Abstract
Colorectal cancer (CRC) is the leading cause of cancer-related mortality worldwide, which makes it urgent to identify novel therapeutic targets for CRC treatment. In this study, DHX9 was filtered out as the prominent proliferation promoters of CRC by siRNA screening. Moreover, DHX9 was overexpressed in CRC cell lines, clinical CRC tissues and colitis-associated colorectal cancer (CAC) mouse model. The upregulation of DHX9 was positively correlated with poor prognosis in patients with CRC. Through gain- and loss-of function experiments, we found that DHX9 promoted CRC cell proliferation, colony formation, apoptosis resistance, migration and invasion in vitro. Furthermore, a xenograft mouse model and a hepatic metastasis mouse model were utilized to confirm that forced overexpression of DHX9 enhanced CRC outgrowth and metastasis in vivo, while DHX9 ablation produced the opposite effect. Mechanistically, from one aspect, DHX9 enhances p65 phosphorylation, promotes p65 nuclear translocation to facilitate NF-κB-mediated transcriptional activity. From another aspect, DHX9 interacts with p65 and RNA polymerase II (RNA Pol II) to enhance the downstream targets of NF-κB (e.g., Survivin, Snail) expression to potentiate the malignant phenotypes of CRC. Together, our results suggest that DHX9 may be a potential therapeutic target for prevention and treatment of CRC patients.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-021-04013-3.
Keywords: Colorectal cancer, DHX9, Outgrowth, Metastasis, NF-κB signaling
Introduction
Colorectal cancer (CRC) is one of the most common malignant tumors worldwide with the second mortality rate and the third highest incidence rate. However, its exact pathogenesis has not been fully clarified [1, 2]. Therefore, it is urgent to explore the molecular mechanism of CRC and unravel new specific cancer therapeutic targets.
Nucleic acid sensing pattern recognition receptors (PRRs) are a group of innate immune receptors recognizing DNA or RNA. They mainly include some members of Toll-like receptors (e.g., TLR3, 7, 8, 9), AIM2-like receptors (e.g., AIM2), cGAS, STING, RIG-I like receptors (e.g., RIG-I, MDA5, LPG2), DExD/H box nucleic acid helicases [e.g., DEAH-box helicase 9 (DHX9), DEAH-box helicase 36 (DHX36) and DEAD-box helicase 21(DDX21), DEAD-box helicase 60 (DDX60)], and Z-DNA binding protein 1(ZBP-1 or DAI) [3]. Some nucleic acid sensing PRRs, such as TLRs, AIM2, and STING, have been demonstrated to be involved in the development of various tumors [4, 5]. Among them, DHX9, also known as nuclear DNA helicase II or RNA helicase A (RHA), is a member of the DExH-box helicase family capable of unwinding RNA and DNA duplexes, as well as aberrant polynucleotide structures [6, 7]. It has been demonstrated that DHX9 has a large number of interacting partners, including protein, circular RNA, and microRNA [8, 9]. Additionally, DHX9 has been found to play an active role in transcription by bridging between transcriptional co-factors and RNA polymerase II (RNA Pol II) or directly binds to specific promoters [10]. Moreover, there is also a strong body of evidence indicating that DHX9 plays significant roles in translational regulation DNA replication and maintenance of genomic stability, microRNA biogenesis and processing, and RNA processing and transport [11].
Since DHX9 participates in a multitude of cellular processes, which also means its deregulation may has serious effects. Compelling evidence has shown that DHX9 is implicated in various human diseases, including cancer, viral infection, and autoimmune diseases [12–14]. As reported previously, DHX9 enhanced transcription of oncogenes to promote tumorigenesis and cancer metastasis through interacting with EGFR or CBP/p300 [15, 16]. Furthermore, DHX9 was also found to be involved in the regulation of cancer stem cells self-renewal via binding with the long non-coding RNA HIF1A-AS2 or protein L1TD1 [17, 18]. Although accumulating evidence indicates that DHX9 functions as a driver of carcinogenesis, there are also some reports implying that it has antitumor activity. Its ability to promote the transcription of the tumor suppressor gene p16INK4A, interact with the tumor suppressor BRCA1, and rescue IRES-mediated p53 translation, suggest that DHX9 may have tumour-suppressive properties [19–21]. Together, the relationship between DHX9 and tumorigenesis is complicated and controversial.
Increasing evidence has demonstrated that nuclear factor kappa B (NF-κB), a proinflammatory transcription factor, plays critical roles in the initiation and propagation of cancer [22]. It has been reported that NF-κB activation is not only involved in development of colitis-associated cancer (CAC), but also in sporadic CRC [23, 24]. Constitutive NF-κB activation results in cellular proliferation, prevents scavenger of pre-neoplastic, and eventually cellular transformation by increasing the anti-apoptosis proteins such as Survivin and Bcl2 [25]. Moreover, NF-κB may contribute to the progression of CRC through mediating the expression of vital target genes that are involved in cell proliferation (e.g., Cyclin D1, D2 and D3) [26], angiogenesis (e.g., VEGF, COX2) [27], and metastasis (e.g., Snail, MMPs) [28]. Of note, DHX9 was associated with p65 and RNA Pol II to enhance NF-κB-mediated transcriptional activation of several cytokines (e.g., CXCL8, IFN-β and TNF) to protect host against DNA virus infection [29, 30]. However, whether these interactions play a role in tumorigenesis and cancer cell metastasis remains to be elucidated.
A study showed that inhibition of DHX9 expression induced fatal damage to cancer cells without affecting the function of normal cells [11], suggesting that it is a safe and effective target for tumor therapy. Furthermore, DHX9 was overexpressed in two cell lines derived from liver xenografts of CRC patients [31], indicating that DHX9 may have a certain promoting effect on CRC. However, the function and mechanism of DHX9 in CRC remains elusive.
This study is to investigate the effects of DHX9 on the malignant phenotype of CRC and uncover the underlying mechanism. Our results showed that DHX9 was filtered out as the prominent proliferation promoter of CRC via siRNA screening. Moreover, DHX9 overexpression was positively associated with shorten overall survival and disease-free survival in patients with CRC. In addition, DHX9 was found to be overexpressed in CRC cell lines, clinical CRC tissues and CAC mouse model. The upregulation of DHX9 was positively correlated with the histopathological differentiation, infiltration depth of intestinal wall and lymph node metastasis in followed-up CRC patients, clinically. Ectopic expression of DHX9 promoted proliferation, colony formation, apoptosis resistance, migration and invasion, and metastasis of CRC cells in vitro and in vivo. However, silence DHX9 reciprocally diminished these characteristics. These effects of DHX9 on CRC are involved in activation of NF-κB pathway via promoting p65 translocation and interacting with p65 and RNA Pol II. Our study identifies DHX9 as a promising treatment target for patients with CRC.
Materials and methods
siRNA screening
The human siRNA libraries used in the screens were purchased from Ribobio (Guangzhou, China). Three independent siRNAs against per gene were contained in the siRNA libraries, which were transfected into HCT116 cells in the 96 plates following the previous study [32]. After 48 h, CRC cells were incorporated with 5-ethynyl-2′-deoxyuridine (EdU) for 2 h according to the manufacturer’s instructions. 4% paraformaldehyde was employed to fix the HCT116 cells following 1 µg/mL DAPI (Sigma) staining at for 10 min. Fluorescent images and the proportion of proliferation of CRC cells were analyzed with high-throughput screening system (MolecularDevices, USA). In the screening system, siRNAs that result in ≥ 50% suppression in the proliferation rate of CRC cells were regarded as candidate targets for inhibiting cancer. Three independent screening experiments were performed. Sequences of siRNAs were listed in Supplementary Table 1.
Human CRC tissue samples
Fresh CRC tissues and corresponding adjacent tissues were obtained after surgical resection. A total of 220 formalin-fixed paraffin-embedded (FFPE) tissues (140 primary cancerous, 80 adjacent noncancerous tissues) were collected at the First Affiliated Hospital of Gannan Medical University from 2017 to 2018 after the informed consent was obtained from each subject or each subject’s guardian according to the institutional guidelines and the Declaration of Helsinki principles. Cancer was diagnosed in these samples based on histological examinations. The average age of patients was 61 years old with a range from 37 to 82.
Immunohistochemistry (IHC)
IHC analysis was performed to detect the protein expression of DHX9 in CRC patient tissues. Briefly, 4 μm FFPE sections were deparaffinized with xylene, rehydrated in a series of graded ethanol, and heated with citrate buffer for antigen retrieval. After endogenous peroxidase blocking in 0.5% H2O2 and unspecific binding site blocking with goat serum for 60 min, the sections were incubated with antibody against DHX9 (Cat# PA5-41,259) from Invitrogen (Carlsbad, CA, USA) with a ratio of 1:500 at 4 ℃ overnight, followed by incubation with secondary antibody (MaxVison, Cat# KIT5004). The slides were stained with DAB and counterstained with hematoxylin. To evaluate the protein level of DHX9, all the slides were observed, photographed and scored according to the staining intensity as described previously [33]: 0 for no staining; 1 for weakly staining; 2 for moderately staining; 3 for strongly staining. The ratio of positive-staining cells was scored as follows: 0% scored 0; < 10% scored 1; 41–70% scored 2; > 70% scored 3. Finally, the scores of DHX9 were determined using the multiplication intensity score and the ratio of positive cell score, and then classified into four groups as follows: negative for a score of 0; low for a score of 1–3; medium for a score of 4–8; and high for a score of 9–12.
CAC mouse model
4-–6-week-old male C57BL/6 mice were purchased from Nanjing Model Animal Research Institute (Nanjing, China). The CAC model was established as described previously [34]. Briefly, mice were injected intraperitoneally with 10 mg/kg of AOM (Azoxymethane, Sigma), PBS was intraperitoneally injected as vehicle control. After 5 days, the mice were subjected to three cycles of 3% DSS (Dextran sodium sulfate, molecular mass 36–40 kDa; MP Biologicals) containing in the drinking water for 6 days with a 14-day recovery period. Mice were sacrificed on day 80 and the intestinal tissues were collected and subjected to RT-qPCR assay and Western blotting.
Xenograft experiments
4-–6-week-old male BALB/c nude mice were purchased from Nanjing Model Animal Research Institute (Nanjing, China). 1 × 107 HCT116 cells stably overexpressing DHX9 or silencing DHX9 were subcutaneously injected into BALB/c nude mice. The tumor volume was measured with calipers and calculated using the formula 0.4 × a × b2 [35], where a is the largest diameter and b is the smallest diameter in millimeters. After 2 weeks, mice were euthanized and tumors were isolated, photographed, and weighed. Tumor tissues were fixed in formalin, embedded in paraffin, and cut into 4 µm sections followed by haematoxylin and eosin (H&E), Ki67 (Maxim, Fuzhou, China), and active caspase-3 (Cell Signaling Technology, Beverly, MA) staining.
Liver metastasis mouse model
4-–6-week-old male BALB/c nude mice were obtained from Nanjing Model Animal Research Institute (Nanjing, China). 2 × 106 DHX9-overexpressed or silenced HCT116 cells resuspended in 50 µl of PBS were injected intrasplenically into the nude mice. At 10 weeks after inoculation, the mice were sacrificed after anesthetized with isoflurane and liver were isolated, followed by fixing with Bouin’’s solution. Livers were embedded in paraffin, cut into 4 µm sections, and subjected to H&E staining. All mice were bred at the animal facility of Gannan Medical University with controlled 40–50% humidity, 20 ± 2 °C temperature and the lighting cycle of 12-h light/darkness. All animal experiments were approved by the Institutional Research Ethics Committees of Gannan Medical University.
Cell culture
Human normal colonic epithelial cells including HCoEpiC and NCM460 were purchased from ScienCell (Carlsbad, CA) and Incell Corporation (San Antonio, TX), respectively. They were cultured in MEM medium supplemented with 10% fetal bovine serum (FBS, Gibco, NY, USA) with 1% penicillin-streptomycin (Solarbio, Beijing, China). The human CRC cell lines HCT116, HCT8 and COLO205 were obtained from the American Type Culture Collection (ATCC, Manassas, VA) and maintained in RPMI 1640 containing 10% FBS. 293T cells from ATCC were cultured in DMEM medium supplemented with 10% FBS.
Generation of stable cell lines
Constructs encoding human DHX9, mutant K417R DHX9, specific shRNAs targeting DHX9 or shRNAs targeting p65 were cloned into the pLVX-puro lentiviral vector [35]. Viral supernatants were collected at 48 h and 72 h after transfection with 293 T cells utilizing pCMV-dR8.2 (packing construct) and the pCMV-VSVG (envelope construct) as previously described [36]. HCT116, HCT8, and COLO205 cell were then infected with lentiviral supernatants 2–3 rounds followed by selection with 1.0 µg/mL puromycin for 1 week.
Reverse-trascription quantitative PCR (RT-qPCR)
Total RNA was extracted from colon tissues or CRC cells using Trizol reagent according to the manufacturer’s instructions. Then, 0.5 μg of total RNA was used to reverse-transcribed into cDNA with ImProm-II™ Reverse Transcription System (Promega, Madison, WI, USA). 10 ng cDNA was used for real-time quantitative PCR with the SYBR™ Green qPCR SuperMix-UDG (Invitrogen, Carlsbad, CA, USA). The primers were synthesized by Sangon Biotech (Shanghai, China) and listed in Supplementary Table 2.
Cell growth curve
Stable DHX9-overexpressed or -silenced CRC cells were seeded in 24-well plate at 1 × 104 per well and allowed growing for 24, 48, 72, and 96 h. Cell numbers in triple wells were determined with trypan blue staining.
Anchorage-independent growth assay
Soft agar assay was performed as previously described [36]. Briefly, 5000 CRC cells each well mixed with 0.5% agarose (Invitrogen, Guangzhou, China) medium containing 10% FBS were plated on a bottom layer of solidified 1% agar in 24-well plates. After theses plates were incubated in a humidified atmosphere containing 5% CO2 at 37 ℃ for 2 weeks, and the colonies consisting of ≥ 50 cells were examined by microscope counting.
Western blotting analysis
Total cellular proteins were extracted by RIPA buffer containing protease inhibitors (Beyotime, Shanghai, China). Nuclear and cytoplasmic proteins for detection of subcellular distribution of p65 were extracted with Nuclear and Cytoplasmic Protein Extraction Kit (Beyotime, Shanghai, China). The proteins were boiled with SDS loading buffer at 100 ℃ for 10 min and then separated by 10% SDS-PAGE. The protein bands were transferred to polyvinylidene difluoride (PVDF) membrane, blocked in 5% milk and probed with primary antibodies overnight at 4 ℃, followed by the corresponding HRP-conjugated secondary antibody against mouse (Cat# 62–6520) and against rabbit (Cat# PA1-28,761) from Invitrogen (Carlsbad, CA, USA). Antibodies against Bcl-XL (Cat# sc-634), p65 (Cat# sc-81622) and Survivin (Cat# sc-17779) were obtained from Santa Cruz Biotech (Santa Cruz, CA). Antibody against DHX9 (Cat# PA5-41,259) was purchased from Invitrogen (Carlsbad, CA, USA). Antibody against cyclin D1 (Cat# ab40754) was purchased from Abcam (Cambridge, MA, USA). Antibodies against phospho-IκBα (S32) (Cat# 2859S), phospho-p65 (S536) (Cat# 3033S), IκBα (Cat# 4814S), Snail (Cat# 3879S), Slug (Cat# 9585S), c-Myc (Cat# 18583S), OCT4 (Cat# 2750S), PTEN (Cat# 9188S) and β-catenin (Cat# 8480S) were from Cell Signaling Technology (Beverly, MA). Antibodies against Bcl-2 (Cat# 551,051) and XIAP (Cat# 610,782) were from BD Biosciences (San Jose, CA). Anti-RNA pol II (Cat# A300-653A) was obtained from Bethyl (Montgomery, TX). Antibody against β-actin (Cat# A1978) was from Sigma-Aldrich (Shanghai, China). Protein bands were visualized by ECL chemiluminescence (Millipore) and analyzed by Image Pro Plus software.
MTS assay
Cell viability was determined by MTS assays. Briefly, 5000 cells were seeded in 96-well plates. After incubation for 0, 24, 48, 72, and 96 h, cells were incubated with MTS (5 mg/ml, 20 μL, Promega) for 4 h. The absorbance was measured at 490 nm using a spectrophotometer (Thermo Fisher Scientific, USA).
Caspase activity assay
The activity of caspase-3, caspase-8, and caspase-9 were detected with caspase-3, caspase-8, and caspase-9 assay kit, respectively (Beyotime, Shanghai, China). According to the manufacturer’s instructions, cells were incubated with lysis on ice for 15 min and subjected to centrifugation. 10 μl substrate of caspase-3, caspase-8 or caspase-9 was added to the supernatant for incubation 1 h at 37 ℃, and then absorbance was measured at 405 nm.
Wound healing assay
The wound healing assay was performed according to the previous study [37]. In short, 3 × 106 CRC cells each well were seeded into six-well plates and grown to 100% confluence. 200 μL sterile pipette tip was employed to scratch straight lines and then cells were replaced with culture medium containing 1% FBS. Images at indicated time were photographed with a phase contrast microscope.
Transwell assays
The ability of CRC cell migration and invasion was detected using transwell chamber (BD Biosciences). For migration assay, 5 × 104 cells were suspended in 200 µL serum-free RPMI-1640 medium and seeded into the upper chamber. Then, 600 µL of medium containing 10% FBS was added in the lower chamber. After incubation for 48 h, cells on the upper chamber were removed, and the cells in the lower chamber were fixed and stained for 10 min in 0.1% Crystal Violet. For cell invasion assay, it was performed as migration assay with an exception. The chambers were uniformly pre-coated with 30% matrigel (BD Biosciences, USA) and incubated at 37 ℃ for 2 h. Migrated or invaded cells in three random fields were captured and counted under a light microscope at 10 × .
Apoptosis analysis by flow cytometry
Cells were collected by centrifugation and suspended to 1 × 106 cells/ml. Cells were double stained with 0.3% Annexin V (Keygen, China) for 30 min at room temperature in dark. PI was added just before subject to flow cytometry analysis (BD Biosciences, San Jose, CA, USA).
RNA sequencing (RNA-Seq)
Total RNA was extracted from vector-transfected or DHX9-silenced HCT116 cells utilizing RNeasy Mini kit (QIAGEN). The RNA library was constructed by TruSeq RNA sample prep kit (Illumina) following the previous study [38]. Gene set enrichment analysis (GSEA) was performed with standalone desktop program. Gene Ontology analysis (GO) was conducted using the default settings of the DAVID tool V6.8 (https://david.ncifcrf.gov).
Co-immunoprecipitation (Co-IP)
CRC cells were washed three times with cold PBS and then lysed on ice in IP lysis buffer [20 mM Tris–HCl (pH 8.0), 100 mM NaCl, 10 mM EDTA, 0.5 NP-40] containing protease inhibitor cocktail [39]. Cell lysate was centrifuged at 12,000 rpm for 10 min and the supernatants were collected to incubate with the antibodies against DHX9 (Cat# A300-855A, Invitrogen), IgG (Cat# 2729S, Cell Signaling Technology) or p65 (Cat# sc-81622, Santa Cruz Biotechnology) overnight at 4 ℃. Protein A/G agarose beads (Cat# sc-2003, Santa Cruz Biotechnology) were used to harvest the immune complexes, and then the proteins were washed with lysis buffer, boiled and subjected to Western blotting analysis.
Immunofluorescence staining
Stable vector-transfected, DHX9-overexpressed or -depleted HCT116 cells were seeded on the coverslips of six-well plates. After 24 h, cells were stimulated with LPS (1 μ1/ml, dissolved in PBS) or PBS for 15 min and then fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton X-100, and blocked using 5% bovine serum albumin. Cells were washed with PBS three times and exposed to anti-p65 (Cat# 6956S, Cell Signaling Technology) with the ratio of 1:100 at 4 °C overnight followed by the Alexa Fluor 488 conjugated secondary antibody (Cat# A0423, Beyotime) for 60 min at room temperature. DAPI was used to stain nucleus for 15 min and images were captured with confocal microscope (LSM 880, Carl-Zeiss).
Luciferase reporter
2 × 105 stable vector-transfected, DHX9-overexpressed or -depleted HCT116 cells per well in the 24-well plates were co-transfected with 10 ng Renilla luciferase reporter plasmid and 0.5 µg NF-κB-TATA-Luc reporter plasmid utilizing polyethylenimine. 36 h after transfection, cells were treated with LPS (200 ng/ml) for 8 h and examined by a Luciferase Assay System (Promega). Renilla expression plasmid was used as an internal control.
Chromatin immunoprecipitation (ChIP)
EZ-ChIP Kit (EMD Millipore) was used for ChIP assay and performed according to the manufacturer’s instructions. Brief, 1 × 107 CRC cells were crosslinked in 1% formaldehyde for 10 min at room temperature and quenched unreacted formaldehyde with glycine. The cell lysates were sonicated to shear DNA to sizes of 200–500 bp and centrifuged at 12,000 g at 4 °C for 10 min. To preclear the chromatin, 60 µL of protein G agarose was added to each tube before immunoprecipitated with 5 µ g anti-p65 (Cat# 6956S, Cell Signaling Technology), anti-DHX9 (Cat# A300-855A, Invitrogen), anti-RNA pol II (Cat# A300-653A) or normal rabbit IgG (Cat# 2729S, Cell Signaling Technology) overnight at 4 °C with rotation. The protein-DNA complexes were reverse crosslinks overnight. RNase A and Proteinase K were used to digest RNA and protein, respectively. The DNA fragments enriched by indicated antibodies were employed for qPCR reaction using specific primers listed in Supplementary Table 3.
Statistical analysis
Statistical analysis was performed by SPSS 23.0 (SPSS, Inc., Chicago, IL) and GraphPad Prism 8 (GraphPad, Inc., La Jolla, CA, USA). All data were represented by mean ± SEM. Statistical significance was assessed using Student’s t test for independent groups or one-way analysis of variance for multiple groups. P < 0.05 was considered as statistically significant.
Results
siRNA screening identifies DHX9 with candidate oncogenic properties in CRC
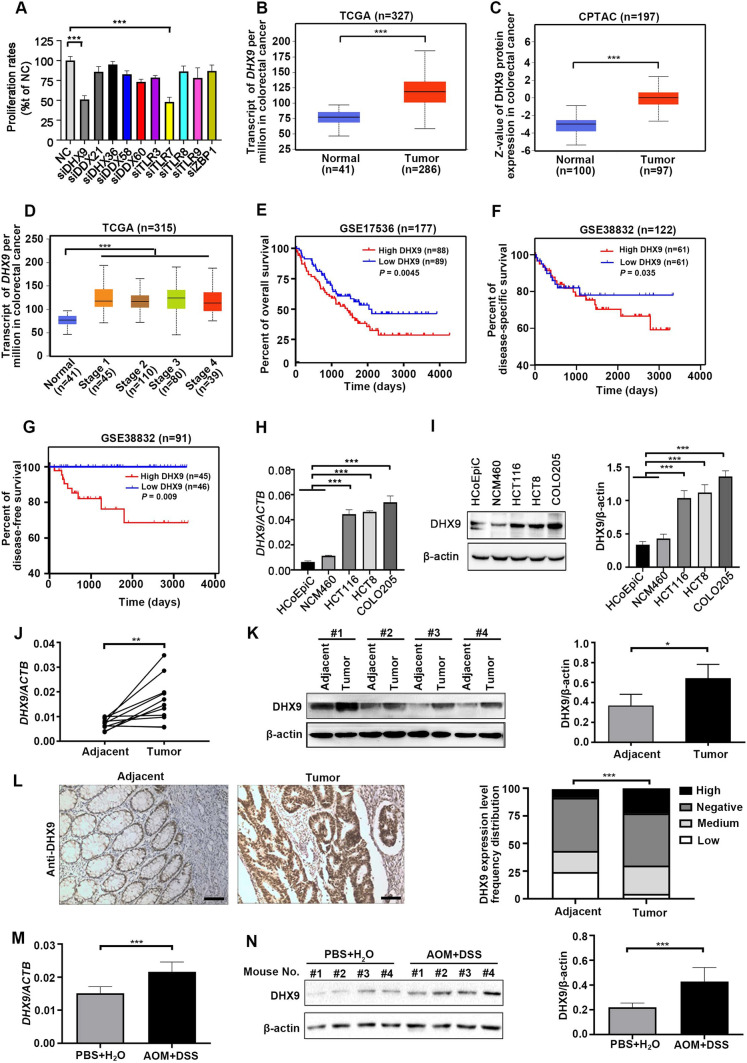
Inflammation is frequently initiated by activation of nucleic acid sensing PRRs which could lead to CRC development [40]. In our previous study, we identified a list of nucleic acid sensing PRRs which were differentially expressed in CRC animal models or patients [4]. To further study their functions, we investigated whether targeting these receptors could suppress tumorigenesis. The overview of high-throughput siRNA screening was displayed in Supplementary Fig. 1A. siRNAs that restricted the EdU proliferation rates of CRC cells by ≥ 50% compared with the ratio of cells infected with non-targeting siRNA (NC) were regarded as candidate targets for CRC inhibition. siRNAs against DHX9 and TLR7 were identified as possible targets for CRC inhibition (Fig. 1A and Supplementary Fig. 1B).
Fig. 1.
siRNA screening identifies DHX9 as an oncogenic molecule in colorectal cancer. A Forty-eight hours after HCT116 cells were transfected with a negative control (NC) siRNA or siRNA against nucleic acid sensing pattern recognition receptors (PRRs), EdU proliferation assay was conducted with high-throughput siRNA screening. ***P < 0.001; Student’s t test. (B-C) The mRNA (B) and protein (C) levels of DHX9 in colorectal cancer (CRC) tissues and normal adjacent tissues in TCGA and CPTAC database, respectively (obtained through UALCAN; http://ualcan.path.uab.edu/index.html). ***P < 0.001; Student’s t test. (D) DHX9 mRNA expression in different CRC stages and normal tissues from the TCGA Dataset. ***P < 0.001; Student’s t test. E Relationship of DHX9 expression with overall survival in patients with CRC in the cohort of GSE17536 database, log-rank test. F The disease-specific survival of patients with low and high DHX9 expression obtained from GSE38832 database, log-rank test. G Association of DHX9 expression with disease free survival of CRC patients in GSE38832 dataset, log-rank test. H The mRNA levels of DHX9 in human normal colonic epithelial cells (HCoEpiC and NCM460) and CRC cells (HCT116, HCT8, and COLO205) were detected by RT-qPCR analysis. ***P < 0.001; one-way ANOVA, post hoc comparisons, Tukey’s test. (I) The protein levels of DHX9 in normal colonic epithelial cells and CRC cells were examined by Western blotting analysis and quantitative data of DHX9 protein levels were shown. ***P < 0.001; one-way ANOVA, post hoc comparisons, Tukey’s test. J The mRNA levels of DHX9 in paired human clinical CRC tissues and corresponding adjacent colorectal tissues (n = 11). **P < 0.01; Student’s t test. K Left, the representative ECL images of DHX9 detected by Western blotting analysis in paired human clinical CRC tissues and corresponding adjacent colorectal tissues. Right, the summary data correspond to Left (n = 3). *P < 0.05; Student’s t test. L Left, representative IHC micrographs of DHX9 staining in paraffin embedded clinical CRC tissues and adjacent normal tissues. Scale bar, 100 μm. Right, rank sum test analysis of IHC results of DHX9 in CRC tissue and adjacent normal tissues (n = 220). ***P < 0.001. M The difference of DHX9 mRNA levels in colorectal tissues of AOM/DSS-treated mice and PBS-treated mice (n = 7). ***P < 0.001; Student’s t test. N Left, the representative ECL images of DHX9 in AOM/DSS-treated mice and PBS-treated mice. Right, the summary data in AOM/DSS-treated group and PBS control group correspond to Left (n = 3). ***P < 0.001; Student’s t test. Data were represented as mean ± SEM from three independent replicate experiments in A, H, I, J, K, M and N
To further investigate the potential oncogenic roles of DHX9 and TLR7 in CRC, we next explored the expression levels of these genes in databases. The results revealed that DHX9 expression instead of TLR7 were tremendously elevated in CRC tissues relative to normal colorectal tissues (Fig. 1B–C and Supplementary Fig. 2A). Moreover, the mRNA levels of DHX9 were upregulated in different stages of CRC compared with normal tissues, which was not observed in TLR7 (Fig. 1D and Supplementary Fig. 2B). In addition, we found that high expression of DHX9 instead of TLR7 was corresponded with a low overall survival rate in the GSE17536 database (Fig. 1E and Supplementary Fig. 2C). Furthermore, elevated DHX9 expression level, but not TLR7, was also strongly correlated with shorter disease-specific survival and disease-free survival (Fig. 1F–G and Supplementary Fig. 2D–E).
We next evaluated the expression of DHX9 in normal colonic epithelial cells and CRC cells. The mRNA levels of DHX9 were remarkedly elevated in CRC cells (HCT116, HCT8, and COLO205) compared with these in human normal colonic epithelial cells (HCoEpiC and NCM460) (Fig. 1H). Consistently, the protein levels of DHX9 were significantly higher in CRC cells (Fig. 1I). The significantly elevated mRNA and protein levels of DHX9 were also found in the clinical CRC samples relative to corresponding adjacent normal colorectal tissues (Fig. 1J–K). The in-situ expression levels of DHX9 in the primary CRC tissues were detected utilizing IHC staining. We found that the expression of DHX9 in the CRC specimens were significantly increased comparing with that in adjacent normal tissue (Fig. 1L and Supplementary Table 4). Notably, the protein expression of DHX9 was positively correlated with histopathological differentiation, infiltration depth of intestinal wall and lymph node metastasis in CRC patients (Supplementary Table 5). We also detected the expression of DHX9 in AOM/DSS-induced CAC model. The result revealed that the DHX9 expression was significantly higher in the colons of AOM/DSS-treated mice than these in PBS-treated group (Fig. 1M–N). Collectively, these results suggest that DHX9 may function as an oncogene to promote the development of CRC.
DHX9 promotes the proliferation of CRC cells
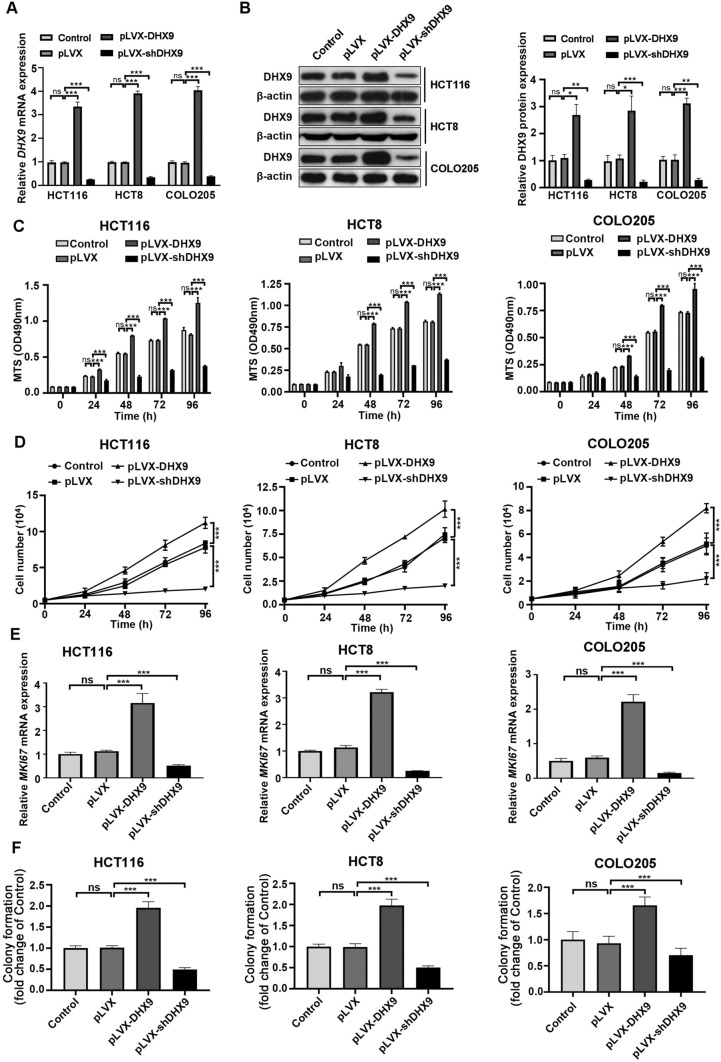
To clarify the effect of DHX9 on the biological behaviors of CRC cells, we established stable DHX9-overexpressed or -silenced CRC cells. The transfection efficiency of three cell lines were detected by RT-qPCR and Western blotting assay, and the results revealed that cells were successfully constructed (Fig. 2A–B). The proliferation rates of DHX9-overexpressed or -silenced CRC cells were determined by MTS assay and trypan blue exclusion assay. We found that there was no significant difference between the blank control CRC cells and the negative group cells with expressing lentiviral vector. DHX9 overexpression markedly aggravated proliferation compared with vector-transfected group cells, while DHX9 knockdown greatly decreased proliferation of CRC cells (Fig. 2C–D), which was confirmed by the mRNA levels of MKi67 encoding proliferation marker Ki67 (Fig. 2E). Furthermore, ectopic expression of DHX9 promoted cell colony formation. In contrast, DHX9 knockdown attenuated colony formation capacity in CRC cells (Fig. 2F and Supplementary Fig. 3A). Therefore, DHX9 is necessary for malignant proliferation of CRC cells.
Fig. 2.
DHX9 promotes the proliferation of colorectal cancer cells in vitro. A CRC cells with untreated (Control), lentiviral vector-transfected (pLVX), stably expressing DHX9-encoding constructs (pLVX-DHX9) or shRNAs against DHX9 (pLVX-shDHX9) were subjected to DHX9 expression analysis by RT-qPCR assay. ns, not significant; ***P < 0.001. B Western blotting assay to analyze DHX9 transfection efficiency in CRC cells. Left, the representative ECL images detected by Western blotting analysis. Right, the summary data in CRC cells correspond to Left (n = 3). ns, not significant; *P < 0.05; **, P < 0.01; ***, P < 0.001. C OD490nm values of CRC cells with untreated, vector-transfected, stably expressing DHX9-encoding constructs or shRNAs against DHX9 for indicated time. ns, not significant; ***P < 0.001. D DHX9-overexpressed and -silenced CRC cells were subjected to trypan blue exclusion assay. ***P < 0.001. E The mRNA levels of MKI67 encoding Ki67 in CRC cells with untreated, vector-transfected, stably expressing DHX9-encoding constructs or shRNAs against DHX9 were detected by RT-qPCR analysis. F Anchorage-independent colony growth of CRC cells with untreated, vector-transfected, stably expressing DHX9-encoding constructs or shRNAs against DHX9 were determined. ns, not significant; ***P < 0.001. All data in bar graphs were shown as mean ± SEM from three independent experiments and assessed by one-way ANOVA, post hoc intergroup comparisons, Tukey’s test
DHX9 contributes to apoptosis resistance in CRC cells
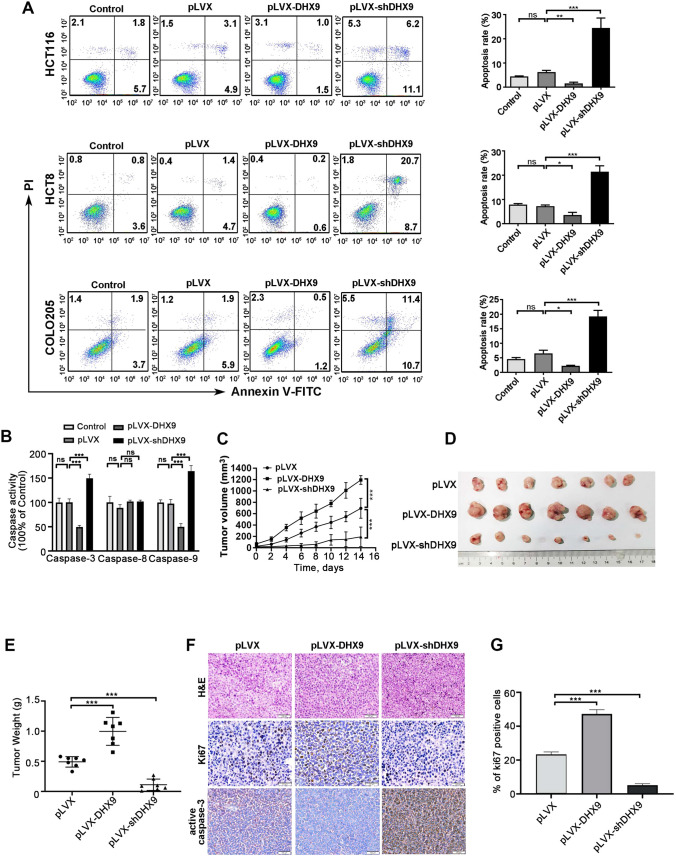
As malignant and immortality potential of tumor cells are associated with apoptosis resistance [41, 42], we investigated whether DHX9 effected on this aspect in DHX9-overexpressed or -silenced CRC cells. Annexin V-FITC/PI dual staining assay results demonstrated that overexpression of DHX9 exhibited lower apoptotic index compared with vector-transfected HCT116 cells, while DHX9-silenced cells revealed higher apoptosis rate. Consistently, similar results were found in HCT8 and COLO205 cells (Fig. 3A).
Fig. 3.
DHX9 depletion induces apoptosis and inhibits outgrowth of xenografted colorectal cancer cells in nude mice. A Annexin V/PI apoptotic assay was performed in CRC cells with untreated, vector-transfected, stably expressing DHX9-encoding constructs or shRNAs against DHX9. Representative flow cytometry dot plots for HCT116, HCT8, and COLO205 cells and corresponding quantitative analysis from three independent experiments were shown. ns, not significant; *P < 0. 05; *P < 0. 01; ***P < 0.001. B The activity of Caspase-3/8/9 were measured in DHX9-overexpressed or -depleted CRC cells and quantitative data were obtained from three independent experiments. ns, not significant;*** P < 0.001. C Xenograft model of tumor growth was used to evaluate the tumorigenesis ability of stable DHX9-overexpressed or -depleted HCT116 cells (n = 6 each group). Tumor growth curve over time was plotted in the nude mice. ***P < 0.001. D Representative images of tumors dissected from mice were presented (n = 7 each group). (E) Weights of tumors in each group were shown (n = 7). F H&E staining and IHC analysis of Ki67 and active caspase-3 in tumor sections from each group were shown. Scale bar, 100 μm. G Quantitative analysis of Ki67 positive cells in tumor sections from each group. n = 3 mice per condition. *P < 0.05; ***P < 0.001. All data in bar graphs were shown as mean ± SEM and analyzed by one-way ANOVA, post hoc intergroup comparisons, Tukey’s test
Moreover, the effects of manipulating DHX9 on the activities of caspase-3, caspase-8, and caspase-9 in HCT116 cells were examined. We found that DHX9 overexpression significantly decreased activities of caspase-3 and caspase-9. However, DHX9 knockdown notably enforced activities of caspase-3 and caspase-9. The caspase-8 activity was not altered in DHX9-overexpressed and -silenced HCT116 cells (Fig. 3B).
DHX9 facilitates outgrowth of the xenografted CRC cells in vivo
To further evaluate the effect of DHX9 on CRC maintenance, a xenograft model of tumor outgrowth was established in nude mice bearing HCT116 cells. We found inoculation of DHX9-overexpressed HCT116 cells appreciably enhanced tumor volume (Fig. 3C), size (Fig. 3D), and weight (Fig. 3E) compared with the injection of vector-transfected HCT116 cells, while knockdown of DHX9 remarkably restrained tumor growth (Fig. 3C–E). IHC analysis in the tumor sections of the three groups further confirmed that DHX9 overexpression dramatically augmented proliferation (marked as Ki67 staining) and abrogated apoptosis (marked as active caspase-3 staining), whereas DHX9 knockdown effectively restrained CRC outgrowth and induced apoptosis (Fig. 3F–G).
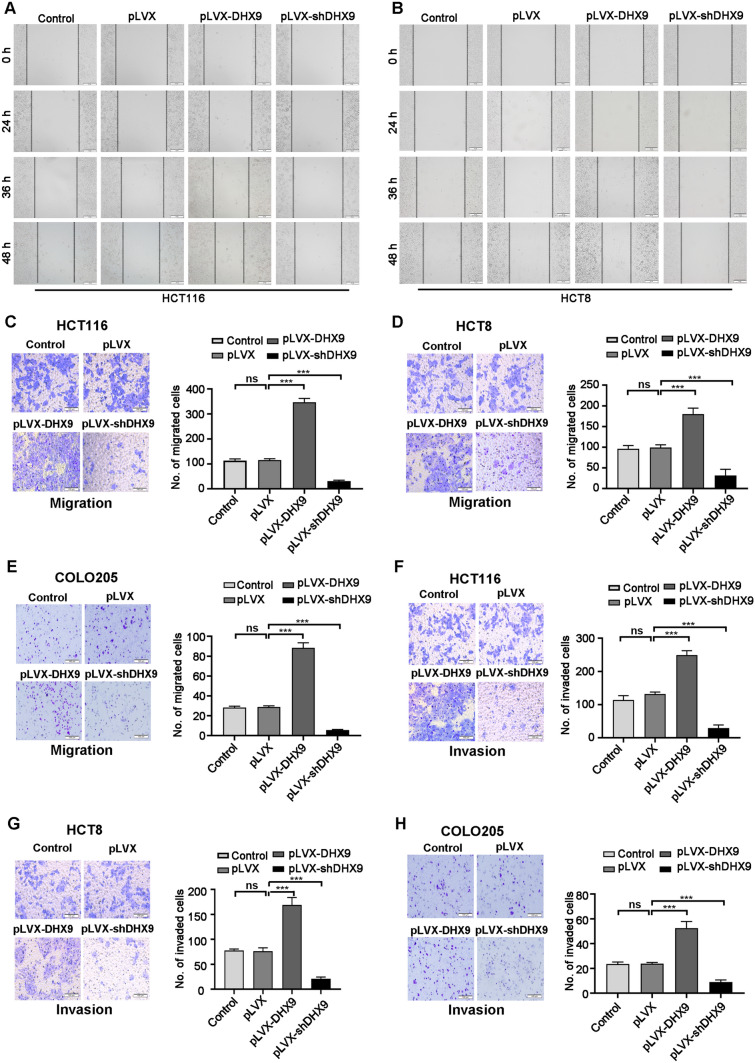
DHX9 augments the capability of migration and invasion in CRC cells
Dissemination of tumor cells is an important malignant feature of CRC, which leads to limited therapeutic options [43]. We therefore examined the motility of CRC cells utilizing the wound healing assay. DHX9 knockdown repressed the motility of CRC cells, while DHX9 overexpression in CRC cells promoted the cell motility (Fig. 4A–B). Next, we detected the effect of DHX9 on migration and invasion of CRC cells. The results showed that compared with the vector-transfected cells, the ability of migration and invasion were significantly elevated in DHX9-overexpressed CRC cells, which was significantly attenuated in DHX9-silenced cells (Fig. 4C–E). Similar results were observed in invasion assay of CRC cells (Fig. 4F–H).
Fig. 4.
DHX9 increases the capability of migration and invasion in colorectal cancer cells. A-B Representative photographs of the wound healing scratch assay from stable DHX9-overexpression or -silenced HCT116 (A) and HCT8 (B) cells (The culture properties of COLO205 cells are mixed, adherent and suspension, which were difficult for wound healing scratch assay). Scale bar, 100 μm. C-E CRC cells with untreated, vector-transfected, stably expressing DHX9-encoding constructs or shRNAs against DHX9 were subjected to transwell chamber assays. Representative images for HCT116, HCT8, and COLO205 cells and quantitative analysis from three random microscopic fields were shown in C, D, E, respectively. Scale bar, 100 μm. ns, not significant; ***P < 0.001. F–H Control, vector-transfected, stably expressing DHX9-encoding constructs or shRNAs against DHX9 CRC cells were underwent matrigel invasion chamber assays. Representative images for HCT116, HCT8, and COLO205 cells and quantitative analysis from three random microscopic fields were shown in F, G, H, respectively. Scale bar, 100 μm. ns, not significant; ***P < 0.001. Data in all bar graphs were represented as mean ± SEM and were assessed by one-way ANOVA, post hoc intergroup comparisons, Tukey’s test
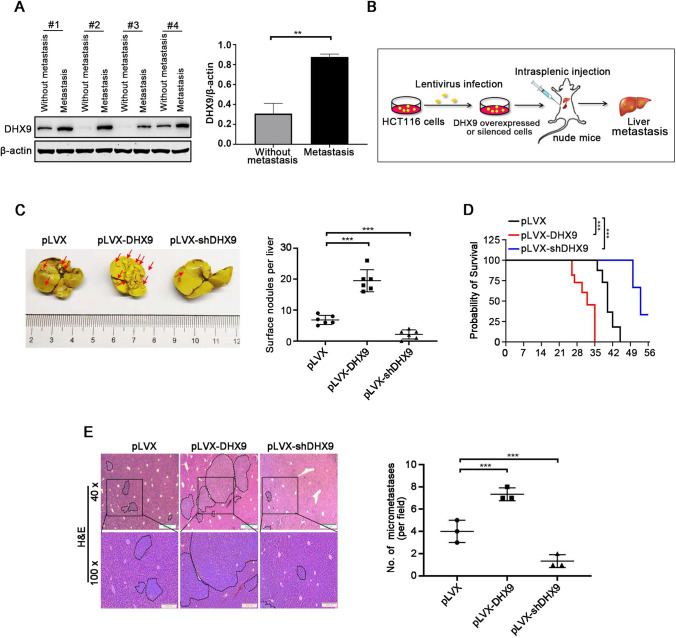
DHX9 increases the liver metastasis of CRC
Approximately fifty percent of CRC patients develop liver metastases, which is the main cause of death [44]. To explore the pro-metastatic role of DHX9 in CRC, we measured primary CRC tumors from patients with or without liver metastasis. Compared with non-metastatic tumor, DHX9 expression was significantly increased in tumor tissues of CRC patients with liver metastases (Fig. 5A). We further evaluated the effect of DHX9 on hepatic metastasis of CRC cells in vivo. As illustrated in Fig. 5B, a hepatic metastasis model utilizing the nude mice intrasplenically injected with DHX9-overexpressed or -silenced CRC cells was employed. As expected, overexpression of DHX9 strongly boosted surface liver metastatic nodules and significantly shorten the overall survival time of nude mice bearing CRC cells (Fig. 5C), whereas DHX9 silence exhibited the contrast effect (Fig. 5D). Consistently, H&E staining analysis showed that forced expression of DHX9 resulted in a robustly increase in number and size of metastatic nodules on the livers of nude mice, while DHX9 knockdown exhibited significantly compromised size and number of metastatic foci on the livers (Fig. 5E). These results suggest that DHX9 promotes liver metastasis of CRC cells in vivo.
Fig. 5.
DHX9 facilitates liver metastasis of colorectal cancer cells. A Western blotting analysis of DHX9 in primary CRC tumors from non-metastatic and liver metastatic patients and quantitative data of DHX9 protein levels were shown Data were represented as mean ± SEM (n = 3). **P < 0.01; Student’s t test. B Graphic illustration for liver metastasis mouse model of CRC. C Representative images of metastatic liver in vector (pLVX), pLVX-DHX9 and pLVX-shDHX9 group were shown (left) and surface nodules were counted (right). Arrows indicated metastatic foci on liver surface. n = 6 mice per condition. Data were represented as mean ± SEM. ***P < 0.001, one-way ANOVA with post hoc intergroup comparison by Tukey's test. D Survival curves analyzed with Kaplan–Meier in the indicated groups of mice were shown (n = 6). Log-rank test, ***P < 0.001. E H&E staining of liver sections, dot plots indicated metastatic nodules. Left, representative images of metastatic nodules in H&E-stained liver section were shown. Scale bar, 500 µm (40 ×), 200 µm (100 ×). Right, quantification of liver metastatic nodules in microscopic fields of 40 × was shown. n = 3 mice per condition. Data were represented as mean ± SEM. ***P < 0.001, one-way ANOVA with post hoc intergroup comparison by Tukey's test
DHX9 regulates the activation of NF-κB signal pathway in CRC
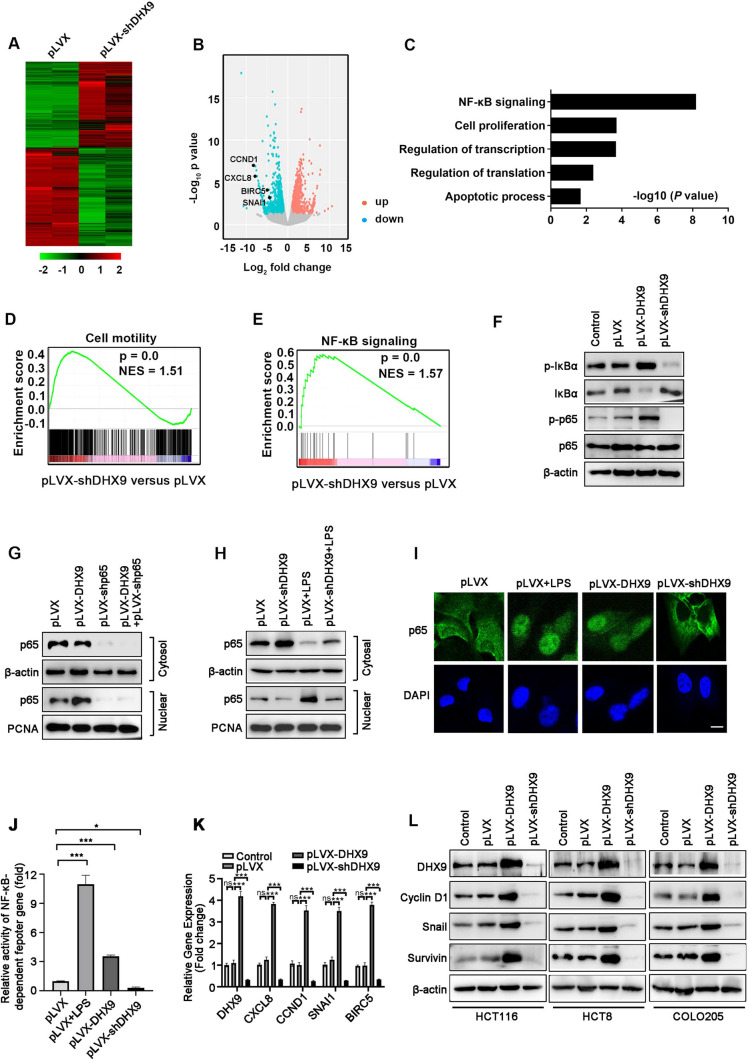
To gain insight into how DHX9 regulates tumorigenesis in CRC, the global transcriptomic change was investigated in DHX9-silenced HCT116 cells by RNA-seq. A total of 3936 genes displayed altered expression in DHX9-silenced HCT116 cells compared with vector-transfected cells (Fig. 6A–B and Supplementary Table 6). In brief, it was found that a total of 2113 genes were up-regulated (≥ twofold). Meanwhile, 1838 genes were down-regulated (≤ twofold). Among them, genes in NF-κB signaling (such as CXCL8, encoding IL-8; CCND1, encoding Cyclin D1; BIRC5, encoding Survivin; SNAI1, encoding Snail) were markedly downregulated by DHX9 ablation (Fig. 6B). GO analysis was performed to dissect the function of down-regulated genes in DHX9-silenced CRC cells, and the results revealed that these genes in NF-κB signaling, cell proliferation, transcription and translation regulation, and apoptosis were notably enriched, highlighting the crucial role of DHX9 in CRC biology (Fig. 6C). In addition, GSEA results showed that the decreased genes in DHX9 knockdown cells were significantly enriched in cell motility (Fig. 6D), which was consistent with our findings that DHX9 ablation resulted in the reduced capacity of migration and invasion in CRC cells in vivo and in vitro. Furthermore, GSEA results demonstrated that NF-κB signaling was markedly enriched in DHX9-silenced cells (Fig. 6E).
Fig. 6.
DHX9 mediates the activation of NF-κB signal pathway in colorectal cancer cells. A Heatmap showed the alteration of gene expression (≥ twofold) obtained from RNA-Seq of vector-transfected and DHX9-silenced HCT116 cells. B Scatter plot displayed fold changes of gene expression (≥ twofold) in DHX9-silenced HCT116 cells compared with vector-transfected cells. C GO analysis of down-regulated genes (≥ twofold) in DHX9-silenced HCT116 cells compared with vector-transfected cells. D-E GSEA revealed that gene sets of cell motility D and NF-κB signaling E were enriched in DHX9-depleted HCT116 cells. F Proteins levels of IKBα, p65 and phosphorylated IKBα and p65 were evaluated by Western blotting in DHX9-overexpressed or -depleted HCT116 cells. G-H Cytosolic and nuclear fractionations were evaluated by Western blotting in DHX9-overexpressed HCT116 cells with or without p65 knockdown G or in DHX9-silenced HCT116 cells with or without LPS (1 μg/ml) pretreatment for 15 min H. I DHX9-overexpressed or -depleted HCT116 cells were subjected to immunofluorescence analysis of p65 localization (green). Vector-transfected (pLVX) HCT116 cells pretreated with LPS (1 μg/ml) for 15 min were served as a positive control. DAPI (blue) was applied to stain the nuclear. Scale bar, 20 μm. J HCT116 cells with stably expressing pLVX, pLVX-DHX9 or shRNAs against DHX9 were co-transfected with 0.5 μg NF-κB-TATA-Luc reporter plasmid and 10 ng Renilla luciferase reporter. 36 h after transfection, pLVX-HCT116 cells were treated with LPS (200 ng/mL) for 8 h. The luciferase activity of cells was measured. The values of firefly luciferase activity were normalized to Renilla luciferase activity. Fold activation were represented as mean ± SEM of three independent transfections. ***P < 0.001, one-way ANOVA with post hoc intergroup comparison by Tukey's test. K RT-qPCR experiment analyzed NF-κB-target genes including CXCL8, encoding IL-8; CCND1, encoding Cyclin D1; BIRC5, encoding Survivin; SNAI1, encoding Snail, in DHX9-overexpressed or -depleted HCT116 cells. Data were from three independent experiments and represented as mean ± SEM. ns, not significant; ***P < 0.001, one-way ANOVA with post hoc intergroup comparison by Tukey's test. L Western blotting analysis verified the protein levels of candidate NF-κB-target genes in DHX9-overexpressed or -depleted cells
To further validate the effect DHX9 on NF-κB signaling, we examined p65 activation and translocation in DHX9-overexpressed and -silenced CRC cells. Western blotting results showed that phosphorylated IκBα and phosphorylated p65 was increased in DHX9-overexpressed HCT116 cells, while decreased in DHX9-silenced cells (Fig. 6F). Concomitantly, the levels of total IκBα were decreased in DHX9-overexpressed cells, whereas increased in DHX9-silenced cells (Fig. 6F). We next examine cytosolic and nuclear p65 distribution in DHX9-overexpressed and -silenced CRC cells. Result showed that overexpression of DHX9 significantly enhanced p65 nuclear translocation, but did not trigger nuclear translocation of p65 in p65-konckdown CRC cells, which serves as a negative control (Fig. 6G). Conversely, knockdown DHX9 notably decreased nuclear protein p65 level, which was rescued by LPS treatment in CRC cells (Fig. 6H). The effect of DHX9 on nuclear translocation of p65 in CRC cells was verified by immunofluorescence assay, in which LPS was used as a positive control (Fig. 6I). Moreover, in DHX9-overexpressed and -silenced HCT116 cells, we transiently co-transfected with NF-κB-TATA-Luc and Renilla luciferase reporter plasmids, and found that DHX9 overexpression significantly enhanced NF-κB reporter activity, which was reversed in DHX9 knockdown cells (Fig. 6J), suggesting that DHX9 can regulate transcription of the NF-κB-dependent genes.
In addition, we measured the expression of NF-κB-target genes in DHX9-overexpressed or -silenced CRC cells. The results revealed that ectopically expression of DHX9 profoundly elevated expression of CXCL8 and Cyclin D1, known proteins that participate in proliferation of cancer [26, 45], while ablation of DHX9 potently disrupted expression of CXCL8 and Cyclin D1 (Fig. 6K-L and Supplementary Fig. 4A-B). Moreover, we evaluated the expression of apoptosis-related proteins after DHX9 overexpression and silence. The results showed that the expression of pro-survival Survivin was obviously upregulated in DHX9-overexpressed cells and decreased in DHX9-silenced CRC cells (Fig. 6K–L and Supplementary Fig. 4A–B). The levels of other apoptosis-related family members (Bcl-XL, Bcl-2, XIAP) did not considerably change (Supplementary Fig. 5A). The results revealed that DHX9 might exert its anti-apoptosis role via Survivin. Furthermore, we detected the expression of metastasis-associated proteins. We found an appreciable increase of Snail in DHX9-overexpressed cells and a significant decrease in DHX9-overexpressed cells without obvious alteration in SLUG, β-catenin, c-Myc, OCT4, and PTEN (Fig. 6K–L and Supplementary Fig. 4–6). The data suggest that DHX9 may promote metastasis via Snail. Consistent with DHX9, the expression of CXCL8, CCND1, BIRC5 and SNAI1 were significantly upregulated in the CRC tumor tissues compared with normal tissues and were associated poor prognosis in CRC patients in the TCGA database (Supplementary Fig. 7A–H). DHX9 mRNA levels were found to be significantly positively correlated with mRNA levels of CXCL8, CCND1, BIRC5 and SNAI1 in the CRC biopsies (Supplementary Fig. 7I–L). These results indicate that DHX9 facilitates proliferation, apoptosis resistance, migration, invasion, and metastasis possibly through modulating the expression of NF-κB-target genes.
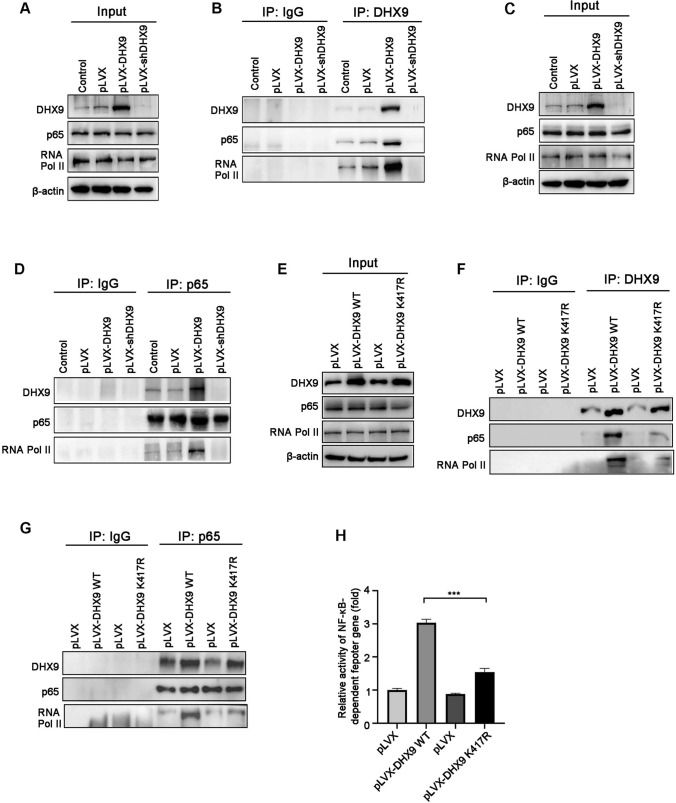
DHX9 interacts with p65 and RNA Pol II to enhance NF-κB-mediated transcription
It has been reported that DHX9 can interact with p65 to enhance transcription of NF-κB-dependent genes in 293T cells [29], we therefore interrogated whether this interaction existed in CRC. Our co-immunoprecipitation (co-IP) results showed that endogenous DHX9 weakly interacted with p65 and RNA Pol II in the immunoprecipitates, whereas these interaction were intensified once forced DHX9 expression. In contrast, these interaction were abrogated in DHX9-silenced CRC cells (Fig. 7A–B). Furthermore, reciprocal co-IP was performed with an antibody against endogenous p65 in DHX9-overexpressed or -silenced cells. We found that endogenous DHX9 and RNA Pol II were readily detectable, while p65 failed to interact with RNA Pol II in DHX9 knockdown cells. Conversely, this interaction was strengthened in DHX9-overxepressed cells (Fig. 7C–D).
Fig. 7.
DHX9 interacts with p65 and RNA Pol II to active NF-κB-mediated transcription in colorectal cancer cells. A-B Lysates of DHX9-overexpressed or -depleted HCT116 cells were subjected to immunoprecipitation with the anti-DHX9 or anti-IgG antibody, and then incubated with the indicated antibodies using Western blotting. C–D Lysates of DHX9-overexpressed or -depleted HCT116 cells were subjected to immunoprecipitation with the anti-p65 or anti-IgG antibody and subjected to Western blotting. E–G Stable HCT116 cells expressing WT DHX9 and K417R DHX9 were harvested and lysates were used to immunoprecipitation with the anti-DHX9 or anti-p65 or anti-IgG antibody, and then subjected to Western blotting. K417R indicates a negative mutant of DHX9, in which Lys 417 is substituted to Arg that abolishes the ATP-dependent helicase activity. H Stable HCT116 cells expressing WT DHX9 or K417R DHX9 were co-transfected with NF-κB-TATA-Luc reporter plasmid and Renilla luciferase reporter. The luciferase activity of cells was measured after 48-h transfection. The values of firefly luciferase activity were normalized to Renilla luciferase activity. Fold activation were represented as mean ± SEM of three independent transfections. ***P < 0.001, one-way ANOVA with post hoc intergroup comparison by Tukey's test
To further determine which domains or amino acids of DHX9 were responsible for interaction with p65 and RNA Pol II, we used stable CRC cell lines expressing WT DHX9 or negative mutant DHX9, which Lys417 of the conserved ATP-binding motif (Gly-Lys-Thr) in catalytic domain was substituted by Arg and resulted in a significant reduction in transcriptional activity [46]. Co-IP results showed that compared with WT DHX9, mutant DHX9 poorly interacted with p65 and RNA Pol II in the immunoprecipitates of HCT116 cells (Fig. 7E–F). Moreover, reciprocal co-IP results indicated that p65 weakly interacted with DHX9 and RNA Pol II in the ATPase/helicase mutant of K417R DHX9-expresssed cells (Fig. 7G). In addition, compared with WT DHX9, mutant DHX9 weakly enhanced the activation of NF-κB reporter activity (Fig. 7H). These results suggest that the ATPase/helicase domain of DHX9 is responsible for the interaction with p65 and RNA Pol II and necessary for activation of NF-κB-dependent transcription.
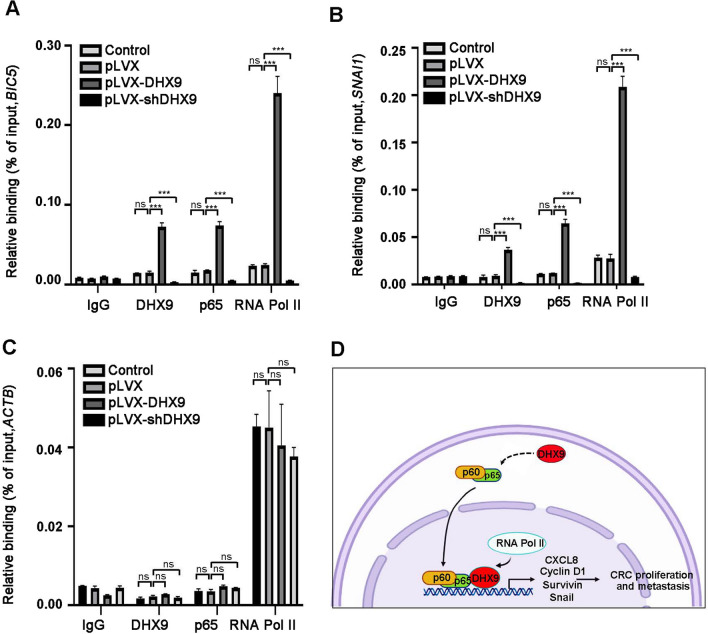
We next investigated how DHX9 affect the transcriptional activity of NF-κB signal pathway in details using ChIP assay. The quantitative ChIP data revealed that DHX9 itself was recruited to the promoters of NF-κB-dependent genes (such as BICR5 and SNAI1), and forced expression of DHX9 enhanced p65’s and RNA Pol II’s occupancy of BICR5 and SNAI1 promoters in CRC cells, while DHX9 ablation diminished p65’s and RNA Pol II’s occupancy of these promoters (Fig. 8A–B). In the same experiment, low levels of DHX9 and p65, while significantly amounts of RNA Pol II, were detected on the ACTB promoter, which served as a negative control, in either DHX9-overexpressed or -silenced CRC cells (Fig. 8C). These ChIP results were consistent with the expression levels of BIRC5 and SNAI1 in DHX9-overexpressed or -knockdown CRC cells and suggest that DHX9 is required for the recruitment of p65 and RNA Pol II to NF-κB-dependent promoters.
Fig. 8.
DHX9 is required for p65 and RNA Pol II recruitment to the promoters of NF-κB-dependent genes. A–C ChIP-qPCR analysis for DHX9, p65 or RNA Pol II occupancy at the promoter of BIRC5 A, SNAI1 B or ACTB C in HCT116 cells stably expressed DHX9 cDNA or shDDR1 constructs. All data in bar graphs were shown as mean ± SEM from three independent experiments and analyzed by one-way ANOVA, post hoc intergroup comparisons, Tukey’s test. D A proposed working model of DHX9 in orchestrating the malignant phenotypes of CRC. On one hand, DHX9 enhances p65 phosphorylation, promotes p65 nuclear translocation to facilitate NF-κB-mediated transcriptional activity. On the other hand, DHX9 interacts with p65 and RNA Pol II and is necessary to recruit p65 and RNA Pol II to the NF-κB-dependent promoters to activate downstream gene transcription, including CXCL8, CCND1, BIRC5 and SNAI1. Enhanced CXCL8, Cyclin D1 and Survivin expression contributes to lower apoptosis and promote proliferation in CRC cells; the strengthened expression of Snail increases the capability of migration and invasion, and ultimately facilities liver colonization and metastasis in CRC
Discussion
Despite a great progress has been made in the development of chemotherapy for CRC, increasing cases show tolerance to such treatments [23]. Hence, there is an urgent need to find more safe and effective molecular targets for the treatment of CRC. In this study, high-throughput siRNA screening was used to identify nucleic acid sensing PRRs that were involved in tumorigenesis of CRC, and DHX9 was found to be a potential oncogene. Moreover, it was shown that DHX9 was predominantly elevated in CRC cells lines and clinical CRC tissues. Additionally, DHX9 also was negatively correlated with poor prognosis in CRC. From a functional aspect, forced overexpression of DHX9 aggravated cell proliferation, anchorage-independent growth, anti-apoptosis, migration and invasion in CRC. On the contrary, DHX9 ablation inhibited these malignant phenotypes. The xenograft mouse model and liver metastasis mouse model validated that ectopic expression of DHX9 promoted CRC growth and metastasis, while DHX9 knockdown exerted converse effect. From the molecular mechanistic perspective, DHX9 enhanced p65 phosphorylation, promoted p65 nuclear translocation, and interacted with p65 and RNA Pol II to stimulate NF-κB-mediated transcription of target genes (such as CXCL8, CCND1, BIRC5 and SNAI1).
DHX9 is an NTP-dependent helicase protein that functions in a variety of cellular processes, indicating that aberrant DHX9 expression may participate in multiple human diseases [12, 14]. Previous studies reported that DHX9 was upregulated in various cancer such as lung cancer [47] and liver cancer [48]. Consistently, we found that DHX9 was highly expressed in CRC cells lines, CAC mouse model and clinical CRC samples at the mRNA and protein level, which was validated in TCGA and CPTAC database. However, the mechanism of DHX9 overexpression needs further investigation. Additionally, it has reported that DHX9 overexpression was associated with poor patient survival in lung cancer [49]. Analogously, we found that elevated DHX9 expression was correlated with aggressive CRC stage, lymph node metastasis and poor prognosis, which was evidenced in clinic CRC samples, GEO, and TCGA database. These findings suggest that DHX9 might perform as an oncogenic factor in CRC.
Sustaining proliferation, apoptosis resistance and enhanced motility are hallmarks of malignant cancer [50]. Our results demonstrated that DHX9 promoted proliferation of CRC cells in vitro and in nude mice. In accordance with our findings, the effect of DHX9 on proliferation has been found in other types of cancer such as myeloma and osteosarcoma [11]. In addition, we observed that DHX9 knockdown induced apoptosis in CRC cells, while ectopic expression of DHX9 enhanced survival. Similarly, loss of DHX9 activating p53-dependent apoptotic program was found in Eμ-myc/Bcl-2 lymphomas [51]. Furthermore, through gain- and loss-of-function experiments, we demonstrated that DHX9 potentiated the capacity of migration and invasion of CRC cells in vitro and in vivo, which identified that DHX9 facilitated hepatic metastasis in CRC. Above all, these results suggest that DHX9 plays a critical role in promoting the growth and metastasis of CRC. However, there are also several studies indicate that DHX9 has tumor-suppressive properties through interacting with the tumor suppressor BRCA1 or upregulating of p53 translation in breast cancer [20] and lung adenocarcinoma [13], respectively. Therefore, whether DHX9 functions as an oncogene or tumor suppressor gene may depend on the tumor type or activity of its interacting partners.
NF-κB activation is constantly observed in CRC and correlated with cellular processes including proliferation, apoptosis, angiogenesis, and metastasis via promoting the expression of related target genes [24]. A previous study demonstrated that DHX9 was required for NF-κB-mediated transcriptional activation of antiviral cytokines [29]. Consistently, it was observed that NF-κB-mediated cytokine, CXCL8, was suppressed in DHX9-silenced CRC cells, and other target genes (e.g., CCND1, BIRC5) involved in proliferation and apoptosis were also inhibited (Fig. 6K–L). Moreover, Snail, as a known downstream target of the NF‐κB signal pathway to promote tumor metastasis [28], was decreased in DHX9-silenced CRC cells (Fig. 6K–L). Interestingly, other target genes of NF‐κB signal pathway, such as Bcl-XL, Bcl-2, XIAP, SLUG, β-catenin, c-Myc, OCT4, and PTEN was unaffected in DHX9-overexpressed or -silenced CRC cells (Supplementary Figs. 5–6). Previous studies demonstrated that a growing list of proteins that interact with NF-κB can affect specific DNA-binding sites and govern selected gene expression [52, 53]. One important addition to this list is Ribosomal Protein S3 (RPS3), a subunit of p65 DNA-binding complexes. It enhances NF-κB-mediated transcription of selected p65 target genes which expression were dependent on RPS3, but not global p65 target genes which expression were not affected by RPS3[54]. In this study, we found that knockdown p65 actually diminished expression of many NF-κB-targeted genes, while DHX9 selectively affected NF-κB-downstream genes (Fig. 6F–G and Supplementary Fig. 8). Therefore, we speculate that DHX9, as a subunit in NF-κB complexes, may induce selective recruitment of p65 to certain promoters of NF-κB-targeted genes in CRC similar to RPS3. Other potential mechanisms for DHX9 selectively regulates NF-κB-targeted expression need to be further clarified.
DHX9 has been demonstrated to interact with many transcription regulatory factors, such as EGFR or CBP/p300 [15, 16]. In this study, we found DHX9 interacted with p65 and RNA Pol II in CRC cells via co-IP assay. Importantly, in mutant study, the conserved ATP-binding motif (Gly-Lys-Thr) in catalytic domain of DHX9 may be responsible for the interaction with p65 and RNA Pol II in CRC cells. In line with previous studies [29, 30], the ATP-binding mutant (K417R) of DHX9 resulted in a great reduction in transcriptional activity of NF‐κB. These results suggest that the ATPase/helicase domain of DHX9 may be responsible for the interaction with p65 and RNA Pol II and be critical for NF-κB-dependent transcription (Fig. 7H). Moreover, ChIP assay indicated that DHX9 itself was recruited to NF-κB-dependent promoters, and modulated p65’s and RNA Pol II’s occupancy of these promoters in CRC cells (Fig. 8A–B). These results suggest that DHX9 may serve as a bridging factor to recruit p65 and RNA Pol II to the NF-κB-specific promoters, which is similar to Akirin2 that bridges NF-κB p50 and the chromatin-remodeling SWI-SNF complex [55].
In summary, our findings reveal that overexpressed DHX9 is associated with aggressive tumor stage, poor prognosis and malignant phenotypes maintenance (e.g., apoptosis resistance, outgrowth and metastasis). Mechanically, on one hand, DHX9 enhances p65 phosphorylation, promotes p65 nuclear translocation to facilitate NF-κB-mediated transcriptional activity. On the other hand, DHX9 interacts with p65 and RNA Pol II, and may serve as a bridging factor to recruit p65 and RNA Pol II to the NF-κB-dependent promoters to activate transcription. This study validate that DHX9 may serve as a diagnostic and therapeutic target for CRC prevention and treatment. However, the definite molecular mechanism underlying how DHX9 regulates NF-κB translocation still need more investigation. Further experiments on the role of DHX9 in tumorigenesis and activation of NF-κB pathway may provide new insights into the effective therapies in CRC.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by funds from National Natural Science Foundation of China (31560260 and 31960163 to Zhiping Liu, Youth Project 82003801 to Shenglan Liu), The Jinggang Scholar Program in Jiangxi Province (to Zhiping Liu), Natural Science Foundation of Jiangxi Province (20171ACB20024 and 20181BAB205032 to Zhiping Liu) and Technological Innovation Team Project of Gannan Medical University (TD201703 to Zhiping Liu).
Author contributions
ZL and FX developed the hypothesis, designed, guided research, and revised manuscript. SL performed the experiments, analyzed data, and wrote the manuscript. LH performed the experiments and analyzed data. JW, XW, LX, WD and LC assisted the experiments. All authors read and approved the final manuscript.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author (Zhiping.Liu@gmu.edu.cn) on reasonable request.
Declarations
Conflict of interest
All authors have no conflict of interest in this paper.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Fuhua Xie, Email: fuhuax2004@163.com.
Zhiping Liu, Email: Zhiping.Liu@gmu.edu.cn.
References
- 1.Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. The Lancet. 2019;394:1467–1480. doi: 10.1016/S0140-6736(19)32319-0. [DOI] [PubMed] [Google Scholar]
- 2.F. Bray, J. Ferlay, I. Soerjomataram, R.L. Siegel, L.A. Torre, A. Jemal, Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries, CA: a cancer journal for clinicians, 68 (2018) 394–424. [DOI] [PubMed]
- 3.He L, Chen Y, Wu Y, Xu Y, Zhang Z, Liu Z. Nucleic acid sensing pattern recognition receptors in the development of colorectal cancer and colitis. Cell Mol Life Sci. 2017;74:2395–2411. doi: 10.1007/s00018-017-2477-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He L, Liu Y, Lai W, Tian H, Chen L, Xie L, Liu Z. DNA sensors, crucial receptors to resist pathogens, are deregulated in colorectal cancer and associated with initiation and progression of the disease, Journal of. Cancer. 2020;11:893–905. doi: 10.7150/jca.34188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He L, Xiao X, Yang X, Zhang Z, Wu L, Liu Z. STING signaling in tumorigenesis and cancer therapy: a friend or foe? Cancer Lett. 2017;402:203–212. doi: 10.1016/j.canlet.2017.05.026. [DOI] [PubMed] [Google Scholar]
- 6.Jain A, Bacolla A, Chakraborty P, Grosse F, Vasquez KM. Human DHX9 helicase unwinds triple-helical DNA structures. Biochemistry. 2010;49:6992–6999. doi: 10.1021/bi100795m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuller-Pace FV. DExD/H box RNA helicases: multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res. 2006;34:4206–4215. doi: 10.1093/nar/gkl460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim S, Kang N, Park SH, Wells J, Hwang T, Ryu E, Kim BG, Hwang S, Kim SJ, Kang S, Lee S, Stirling P, Myung K, Lee KY. ATAD5 restricts R-loop formation through PCNA unloading and RNA helicase maintenance at the replication fork. Nucleic Acids Res. 2020;48:7218–7238. doi: 10.1093/nar/gkaa501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin YC, Yu YS, Lin HH, Hsiao KY. Oxaliplatin-induced DHX9 phosphorylation promotes oncogenic circular RNA CCDC66 expression and development of chemoresistance. Cancers. 2020;12:697. doi: 10.3390/cancers12030697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee T, Pelletier J. The biology of DHX9 and its potential as a therapeutic target. Oncotarget. 2016;7:42716–42739. doi: 10.18632/oncotarget.8446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee T, Paquet M, Larsson O, Pelletier J. Tumor cell survival dependence on the DHX9 DExH-box helicase. Oncogene. 2016;35:5093–5105. doi: 10.1038/onc.2016.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.B. Shen, Y. Chen, J. Hu, M. Qiao, J. Ren, J. Hu, J. Chen, N. Tang, A. Huang, Y. Hu, Hepatitis B virus X protein modulates upregulation of DHX9 to promote viral DNA replication, Cellular microbiology, 22 (2020) e13148. [DOI] [PubMed]
- 13.Yan X, Chang J, Sun R, Meng X, Wang W, Zeng L, Liu B, Li W, Yan X, Huang C, Zhao Y, Li Z, Yang S. DHX9 inhibits epithelial-mesenchymal transition in human lung adenocarcinoma cells by regulating STAT3. Am J Trans Res. 2019;11:4881–4894. [PMC free article] [PubMed] [Google Scholar]
- 14.M. Vázquez-Del Mercado, C.A. Palafox-Sánchez, J.F. Muñoz-Valle, G. Orozco-Barocio, E. Oregon-Romero, R.E. Navarro-Hernández, M. Salazar-Páramo, J. Armendariz-Borunda, J.I. Gámez-Nava, L. Gonzalez-Lopez, J.Y. Chan, E.K. Chan, M. Satoh, High prevalence of autoantibodies to RNA helicase A in Mexican patients with systemic lupus erythematosus, Arthritis research & therapy, 12 (2010) R6. [DOI] [PMC free article] [PubMed]
- 15.Koirala P, Huang J, Ho TT, Wu F, Ding X, Mo YY. LncRNA AK023948 is a positive regulator of AKT. Nat Commun. 2017;8:14422. doi: 10.1038/ncomms14422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.H. Hong, O. An, T.H.M. Chan, V.H.E. Ng, H.S. Kwok, J.S. Lin, L. Qi, J. Han, D.J.T. Tay, S.J. Tang, H. Yang, Y. Song, F. Bellido Molias, D.G. Tenen, L. Chen, Bidirectional regulation of adenosine-to-inosine (A-to-I) RNA editing by DEAH box helicase 9 (DHX9) in cancer, Nucleic acids research, 46 (2018) 7953–7969. [DOI] [PMC free article] [PubMed]
- 17.Mineo M, Ricklefs F, Rooj AK, Lyons SM, Ivanov P, Ansari KI, Nakano I, Chiocca EA, Godlewski J, Bronisz A. The long non-coding RNA HIF1A-AS2 facilitates the maintenance of mesenchymal glioblastoma stem-like cells in hypoxic niches. Cell Rep. 2016;15:2500–2509. doi: 10.1016/j.celrep.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Närvä E, Rahkonen N, Emani MR, Lund R, Pursiheimo JP, Nästi J, Autio R, Rasool O, Denessiouk K, Lähdesmäki H, Rao A, Lahesmaa R. RNA-binding protein L1TD1 interacts with LIN28 via RNA and is required for human embryonic stem cell self-renewal and cancer cell proliferation. Stem cells (Dayton, Ohio) 2012;30:452–460. doi: 10.1002/stem.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Myöhänen S, Baylin SB. Sequence-specific DNA binding activity of RNA helicase A to the p16INK4a promoter. J Biol Chem. 2001;276:1634–1642. doi: 10.1074/jbc.M004481200. [DOI] [PubMed] [Google Scholar]
- 20.Kawai S, Amano A. BRCA1 regulates microRNA biogenesis via the DROSHA microprocessor complex. J Cell Biol. 2012;197:201–208. doi: 10.1083/jcb.201110008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Halaby MJ, Harris BR, Miskimins WK, Cleary MP, Yang DQ. Deregulation of internal ribosome entry site-mediated p53 translation in cancer cells with defective p53 response to DNA damage. Mol Cell Biol. 2015;35:4006–4017. doi: 10.1128/MCB.00365-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamata H, Honda S, Maeda S, Chang L, Hirata H, Karin M. Reactive oxygen species promote TNFalpha-induced death and sustained JNK activation by inhibiting MAP kinase phosphatases. Cell. 2005;120:649–661. doi: 10.1016/j.cell.2004.12.041. [DOI] [PubMed] [Google Scholar]
- 23.Sakamoto K, Maeda S. Targeting NF-kappaB for colorectal cancer. Expert Opin Ther Targets. 2010;14:593–601. doi: 10.1517/14728221003769903. [DOI] [PubMed] [Google Scholar]
- 24.Patel M, Horgan PG, McMillan DC, Edwards J. NF-kappaB pathways in the development and progression of colorectal cancer. Trans Res. 2018;197:43–56. doi: 10.1016/j.trsl.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 25.Pahlavan Y, Kahroba H, Samadi N. Survivin modulatory role in autoimmune and autoinflammatory diseases. J Cell Physiol. 2019;234:19440–19450. doi: 10.1002/jcp.28725. [DOI] [PubMed] [Google Scholar]
- 26.L. Mei, Y.M. Zheng, T. Song, V.R. Yadav, L.C. Joseph, L. Truong, S. Kandhi, Rieske iron-sulfur protein induces FKBP12.6/RyR2 complex remodeling and subsequent pulmonary hypertension through NF-κB/cyclin D1 pathway, Nature communications, 11 (2020) 3527. [DOI] [PMC free article] [PubMed]
- 27.Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441:431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 28.Wu TJ, Chang SS, Li CW, Hsu YH, Chen TC, Lee WC, Yeh CT, Hung MC. Severe hepatitis promotes hepatocellular carcinoma recurrence via NF-κB pathway-mediated epithelial-mesenchymal transition after resection, clinical cancer research : an official journal of the American association for. Can Res. 2016;22:1800–1812. doi: 10.1158/1078-0432.CCR-15-0780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tetsuka T, Uranishi H, Sanda T, Asamitsu K, Yang JP, Wong-Staal F, Okamoto T. RNA helicase A interacts with nuclear factor kappaB p65 and functions as a transcriptional coactivator. Eur J Biochem. 2004;271:3741–3751. doi: 10.1111/j.1432-1033.2004.04314.x. [DOI] [PubMed] [Google Scholar]
- 30.Ng YC, Chung WC, Kang HR, Cho HJ, Park EB, Kang SJ, Song MJ. A DNA-sensing-independent role of a nuclear RNA helicase, DHX9, in stimulation of NF-κB-mediated innate immunity against DNA virus infection. Nucleic Acids Res. 2018;46:9011–9026. doi: 10.1093/nar/gky742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.J. Mi, P. Ray, J. Liu, C.T. Kuan, J. Xu, D. Hsu, B.A. Sullenger, R.R. White, B.M. Clary, In Vivo Selection Against Human Colorectal Cancer Xenografts Identifies an Aptamer That Targets RNA Helicase Protein DHX9, Molecular therapy. Nucleic acids, 5 (2016) e315. [DOI] [PMC free article] [PubMed]
- 32.Meier R, Franceschini A, Horvath P, Tetard M, Mancini R, von Mering C, Helenius A, Lozach PY. Genome-wide small interfering RNA screens reveal VAMP3 as a novel host factor required for Uukuniemi virus late penetration. J Virol. 2014;88:8565–8578. doi: 10.1128/JVI.00388-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin Y, Zhang P, Wang Y, Jin B, Zhou J, Zhang J, Pan J. Neddylation blockade diminishes hepatic metastasis by dampening cancer stem-like cells and angiogenesis in uveal melanoma, clinical cancer research : an official journal of the American association for. Can Res. 2018;24:3741–3754. doi: 10.1158/1078-0432.CCR-17-1703. [DOI] [PubMed] [Google Scholar]
- 34.Man SM, Zhu Q, Zhu L, Liu Z, Karki R, Malik A, Sharma D, Li L, Malireddi RK, Gurung P, Neale G, Olsen SR, Carter RA, McGoldrick DJ, Wu G, Finkelstein D, Vogel P, Gilbertson RJ, Kanneganti TD. Critical role for the DNA sensor AIM2 in stem cell proliferation and cancer. Cell. 2015;162:45–58. doi: 10.1016/j.cell.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J, Liu S, Ye Q, Pan J. Transcriptional inhibition by CDK7/9 inhibitor SNS-032 abrogates oncogene addiction and reduces liver metastasis in uveal melanoma. Mol Cancer. 2019;18:140. doi: 10.1186/s12943-019-1070-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.J. Zhou, S. Liu, Y. Wang, W. Dai, H. Zou, S. Wang, J. Zhang, J. Pan, Salinomycin effectively eliminates cancer stem-like cells and obviates hepatic metastasis in uveal melanoma, Molecular Cancer, 18 (2019). [DOI] [PMC free article] [PubMed]
- 37.Li Y, Chen F, Shen W, Li B, Xiang R, Qu L, Zhang C, Li G, Xie H, Katanaev VL, Jia L. WDR74 induces nuclear beta-catenin accumulation and activates Wnt-responsive genes to promote lung cancer growth and metastasis. Cancer Lett. 2020;471:103–115. doi: 10.1016/j.canlet.2019.12.011. [DOI] [PubMed] [Google Scholar]
- 38.Jiang YY, Lin DC, Mayakonda A, Hazawa M, Ding LW, Chien WW, Xu L, Chen Y, Xiao JF, Senapedis W, Baloglu E, Kanojia D, Shang L, Xu X, Yang H, Tyner JW, Wang MR, Koeffler HP. Targeting super-enhancer-associated oncogenes in oesophageal squamous cell carcinoma. Gut. 2017;66:1358–1368. doi: 10.1136/gutjnl-2016-311818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu C, Nie D, Li J, Du X, Lu Y, Li Y, Zhou J, Jin Y, Pan J. Antitumor effects of blocking protein neddylation in T315I-BCR-ABL leukemia cells and leukemia stem cells. Can Res. 2018;78:1522–1536. doi: 10.1158/0008-5472.CAN-17-1733. [DOI] [PubMed] [Google Scholar]
- 40.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 41.Srivastava SK, Bhardwaj A, Arora S, Singh S, Azim S, Tyagi N, Carter JE, Wang B, Singh AP. MYB is a novel regulator of pancreatic tumour growth and metastasis. Br J Cancer. 2015;113:1694–1703. doi: 10.1038/bjc.2015.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Y, Zhou Y, Li B, Chen F, Shen W, Lu Y, Zhong C, Zhang C, Xie H, Katanaev VL, Jia L. WDR74 modulates melanoma tumorigenesis and metastasis through the RPL5-MDM2-p53 pathway. Oncogene. 2020;39:2741–2755. doi: 10.1038/s41388-020-1179-6. [DOI] [PubMed] [Google Scholar]
- 43.Ceelen W, Ramsay RG, Narasimhan V, Heriot AG, De Wever O. Targeting the tumor microenvironment in colorectal peritoneal metastases. Trends in cancer. 2020;6:236–246. doi: 10.1016/j.trecan.2019.12.008. [DOI] [PubMed] [Google Scholar]
- 44.D.K. Filippiadis, G. Velonakis, The Role of Percutaneous Ablation in the Management of Colorectal Cancer Liver Metastatic Disease, Diagnostics (Basel, Switzerland), 11 (2021). [DOI] [PMC free article] [PubMed]
- 45.Lo MC, Yip TC, Ngan KC, Cheng WW, Law CK, Chan PS, Chan KC, Wong CK, Wong RN, Lo KW, Ng WT, Lee WM, Tsao SW, Kwong LW, Lung ML, Mak NK. Role of MIF/CXCL8/CXCR2 signaling in the growth of nasopharyngeal carcinoma tumor spheres. Cancer Lett. 2013;335:81–92. doi: 10.1016/j.canlet.2013.01.052. [DOI] [PubMed] [Google Scholar]
- 46.Zhang S, Grosse F. Domain structure of human nuclear DNA helicase II (RNA helicase A) J Biol Chem. 1997;272:11487–11494. doi: 10.1074/jbc.272.17.11487. [DOI] [PubMed] [Google Scholar]
- 47.Cao S, Sun R, Wang W, Meng X, Zhang Y, Zhang N, Yang S. RNA helicase DHX9 may be a therapeutic target in lung cancer and inhibited by enoxacin. Am J Trans Res. 2017;9:674–682. [PMC free article] [PubMed] [Google Scholar]
- 48.Y.L. Wang, J.Y. Liu, J.E. Yang, X.M. Yu, Z.L. Chen, Y.J. Chen, M. Kuang, Y. Zhu, S.M. Zhuang, Lnc-UCID Promotes G1/S Transition and Hepatoma Growth by Preventing DHX9-Mediated CDK6 Down-regulation, Hepatology (Baltimore, Md.), 70 (2019) 259–275. [DOI] [PMC free article] [PubMed]
- 49.Sun Z, Wang L, Eckloff BW, Deng B, Wang Y, Wampfler JA, Jang J, Wieben ED, Jen J, You M, Yang P. Conserved recurrent gene mutations correlate with pathway deregulation and clinical outcomes of lung adenocarcinoma in never-smokers. BMC Med Genomics. 2014;7:32. doi: 10.1186/1755-8794-7-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 51.Mills JR, Malina A, Lee T, Di Paola D, Larsson O, Miething C, Grosse F, Tang H, Zannis-Hadjopoulos M, Lowe SW, Pelletier J. RNAi screening uncovers Dhx9 as a modifier of ABT-737 resistance in an Eμ-myc/Bcl-2 mouse model. Blood. 2013;121:3402–3412. doi: 10.1182/blood-2012-06-434365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Natoli G, Saccani S, Bosisio D, Marazzi I. Interactions of NF-kappaB with chromatin: the art of being at the right place at the right time. Nat Immunol. 2005;6:439–445. doi: 10.1038/ni1196. [DOI] [PubMed] [Google Scholar]
- 53.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 54.Wan F, Anderson DE, Barnitz RA, Snow A, Bidere N, Zheng L, Hegde V, Lam LT, Staudt LM, Levens D, Deutsch WA, Lenardo MJ. Ribosomal protein S3: a KH domain subunit in NF-kappaB complexes that mediates selective gene regulation. Cell. 2007;131:927–939. doi: 10.1016/j.cell.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 55.Tartey S, Matsushita K, Vandenbon A, Ori D, Imamura T, Mino T, Standley DM, Hoffmann JA, Reichhart JM, Akira S, Takeuchi O. Akirin2 is critical for inducing inflammatory genes by bridging IκB-ζ and the SWI/SNF complex. EMBO J. 2014;33:2332–2348. doi: 10.15252/embj.201488447. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author (Zhiping.Liu@gmu.edu.cn) on reasonable request.