
Fig. 6.
CCDC146 interacts with CCDC38 and CCDC42. A CCDC146 interacted with CCDC42. pCSII-MYC-Ccdc146 were transfected into HEK293T cells with pEGFP-C1-Ccdc42 48 hours after transfection; cells were collected for immunoprecipitation with anti-MYC, and detected by anti-GFP or anti-MYC antibodies, respectively. B CCDC146 interacted with CCDC38 and CCDC42 in testis. Co-IP of CCDC38 and CCDC42 with FLAG-CCDC146 from testis lysate using anti-FLAG magnetic beads, followed by western blotting with anti-CCDC38, anti-CCDC42, anti-CCDC146 and anti-FLAG (CCDC146) antibodies. C, D Western blots showing CCDC38, CCDC42, ODF2, IFT88, IFT20, and ODF1 protein levels in lysates from Ccdc146+/+ and Ccdc146−/− mice testes. GAPDH served as a loading control. ODF2, IFT88, and IFT20 protein levels were reduced in Ccdc146−/− testes compared with Ccdc146+/+ testes. Data are presented as means ± SEM. two-tailed Student’s t test; ns: no significance; *P < 0·1; **P < 0·01. E Binding mode of CCDC38 on the CCDC146 predicted by docking. Detailed interaction network between CCDC38 and CCDC146. Key residues of CCDC146 (blue) and CCDC38 (light red) are displayed as sticks. H-bonds are displayed in dash lines. F Binding mode of CCDC42 on the CCDC146 predicted by docking. Detailed interaction network between CCDC42 and CCDC146. Key residues of CCDC146 (blue) and CCDC38 (light yellow) are displayed as sticks. H-bonds are displayed in yellow dashed lines. G Alignment of the mode of CCDC38 and CCDC42 on the CCDC146 predicted by docking. H–I Interactions between CCDC38 and WT or D541, or R547 mutants of truncated CCDC146 were detected by Co-IP assays. J–K Co-IP assays detected interactions between CCDC42 and WT or L232, or Y245 mutants of truncated CCDC146
We found that ODF2, IFT88, and IFT20 expression levels were significantly reduced in Ccdc146−/− testicular lysate (Fig. 6C, D) compared to that of the Ccdc146+/+. Notably, we did not observe any significant changes in CCDC38, CCDC42, and ODF1 levels in the Ccdc146−/− testicular lysate. These results indicated that CCDC146 might participate in the ODF2 transport via the IFT pathways.
To further examine the relationship between CCDC146 and CCDC38 or CCDC42, we performed a docking experiment using HDOCK [80] to investigate the binding mode between CCDC146 and CCDC38 or CCDC42. The R547 and D541 of CCDC146 formed hydrogen-bonding interactions with E144 and R24 of CCDC38, respectively (Fig. 6E). Moreover, the Y245 and L232 of CCDC146 were also able to form hydrogen-bonding interactions with L289 and K252 of CCDC42, respectively (Fig. 6F). To explore the relationship between them, we combined the two docking modes using Chimera [81]. We found that the binding region of CCDC146 and CCDC38 is independent of the binding region of CCDC42 and CCDC146. Additionally, with regard to their spatial relationship, the binding of CCDC42–CCDC146 will not interfere with the binding of CCDC38–CCDC146 in the 3D structure (Fig. 6G). To elucidate the role of interaction residues on CCDC146, we changed the properties of the aforementioned binding sites to validate the interactions between CCDC146 and CCDC38 or CCDC42 by generating point mutations in CCDC146. We replaced leucine with arginine (L232R), tyrosine with aspartic acid (Y245D), aspartic acid with arginine (D541R), and arginine with aspartic acid (R547D), respectively. CCDC38 and CCDC146 mutations (D541R, R547D) were expressed in HEK293T cells and followed by immunoprecipitation experiments using the anti-MYC antibody, demonstrating that CCDC146 cannot co-immunoprecipitate with CCDC38 when the hydrogen-bonding interactions were disrupted (Fig. 6H, I). Co-IP assays also showed no interaction between CCDC42 and the CCDC146 mutations (L232R, Y245D), confirming that L232R and Y245D on the CCDC146 were essential for binding CCDC42 (Fig. 6J, K).
Further, we investigated the sequence and structural characteristics of CCDC146. We found that CCDC146 is evolutionarily conserved from bony fish to human beings (Supplement Fig. S6A, B). Only one potential functional domain of CCDC146 (150–920) was predicted by the NCBI’s Conserved Domain Database (CDD), whereas the other parts of CCDC146 remained unknown (Supplement Fig. S6C). For this reason, MEME was employed to predict the conserved motifs of CCDC146 in seven species (Homo sapiens, Xenopus laevis, Salmo salar, Alligator sinensis, Danio rerio, Mus musculus, Acanthaster planci). Though the pairwise similarities of some species were low (Supplement Fig. S6D), 10 motifs were identified. It is worth noting that motif 8 (E213-D271) was conserved in all species and overlapped with the interaction region of CCDC42 and CCDC146 (L232 and Y245), while motif 6 (K493-I591), which contains the interaction region of CCDC38 and CCDC146 (D541R, R547D), was only conserved in Homo sapiens, Xenopus laevis, Alligator sinensis, Mus musculus and Acanthaster planci (Supplement Fig. S6E).
CCDC146 mediated ODF2 transportation by CCDC42 and CCDC38 in different ways
To reveal how CCDC146 influences CCDC42 and CCDC38, we performed immunofluorescence co-staining of them. We used NIH3T3 cells to explore the relationship of CCDC146, CCDC42, and CCDC38 during ciliogenesis because the antibody against CCDC42 does not function for immunofluorescence. Near the centrosome, an overlapping immunostaining pattern was seen in NIH3T3 cells transiently expressing GFP-CCDC38, MYC-CCDC146, and GFP-CCDC42 (Fig. 7A–C, Supplement Fig. S7A–C). After serum starvation, CCDC38 co-localized with Ac-TUBULIN in the cilia, while CCDC146 and CCDC42 still localized to the base of cilia (Fig. 7D–F, Supplement Fig. S7D–F). It demonstrated that CCDC38 would be transported to cilia during ciliogenesis. CCDC146 and CCDC42, on the other hand, would not be transported to the cilia.
Fig. 7.
CCDC146 may facilitate ODF2 transportation by interacting with CCDC42 and CCDC38. A-C CCDC146 co-localized with γ-TUBULIN, CCDC38, and CCDC42 in NIH3T3 cells before serum starvation. pCSII-MYC-Ccdc146 and pEGFP-C1-Ccdc38 or pEGFP-C1-Ccdc42 were co-transfected into NIH3T3 cells. 24 h after transfection, cells were fixed and stained with anti-MYC and anti-GFP antibodies, and the nucleus was stained with DAPI. D CCDC42 cannot co-localize with Ac-TUBULIN in NIH3T3 cells during ciliogenesis. pEGFP-C1-Ccdc42 were transfected into NIH3T3 cells. After serum starvation, cells were fixed and stained with anti-GFP and anti-Ac-TUBULIN antibodies, and the nucleus was stained with DAPI. E CCDC38 co-localized with Ac-TUBULIN in NIH3T3 cells during ciliogenesis. pEGFP-C1-Ccdc38 were transfected into NIH3T3 cells. After serum starvation, cells were fixed and stained with anti-GFP and anti-Ac-TUBULIN antibodies, and the nucleus was stained with DAPI. F CCDC146 cannot co-localize with Ac-TUBULIN in NIH3T3 cells during ciliogenesis. pCSII-MYC-Ccdc146 were transfected into NIH3T3 cells. After serum starvation, cells were fixed and stained with anti-MYC and anti-Ac-TUBULIN antibodies, and the nucleus was stained with DAPI. G The immunofluorescence of CCDC146 in Ccdc146+/+ and Ccdc146−/− mice. Testis germ cells were stained with anti-CCDC146 antibody, and the nucleus was stained with DAPI. H The immunofluorescence of CCDC146 in Ccdc146+/+ and Ccdc146−/− mice. Spermatozoa from cauda of the epididymis were stained with anti-CCDC146 antibody, and the nucleus was stained with DAPI. Scale bars: 2·5 μm (A-F); 5 μm (G); 10 μm (A-F, H). I CCDC146 may interact with microtubule proteins and microtubule inner proteins. Co-IP of sperm flagellum proteins with FLAG-CCDC146 from testis lysate using anti-FLAG magnetic beads, followed by western blotting with anti-α-TUBULIN, anti-β-TUBULIN, anti-TEKT2, anti-SPACA9, anti-DYNLL2, anti-PPIL6, anti-NME5, anti-SPAG6, anti-SPAG16, and anti-FLAG (CCDC146) antibodies. MT microtubule; MIP microtubule inner protein; RS radial spoke; CPA central-pair apparatus
It was reported that CCDC38 localized to the manchette, the sperm connecting piece, and the sperm tail during spermatogenesis [72], while CCDC146 was localized to the sperm connecting piece and sperm tail during spermiogenesis. (Fig. 7G, Supplement Fig. S7G) and the flagellum in spermatozoa from cauda of the epididymis (Fig. 7H, Supplement Fig. S7H). To further investigate the localization of CCDC146 in the sperm flagellum, we purified FLAG-tagged CCDC146 protein and performed Co-IP of sperm flagellum proteins with FLAG-CCDC146 using anti-FLAG magnetic beads. We found that CCDC146 may interact with microtubule proteins (α-TUBULIN and β-TUBULIN) and microtubule inner proteins (TEKT2 and SPACA9) [82] (Fig. 7I). However, CCDC146 can not immunoprecipitated some known radial spoke proteins (DYNLL2, PPIL6, NME5) [83, 84] or central-pair proteins (SPAG6 and SPAG16) [85, 86] (Fig. 7I). Previous studies have shown that the Ccdc42 homolog localizes to the base of motile cilia in T. thermophila [71]. Loss of Ccdc42 in mice is associated with male infertility, and an odd number and positioning of the HTCA, and the mutant sperm lack an attached flagellum [71]. Therefore, we speculate that CCDC146 may play essential roles in gate and cargo transportation during flagellum biogenesis.
Discussion
In this study, we have identified the essential role of CCDC146 in spermiogenesis and male fertility by generating Ccdc146–/– mice. Our study demonstrated that the Ccdc146–/– mice display MMAF phenotype. Ccdc146–/– mice sperm showed abnormal morphology, including a misshapen head and flagella assembly failure. The fact that Ccdc146 is not expressed until about 21 days of age suggests that the gene is not necessary for early cell division but instead plays a role in morphogenesis in forming mature sperms (Fig. 1E).
Proper assembly of the sperm axoneme and accessory structures is essential for sperm motility and fertilization [55, 87–89]. Several genes have been recently reported to be associated with defective sperm flagellum development. For example, subtle ultrastructure disorganizations were reported in Tektin4 knockout mice [90]. Axoneme defects were observed in Odf2 null mice [91]. Akap4, Fsip2, and Cabyr deficiency in mice resulted in a lack of fibrous sheath [92–94]. The ablation of Ropn1 impaired the structure of the sperm principal piece [95]. However, there are few studies on ODF assembly and transport, and the molecular mechanisms causing the failure of ODF transportation remain largely unknown. Previous studies have indicated that CCDC38 interacts with IFT88 and ODF2, and the knockout of Ccdc38 influences the transportation of ODF2 to the flagellum [59]. In addition, it has been reported that CCDC42 can interact with ODF1 and ODF2 and is involved in sperm flagellum biogenesis [71, 73]. It has been reported that CCDC38 localizes to the manchette, the sperm connecting piece, and the sperm tail during spermiogenesis [59]. Loss of Ccdc42 in mice is associated with male infertility due to absent flagellum. Our study found that CCDC146 was located in the sperm connecting piece and sperm tail at step 13–16 during spermatogenesis. We further validated the interaction between CCDC146, CCDC38, and CCDC42 (Fig. 1B, Fig. 6A–B). CCDC146 was also detected to interact with intraflagellar transport (IFT) complexes IFT88 and IFT20. Additionally, we found that the levels of ODF2, IFT88, and IFT20 were significantly reduced (Fig. 6C), while CCDC38, CCDC42, and ODF1 were not affected (Fig. 6D). These results indicated that CCDC146 might participate in the transportations of some proteins via IFT pathways.
Assembly of sperm flagellum structures requires intramanchette transport (IMT) and intraflagellar transport (IFT) to the assembly sites [75, 96, 97]. Cargo proteins are transported through the manchette by IMT to the base of the sperm flagellum, and the developing sperm flagellum by IFT [98]. Formation of HTCA and sperm tail requires many proteins that are presumably transported by manchette. The manchette is crucial to nuclear remodeling and is believed to be the cargo transport rail for assembling the HTCA and sperm tails [71]. The previous study has posed that Ccdc42 may be a passenger protein transported along the manchette from the acrosome-acroplaxome complex to the HTCA. It may then determine the correct number, placement, and stability of the HTCA. Our study found that CCDC38 and CCDC42 could interact with different conserved motifs of CCDC146. In this scenario, we speculate that CCDC146 cooperating with both CCDC38 and CCDC42 facilitates the transportation of some proteins along the intraflagellar transport (IFT) pathway, promoting their assembly into the flagella.
To examine the conservation of CCDC146, we obtained its homologues from several representative species, such as Mammals, Bird, Reptile, Amphibian, Chondrichthyan, and Bony fish, then constructed the phylogenetic tree of CCDC146 with MEGA [99] (Supplement Fig. S6A) and calculated the evolutionary rate (ω) per codon sites of CCDC146 using HyPhy (Supplement Fig. S6B) [100]. According to the phylogenetic tree, we found that CCDC146 is highly conserved in vertebrate species. Thus, the function of CCDC146 might also be conserved in these species. Recently, a study found that there are two patients with homozygous truncating variants in the CCDC146 (coiled-coil domain containing 146) gene [101], which further proved that CCDC146 is conservative in physiological function. Besides, that study also found CCDC146 localized to the flagellum of mature spermatozoa in both humans and mice. However we found that the Ccdc146 gene is highly expressed in the testis in mice. The homologue of this gene (CCDC146) is also expressed in other tissues or organs in human beings, such as the respiratory system, liver, ovary, and other tissues according to the human protein atlas (https://www.proteinatlas.org). The phylogenetic tree analysis also showed that this protein is diversified from mouse to human. These results suggest that the function of human CCDC146 may not be similar to mouse Ccdc146. If there were CCDC146 mutations in human beings, the patients might have more syndromes rather than MMAF only in mice. In addition, the harmful allele had a higher chance of being passed on to the next generation than those genes that affect both male and female fertility, because Ccdc146 knockout female mice showed no defects in fertility (Fig. 1J, H). Our results suggest that the testis highly expressed gene CCDC146 is essential for sperm flagellum biogenesis and male fertility.
Methods
Plasmids
Mouse Ccdc146 were obtained from mouse testis cDNA and were cloned into the pCSII-MYC vector using the Clone Express Ultra One Step Cloning Kit (C115, Vazyme). Mouse Ccdc38, Ccdc42, Odf1, Odf4, and Ift88 were obtained from mouse testis cDNA, and it was cloned into the pEGFP-C1 vector. Mouse Odf2 was obtained from mouse testis cDNA, and it was cloned into the pCDNA-HA-3.0 vector. Mouse Ift20 was obtained from mouse testis cDNA, and it was cloned into the pRK vector. pCDNA-HA-3.0vector was obtained from Addgene (128034). pCSII-MYC, pRK and pEGFP-C1 vectors were obtained from BioVector NTCC.
Animals
The mice Ccdc146 gene is 2934 bp and contains 19 exons. The knockout mice of Ccdc146 were generated by the CRISPR-Cas9 system from Cyagen Biosciences. The genotyping primers for knockout were: F1: 5’-TCTTACCCTCTGTCAGGTCTCC-3’, R1: 5’-TGGGTTTTAGAATCACCACTGCTC-3’, and for WT mice, the specific primers were: F1: 5’-TCTTACCCTCTGTCAGGTCTCC-3’, R2: 5’ GCTGGGTATCACAGAGTAGTTCC-3’.
All the animal experiments were performed according to approved institutional animal care and use committee (IACUC) protocols (# 08–133) of the Institute of Zoology, Chinese Academy of Sciences.
Antibodies
The following primary antibodies were used for immunofluorescence (IF) and western blotting (WB): mouse anti-GFP (M20004, Abmart; 1:1000 for WB), rabbit anti-MYC (BE2011, EASYBIO; 1:1000 for WB), anti-ODF2 (12058-1-AP, Proteintech; 1:500 for WB, 1:200 for IF), anti-ODF1 (24736-1-AP, Proteintech; 1:1000 for WB), mouse anti-α-Tubulin antibody (AC012, ABclonal; 1:1000 for WB, 1:100 for IF), rabbit anti-β-Tubulin antibody (10068-1-AP, Proteintech; 1:1000 for WB), mouse anti-α/β-Tubulin antibody (ab44928, Abcam; 1:100 for IF), mouse anti-GAPDH antibody (AC002, ABclonal; 1:10,000 for WB), mouse anti-Ac-Tubulin antibody (T7451, Sigma-Aldrich; 1:200 for IF), mouse anti-NME5 antibody (12923-1-AP, Proteintech; 1:1000 for WB), rabbit anti-DYNLL2 antibody (16811-1-AP, Proteintech; 1:1000 for WB), rabbit anti-TEKT2 antibody (13518-1-AP, Proteintech; 1:1000 for WB), rabbit anti-SPACA9 antibody (26034-1-AP, Proteintech; 1:1000 for WB), rabbit anti-PPIL6 antibody (17452-1-AP, Proteintech; 1:1000 for WB), rabbit anti-NME5 antibody (12923-1-AP, Proteintech; 1:1000 for WB), mouse anti-SPAG6 antibody (H00008382-M04, Abnova; 1:1000 for WB), mouse anti-SPAG16 antibody (H00079582-M01, Abnova; 1:1000 for WB), rabbit anti-CCDC38 (generated by Dia-an Biotechnology, Wuhan, China; 1:500 for WB), rabbit anti-CCDC146 (generated by Dia-an Biotechnology, Wuhan, China; 1:200 for WB, 1:50 for IF). The Alexa Fluor, 488 conjugate of lectin PNA (1:400, L21409, Thermo Fisher Scientific), was used for immunofluorescence. The following secondary antibodies were used: goat anti-rabbit FITC (1:200, ZF-0311, http://www.zsbio.com/), goat anti-rabbit TRITC (1:200, ZF-0316, Zhong Shan Jin Qiao), goat anti-mouse FITC (1:200, ZF-0312, Zhong Shan Jin Qiao), and goat anti-mouse TRITC (1:200, ZF0313, Zhong Shan Jin Qiao).
Western blotting
As previously reported [102], the tunica albuginea of the testis was peeled and added to RIPA buffer supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF) and protease inhibitor cocktail (PIC; Roche Diagnostics, 04693132001). The solution was sonicated transiently and then placed on ice for 30 min. The samples were centrifuged at 13,523 g for 15 min at 4 °C. Next, the supernatants were collected in new tubes. Protein lysates were electrophoresed and electrotransferred to a nitrocellulose membrane. The membrane was then incubated with a primary antibody followed by a secondary antibody. Finally, the membrane was scanned using an Odyssey infrared imager (LI-COR Biosciences; RRID: SCR_014579).
Co-immunoprecipitation
Transfected cells were lysed in a lysis buffer (50 mM HEPES, PH 7·4, 250 mM NaCl, 0·1% NP-40 containing PIC and PMSF) on ice for 30 min and centrifugated at 12,000 rpm at 4℃ for 15 min, cell lysates were incubated with primary antibody overnight at 4℃, next incubated with protein A for 2 h at 4℃, then washed three times with lysed buffer and subjected to immunoblotting analysis.
Co-immunoprecipitation of endogenous proteins
The purified FLAG-CCDC146 proteins were incubated with Anti-FLAG M2 Magnetic Beads(Sigma, M8823) in ELB IP buffer at 4 °C for 3 h. The beads were washed 3 times with ELB IP buffer. Then, testis lysates were added to the washed FLAG-CCDC146-bound beads and 3xFLAG bound beads, incubating at 4 °C overnight. The beads were washed at least 5 times with ELB IP buffer. The bound proteins were eluted by adding SDS loading buffer, followed by SDS-PAGE and western blotting with antibodies against FLAG, CCDC146, CCDC38, and CCDC42, as well as α-Tubulin, β-Tubulin, TEKT2, SPACA9, DYNLL2, PPIL6, NME5, SPAG6, and SPAG16.
Protein expression and purification
The mouse Ccdc146 open reading frame fusion with FLAG tag on its N-terminal was subcloned between the EcoRI and SalI sites of the pET28a vector, and then the resulting plasmid was introduced into BL21 Rosetta (DE3) competent cells and grown in Terrific Broth at 37 °C to an optical density of 0.8. The temperature of the culture was then shifted to 16 °C and cells were induced with 1 mM isopropyl-D-thiogalactoside (IPTG) for 20 h. Cells were collected by centrifugation at 4℃ and resuspended in ice-cold lysis buffer (pH 7.4, 20 mM Tris, 300 mM NaCl, 10 mM imidazole, and 20% glycerol for hexahistidine (6xHis)-tagged fusion protein) with 1 mM Phenylmethylsulfonyl fluorid (PMSF) and 1 × Protease Inhibitor Cocktail. After lysis by sonication for 10 min on ice and high-speed centrifugation of lysates for 40 min at 4℃, the supernatant was incubated with Ni Sepharose 6 Fast Flow (GE Healthcare) for 2 h at 4 °C. The resin was washed and the protein was eluted using the lysis buffer supplemented with 500 mM imidazole. Purified proteins were dialyzed in the ELB IP buffer (50 mM HEPES, PH 7·4, 250 mM NaCl, 0·1% NP-40).
Sperm motility and sperm count assays
The cauda epididymis was isolated from 8 weeks of mice. Sperm were released in phosphate-buffered saline (PBS, Gibco, C14190500BT) from the incisions of the cauda epididymis for 10 min at 37 ℃. And then the swim-up suspension was used for the analysis of sperm motility with a microscope through a 20 × phase objective. Viewing areas in each chamber were imaged using a CCD camera. The samples were analyzed via computer-assisted semen analysis (CASA) using the Minitube Sperm Vision Digital Semen Evaluation System (12,500/1300, Minitube Group, Tiefenbach, Germany) and were also analyzed by a computer-aided semen analysis system (CASA, Song Jing Tian Lun Biotechnology Co., Ltd., Nanning, China). The incubated sperm solution was then diluted 1:100 and sperm number was counted with a hemocytometer.
Tissue collection and histological analysis
As previously reported [102], the testes were dissected after euthanasia, and fixed with Bouin’s fixative for 24 h at 4 ℃, the testes were dehydrated with graded ethanol and embedded in paraffin. The 5 μm sections were cut and covered on glass slides. Sections were stained with H&E and PAS for histological analysis after deparaffinization.
Transmission electron microscopy
The methods were as reported previously with some modifications [102]. The testis from WT and Ccdc146 depletion mice testis and epididymis were dissected and fixed in 2·5% glutaraldehyde in 0·1 M cacodylate buffer at 4℃ overnight. After washing in 0·1 M cacodylate buffer, samples were cut into small pieces and immersed in 1% OsO4 for 1 h at 4℃. Samples were dehydrated through a graded acetone series and embedded in resin for staining. Ultrathin sections were cut and stained with uranyl acetate and lead citrate; images were acquired and analyzed using a JEM-1400 transmission electron microscope.
Scanning electron microscopy
The sperm were released from epididymis in HTF at 37℃ 15 min, centrifugated for 5 min at 500 g, then washed twice with PB, fixed in 2·5% glutaraldehyde solution overnight, and dehydrated in graded ethanol, subjected to drying and coated with gold. The images were acquired and analyzed using SU8010 scanning electron microscope.
Immunofluorescence
The testis albuginea was peeled and incubated with collagenase IV and hyaluronidase in PBS for 15 min at 37℃, then washed twice with PBS. Next, fixed with 4% PFA for 5 min, then coated on slide glass to dry out. The slides were washed with PBS three times, treated with 0.5% TritonX-100 for 5 min, and blocked with 5% BSA for 30 min. Added the primary antibodies and incubated at 4℃ overnight, followed by incubating with second antibody and DAPI. The images were taken using LSM880 and Sp8 microscopes.
Protein–protein docking
3D structures of CCDC146, CCDC38 and CCDC42 were obtained from UniPort (AF-E9Q9F7-F1, AF-Q8CDN8-F1, and AF-Q5SV66-F1). Protein–protein docking for CCDC146 and CCDC38, as well as CCDC146 and CCDC42, were performed using the HDOCK webserver (http://hdock.phys.hust.edu.cn/) [80]. The details related to the interaction are supplemented by pyMOL [103].
Domain and conserved motifs analysis
The NCBI’s Conserved Domain Database (CDD) (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) was used to predict protein domains of CCDC146. MEME (Multiple EM for Motif Elicitation) motif discovery tools[104] (http://meme-suite.org/tools/meme) were employed to predict conserved motifs of CCDC146 including seven species, Homo sapiens (NP_065930.2), Mus musculus (NP_083471.1), Xenopus laevis (XP_018108171.1), Salmo salar (XP_014010955.1), Alligator sinensis (XP_025048306.1), Danio rerio (NP_001076522.1) and Acanthaster planci (XP_022081518.1). Each motif’s minimum and maximum width were set to 30 aa and 100 aa, respectively. Each motif must be recognized in at least two sequences [105].
Statistical analysis
All data are presented as the mean ± SEM. The statistical significance of the differences between the mean values for the various genotypes was measured by Student’s t-tests with paired, 2-tailed distribution. The data were considered significant when the P-value was less than 0.05(*), 0.01(**), or 0.001(***).
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Jingnan Liang and Pengyan Xia for their help with transmission electron microscopy, Chunli Li for his help with scanning electron microscopy, and Guofeng Zhou for his help with the protein-protein docking experiment. This work was supported by the National Science Fund for Distinguished Young Scholars (81925015) and the National Natural Science Foundation of China (Grant No. 91649202).
Author contribution
All authors contributed to the study conception and design. YM, BW, and YC performed most of the experimental work of the study, analyzed data, and wrote the manuscript. SM, LW, Tingting Han, XL, and FY performed part of the experiments. WL, JZ, and CL designed the experiments, supervised the whole project, and revised the manuscript. All the authors read and approved the final manuscript.
Funding
This work was supported by the National Science Fund for Distinguished Young Scholars (Grant No. 81925015) and the National Natural Science Foundation of China (Grant No. 91649202).
Data availability
All the data supporting the findings of this study are available from the corresponding author upon reasonable request.
Declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
All the animal experiments were approved by the Animal Research Panel of the Committee on Research Practice of the University of the Chinese Academy of Sciences (Approval number: IOZ20180012). All of the animal experiments were performed according to approved institutional animal care and use committee (IACUC) protocols (#08–133) of the Institute of Zoology, Chinese Academy of Sciences. All the 8-week male mice were sacrificed by cervical dislocation before testes collection.
Consent to participate
Informed consent was obtained from all individual participants included in the study.
Consent for publication
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yanjie Ma and Bingbing Wu have contributed equally to this work.
Contributor Information
Chao Liu, Email: liuchao@gwcmc.org.
Jianguo Zhao, Email: zhaojg@ioz.ac.cn.
Wei Li, Email: leways@gwcmc.org.
References
- 1.Hwang YS, et al. Reconstitution of prospermatogonial specification in vitro from human induced pluripotent stem cells. Nat Commun. 2020;11(1):5656. doi: 10.1038/s41467-020-19350-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tüttelmann F, Ruckert C, Röpke A. Disorders of spermatogenesis: perspectives for novel genetic diagnostics after 20 years of unchanged routine. Med Gen. 2018;30(1):12–20. doi: 10.1007/s11825-018-0181-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tuttelmann F, Ruckert C, Ropke A. Disorders of spermatogenesis: perspectives for novel genetic diagnostics after 20 years of unchanged routine. Med Genet. 2018;30(1):12–20. doi: 10.1007/s11825-018-0181-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coutton C, et al. Teratozoospermia: spotlight on the main genetic actors in the human. Hum Reprod Update. 2015;21(4):455–485. doi: 10.1093/humupd/dmv020. [DOI] [PubMed] [Google Scholar]
- 5.Ray PF, et al. Genetic abnormalities leading to qualitative defects of sperm morphology or function. Clin Genet. 2017;91(2):217–232. doi: 10.1111/cge.12905. [DOI] [PubMed] [Google Scholar]
- 6.Jiao SY, Yang YH, Chen SR. Molecular genetics of infertility: loss-of-function mutations in humans and corresponding knockout/mutated mice. Hum Reprod Update. 2021;27(1):154–189. doi: 10.1093/humupd/dmaa034. [DOI] [PubMed] [Google Scholar]
- 7.Fawcett DW. The mammalian spermatozoon. Dev Biol. 1975;44(2):394–436. doi: 10.1016/0012-1606(75)90411-x. [DOI] [PubMed] [Google Scholar]
- 8.Russell LD, Russell J, MacGregor GR, Meistrich ML. Linkage of manchette microtubules to the nuclear envelope and observations of the role of the manchette in nuclear shaping during spermiogenesis in rodents. Am J Anat. 1991;192(2):97–120. doi: 10.1002/aja.1001920202. [DOI] [PubMed] [Google Scholar]
- 9.Eddy EM, Toshimori K, O'Brien DA. Fibrous sheath of mammalian spermatozoa. Microsc Res Tech. 2003;61(1):103–115. doi: 10.1002/jemt.10320. [DOI] [PubMed] [Google Scholar]
- 10.Miyata H, Morohoshi A, Ikawa M. Analysis of the sperm flagellar axoneme using gene-modified mice. Exp Anim. 2020;69(4):374–381. doi: 10.1538/expanim.20-0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao W, et al. Outer dense fibers stabilize the axoneme to maintain sperm motility. J Cell Mol Med. 2018;22(3):1755–1768. doi: 10.1111/jcmm.13457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baltz JM, Williams P, Cone RA. Dense fibers protect mammalian sperm against damage. Biol Reprod. 1990;43(3):485–491. doi: 10.1095/biolreprod43.3.485. [DOI] [PubMed] [Google Scholar]
- 13.Escalier D, Toure A. Morphological defects of sperm flagellum implicated in human male infertility. Med Sci. 2012;28(5):503–511. doi: 10.1051/medsci/2012285015. [DOI] [PubMed] [Google Scholar]
- 14.Touré A, et al. The genetic architecture of morphological abnormalities of the sperm tail. Hum Genet. 2021;140(1):21–42. doi: 10.1007/s00439-020-02113-x. [DOI] [PubMed] [Google Scholar]
- 15.Wang J, et al. Clinical detection, diagnosis and treatment of morphological abnormalities of sperm flagella: a review of literature. Front Genet. 2022;13:1034951. doi: 10.3389/fgene.2022.1034951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X, et al. Homozygous DNAH1 frameshift mutation causes multiple morphological anomalies of the sperm flagella in Chinese. Clin Genet. 2017;91(2):313–321. doi: 10.1111/cge.12857c. [DOI] [PubMed] [Google Scholar]
- 17.Gao Y, et al. Novel bi-allelic variants in DNAH2 cause severe asthenoteratozoospermia with multiple morphological abnormalities of the flagella. Reprod Biomed. 2021;42(5):963–972. doi: 10.1016/j.rbmo.2021.01.011. [DOI] [PubMed] [Google Scholar]
- 18.Lu S, et al. Bi-allelic variants in human WDR63 cause male infertility via abnormal inner dynein arms assembly. Cell Discov. 2021;7(1):110. doi: 10.1038/s41421-021-00327-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tu C, et al. Bi-allelic mutations of DNAH10 cause primary male infertility with asthenoteratozoospermia in humans and mice. Am J Hum Genet. 2021;108(8):1466–1477. doi: 10.1016/j.ajhg.2021.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tu C, et al. Identification of DNAH6 mutations in infertile men with multiple morphological abnormalities of the sperm flagella. Sci Rep. 2019;9(1):15864. doi: 10.1038/s41598-019-52436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu H, et al. DNALI1 deficiency causes male infertility with severe asthenozoospermia in humans and mice by disrupting the assembly of the flagellar inner dynein arms and fibrous sheath. Cell Death Dis. 2023;14(2):127. doi: 10.1038/s41419-023-05653-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen D, et al. A novel CCDC39 mutation causes multiple morphological abnormalities of the flagella in a primary ciliary dyskinesia patient. Reprod Biomed. 2021;43(5):920–930. doi: 10.1016/j.rbmo.2021.07.005. [DOI] [PubMed] [Google Scholar]
- 23.Xu Y, et al. Novel compound heterozygous variants in CCDC40 associated with primary ciliary dyskinesia and multiple morphological abnormalities of the sperm flagella. Pharmacogenomics Pers Med. 2022;15:341–350. doi: 10.2147/PGPM.S359821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cong J, et al. Homozygous mutations in CCDC34 cause male infertility with oligoasthenoteratozoospermia in humans and mice. J Med Genet. 2022;59(7):710–718. doi: 10.1136/jmedgenet-2021-107919. [DOI] [PubMed] [Google Scholar]
- 25.Sha Y, et al. Biallelic mutations of CFAP74 may cause human primary ciliary dyskinesia and MMAF phenotype. J Hum Genet. 2020;65(11):961–969. doi: 10.1038/s10038-020-0790-2. [DOI] [PubMed] [Google Scholar]
- 26.Liu S, et al. CFAP61 is required for sperm flagellum formation and male fertility in human and mouse. Development. 2021 doi: 10.1242/dev.199805. [DOI] [PubMed] [Google Scholar]
- 27.Liu C, et al. Deleterious variants in X-linked CFAP47 induce asthenoteratozoospermia and primary male infertility. Am J Hum Genet. 2021;108(2):309–323. doi: 10.1016/j.ajhg.2021.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He X, et al. Bi-allelic loss-of-function variants in CFAP58 cause flagellar axoneme and mitochondrial sheath defects and asthenoteratozoospermia in humans and mice. Am J Hum Genet. 2020;107(3):514–526. doi: 10.1016/j.ajhg.2020.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tian S, et al. Biallelic mutations in CFAP54 cause male infertility with severe MMAF and NOA. J Med Genet. 2023;60(8):827–834. doi: 10.1136/jmg-2022-108887. [DOI] [PubMed] [Google Scholar]
- 30.Lu Y, et al. The compound heterozygous mutations of CFAP65 cause multiple morphological abnormalities of sperm flagella in infertile men. QJM. 2013 doi: 10.1093/qjmed/hcad205. [DOI] [PubMed] [Google Scholar]
- 31.Beurois J, et al. CFAP70 mutations lead to male infertility due to severe astheno-teratozoospermia. A case report. Hum Reprod. 2019;34(10):2071–2079. doi: 10.1093/humrep/dez166. [DOI] [PubMed] [Google Scholar]
- 32.Martinez G, et al. Biallelic variants in MAATS1 encoding CFAP91, a calmodulin-associated and spoke-associated complex protein, cause severe astheno-teratozoospermia and male infertility. J Med Genet. 2020;57(10):708–716. doi: 10.1136/jmedgenet-2019-106775. [DOI] [PubMed] [Google Scholar]
- 33.Shen Q, et al. Bi-allelic truncating variants in CFAP206 cause male infertility in human and mouse. Hum Genet. 2021;140(9):1367–1377. doi: 10.1007/s00439-021-02313-z. [DOI] [PubMed] [Google Scholar]
- 34.Auguste Y, et al. Loss of calmodulin- and radial-spoke-associated complex protein CFAP251 leads to immotile spermatozoa lacking mitochondria and infertility in men. Am J Hum Genet. 2018;103(3):413–420. doi: 10.1016/j.ajhg.2018.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang S, et al. Biallelic mutations in CFAP43 and CFAP44 cause male infertility with multiple morphological abnormalities of the sperm flagella. Am J Hum Genet. 2017;100(6):854–864. doi: 10.1016/j.ajhg.2017.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dong FN, et al. Absence of CFAP69 causes male infertility due to multiple morphological abnormalities of the flagella in human and mouse. Am J Hum Genet. 2018;102(4):636–648. doi: 10.1016/j.ajhg.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lei C, et al. DRC1 deficiency caused primary ciliary dyskinesia and MMAF in a Chinese patient. J Hum Genet. 2022;67(4):197–201. doi: 10.1038/s10038-021-00985-z. [DOI] [PubMed] [Google Scholar]
- 38.Tu C, et al. Novel mutations in SPEF2 causing different defects between flagella and cilia bridge: the phenotypic link between MMAF and PCD. Hum Genet. 2020;139(2):257–271. doi: 10.1007/s00439-020-02110-0. [DOI] [PubMed] [Google Scholar]
- 39.Xu C, et al. Homozygous SPAG6 variants can induce nonsyndromic asthenoteratozoospermia with severe MMAF. Reprod Biol Endocrinol. 2022;20(1):41. doi: 10.1186/s12958-022-00916-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu Z-J, et al. Novel mutation in ODF2 causes multiple morphological abnormalities of the sperm flagella in an infertile male. Asian J Androl. 2022;24(5):463–472. doi: 10.4103/aja202183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shen Y, et al. Loss-of-function mutations in QRICH2 cause male infertility with multiple morphological abnormalities of the sperm flagella. Nat Commun. 2019;10(1):433. doi: 10.1038/s41467-018-08182-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coutton C, et al. Bi-allelic mutations in ARMC2 lead to severe astheno-teratozoospermia due to sperm flagellum malformations in humans and mice. Am J Hum Genet. 2019;104(2):331–340. doi: 10.1016/j.ajhg.2018.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martinez G, et al. Whole-exome sequencing identifies mutations in FSIP2 as a recurrent cause of multiple morphological abnormalities of the sperm flagella. Hum Reprod. 2018;33(10):1973–1984. doi: 10.1093/humrep/dey264. [DOI] [PubMed] [Google Scholar]
- 44.Liu C, et al. Homozygous variants in AKAP3 induce asthenoteratozoospermia and male infertility. J Med Genet. 2023;60(2):137–143. doi: 10.1136/jmedgenet-2021-108271. [DOI] [PubMed] [Google Scholar]
- 45.Lv M, et al. Homozygous mutations in DZIP1 can induce asthenoteratospermia with severe MMAF. J Med Genet. 2020;57(7):445–453. doi: 10.1136/jmedgenet-2019-106479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu W, et al. Bi-allelic mutations in TTC21A induce asthenoteratospermia in humans and mice. Am J Hum Genet. 2019;104(4):738–748. doi: 10.1016/j.ajhg.2019.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lorès P, et al. Mutations in TTC29, encoding an evolutionarily conserved axonemal protein, result in asthenozoospermia and male infertility. Am J Huma Genet. 2019;105(6):1148–1167. doi: 10.1016/j.ajhg.2019.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ni X, et al. A novel homozygous mutation in WDR19 induces disorganization of microtubules in sperm flagella and nonsyndromic asthenoteratospermia. J Reprod Genet. 2020;37(6):1431–1439. doi: 10.1007/s10815-020-01770-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lorès P, et al. A missense mutation in IFT74, encoding for an essential component for intraflagellar transport of Tubulin, causes asthenozoospermia and male infertility without clinical signs of Bardet-Biedl syndrome. Hum Genet. 2021;140(7):1031–1043. doi: 10.1007/s00439-021-02270-7. [DOI] [PubMed] [Google Scholar]
- 50.Martinez G, et al. New mutations in DNHD1 cause multiple morphological abnormalities of the sperm flagella. Int J Mol Sci. 2023 doi: 10.3390/ijms24032559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lorès P, et al. Homozygous missense mutation L673P in adenylate kinase 7 (AK7) leads to primary male infertility and multiple morphological anomalies of the flagella but not to primary ciliary dyskinesia. Hum Mol Genet. 2018;27(7):1196–1211. doi: 10.1093/hmg/ddy034. [DOI] [PubMed] [Google Scholar]
- 52.Guo T, et al. Bi-allelic BRWD1 variants cause male infertility with asthenoteratozoospermia and likely primary ciliary dyskinesia. Hum Genet. 2021;140(5):761–773. doi: 10.1007/s00439-020-02241-4. [DOI] [PubMed] [Google Scholar]
- 53.Ma H, et al. Novel frameshift mutation in STK33 is associated with asthenozoospermia and multiple morphological abnormalities of the flagella. Hum Mol Genet. 2021;30(21):1977–1984. doi: 10.1093/hmg/ddab165. [DOI] [PubMed] [Google Scholar]
- 54.Liu C, et al. Novel mutations in X-linked, USP26-induced asthenoteratozoospermia and male infertility. Cells. 2021 doi: 10.3390/cells10071594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Toure A, et al. The genetic architecture of morphological abnormalities of the sperm tail. Hum Genet. 2021;140(1):21–42. doi: 10.1007/s00439-020-02113-x. [DOI] [PubMed] [Google Scholar]
- 56.Priyanka PP, Yenugu S. Coiled-coil domain-containing (CCDC) proteins: functional roles in general and male reproductive physiology. Reprod Sci. 2021;28(10):2725–2734. doi: 10.1007/s43032-021-00595-2. [DOI] [PubMed] [Google Scholar]
- 57.Kaczmarek K, et al. Ccdc33, a predominantly testis-expressed gene, encodes a putative peroxisomal protein. Cytogenet Genome Res. 2009;126(3):243–252. doi: 10.1159/000251961. [DOI] [PubMed] [Google Scholar]
- 58.Song M-H, et al. KP-CoT-23 (CCDC83) is a novel immunogenic cancer/testis antigen in colon cancer. Int J Oncol. 2012;41(5):1820–1826. doi: 10.3892/ijo.2012.1601. [DOI] [PubMed] [Google Scholar]
- 59.Zhang R, et al. CCDC38 is required for sperm flagellum biogenesis and male fertility in mice. Development. 2022 doi: 10.1242/dev.200516. [DOI] [PubMed] [Google Scholar]
- 60.Chen J-b, et al. Expression characteristics of the Ccdc70 gene in the mouse testis during spermatogenesis. Zhonghua Nan Ke Xue. 2016;22(1):12–16. [PubMed] [Google Scholar]
- 61.Vandenbrouck Y, Pineau C, Lane L. The functionally unannotated proteome of human male tissues: a shared resource to uncover new protein functions associated with reproductive biology. J Proteome Res. 2020;19(12):4782–4794. doi: 10.1021/acs.jproteome.0c00516. [DOI] [PubMed] [Google Scholar]
- 62.Khan M, et al. The evolutionarily conserved genes: Tex37, Ccdc73, Prss55 and Nxt2 are dispensable for fertility in mice. Sci Reports. 2018;8(1):4975. doi: 10.1038/s41598-018-23176-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang T, et al. Ccdc87 is critical for sperm function and male fertility. Biol Reprod. 2018;99(4):817–827. doi: 10.1093/biolre/ioy106. [DOI] [PubMed] [Google Scholar]
- 64.Imsland F, et al. The Rose-comb mutation in chickens constitutes a structural rearrangement causing both altered comb morphology and defective sperm motility. PLoS Genet. 2012;8(6):e1002775. doi: 10.1371/journal.pgen.1002775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sarkar S, et al. Histone methylation regulates gene expression in the round spermatids to set the RNA payloads of sperm. Reproductive Sci. 2022;29(3):857–882. doi: 10.1007/s43032-021-00837-3. [DOI] [PubMed] [Google Scholar]
- 66.Wang M, et al. CCDC189 affects sperm flagellum formation by interacting with CABCOCO1. Nat Sci Rev. 2023;10(9):nwad181. doi: 10.1093/nsr/nwad181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Burkhard P, Sergei J, Strelkov SV. Coiled coils: a highly versatile protein folding motif. Trends Cell Biol. 2001;11(2):82–88. doi: 10.1016/s0962-8924(00)01898-5. [DOI] [PubMed] [Google Scholar]
- 68.Lupas AN, Bassler J, Dunin-Horkawicz S. The structure and topology of alpha-helical coiled coils. Subcell Biochem. 2017;82:95–129. doi: 10.1007/978-3-319-49674-0_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rose A, et al. Coiled-coil protein composition of 22 proteomes–differences and common themes in subcellular infrastructure and traffic control. BMC Evol Biol. 2005;5:66. doi: 10.1186/1471-2148-5-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Truebestein L, Leonard TA. Coiled-coils: the long and short of it. BioEssays. 2016;38(9):903–916. doi: 10.1002/bies.201600062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pasek RC, et al. Coiled-coil domain containing 42 (Ccdc42) is necessary for proper sperm development and male fertility in the mouse. Dev Biol. 2016;412(2):208–218. doi: 10.1016/j.ydbio.2016.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang R, W B, Liu C, Zhang Z, Wang X, Wang L, Xiao S, Chen Y, Wei H, Jiang H, Gao F, Yuan L, Li W. CCDC38 is required for sperm flagellum biogenesis and male fertility in mice. Development. 2022;149(11):200516. doi: 10.1242/dev.200516. [DOI] [PubMed] [Google Scholar]
- 73.Tapia Contreras C, Hoyer-Fender S. CCDC42 localizes to manchette, HTCA and tail and interacts with ODF1 and ODF2 in the formation of the male germ cell cytoskeleton. Front Cell Dev Biol. 2019;7:151. doi: 10.3389/fcell.2019.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Firat-Karalar EN, Stearns T. The centriole duplication cycle. Philos Trans R Soc Lond B Biol Sci. 2014 doi: 10.1098/rstb.2013.0460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lehti MS, Sironen A. Formation and function of the manchette and flagellum during spermatogenesis. Reproduction. 2016;151(4):R43–54. doi: 10.1530/REP-15-0310. [DOI] [PubMed] [Google Scholar]
- 76.Kierszenbaum AL, Rivkin E, Tres LL. Acroplaxome, an F-actin-keratin-containing plate, anchors the acrosome to the nucleus during shaping of the spermatid head. Mol Biol Cell. 2003;14(11):4628–4640. doi: 10.1091/mbc.e03-04-0226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wei YL, Yang WX. The acroframosome-acroplaxome-manchette axis may function in sperm head shaping and male fertility. Gene. 2018;660:28–40. doi: 10.1016/j.gene.2018.03.059. [DOI] [PubMed] [Google Scholar]
- 78.Piperno GFM. Monoclonal antibodies specific for an acetylated form of a-tubulin recognize the antigen in cilia and flagella from a variety of organisms. J Cell Biol. 1985;101(6):2085–2094. doi: 10.1083/jcb.101.6.2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Symon A, Harley V. SOX9: a genomic view of tissue specific expression and action. Int J Biochem Cell Biol. 2017;87:18–22. doi: 10.1016/j.biocel.2017.03.005. [DOI] [PubMed] [Google Scholar]
- 80.Yan Y, et al. The HDOCK server for integrated protein-protein docking. Nat Protoc. 2020;15(5):1829–1852. doi: 10.1038/s41596-020-0312-x. [DOI] [PubMed] [Google Scholar]
- 81.Pettersen EF, et al. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 82.Gui M, et al. SPACA9 is a lumenal protein of human ciliary singlet and doublet microtubules. Proc Natl Acad Sci United States Am. 2022;119(41):e2207605119. doi: 10.1073/pnas.2207605119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cho EH, et al. A nonsense variant in NME5 causes human primary ciliary dyskinesia with radial spoke defects. Clin Genet. 2020;98(1):64–68. doi: 10.1111/cge.13742. [DOI] [PubMed] [Google Scholar]
- 84.Zhang X, et al. Differential requirements of IQUB for the assembly of radial spoke 1 and the motility of mouse cilia and flagella. Cell Rep. 2022;41(8):111683. doi: 10.1016/j.celrep.2022.111683. [DOI] [PubMed] [Google Scholar]
- 85.Sapiro R, et al. Male infertility, impaired sperm motility, and hydrocephalus in mice deficient in sperm-associated antigen 6. Mol Cell Biol. 2002;22(17):6298–6305. doi: 10.1128/MCB.22.17.6298-6305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang Z, et al. Deficiency of SPAG16L causes male infertility associated with impaired sperm motility. Biol Reprod. 2006;74(4):751–759. doi: 10.1095/biolreprod.105.049254. [DOI] [PubMed] [Google Scholar]
- 87.Gaffney EA, et al. Mammalian sperm motility: observation and theory. Ann Rev Fluid Mech. 2011;43(1):501–528. doi: 10.1146/annurev-fluid-121108-145442. [DOI] [Google Scholar]
- 88.Lehti MS, Sironen A. Formation and function of sperm tail structures in association with sperm motility defects. Biol Reprod. 2017;97(4):522–536. doi: 10.1093/biolre/iox096. [DOI] [PubMed] [Google Scholar]
- 89.Sun X, et al. The role of actin and myosin during spermatogenesis. Mol Biol Rep. 2011;38(6):3993–4001. doi: 10.1007/s11033-010-0517-0. [DOI] [PubMed] [Google Scholar]
- 90.Roy A, et al. Absence of tektin 4 causes asthenozoospermia and subfertility in male mice. FASEB J. 2007;21(4):1013–1025. doi: 10.1096/fj.06-7035com. [DOI] [PubMed] [Google Scholar]
- 91.Donkor FF, et al. Outer dense fibre protein 2 (ODF2) is a self-interacting centrosomal protein with affinity for microtubules. J Cell Sci. 2004;117(20):4643–4651. doi: 10.1242/jcs.01303. [DOI] [PubMed] [Google Scholar]
- 92.Young SAM, et al. CABYR is essential for fibrous sheath integrity and progressive motility in mouse spermatozoa. J Cell Sci. 2016;129(23):4379–4387. doi: 10.1242/jcs.193151. [DOI] [PubMed] [Google Scholar]
- 93.Moretti E, et al. Localization of AKAP4 and tubulin proteins in sperm with reduced motility. Asian J Androl. 2007;9(5):641–649. doi: 10.1111/j.1745-7262.2007.00267.x. [DOI] [PubMed] [Google Scholar]
- 94.Fang X, et al. Hypomorphic and hypermorphic mouse models of Fsip2 indicate its dosage-dependent roles in sperm tail and acrosome formation. Development. 2021 doi: 10.1242/dev.199216. [DOI] [PubMed] [Google Scholar]
- 95.Fiedler SE, et al. Loss of R2D2 proteins ROPN1 and ROPN1L causes defects in murine sperm motility, phosphorylation, and fibrous sheath integrity. Biol Reprod. 2013;88(2):41. doi: 10.1095/biolreprod.112.105262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.San Agustin JT, Pazour GJ, Witman GB. Intraflagellar transport is essential for mammalian spermiogenesis but is absent in mature sperm. Mol Biol Cell. 2015;26(24):4358–4372. doi: 10.1091/mbc.E15-08-0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.O'Donnell L, O’Bryan MK. Microtubules and spermatogenesis. Semin Cell Dev Biol. 2014;30:45–54. doi: 10.1016/j.semcdb.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 98.Kierszenbaum AL, Rivkin E, Tres LL. Cytoskeletal track selection during cargo transport spermatids is relevant to male fertility. Spermatogenesis. 2011;1(3):221–230. doi: 10.4161/spmg.1.3.18018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kumar S, et al. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pond SLK, Frost SDW, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21(5):676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 101.Muroňová J et al (2023) Lack of CCDC146, a ubiquitous centriole and microtubule-associated protein, leads to non-syndromic male infertility in human and mouse. 2023.02.27.530236 [DOI] [PMC free article] [PubMed]
- 102.Liu C, et al. Autophagy is required for ectoplasmic specialization assembly in sertoli cells. Autophagy. 2016;12(5):814–832. doi: 10.1080/15548627.2016.1159377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mooers BH. Shortcuts for faster image creation. Protein Sci. 2020;29(1):268–276. doi: 10.1002/pro.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bailey TL, et al. The MEME suite. Nucleic Acids Res. 2015;43(1):39–49. doi: 10.1093/nar/gkv416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gamallat Y, et al. Bi-allelic mutation in Fsip1 impairs acrosome vesicle formation and attenuates flagellogenesis in mice. Redox Biol. 2021;43:101969. doi: 10.1016/j.redox.2021.101969. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data supporting the findings of this study are available from the corresponding author upon reasonable request.