Abstract
MDM2 is induced by p53 in response to cellular insults such as DNA damage and can have effects upon the cell cycle that are independent or downstream of p53. We used a yeast two-hybrid screen to identify proteins that bind to MDM2 and which therefore might be involved in these effects. We found that MDM2 can bind to the C-terminus of the catalytic subunit of DNA polymerase ɛ (DNA pol ɛ), to a region that is known to be essential in yeast. In an in vitro system we confirmed that MDM2 could bind to the homologous regions of both mouse and human DNA pol ɛ and to full-length human DNA pol ɛ. DNA pol ɛ co-immunoprecipitated with MDM2 from transfected H1299 cells and also from a HeLa cell nuclear extract. We show here that the DNA pol ɛ-interacting domain of MDM2 is located between amino acids 50 and 166. Our studies provide evidence that MDM2 interacts with a region of DNA pol ɛ that plays a critical role in the function of DNA pol ɛ.
INTRODUCTION
MDM2 was originally discovered as one of three genes amplified on double minute chromosomes in a tumourigenic derivative of NIH 3T3 cells (1) and it was later shown that MDM2 has oncogenic potential when overexpressed (2,3). High level expression of MDM2 has also been shown to confer tumourigenic potential upon non-transformed rodent fibroblasts in athymic nude mice (2,3). MDM2 can immortalise rat embryo fibroblasts and can cooperate with activated RAS to transform these cells (2). Elevated levels of MDM2 have been found in a variety of human tumours, most notably in soft tissue sarcomas where 20% of primary tumours contain multiple copies of the MDM2 gene (4). One mechanism by which MDM2 overexpression may lead to tumour development is through its ability to bind to the p53 tumour suppressor (5,6) and block the transcriptional activation function of p53 (5). MDM2 overexpression has been shown to block the transactivation (5,7), cell cycle arrest (8,9) and apoptotic functions of p53 (8,9). In addition, it has been shown that MDM2 regulates the stability of p53 by inducing ubiquitination and thus targeting the protein for degradation by proteosomes (10–12). Studies of primary human tumours have reinforced the idea that MDM2 overexpression inhibits p53 function in vivo. It has been shown that the p53 gene is rarely mutated in tumours in which the MDM2 gene is amplified (13).
MDM2 is itself a transcriptional target of p53 and induction of p53 transcriptional activity leads to increases in MDM2 mRNA and MDM2 protein (14,15). Thus an autoregulatory feedback loop (15) exists between these two proteins. The importance of the MDM2–p53 autoregulatory feedback loop has been confirmed by transgenic animal studies. Mice that possess a homozygous deletion of MDM2 die by day 7 of embryogenesis, whereas mice that possess homozygous deletion of both MDM2 and p53 are viable and develop normally (16,17). These results demonstrate that a primary function of MDM2 during development is to regulate p53 function.
Notwithstanding the critical role MDM2 plays in regulating p53, there is good evidence that high levels of MDM2 alter cellular growth control pathways in a p53-independent manner. For example, MDM2 has been shown to interact with a number of additional growth control molecules, namely RB (18), the E2F-1/DP1 transcription complex (19) and p19ARF (20). Transfection of MDM2 into p53 null cells can alter the growth properties of these cells (21) and MDM2 isoforms that cannot bind to p53 retain the ability to transform NIH 3T3 cells (22). Moreover, MDM2 overexpression can block the growth inhibitory activities of transforming growth factor β1 (TGF-β1) in a p53-independent manner in cultured cells (23). Overexpression of MDM2 has been shown to induce uncoupling of S phase from mitosis in both p53 null and E2F-1 null cells in vivo (24) and p53 null transgenic mice that overexpress MDM2 develop a different spectrum of tumours compared with p53 null mice that do not overexpress MDM2 (25). Taken together these results suggest that MDM2 overexpression can, independent of the interaction with p53, alter cell growth control. In an effort to identify how MDM2 may mediate its effects we have used a yeast two-hybrid screen to identify novel MDM2-binding proteins that may shed some light upon the underlying pathways responsible for these effects.
We have found that MDM2 can bind to the C-terminus of the catalytic subunit of DNA polymerase ɛ (DNA pol ɛ). DNA pol ɛ is an essential molecule in both budding and fission yeast (26,27) and is thought to be involved in a number of cellular processes, including DNA replication (27,28), DNA repair (29–32) and, in budding yeast (but not in fission yeast; 33), the S phase checkpoint (34,35). The MDM2-binding domain on DNA pol ɛ was found to be located within a region of DNA pol ɛ (the C-terminus) that is not required for catalytic activity of the human enzyme in vitro (36) but which is essential for viability in budding yeast (37). Although this region of DNA pol ɛ has not yet been shown to play a role in these processes in multicellular organisms, it is conserved from yeast up to man (38). Our studies provide evidence that MDM2 interacts with a region of DNA pol ɛ that plays a critical role in the function of DNA pol ɛ.
MATERIALS AND METHODS
Plasmids and antibodies
pGAL4-DBD-MDM2 encodes full-length mouse MDM2 cloned in-frame with the GAL4-DNA-binding domain (DBD) and pGAL4-AD-DNA pol ɛ encodes nt 5833–6984 (numbering based on the human cDNA sequence, GenBank accession no. L09561) plus ∼500 bases of the 3′-untranslated region of murine DNA pol ɛ as a GAL4 activation domain (AD) fusion protein in the XhoI site of pACT (Clontech). pGAL4-AD-p53 encodes full-length p53 in the XhoI site of pACT. pQE32-MDM2 was prepared by cloning full-length murine MDM2 as an EcoRV–XhoI fragment from pBBV-MDM2 into the SmaI site of pQE32 (Qiagen). The murine C-terminal fragment of DNA pol ɛ was prepared by PCR amplification from pGAL4AD-DNA pol ɛ using oligonucleotides GAD5 (5′-gag aga gat atc gcc aat ttt aat caa agt ggg aat att-3′) and GAD3 (5′-gag aga gcg gcc gct ttc agt atc tac gat tca tag atc tc-3′) followed by restriction digestion with EcoRV and NotI and cloning into these sites in pBBV. pBBV was prepared by inserting an oligonucleotide containing the black beetle virus ribosomal binding sequences from pBD7 (39) into the HindIII and EcoRV sites of pcDNAI/Neo (Invitrogen). The human C-terminal fragment of DNA pol ɛ was made from cDNA prepared from MCF-7 cells (ATCC clone HTB 22) by amplification with oligonucleotides DNA pol ɛ 5 (5′-gag tct aga gga atc caa cgt gga gga ttt a-3′) and DNA pol ɛ 3 (5′-gag gaa ttc cta atg gcc cag ctg tgg gtt ctt-3′) followed by cloning into the EcoRV and XbaI sites of pBD7. 5′ɛ and 3′ɛ contain cDNAs corresponding to amino acids 1–998 and 996–2287, respectively, of human DNA pol ɛ cloned into in pSK-BBV. p53 contains full-length murine p53 in the pBBV vector prepared from pGAL4-AD-p53 by excising a BamHI–BglII fragment and cloning this into the EcoRV site of pBBV. Full-length human DNA pol ɛ was amplified from MCF-7 cDNA using rTth-XL (Perkin Elmer) and oligonucleotides Pol ɛ 5 (5′-gag agg tac ccc acc ggc tcc atg tct ctg ag-3′) and Pol ɛ 3 (5′-gag aga gtc gac cta atg gcc cag ctg tgg gtt ctt-3′). pCEP-DNA pol ɛ contains full-length human cDNA from DNA pol ɛ cloned into the KpnI and SalI sites of pCEP4 (Invitrogen). Full-length DNA pol ɛ for in vitro transcription/translation reactions was generated by PCR from pCEP-DNA pol ɛ with primers Pol ɛ 5′-XbaI (5′-gag atc tag aca acg gct cca tgt ctc tga g-3′) and Pol ɛ 3′-XbaI (5′-gag atc tag act aat ggc cca gct gtg ggt t-3′) using rTth-XL and subcloned into the XbaI site of pSK-BBV (generated by subcloning the HindIII–BglII fragment from pBBV into the HindIII and BamHI sites of pBluescript SKII+; Stratagene). Plasmids pCMVBamNeo, pCMVBamNeo-MDM2 and p53SN3 were obtained from B. Vogelstein.
The 3C5.1 antibody to DNA pol ɛ for western blotting has been described previously (40). The MDM2-specific antibodies Ab-1 (IF2) and SMP14 were purchased from Oncogene Research Products and Santa Cruz Biotechnology, respectively. The anti-hemagglutinin (HA) antibody 12CA5 was purchased from Roche Molecular Biochemicals.
Yeast two-hybrid analysis and construction of MDM2 deletion mutants
We utilised Clontech’s Matchmaker system to screen a mouse T cell lymphoma library (ML4001AE) for MDM2-interacting clones. The same system was used for analysis of the interaction between the C-terminus of DNA pol ɛ and the various MDM2 deletion mutants. To generate C-terminal deletion mutants in pGAL4-DBD-MDM2, a stuffer fragment (5′-gag act cga gct gca ggt cga cgc ggc cgc ggt acc gca tgc ctg cag ctg gag gag a-3′) was first cloned into the PstI site of this vector. The vector was then restricted with KpnI and NotI and, after purification, DNA was digested with ExoIII for varying periods of time, blunt-ended with Klenow DNA polymerase and religated. Recombinants containing MDM2 deletions were identified after digestion with EcoRI and PstI and sequenced. Transformants were grown overnight in trp–, leu– medium and then streaked onto a trp–, leu–, his– plate. Plates were placed at 30°C for 3 days and the growth of yeast monitored.
In vitro binding assay
MDM2 was expressed in XL-1 bacteria (Stratagene) from the pQE32-MDM2 construct, captured on Ni2+–agarose and washed with buffers B, C and D as described by the manufacturer (Qiagen). Prior to all binding reactions, protein captured on beads was run on a SDS–polyacrylamide gel and Coomassie blue stained to confirm that full-length MDM2, Δ1–49 MDM2 and Δ1–166 MDM2 were successfully purified on the Ni2+–agarose beads. An aliquot of 100 µl of washed beads was then mixed with 10 µl of in vitro translated protein (TNT; Promega) for 3 h at 30°C, followed by washing three times in wash buffer (20 mM HEPES, pH 7.9, 20% glycerol, 0.1 M KCl) supplemented with 75 mM imidazole. Beads were then resuspended in SDS–PAGE loading buffer and analysed by SDS–PAGE and fluorography.
Immunoprecipitation and western blotting
H1299 cells (ATCC clone CRL-5803) were grown and transfected with calcium phosphate as described previously (41). Transfected H1299 cells were then lysed in IP buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 10% glycerol, 0.1% Triton X-100, 0.5 mg/ml BSA, 1 mM PMSF and 1 µg/ml each pepstatin, leupeptin, aprotinin and soybean trypsin inhibitor; Roche Molecular Biochemicals), placed on ice for 10 min, clarified by centrifugation for 10 min and 1–5 mg protein incubated for 1 h at 4°C with 50 µl of protein G–Sepharose (Pharmacia). Beads were removed by centrifugation and the supernatant was then incubated with 1 µg SMP14 antibody or a control antibody of the same isotype for 1 h at 4°C, followed by 2 h at 4°C with 50 µl of protein G–Sepharose. Beads were washed three times in IP buffer and then resuspended in 30 µl of loading buffer prior to SDS–PAGE. Proteins were transferred to Hybond-ECL membrane, probed with antibodies 3C5.1 and IF2 and detected with anti-mouse Ig–horseradish peroxidase and Luminol (Renaissance; NEN).
RESULTS
MDM2 interacts with DNA polymerase ɛ in yeast
To identify novel candidate MDM2-binding proteins we performed a yeast two-hybrid screen using MDM2 as the bait. Numerous candidate clones were identified that appeared to specifically interact with MDM2 in yeast. These cDNAs were subsequently screened using an in vitro binding assay. Four clones were found to encode a peptide that clearly and reproducibly bound to MDM2 in this assay (M.T.Boyd, D.S.Haines and N.Vlatkovic, unpublished results). Sequence analysis of these revealed that each encoded a different protein, namely full-length p53, a C-terminal fragment of the 34 kDa subunit of TFIIE (42), the C-terminus of the catalytic subunit of DNA pol ɛ and a previously unidentified protein (M.T.Boyd, D.S.Haines and N.Vlatkovic, submitted for publication). This report focuses on characterisation of the interaction between MDM2 and DNA pol ɛ.
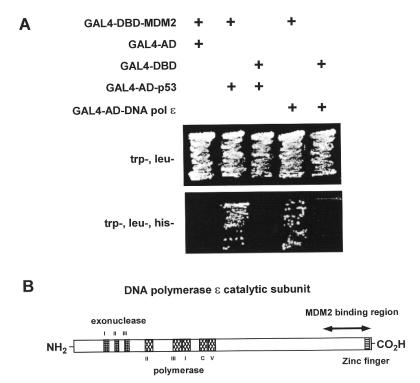
Figure 1A shows that MDM2 interacts with the C-terminus of DNA pol ɛ. All of the yeast transformants were able to grow on the trp–, leu– plate but only those yeast that had been transfomed with MDM2 together with either the C-terminal DNA pol ɛ or p53 fusion constructs were able to grow on the trp–, leu–, his– plate. The region of DNA pol ɛ that was identified in this analysis is shown in Figure 1B.
Figure 1.
MDM2 interacts with the C-terminus of DNA pol ɛ in yeast. (A) Yeast transformed with the indicated plasmids were grown on plates lacking the indicated amino acids. Only yeast containing plasmid constructs for MDM2 together with either p53 or the C-terminus of DNA pol ɛ grew on the trp–, leu–, his– plate. (B) Diagram of the human DNA pol ɛ catalytic subunit with the region identified in our yeast two-hybrid screen and conserved domains indicated.
MDM2 interacts with DNA polymerase ɛ in vitro
We next tested the ability of MDM2 to interact with DNA pol ɛ in vitro. cDNAs encoding the C-terminus of murine DNA pol ɛ (murine 3′ɛ) (the same region present in the GAL4AD-DNA pol ɛ fusion construct), the corresponding region of human DNA pol ɛ (human 3′ɛ) and full-length human DNA pol ɛ were cloned into an in vitro transcription/translation vector. Figure 2 shows that all three forms of DNA pol ɛ bind to recombinant MDM2 (MDM2) prepared from XL-1 bacterial cells and not to protein prepared in the same manner from vector-transformed XL-1 bacteria (XL-1). In vitro synthesised p53 is also shown as a positive control for MDM2 binding. These results demonstrate that MDM2 can bind to the C-terminus of both murine and human DNA pol ɛ and also to full-length human DNA pol ɛ in vitro.
Figure 2.
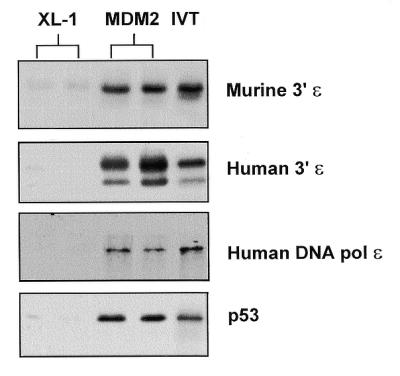
MDM2 interacts with DNA pol ɛ in vitro. Results from an in vitro binding assay in which in vitro translated radiolabelled proteins were mixed with purified protein from XL-1 bacterial cells (XL-1) or recombinant murine MDM2 protein (MDM2). IVT indicates 10% of the input in vitro translated protein. Assays were performed in duplicate as shown.
MDM2 interacts with DNA polymerase ɛ in mammalian cells
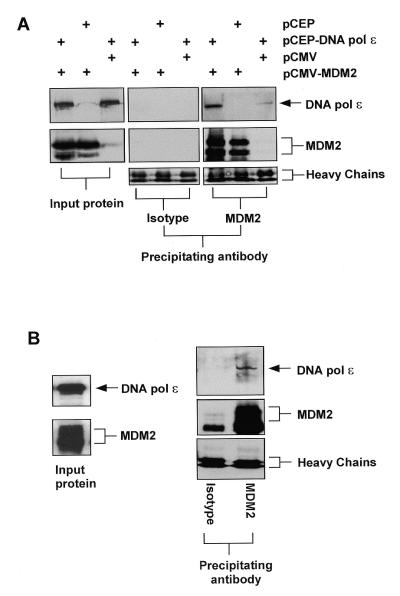
We next wanted to determine whether MDM2 and full-length DNA pol ɛ could interact in mammalian cells. H1299 cells were transfected with constructs expressing either DNA pol ɛ or MDM2 or both as indicated. Following transfection, protein extracts were prepared and immunoprecipitations were performed with either an MDM2-specific antibody or with an isotype-matched control antibody. Western blots were then probed with either an antibody specific for MDM2 or for DNA pol ɛ. Figure 3A shows that after immunoprecipitation with the MDM2-specific antibody, DNA pol ɛ could be detected in extracts from DNA pol ɛ-transfected H1299 cells and in H1299 cells transfected with both MDM2 and DNA pol ɛ, but not in extracts from cells transfected with MDM2 alone. Note that we attempted to perform the reciprocal experiment, id est using an anti-DNA pol ɛ antibody for the immunoprecipitation step, but were thwarted by the presence of a non-specific precipitating protein that co-migrated with MDM2 and which cross-reacted with our secondary antibody (not shown). The ability to detect DNA pol ɛ in the extracts from DNA pol ɛ-transfected cells in the absense of transfected MDM2 is most likely due to the interaction of transfected DNA pol ɛ with endogenous MDM2 present in these cells. The amount of immunoprecipitated DNA pol ɛ was found to be greater in extracts from cells that had been transfected with both DNA pol ɛ and MDM2 expression constructs. This suggests that DNA pol ɛ is the limiting factor in detecting these MDM2–DNA pol ɛ complexes. DNA pol ɛ could not be detected in any of the extracts from the transfected cells that had been immunoprecipitated with an antibody of the same isotype but specific for a heterologous protein. These results demonstrate that MDM2 and DNA pol ɛ can form stable protein complexes in transfected cells.
Figure 3.
MDM2 interacts with DNA pol ɛ in cells. (A) Immunoprecipitations using either an MDM2-specific antibody (SMP14) or a control antibody of the same isotype were performed on extracts of H1299 cells transfected with 10 µg of the indicated plasmid. Following immunoprecipitation and SDS–PAGE, western blot analysis was performed using antibodies specific for either DNA pol ɛ or MDM2 as indicated. The panels on the left are western blots of the protein extract used as the input for the immunoprecipitations probed with the indicated antibodies as for the panels on the right. Also shown is the signal obtained from the heavy chain of the immunoprecipitating antibody which indicates that approximately equal amounts of each were precipitated. (B) Immunoprecipitation and western blot analysis using HeLa cell nuclear extract essentially as described in (A).
In order to determine whether MDM2 and DNA pol ɛ can form stable complexes in untransfected cells, we performed similar precipitation experiments with a HeLa cell nuclear extract. This extract contains an ∼10-fold enrichment of both MDM2 and DNA pol ɛ protein per µg total protein when compared with extracts prepared from unfractionated H1299 cells. Figure 3B shows that DNA pol ɛ can indeed be detected precipitating in a complex with MDM2 in this nuclear extract. As before, DNA pol ɛ could not be detected in immunoprecipitations performed with an isotype control antibody. These results suggest that DNA pol ɛ and MDM2 can form stable protein complexes in mammalian cells.
Amino acids 50–166 of MDM2 are sufficient for binding to DNA polymerase ɛ
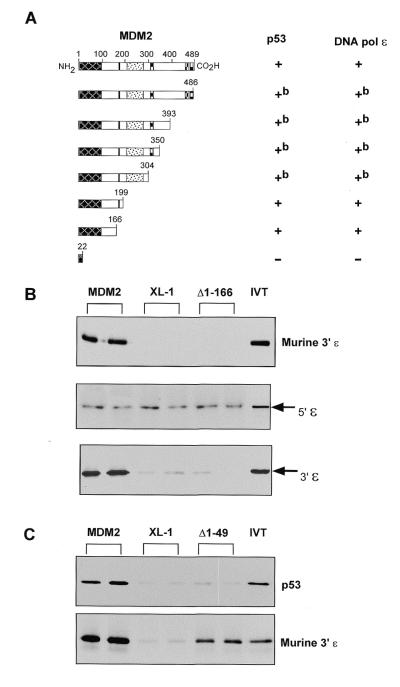
To identify the region of MDM2 that is required for binding to the C-terminal region of DNA pol ɛ, we constructed a series of C-terminal deletion mutants of MDM2 in the pGAL4-DBD-MDM2 construct. Deletion mutants of MDM2 were transformed into yeast containing either pGAL4-AD-p53 or pGAL4-AD-DNA pol ɛ as indicated. Figure 4A demonstrates that an MDM2 deletion mutant containing only the first 166 amino acids of MDM2 is sufficient for interaction with DNA pol ɛ. As expected, this same mutant also retains the ability to interact with p53 (see Fig. 4A; 41). These results demonstrate that the first 166 amino acids of MDM2 are sufficient to interact with the C-terminal region of DNA pol ɛ in yeast.
Figure 4.
Amino acids 50–166 of MDM2 contain the DNA pol ɛ-binding region. (A) Yeast expressing, as indicated, pGAL4-AD-p53 or pGAL4-AD-DNA pol ɛ were transformed with various pGAL4-DBD-MDM2 deletion mutants. Colony growth on trp–, leu–, his– plates is indicated as + and background or no growth is indicated by –. bThese mutants displayed limited intrinsic ability to grow on trp–, leu–, his– plates in the presence of pGAL4-AD (not shown). (B) Results from an in vitro binding assay in which the indicated in vitro translated radiolabelled proteins were mixed with purified protein from XL-1 bacterial cells (XL-1) or recombinant murine MDM2 protein (MDM2) or Δ1–166. 5′ɛ and 3′ɛ contain amino acids 1–998 and 996–2287, respectively, of human DNA pol ɛ. IVT indicates 10% of the input in vitro translated protein. Assays were performed in duplicate as shown. (C) An in vitro binding assay was performed essentially as above with in vitro translated DNA pol ɛ (murine 3′ɛ) or p53 and purified protein from XL-1 bacterial cells (XL-1) or recombinant MDM2 protein (MDM2) or Δ1–49.
To confirm these results using a different system and to further define the DNA pol ɛ binding site on MDM2, two N-terminal deletion mutants of MDM2 were generated. The first mutant (Δ1–166) lacks the region of MDM2 that was found to be necessary for DNA pol ɛ binding in yeast. Figure 4B shows that the C-terminus of murine DNA pol ɛ binds to full-length MDM2 but not to the Δ1–166 mutant. Also shown are binding results for mutants of human DNA pol ɛ that express N- and C-terminal moieties of this molecule. These results indicate that the region of DNA pol ɛ that is necessary to bind to MDM2 is encoded within the C-terminal half of the molecule. Note that the background is higher for the N-terminal moiety of DNA pol ɛ and this may reflect a higher affinity of this portion of the molecule, perhaps because it contains the catalytic region, for the polyanionic bead matrix (agarose). The second MDM2 mutant (Δ1–49) lacks the first 49 amino acids of MDM2 (a region required for p53 binding). Figure 4C shows that the murine C-terminal region of DNA pol ɛ is able to bind to full-length MDM2 in vitro and also to Δ1–49. As expected, p53 does not bind to Δ1–49. Thus we conclude that the minimal DNA pol ɛ binding region of MDM2 is located between amino acids 50 and 166.
DISCUSSION
The pathways by which high level expression of MDM2 alters cellular growth control in a p53-independent manner are unclear. We report here the identification of a novel MDM2-binding protein, DNA pol ɛ. This interaction was identified in a yeast two-hybrid screen and confirmed in vitro and by co-immunoprecipitation. Our results beg the question what is the functional significance of an MDM2–DNA pol ɛ interaction? We have performed numerous experiments to address this, but have not detected any effect of either MDM2 upon DNA pol ɛ or vice versa. MDM2 does not alter the catalytic activity of human DNA pol ɛ in vitro (M.T.Boyd, Y.Li, S.Linn and N.Vlatkovic, unpublished results). This may not be surprising considering that the MDM2-binding region on DNA pol ɛ (the C-terminus) is not contained within the domain that is necessary for catalytic activity of the enzyme in in vitro assays (36). MDM2 can induce proteasome-mediated degradation of p53 (10,11) and Numb (43). Therefore, we also examined whether MDM2 could alter the level of DNA pol ɛ protein, but we found no evidence for this (M.T.Boyd, D.S.Haines and N.Vlatkovic, unpublished results). This is not the first example of an MDM2-binding protein that is not targeted for degradation by binding to MDM2 (44,45).
What other clues are there to this question of function? It has been shown in a transgenic animal model that MDM2 overexpressing cells go through multiple rounds of DNA replication prior to cytokinesis in vivo (24). This phenotype is observed in both p53 null (24) and E2F-1 null (46) backgrounds. The effect appears to be due to the ability of MDM2 to compromise the S phase checkpoint. In yeast this checkpoint has been shown to be dependent upon the DNA polymerase–α-primase complex in fission yeast (47,48) and DNA pol ɛ in budding yeast (34). Moreover, the region of DNA pol ɛ that we have identified as binding to MDM2 overlaps with the region of the yeast gene that is known to be essential for the S phase checkpoint function. This region of DNA pol ɛ contains a zinc finger that is conserved between yeast and humans (38). However, yeast do not possess MDM2 nor do they possess p53. Since MDM2 is up-regulated in a p53-dependent manner during the DNA damage response, the normal function(s) of MDM2 is likely to be dependent upon such a response pathway. From this it seems possible that MDM2 might form part of an S phase checkpoint in metazoan cells and thus may couple the stress response to the normal S phase checkpoint machinery, namely DNA pol ɛ. Given that DNA pol ɛ appears to be dispensable as a polymerase in yeast, it has been suggested that it plays more of a structural role in the replication complex (49). Thus the interaction that we have identified might be part of a recruitment process for MDM2 such that MDM2 may mediate other effects upon the regulation of DNA synthesis. One could reconcile many of the p53-independent effects of MDM2 if it were recruited to the replication complex and activated DNA synthesis. It is noteworthy that RB has been shown to bind to and stimulate the catalytic activity (in vitro) of DNA polymerase α (50). In addition, it has been shown that p53, MDM2 and p19ARF co-localise to distinct nuclear structures and it has been proposed that these structures may be involved in DNA synthesis (51). Clearly there exists considerable uncertainty regarding the function of DNA pol ɛ, particularly in metazoans. Our studies provide evidence that MDM2 interacts with a region of DNA pol ɛ that is essential in yeast and which may well play an important role in the function of DNA pol ɛ in mammalian cells.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported in part by grants from the Breast Health Institute of Philadelphia (to M.T.B.), the W.W. Smith Charitable Trust (to M.T.B. and D.S.H.) and by NIH grants CA70165 (to D.S.H.) and GM30415 (to S.L.).
REFERENCES
- 1.Cahilly-Snyder L., Yang-Feng,T., Francke,U. and George,D.L. (1987) Somat. Cell Mol. Genet., 13, 235–244. [DOI] [PubMed] [Google Scholar]
- 2.Finlay C.A. (1993) Mol. Cell. Biol., 13, 301–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fakharzadeh S.S., Trusko,S.P. and George,D.L. (1991) EMBO J., 10, 1565–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Momand J., Jung,D., Wilczynski,S. and Niland,J. (1998) Nucleic Acids Res., 26, 3453–3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Momand J., Zambetti,G.P., Olson,D., George,D.L. and Levine,A.J. (1992) Cell, 69, 1237–1245. [DOI] [PubMed] [Google Scholar]
- 6.Oliner J.D., Kinzler,K.W., Meltzer,P.S., George,D.L. and Vogelstein,B. (1992) Nature, 358, 80–83. [DOI] [PubMed] [Google Scholar]
- 7.Oliner J.D., Pietenpol,J.A., Thiagalingam,S., Gyuris,J., Kinzler,K.W. and Vogelstein,B. (1993) Nature, 362, 857–860. [DOI] [PubMed] [Google Scholar]
- 8.Chen J., Wu,X., Lin,J. and Levine,A.J. (1996) Mol. Cell. Biol., 16, 2445–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haupt Y., Barak,Y. and Oren,M. (1996) EMBO J., 15, 1596–1606. [PMC free article] [PubMed] [Google Scholar]
- 10.Kubbutat M.H., Jones,S.N. and Vousden,K.H. (1997) Nature, 387, 299–303. [DOI] [PubMed] [Google Scholar]
- 11.Haupt Y., Maya,R., Kazaz,A. and Oren,M. (1997) Nature, 387, 296–299. [DOI] [PubMed] [Google Scholar]
- 12.Fuchs S.Y., Adler,V., Buschmann,T., Wu,X. and Ronai,Z. (1998) Oncogene, 17, 2543–2547. [DOI] [PubMed] [Google Scholar]
- 13.Leach F.S., Tokino,T., Meltzer,P., Burrell,M., Oliner,J.D., Smith,S., Hill,D.E., Sidransky,D., Kinzler,K.W. and Vogelstein,B. (1993) Cancer Res., 53, 2231–2234. [PubMed] [Google Scholar]
- 14.Barak Y., Juven,T., Haffner,R. and Oren,M. (1993) EMBO J., 12, 461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu X., Bayle,J.H., Olson,D. and Levine,A.J. (1993) Genes Dev., 7, 1126–1132. [DOI] [PubMed] [Google Scholar]
- 16.Montes de Oca Luna R., Wagner,D.S. and Lozano G. (1995) Nature, 378, 203–206. [DOI] [PubMed] [Google Scholar]
- 17.Jones S.N., Roe,A.E., Donehower,L. and Bradley,A. (1995) Nature, 378, 206–208. [DOI] [PubMed] [Google Scholar]
- 18.Xiao Z.-X., Chen,J., Levine,A.J., Modjtahedl,N., Xing,J., Sellers,W.R. and Livingston,D.M. (1995) Nature, 375, 694–698. [DOI] [PubMed] [Google Scholar]
- 19.Martin K., Trouche,D., Hagemeier,C., Sorensen,T.S., La Thangue,N.B. and Kouzarides,T. (1995) Nature, 375, 691–694. [DOI] [PubMed] [Google Scholar]
- 20.Pomerantz J., Schreiber-Agus,N., Liegeois,N.J., Silverman,A., Alland,L., Chin,L., Potes,J., Chen,K., Orlow,I., Lee,H.-W., Cordon-Cardo,C. and DePinho,R.A. (1998) Cell, 92, 713–723. [DOI] [PubMed] [Google Scholar]
- 21.Dubs-Poterszman M.C., Tocque,B. and Wasylyk,B. (1995) Oncogene, 11, 2445–2449. [PubMed] [Google Scholar]
- 22.Sigalas I., Calvert,A.H., Anderson,J.J., Neal,D.E. and Lunec,J. (1996) Nature Med., 2, 912–917. [DOI] [PubMed] [Google Scholar]
- 23.Sun P., Dong,P., Dai,K., Hannon,G.J. and Beach,D. (1998) Science, 282, 2270–2272. [DOI] [PubMed] [Google Scholar]
- 24.Lundgren K., Montes de Oca Luna,R., McNeill,Y.B., Emerick,E.P., Spencer,B., Barfield,C.R., Lozano,G., Rosenberg,M.P. and Finlay,C.A. (1997) Genes Dev., 11, 714–725. [DOI] [PubMed] [Google Scholar]
- 25.Jones S.N., Hancock,A.R., Vogel,H., Donehower,L.A. and Bradley,A. (1998) Proc. Natl Acad. Sci. USA, 95, 15608–15612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sugino A., Ohara,T., Sebastian,J., Nakashima,N. and Araki,H. (1998) Genes Cells, 3, 99–110. [DOI] [PubMed] [Google Scholar]
- 27.Morrison A., Araki,H., Clark,A.B., Hamatake,R.K. and Sugino,A. (1990) Cell, 62, 1143–1151. [DOI] [PubMed] [Google Scholar]
- 28.Zlotkin T., Kaufmann,G., Jiang,Y., Lee,M.Y., Syvaoja,J., Dornreiter,I., Fanning,E. and Nethanel,T. (1996) EMBO J., 15, 2298–2305. [PMC free article] [PubMed] [Google Scholar]
- 29.Nishida C., Reinhard,P. and Linn,S. (1988) J. Biol. Chem., 263, 501–510. [PubMed] [Google Scholar]
- 30.Araki H., Ropp,P.A., Johnson,A.L., Johnston,L.H., Morrison,A. and Sugino,A. (1992) EMBO J., 11, 733–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Budd M.E. and Campbell,J.L. (1995) Mol. Cell. Biol., 15, 2173–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Z., Wu,X. and Friedberg,E.C. (1993) Mol. Cell. Biol., 13, 1051–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.D’Urso G. and Nurse,P. (1997) Proc. Natl Acad. Sci. USA, 94, 12491–12496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Navas T.A., Zhou,Z. and Elledge,S.J. (1995) Cell, 80, 29–39. [DOI] [PubMed] [Google Scholar]
- 35.Dua R., Levy,D.L. and Campbell,J.L. (1998) J. Biol. Chem., 273, 30046–30055. [DOI] [PubMed] [Google Scholar]
- 36.Kesti T. and Syvaoja,J.E. (1991) J. Biol. Chem., 266, 6336–6341. [PubMed] [Google Scholar]
- 37.Kesti T., Flick,K., Keranen,S., Syvaoja,J.E. and Wittenberg,C. (1999) Mol. Cell, 3, 679–685. [DOI] [PubMed] [Google Scholar]
- 38.Kesti T., Frantti,H. and Syvaoja,J.E. (1993) J. Biol. Chem., 268, 10238–10245. [PubMed] [Google Scholar]
- 39.Dasmahapatra B., Rozhon,E.J. and Schwartz,J. (1987) Nucleic Acids Res., 15, 3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chui G. and Linn,S. (1995) J. Biol. Chem., 270, 7799–7808. [DOI] [PubMed] [Google Scholar]
- 41.Haines D.S., Landers,J.E., Engle,L.J. and George,D.L. (1994) Mol. Cell. Biol., 14, 1171–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thut C.J., Goodrich,J.A. and Tjian,R. (1997) Genes Dev., 11, 1974–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Juven-Gershon T., Shifman,O., Unger,T., Elkeles,A., Haupt,Y. and Oren,M. (1998) Mol. Cell. Biol., 18, 3974–3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dobbelstein M., Wienzek,S., Konig,C. and Roth,J. (1999) Oncogene, 18, 2101–2106. [DOI] [PubMed] [Google Scholar]
- 45.Zeng X., Chen,L., Jost,C.A., Maya,R., Keller,D., Wang,X., Kaelin,W.G.,Jr, Oren,M., Chen,J. and Lu,H. (1999) Mol. Cell. Biol., 19, 3257–3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reinke V., Bortner,D.M., Amelse,L.L., Lundgren,K., Rosenberg,M.P., Finlay,C.A. and Lozano,G. (1999) Cell Growth Differ., 10, 147–154. [PubMed] [Google Scholar]
- 47.D’Urso G., Grallert,B. and Nurse,P. (1995) J. Cell Sci., 108, 3109–3118. [DOI] [PubMed] [Google Scholar]
- 48.Marini F., Pellicioli,A., Paciotti,V., Lucchini,G., Plevani,P., Stern,D.F. and Foiani,M. (1997) EMBO J., 16, 639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dua R., Levy,D.L. and Campbell,J.L. (1999) J. Biol. Chem., 274, 22283–22288. [DOI] [PubMed] [Google Scholar]
- 50.Takemura M., Kitagawa,T., Izuta,S., Wasa,J., Takai,A., Akiyama,T. and Yoshida,S. (1997) Oncogene, 15, 2483–2492. [DOI] [PubMed] [Google Scholar]
- 51.Zhang Y. and Xiong,Y. (1999) Mol. Cell, 3, 579–591. [DOI] [PubMed] [Google Scholar]