Abstract
Background
Skeletal muscles (SkM) are mechanosensitive, with mechanical unloading resulting in muscle-devastating conditions and altered metabolic properties. However, it remains unexplored whether these atrophic conditions affect SkM mechanosensors and molecular clocks, both crucial for their homeostasis and consequent physiological metabolism.
Methods
We induced SkM atrophy through 14 days of hindlimb suspension (HS) in 10 male C57BL/6J mice and 10 controls (CTR). SkM histology, gene expressions and protein levels of mechanosensors, molecular clocks and metabolism-related players were examined in the m. Gastrocnemius and m. Soleus. Furthermore, we genetically reduced the expression of mechanosensors integrin-linked kinase (Ilk1) and kindlin-2 (Fermt2) in myogenic C2C12 cells and analyzed the gene expression of mechanosensors, clock components and metabolism-controlling genes.
Results
Upon hindlimb suspension, gene expression levels of both core molecular clocks and mechanosensors were moderately upregulated in m. Gastrocnemius but strongly downregulated in m. Soleus. Upon unloading, metabolism- and protein biosynthesis-related genes were moderately upregulated in m. Gastrocnemius but downregulated in m. Soleus. Furthermore, we identified very strong correlations between mechanosensors, metabolism- and circadian clock-regulating genes. Finally, genetically induced downregulations of mechanosensors Ilk1 and Fermt2 caused a downregulated mechanosensor, molecular clock and metabolism-related gene expression in the C2C12 model.
Conclusions
Collectively, these data shed new lights on mechanisms that control muscle loss. Mechanosensors are identified to crucially control these processes, specifically through commanding molecular clock components and metabolism.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-022-04346-7.
Keywords: Atrophy, Hindlimb suspension, Mechanosensing, Molecular clock, Skeletal muscle metabolism
Introduction
Skeletal muscle (SkM) represents the largest metabolically active tissue in the body accounting for approximately 40–50% of total body mass [1]. Hence, maintenance of SkM mass is a fundamental process guaranteeing the management of health. In contrast, substantial loss of SkM mass (SkM atrophy), as caused by injury, inactivity or disease, impairs whole-body metabolism and consequently drives dysfunctional phenotypes [2–4]. SkM metabolism is regulated by autocrine, paracrine and endocrine hormonal signals [5] as well as by myokine-mediated crosstalk with other tissues [6]. In addition to these pathways, we recently identified mechanosensors as potentially novel regulators of SkM metabolism [7]. Another promising SkM metabolism-controlling pathway is the circadian clock machinery since it is critical for both muscle and systems health [8, 9]. However, for both mechanosensors and molecular clock components it remains unknown how loss of SkM mass changes their patterns as well as the expression of metabolism-controlling genes.
SkM has an intrinsic capacity to sense mechanical stimuli (mechanosensing) and to convert these into biochemical events (mechanotransduction) that control muscle growth and metabolism [7]. Mechanosensors constitute important intra-muscular mechanosensitive components [10] and physically link force-generating sarcomeres with the sarcolemma at each Z-disk [7, 10]. Therefore, they control physiological adaptions of individual SkM fibers to environmental stimuli, such as mechanical loading [7, 10]. Among mechanosensors, the integrin-linked kinase (ILK) [7, 10] and Kindlin-2 (Fermt2/KIND2) [11] are crucial and regulate, together with proteins, such as Talin 1 and 2 (TLN1 and TLN2) [12] and Vinculin (VCL) [13], SkM functionality. Recently, the major regulator of integrin-mediated signaling, Ilk1 [14], was demonstrated to play a crucial role in the control of muscle fiber characteristics. Ilk1 knockdown in C2C12 cells resulted in significantly increased slow SkM gene program as well as reduced protein synthesis-enhancing signaling [7]. This identified SkM mechanosensors as muscle fiber type-specific loading management ‘hubs’ that are crucial for SkM fiber metabolic programming.
Further, SkM metabolism is also regulated by internal timing mechanisms that function to predict and prepare the organism for daily environmental changes [15]. These approximate 24-h biological cycles are called circadian rhythms and in SkM they include oscillations in metabolism, transcription and myogenic capacity [9, 16, 17]. The circadian system encompasses a network of molecular clocks found in most cell types, including skeletal muscle, and are controlled and synchronized by the master clock located in the suprachiasmatic nuclei (SCN) of the hypothalamus [9, 15]. Circadian rhythms are regulated by interlocking transcription-translation feedback loops, known as the core molecular clock, that control the expression of rhythmic genes in a tissue-specific manner [9, 18].
At a molecular level, circadian locomotor output cycles kaput (Clock) and brain and muscle arnt-like protein 1 (Bmal1) compose the positive arm of the transcriptional–translational feedback loops, while period 1 and 2 (Per1/2) and cryptochrome 1 and 2 (Cry1/2) drive the negative loop. Clock and Bmal1 heterodimerize and activate the rhythmic transcription of the negative feedback elements Per1, Per2, Cry1 and Cry2. The Cry and Per proteins finally inhibit BMAL1:CLOCK activity and thereby suppress their own expression [9, 15].
In SkM, core molecular clocks control SkM mass, glucose metabolism and insulin sensitivity [18–20]. Furthermore, Clock mutants and Bmal1-deficient mice show reduced SkM force, mitochondrial dysfunction and disrupted myofilament architecture [16, 21]. These findings suggest that molecular clocks modify SkM physiology, which is further supported by the fact that approximately 40% of transcripts with a circadian expression pattern in adult SkM belong to metabolic processes [18]. Interestingly, mechanosensitive integrin signaling contributes to molecular clock regulation in mammary epithelial cells (MECs) [22]. Furthermore, Clock localizes to the sarcomeric Z-disk in cardiomyocytes and acts as a sensor of myofilament cross-bridge activity [23]. These data suggest that the cellular mechano-environment regulates the molecular clock [22, 23], which, however, has not yet been demonstrated.
In the present study, we show that mechanical unloading causes significant alterations in mechanosensing elements as well as metabolism- and molecular clock-regulating genes in a SkM type-dependent manner. Since mechanosensors are involved in the control of muscle fiber characteristics [7], this further underlines their role in SkM fiber type-specific loading management upon altered mechanical loading. We further identified very strong correlations between mechanosensors, metabolism- and circadian clock-controlling genes indicating their close regulatory dependencies. Mechanistically, we highlight that genetically inhibited mechanosensor expression disrupts molecular clock and metabolic equilibria in vitro. Collectively, our data emphasize the impact of SkM-devastating conditions on mechanosensing, metabolism and molecular clock regulation and identify mechanosensors as novel control hubs of the molecular clock machinery.
Materials and methods
Animals and interventions
Eight-week-old male C57BL/6J mice were housed in a conventional animal facility of the KU Leuven at 22–24 °C under a 14 h light/10 h dark cycle. Standard chow and non-wetting water gels (HydroGel; ClearH2O) were supplemented ad libitum on the cage floor to permit easy access. All mice were examined daily and bodyweights were measured every two days to monitor animals’ health and well-being. The experimental procedures were approved by the Animal Ethics Committee of the KU Leuven, Leuven, Belgium (P110/2018). Mice were randomly divided into two groups: (1) sedentary control (CTR, n = 10), mice of this group did not receive any specific intervention; (2) hindlimb suspension (HS; n = 10), mice of this group were subjected to tail suspension for fourteen consecutive days. HS is an established standardized approach to induce SkM atrophy [24]. In brief, a hanger was attached to the tail of the mice. The cages were covered by Plexiglas plates on which curtain rods were fixed. The hanger was then connected to a curtain rod on the top of the cage, thereby preventing the hindlimbs from contacting any supporting surface. The bottom of the cages was covered with cork plates (8 mm thick) providing a robust, but comfortable surface for the forepaws. This combination of both the curtain rods and the cork plates allowed the mice to move freely across the full length of the cage while reducing the external mechanical loading on the hindlimbs. We sacrificed the mice at the same time point after 14 days of HS and then the m. Gastrocnemius (GAS, fast-twitch) and m. Soleus (SOL, slow-twitch) muscles were dissected and frozen in liquid nitrogen.
Immunohistochemistry
Immunohistochemical analysis was conducted as described previously [25]. Briefly, 7-µm-thick cryosections of GAS were warmed to room temperature and subsequently fixed for 10 min in − 20 °C pre-cooled acetone. Next, slides were incubated for 60 min at 37 °C with the antibody recognizing Laminin (dilution: 1:500, Sigma, L9393). Upon rinsing in PBS, slides were then incubated for 60 min at 37 °C with the appropriate polyclonal biotinylated secondary antibody (Donkey Anti-Rabbit IgG H&L Alexa Fluor 488, dilution: 1:500 in PBS; Abcam, ab150073). Finally, the sections were embedded in Dako Fluorescence Mounting Medium (Dako, S3023) and supplied with a coverslip. All slides were examined with a Nikon Eclipse E1000 microscope and compatible NIS Elements software (20× magnification). The MyoVision software (University of Kentucky, Lexington, KY, USA) was used for data acquisition [26].
Western blotting
According to established protocols [27, 28], 6–20 mg of frozen muscle tissue was homogenized 5 × 20 s at 6.0 m/s in ice-cold lysis buffer (Cell Signaling, Boston, MA) using a FastPrep-24™ Classic Instrument (MP Biomedicals, Santa Ana, CA). Homogenates were then centrifuged at 10,000g for 25 min at 4 °C and the supernatant was collected and immediately stored at − 80 °C. The homogenates’ protein contents were determined by using the DC protein assay kit (Bio‐Rad laboratories, Nazareth, Belgium). 10–30 μg of homogenate protein was separated by SDS‐PAGE (8–12% gels) and transferred to a polyvinylidene difluoride (PVDF) membrane. The membranes were blocked in 5% low-fat milk in TBS-T for 60 min and afterwards incubated overnight at 4 °C with the following antibodies: Clock (1:1000, PA1-520, Invitrogen); Bmal1 (1:1000, NB100-2288SS, Novus Biologicals); Ilk1 (1:2000, CST-3862, Cell Signaling); Kind2 (1:1000, CST-13562, Cell Signaling); Tln (1:5000, T3287, Sigma) and Vcl (1:2000, V9131, Sigma) dissolved in 5% low-fat milk in TBS-T. Afterwards, the membranes were incubated for one hour at RT with horseradish peroxidase‐conjugated anti‐mouse or anti‐rabbit secondary antibodies (1:5000 both in 5% low-fat milk in TBS-T for both, Sigma) for chemiluminescent detection. Membranes were scanned and quantified with Genetools and Genesnap softwares (Syngene, Cambridge), respectively. Total GAPDH served as internal loading control and the results are presented as the ratio protein of interest/GAPDH.
RNA extraction and reverse transcription
Total RNA was extracted from 6 to 20 mg of skeletal muscle using TRI Reagent® (MRC, Cincinnati). Quantity and quality of RNA were assessed by a spectrophotometer (SimpliNano, Biochrom). One μg of total RNA was then used for reverse transcription (RT) with a QuantiTect Reverse Transcription Kit (Qiagen, Hilden, Germany). RNA integrity was controlled by RNA gel electrophoresis as described [7, 25, 28].
Real‐time qPCR analysis
Upon reverse transcription of RNA into cDNA using the QuantiTect Reverse Transcription Kit (Qiagen), qPCR was performed by mixing a final reaction volume of 25 μL including the following components: (1) 12.5 μL GoTaq(R) qPCR Master Mix (Promega); (2) 100 nM primer mixes of targeted primers (Table S1) targeting the cDNA of genes of interests; (4) 2 μL of cDNA; (5) 8.5 μL nuclease-free H2O. The reaction was performed on a QuantStudio 3 Real-Time PCR System (Thermo Fisher Scientific, Waltham). The quantity of the genes of interest in each sample was normalized to that of Rpl41 using the comparative (2-∆∆CT) method. Unbiased amplicon generation by qPCR reactions were finally controlled by DNA gel electrophoresis as described [7, 25, 28]. The primer sequences are summarized in Table S1.
Cell culture and transfection
The protocol has been described earlier [7]. Briefly, C2C12 myoblasts were grown in culture medium (high-glucose DMEM containing 10% FBS and 1% P/S) until sub-confluence was reached. Differentiation was initiated by changing the media to high-glucose DMEM containing 2% HS and 1% P/S. For Ilk1 and Fermt2 knockdowns, C2C12 myotubes were transfected after 4 days of differentiation with 10 nM Ilk-SiRNA, Fermt2-SiRNA or respective control SiRNA (TriFECTa RNAi Kit, IDT) using Lipofectamine RNAiMAX (Invitrogen). To examine the transfection efficiency of Lipofectamine RNAiMAX on C2C12 cells we used fluorescent dye–labeled DsiRNA (TYE 563 transfection control dsiRNA). Twenty-four hours post-transfection, the percentage of fluorescent positive cells was determined. Our results revealed that the siRNA transfection efficiency of the C2C12 myotubes was ~ 80%. 48 h after transfection, C2C12 myotubes were harvested for further RNA analysis. All cells were cultured in a 5% CO2 atmosphere at 37 °C.
Statistics
All analyses were performed with GraphPad Prism 8.0.2 (http://www.graphpad.com). Body weight developments and food intake were analyzed by two-way repeated measures ANOVA with time and group as factors. For multiple comparison, Sidak’s post hoc test was used. Gene and protein expression data were log-transformed for statistical analysis. Fiber cross-sectional area (CSA) data as well as gene and protein expression data were analyzed by two-way ANOVA with muscle and group as factors. For multiple comparison, Sidak’s multiple comparison post hoc test was used and within-muscle results are depicted in the figures. Correlations between mechanosensor, circadian clock and metabolism-related genes were assessed using Pearson correlation coefficient analyses. For the gene expression data of the C2C12 siRNA approach, we used a Student’s t test upon log-transformation. For all tests, statistical significance was accepted at p < 0.05.
Results
Hindlimb suspension reduces skeletal muscle fiber size and induces the ubiquitin–proteasome system
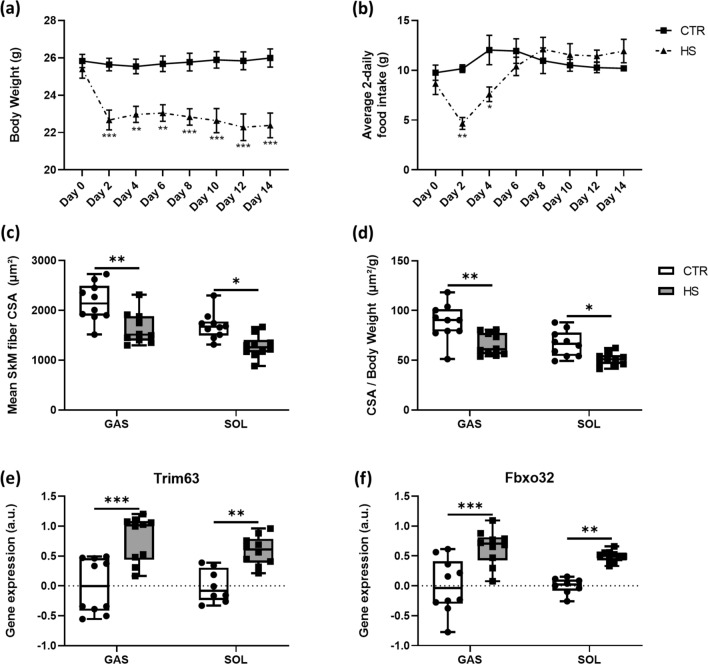
We first evaluated the effect of 14 days HS on body weight and food intake and observed a significantly reduced body weight of HS mice from day 2 until day 14 compared to CTR mice (Fig. 1a). HS mice consumed significantly less food only until day 4 (Fig. 1b) compared to CTR mice, while the food intake normalized after day 4 (Fig. 1b). Next, we analyzed SkM fiber cross-sectional area (CSA) of GAS and SOL muscles. As expected, HS significantly reduced the CSA in GAS and SOL muscles in comparison to the CTR mice (Fig. 1c). In addition, we found a comparable result for the CSA-to-body-weight ratio, which was also reduced in HS compared to CTR mice in both GAS and SOL (Fig. 1d).
Fig. 1.
Skeletal muscle unloading modulates body weights, food intake, fiber cross-sectional area (CSA) and atrophy-related E3 ligases. a Body weight developments and b food intake in CTR (n = 10) and HS (n = 10) mice. c Mean skeletal muscle CSA between CTR (n = 10) and HS (n = 10) mice in GAS and SOL muscle fibers. d CSA/body weight ratios for GAS and SOL of CTR and HS mice. Statistical analysis used was two-way repeated measures ANOVA with Sidak’s multiple comparison post hoc test. Relative gene expression profiles of e Trim63 and f Fbxo32 between CTR and HS mice in GAS and SOL muscles. Statistical analysis used was two-way ANOVA with Sidak’s multiple comparison post hoc test on the following number of animals: GAS 10 CTR and 10 HS animals, SOL 8 CTR and 10 HS animals. *p value < 0.05; **p value < 0.01; ***p value < 0.001
HS causes disuse-related SkM atrophy caused by muscle contractile deficiency [29]. This event is usually accompanied by increased protein degradation mainly mediated by an activated ubiquitin proteasome system (UPS) [30]. Two major SkM-specific E3 ubiquitin ligases, Trim63 (muscle ring finger protein 1 [MuRF1]) and Fbxo32 (atrogin1 or MAFbx), play pivotal roles in this process [3]. We confirmed significant higher levels of Fbxo32 (Fig. 1e) and Trim63 (Fig. 1f) in GAS and SOL of HS compared to CTR mice.
Summarized, these data demonstrate that our HS model induced loss of SkM mass through known pathways indicating a high validity of the model.
Molecular clock genes show a tendency towards upregulation in GAS but are downregulated in SOL upon hindlimb suspension
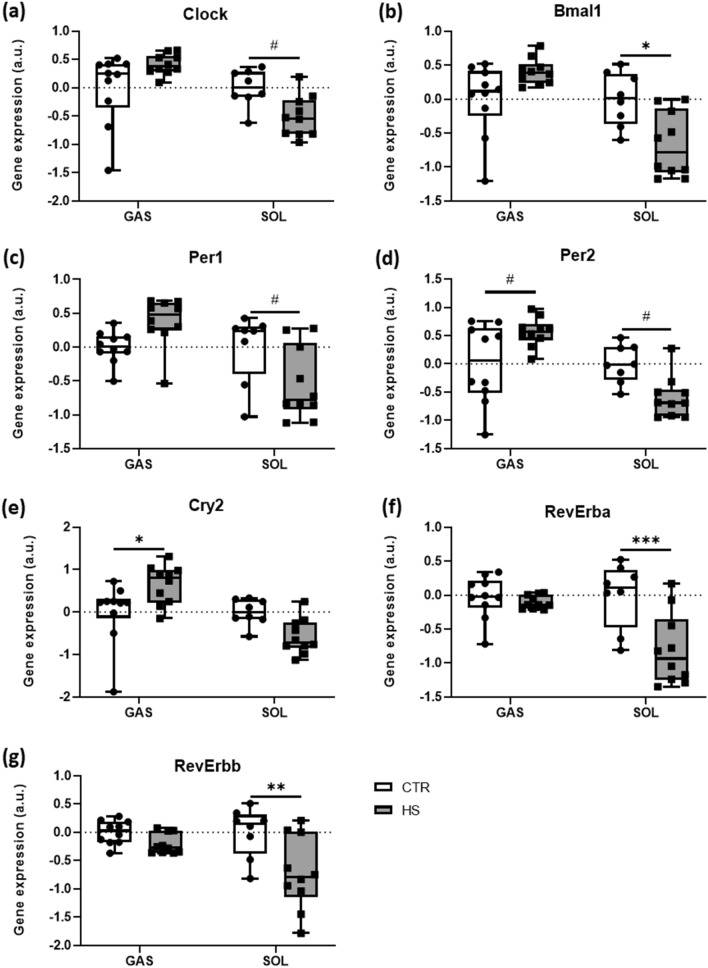
Knockout models demonstrated that molecular clock genes control SkM homeostasis [18]. However, it remains unknown whether physiological models of SkM mass loss alter molecular clock gene expressions. Therefore, we analyzed the effects of HS on molecular clock gene expressions in GAS and SOL muscles.
In GAS, gene expression levels of the two central components of the molecular clock, Clock and Bmal1 [9, 15], did not differ (respectively, Fig. 2a, b) between HS and CTR mice. In addition, expression of Per1 (Fig. 2c) was not different between conditions, whereas other defined target genes of the Clock–Bmal1 heterodimer, such as Per2 (Fig. 2d) or Cry2 (Fig. 2e), were higher in HS compared to CTR. The Clock–Bmal1 complex furthermore controls the expression of the genes coding for the REV-ERB nuclear hormone receptors (RevErba and RevErbb) [22]. Both markers of this auxiliary feedback loop did not significantly change upon the HS intervention (RevErba, Fig. 2f and RevErbb, Fig. 2g).
Fig. 2.
Skeletal muscle unloading regulates expressions of circadian clock genes. Relative gene expression profiles of a Clock, b Bmal1, c Per1, d Per2, e Cry2, f RevErba, and g RevErbb between CTR and HS mice in GAS and SOL muscles. Expression is relative to the housekeeping gene Rpl41. Statistical analysis used was two-way ANOVA with Sidak’s multiple comparison post hoc test on the following number of animals: GAS 10 CTR and 10 HS animals, SOL 8 CTR and 10 HS animals. *p value < 0.05; **p value < 0.01; ***p value < 0.001; #p value = 0.05–0.1
On the other hand, in SOL, the two crucial clock components, Clock and Bmal1 [9, 15], were significantly lower (respectively, Fig. 2a, b) in HS compared to CTR mice. Similarly, we reveal that expression levels of Per1 (Fig. 2c) and Per2 (Fig. 2d) tended to be lower, while Cry2 expression (Fig. 2e) did not differ between conditions. Both markers of the auxiliary feedback loop, RevErba (Fig. 2f) and RevErbb (Fig. 2g), had significantly lower expression levels after the HS intervention.
These data clearly show that molecular clock genes respond to a severe loss of SkM with clear differences between predominantly fast- vs. slow-twitch muscles. Further, the data demonstrate that mechanical loading serves as a crucial regulator of SkM homeostasis, raising the question of whether mechanosensors control molecular clock regulations.
Mechanosensor gene expression is unaltered in GAS but downregulated in SOL upon hindlimb suspension
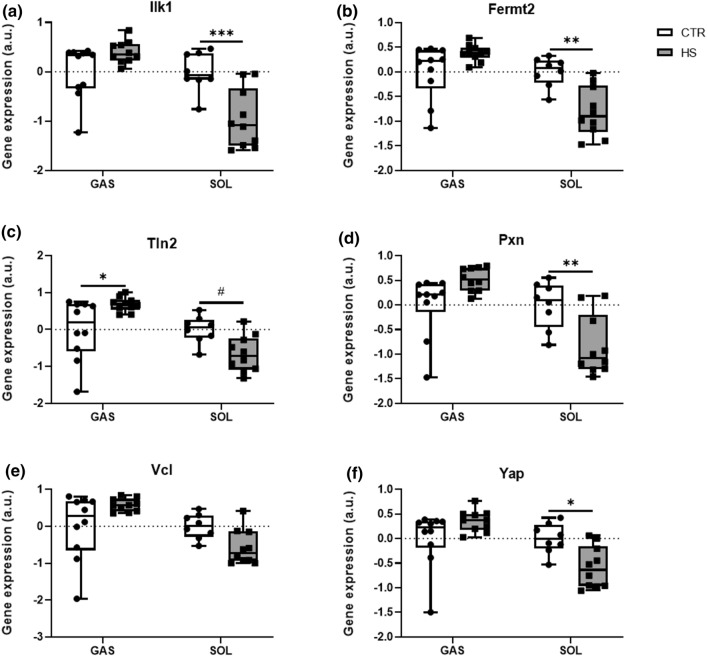
Mechanosensors contribute to SkM adaptation to loading conditions [7, 10]. However, it remains unknown whether loss of SkM mass alters mechanosensors’ gene expressions.
In GAS, gene expression levels of the crucial component Ilk1 [7, 10] and its important binding partner Fermt2 (coding for the protein Kindlin-2) [31], were not different between conditions (Fig. 3a, b). We further studied gene expressions of Tln2, Pxn and Vcl, all accessory molecules of the IRC/IPP complex and involved in integrin activation [10]. While we found significantly higher expression levels of Tln2 (Fig. 3c) in HS mice compared to CTR, there was no difference in the expression levels of Pxn and Vcl (Fig. 3d, e) between conditions. Finally, we found that the gene expression of the Yes-associated Protein (Yap), a transcriptional coactivator downstream of the Hippo pathway and key player in SkM mechanotransduction, myogenesis and SkM homeostasis [32], did not differ between HS and CTR (Fig. 3f). Conversely, in SOL we report significantly downregulated expressions of Ilk1 Fig. 3a) and Fermt2 (Fig. 3b) upon HS. Gene expression levels of Tln2 (Fig. 3c) and Pxn (Fig. 3d) were lower in HS mice compared to CTR, whereas Vcl (Fig. 3e) did not differ. In addition, we identified a significantly lower Yap expression (Fig. 3f) in HS compared to CTR conditions.
Fig. 3.
Mechanosensor gene expression modifications upon skeletal muscle unloading. Relative gene expression profiles of a Ilk, b Fermt2, c Vcl, d Tln2, e Pxn, and f Yap between CTR and HS mice in GAS and SOL muscles. Expression is relative to the housekeeping gene Rpl41. Statistical analysis used was two-way ANOVA with Sidak’s multiple comparison post hoc test on the following number of animals: GAS 10 CTR and 10 HS animals, SOL 8 CTR and 10 HS animals. *p value < 0.05; **p value < 0.01; ***p value < 0.001; #p value = 0.05–0.1
Our data demonstrate that mechanosensors are SkM fiber type-specific loading management hubs that adapt differentially to altered mechanical loading.
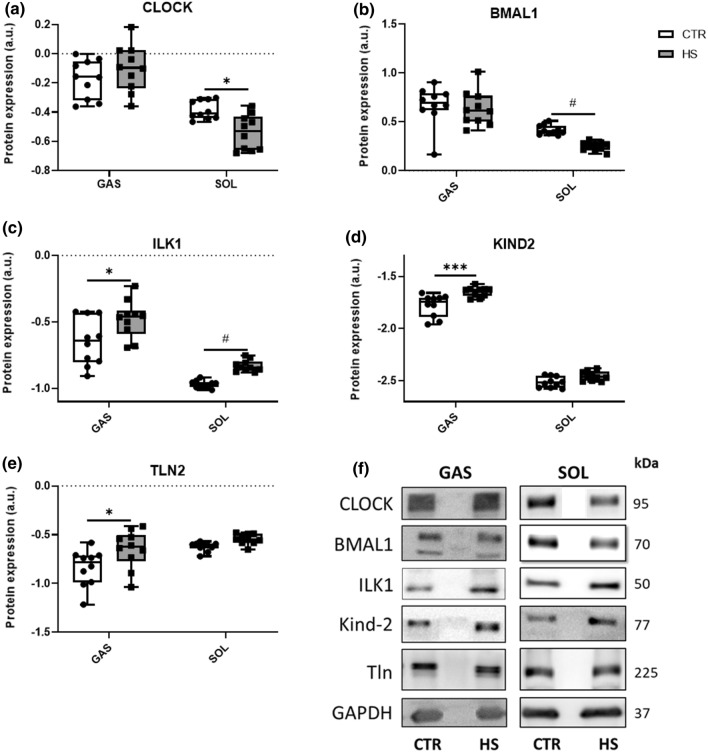
Hindlimb suspension increases protein levels of mechanosensors in GAS but decreases molecular clock components in SOL
As outlined, we found significantly changed genes expressions of molecular clocks and mechanosensors upon HS compared to CTR conditions. We now evaluated potential changes of a selection of these components. In GAS, similar to the gene expression data, the two master clock components Clock and Bmal1 were not differently regulated at the protein level (Fig. 4a, b) between HS and CTR conditions. In line with their gene expression profiles in SOL, we observed significantly lower protein levels of Clock (Fig. 4a) and Bmal1 (Fig. 4b) upon HS. In addition, protein levels of ILK1 (Fig. 4c), KIND2 (Fig. 4d), and TLN2 (Fig. 4e) were significantly higher in GAS of HS compared to CTR mice. Whereas in SOL, protein expression levels of ILK1 (Fig. 4c) tended to be higher upon HS, while protein levels of KIND2 and TLN2 (Fig. 4d, e) did not differ between CTR and HS. Representative western blots are depicted in Fig. 4f.
Fig. 4.
Skeletal muscle unloading does not change circadian clock, but mechanosensor proteins. Relative protein expression profiles of a CLOCK, b BMAL, c ILK, d KIND2, and e TLN2 between CTR and HS mice in GAS and SOL muscles. Expression is relative to the expression of GAPDH. Statistical analysis used was two-way ANOVA with Sidak’s multiple comparison post hoc test on the following number of animals: GAS 10 CTR and 10 HS animals, SOL 10 CTR and 10 HS animals. *p value < 0.05; ***p value < 0.001; #p value = 0.05–0.1. f Representative western blot bands of CLOCK, BMAL1, ILK, KIND2, and TLN2
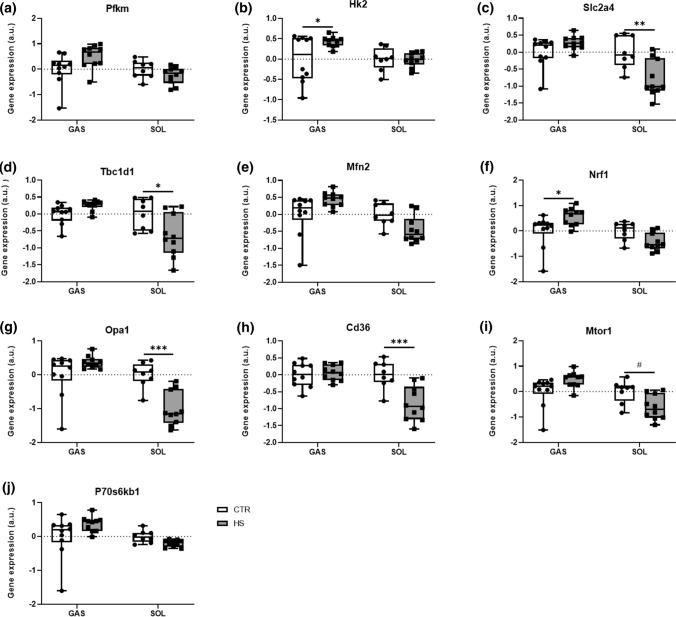
Metabolism- and protein biosynthesis-related genes are moderately upregulated in GAS but downregulated in SOL upon HS
Both mechanosensors [7] and molecular clocks [17, 19] control SkM metabolism. Therefore, we next investigated genes involved in glycolysis as well as oxidative phosphorylation and protein metabolism to gain insights into the question whether SkM metabolism-related gene expression reacts upon unloading and whether they relate to mechanosensors and molecular clock gene expression.
Consequently, we analyzed genes involved in the glucose metabolism (muscle phosphofructokinase [Pfkm] and muscle hexokinase 2 [Hk2]) and its transport (glucose transporter 4 [Slc2a4] and TBC1 domain family member 1 [Tbc1d1]), as glucose is the main metabolic substrate in SkM [33]. Pfkm and Hk2 are rate-limiting enzymes of glycolysis, whereas Slc2a4 and Tbc1d1 control glucose uptake into SkM [34]. In GAS, Pfkm gene expression did not differ between conditions (Fig. 5a), whereas Hk2 expression was significantly higher (Fig. 5b) in HS compared to CTR mice. Furthermore, there was no significant difference in Slc2a4 and Tbc1d1 expression (Fig. 5c, d) between HS and CTR conditions. In SOL, gene expression levels of both Pfkm and Hk2 were not different (Fig. 5a, b) in HS compared to CTR mice. In contrast, Slc2a4 (Fig. 5c) and Tbc1d1 (Fig. 5d) were significantly lower in SOL upon HS compared to CTR conditions.
Fig. 5.
Metabolism-related gene expressions alter upon HS conditions. Relative gene expression profiles of a Pfkm, b Hk2, c Slc2a4, d Tbc1d1, e Mfn2, f Nrf1, g Opa1, h Cd36, i Mtor1, and j P70s6kb1 between CTR and HS mice in GAS and SOL muscles. Expression is relative to the housekeeping gene Rpl41. Statistical analysis used was two-way ANOVA with Sidak’s multiple comparison post hoc test on the following number of animals: GAS 10 CTR and 10 HS animals, SOL 8 CTR and 10 HS animals. *p value < 0.05; **p value < 0.01; ***p value < 0.001; #p value = 0.05–0.1
Mitochondria play pivotal roles as the ‘power plants’ in SkM metabolism and energy homeostasis [35]. Therefore, we studied key regulators of mitochondrial dynamics, including mitofusin-2 (Mfn2), nuclear respiratory factor-1 (Nrf1), optic atrophy 1 (Opa1) as well as the fatty acid transporter CD36 (Cd36) [35]. In GAS, gene expression levels of Mfn2 (Fig. 5e) did not differ between HS and CTR mice, whereas Nrf1 expression (Fig. 5f) was significantly higher in HS. In addition, we found no significant differences in the gene expression levels of both Opa1 and Cd36 (Fig. 5g, h) between both conditions. In SOL, gene expression levels of Mfn2 and Nrf1 (Fig. 5e, f) did not differ between HS and CTR conditions. In contrast, we found significantly lower gene expression levels of Opa1 and Cd36 (Fig. 5g, h) upon HS.
SkM represents the main amino acid-reservoir containing 50–75% of all body proteins and accounts for 30–50% of whole-body protein turnover [1]. The mammalian target of rapamycin (Mtor) and the ribosomal protein S6 kinase beta-1 (P70s6kb1) genes critically regulate protein biosynthesis. Therefore, we investigated the expressions of these two genes in GAS and SOL. In GAS, we found no significant changes in the gene expression of Mtor1 and P70s6kb1 (Fig. 5i, j) between both conditions. Conversely, in SOL Mtor1 gene expression (Fig. 5i) tended to be lower in HS, whereas this was not the case for P70s6kb1 gene expression (Fig. 5j).
Summarized, our data reveal a significant muscle-specific HS-induced alteration of metabolic pathways involving both glycolysis- as well as mitochondria/oxidative metabolism-related genes.
Hindlimb suspension has a limited influence on circadian cycle-dependent E3 ligases, proteasomal activators as well as gluconeogenesis and sarcomeric Z-disk proteins in SOL but not in GAS.
To attain a clearer perspective on further mechanistic details underlying the observed SkM phenotypes upon HS, we analyzed the gene expression levels of the F box E3 ligase Fbxl21, which stabilizes molecular clock components [36] and is further involved in SkM maintenance [37]. Further, we studied a direct target of Fbxl21, namely the core sarcomeric component Titin cap (Tcap) capping titin proteins to manage SkM Z-disk maintenance [38]. In GAS, we found no significant differences in the gene expression levels of Fbxl21 and Tcap (fig. S1a, S1b) between CTR and HS mice. In SOL, Fbxl21 gene expression was not different between conditions (fig. S1a). However, we reported a significantly lower expression of Tcap (fig. S1b).
These data show that severe SkM phenotype alterations induced by HS involve minor changes in genes related to circadian SkM maintenance and sarcomeric Z-disk control in a SkM type-dependent manner.
HS conditions introduce coupled changes in metabolism-related genes, mechanosensor and circadian clock genes
As reported above, we found that HS conditions resulted in marked alterations of mechanosensor, circadian clock and metabolism-related genes.
In GAS, we found that the crucial oxidative metabolism-related gene Nrf1 showed no correlation with Clock under CTR conditions (fig. S2a). Interestingly, in HS conditions, a strong correlation between these genes was observed (fig. S2b). We observed a similar pattern for Nrf1 and the mechanosensor Ilk1 under CTR (fig. S2c) and HS (fig. S2d) conditions. Next, the glucose metabolism-related gene Slc2a4 was correlated with the circadian clock gene Bmal1 as well as with the mechanosensor gene Fermt2. Under CTR conditions, we did not observe a correlation between Slc2a4 and Bmal1 (fig. S2e), whereas the HS intervention caused a strong correlation between these two genes (fig. S2f). We found a comparable pattern for the correlations between Slc2a4 and Fermt2 in CTR (fig. S2g) in contrast to HS (fig. S2h) mice.
In SOL, we report that Nrf1 shows a strong correlation with Clock under CTR conditions (fig. S2i) and HS conditions (S2j). We observed a similar pattern between Nrf1 and the mechanosensor Ilk1 under CTR (fig. S2k) and HS (fig. S2l) conditions. We also correlated Slc2a4 with the circadian clock gene Bmal1 as well as with the mechanosensor gene Fermt2. We observed strong correlations between Slc2a4 and Bmal1 under CTR (fig. S2m) and HS conditions (fig. S2n). We found a comparable pattern for the correlations between Slc2a4 and Fermt2 in CTR (fig. S2o) and HS mice (fig. S2p).
These data demonstrate clearly that metabolism-related genes interrelate with both mechanosensor and circadian clock genes specifically under severe SkM-devastating conditions further pointing to a mechanistic interplay between mechanosensors and circadian clock pathways.
Mechanosensor and circadian clock gene expression profiles are related
We further used correlation analysis to examine the relationship between the two crucial mechanosensors Ilk1 and Fermt2 with the core circadian clock components Clock and Bmal1.
In GAS, we found that Ilk1 and Bmal1 gene expressions show strong correlations under both CTR (fig. S3a) and HS conditions (fig. S3b). Similarly, we observed strong correlations between Ilk1 and Clock under CTR (fig. S3c) and HS conditions (fig. S3d). Similar to Ilk1 and Bmal1, we demonstrate strong correlations between Fermt2 and Bmal1 under both CTR (fig. S3e) and HS (fig. S3f) states. Fermt2 and Clock correlations show a comparable pattern with strong correlations between both genes in both CTR (fig. S3g) and HS (fig. S3h) mice.
In SOL, we reported that Ilk1 and Bmal1 gene expressions showed strong correlations under both CTR (fig. S3i) and HS conditions (fig. S3j). Similarly, Ilk1 and Clock showed strong correlations under CTR (fig. S3k) and HS conditions (fig. S3l). Similar to Ilk1 and Bmal1, we demonstrated strong correlations between Fermt2 and Bmal1 under both CTR (fig. S3m) and HS (fig. S3n) states. Fermt2 and Clock correlations showed a comparable pattern with strong correlations between both genes in both CTR (fig. S3o) and HS (fig. S3p) mice.
Collectively, our data demonstrate that the majority of the tested genes of the mechanosensor clusters are interrelated under both sedentary control as well as under SkM-devastating HS conditions. Hence, these findings could potentially point towards a regulatory connection between these pathways in SkM tissue.
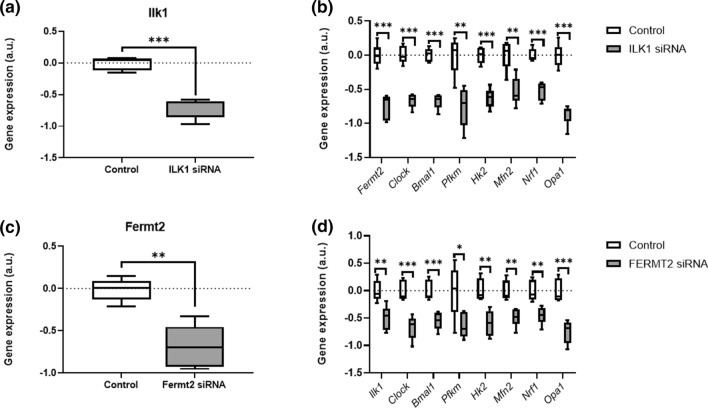
Knockdown of either Ilk1 or Fermt2 in C2C12 myotubes downregulates mechanosensor, molecular clock and metabolism-related gene expression
Given the strong correlations between mechanosensors and molecular clock components found in SkM upon a period of unloading, we aimed to mechanistically establish a potential connection between both pathways in muscle. To this end, we used a siRNA approach to genetically reduce the expressions of the mechanosensors Ilk1 and Fermt2 in C2C12 myotubes.
Relative to control siRNA, the Ilk1 siRNA-treated C2C12 myoblasts showed a 69.49% reduction in the expression levels of Ilk1 (Fig. 6a). We next examined how this knockdown in Ilk1 expression affected the gene expression levels of core costamere (i.e., Fermt2), clock components (i.e., Clock, Bmal1) and metabolic markers. Here, we report that Ilk1 siRNA significantly reduced Fermt2 gene expression levels (Fig. 6b) compared to control siRNA. Moreover, gene expression of Clock and Bmal1 (Fig. 6b) was significantly reduced in Ilk1 siRNA-treated C2C12 myoblasts relative to control. In addition, Ilk1 siRNA-treated C2C12 myoblasts showed a strong decline in the expression levels of metabolic markers Pfkm, Hk2, Mfn2, Nrf1 and Opa1 (all Fig. 6b).
Fig. 6.
Knockdown of either Ilk1 or Fermt2 in C2C12 myotubes alters the expression of mechanosensor, molecular, and metabolism-controlling clock genes. Relative gene expression profiles of a Ilk1, b Fermt2, Clock, Bmal1, Pfkm, Hk2, Mfn2, Nrf1 and Opa1 between Ilk1 SiRNA-treated (n = 6) and control SiRNA-treated (n = 6) C2C12 myotubes. Relative gene expression profiles of c Fermt2, d Ilk1, Clock, Bmal1, Pfkm, Hk2, Mfn2, Nrf1 and Opa1 between Fermt2 siRNA-treated (n = 6) and control siRNA-treated (n = 6) C2C12 myotubes. Expression is relative to the housekeeping gene Rpl41. Statistical analysis used was Student’s t test. *p value < 0.05; **p value < 0.01; ***p value < 0.001
Finally, we used an additional siRNA approach knockdown Fermt2 in C2C12 myotubes to further relate mechanosensors, core clock components and metabolism. Relative to control siRNA, the Fermt2 siRNA-treated C2C12 myoblasts showed a 66.67% reduction in the expression levels of Fermt2 (Fig. 6c). We next examined how this knockdown in Fermt2 expression affected the gene expression levels of core costamere (i.e., Ilk1), clock components (i.e., Clock, Bmal1) and metabolic markers. Here, we report that Fermt2 siRNA significantly reduced Ilk1 gene expression levels (Fig. 6d) compared to control siRNA. Similar to the Ilk1 siRNA approach, Fermt2 siRNA-treated C2C12 myoblasts showed significantly reduced gene expression levels of Clock and Bmal1 relative to control (both Fig. 6d). This was accompanied with a significantly decreased expression of Pfkm, Hk2, Mfn2, Nrf1 and Opa1 (all Fig. 6d) compared to control siRNA.
Collectively, both Ilk1 and Fermt2 siRNA approaches further strengthen our hypothesis of a regulatory connection between both pathways in SkM tissue. This finding unambiguously demonstrates that core mechanosensors causatively regulate SkM molecular clock machineries.
Discussion
The present study demonstrates that SkM-wasting conditions, such as clinically relevant unloading, cause marked alterations of mechanosensor gene expressions. In Gas, these alterations lead to an uncoupling of canonical molecular clock gene expressions (SkM atrophy without any modification of molecular clock gene expression), while expression of these molecular clock genes is concomitant to muscle loss in SOL. Furthermore, we report considerable modifications of metabolism-related gene expressions. We demonstrate strong dependencies between changes in mechanosensors, molecular clock and metabolism-related gene expression profiles and hence identify these genes as potentially novel mechanistic routes that participate in SkM-devastating phenotypes for exploration in follow-up research. Finally, we distinguish, through mechanistic approaches, the mechanosensors Ilk1 and Fermt2 as yet unidentified novel control hubs of the molecular clock and hence metabolism machinery in SkM.
Loss of SkM mass significantly impacts general health since SkM represents the largest metabolically active tissue [1]. Many processes regulate the maintenance of SkM mass and its physiological metabolism [12, 19, 20]. However, the mechanosensor and molecular clock machineries remain underexplored in SkM regulation, including models relevant for clinical conditions with associated modifications in musculoskeletal loading. In this study, we used a hindlimb suspension model to successfully induce a reduction in SkM CSA and an increase in catabolic signaling. These data collectively demonstrate that our HS model optimally reflects clinical phenotypes of severe SkM mass loss and therefore justifies investigating in detail the underlying mechanisms. We demonstrate that molecular clock genes respond in a highly muscle-specific manner to HS. While only a few molecular clock-related genes were upregulated in GAS, we found that the majority of molecular clock-related genes were significantly lower in SOL in HS compared to CTR conditions. Our data are consistent with one previous report, which found higher Clock, Bmal1, and Per2 gene expressions in GAS upon denervation-induced SkM immobilization [21]. In addition, we reported significantly lower expression levels of the nuclear receptors RevErba and RevErbb in SOL but not in GAS between HS and CTR mice. This finding, however, does not explain the downregulation of Clock and Bmal1 in SOL, since RevErba and RevErbb represent the negative transcriptional regulators of Clock and Bmal1 [15]. Together, our data demonstrate a high sensitiveness of the molecular clock gene machinery to SkM unloading and that the type of SkM (predominantly fast- vs. slow-twitch fibers) plays a pivotal role assigning the properties fiber types a leading control herein. This further underlines the necessity for careful discriminations between fiber types and highlights the importance considering SkM fiber type-specific load management to maintain appropriate intrinsic clock system function to preserve SkM functionality.
Mechanical unloading affects the mechanosensor-related gene profiles. Mechanosensors constitute a family of subsarcolemmally located proteins, critically responsible for mechanosensing and related signal transduction in SkM fibers [7]. By this, mechanosensors control transcription processes. Very recently, our group was the first to demonstrate that the mechanosensor Ilk1 controls SkM myosin heavy chain expressions and, thus, SkM metabolic properties [7]. Beside Ilk, the mechanosensors Fermt2 [11], Tln2 [12] or Pxn [39], contribute to SkM integrity and functionality. In GAS, our present data demonstrate that mechanosensor protein expression levels, but not gene expression levels (except for Tln2), were higher in HS compared to CTR mice. In addition, in SOL we observed that both mechanosensor genes and proteins were reduced. Expression of the mechanosignaling molecule Yap did not change in GAS, while we found significantly lower Yap expression in SOL of HS mice. Yap controls focal adhesion assembly [40], the formation of mechanosensor-built costamere structures [10] as well as myogenesis and SkM homeostasis [41]. Further, Yap nuclear localization regulates focal adhesion protein (incl. Tln2, Pxn and Vcl) translation [40]. Hence, the downregulation of Yap in SOL matches regulatory signals of the mechanosensors. With respect to SOL, our data are in line with previous results of downregulated mechanosensor expressions upon one week of botulinum toxin A-induced SkM immobilization [7]. In contrast, we did not observe downregulated mechanosensor expressions in GAS upon unloading despite a significant reduction in SkM CSA. This finding is interesting as it points towards the possibility that mechanosensor and clock genes remain stable under atrophying conditions to stimulate pathways that work against the devastating muscle mass loss (i.e., protein synthesis). Since mechanosensors show specific type 1 or type 2 fiber expressions [7], differences in SkM fiber type distribution between SOL (predominantly slow-twitch type 1 fibers) and GAS (predominant fast-twitch type 2 fibers) could plausibly explain these variational patterns. Different reports showed that type 2 fibers are less susceptible to unloading-induced SkM mass loss compared to type 1 fibers [42, 43]. Hence, the herein detected mechanosensor upregulations could relate to the fiber type characteristics of GAS. Taken together, our mechanosensor-related data highlight the dynamic of this SkM-controlling system and demonstrate that mechanical unloading does not automatically result in mechanosensor downregulation, but that these mechanosensitive components contribute to SkM regulation even under unloading situations.
In line with the eminent role of SkM for whole-body metabolism and SkM metabolism undergoing adaptive remodeling upon unloading [44], we subsequently investigated the impact of HS-induced unloading on metabolism-regulating genes. We found altered expression of genes involved in the regulation of glucose metabolism/glycolysis. More specific, upregulation for Hk2 in GAS, but not in SOL, was reported, which is in line with previous studies demonstrating that genes coding for glycolysis-controlling enzymes, i.e., Hk2 and Pfkm, were upregulated upon unloading [44]. Conversely, in SOL, we demonstrate that key players of glucose uptake [34], i.e., Slc2a4 and Tbc1d1, were lower in HS compared to CTR mice, whereas GAS showed unaltered expression profiles.
Furthermore, the synergistic action of mitochondrial genes, involved in maintaining ATP levels and hence regulation of SkM metabolism, was explored. Upon (un)loading, mitochondrial adaptations comparable to the once reported for glycolysis occur [35]. For instance, Opa1, Nrf1, and Mfn2 are known to regulate mitochondrial fusion and biogenesis, while, e.g., Cd36, is involved in fatty acid transportation [35]. Interestingly, our results demonstrate significant upregulations of Nrf1 in GAS upon unloading, whereas Opa1 and Cd36 were reduced in SOL. This result is striking, because SkM unloading is believed to induce a shift towards glycolytic metabolism [44]. Specifically in SOL, our data seem to indicate that upon HS the overall energy provision through classical pathways is reduced. Furthermore, metabolic pathways also involve protein biosynthesis-related mechanisms, which require Mtor and P70s6kb1 upregulations [45]. We confirm downregulation of these protein biosynthesis-stimulating pathways [2] in SOL with significantly lower gene expression levels of Mtor1 upon unloading, while we report no changes in these muscle anabolic mediators in GAS. Additional analyses showed that SkM-specific Z-disk-maintaining Tcap transcription was lower in SOL, but unaltered in GAS following unloading. In addition, the upstream located circadian rhythm-controlled E3 ligase Fbxl21 and the ATP- and UPS-independent PA28gamma [46], did not change. Altogether, these data highlight that minor SkM-dependent changes in genes involved in sarcomeric Z-disk control and circadian SkM maintenance underlie the unloading-induced alterations in SkM phenotype as reflected in the loss of the CSA, indicative of muscle atrophy.
Strong muscle-specific co-dependencies between mechanosensors, molecular clock and metabolism-related genes under SkM-devastating conditions could be confirmed. This finding is promising as it would allow to identifying potentially novel players involved in the management of SkM-devastating mechanisms. Although we observed clear correlations between mechanosensors and circadian clock pathways at the mRNA level, it should be noted that such correlations could not be found at the protein level. However, studies have shown that correlations between expression levels of mRNA and protein are poor, hovering around 40% explanatory power [47–49]. Hence, regulations at the transcriptional level do not necessarily correlate with respective protein levels.
Our data highlight that reduced mechanosensor profiles, as evident from the reduced the expressions of the mechanosensors Ilk1 and Fermt2 in C2C12 cells, cause significant reductions in Clock and Bmal1 gene expression profiles and, thus, uncouple the canonical molecular clock pathways (in GAS, but not in SOL). In addition, knockdown of Ilk1 and Fermt2 strongly reduced expression levels of metabolism-regulating genes (i.e., Pfkm, Hk2, Mfn2, Nrf1 and Opa1). These alterations in metabolism-controlling genes further support an interplay between mechanosensors, molecular clocks and metabolism in SkM-remodeling. These results together with those from the HS model unambiguously demonstrate that mechanosensors take a regulatory lead in the molecular clock machinery of SkM and control SkM metabolism.
In conclusion, this study demonstrates that unloading induces an uncoupling of canonical mechanosensor and molecular clock gene regulations (in GAS, but not in SOL) paralleled by alterations of metabolism-controlling genes that underlie SkM atrophy (in SOL). Furthermore, mechanosensors are identified as novel, yet unidentified control hubs of the physiological molecular clock and associated metabolism-related pathways in SkM. Herein, we present gene clusters that have currently not directly been associated with the regulation of loss of SkM mass, namely mechanosensors and molecular clock genes. According to our results, these clusters demonstrate promising potentials to being identified as crucial regulators of clinical phenotypes linked to severe SkM mass loss, e.g., cancer cachexia, metabolic disturbances or aging-related SkM mass loss. In these SkM-devastating conditions, the role of the identified metabolism-controlling pathways may bear translational importance and open new avenues to targeted therapeutic interventions.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors would like to thank Monique Ramaekers for technical assistance during the tissue collections.
Author contributions
Project design: FS; Performing experiments: MV, AVRG, AP, SD; Data analysis: FS, MV; Provided reagents: FS, KK; Drafting the manuscript: FS, MV; Final approval of the manuscript: MV, AVRG, AP, SD, KK, FS.
Funding
Mathias Vanmunster is a recipient of an FWO Fundamental Research PhD scholarship (Project Number 1186720N). Sebastiaan Dalle has received a postdoctoral fellowship (12Z8622N) from Research Foundation Flanders (FWO). The project was funded by a KU Leuven grant (Project Number C14/19/092) as well as an FWO grant (Project Number: G056521N) to Frank Suhr.
Data availability
The data will be made available from the corresponding author upon reasonable request.
Declarations
Conflict of interest
The authors declare that no conflict of interest exists.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Frontera WR, Ochala J. Skeletal muscle: a brief review of structure and function. Behav Genet. 2015;45:183–195. doi: 10.1007/s00223-014-9915-y. [DOI] [PubMed] [Google Scholar]
- 2.Bodine SC. Disuse-induced muscle wasting. Int J Biochem Cell Biol. 2013;45:2200–2208. doi: 10.1016/j.biocel.2013.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bodine SC, Latres E, Baumhueter S, Lai VKM, Nunez L, Clarke BA, et al. Identification of ubiquitin ligases required for skeletal Muscle Atrophy. Science (80) 2001;294:1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 4.Otto A, Patel K. Signalling and the control of skeletal muscle size. Exp Cell Res. 2010;316:3059–3066. doi: 10.1016/j.yexcr.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 5.Giudice J, Taylor JM. Muscle as a paracrine and endocrine organ. Curr Opin Pharmacol. 2017;34:49–55. doi: 10.1016/j.coph.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pedersen L, Hojman P. Muscle-to-organ cross talk mediated by myokines. Adipocyte. 2012;1:164–167. doi: 10.4161/adip.20344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mathes S, Vanmunster M, Bloch W, Suhr F. Evidence for skeletal muscle fiber type-specific expressions of mechanosensors. Cell Mol Life Sci. 2019;76:2987–3004. doi: 10.1007/s00018-019-03026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gabriel BM, Zierath JR. Circadian rhythms and exercise—re-setting the clock in metabolic disease. Nat Rev Endocrinol. 2019;15:197–206. doi: 10.1038/s41574-018-0150-x. [DOI] [PubMed] [Google Scholar]
- 9.Harfmann BD, Schroder EA, Esser KA. Circadian rhythms, the molecular clock, and skeletal muscle. J Biol Rhythms. 2015;30:84–94. doi: 10.1177/0748730414561638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pingel J, Suhr F. Are mechanically sensitive regulators involved in the function and (patho)physiology of cerebral palsy-related contractures? J Muscle Res Cell Motil. 2017;38:317–330. doi: 10.1007/s10974-017-9489-1. [DOI] [PubMed] [Google Scholar]
- 11.Dowling JJ, Vreede AP, Kim S, Golden J, Feldman EL. Kindlin-2 is required for myocyte elongation and is essential for myogenesis. BMC Cell Biol. 2008;9:1–16. doi: 10.1186/1471-2121-9-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conti FJ, Monkley SJ, Wood MR, Critchley DR, Müller U. Talin 1 and 2 are required for myoblast fusion, sarcomere assembly and the maintenance of myotendinous junctions. Development. 2009;136:3597–3606. doi: 10.1242/dev.035857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shear CR, Bloch RJ. Vinculin in subsarcolemmal densities in chicken skeletal muscle: localization and relationship to intracellular and extracellular structures. J Cell Biol. 1985;101:240–256. doi: 10.1083/jcb.101.1.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Legate KR, Montañez E, Kudlacek O, Fässler R. ILK, PINCH and parvin: the tIPP of integrin signalling. Nat Rev Mol Cell Biol. 2006;7:20–32. doi: 10.1038/nrm1789. [DOI] [PubMed] [Google Scholar]
- 15.Schroder EA, Esser KA. Circadian rhythms, skeletal muscle molecular clocks, and exercise. Exerc Sport Sci Rev. 2013;41:224–229. doi: 10.1097/JES.0b013e3182a58a70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andrews JL, Zhang X, McCarthy JJ, McDearmon EL, Hornberger TA, Russell B, et al. CLOCK and BMAL1 regulate MyoD and are necessary for maintenance of skeletal muscle phenotype and function. Proc Natl Acad Sci USA. 2010;107:19090–19095. doi: 10.1073/pnas.1014523107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chatterjee S, Barquero N, Li L, Ma K. Circadian clock gene, Bmal1, regulates skeletal muscle metabolism and development. Endocr Rev. 2011;32:P3–434. [Google Scholar]
- 18.Chatterjee S, Nam D, Guo B, Kim JM, Winnier GE, Lee J, et al. Brain and muscle Arnt-like 1 is a key regulator of myogenesis. J Cell Sci. 2013;126:2213–2224. doi: 10.1242/jcs.120519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dyar KA, Ciciliot S, Wright LE, Biensø RS, Tagliazucchi GM, Patel VR, et al. Muscle insulin sensitivity and glucose metabolism are controlled by the intrinsic muscle clock. Mol Metab. 2014;3:29–41. doi: 10.1016/j.molmet.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harfmann BD, Schroder EA, Kachman MT, Hodge BA, Zhang X, Esser KA. Muscle-specific loss of Bmal1 leads to disrupted tissue glucose metabolism and systemic glucose homeostasis. Skelet Muscle. 2016;6:1–13. doi: 10.1186/s13395-016-0082-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakao R, Yamamoto S, Horikawa K, Yasumoto Y, Nikawa T, Mukai C, et al. Atypical expression of circadian clock genes in denervated mouse skeletal muscle. Chronobiol Int. 2015;32:486–496. doi: 10.3109/07420528.2014.1003350. [DOI] [PubMed] [Google Scholar]
- 22.Yang N, Williams J, Pekovic-Vaughan V, Wang P, Olabi S, McConnell J, et al. Cellular mechano-environment regulates the mammary circadian clock. Nat Commun. 2017;8:1–13. doi: 10.1038/ncomms14287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qi L, Boateng SY. The circadian protein Clock localizes to the sarcomeric Z-disk and is a sensor of myofilament cross-bridge activity in cardiac myocytes. Biochem Biophys Res Commun. 2006;351:1054–1059. doi: 10.1016/j.bbrc.2006.10.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morey-Holton ER, Globus RK. Hindlimb unloading rodent model: technical aspects. J Appl Physiol. 2002;92:1367–1377. doi: 10.1152/japplphysiol.00969.2001. [DOI] [PubMed] [Google Scholar]
- 25.Greiwe L, Vinck M, Suhr F. The muscle contraction mode determines lymphangiogenesis differentially in rat skeletal and cardiac muscles by modifying local lymphatic extracellular matrix microenvironments. Acta Physiol. 2016;217:61–79. doi: 10.1111/apha.12633. [DOI] [PubMed] [Google Scholar]
- 26.Wen Y, Murach KA, Vechetti IJ, Fry CS, Vickery C, Peterson CA, et al. MyoVision: Software for automated high-content analysis of skeletal muscle immunohistochemistry. J Appl Physiol. 2018;124:40–51. doi: 10.1152/japplphysiol.00762.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suhr F, Braun K, Vanmunster M, Bloch W. Acute skeletal muscle contractions orchestrate signaling mechanisms to trigger nuclear NFATc1 shuttling and epigenetic histone modifications. Cell Physiol Biochem. 2019;52:633–652. doi: 10.33594/000000045. [DOI] [PubMed] [Google Scholar]
- 28.Dalle S, Van Roie E, Hiroux C, Vanmunster M, Coudyzer W, Suhr F, et al. Omega-3 supplementation improves isometric strength but not muscle anabolic and catabolic signaling in response to resistance exercise in healthy older adults. J Gerontol Ser A. 2020 doi: 10.1093/gerona/glaa309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maki T, Yamamoto D, Nakanishi S, Iida K, Iguchi G, Takahashi Y, et al. Branched-chain amino acids reduce hindlimb suspension-induced muscle atrophy and protein levels of atrogin-1 and MuRF1 in rats. Nutr Res. 2012;32:676–683. doi: 10.1016/j.nutres.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 30.Solomon V, Goldberg AL. Importance of the ATP-ubiquitin-proteasome pathway in the degradation of soluble and myofibrillar proteins in rabbit muscle extracts. J Biol Chem. 1996;271:26690–26697. doi: 10.1074/jbc.271.43.26690. [DOI] [PubMed] [Google Scholar]
- 31.Ussar S, Wang HV, Linder S, Fässler R, Moser M. The Kindlins: subcellular localization and expression during murine development. Exp Cell Res. 2006;312:3142–3151. doi: 10.1016/j.yexcr.2006.06.030. [DOI] [PubMed] [Google Scholar]
- 32.Watt KI, Turner BJ, Hagg A, Zhang X, Davey JR, Qian H, et al. The Hippo pathway effector YAP is a critical regulator of skeletal muscle fibre size. Nat Commun. 2015;6:1–13. doi: 10.1038/ncomms7048. [DOI] [PubMed] [Google Scholar]
- 33.Sinacore DR, Gulve EA. The role of skeletal muscle in glucose transport, glucose homeostasis, and insulin resistance: implications for physical therapy. Phys Ther. 1993;73:878–891. doi: 10.1093/ptj/73.12.878. [DOI] [PubMed] [Google Scholar]
- 34.Stöckli J, Meoli CC, Hoffman NJ, Fazakerley DJ, Pant H, Cleasby ME, et al. The RabGAP TBC1D1 plays a central role in exercise-regulated glucose metabolism in skeletal muscle. Diabetes. 2015;64:1914–1922. doi: 10.2337/db13-1489. [DOI] [PubMed] [Google Scholar]
- 35.Liesa M, Shirihai OS. Mitochondrial dynamics in the regulation of nutrient utilization and energy expenditure. Cell Metab. 2013;17:491–506. doi: 10.1016/j.cmet.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hirano A, Yumimoto K, Tsunematsu R, Matsumoto M, Oyama M, Kozuka-Hata H, et al. FBXL21 regulates oscillation of the circadian clock through ubiquitination and stabilization of cryptochromes. Cell. 2013;152:1106–1118. doi: 10.1016/j.cell.2013.01.054. [DOI] [PubMed] [Google Scholar]
- 37.Wirianto M, Yang J, Kim E, Gao S, Paudel KR, Choi JM, et al. The GSK-3β-FBXL21 axis contributes to circadian TCAP degradation and skeletal muscle function. Cell Rep. 2020;32:108140. doi: 10.1016/j.celrep.2020.108140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gregorio CC, Trombitás K, Centner T, Kolmerer B, Stier G, Kunke K, et al. The NH2 terminus of titin spans the Z-disc: its interaction with a novel 19-kD ligand (T-cap) is required for sarcomeric integrity. J Cell Biol. 1998;143(4):1013–1027. doi: 10.1083/jcb.143.4.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Flück M, Carson JA, Gordon SE, Ziemiecki A, Booth FW. Focal adhesion proteins FAK and paxillin increase in hypertrophied skeletal muscle. Am J Physiol Cell Physiol. 1999;277:C152–162. doi: 10.1152/ajpcell.1999.277.1.C152. [DOI] [PubMed] [Google Scholar]
- 40.Nardone G, Oliver-De La Cruz J, Vrbsky J, Martini C, Pribyl J, Skládal P, et al. YAP regulates cell mechanics by controlling focal adhesion assembly. Nat Commun. 2017;8:15321. doi: 10.1038/ncomms15321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fischer M, Rikeit P, Knaus P, Coirault C. YAP-mediated mechanotransduction in skeletal muscle. Front Physiol. 2016;7:1–12. doi: 10.3389/fphys.2016.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gardetto PR, Schluter JM, Fitts RH. Contractile function of single muscle fibers after hindlimb suspension. J Appl Physiol. 1989;66:2739–2749. doi: 10.1152/jappl.1989.66.6.2739. [DOI] [PubMed] [Google Scholar]
- 43.Augusto V, Padovani CR, Gerson E, Rocha C. Skeletal muscle fiber types in C57BL6J Mice. Braz J Morphol Sci. 2004;21:89–94. [Google Scholar]
- 44.Stein TP, Wade CE. Metabolic consequences of muscle disuse atrophy. J Nutr. 2005;135:1824S–1828S. doi: 10.1093/jn/135.7.1824S. [DOI] [PubMed] [Google Scholar]
- 45.Bodine SC, Stitt TN, Gonzalez M, Kline WO, Stover GL, Bauerlein R, et al. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat Cell Biol. 2001;3:1014–1019. doi: 10.1038/ncb1101-1014. [DOI] [PubMed] [Google Scholar]
- 46.Sun L, Fan G, Shan P, Qiu X, Dong S, Liao L, et al. Regulation of energy homeostasis by the ubiquitin-independent REGγ 3 proteasome. Nat Commun. 2016;7:1–15. doi: 10.1038/ncomms12497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Payne SH. The utility of protein and mRNA correlation. Trends Biochem Sci. 2015;40:1–3. doi: 10.1016/j.tibs.2014.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Sousa AR, Penalva LO, Marcotte EM, Vogel C. Global signatures of protein and mRNA expression levels. Mol Biosyst. 2009;5:1512–1526. doi: 10.1039/b908315d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data will be made available from the corresponding author upon reasonable request.