Abstract
Inflammation and cancerogenesis are strongly interconnected processes, not only because inflammation promotes DNA instability, but also because both processes are driven by pathways such as NF-kB, STAT3, mTOR and MAPKs. Interestingly, these pathways regulate the release of pro-inflammatory cytokines such as IL-6, TNF-α and IL-1β that in turn control their activation and play a crucial role in shaping immune response. The transcription factor p53 is the major tumor suppressor that is often mutated in cancer, contributing to tumor progression. In this overview, we highlight how the interplay between pro-inflammatory cytokines and pro-inflammatory/pro-oncogenic pathways, regulating and being regulated by UPR signaling and autophagy, affects the stability of mutp53 that in turn is able to control autophagy, UPR signaling, cytokine release and the activation of the same oncogenic pathways to preserve its own stability and promote tumorigenesis. Interrupting these positive feedback loops may represent a promising strategy in anticancer therapy, particularly against cancers carrying mutp53.
Keywords: Inflammatory cytokines, Cancer, Mutant p53, Unfolded protein response, Oncogenic pathways, Autophagy
Introduction
Pro-inflammatory and anti-inflammatory cytokines such as IL-6, IL-1β, TNF-α and IL-10 deeply shape immune response; therefore the dysregulation of their production may lead to immune dysfunction, favoring the onset of inflammatory diseases including cancer [1–3]. NF-kB (nuclear factor kappa-light chain- enhancer of activated B cells), MAPKs (mitogen-activated protein kinases), mTOR (mammalian target of rapamycin) and STAT3 (signal transducer and activator of transcription 3) are among the most important pathways that regulate cytokine production and, interestingly, also strongly involved in the control of carcinogenesis. These pathways that bridge inflammation to cancer may be activated in response to cellular stress caused by the presence of oncogenes or by the sensing of PAMPs (pathogens-associated molecular patterns) or DAMPs (damage-associated molecular patterns), molecules that also trigger endoplasmic reticulum (ER) stress and the unfolded protein response (UPR) [4]. UPR is an adaptive response that helps cells to survive in the face of stress whose signaling initiates from proteins that traverse the ER membrane and act as cellular sensors, namely: inositol requiring enzyme 1 (IRE1, also known as ERN1), activating transcription factor 6 (ATF6), and PKR-like ER kinase (PERK, also known as EIF2AK3) [4]. These three sensors activate an integrated transcriptional program that drives multiple processes, including the activation of the oncogenic pathways and the secretion of the cytokines above reported [5, 6]. Interestingly, cytokines released following UPR activation may in turn trigger UPR and through its signaling or directly can reactivate the same oncogenic pathways that promote their production, in a positive feedback loop [7] (Fig. 1).
Fig. 1.
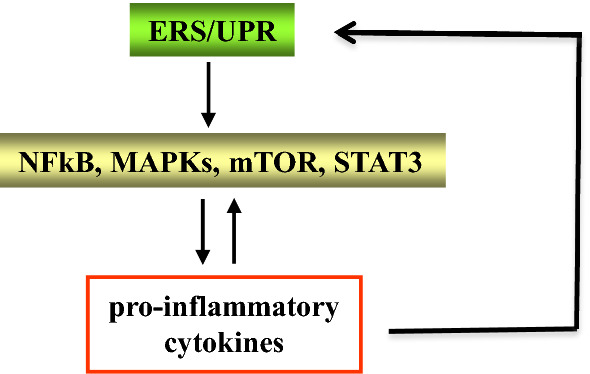
Interplay between oncogenic pathways (NF-kB, mTOR, STAT3 and p38MAPK), ERS (endoplasmic reticulum stress)/UPR (unfolded protein response) and pro-inflammatory cytokines
Mutations in the TP53 oncosuppressor gene are very common in cancers, as a normal functioning p53 does not allow cells to undergo oncogenic transformation [8]. In addition, in cancers in which p53 is not mutated, wild-type (wt) p53 protein may be functionally inactivated by other mechanisms [8, 9]. For example, wtp53 is inhibited or degraded due to the binding to viral proteins, in some virus-associated cancers [10, 11]. Besides losing the wild-type oncosuppressor functions, some mutant (mut) p53 proteins acquire oncogenic properties, defined as gain of function (GOF) [8]. This implies that mutp53 may promote tumor invasion, metastasis, chemoresistance and inflammation [12], e.g., it has been reported that mutp53 may alter cancer cell secretome and consequently modulate the characteristics of the tumor microenvironment [13]. Among other effects, mutp53 has been reported to trigger UPR, in particular the ATF6 branch [14] that shares with the other two arms, IRE1alpha and PERK, the capacity to activate the pro-inflammatory/pro-oncogenic transcription factors NF-κB, STAT3, MAPK and mTOR and to positively regulate pro-inflammatory cytokine production [7], strengthening the link between inflammation and cancer [15]. All the above-mentioned pathways may be activated by mutp53, which earns in turn a greater stability, a distinctive trait of its oncogenic function. Thus, these pathways may prevent mutp53 degradation [14, 16], for example, by negatively regulating macroautophagy, a process known to be involved in mutp53 degradation [17–19] (Fig. 2). As mutp53 is often misfolded, its hyperstability may also depend on the presence of chaperoning molecules such as HSP90 [20, 21] whose expression can be regulated by pathways such as PI3K/AKT/mTOR [22] and STAT3 [23, 24] (Fig. 3).
Fig. 2.
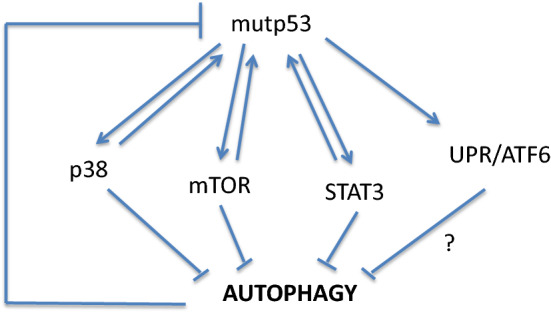
Autophagy induces mutp53 degradation. Mutp53 may activate the oncogenic pathways to inhibit autophagy and therefore prevent its degradation. The results of that interplay further link inflammation to cancerogenesis
Fig. 3.
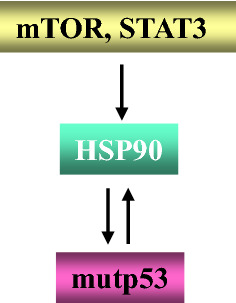
The oncogenic pathways mTOR and STAT3 up-regulate molecular chaperone HSP90 expression level that contributes to maintain mutp53 stability
On the basis of the above background, the aim of this perspective is to discuss how the interplay between UPR, inflammatory cytokines and pro-inflammatory/pro-oncogenic pathways may affect mutp53 expression levels by regulating its stability and/or degradation.
The interplay between UPR, autophagy, pro-inflammatory cytokines and pro-oncogenic pathways regulates mutp53 expression level
When unfolded/misfolded proteins accumulate into the ER, they attract GRP78/BIP detaching it from IRE1α, PERK and ATF6 UPR sensors, resulting in their activation and UPR triggering. This process helps cells to be relieved from stress, although when it is too prolonged or intense it may induce cell death [4]. Among its protective functions, UPR may reduce protein translation, increase ER chaperone transcription, promote mRNA degradation and induce macroautophagy to eliminate misfolded proteins and damaged organelles [25]. Macroautophagy (hereafter referred to as autophagy) is indeed a catabolic process, during which unwanted materials are enclosed in double membrane vesicles that are targeted to lysosomes for degradation. Autophagy has a key role in maintaining essential biological activities during cellular stress and plays also a role in physiological processes; therefore, its dysregulation predisposes to a variety of human diseases, including cancer [26]. Autophagy regulation is highly dependent on rewiring of tumor metabolism, as demonstrated by the fact that mTOR and AMPK cellular sensors are both the master regulators of autophagy [27]. The autophagic process may have a multifacet role in cancer, depending on the cell context, the tumor types and stage, as well as the nature of stress and the metabolic and environmental status of the cancer cells [28]. In this regard, it has been reported that autophagy may inhibit the first steps of cancerogenesis by reducing reactive oxygen species (ROS) and DNA damage, thus preventing oncogenic transformation [29].
On the other side, autophagy promotes survival of established cancers, especially when they undergo chemotherapy/radiotherapy, or even contributes to immunogenicity of cell death in the course of those treatments [28–30]. The ER stress/UPR-dependent autophagy induction has been reported to promote the degradation of mutp53 [31, 32] that is able in turn to counteract this catabolic process to prevent its own elimination [33]. Moreover, mutp53 has been shown to manipulate UPR, inhibiting its pro-death functions while stimulating the pro-survival ones through the activation of ATF6 [14], although the impact of ATF6 activation on autophagy in this context remains to be explored. UPR signaling may contribute to the activation of NF-κB, STAT3, MAPK and mTOR, thus regulating the cytokine release [34] that both influences and is influenced by autophagy. Of note, ATF6 has been shown to synergize with toll-like receptor (TLR) signaling in the activation of NF-κB, stimulating the production of pro-inflammatory cytokines while reducing the anti-inflammatory ones [35] or contribute to the activation of p38 MAPK to regulate cytokine secretion [36]. Intriguingly MKK3, a kinase involved in the phosphorylation of p38 MAPK, is one of mutp53 targets [37] whose inhibition promotes mutp53 degradation through autophagy [31]. These findings allow us to hypothesize that ATF6 activation by mutp53 may contribute to p38 phosphorylation also to counteract autophagy. In addition, ATF6 may activate mTOR [38], the master negative regulator of autophagy, previously reported to be activated by mutp53 to inhibit autophagy [39]. These results point out to a close relationship between UPR, pro-oncogenic pathways regulating the release of pro-inflammatory cytokines, autophagy and mutp53 stability that deserves to be better explored in future studies.
Cross talk between NF-κB, UPR, autophagy and mutp53
The NF-κB family of transcription factors, composed of five members designated as p65 (RelA), RelB, c-Rel, NF-κB1 and NF-κB2, plays an essential role in regulating inflammation. Even if its primary function it to sustain this process by inducing the transcription of pro-inflammatory cytokines and contributing to the activation of NRLP3 inflammosome, NF-κB may also restrain inflammation, for example by promoting the p62/SQSTM1-mediated removal of damaged mitochondria [40]. Prolonged inflammation is associated with an increased risk of cancer, as it favors mismatch repair abnormalities or alters DNA methylation, and DNA hypermethylation has been reported to precede large granular lymphocytic leukemia by activating the NF-κB-Myc axis [15]. NF-κB is activated mainly by mutations of the upstream components of this pathway, as observed in a plethora of hematological as well as solid cancers [41]. Interestingly, NF-κB that promotes the release of pro-inflammatory cytokines may in turn be activated by them, in a regulatory circuit that plays a critical role in cancerogenesis. Indeed, an inflammatory microenvironment paves the way to processes such as endothelial to mesenchymal transition (EMT) and strongly contributes to the impairment of immune response, i.e., by skewing macrophage polarization into M2/TAM, cells that sustain tumor instead of fighting it [20, 42, 43]. Of note, oncogenes such as RAS and Myc have been shown to promote a pro-inflammatory environment and a similar effect may be induced by mutp53, which for that reason may acquire oncogenic properties. While wtp53 interacts with NF-κB and monitors that inflammatory response is properly balanced, maintaining the defense against pathogens and preventing the onset of inflammatory diseases, mutp53 interaction with NF-κB dysregulates its activation to create an inflammatory microenvironment that promotes tumor progression [42]. Mutp53 may induce elevated expression of CXCL5, CXCL8 and CXCL12 [13, 44, 45], increase the expression of NF-κB2 [46], stimulate NF-κB transcriptional activity in cancer cells exposed to TNF-α [47] or interact with NF-κB upon chronic TNF-α signaling to simultaneously activate pro-tumorigenic genes [48]. It has also been reported that mutp53 sustains TNF-α-induced NF-κB signaling through the transcriptional repression of DAB2IP, a cytoplasmic inhibitor of NF-κB, increasing the secretion of inflammatory chemokines which recruit lymphocytes in the tumor bed and coopt them to further promote inflammation [47]. Moreover, mutp53 is able to suppress the anti-inflammatory response by reducing the secreted interleukin-1 receptor antagonist (sIL-1Ra, IL1RN), an effect that contributes to the chronicity of the inflammatory process [49]. NF-κB may be activated through UPR signaling, i.e., by ATF6, the UPR sensor activated by mutp53 [14] that may trigger NF-κB activation through the phosphorylation of AKT [50].
Very complex is the relationship between NF-κB and autophagy. Interestingly, several components of the NF-κB pathway such as IKKa, IKKb and IKKg may be degraded through autophagy, following HSP90 inhibition [51]. NF-κB has been reported to inhibit autophagy in a variety of cancers [52, 53], therefore its activation through ATF6 signaling and by pro-inflammatory cytokines, both induced by mutp53, could contribute to sustain its expression level also by inhibiting autophagy.
Cross talk between STAT3, UPR, autophagy and mutp53
Another pathway playing a crucial role in inflammation, cancerogenesis and immune suppression is STAT3 [54], whose tyrosine phosphorylation, the most critical event for its activation, is mediated by JAK2, even if other kinases, including PERK UPR sensor, are able to do so [55]. JAK2-mediated STAT3 phosphorylation mainly occurs in response to the signaling mediated by pro-inflammatory and immune-suppressive cytokines whose release is also promoted by STAT3 activation, in a positive feedback loop [56]. STAT3 is a very promising target in anticancer therapy, as its inhibition interrupts the release of cytokines that re-activate STAT3 in immune myeloid cells, inducing immune dysfunction, or in B cells, contributing to EBV-driven immortalization, or in established cancer cells, sustaining cell survival [57–60]. While it has been reported that STAT3 and wtp53 negatively regulate each other [61, 62], recent studies have highlighted that STAT3 engages a cross talk with mutp53 in which they sustain each other [24, 63, 64]. Reducing the release of pro-inflammatory cytokines or using monoclonal antibodies able to neutralize their activity could interrupt the harmful alliance between mutp53 and STAT3, in which these cytokines may act as a bridge. In previous studies, we have found that the interplay between STAT3 activation and the production of pro-inflammatory cytokines may reduce autophagy [65], therefore it is possible that such interplay could contribute to the prevention of autophagy-mediated mutp53 degradation.
Cross talk between mTOR, UPR, autophagy and mutp53
mTOR is a serine/threonine protein kinase that belongs to the phosphatidylinositol kinase-related kinase (PIKK) family and constitutes the catalytic subunit of two distinct protein complexes, known as mTORC1 and mTORC2 [66]. A plethora of studies evidenced a crucial role for mTOR pathway in the regulation of fundamental cellular processes, such as protein synthesis [67], EMT [68], Warburg effect [69], autophagy [67], immune response [70] and oxidative stress [71] and demonstrate that deregulated mTOR signaling is implicated in cancer progression as well as aging [72]. Interestingly, mTOR activation may engage a cross talk with ER stress and UPR with important implications in anticancer therapy [73]. A recent report has indicated that ATF6 branch of UPR is directly involved in mTOR activation [38]. Interestingly, mTOR may be also activated by cytokines [74, 75] and, as UPR has a key role in regulating the release of these molecules, UPR signaling could contribute to mTOR activation also by increasing the production of these cytokines. Of note, mutp53, in contrast to its wild-type counterpart, may support mTOR signaling, sustaining an oxidative environment that leads to uncontrolled cancer cell proliferation [76]. Indeed, it has been recently reported that hotspot mutp53 promotes the phosphorylation of the mTORC1 targets S6K1 and 4EBP1 in both colon and non-small carcinoma cells [77]. Considering that mTOR is the master negative regulator of autophagy, it is not surprising that mutp53 may adopt several strategies to maintain mTOR activated in an attempt to prevent its degradation through autophagy. On the other side, the stimulation of mTOR activity by mutp53 represents an Achille’s heel that makes cancer cells carrying mutp53 more sensitive to mTOR inhibitors [39]. This may have particular implications from a therapeutic point of view, since counteracting mTOR pathway may provide new therapeutic openings for clinical studies in cancer patients carrying the mutant TP53 gene.
Cross talk between MAPKs, UPR, autophagy and mutp53
The Ser/Thr mitogen-activated family of protein kinases (MAPKs) includes p38 (α, β, γ, and δ), c-Jun amino-terminal kinases 1–3 (JNK1 to -3), and the extracellular signal-regulated kinases 1 and 2 (ERK1/2). P38 MAPK, one of the best studied MAPKs, may influence a multitude of cellular events, such as cell growth, proliferation, differentiation and inflammation, as it plays an important role in the regulation of cytokines production by the immune cells [78]. P38 as well as the other MAPK are activated through ER stress/UPR signaling, promoting the secretion of pro-inflammatory cytokines [5] that, also in this case, reactivate p38MAPK and UPR, through positive feedback loops [79]. The release of IL-1β, IL-6, and IL-8, the most important cytokines promoting inflammation, is under the direct control of p38 MAPK [80]. MKK3, a kinase involved in p38MAPK activation [81], has been shown to be activated by mutp53 to promote tumor survival [82]. Interestingly, MKK3 depletion may trigger ER stress and autophagy, inducing mutp53 degradation and reducing tumor growth [31].
JNK is another MAPK activated by pro-inflammatory cytokines as well as by UPR signaling [5]. Tumorigenic mutp53 has been reported to disrupt the Daxx-ASK1 circuit that amplifies JNK signaling, making cells more tolerant to stress induced by TNFα [83], differently from what occurs for NF-κB whose activation is sustained by mutp53 [46, 47]. Intriguingly, although in completely different cell contexts, we have previously shown that JNK activation could promote autophagy [84, 85], allowing us to speculate that mutp53 could reduce JNK activation, once again to counteract its degradation through this catabolic route.
Previous findings have suggested that mutp53 may slightly influence ERK1/2 phosphorylation [86], although this molecule may be activated downstream of the mevalonate pathway that is known to be sustained by mutp53 [16] as well as by pro-inflammatory and anti-inflammatory cytokines [87]. More investigations are required to clarify the role of ERK1/2 on autophagy, as it has been reported to either negatively or positively regulate this process in cancer cells [88], depending on the stimuli and the cell types [89]. Also the interplay between mutp53 and ERK1/2 needs to be better elucidated, although we speculate that, given the controversial role ERK1/2 on autophagy, mutp53 could avoid interfering with its activation to preserve its own stability.
Concluding remarks and potential implications for cancer therapy
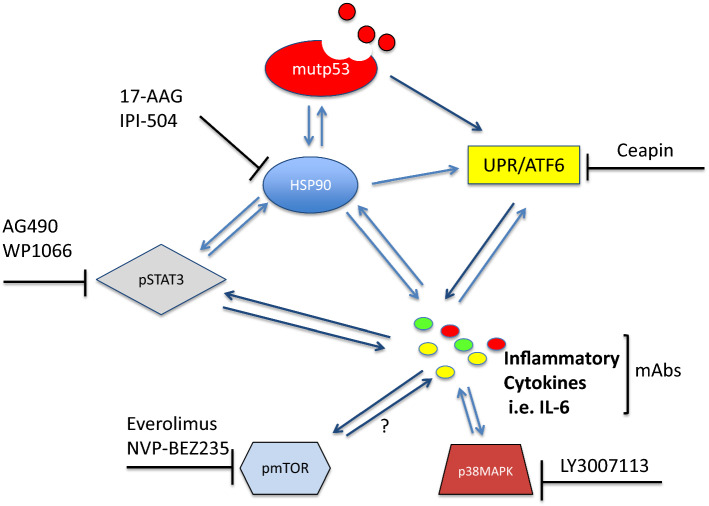
Inflammation may promote cancer onset and progression as well as immune dysfunction. Inflammation is orchestrated by the interplay between several pro-inflammatory cytokines and oncogenic pathways, including NF-kB, MAPK, mTOR and STAT3. These pathways can be activated by oncogenes, PAMPs or DAMPs, also through the triggering of ER stress/UPR signaling that thus contributes to the pro-inflammatory cytokine release. Besides shaping a pro-inflammatory/immune suppressive microenvironment, these cytokines may reactivate the oncogenic pathways that regulate their production, in positive feedback loops that ultimately regulate autophagy and the expression of chaperones such as HSP90. These complex interactions may influence the expression level of mutp53, whose hyperstability is a prerequisite of its GOF. Consequently, interrupting the interplay between UPR, oncogenic pathways, and pro-inflammatory cytokines may affect autophagy and HSP90 expression and concomitantly reduce pro-oncogenic inflammation and the stability of mutp53. Also considering that mutp53 may in turn positively influence the activation of these pathways and thus downregulate it may help to break the bridge between inflammation and cancer. Inhibitors of specific oncogenic pathways such as AG490 or tocilizumab WP1066 for STAT3, everolimus or NVP-BEZ235 for mTOR or LY3007113 for p38MAPK or targeting chaperones such as HSP90 by 17-AAG or derivatives, used alone or in combination, may be promising in anticancer therapy, as they are able to interfere with multiple aspects of cancer biology (Fig. 4). Of note, many of these inhibitors are already in pre-clinical or clinical trials in which they are showing promising results. Antibodies against pro-inflammatory cytokines or their receptors may be also exploited to counteract inflammation and the activation of cancer-promoting molecular pathways that, among other functions, may contribute to mutp53 stability (Fig. 4).
Fig. 4.
Schematic representation of the interplay between pro-inflammatory cytokines, UPR, pro-oncogenic pathways, HSP90, whose targeting may promote mutp53 degradation
Acknowledgements
The research in the laboratory of GD has been supported by Grants from the Italian Association for Cancer Research (AIRC) (IG 2013, 11377; IG 2015, 16742); in the laboratory of Mara Cirone by Grants from the Italian Association for Cancer Research (AIRC) IG 2019 23040) and by Istituto Pasteur Italia Fondazione Cenci Bolognetti. The funding agencies played no role in the concept, design, or writing of this study.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Galdiero MR, Marone G, Mantovani A. Cancer inflammation and cytokines. Cold Spring Harb Perspect Biol. 2018 doi: 10.1101/cshperspect.a028662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Landskron G, De La Fuente M, Thuwajit P, et al. Chronic inflammation and cytokines in the tumor microenvironment. J Immunol Res. 2014 doi: 10.1155/2014/149185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qu X, Tang Y, Hua S. Immunological approaches towards cancer and inflammation: a cross talk. Front Immunol. 2018 doi: 10.3389/fimmu.2018.00563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Senft D, Ronai ZA. UPR, autophagy, and mitochondria crosstalk underlies the ER stress response. Trends Biochem Sci. 2015 doi: 10.1016/j.tibs.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Darling NJ, Cook SJ. The role of MAPK signalling pathways in the response to endoplasmic reticulum stress. Biochim Biophys Acta Mol Cell Res. 2014 doi: 10.1016/j.bbamcr.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 6.Schmitz ML, Shaban MS, Albert BV, et al. The crosstalk of endoplasmic reticulum (ER) stress pathways with NF-κB: complex mechanisms relevant for cancer, inflammation and infection. Biomedicines. 2018 doi: 10.3390/biomedicines6020058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reverendo M, Mendes A, Argüello RJ, et al. At the crossway of ER-stress and proinflammatory responses. FEBS J. 2019 doi: 10.1111/febs.14391. [DOI] [PubMed] [Google Scholar]
- 8.Muller PAJ, Vousden KH. Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell. 2014 doi: 10.1016/j.ccr.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Puca R, Nardinocchi L, Gal H, et al. Reversible dysfunction of wild-type p53 following homeodomain-interacting protein kinase-2Knockdown. Cancer Res. 2008 doi: 10.1158/0008-5472.CAN-07-6776. [DOI] [PubMed] [Google Scholar]
- 10.Chatterjee K, Das P, Chattopadhyay NR, et al. The interplay between Epstein-Barr virus (EBV) with the p53 and its homologs during EBV associated malignancies. Heliyon. 2019 doi: 10.1016/j.heliyon.2019.e02624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lan KH, Sheu ML, Hwang SJ, et al. HCV NS5A interacts with p53 and inhibits p53-mediated apoptosis. Oncogene. 2002 doi: 10.1038/sj.onc.1205589. [DOI] [PubMed] [Google Scholar]
- 12.Mantovani F, Collavin L, Del Sal G. Mutant p53 as a guardian of the cancer cell. Cell Death Differ. 2019 doi: 10.1038/s41418-018-0246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cordani M, Pacchiana R, Butera G, et al. Mutant p53 proteins alter cancer cell secretome and tumour microenvironment: Involvement in cancer invasion and metastasis. Cancer Lett. 2016 doi: 10.1016/j.canlet.2016.03.046. [DOI] [PubMed] [Google Scholar]
- 14.Sicari D, Fantuz M, Bellazzo A, et al. Mutant p53 improves cancer cells’ resistance to endoplasmic reticulum stress by sustaining activation of the UPR regulator ATF6. Oncogene. 2019 doi: 10.1038/s41388-019-0878-3. [DOI] [PubMed] [Google Scholar]
- 15.Greten FR, Grivennikov SI. Inflammation and cancer: triggers, mechanisms, and consequences. Immunity. 2019 doi: 10.1016/j.immuni.2019.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parrales A, Thoenen E, Iwakuma T. The interplay between mutant p53 and the mevalonate pathway. Cell Death Differ. 2018 doi: 10.1038/s41418-017-0026-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garufi A, Pucci D, D’Orazi V, et al. Degradation of mutant p53H175 protein by Zn(II) through autophagy. Cell Death Dis. 2014 doi: 10.1038/cddis.2014.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choudhury S, Kolukula VK, Preet A, et al. Dissecting the pathways that destabilize mutant p53: the proteasome or autophagy? Cell Cycle. 2013 doi: 10.4161/cc.24128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garufi A, Pistritto G, Cirone M, D’Orazi G. Reactivation of mutant p53 by capsaicin, the major constituent of peppers. J Exp Clin Cancer Res. 2016 doi: 10.1186/s13046-016-0417-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilardini Montani MS, Cecere N, Granato M, et al. Mutant P53, stabilized by its interplay with HSP90, activates a positive feed-back loop between NRF2 and P62 that induces chemo-resistance to apigenin in pancreatic cancer cells. Cancers (Basel) 2019 doi: 10.3390/cancers11050703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alexandrova EM, Moll UM. Depleting stabilized GOF mutant p53 proteins by inhibiting molecular folding chaperones: a new promise in cancer therapy. Cell Death Differ. 2017 doi: 10.1038/cdd.2016.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chatterjee M, Andrulis M, Stühmer T, et al. The PI3k/Akt signaling pathway regulates the expression of Hsp70, which critically contributes to Hsp90-chaperone function and tumor cell survival in multiple myeloma. Haematologica. 2013 doi: 10.3324/haematol.2012.066175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Granato M, Chiozzi B, Filardi MR, et al. Tyrosine kinase inhibitor tyrphostin AG490 triggers both apoptosis and autophagy by reducing HSF1 and Mcl-1 in PEL cells. Cancer Lett. 2015 doi: 10.1016/j.canlet.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 24.Romeo MA, Gilardini Montani MS, Benedetti R, Santarelli R, D'Orazi G, Cirone M. STAT3 and mutp53 Engage a Positive Feedback Loop Involving HSP90 and the Mevalonate Pathway. Front Oncol. 2020 doi: 10.3389/fonc.2020.01102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cirone M. Perturbation of bulk and selective macroautophagy, abnormal UPR activation and their interplay pave the way to immune dysfunction, cancerogenesis and neurodegeneration in ageing. Ageing Res Rev. 2020 doi: 10.1016/j.arr.2020.101026. [DOI] [PubMed] [Google Scholar]
- 26.Dikic I, Elazar Z. Mechanism and medical implications of mammalian autophagy. Nat Rev Mol Cell Biol. 2018;19:349–364. doi: 10.1038/s41580-018-0003-4. [DOI] [PubMed] [Google Scholar]
- 27.Dodson M, Darley-Usmar V, Zhang J. Cellular metabolic and autophagic pathways: traffic control by redox signaling. Free Radic Biol Med. 2013;63:207–221. doi: 10.1016/j.freeradbiomed.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kondo Y, Kanzawa T, Sawaya R, Kondo S. The role of autophagy in cancer development and response to therapy. Nat Rev Cancer. 2005 doi: 10.1038/nrc1692. [DOI] [PubMed] [Google Scholar]
- 29.Mathew R, Kongara S, Beaudoin B, et al. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007 doi: 10.1101/gad.1545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Masuelli L, Granato M, Benvenuto M, et al. Chloroquine supplementation increases the cytotoxic effect of curcumin against Her2/neu overexpressing breast cancer cells in vitro and in vivo in nude mice while counteracts it in immune competent mice. Oncoimmunology. 2017 doi: 10.1080/2162402X.2017.1356151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baldari S, Ubertini V, Garufi A, et al. Targeting MKK3 as a novel anticancer strategy: molecular mechanisms and therapeutical implications. Cell Death Dis. 2015 doi: 10.1038/cddis.2014.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garufi A, Federici G, Montani MSG, et al. Interplay between endoplasmic reticulum (ER) stress and autophagy induces mutant p53H273 degradation. Biomolecules. 2020 doi: 10.3390/biom10030392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cordani M, Butera G, Pacchiana R, Donadelli M. Molecular interplay between mutant p53 proteins and autophagy in cancer cells. Biochim Biophys Acta Rev Cancer. 2017 doi: 10.1016/j.bbcan.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 34.Zhang K, Kaufman RJ. From endoplasmic-reticulum stress to the inflammatory response. Nature. 2008 doi: 10.1038/nature07203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rao J, Yue S, Fu Y, et al. ATF6 mediates a pro-inflammatory synergy between ER stress and TLR activation in the pathogenesis of liver ischemia-reperfusion injury. Am J Transplant. 2014 doi: 10.1111/ajt.12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mijošek V, Lasitschka F, Warth A, et al. Endoplasmic reticulum stress is a danger signal promoting innate inflammatory responses in bronchial epithelial cells. J Innate Immun. 2016 doi: 10.1159/000447668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gurtner A, Starace G, Norelli G, et al. Mutant p53-induced up-regulation of mitogen-activated protein kinase kinase 3 contributes to gain of function. J Biol Chem. 2010 doi: 10.1074/jbc.M109.094813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allen D, Seo J. ER stress activates the TOR pathway through Atf6. J Mol Signal. 2018 doi: 10.5334/1750-2187-13-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cordani M, Oppici E, Dando I, et al. Mutant p53 proteins counteract autophagic mechanism sensitizing cancer cells to mTOR inhibition. Mol Oncol. 2016 doi: 10.1016/j.molonc.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhong Z, Umemura A, Sanchez-Lopez E, et al. NF-κB restricts inflammasome activation via elimination of damaged mitochondria. Cell. 2016 doi: 10.1016/j.cell.2015.12.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verzella D, Pescatore A, Capece D, et al. Life, death, and autophagy in cancer: NF-κB turns up everywhere. Cell Death Dis. 2020 doi: 10.1038/s41419-020-2399-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cooks T, Harris CC, Oren M. Caught in the crossfire: p53 in inflammation. Carcinogenesis. 2014;35:1680–1690. doi: 10.1093/carcin/bgu134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strazza M, Mor A. The complexity of targeting chemokines to promote a tumor immune response. Inflammation. 2020 doi: 10.1007/s10753-020-01235-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yeudall WA, Vaughan CA, Miyazaki H, et al. Gain-of-function mutant p53 upregulates CXC chemokines and enhances cell migration. Carcinogenesis. 2012 doi: 10.1093/carcin/bgr270. [DOI] [PubMed] [Google Scholar]
- 45.Scian MJ, Stagliano KER, Anderson MAE, et al. Tumor-derived p53 mutants induce NF-κB2 gene expression. Mol Cell Biol. 2005 doi: 10.1128/mcb.25.22.10097-10110.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weisz L, Damalas A, Liontos M, et al. Mutant p53 enhances nuclear factor κB activation by tumor necrosis factor α in cancer cells. Cancer Res. 2007 doi: 10.1158/0008-5472.CAN-06-2425. [DOI] [PubMed] [Google Scholar]
- 47.Di Minin G, Bellazzo A, DalFerro M, et al. Mutant p53 reprograms TNF signaling in cancer cells through interaction with the tumor suppressor DAB2IP. Mol Cell. 2014 doi: 10.1016/j.molcel.2014.10.013. [DOI] [PubMed] [Google Scholar]
- 48.Rahnamoun H, Lu H, Duttke SH, Benner C, Glass CK, Lauberth SM. Mutant p53 shapes the enhancer landscape of cancer cells in response to chronic immune signaling. Nat Commun. 2017 doi: 10.1038/s41467-017-01117-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ubertini V, Norelli G, D’Arcangelo D, et al. Mutant p53 gains new function in promoting inflammatory signals by repression of the secreted interleukin-1 receptor antagonist. Oncogene. 2015 doi: 10.1038/onc.2014.191. [DOI] [PubMed] [Google Scholar]
- 50.Yamazaki H, Hiramatsu N, Hayakawa K, Tagawa Y, Okamura M, et al. Activation of the Akt-NF-κB pathway by subtilase cytotoxin through the ATF6 branch of the unfolded protein response. J Immunol. 2009 doi: 10.4049/jimmunol.0900017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trocoli A, Djavaheri-Mergny M. The complex interplay between autophagy and NF-κB signaling pathways in cancer cells. Am J Cancer Res. 2011;1(5):629–649. [PMC free article] [PubMed] [Google Scholar]
- 52.Lavieu G, Scarlatti F, Sala G, et al. Regulation of autophagy by sphingosine kinase 1 and its role in cell survival during nutrient starvation. J Biol Chem. 2006 doi: 10.1074/jbc.M506182200. [DOI] [PubMed] [Google Scholar]
- 53.Meijer AJ, Codogno P. AMP-activated protein kinase and autophagy. Autophagy. 2007 doi: 10.4161/auto.3710. [DOI] [PubMed] [Google Scholar]
- 54.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009 doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meares GP, Liu Y, Rajbhandari R, et al. PERK-dependent activation of JAK1 and STAT3 contributes to endoplasmic reticulum stress-induced inflammation. Mol Cell Biol. 2014 doi: 10.1128/mcb.00980-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Agrawal S, Gollapudi S, Su H, Gupta S. Leptin activates human B cells to secrete TNF-α, IL-6, and IL-10 via JAK2/STAT3 and p38MAPK/ERK1/2 signaling pathway. J Clin Immunol. 2011 doi: 10.1007/s10875-010-9507-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Granato M, Gilardini Montani MS, Zompetta C, Santarelli R, Gonnella R, Romeo MA, D'Orazi G, Faggioni A, Cirone M. Quercetin interrupts the positive feedback loop between STAT3 and IL-6, promotes autophagy, and reduces. Biomolecules. 2019 doi: 10.3390/biom9090482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Laudisi F, Cherubini F, Monteleone G, Stolfi C. STAT3 interactors as potential therapeutic targets for cancer treatment. Int J Mol Sci. 2018 doi: 10.3390/ijms19061787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Klionsky DJ, Abdelmohsen K, Abe A et al (2016) Guidelines for the use and interpretation of assays for monitoring autophagy, 3.th edn., Autophagy. 10.1080/15548627.2015.1100356
- 60.Cirone M, Di RL, Lotti LV, et al. Activation of dendritic cells by tumor cell death. Oncoimmunology. 2012 doi: 10.4161/onci.20428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Santarelli R, Carillo V, Romeo MA, et al. STAT3 phosphorylation affects p53/p21 axis and KSHV lytic cycle activation. Virology. 2019 doi: 10.1016/j.virol.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 62.Granato M, Gilardini Montani MS, Santarelli R, et al. Apigenin, by activating p53 and inhibiting STAT3, modulates the balance between pro-apoptotic and pro-survival pathways to induce PEL cell death. J Exp Clin Cancer Res. 2017 doi: 10.1186/s13046-017-0632-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schulz-Heddergott R, Stark N, Edmunds SJ, et al. Therapeutic ablation of gain-of-function mutant p53 in colorectal cancer inhibits Stat3-mediated tumor growth and invasion. Cancer Cell. 2018 doi: 10.1016/j.ccell.2018.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.D'Orazi G, Cirone M. Mutant p53 and cellular stress pathways; a criminal alliancer that promotes cancer progression. Cancers (Basel) 2019 doi: 10.3390/cancers11050614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Santarelli R, Gonnella R, Di Giovenale G, et al. STAT3 activation by KSHV correlates with IL-10, IL-6 and IL-23 release and an autophagic block in dendritic cells. Sci Rep. 2014 doi: 10.1038/srep04241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017 doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang X, Proud CG. The mTOR pathway in the control of protein synthesis. Physiology. 2006 doi: 10.1152/physiol.00024.2006. [DOI] [PubMed] [Google Scholar]
- 68.Gulhati P, Bowen KA, Liu J, et al. mTORC1 and mTORC2 regulate EMT, motility, and metastasis of colorectal cancer via RhoA and Rac1 signaling pathways. Cancer Res. 2011 doi: 10.1158/0008-5472.CAN-10-4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Salmond RJ. mTOR regulation of glycolytic metabolism in T cells. Front Cell Dev Biol. 2018 doi: 10.3389/fcell.2018.00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Powell JD, Pollizzi KN, Heikamp EB, Horton MR. Regulation of immune responses by mTOR. Annu Rev Immunol. 2012 doi: 10.1146/annurev-immunol-020711-075024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Morita M, Gravel SP, Chénard V, et al. MTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell Metab. 2013 doi: 10.1016/j.cmet.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 72.Guertin DA, Sabatini DM. Defining the Role of mTOR in Cancer. Cancer Cell. 2007 doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 73.Appenzeller-Herzog C, Hall MN. Bidirectional crosstalk between endoplasmic reticulum stress and mTOR signaling. Trends Cell Biol. 2012 doi: 10.1016/j.tcb.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 74.Granato M, Rizzello C, Montani MSG, et al. Quercetin induces apoptosis and autophagy in primary effusion lymphoma cells by inhibiting PI3K/AKT/mTOR and STAT3 signaling pathways. J Nutr Biochem. 2017 doi: 10.1016/j.jnutbio.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 75.Bhatt AP, Bhende PM, Sin SH, et al. Dual inhibition of PI3K and mTOR inhibits autocrine and paracrine proliferative loops in PI3K/Akt/mTOR-addicted lymphomas. Blood. 2010 doi: 10.1182/blood-2009-10-251082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cordani M, Butera G, Pacchiana R, et al. Mutant p53-associated molecular mechanisms of ROS regulation in cancer cells. Biomolecules. 2020 doi: 10.3390/biom10030361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Agarwal S, Bell CM, Taylor SM, Moran RG. p53 deletion or hotspot mutations enhance mTORC1 activity by altering lysosomal dynamics of TSC2 and Rheb. Mol Cancer Res. 2016 doi: 10.1158/1541-7786.MCR-15-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhao YW, Pan YQ, Tang MM, Lin WJ. Blocking p38 signaling reduces the activation of pro-inflammatory cytokines and the phosphorylation of p38 in the habenula and reverses depressive-like behaviors induced by neuroinflammation. Front Pharmacol. 2018 doi: 10.3389/fphar.2018.00511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pantouli E, Boehm MM, Koka S. Inflammatory cytokines activate p38 MAPK to induce osteoprotegerin synthesis by MG-63 cells. Biochem Biophys Res Commun. 2005 doi: 10.1016/j.bbrc.2005.01.122. [DOI] [PubMed] [Google Scholar]
- 80.Yeung YT, Bryce NS, Adams S, et al. P38 MAPK inhibitors attenuate pro-inflammatory cytokine production and the invasiveness of human U251 glioblastoma cells. J Neurooncol. 2012 doi: 10.1007/s11060-012-0875-7. [DOI] [PubMed] [Google Scholar]
- 81.Dérijard B, Raingeaud J, Barrett T, et al. Independent human MAP kinase signal transduction pathways defined by MEK and MKK isoforms. Science. 1995 doi: 10.1126/science.7839144. [DOI] [PubMed] [Google Scholar]
- 82.Bossi G, Marampon F, Maor-Aloni R, et al. Conditional RNA interference in vivo to study mutant p53 oncogenic gain of function on tumor malignancy. Cell Cycle. 2008 doi: 10.4161/cc.7.12.6161. [DOI] [PubMed] [Google Scholar]
- 83.Kitamura T, Fukuyo Y, Inoue M, et al. Mutant p53 disrupts the stress MAPK activation circuit induced by ASK1-dependent stabilization of Daxx. Cancer Res. 2009 doi: 10.1158/0008-5472.CAN-09-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Granato M, Santarelli R, Lotti LV, et al. JNK and macroautophagy activation by bortezomib has a pro-survival effect in primary effusion lymphoma cells. PLoS ONE. 2013 doi: 10.1371/journal.pone.0075965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Granato M, Romeo MA, Tiano MS, et al. Bortezomib promotes KHSV and EBV lytic cycle by activating JNK and autophagy. Sci Rep. 2017 doi: 10.1038/s41598-017-13533-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang C, Liu J, Liang Y, et al. Tumour-associated mutant p53 drives the Warburg effect. Nat Commun. 2013 doi: 10.1038/ncomms3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Saud K, Herrera-Molina R, Bernhardi R. Pro- and anti-inflammatory cytokines regulate the ERK pathway: Implication of the timing for the activation of microglial cells. Neurotox Res. 2005 doi: 10.1007/BF03033981. [DOI] [PubMed] [Google Scholar]
- 88.Bryant KL, Stalnecker CA, Zeitouni D, et al. Combination of ERK and autophagy inhibition as a treatment approach for pancreatic cancer. Nat Med. 2019 doi: 10.1038/s41591-019-0368-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kochetkova EY, Blinova GI, Bystrova OA, et al. Suppression of mTORC1 activity in senescent Ras-transformed cells neither restores autophagy nor abrogates apoptotic death caused by inhibition of MEK/ERK kinases. Aging (Albany NY) 2018 doi: 10.18632/aging.101686. [DOI] [PMC free article] [PubMed] [Google Scholar]