Abstract
Microgravity and space radiation (SR) are two highly influential factors affecting humans in space flight (SF). Many health problems reported by astronauts derive from endothelial dysfunction and impaired homeostasis. Here, we describe the adaptive response of human, capillary endothelial cells to SF. Reference samples on the ground and at 1g onboard permitted discrimination between the contribution of microgravity and SR within the combined responses to SF. Cell softening and reduced motility occurred in SF cells, with a loss of actin stress fibers and a broader distribution of microtubules and intermediate filaments within the cytoplasm than in control cells. Furthermore, in space the number of primary cilia per cell increased and DNA repair mechanisms were found to be activated. Transcriptomics revealed the opposing effects of microgravity from SR for specific molecular pathways: SR, unlike microgravity, stimulated pathways for endothelial activation, such as hypoxia and inflammation, DNA repair and apoptosis, inhibiting autophagic flux and promoting an aged-like phenotype. Conversely, microgravity, unlike SR, activated pathways for metabolism and a pro-proliferative phenotype. Therefore, we suggest microgravity and SR should be considered separately to tailor effective countermeasures to protect astronauts’ health.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-021-04025-z.
Keywords: Cytoskeleton, HMEC-1, Immunofluorescence staining, International Space Station, RNA sequencing, Telomeres
Introduction
For the past 60 years, space exploration has offered experimental biology a totally new way of studying life. A robust body of literature now shows that space flight (SF) elicits a strong adaptive response from biological systems of all degrees of complexity, from whole organisms to single cells, and points to microgravity and space radiation (SR) as the two the most influential among the many physical factors affecting humans in space. Since the first reports [1], numerous studies conducted on a variety of cell models document adaptive changes occurring in SF or simulated microgravity, with the most evident being alterations of cell shape, size, volume, and adherence properties that lead to changes in intracellular signaling, gene expression, cell proliferation and differentiation or even cell death [2–9]. While the effects of radiation are well described, regardless of the association with microgravity, the mechanisms by which microgravity disrupts human physiology still remain largely unknown.
Many health problems reported by astronauts are linked to endothelial suffering. Similar health issues are observed in aging or severely sedentary individuals, with a trait d’union represented by the response of healthy individuals to prolonged bed rest treatments [10]. The mechanical properties of endothelial cells (ECs) provide the ability to resist and adapt to shear stress, cyclic stretch, hydrostatic pressure and in general various types of dynamic forces coming from surrounding tissues. In turn, endothelial dysfunction leads to cardiovascular deconditioning and physiological changes that influence angiogenesis, wound healing and inflammation [11].
ECs have been the subject of studies in simulated microgravity (reviewed in [12, 13]); however, getting a general overview of their adaptive responses is a complex matter, for several reasons, first of all, because of the heterogeneity of experimental conditions and lengths of the protocols used.
In addition, very few studies have been performed in space; most were performed using a variety of microgravity simulators on the ground, each with specific technical issues, and all operating around 10–2/10–3 g, (see [14]) for an accurate review of ground-based facilities and terminology). These values are far below weightlessness or real microgravity (10–6 g) that can be experienced on spacecrafts, such as the International Space Station (ISS), orbiting the Earth. Of note, the gravitational field on the ISS is approximately 89% of that on the Earth’s surface. However, on the station, the two main forces at play, i.e., the gravitation force that pulls toward Earth and the centrifugal force that keeps the ISS in orbit, reach a status quo so that the ISS and its content are in constant free fall, which causes apparent zero g and weightlessness. The continuous weightlessness on ISS is key to the ability to perform extended experiments, and the reason why SF cannot be completely replaced by ground simulators. In fact, zero g can be reproduced on Earth only through drop towers or parabolic flights, but only for very limited time frames, seconds to minutes, which are not enough for most biological studies. In addition, other environmental space parameters are not completely reproducible in the laboratory, including for example SR. The other most important reason is intrinsic to the nature of ECs. In fact, this cell type is characterized by marked heterogeneity. Although ECs share the functional roles of lining blood and lymphatic vessels, they also display different phenotypic traits from their different spatial locations within the body. Models of macrovascular ECs have been studied in simulated and real microgravity [12, 13]. Microvascular ECs, have a fundamental role in maintaining homeostasis and represent a highly sensitive target for the environmental conditions of long-term space missions, and as such have been the subject of several studies in simulated microgravity [15–19]. However, up to now, microvascular ECs have never been exposed to real SF and, for this reason, the literature does not provide a systematic distinction of the effect of SR and microgravity on this cell type, which is critically important for maintaining cardiovascular health.
On this basis, we organized a multifaceted, integrated study to explore the molecular basis of the adaptive responses to SF of a model of human dermal, microvascular ECs, HMEC-1 [20]. From this study, we expected a better understanding of these cells’ mechanobiology, to determine why microvascular ECs are so sensitive to SF.
Although within the numerous constraints imposed by the complex logistics of a space experiment, this articulated study was performed thanks to the opportunity, granted by the European Space Agency (ESA) and the Italian Space Agency (ASI), of preparing 40 bioreactors and uploading the exceptional number of 24 of them onto the ISS. Thanks to adequate sample numbers and to proper reference samples on the ground and at 1g in the onboard centrifuge, we could distinguish, using adequate statistics on all sub-experiments performed, the respective contribution of microgravity and SR to the combined response to SF.
In addition to the expected cytoskeletal reorganization, at transcriptional level we unexpectedly found that some gene pathways were controlled in opposite directions by microgravity or radiation. In general, radiation turned on pathways for endothelial activation (hypoxia, cytokines, inflammation), DNA repair and apoptosis, inhibiting the autophagy flux and promoting an aging-like phenotype, while microgravity instead down-regulated them. Microgravity activated pathways for metabolism and a pro-proliferation phenotype, the opposite to radiation. The net effect observed in SF on individual pathways is derived from the sum of both contributions. Remarkably, the MTORC1 gene pathway was among those inhibited by SR and stimulated by microgravity, suggesting that the net effect of inhibition of autophagy (a defense mechanism against stress) might explain EC frailty in SF.
Materials and methods
Sample preparation, space launch and sample retrieval
All space studies are supported by complex logistics, whose planning is an essential part of the experiments [21–23]. The ENDO campaign (mission patch in Supplementary Fig. 1a) was held in September 2015 in Baikonur, Kazakhstan.
For this study, 40 microincubators designed for space cell culture, named Experimental Units (EUs, Kayser Italia, Livorno, Italy; Supplementary Fig. 1b), were prepared and assembled at the Baikonur Cosmodrome (Kazakhstan). Each one housed one single coverslip for cell culture, which represented the samples we refer to throughout the paper.
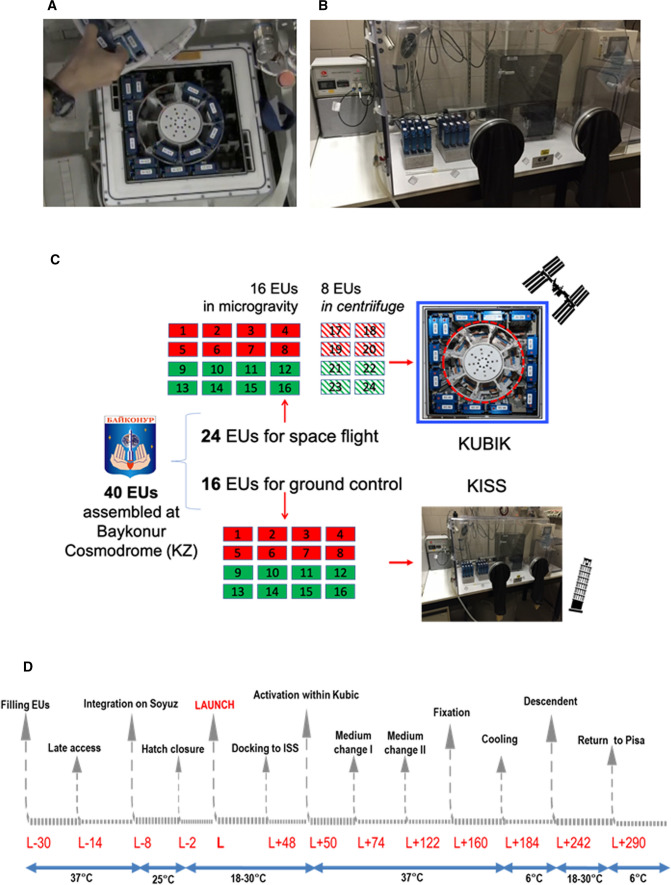
24 EUs out of 40 were brought to the Columbus module of the ISS with the Soyuz TMA-18 M 44S manned mission (Supplementary Fig. 1c), on September 2, 2015, within the IRISS mission of ESA. At rendezvous, the EUs were immediately transferred into the KUBIK facility, an incubator also featuring an internal centrifuge for artificial gravity (Fig. 1a). Of those 24, 16 EUs were exposed to environmental microgravity (μg) and named space-flown-μg samples (SF-μg 1–16), while 8 EUs were set inside the centrifuge reproducing 1g (SF-1g 17–24).
Fig. 1.
A Astronaut Kimiya Yui (Jaxa) extracts the ENDO EUs from KUBIK, at the end of the experiment in the Columbus module of ISS (photo with permission by ESA). Samples aligned against the incubator walls were exposed to μg, while those in the centrifuge (object with circular section in red at the center) were exposed to 1g. B KISS facility hosted in the home laboratory in Pisa, within a thermal incubator. The electronic control of culture procedures allowed running the same protocol, at the same time points, both on ground and on the ISS. C Workflow of the 'Endothelial Cells' study. Of 40 EUs prepared at Baykonur, 24 flew to the ISS. Set inside KUBIC (framed in blue), 16 of them remained in microgravity (SF-μg), and 8 were set in the centrifuge within (circled in red) that can accommodate 8 EUs maximum (SF-1g). The other 16 EUs remained on Earth (GC). Red boxes: half of all EUs were prepared for nucleic acid extraction and fixed with RNAprotect Cell Reagent (Qiagen, USA). Three EUs exposed to μg and two kept in the centrifuge at 1g were used for RNA extraction. Green boxes: half of all EUs were prepared for cell imaging analysis, and fixed with NOTOXHisto (EarthSafe Industries, USA). All coverslips were used up for IF. Coverslips were cut in parts to increase numerosity. For technical replicas, each antibody was hybridized on at least three different parts originated from different coverslips. Diagonal stripes: SF-1g samples. D Timing of the ENDO experiment, including pre-launch (L) and post-L activities in hours, with relevant temperature values, in °C, under the blue arrows
16 EUs, of the 40 prepared in Baikonur, flew to the home laboratory in Pisa with civilian airlines, when the others were flying into space, to be integrated within the KISS facility (Kayser Italia, Livorno, Italy) and to serve as ground control samples (named GC 1–16), (Fig. 1b [23]).
Experiment activation and media exchanges occurred concomitantly in space and on the ground thanks to coordination and communication exchange with the space crew mediated by Biotesc, the Swiss User Support and Operations Centre (USOC) located in Hergiswil (CH).
As always with space rockets, the Soyuz launch phase imposes on the cell samples a complex sequence of accelerations and decelerations [24]. Since, for logistical reasons, it was not possible to fix a set of samples immediately after lift-off, we do not have a control on the condition of cells before docking in the ISS. Therefore, all the consequences of launch were included within the framework of ‘space flight’ conditions.
At the end of the mission, the samples were fixed in space, sent back to Earth with the manned Soyuz TMA-16M 42S on September 12, 2015, retrieved by ESA personnel, brought to the PI’s laboratory in Pisa on September 13, 2015 within an ESA thermal case Yellow Box (Supplementary Fig. 1d), and stored at 6 °C until processed. The experiment flowchart can be viewed in Fig. 1c, and the timeline is represented in Fig. 1d.
Cell line, cell culture, fixation and storage
HMEC-1 (CDC, Atlanta, GA, USA) is a line of human microvascular endothelial cells (ECs) isolated from human foreskins, transfected with pSVT vector, a pBR-322-based plasmid containing the coding region for Simian virus 40A gene product, large T antigen [20]. The cells express von Willebrand’s factor (vWF) and cell adhesion molecules ICAM-1 and are capable of acetylated LDL uptake [20]. The cell line has been well characterized and shown to retain many of the characteristics of ECs. These immortalized cells, because they are a continuously renewable source of human microvascular ECs, can be used as a replacement for primary human dermal ECs for research studies (https://www.lgcstandards-atcc.org/).
HMEC-1 cells were cultured as in [23]. Briefly, 30.000 cells were seeded per coverslip (Cyclic Olefin Copolymer, COC, IBIDI, Martinsried, Germany), measuring 10.5 × 22 mm, coated with 2% gelatin to improve cell adhesion, in MCDB131 medium supplemented with 10% fetal bovine serum (Life Technologies Corporation, Grand Island, NY, USA), hydrocortisone (1 μg/ml), EGF (10 ng/ml), 1 mM sodium bicarbonate, penicillin (1 U/ml), streptomycin (1 μg/ml), and HEPES (20 mM). COC was chosen for its flexibility, to resist the mechanical stress and hard acceleration during Soyuz launch to the ISS, along with it having good optical transparency for subsequent fluorescence microscopy. The medium and all supplements were from Sigma-Aldrich, Saint Louis, MO, USA, unless otherwise specified. Cells were at passage 3, and tested for mycoplasma contamination (DAPI staining, Sigma-Aldrich, Saint Louis, MO, USA, or MycoAlertTM Mycoplasma Detection Kit, Lonza, Allendale, NJ, USA). Cell cultures were incubated at 37 °C in 5% CO2 and 80% humidity before insertion into the EUs. Once inserted, cells remained at room temperature (RT) from launch (L) to L + 48 h, when the EUs were inserted into KUBIK, onboard ISS, or into KISS on ground. The first medium exchange occurred within KUBIK and KISS preheated at 37 °C; this marked the beginning of the experiment. Injection of the first aliquot of fixative or sterile 1X phosphate buffer saline (PBS) marked the end of the culture protocol (Fig. 1d). HMEC-1 cells were then stored at 6 °C as described previously [21–23].
Overall (Fig. 1c), 20 samples prepared for nucleic acid extraction were fixed with RNA Protect Cell Reagent (Qiagen, Germantown MD, USA; 30 s wash in 1X PBS, 1 ml; 30 min in RNAprotect Cell Reagent, 1:6 in 1X PBS, 1 ml, twice). 20 other samples, prepared for morphological analysis were fixed with NoToxHisto (Scientific Laboratory Device, Des Plaines IL, USA; 30 s in NoToxHisto 1:2 in 1X PBS, 1 ml; 30 min NoToxHisto 1:2 in 1X PBS, 1 ml; 30 min, 1X PBS, 1 ml).
The experimental unit
Cells were cultured as in [21, 23], within EUs that were electronically controlled and completed the scientific protocol for both culturing and fixing without human intervention (Kayser Italia, Livorno, Italy). ENDO EUs were used for other space missions with similar configuration [25–28] and, concisely, consist of a main body and a release system unit (Supplementary Fig. 1b). The main body is a passive unit made of biocompatible plastic material (polyetheretherketone, PEEK) that contains the cell culture chamber (CC), connected through a fluidic path to five cylindrical reservoirs with channels and valves for fluid exchange. The release system unit drives the fluid exchange to and from the CC, which contains a volume of 1.3 ml.
When the release system is fully assembled with the main body, the EU is isolated from the environment and allows a bacteriologically sterile cell culture protocol. An electronically controlled, automated procedure injects the fluid contained in each reservoir, at a controlled time and sequence, into the CC, with simultaneous collection of the exhausted solution in the space located above each piston. As the liquid is set in motion, before entering the CC, it flows slowly to avoid turbulences that cause shear stress on the coverslip. Silicon gaskets guarantee the sterile CC isolation from external environment and prevent different fluids from mixing with each other.
Each fully assembled EU was inserted into a KIC (Kayser Italia, Livorno, Italy) and further in soft transportation bags (STB) for integration onboard the Soyuz rocket. Through KIC, the EU was connected to KUBIK once aboard ISS.
In the ENDO configuration, the five reservoirs contained two aliquots of cell culture medium, one 1X PBS wash and two aliquots of a specific cell fixative. At the end of the procedure, fixed cells were preserved in the appropriate solution until each EU was disassembled and the samples processed.
The KUBIK incubator
KUBIK is a small, temperature-controlled incubator or cooler with removable inserts designed for self-contained, automatic microgravity experiment [29]. KUBIK operates from + 6 °C to + 38 °C. Self-contained automatic experiments can be performed using power provided by the facility. Several removable inserts can be used depending on the experiment requirements. For the ENDO study, 16 samples were accommodated on KUBIK interface plate, which provides electrical power to the experiment, to be exposed to microgravity; eight samples were accommodated within the centrifuge insert, to be exposed to 1g on board (Fig. 1a).
Radiation exposure
Dosimetry data for the ‘Endothelial Cells’ experiment were gathered within the ESA DOSIS 3D experiment, whose aim was to monitor the radiation environment inside the Columbus Laboratory of ISS with active and passive radiation detectors at fixed locations, for the support of ongoing biological experiments. All data from the active radiation detectors were provided and evaluated by Christian-Albrechts University (CAU) and German Aerospace Center (DLR).
The location of the experiment in Columbus was COL1D1. The radiation environment inside Columbus is mapped by passive detector packages (PDPs) mounted at 11 locations, and also by 2 active dosimetry telescopes (DOSTEL-1 and DOSTEL-2) that, together with a data and power unit, were mounted within the DOSIS 3D main box at a fixed location beneath the EPM rack, in Columbus. Based on data analysis, the dose absorbed by ‘Endothelial Cells’ experiment was 259 15 μGy/day and the dose equivalent values were 600 22 μSv/day, during the whole short-duration mission (ESA DOSIS-3D-DLR-REP-BIO-001, 1-3-2016, personal communication to the Principal Investigator).
Cell staining and IF analysis
F-actin was stained with Phalloidin–Atto 550 (Sigma-Aldrich, Saint Louis, MO, USA) on cells grown and fixed on COC coverslips, with a procedure modified from [30]. Briefly, HMEC-1 cells were washed in 1X PBS, treated with 0.1% Triton X-100/1X PBS, with 1X PBS, 0.1% Tween-20/1% bovine serum albumin (BSA, Sigma-Aldrich, Saint Louis, MO, USA), and finally labeled with Phalloidin solution (Acto 550-Phalloidin, 1:200 in 1% BSA).
We performed IF as in [22]. Briefly, cells were washed in 1X PBS, permeabilized with 0.1% Triton X in 1X PBS, washed again and blocked in 1% BSA in 1X PBS, 0.1% Tween-20 at RT, incubated with the primary antibody in blocking solution for 1 h at 37 °C, washed twice in 1X PBS and 0.1% Tween-20, and incubated with the secondary antibody in blocking solution at RT. After being washed twice in 1X PBS and 0.1% Tween-20 at RT, nuclei were stained with 4ʹ, 6-diamidino-2-phenylindole (DAPI, 1:1000 in 1X PBS, at RT), washed again, mounted on COC coverslips with Aqua Poly-Mount (Polysciences, Eppelheim, Germany) and stored in the dark at 4 °C. Antibodies and reagents used for fluorescence analysis are listed in Supplementary Table 1.
Fluorescence intensity was calculated using ImageJ1.50i software (http://imagej.nih.gov), with the correct total cell fluorescence parameter (CTCF) with the formula:
To calculate the fluorescence intensity plot, we used the dedicated ImageJ plugin.
For each experimental group (GC; SF-μg; SF-1g), three independent operators counted at least 60 cells (six cells per at least ten fields). Given the paucity of samples, each coverslip was cut in parts and each part was stained with different antibodies, so as to use each antibody on fragments from at least three independent coverslips per experimental group (each coverslip representing a technical replica). Data are indicated as mean ± SEM and statistical analysis was performed with Student’s t test for unpaired samples, two-way Anova test and Bonferroni multiple comparison post-hoc test. Always, the difference between means was judged statistically significant at p ≤ 0.05. All statistical procedures were performed using GraphPad software (GraphPad Software, San Diego, CA USA).
Nuclear and cellular area, circularity, roundness and solidity
Cellular and nuclear shape were analyzed on samples stained with CTNNB and/or DAPI.
Area and different cell descriptors were calculated with ImageJ 1.50i software as in [31]. In each image, single, randomly chosen cells were contoured with the “polygon selection” function, then the selected single cell was analyzed with the 'shape descriptor' function.
Circularity indicates the trend for a cell to assume a circular shape. Roundness does not significantly differ from circularity, except for the introduction in the roundness formula of the major axis’s variable, a parameter that is influenced by cell dimension, which in our case was similar between the GC and FM samples. In geometrical terms, solidity describes the stiffness and deformability of an object and it is related to spatial complexity. High solidity indicates a lower deformability of the cells. The values of cell circularity, cell roundness and cell solidity vary between 0 and 1, with 0 = low level and 1 = high level, according to the formulas:
with A the cell area; p the cell perimeter; CA t cell convex area; ma the cell major axis.
The mitotic index (MI)
MI was calculated as the number of cells undergoing all stages of mitosis, (highlighted with DAPI staining), divided by the total number of cells. Each percentage is the average of N = 17 fields, for each of the three sample types (GC; SF-μg; SF-1g), taken randomly from coverslips of all EUs, for a total of about 700 cells per each sample type. Analysis was performed by two independent operators. Test: one-way ANOVA.
2D Nuclear eccentricity
Nuclear eccentricity describes the position of the nucleus with respect to the cell morphology. Eccentricity varies between 0 and 1. E = 0 means the centers of the nuclei and cell body are coincidental, while E = 1 means the nucleus is in contact with the cell perimeter, according to the formula:
with AC the distance between the center of the cell body and the center of the nucleus. AB the distance between the center of the cell body and its periphery. CD the distance between the center of the nucleus and the nuclear lamina.
We measured nuclear eccentricity on ten images for GS and ten images for FM samples, according to the formula above [32]. We designated in each single cell examined the A and D points along the line from the center of the cell body (A) through the center of the nucleus (C) to the cell periphery (B).
Nuclear volume
Z-stack images of the entire cell were acquired on a Nikon Ti-e microscope interfaced with an A1 scanning head and 405/488/561/630 laser lines, using a 100 × oil immersion objective (Nikon APO TIRF NA 1.49, Nikon Corporation, Tokyo, Japan). Pixel size was set at 246 nm, and Z-step size (z) at 150 nm. Scan speed was set at 4.4 μs/pixel, and frame dimensions at 512 × 512 pixels. 5–15 nuclei were usually present in each field of view.
The resulting stacks were Z-averaged to provide a map of all nuclei in the field, and from this map a set of ROIs (the “nuclei mask”) representing the maximum area (A) of each nucleus was obtained using the “analyze particles” plugin of imageJ (with parameters: min area 50 μm3, circularity 0.3 infinity). Next, the stack was averaged on xy (2 × 2) and thresholded by measuring the intensity of the background signal in the first frames and subtracting an appropriate threshold level (background average + 5 SD) from each image of the stack. The thresholded stack was then measured using the “nuclei mask” obtained in the previous step and measuring for each frame the percentage of nuclear area whose intensity level was over the threshold (f). Nuclear volume for each slice was estimated by multiplying A*f*z. Volumes found in each frame were finally added to provide the overall nuclear volume. Exclusion criteria for the analysis of the nuclei were: a) nucleus on the edge of the imaging field, b) fluorescence intensity greater than threshold in > 2% of the pixels in the first or last frame of the Z-stack, or c) abnormal morphology of the nucleus (e.g., two coalescent nuclei interpreted as one by the “analyze particle” plugin).
Nucleic acid extraction
Total RNA was extracted with ReliaPrep RNA Cell Miniprep System (Promega, Madison WI, USA). RNA integrity was assessed with agarose gel electrophoresis and ethidium bromide staining/UV light (GenoPlex, VWR, USA), concentration and purity with NanoView Spectrophotometer (GE Healthcare, Pittsburgh, PA, USA).
Genomic DNA (gDNA) was extracted with ReliaPrep gDNA Tissue Miniprep System (Promega, Madison WI, USA); gDNA quality was evaluated with spectrophotometric analysis (NanoView Spectrophotometer, GE Healthcare, Pittsburgh, PA, USA).
Telomere length analysis
The ratio of the absorbance at 260 and 280 nm (A260/280) was used to assess the purity of gDNA. An absorbance ratio of 1.7 < A260/280 < 1.9 was considered acceptable.
Telomere length (TL) was measured using the multiplex quantitative real-time PCR method described previously [33]. Briefly, the relative TL (T/S ratio) was reported as the ratio of telomere repeat copy number (T) to single copy globin-beta gene (S). Four biological replicas were analyzed for GCs, three replicas for SF-μg samples, and the only two available, due to space constraint within KUBIK, for SF-1g. All samples were run in triplicate into 384-well plates in a CFX RT-PCR System (Bio-Rad, Hercules, CA, USA), and the whole test was repeated three times.
Telomere Fw: 5ʹACACTAAGGTTTGGGTTTGGGTTTGGGTTTGGGTTAGTGT3ʹ; telomere Rv: 5ʹTGTTAGGTATCCCTATCCCTATCCCTATCCCTATCCCTAACA3ʹ; globin-beta Fw: 5’CGGCGGCGGGCGGCGCGGGCTGGGCGGCTTCATCCACGTTCACCTTG3’; globin-beta Rv: 5ʹGCCCGGCCCGCCGCGCCCGTCCCGCCGGAGGAGAAGTCTGCCGTT3ʹ.
Transcriptome analysis
The RNA-seq libraries for GC, FM-μg and FM-1g samples were prepared using TruSeq Stranded Total RNA with Ribo-Zero Library Prep Kit (Illumina, San Diego, CA). The libraries were sequenced with Illumina HiSeq 2500 platform and CASAVA version 1.8.2 was used to perform base calling. The quality of the 2 × 125 bp reads was assessed with FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/).
Raw reads were aligned using STAR [34] version 2.5.3 to build version hg38 of the human genome defined by GENCODE release 25. Counts for GENCODE basic annotated genes were calculated from the aligned reads using featureCounts function of the Rsubread R package [35]. Normalization was carried out using edgeR R package [36]. After removal of genes encoded on mitochondrial chromosome, raw counts were normalized according to library size to obtain counts per millions (CPM). Only genes with a CPM greater than one in at least test sample were retained for subsequent analysis. Principal component analysis (PCA) was performed using the top 10% variable genes and the function prcomp of R stats package. Differential analysis was performed with voomWithQualityWeights function of limma R package [37], this function models the mean–variance relationship of logCPM with precision weights and sample-specific quality weights in order to down-weight outlier samples. Then, a linear model is fitted for each gene. All pair-wise comparisons between the three defined groups (i.e., GC, SF-1g and SF-μg) were performed. GSEA [38] was used to determine whether the set of genes transcriptionally affected by microgravity or cosmic radiation was enriched in molecular pathways. Gene sets were taken from the Molecular Signatures Database (http://www.broadinstitute.org/gsea/msigdb/index.jsp). GSEA was applied in pre-ranked mode on genes ranked by moderated t-statistic as calculated by limma analysis, using weighted as metric and 1000 permutations of gene sets. R (version 3.3.1) was used for statistical analysis.
Circular plots were generated to display the genomic location of genes with a p value lower that of 0.05 from the limma analysis in the three comparisons: SF-μg vs SF-1g (microgravity), SF-1g vs GC (radiation) and SF-μg vs GC (SF). Red and green bars indicate up- and down-regulated genes, respectively.
Circular plots were generated using R ggbio package [39].
All transcriptome analysis raw data were deposited in GEO, as GSE157937.
Analysis of chromosome territories
Probe preparation
Whole human chromosomes (HSA) were isolated by microdissection and templates were amplified using degenerate oligonucleotide primed polymerase chain reaction (DOP-PCR) as in [40, 41]. Subsequently, templates HSA1 and HSA17 were labeled with biotin-16-dUTP (Boehringer Mannheim). 450 ng biotin-16-dUTP labeled chromosome paint, 7 µg Cot1 (Invitrogen), 30 µg herring sperm (Invitrogen) and 1/10 volume of 3 M sodium acetate pH 5.4 were ethanol precipitated at − 80 °C for at least 1 h prior to dissolving in the hybridization mixture (50% formamide, 10% dextran sulfate, 2 × saline-sodium citrate (SSC) and 1% Tween-20) for 3 h at 50 °C.
2D FISH
FISH was performed using HSA1 and HSA17 chromosome paints on GC and SF-μg samples only, due to paucity of material for further analysis.
After removal of the film from the coverslips’ bottoms, they were briefly washed twice in 1X PBS prior to permeabilization in pre-cooled methanol for 10 min at − 20 °C. Samples were washed in 1X PBS and further permeabilized in 0.5% saponin (w/v) and 0.5% Triton X-100 (v/v) in 1X PBS for 25 min at RT, then were treated with 0.1 N HCl for 12.5 min at RT to improve nuclear DNA accessibility to the probes, and then washed again in 1X PBS. Samples were further treated with 100 µg/ml RNase A in 2X SSC buffer at 37 °C for 1 h and then washed in 1X PBS prior to an incubation in 50% formamide and 2X SSC for 60–90 min at RT.
GC and SF-μg samples probed with HSA1 and HSA17 chromosome paints were denatured at 75 °C in 70% formamide 2X SSC (pH 7.0) for 3 min, and then 50% formamide 2X SSC (pH 7.0) for 1 min. The probe was denatured for 5 min at 75 °C and allowed to reanneal at 37 °C for 10 min before being pipetted onto a slide on a 75 °C hotplate when the coverslip containing the denatured cells was introduced to the probe and sealed with rubber cement. The sample was hybridized for 48 h at 37 °C, in a humidified environment.
After hybridization, coverslips were washed three times in 50% formamide 2X SSC (pH 7.0) for 5 min each at 45 °C, washed again with 0.1X SSC preheated to 60 °C, and placed in a 45 °C water bath three times for 5 min each. Samples were blocked in 3% BSA (Sigma-Aldrich), 0.05% Tween-20, 4X SSC solution for 20 min at 37 °C. Excess block was removed and a mixture containing a 1:200 dilution of streptavidin Cy5 (GE Healthcare Amersham), 4X SSC, 0.05% Tween-20, 3% BSA was applied to the sample and incubated at 37 °C in a humidified environment for 30 min. The coverslips were washed three times, 5 min each, in 4X SSC, 0.05% Tween-20, in darkness, at 42 °C prior to a brief rinse in deionized water and mounting in Vectashield anti-fade mounting media (Vectorlabs) containing DAPI as a counterstain.
Nuclei were examined with a Leica epifluorescence microscope (DM4000) using an HCX PL FLUOTAR 40×/0.75 objective and acquired with a DFC365FX camera. The images were pseudocoloured and analyzed using LAS AF version 4.5.0 software (Leica Microsystems, Danaher, Wetzlar, Germany).
Results
Logistics and samples
The possibility of performing experimental biology in SF is extremely scarce. The preparation of biological samples for space experiments is subject to complicated logistics; in addition, the payload available for each study is usually extremely limited. Therefore, a biology experiment in SF requires careful, highly systematic planning to minimize the possibility of technical failure and loss of data. Also, it is necessary to translate biological requirements of the model system into engineering terms, e.g., in our case to design the proper cell culture conditions taking into account the most likely mission scenarios, which may vary sensibly, and include and test all the possible variables. The preparation phase of the ‘Endothelial Cells’ study that received the operation name ENDO was described in [22], with the quality assessment of the material that returned from space in [23].
The workflow schematic is displayed in Fig. 1c and the experiment timeline is described in Fig. 1d.
Of the 40 samples prepared for the study, 20 were fixed with RNAProtect reagent (Qiagen, USA). Of those, eight were used for RNA-Seq analysis, ten were used for DNA analysis (for genome-wide methylome and telomere length studies). However, two coverslips did not yield sufficient sample for any further analysis. Methylome analysis is still ongoing and is not described here.
Twenty samples were fixed with NoToxHisto (Scientific Laboratory Device, USA), cut in separate sections and prepared for immunofluorescence staining and analysis (modified from [22]).
Fluorescence and Immunofluorescence (IF) analysis
Cell viability, morphology and the cytoskeleton
At opening, all coverslips of the three experimental groups (GC, SF-μg, SF-1g) were almost confluent [23]. The mitotic index (MI) of the cultures could still be scored with the following results: GC = 2.85%; SF-μg = 3.8%; SF-1g = 2.61% with no significant difference between the three samples, implying that proliferation had not been impacted.
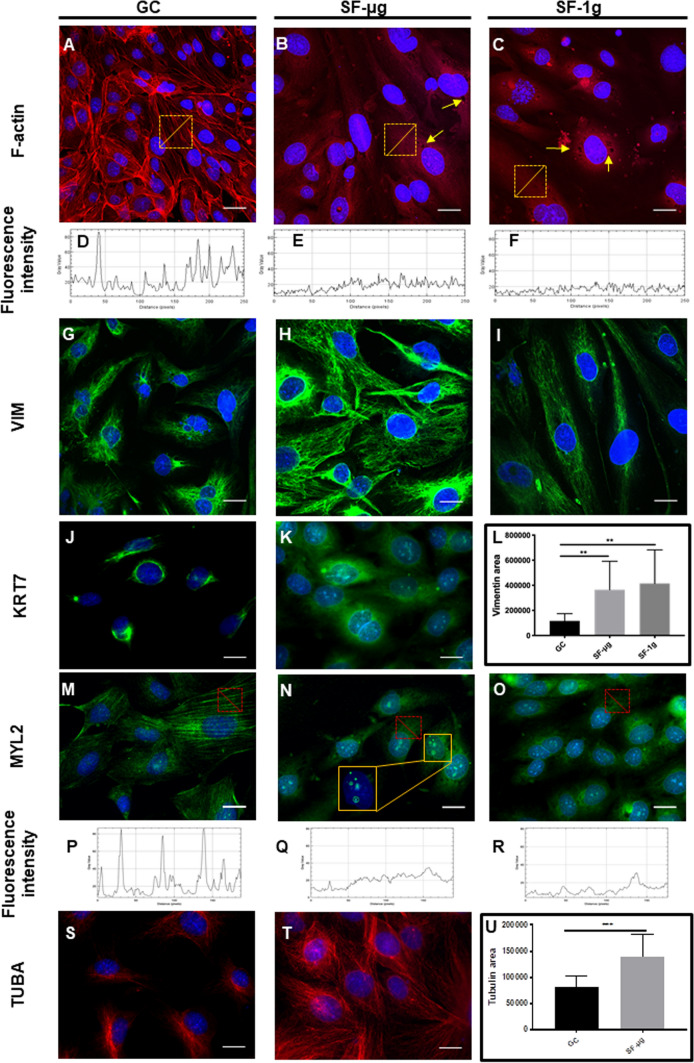
Following mitotic index, we assessed the morphology, presence and distribution of cytoskeletal elements in SF and GC cells beginning with F-actin, a powerful indicator of overall cytoskeletal organization. The GC samples displayed well-defined actin stress fibers and peripheral fibers (Fig. 2a). On the other hand, SF-μg and SF-1g samples displayed a dramatic reorganization of F-actin, with peripheral and stress fibers virtually absent, reduced to a mesh-like structure. On these F-actin meshes, we observed the presence of stress granules, dark accumulations within the cytoplasm, that were often found in all SF samples (Fig. 2b, c, yellow arrows). The analysis of F-actin fluorescence intensity profile revealed a considerable decrease in the fluorescence signal associated with stress fiber peaks in SF HMEC-1 cells (Fig. 2d–f), indicating that not only the distribution had been altered by SF but also the amount of F-actin.
Fig. 2.
Cytoskeletal markers. For analysis of protein markers, cells were fixed with NoToxHisto (Earthsafe Laboratories, USA). Images (A-I) were acquired with confocal laser scanning microscope Nikon C2s (Nikon Corporation, Tokyo, Japan), and (J-P) with Zeiss Axioscope 40 and Zeiss PALM MicroBeam Microscope (Zeiss, Oberkochen, Germany). Magnification is 63× for all. For each experimental group, three independent operators counted at least six cells per at least ten fields. Each coverslip was cut in parts and each part was stained with different antibodies, so as to use each antibody on fragments from at least three independent coverslips per experimental group (N = 3). F-actin showed a remarkable reorganization among the three sample groups. Stress fibers, well distinguishable in GC (A), were no longer evident in SF samples (B, C), as highlighted by the fluorescence intensity plot (D–F). Yellow squares identify the area that was measured. Abundant stress granules, in the form of dark spots, were evident in all SF samples (B, C: yellow arrows). VIM distribution was largely affected, compared to GC (G) in SF, where it showed a more relaxed distribution (H, I), with a statistically significant increase of area (L). KRT7 showed a strong perinuclear, polarized, localization in GC samples (J), as opposed to a largely diffused staining in SF-μg samples (K). Due to KUBIK centrifuge capacity limited to 8 EUs, and the large number of concomitant tests, EUs exposed to 1g onboard were not enough to provide samples for IF with all antibodies otherwise used on SF-μg and GC samples. Therefore, due to lack of material, SF-1g samples were not available for anti-KRT7 staining. MYL2 showed the expected fibers in GC samples (M), which were not present in SF-μg (N), and SF-1g (O) samples, in which we did not observe proper myosin fibers. The lack of proper fibers was highlighted by the fluorescence intensity plot (P–R). SF-μg samples showed well evident dots of nuclear staining (N, inset obtained with digital zoom). TUBA showed tight perinuclear localization in GC samples (S) and cytoplasmic redistribution in SF-μg samples (T) which was significantly enlarged (U), consistently with other cytoskeletal markers. The paucity of samples prevented us from analyzing SF-1g samples. As for KR7 above, due to lack of material, SF-1g samples were not available for anti-TUBA staining
Among intermediate filaments, we analyzed Vimentin (VIM) and Cytokeratin 7 (KRT7). In fact, the disruption of actin fibers, in other models, leads to decreased perinuclear density of VIM and KRT, as a consequence of the functional interplay between the two systems [42]. Indeed, loss of actin dorsal arcs are essential for proper perinuclear accumulation of VIM [43, 44]. The bidirectional transport and establishment of proper arrays of VIM filaments are also organized by microtubules [45]. Therefore, VIM distribution patterns might provide further insights into the overall cytoskeletal configuration alterations by SF. Normally, VIM is predominantly localized in the perinuclear area, often showing a polarized distribution around the nucleus (Fig. 2g). SF cells displayed a diffuse distribution of VIM in the cytoplasm, forming large cotton candy-like meshes, both in SF-1g and SF-μg (Fig. 2h, i) and whose area was significantly larger than in GC (Fig. 2l). Interestingly, in GC samples, anti-KRT7 displayed a tight perinuclear distribution (Fig. 2j), but with a wider, less localized cytoplasmic distribution in SF-μg samples (Fig. 2k).
We then analyzed the distribution of total Myosin Light Chain 2 (MYL2, the sum of phosphorylated and non-phosphorylated MYL2). Myosin is the major cytoskeletal protein in muscle and non-muscle cells, with the ability to self-assemble into bipolar filaments and convert chemical energy of ATP into mechanical work. MYL2-based contractile activity plays important roles in cell spreading, motility, and cell division, in addition to maintenance of cell shape and generation of basal and agonist-induced cytoskeletal tension. This is relevant to ECs, for it contributes to EC function in contexts such as angiogenesis. MYL2 appeared distributed similarly within the cytoplasm of all SF samples, but we did not observe complete myosin fibers as in GCs (Fig. 2m-o). In SF HMEC-1 cells, the analyses of anti-MYL2 fluorescence intensity profile revealed a decrease in the fluorescence signal and a loss of peaks of fluorescence normally associated with fibers (Fig. 2p-r). Of note, we observed discrete dots of staining in the nuclei of SF-μg samples (Fig. 2n, inset) with anti-MYL2 staining.
The anti-tubulin A (TUBA) immunostaining displayed a distribution which was more widely spread-out, with well-defined fibers, in the SF-μg samples when compared to GC samples (Fig. 2s, t). As for the anti-VIM staining, we measured a statistically significant increase of area occupied by anti-TUBA IF signal in SF samples when compared to GC samples (Fig. 2u).
Cell stress, apoptosis and DNA damage
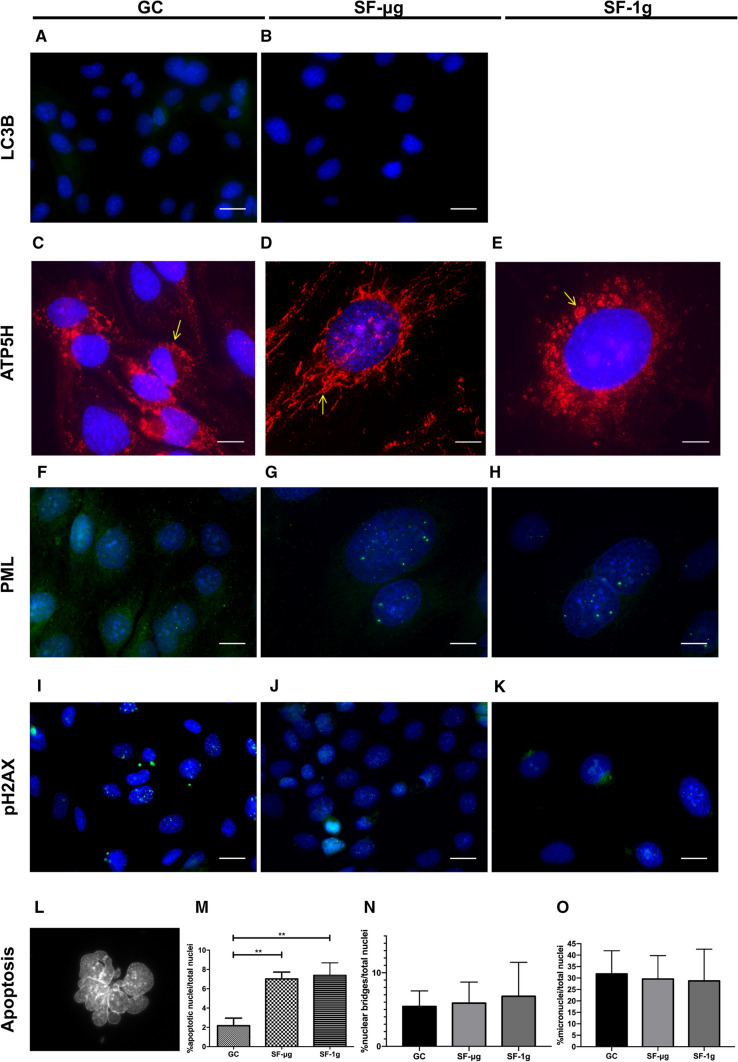
Next, we explored a possible consequence of adaptive stress that could be induced by SF by looking at specific pathways, starting with autophagy. Autophagy is a catabolic process for the autophagosomal–lysosomal degradation of bulk cytoplasmic contents, activated by nutrient deprivation and also by a wide range of other pathophysiological stresses [46]. Specifically, we looked at the light chain 3 isoform B (LC3B) marker protein associated with autophagy. We found an almost undetectable level of LC3B, with no difference between GC and SF-μg samples (Fig. 3a, b). Of note, during the preparation phase of the study, when we challenged HMEC-1 cells with X-rays (1 Gy) to elicit an autophagy response, we obtained a positive result (Supplementary Fig. 2e). This suggested that HMEC-1 cells have a low level of autophagy, which however can be stimulated under certain conditions.
Fig. 3.
Cell stress, apoptosis and DNA damage. LC3B protein was analyzed as an autophagy marker. Both GC (A) and SF-μg (B) samples showed an almost undetectable staining. EUs loaded into the KUBIK centrifuge were not enough to provide SF-1g samples for anti-LC3B IF staining. ATP5H was studied to explore mitochondria subcellular distribution. In GCs, mitochondria showed a perinuclear localization (C), which appeared much more elongated in SF-μg samples (D) and in SF-1g, where we frequently observed aggregates (E). PML was analyzed as marker of DNA damage. GCs were all negative (F), while almost all nuclei of both SF-μg (G) and SF-1g (H) samples showed intense staining characterized by numerous green spots of variable dimension throughout the nucleus. Staining of γH2AX showed abundant foci of various dimensions, also in micronuclei, of almost all GCs nuclei (I), while SF-μg (J) and SF-1g (K) samples showed much fewer positive nuclei, with smaller and less frequent speckles, and negative micronuclei. Apoptotic nuclei were consistently found in SF samples (L, digital zoom), with a statistically significant difference between GCs and SF samples (p < 0.01) but not between SF samples (M). Nuclear bridges (N) and micronuclei (O), although frequently found, were not statistically different among the three groups. Statistical analysis was performed with One way ANOVA and Bonferroni multiple comparison post-hoc tests. Horizontal lines denote mean ± SEM. **p ≤ 0.01. All images were acquired with Zeiss Axioscope 40 and Zeiss PALM MicroBeam Microscope (Zeiss, Oberkochen, Germany). Magnification was 63× for all, except C–H where it was 100×. For each experimental group (SF-μg, SF-1g, GC), three independent operators counted at least 60 cells (six cells per at least ten fields). Each antibody was used on fragments from at least three independent coverslips per experimental group. Each coverslip represented a technical replica
We also analyzed the distribution of a mitochondrial marker to evaluate possible changes in cell energy profiles, also in light of an indication of oxidative stress induced by SF suggested by the transcriptomic study results (see further). We performed IF for the mitochondrial ATP synthase, subunit d (ATP5H), which produces ATP from ADP in the presence of a proton gradient across the membrane generated by electron transport complexes of the respiratory chain. In GCs, mitochondria were distributed within the cytoplasm, mostly in the perinuclear area (Fig. 3c). In the SF-μg samples, mitochondria were assembled in long chains and distributed throughout the cytoplasm (Fig. 3d). In SF-1g samples we found round aggregates of mitochondria (rosettes) in the perinuclear area (Fig. 3e, arrow), resembling aggregates of senescent mitochondria.
DNA damage repair was assessed through well-established markers: promyelocytic leukemia nuclear bodies (PML NBs), and phosphorylated histone γH2AX.
PML NBs are membraneless proteinaceous accumulations of 0.1–1.0 µm in diameter that are part of the nucleoskeleton and regulate a range of nuclear functions, including DNA replication, transcription, stress responses and epigenetic silencing. PML NBs are stress regulated and dynamically recruited to persistent DNA damage lesions by ionizing radiation in primary cell lines, contributing to DNA repair [47–51]. Although the underlying molecular mechanisms remain elusive, PML NBs are found at the sites of DNA damage including double-strand breaks by homologous recombination and are associated with genome maintenance pathways [52]. Remarkably, GCs were all negative for PML bodies (Fig. 3f), while practically 100% of nuclei of both SF-μg and SF-1g samples displayed numerous brightly fluorescent PML bodies throughout the nucleoplasm, although the number and dimension of these bodies varied from nucleus to nucleus (Fig. 3g, h).
γH2AX is a marker of DNA repair found at double-strand breaks that accumulates early in intranuclear spots upon genome damage [53]. Staining of HMEC-1 with anti-γH2AX antibodies yielded an abundance of speckles of various dimensions in GC samples (Fig. 3i), while SF-μg and SF-1g samples displayed a qualitatively different pattern of staining, consisting of fewer positive nuclei with smaller and less frequent foci (Fig. 3j, k). Due to paucity of space-flown samples, we did not apply any established apoptosis assays. However, by scoring cellular morphology, apoptotic cells were statistically more abundant in SFs when compared to GCs (Fig. 3l, m). The frequency of nuclear bridges and micronuclei was similar in all three experimental groups (Fig. 3n, o).
Cell and nuclear shape: mechanotransduction
Mechanotransduction could be described as a multistep process that includes mechanocoupling (transduction of mechanical forces into signals perceived by sensor cells); biochemical coupling (conversion of mechanical signal into a biochemical signal to elicit a cellular response, such as gene activation); transferring of a signal from sensor to effector cells, and then the response of effector cells [54]. The structural link between cytoskeleton and nucleoskeleton, through the nuclear envelope, implies that any change of shape or volume could cause variations in genome transcription.
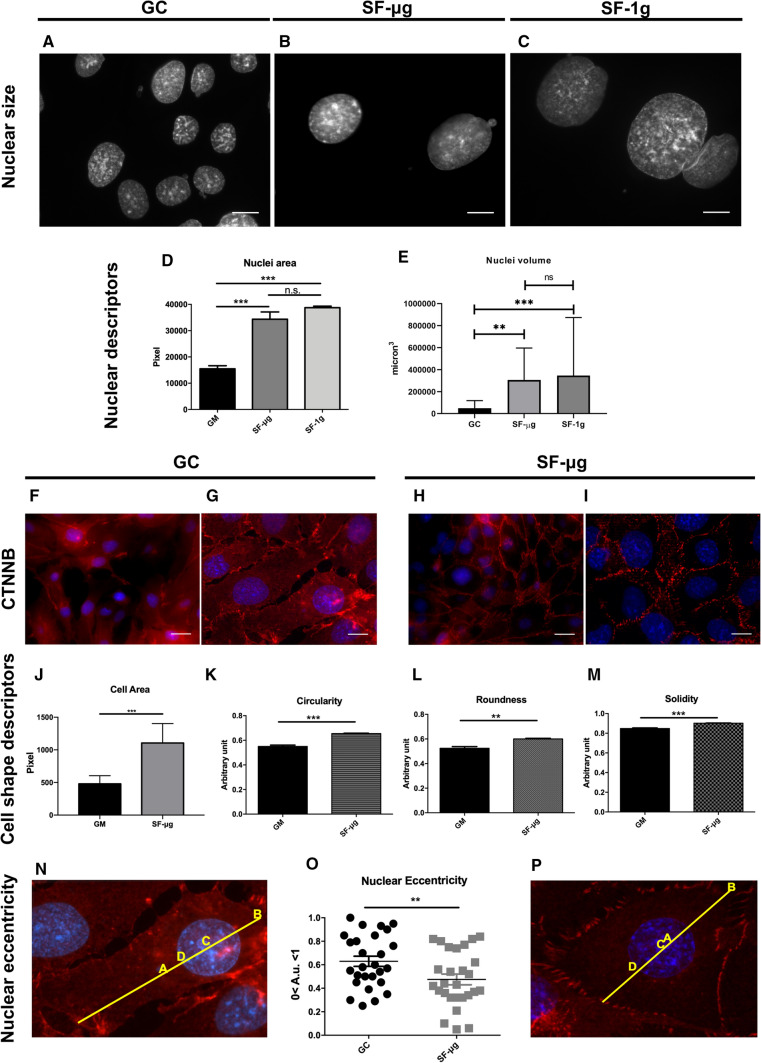
Evaluation of the area of non-apoptotic nuclei displayed significantly larger nuclei in SF samples compared to GCs (Fig. 4a–d, p < 0,001). Interestingly, the volume of nuclei was not significantly different between SF-μg and SF-1g samples with GC nuclei being smaller than both SF samples (Fig. 4e). Other parameters of 2D nuclear shape did not show statistically significant changes (Supplementary Fig. 3a–c).
Fig. 4.
Nuclear and cell shape. Nuclear area was much larger in SF samples (B, C) compared to GCs (A), in a statistically significant way (D). Magnification: 100×. Statistical analysis was performed with One way ANOVA and Bonferroni multiple comparison post-hoc tests. CTNNB was analyzed to evaluate cell–cell junction, cell morphology and nuclear positioning within the cell (F–I). In SF-μg samples the marker was abundantly present at cell junctions (H, I), as opposed to a diffused cytoplasmic localization in GC cells (F, G). Definition of cell borders allowed us to employ cell shape descriptors, i.e., cell area (J), Circularity (K), Roundness (L), Solidity (M) and nuclear eccentricity (N–P). All of them were statistically different between GC and SF-μg samples. Statistical analysis was performed with Student’s t test for unpaired samples. Images were acquired with Zeiss Axioscope 40 and Zeiss PALM MicroBeam Microscope (Zeiss, Oberkochen, Germany). Magnification: 40×: F, H 63×; G, I digital zoom: N, P. For each experimental group, three independent operators counted at least 60 cells (six cells per at least ten fields). Each antibody was used on fragments from at least three independent coverslips per experimental group, each coverslip representing a technical replica. SF-1g samples were not available for IF staining of CTNNB because EUs loaded into the KUBIK centrifuge were not enough for all markers. In all graphs, horizontal lines denote mean ± SEM. **p ≤ 0.01, ***p ≤ 0.001
Catenin-beta (CTNNB)-dependent mechanotransduction is an ancient gene regulatory mechanism, dating back in evolution to at least 600 million years ago [55]. We assessed the CTNNB adaptive response to SF. Regardless of cell density, the marker was distributed diffusely in cytoplasm of the GC cells (Fig. 4f, g). SF-μg cells in contact with each other showed marked aggregation of CTNNB at their cellular boundaries, where it marked well-defined cell–cell junctions (Fig. 4h, i). In general, there seemed to be an increased expression of the marker that allowed us to visualize cell borders and shape (Fig. 4h, i). Images of CTNNB staining were analyzed using different cell shape descriptors, such as area, circularity, roundness and solidity. We found significant changes in the cell area measured in GC and SF-μg cells (Fig. 4j). Circularity, roundness and solidity were also significantly higher in SF-μg than in GC cells (Fig. 4k–m), including nuclear eccentricity (Fig. 4n–p), which could suggest an overall decreased cell motility.
Hippo effectors YAP/TAZ act as an on–off mechanosensing switch by picking up modifications in extracellular matrix (ECM) composition and mechanics. The regulation of their activity has been described by a hierarchical model in which elements of the Hippo pathway are controlled by focal adhesions (FAs). Tuning YAP transcriptional activity leads to the modification of cell mechanics, force development and adhesion strength, and determines cell shape and migration.
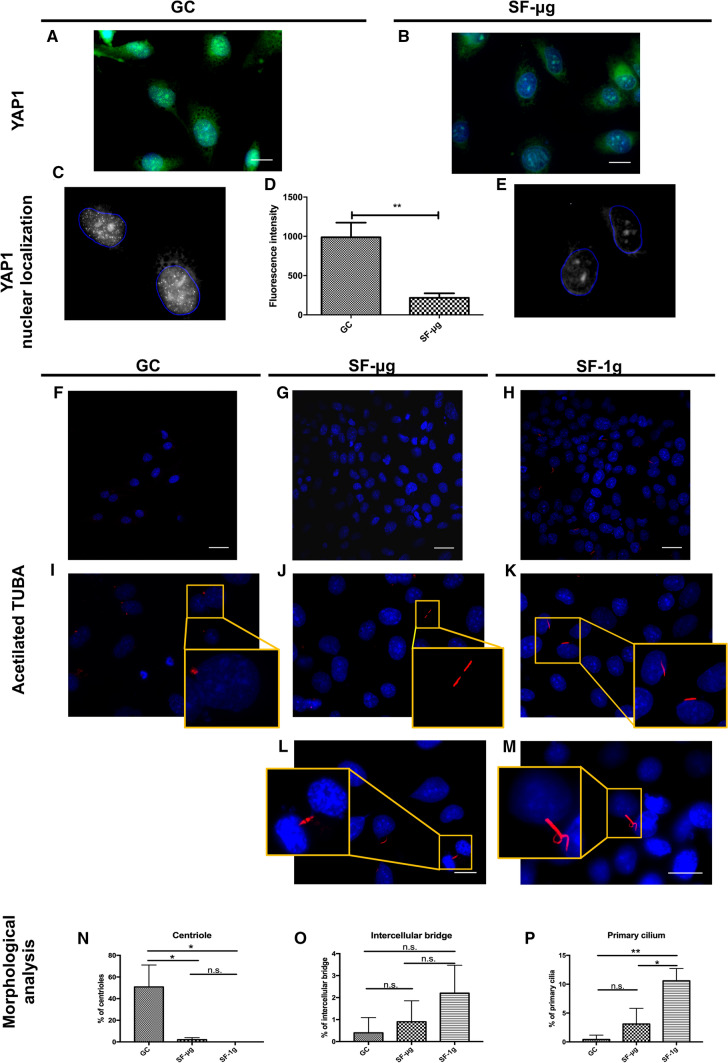
By IF we observed a bright nuclear signal of YAP1 in GC samples, and a very dull signal in SF-μg cells (Fig. 5a–e). Given this result, we explored the status of an important organelle for mechanosensing, i.e., the primary cilium, by employing IF of acetylated tubulin (AcTUBA). Primary cilia are microtubule-based polarized organelles that protrude from the cell surface of many mammalian cell types. On ECs, primary cilia function as calcium-dependent mechanosensors of blood flow, facilitating the regulation of hemodynamic parameters, whereas defects contribute to various vascular dysfunctions [56, 57]. Indeed, with different frequency among the three sample groups, the signal decorated different structures in HMEC-1 (Fig. 5f–h): double dots were identified as centrioles; thin elongated structures as primary cilia; extended structures seemingly connected to each other were identified as interacting primary cilia (cilia-cilia contact) and remnants of mitotic spindles. GC samples featured centrioles only (Fig. 5i). Excitingly, primary cilia were abundantly present in SF-1g samples (Fig. 5k). Primary cilia–cilia contacts were observed in both SF-μg (Fig. 5j) and SF-1g (Fig. 5m), and in the latter sample complex structures were a feature. We observed also labeled microtubules located in intercellular bridges, possibly remnants of late Anaphase bridges (Fig. 5l). Statistical analysis revealed a significantly higher percentage of centrioles in GC samples (Fig. 5n); intercellular bridges were not statistically different among the three groups (Fig. 5o); the primary cilia, however, were much more abundant in SF-1g samples compared to the other groups (Fig. 5p).
Fig. 5.
Elements of signaling and mechanotransduction. YAP1 IF showed nuclear localization in GC samples (A, C), but the signal was not detectable in SF-μg samples (B, E). Fluorescence intensity was statistically different between GC and SF-μg samples (D), with Student’s t test for unpaired samples used for the analysis. Images were acquired with Zeiss Axioscope 40 and Zeiss PALM MicroBeam Microscope (Zeiss, Oberkochen, Germany). Magnification: 63×. For each experimental group (SF-μg, GC), three independent operators counted at least six cells per at least ten fields. Each antibody was used on fragments from at least three independent coverslips per experimental group. Each coverslip represented a technical replica. EUs loaded into the KUBIK centrifuge were not enough to provide SF-1g samples for anti-YAP1 IF staining. AcTUBA is a marker of primary cilium. In GC samples (F, I) the signal highlighted centrioles in different stages of the cell cycle, but no primary cilia (I, digital zoom in the inset). SF samples (G–M) featured other structures, seemingly primary cilia-cilia contact (J, digital zoom in the inset), well-defined primary cilia (K, digital zoom in the inset), overlapping cilia (M), late anaphase bridges (L). Frequency evaluation showed that centrioles were statistically more represented in GC (N) compared with the SF samples. Intercellular bridges (possibly remnants of mitotic spindles) showed a positive trend in SF samples (O). Primary cilia were statistically more represented in SF-1g samples compared with the other two groups (P). Images were acquired with Zeiss Axioscope 40 and Zeiss PALM MicroBeam Microscope (Zeiss, Oberkochen, Germany). Magnification: 63× except L and M (100×). Statistical analysis was performed with one-way ANOVA and Bonferroni multiple comparison post-hoc tests. In all graphs, horizontal lines denote mean ± SEM. *p ≤ 0.05, **p ≤ 0.01
Genome-wide gene expression analysis
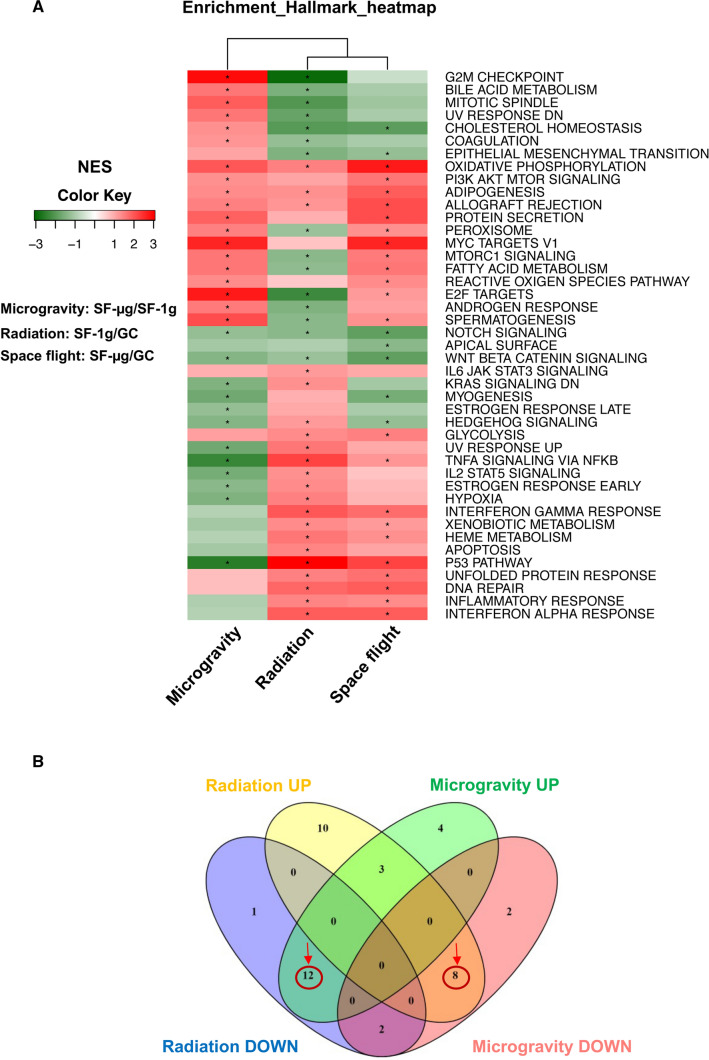
To identify transcriptional changes induced by microgravity, SR, and microgravity and SR combined, we performed RNA-seq and analyzed the transcriptional profiles of three GC, two SF-1g and three SF-μg samples (Supplementary Fig. 4). We performed functional enrichment analysis, using the Gene Set Enrichment Analysis (GSEA) algorithm and the curated Hallmark collections of the Molecular Signature Database, of the following comparisons: i) SF-μg v/s SF-1g (identified as 'Microgravity' in Fig. 6); ii) SF-1g v/s GC ('Radiation' in Fig. 6); and iii) SF-μg v/s GC ('Spaceflight' in Fig. 6). For each gene set, in each comparison, GSEA returned a normalized enrichment score (NES) and a false discovery rate (FDR). The absolute value of the NES represented the degree of the enrichment of the gene set in one of the two groups under comparison, whereas its sign indicated which group was enriched in the gene set (positive if the gene set was enriched in the first group compared to the second, negative vice versa). When using the Hallmark collection, among the most enriched gene sets we found the P53 pathway which was up-regulated in both radiation and spaceflight, while the G2M checkpoint was down-regulated in SF-1g cells v/s GCs (Fig. 6a). Noticeably, assessing the effect of SR or microgravity in the GSEA gene sets, we observed that eight pathways were up-regulated by SR and down-regulated by microgravity (Fig. 6b, intersection of yellow and pink Venn diagrams, red circle). However, 12 other pathways were up-regulated by microgravity, and down-regulated by SR (Fig. 6b, intersection between green and blue Venn diagrams, red circle). The names of gene sets are listed in Table 1.
Fig. 6.
Genome-wide transcriptomic analysis. We performed functional enrichment comparing the following conditions: SF-μg v/s SF-1g (for gene sets affected by microgravity); SF-1g v/s GC (for gene sets affected by SR), and SF-μg v/s GC (for gene sets affected by both microgravity and SR). A GSEA analysis indicated molecular programs activated or repressed in microgravity, radiation and SF. Each row represents a gene set and each column is a comparison. The absolute value of the NES represents the degree of the enrichment of the gene set in one of the two groups under comparison, whereas its sign indicates which group is enriched in the gene set (positive, in red color, if the gene set is enriched in the first group compared to the second; negative, in green, vice versa). Gene sets are significantly enriched with an FDR below 0.25 (*). Row: gene set; column: the comparison between two groups. B To dissect the effect of microgravity from radiation, we analyzed the Venn distribution of the 43 molecular pathways in C. Interestingly, we observed that 12 gene sets up-regulated by microgravity were down-regulated by radiation, whereas eight gene sets down-regulated by microgravity were up-regulated by radiation, highlighting an opposing effect of the two main environmental components of SF
Table 1.
List of molecular pathways included in the Venn diagram of Fig. 6B
| Radiation UP (SF1g vs GC) | Radiation DOWN (SF1g vs GC) | Microgravity UP (SFµg vs SF1g) | Microgravity DOWN (SFµg vs SF1g) |
|---|---|---|---|
| HALLMARK_ADIPOGENESIS | HALLMARK_ANDROGEN_RESPONSE | HALLMARK_ADIPOGENESIS | HALLMARK_ESTROGEN_RESPONSE_EARLY |
| HALLMARK_ALLOGRAFT_REJECTION | HALLMARK_BILE_ACID_METABOLISM | HALLMARK_ALLOGRAFT_REJECTION | HALLMARK_ESTROGEN_RESPONSE_LATE |
| HALLMARK_APOPTOSIS | HALLMARK_CHOLESTEROL_HOMEOSTASIS | HALLMARK_ANDROGEN_RESPONSE | HALLMARK_HEDGEHOG_SIGNALING |
| HALLMARK_DNA_REPAIR | HALLMARK_COAGULATION | HALLMARK_BILE_ACID_METABOLISM | HALLMARK_HYPOXIA |
| HALLMARK_ESTROGEN_RESPONSE_EARLY | HALLMARK_E2F_TARGETS | HALLMARK_CHOLESTEROL_HOMEOSTASIS | HALLMARK_IL2_STAT5_SIGNALING |
| HALLMARK_GLYCOLYSIS | HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION | HALLMARK_COAGULATION | HALLMARK_KRAS_SIGNALING_DN |
| HALLMARK_HEDGEHOG_SIGNALING | HALLMARK_FATTY_ACID_METABOLISM | HALLMARK_E2F_TARGETS | HALLMARK_MYOGENESIS |
| HALLMARK_HEME_METABOLISM | HALLMARK_G2M_CHECKPOINT | HALLMARK_FATTY_ACID_METABOLISM | HALLMARK_NOTCH_SIGNALING |
| HALLMARK_HYPOXIA | HALLMARK_MITOTIC_SPINDLE | HALLMARK_G2M_CHECKPOINT | HALLMARK_P53_PATHWAY |
| HALLMARK_IL2_STAT5_SIGNALING | HALLMARK_MTORC1_SIGNALING | HALLMARK_MITOTIC_SPINDLE | HALLMARK_TNFA_SIGNALING_VIA_NFKB |
| HALLMARK_IL6_JAK_STAT3_SIGNALING | HALLMARK_NOTCH_SIGNALING | HALLMARK_MTORC1_SIGNALING | HALLMARK_UV_RESPONSE_UP |
| HALLMARK_INFLAMMATORY_RESPONSE | HALLMARK_PEROXISOME | HALLMARK_MYC_TARGETS_V1 | HALLMARK_WNT_BETA_CATENIN_SIGNALING |
| HALLMARK_INTERFERON_ALPHA_RESPONSE | HALLMARK_SPERMATOGENESIS | HALLMARK_OXIDATIVE_PHOSPHORYLATION | |
| HALLMARK_INTERFERON_GAMMA_RESPONSE | HALLMARK_UV_RESPONSE_DN | HALLMARK_PEROXISOME | |
| HALLMARK_KRAS_SIGNALING_DN | HALLMARK_WNT_BETA_CATENIN_SIGNALING | HALLMARK_PI3K_AKT_MTOR_SIGNALING | |
| HALLMARK_OXIDATIVE_PHOSPHORYLATION | HALLMARK_PROTEIN_SECRETION | ||
| HALLMARK_P53_PATHWAY | HALLMARK_REACTIVE_OXIGEN_SPECIES_PATHWAY | ||
| HALLMARK_TNFA_SIGNALING_VIA_NFKB | HALLMARK_SPERMATOGENESIS | ||
| HALLMARK_UNFOLDED_PROTEIN_RESPONSE | HALLMARK_UV_RESPONSE_DN | ||
| HALLMARK_UV_RESPONSE_UP | |||
| HALLMARK_XENOBIOTIC_METABOLISM |
Analysis of chromosome territories
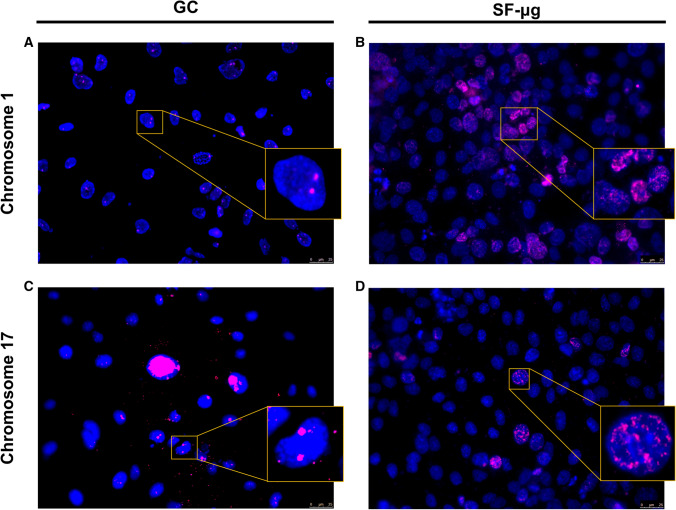
SF was found to have a strong transcriptional impact, with up and down regulation, for genes of the whole genome (Supplementary Fig. 5). Therefore, given the functional link between genome activation and chromosome spatial organization, we asked whether any variations in chromosome territories behavior were detectable in the three experimental conditions. The paucity of material available after the SF narrowed the number of painting probes that could be used for the fluorescence in situ hybridization (FISH) analysis and thus required careful selection. We chose three chromosomes, namely chromosomes 1, 10 and 17, since they represent three different sizes of human chromosomes being large, intermediate and small, respectively, as well as with a range of gene densities from low to high [58]. Due to logistics constraints, the cells were not fixed optimally for FISH protocols. Micro-dissected human chromosomes were used as the initial template to make labeled complementary DNA probes to delineate the chromosome territories. These whole chromosomes paints (WCP) reveal two chromosomes in control human cells routinely [40, 41]. WCP in the control HMEC-1 cells revealed chromosome territories for chromosome 10, but WCP for chromosomes 1 and 17 consistently revealed two foci, not whole territories. However, there being two of these signals in the vast majority of cells, we were convinced they represented the two homologues in the cell nuclei and their relative radial positioning in nuclei fitted with other studies [40, 41, 58]. These chromosomal signals for the two chromosomes 1 and 17 were observed in GCs for chromosomes 1 and 17 (Fig. 7a, c). Most interestingly, specifically in SF-μg it appeared that the genome and the individual chromosomes were affected. The WCP signal was consistently found in speckles and aggregates throughout many of the interphase nuclei (Fig. 7b, d). It is not clear what this unusual distribution of chromosome paint signals are, but they were found in all SF-μg samples processed for FISH. Staining for chromosome 10 in the available experimental samples was unfortunately not successful.
Fig. 7.
Chromosome territories. FISH performed with specific probes for chromosome 1 and 17 showed two signals in ground reference (A, C), and a wide redistribution of territories in microgravity (B, D), with many nuclei featuring abnormal distribution of chromosome painting probes compared to respective ground references. Due to KUBIK centrifuge limited capacity, EUs exposed to 1g onboard were not enough to provide samples for chromosome staining. Nuclei images were acquired with Leica epifluorescence microscope (DM4000) using an HCX PL FLUOTAR objective and a DFC365FX camera. The images were pseudocolored and analyzed using LAS AF version 4.5.0 software. Magnification: 40×/0.75
Analysis of chromosome telomere length
Chromosome telomere length was analyzed on genomic DNA. The relative telomere versus single copy gene (T/S) ratio for GC, SF-1g and SF-μg samples was calculated using the ΔΔCt method with the Pfaffl correction. We found a statistically significant reduction of 54% of the T/S ratio in SF-μg samples (mean value 0.54) compared to the GC samples (mean value 1.17), suggesting that a reduction of telomere had occurred in SF. In the SF-1g samples, we observed a statistically significant increase of 66% (mean value 1.94) compared to the GC samples, possibly induced by SR (p < 0.0001, Fig. 8).
Fig. 8.
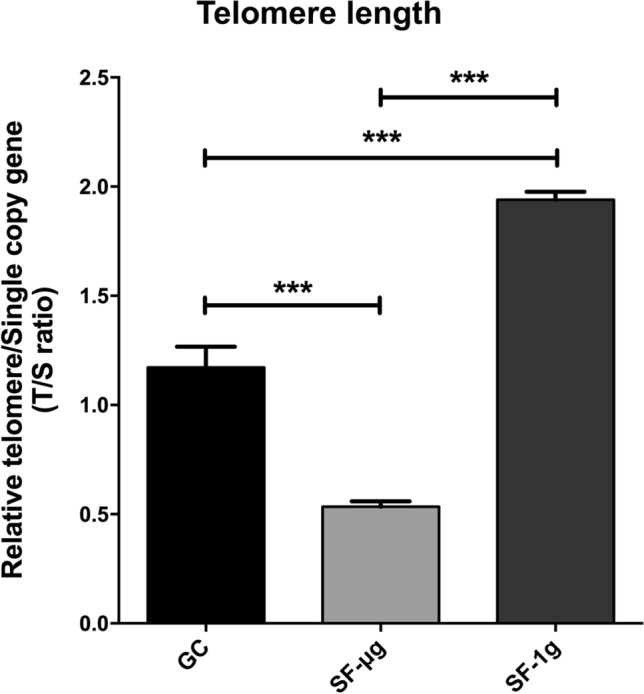
Telomere length analysis. Multiplex PCR showed that the relative telomere length of SF samples changes in a statistically significant way compared to GC samples. Specifically, T/S ratio of SF-μg is about 50% less than GCs. T/S ratio of SF-1g samples is 50% more than GCs. Statistical analysis was performed with one-way ANOVA and Bonferroni multiple comparison post-hoc tests. Horizontal lines denote mean ± SEM. ***p ≤ 0.001
Discussion
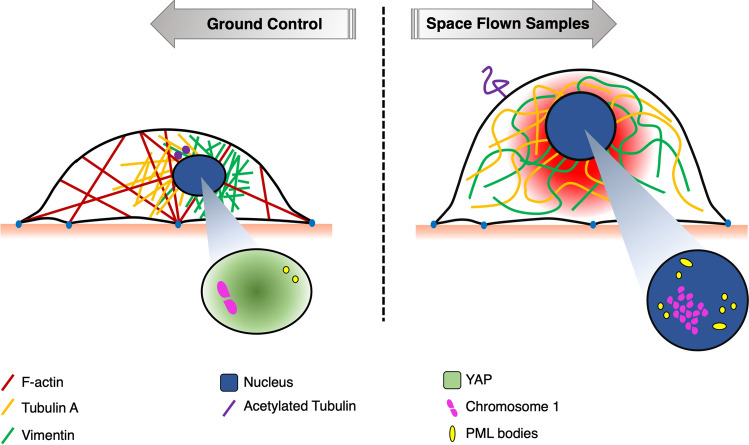
The endothelium is a mechanosensitive, diffuse organ particularly affected by SF. With this work we traced at a cellular and molecular level the basis of the response of ECs to SF, combining morphological observations with transcriptomic and functional cytogenetic data.
Cytoskeleton
The most apparent changes after SF were alterations of cell shape, size and volume, already in part documented after exposure to either real or simulated microgravity [12, 13].
CTNNB staining showed that cell adhesion to the matrix was not particularly affected. Nuclear localization of CTNNB was completely absent in SF-μg, suggesting a minimal Wnt activation in this situation, confirmed by the down-regulation of the Wnt CTNNB gene pathway in SF.
Other analyses of nuclear and cellular parameters suggested important mechanobiological adaptive changes. Regardless of local cell density, nuclear eccentricity of SF samples was smaller than GCs (Fig. 4n–p). Also, the nuclear localization of YAP1 protein was undetectable in SF samples (Fig. 5a–e), and the area of SF cells was significantly enlarged compared to GC (Fig. 4j), along with circularity, roundness and solidity (Fig. 4k–m). Solidity describes in geometric terms the rigidity and deformability of an object. Since cells need to be plastic and pliable to increase their motility, the greater the solidity, the lower the cell deformability, resulting in an increased inability to move. The same applies to circularity and roundness, which represent the tendency of a cell to assume a circular shape, where more circular cells are less spread out and less motile. The data on circularity and roundness for cells in SF strengthened the notion of cells being less motile on a 2D surface in space. In step with morphological observations, transcriptomic data showed the down-regulation by SR of the epithelial–mesenchymal–transition pathway (Fig. 6a). Of note, it was not possible to perform any functional study of motility with SF cells. The equipment available in space did not provide the possibility to do it. Furthermore, to avoid confounding effects linked to the return flight and to ground g, cells were fixed in space and therefore were not exploitable for a live experiments upon return to the home laboratory.
Overall, morphological analysis of microtubules, microfilaments and intermediate filaments showed massive cytoskeletal rearrangements (Fig. 2), consistent with reports in other models [4, 59, 60].
Staining of F-actin revealed a loss of stress fibers and decreased density in all SF samples (Fig. 2). Since early on in history of experimental EC biology in space, F-actin was shown to be affected by SF and simulated microgravity on ground (reviewed in [12, 18, 19]). Here, we were puzzled by the close resemblance of F-actin distribution in SF-μg and SF-1g samples, and thought of two possible explanations. One is that both sample types were exposed to SR and possibly this complex environmental factor affected the cells equally onboard. The other, more likely explanation, is that the 2-day rendezvous flight in μg, experienced by SF-1g samples before integration in the centrifuge onboard, overwhelmed the capacity of HMEC-1 cells to restore F-actin organization at 1g during the following 4 days. In fact, several experiments in simulated microgravity showed a massive reorganization of F-actin, in which cases radiation was certainly not part of the experimental settings (reviewed in [61]).
Actomyosin was also impaired, with some nuclear dots more evident in SF-μg samples (Fig. 2n, inset). Reports of nuclear MYL2 are scarce in literature, but point to a role of the protein, beyond contraction, in transcriptional control of the response to oxidative stress [62]. Indeed, nuclear myosins were recently revealed as integral to genome organization and function [63]. Oxidative phosphorylation was indeed up-regulated by both SR and microgravity, as shown by RNA sequencing (Fig. 6a), which might explain why the nuclear staining was not as strong in SF cells exposed to 1g onboard (Fig. 2o).
IF of TUBA also displayed reorganization of microtubules. Interestingly, an in vitro model of tubulin polymerization showed that microtubule self-organization is gravity sensitive and is inhibited by microgravity [64]. Although in vivo data are missing, this suggests that microtubule disorganization might represent an early event in microgravity-dependent cytoskeletal reorganization. Since transcriptomic results did not show any obvious cytoskeleton-related pathway above the threshold of significance, wider TUBA distribution might originate from increased polymerization or inhibition of depolymerization rate, both highly dynamic phenomena. We would exclude inhibition of depolymerization because it has long been known that inhibition of microtubule depolymerization causes another protein, i.e., VIM, to collapse to the perinuclear region [65]. Indeed, in our system, we observed the opposite, that is a much wider network of VIM fibers in the cytoplasm of SF cells compared to GCs (Fig. 2). In our study, the larger area of VIM staining in HMEC-1 is likely due to a redistribution of the protein and widening of the network, rather than to increased expression (as suggested for other models [66]), as we did not observe transcriptomic change of the corresponding gene. Based on other reports, VIM redistribution happened after several days of spaceflight [66]. Although our data depict the sample at the end of 6-day exposure, we suggest that VIM redistribution might represent a prompt response to microgravity. In fact, treating the same cell type with hypergravity for a few hours also caused a massive dose-dependent change of VIM distribution (in this case, tightening of VIM around the nucleus) again without transcriptional change [67].
In addition to VIM, among intermediate filament proteins providing structural support for the cell to adapt to mechanical stress, we analyzed also KRT7. SF HMEC-1 cells showed increase of the fluorescence signal and a wider network of KRT7 (Fig. 2). Our study is the first to report integrated observations of several cytoskeletal markers in microvascular ECs exposed to real SF. Cumulatively, the reorganized distribution of intermediate filaments and microtubules highlight a theme common to other cell models, that is, the compensation for the loss of mechanical strength caused by F-actin remodeling [5]. Cytoskeleton reorganization can ultimately affect genome function through the Linker of Nucleoskeleton and Cytoskeleton (LINC) complex at the nuclear envelope [68, 69], so that changes of force outside the cell could modulate genome function [70]. This was proved for samples exposed to simulated microgravity [71], and is consistent with our observations of SF samples, both in terms of changes of chromosome territories and changes of genome transcription.
Energy
In light of the robust VIM network rearrangement, we were not surprised to observe that distribution and morphology of mitochondria were also abnormal (Fig. 3). The usual perinuclear distribution observed in GC samples was replaced by different distribution at μg or 1g onboard: thin and elongated fibers in the former group, a seemingly restored perinuclear distribution in the latter, although with enlarged and fragmented mitochondria. The distribution and morphology of mitochondria suggested that the cell energy profile must have changed in space, as highlighted by other authors on the bases of glucose and l-lactate quantification in the medium [66, 72]. A change in mitochondria fusion and/or fission could be ascribed to the observed F-actin redistribution, given actin’s fundamental role in promoting correct fission of mitochrondria [73].
Transcriptomic data has reinforced the suggestion of an altered energy profile, showing increased oxidative phosphorylation gene pathways (Fig. 6).
Signaling
Actin dynamics orchestrate the genesis of primary cilia, a fundamental structure for endothelial function. Disrupted actin polymerization was shown to increase ciliation frequency [74]. On this basis, we explored the status of primary cilia in SF HMEC-1 cells. IF staining of AcTUBA displayed a statistically significant increase of primary cilia in SF-1g samples compared to SF-μg and GC (Fig. 5), in areas of similar cell density. Our interpretation was that the statistically significant increase of primary cilia in SF-1g is due to SR. Indeed, the negative effect of microgravity seems to have been buffered by artificial gravity in the centrifuge.
Unlike other core developmental signaling pathways, vertebrate Hedgehog (Hh) signaling is completely dependent on the primary cilia [57]. Dysregulation of the Hh pathway increases genomic instability and the development of spontaneous and ionizing radiation-induced tumors by an unknown mechanism [75], although in a different cell system, irradiation incidence was shown to correlate positively with increase of primary cilia [76] and with the activation of Hh signaling in cells that survived irradiation [77]. Here, the transcriptomic data demonstrated net down-regulation of the Hedgehog signaling in SF with an up-regulation due to radiation, and a down-regulation with microgravity (Fig. 6). Altogether, the increase of primary cilia suggests a possible deregulation of the whole signaling system, where the increased number of cilia tend to compensate for down-regulation of the Hh signaling pathway.
Cell stress
We then looked at signs of cell stress. The LC3B autophagy marker did not show differences among GC and SF samples, suggesting that either SF did not induce this specific pathway (as other stimulations would do so, see Supplementary Fig. 2), or that the chronic stimulation abated this specific type of response. We propend for the second hypothesis based on the literature, showing in an in vivo model of mouse intestine that radiation persistently activates oxidative stress but down-regulates the autophagy pathway [78].
DNA damage
SF samples showed significantly larger area and volume of nuclei compared to GCs (Fig. 4a–e).
The LINC proteins provide the structural continuum between cytoskeleton and nucleoskeleton, also giving docking sites, with the nuclear lamins, to chromatin on the inner side of the nuclear envelope. For this reason, changes of nuclear morphology have consequences on genome assembly and function. On this basis, we expected to find a different distribution of chromosome territories in SF cells. Indeed, we observed that the signals from the probes for chromosomes 1 and 17, the two sentinels chosen for our study, were often distributed throughout the nuclei in SF exposed to microgravity and not in two territorial accumulations as seen in GC cells (Fig. 7). Due to logistical issues, it has not been possible to gather a full data set for the chromosomes chosen to paint. However, we feel that there are insightful and novel data to present, that for the first time could reveal the effect of SF on the genome in terms of widespread rearrangements of chromosome territories.
Although we did not apply an apoptosis assay (due to scarcity of cell samples from space), the morphological observation showed an increase of apoptotic nuclei in all SF samples compared to GC (Fig. 3l, m), which suggests an increase of DNA damage. There was an increased fraction of formation of nuclear bridges (a sign of affected nuclear division, Fig. 3n), although the latter was not statistically significant in the time span of 6 days in space.
The analysis of DNA damage and repair markers describes a coherent scenario. γH2AX is a marker of DNA double-strand break (DSB) that accumulates in intranuclear spots early upon genome damage. Interestingly, while we observed frequent speckles of various dimensions in GC samples (Fig. 3), SF samples featured fewer positive nuclei, with smaller and less frequent foci.
Genome stress was also highlighted by PML NB, another marker of DSB that were not detected in GC nuclei, but were very obvious in both SF samples (Fig. 3).
To investigate the link between microgravity and SR on genome damage, a wide variety of treatments have been imposed on a broad range of experimental models, either in simulated microgravity or in SF, to respond to different experimental questions (reviewed in [79]). Conflicting results were often reported. As far as HMEC-1 are concerned, we found that the activation of γH2AX became undetectable 24 h after experimental exposure to one gray X-rays on ground (Supplementary Fig. 2f–h). So, it was not surprising that after 4-day exposure to space, SF samples gave negative result for γH2AX, while PML was still detectable. Thus, we propose that in HMEC-1 cells in SF, the DSB repair system involving γH2AX is initially activated, but then later in flight it is saturated by oxidative stress that leads to an undetectable level of H2AX protein. This is in line with [80], reporting a saturation of the repair system in chronic conditions. Instead, PML is recruited to persistent DBS lesions [51] and remains active.
In summary, HMEC-1 are subject, as expected, to DSB in space, showing an articulated response, with activation of different repair systems in time.
Genomic DNA analyzed for telomere length showed shortening in SF, however, with statistically significant lengthening in SF-1g samples (Fig. 8). The shortening effect of microgravity was preponderant over the effect of SR. It is well established that telomere length erodes with aging [81, 82], oxidative stress, inflammation [83, 84], and radiation [85], but the effect of SF on telomere length is still very much controversial, due to data gathered after different lengths and types of exposure, and experimental design and models. In the case of the NASA Twin study [86, 87], the finding of telomere increase was unexpected and surprising. In our case, we were surprised, first, of detecting such differences accumulated in such short time and, second, of finding telomere shortening in SF v/s lengthening in the onboard centrifuge. We stress here that in the studies mentioned above, the time of exposure was much longer compared to our study (1 year or 6 months, v/s total 6 days in space). Therefore, it is possible that we are reporting a transient effect due to short exposure to space. But other elements cannot be ruled out, for instance, the study subject, that is primary lymphocyte blood cells v/s an immortalized EC line. In fact, another cell line, namely embryonic human lens epithelial cells, showed that radiation (on ground) induced telomere length increase, with a dose-dependent response [88]. So, cell culture v/s primary cells might be a significant difference. Further, B and T cells have a peculiar telomere length control compared to other cell types, in that they need to be able to respond with clonal expansion when activated by infections. Even among different B and T cell types, differences exist in telomere length control [89]. Whereas B cells have the highest telomerase activity and longest mean telomere length, CD4+ T cells have slightly higher telomerase activity than CD8+ CD28+ T cells, but similar telomere length; CD8+ CD28− T cells are the most differentiated cell type and have the shortest telomere length and the lowest telomerase activity [89].
The dynamics of telomere length changes observed among pre-, in- and post-flight might reflect a population selection that would be very difficult to see, if at all, in a clonal cell culture. Therefore, telomere elongation observed during SF [87, 90] may reflect changes in lymphocyte cell population dynamics resulting from senescence/apoptosis (critically shortened telomeres), radiosensitivity of lymphocytes, and redistribution of leukocyte subsets in the astronaut blood. Taking into account that telomerase components have activities also independent from telomere lengthening [91], the mechanistic links between exposure to SR and the telomeric and DNA damage responses remain elusive, leaving open the question whether telomere length might be a biomarker of the adaptive response to SF [92]. Here, telomere shortening in SF was interpreted as the consequence of a combination of detrimental environmental factors even after a short-duration flight. Transcriptome changes with respect to signal pathways involved in DNA damage repair complemented the observations (Fig. 6a).
Transcriptome
Transcriptomic data were consistent with cellular observations but provided a much deeper view of the adaptive responses of capillary EC to SF. A number of omics studies conducted in space are reported in the literature, but in most of them the effects of SF are analyzed as a whole, despite space environment being characterized by several parameters (microgravity, cosmic radiation, space electromagnetism, space vibrations), each of which could affect per se the physiology of the model. The configuration of our experiment gave us the rare opportunity to dissect the effect of microgravity and radiation. Therefore, high-throughput RNA sequencing allowed us to identify clearly the genome-wide effects of microgravity, cosmic radiation, and SF on the activation of transcriptional programs in ECs. Functional annotation analysis was applied to explore the expression patterns and regulatory mechanisms during SF using the bioinformatics GSEA tool. In particular, 29 gene sets of the Hallmark collection were deregulated due to SF (22 up-regulated and 7 down-regulated), 36 due to radiation (21 up- and 15 down-regulated) and 32 due to microgravity (19 up- and 12 down-regulated). We would like to remark that through this type of comparative analysis, we have managed to highlight that microgravity and SR do not necessarily exert a synergistic effect on modifying transcriptional programs in SF. In fact, among all the pathways significantly modified by SF, only three (adipogenesis, allograft-rejection and oxidative phosphorylation) were found up-regulated and two other down-regulated (NOTCH-signaling and WNT-Catenin-signaling) in all the three categories of SR, Microgravity and SF.
It was interesting, and somewhat unexpected, to observe that SR and microgravity can even have opposite effect, looking at specific pathways, over the response of ECs to real SF: we found eight gene sets up-regulated by SR and at the same time down-regulated by microgravity (estrogen-response-early, Hedgehog-signaling, hypoxia, IL2-STAT5-signaling, KRAS-signaling-down-regulation, P53-pathway, TNF-signaling-via-NFKB, UV-response-up-regulation) and 12 other pathways up-regulated by microgravity and at the same time down-regulated by SR (androgen-response, bile-acid-metabolism, cholesterol-homeostasis, coagulation, E2F-targets, fatty-acid-metabolism, G2M-checkpoint, mitotic-spindle, mTORC1-signaling, peroxisome, spermatogenesis and UV-response-down-regulation). The most up-regulated pathways in the Radiation group are related to response of inflammation and DNA damage and repair (P53-pathway, TNF-signaling, interferon gamma response, interferon alpha response, DNA repair, inflammation). These same pathways are up-regulated in the SF group (SF-μg vs GC), but not in the microgravity group, where they are down-regulated, suggesting that the overall positive regulation in space is due to the effect of SR. Numerous immune system alterations have been associated with SF in humans and animals [93–95] with the major effects of SF associated with changes in cytokine production, leukocyte subset distribution and antibody production. Different models were used to evaluate the effects of SR on immune function and the activation of inflammatory response based on tissue distribution, length and quality of exposure [96]. Furthermore, nine pathways (E2F-target, fatty-acid-metabolism, mTORC1-signaling, MYC-targets-v1, peroxisome, PI3K-AKT-mTORsignaling, protein-secretion, reactive-oxygen-species-pathway, spermatogenesis) were up-regulated by SF and microgravity but not by SR, where indeed five of these are down-regulated (E2F-target, fatty-acid-metabolism, mTORC1-signaling, peroxisome, spermatogenesis). Most of these pathways are involved in cell growth, proliferation and metabolism. Studies performed in simulated microgravity showed an increase of cell proliferation in HUVEC and BAEC (models of macrovascular ECs), while simulated microgravity inhibited the growth of HMEC-1, a model of microvascular ECs [12]. In our case, the first real SF experiment with HMEC-1, we found a net promotion of the activation of pro-proliferation and anti-apoptotic pathways. Therefore, transcriptomic data show again an opposite effect of concomitant conditions, where microgravity promotes cell proliferation while SR inhibits these same pathways, perhaps to compensate for the negative effects of DNA damage.
A most interesting result of the transcriptome study was the possibility to distinguish at cell level, for the first time in literature to our knowledge, the effect of microgravity from those of SR, the two major stimuli of the space environment. Remarkably, some pathways were controlled in the opposite direction from one or the other condition. Where SR turned on pathways for endothelial activation (hypoxia, cytokines, inflammation), microgravity instead down-regulated them; where microgravity activated pathways for metabolism and cell proliferation, SR exerted the opposite effect. In general, opposite trends were shown by SR, which increased DNA repair and apoptosis promoting an aging phenotype, and microgravity, with mostly a pro-proliferative phenotype.
Lastly, SF increased adipogenesis (with concurring effect of SR and microgravity) and down-regulated myogenesis (with a strong contribution of microgravity), and estrogen response pathways (important for bone integrity), in HMEC-1 cells. Such transcriptome changes strongly suggest that the loss of muscle and bone mass in astronauts are underlined by changes of specific signaling pathways in relevant cell types [97–99].
Cell signaling was also modified in SF, as documented with RNA sequencing by activation of PI3K-AKT-mTOR signaling and inhibition of Hedgehog signaling (Fig. 6), the latter documented also morphologically. The nominal lack of shear stress within the EU along with F-actin reorganization was probably responsible for the increased frequency of primary cilia (Fig. 5).
It is interesting to note that the autophagic marker LC3B was strongly reduced in space (to the limit of undetectability, Fig. 3). This suggests an inhibition of the autophagy flux, also supported by the transcriptomic data of activation of the MTORC1 pathway (Fig. 6), a key negative controller of autophagy, the degradative pathway alternative to proteasome that protects cells under stress [100]. Microgravity and SR acted on the MTORC1 gene pathway with distinct and even opposite signs of effect: SR inhibited the pathway (with consequent enhancement of autophagy as a potential defense mechanism), microgravity strongly stimulated MTORC1 (with consequent inhibition of autophagy).
On this basis, a possible interpretation suggests that HMEC-1 in space, although under stress, has impaired protection due to activation of the MTORC1 pathway. Autophagy protects against environmental and metabolic stress (see the oxidative stress documented in the SF samples by the activation of reactive oxygen species and inflammatory response pathways, Fig. 6). To be activated, autophagy requires an efficient YAP nucleus/cytoplasm shuttling mechanism, in turn guaranteed by the integrity of the cytoskeleton [101], which however was lost in space.
This means that various stimuli, such as oxidative stress, inflammation, and DNA damage that on Earth would increase autophagy, in space are insufficient to activate the autophagic flux. Possibly, cytoskeletal rearrangement and absence of YAP nuclear signaling impair autophagy. On the side, this also suggests that the induction of autophagy in space could protect ECs from the negative effects of microgravity, if not with restoring cytoskeletal integrity at least by counteracting downstream toxic effects through the elimination/recycling of damaged constituents. To our knowledge, this is the first report of inhibition of autophagic flux in ECs in real SF.
It is well known that the cardiovascular function is susceptible to substantial changes in weightlessness, and the altered cardiovascular function in space causes physiological risks in the post-flight period [102]. In addition, the peculiar biological effectiveness of heavy ions in cosmic rays might increase this risk substantially. On this basis, our observations of the impairment of autophagy flux in a model of microvascular ECs in space is important because it might pave the way to pharmacological treatments for preserving the endothelial and the whole cardiovascular function.
As for a link between aging phenotype and SR, similar numbers of apoptotic nuclei, measured in both SF samples, are one of the reasons supporting the hypothesis. In fact, the two sample types were exposed to different g; however, both were exposed to SR. Therefore, the common apoptotic ratio had to be linked to SR. This was also confirmed by the transcriptomic data, showing up-regulation of the apoptosis pathway due to SR (Fig. 6). In addition, the transcriptomic study showed up-regulation of p53 in SF, markedly due to up-regulation by SR (Fig. 6). Since p53 activation is known to be at the crossroads between senescence and aging (reviewed in [103]), our interpretation here was that SR, through activation of p53, caused also up-regulation of the apoptotic pathway (Fig. 3), prodromal to an aging phenotype.
Given the variety of EC models, experimental settings (with all included parameters) and timing of protocols in SF, in simulated microgravity, or exposure to experimental radiation, it is quite complex to compare results across literature. A common theme is definitely represented by a generalized reorganization of the cytoskeleton [13], which seems mostly but not completely due to the effect of microgravity, along with the activation of a pro-inflammatory response mostly due to the effect of SR. Other complex adaptive cell behaviors, like apoptosis or autophagy flux, offer discordant results. Our data show that increase of apoptotic nuclei in SF microvascular ECs is mostly imputable to SR, which could explain why apoptosis was not observed in the same cell type in the condition of simulated microgravity and absence of SR on ground [16].
Autophagy is inhibited by SR, whereas it is activated by microgravity, as indeed seen also in simulated microgravity [104]. As this is the first study to report inhibition of autophagy in real SF, it will be important to wait for other reports on different models to draw general conclusions.
A possible model
From a rheological point of view, the eukaryotic cell can be modeled as a confined compartment, enclosed by the cell membrane. Cytoplasm consists of a water-containing matrix formed by a cytoskeletal network of interconnected protein fibers that might be better described as an aqueous gel. All organelles are not free floating but anchored to the cytoskeleton, a highly dynamic structure that creates an internal positive tension and connects the cell membrane to the nuclear envelope, in a structural continuum. The liquid compartment, the cytosol, has the same surface tension of salt water [105, 106]. Surface tension, internal positive tension, and gravity are basic cell shaping forces. When a drop of polar liquid is released in microgravity, it tends to assume the shape of a sphere, as all forces acting on it tend toward equilibrium. The eukaryotic cell adhering on a stiff surface, as in the case of ENDO EC monolayer, might tend to suddenly ‘dewet’ the adhesion surface when exposed to microgravity, to round up. On ground, cells incur rounding and detachment at specific moments, for example in mitosis, which implies a marked change of internal forces that occurs under strict genetic control. However, unlike liquid drops, metaphase cells do not lose contact and maintain relationships with neighboring cells, actually with a seeming increase of cell–cell junction strength (Fig. 4). We propose that the very first event experienced by ECs in response to sudden gravitational unloading might be cell round up, driven by the liquid compartment (Fig. 9), which in turn would cause nuclear displacement, due to force transmission from the cytoskeleton to the nuclear envelope [8]. This event is supposedly fast. In fact, in a model of glial cells it was observed that 30 min of simulated microgravity were enough to elicit a complex nuclear response to the point of accumulating morphologically apoptotic nuclei [107]. Live fluorescence microscopy observations in SF recently showed that primary macrophages responded to microgravity within seconds to minutes, with nuclei volume and surface that increased up to 150% and 131% before returning to initial values [9]. Interestingly, in the same time frame, actin reorganization was not observed, suggesting it might be a later event. Upon cell round-up and nuclear displacement, cytoskeletal reorganization likely activates a plethora of mechanical and biochemical signaling, pointing to the cytoskeleton as the main gravisensor, affected by the reduction of Rho GTPases activity [108, 109], reduction of F-actin formation and actomyosin stabilization, all leading to cell softening. Tubulin self-assembly into microtubules occurs independent of gravity; however, the assembly and organization of the microtubule network is gravity dependent [64, 110]. This gravity-dependent microtubule network organization was described in multiple cell lines during both real and simulated microgravity exposures [5, 107, 111–113] and could result from poor organization of microtubule organizing center (MTOC) [114]. So, actin reorganization might be an event that follows nuclear repositioning and microtubule disorganization. In our system, after 4 days in SF, we observed an increase of nuclear volume (Fig. 4) and general cytoskeleton remodeling (Fig. 2).
Fig. 9.
A model of subcellular events in space flight. Location of subcellular components at any time depends on the integrity and integration of structural contribution of cytoskeletal compartments (microtubules, intermediate filaments, microfilaments). The position of the nucleus depends on the intermediate filaments network, which together with other scaffold proteins, modifies gene transcription. At 1g, compressive or stretching forces are mechanically balanced by cytoskeletal reorganization to provide the internal resistance to nucleus sedimentation. In space, the response caused by unloading is transmitted likewise to/from the nucleus through the cytoskeleton. If we represent the cell as a nucleated tensegrity structure, we expect exposure to microgravity to cause sudden nuclear repositioning. Later on, chronic exposure elicits adaptive changes, likely initiated by cytoskeleton-bound signaling. In summary, this study reports a general softening of a model of microvascular ECs in SF, with reduction of cell motility. The general reorganization of the cytoskeleton showed remarkable loss of actin stress fibers and compensative enlargement of microtubules and intermediate filaments. Cell signaling was altered, at least concerning Hh, with increased frequency of primary cilia. DNA damage repair mechanisms were activated (PML bodies) and even saturated (γH2AX). Territories of marker chromosomes were found altered compared to GCs
Conclusion
Overall, here we show that SF induced chronic stress on HMEC-1 cells (inferred from increased morphologically apoptotic nuclei in front of stable mitotic index; stress granules; cytoskeletal loosening; cell softening; loss of YAP nuclear localization; mitochondria senescence, and activation of pathways related to oxidative stress, inflammation and DNA damage), with different contributions from microgravity and SR.
Since the mid-1980s, space experiments have begun to examine if microgravity could alter the biological effects of SR. The reported results are still ambiguous; however it is now evident that both stressors induce changes alone, and their combination has a synergistic effect [10]. Oxidative stress appears to be a factor largely underpinning the biological effects of both space phenomena. Recently, a model was proposed, which accounts for their combined effects on cellular responses through (A) damaging and signaling by ROS, (B) damage responses on DNA (repair, replication, transcription, etc.), and (C) changes of gene expression (regulation by chromatin, epigenetic control, etc.) [115]. In line with this model, our data provide deeper insights into the molecular mechanisms activated by SF in microvascular ECs, suggesting that microgravity and SR should be considered separately as well as in their interplay to understand single cell adaptation to space. This will allow targeted and effective countermeasures for protection of space crews’ health in the future, all the more a pressing need considering that the very recent dawn of human launches by commercial ventures theoretically opens the way to a larger human frequentation of space.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file 1 Supplementary Table 1 List of antibodies used for IF, with indication of biological source, commercial provider and dilution used (DOCX 14 KB)
Supplementary file 2 Supplementary Fig. 1 Schematic representation of the ENDO EU. A Mission patch of the ENDO study, supported by ESA and ASI. B 3D rendering of the microincubator (Kayser Italia, Livorno, Italy). Reservoirs containing cell culture media is in red, fixative in green, washing solution in grey. The cell culture support is in red on the incubator proximal side. Further details are in [21–23]. C Launch of Soyuz TMA-18M (manned Mission 44S) from Baikonur Cosmodrome (Kazahstan), at 4:37 GMT, September 2nd 2015. D ESA thermal case Yellow Box, used for maintaining samples while traveling after landing from retrieval site to the PI home laboratory (TIF 114298 KB)
Supplementary file 3 Supplementary Fig. 2 Results of IF preparatory work. HMEC-1 cells were variously stimulated on ground to activate autophagy (A–E) and DNA damage response mechanisms (F–K): culture in serum-free medium (Starvation: 48 h); exposure to oxygen reactive species (50 mM H2O2, 1.5 h prior to fixing); DNA synthesis inhibition (10mM Hydroxyurea, HDU); induction of double strand breaks (exposure to 1 Gray X-rays). L–N labeling with chromosome markers 1, 10 and 17 in ground samples. For each experimental group, three independent operators counted at least 60 cells (six cells per at least ten fields). All tests were repeated at least three times (TIF 104213 KB)
Supplementary file 4 Supplementary Fig. 3 Nuclear and cell shape. Shape descriptors indicated absence of differences in terms of Circularity (A), Roundness (B) and Solidity (C) among the three experimental groups. Statistical analysis was performed with One way ANOVA and Bonferroni multiple comparison post-hoc tests (TIF 90558 KB)
Supplementary file 5 Supplementary Fig. 4 PCA analysis. Unsupervised principal component analysis of the RNA-seq samples (TIF 36294 KB)
Supplementary file6 Supplementary Fig. 5 Integration of genomics and transcriptomics data. Circular plots integrated the gene expression information with physical localization of genes differentially affected by SF (A), showing a different contribution of Microgravity (B) and Radiation (C). All chromosomes were affected. Red and green bars indicate up- and down-regulated genes respectively (TIF 91434 KB)
Acknowledgements
All space biology experiments are supported by complex logistics and require by definition a large range of interventions. The authors wish to acknowledge the contribution of several professionals whose roles were critical for completing this work: Valfredo Zolesi, Alessandro Donati, Aleandro Norfini, Michele Balsamo (Kayser Italia, srl, Livorno, Italy); Jason Hatton, Jutta Krause, Paolo Provasi, Andrea Koheler, Pierfilippo Manieri, Sergio Mugnai (ESA); Sara Piccirillo, Gabriele Mascetti (ASI); Andreas Mogensen (Astronaut Corps, ESA) and Kimiya Yui (Astronaut Corps, Jaxa), Paolo Nespoli (Astronaut Corps, ESA, Ret.); Raimondo Fortezza (Telespazio, Napoli, Italy); Fabienne Wyss, Jeannine Winkler, Bernd Rattenbacher (USOC Hergiswil, CH); Alexander Sverev and all his team of PAO RCS Energia (Russia); Rosa Sapone, Paolo Cergna (Altech spa, Torino, Italy); Thomas Berger (DLR); Sonke Burmeister (CAU); Lucia Giorgetti (IBBA CNR, Pisa, Italy); Edoardo Bertolini (Scuola Superiore Sant’Anna, Pisa, Italy); Daniele Panetta (IFC CNR, Pisa, Italy); Gianni Ciofani, Attilio Marino (IIT, Pontedera, Italy); Gianni Costa (Pierburg Italy); Benedetto Grimaldi (IIT, Genova); Jonny Martini (DHL); Mr. and Mrs. Vittorio E. Angeloni. Last but not least, Riccardo M. Andreazzoli and Matteo and Giulia Meccheri for their cheerful comprehension and forbearance.
Abbreviations
- AcTUBA
Acetylated tubulin alpha
- ATP5H
Mitochondrial ATP synthase, subunit d
- CC
Culture chamber
- COC
Cyclic olefin copolymer
- CTNNB
Catenin-beta
- DEG
Differentially expressed genes
- EC
Endothelial cell
- ESA
European Space Agency
- EU
Experimental Unit
- FISH
Fluorescent in situ hybridization
- GC
Ground controls, samples prepared in parallel with samples sent to space, but grown on Earth
- GSEA
Gene Set Enrichment Analysis
- Hh
Hedgehog
- HMEC-1
Human microvascular endothelial cells-1
- IF
Immunofluorescence
- ISS
International Space Station
- KRT7
Cytokeratin 7
- LC3B
Light chain 3 isoform B
- LINC
Linker of nucleoskeleton and cytoskeleton
- MYL2
Myosin light chain 2
- PBS
Phosphate buffer saline
- γH2AX
Phosphorylated histone H2AX
- PML NB
Promyelocytic leukemia nuclear bodies
- SF
Space flight
- SF-1g
Space-flown samples cultured at 1g within the centrifuge onboard
- SF-μg
Space-flown samples exposed to real microgravity
- SR
Space radiation
- TMA
Transport modified anthropometric
- TUBA
Tubulin alpha
- VIM
Vimentin
- WCP
Whole chromosome probes
- YAP1
Yes-associated protein 1
Author contributions
Conceptualization and overall supervision: DA; methodology: DA, IB, MF, SB, JMB; investigation and validation, data collection, image and statistical analysis: IB, CDC, OVP, MF, SB, JMB, HAF, FS, AB, MA, GS; writing—original draft preparation: DA, IB; writing editing: MF, SB, JMB, GS; project administration: DA; funding acquisition: DA, MA, MEP. All authors have read and agreed to the published version of the manuscript.
Funding
The study was supported by European Space Agency (ESA ILSRA-2009-1026), Agenzia Spaziale Italiana (contract number 5681).
Availability of data and materials
All transcriptome analysis raw data were deposited in GEO as GSE157937.
Code availability
Not applicable.
Declarations
Conflict of interest
The authors declare that there are no conflicts to disclose.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
The manuscript has been approved by all co-authors.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Rijken PJ, De Groot RP, Briegleb W, Kruijer W, Verkleij AJ, Boonstra J, et al. Epidermal growth factor-induced cell rounding is sensitive to simulated microgravity. Aviat Sp Environ Med. 1991;62(1):32–36. [PubMed] [Google Scholar]
- 2.Buken C, Sahana J, Corydon TJ, Melnik D, Bauer J, Wehland M, et al. Morphological and molecular changes in juvenile normal human fibroblasts exposed to simulated microgravity. Sci Rep. 2019;9(1):11882. doi: 10.1038/s41598-019-48378-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dietz C, Infanger M, Romswinkel A, Strube F, Kraus A. Apoptosis induction and alteration of cell adherence in human lung cancer cells under simulated microgravity. Int J Mol Sci. 2019;20(14):3601. doi: 10.3390/ijms20143601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thiel CS, Tauber S, Lauber B, Polzer J, Seebacher C, Uhl R, et al. Rapid morphological and cytoskeletal response to microgravity in human primary macrophages. Int J Mol Sci. 2019;20(10):2402. doi: 10.3390/ijms20102402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vassy J, Portet S, Beil M, Millot G, Fauvel-Lafève F, Karniguian A, et al. The effect of weightlessness on cytoskeleton architecture and proliferation of human breast cancer cell line MCF-7. FASEB J. 2001;15(6):1104–1106. doi: 10.1096/fj.00-0527fje. [DOI] [PubMed] [Google Scholar]
- 6.Infanger M, Kossmehl P, Shakibaei M, Baatout S, Witzing A, Grosse J, et al. Induction of three-dimensional assembly and increase in apoptosis of human endothelial cells by simulated microgravity: impact of vascular endothelial growth factor. Apoptosis. 2006;11(5):749–764. doi: 10.1007/s10495-006-5697-7. [DOI] [PubMed] [Google Scholar]
- 7.Tauber S, Lauber BA, Paulsen K, Layer LE, Lehmann M, Hauschild S, et al. Cytoskeletal stability and metabolic alterations in primary human macrophages in long-term microgravity. PLoS One. 2017;12(4):e0175599. doi: 10.1371/journal.pone.0175599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou L, Zhang C, Zhang F, Lü S, Sun S, Lü D, et al. Theoretical modeling of mechanical homeostasis of a mammalian cell under gravity-directed vector. Biomech Model Mechanobiol. 2018;17(1):191–203. doi: 10.1007/s10237-017-0954-y. [DOI] [PubMed] [Google Scholar]
- 9.Thiel CS, Tauber S, Seebacher C, Schropp M, Uhl R, Lauber B, et al. Real-time 3D high-resolution microscopy of human cells on the international space station. Int J Mol Sci. 2019;20(8):2033. doi: 10.3390/ijms20082033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Demontis GC, Germani MM, Caiani EG, Barravecchia I, Passino C, Angeloni D. Human pathophysiological adaptations to the space environment. Front Physiol. 2017;8:547. doi: 10.3389/fphys.2017.00547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coupé M, Fortrat JO, Larina I, Gauquelin-Koch G, Gharib C, Custaud MA. Cardiovascular deconditioning: from autonomic nervous system to microvascular dysfunctions. Respir Physiol Neurobiol. 2009;169(Suppl):S10–S12. doi: 10.1016/j.resp.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 12.Maier JAM, Cialdai F, Monici M, Morbidelli L. The impact of microgravity and hypergravity on endothelial cells. Biomed Res Int. 2015;2015:434803. doi: 10.1155/2015/434803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin X, Zhang K, Wei D, Tian Y, Gao Y, Chen Z, et al. The impact of spaceflight and simulated microgravity on cell adhesion. Int J Mol Sci. 2020;21:3031. doi: 10.3390/ijms21093031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herranz R, Anken R, Boonstra J, Braun M, Christianen PCM, de Geest M, et al. Ground-based facilities for simulation of microgravity: organism-specific recommendations for their use, and recommended terminology. Astrobiology. 2013;13(1):1–17. doi: 10.1089/ast.2012.0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cotrupi S, Ranzani D, Maier JAM. Impact of modeled microgravity on microvascular endothelial cells. Biochim Biophys Acta. 2005;1746(2):163–168. doi: 10.1016/j.bbamcr.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 16.Mariotti M, Maier JAM. Gravitational unloading induces an anti-angiogenic phenotype in human microvascular endothelial cells. J Cell Biochem. 2008;104:129–135. doi: 10.1002/jcb.21605. [DOI] [PubMed] [Google Scholar]
- 17.Kang CY, Zou L, Yuan M, Wang Y, Li TZ, Zhang Y, et al. Impact of simulated microgravity on microvascular endothelial cell apoptosis. Eur J Appl Physiol. 2011;111(9):2131–2138. doi: 10.1007/s00421-011-1844-0. [DOI] [PubMed] [Google Scholar]
- 18.Tang NP, Hui TT, Ma J, Mei QB. Effects of miR-503-5p on apoptosis of human pulmonary microvascular endothelial cells in simulated microgravity. J Cell Biochem. 2019;120(1):727–737. doi: 10.1002/jcb.27430. [DOI] [PubMed] [Google Scholar]
- 19.Zhao H, Shi Y, Qiu C, Zhao J, Gong Y, Nie C, et al. Effects of simulated microgravity on ultrastructure and apoptosis of choroidal vascular endothelial cells. Front Physiol. 2021;18:11. doi: 10.3389/fphys.2020.577325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ades EW, Candal FJ, Swerlick RA, George VG, Summers S, Bosse DC, et al. HMEC-1: establishment of an immortalized human microvascular endothelial cell line. J Invest Dermatol. 1992;99(6):683–690. doi: 10.1111/1523-1747.ep12613748. [DOI] [PubMed] [Google Scholar]
- 21.Barravecchia I, De CC, Pyankova OV, Scebba F, Mascherpa MC, Vecchione A, et al. Pitting corrosion within bioreactors for space cell-culture contaminated by Paenibacillus glucanolyticus, a case report. Microgravity Sci Technol. 2018;30(3):309–319. [Google Scholar]
- 22.Balsamo M, Barravecchia I, Mariotti S, Merenda A, De Cesari C, Vukich M, et al. Molecular and cellular characterization of space flight effects on microvascular endothelial cell function—preparatorywork for the SFEF project. Microgravity Sci Technol. 2014;26(6):351–363. [Google Scholar]
- 23.Barravecchia I, De Cesari C, Pyankova O, Scebba F, Pè M, Forcato M et al (2018) A comprehensive molecular and morphological study of the effects of space flight on human capillary endothelial cells: sample quality assessment and preliminary results. In: Proceedings, 39th ISGP meeting & ESA life sciences meeting. Front Physiol. section: Environmental, Aviation and Space Physiology. 2018. 10.3389/conf.fphys.2018.26.00050
- 24.Soyuz Users Manual, Issue 3 ST-GTD-SUM-01. Starsem; 2001. 1–204
- 25.Versari S, Longinotti G, Barenghi L, Maier JAM, Bradamante S. The challenging environment on board the International Space Station affects endothelial cell function by triggering oxidative stress through thioredoxin interacting protein overexpression: the ESA-SPHINX experiment. FASEB J. 2013;27(11):4466–4475. doi: 10.1096/fj.13-229195. [DOI] [PubMed] [Google Scholar]
- 26.Tamma R, Colaianni G, Camerino C, Di Benedetto A, Greco G, Strippoli M, et al. Microgravity during spaceflight directly affects in vitro osteoclastogenesis and bone resorption. FASEB J. 2009;23(8):2549–2554. doi: 10.1096/fj.08-127951. [DOI] [PubMed] [Google Scholar]
- 27.Monticone M, Liu Y, Pujic N, Cancedda R. Activation of nervous system development genes in bone marrow derived mesenchymal stem cells following spaceflight exposure. J Cell Biochem. 2010;111(2):442–452. doi: 10.1002/jcb.22765. [DOI] [PubMed] [Google Scholar]
- 28.Franzoso S, Sandonà D, Picard A, Furlan S, Gobbo V, Salvatori S, et al. Cultured adult muscle fibers in the microgravity environment. The MYO experiment in the Foton-M3 space flight mission. Basic Appl Mol. 2009;19:65–76. [Google Scholar]
- 29.ESA document: ESA-HSO-COU-025 2.0 (http://wsn.spaceflight.esa.int/docs/Factsheets/25%20Kubik%20HR_WEB.pdf)
- 30.Mariotti S, Barravecchia I, Vindigni C, Pucci A, Balsamo M, Libro R, et al. MICAL2 is a novel human cancer gene controlling mesenchymal to epithelial transition involved in cancer growth and invasion. Oncotarget. 2014;7(2):1808–1825. doi: 10.18632/oncotarget.6577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marino A, Filippeschi C, Genchi GG, Mattoli V, Mazzolai B, Ciofani G. The Osteoprint: a bioinspired two-photon polymerized 3-D structure for the enhancement of bone-like cell differentiation. Acta Biomater. 2014;10(10):4304–4313. doi: 10.1016/j.actbio.2014.05.032. [DOI] [PubMed] [Google Scholar]
- 32.Barr ML, Hamilton JD. A quantitative study of certain morphological changes in spinal motor neurons during axon reaction. J Comp Neurol. 1948;89(2):93–121. doi: 10.1002/cne.900890203. [DOI] [PubMed] [Google Scholar]
- 33.Sabatino L, Botto N, Borghini A, Turchi S, Andreassi MG. Development of a new multiplex quantitative real-time PCR assay for the detection of the mtDNA4977 deletion in coronary artery disease patients: A link with telomere shortening. Environ Mol Mutagen. 2013;54(5):299–307. doi: 10.1002/em.21783. [DOI] [PubMed] [Google Scholar]
- 34.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liao Y, Smyth GK, Shi W. FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 36.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2009;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu R, Holik AZ, Su S, Jansz N, Chen K, Leong HS, et al. Why weight? Modelling sample and observational level variability improves power in RNA-seq analyses. Nucleic Acids Res. 2015;43(15):e97. doi: 10.1093/nar/gkv412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yin T, Cook D, Lawrence M. ggbio: an R package for extending the grammar of graphics for genomic data. Genome Biol. 2012;13(8):R77. doi: 10.1186/gb-2012-13-8-r77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mehta IS, Amira M, Harvey AJ, Bridger JM. Rapid chromosome territory relocation by nuclear motor activity in response to serum removal in primary human fibroblasts. Genome Biol. 2010;11(1):R5. doi: 10.1186/gb-2010-11-1-r5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Foster HA, Estrada-Girona G, Themis M, Garimberti E, Hill MA, Bridger JM, et al. Relative proximity of chromosome territories influences chromosome exchange partners in radiation-induced chromosome rearrangements in primary human bronchial epithelial cells. Mutat Res Genet Toxicol Environ Mutagen. 2013;756(1–2):66–77. doi: 10.1016/j.mrgentox.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 42.Patteson AE, Pogoda K, Byfield FJ, Mandal K, Ostrowska-Podhorodecka Z, Charrier EE, et al. Loss of vimentin enhances cell motility through small confining spaces. Small. 2019;15(50):1903180. doi: 10.1002/smll.201903180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiu Y, Lehtimäki J, Tojkander S, Cheng F, Jäälinoja H, Liu X, et al. Bidirectional interplay between vimentin intermediate filaments and contractile actin stress fibers. Cell Rep. 2015;11(10):1511–1518. doi: 10.1016/j.celrep.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 44.Jiu Y, Peränen J, Schaible N, Cheng F, Eriksson JE, Krishnan R, et al. Vimentin intermediate filaments control actin stress fiber assembly through GEF-H1 and RhoA. J Cell Sci. 2017;130(5):892–902. doi: 10.1242/jcs.196881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hookway C, Ding L, Davidson MW, Rappoport JZ, Danuser G, Gelfand VI. Microtubule-dependent transport and dynamics of vimentin intermediate filaments. Mol Biol Cell. 2015;26(9):1675–1686. doi: 10.1091/mbc.E14-09-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.D’Arcy MS. Cell death: a review of the major forms of apoptosis, necrosis and autophagy. Cell Biol Int. 2019;43(6):582–592. doi: 10.1002/cbin.11137. [DOI] [PubMed] [Google Scholar]
- 47.Koken MHM, Linares-Cruz G, Quignon F, Viron A, Chelbi-Alix MK, Sobczak-Thepot J, et al. The PML growth-suppressor has an altered expression in human oncogenesis. Oncogene. 1995;10(7):1315–1324. [PubMed] [Google Scholar]
- 48.Dellaire G, Bazett-Jones DP. PML nuclear bodies: dynamic sensors of DNA damage and cellular stress. BioEssays. 2004;26:963–977. doi: 10.1002/bies.20089. [DOI] [PubMed] [Google Scholar]
- 49.Bernardi R, Scaglioni PP, Bergmann S, Horn HF, Vousden KH, Pandolfi PP. PML regulates p53 stability by sequestering Mdm2 to the nucleolus. Nat Cell Biol. 2004;6(7):665–672. doi: 10.1038/ncb1147. [DOI] [PubMed] [Google Scholar]
- 50.Chang FTM, McGhie JD, Chan FL, Tang MC, Anderson MA, Mann JR, et al. PML bodies provide an important platform for the maintenance of telomeric chromatin integrity in embryonic stem cells. Nucleic Acids Res. 2013;41(8):4447–4458. doi: 10.1093/nar/gkt114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vancurova M, Hanzlikova H, Knoblochova L, Kosla J, Majera D, Mistrik M, et al. PML nuclear bodies are recruited to persistent DNA damage lesions in an RNF168-53BP1 dependent manner and contribute to DNA repair. DNA Repair (Amst) 2019;1(78):114–127. doi: 10.1016/j.dnarep.2019.04.001. [DOI] [PubMed] [Google Scholar]
- 52.Chang HR, Munkhjargal A, Kim M-J, Park SY, Jung E, Ryu J-H, et al. The functional roles of PML nuclear bodies in genome maintenance. Mutat Res. 2018;809:99–107. doi: 10.1016/j.mrfmmm.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 53.Mah L-J, El-Osta A, Karagiannis TC. γH2AX: a sensitive molecular marker of DNA damage and repair. Leukemia. 2010;24(4):679–686. doi: 10.1038/leu.2010.6. [DOI] [PubMed] [Google Scholar]
- 54.Van Loon JJWA. Mechanomics and physicomics in gravisensing. Microgravity Sci Technol. 2009;21(1–2):159–167. [Google Scholar]
- 55.Pukhlyakova E, Aman AJ, Elsayad K, Technau U. β-Catenin-dependent mechanotransduction dates back to the common ancestor of Cnidaria and Bilateria. Proc Natl Acad Sci USA. 2018;115(24):6231–6236. doi: 10.1073/pnas.1713682115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Luu VZ, Chowdhury B, Al-Omran M, Hess DA, Verma S. Role of endothelial primary cilia as fluid mechanosensors on vascular health. Atherosclerosis. 2018;275:196–204. doi: 10.1016/j.atherosclerosis.2018.06.818. [DOI] [PubMed] [Google Scholar]
- 57.Kallakuri S, Yu JA, Li J, Li Y, Weinstein BM, Nicoli S, et al. Endothelial cilia are essential for developmental vascular integrity in zebrafish. J Am Soc Nephrol. 2015;26(4):864–875. doi: 10.1681/ASN.2013121314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boyle S, Gilchrist S, Bridger JM, Mahy NL, Ellis JA, Bickmore WA. The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Hum Mol Genet. 2001;10(3):211–219. doi: 10.1093/hmg/10.3.211. [DOI] [PubMed] [Google Scholar]
- 59.Crawford-Young SJ. Effects of microgravity on cell cytoskeleton and embryogenesis. Int J Dev Biol. 2006;50(2–3):183–191. doi: 10.1387/ijdb.052077sc. [DOI] [PubMed] [Google Scholar]
- 60.Corydon TJ, Kopp S, Wehland M, Braun M, Schütte A, Mayer T, et al. Alterations of the cytoskeleton in human cells in space proved by life-cell imaging. Sci Rep. 2016;6:20043. doi: 10.1038/srep20043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bradbury P, Wu H, Choi JU, Rowan AE, Zhang H, Poole K, et al. Modeling the impact of microgravity at the cellular level: implications for human disease. Front Cell Dev Biol. 2020;8:96. doi: 10.3389/fcell.2020.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Y-S, Liu B, Luo X-J, Li T-B, Zhang J-J, Peng J-J, et al. Nuclear cardiac myosin light chain 2 modulates NADPH oxidase 2 expression in myocardium: a novel function beyond muscle contraction. Basic Res Cardiol. 2015;110(4):38. doi: 10.1007/s00395-015-0494-5. [DOI] [PubMed] [Google Scholar]
- 63.Caridi CP, D’agostino C, Ryu T, Zapotoczny G, Delabaere L, Li X, et al. Nuclear F-actin and myosins drive relocalization of heterochromatic breaks. Nature. 2018;559(7712):54–60. doi: 10.1038/s41586-018-0242-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Papaseit C, Pochon N, Tabony J. Microtubule self-organization is gravity-dependent. Proc Natl Acad Sci USA. 2000;97(15):8364–8368. doi: 10.1073/pnas.140029597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goldman RD. The role of three cytoplasmic fibers in BHK-21 cell motility. I. Microtubules and the effects of colchicine. J Cell Biol. 1971;51(3):752–762. doi: 10.1083/jcb.51.3.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li N, Wang C, Sun S, Zhang C, Lü D, Chen Q, et al. Microgravity-induced alterations of inflammation-related mechanotransduction in endothelial cells on board SJ-10 satellite. Front Physiol. 2018;31:9. doi: 10.3389/fphys.2018.01025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.De Cesari C, Barravecchia I, Pyankova OV, Vezza M, Germani MM, Scebba F, et al. Hypergravity activates a pro-angiogenic homeostatic response by human capillary endothelial cells. Int J Mol Sci. 2020;21(7):2354. doi: 10.3390/ijms21072354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Simon DN, Wilson KL. The nucleoskeleton as a genome-associated dynamic “network of networks”. Nat Rev Mol Cell Biol. 2011;12:695–708. doi: 10.1038/nrm3207. [DOI] [PubMed] [Google Scholar]
- 69.Meinke P, Schirmer EC. LINC’ing form and function at the nuclear envelope. FEBS Lett. 2015;589:2514–2521. doi: 10.1016/j.febslet.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 70.Elcock LS, Bridger JM. Exploring the effects of a dysfunctional nuclear matrix. Biochem Soc Trans. 2008;36(6):1378–1383. doi: 10.1042/BST0361378. [DOI] [PubMed] [Google Scholar]
- 71.Neelam S, Richardson B, Barker R, Udave C, Gilroy S, Cameron M, et al. Changes in nuclear shape and gene expression in response to simulated microgravity are linc complex-dependent. Int J Mol Sci. 2020;21(18):1–17. doi: 10.3390/ijms21186762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chakraborty N, Cheema A, Gautam A, Donohue D, Hoke A, Conley C, et al. Gene-metabolite profile integration to understand the cause of spaceflight induced immunodeficiency. npj Microgravity. 2018 doi: 10.1038/s41526-017-0038-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang C, Svitkina TM. Ultrastructure and dynamics of the actin-myosin II cytoskeleton during mitochondrial fission. Nat Cell Biol. 2019;21(5):603–613. doi: 10.1038/s41556-019-0313-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Drummond ML, Li M, Tarapore E, Nguyen TTL, Barouni BJ, Cruz S, et al. Actin polymerization controls cilia-mediated signaling. J Cell Biol. 2018;217(9):3255–3266. doi: 10.1083/jcb.201703196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Leonard JM, Ye H, Wetmore C, Karnitz LM. Sonic Hedgehog signaling impairs ionizing radiation–induced checkpoint activation and induces genomic instability. J Cell Biol. 2008;183(3):385–391. doi: 10.1083/jcb.200804042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Filipová A, Diaz-Garcia D, Bezrouk A, Čížková D, Havelek R, Vávrová J, et al. Ionizing radiation increases primary cilia incidence and induces multiciliation in C2C12 myoblasts. Cell Biol Int. 2015;39(8):943–953. doi: 10.1002/cbin.10462. [DOI] [PubMed] [Google Scholar]
- 77.Qu W, Li D, Wang Y, Wu Q, Hao D. Activation of sonic hedgehog signaling is associated with human osteosarcoma cells radioresistance characterized by increased proliferation, migration, and invasion. Med Sci Monit. 2018;4(24):3764–3771. doi: 10.12659/MSM.908278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Datta K, Suman S, Fornace A. Radiation persistently promoted oxidative stress, activated mTOR via PI3K/Akt, and downregulated autophagy pathway in mouse intestine. Int J Biochem Cell Biol. 2014;57:167. doi: 10.1016/j.biocel.2014.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Moreno-Villanueva M, Wong M, Lu T, Zhang Y, Wu H. Interplay of space radiation and microgravity in DNA damage and DNA damage response. Microgravity. 2017;3:14. doi: 10.1038/s41526-017-0019-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gruosso T, Mieulet V, Cardon M, Bourachot B, Kieffer Y, Devun F, et al. Chronic oxidative stress promotes H2 AX protein degradation and enhances chemosensitivity in breast cancer patients. EMBO Mol Med. 2016;8(5):527–549. doi: 10.15252/emmm.201505891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Aubert G, Lansdorp PM. Telomeres and aging. Physiol Rev. 2008;88:557–579. doi: 10.1152/physrev.00026.2007. [DOI] [PubMed] [Google Scholar]
- 82.Honig LS, Kang MS, Cheng R, Eckfeldt JH, Thyagarajan B, Leiendecker-Foster C, et al. Heritability of telomere length in a study of long-lived families. Neurobiol Aging. 2015;36(10):2785–2790. doi: 10.1016/j.neurobiolaging.2015.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Von Zglinicki T. Role of oxidative stress in telomere length regulation and replicative senescence. Ann New York Acad Sci. 2000;908:99–110. doi: 10.1111/j.1749-6632.2000.tb06639.x. [DOI] [PubMed] [Google Scholar]
- 84.Zhang J, Rane G, Dai X, Shanmugam MK, Arfuso F, Samy RP, et al. Ageing and the telomere connection: an intimate relationship with inflammation. Ageing Res Rev. 2016;25:55–69. doi: 10.1016/j.arr.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 85.Sishc BJ, Nelson CB, McKenna MJ, Battaglia CLR, Herndon A, Idate R, et al. Telomeres and telomerase in the radiation response: Implications for instability, reprograming, and carcinogenesis. Front Oncol. 2015 doi: 10.3389/fonc.2015.00257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Luxton JJ, McKenna MJ, Taylor LE, George KA, Zwart SR, Crucian BE, et al. Temporal telomere and DNA damage responses in the space radiation environment. Cell Rep. 2020;33(10):108435. doi: 10.1016/j.celrep.2020.108435. [DOI] [PubMed] [Google Scholar]
- 87.Luxton JJ, McKenna MJ, Lewis A, Taylor LE, George KA, Dixit SM, et al. Telomere length dynamics and DNA damage responses associated with long-duration spaceflight. Cell Rep. 2020;33(10):108435. doi: 10.1016/j.celrep.2020.108457. [DOI] [PubMed] [Google Scholar]
- 88.Bains SK, Chapman K, Bright S, Senan A, Kadhim M, Slijepcevic P. Effects of ionizing radiation on telomere length and telomerase activity in cultured human lens epithelium cells. Int J Radiat Biol. 2018;95(1):54–63. doi: 10.1080/09553002.2018.1466066. [DOI] [PubMed] [Google Scholar]
- 89.Lin J, Epel E, Cheon J, Kroenke C, Sinclair E, Bigos M, et al. Analyses and comparisons of telomerase activity and telomere length in human T and B cells: insights for epidemiology of telomere maintenance. J Immunol Methods. 2010;352(1–2):71–80. doi: 10.1016/j.jim.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Garrett-Bakelman FE, Darshi M, Green SJ, Gur RC, Lin L, Macias BR, et al. The NASA twins study: a multidimensional analysis of a year-long human spaceflight. Science. 2019;364(6436):127–128. doi: 10.1126/science.aau8650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ali M, Devkota S, Il RJ, Lee J, Lee HW. Telomerase reverse transcriptase induces basal and amino acid starvation-induced autophagy through mTORC1. Biochem Biophys Res Commun. 2016;478(3):1198–1204. doi: 10.1016/j.bbrc.2016.08.094. [DOI] [PubMed] [Google Scholar]
- 92.Welsh J, Bevelacqua JJ, Keshavarz M, Mortazavi SAR, Mortazavi SMJ. Is telomere length a biomarker of adaptive response? Controversial findings of NASA and residents of high background radiation areas. J Biomed Phys Eng. 2019;9(3):381. doi: 10.31661/jbpe.v9i3Jun.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sonnenfeld G, Shearer WT. Immune function during space flight. Nutrition. 2002;18(10):899–903. doi: 10.1016/s0899-9007(02)00903-6. [DOI] [PubMed] [Google Scholar]
- 94.Crucian B, Sams C. Immune system dysregulation during spaceflight: clinical risk for exploration-class missions. J Leukoc Biol. 2009;86(5):1017–1018. doi: 10.1189/jlb.0709500. [DOI] [PubMed] [Google Scholar]
- 95.Guéguinou N, Huin-Schohn C, Bascove M, Bueb J-L, Tschirhart E, Legrand-Frossi C, et al. Could spaceflight-associated immune system weakening preclude the expansion of human presence beyond Earth’s orbit? J Leukoc Biol. 2009;86(5):1027–1038. doi: 10.1189/jlb.0309167. [DOI] [PubMed] [Google Scholar]
- 96.Kennedy AR. Biological effects of space radiation and development of effective countermeasures. Life Sci Space Res. 2014;1:10–43. doi: 10.1016/j.lssr.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gambara G, Salanova M, Ciciliot S, Furlan S, Gutsmann M, Schiffl G, et al. Microgravity-induced transcriptome adaptation in mouse paraspinal longissimus dorsi muscle highlights insulin resistance-linked genes. Front Physiol. 2017;8(MAY):279. doi: 10.3389/fphys.2017.00279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lee SJ, Lehar A, Meir JU, Koch C, Morgan A, Warren LE, et al. Targeting myostatin/activin A protects against skeletal muscle and bone loss during spaceflight. Proc Natl Acad Sci USA. 2020;117(38):23942–23951. doi: 10.1073/pnas.2014716117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Okada R, Fujita S, Suzuki R, Hayashi T, Tsubouchi H, Kato C, et al. Transcriptome analysis of gravitational effects on mouse skeletal muscles under microgravity and artificial 1 g onboard environment. Sci Rep. 2021;11(1):9168. doi: 10.1038/s41598-021-88392-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kroemer G, Mariño G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pavel M, Renna M, Park SJ, Menzies FM, Ricketts T, Füllgrabe J, et al. Contact inhibition controls cell survival and proliferation via YAP/TAZ-autophagy axis. Nat Commun. 2018;9(1):1–18. doi: 10.1038/s41467-018-05388-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hughson RL, Helm A, Durante M. Heart in space: effect of the extraterrestrial environment on the cardiovascular system. Nat Rev Cardiol. 2018;15:167–180. doi: 10.1038/nrcardio.2017.157. [DOI] [PubMed] [Google Scholar]
- 103.Rufini A, Tucci P, Celardo I, Melino G. Senescence and aging: The critical roles of p53. Oncogene. 2013;32:5129–5143. doi: 10.1038/onc.2012.640. [DOI] [PubMed] [Google Scholar]
- 104.Locatelli L, Cazzaniga A, De Palma C, Castiglioni S, Maier JAM. Mitophagy contributes to endothelial adaptation to simulated microgravity. FASEB J. 2020;34(1):1833–1845. doi: 10.1096/fj.201901785RRR. [DOI] [PubMed] [Google Scholar]
- 105.Macis M, Lugli F, Zerbetto F. Modeling living cells response to surface tension and chemical patterns. ACS Appl Mater Interfaces. 2017;9(23):19552–19561. doi: 10.1021/acsami.7b01935. [DOI] [PubMed] [Google Scholar]
- 106.Zhou EH, Martinez FD, Fredberg JJ. Cell rheology: mush rather than machine. Nat Mater. 2013;12(3):184–185. doi: 10.1038/nmat3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Uva BM, Masini MA, Sturla M, Bruzzone F, Giuliani M, Tagliafierro G, et al. Microgravity-induced apoptosis in cultured glial cells. Eur J Histochem. 2002;46(3):209–214. doi: 10.4081/1681. [DOI] [PubMed] [Google Scholar]
- 108.Higashibata A, Imamizo-Sato M, Seki M, Yamazaki T, Ishioka N. Influence of simulated microgravity on the activation of the small GTPase Rho involved in cytoskeletal formation—Molecular cloning and sequencing of bovine leukemia-associated guanine nucleotide exchange factor. BMC Biochem. 2006;28:7. doi: 10.1186/1471-2091-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Louis F, Deroanne C, Nusgens B, Vico L, Guignandon A (2015) RhoGTPases as key players in mammalian cell adaptation to microgravity. Biomed Res Int 747693 [DOI] [PMC free article] [PubMed]
- 110.Tabony J, Glade N, Papaseit C, Demongeot J. Microtubule Self-organisation and its gravity dependence. Adv Space Biol Med. 2002;8:19–58. doi: 10.1016/s1569-2574(02)08014-0. [DOI] [PubMed] [Google Scholar]
- 111.Hughes-Fulford M. Function of the cytoskeleton in gravisensing during spaceflight. Adv Space Res. 2003;32(8):1585–1593. doi: 10.1016/S0273-1177(03)90399-1. [DOI] [PubMed] [Google Scholar]
- 112.Rösner H, Wassermann T, Möller W, Hanke W. Effects of altered gravity on the actin and microtubule cytoskeleton of human SH-SY5Y neuroblastoma cells. Protoplasma. 2006;229(2–4):225–234. doi: 10.1007/s00709-006-0202-2. [DOI] [PubMed] [Google Scholar]
- 113.Janmaleki M, Pachenari M, Seyedpour SM, Shahghadami R, Sanati-Nezhad A. Impact of simulated microgravity on cytoskeleton and viscoelastic properties of endothelial cell. Sci Rep. 2016;1:6. doi: 10.1038/srep32418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lewis ML, Reynolds JL, Cubano LA, Hatton JP, Lawless BD, Piepmeier EH. Spaceflight alters microtubules and increases apoptosis in human lymphocytes (Jurkat) FASEB J. 1998;12(11):1007–1018. doi: 10.1096/fasebj.12.11.1007. [DOI] [PubMed] [Google Scholar]
- 115.Yatagai F, Honma M, Dohmae N, Ishioka N. Biological effects of space environmental factors: a possible interaction between space radiation and microgravity. Life Sci Space Res. 2019;20:113–123. doi: 10.1016/j.lssr.2018.10.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary file 1 Supplementary Table 1 List of antibodies used for IF, with indication of biological source, commercial provider and dilution used (DOCX 14 KB)
Supplementary file 2 Supplementary Fig. 1 Schematic representation of the ENDO EU. A Mission patch of the ENDO study, supported by ESA and ASI. B 3D rendering of the microincubator (Kayser Italia, Livorno, Italy). Reservoirs containing cell culture media is in red, fixative in green, washing solution in grey. The cell culture support is in red on the incubator proximal side. Further details are in [21–23]. C Launch of Soyuz TMA-18M (manned Mission 44S) from Baikonur Cosmodrome (Kazahstan), at 4:37 GMT, September 2nd 2015. D ESA thermal case Yellow Box, used for maintaining samples while traveling after landing from retrieval site to the PI home laboratory (TIF 114298 KB)
Supplementary file 3 Supplementary Fig. 2 Results of IF preparatory work. HMEC-1 cells were variously stimulated on ground to activate autophagy (A–E) and DNA damage response mechanisms (F–K): culture in serum-free medium (Starvation: 48 h); exposure to oxygen reactive species (50 mM H2O2, 1.5 h prior to fixing); DNA synthesis inhibition (10mM Hydroxyurea, HDU); induction of double strand breaks (exposure to 1 Gray X-rays). L–N labeling with chromosome markers 1, 10 and 17 in ground samples. For each experimental group, three independent operators counted at least 60 cells (six cells per at least ten fields). All tests were repeated at least three times (TIF 104213 KB)
Supplementary file 4 Supplementary Fig. 3 Nuclear and cell shape. Shape descriptors indicated absence of differences in terms of Circularity (A), Roundness (B) and Solidity (C) among the three experimental groups. Statistical analysis was performed with One way ANOVA and Bonferroni multiple comparison post-hoc tests (TIF 90558 KB)
Supplementary file 5 Supplementary Fig. 4 PCA analysis. Unsupervised principal component analysis of the RNA-seq samples (TIF 36294 KB)
Supplementary file6 Supplementary Fig. 5 Integration of genomics and transcriptomics data. Circular plots integrated the gene expression information with physical localization of genes differentially affected by SF (A), showing a different contribution of Microgravity (B) and Radiation (C). All chromosomes were affected. Red and green bars indicate up- and down-regulated genes respectively (TIF 91434 KB)
Data Availability Statement
All transcriptome analysis raw data were deposited in GEO as GSE157937.
Not applicable.