Abstract
Autophagy is a lysosome-mediated degradative process that removes damaged proteins and organelles, during which autophagosome–lysosome fusion is a key step of the autophagic flux. Based on our observation that intermediate cytofilament keratin 8 (KRT8) enhances autophagic clearance in cells under oxidative stress condition, we investigated whether KRT8 supports the cytoplasmic architectural networks to facilitate the vesicular fusion entailing trafficking onto filamentous tracks. We found that KRT8 interacts with actin filaments via the cytolinker, plectin (PLEC) during trafficking of autophagosome. When PLEC was knocked down or KRT8 structure was collapsed by phosphorylation, autophagosome–lysosome fusion was attenuated. Inhibition of actin polymerization resulted in accumulation of autophagosomes owing to a decrease in autophagosome and lysosome fusion. Furthermore, myosin motor protein was found to be responsible for vesicular trafficking along the actin filaments to entail autolysosome formation. Thus, the autophagosome–lysosome fusion is aided by PLEC-stabilized actin filaments as well as intermediate cytofilament KRT8 that supports the structural integrity of actin filaments during macroautophagic process under oxidative stress condition.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-022-04144-1.
Keywords: Actin filaments, Autophagy, Autophagosome–lysosome fusion, Keratin 8 (KRT8), Plectin (PLEC)
Introduction
Autophagy is a process that maintains cellular homeostasis through degradation of long-lived proteins and damaged organelles [1, 2]. Macroautophagy, which is the most prevalent form of cellular autophagy [3], begins with formation of a membrane structure called a phagophore. The phagophore then engulfs its cargo, forming a double membrane vesicle called an autophagosome [4]. Autophagosomes may fuse directly with lysosomes or they may first fuse with late endosomes creating amphisomes, which then fuse with lysosomes [5, 6]. Fusion with the lysosome creates a structure called an autolysosome, in which occurs extensive degradation of entrapped cargoes within membranous vesicles, culminating in autophagic flux [7, 8]. Autophagy is linked to several diseases that are likely caused by dysfunctional degradation of cargo proteins and organelles in the autolysosome [9]. Impairment of autophagosome–lysosome fusion causes accumulation of autophagosomes in dystrophic neurites of patients with Alzheimer’s disease and in a mouse model of the disease [10]. Lysosome dysfunction is also responsible for autophagosome accumulation, resulting in an X-linked genetic disorder such as Lowe syndrome [11].
Movement of mature autophagosomes toward lysosomes requires stable microtubules, which is apparent in several diseases possibly caused by dysfunctional autophagosome–lysosome fusion [12]. Autophagosome formation requires microtubules to recruit necessary factors, in which the intact structure of microtubules and microtubule-based motors are required [13, 14]. Participation of the microtubule plus-end kinesin motor in the compartmentalized pathway for autophagosome maturation has been evidenced in primary neurons [15]. The kinesin motor protein is involved in autophagic lysosome reformation through autolysosome tubulation, in which the kinesin motor remains on the microtubule and pulls on the autolysosomal membrane [16]. Another class of cytoskeletal structures, actin filaments, are also known to be implicated in macroautophagy, which was shown by failure of autophagosome formation in the starved cells treated with actin-depolymerizing agents such as cytochalasin D and latrunculin B [14]. The actin cytoskeleton was suggested to be necessary in the early events of autophagosome formation, because actin filaments have been observed colocalized with autophagy initiation markers [17, 18]. Proteomics analysis of autophagy-deficient mouse cells showed that depolymerization of filamentous actin (F-actin) resulted in a failure of autophagosome maturation, suggesting a requirement for an intact actin network in the autophagic flux [19]. Actin-based motor proteins, especially non-muscle myosins, are needed for several stages in autophagy, such as distribution of membranous vesicles and autophagic cargo along the dynamic actin cytoskeleton [20]. For instance, myosin VI (MYO6) and myosin 1C (MYO1C) were known to be involved in autophagosome maturation and lysosome fusion [21]. In particular, MYO6 delivers endocytic cargo to autophagosomes and facilitates autophagosome maturation and lysosomal fusion by binding to autophagy receptors, and an accumulation of autophagosomes was observed in MYO6 knockout mice [22].
Microtubules, actin filaments, and intermediate filaments are components of the main cytoskeletal system and form constant and intimate interactions with each other. Actin filaments and microtubules display a polarity with a fast and a slow growing end, and motor proteins track for unidirectional transport using this polarity. In contrast, intermediate filaments form apolar, flexible, and smooth filaments without tracking motor proteins due to the absence of polarity [23]. Despite the absence of tracking motor proteins, intermediate filaments form immense networks in cytoplasm, extending into all areas of the cytoplasm. Intermediate filaments are important components in mediating the cross talk between microtubules and actin filaments [24]. Recent studies have revealed that interaction of these cytoskeletal components is necessary for structural dynamics in cells, such as maintaining cellular mechanical stability and morphological changes [25, 26]. Intermediate filaments require cytolinker proteins, such as plectin (PLEC), to connect cytoskeletal components. PLEC is a major component of intermediate filaments, which connects intermediate filaments to microtubules, actin filaments, and membrane adhesion sites [27]. PLEC is thus a cross-linking element of the cytoskeleton, which contributes to integration of the cytoplasm [27, 28]. We previously showed that an intermediate filament, keratin 8 (KRT8), facilitates vesicular fusion between autophagosomes and lysosomes [29]. However, as intermediate filaments do not have motor proteins, it remains to be answered how vesicular fusion between autophagosome and lysosome is facilitated by KRT8. Thus, understanding the molecular basis of the cytoskeletal architecture through physical interaction with other cytoskeletal components harboring the tracking motors is warranted to elucidate the dynamics of autophagy flux.
Herein, we investigated the role of KRT8 in autophagosome and lysosome fusion in cells undergoing macroautophagy. When expression of KRT8 was inhibited or its structure was disrupted, autophagosome and lysosome fusion was significantly decreased. In addition, we found that KRT8 interacts with actin filaments via the cytolinker, PLEC. We tested whether inhibition of actin polymerization and disruption of actin filaments decrease vesicular fusion between autophagosomes and lysosomes. We further examined whether the movement of autophagosomes and lysosomes is aided by actin filaments with the support of KRT8 and PLEC through the structural integrity of actin filaments. Our results will contribute to understanding the molecular details of autophagosome–lysosome fusion with an emphasis on the roles of cytofilaments in the dynamic flux of macroautophagy.
Materials and methods
Reagents and antibodies
Methyl viologen dichloride hydrate (paraquat; 856,177), cytochalasin D (C8273), Torin1 (475,991), okadaic acid (O0810) and tetramethylrhodamine B isothiocyanate (phalloidin; P1951) were purchased from Sigma-Aldrich. Dulbecco’s modified Eagle medium: nutrient mixture F-12 (DMEM-F12; 11,330,032) and Opti-MEM (31,985,070) were supplied by GIBCO®. The pBABE-puro mCherry plasmid was purchased from Addgene (22,418), which was originally oriented by Jayanta Debnath (University of California at San Francisco, USA). Plasmid DNAs (pSELECT-GFP-LC3B [psetz-gfplc3b] and pUNO1-hKRT8, coding for Homo sapiens (Hs)/human KRT8 [puno1-hkrt8]) were purchased from InvivoGen. siRNAs targeting KRT8 (sc-35156), PP2A (sc-44033), PLEC (sc-29453), and RAB11FIP2 (sc-90683) were purchased from Santa Cruz Biotechnology. LysoTracker Red DND-99 (L7528), 4′,6-diamidino-2-phenylindole (DAPI; D1306), and Lipofectamine 3000 (L3000015) were purchased from Invitrogen Life Technologies. The following antibodies were used: anti-KRT8 (ab9023), anti-p-KRT8 (ab109452), GAPDH (ab9484), and SQSTM1 (ab56416) were from Abcam; anti-LC3B (2775 s) was purchased from Cell Signaling Technology; anti-RAB11FIP2 (NBP1-57,009) was purchased from Novus Biologicals; anti-PLEC (sc-33649), anti-β-Actin (sc-47778) and horseradish peroxidase-conjugated anti-rabbit and mouse immunoglobulin (sc-2357, sc2031) were from Santa Cruz Biotechnology; Alexa Fluor 488-conjugated anti-mouse immunoglobulin (A11001) was obtained from Invitrogen Life Technologies.
Cell culture and transfection
Retinal pigment epithelial cells, ARPE-19 (ATCC CRL-2302), were purchased from American Type Culture Collection. ARPE-19 cells were maintained in DMEM-F12 supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin and cultured at 37 °C in 5% CO2. To prepare KRT8 overexpressing ARPE-19 cells, cells were seeded in a 60 mm dish and incubated in Opti-MEM medium at 37 °C for 2 h. Then cells were transfected with 10 μg of KRT8 overexpression plasmid (pUNO1-hKRT8) using Lipofectamine 3000 (Invitrogen Life Technologies, L3000015). After 6 h, cells were washed thrice with phosphate-buffered saline (PBS; Welgene, LB 201–02). Then, cells were maintained in DMEM-F12 medium containing 10% fetal bovine serum, 1% penicillin/streptomycin and 20 μg/mL blasticidin (InvivoGen, antbl-1). To knock down the expression of KRT8, PP2A, PLEC and RAB11FIP2, ARPE-19 cells were transfected with anti-KRT8, PP2A, PLEC and RAB11FIP2 siRNAs. Cells were seeded (~ 3 × 105) in a six-well plate. After seeding, cells were incubated in OPTI-MEM medium at 37 °C for 2 h. Then, cells were transfected with 50 nM anti-KRT8, PP2A, PLEC and RAB11FIP2 siRNA using Lipofectamine 3000. After incubating at 37 °C for 9 h, cells were washed thrice with PBS and the medium was changed to DMEM-F12 supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin.
Autophagy assays; mCherry-EGFP-LC3B and GFP-LC3B analysis
ARPE-19 cells were seeded (~ 5 × 104) in a 24-well plate and incubated in Opti-MEM medium at 37 °C for 2 h. After 2 h, cells were transfected with 1 μg of pBABE-puro mCherry-EGFP-LC3B using Lipofectamine 3000. After transfection, cells were treated with 400 μM paraquat, 2 μM Torin1 or 50 nM okadaic acid for 24 h. Cells were fixed with 4% paraformaldehyde for 1 h at room temperature. After fixation, cells were washed three times for 10 min with PBS and nuclei were stained with DAPI (1:1000) for 2 h at room temperature. Cells were then mounted on the coverslip using ProLong Gold antifade reagent (Invitrogen life Technology, p36934). After mounting, cells were observed under a confocal fluorescence microscope (LSM800, Carl Zeiss) at 495–530 nm (EGFP) and 580 to 650 nm (mCherry).
ARPE-19 cells were seeded in a 24-well plate and incubated in Opti-MEM medium at 37 °C for 2 h. Next, cells were transfected with 1 μg of pSELECT-GFP-LC3B for 6 h using Lipofectamine 3000. After transfection, cells were treated with 400 μM paraquat, 2 μM Torin1 or 50 nM okadaic acid for 24 h. Cells were stained with 250 nM Lysotracker Red DND-99 for 2 h in the dark. After staining, cells were fixed in 4% paraformaldehyde at room temperature for 1 h. Then, cells were washed three times for 10 min with PBS and nuclei were stained with DAPI (1:1000) for 2 h. Cells were mounted in ProLong Gold antifade reagent. After mounting, cells were observed under a confocal fluorescence microscope (LSM800, Carl Zeiss).
Western blot analysis
RIPA buffer (50 mM Tris–HCl, pH 8, 150 nM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1.0% Tergitol®, 10 mM 2-mercaptoethanol, 1 mM phenylmethanesulfonylfluoride) and a protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific) were used to lyse the cells. To determine the protein concentration, the Bradford method was used. SDS-polyacrylamide gel electrophoresis (SDS-PAGE) was used to separate proteins (35 μg per lane). After SDS-PAGE, proteins were transferred to an Immobilon-P PVDF membrane (Millipore, IPVH00010). Then, membranes were blocked with 5% skim milk dissolved in TBST buffer (25 mM Tris, 150 mM NaCl, 2 mM KCl, 0.1% Tween 20, pH 7.4) for 1 h at room temperature. Thereafter, membranes were incubated in primary antibodies (1:1000 to 1:2000) at 4 °C overnight. Next, membranes were incubated with horseradish peroxidase-conjugated anti-rabbit and mouse immunoglobulin for 2 h at room temperature. After 2 h, signals were detected using a G:BOX Chemi XL (Syngene, Ozyme, Saint-Quentin-en-Yvelines) with an enhanced chemiluminescence system (Millipore, WBKLS0500). The protein band intensities were normalized to GAPDH which is expressed relative to control using GeneTools (software included in G:BOX Chemi XL).
Analysis of KRT8 and actin filaments morphology
ARPE-19 cells were seeded in a 24-well plate containing coverglasses. Then cells were incubated in the medium with 400 μM paraquat and 20 μM Torin1 at 37 °C for 24 h. After washing, cells were fixed with 4% paraformaldehyde for 1 h at room temperature. Then, cells were permeabilized with 0.2% Triton X-100 for 15 min and blocked with 3% bovine serum albumin for 1 h at room temperature. Next, cells were incubated at 4 °C overnight with anti-KRT8 primary antibody (1:200) and treated with the fluorescence-conjugated secondary antibody (1:1000; Alexa Fluor 488) for 2 h at room temperature in the dark. The nuclei and actin filaments were stained with DAPI (1:1000) and phalloidin (1:1000), respectively. Cells were washed three times with PBS for 10 min after each step. Then, cells were mounted with ProLong Gold antifade reagent. After mounting, the morphology of the KRT8 and actin filaments was observed with a confocal fluorescence microscope (LSM800, Carl Zeiss).
Co-immunoprecipitation
Co-immunoprecipitation (Co-IP) was conducted using the Dynabeads Co-Immunoprecipitation kit (Thermo Fisher Scientific, 14321D) to determine KRT8 binding proteins in ARPE-19 cells and PLEC knockdown ARPE-19 cells. 40 μg of anti-KRT8 antibody was covalently conjugated to 10 mg of Dynabeads and immunoprecipitation was performed according to the manufacturer’s instructions. Cell lysates were incubated with the antibody-coupled Dynabead at 4 °C for 2 h. After incubation, the beads were separated by magnet and washed three times. Captured proteins were eluted twice with the addition of 50 μL of HPH EB buffer (0.5 M NH4OH, 0.5 mM EDTA). Immunoprecipitated proteins that were separated by SDS-PAGE were analyzed by LC–MS/MS peptide mapping and identified by Life Science Laboratory (Seoul, Korea).
Statistical analysis
Replicate data are expressed as means ± standard error of the mean (SEM). Paired data indicate the independent experiments that were evaluated by Student’s t test. One-way analysis of variance (ANOVA) was used for multiple comparisons through Dunnett’s test, in which a value of P < 0.05 was considered statistically significant.
Results
KRT8 facilitates fusion between autophagosomes and lysosomes during ROS-induced autophagy process
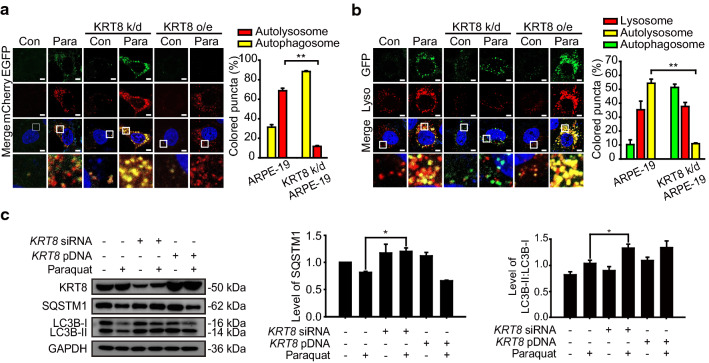
To investigate the role of KRT8 on autophagosome–lysosome fusion, we first examined the effects of different expression levels of KRT8 on the autophagic flux in the cells exposed to a redox-active compound such as paraquat. We transfected cells with mCherry-EGFP-LC3B to label autophagic vacuoles and performed cell imaging analysis after inducing autophagy (Fig. 1a). Autophagosomes are indicated by yellow fluorescence puncta due to the colocalization of mCherry (red) and EGFP (green). After autophagosome–lysosome fusion, EGFP is released from mCherry-EGFP-LC3B and degraded in lysosomes, resulting in a fluorescence change from yellow to red. When cells were exposed to paraquat, the number of LC3B puncta increased as compared with untreated cells, indicating induction of autophagy in cells under oxidative stress. In the KRT8 overexpressing cells under oxidative stress, the merged fluorescence micrographs were dominated with red-colored fluorescence representing autolysosomes. In contrast, merged images of cells in which KRT8 was knocked down and which were under oxidative stress exhibited mainly yellow fluorescence puncta prevalent in the cytoplasm. These results indicate that autophagy was stalled prior to autophagosome–lysosome fusion.
Fig. 1.
KRT8 expression level is associated with autophagy progression in RPE cells under oxidative stress condition. A Confocal microscopic images in mCherry-EGFP-LC3B-expressing ARPE-19 cells. Cells were incubated in the absence or presence of paraquat for 24 h. Enlarged images of the white boxed areas are shown in the lower panels. Scale bar: 5 μm. Bar graph indicates the percentage of each fluorescence level in the merged images of mCherry-EGFP-LC3B under paraquat treatment conditions. Data are presented as the mean ± SEM, n = 3. **P < 0.01. B Confocal microscopic images of Lysotracker Red-stained and GFP-LC3B-transfected ARPE-19 cells under paraquat treatment for 24 h. Scale bar: 5 μm. Bar graph indicates the percentage of each fluorescence level in the merged images of GFP-LC3B and Lysotracker Red under the oxidative stress condition. Data are presented as the mean ± SEM, n = 3. **P < 0.01. C Immunoblots of KRT8, SQSTM1, and LC3B in ARPE-19 cells treated with paraquat for 24 h. Bar graphs indicate SQSTM1 expression level or the ratio of LC3B-II to LC3B-I (triplicate western blot images with ratios of LC3B-II/LC3B-I, shown in Fig. S1). Each protein band intensity was normalized to GAPDH. Data are presented as the mean ± SEM, n = 3. *P < 0.05
To determine whether different expression levels of KRT8 affects the autolysosome formation, we monitored the presence of autophagosomes and lysosomes in the cells under oxidative stress (Fig. 1b). Autophagosome was tracked with GFP-LC3B and lysosome was stained with lysosome-specific fluorescent dye (LysoTracker Red DND-99). ARPE-19 cells under oxidative stress showed accumulation of GFP-LC3B puncta (green) and lysosomal organelles (red), which were colocalized with yellow-colored puncta in the merged image. In the KRT8-overexpressing cells under oxidative stress, yellow-colored fluorescence puncta that indicate the presence of autolysosomes were found in the merged fluorescence image. However, in the KRT8 knocked down cells under oxidative stress, colocalization between GFP-LC3B puncta and lysosomal organelles was rarely observed. Consistent with the result observed in Fig. 1a, this result indicates that KRT8 facilitates the autolysosome formation during the later stage of autophagy.
Under the oxidative stress condition, there was accumulation of LC3B-II and degradation of sequestosome 1 (SQSTM1), showing that the cells underwent autophagy (Figs. 1c & S1). In the KRT8 knockdown cells, the LC3B conversion representing the induction of autophagy was enhanced, but degradation of SQSTM1 was not promoted compared with control cells. This result suggests that autophagic flux was steadily halted before lysosomal fusion of the autophagosome. However, when cells were made to overexpress KRT8, LC3B conversion and subsequent SQSTM1 degradation were increased more than it was in the paraquat-treated control cells. Thus, KRT8 expression efficiently facilitates lysosomal fusion of autophagosomes in cells undergoing extensive autophagy. Taken together, these results indicate that KRT8 facilitates vesicular fusion between the autophagosome and the lysosome during the macroautophagy, likely resulting in more rapid autophagic flux.
Disruption of KRT8 filamentous structure attenuates autophagosome–lysosome fusion under ROS-induced autophagy process
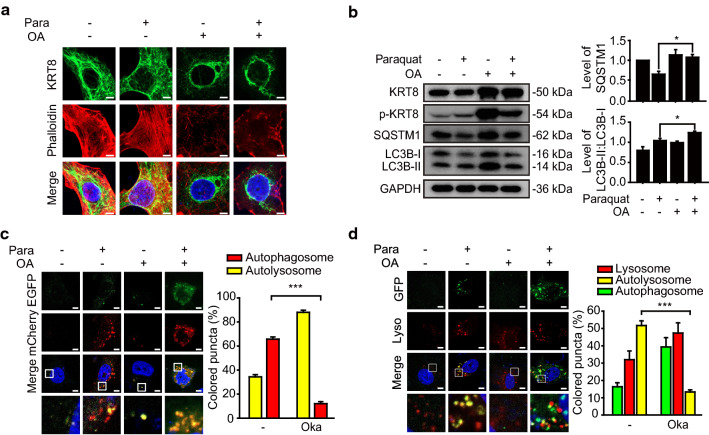
We next hypothesized that disruption of KRT8 filamentous structure may attenuate the fusion between autophagosomes and lysosomes. Phosphorylation of cytoskeletal components has been suggested to regulate intermediate filament polymerization and dynamics [30, 31]. Okadaic acid was used to regulate the filamentous structure of KRT8 by preventing the dephosphorylation of KRT8 through inhibition of phosphatase 2A (PP2A) [32, 33]. We monitored how disruption of KRT8 filamentous structure with okadaic acid treatment affects the shape of KRT8 in the cells under oxidative stress (Fig. 2a). When the cells exposed to paraquat were treated with okadaic acid, KRT8 filaments were disrupted and assembled at perinuclear sites and actin filaments were collapsed. We next investigated whether the fusion between autophagosomes and lysosomes was affected by okadaic acid treatment (Fig. 2b). When cells were treated with okadaic acid, the relative amount of phosphorylated KRT8 was greater than that in untreated cells under oxidative stress. Degradation of SQSTM1 was less pronounced in okadaic acid-treated cells than in untreated cells under oxidative stress. SQSTM1 degradation is a hallmark of autolysosome formation following fusion between autophagosomes and lysosomes. In contrast, the autophagic marker LC3B-II conversion was not affected by okadaic acid treatment of cells under oxidative stress. These results suggest that autophagy was hindered at the stage of vesicular fusion between autophagosomes and lysosomes. Hence, we suggest that disruption of the filamentous structure of KRT8 by okadaic acid halts the progression of autophagy at the vesicular fusion stage.
Fig. 2.
Treatment of RPE cells with okadaic acid suppresses autophagosome–lysosome fusion. ARPE-19 cells were treated with paraquat in the presence or absence of okadaic acid (OA) for 24 h. a Confocal microscopic images of KRT8-immunostained (green) and Alexa 555-Phalloidin-stained (red) ARPE-19 cells. Scale bar: 5 μm. b Immunoblots of KRT8, p-KRT8 and autophagic markers (SQSTM1, LC3B) in ARPE-19 cells. Bar graphs indicate SQSTM1 expression level or the ratio of LC3B-II to LC3B-I. Each protein band intensity was normalized to GAPDH. Data are presented as the mean ± SEM, n = 3. *P < 0.05. c Confocal microscopic images of mCherry-EGFP-LC3B-expressing ARPE-19 cells. Magnified images of the white boxed areas are shown as insets. Scale bar: 5 μm. Bar graph indicates the percentage of each fluorescence level in the merged images of mCherry-EGFP-LC3B under paraquat treatment condition. Data are presented as the mean ± SEM, n = 3. ***P < 0.001. d Confocal microscopic images of LysoTracker Red-stained and GFP-LC3B-transfected ARPE-19 cells. Scale bar: 5 μm. Bar graph indicates the percentage of each fluorescence level in the merged images of GFP-LC3B and Lysotracker Red under oxidative stress condition. Data are presented as the mean ± SEM, n = 3. ***P < 0.001
To test whether the loss of KRT8 filamentous structure caused by okadaic acid treatment hinders the fusion between autophagosomes and lysosomes, we monitored vesicular fusion between autophagosomes and lysosomes in the okadaic acid-treated cells (Fig. 2c). Red fluorescence puncta were predominantly present in the cytoplasm in cells under oxidative stress, indicating a prevalence of autolysosomes. In contrast, the cells treated with okadaic acid lacked red fluorescence and were dominated by yellow fluorescence puncta. This result represents that the autophagic process is paused before the lysosomal fusion of autophagosomes. Thus, consistent with the above result (Fig. 2b), disruption of KRT8 structure with okadaic acid halts the progression of autophagy with an aborted autophagosome–lysosome fusion process.
To further determine whether disruption of KRT8 negatively affects the late stage of autophagic flux, we monitored the colocalization of autophagosomes and lysosomes. We transfected cells with GFP-LC3B to track the autophagosome and stained the lysosome with lysosome-specific fluorescent dye (LysoTracker Red DND-99). ARPE-19 cells under oxidative stress showed accumulation of GFP-LC3B puncta and lysosomal organelles, with yellow puncta showing much colocalization (Fig. 2d). However, the okadaic acid-treated cells did not show colocalization of GFP-LC3B puncta and lysosomal organelles with yellow fluorescence. These results suggest that autophagosome–lysosome fusion was hindered when the filamentous structure of KRT8 was disrupted.
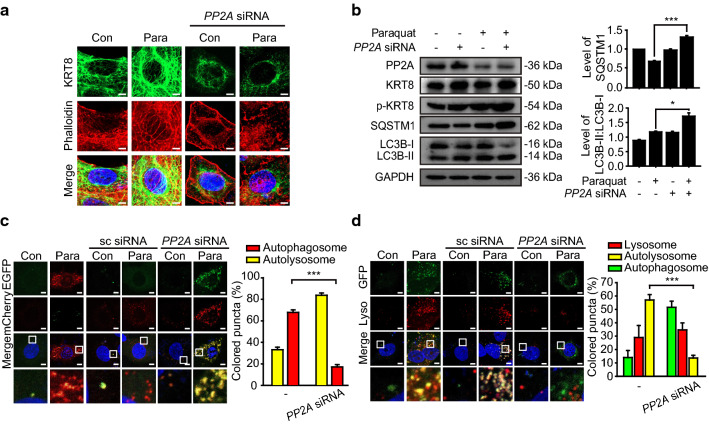
PP2A dephosphorylates the phosphorylated KRT8, by which unphosphorylated KRT8 retains filamentous structure in a branched form in the cytoplasm. To further corroborate that autolysosome formation is diminished when the filamentous structure of KRT8 is disrupted, we examined whether autophagy progression is attenuated by knockdown of PP2A. PP2A knockdown with PP2A-specific siRNA in RPE cells under oxidative stress resulted in perinuclear presence of sparse KRT8 filaments with disrupted actin filaments compared with the control RPE cells (Fig. 3a). In addition, PP2A knockdown with PP2A-specific siRNA resulted in decreased degradation of SQSTM1 with accumulation of LC3B-II in RPE cells under oxidative stress (Fig. 3b). Thus, disruption of the filamentous structure of KRT8 by PP2A knockdown is likely to inhibit the progression of oxidative stress-induced autophagy at a late stage in RPE cells.
Fig. 3.
Knockdown of the PP2A expression level diminishes autophagosome and lysosome fusion. a Confocal microscopic images of anti-PP2A siRNA-transfected ARPE-19 cells with or without paraquat. Cells were stained with Alexa 555-Phalloidin-stained (red) and immunostained for KRT8 (green). Scale bar: 5 μm. b Immunoblots of PP2A, KRT8, p-KRT8, SQSTM1, and LC3B in anti-PP2A siRNA-transfected ARPE-19 cells. Cells were treated with paraquat for 24 h. Bar graphs indicate SQSTM1 expression level or the ratio of LC3B-II to LC3B-I. Each protein band intensity was normalized to GAPDH. Data are presented as the mean ± SEM, n = 3. *P < 0.05 ***P < 0.001. c Confocal microscopic images of mCherry-EGFP-LC3B-expressing ARPE-19 cells. Cells were transfected with anti-PP2A siRNA and incubated with paraquat for 24 h. Scale bar: 5 μm. Bar graph indicates the percentage of each fluorescence level in the merged images of mCherry-EGFP-LC3B under paraquat treatment condition. Data are presented as the mean ± SEM, n = 3. ***P < 0.001. d Confocal microscopic images of LysoTracker Red-stained and GFP-LC3B-transfected ARPE-19 cells. Cells were transfected with anti-PP2A siRNA and incubated with paraquat for 24 h. Scale bar: 5 μm. Bar graph indicates the percentage of each fluorescence level in the merged images of GFP-LC3B and Lysotracker Red under oxidative stress condition. Data are presented as the mean ± SEM, n = 3. ***P < 0.001
Next, we investigated whether the loss of the filamentous architecture of KRT8 caused by PP2A knockdown inhibits autophagy progression during the late stage by hindering fusion between autophagosomes and lysosomes. We monitored vesicular fusion in RPE cells under oxidative stress that were transfected with mCherry-EGFP-LC3B (Fig. 3c). Untreated and scrambled siRNA-transfected RPE cells exhibited accumulation of red fluorescence puncta in the cytoplasm, representing ready formation of autolysosomes under oxidative stress. In contrast, PP2A knockdown using anti-PP2A siRNA resulted in yellow fluorescence puncta in cytoplasm, indicating that autophagy was halted prior to vesicular fusion between autophagosomes and lysosomes. To keep track of the autophagosome–lysosome fusion in ARPE-19 cells with PP2A knockdown, autophagosomes were traced with GFP-LC3B and lysosomes were stained with lysosome-specific fluorescent dye. The ARPE-19 cells under oxidative stress and a negative control group (scrambled siRNA) accumulated GFP-LC3B puncta and lysosomal organelles with significant colocalization, visualized as yellow-colored fluorescence in merged micrographs (Fig. 3d). On the contrary, GFP-LC3B puncta did not colocalize with lysosomes in ARPE-19 cells with PP2A knockdown. Thus, we suggest that maintenance of the filamentous structure of KRT8 in the dephosphorylated state facilitates autophagosome–lysosome fusion during the late stage of autophagy in ARPE-19 cells under oxidative stress.
Unphosphorylated KRT8 is associated with actin filaments through the cytolinker, PLEC
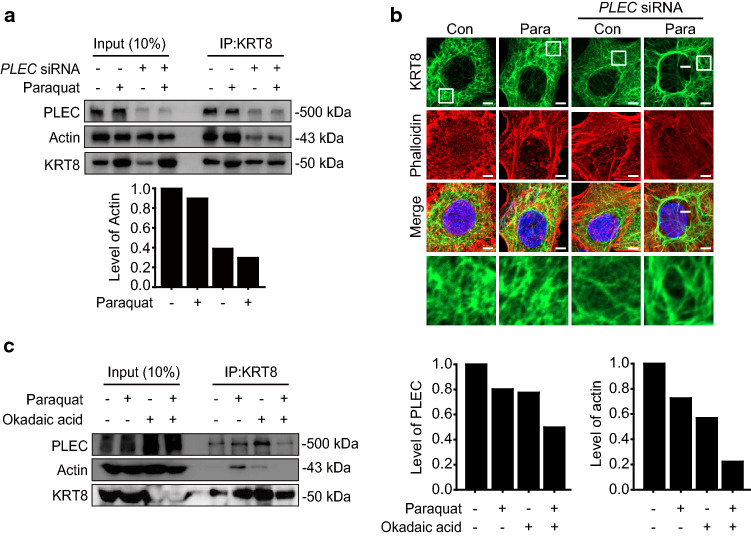
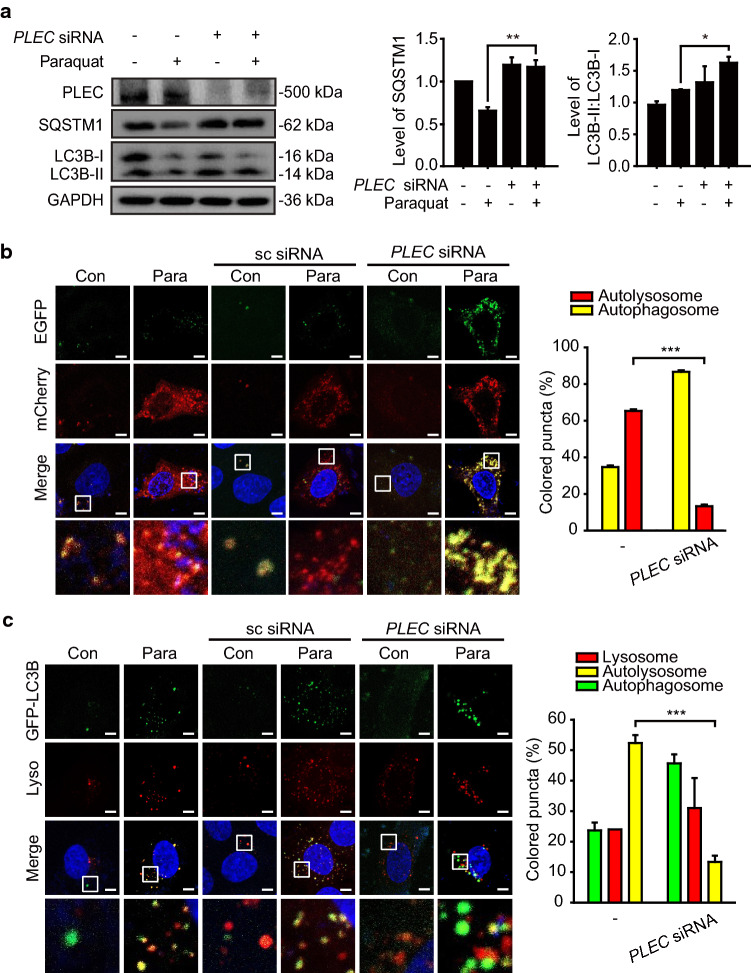
Although KRT8 does not have motor proteins to move autophagosomes and lysosomes along the cytoskeletal track, filamentous KRT8 still facilitates vesicular movement of autophagosomes and lysosomes. To delineate this apparent paradox, we hypothesized that actin filaments harboring motor proteins for vesicular movement are physically associated with KRT8 filaments through a mediating factor. Through the analysis of KRT8 binding proteins by immunoprecipitation, several proteins were found to associate with KRT8 (Table 1). Among them, PLEC was selected as the putative protein that mediates association between KRT8 and actin filaments, because PLEC has been known to play versatile roles such as connecting intermediate filaments to microtubules, actin filaments, and membrane adhesion sites [27, 28]. We used the PLEC knockdown to determine whether PLEC mediates the association of KRT8 with actin filaments. PLEC knockdown with PLEC-specific siRNA resulted in a decreased association of actin filaments with KRT8 compared with untreated RPE cells under oxidative stress (Fig. 4a).
Table 1.
List of proteins found to associate with KRT8 through immunoprecipitation and subsequent LC–MS/MS analysis
| Accession | Protein name | Molecular weight (kDa) | Sequence coverage (%) |
|---|---|---|---|
| XP_005251033.1 | Plectin isoform X3 | 532.145 | 34.72 |
| XP_005251037.1 | Plectin isoform X5 | 516.563 | 35.61 |
| EAW82172.1 | Plectin 1 | 289.821 | 31.58 |
| CAG30412.1 | MYH9 | 226.392 | 16.12 |
| AAA60137.1 | Poly(ADP-ribose) polymerase | 113.011 | 24.85 |
| XP_016882820.1 | Alpha-actinin-4 isoform X3 | 107.175 | 39.55 |
| BAA04656.1 | Motor protein (Mitofilin) | 83.626 | 17.55 |
| EAW68921.1 | Heterogeneous nuclear ribonucleoprotein M | 77.596 | 32.05 |
| P11142.1 | Heat shock cognate 71 kDa protein | 70.854 | 18.42 |
| AAH12295.1 | Lamin B1 | 66.367 | 19.28 |
| AFA52005.1 | Keratin 1 | 66.086 | 12.56 |
| EAW52998.1 | Lamin A/C | 62.105 | 47.77 |
| ACA06101.1 | Vimentin variant 1 | 53.619 | 65.02 |
Fig. 4.
Inhibition of KRT8 dephosphorylation attenuates the interaction between actin filaments, PLEC, and KRT8. a Immunoblots of KRT8 and actin in anti-PLEC siRNA-transfected ARPE-19 cells after co-immunoprecipitation with anti-KRT8 antibody. Cells were incubated with paraquat for 24 h. Bar graph indicates level of co-immunoprecipitated actin with KRT8. Each protein band intensity was normalized to KRT8. b Confocal microscopic images of KRT8-immunostained (green) and Alexa 555-Phalloidin-stained (red) ARPE-19 cells. Cells were treated with paraquat for 24 h. Scale bar: 5 μm. c Immunoblots of KRT8, actin, and PLEC in ARPE-19 cells after co-immunoprecipitation with anti-KRT8 antibody. Cells were incubated with paraquat in the presence or absence of okadaic acid for 24 h. Bar graph indicates the level of co-immunoprecipitated actin and PLEC with KRT8, in which KRT8 was used to normalize each protein band intensity
PLEC has been known to not only act as a cytolinker between actin filaments and intermediate filaments, but also maintain the architecture of intermediate filaments [34]. When the expression of PLEC is inhibited, keratin filament bundles become sparse and there is increased distribution of actin stress fibers [35]. Thus, we investigated the morphology of KRT8 and actin filaments in the PLEC knockdown cells (Fig. 4b). Under oxidative stress, KRT8 filaments held a densely branched shape, while bundles of KRT8 and actin filaments were sparse. These results indicate that PLEC mediates physical association between KRT8 and actin filaments under oxidative stress by maintaining the cytoskeletal architecture composed of KRT8 and actin filaments.
Next, we investigated whether the loss of KRT8 filamentous structure by preventing dephosphorylation of KRT8 may affect cytoskeletal interaction between actin filaments, PLEC, and KRT8 (Fig. 4c). Loss of KRT8 filamentous structure by treating okadaic acid, which inhibits dephosphorylation of KRT8, resulted in a decreased association between KRT8 and actin filaments. Because oxidative stress-induced phosphorylation of KRT8 decreases the association of KRT8 with actin/plectin, inhibition of KRT8 dephosphorylation caused by the okadaic acid treatment under oxidative stress condition (Fig. 2b) is likely to induce attenuation of KRT8 interactions with actin and/or plectin. As expected, both KRT8-actin and KRT8-PLEC interactions were significantly decreased with the okadaic acid treatment under oxidative stress. These results suggest that disruption of KRT8 filamentous structure by preventing the dephosphorylation of KRT8 diminishes the physical interaction between actin filaments and KRT8. Taken together, we suggest that phosphorylation of KRT8 causes disruption of filamentous actin (F-actin) structure and the filamentous structure of unphosphorylated KRT8 is associated with actin filaments via PLEC.
PLEC is required for autophagosome–lysosome fusion by supporting the cytoskeletal architecture
Since PLEC was observed to mediate a physical association between KRT8 and actin filaments in RPE cells under oxidative stress, we examined whether inhibition of association between KRT8 and actin filaments by PLEC knockdown affects progression of autophagy under oxidative stress (Fig. 5). PLEC knockdown using anti-PLEC siRNA resulted in accumulation of LC3B-II and decreased degradation of SQSTM1, compared with untransfected controls (Fig. 5a). To monitor the progression of autophagy in PLEC knockdown RPE cells under oxidative stress, we transfected cells with mCherry-EGFP-LC3B to label autophagic vacuoles and performed cell imaging (Fig. 5b). Untransfected cells showed mainly red fluorescence puncta, indicating formation of autolysosomes under oxidative stress. However, in the PLEC knockdown cells treated with paraquat, the autolysosome formation was significantly decreased, as evidenced by extensive yellow fluorescence puncta. This is consistent with the above result (Fig. 5a). These results indicate that PLEC facilitates the progression of autophagy by enhancing autophagosome–lysosome fusion. To further examine whether PLEC knockdown attenuates vesicular fusion of autophagosomes and lysosomes, autophagosomes were traced with GFP-LC3B and lysosomes were stained with the lysosome-specific fluorescent dye (Fig. 5c). RPE cells under oxidative stress and a control group transfected with scrambled siRNA showed accumulation of GFP-LC3B puncta and lysosomal organelles with extensive colocalization visualized as yellow fluorescence. On the contrary, GFP-LC3B puncta did not colocalize with lysosomes in the PLEC knockdown RPE cells. Taken together, these results suggest that PLEC mediates physical association between actin filaments and KRT8 filaments and plays a supporting role in autolysosome formation during autophagy induced by oxidative stress in RPE cells.
Fig. 5.
PLEC expression is correlated with autophagy progression in RPE cells under oxidative stress. a Immunoblots of PLEC and autophagic markers (SQSTM1, LC3B) in anti-PLEC siRNA-transfected ARPE-19 cells. Cells were treated with paraquat for 24 h. Bar graphs indicate SQSTM1 expression level or the ratio of LC3B-II to LC3B-I. Each protein band intensity was normalized to GAPDH. Data are presented as the mean ± SEM, n = 3. *P < 0.05 **P < 0.01. b Confocal microscopic images of mCherry-EGFP-LC3B-expressing ARPE-19 cells. Cells were transfected with anti-PLEC siRNA and incubated with paraquat for 24 h. Scale bar: 5 μm. Bar graph indicates the percentage of each fluorescence level in the merged images of mCherry-EGFP-LC3B under paraquat treatment condition. Data are presented as the mean ± SEM, n = 3. ***P < 0.001. c Confocal microscopic images of LysoTracker Red-stained and GFP-LC3B-transfected ARPE-19 cells. Cells were transfected with anti-PLEC siRNA and incubated with paraquat for 24 h. Scale bar: 5 μm. Bar graph indicates the percentage of each fluorescence level in the merged images of GFP-LC3B and Lysotracker Red under oxidative stress condition. Data are presented as the mean ± SEM, n = 3. ***P < 0.001
KRT8 and PLEC facilitates autophagosome–lysosome fusion in macroautophagy process
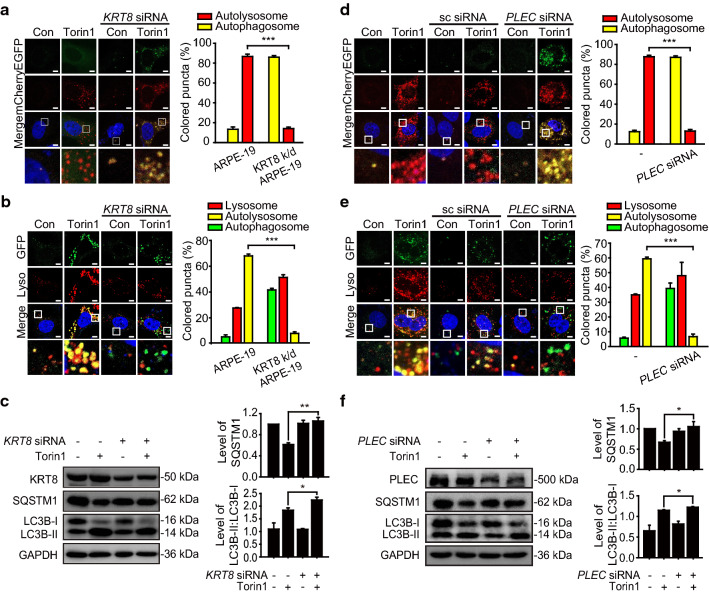
Based on previous observation that KRT8 was phosphorylated and reorganized to a perinuclear place under oxidative stress [29], we substantiated that phosphorylation of KRT8 attenuates the fusion between autophagosome and lysosome in macroautophagy under oxidative stress (Figs. 2,3). These results were explained by disrupted architecture of KRT8-actin filaments, which is mediated by PLEC linker, during the macroautophagy process. Next, we investigated whether shortage of KRT8 would diminish the autophagosome–lysosome fusion in macroautophagy process. To this end, we monitored the role of KRT8 in macroautophagy by treating cells with Torin1, a mechanistic target of rapamycin (MTOR) inhibitor, to induce autophagy by inhibiting mTORC1/2. Unlike the oxidative stress condition, Torin1 treatment maintains the cytoskeletal architecture comprising KRT8-actin filaments in the cells undergoing macroautophagy. Cells harboring mCherry-EGFP-LC3B were treated with Torin1 to monitor the autophagy process (Fig. 6a). Torin1 treatment readily caused autophagy, with cells showing red mCherry fluorescence dominantly. In contrast, merged micrographs of KRT8 knockdown cells were overwhelmed with yellow fluorescence puncta, consistent with the result in cells under oxidative stress (Fig. 1a,b). We next monitored autolysosome formation in the cells undergoing autophagy with Torin1 treatment under the different expression levels of KRT8 condition (Fig. 6b). The cells treated with Torin1 showed accumulation of both GFP-LC3B puncta and lysosomal organelles with extensive colocalization that was visualized as yellow fluorescence in the merged image. Similar to the results shown in the KRT8 knockdown cells under oxidative stress, GFP-LC3B green fluorescence puncta were not colocalized with lysosomes in KRT8 knockdown cells treated with Torin1. Furthermore, cells treated with Torin1 were analyzed for autophagic marker proteins (Fig. 6c). Degradation of SQSTM1 was more pronounced in the cells with Torin1 than the cells with KRT8 knockdown. Thus, macroautophagy caused by Torin1 treatment entails robust autophagosome–lysosome fusion, while shortage of KRT8 significantly decreases autolysosome formation during the macroautophagy process. Therefore, either shortage or phosphorylation of KRT8 causes attenuation of macroautophagy flux.
Fig. 6.
KRT8 and PLEC is required for autolysosome formation in macroautophagy caused by MTOR inhibition condition. a Confocal microscopic images in mCherry-EGFP-LC3B-expressing ARPE-19 cells. Cells were incubated in the absence or presence of Torin1 for 24 h. Enlarged images of the white boxed areas are shown in the lower panels. Scale bar: 5 μm. Bar graph indicates the percentage of each fluorescence level in the merged images of mCherry-EGFP-LC3B under Torin1 treatment condition. Data are presented as the mean ± SEM, n = 3. ***P < 0.001. b Confocal microscopic images of Lysotracker Red-stained and GFP-LC3B-transfected ARPE-19 cells under Torin1 treatment for 24 h. Scale bar: 5 μm. Bar graph indicates the percentage of each fluorescence level in the merged images of GFP-LC3B and Lysotracker Red under the MTOR inhibition condition. Data are presented as the mean ± SEM, n = 3. ***P < 0.001. c Immunoblots of KRT8, SQSTM1, and LC3B in ARPE-19 cells treated with Torin1 for 24 h. Bar graphs indicate SQSTM1 expression level or the ratio of LC3B-II to LC3B-I. Each protein band intensity was normalized to GAPDH. Data are presented as the mean ± SEM, n = 3. *P < 0.05, **P < 0.01. d Confocal microscopic images of mCherry-EGFP-LC3B-expressing ARPE-19 cells. Cells were transfected with anti-PLEC siRNA and incubated with Torin1 for 24 h. Scale bar: 5 μm. Bar graph indicates the percentage of each fluorescence level in the merged images of mCherry-EGFP-LC3B under Torin1 treatment condition. Data are presented as the mean ± SEM, n = 3. ***P < 0.001. e Confocal microscopic images of LysoTracker Red-stained and GFP-LC3B-transfected ARPE-19 cells. Cells were transfected with anti-PLEC siRNA and incubated with Torin1 for 24 h. Scale bar: 5 μm. Bar graph indicates the percentage of each fluorescence level in the merged images of GFP-LC3B and Lysotracker Red under the MTOR inhibition condition. Data are presented as the mean ± SEM, n = 3. ***P < 0.001. f Immunoblots of PLEC, SQSTM1, and LC3B in anti-PLEC siRNA-transfected ARPE-19 cells. Cells were treated with Torin1 for 24 h. Bar graphs indicate SQSTM1 expression level or the ratio of LC3B-II to LC3B-I. Each protein band intensity was normalized to GAPDH. Data are presented as the mean ± SEM, n = 3. *P < 0.05
To verify the role of PLEC on vesicular fusion during macroautophagy caused by Torin1 treatment, we monitored vesicular fusion and the progression of autophagy induced by Torin1 under PLEC knockdown cells. First, we monitored the progression of autophagy in RPE cells treated with Torin1 using mCherry-EGFP-LC3B to label autophagic vacuoles (Fig. 6d). The cells treated with autophagy inducer Torin1 without PLEC knockdown exhibited mainly red fluorescence puncta, indicating formation of autolysosomes. On the contrary, autolysosome formation was inhibited in the PLEC-knockdown cells treated with Torin1, as evidenced by extensive yellow fluorescence puncta, which is consistent with the result observed in paraquat-treated cells (Fig. 5b). Furthermore, autophagosome–lysosome fusion in PLEC knockdown RPE cells treated with Torin1 was monitored (Fig. 6e). Torin1-treated cells showed accumulation of GFP-LC3B puncta and lysosomal organelles with extensive colocalization visualized as yellow fluorescence in the merged micrographs. In contrast, GFP-LC3B green fluorescence puncta did not colocalize with lysosomes in PLEC knockdown cells. In addition, RPE cells treated with Torin1 showed increased hallmarks of autophagy, including LC3B-II conversion and degradation of SQSTM1, compared with control cells (Fig. 6f). When the cells were treated with PLEC-specific siRNA, progression of autophagy in RPE cells was delayed with decreased degradation of SQSTM1. Thus, knockdown of PLEC expression results in failed autolysosome formation in cells undergoing Torin1-activated macroautophagy. Taken together, these results suggest that KRT8 facilitates vesicular fusion between the autophagosome and lysosome during macroautophagy, likely resulting in more rapid autophagic flux. In addition, PLEC mediates physical association between actin filaments and KRT8 filaments and plays a supporting role in autolysosome formation during autophagy induced by mTORC 1/2 inhibition.
Impairment of actin polymerization diminishes autophagosome–lysosome fusion
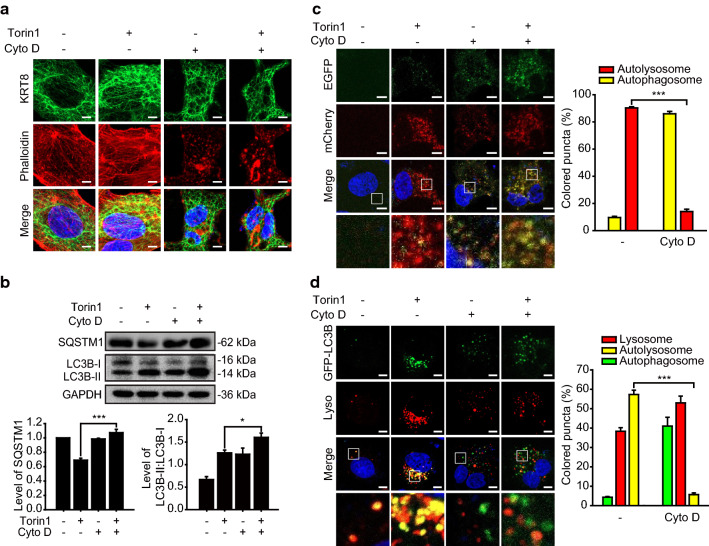
Since we observed that progression of autophagy was facilitated by filamentous KRT8 and PLEC, we investigated whether the structure of the cytoskeletal architecture, including KRT8 and actin filaments, affects the progression of autophagy in RPE cells. We examined whether disruption of actin filaments by inhibition of actin polymerization would affect autophagy in Torin1-treated cells. Disruption of the actin filaments was induced by cytochalasin D, which inhibits polymerization of actin filaments by binding to the ends of actin nuclei and filaments with high affinity [36, 37]. First, we examined the morphology of the KRT8 and actin filaments during autophagy in RPE cells treated with cytochalasin D and Torin1 (Fig. 7a). Actin filaments were broadly disrupted, as evidenced by the presence of small red fluorescence dots when the cells were treated with cytochalasin D under both normal and autophagic conditions. Interestingly, the branched structure of KRT8 filaments was less evident in cells harboring disrupted actin filaments than in cells with intact polymerized actin filaments. It is likely that actin filaments must be intact for maintaining the branched shape of KRT8 filaments in RPE cells.
Fig. 7.
Disruption of actin filaments inhibits autophagosome–lysosome fusion. ARPE-19 cells were treated with Torin1 in the presence or absence of cytochalasin D (Cyto D) for 24 h. a Confocal microscopic images of KRT8-immunostained (green) and Alexa 555-Phalloidin-stained (red) ARPE-19 cells. Scale bar: 5 μm. b Immunoblots of autophagic markers (SQSTM1, LC3B) in ARPE-19 cells. Bar graphs indicate SQSTM1 expression level or the ratio of LC3B-II to LC3B-I. Each protein band intensity was normalized to GAPDH. Data are presented as the mean ± SEM, n = 3. *P < 0.01 ***P < 0.001. c Confocal microscopic images of mCherry-EGFP-LC3B-expressing ARPE-19 cells. Scale bar: 5 μm. Bar graph indicates the percentage of each fluorescence level in the merged images of mCherry-EGFP-LC3B under Torin1 treatment condition. Data are presented as the mean ± SEM, n = 3. ***P < 0.001. d Confocal microscopic images of LysoTracker Red-stained and GFP-LC3B-transfected ARPE-19 cells. Scale bar: 5 μm. Bar graph indicates the percentage of each fluorescence level in the merged images of GFP-LC3B and Lysotracker Red under MTOR inhibition condition. Data are presented as the mean ± SEM, n = 3. ***P < 0.001
Next, we examined the effect of disrupting actin polymerization on the progression of autophagy by monitoring autophagy markers such as LC3B-II conversion and degradation of SQSTM1 (Fig. 7b). When RPE cells were treated with cytochalasin D, progression of autophagy was delayed, illustrated by decreased degradation of SQSTM1 and accumulation of LC3B-II. Thus, disruption of actin polymerization delays progression of Torin1-activated autophagy in RPE cells. We next monitored lysosome formation during Torin1-activated autophagy by using mCherry-EGFP-LC3B to label autophagic vacuoles in RPE cells treated with cytochalasin D (Fig. 7c). Autolysosome formation was inhibited in these cells, with extensive yellow fluorescence puncta in the Torin1-activated autophagic cells. This result indicates that autophagosome–lysosome fusion is inhibited when actin filament polymerization is impaired. In addition, autophagosome–lysosome fusion in RPE cells treated with Torin1 and cytochalasin D was monitored (Fig. 7d). GFP-LC3B (green fluorescence) autophagosome puncta did not colocalize with lysosomes (red fluorescence) in cells treated with Torin1 and cytochalasin D. In contrast, cells treated with only Torin1 showed accumulation of GFP-LC3B puncta and lysosomal organelles with significant colocalization, visualized as yellow fluorescence in the merged micrographs. Therefore, we suggest that disruption of cytoskeletal architecture induced by impairment of actin polymerization diminishes autophagosome–lysosome fusion under macroautophagy induced condition.
RAB11-FIP2 is necessary for facilitation of autophagosome–lysosome fusion
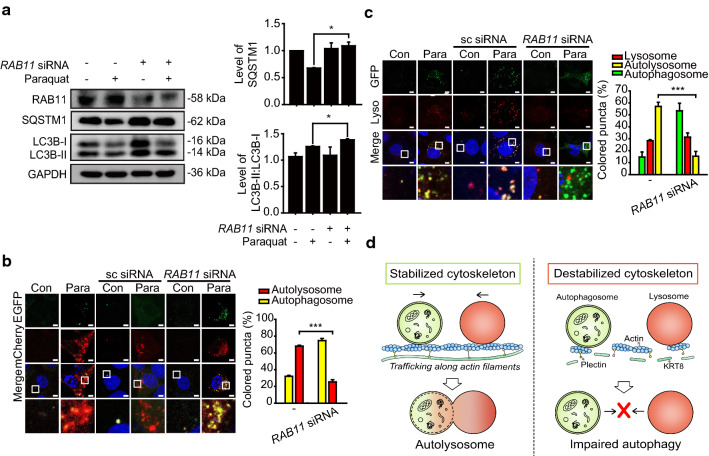
Since we observed that actin filaments must be intact for efficient autolysosome formation in the late stage of autophagy, we postulated that motor proteins working on the actin filaments are responsible for vesicular trafficking along actin filaments during autolysosome formation. Actin filaments have motor proteins that distribute membranous vesicles and autophagic cargoes [20]. Myosins are actin-based cytoskeletal motors that walk along the actin filaments through continuous ATP hydrolysis for directional movement, resulting in trafficking and localization of cellular components [38]. Myosin Va (MYO5A) and Vb (MYO5B) interact with organellar cargoes from the endoplasmic reticulum and recycling endosomes, respectively, in which both MYO5A and MYO5B need the receptor protein Rab11 family-interacting protein 2 (RAB11-FIP2) to interact with their cargoes [39]. Thus, we examined whether disrupting the association of myosin V with autophagic vesicles by knockdown of RAB11-FIP2 diminishes fusion of autophagosomes and lysosomes in RPE cells (Fig. 8).
Fig. 8.
Expression level of RAB11-FIP2 is associated with autophagy progression in RPE cells under oxidative stress. a Immunoblots of RAB11-FIP2 (RAB11), SQSTM1, and LC3B in anti-RAB11-FIP2 siRNA-transfected ARPE-19 cells. Cells were treated with paraquat for 24 h. Bar graphs indicate SQSTM1 expression level or the ratio of LC3B-II to LC3B-I. Each protein band intensity was normalized to GAPDH. Data are presented as the mean ± SEM, n = 3. *P < 0.05. b Confocal microscopic images of mCherry-EGFP-LC3B-expressing ARPE-19 cells exposed to paraquat for 24 h. Scale bar: 5 μm. Bar graph indicates the percentage of each fluorescence level in the merged images of mCherry-EGFP-LC3B under paraquat treatment condition. Data are presented as the mean ± SEM, n = 3. ***P < 0.001. c Confocal microscopic images of LysoTracker Red-stained and GFP-LC3B-transfected ARPE-19 cells. Cells were treated with paraquat for 24 h. Scale bar: 5 μm. Bar graph indicates the percentage of each fluorescence level in the merged images of GFP-LC3B and Lysotracker Red under oxidative stress condition. Data are presented as the mean ± SEM, n = 3. ***P < 0.001. d Graphical summary of the results
Under oxidative stress, RAB11-FIP2 knockdown (with anti-RAB11-FIP2 siRNA) resulted in decreased degradation of SQSTM1, compared with untransfected control cells (Fig. 8a). We next monitored the progression of autophagy in RAB11FIP2 knockdown cells under oxidative stress, where cells were transfected with mCherry-EGFP-LC3B to label autophagic vacuoles (Fig. 8b). Compared with untransfected cells and cells transfected with scrambled siRNA, autolysosome formation was severely inhibited, showing yellow fluorescence puncta in the RAB11-FIP2 knockdown cells treated with paraquat. Thus, the receptor protein RAB11-FIP2, which interacts with myosin motor proteins working on the actin filaments, is responsible for formation of autolysosomes during the late stage of autophagy.
To confirm that the RAB11-FIP2 knockdown results in failed autophagosome–lysosome fusion, autophagosomes and lysosomes were traced with GFP-LC3B and LysoTracker Red, respectively (Fig. 8c). When RPE cells under oxidative stress were transfected with anti-RAB11FIP2 siRNA, GFP-LC3B puncta did not colocalize with lysosomes. In contrast, extensive colocalization between autophagosomes (green fluorescence) and lysosomes (red fluorescence) was observed in untransfected RPE cells and a control group transfected with scrambled siRNA. Importantly, structural disintegration or disruption of KRT8 and actin filaments did not occur under the RAB11-FIP2 knockdown condition; shapes and presence of KRT8 and actin filaments (phalloidin fluorescence) were similar in both absence and presence of the RAB11-FIP2 siRNA treatment (Fig. S2). These results indicate that the receptor protein RAB11-FIP2 is likely required for vesicular trafficking in autolysosome formation in RPE cells. Thus, vesicular trafficking along the actin filaments by myosin motor proteins is presumably responsible for vesicular fusion necessary for autolysosome formation during the late stage of autophagy in RPE cells.
Discussion
In this study, we observed that the intermediate filament KRT8 promotes the progression of autophagy by facilitating vesicular fusion of autophagosomes and lysosomes. Maintenance of actin filament and intermediate filament structure was crucial to autolysosome formation. Vesicular fusion is a seminal event in the late stages of autophagy in cells under oxidative stress. Thus, facilitation of vesicular trafficking with intact cytoskeletal architecture is highly important for uninterrupted autophagic flux. Cellular proteostasis is often achieved by clearance of harmful protein aggregates through autophagy [40]. However, efficiency of autophagy is decreased in aged cells or in pathologically dysfunctional cells, which are often observed in patients with age-related macular degeneration (AMD) [41, 42]. Several studies, including one from our lab, have demonstrated that enhanced autophagy protects cellular function in various diseases such as AMD [29], Huntington’s disease [43], and Mallory–Denk body formation in liver disease [44].
In previous studies, we found that unhampered autophagy could maintain the functional and anatomic integrity of RPE cells despite continuous exposure to oxidative stress [29]. We found that KRT8 was upregulated in cells of patients with neovascular AMD under oxidative stress [45]. We also observed that the upregulated KRT8 facilitated autophagosome–lysosome fusion in RPE cells undergoing autophagy [29, 45]. Other cytoskeletal structures such as microtubules and actin filaments are known to affect the autophagy process [13, 17]. However, the role of intermediate filaments in autophagy has not been explored outside our previous study [29]. Since intermediate filaments do not harbor motor proteins that transport autophagy cargoes [23], it is not well understood how KRT8 facilitates autophagosome–lysosome fusion in RPE cells undergoing autophagy.
In epithelial cells, keratins are one of the dominant intermediate filaments and form cytoplasmic structural networks to link the plasma membrane, nucleus, and other cytoskeletal components by traversing the cell cytoplasm [46, 47]. Keratins are divided into type I (K9-K20) and type II (K1-K8) and they form heterodimers of a type 1 and a type 11 keratin, which then form bundles [48]. KRT8 forms heterodimers with KRT18 (i.e., KRT8/18) and is responsible for regulating the cellular response to apoptotic stimuli [49]. KRT8/18 has been known to affect TNF signaling in liver cells and TNF-dependent apoptotic liver damage is more pronounced in KRT8/18 deficient mice [50]. KRT8/18 also modulates Fas ligand targeting to the cell surface, thereby providing resistance to Fas-mediated apoptosis [51]. In addition to these roles, we demonstrated that KRT8 facilitates vesicular fusion of autophagosomes and lysosomes in stress-induced autophagy.
Macroautophagy is regulated via several pathways; class III PI3K forms a complex with beclin-1 (BECN1) and generates phosphatidylinositol 3‐phosphates to control membrane dynamics, resulting in autophagosome formation [52, 53]. It has been reported that the formation and motility of autophagosomes is influenced by cytofilament microtubules (MTs) but not their fusion with lysosomes [13]. LC3 and ATG proteins, which are involved in the early stages of autophagosome formation, associate with MTs [13, 54]. Motors associated with MTs participate in important steps of autophagosome formation, such as the fusion of ATG16-positive membranes, which then maturate into phagophores [55]. In addition, it has been reported that MTs and their motors are involved in initiation of the autophagic response by regulating MTORC1 and the class III PI3K complex. MTOR activity is also controlled by lysosome localization, which is organized by MTs and kinesin motors. When nutrients are in short supply, MT motors are released from lysosomes, facilitating MTORC1 inactivation and autophagosome formation, and then newly formed autophagosomes can fuse with lysosomes by moving along MTs in perinuclear regions [56, 57]. However, the role of MTs in autophagosome–lysosome fusion has long been controversial due to several reports showing that MTs are dispensable for the fusion between autophagosomes and lysosomes [13, 58, 59].
We observed that fusion of autophagosomes and lysosomes is affected by KRT8 knockdown or disruption. The knockdown or disruption of KRT8 inhibited autophagosome and lysosome fusion during autophagy, resulting in accumulation of autophagosomes. We also found that disruption of KRT8 architecture by KRT8 phosphorylation reduces the efficiency of autophagy. Dephosphorylation of KRT8 by PP2A maintained cytoskeletal architecture for efficient trafficking of vesicles, while the PP2A inhibitor okadaic acid severely attenuated fusion of autophagosomes and lysosomes. Consistent with our result, okadaic acid treatment of neurons elicited disruption of intracellular trafficking with accumulation of several organelles and autophagosomes in neurites [43]. It has been reported that the expression and activity of PP2A is readily declined during aging [60, 61]. This age-related decline of PP2A activity promotes hyperphosphorylation of neurofilaments, which is accompanied with cytoskeleton breakdown [62], leading to neurodegenerative diseases such as Alzheimer’s disease [63, 64]. In addition, okadaic acid suppresses autophagic lysosomal degradation in hepatocytes, possibly due to disruption of the cytoskeleton [65]. We previously reported that the inhibition of ROS-induced KRT8 phosphorylation maintains autophagy and epithelial integrity of the RPE layer in a model of AMD [29]. Lysosomal degradation of damaged or modified proteins may be part of a homeostatic and protective response against oxidative stress exerted on RPEs. Therefore, autophagic flux relies on KRT8 intermediate filaments for the fusion of autophagosomes with lysosomes, which may be therapeutically beneficial in patients with AMD.
We speculated that interaction between KRT8 and actin filaments is essential for autophagosome–lysosome fusion because KRT8 filaments do not bind with any motor proteins. Accordingly, a cytolinker protein PLEC that was known to play a role in connecting intermediate filaments with microtubules and actin filaments [27, 28] was hypothesized to be involved in physical interaction with KRT8 (Table 1). We observed that the interaction between KRT8 and actin filaments was diminished when PLEC was knocked down (Fig. 4a), and vesicular fusion of autophagosomes and lysosomes was inhibited (Fig. 5, Fig. 6). Our results on the architectural role of PLEC are consistent with previous reports on the structure and binding of PLEC. PLEC–IF complexes with irregular networks have been produced by in vitro reconstitution from purified proteins [27]. When PLEC is not available, physical interaction between KRT8 and actin filaments is thus unattainable, resulting in lower frequency of autophagosome–lysosome fusion. We also found that inhibition of actin polymerization negatively affects autophagy by preventing the fusion of autophagosomes and lysosomes (Fig. 7).
In addition, we found that autophagosomes and lysosomes use actin filaments as a tracking lattice for their vesicular fusion by investigating the actin-based motor protein, myosin V. Several factors, including RAB11FIP2, are required as adaptor molecules between cargoes and motor proteins in the process of cargo trafficking by myosin V. It has been known that RAB11FIP2 is involved in endosome recycling and receptor-mediated endocytosis [66], which are targeted processes involving fusion with lysosomes. We have found that knockdown of the myosin V cargo adaptor protein RAB11FIP2 impeded vesicular fusion of autophagosomes and lysosomes (Fig. 8). The actin cytoskeleton actively participates in generation of omegasomes and subsequent elongation of the phagophore to meet the need for increased rate of autophagosome formation in cells under autophagic stimulation [17]. Not mutually exclusive with that report, it is tempting to suggest that the cytoplasmic architecture linking intermediate filaments (IFs) and actin filaments is also important to ensure a tight regulation in the late stage of oxidative stress-induced autophagy. Given that intact cytoplasmic architecture is required for macroautophagy, further studies are necessary to elucidate the roles of both actin modulatory proteins and IFs in the precise regulation of autophagic flux under cellular stress.
In the present study, we have demonstrated a role of KRT8 in the macroautophagy process. It is likely that the movement of autophagosomes and lysosomes proceeds along actin filaments and that KRT8, along with PLEC, provides a structural framework for this process. In contrast, the impairment of autophagy by the attenuation of autophagosome–lysosome fusion is induced by the destabilized cytoskeleton. (Fig. 8D). Thus, IF–PLEC complexes comprise an extensive cross-linking of cytofilament including actin filaments to provide an intact cytoplasmic architecture required for macroautophagy process, which helps in understanding of the pathogenesis of degenerative diseases in retinal epithelium and neurons.
Supplementary Information
Below is the link to the electronic supplementary material.
Author contributions
D-EK and AB formulated the hypothesis, and initiated and organized the study. SS and AB designed and performed experiments with data analysis. SS, AB, JHL and D-EK wrote the manuscript.
Funding
This research was supported by the grant from the National Research Foundation of Korea (2017R1E1A1A01074656) funded by the Korean government.
Data availability
All data generated or analyzed during this study are included in this published article.
Declarations
Conflict of interest
The authors have declared that no competing interests exist.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Sumin Son and Ahruem Baek contributed equally to this work.
References
- 1.Feng Y, He D, Yao Z, Klionsky DJ. The machinery of macroautophagy. Cell Res. 2014;24(1):24–41. doi: 10.1038/cr.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132(1):27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klionsky DJ. The molecular machinery of autophagy: unanswered questions. J Cell Sci. 2005;118(Pt 1):7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klionsky DJ, Ohsumi Y. Vacuolar import of proteins and organelles from the cytoplasm. Ann Rev Cell Dev Biol. 1999;15:1–32. doi: 10.1146/annurev.cellbio.15.1.1. [DOI] [PubMed] [Google Scholar]
- 5.Abada A, Elazar Z. Getting ready for building: signaling and autophagosome biogenesis. Embo Rep. 2014;15(8):839–852. doi: 10.15252/embr.201439076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fader CM, Sanchez D, Furlan M, Colombo MI. Induction of autophagy promotes fusion of multivesicular bodies with autophagic vacuoles in k562 cells. Traffic. 2008;9(2):230–250. doi: 10.1111/j.1600-0854.2007.00677.x. [DOI] [PubMed] [Google Scholar]
- 7.Berg TO, Fengsrud M, Stromhaug PE, Berg T, Seglen PO. Isolation and characterization of rat liver amphisomes. Evidence for fusion of autophagosomes with both early and late endosomes. J Biol Chem. 1998;273(34):21883–21892. doi: 10.1074/jbc.273.34.21883. [DOI] [PubMed] [Google Scholar]
- 8.Nakamura S, Yoshimori T. New insights into autophagosome–lysosome fusion. J Cell Sci. 2017;130(7):1209–1216. doi: 10.1242/jcs.196352. [DOI] [PubMed] [Google Scholar]
- 9.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451(7182):1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu WH, Cuervo AM, Kumar A, Peterhoff CM, Schmidt SD, Lee JH, Mohan PS, Mercken M, Farmery MR, Tjernberg LO et al: Macroautophagy--a novel Beta-amyloid peptide-generating pathway activated in Alzheimer's disease. J Cell Biol (2005), 171(1):87–98. 10.1083/jcb.200505082 [DOI] [PMC free article] [PubMed]
- 11.De Leo MG, Staiano L, Vicinanza M, Luciani A, Carissimo A, Mutarelli M, Di Campli A, Polishchuk E, Di Tullio G, Morra V et al: Autophagosome–lysosome fusion triggers a lysosomal response mediated by TLR9 and controlled by OCRL. Nat Cell Biol (2016), 18(8):839–850. 10.1038/ncb3386 [DOI] [PMC free article] [PubMed]
- 12.Aplin A, Jasionowski T, Tuttle DL, Lenk SE, Dunn WA., Jr Cytoskeletal elements are required for the formation and maturation of autophagic vacuoles. J Cell Physiol. 1992;152(3):458–466. doi: 10.1002/jcp.1041520304. [DOI] [PubMed] [Google Scholar]
- 13.Mackeh R, Perdiz D, Lorin S, Codogno P, Pous C. Autophagy and microtubules—new story, old players. J Cell Sci. 2013;126(Pt 5):1071–1080. doi: 10.1242/jcs.115626. [DOI] [PubMed] [Google Scholar]
- 14.Geeraert C, Ratier A, Pfisterer SG, Perdiz D, Cantaloube I, Rouault A, Pattingre S, Proikas-Cezanne T, Codogno P, Pous C. Starvation-induced hyperacetylation of tubulin is required for the stimulation of autophagy by nutrient deprivation. J Biol Chem. 2010;285(31):24184–24194. doi: 10.1074/jbc.M109.091553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maday S, Holzbaur EL. Autophagosome biogenesis in primary neurons follows an ordered and spatially regulated pathway. Dev Cell. 2014;30(1):71–85. doi: 10.1016/j.devcel.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Du W, Su QP, Chen Y, Zhu Y, Jiang D, Rong Y, Zhang S, Zhang Y, Ren H, Zhang C, et al. Kinesin 1 drives autolysosome tubulation. Dev Cell. 2016;37(4):326–336. doi: 10.1016/j.devcel.2016.04.014. [DOI] [PubMed] [Google Scholar]
- 17.Aguilera MO, Beron W, Colombo MI. The actin cytoskeleton participates in the early events of autophagosome formation upon starvation induced autophagy. Autophagy. 2012;8(11):1590–1603. doi: 10.4161/auto.21459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reggiori F, Monastyrska I, Shintani T, Klionsky DJ. The actin cytoskeleton is required for selective types of autophagy, but not nonspecific autophagy, in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 2005;16(12):5843–5856. doi: 10.1091/mbc.e05-07-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhuo C, Ji Y, Chen Z, Kitazato K, Xiang Y, Zhong M, Wang Q, Pei Y, Ju H, Wang Y. Proteomics analysis of autophagy-deficient Atg7-/- MEFs reveals a close relationship between F-actin and autophagy. Biochem Biophys Res Commun. 2013;437(3):482–488. doi: 10.1016/j.bbrc.2013.06.111. [DOI] [PubMed] [Google Scholar]
- 20.Kruppa AJ, Kendrick-Jones J, Buss F. Myosins, actin and autophagy. Traffic. 2016;17(8):878–890. doi: 10.1111/tra.12410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brandstaetter H, Kishi-Itakura C, Tumbarello DA, Manstein DJ, Buss F. Loss of functional MYO1C/myosin 1c, a motor protein involved in lipid raft trafficking, disrupts autophagosome–lysosome fusion. Autophagy. 2014;10(12):2310–2323. doi: 10.4161/15548627.2014.984272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tumbarello DA, Waxse BJ, Arden SD, Bright NA, Kendrick-Jones J, Buss F. Autophagy receptors link myosin VI to autophagosomes to mediate Tom1-dependent autophagosome maturation and fusion with the lysosome. Nat Cell Biol. 2012;14(10):1024–1035. doi: 10.1038/ncb2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herrmann H, Strelkov SV, Burkhard P, Aebi U. Intermediate filaments: primary determinants of cell architecture and plasticity. J Clin Invest. 2009;119(7):1772–1783. doi: 10.1172/JCI38214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang L, Goldman RD. Intermediate filaments mediate cytoskeletal crosstalk. Nat Rev Mol Cell Biol. 2004;5(8):601–613. doi: 10.1038/nrm1438. [DOI] [PubMed] [Google Scholar]
- 25.Huber F, Boire A, Lopez MP, Koenderink GH. Cytoskeletal crosstalk: when three different personalities team up. Curr Opin Cell Biol. 2015;32:39–47. doi: 10.1016/j.ceb.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 26.Wickstead B, Gull K. The evolution of the cytoskeleton. J Cell Biol. 2011;194(4):513–525. doi: 10.1083/jcb.201102065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wiche G. Plectin: general overview and appraisal of its potential role as a subunit protein of the cytomatrix. Critical Rev Biochem Mol Biol. 1989;24(1):41–67. doi: 10.3109/10409238909082551. [DOI] [PubMed] [Google Scholar]
- 28.Svitkina TM, Verkhovsky AB, Borisy GG. Plectin sidearms mediate interaction of intermediate filaments with microtubules and other components of the cytoskeleton. J Cell Biol. 1996;135(4):991–1007. doi: 10.1083/jcb.135.4.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baek A, Yoon S, Kim J, Baek YM, Park H, Lim D, Chung H, Kim DE. Autophagy and KRT8/keratin 8 protect degeneration of retinal pigment epithelium under oxidative stress. Autophagy. 2017;13(2):248–263. doi: 10.1080/15548627.2016.1256932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eriksson JE, Brautigan DL, Vallee R, Olmsted J, Fujiki H, Goldman RD. Cytoskeletal integrity in interphase cells requires protein phosphatase activity. Proc Natl Acad Sci USA. 1992;89(22):11093–11097. doi: 10.1073/pnas.89.22.11093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goldman RD, Khuon S, Chou YH, Opal P, Steinert PM. The function of intermediate filaments in cell shape and cytoskeletal integrity. J Cell Biol. 1996;134(4):971–983. doi: 10.1083/jcb.134.4.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yatsunami J, Fujiki H, Suganuma M, Yoshizawa S, Eriksson JE, Olson MO, Goldman RD. Vimentin is hyperphosphorylated in primary human fibroblasts treated with okadaic acid. Biochem Biophys Res Commun. 1991;177(3):1165–1170. doi: 10.1016/0006-291x(91)90662-q. [DOI] [PubMed] [Google Scholar]
- 33.Tao GZ, Toivola DM, Zhou Q, Strnad P, Xu B, Michie SA, Omary MB. Protein phosphatase-2A associates with and dephosphorylates keratin 8 after hyposmotic stress in a site- and cell-specific manner. J Cell Sci. 2006;119(Pt 7):1425–1432. doi: 10.1242/jcs.02861. [DOI] [PubMed] [Google Scholar]
- 34.Wiche G, Winter L. Plectin isoforms as organizers of intermediate filament cytoarchitecture. BioArchitecture. 2011;1(1):14–20. doi: 10.4161/bioa.1.1.14630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moch M, Windoffer R, Schwarz N, Pohl R, Omenzetter A, Schnakenberg U, Herb F, Chaisaowong K, Merhof D, Ramms L, et al. Effects of plectin depletion on keratin network dynamics and organization. PLoS ONE. 2016;11(3):e0149106. doi: 10.1371/journal.pone.0149106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Casella JF, Flanagan MD, Lin S. Cytochalasin D inhibits actin polymerization and induces depolymerization of actin filaments formed during platelet shape change. Nature. 1981;293(5830):302–305. doi: 10.1038/293302a0. [DOI] [PubMed] [Google Scholar]
- 37.Schliwa M. Action of cytochalasin D on cytoskeletal networks. J Cell Biol. 1982;92(1):79–91. doi: 10.1083/jcb.92.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hammer JA, 3rd, Sellers JR. Walking to work: roles for class V myosins as cargo transporters. Nat Rev Mol Cell Biol. 2011;13(1):13–26. doi: 10.1038/nrm3248. [DOI] [PubMed] [Google Scholar]
- 39.Kneussel M, Wagner W. Myosin motors at neuronal synapses: drivers of membrane transport and actin dynamics. Nat Rev Neurosci. 2013;14(4):233–247. doi: 10.1038/nrn3445. [DOI] [PubMed] [Google Scholar]
- 40.Mizushima N, Klionsky DJ. Protein turnover via autophagy: implications for metabolism. Ann Rev Nutr. 2007;27:19–40. doi: 10.1146/annurev.nutr.27.061406.093749. [DOI] [PubMed] [Google Scholar]
- 41.Mitter SK, Song C, Qi X, Mao H, Rao H, Akin D, Lewin A, Grant M, Dunn W, Jr, Ding J, et al. Dysregulated autophagy in the RPE is associated with increased susceptibility to oxidative stress and AMD. Autophagy. 2014;10(11):1989–2005. doi: 10.4161/auto.36184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang AL, Lukas TJ, Yuan M, Du N, Tso MO, Neufeld AH. Autophagy and exosomes in the aged retinal pigment epithelium: possible relevance to drusen formation and age-related macular degeneration. PLoS ONE. 2009;4(1):e4160. doi: 10.1371/journal.pone.0004160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoon SY, Choi JE, Kweon HS, Choe H, Kim SW, Hwang O, Lee H, Lee JY, Kim DH. Okadaic acid increases autophagosomes in rat neurons: implications for Alzheimer's disease. J Neurosci Res. 2008;86(14):3230–3239. doi: 10.1002/jnr.21760. [DOI] [PubMed] [Google Scholar]
- 44.Harada M, Hanada S, Toivola DM, Ghori N, Omary MB. Autophagy activation by rapamycin eliminates mouse Mallory-Denk bodies and blocks their proteasome inhibitor-mediated formation. Hepatology. 2008;47(6):2026–2035. doi: 10.1002/hep.22294. [DOI] [PubMed] [Google Scholar]
- 45.Kang GY, Bang JY, Choi AJ, Yoon J, Lee WC, Choi S, Yoon S, Kim HC, Baek JH, Park HS, et al. Exosomal proteins in the aqueous humor as novel biomarkers in patients with neovascular age-related macular degeneration. J Proteome Res. 2014;13(2):581–595. doi: 10.1021/pr400751k. [DOI] [PubMed] [Google Scholar]
- 46.Fuchs E, Weber K. Intermediate filaments: structure, dynamics, function, and disease. Annu Rev Biochem. 1994;63:345–382. doi: 10.1146/annurev.bi.63.070194.002021. [DOI] [PubMed] [Google Scholar]
- 47.Lau AT, Chiu JF. The possible role of cytokeratin 8 in cadmium-induced adaptation and carcinogenesis. Cancer Res. 2007;67(5):2107–2113. doi: 10.1158/0008-5472.CAN-06-3771. [DOI] [PubMed] [Google Scholar]
- 48.Snider NT, Omary MB. Post-translational modifications of intermediate filament proteins: mechanisms and functions. Nat Rev Mol Cell Biol. 2014;15(3):163–177. doi: 10.1038/nrm3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tao GZ, Looi KS, Toivola DM, Strnad P, Zhou Q, Liao J, Wei Y, Habtezion A, Omary MB. Keratins modulate the shape and function of hepatocyte mitochondria: a mechanism for protection from apoptosis. J Cell Sci. 2009;122(Pt 21):3851–3855. doi: 10.1242/jcs.051862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Caulin C, Ware CF, Magin TM, Oshima RG. Keratin-dependent, epithelial resistance to tumor necrosis factor-induced apoptosis. J Cell Biol. 2000;149(1):17–22. doi: 10.1083/jcb.149.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gilbert S, Loranger A, Daigle N, Marceau N. Simple epithelium keratins 8 and 18 provide resistance to Fas-mediated apoptosis. The protection occurs through a receptor-targeting modulation. J Cell Biol. 2001;154(4):763–773. doi: 10.1083/jcb.200102130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290(5497):1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword. Science. 2004;306(5698):990–995. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xie R, Nguyen S, McKeehan K, Wang F, McKeehan WL, Liu L. Microtubule-associated protein 1S (MAP1S) bridges autophagic components with microtubules and mitochondria to affect autophagosomal biogenesis and degradation. J Biol Chem. 2011;286(12):10367–10377. doi: 10.1074/jbc.M110.206532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moreau K, Ravikumar B, Renna M, Puri C, Rubinsztein DC. Autophagosome precursor maturation requires homotypic fusion. Cell. 2011;146(2):303–317. doi: 10.1016/j.cell.2011.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Korolchuk VI, Saiki S, Lichtenberg M, Siddiqi FH, Roberts EA, Imarisio S, Jahreiss L, Sarkar S, Futter M, Menzies FM, et al. Lysosomal positioning coordinates cellular nutrient responses. Nat Cell Biol. 2011;13(4):453–460. doi: 10.1038/ncb2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pous C, Codogno P. Lysosome positioning coordinates mTORC1 activity and autophagy. Nat Cell Biol. 2011;13(4):342–344. doi: 10.1038/ncb0411-342. [DOI] [PubMed] [Google Scholar]
- 58.Fass E, Shvets E, Degani I, Hirschberg K, Elazar Z. Microtubules support production of starvation-induced autophagosomes but not their targeting and fusion with lysosomes. J Biol Chem. 2006;281(47):36303–36316. doi: 10.1074/jbc.M607031200. [DOI] [PubMed] [Google Scholar]
- 59.Scheel J, Matteoni R, Ludwig T, Hoflack B, Kreis TE. Microtubule depolymerization inhibits transport of cathepsin D from the Golgi apparatus to lysosomes. J Cell Sci. 1990;96(Pt 4):711–720. doi: 10.1242/jcs.96.4.711. [DOI] [PubMed] [Google Scholar]
- 60.Veeranna YDS, Lee JH, Vinod KY, Stavrides P, Amin ND, Pant HC, Nixon RA. Declining phosphatases underlie aging-related hyperphosphorylation of neurofilaments. Neurobiol Aging. 2011;32(11):2016–2029. doi: 10.1016/j.neurobiolaging.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sontag E, Luangpirom A, Hladik C, Mudrak I, Ogris E, Speciale S, White CL., 3rd Altered expression levels of the protein phosphatase 2A ABalphaC enzyme are associated with Alzheimer disease pathology. J Neuropathol Exp Neurol. 2004;63(4):287–301. doi: 10.1093/jnen/63.4.287. [DOI] [PubMed] [Google Scholar]
- 62.Strnad P, Windoffer R, Leube RE. In vivo detection of cytokeratin filament network breakdown in cells treated with the phosphatase inhibitor okadaic acid. Cell Tissue Res. 2001;306(2):277–293. doi: 10.1007/s004410100455. [DOI] [PubMed] [Google Scholar]
- 63.Liu R, Wang JZ. Protein phosphatase 2A in Alzheimer's disease. Pathophysiology. 2009;16(4):273–277. doi: 10.1016/j.pathophys.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 64.Sontag JM, Sontag E. Protein phosphatase 2A dysfunction in Alzheimer's disease. Front Mol Neurosci. 2014;7:16. doi: 10.3389/fnmol.2014.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Samari HR, Moller MT, Holden L, Asmyhr T, Seglen PO. Stimulation of hepatocytic AMP-activated protein kinase by okadaic acid and other autophagy-suppressive toxins. Biochem J. 2005;386(Pt 2):237–244. doi: 10.1042/BJ20040609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cullis DN, Philip B, Baleja JD, Feig LA. Rab11-FIP2, an adaptor protein connecting cellular components involved in internalization and recycling of epidermal growth factor receptors. J Biol Chem. 2002;277(51):49158–49166. doi: 10.1074/jbc.M206316200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.