Abstract
Triple-negative breast cancer (TNBC) has an aggressive biological behavior and poor outcome. Our published study showed that PAI-1 could induce the migration and metastasis of TNBC cells. However, the underlying mechanism by which PAI-1 regulates TNBC metastasis has not been addressed. Here, we demonstrated that PAI-1 is high expressed in TNBC and promotes TNBC cells tumorigenesis. Using microarray analysis of lncRNA expression profiles, we identified a lncRNA SOX2-OT, which is induced by PAI-1 and could function as an oncogenic lncRNA in TNBC. Mechanistic analysis demonstrated that SOX2-OT acts as a molecular sponge for miR-942-5p to regulate the expression of PIK3CA, ultimately leading to activating PI3K/Akt signaling pathway and promoting TNBC metastasis. Taken together, our findings suggest that SOX2-OT regulates PAI-1-induced TNBC cell metastasis through miR-942-5p/PIK3CA signaling and illustrate the great potential of developing new SOX2-OT-targeting therapy for TNBC patients.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-021-04120-1.
Keywords: PAI-1/SERPINE1, ceRNA, PAI-039, LY294002, Lung and liver metastases
Introduction
Triple negative breast cancer (TNBC) is identified as lack the expression of estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER-2) [1]. Compared with other subtypes of breast cancer, the prognosis of TNBC patients is worse, which is due to their inherently aggressive clinical behavior and the lack of recognized therapeutic molecular targets [2]. Therefore, the regulatory mechanism of TNBC cell invasion and metastasis process has been a hot topic of research. It has been reported that the tumor microenvironment can also promote metastatic spread by inducing reversible changes in the phenotype of cancer cells [3, 4]. The metastatic potential of breast cancer cells was greatly increased, when mixed with human mesenchymal stem cells [3]. Lymphatic vessel endothelial cells (LECs) are a component of lymphatic vessels (LVs) in the pre-metastatic microhabitats, which can accelerate metastasis and promote angiogenesis when co-cultured with TNBC cells [4]. Our published study has also shown that endothelial cells (ECs) could induce TNBC cells to secrete abundant plasminogen activator inhibitor-1 (PAI-1/SERPINE1) after TGF-β treatment, which caused TNBC cells to undergo epithelial–mesenchymal transition (EMT) and simulated cancer cells metastasis [5]. PAI-1, a major inhibitor of plasminogen activators, is involved in tumor progression and angiogenesis, and high expression of PAI-1 is associated with poor prognosis in breast cancer patients [6–9]. Our study and other literatures had demonstrated that PAI-1 could induce the migration and metastasis of TNBC cells [9, 10]. However, little is known regarding the underlying mechanism by which PAI-1 regulates TNBC metastasis.
Long non-coding RNAs (lncRNAs) are defined as the non-protein-coding transcripts with a length exceeding 200 nucleotides [11]. Accumulating studies have shown that the expression of lncRNAs is dysregulated in various types of cancer, including breast cancer [12], lung cancer [13], cervical cancer [14], pancreatic cancer [15], prostate cancer [16] and hematological malignancies [17]. Recently, numerous lncRNAs have been shown to play important roles in multiple biological processes, such as proliferation, apoptosis, cell cycle arrest, metastasis, and drug resistance [18–21]. In TNBC, many studies have identified a large number of dysregulated lncRNAs that play a critical role in tumor invasion and metastasis through a variety of mechanisms. For instance, lncRNA TROJAN could bind to metastasis-repressing factor ZMYND8 and increase its degradation through the ubiquitin–proteasome pathway, resulting in TNBC proliferation and metastasis [22]. LncRNA NAMPT-AS could promote TNBC cell metastasis and regulate autophagy, through recruiting POU2F2 to activate NAMPT transcription, or serving as a competing endogenous RNA (ceRNA) to rescue NAMPT degradation from miR-548b-3p [23]. Our study also showed that lncRNA ARNILA can act as a ceRNA for miR-204 and promote the expression of its target gene SOX4, which ultimately leads to TNBC EMT, invasion and metastasis [24]. In this study, we aimed to investigate the role of lncRNA in the PAI-1-induced TNBC metastasis. We found that PAI-1 promoted the expression of lncRNA SOX2-OT, which could activate the PI3K/Akt signaling pathway by acting as a molecular sponge for miR-942-5p, ultimately leading to TNBC invasion and metastasis.
Results
PAI-1 is high expressed in TNBC and promotes TNBC cells tumorigenesis
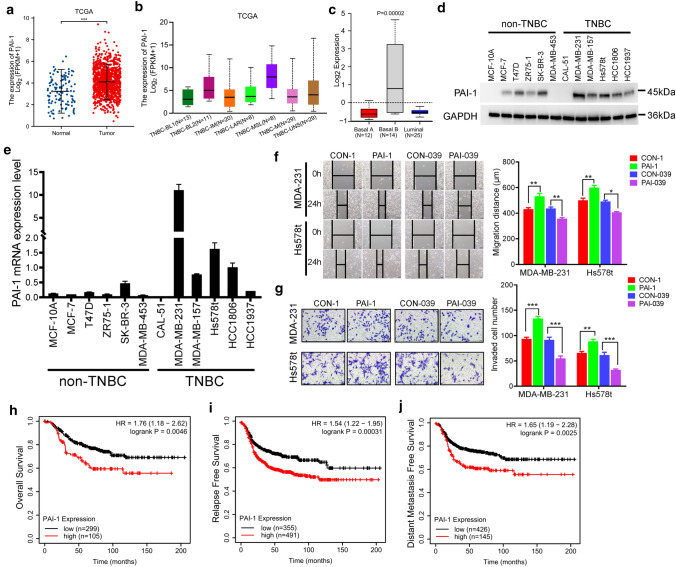
To investigate the expression of PAI-1 in breast cancer, we first analyzed the publically available data set TCGA [25] and found that PAI-1 is significantly overexpressed in breast tumours in comparison with normal breast tissues (Fig. 1a). A significantly elevated PAI-1 expression was also found between the paired primary tumor and normal adjacent tissues (Fig. S1a). The expression levels of PAI-1 were no difference among every PAM50 subtypes of breast cancer, while the mesenchymal stem-like subtype of TNBC showed a higher PAI-1 expression compared with other subtypes (Fig. 1b and S1b) [26]. To examine the above finding in breast cancer cell lines in vitro, we analyzed PAI-1 expression in GOBO database that contains expression data of 51 breast cancer cell lines [27]. A similar result was also obtained that PAI-1 was high expressed in TNBC and Basal B subtype compared with other subtypes (Figs. 1c and S1c, d). Next, we confirmed those results in a panel of normal breast epithelial cell MCF-10A and 11 breast cancer cell lines including five non-TNBC and six TNBC cell lines. Western blotting and reverse transcriptase-PCR analysis showed PAI-1 protein and mRNA expression levels were significantly upregulated in TNBC cell lines as compared with normal breast epithelial cell and non-TNBC cell lines (Figs. 1d, e and S1e). These findings suggested PAI-1 was high expressed in TNBC.
Fig. 1.
PAI-1 is high expressed in TNBC and promotes TNBC cells tumorigenesis. a PAI-1 expression in normal adjacent tissues, and primary tumor tissues from TCGA breast cancer patients. ***P < 0.001 by t test. b PAI-1 expression in different TNBC subtypes of TCGA breast cancer patients from UALCAN cancer database (http://ualcan.path.uab.edu/index.html). c PAI-1 expression in Basal A, Basal B and Luminal subtypes of 51 breast cancer cell lines from GOBO. d Relative PAI-1 protein levels of normal breast epithelial cell MCF-10A and 11 breast cancer cell lines. e Relative PAI-1 mRNA levels of normal breast epithelial cell MCF-10A and 11 breast cancer cell lines. f Wound-healing assay of MDA-MB-231 and Hs578t cell migration treated with recombinant human PAI-1 or PAI-1 small-molecule inhibitor PAI-039. Photographs were obtained immediately (0 h) and at 24 h after wounding. *P < 0.05, **P < 0.01 by t test. g Transwell invasion assay of MDA-MB-231 and Hs578t cells that were treated with recombinant human PAI-1 or PAI-039. **P < 0.01, ***P < 0.001, by t test. h–j Overall survival (OS), Replase free survival (RFS), and Distance metastasis free survival (DMFS) in high and low PAI-1-expressing TNBC patient tissues. P values were calculated with log-rank (Mantel–Cox) test. Patients were stratified into ‘low’ and ‘high’ PAI-1 expression based on auto select best cutoff using Kaplan–Meier plotter database. Results represented the average of three independent experiments, the data represent the mean ± SD
To study a functional role of PAI-1 in TNBC, we treated two TNBC cell lines, MDA-MB-231 and Hs578t, with 10 ng/ml recombinant human PAI-1 and 10 ng/ml PAI-1 inhibitor PAI-039 and then determined the effect of PAI-1 on aggressive growth properties of TNBC. We found recombinant PAI-1 could increase the cell growth of TNBC cells, while PAI-1 small-molecule inhibitor PAI-039 inhibited TNBC cells growth (Fig. S1f, g). Moreover, wound healing assay and transwell invasion assay indicated PAI-1 could increase TNBC cells migration and invasion (Fig. 1f, g). Taken together, those data suggested that PAI-1 could promote TNBC cells tumorigenesis.
To explore the role of PAI-1 in breast cancer patients’ clinical outcomes, we obtained the prognostic information using Kaplan–Meier plotter (http://kmplot.com/analysis/) online breast cancer survival analysis [28]. We observed that high PAI-1 expression positively correlated with reduced overall survival (OS), relapse-free survival (RFS) and distant metastasis-free survival (DMFS) in 1879, 4929, and 2765 breast cancer patients, respectively (Fig. S1i–k). Of interest, we also noticed that high PAI-1 expression was associated with worse OS (HR = 1.76, 95% CI = 1.18–2.62; P = 0.0046; Fig. 1h), RFS (HR = 1.54, 95% CI = 1.22–1.95; P = 0.00031; Fig. 1i) and DMFS (HR = 1.65, 95% CI = 1.19–2.28; P = 0.0025; Fig. 1j) in tissues from 404, 846 and 571 patients with TNBC, respectively. These findings suggested a potential prognostic value of PAI-1 in breast cancer patients.
PAI-1 promotes lncRNA SOX2-OT expression and SOX2-OT functions as an oncogenic lncRNA in TNBC
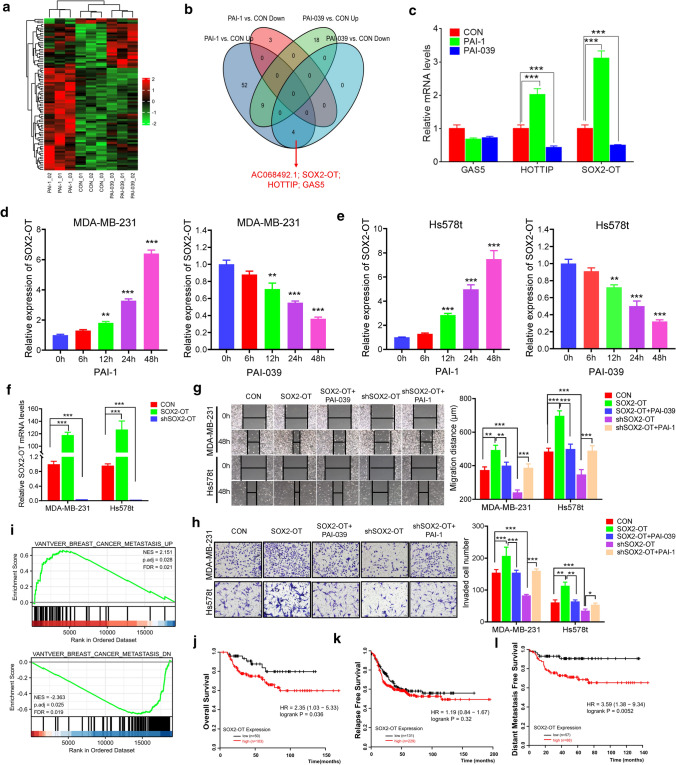
Considering the emerging roles of lncRNAs in the carcinogenesis and development of TNBC [18, 29], we treated MDA-MB-231 cells with or without recombinant human PAI-1 or PAI-1 small-molecule inhibitor PAI-039, then used microarray analysis to obtain the lncRNAs expression profiles (Fig. 2a). We identified four lncRNAs (AC068492.1; SOX2-OT; HOTTIP; GAS5) were upregulated after PAI-1 treatment and downregulated after PAI-039 treatment (Fig. 2b; fold change ≥ 2.0, P < 0.05). We next validated the mRNA expression levels of widely studied lncRNA GAS5, HOTTIP and SOX2-OT. As shown in Fig. 2c, the expression levels of HOTTIP and SOX2-OT were accordant with microarray analysis. Since the role of SOX2-OT in breast cancer was still unknown, we chose the lncRNA SOX2-OT for the further research. We treated two TNBC cell lines MDA-MB-231 and Hs578t with PAI-1 or PAI-039 and measured the expression of SOX2-OT at various timepoints. We found that PAI-1 induced a robust increase of SOX2-OT in both cells, while PAI-039 treatment led to a marked decrease in the expression of SOX2-OT (Fig. 2d, e). Those results suggested that PAI-1 could promote SOX2-OT expression in TNBC.
Fig. 2.
PAI-1 promotes lncRNA SOX2-OT expression and SOX2-OT functions as an oncogenic lncRNA in TNBC. a Heat map representation of microarray data about the lncRNA levels in MDA-MB-231 cells treated with recombinant human PAI-1, or PAI-039. b Venn diagram of the different expressed lncRNAs after PAI-1 or PAI-039 treatment. c Relative mRNA levels of MDA-MB-231 treated with PAI-1 or PAI-039. ***P < 0.001 by t test. d, e SOX2-OT mRNA level of MDA-MB-231 (d) and Hs578t (e) treated with PAI-1 or PAI-039 at various timepoints. **P < 0.01, ***P < 0.001, by t test. f SOX2-OT mRNA level of MDA-MB-231 and Hs578t transfected with SOX2-OT or shSOX2-OT. ***P < 0.001, by t test. g Wound-healing assay of MDA-MB-231 and Hs578t cell migration treated with SOX2-OT or shSOX2-OT transfection, and PAI-039 or PAI-1 treatment. Photographs were obtained immediately (0 h) and at 48 h after wounding. **P < 0.01, ***P < 0.001 by t test. h Transwell invasion assay of MDA-MB-231 and Hs578t cells that were treated with SOX2-OT or shSOX2-OT transfection, and PAI-039 or PAI-1 treatment. *P < 0.05, **P < 0.01, ***P < 0.001 by t test. i GSEA of breast cancer metastasis gene signatures between ‘low’ and ‘high’ SOX2-OT expression in TCGA breast cancer patients. NES, normalized enrichment score. j–l OS, RFS, and DMFS in high and low SOX2-OT-expressing TNBC patient tissues. P values were calculated with log-rank (Mantel–Cox) test. Patients were stratified into ‘low’ and ‘high’ SOX2-OT expression based on auto select best cutoff. Results represented the average of three independent experiments, the data represent the mean ± SD
To explore the functional role of SOX2-OT, we constructed SOX2-OT overexpression plasmid and knocked down SOX2-OT by short hairpin RNA (shRNA) in both MDA-MB-231 and Hs578t cell lines (Fig. 2f). We found overexpression of SOX2-OT could increase cell colony forming ability, while knockdown of SOX2-OT reduced TNBC cell colony forming ability (Fig. S2a). Meanwhile, SOX2-OT knockdown increased, whereas SOX2-OT overexpression reduced, the number of apoptosis cells from both two TNBC cells (Fig. S2b). Wound healing assay and transwell invasion assay indicated SOX2-OT could increase TNBC cells migration and invasion (Fig. 2g, h). Next, we further explored whether PAI-1 could promote TNBC cell migration and invasion through SOX2-OT. Using recombinant PAI-1 or PAI-039 treatment in the SOX2-OT knockdown or overexpressed TNBC cells, we confirmed PAI-1 exert its role through SOX2-OT (Fig. 2g, h). To evaluate whether SOX2-OT expression associated with breast cancer metastasis, we analyzed TCGA database and divided the TCGA breast cancer patients into two groups based on the median expression of SOX2-OT. Gene set enrichment analysis (GSEA) indicated that breast cancer metastasis signature [30] was significantly enriched in the SOX2-OT high-expressed patients (Fig. 2i). Another four published metastasis signaling pathways [31–34] were also mainly enriched upon SOX2-OT upregulation (Fig. S2c). Together, these results suggested that SOX2-OT potentially regulated TNBC metastasis.
Next, we used Kaplan–Meier plotter to investigate the effect of SOX2-OT on the prognosis of TNBC patients. We found that high SOX2-OT expression positively correlated with worse OS (HR = 2.35, 95% CI = 1.03–5.33; P = 0.036; Fig. 2j), and DMFS (HR = 3.59, 95% CI = 1.38–9.34; P = 0.0052; Fig. 2l) in tissues from 153 and 145 patients with TNBC, respectively. We also noticed high SOX2-OT expression had a trend to associate with worse RFS (Fig. 2k). Taken together, our findings suggested that SOX2-OT functions as an oncogenic lncRNA in TNBC.
SOX2-OT acts as a molecular sponge for miR-942-5p
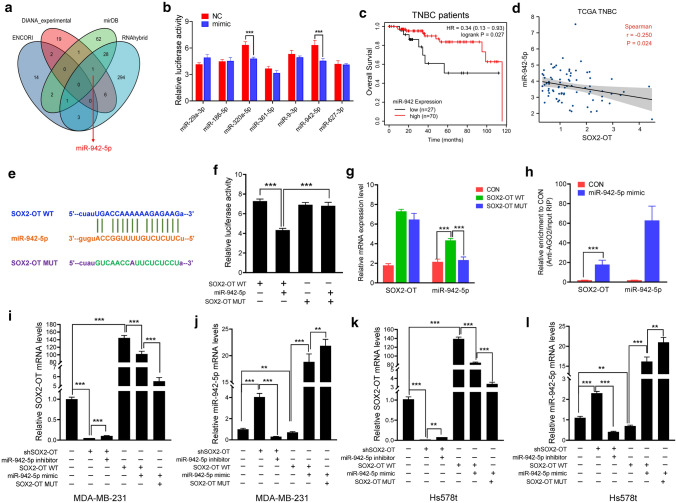
Recently, many lncRNAs have been reported to competitively bind to miRNAs and function as competing endogenous RNAs (ceRNA) in breast cancer [18, 29, 35]. To test the hypothesis that SOX2-OT could act as a ceRNA to regulate TNBC progression, we first performed integrated bioinformatical analysis using four publicly profile data sets and miR-942-5p was predicted to target SOX2-OT in three databases (Fig. 3a). We next performed dual-luciferase reporter assays to confirm the regulatory relationships between SOX2-OT and a total seven tumor suppressor miRNAs, including miR-29a-3p, miR-186-5p, miR-320a-5p, miR-361-5p, miR-9-3p, miR-942-5p and miR-627-3p. The reporter activity was noticeably suppressed by the presence of miR-320a-5p and miR-942-5p mimics (Fig. 3b). Kaplan–Meier plotter showed that both miR-320a and miR-942-5p had a positive correlation with a prolonged OS in systemically untreated TNBC patients (Figs. 3c and S3b), but not all breast cancer patients (Fig. S3a, c). We then analyzed RNA-seq data of TCGA database by comparing the SOX2-OT and those two miRNAs mRNA levels in breast cancer patients and TNBC subpopulation. The results revealed that miR-942-5p, but not miR-320a was negatively correlated with SOX2-OT in both the whole and TNBC populations (Figs. 3d and S3d–f).
Fig. 3.
SOX2-OT acts as a molecular sponge for miR-942-5p. a Venn diagram of the potential binding miRNAs of SOX2-OT according to bioinformatics analysis. b Luciferase activity of SOX2-OT upon transfection of the indicated miRNA mimics in 293 T cells. ***P < 0.001 by t test. c OS in high and low miR-942-expressing TNBC patient tissues. P values were calculated with log-rank (Mantel–Cox) test. Patients were stratified into ‘low’ and ‘high’ expression based on auto select best cutoff using Kaplan–Meier plotter database. d Correlation between SOX2-OT and miR-942-5p expression in TCGA TNBC patients. P values were calculated with Pearson correlation analysis. e Putative miR-942-5p binding site and mutant sequences in SOX2-OT. f Luciferase activity of SOX2-OT WT and SOX2-OT MUT upon transfection of miR-942-5p mimics in 293 T cells. ***P < 0.001 by t test. g Cell lysates of MDA-MB-231 transfected with SOX2-OT or shSOX2-OT were incubated with biotin-labeled SOX2-OT; after pull-down, mRNA expression levels of SOX2-OT and miR-942-5p were detected by qRT-PCR. ***P < 0.001 by t test. h Anti-AGO2 RIP was performed in MDA-MB-231 cells overexpressing miR-942-5p, followed by qRT-PCR to detect SOX2-OT and miR-942-5p associated with AGO2. ***P < 0.001 by t test. i–l Relative SOX2-OT and miR-942-5p mRNA levels in MDA-MB-231 and Hs578t cells transfected with SOX2-OT WT, SOX2-OT MUT or shSOX2-OT, and miR-942-5p mimic or inhibitor. ***P < 0.001 by t test. Results represented the average of three independent experiments, the data represent the mean ± SD
To obtain direct evidence for the interaction between SOX2-OT and miR-942-5p, we subcloned wild-type (SOX2-OT WT) and mutated (SOX2-OT MUT) miR-942-5p binding site into dual-luciferase reporters (Fig. 3e). As shown in Fig. 3f, the relative luciferase activity of SOX2-OT WT was significantly reduced after co-transfection of miR-942-5p mimic, while SOX2-OT mutant vector did not show a response to miR-942-5p mimic. To validate the direct binding between SOX2-OT and miR-942-5p at endogenous levels, we performed SOX2-OT RNA-pull-down experiment in MDA-MB-231 cells transfected with SOX2-OT WT or SOX2-OT MUT vector. qPCR analysis demonstrated that SOX2-OT was able to successfully pull down miR-942-5p in the SOX2-OT WT group compared to the empty vector, and SOX2-OT with mutations in miR-942-5p targeting sites (Fig. 3g). To demonstrate whether SOX2-OT was regulated by miR-942-5p in an AGO2-dependent manner, we conducted anti-AGO2 RNA immunoprecipitation (RIP) in MDA-MB-231 cells transiently overexpressing miR-942-5p. As shown in Fig. 3h, endogenous SOX2-OT pull-down by AGO2 was specifically enriched in miR-942-5p mimic-transfected cells. Furthermore, we found that SOX2-OT knockdown strongly increased miR-942-5p level, which could be downregulated by miR-942-5p inhibitor treatment (Fig. 3i–l). Ectopically expressed SOX2-OT WT reduced the expression level of miR-942-5p, which was reversed by transfecting with miR-942-5p mimics. Notably, mutations in the miR-942-5p-binding site on SOX2-OT abolished this reversed effect, further supporting that SOX2-OT acted as a molecular sponge for miR-942-5p.
SOX2-OT accelerates TNBC cells metastasis via miR-942-5p
miR-942-5p has been reported to inhibit tumorigenesis of several malignant tumors, including colorectal cancer [36, 37], cervical cancer [38] and bone mesenchymal stem cells (BMSCs) [39] and ovarian cancer [40]. However, its function has never been explored in breast cancer and TNBC. To investigate the role of miR-942-5p in the tumorigenesis of TNBC, we treated MDA-MB-231 and Hs578t cells with miR-942-5p mimic or inhibitor and detected the changes of tumor cells biological characteristics (Fig. 4a). We found miR-942-5p inhibitor increased the cell growth of TNBC cells, while miR-942-5p mimic inhibited TNBC cells growth (Fig. S4a, b). We also noticed that miR-942-5p mimic could induce TNBC cells apoptosis, while the effect of miR-942-5p inhibitor on cell apoptosis was negligible (Fig. S4c). Moreover, using wound healing assay and transwell invasion assay, we demonstrated that miR-942-5p could suppress TNBC cells migration and invasion (Fig. 4b, c). Taken together, those data suggested that miR-942-5p could inhibit TNBC cells tumorigenesis.
Fig. 4.
SOX2-OT accelerates TNBC cells metastasis via miR-942-5p. a miR-942-5p mRNA level of MDA-MB-231 and Hs578t transfected with miR-942-5p mimic or inhibitor. ***P < 0.001 by t test. b Wound-healing assay of MDA-MB-231 and Hs578t cells that were treated with miR-942-5p mimic or inhibitor transfection. Photographs were obtained immediately (0 h) and at 48 h after wounding. *P < 0.05, **P < 0.01 by t test. c Transwell invasion assay of MDA-MB-231 and Hs578t cells that were treated with miR-942-5p mimic or inhibitor transfection. **P < 0.01, ***P < 0.001 by t test. d, e Wound-healing assay of MDA-MB-231 and Hs578t cell migration transfected with SOX2-OT WT, SOX2-OT MUT or shSOX2-OT, and miR-942-5p mimic or inhibitor. *P < 0.05, **P < 0.01, ***P < 0.001 by t test. f, g Transwell invasion assay of MDA-MB-231 and Hs578t cells transfected with SOX2-OT WT, SOX2-OT MUT or shSOX2-OT, and miR-942-5p mimic or inhibitor. *P < 0.05, **P < 0.01, ***P < 0.001 by t test. Results represented the average of three independent experiments, the data represent the mean ± SD
Next, we wondered whether SOX2-OT promoted TNBC cells tumorigenesis via miR-942-5p. To address this question, we knocked down or overexpressed SOX2-OT in MDA-MB-231 and Hs578t cells and detected the cell migration and invasion using wound healing assay and transwell invasion assay. As shown in Fig. 4d–g, we found that SOX2-OT knockdown reduced MDA-MB-231 and Hs578t cells migration and invasion, which could be rescued by miR-942-5p inhibitor treatment. Similarly, ectopic expression of SOX2-OT could promote TNBC cells migration and invasion, while adding miR-942-5p mimic could reverse it (Fig. 4d–g). It was noteworthy that SOX2-OT vector treatment with mutations in the miR-942-5p-binding site abolished this reversed effect after transfection with miR-942-5p mimic (Fig. 4d–g). Collectively, SOX2-OT could accelerate TNBC cells metastasis via miR-942-5p.
To address whether the potential pro-proliferative effect of SOX2-OT in TNBC cells was dependent on miR-942-5p, we also detected cell proliferation and apoptosis after SOX2-OT and/or miR-942-5p treatment. We found that inhibition rate of cell growth was increased in the shSOX2-OT cells, and decreased in the SOX2-OT overexpressed cells at various timepoints (Fig. S4d, e). miR-942-5p inhibitor partially reversed shSOX2-OT-induced growth inhibition, and miR-942-5p mimic reversed cell proliferation of SOX2-OT WT treatment (Fig. S4d, e). Notably, mutations in the miR-942-5p-binding site on SOX2-OT abolished this reversed effect. Similarly, colony formation assay and cell apoptosis assay displayed a consistent result with the CCK-8 assay (Fig. S4f–i). Taken together, our results indicated that SOX2-OT promoted cell proliferation and suppressed cell apoptosis in a miR-942-5p-dependent manner.
SOX2-OT activates PI3K/Akt signaling pathway via miR-942-5p
Using four bioinformatical tools, we predicted 1294 potential target genes of miR-942-5p (Fig. S5a). The GO enrichment analysis of these genes showed that PI3K/Akt signaling pathway was the most significant signaling pathways (Fig. S5b). We noticed that PIK3CA was also the potential target of miR-942-5p. To validate the interaction between miR-942-5p and PIK3CA, we subcloned wild-type (PIK3CA WT) and mutated (PIK3CA MUT) miR-942-5p binding site in the 3’-untranslated region (3’UTR) of PIK3CA into dual-luciferase reporters (Fig. 5a). As shown in Fig. 5b, the relative luciferase activity of PIK3CA WT was significantly reduced after co-transfection of miR-942-5p mimic, while PIK3CA mutant vector did not show a response to miR-942-5p mimic. Next, we detected the mRNA expression of PIK3CA in the MDA-MB-231 and Hs578t cells after SOX2-OT overexpression or shSOX2-OT treatment. We found that SOX2-OT overexpression enhanced the mRNA expression level of PIK3CA, while SOX2-OT knockdown significantly diminished PIK3CA mRNA expression (Fig. 5c). We also detected the protein expression of PI3K, p-PI3K, Akt, p-Akt, epithelial marker E-Cadherin, and mesenchymal markers N-Cadherin in the MDA-MB-231 and Hs578t cells after miR-942-5p inhibitor or mimic treatment. As shown in Fig. 5d, miR-942-5p inhibitor promoted the phosphorylation thus the activation of PI3K and Akt (Figs. 5d and S5c, d). To answer the question whether in functional level PIK3CA mutant could restore miR-942-5p overexpression-mediated suppression of tumor cell migration, and invasion, we performed wound healing assay and transwell invasion assay after PIK3CA WT or MUT vector transfection in the miR-942-5p overexpressed MDA-MB-231 and Hs578t cells. We found PIK3CA mutant could promote TNBC cells migration and invasion, compared with PIK3CA WT transfection (Fig. 5e, f). Those data showed that miR-942-5p could target PIK3CA and activate PI3K/Akt signally pathway.
Fig. 5.
SOX2-OT activates PI3K/Akt signaling pathway via miR-942-5p. a Putative miR-942-5p binding site and mutant sequences in the 3’UTR of PIK3CA. b Luciferase activity of PIK3CA WT and PIK3CA MUT upon transfection of miR-942-5p mimics in 293 T cells. ***P < 0.001 by t test. c PIK3CA mRNA level of MDA-MB-231 and Hs578t transfected with miR-942-5p mimic or inhibitor. ***P < 0.001 by t test. d Relative protein levels of MDA-MB-231 and Hs578t transfected with miR-942-5p mimic or inhibitor. e, f Wound-healing assay (e) and Transwell invasion assay (f) of MDA-MB-231 and Hs578t cells that were treated with co-transfection of miR-942-5p mimic and PIK3CA WT or MUT vector. Photographs were obtained immediately (0 h) and at 48 h after wounding. *P < 0.05, **P < 0.01, ***P < 0.001 by t test. g, h PIK3CA mRNA level of MDA-MB-231 and Hs578t transfected with SOX2-OT WT, SOX2-OT MUT or shSOX2-OT, and miR-942-5p mimic or inhibitor. ***P < 0.001 by t test. i Relative protein levels of MDA-MB-231 and Hs578t transfected with SOX2-OT WT, SOX2-OT MUT or shSOX2-OT, and miR-942-5p mimic or inhibitor. Results represented the average of three independent experiments, the data represent the mean ± SD
To investigate whether SOX2-OT could activate PI3K/Akt signaling pathway through targeting miR-942-5p/PIK3CA, we detected the PIK3CA mRNA expression in the MDA-MB-231 and Hs578t cells after SOX2-OT knockdown and SOX2-OT vector transfection. As shown in Fig. 5g, h, we found that SOX2-OT knockdown strongly decreased PIK3CA mRNA level, which could be partially rescued by miR-942-5p inhibitor treatment. Ectopically expressed SOX2-OT WT promoted the expression level of PIK3CA, which was partially reversed by transfecting with miR-942-5p mimics. Significantly, this reversed effect was abolished in the TNBC cells transfected with mutations in the miR-942-5p-binding site on SOX2-OT (Fig. 5g, h), suggesting that SOX2-OT induced PIK3CA expression by competitively binding to miR-942-5p. Next, we detected the change of PI3K/Akt signaling pathway gene expression using western blotting assay. The results displayed a consistent result with the mRNA data (Figs. 5i and S5e, f). Taken together, our findings indicated that SOX2-OT could activate PI3K/Akt signaling pathway via miR-942-5p.
SOX2-OT promotes TNBC metastasis in vivo
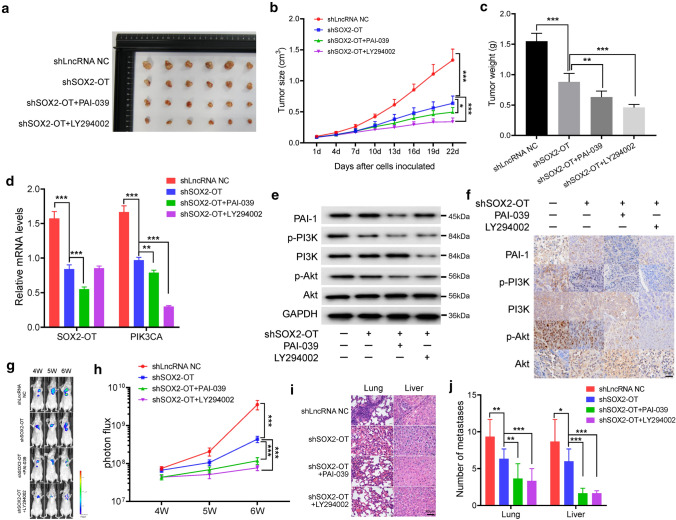
To further demonstrate the oncogenic role of SOX2-OT in TNBC in vivo, we used a xenograft mouse model. Female BALB/c nude mice were subcutaneously injected with shLncRNA NC- or shSOX2-OT-transfected MDA-MB-231 cells and treated intragastrically with PBS, PAI-039, or PI3K inhibitor LY294002 every 3 days. As shown in Fig. 6a–c, SOX2-OT knockdown was shown to strongly inhibit in vivo xenograft tumor growth of MDA-MB-231 cells. Moreover, PAI-039 or LY294002 treatment led to a further inhibition of xenograft tumor growth (Fig. 6a–c). We confirmed that expression levels of SOX2-OT and PIK3CA mRNA were decreased by SOX2-OT knockdown in vivo. Meanwhile, LY294002 treatment caused a lower expression level of PIK3CA (Fig. 6d). Results of western blotting assay and immunohistochemistry (IHC) staining assay demonstrated that the level of PI3K/Akt signaling pathway was decreased in mice injected with shSOX2-OT, whereas the levels of PAI-1 and PI3K were reduced after PAI-039 or LY294002 treatment, respectively (Figs. 6e, f and S6a). Immunofluorescence staining also revealed the change of PAI-1 and p-PI3K expression levels in the xenograft mouse model (Fig. S6b, c).
Fig. 6.
SOX2-OT promotes TNBC metastasis in vivo. a Representative photographs of xenograft tumors were taken 3 weeks after injection. b Tumor sizes were measured at the indicated timepoints. *P < 0.05, ***P < 0.001 by t test. c Excised tumors were weighed. **P < 0.01, ***P < 0.001 by t test. d Relative mRNA expression levels of the excised xenografts. **P < 0.01, ***P < 0.001 by t test. e Relative protein expression levels of the excised xenografts. f Representative IHC staining in the tissue from the excised xenografts. g, h Luciferase signal intensities of the mice in each group after injecting for 4, 5 and 6 weeks. ***P < 0.001 by t test. i Hematoxylin and eosin-stained images of lung and liver tissues isolated from the mice in each group. j Number of lung and liver metastases in the mice. *P < 0.05, **P < 0.01, ***P < 0.001 by t test. Results represented the average of three independent experiments, the data represent the mean ± SD
In addition, we also investigated the function of SOX2-OT in TNBC metastasis in vivo by injection of MDA-MB-231 cells that labeled with firefly luciferase and transfected shLncRNA NC or shSOX2-OT into the tail vein. PBS, PAI-039, or LY294002 was treated intragastrically every 3 days. As shown in Fig. 6g, h, luciferase signal intensity of the mice in the shSOX2-OT group was lower than that in the shLncRNA NC group, further supporting that SOX2-OT promoted TNBC metastasis in vivo. We also found that lung and liver metastases were significantly reduced in the mice treated with PAI-039, or LY294002 (Fig. 6i, j). Immunofluorescence staining confirmed the significantly reduced expression of PAI-1 and p-PI3K after PAI-039, or LY294002 treatment in the lung and liver metastatic tumors (Fig. S6d–g). Collectively, these data suggested that SOX2-OT contributes to TNBC metastasis and that silencing of SOX2-OT inhibits lung and liver metastases in vivo.
Discussion
Our previous study has shown that PAI-1 could induce TNBC cell migration and metastasis [9]. However, the potential regulatory mechanisms by which PAI-1 promotes TNBC metastasis remain unclear. In the present study, we report that lncRNA SOX2-OT, which could be activated by PAI-1, promotes TNBC cell metastasis by competitively binding miR-942-5p, upregulating PIK3CA, and then activating PI3K/Akt signaling pathway. We also found that a higher level of SOX2-OT is associated with worse clinical outcomes in TNBC patients. Thus, SOX2-OT functions as an oncogenic lncRNA and plays a prometastatic role in TNBC.
PAI-1 is a major inhibitor of plasminogen activator and associated with multiple tumor progression, including breast cancer [6–8]. High PAI-1 expression is associated with worse clinical outcomes in breast cancer patients [5, 9]. It is reported that PAI-1 is essential to protect endothelial cells (ECs) from FasL-mediated apoptosis, and PAI-1 knockdown in ECs could enhance the cell-associated plasmin activity and increase spontaneous apoptosis in vitro [41]. Our research also showed that ECs mediates TNBC cells to secrete abundant PAI-1, which increases EC migration and angiogenesis [5]. PAI-1 upregulates CCL5/CCR5 expression and CCL5 secretion in ECs. Meanwhile, CCL5 binds to CCR5 on TNBC cell cytomembrane to induce TNBC invasion, and CCL5, in turn, promotes PAI-1 secretion, forming a positive feedback loop [5]. Another one of our studies found that PAI-1, secreted during the EMT process of TNBC cells, promotes cell growth, migration and invasion in the TNBC cell lines and xenograft mouse model cells [9]. In the present study, we sought to reveal the potential regulatory mechanisms by which PAI-1 promotes TNBC tumorigenesis. We found that PAI-1 promotes lncRNA SOX2-OT expression and SOX2-OT functions as an oncogenic lncRNA through miR-942-5p/PI3K/Akt signaling in TNBC. Consistently, the activation of PI3K/Akt signaling pathway by PAI-1 has also been demonstrated by other investigators. Rømer et al. found that PAI-1 could protect fibrosarcoma cells from etoposide-induced apoptosis through the activation of PI3K/Akt signaling pathway [42]. Recently, ACT001, a novel PAI-1 inhibitor, was reported to inhibit the PI3K/Akt pathway and induce the inhibition of glioma cell proliferation, invasion and migration [43]. Therefore, PAI-1 promotes TNBC cells metastasis by activating PI3K/Akt signaling pathway.
Recently, many lncRNAs have been found to be involved in the regulation of pathological and physiological processes in TNBC cells and have been characterized as diagnostic and prognostic biomarkers of TNBC [18]. Our previous study showed that lncRNA ARNILA could promote TNBC invasion and metastasis as a ceRNA to regulate SOX4 by sponging miR-204 [24]. In this study, we identified that lncRNA SOX2-OT functions as an oncogenic lncRNA and mediates the role of PAI-1 in TNBC metastasis. The lncRNA SOX2-OT gene consists of 10 exons and more than two transcriptional start sites. Aberrant expression of lncRNA SOX2-OT was found in various tumors, and high expression of SOX2-OT was significantly associated with worsening clinical outcomes, suggesting a potential prognostic value of SOX2-OT [44]. Our data also showed that high SOX2-OT expression was positively correlated with worse OS, and DMFS, with a tendency to correlate with worse RFS in tissues from TNBC patients. Several potential regulatory mechanisms of SOX2-OT in cellular biological processes were reported. Such as, Tai et al. showed that SOX2-OT recruits EZH2 to elevate H3K27me3 and epigenetically inhibit PTEN expression, thus facilitating laryngeal squamous cell carcinoma development [45]. Another regulatory mechanism is the competing endogenous RNAs (ceRNA) mechanism, in which lncRNAs act as miRNA ‘sponges’ to sequester miRNAs and spare their protein-coding counterparts from post-translational regulation [35]. Li et al. reported that SOX2-OT competitively binds the miR-200 family to upregulate SOX2, thus inducing EMT and stem features and promoting invasion and metastasis of pancreatic ductal adenocarcinoma (PDAC) [46]. Our results also indicated that SOX2-OT acts as a molecular sponge for miR-942-5p, leading to activating PI3K/Akt signaling pathway and promoting TNBC metastasis in vitro and in vivo. LncRNAs could play important roles in the process of TNBC tumorigenesis through diverse mechanisms [18]. Thus, other potential regulatory mechanisms of SOX2-OT in the processes of TNBC metastasis requires further investigation.
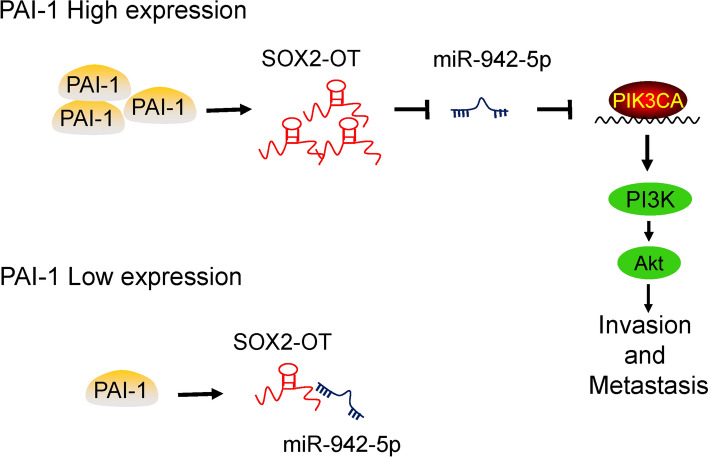
In conclusion, our results to date permit construction of a schematic model demonstrating the critical role of SOX2-OT in the regulation of PAI-1-induced TNBC cell metastasis through miR-942-5p/PIK3CA signaling (Fig. 7). We demonstrate for the first time that SOX2-OT acts as a molecular sponge for miR-942-5p, leading to activating PI3K/Akt signaling pathway and promoting TNBC metastasis. The findings of this study have significant implications for our understanding of the pathogenesis of TNBC metastasis, and illustrate the great potential of the development of new SOX2-OT-targeting therapy for TNBC patients.
Fig. 7.
Schematic model of PAI-1/SOX2-OT/miR-942-5p/PI3K/Akt signaling in the TNBC metastasis. PAI-1 promotes SOX2-OT expression. In patients with a low PAI-1 expression, SOX2-OT blinds to miR-942-5p and the expression level of SOX2-OT is also low. In patients with a high PAI-1 expression, SOX2-OT acts as a molecular sponge for miR-942-5p, leading to activating PI3K/Akt signaling pathway and promoting TNBC invasion and metastasis
Materials and methods
Cell lines and mice
Normal breast epithelial cell line MCF-10A, and breast cancer cell lines HCC1806, HCC1937, CAL-51, MCF-7, ZR75-1, SK-BR-3, T47D, MDA-MB-231, MDA-MB-453, MDA-MB-157, Hs578t were purchased from American Type Culture Collection (ATCC). Cells were cultured in RPMI 1640, McCoy's 5A or DMEM medium (GIBCO, Gaithersburg, MD, USA) supplemented with 10% FBS and 1% penicillin/streptomycin at 37 °C in a humidified atmosphere containing 5% CO2. BALB/c nude mice (female, 4–5 weeks) were purchased from Shanghai SLAC Laboratory animal CO. LTD (Shanghai, China), and maintained in a pathogen-free facility. Animal studies were performed in accordance with institutional guidelines. Experiments were approved by the Ethics Committee of Nanjing First Hospital and were conducted in compliance with the Helsinki Declaration.
Online databases
TCGA breast cancer online database (https://portal.gdc.cancer.gov) was used to obtain the expression of PAI-1 in normal adjacent tissues, and primary breast cancer tissues. UALCAN cancer database (http://ualcan.path.uab.edu/index.html) was used to obtain PAI-1 expression in different TNBC subtypes of TCGA breast cancer patients. GOBO database (http://co.bmc.lu.se/gobo/) was used to analyze the cancer subtype-specific PAI-1 gene expression, which contain 51 breast cancer cell lines including Basal A, Basal B, and Luminal breast cancer cell lines [27]. Kaplan–Meier survival plot (http://kmplot.com/analysis/) was used to obtain the prognostic information of 7830 breast cancer patients and the patient percentile between the upper and lower quartiles were auto-selected based on the computed optimal threshold as cutoff [28].
Reagents
Human recombinant PAI-1 (Topscience #T4254, Shanghai, China) were dissolved in the culture medium, PAI-1 inhibitor PAI-039 (Topscience #T2030, Shanghai, China), PI3K inhibitor LY294002 (Topscience #T2008, Shanghai, China) were dissolved in dimethylsulfoxide (DMSO, Sigma) and stored at − 80 °C as a 1 μg/ml stock solution.
Microarray analysis
MDA-MB-231 cells were treated with 10 ng/ml human recombinant PAI-1 or PAI-039 for 48 h. The total RNA was extracted from above mentioned treated or untreated MDA-MB-231 cells, prepared using Trizol (Invitrogen, USA), and the quality and quantity of the RNA were assessed using NanoDrop ND-1000 (Thermo Fisher Scientific, Waltham, Massachusetts, USA). RNA samples were subjected to Human OneArray v6 (Phalanx Biotech, Hsinchu, Taiwan). The threshold we used to screen upregulated or downregulated lncRNAs were fold change ≥ 2 and a p value ≤ 0.05. Finally, the expression levels of all differentially expressed lncRNAs were plotted on a heatmap. The microarray analysis data have been deposited in the GEO database under accession code GSE181202.
Luciferase assay
293 T cells were cultured in 24-well plates and co-transfected with either the miR-29a-3p/186-5p/320a-5p/361-5p/9-3p/942-5p/627-3p mimics or the control mimics. After 48 h of incubation, firefly and Renilla luciferase activities of the cell lysates were measured using a Dual-Luciferase Reporter Assay System (Promega). The wild-type or mutated SOX2-OT sequences within the predicted binding sites of miR-942-5p, PIK3CA sequences within the predicted binding sites of miR-942-5p in the 3′ UTR of PIK3CA were synthesized. Those sequences were inserted into a luciferase reporter plasmid (GENE, Shanghai, China), and transfected into 293 T cells. The luciferase activity was normalized to Renilla luciferase activity after 48 h of transfection.
RNA immunoprecipitation (RIP) assay
MDA-MB-231 cells were co-transfected with either the miR-942-5p mimics or the control mimics. After 48 h of incubation, the cells were lysed and mixed with treated beads and rotated for 4 h at 4 °C. Then, the AGO2-specific antibody and immunoglobin G (IgG) antibody were used in AGO2 immunoprecipitation according to the manufacturer’s instructions. Beads were subsequently washed 6 times in immunoprecipitation buffer, and the RNA was collected using Trizol (Invitrogen) to perform qRT-PCR to detect the mRNA expression levels of SOX2-OT and miR-942-5p.
RNA pull-down assay
LncRNA SOX2-OT WT, lncRNA SOX2-OT MUT or lncRNA CON were transcribed in vitro, respectively, and biotin-labeled with the Biotin RNA Labeling Mix (Roche) and T7 RNA polymerase (Roche), treated with RNase-free DNase I (Roche), RNeasy Mini Kit (Qiagen, Valencia, CA) for purification. Whole-cell lysate was incubated with purified biotinylated transcripts for 1 h at 25 °C. Then, the complexes were separated with streptavidin agarose beads (Invitrogen), and the RNA complexes binding to the beads were collected. The expression levels of miR-942-5p, and SOX2-OT were analyzed by qRT-PCR.
Gene set enrichment analysis (GSEA)
TCGA breast cancer patients are divided into SOX2-OT high or low expression group based on the median expression of SOX2-OT. Single gene difference analysis was performed using R “DESeq2” package. Fold change ≥ 2, and adjusted P values < 0.01 were considered statistically significant. GSEA v2.0 was used to perform GSEA on various gene signatures. Gene sets were either obtained from the MSigDB Collections or from published gene signatures. Statistical significance was assessed by comparing the enrichment scores with the enrichment results generated by 1000 random permutations of genome to obtain a p value.
Plasmids and transfection
The SOX2-OT Small hairpin RNA and nonspecific control shRNA were all chemically synthesized by KeyGEN Biotech (Nanjing, China). Target sequences for shRNA were shown below: SOX2-OT-1 5'-GGATAGGCCTCACTTACAAGA-3', SOX2-OT-2 5'-GGACTTATCAGCTGGGATAGG-3', SOX2-OT-3, 5'-GACAGCTCTGTTCAGTATTTG-3', Human GAPDH 5'-GTATGACAACAGCCTCAAG-3', Negative Control 5'-GTTCTCCGAACGTGTCACGT-3'. One with the highest targeting efficiency was chosen for further studies. The miR-942-5p mimics, miR-942-5p inhibitor and their respective negative control RNAs (KeyGEN Biotech, Nanjing, China) were introduced into cells at a final concentration of 50 nM. The cells were harvested at 48 h after transfection. Plasmids were transfected into cells using the Lipofectamine 2000 kit (Invitrogen) according to the manufacturer’s protocol.
Cell viability assay
For cell viability analysis, cells were plated in 96-well plates at 4,000 to 6,000 cells per well. The following day, cells were exposed to different concentrations of agents and after 72-h exposure cell survival was assessed with the Cell Counting Kit-8 in accordance with the recommended guideline (KeyGEN Biotech, Nanjing, China). Analysis of the cell viability was performed as previously described [47–49]. Inhibition rate (%) = (Negative control group − Experimental group)/ Negative control group × 100%.
Colony forming assay
To assess effects on colony formation, cells were seeded at 400 cells/mL in 6-well plates. The following day, cells were exposed to agents for 24 h, after which cells were washed with PBS then recovered in fresh medium for 10–14 days. Finally, cells were stained with crystal violet. Colonies containing ≥ 50 cells were counted.
Apoptosis analysis
Cells were treated with plasmids for 48 h, then harvested by trypsinization (no EDTA) and washed with phosphate-buffered saline (PBS). Analysis of the cell apoptosis was performed as previously described [48]. Each sample was tested in triplicate and untreated cells were used as controls.
Wound healing assay
Cell migration was measured using a wound healing assay. In brief, cells were seeded in 12-well plates and cultured to confluence. Wounds of 1.0 mm width were created with a plastic scriber. Then cells were treated with 1 μg/ml mitomycine c for 1 h, and washed, incubated in a serum-free medium. 48 h after wounding, cultures were fixed and observed under a microscope. A minimum of five randomly chosen areas were measured and the distance of cell migration to the wound area was determined.
Transwell invasion assay
Cell invasion was assessed with modified Boyden chamber (Becton Dickinson Labware) assays. Briefly, approximately 1.0 × 105 cells cultured in serum-free medium were added to the upper chamber containing a Matrigel coated membrane with a 24-well insert. The medium containing 10% FBS was added to the lower chamber. After incubation for 24 h, cells that had invaded the opposite side of the membrane were fixed with 4% paraformaldehyde, stained with crystal violet, and counted under a microscope. Cells were counted in five random fields per insert. Three independent experiments were carried out.
Quantitative real-time PCR (qRT-PCR)
Total cellular RNA was extracted from different cell types using TRIzol (Invitrogen, USA) and reversely transcribed according to the Manufacturer’s instruction using the Step One System (Applied Biosystems, Life tech, USA). Primer sequences (forward and reverse, respectively) were as listed below. PAI-1-F 5'-GGTGCTGGTGAATGCCCTCTAC-3', PAI-1-R 5'-TGCTGCCGTCTGATTTGTGGAA-3', SOX2-OT-F 5'-GAGGCTGGTGTAAGGCGATGTG-3', SOX2-OT-R 5'-CATCCAAGGCACCGTGAATCCA-3', HOTTIP-F 5'-TGGGGGAAGGCTTTGGATTG-3', HOTTIP-R 5'-AGCTTTTTCTTGGCGAGAGC-3', GAS5-F 5'-ACCGTTCCATTTTGATTCTGAGG-3', GAS5-R 5'-AAACCCTGAAAGCGAAGCCA-3', PIK3CA-F 5'-CCTGATCTTCCTCGTGCTGCTC-3', PIK3CA-R 5'-TGCCAATGGACAGTGTTCCTCT-3', miR-942-5p-F 5'-TCCAAATTCAAAGAAACAGGGCG-3', miR-942-5p-R 5'-AGTGCAGGGTCCGAGGTATT-3', GAPDH-F 5'-AGATCATCAGCAATGCCTCCT-3', GAPDH-R, 5'-TGAGTCCTTCCACGATACCAA-3', U6-F 5'-CTCGCTTCGGCAGCACA-3', U6-R, 5'-TGGTGTCGTGGAGTCG-3'.
Western blotting
Total protein was extracted using RIPA buffer supplemented with protease and phosphatase inhibitors, with protein concentrations determined using a BCA kit (Thermo Scientific, USA). Analysis of the cell viability was performed as previously described [48, 49]. Antibodies used for western blot were: anti-PAI-1 antibody (ab222754, Abcam, UK), anti-p-PI3K antibody (ab182651, Abcam, UK), anti-PI3K antibody (ab191606, Abcam, UK), anti-p-Akt antibody (ab182651, Abcam, UK), anti-Akt antibody (ab179463, Abcam, UK), anti-E-cadherin antibody (ab40772, Abcam, UK), and anti-N-cadherin antibody (ab98952, Abcam, UK). Bands were normalized to GAPDH expression.
Xenograft transplantation
Approximately 5.0 × 106 MDA-MB-231 shLncRNA NC or shSOX2-OT cells were subcutaneously transplanted into the right side of the posterior flank of nude mice. Tumor growth was examined every 3 days with a vernier caliper. Tumor volumes were calculated by using the equation: V = A × B2/2 (mm3), wherein A is the largest diameter and B is the perpendicular diameter. The tumor-bearing nude mice were treated intragastrically with PBS, PAI-039, or LY294002 every 3 days (n = 6 mice per group). After approximately 3 weeks, mice were euthanized and their tumors harvested.
MDA-MB-231 cells that labeled with firefly luciferase and transfected shLncRNA NC or shSOX2-OT were injected into the tail vein. PBS, PAI-039, or LY294002 was treated intragastrically every 3 days. The metastases were monitored using the IVIS@ Lumina II system (Caliper Life Sciences, Hopkinton, MA) after injecting for 4, 5 and 6 weeks. After 6 weeks, mice were euthanized and their tumors harvested.
Immunohistochemistry
The mice tumors were deparaffinized in xylene, followed by heat-mediated antigen retrieval using citrate buffer (BioGenex Laboratories, San Ramon, CA, USA). Antibody staining was visualized with DAB (Sigma, D-5637) and hematoxylin counterstain. Analysis of immunohistochemistry was performed as previously described [48, 49].
Immunofluorescence
The mice primary tumors and metastatic sites were stained according to immunofluorescence described before [50]. Mitotic and nuclear phenotypes of at least 100 cells per condition were assessed. Immunofluorescence images were obtained with a Zeiss Scope.A1.
Statistical analysis
All statistical analyses were performed using the GraphPad Prism Software (GraphPad Software). Data are presented as means ± SD of three independent experiments. The log-rank (Mantel–Cox) test was used to assess statistical significance of Kaplan–Meier plots. For comparisons, T test, Kruskal–Wallis Test, Dunn’s test, Pearson correlation analysis, and Log-rank test were performed as indicated. A P value of < 0.05 was considered statistically significant. *P < 0.05, **P < 0.01, ***P < 0.001.
Supplementary Information
Below is the link to the electronic supplementary material.
Abbreviations
- TNBC
Triple-negative breast cancer
- ER
Estrogen receptor
- PR
Progesterone receptor
- HER-2
Human epidermal growth factor receptor 2
- LEC
Lymphatic vessel endothelial cell
- LV
Lymphatic vessel
- EC
Endothelial cell
- EMT
Epithelial–mesenchymal transition
- LncRNA
Long non-coding RNA
- ceRNA
Competing endogenous RNA
- RIP
RNA immunoprecipitation
- GSEA
Gene set enrichment analysis
- OS
Overall survival
- RFS
Relapse-free survival
- DMFS
Distant metastasis-free survival
- BMSC
Bone mesenchymal stem cell
- IHC
Immunohistochemistry
Author contributions
WZ and XG designed the study. WZ, SY, DC, DY, JZ, XH and XW performed the experiments and acquired the data. WZ, SY, DC, XH and XW analyzed the data. WZ, SY, XH and XG contributed to discussion and writing of the manuscript.
Funding
This research was supported by National Natural Science Foundation of China (No. 81802667, 81773102), Natural Science Foundation of Jiangsu Province (BK20180133), Nanjing Outstanding Youth Fund (No. JQX20009), Key International Cooperation of the National Natural Science Foundation of China (No. 81920108029), and Key Foundation for Social Development Project of the Jiangsu Province, China (BE2021741).
Availability of data and material
All data and materials in this study are available upon request.
Declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
Studies were performed, according to the ethical guidelines of the Helsinki Declaration and were approved by the Clinical Research Ethics Committee of Nanjing First Hospital.
Consent for publication
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Wenwen Zhang, Shuofei Yang and Datian Chen have contributed equally to this work.
Contributor Information
Wenwen Zhang, Email: wwzhang1022@hotmail.com.
Xin Han, Email: xhan0220@njucm.edu.cn.
Xiaoxiang Guan, Email: xguan@njmu.edu.cn.
References
- 1.Bianchini G, Balko JM, Mayer IA, Sanders ME, Gianni L. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol. 2016;13(11):674–690. doi: 10.1038/nrclinonc.2016.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyle P. Triple-negative breast cancer: epidemiological considerations and recommendations. Ann Oncol. 2012;23(Suppl 6):vi7–12. doi: 10.1093/annonc/mds187. [DOI] [PubMed] [Google Scholar]
- 3.Karnoub AE, Dash AB, Vo AP, Sullivan A, Brooks MW, Bell GW, Richardson AL, Polyak K, Tubo R, Weinberg RA. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449(7162):557–563. doi: 10.1038/nature06188. [DOI] [PubMed] [Google Scholar]
- 4.Lee E, Fertig EJ, Jin K, Sukumar S, Pandey NB, Popel AS. Breast cancer cells condition lymphatic endothelial cells within pre-metastatic niches to promote metastasis. Nat Commun. 2014;5:4715. doi: 10.1038/ncomms5715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang W, Xu J, Fang H, Tang L, Chen W, Sun Q, Zhang Q, Yang F, Sun Z, Cao L, Wang Y, Guan X. Endothelial cells promote triple-negative breast cancer cell metastasis via PAI-1 and CCL5 signaling. FASEB J. 2018;32(1):276–288. doi: 10.1096/fj.201700237RR. [DOI] [PubMed] [Google Scholar]
- 6.Zemzoum I, Kates RE, Ross JS, Dettmar P, Dutta M, Henrichs C, Yurdseven S, Hofler H, Kiechle M, Schmitt M, Harbeck N. Invasion factors uPA/PAI-1 and HER2 status provide independent and complementary information on patient outcome in node-negative breast cancer. J Clin Oncol. 2003;21(6):1022–1028. doi: 10.1200/JCO.2003.04.170. [DOI] [PubMed] [Google Scholar]
- 7.Witzel I, Milde-Langosch K, Schmidt M, Karn T, Becker S, Wirtz R, Rody A, Laakmann E, Schutze D, Janicke F, Muller V. Role of urokinase plasminogen activator and plasminogen activator inhibitor mRNA expression as prognostic factors in molecular subtypes of breast cancer. Onco Targets Ther. 2014;7:2205–2213. doi: 10.2147/OTT.S65344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Declerck PJ, Gils A. Three decades of research on plasminogen activator inhibitor-1: a multifaceted serpin. Semin Thromb Hemost. 2013;39(4):356–364. doi: 10.1055/s-0033-1334487. [DOI] [PubMed] [Google Scholar]
- 9.Xu J, Zhang W, Tang L, Chen W, Guan X. Epithelial-mesenchymal transition induced PAI-1 is associated with prognosis of triple-negative breast cancer patients. Gene. 2018;670:7–14. doi: 10.1016/j.gene.2018.05.089. [DOI] [PubMed] [Google Scholar]
- 10.Humphries BA, Buschhaus JM, Chen YC, Haley HR, Qyli T, Chiang B, Shen N, Rajendran S, Cutter A, Cheng YH, Chen YT, Cong J, Spinosa PC, Yoon E, Luker KE, Luker GD. Plasminogen activator inhibitor 1 (PAI1) promotes actin cytoskeleton reorganization and glycolytic metabolism in triple-negative breast cancer. Mol Cancer Res. 2019;17(5):1142–1154. doi: 10.1158/1541-7786.MCR-18-0836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152(6):1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang M, Wang N, Song P, Fu Y, Ren Y, Li Z, Wang J. LncRNA GATA3-AS1 facilitates tumour progression and immune escape in triple-negative breast cancer through destabilization of GATA3 but stabilization of PD-L1. Cell Prolif. 2020;53(9):e12855. doi: 10.1111/cpr.12855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu T, Han C, Fang P, Zhu H, Wang S, Ma Z, Zhang Q, Xia W, Wang J, Xu L, Yin R. Long non-coding RNAs in lung cancer: implications for lineage plasticity-mediated TKI resistance. Cell Mol Life Sci. 2021;78(5):1983–2000. doi: 10.1007/s00018-020-03691-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu J, Yang B, Wang L, Zhu Y, Zhu X, Xia Z, Zhao Z, Xu L. LncRNA BBOX1-AS1 upregulates HOXC6 expression through miR-361–3p and HuR to drive cervical cancer progression. Cell Prolif. 2020;53(7):e12823. doi: 10.1111/cpr.12823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pandya G, Kirtonia A, Sethi G, Pandey AK, Garg M. The implication of long non-coding RNAs in the diagnosis, pathogenesis and drug resistance of pancreatic ductal adenocarcinoma and their possible therapeutic potential. Biochim Biophys Acta Rev Cancer. 2020;1874(2):188423. doi: 10.1016/j.bbcan.2020.188423. [DOI] [PubMed] [Google Scholar]
- 16.Hua JT, Chen S, He HH. Landscape of noncoding RNA in prostate cancer. Trends Genet. 2019;35(11):840–851. doi: 10.1016/j.tig.2019.08.004. [DOI] [PubMed] [Google Scholar]
- 17.Wong NK, Huang CL, Islam R, Yip SP. Long non-coding RNAs in hematological malignancies: translating basic techniques into diagnostic and therapeutic strategies. J Hematol Oncol. 2018;11(1):131. doi: 10.1186/s13045-018-0673-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang W, Guan X, Tang J. The long non-coding RNA landscape in triple-negative breast cancer. Cell Prolif. 2021;54(2):e12966. doi: 10.1111/cpr.12966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21(11):1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 20.Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2021;22(2):96–118. doi: 10.1038/s41580-020-00315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu SJ, Dang HX, Lim DA, Feng FY, Maher CA. Long noncoding RNAs in cancer metastasis. Nat Rev Cancer. 2021;21(7):446–460. doi: 10.1038/s41568-021-00353-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin X, Xu XE, Jiang YZ, Liu YR, Sun W, Guo YJ, Ren YX, Zuo WJ, Hu X, Huang SL, Shen HJ, Lan F, He YF, Hu GH, Di GH, He XH, Li DQ, Liu S, Yu KD, Shao ZM. The endogenous retrovirus-derived long noncoding RNA TROJAN promotes triple-negative breast cancer progression via ZMYND8 degradation. Sci Adv. 2019;5(3):eaat9820. doi: 10.1126/sciadv.aat9820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang H, Zhang N, Liu Y, Su P, Liang Y, Li Y, Wang X, Chen T, Song X, Sang Y, Duan Y, Zhang J, Wang L, Chen B, Zhao W, Guo H, Liu Z, Hu G, Yang Q. Epigenetic regulation of NAMPT by NAMPT-AS drives metastatic progression in triple-negative breast cancer. Can Res. 2019;79(13):3347–3359. doi: 10.1158/0008-5472.CAN-18-3418. [DOI] [PubMed] [Google Scholar]
- 24.Yang F, Shen Y, Zhang W, Jin J, Huang D, Fang H, Ji W, Shi Y, Tang L, Chen W, Zhou G, Guan X. An androgen receptor negatively induced long non-coding RNA ARNILA binding to miR-204 promotes the invasion and metastasis of triple-negative breast cancer. Cell Death Differ. 2018;25(12):2209–2220. doi: 10.1038/s41418-018-0123-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B, Varambally S. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ringner M, Fredlund E, Hakkinen J, Borg A, Staaf J. GOBO: gene expression-based outcome for breast cancer online. PLoS ONE. 2011;6(3):e17911. doi: 10.1371/journal.pone.0017911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagy A, Munkacsy G, Gyorffy B. Pancancer survival analysis of cancer hallmark genes. Sci Rep. 2021;11(1):6047. doi: 10.1038/s41598-021-84787-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mei J, Hao L, Wang H, Xu R, Liu Y, Zhu Y, Liu C. Systematic characterization of non-coding RNAs in triple-negative breast cancer. Cell Prolif. 2020;53(5):e12801. doi: 10.1111/cpr.12801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 31.Jaeger J, Koczan D, Thiesen HJ, Ibrahim SM, Gross G, Spang R, Kunz M. Gene expression signatures for tumor progression, tumor subtype, and tumor thickness in laser-microdissected melanoma tissues. Clin Cancer Res. 2007;13(3):806–815. doi: 10.1158/1078-0432.CCR-06-1820. [DOI] [PubMed] [Google Scholar]
- 32.Provenzani A, Fronza R, Loreni F, Pascale A, Amadio M, Quattrone A. Global alterations in mRNA polysomal recruitment in a cell model of colorectal cancer progression to metastasis. Carcinogenesis. 2006;27(7):1323–1333. doi: 10.1093/carcin/bgi377. [DOI] [PubMed] [Google Scholar]
- 33.Sung SY, Hsieh CL, Law A, Zhau HE, Pathak S, Multani AS, Lim S, Coleman IM, Wu LC, Figg WD, Dahut WL, Nelson P, Lee JK, Amin MB, Lyles R, Johnstone PA, Marshall FF, Chung LW. Coevolution of prostate cancer and bone stroma in three-dimensional coculture: implications for cancer growth and metastasis. Can Res. 2008;68(23):9996–10003. doi: 10.1158/0008-5472.CAN-08-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rickman DS, Millon R, De Reynies A, Thomas E, Wasylyk C, Muller D, Abecassis J, Wasylyk B. Prediction of future metastasis and molecular characterization of head and neck squamous-cell carcinoma based on transcriptome and genome analysis by microarrays. Oncogene. 2008;27(51):6607–6622. doi: 10.1038/onc.2008.251. [DOI] [PubMed] [Google Scholar]
- 35.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147(2):358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou L, Chen Q, Wu J, Yang J, Yin H, Tian J, Gong L, Kong D, Tao M. miR-942-5p inhibits proliferation, metastasis, and epithelial-mesenchymal transition in colorectal cancer by targeting CCBE1. Biomed Res Int. 2021;2021:9951405. doi: 10.1155/2021/9951405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Du Z, Wang L, Xia Y. Circ_0015756 promotes the progression of ovarian cancer by regulating miR-942-5p/CUL4B pathway. Cancer Cell Int. 2020;20(1):572. doi: 10.1186/s12935-020-01666-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ou R, Mo L, Tang H, Leng S, Zhu H, Zhao L, Ren Y, Xu Y. circRNA-AKT1 sequesters miR-942-5p to upregulate AKT1 and promote cervical cancer progression. Mol Ther Nucleic Acids. 2020;20:308–322. doi: 10.1016/j.omtn.2020.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ouyang Z, Tan T, Zhang X, Wan J, Zhou Y, Jiang G, Yang D, Guo X, Liu T. CircRNA hsa_circ_0074834 promotes the osteogenesis-angiogenesis coupling process in bone mesenchymal stem cells (BMSCs) by acting as a ceRNA for miR-942-5p. Cell Death Dis. 2019;10(12):932. doi: 10.1038/s41419-019-2161-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xie J, Wang S, Li G, Zhao X, Jiang F, Liu J, Tan W. circEPSTI1 regulates ovarian cancer progression via decoying miR-942. J Cell Mol Med. 2019;23(5):3597–3602. doi: 10.1111/jcmm.14260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bajou K, Peng H, Laug WE, Maillard C, Noel A, Foidart JM, Martial JA, DeClerck YA. Plasminogen activator inhibitor-1 protects endothelial cells from FasL-mediated apoptosis. Cancer Cell. 2008;14(4):324–334. doi: 10.1016/j.ccr.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Romer MU, Larsen L, Offenberg H, Brunner N, Lademann UA. Plasminogen activator inhibitor 1 protects fibrosarcoma cells from etoposide-induced apoptosis through activation of the PI3K/Akt cell survival pathway. Neoplasia. 2008;10(10):1083–1091. doi: 10.1593/neo.08486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xi X, Liu N, Wang Q, Chu Y, Yin Z, Ding Y, Lu Y. ACT001, a novel PAI-1 inhibitor, exerts synergistic effects in combination with cisplatin by inhibiting PI3K/AKT pathway in glioma. Cell Death Dis. 2019;10(10):757. doi: 10.1038/s41419-019-1986-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Song X, Yao H, Liu J, Wang Q. The prognostic value of long noncoding RNA Sox2ot expression in various cancers: a systematic review and meta-analysis. Clin Chim Acta. 2018;484:52–59. doi: 10.1016/j.cca.2018.05.038. [DOI] [PubMed] [Google Scholar]
- 45.Tai Y, Ji Y, Liu F, Zang Y, Xu D, Ma S, Qin L, Ma J. Long noncoding RNA SOX2-OT facilitates laryngeal squamous cell carcinoma development by epigenetically inhibiting PTEN via methyltransferase EZH2. IUBMB Life. 2019;71(9):1230–1239. doi: 10.1002/iub.2026. [DOI] [PubMed] [Google Scholar]
- 46.Li Z, Jiang P, Li J, Peng M, Zhao X, Zhang X, Chen K, Zhang Y, Liu H, Gan L, Bi H, Zhen P, Zhu J, Li X. Tumor-derived exosomal lnc-Sox2ot promotes EMT and stemness by acting as a ceRNA in pancreatic ductal adenocarcinoma. Oncogene. 2018;37(28):3822–3838. doi: 10.1038/s41388-018-0237-9. [DOI] [PubMed] [Google Scholar]
- 47.Zhang W, Cao L, Sun Z, Xu J, Tang L, Chen W, Luo J, Yang F, Wang Y, Guan X. Skp2 is over-expressed in breast cancer and promotes breast cancer cell proliferation. Cell Cycle. 2016;15(10):1344–1351. doi: 10.1080/15384101.2016.1160986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang W, Luo J, Yang F, Wang Y, Yin Y, Strom A, Gustafsson JA, Guan X. BRCA1 inhibits AR-mediated proliferation of breast cancer cells through the activation of SIRT1. Sci Rep. 2016;6:22034. doi: 10.1038/srep22034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang W, Luo J, Chen F, Yang F, Song W, Zhu A, Guan X. BRCA1 regulates PIG3-mediated apoptosis in a p53-dependent manner. Oncotarget. 2015;6(10):7608–7618. doi: 10.18632/oncotarget.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Song W, Tang L, Xu Y, Xu J, Zhang W, Xie H, Wang S, Guan X. PARP inhibitor increases chemosensitivity by upregulating miR-664b-5p in BRCA1-mutated triple-negative breast cancer. Sci Rep. 2017;7:42319. doi: 10.1038/srep42319. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and materials in this study are available upon request.